Abstract
Juxtaglomerular (JG) cells, major sources of renin, differentiate from metanephric mesenchymal cells that give rise to JG cells or a subset of smooth muscle cells of the renal afferent arteriole. During periods of dehydration and salt deprivation, renal mesenchymal stromal cells (MSCs) differentiate from JG cells. JG cells undergo expansion and smooth muscle cells redifferentiate to express renin along the afferent arteriole. Gene expression profiling comparing resident renal MSCs with JG cells indicates that the transcription factor Sox6 is highly expressed in JG cells in the adult kidney. In vitro, loss of Sox6 expression reduces differentiation of renal MSCs to renin-producing cells. In vivo, Sox6 expression is upregulated after a low-Na+ diet and furosemide. Importantly, knockout of Sox6 in Ren1d+ cells halts the increase in renin-expressing cells normally seen during a low-Na+ diet and furosemide as well as the typical increase in renin. Furthermore, Sox6 ablation in renin-expressing cells halts the recruitment of smooth muscle cells along the afferent arteriole, which normally express renin under these conditions. These results support a previously undefined role for Sox6 in renin expression.
Keywords: blood pressure, juxtaglomerular cell expansion, kidney, renin-angiotensin-aldosterone system, renin expression, Sox6
INTRODUCTION
The renin-angiotensin-aldosterone system (RAAS) plays a pivotal role in renal development, the control of blood pressure, and fluid homeostasis (6, 8, 9, 20, 42). Inappropriate activation of the RAAS in adult mammals contributes to the development of hypertension, cardiac hypertrophy, and heart failure (9, 18, 34, 36). The first and rate-limiting step in the initiation of the RAAS is catalyzed by the aspartyl protease renin, which is predominantly produced in the kidney (5, 25, 42). The mechanisms underlying renin production have been investigated in the developing kidney as well as during pathological conditions in adult animals (15, 40). In the developing kidney, lineage tracing studies using the Ren1 promoter have shown that renin-producing cells derive from mesenchymal precursors at embryonic day 11.5 (15, 31, 40). These renin precursors are positive for the transcription factor Foxd1 (15, 27, 38). The expression of renin in the embryo is detectable by embryonic day 14.5 (24) and increases thereafter such that by embryonic day 15.5, renin is present in the renal artery, interlobar arteries, and arcuate arteries of the metanephric kidney (32). After birth, renin-expressing cells become restricted to the terminal portion of the afferent arteriole juxtaglomerular (JG) apparatus. In adults, conditions such as chronic ischemia, prolonged adrenergic activation, and Na+ depletion perturb blood pressure homeostasis and increase the number of renin-expressing cells along the afferent arteriole and kidney interstitium and inside the glomerulus, recapitulating the embryonic distribution of renin expression (13, 14, 16). This process of JG cell recruitment occurs via several cellular mechanisms (22, 28, 39). One mechanism involves dedifferentiation and reexpression of renin by renal smooth muscle cells (SMCs) along the afferent arteriole (22, 39). An alternative mechanism is the differentiation of pericytes (3, 32, 38) and adult renal mesenchymal stromal cells (MSCs) into renin-expressing cells (43).
The mechanisms regulating renin expression remain elusive. Various studies have stressed the importance of transcriptional mechanisms in the control of renin expression. Renal SMCs along the afferent arteriole produce renin and participate in JG cell expansion, and, in these cells, the transcription factor named recombining binding protein suppressor of hairless (RBP-J) plays a key role in renin expression and the JG cell identity (15). Moreover, transcription factors such as cAMP response element binding/cAMP response element modulator (CREB/CREM), homeobox/pre-B cell leukemia (Hox/PBX), and liver X receptor-α (LXRα) bind to the renin promoter, in which they exert positive and negative control over renin expression in vitro (5, 21). Also, a recently published study (31) has shown that superenhancers are involved to maintain renin-expressing cell identity and memory to preserve multisystem homeostasis. Here, we show evidence that the transcription factor Sox6 modulates renin expression both in vitro and in vivo. Sox6, which belongs to the sex-determining region Y subfamily Sox D proteins (7, 19, 26), activates or represses gene transcription through association with multiple transcription factors (26). Genecentric arrays and genome-wide association studies have shown a strong association of Sox6 with hypertension (11, 12, 23, 29). Sox6 has a binding site in the renin promoter within the superenhancer region (31). In vitro, we found that Sox6 is necessary for the cAMP-mediated induction of renin expression in renal tissue, specifically stem progenitor or MSCs. In adult mice, low-Na+ diet and furosemide (LowNa/Fu) increases renin expression as well as Sox6 expression. Sox6 colocalizes with renin cells as well as renal MSCs and SMCs in the afferent arteriole. To validate a role for Sox6 in JG cell expansion/renin expression, we used a novel transgenic mouse model in which Sox6 is specifically ablated in renin-expressing cells (Ren1dCre/Sox6fl/fl). Using this model, we found that ablation of Sox6 in renin-expressing cells halts the increase in renin expression after LowNa/Fu. Moreover, in these mice, SMC recruitment to produce renin after LowNa/Fu was halted. Taken together, our findings demonstrate a new role for Sox6 in the control of renin expression and suggest a role for Sox6 in hypertension.
MATERIALS AND METHODS
Animals.
Mice were housed and cared at the Vanderbilt University Medical Center animal facility following National Institutes of Health guidelines and the Guide for the Care and Use of Laboratory Animals. All animal procedures were approved by the Vanderbilt University Medical Center Institutional Animal Care and Use Committee before the experiments were started. Male C57BL/6 wild-type mice (8 wk old) were purchased from Charles River Laboratories, Ren1cYFP and Ren1dCre (39) transgenic mice were kindly provided by Dr. Ariel Gomez (University of Virginia), and Sox6fl/fl (10) mice were kindly provided by Dr. Veronique Lefebvre (Children’s Hospital of Philadelphia). All animals used in in vivo experiments were maintained on a 12:12-h light-dark cycle at an ambient temperature of 24°C and 60% humidity. The sequences of the primers for genotyping were as follows: Ren-Cre, Ren1d Cre primer 5′-GAAGGAGAGCAAAAGGTAAGAG-3′ located in the renin promoter; Cre primer, 5′-TTGGTGTACGGTCAGTAAATTGGAC-3′ located in Cre; Ren1d, Ren1d Cre primer 5′-GAAGGAGAGCAAAAGGTAAGAG-3′ located in the renin promoter of Ren1d and Ren1c; Ren1d and Ren1c and Ren2 Primer, 5′-GTAGTAGAAGGGGGAGTTGTG- 3′ in renin intron 1 of Ren1d and Ren1c and Ren2; and Sox6fl/fl, forward primer 5′-GTCACTCAGAGGTTACTATGGTG-3′ and reverse primer 5′-TTGGAGGCTTTAGCAGCTCTC-3′. DNA gels with PCR products are provided in Supplemental Fig. S1 (Supplemental Data are available online at https://doi.org/10.6084/m9.figshare.8317103.v1). Ren1dCre/Sox6fl/fl transgenic mice (Sox6 is specifically ablated in renin-expressing cells) and Ren1dCre/Sox6wt/wt control littermate mice were used in discerning the role of Sox6 in renin expression control.
Isolation and culture of renal MSCs.
Renal MSC isolation and culture were carried out as previously reported (25). Renal MSCs were differentiated with 3-isobutyl-1-methylxanthine (IBMX; 100 μM) and forskolin (10 μM), as previously described (33). In brief, C57BL/6 mice were euthanized, and kidneys were perfused with PBS, harvested, minced, and digested with 0.1% collagenase type I (catalog no. 17100-017, GIBCO) solution in HBSS (catalog no. 14025, GIBCO) for 30 min at 37°C while being rocked. After tissue digestion, 20 mL (1 volume) warm DMEM (with 10% FBS and 1× penicillin-streptomycin) was added to inactivate collagenase type I. Further procedures were performed on ice. Renal cell suspensions were washed with ice-cold PBS and filtered through 70-μm mesh filters. To remove red blood cells, cell suspensions were treated with ice-cold ACK buffer (0.15 M potassium-ammonium chloride, catalog no. R7757, Sigma). Renal MSCs were isolated using CD44 as a marker (CD44+), MSCs were isolated by two cycles of FACS via specific gates, dead cells were excluded with 7-amino-actinomycin D, and doublets were excluded on the basis of three hierarchical gates (forward/side scatter area, forward scatter height/width, and side scatter height/width). Sorted renal CD44+ cells were cultured in growth medium (MesenPro, ThermoFisher) at 37°C in the presence of 5% CO2. Medium was changed every 2–3 days. Cells were used for experiments during passages 3–5.
Transcript profiling.
Renal MSCs were isolated from adult C57BL/6 Ren1c yellow fluorescent protein (YFP) mice as described above. JG cells were isolated from adult C57BL/6 Ren1c YFP mice by FACS using YFP expression as a surrogate for renin expression. RNA was isolated using the Arcturus RNA PicoPure Isolation Kit (ThermoFisher). cDNA reactions were performed using Ovation Pico WTA from NuGEN. For transcript profiling. Affymetrix Mouse 2.0 was used.
Induction of JG cell expansion and recruitment in vivo.
JG cell expansion was induced with a low-Na+ diet (0.01% NaCl, Envigo) plus furosemide (2.28 mmol/L, Fisher) (LowNa/Fu). Furosemide was dissolved in drinking water (pH 7.4) and provided ad libitum. Since furosemide is light sensitive, amber-colored bottles were used. Furosemide was prepared in the morning and provided fresh to mice every day. Mice were treated with LowNa/Fu for 10 days. After 10 days of treatment, mice were anesthetized using the drop jar method with isoflurane under a fume hood, and kidneys were harvested. Blood was collected directly from heart using a syringe and transferred into EDTA-containing Eppendorf tubes. Blood was centrifuged at 1,500 g for 10 min at room temperature, and plasma was isolated and stored at −80°C until further analysis.
Kidney immunohistochemistry.
