Abstract
Basonuclin (BNC1) is expressed primarily in proliferative keratinocytes and gametogenic cells. However, its roles in spermatogenesis and testicular aging were not clear. Previously we discovered a heterozygous BNC1 truncation mutation in a premature ovarian insufficiency pedigree. In this study, we found that male mice carrying the truncation mutation exhibited progressively fertility loss and testicular premature aging. Genome-wide expression profiling and direct binding studies (by chromatin immunoprecipitation sequencing) with BNC1 in mouse testis identified several spermatogenesis-specific gene promoters targeted by BNC1 including kelch-like family member 10 (Klhl10), testis expressed 14 (Tex14), and spermatogenesis and centriole associated 1 (Spatc1). Moreover, biochemical analysis showed that BNC1 was associated with TATA-box binding protein-associated factor 7 like (TAF7L), a germ cell-specific paralogue of the transcription factor IID subunit TAF7, both in vitro and in testis, suggesting that BNC1 might directly cooperate with TAF7L to regulate spermatogenesis. The truncation mutation disabled nuclear translocation of the BNC1/TAF7L complex, thus, disturbing expression of related genes and leading to testicular premature aging. Similarly, expressions of BNC1, TAF7L, Y-box-binding protein 2 (YBX2), outer dense fiber of sperm tails 1 (ODF1), and glyceraldehyde-3-phosphate dehydrogenase, spermatogenic (GAPDHS) were significantly decreased in the testis of men with non-obstructive azoospermia. The present study adds to the understanding of the physiology of male reproductive aging and the mechanism of spermatogenic failure in infertile men.
Keywords: testicular aging, spermatogenesis, BNC1, TAF7L, gene mutation
Introduction
Premature testicular aging, which mainly results from spermatogenic dysfunction is an important pathogenesis of infertility in elder men. Due to the loss of testicular function, symptoms such as loss of libido, erectile dysfunction, decreased muscle strength, attention deficit, cognitive impairment, and depression may occur in patients with premature testicular aging. The risks of long-term consequences of hypoandrogenism such as osteoporosis, cardiovascular disease, and psychosocial sequelae are not uncommon in patients with premature testicular aging (Zhu et al., 2011). However, there is no effective strategy to stop or slow down the premature testicular reserve depletion process in those patients. Early recognition of testicular aging and prevention of degeneration of spermatogenic function are crucial for preventing the male infertility, especially in consideration of the advanced male reproductive age nowadays.
Premature testicular aging is a heterogeneous disease (Zheng et al., 2019). Potential causes of spermatogenic dysfunction including chromosomal or genetic alterations, environmental factors, infections, metabolic and autoimmune diseases, and iatrogenic factors such as testicular surgery, radiotherapy, or chemotherapy can all lead to premature testicular aging (Zheng et al., 2019). It is usually sporadic and/or simplex, and a genetic basis is considered to be a strong component of the cause for the disorder. Much research has been focused on genes involved in spermatogenesis in recent years, based on animal models and novel approaches such as genome-wide studies. The most commonly known genetic causes of spermatogenic dysfunction are zinc finger (Zf) MYND-type containing 10 (ZMYND10) (Moore et al., 2013), nephrocystin 4 (NPHP4) (Alazami et al., 2014), aurora kinase C (AURKC) (Ben Khelifa et al., 2011), dpy-19 like 2 (DPY19L2) (Harbuz et al., 2011), protein interacting with PRKCA 1 (PICK1) (Liu et al., 2010), cation channel sperm associated 1/2 (CATSPER1/2) (Avenarius et al., 2009; Avidan et al., 2003), spermatogenesis associated 16 (SPATA16) (Dam et al., 2007), ubiquitin-specific peptidase 9 Y-linked (USP9Y) (Sun et al., 1999), and follicle-stimulating hormone receptor (FSHR) (Tapanainen et al., 1997). The list of genetic causes of spermatogenic dysfunction is expanding. Nevertheless, the genetic cause of the majority of spermatogenic dysfunction cases remains unknown.
Basonuclin (BNC1), which contains three pairs of C2H2 Zfs, a nuclear localization signal (NLS) site and a serine stripe (Tseng and Green, 1992), is mainly expressed in gametogenic cells and proliferative keratinocytes and is known as a transcription regulator for both RNA polymerases I and II (Pol I & II). In our previous study (Zhang et al., 2018), we identified a heterozygous 5-bp deletion mutation (c.1153_1157del CCGGG) in the fourth exon of BNC1 gene in a large primary ovarian insufficiency pedigree. The mutated BNC1 protein was expected to lose its three pairs of Zfs and part of NLS (Zhang et al., 2018) and suffer from a nuclear localization disorder (Supplementary Figure S1). Therefore, it cannot function normally, and was proved to be involved in the pathogenesis of premature ovarian insufficiency (Zhang et al., 2018).
A previous study reported that BNC1 existed in spermatogonia, spermatocytes, and spermatids, but was absent in Sertoli cells (Zhang et al., 2012). Bnc1-null male mice were sub-fertile, losing germ cells progressively with age (Zhang et al., 2012). However, the underlying molecular mechanisms remain poorly understood. To investigate the role of the novel mutation described above in testicular aging, we established Bnc1-truncated mutant mice as described in our previous study (Zhang et al., 2018) and found that Bnc1-truncated mutant males were losing fertility progressively with age. Further studies with genome-wide expression profiling and direct binding mapping with BNC1 were conducted to explore the role of BNC1 in testicular function and involved mechanisms. The finding that BNC1 insufficiency may be a cause of testicular premature aging is further supported by the lower expression level of BNC1 in non-obstructive azoospermia (NOA) patients with spermatogenic dysfunction.
Results
Bnc1 truncation mutation leads to progressive fertility loss in male mice
We established a mouse model carrying mutated Bnc1 with a 5-bp deletion (c.1153_1157del CCGGG) in the fourth exon to investigate the role of the mutation in male reproductive aging.
To test the fertility of heterozygous mutant (Het, Bnc1+/T) and homozygous mutant males (Hom, Bnc1T/T), Het males and Hom males were mated respectively with wild-type (Wt, Bnc1+/+) females with normal fertility at 6 weeks old. The Wt littermates were used as controls. During the 5-month testing period, Het males produced a total of 142 pups for a total of five matings and their litter size decreased progressively with age (Figure 1A–C). However, in the five matings with Hom males, only one produced progeny (litter of six) (Figure 1A and B). For Hom males, the single successful mating occurred in younger males (before 12 weeks), whereas no further pup was produced (Figure 1B). By contrast, the mating of Wt males was performed successfully 24 times, producing a total of 213 progeny in five months (Figure 1A). Those results demonstrated a progressive fertility loss in Bnc1 mutant male mice.
Figure 1.
