Abstract
HIV-1 CA is involved in different stages of the viral replication cycle, performing essential roles in both early (uncoating, reverse transcription, nuclear import, integration) and late events (assembly). Recent efforts have demonstrated HIV-1 CA protein as a prospective therapeutic target for the development of new antivirals. The most extensively studied CA inhibitor, PF-3450074 (PF-74, discovered by Pfizer), that targets an inter-protomer pocket within the CA hexamer. Herein we reported the design, synthesis, and biological evaluation of a series of 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as HIV-1 CA inhibitors based on PF-74 scaffold. Most of the analogues demonstrated potent antiviral activities, among them, the anti-HIV-1 activity of 6a-9 (EC50 = 3.13 μM) is particularly prominent. The SPR binding assay of selected compounds (6a-9, 6a-10, 5b) suggested direct and effective interaction with recombinant CA proteins. The mechanism of action studies also demonstrated that 6a-9 displays the effects in both the early and late stages of HIV-1 replication. To explore the potential binding mode of the here presented analogues, 6a-9 was analyzed by MD simulation to predict its binding to the active site of HIV-1 CA monomer. In conclusion, this novel series of antivirals can serve as a starting point for the development of a new generation of HIV-1 treatment regimen and highlights the potentiality of CA as a therapeutic target.
Keywords: HIV-1; CA protein; phenylalanine derivatives; 1,2,3-triazole; SPR assay; MD simulation
Graphical Abstract (for review)
1. Introduction
Acquired immunodeficiency syndrome (AIDS) still remains one of the most important public health challenges worldwide. The human immunodeficiency virus type 1 (HIV-1) is the primary etiological agent responsible for AIDS [1]. Highly active antiretroviral therapy (HAART), combining antivirals with distinct targets and modes of action, has made considerable progress and transformed AIDS into a potentially chronic disease [2]. Nevertheless, the long-term treatment of HAART exhibits severe limitations such as the high costs, the development of drug resistance [3, 4], drug-drug interactions, and a variety of side effects due to serious toxicity [5]. As a result, there is still an urgent and desirable requirement for innovative and potent anti-HIV-1 inhibitors acting on novel targets [6].
The HIV-1 capsid (CA) protein plays multiple roles in both early and late events of the viral replication cycle, all of which are essential for optimal infectivity [7, 8]. The principal functions of the HIV-1 CA protein are to encapsidate and protect the enclosed viral ssRNA genomes [7]. The fullerene-shaped conical capsid, composed of the 1500 CA subunits, is the result of a lattice of approximately 250 hexamers interspersed with 12 pentamers [9]. A single CA protein involves N-terminal domain (NTD, residues 1 to 145) and C-terminal domain (CTD, residues 150 to 231), connected through a flexible linker [10]. Specifically, the CA proteins structural stability and integrity have been indicated to be crucial for the processes of disassembly, reverse transcription, nuclear entry, integration, and assembly. Studies have disclosed that CA-CA interactions affect capsid structural integrity and infectivity [11-13], indicating small changes to the capsid being destructive to the above stages and further infectivity of the virus [14, 15]. Accordingly, the HIV-1 CA protein is an attractive unexploited target for HIV-1 therapy [16-18].
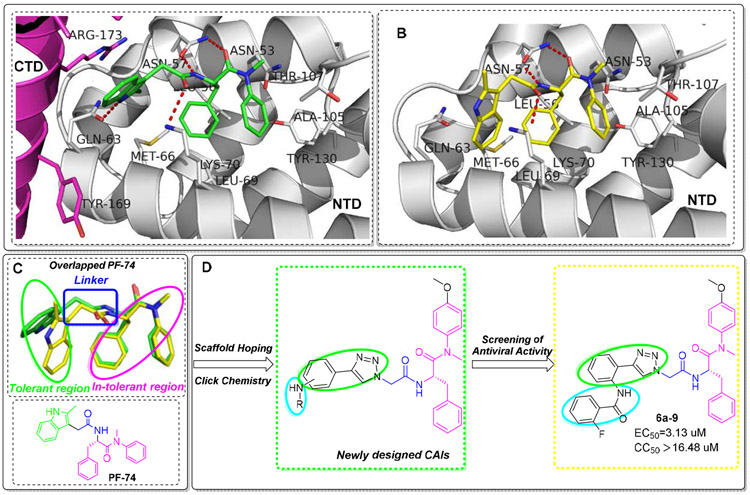
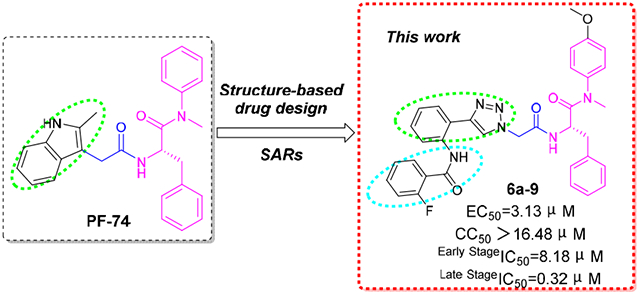
Several chemotypes have been identified as HIV-1 CA inhibitors with distinct binding sites and modes of action [19-29]. The most extensively investigated of these compounds, PF-3450074 (abbreviated to PF-74, discovered by Pfizer, USA) [29], binds to the CA hexamer with more than dozens of times higher affinities compared with isolated or unassembled CA monomer [30]. Nevertheless, the relatively moderate antiviral activity, poor drug-like properties, and especially metabolically lability [19], illustrates the imperative necessity for further structural optimization of PF-74. The X-ray structure of PF-74 in complex with the CA hexamer exhibits that PF-74 sits atop of a preformed binding site enveloping the NTD (of one subunit) – CTD (of the adjacent subunit) interface [11] (Fig.1A), similar to CAPF74 structures (PF-74 complexed with the CA monomer) (Fig.1B) but with the indole moiety arranged in opposite direction than in CAPF74 (Fig.1C). Hence, these co-crystallized structures provide insights into the pharmacophore model of PF-74 (Fig.1C), including an in-tolerant region (phenylalanine scaffold in pink ellipse) and a tolerant region (indole moiety in green ellipse), connected through a linker (in the blue box). In addition, PF-74 shares the same pocket for the host cellular factors NUP153 (nucleoporin 153 kDa) and CPSF6 (cleavage and polyadenylation specific factor 6) [31], suggesting the NTD–CTD interface is very large, and that PF-74 only occupies a small portion of the interface. The flexibility and size of the interface allows sufficient tolerance for further modification with the indole (tolerant) region of PF-74. Our previous work has demonstrated that subtle structural changes to the variable region will have a great impact on the activity [27, 28].
Fig.1.
The co-crystallized structure of PF-74 complexed with CA hexamer (A, PF-74 in green, PDB ID: 5HGL) and monomer (B, PF-74 in yellow, PDB ID: 2XDE), respectively. H-bond interactions are indicated by red dashed lines. Hydrogens (nonpolar) are not shown. Figures were prepared with the PyMOL visualization program (http://www.pymol.org). (C) Chemical structure of PF-74 and overlapped PF-74 conformations from Fig.1 A (PF-74 in green) and B (PF-74 in yellow). (D) The design of novel 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as HIV-1 CA inhibitors.
Owing to the opportunity of abundant crystal structures of PF-74 in complex with HIV-1 CA [10, 30, 32] and its available mechanism of action, in this work, PF-74 was chosen as a lead to focus our attention on its optimization. Considering the asymmetric feature of HIV-1 capsid, it seems that no two CAs are identical in the same capsid [12, 33]. Given the nature of structural pliability and flexibility in the CA, as well as the dynamic process of CA assembly [34], there is a challenge to the rational structure-based drug design. Therefore, the in-depth research of the structure-activity relationships (SARs) of PF-74 derivatives becomes particularly important, especially the diverse modification upon indole moiety (tolerant region). In this study, we choose to retain the privileged (in-tolerant) phenylalanine skeleton (in pink) of the PF-74, while introducing a methoxy group in the para position of the aniline ring which proves to feature more favorable antiviral activity [35, 36] (Fig.1D).
Moreover, in order to further enrich the SAR in this region of our previous work [27], the indole moiety of PF-74 was then replaced with 4-phenyl-1,2,3-triazole (in green ellipse) via scaffold hoping strategy and click chemistry reaction (Fig.1D). In particular, 1,2,3-triazole has been reported to be an important moiety of effective inhibitors of several diseases [37-39], with superior efficacy and diminished toxicity. Thus, exploring this chemical moeity may open new avenues in the treatment of AIDS. Additionally, the SAR of the newly designed compounds were systematically discussed by substituting diversely (in blue ellipse) at different positions on the benzene ring (Fig.1D).
Herein we describe the design, synthesis and biological evaluation of a series of 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as HIV-1 CA inhibitors. The compounds were screened for their antiviral activity in TZM-bl cells, binding was analyzed using surface plasmon resonance (SPR), action stage determination studies were performed, p24 quantification of viral particles was determined and CA assembly was observed in the presence of representative compounds.Moreover, the most potent inhibitor 6a-9 was analyzed by molecular dynamics (MD) simulation to explore the potential binding modes of this series of inhibitors within the CA protein. Finally, the stability assays of 6a-9 were also performed in the presence of human liver microsomes and human plasma, respectively.
2. Results and discussion
2.1. Chemistry
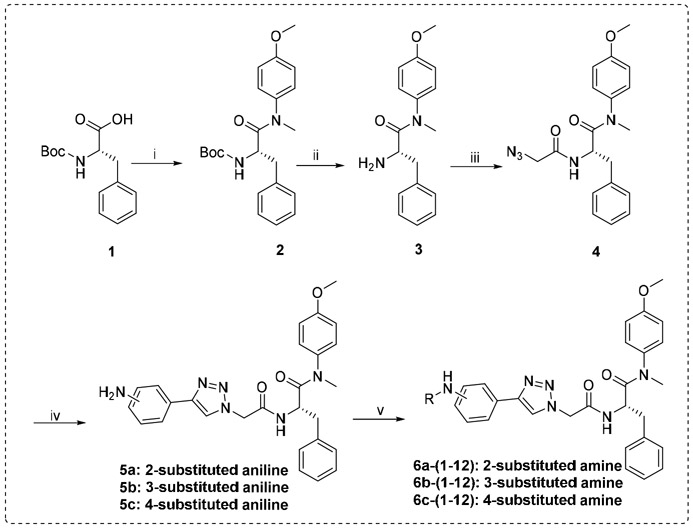
The target compounds were prepared via a concise synthetic route as outlined in Scheme 1. Commercially available (tert-butoxycarbonyl)-L-phenylalanine (1) was treated as the primary starting material to produce all derivatives using well-established methods and treated with 4-methoxy-N-methylaniline and PyBop in DIEA and CH2Cl2 to give 2, followed by removing the Boc protection to produce the free amine 3. Acylation of 3 by reacting with N3CH2COOH in CH3CN solution to produce the azide intermediate 4. This azide scaffold was then reacted with corresponding substituted aminophenylacetylene by CuAAC reaction to obtain correspondingly substituted aniline 5a-5c. Finally, reaction with corresponding acyl chloride to generate the desired compounds 6a-(1-12), 6b-(1-12) and 6c-(1-12).
Scheme 1.
The synthetic route of target compounds. Reagents and Conditions: (i) 4-methoxy-N-methylaniline, PyBop, DIEA, CH2Cl2, 0 °C to r.t.; (ii) CF3COOH, CH2Cl2, r.t.; (iii) N3CH2COOH, HATU, DIEA, CH3CN, 0 ° C to r.t.; (iv) corresponding substituted aminophenylacetylene, L-ascorbic acid sodium salt, CuSO4‧5H2O. THF/H2O (v:v=1:1), r.t.; (v) corresponding acyl chloride, TEA, CH2Cl2, 0 ° C to r.t..
2.2. In vitro anti-HIV assay in TZM-bl cells
All the derivatives were tested for their antiviral activity and cytotoxicity using TZM-bl cells fully infected by HIV-1 NL4.3 virus. EC50 (as measured by a luciferase gene expression assay [40]) and CC50, as well as selectivity index (SI, the ratio of CC50/EC50) values for target compounds 5a-5c, 6a-(1-12), 6b-(1-12) and 6c-(1-12), are shown in Table 1.
Table 1.
Anti-HIV-1 activity and cytotoxicity of the target compounds in TZM-bl cells infected with the HIV-1 NL4-3 virus.
 | ||||
|---|---|---|---|---|
| Compounds | R | EC50a(μM) | CC50b(μM) | SI |
| 5a | H | 3.92 ± 0.72 | >20.64 | >5.27 |
| 6a-1 |  |
3.99 ± 0.65 | >18.99 | >4.76 |
| 6a-2 |  |
3.74 ± 0.70 | >16.99 | >4.54 |
| 6a-3 | 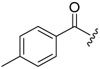 |
>16.59 | >16.59 | |
| 6a-4 | 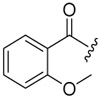 |
8.41 ± 2.42 | >16.16 | >1.92 |
| 6a-5 | 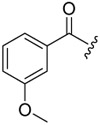 |
12.77 ± 2.91 | >16.16 | >1.27 |
| 6a-6 | 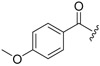 |
>16.16 | >16.16 | |
| 6a-7 | 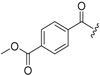 |
>15.46 | >15.46 | |
| 6a-8 | 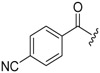 |
>16.30 | >16.30 | |
| 6a-9 | 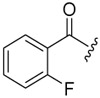 |
3.13 ± 0.91 | >16.48 | >5.27 |
| 6a-10 | 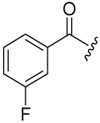 |
3.30 ± 0.63 | >16.48 | >4.99 |
| 6a-11 | 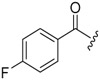 |
3.46 ± 0.59 | >16.48 | >4.76 |
| 6a-12 | 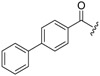 |
>15.04 | >15.04 | |
| 5b | H | 3.30 ± 0.85 | >20.64 | >6.25 |
| 6b-1 | 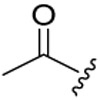 |
14.81 ± 2.66 | >18.99 | >1.28 |
| 6b-2 | 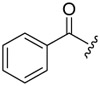 |
12.74 ± 2.72 | >16.99 | >1.33 |
| 6b-3 | 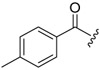 |
>16.59 | >16.59 | |
| 6b-4 | 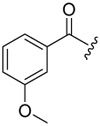 |
>16.16 | >16.16 | |
| 6b-5 | 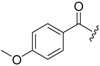 |
>16.16 | >16.16 | |
| 6b-6 | 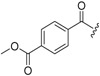 |
>16.16 | >16.16 | |
| 6b-7 | >15.46 | >15.46 | ||
| 6b-8 | 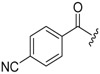 |
12.87 ± 3.10 | >16.30 | >1.27 |
| 6b-9 | 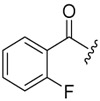 |
13.52 ± 2.47 | >16.48 | >1.22 |
| 6b-10 | 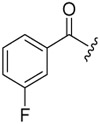 |
9.56 ± 2.31 | >16.48 | >1.72 |
| 6b-11 | 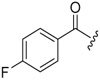 |
>16.48 | >16.48 | |
| 6b-12 | 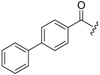 |
>15.04 | >15.04 | |
| 5c | H | 13.62 ± 2.68 | >20.63 | >1.51 |
| 6c-1 | 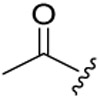 |
>18.99 | >18.99 | |
| 6c-2 | 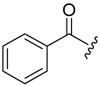 |
12.91 ± 2.89 | >16.99 | >1.32 |
| 6c-3 | 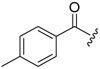 |
>16.59 | >16.59 | |
| 6c-4 | 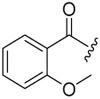 |
>16.16 | >16.16 | |
| 6c-5 | 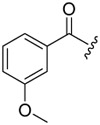 |
>16.16 | >16.16 | |
| 6c-6 | 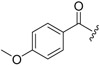 |
>16.16 | >16.16 | |
| 6c-7 | 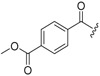 |
>15.46 | >15.46 | |
| 6c-8 | 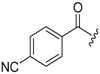 |
12.38 ± 2.28 | >16.30 | >1.32 |
| 6c-9 | 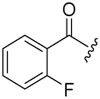 |
11.87 ± 3.13 | >16.48 | >1.39 |
| 6c-10 | 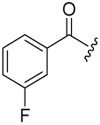 |
12.69 ± 3.46 | >16.48 | >1.30 |
| 6c-11 | 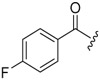 |
13.02 ± 2.97 | >16.48 | >1.27 |
| 6c-12 | 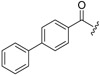 |
12.19 ± 2.56 | >15.04 | >1.23 |
| PF-74 | 0.28 ± 0.06 | >23.50 | >83.93 | |
EC50: the concentration of the compound required to achieve 50% protection of TZM-bl cells against HIV-1-induced cytopathic effect, determined in at least triplicate against HIV-1 in TZM-bl cells.