Immunohistochemistry staining of kidney sections was performed as previously described (43). Briefly, kidneys were perfused fixed with 10% neutral buffered formalin solution, dehydrated in a graduated ethanol series, and embedded in paraffin. Kidney sections were cut at 10 μm thickness. Sections were deparaffinized in Histo-Clear solution (catalog no. HS-202, National Diagnostics) at room temperature and permeabilized with 0.2% Triton X-100. After being blocked with 5% BSA-PBS for 1 h at room temperature, sections were incubated with primary antibodies diluted in 1% BSA-PBS overnight at 4°C. The next day, slides were washed in PBS and incubated with fluorochrome-conjugated secondary antibodies for 1 h at room temperature. The following primary antibodies were used: anti-renin (1/100 dilution, kindly provided by Dr. Tadashi Inagami, Vanderbilt University), anti-renin (1/50 dilution, no. 1206, Innovative Research), anti-α-smooth muscle actin (α-SMA; 1/500 dilution, A5228, Sigma), anti-Sox6 (1/1,000, ab30455, Abcam), and anti-aquaporin 2 (1/1,000, ab15116, Abcam). The specificity of the Sox6 antibody was determined with tissue from Sox6 knockout mice (2). Furthermore, for the specificity of Sox6 antibody and protein signal, a Sox6 peptide competition assay was performed using Sox6 peptide (ab30530, Abcam) with the Sox6 antibody (ab30455, Abcam) (see Supplementary Fig. S2, available online at https://doi.org/10.6084/m9.figshare.8317103.v1). The secondary antibodies were used in 1:500 dilutions and chosen based on primary antibodies and Alexa Fluor fluorophores (ThermoFisher). Nuclei were counterstained with DAPI.
Flow cytometry.
Kidneys were harvested from control and LowNa/Fu-treated mice. Kidneys were minced using surgical scissors and digested with collagenase type I (catalog no. 17100-017, GIBCO) solution in HBSS (catalog no. 14025, GIBCO) and incubated for 30 min at 37°C in a slowly shaking water bath. After tissue digestion, 20 mL (1 volume) warm DMEM (with 10% FBS and 1× penicillin-streptomycin) was added to inactivate collagenase type I. Further procedures were performed on ice. After being strained with a 70-μm strainer to separate nondigested tissue, the cell mixture was centrifuged at 300 g for 5 min at 4°C and the supernatant was discarded. The cell pellet was treated with red blood cell lysis buffer (catalog no. R7757, Sigma) and incubated for 5 min on ice and centrifuged at 300 g for 5 min at 4°C. The cell pellet was washed with cold PBS by repeating the previous step. For extracellular staining, cells were resuspended in ice-cold staining buffer (PBS with 2 mM EDTA and 0.5% BSA) and stained with CD44 antibody (0.4 μg, 2 μL, APC Rat Anti-Mouse CD44, catalog no. 559250, BD Pharmingen) while being rocked for 1 h at 4°C followed with centrifugation of samples at 300 g for 5 min at 4°C. The cell pellet was then washed with staining buffer and centrifuged as above. For intracellular staining, cells were fixed in 4% paraformaldehyde (catalog no. 157-SP50, Electron Microscopy Sciences) by being incubated for 15 min on ice. Cells were washed with staining buffer plus saponin (PBS with 2 mM EDTA, 0.5% BSA, and 0.02% saponin), and the obtained cell pellet was resuspended in saponin buffer at 4°C and considered ready for intracellular staining. Cells were stained with Sox6-Alexa Fluor 647, renin-Alexa Fluor 488, and α-SMA- phycoerythrin (Sigma) and incubated for 1 h at 4°C while being rocked. Cells were centrifuged at 300 g for 5 min at 4°C, and the supernatant was discarded. The cell pellet was washed with staining buffer plus saponin by centrifugation at 300 g for 5 min at 4°C, and the supernatant was discarded. The obtained cell pellet was resuspended in 300 μL PBS and transferred into prelabeled flow cytometry tubes. To identify cell population and draw proper gates, we used flow minus one samples. Flow minus one samples allow us to draw gates identifying multiple cell populations when multiple fluorophores are used. Compensation beads were used to consistently and accurately set flow cytometry compensation for all antibodies used in the experiment. We used antibodies mixed with compensation beads (Biolegend). Samples were analyzed using flow cytometry (BD Biosciences Canto II 3 laser 8 color cytometer). Data analysis was performed using FlowJo (version 10).
Labeling Sox6 and renin antibody with Alexa Fluor.
Sox6 (ab30455, Abcam) and renin (AF4277, R&D Systems) antibodies were labeled with AF647 and AF488, respectively, using an antibody labeling kit (A20181 and A20186, respectively, ThermoFisher). The 1 M sodium bicarbonate solution was prepared by resuspending component B in 1.0 mL deionized water. The antibody concentration was adjusted to 1.0 mg/mL, and 1/10 volume of 1 M sodium bicarbonate solution was added. Next, antibody plus 1 M sodium bicarbonate was added to the vial of Alexa Fluor dye, mixed well, and incubated for 1 h at room temperature. After the purification column had been assembled, resin was added to the column and centrifuged at 1,100 g for 3 min. The reaction mix of antibody and Alexa Fluor was added into the column and centrifuged at 1,100 g for 5 min to collect the labeled antibody.
Western blot analysis.
Kidney tissues were minced with a razor and homogenized with Tissue-Tearor (model no. 985370, BioSpec Products) following the manufacturer’s instructions. Homogenates were lysed with lysis buffer and sonicated for 1.5 min (30 s × 3 times, with one 30-s interval between each sonication) followed by centrifugation for 16,000 g for 20 min at 4°C. The supernatant was collected, and samples were prepared in RIPA buffer, Laemmli buffer, and β-mercaptoethanol (10%) and heated at 95°C for 5 min. Samples (30 μg protein) were loaded in the gels. Western blot analysis of kidney samples was performed as previously described (35). Briefly, tissue homogenate was resolved on 8–16% Tris-glycine gels (catalog no. 456-1063, Bio-Rad) by SDS-PAGE. Gels were transblotted onto polyvinylidene fluoride membranes for 2 h at 4°C. Thereafter, membranes were blocked with a mixture of 5% milk and 3% BSA in Tris-buffered saline-Tween 20 (TBST) at room temperature for 1 h and probed with the respective primary antibodies in TBST overnight at 4°C. After being washed with TBST (3 × 10 min), membranes were incubated with the corresponding secondary antibodies conjugated with horseradish peroxidase (HRP; anti-mouse ref. no. W4028 and anti-rabbit ref. no. W4018) followed by TBST washes (3 × 10 min). Chemiluminescent reagent clarity Western ECL substrate was used (catalog no. 1705062, Bio-Rad) to visualize protein bands using the image station. Protein bands were quantified and normalized with the housekeeping gene β-actin (A1978, Sigma-Aldrich) using software integrated with the image station. The primary antibody dilutions used were 1:500 for Sox6 (ab30455, Abcam), 1:100 for renin (sc-137252, Santa Cruz Biotechnology), and 1:5,000 for β-actin (A1978, Sigma-Aldrich). Secondary antibody (Promega) dilutions were used at 1:500. To determine the specificity and renin band position in Western blot analysis, we used a renin peptide from Santa Cruz Biotechnology (sc-137252p) in a competition assay with the renin antibody (sc-137252). Moreover, to determine the prorenin and renin position in immunoblots, His-tagged mouse recombinant prorenin (AS-72174, AnaSpec) and renin (catalog no. 4277-AS-020, R&D Systems) proteins were run with kidney tissue lysate samples from LowNa/Fu-treated Ren1dcre/Sox6wt/wt mice. Prorenin and renin bands quantified separately and normalized with the housekeeping gene β-actin. Recombinant prorenin and renin proteins have been previously used to localize prorenin and renin proteins in Western blot analysis (17, 37).
Quantitative real-time PCR.
Total RNA was isolated using the Quick-RNA MiniPrep kit (R1054, Zymo Research). RNA was then used to generate cDNA using the high-capacity cDNA reverse transcription kit (catalog no. 4368814, Applied Biosystems). RNA and cDNA final concentrations were measured using NanoDrop One (ThermoFisher). cDNA (200 ng) per quantitative RT-PCR plus 1 μL Taqman primer and the Taqman fast advance master mix (catalog no. 4444554 ThermoFisher) were used. The following validated Taqman primers (ThermoFisher) were used: renin (Mm02342887), Sox6 (Mm00488393), and Gapdh (Mm99999915). Samples were run in a Step One quantitative RT-PCR (ThermoFisher), and threshold cycle values were used to calculate relative expression to Gapdh (housekeeping gene) and fold changes.
In situ hybridization.
Colocalization and amplification of Sox6 and renin mRNA synthesis were examined using the RNAscope 2.5 HD duplex detection assay [Advanced Cell Diagnostics (ACD) Bio-Techne, Newark, CA] for in situ hybridization technology following the manufacturer’s protocols. Briefly, kidneys were perfused fixed with 10% neutral buffered formalin solution, dehydrated in a graduated ethanol series, and embedded in paraffin. Kidney sections were cut at 10 μm thickness. Sections were deparaffinized in Histo-Clear solution (HS-202, National Diagnostics) at room temperature followed by blockade of endogenous peroxidase with RNAscope H2O2. Tissue was retrieved by boiling in target retrieval solution (ACD Bio-Techne) at 100–104°C for 15 min and then treated with protease plus at 40°C for 30 min. Target probes (designed by ACD Bio-Techne: Mm-Sox6, ref. no. 472061-C1, and Mm-Ren1, ref. no. 433461-C2) were hybridized for 2 h at 40°C followed by a series of signal amplification (Amp 1–6) and in between washing with RNAscope wash buffer. Red (renin mRNA) and green (Sox6 mRNA) dyes were used after 6 and 10 signal amplifications, respectively. RNA staining signals were identified as red and green punctuate dots. All hybridization steps at 40°C were performed in a HybEZ Hybridization System (ACD Bio-Techne). After the completion of the RNAscope assay, tissue sections were counterstained with 50% hematoxylin staining solution for 30 s at room temperature followed by mounting with VectaMount mounting medium (H-500, Vector Laboratories, Burlingame, CA) and drying slides in a dry oven at 60°C for 30 min. Tissue sections were viewed with Nikon Eclipse Ti (Software NIS-Elements AR 4.40.00 64-bit). For randomization and blinding, two separate blinded investigators carried out the staining and counting the glomeruli containing renin and Sox6 double-positive cells. Investigators were blinded with respect to animal identifiers and group assignments. Values were averaged between the two blinded investigators.
Fluorescent in situ hybridization.
