Abstract
A trichloroethene (TCE)-dechlorinating community (CANAS) maintained in a completely mixed flow reactor was established from a semi-batch enrichment culture (ANAS) and was monitored for 400 days at a low solids retention time (SRT) under electron acceptor limitation. Around 85% of TCE supplied to CANAS (0.13 mmol d−1) was converted to ethene at a rate of 0.1 mmol d−1, with detection of low production rates of vinyl chloride (6.8 × 10−3 mmol d−1) and cis- dichloroethene (2.3 × 10−3 mmol d−1). Two distinct Dehalococcoides mccartyi strains (ANAS1 and ANAS2) were stably maintained at 6.2 ± 2.8 × 108 cells mL−1 and 5.8 ± 1.2 × 108 cells mL−1, respectively. Electron balance analysis showed 107% electron recovery, in which 6.1% were involved in dechlorination. 16S rRNA amplicon sequencing revealed a structural regime shift between ANAS and CANAS while maintaining robust TCE dechlorination due to similar relative abundances of D. mccartyi and functional redundancy among each functional guild supporting D. mccartyi activity. D. mccartyi transcriptomic analysis identified the genes encoding for ribosomal RNA and the reductive dehalogenases tceA and vcrA as the most expressed genes in CANAS, while hup and vhu were the most critical hydrogenases utilized by D. mccartyi in the community.
Keywords: CMFR, Dechlorination, Community structure, Transcriptomic analysis, Dehalococcoides, Functional robustness
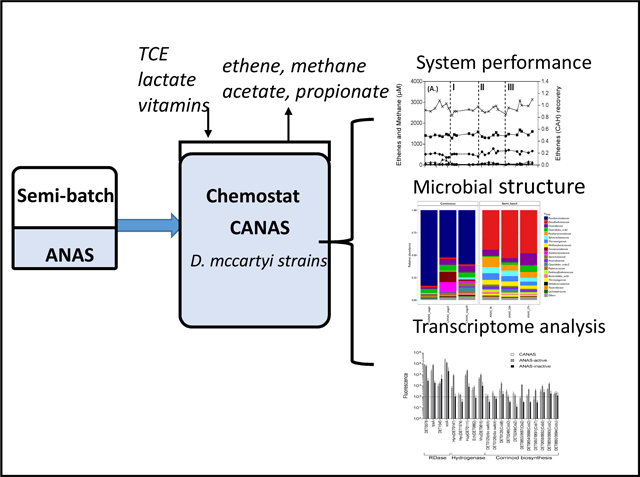
Graphical Abstract
1. Introduction
Perchloroethene (PCE) and trichloroethene (TCE) are common solvents detected in soils and groundwater due to historical improper storage, handling and disposal practices and accidental releases during industrial uses, such as dry cleaning and degreasing operations (Moran et al. 2007, McCarty 2010). In situ bioremediation is a cost-effective and environmentally-friendly approach to reach eco-toxicological safety endpoints at chlorinated ethene-contaminated sites (Stroo et al. 2010). Among all reported dehalogenating microorganisms (Maymo-Gatell et al. 1997, Holliger et al. 1998, Löffler et al. 2000, Adrian et al. 2000, Suyama et al. 2001, Sung et al. 2006, Hug et al. 2013, Dolinova et al. 2017), Dehalococcoides mccartyi is the only known species that metabolically reductively dechlorinates PCE and TCE to non-toxic ethene. Many studies on D. mccartyi-containing microbial communities have been carried out in the past decades (Carr et al. 2000, Cupples et al. 2004, Freeborn et al. 2005, Yu et al. 2005, Daprato et al. 2007, Duhamel et al. 2007, Ziv-El et al. 2012), with the identification of optimal growth conditions supporting D. mccartyi and functional robustness determinants of dechlorinating activity. However, the microbial community assembly processes and inter-species interaction networks that promote successful bioremediation are difficult to predict within the flow-dominated physico-chemical conditions occurring in the subsurface (Kinnunen et al. 2016).
Dechlorinating microbial communities have mainly been studied in batch reactors characterized by high initial concentrations of substrates, nutrients and electron acceptors and dynamic conditions as dechlorination proceeds. The knowledge acquired from such feast-and-famine laboratory incubations has been extremely useful for understanding the dynamics of dechlorinating communities but is difficult to extrapolate to flowing groundwater plumes which are continuous, oligotrophic fluidic ecosystems with site-specific hydraulic retention times and relatively constant and low concentrations of contaminants, substrates and nutrients (Stroo et al. 2010). Continuous-flow bioreactors generate conditions that are more environmentally and ecologically relevant to groundwater plumes than batch systems. In addition, continuous culture systems can be highly reproducible platforms for dynamic perturbation under rigorously defined and controlled conditions relevant to systems-level omics and electron flow studies.
Structural and functional dynamics of dechlorinating communities have been investigated in continuous-flow biofilm and packed column systems (Carr et al. 1998, Azizian et al. 2008, Chung et al. 2008, Maphosa et al. 2010, Sabalowsky et al. 2010, Azizian et al. 2016, Mirza et al. 2016, Mirza et al. 2017, Delgado et al. 2017). Continuous-flow suspended growth systems have also been used to study the impacts of solids retention times (SRTs) and other critical parameters on dechlorination activity of synthetic consortia and more complex communities (Gerritse et al. 1997, Carr et al. 2000, Drzyzga et al. 2001, Zheng et al. 2001, Sabalowsky et al. 2010). Yang and McCarty (1998) also reported stable production of ethene by a methanogenic PCE-dechlorinating enrichment culture maintained in a semi-continuous stirred-tank reactor fed with PCE and benzoate. These studies provided important information on aggregate properties of the system (e.g., functional endpoints, total biomass concentrations, process rates and optimization). What has yet to be reported are quantitative assessments of the dynamics of the dechlorinating populations (e.g., microdiversity and physiological responses of D. mccartyi) and functional guilds supporting them. Here, a functional guild is defined as a group of taxonomically-distinct microorganisms performing the same function (e.g., syntrophic, fermentative bacteria that catalyze the oxidation of lactate) (Wittebolle et al. 2009). Functional redundancy among a functional guild involves the presence of multiple functionally-equivalent taxa that enables functional robustness through compensation by metabolic equivalency and response asynchrony.
In addition, several studies have reported successful and sustainable PCE/TCE dechlorination to ethene in completely mixed flow reactors (CMFRs, i.e. chemostat) with associated population dynamics (Table1) (Berggren et al. 2013, Delgado et al. 2014, Marshall et al. 2014, Mayer-Blackwell et al. 2016, Mayer-Blackwell et al. 2017).
In this study, a TCE-dechlorinating community (CANAS) sustained in a CMFR was established from a long-term dechlorinating enrichment culture (ANAS) which has been functionally maintained in a semi-batch reactor for more than 15 years (Richardson et al. 2002, Lee et al. 2006). The microbial community structure of ANAS and its responses to different environmental conditions have been well characterized (Freeborn et al. 2005, Lee et al. 2011, West et al. 2013). The main objective of this work is to study a continuous-flow mixed culture (CANAS) in the presence of sustained selective pressure (i.e., constant supply of chlorinated ethenes) and low concentrations of electron donor, as frequently observed in contaminated oligotrophic groundwater plumes (Stroo et al. 2010). 16S rRNA gene amplicon sequencing was carried out to assess the effects on the microbial community structure induced by kinetic mode changes, from feast-and-famine conditions (ANAS) to the continuous feeding of low nutrient concentrations (CANAS). A D. mccartyi genus-wide microarray was applied to study the transcriptional dynamics of D. mccartyi populations present in CANAS.
2. Materials and Methods
Inoculum Culture
A 3.0 L CMFR was inoculated with dechlorinating enrichment culture ANAS at 5% v/v. ANAS was originally enriched from contaminated soil obtained from Alameda Naval Air Station (CA) and has been functionally stable for more than 15 years in a continuously stirred semi-batch reactor. Every two weeks, 25 mM (or 2.8 g L−1) lactate was supplied as both electron donor and carbon source and 0.1 mM (or 13 μg L−1) TCE was supplied as the terminal electron acceptor. The growth and maintenance procedures of ANAS have been previously described (Lee et al. 2011, West et al. 2013). Two distinct D. mccartyi strains were identified and isolated from ANAS (strain ANAS1 which contains the TCE reductive dehalogenase gene tceA, and strain ANAS2 which contains the VC reductive dehalogenase gene vcrA) (Lee et al. 2011). ANAS has been extensively studied (Richardson et al. 2002, Lee et al. 2006, West et al. 2008, West et al. 2013) and contains 1,056 bacterial and archaeal taxa on the basis of DNA PhyloChip analysis (Brisson et al. 2012).
