Abstract
Motivation
Much effort has been invested in the identification of protein-protein interactions using text mining and machine learning methods. The extraction of functional relationships between chemical compounds and proteins from literature has received much less attention, and no ready-to-use open-source software is so far available for this task.
Method
We created a new benchmark dataset of 2,613 sentences from abstracts containing annotations of proteins, small molecules, and their relationships. Two kernel methods were applied to classify these relationships as functional or non-functional, named shallow linguistic and all-paths graph kernel. Furthermore, the benefit of interaction verbs in sentences was evaluated.
Results
The cross-validation of the all-paths graph kernel (AUC value: 84.6%, F1 score: 79.0%) shows slightly better results than the shallow linguistic kernel (AUC value: 82.5%, F1 score: 77.2%) on our benchmark dataset. Both models achieve state-of-the-art performance in the research area of relation extraction. Furthermore, the combination of shallow linguistic and all-paths graph kernel could further increase the overall performance slightly. We used each of the two kernels to identify functional relationships in all PubMed abstracts (29 million) and provide the results, including recorded processing time.
Availability
The software for the tested kernels, the benchmark, the processed 29 million PubMed abstracts, all evaluation scripts, as well as the scripts for processing the complete PubMed database are freely available at https://github.com/KerstenDoering/CPI-Pipeline.
Introduction
Interactions of biomolecules are substantial for most cellular processes, involving metabolism, signaling, regulation, and proliferation [1]. Small molecules (compounds) can serve as substrates by interacting with enzymes, as signal mediators by binding to receptor proteins, or as drugs by interacting with specific target proteins [2].
Detailed information about compound-protein interactions is provided in several databases. ChEMBL annotates binding affinity and activity data of small molecules derived from diverse experiments [3]. PDBbind describes binding kinetics of ligands that have been co-crystallized with proteins [4]. DrumPID focuses on drugs and their addressed molecular networks including main and side-targets [5]. DrugBank, SuperTarget, and Matador host information mainly on FDA approved but also experimental drugs and related interacting proteins [6, 7]. However, most of this data was extracted from scientific articles. Given that more than 10,000 new articles are added in the literature database PubMed each week, it is obvious that it requires much effort to extract and annotate these information manually to generate comprehensive datasets. Automatic text mining methods may support this process significantly [1, 8]. Today, only a few approaches exist for this specific task. One of them is the Search Tool Interacting Chemicals (STITCH), developed in its 5th version in 2016, which connects several information sources of compound-protein interactions [2]. This includes experimental data and data derived from text mining methods based on co-occurrences and natural language processing [9, 10]. Similar methods have been applied for developing the STRING 11.0 database, which contains mainly protein-protein interactions [11]. OntoGene is a text mining web service for the detection of proteins, genes, drugs, diseases, chemicals, and their relationships [12]. The identification methods contain rule-based and machine learning approaches, which were successfully applied in the BioCreative challenges, e.g. in the triage task in 2012 [13].
Although STITCH and OntoGene deliver broadly beneficial text mining results, it is difficult to compare their approaches, because no exact statistical measures of their protein-compound interaction prediction methods are reported. Furthermore, no published gold standard corpus of annotated compound-protein interactions could be found to evaluate text mining methods for their detection.
Tikk et al. compared 13 kernel methods for protein-protein interaction extraction on several text corpora. Out of these methods, the SL kernel and APG kernel consistently achieved very good results [14]. To detect binary relationships, the APG kernel considers all weighted syntactic relationships in a sentence based on a dependency graph structure. In contrast, the SL kernel considers only surface tokens before, between, and after the candidate interaction partners.
Both kernels have been successfully applied in different domains, including drug-drug interaction extraction [15], the extraction of neuroanatomical connectivity statements [16], and the I2B2 relation extraction challenge [17].
If two biomolecules appear together in a text or a sentence, they are referred to as co-occurring. A comparably high number of such pairs of biomolecules can be used as a prediction method for functional relationship detection, e.g. between proteins or proteins and chemical compounds [9]. This concept can be refined by requiring interaction words such as specific verbs in a sentence [1, 18].
So far, it was unclear whether machine learning outperforms a rather naive baseline relying on co-occurrences (with or without interaction words) for the detection of functional compound-protein relations in texts. Especially for the identification of newly described interactions in texts, that were not described in any other data source, the annotation of a functional relation is challenging.
In this publication, we evaluated the usability of two diverse machine learning kernels for the detection of functional and non-functional compound-protein relationships in texts, independent of additional descriptors, such as the frequency of co-occurrences of specific pairs. Although the ChemProt-benchmark for the training of the classification of functional compound-protein interactions into different groups (e.g. upregulator, antagonist, etc.) was published before [19], this benchmark is focusing on validated interactions and is therefore not suitable for the separation from functionally unrelated compound-protein pairs that are mentioned in texts. To achieve the goal of benchmarking the task of extracting these pairs from literature, we annotated a corpus of protein and compound names in 2,613 sentences and manually classified their relations as functional and non-functional, i.e. no interaction. Furthermore, the kernels were applied to the large-scale text dataset of PubMed with 29 million abstracts. The approaches have been implemented in an easy-to-use open source software available via GitHub.
