Abstract
Temporal regulation of gene expression is a crucial aspect of metazoan development. In the roundworm Caenorhabditis elegans, the heterochronic pathway controls multiple developmental events in a time-specific manner. The most downstream effector of this pathway, the zinc-finger transcription factor LIN-29, acts in the last larval stage (L4) to regulate elements of the larval-to-adult switch. Here, we explore new LIN-29 targets and their implications for this developmental transition. We used RNA-sequencing to identify genes differentially expressed between animals misexpressing LIN-29 at an early time point and control animals. Among 230 LIN-29-activated genes, we found that genes encoding cuticle collagens were overrepresented. Interestingly, expression of lin-29 and some of these collagens was increased in adults with cuticle damage, suggesting a previously unknown function for LIN-29 in adult cuticle maintenance. On the other hand, genes involved in fat metabolism were enriched among 350 LIN-29-downregulated targets. We used mass spectrometry to assay lipid content in animals overexpressing LIN-29 and observed reduced fatty acid levels. Many LIN-29-repressed genes are normally expressed in the intestine, suggesting cell-nonautonomous regulation. We identified several LIN-29 upregulated genes encoding signaling molecules that may act as mediators in the regulation of intestinally expressed genes encoding fat metabolic enzymes and vitellogenins. Overall, our results support the model of LIN-29 as a major regulator of adult cuticle synthesis and integrity, and as the trigger for metabolic changes that take place at the important transition from rapid growth during larval life to slower growth and offspring production during adulthood.
Keywords: Caenorhabditis elegans, heterochronic, gene expression, collagen, metabolism
FOR successful animal development to occur, a large number of cellular events, including the proper regulation of gene expression, must occur in the right place but also at the right time. While much is known about regulation of metazoan development in the spatial dimension, less is known about the equally important temporal coordination of such events. Here we examine the temporal control of gene expression during the last phase of development in the nematode Caenorhabditis elegans.
After embryogenesis is completed inside an eggshell, this ecdysozoan nematode worm goes through four larval stages (L1–L4), molting its outer, collagen-rich cuticle between stages, before becoming an adult that is capable of laying eggs (Altun and Hall 2009). Genetic and molecular analyses have uncovered the heterochronic pathway as the main regulator of developmental timing in C. elegans. This pathway consists of a network of proteins and microRNAs (miRNAs) that interact to control the expression and stability of key transcription factors that regulate developmental events in a stage-specific manner (Nimmo and Slack 2009; Rougvie and Moss 2013; Moss and Romer-Seibert 2014). Mutations in components of this pathway lead to either the precocious or retarded occurrence of stage-specific events, particularly events involved in the development of the single-layer skin of the worm, the hypodermis. Several members of the heterochronic pathway are conserved in vertebrates and control developmental timing and stem cell fate in those organisms as well (Houbaviy et al. 2003; Moss 2007; Su et al. 2012; Ecsedi and Großhans 2013; Worringer et al. 2014; Tsialikas and Romer-Seibert 2015).
The most downstream heterochronic pathway regulator is LIN-29, a Kruppel-family zinc finger transcription factor (Rougvie and Ambros 1995) with homology to mammalian early growth response (EGR) proteins (Harris and Horvitz 2011). lin-29 function is required for a number of developmental events that take place in the L4 stage in coordination with the worm’s transition from larval to adult life. Some of these LIN-29-regulated events include the formation of the adult cuticle, the terminal differentiation and fusion of the lateral hypodermal cells (also called seam cells), the cessation of the molting cycle, the migration of the developing gonad, and the formation of various somatic reproductive structures in both hermaphrodites and males (Rougvie and Ambros 1995; Bettinger et al. 1996, 1997; Euling et al. 1999; Newman et al. 2000; Inoue et al. 2005; Sternberg 2005; Hayes et al. 2006; Abraham et al. 2007; Fielenbach et al. 2007; Ririe et al. 2008; Harris and Horvitz 2011; Blum et al. 2012; Gupta et al. 2012). According to whole-body RNA-sequencing (RNA-seq) data, lin-29 transcript levels peak in the L3 stage, while immuno-staining and reporter fusions show that a major accumulation of LIN-29 protein takes place in hypodermal cells starting in the L4 (Bettinger et al. 1996; Gerstein et al. 2010; Harris and Horvitz 2011; Aeschimann et al. 2017). In these cells, LIN-29 expression is negatively regulated before the L4 stage by two upstream heterochronic proteins: HBL-1/Hunchback, which presumably acts by repressing lin-29 transcription in the L2; and LIN-41/Trim, which represses by binding the 5′ end of the lin-29a transcript and blocking its translation in the L3 stage (Slack et al. 2000; Lin et al. 2003; Aeschimann et al. 2017). Negative regulation of HBL-1 and LIN-41 by members of the let-7 miRNA family subsequently allows LIN-29 accumulation to occur at the correct time (Slack et al. 2000; Abrahante et al. 2003; Abbott et al. 2005; Aeschimann et al. 2017).
A number of target genes regulated by LIN-29 that may function in stage-specific developmental events have been identified. In the hypodermal seam cells, LIN-29 regulates expression of genes involved in cell division (Hong et al. 1998; Rausch et al. 2015), cell fusion (Friedlander-Shani and Podbilewicz 2011), molting (Harris and Horvitz 2011), and the adult-specific cuticle collagen (col) genes col-7 and col-19 (Liu et al. 1995; Rougvie and Ambros 1995). In recent work, we found that LIN-29 also regulates the L4-expressed col genes col-38, col-49, col-63, and col-138, and showed that mutation of specific LIN-29 binding sites abolished expression of a col-38 reporter transgene in vivo in the L4 hypodermis (Abete-Luzi and Eisenmann 2018). In the anchor cell of the somatic gonad, LIN-29 activates expression of lag-2, a Notch ligand that promotes uterine cell differentiation and the formation of the uterine-seam cell connection (Newman et al. 2000). Finally, LIN-29 was also shown to act nonautonomously to regulate expression of vitellogenin genes vit-1, vit-2, vit-3, and vit-6 in the intestine, promoting an adult-specific event required for fertility (Dowen et al. 2016).