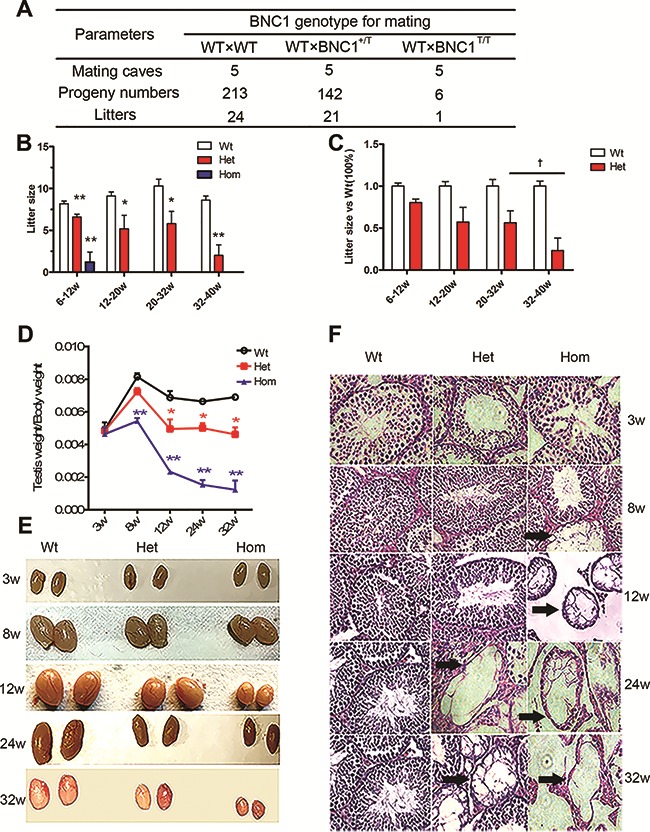
Bnc1 is essential for reproductive success of male mice. (A) Progeny produced over 5 months from five mating cages each with Wt males and Wt females, Het males and Wt females, homozygous males, and Wt females. (B) Numbers of progeny per litter form those types of matings described above at the age of different weeks. (C) Percentage of progeny numbers per litter for matings in Het males compared with Wt males at the age of different weeks. (D) Testis/body weight ratio of Wt, Het, and Hom mice. (E) General appearances of testis from Wt, Het, and Hom male mice at different ages (weeks). (F) Testes slides from Wt (column 1), Het (column 2), and Hom (column 3) mice of various ages (3 weeks, 8 weeks, 12 weeks, 24 weeks, and 32 weeks) were stained with hematoxylin and eosin. Disrupted tubules in testis are indicated by black arrows. Values in Figure 1 are expressed as mean ± SEM (n = 5). Asterisks denote statistically significant differences compared with Wt male mice. *P < 0.05; **P < 0.01; † denotes statistically significant differences compared with the group of Het male mice at the age of 20–32 weeks. †P < 0.05 (Student’s t-test).
In addition, we measured sperm count, sperm motility, testis weight, and serum levels of testosterone, luteinizing hormone (LH), and follicle-stimulating hormone (FSH) in the mouse model. The results showed that decreased testes sizes and a decline in testes/body weight ratio occurred in 8-week-old Hom males and in 12-week-old Het males (Figure 1D and E). Similarly, histological examination revealed a massive germ cell loss in 8-week-old Hom testes and a progressive germ cell loss in Het testes since the age of 24 weeks (Figure 1F). Hormone assay also showed increased FSH and LH levels in 8-week-old Hom males and increased FSH level in 24-week-old Het males (Supplementary Figure S2A and B). Meanwhile, significantly lower serum level of testosterone was also observed in 8-week-old Hom males and in 12-week-old Het males (Supplementary Figure S2C). Consistent with the changes in hormone levels, computer-aided sperm analysis (CASA assay) showed significantly decreased sperm count and sperm motility in 8-week-old Hom males and progressively decreased sperm motility in Het males since the age of 24 weeks (Supplementary Figure S2D–F).
Bnc1 truncation mutation disrupts normal expressions of spermatogenic genes in mouse testis
To gain a better understanding of the phenotypes observed above, we compared the genome-wide expression profiles of 8-week-old Wt and Bnc1+/T testes by gene chip. RNAs were extracted from nine Wt and nine Bnc1+/T testes, which were mixed separately to refrain from individual differences. Microarrays were generated for both Wt and Bnc1+/T samples and then analyzed by ingenuity pathway analysis (IPA). According to the expression data, we identified 1611 downregulated genes and 1276 upregulated genes in mouse testes carrying a Bnc1 mutation (fold change > 2, P < 0.01, Figure 2A and B). Gene ontology (GO) analysis showed that a handful of genes involved in spermatogenesis and fertility were significantly downregulated in the testes of Bnc1+/T males. We further subdivided the relevant genes downregulated by the mutation of Bnc1 into three groups. One prominent group of genes included well-known spermatogenic activators and markers such as F-box/WD-40 repeat-containing protein 7 (Fbwx7), spermatogenic leucine zipper 1 (Spz1), cAMP responsive element modulator (Crem), POU domain, class 3, transcription factor 1 (Pou3f1), spermatogenesis and centriole associated 1 (Spatc1), and spermatid associated (Spert) (Goto et al., 2010; Hsu et al., 2004; Kanatsu-Shinohara et al., 2014; Nantel et al., 1996; Wu et al., 2011). Other two groups of Bnc1-regulated genes including outer dense fiber of sperm tails 1 (Odf1), testis-specific serine kinase 6 (Tssk6), four and a half LIM domains 5 (Fhl5), glyceraldehyde-3-phosphate dehydrogenase, spermatogenic (Gapdhs), serpin family A member 5 (Serpina5), and H1 histone family, member N, testis-specific (H1fnt) were associated with sperm motility and morphology (Figure 2C) (Lardenois et al., 2009; Li et al., 2011; Margaryan et al., 2015; Nair et al., 2008; Uhrin et al., 2000; Yang et al., 2014). To confirm the microarray results, quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR) was carried out on a number of representative genes from these three groups (Supplementary Figure S3). Those results provided evidence that Bnc1 regulates a number of spermatogenic genes in testis. Consistently, downstream analysis via IPA revealed that the upregulated downstream effects by Bnc1 mutation include sperm disorder, asthenozoospermia, teratozoospermic, and oligospermia (Figure 2D).
Figure 2.
