Abstract
Age-related macular degeneration (AMD) is a chronic degenerative disease of the retina. Recent reports have highlighted the potential role of mucosal surface microbes in the pathogenesis of AMD. In this case-control study, the composition of the nasal and oral microbiota in newly diagnosed neovascular age-related macular degeneration cases (6 male, 7 female) was compared to controls without retinal diseases (2 male, 3 female). PCR amplification of 16S rRNA genes was performed with universal primers amplifying the V4 variable region (515F-806R). Distinct microbial community characterization was achieved using Principal Coordinates Analysis (PCoA) of the Bray-Curtis index with comparative analysis between cases and controls performed within QIIME 2. Sequencing of all cases and controls revealed clear separation with strong beta diversity between oral and nasal microbial communities (p < 0.001). Microbial composition differed between cases and controls in both oral and nasal samples. The top three oral microbes identified as different compared to controls included Burkholderiales (7.41 log2fold change, p = 3.29E-05), Actinomyceataceae (6.22 log2fold change, p = 3.73E-06) and Gemella (5.28 log2fold change, p = 0.0002). The top three nasal microbes identified as different compared to controls included Rothia (13.6 log2fold change, p = 3.63E-18), Actinobacteria (10.29 log2fold change, p = 9.81E-10) and Propionibacteriales (8.73 log2fold change, p = 6.74E-09). These relative shifts in communities of bacteria detected in newly diagnosed neovascular AMD patients may suggest additional mechanistic links in disease pathogenesis.
Subject terms: Applied microbiology, Metagenomics, Retinal diseases
Introduction
Chronic inflammatory diseases including peptic ulcer disease, inflammatory bowel disease, arthritis and coronary artery disease have been linked to changes in the local gut and oral microbiome1–3. Microbes have been implicated in the induction of local inflammation, leading to increased mucosal permeability, thereby allowing inflammogenic material direct access to the systemic circulation. This material up-regulates immune mediators at distal sites, inducing local inflammatory pathways in end organs. Age-related macular degeneration (AMD) is considered a disease of chronic inflammation. Amongst the numerous factors implicated in the pathogenesis of AMD, oxidative stress and inflammation are two key themes. Consistent with these paradigms, inflammatory proteins, complement by-products and other immune-associated responses are implicated in the formation and progression of choroidal neovascularization, the form AMD associated with the majority of vision loss experienced by patients4.
The aberrant immune response in AMD patients is at least partially genetically mediated by polymorphisms at two genetic loci, complement factor H (CFH) and age-related maculopathy susceptibility 2 (ARMS2), both of which have been strongly implicated in disease initiation and progression5–7. Gene-environment interactions have also been implicated in the pathogenesis of AMD. Patients with the CFH gene risk variant also exposed to Chlamydia pneumonia (as measured by titres of antibodies against C. pneumonia) have been found to be at increased risk of AMD disease progression8. The role of microbes in AMD is in its infancy, but recent studies have shed light on particular gut and pharyngeal microbes present in patients with AMD, suggesting the diversity in microbial composition as a potential contributor to AMD pathophysiology9,10. To our knowledge, two studies have sought to characterize the microbiome in clinical cases of AMD. Zinkernagel, M. S. et al. sequenced the gut metagenome using fecal samples from 12 patients with recent signs of neovascular AMD in comparison to 11 controls9. The investigators found an increased abundance of Ruminococcus torques, Oscillibacter, Anaerotruncus and Eubacterium ventriosum9. Ho, E. X. P. et al. examined the pharyngeal microbiome in 245 AMD cases and 368 controls. Gemella was present in a higher abundance of AMD cases while Prevotella was reduced10.
