Abstract
Our understanding is limited concerning the tumor immune microenvironment of inflammatory breast cancer (IBC), an aggressive form of primary cancer with low rates of pathologic complete response to current neoadjuvant chemotherapy (NAC) regimens. We retrospectively identified pre-treatment (N = 86) and matched post-treatment tissue (N = 27) from patients with stage III or de novo stage IV IBC that received NAC followed by a mastectomy. Immune profiling was performed including quantification of lymphoid and myeloid infiltrates by immunohistochemistry and T-cell repertoire analysis. Thirty-four of 86 cases in this cohort (39.5%) achieved a pathologic complete response. Characterization of the tumor microenvironment revealed that having a lower pre-treatment mast cell density was significantly associated with achieving a pathologic complete response to NAC (p = 0.004), with responders also having more stromal tumor infiltrating lymphocytes (p = 0.035), CD8+ T cells (p = 0.047), and CD20+ B cells (p = 0.054). Spatial analysis showed close proximity of mast cells to CD8+ T cells, CD163+ monocytes/macrophages, and tumor cells when pathologic complete response was not achieved. PD-L1 positivity on tumor cells was found in less than 2% of cases and on immune cells in 27% of cases, but with no correlation to response. Our results highlight the strong association of mast cell infiltration with poor response to NAC, suggesting a mechanism of treatment resistance and a potential therapeutic target in IBC. Proximity of mast cells to immune and tumor cells may suggest immunosuppressive or tumor-promoting interactions of these mast cells.
Keywords: inflammatory breast cancer, breast cancer, immune microenvironment, mast cells, pathologic complete response
Introduction
Inflammatory breast cancer (IBC) is an aggressive form of the disease characterized by dermal lymphatic blockage by the tumor mass or tumor emboli, leading to the clinical appearance of inflammation on initial presentation (1). Locally advanced IBC is routinely treated with neoadjuvant chemotherapy (NAC) followed by surgery and radiation therapy; however, these patients experience low pathologic complete response (pCR) rates (~15%) (2) and poor survival outcomes compared to stage matched non-IBC cases (3,4). De novo stage IV patients may also receive NAC followed by aggressive local therapy for palliative purposes given the significant breast discomfort associated with IBC; investigations are ongoing into therapeutic benefit of this strategy that emphasizes local disease control (5,6). Unfortunately, targeted genomic profiling has provided limited insight into what distinguishes IBC from non-IBC and molecularly targeted trials have shown limited success.
Immunotherapeutic agents such as monoclonal antibodies targeting immune checkpoints have revolutionized treatment of many types of solid tumors; however, they have shown modest efficacy thus far in breast cancer (7). There remains significant interest in evaluating the role of immune checkpoint blockade in breast cancer, including IBC. Available data have shown that IBC is associated with upregulated inflammatory pathways (e.g. NF-ĸB, JAK/STAT, IL6, COX-2) (8) and that defects in antitumor immunity may contribute to the tumorigenesis and evolution of IBC (9,10). In contrast, cytotoxic T-cell responses and associated exhaustion markers such as PD-L1 have been correlated with favorable outcomes in IBC (11,12). Unfortunately, the majority of published data is based on gene expression profiling. Therefore, there is a critical need for comprehensive morphologic characterization of immunologic aspects of the IBC tumor microenvironment (TME) and the relation to clinical outcomes.
To address this gap in understanding, this study was undertaken to morphologically characterize immunologic aspects of the IBC tumor microenvironment in a prospectively collected cohort of treatment-naïve locally-advanced and de novo stage IV IBC patients, and to correlate these features with pathologic response to NAC. In addition to confirming biomarkers of response previously reported in non-IBC patients (tumor infiltrating lymphocytes, CD8+ T cells), we identified mast cells, a critical regulator of several inflammatory conditions, as strongly associated with poor treatment outcomes. Mast cells represent a potential therapeutic target against which pharmacologic agents already exist, making translation readily accessible for future clinical studies. In addition, these findings provide additional evidence for the ‘inflammatory’ underpinnings of IBC and its poor prognosis.
Methods:
Study Oversight
This study was approved by the MD Anderson Cancer Center Institutional Review Board. The study was conducted in accordance with the ethical principles of the Declaration of Helsinki, and the protocol was conducted with compliance with all relevant ethical regulations. Written informed consent was obtained from all participants.
Cohort and Tissue Selection
Patients with an available hematoxylin and eosin (H&E) slide from pre-treatment breast core biopsy tissue were identified in the MD Anderson IBC Tissue Bank. The MD Anderson IBC Tissue Registry was searched to identify which of these patients had stage III or de novo stage IV disease and received NAC followed by a mastectomy. The immune analyses of de novo stage IV disease were performed on core biopsy tissue from the primary lesion in the breast as opposed to metastatic tissue, facilitating comparisons with stage III disease. Clinicopathologic data was recorded, including age, race, stage, grade, histology, and receptor subtype. Treatment information (including use of chemotherapy, anti-HER2 targeted therapy, and endocrine therapy) and outcome data (including pathologic responses, progression-free survival, and overall survival) were also recorded. Progression-free survival (PFS) was computed from time of surgery to time of relapse if stage III disease or clinical progression as deemed by the treating physician if stage IV disease or death, and overall survival (OS) from time of surgery to time of death. Patients were categorized into responder and non-responder groups for immune profiling (Supplementary Fig. S1). The use of the tissue and clinical data for these studies were covered under a protocol approved by the MD Anderson Institutional Review Board.
Immune Profiling
Tumor-infiltrating lymphocyte assessment
From each case, an H&E slide was assessed for tumor infiltrating lymphocytes (TILs). TILs were enumerated on H&E slides from pre-treatment core biopsies according to the International TIL Working Group consensus guidelines (13). Tumor-infiltrating lymphocytes were quantified by a breast cancer pathologist as percentage of stromal area occupied by mononuclear inflammatory cells based on H&E slides.
Singlet immunohistochemical (IHC) staining and quantification
Immunohistochemistry was performed using an automated stainer (Leica Bond Max, Leica Biosystems) staining for CD8 (Thermo, clone C8/144B, catalog no. MS-457-S, 1:20 dilution) and PD-L1 (pharmDx, clone 22C3, 1:40 dilution). Slides were stained using previously optimized conditions with appropriate positive and negative controls. Leica Bond Polymer Refine detection kit and DAB chromagen were used and counterstained with hematoxylin. Slides were scanned into an Aperio slide scanner (Aperio AT Turbo, Leica Biosystems) and analyzed with Aperio Image Toolbox software for CD8+ cell staining. Regions of invasive tumor and associated stroma were chosen for digital analysis, and quantification of cell density (cells/mm2) was performed. PD-L1 expression was scored as the percentage of positive cells with membranous or cytoplasmic staining pattern (0–100).
Multiplex IHC staining and singlet quantification
Sequential multiplex IHC was performed as previously described (14) using a myeloid panel and a lymphoid panel. Briefly, FFPE tissue sections were deparaffinized, counterstained in hematoxylin, and then the whole slide scanned into an Aperio slide scanner. After blocking with endogenous peroxidase and heat based antigen retrieval, the slides underwent sequential iterative cycles of blocking with 5% normal goat serum / 2.5% BSA / 1x PBS, primary antibody incubation, secondary horseradish conjugated antibody incubation with HistoFine (mouse or rabbit) Simple Stain MAX PO (Nichirei Bioscience Inc) for 30 minutes, and then visualization with 3-amino-9-ethylcarbazole (AEC) detection. The slides were subsequently scanned by Aperio slide scanner after which they were destained with an alcohol gradient, followed by heat-based antibody stripping and antigen retrieval. This cycle was repeated for sequential staining on a single FFPE slide.
