Abstract
Plasma lipoprotein (a) [Lp(a)] levels are largely determined by variation in the LPA gene, which codes for apo(a). Genome-wide association studies (GWASs) have identified nonsynonymous variants in LPA that associate with low Lp(a) levels, although their effect on apo(a) function is unknown. We investigated two such variants, R990Q and R1771C, which were present in four null Lp(a) individuals, for structural and functional effects. Sequence alignments showed the R990 and R1771 residues to be highly conserved and homologous to each other and to residues associated with plasminogen deficiency. Structural modeling showed both residues to make several polar contacts with neighboring residues that would be ablated on substitution. Recombinant expression of the WT and R1771C apo(a) in liver and kidney cells showed an abundance of an immature form for both apo(a) proteins. A mature form of apo(a) was only seen with the WT protein. Imaging of the recombinant apo(a) proteins in conjunction with markers of the secretory pathway indicated a poor transit of R1771C into the Golgi. Furthermore, the R1771C mutant displayed a glycosylation pattern consistent with ER, but not Golgi, glycosylation. We conclude that R1771 and the equivalent R990 residue facilitate correct folding of the apo(a) kringle structure and mutations at these positions prevent the proper folding required for full maturation and secretion. To our knowledge, this is the first example of nonsynonymous variants in LPA being causative of a null Lp(a) phenotype.
Keywords: apolipoprotein (a), endoplasmic reticulum, Golgi, kringle structure, null allele, protein modeling
Lipoprotein (a) [Lp(a)] is an atherogenic and prothrombotic molecule comprised of a LDL covalently linked to apo(a) (1–3). Plasma concentrations of Lp(a) are highly variable and elevated levels are an independent genetic risk factor for CVD (4). Up to 90% of the variation in plasma Lp(a) concentrations can be attributed to variation in the LPA gene, which codes for apo(a) (5, 6). apo(a) has evolved from duplication of multiple copies of the kringle IV (KIV) domain and the kringle V (KV) and protease domains of the PLG gene, which codes for the serine protease, plasminogen (7). A distinctive feature of the LPA gene is a unique size polymorphism caused by a copy number variation from 2 to >40 copies of the KIV-2 domain (8). The KIV-2 copy number variation results in a range of differently sized apo(a) isoforms and has been shown to have a significant influence on Lp(a) levels (4, 5).
The liver is the major site of LPA expression (7, 9). Once translated, apo(a) undergoes extensive processing and glycosylation modification prior to its secretion from hepatocytes and subsequent association with LDL (10–12). Multiple studies have shown that small apo(a) isoforms exhibit a shorter retention time in the ER and are more efficiently secreted than large isoforms (11, 13–15). These observations concur with genetic studies demonstrating the KIV-2 copy number to be inversely correlated with plasma Lp(a) concentration (5, 16–18) and with kinetic studies showing that plasma Lp(a) levels are largely determined by secretion rate (19).
The size of the effect of the KIV-2 copy number variation on Lp(a) levels differs widely between different populations, accounting for between 19% and 77% of the variation (20–22). Regardless of the size of the effect, a common finding is disparity between the Lp(a) levels of individuals with the same KIV-2 copy number (23–25). Remaining differences in levels have been attributed to additional variation in the LPA gene (26, 27). Two SNPs (rs3798220 and rs10455872) linked to small apo(a) isoforms have repeatedly been shown to associate with increased levels in genome-wide association studies (GWASs) (28–30) and rarer SNPs (e.g., rs186696265) that associate with elevated levels independent of apo(a) size do exist (30). Lp(a) null alleles, on the other hand, provide LPA gene variation that associates with decreased plasma Lp(a) levels (31).
Lp(a)-null alleles occur in individuals that have undetectable plasma Lp(a) and negligible apo(a) protein levels and are the result of loss-of-function mutations in the LPA gene (31–33). Of several variants identified in LPA that are predicted to result in a loss-of-function, three have been functionally characterized as causative of a null Lp(a) phenotype. These include a +1 donor splice site mutation in KIV-8, rs41272114 (c.4289+1G>A) (31), and a nonsense mutation in exon 1 of KIV-2 (R20X) (32), which both cause premature stop codons and result in trace amounts of a truncated apo(a) protein in plasma. A rare donor splice variant in exon 1 of KIV-2 associated with undetectable apo(a) in an African individual was also found to be causative of a null phenotype (33). While not defined as a null allele, a common splice variant in exon 2 of KIV-2 (G4925A) was shown to display defective splicing and a marked reduction in apo(a) protein and Lp(a) levels (34). In addition, a predicted loss-of-function splice acceptor variant, rs143431368 (c.4974-2A>G), identified in a Finnish study, was associated with a significant reduction in Lp(a) levels and CVD risk (35). A novel predicted loss-of-function splice acceptor variant (rs199583644) recently identified in African Americans was also significantly associated with decreased Lp(a) (36). A few rare nonsynonymous SNPs have been associated with reduced plasma Lp(a) in GWASs (30, 36–38); however, none have been characterized for their functional effects.
Here, we present structural and functional data for two rare nonsynonymous SNPs (rs41259144 and rs139145675) present in null Lp(a) individuals that have previously been associated with reduced Lp(a) levels. We show that both are in an equivalent position to mutations in PLG that cause plasminogen deficiency and that mutation of that position produces an apo(a) protein that is unable to be secreted. To our knowledge, these SNPs represent the first examples of full-length apo(a)-null alleles.
METHODS
Subjects
The null Lp(a) samples used in this study were selected from two predominantly European cohorts for which Lp(a) levels were available. One cohort (Otago LPA) was a local population being screened for Lp(a) levels and Lp(a) proteomics and genetics (see supplemental Table S1 for demographic data on this population). The other cohort was the Gout in New Zealand Study, for which demographics have been previously published and show similar characteristics to other European gout populations (39). These studies were approved by the Multi-Regional and Lower Southern Regional Ethics Committees, respectively, with participants giving written informed consent. The study abides by the principles of the Helsinki Declaration. The Otago LPA cohort consisted of volunteers recruited from the local community to take part in an Lp(a) screening study. There were no exclusion criteria and the population included both healthy subjects and subjects that had a personal and/or family history of CVD. The European gout population consisted of healthy control and gout subjects recruited from outpatient clinics in Auckland, Wellington, and Christchurch with a diagnosis of gout confirmed by a rheumatologist (39).
Selection criteria for null samples
The six samples that were chosen for further analysis demonstrated a clear apo(a)-null phenotype by three criteria (outlined in supplemental Table S2). First, they had a plasma Lp(a) below the detectable limit of Lp(a) <3 mg/dl. Second, they had no detectable apo(a) bands on phenotyping; third, on sequencing, they were shown to harbor sequence variations that have previously been reported to be associated with a decreased Lp(a). Not all of the samples that met the second criteria were sequenced, and only some samples that were sequenced met the third criteria. We performed next generation sequencing on 11 null samples in total (5 from the Otago LPA and 6 from the NZ Gout population) and only 6 of these met our criteria 3.
Lp(a) measurement and apo(a) phenotyping
Lp(a) levels were measured by a commercial assay [Quantia Lp(a); Abbot Laboratories Inc., Abbot Park, IL] that had a limit of quantification of 3 mg/dl. apo(a) phenotyping was performed on plasma samples by SDS-PAGE on 4% polyacrylamide gels alongside samples of known apo(a) isoform size. Separated plasma proteins were transferred to nitrocellulose membrane and Western blotting was performed using a goat anti-Lp(a) polyclonal antibody to detect apo(a) (Wako Pure Chemical Corporation, Osaka, Japan) and a HRP-labeled anti-goat IgG secondary antibody (Santa Cruz Biotechnology Inc., Dallas, TX). Membranes were imaged using electrochemiluminescence on a Licor Odyssey imager. The isoform designation refers to the total number of all KIV repeats. Samples with an Lp(a) level <3 mg/dl that showed no detectable apo(a) on phenotyping were classified as nulls.
Next generation sequencing of the LPA gene in null individuals
Amplicon primer sets (n = 35) were designed to amplify the promoter and coding regions of LPA. In the case of the nonrepeat kringle domains, primers were designed to anneal in the flanking introns of each exon to provide enough variation between kringle types to prevent nonspecific amplification of highly homologous kringles. The two KIV-2 exons were amplified in a batch-wise manner similar to previous KIV-2 sequencing analyses (33, 34) except that exon 1 and exon 2 were amplified separately. Primers were designed to flank each of the two exons independently and were placed in regions of sequence homology between KIV-2 repeats according to the Ensembl gene reference sequence ENSG00000198670, GRCh38 to facilitate amplification of every repeat within both alleles of a sample simultaneously. Amplicons were cleaned using Agencourt AMPure XP PCR Purification Beads (Beckman Coulter Inc., Indianapolis, IN) and combined at equimolar concentrations to generate one pool for sequencing. Next generation sequencing was performed by New Zealand Genomics Limited (Dunedin, New Zealand) on a single flow cell lane of an Illumina MiSeq sequencer using paired-end chemistries and a 250 bp run. Details of the bioinformatic analysis of the sequencing data can be found in the supplemental Methods.
apo(a) and plasminogen sequence alignments
The amino acid sequence for the single copy kringle domains and the first KIV-2 domain of apo(a) and the kringle domains KI to KV of plasminogen were aligned in Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) and the output file was then processed in Jalview (http://www.jalview.org/). The numbering of residues was according to the NCBI apo(a) precursor protein reference sequence NP_005568.2 for apo(a) and the GenBank protein reference sequence AAA60113.1 for plasminogen.
