Abstract
Mitochondrial DNA (mtDNA) exposed to the extracellular space due to cell death has immunostimulatory properties. Case-control studies reported a positive association between odds of developing preeclampsia and circulating mtDNA. These findings are based on relative quantification protocols that do not allow determination of absolute concentrations of mtDNA and are highly sensitive to nuclear DNA contamination. Furthermore, circulating mtDNA concentrations in response to normal pregnancy, which is an inflammatory state characterized by continuous placental cell apoptosis, have not been established. The main objective of this study was to determine longitudinal changes in circulating mtDNA from preconception to first trimester, third trimester, and postpartum in healthy pregnant women. Absolute real-time PCR quantification of mtDNA and nuclear DNA (nDNA) was performed on whole genomic extracts from serum using TaqMan probes and chemistry. Serum cell-free mtDNA and nDNA concentrations were greater in late pregnancy as compared with early pregnancy and postpartum. Pregnant women carrying neonates at the upper quartile of birth length distribution had higher concentrations of mtDNA in late pregnancy compared with pregnancies carrying neonates at the lower quartile. The correlation between circulating mtDNA and nDNA concentrations varied by sex (i.e., pregnancies carrying female vs. male fetuses). This study is the first to establish temporal patterns of circulating cell-free mtDNA concentrations in normal human pregnancy using absolute DNA quantification techniques. Concentrations of circulating mtDNA in normal pregnancy may be used as reference values for the development of clinical prognostic or diagnostic tests in pregnant women with, or at risk of developing, gestational complications.
Keywords: cell-free DNA, DNA quantification, gestational age, mitochondria
INTRODUCTION
Mitochondria are eukaryotic organelles with an ancestral bacterial origin (27). Because of this ancestral origin, mitochondrial products, including mitochondrial DNA (mtDNA), released into the extracellular space can act as damage-associated molecular patterns to the innate immune system (32). Extracellular mtDNA, which is enriched in unmethylated CpG dinucleotide motifs, similar to bacterial and viral DNA, contributes to inflammatory responses via activation of innate immune receptors (9). Since 1997, when cell-free fetal DNA was first reported by Lo et al. (17), there has been extensive investigation into circulating, cell-free nuclear DNA (nDNA), culminating in the clinical use of noninvasive sequencing of fetal DNA to screen for chromosomal and subchromosomal abnormalities in utero (4). Interestingly, mtDNA is detectable in maternal circulation in normal pregnancy (8) and increases in certain pregnancy complications (6, 8, 25). Yet cell-free mtDNA has received less attention (compared with cell-free nDNA), and currently, there is limited characterization of its temporal changes in the maternal circulation during pregnancy or its biological role during normal gestation and complicated pregnancies.
Case-control studies have reported increased mtDNA copy numbers in serum and whole blood of pregnant women with preeclampsia (19, 25) and a positive association between amount of circulating mtDNA measured during delivery and the odds of having preeclampsia at the time of delivery (25). Recently, Busnelli et al. (6) demonstrated that a reduction in circulating mtDNA early in pregnant women, before the onset of preeclampsia, suggesting that mtDNA may be involved in the pathogenesis of this pregnancy-specific syndrome. Previous research characterizing the levels of mtDNA in maternal circulation during pregnancy included case-control studies (6, 18, 24, 28) and cross-sectional comparisons, studying whole blood as the tissue of interest (6, 8, 24, 28). Importantly, these studies used relative methods of quantification that rely on a nuclear housekeeping gene, instead of absolute quantification protocols that can provide more accurate amounts of mtDNA (6, 8, 24, 28). Concentrations of circulating cell-free mtDNA throughout normal pregnancy have not been determined, and this point-of-reference is important for studies of cell-free mtDNA in disease states. Therefore, the objective of this study was to use absolute DNA quantification techniques to determine the temporal changes in mtDNA concentrations from preconception to first, third, and fourth (postpartum) trimesters in a group of pregnant women followed throughout pregnancy.
METHODS
This is an observational, longitudinal study designed to establish absolute concentrations of cell-free mtDNA in the maternal circulation during pregnancy and postpartum. Data presented herein were collected from healthy women who volunteered for a study protocol assessing autonomic cardiovascular regulation during pregnancy. This study was approved by the Institutional Review Boards of the University of North Texas Health Science Center, the University of Texas Southwestern Medical Center, and Texas Health Presbyterian Hospital Dallas. All subjects provided written informed consent. All procedures were performed in accordance with the guidelines set forth in the Declaration of Helsinki.
Subjects.
Subjects were screened using an in-depth medical history questionnaire and a physical examination. Exclusion criteria included smoking, overt history of chronic disease, prior history of gestational hypertension or preeclampsia, use of recreational drugs or hormonal contraceptives within the previous 6 mo, use of hormonal fertility treatments or supplements, and menstrual irregularities. Details about subject recruitment and screening have been published elsewhere (22).
The study group consisted of 21 pregnant women who were tested at early pregnancy (5–8 wk of gestation), late pregnancy (33–36 wk of gestation), and postpartum (6–10 wk after birth) and 19 nonpregnant women tested at the mid-luteal phase of their menstrual cycle. All pregnancies were singleton, and all pregnant women delivered vaginally at term (37–42 wk of gestation). Postpartum subjects were breastfeeding. Subject characteristics, including maternal and neonatal parameters, are presented in Table 1. Maternal parameters include race, chronological age, height, weight, body mass index, parity, and gestational age. Neonatal characteristics include sex, length, weight, and ponderal index.
Table 1.