Both kernels achieved a much better performance on the benchmark dataset than simply using the concept of co-occurrences. These findings imply that a relatively high classification threshold can be used to automatically identify and extract functional compound-protein interactions from publicly available literature with high precision.
Dataset and methods
Generation of the benchmark dataset
Chemical compounds are referred to as small molecules up to a molecular weight of about 1,000 Da, for which a synonym and a related ID are contained in the PubChem database [20]. Similarly, genes and proteins must have UniProt synonyms and were assigned to related UniProt IDs [21].
PubChem synonyms were automatically annotated with the approach described in the manuscripts about the web services CIL [22] and prolific [23], by applying the rules described by Hettne et al. [24]. Proteins were annotated using the web service Whatizit [25]. Synonyms that were assigned wrongly by the automatic named entity recognition approach were manually removed.
The complete compound-protein-interaction benchmark dataset (CPI-DS) was generated from the first 40,000 abstracts of all PubMed articles published in 2009, using PubMedPortable [26].
All pairs of proteins and compounds co-occurring in a sentence are considered as potential functionally related or putative positive instances. Pairs with no functional relation were subsequently annotated as negative instances. Positive instances must be described as directly interacting, up- or down-regulating each other (directly or indirectly), part of each other, or as cofactors of the related proteins. If a named entity exists as a long-form synonym and an abbreviated form in brackets, both terms are considered as individual entities. The result is a corpus of 2,613 sentences containing at least one compound and protein name (CPI-pair).
For further manual annotation, all sentences were transferred to an HTML form. Verbs that belong to a list of defined interaction verbs, defined by Senger et al. [23] and which are enclosed by a protein-compound pair, were annotated, too. The sentences were annotated by 8 different annotators. All pairs were at least proven by two different annotators. If a pair was either classified as “unclear” by one of the annotators or both annotators classified a pair differently, the annotation instructor made the final decision.
Interaction verbs
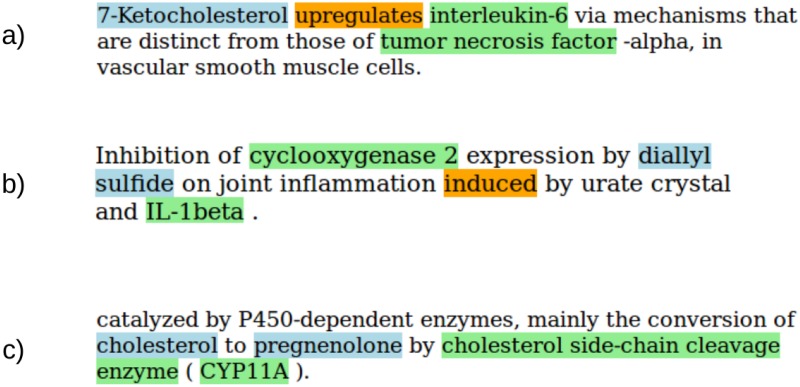
To analyze how much specific verbs, enclosed by a compound and protein name, affect the precision of functional relationships, we further differentiated between sentences with or without this structure. Fig 1 shows detailed examples.
Fig 1.
a) Direct functional relation with interaction verb. The orange-coloured verb is enclosed by the compound 7-ketocholesterol, shown in blue, and the protein interleukin-6, shown in green. The pair was annotated as functional. b) Indirect functional relation with interaction verb. Diallyl sulfide is influencing cyclooxygenase 2 indirectly by inhibiting its expression. The pair was annotated as functional. The compound diallyl sulfide and the protein IL-1beta enclose an interaction verb, but do not describe a functional relation. c) Direct functional relation without interaction verb. The molecule cholesterol is metabolised to pregnenolone by CYP11A. This is indicated by the word conversion. The pair was annotated as functional.
Kernel methods
Shallow linguistic kernel
Giuliano et al. developed this kernel to perform relation extraction from biomedical literature [27]. It is defined as the sum of a global and local context kernel. These customized kernels were implemented with the LIBSVM framework [28]. Tikk et al. adapted the LIBSVM package to compute the distance to the hyperplane, which allowed us to calculate an area under the curve value.
The global context kernel works on unsorted patterns of words up to a length of n = 3. These n-grams are implemented using the bag-of-words approach. The method counts the number of occurrences of every word in a sentence including punctuation, but excludes the candidate entities. The patterns are computed regarding the phrase structures before-between, between, and between-after the considered entities.
The local context kernel considers tokens with their part-of-speech tags, capitalisation, punctuation, and numerals [1, 27]. The left and right ordered word neighborhoods up to window size of w = 3 are considered in two separated kernels, which are summed up for each relationship instance.
All-paths graph kernel
The APG kernel is based on dependency graph representations of sentences, which are gained from dependency trees [29]. In general, the nodes in the dependency graph are the text tokens in the text (including the part-of-speech tag). The edges represent typed dependencies, showing the syntax of a sentence. The highest emphasis is given to edges which are part of the shortest path connecting the compound-protein pair in question.
A graph can be represented in an adjacency matrix. The entries in this matrix determine the weights of the connecting edges. A multiplication of the matrix with itself returns a new matrix with all summed weights of path length two.
All possible paths of all lengths can be calculated by computing the powers of the matrix. Matrix addition of all these matrices results in a final adjacency matrix, which consists of the summed weights of all possible paths [31]. Paths of length zero are removed by subtracting the identity matrix.
