Abstract
The rapid development in sequencing technology is creating an increase in demand for largely intact DNA as starting material as very long strands of DNA are sequenced directly to generate reads that are thousands of bases long. Organisms with thick cell walls are difficult to lyse, often impacting both DNA recovery and quality. Consequently, most mycobacterial DNA extraction methods require bead-beating steps or toxic chemicals. Here we present an updated method that yields abundant, high quality genomic DNA from M. tuberculosis and diverse nontuberculous mycobacterial (NTM) species, in addition to complex biological communities from a variety of sources. This method eliminates the time-consuming phenol and chloroform extraction and ethanol precipitation steps, and high quality DNA from up to 96 samples can be extracted in about 2–3 h of hands-on time. This DNA is suitable for long and short read sequencing technologies as well as PCR and qPCR amplification.
Keywords: NTM, Phylogenomic, Outbreak, Next generation sequencing, Molecular diagnostics
1. Introduction
Infections caused by Mycobacterium tuberculosis complex (MTBC) continue to kill more than 1.2 million people each year and approximately 10 million people develop active tuberculosis (TB) disease each year [1]. Among the greatest concerns pertinent to TB disease is the rise of drug resistant strains including multidrug-resistant (MDR) and extensively drug-resistant (XDR) M. tuberculosis [2]. Nontuberculous mycobacteria are an emerging infectious disease threat [3]; they are innately resistant to many antibiotics [4] and pose an increasing challenge to human populations, including patients with cystic fibrosis [5]. The host characteristics that make particular individuals more susceptible to NTM are not fully understood, but immune system related genes and ciliary genes are likely to impart influence on susceptibility [6], [7], [8]. In order to rapidly identify and elucidate mechanisms of resistance in MTBC and NTM, to follow outbreak and transmission dynamics, to identify newly arising single nucleotide polymorphisms (SNPs) and insertions/deletions (INDELs), and to investigate mycobacterial plasmids, many researchers are implementing methods of mycobacterial whole genome sequencing (WGS) [5,[9], [10], [11], [12], [13]]. Long read WGS can generate sequence reads that are thousands of bases long, using platforms such as PacBio and Oxford Nanopore, and enables the characterization of plasmids and challenging repeat regions of MTB. These methods in particular depend on high quality, very long pieces of input DNA as starting material, as close to full-length chromosomal and plasmid templates as possible.
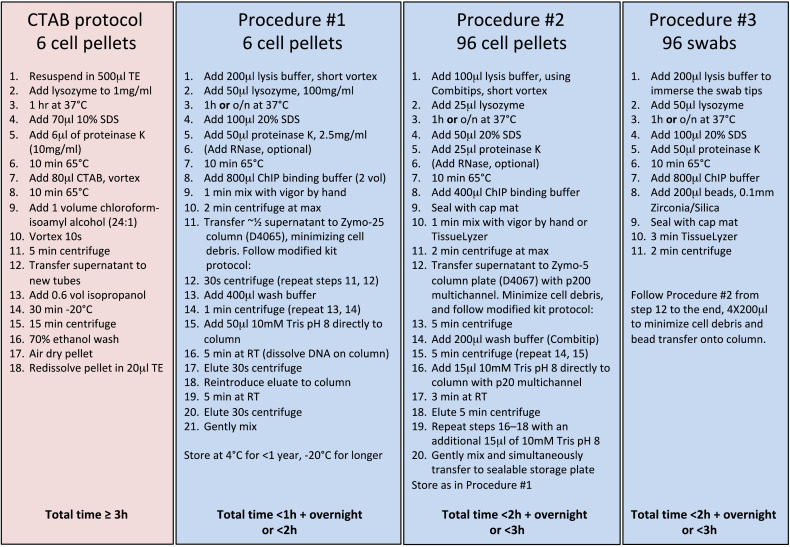
A number of factors play a role in the quality and quantity of extracted DNA in a given sample including growth time of the cultures (which when extended to months results in the presence of dead cells and likely degraded DNA), duration and temperature of heat kill when that is required, storage of pellets before extraction, and reagents and timing of incubations during the extraction itself ([14] and Matthias Merker, personal communication). Most current methods for recovering intact DNA from mycobacteria depend on chemical lysis, bead-beating, extraction of DNA using Cetyltrimethylammonium Bromide (CTAB) and/or phenol, both of which normally co-utilize chloroform along with salt and alcohol for precipitation of the final DNA, from which the nucleic acid is pelleted, dried and resuspended in an aqueous solvent [14,15]. We selected aspects of current methods and kits to generate a single enabling protocol that requires minimal hands-on time starting from the cell pellet for up to 96 samples with the use of multichannel pipettes, is scalable in both DNA quantity and sample number, flexible for sample type (including direct respiratory samples and environmental swabs), and avoids the use of