Neovascular AMD represents a discrete event in the pathogenesis of disease and a time point where unique biological pathways can be sampled experimentally. Considering the etiological agent responsible for the initiation of neovascular AMD remains unknown, it seems plausible that microbial-host interactions may play a role. Therefore, we hypothesized that patients with newly diagnosed neovascular AMD are associated with a distinct oral and nasal microbiome. The diverse microbial environment surrounding the eye, namely the nose and mouth, represents a unique region where microbial diversity could lead to distant chronic inflammatory changes in the eye.
Results
Characterizing oral and nasal bacterial communities in neovascular AMD
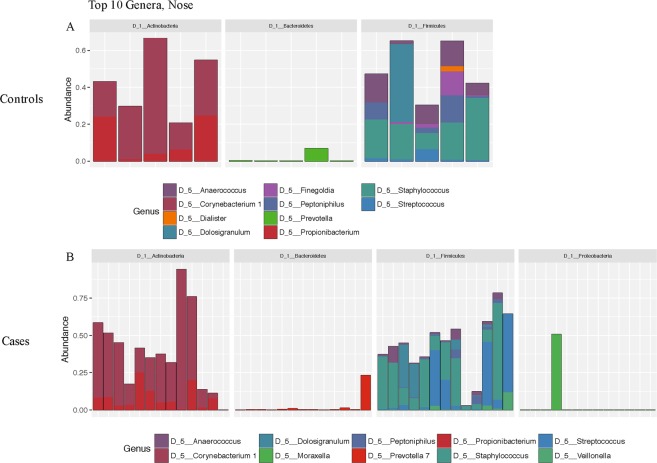
In order to determine whether oral and nasal microbiota were associated with newly developed neovascular AMD, 16S rRNA sequences were compared between 13 treatment naïve, newly diagnosed cases of neovascular AMD and 5 controls without of any evidence of retinal disease. Demographic data of study participants are presented in Table 1. Cases and controls did not differ with respect to gender, smoking status, last cigarette inhaled and antibiotic use (Table 1). Principle coordinates analysis plots of Bray-Curtis distances was plotted for all oral and nasal samples analyzed. Axes 1 and 2, representing 42.2% and 13.1% of the variance, respectively, are displayed in Fig. 1. The difference between all sampled oral and nasal microbiota was statistically significant (P = 0.001). The bacterial communities clustered together based on site sampled, with nasal communities more similar to each other than oral. Oral communities of cases and controls appeared to cluster together. Greater variance was observed within all nasal samples sequences in both cases and controls (Fig. 1). Oral and nasal microbiome composition for cases and controls as well as their relative abundances for the top ten most abundant microbes from major taxonomic groups is presented in Figs. 2 and 3.
Table 1.
Demographics table of cases and controls.
| Demographics | |||
|---|---|---|---|
| Cases | Controls | ||
| No. of Participants | 13 | 5 | |
| Age | <59 | 0 | 1 |
| 60–69 | 2 | 1 | |
| 70–79 | 4 | 1 | |
| 80–89 | 6 | 2 | |
| >89 | 1 | 0 | |
| Gender (%) | Male | 6 (46) | 2 (40) |
| Female | 7 (54) | 3 (60) | |
| Current Smoker (%) | Yes | 2 (15) | 1 (20) |
| No | 11 (85) | 4 (80) | |
| Last Cigarette | 1–2 hours | 1 | 0 |
| 2–4 hours | 1 | 1 | |
| Current Antibiotic Use | Yes | 0 | 0 |
| No | 13 | 5 | |
| Past Antibiotic Use (%) | 1–2 months | 1 (8) | 0 (0) |
| 2–4 months | 1 (8) | 0 (0) | |
| 4–6 months | 0 (0) | 0 (0) | |
| 6–12 months | 2 (15) | 1 (20) | |
| >12 months | 9 (69) | 4 (80) | |
| Current cold/runny nose (%) | Yes | 2 (15) | 1 (20) |
| No | 11 (85) | 4 (80) | |
| Use of nasal sprays (%) | Yes | 1 (8) | 1 (20) |
| No | 12 (92) | 4 (80) | |
Figure 1.
