ABSTRACT
Cool-associated tyrosine phosphorylated protein 1 (Cat1), also referred to as GPCR-kinase interacting protein 1 (Git1), is a ubiquitously expressed, multi-domain protein that is best known for regulating cell shape and migration. Cat1/Git1 functions as a GTPase activating protein (GAP) that inactivates certain members of the ADP-ribosylation factor (Arf) family of small GTPases. It is also a scaffold that brings together several signaling proteins at specific locations within the cell, ensuring their efficient activation. Here we will discuss what is known regarding the classical role of Cat1/Git1 in the regulation of cell morphology and migration, as well as highlight some more recent findings that suggest this interesting signaling/scaffolding protein may also contribute in unexpected ways to oncogenic transformation.
KEYWORDS: Cat1, Git1, Arf, GAP, Cool, Pix, cancer, migration
Cat1/Git1 negatively regulates Arf small GTPase activity
Cells respond to cues that they receive from their surroundings by inducing signaling events that often culminate in the activation of small GTP-binding proteins (GTPases), a diverse family of proteins that are defined by their ability to cycle between an inactive GDP-bound state, and an active GTP-bound state. When properly regulated, small GTPases elicit signaling events that mediate a broad spectrum of cellular processes including changes in cell attachment and shape, migration, intracellular vesicle and protein trafficking, gene expression, cell growth, and even apoptosis. However, dysregulation of the signaling capabilities of these same proteins has been shown to hijack “normal” cellular processes, giving rise to pathological conditions such as cancer. Thus, understanding how small GTPases are regulated and function in different contexts continues to be a major research emphasis.
There are 2 major classes of proteins that directly impact small GTPase activity; specifically, guanine nucleotide exchange factor proteins (GEFs) and GTPase activating proteins (GAPs). GEFs function to activate small GTPases by catalyzing GDP-GTP exchange, while GAPs terminate their signaling activity by stimulating the hydrolysis of GTP to GDP. A rather interesting example of a GAP is Cat1/Git1. This ∼95 kDa protein has an N-terminal GAP-domain containing a consensus zinc finger motif (Fig. 1, GAP Domain) that is required for the ability of Cat1/Git1 to catalyze the hydrolysis of GTP to GDP on certain members of the Arf family of small GTPases, namely Arf1 and Arf6.1,2
Figure 1.
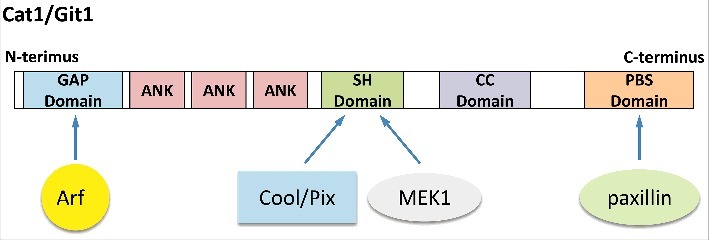
Diagram showing the different structural domains in Cat1/Git1. Starting at its N-terminus, Cat1/Git1 has a GAP domain, followed by 3 ankyrin repeats (ANK), a Spa-2 homology (SH) domain, a coiled-coil (CC) domain, and a paxillin binding sequence (PBS) domain. Also, included on the diagram are some of the major protein partners of Cat1/Git1 (i.e., Arf, Cool/Pix, MEK1, and paxillin) and where they bind.
Arf1 is localized to the Golgi apparatus where it controls the trafficking of transport vesicles containing proteins that will be secreted between the different compartments of this organelle, as well as between the endoplasmic reticulum and the Golgi apparatus. The GTP-bound form of Arf1 recruits coat protein complex 1 (COP1), adaptor proteins (APs)-1,3, and 4, and Golgi-adaptin ear domain homology, Arf-binding domain (GGA), to initiate the budding of transport vesicles, promoting their movement within Golgi compartments.3,4 GAP-mediated GTP hydrolysis of Arf1 terminates these trafficking events by causing the dissociation, or uncoating, of COP1 from the vesicles.3 On the other hand, Arf6 is expressed primarily along discrete regions of the plasma membrane and is associated with some endosomes.3,4 Consistent with these locations, Arf6 has been shown to play an important role in promoting the endocytosis of cell surface receptors.3 GTP-bound Arf6 stimulates endocytosis by increasing the phosphatidylinositol-4-phosphate 5-Kinase (PIP5K) dependent production of phosphatidylinositol 4,5-bisphosphate (PIP2) at the cell surface,3,5 which promotes the recruitment of endocytic proteins such as AP-2 and clathrin to newly formed sites of endocytosis.3 Inactivation of Arf6 limits endocytosis and even promotes the recycling of some endosomes, together with their corresponding cargo, back to the plasma membrane.3
Increases in Arf1 and Arf6 expression and activation have been observed in several different types of human cancer.6-9 For example, Arf1 expression is elevated in high grade and aggressive gastric tumor samples, compared with lower grade tumors or normal tissue.6 The ectopic expression of Arf1 in lower grade gastric cancer cell lines that express relatively low amounts of Arf1 was sufficient to cause these cells to acquire an aggressive phenotype, including enhanced growth, migration, and invasive activities. Similar findings have been reported for Arf6. The levels of this small GTPase were found to be highly upregulated in several aggressive and metastatic cancer cells such as breast, brain and prostate cancer cells.10-12 Knocking-down its expression in these cells using siRNAs, or introducing a mutant form of Arf6 that is unable to bind GTP, suppressed their invasive and metastatic behaviors.10,11 These findings, when combined with the fact that ectopically expressing activating mutants of Arf1 and Arf6 together in non-transformed NIH3T3 fibroblasts induces their ability to form colonies in soft agar (i.e. anchorage-independent growth),13 an in vitro read-out of tumorigenicity, suggest that increasing the expression and/or activation of at least some Arf family members potentially play important roles in several different aspects of cancer progression.
