ABSTRACT
Bacterial biofilms can cause medical problems and issues in technical systems. While a large body of knowledge exists on the phenotypes of planktonic and of sessile cells in mature biofilms, our understanding of what happens when bacteria change from the planktonic to the sessile state is still very incomplete. Fundamental questions are unanswered: for instance, how do bacteria sense that they are in contact with a surface, and what are the very initial cellular responses to surface contact. Here, we review the current knowledge on the signals that bacteria could perceive once they attach to a surface, the signal transduction systems that could be involved in sensing the surface contact and the cellular responses that are triggered as a consequence to surface contact ultimately leading to biofilm formation. Finally, as the main obstacle in investigating the initial responses to surface contact has been the difficulty to experimentally study the dynamic response of single cells upon surface attachment, we also review recent experimental approaches that could be employed to study bacterial surface sensing, which ultimately could lead to an improved understanding of how biofilm formation could be prevented.
Keywords: surface sensing, planktonic cells, sessile cells, signal transduction, adhesion, Escherichia coli
This review describes the current knowledge on bacterial surface sensing, as the very initial step in bacterial biofilm formation.
INTRODUCTION
Bacterial biofilms generate significant technological and therapeutic problems. These issues range from increased fuel consumption of ships due to higher flow resistance (Callow and Callow 2011; Schultz et al. 2011), via fouling of membranes in water treatment facilities (Subramani and Hoek 2010), to serious medical problems. For instance, it is estimated that two-thirds of human infections involve biofilm formation, including infections of the urinary tract, lungs and ears, dental plaque and fouling of implants and contact lenses (Potera 1999). The increased tolerance of bacteria in biofilms towards antimicrobial compounds and the host immune system constitutes a central issue in treatment of bacterial infections (Mah and O'Toole 2001).
While the developmental steps leading to a mature biofilm are reasonably well-characterised, at least in laboratory conditions (Costerton et al. 1995; Monds and O'Toole 2009; Laverty, Gorman and Gilmore 2014), little is known about the first step of biofilm development. Specifically, it is unclear how cells initially sense the surface, which eventually leads to phenotypic adjustment from the planktonic (suspended) to the sessile (surface-attached) state, involving substantial changes in gene expression (Prigent-Combaret et al. 1999; Kuchma and O'Toole 2000; Beloin et al. 2004; Domka et al. 2007). The main reason for this limited knowledge possibly lies in the challenge to experimentally investigate the dynamic response of single cells on a very short timescale during the transition between these two states.
Here, towards pointing out the existing gaps in our understanding of surface sensing, we review the current knowledge on how bacteria recognise and respond to surface attachment with a special emphasis on the model organism Escherichia coli. Although we also describe the attachment process, the sensing of this attachment and the subsequent downstream effects are the focus of this review. For more extensive descriptions of the bacterial attachment process, the reader is referred to another recent review (Berne et al. 2018a). Specifically, we will first review the signals a cell might perceive in close proximity to, or in contact with, a surface and describe the mechanisms that bacteria employ to perceive the presence of and attachment to a surface. While not all biofilms are surface-attached and may instead be formed by bacterial aggregates (Kragh et al. 2016; Melaugh et al. 2016; Sønderholm et al. 2017), such non-surface-connected biofilms are beyond the scope of this review. Second, we report the initial downstream effects that are triggered in response to surface attachment. Third, as the limited understanding of surface sensing and of the very first steps of biofilm formation is likely connected with the fact that the respective processes are difficult to study, we highlight recent technological developments that might support future research on further elucidating the process of surface sensing. Closing the knowledge gaps will offer valuable insights on how to combat biofilm formation.
SURFACE SENSING
Bacteria can adhere to a large variety of surfaces, including glass, metals, many different polymers, as well as to other bacteria and eukaryotic cells (for a review, see Tuson and Weibel 2013; Berne et al. 2018a). In fact, it is practically impossible to develop a surface that cannot be colonized by bacteria, making it resistant towards biofouling, while at the same time being harmless to humans and the environment (Callow and Callow 2011). When a planktonic bacterial cell advances towards a surface from the bulk of a liquid, there are three different cues that might be sensed: (i) changes in physicochemical properties, (ii) attachment of cell appendages and (iii) attachment of the cell body (Fig. 1).
Figure 1.
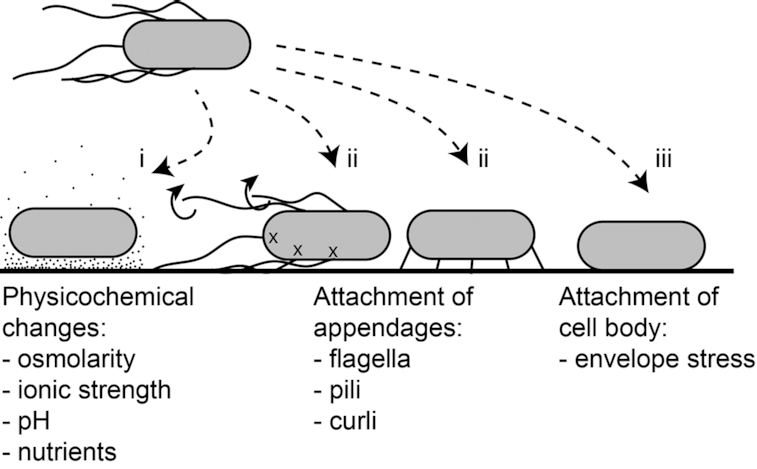
Schematic overview of the different signals that a surface may present to an approaching cell. When a cell approaches and contacts the surface, it may sense (i) different physicochemical properties compared to the bulk liquid, (ii) attachment of cell appendages and (iii) envelope stress due to attachment of the cell body.
Physicochemical changes
The microenvironment close to the surface differs from the bulk liquid in terms of ionic strength, osmolarity, pH and nutrient availability (for reviews, see Goodman and Marshall 1995; Berne et al. 2018a). Since many surfaces are charged, counter-ions accumulate at the solid-liquid interface. In the case of a negatively charged surface, which is the most common, these counter-ions include protons, causing a lowered pH at the surface. It has been suggested that close to a glass surface, the pH may decrease by as much as two units (Hong and Brown 2010).
Furthermore, organic molecules, present in the bulk liquid or secreted by bacteria, can be adsorbed onto surfaces and form a conditioning film (reviewed in Tuson and Weibel 2013; Berne et al. 2018a). Such films can consist of a mixture of (macro)molecules, including proteins, amino acids, lipids and polysaccharides (Loeb and Neihof 1975; Taylor et al. 1997; Garg, Jain and Bhosle 2009) – nutrients that can be metabolised by attached bacteria (Marshall 1988; Samuelsson and Kirchman 1990). Sessile bacteria may therefore be able to grow, even when the nutrient concentration in the bulk liquid is insufficient to sustain growth of planktonic cells (Zobell 1943; Kjelleberg, Humphrey and Marshall 1982).
To sense changes in pH, ionic strength, osmolarity and nutrient availability, bacteria typically employ two-component signal transduction, consisting of a membrane-bound histidine kinase, which senses the stimulus and a cytoplasmic response regulator mediating the cellular response. As physicochemical properties can vary also in the bulk liquid, the stimuli of respective systems are not specific for surfaces. To the best of our knowledge, direct involvement of these signals in surface sensing also has not been demonstrated. However, as we describe below, multiple systems that are responsive to these signals have downstream target genes linked to biofilm formation, which indicates that these systems could be part of the surface sensing machinery.
In E. coli, for instance, systems that sense physicochemical properties and have downstream targets with a role in biofilm formation are CpxAR (Danese and Silhavy 1998; Jubelin et al. 2005; Clarke and Voigt 2011; Raivio 2014), EnvZ/OmpR (Hall and Silhavy 1981; Heyde and Portalier 1987; Mizuno and Mizushima 1990; Thomas and Booth 1992; Pratt et al. 1996; Sato et al. 2000; Clarke and Voigt 2011), RcsCDB (Sledjeski and Gottesman 1996; Francez-Charlot et al. 2005) and possibly BasSR (Hagiwara, Yamashino and Mizuno 2004; Lee, Barrett and Poole 2005; Perez and Groisman 2007). As will also be described below, these systems regulate biofilm-related genes (Table 1), with for instance motility being regulated by CpxAR (Raivio, Leblanc and Price 2013), EnvZ/OmpR (Shin and Park 1995) and RcsCDB (Francez-Charlot et al. 2003). The downstream effects are discussed in more detail below.
Table 1.
An overview of the sensing systems in E. coli that can sense properties associated with surface proximity or attachment. Only the inducing signals that may be relevant for surface sensing, as described in the section 'Surface sensing', are shown. Also, the downstream targets are limited to those that are related to biofilm formation and described in the section 'Downstream effects of the potential surface sensing pathways’.
| Sensing system | Adhesion-related inducing signals | Biofilm-related targets |
|---|---|---|
| BaeSR | Envelope stress (Raffa and Raivio 2002; Leblanc, Oates and Raivio 2011) | — |
| BarA/UvrY | Attached pili (Zhang and Normark 1996) | Motility (Suzuki et al. 2002; Weilbacher et al. 2003; Jonas et al. 2008), pili (Mitra et al. 2013) and matrix (Wang et al. 2005) |
| BasSR | pH (Perez and Groisman 2007) | Curli (Ogasawara et al. 2012) and pili (Ogasawara et al. 2012) |
| CpxAR | Osmolarity (Prigent-Combaret et al. 2001; Jubelin et al. 2005), pH (Danese and Silhavy 1998; Clarke and Voigt 2011), pili subunits (Jones et al. 1997) and envelope stress (Otto and Silhavy 2002; Hirano et al. 2007) | Motility (Raivio, Leblanc and Price 2013) and curli (Evans and Chapman 2014) |
| EnvZ/OmpR | Osmolarity (Hall and Silhavy 1981; Mizuno and Mizushima 1990; Pratt et al. 1996), pH (Heyde and Portalier 1987; Thomas and Booth 1992; Sato et al. 2000; Clarke and Voigt 2011) | Motility (Shin and Park 1995) and curli (Evans and Chapman 2014) |
| PSP | Envelope stress (Jovanovic et al. 2006) | Motility (Jovanovic et al. 2006) |
| RcsCDB | Osmolarity (Sledjeski and Gottesman 1996; Francez-Charlot et al. 2005) and envelope stress (Parker et al. 1992; Chen et al. 2001; Majdalani et al. 2005; Farris et al. 2010) | Motility (Francez-Charlot et al. 2003), curli (Evans and Chapman 2014), pili (Ferrières and Clarke 2003), Ag43 (Ferrières and Clarke 2003) and matrix (Gottesman, Trisler and Torres-Cabassa 1985) |
| Sigma factor σE | Envelope stress (Mecsas et al. 1993; Walsh et al. 2003; Lima et al. 2013) | — |
| Flagella | Hindered rotation (Lele, Hosu and Berg 2013; Tipping et al. 2013; Nord et al. 2017) | Unknown in E. coli |
Alternatively, close to surfaces, bacteria could sense increased nutrient concentrations via changes in metabolism. The uptake of a compound may in some cases act as a signal that is transmitted by the respective transporter to a sensor protein (for a review, see Tetsch and Jung 2009). If the adsorbed nutrients are metabolised, this will change the intracellular metabolic fluxes and may in turn affect gene expression (Kotte, Zaugg and Heinemann 2010; Kochanowski et al. 2013). The metabolic state of the cell is also influenced by alterations in pH, as a local drop in pH close to a negatively charged surface can directly affect the proton motive force (Hong and Brown 2009).
Overall, the physicochemical properties in the vicinity of a surface may trigger a cellular response via two-component systems. However, because these properties can also vary in the bulk liquid, these stimuli are not specific for surfaces and are therefore unlikely to be the main cue indicating surface contact.
Attachment of cell appendages
After describing the physicochemical changes that a bacterium experiences upon approaching a surface, we will next describe the next process that occurs when a cell reaches a surface, which is the attachment of cell appendages. We review how these attachment processes can be sensed.
