Abstract
Live-cell imaging of mitochondrial function and dynamics can provide vital insights into both physiology and pathophysiology, including of metabolic diseases like type 2 diabetes. However, without super-resolution microscopy and commercial analysis software, it is challenging to accurately extract features from dense multilayered mitochondrial networks, such as those in insulin-secreting pancreatic β-cells. Motivated by this, we developed a comprehensive pipeline and associated ImageJ plugin that enables 2D/3D quantification of mitochondrial network morphology and dynamics in mouse β-cells and by extension other similarly challenging cell types. The approach is based on standard confocal microscopy and shareware, making it widely accessible. The pipeline was validated using mitochondrial photolabeling and unsupervised cluster analysis and is capable of morphological and functional analyses on a per-organelle basis, including in 4D (xyzt). Overall, this tool offers a powerful framework for multiplexed analysis of mitochondrial state/function and provides a valuable resource to accelerate mitochondrial research in health and disease.
Keywords: cell metabolism, diabetes, fluorescence microscopy, image analysis, live-cell imaging
INTRODUCTION
Mitochondria are the main energy-producing organelles of eukaryotic cells and are essential for a diverse range of cellular functions, including ATP synthesis, Ca2+ homeostasis, ROS signaling, and the control of apoptotic cell death (12, 36). Microscopy has been instrumental in unraveling intricacies of mitochondrial biology and their diverse roles in cellular physiology and pathophysiology. Electron microscopy has provided fundamental insights into mitochondrial ultrastructure and cellular distribution in health and disease but requires cell fixation and provides only a static snapshot. In contrast, fluorescence microscopy of live cells labeled with mitochondria-targeted fluorescent proteins or dyes has revealed that mitochondria are highly dynamic and motile organelles that undergo frequent fusion and fission events (23, 51, 52). Mitochondrial dynamics and network morphology vary in different cellular states and are important for the function and quality control of the organelle as well as overall cell health and adaptation to stress (12). Healthy mitochondria are generally mobile and tubular in shape and exist in complex networks, whereas cells undergoing profound stress or entering apoptosis often display swollen and fragmented mitochondria, marked by concurrent disruption of metabolism, membrane potential, ROS levels, and Ca2+ signaling (4, 6, 31). Therefore, quantitative imaging-based assessment of mitochondrial morphology and dynamics can provide valuable insights into cellular physiology and pathophysiology.
In pancreatic β-cells, mitochondria play an essential role in insulin secretion, which relies on ATP and other mitochondria-derived metabolites to both trigger and amplify insulin granule exocytosis in response to glucose and other nutrient stimuli (38). Therefore, dysfunction of β-cell mitochondria results in loss of glucose-stimulated insulin secretion (46). Perturbations to mitochondria are also a common feature in insulin target tissues with impaired insulin signaling (21, 41). Mitochondria thus take center stage in both β-cell failure and insulin resistance and are an area of significant focus in efforts to understand the pathophysiology of type 2 diabetes (13, 17, 45). Mitochondria also exist as dynamic networks in β-cells. Fusion within the network may help protect β-cells from nutrient stress-induced apoptosis (31), and mitochondrial fragmentation, swelling, and dysfunction are seen in β-cells from patients with type 2 diabetes and rodent models of diabetes (3, 11, 17, 29). Normal insulin secretion may also be influenced by β-cell mitochondrial dynamics (18, 22, 34), but exactly how networking of the organelle relates to its metabolic capacity in healthy β-cells or during conditions of moderate nutrient excess remains unclear and warrants further investigation. Therefore, improved methods for analysis of mitochondrial morphology and dynamics in live β-cells may help provide new insights into important aspects of β-cell biology.
Most types of microscopy can detect the prominent morphological differences between healthy and severely stressed mitochondria with relative ease. However, it is much more challenging to accurately quantify subtle changes in mitochondrial dynamics or perform three-dimensional (3D) analysis of the full mitochondrial network. This is particularly difficult in cells with a dense mitochondrial network that spans several layers, such as β-cells (11, 31). Although methods have been published that integrate 3D confocal imaging and analysis of mitochondria, these generally use commercial software packages and/or are optimized for relatively flat cell types (32, 47). This is likely one reason why there are only few quantitative analyses of β-cell mitochondrial dynamics and why full 3D investigations of β-cell mitochondria are limited to a small number of examples using super-resolution approaches such as 4Pi-microscopy (11, 37).
To facilitate progress in the important area of mitochondrial biology and dynamics, we present here a pipeline for quantitative multidimensional analysis of mitochondria that is based on standard confocal fluorescence microscopy and the open-source image analysis platform ImageJ/Fiji (43, 44). In this, we identify a superior method for accurate identification of individual mitochondria within dense networks, and we outline a framework for quantitative description of mitochondrial morphology and network characteristics. Applying this pipeline to clonal MIN6 β-cells and primary mouse β-cells, we quantitatively distinguish mitochondrial morphologies, including the functional and morphological changes to physiological and pathophysiological stimuli. Additionally, we discuss the pros and cons of two-dimensional (2D) and 3D imaging approaches, identify image-processing steps required for accurate mitochondrial analysis in 3D, and apply these to quantitate distinct 3D β-cell network morphologies. Finally, we extend our analysis to four-dimensional (4D) imaging by including time lapse data, and we demonstrate the feasibility of using the pipeline to quantitate the dynamics of the entire three-dimensional mitochondrial network in live cells.
MATERIALS AND METHODS
Reagents.
Collagenase type XI (no. C7657), tetramethylrhodamine ethyl ester (TMRE; no. 87917), d-glucose (no. G7528), bovine serum albumin (BSA; no. A7030), FCCP (no. C2920), and palmitic acid (no. P5585) were purchased from Sigma-Aldrich (St. Louis, MO). MitoTracker Deep Red FM (no. M224726), MitoTracker Green FM (MTG, no. M7514), Hoechst 33342 (no. H3570), RPMI 1640 (no. 11879), Dulbecco’s modified Eagle’s medium (DMEM; no. 11995), fetal bovine serum (FBS; no. 10438), trypsin-EDTA (no. 25300), penicillin-streptomycin (10,000 U/mL; no. 15140), and HBSS (no. 14185) were purchased from Life Technologies/Thermo Fisher Scientific (Carlsbad, CA). Dimethyl sulfoxide (DMSO; no. BP231) was purchased from Fisher Scientific (Waltham, MA). Minimum essential media (MEM; no. 15-015-CV) was purchased from Corning (Corning, NY). The mitochondria-targeted YFP (mito-YFP) and mitochondria-targeted photoactivatable GFP (mito-PAGFP) plasmids were gifts from Dr. Mark Cookson (42) and Dr. Richard Youle (Addgene; no. 23348) (23), respectively.
Cell isolation and culture.
MIN6 cells were cultured at 37°C and 5% CO2 using complete DMEM supplemented with 10% FBS and 2% penicillin-streptomycin. Culture media were replaced every 2 days, and cells were passaged upon reaching 70–80% confluency. To assess the effects of palmitate on mitochondrial networks, cells were cultured for 6 h in complete DMEM supplemented with either 1.5 mM palmitate complexed to BSA in a 6:1 ratio or BSA-only vehicle control.
Pancreatic islets were isolated from wild-type male mice of a mixed C57BL/6 and CD1 background using collagenase digestion and filtration-based purification, as previously described (1). The isolated islets were hand-picked and allowed to recover overnight before being dispersed into single cells and seeded on 25-mm glass coverslips (30). The islet cells were cultured in RPMI complete with 10% FBS and 2% penicillin-streptomycin at 37°C and 5% CO2 for 4 days before imaging. All animal procedures were approved by the University of British Columbia Animal Care Committee.
Cell transfection and mitochondrial labeling.
MIN6 cells were seeded at a density of 2.0 × 105 on 25-mm glass coverslips (0.13- to 0.16-mm thickness, VWR no. 16004-310) and incubated for 24 h before being transfected with mito-YFP, mito-dsRed, or mito-PAGFP plasmids using Lipofectamine 2000 (no. 11668; Life Technologies) as per the manufacturer’s protocol. All plasmids were expressed for ≥24 h before confocal microscopy.
To assess the effect of acute glucose exposure on mitochondrial morphology and membrane potential, primary mouse islet cells were cultured for 60 min in complete RPMI media containing 3 or 17 mM glucose and then stained with 0.1 µg/mL Hoechst 33342, 50 nM MTG, and 25 nM TMRE for 30 min, followed by a wash with complete RPMI immediately before imaging.
Image acquisition by confocal microscopy.
Live cells were imaged in a Tokai Hit INUBTFP-WSKM stage-top incubator at 37°C on a Leica SP8 Laser Scanning Confocal Microscope (Concord, ON, Canada). For 2D, images were acquired using a ×63 HC Plan Apochromatic water immersion objective (1.2 NA). Pixel size was adjusted using the “Optimize” function in the Leica LASX Software, resulting in a consistent pixel width of ∼80 nm. The pinhole size was 1.0 Airy unit. For 3D acquisition, z-stacks were obtained using a ×63 oil immersion objective (1.4 NA). Pixel size (x, y) and z-spacing were adjusted as per the calculated optimal Nyquist sampling parameters (44a), and pinhole size was reduced to 0.75 Airy units. The z-step size generally varied between 170 and 220 nm. Bidirectional scanning was enabled, and all images were acquired using at least three frame averages. Laser power, detector filtering/gating, and gain were adjusted to maximize signal without saturation while also minimizing background signal, cross-fluorescence, and photobleaching.
Time lapse 3D Imaging.
Time lapse 3D (xyzt) imaging was performed on MIN6 cells transfected with mito-YFP plasmid. Acquisition settings were established as above, and z-stacks were acquired every 45 s for 30 min. The acquisition time for each stack was ∼30 s. At the 13 min-mark, FCCP was added to the chamber for a final concentration of 25 µM.
