Abstract
Background
Since their emergence in Indonesia in 2005, 200 human infections with clade 2.1 highly pathogenic avian influenza A/H5N1 virus have been reported, associated with exceptionally high mortality (84%) compared to regions affected by other genetic clades of this virus. To provide potential clues towards understanding this high mortality, detailed clinical virological analyses were performed in specimens from 180 H5N1 patients, representing 90% of all Indonesian patients and 20% of reported H5N1-infected patients globally.
Methods
H5N1 RNA was quantified in available upper- and lower-respiratory tract specimens as well as fecal and blood samples from 180 patients with confirmed infection between 2005 and 2017. Mutations in the neuraminidase and M2 genes that confer resistance to oseltamivir and adamantanes were assessed. Fatal and nonfatal cases were compared.
Results
High viral RNA loads in nasal and pharyngeal specimens were associated with fatal outcome. Mortality increased over time during the study period, which correlated with increasing viral RNA loads on admission. Furthermore, the prevalence of amantadine resistance–conferring M2 mutations increased over time, and viral loads were higher in patients infected with viruses that harbored these mutations. Compared to observations from other regions, viral RNA was detected more frequently in feces (80%) and particularly in blood (85%), and antiviral responses to oseltamivir appeared less pronounced.
Conclusions
These observations confirm the association of viral load with outcome of human H5N1 infections and suggest potential differences in virulence and antiviral responses to oseltamivir that may explain the exceptionally high mortality related to clade 2.1 H5N1 infections in Indonesia.
Keywords: highly pathogenic avian influenza virus, H5N1, resistance, viral load, mortality
Detailed virological analyses of 180 Indonesian patients infected with avian influenza A/H5N1 virus point to potential differences in virulence and antiviral responses to oseltamivir that may explain the exceptionally high mortality related to these infections in Indonesia compared to other regions.
Since their emergence as zoonotic pathogens in 1997, highly pathogenic avian influenza A (H5N1) viruses have spread across Asia, Europe, and Africa, posing a persisting threat to animal and human health globally [1]. During 2017 and 2018 alone, outbreaks in poultry and wild birds were reported in 17 Asian and African countries, including 8 countries during the first 6 months of 2018, with millions of birds succumbing to the disease or culled to control outbreaks [2].
Sporadic transmission to humans occurs during poultry outbreaks. Since 2003, 860 human H5N1 infections have been reported to the World Health Organization (WHO), with nearly two-thirds of cases occurring in Egypt and Indonesia. While the number of reported human infections has declined substantially in recent years despite continued circulation in birds, underreporting due to reduced awareness and limited diagnostic capacity in affected African and Asian countries cannot be excluded.
The overall mortality related to reported human infections is high (53%), with remarkable differences among countries and regions, ranging from 33% in Egypt to as high as 84% in Indonesia, where fewer than one-quarter of all reported cases but more than one-third of all deaths occurred [3, 4]. These regional differences in outcome might be related to differences in access to healthcare and clinical management among countries, differences in virulence of circulating viruses, or both. The relatively low mortality in Egypt compared to south and southeast Asian countries, where the case fatality rate is more than twice as high (69%), may, in part, be attributable to earlier identification, hospitalization, and antiviral treatment of patients in Egypt [5]. However, the timing of hospitalization and treatment cannot readily explain the mortality differences among Asian countries. For example, despite similarly late treatment of patients in Vietnam and Indonesia (median, 6 days after illness onset), reported mortality in Vietnam is substantially lower (50% in Vietnam vs 84% in Indonesia) [6, 7]. Although genetically distinct H5N1 virus lineages circulate in different regions and countries, insights into possible differences in virulence or other viral characteristics that could impact clinical outcome are very limited. Studies that relate virological determinants to clinical outcome are largely limited to observations in small numbers of Vietnamese and Cambodian patients infected by clade 1 H5N1 viruses. These studies showed that a fatal outcome is associated with high pharyngeal viral loads, the presence of viremia, and a poor response to oseltamivir treatment due to antiviral resistance [7–13]. Comparable data from patients infected with other H5N1 virus clades may contribute to improved understanding of the differences in mortality but are currently lacking.
In the present study, we report detailed virological analyses in a range of clinical specimens from 180 Indonesian patients with laboratory-confirmed H5N1 infection, representing the first comprehensively studied series of human clade 2.1 H5N1 infections and the largest cohort of human H5N1 infections overall. In addition to confirming that high nasopharyngeal viral loads are associated with poor clinical outcome, the virus was more commonly detected in blood, and early antiviral responses to treatment with oseltamivir appeared less pronounced when compared to Vietnamese H5N1 clade 1–infected patients. Strikingly, the case fatality rate in Indonesia increased over time and was associated with increasing viral loads and the emergence of mutations in the matrix protein that confers adamantane resistance. These observations suggest that differences in virulence and other viral characteristics may contribute to the exceptionally high mortality related to human clade 2.1 H5N1 infections in Indonesia.