Flagella
When bacteria approach a surface, cell appendages will stick to it. Adhesion is supported by flagella, which due to their hydrophobic nature particularly adhere to hydrophobic surfaces (Pratt and Kolter 1998; van Houdt and Michiels 2005; Wood et al. 2006; Bruzaud et al. 2015; Friedlander, Vogel and Aizenberg 2015; Berne et al. 2018a). Not only the presence of flagella but also the ability to rotate them is important for adhesion, as E. coli mutants with non-functional flagella are impaired in biofilm formation and detach more readily compared to the wild-type (Wood et al. 2006; Yoshihara et al. 2015). In contrast, possessing flagella was found to reduce adhesion in Caulobacter crescentus (Berne et al. 2018b), indicating the complexity of the adhesion process.
Once attached, flagella can provide signals to the cell indicating surface contact, which originate from hindered rotation (for a review, see Belas 2014). It was recently shown that when flagellar rotation is blocked, either by mutations in flagellar motor genes or by addition of anti-flagellin antibodies, the DegS-DegU signal transduction pathway, controlling biofilm formation, is activated in Bacillus subtilis (Cairns et al. 2013). Furthermore, in Vibrio parahaemolyticus, an organism that has been long known to sense flagellar inhibition as a signal to initiate swarming (McCarter, Hilmen and Silverman 1988), a transcriptomics study both on mutant strains defective in flagellar rotation and on wild-type cells treated with flagellum-inhibiting drugs, found a gene expression pattern similar to sessile cells (Gode-Potratz et al. 2011). Specifically, about half of the genes that were differentially expressed in surface-attached cells also had altered expression levels when rotation was impaired, suggesting that the flagella are a main surface sensor in this organism. The E. coli flagellum has also been shown to be sensitive towards mechanical forces: the number of stators, i.e. the force-generating protein complexes of the flagellar motor, increases within minutes when the load of rotation is increased by binding a microbead to truncated flagella (Lele, Hosu and Berg 2013; Tipping et al. 2013; Nord et al. 2017).
How hindered flagellar rotation is sensed on the molecular level seems to vary between bacterial species and the mechanism has not been resolved in all cases. In B. subtilis, the interaction affinity of the flagellar motor and the cytoplasmic histidine kinase DegS may be affected by the rotation of the motor, such that halted rotation leads to activation of the DegS-DegU two-component system (Cairns et al. 2013). In other cases, signal transduction of flagellar attachment might go via the flagellar stator-associated FliL protein (Cairns et al. 2013). In one study, FliL was shown to play an important role in sensing the presence of a surface, as respective deletion mutants of Proteus mirabilis and E. coli responded differently to soft agar surfaces than wild-type cells, in terms of motility and gene expression (Lee and Belas 2015). However, another study reported that FliL plays no significant role in mechanosensitivity of the E. coli flagellum, instead suggesting that higher torque in hindered flagella results in exposure of binding sites on the flagellar rotor (Chawla, Ford and Lele 2017). Other explanations for surface sensing by flagella are that, after rotation has stopped, a reduced ion flux through the flagellar motor may impact the membrane potential and energy state of the cell (Cairns et al. 2013) or that blocked rotation could result in an increased torque of the motor, leading to perturbations in the cell envelope (Belas 2013). Sensing surface contact via flagella may also be unrelated to the rotation of this appendage, as is the case in C. crescentus. Here, it was found that arresting flagellar rotation does not lead to the expected downstream response, namely, synthesis of the holdfast structure (Berne et al. 2018b). Instead, the flagellar motor senses the surface by direct contact, via an unresolved mechanism that might involve altered intracellular pH due to changes in ion fluxes (Hug et al. 2017). Sensing via the motor results in synthesis of the second messenger molecule cyclic diguanylate (c-di-GMP) by the motor-associated diguanylate cyclase DgcB (Hug et al. 2017). A complex regulatory mechanism then reinforces the switch to the sessile phenotype by inhibition of motility (Nesper et al. 2017). Thus, while flagella are known to contribute to the perception of surface contact in a range of bacterial species, they might employ different sensing mechanisms.
Pili
In addition to flagella, also pili (fimbriae) attach to surfaces and support biofilm development (Berne et al. 2018a). Their importance is illustrated by a genome-wide study in E. coli, which revealed that loss of genes encoding type I pili had the most detrimental effect out of all single-gene deletions on the formation of biofilms (Niba et al. 2007). The importance of type I pili for adhesion is also illustrated by the finding that attachment to a variety of abiotic surfaces can be greatly reduced by addition of mannose to the medium (Pratt and Kolter 1998). As the FimH subunit on the tip of type I pili is known to bind to mannose, which is also present on eukaryotic cells (Old 1972; Ofek, Mirelman and Sharon 1977), the decreased adhesion is likely due to a reduced interaction of the mannose-saturated pili with the surface (Old 1972; Pratt and Kolter 1998).
Also E. coli’s curli (a type of pili) stick to surfaces and are highly beneficial for adhesion (Pratt and Kolter 1998; Vidal et al. 1998; van Houdt and Michiels 2005; Niba et al. 2007). Increased production of curli through a point mutation in the ompR gene has been shown to enhance surface attachment, while mutations generating curli-deficient cells were found to result in a more than 50% reduction in biofilm formation (Vidal et al. 1998; Niba et al. 2007). A wide range of pili are known to exist in various bacterial species but also within single species generally multiple types of pili are expressed. For example, in addition to type I pili and curli, E. coli may also carry P-pili, type IV pili and several others (Wurpel et al. 2013; Berne et al. 2015).
Mediating adhesion to the surface is a main role of pili but they may have other functions as well. In the case of type IV pili, that are present in many organisms, a continuous process of extension, eventual attachment and retraction facilitates twitching motility on the surface (Skerker and Berg 2001; Xicohtencatl-Cortes et al. 2009). In Pseudomonas aeruginosa, another type of pilus, Cup, does not only mediate cell-surface interactions but seems to also be involved in cell–cell aggregation (D'Argenio et al. 2002; Ruer et al. 2007), underscoring the importance of pili also during subsequent stages of biofilm formation. In C. crescentus, one role of the polar Tad pili might be to bring the cell pole into close proximity of the surface, such that the flagellar motor can sense the contact (Hug et al. 2017; Sangermani et al. 2019). Countless other cell appendages exist, but listing them all is beyond the scope of this review and the reader is referred to a comprehensive review (Berne et al. 2015). Overall, bacteria use a wide range of pili mainly to mediate adhesion to surfaces but also for a variety of other functions.
The attachment of pili is also known to be sensed. For instance, in E. coli attachment of pili was found to result in altered gene expression (Zhang and Normark 1996; Otto et al. 2001; Bhomkar et al. 2010). While here the molecular mechanism is not fully clear, for P. aeruginosa, it has been suggested that the continuously extending and retracting type IV pili perceive tension when attached pili are retracted and that these forces may lead to depolymerisation of the pili and/or conformational changes in pilus subunits, which then enable the interaction between the major pilus subunit PilA and the sensor protein PilJ (Persat et al. 2015). As a result, a signalling pathway is activated that produces the second messenger cyclic AMP to initiate biofilm formation (Persat et al. 2015; Inclan et al. 2016). In C. crescentus, a different type of pilus, Tad, using a comparable mechanism, senses the inability to retract once it is attached to a surface and then possibly stimulates signalling via c-di-GMP, leading to synthesis of a holdfast structure that mediates adhesion (Ellison et al. 2017). Besides sensing adhesion, these Tad pili also play a role in bringing the polar flagellar motor into contact with the surface, which is another cue for adhesion (Hug et al. 2017; Sangermani et al. 2019). Alternatively, pili-mediated attachment could be sensed as an accumulation of mislocalised pilus subunits in the periplasm of attached cells (Mulvey et al. 1998), known to induce the CpxAR two-component system in E. coli (Jones et al. 1997). A second signalling system in E. coli that has been implicated in pili-mediated sensing of the surface is the BarA/UvrY two-component system: the transcription of barA is stimulated by P-pilus attachment in uropathogenic E. coli by a yet unknown mechanism (Zhang and Normark 1996). For a recent review that covers surface sensing via type IV pili, see O'Toole and Wong (2016). Thus, bacteria have different mechanisms in place to sense the adhesion of pili, leading to altered gene expression.
In summary, flagella and pili do not only facilitate adhesion to the surface but also transmit signals that allow bacteria to respond to this adhesion. Surface sensing via cell appendages has been found in multiple species, indicating that it is a common mechanism to perceive surface contact in bacteria.
Cell body attachment
In addition to surface attachment via cell appendages, also the cell body can attach to the surface. We will next first describe the attachment process of the cell body and the forces involved, where the reader is also referred to another recent review (Berne et al. 2018a), and we will then review potential mechanisms to sense attachment of the cell body. Here, adhesion to a surface is mainly mediated by Van der Waals, electrostatic and acid-base interactions between the bacterium and the surface (for reviews see (Hermansson 1999; Renner and Weibel 2011; Berne et al. 2018a)). Upon approaching a surface, a bacterium will initially be attracted by the long-range Van der Waals forces, but short-range repulsive electrostatic forces, for instance provided by the fact that most bacteria and material surfaces are both negatively charged (Jucker, Harms and Zehnder 1996), may subsequently prevent close contact to the surface. This phenomenon is the basis of the Derjaguin-Landau-Verwey-Overbeek theory, which has provided insight in explaining cellular adhesion, although it is a highly simplified representation (Derjaguin and Landau 1941; Verwey and Overbeek 1948). Due to the oppositely oriented forces, the energy minimum may lie at a distance on the order of tens of nanometers from the surface (Figs 2A and 2B) (Simoni et al. 1998). Shielding of the charges on the surface by a conditioning film or high ionic strength of the medium can decrease the contribution of the electrostatic forces, allowing a smaller distance between cell and surface (Marshall, Stout and Mitchell 1971; Renner and Weibel 2011). Physical contact of the cell body with the surface during adhesion can furthermore be facilitated by the long O-antigen part of lipopolysaccharides (LPS) or by cell appendages, while also polar adhesion can be a way to reach surface contact before transitioning to a flat orientation (Fig. 2C) (Feldner, Bredt and Kahane 1983; Jucker et al. 1997; Pratt and Kolter 1998; Prigent-Combaret et al. 2000; Jones et al. 2003; van Houdt and Michiels 2005; Chao and Zhang 2011; DeBenedictis, Liu and Keten 2016; Berne et al. 2018a). Here, it should be noted though, that some organisms do not transition to a flat orientation on the surface, instead remaining solely attached by the cell pole (Ellison et al. 2017; Hug et al. 2017).
Figure 2.
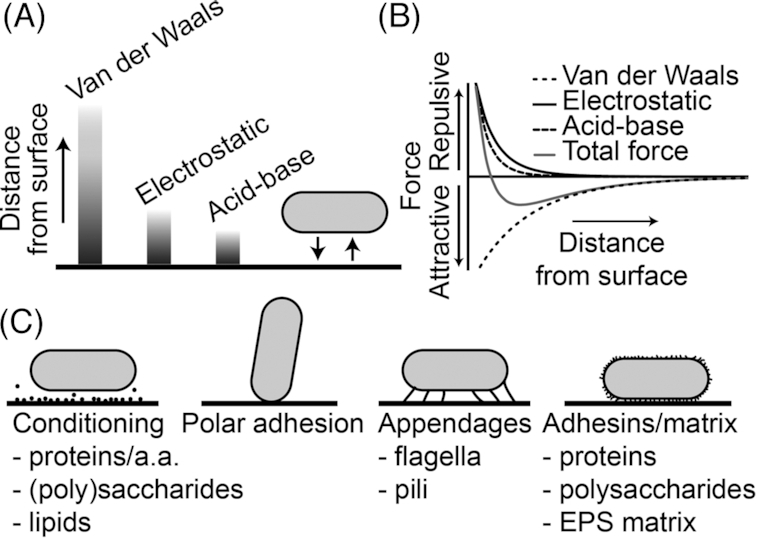
Summary of the forces involved in adhesion and strategies to overcome unfavourable surface interactions. (A), Schematic representation of forces acting on a bacterium when it approaches a surface, with the balance between attractive and repulsive forces keeping the bacterium at a small distance from the surface. (B), Adhesion forces as a function of distance from the surface. Van der Waals forces are attractive, electrostatic forces are generally repulsive and acid-base forces might be either. The total force reaches an energy minimum, which determines the separation of the bacterium from the surface (Busscher et al. 2010). (C), Strategies employed by bacteria to achieve a smaller separation from the surface than allowed by the initial energy minimum, which include (i) conditioning of the surface with e.g. proteins, amino acids, (poly)saccharides and lipids, (ii) polar adhesion, (iii) attachment of appendages and (iv) production of adhesive proteins and polysaccharides that are exposed on the cell surface.