All labels are represented as a feature vector. The feature vector is encoded for every vertex, containing the value 1 for labels that are presented within this particular node. This results in a label allocation matrix.
A feature matrix as defined by Gärtner et al. sums up all weighted paths with all presented labels [30]. This calculation combines the strength of the connection between two nodes with the encoding of their labels. In general, it can be stated that the dependency weights are higher the shorter their distance to the shortest path between the candidate entities is [1]. The similarity of two feature matrix representations can be computed by summing up the products of all their entries [31].
In the implementation used here [1, 31], the regularized least squares classifier algorithm is applied to classify compound-protein interactions with the APG kernel. This classifier is similar to a standard support vector machine, but the underlying mathematical problem does not need to be solved with quadratic programming [31, 32].
Analysis of predictive models
Baseline of the kernel models
We considered co-occurrences as a simple approach to calculate the baseline in the way that every appearance of a compound and a protein in a sentence is classified as a functional relationship (recall 100%, specificity 0%), taking into account the number of all true functional relationships.
Benchmark dataset-based analysis
The evaluation was calculated by document-wise 10-fold cross-validation, as an instance-wise cross-validation leads to overoptimistic performance estimates [33]. Each compound-protein pair was classified as functionally related or not related using the previously described kernel methods and resulting in an overall recall, precision, F1 score, and AUC value for a range of kernel parameters. Furthermore, we have performed a nested-cross validation analysis to check if the selection of the hyperparameters has an influence of the overall-performance of both kernels.
Subsequently, the kernels were applied solely to sentences which contain an interaction verb and sentences containing no interaction verb. The analyses compare each of the three baselines with the kernel method results.
Combination of the APG and SL kernel
To analyze if the combination of both kernels yields a higher precision and F1 score than each individual kernel, we combined them by applying majority voting, with the definition that only those relations were classified as functional for which both kernels predicted a functional relation. As we are considering the benchmark dataset, the values for recall, specificity, precision, accuracy, and F1 score could be calculated for the new majority outcome. Based on what is known from the AUC analysis, we can identify the classification threshold for which each of the two kernels reaches the same precision as resulting from the majority voting with default thresholds. By recalculating the above mentioned parameters for each kernel with the new classification threshold, we can compare the single kernel performance with the majority voting outcome.
Large-scale dataset analysis
We applied both kernels on all PubMed abstracts before 2019, including titles. For the annotation of proteins and small molecules the PubTator web service was applied [34, 35]. The web server annotates genes, proteins, compounds, diseases, and species in uploaded texts. Furthermore, it provides all PubMed abstracts and titles as preprocessed data.
Processing these annotations with PubMedPortable [26] in combination with the CPI pipeline, as explained in the GitHub project documentation, allows for a complete automatic annotation of functional compound-protein relations in texts.
Results and discussion
Baseline analysis
Within all sentences, a total number of 5,562 CPI-pairs was curated and separated into 2,931 functionally related (positive instances) and 2,631 non-functionally related CPI-pairs (negative instances). Considering the prediction approach of co-occurrences, this results in a precision (equal to accuracy in this case) of 52.7% and an F1 score of 69.0% (Table 1).
Table 1. Analysis of the CPI benchmark dataset.
| DS | #Sent. | #CPIs | #No-CPIs | Total | Rec. | Spec. | Prec. | F1 |
|---|---|---|---|---|---|---|---|---|
| CPI-DS | 2613 | 2931 | 2631 | 5562 | 100.0 | 0.0 | 52.7 | 69.0 |
Baseline results for precision, recall, and F1 score based on simple co-occurrences. Results are shown in percent (DS—dataset, Sent.—sentences, Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score).
Shallow linguistic kernel
All parameter combinations in the range 1-3 for n-gram and window size of the SL kernel were evaluated. The selection of n-gram 3 and window size 1 shows the best AUC value and the highest precision in comparison to all other models. In general, a lower value of window size leads to a higher precision and a lower recall (Table 2).
Table 2. Shallow linguistic kernel results on the datasets CPI-DS.
| n | w | Rec. | Spec. | Prec. | F1 | AUC |
|---|---|---|---|---|---|---|
| 1 | 1 | 76.6 | 69.8 | 74.0 | 75.2 | 80.7 |
| 1 | 2 | 85.1 | 61.0 | 71.1 | 77.4 | 80.5 |
| 1 | 3 | 87.2 | 56.3 | 69.1 | 77.0 | 80.3 |
| 2 | 1 | 78.5 | 70.8 | 75.1 | 76.7 | 81.8 |
| 2 | 2 | 85.6 | 62.7 | 72.1 | 78.2 | 81.5 |
| 2 | 3 | 87.0 | 59.8 | 70.9 | 78.1 | 81.3 |
| 3 | 1 | 79.5 | 70.2 | 75.0 | 77.2 | 82.5 |
| 3 | 2 | 86.6 | 62.8 | 72.4 | 78.8 | 82.2 |
| 3 | 3 | 87.3 | 60.0 | 71.1 | 78.3 | 82.1 |
| nes. | cr. val. | 82.3 | 66.2 | 73.4 | 77.5 | 82.2 |
The first parameter shows the n-gram value, the second represents the window size. Values in percent: Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score, AUC—area under the curve, nes. cr. val.—nested cross validation.