Mitochondrial labeling by photoactivatable green fluorescent protein.
MIN6 cells were co-transfected with mito-PAGFP and mito-dsRed 24 h before imaging, as described above. Mito-dsRed was visualized using a 561-nm excitation laser, with emission detected between 585 nm to 650 nm. Individual mitochondria were marked for photolabeling using the “Bleach Point” function in the Leica LasX software, and the PAGFP was activated using a 405-nm laser pulse of 150-ms duration. The activated PAGFP was then imaged using a 488-nm excitation laser with an emission range between 505 nm and 550 nm. All other imaging settings were as described above.
Image deconvolution.
Deconvolution of 3D and 4D stacks was performed in Huygens Professional version 16.10 (SVI) using the Classic Maximum Likelihood Estimation algorithm with a signal-to-noise ratio of 7.0, maximum iterations of 40, and quality threshold of 0.001. For deconvolution in ImageJ/Fiji, the “PSF Generator” plugin (24) was used to generate a theoretical PSF based on our microscope parameters, and deconvolution was performed with the “DeconvolutionLab2” module utilizing the Richardson-Lucy TV algorithm with regularization set to 0.0001 and maximum iterations of 30 (9, 40).
Image processing and thresholding.
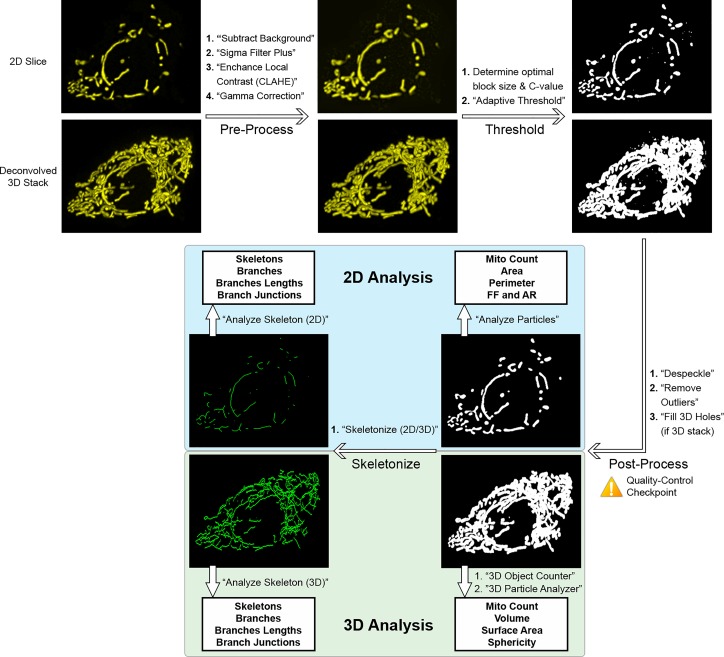
The workflow and procedures for image processing and thresholding are summarized in Fig. 6. Using ImageJ/Fiji, 2D images or deconvolved 3D image stacks (operating on each slice in the stack) were preprocessed using the following commands: 1) “subtract background” (radius = 1 μm) to remove background noise; 2) “sigma filter plus” (radius = 0.1 μm, 2.0 sigma) to reduce noise and smooth object signal while preserving edges; 3) “enhance local contrast” (block size = 64, slope = 2.0 for 2D and 1.25 for 3D stacks) to enhance dim areas while minimizing noise amplification; and 4) “gamma correction” (value = 0.80 for 2D and 0.90 for 3D) (16) to correct any remaining dim areas. To identify mitochondria in the images, we evaluated multiple global and local thresholding algorithms (Fig. 1 and Supplemental Figs. S1–S3; Supplemental Materials for this article are available at https://doi.org/10.6084/m9.figshare.9994088). Based on our comparisons, we elected to use the “adaptive threshold” method (50). In the adaptive threshold plugin, block size was set to an equivalent of 1.25 μm and the optimal C value was empirically determined for each image set (see Supplemental Fig. S2 for additional details). The thresholded images were then postprocessed using “despeckle” and then “remove outliers” (radius = 0.15 μm) to remove residual noise. For 3D stacks, we additionally applied the “fill 3D holes” command from the “3D ROI manager” plugin (33).
Fig. 6.
Summary of pipeline for 2-dimensional (2D) and 3-dimensional (3D) mitochondrial analysis in ImageJ/Fiji. For illustration, an image stack was acquired from a MIN6 cell expressing mitochondria-targeted yellow fluorescent protein (mito-YFP); a representative slice is shown as the 2D input and the entire stack (after deconvolution) as the 3D input. 3D stacks are represented as maximum projections here. Scale bars, 5 μm. See materials and methods and results for additional details and parameter values. AR, aspect ratio; FF, form factor.
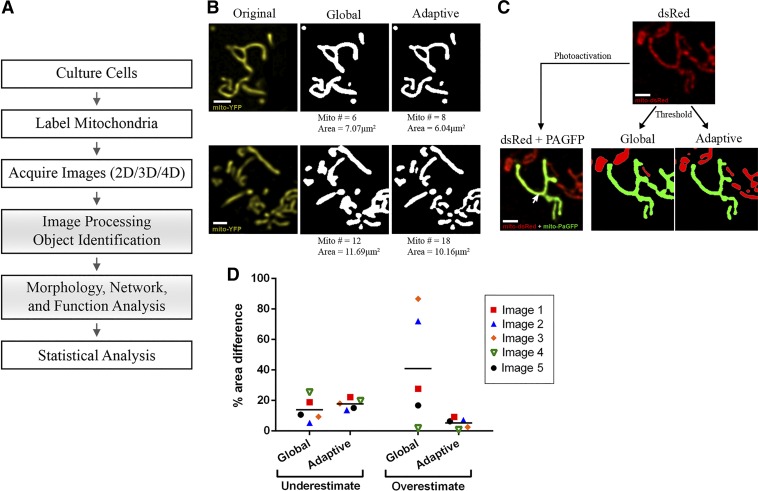
Fig. 1.
General workflow and comparison of mitochondrial identification using global vs. adaptive thresholding methods. A: schematic of the general workflow required for mitochondrial analysis by confocal microscopy. Shaded boxes represent the steps that are addressed and detailed in this paper. B: 2 representative examples of object identification using global thresholding (“default” method) vs. adaptive thresholding (radius = 1.25 μm, C = 11) on images of MIN6-cell mitochondria labeled with mitochondria-targeted yellow fluorescent protein (mito-YFP). The number of identified objects (mitochondria) and their total area are indicated below the images. Scale bar, 1 μm. C: part of the mitochondrial network in a MIN6 cell co-transfected with mito-dsRed and mitochondria-targeted photoactivatable green fluorescent protein (mito-PAGFP). Top: all mitochondria imaged in the mito-dsRed channel. Bottom left: a single mitochondrion (green) was labeled by laser-based mito-PAGFP activation at the point indicated by the arrow. Bottom right: object identification using global vs. adaptive threshold algorithms applied to the dsRed channel; in each image, the object that is identified as contiguous with the PAGFP-labeled mitochondrion is shown in green. Comparison with the original image shows that the adaptive method more accurately distinguished the photo-labeled mitochondrion, whereas global thresholding artificially merged it with adjacent mitochondria. Scale bar, 1 μm. D: quantitative comparison of the degree to which global and adaptive thresholding under- or overestimated the PAGFP-labeled mitochondrion in 5 test images. The corresponding images and details of the estimation algorithm are shown in Supplemental Fig. S3. 2D, 2-dimensional; 3D, 3-dimensional; 4D, 4-dimensional.
2D analysis of mitochondrial function, morphology, and network characteristics.
The approach for quantification of mitochondrial characteristics is summarized in Fig. 6. For 2D analysis, the image was first processed and thresholded (see above), and the resulting binary image was used as the input for the “analyze particles” command (size = 0.06 μm2-infinity, circularity = 0.00–1.00), measuring for “area,” “perimeter,” and “shape descriptors.” Form Factor (FF) was derived as the inverse of the “circularity” output value. For network connectivity analysis, the “skeletonize 2D/3D” command was applied to the thresholded image to produce a skeleton map, and the “analyze skeleton” command was used to calculate the number of branches, branch lengths, and branch junctions in the skeletonized network.
To simultaneously measure mitochondrial polarization and morphology, islet cells were co-stained with MTG and TMRE. Our threshold method was first applied to the MTG channel, and morphological analysis was done on the identified objects. Additionally, the analyze particles command (“add to manager” option enabled) was used to convert the identified objects (mitochondria) into regions of interest (ROIs). These ROIs were then superimposed onto the raw images of the MTG and TMRE channels, and the MTG and TMRE intensities of each individual mitochondrion were measured as the “mean gray value” obtained via the analyze particles command. The degree of mitochondrion polarization was then expressed as the ratio of TMRE to MTG intensity and correlated with mitochondrial morphology on a per-organelle basis.
3D analysis of mitochondrial morphology and network characteristics.
For 3D analysis, the image stacks were first deconvolved, preprocessed, and thresholded as described above and summarized in Fig. 6. Next, the “3D object counter” command (size = 0.6 μm3-Infinity) was used to calculate the number of mitochondrial objects and produce a labeled object map. The object map was subsequently used as an input for the “particle analyzer 3D” command (part of the MorphoLibJ package) (26) to calculate the volume, sphericity (weighted by volume of the object), and corrected surface area (“Crofton 13 directions” method) of each mitochondrial object. Network connectivity analysis was performed on the skeletonized 3D network using the same commands as 2D analysis. For 4D (xyzt) analysis, these 3D analysis steps were performed on each stack obtained in the time course acquisition.
Unsupervised categorization of mitochondrial morphology using spanning tree progression analysis of density-normalized events.