METHODS
Specimen and Data Collection
The analyses were conducted on anonymized available specimens and associated demographic and clinical data from Indonesian patients with laboratory-confirmed H5N1 infection during the period 2005–2017. Specimens and data were collected and stored at the National Institute of Health Research and Development (NIHRD), Ministry of Health of Indonesia, as part of the national procedure for avian influenza case investigation. This national procedure prescribed to send clinical specimens to NIHRD in case of suspected H5N1 infection, including repeat respiratory specimens during hospitalization, for diagnostic and confirmatory testing [14]. Specimens were stored at –80°C after laboratory confirmation was performed using H5-specific reverse-transcriptase polymerase chain reaction (RT-PCR) [15]. The laboratory analyses of the current study were conducted on all available respiratory (nasal, pharyngeal, pleural, tracheal, bronchial), gastrointestinal (rectal, fecal), and blood specimens. Specimen and data collections were approved by the Ministry of Health of Indonesia as part of the national investigation of avian influenza [14]. The current investigations were approved by the ethical and scientific review board of the NIHRD, Ministry of Health.
Virological Analyses
Viral RNA was isolated from clinical specimens using PureLink (Invitrogen/Thermo Fisher, Carlsbad, CA) or QIAamp (Qiagen, Hilde, Germany) RNA extraction kits, according to the manufacturers’ instructions. Quantitative detection of H5N1 RNA was done batch-wise using real-time RT-PCR as described previously [11, 12]. The presence of mutations in the viral neuraminidase and matrix 2 (M2) genes that confer resistance to oseltamivir and adamantanes, respectively, was analyzed using pyrosequencing or Sanger sequencing [16, 17]. Viral RNA was reverse transcribed as described previously [8, 11, 12] and amplified using gene-specific primers. Primer sequences are available upon request. Amplification was performed using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) or HotStar Taq (Qiagen, Hilden, Germany) according to the manufacturers’ instructions. The ExoSAP-IT (Affymetrix, Inc., Santa Clara, CA) purification kit was used to purify the PCR products. Pyrosequencing was performed on the PyroMark ID instrument using Pyro Gold Reagents kits (Biotage AB, Uppsala, Sweden). For Sanger sequencing, DNA was sequenced using the Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Inc., Foster City, CA) according to the manufacturer’s instructions using a 16-capillary 3130xl Genetic Analyzer (Applied Biosystems).
Statistical Analyses
The χ2 or Fisher exact test was used to compare categorical variables, and the Mann-Whitney test was used for group comparisons of continuous variables. Viral RNA loads were analyzed after log transformation. For statistical purposes, the lower detection limit of the assay (10 cDNA copies/mL) was used for correlation analyses in case no viral RNA could be detected. Correlations between variables were calculated using the Spearman rank correlation test. All statistical analyses were done using SPSS v14.0 (SPSS Inc., Chicago, IL) or Graphpad Prism 7.02 (GraphPad Software, La Jolla, CA). A P value < .05 was considered statistically significant.
RESULTS
Patient Demographics and Outcome
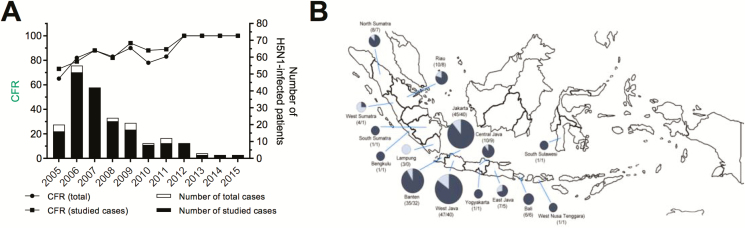
Clinical specimens and associated demographic and clinical outcome data were available for analysis from 180 patients with laboratory-confirmed H5N1 infection, representing 90% of all Indonesian patients reported to WHO since the emergence of H5N1 influenza in Indonesia in 2005 until 2017 (Figure 1A, Table 1). Patients originated from 15 of 34 Indonesian provinces, with the majority of cases from the province of Java (Figure 1B). The median age of patients was 19 years (range, 1–67), and 54% were female (Table 1).
Figure 1.
Reported human H5N1 infections in Indonesia, 2005–2015. A, Number of human H5N1 infections in Indonesia reported to the World Health Organization (white bars) and cases included in this study (black bars). Lines depict case fatality proportions of reported infections (circles) and cases included in this study (squares). B, Geographical origin of cases. Size of pie diagrams reflect number of cases, with the dark fraction representing fatalities. Abbreviation: CFR, case fatality rate.