Colocalization and amplification of Sox6 and renin mRNA synthesis were studied using RNAscope Multiplex Fluorescent Reagent Kit v2 (ACD Bio-Techne) for fluorescent in situ hybridization technology following the manufacturer’s protocols. Briefly, kidneys were perfused fixed with 10% neutral buffered formalin solution, dehydrated in a graduated ethanol series, and embedded in paraffin. Kidney sections were cut at 10 μm thickness. Sections were deparaffinized in Histo-Clear solution (HS-202, National Diagnostics) at room temperature followed by blockade of endogenous peroxidase with RNAscope H2O2. Tissue was retrieved by boiling in target retrieval solution (ACD Bio-Techne) at 100–104°C for 15 min and then treated with protease plus at 40°C for 30 min. Target probes (designed by ACD Bio-Techne: Mm-Ren1-O1, ref. no. 558571, and Mm-Sox6-C2, ref. no. 472061-C2) were hybridized for 2 h at 40°C followed by a series of signal amplification (Amp 1–3) and in between washing with RNAscope wash buffer. Renin and Sox6 mRNA probes were assigned to channels HRP-C1 and HRP-C2, respectively. The HRP-C1 signal was developed using RNAscope Multiplex FL v2 HRP-C1 with Opal 520 (ref. no. FP1487001KT, Perkin-Elmer) followed by blockade with RNAscope Multiplex FL v2 HRP blocker and in between washing with RNAscope wash buffer. Similarly, the HRP-C2 signal was developed using RNAscope Multiplex FL v2 HRP-C2 with Opal 650 (ref. no. FP1496001KT) followed by blockade with RNAscope Multiplex FL v2 HRP blocker and in between washing with RNAscope wash buffer. All hybridization steps at 40°C were performed in a HybEZ Hybridization System (ACD Bio-Techne). After completion of the RNAscope assay, tissue sections were counterstained with DAPI and incubated for 30 s at room temperature followed by mounting with VectaMount mounting medium (H-500, Vector Laboratories). Slides were dried overnight in the dark at room temperature. Tissue sections were viewed with Nikon Eclipse Ti (Software NIS-Elements AR 4.40.00 64-bit). For randomization and blinding, two separate blinded investigators carried out counting of the glomeruli containing renin and Sox6 double-positive cells. Investigators were blinded with respect to animal identifiers and group assignments. Values were averaged between the two blinded investigators.
Plasma renin concentration.
To determine plasma renin activity, blood samples were obtained from mice using EDTA as an anticoagulant and assayed for angiotensinogen I generation as previously described (30). From each sample, 2 μL plasma was added to an incubation cocktail containing 158 μL modified phosphate incubation buffer (0.1 M phosphate buffer, 0.06 M disodium EDTA, and 0.2% gelatin, pH 6.5), 2 μL PMSF, and 138 μL substrate (containing 250 ng angiotensinogen, obtained from nephrectomized rats). Substrate consumption had to be less than 15% of the total added to ensure first-order kinetics of the enzymatic reaction. This sample was incubated at 37°C for 90 min, boiled for 10 min to stop the reaction, and centrifuged for 10 min at 2,500 g, and the supernatant was removed. The sample was frozen for later analysis (typically in <48 h). Angiotensin I generation was analyzed by radioimmunoassay using a GammaCoat Plasma Renin Activity Kit (DiaSorin, Stillwater, MN) according to the manufacturer’s instructions. The assay was controlled using buffer blanks and internal quality controls.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) assays were performed according the manufacturer’s instructions (SimpleChIP Enzymatic Chromatin IP kit no. 9003, Cell Signaling). C57BL/6 wild type (6–8 wk old) were subjected to LowNa/Fu for 10 days. After treatment, mouse kidneys were digested with collagenase as described above, and single cell suspension nuclei were digested with 0.1 μL micrococcal nuclease per 4 × 106 cells (amount of micrococcal nuclease was empirically determined according to the manufacturer’s instructions). Immunoprecipitation was performed with ChIP-validated antibodies: rabbit IgG control (catalog no. 2729, Cell Signaling) and Sox6 (PA5-34616, ThermoFisher). Immunoprecipitated DNA was quantified by quantitative PCR (Power SYBR Green PCR Master Mix catalog no. 4367659, ThermoFisher) with primers for the renin promoter [EpiTect ChIP qPCR Primer Assay for Mouse Ren1, NM_031192.2(-)01Kb): GPM1028245(-)01A, Qiagen]. A serial dilution of the 2% input chromatin DNA (undiluted, 1:5, 1:25, 1:125) was used to create a standard curve and determine the efficiency of amplification. Percent input was calculated, and negative control IgG values were subtracted. Data are presented as fold changes of the percent input between IgG and Sox6 samples. Binding of Sox6 to renin promoter ChIP is presented as fold enrichment.
Statistics.
A t test was performed for experiments containing two conditions. One-way or two-way ANOVA was used for experiments with three or more conditions followed by Bonferroni post hoc tests for comparisons between individual groups. P values of ≤0.05 was considered significant. All statistical analysis was performed using GraphPad Prism 7.03.
RESULTS
Identification of JG-specific transcription factors.
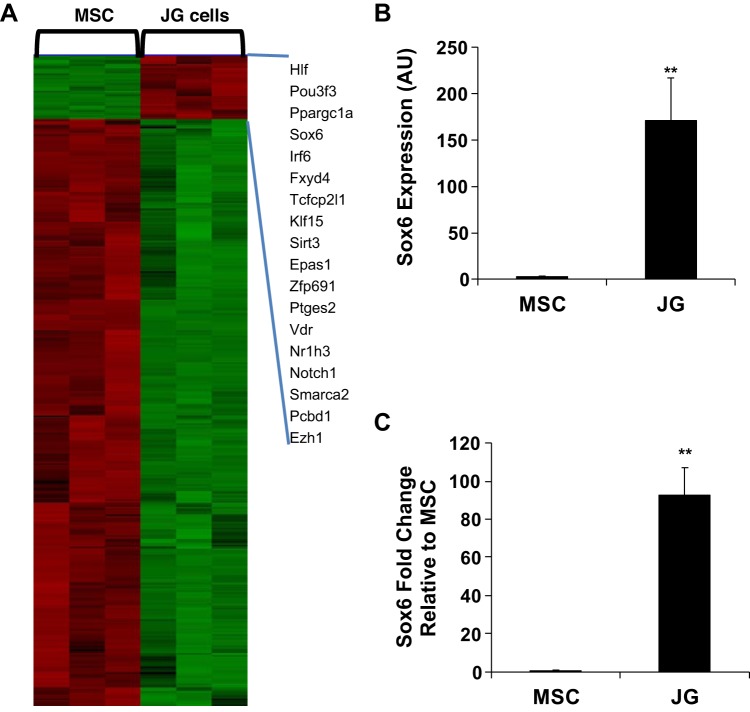
To identify specific transcription factors that are highly expressed in JG cells, we performed a microarray screen comparing renal MSCs and JG cells. As expected, renin and other JG cell markers were highly expressed in JG cells and absent from MSCs (data not shown). The array results showed that 5,573 genes were differentially expressed in JG cells compared with renal CD44+ MSCs: 1,223 transcripts were upregulated (20%) and 4,350 (80%) were downregulated. Of these 5,573 genes, 643 transcripts were involved in transcription, 73 transcripts were upregulated, and 570 transcripts were downregulated. Genes upregulated more than fivefold are shown (Fig. 1A). Sox6 was chosen for its function in controlling cell fate in other systems (2, 19, 26). Microarray fluorescence intensity data (in arbitrary units) from MSC and JG cell samples were plotted. We found that Sox6 was highly expressed in JG cells compared with renal MSCs (Fig. 1B). Sox6 microarray data were validated by quantitative RT-PCR (Fig. 1C).
Fig. 1.
Sox6 expression is upregulated in juxtaglomerular (JG) cells compared with renal mesenchymal stromal cells (MSCs). A: heat map showing the microarray data comparison between renal CD44+ MSCs and JG cells. Data were analyzed using d-Chip, which focuses on transcriptional regulators. Genes upregulated ≥5-fold in JG cells are presented on the right side of the heat map. B: microarray expression data of Sox6 in renal MSCs and JG cells. C: Sox6 quantitative real-time PCR validation of microarray data. Expression values for Sox6 are shown relative to GAPDH. There were at least 4 independent samples/group. P values were calculated using a Student’s unpaired t test. **P < 0.01. AU, arbitrary units.
Sox6 is involved in renin expression in vitro.
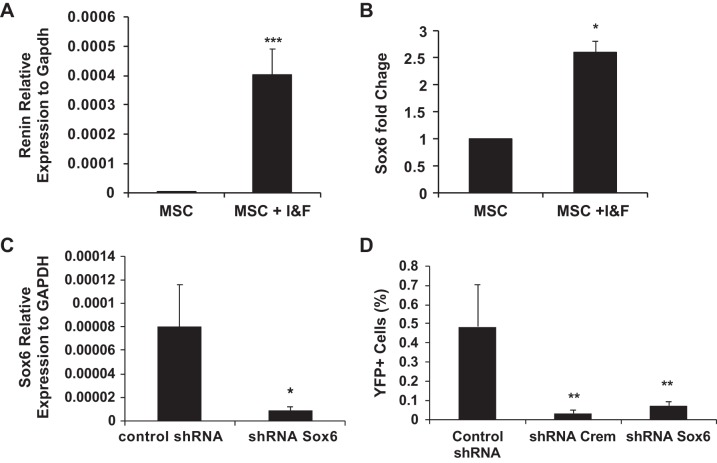
Our gene expression array data indicated that Sox6 is highly expressed in cells that produce renin. To validate this in a model that induces renin expression, CD44+ tissue-specific MSCs were isolated from C57BL6 Ren1c YFP adult kidneys and treated with IBMX (100 μM) and forskolin (10 μM) to differentiate into renin-expressing cells, as previously described (33). This treatment concomitantly increased both renin and Sox6 expression (Fig. 2, A and B), suggesting that Sox6 may control renin expression. We investigated this possibility by reducing Sox6 levels with a specific shRNA in renal MSCs isolated from Ren1c YFP adult mice (Fig. 2, C and D). Control nontargeted shRNA and CREM-specific shRNA were used as negative and positive controls, respectively. Knockdown of Sox6 prevented MSC differentiation into renin-producing cells in response to IBMX and forskolin to the same levels as the positive control CREM (Fig. 2D). This indicates that Sox6 is necessary for renin expression by renal MSCs induced by cAMP.