the more noxious chemicals. We expressly optimized this protocol for the purpose of whole genomic sequencing, and have used it to prepare high quality genomic DNA for short read Illumina 2 × 250 bp and 2 × 300 bp sequencing of hundreds of NTM isolates [3,10] in addition to long read PacBio sequencing of NTM [16,17] and TB [18] genomes. We have also used genomic DNA from this protocol for MinION nanopore sequencing, yielding reads averaging about 6500 bp which, when assembled with Illumina reads, result in closed mycobacterial genomes (unpublished). Fig. 1 summarizes the steps and duration of this procedure relative to the CTAB protocol.
Fig. 1.
Comparison of and estimated times for four procedures. The left-most column is the CTAB procedure [15], often used by mycobacterial researchers for DNA extraction. The next three columns detail variations of the present protocol. Total time estimates are listed at the bottom of the figure. The first three columns begin at the step of bacterial cell pellets. The last column begins with swabs as described in the text.
2. Methods and results
2.1. Mycobacterial culture
Rapidly growing mycobacteria (RGM), including the Mycobacterium abscessus group of species, are grown from single colony in broth (Middlebrook 7H9) or on solid media (7H11) for seven to ten days at 37 °C to procure one to three loopfuls of cells to pellet and extract DNA, approximately equivalent to 50–100 μl in volume. Slowly growing mycobacteria (SGM), including M. tuberculosis, routinely require two to four weeks of incubation to achieve the desired biomass. For sequencing platforms that require only 1–10 ng of DNA, very little bacterial growth is necessary and enough cells may be obtained in much shorter incubation times. If additional cells are needed, as in the case of the microgram amounts of DNA required for PacBio sequencing, our preference is to increase the number of inoculated plates or broths as opposed to extending the growth time because the DNA preparations from older cultures appear to contain more degraded DNA, possibly due to the presence of dead and deteriorating cells. If heat killing of the bacteria is necessary, as is the case for biosafety level 3 (BSL3) organisms such as M. tuberculosis, a shorter and moderate incubation of 80 °C for 20 min is effective to kill the mycobacteria without degrading the DNA [19,20]. After removal of supernatant, the enriched cell pellet should be subjected to DNA extraction soon after or stored frozen until extraction begins.
2.2. Cell lysis
Cell pellets are immersed and briefly vortexed in 100 μl lysis buffer (15% sucrose, 0.05 M Tris-Cl pH 8.0, 0.05 M EDTA, pH 8.0, filtered and stored at room temperature [14]). The cell pellet is often waxy, adhering to the tube and to itself. Rather than losing cells with extensive pipetting, the pellet is allowed to remain in clumps. 25 μl of lysozyme is added (frozen powder aliquots, resuspended by vortex to 100 mg/ml in lysis buffer immediately before use) and samples are incubated overnight at 37 °C on an Eppendorf ThermoMixer® C at 500 rpm. A water bath or other incubator without agitation is also suitable. Lysozyme dismantles the cell wall peptidoglycans in this overnight step. Although we have obtained qualitatively higher yields with some samples with this overnight lysozyme step, the overnight incubation in lysozyme can be replaced with a 1 h lysozyme incubation at 37 °C under the following conditions: 1) DNA yield requirements are relatively minimal, i.e. 1–10 ng of genomic DNA needed for downstream processing, and 2) sample contains a substantial visible pellet of isogenic material rather than a mixed sample of different mycobacterial cell types that may lyse differentially. For large scale preparation of DNAs useful for sequencing with Illumina chemistry, for example, a one hour lysis is normally sufficient if starting with a pellet of isolated cells that is visible in a microfuge tube.
