Abstract
Liver-derived serum amyloid A (SAA) is present in plasma where it is mainly associated with HDL and from which it is cleared more rapidly than are the other major HDL-associated apolipoproteins. Although evidence suggests that lipid-free and HDL-associated forms of SAA have different activities, the pathways by which SAA associates and disassociates with HDL are poorly understood. In this study, we investigated SAA lipidation by hepatocytes and how this lipidation relates to the formation of nascent HDL particles. We also examined hepatocyte-mediated clearance of lipid-free and HDL-associated SAA. We prepared hepatocytes from mice injected with lipopolysaccharide or an SAA-expressing adenoviral vector. Alternatively, we incubated primary hepatocytes from SAA-deficient mice with purified SAA. We analyzed conditioned media to determine the lipidation status of endogenously produced and exogenously added SAA. Examining the migration of lipidated species, we found that SAA is lipidated and forms nascent particles that are distinct from apoA-I-containing particles and that apoA-I lipidation is unaltered when SAA is overexpressed or added to the cells, indicating that SAA is not incorporated into apoA-I-containing HDL during HDL biogenesis. Like apoA-I formation, generation of SAA-containing particles was dependent on ABCA1, but not on scavenger receptor class B type I. Hepatocytes degraded significantly more SAA than apoA-I. Taken together, our results indicate that SAA’s lipidation and metabolism by the liver is independent of apoA-I and that SAA is not incorporated into HDL during HDL biogenesis.
Keywords: hepatocyte, adenosine 5′-triphosphate binding cassette transporter A1, nascent high density lipoprotein, lipid metabolism, lipidation, liver metabolism, inflammatory disease, lipoprotein
The serum amyloid A (SAA) family of secreted proteins is encoded by multiple genes that display a high degree of homology in mammals. SAAs can be divided into two main classes based on their responsiveness to inflammatory stimuli. The “acute phase” SAAs are major acute phase reactants whose serum concentrations increase by 1,000-fold or more in response to tissue injury and infection. In contrast, “constitutive” SAA is minimally induced and is present in relatively low concentrations in plasma (1, 2). The genes for the two major acute phase human SAAs, Saa1 and Saa2, correspond to mouse Saa1.1 and Saa2.1. Mice express a third acute phase SAA, SAA3, which is not expressed in humans due to the presence of a premature stop codon. Both human SAA4 and mouse SAA4 are constitutive SAAs. Circulating SAA is produced and secreted primarily by the liver and is present in plasma mainly associated with HDL (1, 2). However, expression of extrahepatic SAA is also found in inflamed tissues, such as adipose tissue in obesity (3), atherosclerotic lesions (4, 5), synovial tissue in rheumatoid arthritis (6), various carcinomas (7), and lung tissue in chronic obstructive pulmonary disease (8).
Increased circulating SAA has been associated with several inflammatory diseases, including obesity (3), type 2 diabetes (9), atherosclerosis (10, 11), abdominal aortic aneurysms (12), pulmonary disease (13), and cancer (7), and emerging research implies that SAA plays a pathogenic role in the development of these conditions. Thus, understanding the pathways by which SAA levels and activity are regulated is critical. SAA has been shown to exert a number of biological activities consistent with a role in innate immunity, including leukocyte chemotaxis (14, 15), inflammatory cytokine induction (16–18), activation of the NLRP3 inflammasome (19), and upregulation of genes involved in extracellular matrix remodeling, including transforming growth factor-β (TGF-β) (20) and matrix metalloproteinases (21).
The majority of SAA in the circulation is lipoprotein-associated and not in a lipid-free form (22), and accumulating evidence indicates that the lipidation status of SAA is a critical factor governing its function. For example, only lipid-free SAA, but not HDL-associated SAA, stimulates macrophages to express granulocyte colony stimulating factor (G-CSF) in vitro (23). The chemoattractant activity of SAA is also dampened by its association with HDL (24). Similarly, association with HDL inhibits the ability of SAA to stimulate reactive oxygen species generation and NLRP3 inflammasome activation in macrophages (19). Plasma SAA is cleared more rapidly than the other major HDL apolipoproteins, apoA-I and apoA-II (25), suggesting that SAA dissociates from HDL more readily compared with apoA-I and apoA-II. Despite the growing recognition that SAA lipidation and delipidation may be key regulators of its biological activity, the processes involved in determining its lipidation state have not been well defined. In this study, we investigated SAA lipidation by hepatocytes and how such lipidation relates to the formation of nascent HDL. We also examined the role of hepatocytes in the clearance of HDL-associated SAA.
MATERIALS AND METHODS
Animals
C57BL/6 mice and apoA-I-deficient mice were purchased from the Jackson Laboratory. Mice deficient in both of the two major acute phase SAA isoforms, SAA1.1 and SAA2.1 (SAA1.1/2.1-DKO) were bred to obtain a >99.9% C57BL/6 background using the Jackson Laboratory Speed Congenic Service as previously described (26). Mice deficient in SAA1.1, SAA2.1, and SAA3 (SAA1.1/2.1/3-TKO) were obtained by deleting the Saa3 gene in SAA1.1/2.1-DKO mice using CRISPR Genome Editing technology (27). Liver-specific ABCA1-KO mice (albumin Cre+, ABCA1flox/flox) were provided by Dr. John S. Parks, Wake Forest University (28). Scavenger receptor class B type I (SR-BI)−/− and SR-BI+/+ mice were obtained from Dr. Xiang-An Li, University of Kentucky (29). Induction of hepatic SAA expression was achieved by injecting Escherichia coli lipopolysaccharide (LPS; 0111:B4; Sigma-Aldrich, St. Louis, MO) intraperitoneally into C57BL/6 mice (1 μg/g body weight) or by intravenous injection of 1 × 1011 particles of an adenoviral vector expressing SAA (AdSAA) into SAA1.1/2.1/3-TKO mice. Where indicated, an empty vector, AdNull, was used as negative control for adenovirus studies. Animals were housed in micro-isolator cages and maintained on a 14 h light/10 h dark cycle. Mice were provided with normal chow diet and water ad libitum. All studies were performed in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals and with the approval of the University of Kentucky Institutional Animal Care and Use Committee.
Primary hepatocyte culture
Primary hepatocytes were isolated from untreated C57BL/6 mice at 6 or 24 h after LPS or adenoviral vector injection, respectively, using a two-step perfusion method as described previously (30). Briefly, the liver was first perfused with Ca2+/Mg2+-free HBSS containing 10 mM glucose, 10 mM HEPES, and 0.3 mM EDTA and then with HBSS containing 0.05% collagenase type IV (#C5138; Sigma-Aldrich), 1.3 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, and 10 mM HEPES. Hepatocytes were washed three times by low speed centrifugation (50 g for 2 min) and cell viability, assessed by trypan blue exclusion, was confirmed to be >94% prior to plating cells. Cells were plated onto 12-well plates precoated with rat tail collagen (#354236; BD Biosciences, Bedford, MA) at a density of 2 × 105 cells/well and cultured at 37°C under 5% CO2 in Williams’ Medium E (GIBCO) containing 10% fetal bovine serum (GIBCO), 2% penicillin-streptomycin, 1% sodium pyruvate, 1% l-glutamine, and 1% insulin-transferrin-selenium (GIBCO) overnight prior to lipidation assays.
HDL preparation
HDL (d = 1.063–1.21 g/ml) was isolated from mouse plasma by density gradient ultracentrifugation as described previously (31). HDL was dialyzed against 150 mM NaCl and 0.01% EDTA, sterile filtered, and stored under argon gas at 4°C. Protein concentrations were determined by the method of Lowry et al (32). HDL purity and integrity were confirmed by SDS-PAGE (4–20% polyacrylamide SDS gels; Bio-Rad Laboratories, Hercules, CA) and nondenaturing gradient gel electrophoresis (NDGGE) in which HDL was electrophoresed on a 4–20% nondenaturing polyacrylamide gel (Bio-Rad Laboratories) for 3.5 h at 200 V and 4°C.
Mouse SAA and apoA-I purification
Mouse SAA and apoA-I were purified as described (12) from HDL isolated from mouse plasma collected 24 h after injection with 1.0–2.5 μg/g body weight LPS. Briefly, ∼20 mg of HDL were delipidated with ethanol:ether (3:2) and the delipidated proteins were separated by gel filtration on a Sephacryl S-200 column in buffer containing 7 M urea, 20 mM Tris, 150 mM NaCl, and 1 mM EDTA (pH 8.4). apoA-I- and SAA-containing fractions were identified by SDS-PAGE, pooled separately, and dialyzed against 2 mM Tris, 15 mM NaCl, and 0.1 mM EDTA (pH 8.4) prior to 10-fold concentration. LPS contamination in purified apolipoprotein preparations was below the level of detection (ToxinSensorTM Chromogenic LAL Endotoxin assay kit, #L00350C; GenScript, Piscataway, NJ).