CC50: the concentration of the compound required to reduce the viability of uninfected cells by 50%, determined in at least triplicate against HIV-1 in TZM-bl cells; values were averaged from at least three independent experiments.
Generally, antiviral results showed that most of the target compounds exhibited from moderate to potent activity against HIV-1 NL4-3 virus with EC50 values ranging from 3.13 μM to 14.81 μM (Table 1). Among them, 6a-9 (EC50 = 3.13 ± 0.91 μM, CC50> 16.48 μM), 6a-10 (EC50 = 3.30 ± 0.63 μM, CC50> 16.48 μM), 6a-11 (EC50 = 3.46 ± 0.59 μM, CC50> 16.48 μM) and 5b (EC50 = 3.30 ± 0.85 μM, CC50> 20.64 μM) were considered to be the most potent HIV-1 inhibitors.
Among the ortho-substituted aniline derivatives (5a, 6a-(1-12)), taking the non-substituted aniline compound 5a (EC50 value of 3.92 μM) as reference, substitution on the NH2- of the benzene ring (5a) with a CH3CO- (6a-1) or PhCO-group (6a-2), no statistically significant difference was found in antiviral activity (EC50 values of 3.99 μM and 3.74 μM, respectively). However, the substitution on the left terminal benzene ring of 6a-2 with different moieties have diverse effects on antiviral activity: Substitution with 2-CH3O- (6a-4, EC50 value of 8.41 μM), 3-CH3O- (6a-5, EC50 value of 12.77 μM), resulted in 2.2 and 3.4-fold decreases in antiviral activity, respectively; while substitution with F- moiety (6a-9, 6a-10, 6a-11), increased activity slightly with EC50 values of 3.13, 3.30 and 3.46 μM, respectively. It proved that the substitution position of F has favorable tolerance on the activity, and the introduction of F can help maintain or even increase potency. In addition, 6a-9 was also found to be the most active compound among the whole newly synthesized series. However, the substitution with 4-CH3- (6a-3, EC50 value of > 16.59 μM), 4-CH3O- (6a-6, EC50 value of > 16.16 μM), 4-CH3OCO- (6a-7, EC50 value of > 15.46 μM), 4-CN- (6a-8, EC50 value of > 16.30 μM) and 4-Ph- (6a-12, EC50 value of >15.04 μM) were considered to be inactive, indicating that these modifications (especially aliphatic and unsaturated moieties) were inappropriate for antiviral activity.
Within the meta-substituted aniline derivatives (5b, 6b-(1-12)), the most active one was the non-substituted aniline 5b (EC50 value of 3.30 μM), substitution on the NH2-of the benzene ring (5b) with a CH3CO- (6b-1, EC50 value of 14.81 μM) or PhCO-group (6b-2, EC50 value of 12.74 μM), leading to 4.5 and 3.9-fold decreases in antiviral activity, respectively. However, the substitutions on the left terminal benzene ring of 6b-2 have significant impacts on antiviral activity: Most of the substitutions (especially aliphatic and aromatic groups) were considered to be detrimental for inhibition, except for 4-CN- (6b-8, EC50 value of 12.87 μM), 2-F- (6b-9, EC50 value of 13.52 μM) and 3-F (6b-10, EC50 value of 9.56 μM) counterparts. 6b-8 and 6b-9 demonstrated comparable potencies with 6b-2, while 6b-10 was 1.3-fold better active with respect to 6b-2. In this group, although the inhibition of the derivatives containing F- (6b-9, 6b-10, 6b-11) were not ideal, 6b-10 demonstrated the best activity among the benzene ring-substituted derivatives (6b-(2-12)), proved that the 2-F- seems to favor the antiviral potency.
Among the para-substituted aniline derivatives (5c, 6c-(1-12)), taking the non-substituted aniline compound 5c (EC50 value of 13.62 μM) as reference, substitution on the NH2- of the benzene ring (5c) with a CH3CO- (6c-1, EC50 value of > 18.99 μM) was considered to be inactive, while substitution with PhCO- group (6c-2, EC50 value of 12.91 μM) exhibited no major changes in antiviral activity. The substitution on the left terminal benzene ring of 6c-2 with 4-CN- (6c-8, EC50 value of 12.38 μM), 2-F-PhCO- (6c-9, EC50 value of 11.87 μM), 3-F-PhCO- (6c-10, EC50 value of 12.69 μM), 4-F-PhCO- (6c-11, EC50 value of 13.02 μM) and 4-Ph-PhCO- (6c-12, EC50 value of 12.19 μM) were found to maintain a same potency level with unmodified counterpart (6c-2). Unfortunately, the remaining substituted benzene derivatives (mainly aliphatic moieties) were observed to be inactive. F- substituted derivatives (6c-9, 6c-10, 6c-11) all demonstrated moderate antiviral activity, indicating that the F- was essential for the inhibition.
The preliminary SARs analysis based on the above results obtained showed that: i) the ortho-substituted aniline derivatives proved to be much more effective than meta- and para- ones; ii) among the whole series, the aliphatic and aromatic substitutions on the left terminal benzene ring were considered to be detrimental for inhibition, for example, the 4-CH3-PhCO-, 4-CH3O-PhCO- substituted aniline derivatives were all found to be inactive across three groups; iii) F- has proven to be a beneficial group for antiviral activities of benzoyl substituted aniline derivatives, exemplified by the best inhibitor 6a-9; iv) 5a, 5b and 5c were compounds with good cell-based activity in their respective groups, indicating that hydrogen-bond donors play an essential role in inhibition and substitutions on -NH2 should be cautious.
2.3. Surface plasmon resonance (SPR) assay on CA protein
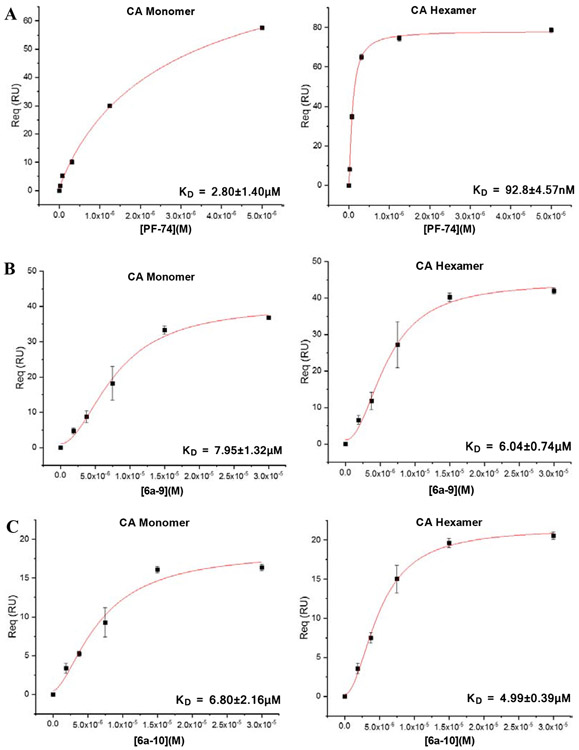
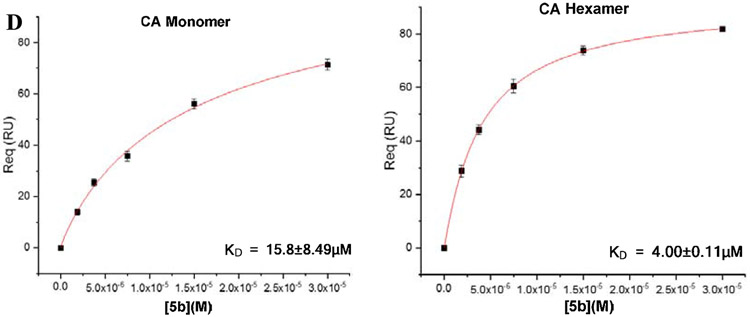
Most of the compounds have demonstrated moderate to potent antiviral activities, we then sought to ascertain whether the newly synthesized compounds bind to the CA protein. In this work, we selected two different CA protein constructs: CA monomer and hexamer, to determine their binding affinity with selected compounds, using an SPR based method as previously reported [19-21]. Among this series, the three most potent antiviral compounds, 6a-9, 6a-10 and 5b, were chosen to determine their direct interactions with the two CA constructs. In this assay, PF-74 serves as an in-line control to ensure the validity of experimental data. The SPR binding results are shown in Fig.2.
Fig. 2.
SPR isotherms of target compounds 6a-9 (B), 6a-10 (C), 5b (D) and PF-74 (A) binding to CA proteins (monomer left, hexamer right), respectively.
As shown in Fig.2B and Fig.2C, both 6a-9 and 6a-10 displayed no significant differences in the equilibrium dissociation constant (KD) values for hexameric and monomeric CA. However, the binding affinity of compound 5b to the hexamer is approximately four times higher compared to the monomer, with KD values of 4.00 and 15.8 μM, respectively (Fig.2D). In addition, the KD values of three newly synthesized compounds binding to CA hexamer are close to each other, this is consistent with the trend of their antiviral activity level. It is worth noting that the KD values of four tested compounds to CA monomer are all in the micromolar level, however, as depicted in Fig.2A, PF-74 demonstrated the tightest interaction and highest selectivity and specificity to CA hexamer, with KD value for CA hexamer of 92.8 nM, suggesting that the achievement of potent antiviral activities of these inhibitors depends on their binding to CA hexamer. In conclusion, these results indicated that this series of inhibitors have high affinities to CA protein and preferentially bind to the hexameric CA, consistent with the performance of PF-74.
2.4. Determination of the action stage of 6a-9
The above SPR results showed that these inhibitors demonstrated high affinities with CA proteins, characterizing their target engagement. Given that CA exerts essential roles in both early and late stages, we then sought to determine which stage(s) of the HIV-1 lifecycle can be inhibited by these inhibitors. Therefore, the modular nature of the single-round infection assay was performed herein to achieve this goal, the infective HIV-1 particles are generated recombinantly in 293T cells, and next utilized to infect U87.CD4.CCR5 cells: early and late stages of the HIV-1 lifecycle can be separated effectively [41]. In this assay, the most potent representative compound 6a-9 was chosen to focus the mechanisms of action studies on.
As depicted in Table 2, the dual-stage inhibition profile of 6a-9 is in accordance with the parental drug PF-74. Specifically, 6a-9 demonstrated the moderate antiviral activities in the early event of the HIV-1 lifecycle (IC50 = 8.18 μM), while in the late stage, the potent inhibitory activity of the 6a-9 is almost the same as that of PF-74 with IC50 values of 0.32 and 0.23 μM, repectively. These results combined with the above SPR data indicated that 6a-9 possibly prefer binding to one of the drug targets of PF-74 being the assembled capsid to exert its inhibitory activity.
Table 2.
Early and late stage infection results of control PF-74 and 6a-9 against HIV-1NL4-3 single-round infectious pseudoviruses.
| Compound | Early Stage IC50 (μM) |
Late Stage IC50 (μM) |
|---|---|---|
| PF-74 | 0.056 ± 0.017 | 0.23 ± 0.17 |
| 6a-9 | 8.18 ± 1.80 | 0.32 ± 0.11 |
2.5. Quantification of capsid (p24) content on 6a-9
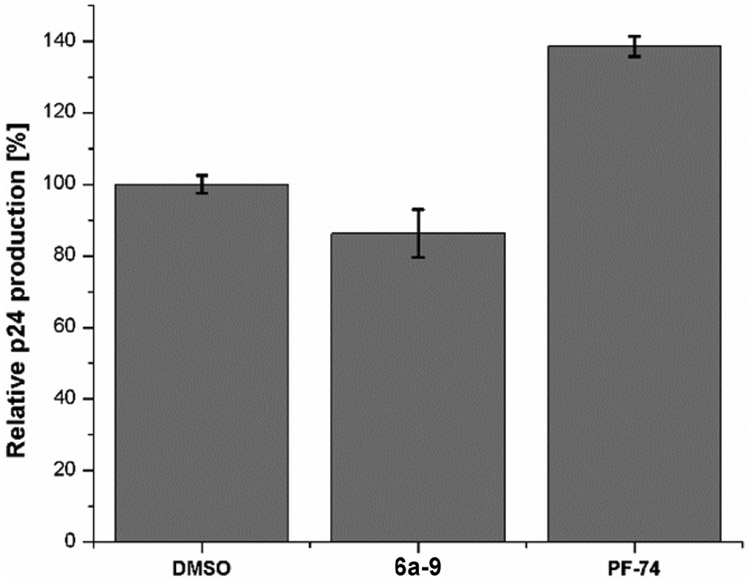
Having demonstrated that 6a-9 displays a more potent inhibition upon the late stage of HIV-1 replication, we then attempted to determine its mechanism of action. Firstly, we decided to test the amount of virus (p24/CA) produced in the presence of the compound. In this assay, p24 quantification of the virus produced in the presence of 10 μM of 6a-9 was performed by lysing the virus with a detergent. The p24 was then captured to an anti-p24 antibody coated ELISA plate and calorimetrically quantified (with a secondary antibody) and compared to a virus produced in the presence of PF-74.
As depicted in Fig.3, the p24 quantification of the produced virus in the presence of 6a-9 showed only a slight reduction (about 15%) of p24 compared to the DMSO control. No obvious inhibition nor stimulation of CA assembly was observed for 6a-9, while PF-74 stimulates the CA assembly to make the virus non-infectious by dramatic effects on the morphology of the immature virion. This assay may also provide evidence that the target of 6a-9 is the assembled capsid.
Fig.3.
The effect of compound 6a-9 on viral production. Standard deviation represents 2 replicates performed in duplicate. Performed with NL4-3 Env pseudo-typed HIV-I virus at 10 μM compound concentration.
2.6. In vitro capsid assembly assay in the presence of 6a-9
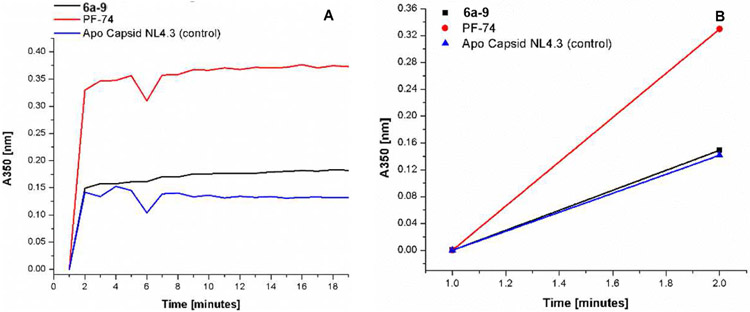
Since the few changes in the p24 content of 6a-9 are not significant enough to be considered a part of the mechanism of action, we next sought to look at the effects of 6a-9 on the in vitro assembly of the HIV-1 CA. As shown in Fig.4, PF-74 accelerated the assembly of HIV-1 CA in the in vitro assay as compared to DMSO control, while 6a-9 neither accelerates nor reduces CA assembly. This could explain the almost unchanged amount of virions produced (p24/CA) in the presence of 6a-9.
Fig.4.
The effect of 6a-9 on the NL4-3 capsid assembly in vitro at 3M NaCl. (A) Capsid assembly was monitored by an increase in turbidity using a spectrophotometer at 350 nm over 19 minutes. Capsid was used at a final concentration of 30 μM, and compounds 6a-9 and PF-74 at a final concentration of 50 μM. (B) Slope/velocity quantification of capsid assembly during the first 2 minutes. Experiments were performed in triplicate. (AU) Absorption unit.
Taking together the results of p24 content and CA assembly assay on 6a-9, we can now speculate that 6a-9 performed its inhibitory effect by binding the assembled CA to alter the entire morphology of the conical CA core in the virus, hence the inhibition of the virus in the late stages (IC50 = 0.32 μM). These preliminary mechanism-of-action studies will definitely lay the foundation for more in-depth research with these and higher potency inhibitors.
2.7. Molecular dynamics (MD) simulation on 6a-9
For a better interpretation of SAR of 6a-9, considered the best CA inhibitor of the series, 6a-9 was simulated for 1 μs to find its binding to the active site of HIV-1 CA monomer using the software Autodock 4.2.6 using default settings [42].