Spanning tree progression analysis of density-normalized events (SPADE; version 3.0) (2, 39) was used to automatically classify 2D mitochondrial images into three different categories based exclusively on their calculated morphological and network parameters. Briefly, mitochondrial parameter data were transformed into a flow cytometry standard (FCS) file using FlowJo version 10 and loaded into SPADE. A SPADE tree was created using default settings, without application of arcsin transformation or removal of outliers. The number of desired clusters was set equal to the total number of images in the data set. The “auto suggest annotation” function was then used to partition the SPADE tree into two subgroups, and the larger of these was subsequently auto-partitioned again, resulting in a total of three subgroups. The data in these SPADE-identified groups were then exported as CSV formatted files for statistical comparison.
Statistical analysis.
All data were represented as means ± SE. Data were analyzed in GraphPad Prism 6.0 software (La Jolla, CA) using Student’s t-test or one-way ANOVA followed by a Sidak multiple-comparison test as appropriate. Statistical significance was set at a threshold of P < 0.05.
RESULTS
Overall workflow and general considerations.
Fluorescence confocal analysis of mitochondria in live cells involves several general steps, each of which is important for high-quality results (Fig. 1A). As a starting point, the cells must be cultured on glass coverslips, or other vessels, that are appropriate for confocal microscopy. The mitochondria should then be labeled using carefully chosen mitochondria-targeted fluorescent proteins or organic dyes (28), and the image acquisition should be optimized and carried out in a manner that provides sufficiently high resolution and image quality for accurate analysis. Because these factors and general steps can vary between specific experiments and microscope systems, an extensive discussion falls beyond the scope of this paper. The imaging parameters and conditions we have used are detailed in Materials and Methods. Our focus in the following will be on the post-acquisition steps that are critical for accurate morphological analysis of mitochondria in the confocal images.
Image acquisition and analysis can be done in 2D or 3D, and by further extending this to include time lapse capture, important information can be extracted about mitochondrial dynamics. The choice between these imaging modes may be influenced by several considerations, including the type and thickness of the cell, the specific parameters to be quantified, and the biological questions being asked. For instance, we will discuss later how some 2D analyses of relatively thick cells, such as pancreatic β-cells, can be associated with inaccuracies that may be mitigated by a full 3D analysis of the mitochondrial network. In all cases, accurate quantification of mitochondrial features involves image-processing steps and identification of the mitochondrial objects in the image. Morphological features can then be extracted using appropriate 2D or 3D shape descriptors, whereas mitochondrial networking can be assessed through skeletonization analysis. In this latter process, the binarized mitochondria are converted into topological skeletons (the thinnest form that is equidistant to its edges), and the branches of the skeleton are analyzed. In the following, we describe each of these postacquisition steps and identify a number of “best approaches” to build a pipeline for accurate multidimensional analysis of mitochondria that we also implement and make available in a comprehensive Mitochondria Analyzer plugin for ImageJ/Fiji (7).
Image thresholding and identification of mitochondria.
Before accurate morphological analysis of fluorescently-labeled mitochondria can be done, it is essential that 1) the mitochondrial population is correctly identified in the images and 2) the individual mitochondrial units can be distinguished within the dense mitochondrial network. For this critical step, a thresholding process based on analysis of the intensity histogram is used to distinguish pixels with “true” fluorescent signal from background signal. This process also groups any identified positive pixels that are connected into discrete objects (i.e., mitochondria) that can be analyzed further. Thresholding approaches can be broadly categorized as either “global” or “local,” which identify positive pixels based on the histogram of the entire image or on dynamic analyses of image subregions, respectively (16, 32). Global thresholding tends to be the most commonly used approach, but this may reflect its relative ease of use rather than accuracy.
To identify the most suitable thresholding strategy for mitochondrial identification, we compared the performance of the global and local threshold methods available for ImageJ/Fiji on images of primary islet cells stained with MitoTracker dye. This was judged on the ability to preserve mitochondrial structural detail while minimizing capture of background signal. For optimal results, all images were pre- and postprocessed to reduce noise (see materials and methods). Among the global-based algorithms in the ImageJ/Fiji “auto threshold” command, we qualitatively estimated that the default method performs similar to, or in several cases better than, the other global algorithms (Supplemental Fig. S1).
The local thresholding methods we tested included the mean, median, and mid-gray algorithms (part of the “auto local threshold” command), as well as the weighted mean method (also called adaptive threshold), which is available through a separate plugin (50) (Supplemental Fig. S2). These local methods compute a threshold for each pixel in the image and require the definition of two parameters: a block size and a C value. The block size specifies the size of the region around each pixel for which the histogram is analyzed and should be chosen based on the size of the objects of interest for the best results (50). The C value provides an offset to the threshold and helps strike a balance between minimizing noise detection and incorrectly splitting objects into smaller pieces (25, 50). Using the adaptive threshold method for optimization, we found that the ideal C value depended on the image’s signal-to-noise contrast and needed to be empirically determined. Therefore, for each set of images that has been acquired and processed in a similar manner, we recommend that various combinations of block size and C values should be tested on a representative image to determine the best combination. The optimized parameters can then be used to threshold all images in the group similarly (see Supplemental Fig. S2 for details). Among the local threshold approaches, our assessment was that the mean and adaptive threshold methods best captured mitochondrial structure and that the adaptive threshold further tended to identify less noise (Supplemental Fig. S2B).
A side-by-side comparison indicated that local (adaptive) thresholding resolves mitochondria better than global thresholding, which appears to capture more out-of-focus signal and/or noise, and therefore, it erroneously merges adjacent objects (Fig. 1, B and C). For a more stringent and quantitative evaluation, we used mitochondria-targeted photoactivatable GFP (mito-PAGFP) to selectively photo label single mitochondria and identify truly contiguous organelles within dense regions of the network (23, 52). As exemplified in Fig. 1C and quantified in Fig. 1D (see also Supplemental Fig. S3), adaptive thresholding was indeed better at delineating photo-labeled mitochondria and distinguishing closely adjacent parts of the network that are physically separate. In contrast, the global threshold algorithm consistently overestimated the mitochondrial size. Taken together, these comparisons established that using adaptive thresholding, with empirically optimized parameter values, is a superior approach for accurate identification of fluorescently labeled β-cell mitochondria.
Two-dimensional analysis of mitochondrial morphology and network connectivity.
After careful image thresholding, the next step is to quantify the morphological features of the identified mitochondrial objects. Therefore, we identified a comprehensive set of parameters to capture and mathematically describe key aspects of the mitochondrial morphology. For 2D analysis, we characterize mitochondrial size by area and perimeter, whereas mitochondrial shape is defined by form factor (FF) and aspect ratio (AR). We evaluate the overall connectivity and morphological complexity of the mitochondrial network based on the skeletonized network and quantify this by the number of branches and the number of branch junctions as well as total (accumulated) length of branches in the skeleton. Supplemental Figure S4 summarizes the various parameters and indicates how they change with various morphologies.
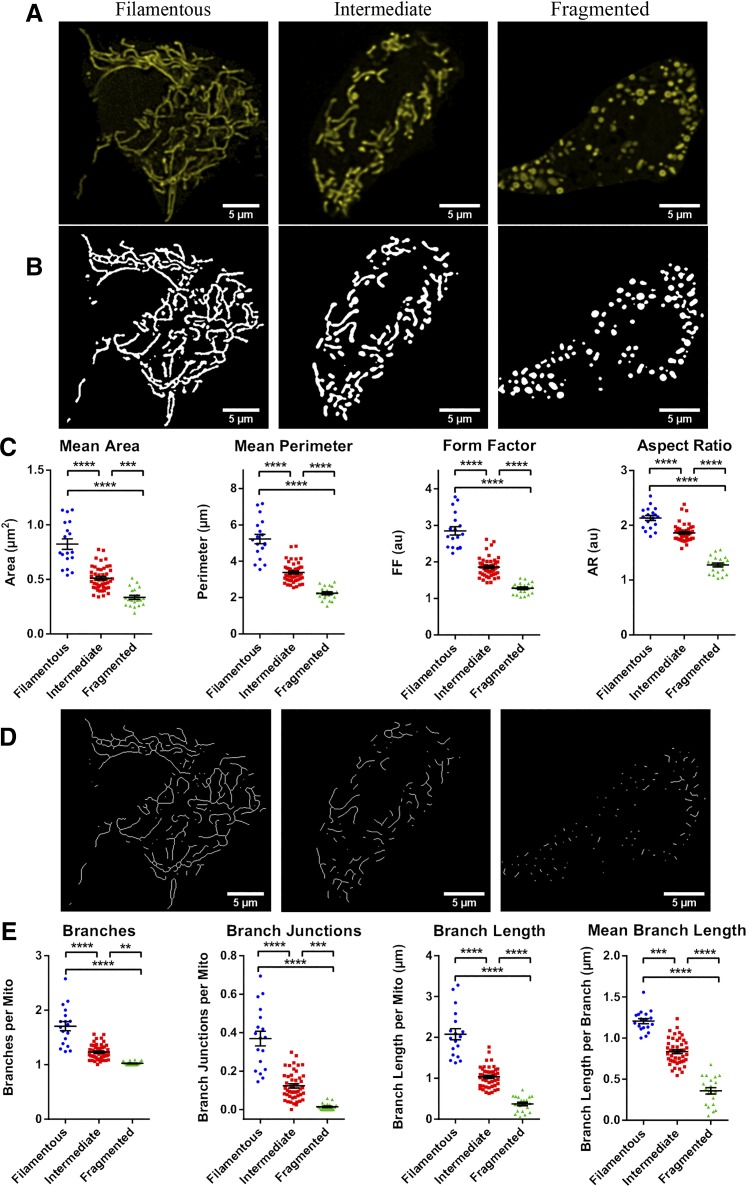
To evaluate the ability of this approach to measure and distinguish mitochondrial morphologies, we transfected MIN6 cells with mito-YFP and generated an image set consisting of 2D slices from 84 cells. We then divided the cells into three different categories based on visual inspection of their mitochondria: 1) a “fragmented” group, characterized by small round mitochondria and little branching; 2) a “filamentous” group, with highly connected networks of long/filamentous mitochondria; and 3) an “intermediate” group of cells containing a mixture of punctate and longer tubular mitochondria. As shown in Fig. 2, analysis of the 2D images resulted in quantitative morphological and networking parameters that differed significantly between the three groups. Of note is that a more in-depth comparison of the two shape descriptors revealed that FF required smaller sample sizes than AR to detect differences between the three morphological subtypes and seemed particularly well-suited for distinguishing between cells with filamentous and intermediate mitochondrial morphologies (Supplemental Fig. S5). This is likely because AR only measures elongation, whereas FF incorporates the perimeter, and therefore it is more sensitive to curvature and the irregular shapes of filamentous mitochondria (Supplemental Fig. S5B). Collectively, these results demonstrate that our combined approach for image processing, thresholding, and analysis enables quantitative identification and comparison of mitochondrial morphological subtypes.