The myeloid panel was modified from the original published protocol (14) with the following cycle order, primary antibodies, and incubation times: Hematoxylin (Dako S3301, 1 min), tryptase (AA1, Abcam, 1:20000, blocking 10 min, primary 30 min, AEC 10 min), CD68 (PG-M1, Abcam, 1:50, blocking 10 min, primary 30 min, AEC 10 min), CD8 (C8/144B, Thermo Scientific, 1:25, blocking 20 min, primary 30 min, AEC 25 min), CD163 (10D6, Thermo Scientific, 1:100, blocking 10 min, primary 30 min, AEC 10 min), HLA-DR (SPM288, Novus Biological, 1:100, blocking 10 min, primary 30 min, AEC 10 min), CD3 (SP7, Thermo Scientific, 1:150, blocking 20 min, primary 60 min, AEC 15 min), and cytokeratin (AE1/AE3 1:50, MNF116 1:50, 8&18 1:25, blocking 10 min, primary 30 min, AEC 5 min). The lymphoid panel was modified with the following cycle order, primary antibodies, and incubation times: Hematoxylin (Dako S3301, 1 min), CD3 (SP7, Thermo Scientific, 1:150, blocking 20 min, primary 60 min, AEC 15 min), FoxP3 (236A/E7, eBioscience, 1:40, blocking 10 min, primary 60 min, AEC 20 min), CD8 (C8/144B, Thermo Scientific, 1:25, blocking 20 min, primary 30 min, AEC 25 min), CD20 (L26, Agilent DAKO, 1:300, blocking 20 min, primary 30 min, AEC 10 min), CD56 (123C3, DAKO, 1:100, blocking 10 min, primary 60 min, AEC 15 min), and cytokeratin (AE1/AE3 1:50, MNF116 1:50, 8&18 1:25, blocking 10 min, primary 30 min, AEC 5 min). Singlet quantification of stains was performed as described above.
DNA extraction
Sections from paraffin embedded tissue were reviewed for pathologic diagnosis and dissected if necessary to ensure that ≥90% of the sample represented tumor. Total cellular DNA was isolated from tissue sections using the QIAmp FFPE DNA isolation kit according to the manufacturer’s protocol (Qiagen Inc, Hilden, Germany) following de-paraffinization and proteinase K treatment.
T-cell receptor (TCR) sequencing and analysis
TCR beta chain CDR3 regions were sequenced by immunoSEQ™ (Adaptive Biotechnologies, Seattle, WA), with primers annealing to V and J segments, resulting in amplification of rearranged VDJ segments from each cell. Clonality and richness values were obtained through the Analyzer website. Clonality was measured as 1-(entropy)/log2(# of productive unique sequences), with entropy taking into account clone frequency.
Cell localization and spatial analysis
Spatial localization
As previously described (14), the images acquired from the multiplex IHC staining were aligned by calculation of xy coordinates of fixed structures within each image and then adjustment made through CellProfiler. Using Image J, these images were converted to gray scale and contrast adjusted to discriminate positive staining from background staining. To quantify the spatial proximities of cells exhibiting the queried markers, candidate nuclei in the hematoxylin IHC stained slides were identified using the connected component analysis algorithm (15) with the MATLAB function ‘bwconncomp’ (16). Candidates whose diameters were too small or too large to be a cell (<4 uM and > 16 uM respectively) were rejected, and IHC stains of markers were overlayed over the nuclei candidates to determine which cells were positive for each marker.
Spatial G-function
The spatial G-function was used to quantify infiltration of cells of one type into another (17). This technique was used in non-small cell lung cancer and intraductal papillary mucinous neoplasms to quantify the proximities of immune cell types to cancer cells, and shown to have a significant association with overall survival and risk of tumor progression (18,19). Used previously in modeling spatial statistics in ecology (20), the G-function G(r) is a nearest neighbor distribution function that indicates the probability of a cell of type “i” having at least one cell of type “j” within a distance "r"μm. If we denote all cells of type ‘i’ as Xi and all cells of type ‘j’ as Xj, then the following equation describes the G-function mathematically:
Where, ρ(xi, Xj) = min(∥xi − xj∥2) : xj ∈ Xj) is the minimum distance between a cell xi and cells xj, and Prob(.) indicates the probability distribution function of the quantity within the bracket.
Consequently, this function rises sharply when cells of type “j” and type “i” are clustered together, and sluggishly when the two groups of cells are further away from each other. Thus, the shape of the G-function provides a signature of the infiltration pattern. The G-function can be efficiently and compactly represented by computing (18), the area under the G-function curve (=G-function AUC). The G-function AUC is computed between a distance r = 0 to r = rmax, as a quantitative surrogate of the amount of infiltration of one cell type into another. Various values of rmax between 5 μm to 50 μm were experimented with. The spatial analysis was performed using R’s spatstat package (21).
Statistical Design
Associations between categorical measures were evaluated using the Chi-Square or Fisher’s exact test as appropriate, whereas differences in continuous measures between groups were tested using the Mann-Whitney U test (if only 2 groups) or Kruskal-Wallis test (if more than 2 groups). Correlations between continuous variables were calculated using Spearman’s rho. Fold change in immune markers with treatment were assessed within each response group using one sample Student t test if sample had normal distribution by Shapiro-Wilk’s test or Wilcoxon signed-rank test if sample was not normally distributed, with a null hypothesis of 1. PFS and OS were estimated using the Kaplan-Meier method and differences between groups were assessed using the log-rank test. Comparison of PFS by immune cell infiltration in post-treatment tissue was made between highest and lowest quartiles of immune cell infiltration. All analyses were performed using GraphPad Prism 7. All statistical tests used an alpha value of 0.05 and were two-sided.
Results:
Cohort Characteristics
A total of 86 IBC patients with available pre-treatment H&E stained slides and who underwent mastectomy after NAC were identified in the IBC tumor registry and tissue bank. Baseline demographics, tumor characteristics, treatment regimens, and pathologic outcome data are shown in Table 1. Approximately 70% of cases had stage III disease with the rest having de novo stage IV disease; 90% had pure ductal histology with the remainder largely having mixed histology, and 70% of cases had grade III disease. HER2/neu amplified (HER2+) and hormone receptor positive, HER2/neu non-amplified (HR+/HER2−) breast cancer receptor subtypes each represented 40% of the cohort population, and treatment regimens largely reflected this with 40% receiving anti-HER2 targeted regimens in combination with chemotherapy. During the time period over which this cohort was assembled, a clinical trial of carboplatin and panitumumab in combination with standard anthracycline and taxane based therapy (NCT01036087) was ongoing for patients with triple negative breast cancer (TNBC) or HR+/HER2− disease (22), and 25% of patients in this cohort received this treatment. Approximately 40% of patients experienced a pCR and comparison of responders and non-responders showed that the responder cohort was enriched with stage III patients and HER2+ disease (Table 1). PFS was improved in patients experiencing a pCR in both the full cohort as well as when analyses were limited to patients with stage III disease (Supplementary Fig. S2A-B).
Table 1.