Radioiodination of HDL and apolipoproteins
Mouse HDL, purified mouse SAA and apoA-I, human SAA (BioVision; #4324), human apoA-I (Meridian Life Science; #A95120H), and human apoA-II (Meridian Life Science; #A50216H) were radiolabeled using the iodine monochloride method (33). The specific activity of 125I-SAA, 125I-apoA-I, 125I-apoA-II, and 125I-HDL ranged from 300 to 400 cpm/ng protein.
Ex vivo lipidation assays
To investigate the lipidation of endogenously produced SAA, primary hepatocytes were prepared from C57BL/6 mice 6 h after administration of 1 μg/g LPS or 24 h after injection of 1 × 1011 particles of an adenoviral vector expressing SAA (AdSAA) into SAA1.1/2.1/3-TKO mice. After an overnight culture, cells were treated with the LXR agonist T0901317 (5 μM, #71810; Cayman Chemical) in Williams’ Medium E containing 0.2% fatty acid-free BSA for 8 h, as stated in the figure legends, to induce the expression of ABCA1. Cells were then cultured in medium containing 0.2% fatty acid-free BSA for 4, 8, and 18 h in the absence of T0901317. To study exogenous SAA lipidation, primary hepatocytes were prepared from SAA1.1/2.1/3-TKO mice or apoA-I-deficient mice, treated with 5 μM T0901317 for 8 h, and then incubated for 4, 8, and 18 h with 10 μg/ml purified mouse SAA in medium containing 0.2% fatty acid-free BSA. Cell lysates and culture media were harvested for analysis by SDS-PAGE and NDGGE followed by Western blotting. Alternatively, primary hepatocytes from C57BL/6 mice, SAA1.1/2.1/3-TKO mice, or apoA-I-deficient mice were treated with T0901317 for 8 h and then incubated with 125I-radiolabeled human apoA-I (5 μg/ml, 0.177 μM), apoA-II (10 μg/ml, 0.575 μM), or SAA (10 μg/ml, 0.870 μM) for 16 h. Aliquots of cell media were separated by NDGGE, and dried gels were subjected to autoradiography.
Gel electrophoresis and Western blot analysis
Total cell lysates (10 μg protein) or culture media (7 μl) were separated by 4–20% SDS-PAGE and transferred to PVDF membranes for immunoblotting. The amount of cell lysate and conditioned media typically corresponded to approximately 3.8% and 1.4% of the total amount in the dish, respectively. The following primary antibodies were used: rabbit anti-mouse SAA (1:2,000) (#ab199030; Abcam); rabbit anti-mouse SAA (12) (1:2,000); rabbit anti-mouse apoA-I (1:2,000) (#K23001R; Meridian Life Science); rabbit anti-mouse SR-BI (1:1,000) (#NB400-104; Novus Biologicals, Littleton, CO); or rabbit anti-mouse ABCA1 (1:1,000) (#ab7360; Abcam). For loading controls, membranes were immunoblotted with anti-β-actin antibody (#A5441; Sigma-Aldrich). Secondary antibodies were peroxidase-conjugated anti-rabbit IgG (#A6154; Sigma-Aldrich) or anti-mouse IgG (#A4416; Sigma-Aldrich). Immunoblots were visualized by the Amersham™ ECL™ Western blotting detection reagents (#RPN2106; GE Healthcare, Pittsburgh, PA).
To assess the lipidated species generated by primary mouse hepatocytes, aliquots of culture media were electrophoresed on 4–20% nondenaturing gradient gels for 3.5 h at 200 V and 4°C and transferred to PVDF membranes (40 min at 100 V, 4°C) for subsequent Western blotting or autoradiography. A protein standard mix containing proteins with known diameter (7.1, 8.2, 10.4, 12.2, and 17.0 nm) was obtained from GE Healthcare (HMW Calibration Kit for Native Electrophoresis, #17-0445-01) and used to estimate the sizes of lipidated species after separation by NDGGE.
Cell association and degradation of HDLs and SAA
Cell association of iodinated mouse HDLs, SAA, and apoA-I was assessed as described previously (34). Hepatocytes were seeded into 12-well cell culture clusters at a density of 2 × 105 cells/well and then cultured overnight. After 8 h without or with T0901317 treatment, cells were washed with PBS and incubated at 37°C for the indicated times with 5 μg/ml 125I-SAA, 5 μg/ml 125I-apoA-I, or 10 μg/ml 125I-HDL in Williams’ Medium E containing 0.2% fatty acid-free BSA. At the end of the experiments, media were collected into glass tubes for degradation analysis. The cells were washed four times with cold PBS containing BSA (1 mg/ml) followed by two washes in the same buffer without BSA. The cells were then solubilized in 0.1 N NaOH for 60 min at room temperature, and the protein concentration was determined by the method of Lowry et al. (32). The 125I content of cell lysates was determined in a γ counter.
For analysis of cell-mediated 125I-HDL degradation, 500 μl of BSA in PBS (1 mg/ml) and 400 μl of 50% trichloroacetic acid were added to 500 μl of media in glass tubes and mixed thoroughly. The mixture was stored at 4°C for at least 30 min or up to 3 days. After centrifugation at 800 g and 4°C for 20 min, 1 ml of supernatant was moved to a new glass tube and 20 μl of freshly made 20% KI were added and mixed, and then 50 μl of 30% H2O2 were added and mixed. After incubation at room temperature for 5 min, 2 ml of chloroform were added to the mixture and vortexed thoroughly until the aqueous layer was clear. After centrifugation at 800 g for 5 min, 750 μl of the top layer was placed in a new tube and counted in a γ counter.
For analysis of cell-mediated 125I-SAA and 125I-apoA-I degradation, culture media collected at the indicated times were applied to an Amicon Ultra 3K device and the flow-through samples containing the degradation products were collected and counted in a γ counter. It is possible that degradation products of SAA and apoA-I larger than 3,000 were retained in the column, resulting in an underestimate of apolipoprotein degradation. We opted to use this method of assessing degradation because the conventional method [i.e., quantifying trichloroacetic acid-soluble 125I counts] failed to produce meaningful results due to substantial precipitation of the 125I-SAA peptide fragments.
Statistical analysis
Results are expressed as the mean ± SEM as indicated in the figure legends. Statistics were calculated with GraphPad’s Prism 5 software. Comparisons of multiple groups were performed using one-way ANOVA or two-way ANOVA with Bonferroni’s multiple comparison tests. P-values less than 0.05 were considered statistically significant and denoted in figures with a single asterisk (*); P-values less than 0.01 and 0.001 were denoted with two (**) and three asterisks (***), respectively.
RESULTS
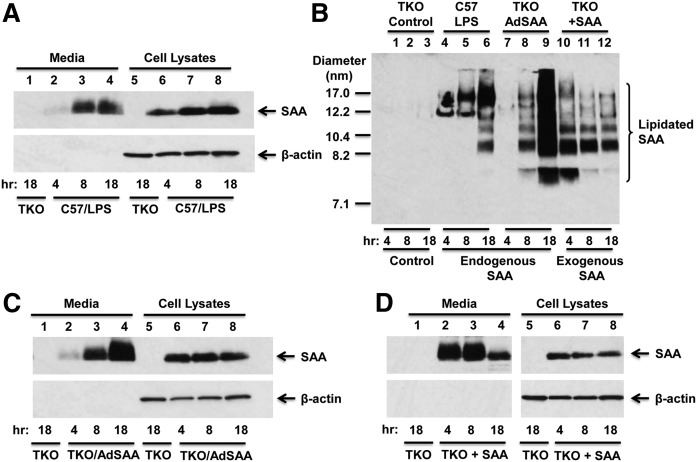
During an acute inflammatory response, SAA is synthesized primarily in the liver and secreted into the circulation, where it is found mainly associated with HDL. Whether SAA incorporates into HDL during HDL biogenesis or through a separate process is not known, and was the focus of this study. SAA expression was induced in C57BL/6 mice by injecting LPS 6 h prior to the preparation of hepatocytes, which allowed for robust SAA expression and secretion throughout the 18 h ex vivo experiments (Fig. 1A). As expected, no immunoreactive SAA was detected in conditioned media or cell lysates of hepatocyte cultures prepared from SAA1.1/2.1/3-TKO mice (Fig. 1A, lanes 1 and 5). Cell culture media were subjected to NDGGE and immunoblotted to assess the lipidation state of the secreted SAA (Fig. 1B; lanes 1–6). This analysis indicated that SAA is efficiently lipidated to form a number of discrete species ranging from ∼8.2 to ∼17 nm in diameter; no lipid-free SAA was detected in the culture media at any of the time points. Notably, the larger (∼12 and 17 nm) particles were predominant during the earlier time intervals, suggesting that smaller species are not converted to larger species during the course of the incubations.
