Abstract
Zinc metallopeptidase STE24 (ZMPSTE24) is essential for the conversion of farnesyl–prelamin A to mature lamin A, a key component of the nuclear lamina. In the absence of ZMPSTE24, farnesyl–prelamin A accumulates in the nucleus and exerts toxicity, causing a variety of disease phenotypes. By ∼4 months of age, both male and female Zmpste24−/− mice manifest a near-complete loss of adipose tissue, but it has never been clear whether this phenotype is a direct consequence of farnesyl–prelamin A toxicity in adipocytes. To address this question, we generated a conditional knockout Zmpste24 allele and used it to create adipocyte-specific Zmpste24–knockout mice. To boost farnesyl–prelamin A levels, we bred in the “prelamin A–only” Lmna allele. Gene expression, immunoblotting, and immunohistochemistry experiments revealed that adipose tissue in these mice had decreased Zmpste24 expression along with strikingly increased accumulation of prelamin A. In male mice, Zmpste24 deficiency in adipocytes was accompanied by modest changes in adipose stores (an 11% decrease in body weight, a 23% decrease in body fat mass, and significantly smaller gonadal and inguinal white adipose depots). No changes in adipose stores were detected in female mice, likely because prelamin A expression in adipose tissue is lower in female mice. Zmpste24 deficiency in adipocytes did not alter the number of macrophages in adipose tissue, nor did it alter plasma levels of glucose, triglycerides, or fatty acids. We conclude that ZMPSTE24 deficiency in adipocytes, and the accompanying accumulation of farnesyl–prelamin A, reduces adipose tissue stores, but only modestly and only in male mice.
Keywords: animal models, farnesylation, nuclear lamins, fluorescence microscopy, lipodystrophies, zinc metallopeptidase STE24
Zinc metallopeptidase STE24 (ZMPSTE24), an integral membrane zinc metalloprotease (1), is required for the biogenesis of mature lamin A, a key component of the nuclear lamina (2, 3). Lamin A is produced from a precursor protein, prelamin A, by four enzymatic processing steps (4). The cysteine in prelamin A’s carboxyl-terminal CaaX motif (–CSIM) is farnesylated by protein farnesyltransferase. Next, the last three amino acids of the protein (–SIM) are clipped off by Ras-converting enzyme 1 (RCE1) or ZMPSTE24. The newly exposed farnesylcysteine is then methylated by isoprenylcysteine methyltransferase (ICMT). Finally, the last 15 amino acids of prelamin A (including the farnesylcysteine methyl ester) are clipped off by ZMPSTE24, releasing mature lamin A. Prelamin A-to-mature lamin A processing is normally very efficient, such that prelamin A is virtually undetectable in cells and tissues. However, prelamin A-to-lamin A processing is blocked by ZMPSTE24 deficiency (2, 3). In the absence of ZMPSTE24, farnesyl–prelamin A accumulates in the cell nucleus, and the biogenesis of mature lamin A is completely abolished.
The accumulation of farnesyl–prelamin A in Zmpste24−/− mice is toxic, resulting in a variety of disease phenotypes (e.g., reduced growth, nonhealing bone fractures, sclerodermatous changes in the skin, and loss of adipose tissue) (2, 5). The extent of disease depends on the level of prelamin A expression. When farnesyl–prelamin A production in Zmpste24−/− mice is reduced by 50% (by introducing a single knockout allele for Lmna), disease phenotypes are completely abolished (5).
The loss of adipose tissue in Zmpste24−/− mice is profound, such that white adipose tissue (WAT) is nearly undetectable in both male and female Zmpste24−/− mice by ∼5 months of age (2, 5). However, the mechanism for the loss of adipose tissue has been unclear. One possibility is that the loss of adipose tissue is a direct consequence of farnesyl–prelamin A toxicity in adipocytes. Such a mechanism is plausible—for several reasons. Missense mutations in LMNA cause partial lipodystrophy in humans (6–8). Also, patients with mandibuloacral dysplasia type B, a disease resulting from loss-of-function mutations in ZMPSTE24, have reduced adipose tissue stores (9, 10). Finally, human immunodeficiency virus (HIV) protease inhibitors (HIV-PIs) that have been linked to the side effect of partial lipodystrophy (e.g., lopinavir) inhibit ZMPSTE24 in cultured fibroblasts, resulting in an accumulation of farnesyl–prelamin A (11, 12). Darunavir, an HIV-PI that is largely free of the lipodystrophy side effect, does not inhibit ZMPSTE24 or lead to an accumulation of farnesyl–prelamin A in fibroblasts (12). Despite these observations, there are ample reasons to be cautious about ascribing the loss of adipose tissue to the toxic effects of farnesyl–prelamin A. First, no one has actually tested the impact of farnesyl–prelamin A accumulation in adipocytes, and it is entirely conceivable that adipose tissue is resistant to the toxicity of farnesyl–prelamin A. For example, Zmpste24-deficient mice are free of liver disease despite a substantial expression of prelamin A in hepatocytes (2, 5). Also, Zmpste24−/− mice have nonhealing bone fractures, most prominently in the ribs and the zygomatic arch (2, 5), and it is conceivable that the loss of adipose tissue is secondary to these bone fractures (and reduced food intake) rather than being a direct result of farnesyl–prelamin A accumulation in adipose tissue.
In the current study, our goal was to determine whether the loss of adipose tissue is a direct consequence of ZMPSTE24 inactivation in adipocytes (and the resulting accumulation of farnesyl–prelamin A). To pursue this goal, we created a conditional knockout allele for Zmpste24 (Zmpste24fl) and used it to create mice lacking ZMPSTE24 specifically in adipocytes. To minimize the possibility of overlooking a small effect of farnesyl–prelamin A on adipocyte biology, we generated adipocyte-specific Zmpste24 knockout mice that were homozygous for the prelamin A–only Lmna (LmnaPLAO) allele (13, 14). Prelamin A production from the LmnaPLAO allele is approximately twice-normal; thus, we were able to examine whether an exaggerated accumulation of farnesyl–prelamin A in adipocytes alters adipose tissue stores in mice.
