ABSTRACT
Protein aggregation occurs as a consequence of perturbations in protein homeostasis that can be triggered by environmental and cellular stresses. The accumulation of protein aggregates has been associated with aging and other pathologies in eukaryotes, and in bacteria with changes in growth rate, stress resistance and virulence. Numerous past studies, mostly performed in Escherichia coli, have led to a detailed understanding of the functions of the bacterial protein quality control machinery in preventing and reversing protein aggregation. However, more recent research points toward unexpected diversity in how phylogenetically different bacteria utilize components of this machinery to cope with protein aggregation. Furthermore, how persistent protein aggregates localize and are passed on to progeny during cell division and how their presence impacts reproduction and the fitness of bacterial populations remains a controversial field of research. Finally, although protein aggregation is generally seen as a symptom of stress, recent work suggests that aggregation of specific proteins under certain conditions can regulate gene expression and cellular resource allocation. This review discusses recent advances in understanding the consequences of protein aggregation and how this process is dealt with in bacteria, with focus on highlighting the differences and similarities observed between phylogenetically different groups of bacteria.
Keywords: protein aggregation, molecular chaperones, disaggregases, aggregate inheritance, cellular aging, stress adaptation
This review discusses recent advances in understanding the consequences of protein aggregation and how this process is dealt with in bacteria, with focus on highlighting the differences and similarities observed between phylogenetically different groups of bacteria.
INTRODUCTION
Proteins are responsible for fulfilling the vast majority of cellular functions. To achieve their functional or native state, most newly synthesized proteins fold into specific three-dimensional structures and sometimes further assemble into multimeric protein complexes. Protein folding depends on a network of non-covalent interactions involving both the polypeptide's backbone and amino acid side chains (Dill et al. 2008). Hydrophobic effects are particularly important in driving this process, burying hydrophobic amino acids in the interior of the native protein and shielding them from the aqueous environment inside the cell (Dill et al. 2008). The native state of proteins is a thermodynamically favored conformation and many proteins reach it in a spontaneous and rapid process (Anfinsen and Scheraga 1975). However, especially large or complex proteins have to cross kinetic energy barriers and form folding intermediates before reaching their native fold, a process that often requires the assistance of molecular chaperones (Brockwell and Radford 2007; Balchin, Hayer-Hartl and Hartl 2016). The life cycle of a protein ends with its degradation, which is carried out by proteolytic machineries (Sauer and Baker 2011). Maintaining proteome integrity requires that protein synthesis, folding, transport and degradation are in a dynamic equilibrium, called protein homeostasis (proteostasis), which is adjusted in response to changing environments (Fig. 1) (Powers et al. 2009; Richter, Haslbeck and Buchner 2010).
Figure 1.
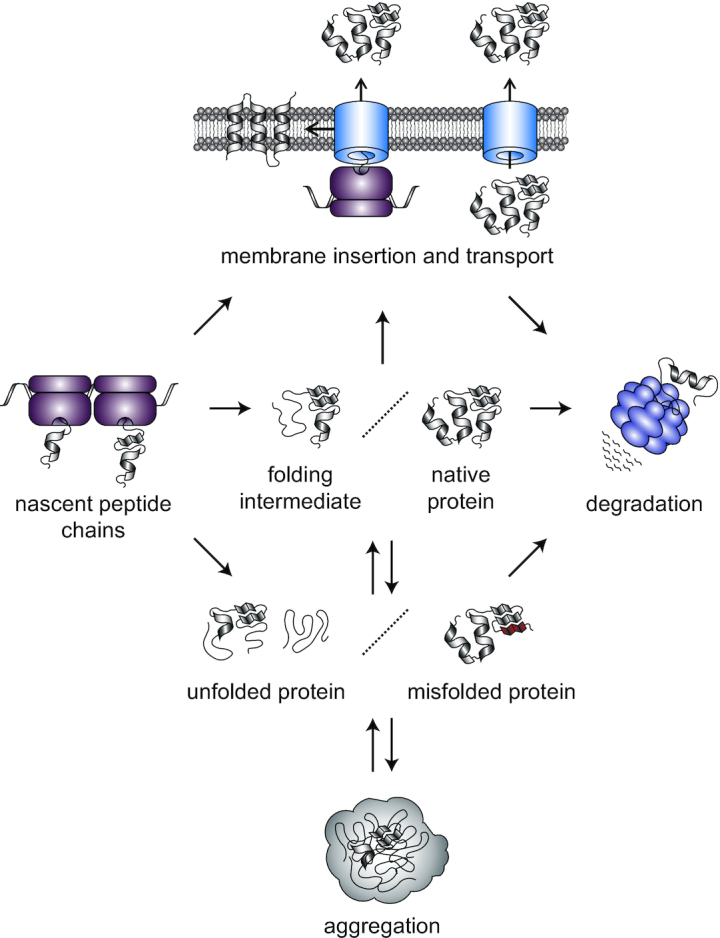
Overview of the major protein homeostatic processes in bacteria. As they exit the ribosome, most peptide chains reach their functional or native state by folding into a specific three-dimensional structure. In the case of larger proteins, this can entail the preceding formation of folding intermediates along the folding pathway. In addition to cytosolic proteins, a fraction of the proteome is inserted into or transported through the membrane. Protein function is threatened by stress conditions that affect protein folding. During stress, noncovalent interactions within the protein can be disrupted, leading to local or global loss of secondary and tertiary structure and the unfolding of a protein (represented as unstructured threads). Through the formation of non-native intramolecular interactions a protein can misfold and assume a structure deviating from its functional state (represented as a red fold). When folding intermediates, unfolded and misfolded proteins are abundant, they can associate with one another through non-native intermolecular interactions and co-aggregate to form larger aggregate structures. In order to maintain a functional proteome, natively folded as well as un/misfolded proteins can be degraded.
During the folding process, folding intermediates are vulnerable to forming aberrant non-native interactions causing the formation of misfolded protein species. Additionally, proteins can unfold resulting in partially or fully disordered polypeptides that lack a secondary or tertiary structure. Through hydrophobic interactions involving exposed residues or intermolecular β-sheet formation, un- and misfolded proteins have a tendency to associate in the crowded intracellular environment, leading to the formation of non-functional protein assemblies called protein aggregates (Fig. 1) (Balchin, Hayer-Hartl and Hartl 2016; Mogk, Bukau and Kampinga 2018). Aggregates generally start out as small soluble oligomers and can grow into large insoluble structures visible at cellular or even tissue scale (Mogk, Bukau and Kampinga 2018). These microscopically observable aggregate assemblies often are a final result of disturbances in protein homeostasis.
Although all organisms have systems in place to cope with protein un/misfolding, acute stress can overwhelm the protein quality control machinery leading to global protein aggregation. Protein aggregation generally results in a loss of protein function and can thus impair critical cellular functions that are required for growth and survival. In particular small soluble aggregate species and misfolded proteins can have cytotoxic effects by binding to and interfering with functional proteins and folding intermediates (Mogk, Bukau and Kampinga 2018). As free-living organisms, bacteria frequently encounter stress conditions inducing protein aggregation, and they have a remarkable ability to recover from such conditions. Importantly, protein aggregation can be induced by various antibiotics and recent research suggests that bacterial pathogenicity is tightly linked to protein quality control mechanisms (Lee et al. 2016). Hence, research addressing protein aggregation and its consequences in bacteria is a relevant research topic with potential impact on infectious disease control.
In this review, we discuss the current state of knowledge of protein aggregation in bacteria. We first highlight the diversity of aggregate types and the conditions that induce their formation. We then briefly describe the general roles of bacterial chaperones and proteases in maintaining proteostasis and discuss the diversity of disaggregation machineries employed amongst phylogenetically unrelated bacteria. We next discuss in detail how persistent aggregates are distributed to progeny in growing populations, and the cellular consequences of aggregate carriage. Finally, we highlight the possibility that protein aggregation can fulfill regulatory functions in stress adaptation.
BACTERIAL AGGREGATE DIVERSITY
Depending on the type of stress, as well as its duration and intensity, different types of aggregates can form that vary in composition, structure, size and their impact on cell function and viability. In bacteria, protein aggregates can be grouped into those that form as a result of environmental stress, and those that form as a result of heterologous protein expression.
Environmental stress-induced aggregation of the susceptible proteome
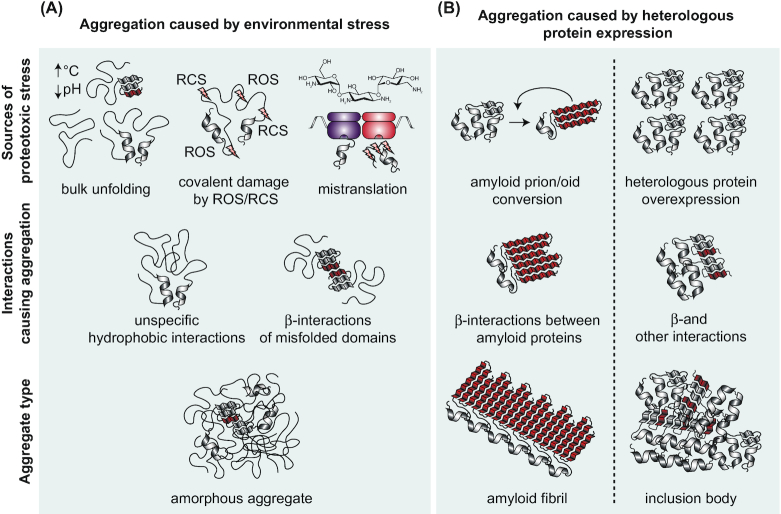
The native fold of a protein is sensitive to many conditions including temperature, osmolarity, ionic strength, pH and macromolecular crowding (Anfinsen and Scheraga 1975). Changes in any of these conditions in the cytosol and envelope of bacteria can impact the folding of the proteome, provoking protein un/misfolding that seeds aggregation (Fig. 2A) (Zhou 2013; Stull et al. 2018; Hantke et al. 2019). Investigation into how this occurs has come primarily from studying aggregation caused by exposure to high temperature. Increased temperature disrupts the weak intra-molecular forces holding a protein in its secondary and/or tertiary structure, causing denaturation (Anfinsen and Scheraga 1975). The effects of heat stress are highly scalable, where exposure to moderate heat stress transiently unfolds or inhibits correct folding of a small percentage of highly susceptible proteins, whereas exposure to extreme heat stress can cause un/misfolding of hundreds of protein species followed by their co-aggregation. Co-aggregation occurs through nonspecific hydrophobic interactions, and to a lesser extent through sequence-specific β-strand interactions, leading to the development of globular amorphous aggregates (Fig. 2A) (Mogk et al. 1999; Balchin, Hayer-Hartl and Hartl 2016; Khodaparast et al. 2018; Mogk, Bukau and Kampinga 2018; Pu et al. 2019; Schramm et al. 2019).
Figure 2.
Summary of proteotoxic stresses, aggregation causing interactions and protein aggregate types. Bacterial protein aggregates can be broadly categorized as those caused by environmental stress and those caused by heterologous protein expression. (A), Different types of environmental stresses lead to protein aggregation. Changes in temperature, osmolarity, ionic strength, pH and macromolecular crowding cause global protein un- and misfolding in a dose-dependent manner. Exposure to substances like hydrogen peroxide and hypochlorous acid cause a surge of reactive oxygen (ROS) and chlorine species (RCS), covalently damaging proteins and irreversibly changing their folding properties. Aminoglycoside antibiotics like kanamycin and streptomycin cause mRNA mistranslation. Incorporation of the wrong amino acids into nascent chains results in aberrant peptides with altered folding properties. Different species of un- and misfolded proteins can interact with one another through unspecific hydrophobic interactions or in a sequence-specific manner through contacting β-strands (β-interactions). Co-aggregation of many protein species results in the formation of globular amorphous aggregates that lack ordered intermolecular interactions. (B), Heterologous protein expression in bacteria can result in the formation of amyloid fibrils and inclusion bodies. Heterologously expressed prion/oid proteins switch between native and misfolded prion conformations, with the prion conformation driving conversion of the same protein species also into prion conformation. Prion/oid proteins can form highly structured amyloid aggregates where monomers of the same protein contact each other through β-sheet interactions running perpendicular to the long axis of the aggregate fibril. In heterologous overexpression for protein production, highly abundant protein can aggregate into globular inclusion bodies containing both amyloid and natively folded structure.
While heat-induced protein unfolding and aggregation is often reversible, some proteotoxic stresses irreparably damage proteins (Santra et al. 2018). Oxidative stress, which occurs for example during exposure to hydrogen peroxide or hypochlorous acid, damages proteins through covalent modification of specific amino acid side chains, altering the folding chemistry and causing un/misfolding that leads again to aggregation (Fig. 2A) (Dahl, Gray and Jakob 2015). Further, accumulation of oxidized amino acid side chains on a protein may also render it resistant to proteolysis (Grune et al. 2004; Maisonneuve et al. 2008a). Exposure to sublethal concentrations of the antibiotic kanamycin or heavy metals affects the fidelity of the ribosome, causing the creation of folding-deficient mistranslated protein species which can induce aggregation (Dukan et al. 2000; Kohanski et al. 2008; Tamás et al. 2018; Schramm et al. 2019). These mistranslated species, particularly those proteins that are incorporated into the membrane, can further exacerbate aggregation by fostering the creation of reactive oxygen species, which leads to damage of oxidation-sensitive proteins (Fig. 2A) (Kohanski et al. 2007; Ling et al. 2012).