Modeling of apo(a) and plasminogen mutants
The X-ray crystal structure for KV of apo(a) [Protein Data Bank (PDB) accession 4BVV] and the X-ray and nuclear magnetic resonance structures for KI and KII of plasminogen (PDB accession 4cik and 1b2i, respectively) were downloaded for use in PyMOL (Schrödinger LLC, New York, NY). As a structure for KIV-4 of apo(a) was not available, a homology model based on the crystal structure of apo(a) KIV-7 (PDB accession 4BVW) was generated using the SCRWL modeling method within the Fold and Function Assignment System (FFAS) (40). Within PyMOL, the structure backbone was visualized as a cartoon and the variant residue (and interacting groups) in each structure was visualized as sticks. Evidence of bonding between atoms of the variant residue and any other atoms in the structure were identified by finding polar contacts within a distance of 3.2 Å. The mutagenesis wizard tool of PyMOL was used to alter the residues to their respective mutants. Sidechain rotamers for variant residues were chosen to minimize steric hindrance.
apo(a) cDNA vector and mutagenesis
A mammalian expression vector containing an apo(a) cDNA insert with six KIV-2 repeats (giving an isoform size of 15) under the control of the cytomegalovirus (CMV) promoter and tagged with GFP was purchased from Origene (Origene Technologies Inc., Rockville, MD). A mutant form of the apo(a)-GFP vector containing the R1771C substitution was created by site-directed mutagenesis of the WT apo(a)-GFP vector using a modified version of the Quikchange protocol and high-fidelity Pfu polymerase (Promega, Madison, WI). The sequence of the mutagenic primers and conditions used for the mutagenesis reaction are provided in supplemental Tables S3 and S4. Successful mutagenesis was verified by Sanger sequencing.
Sequencing of apo(a) cDNA vectors
All WT and R1771C apo(a)-GFP vectors were sequenced using the Oxford Nanopore MinION device and reagents (Oxford Nanopore Technologies Ltd, Oxford, UK). Both plasmids (7 μg) were linearized overnight with EcoRI (New England Biolabs, Ipswich, MA). Digested products were purified using Agencourt AMPure XP beads (Beckman Coulter Inc.). End repair of DNA, dA-tailing, and sequencing adaptor ligation were performed using the SQK-LSK108 ligation sequencing kit (Oxford Nanopore Technologies) as per the manufacturer’s instructions. Prepared libraries for both plasmids (75 μl) were sequenced sequentially for 1 h on the same FLO-MIN106 (R9.4) flowcell (Oxford Nanopore Technologies) with washing in between using the EXP-WSH002 wash kit. Base calling was performed using Albacore 2.1.7 (Oxford Nanopore Technologies) and read assembly was performed using CANU (41) with the default parameters. Sequence alignment with NCBI cDNA reference sequence NM_005577 and the pAC-CMV-GFP vector backbone sequences (Origene Technologies) was performed in Geneious (42).
Recombinant expression of apo(a)
The recombinant apo(a)-GFP proteins were expressed in both HEK293 human embryonic kidney cells (American Type Culture Collection, Manassas, VA) and Huh7 human hepatoma cells (Japanese Collection of Research Bioresources Cellbank, Osaka, Japan). Neither cell line displayed endogenous expression of apo(a) as assessed by Western blot analysis. Cells were grown in T-75 cell culture flasks (Greiner Bio-One, Kremsmünster, Austria) in DMEM supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 μg/ml streptomycin, 100 U/ml penicillin, and 0.25 μg/ml amphotericin B (all from Invitrogen, Carlsbad, CA) in a humidified environment at 37°C with 5% CO2. Cells were seeded in CELLSTAR® 6-well plates (Greiner Bio-One) and transfected for 24 h with 2.5 μg of the apo(a)-GFP expression vectors using Lipofectamine®3000 (Thermo Fisher Scientific) and then maintained in fresh DMEM for a further 48 h. Total mRNA was extracted from mock and transfected cells and mRNA levels of both apo(a)-GFP vectors analyzed by quantitative (q)PCR. Intracellular apo(a) was investigated by Western blotting of cell lysates prepared in RIPA buffer (50 mM Tris-HCl pH 7.8, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) supplemented with mini protease inhibitor cocktail tablets (Hoffman-La Roche, Basel, Switzerland). apo(a) secreted in the media (500 μl) was immunoprecipitated using a goat anti-Lp(a) polyclonal antibody (Wako Pure Chemical Corporation) attached to Dynabeads Protein G (Thermo Fisher Scientific) and the immunoprecipitated proteins were subjected to Western blotting after elution in SDS-PAGE buffer.
qRT-PCR
Total RNA was isolated from HEK293 cells transfected with WT and R1771C apo(a)-GFP vectors using the Quick-RNA miniprep kit (Zymo Research). Isolated RNA was quantified via Nanodrop spectrophotometer and aliquots were immediately stored at −80°C. Total RNA (1 μg) was reversed transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis kit (Roche). Diluted cDNA was used as template for quantitative (q)RT-PCR reactions in a LightCycler 480 Instrument II (Roche) using LightCycler 480 SYBR Green I Master (Roche) and apo(a)-specific primers (see supplemental Table S3). Primers targeting neomycin (supplemental Table S3) were used to amplify the neomycin resistance cassette that is co-expressed on the apo(a)-GFP vector for normalization of apo(a) target Ct values obtained for WT and R1771C samples. Ct values for apo(a) targets and neomycin were used to calculate relative expression of R1771C apo(a)-GFP to WT via the 2−ΔΔCt method (User Bulletin #2 ABI PRISM 7700 Sequence Detection System; http://tools.thermofisher.com/content/sfs/manuals/cms_040980.pdf).
Western blots of recombinant apo(a)
Cell lysates (80 μg protein) or the immunoprecipitated proteins were subjected to SDS-PAGE on 4% polyacrylamide gels. A plasma sample from a subject with 18 KIV repeats was included as an apo(a)-positive control. Separated proteins were transferred to nitrocellulose and probed with a goat-anti-Lp(a) polyclonal antibody (Wako) and a HRP-labeled anti-goat IgG secondary antibody (Santa Cruz Biotechnology Inc.). Actin was used as a loading control for cell lysates, while IgG was used as a loading control for immunoprecipitated proteins, both on 10% polyacrylamide gels. Actin was detected with a goat anti-actin antibody (Sigma-Aldrich, St. Louis, MO) and a HRP-labeled anti-goat IgG secondary antibody (Santa Cruz), while IgG was detected by an anti-goat-HRP secondary antibody (Santa Cruz). Bands were imaged on a LICOR odyssey imager using electrochemiluminescence.
Immunocytochemistry
The human hepatoma cell line, HepG2 (American Type Culture Collection), was used for immunocytochemistry experiments. Cells were maintained in advanced DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 0.25 μg/ml amphotericin B, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen) at 37°C in a humidified environment with 5% CO2. Cells were seeded for 24 h on 0.01% poly l-ornithine-coated (Sigma-Aldrich) 13 mm glass coverslips (Global Science and Technology Ltd, Auckland, New Zealand) and transfected the next day with 500 ng of either WT or R1771C apo(a)-GFP DNA. Transfections were performed using Lipofectamine®3000 transfection reagent (Thermo Fisher Scientific) according to manufacturer’s instructions. Cells were washed in cold phosphate-buffered saline and fixed with 4% paraformaldehyde solution at either 24 or 72 h posttransfection, and then immunohistochemistry was performed.
Cells were permeabilized and blocked with 0.2% Triton X-100 (Bio-Rad Laboratories, Hercules, CA) and 3% normal goat serum (Thermo Fisher Scientific), respectively. Cells were incubated with a rabbit anti-calnexin antibody (ab22595; Abcam, Cambridge, UK), which was subsequently detected with an anti-rabbit Alexa Fluor® 594 antibody (Invitrogen). In a separate experiment, cells were incubated with a mouse anti-apo(a) antibody (LPA4, MABS1284; Sigma-Aldrich) and either a rabbit anti-calnexin antibody, a rabbit anti-Sec31A antibody (ab86600; Abcam), or a rabbit anti-trans-Golgi network 2 (TGOLN2) antibody (Sigma). The anti-apo(a) antibody was subsequently detected with an anti-mouse Alexa Fluor® 647 antibody (Invitrogen). The anti-calnexin, anti-Sec31A, and anti-TGOLN2 were subsequently detected with an anti-rabbit Alexa Fluor® 555 antibody (Invitrogen). Stained coverslips were mounted using ProLong® Gold Antifade reagent with DAPI (Life Technologies, Carlsbad, CA). Images were obtained using an Olympus FluoView™ FV1000 confocal microscope. Images were compiled and analyzed using FIJI (http://fiji.sc/Fiji). Images were quantified using ImageJ.
Proteasomal inhibition of recombinant apo(a)
Huh7 cells were transfected with the apo(a)-GFP vectors for 24 h as described above, and cells were allowed to recover for 12 h. Cells were treated with either DMSO or 20 μM of the proteasomal inhibitor, clasto-lactacystin β-lactone (Merck, Kenilworth, NJ), in DMSO for an additional 12 h at which point cell lysates were harvested in RIPA buffer and 500 μl of cellular media were immunoprecipitated as described above. Harvested lysates (60 μg) or immunoprecipitated media were subjected to SDS-PAGE and Western blotting as described above.
