Abstract
Olfaction, the sense of smell detects and discriminate odors as well as social cues which influence our innate re-sponses. The olfactory system in human beings is found to be weak as compared to other animals; however, it seems to be very precise. It can detect and discriminate millions of chemical moieties (odorants) even in minuscule quantities. The pro-cess initiates with the binding of odorants to specialized olfactory receptors, encoded by a large family of Olfactory Receptor (OR) genes belonging to the G-protein-coupled receptor superfamily. Stimulation of ORs converts the chemical information encoded in the odorants, into respective neuronal action-potentials which causes depolarization of olfactory sensory neurons. The olfactory bulb relays this signal to different parts of the brain for processing. Odors are encrypted using a combinatorial approach to detect a variety of chemicals and encode their unique identity. The discovery of functional OR genes and pro-teins provided an important information to decipher the genomic, structural and functional basis of olfaction. ORs constitute 17 gene families, out of which 4 families were reported to contain more than hundred members each. The olfactory machin-ery is not limited to GPCRs; a number of non-GPCRs is also employed to detect chemosensory stimuli. The article provides detailed information about such olfaction machinery, structures, transduction mechanism, theories of odor perception, and challenges in the olfaction research. It covers the structural, functional and computational studies carried out in the olfaction research in the recent past.
Keywords: GPCR, olfactory bulb, odor, receptor, signal transduction
1. INTRODUCTION
The sensory systems sense the information in our surroundings and transfer it to the specific parts of the brain for processing, by stimulating a series of reactions which causes a neuron to fire an electric signal. Among all the senses possessed by humans (approximately 25), sense of smell/ olfaction is the most primeval and volatile sense [1]. It detects, encodes and discriminates among thousands of small airborne chemicals (odorants); which are volatile, lighter, largely hydrophobic organic compounds possessing varied chemical structure and properties, often at very small concentrations (few parts per trillion) [2-4]. However, olfaction is vital not just to identify and discriminate odorants, it adds an emotional attribute to the objects or events, influences our mood and thoughts, acts as a catalyst in social interactions (modulates behavior and interpersonal relationships) and has played a significant role in the evolution of human habitats [5]. Among all species, the olfactory system has evolved in
response to the two problems; to detect and discriminate between the array of chemical compounds and to the unique sensory challenges faced by them. Different species possess different sets of olfactory receptors, governed by two processes: an evolutionary version of birth and death, and a relaxed selection process [6]. In humans, olfactory genes are located in regions of a chromosome which are prone to copying mistakes. The OR gene undergoes duplication, wherein one of the two identical genes may mutate. The mutation can be fatal, turning a functional gene into pseudogene or non-fatal i.e. continue making the same OR with slight changes in its molecular structure. The change in the structure of OR alters its grip on odorants and hence brings about a delicate shift in the perception of odors and behaviors driven by odorants. The subtle adaptations in the lifestyle, habitat and behavior correspond to increased fine-scale growth and death of OR genes. The olfactory system takes advantage of these changes in OR repertoire to generate distinct and learned olfactory behaviors [6]. As a result, humans possess quite a diverse olfactory repertoire of genes, within and between populations, which relates to a variation in cross-cultural preferences and perception of odors.
The sense of smell works standalone as well in conjunction with the sense of taste. When we chew and swallow food, organic compounds make an access to the olfactory epithelium from the nostrils as well from the mouth. If the sense of smell is knocked out (during cold), one relies only on the sense of taste, hence eatables are not experienced as rich as they should. The smell does not seem to be satisfactorily ‘real’ as it does not exist without a perceiving material. The odors are too brief in their appearances, insubstantial, have a complex molecular basis and are perceived individually.
Earlier in comparison to other senses, sense of smell was considered as unimportant, even dismissed as the most ‘ungrateful’ sense and neglected for the research pursuits. The interest in how humans perceive smell is fairly recent and is established by two important scientific endeavors; discovery of olfactory-receptor (OR) superfamily of proteins by Buck and Axel [7] and Human Genome Project [8-10], which shifted the focus to research on olfactory genes, proteins, transduction mechanism and development of olfactory repertories [8, 11-15]. However, because of its emotive and deceptive nature, yet there is no appropriate scientific justification of how smell is actually perceived, especially in humans. The phenomena involve various mechanisms and answer to the question of how we actually perceive odors must be sought at different levels.
2. THE NOSE: NATURAL CHEMO-DETECTOR
The nose continuously monitors the dynamic chemical composition of the environment and perceives it as distinct aromas Fig. (1). With a sniff, airborne, volatile chemicals are inhaled into the nasal cavity by the respiratory airflow. In the rooftop of the two nasal cavities, located in between the eyes, there is a region known as nasal/ olfactory epithelium (OE), i.e. primary interaction site, which leads to the succeeding signal transduction and processing of signals at the neurological level. The area of OE of each nasal passage is about 2.5 square centimeter, which contains approximately 50 million distinct olfactory sensory cells/ neurons (OSNs) arranged in structurally/anatomically and functionally divergent order [2, 16]. Olfactory cells are focused, and neurons turnover approximately every 40 days. Separating an OE from the brain is a piece of bone known as a cribriform plate. Sitting right above the cribriform plate is an extension from the brain, which looks like a bulb known as the olfactory bulb (OB). The cribriform plate is a bone with several holes in it, through which the sensory cells send their projection into the OB, which is basically a bundle of nerves (cranial nerves) that exit the brain. This bundle of nerves sends little projections through the cribriform plate into the OE where they differentiate. Likewise, there are thousands of different types of cells (OSNs) embedded among the rest of the epithelial cells, sending little connections (dendrites) into the OE. The OSNs are bipolar in nature, projecting an axon to the OB and extending several dendrites in the epithelium. The apical dendrite contains 5-10 immotile cilia, regarded as the actual chemosensory structures, embedded in the nasal mucus of about 60 microns thickness. Olfactory cilia provide sites of primary olfactory processes i.e. the large surface area where odorous molecules can interact. The membrane of cilia contains olfactory receptor proteins (ORs) i.e. the elements of the olfaction transduction machinery [2, 16-18]. Each type of OSNs has a particular olfactory receptor (OR) which binds to a specific odorous molecule. The interaction of odorants with the ORs triggers a cascade of signaling events which causes the cell to fire, i.e. increases the membrane conductance due to change in membrane potential. The generated potential is converted into action potential of distinct frequency and passed on to the OB. The whole bunch of OSNs, sensitive to a particular odorous molecule, fires an action potential to a particular location in the OB known as glomerulus, an anatomical structure primarily consisting of neutrophil. The glomerulus acts as a destination point for various OSNs (sensitive to the same molecule). Each OSN expressing the same ORs converges meticulously onto the same glomerulus; generally, there are two glomeruli situated at the lateral and the medial hemisphere of OB thereby maintaining a spatial order. This odorant-specific particular axonal convergence pattern forms the anatomical basis of olfactory sensory map [19, 20]. In the glomerulus, OSNs synapse onto other cells are known as mitral or tufted cells, which further send neuronal signals to the primary olfactory cortex. For every odorant, a specific pattern of neuronal signals is generated comprising of the strength of the signal, time period, and quality of odorant stimuli. Each odorant produces a unique pattern of neuronal signals as it stimulates only a certain specific population of OSNs and each cell/ neuron responds differently to different odors [2]. From primary olfactory cortex, neuronal signals are sent to the higher cortical areas and the limbic system. The higher cortical area allows the conscious perception of odors and the limbic system governs analogous emotions, memory storage, behavioral and sensational effects.
Fig. (1).
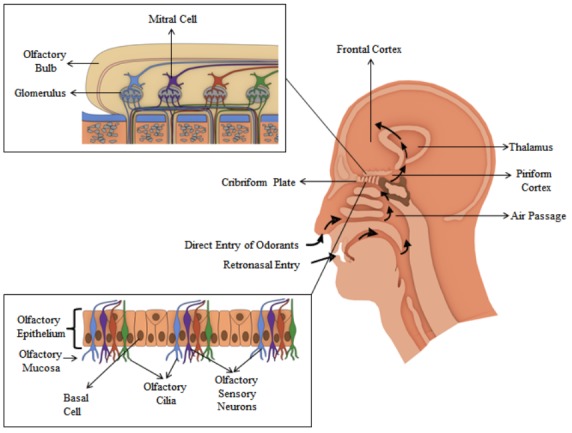
The organization of various olfactory sensory neurons (OSNs) in OE. There are hundreds of OSNs that are sensitive to a particular odorous molecule and all these hundreds of OSNs send their projection to one glomerulus. Every glomerulus has several mitral and tufted cells that send synapse to the brain. Odor molecule binds and activates specific ORs. The activation, possibly causes the structural changes in OR’s, triggering a signaling cascade wherein the chemical signal is converted into electrical signals which generate an olfactory response. (The color version of the figure is available in the electronic copy of the article).
Thus, the answer to how humans perceive odors lies in another query i.e. how the brain interprets neuronal activity patterns generated due to precise interaction of odorants with distinct OSNs.
3. THE HUMAN VOMERONASAL ORGAN
The vomeronasal organ (VNO) is an accessory system to the main olfactory system (olfactory epithelium) and considered as ‘a specialized nose’, which can detect the chemical signals emitted by animals to determine species, identity and gender. It is usually located at the base of the nasal septum or in the roof of the mouth. These organs are primarily found in snakes, insects, rodents, etc. where pheromones are involved in attraction and reproduction. In rodents, the VNO contains vomeronasal receptor neurons located in a sensory epithelium of the vomeronasal duct; their afferent axons connect the duct with the accessory olfactory bulb, allied glands and ganglionic cells in the nasal septal mucosa [21]. VNO contains bipolar cells similar to the developing vomeronasal sensory neurons (VSNs) of other species. There are many elongated cells having a microvillar surface to the lumen of the organ in humans but most are not similar to microvillar vomeronasal sensory organs (VSNs) of other species. They have not been shown to have axons leaving the epithelium nor to make synaptic contact with axons in the epithelium, therefore, they are unable to directly communicate with the brain [22].
VSNs contain two types of receptors; Vomeronasal type 1 receptor (V1R) and Vomeronasal type 2 receptor (V2R). V1R is a multigene family with topographically restricted expression in the apical, Gαi2-expressing layer of the VNO neuroepithelium. They belong to Class-A (rhodopsin-like) GPCRs [23] but shares no sequence homology with ORs, however, there are certain common features between the two like no introns in coding region, clustered chromosomal organization [24], monogenic [25] and monoallelic expression [26] and follows one neuron-one (or a few) receptors(s) hypothesis given by Mombaerts et al. [27]. V1R superfamily is highly diverse and is subdivided into 12 extremely isolated gene families which share 40% minimum sequence identity and less than 15% interfamily homology. Intact V1R genes are conserved in teleost species [24, 28, 29], however high cross-species variability is found in mammals in both V1R gene count and primary sequence [30]. V2R receptors represent the second multigene family of VNO-specific GPCRs exclusively expressed in Gαo –positive VSNs and shows no similarity with ORs [31-33]. V2Rs clustered on most chromosomes are divided into four distinct subfamilies (A, B, C, and D) [34, 35]. Family A represents 80% of the V2R genes (more than 100 members) while family D has only four members [34, 35]. In humans, functional repertoires of V1r are substantially degenerated while V2r repertoires are completely degenerated [24, 34]. Presence of the VNO in humans is still doubtful and of debate, however, they have been reported in human fetuses and infrequently in adults only after the eighteenth century. A study by Mombaerts et al. [36] identified a gene (V1RL1 receptor) in the epithelial tissue, which closely resembles a mouse pheromone receptor. An indirect evidence of VNO-like discrimination in humans was also reported which observed the activation of the hypothalamus, by women, on smelling an androgen-like compound and by men on smelling an estrogen-compound [37]. These findings are just the beginning of re-considering the functional organization of a matter human ORs and in the near future, there may be the inclusion of a few more genes.
4. OLFACTORY BULB
OB is situated in the foremost part of the brain. It acts as a first relay center, which receives olfactory stimuli from OSNs, processes and transmits them to different regions of the olfactory cortex. OB is made up of seven layers [38] Fig. (2); a) Olfactory Nerve Layer (ONL) is the outermost layer comprising fascicles of unmyelinated olfactory axons which penetrate the cribriform plate. The olfactory nerve bundles are enclosed by unsheathing glia and penetrate into glomeruli in the Glomerular layer. b) Glomerular layer (GL): Glomeruli are unique compactly packed structures in the brain. Each glomerulus consists of neutrophil-rich spheroidal structures, surrounded by different small to medium-sized neurons. In GL, terminals of olfactory neurons make synapse with dendritic tufts of different types of neurons (mitral cells, tufted cells and intrinsic local circuit neurons). Neurons around and subjacent to glomeruli are heterogeneous and are termed as juxtaglomerular neurons. They are further classified into periglomerular cells, external tufted cells and superficial short-axon cells. The dendrites of periglomerular cells and external tufted cells enter glomeruli while superficial short-axon cells send dendrites only into the juxtaglomerular region. c) External plexiform layer (EPL) contains soma of tufted cells and of few short-axon cells. Secondary dendrites of both mitral and tufted cells make synapse in the EPL with granule cell dendrites and EPL anaxonic multipolar neuron dendrites. d) Mitral cell layer (MCL) is a thin layer, made up of large somas of mitral cells and smaller somas of other local circuit neurons. e) Internal plexiform layer (IPL) is a cell-free thin layer. f) Granule cell layer (GCL) consists of several rows of compactly packed granule cell somata and larger somas of deep short-axon cells. g) Subependymal-ependymal layer (SEL) is the core region of the OB.
Fig. (2).
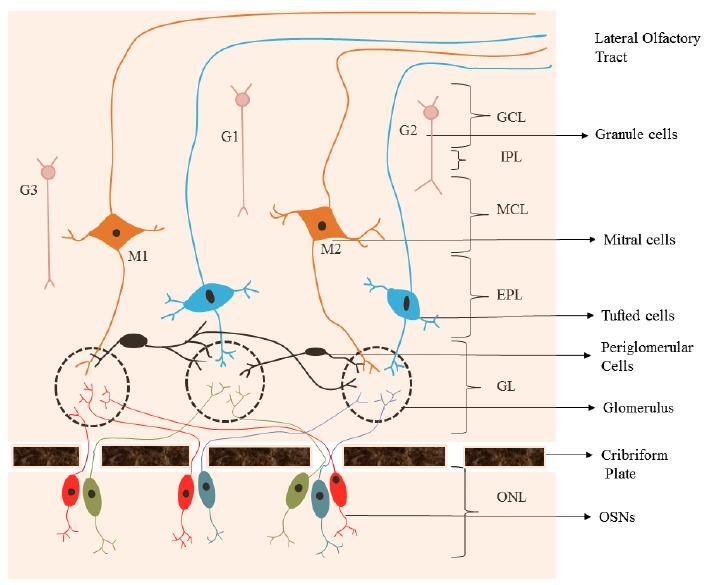
Olfactory Bulb [38]. (The color version of the figure is available in the electronic copy of the article).
There are two types of heterogeneous neurons in OB- principal neurons and non-principal neurons Fig. (3). The principal neurons are heterogeneous in structural, physiological and chemical properties, example mitral and tufted cells. They send single primary dendrites into lone glomeruli to build a characteristic cluster and many secondary dendrites in the EPL. Some of the tufted cells do not have secondary dendrites. On the basis of distribution of secondary dendrites in EPL, mitral cells are of two types, type 1 (M1) and type 2 (M2). Based on the location of their somas, tufted cells are of three types, namely deep, middle and external tufted cells. The non-principal neurons comprise local circuit neuron and projection neuron. The local circuit neuron (inhibitory neurons) extends its axons within the OB and is classified into periglomerular cells, granule cells, and short-axon cells. Periglomerular cells are found in the juxtaglomerular regions. They send dendrites into glomeruli where they intermingle with mitral/tufted cells dendritic tufts. Granule cells make reciprocal pairs of synapses with secondary dendrites of mitral/tufted cells. There are three subtypes of granule cells based on the location of soma and spreading of peripheral dendrites in the EPL: Type 1 (G1) having dendrites throughout EPL, type 2 (G2) dendrites and somata in the deep half of the EPL, type 3 (G3) dendrites and somata in the superficial half of the EPL. Granule cells modify the effects of GABA in the body (GABAergic). Short-axon cells are present in all seven layers of OB and include diverse types of neurons [38].
Fig. (3).
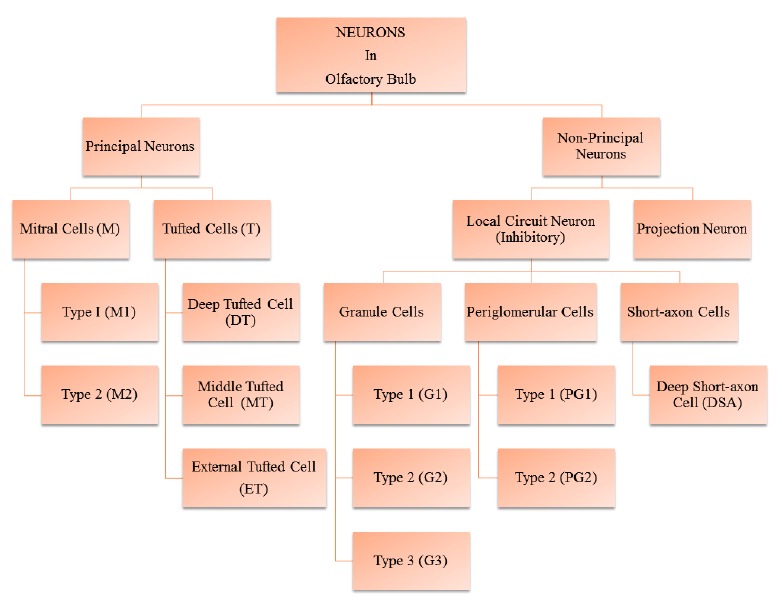
Types of Neurons in Olfactory Bulb. (The color version of the figure is available in the electronic copy of the article).