In addition to stresses that directly disrupt the folding of susceptible proteins, un/misfolding and aggregation can be induced through disabling of the chaperone machinery. The entry into stationary phase is associated with the un/misfolding of many protein species and the development of protein aggregates, which in Escherichia coli is thought to stem primarily from the decline in ATP availability (Pu et al. 2019). ATP is required to power the major chaperones maintaining and helping proteins to achieve their native form, however, it also acts as a hydrotrope that supports protein solubility in the crowded intracellular environment (Patel et al. 2017; Pu et al. 2019). As levels of ATP decrease during the transition to stationary phase hundreds of proteins were found to aggregate in E. coli (Pu et al. 2019). Some proteotoxic stresses affect proteostasis at multiple points, such as how hypochlorous acid or hydrogen peroxide exposure directly causes protein un/-misfolding by oxidative damage but also disable chaperone-mediated resolubilization by damaging the chaperones and depleting ATP from the cell (Khor, Fisher and Schöneich 2004; Melkani et al. 2004; Winter et al. 2005). In this way, the composition of aggregates depends on the mechanism of the original unfolding stress, its intensity, as well as the available chaperoning capacity.
Numerous discrete protein species may aggregate in response to the proteotoxic stresses commonly encountered by bacteria, and the aggregation of many essential and non-essential proteins may threaten or impair a wide array of cellular processes (Mogk et al. 1999; Tomoyasu et al. 2001; Pu et al. 2019; Schramm et al. 2019). Some proteins are particularly sensitive to aggregation only in specific conditions, such as hypochlorous acid or antibiotic stress, however, others are prone to aggregation in many conditions (Weids et al. 2016; Santra et al. 2018; Pu et al. 2019; Schramm et al. 2019). This propensity to aggregate may be related to the functional domains of these proteins and their conserved chaperone dependency, as homologues of aggregation-prone proteins have been demonstrated to aggregate in different species. A large overlap exists between proteins that aggregate in stationary phase and heat shock in E. coli, and many of these proteins were also identified in heat shock-induced aggregates in Caulobacter crescentus (Mogk et al. 1999; Tomoyasu et al. 2001; Pu et al. 2019; Schramm et al. 2019). Proteins that have been identified in protein aggregates include those associated with carbon metabolism, oxidative phosphorylation, translation, as well as those involved in DNA replication and repair (Mogk et al. 1999; Tomoyasu et al. 2001; Pu et al. 2019; Schramm et al. 2019). The temporary sequestration of certain aggregation-prone proteins may function as regulatory nodes that shut down critical cellular processes in the interest of survival when mild stress is encountered, while more severe stress renders a significant fraction of the proteome non-functional, rapidly arresting growth and replicative processes (see Section 5).
Bacterial amyloid and heterologous overexpression-induced inclusions
In addition to relatively unstructured amorphous aggregates that form in response to environmental stress, other types of aggregates exist that assume a more ordered structure. Certain proteins can adopt a conformation allowing them to associate with one another in a sequence and stereospecific manner to form highly stable amyloid fibrils where monomers contact each other through β-sheets running perpendicular to the long axis of the fibril (Fig. 2B) (Eisenberg and Sawaya 2017). When resulting from protein misfolding, these amyloids represent aggregates that have been strongly associated with dysfunction and toxicity especially in eukaryotes (Aguzzi, Lakkaraju and Frontzek 2018). In contrast, bacterial amyloid in its endogenous context has mainly been linked to functional protein assemblies, making these amyloids distinct from the nonfunctional amyloid aggregates. Endogenous bacterial amyloids can be found both intra and extracellularly. The intracellular replication initiation factor RepA was shown to form amyloid oligomers when inhibiting plasmid replication, while the role of the amyloid formation of the RNA chaperone Hfq on DNA is less clear (Molina-García et al. 2016; Malabirade et al. 2018). Extracellular amyloid fibrils serve as structural elements and adhesion factors of the extracellular matrix and are often involved in host colonization. A well-known example is the amyloidogenic CsgA protein responsible for the formation of E. coli curli fibers (Chapman et al. 2002), however, many other proteins have been identified to form extracellular amyloid fibrils across the bacterial domain (recently reviewed by Marcoleta et al. 2019).
Some proteins can misfold into conformations that not only exhibit a strong tendency to form amyloid but also strongly encourage the misfolding of other proteins of the same species (Fig. 2B) (Aguzzi, Lakkaraju and Frontzek 2018). These amyloid prion (termed prionoid if only vertical and not horizontal transmission is observed) proteins have not as of yet been observed in endogenous bacterial contexts, however, heterologously expressed intracellular proteins have been documented to exhibit prionoid behavior in bacteria. It was demonstrated that the amyloidogenic yeast prion Sup35 could achieve its prionoid conformation and also cause the formation of self-perpetuating amyloid aggregates in E. coli (Garrity et al. 2010; Yuan et al. 2014). Additionally, the transcription termination factor Rho of Clostridium botulinum E3 strain Alaska E43 was shown to contain a domain that was able to substitute the function of the Sup35 prion-forming domain in yeast, and that exhibited prionoid behavior when expressed in E. coli (Yuan and Hochschild 2017). When the WH1 domain of the bacterial RepA protein was expressed to induce artificial intracellular prionoid aggregates in bacteria, a strong conformation-dependent toxicity was observed (Gasset-Rosa et al. 2014). Oligomeric amyloid aggregates formed by this protein created membrane pores that reduced membrane potential and induced oxidative stress and diverse aggregation of other proteins (Molina-García et al. 2016, 2017). As with small un/misfolded species caused by environmental proteotoxic stress, it is thought that misfolded or oligomeric amyloid species that remain in a soluble state are most disruptive to the cell, and co-aggregation of other protein species with the amyloidogenic protein is thought to confer much of the observed toxicity (Olzscha et al. 2011). Sequestration of the most detrimental misfolded oligomers into amyloid fibrils has therefore been proposed to reduce amyloid toxicity (Ross and Poirier 2004; Chiti and Dobson 2006; Rambaron and Serpell 2008). For example, it was found that the secreted antibacterial pore-forming bacteriocin microcin E492 from Klebsiella pneumoniae can form intracellular amyloid aggregates when expressed in E. coli which may sequester the toxic form (Aguilera et al. 2016).
Amyloid structures may also be incorporated into other types of aggregated protein structures. The microscopically observable intracellular inclusion bodies arising from heterologous protein overexpression in bacteria such as E. coli are often composed mainly of the recombinant protein and contain both amyloid and native or native-like structures (Fig. 2B) (de Marco et al. 2019). Accordingly, inclusion bodies can form globular structures containing a fibrillar interior and protruding fibrillar portions (Morell et al. 2008). Overexpression-induced inclusion bodies are mostly benign to the host cell and a fraction of the protein of interest can maintain its activity in the aggregated state (de Marco et al. 2019). These examples of native bacterial amyloids and inclusion bodies demonstrate that the formation of amyloid can represent a functional state of a protein, and that protein insolubility is not always associated with protein loss of function.
PROTEIN QUALITY CONTROL SYSTEMS COPING WITH PROTEIN AGGREGATION
Protein misfolding and aggregation is a result of disturbances in proteostasis which can lead to a functional deficit in cells. Like most other cells, bacteria deploy a plethora of chaperones and proteases in the cytosol, the cytoplasmic membrane and the cellular envelope, which are crucial to maintain a functional proteome. The function and mechanisms of these systems have been extensively studied and are reviewed in detail by others (Goemans, Denoncin and Collet 2014; Balchin, Hayer-Hartl and Hartl 2016; Mogk, Bukau and Kampinga 2018). In the following section, we provide a brief overview of how major components of the bacterial protein quality network collaborate to control the threat of protein aggregation and in particular discuss the different machineries employed in bacterial protein disaggregation.
Protein quality control systems preventing and resolving protein aggregation
The chaperone and protease repertoire of individual species can vary, however, the core elements of this network are highly conserved and widespread amongst the domains of life (Wong and Houry 2004; Powers and Balch 2013). The major chaperones which are constitutively expressed to handle the de novo folding of proteins in the bacterial cytosol typically include the ribosome-associated trigger factor (TF), the Hsp70 (70-kDa heat shock protein (HSP)) family chaperone DnaK with its J-domain containing Hsp40 co-chaperones such as DnaJ and nucleotide exchange factors such as GrpE (together referred to as DnaKJE), the Hsp60 chaperonin GroEL and co-chaperone GroES (GroESL) and in some bacteria the Hsp90 chaperone HtpG. These chaperones recognize and bind to hydrophobic residues exposed in their non-native substrates and aid the folding process of the substrate by protecting these residues from forming aberrant interactions (Balchin, Hayer-Hartl and Hartl 2016). While the monomeric TF functions independently of ATP (Maier, Scholz and Schmid 2001), DnaKJE, GroESL and HtpG substrate binding and chaperoning function is coupled to ATP binding and ATP hydrolysis-dependent conformational changes. Structural studies revealed that DnaK and HtpG function as monomeric or dimeric molecular clamps, respectively (Pearl and Prodromou 2006; Mayer 2013; Mayer and Gierasch 2019) and that the GroESL machinery is oligomeric and forms two chambers in which it can enclose substrates, providing a beneficial environment for folding (Hayer-Hartl, Bracher and Hartl 2016). In addition to their role in the folding of cytosolic proteins TF, DnaKJE and GroESL can also facilitate post- and co-translational protein secretion and membrane insertion involving dedicated chaperone and targeting machineries such as SecAB, the signal recognition particle (SRP), redox enzyme maturation proteins and the membrane insertase YidC (Castanié-Cornet, Bruel and Genevaux 2014). The folding and membrane insertion of translocated periplasmic and outer membrane proteins in Gram-negative bacteria is subsequently taken over by an ATP-independent periplasmic chaperone network (Goemans, Denoncin and Collet 2014).
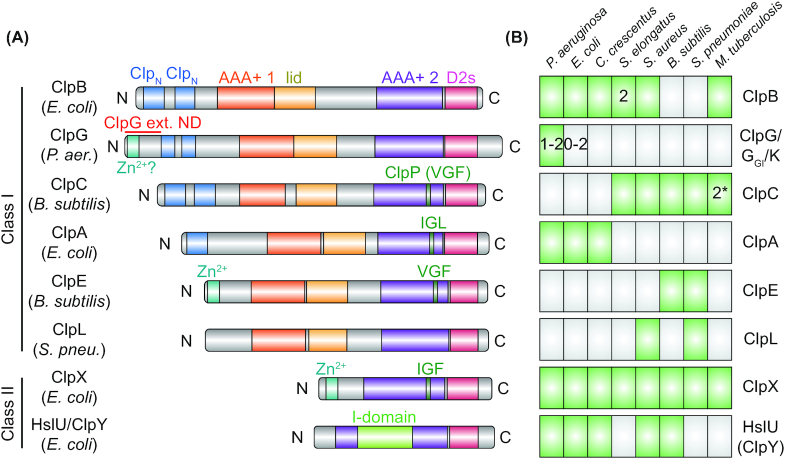
In addition to the housekeeping chaperones involved in protein synthesis and transport, proteases participate in proteostasis maintenance by degrading damaged and unneeded proteins. Among the major proteases are ATP-dependent proteases. These proteases which contain AAA+ (ATPases associated with a variety of cellular activities) domains oligomerize to form a ring structure which unfolds and processively transfers protein substrates into a peptidase ring via conformational changes induced by ATP binding and hydrolysis (Sauer and Baker 2011; Bittner, Arends and Narberhaus 2016). Highly conserved and widely distributed bacterial ATP-dependent proteases with housekeeping functions include the hexameric FtsH and Lon proteases, in which unfolding and proteolytic activities are provided by the same protein, as well as the two-component proteases, where unfolding and degradation are performed by different subunits. The two-component proteases include the hexameric HslU unfoldase ring, which associates with the hexameric HslV peptidase ring and the hexameric ClpA, ClpC, ClpE or ClpX unfoldase rings that associate with the heptameric ClpP peptidase ring via ClpP interaction loops located in the AAA+ domains. Although HslV and ClpP are not phylogenetically related, the associating unfoldase rings all belong to the large phylogenetic group of Hsp100 proteins with generally conserved domain structure (Fig. 3A) (Duran, Weaver and Lucius 2017). These may be widely distributed amongst bacterial phylogenetic groups, such as ClpX, or more phylum specific (Fig. 3B).
Figure 3.
