Abstract
Human cytomegalovirus (HCMV) components are often found in tumors, but the precise relationship between HCMV and cancer remains a matter of debate. Pro-tumor functions of HCMV were described in several studies, but an association between HCMV seropositivity and reduced cancer risk was also evidenced, presumably relying on recognition and killing of cancer cells by HCMV-induced lymphocytes. This study aimed at deciphering whether CMV influences cancer development in an immune-independent manner. Using immunodeficient mice, we showed that systemic infection with murine CMV (MCMV) inhibited the growth of murine carcinomas. Surprisingly, MCMV, but not HCMV, also reduced human colon carcinoma development in vivo. In vitro, both viruses infected human cancer cells. Expression of human interferon-β (IFN-β) and nuclear domain (ND10) were induced in MCMV-infected, but not in HCMV-infected human colon cancer cells. These results suggest a decreased capacity of MCMV to counteract intrinsic defenses in the human cellular host. Finally, immunodeficient mice receiving peri-tumoral MCMV therapy showed a reduction of human colon cancer cell growth, albeit no clinical sign of systemic virus dissemination was evidenced. Our study, which describes a selective advantage of MCMV over HCMV to control human colon cancer, could pave the way for the development of CMV-based therapies against cancer.
Keywords: mCMV, hCMV, Anti-tumor potential, Decreased host defense, Human Colon cancer, Cross-species, Virotherapy, IFN-B
Introduction
Cytomegaloviruses (CMVs) are large, enveloped, double-stranded DNA herpesviruses that establish a lifelong latent infection. Seroprevalence of human cytomegalovirus (HCMV) ranges from 50% to 100% in the general adult population. CMVs have co-evolved with their host following co-speciation resulting in highly species-specific viruses. Murine CMV (MCMV) shares a high degree of sequence homology and biology with HCMV but is unable to replicate efficiently in humans. CMVs’ restricted host range is not due to the absence of appropriate entry receptors on the cell surface but results from the inability of the virus to prevent apoptosis1 or to subvert host defense mechanisms2 in a distant host.
Primary CMV infection is usually subclinical thanks to robust intracellular and systemic defense mechanisms, which ultimately lead to the control of viral replication and establishment of latency. Type I interferons (IFNs) are produced as a first line of defense by stromal and endothelial cells and innate lymphocytes, upon sensing of CMV DNA by Toll-like receptors (TLRs), and more recently identified cyclic GMP-AMP synthase (cGAS) and interferon-inducible nucleoprotein (IFI) 16 intracellular DNA sensors.3, 4, 5, 6 Type I IFNs induce an antiviral state in their cellular host and possess indirect anti-pathogen activity via the induction of IFN-stimulated genes (ISGs). Among IFN-stimulated factors are the constitutively expressed cellular proteins that form the nuclear domain (ND10), a macromolecular complex that creates a condensed chromatin environment around the major immediate early (IE) promoter of HCMV, resulting in the inhibition of IE gene transcription and subsequent productive viral infection.7, 8, 9 Type I IFNs operate as autocrine and paracrine factors and orchestrate innate and adaptive immune responses.
The immune response against CMV relies on multiple and redundant immune effector functions from the innate and adaptive immune systems. While the acute phase of infection is dominated by the triptych natural killer and dendritic cell (DC-NK)-αβ T cell responses, long-term control of CMV is primarily attributed to αβ T cells, although CMV-reactive memory NK cells have been described more recently (reviewed in O’Sullivan et al.10). We also described that γδ T cells participate to the immune response against CMV in human and in mouse (reviewed in Khairallah et al.11).
The relationship between CMV and cancer has been investigated for decades but remains a matter of debate. In the 1970s, the group of Rapp reported the transformation of embryo lung fibroblasts upon in vitro infection with a clinical isolate of HCMV.12 However, the notion that HCMV could be oncogenic was superseded by the concept of oncomodulation,13 due to the reported controversies about the presence of HCMV in tumors.14, 15, 16 Supporting an oncomodulatory role of HCMV, several research groups have described an increased malignancy of human tumor cell lines infected by HCMV.17, 18, 19 More recently, the group of Herbein reconsidered the oncogenic potential of HCMV and showed that long-term culture of human mammary epithelial cells (HMEC) in presence of HCMV strain DB induced their transformation20 (reviewed in Herbein21). Concerning colorectal cancer, a pro-tumor role of HCMV has been put forward.22,23 However, HCMV may influence the outcome of colorectal cancer in an age-dependent manner. Indeed, the presence of HCMV in colorectal tumors was associated with shorter disease-free survival in ≥65-year-old patients24 and a favorable outcome in non-elderly patients.25
While a pro-tumor role of HCMV has been predominantly evoked, a recent report described an inhibitory role of HCMV on the development of human hepatocellular carcinoma xenografted in non-obese diabetic (NOD) scid gamma (NSG) mice.26 An anti-tumor role of CMV was also reported in mouse models, after systemic infection of MCMV in the case of a liver lymphoma27 and after intra-tumoral injection of MCMV in the case of melanomas.28,29 In human, HCMV reactivation after hematopoietic stem cell transplantation (HSCT) or kidney transplantation has been associated with a decreased rate of relapse for acute myeloid leukemia (AML)30, 31, 32, 33 and a reduced risk of skin cancer,34 respectively. The mechanism underpinning this beneficial effect of HCMV was suggested to rely on the reported recognition of cancer cells by donor-derived, HCMV-stimulated non-Vδ2Vγ9 T cells35, 36, 37, 38, 39 and NKG2Cpos NK cytotoxic effector cells (for reviews see Litjens et al.40 and Bigley et al.41). Yet, Koldehof et al.42 showed a direct pro-apoptotic effect of HCMV on acute leukemia cell lines that could explain, at least in part, the decreased leukemic relapse rate in AML patients with HCMV reactivation.
The reported discrepancies about the role of CMV in cancer might be due to variable factors including the state of cytomegalovirus infection (acute versus latent) and the host immune status, as well as the tumor origin and microenvironment. The present study aimed at investigating whether and how CMV would affect cancer cell growth without the influence of major immune effectors in highly immunodeficient mice.
Results
Dose-Dependent Inhibition of Mouse Cancer Cell Growth in Immunodeficient Mice
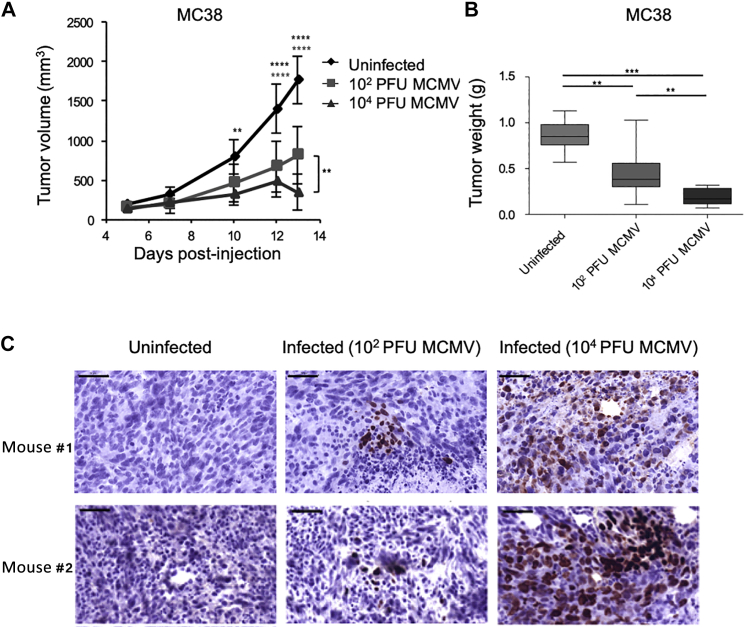
In order to test the effect of MCMV on tumors without the impact of main anti-tumor immune effectors, we used the most highly immunodeficient mice available (NSG). MC38 colon cancer cells were injected subcutaneously (s.c.) in NSG mice that concomitantly received MCMV intraperitoneally (i.p.) or were left uninfected. Two different doses of virus were used (104 and 102 plaque-forming units [PFUs]). As shown in Figure 1, the growth of MC38 cells was inhibited in infected mice in a dose-dependent manner. MCMV was also able to inhibit in a dose-dependent manner the growth of another type of tumor, i.e., the B16 melanoma in a dose-dependent manner (data not shown). At the end of the experiment, a significant difference was observed between the two groups of infected mice (102 versus 104 PFUs) for both tumor volumes (Figure 1A) and tumor weight (Figure 1B). We next tested whether MCMV was able to infect tumor cells in vivo and whether the dose-dependent intensity of tumor growth inhibition was dependent on the number of infected tumor cells in the host. The hypothesis that MC38 cells were infected by MCMV was confirmed by detection of immediate early (IE-1) proteins within MC38 tumors (Figure 1C). As depicted, the number of IE-1+ cells appeared to be higher in mice that had received 104 versus 102 PFUs of viral inoculum. Altogether these results demonstrated that MCMV limits murine tumor growth in vivo, which could, at least partially, be attributed to the virus’s ability to infect tumor cells.
Figure 1.
