ABSTRACT
VirE2-INTERACTING PROTEIN1 (VIP1) is a basic leucine zipper protein in Arabidopsis thaliana. VIP1 changes its subcellular localization from the cytoplasm to the nucleus when cells are exposed to mechanical or hypo-osmotic stress. The nuclear localization of VIP1 is inhibited either by inhibitors of calcium signaling or by inhibitors of protein phosphatases 1, 2A and 4 (PP1, PP2A and PP4, respectively). VIP1 binds to the PP2A B”-family subunits, which have calcium-binding EF-hand motifs and which act as the regulatory, substrate-recruiting B subunit of PP2A. The VIP1 de-phosphorylation can therefore be mediated by PP2A. However, details of the PP2A-mediated de-phosphorylation of VIP1 are unclear. Here, with yeast two-hybrid assays and in-vitro pull-down assays, we show that VIP1 does not interact with the scaffolding A subunit of PP2A, but that VIP1 does interact with the catalytic C subunits. Our data raise the possibility that not only the B”-family B subunit of PP2A but also its C subunit contributes to the PP2A-mediated de-phosphorylation of VIP1.
KEYWORDS: Arabidopsis thaliana, protein phosphatase 2A, protein-protein interaction, VIP1
VIP1 (VirE2-INTERACTING PROTEIN 1) is a basic leucine zipper (bZIP) protein in Arabidopsis thaliana and one of the plant group I bZIP proteins.1 VIP1 plays roles in plant development and responses to various stimuli such as hypo-osmotic stress and mechanical stress.2–9 VIP1 is present in the cytoplasm under a stable condition but is transiently localized to the nucleus when Arabidopsis cells are either hypo-osmotically or mechanically stressed.8–10 The nuclear localization of VIP1 requires calcium signaling, VIP1 de-phosphorylation, and dissociation from 14-3-3 proteins.11–13 Inhibitors of protein phosphatases 1, 2A and 4 (PP1, PP2A and PP4, respectively) inhibit both the de-phosphorylation and the nuclear localization of VIP1.13 VIP1 does not bind either B-family or B’-family B subunits of PP2A. However, VIP1 does bind its B”-family B subunits, which have calcium-binding EF-hand motifs.13 These findings raise the possibility that the VIP1 de-phosphorylation is mediated by PP2A.13 However, mechanisms of the PP2A-mediated de-phosphorylation of VIP1 remain to be elucidated.
PP2A consists of the scaffolding A subunit, the regulatory, substrate-recruiting B subunit, and the catalytic C subunit.14–17 To further characterize the interactions between VIP1 and PP2A, one of the Arabidopsis PP2A A subunit isoforms, PP2A-A2, and two of the PP2A C subunit isoforms, PP2A-C3 and PP2A-C5, were subjected to yeast two-hybrid (Y2H) and in-vitro pull-down assays. In the Y2H assays, GAL4 DNA-binding domain (BD)-fused PP2A-A2 did not enable yeast cells to survive on the highly stringent quadruple-dropout (QDO) selection medium, which lacks leucine, tryptophan, histidine and adenine, when co-transformed with GAL4 activation domain (AD)-fused VIP1, but BD-fused PP2A-C3 did enable it (Figure 1a, left panels). Co-expression of AD-fused VIP1 and BD-fused PP2A-C5 did not enable yeast cells to survive on the QDO medium, but did enable them to survive on the moderately stringent triple-dropout (TDO, leucine-, tryptophan- and histidine-deficient) medium supplemented with 3-amino-1,2,4-triazole (3-AT, a histidine biosynthesis inhibitor that can suppress false-positive results in Y2H assays) (Figure 1a, right panels). In similar Y2H assays, co-expression of BD-fused PP2A-C3 with an AD-fused truncated version of VIP1 (either AD-fused VIP1 N (1–186 amino acids of VIP1, which contain putative phosphorylation sites) or AD-fused VIP1 C (165–341 amino acids of VIP1, which contain the bZIP domain)) enabled yeast cells to survive on the QDO medium (Figure 1b, left panel). Neither AD-fused VIP1 N nor AD-fused VIP1 C enabled yeast cells to survive on the QDO medium when co-expressed with BD-fused PP2A-C5 (Figure 1b, middle). However, co-expression of BD-fused PP2A-C5 and one of the AD-fused VIP1 variants did enable yeast cells to survive on the TDO medium supplemented with 3-AT (Figure 1b, right).
Figure 1.
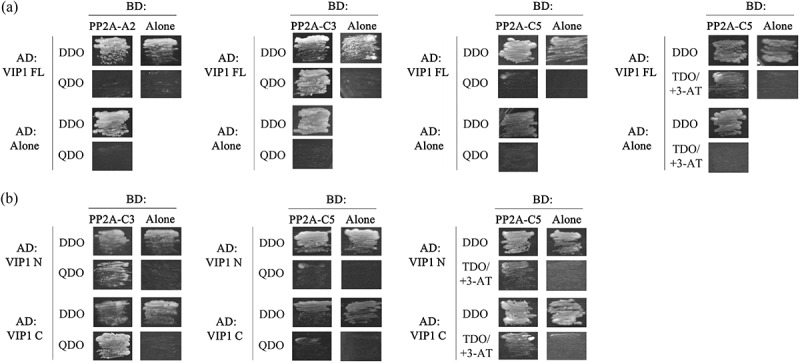
VIP1 interacts with PP2A-C subunits in a yeast two-hybrid system.