Endoglycosidase digestion of recombinant apo(a)
Huh7 cells were seeded in 10 cm CELLSTAR® dishes (Greiner Bio-One) and transfected for 24 h with 14.5 μg of WT or R1771C apo(a)-GFP vector and cells allowed to recover for 48 h. Cells were lysed in RIPA buffer and lysates were concentrated in Amicon Ultra 0.5 ml 100K centrifugal filters (Merck) at 14,000 g for 10 min. Glycoprotein denaturing buffer (NEB) and water were added to the concentrated lysates (60 μg) and heated for 10 min at 100°C, and then chilled and briefly centrifuged. For PNGase F reactions, Glycobuffer 2 (NEB), 10% NP-40, water, and PNGase F (NEB) were added to the denatured lysates. For Endo H reactions, Glycobuffer 3 (NEB), water, and Endo H (NEB) were added to the denatured lysates. For untreated samples, denatured lysates were combined with Glycobuffer 2 (NEB), 10% NP-40, and water. All samples were incubated at 37°C for 1 h and then mixed with SDS-PAGE buffer and subjected to SDS-PAGE and Western blotting as described above.
Translation inhibition of recombinant apo(a)
Huh7 cells were transfected with WT and R1771C apo(a)-GFP vectors for 24 h as for the proteasomal inhibition experiment. Lysates and media were immediately harvested (time 0). Alternatively, cells were supplemented with fresh DMEM for untreated cells or fresh DMEM containing 50 μg/ml of cycloheximide (Abcam) for treated cells for either 12 or 24 h, after which lysates and media were harvested. Media (500 μl) from the three different time points was immunoprecipitated as previously described. Cell lysates (60 μg) and immunoprecipitated media were then subjected to SDS-PAGE and Western blotting as described above.
RESULTS
Sequencing of null individuals
The LPA genes of eleven European individuals classed as phenotypically null for Lp(a), i.e., Lp(a) undetectable by assay and no apo(a) bands on phenotyping (supplemental Fig. S1), were subjected to next generation sequencing. Six individuals who carried variations known to be associated with low Lp(a) were chosen for further study. The characteristics and genotypes of these individuals are shown in Table 1 with the associated sequencing data shown in supplemental Figs. S2–S8. One individual (G5788) was identified as heterozygous for the known null allele, rs41272114 (31) (supplemental Fig. S2), and two individuals (G5792 and LPA114) were heterozygous for rs41272112 and another known null allele, R20X (32) (supplemental Figs. S3, S4). Another three individuals (G5780, G5591, and LPA089) were found to carry the nonsynonymous SNP, rs41259144 (c.2926G>A), in heterozygous form, which results in the amino acid substitution R990Q in KIV-4 of apo(a) (Table 1, supplemental Figs. S5–S7). One of these individuals (LPA089) also carried the nonsynonymous SNP, rs139145675 (c.5311C>T), in heterozygous form, which results in a R1771C substitution in KV of apo(a) (Table 1) (supplemental Fig. S7). A seventh individual (G5751) who had no detectable Lp(a) but displayed a trace amount of a single apo(a) isoform was also sequenced. This individual was found to be heterozygous for the R990Q substitution and a common variant, G4925A, in KIV-2 associated with low Lp(a) levels (34, 43) (supplemental Fig. S8). Other nonsynonymous variants found in these individuals are listed in Table 1 and include one novel SNP (W52G) in KIV-1 and four previously described SNPs in KIV-2 (M38L and T46N) and KIV-8 (T1399P and P1428L) (supplemental Figs. S2–S8).
TABLE 1.
Characteristics and genotypes of null Lp(a) subjects selected for study
| Participant | G5780 | G5591 | G5788 | G5792 | LPA089 | LPA114 | G5751 |
| Age | 50 | 27 | 62 | 72 | 79 | 52 | 64 |
| Gender | Male | Male | Male | Male | Male | Male | Male |
| CVD history | Yes | No | Yes | Yes | Noa | Noa | No |
| TC (mmol/l) | 5.5 | 2.8 | 7.4 | 5.8 | 5.6 | 6.2 | 6.2 |
| TG (mmol/l) | 3.11 | 1.24 | 4.02 | 4.39 | 0.6 | 2.5 | 1.73 |
| HDL-C (mmol/l) | 1 | 1 | 0.76 | 1.43 | 1.67 | 1.48 | 1.53 |
| LDL-C (mmol/l) | 3.09 | 1.24 | 4.81 | 2.37 | 3.7 | 3.6 | 3.88 |
| Lp(a) (mg/dl) | <3 | <3 | <3 | <3 | <3 | <3 | <3 |
| apo(a) phenotype | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 0/0 | 21/0 |
| Genotypes | |||||||
| KIV-1 | |||||||
| W52G | Yes | — | — | — | — | — | — |
| KIV-2 | |||||||
| R20X | — | — | — | Yes | — | Yes | — |
| M38L | Yes | — | — | — | — | — | — |
| T46N | — | — | Yes | — | — | — | — |
| A114T (G4925A) | — | — | — | — | — | — | Yes |
| KIV-4 | |||||||
| R990Q rs41259144 | Yes | Yes | — | — | Yes | — | Yes |
| KIV-8 | |||||||
| +1splice rs41272114 | — | — | Yes | Yes | — | Yes | — |
| T1399P rs41272110 | — | — | — | — | — | — | Yes |
| P1428L rs76144756 | — | — | — | — | — | — | Yes |
| KV | |||||||
| R1771C rs139145675 | — | — | — | — | Yes | — | — |
| Protease | |||||||
| R2016C rs3124784 | — | — | — | — | Yes | — | — |
The KIV-2 variants are numbered as per Coassin et al. (43), which numbers the first amino acid. Variants in bold are already known null variants or variants associated with significantly decreased Lp(a). For participants with multiple variants, the heterozygosity status could not be definitely determined.
Family history of CVD data was also available for these two participants, which showed no family history.
The R990Q and R1771C variants are predicted to be damaging and probably damaging by SIFT and PolyPhen2, respectively. Large GWASs have identified both as rare (global mean allele frequency of 0.004–0.005) but significantly associated with decreased Lp(a) levels (supplemental Tables S5. S6). Indeed, some of these studies show that the allelic effect of R990Q and R1771C on plasma Lp(a) are comparable to the effect of the most common null allele, rs41272114 (+1 splice donor in KIV-8) (supplemental Table S7). This prompted us to further investigate these two variants for their potential as null alleles, which included an initial Sanger sequencing approach to confirm their presence in subject LPA089 (supplemental Fig. S9). Interestingly, a further five individuals with varying Lp(a) levels (supplemental Table S8) were identified by genotyping of the LPA Otago population to harbor the R990Q mutation. Sanger sequencing (supplemental Fig. S10) confirmed these five subjects as being heterozygous for the R990Q SNP. Interestingly, all five subjects exhibited only one isoform on Western blotting (supplemental Fig. S11), consistent with the R990Q allele being null and the other allele being WT.
Sequence alignment and structural modeling of R990Q and R1771C mutants
An alignment of apo(a) and plasminogen kringle domains shows significant sequence similarity between all apo(a) KIV domains and significant but slightly less similarity with apo(a) KV and all five plasminogen domains (Fig. 1). The alignment shows that R990Q in KIV-4 and R1771C in KV (highlighted in blue) are homologous arginine residues that are highly conserved in both apo(a) and plasminogen. Interestingly, four previously described PLG variants associated with plasminogen deficiency [R153K in KI (44), R235H in KII (45), R325H KIII (46), and R532H in KV (47), highlighted in blue] involve this arginine residue.
Fig. 1.
Sequence alignment of apo(a) and plasminogen kringle domains show the R990 and R1771 residues to be homologous to plasminogen deficiency mutations. Alignment of single copy kringle domains and the first KIV-2 domain of apo(a) as well as kringle domains of plasminogen performed using Clustal Omega. Shading was performed in Jalview with residue sequence identity set to 50%. Conserved cysteine residues involved in intra-kringle disulfide linkages are shaded yellow and surrounded by blue boxes. The R990Q and R1771C residues and those mutated in cases of plasminogen deficiency (R153K, R235H, R325H, and R532H) are shaded blue, and this position is surrounded by blue boxes. The numbering scheme for apo(a) kringle domains is based on the NCBI apo(a) precursor protein sequence (NP_005568.2). The numbering scheme for amino acids in the plasminogen kringle domains is based on GenBank plasminogen sequence AAA60113.1. This plasminogen sequence contains the signal peptide hence the numbering of the mutant plasminogen residues shown in blue here is +19 to the positions previously described as R134K, R216H, R306H, and R513H.