Dose-Dependent Inhibition of Mouse Cancer Cell Growth in Immunodeficient Mice
NSG mice received s.c. injection of 5 × 105 MC38 tumor cells and were left untreated or infected with 102 or 104 PFUs of MCMV. (A) Tumor growth was monitored 3 times a week. (B) Tumors were weighted 2 weeks post-infection, after sacrifice. The data represent the mean ± SEM of tumor volumes (A) and weights (B) from 8–10 mice per group for 1 representative experiment out of 2. Significant differences between control and infected mice are shown at different time points, while significant differences between the 2 groups of infected mice (102 or 104 PFUs) are shown at the end of experiment. Statistical tests were two-way ANOVA (A) and Mann-Whitney (B) (*p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001). (C) IE-1 protein expression was evaluated by immunohistochemistry on tumor biopsies collected at the end of the experiment, from uninfected, 102 and 104 PFUs infected mice. Images are from one representative mouse for each experiment. Scale bar represents 50 μm.
Cancer Cells Show Different Permissiveness to MCMV
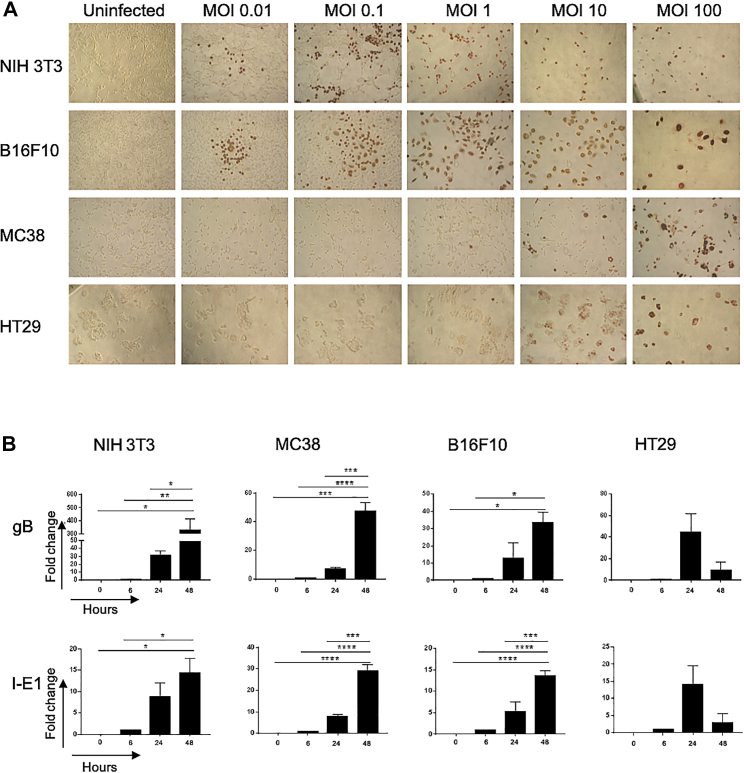
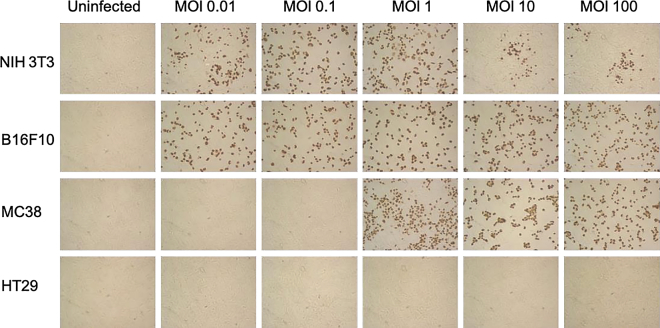
Permissiveness of mouse cancer cells to MCMV was further analyzed in vitro, relative to highly permissive murine fibroblasts (3T3). Because MCMV replicates very poorly in human cells due to high species specificity, we also tested permissiveness of human cancer cells. The results from these analyses are shown in Figure 2. Staining of cells with anti-IE-1 monoclonal antibody (mAb) showed that a low viral dose (multiplicity of infection or MOI = 0.01) was sufficient to infect B16F10 and 3T3 (Figure 2A). In contrast, MC38 and HT29 colon cancer cells required higher doses of virus (MOI ≥ 1) to be infected. Transcripts for MCMV I-E1 and glycoprotein B (gB) were detected in all cancer cells infected with MCMV (MOI = 10, Figure 2B). The presence of the “late” gB protein was also evidenced by immunohistochemistry, although in low proportion even at high MOI in colon cancer cells, especially for human HT29 cells (Figure S1). We then evaluated whether cancer cells can support productive viral replication. After contact with increasing doses of MCMV and inactivation of residual viral inoculum, cancer cells were incubated for 5 days, supernatants were transferred onto 3T3 fibroblasts for 2 days, and the initiation of viral replication assessed by IE-1 staining. As evidenced by the presence of IE-1+ fibroblasts, infectious viral particles were released from cancer cells, as long as the initial MOI was ≥0.01 for B16F10 and 3T3 and ≥1 for MC38 (Figure 3). In contrast, although MCMV IE-1 antigens could be detected in human HT29 colon cancer cells infected with MCMV (MOI > 1, Figure 2A), no infectious viral particles seemed to be present in the supernatant (Figure 3). These results suggest that MCMV can replicate in mouse cancer cells but not in human cancer cells. However, mouse cancer cell lines show different permissiveness to MCMV, MC38 colon cancer cells being less permissive than B16F10 melanomas.
Figure 2.
Cancer Cells Show Different Permissiveness to MCMV
(A) The different cell lines were left uninfected or were infected with variable doses of MCMV; the expression of IE-1 proteins was evaluated after 48 h. (B) Murine (3T3, MC38, B16F10) and human (HT29) cancer cells were infected with MCMV (MOI 10). RNA was extracted from cells 6 h, 24 h, and 48 h after infection. Gene expression at 24 and 48 h is expressed as fold change relative to expression of gB and IE-1 at 6 h. Error bars represent the mean ± SEM of 3 independent experiments. One-way ANOVA statistical tests were used; *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001.
Figure 3.
Human Colon Cancer Cells Do Not Support MCMV Replication
Indicated cells were seeded and let to adhere for 24 h prior to infection with increasing doses of MCMV. Viral particles that were not internalized after 1 h incubation were removed by acidic treatment. After 5 days, culture supernatants were collected and transferred onto freshly seeded 3T3 fibroblasts. IE-1 expression was analyzed 48 h after transfer. Images are from 1 experiment representative of 2.
MCMV Affects Survival of Both Mouse and Human Cancer Cells
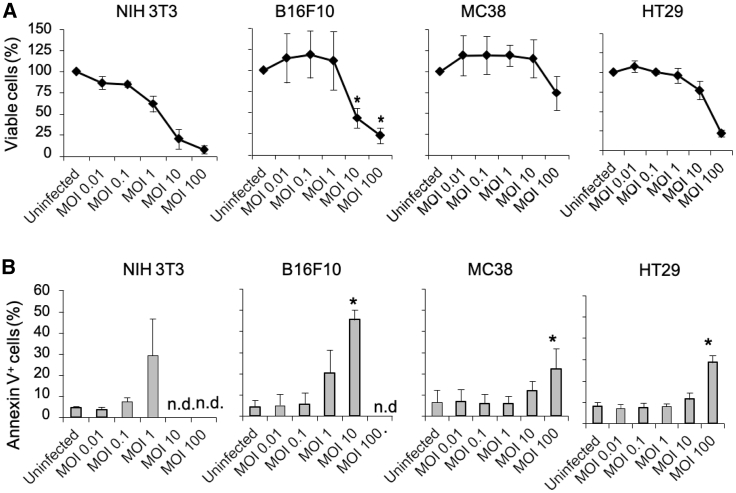
We next assessed cell survival of MCMV-infected cancer cells. As measured by dye compound 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromidefo (MTT) assay, all cells tested were sensitive to MCMV-mediated growth inhibition (Figure 4A). The viral dose inhibiting 50% of growth (IC50) 3 days post-infection related to 1 < MOI < 10 for B16F10 and 3T3, 10 < MOI < 100 for HT29, and MOI > 100 for MC38 (Figure 4A). The concomitant assessment of apoptosis by flow cytometry revealed that at MOI = 1, 30% of 3T3, 20% of B16F10, 5% of MC38, and 8% of HT29 cells were annexin V+ after 3 days of infection. MOI = 100 was 100% lethal for B16F10 and 3T3 cells, while 20% of MC38 and 30% of HT29 were annexin V+ (Figure 4B). These data confirm that B16F10 cells are more sensitive to MCMV infection than MC38 cells. They also unexpectedly show that, despite the absence of a productive viral cycle, MCMVs inhibit proliferation and induce apoptosis of human HT29 colon cancer cells at similar or even lower doses than for the permissive MC38 cells.
Figure 4.
MCMV Affects Survival of Both Mouse and Human Cancer Cells
Cell lines were infected with MCMV at different MOI for 72 h. (A) Cell viability of cells was assessed using an MTT assay. (B) The proportion of annexin V+ cells was measured by flow cytometry. Data represent the mean ± SEM of 3 or 4 independent experiments. The proportions of viable cells (A) and annexin V+ cells (B) between control and infected cells were compared and statistics analyzed using two-way ANOVA (*p < 0.05).