Fig. 2.
Sox6 is essential for cAMP induction of renin expression in vitro. Renal CD44+ mesenchymal stromal cells (MSCs) were isolated from adult wild-type mice. Quantitative RT-PCR analysis of renin and Sox6 expression after 3-isobutyl-1-methylxanthine and forskolin (I&F) treatment was performed after 7 days of treatment. Data are presented as means ± SE; n = 4. P values were calculated with an unpaired t test. *P < 0.05; ***P < 0.001. A: renin. B: Sox6. Renal CD44+ MSCs were isolated from C57BL6 Ren1c yellow fluorescent protein (YFP) adult mice and cultured in growth medium for three to five passages before use, and YFP expression was used as a surrogate for renin expression. C: quantitative RT-PCR analysis of Sox6 knockdown in MSCs. Data are presented as means ± SE of three independent experiments. P values were calculated with an unpaired t test. *P < 0.05. D: downregulation of Sox6 affects the differentiation of renal MSCs to renin-expressing cells. Data are presented as means ± SE of three independent experiments. P values were calculated using an unpaired t test. **P < 0.01 comparing Sox6 shRNA with control shRNA or cAMP-responsive element modulator (Crem) shRNA with control shRNA.
Sox6 expression increases during LowNa/Fu treatment.
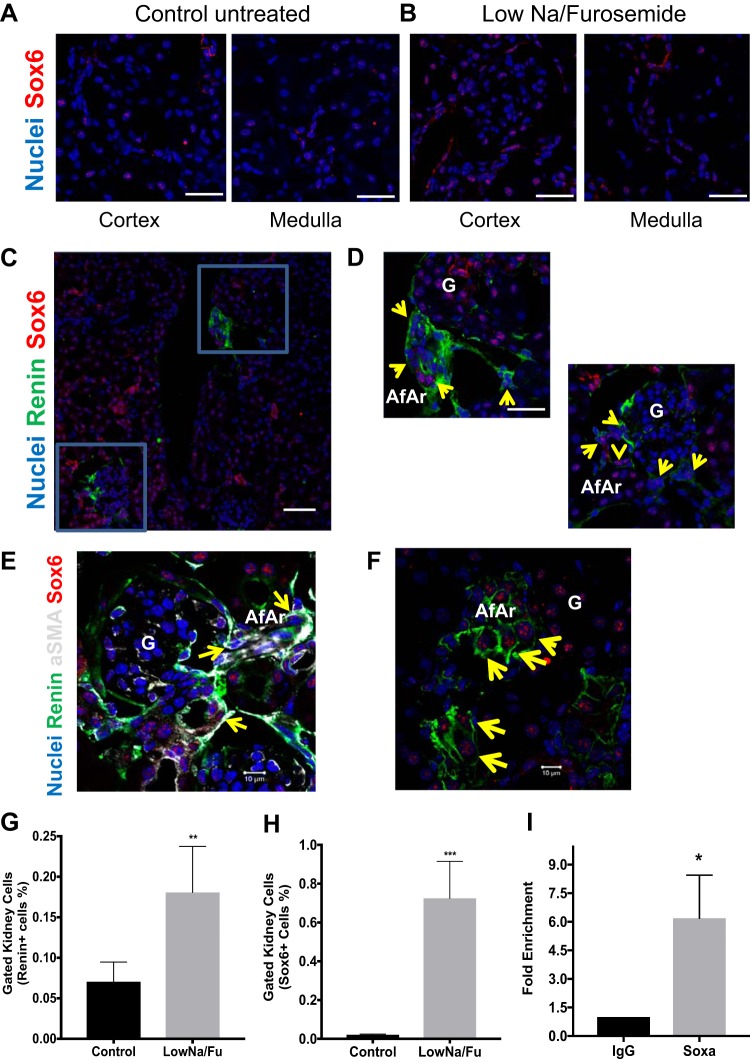
To unravel the role of Sox6 in the kidney and its contribution to the regulation of blood pressure homeostasis, we initially analyzed Sox6 expression in adult mice. We found that Sox6 is expressed in the cortex and medulla of the adult kidney (Fig. 3, A and B). The specificity of the Sox6 antibody was determined using tissue from Sox6 knockout mice (2) and a specific peptide competition assay (Supplementary Fig. S2, available online at https://doi.org/10.6084/m9.figshare.8317103.v1). After LowNa/Fu treatment, Sox6 was colocalized with renin-expressing cells in the glomeruli and afferent arteriole (Fig. 3, C and D). Colocalization of Sox6 staining was also observed with cells involved in JG recruitment, such as α-SMA+ SMCs in the afferent arteriole (Fig. 3E) (33) as well as vascular and perivascular MSCs that differentiate into renin-expressing cells (Fig. 3F) (43). Confirming our previous study, renin and Sox6 expression was determined using flow cytometry. Renin expression was significantly upregulated after LowNa/Fu, which also induces JG cell recruitment (Fig. 3G) (43). Concomitant with a 3-fold increase in the number of renin-expressing cells (renin+ cells), Sox6-expressing cells (Sox6+ cells) increased 36-fold (Fig. 3, G and H). To further support a role for Sox6 in controlling renin expression, we performed a ChIP assay on freshly isolated kidney cells from LowNa/Fu-treated mice. From in silico analysis of the renin promoter, we found four putative Sox6-binding sites within −5 kb from the transcription start site. Analysis of this region by ChIP indicated that Sox6 was bound to the renin promoter in renal cells during JG cell expansion (Fig. 3I). These results support the notion that Sox6 modulates renin expression during JG cell expansion, controlling renin expression during this process.
Fig. 3.
Sox6 expression is upregulated during juxtaglomerular cell expansion. C57BL/6 wild-type mice (6–8 wk old) were administered a low-Na+ diet (0.02% NaCl) plus furosemide (LowNa/Fu) (drinking water: 2.28 mmol/L) for 10 days. Control animals received normal chow (0.6% NaCl). Representative ×63 confocal images of kidney medulla and cortex sections stained with Sox6 from LowNa/Fu-treated and control untreated mice (4 mice/condition with three 10-μm kidney sections/kidney and multiple confocal images per section) are shown. Sox6 is shown in red; nuclei are shown in blue. A: Sox6 expression in the kidney cortex and medulla of untreated control mice. Scale bars = 20 μm. B: Sox6 expression in the kidney cortex and medulla of mice after of LowNa/Fu treatment. Scale bars = 20 μm. C: representative ×20 confocal image of a kidney section stained for Sox6 (red), renin (green), and nuclei (blue) from a LowNa/Fu-treated mouse. Scale bar = 50 μm. Areas enclosed in the blue boxes were enlarged with a ×63 objective. D: magnified ×63 images of the glomeruli (G) showing the coexpression of renin and Sox6 in the afferent arteriole (AfAr). Yellow arrows point to Sox6+ renin+ double-positive cells. Scale bar = 20 μm. E: representative ×63 confocal images of kidney sections stained with renin (green), Sox6 (red), α-smooth muscle actin (aSMA; gray), and nuclei (blue) from LowNa/Fu-treated mice. Yellow arrows point to renin+ Sox6+ α-smooth muscle actin+ triple-positive cells. F: representative ×63 confocal image of a kidney section stained for Sox6 (red), CD44 (green), and nuclei (blue) from a LowNa/Fu-treated mouse. Yellow arrows point to double-positive Sox6+ CD44+ cells. Scale bar = 10 μm. Flow cytometry analysis of isolated mouse kidney cells used specific antibodies to renin and Sox6 to quantify renal cells expressing these proteins. G: renin expression in mice before and after LowNa/Fu treatment. H: Sox6 expression in mice before and after LowNa/Fu treatment. Data are presented as means ± SE; n = 4–5. P values were calculated with an unpaired t test. **P < 0.01 and ***P < 0.001, LowNa/Fu vs. no treatment. I: chromatin immunoprecipitation assay showing binding of Sox6 to the renin promoter with IgG and specific Sox6 antibody. Data are presented as fold changes of the percent input between IgG and Sox6 samples. Data are presented as means ± SE; n = 3. P values were calculated with an unpaired t test. *P < 0.05.
Renin expression in LowNa/Fu and nontreated animals.