Cohort Characteristics.
| Full cohort (n=86) |
pCR (n=34) |
Non-pCR (n=52) |
p value | |
|---|---|---|---|---|
| Age at Diagnosis, mean years (range) | 50 (23-75) | 48 (23-64) | 51 (27-75) | 0.092 |
| Race/Ethnicity, n (%) | 0.020 | |||
| White | 72 (83.7) | 26 (76.5) | 46 (88.5) | |
| Black | 6 (7.0) | 1 (2.9) | 5 (9.6) | |
| Hispanic | 7 (8.1) | 6 (17.6) | 1 (1.9) | |
| Asian | 1 (0.1) | 1 (2.9) | 0 (0.0) | |
| Stage, n (%) | <0.001 | |||
| Stage IIIB | 32 (37.2) | 12 (35.3) | 20 (38.5) | |
| Stage IIIC | 30 (34.9) | 19 (55.9) | 11 (21.2) | |
| Stage IV | 24 (27.9) | 3 (8.8) | 21 (40.4) | |
| Receptor Subtype, n (%) | 0.019 | |||
| TNBC | 20 (23.2) | 8 (23.5) | 12 (23.1) | |
| HER2+ | 34 (39.5) | 19 (55.9) | 15 (28.8) | |
| HR+/ HER2− | 32 (37.2) | 7 (20.6) | 25 (48.1) | |
| Histology, n (%) | 1 | |||
| Ductal | 76 (88.4) | 30 (88.2) | 46 (88.5) | |
| Lobular | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Mixed | 9 (10.5) | 4 (11.8) | 5 (9.6) | |
| Poorly differentiated | 1 (0.1) | 0 (0.0) | 1 (1.9) | |
| Grade, n (%) | ||||
| I | 0 (0) | 0 (0.0) | 0 (0.0) | 0.358 |
| II | 28 (32.6) | 9 (26.5) | 19 (36.5) | |
| III | 58 (67.4) | 25 (73.5) | 33 (63.5) | |
| Neoadjuvant Treatment Received | 0.004 | |||
| Anthracycline + taxane | 29 (33.7) | 4 (11.8) | 24 (46.2) | |
| Taxane + anti-HER2 +/− anthracycline or carboplatin | 34 (39.5) | 19 (55.9) | 16 (30.8) | |
| Anthracycline + carboplatin + taxane + panitumumab | 21 (24.4) | 10 (29.4) | 11 (21.2) | |
| Other (T-DM1, taxol+avastin) | 2 (2.3) | 1 (2.9) | 1 (1.9) |
pCR, pathologic complete response; TNBC, triple negative breast cancer; HER2+, HER2/neu amplified breast cancer; HR+/HER2-, hormone receptor (estrogen receptor or progesterone receptor) positive, HER2/neu non-amplified breast cancer
Characterization of Immunologic Aspects of the Tumor Microenvironment
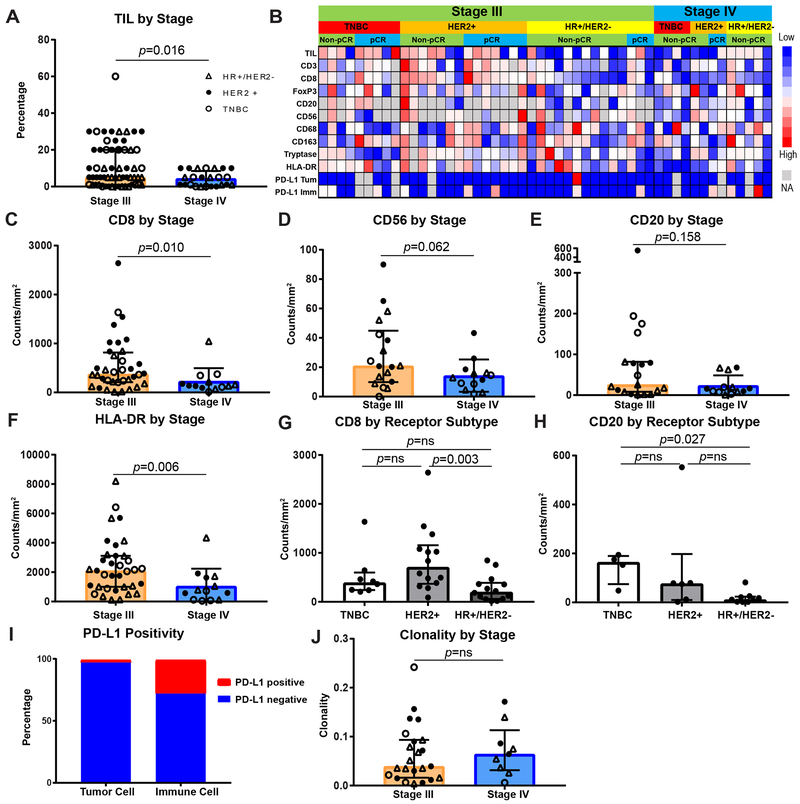
The IBC TME was first interrogated by presenting stage and by breast cancer receptor status (Fig. 1A-J, Supplementary Fig. S3A-Q, Supplementary Fig. S4A-L, Supplementary Fig. S5A-D, Supplementary Fig. S6A-J). Representative H&Es and stains are shown in Supplementary Fig. S3A-Q. Greater TIL frequencies were found in stage III disease compared to de novo stage IV disease (p=0.016) and this was largely driven by the HER2+ and TNBC subtypes (Fig. 1A, Supplementary Fig. S4H). Singlet IHC staining as well as multiplex IHC were performed to further characterize lymphoid and myeloid subsets (Fig. 1B-H, Supplementary Fig. S4A-E). Tumors from patients with stage III disease demonstrated higher lymphoid infiltrates (CD8+, CD56+, and CD20+ cells) as well as HLA-DR, which is often seen on antigen presenting cells as well as activated lymphoid cells compared to de novo stage IV disease (p = 0.010, 0.062, 0.158, and 0.006, respectively). Differences in numbers of CD8+ and HLA-DR+ cells between IBC stages were largely seen in HER2+ and TNBC subtypes; differences in CD56+ cells were predominantly seen in HR+/HER2− disease, and differences in CD20+ cells primarily in TNBC (Supplementary Fig. S4I-L). Density of cells staining double positive for HLA-DR and CD68, CD163, or CD8 did not reveal any particular immune cell subset was accounting for HLA-DR+ cells being overrepresented in stage III disease (Supplementary Fig. S5A-D). Comparisons of receptor subtypes within stage III disease showed that HER2+ disease had significantly higher numbers of CD8+ cells compared to HR+/HER2− disease (p = 0.003), and that TNBC had higher CD20+ cells compared to HR+/HER2− disease (p = 0.027) (Fig. 1G-H, Supplementary Fig. S6A-H).
Figure 1. Stage III IBC has a higher infiltrate of lymphoid cells and MHC class II presentation, whereas hormone receptor positive disease has a lower lymphoid infiltrate.