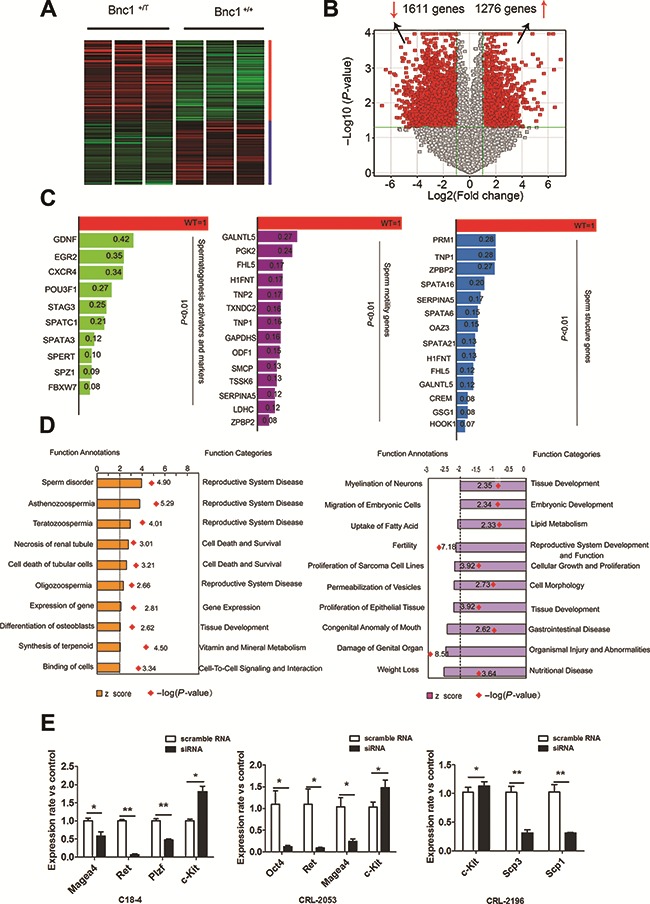
Bnc1 insufficiency dramatically deregulates testis gene expression in vivo and in vitro. (A) Global gene expression of Bnc1+/T and Wt testis were analyzed via gene chip. Green column represents genes downregulated (1611 genes); red column represents genes upregulated (1276 genes). (B) Volcano plots of genes with differential expression in Bnc1+/T versus Wt testes. The x axis represents the log2 of the fold change, and the y axis represents the −log10 of the P-value from a student’s t-test. The red points in the plot represent the differentially expressed genes with statistical significance (with a fold change ≥ 2, and P-value < 0.05). (C) Relative expression levels of representative spermatogenic activators and markers, sperm motility-related genes, and sperm structure-related genes. (D) Downstream effects of deregulated testis gene expression by mutated Bnc1. The orange column shows the upregulated effects including asthenozoospermia, teratozoospermic, and oligospermia, and the purple column shows the downregulated effects including fertility. Red dots denote –log(P-value) of each effect. (E) Knocking down Bnc1 disrupts the expressions of several genes in spermatogonia cells. Several spermatogonial stem cell markers such as Magea4, Ret, and Plzf were downregulated, whereas the differentiation marker Kit was upregulated in C18-4 cell and CRL-2053 cell after knocking down Bnc1. *P < 0.05; **P < 0.01.
Furthermore, knocking down Bnc1 in spermatogenic cells (C18-4 indicating type-A spermatogonial stem cell, CRL-2053 indicating type-B spermatogonial stem cell, and CRL-2196 indicating spermatocyte) via siRNA:small interfering RNA (siRNA) also led to the downregulation of several markers for spermatogonial stem cells including MAGE family member A4 (Magea4), ret proto-oncogene (Ret), promyelocytic leukemia Zf (Plzf) and the upregulation of kit proto-oncogene receptor tyrosine kinase (c-kit), which is required for differentiation of spermatogonia cells (Figure 2E). Those data also suggested an important role of Bnc1 in the regulation of early stage spermatogenesis.
BNC1 directly binds to promoters of many target genes in mouse testis
Given that hundreds of genes are up- or downregulated by >2-fold in Bnc1+/T testis, we further determined whether BNC1 directly bound to the promoters of those differently expressed genes in testis. Chromatin immunoprecipitation sequencing (ChIP-seq) mapping of BNC1 binding sites was carried out in Wt testes with histone H3 tri methyl K4 (H3K4Me3) binding sites as positive controls and input as negative controls. Firstly, we use intersecting magnetic cell separation and Grizzly Peak algorithms to analyze the mapped ChIP tags from deep sequencing with Bowtie. The analysis identified 11743 significant peaks for H3, 4922 significant peaks for BNC1, and no significant peak for input control. The ChIP-seq-detected binding sites of BNC1 contain known BNC1 binding sites proposed by Tseng and Green (1992) and Wang et al. (2006). Next, the distances of the BNC1 binding peaks to transcription start sites (TSS) were analyzed, which showed that 56% of BNC1 peaks were within 1 kb of TSS (Figure 3A and B). Co-localization analysis revealed that nearly half of the BNC1 peaks overlap H3 peaks (Figure 3C), suggesting that BNC1 is largely associated with the promoters of actively transcribed genes in testis. Furthermore, we found that 132 genes downregulated by Bnc1 mutation including many spermatogenic genes bore direct binding sites for BNC1. In particular, co-localization analysis showed that BNC1 co-localizes with H3 at promoter sites of hook microtubule tethering protein 1 (Hook1) (Figure 3D), germ cell-specific gene 1 (Gsg1) (Figure 3E), and glial cell-derived neurotrophic factor (Gdnf) (Figure 3F). ChIP-qPCR analysis with Wt and Bnc1T/T testes confirmed that BNC1 binding was dramatically decreased in Bnc1T/T testes at target promoters, including kelch-like family member 10 (Klhl10), testis expressed 14 (Tex14), Tssk6, Spatc1, transition protein 2 (Tnp2), vascular endothelial growth factor A (Vegfa), Gdnf, and Hook1 (Figure 3G), suggesting that Bnc1 truncation mutation impeded the effects of BNC1 on regulating spermatogenic genes expression in testis.
Figure 3.
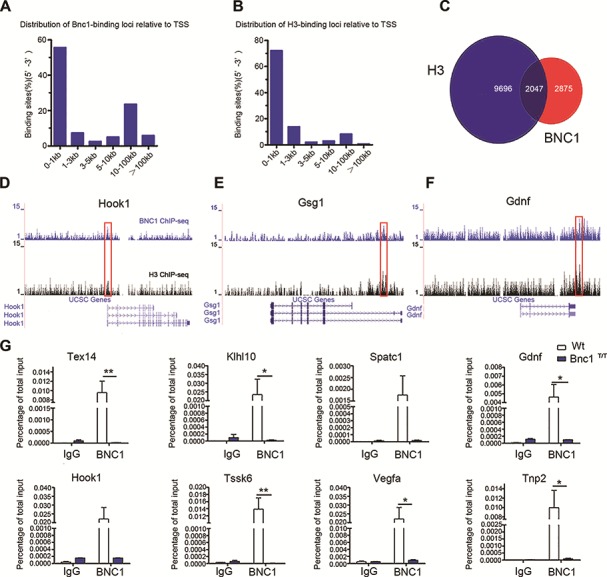
ChIP-seq analysis identifies BNC1 and H3K4Me3 binding sites in testis. (A and B) Percentages of BNC1 (A) and H3K4Me3 (B) binding peaks versus the distance of the peaks to TSS on genome-wide scale in testis. (C) Overlapping between BNC1 and H3K4Me3 binding peaks in testis. (D–F) Read accumulation of BNC1 and H3K4Me3 are shown in Hook1 (D), Gsg1 (E) and Gdnf (F) gene loci. Co-localized peaks are marked with red solid boxes. Input served as negative control for ChIP-seq analysis. (G) ChIP-qPCR analysis of IgG, and BNC1 on Tex14, Klhl10, Spatc1, Gdnf, Hook1, Tssk6, Vegfa, and Tnp2 promoters in Wt and Bnc1T/T testis; values are expressed as mean ± SEM of three independent experiments. Asterisks denote statistically significant differences of Bnc1T/T compared with Wt. *P < 0.05; **P < 0.01 (Student’s t-test).