5. OLFACTORY RECEPTOR GENES
Previously [39-41], OR genes were believed to constitute roughly 3% of the 30,000 genes comprising the human genome. A study by Rouquier et al. [42, 43] found out that 72% of human OR genes are pseudogenes. Since humans require less olfactory acuity as compared to the other primates, molecular disruptions were accumulated during evolution followed by eradication of the functionality of OR genes in humans. The possible reasons for the transformation of functional genes into pseudogenes are frame shifts, single nucleotide polymorphism (SNP) and stop codon.
One particular OR gene is expressed in a small set of OSNs and, each OSN expresses only one OR protein [44]. The monoallelic/ monogenic expression of OR genes in OSNs is mainly a stochastic process, regulated by a negative feedback mechanism which is mediated by OR protein themselves [28, 45]. Consistent with previous predictions [42], two studies reported 347 [11] and 368 [13] full-length, functional OR genes. The number of genes and pseudogenes identified in the three studies is almost the same. The human genome approximately contains 900 OR genes, of which 396 are functional (protein-coding) OR genes and 468 are pseudogenes [13, 28, 46, 47]. The detailed analysis of human OR family carried out by Glusman et al. [11] explained diversification events and a rate (‘molecular clock’) at which mutations become fixed during human evolution. They performed comprehensive data mining using gene discovery algorithms and detected 601 new ORs. In addition to the detection, they also explained the localization, classification, isochore analysis and potential orthologs for OR genes.
The genes encoding OR do not contain introns within their coding region, however experimental and computational comparison of cDNA and genomic sequences [48, 49], and transcription analysis [50, 51] revealed the presence of a long intron splitting in the 5’ un-translated region. The in situ hybridization experiments [11, 13, 42-43, 46, 52-54] confirmed the distribution of human OR genes in multiple clusters of variable sizes throughout all chromosomes (Table 1), except chromosome 20 and Y. Chromosome 11 is the richest in OR genes as it contains the maximum number of OR genes (in two super-clusters, repeated cluster reflect the evolutionary origin of the family), followed by chromosomes 1, 9, 6, and 14 Fig. (4). Chromosomes 10, 22 and X contain single OR gene. Of all the functional OR genes identified so far, none of them were observed in chromosomes 4, 18 and 21 [11, 13]. The OR gene superfamily is the largest in the human genome. It comprises 18 families and 301 subfamilies (Table 1, Supplementary Data). The individual members of the subfamily are often located on different (two or more) chromosomes.
Table 1. Human olfactory receptor family.
| Sr. No. | Family | Subfamily | Genes | Pseudo-genes | Chromosome |
|---|---|---|---|---|---|
| 1 | OR1 | 21 | 28 | 11 | 1,5,9,11,16,17,19, X |
| 2 | OR2 | 41 | 67 | 46 | 1,5,6,7,9,11,12,16,19, X |
| 3 | OR3 | 3 | 4 | 2 | 1,17, X |
| 4 | OR4 | 24 | 57 | 80 | 1,5,6,8,11,14,15,18,19,21, X |
| 5 | OR5 | 49 | 47 | 64 | 2,3,6,9,11, X |
| 6 | OR6 | 21 | 30 | 21 | 1,2,7,8,10,11,12,14 |
| 7 | OR7 | 9 | 11 | 102 | 2,3,4,5,7,8,9,10,11,12,13,14,19,21 |
| 8 | OR8 | 18 | 24 | 23 | 11,12 |
| 9 | OR9 | 12 | 9 | 14 | 1,2,7,11,12 |
| 10 | OR10 | 28 | 36 | 28 | 1,6,7,11,12,14,19 |
| 11 | OR11 | 11 | 8 | 17 | 1,5,12,14,15, X |
| 12 | OR12 | 1 | 2 | 1 | 6 |
| 13 | OR13 | 11 | 12 | 10 | 1,9,10, X |
| 14 | OR14 | 6 | 6 | 1 | 1,6 |
| 15 | OR51 | 21 | 23 | 21 | 11 |
| 16 | OR52 | 22 | 26 | 23 | 11 |
| 17 | OR55 | 1 | 0 | 1 | 11 |
| 18 | OR56 | 2 | 6 | 3 | 11 |
Fig. (4).
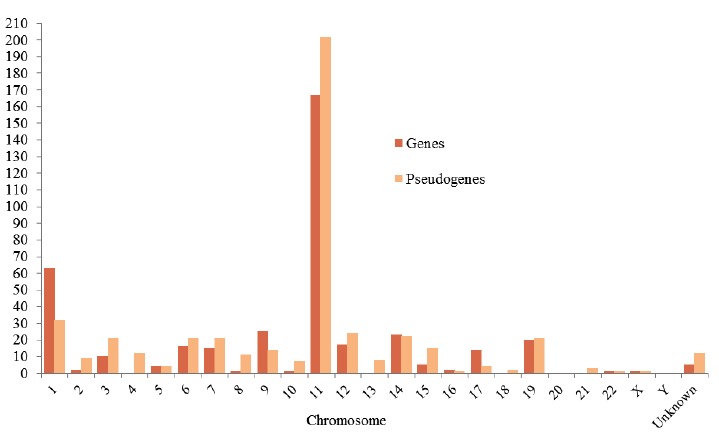
Chromosomal location of genes. (The color version of the figure is available in the electronic copy of the article).
The nomenclature for human OR genes and Cytochrome (CYP) gene family is similar [55-60]. OR gene sequences are checked for sequence identity. A sequence identity of 40% or higher is observed between any two randomly chosen OR genes, while the lowest reported sequence identity is about 20% [11, 13]. Based on sequence identity, different classification and nomenclature for the massive OR gene family are suggested. The Human Olfactory Receptor Data Explorer (HORDE) [46], a repository of OR genes, is based on the convention; family comprises of sequences with 40% or more sequence identity and subfamily comprises sequences with 60% or more sequence identity [61, 62]. In another classification scheme, minimum sequence identity required to be a member of a family is 43% and nomenclature is done on the basis of the chromosome number, the family number, and a unique member identifier [13]. The Olfactory Receptor Database, maintained and supported by the Human Brain Project, and the U.S. National Institute on Deafness and other Communication Disorders, classifies OR genes according to the chronological order of their publication [53]. Phylogenetically, OR genes are classified into two classes: Class I and Class II OR genes. Mammals contain both the classes [63].
6. OLFACTORY RECEPTOR PROTEINS
The molecular basis of odor detection and discrimination remained speculative for almost the entire 20th century because the receptors were not known. The discovery of the ORs happened to be the turning point in the research on the olfactory mechanism. OR proteins are the physical barriers between the environment and the brain. In order to understand the molecular basis of olfaction, it is vital to understand the nature, diversity and specificity of ORs [2]. The odor perception was assumed to be based on shape-sensitive mechanism. Various studies highlighted the role of G protein-coupled, cAMP –mediated transduction mechanism in odor recognition and discrimination [64-68]. These studies suggested the structural and functional belongingness of ORs to the family of membrane receptors known as Guanosine Tri Prophosphate (GTP) - binding Protein Coupled Receptors (GPCRs) family [2, 69]. OR proteins possess appreciable sequence similarity to other members of GPCR family and contain the characteristics of GPCR i.e. seven helical trans-membrane structure [7].
The classical work revealed that each OSN expresses only one type of OR. Interactions between ORs and odors are perplexed: A single OR can bind to multiple odorants, on the other hand, a single odorant can provoke a response from more than one OR, thus leading to a unique combination of ORs for each odorant [70-72]. The OSNs expressing specific ORs may intermingle with OSNs expressing other ORs, but the group of OSN expressing one type of OR is restricted to a specific anatomical zone within the main OE [44]. The functional significance of these OR-expression zones remains unknown. Both in-vitro and in-vivo functional studies have confirmed the chemosensory role of ORs i.e. they are activated by diverse chemical compounds, dictate tuning of OSNs, confer selective odor response on either OSNs or heterologous cells, and respond with different efficacies to the same odorant [44, 63, 70-76].
6.1. Structural Features of Olfactory Receptors
ORs belong to the GPCRs-rhodopsin family, which plays a key role in cell recognition, activating signal transduction, mediating senses (smell, taste, pain, sight) [7, 44, 70, 77, 78]. GPCRs are ubiquitous proteins which traverse the cell membranes and are included in metabotropic receptor family. Structurally, OR contains seven trans-membrane (TM) helical domains, connected by three putative extracellular loops (EL) and three putative intracellular loops (IL), an extracellular N-terminus, an intracellular C-terminus, similar to GPCR's [79]. OR possess certain sequence features which are unique Fig. (5). The intracellular loop contains a conserved sequence motif, at the intersection of TM3 and IL2, aspartate-arginine-tyrosine (DRY) amino acid motif, a hallmark of GPCRs [2-3, 7, 63]. TM1, TM2 and TM7 are conserved [2]. The central TM domains are structurally diverse and their amino acid side-chains determine the specificity of the binding site. Various molecular modeling and mutagenesis studies reported the hyper-variable regions of 20 amino acids in TM3, TM4 and TM5 which contribute to selective binding of different odor molecules [2, 7, 41, 65, 73, 80]. The spatial location of OR-binding pocket is similar to that of GPCRs (β-adrenergic receptor) but differs in the environment. The binding pocket of ORs has a hydrophobic environment, which indicates hydrophobic interactions between odorants and ORs, while in the GPCRs (in case of β-adrenergic receptor) binding site, ligands form ionic bonds, including hydrogen bonds and electrostatic interactions [81, 82]. However, the binding specificity of the ORs is not only determined by central TM-domains hyper-variable region, both N-termini and C-termini also influence the binding specificities of ORs. Both terminals are short, containing approximately 20 amino acids each. As in other GPCRs, N-terminal contains consensus sequences for N-glycosylation sites, while the C-terminal and IL3-loop contain phosphorylation sites. The kinases and second messenger (cAMP)-activated kinases induce phosphorylation thus uncoupling the signaling cascade. The EL1 and EL2-loop contain conserved cysteine residues which are involved in inter- and intra-molecular disulfide linkages. Of the three cysteine residues in EL2-loop, one cysteine has a particular function. The sequence motif (HXXC[DE]) in EL2-loop constitutes a metal binding site. The EL2-loop is transformed into α-helical structure confirmation (eighth helix) on binding with Zn(II) or Cu(II). Therefore, ORs undergo structural rearrangement when an odorant with high affinity to the metal ions replaces one of the metal-ligated amino acid in EL2-loop, a phenomenon required for activation of Gα-olf proteins attached to OR receptors. ORs do not contain a fourth cytoplasmic loop, a typical feature of GPCRs [83]. The sequence motifs KAFSTC, PMYFFL and YRDYAM found on the cytoplasmic side of the TM domains Fig. (5) contribute to the normal folding and activation of OR. The amino acid residues in the extracellular side of TM domains are variable thereby signifying numerous probable recognition sites for a myriad of odorants [81, 84].
Fig. (5).
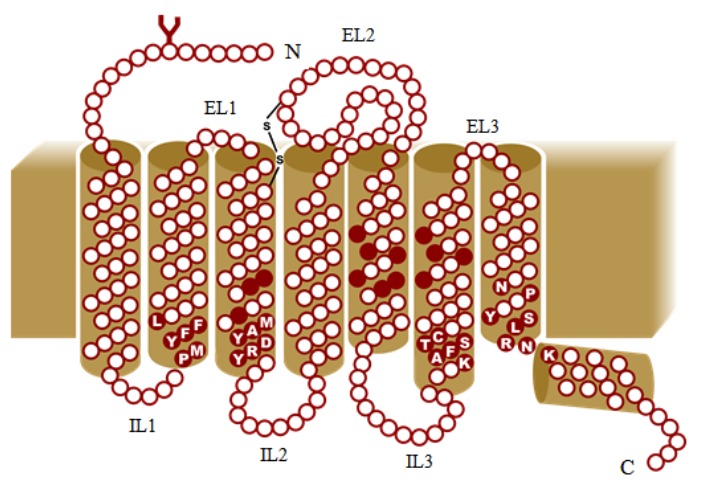
Trans-membrane topology of Odorant Receptors (OR). (The color version of the figure is available in the electronic copy of the article).
The existence of GPCRs as self-associated dimers or higher-order oligomers is well documented [85-88]. Kaupmann et al [89] demonstrated the crucial role of oligomerization for receptor localization and intracellular signaling.
A contrasting study reported that the binding of ligands on the GPCR surface activates it and thereby facilitates oligomerization [90]. Earlier there was no certain evidence of the precise relation between GPCRs oligomerization and activation. Although heterodimerization of ORs have been reported in other species [91, 92]. However, few ORs, including human OR1740, do not heterodimerize with non-olfactory GPCR. Fallou et al., unambiguously demonstrated homo-dimerization by human OR (OR1740) at the plasma membrane [85]. This study suggests that binding of odorant induces conformational changes in OR dimers and activity of potential OR dimers could depend on the number of bound odorant ligands. They also proposed a model for OR-OBP-ligand interactions which is based on two hypotheses. One hypothesis suggests the competitive binding of OBP and odorants to the OR. The second hypothesis deals with homo-dimerization of ORs [93, 94].
7. OLFACTORY SIGNAL TRANSDUCTION MECHANISM
G-proteins contain three subunits; an alpha subunit, namely Gα-olf which is specific to the ORs, a beta subunit and a gamma subunit. G-alpha is considered as the active unit while beta and gamma subunits regulate the activity of the alpha subunit. While in inactive (OFF) state, alpha subunit binds to Guanosine-DiPhosphate (GDP) [95]. GPCR activates on binding to an odorant and initiates a transduction cascade. The activation causes conformational changes in GPCR, both tilting and rotation of TM6 relative to TM3 [96-99]. Highly conserved serine (Ser) residue in the KAFSTC motif located in the cytoplasmic domain of TM6 has a significant role in receptor efficacy as it regulates the conformational changes of ORs (inactive to active forms). It is speculated that mutation of Ser to Ala or Val disrupts the ionic network, consequently, the receptor remains in an inactive state, thus making it easier for the receptor to move back to its active state [100]. The movement of TM6 exposes (technically buried) amino acids in ILs and in the C-terminal domain, thereby ‘unlocking’ a network of specific ionic interactions at the cytoplasmic ends of TMs [101]. Coupling of stimulated OR and G-protein, leads to a replacement of GDP in the alpha subunit by GTP, thereby attainment of activation (ON) state Fig. (6).
Fig. (6).
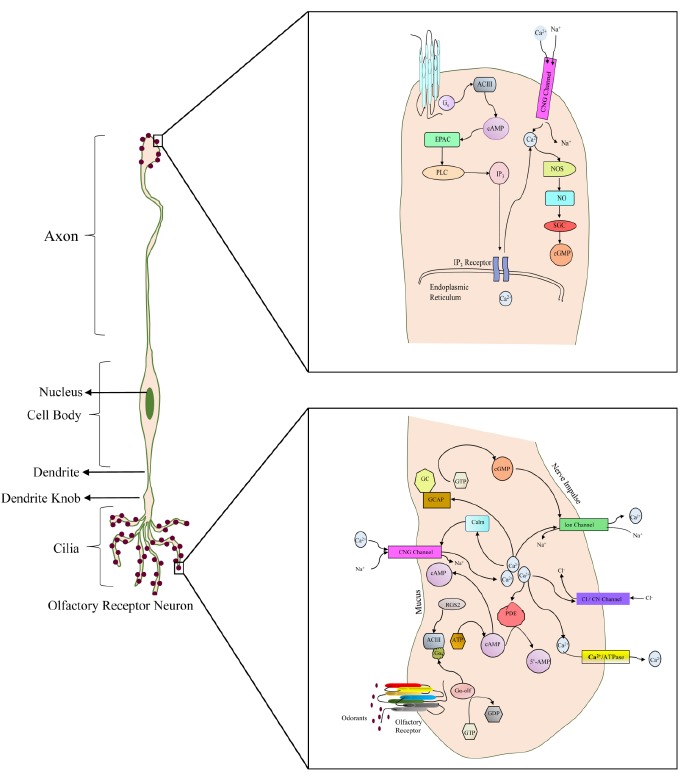
Olfactory transduction mechanism. (The color version of the figure is available in the electronic copy of the article).