A, Quantification by TIL percentage of stromal area of primary breast tissue from stage III or de novo stage IV disease (n = 62 stage III and 24 stage IV patients). Receptor subtypes are denoted with triangle indicating hormone receptor positive, HER2/neu non-amplified (HR+/HER2−) disease, closed circle indicating HER2/neu amplified (HER2+) disease, and open circle indicating triple negative breast cancer (TNBC). B, Supervised clustering by stage then receptor status and chemotherapy response of cases that received additional immunohistochemical (IHC) immune assessment (n = 37 stage III and 13 stage IV patients). TIL percentage, CD3, CD8, FoxP3, CD20, CD56, CD68, CD163, tryptase, HLA-DR, and PD-L1 expression on tumor and immune cells are shown, with red indicating higher infiltration and blue lower infiltration. C-F, Quantification of CD8+, CD56+, CD20+, and HLA-DR+ cells by stage (n = 36 stage III and 13 stage IV, 18 stage III and 13 stage IV, 19 stage III and 13 stage IV, 35 stage III and 13 stage IV, respectively). G, H, Quantification of CD8+ and CD20+ cells by tumor receptor status (n = 8 TNBC, 14 HER2+, 14 HR+/HER2+ for CD8+, 4 TNBC, 6 HER2+, 9 HR+/HER2− for CD20+ cells). I, Bar graph representing percentage of cases with PD-L1+ tumors and immune cells (n = 43). J, T-cell receptor clonality by stage (n = 23 stage III and 9 stage IV). A, C-H, J, Bar heights indicate median values, and interquartile ranges are presented in addition to individual data points. All comparisons were made using two-sided Mann-Whitney U test (if 2 groups) or Kruskal-Wallis test with adjustment for multiple testing (if more than 2 groups).
PD-L1 expression at ≥ 1% tumor cell positivity was demonstrated in only 1 of 43 cases (Supplementary Fig. 3E), and this case displayed only 1% staining. In contrast, approximately 30% of immune cells were positive for PD-L1 staining in this cohort but with no differences by stage or receptor subtype (Fig. 1I, Supplementary Fig. S4F).
To further characterize the specificity of the T-cell infiltrate, T-cell repertoire analysis was performed. No differences in T-cell clonality (reactivity) or T-cell richness (diversity) were seen by IBC stage or receptor subtype (Fig. 1J, Supplementary Fig. S4G, Supplementary Fig. S6I-J).
TILs and Mast cells are Associated with Response to Neoadjuvant Chemotherapy
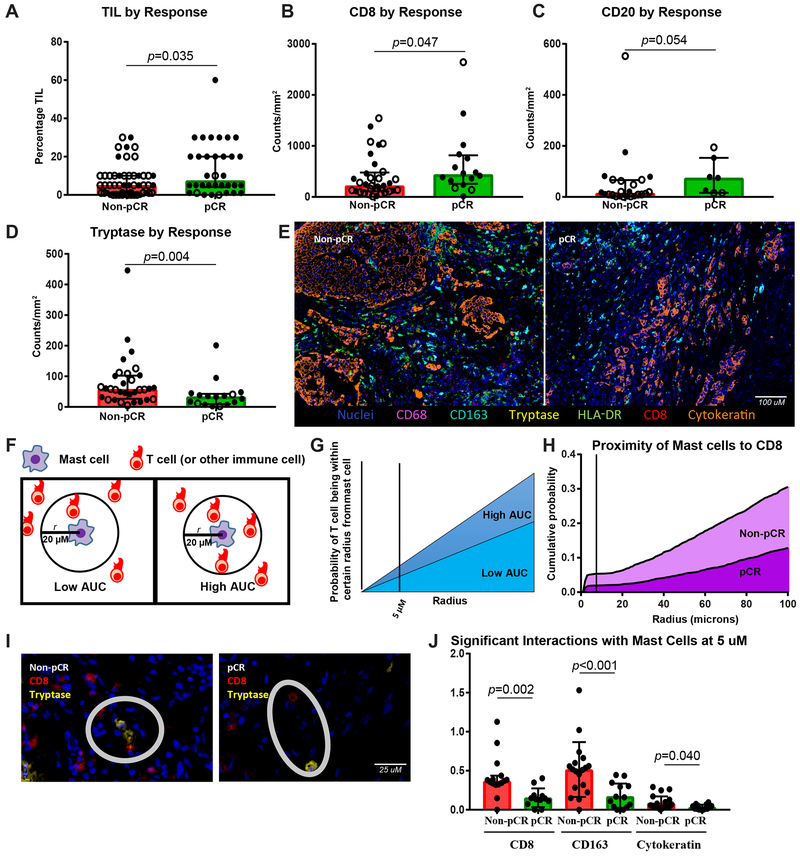
We next asked if immune cell subsets were different between responders, defined as those experiencing pCR, and non-responders (Fig. 2A-G, Supplementary Fig. S7A-J, Supplementary Fig. S8A-M). Consistent with prior reports in non-IBC breast cancer, the presence of TILs was increased in the tumors of patients experiencing pCR (p = 0.035) with higher subsets of CD8+ (p = 0.047) and CD20+ cells (p = 0.054) (Fig. 2A-C).
Figure 2. Mast cells are the strongest immune predictor of poor response to chemotherapy, with higher TILs seen in responders than nonresponders.
A-D, TIL percentage and densities of CD8+, CD20+, and tryptase+ cells by response, with response defined as achieving a pathologic complete response (pCR) or not (non-pCR) (n = 52 non-pCR and 34 pCR, 33 non-pCR and 16 pCR, 24 non-pCR and 7 pCR, 32 non-pCR and 17 pCR, respectively). Closed circle indicates stage III cases, and open circle stage IV cases. E, Representative multiplex immunohistochemistry image of a non-pCR and pCR case with nuclei (blue), CD68 (purple), CD163 (turquoise), tryptase (yellow), HLA-DR (green), CD8 (red), and cytokeratin (orange) stains depicted. F, G, Depiction of methodology for spatial analyses performed. Probabilities of a cell of interest being within a certain radius to another cell of interest were computed and area under the curve (AUC) calculated, with higher AUC indicating closer proximity. H, Non-pCR cases (n = 15) demonstrated higher AUC of mast cells to CD8+ T cells than pCR cases (n=12). I, Representative non-pCR and pCR cases showing distance between CD8+ T cell (red) and tryptase+ mast cell (yellow). J, Comparisons by response of AUC between mast cells and CD8+, CD163+, and cytokeratin+ (tumor) cells (n = 15 non-pCR and 12 pCR, 18 non-pCR and 14 pCR, 21 non-pCR and 14 pCR, respectively). A-D, J, Bar heights indicate median values, and interquartile ranges are presented in addition to individual data points. Comparisons were made using two-sided Mann-Whitney U test.
The immune marker most significantly associated with response was tryptase, a mast cell marker. Numbers of tryptase+ cells were significantly higher in non-responders than responders (p = 0.004) (Fig. 2D). Representative tryptase staining in non-responder and responder cases are shown in Supplementary Fig. S3N-O, respectively. This association was seen in the full cohort, in stage III cases only, and also within each receptor subtype among the stage III cohort (Supplementary Fig. S8G). All other lymphoid and myeloid markers tested as well as PD-L1 and T-cell clonality and diversity metrics were not associated with response.
To further understand what immune interactions may account for this inverse relationship between mast cells and response, spatial assessment was performed on the multiplex myeloid panel (Fig. 2E-G). Focusing on 5 µM, 10 µM, and 20 µM distances between cells, within presumed range of interaction, we found that mast cells were within close proximity to CD8+ T cells, CD163+ macrophages, and tumor cells (as assessed by cytokeratin) in non-responders compared to responders (Fig. 2H-J, Supplementary Fig. S7I-J), suggesting mast cells may be exerting immunosuppressive effects by interaction with these cell types in particular.