Principal coordinate analysis (PCoA) plot with Bray-Curtis dissimilarity of all nose and mouth samples. Nose samples clustered separately from mouth samples. Blue symbols denote controls. Red symbols denote cases. Circle symbols denote mouth samples. Triangle symbols denote nasal samples. Ellipses represent 95% confidence intervals around the centroid of cases and controls respectively. Clustering of cases and showed shifts between cases and controls in both nose and mouth sites.
Figure 2.
Relative abundance of the top ten bacterial communities generated by 16S rRNA sequencing of the nasal cavity swabs in (A) controls and (B) cases. Sequences were classified as described in the methods section.
Figure 3.
Relative abundance of the top ten bacterial communities generated by 16S rRNA sequencing of the oral cavity swabs in (A) controls and (B) cases. Sequences were classified as described in the methods section.
Comparison of microbial composition between cases and controls
Log2fold differences in the relative abundance of microbial taxa of cases and controls for oral and nasal communities is presented in Table 2. Eight nasal microbial communities showed a statistical difference between cases and controls. Nasal streptococcus showed the highest relative shift in abundance in cases as compared to controls (3.1 log2 fold increase (P < 0.01). Nasal Burkholderiales was not relatively abundant, but increased 7.4 log2 fold (P < 3.29E-05) over controls. Actinomycetaceae (p = 3.73E-06), Gemella (p = 0.0002), Proteobacteria (p = 0.004), Actinomyces (p = 0.002) and Veillonella (p = 0.005) all were significantly higher in cases as compared to controls (Table 2). Members of the clostridia class were 4-fold higher in controls as compared to cases (p = 0.007). In the oral mucosa, 15 of the detected microbial communities were statistically different between cases and controls (Table 2). Oral Propionibacteriales and Rothia increased by 8.6 (p = 6.74E-09) and 13.6 log2 fold (p = 3.63E-18) respectively over controls. Staphylococcus and Cornyebacteriaceae in the mouth had a 7.5 (p = 6.96E-05) and 7.2 (p = 2.33E-05) log2fold increase respectively over controls. Fusobacterium (p = 1.00E-10) and Bacilli (p = 0.007) were statistically higher in controls as compared to cases.
Table 2.
Log2fold change in the relative abundance of operational taxonomic units of cases over controls.
| Nasal cases as compared to controls | |||
|---|---|---|---|
| Log2FC | P value | Taxonomy | Taxonomic Rank |
| −4.16 | 0.007 | Clostridia | Class |
| 2.92 | 0.005 | Veillonella | Species |
| 3.10 | 0.011 | Streptococcus | Species |
| 3.14 | 0.002 | Actinomyces | Species |
| 4.75 | 0.004 | Proteobacteria | Family |
| 5.28 | 0.0002 | Gemella | Genus |
| 6.23 | 3.73E-06 | Actinomycetaceae | Family |
| 7.42 | 3.29E-05 | Burkholderiales | Order |
| Oral cases as compared to controls | |||
| −10.53 | 1.00E-10 | Fusobacterium | Genus |
| −3.91 | 0.007 | Bacilli | Class |
| 2.20 | 0.032 | Actinomyces | Species |
| 2.73 | 0.022 | Prevotella | Genus |
| 4.42 | 0.007 | Anaerococcus | Species |
| 5.27 | 7.97E-05 | Finegoldia | species |
| 6.04 | 0.0004 | Clostrida | Phylum |
| 6.30 | 6.23E-05 | Propionibacterium | Species |
| 6.57 | 5.87E-05 | Campylobactor | Species |
| 7.16 | 2.33E-05 | Corynebacteriaceae | Genus |
| 7.36 | 4.31E-06 | Peptoniphilus | Genus |
| 7.51 | 6.96E-06 | Staphylococcus | Species |
| 8.73 | 6.74E-09 | Propionibacteriales | Family |
| 10.29 | 9.81E-10 | Actinobacteria | Phylum |
| 13.57 | 3.63E-18 | Rothia | Genus |
Abbreviations: Fold change (FC). Positive log2 fold change is relative abundance in cases compared to controls.