Distribution of Hsp100 proteins among bacteria and Hsp100 domain structure. (A), Domain organization of bacterial Hsp100 homologs from different bacterial species. Domains annotated using InterPro (http://www.ebi.ac.uk/interpro/) show the respective identifiers. Hsp100 proteins can be categorized as two classes according to the number of AAA+ domains they contain: Class I Hsp100s ClpA, ClpB, ClpC, ClpE, GlpG (or ClpK) and ClpL have two while class II Hsp100s ClpX and HslU (also called ClpY) have only one. The AAA+ core domains (AAA+ 1/2; IPR003959) are C-terminally bordered by the AAA+ lid domain (lid; IPR041546) or the D2 small domain (D2s; IPR019489), respectively. HslU (ClpY) has an intermediate domain (I-domain, residues 108–243) inserted into its AAA+ core domain. The remaining domain organization of Hsp100s is protein specific, with differences in the length of the region between the AAA+ domains as well as in the N-termini, which may contain one or several conserved Clp N-terminal domains (ClpN; IPR004176) or a zinc-binding motif (Zn2+; IPR010603 in ClpX). The stand-alone disaggregases ClpG and ClpGGI possess an N-terminal extension with a putative zinc-binding motif (Zn2+?), which is involved in protein aggregate interaction (ClpG), or ATPase activity regulation (ClpGGI). HslU (ClpY) associates with the peptidase HslV (ClpQ) while ClpA, ClpC, ClpE and ClpX associate with the peptidase ClpP. ClpP interaction involves a tripeptide motif interaction loop in an AAA+ domain of the Hsp100s (tripeptide sequence indicated). ClpL lacks a known ClpP interacting motif and it is unknown if it interacts with a peptidase subunit. (B), Distribution of Hsp100 proteins in selected bacterial species belonging to the Proteobacteria (Pseudomonas aeruginosa, Escherichia coli and Caulobacter crescentus), Cyanobacteria (Synechococcus elongatus), Firmicutes (Staphylococcus aureus, Bacillus subtilis and Streptococcus pneumoniae) and Actinobacteria (Mycobacterium tuberculosis). ClpG belongs to the Pseudomonas aeruginosa core genome but a second homolog (ClpGGI) can be found on genomic island 1 of P. aeruginosa clone C. Most E. coli species lack ClpG, however, some E. coli strains harbor a ClpG or ClpGGIhomolog on a genomic island or on mobile genetic elements. Synechococcus elongatus possesses two ClpB homologs (ClpB1 and ClpB2) and two potential ClpC homologs (ClpC1 and the unusual truncated ClpC2/ClpX’) can be found in Mycobacterium tuberculosis.
In response to proteotoxic stress, bacteria employ stress-adaptive transcriptional programs, such as the heat shock response, where cellular resources become redirected toward the chaperone and protease network in order to prevent further un/misfolding and aggregation as well as to facilitate refolding, protein removal and aggregate dissolution (Richter, Haslbeck and Buchner 2010). Proteotoxic stress-induced transcriptional reprogramming involves activators and repressors whose activity is regulated by chaperones and proteases (Roncarati and Scarlato 2017) (further discussed in Section 5.1). The majority of chaperones and proteases that are central to protein homeostasis under optimal conditions are strongly upregulated as part of the heat shock response to cope with the increased folding demand (Richter, Haslbeck and Buchner 2010). In E. coli, both GroESL and DnaKJE are upregulated following temperature upshifts and play critical roles in preventing global protein aggregation by folding and re-folding a large fraction of the proteome (Tomoyasu et al. 2001; Mogk et al. 2003; Chapman et al. 2006). Among the proteases in E. coli, Lon has a well-documented role in degrading un/misfolded proteins that accumulate under non-optimal conditions (Tomoyasu et al. 2001; Gur and Sauer 2008), a role that seems to be fulfilled mainly by ClpP protease complexes in B. subtilis (Krüger et al. 2000; Kock, Gerth and Hecker 2004). In addition to the housekeeping chaperones and proteases that operate under both optimal and stress conditions in the cytosol, proteotoxic stress responses also induce chaperones that prevent aggregate formation under specific stress conditions or in other cellular compartments. Well-studied examples are the cytosolic redox-regulated chaperone Hsp33 and the hypochlorous acid-activated protein CnoX, both of which function independently of ATP and are important for resisting oxidative stress which can temporarily inactivate ATP-dependent chaperones (Dahl, Gray and Jakob 2015; Goemans et al. 2018). Additionally, stress-induced ATP-independent chaperones such as HdeA/B and Spy operate in the periplasm, a compartment which lacks ATP and in addition to general proteotoxic stress is more exposed to environmental changes in pH and ionic strength (Goemans, Denoncin and Collet 2014; Lee, Kim and Bardwell 2018; Stull et al. 2018).
Different types of disaggregation machineries in bacteria
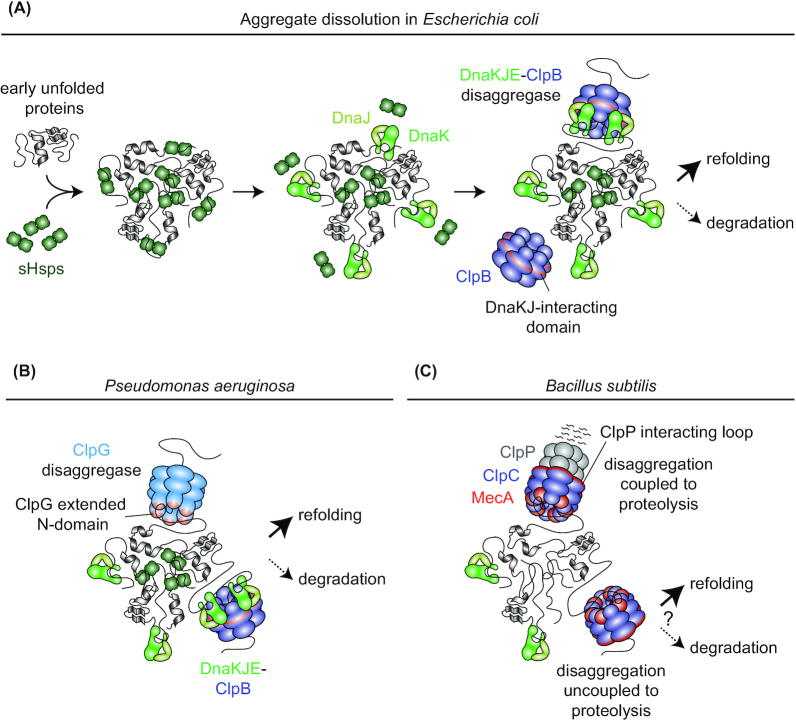
The protein quality control network is not only involved in preventing protein aggregation but also has dedicated systems to undo protein aggregation once it has occurred. Disaggregation machineries are able to dissolve protein aggregates and in this way ensure recovery from acute stress and continued survival. In bacteria, ClpB is a highly conserved representative of this group of chaperones. ClpB belongs to the phylogenetic group of Hsp100 proteins (Fig. 3A) that form a hexameric ring capable of processively threading unfolded proteins through its central pore under ATP consumption (Duran, Weaver and Lucius 2017; Rizo et al. 2019). However, unlike the related HslU, ClpA, ClpC, ClpE or ClpX Hsp100 proteins, it does not associate with a peptidase ring and E. coli proteins recovered from aggregates by ClpB activity are usually subsequently refolded to regenerate functional proteins (Weibezahn et al. 2004). ClpB cannot prevent protein aggregation and has no disaggregation activity of its own but instead cooperates with DnaKJE to form the DnaKJE-ClpB bichaperone system (Fig. 4A) (Mogk, Bukau and Kampinga 2018). Similarly to its eukaryotic homolog Hsp104, which strictly depends on Hsp70 (Glover and Lindquist 1998), the bacterial ClpB chaperone requires DnaKJE for its association with protein aggregates, for substrate transfer and for modulation of its ATPase activity (Ziȩtkiewicz, Krzewska and Liberek 2004; Acebrón et al. 2009; Deville et al. 2017; Hayashi et al. 2017). Work in different bacteria showed that clpB expression is strongly upregulated in response to heat shock and other stress conditions and that ∆clpB mutants are strongly compromised in heat resistance and unable to resolve protein aggregates following stress release (Eriksson and Clarke 1996; Allan, Mullany and Tabaqchali 1998; Ekaza et al. 2001; Mogk et al. 2003; Winkler et al. 2010; Vaubourgeix et al. 2015; Schramm et al. 2019). Furthermore, elevating the levels of the DnaKJE-ClpB machinery was shown to be sufficient to almost completely revert protein aggregation after heat shock in an E. coli strain deficient in heat shock response regulation (Tomoyasu et al. 2001), demonstrating that DnaKJE-ClpB constitutes the major system for preventing and reversing protein aggregation in E. coli. In E. coli and other bacteria, protein aggregate dissolution by the DnaKJE-ClpB bichaperone is assisted by the small HSPs (sHSPs). sHSPs can bind proteins at an early stage of unfolding and stabilize them in a native-like conformation inside aggregate assemblies that are amenable to disaggregation and subsequent refolding (Fig. 4A) (Mogk, Ruger-Herreros and Bukau 2019). In different bacteria sHSPs have been shown to represent the most abundant protein species in aggregates (Laskowska, Wawrzynów and Taylor 1996; Cashikar, Duennwald and Lindquist 2005; LeThanh, Neubauer and Hoffmann 2005; Gasset-Rosa et al. 2014; Schramm et al. 2019).
Figure 4.
Protein disaggregation in different bacterial species. (A), Protein disaggregation via the DnaKJE-ClpB bichaperone machinery in E. coli (GrpE not shown). sHSPs (IbpA and IbpB in E. coli) bind recently unfolded proteins and are incorporated into aggregates while maintaining proteins in a near-native state that facilitates eventual disaggregation by DnaKJE-ClpB. Together with the co-chaperone DnaJ, DnaK binds to aggregated protein and displaces sHSPs from the aggregate surface. DnaK recruits the ClpB disaggregase to the aggregate and this interaction stimulates high ClpB ATPase activity. Substrates are threaded through the central pore of ClpB and, after extraction from the aggregate, can be refolded (preferred) or degraded. (B), Protein disaggregation machineries in Pseudomonas aeruginosa. In addition to the DnaKJE-ClpB disaggregation machinery, P. aeruginosa employs ClpG which does not require stimulating factors for high disaggregation activity. Proteins extracted by both machineries can subsequently be refolded or degraded. (C), Potential protein disaggregation mechanisms in B. subtilis. B. subtilis lacks ClpB or another stand-alone disaggregase. Instead, the ClpP interacting loop containing Hsp100 ClpC interacts with the adaptor MecA and binds to aggregated proteins to drive their disaggregation in vitro. Although protein degradation-coupled disaggregation by MecA-ClpCP has been shown to be more efficient, MecA-ClpC can also disaggregate proteins without associating with ClpP. Thus, potential disaggregation through MecA-ClpC not associated with ClpP could allow for the refolding (or the downstream degradation by another protease) of extracted proteins in vivo.
In addition to ClpB, some bacteria use other Hsp100-type proteins for disaggregation. The opportunistic pathogen Pseudomonas aeruginosa employs the stand-alone ClpG disaggregase in addition to the DnaKJE-ClpB bichaperone to ensure heat tolerance (Fig. 4B) (Lee et al. 2017). ClpG exhibits high basal ATPase activity and directly binds to aggregates without requiring the assistance of DnaKJE or other accessory factors, a process which involves an extended N-terminal domain essential for aggregate targeting (Fig. 4B). Interestingly, the medically important and environmentally widespread P. aeruginosa clone C harbors two copies of clpG, one of them on the P. aeruginosa core genome and the other one (ClpGGI) on a clone C specific genomic island that harbors several other genes encoding protein quality control proteins. Although ClpGGI is like ClpG in that it is a stand-alone disaggregase featuring an extended N-terminal domain, this domain is not essential for aggregate binding but rather involved in the regulation of the ATPase activity. Both ClpG proteins work independently from one another and exert overlapping and compensatory activities. ClpGGI (also called ClpK in some species (Bojer et al. 2010)) is widespread among other pathogenic bacteria suggesting that it might be important for virulence or antibiotic resistance (Lee et al. 2016, 2017, Boll et al.2017). Furthermore, some Gram-positive bacteria harbor the ClpL protein which like ClpB and ClpG lacks a ClpP peptidase interacting loop (Fig. 3A and B). ClpL confers disaggregation activity in vitro and is involved in heat stress resistance in several species suggesting that this protein might represent yet another bacterial disaggregation machinery (Kwon et al. 2003; Frees et al. 2004; Suokko et al. 2008; Park et al. 2015).
In addition to ClpB, ClpG and ClpL, there are indications that the ClpP peptidase interaction loop containing ClpA, ClpC, ClpE and ClpX also disaggregate proteins in vivo. It was shown that these Hsp100s from the phylogenetically distant species E. coli, B. subtilis and the cyanobacterium Synechococcus elongatus can disaggregate protein aggregates with varying efficiency in vitro (Dougan et al. 2002; Schlothauer et al. 2003; Andersson et al. 2006; LaBreck et al. 2017) and that they associate with large microscopically detectable aggregates in E. coli and B. subtilis (Krüger et al. 2000; Schlothauer et al. 2003; Miethke, Hecker and Gerth 2006; Winkler et al. 2010; Kumar and Sourjik 2012). Furthermore, some bacteria, including Gram-positive B. subtilis, lack ClpB, ClpG or ClpL homologs (Fig. 3B) and are less dependent on DnaKJE for thermal stress adaptation, suggesting that ClpP peptidase interaction loop containing unfoldases might play a more prominent role in protein disaggregation in such species (Fig. 4C). Whether in vivo this activity is performed while associated with ClpP, and thus coupled to the degradation of the extracted proteins, is still unclear. In vitro data indicate that ClpC in complex with the adaptor MecA from B. subtilis also possesses disaggregation activity independent of ClpP (Schlothauer et al. 2003). ClpP-independent disaggregation by ClpC and other related unfoldase rings could potentially provide a way to facilitate refolding of proteins following their disaggregation (Fig. 4C).