GTP bound Gα-olf dissociates itself from beta- and gamma- subunits and moves on to stimulate adenylyl cyclase III (ACIII), which cyclizes Adenosine TriPhosphate (ATP) into second messenger, cyclic-3’5’-AdenosylMonoPhosphate (cAMP). Various studies have indicated the vital role of cAMP in olfactory signal transduction [102-107]. Using calcium imaging technique, it was observed that the increased intracellular concentration of cAMP moves throughout the cell cytoplasm and activates olfactory-specific cyclic nucleotide-gated channel (CNG) (ion-channels). CNG channel comprises the assembly of CNGA2, CNGA4 and CNGB1b subunits, which accomplishes ligand sensitivity and selectivity [108-110], leading to an influx of extracellular inorganic ions (Ca2+ and Na+) into the OSN. Calcium influx through CNG channels activates Ca2+-gated Cl- channel, causing efflux of Cl- from the OSN via cilia. Ion channels usually maintain the cell in a stable state, i.e. polarized state, with a potential of 90mV across the cell membrane. The organized calcium influx and chloride efflux cause depolarization of membrane potential in OSNs [110-113] which generate an electrical signal, representative of transfer of chemical signal to OB via axons. The cAMP concentration lowers as it hydrolyzes to AMP. Meanwhile, the alpha subunit of G-Protein effectively terminates its own activity, hydrolyzes GTP to GDP, rejoins to the beta and gamma subunits and retains its resting (OFF) state [114, 115]. The cell regains its electrical neutrality and maintains Ca2+ homeostasis by pumping Ca2+ out of OSNs by sodium-calcium (Na+/Ca2+) exchangers, voltage-gated chloride channel (ClCn) and transport proteins Ca2+ ATPase present in the cilia and dendritic knobs.
OR proteins present at axon terminal work as autonomous compartment which not only increases the local concentration of cAMP and Ca2+ through CNG channels, but also guides the convergence location of axons in OB. The concentration of Ca2+ is regulated by two mechanisms; first by usual CNG channel, and second by immobilization of Ca2+ from inside the nucleus by guanine nucleotide exchange factor (EPAC) which plays an important role in cGMP production. Increased concentration of Ca2+ activates nitric oxide (NOS) synthase, thereby generating nitric oxide (NO). Nitric oxide utilizing cAMP activates soluble guanylyl cyclase (SGC) and locally produces cGMP. cGMP thus produced is not directly involved in initial stimulus detection, but influences axon guidance and regulates axonic gene expression at local as well as nuclear level [19].
8. OLFACTORY ADAPTATION:
Adaptation / Short-term adaptation (STA) refers to a decrease in the response to the odors by OSNs. STA has a recovery time of seconds, and is induced by a very brief odor pulse. Once the odorant is captured and a training stimulus is relayed across OB to the brain, OSN adapts itself to previous exposure within a few seconds. The removal of intracellular Ca2+ abolishes STA [116]. Within OSN, there are other modulators such as calcium-binding protein Calmodulin (Calm), which regulates changes in intracellular Ca2+ concentrations by forming a complex with Ca2+. STA occurs at the level of CNG [117], which can be comprehended by the fact that the CNG channel has a binding site for Ca2+/Calm complex. Binding of Ca2+/Calm complex to CNG channel reduces channels sensitivity to cAMP, thus CNG channel closes. Ca2+/Calm activates enzyme Phosphodiesterase (PDE) which transforms cAMP to 5’AMP. This transformation reduces the excitation of the cell even though odorant is still present [118, 119].
In addition to the cAMP, cyclic Guanosine Monophosphate (cGMP) is also produced during odorant stimulation. As the concentration of cGMP decreases, CNG channels closes. It brings about two effects: Ca2+ influx is reduced with hyperpolarization of the membrane potential. The decrease in intracellular Ca2+ concentration is important for olfactory adaptation. Dropped intracellular Ca2+ concentration disinhibits Guanylate-Cyclase-Activating Protein (GCAP), which activates Guanylate Cyclase (GC) and leads to re-synthesis of cGMP. Another element regulated by cGMP has been discovered in olfactory neurons known as cGMP-activated PDE2 which hydrolyzes both cAMP and cGMP, hence terminating their action. Regulators of G-protein Signaling (RGS) also suppress GPCR mediated signals by accelerating the hydrolysis of GTP bound to the Gα subunit [14, 120] All the findings mentioned above indicate that STA is regulated by not only CNG channel desensitization but also by other signaling pathways which control cycling of GPCR, and tune the levels of secondary messengers in OSNs [116].
9. DESENSITIZATION OF OLFACTORY RECEPTORS
Desensitization involves loss of responsiveness by ORs in the repetitive presence of odorants or stimulus. The mechanism includes phosphorylation of the ORs resulting in uncoupling from its heterodimeric Gα-olf protein. It causes internalization of membrane-bound ORs to the cytosol and down-regulation of the cellular component of ORs. The intracellular kinases, both second messenger-dependent protein kinases (cAMP-dependent protein kinase A (PKA) and calcium-dependent protein kinase C (PKC)) and GPCR specific kinases (GRKs), phosphorylate the serine and threonine residues present in the intercellular loops and C-terminal regions of ORs. GRKs promote binding of arrestins, cytosolic co-factor proteins, causing steric uncoupling of the receptor and G-protein [121, 122]. The role of GRK and arrestins in the down- regulation of GPCR has been explored to a large extent, however, the precise mechanism of ORs desensitization is still elusive. Few studies demonstrated the role of GRK3, member of GRK family and β-arrestin2 in OR desensitization [123-126].
10. OLFACTORY BINDING PROTEINS (OBPS)
The entry, residence, and exit time period of odorants are crucial for olfaction recognition mechanism. The process of signal transduction is believed to be triggered by the entry of volatile molecules in the nasal cavity, followed by traversal in the mucus layer, which covers the nasal epithelium where they are recognized by OSNs cilia. However, the discovery of small, globular, water-soluble, ligand-specific proteins in the mucus fluid produced by nasal glands has steered a new concept of odorants being “carried away/ transferred” across the hydrophilic mucus layer Fig. (7a) towards their respective ORs via these proteins [2, 127-130]. It is believed that OBPs bind reversibly to accommodate hydrophobic odorants and enhance their access to OR binding sites [131, 132]. OBP belongs to the family of lipocalin proteins which include carrier proteins such as retinol-binding protein, β-lactoglobulin. There are two classes of OBP; one in vertebrate and other in insects, both perform the same function but are structurally different. Structurally, OBP consists of eight antiparallel β-sheets with an α-helical domain at the carboxyl terminal Fig. (7b). The β-sheets are folded into a continuous hydrogen-bonded β-barrel. OBPs are very stable to organic solvents, temperature, and proteolytic digestion [133]. OBPs do not undergo any chemical reaction with odorants in the initial stage of smell perception [134]. The reaction is equilibrated and occurs backward, i.e. when the system is completely hydrated. The hydrophobic ligand is accommodated inside the barrel cavity of and is protected from water, this OBP-odorant complex resembles the Schiff base. In a study, human OBPn (OBPIIa) has shown a strong affinity for aldehydes and fatty acids due to the formation of a Schiff base between a Lysine 127 residue, located in the binding cavity, and the aldehyde function [135]. To activate OR, odorant has to be released in its original form (aldehyde), the binding pocket of OBP opens up and is filled with water to recover the aldehyde function. Apart from solvation, the solvent plays an important role of chemical protagonist, which in absence induces the formation of Schiff base and in presence aids in the recovery of the aldehyde group near OSN membrane.
Fig. (7).
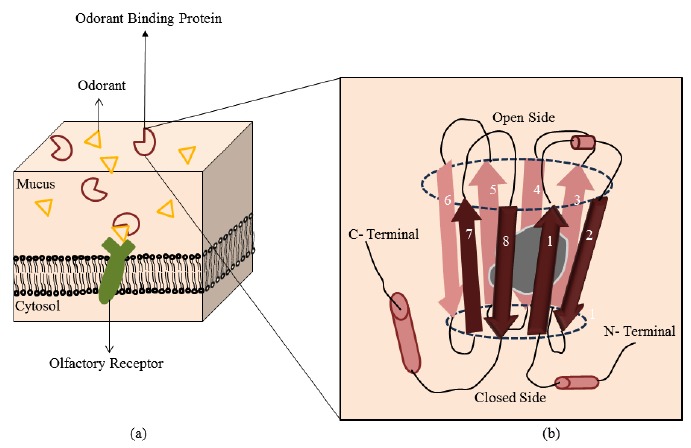
(a) Working of OBP and (b) Structure of OBP [2]. (The color version of the figure is available in the electronic copy of the article).
OBPs are highly diverse which led to the belief that each subtype is specialized to identify a distinct repertoire of odorants. The mechanism of odorant and OBP interaction is poorly understood. OBPs possess a unique ligand binding profile and act as a selective filter in odor pre-selection rather than just a passive carrier of odorants in the mucus layer [2, 131, 136-139]. Another study demonstrated OBP’s direct involvement in the direct activation of receptor [140] but it requires more testing.
11. OTHER CHEMOSENSORY RECEPTORS
The responsiveness of olfactory epithelium to diverse stimulus ranging from general odors to semiochemicals suggested the presence of other chemosensory receptors apart from canonical ORs [141-143]. Various studies have reported the existence of other GPCRs and non-GPCRs in the main olfactory systems [144-148].
11.1. Trace amine-associate Receptors (TAARs)
Discovered in 2001, TAARs, distantly related to biogenic amine GPCRs [149, 150], recognize low abundance neurotransmitters (trace amines) [151] via a key salt bridge involving a conserved trans-membrane three aspartic acid [152] and evoke stereotyped behaviors [146]. Except for TAAR1, all TAARs function as olfactory receptors [144, 153] however; TAARs are not phylogenetically related to ORs. Their sequence is generally conserved across species, but the number varies from species to species. The human repertoire consists of 6 full-length TAARs [153- 156]. Taar genes contain one translated exon and are found in a single genomic cluster, except Taar2. Each Taar allele defines a unique sensory neuron population that does not express other Taars or Ors. Like ORs, TAAR proteins are expressed in olfactory cilia to detect odor as well as at the axon terminal to participate in axon guidance and are involved in similar intracellular pathways [144, 157]. The expression of ORs and TAARs is mutually exclusive as OSNs expressing both have not been reported [144].
11.2. Non-GPCRs Chemosensors
The repertoire of olfactory receptors is not restricted to GPCRs; non-GPCRs have been identified in a small number of OSNs of the “necklace” subsystem within the olfactory epithelium [148, 158], capable of detecting diverse range of stimuli, like gases (carbon dioxide and carbon difsulfie), pheromones, urinary peptides and plant-derived odorants [147, 159-162]. The subsystem is so called because OSNs project axons to a ring of approximately 12-40 interconnected glomeruli, which encircle the caudal olfactory bulb like beads on a necklace [149]. The necklace neurons specifically contain single-pass transmembrane protein guanylate cyclase-D (GC-D) which detects and responds to diverse chemical stimuli. [152, 161-164]. The necklace OSNs also express one or more chemoreceptor encoded by Ms4a gene family, which encodes a four-pass transmembrane protein namely MS4A. The MS4A protein effectively detects specific odors like fatty acids, mouse pheromone [158].
12. THEORIES OF ODOR PERCEPTION
The sense of smell is important in medical and biological studies as it performs a dynamic range of very crucial functions in organisms. The mechanisms by which myriad of structurally diverse odorants are readily recognized and differentiated is still a puzzle. The molecular basis of odors is divided into two domains; molecular biology and fragrance chemistry. The fragrance chemistry looks for relationships between the odorants and odor quality and develops rules for structure-odor relations (SOR) [165]. While molecular biology focuses on the interaction between odorants and ORs and study relationship in their structure-activity patterns (SARs) [166]. The fundamental odor perception mechanism is a combination of both domains i.e. determining SARs, which must correspond to SORs to a certain degree [167]. The structural hypothesis emphasizes the role of the features (steric, nucleophilic, electrophilic characteristics) of odorants in the perception of smell [168]. Another study states that the odorant must possess certain molecular properties like water solubility, low polarity, surface activity, high vapor pressure (volatility), lipophilicity (to be able to dissolve in fat), in order to provide sensory properties. From various studies, it is observed that the odorant known till date possesses a molecular weight ≤ 294 [169]. However, odor is not an intrinsic property of a molecule rather it refers to a precise mechanism of recognition i.e. sensory response which is generated when the odorant binds to the appropriate receptor ORs, expressed in OSNs located in OE [7, 63, 170, 171] and the accuracy with which odors are discriminated depends on the specificity of the ORs to a particular odorant. Many theories related to olfaction have been proposed from time to time; few are described in the paper.
12.1. The Steric Theory of Odor
The most sophisticated theory for olfaction given by Troland is based on the steric factors. According to the theory, molecules adsorbed on the surfaces of olfactory cells, depolarize the cells and generate a neural impulse. The odor quality is determined by the nerve impulse’s timings, while odor intensity is perceived by the total number of similarly excited cells or by the total number of impulses in the nerve [172]. Moncrieff [173-175] proposed a “site filling” theory, which states that an odorant is smelled when it binds to the complimentary binding site on ORs. Binding follows “lock and key” hypothesis as in the case of enzyme-substrate interaction. Therefore, it is the molecular shape, volume and size of odorant, which determines the odor and its quality [176]. This steric theory explains the specificity and sensitivity of OR towards a certain set of odorants. Davies et al. proposed 'penetration theory' i.e. olfactory stimuli goes inside the OSNs. After strong criticism, they gave a new theory, which tried to explain olfaction mechanism on the basis of odorant size, shape and partition coefficient, but failed to address odor quality and the relation between the intensity of stimuli and odor concentration [177].
12.2. The Radiation Theory
Every atom or molecule has associated electron vibrations which set up the vibrations in the surrounding medium, and are reinforced by resonance. Jones et al. proposed that the olfactory nerves possess similar electrical vibrations with a minor variation among nerves, and molecules of odorous substances will reinforce those whose period of vibration relates to their own. The different rates of vibration are perceived as different smell characteristics [172]. However, this theory was refused.
12.3. The Vibrational Theories of Odor
12.3.1. Infra-red Theories
The theory associates odor with infrared resonance (IR), measurements of molecule vibration [178, 179]. The infrared theory of olfaction, first formulated by Ogle [180], hypothesized that analogous to audio and visual receptors which respond to waves, pigmentation in the nose also responds to radiation which lies in the infra-red spectrum. The theory faced many criticisms and most famous was by Beck et al. They reported that olfactory receptors radiate selectively, depending upon the size and shape. When an odorant comes in the radiation field of receptors, due to infra-red absorption characteristics of the odorant, receptors lose energy. The loss of energy generates the neural impulse. Different odorants have different infra-red absorption spectra and they stimulate the different set of receptors [173, 181]. Another study stated, the existence of substances (carbon disulfide) having odor, but their absorption spectra curves do not lie within the infra-red spectrum and odorless substances (carbon dioxide) exhibiting absorption of infra-red radiation [182]. The theory also fails in the case of enantiomers (Menthol and Carvone) that have identical infrared spectra, but distinctly different smell [183, 184].
12.3.2. Ultraviolet Theory
Given by Heyninx [179], the theory states that ultraviolet absorption bands of odorous molecules are due to the constituent molecules which vibrate with a frequency equivalent to that of the absorbed light. Based on the ultraviolet absorption bands, different frequencies of vibrations can be determined, which can define the differences in the qualities of different odors. Due to many inconsistencies, the ultraviolet theory was negated.
12.3.3. Raman Shift Theory
When a substance is radiated by laser light of particular wavelength, the energy of laser photons is shifted either up or down due to which substance emits shorter or longer wavelengths than the original light. This shift/ difference in energy, known as Raman shift, gives information about the vibrational modes of the system. Dyson in 1928 attempted to explain the olfaction mechanism using the Raman shift. He believed that the intra-molecular vibrations of odorants are responsible for odors and can be measured by utilizing Raman spectrum. The Raman shift of all odorous compounds lies between 140-130 millimicrons and odorants with similar Raman Shifts have similar odors [182, 185]. Though the theory was more convincing than the previous ones, however, exceptions have been found, like various compounds having the same odor, but different Raman shift [186], odorants with different odors, having same Raman Shift, odorless molecules have absorption bands in the Raman Spectrum [172].
12.3.4. Vibrational Induced Electron Tunneling Spectroscopy Theory
The electron tunneling involves the transfer of electrons down the backbone of the protein. Turin proposed a model in which zinc-binding motif is present in both OR and the G-protein, to explain electron transfer during olfaction [187]. The ability of zinc to form bridges between proteins and the presence of redox-active amino acid Cysteine in OR’s binding site establishes the link between electron flow and signal transduction via G-Protein. When OR binding site is empty, the disulfide bridge between OR and associated G-proteins remains in oxidized state and electrons are not able to tunnel across the binding site. After odorant binds to the OR, electrons excite to their vibrational mode. If vibrational mode energy is equal to the energy gap between the bound and unbound state, electron loses its energy and moves through the protein, reducing the disulfide bridge formed via zinc ion and leading to a release of G-protein from the ORs. The receptor acts as a spectrometer here, which detects a single well-defined energy, E. If the energy difference between sink and source is sufficiently larger, then the electron will move (current flow) across the receptor only if the odorant with required vibrational energy is present in the binding site. In the case of several vibrational modes, atoms partial charges and their relative orientation define the relative strength of the coupling which further determines which mode will be excited [188]. The theory is not validated, but appears to be reasonably sound.