Following the lysozyme step, samples are removed from the incubator and two reagents are added (or three, if RNA removal is deemed necessary): 25 μl of 2.5 mg/ml proteinase K (resuspended by pipet in nuclease-free water and stored frozen at −20 °C in aliquots), 50 μl 20% SDS (made from solid to achieve this high concentration, solubilized overnight, filtered, and stored at room temperature), and 4 μl RNaseA/T1 if removal of RNA is desired (Thermo part EN0551). The samples are then incubated for 10 min at 65 °C. The high concentration of SDS is a critical step in the extraction [14], but would interfere with lysozyme activity. For this reason, this detergent step is temporally separated from and after the lysozyme step [14].
2.2.1. Cell lysis for environmental swabs, clinical specimens, and other non-isolate sample types
This protocol has been used successfully for DNA preparation from numerous complex samples such as sputum, bronchoalveolar lavage (BAL), and swabs from home dust samples. For these samples, a bead-beating step is added for recovery of fungal and plant DNA, and other DNA originating from difficult to lyse cells. In the case of cells in suspension, such as BAL, cells are pelleted and supernatant removed from the pellet, which is resuspended in lysis buffer as stated above. For sputum, 100–200 μl of sputum is used in place of the resuspended pellet, adding lysozyme directly to the sputum for the initial lysis step. Swabs must be made from synthetic, DNA-free material; we utilize double tip swabs in order to have a duplicate for culture-based experiments and microbiome analysis (BD BBL CultureSwab EZ sterile polyurethane, dual foam swab, part 220145.) One of the two swab tips is cleaved using an ethanol-wiped pet nail clipper directly into a 2 ml deepwell 96-well plate (Nunc™ Thermo part 278743), 200μl of lysis buffer is added and 50 μl lysozyme for overnight incubation. If 100 μl of lysis buffer is used, 100 μl of zirconium beads (0.1 mm Zirconia/Silica Beads, BioSpec part 11079101z) are added after the proteinase K and detergent step. Likewise, if 200 μl of lysis buffer is used, 200 μl of beads are added after the proteinase K and detergent step. If samples are in individual tubes, beads are dispensed by hand into separate microfuge tubes and then added. For the 96-well format, we use a 96-well powder dispenser tailored for us by LabTIE (Leiden, NL), and seal the plate with a cap mat (Nunc™ Thermo part 276000). ChIP binding buffer from the applicable Zymo kit (see below) is added using a Combitip repeat pipettor (Eppendorf) before bead beating, which is done on a Qiagen TissueLyzer II for 3 min at 30 Hz. The tubes or plate are centrifuged for 2 min at maximum speed to pellet cell debris. All plate centrifugation steps are done in a Sorvall ST16. The supernatant is then transferred to the column using a p200 multichannel pipette to transfer 800 μl (see below), and the rest of the procedure is followed according to the manufacturer or modified as described below.
