Abstract
Scutellarein has been identified to serve an anti-tumor function in human colon cancer, but the underlying mechanisms remain largely unclear. The present study further investigated the effect and mechanism of scutellarein, extracted from wild chrysanthemum, in the progression of colon cancer. MTT, clone formation, flow cytometry and tumor-bearing mice assays were used to detect cell viability, clone formation, apoptosis and tumorigenesis, respectively. Western blot and quantitative PCR assays were performed for protein and mRNA expression detection. The results revealed that, compared with the control group, scutellarein treatment significantly inhibited the viability and induced the apoptosis of colon cancer cells (P<0.05), with significant decreases in receptor for advanced glycosylation end products (RAGE) protein expression and stability and an increase in RAGE ubiquitination (P<0.05). However, the effects of scutellarein exerted in cell apoptosis and viability were rescued by RAGE overexpression, and accelerated by RAGE knockdown. Additionally, it was observed that scutellarein treatment induced a significant increase in the expression of cell division control protein 4 (CDC4) compared with the control group (P<0.05), which was then verified to interact with RAGE protein and mediate its ubiquitination. Overexpression of CDC4 inhibited colon cancer cell viability and promoted the apoptosis of SW480 and T84 cells, whereas this function was weakened when RAGE was overexpressed. Furthermore, CDC4 downregulation significantly neutralized scutellarein functions in promoting cell apoptosis and inhibiting cell viability and tumorigenesis in colon cancer cells compared with the scutellarein group (P<0.05). In conclusion, the present study revealed that scutellarein inhibited the development of colon cancer through upregulating CDC4-mediated RAGE ubiquitination.
Keywords: scutellarein, wild chrysanthemum, colon cancer, cell division control protein 4, receptor for advanced glycosylation end products, ubiquitination
Introduction
Colon cancer is one of the most common malignant alimentary canal tumor types, and mainly occurs on the joint of the rectum and colon sigmoideum (1). A total of 1.2 million patients are diagnosed annually and ~0.6 million mortalities occur each year globally (1). Nearly 60% patients with colon cancer will develop metastasis, which results in a high mortality of patients with colon cancer (2,3). Colon cancer is the fifth highest cause of cancer-associated mortalities in China, and the morbidity continues to rapidly rise, with a 5-year survival rate of 40-60% (4). Therefore, it is important to further reveal the underlying mechanisms of colon cancer.
An increasing number of herbs have been used in medicines and cosmetics for centuries. Among them, wild chrysanthemum (Chrysanthemum indicum L.) has a range of biological activities. In China, wild chrysanthemum is used to cure numerous types of inflammatory diseases (5). Studies have documented that flavonoids are the active ingredients in wild chrysanthemum, including hispidulin, isorhoifolin and scutellarein (6-8). Scutellarein, a flavone present in the perennial herb Scutellaria baicalensis, is the aglycone of scutellarin with a free hydroxyl in 7 position, and exerts a higher bioavailability compared with scutellarin (9). A variety of studies have demonstrated that scutellarein may inhibit the viability of human lung cancer cells and fibrosarcoma cells (9-11), in addition to serving an anti-tumor function in human colon cancer (12,13). However, the underlying mechanisms of scutellarin, derived from the wild chrysanthemum, in the repression of colon cancer progression remain largely unknown.
Cluster of differentiation (CD) family proteins, including CD44, CD50 and CD74, are all members of the immuno-globulin superfamily, and are associated with cancer cell migration and immune response (14,15). Melanoma cell adhesion molecule (MCAM), also known as CD146, is a cell adhesion molecule and is involved in several cellular processes, including cell invasion, migration, immune response and signal transduction (16). The increased expression of MCAM in the primary tumor types is associated with metastasis and prognosis in several cancer types (16-18). Mucosal addressin cell adhesion molecule 1 (Madcam1) is known as a type of adhesion molecule, which is involved in inflammatory responses with no expression in the majority of normal tissues (19,20). Receptor for advanced glycation end products (RAGE) is a signal transduction receptor that is involved in multiple pathologic conditions, including carcinogenesis and inflammation relying on its diverse ligands (21,22). It is well acknowledged that RAGE expression is associated with multifarious cancer types, including colon cancer (23). These results suggest that all of the aforementioned proteins serve a function in carcinogenesis. However, whether scutellarein inhibits the development of colon cancer through modulating these oncoproteins (CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206, RAGE and Madcam1) is not yet known.
Ubiquitylation is considered to be an important modification of nuclear and cytoplasmic proteins at the post-translational level, as it is known that ubiquitylation serves an important function in cell apoptosis, cell cycle and DNA damage repair (24,25). A number of studies have demonstrated that ubiquitylation is strongly associated with the initiation and progression of carcinogenesis (26,27). Notably, numerous ubiquitin-associated proteins have been revealed as tumor suppressors/oncogenes and therapeutic targets in different tumor types (28,29). However, whether ubiquitylation is involved in scutellarein-mediated colon cancer repression remains unknown.
The present study aimed to investigate the function of scutellarein in ubiquitylation, and to determine the expression of a number of ubiquitin-associated proteins in the presence of scutellarein. In order to elucidate the functions of scutellarein for the potential treatment of colon cancer, the anticancer function of scutellarein from wild chrysanthemum in the development of colon cancer cells and its associated mechanisms were examined.
Materials and methods
Extraction of scutellarein
A whole plant of wild chrysanthemum was gathered from the wild, dried in the air and then ground, followed by extraction using 99.5% ethanol three times. Subsequently, the ethanol was removed and the extract was suspended in water and partitioned with petroleum ether and acetyl acetate. Under reduced pressure, the acetyl acetate was evaporated and obtained. This was handled with column chromatography on silica gel eluted with gradient dichloromethane and purified by Sephadex LH-20 to obtain a yellow powder. The structure of the compound was determined using 1H and 13C nuclear magnetic resonance (NMR) spectra and electrospray ionization mass spectrometry analysis. The results revealed the peaks to be at m/z 287[M+H]+ and 285[M−H]−, demonstrating that the molecular weight was 286 g/mol. On the other hand, two single peaks at δ 6.77 (1H, s) and 6.57 (1H, s) were observed on the 1H NMR spectrum. Furthermore, on the 13C NMR spectrum, 13 carbon signals were detected. The purity of the scutellarein was identified by using high performance liquid chromatography on an Agilent 1,100 high performance liquid chromatography system (Agilent Technologies, Inc.), and was determined to be 96.5%. The chromatographic column was Waters Xterra Rp-C18 (3.0×100 mm; 3.5 µm), the mobile phase A was acetonitrile solution, and the mobile phase B was 0.1% formic acid water. The gradient elution conditions were 10-20% A (0-10 min), 20-25% A (10-30 min) and 25-45% A (30-60 min), and the volume flow rate was 0.4 ml/min with a column temperature of 20°C and an injection volume of 5 µl. The UV detection wavelength was 275 nm, and the running time was 60 min.
Cell culture
Colon cell lines CL-40, SW480 and T84 were all obtained from the American Type Culture Collection. T84 cells were maintained in high glucose-DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) with the presence of 10% fetal bovine serum (FBS; Biological Industries) and 1% (v/v) penicillin-streptomycin, while SW480 cells were cultured in DMEM/F-12 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin, and CL-40 cells were incubated with RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% penicillin-streptomycin. All cells were maintained in a humidified condition of 5% CO2 at 37°C.
MTT analysis
MTT analysis was performed to investigate the half maximal inhibitory concentration (IC50) of scutellarein in colon cancer cells and cell viability. SW480 and T84 cells (3,000 cells/well) were cultured in 96-well plates for 24 h following adherence, and were then exposed to scutellarein at a gradient concentration for 36 h. Subsequently, 10 µl MTT solution (Beyotime Institute of Biotechnology) were added per well and the cells were incubated for another 4 h at 37°C. DMSO was used to dissolve the purple formazan. Then, the optical density (OD) values were measured at 570 nm, and the IC50 value was calculated. In addition, cell viability was also detected using MTT. Cells were cultured in 96-well plates, and underwent different treatments, including scutellarein, scutellarein+RAGE overexpression (OE-RAGE), scutellarein+RAGE short hairpin RNA (sh-RAGE), OE-CDC4, OE-CDC4+OE-RAGE, following adherence. Following incubation at 37°C for 1, 2, 3, 4 and 5 days, the cells were treated with MTT solution for 4 h, and the OD values were measured at 570 nm.