Subject demographics
| Maternal Characteristics |
|||||
|---|---|---|---|---|---|
| All Subjects | Nonpregnant | Early | Late | Post | |
| n | 40 | 19 | 21 | 21 | 21 |
| Race, % | |||||
| Asian | 13/40 (33) | 6/19 (32) | 7/21 (33) | 7/21 (33) | 7/21 (33) |
| Black | 11/40 (28) | 8/19 (42) | 3/21 (14) | 3/21 (14) | 3/21 (14) |
| Caucasian | 15/40 (38) | 5/19 (26) | 10/21 (48) | 10/21 (48) | 10/21 (48) |
| Hispanic | 1/40 (3) | – | 1/21 (5) | 1/21 (5) | 1/21 (5) |
| Age, yr (min, max) | 31 (20, 43) | 33 (23, 43) | 30 (20, 37) | 31 (20, 38) | 31 (21, 38) |
| Height, cm | 164 (149, 178) | 163 (150, 178) | 164 (149, 177) | 164 (150, 178) | 164 (149, 177) |
| Weight, kg | 66 (44, 99) | 68 (46, 91) | 61 (44, 94) | 72 (56, 99) | 65 (47, 96) |
| BMI, kg/m2 | 25 (18, 39) | 25 (18, 33) | 23 (18, 36) | 27 (22, 39) | 24 (21, 38) |
| Parity, % | |||||
| 0 | 21/39 (54)* | 10/18 (56)1 | 11/21 (52) | 11/21 (52) | |
| 1 | 14/39 (36) | 6/18 (33) | 8/21 (38) | 8/21 (38) | 11/21 (52) |
| 2 | 2/39 (5) | 1/18 (6) | 1/21 (5) | 1/21 (5) | 8/21 (38) |
| 3 | 1/39 (3) | 0/18 (0) | 1/21 (5) | 1/21 (5) | 1/21 (5) |
| 4 | 1/39 (3) | 1/18 (6) | 1/21 (5) | ||
| Gestational age at blood sampling, wk (min, max) | 6.7 (4.5, 8.1) | 34.1 (32, 36.5) | |||
| Time postpartum, wk | 8.2 (6, 10) | ||||
| Neonatal characteristics | |||||
| Sex, % | |||||
| Female | 10/21 (48) | ||||
| Male | 11/21 (52) | ||||
| Birth length, cm (min, max) | 50 (43, 56)† | ||||
| Birth weight, kg | 3.2 (2.8, 3.8) | ||||
Characteristics of two maternal cohorts are presented: 1) nonpregnant subjects (data from this group were used for cross-sectional comparisons) and 2) pregnant subjects (data from these subjects were collected at early and late pregnancy and postpartum). Maternal race, maternal parity, and neonatal sex are presented as a ratio to the cohort with a percentage in parenthesis. Maternal age, height, weight, body mass index (BMI), gestational age, time postpartum, neonatal length, weight, and ponderal index are all presented as mean with range in parenthesis. For “All Subjects” parity, parity status for the longitudinal pregnant cohort was determined before parturition in the current study.
Missing one cross-sectional nonpregnant subject’s parity status;
Missing one fetal length, calculated from n = 20 rather than 21.
Experimental design and methodology.
Starting 2 days before blood collection, subjects consumed an isocaloric constant diet consisting of 200 mEq sodium, 100 mEq potassium, and 1,000 mg calcium daily. Fluid intake was ad libitum. On the day of blood collection, subjects had a light breakfast at least 2 h previously, no caffeine or alcohol for at least 48 h, and no strenuous exercise at least 24 h before blood sample collection. Preexperimental conditions have been described in detail elsewhere (22). Blood samples were collected in the morning in the supine position via an intravenous catheter inserted into the antecubital vein of the left arm. Blood was collected in serum collection tubes (cat. no. 366430, Becton Dickinson) and was allowed to clot for 30 min at room temperature before being centrifuged at 3,500 revolutions/min for 15 min. Serum was then stored at −80°C.
DNA measurements: absolute quantification polymerase chain reaction.
DNA was isolated from serum samples (250 µL) using Mag-Bind Blood & Tissue DNA HDQ 96 kit (cat. no. M6399, Omega Bio-tek, Norcross, GA;) using an automated liquid handling system (Microlab STARlet, Hamilton, Reno, NV) according to the manufacturer instructions. Proteinase K (20 µL) and lysis buffer (290 µL) were added to 250 µL of serum, mixed, and incubated at 70°C for 10 min. Then, magnetic particles with DNA binding moieties were added to this solution, mixed for 10 min, and separated from the supernatant using a strong magnet. DNA bound to magnetic particles underwent 3 wash cycles, eluted from magnetic particles with 100 µL of elution buffer, and were subsequently stored in −20°C until ready for analysis.
Nuclear DNA was quantified using Quantifiler Trio DNA Quantification kit (cat. no. 4482910, Applied Biosystems, Waltham, MA) and the 7500 Real-Time PCR System (Applied Biosystems). Results were quantified using standard reference DNA (Quantifiler THP DNA Standard) over five 10× dilutions (50 to 0.005 ng/µL). This TaqMan-based kit allows for quantification of autosomal as well as Y-chromosomal DNA. DNA isolate (2 µL) was added to 18 µL of master mix, resulting in a 20-µL reaction mixture. MicroAmp optical 96-well reaction plates (cat. no. N8010560, Applied Biosystems) were sealed with MicroAmp optical adhesive cover (cat. no. 4311971, Applied Biosystems). PCR settings were as follows: 95°C for 2 min and 40 cycles of 95°C for 9 s with 60°C at 30 s. Cycle threshold (CT) was determined automatically.
Mitochondrial DNA was quantified using a method similar to the previously published technique by Kavlick et al. (14). This TaqMan chemistry-based method of absolute quantification PCR employed primers (sense and anti-sense; 900 nM), TaqMan MGB probe (900 nM) (Table 2), and TaqMan Universal PCR Master Mix (12.5 µL) (cat. no. 4324018, Applied Biosystems). Primers target mitochondrial NADH:ubiquinone oxidoreductase core subunit 5 (MT-ND5) (GenBank Gene ID: 4540) at position 13,288–12,392 of the mitochondrial genome (based on revised Cambridge Reference Sequence positions) (1, 2, 7). The amplified region (13,288–12,392) has few known mutations and has been demonstrated to be absent from the nuclear genome (13, 14), providing protection from both the effects of hypervariability and pseudogene contamination, respectively. Finally, it is a very sensitive method of mtDNA concentration analysis, able to detect even minute quantities of mtDNA (14). The quantitative standard employed was a double stranded gBlocks gene fragment (Integrated DNA Technologies, Coralville, IA) (Table 2) diluted over eight 10× dilutions (1,000 to 0.0001 pg/µL). DNA isolate (2 µL) was added to 23 µL of master mix, resulting in a 25-µL reaction mixture. This reaction was performed using the same reaction plates and covers as above. A 7500 Real-Time PCR instrument (Applied Biosystems) was used to amplify and analyze with the following settings: 9600 emulation, 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s with 60°C for 1 min. For analysis, CT was set at 0.2. Quantification was achieved by comparing ΔRn (normalized reporter signal) values of samples with those of a standard.
