Abstract
Background and purpose:
Breast cancer (BC) is one of the major causes of female cancer-related death. It has recently been demonstrated that metabolic reprogramming including alteration in lipid metabolism is indicated in various types of cancer. The enzymes of the acyl-coenzyme A synthetase long-chain family (ACSLs) are responsible for converting fatty acids to their corresponding fatty acyl-coenzyme A esters which are essential for some lipid metabolism pathways. ACSL4 is one of the isoforms of ACSLs and has a marked preference for arachidonic and eicosapentaenoic acids. The objective of this study was to evaluate ACSL4 expression, its prognostic significance, and its correlation with p53 tumor suppressor in BC patients.
Experimental approach:
In this study 55 pairs of fresh samples of BC and adjacent non-cancerous tissue were used to analyze ACSL4 expression, using real-time polymerase chain reaction and immunohistochemistry (IHC) staining. The expression of other studied variables was also examined using the IHC technique.
Findings / Results:
ACSL4 expression was significantly higher in BC tissues compared to the adjacent normal tissue. This upregulation was negatively correlated with Ki-67 and age, and positively correlated with p53 status. The correlation between ACSL4 and p53 may indicate the role of p53 in the regulation of lipid metabolism in cancer cells, in addition to its role in the regulation of ferroptosis cell death.
Conclusion and implications:
Our results indicated that the expression of ACSL4 may be considered as a prognostic indicator and potential therapeutic target in BC. However, further studies are needed to confirm the significance of these findings.
Keywords: ACSL4, Breast cancer, p53, Tumor suppressor
INTRODUCTION
Breast cancer (BC) is one of the major causes of female cancer-related death (1). It is a global concern and each year nearly 440,000 patients die from BC. Also, it is estimated that one in every nine females will be affected once during their lifetime (2). Various physiological conditions including hypoxia, reactive oxygen species, and metabolic dysfunction promote cancer progression (3). Metabolic reprogramming in cancer cells leads to promoting growth, survival, proliferation, and long-term maintenance in cancer cells (3,4). Alteration in lipid metabolism is also indicated in cancer and is essential for the increase of proliferation, progression, and metastasis (3).
Activation of fatty acids by esterification with a molecule of coenzyme A (CoA) is prerequisite for the metabolism of fatty acids in a number of lipid metabolism pathways including lipogenesis, glycerolipid synthesis, lipidation of proteins, and β-oxidation (5,6).
The enzymes responsible for the conversion of fatty acids to fatty acyl-CoA esters are a family of proteins known as fatty acyl-CoA synthetases (ACS) (7). ACSs are classified based upon the chain length of their preferred substrates (short, medium, long, and very long) (5,6). The long-chain acyl-CoA synthetase family (ACSLs) is active on C12-C22 fatty acids. ACSLs contain five isoforms, ACSL1, ACSL3, ACSL4, ACSL5, and ACSL6 (6). These isoforms differ in subcellular localization and substrate specificity. For example, ACSL4 has a marked preference for arachidonic acid (20:4) and eicosapentaenoic acid (20:5) (5,7). In addition, ACSL4 catalyzes the esterification of arachidonoyl and adrenoyl into phosphatidylethanolamines, thereby, has a central role in lipid peroxidation in ferroptosis related cell death (8).
Furthermore, it has been demonstrated that ACSL4 is overexpressed in several cancer cell lines including breast, prostate, colon, gastric, and liver. Inasmuch as its inhibition reduces cell invasion and migration and induces apoptotic cell death, therefore ACSL4 may be considered as an interesting therapeutic target in cancer (9).
There is evidence to indicate that metabolic alterations in cancer are promoted by activation of oncogenes and loss of tumor suppressors that leads to an increased nutrient uptake to provide energetic and biosynthetic pathways (10). For example, it has been reported that p53 tumor suppressor, also dubbed as the “guardian of the genome”, regulates lipid metabolism through gene expression or protein-protein interactions (11). Some studies have also shown that p53, through increased expression of miR-34 (a tumor suppressor miRNA family), leads to inhibition of ACSL4 expression (12).