Western blot analysis
The colon cancer SW480 and T84 cells were seeded into 6-well plates and cultured for 12 h until adherence. Then, cells underwent different treatments (scutellarein, scutellarein+OE-RAGE, scutellarein+sh-RAGE, OE-CDC4 and OE-CDC4+OE-RAGE) for 48 h, and then the cells were gathered. The total protein was isolated using RIPAIII (Beijing Solarbio Science & Technology Co., Ltd.) and centrifuged at 12,000 × g at 4°C for 20 min. Proteins from supernatants were quantified using bicinchoninic acid analysis (EMD Millipore) and boiled with 1x loading buffer. A total of 20 mg protein was separated using 10% SDS-PAGE, and was subsequently transferred onto polyvinylidene fluoride membranes (EMD Millipore). The membranes were immersed into 5% non-fat milk for 1 h at room temperature to block, following by being washed with TBS with 0.1% (v/v) Tween-20 a total of 3 times. Then, the membranes were incubated with primary antibodies at 4°C overnight. Next day, the membranes were incubated with the corresponding horseradish peroxidase (HRP)-conjugated anti-mouse (cat. no. 7076) or anti-rabbit (cat. no. 7074) secondary antibody (1:10,000 dilution; Cell Signaling Technology, Inc.) at room temperature for 2 h. Subsequently, the complexes were tested using the electrochemical luminescence of horseradish peroxidase substrate (EMD Millipore) and analyzed using ImageJ software (version 1.48; National Institutes of Health). The primary antibodies were as follows: Anti-CD44 (1:1,000; cat. no. #3578), anti-ring finger and CHY zinc finger domain containing 1 (RCHY1; 1:1,000; cat. no. #5754), anti-cleaved caspase3 (1:1,000; cat. no. #9661), anti-caspase3 (1:1,000; cat. no. #9662), anti-cleaved caspase7 (1:1,000; cat. no. #9492), anti-caspase7 (1:1,000, no. #9491), anti-phosphorylated (p-) p65 (1:1,000; cat. no. #3033), anti-p65 (1:1,000; cat no. #8242) and anti-protein kinase C (PKC; 1:1,000; cat. no. #2056) antibodies were all obtained from Cell Signaling Technology, Inc. Anti-RAGE (1:1,000; cat. no. sc365154), anti-cell division control protein 4 (CDC4; 1:1,000; cat. no. sc293423), anti-CD50 (1:1,000; cat. no. sc71307), anti-CD74 (1:1,000; cat. no. sc166047), anti-CD138 (1:1,000; cat. no. sc390791), anti-CD151 (1:1,000; cat. no. sc271216), anti-CD166 (1:1,000; cat. no. sc74558), anti-CD206 (1:1,000; cat. no. sc70586), anti-MCAM (1:1,000; cat. no. sc376762), anti-Madcam1 (1:1,000; cat. no. sc374398), anti-Cbl proto-oncogene (CBL; 1:1,000; cat. no. sc1651), anti-Ubiquitin (1:1,000; cat. no. sc166553), anti-SMAD specific ubiquitin protein ligase 1 (Smurf1; 1:1,000; cat. no. sc100616), anti-MDM2 proto-oncogene (MDM2; 1:1,000; cat. no. sc5304) and anti-GAPDH (1:1,000; cat. no. sc47724) were all purchased from Santa Cruz Biotechnology, Inc., and anti-vascular endothelial growth factor (VEGF; 1:1,000; cat. no. ab46154) was purchased from Abcam. The corresponding secondary antibodies were obtained from OriGene Technologies, Inc.
Cycloheximide (CHX) analysis
The colon cancer SW480 and T84 cells were routinely cultured and underwent different treatments. Then, the cells were treated with CHX (100 µg/ml) for 1, 2, 4, 8 and 24 h at 37°C, following which the total protein was extracted.
Colony formation assay
Colon cancer SW480 and T84 cells (1,000 cells/dish) were seeded in 3.5 cm dishes and cultured in a humidified environment with 5% CO2 at 37°C for 24 h. Then, different concentrations of scutellarein (10, 20, 40, 60 and 80 µM) were added into each dish and the cells were incubated with scutellarein at 37°C for 15 days. Cells treated with DMSO were used as the negative control. Subsequent to the incubation at 37°C for 15 days, the cells were washed with phosphate buffered saline (PBS) 3 times, following by staining with 1 ml crystal violet solution for 10 min at room temperature. The cells were then washed with PBS prior to the colonies being counted.
Reverse transcription-quantitative PCR (RT-qPCR)
Colon cancer SW480 and T84 cells with different treatments were obtained and mRNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Subsequently, reverse transcription (42°C for 30 min followed by 85°C for 5 min) was performed using EasyScript Reverse Transcriptase (Beijing Transgen Biotech Co., Ltd.) followed by qPCR on a DA7600 Real-time Nucleic Acid Amplification Fluorescence Detection System (Bio-Rad Laboratories, Inc.) in a 25 µl volume system using the TransStart Green qPCR SuperMix (Beijing Transgen Biotech Co., Ltd.). The thermocycling conditions were as follows: Initial denaturation for 3 min at 95°C, followed by 35 cycles of denaturation at 95°C for 10 sec and annealing/extension at 55°C for 30 sec.
The primers were obtained from Sangon Biotech Co., Ltd. Melting-curve analysis was conducted to recognize the reaction specificity. All experiments were performed in triplicate and the data were analyzed using the 2−ΔΔCq method (30). Primers of RAGE, CDC4 and β-catenin were as follows: RAGE forward, 5′-GGG GTA CCA AGG AAG CAG GTA GGC AGC C-3′ and reverse, 5′-CCG CTC GAA TCC ATT CCT GTT CAT CTG C-3′; CDC4 forward, 5′-GTG GGA CAT ACA GGT GGA-3′ and reverse, 5′-CAA CGC ACA GTG GAA CTA-3′; β-actin forward, 5′-CAT GTA CGT TGCT ATC CAG GC-3′ and reverse, 5′-CTC CTT AAT GTC ACG CAC GAT-3′.
Immunoprecipitation (IP)
An IP assay was performed using protein G plus A-agarose beads as established, with CDC4 or RAGE antibodies obtained from Santa Cruz Biotechnology, Inc. In brief, SW480 and T84 cells were first rinsed with cold PBS and lysed in IP lysis buffer (Thermo Fisher Scientific, Inc.); the total proteins were obtained after being centrifuged at 20,000 × g for 30 min at 4°C and served as the 'Input' sample. Then, the cell lysate containing 200 µg protein was incubated with Dynabeads® protein G (Thermo Fisher Scientific, Inc.) for 1 h at room temperature, and incubated with 2 µg of antibody against RAGE (cat. no. sc365154; Santa Cruz Biotechnology, Inc.) or IgG (cat. no. 5946, Cell Signaling Technology, Inc.; negative control) overnight at 4°C, followed by incubation with Dynabeads® protein G for another 1 h at room temperature to form the immune complex. Then, the protein samples were loaded onto gels for western blot analysis.
Cell transfection
T84 and SW480 cells (3×105 cells/well) were seeded into 6-well plates and cultured with antibiotic-free standard growth medium for 24 h until the confluence reached 60-80%. Subsequently, the cells were infected with the following lentivirus vectors using 7 µg/ml polybrene (Hanbio Biotechnology Co., Ltd.), including RAGE shRNA lentivirus vector (cat. no. sc-36374-SH; Santa Cruz Biotechnology, Inc.), CDC4 shRNA lentivirus vector (sh-CDC4; cat. no. sc-37547-SH; Santa Cruz Biotechnology, Inc.) and the control shRNA plasmid-A (sh-NC; cat. no. sc-108060; Santa Cruz Biotechnology, Inc.). In addition, the RAGE clustered regularly interspaced short palindromic repeats (CRISPR) Activation Plasmid (cat. no. sc-400284-ACT; Santa Cruz Biotechnology, Inc.), and CDC4 CRISPR Activation Plasmid (cat. no. sc-401257-ACT; Santa Cruz Biotechnology, Inc.) were used to upregulate the expression of RAGE (OE-RAGE) and CDC4 (OE-CDC4) in colon cancer, and were transfected into cells using Lipo3000 (cat. no. L3000008; Thermo Fisher Scientific, Inc.) at 37°C for 48 h, in addition to the Control CRISPR/dCas9 Activation Plasmid (OE-NC; cat. no. sc-437275; Santa Cruz Biotechnology, Inc.). Subsequent to 48 h cell infection/transfection, the mRNA and protein samples were extracted from the cells and submitted to RT-qPCR and western blot analysis.
Flow cytometry
T84 and SW480 cells were seeded into 6-well plates and cultured for 24 h. Following different treatments, the cells were gathered into individual centrifuge tubes and suspended in Annexin-binding buffer. Subsequently, the cells were incubated with Annexin V-fluorescein isothiocyanate and propidium iodide solution (BD Biosciences) at room temperature in the shade for 15 min, according to the manufacturer's instructions. The apoptotic percentages of all samples were analyzed using flow cytometry (BD Biosciences). Cell apoptotic rates were analyzed using FlowJo v.10 (FlowJo, LLC).