MATERIALS AND METHODS
A conditional knockout allele for Zmpste24
A recombineering-ready mouse genomic BAC library (library ID: CHORI-38) gridded on eleven 22 × 22 cm high-density nylon filters was screened for Zmpste24 with an oligonucleotide hybridization probe (GCTGACTATATTGCCCCTCTGTTTGACAAATTCACACCTC). Positive BACs were used to construct a targeting vector by λ-Red recombination with FRT-Cre and Gateway (Invitrogen) reactions. Exon 6 of Zmpste24 was “floxed” by introducing loxP sites on either side of exon 6 with three sequential recombineering reactions. First, Gateway attR1 and attR2 sites were introduced upstream of exon 6 between sequences GTTTCTCATATTCTAATCTGCTTCATCACAGACAATCATACAGGCAAGGG and TGAGAGATAAAGCAACAAACGCCACACAGTTCAGGTGGAAATAACCCTCA. Second, a floxed kanamycin-resistant cassette was introduced downstream of exon 6 between sequences GCCTGGCTCCTTTGGGTAATTGAATTGGACTCTTGAATGAATGATTAGCT and AAACAGTTTAATAATGTGAGGTGTCTGATGGTGCCACAGATTAGAGTCCC. Third, an ∼9.5 kb genomic fragment was introduced into a pBR-based plasmid flanked by attR3 and attR4 sites. The resultant clones were transformed into Cre-expressing bacteria to excise the floxed kanamycin-resistant cassette, leaving behind a single 3′ loxP site in intron 6. Gateway reactions were performed to introduce a 5′ loxP site (as a component of a β-galactosidase–neomycin trapping cassette) at the attR1 to attR2 sites and a diphtheria toxin fragment A negative-selection marker at the attR3 to attR4 sites. The targeting vector was sequenced, verified, linearized with AsiSI, and electroporated into mouse embryonic stem cells.
Mice
Zmpste24−/− and LmnaPLAO/PLAO mice have been described previously (1, 2, 14, 15). Transgenic adiponectin-Cre (Adipoq-Cre) (stock no. 028020) and ROSAmT/mG reporter (stock no. 007676) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The Adipoq-Cre mouse strain was genotyped by PCR with forward primer 5′-GGATGTGCCATGTGAGTCTG-3′ and reverse primer 5′-ACGGACAGAAGCATTTTCCA-3′ (yielding an ∼200 bp product). The ROSAmT/mG mouse strain was genotyped with a mutant forward primer 5′-TAGAGCTTGCGGAACCCTTC-3′, a wild-type forward primer 5′-AGGGAGCTGCAGTGGAGTAG-3′, and a common reverse primer 5′-CTTTAAGCCTGCCCAGAAGA-3′ (yielding a 128 bp product for the mutant allele and a 212 bp product for the wild-type allele). The Zmpste24fl allele was genotyped by PCR with forward primer 5′-GGTAGCCTGATGCCAAATCC-3′ and reverse primer 5′-CACACGGTTGAAAGGTAGAG-3′. The 550 bp product was incubated with NotI-HF (New England Biolabs #R3189L) at 37°C for 2 h. The mutant allele yields a 330 bp and 220 bp product. The PCR product from the wild-type allele is not cleaved by NotI. Mice were housed in a specific pathogen–free barrier facility with a 12 h light/dark cycle. The mice were fed pelleted mouse chow (NIH31) and water ad libitum, and nutritional food cups (ClearH2O, Westbrook, ME) as required for supportive care. All animal studies were approved by Univerisity of California, Los Angeles (UCLA)’s Animal Research Committee.
Echo-MRI
Body composition in live mice was measured using an EchoMRI 3-in-1 analyzer (EchoMRI Corp., Houston, TX), which assesses lean mass, fat mass, free water (mostly urine), and total water.
Plasma glucose, triglyceride, and free fatty acid levels
A blood sample (100 μl) was collected from anesthetized mice by retro-orbital puncture with a heparinized capillary tube (Kimble Chase). Plasma was separated from red blood cells by centrifugation (13,000 g for 30 s) and stored at −80°C until analysis. Plasma triglycerides (Sigma; TR0100), free fatty acids (Abcam; ab65341), and glucose (Cayman Chemical; 10009582) were measured according to kit instructions.
Measurement of macrophage content in WAT by fluorescence activated cell sorting
The stromal vascular fraction from gonadal WAT was prepared as described (16). Briefly, gonadal WAT was minced on ice, digested with collagenase type II (Worthington; LS004176) in PBS containing 0.5% BSA at 37°C and filtered through a 100 μm filter. After centrifugation at 300 g for 10 min at 4°C, the cell pellet was incubated with RBC lysis solution (Caprico Biotech), centrifuged, and the resuspended cell pellet filtered a second time through a 100 μm filter. The single-cell suspension was counted and immediately stained with fluorescently labeled antibodies. Anti-mouse CD16/CD32 antibody (Fc block) was added to the cells (1 μg/106 cells) and incubated on ice for 10 min. An FITC-labeled Cd11b antibody (eBioscience; #11-0112-41) and an APC-labeled F4/80 antibody (eBioscience; #17-4801-82) were added to the samples (1:100 dilution) and incubated in the dark at 4°C for 30 min. The cells were washed with 3 ml of fluorescence activated cell sorting (FACS) buffer (PBS containing 0.5% BSA and 0.2 mM EDTA) and centrifuged at 500 g for 5 min at 4°C. The cell pellets were resuspended in 300–400 μl of FACS buffer and stored at 4°C until analysis by FACS. Cells were stained with propidium iodide (Invitrogen; P3566) ∼15 min before FACS analysis.
Western blotting
Urea-soluble protein extracts from tissues were prepared as described (5, 17). Proteins were size fractionated on 4–12% gradient polyacrylamide Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membranes. The membranes were blocked with Odyssey blocking solution (LI-COR Bioscience, Lincoln, NE) for 1 h at room temperature and incubated with primary antibodies at 4°C overnight. Primary antibodies included anti-prelamin A (clone 3C8; 2 μg/ml) (18), anti-lamin A/C (Santa Cruz Biotech, SC376248; 1:1,500), and anti-tubulin (Novus Bio, NB600-506; 1:3,000) antibodies. After washing the membranes with PBS containing 0.2% Tween-20, they were incubated with infrared (IR) dye–labeled secondary antibodies at room temperature for 1 h. The IR signals were quantified with an Odyssey IR scanner (LI-COR Biosciences).