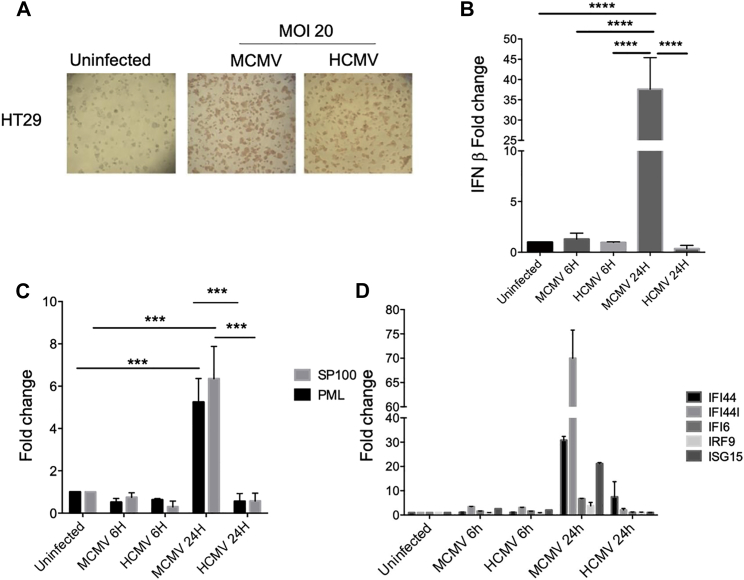
Increased Host Defense Mechanisms in MCMV versus HCMV-Infected HT29 Cells
CMV replication in cells from a different species might be prohibited due to the inability of the virus to overcome host defense mechanisms. The data presented in Figure 4 are in agreement with this idea. A similar level of viral IE-1 proteins was evidenced in HT29 cells infected with either murine or human CMV at a high MOI (Figure 5A). However, 24 h post-infection, IFN-β expression was highly increased in MCMV- but not HCMV-infected cells, relative to uninfected cells (Figure 5B). Similar results were obtained at day 7 (data not shown). Because IFN-β possesses indirect anti-pathogen activity via the induction of various ISGs and ND10, we examined in infected versus control HT29 cells: (1) the expression of promyelocytic leukemia (PML) and speckled protein of 100 kDa (SP100), two major components of ND10, and (2) the transcription of a panel of ISGs that were previously shown to be regulated in cancer.43, 44, 45 As observed in Figure 5C, the expression of PML and SP100 was upregulated 24 h post-MCMV infection, while HCMV had no effect on the expression of these genes. Similarly, the transcription of ISGs was differently affected by the two viruses, with a reproducible (although non-significant) increase of IFI44, IFI44L, and ISG15 in 24 h-MCMV infected cells only, relative to uninfected cells (Figure 5D). As a whole, these results indicate that MCMV, by contrast to HCMV, is able to stimulate the type I IFN response in human tumor cells.
Figure 5.
Increased Host Defense Mechanisms in MCMV- versus HCMV-Infected HT29 Cells
(A) HT29 cells were left uninfected or subjected to MCMV or HCMV infection (MOI = 20); staining of murine (MCMV) and human (HCMV) IE-1 proteins was performed after 48 h. Images are representative of 2 to 3 independent experiments. (B–D) HT29 cells were uninfected or infected with MCMV or HCMV (MOI = 20). IFN-β (B), ND10 (C), and ISG (D) mRNA levels were evaluated 6 h and 24 h post-infection by qRT-PCR. Gene expression in infected cells is expressed as fold change relative to expression in control HT29 cells. Error bars represent the mean ± SEM of at least 3 independent experiments. Two-way ANOVA statistical tests were used (*p < 0.05, **p < 0.005, ****p < 0.0001).
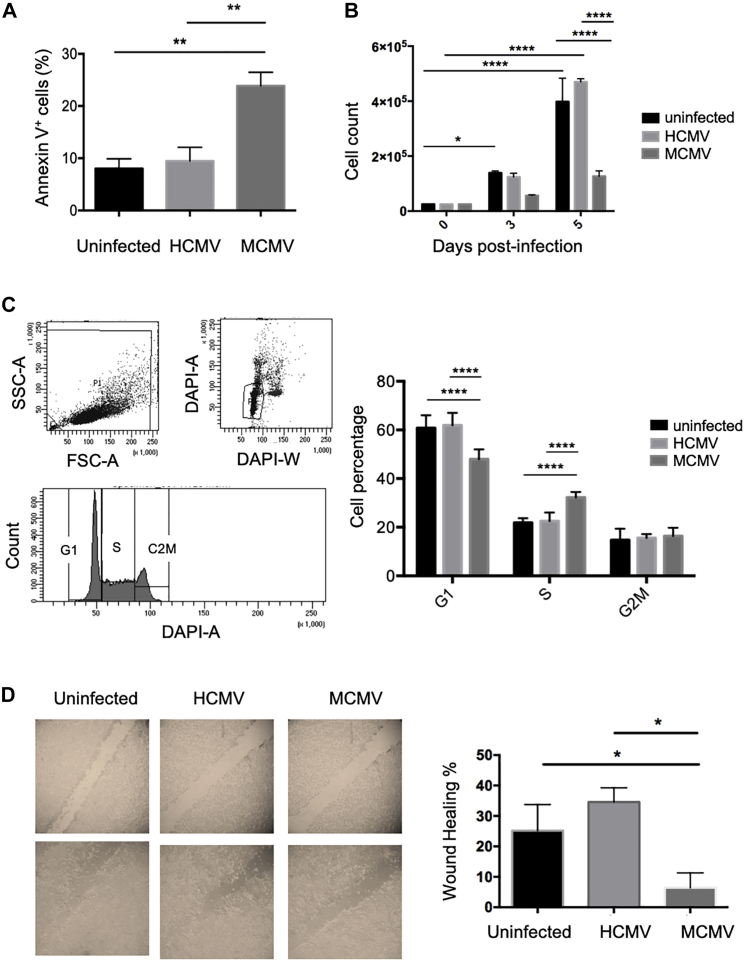
MCMV, but Not HCMV, Affects Human Colon Cancer Cell Properties In Vitro
Our next in vitro experiments aimed at comparing MCMV versus HCMV impact on diverse properties of human colon cancer cells. 3 days post-infection, 8%–10% cells stained positive for annexin V among control and HCMV-infected HT29 cells, while 22% of MCMV-infected HT29 cells were annexin V+ (Figure 6A). Moreover, HT29 cell counts were much lower when the cells were infected with MCMV, relative to control cells, or to HCMV-infected cells after 5 days of culture (Figure 6B). MCMV was also able to target a metastatic human colon cancer cell line (the SW480 cells), as evidenced by lower cell counts in culture (Figure S2A), and increased expression of annexin V in MCMV-infected cells only (Figure S2B). We next tested the effect of MCMV versus HCMV infection on HT29 cell cycle and observed a slight decrease in the proportion of cells in G1 phase concomitant to an increase in S phase in MCMV-infected cells only (Figure 6C). Finally, wound-healing inhibition by MCMV but not HCMV was shown using a scratch assay, suggesting an impaired migratory phenotype induced by MCMV but not HCMV (Figure 6D).
Figure 6.
MCMV Affects Human Colon Cancer Cells In Vitro, in Contrast to HCMV
HT29 cells were left uninfected or infected with MCMV or HCMV (MOI = 20). (A–D) The impact of MCMV and HCMV on apoptosis (A), culture cell counts (B), cell cycle (C), and wound healing (D) was determined (for each technique see the Materials and Methods section). Error bars represent the mean ± SEM of 2–4 independent experiments. Two-way ANOVA statistical tests were used (*p < 0.05, **p < 0.005, ****p < 0.0001).
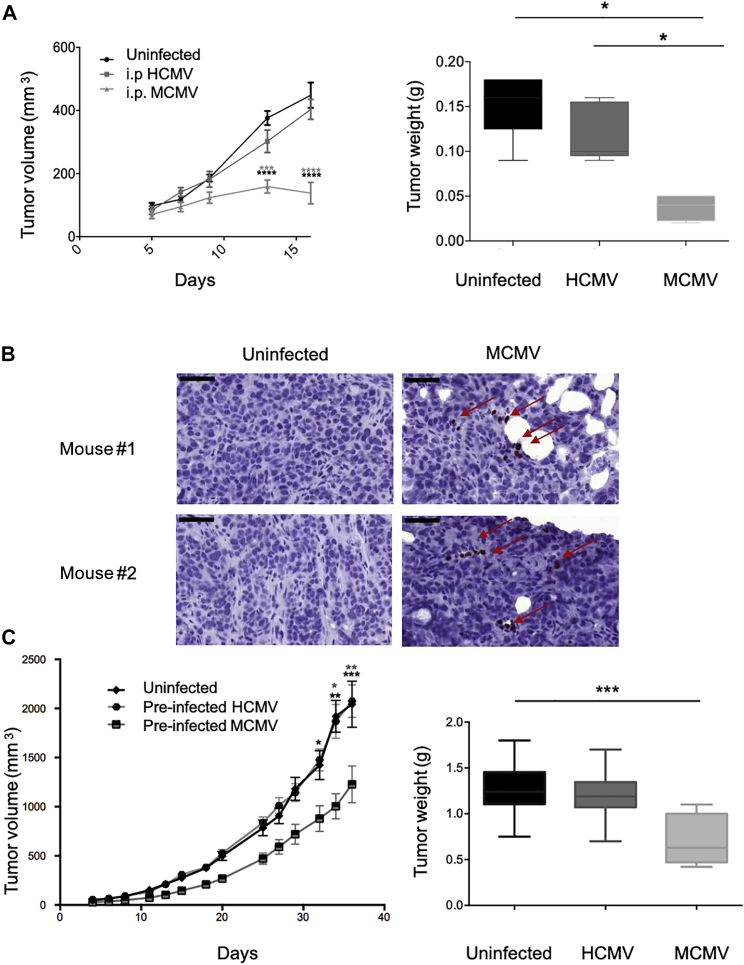
MCMV-Restricted Inhibition of Human Colon Cancer Cell Growth in Mice
As a first approach to test the impact of MCMV on human colon cancer cell growth in vivo, HT29 cells were implanted s.c. in immune-deficient mice that were concomitantly infected i.p. with MCMV (104 PFUs). HT29 tumors barely developed in MCMV-treated mice and tumor weights were very low compared to tumors from untreated mice (Figure 7A). The presence of MCMV IE-1 antigens was evidenced in HT29 tumors, although few cells stained positive relative to what we observed previously with murine colon cancer cells (Figures 7B and 1C). HCMV had no effect on HT29 tumor growth in the same setting but this might be because HCMV is unable to produce a systemic infection in mice. For this reason, in another set of experiments, HT29 cells were pre-infected with MCMV or HCMV before s.c. implantation in NSG mice. Tumor growth was significantly inhibited by MCMV pre-infection in contrast to HCMV and, at the end of the experiment, tumor masses were much lower in MCMV-infected mice than in HCMV-infected or control mice (Figure 7C). Thus, our results demonstrate that despite species-specific barriers, MCMV can inhibit human tumor growth.