Animal experiments
The function of scutellarein in inhibiting tumor progression was assessed using a mouse model. A total of 30 male BALB/c nude mice aged 5 weeks old (weight 18-20 g) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai, China) and maintained under specific pathogen-free conditions at 20-26°C with 55±5% humidity, a 12 h light/dark cycle and ad libitum access to food and water. The experiment protocols were ethically approved by the Institutional Animal Care and Use Committee of the Affiliated Hospital of Southwest Medical University (Sichuan, China). The T84 cells (5×106 in 200 µl PBS) were tail-vein injected into the mice to generate a colon cancer-bearing model. Once the tumors had grown, the mice were randomly separated into 3 groups and intraperitoneally injected with scutellarein (0.5 µg/g body weight) and shRNA-CDC4 (0.1 µg/g body weight). Mice treated with PBS alone were considered to be the control group. The animal health and behavior, in addition to the tumor sizes, were monitored every 3 days. A total of 20 days later, the mice were euthanized by cervical dislocation and the heads were broken and the tumor weight of each mouse was evaluated. Then, the tumors were removed from the mice and subjected to immunohistochemical staining with an anti-Ki-67 antibody (1:100 dilution; cat. no. 12202; Cell Signaling Technology, Inc.) and hematoxylin (100%, 3 min) and eosin (0.5%, 30 sec) staining at room temperature to assess the effect of scutellarein/CDC4 on the expression of Ki-67 and pathological alteration. The diameter for the maximum tumor was 1.8 cm and all mice were euthanized at the end of the experiment.
For the immunohistochemical staining, tissue sections were deparaffinized and rehydrated with xylene (100%) and ethanol (100, 95, 80 and 70%). Subsequently, the antigens were retrieved using citrate antigen retrieval solution at 95°C (Beyotime Institute of Biotechnology) and blocked with 5% goat serum (AmyJet Scientific, Inc.) diluted in TBS + 0.5% Tween-20 at room temperature. The sections were probed with the primary antibody against Ki-67 (1:100 dilution; cat. no. 12202; Cell Signaling Technology, Inc.) at 4°C overnight. Following the primary antibody incubation, sections were incubated with a HRP-conjugated secondary antibody (1:500; cat. no. 8114; Cell Signaling Technology, Inc.) at room temperature for 30 min, followed by incubation with the chromogen 3,30-diaminobenzidine tetrachloride (R&D Systems, Inc.) for 2-3 sec at room temperature. Cell nuclei were stained with 1% Harris hematoxylin solution for 30 sec at room temperature. The protein expression of Ki-67 was evaluated by two pathologists on the base of the positive staining proportion and the staining intensity, as previously described (31). The positive staining percentage was scored as 0 for ≤5%, 1 for 6-25%, 2 for 26-50%, 3 for 51-75% and 4 for >75%. Intensity was marked as follows: 0 represented no staining, 1 represented weak staining, 2 represented moderate staining and 3 represented strong staining. The final score was obtained by multiplying the percentage score and intensity score.
Statistical analysis
The results are representative of independent experiments, and the data were presented as the mean ± standard error of the mean. SPSS22.0 statistical software (IBM Corp.) was used to analyze the data between groups with different treatments using paired Student's t-tests for 2 groups and one way analysis of variance followed by a Tukey's test for multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Scutellarein treatment induces cell apoptosis and inhibits cell viability in CL-40, T84 and SW480 cells
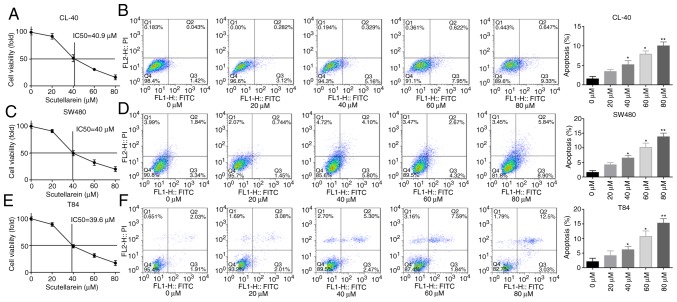
First, the present study investigated the effect of scutellarein treatment on cell apoptosis and viability in colon cancer. The CL-40, T84 and SW480 cells were exposed to gradient concentrations of scutellarein, and the cytotoxic activity of scutellarein was evaluated using an MTT assay. The IC50 values of scutellarein in CL-40, T84 and SW480 cell lines were determined to be 40.9, 40 and 39.6 µM, respectively (Fig. 1A, C and E). In addition, the ratios of cleaved caspase3/total caspase3 and cleaved/total caspase7 were significantly increased when the cells were treated with scutellarein in a dose-dependent manner (Fig. S1A-C), in addition to the apoptosis rates detected using a flow cytometry assay in CL-40, T84 and SW480 cell lines (Fig. 1B, D and F) compared with the control group (P<0.05). Furthermore, the function of scutellarein treatment in the clone formation ability of CL-40, T84 and SW480 cells was examined using a colony formation assay. As presented in Fig. S1D, compared with the control group, the plates with scutellarein-treated cells demonstrated fewer cell clones, and ≥40 µM scutellarein induced a significant inhibition in the clone formation ability in a dose-dependent manner compared with the 0 µM group (P<0.05). These results illustrated that scutellarein treatment was able to efficiently inhibit colon cancer cell viability and induce cell apoptosis.
Figure 1.
Scutellarein treatment inhibited cell viability and promoted cell apoptosis in colon cancer cells (CL-40, SW480 and T84). The cells were exposed to scutellarein (0, 20, 40, 60 and 80 µM) for 24 h, and then assays were performed. (A) Cytotoxic effect of scutellarein on CL-40 cells were determined using an MTT assay, and the IC50 value was determined to be 40.9 µM. (B) Effect of scutellarein treatment on the apoptosis rate of CL-40 cells was evaluated by using a flow cytometry assay. (C) Cell viability of SW480 cells was determined using an MTT assay, and the IC50 value was determined to be 40 µM. (D) SW480 cell apoptosis was detected by using flow cytometry technology. (E) Cytotoxic effect of scutellarein on T84 cells was assessed using an MTT assay, and the IC50 value was determined to be 39.6 µM. (F) Cell apoptosis in T84 cells was measured using a flow cytometry assay. The data were represented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01 vs. the control group. IC50, half maximal inhibitory concentration.
Scutellarein inhibits cell proliferation and promotes cell apoptosis via reducing RAGE expression in T84 and SW480 cells
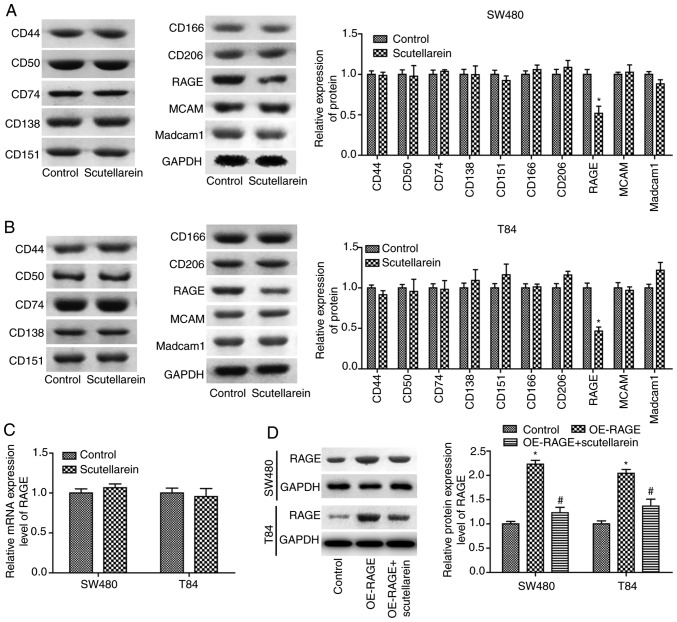
To reveal the mechanisms underlying scutellarein in colon cancer inhibition, the present study then investigated the effects of scutellarein on the expression of numerous oncoproteins, including CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206, RAGE and Madcam1 (14,15). The results revealed that scutellarein treatment (40 µM) significantly inhibited the protein expression levels of RAGE compared with the control group (P<0.05) in T84 (Fig. 2A) and SW480 cells (Fig. 2B), with no significant influence in the protein expression levels of CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206 and Madcam1 (P>0.05). However, scutellarein treatment exerted no significant effect on the mRNA levels of RAGE compared with the control group (P>0.05; Fig. 2C). Furthermore, the present study revealed that the significantly increased expression of RAGE induced by OE-RAGE transfection was significantly reduced when T84 and SW480 cells were treated with scutellarein (40 µM; P<0.05; Fig. 2D). These results demonstrated that scutellarein treatment negatively regulated RAGE expression at the protein level.
Figure 2.