Macrophages and MHC class II molecules are associated with improved PFS
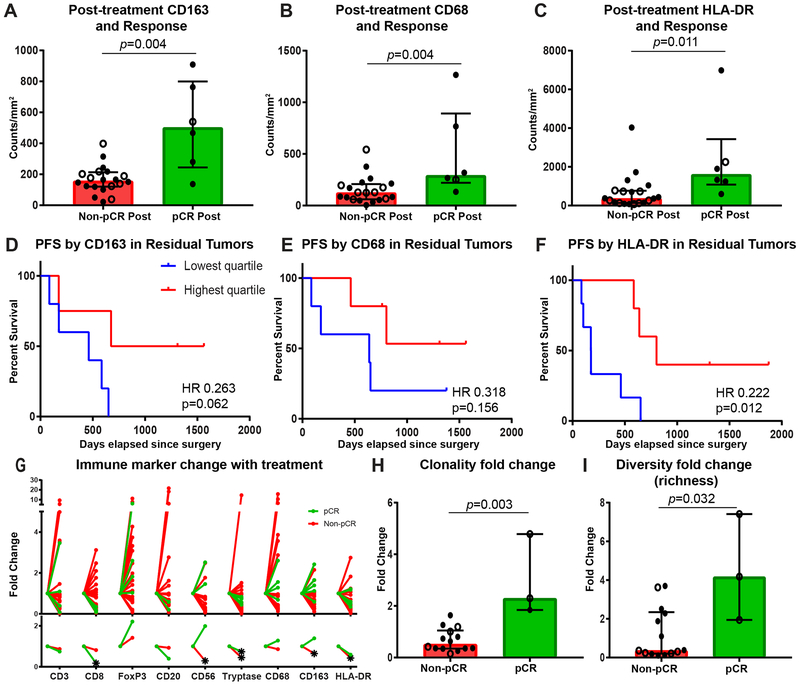
On a subset of cases with available post-treatment tissue from their surgical specimen (n = 27), immune profiling was performed to characterize immune infiltrates in residual tumor as well as areas of treatment effect and to study the change in immune cells with therapy (Fig. 3A-F, Supplementary Fig. S9A-O, Supplementary Fig. S10A-I). Pathologic response, stage, receptor subtype, and treatment characteristics of this cohort are provided in Supplementary Table S1. The cohort included tissue retrieved from the previous tumor bed in 6 patients that experienced a pCR and residual tumor in 21 patients that did not experience a pCR. Analysis of post-treatment tissue showed that myeloid infiltrates including macrophages, assessed by CD163 (p = 0.004) and CD68 (p = 0.004), as well as HLA-DR (p = 0.011), were present in significantly higher densities in the prior tumor bed of cases that achieved a pCR compared to in the residual tumor of non-pCR cases (Fig. 3A-C). In addition, higher infiltration of these cell types was associated with improved PFS (p = 0.062, 0.156, and 0.012 for CD163, CD68, and HLA-DR, respectively) and a trend in improved OS among the non-pCR cases (Fig. 3D-F, Supplementary Fig. S10A-C). FoxP3+ and CD56+ cells were also elevated in pCR mastectomy tissue, whereas lower numbers of FoxP3+ cells were associated with trend in improved OS among non-pCR cases (Supplementary Fig. S9C,E, Supplementary Fig. S10G).
Figure 3. Higher post-treatment myeloid cells are associated with improved outcomes.
A-C, Densities of CD163+, CD68+, and HLA-DR+ cells in the tumor bed at time of mastectomy by response, with response defined as achieving a pathologic complete response (pCR) or not (non-pCR) (n = 20 non-pCR and 6 pCR). D-F, Kaplan-Meier estimates of progression-free survival from time of surgery using two-sided log rank test by residual densities of CD163+, CD68+, and HLA-DR+ cells in residual tumor of non-pCR cases (n = 20). G, Immune marker fold change from baseline to mastectomy tumor bed of non-pCR (red) and pCR (green) cases for density of CD3+ (n=19 non-pCR and 4 pCR), CD8+ (n=20 non-pCR and 5 pCR), FoxP3+ (n=19 non-pCR and 4 pCR), CD20+ (n=15 non-pCR and 2 pCR), CD56+ (n=14 non-pCR and 2 pCR), tryptase+ (n=18 non-pCR and 5 pCR), CD68+ (n=19 non-pCR and 5 pCR), CD163+ (n=19 non-pCR and 5 pCR), and HLA-DR+ (n=19 non-pCR and 5 pCR) cells. Individual data points are depicted in top panel and median fold change on bottom. Star indicates significant fold change from baseline, determined by one sample t test if variable normally distributed or Wilcoxon Signed Rank Test if not normally distributed by Shapiro-Wilk normality test. H, I, T-cell repertoire clonality and diversity fold changes from baseline to mastectomy tumor bed by response (n = 14 non-pCR and 3 pCR). A-C, H, I, Closed circle indicates stage III cases, and open circle indicates stage IV cases. Bar heights indicate median values, and interquartile ranges are presented in addition to individual data points. All comparisons were made using two-sided Mann-Whitney U test except where indicated.
When looking at fold change from pre-treatment biopsy specimens to mastectomy specimens (Fig. 3G), there was a significant decrease in CD8+ cells (p < 0.001) as well as tryptase+ cell density (p = 0.012) with treatment in patients that experienced a pCR. Non-pCR cases also demonstrated a significant decrease in tryptase+ cell density as well as CD56, HLA-DR, and CD163 expression (p = 0.021, 0.049, 0.020, and 0.020, respectively). Fold change by treatment type is shown in Supplementary Fig. S11A, though it is difficult to draw conclusions on differential changes by therapy given the limited size of the cohort and that the pCR cases consisted of 5 patients who received anti-HER2 targeted therapy and 1 that received an EGFR inhibitor along with chemotherapy.
Treatment induces higher clonality as well as higher T-cell diversity in responders
T-cell repertoire analysis using T-cell receptor sequencing overall showed a significant change in repertoire with very low Morisita overlap values in both pCR and non-pCR cases (Supplementary Fig. S11B). Responders showed an increase in both clonality and diversity (richness) compared to non-responder cases (p = 0.003 and 0.032, respectively) (Fig. 3H-I).
Discussion:
This study provides a large and comprehensive characterization of the IBC tumor immune microenvironment that has been performed using reliable methodology such as IHC to quantify immune cell subsets and determine their relationship to response. Mast cell infiltration, as identified by tryptase staining, was significantly associated with poor response to neoadjuvant chemotherapy in all stage and receptor subtypes of IBC and presents a possible therapeutic target.
Mast cells, which are known regulators of allergic and non-allergic inflammatory responses (23), are known to infiltrate breast cancer, but the literature has been inconsistent as to whether they are a favorable (24,25) or poor prognostic indicator (26), with some data suggesting they may play different roles by breast cancer receptor subtype and grade (27,28). This functional heterogeneity of mast cells may be a result of their ability to secrete a wide range of chemokines, growth factors, and other soluble mediators in a context-dependent fashion (29). Downstream effects therefore range from tumor promoting (stimulating angiogenesis / lymphangiogenesis (30,31), extracellular matrix degradation, direct immunosuppression via secretion of TGFβ and IL10, and immune cell recruitment and activation of inhibitory cells such as myeloid derived suppressor cells and T regulatory cells) to tumor inhibiting (direct cytotoxic activity and immune cell recruitment and activation of cytotoxic T lymphocytes and other antitumor immune cells) (32). In this cohort, in which mast cells were inversely associated with pCR across IBC subtypes, we explored potential immune interactions contributing to treatment resistance by analysis of spatial relationships with a multiplex IHC platform. By focusing on distances up to 20 µM (33), we found that mast cells were within range for direct or paracrine interactions with CD8+ T cells, the primary effectors of antitumor responses, as well as CD163+ macrophages and tumor cells. Mast cells may therefore at least partially be exerting their inhibitory effect in IBC through suppressing CD8+ T cells, enhancing immunosuppressive CD163+ macrophages, and directly promoting tumor cell growth. Prior studies support such interactions (34–39). Though this work will need to be validated with mechanistic studies, these data suggest mast cells may be a therapeutic target to enhance responses to NAC, and already existent agents such as mast cell stabilizers and c-Kit inhibitors make this readily translatable. In addition, these findings give further support that inflammation in ‘inflammatory’ breast cancer extends beyond clinical appearance to underlying pathophysiology.