CMFR operation and sampling
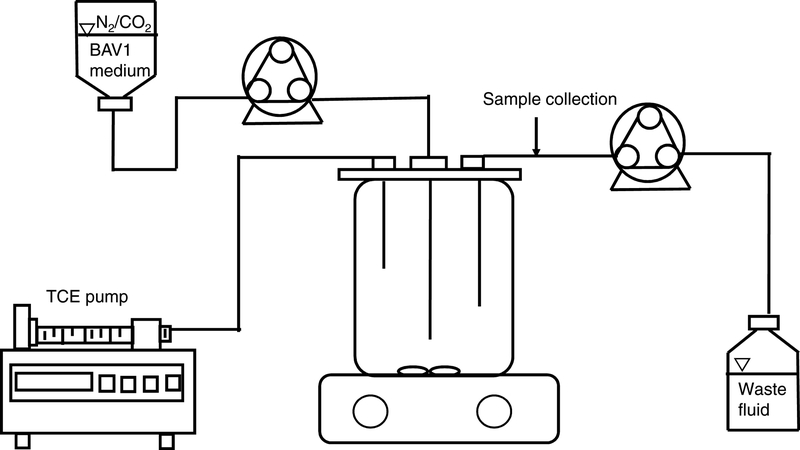
The CANAS culture was maintained in the 3.0 L CMFR with no headspace represented in Figure 1 (New Brunswick Eppendorf, Enfield, CT) fitted with Teflon caps and viton tubing (Masterflex, Cole-Parmer Instrument Co., Vernon Hills, IL) (Supporting material, Fig. S1). Pure TCE was stored in a 1.0 mL gastight syringe with a PTFE plunger tip (Hamilton, Reno, NV) and was continuously fed into the glass reactor using a syringe pump (NE-300 programmable syringe pump, New Era Pump Systems, Farmingdale, NY) at a flow rate of 12 μL d−1 (0.133 mmol d−1). The composition of the influent (flow rate = 75 mL d−1) was modified from the defined medium used for the growth of D. mccartyi strain 195 (He et al. 2007) by including 10 mM lactate as the electron donor/carbon source and by increasing the concentration of L-cysteine and sodium sulfide (Na2S) reducing agent to 0.4 mM of each to ensure anoxic conditions inside the influent tubing and reactor (Supplemental material, Table S1)(Johnson et al. 2008). Lactate was supplied to the reactor at a rate of 750 μmol d−1, i.e., 3 meeq d−1 taking into account 4 eeq per mole of lactate on the basis of H2 production (Supporting material, equation S1) which is in excess of electron equivalents required for dechlorination of TCE to ethene (133 μmol d−1, i.e., 0.8 meeq d−1).
Figure 1.
Diagram of the CMFR system used for cultivation of CANAS.
The liquid flow rate of 0.075 L d−1 resulted in a solids retention time (SRT) of 40 days (dilution rate of 0.025 d−1), theoretically equal to the hydraulic retention time in the CMFR. Influent pH was adjusted to 7.0–7.3 using 30 mM sodium bicarbonate and 10 mM TES buffer. The CMFR was continuously stirred with a magnetic stir bar (~10 rpm) at room temperature (23 ± 2 °C) to ensure rapid dissolution of TCE and to minimize mass-transfer limitations (detailed information of operation is summarized in Supporting material, Table S2 and Fig. S2). 1.5 mL culture samples were collected frequently throughout the experimental period for quantification of metabolites and cell numbers. Cell pellets were harvested after centrifugation at 14,000 rpm (4 °C) for 10 min. After the CMFR maintained steady-state for five SRTs (i.e., stable levels of chlorinated ethenes, ethene, methane, hydrogen, lactate, volatile fatty acids, and specific biomarkers for 200 days), duplicate culture samples were collected during three consecutive SRTs (at stages I, II, III) to generate biological triplicates for subsequent DNA/RNA studies. Briefly, 100 mL culture samples were collected in duplicate at the sample port and filtered on a 0.2 μm autoclaved GVWP filter (Durapore membrane, Millipore, Billerica, MA). Each filter was immediately placed in a 2 mL attached screw cap micro-centrifuge tube (Corning, Lowell, MA), frozen with liquid nitrogen and stored at −80 °C until further processing. The CMFR design, configuration and maintenance strategies, as well as routine sample collection and preparation procedures, are summarized in the supporting material.
Chemical and molecular methods
Chloroethenes, ethene and methane were measured by injecting 100 μL headspace sample to a FID-gas chromatograph (Agilent Technologies, Santa Clara, CA), and 300 μL headspace was injected into a RGD-gas chromatograph (Trace Analytical, Menlo Park, CA) for hydrogen quantification as previously described (Freeborn et al. 2005, Lee et al. 2006). Organic acids, including lactate, acetate and propionate were analyzed in 2 mL liquid samples by HPLC equipped with a photodiode array detector set at 210 nm (Waters 2996 photodiode array detector, Milford, MA) as previously described (Freeborn et al. 2005).
Quantification of Bacteria, Archaea and specific microbial populations or functional groups present in CANAS was performed at regular interval times using real-time quantitative PCR (qPCR) targeting tceA and vcrA genes and different regions of 16S rRNA genes as previously reported (Men et al. 2012, West et al. 2013) with primers summarized in Table S3. Cell lysis was carried out by mechanical and chemical disruption and nucleic acids were purified using a previously described phenol extraction (West et al. 2008). DNA and RNA were separated using AllPrep DNA/RNA Mini Kit (Qiagen, Valencia, CA) according to the manufacturer’s instruction. RNA was eluted from the spin column with 100 μL of nuclease-free water and was further purified by using Turbo DNA-free kit (Ambion, Austin, TX) to remove additional DNA contamination according to the manufacturer’s instructions. The quality of RNA samples was checked by electrophoresis, and the concentration of RNA samples was quantified using a nanophotometer P-300 (Implen, Westlake Village, CA, USA). The ratio of A260/A280 for all samples was between 1.80 and 2.0. Purified RNA was stored in nuclease-free water at −80 °C prior to further analysis.
Microbial community structure analysis using targeted sequencing of 16S rRNA genes
Sequencing of the V4 hypervariable region amplicons was performed on a HiSeq 2000 in a 2 × 150 bp configuration. Due to quality issues, single reads instead of assembled read pairs were kept for downstream analyses. Sequences were analyzed through our internal rRNA short amplicon analysis pipeline as previously described (Tremblay et al. 2015, Yergeau et al. 2015). Briefly, reads were quality-filtered and quality-trimmed, and clustered at 97% similarity. Resulting OTUs were assigned to a taxonomic lineage using the RDP classifier with a Greengenes (v13_5) training set. OTU tables were generated, filtered to exclude eukaryotes and chloroplasts and normalized using the trimmed mean of M-values (TMM) method in the edgeR package (Robinson et al. 2010, McMurdie et al. 2014). TMM-normalized OTU tables were used for assessing relative abundances of OTUs for every taxonomic level and computing beta-diversity metrics. Repeated rarefactions at 5,000 reads (performing 10 iterations at each sampling depth) were generated from the raw (un-normalized) OTU table to generate alpha diversity metrics. Taxonomic and phylogenetic alpha and beta diversity metrics were computed using Qiime and downstream analyses were done with in-house Perl and R scripts. OTU tables and further details on 16S amplicon sequencing are included in supplemental material.
Nucleotide sequence accession numbers
Raw sequence reads of the 16S rRNA gene amplicon data for this project are available through the NCBI Sequence Read Archive (SRA) (study accession: SRP090533; Bioproject PRJNA309107).
Transcriptomic analysis
The samples selected for transcriptomic analysis were 1) ANAS-0h (before a new feeding cycle when the culture was in starvation condition; 2) ANAS-20h (the culture was metabolically active after 20 hours of feeding, all dechlorination metabolites were present); 3) CANAS (biological triplicate samples were collected from stage I, II and III at steady state) (Figure 2). Microarray sample preparation and application procedures were previously described (West et al. 2013). Briefly, 1 μg of community genomic DNA (gDNA) was used for DNA analysis and 10 μg of community total RNA was used as starting material for RNA analysis as previously described (West et al. 2008). All probe sets that were detected as “present” by DNA microarray analysis were used in the RNA analysis. DNA for any particular ORF was deemed “present” if each replicate probe set for that ORF had signal intensity greater than 140 and a P value less than 0.05 in the DNA chips. RNA for any particular ORF was considered “present” if the average signal intensity of the probe sets for that ORF was greater than 120. The criteria applied in this study are the same as previously described for the ANAS transcriptomic study (West et al. 2013).
Figure 2.
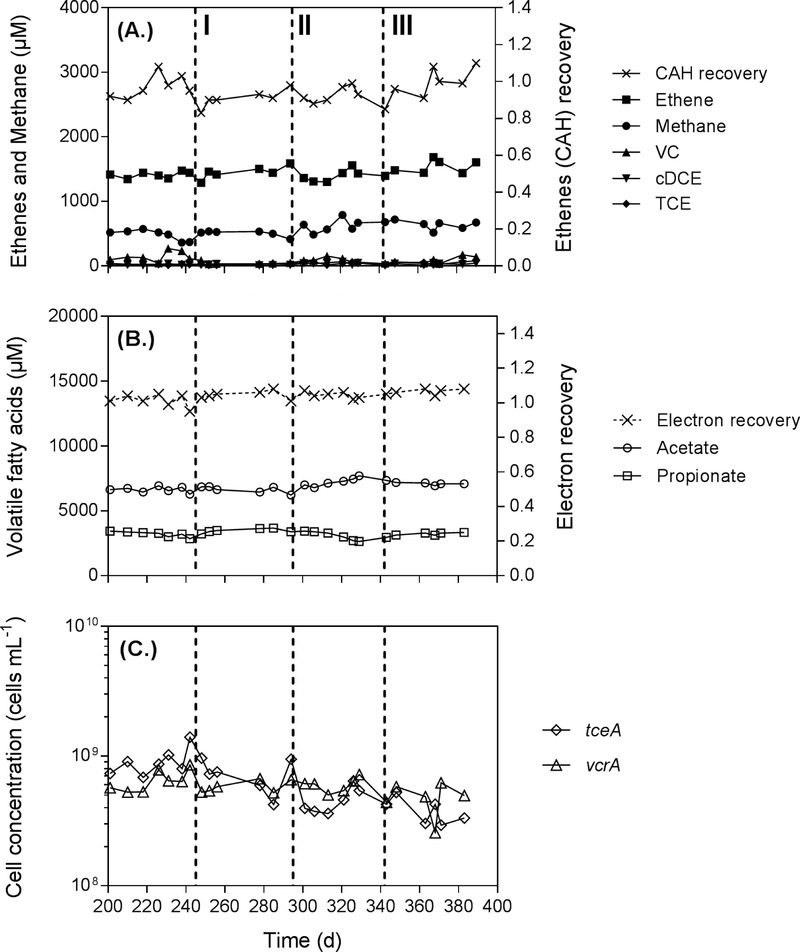
Time course of (A.) TCE dechlorination, production of TCE metabolites and methane concentrations measured in the effluent samples and chlorinated aliphatic hydrocarbons (CAHs) recovery (×100%); (B.) Volatile fatty acid concentrations (lactate was below the detection limit) and electron balance (see Table 2); and (C.) D. mccartyi strains ANAS1 (tce A) and ANAS2 (vcr A) cell concentrations at steady-state in CANAS during the monitored experimental period (stages I, II and III).