Immunofluorescence microscopy
Formaldehyde-fixed paraffin–embedded sections (4–6 μm thick) were deparaffinized and exposed to heat and pressure in R-Universal buffer using the Antigen-Retriever 2100 (Aptum Biologics, UK). Frozen sections (10 μm thick) were prepared from OCT-embedded–fixed tissues. The tissue sections were then processed for immunofluorescence microscopy (17, 19). Primary antibodies included a rat anti-prelamin A monoclonal antibody (clone 3C8; 12 μg/ml) (18), a goat anti-CD31 antibody (Novus, AF3628; 4 μg/ml), a rabbit anti-collagen IV antibody (Abcam, ab19808; 1:300), and a mouse anti-lamin A/C antibody (Santa Cruz Biotechnology, sc-375248; 1:500) labeled with Alexa 647 (Thermo Fisher). Secondary antibodies included Alexa 488-labeled donkey anti–rat IgG, Alexa 555-labeled donkey anti–goat IgG, and Alexa 555-labeled goat anti–rabbit IgG, all used at 1:1,000 dilution. Confocal fluorescence microscopy images were obtained with a Zeiss LSM700 laser–scanning microscope and processed by Zen 2010 software (Zeiss) to generate maximum image projections.
Histological studies
Mice were perfused in situ with PBS followed by fixative solution (3% paraformaldehyde in PBS). Excised tissues were stored in fixative for 1–3 days at 4°C and then embedded in paraffin. Tissue sections (4–6 μm thick) were prepared and stained with H&E or processed for immunohistochemical detection of F4/80 by UCLA’s Translational Pathology Core Laboratory. The stained sections were coded, and photographs captured on a Nikon E600 light microscope with 20× and 40× objectives using a Nikon DS-Fi2 camera operated by NIS Elements software (version 4.0). To quantify crown-like structures (CLSs), six representative images of H&E-stained sections were recorded for each sample by a microscopist blinded to genotype. Coded images were scored by three trained observers (the variation between observers was <9%). Numbers of CLSs in each photograph were counted and expressed relative to the total number of adipocytes examined.
Quantitative real time-PCR
Total RNA was isolated and treated with DNase I (Ambion, Life Technologies). RNA was reverse-transcribed with random primers, oligo(dT), and SuperScript III (Invitrogen). Quantitative PCR reactions were performed on a 7900HT Fast Real-Time PCR system (Applied Biosystems) with SYBR Green PCR Master Mix (Bioline, Taunton, MA). Transcript levels were determined with the comparative cycle threshold method and normalized to levels of cyclophilin A. Primer sequences are listed in supplemental Table S3.
Statistical analysis
Statistical analyses were performed with Microsoft Excel for Mac 2011. Unless indicated, experimental groups were analyzed by a two-tailed Student’s t-test. Groups were considered different at P < 0.05.
RESULTS
A conditional knockout allele for Zmpste24
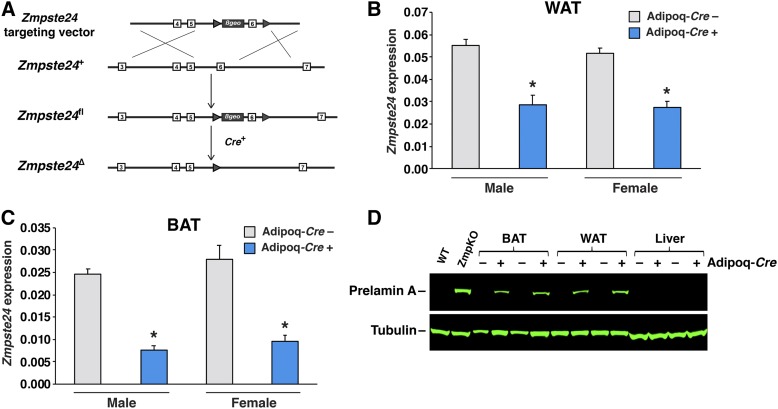
A gene-targeting vector to produce a conditional knockout allele for Zmpste24 (Zmpste24fl) was constructed by BAC recombineering (20–22). Exon 6 of Zmpste24, which encodes transmembrane domain 5 and part of ZMPSTE24’s protease domain (23), was flanked by loxP sites, and a β-galactosidase–neomycin (βgeo) cassette was introduced into intron 5 (Fig. 1A). Cre-mediated excision of exon 6 is predicted to yield a frameshift. Targeted mouse embryonic stem cells were identified by long-range PCR and used to generate chimeric mice, which were bred with C57BL/6 females to create Zmpste24fl/+ mice.
Fig. 1.
Creating adipocyte-specific Zmpste24 knockout mice. A: Strategy to create a conditional Zmpste24 knockout allele (Zmpste24fl). A gene-targeting vector was used to introduce a β-galactosidase–neomycin (βgeo) selection cassette into intron 5 and loxP sites (arrowheads) flanking exon 6 of Zmpste24, creating the Zmpste24fl allele. Exons are depicted as boxes. Cre-mediated excision of exon 6 creates a frameshift and inactivates ZMPSTE24. B, C: Quantitative RT-PCR studies of Zmpste24 expression in gonadal WAT (gWAT) and interscapular BAT (BAT) from male and female LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray) and littermate LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue) mice. Data were normalized to cyclophilin A. Data are expressed as mean ± SEM for 5–7 male mice and 4–5 female mice per genotype. *P < 0.01. D: Western blots of tissue extracts from LmnaPLAO/PLAOZmpste24fl/fl mice with and without the Adipoq-Cre transgene. Liver extracts from a Zmpste24+/+ (WT) and Zmpste24−/− (ZmpKO) mice were included as controls. Prelamin A was detected with a prelamin A–specific antibody (top). Tubulin was assessed as a loading control (bottom).