Fig. 2.
Quantitative comparison of mitochondrial morphology and network connectivity in 2D (2-dimensional). Based on visual inspection of their mitochondria, 84 images of mitochondria-targeted yellow fluorescent protein (mito-YFP)-expressing MIN6 cells were categorized into 3 morphological groups: fragmented (20 cells), intermediate (46 cells), or filamentous (18 cells). A and B: examples of the YFP-labeled mitochondria in representative cells from each group (A) and the objects identified by application of adaptive thresholding to the images (B). C: 2D morphological analysis of all cells in each of the categories. D: skeletonization of the mitochondrial objects identified in B. E: quantitative analysis and comparison of mitochondrial network connectivity performed on all cells in each morphological category. Data are represented by means ± SE. One-way ANOVA with Sidak post hoc test was used to compare the groups; **P < 0.01; ***P < 0.001; ****P < 0.0001. AR, aspect ratio; AU, arbitrary units; FF, form factor.
Validation of morphometric quantifications and classifications by unsupervised clustering.
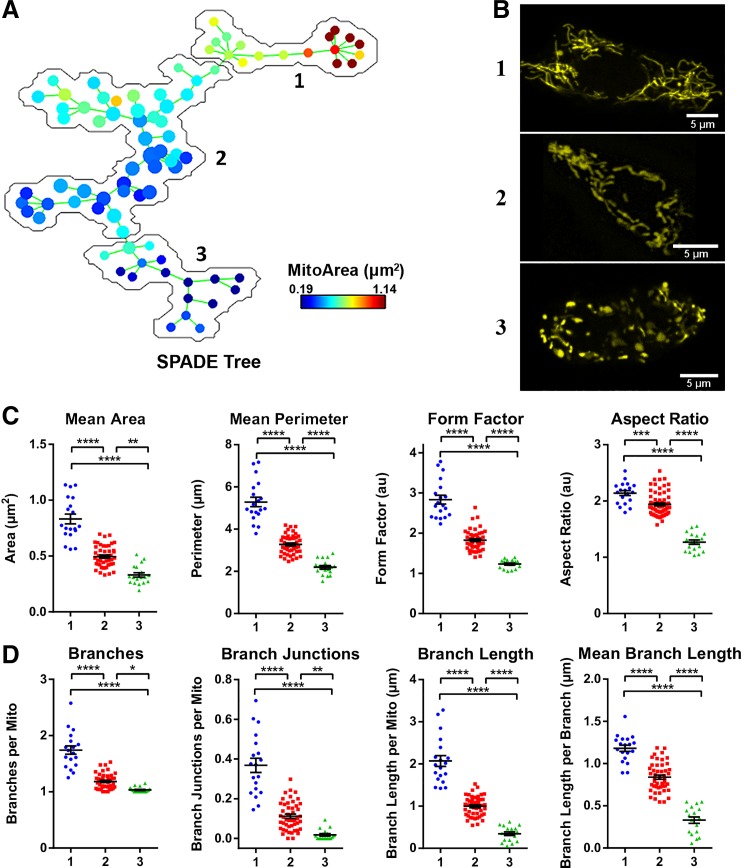
Next, we further tested our pipeline by using spanning tree progression analysis of density-normalized events (SPADE) (2, 39) to obtain an unbiased clustering of our test images. The morphological parameters that had been calculated from our image set of 84 mito-YFP-expressing MIN6 cells (shown in Fig. 2) were loaded into SPADE, which used these to generate a population tree in which each node represents a cell (Fig. 3A). This SPADE tree was then subdivided into three cell populations based on automatic clustering of their mitochondrial features (Fig. 3A; see materials and methods for details). When images from each of the three SPADE-identified groups were subsequently examined, the mitochondria in each group were noticeably dissimilar in appearance (Fig. 3B), and comparative analysis revealed that there were significant differences in all the morphological descriptors (Fig. 3, C and D). The morphometric data indicated that SPADE subgroups 1, 2, and 3 corresponded to cells with filamentous, intermediate, and fragmented mitochondria, respectively. This was confirmed by an 88% match between the unsupervised SPADE clustering and our manual grouping of the cells. Together, these results provide an unbiased validation of the applicability and robustness of our 2D pipeline for analysis of mitochondrial network structure and complexity.
Fig. 3.
Unsupervised categorization of mitochondrial features using spanning tree progression analysis of density-normalized events (SPADE). A: a SPADE tree was generated based on the same set of 84 images used in Fig. 2 and then automatically subdivided into 3 groups; group 1 contains 19 nodes/cells, group 2 contains 47 nodes/cells, and group 3 contains 18 nodes/cells. B: representative images extracted from each of the 3 SPADE-generated groups. C and D: comparison of the mitochondrial morphology and network parameters between the 3 SPADE-identified cell groups. All data are represented by means ± SE. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 as determined by 1-way ANOVA with Sidak post hoc test; n = 84 images.
Limitations of 2D mitochondrial analysis.
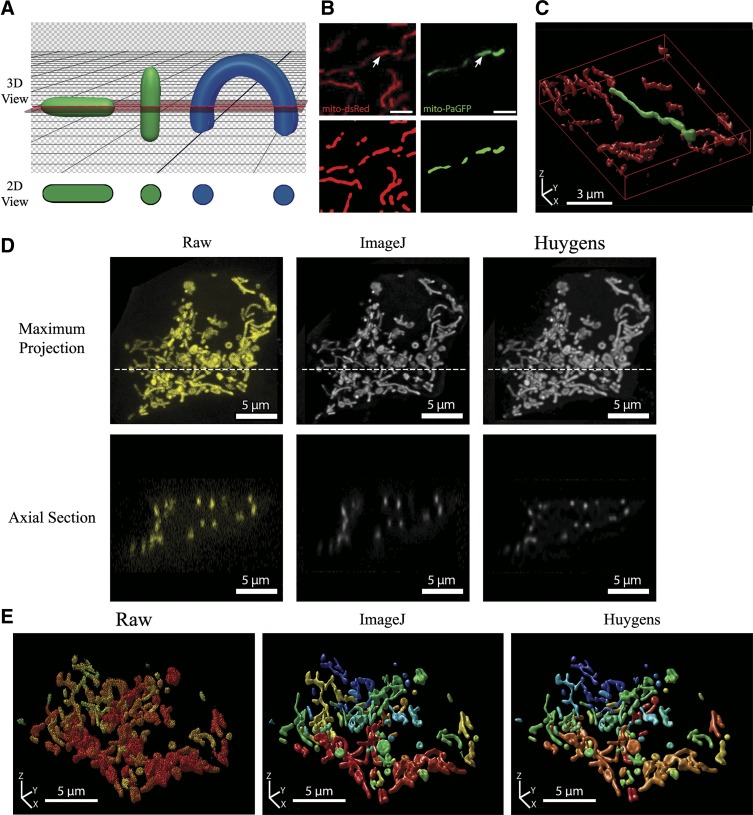
Our 2D analyses reliably measure mitochondrial morphology in an optical cross-section and can provide valuable information regarding the state of the organelle. However, when cells are relatively thick and tend to have a mitochondrial network that spans several layers, this approach has its challenges and limitations. It is difficult to know whether a given plane in a cell is truly representative, and as illustrated by the green objects in Fig. 4A, the 2D appearance of a mitochondrion will also depend on its orientation relative to the optical cross-section. Moreover, a 2D image is unlikely to reveal the actual interconnectedness of the mitochondrial network. When a mitochondrion spans multiple planes and intersects the focal cross-section at several points, it can result in a notable misrepresentation of the morphology, as illustrated by the blue schematic object in Fig. 4A. That this also occurs in situ is demonstrated in the side-by-side 2D and 3D visualization of a photo-labeled mitochondrion in Fig. 4, B and C. When viewed in 2D, the localized photoactivation of mito-PAGFP seemed to label four small and distinct mitochondria (Fig. 4B; shown in green), but a full 3D reconstruction revealed that it was in fact one continuous organelle (Fig. 4C), consistent with diffusion-mediated distribution of GFP within the lumen.
Fig. 4.