2.2.2. Additional recommendations for reagents and consumables
Lysis buffer is mixed and filtered into 50 ml aliquots at room temperature for 2 months or 250 ml to 500 ml filtered bottles at 4 °C for up to one year, being careful to dispense by pouring rather than pipetting from the bottle or tube to minimize the chance of contamination. Lysozyme solid is stored at −20 °C in aliquots of practical size; for example, 32 mg of lysozyme in a 1.5 ml microfuge tube can be resuspended by vortex in 320 μl of lysis buffer to extract 6 large cell pellets using 50 μl apiece or 12 smaller cell pellets using 25 μl apiece. All reagents are available in bulk from Sigma, Fisher Scientific, VWR, and other major chemical distributors. Hundreds to thousands of samples can be processed with single purchases of these items in solid or bulk when stored properly. For 96 well plate format, multichannel pipetting and repeat pipettors (Eppendorf Combitip) are used with reagent reservoirs and strip tubes for the steps throughout the protocol. The major expense for our method is the column clean up. In the 96 well column format that yields a maximum of 5 μg per column, the current cost is $1.30 per sample. Because the reagents can be purchased in bulk, we estimate that the cost is well under $2.00 per sample for the entire DNA extraction, including plastic consumables and reagents.
2.3. Scalable DNA extraction and cleanup
The next step transitions to a commercial kit for column purification of the DNA, completely replacing the phenol/chloroform extraction and ethanol precipitation steps. We use the Zymo Genomic DNA Clean and Concentrator kits. 400 μl of the Zymo ChIP DNA binding buffer is added and the samples are mixed vigorously by hand for at least 60 s. The cell debris is pelleted for 2 min at maximum in a microfuge and the supernatants transferred to the Zymo column. Individual kits offer a variety of options for scalability. For large quantities of DNA, we double all of the volumes stated above and use the tube format Genomic DNA Clean and Concentrator-25. For smaller preparations and to process many samples at one time, we use the Genomic DNA Clean and Concentrator-5 columns in 96 well plate format for up to 96 samples at a time. For fewer than 96 samples, the wells/columns are covered with a plate seal to prevent settling of dust, and the seal is cut to expose the desired number of wells/columns. The kits are used according to the manufacturer's protocol with two modifications. First, instead of the kit's EDTA-containing elution buffer, 10 mM Tris, pH7-8 is normally used for the final elution, particularly for samples that are to be sequenced with PacBio chemistry, since even small quantities of EDTA can impede downstream processes included in sequencing library preparation. Nuclease-free water with pH above 6 can also be used for the elution. Second, we perform an additional elution step in order to maximize DNA recovery. We measure approximately 50% additional DNA on the second elution. If highly concentrated DNA is desired, the first eluate is reintroduced onto the column, incubated per protocol again, and re-eluted. Large preparations and less concentrated DNA samples can be obtained by adding a second portion of elution buffer rather than re-eluting in the same. We normally elute in two 15μl aliquots (Figure 1, Procedure #2 step 19), using a multichannel p20 to carefully dispense directly onto the column. The room temperature column incubation in elution buffer is also critical for success.