(a) The pGBKT7 vector (TaKaRa Bio, Kusatsu, Japan) containing no insert (BD: Alone), PP2A-A2 (BD: PP2A-A2), PP2A-C3 (BD: PP2A-C3), or PP2A-C5 (BD-PP2A-C5) was co-introduced with the pGADT7-Rec vector (TaKaRa Bio) containing no insert (AD: Alone) or VIP1 (AD: VIP1 FL) into the yeast strain AH109. (B) The pGBKT7 vector containing no insert (BD: Alone), PP2A-C3 (BD: PP2A-C3), or PP2A-C5 (BD-PP2A-C5) was co-introduced with the pGADT7-Rec vector containing no insert (AD: Alone) or truncated forms of VIP1, VIP1 N (amino acids 1–186; AD: VIP1 N) and VIP1C (amino acids: 165–341; AD: VIP1 C) into the yeast strain AH109. The transformed yeast cells were cultured on the double-dropout medium (DDO: synthetic dextrose (SD) medium lacking tryptophan and leucine), the quadruple-dropout medium (QDO: SD medium lacking tryptophan, leucine, histidine, and adenine), and the triple-dropout medium supplemented with 3-amino-1,2,4-triazole (3-AT) (TDO/+3-AT: SD medium that lacks tryptophan, leucine, and histidine, and that contains 5mM 3-AT) to examine activation of the reporter genes HIS3 and ADE2. Experiments were performed four times and a representative result is presented.
Glutathione S-transferase (GST)-fused VIP1 and His-tagged forms of PP2A-A2 and PP2A-C3 were expressed in Escherichia coli cells and used for in-vitro pull-down assays. GST-fused VIP1 did not pull down His-tagged PP2A-A2 (Figure 2a, left panel), but did pull down His-tagged PP2A-C3 (Figure 2a, middle). His-tagged PP2A-C5 could not be expressed well in Escherichia coli but could be expressed as Myc-tagged protein with in-vitro transcription and translation. GST-fused VIP1 also pulled down Myc-tagged PP2A-C5 (Figure 2a, right). GST-fused VIP1 C pulled down His-tagged PP2A-C3 and Myc-tagged PP2A-C5 more strongly than GST-fused VIP1 N (Figure 2b).
Figure 2.
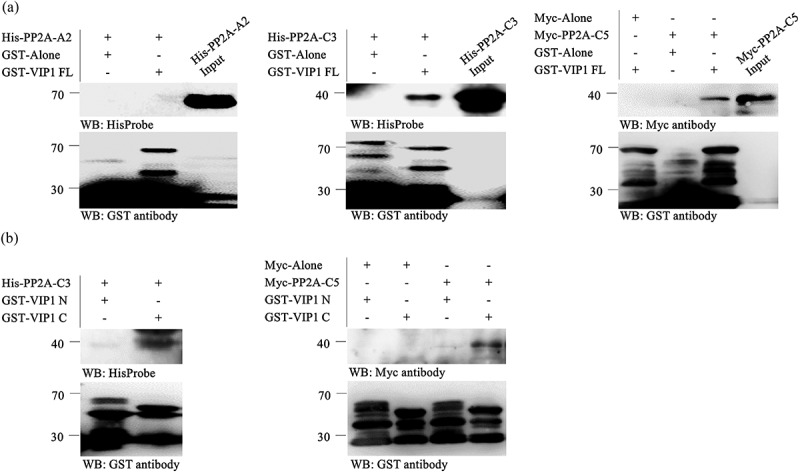
VIP1 interacts with PP2A-C subunits in vitro.
(a) Pull-down assay with full-length VIP1 (VIP1 FL), His-tagged PP2A-A2 (His-PP2A-A2), His-tagged PP2A-C3 (His-PP2A-C3), and Myc-tagged PP2A-C5 (Myc-PP2A-C5). Either GST alone (GST-Alone) or the GST-VIP1 FL fusion protein was bound to the Glutathione Sepharose 4B resin (GE Healthcare, Little Chalfont, UK), reacted with His-PP2A-A2, His-PP2A-C3, Myc-Alone, or Myc-PP2A-C5, and then eluted from the resin. GST-fused proteins, His-tagged proteins and Myc-tagged proteins in the resulting solutions were detected by western blotting with a horseradish peroxidase (HRP)-conjugated GST antibody (Fujifilm Wako Pure Chemical Co., Osaka, Japan), HisProbe-HRP (Thermo Fisher Scientific Inc., Waltham, MA, USA), and anti-Myc-tag pAb (Medical & Biological Laboratories CO., Nagoya, Japan), respectively. Experiments were performed three times and a representative result is presented. Protein masses (kDa) are indicated on the left. (B) Pull-down assay with the N-terminal region (1–186 amino acids) of VIP1 (VIP1 N) and its C-terminal region (165–341 amino acids, VIP1 C). Either GST-fused VIP1 N (GST-VIP1 N) or GST-fused VIP1 C (GST-VIP1 C) was bound to the resin and reacted with either His-PP2A-C3 (left panel) or Myc-PP2A-C5 (right). These proteins were then detected as in the panel A. Experiments were performed three times and a representative result is presented. Protein masses (kDa) are indicated on the left.