Limitations of 2-dimensional (2D) morphometric analysis and the importance of deconvolution for the quality and accuracy of 3-dimensional (3D) mitochondrial analysis. A: schematic illustrating the effect of object orientation in 3D space on the image capture in a horizontal 2D slice. The apparent 2D morphology of the same tubular object (shown in green) will depend on its orientation relative to the confocal plane. If a curved object (shown in blue) intersects the confocal plane at several locations, it will erroneously be identified as separate objects. B: MIN6 cells were co-transfected with mito-dsRed and mitochondria-targeted photoactivatable green fluorescent protein (mito-PAGFP) and photoactivation induced at the point indicated by an arrowhead. Scale bars, 3 µm. Top: 2D image of Mito-dsRed and mito-PAGFP channels after photoactivation. Bottom: objects identified after preprocessing and thresholding of the 2D cross-section. C: full 3D imaging and reconstruction (rendered using Huygens Professional software) of the same mitochondrial population shown in B. Note that the photo-labeled mitochondrion in 2D appears as a series of separate mitochondria, whereas 3D visualization correctly identifies it as 1 contiguous organelle. D: a full z-stack was acquired from a mitochondria-targeted yellow fluorescent protein (mito-YFP)-expressing MIN6 cell that was 11 μm in height. Top: maximum projection views of the z-stack before and after deconvolution. The confocal image stack was deconvolved using either ImageJ DeconvolutionLab (Richardson-Lucy algorithm) or Huygens Professional (Classical Maximum Likelihood Estimation) software for 40 iterations. Dotted line indicates the position of the axial section shown below. Bottom: axial sections (xz-plane) of the raw and deconvolved image stacks. The reduction in axial stretching of objects can be seen in the deconvolved stacks, with the best improvement achieved using the Huygens algorithm (see additional details in Supplemental Fig. S6 and Supplemental Table S1). E: 3D renderings of the z-stack before and after deconvolution with ImageJ or Huygens Professional. All 3D visualizations were generated using the Huygens 3D object renderer, with a unique color assigned to separate objects.
Another inherent limitation of 2D analysis is that it does not allow direct quantitation of the total mitochondrial mass. Cross-sectional area has been used to estimate mass in relatively flat cells like neurons and fibroblasts, where mitochondria are confined to a limited number of planes (5, 32). However, this approximation is less appropriate for thicker cells, including β-cells. A common alternative, intended to capture as much of the mitochondrial network as possible, involves acquiring a stack of z-slices and projecting these into a single plane for faster and simpler analysis (31, 47). Such projections contain information from the whole network, but in voluminous cells this will erroneously merge overlapping mitochondria and produce indiscriminate clusters in the resulting image.
Because the importance of mitochondrial dynamics and its implication for cellular health and disease has become more apparent, there is also an increasing need for more comprehensive characterization of the organelle. Accordingly, there will inevitably be instances where the caveats of 2D analysis we have discussed above become restricting. Therefore, to enable more precise quantification of mitochondrial volume and network structure, we expanded our pipeline to include a complete 3D representation and analysis.
Three-dimensional imaging and analysis of mitochondria.
Full 3D reconstruction of mitochondria can be accomplished by taking a stack of serial slices throughout the volume of the cell and integrating them with software such as ImageJ/Fiji. However, there are technical challenges and constraints specifically associated with 3D imaging. Foremost of these is that the maximum axial resolution (z-axis) of confocal microscopes is ∼500–800 nm, which is almost three times worse than the lateral (xy-plane) (8, 14). Because mitochondria are often <1 μm in diameter, they approach this limit (11). This can lead to a distorted appearance of imaged mitochondria, particularly in the z-axis, where it causes artificial stretching and blending of signal from objects in close vertical proximity to each other. In the following section, we discuss steps that can be taken to mitigate some of these caveats and improve 3D results.
Image acquisition and processing requirements for accurate 3D analysis.
An important first consideration when acquiring a stack of images for 3D analysis is the z-distance between adjacent imaging planes. If the spacing is too large, the final reconstruction will be inaccurate. On the other hand, oversampling will take unnecessary time, increase phototoxicity, and require additional resources for image storage and analysis. Therefore, the distance between serial sections should be set according to the optimal Nyquist sampling rate, which provides the ideal density of information to permit accurate digital reconstruction of an object (35). The Nyquist distance can be calculated using online resources (44a).
Even under optimal conditions, a confocal image will be affected by inherent diffraction-induced distortion of the imaged object. This distortion can be represented by a point-spread function (PSF) and then computationally corrected by using deconvolution algorithms. By removing the effects of the PSF, the deconvolution process provides a more correct representation of the underlying object and also helps eliminate out-of-focus light and/or noise in the image (8). In Fig. 4, we illustrate this and use the free DeconvolutionLab2 module for ImageJ/Fiji and the commercial deconvolution software Huygens Professional (SVI) to test the effect of deconvolution on 3D stacks of mitochondria (see materials and methods for details). As seen in Fig. 4D, mitochondria in the raw image stack have approximately two to three times greater diameter in the xz-view than in the xy-view, which illustrates the z-stretching. The deconvolution algorithms help reduce this distortion, remove noise, and improve the contrast and separation of adjacent objects (Fig. 4D and Supplemental Fig. S6). In general, we found that the Huygens deconvolution package reduced axial stretching more effectively than the ImageJ DeconvolutionLab2 module. By and large, however, both deconvolution algorithms significantly increased the quality of 3D mitochondrial network reconstructions compared with the raw confocal images (Fig. 4E). Deconvolution also affected subsequent 3D quantifications of mitochondrial number, shape, and size in a way that indicated superior separation of individual mitochondria within the full population (Supplemental Table S1; see discussion of the 3D analysis parameters below). In summary, these results demonstrate that deconvolution of the raw confocal image stacks helps mitigate limitations of 3D imaging and is a necessary step for accurate reconstruction and quantification of the full mitochondrial network.
Three-dimensional quantification of mitochondrial morphology and network connectivity.
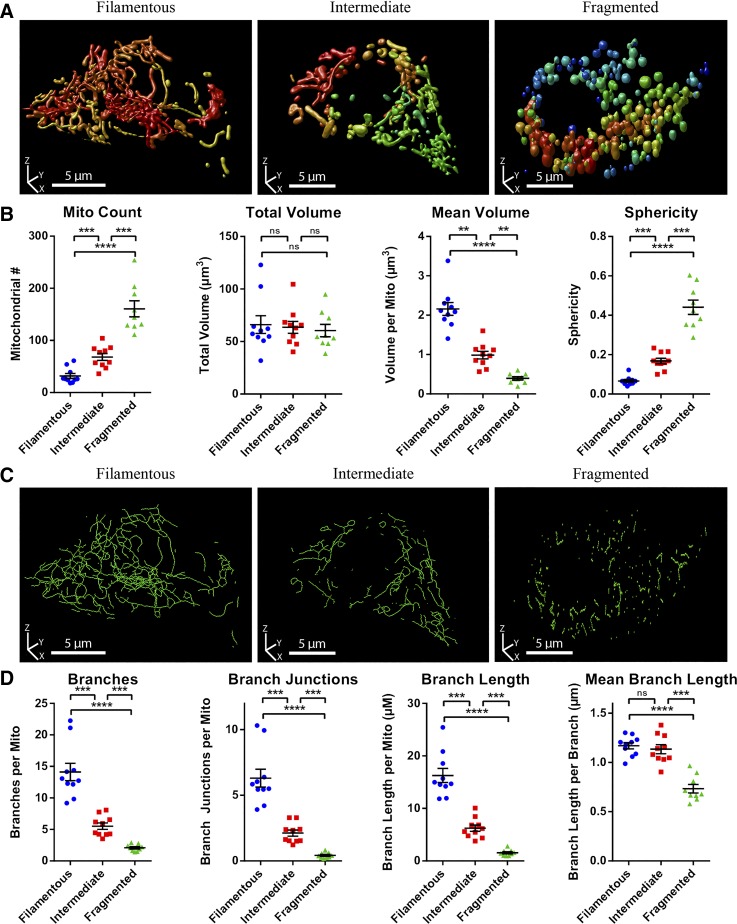
When a high-quality representation of the full mitochondrial network has been generated, ImageJ/Fiji can be used to extract information about the 3D morphology and connectivity by the same general principles previously discussed for 2D. Mitochondrial size in 3D is represented by volume and surface area, whereas shape is characterized by the sphericity of the mitochondrial object. The complexity of the 3D network is quantified by the same branch parameters used for 2D (see Supplemental Fig. S4 for a summary). Analogous to our 2D analyses, we evaluated our 3D approach by generating a set of image stacks from mito-YFP-expressing MIN6 cells and grouping these as fragmented, filamentous, or intermediate based on the visual appearance of the reconstructed mitochondrial networks (Fig. 5A). Quantification using ImageJ/Fiji (see Fig. 6 and materials and methods for details) showed that the number of mitochondria per cell and their average sphericity progressively increased, whereas the average mitochondrial volume decreased, as we moved from filamentous to intermediate to fragmented morphologies (Fig. 5B). In contrast, the total mitochondrial volume of each cell remained constant, highlighting that significant morphological heterogeneity can occur independent of changes to mitochondrial mass (Fig. 5B). In the skeletonized network, the number of branches and branch junctions progressively decreased, illustrating that mitochondrial fragmentation, not surprisingly, is associated with a reduction in overall network complexity (Fig. 5, C and D). Together, the above results and discussions demonstrate how standard confocal imaging can be combined with ImageJ/Fiji-based processing and analysis to quantify volume, morphology, and connectivity of the entire mitochondrial network in live pancreatic β-cells. To our knowledge, 3D investigation of β-cell mitochondria at this level has previously been done only on fixed INS-1 cells using specialized high-resolution 4Pi imaging techniques (11, 37) or super-resolution microscopy (10).
Fig. 5.
Quantitative comparison of mitochondrial morphology and network connectivity in 3D (3-dimensional). Image stacks of mitochondria-targeted yellow fluorescent protein (mito-YFP)-expressing MIN6 cells were visualized in 3D and their mitochondria manually categorized as fragmented, intermediate, or filamentous. A: 3D renderings (produced using Huygens Professional) of representative mito-YFP-expressing MIN6 cells from each of the morphological categories. B: quantitative 3D analysis and comparison of mitochondrial morphology between cells in each category. C: 3D renderings of the skeletonized mitochondrial network of the cells depicted in A. D: quantitative 3D analysis and comparison of mitochondrial network connectivity between cells in each category. All data are represented by means ± SE. **P < 0.01, ***P < 0.001, and ****P < 0.0001, as determined by 1-way ANOVA with Sidak post hoc test; n = 10 cells in each category.