The transition to adulthood is a fundamental life history event for all animals and it involves at least three major changes: the conclusion of a period of rapid somatic growth and differentiation, the acquisition of reproductive capabilities (e.g., sexual organogenesis), and the associated metabolic adjustment underlying a switch in energy investment from somatic to germinal functions. To further explore the network of events coordinated by LIN-29, and to uncover potential new roles for this heterochronic protein, we temporally misexpressed LIN-29 and examined changes in development and gene expression. Using RNA-seq analysis, we identified several hundred genes for which expression was up- or downregulated upon temporal misexpression of LIN-29. These include 33 upregulated genes encoding cuticle collagens, suggesting a rather preponderant cell-autonomous role for LIN-29 in cuticle production at the last molt. Interestingly, our data suggests that LIN-29 and most likely its upstream regulators are also used to upregulate collagen gene expression in the adult in response to defects in cuticle integrity. Among target genes with decreased expression upon LIN-29 overexpression, we identified genes encoding enzymes involved in lipid metabolism, many of which are normally expressed in the intestine and are downregulated in the L4 stage. We found that several signaling molecules encoded by LIN-29-upregulated targets are required for both the positive expression of intestinal vitellogenin genes, and for the repression of some intestinal metabolic enzyme genes. Together, these results indicate that in addition to its roles in hypodermal developmental events, including cuticle collagen gene expression, LIN-29 may play a broader, cell-nonautonomous role in the regulation of fat metabolism perhaps contributing to a metabolic restructuring at the larval-to-adult transition.
Materials and Methods
C. elegans growth and strains used
C. elegans animals were cultured using standard methods (Brenner 1974). Worms were grown on NGM plates and fed with Escherichia coli OP50, or HT115 in the case of RNA interference (RNAi) experiments. Experiments were performed at 20° unless indicated otherwise. Bristol strain N2 of C. elegans was the wild-type strain. The following strains and alleles were used in this work:
NR222: rde-1(ne219) V; kzIs9[pKK1260(lin-26p::nls::gfp), pKK1253(lin-26p::rde-1), pRF4(rol-6(su1006)] (Qadota et al. 2007)
CB6147: bus-8(e2882) X (Partridge et al. 2008)
SV1009: heIs63 [wrt-2p::gfp::ph + wrt-2p::gfp::H2B + lin-48p::mCherry] V
HW1692: lin-29(xe37) II (Aeschimann et al. 2019)
HW1695: lin-29(xe40) II (Aeschimann et al. 2019)
hs::lin-29: deSi5[pPA5 = hsp-16.2p::lin-29a::unc-54-3′UTR; unc-119(+)] II; unc-119(ed3) III (Abete-Luzi and Eisenmann 2018)
hs::control: deSi6[pPA4 = hsp-16.2p::unc-54-3′UTR; unc-119(+)] II; unc-119(ed3) III (Abete-Luzi and Eisenmann 2018)
Ectopic induction of LIN-29 via heat shock
Embryos obtained from bleaching hs::lin-29 and hs::control strains were hatched overnight in liquid in the absence of food, the resulting synchronized early L1 stage animals were grown for a given amount of time with food at 20° (or at 25° when indicated), induced by heat-shock exposure for 30 min at 37°, then returned to growing temperature until scoring, imaging, or collection for RNA preparations. Specific developmental stages were determined by time in hours postfeeding (hpf) and verified by the extent of gonad migration and/or vulval cell division/morphology.
Induction protocol for analyses of body morphology and vulva phenotypes:
Animals were grown at 20°. Heat shocks corresponding to the late L2, late L3, and mid L4 stages were done at 23, 33, and 43 hpf, respectively.
Induction protocol for analysis of precocious seam cell fusion:
Strains also carried the heIs63 array. For late L2 induction, animals were grown at 20°, heat-shocked at 23 hpf, and scored at 28 hpf. For single L3 induction animals were grown at 25°, heat-shocked at 22 hpf (early L3), and scored at 25 hpf. For double L3 induction, animals were grown at 25°, heat-shocked at 22 and 25 hpf (mid L3), and scored at 27 hpf.
Induction protocols for analyses of precocious alae and gonad migration defects:
Worms were grown at 25°, heat-shocked at 22 and 25 hpf, and then scored at the early-to-mid L4 stage (29–32 hpf).
Induction protocols for assessment of LIN-29 target gene expression:
Animals were grown at 20°. Induction for RNA-seq analysis was done by heat shock in the early L3 at 28 hpf. Adult induction for quantitative RT-PCR (RT-qPCR) assessment of intestinal targets was carried out in gravid adults (66 hpf).
“Young adult animals” indicates pregravid adult animals that have yet to accumulate or lay eggs; “day 1 adult animals” indicates animals in the first day of egg laying.
Imaging
Animals were mounted on 2.5% agarose pads and suspended in anesthetic solution (5 mM levamisole in M9). Nomarski (DIC) and epifluorescence microscopy was performed on a Zeiss Axioplan 2 and recorded with a Lumenera Infinity 3 camera and Infinity Analyze software.
RNA-seq and target gene identification
hs::lin-29 and hs::control worms grown at 20° were induced in the early L3 (28 hpf), given a 1-hr recovery at 20°, collected, and frozen at −80° for a minimum of 15 min. Pellets (50–100 µl) were washed three times, resuspended in DEPC water (600 μl), and homogenized with a gentleMAC dissociator (Miltenyi Biotec). RNA preparations were performed with Quick-RNA MiniPrep kit (Zymo Research). A total of six samples (three biological replicates for hs::lin-29 and hs::control each) were sequenced with single-end 50 base reads on an Illumina HiSeq 2500. Bioinformatics quality controls were done using FastQC, version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc). The ce10 reference genome was aligned using STAR, version 2.5.1b. The number of reads mapped to genes were counted using htseq, version 0.6.1p1. Differentially expressed genes were determined using DESeq2, version 1.12.3 with the cut-off of 0.05 on false discovery rate. Transcriptomic data from this work has been deposited in the Gene Expression Omnibus archive under accession number GSE118433.
RNAi treatments
In most cases, synchronized L1 stage animals were incubated at 20° [except for lin-29(RNAi) and bli-1(RNAi) experiments, in which worms were grown at 25°] and fed with HT115 E. coli previously transformed with specific RNAi clones (Kamath et al. 2000). The RNAi control was empty “feeding” vector L4440 (gift from Andrew Fire; plasmid 1654; Addgene). RNAi clones used in this work were from the Ahringer RNAi library (Kamath and Ahringer 2003) (lin-29, wrt-6, grd-11), the Vidal RNAi library (Rual et al. 2004) (ins-37, grl-14), or previous work (Jackson et al. 2014) (bli-1).