Fig. 1.
Hepatocytes efficiently lipidate endogenous and exogenous SAA. A, C, D: SAA content of conditioned media and cell lysates from primary hepatocyte cultures. Mouse primary hepatocytes were prepared as described in the Materials and Methods and, after overnight culture, were treated with 5 μM T0901317 to induce ABCA1 expression, and then cultured in medium containing 0.2% fatty acid-free BSA. Conditioned media (7 μl) and cell lysates (10 μg protein) were collected at the indicated times were separated by SDS-PAGE followed by Western blotting to detect SAA with β-actin as loading control for cell lysates. A: Hepatocytes were prepared from untreated SAA1.1/2.1/3-TKO mice and C57BL/6 (C57) mice 6 h after injection of 1 μg/g body weight LPS. The results shown are representative of three immunoblots. B: Cell media from A, C, and D were subjected to NDGGE followed by Western blotting to detect SAA. The migration of size standards and lipidated SAA are indicated. The results shown are representative of two separate immunoblots. Note that a lower exposure of this immunoblot is presented in supplemental Fig. S1. C: Hepatocytes were prepared from untreated SAA1.1/2.1/3-TKO mice or SAA1.1/2.1/3-TKO mice 24 h after administration of 1 × 1011 AdSAA. The results are representative of three immunoblots. D: Hepatocytes from untreated SAA1.1/2.1/3-TKO mice were incubated without or with 10 μg/ml purified mouse SAA; the results are representative of three immunoblots.
To study endogenous SAA lipidation in the absence of inflammation, an adenoviral vector expressing SAA (AdSAA) was injected into SAA1.1/2.1/3-TKO mice 24 h prior to the preparation of hepatocyte cultures. As shown in Fig. 1C, SAA expression was sustained in primary hepatocytes from AdSAA-administered mice and accumulated in the culture media in a time-dependent fashion. Most of the SAA secreted by AdSAA-transduced hepatocytes was lipidated (Fig. 1B; supplemental Fig. S1, lanes 7–9), forming species that appeared to be qualitatively similar to the lipidated SAA species produced by hepatocytes undergoing an acute inflammatory response. At the doses administered, AdSAA provided markedly higher levels of secreted SAA compared with LPS, and resulted in the appearance of a portion of lipidated SAA that was smaller in size (∼7.5 nm) and thus likely relatively lipid poor. Taken together, these results suggest that the lipidation of endogenously expressed SAA is not markedly influenced by inflammatory signaling in the liver, but can be overwhelmed in circumstances where SAA secretion is extremely high.
We next investigated to determine whether the lipidation of exogenous SAA by mouse hepatocytes is distinctly different from the lipidation of secreted endogenously expressed SAA. Primary hepatocytes from SAA1.1/2.1/3-TKO mice were incubated with purified mouse SAA. Cell lysates and culture media were collected after 4, 8, or 18 h, separated by SDS-PAGE, and analyzed by Western blotting. Interestingly, exogenously added SAA was readily detected in cell lysates at each of the time points, whereas SAA in the media disappeared during the 18 h time course (Fig. 1D). To determine the lipidation status of extracellular SAA, aliquots of culture media were separated by NDGGE followed by Western blotting (Fig. 1B, lanes 10–12). Based on the migration of immunopositive species, exogenously added SAA was lipidated to form species corresponding to endogenously expressed SAA, ranging from ∼7.5 to ∼17.0 nm in diameter. However, unlike secreted SAA, which accumulated in the culture media over time, exogenously added SAA appeared to disappear from the media as both lipidated and lipid-poor forms.
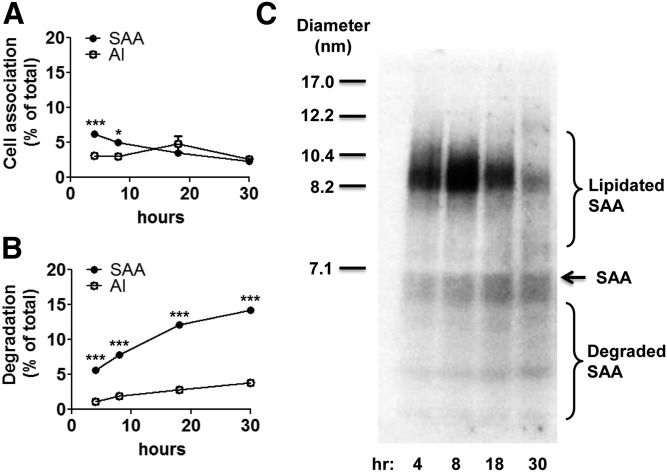
To investigate whether the disappearance of exogenous SAA from conditioned media is due to cell uptake, hepatocytes were incubated with 5 μg/ml 125I-SAA for cell association and degradation assays. An additional set of hepatocytes was incubated with 5 μg/ml 125I-apoA-I for comparison. In the case of SAA, ∼6.3% and ∼2.4% of the apolipoprotein added to the dish was found cell-associated after 4 and 30 h, respectively (Fig. 2A), and this decrease in cell-associated SAA was accompanied by an accumulation of degraded SAA in the culture media (Fig. 2B). In contrast, only 3.1% of apoA-I added to the dish was cell associated at 4 h, and the amount of cell-associated apoA-I remained relatively constant throughout the 30 h incubation (Fig. 2A). The amount of apoA-I degradation products in the media after 30 h was ∼4-fold lower compared with SAA (Fig. 2B). Taken together, these results suggest that hepatocytes take up and degrade SAA to a significantly greater extent compared with apoA-I. To visualize the lipidation and degradation status of extracellular 125I-SAA, conditioned media were separated by NDGGE followed by autoradiography (Fig. 2C). At the 4 and 8 h time points, a predominant ∼8.2 nm species was apparent that likely corresponds to a major lipidated SAA species detected by immunoblotting (Fig. 1B). At the later time points (18 and 30 h), this 8.2 nm species disappeared coincidentally with the accumulation of SAA degradation products (Fig. 2B, C). Our results suggest that exogenous SAA is efficiently lipidated by hepatocytes, but the lipidated products are labile, due at least in part to hepatocyte uptake and degradation.
Fig. 2.
Exogenous SAA is degraded by hepatocytes to a greater extent than apoA-I. Primary hepatocytes were prepared from SAA1.1/2.1/3-TKO mice as described in the Materials and Methods and incubated at 37°C for the indicated times with 5 μg/ml 125I-SAA or 125I-apoA-I. A: After removing the culture media, cells were solubilized in 0.1 N NaOH, and the protein and 125I contents of the lysates were determined and expressed as the percentage of the total amount of 125I added to the dish. B: The cell-mediated degradation of SAA and apoA-I was measured in the culture media and expressed as the percentage of the total amount of 125I added to the dish. Statistical analysis was performed using two-way ANOVA with Bonferroni’s multiple comparison tests. P-values less than 0.05 and 0.001 are denoted with one (*) and three asterisks (***), respectively. C: Aliquots of the cell media at each time point were separated by NDGGE followed by autoradiography to visualize lipidated and degraded SAA. The migration of size standards, lipidated SAA, lipid-free SAA, and SAA degradation products are indicated.