Together, although the bacterial ClpB-DnaKJE bichaperone system is the primary disaggregation machine in many bacteria (Mogk, Bukau and Kampinga 2018), there is increasing evidence that bacteria can also make use of other Hsp100-dependent systems to cope with protein aggregates. Interestingly, while plants and fungi also make use of Hsp100/Hsp70 disaggregation machineries for cytoplasmic protein disaggregation, animals lack a Hsp100-based disaggregation machine (Mogk, Bukau and Kampinga 2018; Nillegoda, Wentink and Bukau 2018). Instead, disaggregation is achieved by collaboration of Hsp70 with specific J-domain proteins and Hsp110 nucleotide exchange factors, also part of the Hsp70 family (Rampelt et al. 2012; Nillegoda et al. 2015; Nillegoda, Wentink and Bukau 2018). Whether disaggregation in bacteria can also take place in an Hsp100-independent way remains unclear.
PROTEIN AGGREGATES AS INHERITED DAMAGE AND AGING FACTOR
When stress causes protein un- and misfolding above what the proteostasis network can prevent or repair, protein aggregates persist in the cell. If under such conditions growth and division continue, these persistent aggregates are distributed between the progeny at each cell division. Where aggregates form and how they move are not universal among bacteria and these parameters constrain how aggregates can be distributed during cell division with consequences for how persistent protein aggregates are shared by the population. Unequal distribution of un/misfolded protein and aggregates has been proposed to drive population heterogeneity and to underpin replicative decline or aging, in the population segment inheriting aggregates. However, the impact of aggregate carriage remains a controversial topic. This section describes the spatiotemporal constraints on how aggregates form and move in bacteria, how aggregates are distributed during cell division and what impact they might have in bacteria.
Visualization of protein aggregates in bacteria
As described in Section 1, larger protein aggregates are seeded by individual un/misfolded proteins. The process of protein un/misfolding and aggregation can be monitored in bacterial populations directly by using biochemical techniques to isolate and interrogate insoluble proteins (Mogk et al. 1999; Maisonneuve et al. 2008b; Fay and Glickman 2014; Schramm et al. 2017). These techniques have provided information on which protein species are prone to aggregation, and in which stress conditions these are present or absent from aggregates. In order to investigate aggregate formation and resolution in individual cells, standard fluorescence microscopy and more advanced techniques such as super-resolution fluorescence microscopy and cryo-tomography are employed, the former relying on fluorescently tagged proteins that aggregate or localize to sites of aggregation (Winkler et al. 2010; Vaubourgeix et al. 2015; Schramm et al. 2019). Fluorescence microscopy in combination with time-lapse microscopy can provide time-resolved information on the location and trajectories of aggregates within the limits of detection, and give insight into how aggregates build, move, and are resolved over the lifespan of individual cells (Coquel et al. 2013; Gupta et al. 2014; Schramm et al. 2019). Fluorescently tagged endogenous sHSPs have frequently been used as aggregate reporters in E. coli, as they are highly upregulated in response to proteotoxic stress and are abundantly incorporated into aggregates (Lindner et al. 2008; Kumar and Sourjik 2012; Govers et al. 2018; Hantke et al. 2019; Mogk, Ruger-Herreros and Bukau 2019). However, other components of the protein homeostasis network also localize to protein aggregates and have been used to label these when fluorescently tagged, including DnaK and ClpB (Kirstein et al. 2008; Winkler et al. 2010; Kumar and Sourjik 2012; Schramm et al. 2019).
In addition to components of the proteostasis network, aggregation-prone endogenous proteins identified through mass spectrometry of isolated protein aggregates have been used as reporters (Winkler et al. 2010; Hantke et al. 2019; Schramm et al. 2019). Finally, the use of unstable exogenous proteins such as firefly luciferase or aggregation-prone sequences fused to fluorescence reporters allow tracking the process of protein aggregation in bacteria without altering endogenous proteins (Winkler et al. 2010; Fay and Glickman 2014; Schramm et al. 2019). As the addition of fluorescent tags can influence the stability of proteins and increase or decrease their tendency to aggregate, the choice of fluorescent protein and validation of the functionality of the resulting fusion protein are critical. In particular, the use of non-monomeric fluorescent proteins on sHSPs is known to increase the number of aggregates detected during non-stress conditions and can influence the size of aggregates formed during stress (Landgraf et al. 2012; Govers et al. 2018; Schramm et al. 2019). These concerns are currently addressed through the use of monomeric fluorescent protein variants together with careful verification that the fusion protein retains functionality when using endogenous proteins (Winkler et al. 2010; Landgraf et al. 2012; Govers et al. 2018; Schramm et al. 2019).
Subcellular localization of protein aggregates in bacteria
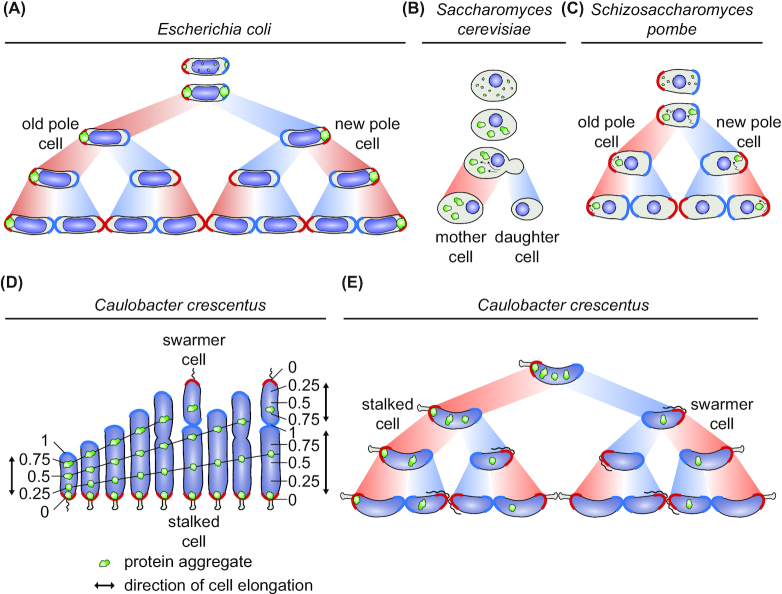
Large intracellular structures, such as DNA, impose spatial constraints on where aggregate particles are able to develop and influence how they are able to move from these positions inside a cell. Differences in intracellular organization between bacteria therefore give rise to different aggregation formation and movement patterns. In E. coli, aggregates demonstrate a clear preference for forming at the poles in response to many different stresses, as well as at midcell in predivisional cells (Fig. 5A) (Lindner et al. 2008; Winkler et al. 2010; Kumar and Sourjik 2012; Coquel et al. 2013; Govers, Dutré and Aertsen 2014; Gupta et al. 2014; Neeli-Venkata et al. 2016). These regions correspond to the area of the cytoplasm that is unoccupied by the nucleoid and perturbing the arrangement and number of chromosomes influences how aggregates are able to form. Creation of anucleoid E. coli cells, such as by deletion of the gene encoding the chromosome partitioning protein MukB, results in the formation of a single large aggregate in the center of the cell (Winkler et al. 2010; Govers, Dutré and Aertsen 2014), and conversely, a filamentous minCD mutant with multiple chromosomes forms larger numbers of regularly spaced aggregates located between the nucleoids (Lindner et al. 2008). Changing the conformation of the chromosome, such as by reducing or increasing the size of the nucleoid, also affects the fraction of aggregates that occur away from the poles, with a larger nucleoid resulting in more aggregates forming within the nucleoid space (Neeli-Venkata et al. 2016). Furthermore, it was shown that increasing the viscosity of the cytoplasm also increases the likelihood of aggregates existing in positions within the nucleoid instead of at the poles (Oliveira et al. 2016). Therefore, aggregates are not formed actively at the poles, a finding confirmed by biophysical measurements showing aggregate movement trajectories that are consistent with free diffusion, or Brownian motion (Coquel et al. 2013; Gupta et al. 2014). But rather, these experiments collectively indicate that molecular crowding from the nucleoid prevents formation of larger aggregates within this space, and that diffusion together with aggregation are sufficient to drive polar (or midcell when two nuceloids are present) aggregate formation in E. coli. A similar situation occurs in B. subtilis exposed to low intensity stress, where a few protein aggregates preferentially form in the DNA-free spaces at the poles and between nucleoids, while additional distributed aggregates are observable with increasing stress intensity (Kirstein et al. 2008; Runde et al. 2014; Hantke et al. 2019).
Figure 5.
Aggregate formation and inheritance in unicellular organisms. (A), In E. coli, cells are born with an old pole (labeled in red) present in the progenitor cell and a new pole (labeled in blue) built during division. The cell inheriting the old pole of the progenitor cell is termed the old pole daughter cell and the one inheriting the new pole the new pole daughter cell. An E. coli population consists of cell lineages with many different pole inheritance histories (red areas connecting cells indicate old pole inheritance, blue areas new pole inheritance). In E. coli, aggregating proteins (green particles) are occluded from the central nucleoid (violet region), enforcing aggregate localization at the nucleoid-free poles from which they rarely move. This pattern creates a strong asymmetry of aggregate inheritance as aggregates are specifically retained in the cell lineages consecutively inheriting the progenitor cell's old pole. (B), During asymmetric cell division of S. cerevisiae a smaller cell buds off from a larger mother cell. Heat shock-induced aggregates are collected and deposited at different sites in the cell and are actively and passively retained in the mother cell, creating strong asymmetry of aggregate distribution. (C), In the symmetrically dividing S. pombe stress-induced aggregates form and fuse together either in the space between old pole and the nucleus (violet circle) or new pole and the nucleus. Aggregates are mobile however the nucleus impedes frequent movement to the opposite pole half. This leads to preferential aggregate retention in cell lineages consecutively inheriting the progenitor cell's old pole. (D), Schematic showing how growth along the length of the cell outside the pole regions determines aggregate localization and inheritance in C. crescentus. In this organism, cell division is asymmetric and yields a larger stalked cell (the old pole cell) and a smaller swarmer cell (the new pole cell). The C. crescentus nucleoid expands through the entire cell and aggregates form as distributed foci throughout the length of the cell. Numbers represent relative cell positions between the old (0) and new pole (1). Lines depict these relative positions as the cell elongates and divides. With each division event, aggregates will gradually assume a position closer to the new pole until they are inherited by the other cell type. A minority of aggregates remain trapped after forming at the pole. (E), C. crescentus aggregate inheritance pattern resulting from the process described in (D). Aggregates are not retained in the lineage consecutively inheriting the old pole, but instead are constantly distributed between old and new pole cells (with the exception of the minor pole aggregate fraction).
A more numerous and distributed pattern of protein aggregation localization has been demonstrated both in Mycobacteria and C. crescentus. In Mycobacteria, multiple punctate foci of protein aggregation form throughout the entire cytoplasm in response to antibiotic treatment and heat stress (Fay and Glickman 2014; Vaubourgeix et al. 2015), contrasting with the localization of aggregates in E. coli. However, also in contrast to E. coli and B. subtilis, during log phase the Mycobacterial chromosome is organized into multiple small nucleoids that condense upon entry into stationary phase (Fay and Glickman 2014; Scutigliani et al. 2018). These differences in intracellular crowding may drive the different formation pattern of protein aggregates in Mycobacteria, where aggregates initially form as distributed foci which are then collected into larger deposits sitting preferentially at a pole (Fay and Glickman 2014; Vaubourgeix et al. 2015). In C. crescentus, protein aggregation also occurs in multiple punctate foci that are distributed throughout the entire cell volume in response to heat and antibiotic stress (Schramm et al. 2019). However, whereas the aggregates of Mycobacteria continue to move after formation, those of C. crescentus are mostly stationary (Vaubourgeix et al. 2015; Schramm et al. 2019). One important structural difference between these organisms is that the nucleoid of C. crescentus extends throughout the entire cell volume (Ward and Newton 1997; Gray et al. 2019). In C. crescentus, protein aggregates may even displace the chromosome, as shown by reduced DNA staining in areas occupied by aggregates (Schramm et al. 2019). Measurements of the ratio of the nucleoid to total cytoplasm (NC ratio) across many bacterial species has demonstrated that protein complexes in organisms with a low NC ratio, such as E. coli, can diffuse and move freely once the DNA mesh is escaped, whereas mobility is very limited in organisms with a high NC ratio, such as C. crescentus (Gray et al. 2019). These differences have important implications for the mobility of protein aggregates in different species, as well as for the cellular locations they can collect in.