Adipocyte-specific Zmpste24 knockout mice
To inactivate Zmpste24 in adipocytes, we bred Zmpste24fl/fl mice harboring a Cre transgene driven by the adiponectin promoter (Adipoq-Cre) (24, 25). Quantitative RT-PCR studies revealed that the Adipoq-Cre transgene was expressed in adipose tissue but not in liver. Also, fluorescence microscopy studies on tissues of RosamT/mG transgenic mice (26) carrying the Adipoq-Cre revealed recombination in adipose tissue but not in kidney or peritoneal macrophages (supplemental Fig. S1A–C). In Zmpste24fl/flAdipoq-Cre+ mice, Zmpste24 transcript levels were reduced by ∼50% in WAT (Fig. 1B) and ∼70% in brown adipose tissue (BAT) (Fig. 1C), whereas transcript levels were not altered in liver and kidney and reduced by only 9% in peritoneal macrophages (supplemental Fig. S1D–F).
We were uncertain whether the levels of farnesyl–prelamin A accumulation in adipocytes of Zmpste24fl/flAdipoq-Cre+ mice would be sufficient to elicit disease phenotypes. For that reason, we bred Zmpste24fl/flAdipoq-Cre+ mice homozygous for the LmnaPLAO (14, 15). All of the output from the LmnaPLAO allele is channeled into prelamin A (rather than into both lamin C and prelamin A) (14, 15), resulting in an ∼2-fold increase in prelamin A expression. We showed previously that prelamin A in LmnaPLAO/PLAO mice is fully processed to mature lamin A and that twice-normal amounts of lamin A have no effect on the vitality of mice or body weight (14, 15). Also, the levels of farnesyl–prelamin A in Zmpste24−/−LmnaPLAO/PLAO mice are double those in Zmpste24−/−Lmna+/+ mice (15). Not surprisingly, the disease phenotypes in Zmpste24−/−LmnaPLAO/PLAO mice (e.g., reduced weight gain, bone fractures) occur earlier and are more severe than in Zmpste24−/−Lmna+/+ mice (15).
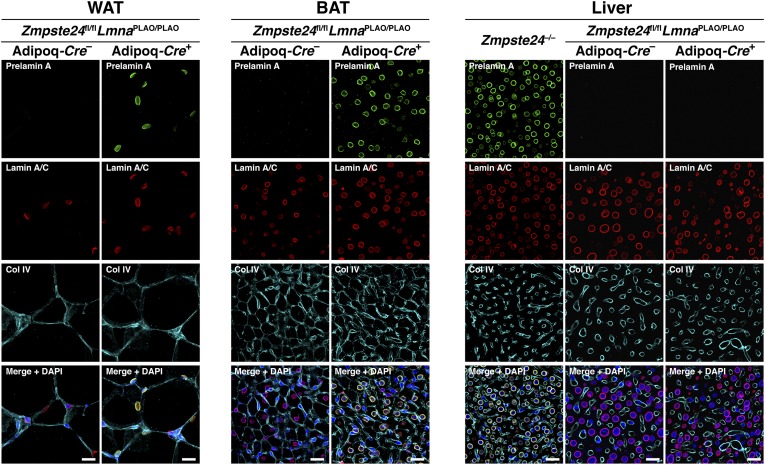
Western blots of BAT and WAT extracts from LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice revealed an accumulation of farnesyl–prelamin A (Fig. 1D). There was no prelamin A accumulation in the liver of these mice. The accumulation of prelamin A in the WAT and BAT of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice was located in adipocytes, as judged by immunohistochemistry (Fig. 2). As expected, the prelamin A was located at the nuclear rim (Fig. 2). No prelamin A accumulation was observed in littermate mice lacking the Adipoq-Cre transgene (Fig. 2). Also, no prelamin A was detected in the endothelial cells of adipose tissue (supplemental Fig. S2).
Fig. 2.
Prelamin A accumulates in adipocytes of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice, as judged by immunofluorescence microscopy. Tissue sections of gonadal WAT, BAT, and liver from a LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– mouse and a LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mouse were stained with antibodies against prelamin A (green), lamin A/C (red), and collagen IV (cyan). DNA was stained with DAPI (blue). Liver from a Zmpste24−/− mouse was included as a positive control for the prelamin A antibody. Images were recorded with a confocal microscope using a 20× objective and a 2× digital zoom. Identical microscope settings were used for all three tissue samples. Scale bar, 20 μm.