Table 1.
Overview of Demographic and Clinical Characteristics of H5N1-Infected Patients
| Median age (range), y | 19 (1–67) |
| Gender | |
| Female | 97 |
| Male | 83 |
| Year of infection | |
| 2006 | 16 |
| 2007 | 51 |
| 2008 | 42 |
| 2009 | 22 |
| 2010 | 17 |
| 2011 | 8 |
| 2012 | 9 |
| 2013 | 2 |
| 2014 | 2 |
| 2015 | 2 |
| Geographical origin | |
| Bali | 6 |
| Banten | 35 |
| Bengkulu | 1 |
| Jakarta | 45 |
| West Java | 47 |
| Central Java | 10 |
| East Java | 7 |
| Lampung | 3 |
| West Nusa Tenggara | 1 |
| Riau | 10 |
| South Sulawesi | 1 |
| West Sulawesi | 4 |
| South Sumatra | 1 |
| North Sumatra | 8 |
| Yogyakarta | 1 |
| Median days after symptom onset (range) | 9 (–1 to 27) |
| Antiviral treatment | |
| Yes | 123 |
| No | 57 |
| Clinical outcome | |
| Survival | 27 |
| Death | 153 |
| Type of specimen | |
| Upper respiratory tract | 699 |
| Nasal swabs | 346 |
| Pharyngeal swabs | 346 |
| Other | 7 |
| Lower respiratory tract | 141 |
| Bronchial washes | 22 |
| Tracheal aspirates | 66 |
| Pleural fluids | 44 |
| Other | 9 |
| Blood | 26 |
| Gastrointestinal tract | 44 |
| Central nervous system | 1 |
| Other | 3 |
| Total | 914 |
Of the 180 studied cases, 153 (85 %) were fatal. The case fatality rate increased from 73% in 2005 to 100% in 2012 and subsequent years (Figure 1A). There were no significant differences in age, gender, or days since illness onset before presentation between surviving and fatal cases (data not shown). Over time, the days since illness onset were also similar each year until 2012 but appeared longer during the period 2013–2015, although numbers are very small (Supplementary Figure 1).
H5N1 RNA Load in Clinical Specimens
Available stored clinical specimens from the 180 patients included nasal swabs from 164 (91%), pharyngeal swabs from 157 (88%), lower respiratory tract specimens (bronchial washes, tracheobronchial aspirates, pleural fluids) from 92 (52%), serum or plasma from 26 (14%), and rectal swabs or feces from 25 (14%). Quantitative detection of H5N1 RNA was performed in all 914 available specimens (Table 1).
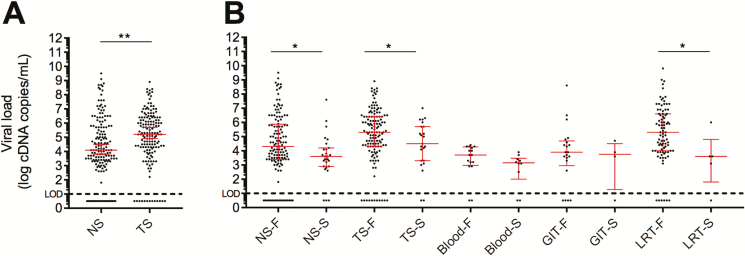
Viral RNA was detected in 144 of 157 pharyngeal and 142 of 163 nasal specimens obtained at admission, with significantly higher H5N1 RNA levels observed in throat swabs than in nasal swabs (Figure 2A). To estimate the natural course of viral RNA load during human H5N1 infection, viral loads in initial nasal and pharyngeal specimens obtained from individual patients prior to initiation of treatment were plotted against the days since reported onset of symptoms at the time of sample collection (Supplementary Figure 2). This showed that H5N1 RNA could be detected up to 19 days after onset of symptoms with no clear trend toward lower viral RNA levels in specimens collected later during the course of infection.
Figure 2.
Viral RNA load in clinical specimens. A, H5N1 RNA levels in admission NS and TS. B, Comparison of H5N1 RNA levels in NS and TS of F and S cases. H5N1 RNA levels in LRT, GIT, and blood specimens. Median value and interquartile range are depicted in red. Abbreviations: F, fatal; GIT, gastrointestinal tract; LRT, lower respiratory tract; NS, nasal swabs; S, surviving; TS, throat swabs. *P < .05; **P < .005.