2.4. DNA quantitation and quality assessment
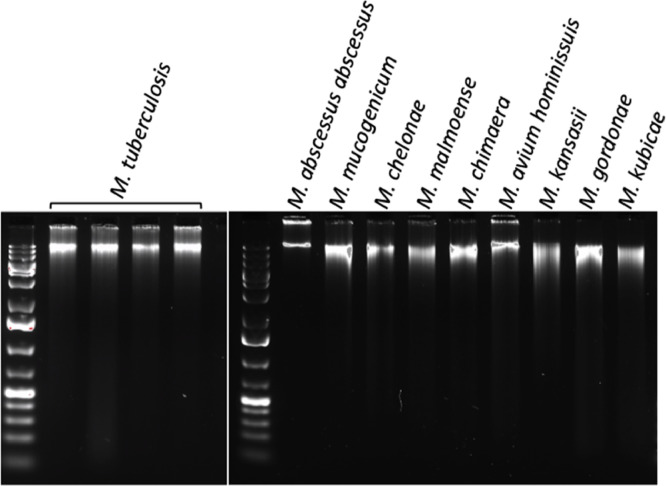
The eluted DNAs are gently pipet-mixed to assure homogeneity before fluorescent (Qubit) quantitation. Freezing and thawing of the DNA can result in fragmentation, so DNA is stored at 4 °C during repeated use and then −20 °C for archive storage. From a large cell pellet (equivalent to 100 μl) we recover yields ranging from 100–200 ng of DNA up to 8–10 μg. In almost every case, if a cell pellet is visible, we recover enough DNA for successful Illumina library preparation using the Nextera DNA Flex Library Kit starting with 10 ng. Absorbance ratios of 260/280 reproducibly obtain a median above 1.6 according to NanoDrop (Thermo.) An agarose gel can be used to evaluate sample quality (Fig. 2). The presence of smearing in this gel is minimal, and all bands indicate the dominant presence of long, intact chromosomal genomic DNA. Of the samples shown on the gel, seven of the genomic DNA preparations, namely M. tuberculosis, M. mucogenicum, M. chelonae, M. chimaera, M. avium hominissuis, M. gordonae, and M. kubicae have been used to generate very long sequence reads using either PacBio or Nanopore chemistries ([16], [17], [18] and unpublished results).
Fig. 2.
Quality assessment of mycobacterial genomic DNA. DNA was extracted from four strains of M. tuberculosis and nine NTM species as indicated using the present protocol and assessed on two distinct 0.8% agarose gels in 1X TAE stained with GelRed, 1 kb Plus ladder (Thermo Scientific), with separation for 90 min at 80 V. Quantities range from 15 to 36 ng per lane. The large DNA migrates with the highest marker band at about 20 kbases.
3. Discussion
The described protocol represents several improvements to current widely-used mycobacterial DNA protocols: we eliminate a number of noxious chemicals, reduce the hands-on time and number of steps needed, and increase reproducibility among samples and researchers by replacing extraction and alcohol precipitation with a column. We also reduce the reliance on specialized equipment, including bead beating equipment and reagents, and the corresponding time needed for the often-laborious steps such as bead dispensing. As the fields of mycobacterial genomic research and mycobacterial molecular diagnostics [11] continue to advance, it has become even more essential to have an optimized, streamlined, nontoxic, time-efficient protocol in place in order to purify high quality NTM and MTB DNA in a timely manner for short read and long read next generation genomic sequencing. This protocol addresses these needs, as we have demonstrated in the use of this protocol to prepare the DNA from hundreds of mycobacterial samples for short read and long read genomic sequencing [3,10,17,18].
The steps in this procedure are also amenable to scaling up sample numbers, allowing DNA to be prepared in plate format. For instance, our current implementations include preparation in 96 well plate format to facilitate Illumina HiSeq runs of 288 multiplexed, taxonomically diverse mycobacterial samples (3 × 96 well plates) per HiSeq sequencing run. This method has been used successfully to prepare DNA from diverse mycobacteria including M. tuberculosis, M. abscessus, M. avium, M. intracellulare, M. chimaera, M. chelonae and many other NTM species, making it a robust protocol for diverse mycobacterial DNA preparation for genomic sequencing.
Another major advantage of this protocol is that it eliminates the need for a phenol: chloroform extraction and alcohol precipitation, which have long been standard in mycobacterial DNA work. Our method demonstrates that these steps are not necessary, eliminating exposure to noxious reagents like phenol and chloroform. This improvement increases the safety of the lab environment, but also enables scaling up to larger sample numbers that can be processed in a time-efficient manner. There is no nucleic acid pelleting, drying, and resuspension, which means that there is no opportunity for loss of such a pellet during the procedure. Instead, the DNA is eluted directly from the column into solution at an appropriate concentration. In addition to the safety advantages, elimination of phenol/chloroform extraction and ethanol precipitation and resuspension results in much greater reproducibility among researchers.