12.4. Mechanical Theories
It states that the air movement in nose leads to vibration of olfactory hairs and the vibrations were modulated by odorants according to their molecular weight and momentum. The theory was abandoned as the odor quality was not correlated with the odorant molecular weight [172].
12.5. Stimulus Pattern Theories
An extensive work was carried out by Adrian in the electrophysiology of olfaction [172]. It was proposed that different odors stimulate different regions of the olfactory membrane due to their different physical properties and the eddy currents in the nasal passage [189]. The revised theory correlates the quality of odor to the membrane spatial patterning, coupled with temporal differences in arousal time and response decay. The theory was criticized due to unaccountability for olfactory stimulation and inability of replicating the experiments [172, 173].
12.6. Phase Boundary Theories
These theories deal with the mechanism of receptor stimulation. It was believed that the odorants dissolve in the mucus layer first, followed by absorption in the OSNs or adsorption on the surface. In both the events (absorption and adsorption) cellular metabolism would be disturbed, resulting in neural impulse. However, nothing was said about the olfactory quality [172, 173].
12.7. Chemical Theories
Various diverse chemical theories were given time to time, which relates the chemical properties of the receptors to olfaction and/or transduction mechanism [172, 179]. Mullins et al. proposed the existence of a minimum of two types of receptors in the olfactory epithelium which possesses a different solubility parameter and encompasses on the determinant role of odorant molecular shape [190]. The surface of receptors contains numerous, randomly arranged pores of variable sizes wherein odorants of similar size can bind. The binding excites only that particular surface of the receptor while the rest part remains insensitive. Large molecules are not able to excite the membrane due to insufficient number of suitable pores. The theory was rejected because the existence of hypothetical pores was not established [190]. Stoll et al reported decreases in the intensity of odor with the decrease in the molecular weight of the odorant, in the case of homologous series of bicyclic farnesyl synthetic compounds [173].
12.8. Enzyme Theories
The modern theories of olfactory stimulation revolve around the presence of various active enzymes in olfactory epithelium which are selectively inhibited by odorants. The inhibition alters the relative concentrations of some compounds in the receptor, resulting in the generation of nerve impulse [174]. The odorant-enzyme complex brings out conformation changes in odorant resulting in the exposure of buried points. The most significant theory is proposed by Amoore in which he correlated the odor quality with the shape and size of an odorant [191]. He characterized odors into seven primary classes (floral, pungent, peppermint, putrid, musky, camphoraceous and ethereal), postulated five hypothetical receptor sites for five of the primary odors and built a three-dimensional model of their atomic units. Amoore theory faced no serious objections and has been successfully able to predict and explain the olfaction mechanism. The only drawback with the theory is its inability to explain the actual stimulation processes which involve receptor.
The extensive review of all the theories is available in published works of Jones et al [172] and Moncrieff et al [175].
13. COMPUTATIONAL BIOLOGY OF OLFACTORY RECEPTORS
In the face of advancement in science and technology, insight into the specifics of the odorant recognition process has not improved significantly. There is no conclusive, concrete evidence as to how odorants bind or dock into a specific OR, which is a shape-sensitive mechanism. The reason is the notoriously difficult family to which OR belongs to. Despite their importance, limited structural information on GPCRs is available. The standard method of structure prediction by crystallizing the protein (X-ray crystallography) is not feasible with trans-membrane proteins, as a result, its binding site is experimentally inaccessible. Nuclear Magnetic Resonance (NMR) studies also sometimes fail to predict structures for GPCR [192, 193]. However, recently, there have been some decent efforts to solve the structure of many GPCRs with X-ray diffraction or other techniques. 5-HT2C receptor structural studies reveal the structural basis of GPCR polypharmacology [194], structural analysis of D2 dopamine receptor bound to the risperidone [195], ligand binding studies of neuropeptide Y Y1receptor [196] to name a few. Nevertheless, it is difficult to figure out the structural changes OR undergo at the molecular level. The atomic-level structure has been solved for bovine rhodopsin [197, 198] and beta-adrenergic receptor [196, 199, 200] by x-ray diffraction studies. Considering the importance, diversity and the vital role of GPCRs, it is important to develop theoretical methods to predict their structure and function [201, 202]. Nagarajan et al developed computational strategies and techniques to predict the structure of GPCRs (MembStruck protocol) and ligand binding sites (HierDock protocol), which were validated by comparing the predicted models to the experimental data of rhodopsin and bacteriorhodopsin [203].
MembStruck protocol for GPCR structure prediction begins with a prediction of trans-membrane region using hydropathicity analysis [204] in combination with input from multi-sequence profiles. Based on predicted TM regions, canonical right-handed α-helices are constructed to allow the sequence-specific distortions (because of proline). They are optimized with fixed bonds and angles. Once the TM scaffold is built, the axis of each helix in seven-helical TM bundle is rotated for hydrophobic-based positioning so that the net hydrophobic moment of each helix points outward, towards the membrane. The loops and termini are added, followed by coarse-grain optimization while simulating the surrounding lipid bi-layer using lipid molecules [203]. HierDock protocol [205] has been applied successfully to predict ligand binding sites of both globular and membrane proteins [206, 207]. Since the ligand binding site of GPCRs is not known beforehand, therefore coarse grain docking technique is applied for scanning entire protein to identify probable sites. Often used in conjunction with Membstruck protocol, it involves the progression of steps which discards ligand configurations failing to meet the criteria established while coarse grain docking. For each site, relative energies of ligands are determined as a difference between potential energy of ligand in the solvent and ligand in the protein. Low energy structures are further refined. The final protein-ligand complex could be used to explore binding mechanisms.
The atomic-resolution structures of only two receptors (rhodopsin and β-andregenic), having differently shaped ligand binding sites as compared to the ORs, are used to model ORs. Anselmi et al. used homology modeling and molecular dynamics to predict the binding site residues of human olfactory receptor OR3A1 followed by docking of few odorant molecules into the predicted binding site. Moreover, they proposed a correlation with the odorous properties of the ligands and investigated the residues involved in the binding. The study also highlights the olfactive stimulation of the OR with odorous molecules using calcium imaging or electrophysiological recordings [208].
14. OLFACTORY DATABASES
In the absence of experimentally derived structures of OR's, computational methods are used to model and to simulate interactions with odorants using static/ dynamic methods [206, 207, 209-211]. The results of potentially challenging and complicated modeling strategies are required to be stored and disseminated, so that they can be used for better understanding. With this idea, the information related to ORs and odorants is stored in well established, web-based resources, namely: the Human Olfactory Data Explorer (HORDE) [46], the Olfactory Receptor Database (ORDB) [47], Olfactory Receptor Microarray Database (ORMD) [212] and ODORactor [213], OlfactionDB [214]. All databases and web servers’ aims to store and assist experimental research related to olfaction.
HORDE is a complete repertoire of human OR genes and pseudogenes. It provides insights into the structure, function, and evolution of ORs. It contains OR orthologs from six mammalian species, namely, mouse, rat, chimpanzee, cow, dog, platypus, and opossum. The information is stored using an automated computational pipeline, which mines the relevant genes out of complete genome [215]. The information involves genomic organization of ORs into clusters, identification of clusters, gene models, Microarray and ESTs data. HORDE gene nomenclature is standardized using sequence comparisons that reflects OR evolution. The convention used in HORDE for instance; hOR4H11 means that the receptor belongs to family 4 and is the 11th gene of subfamily H Fig. (8a).
Fig. (8).
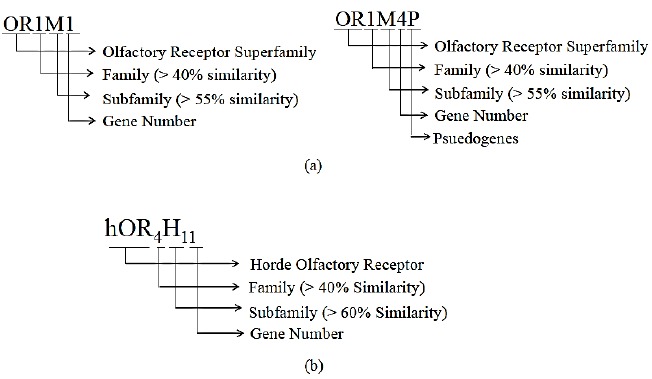
Nomenclature used by OR databases; (a) ORDB Nomenclature, (b) HORDE Nomenclature.
ORDB contains genomics and proteomics information related to ORs and other chemosensory receptors [49]. Proposed nomenclature for ORDB includes receptor superfamily i.e. olfactory (OR) followed by family, subfamily and gene information. For example, OR1M1, where OR represents olfactory superfamily, 1 is for the family, M for subfamily, 1 represents gene number within the subfamily. Similarly OR1M4P, here represents pseudogene Fig. (8b).
ORDB is integrated with three companion databases OdorDB, ORModelDB, and OdorMapDB which are also a part of the Sense Lab suite of databases. OdorDB provides information about the functional aspect of ORs i.e. with which the odorant, particularly OR can interact. At present, it stores information of approximately 257 odorants, for 75 odorants, their interacting ORs (experimental studies) is known. ORModelDB stores computationally predicted structures of
ORs based on ab initio or semi-empirical methods with the aim to decipher the OR-odorant interaction mechanism at a molecular level. The number of models is less (08 models) indicating gaps in in-silico aspect of OR-odorant interaction. OdorMapDB, another database associated with ORDB, contains information regarding OB’s molecular and functional organization from high-resolution fMRI, c-fos and 2-deoxyglucose studies. All four databases are cross-linked.
ORMD is a repository and management system for microarray experiments related to ORs. It stores Affymetrix gene-chip data in the olfactory epithelium as well as other tissues of rodents in minimum information about a microarray experiment (MIAME) format. ORMD contains both private as well as public gene expression data. It allows users to not only deposit gene expression data, but also manage their experiments. ORMD is cross-linked with ORDB, i.e. it connects the expression data with corresponding genes.
ODORactor is a MySQL based web server [215] to search existing OR-odorant pairs and uses SVM-based prediction to identify ORs for chemical compounds. ORs for a query chemical compound are predicted based on two functions, namely, odorant verification and OR identification. It houses manually curated information from literature about odorants (3038) and ORs (1608) in both humans as well as mouse. It is an effective platform for identifying probable ORs, odorants, Odorant-OR interaction and for basic olfaction research [216].
OlfactionDB is another free, manually curated, comprehensive, and publicly available database storing information of approximately 400 odorant-receptor interactions. It is developed for managing information about odorants and their receptors. It is integrated with a variety of online tools to carry out keyword-based search, sequence or ligand similarity search, Uniprot/PubChem accession number search [214].
CONCLUSION
Olfaction is the oldest sensory modality known in evolution. Odor detection is accomplished by an array of different ORs expressed by olfactory sensory neurons in the nasal olfactory epithelium. Olfaction signal transduction is crucial for sensing our environment. There are unanswered questions, however. Now, we have some data; the advancement in biological techniques has accelerated research in olfaction. With the knowledge of ORs, as multifunctional signaling molecules, their belongingness to GPCR family, OBPs working as a pre-selection filter, various experimental evidences of OR-odorant interactions, a lot of efforts have been made to get insights into structural and functional aspects of ORs. GPCR research is confined due to the limited availability of experimentally derived structures, which is a challenging task both experimentally and computationally. For databases like ORDB house models for ORs, the number is very small as compared to the actual OR receptors known. The mechanism underlying the expression of even a single allele in any given OSN is not yet understood. Studies to identify conserved motifs in the transcription factor binding site or promoter site, residue motifs specific to binding site have failed time and again thereby marking a new area of research. ORs structural and functional diversity is consistent with their ability to recognize structurally diverse chemical compounds. Odors are encrypted using a combinatorial approach i.e. structurally similar odorants bind to altogether different but overlapping ORs, which increase the complexity of the problem many folds. Also, the perception quality of odorant varies with concentration. Diverse approaches have yielded largely convergent results. Mechanistics of odorant-OR interactions, ORs activation and desensitization are still a black box. It is also not known as to how different areas of the olfactory cortex receive signals; from different subsets of ORs or from all the ORs. Further, how are the signals organized in the cortex? is it random scattering similar to that in the epithelium or are they mapped onto unique locations analogous to the OB or organization is completely different. There are several such queries which require more experimentation and in-depth research in olfaction. Computational analysis (structure modeling, docking and simulation protocols) offers a view of the OR-odorant interactions for a better understanding of what leads to olfaction. By modeling ORs, one can predict the underlying structural and functional relationship of odorous compounds coupling to their corresponding receptors. In the absence of experimental studies, site-directed mutagenesis could be used in modeling and docking studies to decipher the 3D structure of ORs. They can be used to generate hypothesis for OR structure’s and OR-odorant interaction prediction which needs to be validated through experiments. Computational databases storing complete human OR universe known till date are a fundamental asset for future research in olfaction. Twenty-seven years after the discovery of ORs, there is still much more to research and learn about their structure, function and mechanism.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Harel D., Carmel L., Lancet D. Towards an odor communication system. Comput. Biol. Chem. 2003;27(2):121–133. doi: 10.1016/s1476-9271(02)00092-0. [http://dx.doi. org/10.1016/S1476-9271(02)00092-0]. [PMID: 12821309]. [DOI] [PubMed] [Google Scholar]