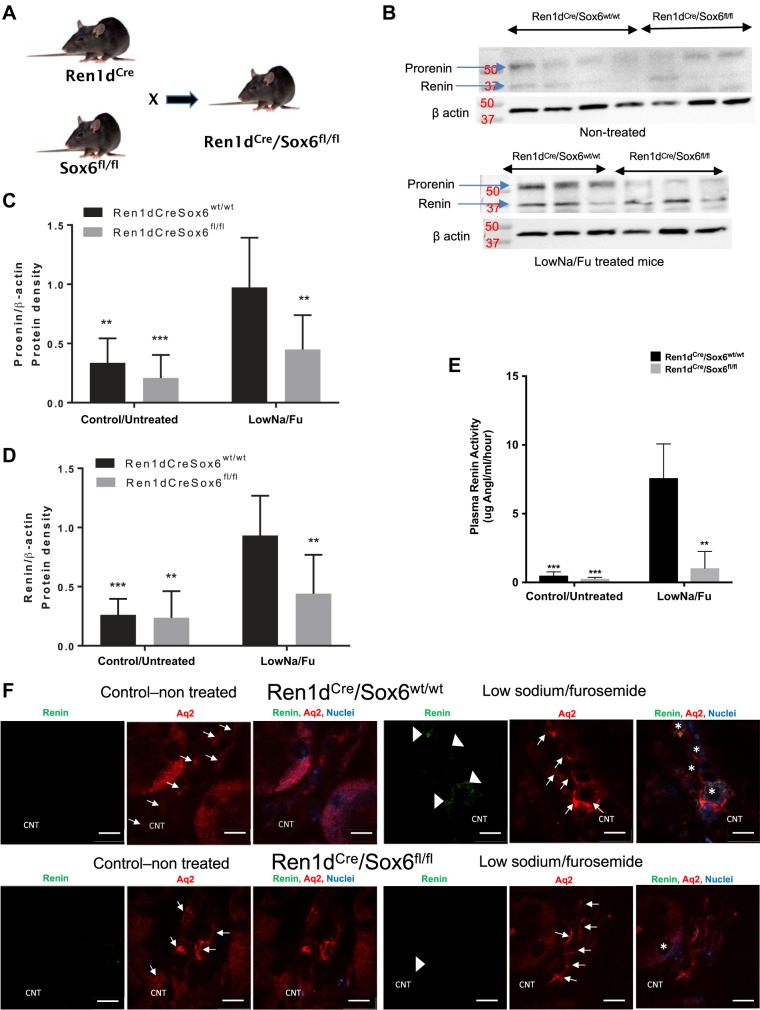
To further define a role for Sox6 in controlling renin expression during physiological conditions, we developed a new transgenic mouse by crossing the Ren1dCre transgenic mouse (39) with the Sox6fl/fl mouse (10) (Fig. 4A). The resulting Ren1dCre/Sox6fl/fl mice lack Sox6 specifically in renin-expressing cells. To elaborate a role of Sox6 in renin expression, we measured renin protein by Western blot analysis after LowNa/Fu treatment. By Western blot analysis, we detect two renin bands running at ~38 and 52 kDa. We run His-tagged recombinant renin and His-tagged recombinant prorenin proteins with LowNa/Fu samples from Ren1dcre/Sox6fl/fl and Ren1dcreSox6wt/wt mice to establish the effects of knockout of Sox6 in renin-expressing cells over prorenin and renin expression. We determine that the upper band corresponds to prorenin at ~52 kDa (recombinant His-prorenin band appear near 50 kDa) and the lower band that appears at ~40 kDa corresponds to renin. Moreover, the renin peptide competition assay shows that the renin band, which appears near 37 kDa, completely disappears, and the band near 50 kDa is faint, showing that this antibody (sc-137252, Santa Cruz Biotechnology) recognizes prorenin and renin. In the LowNa/Fu-treated group, levels of renin and prorenin protein expression were significantly higher in wild-type (Ren1dcre/Sox6wt/wt) mice compared with Sox6-specific knockout (Ren1dcre/Sox6fl/fl) mice (Fig. 4B). Quantitatively, both prorenin and renin expression in Ren1dcre/Sox6fl/fl mice did not increase after stimulation with LowNa/Fu; moreover, their expression was significantly lower compared with wild-type mice. In the nontreated group, levels of prorenin and renin expression decreased (prorenin decrease ~40% and renin ~12%) in Sox6-specific knockout compared with wild-type mice. Within the wild-type groups, there was a significant difference in the levels of prorenin and renin expression in nontreated compared with LowNa/Fu-treated mice (Fig. 4, B–D). As another parameter of JG cell expansion, we measured plasma renin activity after LowNa/Fu treatment. Mice with specific ablation of Sox6 in renin-expressing cells had significantly lower plasma renin activity compared with wild-type mice (Fig. 4E). To assess the expression of renin in the collecting duct (CD) and connecting tubule (CNT), aquaporin 2 was used as a marker. Renin-expressing cells were found around the CNT and CD in wild-type mice treated with LowNa/Fu (Fig. 4F). However, in mice with specific ablation of Sox6 in renin-expressing cells, renin expression was not detected in CNT or CD cells (Fig. 4F). Renin expression in CNT and CD cells was not detected in control nontreated wild-type or Sox6 knockout mice. Figure 4F only shows the data in CNT, although we found a similar pattern of renin expression in CDs as in CNT in all four groups of mice (data not shown). These results indicate that Sox6 has a new function in renin expression during LowNa/Fu treatment known to stimulate an increase in renal renin expression.
Fig. 4.
Specific knockout of Sox6 in renin-expressing cells inhibits the increase in renin expression during juxtaglomerular cell expansion. A: schematic representation of the development of Ren1dCre/Sox6fl/fl mice. Mice were administered a low-Na+ diet (0.02% NaCl) plus furosemide (LowNa/Fu) (drinking water: 2.28 mmol/L) for 10 days. Control animals received normal chow (0.6% NaCl). B: after 10 days of LowNa/Fu treatment, kidneys were isolated and Western blot analysis was performed. Representative Western blot images show levels of prorenin/renin expression in nontreated (top) and LowNa/Fu-treated (bottom) mice. β-Actin was used as a loading control. To determine the specificity of the renin antibody, we used recombinant mouse renin in a competition assay. C: quantification of prorenin expression immunoblots. D: quantification of renin expression immunoblots. Data are presented as means ± SE; n = 5–10. P values were calculated with two-way ANOVA followed by a Tukey post hoc test. **P < 0.01; ***P < 0.001. E: plasma was isolated from nontreated and LowNa/Fu-treated Ren1dCre/Sox6fl/fl and Ren1dCre/Sox6wt/wt mice (6–8 wk old), and plasma renin activity determined as μg ANG I·mL−1·h−1. Data are presented as means ± SE; n = 3 per group. P values were calculated with one-way ANOVA followed by a Tukey post hoc test. **P < 0.01; ***P < 0.001. F: after 10 days of LowNa/Fu treatment, kidneys were isolated and the expression of renin (green) and aquaporin 2 (Aq2; red) was used to establish renin expression in the collecting duct (CD) and connecting tubule (CNT). Aquaporin 2 is a known marker of the CD and CNT. Representative ×90 confocal microscopy images of kidney sections (cortex part) were stained for renin (green) and aquaporin 2 (red) from LowNa/Fu-treated mice and control untreated mice. Arrows and arrowheads indicate aquaporin 2 and renin expression, respectively, in the CNT. *Expression of renin and aquaporin 2 together in the same cell of the CNT. Scale bars = 20 μm.
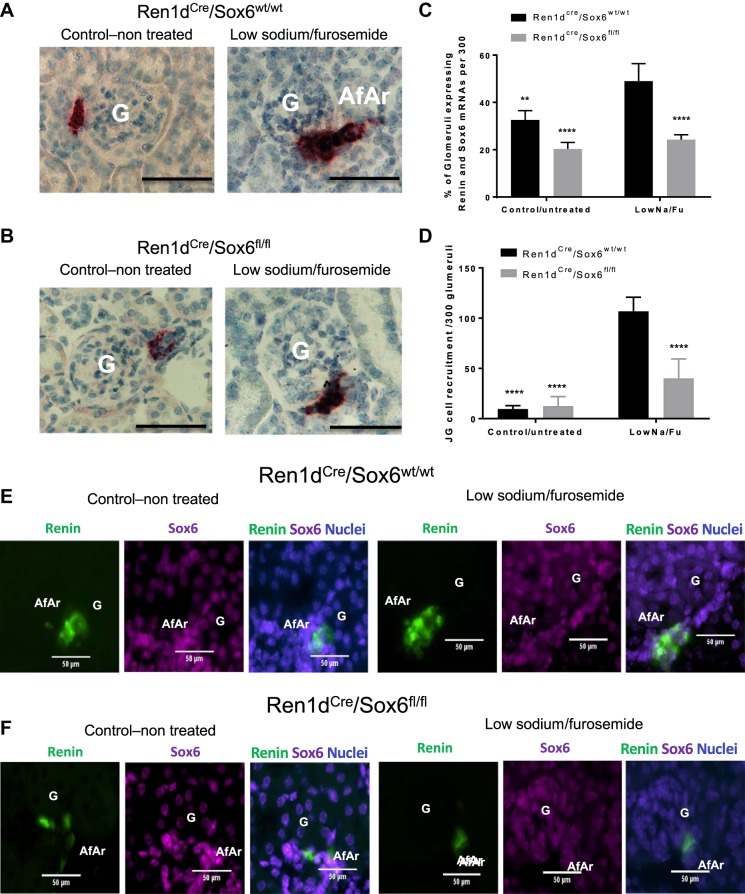
To define if Sox6 affects renin transcription after LowNa/Fu, RNA expression levels were measured by in situ hybridization using specific probes to renin and Sox6 mRNAs. At baseline, the number of renin mRNA-positive cells in the JG area was significantly lower in Ren1dCre/Sox6fl/fl mice compared with Ren1dCre/Sox6wt/wt mice (Fig. 5, A–C). After LowNa/Fu treatment, renin mRNA levels and in situ signal intensity increased in Ren1dCre/Sox6wt/wt mice and the number of glomeruli with renin mRNA-positive cells increased (Fig. 5, A–C). However, there was no increase in renin mRNA levels in Ren1dCre/Sox6fl/fl mice as well as Sox6 and renin double-positive cells (Fig. 5, A–C). Likewise, to demonstrate the colocalization of Sox6 and renin mRNAs as well as the JG cell recruitment along the afferent arteriole, specific fluorescent probes were used to detect Sox6 and renin mRNAs. After LowNa/Fu, the number of renin-expressing cells along the afferent arteriole was higher in Ren1dCre/Sox6wt/wt mice (Fig. 5, D–F) compared with Ren1dCre/Sox6fl/fl mice. There was coexpression of renin and Sox6 mRNAs in cells along the afferent arteriole (Fig. 5E). Notably, specific ablation of Sox6 in renin-expressing cells in Ren1dCre/Sox6fl/fl mice significantly decreased the recruitment of cells to express renin along the afferent arteriole (Fig. 5, E and F). These results indicate that Sox6 plays a role in renin expression control and in the recruitment of cells along the afferent arteriole to express renin during JG cell recruitment, influencing both mRNA and protein levels.
Fig. 5.