Therefore, we studied the expression of ACSL4 in pairs of BC and adjacent normal tissues to investigate the correlation between p53 (wild-type and mutant) status and ACSL4 expression to further the understanding of p53 influence in metabolic regulation of cancer.
MATERIALS AND METHODS
Human breast cancer specimens
Fifty-five pairs of human BC and adjacent normal tissue were obtained from the Ordibehesht Hospital in Isfahan, I.R. Iran from patients undergoing surgical resection during 2016 to 2017. After resection, all the samples were frozen immediately in liquid nitrogen and stored at -80 °C prior to RNA extraction. The histological data of tumors including histology, tumor size, grade, and stage were confirmed by a pathologist. This study was approved by the Ethics Committee of Isfahan University of Medical Sciences under the ethical code: 396510 and informed consent was obtained from all patients.
Quantitative real-time polymerase chain reaction
Total RNA of fresh frozen samples was extracted using the BioFACT™ Total RNA Prep Kit (Ver.2.0, BioFACT, Daejeon, Korea) according to the manufacturer’s protocol. Thereafter, the quality and quantity of mRNA were confirmed by gel electrophoresis and Nanodrop (ODs 260 and 280 nm), respectively. At the reverse transcription step, the RNA was used for cDNA synthesis using a BioFact™ RT-Kit (BioFACT, Daejeon, Korea) as directed by the manufacturer in a 20 μL reaction mixture including 1 μL oligo dt primer, 1 μL random hexamer primer, 9 μL RNA, and 9 μL master mix, this mixture was then incubated at 50 °C for 60 min and 95 °C for 5 min. The generated cDNA was subjected to SYBR green-based standard quantitative real-time polymerase chain reaction (RT-PCR) analysis (BioFACT™ 2X Real-Time PCR master mix, For SYBR green I; BioFACT, Daejeon, Korea) on an ABI StepOnePlus RT- PCR system (Applied Biosystems, USA). Beta-actin was used as an internal control to normalize the RNA input. The sequences of the primers used indicated in Table 1. Primers were designed with Allele ID software version 7.6 (Premier Biosoft, USA) and NCBI BLAST was used to check for Primer Specificity.
Table 1.
The sequences of the primers used in the study.
| Genes | Forward sequences | Reverse sequences |
|---|---|---|
| ACSL4 | 5´-AGAATACCTGGACTGGGACCGAAG -3 | 5´-TGCTGGACTGGTCAGAGAGTGTAA-3´ |
| β-actin | 5´-GTTGTCGACGACGAGCG-3´ | 5´-GCACAGAGCCTCGCCTT-3´ |
The RT-PCR reaction was performed in three steps. Step one included the activation of the enzyme at 95 °C for 15 min. Step two consisted of 40 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The melting curve was generated in step three and at the end of each examination to confirm if a single product was amplified and included the range of 60 °C to 95 °C at 0.3 °C per 5 s increments. All the examinations were carried out in triplicate for every sample. The cycle threshold value is determined as the cycle number at which the fluorescence intensity passes a fixed threshold above the background fluorescence (13).
The gene expression was relatively quantified by ΔCt method compared to the expression of the internal control gene β-ACTIN (ΔCt = Ct of target gene - Ct of internal control). -ΔCt method was used for calculating the Box plot and the fold change was calculated using the 2-ΔΔCt method for comparison between tissues. Values are reported as mean ± standard error of mean (SEM).
Immunohistochemistry
Slices of 5 μm thickness were cut from paraffin-embedded tissue samples using a microtome and placed on a slide coated with poly-L-lysine. Slices were then dewaxed in xylene, rehydrated in ethanol, and washed with phosphate-buffered saline. After antigen retrieval of sections in 10 mM citrate buffer in a microwave oven, endogenous peroxidase was blocked with H2O2. Tissue sections were, then, stained with primary antibodies; p53 antibody (DO-7; Diagnostic BioSystems, Pleasanton, CA, USA); ACSL4 antibody (sc-271800; Santa Cruz, USA) and incubated with horseradish peroxidase-linked secondary antibody. Finally, these sections were visualized using chromogenic 3’-diaminobenzidine tetrahydrochloride (14), as a substrate. The hematoxylin was used for background staining. In this method, sections with overexpression of ACSL4 exhibited staining of brown. ImageJ software (version 1.52h) was used for the quantification of photomicrographs taken by Olympus light microscope with ×40 amplification, and results were reported as Pix/μm2.