3. Results and Discussion
Steady state performance of the community
The 40-day SRT selected for this study was lower than that of ANAS (Table S1) and was based on two objectives: To avoid washing out the slow growing microorganisms (e.g., syntrophs) and to maintain a sufficient steady-state volumetric biomass productivity (cells L−1 d−1) for molecular analyses. The basic maintenance parameters for CANAS are compared with previous CMFR PCE/TCE dechlorination to ethene studies in Table 1. The donor to acceptor ratio in the influent supplied to CANAS was about 19:1 (based on 12 equivalents of available electrons per mole of lactate and 6 eeq per mole of TCE reduced), ensuring TCE to be the limiting reactant for dechlorination (as observed in TCE-contaminated plumes) with the electron donor concentration in excess. Samples were collected at steady-state for both microbial community structure and transcriptomic analyses during the experimental period from day 201 to day 400 (Figure 2). During this time, around 92 % of TCE in the influent (1.8 mM (0.13 mmol TCE d−1 / 0.075 L d−1 influent)) was converted to 1.5 ± 0.10 mM ethene, 0.09 ± 0.06 mM vinyl chloride (VC) and 0.03 ± 0.01 mM cis-DCE (Figure 2A) based on effluent sample measurement. The chlorinated solvent recovery rate ranged from 83 to 108% during the experimental period (Figure 2A). The dechlorination rate of the reactor was 120 μmol Cl− L−1 d−1. Methane concentration in the reactor was maintained at a low level of 0.56 ± 0.10 mM (with a production rate of 14 ± 2.5 μmol L−1 d−1).
ANAS, the inoculum of CANAS, consistently exhibited faster growth of D. mccartyi strain ANAS1 (with one copy of tceA) than ANAS2 (with one copy of vcrA), resulting in a ANAS1:ANAS2 ratio of 4:1 (Lee et al. 2011). In CANAS, this ratio was stably maintained near 1:1 (6.2 ± 2.8 × 108 cell mL−1 vs. 5.8 ± 1.2 × 108 cell mL−1) (Figure 2C), demonstrating that sustained exposure to low TCE concentrations (electron acceptor limitation) and constant supplies of electron donor, nutrients and growth factors can shift the relative abundances of D. mccartyi populations as has previously been shown under electron donor limitation (Mayer-Blackwell et al. 2017). The abundance of D. mccartyi strain ANAS2, which is responsible for VC reduction in the microbial community, could be increased in the completely mixed flow reactor and thus could facilitate more complete dechlorination.
Complete fermentation of 10 mM lactate in the influent was observed with the accumulation of propionate and acetate in the reactor effluent at concentrations of 3.1 ± 0.4 mM and 6.7 ± 0.7 mM, respectively (Figure 2B). H2 concentrations were maintained at around 0.2 μM (data not shown), which was greater than the reported minimum H2 threshold (2 nM) to support dechlorination (Yang et al. 1998). Recovery of electron equivalents was 95–108% throughout the steady-state period (Table 2), a result similar to previous bioreactor studies (Drzyzga et al. 2001). Only 6.1% of the electrons were consumed for reductive dechlorination while 75.2% accumulated as propionate and acetate, and 2.4 % as methane and trace H2. Biomass production accounts for a large portion of electron flow in this reactor (15.8 %), compared to previous studies (7.5 to 9.2%) (Azizian et al. 2010, Berggren et al. 2013). Biomass calculations assumed an elemental microbial cell composition of C5H7O2N, an average dry cell weight of 1.1 × 10−14 g cell−1 (with the assumption: the average cell size is 0.2μm in diameter, 2 μm in length, 20% of the cell is in the solid form, 90% of the solid is in organic form, and an average of two 16S rRNA copies are present in each bacteria cell), and total cell concentrations of Bacteria (2.3 ± 0.8 ×1010 16S rRNA copies mL−1) and Archaea (0.7 ± 0.3 ×107 16S rRNA copies mL−1) based on average qPCR of 16S rRNA copy numbers (Figure S3) (Duhamel et al. 2007). The ratios of D. mccartyi 16S rRNA copies to total 16S rRNA copies (including Bacteria and Archaea) were 4.2 ± 0.4 %, 5.8 ± 0.3%, and 5.3 ± 0.4% for CANAS at stages I, II and III and 4.1 ± 0.2 % and 4.7 ± 0.1% for ANAS during active and inactive dechlorination (Fig S3 B) (West et al. 2013).
To our knowledge, only one other study has reported cell densities and production rates of D. mccartyi in a CMFR with sustainable dechlorination to ethene (Table 1) (Delgado et al. 2014). Here, 85 % of TCE in the influent was converted to ethene with a dechlorination rate of 120 μmol Cl−1 L−1 d−1 which is lower than the highest dechlorination rate reported in a chemostat (130 μmol Cl−1 L−1 d−1; SRT = 3 d)(Delgado et al. 2014) but higher than a previous study with a 55-day SRT exhibiting a dechlorination rate of 80 μmol Cl− L−1 d−1 (Table 1)(Berggren et al. 2013). The relatively high D. mccartyi cell concentration (1.2 ± 0.3 × 109 cells mL−1 (Figure S3A)) observed in CANAS is comparable to the previously reported maximum D. mccartyi cell concentrations obtained in laboratory studies(Vainberg et al. 2009, Delgado et al. 2014). The D. mccartyi production rate in CANAS (3 × 1010 cells L−1 d−1, SRT 40 days) was about 10% of that observed in the 3-day SRT chemostat (Delgado et al. 2014) (Table 1), which is explained by the dilution rate (SRT−1) of CANAS that is much lower than the reported maximum specific growth rates of D. mccartyi (0.21–0.4d−1) (Cupples et al. 2003, Cupples et al. 2004, Johnson et al. 2008). The cell yield of D. mccartyi relative to TCE dechlorination in CANAS reached 2.5 ± 0.7 ×108 cells per μmol Cl− released (Table 1), which is in the range of previously reported cell yields of D. mccartyi isolates (He et al. 2007, Cheng et al. 2009, Men et al. 2012, Yan et al. 2012, Yan et al. 2013, Mao et al. 2015, Cooper et al. 2015), but lower than the yield reported in the 3-day SRT chemostat (5.3 × 108 cells per μmol Cl− released) (Delgado et al. 2014) likely due to enhanced endogenous cell decay within CANAS and the ability of D. mccartyi to decouple dechlorination from growth for maintenance activities (Cupples et al. 2003; Cupples et al. 2004; Johnson et al. 2008).