Reduced body weight and fat mass in male adipocyte-specific Zmpste24 knockout mice
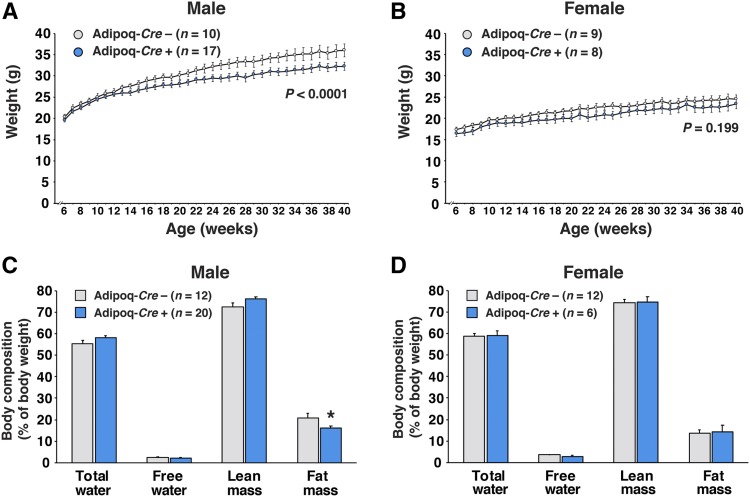
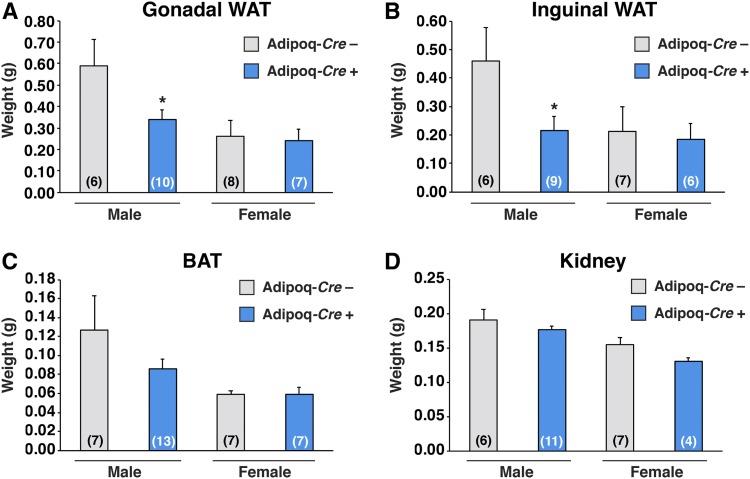
Body weights in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice were lower than in littermate controls lacking the Adipoq-Cre transgene (P < 0.0001); however, no significant changes were observed in the female mice (P = 0.199) (Fig. 3A, B). Reduced fat mass (as judged by Echo-MRI) was observed in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice (P < 0.05) but not in the female mice (Fig. 3C, D). Both gonadal and inguinal WAT fat pad weights were lower in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice than in littermates lacking the Adipoq-Cre transgene (P < 0.05) (Fig. 4A, B). There was a trend for lower BAT weights in male mice, but the difference did not achieve statistical significance (Fig. 4C) (P = 0.24). There were no differences in kidney weights (Fig. 4D). Fat pad weights in female LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice were no different than in littermate controls (Fig. 4A–C). The adipose tissue phenotypes in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice did not appear to be explained by changes in food consumption (supplemental Fig. S3).
Fig. 3.
Inactivating Zmpste24 expression in adipocytes leads to reduced body weight gain and reduced body fat mass in male mice. A, B: Body weight curves for male (A) and female (B) LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray circles) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue circles) mice. Body weights were measured weekly; graphs show the mean ± SEM. The weight curves were analyzed by repeated measures ANOVA. C, D: Whole-body composition was measured by Echo-MRI in male (C) and female (D) LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue) mice. Measurements of total water, free water, lean mass, and fat mass were expressed relative to body weight; graphs depict the mean ± SEM. *P < 0.05.
Fig. 4.
Inactivating Zmpste24 in adipocytes decreases adipose tissue weight in male mice. Gonadal WAT (A), inguinal WAT (B), BAT (C), and kidney tissue (D) from male and female LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue) mice were weighed. Graphs show the mean ± SEM. Numbers of mice per group are shown in parentheses. *P < 0.05.
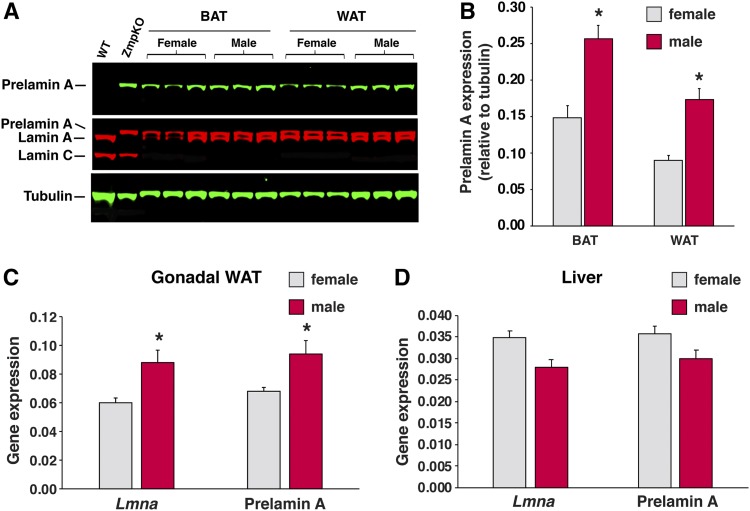
Because the severity of disease phenotypes in conventional Zmpste24-deficient mice depends on the level of prelamin A expression (5, 15), we hypothesized that the more prominent adipose tissue findings in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice might relate to greater amounts of farnesyl–prelamin A accumulation in adipose tissue. Indeed, as judged by Western blotting, the prelamin A levels in BAT and WAT extracts were ∼60% greater in male mice than in female mice (P < 0.02) (Fig. 5A, B). The higher prelamin A protein levels in the male mice are likely due to increased expression of the Lmna gene. Prelamin A transcripts in adipose tissue were 46% higher in male mice than in female mice (P < 0.05) (Fig. 5C). Prelamin A transcripts in the liver were similar in male and female mice (Fig. 5D). The modest decrease in adiposity in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice was not accompanied by perturbations in free fatty acid, triglyceride, or glucose levels (supplemental Fig. S4).
Fig. 5.
Levels of prelamin A protein and Lmna gene expression in WAT are higher in male mice than female mice. A: Western blots on tissue extracts from female and male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice. Tissue extracts from a Zmpste24+/+ (WT) and Zmpste24−/− (ZmpKO) mice were included as controls. Prelamin A was detected with a prelamin A–specific antibody (green, top). Lamin A, lamin C, and prelamin A were detected with a lamin A/C antibody (red, middle). Tubulin was measured as a loading control (green, bottom). B: Quantification of prelamin A expression, normalized to tubulin, in the Western blots shown in panel A. Graph shows the mean ± SEM; n = 3 mice per group). *P < 0.02. C, D: Quantitative RT-PCR studies of total Lmna expression (prelamin A and lamin C) and prelamin A expression in gonadal WAT (C) and liver (D) tissue from female (gray) and male (red) LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– mice. Data were normalized to cyclophilin A. Data show the mean ± SEM for 9 female mice and 12 male mice. *P < 0.05.