RNA isolation and RT-qPCR
For each experiment, relative transcript levels were assessed by two-step RT-qPCR with three-to-four independent biological replicates. RNAi-treated, heat-shocked, and control animals were collected and stored at −80° for a minimum of 15 min. Worm pellets (50–100 µl) were washed three times, resuspended in DEPC water (∼600 µl), homogenized with a gentleMAC dissociator (Miltenyi Biotec) and used for RNA preparations via Quick-RNA MiniPrep kit (Zymo Research). Total RNA was reverse transcribed with a mix of oligo(dT) and random primers using iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Real-time PCRs were performed with exon-exon spanning primers (Supplemental Material, Table S5) and the iTaq Universal SYBR Green Supermix system (Bio-Rad). All Ct values were normalized to housekeeping gene gpd-2 and data were analyzed by the 2ΔΔCt method (Livak and Schmittgen 2001).
Protein category (Gene Ontology term) and tissue enrichment analyses
Analysis of target gene lists for protein function was performed using UniProt Knowledgebase (www.uniprot.orgl); Gene Ontology term enrichment was performed using DAVID (https://david.ncifcrf.gov; Huang et al. 2009) and AmiGO 2/PANTHER (http://amigo.geneontology.org; Carbon et al. 2009; Munoz-Torres and Carbon 2017); and tissue enrichment analysis (TEA) was performed using the WormBase Enrichment Analysis tool (www.wormbase.org; Angeles-Albores et al. 2016). Enrichment analyses were done using default parameters. Published data on target genes (IDs; RNAi phenotypes, sites of expression, times of expression, etc.) from Table S2 was retrieved using the WormBase Simplemine tool (www.wormbase.org; Lee et al. 2018).
Fatty acid gas chromatography–mass spectrometry analysis
For animals overexpressing LIN-29, synchronized L1 stage hs::lin-29 and hs::control worms were grown at 20°, induced by heat shock (30 min 37°) twice in the L3 stage (28 and 33 hpf) and once in the L4 stage (43 hpf), and returned to 20° until 65 hpf. For animals with reduction of lin-29 function only in the hypodermis, synchronized L1 larvae of strain NR222 were grown at 20° and fed HT115 bacteria containing either lin-29(RNAi) construct or empty-vector control. In all cases, adults (65 hpf) were washed from plates with water (four to six 60-mm plates per biological replicate) and transferred to preweighed glass vials. Worm samples were processed for fatty acid methyl ester analysis as described (Watts and Browse 2002) with the modification that naphthalene d8 (1 ng/μl final in injection mix) was added as an internal loading standard. Samples (1 μl) of the organic phase were analyzed by gas chromatography using a PerkinElmer Clarus 680 Gas Chromatograph equipped with a PerkinElmer (Norwalk, CT) Elite 5-MS column and helium as the carrier gas at 1.5 ml/min. Samples were injected without splitting at 250° and the following temperature program was used: 100° hold 2 min, 4°/min to 150° hold 4 min, 6°/min to 320° hold 4 min. Fatty acid methyl esters were identified by EI+ using a PerkinElmer Clarus SQ 8C Mass Spectrometer and TurboMass Ver6.0.0 software in the range 50.00–200.0 m/z. All biological replicates were processed and analyzed on the same day. For the hs::lin-29 vs. hs::control study, a total of four biological trials of each strain were analyzed on two separate dates [two trials per gas chromatography–mass spectrometry (GC-MS) run for each strain]. Trials performed on different dates were not averaged. For the lin-29(RNAi) vs. control RNAi analysis, four biological replicates of each treatment were assessed together in a single GC-MS run (all trials averaged). For each fatty acid, the quantities determined by GC-MS were successively normalized to the naphthalene internal standard and to the weight of the sample.
Survival analysis
L1-synchronized hs::lin-29 and hs::control worms were grown at 20° until the first eggs were laid, then transferred to 5-fluoro-2′-deoxyuridine (FUdR) solid media (to induce sterility) at 25°. It was previously shown that life span tends to be intrinsically shorter when animals are fed proliferating bacteria (most likely due to an age-related susceptibility to infection) and one recommended alternative is to use UV-killed bacteria as source of food for survival analysis (Garigan et al. 2002; Sutphin and Kaeberlein 2009). In this study, we tested both conditions and animals were fed with either live bacteria (E. coli OP50) or dead bacteria (UV-killed E. coli OP50 on 50 µg/ml carbenicillin NGM plates). Heat-shock inductions of LIN-29 started in day 1 adults and were repeated either daily or every other day. All cohorts were followed until 100% mortality, and survival curves were determined with OASIS 2 using the Kaplan–Meier method and statistically analyzed with the log rank test (https://sbi.postech.ac.kr/oasis2; Oncotarget 11269; Han et al. 2016).
Data availability statement
Original gene expression data underlying this work are fully available without restriction from the Gene Expression Omnibus archive (www.ncbi.nlm.nih.gov/geo/) under accession number GSE118433. After publication, some of the data from this paper will be available in the publicly accessible, curated database WormBase (wormbase.org). Any reagents and strains utilized in this work will either be available from a publicly accessible strain repository (the Caenorhabditis Genetic Center) or freely available upon request from the corresponding author. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11663883.
Results
hs::lin-29 induction before the L4 stage leads to defects in body morphology, vulval development, and gonad migration
Loss-of-function mutations in upstream heterochronic pathway regulators precociously express LIN-29 earlier in development (Slack et al. 2000; Aeschimann et al. 2017); however, the consequences of direct misexpression of LIN-29 have not previously been assayed. To that end we used a strain containing a single-copy, integrated transgene containing a full-length lin-29a complementary DNA downstream of a heat-shock promoter (referred to as hs::lin-29) and a control strain with the identical heat-shock promoter and no insert (hs::control) (Abete-Luzi and Eisenmann 2018). We previously showed that this reagent was sufficient to drive ectopic expression of a reporter for the LIN-29 target col-38 (which is normally expressed in the L4 stage) in either the L2/L3 stage or in the adult, when induced at those respective times of development (Abete-Luzi and Eisenmann 2018). In these cases, the col-38 reporter expression was only observed in the hypodermal cells that normally express col-38, indicating that temporal but not spatial expression was affected. Here, we used this reagent to test whether temporal misexpression of LIN-29 during development was sufficient to cause phenotypes in processes associated with lin-29.