All-paths graph kernel
We evaluated the APG kernel using the same cross-validation splits as for the SL kernel. Results shown in Table 3 indicate that experiments achieve similar performance independent of the hyperplane optimization parameter c. Mathematically, a larger generalization parameter c represents a lower risk of overfitting [31, 32].
Table 3. APG kernel results on the datasets CPI-DS.
| c | Rec. | Spec. | Prec. | F1 | AUC |
|---|---|---|---|---|---|
| 0.25 | 81.7 | 71.8 | 76.6 | 79.0 | 84.6 |
| 0.50 | 82.7 | 70.2 | 75.8 | 79.0 | 84.6 |
| 1.00 | 81.4 | 72.0 | 76.6 | 78.8 | 84.4 |
| 2.00 | 79.7 | 73.2 | 77.0 | 78.2 | 84.1 |
| nes. cr. val. | 81.7 | 71.7 | 76.4 | 78.9 | 84.5 |
c is hyperplane optimization parameter. Values in percent: Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score, AUC—area under the curve, nes. cr. val.—nested cross validation.
Both kernels in comparison
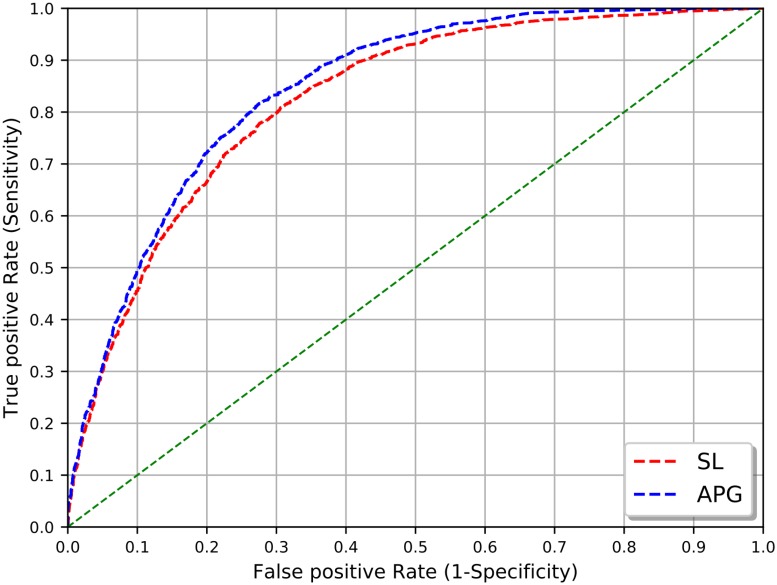
In general, the APG kernel achieved slightly better results than the SL kernel in terms of the resulting AUC value (Fig 2), which is inline with previous findings for other domains, e.g. protein-protein interactions [1]. Considering the models with the highest AUC and precision values for SL (n = 3, w = 1), and APG (c = 0.25) kernel, the results clearly outperform the baseline approach of simple co-occurrences.
Fig 2. SL and APG kernel comparison.
Area under the curve (AUC) of SL kernel (n = 3, w = 1) and APG kernel (c = 0.25).
The complete cross-validation procedure for all parameter settings (including linguistic preprocessing) required almost 5.3 h for the APG kernel and about 34 min for the SL kernel on an Intel Core i5-3570 (4x 3.40GHz). For the APG kernel, a substantial amount of the time is required for dependency parsing. This aspect has to be considered within the scenario of applying a selected model to all PubMed articles (see section Large-scale dataset application).
Both kernels in combination
The combination of both kernels by majority voting yielded a precision of 80.5% and an F1 score of 74.0%. As described in the methods section, the precision was set to the same value (80.5%) for each individual kernel and the appropriate classification threshold was derived from the AUC analysis of each kernel. The resulting F1 score was slightly lower for the APG kernel (73.9%) and considerably lower for the SL kernel (69.4%). This indicates that the combination of both kernels by majority voting leads to a small improvement of the performance (Table 4).
Table 4. Comparison of the combined kernels to each individual kernel.
The precision of each kernel was set to the same level as in the combination by majority voting.
| Kernel | Rec. | Spec. | Prec. | Acc. | F1 |
|---|---|---|---|---|---|
| SL | 61.0 | 83.6 | 80.5 | 71.7 | 69.4 |
| APG | 68.4 | 81.6 | 80.5 | 74.6 | 73.9 |
| Majority voting | 68.5 | 81.6 | 80.5 | 74.7 | 74.0 |
Values in percent: Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score.