Table 2.
Nucleotide sequences for absolute qPCR of mitochondrial DNA
| Forward primer | 5′-GGC ATC AAC CAA CCA CAC CTA-3′ |
| Reverse primer | 5′-ATT GTT AAG GTT GTG GAT GGA-3′ |
| TaqMan probe | 5′-6FAM CAT TCC TGC ACA TCT G MGBNFQ-3′ |
| Synthetic standard (gBlock) | 5′-TG TTC TGT TCA TTG TTA AGG TTG TGG ATG GAC CCG GAG CAC ATA AAT AGT CGT TAT TTG AAG GCG TGG GTA CAG ATG TGC AGG AAT GCT AGG TGT GGT TGG TTG ATG CCG ATT GGA TTG-3′ 5′-CAA TCC AAT CGG CAT CAA CCA ACC ACA CCT AGC ATT CCT GCA CAT CTG TAC CCA CGC CTT CAA ATA ACG ACT ATT TAT GTG CTC CGG GTC CAT CCA CAA CCT TAA CAA TGA ACA GAA CA-3′ |
6FAM, 6-Carboxyfluorescein; MGBNFQ, minor groove binder nonfluorescent quencher; qPCR, quantification polymerase chain reaction.
For both mtDNA and nDNA measurements, amplification efficiency, y-intercept, and R2 were analyzed before absolute quantification. Efficiency >80% and R2 >99% were considered adequate. Amplifications were performed in duplicate unless otherwise noted. Using known volumes of subject serum (250 µL) and final elution volumes (100 µL), nDNA and mtDNA concentrations were calculated and expressed as picograms (pg) per milliliter of serum. Total DNA was calculated as the sum of mtDNA and nDNA.
Statistics.
Statistical analyses were performed using JMP (Pro 12; SAS Institute, Cary, NC) and Prism (Version 7, Graphpad, San Diego, CA) software. The D’Agostino–Pearson omnibus test was used to assess data distribution. Data that did not fit a normal distribution were log-transformed before parametric statistics were used. All figures present untransformed data for clarity. The robust regression and outlier removal method was used to identify and remove outliers (Prism). DNA outcomes were compared across trimesters and postpartum using a one-way mixed model analysis of variance (ANOVA) followed by Tukey’s post hoc multiple comparisons. In a separate analysis, we compared DNA outcomes in nonpregnant subjects with those in various pregnancy stages using Student’s t tests with Bonferroni correction for multiple comparisons. DNA outcomes for these cohorts are presented as medians with interquartile range (IQR). A subset of subjects was tested before pregnancy, at early pregnancy, late pregnancy, and postpartum. Data from this group were analyzed using a one-way mixed model ANOVA followed by Tukey’s post hoc multiple comparisons. A two-way mixed model ANOVA followed by Tukey’s post hoc test was used to assess the effects of fetal sex on circulating mtDNA across gestational stages. To assess the association between maternal and neonatal characteristics with circulating mtDNA, we compared circulating mtDNA between subjects in the lower quartile (Q1) with those in the upper quartile (Q3) for maternal age, height, and gestational weight gain and neonatal weight, length, and ponderal index. Neonatal quartiles (Q1 and Q3) were compared with World Health Organization child growth standards (12). DNA outcomes for these analyses are presented as means ± SE. Pearson correlation was used to measure the strength and direction of association between mtDNA and nDNA at each reproductive stage. Subject characteristics are presented as means (range), unless otherwise indicated. Exact P values are presented for each analysis, and rejection of statistical test null hypothesis was set to P ≤ 0.05.
RESULTS
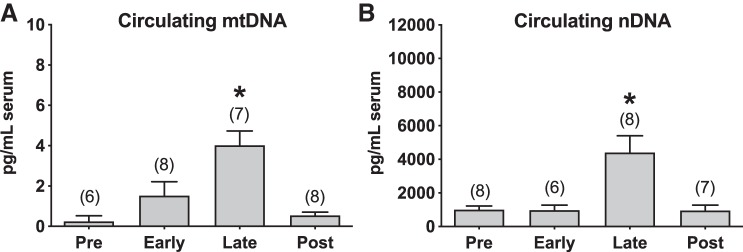
Concentrations of serum cell-free mtDNA were greater in late pregnancy compared with early pregnancy and postpartum, and there were no differences in mtDNA between early pregnancy and postpartum (Fig. 1A). The same pattern was observed in concentrations of cell-free serum nDNA, with samples collected at late pregnancy exhibiting a sixfold higher concentration compared with all other gestational stages (Fig. 1B). Cross-sectional comparisons revealed lower serum concentrations of mtDNA and nDNA from nonpregnant women compared with women tested at late pregnancy (Fig. 1, A and B). When normalized to total cell-free DNA, concentrations of mtDNA were higher in late pregnancy and postpartum compared with nonpregnant state, and there were no differences among gestational stages (Fig. 1C). The results from the longitudinal and cross-sectional comparisons were reproducible in a subset of subjects who had been tested before pregnancy, during pregnancy, and postpartum (Fig. 2, A and B).
Fig. 1.