It should be noted that we and others have observed variability, sometimes extreme variability, in DNA yield from samples extracted in parallel and from equally large cell pellets. For example, in one batch we extracted two M. abscessus isolates, one strain of which demonstrated a smooth colony morphology, and the other rough, but both derived from the same patient sputum sample. One of the rough pellets yielded 1.4 μg of DNA and the smooth pellet yielded 11.8 ng, in spite of approximately equally large pellet sizes. Both extractions yielded intact DNA, and both were successfully used for Illumina WGS. Similar disparity in yields was obtained in three successive DNA extractions from the same strains, indicating highly reproducible DNA recovery that was strain-specific.
Acceleration of the diagnostic process for mycobacteria could dramatically improve treatment of these diseases and infections. Methods have already been developed to generate informative whole genome sequence data from Mycobacteria Growth Indicator Tube (MGIT) cultures [21] and from direct respiratory specimens, the latter technique being most impeded by the dominant presence of human and other eukaryotic DNA, which was overcome with an effective eukaryotic DNA depletion before mycobacterial DNA extraction [22]. Another group bypassed the extensive time to regrow these very slowly growing bacteria by extracting DNA from frozen glycerol stocks [23]. In all of these cases, the steps beginning with overnight (or one hour) lysis in this paper should be capable of replacing the steps that follow human DNA depletion from the point of bacterial cell pelleting. Furthermore, with an effective human DNA depletion and beginning with direct specimens, this method will allow scaling up in diagnostic and research laboratories to bypass the cultural decontamination step that kills up to 90% of the mycobacteria in a sample [24], in addition to eliminating the extensive growth time that follows.
In summary, this enabling protocol facilitates a straightforward and accelerated preparation of high quality mycobacterial DNA for short read and long read genomic sequencing, is amenable to scaling-up to plate format, increases reproducibility, and greatly reduces exposure to toxic chemicals in the preparation of the DNA.
Ethical statement
Samples used for development of these methods and reported herein have been removed from any links to their sources; therefore, institutional ethics committee approval is not required for this publication.
Author contributions
LEE optimized and evaluated the refinements of the method and ran the gels shown in Fig. 2. Both authors wrote, edited, and approved the manuscript.
CRediT authorship contribution statement
L. Elaine Epperson: Methodology, Validation, Writing - original draft, Writing - review & editing. Michael Strong: Supervision, Formal analysis, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Funding and Acknowledgments
We thank the Cystic Fibrosis Foundation (grant number NICK15R0) and National Science Foundation (grant number 1743587) for support, including funding to establish a cross-disciplinary, collaborative infrastructure. We gratefully acknowledge the National Jewish Health Mycobacteriology Laboratory and Jennifer Honda and her Laboratory, including Stephanie Dawrs, Grant Norton, and Ravleen Virdi for providing a wide range of carefully-grown mycobacterial species with which to refine this protocol. We thank the many researchers who published or shared their mycobacterial DNA preparation protocols, providing the pillars of the present protocol, including Stefan Niemann, Thomas Kohl, and Matthias Merker at Forschungszentrum Borstel in Germany. We thank David Durbin, Maha Farhat, Max Salfinger, Theresa Savidge, and Alyssa Sherwood for giving us access to slowly growing strains of M. tuberculosis, and the Colorado CF RDP for access to NTM strains. We thank Josh Hunkins, Vinicius Calado, Nabeeh Hasan, Rebecca Davidson, Sean Beagle, Jo Hendrix, and Sara Kammlade for ongoing genomic and collaborative discussions.
Contributor Information
L. Elaine Epperson, Email: eppersone@njhealth.org.
Michael Strong, Email: strongm@njhealth.org.