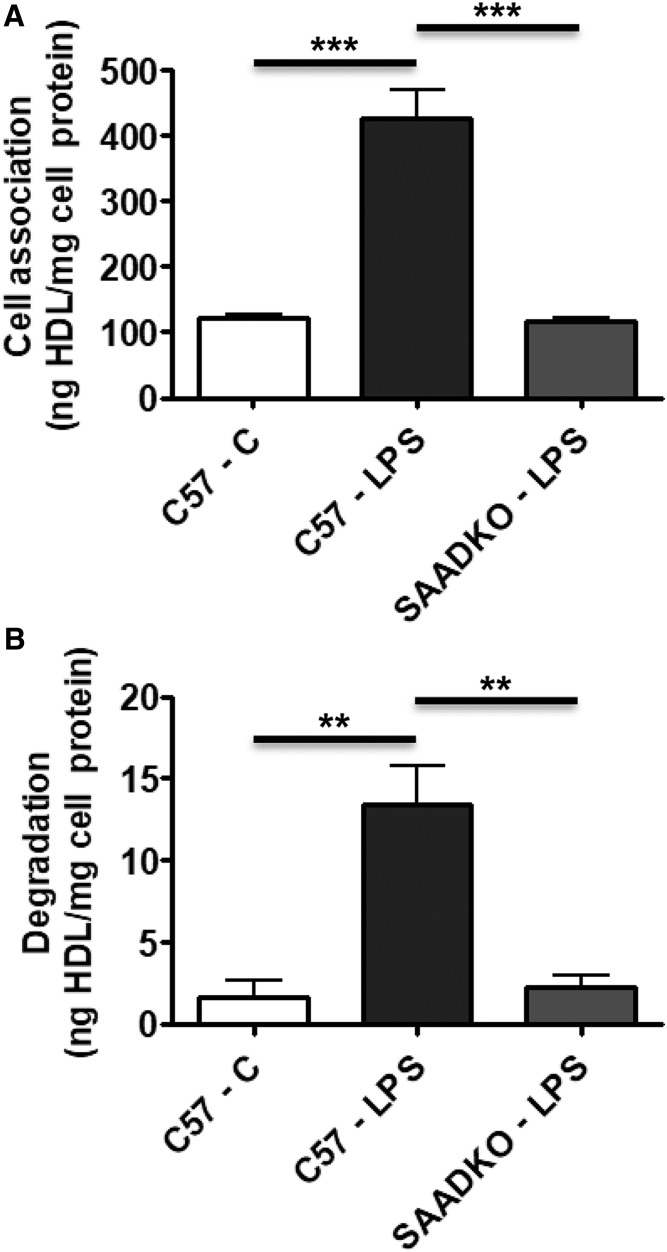
Because plasma SAA is mainly associated with HDL and not lipid free, the finding that SAA appears to be taken up and degraded more readily by hepatocytes than apoA-I prompted us to assess hepatocyte metabolism of 125I-HDL with and without SAA enrichment. 125I-HDLs were prepared using plasma collected from untreated C57BL/6 mice (C57-C HDL) and from C57BL/6 and SAA1.1/2.1-DKO mice 24 h after LPS injection (C57-LPS HDL and SAADKO-LPS HDL, respectively). Based on Coomassie staining after SDS-PAGE, we estimated that SAA comprised ∼30% of total apolipoproteins in C57-LPS HDLs, whereas SAA was undetec in C57-C and SAADKO-LPS HDLs (data not shown). Primary hepatocytes from C57BL/6 mice were incubated at 37°C for 4 h with the 125I-labeled HDLs. As shown in Fig. 3A, there was a significant increase (3.5-fold) in cell association of 125I-C57-LPS HDL compared with C57-C HDL, which was accompanied by a significant 8-fold increase in degradation (Fig. 3B). Notably, this increase in cell association and degradation was totally abrogated for 125I-SAADKO-LPS HDL, suggesting that SAA largely accounts for the observed increase in the association with hepatocytes and subsequent degradation of C57-LPS HDL compared with C57-C HDL. The increase in the cell association and degradation of 125I-C57-LPS HDL compared with the other HDLs could reflect either uptake and degradation of the entire 125I-labeled particle or selective uptake and degradation of the HDL-associated SAA.
Fig. 3.
SAA increases HDL association and degradation in hepatocytes. HDLs were prepared from untreated C57BL/6 mice and C57BL/6 and SAA1.1/2.1-DKO mice 24 h after LPS injection. The HDLs were radioiodinated and then incubated with primary hepatocytes from C57BL/6 mice for 4 h at 37°C (10 μg HDL protein per milliliter). Cell association (A) and degradation (B) were determined as described in the Materials and Methods and expressed as nanograms of HDL protein per milligram of cell protein. Values shown are the mean and SEM of triplicate determinations. Statistical analysis was performed using one-way ANOVA with Bonferroni’s multiple comparison tests. P-values less than 0.01 and 0.001 are denoted with two (**) and three asterisks (***), respectively.
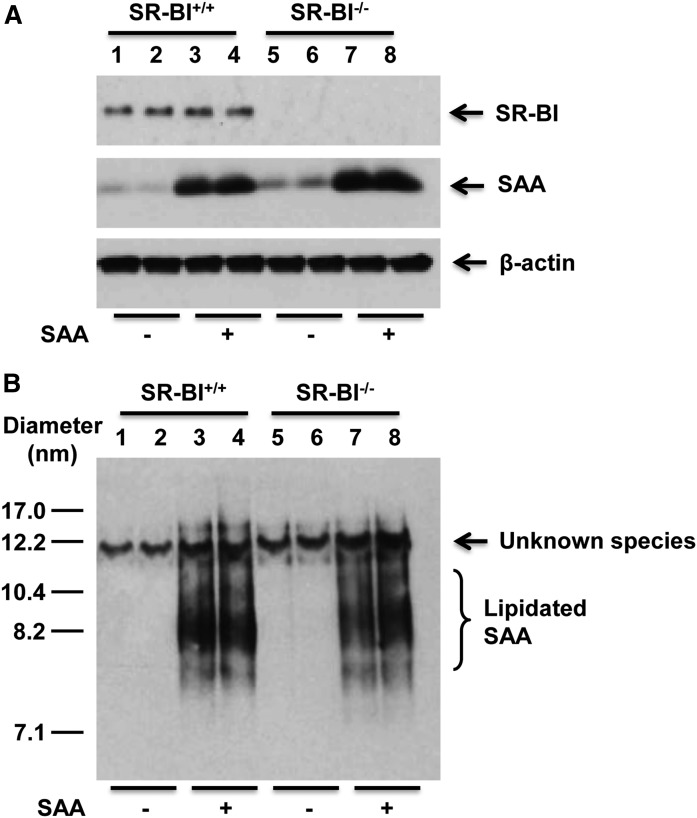
Several studies have implicated SR-BI in mediating cholesterol efflux to SAA (35, 36), as well as SAA uptake and clearance (25, 37). To investigate whether SAA lipidation is dependent on SR-BI, primary hepatocytes were prepared from SR-BI+/+ and SR-BI−/− mice and then incubated with exogenously added mouse SAA. Western blotting confirmed SR-BI expression only in cells expressing SR-BI, and there was no evidence that incubations with SAA altered hepatocyte expression of SR-BI (Fig. 4A, top panel). It also did not appear that the lack of SR-BI reduced the amount of exogenous SAA associated with hepatocytes as assessed by Western blotting of cell lysates after the 18 h incubation (Fig. 4A, middle panel; compare lanes 3–4 and 7–8). To investigate the impact of SR-BI deficiency on SAA lipidation, culture media were separated by NDGGE and immunoblotted with anti-SAA antibody (Fig. 4B). In this and some other studies, an immunoreactive species with an apparent diameter ∼12 nm was detected even in samples lacking SAA. The identity of this species is unknown. As shown in Fig. 4B, the lipidation of exogenous SAA by SR-BI−/− hepatocytes was not markedly altered compared with SR-BI+/+ hepatocytes, suggesting that SAA lipidation is not dependent on SR-BI.
Fig. 4.
Hepatocyte lipidation of SAA is not dependent on SR-BI. Primary hepatocytes from SR-BI+/+ and SR-BI−/− mice were incubated without or with 10 μg/ml purified mouse SAA for 18 h in the absence of T0901317 treatment. A: Cell lysates (20 μg protein) were separated by SDS-PAGE followed by Western blotting to detect SAA and SR-BI; β-actin was assessed for loading control. B: Cell media (18 μl) were separated by NDGGE followed by Western blotting to determine the lipidation status of SAA. The results shown are representative of two independent experiments and two separate immunoblots. The migration of size standards and lipidated SAA species are indicated.