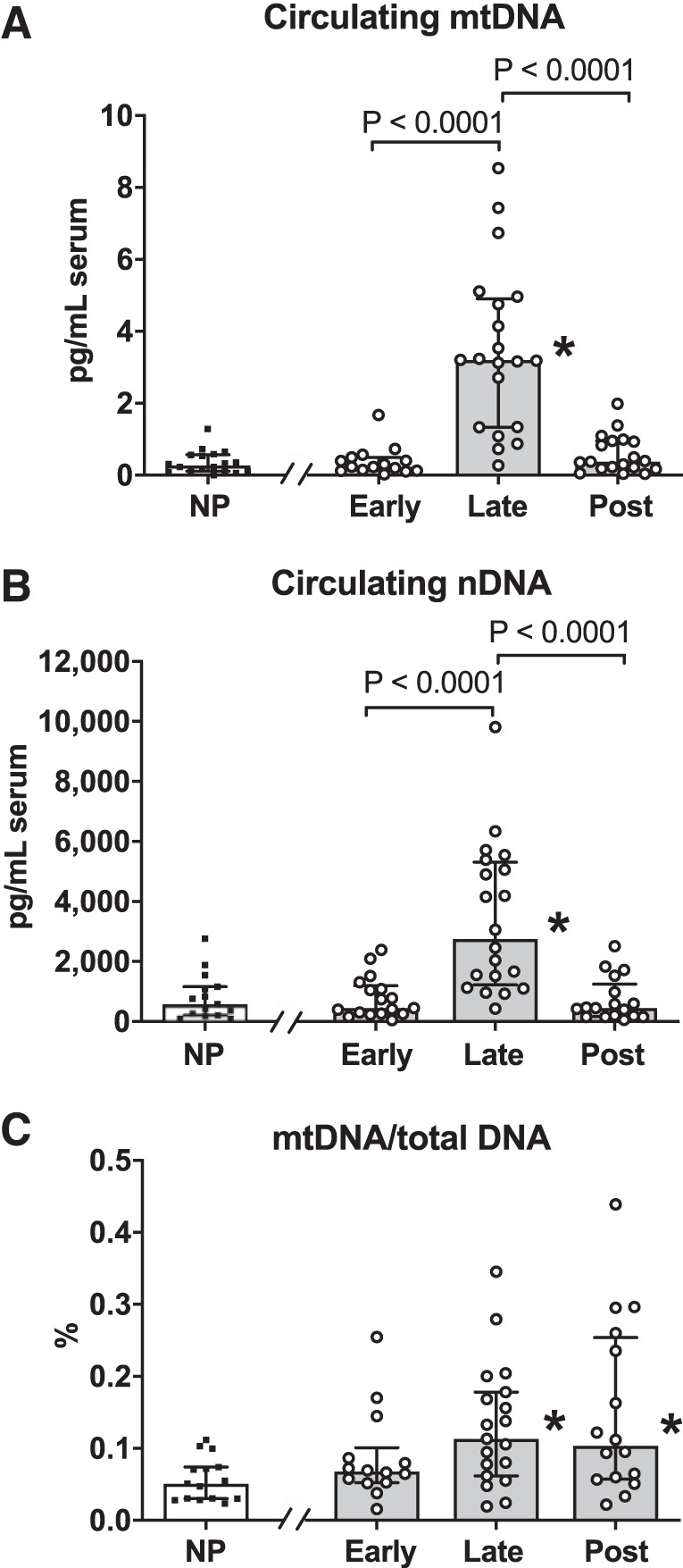
Circulating cell-free mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) in healthy nonpregnant (NP), pregnant, and postpartum women. A: mtDNA concentrations (pg/mL) in serum. B: nDNA concentrations (pg/mL) in serum. C: mtDNA as a percentage of total cell-free DNA in serum. A mixed-model analysis of variance followed by Tukey’s post hoc was used to analyze data from pregnant women tested at early pregnancy, late pregnancy, and postpartum. Unpaired t tests with Bonferroni correction were used to compare data between NP women vs. each stage of gestation and postpartum. *P < 0.05 vs. NP. All values presented as median and interquartile range. Early, gestational weeks 5–8; late, gestational weeks 33–36; and post, 6–10 wk after parturition. Closed squares, NP group; open circles, pregnant women at early and late pregnancy and postpartum.
Fig. 2.
Circulating cell-free mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) in a subset of subjects tested before pregnancy, during pregnancy, and postpartum. A: mtDNA concentrations (pg/mL) in serum. B: nDNA concentrations (pg/mL) in serum. Mixed-model analysis of variance followed by Tukey’s post hoc. *P ≤ 0.027 late vs. pre, early, post. All values presented as means ± SE. Numbers in parentheses indicate number of observations used in the statistical analysis. Early, gestational weeks 5–8; late, gestational weeks 33–36; and post, 6–10 wk after parturition.
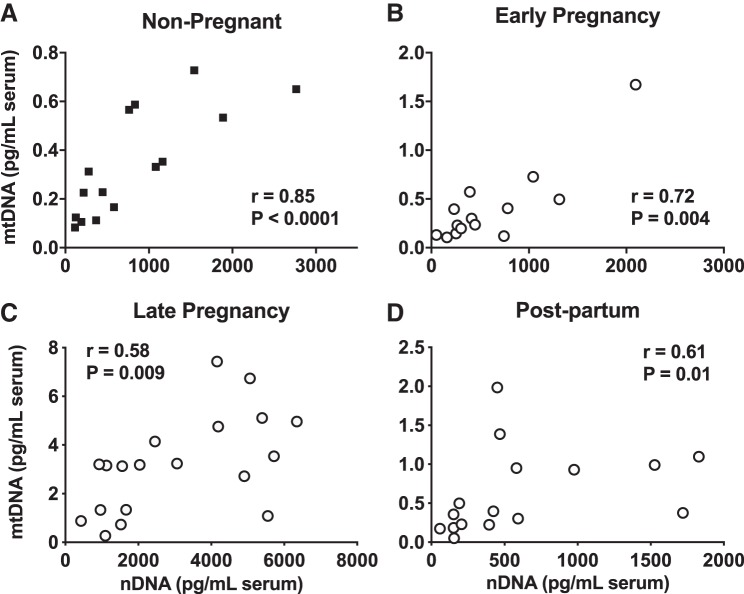
There were no differences in concentrations of serum mtDNA between subjects at Q1 compared with subjects at Q3 for maternal age (Fig. 3A), height (Fig. 3B), gestational weight gain (Fig. 3C), ponderal index (Fig. 3D), or neonatal weight (Fig. 3E). Pregnant women carrying neonates at Q3 for birth length had higher concentrations of mtDNA in late pregnancy (Fig. 3F). Neonates at Q3 were long (54.03 ± 0.61 cm) when compared with the World Health Organization child growth standards (12), whereas neonates at Q1 had normal length (44.66 ± 0.61 cm). There were moderate and strong positive correlations between concentrations of mtDNA and nDNA in serum from nonpregnant women (Fig. 4A) and women tested at early pregnancy (Fig. 4B), late pregnancy (Fig. 4C), and postpartum (Fig. 4D).