Pipeline summary.
Figure 6 illustrates the overall pipeline for 2D and 3D mitochondrial analysis. In summary, 2D image slices or 3D image stacks are first acquired, with the latter deconvolved before analysis. In ImageJ/Fiji, deconvolution of 3D stacks is done using the DeconvolutionLab2 module (40), and if desired, the 3D stack can be visualized using the “3D viewer” or “volume viewer” functions. Alternatively, 3D deconvolution and visualization can be done using commercial software, such as Huygens, if available to the user (Fig. 4). For analysis, all images are then preprocessed using the commands subtract background, sigma filter plus, enhance local contrast, and gamma correction. We then empirically test a range of block sizes and C values for the adaptive threshold command to establish the optimal values and use these as input when applying the threshold algorithm. The resulting binarized images are postprocessed using the despeckle, remove outliers, and fill 3D holes commands. At this stage, we recommend comparing the final thresholded image to the original images as a quality control check of the object identification and segmentation. The identified mitochondrial objects are then analyzed in 2D using analyze particles command, which provides mitochondrial count, area, perimeter, form factor (FF), and aspect ratio (AR). For 3D analysis, we use the 3D object counter and “3D particle analyzer” (from the MorphoLibJ package) commands to quantify count, volume, surface area, and sphericity. The thresholded objects are then converted into skeletons using “skeletonize (2D/3D)”, and we apply the analyze skeleton command to obtain the number of skeletons, number of branches, length of branches, and number of branch junctions in the 2D or 3D network. Additional details and parameter values can be found in materials and methods.
Quantifying physiological and pathophysiological changes to mitochondrial morphology and networking.
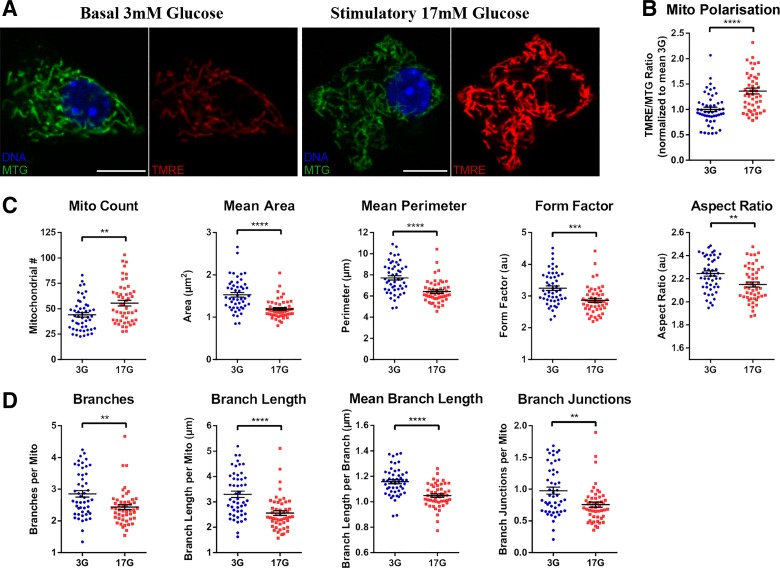
Having established and validated the mitochondrial analysis pipeline, we next tested its ability to characterize mitochondrial changes under relevant physiological and pathophysiological conditions. As a test of acute functional responses, primary mouse islet cells were cultured in either basal (3 mM) or stimulatory (17 mM) glucose for 1 h and co-stained with MitoTracker green (MTG) and the mitochondrial membrane potential-sensitive dye TMRE (Fig. 7A). The MTG fluorescence is insensitive to changes in mitochondrial polarization and served as the signal for mitochondrial detection and morphological characterization (54). The TMRE intensity provided a simultaneous readout of the activity of the individual mitochondrial units, and as expected, stimulatory glucose increases the TMRE/MTG intensity ratio (Fig. 7B). By visual inspection there were no obvious differences in mitochondrial morphology between the cells in low and high glucose (Fig. 7A), but quantitative analysis revealed a number of significant effects (Fig. 7, C and D). Despite no change to total mitochondrial area, glucose stimulation increased the number of mitochondria, reduced their average size (area and perimeter), and made them more round (decreased form factor), all of which suggests increased mitochondrial fission (Fig. 7C). This was further supported by skeletonization analysis, which showed that stimulatory glucose caused an overall reduction in mitochondrial network connectivity (decreased branch parameters) (Fig. 7D). This experiment agrees with previous reports linking Drp1-dependent mitochondrial fission to glucose-stimulated insulin secretion (18, 22) and demonstrates that our analysis pipeline is sensitive enough to allow quantitative detection of subtle physiological changes to mitochondrial morphology and networking.
Fig. 7.
Two-dimensional (2D) analysis shows that glucose stimulation is associated with mitochondrial fission in pancreatic islet cells. Dispersed mouse islet cells were treated in either 3 (3G) or 17 mM glucose (17G) for 60 min and then labeled with Hoechst 33342, MitoTracker Green FM (MTG), and tetramethylrhodamine (TMRE) before 2D imaging. A: representative images of an MTG- and TMRE-stained islet cell in 3G and 17G. B: TMRE/MTG ratio (normalized to average 3G), indicating the degree of mitochondrial hyperpolarization. Mitochondrial morphology and polarization were quantified using our 2D analysis pipeline in Fiji/ImageJ (see materials and methods). C and D: comparison of mitochondrial morphometry (C) and mitochondrial network connectivity (D) demonstrates significant differences between cells acutely treated with low and stimulatory glucose. All data are represented by means ± SE. **P < 0.01, ***P < 0.001, and ****P < 0.0001 as determined by Student’s t-test; n = 49 cells in each glucose treatment from 4 mice.
As an example of a full 3D application, we quantified the mitochondrial changes in palmitate-treated MIN6 cells, an in vitro model of the β-cell lipotoxicity associated with obesity and type 2 diabetes. As expected from previous 2D analyses (31), we observed a fragmentation of the mitochondrial network following treatment with a high concentration of palmitate (Supplemental Fig. S7). This pathophysiological stress response did not affect total mitochondrial volume but was clearly reflected in all parameters describing the 3D shape and size of individual mitochondrial units (Supplemental Fig. S7). Comparing the 2D morphological changes associated with 1 h of glucose stimulation and 6 h of palmitate exposure, it is interesting to note that palmitate treatment reduced AR by 26% and FF by 29%, whereas glucose stimulation decreased AR and FF by only 4 and 12%, respectively. This suggests that physiological fission generates daughter mitochondria that largely retain their shape, in contrast to the more pronounced stress-induced fragmentation, which also causes a striking rounding of the smaller organelles.
Four-dimensional analysis of mitochondrial dynamics.
At any given time, the overall structure of a mitochondrial network reflects the net balance of fusion and fission between individual mitochondria. These are dynamic, energy-dependent processes that involve mitochondrial movement and coordinated actions of proteins that mediate fusion of the outer and inner membranes or constriction and splitting of the organelle (36). Based on static image analysis alone, it can be difficult to know the reason for a change in morphometry. For instance, a more connected and elongated network can be the result of an increase in fusion events, a decrease in fission activity, or a combination of both. Therefore, to further understand the underlying changes, it can be valuable to monitor mitochondrial movement, morphological changes, and organelle interactions in real time. In practical terms, this requires that image acquisition can be repeated at sufficiently frequent intervals and that the analysis is extended to the time domain. Previous studies have applied these principles to 2D images to provide important insights regarding mitochondrial dynamics and turnover in pancreatic β-cells (31, 51).
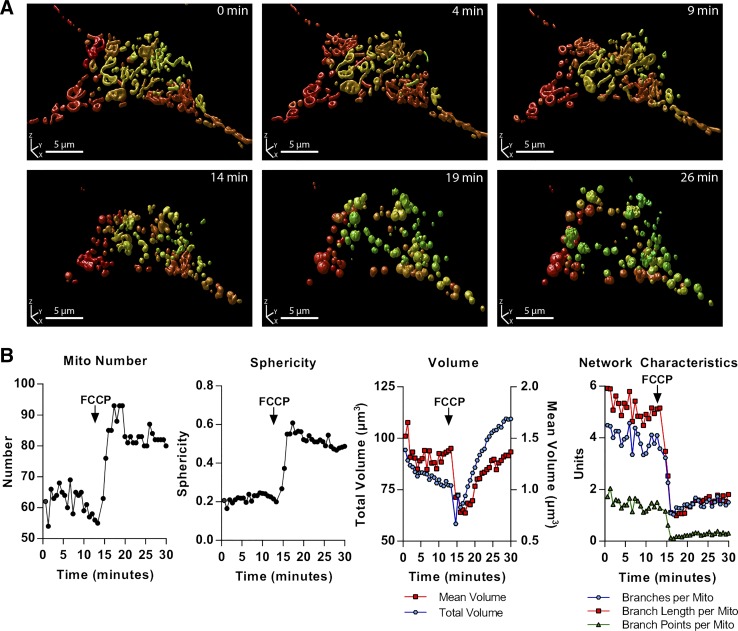
Here, we tested the feasibility of recording and quantifying the time-dependent dynamics of the full 3D mitochondrial network (i.e., an extension to 4D analysis). For this, we expressed mito-YFP in MIN6 cells and imaged these in a stage top incubator on the confocal microscope. 3D time lapse data were generated by acquiring z-stacks of the cells at regular time intervals (every 45 s) for a period of 30 min. At the 13-min mark, we added a high concentration of the mitochondrial uncoupling agent FCCP, with the purpose of inducing a relatively rapid change in mitochondrial dynamics and architecture. Quantitative analyses showed that the distribution of mitochondrial morphologies did not change before the addition of FCCP, indicating that there was no photodamage to the cell (data not shown). As seen in Fig. 8 and Supplemental Video S1, the FCCP triggered a rapid and dramatic loss of mitochondrial connectivity along with an increase in the number of organelles. Interestingly, there was also a transient decrease in both average and total mitochondrial volume, which indicates an initial contraction and shrinking of the mitochondrial fragments, followed by significant swelling, a known response to stress and osmotic shock (48). The abrupt and severe deterioration of the mitochondrial network likely reflects the induction of apoptosis due to profound damage from high levels of FCCP.