Microbial community structure analysis
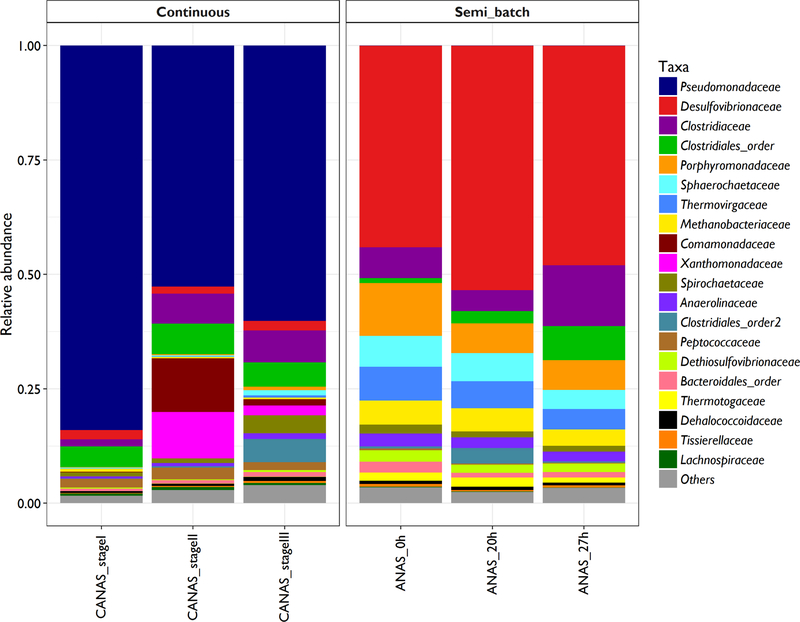
No significant difference in alpha diversity was observed between CANAS and ANAS using the Faith’s phylogenetic diversity index (PD = 21.6 ± 3.1 and 23.7 ± 1.7, respectively (p = 0.3709; unpaired, two-tailed t-test with Welch’s correction)) or the non-phylogenetic Shannon’s index (3.8 ± 1.4 for CANAS and 6.0 ± 0.2 for ANAS, p = 0.1163) and Chao1 estimators (551.9 ± 166.1 for CANAS and 717.9 ± 22.4 for ANAS, p = 0.2241). However, the composition and the relative abundance of OTUs identified in ANAS and CANAS demonstrated a microbial community structural shift along with the maintenance of TCE dechlorination performance. Archaea and Bacteria in CANAS represented 0.44% ± 0.19 and 99.56% ± 0.19 of the community respectively, with a significant difference in relative abundances compared to ANAS (4.86 ± 0.95 (p = 0.0014) and 95.1 ± 0.94 (p = 0.0014)). The lower relative abundance of Archaea in CANAS is consistent with the decreased methanogenic activity observed in the CMFR (Figure 2A; 0.04 mol methane / mol lactate for CANAS and 0.23 mol methane / mole lactate for ANAS). At the phylum and class levels, CANAS is dominated by members of Proteobacteria (77% ± 9.7 of which 91% ± 8.8 belong to the class Gammaproteobacteria) and Firmicutes (16% ± 6.0, 99.9% ± 0.02 of which belong to the class Clostridia) (Figures S4 and S5). ANAS is characterized by a more even community with Proteobacteria (49% ± 4.7) identified as the prominent phylum but with a dominance of the class Deltaproteobacteria (99.8% ± 0.03) (Figures S4 and S5). The Gini coefficients (G) calculated on the basis of the taxonomy tables (phylum (L2) and class (L3) levels) demonstrated a lower evenness in CANAS than in ANAS (GL2 = 0.94 ± 0.01, GL3 = 0.96 ± 0.01 and GL2 = 0.86 ± 0.02, GL3 = 0.94 ± 0.01, respectively) (Figure S6). At the taxonomic family level, twelve families with a relative abundance higher than 1% were maintained during the feast and famine feeding cycle of ANAS (i.e., Desulfovibrionaceae, Clostridiaceae, members from an unidentified family of Clostridiales (Clostridiales_order), Porphyromonadaceae, Sphaerochaetaceae, Thermovirgaceae, Methanobacteriaceae, Spirochaetaceae, Anaerolinaceae, Dethiosulfovibrionaceae, members from an unidentified family of Bacteroidales (Bacteroidales _order) and Thermotogaceae) (Figure 3). In CANAS, only five dominant families persisted at a relative abundance higher than 1% (i.e., Pseudomonadaceae, Desulfovibrionaceae, Clostridiaceae, members from an unidentified family of Clostridiales (Clostridiales_order) and Peptococcaceae) (Figure 3).
Figure 3.
Microbial community composition based on family-level taxonomic classification of 16S rRNA gene tags for CANAS during three consecutive stages (I, II and III, roughly 50 days apart) and ANAS at three distinct time-points in a single feeding cycle (ANAS-0h, after a 14-day batch incubation (starved culture), ANAS-20h, 20 hours after lactate and TCE feeding (all lactate consumed) and ANAS-27h, 27 hours after lactate and TCE feeding (all TCE was converted to ethene)). The stacked bar charts present the nineteen most abundant taxa detected in ANAS and CANAS.
Compared to ANAS, the microbial community in CANAS shifted to a new composition with the identification of two dominant families that exhibited a relative abundance (to bacteria and archaea) lower than 0.3% in ANAS (Pseudomonadaceae (66% ± 16 of CANAS) and Peptococcaceae (2.1% ± 0.5 of CANAS)). The high abundance of Pseudomonas was confirmed by qPCR of 16S rRNA to exclude gene tag sequencing biases (data not shown). Although Pseudomonadaceae is mostly associated with aerobic and facultative anaerobic bacteria, all members of Peptococcaceae of the order Clostridiales are obligate anaerobes commonly involved in syntrophic associations (Kleinsteuber et al. 2012, Stackebrandt 2014). The relative abundance of Clostridiaceae increased from 1.5 to 7.0% in CANAS from stage I to III which is similar to ANAS (8.2% ± 4.5). Desulfovibrionaceae was relatively stable in CANAS (1.9% ± 0.28), which is much lower than the relative abundance observed in ANAS (48% ± 4.7). Methanobacteriaceae was the most abundant archaeal family detected in both CANAS and ANAS and accounted for 0.43% ± 0.19 and 4.6% ± 0.91 of the total community, respectively. In CANAS, 98% ± 2.3 of archaeal OTUs belong to the Methanobacteriaceae (96% ± 6.0 of Methanobacterium sp.) of which most members are hydrogenotrophic methanogens (Oren 2014). In ANAS, 95% ± 0.5 of archaeal OTUs belong to the Methanobacteriaceae and 4.6% ± 0.5 belong to the Methanospirillaceae which mainly catalyzes methanogenesis from H2/CO2 (Oren 2014). The relative abundances of Dehalococcoidaceae in CANAS (0.59% ± 0.26 for stages I, II, III) were similar to ANAS (0.69% ± 0.04 for ANAS-0h, ANAS-20h and ANAS-27h) with the maintenance of robust dechlorination rates (i.e., 12 μmol Cl−1 L−1 d−1 for CANAS and 59 μmol Cl−1 L−1 d−1 for ANAS). In all conditions, the relative abundance of Dehalococcoides sp. detected by gene tag sequencing was lower than the percentage calculated from qPCR analysis (4–6%, Figure S3).
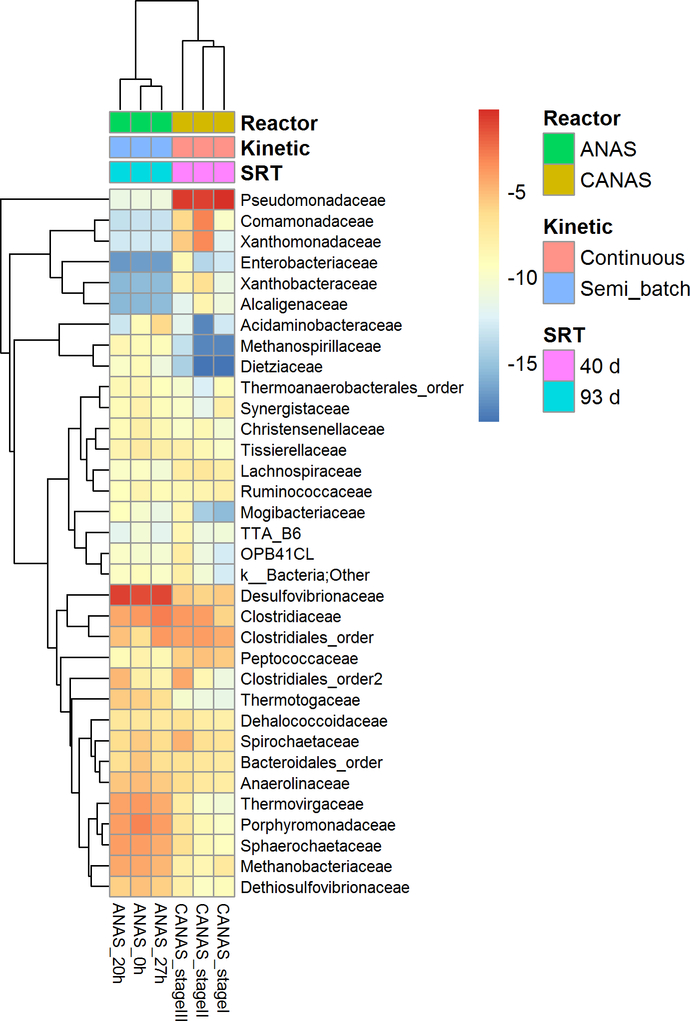
The structural regime shift observed between CANAS and ANAS (i.e., change in the microbial community composition and structure (Bürgmann et al. 2011, Engel et al. 2014, Andersen et al. 2008, Scheffer and Carpenter, 2003)) was confirmed using hierarchical cluster and PCoA analyses. Hierarchical cluster analysis of the most abundant OTUs (at the class level (Figure S7) and family level (Figure 4)) identified in ANAS and CANAS indicates that samples segregate according to their reactor type. Pairwise (intra- and inter-cluster) distances analysis for ANAS and CANAS (Figure S8 A) showed that intra-cluster comparisons (ANAS-ANAS and CANAS-CANAS) and inter-cluster distances (ANAS-CANAS) were significantly greater than intra-cluster distances (p < 0.0001, Figure S8 A). In addition, the Weighted UniFrac-based PCoA demonstrated ANAS and CANAS formed two distinct clusters along PCo1 (78% variation), supporting their separation (Figure S8 B).
Figure 4.
Taxonomy heatmap of 16S rRNA gene tags (family-level taxonomic classification) of ANAS and CANAS microbial communities at different time-points. Microbial families accounting for a relative abundance greater than or equal to 0.2% in at least one sample were included in the heatmap. The log2 values of relative abundance (in %) of each taxon were used for hierarchical cluster analysis using the pheatmap R package.