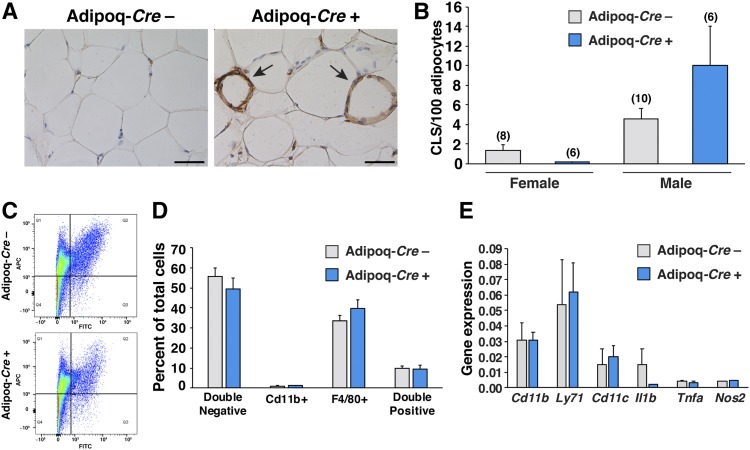
We examined gene expression related to adipocyte differentiation, triglyceride metabolism, extracellular matrix synthesis, and the p53 pathway in the adipose tissue of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice and littermate controls, but we did not find significant changes (supplemental Tables S1, S2). We also quantified CLSs (macrophages in the process of engulfing dead or dying adipocytes) in the adipose tissue of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice (Fig. 6A). CLSs were more numerous in male mice than in female mice (P < 0.05), but we found no differences in CLSs in the adipose tissue of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice and littermate controls lacking the Adipoq-Cre transgene (P = 0.11) (Fig. 6B). We also performed FACS analysis on the stromal vascular fraction of adipose tissue from 40-week-old male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice and littermate controls, with the goal of quantifying numbers of macrophages (CD11b and F4/80 double-positive cells) (Fig. 6C); however, we found no differences in the macrophage content of adipose tissue in the two groups of mice (Fig. 6D). Consistent with these findings, we did not observe changes in the expression of macrophage-related genes (Cd11b, Ly71, Cd11c, Il1b) or changes in the expression of genes reflecting macrophage activation state (Tnfa, Nos2) in LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice and littermate controls (Fig. 6E).
Fig. 6.
Macrophage content in gonadal WAT of adipocyte-specific Zmpste24 knockout mice. A: Images of sections of gonadal WAT, stained with antibodies against F4/80, from a male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– mouse (left) and a littermate male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mouse (right). Antibody binding was detected by peroxidase staining (brown) and identified macrophages surrounding adipocytes (arrows). Scale bar, 50 μm. B: CLSs in gonadal WAT were quantified in H&E–stained paraffin-embedded tissue sections. Photographs of tissue sections (three fields per tissue) were recorded by an individual blinded to genotype, and the number of CLSs counted by three trained observers. The average score for each mouse was calculated and expressed relative to the total number of adipocytes. The mean ± SEM are plotted. Numbers of mice per group are shown in parentheses. C: The stromal vascular fraction from gonadal WAT was isolated by differential centrifugation and analyzed by FACS after staining with APC-labeled anti-F4/80 and FITC-labeled anti-CD11b antibodies. Representative distributions for F4/80 positive (Q1), F4/80 and CD11b double positive (Q2), CD11b positive (Q3), and double negative (Q4) cells are shown for a LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (top) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (bottom) mice. A total of 5,000 single cell events were collected for each experiment. D: The number of double negative, CD11b positive, F4/80 positive, and F4/80 and CD11b double positive cells as a percentage of total cells analyzed are plotted for LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue) male mice. The mean ± SEM is shown for Adipoq-Cre– mice (n = 8) and Adipoq-Cre+ mice (n = 4). E: Quantitative RT-PCR studies of Cd11b, Ly71, Cd11c, Ifng, Il1b, Tnf, and Nos2 expression in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre– (gray) and LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ (blue) mice. The mean ± SEM is shown for Adipoq-Cre– mice (n = 5) and Adipoq-Cre+ (n = 7) mice.
DISCUSSION
We used a newly developed Zmpste24 conditional knockout allele and the Adipoq-Cre transgene to create adipocyte-specific Zmpste24 knockout mice. Our goal was to examine the impact of farnesyl–prelamin A accumulation in adipocytes. We created adipocyte-specific Zmpste24 knockout mice that were homozygous for the LmnaPLAO allele, reasoning that twice-normal amounts of prelamin A expression would make farnesyl–prelamin A toxicity more pronounced and easier to detect.
Our a priori expectation was that we would encounter substantial loss of adipose tissue in LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice. This expectation was based on the fact that adipose tissue is virtually undetectable in 4–5-month-old conventional Zmpste24 knockout mice (2, 5) and on the fact that the levels of farnesyl–prelamin A accumulation in our mouse model would be twice those in conventional Zmpste24 knockout mice. However, despite an exaggerated accumulation of farnesyl–prelamin A expression in adipocytes, the body weight, body fat content, and adipose tissue depot weights were normal in female LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice. In male mice, body weight, body fat content, and adipose tissue depot weights were lower but the differences were modest—never approaching the striking loss of adipose tissue observed in conventional Zmpste24 knockout mice. We did not identify significant changes in p53-related gene expression (27) in the adipose tissue of LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice, nor did we find more CLSs in LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice than in littermate mice lacking the Adipoq-Cre transgene. Thus, the impact of ZMPSTE24 deficiency in adipocytes (and the accompanying accumulation of farnesyl–prelamin A) is modest. Adipocytes, like hepatocytes in the liver, do not appear to be particularly sensitive to farnesyl–prelamin A toxicity. In view of these findings, it seems likely that the profound loss of adipose tissue in conventional Zmpste24−/− mice is secondary to disease in other tissues [e.g., the result of poor nutrition as a result of nonhealing bone fractures of the zygomatic arch (5)] or the inhibition of ZMPSTE24 in cell types important for adipose development (e.g., adipocyte stem cells).