Discussion
In this brief report, we characterized local shifts in the oral and nasal microbiome in patients with newly diagnosed neovascular AMD compared to controls without retinal pathology. The nasal mucosa of cases showed a shift in the dominant species, with streptococcus being most prevalent. Burkholderiales, Gemella and Actinomycetaceae showed the greatest fold change compared to controls. In the oral cavity, cases showed a shift in a composition of microbial communities, with members of the Actinobacteria phyla being the greatest: Rothia (genus), Proprionbacteriaceae (family), Corynebacteriaceae (genus). Considering the pathogenesis of AMD is partly regulated by the innate immune system, it is therefore possible that local changes in microbial composition are driving the up-regulation of pro-inflammatory pathways at distant organ sites, including the choroid/RPE complex1,2,11.
Microbiota trends identified in this report in the context of the published literature are intriguing. Rothia, Proprionbacteriaceae and Corynebacterium, have all been found simultaneously within the oral and atherosclerotic plaques of patients with coronary artery disease3. Considering the finding of characteristic pathogenic oral microbes in patients with newly diagnosed neovascular AMD, these bacteria may represent pathogenic species linked to promoting inflammation. Further, 16S rRNA of Gemella and Streptococcus are part of the pharyngeal microbiome recently described in late AMD patients as opposed to early AMD or controls10. Our report confirms the finding of Gemella and Streptococus as part of the nasal microbiome in newly diagnosed cases of neovascular AMD. Taken together, these species may hold pathogenic links to AMD progression and shed light onto previously unrecognized areas of research.
Connections have been established between the microbiome, metabolism and various chronic inflammatory diseases. Helicobacter pylori infection and peptic ulcer disease established the prototypical relationship between infection and chronic disease2. Mycobacterium avium species have been strongly linked to patients with Crohn’s disease1. Gut microbes can promote the production of pro-atherogenic metabolites such as trimethylamine-N-oxide12. In addition, alterations in a host’s metabolism have been shown to shift the gut microbiome and predispose mice to choroidal neovascularization13. Therefore, a role for bacteria in AMD should not be surprising, but its complex nature requires more intensive and controlled research.
Cumulative exposure to pathogens, or pathogen burden, has been implicated in the pathogenesis of chronic inflammatory diseases. In atherosclerosis, the amount of bacterial DNA has been linked to arterial inflammation and plaque stability14. In ophthalmology, overproduction of lipopolysaccharides, a known initiator of inflammation, has recently been implicated in neurodegeration and glaucoma15. In light of these new areas of research, shifts in an individuals’ oral or nasal microbiome may have significant pathogenic consequences. A causal link has yet to be clearly established, but mechanistically, this may occur either from direct transmission of bacteria or the dissemination of pro-inflammatory cytokines from focal inflammation within the oral or nasal cavities. Considering the choroid receives the greatest blood flow per unit tissue in the entire body, it is plausible either microbes or inflammatory products circulate in this vascular bed and the area with the greatest metabolism, oxidative stress and cellular turnover would be most susceptible (i.e the macula). Additional studies are required to further elucidate whether bacterial DNA is present in neovascular membranes or within drusen of eyes with AMD.
The generalizability of this study remains to be defined. The small number of participants, sequencing depth and number of comparisons may allow for spurious associations to be made. Further, oral and nasal microbiomes may be transiently affected by daily routines and dietary practices, that may alter microbial relative abundances and species leading to sampling error. To better understand the role of microbes in newly diagnosed neovascular AMD, a greater number of intra- and inter-day comparisons need to be made, but as well in non-neovascular AMD patients, AMD mimickers, other retinal diseases and non-retinal disease controls. Co-morbid disease, medication use, dietary habits and numerous other factors that were not controlled for may confound our findings.