While sublethal stresses are typically used to monitor aggregate formation and recovery, the intensity of the stress also affects the number and position of aggregates present during the cell cycle. Higher stress temperatures or antibiotic concentrations induce additional, more stable and sometimes larger foci of protein aggregation in most bacteria (Runde et al. 2014; Vaubourgeix et al. 2015; Govers et al. 2018; Hantke et al. 2019; Schramm et al. 2019). In C. crescentus, mild heat shock or low antibiotic concentrations results in short-lived aggregates that are dissolved before completion of one cell cycle, however, higher intensity stress or genetic mutation of the protein quality control machinery results in a fraction of long-lived aggregates that can persist for several generations (Schramm et al. 2019). The number and position of these persistent aggregates prior to division is tightly linked to how aggregates are distributed to different segments of the population.
Aggregate distribution in dividing bacteria
When the load of un/misfolded and aggregated protein persists for longer than the time needed for cell cycle progression and division, it will somehow be distributed between the emerging two progeny. In bacteria this aggregate distribution was first described in E. coli, where polar aggregate localization drives a characteristic inheritance pattern during subsequent generations (Fig. 5A) (Lindner et al. 2008). As aggregates localize and remain preferentially at the poles in the progenitor cell, a division event results in two daughter cells that each contain one aggregate at the oldest cell pole (Coquel et al. 2013; Gupta et al. 2014). A second division event results in four daughter cells, two of which contain only new poles built during division and thus escaping entirely the inheritance of an aggregate and two cells that contain the original and oldest poles from the progenitor cell and therefore the associated aggregates (Fig. 5A). This distribution of aggregates has historically been referred to as ‘asymmetric inheritance’ of protein aggregates, where one daughter cell type inherits all, or the majority, of the aggregated protein present in the progenitor cell, and the other escapes aggregate carriage (Nyström and Liu 2014). Crucially, as persistent aggregates only rarely move away from their position proximal to the pole, in this pattern aggregates are retained in the lineage of cells that consecutively inherits the old pole (Fig. 5A) (Lindner et al. 2008; Winkler et al. 2010). The phenomenon of asymmetric aggregate inheritance was originally described in the budding yeast S. cerevisiae, and later also in the fission yeast Schizosaccharomyces pombe. In budding yeast, the larger and older mother cell lineage preferentially retains aggregates by both active and passive mechanisms during division (Fig. 5B) (Erjavec et al. 2007; Tessarz et al. 2009; Liu et al. 2010; Zhou et al. 2011, 2014; Spokoini et al. 2012; Song et al. 2014). Similarly to E. coli, in S. pombe aggregates form on either or both sides of the central nucleus and are inherited during subsequent division events as a part of the cell half containing the oldest pole (Fig. 5C) (Coelho et al. 2013, 2014; Nakaoka and Wakamoto 2017).
The similarity in the pattern of aggregate inheritance between yeast and E. coli as well as the observation of polar aggregate localization in other bacteria led to the prediction that asymmetric aggregate inheritance might take place in most prokaryotes, however, a recent study of aggregate inheritance in C. crescentus has challenged this view. C. crescentus divides asymmetrically and produces a smaller swarmer cell and a larger stalked cell at the end of each division cycle, and owing to swarmer to stalked cell differentiation the stalked cell population always maintains the oldest poles (Fig. 5D) (Curtis and Brun 2010). As described above, C. crescentus develops multiple protein aggregates that are dispersed throughout the cell volume and without preference for the cell poles. Tracking aggregates by time-lapse microscopy showed that these are distributed to both daughter cells in the same ratio at each division, and that this inheritance pattern is driven by the elongation of the growing progenitor cell (Fig. 5D) (Schramm et al. 2019). As the stalked cell is larger than the swarmer cell this segment of the population consistently inherits a larger percentage of the aggregate load compared to swarmer cells. However, with successive divisions individual stalked cells gradually decrease their carried aggregate load. Furthermore, in the case where only one aggregate is carried by a progenitor cell it is not necessarily passed on to the old pole stalked cell, but can be distributed to either daughter cell type, thereby deviating from asymmetric aggregate inheritance as the term has been historically used (Fig. 5E) (Schramm et al. 2019).
Consequences of aggregate carriage and the link to aging
Conflicting evidence exists about the impact aggregate inheritance has on a bacterial cell, with results indicating aggregates may be detrimental, beneficial or of no consequence depending on the environmental condition. The first example of a unicellular organism that might experience a disadvantage from inheriting aggregates was budding yeast, where the replicative output of the aggregate retaining mother cell is reduced compared to the aggregate evading daughter cell (Erjavec et al. 2007; Denoth Lippuner, Julou and Barral 2014). The decrease in reproductive capacity and growth rate over generations associated with accumulation of aggregated protein was termed ‘replicative aging’ (recently reviewed by Florea 2017 and Moger-Reischer and Lennon 2019). As protein aggregates are also inherited asymmetrically in some bacteria, studies followed to determine if aggregate inheritance might also be linked to aging in bacteria. Compared to aggregate-evading cells, retention of a protein aggregate was shown to contribute to a reduced growth rate when monitored by a model aggregating protein during recovery from heat stress (Winkler et al. 2010) or by a sHSP during spontaneous aggregate formation (Lindner et al. 2008), findings which have since been extended to entire stressed populations (Vedel et al. 2016). In another study, it was observed that ejection of aggregates by strains possessing the ability to create minicells from the pole regions was able to confer a growth advantage (Rang et al. 2018). In M. smegmatis a clear correlation was established between aggregate inheritance and mortality, where higher intensity stress correlated with more unequal distribution of the aggregate load between daughter cells and inheritance of more of the aggregate load reduced growth rate or led to cell death (Vaubourgeix et al. 2015). Since the original suggestion of bacterial aging, several studies have sought to understand and model how unequal distribution of protein aggregates could benefit bacterial populations. Collectively, these studies suggest that under high-intensity stress when repair processes become inefficient, unequal segregation can be beneficial as it allows faster growth rates as well as higher stress tolerance of damage-evading cells (Clegg, Dyson and Kreft 2014; Chao et al. 2016; Vedel et al. 2016; Proenca et al. 2019).
While the paradigm constructed from these studies has associated aggregate carriage with growth disadvantage, newer studies have called this connection into question. Re-analysis of the impact of aggregates on cell growth in E. coli indicated that carrying an aggregate had no detrimental effect on growth rate (Govers et al. 2018). Aggregate carriage in this system instead even resulted in increased resistance to subsequent stresses, attributed to elevated local concentrations of proteostasis network proteins, suggesting that under certain conditions harboring aggregates could enhance survival (Govers et al. 2018). In C. crescentus, abnormally large aggregates in a strain with altered sHsp activity reduced growth rate (Schramm et al. 2019) but no obvious growth defect could be detected in cells carrying regularly sized aggregates. Previous work in C. crescentus demonstrated that over many generations stalked cells show a decline in reproductive output (Ackermann, Stephen and Jenal 2003). This observation provoked the hypothesis that retention of protein aggregates in the stalked cell might cause aging in this organism (Ackermann, Stephen and Jenal 2003; Ackermann et al. 2007; Lindner et al. 2008). The more recent data, including the lack of stalked-cell specific aggregate retention, questions the hypothesized role of protein aggregates as aging factors in C. crescentus, instead suggesting that this form of stalked cell senescence could be due to other factors that may accumulate in the old pole stalked cell, such as older membrane components, or that distribution of large stress-induced aggregates is a separate process from replicative aging. In support for the former alternative, in E. coli it was shown that simply the age of the pole independent of aggregate presence can affect growth rate (Stewart et al. 2005; Lindner et al. 2008; Winkler et al. 2010; Bergmiller et al. 2017; Proenca et al. 2018, 2019) and aging factors other than damaged proteins have also been suggested to be present in S. cerevisiae (Denoth Lippuner, Julou and Barral 2014).
Taken together, how aggregate carriage impacts bacterial growth and population fitness appears to be influenced both by the type and size of the aggregates present, as well as the environment that bacteria are exposed to. Future research, including studies in other bacterial species, will be necessary to fully understand the parameters governing the effects of protein aggregation and how this is linked to phenotypic and population outcomes.
Prionoid propagation in bacteria
Aggregate inheritance and its impact has mainly been studied under conditions inducing the formation of amorphous aggregates composed of many protein species, such as after heat shock or antibiotic treatment. To better understand the contribution of the conformation of protein species to toxicity and damage inheritance in bacteria, the behavior of prionoid proteins and aggregates formed by these have been studied. Heterologous expression of the toxic prionoid protein RepA-WH1 in E. coli induced amyloid aggregates that took either globular or ‘comet-like’ forms, which were distributed to daughter cells according to the formed conformation (Gasset-Rosa et al. 2014). The elongated ‘comet-like’ structures, spanning the length of the cell, may be split during a division event and inherited by both daughter cells in this conformation. However, daughter cells escaping inheritance of a prionoid aggregate were also more likely to reform aggregates in the same conformation present in the progenitor cell (Gasset-Rosa et al. 2014). Importantly, carriage of a ‘comet-like’ structure was associated with milder growth defect than the globular form, indicating that the conformation of aggregating protein can influence its toxicity (Gasset-Rosa et al. 2014). These findings indicate that the conformation of un/misfolded proteins below the limit of microscopic detection is able to influence the aggregation of other proteins, and that alternate aggregation-prone conformations of a protein species can disrupt cellular processes differently.
PROTEIN AGGREGATION AS A REGULATORY MECHANISM
Although stress-induced protein un- and misfolding and aggregation generally perturbs cellular function, recent work suggests that the stress-induced aggregation of specific regulatory proteins constitutes an important regulatory mechanism in bacteria. Furthermore, in analogy to the phase separation of specific groups of proteins in eukaryotic stress granules, the sequestration of certain proteins in bacterial protein aggregates could potentially provide a means to reallocate cellular resources from growth-promoting to cytoprotective functions. Here, we discuss recent work suggesting that protein aggregation in bacteria represents an integral part of stress response regulation and cellular resource allocation.
Transcriptional control through protein aggregation in bacteria
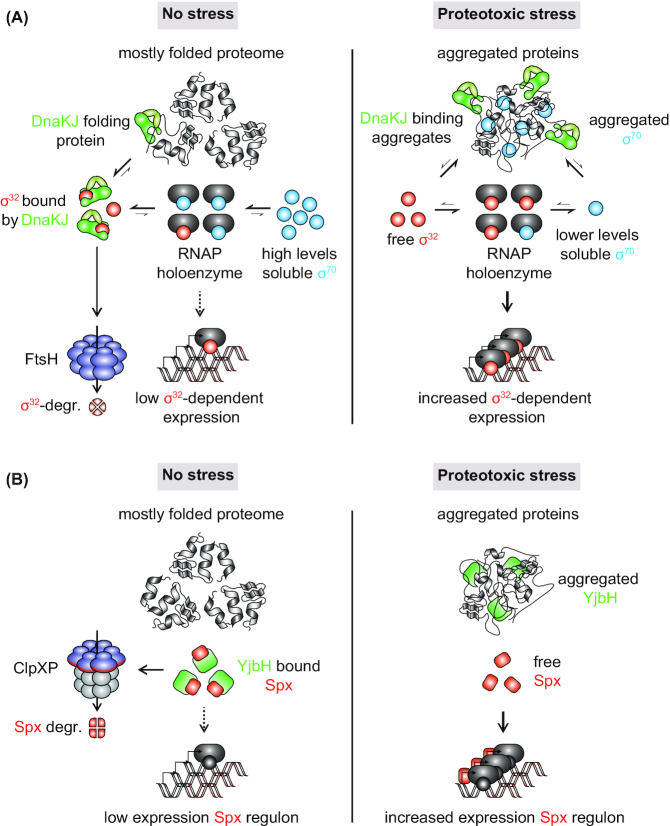
Bacteria utilize a range of different transcriptional activators and repressors for regulating gene expression in response to proteotoxic stress (Narberhaus 1999; Roncarati and Scarlato 2017). In many cases, these regulators are directly linked to sensing the intracellular protein folding status and their activity can either be directly or indirectly regulated through protein un-, misfolding and aggregation events. In E. coli and other proteobacteria the heat shock sigma factor σ32 induces transcriptional changes in response to proteotoxic stress (Reisenauer, Mohr and Shapiro 1996; Arsène, Tomoyasu and Bukau 2000; Permina and Gelfand 2003). In the absence of proteotoxic stress σ32 activity and levels are kept low by DnaKJE and SRP that inactivate and target it for degradation by the protease FtsH (Fig. 6A) (Lim et al. 2013; Schumann 2016; Roncarati and Scarlato 2017). In response to stress most DnaKJE binds to un/misfolded and aggregated proteins, which liberates and stabilizes σ32, which then redirects RNA polymerase to heat shock promoters (Roncarati and Scarlato 2017). σ32 competes with the housekeeping sigma factor σ70 for a common pool of RNA polymerase (RNAP) (Jishage et al. 2002). Interestingly, σ70 is thermally unstable and temperature upshift leads to σ70 unfolding and sequestration into aggregates, which was suggested to further enhance association of σ32 with RNAP and consequently induction of σ32-dependent genes (Fig. 6A) (Blaszczak et al. 1995). Disaggregation and refolding of σ70 after upregulation of the disaggregation machinery could then help in shutting off the heat shock response. In C. crescentus, σ70 was also found to be enriched in insoluble aggregate fractions upon heat stress (Schramm et al. 2019). Furthermore, it was shown that an upregulation of σ70 levels reduces σ32 activity, probably through competition for a common pool of RNAP (Schramm et al. 2017). Competition between σ70 and σ32 has been proposed as an important mechanism to downregulate heat shock response induction during the heat stress recovery phase in C. crescentus (da Silva et al.2003).