Based on these findings, it is reasonable to speculate that proteins that inactivate Arf1 and Arf6, such as GAPs like Cat1/Git1, would function to limit the cancer promoting actions of these small GTPases. Indeed, this seems to be at least partially correct, as expression of a point mutant of Cat1/Git1 that interferes with its ability to act as a GAP (Cat1/Git1 R39A) in HeLa cervical carcinoma cells not only led to increased levels of activated, GTP-bound Arf1 and Arf6, but also strongly enhanced their ability to form colonies in soft agar, compared with the parental HeLa cell line.13 However, there also appears to be more to the story, as immunohistochemistry performed on 80 cervical cancer sections and 20 normal cervical tissue samples revealed that an overwhelming majority (i.e., ∼95%) of the tumor samples overexpressed Cat1/Git1.13 Increases in the levels of Cat1/Git1 has also been observed in several different advanced-stage breast cancer cell lines.14 We discovered that depleting Cat1/Git1 expression in cervical cancer cells, or in fibroblasts that have been transformed through the expression of an activated form of the small GTPase Cdc42 (Cdc42 F28L), using siRNAs, caused both cell types to lose their colony forming capabilities.13 Moreover, knocking-down Cat1/Git1 expression in several cancer cells that were selected for their highly invasive and metastatic activity was shown to revert these phenotypes.14-19 Thus, these findings highlight 2 critical points that will be the focus of the remainder of this review. First, Cat1/Git1 must have roles other than simply functioning as an Arf GAP. Second, the regulation and functional consequences of Cat1/Git1 in malignant transformation is currently not well understood and will likely be complex and context dependent. At least in some cases, Cat1/Git1 helps maintain oncogenic transformation by potentially impacting several different aspects of cancer progression, such as promoting invasive and metastatic activity, as well as enhancing cell growth and survival. These findings will be highlighted. However, other lines of evidence suggest that Cat1/Git1 may also act to limit transformed characteristics, i.e., possibly when functioning as Arf GAP. Determining whether Cat1/Git1 helps to maintain or prevents the formation of different types of cancers, as well as identifying the mechanisms through which these effects are mediated, will undoubtedly be a point of emphasis going forward.
Cat1/Git1 is a protein scaffold
In addition to its N-terminal GAP domain, several other domains have been identified in Cat1/Git1 (Fig. 1), including ankyrin repeats (denoted as ANK), a Spa2-homology domain (denoted as the SH Domain), a coiled-coil domain (denoted as the CC Domain), and a C-terminal paxillin binding sequence domain (denoted as the PBS Domain). Each domain has been shown to carry-out out a distinct function. The ankyrin repeats help localize Cat1/Git1 to endosomes,20 while the coiled-coil domain allows for the formation of Cat1/Git1 homodimers.21,22 However, the Spa-2 homology and paxillin binding sequence domains, which have been extensively studied, serve as binding sites for proteins.20,23-26 The identification of these distinct regions and the proteins that bind them, has shed some light on the mechanism through which Cat1/Git1 mediate its effects on cellular transformation. In particular, it is now appreciated that Cat1/Git1 can also function as a protein scaffold that brings together several different proteins, ensuring their proper localization and efficient activation. To date, at least 100 Cat1/Git1 associating proteins have been reported, underscoring the potential of this protein to influence several different signaling pathways and cellular processes.27 Here, we will highlight what is currently known regarding some of the most prominent roles played by Cat1/Git1 as a scaffold. It is important to keep in mind that, although each of the distinct functions of Cat1/Git1 will be described separately, some of them are most likely occurring simultaneously, while others act antagonistically, to mediate specific biologic effects.