BNC1 and TAF7L target similar spermatogenic genes
Based on IPA analysis, we found that many target genes of BNC1 that play key roles in spermatogenesis were also regulated by TATA-box binding protein associated factor 7 like (TAF7L). We further compared all of the target genes regulated by BNC1 and TAF7L in testis identified by RNA-seq or microarray (Zhou et al., 2013) and found that BNC1 and TAF7L co-regulated a series of genes directing post-meiotic spermatogenesis, such as Gapdhs (Margaryan, et al., 2015), Tnp1/2 (Zhao et al., 2004), protamine 1/2 (Prm1/2) (Steger et al., 2000), fascin actin-bundling protein1/2 (Fscn1/2) (Zhou et al., 2013), sperm mitochondria-associated cysteine-rich protein (Smcp) (Cullinane et al., 2015), Y-box-binding protein 2 (Ybx2) (Najafipour et al., 2015), and poly(A) polymerase beta (Papolb) (Kashiwabara et al., 2018) (Figure 4A and B). Both BNC1 and TAF7L are required for proper expression of those spermatogenic genes. The results were also in accordance with the spermatogenesis defects observed in both Bnc1T/T and Taf7l−/Y testes, which showed similar reduced sperm count and motility (Cheng et al., 2007). ChIP-qPCR analysis with Wt and Bnc1T/T testes confirmed that TAF7L binding at target promoters (Klhl10, Tssk3, Prm1/2, Tex14, and Spatc1) also significantly decreased in Bnc1T/T testes (Figure 4C). Furthermore, according to CHIP-seq mapping of binding sites, a few representative gene loci such as Klhl10, Spatc1, and Tex14 clearly show co-localization of BNC1, TAF7L, and H3 at their promoter sites (Figure 4D and F). These findings suggest that BNC1 might cooperate with TAF7L to control the expression of many spermatogensis-related genes, as deduced from microarray, RT-qPCR, and ChIP-seq analysis.
Figure 4.
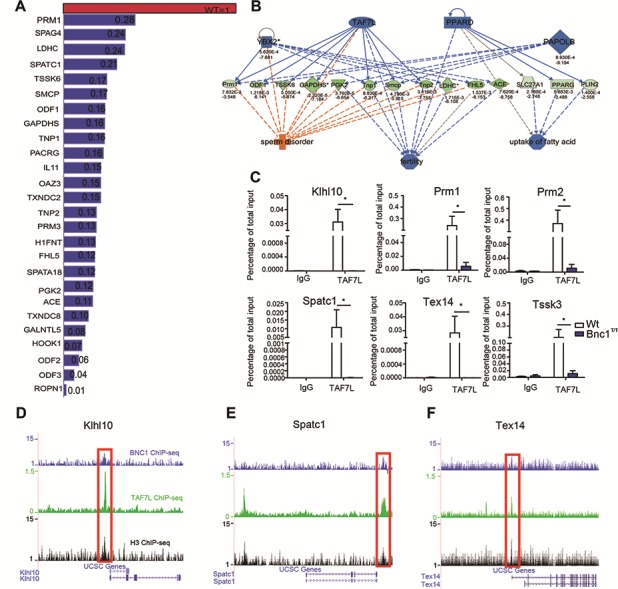
Bnc1 mutation impairs the expression of spermatogenetic genes in a similar way to Taf7l−/Y in testis. (A) Expression of Taf7l-regulated spermatogenic genes in Bnc1+/T testis were compared with those in Wt testis. A value of one was arbitrarily assigned to expression levels of genes in Wt testis and their corresponding expression levels in Bnc1+/T testis were relative to this value. The expression levels of genes were from the expression profile data. (B) Genes downregulated by Bnc1 mutation and involved in sperm disorder and testicular aging were also regulated by TAF7L. Genes in blue were downregulated in Bnc1+/T testes. Genes in green were also downregulated in Bnc1+/T testes, which can be regulated by upstream genes in blue. The deeper the color, the higher the degree of downregulation. Imaginary in blue indicated that the decreased expression level of the upstream genes might suppress the expression of the downstream genes. Imaginary in orange means the decreased expression level of the upstream genes may lose their function as inhibitors of downstream effects. Full line in blue means the upstream genes can definitely regulate the expression of the downstream genes and effects. Lines in gray mean the upstream genes may have no effects on the regulation of downstream genes and effects. (C) ChIP-qPCR analysis of IgG, and TAF7L on Klhl10, Prm1/2, Spatc1, Tex14, and Tssk3 promoters in Wt and Bnc1T/T testis showed that Bnc1 mutation greatly diminished the binding of TAF7L to target gene promoters. Values are expressed as mean ± SEM of three independent experiments. Asterisks denote statistically significant differences of Bnc1T/T compared with Wt. *P < 0.05; **P < 0.01 (Student’s t-test). (D–F) Read accumulation of BNC1, TAF7L, and H3K4Me3 are shown on the Klhl10 (D), Spatc1 (E), and Tex14 (F) gene loci. Co-localized peaks are marked with red solid boxes. Input served as negative control for ChIP-seq analysis.
Co-localization and interaction between proteins of BNC1 and TAF7L
To further explore the possibility of cooperation between BNC1 and TAF7L in regulation of spermatogenic gene expression and the effects of mutated BNC1 on the interaction, we co-transferred FLAG-tagged TAF7L plasmid with CFP-tagged wild-type BNC1 plasmid (BNC1 WT) or CFP-tagged truncated mutated BNC1 plasmid (BNC1 MT) in HEK293T cells, and used FLAG antibody to co-IP with BNC1 WT and BNC1 MT proteins or CFP antibody to co-immunoprecipitation (IP) with FLAG-tagged TAF7L protein, respectively. The results show that TAF7L can be efficiently pulled down by both BNC1 WT and BNC1 MT proteins and vice versa (Supplementary Figure S4). In consideration of the nuclear localization disorder of BNC1 MT protein, we carried out co-IP experiments in cytosolic protein and nuclear protein, respectively. The results showed that co-precipitation of BNC1 WT protein and TAF7L mainly occurred in nuclei while the co-precipitation of BNC1 MT protein and TAF7L occurred only in cytoplasm (Figure 5A). Similarly, the same phenomenon was observed in confocal scanning of HEK293T cell co-expressed with RFP-tagged TAF7L and CFP-tagged BNC1 WT or BNC1 MT plasmids (Figure 5C). In addition, we used affinity-purified BNC1 antibody to co-immunoprecipitate endogenous TAF7L in CRL-2196 cell line. The results indicate that BNC1 could pull down endogenous TAF7L in the CRL-2196 cell (Figure 5B). Finally, co-localization of BNC1 and TAF7L was confirmed via immunofluorescence in vivo. BNC1 co-localized with TAF7L mainly in post-meiotic germ cells in mouse seminiferous tubule (Supplementary Figure S5). Those data suggested that BNC1, a germ cell-specific transcription factor, might cooperate with an atypical testis-specific core promoter recognition factor TAF7L to regulate a subset of genes necessary for spermatogenesis.
Figure 5.