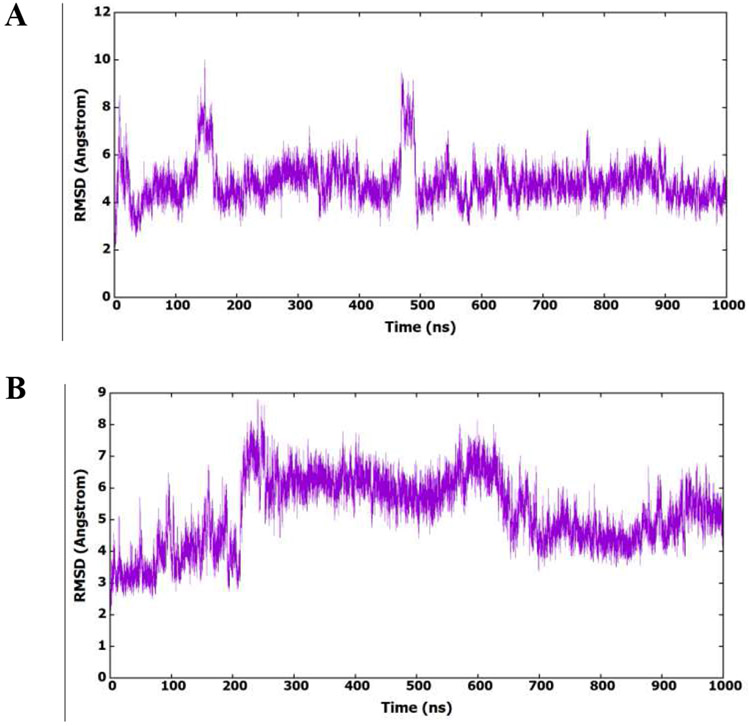
Fig.5A shows the root mean square deviation (RMSD) of amino acids (heavy atoms) during the simulation. The figure shows that the protein structure exists in different conformational ensembles with a highly abundant conformation. The presence of the protein in different conformational forms could be accompanied with different binding modes of the inhibitor. The RMSD of 6a-9 was calculated and plotted in Fig.5B to find its conformational existence and binding to the capsid protein. It is clear from the figure that 6a-9 exists in different conformational forms which shows different binding modes to the active site.
Fig.5.
(A) RMSD (heavy atoms) of amino acids of CA HIV-1 monomer in reference to the first frame of the MD simulation. (B) RMSD (heavy atoms) of the bound 6a-9 in reference to the docked conformer.
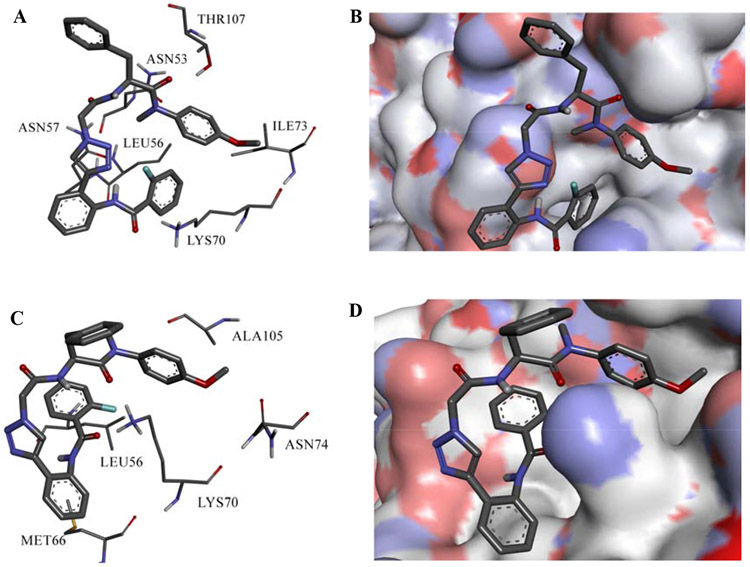
Results of RMSD of the protein and the inhibitor show that the inhibitor binds with different modes, therefore the entire trajectory has been clustered based on 6a-9 (no fit). Clustering resulted in four different structural clusters with two most populated. Fig.6A and C show representative structure interactions of the first (57.6%) and second (13.0%) clusters respectively: expanded views for 6a-9 binding to the active site of both clusters are represented in Fig.6B and D. According to the clustering results, it is clear that 6a-9 has different binding modes. The binding of 6a-9 to the CA monomer is similar to that of PF-74 (the prototype inhibitor) and 13m (our previously synthesized inhibitor [27]), where the core scaffold is oriented to the inside of the active site and the substituent is oriented to the outside of the active site.
Fig.6.
Binding interactions of 6a-9 in the first (A) and second (C) clusters. Expanded views of the representative structures of the first (B) and second (D) clusters.
The phenyl ring of the core region of 6a-9 forms hydrophobic interaction with LYS70 in the second cluster binding mode, and it could form an ion-induced dipole interaction with the positively charged nitrogen atom of LYS70. This is in contrast to the binding mode in the first cluster where the phenyl ring does not form any contact with the protein. Furthermore, 6a-9 forms aliphatic-aromatic hydrophobic interactions with LEU56 with its fluorobenzene ring in the second cluster similar to the binding of PF-74. LEU56 is far from binding to the aromatic ring in the most populated cluster. 6a-9 could form hydrophobic interactions with ILE73 in the first cluster and ALA105 in the second cluster. There is no proper hydrogen bond formation with the methoxy group: however, an aliphatic hydrogen bond could be formed between the methoxy group and LYS70 backbone in the first cluster and ASN74 backbone in the second cluster. The phenyl ring next to the triazole ring forms hydrophobic interactions with MET66 in the second cluster and no interaction of this ring in the first cluster binding mode. Finally, 6a-9 forms a hydrogen bond with THR107 in the first cluster binding mode and with LYS70 in the second binding mode. However, hydrogen bond analysis showed very low frequency of forming hydrogen bonds. Results of MD simulation analysis shows that the binding is weak in the most predominant binding mode, however more interactions are present in the less populated cluster.
2.8. Metabolic stabilities in the presence of human liver microsomes and human plasma
The metabolic stability of 6a-9 and PF-74 was measured in a human liver microsomes (HLM) assay. Testosterone, diclofenac, and propranolol were used as control with moderate metabolic stability. As shown in Table 3, PF-74 was rapidly metabolized with a half-life of 0.6 min, while t1/2 of 6a-9 was 0.9 min. CLint(liver) of 6a-9 was also much lower than that of PF-74.
Table 3.
Metabolic stability assay in human liver microsomes
| HLM (Final concentration of 0.5 mg protein/mL) | ||||||
|---|---|---|---|---|---|---|
| Sample | R2 a | T1/2b (min) |
CLint(mic)c (μL/min/mg) |
CLint(liver)d (mL/min/kg) |
Remaining (T=60min) |
Remaining (NCFe=60min) |
| 6a-9 | 1.0000 | 0.9 | 1559.4 | 1403.5 | 0.3% | 122.1% |
| PF-74 | 1.0000 | 0.6 | 2403.0 | 2162.7 | 0.0% | 95.4% |
| Testosterone | 0.9239 | 7.4 | 188.3 | 169.4 | 3.6% | 74.4% |
| Diclofenac | 0.9924 | 5.6 | 248.2 | 223.4 | 0.5% | 95.9% |
| Propafenone | 0.9616 | 5.4 | 257.7 | 231.9 | 0.1% | 97.9% |
R2 is the correlation coefficient of the linear regression for determination of the kinetic constant.
T1/2 is half-life and CLint(mic) is the intrinsic clearance.
CLint(mic) = (0.693/half-life)/mg microsome protein per mL.
CLint(liver) = CLint(mic) × mg microsomal protein/g liver weight × g liver weight/kg body weight.
NCF: no cofactor. No NADPH regenerating system was added to the NCF sample (replaced by buffer) during the 60 min incubation. If the remaining amount is less than 60%, then non-NADPH dependent reaction occurs.
The stabilities of 6a-9 and PF-74 were also evaluated in a human plasma assay. Propantheline bromide was tested for control. As shown in Table 4, 6a-9 remained intact (measured as 107.4% of the initial amount) after incubation for 120 min, and it also showed better stability than PF-74 (85.6% after 120 min).
Table 4.
Result of human plasma stability assay
| Compound | Batch | Time Point | % Remaininga |
|---|---|---|---|
| (min) | Human | ||
| 6a-9 | / | 0 | 100.0 |
| 10 | 91.3 | ||
| 30 | 94.6 | ||
| 60 | 86.9 | ||
| 120 | 107.4 | ||
| PF-74 | / | 0 | 100.0 |
| 10 | 87.3 | ||
| 30 | 83.4 | ||
| 60 | 86.5 | ||
| 120 | 85.6 | ||
| Propantheline bromide | ODR-4547 | 0 | 100.0 |
| 10 | 47.7 | ||
| 30 | 9.1 | ||
| 60 | 0.6 | ||
| 120 | 0.1 |
% remaining = 100 × (PAR at appointed incubation time/PAR at time T0). PAR is the peak area ratio of test compound to internal standard. Accuracy should be within 80–120% of the indicated value.
3. Conclusions
In this study, we have designed, synthesized, and evaluated a novel series of 4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as HIV-1 CA inhibitors based on PF-74 skeleton. Most of the analogues demonstrated potent antiviral activities. Among them, the anti-HIV activity of 6a-9 (EC50 = 3.13 μM) is particularly prominent. The SPR binding assay demonstrated that this novel series of inhibitors could form direct and tight interactions with recombinant CA proteins. Inspired by its high efficiency, SPR can be utilized as a rapid screening method to discover potent CA inhibitors in due course. The action stage determination studies also demonstrated that 6a-9 displays the dual-stage inhibition profile with the more potent inhibition in the late stage of HIV replication than early stage. The mechanism of action studies (p24/CA content and CA assembly assay) on 6a-9 showed no obvious changes in p24 content or assembly rate compared to DMSO control. Taking together the above results, we can now speculate that 6a-9 performed its inhibitory effect by binding the assembled CA to disturb the normal morphology of the conical CA core in the virus, thus inhibiting the late event of the virus. To predict the potential binding patterns of this kind of analogues, we also conducted MD simulation on 6a-9, results showed that the binding of 6a-9 to the CA is similar to that of PF-74, however, the calculated binding affinity is relatively weak in the most predominant binding mode, providing a reasonable explanation for its weaker antiviral activity than PF-74. The human plasma and liver microsomes stability assays also demonstrated that the stability of 6a-9 was slightly improved compared with the PF-74.
Given the nature of CA structural pliability and flexibility as well as the dynamic process of CA assembly [34], a dynamical perspective on inhibitory mechanisms of PF-74 derivatives should be adopted, thus representing a barrier for rational structure-based drug design. In the circumstances, the in-depth study of the SARs becomes particularly essential. In this work, the systematic analyses of SARs on newly synthesized compounds are expected to offer beneficial guidance for further rational optimization. We firmly believe that this novel series of capsid inhibitors can act as a starting point for the advancement of a next generation of HIV-1 treatment regimen and highlights the potentiality of CA as a therapeutic target. The research of development on the PF-74 scaffold is still in progress within our team to find CA inhibitors with enhanced potency and better drug-like properties.
4. Experimental Section
4.1. Chemistry
1H NMR and 13C NMR spectra were recorded on a Bruker AV-400 spectrometer using solvents as indicated (DMSO-d6, Methanol-d4). Chemical shifts were reported in δ values (ppm) with tetramethylsilane (TMS) as the internal reference, and J values were reported in hertz (Hz). Melting points (mp) were determined on a micromelting point apparatus and were uncorrected. TLC was performed on Silica Gel GF254 for TLC (Merck) and spots were visualized by iodine vapor or by irradiation with UV light (λ = 254 nm). Flash column chromatography was performed on column packed with Silica Gel60 (200-300 mesh).
Thin layer chromatography was performed on pre-coated HUANGHAI_HSGF254, 0.15-0.2 mm TLC-plates. Solvents were of reagent grade and were purified and dried by standard methods when necessary. Concentration of the reaction solutions involved the use of rotary evaporator at reduced pressure. The solvents of CH2Cl2, Et3N and methanol etc. were obtained from Sinopharm Chemical Reagent Co., Ltd (SCRC), which were of AR grade. The key reactants including 4-methoxy-N-methylaniline, N-(tert-Butoxycarbonyl)-L-phenylalanine, 2-azidoacetic acid etc. were purchased from Bide Pharmatech Co. Ltd.
4.1.1. Tert-butyl(S)-(1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)carb amate (2)
A solution of (tert-butoxycarbonyl)-L-phenylalanine (1, 2.90 g, 10.93 mmol, 1.5 eq.) in 20 mL dichloromethane was added PyBop (5.69 g, 10.93 mmol, 1.5 eq.) at 0°C, and the mixture stirred for 0.5 h. Subsequently, DIEA (3.61mL, 21.87 mmol, 3 eq.) and 4-methoxy-N-methylaniline (1.0 g 7.29 mmol, 1 eq.) were added to the mixture and then stirred at room temperature for another 6 h (monitored by TLC). The resulting mixture was evaporated under reduced pressure and the residue was initially washed by 1N HCl and extracted with ethyl acetate (3 × 20 mL). Then, the combined organic layer was washed with saturated sodium bicarbonate (3 × 20 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford corresponding crude product, which was purified by flash column chromatography to afford intermediate 2 as yellow oil with a yield of 88%. 1H NMR (400 MHz, DMSO-d6) δ 7.22 (d, J = 8.3 Hz, 2H, Ph-H), 7.20 – 7.11 (m, 3H, Ph-H), 7.09 (d, J = 8.2 Hz, 1H, NH), 7.03 (d, J = 8.6 Hz, 2H, Ph-H), 6.79 (d, J = 7.3 Hz, 2H, Ph-H), 4.27 – 4.06 (m, 1H, CH), 3.81 (s, 3H, OCH3), 3.13 (s, 3H, NCH3), 2.75 (dd, J = 13.4, 3.8 Hz, 1H, PhCH), 2.61 (dd, J = 13.3, 10.3 Hz, 1H, PhCH), 1.30 (s, 9H, C(CH3)3). 13C NMR (100 MHz, DMSO-d6) δ 172.22 (C=O), 158.98, 155.75 (C=O), 138.53 (2×C), 136.12 (2×C), 129.28 (2×C), 128.47 (2×C), 126.70, 115.21 (2×C), 78.33, 55.94, 53.55, 37.86, 37.07, 28.65 (3×C). ESI-MS: m/z 385.4 (M+1), 407.5 (M+23). C22H28N2O4 [384.5]
4.1.2. (S)-2-amino-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (3)
Trifluoroacetate (3.86 mL, 52.02 mmol, 5.0 eq.) was added dropwise to intermediate 2 (4.0 g, 10.40 mmol, 1.0 eq.) in 30 mL dichloromethane and stirred at room temperature for 1 h. Then, the resulting mixture solution was alkalized to pH ~ 7 with saturated sodium bicarbonate solution, and then extracted with dichloromethane (40 mL). Then, the combined organic layer was washed with saturated sodium bicarbonate (3 × 20 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford corresponding crude product 3 as yellow oil with the yield of 80%. 1H NMR (400 MHz, DMSO-d6) δ 7.29 – 7.13 (m, 3H, Ph-H), 7.03 – 6.75 (m, 6H, Ph-H), 3.77 (s, 3H, OCH3), 3.44 – 3.35 (m, 1H, CH), 3.06 (s, 3H, NCH3), 2.75 (dd, J = 12.8, 6.7 Hz, 1H, PhCH), 2.45 (dd, J = 12.9, 7.1 Hz, 1H, PhCH), 1.87 (s, 2H, NH2). 13C NMR (100 MHz, DMSO-d6) δ 174.89 (C=O), 158.75, 139.00, 136.35, 129.51 (2×C), 128.93 (2×C), 128.47 (2×C), 126.55, 115.04 (2×C), 55.85, 53.35, 42.19, 37.45. ESI-MS: m/z 285.05 (M+1). C17H20N2O2 [284.36].
4.1.3. (S)-2-(2-azidoacetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (4)
N3CH2COOH (40 mg, 0.40 mmol, 1.2 eq.) and HATU (188 mg, 0.50 mmol, 1.5 eq.) were mixed in15 mL CH3CN and stirred in an ice bath for 1 h. Then, the intermediate 3 (94 mg, 0.33 mmol, 1 eq.) and DIEA (109 μL, 0.66 mmol, 2 eq.) were added to the above solution slowly at 0 °C. The reaction system was then stirred at room temperature for additional 12 h. The solvent was removed under reduced pressure and then saturated sodium bicarbonate solution (20 mL) was added, extracted with dichloromethane (20 mL). The combined organic layer was washed with 1N HCl (10 mL), the resulting organic layer was then washed with saturated salt water, dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to afford corresponding crude product, which was purified by flash column chromatography to provide compound 4 as yellow oil with the yield of 39%. 1H NMR (400 MHz, Methanol-d4) δ 7.27 – 7.13 (m, 3H, Ph-H), 7.11 – 6.59 (m, 6H, Ph-H), 4.67 (t, J = 7.4 Hz, 1H, NH), 3.82 (s, 3H, OCH3), 3.81 – 3.74 (m, 1H, CH), 3.31 (dt, J = 3.1, 1.6 Hz, 2H, N3CH2), 3.16 (s, 3H, NCH3), 2.97 (dd, J = 13.3, 7.0 Hz, 1H, PhCH), 2.73 (dd, J = 13.3, 7.9 Hz, 1H, PhCH). 13C NMR (100 MHz, Methanol-d4) δ 171.05 (C=O), 167.29 (C=O), 158.67, 135.77, 134.23, 128.05 (2×C), 127.49, 127.30 (2×C), 125.75 (2×C), 113.66 (2×C), 53.79, 51.03, 50.30, 36.99, 36.05. ESI-MS: m/z 368.3 (M+1), 390.3 (M+23), 406.5 (M+39). C19H21N5O3 [367.4],
4.1.4. General procedure for the synthesis of 5a-5c
The key intermediate 4 (110 mg, 0.30 mmol, 1 eq.), corresponding substituted aminophenylacetylene (42 mg, 0.36 mmol, 1.2 eq.), L-ascorbic acid sodium salt (7.5 mg, 0.03 mmol, 0.1 eq.), CuSO4‧5H2O (17.8 mg, 0.09 mmol, 0.3 eq.) were dissolved in the solution of tetrahydrofuran/water (v:v = 1:1, 6 mL). The resulting mixture was stirred at room temperature for 12 h. Then the reaction mixture was extracted with dichloromethane (20 mL), and the combined organic phase was washed with saturated salt water (20 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to give the corresponding crude target product, which was purified by flash column chromatography to afford product 5a-5c.