Scutellarein treatment decreased RAGE protein levels in SW480 and T84 cells. Western blot analysis was performed to determine the effects of scutella-rein on the expression of CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206, RAGE and Madcam1 once the cells were treated with scutellarein (40 µM) in (A) SW480 cells and (B) T84 cells, and GAPDH was used as a loading control. (C) mRNA expression levels of RAGE were determined by using a reverse transcription-quantitative PCR assay once the cells were treated with scutellarein (40 µM). (D) Western blot analysis was performed to investigate the expression of RAGE protein once cells were transfected with the OE-RAGE plasmid together with scutellarein (40 µM) or not. The data demonstrated are representative of three independent experiments. n=3. *P<0.05 vs. the control group; #P<0.05 vs. the OE-RAGE group. CD, cluster of differentiation; MCAM, melanoma cell adhesion molecule; RAGE, receptor for advanced glycation end products; Madcam1, mucosal addressin cell adhesion molecule 1; OE, overexpression.
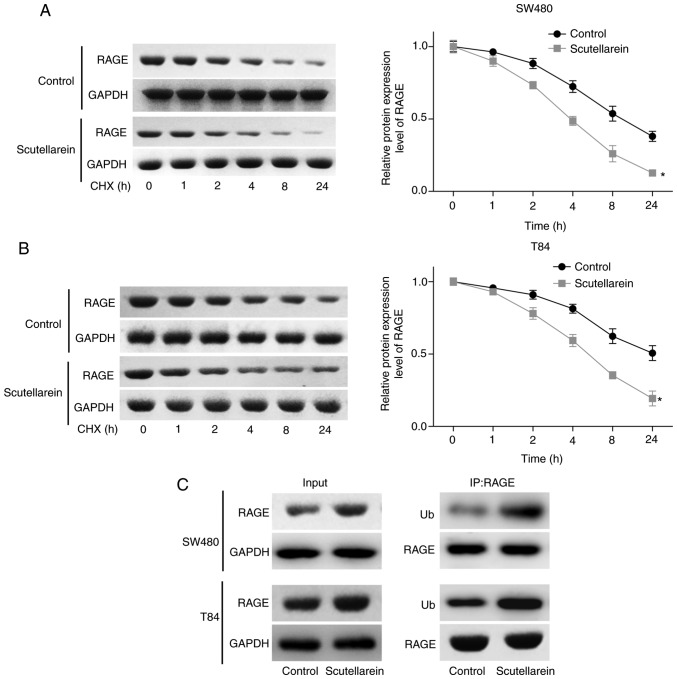
To reveal the underlying mechanism of the scutellarein treatment-induced reduction in the RAGE protein expression level, the present study then investigated the effect of scutellarein treatment on the protein stability and ubiquitination of RAGE. The CHX analysis demonstrated that scutellarein treatment significantly reduced the stability of RAGE protein in SW480 (P<0.05; Fig. 3A) and T84 cell lines compared with the control (P<0.05; Fig. 3B). In addition, scutellarein treatment significantly enhanced the ubiquitination of the RAGE protein (Fig. 3C). These results revealed that scutellarein treatment may reduce RAGE protein expression via enhancing its ubiquitination and impairing its stability.
Figure 3.
Scutellarein treatment weakened the protein stability of RAGE and increased its ubiquitination. (A) SW480 and (B) T84 cells were exposed to 40 µM scutellarein for 24 h, and then the cells were treated with CHX (100 µg/ml) for 1, 2, 4, 8 or 24 h, followed by a western blot assay to detect the expression levels of RAGE. (C) An IP assay was used to detect the effect of scutellarein (40 µM) treatment on the ubiquitination of the RAGE protein. The data presented are representative of three independent experiments. *P<0.05 vs. the control group. RAGE, receptor for advanced glycation end products; CHX, cycloheximide; IP, immunoprecipitation; Ub, ubiquitin.
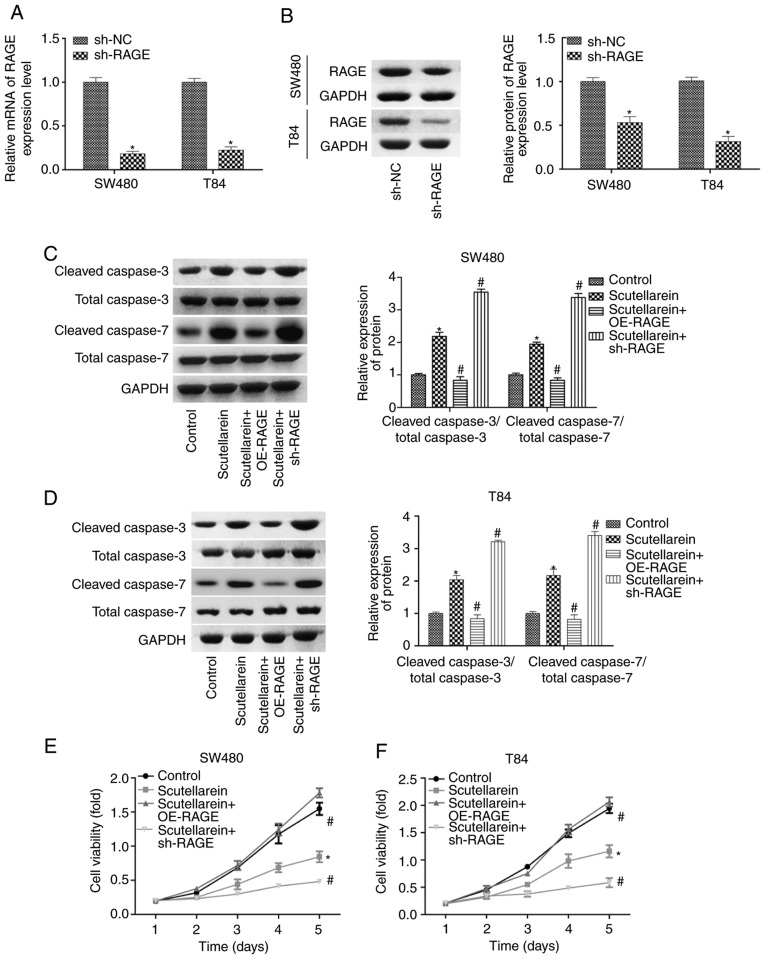
The present study then investigated the effects of RAGE in scutellarein-mediated cell proliferation repression and apoptosis promotion in T84 and SW480 cells. The expression levels of RAGE were significantly reduced when the cells were infected with shRNA targeting the RAGE gene at the mRNA and protein levels when compared with the sh-NC group (P<0.05; Fig. 4A and B). As demonstrated in Fig. 4C and D, the overexpression of RAGE significantly blocked the function of scutellarein in promoting the expression levels of cleaved caspase3/7, while the knockdown of RAGE enhanced the function of scutellarein, with a statistically significant difference compared with the scutellarein-treated group (P<0.05; Fig. 4C and D). Additionally, the proliferation of T84 and SW480 cells in the presence of scutellarein with or without RAGE overexpression or knockdown was also assessed. Compared with the control group, cell proliferation was significantly repressed following scutellarein treatment, and RAGE overexpression significantly weakened this function while RAGE downregulation enhanced this function in SW480 and T84 cell lines (P<0.05; Fig. 4E and F). In addition, the present study detected the effect of the scutellarein/RAGE axis on the activation of signals which have been reported to be under the regulation of scutellarein, including nuclear factor-κβ (32), PKC (33) and VEGF (34). The results revealed that scutellarein treatment significantly decreased the expression levels of p-p65, PKC and VEGF compared with the control group (P<0.05), whereas the expression levels of p-p65 and VEGF were neutralized when RAGE was overexpressed, while RAGE downregulation further decreased p-p65 and VEGF expression levels compared with the scutellarein group (P<0.05; Fig. S2). However, the deregulation of RAGE exhibited no significant influence in PKC expression in SW480 and T84 cell lines (P>0.05; Fig. S2). These results demonstrated that scutellarein promoted colon cancer cell apoptosis and repressed cell viability via downregulating the expression of RAGE.
Figure 4.
Effects of RAGE on scutellarein-mediated apoptosis promotion and proliferation repression in SW480 and T84 cells. Knockdown efficiency of sh-RAGE was detected using (A) reverse transcription-quantitative PCR and (B) western blot assays in SW480 and T84 cells. Then, the SW480 and T84 cells were treated with scutellarein, scutellarein+OE-RAGE or scutellarein+sh-RAGE. Relative protein levels of cleaved caspase3/7 and total caspase3/7 in (C) SW480 and (D) T84 cells were determined by western blot analysis. Proliferation of (E) SW480 and (F) T84 cells was assessed using an MTT assay. n=3. *P<0.05 vs. the control group; #P<0.05 vs. the scutellarein group. RAGE, receptor for advanced glycation end products; sh-, short hairpin RNA; OE, overexpression; NC, negative control.