Evaluation of p53
In this study, p53 expression was grouped into wild-type and mutant form using immunohistochemistry (IHC) method in tissue samples. This method is based on the stability of p53 since wild-type p53 is an unstable protein compared to the mutant p53 protein that is more stable. Conformational changes in mutant p53 leads to its stabilization and accumulation in cancer cells that is detectable by the IHC method. Thus, p53-positive is indicative of mutant p53, while p53-negative indicates the wild-type.
Evaluation of ER, PR, and HER
The expression of ER, PR, and HER protein was evaluated using IHC method in paraffin- embedded tissue samples as describe above using standard routine laboratory procedure.
Statistical analysis
Statistical analysis was performed using the SPSS software (v.21, IBM Corporation). Data are presented as mean ± SEM. For the gene expression studies the results were analyzed using Student’s t-tests between the tumor and control groups. Correlations between ACSL4 expression, p53 status and Ki- 67 with clinicopathological parameters were presented using multiple linear regression.
RESULTS
ACSL4 mRNA expression in breast cancer and adjacent normal tissue
To assess the potential effect of ACSL4 mRNA expression on BC, the relative transcription levels of ACSL4 gene were evaluated in 55 paired human BC specimens and adjacent normal tissue by RT-PCR.
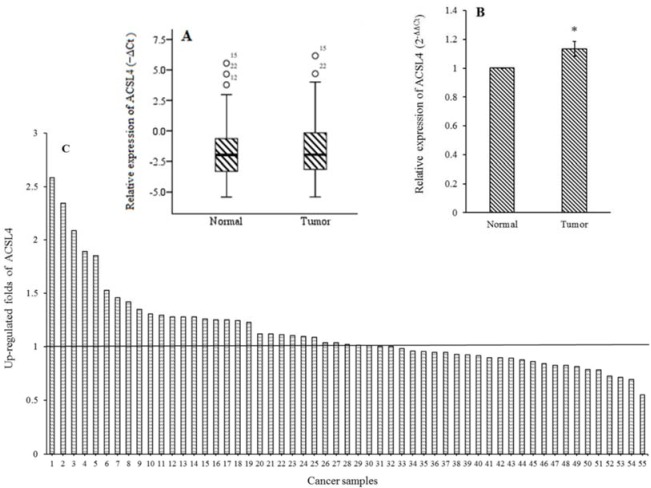
The data indicated that the expression of ACSL4 in BC tissue was significantly higher in comparison with adjacent normal tissue (P < 0.001, Fig. 1A). The analyses of the results by 2-ΔΔCt method display that the relative mRNA expression of ACSL4 was upregulated in 30 of 55 BC samples at more than 1.13-fold change, in tumor tissues compared with adjacent normal tissues (P < 0.05, Fig. 1B and C).
Fig. 1.
Expression of ACSL4 mRNA in breast cancer tissues and adjacent normal tissues. Real-time polymerase chain reaction was performed to evaluate the expression level of ACSL4. The mRNA expression data were normalized to the beta-actin (ACTB) signal and (A) the comparison of the mRNA expression of ACSL4 was shown as Box plot; (B and C) the fold change of ACSL4 expression in each sample pair was calculated using 2-ΔΔCT as columns, mean ± SEM. *P < 0.01 Indicates significant difference with normal group.
ACSL4 protein expression in BC and adjacent normal tissue
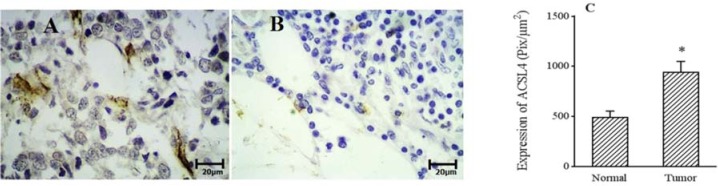
To confirm the RT-PCR data, 20 pairs of BC samples were randomly selected and examined by IHC staining with a specific antibody against ACSL4. Consistent with the RT-PCR results, the positive ratio of ACSL4 staining was significantly higher in BC tissues compared to that in adjacent normal tissues (Fig. 2).