Fig. 3.
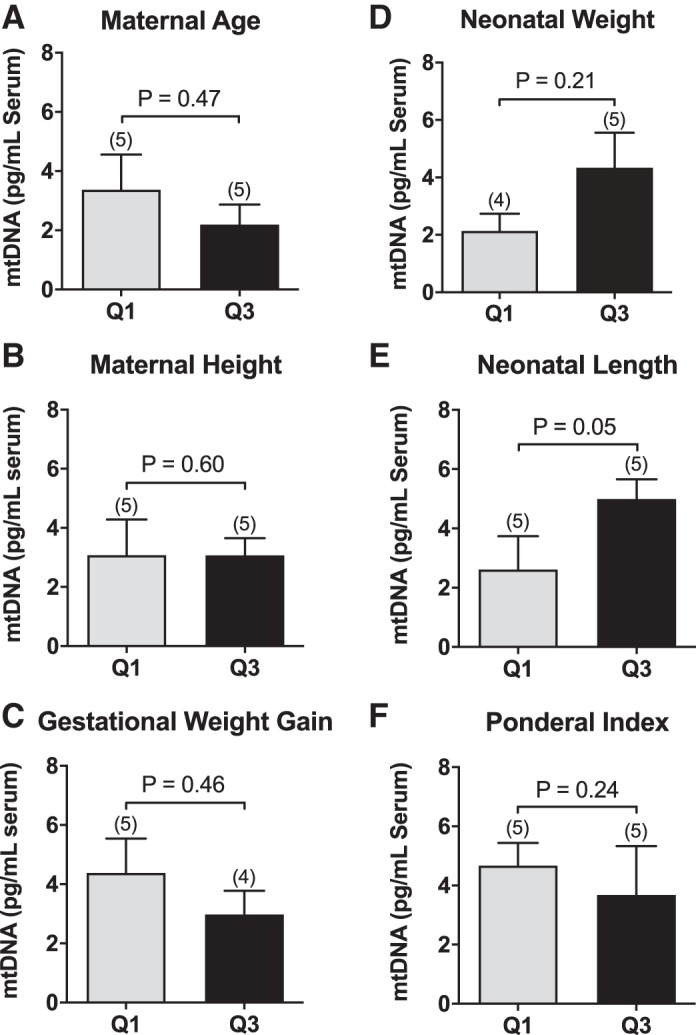
Circulating cell-free mitochondrial DNA (mtDNA) in the lower and upper quartiles for maternal and neonatal characteristics. Concentrations of mtDNA (pg/mL) measured in late pregnancy in maternal serum from pregnancies at the lower quartile (Q1) vs. upper quartile (Q3) for maternal chronological age (A), maternal height (B), maternal gestational weight gain (C), ponderal index (D), neonatal weight(E), and neonatal length (F). Unpaired t tests. Values are presented as means ± SE.
Fig. 4.
Correlations between circulating cell-free mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) measured in nonpregnant state, at early and late pregnancy, and postpartum. A: nonpregnant. B: early pregnancy (gestational weeks 5–8). C: late pregnancy (gestational weeks 33–36). D: postpartum (6–10 wk after parturition). Pearson correlation coefficient r and P values are presented for each analysis. Closed squares, nonpregnant group; open circles, pregnant group tested at early and late pregnancy and postpartum.
In a separate analysis, we determined the effects of neonatal sex on circulating serum mtDNA and found no significant sex-by-gestational stage interaction (Fig. 5). The relationship between circulating mtDNA and nDNA concentrations varied by sex. In blood samples from pregnant women carrying male fetuses, there was no correlation between mtDNA and nDNA at early pregnancy (r = 0.68, P = 0.09) or postpartum (r = 0.59, P = 0.09), but there was a moderate correlation between mDNA and nDNA at late pregnancy (r = 0.65, P = 0.04) (Fig. 6). There was no correlation between mtDNA and nDNA concentrations in samples collected from women carrying female fetuses at late pregnancy (r = 0.43, P = 0.24) or postpartum (r = 0.69, P = 0.08) (Fig. 6). The correlation between mtDNA and nDNA was strong in samples collected at early pregnancy (r = 0.81, P = 0.03) from women carrying female fetuses (Fig. 6).
Fig. 5.
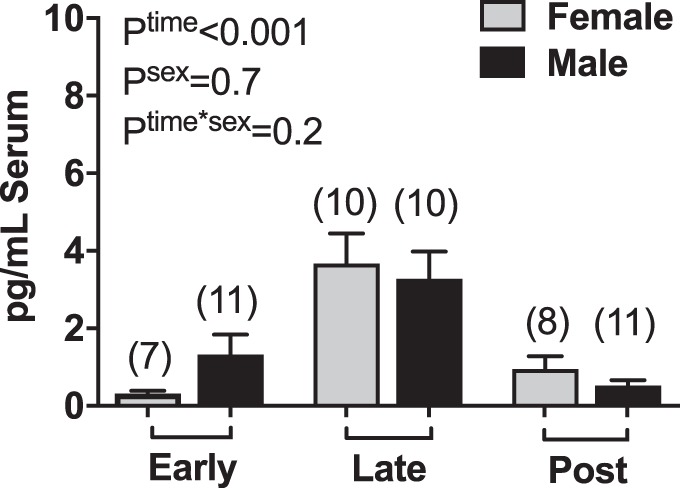
Circulating cell-free mitochondrial DNA (mtDNA) in healthy pregnant women carrying male and female fetuses. Concentrations of mtDNA in maternal serum at early pregnancy, late pregnancy, and postpartum (two-way mixed model analysis of variance). Values are presented as means ± SE. Numbers in parentheses indicate number of observations used in the statistical analysis. Early, gestational weeks 5–8; late, gestational weeks 33–36; and post, 6–10 wk after parturition.
Fig. 6.