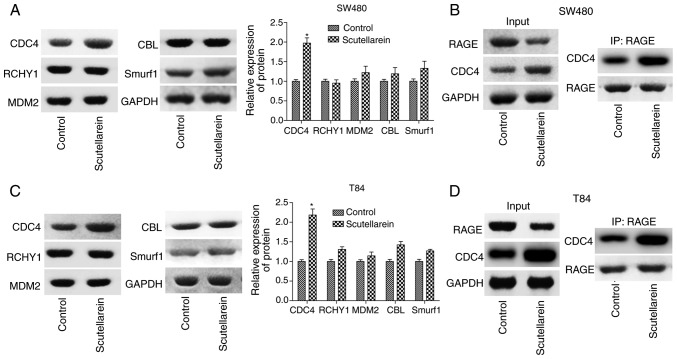
Scutellarein increases the combination of CDC4 and RAGE
To investigate the mechanism of the suppressant effects of scutellarein on RAGE levels, the present study detected the expression of a series of ubiquitination-associated proteins, including CDC4, RCHY1, MDM2, CBL and Smurf1 in T84 and SW480 cells using western blot analysis. It was revealed that only the expression levels of CDC4 were significantly increased in presence of scutellarein compared with the control (P<0.05), with no notable change in the expression levels of RCHY1, MDM2, CBL and Smurf1 (Fig. 5A and C). As CDC4 was considered as one of the ubiquitin ligases, the increase of CDC4 may have induced the degradation of downstream molecules. IP was performed to investigate the combination of CDC4 and RAGE and the results revealed that scutellarein enhanced the direct or indirect combination of CDC4 and RAGE protein expression in SW480 cells (Fig. 5B) and T84 cells (Fig. 5D). These results indicated that CDC4 may serve a function in the scutellarein-mediated reduction of RAGE expression.
Figure 5.
Effects of scutellarein on the combination of CDC4 and RAGE proteins. (A) Western blot analysis was performed to investigate the effects of scutellarein (40 µM) on the expression of ubiquitin-associated proteins, including CDC4, RCHY1, MDM2, CBL and Smurf1 in SW480 cells. (B) An IP assay was performed to detect the effects of scutellarein on the combination of CDC4 and RAGE in SW480 cells. (C) Representative data presenting the levels of ubiquitin-associated proteins (CDC4, RCHY1, MDM2, CBL and Smurf1) in T84 cells treated with scutellarein. (D) An IP assay was used to assess the effects of scutellarein on the combination of CDC4 and RAGE proteins in T84 cells. n=3. *P<0.05 vs. the control group. RAGE, receptor for advanced glycation end products; CDC4, cell division control protein 4; IP, immunoprecipitation; RCHY1, ring finger and CHY zinc finger domain containing 1; MDM2, MDM2 proto-oncogene; CBL, Cbl proto-oncogene; Smurf1, SMAD specific ubiquitin protein ligase 1.
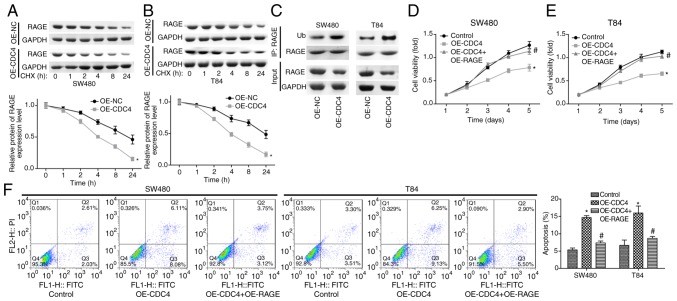
Scutellarein treatment inhibits cell proliferation and promotes cell apoptosis via CDC4-mediated RAGE downregulation
To further study the function CDC4 served in the scutellarein-mediated reduction of RAGE expression and colon cancer inhibition, the present study then performed CHX analysis to investigate the effect of CDC4 on the stability of RAGE. The results demonstrated that the overexpression of CDC4 significantly accelerated the degradation of the RAGE protein in SW480 cells (Fig. 6A) and T84 cells (Fig. 6B) compared with the control (P<0.05). In addition, it was revealed that the over-expression of CDC4 increased the expression of ubiquitinated RAGE compared with the control (Fig. 6C). Cell proliferation was significantly inhibited (P<0.05; Fig. 6D and E) while cell apoptosis (P<0.05; Fig. 6F) was significantly increased when CDC4 was overexpressed in SW480 and T84 cells compared with the control, whereas these effects were significantly abolished when RAGE was overexpressed at the same time (P<0.05). These results indicated that CDC4 promoted colon cancer cell apoptosis and inhibited cell proliferation via increasing the RAGE ubiquitination levels.
Figure 6.
Effects of CDC4/RAGE on the proliferation and apoptosis of SW480 and T84 cells. Following 24 h of cell transfection with OE-NC or OE-CDC4, (A) SW480 and (B) T84 cells were treated with CHX (100 µg/ml) for 1, 2, 4, 8 or 24 h, and then a western blot assay was performed to detect the protein levels of RAGE. (C) A western blot assay was performed to investigate the ubiquitination of RAGE subsequent to SW480 and T84 cells being transfected with OE-CDC4 or OE-NC. Cell proliferation of (D) SW480 and (E) T84 cells was detected using MTT analysis once the cells were transfected with OE-CDC4, OE-CDC4+OE-PAGE or their negative control vector. (F) Effects of CDC4 and RAGE overexpression on the apoptosis of SW480 and T84 cells were analyzed using flow cytometry. n=3. *P<0.05 vs. control group; #P<0.05 vs. the OE-CDC4 group. RAGE, receptor for advanced glycation end products; CDC4, cell division control protein 4; OE, overexpression; NC, negative control; CHX, cycloheximide.
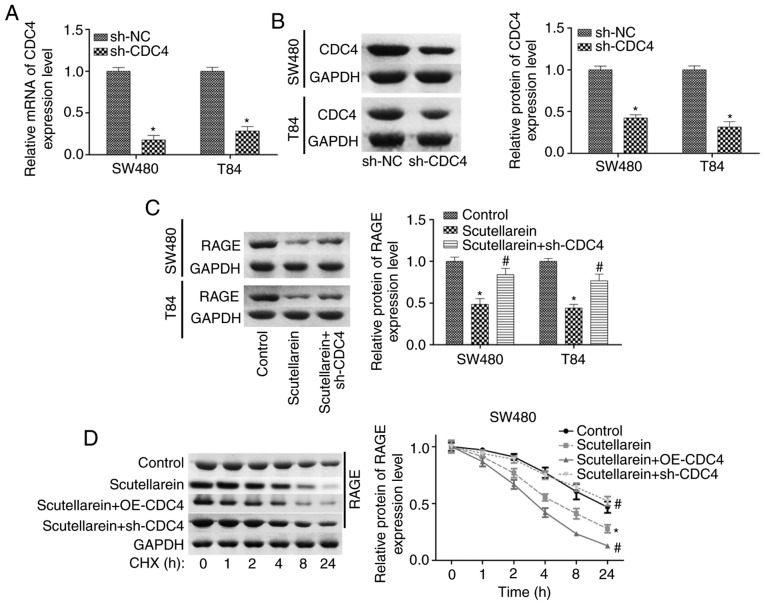
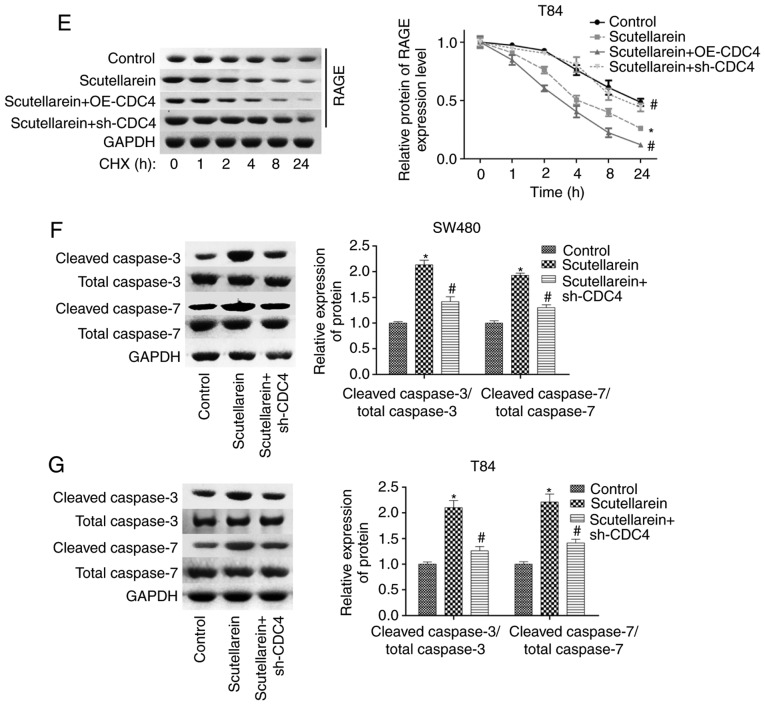
Furthermore, the present study investigated the function of CDC4 in scutellarein-mediated proliferation inhibition and apoptosis promotion in colon cancer. The expression of CDC4 was significantly reduced when the cells were infected with sh-CDC4 at the mRNA and protein levels compared with the negative control (P<0.05; Fig. 7A and B). Knockdown of CDC4 significantly rescued the reduction of RAGE in SW480 and T84 cells mediated by scutellarein treatment (P<0.05; Fig. 7C). CHX analysis revealed that the overexpression of CDC4 significantly enhanced the effect of scutellarein in reducing RAGE stability, while the knockdown of CDC4 significantly weakened the effect of scutellarein (P<0.05; Fig. 7D and E). Furthermore, it was revealed that the increased expression levels of cleaved caspase3/7 caused by scutellarein treatment were significantly reduced when CDC4 was downregulated (P<0.05; Fig. 7F and G). In addition, the knockdown of CDC4 significantly rescued scutellarein-mediated reductions in the expression levels of p-p65 and VEGF (P<0.05), with no notable change in the expression of PKC (P>0.05; Fig. S3). These results demonstrated that the increased level of CDC4 served a vital function in scutellarein-mediated proliferation inhibition and apoptosis promotion in colon cancer.