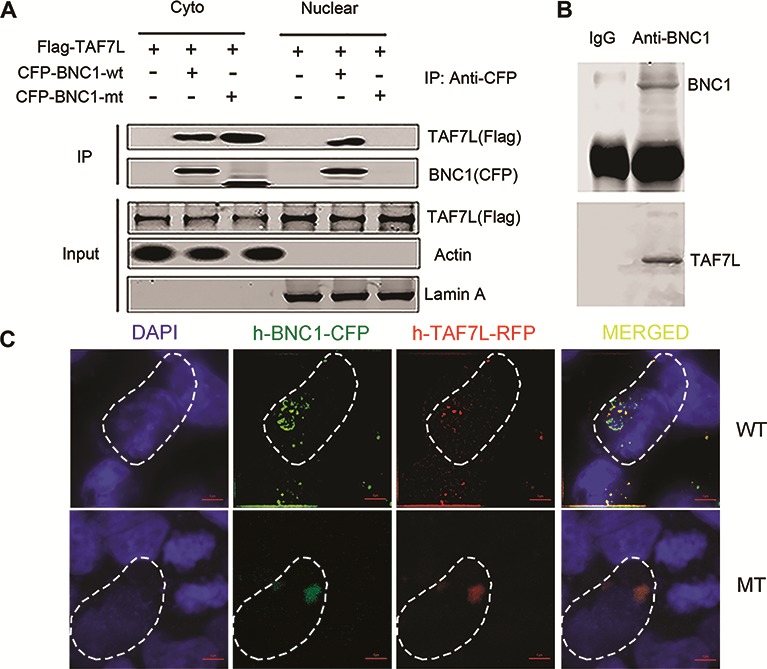
BNC1 associates with TAF7L both in vitro and in vivo. (A) FLAG-TAF7L and CFP-BNC1 fusion plasmids were over-expressed in HEK293T cells, and IPs were performed using both FLAG and CFP antibodies in cytoplasmic and nucleoplasmic proteins, respectively, followed by western blotting analysis with FLAG and CFP antibodies. (B) IPs were performed on CRL-2196 cell lysates using BNC1 antibody, followed by western blotting with TAF7L and BNC1 antibodies. (C) Subcellular localization of BNC1 WT protein and BNC1 MT protein and their associations with TAF7L in HEK293T cells co-transfected with BNC1-CFP and TAF7L-RFP fusion plasmids. When the two plasmids were co-transfected into HEK293T cells, MT BNC1-CFP protein was co-localized with TAF7L-RFP protein in cytoplasm, whereas WT BNC1-CFP protein was co-localized with TAF7L-RFP protein in nucleoplasm. Nuclei of cultured cells were stained with DAPI. Contour of the cell was marked by the fusiformises in white dashed lines. Scale bar, 5 μm.
BNC1 is associated with spermatogenic dysfunction in human
To further confirm the role of BNC1 in human spermatogenesis, we validated the mRNA level of BNC1 and other selected important genes, which were found in Het male mice, on testicular biopsies of infertile patients with OA (n = 10) and NOA (n = 10) (Figure 6A) by q-PCR. Expressions of BNC1, TAF7L, GAPDHS,ODF1, and YBX2 were significantly decreased in NOA group (Figure 6B and C). These results suggested that NOA patients had a pattern of molecular markers similar to that of male mice with truncated Bnc1 and further confirmed the important role of BNC1 in human spermatogenesis.
Figure 6.
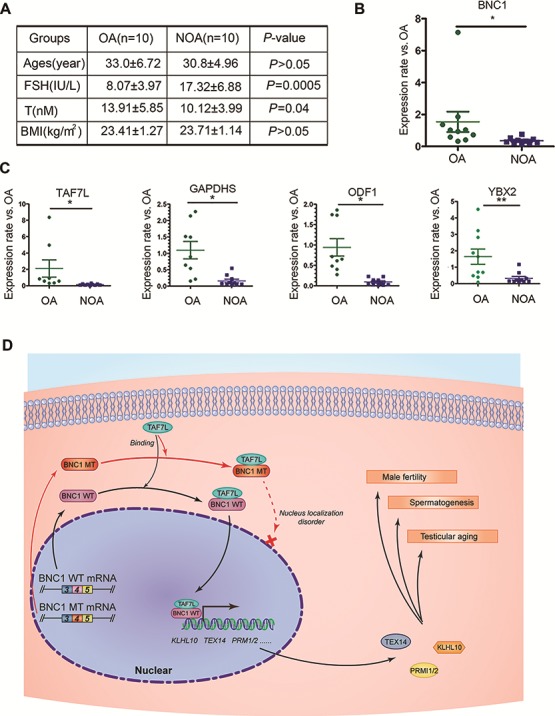
BNC1 is associated with spermatogensis disorder in human male individuals. (A) The base line characters of involved subjects for screening the expression levels of BNC1 and its related genes. OA, obstructive azoospermia; NOA, non-obstructive azoospermia. (B) qPCR showed the different expression levels of BNC1 in OA and NOA groups. (C) qPCR showed the different expression levels of BNC1-related genes including TAF7L, GAPDHS, ODF1, and YBX2 in OA and NOA groups. (D) A model to explain the mechanism of spermatogenesis regulation by BNC1 and the mechanism of testicular aging caused by mutated BNC1. *P < 0.05; **P < 0.01.
Discussion
Spermatogenesis is a cyclic process when diploid spermatogonia differentiate into mature haploid spermatozoa, which is mainly driven by two pre- and post-meiotic transcription waves. In the pre-meiotic transcription phase, individual spermatogonia differentiate into primary spermatocytes, which undergo two meiotic divisions to generate haploid round spermatids later (Greenbaum et al., 2011; Mahoney et al., 1998). With dramatic biochemical and morphological changes, haploid round spermatids turn into the elongated shape of mature spermatozoa later during the post-meiotic transcription phase of spermatogenesis. Exploring the intricate mechanisms of spermatogenesis has important implications for human health and reproduction. Our data demonstrated that Bnc1 insufficiency promoted the age-related loss of testicular functions including spermatogenesis and steroid hormone synthesis. Given that Bnc1 mainly expressed in germ cells and not expressed in somatic cells in testis, the changes of reproductive hormones levels including lower levels of testosterone and higher level of FSH in Bnc1 mutated mice might result from the degeneration of the testes characterized by loss of germ cells. To further determine the underlying mechanisms involved in Bnc1’s role in testes, we analyzed the global influence of Bnc1 mutation in the whole mouse testis via gene chip analysis. Bnc1 was expressed in germ cells exclusively in testis. Therefore, both down- and upregulated genes found by microarray and RT-qPCR in this study were probably direct consequences of Bnc1 mutation.
Transcription, beginning with the utilization of distinct promoter elements (Kimmins et al., 2004) in uniquely reorganized chromatin (Sassone-Corsi, 2002), is a key step in regulating spermatogenesis. Transcription in testis is driven by several testis-specific transcription factors such as TATA-binding protein (TBP)-associated factor 4b (TAF4B, a testis-specific homolog of TAF4) (Falender et al., 2005), TBP-related factor 2 (TRF2) (Zhang et al., 2001), TAF7L (Cheng et al., 2007; Pointud et al., 2003), and so on. For example, mice bearing deficient Taf4b were initially normal, while underwent progressive germ-cell loss and even became infertile at the age of three months with Sertoli cells only in seminiferous tubules (Falender et al., 2005). Taf7l-deficient mice showed defects in fertility, aberrant sperm structure, reduced sperm count, and weakened motility (Cheng et al., 2007).