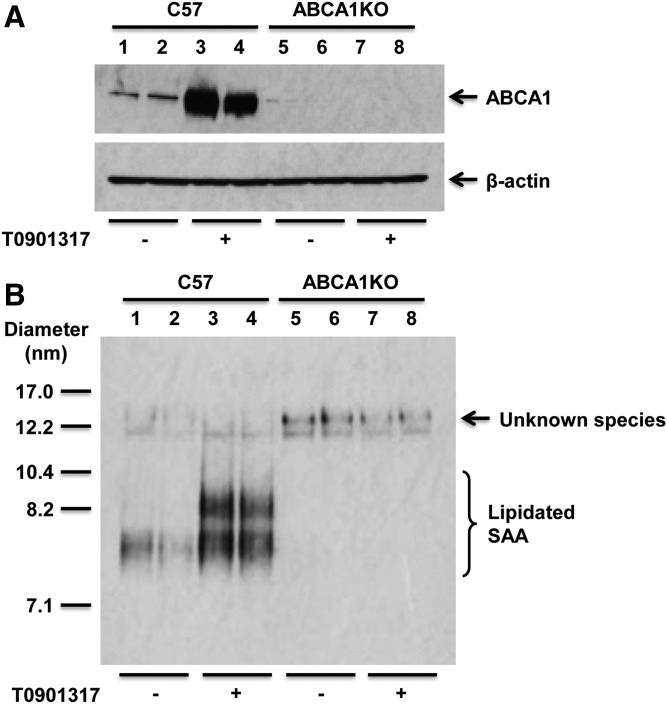
As indicated in the figure legends, hepatocyte cultures were preincubated with the LXR agonist T0901317 to induce the expression of ABCA1, which we have shown significantly increases cholesterol efflux and lipidation of apoA-I by primary mouse hepatocytes (38). To investigate whether SAA lipidation is dependent on ABCA1, primary hepatocytes were prepared from C57BL/6 and ABCA1-KO mice and incubated with purified mouse SAA without and with pretreatment with LXR agonist. As expected, ABCA1 expression was detected only in C57BL/6 cells, where it was dramatically induced by LXR agonist (Fig. 5A). As shown in Fig. 5B, exogenous SAA was efficiently lipidated by hepatocytes from C57BL/6 mice but not ABCA1-KO mice, and induction of ABCA1 by LXR agonist was associated with an increase in the number and size of lipidated SAA species. Thus, the lipidation of SAA is clearly dependent on ABCA1.
Fig. 5.
Hepatocyte lipidation of SAA is dependent on ABCA1. Primary hepatocytes from C57BL/6 (C57) and ABCA1-deficient (ABCA1KO) mice were treated without or with T0901317 (5 μM) for 8 h, and then incubated in the presence or absence of 10 μg/ml mouse SAA for 18 h, as indicated. A: Cell lysates were analyzed by SDS-PAGE followed by Western blotting for ABCA1 expression; β-actin was assessed for loading control. B: Cell media were separated by NDGGE and then analyzed by Western blotting to determine the lipidation status of SAA. The results shown are representative of three independent experiments and three separate immunoblots. The migration of size standards and lipidated SAA species are indicated.
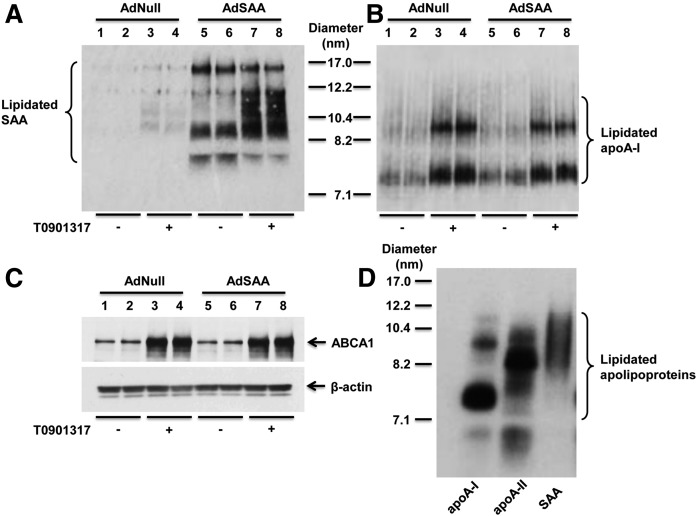
We next investigated whether SAA expression alters the lipidation of mouse apoA-I. In particular, we were interested in determining whether both SAA and apoA-I incorporate into the same nascent particles. For these studies, primary hepatocytes were prepared from C57BL/6 mice 24 h after injection of AdSAA or Adnull as negative control. After 8 h incubation with T0901317, cells were incubated with mouse apoA-I for 16 h. Cell media were separated by NDGGE followed by Western blotting to determine the lipidation status of SAA and apoA-I (Fig. 6A, B). While lipidated SAA was more readily detected in conditioned media from hepatocytes transduced with AdSAA compared with Adnull, the size of the lipidated species generated by SAA-overexpressing cells compared with control cells appeared to be qualitatively similar (Fig. 6A). Treatment with the LXR agonist enhanced the generation of lipidated SAA species by both sets of cells, consistent with a role for ABCA1 in this process. Importantly, overexpression of SAA in hepatocytes did not alter the amount or size distribution of lipidated species containing apoA-I (Fig. 6B). This finding along with the observation that the lipidated products containing apoA-I compared with SAA appear to be distinct in size (compare Fig. 6A and B) strongly indicate that the two apolipoproteins are not present on the same nascent particles.
Fig. 6.
Endogenous SAA expression does not alter the lipidation of apoA-I. A, B: Primary hepatocytes were prepared from C57BL/6 mice 24 h after injection of 1 × 1011 particles AdNull or AdSAA. After 8 h incubation without or with T0901317 (5 μM), cells were incubated with mouse apoA-I (10 μg/ml) for 16 h. Cell media were separated by NDGGE followed by Western blotting to determine the lipidation status of SAA (A) and apoA-I (B). The migration of size standards and lipidated apoA-I and SAA species are indicated. C: Cell lysates (20 μg protein) were analyzed by SDS-PAGE followed by Western blotting for ABCA1 expression; β-actin was assessed for loading control. D: Primary hepatocytes from C57BL/6 mice were treated with 5 μM T0901317 for 8 h and then incubated with 125I-radiolabeled human apoA-I (5 μg/ml), apoA-II (10 μg/ml), or SAA (10 μg/ml) for 16 h. Cell media were separated by NDGGE followed by autoradiography. The migration of size standards and lipidated species are indicated.
As another approach to investigate the relationship between the lipidation of SAA and other HDL apolipoproteins, primary hepatocytes prepared from C57BL/6 mice were incubated with 125I-radiolabeled human apoA-I, apoA-II, or SAA for 16 h. Cell media were then separated by NDGGE followed by autoradiography to allow for direct comparison of the lipidated species containing each apolipoprotein. As shown in Fig. 6D, 125I-apoA-I was associated with particles primarily ∼7.4 and ∼10.0 nm in diameter, whereas the major species containing 125I-apoA-II was ∼8.2 nm in diameter. In contrast, SAA-containing lipidated species comprised particles with sizes ranging from ∼8.2 to 12.2 nm in size. The discrepancy in the size distribution of the apoA-I, apoA-II, and SAA species indicates that the apolipoproteins are lipidated independently of one another, and provides evidence that SAA is not incorporated into apoA-I-containing HDL during HDL biogenesis.
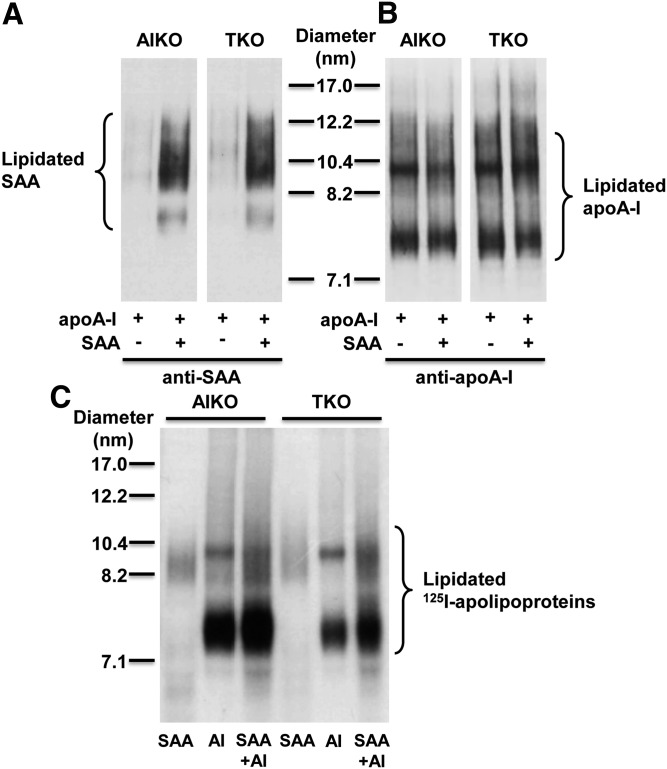
To provide additional evidence that hepatocytes do not generate nascent particles containing both apoA-I and SAA, we performed experiments using hepatocytes isolated from apoA-I-deficient (AIKO) and SAA-deficient (TKO) mice that lack the ability to form lipidated particles containing both SAA and apoA-I endogenously produced. After 8 h incubation with T0901317, cells were incubated for 16 h with mouse apoA-I (10 μg/ml) in the absence or presence of mouse SAA (10 μg/ml), and then aliquots of conditioned media were separated by NDGGE followed by Western blotting to visualize lipidated species containing SAA and/or apoA-I (Fig. 7A, B). In both AIKO and TKO hepatocytes, the presence of exogenous SAA did not alter the amount or size distribution of lipidated species containing apoA-I (Fig. 7B), as would be expected if the two apolipoproteins were incorporated on the same nascent HDLs. As another approach, we also incubated AIKO and TKO hepatocytes with 125I-radiolabeled mouse SAA (10 μg/ml), mouse apoA-I (10 μg/ml), or a mixture of both 125I-SAA and 125I-apoA-I for 16 h, and then visualized lipidated species by autoradiography (Fig. 7C). Strikingly, the major lipidated species observed when only apoA-I was present (∼7.4 and ∼10 nm) were not shifted in size with the addition of SAA. Similarly, the polydisperse species containing only SAA (∼8.2–12.2 nm) appeared to be unaltered in the presence of apoA-I. These data strongly indicate that the two apolipoproteins are not present on the same nascent particles.