Figure 7.
Scutellarein treatment decreased RAGE expression and induced cell apoptosis via increasing CDC4 expression. (A) Reverse transcription-quantitative PCR and (B) western blot assays were used to determine the knockdown efficiency of sh-CDC4 in SW480 and T84 cells. *P<0.05 vs. the sh-NC group. Next, SW480 and T84 cells were divided into three groups (control, scutellarein and scutellarein+sh-CDC4), and then submitted to the following assays. (C) Expression of RAGE was detected using a western blot assay. Protein stability of RAGE was detected by using the CHX reagent together with a western blot assay in (D) SW480 and (E) T84 cells. Levels of cleaved caspase3/7 and total caspase3/7 were determined using a western blot assay in (F) SW480 and (G) T84 cells. *P<0.05 vs. the control group; #P<0.05 vs. the scutellarein group. RAGE, receptor for advanced glycation end products; CDC4, cell division control protein 4; sh-, short hairpin RNA; NC, negative control; CHX, cycloheximide.
Scutellarein suppresses the progression of colon cancer via increasing CDC4 expression in vivo
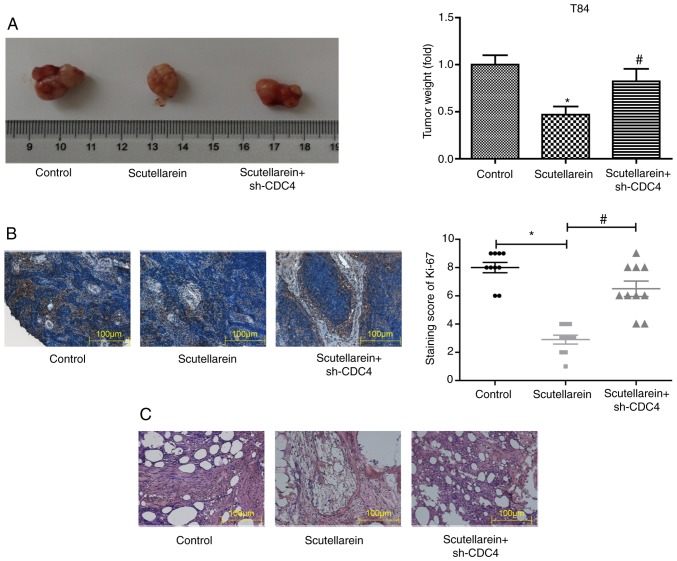
Next, the present study further investigated whether scutellarein inhibited the progression of colon cancer in vivo via upregulating CDC4 expression. A colon cancer-bearing mice model was established by the injection of the colon cancer cell line T84. The solid tumor types were removed and weighed once the mice were sacrificed by cervical dislocation. In comparison with the control group, scutellarein treatment significantly reduced the tumor volume by nearly 50%, while the knockdown of CDC4 significantly increased the tumor volume compared with the scutellarein alone treated group (P<0.05; Fig. 8A). In addition, the knockdown of CDC4 also significantly neutralized the functions of scutellarein treatment in reducing the expression levels of Ki-67 compared with the control (P<0.05; Fig. 8B) and the alleviation in the histological change of tumor tissues (Fig. 8C). These results revealed that scutellarein suppressed the in vivo tumor formation of colon cancer via increasing CDC4 expression.
Figure 8.
Effects of scutellarein/CDC4 on the tumor growth in a mice xenograft model with a T84 cell injection. (A) Images of tumors removed from the mice treated with scutellarein and scutellarein+sh-CDC4. Bar graph represents the relative weight of the solid tumor types. (B) Immunohistochemical staining was used to detect the expression of Ki-67 in the tumor tissues derived from different groups. (C) Hematoxylin and eosin staining was used to assess the pathological alteration of the tumor tissues derived from different groups. Data are presented as the mean ± standard error of the mean. n=10. *P<0.05 vs. the control group; #P<0.05 vs. the scutellarein group. CDC4, cell division control protein 4; sh-, short hairpin RNA.
Discussion
As is well known, colon cancer is one of the most malignant carcinoma types (35). Wild chrysanthemum is the flower head of Chrysanthemum indicum L., which may be obtained wildly in the majority of natural habitats in China (5,36). The present study was designed to investigate the effects and mechanism of scutellarein in the development of colon cancer. The present study demonstrated that scutellarin, derived from wild chrysanthemum, notably inhibited the development of colon cancer in vitro and in vivo via CDC4-mediated RAGE ubiquitination enhancement.
As early as 2005, Goh et al (13) reported that the chemically standardized extract from Scutellaria barbata induced a significant increase in the sub G1 phase and promoted cell apoptosis in colon cancer LoVo cells. Subsequently, a study by Wang et al (37) also demonstrated the anti-tumor activity of scutellarein in colon cancer. Recently, Guo et al (12) demonstrated that scutellarein identified from Scutellaria barbata significantly increased the apoptosis of colon cancer HCT116 cells via increasing the production of intracellular reactive oxygen species. As CL-40, T84 and SW480 cell lines are from different types of colon cancer, in that CL-40 cells are derived from the colon adenocarcinoma, T84 cells are derived from the lung metastatic site and SW480 cells are derived from the primary site of colon cancer of Dukes' type B, they were selected for the present study to investigate the effects of scutellarein in the viability and apoptosis of colon cancer cells. Consistent with previous results (12,13,37), the present study also observed that scutellarein treatment significantly inhibited colony formation and cell proliferation and accelerated cell apoptosis in colon cancer CL-20, SW480 and T84 cells (P<0.05). In addition, it was demonstrated that scutellarein significantly inhibited the in vivo tumor formation ability of T84 cells using colon cancer-bearing mice (P<0.05). The present results further illustrate the inhibitory function of scutellarein in colon cancer progression.
To investigate the underlying mechanism of scutellarein-mediated repression in colon cancer, the present study investigated the effects of scutellarein treatment on the expression of numerous oncoproteins, including CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206, RAGE and Madcam1. As CL-40 and SW480 are primary cell lines of colon cancer, one was selected (SW480) together with T84 (a metastasis cell line) for the subsequent experiments. The present results demonstrate that scutellarein treatment significantly inhibited the expression of RAGE compared with the control (P<0.05), with no substantial influence in the expression levels of CD44, CD50, CD74, CD138, MCAM, CD151, CD166, CD206 and Madcam1. An increasing number of studies have demonstrated the importance of RAGE in the pathogenesis of multiple human disease types, including cancer (38-41). The deregulation of RAGE is considered to be an important factor in tumorigenesis, and the increase of RAGE is associated with a diverse range of malignancies (22,42). In the present study, the results demonstrated that scutellarein treatment significantly decreased the protein levels of RAGE, but not the mRNA levels, compared with the control (P<0.05). Thus, the present study investigated the effect of scutellarein on RAGE protein stability and ubiquitination, and the results revealed that scutellarein treatment decreased the protein stability of RAGE and enhanced its ubiquitination. Similarly, RAGE was also reported to be regulated by the ubiquitination-degradation pathway by the ubiquitin E3 ligase subunit F-box protein O10 (43). Furthermore, the present study observed that the overexpression of RAGE weakened while the knockdown of RAGE enhanced scutellarein functions in promoting cell apoptosis and inhibiting cell proliferation in colon cancer, suggesting that scutellarein inhibited colon cancer progression via downregulating RAGE.