Figure 7.
MCMV-Restricted Inhibition of Human Colon Cancer Cell Growth in Mice
(A) NSG mice received s.c. injection of 5 × 105 HT29 tumor cells and were left untreated, or i.p. infected with 104 PFUs of MCMV or HCMV. Tumor growth was monitored three times a week and tumors were weighted at the end of experiment. Data represent the mean ± SEM of tumor volumes and weights from 5–8 mice for 1 representative experiment out of 2. Significant differences between control and infected mice are shown at different time points. (B) IE-1 staining was performed on tumor biopsies collected at the end of the experiment. Images are from one representative mouse for each experiment. Scale bar represents 50 μm. (C) NSG mice received s.c. injection of 5 × 105 HT29 control cells or 5 × 105 HT29 cells pre-infected in vitro with HCMV or MCMV (MOI = 10). Tumor growth was monitored three times a week and tumors were weighted 38 days post-infection, after sacrifice. Data represent the mean ± SEM of tumor volumes and weight from 10 mice for 1 representative experiment out of 2. (A) and (C) Statistical tests were two-way ANOVA for tumor growth and Mann-Whitney for tumor weights (*p < 0.05, ***p < 0.001, ****p < 0.0001).
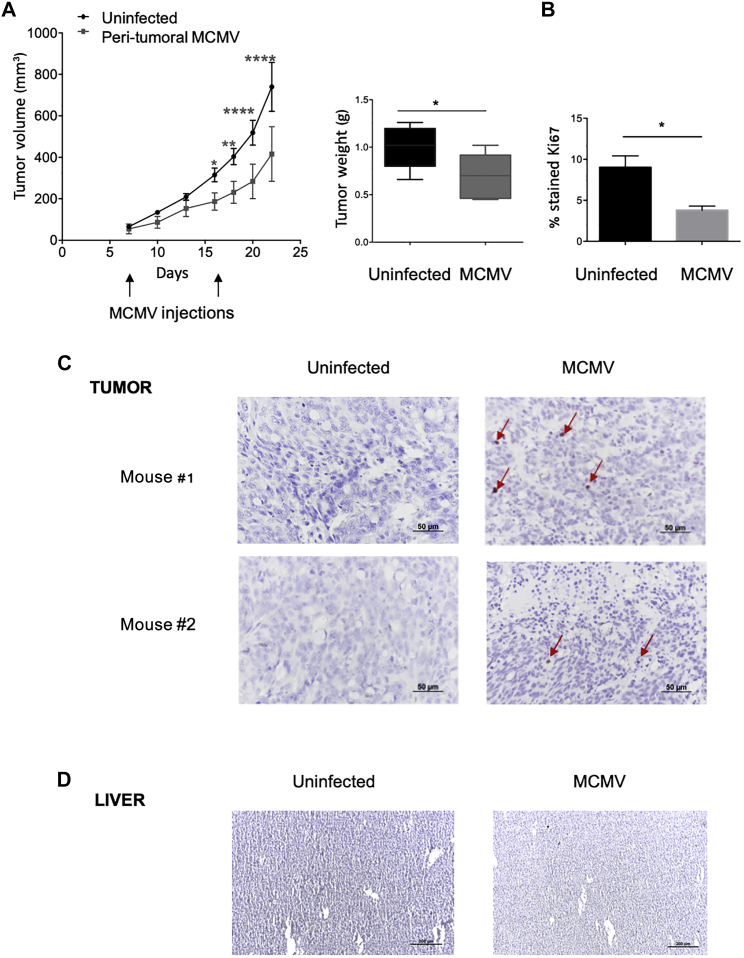
Inhibition of Human Cancer Cell Growth by Local MCMV Therapy
A third set of experiments was carried out to evaluate the effect of a therapy based on local MCMV injections. Two peri-tumoral injections were performed in the course of the experiment (Figure 8A). Tumor development in mice receiving local MCMV therapy and control uninfected mice was followed in parallel. Human cancer cell growth was inhibited in mice challenged with MCMV and resulted in a reduction in tumor weights (Figure 8A). Furthermore, tumors from mice treated with local MCMV therapy showed a decreased proportion of Ki67+ cells (Figure 8B; Figure S3). Finally, IE-1+ cells were visualized in human tumors in mice receiving MCMV therapy at the end of the experiment, although at low proportion (Figure 8C). In contrast, IE-1 was undetectable in liver tissues, suggesting negligible viral dissemination (Figure 8D). These results indicate that MCMV could be used as a local treatment to reduce human tumor growth.
Figure 8.
Inhibition of Human Cancer Cell Growth by Local MCMV Therapy
20 NSG mice received s.c. injection of 5 × 105 HT29 uninfected cells. Half of mice bearing HT29 untreated tumors (10) received local MCMV therapy at days 8 and 17 (103 PFUs). (A) Tumor growth was monitored three times a week and tumors were weighted 33 days post-infection, after sacrifice. Data represent the mean ± SEM of tumor volumes and weights from 10 mice of 1 representative experiment out of 2. (B) Ki67 staining was done on tumor biopsies, at the end of experiment. Statistical tests were two-way ANOVA for tumor growth and Mann-Whitney for tumor weights and Ki67 staining (*p < 0.05, **p < 0.005, ***p < 0.001). (C and D) IE-1 staining was performed on tumor biopsies (C) and liver (D). Scale bar represents 50 μm for tumors and 200 μm for liver. Arrows indicate IE-1-positive cells.
Discussion
Our results demonstrate that both murine and human tumor cells can be infected with MCMV, which, even when not replicating in these cells, induces a type I IFN response and decrease in tumor cell growth.
First, we showed a viral dose-dependent inhibition of B16F10 and MC38 murine cancer cell growth in highly immunodeficient mice infected with MCMV. The presence of IE-1+ cells in MC38 tumors in vivo, and the results of our in vitro analyses, suggest a direct viral anti-tumor effect through cancer cell killing, although other mechanisms might take place. In a recent study by Erkes et al.,28 a CD8+ αβ T cell-dependent inhibition of tumor growth was observed in immunocompetent mice, after intra-tumoral injection of MCMV in B16F0 s.c. tumors. In our study, the absence of immune control in NSG mice probably allows the virus (injected i.p.) to disseminate to and reach distant sites and cancer cells. Although this scenario probably only rarely occurs in physiological situations, it might take place with specific tumors and/or in particular contexts of immunosuppression. However, when using MCMV as a therapeutic agent to target human cancer, using relatively high MOI appears necessary to affect survival of human cancer cells and thus constitutes a guarantee of success with limited risk of dissemination due to the species specificity barrier.