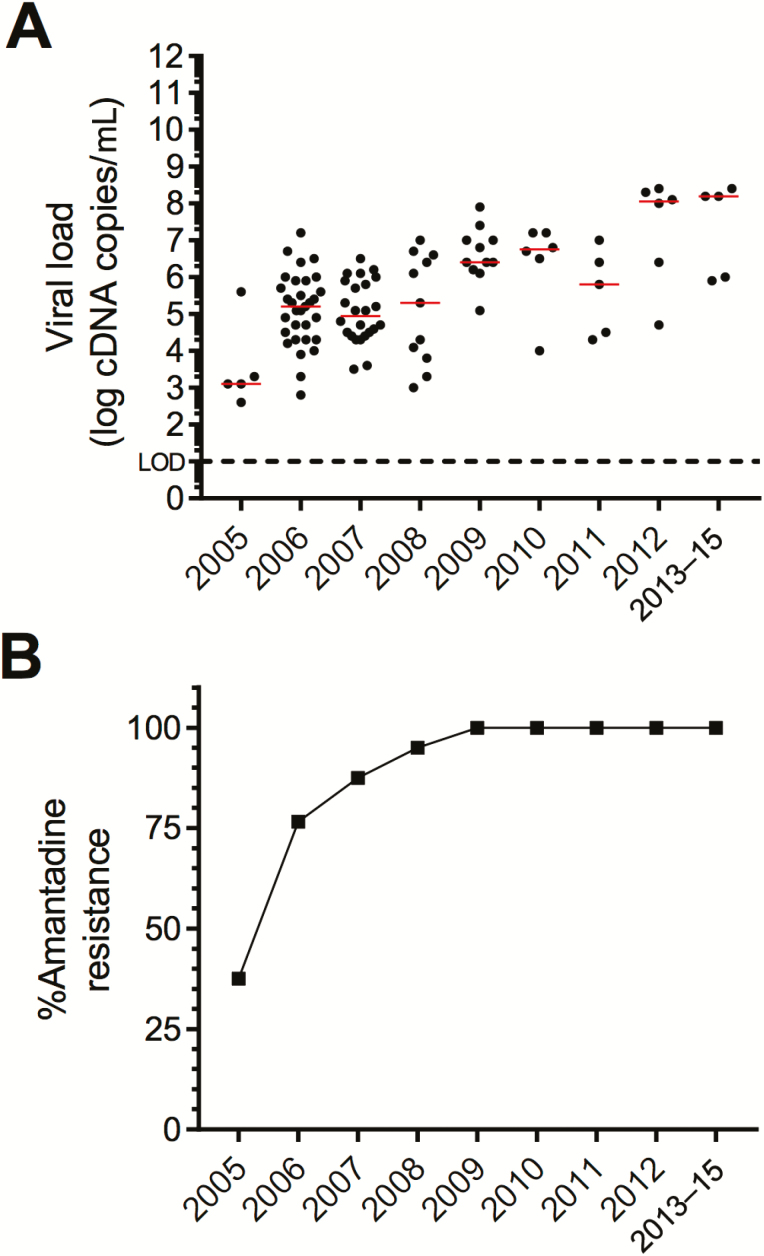
Both in nasal and pharyngeal specimens, H5N1 RNA levels were significantly higher in patients who died than in those who survived (nasal: median, 4.3 log vs 3.6 log cDNA copies/mL, respectively; P = .0135 and pharyngeal; mean, 5.3 log vs 4.5 log cDNA copies/mL, respectively; P = .041; Figure 2B). During the study period, median viral RNA loads in pretreatment specimens gradually increased, which correlated significantly with the rising mortality rates during the same period (Spearman ρ = .86, P = .0014; Figure 3).
Figure 3.
Viral RNA load and prevalence of amantadine resistance over time. A, H5N1 RNA levels in admission throat swabs per year of infection. Median value indicated in red. B, Prevalence of amantadine resistance-conferring M2 mutations over time.
Viral RNA was also detected in 85 of 92 lower respiratory tract specimens, of which all except 4 were collected from fatal cases (Figure 2C). In 20 of 25 (80%) patients, viral RNA could be detected in rectal swabs or feces. Detection rates and viral loads were similar between fatal and surviving cases, but the number of surviving cases from whom specimens were available was small (n = 4). In available serum or blood specimens, viral RNA was detected in 22 of 26 (85%) patients, with similar detection rates observed in fatal and surviving cases (14 of 16 [88%] and 8 of 10 [80%] patients, respectively). Viral RNA levels in blood were higher in fatal cases, but this difference did not reach statistical significance (median, 3.7 log cDNA copies/mL [range, 2.9–4.4] vs 3.2 [range 2.5–3.9], respectively; P = .08).