Ubiquitination is a process in which ubiquitin alters and specifically modifies the target proteins with the help of a series of particular enzymes, including ubiquitin activating enzyme, ubiquitin conjugating enzyme and ubiquitin ligase (44). An increasing number of studies have demonstrated that ubiquitination is strongly implicated in the pathogenesis of carcinogenesis (45,46). In the present study, the underlying mechanism of the scutellarein-mediated ubiquitination of RAGE protein was examined. An increase in CDC4 expression was observed in the presence of scutellarein. CDC4/F-box and WD repeat domain containing 7, located on the 4q, is considered to be a vital anti-oncogene in various cancer types, in addition to a potential therapeutic target, with mutations observed in pancreatic, colorectal and ovarian tumor types (47). The results of the present study confirmed that CDC4 may combine with the RAGE protein and then induce its ubiquitination and the subsequent degradation. Furthermore, a significant decrease in cell proliferation and an increase in cell apoptosis rates were observed in SW480 and T84 cells with CDC4 overexpression compared with the control (P<0.05), while this effect was impaired when RAGE was downregulated, suggesting that CDC4 inhibited colon cancer progression via decreasing the expression of RAGE. Furthermore, the present results also revealed the important function of CDC4 in scutellarein-mediated increases in cell apoptosis and inhibition in cell proliferation and tumorigenesis in colon cancer.
However, there are a number of limitations to the present study. In the present study it was mainly revealed that scutellarein inhibited the expression of the oncoprotein RAGE via increasing CDC4-mediated ubiquitination enhancement, which then inhibited cell proliferation and induced cell apoptosis. However, the present study did not investigate the molecular mechanism underlying the scutellarein-mediated CDC4 upregulation. In addition, the results revealed that scutellarein inhibited cell proliferation and induced cell apoptosis in T84 and SW480 cells in a similar manner. However, T84 cells are derived from a lung metastatic site, while SW480 cells are derived from a primary site of colon cancer of Dukes' type B, indicating that these are two different types of cells with different malignant behaviors. It was hypothesized that this may be induced by the same protein which is located at the upstream of CDC4, but this was not further investigated. These will be investigated in future studies through performing sequencing to determine the gene which is modulated by scutellarein in T84 and SW480 cell lines. Furthermore, the present study did not examine the effect of scutellarein on the function of CL-40 cells due to the limitations in funding and time. The present study only examined the effects and underlying mechanisms of scutellarein, an active ingredient of wild chrysanthemum, on the progression of colon cancer. However, it is not clear whether chamomile and other chrysanthemums have similar effects. In future studies, the effects of chamomile and other chrysanthemums on colon cancer progression will be examined.
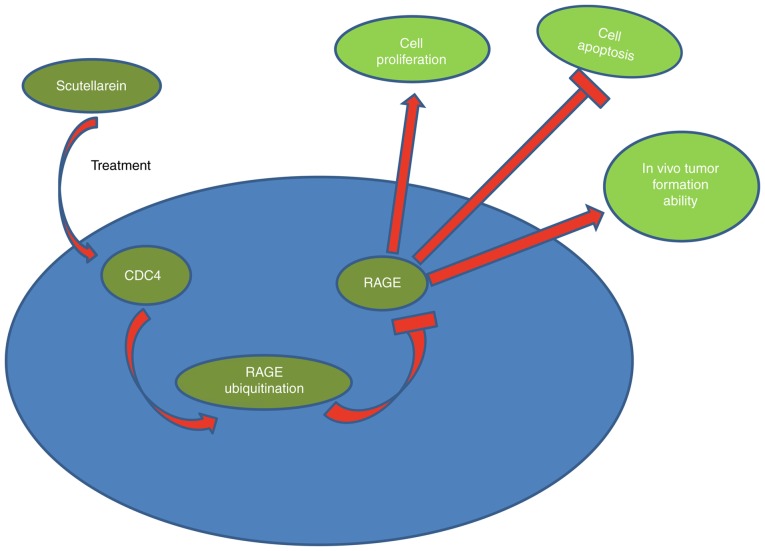
In conclusion, the present investigation revealed that scutellarein derived from wild chrysanthemum may effectively suppress the proliferation and tumorigenesis and induce the apoptosis of colon cancer cells through downregulating RAGE expression mediated by CDC4 upregulation (Fig. 9). The present results may provide more useful information for the application of scutellarein in the treatment of colon cancer.
Figure 9.
Schematic diagram of the anti-tumor function of scutellarein in colon cancer. Scutellarein treatment reduced the expression of RAGE via the CDC4-mediated ubiquitination degradation pathway, which then inhibited cell proliferation, tumorigenesis and induced cell apoptosis in colon cancer. RAGE, receptor for advanced glycation end products; CDC4, cell division control protein 4.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was supported by a research grant for Doctors of the Affiliated Hospital of Southwest Medical University and Luzhou People's Government-Southwest Medical University Cooperative Scientific Research Project (grant no. 2019LZXNDJ26).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YL and JW provided the idea and performed the experiments, in addition to writing the manuscript. SZ and JL performed the data analyses and parts of the experiments. WD performed parts of the data analyses. YL revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Athymic nude mouse assays were performed in accordance with the institutional principles for the concern and use of animals and the protocol was ethically approved by the ethical committee of the Affiliated Hospital of Southwest Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Labianca R, Beretta GD, Kildani B, Milesi L, Merlin F, Mosconi S, Pessi MA, Prochilo T, Quadri A, Gatta G, et al. Colon cancer. Crit Rev Oncol Hematol. 2010;74:106–133. doi: 10.1016/j.critrevonc.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Freeman HJ. Early stage colon cancer. World J Gastroenterol. 2013;19:8468–8473. doi: 10.3748/wjg.v19.i46.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orangio GR. The economics of colon cancer. Surg Oncol Clin N Am. 2018;27:327–347. doi: 10.1016/j.soc.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Shi J, Huang H, Ren J, Li N, Dai M. Burden of colorectal cancer in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2015;36:709–714. In Chinese. [PubMed] [Google Scholar]
- 5.Sun S, Jiang P, Su W, Xiang Y, Li J, Zeng L, Yang S. Wild chrysanthemum extract prevents UVB radiation-induced acute cell death and photoaging. Cytotechnology. 2016;68:229–240. doi: 10.1007/s10616-014-9773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang WS, Kim D, Yi YS, Kim JH, Jeong HY, Hwang K, Kim JH, Park J, Cho JY. AKT-targeted anti-inflammatory activity of the methanol extract of Chrysanthemum indicum var. albescens J Ethnopharmacol. 2017;201:82–90. doi: 10.1016/j.jep.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Deng Y, Jiang J, Chen S, Teng N, Song A, Guan Z, Fang W, Chen F. Combination of multiple resistance traits from wild relative species in Chrysanthemum via trigeneric hybridization. PLoS One. 2012;7:30. doi: 10.1371/journal.pone.0044337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassanpouraghdam MB. Flowerhead volatile oil composition of soilless culture-grown Chrysanthemum balsamita L. Nat Prod Res. 2009;23:672–677. doi: 10.1080/14786410802591182. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Chen G, Liu X, Qiu Y, Yang S, Zhang Y, Fang X, Zhang C, Liu X. Scutellarein inhibits cancer cell metastasis in vitro and attenuates the development of fibrosarcom in vivo. Int J Mol Med. 2015;35:31–38. doi: 10.3892/ijmm.2014.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng CY, Hu CC, Yang HJ, Lee MC, Kao ES. Inhibitory effects of scutellarein on proliferation of human lung cancer A549 cells through ERK and NFκB mediated by the EGFR pathway. Chin J Physiol. 2014;57:182–187. doi: 10.4077/CJP.2014.BAC200. [DOI] [PubMed] [Google Scholar]
- 11.Parajuli P, Joshee N, Rimando AM, Mittal S, Yadav AK. In vitro antitumor mechanisms of various Scutellaria extracts and constituent flavonoids. Planta Med. 2009;75:41–48. doi: 10.1055/s-0028-1088364. [DOI] [PubMed] [Google Scholar]
- 12.Guo F, Yang F, Zhu YH. Scutellarein from Scutellaria barbata induces apoptosis of human colon cancer HCT116 cells through the ROS-mediated mitochondria-dependent pathway. Nat Prod Res. 2019;33:2372–2375. doi: 10.1080/14786419.2018.1440230. [DOI] [PubMed] [Google Scholar]