Decreased murine cancer cell viability was shown upon MCMV infection of B16F10 and MC38 cancer cells in vitro. Moreover, murine cancer cells infected by MCMV produced infectious viral particles. In both cancer cell lines, cell death was triggered as soon as IE-1+ cells were detected, but a 100-fold lower viral dose was required to induce the release of viral particles from B16F10 melanomas, relative to MC38 colon carcinomas. This was unexpected since CMV was shown to poorly replicate in cancer cells, due to the expression of oncogenic alleles that induce multiple blocks including inhibition of viral entry, expression of IE genes and viral DNA replication.46
Increased apoptosis of murine cancer cells upon MCMV infection in vitro did not preclude the possibility that MCMV could also operate through an indirect mechanism to inhibit cancer cell growth in vivo, such as the release of soluble inhibitory factors by non-tumoral cells. Interestingly, inhibition of human HT29 tumors was also evidenced after i.p. infection with MCMV (Figure 6A), despite the fact that most mouse cytokines do not cross-react with human cells. In vivo infection of human colon cancer cells with MCMV was suggested by the presence of IE-1+ (Figure 7B) and HLA-I+ (data not shown) cells in serial sections of HT29 tumors. In vitro, apoptosis of MCMV-infected HT29 cells correlated with IE-1+ staining; however, no viral particles were detected in the supernatants, indicating that the viral lytic cycle was aborted in human tumor cells, contrarily to murine cancer cells (Figure 3). Finally, few IE-1+ cells were detected within HT29 tumors collected from MCMV-treated mice (Figures 7B and 8C), even when human cancer cells were pre-infected with MCMV (data not shown). This could be due to the preferential proliferation of remaining non-infected cells in vivo or to an inhibition of murine viral transcription/replication in the human cancer cell line.47
In addition to inducing HT29 cell apoptosis, MCMV was able to act on various cellular functions, supporting its anti-tumor potential on human colon cancer cells. MCMV infection of HT29 cells affected wound healing, triggered a cell-cycle arrest in S-phase and inhibited cell proliferation. Furthermore, HT29 cells infected by MCMV expressed IFN-β mRNA and, most likely, IFN-β protein, as suggested by the induction of ISGs. This cytokine is probably involved in HT29 growth inhibition since it was shown to activate caspase-dependent apoptosis of HT29 cells,48 as well as to trigger the prolongation of S-phase and to block subsequent transition into G2/M-phase.49 In vivo, anti-angiogenic activity of IFN-β has also been reported.50 Presumably, IFN-β independent inhibitory mechanisms also operate in MCMV-induced tumor cell control. Increased apoptosis and decreased cell counts in culture were shown in the infected SW480 colon cancer cell line as well, despite its reported defectiveness in cGAS expression,51 a DNA-sensor involved in the IFN-response in CMV-infected cells.4,5
In contrast to MCMV, HCMV (strain TB42-E) did not have a significant impact on HT29 colon cancer cells in vitro (Figure 6) and in vivo (Figure 7). Using a different strain of HCMV (AD169), Teo et al.52 showed a significant increase in cell proliferation and viability, 48 h post in vitro infection of HT29 cells. We also tested whether HT29 cells can be infected by the Merlin strain, but no HCMV IE-1 Ag were detected, in contrast to infection with TB42-E (Figure S4). While the relationship between HCMV and cancer remains puzzling, our results describe a selective advantage of MCMV over HCMV in fighting human cancer. Peri-tumoral injection of MCMV inhibited of HT29 tumor growth. MCMV did not seem to disseminate to the liver of mice. Although highly sensitive to MCMV, NSG mice did not show clinical signs of infection along the course of the experiment, and IE-1 proteins were not detected in the livers (Figure 8D).
During cross-species infection, a post-penetration block of viral replication occurs and limits viral spread.53,54 This block might be due to the inability of the virus to prevent apoptosis1 or to subvert host defense mechanisms. Indeed, IE-1 from MCMV was unable to counteract human ND10 components, in contrast to IE-1 from HCMV.2 In line with these findings in non-tumoral cells, we found an increased transcription of IFN-β, SP100, and PML in HT29 cells infected by MCMV, but not HCMV. Therefore, we propose that MCMV is unable to counteract cell intrinsic responses in human cancer cells.
The peculiar features of CMV (large DNA genome facilitating recombinant gene cloning, possible re-infection, induction of strong and life-long immune surveillance), have led us to propose the use of CMV as a viral vaccine vector.55, 56, 57 CMV-based cancer vaccines are also under consideration and have shown some protection in murine models of prostate cancer58 and melanoma,59 although different efficacies were evidenced, depending on the antigenic epitope and to the route of infection28,60 (reviewed in Qiu et al.61 and Quinn et al.62). However, therapeutic vaccination for late-stage cancer patients could be dangerous because of their immunocompromised status and must require the use of spread defective viral variants.63 Alternatively, recombinant MCMV has been suggested as a potential antigen delivery vector in humans, due to its ability to target dendritic cells without compromising their antigen-presenting ability.64,65 Yet, CMV is not considered as an oncolytic virus and has never been evoked as a potential virotherapeutic agent to target tumors. Our study paves the way for the development of undescribed therapeutic protocols based on the local tumor delivery of MCMV, a strategy that could be of relevance for treating cancer in immunocompromised recipients. Using MCMV (or HCMV-based vectors containing MCMV proteins) could activate cancer cell intrinsic responses that will ultimately lead to cell death and could avoid uncontrolled and harmful viral multiplication in human surrounding tissues. The induction of IFN-β production by MCMV in cancer cells could also trigger innate immune responses,66 limit neoangiogenesis,50 and increase the control of tumor growth.
Materials and Methods
Cell Lines and Viral Stocks
We used 8- to 12-week-old NSG male or female mice from the Jackson Laboratories. The mice were housed in an appropriate mouse facility and kept under pathogen-free conditions (Animalerie A2, University of Bordeaux, France, approval n° B33-063-916). Our laboratory is accredited for the manipulation of genetically modified organisms (declaration no. 3153). This study was approved by the Ethics Review Committee of Bordeaux (reference no. 2016092917471799-V2, APAFiS #7022).
C57BL/6-derived MC38 colon adenocarcinoma and B16F10 murine melanoma cell lines were generously donated by Dr. B. Robert (Cancer Research Institute, Montpellier, France) and Dr. P. Voisin (Centre de Résonance Magnétique des Sytèmes Biologiques, Bordeaux, France). NIH 3T3 murine fibroblasts, HT29, and SW480 human colon adenocarcinoma cell lines were from the American Type Culture Collections (ATCC, LGC Standards, Molsheim, France). Cells were cultivated at 37°C, 5% CO2 in DMEM (GIBCO, Thermo Fisher Scientific, Paisley, UK) supplemented with 8% of heat inactivated fetal bovine serum (FBS) (HyClone Laboratories, GE Healthcare, Logan, UT, USA). All cell lines used were regularly tested for the absence of mycoplasma.
MCMV infection was performed with the Smith strain (ATCC VR-194, LGC Standards, Molsheim, France). For in vivo experiments, the virus stock was prepared by homogenizing salivary glands (SVGs) harvested from BALB/c mice (Charles Rivers Laboratory, Saint-Germain-sur-l’Arbresles, France), which had been infected 3 weeks earlier with MCMV. For in vitro experiments, the virus stock was prepared by successive passages on a NIH 3T3 cell layer. Virus titer was determined by the standard plaque assay method on murine embryonic fibroblasts (MEFs). For HCMV infection, we used the clinical strain of HCMV TB42-E (generously donated by C. Sinzger, University of Tübingen, Germany). Regarding the genomic integrity of TB42-E strain, the sequence used was shown to be intact apart from two genes: RL13 and UL1 (A. Davison, personal communication). The HCMV Merlin strain was generously donated by C. Fielding (Cardiff University, UK). HCMV stocks were prepared by successive passages on a fibroblast cell layer.
Implantation of Tumor Cells and Infection of Mice
For systemic MCMV infections (Figures 1A and 7A), the mice were anesthetized and subcutaneously given 5 × 105 tumor cells (MC38, B16F10, or HT29) in the right flank. The mice were infected the same day by i.p. injection of 102 or 104 PFUs of MCMV or HCMV. In the experiments described in Figure 7C, the mice were anesthetized and subcutaneously given 5 × 105 HT29 control cells or HT29 cells that had been pre-infected in vitro for 4 days with MCMV or HCMV, at a MOI = 10. For local MCMV therapy (Figure 8A), mice received two peri-tumoral injections of 103 PFUs of MCMV (SGV) at day 8 and at day 17. Tumor growth was monitored by measuring the length and width of tumors with a caliper two to three times per week. The tumor volume was estimated using the following formula: tumor volume (mm3) = [length (mm) x width2 (mm)] / 2.
Infection of Cells and Detection of Viral Proteins In Vitro
NIH 3T3, MC38, B16F10, and HT29 cells were seeded at a density of 3,000 cells per well in a 96-well plate in 100 μL of complete medium. After 24 h, cells were infected at various MOI (0.01, 0.1, 1, 10, and 100). After 48 h of incubation at 37°C, 5% CO2, the cells were washed with phosphate buffered saline (PBS) and fixed. Endogenous peroxidases were inhibited with an H2O2 solution for 20 min at −20°C. Staining was performed using monoclonal antibody (mAb) against MCMV IE-1 (clone Croma 101 from CapRi), mAb against MCMV gB (clone Croma 7 from CapRi, University of Rijeka, Croatia), mAb against HCMV IE-1 (Argene, Vernoile, France), and Vectastain Universal Elite ABC Kit (Vector Laboratories, Peterborough, UK) following the manufacturer’s recommendations. The revelation was done using the AEC Kit (Vector Laboratories, Peterborough, UK).
Supernatant Transfer Experiments
NIH 3T3, MC38, B16F10, and HT29 cells were seeded at a density of 3,000 cells per well in a 96-well plate in 100 μL of complete medium. After 24 h, cells were infected with various MOIs (0.01, 0.1, 1, 10, and 100). After centrifugation and incubation for 1 h at 37°C, cells were washed with DMEM. Remaining viral inoculum was eliminated using citric buffer at pH = 3. After 5 days of incubation at 37°C, 5% CO2, supernatant was transferred on NIH 3T3 (3,000 per well) in a 96-well plate. IE-1 staining was performed after 48 h.