This study also demonstrated known lymphoid biomarkers of response to NAC as seen in non-IBC, including TILs (40), CD8+ T-cell infiltrate (41), and CD20+ B cell infiltrate (42,43), though notably these lymphoid infiltrates were less significantly associated with response compared to mast cell infiltration. Differences were noted in density of these lymphoid infiltrates by stage and by receptor status, with stage IV disease and hormone receptor positive disease having a ‘colder’ TME, therefore perhaps less immunogenic tumors than other subsets, potentially accounting for differential responses to therapy in these IBC subsets. Although lower TIL and specific lymphocyte subsets have previously been seen in matched primary metastases compared to primary breast cancer sites, this current study is unique in comprehensively characterizing primary breast tissue of de novo metastatic disease and showed that a less inflamed TME is seen at the site of the primary tumor and likely precedes development of distant metastases. PD-L1 positivity (≥1%) was seen on tumor cells in only 1 of 43 pre-treatment cases assessed and on immune cells in ~30% of cases. Low PD-L1 positivity, in particular tumor cell positivity, is lower than seen in non-IBC even with the same antibody clone, suggesting this is not a primary mechanism for immune evasion in IBC (7,44–49). Consistent with this, neither tumor nor immune cell PD-L1 positivity was correlated to response in this cohort. This rate of PD-L1 expression is consistent with prior RNA expression based studies in IBC (11), and our IHC methodology additionally provides an accurate quantification of number of cells with positive PD-L1 expression and further subdivides into tumor versus immune cell positivity. Unlike the prior report, our finding that PD-L1 is not predictive of pCR may reflect a more accurate characterization of PD-L1 expression though limited sample size may also be contributing.
Post-treatment tissue assessment revealed greater macrophage infiltration and HLA-DR staining in the prior tumor bed of patients experiencing a complete response to NAC as well as in residual tissue of non-pCR cases that had improved progression-free survival. Interpretation of these findings is confounded by the overrepresentation of anti-HER2 and anti-EGFR targeted treatments combined with NAC in the responders for the post-treatment cases available. Therefore this may reflect that monoclonal antibody–treated patients demonstrate evidence of antibody-dependent phagocytosis, in which macrophages are critical effectors, and highlight the importance of this mechanism in addition to NK cells in mediating improved outcomes (50,51). As expected, higher NK cell infiltration was also seen with therapy in responders. In this limited cohort, no significant relationship was seen between lymphoid infiltration in residual tumor and progression-free survival outcome, though greater numbers of FoxP3+ cells were seen in pCR compared to non-pCR cases, perhaps again reflective of a trastuzumab treatment effect, as has been previously demonstrated (52); among the non-pCR cases, lower FoxP3+ cell infiltration was associated with improved overall survival as previously reported in non-IBC (53). Analysis of change in T-cell repertoire dynamics showed that response was associated with an increase in clonality but also an increase in diversity metrics. This suggests that NAC (and again in this case monoclonal antibody and specifically HER2 targeted therapy) led to an increase in overall tumor reactive T cells as well as an increase in specific expanded T-cell clones accounting for increase in clonality. Similar to what was seen in pre-treatment tissue, our cases were largely PD-L1 negative on tumor cells and positive in õne third of cases on immune cells, neither with correlation to survival outcome. Another study using tissue microarrays of 68 mastectomy IBC cases diagnosed prior to 2004 found that 37% of non-pCR cases were positive for PD-L1 on tumor cells and that this predicted worse survival (54); differences in patient population (earlier stage patients and unknown neoadjuvant treatment characteristics in the other cohort), PD-L1 antibody clone used, and a limited sample size in our post-treatment cohort limit meaningful comparisons between these 2 studies. When comparing pCR and non-pCR cases, differences between cases in the area occupied by tumor cells in the residual tumor bed of non-pCR cases (versus pCR cases for which there are no tumor cells present) may have added bias to our post-treatment results. However, the interspersed nature of IBC infiltration into the tissue made this difficult to account for. Nevertheless, the presence of consistent findings in our study between the pCR versus non-pCR tumor bed analysis and the survival analysis among non-pCR cases lends support for the accuracy and clinical relevance of our findings regarding higher macrophage infiltration and HLA-DR+ cells in post-treatment tissue of responders. Our results will need to be validated in larger datasets with more homogenous populations by stage, receptor status, and associated therapies.
The primary limitation of this study is the relatively small size of the cohort and heterogeneity with respect to stage, receptor status, and associated differences in neoadjuvant treatments given. These mixed classes and treatment statuses therefore limit reliability of pooling data for pre- and post-treatment analyses. However, given the relative rarity of IBC and the comprehensive assessment of primary breast tissue from de novo IBC, the work presented here provides thorough immune characterization of IBC. To ensure accurate and robust conclusions from this data, all analyses were performed for the full cohort as well as within stage and receptor subsets when possible to determine both generalizable and subset specific findings. These will need to be validated in additional IBC cohorts as well as compared to matched non-IBC to better understand the unique pathophysiology of IBC. In addition, with respect to our findings on mast cell infiltration predicting poor response to chemotherapy, mechanistic studies in addition to functional and activation profiling of the immune subsets (such as mast cells and CD8+ T cells) are needed to further validate our findings and identify optimal modalities of therapeutically targeting IBC tumors.
In summary, this study provides a comprehensive immune profiling of IBC from which future studies can build. We showed expected biomarkers of response to NAC such as TILs and CD8+ T-cell infiltrate, which have a known role across solid tumors in shaping response to therapy. We additionally identified mast cells as significantly associated with poor responses to therapy and a potential therapeutic target, highlighting the role of non-lymphoid subsets in IBC and perhaps even more generally the breast cancer TME. Together, these analyses provide an integrated profile of IBC to inform future design of IBC specific clinical trials.
Supplementary Material
Acknowledgments
Financial Support: This research was funded by the Breast Cancer Research Foundation / Conquer Cancer Foundation Young Investigator Award (to SMR), Breast Cancer Research Foundation (to NU), Morgan Welch Inflammatory Breast Cancer Research Program and Clinic, State of Texas Rare and Aggressive Breast Cancer Research Program Grant, and the MD Anderson Cancer Center Support grant (NCI #CA16672). SMR received support from National Institutes of Health T32 Training Grant T32 CA 009666. AR (Reuben) received support from the Kimberley Clark Foundation Award for Scientific Achievement provided by MD Anderson’s Odyssey Fellowship Program. AR (Rao) received support from the CCSG Bioinformatics Shared Resource P30 CA016672, an Institutional Research Grant from The University of Texas MD Anderson Cancer Center, CPRIT RP170719, CPRIT RP150578, NCI R01CA214955–01A1, a Career Development Award from the MD Anderson Brain Tumor SPORE, a gift from Agilent technologies, and a Research Scholar Grant from the American Cancer Society (RSG-16–005-01). EAM received support from the Pink Ribbons Project.