In this study, the microbial community structure of a TCE-dechlorinating enrichment culture, maintained functionally active for more than 15 years in a semi-batch reactor, significantly shifted after inoculation and maintenance in a CMFR. Despite the observed community structural shift, CANAS demonstrated a high functional robustness with the maintenance of efficient TCE dechlorination to ethene. Although large proportions of facultative anaerobic microorganisms were detected in CANAS (e.g., Pseudomonadaceae) with a significant decrease in relative abundance of the strictly anaerobic Desulfovibrionaceae family, the dechlorination function was stably preserved suggesting that ANAS1 and ANAS2 could take advantage of the functional redundancy of microbial functional guilds that support their activities.
Using qPCR measurements normalized to the bacterial and archaeal 16S rRNA copies detected in the whole community, D. mccartyi accounted for around 4 to 6% of the community in both ANAS and CANAS, as reported in other TCE-dechlorinating D. mccartyi-containing communities (Azizian et al. 2008, Behrens et al. 2008, West et al. 2013). In the EV-5L and VS-5L CMFRs (Table 1) (Mayer-Blackwell et al. 2016, Mayer-Blackwell et al. 2017), the relative abundance of D. mccartyi was higher than 90% of the total bacterial community using targeted sequencing of the hypervariable V1–V2 or V2–V3 16S rRNA regions. In the CMFR PM-2L (2-II) used to inoculate the CMFR PM-5L (Table 1), Dehalococcoides sp. accounted for around 35% of the bacterial community using 16S rRNA-based qPCR assays (Berggren et al. 2013). In this study, we used the primer pair 515F-806R (Caporaso et al. 2011, Tremblay et al. 2015) for targeted amplicon sequencing which shows a single primer mismatch at the eleventh position from the 5’-end of the reverse primer 806R (position 10 from the 3’ end; 5’- GGACTACHVG(G/A)GTWTCTAAT-3’) with the 16S rRNA sequence of D. mccartyii strain 195. In silico analysis of the primer pair 515F/806R with Primer-BLAST (Ye et al. 2012) revealed the same single base mismatch for all D.s mccartyi 16S rRNA sequences. This can explain the lower relative abundance values of D. mccartyi sp. in CANAS measured by 16S amplicon sequencing compared to qPCR assays. Apprill et al. (2015) (Apprill et al. 2015) reported the underestimation of SAR11 bacterioplankton due to the effect of a single internal primer-template mismatch at the position 8 from the 5’ end of the reverse primer 806R. It is also possible that different sample preparation methods (cells were collected by filtration or by centrifugation) could lead to the discrepancy between the amplicon sequencing and the qPCR results.
Using qPCR measurements, the 16S gene copies of methanogens in CANAS were below 30% of those measured in ANAS (Figure S3A) (West et al. 2013). Similar results were obtained based on 16S amplicon sequencing with a ratio of Methanobacteriaceae in CANAS vs. ANAS of 0.093 ± 0.045, indicating that CANAS has about 10% of the abundance of Methanobacteriaceae compared to ANAS. H2 levels measured in CANAS were maintained at about 0.2 μM which is higher than the required methanogenic threshold (Yang et al. 1998, Loffler et al. 1999). However, methane production was only about 30% compared to ethene production based on electron equivalents (Table 2). This contrasts with the ANAS bioreactor in which methane production was about 500% of ethene production (West et al., 2013). Since methanogenesis requires more reduced redox conditions (EH < −200 mV) than reductive dechlorination and oxygen reduction(Le Mer et al. 2001), methane production in CANAS might be limited by the increase of the redox potential as observed by the dominance of facultative anaerobic microorganisms.
Transcriptional analysis
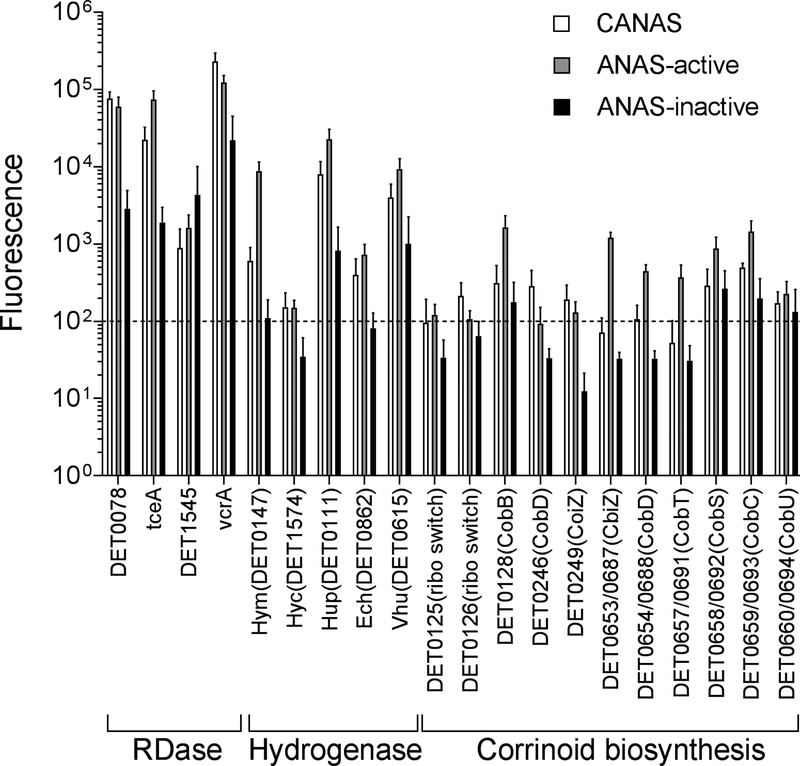
Gene expression profiles of D. mccartyi in ANAS during active (ANAS-20h) and inactive (ANAS-0h) dechlorination were compared with those obtained in CANAS under sustained dechlorination at steady-state. In order to make valid comparisons of transcriptomic data between the two consortia, all expression levels were normalized to amended standard samples because Dehalococcoides species were present at similar relative abundances in both ANAS and CANAS (Figure S3B). Unsurprisingly, the gene expression profiles of D. mccartyi in CANAS remained relatively constant in the three consecutive stages (Figure 5), while the expression profile of ANAS varied significantly as expected in a semi-batch reactor. Most highly expressed genes in CANAS throughout the experimental period were genes encoding for ribosomal RNA (5S, 16S and 23S rRNA) and reductive dehalogenases tceA and vcrA. Interestingly, the expression levels of tceA (DET0079) were significantly lower in CANAS (p = 0.03), while vcrA was significantly higher (p = 0.03) compared to ANAS during active dechlorination. The transcriptomic results agree with the higher cell ratio of ANAS2 to ANAS1 observed in CANAS compared to ANAS. The tceA anchor protein gene (DET0078), and two other RDase genes (DET0173 and DET 1545) in CANAS were similar to those detected in ANAS-active (Figure 5). In contrast, the expression levels of putative hydrogenases (hym, hup, vhu, hyc, ech, hyp) and several oxidoreductase genes (fdh, mod, nuo, fpoF, por) were significantly lower compared to ANAS-active, but higher than expression levels detected in ANAS-starved (Figure 5, supporting material Table S4). Specifically, the most highly expressed hydrogenases in CANAS were hup (DET0111), hym (DET0728), vhu, which were expressed to the same extent or slightly lower as detected in ANAS-active, but were significantly higher than ANAS-starved (Table S4).
Figure 5.
Functional gene expression profiles of reductive dehalogenases (RDase) genes, oxidoreductase genes and corrinoid transport/biosynthesis genes in CANAS and ANAS during active dechlorination (20 hours after feeding substrate, ANAS-active (ANAS-20h)) and starvation (14 days after feeding substrate, ANAS-inactive (ANAS-0h)). Dashed line indicates significant fluorescence level (fluorescence signal > 100). Error bars represent the SD of biological triplicates.
Other oxidoreductases followed the same trend as the hydrogenase expression (Table S4), with the expression of the putative formate dehydrogenase (fdh DET 0185–0187) in CANAS at about 20% of that measured in ANAS-active, but 8 to 10 times higher than in ANAS-starved. A previous study had shown that the expression levels of DET 0187 in dechlorinating microbial communities were similar or higher than those of the hydrogenases under batch growth conditions (Morris et al. 2007). However, a recent transcriptomic study revealed that DET0187 was expressed at similar levels to the Hup subunit (DET 0112) in multiple D.mccartyi strains (Mansfeldt et al. 2014). The Fdh-like oxidoreductase (DET0187) was shown to be associated with the Fdh-like protein DET0112 from the hup operon (Mansfeldt et al. 2014) and a recent transcriptomic study of D. mccartyi-containing microbial communities indicated the expression pattern of the Hup and the Fdh-like oxidoreductase are strongly linked (Mansfeldt et al. 2016). Although the exact function of the Fdh-like oxidoreductase in D. mccartyi remains unknown, protein analysis of D. mccartyi strain CBDB1 suggested it is involved in the enzyme complexes that are critical to the electron transport chain of the D. mccartyi species (Kublik, et al. 2016; Türkowsky et al. 2018). In our study, the expression levels of fdh (DET0186-DET 0187) in both ANAS and CANAS were found to be at similar levels to the hydrogenases hup, hym, and vhu (Supplemental material Table S4), consistent with the previous findings (Mansfeldt et al. 2014, Mansfeldt et al. 2016, Heavner et al. 2018).