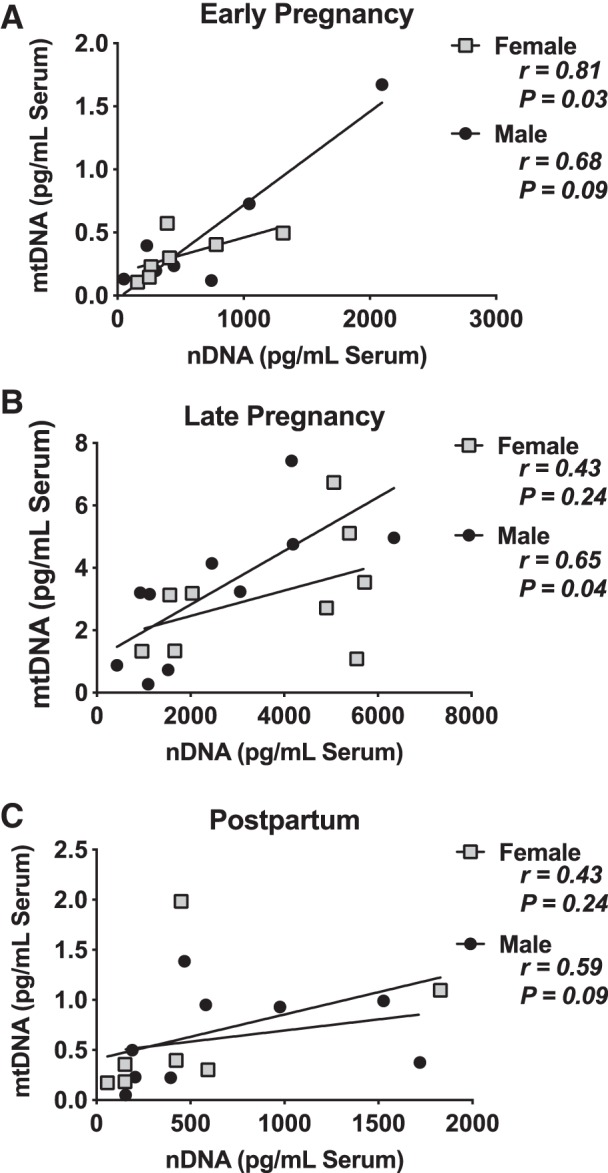
Correlations between circulating cell-free mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) measured at early (A), late pregnancy (B), and postpartum (C) by fetal sex. Early, gestational weeks 5–8; late, gestational weeks 33–36; and post, 6–10 wk after parturition. Pearson correlation coefficient r and P values are presented for each analysis. Line of best fit is provided.
DISCUSSION
The main finding of this study is that in normal pregnancy, absolute concentrations of circulating cell-free mtDNA and nDNA increase with advancing gestational age and return to nonpregnant concentrations within approximately 2 mo from parturition. The relationship between mtDNA and nDNA, an indirect assessment of mechanisms of mtDNA release into the maternal circulation, differs across gestational stages and is affected by fetal sex. To the best of our knowledge, this is the first study to measure absolute quantities of circulating mtDNA in pregnant women and establish patterns of mtDNA changes in normal pregnancy.
We demonstrated for the first time that mtDNA concentrations increase during late pregnancy. Our findings are in direct opposition to those of Colleoni et al. (8), who reported reduced whole blood mtDNA content in pregnant, compared with nonpregnant, women. To put our results in perspective, it is important to note that our study had several key differences from that of Colleoni et al. We measured mtDNA in the same women before and across trimesters of pregnancy (within-subject design), whereas the experimental design in the study by Colleoni et al. was cross-sectional. Additionally, we investigated cell-free mtDNA in serum rather than cell-containing whole blood. The fraction of blood used would be expected to alter mtDNA concentrations, as some fractions (whole blood and buffy coat) contain cells, which have their own complement of nDNA (leukocytes) and mtDNA (leukocytes and platelets) separately from a cell-free fraction (plasma or serum). Another difference between these studies is that rather than relying on a nuclear-genome-located housekeeping gene to calculate a relative change in mtDNA, we analyzed mtDNA and nDNA separately using synthetic dsDNA as a standard. This allows us to interpret changes in mtDNA separately from underlying changes in nDNA as well as compare them to each other.
The observed rise in serum mtDNA concentrations during pregnancy mirrors placental and fetal development. Placental thickness is linearly correlated with gestational age in normal pregnancy (11). Using data from a cross-sectional study by Elchalal et al. (10), we can estimate that our “late pregnancy” group (mean gestational age = 34.1 wk) had placentas ~40 mm thick, whereas those of our “early pregnancy” group (mean gestational age = 6.7 wk) would have had placentas less than 25 mm thick. In our study, we found an increase in late pregnancy serum mtDNA from women who delivered longer neonates. Although further studies on the relationship between placental size and circulating mtDNA would need to be undertaken, our findings suggest that in normal pregnancy, a larger placenta, and thus greater trophoblastic mass, may be related to greater concentrations of circulating mtDNA. This is further supported by the herein observed return to baseline concentrations of serum mtDNA after delivery of the fetoplacental unit.
The observed correlations between mtDNA and nDNA during all pregnancy stages may be related to apoptosis or programmed cell death. Apoptosis increases throughout gestation as well, peaking in the third trimester (28). Correlations between cell-free nDNA and apoptosis of placental trophoblasts have been noted previously (5, 24). Because apoptosis is a known mechanism of release of extracellular nDNA and mtDNA (3), this is a likely cause of the correlations observed between mtDNA and nDNA in the study at present. In late pregnancy, the correlation between mtDNA and nDNA dissipates when female pregnancies are considered separately from male pregnancies. A possible explanation is that male placentas invade more deeply into the spiral arteries of the uterine wall and express proinflammatory transcripts to a greater extent than female pregnancies (26). If these male-specific processes result in cell lysis and extrusion of mtDNA into the extracellular space, one would expect greater concentrations of circulating mtDNA in pregnancies carrying male fetuses, rather than the observed sex differences in the relationship between mtDNA and nDNA. The lack of (fetal) sex differences in concentrations of mtDNA during pregnancy, in the presence of sex differences in the relationship between mtDNA and nDNA, posits novel questions for investigation of mtDNA release mechanisms.