The Cool-Cat1/Git1 complex- Cat1/Git1 was initially identified in our laboratory almost 20 y ago as a tyrosine phosphorylated binding partner for a GEF called cloned-out-of-library (Cool), hence the name Cool-associated tyrosine phosphorylated protein, or Cat.23 The same protein and additional family members were identified by Premont and colleagues (Git),28 Turner and colleagues (Pkl for paxillin kinase linker),26 and Di Cesare and colleagues (p95-APP for p95-ADP ribosylation factor GTPase-activating protein).20 Cool, which is also referred to as p21-activated kinase-interacting exchange factor (Pix), is a GEF for Cdc42 and Rac,29,30 2 members of the Rho family of small GTPases. It binds to Cat1/Git1 through the Spa2-homology domain,24 and Cool-Cat1/Git1 complexes are localized throughout the cytosol, along the cell membrane, and especially to focal adhesions, distinct structures that physically connect a cell to its extracellular environment (focal adhesions will be discussed in more detail later).31 Because both Cool and Cat1/Git1 can form homodimers, it is thought that Cool-Cat1/Git1 complexes assemble into large oligomeric complexes in cells as a mechanism to promote signal amplification.27,32,33
The localization of Cool-Cat1/Git1 complexes to the cell surface has been shown to be important for mediating G-protein coupled receptor- and growth factor receptor-induced activation of Cdc42 and Rac, as well as for the regulation of Arf GTPase activation.27,31 Activation of these signaling pathways lead to cytoskeletal rearrangements which are required for promoting cancer cell migration and invasion, and enhancement of cell growth.27,31 The ectopic expression of a mutant form of Cat1/Git1 that is unable to associate with Cool abolishes these effects, highlighting the importance of this protein-protein interaction.34,35
The MEK1-ERK1/2-Cat1/Git1 complex- Cat1/Git1 has also been shown to promote the activation of the mitogen activated protein kinase ERK1/2.36 This is an outcome of a complex that can form at focal adhesions between Cat1/Git1, MEK1.36 (i.e., the kinase that phosphorylates ERK1/2), and ERK1/2.37 Similar to Cool, MEK1 binds to the Spa-2 homology domain of Cat1/Git1, an interaction that is enhanced when Cat1/Git1 is phosphorylated by the non-receptor tyrosine kinase c-Src.36,37 While it is not entirely clear how Cat1/Git1 binds to ERK1/2, this interaction is augmented following the treatment of cells with epidermal growth factor (EGF).36 Interestingly, knocking-down Cat1/Git1 expression levels in vascular smooth-muscle cells, or human embryonic kidney 293 cells, using siRNAs was sufficient to block EGF- and c-Src-mediated ERK activation and cell migration.36,37 On the other hand, the ectopic expression of Cat1/Git1 in these same cells was found to extend EGF-induced ERK activation.36 These findings suggested for the first time that Cat1/Git1, at least in certain contexts, could function as a signaling scaffold that connects growth factor receptor signaling to the ERK pathway. They also raised the intriguing possibility that the ability of Cat1/Git1 to promote ERK activation might also contribute to tumor expansion, since the Ras-ERK pathway is known to stimulate cell growth. Indeed, it was recently shown that Cat1/Git1 mediated activation of ERK was responsible for increasing the rates of tumor formation in a liver cancer model.16,38
The Paxillin-Cat1/Git1 complex- Focal adhesions are unique and dynamic structures found along the plasma membrane.39 They use integrins to physically connect the cell to the extracellular matrix and consist of several additional proteins which are important for helping maintaining cell attachment.39 Focal adhesions also can assemble and disassemble, or turnover, in a coordinated manner. This is one of the principal mechanisms underlying cell migration.39 However, focal adhesions also serve as signaling hubs that transduce signals initiated by cellular interactions with the extracellular environment.40 In addition to regulating cell attachment and motility, these signaling events have been shown to increase the rates of cell growth and survival.41
Paxillin is one of the major constituents of focal adhesions and is known to regulate cell spreading and cell migration.42 It functions as a scaffold/adaptor protein and is responsible for the recruitment of several different proteins to focal adhesions via their association with one of paxillin's 5 N-terminal leucine-rich (LD) motifs and 4 C-terminal double-zinc finger motifs, referred to as LIM domains.42 The fourth LD domain of paxillin (i.e., the LD4 domain) was shown to bind to the C-terminal region of Cat1/Git1.26 The resulting paxillin-Cat1/Git1 interaction causes Cat1/Git1 to localize to focal adhesions, while ectopic expression of mutant forms of Cat1/Git1 that lack the paxillin binding domain are no longer enriched in these structures.43,44
In a series of eloquent studies performed by several different groups, it was shown how interfering with the ability of Cat1/Git1 to localize to focal adhesions had a dramatic effect on cell attachment and migration. In one such study, the ectopic expression of a Cat1/Git1 mutant defective in its ability to bind paxillin, while retaining its other activities, in CHO.K1 fibroblasts, was found to be sufficient to cause aberrant cell spreading43 and blocked the ability of cells to migrate into a wound.45 Expression of a mutant form of paxillin that is unable to bind Cat1/Git1 in cells yielded similar results.43,45 Based on these findings, it was thought that the exclusion of Cat1/Git1 from focal adhesions might affect localized signaling events that are important for mediating these effects. This idea turned out to be correct, as subsequent discoveries showed that by failing to associate with paxillin, Cat1/Git1 is not able to help mediate the Cool/Pix dependent activation of Cdc42 and Rac1.43,46-49 This would in turn compromise the activation of the serine-threonine kinase p21-activated kinase (PAK) at focal adhesions,50,51 which is a key event for triggering cytoskeletal rearrangements and promoting cell motility.50-54 It is also worth noting that immunofluorescent experiments have revealed that both the Cool-Cat1/Git1 complex and the MEK1-ERK1/2-Cat1/Git1 complex are enriched in focal adhesions.37 Thus, it is interesting to consider the possibility that the localization of Cat1/Git1 to these structures enables it to mediate the efficient and localized activation of signaling events needed to elicit biologic effects (i.e., cell attachment and migration).