Following the initial adhesion of the cell body, gradually the attachment becomes stronger over a time window of seconds to minutes, which is facilitated by rearrangements at the interface between cell envelope and surface that progressively maximise attractive interactions, e.g. by removal of interfacial water, protein conformational changes and an increase of favourable acid-base and hydrophobic interactions between cell and surface (Boks et al. 2008; Busscher et al. 2010). Indeed, establishing initial stable attachment to the surface does not require biological activity, as the adhesion force of polystyrene particles to glass has also been found to strongly increase within minutes on a surface (Meinders and Busscher 1993).
At this stage, the adhesive strength could still be reinforced through production of adhesins. Irreversible adhesion in fact does not require a large contact area between cell and surface. For instance, C. crescentus can irreversibly attach in a polar orientation due to the production of adhesins at the cell pole (Ellison et al. 2017; Hug et al. 2017). Similar results have been found for Asticcacaulis biprosthecum and Agrobacterium tumefaciens, where also polar adhesins were produced within minutes following surface contact, which shows that rapid surface-induced strengthening of polar adhesion is a common strategy in multiple genera (Li et al. 2012). However, other bacterial species preferentially assume a flat orientation on the surface. For example, in P. aeruginosa, production of the Pel polysaccharide, a component of the exopolymeric matrix, stimulates the transition from polar adhesion to a flat orientation by increasing short-range attractive interactions (Cooley et al. 2013). Pseudomonas fluorescens uses a similar mechanism to transition to irreversible attachment, secreting the large adhesive protein LapA, which remains associated with the cell-surface, to assume a flat orientation (Hinsa et al. 2003). Initial adhesion does not necessarily result in irreversible attachment within a short time span, as bacteria may first explore the surface by moving (or swarming) over it, using Type IV pili or flagella (Mobley and Belas 1995; Gibiansky et al. 2010; Lee and Belas 2015). Also, some bacteria possess detachment programs to allow one cell to leave the surface following division (Conrad et al. 2011; Laventie et al. 2019). Recently, it was found that detached P. aeruginosa cells retain a multigenerational memory, mediated by oscillations of cAMP levels and Type IV pili activity, that prepares them for a stronger adhesion when a new surface is encountered (Lee et al. 2018). These examples illustrate that the adhesion response upon surface contact can differ between bacterial species and even between single genetically identical cells.
Following irreversible adhesion, bacteria stick together mainly by production of an exopolymeric matrix and via their appendages (van Houdt and Michiels 2005), but also by synthesizing adhesins (e.g. Ag43 for E. coli) that promote aggregation of bacteria (van der Woude and Henderson 2008).
The importance of understanding adhesion forces is exemplified by the effect that combined adhesion forces between cell and surface have on the fate of the attached cell (Busscher and van der Mei 2012). Weak adhesion forces (e.g. on polymer brush-coatings) can lead to the formation of unstable biofilms with aberrant structure and smaller thickness (Nejadnik et al. 2008; Gu et al. 2017). Possibly, such weak interactions do not provide sufficient signal for the adhered bacteria to transition to subsequent steps in the development of the biofilm, such that they fail to form a robust matrix-enclosed structure. On the other hand, in the case of too strong adhesion forces, which bacteria especially encounter on positively charged surfaces, extensive envelope stress and loss of viability may occur (Liu, Strauss and Camesano 2008; Busscher and van der Mei 2012). In the intermediate regime, corresponding to interactions between bacteria and many common materials, the forces are sufficiently strong to trigger biofilm formation, without affecting viability (Busscher and van der Mei 2012). Generally, surfaces are not perfectly homogeneous but instead contain patches with different properties, in terms of surface charge, hydrophobicity, roughness and composition of the conditioning film (Ren et al. 2018). This means that attached bacteria may experience different interactions with the same surface, which, together with the limited range of interbacterial communication, was proposed to be the cause of commonly observed heterogeneous micro-environments within biofilms where bacteria have distinct phenotypes (Ren et al. 2018). Thus, understanding the forces that govern bacteria-surface interactions not only allows us to predict whether irreversible adhesion will occur but might also explain emergent properties in mature biofilms.
Physical contact of the cell body with a surface is widely thought to be sensed as envelope stress (Otto and Silhavy 2002; Ferrières and Clarke 2003; Lejeune 2003; Hirano et al. 2007; Dorel 2010; Busscher and van der Mei 2012; Morgenstein and Rather 2012; Harapanahalli et al. 2015b; O'Toole and Wong 2016), which is often defined as the presence of unfolded proteins (Jones et al. 1997; DiGiuseppe and Silhavy 2003) and LPS (Lima et al. 2013) in the periplasm. However, to the best of our knowledge, there is no direct experimental evidence for such surface-induced protein unfolding or LPS mislocalization. If a broader definition of envelope stress is used, including all perturbations of the extracytoplasmic space of the cell, such as altered membrane curvature (Conter et al. 2002; Mcdonald et al. 2015) and loss of membrane integrity (Guilvout et al. 2006), then adhesion of the cell body can indeed be seen as a cause of envelope stress. Specifically, there is evidence, obtained in Staphylococcus aureus with three different experimental techniques, that adhesion causes deformations of the cell shape, and thus altered membrane curvature (Chen et al. 2014; Harapanahalli et al. 2015a), and compresses the thickness of the cell envelope near the site of surface contact (Gu et al. 2017). Such attachment-related membrane tension has also been proposed to cause mechanosensitive channels to open (Harapanahalli et al. 2015b), potentially having large effects on cellular homeostasis.
Usually, cellular responses to envelope stress are studied after invoking the stress by chemical means (e.g. treatment with compounds that insert into or disrupt the lipid bilayer (Conter et al. 2002; Farris et al. 2010)), or by genetic modifications (e.g. overexpression of membrane proteins (DiGiuseppe and Silhavy 2003) or mutations in the synthesis pathways of LPS or phospholipids (Parker et al. 1992; Mileykovskaya and Dowhan 1997; Keller et al. 2015)). Since a number of signalling systems that respond to such laboratory-induced envelope stress regulate biofilm-associated genes, it is plausible that surface-induced envelope stress upon physical contact of the cell body may be a way to sense adhesion. While in the following, we will describe the envelope stress response systems with a focus on E. coli, it should be noted that in many cases homologous systems exist in other species. For instance, as described below, the Rcs system is also found in P. mirabilis (Morgenstein and Rather 2012) and P. aeruginosa (Mikkelsen et al. 2009), and the Cpx system is also found in multiple organisms (Raivio 2014).
Besides the above-mentioned physicochemical parameters and attachment of pili, envelope stress can activate the E. coli CpxAR two-component system. Activation can happen when there are defects in LPS assembly (Klein et al. 2009, 2014). Also, unfolded proteins can induce the system, by binding to the auxiliary regulator CpxP, which lifts its inhibition of the histidine kinase CpxA (Raivio, Popkin and Silhavy 1999; Zhou et al. 2011; Vogt and Raivio 2012; Tschauner et al. 2014). Once bound to an unfolded protein, the direct interaction of CpxP with CpxA is released and CpxP is proteolysed (Isaac et al. 2005). It has also been suggested that, in response to physical contact with a surface, the Cpx system gets activated via the outer membrane lipoprotein NlpE (Otto and Silhavy 2002). Its tertiary structure might predispose NlpE to partial unfolding by membrane perturbations, leading to activation of the kinase functionality of the inner membrane protein CpxA, supposedly by direct interactions between these proteins (Hirano et al. 2007). If this model is correct, then it exemplifies how envelope stress can be exploited by the cell as a signal for surface attachment. However, recent findings indicate that the CpxAR system is not activated by NlpE-mediated surface sensing, as the previously reported results could not be reproduced, neither by employing the original population-level assay, nor by a novel single-cell experimental approach (Kimkes and Heinemann 2018), consistent with recent findings on the interaction between NlpE and CpxA (Delhaye, Laloux and Collet 2019). Thus, more research is needed to clarify if and how CpxAR is involved in surface sensing.
Envelope stress can also activate the RcsCDB phosphorelay. The activity of this system was found to be altered upon deletion of genes involved in LPS synthesis (Parker et al. 1992; Majdalani et al. 2005), mutation or overexpression of genes encoding envelope-localised proteins (Chen et al. 2001; Majdalani and Gottesman 2006), and upon membrane damage caused by the action of antimicrobial peptides (Farris et al. 2010). When E. coli is grown on a solid surface, the expression of genes controlled by the Rcs phosphorelay has been shown to increase rapidly, but the underlying mechanism had not been solved (Ferrières and Clarke 2003). Also in P. mirabilis, it was found that the Rcs system is responsive to surface contact, but contrary to what was found in E. coli, surfaces inhibit the Rcs system in P. mirabilis (Morgenstein and Rather 2012). Likely, the outer membrane lipoprotein RcsF is involved in surface sensing. RcsF is inserted into the outer membrane via the major subunit of the β-barrel assembly machinery BamA and forms complexes with several abundant β-barrel proteins (Fig. 3) (Cho et al. 2014; Konovalova et al. 2014). Here, part of RcsF is exposed on the cell surface, which was proposed to enable RcsF to sense perturbations of the LPS layer (Konovalova et al. 2014). Upon envelope stress, BamA may fail to generate the complexes between RcsF and other outer membrane proteins, thereby enabling RcsF to activate the Rcs phosphorelay via the inner membrane receptor IgaA (Cho et al. 2014). This model is in agreement with findings that inner membrane anchored RcsF or truncated forms that localise to the periplasm, constitutively activate the Rcs phosphorelay (Farris et al. 2010). A recent study found that it is crucial for activation of the pathway that RcsF has the correct length to span the periplasm (Asmar et al. 2017). Possibly, the activation of the Rcs system on a surface might be facilitated by small deformations in the cell envelope that affect the distance between the membranes, enabling the direct contact.
Figure 3.
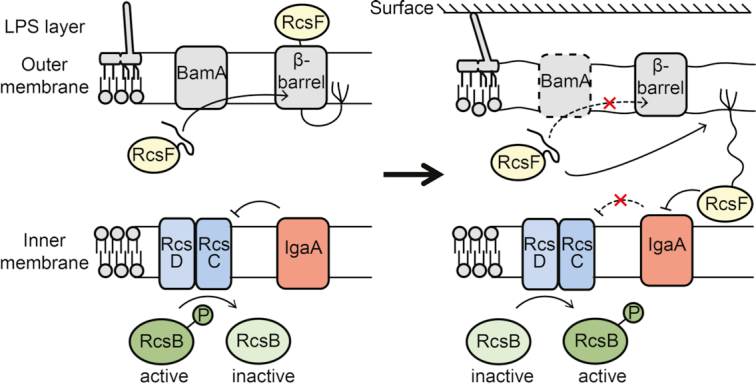
Proposed method of activation of the Rcs system via the outer membrane sensor RcsF. Left: Under non-inducing conditions, the lipoprotein RcsF is tethered to the outer membrane and threads through the lumen of β-barrel proteins (Cho et al. 2014; Konovalova et al. 2014) (the two studies reported opposite orientations of RcsF in the membrane). In this state, RcsF is unable to contact its inner membrane receptor IgaA, and can therefore not activate the Rcs system, i.e. the dephosphorylation activity of RcsC predominates. Right: By a yet unclear mechanism, envelope stress can interfere with the ability of periplasmic chaperones or BamA to localize RcsF to its regular location and thereby enables it to activate the Rcs system (Cho et al. 2014).
Three other systems in E. coli that are or might be responsive to envelope stresses, although without any clear involvement in surface sensing or role in biofilm formation, are the sigma factor σE (Mecsas et al. 1993; Lima et al. 2013), the phage shock protein (PSP) response system (Brissette et al. 1991; Jovanovic, Weiner and Model 1996; Lloyd et al. 2004; Jovanovic et al. 2006) and the BaeSR two-component system (Nagasawa, Ishige and Mizuno 1993; Raffa and Raivio 2002).