Fig. 2.
Photomicrograph images of immunohistochemistry staining of ACSL4 in sections of (A) breast tumor tissue and (B) the adjacent normal breast tissue. Brown staining shows the expression of ACSL4 in breast tumor tissues. The quantification of ACSL4 protein was evaluated using ImageJ (ver. 1.52h) software by Olympus light microscope with ×40 amplification (Pix/μm2). *P < 0.05 Indicates significant difference with normal group.
The relationship between expression of ACSL4 mRNA, p53, Ki-67 and clinicopathological characteristics of BC
The association between expression of ACSL4 mRNA, p53, Ki-67, and the clinicopathological characteristics in BC patients is shown in Table 2, Figs. 3 and 4. We used backward stepwise regression for evaluation relationship between ACSL4 with the other variables. It is a stepwise regression approach that begins with a full (saturated) model and at each step gradually eliminates variables from the regression model to find a reduced model that best explains the data. This is also known as backward elimination regression. Table 2 shows the first step and final step of multiple linear regressions.
Table 2.
Multiple linear regression between ACSL4 mRNA expression (dependent variable) and other variables. The ACSL4 mRNA expression was measured based on ACTB in tumor and adjacent normal tissues with 2-ΔΔct from at least 2 experiments. Estrogen and progesterone receptors and human epidermal growth factor receptor-2 expressions were measured using immunohistochemistry (IHC) method, and the results are reported as expression of positive (+) or negative (-). Ki-67 nuclear expression was measured using IHC method, and the results are reported as percent of expression. p53 expression was measured based on IHC, and the results were reported as the mutant p53 (+) and wild-type p53 (-). * P ≤ 0.05 indicate significant differences between variables and expression of ACSL4 group. The following linear regression (backward) equation was used for calculating the mRNA expression of gene of interest; ACSL4 mRNA expression = 0.386 p35 - 0.560 ki - 67 - 0.416 age
| ACSL4 mRNA expression | Unstandardized coefficients | Standardized coefficients | P values | |
|---|---|---|---|---|
| Models | Variables | B | Beta | |
| 1 | Constant | 1.796 | 0.001 | |
| age | -0.012 | -0.410 | 0.019* | |
| grade | 0.111 | 0.186 | 0.308 | |
| stage | -0.014 | -0.020 | 0.912 | |
| tumor size | 0.000 | 0.000 | 0.998 | |
| Estrogen receptors | 0.012 | 0.013 | 0.962 | |
| Progesterone receptors | -0.040 | -0.049 | 0.837 | |
| human epidermal growth factor | 0.030 | 0.097 | 0.583 | |
| Ki67 | -0.017 | -0.679 | 0.003* | |
| p53 | 0.252 | 0.343 | 0.056 | |
| 6 | Constant | 1.945 | 0.000 | |
| age | -0.012 | -0.416 | 0.006* | |
| Ki67 | -0.014 | -0.560 | 0.001* | |
| p53 | 0.284 | 0.386 | 0.014* | |
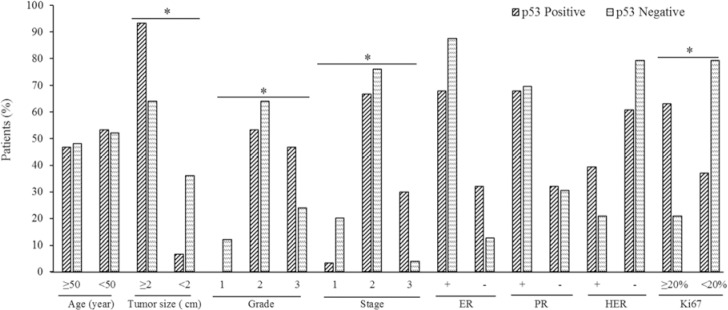
Fig. 3.
Relationship between p53 and clinicopathological variables of breast cancer. Results are according to chi-square test. Marked groups have significant association with p53 status, *P < 0.05. p53 positive was considered as mutant and p53 negative as wild-type. ER, Estrogen receptor; PR, progesterone receptor; HER, human epidermal growth factor receptor-2.