4.1.4.1. (S)-2-(2-(4-(2-aminophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (5a).
White solid, yield: 83%. mp: 94-95°C. 1H NMR (400 MHz, DMSO-d6) δ 8.90 (d, J = 7.6 Hz, 1H, NH), 8.33 (s, 1H, triazole-H), 7.42 (d, J = 7.5 Hz, 1H, Ph-H), 7.29 – 7.14 (m, 3H, Ph-H), 7.14 – 6.98 (m, 3H, Ph-H), 6.91 (dd, J = 16.8, 7.5 Hz, 4H, Ph-H), 6.76 (d, J = 8.1 Hz, 1H, Ph-H), 6.59 (t, J = 7.3 Hz, 1H, Ph-H), 6.16 (s, 2H, NH2), 5.25 – 4.99 (m, 2H, triazoleCH2), 4.46 (q, J = 8.1 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.2, 4.7 Hz, 1H, PhCH), 2.69 (dd, J = 13.1, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.40 (C=O), 159.02, 147.54, 145.05, 137.68, 135.78, 129.33 (2×C), 129.10 (2×C), 128.88, 128.70 (2×C), 127.93, 127.03, 123.22, 116.40, 116.25, 115.10 (2×C), 113.02, 55.85, 52.10, 51.85, 37.87, 37.77. ESI-MS: m/z 485.5 (M+1), 507.4 (M+23). C27H28N6O3 [484.6].
4.1.4.2. (S)-2-(2-(4-(3-aminophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (5b).
White solid, yield: 87%. mp: 90-91°C. 1H NMR (400 MHz, DMSO-d6) δ 8.88 (d, J = 7.8 Hz, 1H, NH), 8.20 (s, 1H, triazole-H), 7.29 – 7.14 (m, 3H, Ph-H), 7.13 – 7.02 (m, 4H, Ph-H), 6.91 (dd, J = 17.6, 8.2 Hz, 5H, Ph-H), 6.52 (d, J = 7.6 Hz, 1H, Ph-H), 5.17 (s, 2H, PhNH2), 5.13 (d, J = 16.4 Hz, 1H, triazoleCH), 5.02 (d, J = 16.2 Hz, 1H, triazoleCH), 4.45 (q, J = 8.2 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.10 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 4.8 Hz, 1H, PhCH), 2.69 (dd, J = 13.2, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.20 (C=O), 165.53 (C=O), 159.01, 149.51, 147.15, 137.69, 135.79, 131.59, 129.81, 129.33 (2×C), 129.11, 128.71 (2×C), 127.03, 122.90, 115.09 (2×C), 113.97, 113.36, 110.80 (2×C), 55.85, 52.10, 51.73, 37.85, 37.77. ESI-MS: m/z 485.5 (M+1), 507.5 (M+23). C27H28N6O3 [484.6]
4.1.4.3. (S)-2-(2-(4-(4-aminophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (5c).
White solid, yield: 48%. mp: 105-106°C. 1H NMR (400 MHz, DMSO-d6) δ 8.86 (d, J = 7.8 Hz, 1H, NH), 8.07 (s, 1H, triazole-H), 7.47 (d, J = 8.4 Hz, 2H, Ph-H), 7.27 – 7.13 (m, 3H, Ph-H), 7.06 (d, J = 6.6 Hz, 2H, Ph-H), 6.93 (d, J = 9.0 Hz, 2H, Ph-H), 6.91 – 6.82 (m, 2H, Ph-H), 6.60 (d, J = 8.4 Hz, 2H, Ph-H), 5.23 (s, 2H, NH2), 5.09 (d, J = 16.3 Hz, 1H, triazoleCH), 4.98 (d, J = 16.3 Hz, 1H, triazoleCH), 4.45 (td, J = 8.5, 5.5 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.10 (s, 3H, NCH3), 2.91 (dd, J = 13.6, 5.2 Hz, 1H, PhCH), 2.68 (dd, J = 13.4, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.58 (C=O), 159.01, 149.03, 147.48, 137.69, 135.80, 129.33 (2×C), 129.10 (2×C), 128.69 (2×C), 127.02, 126.55 (2×C), 121.07, 118.78, 115.09 (2×C), 114.39 (2×C), 55.85, 52.06, 51.71, 37.87, 37.77. ESI-MS: m/z 485.5 (M+1), 507.4 (M+23), 523.5 (M+39). C27H28N6O3 [484.6].
4.1.5. General procedure for the synthesis of target compounds 6a-(1-12), 6b-(1-12), 6c-(1-12)
The compound 5a-5c (0.12 g, 0.25 mmol, 1eq.), TEA (69 μL, 0.50 mmol, 2 eq.), corresponding acyl chloride (0.37 mmol, 1.5 eq.) were dissolved in the solution of dichloromethane (10 mL) under ice cooling. The resulting mixture was stirred at room temperature for 10 h. Then the reaction mixture was extracted with dichloromethane (10 mL), and the combined organic phase was washed with saturated salt water (10 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to give the corresponding crude target product, which was purified by flash column chromatography to afford product 6a-(1-12), 6b-(1-12), 6c-(1-12).
4.1.5.1. (S)-2-(2-(4-(2-acetamidophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (6a-1).
White solid, yield: 47%. mp: 95-96°C. 1H NMR (400 MHz, DMSO-d6) δ 10.71 (s, 1H, PhNH), 8.95 (d, J = 7.8 Hz, 1H, NH), 8.47 (s, 1H, triazole-H), 8.10 (d, J = 8.1 Hz, 1H, Ph-H), 7.77 (d, J = 7.6 Hz, 1H, Ph-H), 7.33 (t, J = 7.4 Hz, 1H, Ph-H), 7.29 – 7.15 (m, 4H, Ph-H), 7.14 – 7.01 (m, 2H, Ph-H), 6.93 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.6 Hz, 2H, Ph-H), 5.23 (d, J = Hz, 1H, triazoleCH), 5.12 (d, J = 16.3 Hz, 1H, triazoleCH), 4.54 – 4.38 (m, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.3 Hz, 1H, PhCH), 2.12 (s, 3H, COCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 168.89 (C=O), 165.29 (C=O), 159.03, 145.45, 137.66, 135.86, 135.78, 129.32 (2×C), 129.10 (2×C), 128.77, 128.71 (2×C), 128.07, 127.04, 125.03, 124.76, 123.43, 120.94, 115.09 (2×C), 55.86, 52.15, 51.94, 37.85, 37.77, 24.89. ESI-MS: m/z 527.4 (M+1), 544.5 (M+18), 549.4 (M+23), 565.4 (M+39). C29H30N6O4 [526.6].
4.1.5.2. (S)-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)a mino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-2).
White solid, yield: 53%. mp: 103-104°C. 1H NMR (400 MHz, DMSO-d6) δ 12.11 (s, 1H, PhNH), 8.98 (d, J = 7.8 Hz, 1H, NH), 8.65 (s, 1H, triazole-H), 8.61 (d, J = 8.3 Hz, 1H, Ph-H), 8.06 (d, J = 7.0 Hz, 2H, Ph-HH), 7.81 (d, J = 7.8 Hz, 1H, Ph-H), 7.70 – 7.50 (m, 3H, Ph-H), 7.41 (t, J = 7.8 Hz, 1H, Ph-H), 7.30 – 7.14 (m, 4H, Ph-H), 7.06 (d, J = 6.5 Hz, 2H, Ph-H), 6.93 (d, J = 9.0 Hz, 2H, Ph-H), 6.88 (d, J = 6.7 Hz, 2H, Ph-H), 5.26 (d, J = Hz, 1H, triazoleCH), 5.15 (d, J = 16.4 Hz, 1H, triazoleCH), 4.46 (td, J = 8.5, 5.5 Hz, 1H, CH), 3.74 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ (C=O), 165.34 (C=O), 165.15 (C=O), 159.02, 146.34, 137.65, 136.27, 135.76, 135.16, 132.45, 129.35 (2×C), 129.32 (2×C), 129.09 (2×C), 129.06, 128.70 (2×C), 128.01, 127.63 (2×C), 127.02, 125.07, 124.58, 121.91 , 119.22, 115.10 (2×C), 55.84, 52.14, 52.12, 37.87, 37.77. ESI-MS: m/z 589.4 (M+1), 606.4 (M+18), 611.3 (M+23), 627.4 (M+39). C34H32N6O4 [588.7].
4.1.5.3. (S)-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)a mino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-4-methylbenzamide (6a-3).
White solid, yield: 70%. mp: 118-119°C. 1H NMR (400 MHz, DMSO-d6) δ 12.07 (s, 1H, PhNH), 8.99 (d, J = 7.8 Hz, 1H, NH), 8.64 (s, 1H, triazole-H), 8.61 (d, J = 8.3 Hz, 1H, Ph-H), 7.96 (d, J = 8.1 Hz, 2H, Ph-H), 7.80 (d, J = 7.7 Hz, 1H, Ph-H), 7.40 (t, J = 6.7 Hz, 3H, Ph-H), 7.20 (tt, J = 14.2, 7.3 Hz, 4H, Ph-H), 7.13 – 6.99 (m, 2H, Ph-H), 6.93 (d, J = 8.9 Hz, 2H, Ph-H), 6.88 (d, J = 6.8 Hz, 2H, Ph-H), 5.26 (d, J = 16.4 Hz, 1H, triazoleCH), 5.15 (d, J = 16.4 Hz, 1H, triazoleCH), 4.61 – 4.34 (m, 1H, CH), 3.74 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.2 Hz, 1H, PhCH), 2.40 (s, 3H, PhCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.14 (C=O), 165.27 (C=O), 165.16 (C=O), 159.02, 146.39 , 142.52 , 137.65 , 136.37, 135.76, 132.40 , 129.88 (2×C), 129.32 (2×C), 129.09 (2×C), 129.04 , 128.70 (2×C), 127.99 , 127.64 (2×C), 127.02 , 125.04 , 124.42 , 121.80 , 119.04, 115.09 (2×C), 55.84, 52.12, 52.11, 37.87, 37.77, 21.49. ESI-MS: m/z 603.4 (M+1), 620.5 (M+18), 625.4 (M+23), 641.3 (M+39). C35H34N6O4 [602.7].
4.1.5.4. (S)-2-methoxy-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpro pan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-4).
White solid, yield: 65%. mp: 99-100°C. 1H NMR (400 MHz, DMSO-d6) δ 11.66 (s, 1H, PhNH), 8.95 (d, J = 7.9 Hz, 1H, NH), 8.55 (d, J = 8.3 Hz, 1H, Ph-H), 8.47 (s, 1H, triazole-H), 7.93 (d, J = 6.9 Hz, 1H, Ph-H), 7.64 (d, J = 7.6 Hz, 1H, Ph-H), 7.60 – 7.50 (m, 1H, Ph-H), 7.40 (t, J = 7.8 Hz, 1H, Ph-H), 7.28 – 7.13 (m, 5H, Ph-H), 7.15 – 6.99 (m, 3H, Ph-H), 6.93 (d, J = 8.9 Hz, 2H, Ph-H), 6.88 (d, J = 6.6 Hz, 2H, Ph-H), 5.23 (d, J = 16.3 Hz, 1H, triazoleCH), 5.13 (d, J = 16.3 Hz, 1H, triazole-CH), 4.47 (td, J = 8.5, 5.4 Hz, 1H, CH), 3.87 (s, 3H, OCH3), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.16 (C=O), 165.27 (C=O), 164.08 (C=O), 159.02, 157.36, 145.64, 137.66, 136.09, 135.77, 133.66, 131.58, 129.32 (2×C), 129.10, 128.89, 128.82, 128.69 (2×C), 127.02, 125.08, 124.45, 123.07, 122.82, 121.13 (2×C), 120.14, 115.10 (2×C), 112.51, 56.29, 55.85, 52.12, 51.97, 37.85, 37.77. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.4 (M+23), 657.4 (M+39). C35H34N6O5 [618.7].
4.1.5.5. (S)-3-methoxy-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpro pan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-5).
White solid, yield: 79%. mp: 99-100°C. 1H NMR (400 MHz, DMSO-d6) δ 12.13 (s, 1H, PhNH), 8.97 (d, J = 7.8 Hz, 1H, NH), 8.65 (s, 1H, triazole-H), 8.62 (d, J = 8.3 Hz, 1H, Ph-H), 7.82 (d, J = 7.5 Hz, 1H, Ph-H), 7.65 (d, J = 7.8 Hz, 1H, Ph-H), 7.62 (s, 1H, Ph-H), 7.51 (t, J = 7.9 Hz, 1H, Ph-H), 7.42 (t, J = 7.6 Hz, 1H, Ph-H), 7.21 (dp, J = 16.5, 7.4 Hz, 5H, Ph-H), 7.06 (d, J = 6.6 Hz, 2H, Ph-H), 6.93 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.8 Hz, 2H, Ph-H), 5.26 (d, J = 16.4 Hz, 1H, triazoleCH), 5.16 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.5 Hz, 1H, CH), 3.87 (s, 3H, OCH3), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.5, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.14 (C=O), 165.15 (C=O), 165.05 (C=O), 159.96, 159.02, 146.36, 137.65, 136.59, 136.24, 135.76, 130.51, 129.32 (2×C), 129.10 (2×C), 128.71 (2×C), 128.01, 127.03, 125.10, 124.57, 121.72, 119.80 (2×C), 119.08, 118.43, 115.09 (2×C), 112.60, 55.84, 55.77 (2×C), 52.14, 37.86, 37.77. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.4 (M+23), 657.4 (M+39). C35H34N6O5 [618.7].
4.1.5.6. (S)-4-methoxy-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-6).
White solid, yield: 68%. mp: 110-111°C. 1H NMR (400 MHz, DMSO-d6) δ 12.03 (s, 1H, PhNH), 8.97 (d, J = 7.8 Hz, 1H, NH), 8.64 (s, 1H, triazole-H), 8.62 (d, J = 8.1 Hz, 1H, Ph-H), 8.04 (d, J = 8.8 Hz, 2H, Ph-H), 7.85 – 7.76 (m, 1H, Ph-H), 7.45 – 7.36 (m, 1H, Ph-H), 7.26 – 7.16 (m, 4H, Ph-H), 7.14 (d, J = 8.9 Hz, 2H, Ph-H), 7.06 (d, J = 6.8 Hz, 2H, Ph-H), 6.93 (d, J = 9.0 Hz, 2H, Ph-H), 6.89 (d, J = 6.6 Hz, 2H, Ph-H), 5.26 (d, J = Hz, 1H, triazoleCH), 5.16 (d, J = 16.4 Hz, 1H, triazoleCH), 4.47 (td, J = 8.5, 5.3 Hz, 1H, CH), 3.86 (s, 3H, OCH3), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.5, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.15 (C=O), 165.18 (C=O), 164.85 (C=O), 162.62, 159.02, 146.45, 137.66, 136.51, 135.76, 129.53 (2×C), 129.32 (2×C), 129.10 (2×C), 129.04, 128.71 (2×C), 127.96, 127.26, 127.03, 125.04, 124.25, 121.69, 118.87, 115.09 (2×C), 114.59 (2×C), 55.95, 55.84, 52.16, 52.11, 37.85, 37.77. ESI-MS: m/z 619.5 (M+1), 641.3 (M+23). C35H34N6O5 [618.7].