Also in P. aeruginosa, surface-induced envelope stress could be a trigger for biofilm initiation. Specifically, contact with a surface can lead to the activation of the Wsp pathway (Güvener and Harwood 2007; Francis, Stevenson and Porter 2017). It has been proposed that deformation of the cell envelope is the signal that is sensed by this pathway, as the WspA protein forms weakly interacting clusters in the inner membrane that might be affected by mechanical forces originating from bacteria-surface interactions (O'Connor et al. 2012). Activation of this system leads to the phosphorylation of the diguanylate cyclase WspR and, in turn, the produced c-di-GMP inhibits the transcription factor FleQ (Hickman and Harwood 2008). Thereby, activation of the Wsp system on a surface represses flagella biosynthesis genes and induces the production of biofilm matrix components. Very recently, it was found that activation of the Wsp system on a surface is highly heterogeneous, resulting in two subpopulations on a surface: one that colonises the surface by initiating microcolonies and one that remains motile to explore the surface (Armbruster et al. 2019).
Recently it was shown that mechanical forces on surface-attached cells could, besides causing envelope stress, could also be sensed by other means. One of these means might be sensing shear forces. This has been shown in P. aeruginosa cells that were attached via type IV pili, where flow caused the production of c-di-GMP and subsequent biofilm initiation (Rodesney et al. 2017). Similar results have been obtained in enterohemorrhagic E. coli, where increasing flow rates on attached cells led to increased induction of a pathogenicity island, although here the involvement of pili was less clear (Alsharif et al. 2015). Although it is often assumed that bacteria that are responsive to flow, sense the shear force, a recent study showed that P. aeruginosa can in fact sense the shear rate, i.e. the speed at which the liquid moves along the bacterium (Sanfilippo et al. 2019). The mechanism for sensing the shear rate is, however, yet unsolved. Another means to sense mechanical forces on surface-attached cells could be via induction of voltage and calcium transients in E. coli (Bruni et al. 2017). Changes in calcium concentration were found to result in alterations in protein levels. The mechanisms by which these calcium transients are generated and regulate gene expression remain unsolved but may prove to be an interesting novel sensing system for bacteria attached via the cell body.
In summary, attachment of the cell body can give rise to envelope deformations and maybe other forms of envelope stress. These perturbations are sensed by at least the Rcs system in E. coli and P. mirabilis, and the Wsp system in P. aeruginosa. It is very plausible that other bacteria also recognise surface-induced envelope deformations as a signal for adhesion.
DOWNSTREAM EFFECTS OF THE POTENTIAL SURFACE SENSING PATHWAYS
In the following, we outline how the above-mentioned systems that may sense the proximity to or contact with a surface could induce the changes necessary to switch from a mobile to a sessile lifestyle. Here, we will focus on how those potential surface sensing systems that specifically affect motility and adhesion, as key phenotypic changes for the lifestyle switch.
Central role of c-di-GMP
Several of the above sensing systems ultimately lead to increased cyclic-diguanylate (c-di-GMP) levels, which plays a central role in signal transduction that leads to the switch to the sessile phenotype. At high concentrations, c-di-GMP leads to enhanced synthesis of pili and matrix and reduced motility, and thereby plays an important role in the switch between mobile and biofilm lifestyles (for reviews, see Wolfe and Visick 2008; Jenal, Reinders and Lori 2017). Regulation of biofilm initiation via c-di-GMP is highly conserved in the bacterial kingdom, and species that do not synthesize this compound, e.g. S. aureus, use similar second messengers, e.g. c-di-AMP, to fulfill the same functions (Corrigan et al. 2011). The E. coli genome contains 29 genes that are proposed to synthesize or degrade c-di-GMP (Weber et al. 2006), a number of them known to be induced by the above described potential surface sensing pathways. For instance, the CpxAR system upregulates the expression of the diguanylate cyclase dgcZ (ydeH) (Raivio, Leblanc and Price 2013; Lacanna et al. 2016). Furthermore, the production of a number of diguanylate cyclases is regulated by the carbon storage protein CsrA (Jonas et al. 2008), whose activity is indirectly controlled by the BarA/UvrY two-component system via regulation of the expression of the small RNAs CsrB and CsrC (Suzuki et al. 2002; Weilbacher et al. 2003). In P.aeruginosa, c-di-GMP production is regulated by surface contact via the Wsp system (Güvener and Harwood 2007; Hickman and Harwood 2008; Francis, Stevenson and Porter 2017). Thus, c-di-GMP links the activation of some potential surface sensing systems to biofilm initiation.
Another second messenger molecule that is induced upon surface contact, is cyclic AMP (Persat et al. 2015). This signaling molecule is important for the regulation of virulence factors, such as Type II and III secretion systems, motility systems and adhesive appendages in P. aeruginosa (Wolfgang et al. 2003).
Downregulation of motility
Some of the above-mentioned surface sensing systems downregulate motility, which is no longer needed for sessile cells in a biofilm. Although the biosynthesis and rotation of flagella are decreased on a surface, there are generally microenvironments within biofilms where flagella are still expressed to mediate cell–cell and cell–surface interactions (Serra et al. 2013). With constitutive expression of the flagellar regulator FlhDC, biofilm formation by E. coli is significantly impaired, indicating the importance of their timely regulation (Prüß et al. 2010), even though the reduced biofilm formation might also have been due to the constitutive expression of a coregulated phosphodiesterase in this case. The above outlined, putative surface sensing pathways CpxAR (Raivio, Leblanc and Price 2013), RcsCDB (Francez-Charlot et al. 2003), EnvZ/OmpR (Shin and Park 1995) and BarA/UvrY (Suzuki et al. 2002; Weilbacher et al. 2003; Jonas et al. 2008) are all involved in controlling flagellar expression and activity in E. coli, for instance by repressing the flagellar regulator flhDC (Soutourina and Bertin 2003) or by increasing the production of c-di-GMP (Wolfe and Visick 2008), which activates YcgR, which in turn binds to and slows down the rotation of the E. coli flagella (Boehm et al. 2010). The c-di-GMP level affects motility not only in E. coli, but also in e.g. P. aeruginosa (Petrova, Cherny and Sauer 2014) (Hickman and Harwood 2008), C. crescentus (Abel et al. 2013) and V. cholerae (Krasteva et al. 2010). For a review about two-component-system-based regulation of motility, see (Prüß 2017).
Control over adhesive appendages
Some of the surface sensing systems exert control over expression of adhesive appendages. For switching to the sessile lifestyle, it is essential to control attachment to the surface and other cells, which can be mediated in part by the synthesis of pili. A number of the above mentioned pathways are involved in expression of the E. coli curli (for a review see (Evans and Chapman 2014)). Activators are the two-component systems EnvZ/OmpR and BasSR, while the CpxAR and RcsCDB systems have been found to repress the curli genes (Evans and Chapman 2014). Probably connected to the loss of curli, deletion of the gene encoding the response regulator OmpR has been found to result in complete loss of adhesion (Vidal et al. 1998), although other studies only reported a moderate reduction in biofilm formation (Niba et al. 2007; Prüß et al. 2010). The genes for curli synthesis are encoded by two operons in E. coli, csgBAC and csgDEFG, both of which are dependent on the general stress response sigma factor σS (Hammar et al. 1995). The csgD gene encodes a transcription factor that plays a major role in reduction of motility and is considered as a master regulator of the switch to the sessile phenotype (Ogasawara, Yamamoto and Ishihama 2011).
Different types of pili in a variety of species are dynamically regulated by c-di-GMP, such as MshA pili in Vibrio cholerae (Jones et al. 2015), Type IV pili in multiple species including Myxococcus xanthus (Skotnicka et al. 2016), P. aeruginosa (Jain, Sliusarenko and Kazmierczak 2017) and Xanthomonas citri (Guzzo et al. 2013) and Tad pili in C. crescentus (Sangermani et al. 2019). Also in E. coli, the production of pili has been shown to be regulated by the second messenger c-di-GMP (Claret et al. 2007). The BarA/UvrY two-component system also plays a role in the expression of pili; in a uvrY deletion strain fewer cells express pili, while the opposite is true for a strain missing the csrA gene, whose product is negatively regulated by BarA/UvrY (Mitra et al. 2013). However, since CsrA regulates the expression of a number of diguanylate cyclases (Jonas et al. 2008), the effect of the BarA/UvrY system on pili expression could also be indirect, via altered levels of c-di-GMP. Furthermore, the two-component system BasSR was found to directly control expression of pilus genes (Ogasawara et al. 2012). Biosynthesis of the CupD fimbriae of P. aeruginosa, which play an important role in biofilm formation in more virulent strains, is regulated by the Rcs system (Mikkelsen et al. 2009). As described above, this system is highly responsive to surface contact in E. coli and P. mirabilis (Ferrières and Clarke 2003; Morgenstein and Rather 2012), however, it has not been tested for surface sensing in P. aeruginosa.
Production of the exopolymeric matrix
Another important aspect of the sessile lifestyle is the synthesis of the exopolymeric matrix, supporting bacteria to stick together and shielding them to some extent from influences from outside, both physical and chemical. In E. coli, the RcsCDB system positively regulates the expression of the wca (also called cps) genes, which are responsible for the production of the polysaccharide colanic acid (Gottesman, Trisler and Torres-Cabassa 1985). Colanic acid is essential for the development of the three-dimensional biofilm structure (Danese, Pratt and Kolter 2000). The BarA/UvrY system can increase the production of another polysaccharide, poly-β-1,6-N-acetyl-D-glucosamine (PGA), via inhibition of CsrA (Wang et al. 2005). PGA is important for sessile E. coli cells, as indicated by the finding that enzymatic hydrolysis of this compound greatly reduces the ability to form biofilms (Wang, Preston III and Romeo 2004; Itoh et al. 2005).
Also in other species, matrix production is controlled upon arriving at a surface. In V. cholerae matrix production, specifically the synthesis of the Vibrio polysaccharide, is controlled by the c-di-GMP level (Krasteva et al. 2010). In P. aeruginosa, the biofilm matrix is formed by nucleic acids, proteins and the polysaccharides alginate, Psl and Pel (Ma et al. 2009; Franklin et al. 2011). Biosynthesis of Pel is regulated by c-di-GMP (Ueda and Wood 2009) (Hickman and Harwood 2008). Alginate production is under control of the sigma factor AlgT (Wozniak, Sprinkle and Baynham 2003), which is homologous to the E. coli σE (Hershberger et al. 1995). Thus, also in P. aeruginosa, production of exopolymeric matrix components depends on surface sensing.
EXPERIMENTAL DEVELOPMENTS FOR STUDYING SURFACE SENSING
Investigation of surface sensing and the corresponding initial responses is complicated due to several inherent and experimental challenges. First, cells simultaneously encounter multiple changes once they approach a surface, i.e. variation in pH, osmolarity, nutrient availability, forces on the flagella and pili, and potentially envelope stress, which makes it difficult to trace the response to a single stimulus. Also, biofilm-related genes are not solely regulated by surface-induced stimuli. For instance, it has been found that pH affects motility and the ability of planktonic E. coli to adhere, as exemplified by the presence of fewer and shorter flagella and more pili when cells are cultured in non-neutral pH (Chang et al. 2013). Thus, identifying the molecular mechanisms activating surface-induced systems requires either ‘isolation’ of the phenomena that can be sensed at a surface or requires solving of an intertwined, multivariate problem.
Second, the different potential surface sensing systems overlap in both their activating signals and downstream functions. For instance, the activity of the EnvZ/OmpR system depends on at least three other systems implicated in surface sensing: CpxAR regulates transcription of ompR itself (Dudin et al. 2014), both CpxAR and σE control transcription of mzrA (Dartigalongue, Missiakas and Raina 2001; Raivio, Leblanc and Price 2013), which in turn influences the activity of the EnvZ/OmpR system (Gerken et al. 2009), and there appears to be crosstalk between histidine kinase BarA and response regulator OmpR (Nagasawa et al. 1992). Also for the downstream targets there is overlap, as both CpxAR and EnvZ/OmpR regulate the expression of the membrane proteins TppB, OmpC and OmpF (Goh, Siino and Igo 2004; Batchelor et al. 2005; Raivio, Leblanc and Price 2013) and both RcsCDB and EnvZ/OmpR may control the colanic acid synthesis genes (Hagiwara et al. 2003). Similar overlaps in inputs and outputs exist also for other proposed surface sensing systems. Thus, together the systems form an entangled network that obscures investigation of individual pathways.
Third, a more practical limitation with respect to the study of initial surface contact is that most current experimental approaches require relatively large numbers of cells. However, if the initial responses to spontaneous cellular adhesion to surfaces is studied, generally only very few cells will be attached to the surface initially. Additionally, not all cells get in contact with the surface at the same time, implying that in the earliest stages of surface attachment there will be significant heterogeneity within the population.