Specific knockout of Sox6 in renin-expressing cells inhibits juxtaglomerular (JG) cell recruitment during low-Na+ and furosemide (LowNa/Fu) treatment. After 10 days of LowNa/Fu treatment, kidneys were isolated, perfusion fixed with 10% neutral buffered formalin solution, dehydrated in a graduated ethanol series, and embedded in paraffin. Renin (red) mRNA amplification was performed following the manufacturer’s instructions. Red staining represents renin mRNA expression. Specific knockout of Sox6 in JG cells inhibited the increase in renin mRNA expression in cells along the afferent arteriole during JG cell recruitment. A: control untreated and LowNa/Fu-treated Ren1dCre/Sox6wt/wt mice. B: control untreated and LowNa/Fu-treated Ren1dCre/Sox6fl/fl mice. Representative microscopy images are shown. G, glomerulus; AfAr, afferent arteriole. Magnification: ×60. Scale bars = 20 μm. C: quantification of at least 300 glomeruli/sample expressing renin mRNA along the afferent arteriole from the in situ hybridization experiments. Specific probes for renin (red) were used to detect renin mRNA. Data are presented as means ± SE; n = 4. P values were was calculated with one-way ANOVA followed by a Tukey post hoc test. **P < 0.01; ****P < 0.0001. Specific fluorescent probes for renin (opal 520, green) and Sox6 (opal 650, magenta) were used to detect renin and Sox6 mRNA, respectively. DAPI was used as a nuclear stain. D: quantification of JG cell recruitment along the afferent arteriole in at least 300 glomeruli/sample expressing renin and Sox6 mRNA from the in situ hybridization experiments. Specific probes for renin (green) and Sox6 (magenta) were used to detect renin and Sox6 mRNAs. Data are presented as means ± SE; n = 4. P values were was calculated with one-way ANOVA followed by a Tukey post hoc test. ****P < 0.0001. E: control untreated and LowNa/Fu-treated Ren1dCre/Sox6wt/wt mice. F: control untreated and LowNa/Fu-treated Ren1dCre/Sox6fl/fl mice. Representative microscopy images are shown. Magnification: ×30. Scale bars = 50 μm.
Knockout of Sox6 in renin-expressing cells prevents the increase in renin expression during JG cell recruitment.
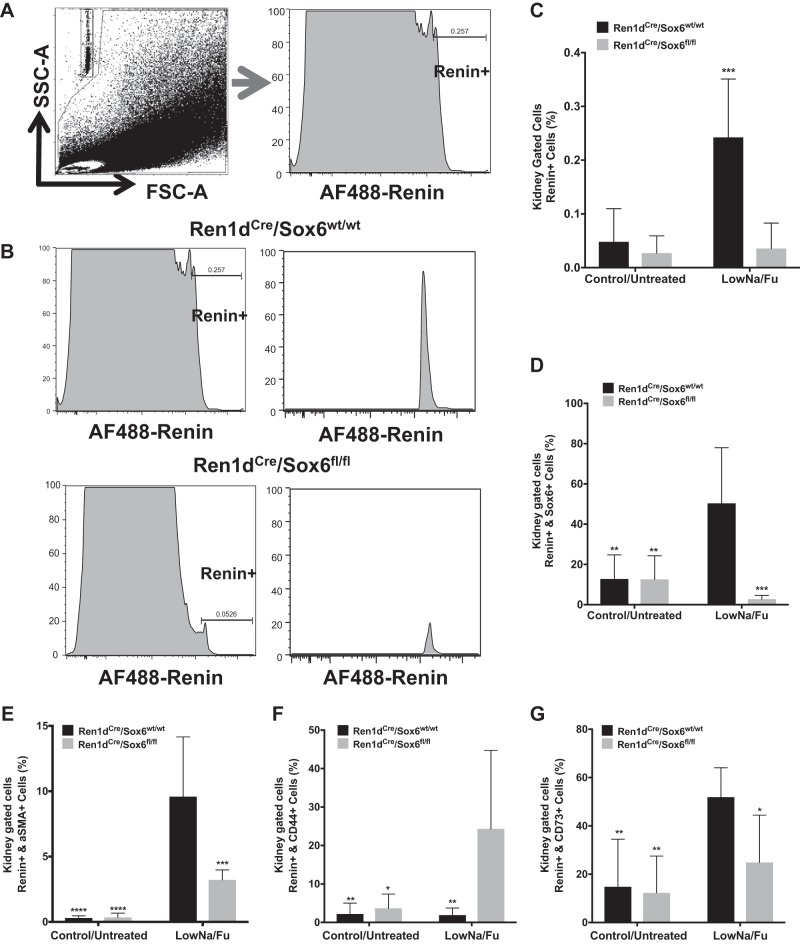
Next, we measured the number of renin-expressing cells in vivo after LowNa/Fu treatment using flow cytometry. Compared with control mice, Ren1dCre/Sox6fl/fl mice failed to increase the number of renin-expressing cells in response to LowNa/Fu (Fig. 6, A–G). Expression of Sox6 was upregulated after changes in renin expression induced by LowNa/Fu treatment (Fig. 3), and specific ablation of Sox6 in renin-expressing cells inhibited the increase in renin-producing cells during JG cell expansion and recruitment (Fig. 6, B and C) as well as the number of Sox6+ and renin+ double-positive cells (Fig. 6D). Furthermore, our results showed that Sox6 is important for renin expression by SMCs during JG cell recruitment (Fig. 6E). Specific ablation of Sox6 in renin-expressing cells inhibited the recruitment of SMCs (renin+ and α-SMA+ cells) along the afferent arteriole normally seen after LowNa/Fu treatment (Fig. 6E). CD44 and CD73 were used as markers of renal MSCs or renal stem progenitor cells. We found that there was an increase in the number of renin+ and CD44+ double-positive cells and a decrease in the number of renin+ and CD73+ cells in mice lacking Sox6 in renin-expressing cells (Fig. 6, F and G). This suggests that ablation of Sox6 in vivo decreases the differentiation of MSCs to renin-expressing cells and may affect the maintenance of the progenitor phenotype after differentiation to renin-expressing cells (Fig. 6, F and G). Taken together, the results presented here allow us to conclude that Sox6 has a new function in renin expression control.
Fig. 6.
Knockout of Sox6 in juxtaglomerular (JG) cells inhibits the increase in renin-expressing cells during JG cell expansion. Ren1dCre/Sox6fl/fl and Ren1dCre/Sox6wt/wt mice (6–8 wk old) were administered a low-Na+ diet (0.02% NaCl) plus furosemide (LowNa/Fu) (drinking water: 2.28 mmol/L) for 10 days. Flow cytometry analysis of isolated mouse kidney cells used specific antibodies to renin and Sox6 to quantify renal cells expressing these proteins. A, left: representative flow cytometry dot plots of kidneys from Ren1dCre/Sox6fl/fl and Ren1dCre/Sox6wt/wt mice showing the gating strategy. A, right: representative flow cytometry histograms of kidneys from Ren1dCre/Sox6fl/fl and Ren1dCre/Sox6wt/wt mice showing gating for renin-expressing cells [Alexa Fluor (AF)488-renin+]. B: histograms of renin-expressing cells in kidneys from Ren1dCre/Sox6wt/wt (top) and Ren1dCre/Sox6fl/fl (bottom) mice after LowNa/Fu treatment. C−G: bar graphs show kidney cells positive for the expression of renin (C), renin and Sox6 double-positive cells (D), renin and α-smooth muscle actin (aSMA) double-positive cells (E), renin and CD44 double-positive cells (F), and renin and CD73 double-positive cells (G). Data are presented as means ± SE; n = 6–7. P values were calculated with an unpaired t test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
DISCUSSION
Renin is the rate-limiting enzyme in the RAAS and thus plays a pivotal role. The RAAS activity is essential for water and electrolyte balance and blood pressure control. The RAAS exerts its blood pressure control through plasma circulating renin. Although renin is produced in different tissues, the predominant site of production is the kidney, where it is produced and stored by JG cells (1). Control of renin expression and secretion is of great importance for human health. Renin expression and secretion are carefully coordinated by several proteins that work in a harmonized manner maintaining blood pressure and fluid homeostasis.
In the present study, we demonstrated that Sox6 has a new function in the control of renin expression. Under physiological conditions, JG cells arise from several cellular sources. Previously, we reported that resident renal MSCs play a role in JG cell recruitment and that a subset of these MSCs differentiates into renin-expressing cells (43). Other studies have stressed the importance of arteriolar SMCs in the afferent arteriole and after LowNa/Fu or captopril, SMCs along the afferent arteriole are recruited to produce renin (13, 31, 39). In our study, we show that not only does Sox6 colocalize with renin, but it also colocalizes with markers of both renal MSCs and SMCs after LowNa/Fu treatment, which increases renin expression. For example, colocalization of Sox6 and renin mRNAs from in situ hybridization and the flow cytometry quantification of renin+, Sox6+, α-SMA+ SMCs showed that Sox6-specific ablation in renin-expressing cells inhibits the increase in renin+ and α-SMA+ cells during JG cell recruitment. Although JG cells are the major producers of renin in the adult kidney, cells in the CD and CNT can express renin with a minor contribution as well. There were no renin-positive cells in CD or CNT cells after LowNa/Fu in mice with specific ablation of Sox6 in renin-expressing cells. Moreover, in vitro, we found that Sox6 controls the expression of renin in MSCs after cAMP stimulus. Collectively, in vitro and in vivo studies have supported our hypothesis that Sox6 contributes in the regulation of renin expression in the kidney.
Genecentric array and genome-wide association studies have identified an association between hypertension in various ethnic groups and single-nucleotide polymorphisms (SNPs) in the Sox6 gene (11, 12, 23, 29). It is currently unknown how these SNPs affect Sox6 function. In light of our study, it is possible that these SNPs affect the ability of Sox6 to control renin expression, dysregulating the RAAS. Further studies in our laboratory will define the role of Sox6 SNPs in renin expression and hypertension.
The second messenger cAMP is important for renin expression. The renin promoter contains cAMP-responsive elements in the proximal and distal promoters in which cAMP-responsive transcription factors bind promoting renin expression (5). In silico analysis of the renin promoter has shown that there is a Sox6-binding site in the proximal promoter and not within any of the cAMP-responsive elements. However, Sox6 expression itself is stimulated by cAMP, and its promoter contains binding sites for cAMP-responsive transcription factors, such as activating transcription factor (ATF)1, ATF2, and ATF7, which may bind and stimulate Sox6 expression. Future experiments defining the role of cAMP in Sox6 expression and its relationship to renin expression will be of interest. In vitro, downregulation of Sox6 in renal MSCs inhibited the expression of renin induced by cAMP in these cells, showing that Sox6 is necessary for the cAMP-stimulated differentiation of renal MSCs into renin-expressing cells. In vivo, prorenin and renin protein expression as well as renin mRNA expression increased in wild-type mice after 10 days of LowNa/Fu. Sox6-specific ablation in renin-expressing cells inhibited the increase in prorenin and renin protein and renin mRNA expressions observed in wild-type mice during JG cell recruitment.