We exposed hs::lin-29 animals to a heat-shock pulse either once in the L2, L3, or L4 stage, or twice (in the L2 and L3 stages, or in the L2 and L4 stages), and looked for morphological defects in these same animals as young adults. Adults that were subject to early temporal overexpression of LIN-29 displayed three morphological phenotypes: whereas a few animals displayed a Dumpy phenotype (Dpy), many more were egg-laying defective (Egl) or showed a substantial decrease in body size (Small), or both (Figure 1B, Figure S1, and Table 1). We noted that in many of the Small animals the pharynx was bent inside the head of the animal, as if the pharynx was too large to fit inside a smaller body (penetrance = 38% of Small adults, n = 35; Figure S2). No morphological phenotypes were observed when LIN-29 expression was induced only in the L4 stage when LIN-29 is normally present. We did note that the penetrances of both the Egl and Small phenotypes in animals subjected to a heat shock in the L2 were increased with an additional L4 induction, suggesting that excess LIN-29 in the L4 stage can contribute to these phenotypes. The morphological phenotypes we observed in lin-29 gain-of-function conditions are similar to those seen in lin-41 loss-of-function mutants, in which there is early accumulation of LIN-29 in the L3 stage: these animals also show Dpy, Small, and slightly Egl phenotypes (Slack et al. 2000; Tocchini et al. 2014).
Figure 1.
Early expression of LIN-29 results in body morphology and vulval defects. Nomarski images of hs::control (A, C, and E) or hs::lin-29 (B, D, and F) animals that were subjected to heat shock early in development. (A and B) Adult animals that were given a single heat shock in both the L2 and L4 stages. (C–F) Animals were given a single heat shock in both the L2 and L3 stages, and scored in the L4 (C and D) or the adult (E and F) stage. Single arrowhead in D indicates an underinduced vulva in the L4 stage. Double arrowhead in F indicates an L4 stage vulva (compare to hs::control L4, C) in an adult hs::lin-29 animal (note the presence of unlaid, late-stage embryos in the uterus). Bar, 50 µm.
Table 1. Adult phenotypes following lin-29 misexpression in larval life.
| Time of heat shock | Strain | % WT | % Dpy | % Egl | % Small | % Small-Egl | N |
|---|---|---|---|---|---|---|---|
| No heat shock | hs::lin-29 | 98 | 0 | 0 | 2 | 0 | 130 |
| L2 | hs::control | 99 | 0 | 0 | 1 | 0 | 73 |
| hs::lin-29 | 55 | 7 | 1 | 16* | 21* | 73 | |
| L3 | hs::control | 94 | 0 | 2 | 2 | 0 | 48 |
| hs::lin-29 | 22 | 8 | 38* | 8 | 23* | 86 | |
| L4 | hs::control | 100 | 0 | 0 | 0 | 0 | 72 |
| hs::lin-29 | 100 | 0 | 0 | 0 | 0 | 56 | |
| L2 + L3 | hs::control | 98 | 0 | 0 | 2 | 0 | 111 |
| hs::lin-29 | 12 | 5 | 55* | 8 | 20* | 145 | |
| L2 + L4 | hs::control | 99 | 0 | 0 | 1 | 0 | 224 |
| hs::lin-29 | 17 | 11* | 7 | 28* | 43* | 161 |
Strains carrying hs::lin-29 or hs::control were submitted to different protocols of heat shock (column 1) to test the consequences of LIN-29 induction at different times during development. Day 1 adults were assessed for body morphology phenotypes by direct observation. * P < 0.001 (Fisher’s exact test) compared to the corresponding hs::control. WT, wild type; Dpy, Dumpy, Egl, Egg-laying defective; Small, substantial decrease in body size.
To investigate the basis for the Egl phenotype observed upon misexpression of LIN-29, we assessed the L2 + L3 heat-shocked animals for vulval defects at the L4 and young adult stages; we saw vulval abnormalities with significant penetrance at both times (see Figure 1D, Figure S1, and Table 2). In the L4 stage, heat-shock-treated animals showed too few cells adopting vulval fates (“underinduced”) or vulval inductions that were abnormal in morphology (“abnormal”). lin-29 is known to be required for development of the egg-laying apparatus: lin-29 mutants were first identified based on their loss-of-function Egl and protruding vulva (Pvl) phenotypes, and lin-29 was later shown to be required for the formation of the connection between the uterus and vulva and for expression of certain genes in vulval cells (Ambros and Horvitz 1984; Bettinger et al. 1996, 1997; Newman et al. 2000; Inoue et al. 2005). However, one vulval phenotype we observed deserves comment: when examined as gravid adults, one-third of heat-shocked hs::lin-29 animals showed a vulval morphology that resembled that found in mid-to-late L4 stage animals (Figure 1F, Figure S1, and Table 2). To our knowledge, this type of “arrested L4 vulva in an adult” phenotype has not been observed before in other heterochronic or vulval mutants. A possible explanation for both the “arrested vulva” phenotype and the “bent pharynx” phenotype in Small animals is that in animals experiencing an earlier than normal pulse of LIN-29, the hypodermis may have delayed or arrested development instead of progressing to adulthood, resulting in a hypodermis that is temporally out of sync with other body tissues. Thus, in some animals we observed an L4 vulva in an adult animal with embryos, while in other animals we found an adult-sized pharynx in an L4-sized body.
Table 2. Vulva defects induced by early expression of lin-29.
| Vulva phenotypes | ||||||
|---|---|---|---|---|---|---|
| Stage | Strain | % Wild type | % Abnormal | % Underinduced | % L4-like lumen | N |
| L4 | hs::control | 100 | 0 | 0 | n.d. | 73 |
| L4 | hs::lin-29 | 66 | 18 | 15 | n.d. | 110 |
| Adult | hs::control | 100 | 0 | n.d. | 0 | 30 |
| Adult | hs::lin-29 | 21 | 46 | n.d. | 33 | 67 |
Vulva developmental defects were assessed in the indicated strains after early induction of LIN-29 by a single heat shock in both the L2 and L3 stages. Phenotypes were scored first in the L4 stage, then in adults of the same cohorts. In all cases P < 0.001 (Fisher’s exact test) compared to the corresponding hs::control.
Finally, we also observed that overexpression of LIN-29 in the L3 stage was sufficient to cause a mild gonad migration phenotype. Although reduction of lin-29 function does not cause a gonad migration defect on its own, genetic and molecular analyses indicate that lin-29 acts on the migrating distal tip cell to control the timing of its turning event (Fielenbach et al. 2007; Huang et al. 2014). We found that 13% of hs::lin-29 animals given two heat-shock pulses in the L3 stage had defects in gonad migration (n = 48, compared to 0% for hs::control animals, n = 32). In these animals the elongating gonad arms turned dorsalward correctly, but then one arm migrated in the incorrect direction along the anterior-posterior axis.