Figure 6.
Protein aggregation in stress adaptation. (A), Possible role of σ70 aggregation in the regulation of the E. coli heat shock response. In the absence of stress, free DnaKJ (GrpE not shown) will bind to the heat shock sigma factor σ32 and facilitate its degradation by the membrane-bound protease FtsH (contribution of SRP to σ32 regulation not shown). High levels of the housekeeping sigma factor σ70 could outcompete residual free σ32 for binding to the RNA polymerase (RNAP), further inhibiting inappropriate heat shock response induction. During heat shock, DnaKJ will largely relocalize to aggregating protein and liberate σ32. The thermosensitive σ70 will aggregate, potentially reducing the levels of soluble molecules capable of competing with σ32 and further enhancing heat shock response induction. (B), Protein aggregation in the regulation of the B. subtilis heat and oxidative stress response. In the absence of stress, levels of the heat and oxidative stress master regulator Spx are kept low through YjbH adaptor-mediated degradation by the protease ClpXP. Aggregation induced through heat and oxidative stress induces co-aggregation of YjbH. Liberated Spx can induce the expression of stress-adaptive genes and the repression of proliferative genes.
In many bacteria, heat shock gene regulation also involves repressor proteins that prevent heat shock gene induction in the absence of stress. The proteins CtsR, HrcA and HspR are widespread repressors and bind to inverted or direct repeat DNA motifs in the operons of heat shock genes like dnaKJ, grpE, groESL and clps (recently reviewed by Roncarati and Scarlato 2017). CtsR directly functions as a thermosensor and genes controlled by this repressor are expressed when increases in temperature induce conformational changes that reduce its ability to bind DNA (Elsholz et al. 2010). While HrcA has also been proposed to directly function as a thermosensor at least in some organisms, it is less clear if this is also the case for HspR (Bucca et al. 2000; Hitomi et al. 2003; Roncarati, Danielli and Scarlato 2014; Roncarati and Scarlato 2017). Both HrcA and in some cases HspR activity has been shown to be regulated indirectly through the availability of chaperones. HrcA depends on GroESL both for de novo folding and reactivation after denaturation (Mogk et al. 1997; Reischl, Wiegert and Schumann 2002; Wilson et al. 2005; Hanson and Tan 2015), while HspR was shown to require an association with DnaK to bind DNA and repress its target genes (Bucca et al. 2000, 2003). Increasing folding demands under stress conditions titrate the chaperones away from these repressors, rendering them nonfunctional and leading to the induction of heat shock genes.
The stress-induced aggregation of specific regulatory proteins has also been shown to play a direct role in stress response activation in Bacillus subtilis (Fig. 6B). In this organism the heat shock and oxidative stress response master regulator Spx is subject to regulated proteolysis by ClpXP, which requires the adaptor protein YjbH (Larsson, Rogstam and Wachenfeldt 2007; Garg et al. 2009; Chan, Hahn and Zuber 2014; Awad et al. 2019; Schäfer and Turgay 2019). YjbH is a relatively unstable protein that unfolds and aggregates in response to stresses like diamide, heat or ethanol exposure (Engman and Wachenfeldt 2015). Aggregation of YjbH under such conditions causes the stabilization of Spx, allowing it to repress genes important for proliferation and activate the expression of stress adaptive genes (Fig. 6B) (Nakano et al. 2003; Reyes and Zuber 2008; Rochat et al. 2012; Gaballa et al. 2019; Schäfer et al. 2019). It has been proposed that YjbH aggregation is either driven by the property of YjbH to interact with other un- and misfolded proteins or by improper folding of YjbH due to the reduced availability of chaperones under such conditions (Engman and Wachenfeldt 2015). Once aggregated, YjbH is not returned to its folded cytosolic form but is instead degraded. Shutdown of the Spx-dependent stress response following stress relief takes place when the intracellular environment allows for proper folding of newly synthesized YjbH (Engman and Wachenfeldt 2015).
Sequestration of proteins with growth-promoting functions by phase separation and protein aggregation
In eukaryotes, stress granules are a class of phase separated structures that form under various stresses, including heat shock and usually contain poly(A)-mRNA, translation initiation factors and other RNA-binding proteins (Protter and Parker 2016). Stress granules are insoluble large assemblies that can be dissolved by the same chaperone disaggregation machineries as protein aggregates. Proteins collecting in stress granules were shown to contain specific domains that drive their stress-dependent assembly (Riback et al. 2017). In contrast to protein aggregation that is traditionally seen as a detrimental event, the reversible sequestration of translation preinitiation complexes into stress granules was proposed to serve a stress-adaptive resource reallocation from growth-promoting to cytoprotective function (Cherkasov et al. 2013; Wallace et al. 2015; Riback et al. 2017). Stress granules have not as of yet been described in bacteria, however, ribosomal proteins and translation factors like EF-G and EF-Tu are often part of protein aggregates (Mogk et al. 1999; Tomoyasu et al. 2001; Maisonneuve et al. 2008a; Schramm et al. 2019), and their sequestration in aggregates could constitute a way to globally reduce protein synthesis under proteotoxic stress conditions under which nascent chains are particularly aggregation prone (Hartl and Hayer-Hartl 2009). Consistent with this idea, downregulation of EF-Tu is sufficient to reduce protein aggregation in E. coli mutants lacking DnaK and TF (Bruel et al. 2012).
The aggregation and sequestration of other proteins involved in distinct cellular processes could temporarily arrest processes vulnerable to stress in a similar manner and liberate resources for cytoprotective functions. Recent work also suggests a link between protein aggregation and antibiotic tolerance in E. coli (Fig. 7). It was shown that the presence of protein aggregates formed as a consequence of long-term stationary phase, heat shock, streptomycin or hydrogen peroxide exposure positively correlated with metabolic inactivity and antibiotic tolerance and that metabolic inactivity was linked to the depletion of proteins involved in translation, DNA replication, carbon metabolism and oxidative phosphorylation (Leszczynska et al. 2013; Mordukhova and Pan 2014; Pu et al. 2019). Exit from this metabolically inactive state correlates with ATP-dependent protein disaggregation by the DnaKJE-ClpB machinery (Pu et al. 2019), although it is currently unclear if disaggregation is a prerequisite for or rather a result of growth resumption.
Figure 7.
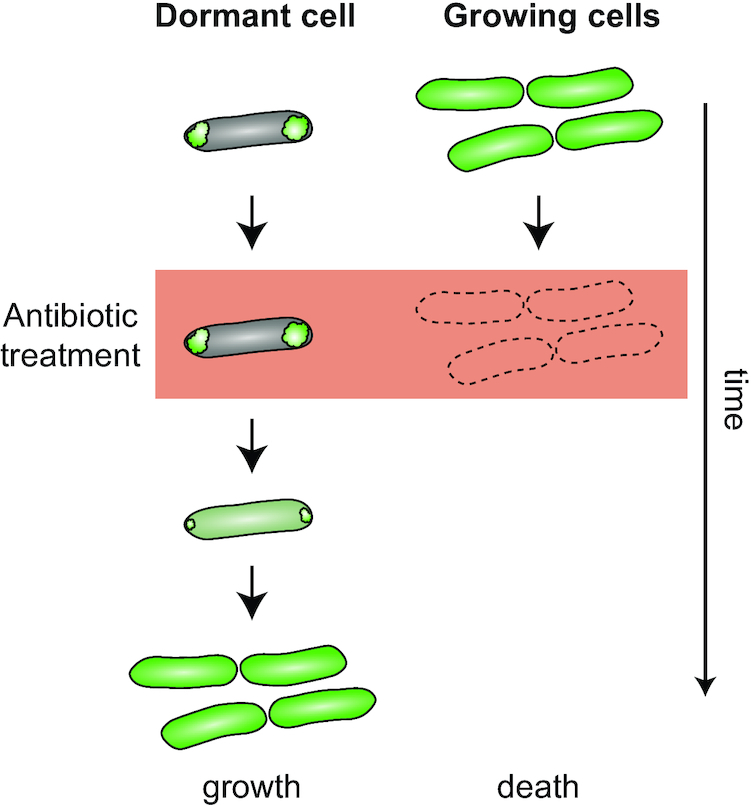
The potential impact of protein aggregation on bacterial antibiotic tolerance. After proteotoxic stress exposure, a population of E. coli cells is heterogeneous in its capability to resume growth. The aggregation of a large fraction of the proteome (green signal localized in foci instead of distributed throughout the cell as in growing cells) can render a cell inactive, or dormant, without causing its death. Dormant cells survive antibiotic treatment that otherwise kills metabolically active and growing cells. The exit from dormancy and resumption of the growth program is correlated with ATP-dependent protein disaggregation.
CONCLUSIONS AND FUTURE PERSPECTIVES
All life requires the maintenance of protein homeostasis, a feat that is particularly challenging when exposed to fluctuating environmental conditions or when interacting with other species. Because of their comparably lower complexity, short generation times and genetic tractability, bacteria continue to be important models for addressing fundamental questions regarding protein homeostasis maintenance as well as to discover biological principles that are valid across the domains of life. The diversity in the composition and utilization of proteostasis networks that we describe in this review holds promise for the identification of new factors and mechanisms bacteria use to cope with protein aggregation, and will shed new light into how different solutions can meet the universal challenge of maintaining proteome integrity. For example, the finding that the well-conserved ClpB disaggregase is absent in many bacteria suggests that alternative ways evolved to deal with protein aggregation. Future research will clarify what role other Hsp100 proteins may play in protein disaggregation in vivo, and how potential triage decisions between refolding and degradation take place mechanistically.
In addition to the strategies bacteria employ to prevent and revert aggregation, the patterns by which persistent protein aggregates are distributed during cell division are also more diverse than previously anticipated. Contrary to the general paradigm of asymmetric aggregate inheritance constructed from studies in yeast and E. coli, in the asymmetrically dividing bacterium C. crescentus most aggregates are not retained in a specific daughter cell type, but are rather distributed to both daughter cells. It will be interesting to study aggregate inheritance in species with unusual morphology and internal structures, such as budding Hyphomonas or filamentous Streptomyces. If and how differences in bacterial protein aggregate inheritance patterns are connected to different adaptive traits is still a topic of ongoing debate, and more research is required to address the consequences connected to aggregate carriage. Continued research in this field is expected to lead to a more complete picture of how bacteria cope with persistent protein aggregates.
While protein aggregation has mostly been seen as a symptom of stress and damage, newer research suggests that it could constitute an important part of the regulation of stress adaptation. Transcriptional regulation of stress responses or the reallocation of cellular resources between proliferative and protective functions through conditional aggregation of specific proteins could help cells to cope with stresses that threaten the proteome. Recent research on eukaryotic protein phase separation in the process of stress granule formation has suggested that protein sequestration into larger assemblies could be an evolved trait of many proteins. Further analysis of the bacterial aggregating proteome and its diversity is required to assess if aggregation propensity could be a selectable trait.
Answering fundamental questions about bacterial protein homeostasis maintenance and comparing the different strategies that are employed by bacteria remains a fascinating topic in cell biology. In addition to important insights into the fundamental aspects of cellular life, a detailed molecular understanding of bacterial protein homeostasis is also of critical importance when combating bacterial infections. It is increasingly recognized that the success and survival of many pathogens is tightly linked to the ability to cope with proteotoxic stresses, and several pathogens boast extended proteostasis networks that increase their resilience.
Contributor Information
Frederic D Schramm, Science for Life Laboratory and Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Svante Arrhenius väg 20C, Stockholm 10691, Sweden.
Kristen Schroeder, Science for Life Laboratory and Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Svante Arrhenius väg 20C, Stockholm 10691, Sweden.
Kristina Jonas, Science for Life Laboratory and Department of Molecular Biosciences, The Wenner-Gren Institute, Stockholm University, Svante Arrhenius väg 20C, Stockholm 10691, Sweden.
FUNDING
Research in the Jonas laboratory is financially supported by the Swedish Research Council (2016-03300), the Swedish Foundation for Strategic Research (FFL15-0005) and funding from the Science for Life Laboratory and Stockholm University.
Conflict of interest. None declared.