Functional relationships with and without an enclosed interaction verb
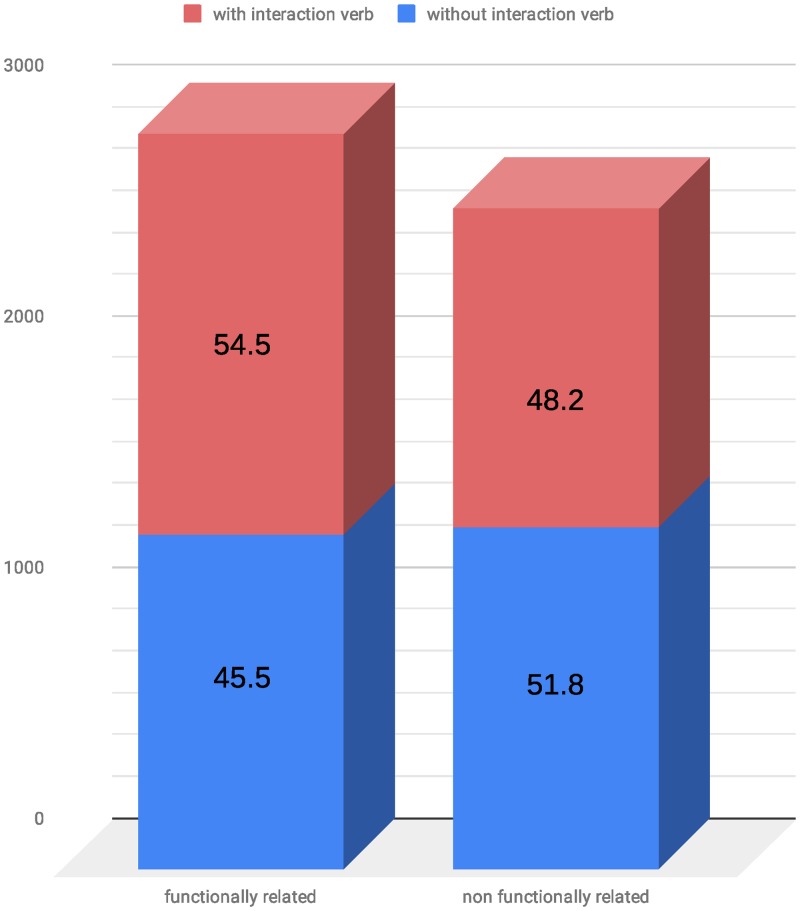
Subsequently, we analyzed the impact of interaction words on the classification of both kernels. The independence of functional relationships and the existence of an interaction verb was tested with a chi-squared test. Both characteristic features are not independent from each other (p<0.01). The fraction of sentences containing an interaction verb is higher in the functionally related CPI-pairs (Fig 3).
Fig 3. Ratios of CPI-pairs in sentences with and without interaction verbs.
To see if and how the two different kernel functions make use of this correlation, we divided the CPI-DS into two groups, considering compound-protein pairs which contain an interaction verb (CPI-DS_IV) and pairs of compounds and proteins that do not show this structure, i.e. no interaction verb enclosed (CPI-DS_NIV). Table 5 shows the baseline results by using simple co-occurrences. In both datasets, the baseline achieves an F1 score of 71.6% and 66.2% for APG and SL kernel respectively. Regarding the analyses of the kernels, we recalculated the results from the complete CPI-DS cross-validation run on CPI-DS_IV and CPI-DS_NIV.
Table 5. Basic statistics of the two compound-protein interaction corpora.
| DS | #Sent. | #CPIs | #No-CPIs | Total | Rec. | Spec. | Prec. | F1 |
|---|---|---|---|---|---|---|---|---|
| CPI-DS_IV | 1209 | 1598 | 1269 | 2867 | 100.0 | 0.0 | 55.7 | 71.6 |
| CPI-DS_NIV | 1404 | 1333 | 1362 | 2695 | 100.0 | 0.0 | 49.5 | 66.2 |
Baseline results for precision, recall, and F1 score are derived by using co-occurrences. Values in percent (DS—dataset, Sent.—sentences, Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score).
Shallow linguistic kernel
For both datasets (CPI-DS_IV and CPI-DS_NIV), the parameter selection n-gram 3 and window size 1 shows the highest area under the curve value (Table 6). Again, a lower value of window size leads to a higher precision and a lower recall. The SL kernel performs slightly better in distinguishing between functional and non-functional relations on dataset CPI-DS NIV but the overall results between the two datasets are very similar.
Table 6. SL kernel results on the datasets CPI-DS_IV and CPI-DS_NIV.
| Param. | CPI-DS_IV | CPI-DS_NIV | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | w | Rec. | Spec. | Prec. | F1 | AUC | Rec. | Spec. | Prec. | F1 | AUC |
| 1 | 1 | 77.5 | 67.4 | 75.7 | 76.5 | 79.4 | 78.7 | 70.7 | 73.1 | 75.6 | 80.6 |
| 1 | 2 | 81.3 | 65.0 | 75.3 | 78.1 | 79.9 | 82.2 | 64.7 | 70.2 | 75.6 | 79.7 |
| 1 | 3 | 80.6 | 64.3 | 74.6 | 77.4 | 79.6 | 84.0 | 63.0 | 69.5 | 75.9 | 79.2 |
| 2 | 1 | 78.1 | 70.0 | 77.3 | 77.6 | 80.8 | 78.4 | 71.7 | 73.9 | 75.9 | 81.7 |
| 2 | 2 | 80.5 | 66.3 | 75.6 | 77.9 | 80.8 | 84.0 | 63.9 | 70.3 | 76.4 | 80.9 |
| 2 | 3 | 80.2 | 65.8 | 75.1 | 77.5 | 80.2 | 85.0 | 63.7 | 70.3 | 76.8 | 80.4 |
| 3 | 1 | 77.9 | 71.1 | 78.0 | 77.8 | 81.3 | 80.3 | 70.5 | 73.4 | 76.6 | 82.5 |
| 3 | 2 | 81.2 | 66.5 | 76.0 | 78.4 | 81.1 | 85.7 | 63.4 | 70.4 | 77.2 | 81.8 |
| 3 | 3 | 80.1 | 66.9 | 75.9 | 77.8 | 80.8 | 86.7 | 62.0 | 69.7 | 77.2 | 81.4 |
The first parameter shows the n-gram value, and the second number represents the window size. Values in percent: Rec.—recall, Spec.—specificity, Prec.—precision, F1—F1 score, AUC—area under the curve.