Early hs::lin-29 induction is sufficient to promote precocious seam cell fusion, but not precocious alae formation
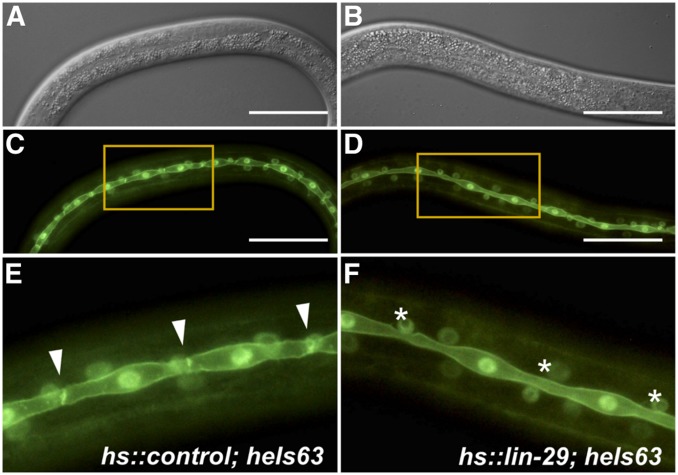
LIN-29 is also known to regulate the fusion of the hypodermal seam cells with each other at the end of the L4 stage via expression of the fusogen aff-1 (Friedlander-Shani and Podbilewicz 2011). Therefore, we looked at seam cell behavior when precociously inducing LIN-29 in the late L2 and in the L3 stage. We used the heIs63 transgene, which expresses nuclear- and membrane-localized GFP from a seam-cell-specific promoter (Wildwater et al. 2011), to examine seam cell morphology in hs::lin-29 and hs::control animals. We found that a single heat shock is enough to induce precocious seam cell fusion at high penetrance (Figure 2 and Table 3). We noted that the timing of the heat shock relative to the timing of the seam cell division affected the penetrance of the phenotype: cells that were newly divided and had not yet restored cell-cell contact upon heat shock did not display a precocious fusion phenotype, while single seam cells in contact with neighboring cells usually did show precocious fusion when LIN-29 was induced. Our results indicate that LIN-29 is not only required but also sufficient for seam cell fusion, at least in the L3 stage.
Figure 2.
Early expression of LIN-29 is sufficient to cause precocious seam cell fusion. Shown here are synchronized L3 stage animals expressing nuclear and plasma membrane localized GFP in the hypodermal seam cells (from array heIs63) and carrying either hs::control (A, C, and E) or hs::lin-29 (B, D, and F). Populations of animals were given a heat shock in the late L2 (see Table 3; Materials and Methods). Nomarski (A and B) and epifluorescence (C and D) microscopy of larvae 5 hr after heat shock. Precocious seam cell fusion is observed, as seen in the magnified view (E and F; from insets shown in C and D): cell junctions between seam cells are still present in the hs::control strain (E; arrowheads) but are absent in animals carrying hs::lin-29 (F; asterisks). Bar, 50 µm.
Table 3. LIN-29-induced seam cell fusion in earlier developmental stages.
| Time of heat shock | Time observed | Strain | % Multiple or all seam cells fused | % Only one fusion between two cells | N |
|---|---|---|---|---|---|
| Late L2 | Early L3 | hs::control; heIs63 | 0 | 7 | 42 |
| hs::lin-29; heIs63 | 45 | 0 | 42 | ||
| Early L3 | Mid L3 | hs::control; heIs63 | 0 | 0 | 30 |
| hs::lin-29; heIs63 | 82 | 3 | 60 | ||
| Early L3 + mid L3 | Late L3 | hs::control; heIs63 | 0 | 0 | 18 |
| hs::lin-29; heIs63 | 100 | 0 | 30 |
Animals expressing GFP in the nucleus and at the plasma membrane of the seam cells (from heIs63; see Materials and Methods) and carrying either hs::lin-29 or hs::control were heat-shocked as indicated and observed for precocious seam cell fusion at the time shown. Multiple and single fusion events were scored with epifluorescence microscopy. In all cases P ≤ 0.0001 (Fisher’s exact test) compared to the corresponding hs::control.
Unlike seam cell fusion, we found that early overexpression of LIN-29 was not sufficient to induce adult alae formation. It has long been known that lin-29 mutants lack adult alae, indicating lin-29 is necessary for production of these adult cuticular structures in the L4 stage (Ambros and Horvitz 1984). We gave hs::lin-29 animals two heat-shock treatments in the L3 stage and observed them from 2 to 5 hr after the second heat-shock period. Although we observed short, disorganized striations in rare animals, in no case did we observed the presence of unambiguous adult alae, even in small amounts. Precocious adult alae have been observed at the L3 molt in lin-41(lf) mutants and in lin-41 or hbl-1 RNAi-treated animals (Slack et al. 2000; Lin et al. 2003; Fielenbach et al. 2007) in which LIN-29 accumulated early. These results indicate that overexpression of the LIN-29a isoform under the conditions we assayed is not sufficient for production of adult alae in the L3 stage, suggesting that perhaps another LIN-29 isoform, or the repression of other upstream heterochronic regulators is necessary for this phenotype.