Together, these results support the idea that VIP1 binds not only the B”-family B subunit of PP2A but also its C subunit. The PP2A C subunit may contribute to stabilizing the interaction between VIP1 and the PP2A holoenzyme containing the B”-family B subunit. PP2A-C3 and PP2A-C4 regulate auxin responses,18 whereas PP2A-C5 binds Arabidopsis chloride channel proteins, AtCLCc and AtCLCb, and regulates salt stress responses.19 VIP1 also regulates auxin responses and salt stress responses.7,9 It should be interesting to examine the physiological relevance of the interaction between VIP1 and the PP2A C subunit.
Funding Statement
This work was supported by the Kato Memorial Bioscience Foundation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Jakoby M, Weisshaar B, Drandouml;ge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F.. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7:1–3. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 2.Tzfira T, Vaidya M, Citovsky V.. VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. Embo J. 2001;20:3596–3607. doi: 10.1093/emboj/20.13.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc Natl Acad Sci U S A. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Krichevsky A, Vaidya M, Tzfira T, Citovsky V. Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc Natl Acad Sci U S A. 2005;102:5733–5738. doi: 10.1073/pnas.0404118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pitzschke A, Djamei A, Teige M, Hirt H. VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc Natl Acad Sci U S A. 2009;106:18414–18419. doi: 10.1073/pnas.0905599106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Y, Zhao Q, Gao L, Yu XM, Fang P, Oliver DJ, Xiang CB. Isolation and characterization of low-sulphur-tolerant mutants of Arabidopsis. J Exp Bot. 2010;61:3407–3422. doi: 10.1093/jxb/erq161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapham R, Lee LY, Tsugama D, Lee S, Mengiste T. Gelvin SB VIP1 and its homologs are not required for agrobacterium-mediated transformation, but play a role in botrytis and salt stress responses. Front Plant Sci. 2018;9:749. doi: 10.3389/fpls.2018.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsugama D, Liu S, Takano T. A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol. 2012;159:144–155. doi: 10.1104/pp.112.197020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsugama D, Liu S, Takano T. The bZIP protein VIP1 is involved in touch responses in Arabidopsis roots. Plant Physiol. 2016;171:1355–1365. doi: 10.1104/pp.16.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsugama D, Liu S, Takano T. VIP1 is very important/interesting protein 1 regulating touch responses of Arabidopsis. Plant Signal Behav. 2016;11:e1187358. doi: 10.1080/15592324.2016.1187358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsugama D, Liu S, Fujino K, Takano T. Calcium signalling regulates the functions of the bZIP protein VIP1 in touch responses in Arabidopsis thaliana. Ann Bot. 2018;122:1219–1229. doi: 10.1093/aob/mcy125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeo K, Ito T. Subcellular localization of VIP1 is regulated by phosphorylation and 14-3-3 proteins. FEBS Lett. 2017;591:1972–1981. doi: 10.1002/1873-3468.12686. [DOI] [PubMed] [Google Scholar]
- 13.Tsugama D, Yoon HS, Fujino K, Liu S, Takano T. Protein phosphatase 2A regulates the nuclear accumulation of the Arabidopsis bZIP protein VIP1 under hypo-osmotic stress. J Exp Bot. 2019. doi: 10.1093/jxb/erz384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31:853–873. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E, Yu JW, Strack S, Jeffrey PD, Shi Y. Structure of the protein phosphatase 2A holoenzyme. Cell. 2006;127:1239–1251. doi: 10.1016/j.cell.2006.11.033. [DOI] [PubMed] [Google Scholar]
- 16.Cho US, Xu W. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature. 2007;445:53–57. doi: 10.1038/nature05351. [DOI] [PubMed] [Google Scholar]
- 17.Wlodarchak N, Guo F, Satyshur KA, Jiang L, Jeffrey PD, Sun T, Stanevich V, Mumby MC, Xing Y. Structure of the Ca2+-dependent PP2A heterotrimer and insights into Cdc6 dephosphorylation. Cell Res. 2013;23:931–946. doi: 10.1038/cr.2013.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ballesteros I, Domínguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartin M, Sánchez‐Serrano JJ. Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J. 2013;73:862–872. doi: 10.1111/tpj.12078. [DOI] [PubMed] [Google Scholar]
- 19.Hu R, Zhu Y, Wei J, Chen J, Shi H, Shen G, Zhang H. Overexpression of PP2A‐C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions. Plant Cell Environ. 2017;40:150–164. doi: 10.1111/pce.12837. [DOI] [PubMed] [Google Scholar]