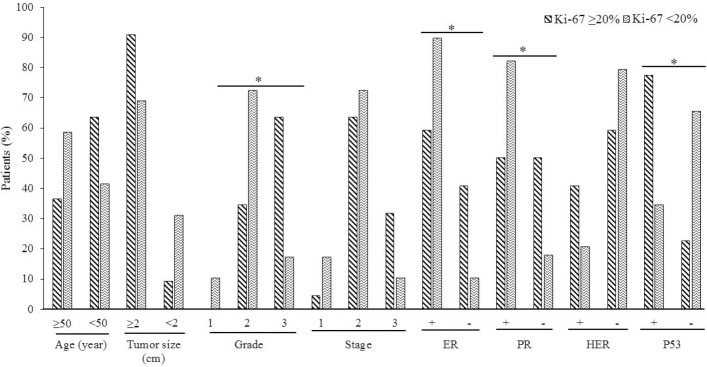
Fig. 4.
Relationship between Ki-67 and clinicopathological variables of breast cancer. Results are according to chi-square test. Marked groups have significant association with ki-67 expression. *P < 0.05. Ki-67 is reported as percent of expression. ER, Estrogen receptor; PR, progesterone receptor; HER, human epidermal growth factor receptor-2.
These analyses showed that by adjusting other variables in the model, ACSL4 expression in p53 positive patients was on an average 0.386 fold higher in comparison to individuals of p53 negative. Conversely, ACSL4 mRNA expression showed negative correlations with Ki-67 (-0.560) and age (-0.416), P < 0.05.
The expression of p53 protein was positively associated with tumor size (P = 0.007), pathology grade (P = 0.041), tumor stage (P = 0.010), and Ki-67 (P = 0.002, Fig. 3). In addition, the proliferation index expression, Ki-67, was positively associated with pathology grade (P = 0.001), and negatively correlated with estrogen receptor (ER) and progesterone receptor (PR) expression (P = 0.011, P = 0.016, respectively; Fig. 4).
DISCUSSION
Dysregulation in fatty acid metabolism has been indicated in several types of cancers including breast cancer (3,15), and suggests that increased fatty acid metabolism promotes tumor progression in various types of cancers (3). ACSL enzymes family catalyze the activation of long-chain fatty acids to the corresponding acyl-CoA esters, which is pre-requisite for the synthesis of triacylglycerols and phospholipids, or entering the β oxidation pathway (5). It has been shown that ACSL4, as one of the isoforms of ACSLs, overexpressed in various types of cancer (16).
In this study, we demonstrated that ACSL4 expression was enhanced in BC tumor tissue compared to the adjacent normal tissue. In agreement with our result, Chiyen et al. have reported high ACSL4 expression in breast cancer cell lines (17). In addition, the data presented in Table 2 showed that the expression of ACSL4 was negatively correlated with Ki-67 and age, and positively correlated with p53 status. Meanwhile, ACSL4 expression was not associated with the expression of ER, PR, human epidermal growth factor receptor-2 (HER2/neu) and other clinicopathological parameters in BC.
The expression of ER, PR, and HER2/neu is routinely assessed in BC patients as prognostic and predictive biomarkers in breast carcinoma (18). BC patients with tumor positive ER and/or PR have a reduction in the risk of mortality (19). While tumors with HER2/neu overexpression are more aggressive and have poor prognosis (18,20). Monaco and Chiyen et al. by analysis of the published expression databases, have reported that high expression of ACSL4 is negatively correlated with sex steroid (androgen and estrogen) receptors status in both BC and prostate cancers (17,21). Since estrogen regulates ER and PR, the correlation between ACSL4 and age may be modulated by estrogen levels.
The nuclear protein Ki-67, as a marker of cellular proliferation, is one of the main characteristics of cancer. According to several studies, high Ki-67 expression is significantly associated with poor prognosis, higher risk of recurrence, and lower survival rate (22). The negative correlation of Ki-67 with PR and ER, and its positive correlation with tumor grade (as poor prognostic index) (23) observed in this study confirm previous reports regarding the association of Ki-67 with poor prognosis in BC. Although, the mechanism of association of ACSL4 with Ki-67 is unknown, these data may suggest that ACSL4 may be a useful prognostic indicator in BC.