Here, we describe the novel function of the transcription factor Sox6 in renin expression using a new transgenic mouse in which Sox6 is ablated only in renin-expressing cells. Several transcription factors bind the renin promoter and control its expression [for reviews, see Castrop et al. (5), Sigmund and Gross (41), and Lopez and Gomez (28)]. The Cre-LoxP system was used to identify RBP-J as a transcriptional regulator important for renin cell phenotype and plasticity (4); in this animal, blood pressure at baseline is lower and after LowNa/Fu or captopril, there is no recruitment of SMCs along the afferent arteriole to produce renin. Here, we used the same system to specifically knock out Sox6 in renin-expressing cells (Ren1dCre/Sox6fl/fl mice). At baseline, blood pressure is similar in both wild-type and Ren1dCre/Sox6fl/fl mice. However, ablation of Sox6 in renin-expressing cells halts the increase in prorenin and renin expression after LowNa/Fu treatment as well as the recruitment of SMCs along the afferent arteriole, indicating that Sox6 has a new function in the control of renin expression. This indicates that Sox6 ablation impairs the reexpression and recruitment of SMCs along the afferent arteriole to express renin.
Taken together, our findings indicate that Sox6 has a previously undefined role in modulating renin expression in response to Na+ and volume deprivation. Given this critical function, Sox6 might also be a therapeutic target for the treatment of hypertension.
GRANTS
This work was supported by American Heart Association Scientist Development Award 16SDG29880007 (to J. A. Gomez), the Vanderbilt University Medical Center Faculty Research Scholars Program (to J. A. Gomez), and National Heart, Lung, and Blood Institute Research Scientist Development Grant 1-K01-HL-135461-01 (to J. A. Gomez).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
S.M. and J.A.G. conceived and designed research; M.S., C.P.H., L.X., J.A.G.-B., M.L.R., J.F., A.J.P., M.M., and J.A.G. performed experiments; M.S., C.P.H., L.X., J.A.G.-B., M.L.R., J.F., A.J.P., M.M., and J.A.G. analyzed data; A.J.P., M.M., V.G., V.J.D., and J.A.G. interpreted results of experiments; S.M. and J.A.G. prepared figures; M.S., C.P.H., and J.A.G. drafted manuscript; J.A.G. edited and revised manuscript; J.A.G. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. R. Ariel Gomez for kindly providing the Ren1dCre mice, Dr. Monique Lefebvre (Cleveland Clinic) for generously providing the Sox6fl/fl transgenic mice, Ela Contreras for technical support with Fig. 5, A–C. We thank Dr. David G. Harrison for comments on the manuscript.
REFERENCES
- 1.Anderson LM, Choe SE, Yukhananov RY, Hopfner RL, Church GM, Pratt RE, Dzau VJ. Identification of a novel set of genes regulated by a unique liver X receptor-alpha-mediated transcription mechanism. J Biol Chem 278: 15252–15260, 2003. doi: 10.1074/jbc.M208644200. [DOI] [PubMed] [Google Scholar]
- 2.Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci 12: 1238–1247, 2009. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg AC, Chernavvsky-Sequeira C, Lindsey J, Gomez RA, Sequeira-Lopez ML. Pericytes synthesize renin. World J Nephrol 2: 11–16, 2013. doi: 10.5527/wjn.v2.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellanos Rivera RM, Monteagudo MC, Pentz ES, Glenn ST, Gross KW, Carretero O, Sequeira-Lopez ML, Gomez RA. Transcriptional regulator RBP-J regulates the number and plasticity of renin cells. Physiol Genomics 43: 1021–1028, 2011. doi: 10.1152/physiolgenomics.00061.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castrop H, Höcherl K, Kurtz A, Schweda F, Todorov V, Wagner C. Physiology of kidney renin. Physiol Rev 90: 607–673, 2010. doi: 10.1152/physrev.00011.2009. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Lasaitiene D, Friberg P. The renin-angiotensin system in kidney development. Acta Physiol Scand 181: 529–535, 2004. doi: 10.1111/j.1365-201X.2004.01327.x. [DOI] [PubMed] [Google Scholar]
- 7.Cohen-Barak O, Hagiwara N, Arlt MF, Horton JP, Brilliant MH. Cloning, characterization and chromosome mapping of the human SOX6 gene. Gene 265: 157–164, 2001. doi: 10.1016/S0378-1119(01)00346-8. [DOI] [PubMed] [Google Scholar]
- 8.Crowley SD, Coffman TM. Recent advances involving the renin-angiotensin system. Exp Cell Res 318: 1049–1056, 2012. doi: 10.1016/j.yexcr.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowley SD, Gurley SB, Herrera MJ, Ruiz P, Griffiths R, Kumar AP, Kim HS, Smithies O, Le TH, Coffman TM. Angiotensin II causes hypertension and cardiac hypertrophy through its receptors in the kidney. Proc Natl Acad Sci USA 103: 17985–17990, 2006. doi: 10.1073/pnas.0605545103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumitriu B, Dy P, Smits P, Lefebvre V. Generation of mice harboring a Sox6 conditional null allele. Genesis 44: 219–224, 2006. doi: 10.1002/dvg.20210. [DOI] [PubMed] [Google Scholar]
- 11.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, Tayo BO, Sun YV, Gottesman O, Adeyemo A, Johnson AD, Young JH, Rice K, Duan Q, Chen F, Li Y, Tang H, Fornage M, Keene KL, Andrews JS, Smith JA, Faul JD, Guangfa Z, Guo W, Liu Y, Murray SS, Musani SK, Srinivasan S, Velez Edwards DR, Wang H, Becker LC, Bovet P, Bochud M, Broeckel U, Burnier M, Carty C, Chasman DI, Ehret G, Chen WM, Chen G, Chen W, Ding J, Dreisbach AW, Evans MK, Guo X, Garcia ME, Jensen R, Keller MF, Lettre G, Lotay V, Martin LW, Moore JH, Morrison AC, Mosley TH, Ogunniyi A, Palmas W, Papanicolaou G, Penman A, Polak JF, Ridker PM, Salako B, Singleton AB, Shriner D, Taylor KD, Vasan R, Wiggins K, Williams SM, Yanek LR, Zhao W, Zonderman AB, Becker DM, Berenson G, Boerwinkle E, Bottinger E, Cushman M, Eaton C, Nyberg F, Heiss G, Hirschhron JN, Howard VJ, Karczewsk KJ, Lanktree MB, Liu K, Liu Y, Loos R, Margolis K, Snyder M, Psaty BM, Schork NJ, Weir DR, Rotimi CN, Sale MM, Harris T, Kardia SL, Hunt SC, Arnett D, Redline S, Cooper RS, Risch NJ, Rao DC, Rotter JI, Chakravarti A, Reiner AP, Levy D, Keating BJ, Zhu X, Go MJ, Kim YJ, Lee J-Y, Jeon J-P, Kim SS, Han B-G, Cho YS, Sim X, Tay WT, Ong RTH, Seielstad M, Liu JJ, Aung T, Wong TY, Teo YY, Tai ES, Chen C-H, Chang L, Chen Y-T, Wu J-Y, Kelly TN, Gu D, Hixson JE, Sung YJ, He J, Tabara Y, Kokubo Y, Miki T, Iwai N, Kato N, Takeuchi F, Katsuya T, Nabika T, Sugiyama T, Zhang Y, Huang W, Zhang X, Zhou X, Jin L, Zhu D; Asian Genetic Epidemiology Network Consortium . Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. Am J Hum Genet 93: 545–554, 2013. doi: 10.1016/j.ajhg.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganesh SK, Tragante V, Guo W, Guo Y, Lanktree MB, Smith EN, Johnson T, Castillo BA, Barnard J, Baumert J, Chang YP, Elbers CC, Farrall M, Fischer ME, Franceschini N, Gaunt TR, Gho JM, Gieger C, Gong Y, Isaacs A, Kleber ME, Mateo Leach I, McDonough CW, Meijs MF, Mellander O, Molony CM, Nolte IM, Padmanabhan S, Price TS, Rajagopalan R, Shaffer J, Shah S, Shen H, Soranzo N, van der Most PJ, Van Iperen EP, Van Setten J, Vonk JM, Zhang L, Beitelshees AL, Berenson GS, Bhatt DL, Boer JM, Boerwinkle E, Burkley B, Burt A, Chakravarti A, Chen W, Cooper-Dehoff RM, Curtis SP, Dreisbach A, Duggan D, Ehret GB, Fabsitz RR, Fornage M, Fox E, Furlong CE, Gansevoort RT, Hofker MH, Hovingh GK, Kirkland SA, Kottke-Marchant K, Kutlar A, Lacroix AZ, Langaee TY, Li YR, Lin H, Liu K, Maiwald S, Malik R, Murugesan G, Newton-Cheh C, O’Connell JR, Onland-Moret NC, Ouwehand WH, Palmas W, Penninx BW, Pepine CJ, Pettinger M, Polak JF, Ramachandran VS, Ranchalis J, Redline S, Ridker PM, Rose LM, Scharnag H, Schork NJ, Shimbo D, Shuldiner AR, Srinivasan SR, Stolk RP, Taylor HA, Thorand B, Trip MD, van Duijn CM, Verschuren WM, Wijmenga C, Winkelmann BR, Wyatt S, Young JH, Boehm BO, Caulfield MJ, Chasman DI, Davidson KW, Doevendans PA, Fitzgerald GA, Gums JG, Hakonarson H, Hillege HL, Illig T, Jarvik GP, Johnson JA, Kastelein JJ, Koenig W, März W, Mitchell BD, Murray SS, Oldehinkel AJ, Rader DJ, Reilly MP, Reiner AP, Schadt EE, Silverstein RL, Snieder H, Stanton AV, Uitterlinden AG, van der Harst P, van der Schouw YT, Samani NJ, Johnson AD, Munroe PB, de Bakker PI, Zhu X, Levy D, Keating BJ, Asselbergs FW CARDIOGRAM, METASTROKE; LifeLines Cohort Study . Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet 22: 1663–1678, 2013. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez RA, Chevalier RL, Everett AD, Elwood JP, Peach MJ, Lynch KR, Carey RM. Recruitment of renin gene-expressing cells in adult rat kidneys. Am J Physiol 259: F660–F665, 1990. doi: 10.1152/ajprenal.1990.259.4.F660. [DOI] [PubMed] [Google Scholar]