The mTOR-Cat1/Git1 complex- Deregulation of the phosphatidylinositol-3-kinase (PI3 kinase)/AKT signaling pathway is a common occurrence in human malignancies.55,56 This pathway is initiated when the activated form of PI3 kinase generates phosphatidylinositol 3,4,5-trisphosphate (PIP3) from phosphatidylinositol 4,5-bisphosphate (PIP2) along the cell surface. Phosphoinositide-dependent kinase 1 (PDK1) is then recruited to the plasma membrane where it activates AKT. AKT, in turn, prevents tuberous sclerosis complex 2 (TSC2) from acting as a GAP for the small GTPase Ras homolog enriched in brain (Rheb), leading to increases in the levels of the GTP-bound form of Rheb. Activated Rheb binds and activates the mechanistic target of rapamycin (mTOR), specifically, when it is interacting with Raptor and PRAS40 as part of a protein complex referred to as mTOR complex 1 (mTORC1). Activation of mTORC1 induces the phosphorylation of p70S6 kinase, resulting in the synthesis of proteins that stimulate cell growth and survival. It is also worth noting that mTOR can interact with an additional set of proteins, including Rictor and mSIN1, to form mTOR complex 2 (mTORC2).56 This protein complex is functionally distinct from mTORC1 and is best known for its ability to mediate cytoskeletal rearrangements through the activation of protein kinase C-α (PKCα), as well as members of the Rho family of small GTPases. mTORC2 has also been shown to promote cell survival by phosphorylating AKT.56,57
Cat1/Git1 was recently identified in a screen as a novel mTOR-binding partner in astrocytes.58 The authors showed that the ability of Cat1/Git1 to co-immunoprecipitate with mTOR was dependent on AKT activity, and that the interaction between these 2 proteins was important for mediating the survival of astrocytes. However, what made the findings from this study especially intriguing was the condition under which the interaction between Cat1/Git1 and mTOR appears to occur. Rather than co-immunoprecipitating with mTOR as part of either mTORC1 or mTORC2, Cat1/Git1 interacts with mTOR to form a unique complex that does not include the core components of either mTORC1 or mTORC2, specifically Raptor and Rictor. These findings suggest that Cat1/Git1 can play an important role in regulating the PI3 kinase/AKT/mTOR signaling pathway by forming a novel protein complex in at least some cell types (i.e., astrocytes).
Cat1/Git1 in cancer progression
The protein levels of Cat1/Git1 are frequently upregulated in high grade and aggressive forms of several different types of cancer cells including cervical,13 breast,14,18 liver,59 lung,15 kidney60 and oral cancers.17 At least one mechanism which can account for these changes in Cat1/Git1 expression has been discovered and it involves microRNAs. To date, 4 distinct microRNAs that target Cat1/Git1 have been identified. Each of these microRNAs is expressed at relatively high levels in normal cells and low-grade cancer cells, and function to keep Cat1/Git1 levels in check. However, in high grade cancer cells with invasive and metastatic activities, the microRNAs targeting Cat1/Git1 are downregulated and lead to increases in Cat1/Git1 expression.14,17-19 Reversing this effect in some highly aggressive breast cancer, oral squamous cancer and lung cancer cell lines, through the introduction of siRNAs or shRNAs that specifically target Cat1/Git1, caused the cells to lose their invasive and metastatic capabilities.14,17-19 Since cancer cell invasion and metastasis rely heavily on cell migration, and because Cat1/Git1 is a well-established promoter of cell motility, one can appreciate how this protein exerts a major influence on the aggressive behavior of cancer cells.
However, there is also a growing number of recent findings that implicate Cat1/Git1 as an important contributor to cancer cell growth and survival. For example, Cat1/Git1, functioning as a scaffold for MEK1 and ERK1/2, was recently shown to be essential for mediating the activation of ERK1/2 by methionine adenosyltransferase 2B (MAT2B) in liver and colon cancer models.16,38 The authors went on to show that the formation of this complex had important consequences on tumor growth. Namely, liver tumors with increased levels of Cat1/Git1 and MAT2B had more ERK1/2 activity and grew faster, compared with those tumors without elevated levels of these proteins.16,38
We also have discovered Cat1/Git1 plays an essential role in promoting the transformed phenotype of HeLa cervical carcinoma cells and fibroblasts transformed through the expression of an activated Cdc42 mutant, i.e., Cdc42 F28L.13 Knocking-down Cat1/Git1 expression from either of these cell types using siRNAs strongly inhibited their ability to form colonies in soft agar. While introduction of an siRNA-insensitive form of wild-type Cat1/Git1 in HeLa cells expressing Cat1/Git1 siRNA completely rescued this effect, a siRNA-insensitive mutant form of Cat-/Git1, defective in its ability to bind paxillin, was ineffective. Because paxillin has been shown to mediate the growth of some cancer and transformed cell lines,61 we initially suspected this meant that the formation of paxillin-Cat1/Git1 complexes, most likely at focal adhesions, might recruit and activate a unique set of proteins essential for the growth of cancer/transformed cells.