Fig. 7.
The presence of SAA does not alter the lipidation of apoA-I. Primary hepatocytes were prepared from apoA-I-deficient (AIKO) and SAA1.1/2.1/3-deficient (TKO) mice and treated with T0901317 (5 μM) for 8 h. A, B: Cells were incubated with mouse apoA-I (10 μg/ml) in the absence or presence of mouse SAA (10 μg/ml) for 16 h, as indicated. Cell media were separated by NDGGE followed by Western blotting to determine the lipidation status of SAA (A) and apoA-I (B). The results shown are representative of two immunoblots. The migration of size standards and lipidated SAA and apoA-I species are indicated. C: Cells were incubated with 125I-radiolabeled mouse SAA (10 μg/ml), 125I-radiolabeled mouse apoA-I (10 μg/ml), or a mixture of both (each at 10 μg/ml) for 16 h. Cell media were separated by NDGGE followed by autoradiography. The results shown are representative of three gels. The migration of size standards and lipidated species are indicated.
DISCUSSION
The liver undergoes major reprogramming during an acute inflammatory response with a number of genes either suppressed or induced. The most sensitive acute phase reactant is SAA, which may be transiently upregulated 1,000-fold or more within 12–48 h after infection or tissue injury (1, 2). Acute phase SAA is present in virtually all vertebrates studied and is highly conserved through at least 500 million years of evolution, indicating that the induction of SAA plays an important survival role in a systemic inflammatory response (39, 40). The vast majority of SAA secreted by the liver is typically found associated with the HDL fraction in plasma; little or none is in a lipid-free form (22). Although it is now recognized that lipid-free and HDL-associated SAA are functionally distinct (23, 35, 36), the pathways leading to the association/disassociation of SAA with HDL are not clearly understood. Whether SAA-containing HDL is formed through the modification of preexisting HDL or during the assembly of new HDL particles has not been conclusively established. In the present study, we demonstrate that HDL does not acquire SAA during nascent HDL biogenesis. This data, coupled with our prior study demonstrating that SAA is an exchangeable apolipoprotein (22), provide new insights into SAA metabolism and the regulation of its function.
We focused on the interaction of SAA with hepatocytes, the primary site of HDL biogenesis. An important finding of our study is that SAA is lipidated by hepatocytes independent of apoA-I. Based on their migration on nondenaturing gradient gels, these two apolipoproteins do not appear to be present on the same nascent particles. Whereas lipid-poor apoA-I generated two distinct lipidated species ∼7.4 and ∼10.0 nm in diameter, the lipidated SAA species were much more poly-disperse, ranging from ∼8.2 to 12.2 nm in size. Moreover, the apparent size of apoA-I-containing species was not altered when SAA was also present, supporting the conclusion that the vast majority of SAA is not incorporated into apoA-I-containing particles during HDL biogenesis. Our results are similar to those of Abe-Dohme et al. (41), who characterized the size distribution of cholesterol- and phosphatidylcholine-containing particles present in conditioned media from HEK293 cells stably transfected to express ABCA1. Based on HPLC elution profiles, cells incubated with apoA-I for 48 h formed two major species of lipidated particles that differed in size compared with the dominant species formed when the cells were incubated with lipid-poor SAA. In another study, Cabana et al. (42) determined that apoA-I and SAA recovered from conditioned media of McA RH777 rat hepatoma cell cultures by density gradient centrifugation were distributed in different density fractions. Our novel finding that the initial lipidation of SAA does not give rise to particles containing both apoA-I and SAA raises interesting questions as to how SAA incorporates into the apoA-I-containing HDL fraction in vivo. It is known that SAA-containing HDL particles can be produced by the displacement of apoA-I on HDL by lipid-free SAA (42), and that SAA can be exchanged between HDL and apoB-containing lipoproteins (22). Whether the lipidated species generated by ABCA1 efficiently incorporate into mature HDL particles, and whether this leads to the dissociation of apoA-I, requires further investigation.
We determined that SAA lipidation by hepatocytes does not occur in the absence of ABCA1. SAA contains two α-helical regions and a short amphipathic α-helix within the N-terminal portion that is thought to be involved in lipid binding, and thus appears to fulfill the structural requirements for ABCA1 efflux (39). The finding in the current study that ABCA1 is required for SAA lipidation is consistent with previous studies in stably transfected HEK293 cells (41) and primary hepatocytes isolated from mice after LPS injection (43). Furthermore, Hu et al. (43) showed that in the absence of ABCA1, SAA biosynthesis is induced in the liver after LPS administration, but it is not recovered in the plasma in a lipidated form, suggesting that there is no alternative pathway for SAA-HDL production in the absence of ABCA1. On the other hand, SAA “only” HDL can be detected in apoA-I-deficient mice injected with LPS (44, 45), consistent with the biogenesis of SAA-containing particles that lack apoA-I. However, the formation of such particles appears to rely on an unidentified component of the inflammatory response, because SAA expressed in the livers of apoA-I-deficient mice via adenoviral vector was found almost exclusively in the lipoprotein-free fraction (46).
Our studies indicate that SAA is not lipidated by SR-BI, a high affinity receptor for SAA (37). However, we cannot rule out the possibility that SR-BI plays a role in the formation of HDL-associated SAA. According to a published study, lipid-free and lipid-poor SAA failed to stimulate cholesterol efflux via SR-BI. However, SR-BI-mediated cholesterol efflux did occur following extensive lipidation of SAA (35). Thus, the impact of the lipidation status of SAA on cholesterol mobilization by SR-BI appears to be similar to what has been observed for other amphipathic apolipoproteins, including apoA-I (47).
Kinetic studies in mice determined that SAA on HDL is cleared more rapidly from plasma than apoA-I (25, 48, 49). In the current study, we showed that exogenously added SAA associates with hepatocytes and is degraded by hepatocytes to a significantly greater extent compared with apoA-I. Thus, SAA is efficiently lipidated by hepatocytes, but the lipidated products are labile, possibly due to hepatocyte uptake and degradation. It is also possible that hepatocytes secrete a soluble protease capable of degrading extracellular SAA. However, results from mixing experiments in which SAA was incubated with hepatocyte-conditioned media indicate that the degradation of SAA is cell dependent (data not shown). Thus, available data suggests that SAA is either degraded inside the cell and resecreted, or is degraded by a protease that is tethered on the hepatocyte cell surface. We also showed that the presence of SAA increases the association and subsequent degradation of radioiodinated HDL by hepatocytes, consistent with the notion that SAA is selectively dissociated from the HDL particle by hepatocytes.
Notably, the uptake/degradation of SAA was not markedly altered in hepatocytes lacking SR-BI. This finding aligns with our previous report demonstrating that although the in vivo clearance rate of SAA was reduced in mice lacking SR-BI, it remained significantly faster compared with that of apoA-I, indicating a relatively minor role of SR-BI in SAA’s rapid clearance (25). However, the current studies appear to be in conflict with a previous study identifying human SR-BII as a functional SAA receptor that mediates uptake of lipid-free SAA (50), because the SR-BI−/− mice used in the present study also lack SR-BII, an mRNA splicing variant of SR-BI that contains an alternative C-terminal cytoplasmic tail (51, 52). The discrepant conclusions regarding the role of SR-BII in SAA metabolism from the two studies may be due to differences in experimental approaches (loss-of-function versus gain-of-function), cell systems (primary mouse hepatocytes versus transfected HeLa cells), or species differences (mouse versus human SR-BII). Clearly, additional studies are needed to investigate the pathway(s) by which hepatocytes take up and degrade SAA in all of its lipidated forms.