In conclusion, newly diagnosed neovascular AMD in this small case series is accompanied by a shift in nasal and oral microbiota composition which may be associated with disease pathogenesis. Future studies are needed to elucidate the mechanism of this shift in microbial communities and aim to modulate it as a treatment modality for patients.
Methods
This case-control study was approved by the institutional ethics committee (Queen’s University Health Sciences) in accordance to the tenets of the Declaration of Helsinki. Oral and written informed consent was obtained from all participants prior to the collection of demographic information or patient samples.
Patient recruitment
Treatment-naïve patients with newly diagnosed neovascular AMD were recruited as cases. All cases with neovascular AMD were screened by an ophthalmologist with subspecialty training in retina. Neovascular AMD was confirmed on clinical biomicroscopic examination, spectral domain-optical coherence tomography and intravenous fluorescein angiography. Patients with geographic atrophy or exclusive dry AMD were excluded from study participation. Controls consisted of patients presenting for cataract extraction. Controls were excluded if there was a past history of retinal co-morbidity. Cases and controls were excluded if they were actively taking antibiotics. All participants were from single family dwellings.
Patient demographics
Age, gender, current smoking status, last cigarette consumed, current or past use of antibiotics, current cold or runny nose status and use of nasal sprays were all collected from consented participants.
Biospecimen collection
Swabs of buccal and nasal mucosa were taken in all participants. Buccal mucosa was swabbed using standard sterile uBiome (CA, USA) RNA/DNA free swabs. To standardize swabbing, the involved eye in cases or controls was used to designate which side of the nose or mouth to swab. Swabbing occurred after 1300 hours in all cases and controls. The buccal mucosa was swabbed with a forward and back motion for 30 seconds. Swabs were discarded if any contact with dental material or skin was made. For nasal swabs, the anterior nares were swabbed in a rotating fashion for 30 seconds. Nasal swabs were discarded if any nostril or upper lip skin was contacted. Buccal and Nasal mucosa were swabbed by a trained technician using a standardized procedure. All swabs were placed into a uBiome (CA, USA) storage container at −20 °C until further processing. Storage containers were barcoded without patient or disease identifying information. Sequencing and taxonomic annotation methods were performed blinded to disease status.
16S rRNA gene sequencing
16S rRNA sequencing was performed by uBiome (CA, USA) and performed according to previously published methodology outlined below. Oral and nasal samples were collected by study investigators blinded by patient disease status. Participants were collected using commercially available uBiome microbiome sampling kits consisting of sterile swabs, which follow the requirements by the NIH Human Microbiome Project16. DNA was extracted in a class 1000 clean room by a guanidine thiocyanate silica column-based purification method using a liquid-handling robot17,18. PCR amplification of the 16S rRNA genes was performed with universal primers amplifying the V4 variable region (515F: GTGCCAGCMGCCGCGGTAA and 806R: GGACTACHVGGGTWTCTAAT)19. In addition, the primers contained Illumina tags and barcodes. Samples were barcoded with a unique combination of forward and reverse indexes allowing for simultaneous processing of multiple samples. PCR products were pooled, column-purified, and size-selected through microfluidic DNA fractionation20. Consolidated libraries were quantified by quantitative real-time PCR using the Kapa Bio-Rad iCycler qPCR kit on a BioRad MyiQ before loading into the sequencer. Sequencing was performed in a pair-end modality on the Illumina NextSeq500 platform rendering 2 × 150 bp pair-end sequences.
Taxonomic annotation and reference database generation
Demultiplexing of samples was performed using Illumina’s BCL2FASTQ algorithm. Reads were filtered using an average Q-score >30 as previously published21 The most abundant sequence per cluster was considered the real and assigned the count of all reads in the cluster. The remainder were considered to contain errors as a by-product of sequencing. Chimera removal was performed on the representative reads from all clusters using the VSEARCH algorithm22.