- 14.Gomez RA, Pentz ES, Jin X, Cordaillat M, Sequeira Lopez ML. CBP and p300 are essential for renin cell identity and morphological integrity of the kidney. Am J Physiol Heart Circ Physiol 296: H1255–H1262, 2009. doi: 10.1152/ajpheart.01266.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez RA, Sequeira-Lopez MLS. Renin cells in homeostasis, regeneration and immune defence mechanisms. Nat Rev Nephrol 14: 231–245, 2018. doi: 10.1038/nrneph.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez RA, Sequeira Lopez ML. Who and where is the renal baroreceptor?: the connexin hypothesis. Kidney Int 75: 460–462, 2009. doi: 10.1038/ki.2008.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez AA, Zamora L, Reyes-Martinez C, Salinas-Parra N, Roldan N, Cuevas CA, Figueroa S, Gonzalez-Vergara A, Prieto MC. (Pro)renin receptor activation increases profibrotic markers and fibroblast-like phenotype through MAPK-dependent ROS formation in mouse renal collecting duct cells. Clin Exp Pharmacol Physiol 44: 1134–1144, 2017. doi: 10.1111/1440-1681.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granger JP, Schnackenberg CG. Renal mechanisms of angiotensin II-induced hypertension. Semin Nephrol 20: 417–425, 2000. [PubMed] [Google Scholar]
- 19.Hagiwara N. Sox6, jack of all trades: a versatile regulatory protein in vertebrate development. Dev Dyn 240: 1311–1321, 2011. doi: 10.1002/dvdy.22639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall JE. Control of blood pressure by the renin-angiotensin-aldosterone system. Clin Cardiol 14, Suppl 4: 6–IV21, 1991. doi: 10.1002/clc.4960141802. [DOI] [PubMed] [Google Scholar]
- 21.House MA. Thrombolytic therapy for acute myocardial infarction: the elderly population. AACN Clin Issues Crit Care Nurs 3: 106–113, 1992. doi: 10.4037/15597768-1992-1013. [DOI] [PubMed] [Google Scholar]
- 22.Humphreys BD, Lin SL, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. Fate tracing reveals the pericyte and not epithelial origin of myofibroblasts in kidney fibrosis. Am J Pathol 176: 85–97, 2010. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O’Brien ET, Poulter NR, Sever P, Shields DC, Thom S, Wannamethee SG, Whincup PH, Brown MJ, Connell JM, Dobson RJ, Howard PJ, Mein CA, Onipinla A, Shaw-Hawkins S, Zhang Y, Davey Smith G, Day IN, Lawlor DA, Goodall AH, Fowkes FG, Abecasis GR, Elliott P, Gateva V, Braund PS, Burton PR, Nelson CP, Tobin MD, van der Harst P, Glorioso N, Neuvrith H, Salvi E, Staessen JA, Stucchi A, Devos N, Jeunemaitre X, Plouin PF, Tichet J, Juhanson P, Org E, Putku M, Sõber S, Veldre G, Viigimaa M, Levinsson A, Rosengren A, Thelle DS, Hastie CE, Hedner T, Lee WK, Melander O, Wahlstrand B, Hardy R, Wong A, Cooper JA, Palmen J, Chen L, Stewart AF, Wells GA, Westra HJ, Wolfs MG, Clarke R, Franzosi MG, Goel A, Hamsten A, Lathrop M, Peden JF, Seedorf U, Watkins H, Ouwehand WH, Sambrook J, Stephens J, Casas JP, Drenos F, Holmes MV, Kivimaki M, Shah S, Shah T, Talmud PJ, Whittaker J, Wallace C, Delles C, Laan M, Kuh D, Humphries SE, Nyberg F, Cusi D, Roberts R, Newton-Cheh C, Franke L, Stanton AV, Dominiczak AF, Farrall M, Hingorani AD, Samani NJ, Caulfield MJ, Munroe PB. Cardiogenics Consortium; Global BPgen Consortium . Blood pressure loci identified with a gene-centric array. Am J Hum Genet 89: 688–700, 2011. doi: 10.1016/j.ajhg.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones CA, Sigmund CD, McGowan RA, Kane-Haas CM, Gross KW. Expression of murine renin genes during fetal development. Mol Endocrinol 4: 375–383, 1990. doi: 10.1210/mend-4-3-375. [DOI] [PubMed] [Google Scholar]
- 25.Kurtz A. Control of renin synthesis and secretion. Am J Hypertens 25: 839–847, 2012. doi: 10.1038/ajh.2011.246. [DOI] [PubMed] [Google Scholar]
- 26.Lefebvre V. The SoxD transcription factors–Sox5, Sox6, and Sox13–are key cell fate modulators. Int J Biochem Cell Biol 42: 429–432, 2010. doi: 10.1016/j.biocel.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin EE, Sequeira-Lopez ML, Gomez RA. RBP-J in FOXD1+ renal stromal progenitors is crucial for the proper development and assembly of the kidney vasculature and glomerular mesangial cells. Am J Physiol Renal Physiol 306: F249–F258, 2014. doi: 10.1152/ajprenal.00313.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez ML, Gomez RA. The renin phenotype: roles and regulation in the kidney. Curr Opin Nephrol Hypertens 19: 366–371, 2010. doi: 10.1097/MNH.0b013e32833aff32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Wang L, Lin X, Huang J, Charles Gu C, He M, Shen H, He J, Zhu J, Li H, Hixson JE, Wu T, Dai J, Lu L, Shen C, Chen S, He L, Mo Z, Hao Y, Mo X, Yang X, Li J, Cao J, Chen J, Fan Z, Li Y, Zhao L, Li H, Lu F, Yao C, Yu L, Xu L, Mu J, Wu X, Deng Y, Hu D, Zhang W, Ji X, Guo D, Guo Z, Zhou Z, Yang Z, Wang R, Yang J, Zhou X, Yan W, Sun N, Gao P, Gu D. Genome-wide association study in Chinese identifies novel loci for blood pressure and hypertension. Hum Mol Genet 24: 865–874, 2015. doi: 10.1093/hmg/ddu478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lum C, Shesely EG, Potter DL, Beierwaltes WH. Cardiovascular and renal phenotype in mice with one or two renin genes. Hypertension 43: 79–86, 2004. doi: 10.1161/01.HYP.0000107401.72456.50. [DOI] [PubMed] [Google Scholar]
- 31.Martinez MF, Medrano S, Brown EA, Tufan T, Shang S, Bertoncello N, Guessoum O, Adli M, Belyea BC, Sequeira-Lopez MLS, Gomez RA. Super-enhancers maintain renin-expressing cell identity and memory to preserve multi-system homeostasis. J Clin Invest 128: 4787–4803, 2018. doi: 10.1172/JCI121361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neubauer B, Machura K, Chen M, Weinstein LS, Oppermann M, Sequeira-Lopez ML, Gomez RA, Schnermann J, Castrop H, Kurtz A, Wagner C. Development of vascular renin expression in the kidney critically depends on the cyclic AMP pathway. Am J Physiol Renal Physiol 296: F1006–F1012, 2009. doi: 10.1152/ajprenal.90448.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pentz ES, Lopez ML, Cordaillat M, Gomez RA. Identity of the renin cell is mediated by cAMP and chromatin remodeling: an in vitro model for studying cell recruitment and plasticity. Am J Physiol Heart Circ Physiol 294: H699–H707, 2008. doi: 10.1152/ajpheart.01152.2007. [DOI] [PubMed] [Google Scholar]
- 34.Raman VK, Lee YA, Lindpaintner K. The cardiac renin-angiotensin-aldosterone system and hypertensive cardiac hypertrophy. Am J Cardiol 76: 18D–23D, 1995. doi: 10.1016/S0002-9149(99)80487-1. [DOI] [PubMed] [Google Scholar]
- 35.Saleem M, Pokkunuri I, Asghar M. Superoxide increases angiotensin II AT1 receptor function in human kidney-2 cells. FEBS Open Bio 6: 1273–1284, 2016. doi: 10.1002/2211-5463.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saleem M, Wang X, Pokkunuri I, Asghar M. Superoxide via Sp3 mechanism increases renal renin activity, renal AT1 receptor function, and blood pressure in rats. Am J Physiol Renal Physiol 315: F1478–F1483, 2018. doi: 10.1152/ajprenal.00194.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salinas-Parra N, Reyes-Martínez C, Prieto MC, Gonzalez AA. Prostaglandin E2 induces prorenin-dependent activation of (pro)renin receptor and upregulation of cyclooxygenase-2 in collecting duct cells. Am J Med Sci 354: 310–318, 2017. doi: 10.1016/j.amjms.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sequeira Lopez ML, Gomez RA. Development of the renal arterioles. J Am Soc Nephrol 22: 2156–2165, 2011. doi: 10.1681/ASN.2011080818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sequeira López ML, Pentz ES, Nomasa T, Smithies O, Gomez RA. Renin cells are precursors for multiple cell types that switch to the renin phenotype when homeostasis is threatened. Dev Cell 6: 719–728, 2004. doi: 10.1016/S1534-5807(04)00134-0. [DOI] [PubMed] [Google Scholar]
- 40.Sequeira Lopez ML, Pentz ES, Robert B, Abrahamson DR, Gomez RA. Embryonic origin and lineage of juxtaglomerular cells. Am J Physiol Renal Physiol 281: F345–F356, 2001. doi: 10.1152/ajprenal.2001.281.2.F345. [DOI] [PubMed] [Google Scholar]
- 41.Sigmund CD, Gross KW. Structure, expression, and regulation of the murine renin genes. Hypertension 18: 446–457, 1991. doi: 10.1161/01.HYP.18.4.446. [DOI] [PubMed] [Google Scholar]
- 42.Sparks MA, Crowley SD, Gurley SB, Mirotsou M, Coffman TM. Classical renin-angiotensin system in kidney physiology. Compr Physiol 4: 1201–1228, 2014. doi: 10.1002/cphy.c130040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Gomez JA, Klein S, Zhang Z, Seidler B, Yang Y, Schmeckpeper J, Zhang L, Muramoto GG, Chute J, Pratt RE, Saur D, Mirotsou M, Dzau VJ. Adult renal mesenchymal stem cell-like cells contribute to juxtaglomerular cell recruitment. J Am Soc Nephrol 24: 1263–1273, 2013. doi: 10.1681/ASN.2012060596. [DOI] [PMC free article] [PubMed] [Google Scholar]