Response to Antiviral Treatment
Treatment with oseltamivir had a statistically significant impact on clinical outcome: 22 of 123 patients (18%) who received treatment survived vs none of the remaining 57 untreated patients (P < .001). The median duration from onset of illness to initiation of treatment was 7 days (range, 0–18), with no statistically significant differences between surviving and fatal cases (median, 6 days [range 0–18] vs 7 days [range 0–14], respectively; P = .15). However, the survival rate was significantly higher in patients treated earlier than the median of 7 days compared to those treated on day 7 or later (16/51 [31%] vs 11/71 [15%], respectively; P = .037). This difference in survival was more pronounced when patients treated on or before 4 days after illness onset were compared to those treated later (8/16 [50%] vs 19/106 [18%], respectively; P = .004).
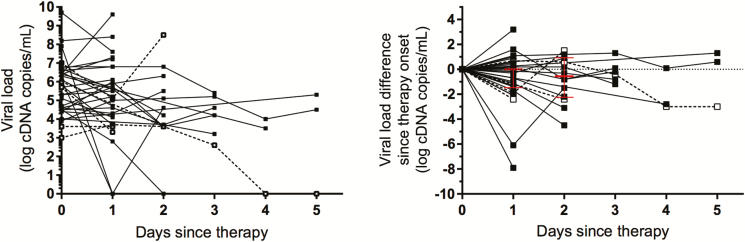
Serial pharyngeal specimens collected before and during antiviral treatment were available from 34 patients. The median reduction in viral RNA load after 2 days of treatment was 0.55 log cDNA copies/mL (range, –4.5 to 1.5), with only 2 patients reaching undetectable levels within 2 days (Figure 4). During subsequent days, viral RNA remained detectable in 6 of 6 patients at day 3 and in 2 of 3 patients at days 4 and 5.
Figure 4.
Viral RNA load during oseltamivir treatment. A, H5N1 RNA levels in serial throat swabs collected from fatal (black line) and surviving (dashed line) cases before and during oseltamivir treatment. B, Changes in H5N1 RNA load from pretreatment baseline levels. Median value and interquartile range are depicted in red.
Oseltamivir and Adamantane Resistance Mutations
The presence of the oseltamivir resistance–conferring His275Tyr (H275Y) substitution in neuraminidase (NA) was investigated in admission respiratory specimens from 155 patients and in 168 pharyngeal samples collected from 78 patients after initiation of oseltamivir treatment (Supplementary Table 1) and was not observed in any of the specimens tested.
While detection of the H275Y substitution was based on residue-specific pyrosequencing methods in the majority of specimens, Sanger sequencing was used in a proportion of samples, which allowed detection of 2 other resistance-conferring substitutions, that is, R293K and N295S (N1 numbering) [18]. Neither of these substitutions was found in 29 specimens from 14 patients after initiation of treatment (Supplementary Table 2) and in specimens from 14 patients with no treatment.
Sequence analyses of the M2 gene in admission specimens from 153 patients revealed the presence of mutations that confer resistance to adamantanes in 131 (85.6%) patients. The mutations identified were Val27Ala (n = 109), Val27Thr (n = 1), Ser31Asn (n = 7), Val27Ala + Ser31Asn (n = 3), Val27Ala + Ser31Gly (n = 7), and Leu26Ile + Ser31Asn (n = 4). M2 resistance mutations were detected more frequently in fatal cases compared to survivors (117/132 [89%] vs 14/21 [67%], respectively; P = .015). Over time, the prevalence of M2 resistance mutations increased gradually from 37.5% in 2005 to 95% in 2009 and to 100% during subsequent years (Figure 3B). These increases correlated with increasing median pharyngeal viral RNA loads during the same period (Spearman ρ = 0.83; P = .0019). As a result, admission viral RNA loads in throat specimens from patients who harbored virus with M2 resistance mutations were higher than those from patients infected with virus without these mutations (median, 5.3 log cDNA copies/mL vs 4.3 log cDNA copies/mL; P = .0042).
DISCUSSION
In a large cohort of 180 Indonesian patients with influenza A/H5N1 infection, representing one-fifth of all globally reported cases and 90% of those reported from Indonesia, we provide information to explain the exceptionally high and increasing mortality related to human H5N1 clade 2.1 infections in Indonesia.