However, in our most recent study,62 where we set out to learn more about how paxillin-Cat1/Git1 interactions mediate cellular transformation, we discovered that our initial idea was incorrect. Instead of the formation of a complex between paxillin and Cat1/Git1 helping to induce a stimulatory signal that promotes colony formation in soft agar, it appears that the ability of Cat1/Git1 to associate with paxillin prevents it from exerting a negative regulatory effect on this transformed phenotype. The first indication that this was likely to be the case came from an experiment where it was shown that HeLa cells transfected with siRNAs targeting paxillin consistently performed better in soft agar assays (i.e., they formed more colonies), compared with the same cells expressing a control siRNA.62 Moreover, the inability of HeLa cells, depleted of Cat1/Git1 expression using siRNAs, to form colonies in soft agar could be restored if paxillin expression was also knocked-down in the cells.
We then went on to uncover the mechanism through which Cat1/Git1 helps maintain this transformed phenotype. It involves Cat1/Git1 binding paxillin and, presumably functioning as a GAP, to counteract the ability of paxillin to increase the levels of active, GTP-bound Arf1 in cells (Fig. 2A). As long as paxillin-mediated Arf1 activity is kept in check by Cat1/Git1, it is unable to stimulate the activation of mTOR complex 1 (mTORC1) and its downstream effector p70S6 kinase.55,56 This, in turn, prevents p70S6 kinase from negatively regulating the PI3-kinase/AKT pathway, thereby ensuring that enough AKT activity is maintained in the cancer/transformed cells to allow them to form colonies in soft agar.62 However, when Cat1/Git1 expression is knocked-down in cancer cells, a delicate balance is disrupted and paxillin is now capable of activating the Arf1-mTORC1-P70S6 kinase pathway (Fig. 2B). In this situation, sufficient levels of PI3-kinase/AKT activity are not achieved, causing the cancer cells to lose their transformed characteristics. For additional information regarding these findings, please see our manuscript.62
Figure 2.
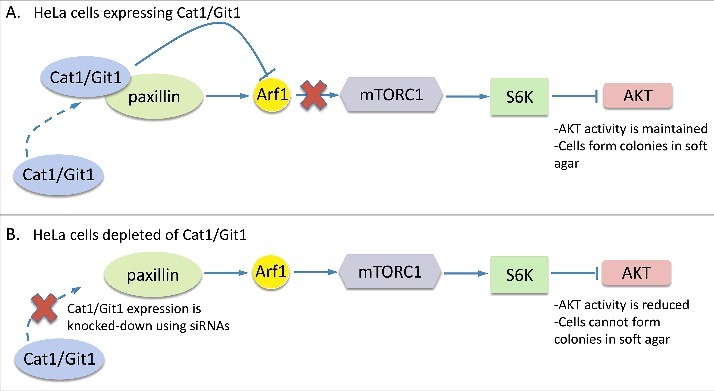
Diagram showing the mechanism through which Cat1/Git1 promotes HeLa cervical carcinoma cell transformation. A) When expressed in HeLa cells, Cat1/Git1 associates with paxillin to prevent it from functioning as a negative regulator of cellular transformation. Specifically, the interaction between Cat1/Git1 and paxillin inhibits the ability of paxillin to activate the Arf1-mTORC1-P70S6 kinase (S6K) pathway, which ensures that the necessary level of AKT activation required for supporting soft agar colony formation is met. B) However, under conditions where Cat1/Git1 expression is knocked-down in HeLa cells using siRNAs, paxillin is now able to stimulate the activation of the Arf1-mTORC1-P70S6 kinase pathway and inhibit AKT activation. These cells are no longer capable of forming colonies in soft agar.
Concluding remarks
Cat1/Git1 is a GAP for Arf1 and Arf6, as well as a protein scaffold/adaptor that binds several different protein partners. It is best known for its roles in promoting cell attachment and migration. However, during the past few years, Cat1/Git1 has been attracting a good deal of attention for its ability to impact several unique aspects of cancer progression. For example, Cat1/Git1 is frequently overexpressed in advanced-stage and highly aggressive forms of several types of cancer, where it promotes their invasive and metastatic activities. It is also becoming increasingly clear that Cat1/Git1, at least in certain contexts, contributes to cellular transformation by stimulating the activation of ERK1/2 and AKT, and promoting the anchorage-independent growth and survival of cancer cells.
While our understanding of the contribution of Cat1/Git1 to cancer progression is still in its infancy, it seems clear that, at least in certain contexts, it plays important roles in the development of this disease. Because of the ability of Cat1/Git1 to interact with several different proteins, as well as regulate a wide range of cellular processes, it seems likely that this interesting signaling/scaffold protein will emerge as a major player in cancer biology.
Disclosure of potential conflicts of interest
The authors report no conflict of interest.
Funding
This work was supported by the NIH under grants GM040654 and GM047458.