REFERENCES
- Acebrón SP, Martín I, del Castillo Uet al.. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009;583:2991–6. [DOI] [PubMed] [Google Scholar]
- Ackermann M, Chao L, Bergstrom CTet al.. On the evolutionary origin of aging. Aging Cell. 2007;6:235–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M, Stephen CS, Jenal U. Senescence in a bacterium with asymmetric division. Science. 2003;300:1920. [DOI] [PubMed] [Google Scholar]
- Aguilera P, Marcoleta A, Lobo-Ruiz Pet al.. Identification of key amino acid residues modulating intracellular and in vitro microcin E492 amyloid formation. Front Microbiol. 2016;7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A, Lakkaraju AKK, Frontzek K. Toward therapy of human prion diseases. Annu Rev Pharmacol Toxicol. 2018;58:331–51. [DOI] [PubMed] [Google Scholar]
- Allan E, Mullany P, Tabaqchali S. Construction and characterization of a Helicobacter pylori clpB mutant and role of the gene in the stress response. J Bacteriol. 1998;180:426–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson FI, Blakytny R, Kirstein Jet al.. Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J Biol Chem. 2006;281:5468–75. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB, Scheraga HA. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. [DOI] [PubMed] [Google Scholar]
- Arsène F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. [DOI] [PubMed] [Google Scholar]
- Awad W, Al-Eryani Y, Ekström Set al.. Structural basis for YjbH adaptor-mediated recognition of transcription factor Spx. Structure. 2019;27:1–14. [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. [DOI] [PubMed] [Google Scholar]
- Bergmiller T, Andersson AMC, Tomasek Ket al.. Biased partitioning of the multidrug efflux pump AcrAB-TolC underlies long-lived phenotypic heterogeneity. Science. 2017;356:311–5. [DOI] [PubMed] [Google Scholar]
- Bittner L-M, Arends J, Narberhaus F. Mini review: ATP-dependent proteases in bacteria. Biopolymers. 2016;105:505–17. [DOI] [PubMed] [Google Scholar]
- Blaszczak A, Zylicz M, Georgopoulos Cet al.. Both ambient temperature and the DnaK chaperone machine modulate the heat shock response in Escherichia coli by regulating the switch between σ70 and σ32 factors assembled with RNA polymerase. EMBO J. 1995;14:5085–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojer MS, Struve C, Ingmer Het al.. Heat resistance mediated by a new plasmid encoded Clp ATPase, ClpK, as a possible novel mechanism for nosocomial persistence of Klebsiella pneumoniae. PLoS One. 2010;5:e15467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll EJ, Marti R, Hasman Het al.. Turn up the heat - Food and clinical Escherichia coli isolates feature two transferrable loci of heat resistance. Front Microbiol. 2017;8:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockwell DJ, Radford SE. Intermediates: ubiquitous species on folding energy landscapes?. Curr Opin Struct Biol. 2007;17:30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruel N, Castanié-Cornet M-P, Cirinesi A-Met al.. Hsp33 controls elongation factor-Tu stability and allows Escherichia coli growth in the absence of the major DnaK and trigger factor chaperones. J Biol Chem. 2012;287:44435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucca G, Brassington AME, Hotchkiss Get al.. Negative feedback regulation of dnaK, clpB and lon expression by the DnaK chaperone machine in Streptomyces coelicolor, identified by transcriptome and in vivo DnaK-depletion analysis. Mol Microbiol. 2003;50:153–66. [DOI] [PubMed] [Google Scholar]
- Bucca G, Brassington AME, Schönfeld H-Jet al.. The HspR regulon of Streptomyces coelicolor: a role for the DnaK chaperone as a transcriptional co-repressor. Mol Microbiol. 2000;38:1093–103. [DOI] [PubMed] [Google Scholar]
- Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. J Biol Chem. 2005;280:23869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanié-Cornet M-P, Bruel N, Genevaux P. Chaperone networking facilitates protein targeting to the bacterial cytoplasmic membrane. Biochim Biophys Acta. 2014;1843:1442–56. [DOI] [PubMed] [Google Scholar]
- Chan CM, Hahn E, Zuber P. Adaptor bypass mutations of Bacillus subtilis spx suggest a mechanism for YjbH-enhanced proteolysis of the regulator Spx by ClpXP. Mol Microbiol. 2014;93:426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Rang CU, Proenca AMet al.. Asymmetrical damage partitioning in bacteria: a model for the evolution of stochasticity, determinism, and genetic assimilation. PloS Comput Biol. 2016;12:e1004700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E, Farr GW, Usaite Ret al.. Global aggregation of newly translated proteins in an Escherichia coli strain deficient of the chaperonin GroEL. Proc Natl Acad Sci USA. 2006;103:15800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JSet al.. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin Set al.. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–62. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. [DOI] [PubMed] [Google Scholar]
- Clegg RJ, Dyson RJ, Kreft J-U. Repair rather than segregation of damage is the optimal unicellular aging strategy. BMC Biol. 2014;12:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Dereli A, Haese Aet al.. Fission yeast does not age under favorable conditions, but does so after stress. Curr Biol. 2013;23:1844–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho M, Lade SJ, Alberti Set al.. Fusion of protein aggregates facilitates asymmetric damage segregation. PLoS Biol. 2014;12:e1001886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel A-S, Jacob J-P, Primet Met al.. Localization of protein aggregation in Escherichia coli is governed by diffusion and nucleoid macromolecular crowding effect. PLoS Comput Biol. 2013;9:e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD, Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl J-U, Gray MJ, Jakob U. Protein quality control under oxidative stress conditions. J Mol Biol. 2015;427:1549–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva ACA, Simão RCG, Susin MFet al.. Downregulation of the heat shock response is independent of DnaK and σ32 levels in Caulobacter crescentus. Mol Microbiol. 2003;49:541–53. [DOI] [PubMed] [Google Scholar]
- de Marco A, Ferrer-Miralles N, Garcia-Fruitós Eet al.. Bacterial inclusion bodies are industrially exploitable amyloids. FEMS Microbiol Rev. 2019;43:53–72. [DOI] [PubMed] [Google Scholar]
- Denoth Lippuner A, Julou T, Barral Y. Budding yeast as a model organism to study the effects of age. FEMS Microbiol Rev. 2014;38:300–25. [DOI] [PubMed] [Google Scholar]
- Deville C, Carroni M, Franke KBet al.. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv. 2017;3:e1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill KA, Ozkan SB, Shell MSet al.. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Reid BG, Horwich ALet al.. ClpS, a substrate modulator of the ClpAP Machine. Mol Cell. 2002;9:673–83. [DOI] [PubMed] [Google Scholar]
- Dukan S, Farewell A, Ballesteros Met al.. Protein oxidation in response to increased transcriptional or translational errors. Proc Natl Acad Sci USA. 2000;97:5746–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran EC, Weaver CL, Lucius AL. Comparative analysis of the structure and function of AAA+ motors ClpA, ClpB, and Hsp104: common threads and disparate functions. Front Mol Biosci. 2017;4:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg DS, Sawaya MR. Structural studies of amyloid proteins at the molecular level. Annu Rev Biochem. 2017;86:69–95. [DOI] [PubMed] [Google Scholar]
- Ekaza E, Teyssier J, Ouahrani-Bettache Set al.. Characterization of Brucella suis clpB and clpAB mutants and participation of the genes in stress responses. J Bacteriol. 2001;183:2677–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz AKW, Michalik S, Zühlke Det al.. CtsR, the Gram-positive master regulator of protein quality control, feels the heat. EMBO J. 2010;29:3621–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engman J, von Wachenfeldt C. Regulated protein aggregation: a mechanism to control the activity of the ClpXP adaptor protein YjbH. Mol Microbiol. 2015;95:51–63. [DOI] [PubMed] [Google Scholar]
- Eriksson M-J, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erjavec N, Larsson L, Grantham Jet al.. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay A, Glickman MS. An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet. 2014;10:e1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea M. Aging and immortality in unicellular species. Mech Ageing Dev. 2017;167:5–15. [DOI] [PubMed] [Google Scholar]
- Frees D, Chastanet A, Qazi Set al.. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–62. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Antelmann H, Hamilton CJet al.. Regulation of Bacillus subtilis bacillithiol biosynthesis operons by Spx. Microbiology. 2019;159:2025–35. [DOI] [PubMed] [Google Scholar]
- Garg SK, Kommineni S, Henslee Let al.. The YjbH protein of Bacillus subtilis enhances ClpXP-catalyzed proteolysis of Spx. J Bacteriol. 2009;191:1268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity SJ, Sivanathan V, Dong Jet al.. Conversion of a yeast prion protein to an infectious form in bacteria. Proc Natl Acad Sci USA. 2010;107:10596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasset-Rosa F, Coquel A-S, Moreno-del Álamo Met al.. Direct assessment in bacteria of prionoid propagation and phenotype selection by Hsp70 chaperone. Mol Microbiol. 2014;91:1070–87. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. [DOI] [PubMed] [Google Scholar]
- Goemans C, Denoncin K, Collet J-F. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta. 2014;1843:1517–28. [DOI] [PubMed] [Google Scholar]
- Goemans CV, Vertommen D, Agrebi Ret al.. CnoX is a chaperedoxin: a holdase that protects its substrates from irreversible oxidation. Mol Cell. 2018;70:614–27. [DOI] [PubMed] [Google Scholar]
- Govers SK, Dutré P, Aertsen A. In vivo disassembly and reassembly of protein aggregates in Escherichia coli. J Bacteriol. 2014;196:2325–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govers SK, Mortier J, Adam Aet al.. Protein aggregates encode epigenetic memory of stressful encounters in individual Escherichia coli cells. PLoS Biol. 2018;16:e2003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WT, Govers SK, Xiang Yet al.. Nucleoid size scaling and intracellular organization of translation across bacteria. Cell. 2019;177:1632–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker Ket al.. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and “aggresomes” during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–30. [DOI] [PubMed] [Google Scholar]
- Gupta A, Lloyd-Price J, Neeli-Venkata Ret al.. In vivo kinetics of segregation and polar retention of MS2-GFP-RNA complexes in Escherichia coli. Biophys J. 2014;106:1928–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA+ protease. Genes Dev. 2008;22:2267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson BR, Tan M. Transcriptional regulation of the Chlamydia heat shock stress response in an intracellular infection. Mol Microbiol. 2015;97:1158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke I, Schäfer H, Janczikowski Aet al.. YocM a small heat shock protein can protect Bacillus subtilis cells during salt stress. Mol Microbiol. 2019;111:423–40. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol. 2009;16:574–81. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Nakazaki Y, Kagii Ket al.. Fusion protein analysis reveals the precise regulation between Hsp70 and Hsp100 during protein disaggregation. Sci Rep. 2017;7:8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M, Bracher A, Hartl FU. The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem Sci. 2016;41:62–76. [DOI] [PubMed] [Google Scholar]
- Hitomi M, Nishimura H, Tsujimoto Yet al.. Identification of a helix-turn-helix motif of Bacillus thermoglucosidasius HrcA essential for binding to the CIRCE element and thermostability of the HrcA-CIRCE complex, indicating a role as a thermosensor. J Bacteriol. 2003;185:381–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M, Kvint K, Shingler Vet al.. Regulation of σ factor competition by the alarmone ppGpp. Genes Dev. 2002;70:1260–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaparast L, Khodaparast L, Gallardo Ret al.. Aggregating sequences that occur in many proteins constitute weak spots of bacterial proteostasis. Nat Commun. 2018;9:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor HK, Fisher MT, Schöneich C. Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO−). J Biol Chem. 2004;279:19486–93. [DOI] [PubMed] [Google Scholar]
- Kirstein J, Strahl H, Molière Net al.. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol. 2008;70:682–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock H, Gerth U, Hecker M. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J Bacteriol. 2004;186:5856–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete Bet al.. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski Jet al.. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger E, Witt E, Ohlmeier Set al.. The Clp proteases of Bacillus subtilis are directly involved in degradation of misfolded proteins. J Bacteriol. 2000;182:3259–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Sourjik V. Physical map and dynamics of the chaperone network in Escherichia coli. Mol Microbiol. 2012;84:736–47. [DOI] [PubMed] [Google Scholar]
- Kwon H-Y, Kim S-W, Choi M-Het al.. Effect of heat shock and mutations in ClpL and ClpP on virulence gene expression in Streptococcus pneumoniae. Infect Immun. 2003;71:3757–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBreck CJ, May S, Viola MGet al.. The protein chaperone ClpX targets native and non-native aggregated substrates for remodeling, disassembly, and degradation with ClpP. Front Mol Biosci. 2017;4:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf D, Okumus B, Chien Pet al.. Segregation of molecules at cell division reveals native protein localization. Nat Methods. 2012;9:480–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson JT, Rogstam A, von Wachenfeldt C. YjbH is a novel negative effector of the disulphide stress regulator, Spx, in Bacillus subtilis. Mol Microbiol. 2007;66:669–84. [DOI] [PubMed] [Google Scholar]
- Laskowska E, Wawrzynów A, Taylor A. IbpA and IbpB, the new heat-shock proteins, bind to endogenous Escherichia coli proteins aggregated intracellularly by heat shock. Biochimie. 1996;78:117–22. [DOI] [PubMed] [Google Scholar]