Analyses of Biopsies
The tumors (MC38 and HT29) and eventually the livers were taken at the end of the experiments, weighted, placed in cassettes, and fixed with 4% paraformaldehyde for 4 h. The samples were transferred to a 70% ethanol solution and included in paraffin. Antigens were unmasked on sections of 4 μm thick, by 30 min incubation in Tris buffer (10 mM) EDTA (1 mM) at pH 9. After inhibition of endogenous peroxidases with 3% H2O2 solution, nonspecific binding sites and avidin were blocked by the blocking solutions of Vectastain Universal Elite ABC Kit and Avidin/Biotin Blocking Kit, respectively (Vector Laboratories, Peterborough, UK). Staining was performed using mAb against Ki67 (clone sp6 Invitrogen, Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA), mAbs against MCMV IE-1 (clone Croma 101), and Vectastain Universal Elite ABC Kit. The revelation was done using Novared (Vector Laboratories, Peterborough, UK). A counterstaining was done in Mayer hemalun before the analysis. The slides were scanned with the Pannoramic Scan and analyzed with Pannoramic Viewer or Nikon camera.
Viability Assay
The viability of tumor cells and 3T3 post-infection with MCMV was assessed by an MTT assay. The cells were seeded at a rate of 500 to 2,500 cells per well according to the cell line in 96-well plate in 100 μL of complete medium. After 24 h of adhesion, the cells were infected with various MOI (0.01, 0.1, 1, 10, and 100). After 3 days of infection, the cells were incubated for 2 to 4 h at 37°C, 5% CO2 with 15 μL per well of a solution of 3- (4,5-dimethylthiazol-2-yl) −2,5 bromide-diphenyl tetrazolium at 5 mg/mL (Sigma Aldrich, Merck, St. Louis, MO, USA). The formazan crystals were then solubilized by the addition of 105 μL per well of isopropanol containing 5% of formic acid. The optical density was read at 570 nm with a Varioskan Flash (Thermo Fisher Scientific, Waltham, MA, USA). The optical density absorbance of the medium alone was then subtracted from that of the conditions tested to evaluate the specific absorbance of each condition.
Quantitative Real-Time PCR and qRT-PCR
HT29 cells were seeded at a rate of 2 × 105 cells per well in a 24-well plate in 1 mL of complete medium. After 24 h of adhesion, the cells were infected at MOI = 20 and incubated for 6 h, 24 h, or 48 h at 37°C, 5% CO2. RNA was prepared with the NucleoSpin RNA Plus Kit (Macherey-Nagel Hoerdt, France). RNA quality and quantity were analyzed using Agilent TapeStation System and DS-11 DeNovix spectrometer, respectively. RNA was converted to cDNA with the Go-Script Reverse Transcription Kit (Promega, La Farlede, France). The qRT-PCR was realized using the GoTaq Master Mix from Promega, La Farlede, France. Samples were distributed by means of Eppendorf epMotion 5073 automated pipetting. PCR was performed with the Bio-Rad CFX384 real-time PCR detection system. All procedures were performed according to the manufacturer’s instructions. The following targets were analyzed: IFN-β, IFI6, IFI44, IFI44L, IRF9, ISG15, PML, and SP100, as well as MCMV I-E1 and gB. Primers sequences are depicted in Table 1 and were provided by Sigma Genosys (The Woodlands, USA). Fold change gene expression was calculated using the 2−ΔΔCt method.
Table 1.
List of Primers Used for RT-PCR Analysis
| Gene | Sequence (5′ to 3′) | GenBank Access Number |
|---|---|---|
| IRF9 | 5′-GCCCTACAAGGTGTATCAGTTG-3′ | NM_006084 |
| 5′-TGCTGTCGCTTTGATGGTACT-3′ | ||
| ISG15 | 5′-GAGGCAGCGAACTCATCTTT-3′ | NM_005101 |
| 5′-CTTCAGCTCTGACACCGACA-3′ | ||
| SP100 | 5′-TCCATGACAAATTGCCTCTCC-3′ | NM_001206702 |
| 5′-GAGATGGGGAACCCGAAGG-3′ | ||
| PML | 5′-CTTCTGCTCCAACCCCAAC-3′ | NM_033239 |
| 5′-AAGGCACTATCCTGCTCCTG-3′ | ||
| IFI44 | 5′-GGTGGGCACTAATACAACTGG-3′ | NM_006417 |
| 5′-CACACAGAATAAACGGCAGGTA-3′ | ||
| IFI44L | 5′-TCTGCCATTTATGTTGTGTGACA-3′ | NM_006820 |
| 5′-CAGGTGTAATTGGTTTACGGGAA-3′ | ||
| IFI6 | 5′-GGTCTGCGATCCTGAATGGG-3′ | NM_022873 |
| 5′-TCACTATCGAGATACTTGTGGGT-3′ | ||
| IFN-β | 5′-ATGACCAACAAGTGTCTCCTCC-3′ | NM_002176 |
| 5′-GGAATCCAAGCAAGTTGTAGCTC-3′ | ||
| MCMV I-E1 | 5′-CTCATGGACCGCATCGCTGACCAC GTGGG-3′ |
M11788.1 |
| 5′-TGGCTGATTGATAGTTCTGTTTT ATCA-3′ | ||
| MCMV gB | 5′-GGTAAGGCGTGGACTAGCGAT-3′ | M86302.1 |
| 5′-CTAGCTGTTTTAACGCGCGG-3′ |
Cell-Cycle Analysis
HT29 cells were seeded at a density of 2 × 105 cells per well in a 24-well plate in 1 mL complete medium. After 24 h of adhesion, the cells were infected at MOI = 20 and incubated for 48 h at 37°C, 5% CO2. After washing, the cells were fixed with 70% ice cold ethanol for 1 h at 4°C. After centrifugation for 5 min at 400 g, cells were washed once using PBS, 1% BSA. Cells were then stained with 0.5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Aldrich, St. Louis, MO, USA) for 30 min. Acquisition was made using BD LSRFortessa and analysis with BD-DIVA software.
Annexin V Detection
Murine NIH 3T3, MC38, B16F10, and the human HT29 and SW480 colon cancer cells were seeded at a density of 3,000 cells per well in a 96-well plate in 100 μL of complete medium. After 24 h of adhesion, the cells were infected at different MOI and incubated for 72 h at 37°C, 5% CO2. After PBS washing, cell apoptosis was determined with the BD PharMingen FITC Annexin V Apoptosis Detection Kit I (BD, Allschwil, Switzerland). Staining of annexin V was carried out according to the manufacturer’s instructions. Acquisition was made on BD LSRFortessa and analysis was done with the FlowJo (Tree Star) software.
Wound Healing Assay
For wound-healing assays, HT29 (1 × 106) were cultured in 6-well plates for 24 h to obtain a confluent monolayer. Vertical artificial scratches were made in the wells by scraping the cell monolayer along the diameter of each well using a 10 μL pipette tip. Cells that separated from the monolayer were removed. Then, medium with or without MCMV at MOI = 20 was added. After 48 h in culture, the microscopic images of the scratched area before and after treatment were obtained. The percentage of wound healing was analyzed using ImageJ software.
Statistical Analyses
Statistical studies were performed using GraphPad Prism 5 software. For in vivo experiments, statistical tests were two-way ANOVA (comparison of tumor volumes) and Mann-Whitney (comparison of tumor weights). For in vitro experiments, statistical tests were one-way or two-way ANOVA, as indicated in figure legends.
Author Contributions
Project administration and supervision: M.C. and J.D.-M. Conceptualization and validation: L.M., C.K., M.C., and J.D.-M. Investigation and Methodology: L.M., C.K., N.Y., V.P., B.R., J.I., A.G., and X.G. Formal analysis: L.M., C.K., and N.Y. Resources: B.R. and J.I. (animals) V.P., X.G., P.D., and A.G. (instrumentations). Visualization: L.M., C.K., M.C., and J.D.-M. Writing: M.C. Funding acquisition: M.C. and J.D.-M.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported in part by grants from the Centre National de la Recherche Scientifique (CNRS, France); the Fondation pour la Recherche Médicale (DEQ20110421287) and the Victor and Ermina Mescle price 2017; the Agence National de la Recherche (ANR-12-BSV3-0024-02); and the Ligue Contre le Cancer (15454) and SIRIC BRIO. C.K.’s doctoral funding was from the Conseil Régional d’Aquitaine and the Fondation pour la Recherche Médicale (FDT20140931227). L.M.’s doctoral funding was from the Centre National de la Recherche Scientifique (CNRS, France). We thank Anaëlle Stum for technical assistance at the flow cytometry facility.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.01.007.