4.1.5.7. Methyl (S)-4-((2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)carbamoyl)benzoate (6a-7).
White solid, yield: 76%. mp: 130-131°C. 1H NMR (400 MHz, DMSO-d6) δ 12.04 (s, 1H, PhNH), 8.95 (d, J = 7.8 Hz, 1H, NH), 8.61 (s, 1H, triazole-H), 8.50 (d, J = 8.2 Hz, 1H, Ph-H), 8.22 – 8.10 (m, 4H, Ph-H), 7.83 (d, J = 7.2 Hz, 1H, Ph-H), 7.43 (t, J = 7.4 Hz, 1H, Ph-H), 7.28 (t, J = 7.5 Hz, 1H, Ph-H), 7.19 (q, J = 8.7, 7.5 Hz, 3H, Ph-H), 7.06 (d, J = 7.4 Hz, 2H, Ph-H), 6.92 (d, J = 8.9 Hz, 2H, Ph-H), 6.88 (d, J = 6.6 Hz, 2H, Ph-H), 5.25 (d, J = 16.3 Hz, 1H, triazoleCH), 5.15 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.5, 5.5 Hz, 1H, CH), 3.90 (s, 3H, COOCH3), 3.74 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.1 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.12 (C=O), 166.06 (C=O), 165.14 (C=O), 164.61 (C=O), 159.03, 146.07, 139.27, 137.64 (2×C), 135.89 (2×C), 135.78, 132.84, 130.05 (2×C), 129.32 (2×C), 129.06 (2×C), 128.69 (2×C), 128.09 (2×C), 127.00, 125.08 (2×C), 122.58, 120.11, 115.10 (2×C), 55.84 (2×C), 52.91, 52.12, 37.91, 37.76. ESI-MS: m/z 647.5 (M+1), 664.5 (M+18), 669.4 (M+23), 685.5 (M+39). C35H34N6O5 [646.7].
4.1.5.8. (S)-4-cyano-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-8).
White solid, yield: 61%. mp: 142-143°C. 1H NMR (400 MHz, DMSO-d6) δ 12.04 (s, 1H, PhNH), 8.95 (d, J = 7.8 Hz, 1H, NH), 8.61 (s, 1H, triazole-H), 8.47 (d, J = 8.2 Hz, 1H, Ph-H), 8.18 (d, J = 8.2 Hz, 2H, Ph-H), 8.07 (d, J = 8.3 Hz, 2H, Ph-H), 7.83 (d, J = 7.7 Hz, 1H, Ph-H), 7.43 (t, J = 7.7 Hz, 1H, Ph-H), 7.28 (t, J = 7.5 Hz, 1H, Ph-H), 7.20 (q, J = 8.8, 7.6 Hz, 3H, Ph-H), 7.06 (d, J = 7.3 Hz, 2H, Ph-H), 6.92 (d, J = 8.8 Hz, 2H, Ph-H), 6.88 (d, J = 6.8 Hz, 2H, Ph-H), 5.25 (d, J = 16.3 Hz, 1H, triazoleCH), 5.15 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.4 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.3, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.14 (C=O), 165.17 (C=O), 164.02 (C=O), 159.04, 146.01, 139.17, 137.65, 135.78, 135.71, 133.36 (2×C), 129.31 (2×C), 129.07 (2×C), 128.70 (2×C), 128.53 (2×C), 128.12, 127.02, 125.28, 125.12, 122.71, 120.32, 118.69 (2×C), 115.11 (2×C), 114.68, 55.85, 52.16, 52.10, 37.90, 37.77. ESI-MS: m/z 614.3 (M+1), 631.4 (M+18), 636.4 (M+23), 652.5 (M+39). C35H31N7O4 [613.7].
4.1.5.9. (S)-2-fluoro-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-9).
White solid, yield: 76%. mp: 105-106°C. 1H NMR (400 MHz, DMSO-d6) δ 11.76 (s, 1H, PhNH), 8.95 (d, J = 7.9 Hz, 1H, NH), 8.58 (s, 1H, triazole-H), 8.48 (d, J = 8.1 Hz, 1H, Ph-H), 7.87 (t, J = 7.1 Hz, 1H, Ph-H), 7.78 (d, J = 7.7 Hz, 1H, Ph-H), 7.64 (q, J = 6.0 Hz, 1H, Ph-H), 7.50 – 7.33 (m, 3H, Ph-H), 7.27 (t, J = 7.5 Hz, 1H, Ph-H), 7.19 (q, J = 8.3, 7.3 Hz, 3H, Ph-H), 7.11 – 6.99 (m, 2H, Ph-H), 6.93 (d, J = 9.0 Hz, 2H, Ph-H), 6.88 (d, J = 6.5 Hz, 2H, Ph-H), 5.22 (d, J = 16.4 Hz, 1H, triazoleCH), 5.12 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.5, 5.4 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.13 (C=O), 165.14 (C=O), 162.58 (C=O), 159.69 (d, 1JCF = 248.4 Hz), 159.02, 145.90, 137.64, 135.72 (d, 3JCF = 9.7 Hz), 133.94 (d, 3JCF = 8.6 Hz), 130.84 (d, 4JCF = 2.1 Hz), 129.32 (2×C), 129.08 (2×C), 128.99, 128.69 (2×C), 128.21, 127.01, 125.41 (d, 4JCF = 3.4 Hz), 125.07, 124.98, 124.13 (d, 2JCF = 13.4 Hz), 122.63, 119.87, 117.13, 116.90, 115.10 (2×C), 55.84, 52.11, 52.03, 37.89, 37.77. HRMS: m/z 607.2466 (M+1)+, 629.2273 (M+23)+. C34H31FN6O4 [606.2391].
4.1.5.10. (S)-3-fluoro-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-10).
White solid, yield: 59%. mp: 109-110°C. 1H NMR (400 MHz, DMSO-d6) δ 12.06 (s, 1H, PhNH), 8.97 (d, J = 7.8 Hz, 1H, NH), 8.64 (s, 1H, triazole-H), 8.52 (d, J = 8.2 Hz, 1H, Ph-H), 7.91 (d, J = 7.7 Hz, 1H, Ph-H), 7.83 (t, J = 7.8 Hz, 2H, Ph-H), 7.66 (q, J = 7.8 Hz, 1H, Ph-H), 7.51 (dt, J = 8.3, 4.4 Hz, 1H, Ph-H), 7.42 (t, J = 7.6 Hz, 1H, Ph-H), 7.27 (t, J = 7.5 Hz, 1H, Ph-H), 7.20 (q, J = 9.3, 7.8 Hz, 3H, Ph-H), 7.13 – 6.99 (m, 2H, Ph-H), 6.93 (d, J = 8.8 Hz, 2H, Ph-H), 6.89 (d, J = 6.8 Hz, 2H, Ph-H), 5.26 (d, J = Hz, 1H, triazoleCH), 5.16 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 8.3 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.3, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.13 (C=O), 165.16 (C=O), 164.02 (d, 4JCF = 2.3 Hz, C=O), 162.67 (d, 1JCF = 243.5 Hz), 159.02, 146.17, 137.65 (2×C), 137.58 (d, 3JCF = 6.7 Hz), 135.92, 135.76, 131.56 (d, 3JCF = 7.7 Hz), 129.32 (2×C), 129.08 (2×C), 128.70 (2×C), 128.05, 127.01, 125.10, 124.96, 123.69 (d, 4JCF = 2.3 Hz), 122.32, 119.78, 119.37 (d, 2JCF = 20.8 Hz), 115.09 (2×C), 114.63 (d, 2JCF = 22.8 Hz), 55.84, 52.13, 52.11, 37.88, 37.76. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23), 645.4 (M+39). C34H31FN6O4 [606.7].
4.1.5.11. (S)-4-fluoro-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6a-11).
White solid, yield: 67%. mp: 123-124°C. 1H NMR (400 MHz, DMSO-d6) δ 12.01 (s, 1H, PhNH), 8.97 (d, J = 7.8 Hz, 1H, NH), 8.63 (s, 1H, triazole-H), 8.54 (d, J = 8.2 Hz, 1H, Ph-H), 8.12 (dd, J = 8.7, 5.5 Hz, 2H, Ph-H), 7.88 – 7.78 (m, 1H, Ph-H), 7.43 (q, J = 8.3, 7.7 Hz, 3H, Ph-H), 7.32 – 7.23 (m, 1H, Ph-H), 7.23 – 7.15 (m, 3H, Ph-H), 7.06 (d, J = 6.8 Hz, 2H, Ph-H), 6.93 (d, J = 9.0 Hz, 2H, Ph-H), 6.88 (d, J = 6.7 Hz, 2H, Ph-H), 5.26 (d, J = 16.4 Hz, 1H, triazoleCH), 5.15 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.5, 5.4 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.15 (C=O), 165.18 (C=O), 164.72 (d, 1JCF = 243.5 Hz), 164.32 (C=O), 159.03, 146.23, 137.65, 136.12, 135.76, 131.67 (d, 4JCF = 2.3 Hz), 130.41, 130.32, 129.31 (2×C), 129.09, 129.04, 128.70 (2×C), 128.03, 127.02, 125.08, 124.74, 122.22, 119.61, 116.44 (2×C), 116.22, 115.09 (2×C), 55.84, 52.16, 52.09, 37.86, 37.76. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23), 645.4 (M+39). C34H31FN6O4 [606.7].
4.1.5.12. (S)-N-(2-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-[1,1'-biphenyl]-4-carboxamide (6a-12).
White solid, yield: 58%. mp: 120-121°C. 1H NMR (400 MHz, DMSO-d6) δ 12.16 (s, 1H, PhNH), 8.98 (d, J = 7.8 Hz, 1H, NH), 8.67 (s, 1H, triazole-H), 8.63 (d, J = 8.3 Hz, 1H, Ph-H), 8.16 (d, J = 8.2 Hz, 2H, Ph-H), 7.91 (d, J = 8.2 Hz, 2H, Ph-H), 7.83 (d, J = 7.7 Hz, 1H, Ph-H), 7.79 (d, J = 7.6 Hz, 2H, Ph-H), 7.52 (t, J = 7.5 Hz, 2H, Ph-H), 7.30 – 7.15 (m, 5H, Ph-H), 7.11 – 7.04 (m, 2H, Ph-H), 6.95 – 6.85 (m, 5H, Ph-H), 5.27 (d, J = 16.4 Hz, 1H, triazoleCH), 5.18 (d, J = 13.5 Hz, 1H, triazoleCH), 4.51 – 4.42 (m, 1H, CH), 3.73 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.3, 9.3 Hz, 1H, PhCH). 13C NMR (101 MHz, DMSO-d6) δ 171.15 (C=O), 165.18 (C=O), 165.00 (C=O), 159.02, 146.36, 146.05, 143.92, 139.41, 137.65, 136.30, 135.76, 133.90, 129.54 (2×C), 129.32 (2×C), 129.09 (2×C), 128.70 (2×C), 128.34 (2×C), 127.53 (2×C), 127.42 (2×C), 127.02, 125.10, 124.60, 132.22, 121.99, 119.27, 116.26, 115.09 (2×C), 55.83, 52.15, 52.11, 37.87, 37.76. ESI-MS: m/z 665.4 (M+1), 682.5 (M+18). C40H36N6O4 [664.8].
4.1.5.13. (S)-2-(2-(4-(3-acetamidophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (6b-1).
White solid, yield: 52%. mp: 206-207°C. 1H NMR (400 MHz, DMSO-d6) δ 10.05 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.34 (s, 1H, triazole-H), 8.12 (s, 1H, Ph-H), 7.56 (d, J = 8.0 Hz, 1H, Ph-H), 7.45 (d, J = 7.7 Hz, 1H, Ph-H), 7.36 (t, J = 7.9 Hz, 1H, Ph-H), 7.27 – 7.14 (m, 3H, Ph-H), 7.14 – 7.00 (m, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.4 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.5, 5.4 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.5, 9.2 Hz, 1H, PhCH), 2.07 (s, 3H, COCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 168.87 (C=O), 165.46, 159.02, 146.43, 140.33, 137.67, 135.79, 131.54, 129.75, 129.33 (2×C), 129.10, 128.70 (2×C), 127.03, 123.36 (2×C), 120.40, 118.85, 115.92, 115.10 (2×C), 55.85, 52.09, 51.80, 37.88, 37.77, 24.53. ESI-MS: m/z 527.4 (M+1), 544.5 (M+18), 549.4 (M+23), 565.5 (M+39). C29H30N6O4 [526.6].
4.1.5.14. (S)-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-2).
White solid, yield: 73%. mp: 135-136°C. 1H NMR (400 MHz, DMSO-d6) δ 10.38 (s, 1H, PhNH), 8.93 (d, J = 7.8 Hz, 1H, NH), 8.37 (s, 1H, Ph-H), 8.34 (s, 1H, triazole-H), 8.00 (d, J = 7.1 Hz, 2H, Ph-H), 7.78 (d, J = 8.0 Hz, 1H, Ph-H), 7.62-7.52 (m, 4H, Ph-H), 7.42 (t, J = 7.9 Hz, 1H, Ph-H), 7.23-7.18 (m, 3H, Ph-H), 7.08 – 7.06 (m, 2H, Ph-H), 6.95-6.88 (m, 4H, Ph-H), 5.18 (d, J = 16.3 Hz, 1H, triazoleCH), 5.07 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.5, 5.5 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 5.0 Hz, 1H, PhCH ), 2.69 (dd, J = 13.5, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 166.06 (C=O), 165.48 (C=O), 159.02, 146.45, 140.20, 137.68, 135.79, 135.30, 132.10, 131.52, 129.70, 129.34 (2×C), 129.11, 128.87 (2×C), 128.71 (2×C), 128.16 (2×C), 127.04, 123.43 (2×C), 121.04, 120.22, 117.40, 115.10 (2×C), 55.85, 52.11, 51.82, 37.87, 37.78. ESI-MS: m/z 589.5 (M+1), 606.4 (M+18), 611.4 (M+23), 627.4 (M+39). C34H32N6O4 [588.7].
4.1.5.15. (S)-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-4-methylbenzamide (6b-3).
White solid, yield: 63%. mp: 212-213°C. 1H NMR (400 MHz, DMSO-d6) δ 10.28 (s, 1H, PhNH), 8.92 (d, J = 7.8 Hz, 1H, NH), 8.37 (s, 1H, Ph-H), 8.34 (s, 1H, triazole-H), 7.92 (d, J = 8.0 Hz, 2H, Ph-H), 7.78 (d, J = 7.9 Hz, 1H, Ph-H), 7.53 (d, J = 7.7 Hz, 1H, Ph-H), 7.42 (t, J = 7.9 Hz, 1H, Ph-H), 7.35 (d, J = 8.0 Hz, 2H, Ph-H), 7.21 (q, J = 7.9, 7.0 Hz, 3H, Ph-H), 7.15 – 7.01 (m, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.90 (d, J = 6.6 Hz, 2H, Ph-H), 5.18 (d, J = 16.3 Hz, 1H, triazoleCH), 5.07 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.3 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH), 2.40 (s, 3H, PhCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.85 (C=O), (C=O), 159.02, 146.47, 142.11, 140.27, 137.68, 135.79, 132.40, 131.49, 129.67, 129.39 (2×C), 129.34 (2×C), 129.12, 128.71 (2×C), 128.19 (2×C), 127.04, 123.41 (2×C), 120.92, 120.21, 117.39, 115.10 (2×C), 55.85, 52.10, 51.81, 37.87, 21.50. ESI-MS: m/z 603.4 (M+1), 620.5 (M+18), 625.4 (M+23), 641.3 (M+39). C35H34N6O4 [602.7].
4.1.5.16. (S)-2-methoxy-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-4).
White solid, yield: 65%. mp: 125-126°C. 1H NMR (400 MHz, DMSO-d6) δ 10.23 (s, 1H, PhNH), 8.93 (d, J = 7.8 Hz, 1H, NH), 8.37 (s, 1H, Ph-H), 8.29 (s, 1H, triazole-H), 7.68 (dd, J = 11.9, 8.0 Hz, 2H, Ph-H), 7.52 (t, J = 9.4 Hz, 2H, Ph-H), 7.41 (t, J = 7.9 Hz, 1H, Ph-H), 7.30 – 7.15 (m, 4H, Ph-H), 7.08 (t, J = 7.3 Hz, 3H, Ph-H), 6.94 (d, J = 8.8 Hz, 2H, Ph-H), 6.90 (d, J = 6.6 Hz, 2H, Ph-H), 5.18 (d, J = 16.3 Hz, 1H, triazoleCH), 5.07 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.4 Hz, 1H, CH), 3.92 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.46 (C=O), 165.10 (C=O), 159.02, 156.96, 146.43, 140.06, 137.68, 135.79, 132.52, 131.62, 130.13, 129.76, 129.34 (2×C), 129.11, 128.71 (2×C), 127.03, 125.41, 123.45, 120.95 (2×C), 120.88, 119.65, 116.75, 115.10 (2×C), 112.46, 56.37, 55.85, 52.10, 51.83, 37.89, 37.77. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.3 (M+23), 657.3 (M+39). C35H34N6O5 [618.7].
4.1.5.17. (S)-3-methoxy-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-5).