All-paths graph kernel
Table 7 shows that experiments within the same dataset achieve similar performances, independent of the hyperplane optimization parameter c. For both datasets, the AUC values do not differ by more than 0.6%, indicating a high robustness of the classifier. Furthermore, the AUC values of dataset CPI-DS_IV are about 1-2% better than on dataset CPI-DS_NIV, due to clearly higher recall and precision values. Therefore, the APG kernel performes slightly better in distinguishing between functional and non-functional relations on dataset CPI-DS_IV.
Table 7. CPI-DS_IV and CPI-DS_NIV results for the APG kernel pipeline.
| Param. | CPI-DS_IV | CPI-DS_NIV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| c | Rec. | Spec. | Prec. | F1 | AUC | Rec. | Spec. | Prec. | F1 | AUC |
| 0.25 | 82.8 | 70.0 | 77.9 | 80.1 | 84.4 | 80.4 | 69.4 | 73.4 | 76.4 | 82.2 |
| 0.50 | 82.0 | 70.0 | 78.0 | 79.8 | 84.5 | 77.9 | 70.6 | 73.4 | 75.1 | 82.4 |
| 1.00 | 81.2 | 72.0 | 78.9 | 79.8 | 84.3 | 75.1 | 74.3 | 75.1 | 74.9 | 82.6 |
| 2.00 | 80.5 | 71.3 | 78.7 | 79.4 | 83.8 | 74.9 | 74.2 | 74.7 | 74.6 | 82.8 |
Values in percent: Rec.—recall, Prec.—precision, F1—F1 score, AUC—area under the curve.
Large-scale dataset application
The kernels have been successfully applied to all PubMed titles and abstracts that were published before July 2019, comprising about 29M references to biomedical articles. The dataset consists of more than 120M sentences, with around 16M containing at least one compound-protein pair. Table 8 shows that the APG kernel predicts 54.9% of the potential candidate pairs to be functionally related, while the SL kernel predicts 56.2% as functional. On an Intel Core i5-3570 (4x 3.40GHz), the total run-time of the SL kernel was moderately less than the one of the APG kernel.
Table 8. Application of CPI-Pipeline on PubMed dataset.
| Kernel | APG | SL |
|---|---|---|
| PubMed articles | 29M | |
| Number of sentences | 125M | |
| Number of sentences with candidate pairs | 6.1M | |
| Number of candidate pairs | 14.5M | |
| Functional relations | 7.9M = 54.9% | 8.1M = 56.2% |
| Non-functional relations | 6.5M = 45.1% | 6.3M = 43.8% |
| Number of identical predictions | 11M = 76.6% (43.9% functional, 32.7% non-functional) |
|
| Number of predicted distinct functional relations | 2.1M | |
| Pre-processing elapsed time | 21.0 days | 14.4 days |
| Kernel elapsed time | 4.3 days | 1.6 days |
| Total elapsed time | 25.3 days | 16.0 days |
PubMed dataset statistics for the selected APG (c = 0.25) and SL (n = 3, w = 1) models.
Conclusion
The SL and APG kernels were already applied in different domains, e.g. protein-protein interactions, drug-drug interactions, and neuroanatomical statements. The approach presented herein is focusing on the extraction of functional compound-protein relationships from literature. A benchmark dataset was developed to evaluate both kernels with a range of parameters. This corpus consists of 2,613 sentences, manually annotated with 5,562 compound-protein relationships after automatic named entity recognition. Both kernels are dependent on successfully recognizing protein and compound names in texts. Considering PubMed articles, reliable annotations of these entities are provided by PubTator [34], and publications can be locally processed with PubMedPortable [26].
Cross-validation results with an AUC value of around 82% for the SL kernel and 84% for the APG kernel represent a remarkable performance within the research area of relation extraction.
Considering the filtering of specific interaction verbs, the SL kernel performance is almost the same, while the APG kernel performs slightly better on sentences with an interaction verb. This seems to be intuitive because the APG kernel uses a dependency graph structure and the SL kernel evaluates word neighborhoods. However, since the filtering by interaction verbs does not yield a clearly better precision for both datasets and kernels, this approach can be ignored for the development of an automatic classification method.
The combination of both kernels could slightly increase the overall performance of the classification compared to the single APG kernel. Since both kernels are quite different regarding their classification approach, their combination is supposed to result in a high robustness of the predictions. For fully automatic methods the classification threshold for both kernels can be adjusted to a relatively high precision. The models for predicting functional relationships between compounds and proteins can then be considered e.g. as a filter to decrease the number of sentences a user has to read for the identification of specific relationships.
The selected procedure of training a model with the SL and APG kernels might also come into question for the identification of other types of relationships, such as gene-disease or compound-compound relationships.