- 2.Breer H. Olfactory receptors: molecular basis for recognition and discrimination of odors. Anal. Bioanal. Chem. 2003;377(3):427–433. doi: 10.1007/s00216-003-2113-9. [http://dx.doi.org/10.1007/s00216-003-2113-9]. [PMID: 12898108]. [DOI] [PubMed] [Google Scholar]
- 3.Spehr M., Munger S.D. Olfactory receptors: G protein-coupled receptors and beyond. J. Neurochem. 2009;109(6):1570–1583. doi: 10.1111/j.1471-4159.2009.06085.x. [http://dx.doi.org/10.1111/j.1471-4159.2009.06085.x]. [PMID: 19383089]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touhara K., Vosshall L.B. Sensing odorants and pheromones with chemosensory receptors. Annu. Rev. Physiol. 2009;71:307–332. doi: 10.1146/annurev.physiol.010908.163209. [http://dx.doi.org/10.1146/annurev.physiol.010908.163209]. [PMID: 19575682]. [DOI] [PubMed] [Google Scholar]
- 5.Sarafoleanu C., Mella C., Georgescu M., Perederco C. The importance of the olfactory sense in the human behavior and evolution. J. Med. Life. 2009;2(2):196–198. [PMID: 20108540]. [PMC free article] [PubMed] [Google Scholar]
- 6.Bear D.M., Lassance J.M., Hoekstra H.E., Datta S.R. Evolution of the genetic and neural architecture for vertebrate odor perception. Curr. Biol. 2016;26:R1039–R1049. doi: 10.1016/j.cub.2016.09.011. [http://dx.doi.org/10. 1016/j.cub.2016.09.011]. [PMID: 27780046]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck L., Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [http://dx.doi.org/10.1016/0092-8674(91)90418-X]. [PMID: 1840504]. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X., Firestein S. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 2002;5(2):124–133. doi: 10.1038/nn800. [http://dx.doi.org/ 10.1038/nn800]. [PMID: 11802173]. [DOI] [PubMed] [Google Scholar]
- 9.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J., Szustakowki J. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [http://dx.doi.org/10.1038/ 35057062]. [PMID: 11237011]. [DOI] [PubMed] [Google Scholar]
- 10.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., Gocayne J.D., Amanatides P., Ballew R.M., Huson D.H., Wortman J.R., Zhang Q., Kodira C.D., Zheng X.H., Chen L., Skupski M., Subramanian G., Thomas P.D., Zhang J., Gabor Miklos G.L., Nelson C., Broder S., Clark A.G., Nadeau J., McKusick V.A., Zinder N., Levine A.J., Roberts R.J., Simon M., Slayman C., Hunkapiller M., Bolanos R., Delcher A., Dew I., Fasulo D., Flanigan M., Florea L., Halpern A., Hannenhalli S., Kravitz S., Levy S., Mobarry C., Reinert K., Remington K., Abu-Threideh J., Beasley E., Biddick K., Bonazzi V., Brandon R., Cargill M., Chandramouliswaran I., Charlab R., Chaturvedi K., Deng Z., Di Francesco V., Dunn P., Eilbeck K., Evangelista C., Gabrielian A.E., Gan W., Ge W., Gong F., Gu Z., Guan P., Heiman T.J., Higgins M.E., Ji R.R., Ke Z., Ketchum K.A., Lai Z., Lei Y., Li Z., Li J., Liang Y., Lin X., Lu F., Merkulov G.V., Milshina N., Moore H.M., Naik A.K., Narayan V.A., Neelam B., Nusskern D., Rusch D.B., Salzberg S., Shao W., Shue B., Sun J., Wang Z., Wang A., Wang X., Wang J., Wei M., Wides R., Xiao C., Yan C., Yao A., Ye J., Zhan M., Zhang W., Zhang H., Zhao Q., Zheng L., Zhong F., Zhong W., Zhu S., Zhao S., Gilbert D., Baumhueter S., Spier G., Carter C., Cravchik A., Woodage T., Ali F., An H., Awe A., Baldwin D., Baden H., Barnstead M., Barrow I., Beeson K., Busam D., Carver A., Center A., Cheng M.L., Curry L., Danaher S., Davenport L., Desilets R., Dietz S., Dodson K., Doup L., Ferriera S., Garg N., Gluecksmann A., Hart B., Haynes J., Haynes C., Heiner C., Hladun S., Hostin D., Houck J., Howland T., Ibegwam C., Johnson J., Kalush F., Kline L., Koduru S., Love A., Mann F., May D., McCawley S., McIntosh T., McMullen I., Moy M., Moy L., Murphy B., Nelson K., Pfannkoch C., Pratts E., Puri V., Qureshi H., Reardon M., Rodriguez R., Rogers Y.H., Romblad D., Ruhfel B., Scott R., Sitter C., Smallwood M., Stewart E., Strong R., Suh E., Thomas R., Tint N.N., Tse S., Vech C., Wang G., Wetter J., Williams S., Williams M., Windsor S., Winn-Deen E., Wolfe K., Zaveri J., Zaveri K., Abril J.F., Guigó R., Campbell M.J., Sjolander K.V., Karlak B., Kejariwal A., Mi H., Lazareva B., Hatton T., Narechania A., Diemer K., Muruganujan A., Guo N., Sato S., Bafna V., Istrail S., Lippert R., Schwartz R., Walenz B., Yooseph S., Allen D., Basu A., Baxendale J., Blick L., Caminha M., Carnes-Stine J., Caulk P., Chiang Y.H., Coyne M., Dahlke C., Mays A., Dombroski M., Donnelly M., Ely D., Esparham S., Fosler C., Gire H., Glanowski S., Glasser K., Glodek A., Gorokhov M., Graham K., Gropman B., Harris M., Heil J., Henderson S., Hoover J., Jennings D., Jordan C., Jordan J., Kasha J., Kagan L., Kraft C., Levitsky A., Lewis M., Liu X., Lopez J., Ma D., Majoros W., McDaniel J., Murphy S., Newman M., Nguyen T., Nguyen N., Nodell M., Pan S., Peck J., Peterson M., Rowe W., Sanders R., Scott J., Simpson M., Smith T., Sprague A., Stockwell T., Turner R., Venter E., Wang M., Wen M., Wu D., Wu M., Xia A., Zandieh A., Zhu X. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [http://dx.doi.org/10.1126/science.1058040]. [PMID: 11181995]. [DOI] [PubMed] [Google Scholar]
- 11.Glusman G., Yanai I., Rubin I., Lancet D. The complete human olfactory subgenome. Genome Res. 2001;11(5):685–702. doi: 10.1101/gr.171001. [http://dx.doi.org/10.1101/gr.171001]. [PMID: 11337468]. [DOI] [PubMed] [Google Scholar]
- 12.Niimura Y., Nei M. Comparative evolutionary analysis of olfactory receptor gene clusters between humans and mice. Gene. 2005;346:13–21. doi: 10.1016/j.gene.2004.09.025. [http://dx.doi.org/10.1016/j.gene.2004.09.025]. [PMID: 15716120]. [DOI] [PubMed] [Google Scholar]
- 13.Zozulya S., Echeverri F., Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001 doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malnic B., Godfrey P.A., Buck L.B. The human olfactory receptor gene family. Proc. Natl. Acad. Sci. USA. 2004;101(8):2584–2589. doi: 10.1073/pnas.0307882100. [http://dx.doi.org/10.1073/pnas.0307882100]. [PMID: 14983052]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godfrey P.A., Malnic B., Buck L.B. The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. USA. 2004;101(7):2156–2161. doi: 10.1073/pnas.0308051100. [http://dx.doi.org/10.1073/pnas.0308051100]. [PMID: 14769939]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breer H., Fleischer J., Strotmann J. The sense of smell: multiple olfactory subsystems. Cell. Mol. Life Sci. 2006;63(13):1465–1475. doi: 10.1007/s00018-006-6108-5. [http://dx.doi.org/10.1007/s00018-006-6108-5]. [PMID: 16732429]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strotmann J., Levai O., Fleischer J., Schwarzenbacher K., Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J. Neurosci. 2004;24(35):7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [http://dx.doi.org/ 10.1523/JNEUROSCI.2588-04.2004]. [PMID: 15342743]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menco B.P., Bruch R.C., Dau B., Danho W. Ultrastructural localization of olfactory transduction components: the G protein subunit Golf alpha and type III adenylyl cyclase. Neuron. 1992;8(3):441–453. doi: 10.1016/0896-6273(92)90272-f. [http://dx.doi.org/10.1016/0896-6273(92)90272-F]. [PMID: 1550671]. [DOI] [PubMed] [Google Scholar]
- 19.Mombaerts P. How smell develops. Nat. Neurosci. 2001;4(Suppl.):1192–1198. doi: 10.1038/nn751. [http://dx.doi.org/10.1038/nn751]. [PMID: 11687829]. [DOI] [PubMed] [Google Scholar]
- 20.Lodovichi C., Belluscio L. Odorant receptors in the formation of the olfactory bulb circuitry. Physiology (Bethesda) 2012;27(4):200–212. doi: 10.1152/physiol.00015.2012. [http://dx.doi.org/10.1152/physiol.00015.2012]. [PMID: 22875451]. [DOI] [PubMed] [Google Scholar]
- 21.Witt M., Woźniak W. Structure and function of the vomeronasal organ. Adv. Otorhinolaryngol. 2006;63:70–83. doi: 10.1159/000093751. [http://dx.doi.org/ 10.1159/000093751]. [PMID: 16733333]. [DOI] [PubMed] [Google Scholar]
- 22.Meredith M. Human vomeronasal organ function: a critical review of best and worst cases. Chem. Senses. 2001;26(4):433–445. doi: 10.1093/chemse/26.4.433. [http://dx.doi.org/10.1093/chemse/26.4.433]. [PMID: 11369678]. [DOI] [PubMed] [Google Scholar]
- 23.Dulac C., Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83(2):195–206. doi: 10.1016/0092-8674(95)90161-2. [http://dx.doi.org/10.1016/0092-8674(95)90161-2]. [PMID: 7585937]. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez I., Mombaerts P. Novel human vomeronasal receptor-like genes reveal species-specific families. Curr. Biol. 2002;12(12):R409–R411. doi: 10.1016/s0960-9822(02)00909-0. [http://dx.doi.org/10.1016/S0960-9822(02) 00909-0]. [PMID: 12123587]. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez I., Feinstein P., Mombaerts P. Variable patterns of axonal projections of sensory neurons in the mouse vomeronasal system. Cell. 1999;97(2):199–208. doi: 10.1016/s0092-8674(00)80730-8. [http://dx.doi.org/10.1016/ S0092-8674(00)80730-8]. [PMID: 10219241]. [DOI] [PubMed] [Google Scholar]
- 26.Roppolo D., Vollery S., Kan C.D., Lüscher C., Broillet M.C., Rodriguez I. Gene cluster lock after pheromone receptor gene choice. EMBO J. 2007;26(14):3423–3430. doi: 10.1038/sj.emboj.7601782. [http://dx.doi.org/10. 1038/sj.emboj.7601782]. [PMID: 17611603]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited. Curr. Opin. Neurobiol. 2004;14(1):31–36. doi: 10.1016/j.conb.2004.01.014. [b]. [DOI] [PubMed] [Google Scholar]
- 28.Dulac C., Wagner S. Genetic analysis of brain circuits underlying pheromone signaling. Annu. Rev. Genet. 2006;40:449–467. doi: 10.1146/annurev.genet.39.073003.093937. [http://dx.doi.org/10.1146/annurev.genet.39.073003.093937]. [PMID: 16953793]. [DOI] [PubMed] [Google Scholar]
- 29.Shi P., Zhang J. Comparative genomic analysis identifies an evolutionary shift of vomeronasal receptor gene repertoires in the vertebrate transition from water to land. Genome Res. 2007;17(2):166–174. doi: 10.1101/gr.6040007. [http://dx.doi.org/10.1101/gr.6040007]. [PMID: 17210926]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grus W.E., Shi P., Zhang Y.P., Zhang J. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl. Acad. Sci. USA. 2005;102(16):5767–5772. doi: 10.1073/pnas.0501589102. [http://dx.doi.org/10.1073/pnas. 0501589102]. [PMID: 15790682]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrada G., Dulac C. A novel family of putative pheromone receptors in mammals with a topographically organized and sexually dimorphic distribution. Cell. 1997;90(4):763–773. doi: 10.1016/s0092-8674(00)80536-x. [http://dx. doi.org/10.1016/S0092-8674(00)80536-X]. [PMID: 9288755]. [DOI] [PubMed] [Google Scholar]
- 32.Matsunami H., Buck L.B. A multigene family encoding a diverse array of putative pheromone receptors in mammals. Cell. 1997;90(4):775–784. doi: 10.1016/s0092-8674(00)80537-1. [http://dx.doi.org/10.1016/S0092-8674(00)80537-1]. [PMID: 9288756]. [DOI] [PubMed] [Google Scholar]
- 33.Ryba N.J., Tirindelli R. A new multigene family of putative pheromone receptors. Neuron. 1997;19(2):371–379. doi: 10.1016/s0896-6273(00)80946-0. [http://dx. doi.org/10.1016/S0896-6273(00)80946-0]. [PMID: 9292726]. [DOI] [PubMed] [Google Scholar]
- 34.Young J.M., Trask B.J. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23(5):212–215. doi: 10.1016/j.tig.2007.03.004. [http://dx.doi.org/10.1016/j.tig.2007.03.004]. [PMID: 17382427]. [DOI] [PubMed] [Google Scholar]
- 35.Silvotti L., Moiani A., Gatti R., Tirindelli R. Combinatorial co-expression of pheromone receptors, V2Rs. J. Neurochem. 2007;103(5):1753–1763. doi: 10.1111/j.1471-4159.2007.04877.x. [http://dx.doi.org/10.1111/j.1471-4159.2007. 04877.x]. [PMID: 17854397]. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez I., Greer C.A., Mok M.Y., Mombaerts P. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat. Genet. 2000;26(1):18–19. doi: 10.1038/79124. [http://dx.doi.org/10.1038/79124]. [PMID: 10973240]. [DOI] [PubMed] [Google Scholar]
- 37.Savic I., Berglund H., Gulyas B., Roland P. Smelling of odorous sex hormone-like compounds causes sex-differentiated hypothalamic activations in humans. Neuron. 2001;31(4):661–668. doi: 10.1016/s0896-6273(01)00390-7. [http:// dx.doi.org/10.1016/S0896-6273(01)00390-7]. [PMID: 11545724]. [DOI] [PubMed] [Google Scholar]
- 38.Kosaka T., Kosaka K. Olfactory Anatomy.
- 39.Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286(5440):707–711. doi: 10.1126/science.286.5440.707. [http:// dx.doi.org/10.1126/science.286.5440.707]. [PMID: 10531047]. [DOI] [PubMed] [Google Scholar]
- 40.Mombaerts P. Molecular biology of odorant receptors in vertebrates. Annu. Rev. Neurosci. 1999;22:487–509. doi: 10.1146/annurev.neuro.22.1.487. [http://dx.doi.org/ 10.1146/annurev.neuro.22.1.487]. [PMID: 10202546]. [DOI] [PubMed] [Google Scholar]
- 41.Pilpel Y., Sosinsky A., Lancet D. Molecular biology of olfactory receptors. Essays Biochem. 1998;33:93–104. doi: 10.1042/bse0330093. [http://dx.doi.org/ 10.1042/bse0330093]. [PMID: 10488444]. [DOI] [PubMed] [Google Scholar]
- 42.Rouquier S., Taviaux S., Trask B.J., Brand-Arpon V., van den Engh G., Demaille J., Giorgi D. Distribution of olfactory receptor genes in the human genome. Nat. Genet. 1998;18(3):243–250. doi: 10.1038/ng0398-243. [http://dx.doi.org/10.1038/ng0398-243]. [PMID: 9500546]. [DOI] [PubMed] [Google Scholar]
- 43.Rouquier S., Blancher A., Giorgi D. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl. Acad. Sci. USA. 2000;97(6):2870–2874. doi: 10.1073/pnas.040580197. [http://dx.doi.org/10.1073/pnas.040580197]. [PMID: 10706615]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Serizawa S., Miyamichi K., Nakatani H., Suzuki M., Saito M., Yoshihara Y., Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302(5653):2088–2094. doi: 10.1126/science.1089122. [http://dx.doi.org/10.1126/science.1089122]. [PMID: 14593185]. [DOI] [PubMed] [Google Scholar]
- 45.Lewcock J.W., Reed R.R. A feedback mechanism regulates monoallelic odorant receptor expression. Proc. Natl. Acad. Sci. USA. 2004;101(4):1069–1074. doi: 10.1073/pnas.0307986100. [http://dx.doi.org/10.1073/pnas. 0307986100]. [PMID: 14732684]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. bioinformatics.weizmann.ac.il/HORDE (Accessed June 19, 2018)bioinformatics.weizmann.ac.il/HORDE
- 47. senselab.med.yale.edu/ordb
- 48.Glusman G., Clifton S., Roe B., Lancet D. Sequence analysis in the olfactory receptor gene cluster on human chromosome 17: recombinatorial events affecting receptor diversity. Genomics. 1996;37(2):147–160. doi: 10.1006/geno.1996.0536. [http://dx.doi.org/10.1006/geno.1996.0536]. [PMID: 8921386]. [DOI] [PubMed] [Google Scholar]
- 49.Walensky L.D., Ruat M., Bakin R.E., Blackshaw S., Ronnett G.V., Snyder S.H. Two novel odorant receptor families expressed in spermatids undergo 5′-splicing. J. Biol. Chem. 1998;273(16):9378–9387. doi: 10.1074/jbc.273.16.9378. [http://dx.doi.org/10.1074/jbc.273.16.9378]. [PMID: 9545261]. [DOI] [PubMed] [Google Scholar]