The p53 tumor suppressor status was evaluated in the study samples using IHC staining and samples were divided into mutant and wild-type groups. The mutant p53 is stable and is detectable by IHC, while the wild-type is unstable and degrades quickly, thus, not detectable by IHC (24,25). Our results showed that 54.5% of the study patients were p53 mutant and the remaining wild-type. The p53 expression has a significant association with the tumor size, stage, pathology grade, and Ki-67. Therefore, it may be considered as a biomarker of prognostic in BC patients. Accordingly, some studies have considered it as an independent predictor of survival (26) and a favorable prognostic indicator (27). On the other hand, other studies have considered p53 as a poor prognosis in triple-negative breast cancer (28). Therefore, further studies are needed to confirm the prognostic significance of p53 status in BC.
In addition to the ability of p53 in inducing senescence and apoptosis, it is widely believed that p53 regulation of lipid metabolism is via transcriptional control or protein-protein interaction. Therefore, p53 wild-type increases fatty acid oxidation while inhibiting glycolysis. This appears to play a key role in its underlying tumor suppressor functions. p53 can also inhibit fatty acid synthesis. Conversely, many p53 mutant cells have a different impact upon lipid metabolism and enhance fatty acid synthesis, via their gain-of- function activities (29,30).
Several mechanisms may contribute to the correlation between p53 and ACSL4. For example, miR-34, a tumor suppressor miRNA, is activated by p53 transcriptionally. In turn, overexpression of miR34 down-regulates ACSL4 expression (31,32). In this study, we have shown a positive association between p53 and ACSL4, which is in accordance with this mechanism and further confirming the role of p53 in the regulation of lipid metabolism.
Furthermore, some studies have indicated that mutant p53 sensitizes cells to ferroptosis, a form of regulated cell death, more than wild-type p53 in cancer cells (8,11). Since ACSL4 is an essential component for ferroptosis execution (33), the positive correlation observed between the mutant p53 and ACSL4 in our study may represent an increased tendency to ferroptosis in these samples. This function of ACSL4 can support the role of tumor-suppressive of ACSL4 that has been reported in gastric cancer (16), and also may confirm the results of a Kaplan-Meier plotter database that indicated a significant positive correlation between high ACSL4 expression and the patients’ survival rate in BC (3). Although, there are reports to indicate that ACSL4 expression may be associated with aggressive BC type (34), nevertheless, evidence regarding the role of tumor-suppressive of ACSL4 seems more convincing. It may be speculated that the increase of ACSL4 in p53 mutant tumors can be considered as the defense mechanisms of the body which has been created as a result of wild-type p53 elimination, as a tumor suppressor.
CONCLUSION
In conclusion, ACSL4 expression is upregulated in BC tumor tissue samples compared to the adjacent normal tissue in an Iranian population. The positive correlation observed between ACSL4 expression and mutant p53 status, on one hand, may provide evidence for the effect of p53 on the metabolism of lipids in tumors and on the other hand suggests the role of mutant p53 and ACSL4 in the onset of ferroptosis.
Therefore, this finding along with a negative significant association of ACSL4 with ki-67 suggests that the expression of ACSL4 may be considered as a prognostic indicator and as a tumor suppressor for BC patients. Therefore, it may be considered as a potential therapeutic target. However, further studies are needed to confirm the significance of these findings.
CONFLICT OF INTEREST STATMENT
The authors declare no conflict of interest for this study.
AUTHORS’ CONTRIBUTION
M. Pourfarzam conceived and supervised the study. M. Pourfarzam and N. Dinarvand designed experiments. N. Dinarvand performed experiments and analyzed data. S.M. Hakimian performed biopsies. H. Khanahmad ., A. Sheikhi, and B. Rashidi assisted with experimental design and data analysis. M. Pourfarzam and N. Dinarvand wrote the paper. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
This work was financially supported by Isfahan University of Medical Sciences, I.R. Iran (Grant No. 396510) and Dezful University of Medical Sciences, I.R. Iran (Grant No. IR.DUMS.REC.1396.35). The authors wish to thank Dr. S edighe Rastaghi and Dr. Shahrzad Baradaran for their excellent technical support.