Misexpression of LIN-29 in the adult shortens life span
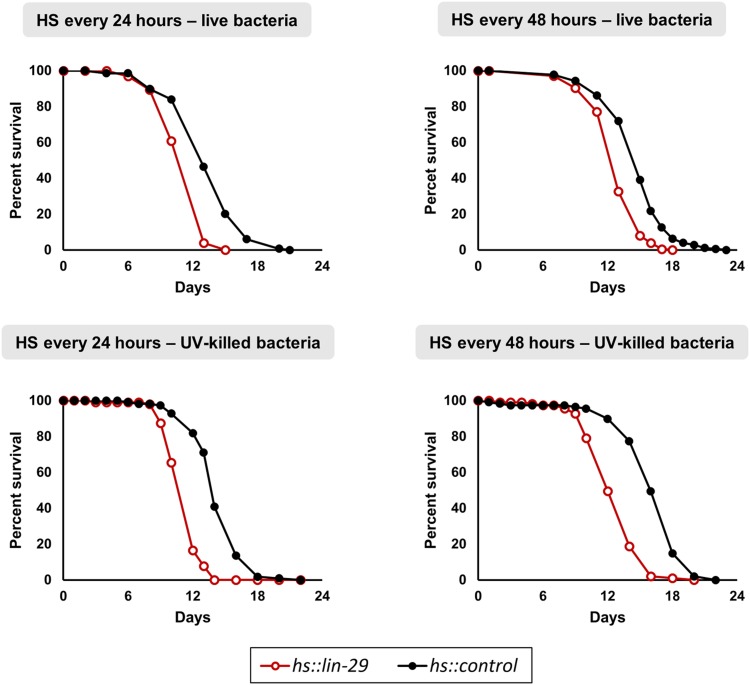
Previous work showed that two heterochronic genes that function early in the larval life, lin-14 and lin-4, can affect life span even when their expression is manipulated solely in the adult (Boehm and Slack 2005). This was surprising because both lin-14 and lin-4 were known as key regulators of the L1 to L2 transition and any effects in the adult were unknown. We know that the expression of lin-29 in the L4 promotes the developmental transition to the adult in some tissues and were curious whether this LIN-29 “maturing” instruction could have any beneficial or adverse effect on the aging of the animal if overexpressed during adulthood. We tested multiple pulses of LIN-29 only in the adult stage and assessed survival rates in four different conditions: a single heat shock every 24 hr or every 48 hr, and feeding with either dead bacteria or live bacteria. Worms subjected to these protocols showed no change in foraging behavior, no altered pharyngeal pumping, and no other visible phenotype or sickness, yet all four experiments showed significantly shorter mean life span and maximum life span (see Figure 3 and Table S1). Although we do not know the cause of this effect on life span, this result indicates that the disruption of normal temporal gene expression patterns by the misexpression of LIN-29 later during adulthood is detrimental to the animal. The observation that LIN-29 regulates cuticle collagen genes and genes involved in energy metabolism (this work, see below; Liu et al. 1995; Rougvie and Ambros 1995; Dowen et al. 2016; Abete-Luzi and Eisenmann 2018), and that both of these types of genes have effects on life span (Ewald et al. 2015; Duffy et al. 2016; Bustos and Partridge 2017), may be relevant to this observation.
Figure 3.
Periodic adult overexpression of LIN-29 shortens life span. Synchronized adult animals carrying either hs::lin-29 or hs::control were periodically exposed to heat shock either every 24 or every 48 hr, and fed with either live or dead bacteria. Cohorts were FUdR-sterilized and followed until the last individual died. Survival curves were computed using the Kaplan–Meier estimator and statistical differences between hs::lin-29 and hs::control groups were calculated with the log-rank test (in all cases P < 0.0001). In all four conditions both mean and maximum life span were shorter in hs::lin-29 animals (see Table S1).
Identification of genes regulated after LIN-29 temporal misexpression
To identify target genes regulated by LIN-29 we used a gain-of-function approach in which we examine global changes in gene expression following overexpression of wild-type LIN-29 using the heat-shock promoter, as we did for the transcription factor BAR-1 (Jackson et al. 2014; Gorrepati et al. 2015). To our knowledge, most work previously done on LIN-29 has been done using lin-29 reduction-of-function mutation or RNAi-treated strains. For example, while investigating targets regulated by the miRNA let-7, Hunter et al. conducted microarray analyses of L4 stage lin-29(n333) mutant animals vs. wild type (Hunter et al. 2013). However, a caveat of looking for target genes with a reduction-of-function approach is that observed changes in gene expression or phenotype may be an indirect, downstream consequence of changes in cell fate or other defects caused by the loss of a regulatory factor during development. Although there are caveats to the gain-of-function approach as well (see Discussion), we believe the approach of expressing LIN-29 at a discrete time in otherwise normally developed animals and then examining changes in gene expression a short time later, may be more likely to avoid such secondary downstream effects. We believe that genes showing altered regulation shortly after LIN-29 overexpression at an earlier time in development are likely to represent targets of LIN-29 during its normal role in the L4 stage.
Strains carrying either hs::lin-29 or hs::control were given a single heat shock in the early L3 stage and RNA-seq analysis was performed on triplicate samples collected 1 hr after the end of the heat-shock period. We chose this time since it is close to but earlier than the normal peak of LIN-29 gene expression, so other aspects necessary for LIN-29 function such as the presence of other transcription factors or a permissible chromatin state, may be present. We found 1101 genes that were differently expressed (P < 0.05) between the two heat-shocked strains.
Using an arbitrary cut-off of ≥1.7-fold, we narrowed our target list to 230 and 350 genes that were upregulated and downregulated respectively, upon early overexpression of LIN-29 (Table S2). To gain insight into the role of these differentially regulated genes we characterized them based on their molecular function (Table 4 and Table S2). The major categories of upregulated genes included those encoding proteins of unknown function (n = 104), cuticle collagens (n = 33), seven transmembrane receptors (n = 12), transcription factors (n = 12), zinc metalloproteases (n = 10), and C-type lectins (n = 9). For the downregulated genes, the major categories were proteins of unknown function (n = 180), enzymes functioning in fatty acid metabolism (n = 25), F-box proteins (n = 18), and transcription factors (n = 17). In addition to these gene classes, other types of genes that were found in large numbers in the set of all 1101 LIN-29-regulated genes were those encoding nuclear hormone receptor transcription factors (25 genes), cytochrome P450 enzymes (11 genes), UDP glycosyl transferase genes (11 genes), and proteins with transthyretin domains (11 genes) or prion-like (polyQ/N) domains (10 genes).
Table 4. Main categories of gene products among LIN-29-regulated genes.