FASTQ files
16S amplicons from each sample were individually barcoded and sequenced in multiplex in the NextSeq500 platform in a 150 bp paired-end modality. Raw data was demultiplexed, and the forward and reverse reads obtained in each of the 4 lanes per sample were filtered using the following criteria: (1) forward and reverse reads in a pair must have an average Q-score >30, (2) primers, and any leading random nucleotides are trimmed, and forward reads are capped at 125 bp and reverse reads are capped at 124 bp, (3) forward and reverse reads of each pair are appended, and sequences with more than 8 consecutive same nucleotide repeats are discarded, (4) the remaining sequences are clustered using a distance of 1 nucleotide using the Swarm algorithm, and the most abundant sequence per cluster is considered the representative of the cluster and assigned a count corresponding to the sum of sequences that compose the cluster, (5) a chimera removal using these centroid representative sequences is performed using the VSEARCH uchime_denovo algorithm21,22. Singletons that remain after chimera removal are also discarded, (6) both forward and reverse reads that match with at least 77% sequence identity to the same sequence in version 123 of the SILVA database are assumed to be 16S sequences23.
Taxonomy annotation table
The most abundant forward-reverse read pair per Swarm cluster is assigned taxonomic annotation according to the following similarity thresholds: >97% sequence identity, the reads are annotated to the same species of the hit in SILVA; >95% sequence identity, the sequence is annotated to the same genus of the hit in SILVA; >90% sequence identity, the sequence is annotated to the same family of the hit in SILVA; >85% sequence identity, the sequence is annotated to the same order of the hit in SILVA; >80% sequence identity, the sequence is annotated to the same class of the hit in SILVA; >77% sequence identity, the sequence is annotated to the same phylum of the hit in SILVA
Sequence processing, feature table construction and analysis
Sequencing reads were processed by Metagenom Bio (Toronto, Canada). Raw 16S sequence data were collapsed into a count (abundance) table, traditionally called an Operational Taxnomic Unit (OTU) table or feature table. Briefly, sequences were processed (filtered, assembled, collapsed into sequence variants) using the dada2 plugin within QIIME 224,25. This workflow de-noises Illumina sequences and attempts to collapse reads into variants representing the original amplicon sequence and its abundance. Taxonomic classification of representative sequences was performed using a scikit-learn naive Bayesian classifier trained against SILVA v.128 trimmed to the amplicon region. Feature table (OTU table) normalization was performed through rarefication (subsampling to equal depth)23.
Statistical analysis of the microbiome
Distinct microbial community characterization was achieved using Principal Coordinates Analysis (PCoA) of the Bray-Curtis index. Comparative analysis between cases and controls was performed with the DESeq. 2 plugin within QIIME. The difference in abundance of a given bacterial taxon between cases and controls was quantified as the log2 fold change in counts for the corresponding OTU. Relative abundance of a given taxon across all samples was calculated as mean count for the corresponding OTU divided by the sum of the mean counts of all OTUs with counts greater than 1000; this threshold was chosen to exclude noise related to sampling depth.
Acknowledgements
We would like to acknowledge Metagenome Bio Inc. for their assistance in the methodological interpretation and statistical analysis of the 16S rRNA data. We would like to acknowledge uBiome Inc for their involvement in the 16S rRNA methodology and sequencing of participant samples. Jacob Rullo was funded by a Research Trainee Award by Physicians Services Incorporated.