REFERENCES
- 1.Wang D, Yin L, Wei J, Yang Z, Jiang G. ATP citrate lyase is increased in human breast cancer, depletion of which promotes apoptosis. Tumor Biol. 2017;39(4):1–10. doi: 10.1177/1010428317698338. [DOI] [PubMed] [Google Scholar]
- 2.Azordegan N, Fraser V, Le K, Hillyer LM, Ma DW, Fischer G, et al. Carcinogenesis alters fatty acid profile in breast tissue. Mol Cell Biochem. 2013;374(1-2):223–232. doi: 10.1007/s11010-012-1523-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen WC, Wang CY, Hung YH, Weng TY, Yen MC, Lai MD. Systematic analysis of gene expression alterations and clinical outcomes for long-chain acyl-coenzyme a synthetase family in cancer. PLoS One. 2016;11(5):1–23. doi: 10.1371/journal.pone.0155660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Yin L, Wei J, Yang Z, Jiang G. ATP citrate lyase is increased in human breast cancer, depletion of which promotes apoptosis. Tumour Biol. 2017;39(4) doi: 10.1177/1010428317698338. DOI: 101177/1010428317698338. [DOI] [PubMed] [Google Scholar]
- 5.Wu X, Deng F, Li Y, Daniels G, Du X, Ren Q, et al. ACSL4 promotes prostate cancer growth, invasion and hormonal resistance. Oncotarget. 2015;6(42):44849–44863. doi: 10.18632/oncotarget.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi Sebastiano M, Konstantinidou G. Targeting long chain acyl-CoA synthetases for cancer therapy. Int J Mol Sci. 2019;20(15):1–16. doi: 10.3390/ijms20153624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooke M, Orlando U, Maloberti P, Podestá EJ, Maciel FC. Tyrosine phosphatase SHP2 regulates the expression of acyl-CoA synthetase ACSL4. J Lipid Res. 2011;52(11):1936–1948. doi: 10.1194/jlr.M015552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20(7):1692–1704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 9.Belkaid A, Ouellette RJ, Surette ME. 17β-estradiol- induced ACSL4 protein expression promotes an invasive phenotype in estrogen receptor positive mammary carcinoma cells. Carcinogenesis. 2017;38(4):402–410. doi: 10.1093/carcin/bgx020. [DOI] [PubMed] [Google Scholar]
- 10.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17(4):351–369. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnanapradeepan K, Basu S, Barnoud T, Budina-Kolomets A, Kung CP, Murphy ME. The p53 tumor suppressor in the control of metabolism and ferroptosis. Front Endocrinol (Lausanne) 2018;9:124–130. doi: 10.3389/fendo.2018.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, et al. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10(8):1–16. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reisi P, Eidelkhani N, Rafiee L, Kazemi M, Radahmadi M, Alaei H. Effects of doxepin on gene expressions of Bcl-2 family, TNF-α, MAP kinase 14, and Akt1 in the hippocampus of rats exposed to stress. Res Pharm Sci. 2017;12(1):15–20. doi: 10.4103/1735-5362.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saadatian-Elahi M, Norat T, Goudable J, Riboli E. Biomarkers of dietary fatty acid intake and the risk of breast cancer: a meta-analysis. Int J Cancer. 2004;111(4):584–591. doi: 10.1002/ijc.20284. [DOI] [PubMed] [Google Scholar]
- 15.Wang YP, Lei QY. Perspectives of reprogramming breast cancer metabolism. Adv Exp Med Biol. 2017;1026:217–232. doi: 10.1007/978-981-10-6020-5_10. [DOI] [PubMed] [Google Scholar]
- 16.Ye X, Zhang Y, Wang X, Li Y, Gao Y. Tumor-suppressive functions of long-chain acyl-CoA synthetase 4 in gastric cancer. IUBMB Life. 2016;68(4):320–327. doi: 10.1002/iub.1486. [DOI] [PubMed] [Google Scholar]
- 17.Yen MC, Kan JY, Hsieh CJ, Kuo PL, Hou MF, Hsu YL. Association of long-chain acyl-coenzyme A synthetase 5 expression in human breast cancer by estrogen receptor status and its clinical significance. Oncol Rep. 2017;37(6):3253–3260. doi: 10.3892/or.2017.5610. [DOI] [PubMed] [Google Scholar]