We were initially puzzled by the fact that changes in adipose tissue mass were evident only in male LmnaPLAO/PLAOZmpste24fl/flAdipoq-Cre+ mice, but we uncovered a likely explanation. The expression of prelamin A transcripts was ∼40% higher in male mice than in female mice, and by Western blotting, the level of prelamin A accumulation in adipose tissue was ∼60% higher in male mice. These 40–60% differences are obviously not enormous, but it is important to note that modest differences in farnesyl–prelamin A accumulation have a huge effect on disease phenotypes. Reducing prelamin A expression levels by 50% in Zmpste24−/− mice completely eliminates disease phenotypes (5), whereas doubling prelamin A expression levels with the LmnaPLAO allele markedly increased the severity of disease (15). However, we cannot exclude the contribution of other mechanisms. For example, differences in genetic background have been suggested to explain the more severe lipodystrophy in male R482Q-lamin A transgenic mice (28), whereas androgen synthesis has been suggested to account for the earlier onset of cardiomyopathy in male LmnaH222P/H222P mice (29).
The fact that a deficiency of ZMPSTE24 in adipocytes and the accompanying accumulation of farnesyl–prelamin A did not induce lipodystrophy and lipodystrophy-related metabolic abnormalities will likely raise doubts about the relevance of ZMPSTE24 inhibition to the lipodystrophy observed in patients treated with HIV-PIs (e.g., lopinavir). The fact that therapeutic concentrations of lopinavir bind to ZMPSTE24 (30) and inhibit ZMPSTE24 activity in cultured fibroblasts is well documented (11, 12, 31), but it is important to note that the level of inhibition is far from complete. Even in the presence of high levels of lopinavir, more than half of the prelamin A in fibroblasts is cleaved by ZMPSTE24 and processed to mature lamin A (11, 12). Also, it is not clear that the lopinavir-induced accumulation of farnesyl–prelamin A observed in cultured fibroblasts occurs in the tissues of patients. In one study (32), prelamin A was detected by Western blot in the adipose tissue of patients undergoing treatment for HIV, but the amount of prelamin A, relative to mature lamin A, was extremely low. Another study failed to detect any prelamin A in leukocytes from HIV-PI–treated patients (33). Those observations, combined with the findings in the current studies (i.e., modest effects of complete ZMPSTE24 inactivation on adipocyte biology), it seems unlikely that the lipodystrophy in HIV-PI–treated patients can be ascribed to drug-induced inhibition of ZMPSTE24 or an accumulation of farnesyl–prelamin A.
Supplementary Material
Acknowledgments
The authors acknowledge the assistance of the Translational Pathology Core Laboratory and FACS cores at UCLA.
Footnotes
Abbreviations:
- BAT
- brown adipose tissue
- CLS
- crown-like structure
- FACS
- fluorescence activated cell sorting
- HIV
- human immunodeficiency virus
- HIV-PI
- human immunodeficiency virus protease inhibitor
- IR
- infrared
- LmnaPLAO
- prelamin A–only Lmna
- UCLA
- University of California, Los Angeles
- WAT
- white adipose tissue
- ZMPSTE24
- zinc metallopeptidase STE24
This work was supported by the National Institutes of Health Grants AG047192 (to L.G.F.), UL1TR001881 (to N.J.), and HL126551 (to S.G.Y.), and the National Institutes of Health Ruth L. Kirschstein National Research Service Award T32GM065823 (to N.Y.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Leung G. K., Schmidt W. K., Bergo M. O., Gavino B., Wong D. H., Tam A., Ashby M. N., Michaelis S., and Young S. G.. 2001. Biochemical studies of Zmpste24-deficient mice. J. Biol. Chem. 276: 29051–29058. [DOI] [PubMed] [Google Scholar]
- 2.Bergo M. O., Gavino B., Ross J., Schmidt W. K., Hong C., Kendall L. V., Mohr A., Meta M., Genant H., Jiang Y., et al. 2002. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc. Natl. Acad. Sci. USA. 99: 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pendás A. M., Zhou Z., Cadiñanos J., Freije J. M. P., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodríguez F., Tryggvason K., et al. 2002. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice. Nat. Genet. 31: 94–99. [DOI] [PubMed] [Google Scholar]
- 4.Young S. G., Fong L. G., and Michaelis S.. 2005. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—New evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 46: 2531–2558. [DOI] [PubMed] [Google Scholar]
- 5.Fong L. G., Ng J. K., Meta M., Cote N., Yang S. H., Stewart C. L., Sullivan T., Burghardt A., Majumdar S., Reue K., et al. 2004. Heterozygosity for Lmna deficiency eliminates the progeria-like phenotypes in Zmpste24-deficient mice. Proc. Natl. Acad. Sci. USA. 101: 18111–18116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao H., and Hegele R. A.. 2000. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 9: 109–112. [DOI] [PubMed] [Google Scholar]
- 7.Shackleton S., Lloyd D. J., Jackson S. N., Evans R., Niermeijer M. F., Singh B. M., Schmidt H., Brabant G., Kumar S., Durrington P. N., et al. 2000. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 24: 153–156. [DOI] [PubMed] [Google Scholar]
- 8.Speckman R. A., Garg A., Du F., Bennett L., Veile R., Arioglu E., Taylor S. I., Lovett M., and Bowcock A. M.. 2000. Mutational and haplotype analyses of families with familial partial lipodystrophy (Dunnigan variety) reveal recurrent missense mutations in the globular C-terminal domain of lamin A/C. Am. J. Hum. Genet. 66: 1192–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal A. K., Fryns J-P., Auchus R. J., and Garg A.. 2003. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum. Mol. Genet. 12: 1995–2001. [DOI] [PubMed] [Google Scholar]