Fig. 8.
Time lapse 3-dimensional (3D) imaging (xyzt) and analysis of mitochondrial dynamics. Mitochondria-targeted yellow fluorescent protein (mito-YFP)-expressing MIN6 cells were imaged in a stage top incubator, with 1 full image stack acquired every 45 s. A: 3D renderings (produced in Huygens Professional) of the mitochondrial network in a single cell at different time points. A high concentration FCCP (25 µM) was added to the incubation media around the 13-min mark. B: quantitative analysis of the time-dependent effects of FCCP on mitochondrial number, sphericity, total and mean volume, and network characteristics in the cell.
With this proof-of-principle experiment, we have established the feasibility of analyzing the temporal dynamics of a full mitochondrial network using standard confocal microscopy. A powerful next step could be to combine this 4D approach with other tools such as photo labeling and tracking of individual organelles to generate even more complete and in-depth knowledge of the events that shape the mitochondrial network in health and disease.
DISCUSSION
Most aspects of cellular function and survival are linked to mitochondrial physiology or signals originating from the organelle, and in these contexts the importance of mitochondrial morphology and dynamics has become evident (12, 36). The integrity of the organelle itself, and by extension the metabolic health of the cell, depends on the capacity for mitochondrial adaptation to stress and on selective turnover of damaged parts of the network by mitophagy (51). These processes rely on mitochondrial fusion and fission dynamics, which require sensitive live-cell imaging approaches to study (20). Our current understanding of mitochondrial dynamics in pancreatic β-cells has also been based largely on such imaging approaches (31, 51). However, it is challenging to accurately quantify β-cell mitochondrial morphometry and dynamics by fluorescence microscopy, and many important questions remain unanswered.
In the previous sections, we established a comprehensive set of methods for quantitative image analysis and “morphofunctional” characterization of mitochondria based on standard confocal microscopy and the ImageJ/Fiji shareware. The robustness of these approaches was validated in several ways, including by unsupervised data clustering. We demonstrated the applicability of the resulting pipeline for cells with dense multilayered mitochondria by conducting detailed 2D and 3D morphometric analyses of β-cells and further extended these to 4D time lapse imaging with the accuracy needed for quantitative assessment of network dynamics. To help researchers implement these methods, we have also built our analysis pipeline into a plugin for ImageJ/Fiji called Mitochondria Analyzer. The plugin is publicly available (7) and includes a graphical user interface to facilitate preprocessing, parameter optimization, image thresholding, and automated morphofunctional analysis of mitochondrial images or image stacks according to the workflow we have presented (Fig. 6).
When testing the pipeline, we demonstrated the capability for multiparameter characterization by performing 2D analyses of islet cells co-stained with MTG and TMRE for simultaneous recordings of changes to mitochondrial morphology and membrane potential. However, the pipeline can in principle be applied to any number of mitochondrial parameters, provided they can be jointly imaged and then quantified using shape- and intensity-based descriptors. Therefore, we predict that the same type of analysis using a stable mitochondrial label combined with one or more spectrally distinct fluorescent biosensors, e.g., for mitochondrial redox state, matrix Ca2+, or pH, could provide valuable insights into physiological and pathophysiological structure-function relationships in mitochondria. Importantly, the analysis pipeline treats all identified objects separately and can, therefore, extract the morphological and functional descriptors on a per-mitochondria basis. In the previous sections, we presented our results based on the cellular averages, but the same data sets contain the descriptors associated with thousands of individual mitochondria and can be mined for a wealth of information about morphometry-physiology correlations and heterogeneity at the organelle level (27, 55). It should also be emphasized that the practical considerations and best-practices we have discussed and incorporated into our pipeline are not restricted to β-cell analyses but can also be applied to other cell types.
Finally, the pipeline can in principle also be used to investigate fluorescently labeled organelles other than mitochondria, provided that appropriate thresholding/analysis parameter adjustments can be made. The importance of inter-organelle contacts for cellular function and health are becoming clear, as is the highly complex and dynamic nature of the “organelle interactome” (15, 49, 53). Within the technical boundaries associated with standard confocal microscopy, the analysis approaches we have described here can help most research laboratories achieve the level of accuracy needed to explore internal mitochondrial network interactions, and likely also the mitochondrial relationship with other organelles, as we work to clarify the subcellular basis of diabetes and other diseases.
GRANTS
This work was supported by operating funds from the Canadian Institutes of Health Research (CIHR; MOP-119537) and JDRF (2-2013-50). Research infrastructure was funded by the Canadian Foundation for Innovation (no. 30214). D. S. Luciani received salary support from BCCHRI and Diabetes Canada. R. Shi was supported by a Canada Graduate Scholarship from CIHR.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
A.C., R.S., and D.S.L. conceived and designed research; A.C. and R.S. performed experiments; A.C., R.S., and D.S.L. analyzed data; A.C., R.S., and D.S.L. interpreted results of experiments; A.C. and R.S. prepared figures; A.C., R.S., and D.S.L. drafted manuscript; A.C., R.S., and D.S.L. edited and revised manuscript; A.C., R.S., and D.S.L. approved final version of manuscript; A.C. developed the Mitochondria Analyzer plugin for ImageJ.
ACKNOWLEDGMENTS
We thank Dr. Jingsong Wang, Mei Tang, and Mitsuhiro Komba at BC Children’s Hospital Research Institute (BCCHRI) for technical assistance and Daniel J. Pasula for constructive feedback and discussion.
REFERENCES
- 1.Aharoni-Simon M, Shumiatcher R, Yeung A, Shih AZ, Dolinsky VW, Doucette CA, Luciani DS. Bcl-2 regulates reactive oxygen species signaling and a redox-sensitive mitochondrial proton leak in mouse pancreatic β-Cells. Endocrinology 157: 2270–2281, 2016. doi: 10.1210/en.2015-1964. [DOI] [PubMed] [Google Scholar]
- 2.Anchang B, Hart TD, Bendall SC, Qiu P, Bjornson Z, Linderman M, Nolan GP, Plevritis SK. Visualization and cellular hierarchy inference of single-cell data using SPADE. Nat Protoc 11: 1264–1279, 2016. doi: 10.1038/nprot.2016.066. [DOI] [PubMed] [Google Scholar]
- 3.Anello M, Lupi R, Spampinato D, Piro S, Masini M, Boggi U, Del Prato S, Rabuazzo AM, Purrello F, Marchetti P. Functional and morphological alterations of mitochondria in pancreatic beta cells from type 2 diabetic patients. Diabetologia 48: 282–289, 2005. doi: 10.1007/s00125-004-1627-9. [DOI] [PubMed] [Google Scholar]
- 4.Arnoult D. Mitochondrial fragmentation in apoptosis. Trends Cell Biol 17: 6–12, 2007. doi: 10.1016/j.tcb.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Berman SB, Chen YB, Qi B, McCaffery JM, Rucker EB III, Goebbels S, Nave KA, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-x L increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol 184: 707–719, 2009. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brookes PS, Yoon Y, Robotham JL, Anders MW, Sheu SS. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am J Physiol Cell Physiol 287: C817–C833, 2004. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- 7.Chaudhry A. Mitochondria Analyzer (Online) https://github.com/AhsenChaudhry/Mitochondria-Analyzer [15 July 2019].