As shown previously in small case series of patients infected with clade 1 H5N1 viruses, viral RNA could be detected in respiratory specimens at high levels for more than 2 weeks after illness onset, indicating prolonged virus replication and shedding during the course of the disease [7–13]. Furthermore, a fatal outcome was not only associated with higher viral loads in the throat, as has been reported previously, but also in nasal specimens. The limited number of patients investigated in previous series may explain why the latter was not observed before. The detection rates of viral RNA in gastrointestinal specimens and particularly in blood were higher in Indonesian patients than previously reported in Vietnamese H5N1-infected patients (gastrointestinal, 80% vs 71% and blood, 85% vs 56%, respectively) [8]. It remains unclear whether the detection of viral RNA in these specimens reflects infectious virus since no attempts were made to isolate virus due to sample volume restrictions. However, isolation of infectious virus from feces and blood during human H5N1 infections has previously been reported [12, 19]. Hence, although numbers of patients in this and previous series were small, it is tempting to speculate that higher rates of viremia in our patients and consequent increased risk of systemic spread of clade 2.1 viruses may have contributed to the higher mortality among Indonesian patients.
Treatment with oseltamivir was associated with a survival benefit. This benefit was greater when treatment was started earlier in the course of infection, confirming earlier observations and emphasizing the need for early recognition and diagnosis [7, 20]. Although differences in survival among countries may be difficult to interpret in view of possible differences in healthcare access and medical practices among H5N1-affected regions, reported survival in oseltamivir-treated patients infected by clade 1 H5N1 viruses is substantially higher than in our clade 2.1-infected patients (52.5% vs 18%), despite late treatment initiation in both [21]. Common oseltamivir resistance–conferring neuraminidase mutations, including H275Y, R293K, and N295S [18], were not detected during treatment in any of the specimens tested. In addition, we looked for a range of other neuraminidase mutations reportedly associated with reduced oseltamivir susceptibility in recently published whole-genome, next-generation sequencing data that we generated from 44 Indonesian patients to study viral quasispecies evolution during human infection (Supplementary Table 3) [22]. Except for 2 variants present as minority viral populations in 4 patients (V116A in 3 patients at 3.6%–6.0% of the viral population, N295S in 1 at 1.6%), other substitutions were not observed [23–26].
The above observations argue against the emergence of known resistance mutations to explain the apparent limited benefit of oseltamivir treatment in Indonesian patients. However, in vitro susceptibility testing of H5N1 viruses has shown that clade 2.1 viruses are intrinsically up to 30-fold less susceptible to oseltamivir than clade 1 viruses [27]. Similar differences in oseltamivir susceptibility between human seasonal influenza A and B viruses have been linked to a longer duration of virus shedding and fever during treatment of influenza B virus–infected patients [28, 29]. Along the same lines, it cannot be excluded that similar differences in drug susceptibility between clade 1 and clade 2.1 H5N1 viruses are associated with differences in antiviral responses and consequently clinical benefit. In a small case series of Vietnamese H5N1 clade 1–infected patients, rapid reductions in viral RNA to undetectable levels, mostly within 2 days of treatment, were observed in surviving patients, while viral RNA remained detectable at high levels in fatal cases and was associated with the emergence of resistance [11]. These observations appear to contrast with our findings in Indonesian patients that show that, despite an absence of oseltamivir resistance mutations, viral RNA was still detectable after 2 days of treatment in 42 of 44 patients and remained detectable throughout the course of treatment in the vast majority of patients who were followed. Although the numbers of evaluated patients are limited, this might be explained by the aforementioned differences in oseltamivir susceptibility.
Changes in mortality were observed during the study period, increasing from 69% overall to 100% in 2012 and beyond. These changes in mortality over time correlated with increasing admission viral loads and with an increasing prevalence of viruses that contain adamantane resistance–conferring M2 mutations during the same period. In turn, admission viral loads were higher in the presence of M2 mutations.
Time from illness onset to presentation of patients and initiation of treatment remained similar until 2012 but appeared longer thereafter, which may have contributed to the high mortality during that period. However, the increasing viral loads during the same period remain striking given that no correlation between viral RNA load and illness duration was observed overall (Supplementary Figure 2). In addition to supporting an important role of viral load in determining outcome, these observations suggest an increasing virulence of circulating H5N1 viruses over time, possibly associated with the presence of M2 substitutions. Similar to observations in seasonal human influenza viruses [30, 31], the increasing prevalence of adamantane resistance M2 mutations in Indonesian H5N1 viruses occurred in the absence of apparent widespread use of amantadine, neither in poultry nor in humans. This suggests that the presence of these mutations is associated with a natural fitness advantage, either indirectly through a genetic “hitchhiking” effect in association with changes elsewhere in the viral genome or directly through potential advantageous effects on the biological function of the M2 protein. The latter direct biological role of observed M2 mutations in enhancing virulence cannot be excluded and deserves further study. In murine experiments, mortality in mice infected with isogenic recombinant H1N1 viruses that harbored amantadine resistance mutations (Val27Ala, Ser31Gly, or both) was higher than in mice infected with wild-type H1N1 viruses [32, 33]. Furthermore, the Val27Ala change in M2, representing the most prevalent mutation observed in our patients, has been associated with increased M2 activity in avian influenza A/H7N1 viruses, which suggests that this particular substitution may have functional significance [34].