The analysis of serum mtDNA separately from an nDNA housekeeper sets the foundation for future studies to investigate the mechanisms of mtDNA clearance and its function in normal pregnancy and pregnancy complications. mtDNA itself is of interest due to its immunostimulatory nature, first noted by Collins et al. in 2004 (9). mtDNA is capable of activating the innate immune system, as reviewed by West et al. (30). Our laboratory has previously demonstrated that activation of Toll-like receptor 9 by a synthetic mtDNA mimetic during rat pregnancy recapitulates some features of human preeclampsia by increasing maternal blood pressure, vascular tone, and vascular oxidative stress (23). In the present study, we demonstrated a positive correlation between mtDNA and nDNA in all stages. This finding sets the foundation for experimental studies to test the hypothesis that this relationship reflects activation of apoptotic pathways during late pregnancy in normal pregnant women and whether this mechanism may change in pregnancy complications. Interestingly, extracellular mtDNA is a damage-associated molecular pattern participating in inflammatory responses. Yet in our studies, all subjects were healthy, nonobese, and experienced uncomplicated pregnancies. In this context, our findings are intriguing as they suggest that circulating mtDNA may play a functional role in late pregnancy (e.g., labor).
Study limitations include a small sample size, which did not allow us to account for potential racial differences, and lack of validation of our findings in additional cohorts/sites. Additionally, the present study did not assess experimentally the origin of cell-free mtDNA; thus, it is currently uncertain whether cell-free mtDNA is released in the maternal circulation during pregnancy by maternal or fetal tissues. Until recently, it was accepted that mtDNA genetics involve uniparental inheritance, transmitted from the mother to the offspring (29). Recent studies, however, have challenged this notion, suggesting that paternal inheritance of mtDNA may be possible (18). An additional limitation of the current work is that the role of fetal sex in circulating mtDNA changes during pregnancy was assessed in a post hoc analysis. On the other hand, the study design had high internal validity, as we controlled for confounding factors known to alter mtDNA (as measured in nonpregnant adults) such as time of blood collection, chronic disease, smoking, age, recent surgery, systemic hormonal use, and sex (15, 16, 20, 21). An additional strength of this study is that subjects were followed postpartum, emphasizing the clinical importance of this time period. Furthermore, we studied pregnant women longitudinally (within-subject design) and used an absolute quantification protocol of mtDNA. One of the strengths of this method is that the synthetic double-stranded standard used is less likely to be contaminated by DNA, RNA, or protein as compared with a plasmid-derived standard and the quantification of mtDNA is not affected by changes in nuclear DNA.
Perspectives and Significance
The present study provides initial concentrations of mtDNA in normal pregnancy and sets the foundation for longitudinal cohort studies to establish normative mtDNA values. Circulating mtDNA has been reported to change in pregnancy complications such as preeclampsia and intrauterine growth restriction (8, 19, 25, 31). More recent studies have suggested that a change in mtDNA in early pregnancy may be a predictor of risk for developing preeclampsia (6). Absolute quantification of mtDNA in blood samples may aid in providing the resolution needed for the development of a clinical test to predict risk of preeclampsia in pregnant women and perform accurate cross-sectional comparisons among different clinical studies. Although the current study does not provide any information on the origin or function of cell-free mtDNA, it provides a platform for experimental studies to determine the mechanisms underlying the release and clearance of mtDNA in the maternal circulation in normal pregnancy and pregnancy complications. Thus, the present study lays the foundation for future research to determine whether measuring mtDNA during pregnancy can be used for monitoring human placenta development and maternal health risk.
GRANTS
This research was supported in part by the American Heart Association Grant Nos. 13SDG17050056 (to S. Goulopoulou), 13GRNT16990064 (to Q. Fu), 18PRE33960162 (to S. C. Cushen), and 19TPA34850131 (to S. Goulopoulou) and the National Institutes of Health Grant No. HL088184 (to Q. Fu).
DISCLAIMERS
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Q.F., N.R.P., and S.G. conceived and designed research; S.C.C., M.L.S., A.B., J.S., S.S.J., Y.O., and N.R.P. performed experiments; S.C.C., M.L.S., S.S.J., Y.O., S.A.R., N.R.P., and S.G. analyzed data; S.C.C., M.L.S., Q.F., S.A.R., and S.G. interpreted results of experiments; S.C.C. and S.G. prepared figures; S.C.C. and S.G. drafted manuscript; S.C.C., M.L.S., A.B., J.S., S.S.J., Y.O., Q.F., S.A.R., N.R.P., and S.G. edited and revised manuscript; S.C.C., M.L.S., A.B., J.S., S.S.J., Y.O., Q.F., S.A.R., N.R.P., and S.G. approved final version of manuscript.
ACKNOWLEDGMENTS
Present addresses: M. L. Sprouse, Dept. of Nanomedicine, Houston Methodist Research Institute, Houston, TX; S. S. Jarvis, Dept. of Biological Sciences, Northern Arizona University, Flagstaff, AZ; Y. Okada, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan.