- 18.Iqbal BM, Buch A. Hormone receptor (ER, PR, HER2/neu) status and proliferation index marker (Ki-67) in breast cancers: Their onco-pathological correlation, shortcomings and future trends. Med J D Y Patil Univ. 2016;9(6):674–679. [Google Scholar]
- 19.Dunnwald LK, Rossing MA, Li CI. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res. 2007;9(1):1–10. doi: 10.1186/bcr1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist. 2004;9(6):606–616. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 21.Monaco ME, Creighton CJ, Lee P, Zou X, Topham MK, Stafforini DM. Expression of long-chain fatty Acyl-CoA synthetase 4 in breast and prostate cancers is associated with sex steroid hormone receptor negativity. Transl Oncol. 2010;3(2):91–98. doi: 10.1593/tlo.09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ragab HM, Samy N, Afify M, El Maksoud NA, Shaaban HM. Assessment of Ki-67 as a potential biomarker in patients with breast cancer. J Genet Eng Biotechnol. 2018;16(2):479–84. doi: 10.1016/j.jgeb.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chia SK, Wykoff CC, Watson PH, Han C, Leek RD, Pastorek J, et al. Prognostic significance of a novel hypoxia-regulated marker, carbonic anhydrase IX, in invasive breast carcinoma. J Clin Oncol. 2001;19(16):3660–3668. doi: 10.1200/JCO.2001.19.16.3660. [DOI] [PubMed] [Google Scholar]
- 24.Milicevic Z, Bajic V, Živkovic L, Kasapovic J, Andjelkovic U, Spremo-Potparevic B. Identification of p53 and its isoforms in human breast carcinoma cells. ScientificWorldJournal. 2014;2014:1–10. doi: 10.1155/2014/618698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang P, Du C W, Kwan M, Liang SX, Zhang G J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci Rep. 2013;3:1–6. doi: 10.1038/srep02246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dookeran KA, Dignam JJ, Ferrer K, Sekosan M, McCaskill-Stevens W, Gehlert S. p53 as a marker of prognosis in African-American women with breast cancer. Ann Surg Oncol. 2010;17(5):1398–1405. doi: 10.1245/s10434-009-0889-3. [DOI] [PubMed] [Google Scholar]
- 27.Jin MS, Park IA, Kim JY, Chung YR, Im SA, Lee KH, et al. New insight on the biological role of p53 protein as a tumor suppressor: re-evaluation of its clinical significance in triple-negative breast cancer. Tumour Biol. 2016;37(8):11017–11024. doi: 10.1007/s13277-016-4990-5. [DOI] [PubMed] [Google Scholar]
- 28.Pan Y, Yuan Y, Liu G, Wei Y. P53 and Ki-67 as prognostic markers in triple-negative breast cancer patients. PLoS One. 2017;12(2):1–13. doi: 10.1371/journal.pone.0172324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parrales A, Iwakuma T. p53 as a regulator of lipid metabolism in cancer. Int J Mol Sci. 2016;17(12):1–11. doi: 10.3390/ijms17122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356(2 Pt A):197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai C, Gao Y, Zhang X, Yang W, Guan W. MicroRNA-34c acts as a bidirectional switch in the maturation of insulin-producing cells derived from mesenchymal stem cells. Oncotarget. 2017;8(63):106844–106857. doi: 10.18632/oncotarget.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro F, Lieberman J. miR-34 and p53: new insights into a complex functional relationship. PLoS One. 2015;10(7):1–23. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13(1):91–115. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, Li Y, Wang J, Wen X, Marcus MT, Daniels G, et al. Long chain fatty Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone resistance in human breast cancer. PLoS One. 2013;8(10):1–20. doi: 10.1371/journal.pone.0077060. [DOI] [PMC free article] [PubMed] [Google Scholar]