SAA has been shown to exert a number of biological effects consistent with its role in innate immunity and tissue repair, including leukocyte chemotaxis, inflammatory cytokine induction, and upregulation of genes involved in extracellular matrix remodeling (15, 16, 20). Notably, these activities have been attributed to lipid-free SAA and are markedly abrogated when SAA is associated with HDL (19, 23, 24). Like lipid-free apoA-I, lipid-free SAA seems to be removed rapidly from the extracellular space by an unknown mechanism in vivo, as it does not accumulate in plasma. Because lipid-free SAA is cleared more rapidly than HDL-bound SAA (48), it is possible that the main purpose for SAA’s association with HDL is to prolong its life in the circulation and to regulate its pro-inflammatory effects during an acute phase response.
In summary, SAA is lipidated by hepatocytes in an ABCA1-dependent manner to form nascent particles that are distinct from apoA-I-containing particles, indicating that SAA is not incorporated into HDL during HDL biogenesis. These findings provide new insights into SAA metabolism and the regulation of its function.
Supplementary Material
Footnotes
Abbreviations:
- LPS
- lipopolysaccharide
- NDGGE
- nondenaturing gradient gel electrophoresis
- SAA
- serum amyloid A
- SR-BI
- scavenger receptor class B type I
This work was supported by National Institutes of Health Grant R01 HL134731 (to N.R.W. and F.C.d.B.) and U.S. Department of Veterans Affairs Grants CX000773 (to N.R.W.), and CX000975 (to L.R.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Uhlar C. M., Burgess C. J., Sharp P. M., and Whitehead A. S.. 1994. Evolution of the serum amyloid A (SAA) protein superfamily. Genomics. 19: 228–235. [DOI] [PubMed] [Google Scholar]
- 2.Uhlar C. M., and Whitehead A. S.. 1999. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265: 501–523. [DOI] [PubMed] [Google Scholar]
- 3.Sjöholm K., Palming J., Olofsson L. E., Gummesson A., Svensson P. A., Lystig T. C., Jennische E., Brandberg J., Torgerson J. S., Carlsson B., et al. 2005. A microarray search for genes predominantly expressed in human omental adipocytes: adipose tissue as a major production site of serum amyloid A. J. Clin. Endocrinol. Metab. 90: 2233–2239. [DOI] [PubMed] [Google Scholar]
- 4.Meek R. L., Urieli-Shoval S., and Benditt E. P.. 1994. Expression of apolipoprotein serum amyloid A mRNA in human atherosclerotic lesions and cultured vascular cells: implications for serum amyloid A function. Proc. Natl. Acad. Sci. USA. 91: 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamada T., Kakihara T., Kamishima T., Fukuda T., and Kawai T.. 1996. Both acute phase and constitutive serum amyloid A are present in atherosclerotic lesions. Pathol. Int. 46: 797–800. [DOI] [PubMed] [Google Scholar]
- 6.O’Hara R., Murphy E. P., Whitehead A. S., FitzGerald O., and Bresnihan B.. 2004. Local expression of the serum amyloid A and formyl peptide receptor-like 1 genes in synovial tissue is associated with matrix metalloproteinase production in patients with inflammatory arthritis. Arthritis Rheum. 50: 1788–1799. [DOI] [PubMed] [Google Scholar]
- 7.Malle E., Sodin-Semrl S., and Kovacevic A.. 2009. Serum amyloid A: an acute-phase protein involved in tumour pathogenesis. Cell. Mol. Life Sci. 66: 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozinovski S., Vlahos R., Anthony D., McQualter J., Anderson G., Irving L., and Steinfort D.. 2016. COPD and squamous cell lung cancer: aberrant inflammation and immunity is the common link. Br. J. Pharmacol. 173: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marzi C., Huth C., Herder C., Baumert J., Thorand B., Rathmann W., Meisinger C., Wichmann H. E., Roden M., Peters A., et al. 2013. Acute-phase serum amyloid A protein and its implication in the development of type 2 diabetes in the KORA S4/F4 study. Diabetes Care. 36: 1321–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Getz G. S., Krishack P. A., and Reardon C. A.. 2016. Serum amyloid A and atherosclerosis. Curr. Opin. Lipidol. 27: 531–535. [DOI] [PubMed] [Google Scholar]
- 11.Shridas P., and Tannock L. R.. 2019. Role of serum amyloid A in atherosclerosis. Curr. Opin. Lipidol. 30: 320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb N. R., De Beer M. C., Wroblewski J. M., Ji A., Bailey W., Shridas P., Charnigo R. J., Noffsinger V. P., Witta J., Howatt D. A., et al. 2015. Deficiency of endogenous acute-phase serum amyloid A protects apoE-/- mice from angiotensin II-induced abdominal aortic aneurysm formation. Arterioscler. Thromb. Vasc. Biol. 35: 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anthony D., Seow H. J., Uddin M., Thompson M., Dousha L., Vlahos R., Irving L. B., Levy B. D., Anderson G. P., and Bozinovski S.. 2013. Serum amyloid A promotes lung neutrophilia by increasing IL-17A levels in the mucosa and gammadelta T cells. Am. J. Respir. Crit. Care Med. 188: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badolato R., Johnston J. A., Wang J. M., McVicar D., Xu L. L., Oppenheim J. J., and Kelvin D. J.. 1995. Serum amyloid A induces calcium mobilization and chemotaxis of human monocytes by activating a pertussis toxin-sensitive signaling pathway. J. Immunol. 155: 4004–4010. [PubMed] [Google Scholar]
- 15.Lee H. Y., Kim S. D., Shim J. W., Lee S. Y., Lee H., Cho K. H., Yun J., and Bae Y. S.. 2008. Serum amyloid A induces CCL2 production via formyl peptide receptor-like 1-mediated signaling in human monocytes. J. Immunol. 181: 4332–4339. [DOI] [PubMed] [Google Scholar]
- 16.Dong Z., Wu T., Qin W., An C., Wang Z., Zhang M., Zhang Y., Zhang C., and An F.. 2011. Serum amyloid A directly accelerates the progression of atherosclerosis in apolipoprotein E-deficient mice. Mol. Med. 17: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furlaneto C. J., and Campa A.. 2000. A novel function of serum amyloid A: a potent stimulus for the release of tumor necrosis factor-alpha, interleukin-1beta, and interleukin-8 by human blood neutrophil. Biochem. Biophys. Res. Commun. 268: 405–408. [DOI] [PubMed] [Google Scholar]
- 18.Song C., Hsu K., Yamen E., Yan W., Fock J., Witting P. K., Geczy C. L., and Freedman S. B.. 2009. Serum amyloid A induction of cytokines in monocytes/macrophages and lymphocytes. Atherosclerosis. 207: 374–383. [DOI] [PubMed] [Google Scholar]
- 19.Shridas P., De Beer M. C., and Webb N. R.. 2018. High-density lipoprotein inhibits serum amyloid A-mediated reactive oxygen species generation and NLRP3 inflammasome activation. J. Biol. Chem. 293: 13257–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson P. G., Thompson J. C., Webb N. R., de Beer F. C., King V. L., and Tannock L. R.. 2008. Serum amyloid A, but not C-reactive protein, stimulates vascular proteoglycan synthesis in a pro-atherogenic manner. Am. J. Pathol. 173: 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H. Y., Kim M. K., Park K. S., Bae Y. H., Yun J., Park J. I., Kwak J. Y., and Bae Y. S.. 2005. Serum amyloid A stimulates matrix-metalloproteinase-9 upregulation via formyl peptide receptor like-1-mediated signaling in human monocytic cells. Biochem. Biophys. Res. Commun. 330: 989–998. [DOI] [PubMed] [Google Scholar]