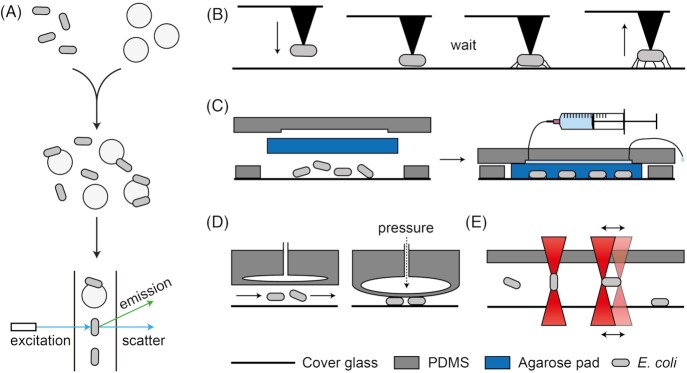
The low number of attached cells and heterogeneity in the population, could be tackled with single-cell methods. In one study, it was demonstrated that incubating bacteria with microbeads to which they can adhere, followed by flow cytometric analysis of this mixture, allows for the observation of both planktonic and sessile cells at the same time (Fig. 4A) (Beloin et al. 2008). Using this method, it was shown that there is a rapid decrease in respiration when E. coli cells adhere to polystyrene microbeads (Geng et al. 2014).
Figure 4.
Experimental approaches for the study of surface sensing. (A), Mixing of bacteria with microbeads leads to a mixed population, in which both planktonic and sessile cells are present. Subsequent measurements with flow cytometry will therefore allow for a comparison of both phenotypes in the same experiment, such that surface-specific induction can be fully isolated from any potentially confounding effects (Beloin et al. 2008). (B), Atomic force microscopy could be employed to measure changes in adhesion forces. Shown here is a bacterium that is brought into contact with a surface for an extended time, which may induce production of adhesins or appendages, followed by retraction of the cantilever and measurement of the required force. (C), A typical microfluidic setup for continuous microscopic observation of sessile cells. Pictured here is the immobilization of bacteria under an agar pad. However, with a surface coating such as polylysine or (3-aminopropyl)triethoxysilane, the bacteria can also be immobilised directly to the glass without the need for an agar pad, in which case the flow of medium is directly over the cells. This kind of experimental approach is not suitable for obtaining quantitative fluorescence data for planktonic cells. (D), A specially designed microfluidic setup that allows for invoking temporary surface contact of bacteria in a flow channel, by reversibly collapsing the channel (Okumus et al. 2016, 2018). (E), Side view of a microfluidic channel, showing a focussed infrared laser beam (‘optical tweezers’) that enables the manipulation of individual bacteria. Once trapped, the bacteria can be moved freely through a microfluidic device. Oscillating the laser trap gives control over the orientation of the bacteria, such that well-focussed images of planktonic cells can be obtained (Zhang, Kimkes and Heinemann; Carmon and Feingold 2011).
Moreover, recent improvements of RNA-seq techniques towards single cell sensitivity (Wu et al. 2014) will enable transcriptome analyses on the single attached cell level. For transcriptomic profiling of individual bacterial cells, single-cell RNA-seq is still plagued by a number of problems, i.e. handling of individual cells, low amounts of mRNA and the absence of polyadenylated tails (Zhang et al. 2018). Microfluidics-based platforms for isolation of DNA or mRNA from single cells in micro-chambers exist, that also allow for microscopic observation prior to cell lysis (e.g. Fluidigm C1). While such devices are generally aimed at research on eukaryotic cells, they have been successfully applied also to a bacterial study (Yu et al. 2017). If adhesion in the micro-chambers could be well-controlled, for instance by application of surface coatings, and the time point of initial surface contact of each cell would be known from microscopic observation, then the transcriptional response to adhesion could be studied in bacteria with single-cell sensitivity.
For transcriptomics of surface-attached cells, instead of RNA-seq, a recent adaptation to single-molecule fluorescence in situ hybridization (smFISH) could be employed, which greatly increased the throughput and number of detectable transcripts. This technique, called multiplexed error-robust FISH (MERFISH), allows for the detection and quantification of hundreds to thousands of individual mRNA species in single-cells (Chen et al. 2015; Moffitt et al. 2016a). As MERFISH uses fixed cells that are immobilised to a cover glass, it should be possible to get transcriptomic data of individual cells at several time points after adhesion, by varying the time between surface attachment and fixation. While mainly designed for eukaryotic cells, MERFISH has been successfully applied in an E. coli study (Moffitt et al. 2016b). Also in the field of proteomics, there are experimental developments that may in the future prove promising for the analysis of single surface-attached cells, with the sensitivity now approaching the level of single mammalian cells (Budnik et al. 2018; Swaminathan et al. 2018).
To study the adhesion strength of single cells to surfaces, atomic force microscopy (AFM) is a well-established method (Camesano, Liu and Datta 2007). Briefly, in AFM the deflection of a cantilever is detected while a sharp tip connected to the cantilever interacts with a surface. From the deflection and spring constant of the cantilever, the force of interaction can then be calculated. By binding a single bacterium to the tip, its interaction with a variety of surfaces can be determined, or alternatively, the tip can be modified with different surface properties (i.e. by attaching a microbead) and sample a surface containing a confluent layer of attached cells (Razatos et al. 1998a, 1998b; Ong et al. 1999). Surface-induced cellular responses that can be detected by AFM are limited to those that affect the interaction strength with the surface, such as the regulation of cell appendages (Fig. 4B). With AFM it has been found that Shewanella oneidensis produced an iron reductase that affected its adhesion to an iron mineral surface within 30 min of surface contact (Lower, Hochella and Beveridge 2001). While this response was probably not due to physical contact sensing per se, but rather due to the chemical signal iron, AFM should be a suitable technique to study surface sensing. Indeed, AFM was used to show that attachment of Staphylococcus aureus led to higher abundance of an adhesin on the cell surface (Lower et al. 2010). AFM has also been used to study surface-induced cell envelope deformation in single bacteria (Chen et al. 2014). A very recent review describes the possibilities of AFM as a tool for the study of microbial adhesion (Mathelié-Guinlet et al. 2019).
Ultimately, microscopic techniques might be the method of choice to observe changes upon surface contact. Nearly all currently used techniques for microscopic time-lapse analyses, such as immobilisation of cells under an agar pad or binding of cells to a glass slide (Fig. 4C; for a comparison of methods, see e.g. (Louise Meyer et al. 2010)), are in fact techniques to investigate cells while they are attached to a surface. In contrast, only very few microscopic techniques allow for tracking single planktonic cells over time (e.g. (Johnson-Chavarria et al. 2014), where single bacteria are hydrodynamically trapped at the junction of two flow channels, or (Taute et al. 2015), which uses phase-contrast microscopy to follow the position of freely moving bacteria over time in 3D, for which the Z-position is determined from the diffraction pattern of out-of-focus cells). Generally, microscopic methods for planktonic cells are either very low-throughput or not suitable for quantitative fluorescence measurements, as freely moving cells will rarely be located perfectly within the focal plane.
Although microscopic investigation of planktonic cells is problematic, as it is hindered by lack of focus, it is possible to enforce a temporary surface contact and immediately image the cells. Planktonic cells can be forced into contact with the surface by reversibly collapsing the flow channel in a specially designed PDMS-based microfluidic device (Fig. 4D) (Okumus et al. 2016, 2018). This setup should also enable the observation of the response to attachment, by collapsing the channel for an extended time.
Being able to observe cell appendages, such as flagella and pili, might yield a deeper understanding of their role in surface sensing. Recent work described the labelling of pili by a click chemistry approach (Ellison et al. 2017, 2019). Cell appendages can also be observed under the microscope in a label-free way, namely, by interferometric scattering microscopy (iSCAT) (Ortega Arroyo, Cole and Kukura 2016; Talà et al. 2019).
The possibility to handle individual cells during continued microscopic observation, such that surface attachment could be induced in a controlled manner, would facilitate investigations of the initial response to surface contact. Here, optical tweezers, an instrumentation that uses focussed light to hold objects, might offer exciting possibilities, specifically the manipulation of single cells in a microfluidic setup (Fig. 4E) (Ashkin and Dziedzic 1989; Zhang and Liu 2008). Use of an oscillating optical trap allows for imaging of rod-shaped cells with the long cell axis along the focal plane (Carmon and Feingold 2011). Optimization of this approach enabled stable holding of bacteria for tens of minutes without affecting their viability (Zhang, Kimkes and Heinemann 2019), such that planktonic cells could be investigated under the microscope and subsequently be brought into contact with the surface in a controlled and dynamic manner to observe their initial response. Apart from controlling attachment, it is conceivable that with two tweezers forces could be applied to two different points on the cell surface, which might induce tension in the membrane and therefore enable controlled generation of envelope stress. So far, however, optical tweezers have been applied to investigate the effect of spatial organisation in a multispecies biofilm (Hutchison et al. 2014) and to inhibit rotation of E. coli flagella (Lele, Hosu and Berg 2013), but not to investigate the cellular response to induced surface contact. Combined with high-resolution microscopy and fluorescence microscopy techniques, such as fluorescence resonance energy transfer (FRET) (Kentner and Sourjik 2010), optical tweezers might ultimately allow for the investigation of conformational changes and protein-protein interactions that eventually are responsible for surface sensing.
CONCLUSION
In this review, we provided an overview of the current knowledge of surface sensing mechanisms and the very initial steps of biofilm formation. The phenomena that can occur when a cell approaches a surface (i.e. physicochemical changes, attachment of surface appendages and envelope stress) are mostly well characterised. Also, the global phenotypic changes that cells undergo when switching from planktonic to sessile lifestyle are known. However, much less is understood about how contact with a surface is perceived and how the actual biofilm initiation is regulated. Thus, the complete picture of the switch from planktonic to sessile lifestyle remains elusive.
For most of the discussed sensing systems, even those that are extensively investigated, involvement in surface sensing has not been confirmed and the precise molecular mechanisms are still unknown. For example, upon attachment to a surface, does the cell envelope slightly compress, allowing outer membrane-localised RcsF to span the periplasm to transduce the signal to the inner membrane, or does attachment-induced envelope stress prevent insertion of RcsF into the outer membrane, thereby facilitating interaction with its inner membrane receptor IgaA? How does E. coli sense the attachment of its flagella and pili and does the former regulate gene expression? Further, for many pathways that have a biofilm-related downstream effect, the primary stimulus of surface sensing remains unsolved.
Another key question is why E. coli has multiple pathways that may sense adhesion, and how these systems are interconnected with each other. The decision to switch to a sessile lifestyle has important implications for the fate of the cell. Therefore, the presence and usage of multiple sensing systems, each responding to different inputs, likely ensures that the adaptation to the sessile lifestyle is only initiated if all conditions indicating surface attachment are met. However, the advantage of sensing the same input via multiple systems is difficult to understand (e.g. osmolarity can be sensed by EnvZ/OmpR, CpxAR and RcsCDB). Are these seemingly redundant sensors all activated under the same conditions or do they respond differently to specific surface properties? It has been proposed that the EnvZ/OmpR, CpxAR and RcsCDB systems form a combinatorial sensor, enabling a cell to distinguish between different inputs by the ratio of induction of these pathways (Clarke and Voigt 2011). Even though this has not been shown in relation to surface sensing, such combinatorial sensing could be relevant in this case as well.
Until now, the multiple and simultaneous changes occurring when a cell approaches a surface, the similar inputs for multiple surface-related sensing systems and the overlap in their target genes tremendously obscures a systems-level picture of the first steps in the initiation of biofilm formation. However, novel single-cell technologies could generate valuable insights into time-dependent cellular responses after surface contact. Improved understanding of surface sensing will greatly contribute to better prevention of biofilm formation.
ACKNOWLEDGEMENTS
We would like to thank Hannah Schramke for valuable feedback on the manuscript and Bert Poolman for helpful discussions.
Notes
This review describes the current knowledge of surface sensing and experimental developments for the study of this process, which is the initial step in bacterial biofilm formation.
Contributor Information
Tom E P Kimkes, Molecular Systems Biology, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 4, 9747 AG Groningen, the Netherlands.
Matthias Heinemann, Molecular Systems Biology, Groningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Nijenborgh 4, 9747 AG Groningen, the Netherlands.