- 8.Cole RW, Jinadasa T, Brown CM. Measuring and interpreting point spread functions to determine confocal microscope resolution and ensure quality control. Nat Protoc 6: 1929–1941, 2011. doi: 10.1038/nprot.2011.407. [DOI] [PubMed] [Google Scholar]
- 9.Dey N, Blanc-Feraud L, Zimmer C, Roux P, Kam Z, Olivo-Marin JC, Zerubia J. Richardson-Lucy algorithm with total variation regularization for 3D confocal microscope deconvolution. Microsc Res Tech 69: 260–266, 2006. doi: 10.1002/jemt.20294. [DOI] [PubMed] [Google Scholar]
- 10.Dlasková A, Engstová H, Špaček T, Kahancová A, Pavluch V, Smolková K, Špačková J, Bartoš M, Hlavatá LP, Ježek P. 3D super-resolution microscopy reflects mitochondrial cristae alternations and mtDNA nucleoid size and distribution. Biochim Biophys Acta Bioenerg 1859: 829–844, 2018. doi: 10.1016/j.bbabio.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 11.Dlasková A, Spacek T, Santorová J, Plecitá-Hlavatá L, Berková Z, Saudek F, Lessard M, Bewersdorf J, Jezek P. 4Pi microscopy reveals an impaired three-dimensional mitochondrial network of pancreatic islet beta-cells, an experimental model of type-2 diabetes. Biochim Biophys Acta 1797: 1327–1341, 2010. doi: 10.1016/j.bbabio.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Eisner V, Picard M, Hajnóczky G. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol 20: 755–765, 2018. doi: 10.1038/s41556-018-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fex M, Nicholas LM, Vishnu N, Medina A, Sharoyko VV, Nicholls DG, Spégel P, Mulder H. The pathogenetic role of β-cell mitochondria in type 2 diabetes. J Endocrinol 236: R145–R159, 2018. doi: 10.1530/JOE-17-0367. [DOI] [PubMed] [Google Scholar]
- 14.Fouquet C, Gilles JF, Heck N, Dos Santos M, Schwartzmann R, Cannaya V, Morel MP, Davidson RS, Trembleau A, Bolte S. Improving axial resolution in confocal microscopy with new high refractive index mounting media. PLoS One 10: e0121096, 2015. doi: 10.1371/journal.pone.0121096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y, Li M, Qiu F, Zhang M, Zhang YH. Cell-permeable organic fluorescent probes for live-cell long-term super-resolution imaging reveal lysosome-mitochondrion interactions. Nat Commun 8: 1307, 2017. doi: 10.1038/s41467-017-01503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartig SM. Basic image analysis and manipulation in ImageJ. Curr Protoc Mol Biol Chapter 14: Unit14.15, 2013. doi: 10.1002/0471142727.mb1415s102. [DOI] [PubMed] [Google Scholar]
- 17.Haythorne E, Rohm M, van de Bunt M, Brereton MF, Tarasov AI, Blacker TS, Sachse G, Silva Dos Santos M, Terron Exposito R, Davis S, Baba O, Fischer R, Duchen MR, Rorsman P, MacRae JI, Ashcroft FM. Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic β-cells. Nat Commun 10: 2474, 2019. doi: 10.1038/s41467-019-10189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennings TG, Chopra DG, DeLeon ER, VanDeusen HR, Sesaki H, Merrins MJ, Ku GM. In vivo deletion of β-cell Drp1 impairs insulin secretion without affecting islet oxygen consumption. Endocrinology 159: 3245–3256, 2018. doi: 10.1210/en.2018-00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakobs S. High resolution imaging of live mitochondria. Biochim Biophys Acta 1763: 561–575, 2006. doi: 10.1016/j.bbamcr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Jheng HF, Tsai PJ, Guo SM, Kuo LH, Chang CS, Su IJ, Chang CR, Tsai YS. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol 32: 309–319, 2012. doi: 10.1128/MCB.05603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabra UD, Pfuhlmann K, Migliorini A, Keipert S, Lamp D, Korsgren O, Gegg M, Woods SC, Pfluger PT, Lickert H, Affourtit C, Tschöp MH, Jastroch M. Direct substrate delivery into mitochondrial fission-deficient pancreatic islets rescues insulin secretion. Diabetes 66: 1247–1257, 2017. [Erratum in: Diabetes 66: 3144, 2017.] doi: 10.2337/db16-1088. [DOI] [PubMed] [Google Scholar]
- 23.Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the Bax activation phase of apoptosis. J Cell Biol 164: 493–499, 2004. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirshner H, Aguet F, Sage D, Unser M. 3-D PSF fitting for fluorescence microscopy: implementation and localization application. J Microsc 249: 13–25, 2013. doi: 10.1111/j.1365-2818.2012.03675.x. [DOI] [PubMed] [Google Scholar]
- 25.Landini G; ImageJ . Auto Local Threshold (Online) https://imagej.net/Auto_Local_Threshold [2 September 2018].
- 26.Legland D, Arganda-Carreras I, Andrey P. MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32: 3532–3534, 2016. doi: 10.1093/bioinformatics/btw413. [DOI] [PubMed] [Google Scholar]
- 27.Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, Beeson CC, Rohrer B. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim Biophys Acta 1853: 348–360, 2015. doi: 10.1016/j.bbamcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Yang L, Long Q, Weaver D, Hajnóczky G. Choosing proper fluorescent dyes, proteins, and imaging techniques to study mitochondrial dynamics in mammalian cells. Biophys Rep 3: 64–72, 2017. doi: 10.1007/s41048-017-0037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu H, Koshkin V, Allister EM, Gyulkhandanyan AV, Wheeler MB. Molecular and metabolic evidence for mitochondrial defects associated with beta-cell dysfunction in a mouse model of type 2 diabetes. Diabetes 59: 448–459, 2010. doi: 10.2337/db09-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luciani DS, Ao P, Hu X, Warnock GL, Johnson JD. Voltage-gated Ca(2+) influx and insulin secretion in human and mouse beta-cells are impaired by the mitochondrial Na(+)/Ca(2+) exchange inhibitor CGP-37157. Eur J Pharmacol 576: 18–25, 2007. doi: 10.1016/j.ejphar.2007.07.055. [DOI] [PubMed] [Google Scholar]
- 31.Molina AJ, Wikstrom JD, Stiles L, Las G, Mohamed H, Elorza A, Walzer G, Twig G, Katz S, Corkey BE, Shirihai OS. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes 58: 2303–2315, 2009. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaisen J, Nilsson LI, Pettersen IK, Willems PH, Lorens JB, Koopman WJ, Tronstad KJ. Automated quantification and integrative analysis of 2D and 3D mitochondrial shape and network properties. PLoS One 9: e101365, 2014. doi: 10.1371/journal.pone.0101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ollion J, Cochennec J, Loll F, Escudé C, Boudier T. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29: 1840–1841, 2013. doi: 10.1093/bioinformatics/btt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park KS, Wiederkehr A, Kirkpatrick C, Mattenberger Y, Martinou JC, Marchetti P, Demaurex N, Wollheim CB. Selective actions of mitochondrial fission/fusion genes on metabolism-secretion coupling in insulin-releasing cells. J Biol Chem 283: 33347–33356, 2008. doi: 10.1074/jbc.M806251200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawley JB. Points, pixels, and gray levels: digitizing imaging data. In: Handbook of Biological Confocal Microscopy, edited by Pawley JB. Basel, Switzerland: Springer, 2006, p. 59–79. [Google Scholar]
- 36.Pernas L, Scorrano L. Mito-Morphosis: Mitochondrial Fusion, Fission, and Cristae Remodeling as Key Mediators of Cellular Function. Annu Rev Physiol 78: 505–531, 2016. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 37.Plecitá-Hlavatá L, Lessard M, Santorová J, Bewersdorf J, Jezek P. Mitochondrial oxidative phosphorylation and energetic status are reflected by morphology of mitochondrial network in INS-1E and HEP-G2 cells viewed by 4Pi microscopy. Biochim Biophys Acta 1777: 834–846, 2008. doi: 10.1016/j.bbabio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Prentki M, Matschinsky FM, Madiraju SR. Metabolic signaling in fuel-induced insulin secretion. Cell Metab 18: 162–185, 2013. doi: 10.1016/j.cmet.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 39.Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29: 886–891, 2011. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sage D, Donati L, Soulez F, Fortun D, Schmit G, Seitz A, Guiet R, Vonesch C, Unser M. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 115: 28–41, 2017. doi: 10.1016/j.ymeth.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. J Clin Invest 126: 12–22, 2016. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandebring A, Thomas KJ, Beilina A, van der Brug M, Cleland MM, Ahmad R, Miller DW, Zambrano I, Cowburn RF, Behbahani H, Cedazo-Mínguez A, Cookson MR. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS One 4: e5701, 2009. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682, 2012. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675, 2012. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Scientific Volume Imaging Microscopy Nyquist rate and PSF calculator (Online) https://svi.nl/NyquistCalculator [29 March 2019].
- 45.Sharoyko VV, Abels M, Sun J, Nicholas LM, Mollet IG, Stamenkovic JA, Göhring I, Malmgren S, Storm P, Fadista J, Spégel P, Metodiev MD, Larsson NG, Eliasson L, Wierup N, Mulder H. Loss of TFB1M results in mitochondrial dysfunction that leads to impaired insulin secretion and diabetes. Hum Mol Genet 23: 5733–5749, 2014. doi: 10.1093/hmg/ddu288. [DOI] [PubMed] [Google Scholar]
- 46.Silva JP, Köhler M, Graff C, Oldfors A, Magnuson MA, Berggren PO, Larsson NG. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet 26: 336–340, 2000. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 47.Song W, Bossy B, Martin OJ, Hicks A, Lubitz S, Knott AB, Bossy-Wetzel E. Assessing mitochondrial morphology and dynamics using fluorescence wide-field microscopy and 3D image processing. Methods 46: 295–303, 2008. doi: 10.1016/j.ymeth.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, Ellisman MH, Green DR, Frey TG. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat Cell Biol 9: 1057–1065, 2007. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- 49.Thivolet C, Vial G, Cassel R, Rieusset J, Madec AM. Reduction of endoplasmic reticulum- mitochondria interactions in beta cells from patients with type 2 diabetes. PLoS One 12: e0182027, 2017. doi: 10.1371/journal.pone.0182027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tseng Q. AdaptiveThreshold - ImageJ Plugin (Online) https://sites.google.com/site/qingzongtseng/adaptivethreshold#use [2 September 2018].
- 51.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27: 433–446, 2008. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Twig G, Graf SA, Wikstrom JD, Mohamed H, Haigh SE, Elorza A, Deutsch M, Zurgil N, Reynolds N, Shirihai OS. Tagging and tracking individual networks within a complex mitochondrial web with photoactivatable GFP. Am J Physiol Cell Physiol 291: C176–C184, 2006. doi: 10.1152/ajpcell.00348.2005. [DOI] [PubMed] [Google Scholar]
- 53.Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167, 2017. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wikstrom JD, Katzman SM, Mohamed H, Twig G, Graf SA, Heart E, Molina AJ, Corkey BE, de Vargas LM, Danial NN, Collins S, Shirihai OS. beta-Cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes 56: 2569–2578, 2007. doi: 10.2337/db06-0757. [DOI] [PubMed] [Google Scholar]
- 55.Zahedi A, On V, Phandthong R, Chaili A, Remark G, Bhanu B, Talbot P. Deep analysis of mitochondria and cell health using machine learning. Sci Rep 8: 16354, 2018. doi: 10.1038/s41598-018-34455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]