White solid, yield: 48%. mp: 115-116°C. 1H NMR (400 MHz, DMSO-d6) δ 10.34 (s, 1H, PhNH), 8.92 (d, J = 7.6 Hz, 1H, NH), 8.37 (s, 1H, Ph-H), 8.33 (s, 1H, triazole-H), 7.79 (d, J = 7.7 Hz, 1H, Ph-H), 7.59 (d, J = 7.5 Hz, 1H, Ph-H), 7.54 (s, 2H, Ph-H), 7.45 (dt, J = 13.0, 7.9 Hz, 2H, Ph-H), 7.29 – 7.13 (m, 4H, Ph-H), 7.08 (d, J = 6.6 Hz, 2H, Ph-H), 6.94 (d, J = 8.5 Hz, 2H, Ph-H), 6.90 (d, J = 6.5 Hz, 2H, Ph-H), 5.19 (d, J = 16.3 Hz, 1H, triazoleCH), 5.08 (d, J = 16.3 Hz, 1H, triazoleCH), 4.48 (q, J = 7.9 Hz, 1H, CH), 3.86 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.2, 4.6 Hz, 1H, PhCH), 2.71 (dd, J = 13.0, 9.4 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.75 (C=O), 165.47 (C=O), 159.65, 159.02, 146.44, 140.12, 137.68, 136.68, 135.79, 131.52, 130.03, 129.69, 129.34 (2×C), 129.10, 128.71 (2×C), 127.03, 123.42, 121.09, 120.38 (2×C), 120.31, 117.87, 117.49, 115.11 (2×C), 113.36, 55.85, 55.82, 52.10, 51.83, 37.89, 37.78. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.3 (M+23), 657.5 (M+39). C35H34N6O5 [618.7].
4.1.5.18. (S)-4-methoxy-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-6).
White solid, yield: 63%. mp: 199-200°C. 1H NMR (400 MHz, DMSO-d6) δ 10.20 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.36 (s, 1H, Ph-H), 8.32 (s, 1H, triazole-H), 8.01 (d, J = 8.7 Hz, 2H, Ph-H), 7.78 (d, J = 8.1 Hz, 1H, Ph-H), 7.52 (d, J = 7.7 Hz, 1H, Ph-H), 7.41 (t, J = 7.9 Hz, 1H, Ph-H), 7.26 – 7.16 (m, 3H, Ph-H), 7.08 (d, J = 8.7 Hz, 4H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.6 Hz, 2H, Ph-H), 5.18 (d, J = 16.3 Hz, 1H, triazoleCH), 5.07 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.6 Hz, 1H, CH), 3.85 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.5, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.47 (C=O), 165.40 (C=O), 162.40, 159.02, 146.50, 140.37, 137.68, 135.79, 131.47, 130.10 (2×C), 129.63, 129.34 (2×C), 129.11, 128.71 (2×C), 127.30, 127.03, 123.38 (2×C), 120.80, 120.19, 117.38, 115.11 (2×C), 114.08 (2×C), 55.91, 55.85, 52.10, 51.83, 37.89, 37.78. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.4 (M+23), 657.5 (M+39). C35H34N6O5 [618.7].
4.1.5.19. Methyl (S)-4-((3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)carbamoyl)benzoate (6b-7).
White solid, yield: 58%. mp: 153-154°C. 1H NMR (400 MHz, DMSO-d6) δ 12.04 (s, 1H, PhNH), 8.95 (d, J = 7.8 Hz, 1H, NH), 8.61 (s, 1H, triazole-H), 8.49 (d, J = 8.2 Hz, 1H, Ph-H), 8.21 – 8.09 (m, 4H, Ph-H), 7.83 (d, J = 7.4 Hz, 1H, Ph-H), 7.43 (t, J = 7.5 Hz, 1H, Ph-H), 7.28 (t, J = 7.5 Hz, 1H, Ph-H), 7.19 (q, J = 8.7, 7.5 Hz, 3H, Ph-H), 7.06 (d, J = 6.9 Hz, 2H, Ph-H), 6.92 (d, J = 8.9 Hz, 2H, Ph-H), 6.88 (d, J = 6.7 Hz, 2H, Ph-H), 5.25 (d, J = 16.3 Hz, 1H, triazoleCH), 5.15 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.6 Hz, 1H, CH), 3.90 (s, 3H, COOCH3), 3.74 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.4, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.13 (C=O), 166.05 (C=O), 165.16 (C=O), 164.61 (C=O), 159.02, 146.05, 139.26, 137.65, 135.86, 135.76, 132.82, 130.06 (2×C), 129.31 (2×C), 129.07 (2×C), 128.70 (2×C), 128.11 (2×C), 127.01 (2×C), 125.11 (2×C), 122.60 (2×C), 120.12, 115.09 (2×C), 55.84, 52.94, 52.14, 52.08, 37.86, 37.76. ESI-MS: m/z 647.4 (M+1), 664.4 (M+18), 669.4 (M+23), 685.5 (M+39). C36H34N6O6 [646.7].
4.1.5.20. (S)-4-cyano-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-8).
White solid, yield: 36%. mp: 224-225°C. 1H NMR (400 MHz, DMSO-d6) δ 10.61 (s, 1H, PhNH), 8.93 (d, J = 7.8 Hz, 1H, NH), 8.39 (s, 1H, Ph-H), 8.34 (s, 1H, triazole-H), 8.15 (d, J = 8.3 Hz, 2H, Ph-H), 8.05 (d, J = 8.3 Hz, 2H, Ph-H), 7.78 (d, J = 8.1 Hz, 1H, Ph-H), 7.57 (d, J = 7.7 Hz, 1H, Ph-H), 7.45 (t, J = 7.9 Hz, 1H, Ph-H), 7.21 (q, J = 7.8, 7.0 Hz, 3H, Ph-H), 7.13 – 7.02 (m, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.6 Hz, 2H, Ph-H), 5.19 (d, J = 16.3 Hz, 1H, triazoleCH), 5.08 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.5, 5.1 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.46 (C=O), 164.67 (C=O), 159.02, 146.33, 139.79, 139.31, 137.68, 135.79, 132.95 (2×C), 131.61, 129.81, 129.33 (2×C), 129.11, 129.03 (2×C), 128.71 (2×C), 127.03, 123.48 (2×C), 121.47, 120.27, 118.80, 117.43, 115.10 (2×C), 114.36, 55.85, 52.11, 51.83, 37.88, 37.77. ESI-MS: m/z 614.3 (M+1), 631.5 (M+18), 636.3 (M+23), 652.4 (M+39). C35H31N7O4 [613.7].
4.1.5.21. (S)-2-fluoro-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-9).
White solid, yield: 61%. mp: 114-115°C. 1H NMR (400 MHz, DMSO-d6) δ 10.55 (s, 1H, PhNH), 8.93 (d, J = 7.9 Hz, 1H, NH), 8.38 (s, 1H, Ph-H), 8.30 (s, 1H, triazole-H), 7.70 (t, J = 7.7 Hz, 2H, Ph-H), 7.65 – 7.57 (m, 1H, Ph-H), 7.55 (d, J = 7.8 Hz, 1H, Ph-H), 7.43 (t, J = 7.9 Hz, 1H, Ph-H), 7.36 (q, J = 9.0, 7.4 Hz, 2H, Ph-H), 7.26 – 7.16 (m, 3H, Ph-H), 7.14 – 7.01 (m, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.90 (d, J = 6.4 Hz, 2H, Ph-H), 5.18 (d, J = 16.3 Hz, 1H, triazoleCH), 5.07 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.5 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.46 (C=O), 163.35 (C=O), 159.36 (d, 1JCF = 247.0 Hz), 159.02, 146.34, 139.88, 137.68, 135.79, 133.02 (d, 3JCF = 8.4 Hz), 131.67, 130.38 (d, 4JCF = 2.7 Hz), 129.86, 129.34 (2×C), 129.11, 128.71 (2×C), 127.03, 125.44 (d, 2JCF = 15.0 Hz), 125.04 (d, 4JCF = 3.4 Hz), 123.47, 121.25, 119.63, 116.75 (2×C), 116.53, 115.10 (2×C), 55.85, 52.10, 51.82, 37.88, 37.77. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23), 645.4 (M+39). C34H31FN6O4 [606.7].
4.1.5.22. (S)-3-fluoro-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-10).
White solid, yield: 75%. mp: 167-168°C. 1H NMR (400 MHz, DMSO-d6) δ 10.44 (s, 1H, PhNH), 8.93 (d, J = 7.8 Hz, 1H, NH), 8.39 (s, 1H, Ph-H), 8.34 (s, 1H, triazole-H), 7.83 (dt, J = 24.4, 8.0 Hz, 3H, Ph-H), 7.61 (q, J = 7.9 Hz, 1H, Ph-H), 7.56 (d, J = 7.7 Hz, 1H, Ph-H), 7.46 (dt, J = 16.2, 7.3 Hz, 2H, Ph-H), 7.29 – 7.15 (m, 3H, Ph-H), 7.15 – 7.01 (m, 2H, Ph-H), 6.94 (d, J = 8.8 Hz, 2H, Ph-H), 6.90 (d, J = 6.5 Hz, 2H, Ph-H), 5.19 (d, J = 16.3 Hz, 1H, triazoleCH), 5.08 (d, J = 16.3 Hz, 1H, triazoleCH), 4.54 – 4.41 (m, 1H, CH), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.47 (C=O), 164.64 (d, 4JCF = 2.4 Hz, C=O), 162.39 (d, 1JCF = 242.7 Hz), 159.02, 146.39, 139.92, 137.68, 137.57 (d, 3JCF = 6.8 Hz), 135.79, 131.56, 131.11, 131.03, 129.75, 129.34 (2×C), 129.11, 128.71 (2×C), 127.03, 124.40 (d, 4JCF = 2.6 Hz), 123.45, 121.28, 120.28, 119.01 (d, 2JCF = 21.4 Hz), 117.46, 115.10 (2×C), 114.88, 55.85, 52.11, 51.83, 37.88, 37.77. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23), 645.4 (M+39). C34H31FN6O4 [606.7].
4.1.5.23. (S)-4-fluoro-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6b-11).
White solid, yield: 30%. mp: 173-174°C. 1H NMR (400 MHz, DMSO-d6) δ 10.44 (s, 1H, PhNH), 8.95 (d, J = 7.8 Hz, 1H, NH), 8.37 (s, 1H, Ph-H), 8.34 (s, 1H, triazole-H), 8.11 (dd, J = 8.1, 5.7 Hz, 2H, Ph-H), 7.79 (d, J = 7.8 Hz, 1H, Ph-H), 7.53 (d, J = 7.6 Hz, 1H, Ph-H), 7.48 – 7.31 (m, 3H, Ph-H), 7.21 (q, J = 7.2, 6.4 Hz, 3H, Ph-H), 7.12 – 6.99 (m, 2H, Ph-H), 6.94 (d, J = 8.7 Hz, 2H, Ph-H), 6.90 (d, J = 6.7 Hz, 2H, Ph-H), 5.19 (d, J = 16.3 Hz, 1H, triazoleCH), 5.08 (d, J = 16.2 Hz, 1H, triazoleCH), 4.46 (q, J = 8.1 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 4.8 Hz, 1H, PhCH), 2.70 (dd, J = 13.3, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.17 (C=O), 165.47 (C=O),165.00 (d, 1JCF = 245.0 Hz), 164.93 (C=O), 159.02, 146.43, 140.13, 137.71, 135.82, 130.99 (2×C), 130.90, 129.67, 129.35 (2×C), 129.10, 128.69 (2×C), 127.01, 123.40, 121.09, 120.31, 117.50, 115.89 (2×C), 115.67 (2×C), 115.11 (2×C), 55.87, 52.13, 51.85, 37.88, 37.78. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23), 645.3 (M+39). C34H31FN6O4 [606.7].
4.1.5.24. (S)-N-(3-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-[1,1'-biphenyl]-4-carboxamide (6b-12).
White solid, yield: 58%. mp: 170-171°C. 1H NMR (400 MHz, DMSO-d6) δ 10.43 (s, 1H, PhNH), 8.94 (d, J = 7.8 Hz, 1H, NH), 8.39 (s, 1H, Ph-H), 8.38 (s, 1H, triazole-H), 8.12 (d, J = 8.2 Hz, 2H, Ph-H), 7.86 (d, J = 8.3 Hz, 2H, Ph-H), 7.82 (d, J = 8.4 Hz, 1H, Ph-H), 7.78 (d, J = 7.6 Hz, 2H, Ph-H), 7.53 (q, J = 7.4 Hz, 3H, Ph-H), 7.44 (dt, J = 7.8, 4.3 Hz, 2H, Ph-H), 7.22 (q, J = 7.8, 7.0 Hz, 3H, Ph-H), 7.14 – 7.01 (m, 2H, Ph-H), 6.95 (d, J = 8.8 Hz, 2H, Ph-H), 6.90 (d, J = 6.6 Hz, 2H, Ph-H), 5.19 (d, J = 16.3 Hz, 1H, triazoleCH), 5.08 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 8.4 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.71 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.66 (C=O), 165.48 (C=O), 159.02, 146.47, 143.62, 140.23, 139.57, 137.69, 135.79, 134.03, 131.54, 129.71, 129.54 (2×C), 129.34 (2×C), 129.11 (2×C), 128.88 (2×C), 128.71 (2×C), 128.63, 127.40 (2×C), 127.07 (2×C), 123.43 (2×C), 121.05, 120.24, 117.41, 115.11 (2×C), 55.85, 52.11, 51.83, 37.88, 37.78. ESI-MS: m/z 665.4 (M+1), 682.5 (M+18), 687.4 (M+23). C40H36N6O4 [664.8].
4.1.5.25. (S)-2-(2-(4-(4-acetamidophenyl)-1H-1,2,3-triazol-1-yl)acetamido)-N-(4-methoxyphenyl)-N-methyl-3-phenylpropanamide (6c-1).
White solid, yield: 47%. mp: 134-135°C. 1H NMR (400 MHz, DMSO-d6) δ 10.04 (s, 1H, PhNH), 8.89 (d, J = 7.8 Hz, 1H, NH), 8.30 (s, 1H, triazole-H), 7.75 (d, J = 8.5 Hz, 2H, Ph-H), 7.65 (d, J = 8.5 Hz, 2H, Ph-H), 7.21 (q, J = 7.2, 6.7 Hz, 3H, Ph-H), 7.06 (d, J = 6.6 Hz, 2H, Ph-H), 6.93 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.5 Hz, 2H, Ph-H), 5.15 (d, J = 16.4 Hz, 1H, triazoleCH), 5.04 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (q, J = 8.3 Hz, 1H, CH), 3.75 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.5, 5.0 Hz, 1H, PhCH), 2.69 (dd, J = 13.3, 9.2 Hz, 1H, PhCH), 2.06 (s, 3H, COCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 168.78 (C=O), 165.48 (C=O), 159.02, 146.40, 139.44, 137.68, 135.80, 129.33 (2×C), 129.10, 128.70 (2×C), 127.02, 125.97 (2×C), 125.89, 122.70 (2×C), 119.67 (2×C), 115.10 (2×C), 55.85, 52.09, 51.80, 37.89, 37.77, 24.51. ESI-MS: m/z (M+1), 544.5 (M+18), 549.4 (M+23), 565.4 (M+39). C29H30N6O4 [526.6].
4.1.5.26. (S)-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)a mino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-2).
White solid, yield: 48%. mp: 171-172°C. 1H NMR (400 MHz, DMSO-d6) δ 10.36 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 7.98 (d, J = 7.2 Hz, 2H, Ph-H), 7.89 (d, J = 8.7 Hz, 2H, Ph-H), 7.83 (d, J = 8.6 Hz, 2H, Ph-H), 7.61 (t, J = 7.2 Hz, 1H, Ph-H), 7.55 (t, J = 7.3 Hz, 2H, Ph-H), 7.22 (q, J = 7.3, 6.7 Hz, 3H, Ph-H), 7.07 (d, J = 6.5 Hz, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.90 (d, J = 6.5 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.4, 5.6 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.1 Hz, 1H, PhCH), 2.71 (dd, J = 13.6, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 166.03 (C=O), 165.49 (C=O), 159.03, 145.38, 139.29, 137.69, 135.80, 135.39, 132.06, 129.34 (2×C), 129.10 (2×C), 128.86 (2×C), 128.71 (2×C), 128.13 (2×C), 127.03, 126.53, 125.87 (2×C), 122.87, 121.05 (2×C), 115.11 (2×C), 55.86, 52.09, 51.83, 37.90, 37.78. ESI-MS: m/z 589.4 (M+1), 606.4 (M+18), 611.3 (M+23), 627.4 (M+39). C34H32N6O4 [588.7].
4.1.5.27. (S)-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-4-methylbenzamide (6c-3).