In summary, our study of a large series of H5N1-infected Indonesian patients confirms the association between high upper respiratory viral loads and fatal outcome and provides information to explain the exceptionally high mortality related to H5N1 disease in Indonesia, including a high proportion of patients with viremia, potential suboptimal antiviral efficacy of oseltamivir, and an increasing virulence of circulating viruses, possibly associated with the emergence of M2 mutations. These observations deserve consideration in future further research efforts in human infections with H5N1 and other avian influenza viruses.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. H. A. P., V. S., H. R. v. D., and M. D. d. J. contributed to the conception and design of the study; H. A. P. and V. S. participated in acquisition of data; H. A. P. and T. T. T. conducted the laboratory analyses; H. A. P., D. E., S. I., V. S., P. Q. T., H. R. v. D., and M. D. d. J. were responsible for data analyses and interpretation; and H. A. P., D. E., H. R. v. D., and M. D. d. J. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Acknowledgments. This article is dedicated to the memory of the late Dr Endang R Sedyaningsih, former Minister of Health of Indonesia, who initiated and inspired the research reported here. The authors thank their many colleagues at the Directorate General of Disease Control and Environmental Health and the National Institute of Health Research and Development, Ministry of Health, Jakarta, Indonesia; the provincial, district, and subdistrict health offices; and healthcare providers at Indonesian hospitals who attended H5N1 patients. They thank Phung Quoc Tuan from Oxford University Clinical Research Unit, Ho Chi Minh City, Vietnam, for advice regarding statistical analyses.
Disclaimer. The Ministry of Health of Indonesia and the funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. Financial support was provided by the Indonesia Ministry of Health (to H. A. P., S. I., and V. S.), the UK Wellcome Trust (to H. R. v. D), the European Union FP7 programs European Management Platform for Emerging and Re-emerging Infectious disease Entities (223498; to M. D. J.) and ANTIcipating the Global Onset of Novel Epidemics (278976; to M. D. J. and D. E.), a SPIN Challenges Exploration grant from the Royal Netherlands Academy of Arts and Sciences (to M. D. J.), a doctorate scholarship from the Dutch Organisation for Internationalisation in Education (Nederlandse organisatie voor internationalisering in onderwijs; to H. A. P.), and the European Union H2020 Marie Curie International Incoming Fellowship (to D. E.).
Potential conflicts of interest. The authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Sedyaningsih ER, Isfandari S, Setiawaty V, et al. Epidemiology of cases of H5N1 virus infection in Indonesia, July 2005–June 2006. J Infect Dis 2007; 196:522–7. [DOI] [PubMed] [Google Scholar]
- 2. influenza OSRfa. Update on avian influenza in animals (types H5 and H7) Available at: www.oie.int/animal-health-in-the-world/update-on-avian-influenza/. Accessed December 2018.
- 3. World Health Organization. Cumulative number of confirmed human cases of avian influenza A/(H5N1) reported to WHO Available at: http://www.who.int/influenza/human_animal_interface/2016_12_19_tableH5N1.pdf?ua=1. Accessed 19 December 2018.
- 4. Kandun IN, Wibisono H, Sedyaningsih ER, et al. Three Indonesian clusters of H5N1 virus infection in 2005. N Engl J Med 2006; 355:2186–94. [DOI] [PubMed] [Google Scholar]
- 5. Lai S, Qin Y, Cowling BJ, et al. Global epidemiology of avian influenza A H5N1 virus infection in humans, 1997–2015: a systematic review of individual case data. Lancet Infect Dis 2016; 16:e108–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liem NT, Tung CV, Hien ND, et al. Clinical features of human influenza A (H5N1) infection in Vietnam: 2004–2006. Clin Infect Dis 2009; 48:1639–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kandun IN, Tresnaningsih E, Purba WH, et al. Factors associated with case fatality of human H5N1 virus infections in Indonesia: a case series. Lancet 2008; 372:744–9. [DOI] [PubMed] [Google Scholar]
- 8. de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12:1203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buchy P, Mardy S, Vong S, et al. Influenza A/H5N1 virus infection in humans in Cambodia. J Clin Virol 2007; 39:164–8. [DOI] [PubMed] [Google Scholar]
- 10. Tran TH, Nguyen TL, Nguyen TD, et al. ; World Health Organization International Avian Influenza Investigative Team Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 2004; 350:1179–88. [DOI] [PubMed] [Google Scholar]
- 11. de Jong MD, Tran TT, Truong HK, et al. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N Engl J Med 2005; 353:2667–72. [DOI] [PubMed] [Google Scholar]
- 12. de Jong MD, Bach VC, Phan TQ, et al. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N Engl J Med 2005; 352:686–91. [DOI] [PubMed] [Google Scholar]
- 13. Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza AV , Abdel-Ghafar AN, Chotpitayasunondh T, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med 2008; 358:261–73. [DOI] [PubMed] [Google Scholar]
- 14. Development, NIoHRa. Guidelines for taking, handling, and shipping of specimens from avian influenza cases (Pedoman pengambilan, penatalaksanaan, dan pengiriman spesimen Flu Burung). 2nd ed. Jakarta, Indonesia: NIHRD, 2007. [Google Scholar]
- 15. World Health Organization. WHO case definition for human infection with influenza A(H5N1) virus Available at: http://www.who.int/influenza/resources/documents/case_definition2006_08_29/en/. Accessed 29 August 2018.