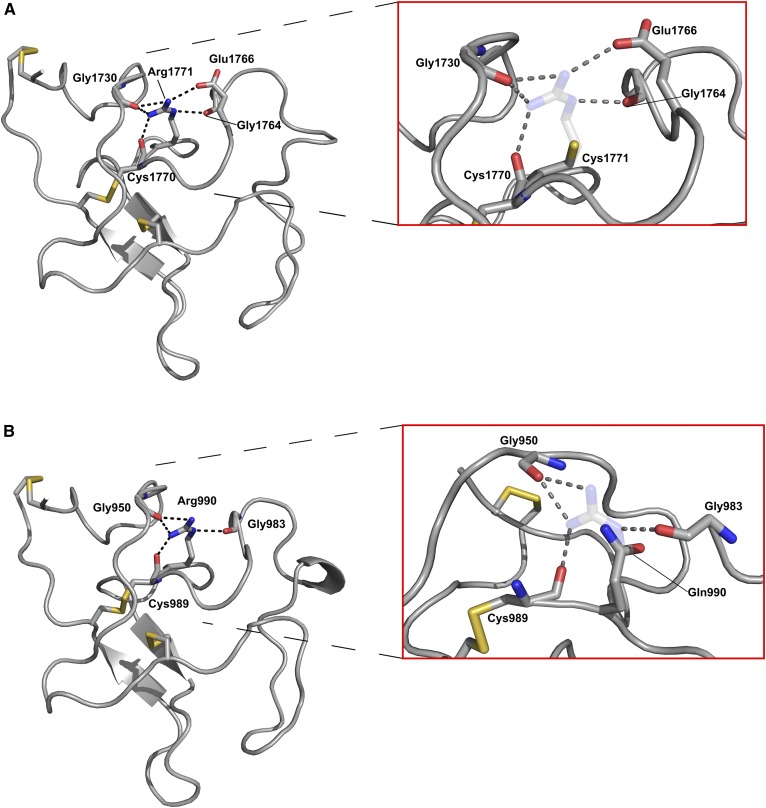
The PDB protein structure for apo(a) KV was used to model the structural consequences of R1771C, and a homology model for KIV-4 was used to model the effect of R990Q. Figure 2A shows the 1.8 Å X-ray crystal structure of KV with the cysteine residues predicted to participate in intramolecular disulfide bonds in yellow. The delta carbon-bonded amine of the guanidinium group of R1771 is positioned to make a 2.8 Å polar contact with the main chain carbonyl of G1764. One of the amine groups in the R1771 guanidinium sidechain appears to make a 2.9 Å polar contact with a carbonyl group on the E1766 sidechain as well as a 3.1 Å polar contact with the main chain carbonyl of G1730. The other amine in the R1771 guanidinium group is positioned to make a 2.9 Å polar contact with the carbonyl main chain of G1730 as well as a 2.8 Å polar contact with the carbonyl main chain of C1770. When R1771 is mutated to C1771, all of these putative polar contacts are predicted to be destroyed (inset). A homology model of apo(a) KIV-4 (Fig. 2B) showed that the R990 residue was predicted to make similar polar contacts as predicted for R1771, although the equivalent interaction with residue 1766 was not present, as this position is occupied by an isoleucine (I985) in KIV-4. As with R1771C, all putative contacts were predicted to be lost upon mutation of R990 to Q990 (inset). Furthermore, modeling of plasminogen kringles I and II predicted the equivalent arginine residues, R153 and R235, to make similar contacts to R1771 and R990 (supplemental Fig. S12A, B). The mutation of both these plasminogen residues is associated with severely impaired plasminogen secretion (46).
Fig. 2.
Protein structures of apo(a) KIV-4 and KV highlighting the R990 and R1771 residues and their interactions. A: X-ray crystal structure of apo(a) KV (PDB 4BVV). Left, WT residue R1771 with interactions predicted to be made between the arginine sidechain and chemical groups of neighboring residues shown in black. Right, predicted effects of the R1771C substitution with the putative side chain interactions predicted to be abolished by the change to cysteine shown in gray with sulfur in yellow. The WT arginine residue is shown as transparent. B: Homology model of KIV-4 based on the X-ray crystal structure for KIV-7 (PDB 4BVW). Left, WT residue R990 with interactions predicted to be made between the arginine sidechain and chemical groups of neighboring residues shown in black. Right, predicted effects of the R990Q substitution with the putative side chain interactions predicted to be abolished by the change to glutamine shown in gray. The WT arginine residue shown as transparent, and view is rotated approximately 90° around a vertical axis relative to left panel.
Recombinant expression of the R1771C mutant
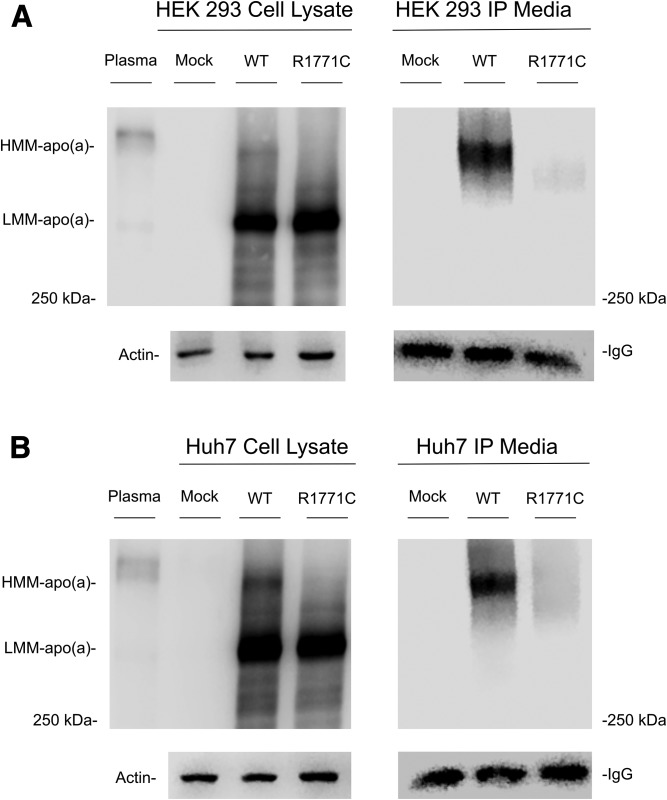
To determine the effects of changes at this position on synthesis and secretion of apo(a), a GFP-tagged apo(a) cDNA expression construct was subjected to directed mutagenesis and recombinantly expressed. The very high sequence homology surrounding the R990 residue precluded the introduction of the R990Q mutation; however, the R1771C mutation was successfully introduced. A qPCR analysis showed similar mRNA expression levels between the WT and R1771C apo(a)-GFP vectors in both cell lines (supplemental Table S9). Western blots of cell lysates from transfected HEK293 (Fig. 3A) and Huh7 (Fig. 3B) cells showed both the WT and R1771C apo(a)-GFP proteins to be expressed at similar levels in a low molecular mass form that corresponds to the immature form of the protein. The WT apo(a)-GFP also showed a less abundant higher molecular mass form of apo(a), which represents the mature form of apo(a). The mature protein migrated close to an apo(a) isoform in human plasma predicted to be slightly bigger than the apo(a)-GFP proteins. The mature form was not apparent in the R1771C apo(a)-GFP transfected cells. Western blots of apo(a) immunoprecipitants from HEK293 (Fig. 3A) and Huh7 (Fig. 3B) cell culture media showed that the WT apo(a)-GFP was secreted in the mature form. This was in contrast, to the R1771C apo(a)-GFP protein, which showed limited secretion into the culture media.
Fig. 3.
Western blots of cell lysates and immunoprecipitated culture media from HEK293 cells transfected with WT and R1771C apo(a)-GFP vectors. HEK293 (A) and Huh7 (B) cells were seeded in 6-well plates and either mock-transfected or transfected with WT or R1771C apo(a)-GFP vectors. At 72 h posttransfection, cell lysates and media were harvested. apo(a) was isolated from culture media by immunoprecipitation on Dynabeads protein G with an anti-Lp(a) antibody and eluted under reducing conditions. Cell lysates and immunoprecipitants (IPs) were run on SDS 4% polyacrylamide gels. Plasma from an individual with apo(a) KIV isoform size of 18 was run as a positive control on lysate gels. After transfer to nitrocellulose, membranes were probed with a goat anti-Lp(a) antibody and a HRP-labeled anti-goat IgG secondary antibody. For loading controls, lysates or immunoprecipitants were run on SDS 10% polyacrylamide gels, transferred to nitrocellulose, and probed with either an anti-actin antibody for lysates or an HRP-labeled anti-goat IgG antibody for immunoprecipitants. Gels were loaded with molecular mass standard labeled in kilodaltons. HMM, high molecular mass; LMM, low molecular mass.