Recent work on biomedical relation classification using deep learning approaches outperformed the overall results of kernel methods for related problems [36–43]. We are looking forward to the deep learning achievements on the presented benchmark data set of functionally related and unrelated compound-protein pairs and the comparison to the results of the kernel methods presented here.
Acknowledgments
We want to thank Kevin Selm for modifying the jSRE software implementation of Giuliano et al. to enable parameter selection. We also want to thank Elham Abassian for her support in programming.
Data Availability
The software for the tested kernels, the benchmark, the processed 29 million PubMed abstracts, all evaluation scripts, as well as the scripts for processing the complete PubMed database are freely available at: https://github.com/KerstenDoering/CPI-Pipeline.
Funding Statement
KD received funding for this project by the German National Research Foundation (DFG, Lis45). • AFAM was funded by a doctoral research grant from the German Academic Exchange Service [DAAD, Award No. 91653768] • MG was funded by China Scholarship Council [Award No. 2 201908080143] • JL was supported by the German National Research Foundation [DFG, Research Training Group 1976] and funded by the Baden-Württemberg Foundation [BWST_WSF-043] • PT received funding from the German Research Center for Artificial Intelligence (DFKI). The company DKFI is a non-profit public-private partnership. • Calculations were partly performed on a cluster hosted by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen. The cluster is funded by the state of Baden-Württemberg through bwHPC and the German Research Foundation (DFG) through grant no INST 37/935-1 FUGG. The article processing charge was funded by the German Research Foundation (DFG) and the Albert Ludwigs University Freiburg in the funding programme Open Access Publishing. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Tikk D, Thomas P, Palaga P, Hakenberg J, Leser U. A comprehensive benchmark of kernel methods to extract protein-protein interactions from literature. PLoS Comput Biol. 2010;6:e1000837 10.1371/journal.pcbi.1000837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44(D1):D380–4. 10.1093/nar/gkv1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, et al. The ChEMBL database in 2017. Nucleic Acids Res. 2017;45(D1):D945–D54. 10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang R, Fang X, Lu Y, Yang CY, Wang S. The pdbbind database: Methodologies and updates. J Med Chem. 2005;48(12):4111–9. 10.1021/jm048957q [DOI] [PubMed] [Google Scholar]
- 5. Kunz M, Liang C, Nilla S, Cecil A, Dandekar T. The drug-minded protein interaction database (drumpid) for efficient target analysis and drug development. Database (Oxford). 2016;2016 10.1093/database/baw041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. Drugbank 5.0: A major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–D82. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Günther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, et al. Supertarget and matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36(Database issue):D919–22. 10.1093/nar/gkm862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100(21):12123–8. 10.1073/pnas.2032324100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet. 2006;7(2):119–29. 10.1038/nrg1768 [DOI] [PubMed] [Google Scholar]
- 10. Saric J, Jensen LJ, Ouzounova R, Rojas I, Bork P. Extraction of regulatory gene/protein networks from Medline. Bioinformatics. 2006;22(6):645–50. 10.1093/bioinformatics/bti597 [DOI] [PubMed] [Google Scholar]
- 11. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The string database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D8. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rinaldi F, Clematide S, Marques H, Ellendorff T, Romacker M, Rodriguez-Esteban R. OntoGene web services for biomedical text mining. BMC Bioinformatics. 2014;15 Suppl 14:S6 10.1186/1471-2105-15-S14-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinaldi F, Clematide S, Hafner S, Schneider G, Grigonyte G, Romacker M, et al. Using the OntoGene pipeline for the triage task of BioCreative 2012. Database (Oxford). 2013;2013:bas053 10.1093/database/bas053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tikk D, Solt I, Thomas P, Leser U. A detailed error analysis of 13 kernel methods for protein-protein interaction extraction. BMC Bioinformatics. 2013;14:12 10.1186/1471-2105-14-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas P, Neves M, Rocktäschel T, Leser U. WBI-DDI: Drug-Drug Interaction Extraction using Majority Voting. In: Proceedings of the 7th International Workshop on Semantic Evaluation (SemEval 2013), 2013, pp. 628–635.
- 16. French L, Lane S, Xu L, Siu C, Kwok C, Chen Y, et al. Application and evaluation of automated methods to extract neuroanatomical connectivity statements from free text. Bioinformatics. 2012;28(22):2963–70. 10.1093/bioinformatics/bts542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solt I, Szidarovszky FP, Tikk D. Concept, Assertion and Relation Extraction at the 2010 i2b2 Relation Extraction Challenge using parsing information and dictionaries. In: Proceedings of the 2010 i2b2/VA Workshop on Challenges in Natural Language Processing for Clinical Data, Boston, MA, 2010.
- 18. Kabiljo R, Clegg AB, Shepherd AJ. A realistic assessment of methods for extracting gene/protein interactions from free text. BMC Bioinformatics. 2009;10:233 10.1186/1471-2105-10-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krallinger M, Rabal O, Akhondi SA. Overview of the BioCreative VI chemical-protein interaction Track. In: Proceedings of the sixth BioCreative challenge evaluation workshop (Vol. 1, pp. 141–146).