White solid, yield: 48%. mp: 138-139°C. 1H NMR (400 MHz, DMSO-d6) δ 10.27 (s, 1H, PhNH), 8.91 (d, J = 7.7 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 7.99 – 7.85 (m, 4H, Ph-H), 7.82 (d, J = 8.3 Hz, 2H, Ph-H), 7.35 (d, J = 7.6 Hz, 2H, Ph-H), 7.22 (q, J = 8.0, 6.9 Hz, 3H, Ph-H), 7.07 (d, J = 7.0 Hz, 2H, Ph-H), 6.94 (d, J = 8.4 Hz, 2H, Ph-H), 6.90 (d, J = 6.4 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 7.7 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.2, 4.5 Hz, 1H, PhCH), 2.76 – 2.64 (m, 1H, PhCH), 2.40 (s, 3H, PhCH3). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.82 (C=O), 165.49 (C=O), 159.02, 146.40, 142.08, 139.36, 137.68, 135.80, 132.48, 129.38 (2×C), 129.34 (2×C), 129.10 (2×C), 128.70 (2×C), 128.17 (2×C), 127.03, 126.42, 125.85 (2×C), 122.84, 121.03 (2×C), 115.10 (2×C), 55.86, 52.09, 51.83, 37.90, 37.78, 21.49. ESI-MS: m/z 603.4 (M+1), 620.5 (M+18), 625.5 (M+23), 641.4 (M+39). C35H34N6O4 [602.7].
4.1.5.28. (S)-2-methoxy-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-4).
White solid, yield: 34%. mp: 146-147°C. 1H NMR (400 MHz, DMSO-d6) δ 10.23 (s, 1H, PhNH), 8.91 (d, J = 7.7 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 7.92 – 7.77 (m, 4H, Ph-H), 7.66 (d, J = 7.3 Hz, 1H, Ph-H), 7.52 (t, J = 7.6 Hz, 1H, Ph-H), 7.21 (t, J = 8.2 Hz, 4H, Ph-H), 7.08 (t, J = 7.2 Hz, 3H, Ph-H), 6.94 (d, J = 8.6 Hz, 2H, Ph-H), 6.90 (d, J = 6.7 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 8.2 Hz, 1H, CH), 3.92 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), (s, 3H, NCH3), 2.93 (dd, J = 13.4, 4.9 Hz, 1H, PhCH), 2.71 (dd, J = 13.4, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.49 (C=O), 164.97 (C=O), 159.03, 156.95, 146.38, 139.15, 137.69, 135.81, 132.52, 130.13, 129.34 (2×C), 129.10, 128.70 (2×C), 127.03, 126.39, 125.96 (2×C), 125.39, 122.83, 120.96 (2×C), 120.42 (2×C), 115.11 (2×C), 112.47, 56.37, 55.86, 52.09, 51.82, 37.91, 37.78. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.4 (M+23). C35H34N6O5 [618.7].
4.1.5.29. (S)-3-methoxy-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-5).
White solid, yield: 52%. mp: 141-142°C. 1H NMR (400 MHz, DMSO-d6) δ 10.32 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 7.87 (d, J = 8.7 Hz, 2H, Ph-H), 7.83 (d, J = 8.7 Hz, 2H, Ph-H), 7.56 (d, J = 7.6 Hz, 1H, Ph-H), 7.51 (s, 1H, Ph-H), 7.46 (t, J = 7.9 Hz, 1H, Ph-H), 7.26 – 7.15 (m, 4H, Ph-H), 7.07 (d, J = 6.5 Hz, 2H, Ph-H), 6.94 (d, J = 8.8 Hz, 2H, Ph-H), 6.90 (d, J = 6.5 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 8.3 Hz, 1H, CH), 3.85 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.12 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.74 (C=O), 165.48 (C=O), 159.65, 159.03, 146.38, 139.20, 137.68, 136.79, 135.81, 130.03, 129.34 (2×C), 129.10, 128.70 (2×C), 127.02, 126.58, 125.86 (2×C), 122.87, 121.13 (2×C), 120.35 (2×C), 117.81, 115.11 (2×C), 113.39, 55.82 (2×C), 52.09, 51.84, 37.91, 37.78. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.3 (M+23), 657.4 (M+39). C35H34N6O5 [618.7].
4.1.5.30. (S)-4-methoxy-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-6).
White solid, yield: 55%. mp: 141-142°C. 1H NMR (400 MHz, DMSO-d6) δ 10.20 (s, 1H, PhNH), 8.92 (d, J = 7.1 Hz, 1H, NH), 8.34 (s, 1H, triazole-H), 7.98 (d, J = 7.8 Hz, 2H, Ph-H), 7.86 (d, J = 7.5 Hz, 2H, Ph-H), 7.81 (d, J = 7.6 Hz, 2H, Ph-H), 7.26 – 7.15 (m, 3H, Ph-H), 7.08 (d, J = 7.2 Hz, 4H, Ph-H), 6.94 (d, J = 8.1 Hz, 2H, Ph-H), 6.89 (d, J = 6.0 Hz, 2H, Ph-H), 5.16 (d, J = 16.1 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (q, J = 8.5 Hz, 1H, CH), 3.85 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.99 – 2.87 (m, 1H, PhCH), 2.76 – 2.62 (m, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.50 (C=O), 165.36 (C=O), 162.39, 159.02, 146.42, 139.47, 137.69, 135.80, 130.08 (2×C), 129.34 (2×C), 129.11 (2×C), (2×C), 127.37, 127.03, 126.29, 125.83 (2×C), 122.82, 120.99 (2×C), 115.10 (2×C), 114.08 (2×C), 55.91, 55.85, 52.10, 51.82, 37.88, 37.78. ESI-MS: m/z 619.5 (M+1), 636.4 (M+18), 641.4 (M+23), 657.4 (M+39). C35H34N6O5 [618.7].
4.1.5.31. Methyl (S)-4-((4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)carbamoyl)benzoate (6c-7).
White solid, yield: 77%. mp: 129-130°C. 1H NMR (400 MHz, DMSO-d6) δ 10.54 (s, 1H, PhNH), 8.90 (d, J = 7.7 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 8.10 (s, 4H, Ph-H), 7.89 (d, J = 8.6 Hz, 2H, Ph-H), 7.84 (d, J = 8.6 Hz, 2H, Ph-H), 7.28 – 7.14 (m, 3H, Ph-H), 7.07 (d, J = 6.6 Hz, 2H, Ph-H), 6.94 (d, J = 8.7 Hz, 2H, Ph-H), 6.90 (d, J = 6.7 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.48 (q, J = 8.2 Hz, 1H, CH), 3.91 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.3, 9.1 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 166.15 (C=O), 165.49 (C=O), 165.17 (C=O), 159.03, 146.33, 139.47, 139.00, 137.68, 135.80, 132.51, 129.66 (2×C), 129.34 (2×C), 129.10 (2×C), 128.70 (2×C), 128.58 (2×C), 127.02, 126.83, 125.92 (2×C), 122.94, 121.15 (2×C), 115.10 (2×C), 55.86, 52.91, 52.10, 51.84, 37.90, 37.77. ESI-MS: m/z 647.5 (M+1), 664.4 (M+18), 669.4 (M+23). C36H34N6O6 [646.7].
4.1.5.32. (S)-4-cyano-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-8).
White solid, yield: 71%. mp: 140-141°C. 1H NMR (400 MHz, DMSO-d6) δ 10.60 (s, 1H, PhNH), 8.92 (d, J = 7.8 Hz, 1H, NH), 8.36 (s, 1H, triazole-H), 8.13 (d, J = 8.3 Hz, 2H, Ph-H), 8.04 (d, J = 8.3 Hz, 2H, Ph-H), 7.88 (d, J = 8.9 Hz, 2H, Ph-H), 7.84 (d, J = 8.9 Hz, 2H, Ph-H), 7.28 – 7.13 (m, 3H, Ph-H), 7.13 – 7.00 (m, 2H, Ph-H), 6.94 (d, J = 8.8 Hz, 2H, Ph-H), 6.90 (d, J = 6.5 Hz, 2H, Ph-H), 5.17 (d, J = 16.4 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (q, J = 8.4 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.6, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.49 (C=O), (C=O), 159.02, 146.28, 139.39, 138.84, 137.69, 135.80, 132.95 (2×C), 129.33 (2×C), 129.10 (2×C), 129.02 (2×C), 128.70 (2×C), 127.02, 126.96, 125.93 (2×C), 122.98, 121.16 (2×C), 118.80, 115.10 (2×C), 114.33, 55.85, 52.10, 51.82, 37.89, 37.77. ESI-MS: m/z 614.3 (M+1), 631.5 (M+18), 636.4 (M+23). C35H31N7O4 [613.7].
4.1.5.33. (S)-2-fluoro-N-(4-(1-(2-(( 1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-9).
White solid, yield: 42%. mp: 122-123°C. 1H NMR (400 MHz, DMSO-d6) δ 10.53 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 7.82 (s, 4H, Ph-H), 7.73 – 7.65 (m, 1H, Ph-H), 7.64 – 7.55 (m, 1H, Ph-H), 7.38 (d, J = 10.4 Hz, 1H, Ph-H), 7.33 (d, J = 7.0 Hz, 1H, Ph-H), 7.26 – 7.15 (m, 3H, Ph-H), 7.12 – 7.01 (m, 2H, Ph-H), 6.94 (d, J = 9.0 Hz, 2H, Ph-H), 6.89 (d, J = 6.3 Hz, 2H, Ph-H), 5.17 (d, J = 16.3, Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.4, 5.5 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.92 (dd, J = 13.4, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ (C=O), 165.48 (C=O), 163.23 (C=O), 159.33 (d, 1JCF = 247.2 Hz), 159.02, 146.30, 138.95, 137.69, 135.80, 133.00 (d, 3JCF = 8.5 Hz), 130.37 (d, 4JCF = 2.9 Hz), 129.33 (2×C), 129.11, 128.71 (2×C), 127.03, 126.73, 126.01 (2×C), 125.46 (d, 2JCF = 15.2 Hz), 125.04 (d, 4JCF = 3.4 Hz), 122.93, 120.45 (2×C), 116.75, 116.53, 115.10 (2×C), 55.86, 52.10, 51.82, 37.89, 37.78. ESI-MS: m/z 607.4 (M+1), 624.5 (M+18), 629.4 (M+23). C34H31FN6O4 [606.7].
4.1.5.34. (S)-3-fluoro-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-10).
White solid, yield: 46%. mp: 120-121°C. 1H NMR (400 MHz, DMSO-d6) δ 10.43 (s, 1H, PhNH), 8.92 (d, J = 7.8 Hz, 1H, NH), 8.36 (s, 1H, triazole-H), 7.85 (q, J = 8.6 Hz, 5H, Ph-H), 7.79 (d, J = 9.7 Hz, 1H, Ph-H), 7.61 (q, J = 7.9 Hz, 1H, Ph-H), 7.46 (td, J = 8.6, 2.2 Hz, 1H, Ph-H), 7.21 (q, J = 7.4, 6.8 Hz, 3H, Ph-H), 7.13 – 7.02 (m, 2H, Ph-H), 6.94 (d, J = 8.8 Hz, 2H, Ph-H), 6.89 (d, J = 6.6 Hz, 2H, Ph-H), 5.17 (d, J = 16.4 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.4, 5.5 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.0 Hz, 1H, PhCH), 2.70 (dd, J = 13.5, 9.3 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.49 (C=O), 164.61 (d, 4JCF = 2.4 Hz, C=O), 162.38 (d, 1JCF = 242.7 Hz), 159.02, 146.32, 138.98, 137.69 (2×C), 137.62, 135.80, 131.07 (d, 3JCF = 7.9 Hz), (2×C), 129.10, 128.71 (2×C), 127.03, 126.77, 125.90 (2×C), 124.38 (d, 4JCF = 2.7 Hz), 122.94, 121.14 (2×C), 118.98 (d, 2JCF = 21.1 Hz), 115.10 (2×C), 114.87, 55.85, 52.10, 51.82, 37.88, 37.78. ESI-MS: m/z 607.4 (M+1), 624.5 (M+18), 629.4 (M+23). C34H31FN6O4 [606.7].
4.1.5.35. (S)-4-fluoro-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)benzamide (6c-11).
White solid, yield: 35%. mp: 125-126°C. 1H NMR (400 MHz, DMSO-d6) δ 10.37 (s, 1H, PhNH), 8.91 (d, J = 7.8 Hz, 1H, NH), 8.35 (s, 1H, triazole-H), 8.06 (dd, J = 8.6, 5.6 Hz, 2H, Ph-H), 7.84 (q, J = 8.8 Hz, 4H, Ph-H), 7.38 (t, J = 8.8 Hz, 2H, Ph-H), 7.21 (q, J = 7.3, 6.7 Hz, 3H, Ph-H), 7.07 (d, J = 6.6 Hz, 2H, Ph-H), 6.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.89 (d, J = 6.5 Hz, 2H, Ph-H), 5.17 (d, J = 16.3 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.46 (td, J = 8.5, 5.6 Hz, 1H, CH), 3.76 (s, 3H, OCH3), (s, 3H, NCH3), 2.93 (dd, J = 13.5, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.4, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.18 (C=O), 165.49 (C=O), 164.90 (C=O), 164.55 (d, 1JCF = 247.6 Hz), 159.02, 146.35, 139.18, 137.69, 135.80, 131.80 (d, 4JCF = 2.8 Hz), 130.93 (2×C), 130.84, 129.33 (2×C), 129.10, 128.70 (2×C), 127.02, 126.60, 125.88 (2×C), 122.89, 121.08 (2×C), 115.92 (2×C), 115.71, 115.10, 55.86, 52.10, 51.82, 37.89, 37.78. ESI-MS: m/z 607.4 (M+1), 624.4 (M+18), 629.4 (M+23). C34H31FN6O4 [606.7].
4.1.5.36. (S)-N-(4-(1-(2-((1-((4-methoxyphenyl)(methyl)amino)-1-oxo-3-phenylpropan-2-yl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)phenyl)-[1,1'-biphenyl]-4-carboxamide (6c-12).
White solid, yield: 41%. mp: 160-161°C. 1H NMR (400 MHz, DMSO-d6) δ 10.41 (s, 1H, PhNH), 8.92 (d, J = 7.8 Hz, 1H, NH), 8.36 (s, 1H, triazole-H), 8.09 (d, J = 8.3 Hz, 2H, Ph-H), 7.91 (d, J = 8.7 Hz, 2H, Ph-H), 7.85 (t, J = 8.1 Hz, 4H, Ph-H), 7.78 (d, J = 7.5 Hz, 2H, Ph-H), 7.52 (t, J = 7.6 Hz, 2H, Ph-H), 7.43 (t, J = 7.3 Hz, 1H, Ph-H), 7.27 – 7.16 (m, 3H, Ph-H), 7.07 (d, J = 6.4 Hz, 2H, Ph-H), 6.94 (d, J = 9.0 Hz, 2H, Ph-H), 6.90 (d, J = 6.3 Hz, 2H, Ph-H), 5.17 (d, J = 16.4 Hz, 1H, triazoleCH), 5.06 (d, J = 16.3 Hz, 1H, triazoleCH), 4.47 (td, J = 8.5, 5.4 Hz, 1H, CH), 3.76 (s, 3H, OCH3), 3.11 (s, 3H, NCH3), 2.93 (dd, J = 13.4, 5.1 Hz, 1H, PhCH), 2.70 (dd, J = 13.5, 9.2 Hz, 1H, PhCH). 13C NMR (100 MHz, DMSO-d6) δ 171.19 (C=O), 165.63 (C=O), 165.50 (C=O), 159.02, 146.39, 143.60, 139.56, 139.31, 137.69, 135.80, 134.11, 129.54 (2×C), 129.34 (2×C), 129.11 (2×C), 128.86 (2×C), 128.71 (2×C), 128.63, 127.40 (2×C), 127.07 (2×C), 127.04, 126.53, 125.88 (2×C), 122.89, 121.05 (2×C), 115.10 (2×C), 55.86, 52.10, 51.82, 37.88, 37.78. ESI-MS: m/z 665.4 (M+1), 682.5 (M+18), 687.4 (M+23), 703.5 (M+39). C40H36N6O4 [664.8].
4.2. In vitro anti-HIV assay in TZM-bl Cells [40]
Inhibition of HIV-1 infection was measured as a reduction in the level of luciferase gene expression after a single round of virus infection of TZM-bl cells as described previously. Briefly, 200 TCID50 of virus (NL4-3) was used to infect TZM-bl cells in the presence of various concentrations of compounds. Two days after infection, the culture medium was removed from each well, and 100 μL of Bright Glo reagent (Promega, San Luis Obispo, CA) was added to the cells to measure luminescence using a Victor 2 luminometer. The effective concentration (EC50) against HIV-1 strains was defined as the concentration that caused a 50% decrease in luciferase activity (relative light units) compared to that of virus control wells.
4.3. Cytotoxicity Assay [43]
A CytoTox-Glo™ cytotoxicity assay (Promega) was used to determine the cytotoxicity of the synthesized compounds. Parallel to the antiviral assays, TZM-bl cells were cultured in the presence of various concentrations of the compounds for 2 days. The percent of viable cells was determined by following the protocol provided by the manufacturer. The 50% cytotoxic concentration (CC50) was defined as the concentration that caused a 50% reduction in cell viability.