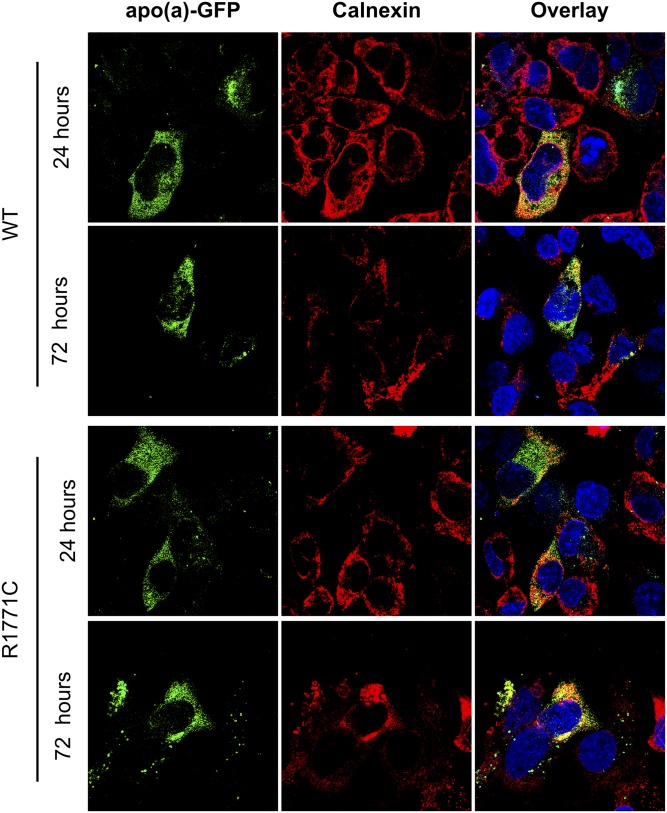
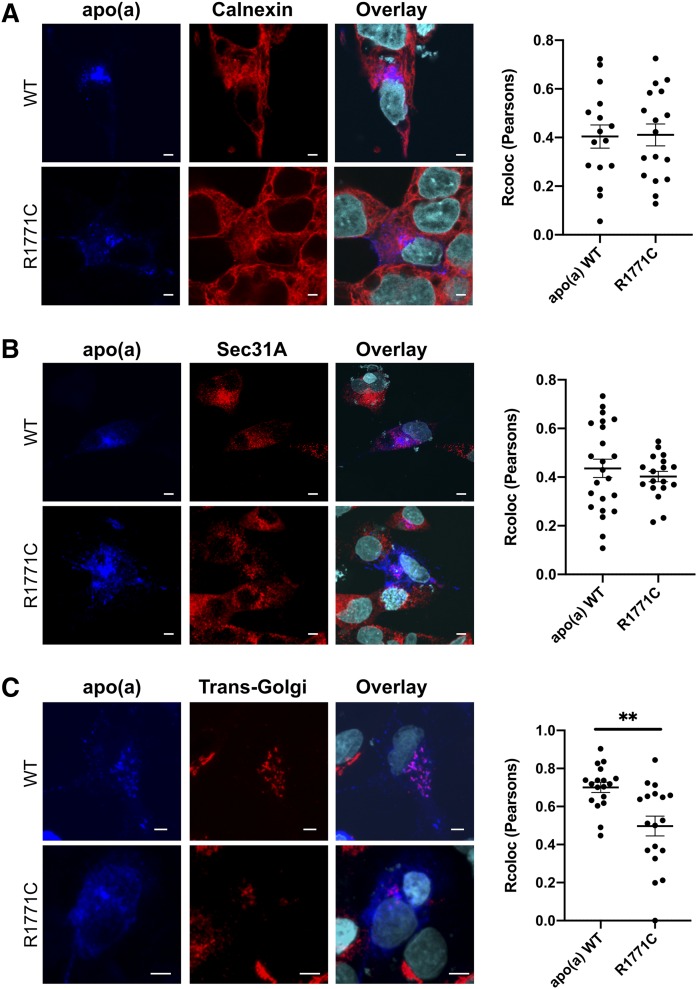
Visualization of the WT apo(a)-GFP protein inside HepG2 liver cells (Fig. 4) showed colocalization (yellow signal) with the ER-resident protein, calnexin, 24 and 72 h posttransfection. The R1771C apo(a)-GFP protein also showed a similar colocalization in the ER at 24 and 72 h. Quantification of the trafficking of the apo(a)-GFP proteins through the secretory pathway at 24 h (Fig. 5) showed both the WT and R1771C apo(a)-GFP proteins to display similar levels of colocalization (pink signal) with calnexin and Sec31A, a CopII complex protein regulating ER to Golgi trafficking. However, colocalization with TGOLN2, a Golgi resident, was significantly reduced in the R1771C mutant compared with WT apo(a) consistent with a lack of progression through the secretory pathway.
Fig. 4.
The R1771C apo(a) protein persists in the ER. HepG2 cells were transfected with 500 ng of either WT or R1771C apo(a)-GFP cDNA and fixed and imaged by confocal microscopy at 24 or 72 h. Cells were permeabilized and stained for the ER resident marker, calnexin, which was detected with a Alexa Fluor® 594 secondary antibody. Intracellular WT and R1771C apo(a)-GFP were imaged for the presence of transfected apo(a)-GFP (green) and calnexin compartment marker (red). Multiple fields of view were visualized and representative images are shown. Images are also shown as overlays with a DAPI nuclear stain (blue).
Fig. 5.
The R1771C apo(a) shows a reduced progression through the secretory pathway. HepG2 cells were transfected with 500 ng of either WT or R1771C apo(a)-GFP cDNA and fixed and imaged by confocal microscopy at 24 h. Cells were permeabilized and stained with antibodies for apo(a), the ER protein, calnexin, the CopII protein, Sec31A, and the Golgi marker protein, TGOLN2. The apo(a) antibody was detected with an anti-mouse Alexa Fluor® 647 secondary antibody (blue), the calnexin, the Sec31A and TGOLN2 antibodies were detected with an anti-rabbit Alexa Fluor® 555 secondary antibody (red). Multiple fields of view were visualized and representative images are shown. Images are also shown as overlays with a DAPI nuclear stain (cyan). Colocalization of the Alexa Fluor® 647 signal detecting apo(a) with the Alexa Fluor® 555 signal detecting the various compartment markers was assessed in confocal images of transfected cells by calculating the Pearson’s correlation coefficient using the Colocalization Finder plugin of ImageJ. Data represents mean ± SEM for at least fifteen individual cells. **P < 0.01 for R1771C versus WT apo(a).
Effect of various treatments on recombinant apo(a)
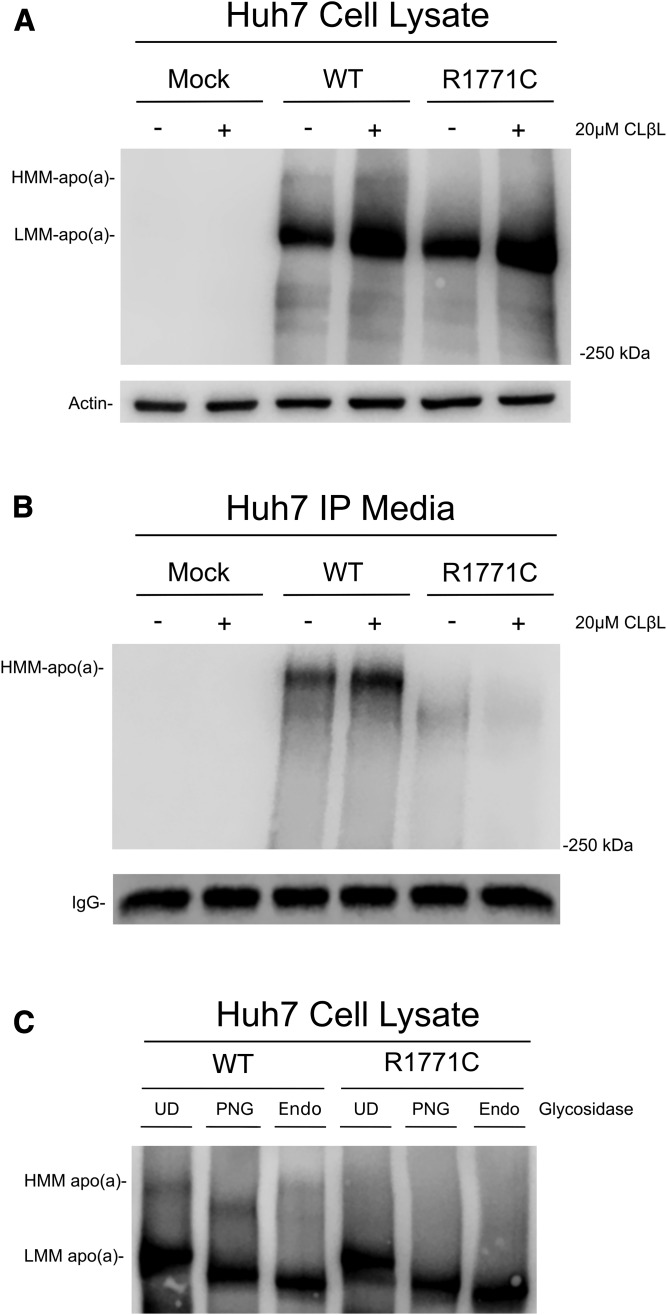
To determine whether the R1771C mutant was being degraded via the proteasome, cells expressing the recombinant apo(a) proteins were treated with a proteasomal inhibitor, clasto-lactacystin β-lactone. This caused an accumulation of the immature apo(a) in both WT and R1771C apo(a)-GFP cell lysates, indicating that both the WT and mutant apo(a) protein are subject to proteasomal degradation (Fig. 6A). The treatment also caused an increase in the WT apo(a) appearing in the media; however, there was no increase in the secretion of the R1771C apo(a) (Fig. 6B).
Fig. 6.
Treatment of recombinant apo(a) cells with proteasomal inhibitors and glycosidases. A: Huh7 cells were mock-transfected or transfected for 24 h with WT or R1771C apo(a)-GFP vectors and then incubated for 12 h in fresh DMEM. Cells were then treated for a further 12 h in the presence of 20 μM of clasto-lactacystin β-lactone after which cells were lysed in RIPA buffer and lysates (60 μg) subjected to SDS-PAGE and Western blotting as previously described. B: Culture media (500 μl) from Huh7 cells transfected and treated with clasto-lactacystin β-lactone as above was immunoprecipitated and subjected to SDS-PAGE and Western blotting as previously described. C: Huh7 cells were transfected for 24 h with WT and R1771C apo(a) vectors. Cell lysates were harvested in RIPA buffer and concentrated using Amicon Ultra centrifugal filters. Concentrated lysates were digested with PNGase F (PNG), Endo H (Endo), or undigested (UD). Reactions were added to SDS-PAGE buffer, electrophoresed on 4% SDS-PAGE gels, and subjected to Western blotting as described previously.