Previous studies have shown that D. mccartyi must rely on exogenous corrinoids for RDase activity (Magnuson et al. 1998, Banerjee and Ragsdale. 2003, Johnson et al. 2008, West et al. 2013, Yan et al. 2013, Men et al. 2015). Genes associated with corrinoid salvaging and transport were most highly expressed in ANAS-active with lower expression levels of both corrinoid transport and synthesis genes throughout the different monitored stages of CANAS supplemented with higher concentrations of vitamin B12 (Figure 5), consistent with a previous study of Dehalococcoides mccartyi 195 grown with excess or limiting vitamin B12 (Johnson et al. 2008). The different expression patterns of corrinoid-related genes compared to genes for RDases indicate the expression of corrinoid-related genes may not be appropriate biomarkers for chlorinated ethene dechlorination activity. Previous studies have proposed hydrogenase genes (hupL) as biomarkers to indicate the activity of reductive dechlorination (Morris et al. 2007, Rowe et al. 2008, Berggren et al. 2013). In this study, we found similar positive correlation between hupL and tceA gene expression. Previous modeling studies have indicated that transcripts instead of DNA/proteins/VSS may serve as better biomarkers to predict reductive dechlorination performance (Baelum et al. 2013, Heavner et al. 2013), however, the consistent high levels of expression demonstrated for active and starving conditions of ANAS suggest that this approach is problematic.
4. Conclusion
Dechlorinating microbial communities have been extensively studied in batch reactors to reveal their dynamics as dechlorination proceeds and to construct kinetic models of reductive dechlorination pathways of chlorinated ethenes. However, it is difficult to extrapolate the knowledge acquired from such transient and fluctuating systems to flowing groundwater plumes with continuous and sustained supply of low concentrations of contaminants, nutrients and substrates. It is critical to understand the microbial community structure and dynamics in such continuous flow conditions, in order to validate the applications of molecular biomarkers or mathematical models during the bioremediation practices. In this study, we developed a completely mixed flow reactor containing the TCE-dechlorinating community (CANAS) from a long-term dechlorinating enrichment culture (ANAS) maintained in a semi-batch reactor. Based on the reactor performance assessment, microbial community structure analysis and the transcriptome dynamic analysis, the findings present in this study are as follows:
The reductive dechlorinating microbial community CANAS developed in the completely mixed flow reactor showed effective TCE reduction at the selected SRT (40 days) with the major end-product of ethene (~85%) and a very low production of other intermediates (VC, cis-DCE) at the steady-state condition.
The microbial community structure shifted significantly from semi-continuous flow condition to completely mixed flow condition while robust reductive dechlorination performance was stably maintained, suggesting that ANAS1 and ANAS2 could take advantage of the functional redundancy of microbial functional guilds that support their activities.
The ratio of D.mccartyi strains ANAS1 to ANAS2 was stably maintained near 1:1 in the CANAS compared with 4:1 in the ANAS, demonstrating that sustained exposure to low TCE concentrations can shift the relative abundance of D. mccartyi populations.
This is the first study to examine global gene expression of Dehalococcoides species under continuous flow conditions. The most highly expressed genes encoded for ribosomal RNA and reductive dehalogenases tceA and vcrA, while the expression of putative hydrogenases and several oxidoreductases genes were maintained at the levels between the ANAS-active and ANAS-inactive conditions, suggesting that we need to be careful when selecting biomarkers to indicate reductive dechlorination activity.
Supplementary Material
Highlights.
Reductive dechlorination of TCE to ethene by a steady-state chemostat culture.
Shift of microbial community from batch-fed to continuous exposure to low TCE.
Community functional robustness ensures stable TCE-to-ethene reduction.
Reductive dehalogenases tceA and vcrA show the highest functional gene expression.
Selection of bio-marker genes for monitoring of TCE-contaminated groundwater plumes.
Acknowledgdements
This work was supported by research grants from the NIEHS (P42-ES04705-14) and the NSF (CBET-1336709). This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668 and S10RR027303.
The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231
Abbreviations
- CMFR
completely mixed flow reactors
- PCE
perchloroethene
- SRA
Sequence Read Archive
- SRT
sludge retention time
- TCE
trichloroethene
- TMM
trimmed mean of M-values
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Reference
- Adrian L, Szewzyk U, Gorisch H, 2000Bacterial growth based on reductive dechlorination of trichlorobenzenes. Biodegradation 11(1): p. 73–81. [DOI] [PubMed] [Google Scholar]
- Andersen T, Carstensen J, Hernandez-Garcia E, Duate CM, 2009. Ecological thresholds and regime shifts: approaches to identification. Trends in Ecology & Evolution. 18, 49–57, DOI: 10.1016/j.tree.2008.07.014 [DOI] [PubMed] [Google Scholar]
- Apprill A, McNally S, Parsons R, Weber. L, 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology 75(2), 129–137. [Google Scholar]
- Azizian MF, Behrens S, Sabalowsky A, Dolan ME, Spormann AM, Semprini. L, 2008. Continuous-flow column study of reductive dehalogenation of PCE upon bioaugmentation with the Evanite enrichment culture. Journal of Contaminant Hydrology 100(1–2), 11–21.DOI: 10.1016/j.jconhyd.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Azizian MF, Marshall IPG, Behrens S, Spormann AM, Semprini L, 2010. Comparison of lactate, formate, and propionate as hydrogen donors for the reductive dehalogenation of trichloroethene in a continuous-flow column. Journal of Contaminant Hydrology 113(1–4), 77–92.DOI: 10.1016/j.jconhyd.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Azizian MF, Semprini L, 2016. Simultaneous anaerobic transformation of tetrachloroethene and carbon tetrachloride in a continuous flow column. Journal of Contaminant Hydrology 190, 58–68.DOI: 10.1016/j.jconhyd.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Baelum J, Chambon JC, Scheutz C, Binning PJ, Laier T, Bjerg PL. Jacobsen CS, 2013. A conceptual model linking functional gene expression and reductive dechlorination rates of chlorinated ethenes in clay rich groundwater sediment. Water Research 47(7), 2467–2478.DOI: 10.1016/j.watres.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Ragsdale SW, 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annual Review of Biochemistry 72: 209–247. [DOI] [PubMed] [Google Scholar]
- Behrens S, Azizian MF, McMurdie PJ, Sabalowsky A, Dolan ME, Semprini L, Spormann AM, 2008. Monitoring abundance and expression of “Dehalococcoides” species chloroethene-reductive dehalogenases in a tetrachloroethene-dechlorinating flow column. Applied and Environmental Microbiology 74(18), 5695–5703.DOI: 10.1128/aem.00926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren DRV, Marshall IPG, Azizian MF, Spormann AM, Semprini L, 2013. Effects of Sulfate Reduction on the Bacterial Community and Kinetic Parameters of a Dechlorinating Culture under Chemostat Growth Conditions. Environmental Science & Technology 47(4), 1879–1886.DOI: 10.1021/es304244z. [DOI] [PubMed] [Google Scholar]
- Brisson VL, West KA, Lee PKH, Tringe SG, Brodie EL, Alvarez-Cohen L, 2012. Metagenomic analysis of a stable trichloroethene-degrading microbial community. ISME Journal 6(9), 1702–1714.DOI: 10.1038/ismej.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürgmann H, Jenni S, Vazquez F, Udert KM. 2011. Regime shift and microbial dynamics in a sequencing batch reactor for nitrification and anammox treatment of urine. Applied Environmental Microbiology 77, 5897–5907, DOI: 10.1128/AEM.02986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N,Knight R, 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences 108(Supplement 1), 4516–4522.DOI: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr CS, Garg S, Hughes JB, 2000. Effect of dechlorinating bacteria on the longevity and composition of PCE-containing nonaqueous phase liquids under equilibrium dissolution conditions. Environmental Science & Technology 34(6), 1088–1094.DOI: 10.1021/es990989t. [DOI] [Google Scholar]
- Carr CS, Hughes JB, 1998. Enrichment of high-rate PCE dechlorination and comparative study of lactate, methanol, and hydrogen as electron donors to sustain activity. Environmental Science & Technology 32(12), 1817–1824.DOI: 10.1021/es970985t. [DOI] [Google Scholar]
- Cheng D, Chow WL, He J, 2009. A Dehalococcoides-containing co-culture that dechlorinates tetrachloroethene to trans-1,2-dichloroethene. The ISME Journal 4, 88DOI: 10.1038/ismej.2009.90 [DOI] [PubMed] [Google Scholar]
- Chung J, Krajmalnik-Brown R, Rittmann BE, 2008. Bioreduction of trichloroethene using a hydrogen-based membrane biofilm reactor. Environmental Science & Technology 42(2), 477–483.DOI: 10.1021/es702422d. [DOI] [PubMed] [Google Scholar]