References
- [1].Vitale N, Patton WA, Moss J, Vaughan M, Lefkowitz RJ, Premont RT. GIT proteins, a novel family of phosphatidylinositol 3,4,5- trisphosphate-stimulated GTPase-activating proteins for ARF6. J Biol Chem. 2000;275(18):13901-6. doi: 10.1074/jbc.275.18.13901. PMID:10788515 [DOI] [PubMed] [Google Scholar]
- [2].Mazaki Y, Hashimoto S, Okawa K, Tsubouchi A, Nakamura K, Yagi R, Yano H, Kondo A, Iwamatsu A, Mizoguchi A, et al.. An ADP-ribosylation factor GTPase-activating protein Git2-short/KIAA0148 is involved in subcellular localization of paxillin and actin cytoskeletal organization. Mol Biol Cell. 2001;12(3):645-62. doi: 10.1091/mbc.12.3.645. PMID:11251077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].D'Souza-Schorey C, Chavrier P. ARF proteins: Roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7(5):347-58. doi: 10.1038/nrm1910. PMID:16633337 [DOI] [PubMed] [Google Scholar]
- [4].Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005;33(Pt 4):639-42. doi: 10.1042/BST0330639. PMID:16042562 [DOI] [PubMed] [Google Scholar]
- [5].Martin TF. PI(4,5)P(2) regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13(4):493-9. doi: 10.1016/S0955-0674(00)00241-6. PMID:11454457 [DOI] [PubMed] [Google Scholar]
- [6].Tsai MM, Lin PY, Cheng WL, Tsai CY, Chi HC, Chen CY, Tseng YH, Cheng YF, Chen CD, Liang Y, et al.. Overexpression of ADP-ribosylation factor 1 in human gastric carcinoma and its clinicopathological significance. Cancer Sci. 2012;103(6):1136-44. doi: 10.1111/j.1349-7006.2012.02243.x. PMID:22348287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Xie X, Tang SC, Cai Y, Pi W, Deng L. Suppression of breast cancer metastasis through the inactivation of ADP-ribosylation factor 1. Oncotarget. 2016;7(36):58111-20. doi: 10.18632/oncotarget.11185. PMID:27517156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Davis JE, Xie X, Guo J, Huang W, Chu WM, Huang S, Teng Y, Wu G, Davis JE, Xie X, et al.. ARF1 promotes prostate tumorigenesis via targeting oncogenic MAPK signaling. Oncotarget. 2016;7(26):39834-45. doi: 10.18632/oncotarget.9405. PMID:27213581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gu G, Chen Y, Duan C, Zhou L, Chen C, Chen J, Cheng J, Shi N, Jin Y, Xi Q, et al.. Overexpression of ARF1 is associated with cell proliferation and migration through PI3K signal pathway in ovarian cancer. Oncol Rep. 2017;37(3):1511-20. doi: 10.3892/or.2017.5388. PMID:28098897 [DOI] [PubMed] [Google Scholar]
- [10].Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H. Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci U S A. 2004;101(17):6647-52. doi: 10.1073/pnas.0401753101. PMID:15087504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hu B, Shi B, Jarzynka MJ, Yiin JJ, D'Souza-Schorey C, Cheng SY. ADP-ribosylation factor 6 regulates glioma cell invasion through the IQ-domain GTPase-activating protein 1-Rac1-mediated pathway. Cancer Res. 2009;69(3):794-801. doi: 10.1158/0008-5472.CAN-08-2110. PMID:19155310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morgan C, Lewis PD, Hopkins L, Burnell S, Kynaston H, Doak SH. Increased expression of ARF GTPases in prostate cancer tissue. Springerplus. 2015;4:342. doi: 10.1186/s40064-015-1136-y. PMID:26185744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yoo SM, Antonyak MA, Cerione RA. The adaptor protein and Arf GTPase-activating protein Cat1/Git1 is required for cellular transformation. J Biol Chem. 2012;287(37):31462-70. doi: 10.1074/jbc.M112.353615. PMID:22807447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chan SH, Huang WC, Chang JW, Chang KJ, Kuo WH, Wang MY, Lin KY, Uen YH, Hou MF, Lin CM, et al.. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33(36):4496-507. doi: 10.1038/onc.2014.10. PMID:24608434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chang JS, Su CY, Yu WH, Lee WJ, Liu YP, Lai TC, Jan YH, Yang YF, Shen CN, Shew JY, et al.. GIT1 promotes lung cancer cell metastasis through modulating Rac1/Cdc42 activity and is associated with poor prognosis. Oncotarget. 2015;6(34):36278-91. doi: 10.18632/oncotarget.5531. PMID:26462147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Peng H, Li TW, Yang H, Moyer MP, Mato JM, Lu SC. Methionine adenosyltransferase 2B-GIT1 complex serves as a scaffold to regulate Ras/Raf/MEK1/2 activity in human liver and colon cancer cells. Am J Pathol. 2015;185(4):1135-44. doi: 10.1016/j.ajpath.2014.12.016. PMID:25794709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Huang WC, Chan SH, Jang TH, Chang JW, Ko YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al.. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74(3):751-64. doi: 10.1158/0008-5472.CAN-13-1297. PMID:24335959 [DOI] [PubMed] [Google Scholar]