FUNDING
This work was supported by the Netherlands Organisation for Scientific Research (NWO) through a VIDI Grant to MH [project number 864.11.001].
Conflicts of interests. None declared.
REFERENCES
- Abel S, Bucher T, Nicollier Met al.. Bi-modal Distribution of the Second Messenger c-di-GMP Controls Cell Fate and Asymmetry during the Caulobacter Cell Cycle. PLoS Genet. 2013;9:e1003744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsharif G, Ahmad S, Islam MSet al.. Host attachment and fluid shear are integrated into a mechanical signal regulating virulence in Escherichia coli O157:H7. Proc Natl Acad Sci. 2015;112:5503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster CR, Lee CK, Parker-Gilham Jet al.. Heterogeneity in surface sensing suggests a division of labor in Pseudomonas aeruginosa populations. Elife. 2019;8:e45084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A, Dziedzic JM. Optical trapping and manipulation of single living cells using infra-red laser beams. Berichte der Bunsengesellschaft für Phys Chemie. 1989;93:254–60. [Google Scholar]
- Asmar AT, Ferreira JL, Cohen EJet al.. Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol. 2017;15:e2004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor E, Walthers D, Kenney LJet al.. The Escherichia coli CpxA-CpxR envelope stress response system regulates expression of the porins ompf and ompC. J Bacteriol. 2005;187:5723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol. 2014;22:517–27. [DOI] [PubMed] [Google Scholar]
- Belas R. When the swimming gets tough, the tough form a biofilm. Mol Microbiol. 2013;90:1–5. [DOI] [PubMed] [Google Scholar]
- Beloin C, Houry A, Froment Met al.. A short-time scale colloidal system reveals early bacterial adhesion dynamics. PLoS Biol. 2008;6:e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloin C, Valle J, Latour-Lambert Pet al.. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol. 2004;51:659–74. [DOI] [PubMed] [Google Scholar]
- Berne C, Ducret A, Hardy GGet al.. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbiol Spectr. 2015;3:MB–0018-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Ellison CK, Agarwal Ret al.. Feedback regulation of Caulobacter crescentus holdfast synthesis by flagellum assembly via the holdfast inhibitor HfiA. Mol Microbiol. 2018b;110:219–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berne C, Ellison CK, Ducret Aet al.. Bacterial adhesion at the single-cell level. Nat Rev Microbiol. 2018a;16:616–27. [DOI] [PubMed] [Google Scholar]
- Bhomkar P, Materi W, Semenchenko Vet al.. Transcriptional Response Of E. coli Upon FimH-mediated Fimbrial Adhesion. Gene Regul Syst Bio. 2010;4:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A, Kaiser M, Li Het al.. Second messenger-mediated adjustment of bacterial swimming velocity. Cell. 2010;141:107–16. [DOI] [PubMed] [Google Scholar]
- Boks NP, Busscher HJ, van der Mei HCet al.. Bond-Strengthening in staphylococcal adhesion to hydrophilic and hydrophobic surfaces using atomic force microscopy. Langmuir. 2008;24:12990–4. [DOI] [PubMed] [Google Scholar]
- Brissette JL, Weiner L, Ripmaster TLet al.. Characterization and Sequence of the Escherichia coli stress-induced psp operon. J Mol Biol. 1991;220:35–48. [DOI] [PubMed] [Google Scholar]
- Bruni GN, Weekley RA, Dodd BJTet al.. Voltage-gated calcium flux mediates Escherichia coli mechanosensation. Proc Natl Acad Sci. 2017;114:9445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruzaud J, Tarrade J, Coudreuse Aet al.. Flagella but not type IV pili are involved in the initial adhesion of Pseudomonas aeruginosa PAO1 to hydrophobic or superhydrophobic surfaces. Colloids Surfaces B Biointerfaces. 2015;131:59–66. [DOI] [PubMed] [Google Scholar]
- Budnik B, Levy E, Harmange Get al.. Mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher HJ, Norde W, Sharma PKet al.. Interfacial re-arrangement in initial microbial adhesion to surfaces. Curr Opin Colloid Interface Sci. 2010;15:510–7. [Google Scholar]
- Busscher HJ, van der Mei HC. How do bacteria know they are on a surface and regulate their response to an adhering state? PLoS Pathog. 2012;8:e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns LS, Marlow VL, Bissett Eet al.. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol Microbiol. 2013;90:6–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callow JA, Callow ME. Trends in the development of environmentally friendly fouling-resistant marine coatings. Nat Commun. 2011;2:244. [DOI] [PubMed] [Google Scholar]
- Camesano TA, Liu Y, Datta M. Measuring bacterial adhesion at environmental interfaces with single-cell and single-molecule techniques. Adv Water Resour. 2007;30:1470–91. [Google Scholar]
- Carmon G, Feingold M. Controlled alignment of bacterial cells with oscillating optical tweezers. J Nanophotonics. 2011;5:051803. [Google Scholar]
- Chang K-C, Cheng S-J, Chen Y-Cet al.. Nanoscopic analysis on pH induced morphological changes of flagella in Escherichia coli. J Microbiol Immunol Infect. 2013;46:405–12. [DOI] [PubMed] [Google Scholar]
- Chao Y, Zhang T. Probing roles of lipopolysaccharide, type 1 fimbria, and colanic acid in the attachment of Escherichia coli strains on inert surfaces. Langmuir. 2011;27:11545–53. [DOI] [PubMed] [Google Scholar]
- Chawla R, Ford KM, Lele PP. Torque, but not FliL, regulates mechanosensitive flagellar motor-function. Sci Rep. 2017;7:5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JRet al.. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Takeda S, Yamada Het al.. Characterization of the RcsC → YojN → RcsB phosphorelay signaling pathway involved in capsular synthesis in escherichia coli. Biosci Biotechnol Biochem. 2001;65:2364–7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Harapanahalli AK, Busscher HJet al.. Nanoscale cell wall deformation impacts long-range bacterial adhesion forces on surfaces. Appl Environ Microbiol. 2014;80:637–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Szewczyk J, Pesavento Cet al.. Detecting envelope stress by monitoring beta-barrel assembly. Cell. 2014;159:1652–64. [DOI] [PubMed] [Google Scholar]
- Claret L, Miquel S, Vieille Net al.. The flagellar sigma factor flia regulates adhesion and invasion of crohn disease-associated escherichia coli via a cyclic dimeric gmp-dependent pathway. J Biol Chem. 2007;282:33275–83. [DOI] [PubMed] [Google Scholar]
- Clarke EJ, Voigt CA. Characterization of combinatorial patterns generated by multiple two-component sensors in E. coli that respond to many stimuli. Biotechnol Bioeng. 2011;108:666–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad JC, Gibiansky ML, Jin Fet al.. Flagella and pili-mediated near-surface single-cell motility mechanisms in P. aeruginosa. Biophys J. 2011;100:1608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conter A, Sturny R, Gutierrez Cet al.. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J Bacteriol. 2002;184:2850–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley BJ, Thatcher TW, Hashmi SMet al.. The extracellular polysaccharide Pel makes the attachment of P. aeruginosa to surfaces symmetric and short-ranged. Soft Matter. 2013;9:3871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan RM, Abbott JC, Burhenne Het al.. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DEet al.. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Calfee MW, Rainey PBet al.. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Pratt LA, Kolter R. Exopolysaccharide Production is required for development of Escherichia coli K-12 biofilm architecture. J Bacteriol. 2000;182:3593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danese PN, Silhavy TJ. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol. 1998;180:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dartigalongue C, Missiakas D, Raina S. Characterization of the Escherichia coli sigma(E) regulon. J Biol Chem. 2001;276:20866–75. [DOI] [PubMed] [Google Scholar]
- DeBenedictis EP, Liu J, Keten S. Adhesion mechanisms of curli subunit CsgA to abiotic surfaces. Sci Adv. 2016;2:e1600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaye A, Laloux G, Collet J-F. The lipoprotein NlpE is a Cpx sensor that serves as a sentinel for protein sorting and folding defects in the Escherichia coli envelope. J Bacteriol. 2019;201:e00611–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derjaguin B, Landau L. Theory of the stability of strongly charged lyophobic sols and of the adhesion of strongly charged particles in solutions of electrolytes. Acta Physicochim URSS. 1941;14:633–62. [Google Scholar]
- DiGiuseppe PA, Silhavy TJ. Signal detection and target gene induction by the CpxRA two-component system. J Bacteriol. 2003;185:2432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal Tet al.. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–46. [DOI] [PubMed] [Google Scholar]
- Dorel C. Manipulating bacterial cell fate: key role of surface-sensing and signal transduction. In: Méndez-Vilas A (ed). Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology. Badajoz: Formatex, 2010, 791–800. [Google Scholar]
- Dudin O, Geiselmann J, Ogasawara Het al.. Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J Bacteriol. 2014;196:707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Dalia TN, Dalia ABet al.. Real-time microscopy and physical perturbation of bacterial pili using maleimide-conjugated molecules. Nat Protoc. 2019;14:1803–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison CK, Kan J, Dillard RSet al.. Obstruction of pilus retraction stimulates bacterial surface sensing. Science. 2017;358:535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ML, Chapman MR. Curli biogenesis: Order out of disorder. Biochim Biophys Acta. 2014;1843:1551–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris C, Sanowar S, Bader MWet al.. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J Bacteriol. 2010;192:4894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldner J, Bredt W, Kahane I. Influence of cell shape and surface charge on attachment of Mycoplasma pneumoniae to glass surfaces. J Bacteriol. 1983;153:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrières L, Clarke DJ. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on a solid surface. Mol Microbiol. 2003;50:1665–82. [DOI] [PubMed] [Google Scholar]
- Francez-Charlot A, Castanié-Cornet M-P, Gutierrez Cet al.. Osmotic Regulation of the Escherichia coli bdm (Biofilm-Dependent Modulation) Gene by the RcsCDB His-Asp Phosphorelay. J Bacteriol. 2005;187:3873–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francez-Charlot A, Laugel B, Van Gemert Aet al.. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol. 2003;49:823–32. [DOI] [PubMed] [Google Scholar]
- Francis VI, Stevenson EC, Porter SL. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2017;364:fnx104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MJ, Nivens DE, Weadge JTet al.. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol. 2011;2:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander RS, Vogel N, Aizenberg J. Role of flagella in adhesion of escherichia coli to abiotic surfaces. Langmuir. 2015;31:6137–44. [DOI] [PubMed] [Google Scholar]
- Garg A, Jain A, Bhosle NB. Chemical characterization of a marine conditioning film. Int Biodeterior Biodegrad. 2009;63:7–11. [Google Scholar]
- Geng J, Beloin C, Ghigo J-Met al.. Bacteria hold their breath upon surface contact as shown in a strain of escherichia coli, using dispersed surfaces and flow cytometry analysis. PLoS One. 2014;9:e102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken H, Charlson ES, Cicirelli EMet al.. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol Microbiol. 2009;72:1408–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibiansky ML, Conrad JC, Jin Fet al.. Bacteria use type iv pili to walk upright and detach from surfaces. Science. 2010;330:197. [DOI] [PubMed] [Google Scholar]
- Gode-Potratz CJ, Kustusch RJ, Breheny PJet al.. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh E-B, Siino DF, Igo MM. The Escherichia coli tppB (ydgR) gene represents a new class of OmpR-regulated genes. J Bacteriol. 2004;186:4019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AE, Marshall KC. Genetic responses of bacteria at surfaces. In: Costerton JW, Lappin-Scott HM (eds). Microbial Biofilms. Cambridge: Cambridge University Press, 1995, 80–98. [Google Scholar]
- Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J Bacteriol. 1985;162:1111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilvout I, Chami M, Engel Aet al.. Bacterial outer membrane secretin PulD assembles and inserts into the inner membrane in the absence of its pilotin. EMBO J. 2006;25:5241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Valdevit A, Chou TMet al.. Substrate effects on cell-envelope deformation during early-stage Staphylococcus aureus biofilm formation. Soft Matter. 2017;13:2967–76. [DOI] [PubMed] [Google Scholar]
- Guzzo CR, Dunger G, Salinas RKet al.. Structure of the PilZ-FimXEAL-c-di-GMP complex responsible for the regulation of bacterial type IV pilus biogenesis. J Mol Biol. 2013;425:2174–97. [DOI] [PubMed] [Google Scholar]
- Güvener ZT, Harwood CS. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol. 2007;66:1459–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Sugiura M, Oshima Tet al.. Genome-wide analyses revealing a signaling network of the rcsc-yojn-rcsb phosphorelay system in Escherichia coli. J Bacteriol. 2003;185:5735–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D, Yamashino T, Mizuno T. A genome-wide view of the Escherichia coli bass-basr two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 2004;68:1758–67. [DOI] [PubMed] [Google Scholar]
- Hall MN, Silhavy TJ. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981;146:23–43. [DOI] [PubMed] [Google Scholar]
- Hammar M, Arnqvist A, Bian Zet al.. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–70. [DOI] [PubMed] [Google Scholar]