References
- 1.WHO . World Health Organization; Geneva: 2019. Global tuberculosis report 2019. [Google Scholar]
- 2.Sandgren A. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6(2):e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasan N.A. Genomic analysis of cardiac surgery-associated Mycobacterium chimaera infections, United States. Emerg Infect Dis. 2019;25(3):559–563. doi: 10.3201/eid2503.181282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davidson R.M. Genome sequencing of Mycobacterium abscessus isolates from patients in the United States and comparisons to globally diverse clinical strains. J Clin Microbiol. 2014;52(10):3573–3582. doi: 10.1128/JCM.01144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant J.M. Emergence and spread of a human-transmissible multidrug-resistant nontuberculous mycobacterium. Science. 2016;354(6313):751–757. doi: 10.1126/science.aaf8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipner E.M., Garcia B.J., Strong M. Network analysis of human genes influencing susceptibility to mycobacterial infections. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fowler C.J. Abnormal nasal nitric oxide production, ciliary beat frequency, and toll-like receptor response in pulmonary nontuberculous mycobacterial disease epithelium. Am J Respir Crit Care Med. 2013;187(12):1374–1381. doi: 10.1164/rccm.201212-2197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski E.P. Pulmonary nontuberculous mycobacterial infection. a multisystem, multigenic disease. Am J Respir Crit Care Med. 2015;192(5):618–628. doi: 10.1164/rccm.201502-0387OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caverly L.J., Spilker T., LiPuma J.J. Complete genome sequences of 17 rapidly growing nontuberculous mycobacterial strains. Genome Announc. 2016;4(5) doi: 10.1128/genomeA.01009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hasan N.A. 2019. Population genomics of nontuberculous mycobacteria recovered from United States cystic fibrosis patients. bioRxiv 663559. [Google Scholar]
- 11.Consortium C.R. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379(15):1403–1415. doi: 10.1056/NEJMoa1800474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedrizzi T. Genomic characterization of nontuberculous mycobacteria. Sci Rep. 2017;7:45258. doi: 10.1038/srep45258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohl T.A. Whole-genome-based Mycobacterium tuberculosis surveillance: a standardized, portable, and expandable approach. J Clin Microbiol. 2014;52(7):2479–2486. doi: 10.1128/JCM.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Käser M. Optimized DNA preparation from mycobacteria. Cold Spring Harb Protoc. 2010;2010(4):1–7. doi: 10.1101/pdb.prot5408. [DOI] [PubMed] [Google Scholar]
- 15.van Soolingen D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29(11):2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasan N.A. Complete genome sequence of Mycobacterium chimaera strain AH16. Genome Announc. 2016;4(6):1–2. doi: 10.1128/genomeA.01276-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao X. Complete genome sequence of Mycobacterium avium subsp. hominissuis strain H87 isolated from an indoor water sample. Genome Announc. 2017;5(16) doi: 10.1128/genomeA.00189-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vargas R. 2019. In-host population dynamics of M. tuberculosis during treatment failure. bioRxiv 726430. [Google Scholar]
- 19.van Embden J.D. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31(2):406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doig C. The efficacy of the heat killing of Mycobacterium tuberculosis. J Clin Pathol. 2002;55(10):778–779. doi: 10.1136/jcp.55.10.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Votintseva A.A. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53(4):1137–1143. doi: 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Votintseva A.A. Same-Day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55(5):1285–1298. doi: 10.1128/JCM.02483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjorn-Mortensen K. Direct DNA extraction from Mycobacterium tuberculosis frozen stocks as a reculture-independent approach to whole-genome sequencing. J Clin Microbiol. 2015;53(8):2716–2719. doi: 10.1128/JCM.00662-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diagnostic Standards and Classification of Tuberculosis in Adults and Children This official statement of the American thoracic society and the centers for disease control and prevention was adopted by the ATS board of directors, July 1999. this statement was endorsed by the council of the infectious disease society of America, September 1999. Am J Respir Crit Care Med. 2000;161(4 Pt 1):1376–1395. doi: 10.1164/ajrccm.161.4.16141. [DOI] [PubMed] [Google Scholar]