- [18].Tao WY, Wang CY, Sun YH, Su YH, Pang D, Zhang GQ. MicroRNA-34c suppresses breast cancer migration and invasion by targeting GIT1. J Cancer. 2016;7(12):1653-62. doi: 10.7150/jca.14762. PMID:27698902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19(12):2793-805. doi: 10.1111/jcmm.12666. PMID:26283050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Di Cesare A, Paris S, Albertinazzi C, Dariozzi S, Andersen J, Mann M, Longhi R, de Curtis I. p95-APP1 links membrane transport to Rac-mediated reorganization of actin. Nat Cell Biol. 2000;2(8):521-30. doi: 10.1038/35019561. PMID:10934473 [DOI] [PubMed] [Google Scholar]
- [21].Kim S, Ko J, Shin H, Lee JR, Lim C, Han JH, Altrock WD, Garner CC, Gundelfinger ED, Premont RT, et al.. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J Biol Chem. 2003;278(8):6291-300. doi: 10.1074/jbc.M212287200. PMID:12473661 [DOI] [PubMed] [Google Scholar]
- [22].Paris S, Longhi R, Santambrogio P, de Curtis I. Leucine-zipper-mediated homo- and hetero-dimerization of GIT family p95-ARF GTPase-activating protein, PIX-, paxillin-interacting proteins 1 and 2. Biochem J. 2003;372(Pt 2):391-8. doi: 10.1042/bj20030047. PMID:12611588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bagrodia S, Bailey D, Lenard Z, Hart M, Guan JL, Premont RT, Taylor SJ, Cerione RA. A tyrosine-phosphorylated protein that binds to an important regulatory region on the cool family of p21-activated kinase-binding proteins. J Biol Chem. 1999;274(32):22393-400. doi: 10.1074/jbc.274.32.22393. PMID:10428811 [DOI] [PubMed] [Google Scholar]
- [24].Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol. 2000;20(17):6354-63. doi: 10.1128/MCB.20.17.6354-6363.2000. PMID:10938112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Premont RT, Claing A, Vitale N, Perry SJ, Lefkowitz RJ. The GIT family of ADP-ribosylation factor GTPase-activating proteins. Functional diversity of GIT2 through alternative splicing. J Biol Chem. 2000;275(29):22373-80. doi: 10.1074/jbc.275.29.22373. PMID:10896954 [DOI] [PubMed] [Google Scholar]
- [26].Turner CE, Brown MC, Perrotta JA, Riedy MC, Nikolopoulos SN, McDonald AR, Bagrodia S, Thomas S, Leventhal PS. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: A role in cytoskeletal remodeling. J Cell Biol. 1999;145(4):851-63. doi: 10.1083/jcb.145.4.851. PMID:10330411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhou W, Li X, Premont RT. Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J Cell Sci. 2016;129(10):1963-74. doi: 10.1242/jcs.179465. PMID:27182061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Premont RT, Claing A, Vitale N, Freeman JL, Pitcher JA, Patton WA, Moss J, Vaughan M, Lefkowitz RJ. β2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci U S A. 1998;95(24):14082-7. doi: 10.1073/pnas.95.24.14082. PMID:9826657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1(2):183-92. doi: 10.1016/S1097-2765(00)80019-2. PMID:9659915 [DOI] [PubMed] [Google Scholar]
- [30].Bagrodia S, Taylor SJ, Jordon KA, Van Aelst L, Cerione RA. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273(37):23633-6. doi: 10.1074/jbc.273.37.23633. PMID:9726964 [DOI] [PubMed] [Google Scholar]
- [31].Frank SR, Hansen SH. The PIX–GIT complex: A G protein signaling cassette in control of cell shape. Semin Cell Dev Biol. 2008;19(3):234-44. doi: 10.1016/j.semcdb.2008.01.002. PMID:18299239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: An oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16(9):1001–11. doi: 10.1016/S0898-6568(04)00023-3. PMID:15212761 [DOI] [PubMed] [Google Scholar]
- [33].Schlenker O, Rittinger K. Structures of dimeric GIT1 and trimeric β-PIX and implications for GIT–PIX complex assembly. J Mol Biol. 2009;386(2):280–9. doi: 10.1016/j.jmb.2008.12.050. PMID:19136011 [DOI] [PubMed] [Google Scholar]
- [34].Feng Q, Baird D, Yoo S, Antonyak M, Cerione RA. Phosphorylation of the Cool-1/β-Pix protein serves as a regulatory signal for the migration and invasive activity of Src-transformed cells. J Biol Chem. 2010;285(24):18806–16. doi: 10.1074/jbc.M109.098079. PMID:20375009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Loo TH, Ng YW, Lim L, Manser E. GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol Cell Biol. 2004;24(9):3849–59. doi: 10.1128/MCB.24.9.3849-3859.2004. PMID:15082779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yin G, Haendeler J, Yan C, Berk BC. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol Cell Biol. 2004;24(2):875–85. doi: 10.1128/MCB.24.2.875-885.2004. PMID:14701758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang N, Cai W, Yin G, Nagel DJ, Berk BC. GIT1 is a novel MEK1-ERK1/2 scaffold that localizes to focal adhesions. Cell Biol Int. 2009;34(1):41–7. doi: 10.1042/CBI20090016. PMID:19947948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peng H, Dara L, Li TW, Zheng Y, Yang H, Tomasi ML, Tomasi I, Giordano P, Mato JM, Lu SC. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology. 2013;57(6):2299–313. doi: 10.1002/hep.26258. PMID:23325601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: Integrating signals from front to back. Science. 2003;302(5651):1704–9. doi: 10.1126/science.1092053. PMID:14657486 [DOI] [PubMed] [Google Scholar]