Interestingly, in the present study, we found that Bnc1+/T mice resembled Taf7l−/Y mice in most of the phenotype, and most of the downregulated genes directing spermatogenesis upon mutation of Bnc1 were also downregulated by Taf7l-null mutations such as Gapdhs, Tnp1/2, Prm1/2, Fscn1/2, H1fnt, and Smcp (Zhou et al., 2013). Taf7l, firstly identified in spermatogonia, is a specific X-linked gene in testis and adipose (Wang et al., 2005). It is expressed throughout the whole process of male germ cell differentiation (Pointud et al., 2003). Previous study indicated that Taf7l was reactivated from meiotic sex chromosome inactivation and relocated into the nuclei of pachytene/round spermatids where Bnc1 was highly expressed during the post meiotic wave of spermatogenesis (Pointud et al., 2003). We postulated that a testis-specific transcription complex including both TAF7L and BNC1 was formed to target to regulate BNC1-dependent genes expression (Ohbayashi et al., 2003; Teichmann et al., 1999). Actually, our biochemical analysis showed that BNC1 was associated with TAF7L both in vitro and in mouse testis. Furthermore, a few representative gene loci such as Klhl10, Tex14, and Spatc1 clearly showed co-localization of BNC1, TAF7L, and H3 at their promoter sites according to ChIP-seq mapping of binding sites and Bnc1 mutation greatly diminished the binding of TAF7L to target gene promoters. We believe that BNC1/TAF7L complex is essential for regulation of spermatogenesis and testes function. The truncation mutation of BNC1, which disabled nuclear localization of BNC1 protein, might prevent BNC1/TAF7L complex from binding with the promoters of target genes and lead to impaired spermatogenesis and testicular aging in male mice. The present study provided an important example of functional cross-talk between two atypical core promoter recognition factors performing coordinately to direct tissue-specific gene transcription. However, further studies are needed to clarify the way in which BNC1 is associated with TAF7L.
Zhang et al. (2012) first proposed that BNC1 may have more than one function in spermatogenesis, based on a gene-dosage effect. Similarly, an important testis phenotype of Bnc1 mutant mice is the progressive germ cell loss, which is different from Taf7l−/Y mice but similar to a null mutation of Taf4b, another gonad-specific subunit of transcription factor IID (Falender et al., 2005). In a previous study, TAF7L was reported to be expressed in cytoplasm in spermatogonia, while was expressed in nucleus in spermatocytes (Pointud et al., 2003). This indicates that the interaction between TAF7L and BNC1 may not occur in spermatogenic stem cells. Apart from this, we still suppose that the TAF7L–BNC1 interaction may account for some effects of BNC1, but not on maintaining stem cells. Further study is needed in germ cell lineage to better clarify this question, since our co-IP experiment was only done in HEK cells. Genome-wide expression analysis and genome-wide binding analysis suggest a role of BNC1 in the regulation of Gdnf, stromal antigen 3 (Stag3), Pou3f1, and chemokine (C-X-C motif) receptor 4 (Cxcr4), which are required for spermatogonial stem cells maintaining (Chen et al., 2016). Furthermore, knocking down Bnc1 in spermatogonial stem cells via siRNA also leads to downregulation of Magea4, Ret, and Plzf (Figure 2E). Most of those factors are known signal transducers of GDNF/RET/AKT pathway. Thus, those data suggest an important role for BNC1 in GDNF/RET/AKT signal transduction, which is involved in the effects of TAF4B on spermatogenesis and spermatogonial stem cell maintaining (Falender et al., 2005). Moreover, BNC1 appears to be a transcription repressor for SCF/kit (Figure 2E). Loss of Bnc1 therefore may lead to further stem cell differentiation. This analysis suggested that in early stage of spermatogenesis including self-renewal of spermatogonial stem cells, BNC1 might also play an important role. Zhang et al. (2012) has also reported that the testis phenotypes of Bnc1 null mutation and Taf4b null mutation are highly similar in spermatogonia stem cell maintenance. Thus, BNC1’s role in spermatogenesis is not confined to co-regulation with TAF7L but may also co-regulate with other TAFs such as TAF4B.
In conclusion, we found a Bnc1 insufficiency mutation promoted age-related loss of testicular functions in the present study. BNC1, which co-regulates a subset of spermatogenic genes together with TAF7L, appears to be essential for spermatogenesis. The truncation mutation of BNC1, which was found in the premature ovarian insufficiency family described above, disabled nuclear localization of BNC1 protein, thus, preventing the BNC1/TAF7L complex from binding with the promoters of target genes such as Prm1/2, Tex14, and Klhl10 (Figure 6D). It may represent the possible mechanisms for the involvement of Bnc1 truncation mutation in premature testicular aging. Further studies in larger premature testicular aging cohorts are recommended to clarify better the BNC1 mutation rate and its importance in patients with premature testicular aging. Meanwhile, further studies performed with germ cells of different stages may help to explore the entire molecular mechanisms involved in the function of BNC1 in spermatogenesis and testicular aging. Nevertheless, our results contribute to expanding the list of genetic causes of premature testicular aging and provide insight into a new target gene for early diagnosis, intervention and future treatment of premature testicular aging. Based on our study, we recommend that men with BNC1 mutations should be monitored for their testicular function and life quality by regular interview, blood hormone assay, and sperm quality investigation. Early intervention including pro-active measures of fertility preservation could be initiated for affected men if necessary. Furthermore, pre-implantation diagnosis could be performed to block transmission of the mutation to offspring.
Materials and methods
Fertility assay
Mice with mutated Bnc1 were generated by insertion of a neomycin-resistance cassette in reverse orientation into the fourth exon (c.1153_1157del CCGGG) of Bnc1 as described in our previous study (Zhang et al., 2018) at the animal model research center of Nanjing University. Het mice were backcrossed to the inbred C57BL/6 strain. Mice were maintained on a 12L:12D cycle with free access to food and water in the vivarium at Zhejiang University. For natural breeding experiments, males were mated with one female. Neonatal males were collected from timed pregnancies. Tissues were frozen for RNA isolation. Mice were maintained in accordance to the NIH Guide for the Care and Use of Laboratory Animals with institutional oversight by Zhejiang University.
Fifteen mating cages of three mating pattern were used as follows: Wt females with Wt males, Het Bnc1+/T males, or Hom Bnc1T/T males for over 7 months, initiated at 6 weeks of age; mating cage numbers, total progeny produced, and total litter production times were recorded for 5 months.
Sperm assays
Uncapacitated cauda epididymal sperm were collected by placing minced cauda epididymis from Wt, Bnc1+/T, and Bnc1T/T mice in HTF medium (Irvine scientific, 90126). One drop of the sperm suspension was transferred to the incubation chamber at 37°C and incubated for 15 min in a 5% CO2–95% O2 incubator. Aliquots of each sperm suspension were loaded into a 100-mm deep chamber pre-warmed at 37°C. Sperm motility parameters were evaluated via a computer-assisted semen analysis system running IVOS (Hamilton Thorne Research, version 12.2L). Eight motion parameters including motility, average path velocity, straight-line velocity, curvilinear velocity (VCL), amplitude of lateral head displacement, beat-cross frequency, straightness, and linearity, were examined. For statistical testing, each parameter was grouped for each genotype and for age of observation. Student’s t-test for independent observations was used to define differences between the Wt and the mutant in VCL means (normalized by natural logarithms). Statistical analyses were performed using the Graph Pad software.
Microarray
Total RNA was prepared from 8-week-old testes by the way described in Supplementary Materials and methods. Both Wt and Bnc1+/T testes were analyzed in three duplicates. For each sample five micrograms of total RNA were used to generate biotinylated cRNA. The cRNA samples were hybridized to MouseWG-6 v2.0 Gene Chips (Illumina) at the Genergy Biotechnology Microarray Core Facility according to the manufacturer’s technical manual (Illumina). The resulting bead chips were scanned and individual hybridization signals were quantified using an Illumina Bead Array Reader. To identify the differentially expressed genes, Student’s t-test analysis was performed. The threshold we used to screen up- or downregulated genes is fold change ≥2 with a P-value cut-off of <0.01. GO and annotation analysis was conducted using DAVID Bioinformatics Resources 6.7 for function analysis of the screened differentially expressed genes. We referred to the National Center for Biotechnology Information’s GEO database (www.ncbi.nlm.nih.gov/geo) under accession no. GSE50807 for RNA-Seq data of Taf7l−/Y testes.