| Category | Genes with significant differential expression | 1.7-Fold upregulated genes | 1.7-Fold downregulated genes | |||
|---|---|---|---|---|---|---|
| Count | % | Count | % | Count | % | |
| Unknown | 502 | 45.6 | 104 | 45.2 | 180 | 51.4 |
| Lipid metabolism (Zhang) | 65 | 5.9 | 2 | 0.9 | 25 | 7.1 |
| Cuticle collagen | 46 | 4.2 | 33 | 14.3 | 3 | 0.9 |
| F-box protein | 26 | 2.4 | 3 | 1.3 | 18 | 5.1 |
| Nuclear hormone receptor | 25 | 2.3 | 4 | 1.7 | 11 | 3.1 |
| Other transcription factor | 21 | 1.9 | 8 | 3.5 | 6 | 1.7 |
| C-lectin | 14 | 1.3 | 9 | 3.9 | 1 | 0.3 |
| 7TM receptor | 13 | 1.2 | 12 | 5.2 | 1 | 0.3 |
| Cytochrome P450 | 11 | 1.0 | 3 | 1.3 | 6 | 1.7 |
| Transthyretin-domain | 11 | 1.0 | 0 | 0.0 | 6 | 1.7 |
| UDP-glucuronosyltransferase | 10 | 0.9 | 3 | 1.3 | 3 | 0.9 |
| Prion-like-(Q/N-rich)-domain | 10 | 0.9 | 3 | 1.3 | 2 | 0.6 |
| Noncoding RNA | 8 | 0.7 | 2 | 0.9 | 0 | 0.0 |
| O-acyltransferase | 8 | 0.7 | 2 | 0.9 | 5 | 1.4 |
| Nematode specific protein B | 6 | 0.5 | 5 | 2.2 | 0 | 0.0 |
| Zinc metalloprotease | 6 | 0.5 | 10 | 4.3 | 2 | 0.6 |
| Protein phosphatase | 4 | 0.4 | 4 | 1.7 | 2 | 0.6 |
| Extracellular signaling protein | 4 | 0.4 | 4 | 1.7 | 0 | 0.0 |
| Neuropeptide like protein | 4 | 0.4 | 0 | 0.0 | 2 | 0.6 |
| Peroxisomal assembly factor | 3 | 0.3 | 0 | 0.0 | 3 | 0.9 |
| Solute carrier protein | 3 | 0.3 | 0 | 0.0 | 3 | 0.9 |
“Genes with significant differential expression” are those with a differential change between hs::lin-29 and hs::control strains; P < 0.05 (n = 1101). The other gene sets are those genes showing ≥1.7-fold upregulation (n = 230) or downregulation (n = 350) in hs::lin-29 compared to hs::control and P < 0.05.
Genes encoding cuticle collagens are major upregulated targets of LIN-29 and are activated in response to cuticle defects in the adult
We performed Gene Ontology analyses to search for overrepresented categories of genes in three data sets: all significant regulated genes (1101 genes), genes upregulated ≥1.7-fold (230 genes), and genes downregulated ≥1.7-fold (350 genes) (Table 5). The sole significantly enriched category among the upregulated genes was “structural constituent of cuticle,” which consists of 33 cuticle collagen genes (Table 6). This group includes col-38, col-49, col-63, and col-138, which we previously showed by qPCR were upregulated in hs::lin-29 animals and downregulated in lin-29(RNAi) animals, and col-19, which was previously shown to be regulated by LIN-29 (Liu et al. 1995; Abrahante et al. 1998; Abete-Luzi and Eisenmann 2018). The fact that 33 of the 187 col genes in C. elegans were found to be upregulated in our analysis suggests that col genes are a major target of regulation by LIN-29.
Table 5. Enrichment analysis of LIN-29 target genes.
| All LIN-29-regulated targets (n = 1101) | Gene count | |||||
|---|---|---|---|---|---|---|
| Category/GO term | In genome | Found | Expected | Fold | P value | |
| GO biological process | Peroxisome organization | 18 | 7 | 0.97 | 7.23 | 1.89E−04 |
| Fatty acid metabolic process | 105 | 21 | 5.65 | 3.72 | 1.56E−06 | |
| Defense response to Gram-positive bacterium | 60 | 12 | 3.23 | 3.72 | 2.65E−04 | |
| Innate immune response | 345 | 57 | 18.56 | 3.07 | 2.55E−12 | |
| Oxidation-reduction process | 618 | 60 | 33.25 | 1.8 | 3.76E−05 | |
| Cellular response to chemical stimulus | 612 | 57 | 32.92 | 1.73 | 1.57E−04 | |
| GO molecular function | Structural constituent of cuticle | 168 | 48 | 9.04 | 5.31 | 2.90E−18 |
| Iron ion binding | 118 | 20 | 6.35 | 3.15 | 2.38E−05 | |
| Oxidoreductase activity | 539 | 56 | 29 | 1.93 | 1.08E−05 | |
| GO cellular component | Peroxisomal membrane | 18 | 8 | 0.97 | 8.26 | 3.08E−05 |
| Membrane raft | 80 | 14 | 4.3 | 3.25 | 2.87E−04 | |
| Extracellular space | 294 | 37 | 15.82 | 2.34 | 7.31E−06 | |
| Lipid metabolisma | N/A | 471 | 65 | 25.33 | 2.56 | 1.72E−12 |
| 1.7-fold upregulated targets (n = 230) | Gene count | |||||
|---|---|---|---|---|---|---|
| Category/GO term | In genome | Found | Expected | Fold | P value | |
| GO molecular function | Structural constituent of cuticle | 168 | 33 | 1.9 | 17.36 | 5.46E−29 |
| Lipid metabolisma | N/A | 471 | 2 | 5.29 | 0.94 | 6.84E−02 |
| 1.7-fold downregulated targets (n = 350) | Gene count | |||||
| Category/GO term | In genome | Found | Expected | Fold | P value | |
|---|---|---|---|---|---|---|
| GO biological process | Fatty acid metabolic process | 105 | 11 | 1.79 | 6.16 | 3.99E−06 |
| Innate immune response | 345 | 18 | 5.87 | 3.07 | 4.40E−05 | |
| GO cellular component | Peroxisomal membrane | 18 | 5 | 0.31 | 16.34 | 3.33E−05 |
| Lipid metabolisma | N/A | 471 | 25 | 8.05 | 3.11 | 4.61E−07 |
Enrichment analyses were done for all LIN-29 significant targets and for both 1.7-fold LIN-29 up- and downregulated subsets using Gene Ontology (GO) Consortium (see Materials and Methods).
Genes were also compared to the list of 471 C. elegans metabolic genes from Y. Zhang et al. (2013).
Table 6. 33 cuticle collagen genes upregulated ≥1.7-fold upon lin-29 overexpression.
| Gene name | Fold change |
|---|---|
| col-49* | 48.1 |
| col-38* | 21.1 |
| col-124 | 16.5 |
| col-140 | 15.8 |
| col-178 | 15.4 |
| col-139 | 11.5 |
| col-71 | 10.4 |
| col-120 | 9.8 |
| col-20 | 8.9 |
| col-129 | 8.4 |
| bli-6 | 7.3 |
| rol-1 | 6.8 |
| col-81 | 6.0 |
| col-79 | 5.9 |
| col-88 | 5.7 |
| col-138* | 5.0 |
| col-19§ | 4.6 |
| bli-1* | 4.4 |
| col-77 | 4.1 |
| col-60 | 3.7 |
| col-176 | 3.3 |
| col-101 | 3.1 |
| col-63* | 2.7 |
| lon-3 | 2.1 |
| col-150 | 2.1 |
| col-182 | 2.0 |
| col-109 | 2.0 |
| col-91 | 2.0 |
| col-73 | 1.8 |
| col-8 | 1.8 |
| col-179 | 1.8 |
| col-142 | 1.8 |
| col-48 | 1.7 |
Cuticle collagen genes which were previously shown to be regulated by LIN-29 are indicated as * (Abete-Luzi and Eisenmann 2018) and § (Liu et al. 1995; Rougvie and Ambros 1995).