- 16. Lackenby A, Democratis J, Siqueira MM, Zambon MC. Rapid quantitation of neuraminidase inhibitor drug resistance in influenza virus quasispecies. Antivir Ther 2008; 13:809–20. [PubMed] [Google Scholar]
- 17. Deyde VM, Nguyen T, Bright RA, et al. Detection of molecular markers of antiviral resistance in influenza A (H5N1) viruses using a pyrosequencing method. Antimicrob Agents Chemother 2009; 53:1039–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. World Health Organization. Summary of neuraminidase amino acid substitutions associated with reduced inhibition by neuraminidase inhibitors Available at: https://www.who.int/influenza/gisrs_laboratory/antiviral_susceptibility/NAI_Reduced_Susceptibility_Marker_Table_WHO.pdf?ua. Accessed December 2018.
- 19. de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 2006; 12:1203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Adisasmito W, Chan PK, Lee N, et al. Effectiveness of antiviral treatment in human influenza A(H5N1) infections: analysis of a global patient registry. J Infect Dis 2010; 202:1154–60. [DOI] [PubMed] [Google Scholar]
- 21. Chan PK, Lee N, Zaman M, et al. Determinants of antiviral effectiveness in influenza virus A subtype H5N1. J Infect Dis 2012; 206:1359–66. [DOI] [PubMed] [Google Scholar]
- 22. Welkers MRA, Pawestri HA, Fonville JM, et al. Genetic diversity and host adaptation of avian H5N1 influenza viruses during human infection. Emerg Microbes Infect 2019; 8:262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boltz DA, Douangngeun B, Phommachanh P, et al. Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People’s Democratic Republic. J Gen Virol 2010; 91:949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hurt AC, Selleck P, Komadina N, Shaw R, Brown L, Barr IG. Susceptibility of highly pathogenic A(H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antiviral Res 2007; 73:228–31. [DOI] [PubMed] [Google Scholar]
- 25. Ilyushina NA, Seiler JP, Rehg JE, Webster RG, Govorkova EA. Effect of neuraminidase inhibitor-resistant mutations on pathogenicity of clade 2.2 A/Turkey/15/06 (H5N1) influenza virus in ferrets. PLoS Pathog 2010; 6:e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le QM, Kiso M, Someya K, et al. Avian flu: isolation of drug-resistant H5N1 virus. Nature 2005; 437:1108. [DOI] [PubMed] [Google Scholar]
- 27. McKimm-Breschkin JL, Selleck PW, Usman TB, Johnson MA. Reduced sensitivity of influenza A (H5N1) to oseltamivir. Emerg Infect Dis 2007; 13:1354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kawai N, Ikematsu H, Iwaki N, et al. Longer virus shedding in influenza B than in influenza A among outpatients treated with oseltamivir. J Infect 2007; 55:267–72. [DOI] [PubMed] [Google Scholar]
- 29. Kawai N, Ikematsu H, Iwaki N, et al. A comparison of the effectiveness of oseltamivir for the treatment of influenza A and influenza B: a Japanese multicenter study of the 2003–2004 and 2004–2005 influenza seasons. Clin Infect Dis 2006; 43:439–44. [DOI] [PubMed] [Google Scholar]
- 30. Simonsen L, Viboud C, Grenfell BT, et al. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 2007; 24:1811–20. [DOI] [PubMed] [Google Scholar]
- 31. Deyde VM, Xu X, Bright RA, et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis 2007; 196:249–57. [DOI] [PubMed] [Google Scholar]
- 32. Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother 2005; 49:556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pizzorno A, Bouhy X, Abed Y, Boivin G. Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis 2011; 203:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grambas S, Bennett MS, Hay AJ. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology 1992; 191:541–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.