- [40].Schwartz MA, Schaller MD, Ginsberg MH. Integrins: Emerging paradigms of signal transduction. Annu Rev Cell Dev Biol. 1995;11:549–99. doi: 10.1146/annurev.cb.11.110195.003001. PMID:8689569 [DOI] [PubMed] [Google Scholar]
- [41].Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285(5430):1028–32. doi: 10.1126/science.285.5430.1028. PMID:10446041 [DOI] [PubMed] [Google Scholar]
- [42].Deakin NO, Turner CE. Paxillin comes of age. J Cell Sci. 2008;121(Pt 15):2435–44. doi: 10.1242/jcs.018044. PMID:18650496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].West KA, Zhang H, Brown MC, Nikolopoulos SN, Riedy MC, Horwitz AF, Turner CE. The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL). J Cell Biol. 2001;154(1):161–76. doi: 10.1083/jcb.200101039. PMID:11448998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brown MC, West KA, Turner CE. Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol Biol Cell. 2002;13(5):1550–65. doi: 10.1091/mbc.02-02-0015. PMID:12006652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu JA, Deakin NO, Turner CE. Paxillin-kinase-linker tyrosine phosphorylation regulates directional cell migration. Mol Biol Cell. 2009;20(22):4706–19. doi: 10.1091/mbc.E09-07-0548. PMID:19776348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nishiya N, Kiosses WB, Han J, Ginsberg MH. An α4 integrin–paxillin–Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat Cell Biol. 2005;7(4):343–52. doi: 10.1038/ncb1234. PMID:15793570 [DOI] [PubMed] [Google Scholar]
- [47].Jones CA, Nishiya N, London NR, Zhu W, Sorensen LK, Chan AC, Lim CJ, Chen H, Zhang Q, Schultz PG, et al.. Slit2–Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat Cell Biol. 2009;11(11):1325–31. doi: 10.1038/ncb1976. PMID:19855388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D'Souza-Schorey C, Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18(6):1480–91. doi: 10.1093/emboj/18.6.1480. PMID:10075920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154(3):599–610. doi: 10.1083/jcb.200104019. PMID:11481345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sells MA, Knaus UG, Bagrodia S, Ambrose DM, Bokoch GM, Chernoff J. Human p21-activated kinase (Pak1) regulates actin organization in mammalian cells. Curr Biol. 1997;7(3):202–10. doi: 10.1016/S0960-9822(97)70091-5. PMID:9395435 [DOI] [PubMed] [Google Scholar]
- [51].Bokoch GM. Biology of the p21-Activated Kinases. Annu Rev Biochem. 2003;72:743–81. doi: 10.1146/annurev.biochem.72.121801.161742. PMID:12676796 [DOI] [PubMed] [Google Scholar]
- [52].Nayal A, Webb DJ, Brown CM, Schaefer EM, Vicente-Manzanares M, Horwitz AR. Paxillin phosphorylation at Ser273 localizes a GIT1–PIX–PAK complex and regulates adhesion and protrusion dynamics. J Cell Biol. 2006;173(4):587–9. doi: 10.1083/jcb.200509075. PMID:16717130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol. 2003;161(1):131–42. doi: 10.1083/jcb.200211002. PMID:12695502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25(13):3379–88. doi: 10.1523/JNEUROSCI.3553-04.2005. PMID:15800193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25(9):545–55. doi: 10.1016/j.tcb.2015.06.002. PMID:26159692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–71. doi: 10.1016/j.cell.2017.03.035. PMID:28388417 [DOI] [PubMed] [Google Scholar]
- [57].Vadlakonda L, Dash A, Pasupuleti M, Anil Kumar K, Reddanna P. The paradox of Akt-mTOR interactions. Front Oncol. 2013;3:165. doi: 10.3389/fonc.2013.00165. PMID:23802099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Smithson LJ, Gutmann DH. Proteomic analysis reveals GIT1 as a novel mTOR complex component critical for mediating astrocyte survival. Genes Dev. 2016; 30(12):1383–8. doi: 10.1101/gad.279661.116. PMID:27340174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen J, Yang P, Yang J, Wen Z, Zhang B, Zheng X. GIT1 is a novel prognostic biomarker and facilitates tumor progression via activating ERK/MMP9 signaling in hepatocellular carcinoma. Onco Targets Ther. 2015;8:3731–42. doi: 10.2147/OTT.S96715. PMID:26719701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lu X, Wan F, Zhang H, Shi G, Ye D. ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumor Biol. 2016;37(1):253–62. doi: 10.1007/s13277-015-3792-5. PMID:26198048 [DOI] [PubMed] [Google Scholar]
- [61].Deakin NO, Pignatelli J, Turner CE. Diverse roles for the paxillin family of proteins in cancer. Genes Cancer. 2012;3(5–6):362–70. doi: 10.1177/1947601912458582. PMID:23226574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yoo SM, Latifkar A, Cerione RA, Antonyak MA. Cool-associated tyrosine-phosphorylated protein 1 is required for the anchorage-independent growth of cervical carcinoma cells by binding paxillin and promoting AKT activation. J Biol Chem. 2017;292(9):3947–57. doi: 10.1074/jbc.M116.769190. PMID:28100775 [DOI] [PMC free article] [PubMed] [Google Scholar]