- Harapanahalli AK, Chen Y, Li Jet al.. Influence of adhesion force on icaA and cidA gene expression and production of matrix components in Staphylococcus aureus biofilms. Appl Environ Microbiol. 2015a;81:3369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapanahalli AK, Younes JA, Allan Eet al.. Chemical Signals and Mechanosensing in Bacterial Responses to Their Environment. PLoS Pathog. 2015b;11:e1005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermansson M. The DLVO theory in microbial adhesion. Colloids Surfaces B Biointerfaces. 1999;14:105–19. [Google Scholar]
- Hershberger CD, Ye RW, Parsek MRet al.. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative σ factor (σE). Proc Natl Acad Sci USA. 1995;92:7941–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyde M, Portalier R. Regulation of major outer membrane porin proteins of Escherichia coli K 12 by pH. Mol Gen Genet. 1987;208:511–7. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsa SM, Espinosa-Urgel M, Ramos JLet al.. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–18. [DOI] [PubMed] [Google Scholar]
- Hirano Y, Hossain MM, Takeda Ket al.. Structural Studies of the Cpx Pathway Activator NlpE on the Outer Membrane of Escherichia coli. Structure. 2007;15:963–76. [DOI] [PubMed] [Google Scholar]
- Hong Y, Brown DG. Alteration of bacterial surface electrostatic potential and pH upon adhesion to a solid surface and impacts to cellular bioenergetics. Biotechnol Bioeng. 2010;105:965–72. [DOI] [PubMed] [Google Scholar]
- Hong Y, Brown DG. Variation in bacterial ATP level and proton motive force due to adhesion to a solid surface. Appl Environ Microbiol. 2009;75:2346–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Deshpande S, Sprecher KSet al.. Second messenger-mediated tactile response by a bacterial rotary motor. Science. 2017;358:531–4. [DOI] [PubMed] [Google Scholar]
- Hutchison JB, Rodesney CA, Kaushik KSet al.. Single-cell control of initial spatial structure in biofilm development using laser trapping. Langmuir. 2014;30:4522–30. [DOI] [PubMed] [Google Scholar]
- Inclan YF, Persat A, Greninger Aet al.. A scaffold protein connects type IV pili with the Chp chemosensory system to mediate activation of virulence signaling in Pseudomonas aeruginosa. Mol Microbiol. 2016;101:590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac DD, Pinkner JS, Hultgren SJet al.. The extracytoplasmic adaptor protein CpxP is degraded with substrate by DegP. PNAS. 2005;102:17775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Wang X, Hinnebusch BJet al.. Depolymerization of beta-1,6-N-Acetyl-D-Glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Sliusarenko O, Kazmierczak BI. Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly. PLoS Pathog. 2017;13:e1006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol. 2017;15:271–84. [DOI] [PubMed] [Google Scholar]
- Johnson-Chavarria EM, Agrawal U, Tanyeri Met al.. Automated single cell microbioreactor for monitoring intracellular dynamics and cell growth in free solution. Lab Chip. 2014;14:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm Ret al.. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Danese PN, Pinkner JSet al.. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Utada A, Davis KRet al.. C-di-GMP regulates motile to sessile transition by modulating msha pili biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog. 2015;11:e1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JF, Feick JD, Imoudu Det al.. Oriented adhesion of Escherichia coli to polystyrene particles. Appl Environ Microbiol. 2003;69:6515–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G, Lloyd LJ, Stumpf MPHet al.. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J Biol Chem. 2006;281:21147–61. [DOI] [PubMed] [Google Scholar]
- Jovanovic G, Weiner L, Model P. Identification, nucleotide sequence, and characterization of pspf, the transcriptional activator of the Escherichia coli stress-induced psp operon. J Bacteriol. 1996;178:1936–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Vianney A, Beloin Cet al.. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol. 2005;187:2038–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker BA, Harms H, Hug SJet al.. Adsorption of bacterial surface polysaccharides on mineral oxides is mediated by hydrogen bonds. Colloids Surfaces B Biointerfaces. 1997;9:331–43. [Google Scholar]
- Jucker BA, Harms H, Zehnder AJB. Adhesion of the Positively Charged Bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and teflon. J Bacteriol. 1996;178:5472–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Ariöz C, Hansmeier Net al.. The Escherichia coli Envelope Stress Sensor CpxA Responds to Changes in Lipid Bilayer Properties. Biochemistry. 2015;54:3670–6. [DOI] [PubMed] [Google Scholar]
- Kentner D, Sourjik V. Use of fluorescence microscopy to study intracellular signaling in bacteria. Annu Rev Microbiol. 2010;64:373–90. [DOI] [PubMed] [Google Scholar]
- Kimkes TEP, Heinemann M. Reassessing the role of the Escherichia coli CpxAR system in sensing surface contact. PLoS One. 2018;13:e0207181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelleberg S, Humphrey BA, Marshall KC. Effect of interfaces on small, starved marine bacteria. Appl Environ Microbiol. 1982;43:1166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Kobylak N, Lindner Bet al.. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem. 2014;289:14829–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Lindner B, Brabetz Wet al.. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases. Minimal lipopolysaccharide structure and induction of envelope stress response. J Biol Chem. 2009;284:15369–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanowski K, Volkmer B, Gerosa Let al.. Functioning of a metabolic flux sensor in Escherichia coli. Proc Natl Acad Sci. 2013;110:1130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konovalova A, Perlman DH, Cowles CEet al.. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of β-barrel proteins. PNAS. 2014;111:E4350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotte O, Zaugg JB, Heinemann M. Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol. 2010;6:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh KN, Hutchison JB, Melaugh Get al.. Role of multicellular aggregates in biofilm formation. MBio. 2016;7:e00237–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva PV, Fong JCN, Shikuma NJet al.. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327:866–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, O'Toole GA. Surface-induced and biofilm-induced changes in gene expression. Curr Opin Biotechnol. 2000;11:429–33. [DOI] [PubMed] [Google Scholar]
- Lacanna E, Bigosch C, Kaever Vet al.. Evidence for escherichia coli diguanylate cyclase dgcz interlinking surface sensing and adhesion via multiple regulatory routes. O'Toole GA (ed). J Bacteriol. 2016;198:2524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laventie B-J, Sangermani M, Estermann Fet al.. A surface-induced asymmetric program promotes tissue colonization by pseudomonas aeruginosa. Cell Host Microbe. 2019;25:1–13. [DOI] [PubMed] [Google Scholar]
- Laverty G, Gorman SP, Gilmore BF. Biomolecular Mechanisms of Pseudomonas aeruginosa and Escherichia coli Biofilm Formation. Pathogens. 2014;3:596–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc SKD, Oates CW, Raivio TL. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol. 2011;193:3367–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, De Anda J, Baker AEet al.. Multigenerational memory and adaptive adhesion in early bacterial biofilm communities. Proc Natl Acad Sci U S A. 2018;115:4471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LJ, Barrett JA, Poole RK. Genome-Wide Transcriptional Response of Chemostat-Cultured Escherichia coli to Zinc. J Bacteriol. 2005;187:1124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y-Y, Belas R. Loss of flil alters proteus mirabilis surface sensing and temperature-dependent swarming. J Bacteriol. 2015;197:159–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune P. Contamination of abiotic surfaces: what a colonizing bacterium sees and how to blur it. Trends Microbiol. 2003;11:179–84. [DOI] [PubMed] [Google Scholar]
- Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. PNAS. 2013;110:11839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Brown PJB, Tang JXet al.. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima S, Guo MS, Chaba Ret al.. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science. 2013;340:837–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Strauss J, Camesano TA. Adhesion forces between Staphylococcus epidermidis and surfaces bearing self-assembled monolayers in the presence of model proteins. Biomaterials. 2008;29:4374–82. [DOI] [PubMed] [Google Scholar]
- Lloyd LJ, Jones SE, Jovanovic Get al.. Identification of a new member of the phage shock protein response in Escherichia coli, the phage shock protein G (PspG). J Biol Chem. 2004;279:55707–14. [DOI] [PubMed] [Google Scholar]
- Loeb GI, Neihof RA. Marine conditioning films. In: Baier RE (ed). Applied Chemistry at Protein Interfaces. Washington D.C.: American Chemical Society, 1975, 319–35. [Google Scholar]
- Louise Meyer R, Zhou X, Tang Let al.. Immobilisation of living bacteria for AFM imaging under physiological conditions. Ultramicroscopy. 2010;110:1349–57. [DOI] [PubMed] [Google Scholar]
- Lower SK, Hochella MF, Beveridge TJ. Bacterial recognition of mineral surfaces: Nanoscale interactions between Shewanella and α-FeOOH. Science. 2001;292:1360–3. [DOI] [PubMed] [Google Scholar]
- Lower SK, Yongsunthon R, Casillas-Ituarte NNet al.. A tactile response in Staphylococcus aureus. Biophys J. 2010;99:2803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah T-FC, O'Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9:34–9. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2006;59:379–405. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Heck M, Stout Vet al.. Role of RcsF in signaling to the rcs phosphorelay pathway in escherichia coli. J Bacteriol. 2005;187:6770–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Conover M, Lu Het al.. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall KC, Stout R, Mitchell R. Mechanism of the Initial Events in the Sorption of Marine Bacteria to Surfaces. J Gen Microbiol. 1971;68:337–48. [Google Scholar]
- Marshall KC. Adhesion and growth of bacteria at surfaces in oligotrophic habitats. Can J Microbiol. 1988;34:503–6. [Google Scholar]
- Mathelié-Guinlet M, Viela F, Viljoen Aet al.. Single-molecule atomic force microscopy studies of microbial pathogens. Curr Opin Biomed Eng. 2019;12:1–7. [Google Scholar]
- McCarter L, Hilmen M, Silverman M. Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell. 1988;54:345–51. [DOI] [PubMed] [Google Scholar]
- Mcdonald C, Jovanovic G, Ces Oet al.. Membrane stored curvature elastic stress modulates recruitment of maintenance proteins pspa and vipp1. MBio. 2015;6:e01188–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J, Rouviere PE, Erickson JWet al.. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 1993;7:2618–28. [DOI] [PubMed] [Google Scholar]
- Meinders JM, Busscher HJ. Influence of ionic strength and shear rate on the desorption of polystyrene particles from a glass collector as studied in a parallel-plate flow chamber. Colloids Surfaces A Physicochem Eng Asp. 1993;80:279–85. [Google Scholar]
- Melaugh G, Hutchison J, Kragh KNet al.. Shaping the growth behaviour of biofilms initiated from bacterial aggregates. PLoS One. 2016;11:e0149683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Ball G, Giraud Cet al.. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4:e6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol. 1997;179:1029–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Palaniyandi S, Herren CDet al.. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One. 2013;8:e55492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Mizushima S. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol Microbiol. 1990;4:1077–82. [DOI] [PubMed] [Google Scholar]
- Mobley HLT, Belas R. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 1995;3:280–4. [DOI] [PubMed] [Google Scholar]
- Moffitt JR, Hao J, Wang Get al.. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci. 2016a;113:11046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Pandey S, Boettiger ANet al.. Spatial organization shapes the turnover of a bacterial transcriptome. Elife. 2016b;5:e13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. [DOI] [PubMed] [Google Scholar]
- Morgenstein RM, Rather PN. Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol. 2012;194:669–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey MA, Lopez-Boado YS, Wilson CLet al.. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–7. [DOI] [PubMed] [Google Scholar]
- Nagasawa S, Ishige K, Mizuno T. Novel members of the two-component signal transduction genes in escherichiacoli. J Biochem. 1993;114:350–7. [DOI] [PubMed] [Google Scholar]
- Nagasawa S, Tokishita S, Aiba Het al.. A novel sensor-regulator protein that belongs to the homologous family of signal-transduction proteins involved in adaptive responses in Escherichia coli. Mol Microbiol. 1992;6:799–807. [DOI] [PubMed] [Google Scholar]
- Nejadnik MR, van der Mei HC, Norde Wet al.. Bacterial adhesion and growth on a polymer brush-coating. Biomaterials. 2008;29:4117–21. [DOI] [PubMed] [Google Scholar]