Supplemental Information
References
- 1.Jurak I., Brune W. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 2006;25:2634–2642. doi: 10.1038/sj.emboj.7601133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang Q., Maul G.G. Mouse cytomegalovirus crosses the species barrier with help from a few human cytomegalovirus proteins. J. Virol. 2006;80:7510–7521. doi: 10.1128/JVI.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa H., Ma Z., Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lio C.W., McDonald B., Takahashi M., Dhanwani R., Sharma N., Huang J., Pham E., Benedict C.A., Sharma S. cGAS-STING Signaling Regulates Initial Innate Control of Cytomegalovirus Infection. J. Virol. 2016;90:7789–7797. doi: 10.1128/JVI.01040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paijo J., Döring M., Spanier J., Grabski E., Nooruzzaman M., Schmidt T., Witte G., Messerle M., Hornung V., Kaever V., Kalinke U. cGAS Senses Human Cytomegalovirus and Induces Type I Interferon Responses in Human Monocyte-Derived Cells. PLoS Pathog. 2016;12:e1005546. doi: 10.1371/journal.ppat.1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diner B.A., Lum K.K., Toettcher J.E., Cristea I.M. Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection. MBio. 2016;7:e01553-16. doi: 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woodhall D.L., Groves I.J., Reeves M.B., Wilkinson G., Sinclair J.H. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 2006;281:37652–37660. doi: 10.1074/jbc.M604273200. [DOI] [PubMed] [Google Scholar]
- 8.Adler M., Tavalai N., Müller R., Stamminger T. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 2011;92:1532–1538. doi: 10.1099/vir.0.030981-0. [DOI] [PubMed] [Google Scholar]
- 9.Ashley C.L., Glass M.S., Abendroth A., McSharry B.P., Slobedman B. Nuclear domain 10 components upregulated via interferon during human cytomegalovirus infection potently regulate viral infection. J. Gen. Virol. 2017;98:1795–1805. doi: 10.1099/jgv.0.000858. [DOI] [PubMed] [Google Scholar]
- 10.O’Sullivan T.E., Sun J.C., Lanier L.L. Natural Killer Cell Memory. Immunity. 2015;43:634–645. doi: 10.1016/j.immuni.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khairallah C., Déchanet-Merville J., Capone M. γδ T Cell-Mediated Immunity to Cytomegalovirus Infection. Front. Immunol. 2017;8:105. doi: 10.3389/fimmu.2017.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geder K.M., Lausch R., O’Neill F., Rapp F. Oncogenic transformation of human embryo lung cells by human cytomegalovirus. Science. 1976;192:1134–1137. doi: 10.1126/science.179143. [DOI] [PubMed] [Google Scholar]
- 13.Cinatl J., Jr., Cinatl J., Vogel J.U., Rabenau H., Kornhuber B., Doerr H.W. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 14.Michaelis M., Doerr H.W., Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnsen J.I., Baryawno N., Söderberg-Nauclér C. Is human cytomegalovirus a target in cancer therapy? Oncotarget. 2011;2:1329–1338. doi: 10.18632/oncotarget.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawler S.E. Cytomegalovirus and glioblastoma; controversies and opportunities. J. Neurooncol. 2015;123:465–471. doi: 10.1007/s11060-015-1734-0. [DOI] [PubMed] [Google Scholar]
- 17.Lepiller Q., Abbas W., Kumar A., Tripathy M.K., Herbein G. HCMV activates the IL-6-JAK-STAT3 axis in HepG2 cells and primary human hepatocytes. PLoS ONE. 2013;8:e59591. doi: 10.1371/journal.pone.0059591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price R.L., Song J., Bingmer K., Kim T.H., Yi J.Y., Nowicki M.O., Mo X., Hollon T., Murnan E., Alvarez-Breckenridge C. Cytomegalovirus contributes to glioblastoma in the context of tumor suppressor mutations. Cancer Res. 2013;73:3441–3450. doi: 10.1158/0008-5472.CAN-12-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soroceanu L., Matlaf L., Khan S., Akhavan A., Singer E., Bezrookove V., Decker S., Ghanny S., Hadaczek P., Bengtsson H. Cytomegalovirus Immediate-Early Proteins Promote Stemness Properties in Glioblastoma. Cancer Res. 2015;75:3065–3076. doi: 10.1158/0008-5472.CAN-14-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A., Tripathy M.K., Pasquereau S., Al Moussawi F., Abbas W., Coquard L., Khan K.A., Russo L., Algros M.P., Valmary-Degano S. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. EBioMedicine. 2018;30:167–183. doi: 10.1016/j.ebiom.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herbein G. The Human Cytomegalovirus, from Oncomodulation to Oncogenesis. Viruses. 2018;10:E408. doi: 10.3390/v10080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H.P., Chan Y.J. The oncomodulatory role of human cytomegalovirus in colorectal cancer: implications for clinical trials. Front. Oncol. 2014;4:314. doi: 10.3389/fonc.2014.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai B., Wang X., Chen E., Zhu H. Human cytomegalovirus infection and colorectal cancer risk: a meta-analysis. Oncotarget. 2016;7:76735–76742. doi: 10.18632/oncotarget.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H.P., Jiang J.K., Lai P.Y., Chen C.Y., Chou T.Y., Chen Y.C., Chan C.H., Lin S.F., Yang C.Y., Chen C.Y. Tumoral presence of human cytomegalovirus is associated with shorter disease-free survival in elderly patients with colorectal cancer and higher levels of intratumoral interleukin-17. Clin. Microbiol. Infect. 2014;20:664–671. doi: 10.1111/1469-0691.12412. [DOI] [PubMed] [Google Scholar]
- 25.Chen H.P., Jiang J.K., Chen C.Y., Yang C.Y., Chen Y.C., Lin C.H., Chou T.Y., Cho W.L., Chan Y.J. Identification of human cytomegalovirus in tumour tissues of colorectal cancer and its association with the outcome of non-elderly patients. J. Gen. Virol. 2016;97:2411–2420. doi: 10.1099/jgv.0.000558. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A., Coquard L., Pasquereau S., Russo L., Valmary-Degano S., Borg C., Pothier P., Herbein G. Tumor control by human cytomegalovirus in a murine model of hepatocellular carcinoma. Mol. Ther. Oncolytics. 2016;3:16012. doi: 10.1038/mto.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlach K.C., Böhm V., Seckert C.K., Reddehase M.J., Podlech J. Lymphoma cell apoptosis in the liver induced by distant murine cytomegalovirus infection. J. Virol. 2006;80:4801–4819. doi: 10.1128/JVI.80.10.4801-4819.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erkes D.A., Xu G., Daskalakis C., Zurbach K.A., Wilski N.A., Moghbeli T., Hill A.B., Snyder C.M. Intratumoral Infection with Murine Cytomegalovirus Synergizes with PD-L1 Blockade to Clear Melanoma Lesions and Induce Long-term Immunity. Mol. Ther. 2016;24:1444–1455. doi: 10.1038/mt.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erkes D.A., Wilski N.A., Snyder C.M. Intratumoral infection by CMV may change the tumor environment by directly interacting with tumor-associated macrophages to promote cancer immunity. Hum. Vaccin. Immunother. 2017;13:1778–1785. doi: 10.1080/21645515.2017.1331795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elmaagacli A.H., Steckel N.K., Koldehoff M., Hegerfeldt Y., Trenschel R., Ditschkowski M., Christoph S., Gromke T., Kordelas L., Ottinger H.D. Early human cytomegalovirus replication after transplantation is associated with a decreased relapse risk: evidence for a putative virus-versus-leukemia effect in acute myeloid leukemia patients. Blood. 2011;118:1402–1412. doi: 10.1182/blood-2010-08-304121. [DOI] [PubMed] [Google Scholar]
- 31.Green M.L., Leisenring W.M., Xie H., Walter R.B., Mielcarek M., Sandmaier B.M., Riddell S.R., Boeckh M. CMV reactivation after allogeneic HCT and relapse risk: evidence for early protection in acute myeloid leukemia. Blood. 2013;122:1316–1324. doi: 10.1182/blood-2013-02-487074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manjappa S., Bhamidipati P.K., Stokerl-Goldstein K.E., DiPersio J.F., Uy G.L., Westervelt P., Liu J., Schroeder M.A., Vij R., Abboud C.N. Protective effect of cytomegalovirus reactivation on relapse after allogeneic hematopoietic cell transplantation in acute myeloid leukemia patients is influenced by conditioning regimen. Biol. Blood Marrow Transplant. 2014;20:46–52. doi: 10.1016/j.bbmt.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inagaki J., Noguchi M., Kurauchi K., Tanioka S., Fukano R., Okamura J. Effect of Cytomegalovirus Reactivation on Relapse after Allogeneic Hematopoietic Stem Cell Transplantation in Pediatric Acute Leukemia. Biol. Blood Marrow Transplant. 2016;22:300–306. doi: 10.1016/j.bbmt.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Couzi L., Lafarge X., Pitard V., Neau-Cransac M., Dromer C., Billes M.A., Lacaille F., Moreau J.F., Merville P., Déchanet-Merville J. Gamma-delta T cell expansion is closely associated with cytomegalovirus infection in all solid organ transplant recipients. Transpl. Int. 2011;24:e40–e42. doi: 10.1111/j.1432-2277.2010.01181.x. [DOI] [PubMed] [Google Scholar]