To determine whether the R1771C mutant was being glycosylated, the recombinant apo(a) proteins were subjected to digestion with various endoglycosidases. Results showed that digestion with PNGase F, which cleaves all N-linked sugars, reduced the size of both the WT and mutant immature apo(a), indicative of N-linked glycosylation having taken place in the ER (Fig. 6C). This was also the case for the mature form of apo(a) in the WT. The immature apo(a) for both WT and R1771C also exhibited a size decrease upon treatment with Endo H (Fig. 6C), indicating that this form of apo(a) had not yet been processed in the Golgi. The mature form of apo(a) in the WT, in contrast, did not alter size with Endo H treatment, indicative that glycolytic modifications had taken place in the Golgi.
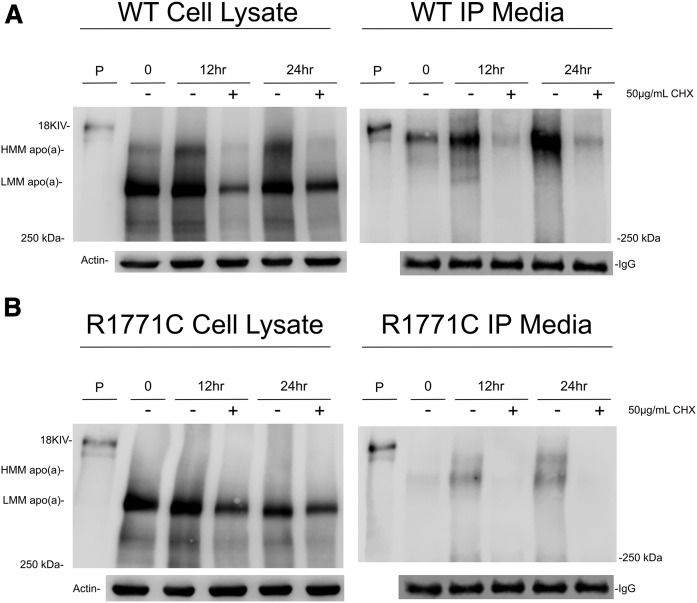
To investigate the effect of translation inhibition on the progression of the WT and R1771C apo(a)-GFP recombinants through the secretory pathway, we treated cells with cycloheximide. Results showed a low ratio of mature to immature protein in untreated WT apo(a) recombinant cells at 0 h (Fig. 7A) with the level of the immature protein remaining constant and the mature form increasing with time (24 h). This corresponded to an increasing appearance of the mature apo(a) in the media. Cycloheximide treatment effectively decreased the levels of both the immature and mature forms in cell lysates and severely reduced the secretion of mature WT apo(a) into the media. For the R1771C mutant apo(a) (Fig. 7B), as with the WT apo(a), the amount of immature apo(a) remained constant with time with the cycloheximide treatment causing a decrease. Interestingly, there appeared to be secretion of a small amount of a mature apo(a) form and an intermediate form migrating between the immature and mature forms at 12 and 24 h (Fig. 7B), which was blocked by cycloheximide treatment.
Fig. 7.
Translation inhibition decreases the amounts of WT and R1771C apo(a) in transfected cells. Huh7 cells were transfected with WT (A) and R1771C (B) apo(a)-GFP vectors. Cell lysates and media were harvested after 24 h of transfection (time 0) or incubated with fresh DMEM or DMEM containing 50 μg/ml of cycloheximide for either 12 or 24 h and subsequently harvested. apo(a) was immunoprecipitated from media (500 μl) as described in the Methods. Cell lysates (60 μg) and immunoprecipitated media were subjected to SDS-PAGE and Western blotting as described previously. An 18 KIV apo(a) isoform from plasma was loaded as a positive control.
DISCUSSION
Here we present evidence that the nonsynonymous SNP, rs139145675, which causes a R1771C substitution in apo(a) KV, is causal of a null Lp(a) phenotype. In addition, we provide data in support of another SNP, rs41259144, resulting in a R990Q substitution in apo(a) KIV-4 also being associated with a null Lp(a) phenotype. All known null apo(a) alleles reported to date are a result of either splice site or nonsense mutations that give rise to truncated apo(a) proteins. To our knowledge, the R1771C and R990Q variants are the first examples of full-length apo(a) variants resulting in null apo(a) alleles.
Both the R990Q and R1771C variants have previously been identified in GWASs as being associated with decreased plasma Lp(a) (30, 36–38), where their effect was shown to be comparable to that of the most common null allele, rs41272114. Compared with rs41272114, however, the allele frequencies of R990Q and R1771C are low (36–38), making homozygotes or compound heterozygotes rare. Even heterozygotes for either variant are unlikely to be identified based on Lp(a) levels because their effect will be masked by the other LPA allele, particularly in the case of a small apo(a) isoform. We were fortunate enough to identify both variants in an individual selected for study. The null phenotype of this individual suggested that the mutations were in the compound heterozygous state prompting us to investigate the effect of these two variants further. The lack of presence of the R1771C mutation in five identified R990Q carriers supported our conclusion of compound heterozygosity.
apo(a) sequence alignments showed the R990Q and R1771C variants to be equivalent to one another in terms of position in the apo(a) kringle sequence and located adjacent to a highly conserved cysteine residue involved in intramolecular cross-linking. They also appeared to be in the same position as variants in the PLG gene that had been identified as the cause of plasminogen deficiency (Fig. 1). Protein prediction software predicted both the R990Q and R1771C variants to be damaging to apo(a) structure, a result we further investigated by the modeling of the protein structures for the apo(a) KV and KIV-4 domains. Protein modeling indicated that both arginine residues made multiple polar contacts with neighboring residues forming a hydrogen-bonding network centered around a conserved cysteine involved in one of three intramolecular disulfide bonds (Fig. 2). The predicted interactions were in reasonable agreement with those identified in the original crystal structure of the plasminogen KIV domain (48).
Our protein modeling suggests that mutation of either of the R990 and R1771 residues would abolish all contacts made by these residues. Predicting the effect of the removal of these contacts is difficult; however, an early study examining the folding properties of isolated plasminogen KIV domains may give some insight. This study demonstrated that the early formation of two of the three disulfide bonds involved in intramolecular cross-linking was crucial for proper kringle folding (49). Sequence alignments of the plasminogen kringle domains showed that the most highly conserved residues were ones clustered around the cysteine residues participating in intramolecular disulfide bonds. The authors concluded that these highly conserved residues facilitate the correct folding of plasminogen (and other kringle containing proteins) to obtain the conformation required for disulfide bond formation. As the R990 and R1771 residues fall into this category, it is highly likely that mutation of either of these residues will circumvent the proper folding of apo(a) leaving it unable to be correctly processed for secretion. These predictions are supported by studies of PLG mutants associated with plasminogen deficiency, which involve the mutation of highly conserved arginine residues. These include the plasminogen variants R153K in KI (44), R235H in KII (45), R325H in KIII (46), and R532H in KV (47), all of which are found in an equivalent location within plasminogen kringles to that of R990 and R1771 in apo(a) (see Fig. 1). The in vitro studies of the R153K and R235H plasminogen variants showed a markedly reduced secretion of these mutants compared with the WT plasminogen protein despite similar intracellular expression levels (46).
The above findings with plasminogen mutants were analogous to what we found with the R1771 variant in our in vitro studies using recombinant expression of apo(a)-GFP tagged constructs. Here, we showed that while the mutant R1771C protein was expressed in both liver and kidney cells, it was unable to be recovered in the media, indicating a lack of secretion. This was in contrast to the WT apo(a) protein, which was recovered in the media on apo(a) immunoprecipitation. Our results show that while the introduced R1771C mutation does not alter apo(a) expression, the mutant protein displays a secretion-deficient phenotype indicative of a null allele. An initial clue that the R1771C mutant was defective was evident in the intracellular protein profiles of transfected cells (Fig. 3). While the WT showed a less abundant mature apo(a)-GFP species in conjunction with a more abundant immature species, cells expressing the R1771C apo(a)-GFP protein only appeared to contain the immature form. Only the mature form of WT apo(a)-GFP was detected in the media of transfected cells. The secretion deficit of the mutant might be expected to lead to an accumulation of the mutant protein in liver; however, treatment with a proteasomal inhibitor indicated that both the WT and R1771C proteins were degraded by the proteasome (Fig. 6A), which likely offsets accumulation of the mutant protein. In the case of the WT apo(a)-GFP, proteasomal inhibition also promoted an increased secretion into the media (Fig. 6B) as expected given the increase in intracellular levels.
Imaging of the WT and R1771C apo(a) proteins indicated differential patterns of colocalization with ER and Golgi proteins in transfected cells (Fig. 5). While there was a similar level of colocalization of the WT and R1771C proteins with calnexin and Sec21A (Fig. 5A, B) showing ER to Golgi trafficking, the R1771C mutant displayed significantly less colocalization with TGOLN2 (Fig. 5C), consistent with a limited flux through the Golgi into the secretory pathway. These results were supported by endoglycosidase digests of the recombinant apo(a) proteins (Fig. 6C) that showed the immature form, the only form present in R1771C recombinant cells, had been glycosylated in the ER but not in the Golgi. These results suggested that the poor secretion of R1771C was unlikely due to defective glycosylation but rather due to a failure of the glycosylated protein to fold into a conformation capable of being processed in the Golgi. It is possible that additional regulatory systems associated with the Golgi could play a role in the fate of any R1771C apo(a) escaping proteasomal degradation, with lysosomal degradation directed by mannose-mediated shuttling from the Golgi, a potential mechanism worth considering in future analyses.
