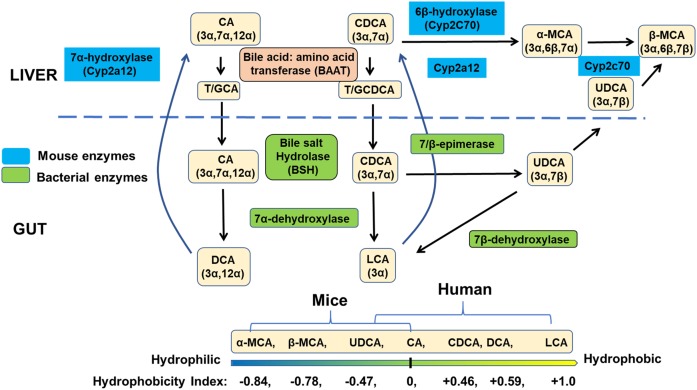
Humans and mice have substantially different bile acid (BA) pool compositions (1). As the major primary BAs, humans synthesize cholic acid (CA) and chenodeoxycholic acid (CDCA), whereas mice have mainly CA and 6-hydroxylated muricholic acids (MCAs) that are made from CDCA (Fig. 1). Hydroxylation at the C-6 position significantly affects the physicochemical properties of BAs, making the BA pool more hydrophilic, less potent as detergents, and less injurious. In addition, 6-hydroxylation dramatically changes BA signaling properties, converting the most potent endogenous FXR agonist (CDCA) to antagonists (MCAs). These human-mouse BA species differences require us to be extremely prudent when extrapolating discoveries from mouse studies to humans for BA signaling or BA-related liver and metabolic diseases.
Fig. 1.
BA synthesis in the liver and biotransformation by gut bacteria in humans and mice. Cholic acid (CA) and chenodeoxycholic acid (CDCA) are two primary BAs synthesized in human livers. In mice, CDCA is 6β-hydroxylated to α-muricholic acid (αMCA), which is epimerized to βMCA by CYP2C70. CA and MCAs are primary BAs synthesized in mouse livers. In human livers, primary BAs are conjugated to amino acid glycine (G) and taurine (T) in a ratio about 3–1 for biliary secretion. In mouse livers, most BAs are conjugated to T. In the colon, bacterial bile salt hydrolase (BSH) deconjugates BAs, which is followed by bacterial 7a-dehydroxylation to remove a 7α-hydroxyl-group from CA and CDCA to form deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. CDCA also can be epimerized to UDCA. UDCA is a primary BA in mice, but most of the UDCA is likely produced by bacterial metabolism. In humans, UDCA is a secondary BA generated by the gut microbiota from CDCA. In mice, UDCA can be converted to βMCA by CYP2C70 or to LCA by bacterial 7β-dehydroxylase. In the mouse liver, DCA and LCA can be converted back to CA and CDCA, respectively, by CYP2A12. Thus, CA, βMCA, and αMCA are predominant BAs in mice, leading to a more hydrophilic circulating BA pool, whereas CA, CDCA, and DCA are predominant BAs in humans, generating a more hydrophobic BA pool. In Cyp2c70 KO mice, MCAs are absent and CDCA levels are increased, whereas in Cyp2a12 deficient mice, DCA is accumulated. In Cyp2c70 and Cyp2a12 double deficient (DKO) mice, DCA, CDCA, and LCA are increased, similar to human BA composition. The bottom diagram shows BA hydrophobicity index of major primary and secondary BAs in humans and mice.
Therefore, there has been considerable interest in the identity of the enzyme(s) responsible for converting CDCA to MCAs, the reason why rodents need this conversion for their physiology and metabolism, and ultimately, whether we can make the mouse a better model to understand human BA metabolism and signaling. We know that synthesis of 6-hydroxylated BAs is mediated by a cytochrome P450, but identification of this enzyme remained a mystery until 2016, when Takahashi et al. (2) reported that mice lacking the CYP2C family of enzymes were unable to synthesize MCAs, and consequently, CYP2C70 was identified as the likely CYP2C isoform responsible for 6-hydroxylation of CDCA to αMCA and ursodeoxycholic acid (UDCA) to βMCA. In contrast, the major human CYP2C enzyme, CYP2C9, is unable to hydroxylate BAs, in agreement with the relative paucity of 6-hydroxylated BAs in humans (2).
Two recent studies in the Journal of Lipid Research confirm the importance of CYP2C70 in the synthesis of 6-hydroxylated BAs and provide novel insights to the pathway for BA biosynthesis in mice and how BA composition impacts BA homeostasis and signaling (3, 4). In the first study, de Boer et al. (3) used CRISPR/Cas9 technology that includes a liver-specific Cas9-transgenic mouse and adenoviral delivery of a guide RNA to target Cyp2c70 to generate an adult-onset hepatic Cyp2c70 “knock-out” mouse model (Cyp2c70ako). As expected, the CYP2C70 deficiency resulted in a more human-like and hydrophobic BA profile in the Cyp2c70ako mice. However, the presence of residual levels of MCAs in the BA pool suggests that genome editing of Cyp2c70 may have been incomplete using this approach or possibly other members of the CYP2C family may possess overlapping activity with CYP2C70. Takahashi et al. (2) had previously reported that CYP2C70 could convert CDCA to αMCA but not to βMCA, raising the question of which additional enzyme(s) are required for synthesis of this major murine 6-hydroxylated BA species. Through the innovative use of deuterated BA tracers and their Cyp2c70ako model, de Boer et al. were able to answer this vexing question and demonstrate that CYP2C70 catalyzes the formation of βMCA primarily by sequential 6β-hydroxylation and C-7-epimerization of CDCA, generating αMCA as an intermediate metabolite that cycles in the enterohepatic circulation. In addition, fecal cholesterol excretion was reduced with FXR activation. These results suggest that BA composition plays an important role in cholesterol homeostasis. Surprisingly, the fecal microbiome was not altered even with these changes in BA composition, which may be due to the acute nature of hepatic-only gene deletion in adult mice, the relative short duration of the study, or the selective sampling of only fecal but not cecum or small intestine microbiome.