Author contributions
J.R. designed the study, collected samples, analyzed data and prepared the manuscript. P.M.F. prepared the manuscript. M.Q. analyzed data, prepared the manuscript. N.S. collected samples. S.B. collected samples. I.I. designed the study. S.S. designed the study and prepared the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Timms VJ, Daskalopoulos G, Mitchell HM, Neilan BA. The association of mycobacterium avium subsp. paratuberculosis with inflammatory bowel disease. PLoS One. 2016;11(2):e0148731. doi: 10.1371/journal.pone.0148731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang JQ, Sridhar S, Hunt RH. Role of Helicobacter pylori infection and non-steroidal anti-inflammatory drugs in peptic-ulcer disease: A meta-analysis. Lancet. 2002;359(9300):14–22. doi: 10.1016/S0140-6736(02)07273-2. [DOI] [PubMed] [Google Scholar]
- 3.Koren O, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc. Natl Acad. Sci. 2011;108(Supplement_1):4592–8. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman HR, Chan CC, Ferris FL, Chew EY. Age-related macular degeneration. Lancet. 2008;372(9652):1835–45.. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vavvas DG, et al. CFH and ARMS2 genetic risk determines progression to neovascular age-related macular degeneration after antioxidant and zinc supplementation. Proc. Natl Acad. Sci. USA. 2018;115(4):E696–704. doi: 10.1073/pnas.1718059115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jabbarpoor Bonyadi MH, Yaseri M, Nikkhah H, Bonyadi M, Soheilian M. Association of risk genotypes of ARMS2/LOC387715 A69S and CFH Y402H with age-related macular degeneration with and without reticular pseudodrusen: a meta-analysis. Acta Ophthalmol. 2018;96(2):e105–10. doi: 10.1111/aos.13494. [DOI] [PubMed] [Google Scholar]
- 7.DeAngelis MM, et al. Genetics of age-related macular degeneration (AMD) Hum. Mol. Genet. 2017;26(R1):R45–50. doi: 10.1093/hmg/ddx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baird PN, et al. Gene-environment interaction in progression of AMD: The CFH gene, smoking and exposure to chronic infection. Hum. Mol. Genet. 2008;17(9):1299–305. doi: 10.1093/hmg/ddn018. [DOI] [PubMed] [Google Scholar]
- 9.Zinkernagel MS, et al. Association of the intestinal microbiome with the development of neovascular age-related macular degeneration. Sci. Rep. 2017;7(1):40826. doi: 10.1038/srep40826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho EXP, et al. Human pharyngeal microbiota in age-related macular degeneration. PLoS One. 2018;13(8):e0201768. doi: 10.1371/journal.pone.0201768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Sci. 2005;308(5720):385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koeth RA, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013;19(5):576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Andriessen EM, et al. Gut microbiota influences pathological angiogenesis in obesity‐driven choroidal neovascularization. EMBO Mol. Med. 2016;8(12):1366–79.. doi: 10.15252/emmm.201606531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein SE, et al. Infection and atherosclerosis: potential roles of pathogen burden and molecular mimicry. Arterioscler. Thromb. Vasc. Biol. 2000;20(6):1417–20. doi: 10.1161/01.ATV.20.6.1417. [DOI] [PubMed] [Google Scholar]
- 15.Astafurov K, et al. Oral microbiome link to neurodegeneration in glaucoma. PLoS One. 2014;9(9):e104416. doi: 10.1371/journal.pone.0104416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mcinnes P. Manual of Procedures for Human Microbiome Project Core Microbiome Sampling Protocol A HMP Protocol # 07-001 (2010).
- 17.Hummel W, Kula MR. Simple method for small-scale disruption of bacteria and yeasts. J. Microbiol. Methods. 1989;9(3):201–9. doi: 10.1016/0167-7012(89)90037-7. [DOI] [Google Scholar]
- 18.Cady NC, Stelick S, Batt CA. Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 2003;19(1):59–66. doi: 10.1016/S0956-5663(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 19.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl Acad. Sci. USA. 2011;108(Suppl(Supplement 1)):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minalla, A. R., Dubrow, R. & Bousse, L. J. Feasibility of high-resolution oligonucleotide separation on a microchip. In: Mastrangelo, C. H. and Becker, H., editors. International Society for Optics and Photonics, 90–7 (2001).
- 21.Mahé F, Rognes T, Quince C, de Vargas C, Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(D1):D590. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]