Lastly, a time course experiment showed the secretion of the WT apo(a) to increase with time with secretion being inhibited by cycloheximide through a reduction in the amount of precursor and mature protein (Fig. 7). Interestingly, the time course experiment showed the appearance of the R1771C mutant apo(a) in the media in two forms, a mature form and an intermediate form indicative of a partially processed form being secreted, both of which were blocked by cycloheximide. The significance of this is uncertain because the amount of mutant protein compared with the WT was very small. Furthermore, the apo(a) isoform expressed in our system is quite small (15 KIV repeats) and expected to be more readily secreted than larger isoforms allowing for some processing of folding defective mutants like R1771C. It would be of interest to express the R1771C mutant in the setting of a larger isoform to establish whether the small amount of secretion is lost in the context of a more common apo(a) isoform.
Our in vitro results are in line with studies of recombinantly expressed WT apo(a) in liver and kidney cells (15, 50). In these studies, two intracellular forms of apo(a) protein were apparent, an immature form of the apo(a) protein and a mature form that was secreted (15, 50). These observations are also in concordance with studies of apo(a) in baboon hepatocytes in which an immature precursor and a mature form that was secreted were apparent (11). Interestingly, this study identified a null allele in baboons with no plasma Lp(a) that displayed a similar profile to that of our R1771C mutant, i.e., the allele produced a detectable intracellular apo(a) protein that was not secreted; however, this null allele was not genetically characterized (11). The response of the WT and R1771C proteins to glycosidase treatments also mirrored that observed by these studies (10, 11). Furthermore, the response of the WT and R1771C apo(a) proteins to proteasomal inhibition was similar to that previously observed in hepatocytes with both WT and a mutant secretion-defective form of apo(a) (51).
While efforts to thoroughly interrogate the R990Q variant in vitro were hampered, there is much circumstantial evidence to support the hypothesis that this is also a null allele. The R1771C variant serves as a reasonable proxy for the R990Q variant due to the high level of structural and sequence similarity between the apo(a) kringle KV and KIV-4 domains in which they are found. Most importantly it was predicted from protein modeling that R990Q displayed similar protein contacts as R1771 that would also be ablated on mutation. The presence of R990Q in an individual also harboring the proven null R1771C who displayed undetectable Lp(a) and no apo(a) is indicative of a null-like effect. In addition, five R990Q heterozygotes were identified that displayed a range of Lp(a) levels but who showed only one single apo(a) isoform on apo(a) phenotyping.
This study presents the first evidence for nonsynonymous mutations in apo(a) being causative of a null apo(a) phenotype and for the first time provides a genuine biochemical link between null Lp(a) and null plasminogen phenotypes. The mutations, which involve residues that are highly conserved in all apo(a) kringle domains, are hypothesized to impair the ability of the protein to fold, circumventing its processing to maturity for secretion. It is possible that other mutations occurring in these highly conserved regions give a similar phenotype and may collectively contribute to the well-known variation in Lp(a) levels. It is also possible that other nonsynonymous mutations could result in an increased propensity to be secreted. Our study is timely given that large LPA sequencing studies are now providing fertile ground for such discoveries.
Supplementary Material
Acknowledgments
The authors thank Carolyn Porteous and Malcolm Rutledge for the apo(a) phenotyping gels and Lamia Khaled for providing the demographic data on the Otago LPA population. They also thank Nicola Dalbeth and Lisa Stamp for recruitment of gout patients, and Mandy Phipps-Green for providing demographic data on the gout patients. The authors thank Dr. Gregory Redpath for reviewing and commenting on the manuscript.
Footnotes
Abbreviations:
- GWAS
- genome-wide association study
- KIV
- kringle IV
- KV
- kringle V
- Lp(a)
- lipoprotein (a)
- PDB
- Protein Data Bank
- TGOLN2
- trans-Golgi network 2
This work was supported by funding from the Maurice Wilkins Centre, the Dunedin School of Medicine Deans Bequest fund, and a Marjorie McCallum Award to B.M.M. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Utermann G. 1989. The mysteries of lipoprotein(a). Science. 246: 904–910. [DOI] [PubMed] [Google Scholar]
- 2.Nordestgaard B. G., Chapman M. J., Ray K., Boren J., Andreotti F., Watts G. F., Ginsberg H., Amarenco P., Catapano A., Descamps O. S., et al. ; European Atherosclerosis Society Consensus Panel. 2010. Lipoprotein(a) as a cardiovascular risk factor: current status. Eur. Heart J. 31: 2844–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boffa M. B., and Koschinsky M. L.. 2016. Lipoprotein (a): truly a direct prothrombotic factor in cardiovascular disease? J. Lipid Res. 57: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamstrup P. R., Tybjaerg-Hansen A., Steffensen R., and Nordestgaard B. G.. 2009. Genetically elevated lipoprotein(a) and increased risk of myocardial infarction. JAMA. 301: 2331–2339. [DOI] [PubMed] [Google Scholar]
- 5.Kraft H. G., Kochl S., Menzel H. J., Sandholzer C., and Utermann G.. 1992. The apolipoprotein (a) gene: a transcribed hypervariable locus controlling plasma lipoprotein (a) concentration. Hum. Genet. 90: 220–230. [DOI] [PubMed] [Google Scholar]
- 6.Boerwinkle E., Leffert C. C., Lin J., Lackner C., Chiesa G., and Hobbs H. H.. 1992. Apolipoprotein(a) gene accounts for greater than 90% of the variation in plasma lipoprotein(a) concentrations. J. Clin. Invest. 90: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean J. W., Tomlinson J. E., Kuang W. J., Eaton D. L., Chen E. Y., Fless G. M., Scanu A. M., and Lawn R. M.. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330: 132–137. [DOI] [PubMed] [Google Scholar]
- 8.Lackner C., Cohen J. C., and Hobbs H. H.. 1993. Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2: 933–940. [DOI] [PubMed] [Google Scholar]
- 9.Kraft H. G., Menzel H. J., Hoppichler F., Vogel W., and Utermann G.. 1989. Changes of genetic apolipoprotein phenotypes caused by liver transplantation. Implications for apolipoprotein synthesis. J. Clin. Invest. 83: 137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White A. L., Rainwater D. L., Hixson J. E., Estlack L. E., and Lanford R. E.. 1994. Intracellular processing of apo(a) in primary baboon hepatocytes. Chem. Phys. Lipids. 67–68: 123–133. [DOI] [PubMed] [Google Scholar]
- 11.White A. L., Hixson J. E., Rainwater D. L., and Lanford R. E.. 1994. Molecular basis for “null” lipoprotein(a) phenotypes and the influence of apolipoprotein(a) size on plasma lipoprotein(a) level in the baboon. J. Biol. Chem. 269: 9060–9066. [PubMed] [Google Scholar]
- 12.Bonen D. K., Nassir F., Hausman A. M., and Davidson N. O.. 1998. Inhibition of N-linked glycosylation results in retention of intracellular apo[a] in hepatoma cells, although nonglycosylated and immature forms of apolipoprotein[a] are competent to associate with apolipoprotein B-100 in vitro. J. Lipid Res. 39: 1629–1640. [PubMed] [Google Scholar]
- 13.Brunner C., Lobentanz E. M., Petho-Schramm A., Ernst A., Kang C., Dieplinger H., Muller H. J., and Utermann G.. 1996. The number of identical kringle IV repeats in apolipoprotein(a) affects its processing and secretion by HepG2 cells. J. Biol. Chem. 271: 32403–32410. [DOI] [PubMed] [Google Scholar]
- 14.White A. L., Guerra B., and Lanford R. E.. 1997. Influence of allelic variation on apolipoprotein(a) folding in the endoplasmic reticulum. J. Biol. Chem. 272: 5048–5055. [DOI] [PubMed] [Google Scholar]
- 15.Lobentanz E. M., Krasznai K., Gruber A., Brunner C., Muller H. J., Sattler J., Kraft H. G., Utermann G., and Dieplinger H.. 1998. Intracellular metabolism of human apolipoprotein(a) in stably transfected Hep G2 cells. Biochemistry. 37: 5417–5425. [DOI] [PubMed] [Google Scholar]
- 16.Kraft H. G., Lingenhel A., Kochl S., Hoppichler F., Kronenberg F., Abe A., Muhlberger V., Schonitzer D., and Utermann G.. 1996. Apolipoprotein(a) kringle IV repeat number predicts risk for coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 16: 713–719. [DOI] [PubMed] [Google Scholar]
- 17.Gaw A., Boerwinkle E., Cohen J. C., and Hobbs H. H.. 1994. Comparative analysis of the apo(a) gene, apo(a) glycoprotein, and plasma concentrations of Lp(a) in three ethnic groups. Evidence for no common “null” allele at the apo(a) locus. J. Clin. Invest. 93: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Utermann G. 1999. Genetic architecture and evolution of the lipoprotein(a) trait. Curr. Opin. Lipidol. 10: 133–141. [DOI] [PubMed] [Google Scholar]