- 10.Barrowman J., Wiley P. A., Hudon-Miller S. E., Hrycyna C. A., and Michaelis S.. 2012. Human ZMPSTE24 disease mutations: residual proteolytic activity correlates with disease severity. Hum. Mol. Genet. 21: 4084–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffinier C., Hudon S. E., Farber E. A., Chang S. Y., Hrycyna C. A., Young S. G., and Fong L. G.. 2007. HIV protease inhibitors block the zinc metalloproteinase ZMPSTE24 and lead to an accumulation of prelamin A in cells. Proc. Natl. Acad. Sci. USA. 104: 13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffinier C., Hudon S. E., Lee R., Farber E. A., Nobumori C., Miner J. H., Andres D. A., Spielmann H. P., Hrycyna C. A., Fong L. G., et al. 2008. A potent HIV protease inhibitor, darunavir, does not inhibit ZMPSTE24 or lead to an accumulation of farnesyl-prelamin A in cells. J. Biol. Chem. 283: 9797–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies B. S., Coffinier C., Yang S. H., Barnes R. H. II, Jung H. J., Young S. G., and Fong L. G.. 2011. Investigating the purpose of prelamin A processing. Nucleus. 2: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffinier C., Jung H. J., Li Z., Nobumori C., Yun U. J., Farber E. A., Davies B. S., Weinstein M. M., Yang S. H., Lammerding J., et al. 2010. Direct synthesis of lamin A, bypassing prelamin A processing, causes misshapen nuclei in fibroblasts but no detectable pathology in mice. J. Biol. Chem. 285: 20818–20826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies B. S., Barnes R. H. II, Tu Y., Ren S., Andres D. A., Spielmann H. P., Lammerding J., Wang Y., Young S. G., and Fong L. G.. 2010. An accumulation of non-farnesylated prelamin A causes cardiomyopathy but not progeria. Hum. Mol. Genet. 19: 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr J. S., Kennedy A. J., and Hasty A. H.. 2013. Isolation of adipose tissue immune cells. J. Vis. Exp. 75: e50707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim P. H., Luu J., Heizer P., Tu Y., Weston T. A., Chen N., Lim C., Li R. L., Lin P. Y., Dunn J. C. Y., et al. 2018. Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci. Transl. Med. 10: eaat7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee R., Chang S. Y., Trinh H., Tu Y., White A. C., Davies B. S., Bergo M. O., Fong L. G., Lowry W. E., and Young S. G.. 2010. Genetic studies on the functional relevance of the protein prenyltransferases in skin keratinocytes. Hum. Mol. Genet. 19: 1603–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies B. S. J., Beigneux A. P., Barnes R. H. II, Tu Y., Gin P., Weinstein M. M., Nobumori C., Nyrén R., Goldberg I. J., Olivecrona G., et al. 2010. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., Iyer V., Mujica A. O., Thomas M., Harrow J., Cox T., et al. 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S. H., Chang S. Y., Yin L., Tu Y., Hu Y., Yoshinaga Y., de Jong P. J., Fong L. G., and Young S. G.. 2011. An absence of both lamin B1 and lamin B2 in keratinocytes has no effect on cell proliferation or the development of skin and hair. Hum. Mol. Genet. 20: 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park C. Y., Jeker L. T., Carver-Moore K., Oh A., Liu H. J., Cameron R., Richards H., Li Z., Adler D., Yoshinaga Y., et al. 2012. A resource for the conditional ablation of microRNAs in the mouse. Cell Reports. 1: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quigley A., Dong Y. Y., Pike A. C., Dong L., Shrestha L., Berridge G., Stansfeld P. J., Sansom M. S., Edwards A. M., Bountra C., et al. 2013. The structural basis of ZMPSTE24-dependent laminopathies. Science. 339: 1604–1607. [DOI] [PubMed] [Google Scholar]
- 24.Wang F., Mullican S. E., DiSpirito J. R., Peed L. C., and Lazar M. A.. 2013. Lipoatrophy and severe metabolic disturbance in mice with fat-specific deletion of PPARgamma. Proc. Natl. Acad. Sci. USA. 110: 18656–18661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K. Y., Russell S. J., Ussar S., Boucher J., Vernochet C., Mori M. A., Smyth G., Rourk M., Cederquist C., Rosen E. D., et al. 2013. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 62: 864–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muzumdar M. D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45: 593–605. [DOI] [PubMed] [Google Scholar]
- 27.Varela I., Cadinanos J., Pendas A. M., Gutierrez-Fernandez A., Folgueras A. R., Sanchez L. M., Zhou Z., Rodriguez F. J., Stewart C. L., Vega J. A., et al. 2005. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 437: 564–568. [DOI] [PubMed] [Google Scholar]
- 28.Wojtanik K. M., Edgemon K., Viswanadha S., Lindsey B., Haluzik M., Chen W., Poy G., Reitman M., and Londos C.. 2009. The role of LMNA in adipose: a novel mouse model of lipodystrophy based on the Dunnigan-type familial partial lipodystrophy mutation. J. Lipid Res. 50: 1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimura T., Onoue K., Takahashi-Tanaka Y., Ishikawa T., Kuwahara M., Setou M., Shigenobu S., Yamaguchi K., Bertrand A. T., Machida N., et al. 2013. Nuclear accumulation of androgen receptor in gender difference of dilated cardiomyopathy due to lamin A/C mutations. Cardiovasc. Res. 99: 382–394. [DOI] [PubMed] [Google Scholar]
- 30.Mehmood S., Marcoux J., Gault J., Quigley A., Michaelis S., Young S. G., Carpenter E. P., and Robinson C. V.. 2016. Mass spectrometry captures off-target drug binding and provides mechanistic insights into the human metalloprotease ZMPSTE24. Nat. Chem. 8: 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudon S. E., Coffinier C., Michaelis S., Fong L. G., Young S. G., and Hrycyna C. A.. 2008. HIV-protease inhibitors block the enzymatic activity of purified Ste24p. Biochem. Biophys. Res. Commun. 374: 365–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caron M., Auclair M., Donadille B., Bereziat V., Guerci B., Laville M., Narbonne H., Bodemer C., Lascols O., Capeau J., et al. 2007. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 14: 1759–1767. [DOI] [PubMed] [Google Scholar]
- 33.Perrin S., Cremer J., Faucher O., Reynes J., Dellamonica P., Micallef J., Solas C., Lacarelle B., Stretti C., Kaspi E., et al. 2012. HIV protease inhibitors do not cause the accumulation of prelamin A in PBMCs from patients receiving first line therapy: the ANRS EP45 “aging” study. PLoS One. 7: e53035 [Erratum. 2013. PLoS One. 8: doi:10.1371/annotation/f02cdfd3-b271-46fd-a78f-5a030f0b416d.] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.