4.4. Binding to CA Proteins Analysis via Surface Plasmon Resonance (SPR)
All binding assays were performed on a ProteOn XPR36 SPR Protein Interaction Array System (Bio-Rad Laboratories, Hercules, CA). The instrument temperature was set at 25°C for all kinetic analyses. ProteOn GLH sensor chips were preconditioned with two short pulses each (10 seconds) of 50 mM NaOH, 100 mM HCl, and 0.5% sodium dodecyl sulfide. Then the system was equilibrated with PBS-T buffer (20 mM sodium phosphate, 150 mM NaCl, and 0.005% polysorbate 20, pH 7.4). The surface of a GLH sensorchip was activated with a 1:100 dilution of a 1:1 mixture of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (0.2 M) and sulfo-N-hydroxysuccinimide (0.05 M). Immediately after chip activation, the HIV-1NL4-3 capsid protein constructs, purified as in Xu et al. [22], were prepared at a concentration of 100 μg/ml in 10 mM sodium acetate, pH 5.0 and injected across ligand flow channels for 5 min at a flow rate of 30 μl/min. Then, after unreacted protein had been washed out, excess active ester groups on the sensor surface were capped by a 5 minute injection of 1 M ethanolamine HCl (pH 8.0) at a flow rate of 5 μl/min. A reference surface was similarly created by immobilizing a non-specific protein (IgG b12 anti HIV-1 gp120; was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Anti-HIV-1 gp120 Monoclonal (IgG1 b12) from Dr. Dennis Burton and Carlos Barbas) and was used as a background to correct non-specific binding.
To prepare a compound for direct binding analysis, compound stock solutions, along with 100% DMSO, and totaling 30μl was made to a final volume of 1 ml by addition of sample preparation buffer (PBS, pH 7.4). Preparation of analyte in this manner ensured that the concentration of DMSO was matched with that of running buffer with 3% DMSO. Serial dilutions were then prepared in the running buffer (PBS, 3% DMSO, 0.005% polysorbate 20, pH 7.4) and injected at a flow rate of 100 μl/min, for a 1 minute association phase, followed by up to a 5 minutes dissociation phase using the “one shot kinetics” capability of the Proteon instrument [44]. Data were analyzed using the ProteOn Manager Software version 3.0 (Bio-Rad). The responses from the reference flow cell were subtracted to account for the nonspecific binding and injection artifacts. The equilibrium dissociation constant (KD) for the interactions, and derived from a minimum of three experiments, were calculated in ProteOn Manager Version 3.1.0.6 (Bio-Rad, Hercules, CA), using the equilibrium analysis function.
4.5. Action Stage Determination of 6a-9
Please refer to our previously published research for methodology [27].
4.6. ELISA-based Quantification of Capsid (p24) Content [19]
An ELISA plate was coated with 50 ng of mouse anti-p24 (Abcam, ab9071) per well for 2 hours at room temperature, blocked with 3% BSA for 2 hours at room temperature and washed with PBST buffer (0.1% Tween-20 in PBS). Pseudovirus stocks were lysed with 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO) at 37°C for 1 hour and added to the plate overnight at 4°C. Simultaneously, p24 protein was added for the generation of a standard curve. Following the overnight incubation, the plate was washed with PBST buffer and 1:5000 dilution of rabbit anti-p24 (Abcam, ab63913) was added for 2 hours at room temperature. After washing the unbound rabbit anti-p24 off the plate with PBST buffer, goat anti-rabbit-HRP at a 1:5000 dilution was added for 1 hour at room temperature. The plate was then extensively washed with PBST buffer. Subsequently, a solution of 0.4 mg/ml o-phenylenediamine in a phosphate-citrate buffer with sodium perborate (Sigma-Aldrich) was added and incubated in the dark for 30 minutes. Optical densities were then obtained at 450 nm in a Multiskan™ GO Microplate Spectrophotometer (Thermo Scientific).
4.7. In Vitro Capsid Assembly Assay [19]
Briefly, 1.0 μl of concentrated compound (10 mM) in 100% DMSO was added to a 74-μl aqueous solution (solution was made by mixing 2 ml of 5 M NaCl with 1 ml of 200 mM NaH2PO4, pH 8.0). To initiate the assembly reaction, 25 μl of purified capsid protein (120 μM) was added. An identical reaction mixture was prepared, omitting the compound (i.e., just DMSO as a vehicle control). Readings were taken at 350 nm every one minute for 19 minutes. Capsid was used at a final concentration of 30 μM. PF-74 (positive control) and compounds at 50μM (3% DMSO final).
4.8. Molecular Dynamics Simulation
The most active compound 6a-9 was docked by Autodock 4.2.6 using default settings [42]. Also, to keep consistency of the MD simulation for different series of CA HIV-1 monomer inhibitors, we used the same procedure for MD simulation and its analysis, please refer to our previously published research for methodology [27].
4.9. Metabolic Stabilities in Human Liver Microsomes and Human Plasma
Please refer to our previously published research for methodology [45].
Highlights (for review).
4-phenyl-1H-1,2,3-triazole phenylalanine derivatives as HIV-1 CA inhibitors were first reported.
6a-9 showed the best anti-HIV-1 activity among this series.
6a-9 exhibited the dual-stage inhibitory activity like the lead PF-74.
Acknowledgements
Financial support from the National Natural Science Foundation of China (NSFC Nos. 81273354, 81573347), Key Project of NSFC for International Cooperation (No. 81420108027), Young Scholars Program of Shandong University (YSPSDU, No. 2016WLJH32), the Fundamental Research Funds of Shandong University (No. 2017JC006), Key research and development project of Shandong Province (Nos. 2017CXGC1401, 2019JZZY021011), Major Project of Science and Technology of Shandong Province (No. 2015ZDJS04001), NIH grants R01GM125396 (transitioning to R01AI150491) (Cocklin, PI) and T32-MH079785 are gratefully acknowledged.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- [1].De Clercq E, Antivirals: past, present and future, Biochem. Pharmacol. 85 (2013) 727–744. [DOI] [PubMed] [Google Scholar]
- [2].Kang D, Huo Z, Wu G, Xu J, Zhan P, Liu X, Novel fused pyrimidine and isoquinoline derivatives as potent HIV-1 NNRTIs: a patent evaluation of WO2016105532A1, WO2016105534A1 and WO2016105564A1, Expert. Opin. Ther. Pat 27 (2017) 383–391. [DOI] [PubMed] [Google Scholar]
- [3].Menendez-Arias L, Molecular basis of human immunodeficiency virus type 1 drug resistance: overview and recent developments, Antiviral Res. 98 (2013) 93–120. [DOI] [PubMed] [Google Scholar]
- [4].Menendez-Arias L, Richman DD, Editorial overview: antivirals and resistance: advances and challenges ahead, Curr. Opin. Virol 8 (2014) iv–vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhan P, Pannecouque C, De Clercq E, Liu X, Anti-HIV Drug Discovery and Development: Current Innovations and Future Trends, J. Med. Chem 59 (2016) 2849–2878. [DOI] [PubMed] [Google Scholar]
- [6].Costi R, Metifiot M, Esposito F, Cuzzucoli Crucitti G, Pescatori L, Messore A, Scipione L, Tortorella S, Zinzula L, Novellino E, Pommier Y, Tramontano E, Marchand C, Di Santo R, 6-(1-Benzyl-1H-pyrrol-2-yl)-2,4-dioxo-5-hexenoic acids as dual inhibitors of recombinant HIV-1 integrase and ribonuclease H, synthesized by a parallel synthesis approach, J. Med. Chem 56 (2013) 8588–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Le VS, Mouland AJ, Valienteecheverría F, Roles of HIV-1 capsid in viral replication and immune evasion, Virus. Res 193 (2014) 116–129. [DOI] [PubMed] [Google Scholar]
- [8].Perrier M, Bertine M, Le Hingrat Q, Joly V, Visseaux B, Collin G, Landman R, Yazdanpanah Y, Descamps D, Charpentier C, Prevalence of gag mutations associated with in vitro resistance to capsid inhibitor GS-CA1 in HIV-1 antiretroviral-naive patients, J. Antimicrob. Chemother 72 (2017) 2954–2955. [DOI] [PubMed] [Google Scholar]
- [9].Sundquist WI, Kräusslich HG, HIV-1 assembly, budding, and maturation, Cold. Spring. Harb. Perspect. Med 7 (2012) a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG, STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability, Science. 349 (2015) 99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua Y, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M, X-ray structures of the hexameric building block of the HIV capsid, Cell. 137 (2009) 1282–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao G, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning J, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang P, Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics, Nature. 497 (2013) 643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pornillos O, Ganser-Pornillos BK, Yeager M, Atomic-level modelling of the HIV capsid, Nature. 469 (2011) 424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI, Assembly properties of the human immunodeficiency virus type 1 CA protein, J. Virol 78 (2004) 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].López CS, Eccles JD, Still A, Sloan RE, Barklis RL, Tsagli SM, Barklis E, Determinants of the HIV-1 core assebly pathway, Virology. 417 (2011) 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Adamson CS, Freed EO, Novel approaches to inhibiting HIV-1 replication, Antiviral. Res 85 (2010) 119–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mascarenhas AP, Musier-Forsyth K, The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors. FEBS. J 276 (2009) 6118–6127. [DOI] [PubMed] [Google Scholar]
- [18].Neira JL, The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly, FEBS. J 276 (2009) 6110–6117. [DOI] [PubMed] [Google Scholar]
- [19].Xu JP, Francis AC, Meuser ME, Mankowski M, Ptak RG, Rashad AA, Melikyan GB, Cocklin S, Exploring modifications of an HIV-1 capsid inhibitor: design, synthesis, and mechanism of action, J. Drug. Des. Res 5 (2018) pii: 1070. [PMC free article] [PubMed] [Google Scholar]
- [20].Kortagere S, Madani N, Mankowski MK, Schon A, Zentner I, Swaminathan G, Princiotto A, Anthony K, Oza A, Sierra LJ, Passic SR, Wang X, Jones DM, Stavale E, Krebs FC, Martin-Garcia J, Freire E, Ptak RG, Sodroski J, Cocklin S, Smith AB 3rd, Inhibiting early-stage events in HIV-1 replication by small-molecule targeting of the HIV-1 capsid, J. Virol 86 (2012) 8472–8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kortagere S, Xu JP, Mankowski MK, Ptak RG, Cocklin S, Structure-activity relationships of a novel capsid targeted inhibitor of HIV-1 replication, J. Chem. Inf. Model 54 (2014) 3080–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xu JP, Branson JD, Lawrence R, Cocklin S, Identification of a small molecule HIV-1 inhibitor that targets the capsid hexamer, Bioorg. Med. Chem. Lett 26 (2016) 824–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Curreli F, Zhang H, Zhang X, Pyatkin I, Victor Z, Altieri A, Debnath AK, Virtual screening based identification of novel small-molecule inhibitors targeted to the HIV-1 capsid, Bioorg. Med. Chem 19 (2011) 77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lamorte L, Titolo S, Lemke CT, Goudreau N, Mercier JF, Wardrop E, Shah VB, von Schwedler UK, Langelier C, Banik SS, Aiken C, Sundquist WI, Mason SW, Discovery of novel small-molecule HIV-1 replication inhibitors that stabilize capsid complexes, Antimicrob. Agents. Chemother 57 (2013) 4622–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kelly BN, Kyere S, Kinde I, Tang C, Howard BR, Robinson H, Sundquist WI, Summers MF, Hill CP, Structure of the antiviral assembly inhibitor CAP-1 complex with the HIV-1 CA protein, J. Mol. Biol 373 (2007) 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tang C, Loeliger E, Kinde I, Kyere S, Mayo K, Barklis E, Y Sun M Huang, M.F. Summers, Antiviral inhibition of the HIV-1 capsid protein, J. Mol. Biol 327 (2003)1013–1020. [DOI] [PubMed] [Google Scholar]
- [27].Wu G , Zalloum WA, Meuser ME, Jing L, Kang D, Chen CH, Tian Y, Zhang F, Cocklin S, Lee KH, Liu X, Zhan P, Discovery of phenylalanine derivatives as potent HIV-1 capsid inhibitors from click chemistry-based compound library, Eur. J. Med. Chem 158 (2018) 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jiang X, Wu G , Zalloum WA, Meuser ME, Dick A, Sun L, Chen CH, Kang D, Jing L, Jia R, Cocklin S, Lee KH, Liu X, Zhan P, Discovery of novel 1,4-disubstituted 1,2,3-triazole phenylalanine derivatives as HIV-1 capsid inhibitors, RSC. Adv 9 (2019) 28961–28986. 10.1039/C9RA05869A [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL, HIV capsid is a tractable target for small molecule therapeutic intervention, PLoS. Pathog 6 (2010) e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M, Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl. Acad. Sci. USA 111 (2014) 18625–18630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Henning MS, Dubose BN, Burse MJ, Aiken C, Yamashita M, In vivo functions of CPSF6 for HIV-1 as revealed by HIV-1 capsid evolution in HLA-B27-positive subjects. PLoS. Pathog 10 (2014) e1003868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Price AJ, Jacques DA, McEwan WA, Fletcher AJ, Essig S, Chin JW, Halambage UD, Aiken C, James LC, Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly, PLoS. Pathog 10 (2014)e1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ganser-Pornillos BK, Cheng A, Yeager M, Structure of full-length HIV-1 CA: a model for the mature capsid lattice, Cell. 131 (2007) 70–79. [DOI] [PubMed] [Google Scholar]
- [34].Pak AJ, Grime JMA, Yu A, Voth GA, Off-Pathway Assembly: A Broad-Spectrum Mechanism of Action for Drugs That Undermine Controlled HIV-1 Viral Capsid Formation, J. Am. Chem. Soc (2019). doi: 10.1021/jacs.9b01413. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bondy SS, Chou CH, Link JO, TSE WC, Antiviral agents, WO2015/130966A1, (2015). [Google Scholar]
- [36].Bender JA, Lopez OD, Nguyen VN, Yang Z, Wang AX, Wang G, Meanwell NA, Beno BR, Fridell RA, Belema M, Thangathirupathy S, Inhibitors of human immunodeficiency virus replication, WO2016/172425A1, (2016). [Google Scholar]
- [37].Jiang X, Hao X, Jing L, Wu G, Kang D, Liu X, Zhan P, Recent applications of click chemistry in drug discovery, Expert. Opin. Drug. Discov (2019) 1–11. [DOI] [PubMed] [Google Scholar]
- [38].Gao P, Sun L, Zhou J, Li X, Zhan P, Liu X, Discovery of novel anti-HIV agents via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) click chemistry-based approach, Expert. Opin. Drug. Discov 11 (2016) 857–871. [DOI] [PubMed] [Google Scholar]
- [39].Wang X, Huang B, Liu X, Zhan P. Discovery of bioactive molecules from CuAAC click-chemistry-based combinatorial libraries, Drug. Discov. Today 21 (2016) 118–132. [DOI] [PubMed] [Google Scholar]
- [40].Lai W, Huang L, Ho P, Li ZJ, Montefiori D, Chen CH, Betulinic acid derivatives that target gp120 and inhibit multiple genetic subtypes of HIV-1, Antimicrob. Agents Chemother 52 (2008) 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zentner I, Sierra LJ, Fraser AK, Maciunas L, Mankowski MK, Vinnik A, Fedichev P, Ptak RG, Martin-Garcia J, Cocklin S, Identification of a small-molecule inhibitor of HIV-1 assembly that targets the phosphatidylinositol (4,5)-bisphosphate binding site of the HIV-1 matrix protein, ChemMedChem. 8 (2013) 426–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ, Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility, J. Comput. Chem 16 (2009) 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dang Z, Lai W, Qian K, Ho P, Lee KH, Chen CH, Huang L, Betulinic acid derivatives as human immunodeficiency virus type 2 (HIV-2) inhibitors, J. Med. Chem 52 (2009) 7887–7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bravman T, Bronner V, Lavie K, Notcovich A, Papalia GA, Myszka DG, Exploring "one-shot" kinetics and small molecule analysis using the ProteOn XPR36 array biosensor, Anal. Biochem 358 (2006) 281–288. [DOI] [PubMed] [Google Scholar]
- [45].Zhang J, Poongavanam V, Kang DW, Bertagnin C, Lu HM, Kong XJ, Ju H, Lu XY, Gao P, Tian Y, Jia HY, Desta S, Ding X, Sun L, Fang ZJ, Huang BS, Liang XW, Jia RF, Ma XL, Xu WF, Murugan NA, Loregian A, Huang B, Zhan P, Liu XY, Optimization of N-substituted oseltamivir derivatives as potent inhibitors of group-1 and -2 influenza A neuraminidases, including a drug-resistant variant, J. Med. Chem 61 (2018) 6379–6397. [DOI] [PubMed] [Google Scholar]