- 50.Asai H., Kasai H., Matsuda Y., Yamazaki N., Nagawa F., Sakano H., Tsuboi A. Genomic structure and transcription of a murine odorant receptor gene: differential initiation of transcription in the olfactory and testicular cells. Biochem. Biophys. Res. Commun. 1996;221(2):240–247. doi: 10.1006/bbrc.1996.0580. [http://dx.doi.org/10.1006/bbrc.1996.0580]. [PMID: 8619840]. [DOI] [PubMed] [Google Scholar]
- 51.Qasba P., Reed R.R. Tissue and zonal-specific expression of an olfactory receptor transgene. J. Neurosci. 1998;18(1):227–236. doi: 10.1523/JNEUROSCI.18-01-00227.1998. [http://dx.doi.org/10.1523/JNEUROSCI.18-01-00227.1998]. [PMID: 9412503]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E., Adato A., Peter I., Khen M., Atarot T., Groner Y., Lancet D. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31(1):142–146. doi: 10.1093/nar/gkg050. [http://dx.doi.org/10.1093/nar/gkg050]. [PMID: 12519968]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Olender T., Feldmesser E., Atarot T., Eisenstein M., Lancet D. The olfactory receptor universe--from whole genome analysis to structure and evolution. Genet. Mol. Res. 2004;3(4):545–553. [PMID: 15688320]. [PubMed] [Google Scholar]
- 54.Olender T., Lancet D., Nebert D.W. Update on the olfactory receptor (OR) gene superfamily. Hum. Genomics. 2008;3(1):87–97. doi: 10.1186/1479-7364-3-1-87. [http://dx.doi.org/10.1186/1479-7364-3-1-87]. [PMID: 19129093]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nebert D.W., Adesnik M., Coon M.J., Estabrook R.W., Gonzalez F.J., Guengerich F.P., Gunsalus I.C., Johnson E.F., Kemper B., Levin W. The P450 gene superfamily. Recommended nomenclature. DNA Cell Biol. 1987;6:1–11. doi: 10.1089/dna.1987.6.1. [PMID: 1991046]. [DOI] [PubMed] [Google Scholar]
- 56.Nebert D.W., Gonzalez F.J. P450 genes: structure, evolution, and regulation. Annu. Rev. Biochem. 1987;56:945–993. doi: 10.1146/annurev.bi.56.070187.004501. [http://dx. doi.org/10.1146/annurev.bi.56.070187.004501]. [PMID: 3304150]. [DOI] [PubMed] [Google Scholar]
- 57.Nebert D.W., Nelson D.R., Adesnik M., Coon M.J., Estabrook R.W., Gonzalez F.J., Guengerich F.P., Gunsalus I.C., Johnson E.F., Kemper B. The P450 superfamily: updated listing of all genes and recommended nomenclature for the chromosomal loci. DNA. 1989;8(1):1–13. doi: 10.1089/dna.1.1989.8.1. [http://dx.doi.org/10.1089/dna.1.1989.8.1]. [PMID: 2651058]. [DOI] [PubMed] [Google Scholar]
- 58.Nebert D.W., Nelson D.R., Coon M.J., Estabrook R.W., Feyereisen R., Fujii-Kuriyama Y., Gonzalez F.J., Guengerich F.P., Gunsalus I.C., Johnson E.F. The P450 superfamily: update on new sequences, gene mapping, and recommended nomenclature. DNA Cell Biol. 1991;10(1):1–14. doi: 10.1089/dna.1991.10.1. [http://dx.doi.org/10.1089/dna. 1991.10.1]. [PMID: 1991046]. [DOI] [PubMed] [Google Scholar]
- 59.Nelson D.R., Kamataki T., Waxman D.J., Guengerich F.P., Estabrook R.W., Feyereisen R., Gonzalez F.J., Coon M.J., Gunsalus I.C., Gotoh O. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12(1):1–51. doi: 10.1089/dna.1993.12.1. [http://dx.doi.org/10.1089/dna.1993.12.1]. [PMID: 7678494]. [DOI] [PubMed] [Google Scholar]
- 60.Nelson D.R., Koymans L., Kamataki T., Stegeman J.J., Feyereisen R., Waxman D.J., Waterman M.R., Gotoh O., Coon M.J., Estabrook R.W., Gunsalus I.C., Nebert D.W. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics. 1996;6(1):1–42. doi: 10.1097/00008571-199602000-00002. [http://dx.doi. org/10.1097/00008571-199602000-00002]. [PMID: 8845856]. [DOI] [PubMed] [Google Scholar]
- 61.Glusman G., Bahar A., Sharon D., Pilpel Y., White J., Lancet D. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm. Genome. 2000;11(11):1016–1023. doi: 10.1007/s003350010196. [http://dx.doi.org/10.1007/s003350010196]. [PMID: 11063259]. [DOI] [PubMed] [Google Scholar]
- 62.Lancet. D.; Ben-Arie, N. Olfactory receptors. Curr. Biol. 1993;3(10):668–674. doi: 10.1016/0960-9822(93)90064-u. [http://dx.doi.org/10.1016/0960-9822(93)90064-U]. [PMID: 15335857]. [DOI] [PubMed] [Google Scholar]
- 63.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5(4):263–278. doi: 10.1038/nrn1365. [a]. [DOI] [PubMed] [Google Scholar]
- 64.Pace U., Hanski E., Salomon Y., Lancet D. Odorant-sensitive adenylate cyclase may mediate olfactory reception. Nature. 1985;316(6025):255–258. doi: 10.1038/316255a0. [http://dx.doi.org/10.1038/316255a0]. [PMID: 3927168]. [DOI] [PubMed] [Google Scholar]
- 65.Abaffy T., Matsunami H., Luetje C.W. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006;97(5):1506–1518. doi: 10.1111/j.1471-4159.2006.03859.x. [http://dx.doi.org/10.1111/j.1471-4159.2006. 03859.x]. [PMID: 16606354]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pace U., Lancet D. Olfactory GTP-binding protein: signal-transducing polypeptide of vertebrate chemosensory neurons. Proc. Natl. Acad. Sci. USA. 1986;83(13):4947–4951. doi: 10.1073/pnas.83.13.4947. [http://dx.doi.org/ 10.1073/pnas.83.13.4947]. [PMID: 3088569]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nakamura T., Gold G.H. A cyclic nucleotide-gated conductance in olfactory receptor cilia. Nature. 1987;325(6103):442–444. doi: 10.1038/325442a0. [http://dx.doi.org/10.1038/325442a0]. [PMID: 3027574]. [DOI] [PubMed] [Google Scholar]
- 68.Sklar P.B., Anholt R.R., Snyder S.H. The odorant-sensitive adenylate cyclase of olfactory receptor cells. Differential stimulation by distinct classes of odorants. J. Biol. Chem. 1986;261(33):15538–15543. [PMID: 3536906]. [PubMed] [Google Scholar]
- 69.Bockaert J., Pin J.P. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18(7):1723–1729. doi: 10.1093/emboj/18.7.1723. [http://dx.doi.org/10.1093/emboj/18.7.1723]. [PMID: 10202136]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malnic B., Hirono J., Sato T., Buck L.B. Combinatorial receptor codes for odors. Cell. 1999;96(5):713–723. doi: 10.1016/s0092-8674(00)80581-4. [http://dx.doi.org/ 10.1016/S0092-8674(00)80581-4]. [PMID: 10089886]. [DOI] [PubMed] [Google Scholar]
- 71.Krautwurst D., Yau K.W., Reed R.R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95(7):917–926. doi: 10.1016/s0092-8674(00)81716-x. [http://dx.doi.org/10.1016/S0092-8674(00)81716-X]. [PMID: 9875846]. [DOI] [PubMed] [Google Scholar]
- 72.Floriano W.B., Vaidehi N., Goddard W.A. III Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptors. Chem. Senses. 2004;29(4):269–290. doi: 10.1093/chemse/bjh030. [http://dx. doi.org/10.1093/chemse/bjh030]. [PMID: 15150141]. [DOI] [PubMed] [Google Scholar]
- 73.Shirokova E., Schmiedeberg K., Bedner P., Niessen H., Willecke K., Raguse J.D., Meyerhof W., Krautwurst D. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. J. Biol. Chem. 2005;280(12):11807–11815. doi: 10.1074/jbc.M411508200. [http://dx.doi.org/10.1074/jbc. M411508200]. [PMID: 15598656]. [DOI] [PubMed] [Google Scholar]
- 74.Abaffy T., Matsunami H., Luetje C.W. Functional analysis of a mammalian odorant receptor subfamily. J. Neurochem. 2006;97(5):1506–1518. doi: 10.1111/j.1471-4159.2006.03859.x. [http://dx.doi.org/10.1111/j.1471-4159.2006. 03859.x]. [PMID: 16606354]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grosmaitre X., Vassalli A., Mombaerts P., Shepherd G.M., Ma M. Odorant responses of olfactory sensory neurons expressing the odorant receptor MOR23: a patch clamp analysis in gene-targeted mice. Proc. Natl. Acad. Sci. USA. 2006;103(6):1970–1975. doi: 10.1073/pnas.0508491103. [http://dx.doi.org/10.1073/pnas.0508491103]. [PMID: 16446455]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Touhara K., Sengoku S., Inaki K., Tsuboi A., Hirono J., Sato T., Sakano H., Haga T. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc. Natl. Acad. Sci. USA. 1999;96(7):4040–4045. doi: 10.1073/pnas.96.7.4040. [http://dx.doi.org/10. 1073/pnas.96.7.4040]. [PMID: 10097159]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dong X., Han S., Zylka M.J., Simon M.I., Anderson D.J. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106(5):619–632. doi: 10.1016/s0092-8674(01)00483-4. [http://dx.doi. org/10.1016/S0092-8674(01)00483-4]. [PMID: 11551509]. [DOI] [PubMed] [Google Scholar]
- 78.Wilson S., Bergsma D. Orphan G-protein coupled receptors: novel drug targets for the pharmaceutical industry. Drug Des. Discov. 2000;17(2):105–114. [PMID: 11045900]. [PubMed] [Google Scholar]
- 79.Baldwin J.M. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12(4):1693–1703. doi: 10.1002/j.1460-2075.1993.tb05814.x. [http:// dx.doi.org/10.1002/j.1460-2075.1993.tb05814.x]. [PMID: 8385611]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boekhoff I., Breer H. Differential stimulation of second messenger pathways by distinct classes of odorants. Neurochem. Int. 1990;17(4):553–557. doi: 10.1016/0197-0186(90)90043-s. [http://dx.doi.org/10.1016/0197-0186(90) 90043-S]. [PMID: 20504658]. [DOI] [PubMed] [Google Scholar]
- 81.Katada S., Hirokawa T., Oka Y., Suwa M., Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J. Neurosci. 2005;25(7):1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [http://dx.doi.org/10.1523/JNEUROSCI. 4723-04.2005]. [PMID: 15716417]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katada S., Hirokawa T., Touhara K. Exploring the odorant binding site of a G-protein-coupled olfactory receptor. Curr. Comput. Aided Drug Des. 2008;4:123–131. [http://dx.doi.org/10.2174/ 157340908784533247]. [Google Scholar]
- 83.Wang J., Luthey-Schulten Z.A., Suslick K.S. Is the olfactory receptor a metalloprotein? Proc. Natl. Acad. Sci. USA. 2003;100(6):3035–3039. doi: 10.1073/pnas.262792899. [http://dx.doi.org/10.1073/pnas.262792899]. [PMID: 12610211]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abaffy T., Malhotra A., Luetje C.W. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J. Biol. Chem. 2007;282(2):1216–1224. doi: 10.1074/jbc.M609355200. [http://dx.doi.org/10.1074/jbc.M609355200]. [PMID: 17114180]. [DOI] [PubMed] [Google Scholar]
- 85.Wade F., Espagne A., Persuy M.A., Vidic J., Monnerie R., Merola F., Pajot-Augy E., Sanz G. Relationship between homo-oligomerization of a mammalian olfactory receptor and its activation state demonstrated by bioluminescence resonance energy transfer. J. Biol. Chem. 2011;286(17):15252–15259. doi: 10.1074/jbc.M110.184580. [http://dx. doi.org/10.1074/jbc.M110.184580]. [PMID: 21454689]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park P.S., Filipek S., Wells J.W., Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43(50):15643–15656. doi: 10.1021/bi047907k. [http://dx.doi.org/10.1021/ bi047907k]. [PMID: 15595821]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duncan R.R., Bergmann A., Cousin M.A., Apps D.K., Shipston M.J. Multi-dimensional time-correlated single photon counting (TCSPC) fluorescence lifetime imaging microscopy (FLIM) to detect FRET in cells. J. Microsc. 2004;215(Pt 1):1–12. doi: 10.1111/j.0022-2720.2004.01343.x. [http://dx. doi.org/10.1111/j.0022-2720.2004.01343.x]. [PMID: 15230870]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfleger K.D., Eidne K.A. Monitoring the formation of dynamic G-protein-coupled receptor-protein complexes in living cells. Biochem. J. 2005;385(Pt 3):625–637. doi: 10.1042/BJ20041361. [http://dx.doi.org/10.1042/ BJ20041361]. [PMID: 15504107]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kaupmann K., Malitschek B., Schuler V., Heid J., Froestl W., Beck P., Mosbacher J., Bischoff S., Kulik A., Shigemoto R., Karschin A., Bettler B. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature. 1998;396(6712):683–687. doi: 10.1038/25360. [http://dx.doi.org/10.1038/25360]. [PMID: 9872317]. [DOI] [PubMed] [Google Scholar]
- 90.Milligan G., Wilson S., López-Gimenez J.F. The specificity and molecular basis of α1-adrenoceptor and CXCR chemokine receptor dimerization. J. Mol. Neurosci. 2005;26(2-3):161–168. doi: 10.1385/JMN:26:2-3:161. [http://dx.doi.org/10.1385/JMN:26:2-3:161]. [PMID: 16012189]. [DOI] [PubMed] [Google Scholar]
- 91.Hague C., Hall R.A., Minneman K.P. Olfactory receptor localization and function: an emerging role for GPCR heterodimerization. Mol. Interv. 2004;4(6):321–322. doi: 10.1124/mi.4.6.4. [http://dx.doi.org/10.1124/mi.4.6.4]. [PMID: 15616160]. [DOI] [PubMed] [Google Scholar]
- 92.Neuhaus E.M., Gisselmann G., Zhang W., Dooley R., Störtkuhl K., Hatt H. Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat. Neurosci. 2005;8(1):15–17. doi: 10.1038/nn1371. [http://dx.doi.org/10.1038/nn1371]. [PMID: 15592462]. [DOI] [PubMed] [Google Scholar]
- 93.Minic J., Persuy M.A., Godel E., Aioun J., Connerton I., Salesse R., Pajot-Augy E. Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J. 2005;272(2):524–537. doi: 10.1111/j.1742-4658.2004.04494.x. [http://dx.doi.org/10.1111/ j.1742-4658.2004.04494.x]. [PMID: 15654890]. [DOI] [PubMed] [Google Scholar]
- 94.Pajot-Augy E., Crowe M., Levasseur G., Salesse R., Connerton I. Engineered yeasts as reporter systems for odorant detection. J. Recept. Signal Transduct. Res. 2003;23(2-3):155–171. doi: 10.1081/rrs-120025196. [http://dx. doi.org/10.1081/RRS-120025196]. [PMID: 14626444]. [DOI] [PubMed] [Google Scholar]
- 95.John C. Olfaction. Leffingwell Reports. 2002;2(1):1–34. [Google Scholar]
- 96.Breer H., Boekhoff I., Tareilus E. Rapid kinetics of second messenger formation in olfactory transduction. Nature. 1990;345(6270):65–68. doi: 10.1038/345065a0. [http://dx.doi.org/10.1038/345065a0]. [PMID: 2158631]. [DOI] [PubMed] [Google Scholar]
- 97.Farrens D.L., Altenbach C., Yang K., Hubbell W.L., Khorana H.G. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274(5288):768–770. doi: 10.1126/science.274.5288.768. [http://dx.doi.org/10.1126/science.274.5288.768]. [PMID: 8864113]. [DOI] [PubMed] [Google Scholar]
- 98.Sheikh S.P., Zvyaga T.A., Lichtarge O., Sakmar T.P., Bourne H.R. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383(6598):347–350. doi: 10.1038/383347a0. [http://dx.doi.org/10.1038/383347a0]. [PMID: 8848049]. [DOI] [PubMed] [Google Scholar]
- 99.Ballesteros J.A., Jensen A.D., Liapakis G., Rasmussen S.G., Shi L., Gether U., Javitch J.A. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem. 2001;276(31):29171–29177. doi: 10.1074/jbc.M103747200. [http://dx.doi.org/10.1074/jbc.M103747200]. [PMID: 11375997]. [DOI] [PubMed] [Google Scholar]
- 100.Salom D., Lodowski D.T., Stenkamp R.E., Le Trong I., Golczak M., Jastrzebska B., Harris T., Ballesteros J.A., Palczewski K. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. USA. 2006;103(44):16123–16128. doi: 10.1073/pnas.0608022103. [http://dx.doi.org/10.1073/pnas.0608022103]. [PMID: 17060607]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kato A., Katada S., Touhara K. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. J. Neurochem. 2008;107(5):1261–1270. doi: 10.1111/j.1471-4159.2008.05693.x. [http://dx.doi.org/10.1111/j.1471-4159.2008.05693.x]. [PMID: 18803693]. [DOI] [PubMed] [Google Scholar]
- 102.Kaupp U.B. Olfactory signalling in vertebrates and insects: differences and commonalities. Nat. Rev. Neurosci. 2010;11(3):188–200. doi: 10.1038/nrn2789. [http://dx.doi.org/10.1038/nrn2789]. [PMID: 20145624]. [DOI] [PubMed] [Google Scholar]
- 103.Wong S.T., Trinh K., Hacker B., Chan G.C., Lowe G., Gaggar A., Xia Z., Gold G.H., Storm D.R. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27(3):487–497. doi: 10.1016/s0896-6273(00)00060-x. [http://dx.doi. org/10.1016/S0896-6273(00)00060-X]. [PMID: 11055432]. [DOI] [PubMed] [Google Scholar]
- 104.Belluscio L., Gold G.H., Nemes A., Axel R. Mice deficient in G(olf) are anosmic. Neuron. 1998;20(1):69–81. doi: 10.1016/s0896-6273(00)80435-3. [http://dx.doi.org/ 10.1016/S0896-6273(00)80435-3]. [PMID: 9459443]. [DOI] [PubMed] [Google Scholar]