- 20. Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D9. 10.1093/nar/gky1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D69. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grüning BA, Senger C, Erxleben A, Flemming S, Günther S. Compounds In Literature (CIL): screening for compounds and relatives in PubMed. Bioinformatics. 2011;27(9):1341–2. 10.1093/bioinformatics/btr130 [DOI] [PubMed] [Google Scholar]
- 23. Senger C, Grüning BA, Erxleben A, Döring K, Patel H, Flemming S, et al. Mining and evaluation of molecular relationships in literature. Bioinformatics. 2012;28(5):709–14. 10.1093/bioinformatics/bts026 [DOI] [PubMed] [Google Scholar]
- 24. Hettne KM, Stierum RH, Schuemie MJ, Hendriksen PJ, Schijvenaars BJ, Mulligen EM, et al. A dictionary to identify small molecules and drugs in free text. Bioinformatics. 2009;25(22):2983–91. 10.1093/bioinformatics/btp535 [DOI] [PubMed] [Google Scholar]
- 25. Rebholz-Schuhmann D, Arregui M, Gaudan S, Kirsch H, Jimeno A. Text processing through Web services: calling Whatizit. Bioinformatics. 2008;24(2):296–8. 10.1093/bioinformatics/btm557 [DOI] [PubMed] [Google Scholar]
- 26. Döring K, Grüning BA, Telukunta KK, Thomas P, Günther S. PubMedPortable: A Framework for Supporting the Development of Text Mining Applications. PLoS One. 2016;11(10):e0163794 10.1371/journal.pone.0163794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giuliano C, Lavelli A, Romano L. Exploiting shallow linguistic information for relation extraction from biomedical literature. In: Proc. of the 11st Conf. of the European Chapter of the Association for Computational Linguistics (EACL’06). Trento, Italy: The Association for Computer Linguistics. 2006, pp. 401–408.
- 28. Chang C.-C. & Lin C.-J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011; 2, 1–27. 10.1145/1961189.1961199 [DOI] [Google Scholar]
- 29.Marneffe M, Maccartney B, and Manning C. Generating Typed Dependency Parses from Phrase Structure Parses. In Proceedings of the Fifth International Conference on Language Resources and Evaluation (LREC-2006), Genoa, Italy. European Language Resources Association (ELRA). ACL Anthology Identifier: L06–1260.
- 30.Gärtner T, Flach P, Wrobel S. On graph kernels: hardness results and efficient alternatives. In: Proceedings of 16th annual conference on learning theory, Washington, USA. 2003 pp 129–143.
- 31. Airola A, Pyysalo S, Björne J, Pahikkala T, Ginter F, Salakoski T. All-paths graph kernel for protein-protein interaction extraction with evaluation of cross-corpus learning. BMC Bioinformatics. 2008;9 Suppl 11:S2 10.1186/1471-2105-9-S11-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rifkin R, Yeo G, Poggio T. Regularized least-squares classification. Nato Science Series Sub Series III: Computer and Systems Sciences 190, 131–154. IOS PRESS. (2003).
- 33.Sætre R, Sagae K, Tsujii J. Syntactic features for protein-protein interaction extraction. In Christopher J. O. Baker and Jian Su, editors, LBM 2007, volume 319, pages 6.1–6.14, Singapore. CEUR Workshop Proceedings.
- 34. Wei CH, Harris BR, Li D, Berardini TZ, Huala E, Kao HY, et al. Accelerating literature curation with text-mining tools: a case study of using PubTator to curate genes in PubMed abstracts. Database (Oxford). 2012;2012:bas041 10.1093/database/bas041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wei CH, Kao HY, Lu Z. PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res. 2013;41(Web Server issue):W518–22. 10.1093/nar/gkt441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Islamaj Dogan R, Kim S, Chatr-Aryamontri A, Wei CH, Comeau DC, Antunes R, et al. Overview of the BioCreative VI Precision Medicine Track: mining protein interactions and mutations for precision medicine. Database (Oxford). 2019. January 1;2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y, Lin H, Yang Z, Wang J, Zhang S, Sun Y, et al. A hybrid model based on neural networks for biomedical relation extraction. J Biomed Inform. 2018. May;81:83–92. 10.1016/j.jbi.2018.03.011 [DOI] [PubMed] [Google Scholar]
- 38. Peng Y, Rios A, Kavuluru R, Lu Z. Extracting chemical-protein relations with ensembles of SVM and deep learning models. Database (Oxford). 2018. January 1;2018 10.1093/database/bay073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng Y, Lu Z. Deep learning for extracting protein-protein interactions from biomedical literature. BioNLP. 2017. [Google Scholar]
- 40. Lim S, Kang J. Chemical-gene relation extraction using recursive neural network. Database (Oxford). 2018. January 1;2018 10.1093/database/bay060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Antunes R, Matos S. Extraction of chemical-protein interactions from the literature using neural networks and narrow instance representation. Database (Oxford). 2019. January 1;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corbett P, Boyle J. Improving the learning of chemical-protein interactions from literature using transfer learning and specialized word embeddings. Database (Oxford). 2018. January 1;2018 10.1093/database/bay066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warikoo N, Chang YC, Hsu WL. LPTK: a linguistic pattern-aware dependency tree kernel approach for the BioCreative VI CHEMPROT task. Database (Oxford). 2018. January 1;2018 10.1093/database/bay108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The software for the tested kernels, the benchmark, the processed 29 million PubMed abstracts, all evaluation scripts, as well as the scripts for processing the complete PubMed database are freely available at: https://github.com/KerstenDoering/CPI-Pipeline.