- 22.Wilson P. G., Thompson J. C., Shridas P., McNamara P. J., de Beer M. C., de Beer F. C., Webb N. R., and Tannock L. R.. 2018. Serum amyloid A is an exchangeable apolipoprotein. Arterioscler. Thromb. Vasc. Biol. 38: 1890–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M. H., de Beer M. C., Wroblewski J. M., Webb N. R., and de Beer F. C.. 2013. SAA does not induce cytokine production in physiological conditions. Cytokine. 61: 506–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badolato R., Wang J. M., Murphy W. J., Lloyd A. R., Michiel D. F., Bausserman L. L., Kelvin D. J., and Oppenheim J. J.. 1994. Serum amyloid A is a chemoattractant: induction of migration, adhesion, and tissue infiltration of monocytes and polymorphonuclear leukocytes. J. Exp. Med. 180: 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim M. H., de Beer M. C., Wroblewski J. M., Charnigo R. J., Ji A., Webb N. R., de Beer F. C., and van der Westhuyzen D. R.. 2016. Impact of individual acute phase serum amyloid A isoforms on HDL metabolism in mice. J. Lipid Res. 57: 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Beer M. C., Webb N. R., Wroblewski J. M., Noffsinger V. P., Rateri D. L., Ji A., van der Westhuyzen D. R., and de Beer F. C.. 2010. Impact of serum amyloid A on high density lipoprotein composition and levels. J. Lipid Res. 51: 3117–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson J. C., Wilson P. G., Shridas P., Ji A., de Beer M., de Beer F. C., Webb N. R., and Tannock L. R.. 2018. Serum amyloid A3 is pro-atherogenic. Atherosclerosis. 268: 32–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timmins J. M., Lee J. Y., Boudyguina E., Kluckman K. D., Brunham L. R., Mulya A., Gebre A. K., Coutinho J. M., Colvin P. L., Smith T. L., et al. 2005. Targeted inactivation of hepatic Abca1 causes profound hypoalphalipoproteinemia and kidney hypercatabolism of apoA-I. J. Clin. Invest. 115: 1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Z., Ai J., Guo L., Ye X., Bondada S., Howatt D., Daugherty A., and Li X. A.. 2018. SR-BI (scavenger receptor class B type 1) is critical in maintaining normal T-cell development and enhancing thymic regeneration. Arterioscler. Thromb. Vasc. Biol. 38: 2706–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji A., Wroblewski J. M., Webb N. R., and van der Westhuyzen D. R.. 2014. Impact of phospholipid transfer protein on nascent high-density lipoprotein formation and remodeling. Arterioscler. Thromb. Vasc. Biol. 34: 1910–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coetzee G. A., Strachan A. F., van der Westhuyzen D. R., Hoppe H. C., Jeenah M. S., and de Beer F. C.. 1986. Serum amyloid A-containing human high density lipoprotein 3. Density, size, and apolipoprotein composition. J. Biol. Chem. 261: 9644–9651. [PubMed] [Google Scholar]
- 32.Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J.. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275. [PubMed] [Google Scholar]
- 33.Bilheimer D. W., Eisenberg S., and Levy R. I.. 1972. The metabolism of very low density lipoprotein proteins. I. Preliminary in vitro and in vivo observations. Biochim. Biophys. Acta. 260: 212–221. [DOI] [PubMed] [Google Scholar]
- 34.de Beer M. C., Durbin D. M., Cai L., Mirocha N., Jonas A., Webb N. R., de Beer F. C., and van Der Westhuyzen D. R.. 2001. Apolipoprotein A-II modulates the binding and selective lipid uptake of reconstituted high density lipoprotein by scavenger receptor BI. J. Biol. Chem. 276: 15832–15839. [DOI] [PubMed] [Google Scholar]
- 35.Marsche G., Frank S., Raynes J. G., Kozarsky K. F., Sattler W., and Malle E.. 2007. The lipidation status of acute-phase protein serum amyloid A determines cholesterol mobilization via scavenger receptor class B, type I. Biochem. J. 402: 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Westhuyzen D. R., Cai L., de Beer M. C., and de Beer F. C.. 2005. Serum amyloid A promotes cholesterol efflux mediated by scavenger receptor B-I. J. Biol. Chem. 280: 35890–35895. [DOI] [PubMed] [Google Scholar]
- 37.Cai L., de Beer M. C., de Beer F. C., and van der Westhuyzen D. R.. 2005. Serum amyloid A is a ligand for scavenger receptor class B type I and inhibits high density lipoprotein binding and selective lipid uptake. J. Biol. Chem. 280: 2954–2961. [DOI] [PubMed] [Google Scholar]
- 38.Ji A., Wroblewski J. M., Cai L., de Beer M. C., Webb N. R., and van der Westhuyzen D. R.. 2012. Nascent HDL formation in hepatocytes and role of ABCA1, ABCG1, and SR-BI. J. Lipid Res. 53: 446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frame N. M., and Gursky O.. 2017. Structure of serum amyloid A suggests a mechanism for selective lipoprotein binding and functions: SAA as a hub in macromolecular interaction networks. Amyloid. 24: 13–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sack G. H., Jr. 2018. Serum amyloid A - a review. Mol. Med. 24: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe-Dohmae S., Kato K. H., Kumon Y., Hu W., Ishigami H., Iwamoto N., Okazaki M., Wu C. A., Tsujita M., Ueda K., et al. 2006. Serum amyloid A generates high density lipoprotein with cellular lipid in an ABCA1- or ABCA7-dependent manner. J. Lipid Res. 47: 1542–1550. [DOI] [PubMed] [Google Scholar]
- 42.Cabana V. G., Feng N., Reardon C. A., Lukens J., Webb N. R., de Beer F. C., and Getz G. S.. 2004. Influence of apoA-I and apoE on the formation of serum amyloid A-containing lipoproteins in vivo and in vitro. J. Lipid Res. 45: 317–325. [DOI] [PubMed] [Google Scholar]
- 43.Hu W., Abe-Dohmae S., Tsujita M., Iwamoto N., Ogikubo O., Otsuka T., Kumon Y., and Yokoyama S.. 2008. Biogenesis of HDL by SAA is dependent on ABCA1 in the liver in vivo. J. Lipid Res. 49: 386–393. [DOI] [PubMed] [Google Scholar]
- 44.Cabana V. G., Reardon C. A., Wei B., Lukens J. R., and Getz G. S.. 1999. SAA-only HDL formed during the acute phase response in apoA-I+/+ and apoA-I-/- mice. J. Lipid Res. 40: 1090–1103. [PubMed] [Google Scholar]
- 45.Hajri T., Elliott-Bryant R., Sipe J. D., Liang J. S., Hayes K. C., and Cathcart E. S.. 1998. The acute phase response in apolipoprotein A-1 knockout mice: apolipoprotein serum amyloid A and lipid distribution in plasma high density lipoproteins. Biochim. Biophys. Acta. 1394: 209–218. [DOI] [PubMed] [Google Scholar]
- 46.Webb N. R., de Beer M. C., van der Westhuyzen D. R., Kindy M. S., Banka C. L., Tsukamoto K., Rader D. L., and de Beer F. C.. 1997. Adenoviral vector-mediated overexpression of serum amyloid A in apoA-I-deficient mice. J. Lipid Res. 38: 1583–1590. [PubMed] [Google Scholar]
- 47.Ji Y., Jian B., Wang N., Sun Y., Moya M. L., Phillips M. C., Rothblat G. H., Swaney J. B., and Tall A. R.. 1997. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272: 20982–20985. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman J. S., and Benditt E. P.. 1983. Plasma clearance kinetics of the amyloid-related high density lipoprotein apoprotein, serum amyloid protein (apoSAA), in the mouse. Evidence for rapid apoSAA clearance. J. Clin. Invest. 71: 926–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tape C., and Kisilevsky R.. 1990. Apolipoprotein A-I and apolipoprotein SAA half-lives during acute inflammation and amyloidogenesis. Biochim. Biophys. Acta. 1043: 295–300. [DOI] [PubMed] [Google Scholar]
- 50.Baranova I. N., Souza A. C. P., Bocharov A. V., Vishnyakova T. G., Hu X., Vaisman B. L., Amar M. J., Chen Z., Remaley A. T., Patterson A. P., et al. 2017. Human SR-BII mediates SAA uptake and contributes to SAA pro-inflammatory signaling in vitro and in vivo. PLoS One. 12: e0175824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Webb N. R., Connell P. M., Graf G. A., Smart E. J., de Villiers W. J., de Beer F. C., and van der Westhuyzen D. R.. 1998. SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. J. Biol. Chem. 273: 15241–15248. [DOI] [PubMed] [Google Scholar]
- 52.Webb N. R., de Villiers W. J., Connell P. M., de Beer F. C., and van der Westhuyzen D. R.. 1997. Alternative forms of the scavenger receptor BI (SR-BI). J. Lipid Res. 38: 1490–1495. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.