- 35.Halary F., Pitard V., Dlubek D., Krzysiek R., de la Salle H., Merville P., Dromer C., Emilie D., Moreau J.F., Déchanet-Merville J. Shared reactivity of Vdelta2(neg) gammadelta T cells against cytomegalovirus-infected cells and tumor intestinal epithelial cells. J. Exp. Med. 2005;201:1567–1578. doi: 10.1084/jem.20041851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willcox C.R., Pitard V., Netzer S., Couzi L., Salim M., Silberzahn T., Moreau J.F., Hayday A.C., Willcox B.E., Déchanet-Merville J. Cytomegalovirus and tumor stress surveillance by binding of a human γδ T cell antigen receptor to endothelial protein C receptor. Nat. Immunol. 2012;13:872–879. doi: 10.1038/ni.2394. [DOI] [PubMed] [Google Scholar]
- 37.Guerville F., Daburon S., Marlin R., Lartigue L., Loizon S., Pitard V., Couzi L., Moreau J.F., Déchanet-Merville J., Faustin B. TCR-dependent sensitization of human γδ T cells to non-myeloid IL-18 in cytomegalovirus and tumor stress surveillance. OncoImmunology. 2015;4:e1003011. doi: 10.1080/2162402X.2014.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marlin R., Pappalardo A., Kaminski H., Willcox C.R., Pitard V., Netzer S., Khairallah C., Lomenech A.M., Harly C., Bonneville M. Sensing of cell stress by human γδ TCR-dependent recognition of annexin A2. Proc. Natl. Acad. Sci. USA. 2017;114:3163–3168. doi: 10.1073/pnas.1621052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheper W., van Dorp S., Kersting S., Pietersma F., Lindemans C., Hol S., Heijhuurs S., Sebestyen Z., Gründer C., Marcu-Malina V. γδT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia. 2013;27:1328–1338. doi: 10.1038/leu.2012.374. [DOI] [PubMed] [Google Scholar]
- 40.Litjens N.H.R., van der Wagen L., Kuball J., Kwekkeboom J. Potential Beneficial Effects of Cytomegalovirus Infection after Transplantation. Front. Immunol. 2018;9:389. doi: 10.3389/fimmu.2018.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bigley A.B., Baker F.L., Simpson R.J. Cytomegalovirus: an unlikely ally in the fight against blood cancers? Clin. Exp. Immunol. 2018;193:265–274. doi: 10.1111/cei.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koldehoff M., Lindemann M., Opalka B., Bauer S., Ross R.S., Elmaagacli A.H. Cytomegalovirus induces apoptosis in acute leukemia cells as a virus-versus-leukemia function. Leuk. Lymphoma. 2015;56:3189–3197. doi: 10.3109/10428194.2015.1032968. [DOI] [PubMed] [Google Scholar]
- 43.Hallen L.C., Burki Y., Ebeling M., Broger C., Siegrist F., Oroszlan-Szovik K., Bohrmann B., Certa U., Foser S. Antiproliferative activity of the human IFN-alpha-inducible protein IFI44. J. Interferon Cytokine Res. 2007;27:675–680. doi: 10.1089/jir.2007.0021. [DOI] [PubMed] [Google Scholar]
- 44.Zuo C., Sheng X., Ma M., Xia M., Ouyang L. ISG15 in the tumorigenesis and treatment of cancer: An emerging role in malignancies of the digestive system. Oncotarget. 2016;7:74393–74409. doi: 10.18632/oncotarget.11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang W.C., Tung S.L., Chen Y.L., Chen P.M., Chu P.Y. IFI44L is a novel tumor suppressor in human hepatocellular carcinoma affecting cancer stemness, metastasis, and drug resistance via regulating met/Src signaling pathway. BMC Cancer. 2018;18:609. doi: 10.1186/s12885-018-4529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu S., Schafer X., Munger J. Expression of Oncogenic Alleles Induces Multiple Blocks to Human Cytomegalovirus Infection. J. Virol. 2016;90:4346–4356. doi: 10.1128/JVI.00179-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dağ F., Dölken L., Holzki J., Drabig A., Weingärtner A., Schwerk J., Lienenklaus S., Conte I., Geffers R., Davenport C. Reversible silencing of cytomegalovirus genomes by type I interferon governs virus latency. PLoS Pathog. 2014;10:e1003962. doi: 10.1371/journal.ppat.1003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juang S.H., Wei S.J., Hung Y.M., Hsu C.Y., Yang D.M., Liu K.J., Chen W.S., Yang W.K. IFN-beta induces caspase-mediated apoptosis by disrupting mitochondria in human advanced stage colon cancer cell lines. J. Interferon Cytokine Res. 2004;24:231–243. doi: 10.1089/107999004323034105. [DOI] [PubMed] [Google Scholar]
- 49.Katayama T., Nakanishi K., Nishihara H., Kamiyama N., Nakagawa T., Kamiyama T., Iseki K., Tanaka S., Todo S. Type I interferon prolongs cell cycle progression via p21WAF1/CIP1 induction in human colon cancer cells. Int. J. Oncol. 2007;31:613–620. [PubMed] [Google Scholar]
- 50.Spaapen R.M., Leung M.Y., Fuertes M.B., Kline J.P., Zhang L., Zheng Y., Fu Y.X., Luo X., Cohen K.S., Gajewski T.F. Therapeutic activity of high-dose intratumoral IFN-β requires direct effect on the tumor vasculature. J. Immunol. 2014;193:4254–4260. doi: 10.4049/jimmunol.1401109. [DOI] [PubMed] [Google Scholar]
- 51.Xia T., Konno H., Ahn J., Barber G.N. Deregulation of STING Signaling in Colorectal Carcinoma Constrains DNA Damage Responses and Correlates With Tumorigenesis. Cell Rep. 2016;14:282–297. doi: 10.1016/j.celrep.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teo W.H., Chen H.P., Huang J.C., Chan Y.J. Human cytomegalovirus infection enhances cell proliferation, migration and upregulation of EMT markers in colorectal cancer-derived stem cell-like cells. Int. J. Oncol. 2017;51:1415–1426. doi: 10.3892/ijo.2017.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim K.S., Carp R.I. Abortive infection of human diploid cells by murine cytomegalovirus. Infect. Immun. 1972;6:793–797. doi: 10.1128/iai.6.5.793-797.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafemina R.L., Hayward G.S. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J. Gen. Virol. 1988;69:355–374. doi: 10.1099/0022-1317-69-2-355. [DOI] [PubMed] [Google Scholar]
- 55.Hansen S.G., Ford J.C., Lewis M.S., Ventura A.B., Hughes C.M., Coyne-Johnson L., Whizin N., Oswald K., Shoemaker R., Swanson T. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen S.G., Sacha J.B., Hughes C.M., Ford J.C., Burwitz B.J., Scholz I., Gilbride R.M., Lewis M.S., Gilliam A.N., Ventura A.B. Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science. 2013;340:1237874. doi: 10.1126/science.1237874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marzi A., Murphy A.A., Feldmann F., Parkins C.J., Haddock E., Hanley P.W., Emery M.J., Engelmann F., Messaoudi I., Feldmann H., Jarvis M.A. Cytomegalovirus-based vaccine expressing Ebola virus glycoprotein protects nonhuman primates from Ebola virus infection. Sci. Rep. 2016;6:21674. doi: 10.1038/srep21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klyushnenkova E.N., Kouiavskaia D.V., Parkins C.J., Caposio P., Botto S., Alexander R.B., Jarvis M.A. A cytomegalovirus-based vaccine expressing a single tumor-specific CD8+ T-cell epitope delays tumor growth in a murine model of prostate cancer. J. Immunother. 2012;35:390–399. doi: 10.1097/CJI.0b013e3182585d50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grenier J.M., Yeung S.T., Qiu Z., Jellison E.R., Khanna K.M. Combining Adoptive Cell Therapy with Cytomegalovirus-Based Vaccine Is Protective against Solid Skin Tumors. Front. Immunol. 2018;8:1993. doi: 10.3389/fimmu.2017.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu Z., Huang H., Grenier J.M., Perez O.A., Smilowitz H.M., Adler B., Khanna K.M. Cytomegalovirus-Based Vaccine Expressing a Modified Tumor Antigen Induces Potent Tumor-Specific CD8(+) T-cell Response and Protects Mice from Melanoma. Cancer Immunol. Res. 2015;3:536–546. doi: 10.1158/2326-6066.CIR-14-0044. [DOI] [PubMed] [Google Scholar]
- 61.Qiu Z., Grenier J.M., Khanna K.M. Reviving virus based cancer vaccines by using cytomegalovirus vectors expressing modified tumor antigens. OncoImmunology. 2015;5:e1056974. doi: 10.1080/2162402X.2015.1056974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinn M., Erkes D.A., Snyder C.M. Cytomegalovirus and immunotherapy: opportunistic pathogen, novel target for cancer and a promising vaccine vector. Immunotherapy. 2016;8:211–221. doi: 10.2217/imt.15.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snyder C.M., Cho K.S., Bonnett E.L., Allan J.E., Hill A.B. Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS Pathog. 2011;7:e1002295. doi: 10.1371/journal.ppat.1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Messerle M., Sapinoro R., Santos K., Hocknell P.K., Jin X., Dewhurst S. Murine cytomegalovirus abortively infects human dendritic cells, leading to expression and presentation of virally vectored genes. J. Virol. 2003;77:7182–7192. doi: 10.1128/JVI.77.13.7182-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang X., Chen D.G. Recombinant murine cytomegalovirus vector activates human monocyte-derived dendritic cells in a NF-kappaB dependent pathway. Mol. Immunol. 2009;46:3462–3465. doi: 10.1016/j.molimm.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolpert F., Happold C., Reifenberger G., Florea A.M., Deenen R., Roth P., Neidert M.C., Lamszus K., Westphal M., Weller M., Eisele G. Interferon-β Modulates the Innate Immune Response against Glioblastoma Initiating Cells. PLoS ONE. 2015;10:e0139603. doi: 10.1371/journal.pone.0139603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.