- Lee C, Franke KB, Mansour Set al.. Stand-alone ClpG disaggregase confers superior heat tolerance to bacteria. Proc Natl Acad Sci USA. 2017;115:E273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Kim H, Bardwell JCA. Electrostatic interactions are important for chaperone-client interaction in vivo. Microbiology. 2018;164:992–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wigren E, Lünsdorf Het al.. Protein homeostasis - more than resisting a hot bath. Curr Opin Microbiol. 2016;30:147–54. [DOI] [PubMed] [Google Scholar]
- Leszczynska D, Matuszewska E, Kuczynska-Wisnik Det al.. The formation of persister cells in stationary-phase cultures of Escherichia coli is associated with the aggregation of endogenous proteins. PLoS One. 2013;8:e54737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeThanh H, Neubauer P, Hoffmann F. The small heat-shock proteins IbpA and IbpB reduce the stress load of recombinant Escherichia coli and delay degradation of inclusion bodies. Microb Cell Fact. 2005;4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Miyazaki R, Neher Set al.. Heat shock transcription factor σ32 co-opts the signal recognition particle to regulate protein homeostasis in E. coli. PLoS Biol. 2013;11:e1001735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AB, Madden R, Demarez Aet al.. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci USA. 2008;105:3076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Cho C, Guo L-Tet al.. Protein aggregation caused by aminoglycoside action is prevented by a hydrogen peroxide scavenger. Mol Cell. 2012;48:713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero Aet al.. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–67. [DOI] [PubMed] [Google Scholar]
- Maier R, Scholz C, Schmid FX. Dynamic association of trigger factor with protein substrates. J Mol Biol. 2001;314:1181–90. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Fraysse L, Lignon Set al.. Carbonylated proteins are detectable only in a degradation-resistant aggregate state in Escherichia coli. J Bacteriol. 2008a;190:6609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E, Fraysse L, Moinier Det al.. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J Bacteriol. 2008b;190:887–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabirade A, Partouche D, El Hamoui Oet al.. Revised role for Hfq bacterial regulator on DNA topology. Sci Rep. 2018;8:16792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcoleta A, Wien F, Arluison Vet al.. Bacterial amyloids. eLS. 2019, 10.1002/9780470015902.a0028401. [DOI] [Google Scholar]
- Mayer MP, Gierasch LM. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J Biol Chem. 2019;294:2085–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38:507–14. [DOI] [PubMed] [Google Scholar]
- Melkani GC, McNamara C, Zardeneta Get al.. Hydrogen peroxide induces the dissociation of GroEL into monomers that can facilitate the reactivation of oxidatively inactivated rhodanese. Int J Biochem Cell Biol. 2004;36:505–18. [DOI] [PubMed] [Google Scholar]
- Miethke M, Hecker M, Gerth U. Involvement of Bacillus subtilis ClpE in CtsR degradation and protein quality control. J Bacteriol. 2006;188:4610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moger-Reischer RZ, Lennon JT. Microbial ageing and longevity. Nat Rev Microbiol. 2019, DOI:10.1038/s41579-019-0253-y. [DOI] [PubMed] [Google Scholar]
- Mogk A, Bukau B, Kampinga HH. Cellular handling of protein aggregates by disaggregation machines. Mol Cell. 2018;69:214–26. [DOI] [PubMed] [Google Scholar]
- Mogk A, Deuerling E, Vorderwülbecke Set al.. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–95. [DOI] [PubMed] [Google Scholar]
- Mogk A, Homuth G, Scholz Cet al.. The GroE chaperonin machine is the major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Ruger-Herreros C, Bukau B. Cellular functions and mechanisms of action of small heat shock proteins. Annu Rev Microbiol. 2019;73:4.1–4.22. [DOI] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff Pet al.. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-García L, Gasset-Rosa F, Moreno-del Álamo Met al.. Functional amyloids as inhibitors of plasmid DNA replication. Sci Rep. 2016;6:25425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-García L, Moreno-del Álamo M, Botias Pet al.. Outlining core pathways of amyloid toxicity in bacteria with the RepA-WH1 prionoid. Front Microbiol. 2017;8:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordukhova EA, Pan J-G. Stabilization of homoserine-o-succinyltransferase (MetA) decreases the frequency of persisters in Escherichia coli under stressful conditions. PLoS One. 2014;9:e110504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morell M, Bravo R, Espargaró Aet al.. Inclusion bodies: Specificity in their aggregation process and amyloid-like structure. Biochim Biophys Acta. 2008;1783:1815–25. [DOI] [PubMed] [Google Scholar]
- Nakano S, Nakano MM, Zhang Yet al.. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc Natl Acad Sci U S A. 2003;100:4233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaoka H, Wakamoto Y. Aging, mortality, and the fast growth trade-off of Schizosaccharomyces pombe. PLoS Biol. 2017;15:e2001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narberhaus F. Negative regulation of bacterial heat shock genes. Mol Microbiol. 1999;31:1–8. [DOI] [PubMed] [Google Scholar]
- Neeli-Venkata R, Martikainen A, Gupta Aet al.. Robustness of the process of nucleoid exclusion of protein aggregates in Escherichia coli. J Bacteriol. 2016;198:898–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Kirstein J, Szlachcic Aet al.. Crucial Hsp70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;542:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda NB, Wentink AS, Bukau B. Protein disaggregation in multicellular organisms. Trends Biochem Sci. 2018;43:285–300. [DOI] [PubMed] [Google Scholar]
- Nyström T, Liu B. Protein quality control in time and space - links to cellular aging. FEMS Yeast Res. 2014;14:40–8. [DOI] [PubMed] [Google Scholar]
- Oliveira SMD, Neeli-Venkata R, Goncalves NSMet al.. Increased cytoplasm viscosity hampers aggregate polar segregation in Escherichia coli. Mol Microbiol. 2016;99:686–99. [DOI] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner ACet al.. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. [DOI] [PubMed] [Google Scholar]
- Park S-S, Kwon H, Tran TD-Het al.. ClpL is a chaperone without auxiliary factors. FEBS J. 2015;282:1352–67. [DOI] [PubMed] [Google Scholar]
- Patel A, Malinovska L, Saha Set al.. ATP as a biological hydrotrope. Science. 2017;356:753–6. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–94. [DOI] [PubMed] [Google Scholar]
- Permina EA, Gelfand MS. Heat shock (σ32 and HrcA/CIRCE) regulons in β-, γ- and ε-proteobacteria. J Mol Microbiol Biotechnol. 2003;6:174–81. [DOI] [PubMed] [Google Scholar]
- Powers ET, Balch WE. Diversity in the origins of proteostasis networks - a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin Aet al.. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–91. [DOI] [PubMed] [Google Scholar]
- Proenca AM, Rang CU, Buetz Cet al.. Age structure landscapes emerge from the equilibrium between aging and rejuvenation in bacterial populations. Nat Commun. 2018;9:3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proenca AM, Rang CU, Qiu Aet al.. Cell aging preserves cellular immortality in the presence of lethal levels of damage. PLoS Biol. 2019;17:e3000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Li Y, Jin Xet al.. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol Cell. 2019;73:143–56. [DOI] [PubMed] [Google Scholar]
- Rambaron RN, Serpell LC. Amyloid fibrils: abnormal protein assembly. Prion. 2008;2:112–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelt H, Kirstein-Miles J, Nillegoda NBet al.. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31:4221–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rang CU, Proenca A, Buetz Cet al.. Minicells as a damage disposal mechanism in Escherichia coli. mSphere. 2018;3:e00428–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischl S, Wiegert T, Schumann W. Isolation and analysis of mutant alleles of the Bacillus subtilis HrcA repressor with reduced dependency on GroE function. J Biol Chem. 2002;277:32659–67. [DOI] [PubMed] [Google Scholar]
- Reisenauer A, Mohr CD, Shapiro L. Regulation of a heat shock σ32 homolog in Caulobacter crescentus. J Bacteriol. 1996;178:1919–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes DY, Zuber P. Activation of transcription initiation by Spx: formation of transcription complex and identification of a Cis-acting element required for transcriptional activation. Mol Microbiol. 2008;69:765–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear-Scott JLet al.. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–66. [DOI] [PubMed] [Google Scholar]
- Rizo AN, Lin J, Gates SNet al.. Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase. Nat Commun. 2019;10:2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Nicolas P, Delumeau Oet al.. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucleic Acids Res. 2012;40:9571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncarati D, Danielli A, Scarlato V. The HrcA repressor is the thermosensor of the heat-shock regulatory circuit in the human pathogen Helicobacter pylori. Mol Microbiol. 2014;92:910–20. [DOI] [PubMed] [Google Scholar]
- Roncarati D, Scarlato V. Regulation of heat-shock genes in bacteria: from signal sensing to gene expression output. FEMS Microbiol Rev. 2017;41:549–74. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10:S10–7. [DOI] [PubMed] [Google Scholar]
- Runde S, Molière N, Heinz Aet al.. The role of thiol oxidative stress response in heat-induced protein aggregate formation during thermotolerance in Bacillus subtilis. Mol Microbiol. 2014;91:1036–52. [DOI] [PubMed] [Google Scholar]
- Santra M, Dill KA, De Graff AMRet al.. How do chaperones protect a cell's proteins from oxidative damage?. Cell Syst. 2018;6:743–51. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. [DOI] [PubMed] [Google Scholar]
- Schlothauer T, Mogk A, Dougan DAet al.. MecA, an adaptor protein necessary for ClpC chaperone actvity. Proc Natl Acad Sci USA. 2003;100:2306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm FD, Heinrich K, Thüring Met al.. An essential regulatory function of the DnaK chaperone dictates the decision between proliferation and maintenance in Caulobacter crescentus. PLoS Genet. 2017;13:e1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm FD, Schroeder K, Alvelid Jet al.. Growth-driven displacement of protein aggregates along the cell length ensures partitioning to both daughter cells in Caulobacter crescentus. Mol Microbiol. 2019;111:1430–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann W. Regulation of bacterial heat shock stimulons. Cell Stress Chaperones. 2016;21:959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer H, Heinz A, Sudzinová Pet al.. Spx, the central regulator of the heat and oxidative stress response in B. subtilis, can repress transcription of translation-related genes. Mol Microbiol. 2019;111:514–33. [DOI] [PubMed] [Google Scholar]
- Schäfer H, Turgay K. Spx, a versatile regulator of the Bacillus subtilis stress response. Curr Genet. 2019;65:871–6. [DOI] [PubMed] [Google Scholar]
- Scutigliani EM, Scholl ER, Grootemaat AEet al.. Interfering with DNA decondensation as a strategy against mycobacteria. Front Microbiol. 2018;9:2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Yang Q, Yang Jet al.. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet. 2014;10:e1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spokoini R, Moldavski O, Nahmias Yet al.. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2012;2:738–47. [DOI] [PubMed] [Google Scholar]
- Stewart EJ, Madden R, Paul Get al.. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull F, Hipp H, Stockbridge RBet al.. In vivo chloride concentrations surge to proteotoxic levels during acid stres. Nat Chem Biol. 2018;14:1051–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suokko A, Poutanen M, Savijoki Ket al.. ClpL is essential for induction of thermotolerance and is potentially part of the HrcA regulon in Lactobacillus gasseri. Proteomics. 2008;8:1029–41. [DOI] [PubMed] [Google Scholar]
- Tamás MJ, Fauvet B, Christen Pet al.. Misfolding and aggregation of nascent proteins: a novel mode of toxic cadmium action in vivo. Curr Genet. 2018;64:177–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Schwarz M, Mogk Aet al.. The Yeast AAA+ chaperone Hsp104 is part of a network that links the actin cytoskeleton with the inheritance of damaged proteins. Mol Cell Biol. 2009;29:3738–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen Het al.. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol. 2001;40:397–413. [DOI] [PubMed] [Google Scholar]
- Vaubourgeix J, Lin G, Dhar Net al.. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17:178–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel S, Nunns H, Košmrlj Aet al.. Asymmetric damage segregation constitutes an emergent population-level stress response. Cell Syst. 2016;3:187–98. [DOI] [PubMed] [Google Scholar]
- Wallace EWJ, Kear-Scott JL, Pilipenko EV. et al.. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol Microbiol. 1997;26:897–910. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker Cet al.. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the cetral pore of ClpB. Cell. 2004;119:653–65. [DOI] [PubMed] [Google Scholar]
- Weids AJ, Ibstedt S, Tamás MJet al.. Distinct stress conditions result in aggregation of proteins with similar properties. Sci Rep. 2016;6:24554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Wu CC, Yates JR IIIet al.. Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J Bacteriol. 2005;187:7535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Seybert A, König Let al.. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek Aet al.. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–92. [DOI] [PubMed] [Google Scholar]
- Wong P, Houry WA. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J Struct Biol. 2004;146:79–89. [DOI] [PubMed] [Google Scholar]
- Yuan AH, Garrity SJ, Nako Eet al.. Prion propagation can occur in a prokaryote and requires the ClpB chaperone. Elife. 2014;3:e02949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan AH, Hochschild A. A bacterial global regulator forms a prion. Science. 2017;201:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JRet al.. Motility and segregation of Hsp104-associated protein aggregates in budding yeast. Cell. 2011;147:1186–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JRet al.. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H-X. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziȩtkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279:44376–83. [DOI] [PubMed] [Google Scholar]