REFERENCES
- 1.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 2.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147, 1999. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 3.Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol Rev Camb Philos Soc 93: 1649–1683, 2018. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW, Chiu RWK. Sequencing of circulating cell-free DNA during pregnancy. N Engl J Med 379: 464–473, 2018. doi: 10.1056/NEJMra1705345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum Reprod Update 11: 59–67, 2005. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- 6.Busnelli A, Lattuada D, Ferrari S, Reschini M, Colciaghi B, Somigliana E, Fedele L, Ferrazzi E. Mitochondrial DNA copy number in peripheral blood in the first trimester of pregnancy and different preeclampsia clinical phenotypes development: a pilot study. Reprod Sci 26: 1054–1061, 2019. doi: 10.1177/1933719118804410. [DOI] [PubMed] [Google Scholar]
- 7.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 44: D67–D72, 2016. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colleoni F, Lattuada D, Garretto A, Massari M, Mandò C, Somigliana E, Cetin I. Maternal blood mitochondrial DNA content during normal and intrauterine growth restricted (IUGR) pregnancy. Am J Obstet Gynecol 203: 365.e1–365.e6, 2010. doi: 10.1016/j.ajog.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Collins LV, Hajizadeh S, Holme E, Jonsson IM, Tarkowski A. Endogenously oxidized mitochondrial DNA induces in vivo and in vitro inflammatory responses. J Leukoc Biol 75: 995–1000, 2004. doi: 10.1189/jlb.0703328. [DOI] [PubMed] [Google Scholar]
- 10.Elchalal U, Ezra Y, Levi Y, Bar-Oz B, Yanai N, Intrator O, Nadjari M. Sonographically thick placenta: a marker for increased perinatal risk—a prospective cross-sectional study. Placenta 21: 268–272, 2000. doi: 10.1053/plac.1999.0466. [DOI] [PubMed] [Google Scholar]
- 11.Fadl S, Moshiri M, Fligner CL, Katz DS, Dighe M. Placental Imaging: Normal Appearance with Review of Pathologic Findings. Radiographics 37: 979–998, 2017. doi: 10.1148/rg.2017160155. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization (WHO) Multicentre Growth Reference Study Group WHO Child Growth Standards: Length/Height-for-Age, Weight-for-Age, Weight-for-Length, Weight-for-Height and Body Mass Index-for-Age: Methods and Development. Geneva, Switzerland: World Health Organization, 2006. [Google Scholar]
- 13.Kavlick MF. Development of a triplex mtDNA qPCR assay to assess quantification, degradation, inhibition, and amplification target copy numbers. Mitochondrion 46: 41–50, 2019. doi: 10.1016/j.mito.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Kavlick MF, Lawrence HS, Merritt RT, Fisher C, Isenberg A, Robertson JM, Budowle B. Quantification of human mitochondrial DNA using synthesized DNA standards. J Forensic Sci 56: 1457–1463, 2011. doi: 10.1111/j.1556-4029.2011.01871.x. [DOI] [PubMed] [Google Scholar]
- 15.Knez J, Winckelmans E, Plusquin M, Thijs L, Cauwenberghs N, Gu Y, Staessen JA, Nawrot TS, Kuznetsova T. Correlates of peripheral blood mitochondrial DNA content in a general population. Am J Epidemiol 183: 138–146, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, Lee DC, Im JA, Lee JW. Mitochondrial DNA copy number in peripheral blood is independently associated with visceral fat accumulation in healthy young adults. Int J Endocrinol 2014: 586017, 2014. doi: 10.1155/2014/586017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet 350: 485–487, 1997. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 18.Luo S, Valencia CA, Zhang J, Lee NC, Slone J, Gui B, Wang X, Li Z, Dell S, Brown J, Chen SM, Chien YH, Hwu WL, Fan PC, Wong LJ, Atwal PS, Huang T. Biparental inheritance of mitochondrial DNA in humans. Proc Natl Acad Sci USA 115: 13039–13044, 2018. doi: 10.1073/pnas.1810946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marschalek J, Wohlrab P, Ott J, Wojta J, Speidl W, Klein KU, Kiss H, Pateisky P, Zeisler H, Kuessel L. Maternal serum mitochondrial DNA (mtDNA) levels are elevated in preeclampsia – a matched case-control study. Pregnancy Hypertens 14: 195–199, 2018. doi: 10.1016/j.preghy.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Meddeb R, Dache ZAA, Thezenas S, Otandault A, Tanos R, Pastor B, Sanchez C, Azzi J, Tousch G, Azan S, Mollevi C, Adenis A, El Messaoudi S, Blache P, Thierry AR. Quantifying circulating cell-free DNA in humans. Sci Rep 9: 5220, 2019. doi: 10.1038/s41598-019-41593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miliotis S, Nicolalde B, Ortega M, Yepez J, Caicedo A. Forms of extracellular mitochondria and their impact in health. Mitochondrion 48: 16–30, 2019. doi: 10.1016/j.mito.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Okada Y, Best SA, Jarvis SS, Shibata S, Parker RS, Casey BM, Levine BD, Fu Q. Asian women have attenuated sympathetic activation but enhanced renal-adrenal responses during pregnancy compared to Caucasian women. J Physiol 593: 1159–1168, 2015. doi: 10.1113/jphysiol.2014.282277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osikoya O, Jaini PA, Nguyen A, Valdes M, Goulopoulou S. Effects of low-dose aspirin on maternal blood pressure and vascular function in an experimental model of gestational hypertension. Pharmacol Res 120: 267–278, 2017. doi: 10.1016/j.phrs.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Phillippe M. Cell-free fetal DNA, telomeres, and the spontaneous onset of parturition. Reprod Sci 22: 1186–1201, 2015. doi: 10.1177/1933719115592714. [DOI] [PubMed] [Google Scholar]
- 25.Qiu C, Hevner K, Enquobahrie DA, Williams MA. A case-control study of maternal blood mitochondrial DNA copy number and preeclampsia risk. Int J Mol Epidemiol Genet 3: 237–244, 2012. [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenfeld CS. Sex-specific placental responses in fetal development. Endocrinology 156: 3422–3434, 2015. doi: 10.1210/en.2015-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz RM, Dayhoff MO. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science 199: 395–403, 1978. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- 28.Sharp AN, Heazell AE, Crocker IP, Mor G. Placental apoptosis in health and disease. Am J Reprod Immunol 64: 159–169, 2010. doi: 10.1111/j.1600-0897.2010.00837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace DC. Why do we still have a maternally inherited mitochondrial DNA? Insights from evolutionary medicine. Annu Rev Biochem 76: 781–821, 2007. doi: 10.1146/annurev.biochem.76.081205.150955. [DOI] [PubMed] [Google Scholar]
- 30.West AP, Shadel GS. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat Rev Immunol 17: 363–375, 2017. doi: 10.1038/nri.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams MA, Sanchez SE, Ananth CV, Hevner K, Qiu C, Enquobahrie DA. Maternal blood mitochondrial DNA copy number and placental abruption risk: results from a preliminary study. Int J Mol Epidemiol Genet 4: 120–127, 2013. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104–107, 2010. doi: 10.1038/nature08780. [DOI] [PMC free article] [PubMed] [Google Scholar]