- Cooper M, Wagner A, Wondrousch D, Sonntag F, Sonnabend A, Brehm M,Schüürmann G, Adrian L, 2015. Anaerobic Microbial Transformation of Halogenated Aromatics and Fate Prediction Using Electron Density Modeling. Environmetal Science & Technology 49(10), 6018–6028.DOI: 10.1021/acs.est.5b00303 [DOI] [PubMed] [Google Scholar]
- Cupples AM, Spormann AM, McCarty PL, 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Applied and Environmental Microbiology 69(2), 953–959.DOI: 10.1128/aem.69.2.953-959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples AM, Spormann AM, McCarty PL, 2004. Vinyl chloride and cis-dichloroethene dechlorination kinetics and microorganism growth under substrate limiting conditions. Environmental Science & Technology 38(4), 1102–1107.DOI: 10.1021/es0348647. [DOI] [PubMed] [Google Scholar]
- Daprato RC, Löffler FE Hughes JB, 2007. Comparative analysis of three tetrachloroethene to ethene halorespiring consortia suggests functional redundancy. Environmental Science & Technology 41(7), 2261–2269.DOI: 10.1021/es061544p. [DOI] [PubMed] [Google Scholar]
- Delgado AG, Fajardo-Williams D, Popat SC, Torres CI, Krajmalnik-Brown R, 2014. Successful operation of continuous reactors at short retention times results in high-density, fast-rate Dehalococcoides dechlorinating cultures. Applied Microbiology and Biotechnology 98(6), 2729–2737.DOI: 10.1007/s00253-013-5263-5. [DOI] [PubMed] [Google Scholar]
- Delgado AG, Fajardo-Williams D, Bondank E, Esquivel-Elizondo S, Krajmalnik-Brown R, 2017. Coupling Bioflocculation of Dehalococcoides mccartyi to High-Rate Reductive Dehalogenation of Chlorinated Ethenes. Environmental Science & Technology 51(19), 11297–11307. [DOI] [PubMed] [Google Scholar]
- Dolinova I, Strojsova M, Cernik M, Nemecek J, Machackova J, Sevcu A, 2017. MMicrobial degradation of chloroethenes: a review. Environ Science and Pollut Research 24(15), 13262–13283. [DOI] [PubMed] [Google Scholar]
- Drzyzga O, Gerritse J, Dijk JA, Elissen H, Gottschal JC, 2001. Coexistence of a sulphate-reducing Desulfovibrio species and the dehalorespiring Desulfitobacterium frappieri TCE1 in defined chemostat cultures grown with various combinations of sulphate and tetrachloroethene. Environmental Microbiology 3(2), 92–99.DOI: 10.1046/j.1462-2920.2001.00157.x. [DOI] [PubMed] [Google Scholar]
- Duhamel M, Edwards EA, 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environmental Science & Technology 41(7), 2303–2310.DOI: 10.1021/es062010r. [DOI] [PubMed] [Google Scholar]
- Engel AS, Gupta AA., 2014. Regime shift in sandy beach microbial communities following Deepwater Horizon oil spill remediation efforts. PLoS ONE 9: e102934 10.1371/journal.pone.0102934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L, 2005. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environmental Science & Technology 39(21), 8358–8368.DOI: 10.1021/es048003p. [DOI] [PubMed] [Google Scholar]
- Gerritse J, Kloetstra G, Borger A, Dalstra G, Alphenaar A, Gottschal JC, 1997. Complete degradation of tetrachloroethene in coupled anoxic and oxic chemostats. Appl Microbiol Biotechnol 48(4), 553–562.DOI: 10.1007/s002530051096. [DOI] [PubMed] [Google Scholar]
- He J, Holmes VF, Lee PKH, Alvarez-Cohen L, 2007. Influence of Vitamin B12 and Cocultures on the Growth of Dehalococcoides Isolates in Defined Medium. Applied and Environmental Microbiology 73(9), 2847–2853.DOI: 10.1128/aem.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heavner GLW, Rowe AR, Mansfeldt CB, Pan JK, Gossett JM, Richardson RE, 2013. Molecular Biomarker-Based Biokinetic Modeling of a PCE-Dechlorinating and Methanogenic Mixed Culture. Environmental Science & Technology 47(8), 3724–3733.DOI: 10.1021/es303517s. [DOI] [PubMed] [Google Scholar]
- Heavner GLW, Mansfeldt CB, Debs GE, Hellerstedt ST, Rowe AR, Richardson RE, 2018. Biomarkers’ Responses to Reductive Dechlorination Rates and Oxygen Stress in Bioaugmentation Culture KB-1 (TM). Microorganisms 6 (1) UNSP 13 DOI: 10.3390/microorganisms6010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliger C, Hahn D, Harmsen H, Ludwig W, Schumacher W, Tindall B, Vazquez F, Weiss N, Zehnder AJB, 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Archives of Microbiology 169(4), 313–321.DOI: 10.1007/s002030050577. [DOI] [PubMed] [Google Scholar]
- Hug LA. Maphosa F, Leys D, Löffler FE, Smidt H, Edwards EA, Adrian L, 2013. Overview of organohalide-respiring bacteria and a proposal for a classification system for reductive dehalogenases. Philosophical Transactions of the Royal Society B-Biological Science 368: 20120322 10.1098/rstb.2012.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L, 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Applied and Environmental Microbiology 74(9), 2864–2872.DOI: 10.1128/aem.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen M, Dechesne A, Proctor C, Hammes F, Johnson D, Quintela-Baluja M, Graham D, Daffonchio D, Fodelianakis S, Hahn N, Boon N, Smets BF, 2016. A conceptual framework for invasion in microbial communities. Isme Journal 10(12), 2773–2779.DOI: 10.1038/ismej.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinsteuber S, Schleinitz KM, Vogt C 2012. Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers. Applied Microbiology and Biotechnology 94(4), 851–873.DOI: 10.1007/s00253-012-4025-0. [DOI] [PubMed] [Google Scholar]
- Kublik A, Deobald D, Hartwig S, Schiffmann CL, Andrades A, von Bergen M, Sawers RG, Adrian L, 2016. Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement: Respiratory reductive dehalogenase protein complex. Environmental Microbiology 18, 3044–3056. [DOI] [PubMed] [Google Scholar]
- Le Mer J, Roger P, 2001. Production, oxidation, emission and consumption of methane by soils: A review. European Journal of Soil Biology 37(1), 25–50.DOI: 10.1016/s1164-5563(01)01067-6. [DOI] [Google Scholar]
- Lee PKH, Cheng D, Hu P, West KA, Dick GJ, Brodie EL, Andersen GL, Zinder SH, He JZ, Alvarez-Cohen L, 2011. Comparative genomics of two newly isolated Dehalococcoides strains and an enrichment using a genus microarray. ISME Journal 5(6), 1014–1024.DOI: 10.1038/ismej.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PKH, Johnson DR, Holmes VF, He JZ, Alvarez-Cohen L, 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Applied and Environmental Microbiology 72(9), 6161–6168.DOI: 10.1128/aem.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler FE, Sun Q, Li JR, Tiedje JM, 2000. 16S rRNA gene-based detection of tetrachloroethene-dechlorinating Desulfuromonas and Dehalococcoides species. Applied and Environmental Microbiology 66(4), 1369–1374.DOI: 10.1128/aem.66.4.1369-1374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löffler FE, Tiedje JM,Sanford RA, 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Applied and Environmental Microbiology 65(9), 4049–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson JK., Stern RV, Gossett JM, Zinde r SH., Burris DR, 1998. Reductive dechlorination of tetrachloroethene to ethene by a two-component enzyme pathway. Applied Environmental Microbiology 64:1270–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeldt CB, Rowe AR, Heavner GLW, Zinder SH,. Richardson RE, 2014. Meta-Analyses of Dehalococcoides mccartyi Strain 195 Transcriptomic Profiles Identify a Respiration Rate-Related Gene Expression Transition Point and Interoperon Recruitment of a Key Oxidoreductase Subunit. Applied and Environmental Microbiology 80(19), 6062–6072.DOI: 10.1128/aem.02130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeldt CB, Heavner GW, Rowe AR, Hayete B, Church BW, Richardson RE, 2016. Inferring Gene Networks for Strains of Dehalococcoides Highlights Conserved Relationships between Genes Encoding Core Catabolic and Cell-Wall Structural Proteins. PLoS ONE 11, e0166234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao XW, Stenuit B, Polasko A, Alvarez-Cohen L, 2015. Efficient Metabolic Exchange and Electron Transfer within a Syntrophic Trichloroethene-Degrading Coculture of Dehalococcoides mccartyi 195 and Syntrophomonas wolfei. Applied and Environmental Microbiology 81(6), 2015–2024.DOI: 10.1128/aem.03464-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maphosa F, Smidt H, De Vos WM, Roling WFM, 2010. Microbial Community- And Metabolite Dynamics of an Anoxic Dechlorinating Bioreactor. Environmental Science & Technology 44(13), 4884–4890.DOI: 10.1021/es903721s. [DOI] [PubMed] [Google Scholar]
- Marshall IPG, Azizian MF, Semprini L, Spormann AM, 2014. Inferring community dynamics of organohalide-respiring bacteria in chemostats by covariance of rdhA gene abundance. Fems Microbiology Ecology 87(2), 428–440.DOI: 10.1111/1574-6941.12235. [DOI] [PubMed] [Google Scholar]
- Mayer-Blackwell K, Azizian MF, Green JK, Spormann AM, Semprini L, 2017. Survival of Vinyl Chloride Respiring Dehalococcoides mccartyi under Long-Term Electron Donor Limitation. Environmental Science & Technology 51(3), 1635–1642.DOI: 10.1021/acs.est.6b05050. [DOI] [PubMed] [Google Scholar]
- Mayer-Blackwell K, Fincker M, Molenda O, Callahan B, Sewell H, Holmes S, Edwards EA, Spormann AM, 2016. 1,2-Dichloroethane Exposure Alters the Population Structure, Metabolism, and Kinetics of a Trichloroethene-Dechlorinating Dehalococcoides mccartyi Consortium. Environmental Science & Technology 50(22), 12187–12196.DOI: 10.1021/acs.est.6b02957. [DOI] [PubMed] [Google Scholar]