Footnotes
Conflict of Interest Disclosure Statement: VG is an inventor on a US patent application (PCT/US17/53,717) submitted by The University of Texas MD Anderson Cancer Center that covers methods to enhance checkpoint blockade therapy by the microbiome, and is a consultant at MicrobiomeDX and ExpertConnect. JAW has received compensation for speaker’s bureau and honoraria from Dava Oncology, Bristol-Myers Squibb and Illumina and has served on advisory committees for GlaxoSmithKline, Roche/Genentech, Novartis, and AstraZeneca, outside of the submitted work. EAM has received compensation for serving on scientific advisory boards for AstraZeneca, Merck, Roche/Genentech, SELLAS life sciences, and Tapimmune, outside of the submitted work. All other authors declare no competing interests.
References:
- 1.Robertson FM, Bondy M, Yang W, Yamauchi H, Wiggins S, Kamrudin S, et al. Inflammatory breast cancer: the disease, the biology, the treatment. CA Cancer J Clin 2010;60(6):351–75 doi 10.3322/caac.20082. [DOI] [PubMed] [Google Scholar]
- 2.Masuda H, Brewer TM, Liu DD, Iwamoto T, Shen Y, Hsu L, et al. Long-term treatment efficacy in primary inflammatory breast cancer by hormonal receptor- and HER2-defined subtypes. Ann Oncol 2014;25(2):384–91 doi 10.1093/annonc/mdt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouad TM, Kogawa T, Liu DD, Shen Y, Masuda H, El-Zein R, et al. Overall survival differences between patients with inflammatory and noninflammatory breast cancer presenting with distant metastasis at diagnosis. Breast Cancer Res Treat 2015;152(2):407–16 doi 10.1007/s10549-015-3436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Monneur A, Goncalves A, Gilabert M, Finetti P, Tarpin C, Zemmour C, et al. Similar response profile to neoadjuvant chemotherapy, but different survival, in inflammatory versus locally advanced breast cancers. Oncotarget 2017;8(39):66019–32 doi 10.18632/oncotarget.19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss A, Menen RS, Lin HY, Shen Y, Rosso KJ, Shaitelman S, et al. Factors associated with improved outcomes for metastatic inflammatory breast cancer patients. Breast Cancer Res Treat 2018. doi 10.1007/s10549-018-4715-0. [DOI] [PubMed] [Google Scholar]
- 6.Yan Y, Tang L, Tong W, Zhou J. The role and indications of aggressive locoregional therapy in metastatic inflammatory breast cancer. Sci Rep 2016;6:25874 doi 10.1038/srep25874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathan MR, Schmid P. The emerging world of breast cancer immunotherapy. Breast 2018;37:200–6 doi 10.1016/j.breast.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Fouad TM, Kogawa T, Reuben JM, Ueno NT. The Role of Inflammation in Inflammatory Breast Cancer. Adv Exp Med Biol 2014;816:53–73 doi 10.1007/978-3-0348-0837-8_3. [DOI] [PubMed] [Google Scholar]
- 9.Mego M, Gao H, Cohen EN, Anfossi S, Giordano A, Tin S, et al. Circulating tumor cells (CTCs) are associated with abnormalities in peripheral blood dendritic cells in patients with inflammatory breast cancer. Oncotarget 2017;8(22):35656–68 doi 10.18632/oncotarget.10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim B, Woodward WA, Wang X, Reuben JM, Ueno NT. Inflammatory breast cancer biology: the tumour microenvironment is key. Nat Rev Cancer 2018. doi 10.1038/s41568-018-0010-y. [DOI] [PubMed] [Google Scholar]
- 11.Bertucci F, Finetti P, Colpaert C, Mamessier E, Parizel M, Dirix L, et al. PDL1 expression in inflammatory breast cancer is frequent and predicts for the pathological response to chemotherapy. Oncotarget 2015;6(15):13506–19 doi 10.18632/oncotarget.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertucci F, Ueno NT, Finetti P, Vermeulen P, Lucci A, Robertson FM, et al. Gene expression profiles of inflammatory breast cancer: correlation with response to neoadjuvant chemotherapy and metastasis-free survival. Ann Oncol 2014;25(2):358–65 doi 10.1093/annonc/mdt496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 2015;26(2):259–71 doi 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep 2017;19(1):203–17 doi 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shapiro L Connected Component Labeling and Adjacency Graph Construction. Machine Intelligence and Pattern Recognition 1996;19:1–30. [Google Scholar]
- 16.McAndrew A An introduction to digital image processing with matlab notes for scm2511 image processing. . 2004. [Google Scholar]
- 17.Baddeley A Analysing spatial point patterns in R. Technical report C, 2010. Version 4. https://research.csiro.au/software/r-workshop-notes. Accessed 1/25/2018.
- 18.Barua S, Fang P, Sharma A, Fujimoto J, Wistuba I, Rao AUK, et al. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer 2018;117:73–9 doi 10.1016/j.lungcan.2018.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barua S, Solis L, Parra ER, Uraoka N, Jiang M, Wang H, et al. A Functional Spatial Analysis Platform for Discovery of Immunological Interactions Predictive of Low-Grade to High-Grade Transition of Pancreatic Intraductal Papillary Mucinous Neoplasms. Cancer Inform 2018;17:1176935118782880 doi 10.1177/1176935118782880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szmyt J Spatial statistics in ecological analysis: from indices to functions. Silva Fenn 2014;48(1) doi ARTN 100810.14214/sf.1008. [Google Scholar]
- 21.Baddeley A, Turner R. spatstat: An R package for analyzing spatial point patterns. J Stat Softw 2005;12(6):1–42. [Google Scholar]
- 22.Matsuda N, Wang X, Lim B, Krishnamurthy S, Alvarez RH, Willey JS, et al. Safety and Efficacy of Panitumumab Plus Neoadjuvant Chemotherapy in Patients With Primary HER2-Negative Inflammatory Breast Cancer. JAMA Oncol 2018. doi 10.1001/jamaoncol.2018.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Theoharides TC, Alysandratos KD, Angelidou A, Delivanis DA, Sismanopoulos N, Zhang B, et al. Mast cells and inflammation. Biochim Biophys Acta 2012;1822(1):21–33 doi 10.1016/j.bbadis.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajput AB, Turbin DA, Cheang MC, Voduc DK, Leung S, Gelmon KA, et al. Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: a study of 4,444 cases. Breast Cancer Res Treat 2008;107(2):249–57 doi 10.1007/s10549-007-9546-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dabiri S, Huntsman D, Makretsov N, Cheang M, Gilks B, Bajdik C, et al. The presence of stromal mast cells identifies a subset of invasive breast cancers with a favorable prognosis. Mod Pathol 2004;17(6):690–5 doi 10.1038/modpathol.3800094. [DOI] [PubMed] [Google Scholar]