- Nesper J, Hug I, Kato Set al.. Cyclic di-GMP differentially tunes a bacterial flagellar motor through a novel class of CheY-like regulators. Elife. 2017;6:e28842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niba ETE, Naka Y, Nagase Met al.. A genome-wide approach to identify the genes involved in biofilm formation in E. coli. DNA Res. 2007;14:237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AL, Gachon E, Perez-Carrasco Ret al.. Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc Natl Acad Sci. 2017;114:12952–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor JR, Kuwada NJ, Huangyutitham Vet al.. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol. 2012;86:720–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole GA, Wong GCL. Sensational biofilms: Surface sensing in bacteria. Curr Opin Microbiol. 2016;30:139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofek I, Mirelman D, Sharon N. Adherence of Escherichia coli to human mucosal cells mediated by mannose receptors. Nature. 1977;265:623–5. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Shinohara S, Yamamoto Ket al.. Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiol. 2012;158:1482–92. [DOI] [PubMed] [Google Scholar]
- Ogasawara H, Yamamoto K, Ishihama A. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol. 2011;193:2587–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B, Baker CJ, Arias-Castro JCet al.. Single-cell microscopy of suspension cultures using a microfluidics-assisted cell screening platform. Nat Protoc. 2018;13:170–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumus B, Landgraf D, Lai GCet al.. Mechanical slowing-down of cytoplasmic diffusion allows in vivo counting of proteins in individual cells. Nat Commun. 2016;7:11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Old DC. Inhibition of the interaction between fimbrial haemagglutinins and erythrocytes by d-mannose and other carbohydrates. J Gen Microbiol. 1972;71:149–57. [DOI] [PubMed] [Google Scholar]
- Ong Y-L, Razatos A, Georgiou Get al.. Adhesion forces between E. coli bacteria and biomaterial surfaces. Langmuir. 1999;15:2719–25. [Google Scholar]
- Ortega Arroyo J, Cole D, Kukura P. Interferometric scattering microscopy and its combination with single-molecule fluorescence imaging. Nat Protoc. 2016;11:617–33. [DOI] [PubMed] [Google Scholar]
- Otto K, Norbeck J, Larsson Tet al.. Adhesion of type 1-fimbriated Escherichia coli to abiotic surfaces leads to altered composition of outer membrane proteins. J Bacteriol. 2001;183:2445–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto K, Silhavy TJ. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. PNAS. 2002;99:2287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Kloser AW, Schnaitman CAet al.. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J Bacteriol. 1992;174:2525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, Groisman EA. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol. 2007;63:283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persat A, Inclan YF, Engel JNet al.. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci. 2015;112:7563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrova OE, Cherny KE, Sauer K. The Pseudomonas aeruginosa diguanylate cyclase GcbA, a homolog of P. fluorescens GcbA, promotes initial attachment to surfaces, but not biofilm formation, via regulation of motility. J Bacteriol. 2014;196:2827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potera C. Forging a Link Between Biofilms and Disease. Science. 1999;283:1837–9. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Hsing W, Gibson KEet al.. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol. 1996;20:911–7. [DOI] [PubMed] [Google Scholar]
- Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–93. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Brombacher E, Vidal Oet al.. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J Bacteriol. 2001;183:7213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigent-Combaret C, Prensier G, Le Thi TTet al.. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol. 2000;2:450–64. [DOI] [PubMed] [Google Scholar]
- Prigent-Combaret C, Vidal O, Dorel Cet al.. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J Bacteriol. 1999;181:5993–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM, Verma K, Samanta Pet al.. Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch Microbiol. 2010;192:715–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüß BM. Involvement of two-component signaling on bacterial motility and biofilm development. J Bacteriol. 2017;199:e00259–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffa RG, Raivio TL. A third envelope stress signal transduction pathway in Escherichia coli. Mol Microbiol. 2002;45:1599–611. [DOI] [PubMed] [Google Scholar]
- Raivio TL, Leblanc SKD, Price NL. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol. 2013;195:2755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL, Popkin DL, Silhavy TJ. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol. 1999;181:5263–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio TL. Everything old is new again : an update on current research on the Cpx envelope stress response. Biochim Biophys Acta. 2014;1843:1529–41. [DOI] [PubMed] [Google Scholar]
- Razatos A, Ong Y-L, Sharma MMet al.. Evaluating the interaction of bacteria with biomaterials using atomic force microscopy. J Biomater Sci Polym Ed. 1998a;9:1361–73. [DOI] [PubMed] [Google Scholar]
- Razatos A, Ong Y-L, Sharma MMet al.. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. PNAS. 1998b;95:11059–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Wang C, Chen Zet al.. Emergent heterogeneous micro-environments in biofilms: substratum surface heterogeneity and bacterial adhesion force-sensing. FEMS Microbiol Rev. 2018;42:259–72. [DOI] [PubMed] [Google Scholar]
- Rodesney CA, Roman B, Dhamani Net al.. Mechanosensing of shear by Pseudomonas aeruginosa leads to increased levels of the cyclic-di-GMP signal initiating biofilm development. Proc Natl Acad Sci. 2017;114:5906–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Stender S, Filloux Aet al.. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelsson MO, Kirchman DL. Degradation of adsorbed protein by attached bacteria in relationship to surface hydrophobicity. Appl Environ Microbiol. 1990;56:3643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfilippo JE, Lorestani A, Koch MDet al.. Microfluidic-based transcriptomics reveal force-independent bacterial rheosensing. Nat Microbiol. 2019. DOI:10.1038/s41564-019-0455-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangermani M, Hug I, Sauter Net al.. Tad pili play a dynamic role in Caulobacter crescentus surface colonization. MBio. 2019;10:e01237–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Machida K, Arikado Eet al.. Expression of outer membrane proteins in Escherichia coli growing at acid pH. Appl Environ Microbiol. 2000;66:943–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MP, Bendick JA, Holm ERet al.. Economic impact of biofouling on a naval surface ship. Biofouling. 2011;27:87–98. [DOI] [PubMed] [Google Scholar]
- Serra DO, Richter AM, Klauck Get al.. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. MBio. 2013;4:e00103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Park C. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J Bacteriol. 1995;177:4696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni SF, Harms H, Bosma TNPet al.. Population heterogeneity affects transport of bacteria through sand columns at low flow rates. Environ Sci Technol. 1998;32:2100–5. [Google Scholar]
- Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci. 2001;98:6901–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotnicka D, Petters T, Heering Jet al.. Cyclic di-GMP regulates type IV pilus-dependent motility in Myxococcus xanthus. J Bacteriol. 2016;198:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1996;178:1204–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–23. [DOI] [PubMed] [Google Scholar]
- Subramani A, Hoek EMV. Biofilm formation, cleaning, re-formation on polyamide composite membranes. Desalination. 2010;257:73–9. [Google Scholar]
- Suzuki K, Wang X, Weilbacher Tet al.. Regulatory Circuitry of the CsrA/CsrB and BarA/UvrY Systems of Escherichia coli. J Bacteriol. 2002;184:5130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan J, Boulgakov AA, Hernandez ETet al.. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat Biotechnol. 2018;36:1076–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderholm M, Kragh KN, Koren Ket al.. Pseudomonas aeruginosa aggregate formation in an alginate bead model system exhibits in vivo-like characteristics. Appl Environ Microbiol. 2017;83:e00113–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talà L, Fineberg A, Kukura Pet al.. Pseudomonas aeruginosa orchestrates twitching motility by sequential control of type IV pili movements. Nat Microbiol. 2019;4:774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taute KM, Gude S, Tans SJet al.. High-throughput 3D tracking of bacteria on a standard phase contrast microscope. Nat Commun. 2015;6:8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GT, Zheng D, Lee Met al.. Influence of surface properties on accumulation of conditioning films and marine bacteria on substrata exposed to oligotrophic waters. Biofouling J Bioadhesion Biofilm Res. 1997;11:31–57. [Google Scholar]
- Tetsch L, Jung K. The regulatory interplay between membrane-integrated sensors and transport proteins in bacteria. Mol Microbiol. 2009;73:982–91. [DOI] [PubMed] [Google Scholar]
- Thomas AD, Booth IR. The regulation of expression of the porin gene ompC by acid pH. J Gen Microbiol. 1992;138:1829–35. [DOI] [PubMed] [Google Scholar]
- Tipping MJ, Delalez NJ, Lim Ret al.. Load-dependent assembly of the bacterial flagellar motor. MBio. 2013;4:e00551–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschauner K, Hörnschemeyer P, Müller VSet al.. Dynamic interaction between the CpxA sensor kinase and the periplasmic accessory protein CpxP mediates signal recognition in E. coli. PLoS One. 2014;9:e107383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson HH, Weibel DB. Bacteria-surface interactions. Soft Matter. 2013;9:4368–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, Pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 2009;5:e1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Henderson IR. Regulation and function of Ag43 (flu). Annu Rev Microbiol. 2008;62:153–69. [DOI] [PubMed] [Google Scholar]
- van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol. 2005;156:626–33. [DOI] [PubMed] [Google Scholar]
- Verwey EJW, Overbeek JTG. Theory of the Stability of Lyophobic Colloids: The Interaction of Sol Particles Having an Electric Double Layer. New York: Elsevier Inc., 1948. [Google Scholar]
- Vidal O, Longin R, Prigent-combaret Cet al.. Isolation of an escherichia coli k-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompr allele that increases curli expression. J Bacteriol. 1998;180:2442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt SL, Raivio TL. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett. 2012;326:2–11. [DOI] [PubMed] [Google Scholar]
- Walsh NP, Alba BM, Bose Bet al.. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. [DOI] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki Ket al.. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–63. [DOI] [PubMed] [Google Scholar]
- Wang X, Preston JF III, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Pesavento C, Possling Aet al.. Cyclic-di-GMP-mediated signalling within the sigma-S network of Escherichia coli. Mol Microbiol. 2006;62:1014–34. [DOI] [PubMed] [Google Scholar]
- Weilbacher T, Suzuki K, Dubey AKet al.. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol Microbiol. 2003;48:657–70. [DOI] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang MC, Lee VT, Gilmore MEet al.. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev Cell. 2003;4:253–63. [DOI] [PubMed] [Google Scholar]
- Wood TK, González Barrios AF, Herzberg Met al.. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–7. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Sprinkle AB, Baynham PJ. Control of Pseudomonas aeruginosa algZ Expression by the Alternative Sigma Factor AlgT. J Bacteriol. 2003;185:7297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AR, Neff NF, Kalisky Tet al.. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods. 2014;11:41–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurpel DJ, Beatson SA, Totsika Met al.. Chaperone-usher fimbriae of Escherichia coli. PLoS One. 2013;8:e52835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicohtencatl-Cortes J, Monteiro-Neto V, Saldaña Zet al.. The type 4 pili of enterohemorrhagic Escherichia coli O157:H7 are multipurpose structures with pathogenic attributes. J Bacteriol. 2009;191:411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara A, Nobuhira N, Narahara Het al.. Estimation of the adhesive force distribution for the flagellar adhesion of Escherichia coli on a glass surface. Colloids Surfaces B Biointerfaces. 2015;131:67–72. [DOI] [PubMed] [Google Scholar]
- Yu FB, Blainey PC, Schulz Fet al.. Microfluidic-based mini-metagenomics enables discovery of novel microbial lineages from complex environmental samples. Elife. 2017;6:e26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu K-K. Optical tweezers for single cells. J R Soc Interface. 2008;5:671–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JP, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Huang Yet al.. Recent developments in single-cell rna-seq of microorganisms. Biophys J. 2018;115:173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Kimkes TEP, Heinemann M. Manipulating rod-shaped bacteria with optical tweezers. Scientific Reports. DOI:10.1038/s41598-019-55657-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Keller R, Volkmer Ret al.. Structural basis for two-component system inhibition and pilus sensing by the auxiliary CpxP protein. J Biol Chem. 2011;286:9805–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobell CE. The effect of solid surfaces upon bacterial activity. J Bacteriol. 1943;46:39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]