- Mayer HP,Conrad R, 1990. Factors Influencing The Population of Methanogenic Bacteria and the Initiation of Methane Production Upon Flooding of Paddy Soil. Fems Microbiology Ecology 73(2), 103–111. [Google Scholar]
- Maymo-Gatell X, Chien YT, Gossett JM, Zinder SH, 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276(5318), 1568–1571.DOI: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- McCarty PL 2010. Groundwater contamination by chlorinated solvents: History, remediation technologies and strategies In Situ Remediation of Chlorinated Solvent Plumes. Stroo HF and Ward CH. New York, NY, Springer: 1–28. [Google Scholar]
- McMurdie PJ, Holmes S, 2014. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. Plos Computational Biology 10(4).DOI: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men Y, Seth EC, Yi S, Crofts TS, Allen RH, Taga ME, Alvarez-Cohen L, 2015. Identification of specific corrinoids reveals corrinoid modification in dechlorinating microbial communities. Environmental Microbiology 17(12), 4873–4884.DOI: 10.1111/1462-2920.12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men YJ, Feil H, VerBerkmoes NC, Shah MB, Johnson DR, Lee PKH, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L, 2012. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME Journal 6(2), 410–421.DOI: 10.1038/ismej.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza BS, Sorensen DL, Dupont RR, McLean JE, 2016. Dehalococcoides abundance and alternate electron acceptor effects on large, flow-through trichloroethene dechlorinating columns. Applied Microbiology and Biotechnology 100(5), 2367–2379.DOI: 10.1007/s00253-015-7112-1. [DOI] [PubMed] [Google Scholar]
- Mirza BS, Sorensen DL, McGlinn DJ, Dupont RR, McLean JE, 2017. Dehalococcoides and general bacterial ecology of differentially trichloroethene dechlorinating flow-through columns. Applied Microbiology and Biotechnology 101(11), 4799–4813.DOI: 10.1007/s00253-017-8180-1. [DOI] [PubMed] [Google Scholar]
- Moran MJ, Zogorski JS, Squillace PJ, 2007. Chlorinated solvents in groundwater of the United States. Environmental Science & Technology 41(1), 74–81.DOI: 10.1021/es061553y. [DOI] [PubMed] [Google Scholar]
- Morris RM, Fung JM, Rahm BG, Zhang S, Freedman DL, Zinder SH, Richardson RE, 2007. Comparative proteomics of Dehalococcoides spp. reveals strain-specific peptides associated with activity. Applied and Environmental Microbiology 73(1), 320–326.DOI: 10.1128/aem.02129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A 2014. The family Methanobacteriaceae. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. Rosenberg E, DeLong EF, Lory S, Stackebrandt Eand Thompson F. Berlin, Germany, Springer: 165–193. [Google Scholar]
- Oren A 2014. The family Methanospirillaceae. The Prokaryotes: Other Major Lineages of Bacteria and The Archaea. Rosenberg E, DeLong EF, Lory S, Stackebrandt E and Thompson F. Berlin, Germany, Springer: 283–290. [Google Scholar]
- Richardson RE, Bhupathiraju VK, Song DL, Goulet TA, Alvarez-Cohen L, 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environmental Science & Technology 36(12), 2652–2662.DOI: 10.1021/es0157797. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK, 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140.DOI: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe AR, Lazar BJ, Morris RM, Richardson RE, 2008. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated “Dehalococcoides” and Methanospirillum species. Applied Environmental Microbiology 74(21), 6709–6719.DOI: 10.1128/aem.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabalowsky AR,Semprini L, 2010. Trichloroethene and cis-1,2-Dichloroethene Concentration-Dependent Toxicity Model Simulates Anaerobic Dechlorination at High Concentrations. II: Continuous Flow and Attached Growth Reactors. Biotechnology and Bioengineering 107(3), 540–549.DOI: 10.1002/bit.22822. [DOI] [PubMed] [Google Scholar]
- Scheffer M, Carpenter SR, 2003. Catastrophic regime shifts in ecosystems: linking theory to observation. Trends in Ecology & Evolution 18, 648–656, DOI: 10.1016/j.tree.2003.09.002 [DOI] [Google Scholar]
- Stackebrandt E, 2014. The emended family Peptococcaceae and description of the families Desulfitobacteriaceae, Desulfotomaculaceae, and Thermincolaceae The Prokaryotes: Firmicutes and Tenericutes. Rosenberg E, DeLong EF, Lory S, Stackebrandt E and Thompson F. Berlin, Germany, Springer: 285–290. [Google Scholar]
- Stroo HF,Ward CH Eds., 2010. In Situ Remediation of Chlorinated Solvent Plumes SERDP/ESTCP Environmental Remediation Technology New York, NY, Springer. [Google Scholar]
- Sung Y, Fletcher KF, Ritalaliti KM, Apkarian RP, Ramos-Hernandez N, Sanford RA, Mesbah NM, Löffler FE, 2006. Geobacter lovleyi sp nov strain SZ, a novel metal-reducing and tetrachloroethene-dechlorinating bacterium. Applied and Environmental Microbiology 72(4), 2775–2782.DOI: 10.1128/aem.72.4.2775-2782.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama A, Iwakiri R, Kai K, Tokunaga T, Sera N, Furukawa K, 2001. Isolation and characterization of Desulfitobacterium sp strain Y51 capable of efficient dehalogenation of tetrachloroethene and polychloroethanes. Bioscience Biotechnology and Biochemistry 65(7), 1474–1481.DOI: 10.1271/bbb.65.1474. [DOI] [PubMed] [Google Scholar]
- Tremblay J, Singh K, Fern A, Kirton ES, He SM, Woyke T, Lee J, Chen F, Dangl JL, Tringe SG, 2015. Primer and platform effects on 16S rRNA tag sequencing. Frontiers in Microbiology 6DOI: 10.3389/fmicb.2015.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkowsky D, Jehmlich N, Diekert G, Adrian L, von Bergen M, Goris T, 2018. An integrative overview of genomic, transcriptomic and proteomic analyses in organohalide respiration research. FEMS Microbiol Ecology 94 (3) DOI: 10.1093/femsec/fiy013 [DOI] [PubMed] [Google Scholar]
- Vainberg S, Condee CW, Steffan RJ, 2009. Large-scale production of bacterial consortia for remediation of chlorinated solvent-contaminated groundwater. Journal of Industrial Microbiology & Biotechnology 36(9), 1189–1197.DOI: 10.1007/s10295-009-0600-5. [DOI] [PubMed] [Google Scholar]
- Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N, 2009. Initial community evenness favours functionality under selective stress. Nature, 458, 623–626, DOI: 10.1038/nature07840 [DOI] [PubMed] [Google Scholar]
- West KA, Johnson DR, Hu P, DeSantis TZ, Brodie EL, Lee PKH, Feil H, Andersen GL, Zinder SH, Alvarez-Cohen L, 2008. Comparative genomics of “Dehalococcoides ethenogenes” 195 and an enrichment culture containing unsequenced “Dehalococcoides” strains. Applied and Environmental Microbiology 74(11), 3533–3540.DOI: 10.1128/aem.01835-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KA, Lee PKH, Johnson DR, Zinder SH, Alvarez-Cohen L, 2013. Global gene expression of Dehalococcoides within a robust dynamic TCE-dechlorinating community under conditions of periodic substrate supply. Biotechnology and Bioengineering 110(5), 1333–1341.DOI: 10.1002/bit.24819. [DOI] [PubMed] [Google Scholar]
- Yan J, Im J, Yang Y, Löffler FE, 2013. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philosophical Transactions of the Royal Society B-Biological Sciences 368(1616).DOI: 10.1098/rstb.2012.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Ritalahti KM, Wagner DD, Löffler FE, 2012. Unexpected Specificity of Interspecies Cobamide Transfer from Geobacter spp. to Organohalide-Respiring Dehalococcoides mccartyi Strains. Applied and Environmental Microbiology 78(18), 6630–6636.DOI: 10.1128/aem.01535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YR, McCarty PL, 1998. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environmental Science & Technology 32(22), 3591–3597.DOI: 10.1021/es980363n. [DOI] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S and Madden TL., 2012. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. Bmc Bioinformatics 13DOI: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Bell TH, Champagne J, Maynard C, Tardif S, Tremblay J Greer CW, 2015. Transplanting Soil Microbiomes Leads to Lasting Effects on Willow Growth, but not on the Rhizosphere Microbiome. Frontiers in Microbiology 6DOI: 10.3389/fmicb.2015.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SH, Dolan ME, Semprini L, 2005. Kinetics and inhibition of reductive dechlorination of chlorinated ethylenes by two different mixed cultures. Environmental Science & Technology 39(1), 195–205.DOI: 10.1021/es0496773. [DOI] [PubMed] [Google Scholar]
- Zheng D, Carr CS, Hughes JB, 2001. Influence of Hydraulic Retention Time on Extent of PCE Dechlorination and Preliminary Characterization of the Enrichment Culture. Bioremediation Journal 5(2), 159–168.DOI: 10.1080/20018891079384. [DOI] [Google Scholar]
- Ziv-El M, Delgado AG, Yao Y, Kang DW, Nelson KG, Halden RU, Krajmalnik-Brown R, 2012. Development and characterization of DehaloR boolean AND 2, a novel anaerobic microbial consortium performing rapid dechlorination of TCE to ethene (vol 92, pg 1063, 2011). Applied Microbiology and Biotechnology 95(1), 273–274.DOI: 10.1007/s00253-012-4161-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.