- 26.Amini RM, Aaltonen K, Nevanlinna H, Carvalho R, Salonen L, Heikkila P, et al. Mast cells and eosinophils in invasive breast carcinoma. BMC Cancer 2007;7:165 doi 10.1186/1471-2407-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fakhrjou A, Naghavi-Behzad M, Montazeri V, Karkon-Shayan F, Norouzi-Panahi L, Piri R. The relationship between histologic grades of invasive carcinoma of breast ducts and mast cell infiltration. South Asian J Cancer 2016;5(1):5–7 doi 10.4103/2278-330X.179699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glajcar A, Szpor J, Pacek A, Tyrak KE, Chan F, Streb J, et al. The relationship between breast cancer molecular subtypes and mast cell populations in tumor microenvironment. Virchows Arch 2017;470(5):505–15 doi 10.1007/s00428-017-2103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frossi B, Mion F, Sibilano R, Danelli L, Pucillo CEM. Is it time for a new classification of mast cells? What do we know about mast cell heterogeneity? Immunol Rev 2018;282(1):35–46 doi 10.1111/imr.12636. [DOI] [PubMed] [Google Scholar]
- 30.Mukai K, Tsai M, Saito H, Galli SJ. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol Rev 2018;282(1):121–50 doi 10.1111/imr.12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cimpean AM, Tamma R, Ruggieri S, Nico B, Toma A, Ribatti D. Mast cells in breast cancer angiogenesis. Crit Rev Oncol Hematol 2017;115:23–6 doi 10.1016/j.critrevonc.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Varricchi G, Galdiero MR, Loffredo S, Marone G, Iannone R, Marone G, et al. Are Mast Cells MASTers in Cancer? Front Immunol 2017;8:424 doi 10.3389/fimmu.2017.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A, et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun 2017;8:15095 doi 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jachetti E, Cancila V, Rigoni A, Bongiovanni L, Cappetti B, Belmonte B, et al. Cross-Talk between Myeloid-Derived Suppressor Cells and Mast Cells Mediates Tumor-Specific Immunosuppression in Prostate Cancer. Cancer Immunol Res 2018. doi 10.1158/2326-6066.CIR-17-0385. [DOI] [PubMed] [Google Scholar]
- 35.Wasiuk A, Dalton DK, Schpero WL, Stan RV, Conejo-Garcia JR, Noelle RJ. Mast cells impair the development of protective anti-tumor immunity. Cancer Immunol Immunother 2012;61(12):2273–82 doi 10.1007/s00262-012-1276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jutel M, Watanabe T, Klunker S, Akdis M, Thomet OA, Malolepszy J, et al. Histamine regulates T-cell and antibody responses by differential expression of H1 and H2 receptors. Nature 2001;413(6854):420–5 doi 10.1038/35096564. [DOI] [PubMed] [Google Scholar]
- 37.Nagai Y, Tanaka Y, Kuroishi T, Sato R, Endo Y, Sugawara S. Histamine reduces susceptibility to natural killer cells via down-regulation of NKG2D ligands on human monocytic leukaemia THP-1 cells. Immunology 2012;136(1):103–14 doi 10.1111/j.1365-2567.2012.03565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saleem SJ, Martin RK, Morales JK, Sturgill JL, Gibb DR, Graham L, et al. Cutting edge: mast cells critically augment myeloid-derived suppressor cell activity. J Immunol 2012;189(2):511–5 doi 10.4049/jimmunol.1200647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z, Zhang B, Li D, Lv M, Huang C, Shen GX, et al. Mast cells mobilize myeloid-derived suppressor cells and Treg cells in tumor microenvironment via IL-17 pathway in murine hepatocarcinoma model. PLoS One 2010;5(1):e8922 doi 10.1371/journal.pone.0008922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Denkert C, von Minckwitz G, Darb-Esfahani S, Lederer B, Heppner BI, Weber KE, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol 2018;19(1):40–50 doi 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 41.Seo AN, Lee HJ, Kim EJ, Kim HJ, Jang MH, Lee HE, et al. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br J Cancer 2013;109(10):2705–13 doi 10.1038/bjc.2013.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown JR, Wimberly H, Lannin DR, Nixon C, Rimm DL, Bossuyt V. Multiplexed quantitative analysis of CD3, CD8, and CD20 predicts response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res 2014;20(23):5995–6005 doi 10.1158/1078-0432.CCR-14-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song IH, Heo SH, Bang WS, Park HS, Park IA, Kim YA, et al. Predictive Value of Tertiary Lymphoid Structures Assessed by High Endothelial Venule Counts in the Neoadjuvant Setting of Triple-Negative Breast Cancer. Cancer Res Treat 2017;49(2):399–407 doi 10.4143/crt.2016.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol 2016;34(21):2460–7 doi 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat 2018;167(3):671–86 doi 10.1007/s10549-017-4537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cimino-Mathews A, Thompson E, Taube JM, Ye X, Lu Y, Meeker A, et al. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol 2016;47(1):52–63 doi 10.1016/j.humpath.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rugo HS DJ-P, Im S-A, Ott PA, Piha-Paul SA, Bedard PL, Sachdev J, Le Tourneau C, van Brummelen E, Varga A, Saraf S, Pietrangelo D, Karantza V, Tan A. Preliminary efficacy and safety of pembrolizumab (MK-3475) in patients with PD-L1–positive, estrogen receptor-positive (ER+)/HER2-negative advanced breast cancer enrolled in KEYNOTE-028. [abstract]. In: Proceedings of the Thirty-Eighth Annual CTRC-AACR San Antonio Breast Cancer Symposium: 2015 Dec 8–12; San Antonio, TX. Philadelphia (PA): AACR; Cancer Res 2016;76(4 Suppl):Abstract nr S5–07. [Google Scholar]
- 48.Pelekanou V, Barlow WE, Nahleh ZA, Wasserman B, Lo YC, von Wahlde MK, et al. Tumor-Infiltrating Lymphocytes and PD-L1 Expression in Pre- and Posttreatment Breast Cancers in the SWOG S0800 Phase II Neoadjuvant Chemotherapy Trial. Mol Cancer Ther 2018. doi 10.1158/1535-7163.MCT-17-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou Y, Nitta H, Wei L, Banks PM, Parwani AV, Li Z. Evaluation of Immune Reaction and PD-L1 Expression Using Multiplex Immunohistochemistry in HER2-Positive Breast Cancer: The Association With Response to Anti-HER2 Neoadjuvant Therapy. Clin Breast Cancer 2018;18(2):e237–e44 doi 10.1016/j.clbc.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Fan X, Deng H, Brezski RJ, Rycyzyn M, Jordan RE, et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcgamma receptors on macrophages. J Immunol 2015;194(9):4379–86 doi 10.4049/jimmunol.1402891. [DOI] [PubMed] [Google Scholar]
- 51.Weiskopf K, Weissman IL. Macrophages are critical effectors of antibody therapies for cancer. MAbs 2015;7(2):303–10 doi 10.1080/19420862.2015.1011450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muraro E, Comaro E, Talamini R, Turchet E, Miolo G, Scalone S, et al. Improved Natural Killer cell activity and retained anti-tumor CD8(+) T cell responses contribute to the induction of a pathological complete response in HER2-positive breast cancer patients undergoing neoadjuvant chemotherapy. J Transl Med 2015;13:204 doi 10.1186/s12967-015-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ladoire S, Mignot G, Dabakuyo S, Arnould L, Apetoh L, Rebe C, et al. In situ immune response after neoadjuvant chemotherapy for breast cancer predicts survival. J Pathol 2011;224(3):389–400 doi 10.1002/path.2866. [DOI] [PubMed] [Google Scholar]
- 54.He J, Huo L, Ma J, Zhao J, Bassett RL, Sun X, et al. Expression of Programmed Death Ligand 1 (PD-L1) in Posttreatment Primary Inflammatory Breast Cancers and Clinical Implications. Am J Clin Pathol 2018;149(3):253–61 doi 10.1093/ajcp/aqx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.