- 13.Goh D, Lee YH, Ong ES. Inhibitory effects of a chemically standardized extract from Scutellaria barbata in human colon cancer cell lines, LoVo. J Agric Food Chem. 2005;53:8197–8204. doi: 10.1021/jf051506+. [DOI] [PubMed] [Google Scholar]
- 14.Behzad MM, Asnafi AA, Jaseb K, Jalali Far MA, Saki N. Expression of CD markers' in immune thrombocytopenic purpura: Prognostic approaches. APMIS. 2017;125:1042–1055. doi: 10.1111/apm.12755. [DOI] [PubMed] [Google Scholar]
- 15.Bacher P, Scheffold A. Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A. 2013;83:692–701. doi: 10.1002/cyto.a.22317. [DOI] [PubMed] [Google Scholar]
- 16.Tripathi SC, Fahrmann JF, Celiktas M, Aguilar M, Marini KD, Jolly MK, Katayama H, Wang H, Murage EN, Dennison JB, et al. MCAM mediates chemoresistance in small-cell lung cancer via the PI3K/AKT/SOX2 signaling pathway. Cancer Res. 2017;77:4414–4425. doi: 10.1158/0008-5472.CAN-16-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013;330:150–162. doi: 10.1016/j.canlet.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Zhang J, Wang Z, Lu D, Feng J, Yang D, Chen X, Yan X. CD146 is a potential marker for the diagnosis of malignancy in cervical and endometrial cancer. Oncol Lett. 2013;5:1189–1194. doi: 10.3892/ol.2013.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanida S, Mizoshita T, Mizushima T, Sasaki M, Shimura T, Kamiya T, Kataoka H, Joh T. Involvement of oxidative stress and mucosal addressin cell adhesion molecule-1 (MAdCAM-1) in inflammatory bowel disease. J Clin Biochem Nutr. 2011;48:112–116. doi: 10.3164/jcbn.10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Truffi M, Colombo M, Peñaranda-Avila J, Sorrentino L, Colombo F, Monieri M, Collico V, Zerbi P, Longhi E, Allevi R, et al. Nano-targeting of mucosal addressin cell adhesion molecule-1 identifies bowel inflammation foci in murine model. Nanomedicine (Lond) 2017;12:1547–1560. doi: 10.2217/nnm-2017-0004. [DOI] [PubMed] [Google Scholar]
- 21.Logsdon CD, Fuentes MK, Huang EH, Arumugam T. RAGE and RAGE ligands in cancer. Curr Mol Med. 2007;7:777–789. doi: 10.2174/156652407783220697. [DOI] [PubMed] [Google Scholar]
- 22.Palanissami G, Paul SFD. RAGE and Its ligands: Molecular interplay between glycation, inflammation, and hallmarks of cancer-a review. Horm Cancer. 2018;9:295–325. doi: 10.1007/s12672-018-0342-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuentes MK, Nigavekar SS, Arumugam T, Logsdon CD, Schmidt AM, Park JC, Huang EH. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Dis Colon Rectum. 2007;50:1230–1240. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 24.Foot N, Henshall T, Kumar S. Ubiquitination and the regulation of membrane proteins. Physiol Rev. 2017;97:253–281. doi: 10.1152/physrev.00012.2016. [DOI] [PubMed] [Google Scholar]
- 25.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou MJ, Chen FZ, Chen HC. Ubiquitination involved enzymes and cancer. Med Oncol. 2014;31:93. doi: 10.1007/s12032-014-0093-6. [DOI] [PubMed] [Google Scholar]
- 27.Jin X, Yang C, Fan P, Xiao J, Zhang W, Zhan S, Liu T, Wang D, Wu H. CDK5/FBW7-dependent ubiquitination and degradation of EZH2 inhibits pancreatic cancer cell migration and invasion. J Biol Chem. 2017;292:6269–6280. doi: 10.1074/jbc.M116.764407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi: 10.1080/15384101.2017.1288326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan C, Su H, Song X, Cao H, Kong L, Cui W. Smad ubiquitination regulatory factor 1 (Smurf1) promotes thyroid cancer cell proliferation and migration via ubiquitin-dependent degradation of kisspeptin-1. Cell Physiol Biochem. 2018;49:2047–2059. doi: 10.1159/000493715. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Xin B, He X, Wang J, Cai J, Wei W, Zhang T, Shen X. Nerve growth factor regulates CD133 function to promote tumor cell migration and invasion via activating ERK1/2 signaling in pancreatic cancer. Pancreatology. 2016;16:1005–1014. doi: 10.1016/j.pan.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Huang XW, Xu Y, Sui X, Lin H, Xu JM, Han D, Ye DD, Lv GF, Liu YX, Qu XB, Duan MH. Scutellarein suppresses Aβ-induced memory impairment via inhibition of the NF-κB pathway in vivo an in vitro. Oncol Lett. 2019;17:5581–5589. doi: 10.3892/ol.2019.10274. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Tian X, Chang L, Ma G, Wang T, Lv M, Wang Z, Chen L, Wang Y, Gao X, Zhu Y. Delineation of platelet activation pathway of scutellarein revealed its intracellular target as protein kinase C. Biol Pharm Bull. 2016;39:181–191. doi: 10.1248/bpb.b15-00511. [DOI] [PubMed] [Google Scholar]
- 34.Thirusangu P, Vigneshwaran V, Vijay Avin BR, Rakesh H, Vikas HM, Prabhakar BT. Scutellarein antagonizes the tumorigenesis by modulating cytokine VEGF mediated neoangiogenesis and DFF-40 actuated nucleosomal degradation. Biochem Biophys Res Commun. 2017;484:85–92. doi: 10.1016/j.bbrc.2017.01.067. [DOI] [PubMed] [Google Scholar]
- 35.Labianca R, Nordlinger B, Beretta GD, Mosconi S, Mandalà M, Cervantes A, Arnold D, ESMO Guidelines Working Group Early colon cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi64–vi72. doi: 10.1093/annonc/mdt354. [DOI] [PubMed] [Google Scholar]
- 36.Bosman FT, Yan P. Molecular pathology of colon cancer. Pol J Pathol. 2014;65(4 Suppl 1):S1–S11. In Polish. [PubMed] [Google Scholar]
- 37.Wang F, Yang B, Zhao Y, Liao X, Gao C, Jiang R, Han B, Yang J, Liu M, Zhou R. Host-guest inclusion system of scutellarein with 2-hydroxypropyl-beta-cyclodextrin: Preparation, characterization, and anticancer activity. J Biomater Sci Polym Ed. 2014;25:594–607. doi: 10.1080/09205063.2014.884875. [DOI] [PubMed] [Google Scholar]
- 38.van Zoelen MA, Achouiti A, van der Poll T. RAGE during infectious diseases. Front Biosci (Schol Ed) 2011;3:1119–1132. doi: 10.2741/s215. [DOI] [PubMed] [Google Scholar]
- 39.Lee TW, Kao YH, Chen YJ, Chao TF, Lee TI. Therapeutic potential of vitamin D in AGE/RAGE-related cardiovascular diseases. Cell Mol Life Sci. 2019;76:4103–4115. doi: 10.1007/s00018-019-03204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azizan N, Suter MA, Liu Y, Logsdon CD. RAGE maintains high levels of NFκB and oncogenic Kras activity in pancreatic cancer. Biochem Biophys Res Commun. 2017;493:592–597. doi: 10.1016/j.bbrc.2017.08.147. [DOI] [PubMed] [Google Scholar]
- 41.Rahimi F, Karimi J, Goodarzi MT, Saidijam M, Khodadadi I, Razavi AN, Nankali M. Overexpression of receptor for advanced glycation end products (RAGE) in ovarian cancer. Cancer Biomark. 2017;18:61–68. doi: 10.3233/CBM-160674. [DOI] [PubMed] [Google Scholar]
- 42.Tesarova P, Cabinakova M, Mikulova V, Zima T, Kalousova M. RAGE and its ligands in cancer-culprits, biomarkers, or therapeutic targets? Neoplasma. 2015;62:353–364. doi: 10.4149/neo_2015_061. [DOI] [PubMed] [Google Scholar]
- 43.Evankovich J, Lear T, McKelvey A, Dunn S, Londino J, Liu Y, Chen BB, Mallampalli RK. Receptor for advanced glycation end products is targeted by FBXO10 for ubiquitination and degradation. FASEB J. 2017;31:3894–3903. doi: 10.1096/fj.201700031R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rittinger K, Ikeda F. Linear ubiquitin chains: Enzymes, mechanisms and biology. Open Biol. 2017;7 doi: 10.1098/rsob.170026. pii: 170026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ao N, Chen Q, Liu G. The small molecules targeting ubiquitin-proteasome system for cancer therapy. Comb Chem High Throughput Screen. 2017;20:403–413. doi: 10.2174/1386207320666170710124746. [DOI] [PubMed] [Google Scholar]
- 46.Ding F, Xiao H, Wang M, Xie X, Hu F. The role of the ubiquitin-proteasome pathway in cancer development and treatment. Front Biosci (Landmark Ed) 2014;19:886–895. doi: 10.2741/4254. [DOI] [PubMed] [Google Scholar]
- 47.Davis H, Tomlinson I. CDC4/FBXW7 and the 'just enough' model of tumourigenesis. J Pathol. 2012;227:131–135. doi: 10.1002/path.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.