- 105.Brunet L.J., Gold G.H., Ngai J. General anosmia caused by a targeted disruption of the mouse olfactory cyclic nucleotide-gated cation channel. Neuron. 1996;17(4):681–693. doi: 10.1016/s0896-6273(00)80200-7. [http://dx.doi.org/ 10.1016/S0896-6273(00)80200-7]. [PMID: 8893025]. [DOI] [PubMed] [Google Scholar]
- 106.Zheng J., Zagotta W.N. Stoichiometry and assembly of olfactory cyclic nucleotide-gated channels. Neuron. 2004;42(3):411–421. doi: 10.1016/s0896-6273(04)00253-3. [http://dx.doi.org/10.1016/S0896-6273(04)00253-3]. [PMID: 15134638]. [DOI] [PubMed] [Google Scholar]
- 107.Kato A., Touhara K. Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell. Mol. Life Sci. 2009;66(23):3743–3753. doi: 10.1007/s00018-009-0111-6. [http://dx.doi.org/10.1007/s00018-009-0111-6]. [PMID: 19652915]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schroeder B.C., Cheng T., Jan Y.N., Jan L.Y. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134(6):1019–1029. doi: 10.1016/j.cell.2008.09.003. [http://dx.doi.org/10.1016/j.cell. 2008.09.003]. [PMID: 18805094]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang Y.D., Cho H., Koo J.Y., Tak M.H., Cho Y., Shim W.S., Park S.P., Lee J., Lee B., Kim B.M., Raouf R., Shin Y.K., Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008;455(7217):1210–1215. doi: 10.1038/nature07313. [http://dx. doi.org/10.1038/nature07313]. [PMID: 18724360]. [DOI] [PubMed] [Google Scholar]
- 110.Bradley J., Reisert J., Frings S. Regulation of cyclic nucleotide-gated channels. Curr. Opin. Neurobiol. 2005;15(3):343–349. doi: 10.1016/j.conb.2005.05.014. [http://dx.doi.org/10.1016/j.conb.2005.05.014]. [PMID: 15922582]. [DOI] [PubMed] [Google Scholar]
- 111.Liman E.R., Buck L.B. A second subunit of the olfactory cyclic nucleotide-gated channel confers high sensitivity to cAMP. Neuron. 1994;13(3):611–621. doi: 10.1016/0896-6273(94)90029-9. [http://dx.doi.org/10.1016/0896-6273 (94)90029-9]. [PMID: 7522482]. [DOI] [PubMed] [Google Scholar]
- 112.Prasad B.C., Reed R.R. Chemosensation: molecular mechanisms in worms and mammals. Trends Genet. 1999;15(4):150–153. doi: 10.1016/s0168-9525(99)01695-9. [http:// dx.doi.org/10.1016/S0168-9525(99)01695-9]. [PMID: 10203825]. [DOI] [PubMed] [Google Scholar]
- 113.Reisert J., Lai J., Yau K.W., Bradley J. Mechanism of the excitatory Cl- response in mouse olfactory receptor neurons. Neuron. 2005;45(4):553–561. doi: 10.1016/j.neuron.2005.01.012. [http://dx.doi.org/10.1016/j.neuron.2005. 01.012]. [PMID: 15721241]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kurahashi T., Yau K.W. Co-existence of cationic and chloride components in odorant-induced current of vertebrate olfactory receptor cells. Nature. 1993;363(6424):71–74. doi: 10.1038/363071a0. [http://dx.doi.org/ 10.1038/363071a0]. [PMID: 7683113]. [DOI] [PubMed] [Google Scholar]
- 115.Schild D., Restrepo D. Transduction mechanisms in vertebrate olfactory receptor cells. Physiol. Rev. 1998;78(2):429–466. doi: 10.1152/physrev.1998.78.2.429. [http:// dx.doi.org/10.1152/physrev.1998.78.2.429]. [PMID: 9562035]. [DOI] [PubMed] [Google Scholar]
- 116.Antunes G., Sebastião A.M., Simoes de Souza F.M. Mechanisms of regulation of olfactory transduction and adaptation in the olfactory cilium. PLoS One. 2014;9(8):e105531. doi: 10.1371/journal.pone.0105531. [http://dx.doi.org/ 10.1371/journal.pone.0105531]. [PMID: 25144232]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kurahashi T., Menini A. Mechanism of odorant adaptation in the olfactory receptor cell. Nature. 1997;385(6618):725–729. doi: 10.1038/385725a0. [http:// dx.doi.org/10.1038/385725a0]. [PMID: 9034189]. [DOI] [PubMed] [Google Scholar]
- 118.Barnea G., O’Donnell S., Mancia F., Sun X., Nemes A., Mendelsohn M., Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304(5676):1468. doi: 10.1126/science.1096146. [http://dx.doi.org/10.1126/ science.1096146]. [PMID: 15178793]. [DOI] [PubMed] [Google Scholar]
- 119.Buck L.B. The search for odorant receptors. 2004. [DOI] [PubMed]
- 120.Eggan K., Baldwin K., Tackett M., Osborne J., Gogos J., Chess A., Axel R., Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428(6978):44–49. doi: 10.1038/nature02375. [http://dx.doi.org/10.1038/ nature02375]. [PMID: 14990966]. [DOI] [PubMed] [Google Scholar]
- 121.Benovic J.L., Kühn H., Weyand I., Codina J., Caron M.G., Lefkowitz R.J. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc. Natl. Acad. Sci. USA. 1987;84(24):8879–8882. doi: 10.1073/pnas.84.24.8879. [http://dx. doi.org/10.1073/pnas.84.24.8879]. [PMID: 2827157]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lohse M.J., Benovic J.L., Codina J., Caron M.G., Lefkowitz R.J. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248(4962):1547–1550. doi: 10.1126/science.2163110. [http://dx.doi.org/ 10.1126/science.2163110]. [PMID: 2163110]. [DOI] [PubMed] [Google Scholar]
- 123.Boekhoff I., Inglese J., Schleicher S., Koch W.J., Lefkowitz R.J., Breer H. Olfactory desensitization requires membrane targeting of receptor kinase mediated by beta gamma-subunits of heterotrimeric G proteins. J. Biol. Chem. 1994;269(1):37–40. [PMID: 8276821]. [PubMed] [Google Scholar]
- 124.Peppel K., Boekhoff I., McDonald P., Breer H., Caron M.G., Lefkowitz R.J. G protein-coupled receptor kinase 3 (GRK3) gene disruption leads to loss of odorant receptor desensitization. J. Biol. Chem. 1997;272(41):25425–25428. doi: 10.1074/jbc.272.41.25425. [http://dx.doi.org/10.1074/ jbc.272.41.25425]. [PMID: 9325250]. [DOI] [PubMed] [Google Scholar]
- 125.Mashukova A., Spehr M., Hatt H., Neuhaus E.M. Beta-arrestin2-mediated internalization of mammalian odorant receptors. J. Neurosci. 2006;26(39):9902–9912. doi: 10.1523/JNEUROSCI.2897-06.2006. [http://dx.doi.org/10.1523/ JNEUROSCI.2897-06.2006]. [PMID: 17005854]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Neuhaus E.M., Mashukova A., Barbour J., Wolters D., Hatt H. Novel function of beta-arrestin2 in the nucleus of mature spermatozoa. J. Cell Sci. 2006;119(Pt 15):3047–3056. doi: 10.1242/jcs.03046. [http://dx.doi.org/ 10.1242/jcs.03046]. [PMID: 16820410]. [DOI] [PubMed] [Google Scholar]
- 127.Pelosi P., Pisanelli A.M., Badaccini N.E., Gagliardo A. Binding of [3H]-2-isobutyl-3-methoxypyrazine to cow olfactory mucosa. Chem. Senses. 1981;6:77–85. [http://dx.doi.org/10.1093/chemse/ 6.2.77]. [Google Scholar]
- 128.Pes D., Pelosi P. Odorant-binding proteins of the mouse. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995;112(3):471–479. doi: 10.1016/0305-0491(95)00063-1. [http://dx.doi.org/10.1016/0305-0491(95)00063-1]. [PMID: 8529023]. [DOI] [PubMed] [Google Scholar]
- 129.Pelosi P. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 1994;29(3):199–228. doi: 10.3109/10409239409086801. [http://dx.doi.org/10.3109/10409239409086801]. [PMID: 8070277]. [DOI] [PubMed] [Google Scholar]
- 130.Pevsner J., Snynder S.H. Odorant-binding protein; odorant transport function in the vertebrate nasal epithelium. Chem. Senses. 1990;15:217–222. [http://dx.doi.org/10.1093/chemse/15.2.217]. [Google Scholar]
- 131.Bignetti E., Cavaggioni A., Pelosi P., Persaud K.C., Sorbi R.T., Tirindelli R. Purification and characterisation of an odorant-binding protein from cow nasal tissue. Eur. J. Biochem. 1985;149(2):227–231. doi: 10.1111/j.1432-1033.1985.tb08916.x. [http://dx.doi.org/10.1111/j.1432-1033.1985.tb08916. x]. [PMID: 3996407]. [DOI] [PubMed] [Google Scholar]
- 132.Pevsner J., Trifiletti R.R., Strittmatter S.M., Snyder S.H. Isolation and characterization of an olfactory receptor protein for odorant pyrazines. Proc. Natl. Acad. Sci. USA. 1985;82(9):3050–3054. doi: 10.1073/pnas.82.9.3050. [http://dx.doi.org/10.1073/pnas.82.9.3050]. [PMID: 2986147]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mulla M.Y., Tuccori E., Magliulo M., Lattanzi G., Palazzo G., Persaud K., Torsi L. Capacitance-modulated transistor detects odorant binding protein chiral interactions. Nat. Commun. 2015;6:6010. doi: 10.1038/ncomms7010. [http://dx.doi.org/10.1038/ncomms7010]. [PMID: 25591754]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Meierhenrich U.J., Golebiowski J., Fernandez X., Cabrol-Bass D. The molecular basis of olfactory chemoreception. Angew. Chem. Int. Ed. Engl. 2004;43(47):6410–6412. doi: 10.1002/anie.200462322. [http://dx.doi.org/10.1002/ anie.200462322]. [PMID: 15578781]. [DOI] [PubMed] [Google Scholar]
- 135.Charlier L., Cabrol-Bass D., Golebiowski J. How does human odorant binding protein bind odorants? The case of aldehydes studied by molecular dynamics. C. R. Chim. 2009;12:905–910. [http://dx.doi.org/10.1016/j.crci.2008.09.022]. [Google Scholar]
- 136.Tegoni M., Ramoni R., Bignetti E., Spinelli S., Cambillau C. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nat. Struct. Biol. 1996;3(10):863–867. doi: 10.1038/nsb1096-863. [http://dx.doi.org/10.1038/nsb1096-863]. [PMID: 8836103]. [DOI] [PubMed] [Google Scholar]
- 137.Vincent F., Spinelli S., Ramoni R., Grolli S., Pelosi P., Cambillau C., Tegoni M. Complexes of porcine odorant binding protein with odorant molecules belonging to different chemical classes. J. Mol. Biol. 2000;300(1):127–139. doi: 10.1006/jmbi.2000.3820. [http://dx.doi.org/10.1006/jmbi. 2000.3820]. [PMID: 10864504]. [DOI] [PubMed] [Google Scholar]
- 138.Löbel D., Strotmann J., Jacob M., Breer H. Identification of a third rat odorant-binding protein (OBP3). Chem. Senses. 2001;26(6):673–680. doi: 10.1093/chemse/26.6.673. [http://dx.doi.org/10.1093/chemse/26.6.673]. [PMID: 11473933]. [DOI] [PubMed] [Google Scholar]
- 139.Löbel D., Marchese S., Krieger J., Pelosi P., Breer H. Subtypes of odorant-binding proteins--heterologous expression and ligand binding. Eur. J. Biochem. 1998;254(2):318–324. doi: 10.1046/j.1432-1327.1998.2540318.x. [http://dx.doi. org/10.1046/j.1432-1327.1998.2540318.x]. [PMID: 9660186]. [DOI] [PubMed] [Google Scholar]
- 140.Wojnar P., Lechner M., Merschak P., Redl B. Molecular cloning of a novel lipocalin-1 interacting human cell membrane receptor using phage display. J. Biol. Chem. 2001;276(23):20206–20212. doi: 10.1074/jbc.M101762200. [http://dx.doi.org/10.1074/jbc.M101762200]. [PMID: 11287427]. [DOI] [PubMed] [Google Scholar]
- 141.Munger S.D., Leinders-Zufall T., Zufall F. Subsystem organization of the mammalian sense of smell. Annu. Rev. Physiol. 2009;71:115–140. doi: 10.1146/annurev.physiol.70.113006.100608. [http://dx.doi.org/10.1146/annurev.physiol.70.113006. 100608]. [PMID: 18808328]. [DOI] [PubMed] [Google Scholar]
- 142.Spehr M., Spehr J., Ukhanov K., Kelliher K.R., Leinders-Zufall T., Zufall F. Parallel processing of social signals by the mammalian main and accessory olfactory systems. Cell. Mol. Life Sci. 2006;63(13):1476–1484. doi: 10.1007/s00018-006-6109-4. [http://dx.doi.org/10.1007/s00018-006-6109-4]. [PMID: 16732428]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lin W., Arellano J., Slotnick B., Restrepo D. Odors detected by mice deficient in cyclic nucleotide-gated channel subunit A2 stimulate the main olfactory system. J. Neurosci. 2004;24(14):3703–3710. doi: 10.1523/JNEUROSCI.0188-04.2004. [http://dx.doi.org/10.1523/JNEUROSCI.0188-04.2004]. [PMID: 15071119]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liberles S.D., Buck L.B. A second class of chemosensory receptors in the olfactory epithelium. Nature. 2006;442(7103):645–650. doi: 10.1038/nature05066. [http://dx.doi.org/10.1038/nature05066]. [PMID: 16878137]. [DOI] [PubMed] [Google Scholar]
- 145.Wallrabenstein I., Kuklan J., Weber L., Zborala S., Werner M., Altmüller J., Becker C., Schmidt A., Hatt H., Hummel T., Gisselmann G. Human trace amine-associated receptor TAAR5 can be activated by trimethylamine. PLoS One. 2013;8(2):e54950. doi: 10.1371/journal.pone.0054950. [http://dx.doi.org/10.1371/journal.pone.0054950]. [PMID: 23393561]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liberles S.D. Trace amine-associated receptors: ligands, neural circuits, and behaviors. Curr. Opin. Neurobiol. 2015;34:1–7. doi: 10.1016/j.conb.2015.01.001. [http://dx.doi.org/10.1016/j.conb.2015.01.001]. [PMID: 25616211]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fulle H.J. Vassar. R.; Foster, D.C.; Yang, R.B.; Axel, R.; Garbers, D.L. A receptor guanylul cyclase expressed. specifically in olfactory sensory neurons. Proc. Natl. Acad. Sci. USA. 1995;92:3571–3575. doi: 10.1073/pnas.92.8.3571. [http://dx.doi.org/10.1073/pnas.92.8.3571]. [PMID: 7724600]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gibson A.D., Garbers D.L. Guanylyl cyclases as a family of putative odorant receptors. Annu. Rev. Neurosci. 2000;23:417–439. doi: 10.1146/annurev.neuro.23.1.417. [http://dx.doi.org/10.1146/annurev.neuro.23.1.417]. [PMID: 10845070]. [DOI] [PubMed] [Google Scholar]
- 149.Borowsky B., Adham N., Jones K.A., Raddatz R., Artymyshyn R., Ogozalek K.L., Durkin M.M., Lakhlani P.P., Bonini J.A., Pathirana S., Boyle N., Pu X., Kouranova E., Lichtblau H., Ochoa F.Y., Branchek T.A., Gerald C. Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 2001;98(16):8966–8971. doi: 10.1073/pnas.151105198. [http://dx.doi.org/ 10.1073/pnas.151105198]. [PMID: 11459929]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bunzow J.R., Sonders M.S., Arttamangkul S., Harrison L.M., Zhang G., Quigley D.I., Darland T., Suchland K.L., Pasumamula S., Kennedy J.L., Olson S.B., Magenis R.E., Amara S.G., Grandy D.K. Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol. Pharmacol. 2001;60(6):1181–1188. doi: 10.1124/mol.60.6.1181. [http://dx.doi.org/10.1124/ mol.60.6.1181]. [PMID: 11723224]. [DOI] [PubMed] [Google Scholar]
- 151.Lindemann L., Hoener M.C. A renaissance in trace amines inspired by a novel GPCR family. Trends Pharmacol. Sci. 2005;26(5):274–281. doi: 10.1016/j.tips.2005.03.007. [http://dx.doi.org/10.1016/j.tips.2005.03.007]. [PMID: 15860375]. [DOI] [PubMed] [Google Scholar]
- 152.Shi L., Javitch J.A. The binding site of aminergic G protein-coupled receptors: the transmembrane segments and second extracellular loop. Annu. Rev. Pharmacol. Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [http://dx.doi.org/10.1146/annurev.pharmtox.42.091101.144224]. [PMID: 11807179]. [DOI] [PubMed] [Google Scholar]
- 153.Horowitz L.F., Saraiva L.R., Kuang D., Yoon K.H., Buck L.B. Olfactory receptor patterning in a higher primate. J. Neurosci. 2014;34(37):12241–12252. doi: 10.1523/JNEUROSCI.1779-14.2014. [http://dx.doi.org/10.1523/JNEUROSCI. 1779-14.2014]. [PMID: 25209267]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Grus W.E., Zhang J. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol. Biol. Evol. 2008;25(8):1593–1601. doi: 10.1093/molbev/msn107. [http://dx.doi.org/10.1093/molbev/msn107]. [PMID: 18460446]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hashiguchi Y., Nishida M. Evolution of trace amine associated receptor (TAAR) gene family in vertebrates: lineage-specific expansions and degradations of a second class of vertebrate chemosensory receptors expressed in the olfactory epithelium. Mol. Biol. Evol. 2007;24(9):2099–2107. doi: 10.1093/molbev/msm140. [http://dx.doi.org/10.1093/ molbev/msm140]. [PMID: 17634392]. [DOI] [PubMed] [Google Scholar]