Western blot analysis and IP
A total of 500 μg of whole-cell extracts or separated cytoplasmic and nuclear protein from HEK293T cells transfected with CFP-BNC1 (WT) or CFP-BNC1 (MT) and FLAG-TAF7L plasmids. TAF7L and CFP-BNC1 (WT) or CFP-BNC1 (MT) proteins were immunoprecipitated with antibodies against FLAG (EarthOx, E022060) or CFP (EarthOx, E022030) at 4°C overnight incubated with 0.3 M NaCl and 0.2% NP-40. Then they were incubated in 30 μl of protein A/G beads for an additional 2 h at 4°C. After washing with buffer containing 0.3 M NaCl and 0.1% NP-40, 10% SDS/PAGE separation was conducted to the remaining beads, which was followed by western blotting analysis with FLAG or CFP antibodies to detect tagged proteins in the inputs and IPs.
ChIP, ChIP library preparation, and deep sequencing
Ten Wt testes from 8-week-old mice were fixed with 1% (v/v) formaldehyde for in vivo cross-linking for 15 min at room temperature, which were terminated with 0.125 M glycine for an additional 5 min. A Covaris sonicator (Covaris, Covaris S220) was used to fragment the chromatin obtained from Wt testis tissue to sizes ranging from 100 to 500 bp. In each IP reaction with 4 μg of antibodies against H3K4Me3 (Abcam, ab8580) or BNC1 (Huaan), 1 mg of fragmented chromatin was used; input was used as negative control. Preparation of the sequencing libraries of the DNA samples from IP and inputs was precisely followed the instructions of Illumina at Genergy Biotechnology Microarray Core Facility; qualities of the libraries were assessed using 2100 Bioanalyzer, ultra-high-throughput sequencing was conducted on the qualified libraries via Illumina HiSeq 2000 sequencer at Genergy Biotechnology Microarray Core Facility. We referred to the National Center for Biotechnology Information’s GEO database (www.ncbi.nlm.nih.gov/geo/) under accession no. GSE50807 for ChIP-Seq data of TAF7L in testes.
For ChIP-qPCR analysis, three duplicates of sonicated chromatin were prepared separately from Wt testes or Hom testes as described above, which was used in each IP reaction with 4 μg of H3K4Me3 (Abcam, ab8580) or BNC1 (Huaan) antibodies, and 4 μg of rabbit IgG were used as negative control. Handling of beads, including washings and elution, reversed cross-linking, and DNA precipitation were performed as the manual of pierce agarose ChIP kit (Thermo Scientific, 26156). DNA samples of the IP from antibodies and IgG were subjected to qPCR assay.
Testicular biopsy sample collection
Human testis tissues were collected from 20 azoospermic patients referred to the Assisted Reproductive Technology Centre of Women’s Hospital, School of Medicine, Zhejiang University. Azoospermic patients were referred to the Department of Urology for further assessment of the cause of azoospermia and for sperm retrieval after the couple had undergone a complete assessment, including a gynecological evaluation of the female partner. The diagnosis of NOA or OA was based on the patient’s history, physical, and genital examination, semen analysis, the patient’s endocrine profile, and genetic studies. The data analyzed included age, weight and height, smoking and drinking habits, and history of cryptorchidism, mumps orchitis, varicocele, environmental or radiation exposure, prescribed drug use, previous chemotherapy, and surgical procedures. All patients underwent a genital examination and a scrotal Doppler ultrasound scan to detect testicular volume and to exclude the presence of epididymis head or tail dilatation, unilateral or bilateral absence of the vas deferens and varicocele. A transrectal ultrasound scan was performed to rule out the presence of prostate median cysts and of anomalies of the seminal vesicles suggestive of obstruction of the male genital tract. Genetic studies were performed, including a karyotype, an analysis of microdeletions in the Y chromosome, and an analysis of mutations in the CFTR gene. The hormonal profile was also documented during hospitalization prior to testicular sperm extraction (TESE) in all patients. All participants were adequately counseled, signed a detailed consent form prior to surgery, and agreed to have their data anonymously utilized. The use of discarded surgical samples was approved by the Ethics Committee of the Women’s Hospital, School of Medicine, Zhejiang University.
Supplementary Material
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Acknowledgments
We thank Prof. Fei Sun who works in University of Science and Technology of China for supplying cell line C18-4 and Prof. Yibing Han who works in Tongji University for supplying cell line CRL-2053 and CRL-2196 for us. We thank Prof. Jun Shen from Harward University for revising the manuscript for us.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFC1005003, 2017YFC1001003, and 2017YFC1001303), the National Natural Science Foundation of China (81471421, 81401219, and 81701461), The Fundamental Research Funds for the Central Universities, Natural Science Foundation of Zhejiang Province (Q19H040040), Key Research Program of Zhejiang Provincial Natural Science Foundation (LZ18H040001), Municipal Human Resources Development Program for Outstanding Young Talents in Medical and Health Sciences in Shanghai (2018YQ39), and Zhejiang University Education Foundation Global Partnership Fund.
Conflict of interest: none declared.
Author Contributions: H.-F.H. and D.Z. designed and coordinated the whole project. J.-Y.L. and Y.-F.L. performed most of the experiments on animal model. H.-Y.X., P.-P.L., and M.-E.L. were involved in ChIP setup and J.-Y.Z. helped with final data analysis. Y.-Y.Y. was involved in manuscript revision. Y.-Q.Q. and Y.H. designed appropriate primers and performed qPCR after ChIP from testis tissue. K.L. and G.-F.X. performed sperm motility assay. C.L. and G.-L.D. helped with plasmid construction and co-IP analysis. C.-M.X. and Y.-C.M. helped with human testis and blood sample collection. X.-M.L. helped constructing the ChIP-seq library for sequencing. J.-Z.S. was involved in manuscript preparation.