- 19.Rader D. J., Cain W., Ikewaki K., Talley G., Zech L. A., Usher D., and Brewer H. B. Jr.. 1994. The inverse association of plasma lipoprotein(a) concentrations with apolipoprotein(a) isoform size is not due to differences in Lp(a) catabolism but to differences in production rate. J. Clin. Invest. 93: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandholzer C., Hallman D. M., Saha N., Sigurdsson G., Lackner C., Csaszar A., Boerwinkle E., and Utermann G.. 1991. Effects of the apolipoprotein(a) size polymorphism on the lipoprotein(a) concentration in 7 ethnic groups. Hum. Genet. 86: 607–614. [DOI] [PubMed] [Google Scholar]
- 21.Geethanjali F. S., Luthra K., Lingenhel A., Kanagasaba-Pathy A. S., Jacob J., Srivastava L. M., Vasisht S., Kraft H. G., and Utermann G.. 2003. Analysis of the apo(a) size polymorphism in Asian Indian populations: association with Lp(a) concentration and coronary heart disease. Atherosclerosis. 169: 121–130. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt K., Noureen A., Kronenberg F., and Utermann G.. 2016. Structure, function, and genetics of lipoprotein (a). J. Lipid Res. 57: 1339–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perombelon Y. F., Soutar A. K., and Knight B. L.. 1994. Variation in lipoprotein(a) concentration associated with different apolipoprotein(a) alleles. J. Clin. Invest. 93: 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen J. C., Chiesa G., and Hobbs H. H.. 1993. Sequence polymorphisms in the apolipoprotein (a) gene. Evidence for dissociation between apolipoprotein(a) size and plasma lipoprotein(a) levels. J. Clin. Invest. 91: 1630–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puckey L. H., Lawn R. M., and Knight B. L.. 1997. Polymorphisms in the apolipoprotein(a) gene and their relationship to allele size and plasma lipoprotein(a) concentration. Hum. Mol. Genet. 6: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 26.Ogorelkova M., Kraft H. G., Ehnholm C., and Utermann G.. 2001. Single nucleotide polymorphisms in exons of the apo(a) kringles IV types 6 to 10 domain affect Lp(a) plasma concentrations and have different patterns in Africans and Caucasians. Hum. Mol. Genet. 10: 815–824. [DOI] [PubMed] [Google Scholar]
- 27.Chretien J. P., Coresh J., Berthier-Schaad Y., Kao W. H., Fink N. E., Klag M. J., Marcovina S. M., Giaculli F., and Smith M. W.. 2006. Three single-nucleotide polymorphisms in LPA account for most of the increase in lipoprotein(a) level elevation in African Americans compared with European Americans. J. Med. Genet. 43: 917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke R., Peden J. F., Hopewell J. C., Kyriakou T., Goel A., Heath S. C., Parish S., Barlera S., Franzosi M. G., Rust S., et al. 2009. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 361: 2518–2528. [DOI] [PubMed] [Google Scholar]
- 29.Lanktree M. B., Anand S. S., Yusuf S., Hegele R. A., and Investigators S.. 2010. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ Cardiovasc Genet. 3: 39–46. [DOI] [PubMed] [Google Scholar]
- 30.Mack S., Coassin S., Rueedi R., Yousri N. A., Seppala I., Gieger C., Schonherr S., Forer L., Erhart G., Marques-Vidal P., et al. 2017. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J. Lipid Res. 58: 1834–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogorelkova M., Gruber A., and Utermann G.. 1999. Molecular basis of congenital lp(a) deficiency: a frequent apo(a) ‘null’ mutation in caucasians. Hum. Mol. Genet. 8: 2087–2096. [DOI] [PubMed] [Google Scholar]
- 32.Parson W., Kraft H. G., Niederstatter H., Lingenhel A. W., Kochl S., Fresser F., and Utermann G.. 2004. A common nonsense mutation in the repetitive Kringle IV-2 domain of human apolipoprotein(a) results in a truncated protein and low plasma Lp(a). Hum. Mutat. 24: 474–480. [DOI] [PubMed] [Google Scholar]
- 33.Noureen A., Fresser F., Utermann G., and Schmidt K.. 2015. Sequence variation within the KIV-2 copy number polymorphism of the human LPA gene in African, Asian, and European populations. PLoS One. 10: e0121582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coassin S., Erhart G., Weissensteiner H., Eca Guimaraes de Araujo M., Lamina C., Schonherr S., Forer L., Haun M., Losso J. L., Kottgen A., et al. 2017. A novel but frequent variant in LPA KIV-2 is associated with a pronounced Lp(a) and cardiovascular risk reduction. Eur. Heart J. 38: 1823–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim E. T., Wurtz P., Havulinna A. S., Palta P., Tukiainen T., Rehnstrom K., Esko T., Magi R., Inouye M., Lappalainen T., et al. ; Sequencing Initiative Suomi (SISu) Project. 2014. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zekavat S. M., Ruotsalainen S., Handsaker R. E., Alver M., Bloom J., Poterba T., Seed C., Ernst J., Chaffin M., Engreitz J., et al. 2018. Deep coverage whole genome sequences and plasma lipoprotein(a) in individuals of European and African ancestries. Nat. Commun. 9: 2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J., Lange L. A., Sabourin J., Duan Q., Valdar W., Willis M. S., Li Y., Wilson J. G., and Lange E. M.. 2015. Genome- and exome-wide association study of serum lipoprotein (a) in the Jackson Heart Study. J. Hum. Genet. 60: 755–761. [DOI] [PubMed] [Google Scholar]
- 38.Burgess S., Ference B. A., Staley J. R., Freitag D. F., Mason A. M., Nielsen S. F., Willeit P., Young R., Surendran P., Karthikeyan S., et al. ; European Prospective Investigation Into Cancer and Nutrition–Cardiovascular Disease (EPIC-CVD) Consortium. 2018. Association of LPA variants with risk of coronary disease and the implications for lipoprotein(a)-lowering therapies: a Mendelian randomization analysis. JAMA Cardiol. 3: 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollis-Moffatt J. E., Xu X., Dalbeth N., Merriman M. E., Topless R., Waddell C., Gow P. J., Harrison A. A., Highton J., Jones P. B., et al. 2009. Role of the urate transporter SLC2A9 gene in susceptibility to gout in New Zealand Maori, Pacific Island, and Caucasian case-control sample sets. Arthritis Rheum. 60: 3485–3492. [DOI] [PubMed] [Google Scholar]
- 40.Jaroszewski L., Li Z., Cai X. H., Weber C., and Godzik A.. 2011. FFAS server: novel features and applications. Nucleic Acids Res. 39: W38–W44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H., and Phillippy A. M.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28: 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coassin S., Schonherr S., Weissensteiner H., Erhart G., Forer L., Losso J. L., Lamina C., Haun M., Utermann G., Paulweber B., et al. 2019. A comprehensive map of single-base polymorphisms in the hypervariable LPA kringle IV type 2 copy number variation region. J. Lipid Res. 60: 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuster V., Zeitler P., Seregard S., Ozcelik U., Anadol D., Luchtman-Jones L., Meire F., Mingers A. M., Schambeck C., and Kreth H. W.. 2001. Homozygous and compound-heterozygous type I plasminogen deficiency is a common cause of ligneous conjunctivitis. Thromb. Haemost. 85: 1004–1010. [PubMed] [Google Scholar]
- 45.Schuster V., Mingers A. M., Seidenspinner S., Nussgens Z., Pukrop T., and Kreth H. W.. 1997. Homozygous mutations in the plasminogen gene of two unrelated girls with ligneous conjunctivitis. Blood. 90: 958–966. [PubMed] [Google Scholar]
- 46.Tefs K., Gueorguieva M., Klammt J., Allen C. M., Aktas D., Anlar F. Y., Aydogdu S. D., Brown D., Ciftci E., Contarini P., et al. 2006. Molecular and clinical spectrum of type I plasminogen deficiency: A series of 50 patients. Blood. 108: 3021–3026. [DOI] [PubMed] [Google Scholar]
- 47.Schuster V., Seidenspinner S., Zeitler P., Escher C., Pleyer U., Bernauer W., Stiehm E. R., Isenberg S., Seregard S., Olsson T., et al. 1999. Compound-heterozygous mutations in the plasminogen gene predispose to the development of ligneous conjunctivitis. Blood. 93: 3457–3466. [PubMed] [Google Scholar]
- 48.Mulichak A. M., Tulinsky A., and Ravichandran K. G.. 1991. Crystal and molecular structure of human plasminogen kringle 4 refined at 1.9-A resolution. Biochemistry. 30: 10576–10588. [DOI] [PubMed] [Google Scholar]
- 49.Trexler M., and Patthy L.. 1983. Folding autonomy of the kringle 4 fragment of human plasminogen. Proc. Natl. Acad. Sci. USA. 80: 2457–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koschinsky M. L., Tomlinson J. E., Zioncheck T. F., Schwartz K., Eaton D. L., and Lawn R. M.. 1991. Apolipoprotein(a): expression and characterization of a recombinant form of the protein in mammalian cells. Biochemistry. 30: 5044–5051. [DOI] [PubMed] [Google Scholar]
- 51.White A. L., Guerra B., Wang J., and Lanford R. E.. 1999. Presecretory degradation of apolipoprotein [a] is mediated by the proteasome pathway. J. Lipid Res. 40: 275–286. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.