- 156.Nei M., Niimura Y., Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 2008;9(12):951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- 157.Zhang J., Pacifico R., Cawley D., Feinstein P., Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. J. Neurosci. 2013;33(7):3228–3239. doi: 10.1523/JNEUROSCI.4299-12.2013. [http://dx.doi.org/10.1523/ JNEUROSCI.4299-12.2013]. [PMID: 23407976]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Greer P.L., Bear D.M., Lassance J.M., Bloom M.L., Tsukahara T., Pashkovski S.L., Masuda F.K., Nowlan A.C., Kirchner R., Hoekstra H.E., Datta S.R. A Family of non-GPCR Chemosensors Defines an Alternative Logic for Mammalian Olfaction. Cell. 2016;165(7):1734–1748. doi: 10.1016/j.cell.2016.05.001. [http://dx.doi.org/10.1016/j.cell.2016.05.001]. [PMID: 27238024]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hu J., Zhong C., Ding C., Chi Q., Walz A., Mombaerts P., Matsunami H., Luo M. Detection of near-atmospheric concentrations of CO2 by an olfactory subsystem in the mouse. Science. 2007;317(5840):953–957. doi: 10.1126/science.1144233. [http://dx.doi.org/10.1126/science. 1144233]. [PMID: 17702944]. [DOI] [PubMed] [Google Scholar]
- 160.Juilfs D.M., Fülle H.J., Zhao A.Z., Houslay M.D., Garbers D.L., Beavo J.A. A subset of olfactory neurons that selectively express cGMP-stimulated phosphodiesterase (PDE2) and guanylyl cyclase-D define a unique olfactory signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1997;94(7):3388–3395. doi: 10.1073/pnas.94.7.3388. [http://dx.doi.org/ 10.1073/pnas.94.7.3388]. [PMID: 9096404]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leinders-Zufall T., Cockerham R.E., Michalakis S., Biel M., Garbers D.L., Reed R.R., Zufall F., Munger S.D. Contribution of the receptor guanylyl cyclase GC-D to chemosensory function in the olfactory epithelium. Proc. Natl. Acad. Sci. USA. 2007;104(36):14507–14512. doi: 10.1073/pnas.0704965104. [http://dx.doi.org/10.1073/pnas.0704965104]. [PMID: 17724338]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Meyer M.R., Angele A., Kremmer E., Kaupp U.B., Muller F. A cGMP-signaling pathway in a subset of olfactory sensory neurons. Proc. Natl. Acad. Sci. USA. 2000;97(19):10595–10600. doi: 10.1073/pnas.97.19.10595. [http://dx. doi.org/10.1073/pnas.97.19.10595]. [PMID: 10984544]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo D., Zhang J.J., Huang X.Y. Stimulation of guanylyl cyclase-D by bicarbonate. Biochemistry. 2009;48(20):4417–4422. doi: 10.1021/bi900441v. [http:// dx.doi.org/10.1021/bi900441v]. [PMID: 19331426]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sun L., Wang H., Hu J., Han J., Matsunami H., Luo M. Guanylyl cyclase-D in the olfactory CO2 neurons is activated by bicarbonate. Proc. Natl. Acad. Sci. USA. 2009;106(6):2041–2046. doi: 10.1073/pnas.0812220106. [http://dx.doi.org/10.1073/pnas.0812220106]. [PMID: 19181845]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chastrette M. Trends in structure-odor relationships. SAR QSAR Environ. Res. 1997;6(3-4):215–254. doi: 10.1080/10629369708033253. [http://dx.doi.org/10.1080/ 10629369708033253]. [PMID: 9487700]. [DOI] [PubMed] [Google Scholar]
- 166.Laska M., Trolp S., Teubner P. Odor structure-activity relationships compared in human and nonhuman primates. Behav. Neurosci. 1999;113(5):998–1007. doi: 10.1037//0735-7044.113.5.998. [http://dx.doi.org/10.1037/0735-7044. 113.5.998]. [PMID: 10571482]. [DOI] [PubMed] [Google Scholar]
- 167.Ohloff G., Pickenhagen W., Kraft P. Scent and Chemistry The molecular world of Odors. 1st ed. Wiley-VCH; 2001. [Google Scholar]
- 168.Dravnieks A. Current status of Odour Theories Flavour Chemistry. Adv. Chem. 1969;56:29–52. [http://dx.doi.org/10.1021/ba-1966-0056.ch002]. [Google Scholar]
- 169.Demole E., Wuest H. Synthèses stéréosélectives de deuxtrioxydes C18H30O3 stéréoisomères, d’ambréinolide et sclaréol-lactone a partir de derives du (+)-manool. Helv. Chim. Acta. 1969;50:1314–1322. [http://dx.doi.org/10.1002/hlca.19670500514]. [Google Scholar]
- 170.Zhang X., Rogers M., Tian H., Zhang X., Zou D.J., Liu J., Ma M., Shepherd G.M., Firestein S.J. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc. Natl. Acad. Sci. USA. 2004;101(39):14168–14173. doi: 10.1073/pnas.0405350101. [http://dx. doi.org/10.1073/pnas.0405350101]. [PMID: 15377787]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Feldmesser E., Olender T., Khen M., Yanai I., Ophir R., Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [http://dx.doi.org/10.1186/1471-2164-7-121]. [PMID: 16716209]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jones F.N., Margaret H. Modern Theories of Olfaction, a Critical Review. J. Psychol. 1953;36:207–241. [http://dx.doi.org/10. 1080/00223980.1953.9712890]. [Google Scholar]
- 173.Bourgeois A.E., Bourgeois J.O. Theories of Olfaction: A Review. Interam. J. Psychol. 2017;•••:19–31. [Google Scholar]
- 174.Wendt G.R. Somesthesis and the chemical senses. Annu. Rev. Psychol. 1952;3:105–130. doi: 10.1146/annurev.ps.03.020152.000541. [http://dx.doi.org/10.1146/annurev.ps. 03.020152.000541]. [PMID: 12977187]. [DOI] [PubMed] [Google Scholar]
- 175.Moncrieff R.W. The characterization of odours. J. Physiol. 1954;125(3):453–465. doi: 10.1113/jphysiol.1954.sp005172. [http://dx.doi.org/10.1113/jphysiol.1954.sp005172]. [PMID: 13212711]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Amoore J.E. Molecular Basis of Odor. Springfield: C.C. Thomas Pub.; 1970. [Google Scholar]
- 177.Davies J.T., Taylor F.H. The role of adsorption and molecular morphology in olfaction; the calculation of olfactory thresholds. Biol. Bull. 1959;117:222–238. [http://dx.doi.org/10.2307/1538902]. [Google Scholar]
- 178.Wright R.H. The Sense of Smell. Boca Raton: CRC Press; 1982. [Google Scholar]
- 179.Moncrieff R.W. The Chemical Senses. New York: Wiley; 1946. [Google Scholar]
- 180.Gasser H.S. Olfactory nerve fibers. J. Gen. Physiol. 1956;39(4):473–496. doi: 10.1085/jgp.39.4.473. [http://dx.doi.org/10.1085/jgp.39.4.473]. [PMID: 13295549]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Beck L.H., Miles W.R. Some theoretical and experimental relationships between infra-red absorption and olfaction. Science. 1947;106:511. [PMID: 20340842]. [Google Scholar]
- 182.Morgan C.T., Stellar E. Physiological Psychology. New York: McGraw- Hill; 1950. pp. 109–116. [Google Scholar]
- 183.Leffingwell J.C. Comment in Gustation and Olfaction. NY: Academic Press; 1971. [Google Scholar]
- 184.Langenau E.E. Olfaction and Taste. Vol. III New York: Rockefeller University Press; 1967. [Google Scholar]
- 185.Wright R.H. Odour and molecular vibration. Nature. 1961;190:1101–1102. doi: 10.1038/209571a0. [http://dx.doi.org/10.1038/1901101a0]. [PMID: 5921179]. [DOI] [PubMed] [Google Scholar]
- 186.Young C.W., Pletcher D.E., Wright N. On olfaction and infrared radiation theories. Science. 1948;108(2807):411–412. doi: 10.1126/science.108.2807.411-a. [http://dx. doi.org/10.1126/science.108.2807.411-a]. [PMID: 17782696]. [DOI] [PubMed] [Google Scholar]
- 187.Stemp E.D., Barton J.K. Electron transfer between metal complexes bound to DNA: is DNA a wire? Met. Ions Biol. Syst. 1996;33:325–365. [PMID: 8742848]. [PubMed] [Google Scholar]
- 188.Turin L. A spectroscopic mechanism for primary olfactory reception. Chem. Senses. 1996;21(6):773–791. doi: 10.1093/chemse/21.6.773. [http://dx.doi.org/10. 1093/chemse/21.6.773]. [PMID: 8985605]. [DOI] [PubMed] [Google Scholar]
- 189.Geldard F.A. Somesthesis and the chemical senses. Annu. Rev. Psychol. 1950;1:71–86. doi: 10.1146/annurev.ps.01.020150.000443. [http://dx.doi.org/10.1146/annurev.ps.01. 020150.000443]. [PMID: 14771867]. [DOI] [PubMed] [Google Scholar]
- 190.Mullins L.J. Olfaction. Ann. N. Y. Acad. Sci. 1955;62:247–276. [http://dx.doi.org/10.1111/j.1749-6632.1955.tb35354.x]. [Google Scholar]
- 191.Amoore J.E. Stereochemical theory of olfaction. Nature. 1963;198:271–272. doi: 10.1038/198271a0. [http://dx.doi.org/10.1038/198271a0]. [PMID: 14012641]. [DOI] [PubMed] [Google Scholar]
- 192.Topiol S., Sabio M. X-ray structure breakthroughs in the GPCR transmembrane region. Biochem. Pharmacol. 2009;78(1):11–20. doi: 10.1016/j.bcp.2009.02.012. [http://dx.doi.org/10.1016/j.bcp.2009.02.012]. [PMID: 19447219]. [DOI] [PubMed] [Google Scholar]
- 193.Crasto C.J. Computational Biology of Olfactory Receptors. Curr. Bioinform. 2009;4(1):8–15. doi: 10.2174/157489309787158143. [http://dx.doi.org/10.2174/ 157489309787158143]. [PMID: 21984880]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Peng Y., McCorvy J.D., Harpsøe K., Lansu K., Yuan S., Popov P., Qu L., Pu M., Che T., Nikolajsen L.F., Huang X.P., Wu Y., Shen L., Bjørn-Yoshimoto W.E., Ding K., Wacker D., Han G.W., Cheng J., Katritch V., Jensen A.A., Hanson M.A., Zhao S., Gloriam D.E., Roth B.L., Stevens R.C., Liu Z.J. 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. Cell. 2018;172(4):719–730.e14. doi: 10.1016/j.cell.2018.01.001. [http://dx.doi.org/ 10.1016/j.cell.2018.01.001]. [PMID: 29398112]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wang S., Che T., Levit A., Shoichet B.K., Wacker D., Roth B.L. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature. 2018;555(7695):269–273. doi: 10.1038/nature25758. [http://dx.doi.org/10.1038/nature25758]. [PMID: 29466326]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yang Z., Han S., Keller M., Kaiser A., Bender B.J., Bosse M., Burkert K., Kögler L.M., Wifling D., Bernhardt G., Plank N., Littmann T., Schmidt P., Yi C., Li B., Ye S., Zhang R., Xu B., Larhammar D., Stevens R.C., Huster D., Meiler J., Zhao Q., Beck-Sickinger A.G., Buschauer A., Wu B. Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor. Nature. 2018;556(7702):520–524. doi: 10.1038/s41586-018-0046-x. [http://dx.doi.org/10.1038/s41586-018-0046-x]. [PMID: 29670288]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Palczewski K., Kumasaka T., Hori T., Behnke C.A., Motoshima H., Fox B.A., Le Trong I., Teller D.C., Okada T., Stenkamp R.E., Yamamoto M., Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289(5480):739–745. doi: 10.1126/science.289.5480.739. [http://dx.doi.org/10.1126/science.289.5480.739]. [PMID: 10926528]. [DOI] [PubMed] [Google Scholar]
- 198.Teller D.C., Okada T., Behnke C.A., Palczewski K., Stenkamp R.E. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs). Biochemistry. 2001;40(26):7761–7772. doi: 10.1021/bi0155091. [http://dx.doi.org/10.1021/bi0155091]. [PMID: 11425302]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Cherezov V., Rosenbaum D.M., Hanson M.A., Rasmussen S.G., Thian F.S., Kobilka T.S., Choi H.J., Kuhn P., Weis W.I., Kobilka B.K., Stevens R.C. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318(5854):1258–1265. doi: 10.1126/science.1150577. [http://dx.doi.org/10.1126/ science.1150577]. [PMID: 17962520]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rasmussen S.G., Choi H.J., Rosenbaum D.M., Kobilka T.S., Thian F.S., Edwards P.C., Burghammer M., Ratnala V.R., Sanishvili R., Fischetti R.F., Schertler G.F., Weis W.I., Kobilka B.K. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450(7168):383–387. doi: 10.1038/nature06325. [http://dx. doi.org/10.1038/nature06325]. [PMID: 17952055]. [DOI] [PubMed] [Google Scholar]
- 201.Strader C.D., Fong T.M., Tota M.R., Underwood D., Dixon R.A.F. Structure and function of G protein-coupled receptors. Annu. Rev. Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [http://dx.doi.org/10. 1146/annurev.bi.63.070194.000533]. [PMID: 7979235]. [DOI] [PubMed] [Google Scholar]
- 202.Shacham S., Topf M., Avisar N., Glaser F., Marantz Y., Bar-Haim S., Noiman S., Naor Z., Becker O.M. Modeling the 3D structure of GPCRs from sequence. Med. Res. Rev. 2001;21(5):472–483. doi: 10.1002/med.1019. [http://dx.doi.org/10.1002/med.1019]. [PMID: 11579443]. [DOI] [PubMed] [Google Scholar]
- 203.Vaidehi N., Floriano W.B., Trabanino R., Hall S.E., Freddolino P., Choi E.J., Zamanakos G., Goddard W.A., III Prediction of structure and function of G protein-coupled receptors. Proc. Natl. Acad. Sci. USA. 2002;99(20):12622–12627. doi: 10.1073/pnas.122357199. [http://dx.doi.org/ 10.1073/pnas.122357199]. [PMID: 12351677]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Eisenberg D., Weiss R.M., Terwilliger T.C. The hydrophobic moment detects periodicity in protein hydrophobicity. Proc. Natl. Acad. Sci. USA. 1984;81(1):140–144. doi: 10.1073/pnas.81.1.140. [http://dx.doi.org/10.1073/ pnas.81.1.140]. [PMID: 6582470]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Floriano W.B., Vaidehi N., Goddard W.A., III, Singer M.S., Shepherd G.M. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc. Natl. Acad. Sci. USA. 2000;97(20):10712–10716. doi: 10.1073/pnas.97.20.10712. [http://dx.doi.org/10.1073/ pnas.97.20.10712]. [PMID: 11005853]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Floriano W.B., Nagarajan V., Goddard W.A., III 2001.
- 207.Datta D., Vaidehi N., Xu X., Goddard W.A., III Mechanism for antibody catalysis of the oxidation of water by singlet dioxygen. Proc. Natl. Acad. Sci. USA. 2002;99(5):2636–2641. doi: 10.1073/pnas.052709399. [http://dx. doi.org/10.1073/pnas.052709399]. [PMID: 11880618]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Anselmi C., Buonocore A., Centini M., Facino R.M., Hatt H. The human olfactory receptor 17-40: requisites for fitting into the binding pocket. Comput. Biol. Chem. 2011;35(3):159–168. doi: 10.1016/j.compbiolchem.2011.04.011. [http:// dx.doi.org/10.1016/j.compbiolchem.2011.04.011]. [PMID: 21704262]. [DOI] [PubMed] [Google Scholar]
- 209.Lai P.C., Singer M.S., Crasto C.J. Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem. Senses. 2005;30(9):781–792. doi: 10.1093/chemse/bji070. [http://dx.doi.org/10.1093/chemse/ bji070]. [PMID: 16243965]. [DOI] [PubMed] [Google Scholar]
- 210.Lai P.C., Crasto C.J. Beyond modeling: all-atom olfactory receptor model simulations. Front. Genet. 2012;3:61. doi: 10.3389/fgene.2012.00061. [http://dx. doi.org/10.3389/fgene.2012.00061]. [PMID: 22563330]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lai P.C., Guida B., Shi J., Crasto C.J. Preferential binding of an odor within olfactory receptors: a precursor to receptor activation. Chem. Senses. 2014;39(2):107–123. doi: 10.1093/chemse/bjt060. [http://dx.doi.org/10.1093/ chemse/bjt060]. [PMID: 24398973]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu N., Crasto C.J., Ma M. Integrated olfactory receptor and microarray gene expression databases. BMC Bioinformatics. 2007;8:231. doi: 10.1186/1471-2105-8-231. [http://dx.doi.org/10.1186/1471-2105-8-231]. [PMID: 17603910]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. mdl.shsmu.edu.cn/ODORactor
- 214.Modena D. Trentini, M.; Corsini, M.; Bombaci, A.; Giorgetti, A. Olfaction D.B: A database of olfactory receptors and their ligands. Adv. Life Sci. 2011;1:1–5. [http://dx.doi.org/10.5923/j.als. 20110101.01]. [Google Scholar]
- 215.Olender T., Nativ N., Lancet D. HORDE: Comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013;1003:23–38. doi: 10.1007/978-1-62703-377-0_2. [http://dx.doi.org/10.1007/978-1-62703-377-0_2]. [PMID: 23585031]. [DOI] [PubMed] [Google Scholar]
- 216.Liu X., Su X., Wang F., Huang Z., Wang Q., Li Z., Zhang R., Wu L., Pan Y., Chen Y., Zhuang H., Chen G., Shi T., Zhang J. ODO Ractor: a web server for deciphering olfactory coding. Bioinformatics. 2011;27(16):2302–2303. doi: 10.1093/bioinformatics/btr385. [http://dx.doi.org/10.1093/ bioinformatics/btr385]. [PMID: 21700676]. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.