In another comprehensive study in the Journal, Honda et al. (4) demonstrate that CYP2C70 is the major enzyme responsible for the synthesis of the 6-hydroxylated primary BAs. Moreover, the authors also answered a second important question regarding the mouse-human BA species differences by identifying CYP2A12 as the cytochrome P450 enzyme responsible for the 7α-rehydroxylation of DCA and LCA in mice. Although hepatic 7α-rehydroxylation is an important determinant of the concentration of DCA in the BA pool in many species, it is absent in humans. The identity of this enzyme has long remained a mystery. Honda et al. describe the use of CRISPR/Cas9 technology to generate single and double whole-body KO (DKO) mice for Cyp2a12 and Cyp2c70. In humans, the major species of the BA pool consists of CA, CDCA, and DCA in a ratio of about 40:40:20, whereas the DCA levels are very low in mice, likely due to the presence of CYP2C12. This study confirms the role of CYP2C70 as a CDCA 6β-hydroxylase and provides convincing evidence showing the role of CYP2A12 in mouse livers to catalyze the 7α-rehydroxylation of TDCA and TLCA (Fig. 1). This study suggests that CYP2C70 converts CDCA to αMCA and UDCA to βMCA, but the accumulation of UDCA is much less than that of CDCA in Cyp2c70 KO mice, suggesting that in mice, most βMCA is synthesized from CDCA via αMCA but not via UDCA. Cyp2c70 KO and Cyp2c70/Cyp2a12 DKO mice have reduced BA pool size compared with wild-type and Cyp2a12 KO mice, which reflects reduced hepatic expression of Cyp7a1 and Cyp8b1. The particularly dramatic reduction in Cyp8b1 expression agrees with the reduction in CA in the BA pool of these mice.
Increased levels of CDCA as a result of blocking its conversion to MCA in Cyp2c70 and DKO mice should lead to increased FXR activation. Surprisingly, results from the Honda study showed FXR was not further activated in both liver and intestine. Instead, the cytokine and c-Jun N-terminal kinase (JNK) signaling pathway was activated; therefore, the authors suggest that this JNK activation by unconjugated CDCA may be the major mechanism for BA-mediated inhibition of their own synthesis in Cyp2c70 KO mice. It was first reported that CDCA induced the inflammatory cytokines in Kupffer cells to activate cytokine receptors and the mitogen-activated protein kinase (MAPK)-JNK pathway (5). Unconjugated CDCA activation of the MAPK-JNK pathway has been shown to inhibit the transcription of genes encoding key enzymes in BA synthesis, Cyp7a1 and Cyp8b1, by activating c-Jun to inhibit hepatocyte nuclear factor 4α and peroxisome proliferator activated receptor γ-co-activator-1α (6, 7).
These studies of Cyp2c70 KO mice reported higher serum ALT activities and other liver injury markers. It is possible that the presence of “novel” CDCA in the livers of these KO mice causes cellular damage by JNK activation and inflammation. The signs of liver injury could be a concern when using these humanized mice.
In conclusion, long-sought enzymes responsible for major murine-human differences in BA pool composition have finally been unveiled by taking advantage of a wealth of knowledge regarding the human and rodent P450 enzyme families and novel gene editing technologies. However, although Cyp2c70 KO mice possess a humanized BA composition, it remains unclear whether this model will be the key to studying human liver diseases associated with BA dysregulation due to baseline liver injury and inflammation manifested in these mice. More studies and improved mouse models are urgently needed.
Footnotes
This work was supported by National Institutes of Health Grants NIH-GM104037, NIH-ES029258, NIH-ES007148, NIHDK116495 and VA-BX002741 to G.G. and DK44224 and NIH-DK58379 to J.Y.L.C. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of the article.
REFERENCES
- 1.Li J., and Dawson P. A.. 2019. Animal models to study bile acid metabolism. Bioch. Biophys. Acta Mol. Basis Dis. 1865: 895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi S., Fukami T., Masuo Y., Brocker C. N., Xie C., Krausz K. W., Wolf C. R., Henderson C. J.,and Gonzalez F. J.. 2016. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57: 2130–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Boer J. F., Verkade E., Mulder N. L., de Vries H. D., Huijkman N., Koehorst M., Boer T., Wolters J. C., Bloks V. W., van de Sluis B.,and Kuipers F.. 2020. A human-like bile acid pool induced by deletion of hepatic Cyp2c70 modulates effects of farnesoid x receptor activation in mice. J. Lipid Res. 61: 291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honda A., Miyazaki T., Iwamoto J., Hirayama T., Morishita Y., Monma T., Ueda H., Mizuno S., Sugiyama F., Takahashi S., et al. 2020. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 61: 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyake J. H., Wang S-L.,and Davis R. A.. 2000. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha -hydroxylase. J. Biol. Chem. 275: 21805–21808. [DOI] [PubMed] [Google Scholar]
- 6.Li T., Jahan A.,and Chiang J. Y.. 2006. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahan A., and Chiang J. Y. L.. 2005. Cytokine regulation of human sterol 12α-hydroxylase (CYP8B1) gene. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G685–G695. [DOI] [PubMed] [Google Scholar]