References
- Alazami A.M., Alshammari M.J., Baig M., et al. (2014). NPHP4 mutation is linked to cerebello-oculo-renal syndrome and male infertility. Clin. Genet. 85, 371–375. [DOI] [PubMed] [Google Scholar]
- Avenarius M.R., Hildebrand M.S., Zhang Y., et al. (2009). Human male infertility caused by mutations in the CATSPER1 channel protein. Am. J. Hum. Genet. 84, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidan N., Tamary H., Dgany O., et al. (2003). CATSPER2, a human autosomal nonsyndromic male infertility gene. Eur. J. Hum. Genet. 11, 497–502. [DOI] [PubMed] [Google Scholar]
- Ben Khelifa M., Zouari R., Harbuz R., et al. (2011). A new AURKC mutation causing macrozoospermia: implications for human spermatogenesis and clinical diagnosis. Mol. Hum. Reprod. 17, 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., Willis W.D., and Eddy E.M. (2016). Targeting the Gdnf gene in peritubular myoid cells disrupts undifferentiated spermatogonial cell development. Proc. Natl Acad. Sci. USA 113, 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Buffone M.G., Kouadio M., et al. (2007). Abnormal sperm in mice lacking the Taf7l gene. Mol. Cell. Biol. 27, 2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinane D.L., Chowdhury T.A., and Kleene K.C. (2015). Mechanisms of translational repression of the Smcp mRNA in round spermatids. Reproduction 149, 43–54. [DOI] [PubMed] [Google Scholar]
- Dam A.H., Koscinski I., Kremer J.A., et al. (2007). Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am. J. Hum. Genet. 81, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falender A.E., Freiman R.N., Geles K.G., et al. (2005). Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev. 19, 794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., O'Brien D.A., and Eddy E.M. (2010). Speriolin is a novel human and mouse sperm centrosome protein. Hum. Reprod. 25, 1884–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum M.P., Iwamori T., Buchold G.M., et al. (2011). Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 3, a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbuz R., Zouari R., Pierre V., et al. (2011). A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am. J. Hum. Genet. 88, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.H., Hsieh-Li H.M., and Li H. (2004). Dysfunctional spermatogenesis in transgenic mice overexpressing bHLH-zip transcription factor, Spz1. Exp. Cell Res. 294, 185–198. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Onoyama I., Nakayama K.I., et al. (2014). Skp1-Cullin-F-box (SCF)-type ubiquitin ligase FBXW7 negatively regulates spermatogonial stem cell self-renewal. Proc. Natl Acad. Sci. USA 111, 8826–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwabara S.I., Tsuruta S., Yamaoka Y., et al. (2018). PAPOLB/TPAP regulates spermiogenesis independently of chromatoid body-associated factors. J. Reprod. Dev. 64, 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmins S., Kotaja N., Davidson I., et al. (2004). Testis-specific transcription mechanisms promoting male germ-cell differentiation. Reproduction 128, 5–12. [DOI] [PubMed] [Google Scholar]
- Lardenois A., Chalmel F., Demougin P., et al. (2009). Fhl5/act, a CREM-binding transcriptional activator required for normal sperm maturation and morphology, is not essential for testicular gene expression. Reprod. Biol. Endocrinol. 7, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sosnik J., Brassard L., et al. (2011). Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol. Hum. Reprod. 17, 42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Shi Q.W., and Lu G.X. (2010). A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J. Androl. 12, 556–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney M.G., Tang W., Xiang M.M., et al. (1998). Translocation of the zinc finger protein basonuclin from the mouse germ cell nucleus to the midpiece of the spermatozoon during spermiogenesis. Biol. Reprod. 59, 388–394. [DOI] [PubMed] [Google Scholar]
- Margaryan H., Dorosh A., Capkova J., et al. (2015). Characterization and possible function of glyceraldehyde-3-phosphate dehydrogenase-spermatogenic protein GAPDHS in mammalian sperm. Reprod. Biol. Endocrinol. 13, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D.J., Onoufriadis A., Shoemark A., et al. (2013). Mutations in ZMYND10, a gene essential for proper axonemal assembly of inner and outer dynein arms in humans and flies, cause primary ciliary dyskinesia. Am. J. Hum. Genet. 93, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair M., Nagamori I., Sun P., et al. (2008). Nuclear regulator Pygo2 controls spermiogenesis and histone H3 acetylation. Dev. Biol. 320, 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafipour R., Moghbelinejad S., Samimi Hashjin A., et al. (2015). Evaluation of mRNA contents of YBX2 and JHDM2A genes on testicular tissues of Azoospermic men with different classes of spermatogenesis. Cell J. 17, 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel F., Monaco L., Foulkes N.S., et al. (1996). Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380, 159–162. [DOI] [PubMed] [Google Scholar]
- Ohbayashi T., Shimada M., Nakadai T., et al. (2003). Vertebrate TBP-like protein (TLP/TRF2/TLF) stimulates TATA-less terminal deoxynucleotidyl transferase promoters in a transient reporter assay, and TFIIA-binding capacity of TLP is required for this function. Nucleic Acids Res. 31, 2127–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud J.C., Mengus G., Brancorsini S., et al. (2003). The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116, 1847–1858. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. (2002). Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296, 2176–2178. [DOI] [PubMed] [Google Scholar]
- Steger K., Pauls K., Klonisch T., et al. (2000). Expression of protamine-1 and -2 mRNA during human spermiogenesis. Mol. Hum. Reprod. 6, 219–225. [DOI] [PubMed] [Google Scholar]
- Sun C., Skaletsky H., Birren B., et al. (1999). An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat. Genet. 23, 429–432. [DOI] [PubMed] [Google Scholar]
- Tapanainen J.S., Aittomaki K., Min J., et al. (1997). Men homozygous for an inactivating mutation of the follicle-stimulating hormone (FSH) receptor gene present variable suppression of spermatogenesis and fertility. Nat. Genet. 15, 205–206. [DOI] [PubMed] [Google Scholar]
- Teichmann M., Wang Z., Martinez E., et al. (1999). Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells. Proc. Natl Acad. Sci. USA 96, 13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H., and Green H. (1992). Basonuclin: a keratinocyte protein with multiple paired zinc fingers. Proc. Natl Acad. Sci. USA 89, 10311–10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrin P., Dewerchin M., Hilpert M., et al. (2000). Disruption of the protein C inhibitor gene results in impaired spermatogenesis and male infertility. J. Clin. Invest. 106, 1531–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang S., Schultz R.M., et al. (2006). Search for basonuclin target genes. Biochem. Biophys. Res. Commun. 348, 1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.J., Page D.C., and McCarrey J.R. (2005). Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum. Mol. Genet. 14, 2911–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Goodyear S.M., Tobias J.W., et al. (2011). Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol. Reprod. 85, 1114–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Grzmil P., Meinhardt A., and Hoyer-Fender S. (2014). Haplo-deficiency of ODF1/HSPB10 in mouse sperm causes relaxation of head-to-tail linkage. Reproduction 148, 499–506. [DOI] [PubMed] [Google Scholar]
- Zhang D., Liu Y., Zhang Z., et al. (2018). Basonuclin 1 deficiency is a cause of primary ovarian insufficiency. Hum. Mol. Genet. 27, 3787–3800. [DOI] [PubMed] [Google Scholar]
- Zhang D., Penttila T.L., Morris P.L., et al. (2001). Cell- and stage-specific high-level expression of TBP-related factor 2 (TRF2) during mouse spermatogenesis. Mech. Dev. 106, 203–205. [DOI] [PubMed] [Google Scholar]
- Zhang X., Chou W., Haig-Ladewig L., et al. (2012). BNC1 is required for maintaining mouse spermatogenesis. Genesis 50, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Shirley C.R., Mounsey S., et al. (2004). Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol. Reprod. 71, 1016–1025. [DOI] [PubMed] [Google Scholar]
- Zheng W., Zou Z., Lin S., et al. (2019). Identification and functional analysis of spermatogenesis-associated gene modules in azoospermia by weighted gene coexpression network analysis. J. Cell. Biochem. 120, 3934–3944. [DOI] [PubMed] [Google Scholar]
- Zhou H., Grubisic I., Zheng K., et al. (2013). Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc. Natl Acad. Sci. USA 110, (3), 16886–16891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q.X., Meads C., Lu M.L., et al. (2011). Turning point of age for semen quality: a population-based study in Chinese men. Fertil. Steril. 96, 572–576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