As an independent assessment of this result, we used the SPELL search engine, which analyzes 400 data sets covering 6524 C. elegans microarray and RNA-seq experiments, to identify genes with a similar pattern of expression to query genes (Hibbs et al. 2007). We queried SPELL using the three col genes we previously showed were regulated by LIN-29 (col-38, col-49, and col-63; Abete-Luzi and Eisenmann 2018). Among the top 100 genes identified, 48 were cuticle col genes and 24 of these 48 genes were also identified as upregulated targets of LIN-29 in our analysis (Table S3). This result corroborates that a large number of cuticle col genes are coregulated under a variety of normal and experimental conditions. The fact that most of these col genes show a peak of expression in the L4 stage during normal development (see below) suggests they are likely to represent a battery of col gene targets of LIN-29 at the L4 to adult transition for use in synthesis of the adult cuticle.
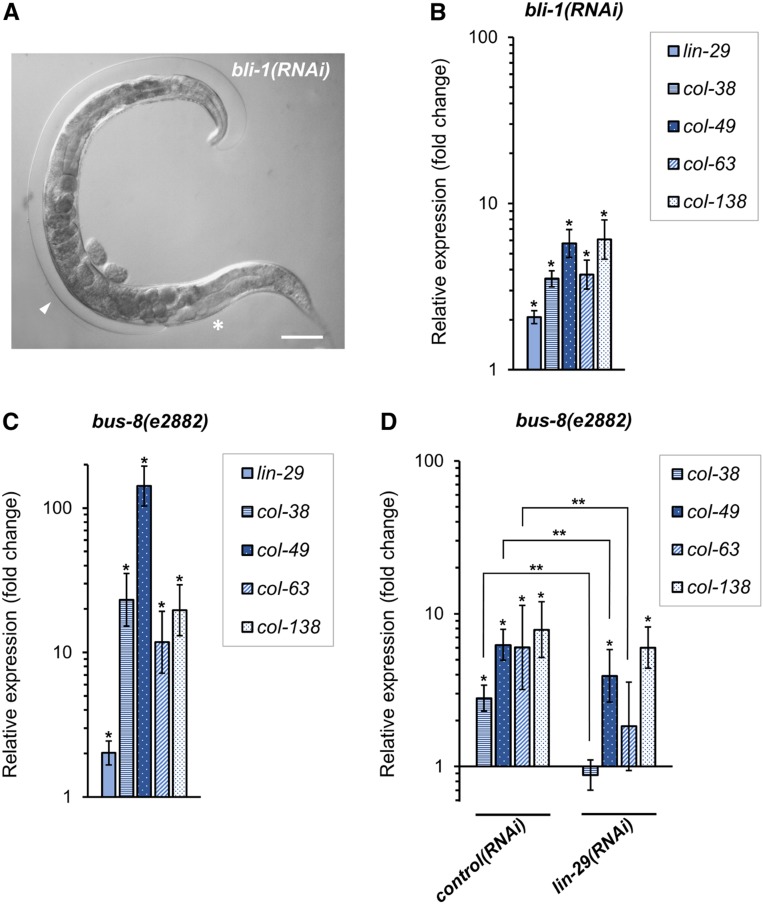
Recent work showed that one of the LIN-29 col gene targets, col-19, is upregulated in adult animals in which cuticle integrity has been damaged via RNAi against the major cuticle collagen gene bli-1 (Zhao et al. 2019). BLI-1 collagen localizes to the medial strut layer of the cuticle, and when bli-1 function is compromised by mutation or RNAi, large fluid-filled blisters cover the surface of the worm (Lints and Hall 2009) (Figure 4A). A mechanism exists within the hypodermis to sense cuticle damage such as that caused by bli-1(RNAi) or physical damage, and alter gene expression to induce an innate immune response (Zhang et al. 2015). Interestingly, bli-1(RNAi) animals also show upregulation of the heterochronic miRNA gene let-7 and downregulation of the heterochronic genes hbl-1 and lin-41 (Zhao et al. 2019). Since LIN-41 is a direct regulator of lin-29 expression (Slack et al. 2000; Aeschimann et al. 2017), we reasoned that lin-29 may be upregulated in response to adult cuticle damage, perhaps to induce expression of col gene targets that were used to synthesize the adult cuticle initially. Consistent with this hypothesis, we found that in day 1 adult bli-1 RNAi-treated animals, lin-29 expression was increased twofold, and the expression of four LIN-29 col gene targets (col-38, col-49, col-63, and col-138), which are normally expressed in the L4 stage, was increased in these bli-1(RNAi)-treated adult animals (Figure 4B). To corroborate this result, we examined the expression of lin-29 and its L4 col gene targets in adult bus-8 mutant animals. bus-8 encodes a hypodermally expressed glycosyltransferase, and reduction-of-function mutants are hyperpermeable to drugs and other reagents due to defects in epidermal and cuticle integrity (Partridge et al. 2008). We found that in bus-8(e2882) mutant adults, lin-29 and the L4 col gene targets were also upregulated (Figure 4C). Furthermore, for three of four col genes tested, the upregulation in the bus-8 background was dependent on lin-29 function (Figure 4D). We were unable to test the lin-29 dependence in the bli-1 background because the Bli phenotype is completely dependent on lin-29 function (Table S4). The result that heterochronic proteins participate in a hypodermal response to cuticle damage suggests that this pathway not only contributes to normal cuticle synthesis before the adult stage, but also functions in cuticle maintenance in adults in response to damage or breaches in integrity.
Figure 4.
Upregulation of lin-29 and col gene targets of LIN-29 in adults in response to defects in cuticle integrity. (A) Adult bli-1(RNAi) hermaphrodite showing Blister phenotype. Asterisk indicates normal cuticle, arrowhead indicates fluid-filled, Blistered cuticle. Bar, 50 µm. (B and C) Endogenous expression of lin-29 and known lin-29-regulated cuticle collagen genes col-38, col-49, col-63, and col-138 assessed by RT-qPCR in synchronized (B) young adults after bli-1(RNAi) feeding treatment (quantification was relative to expression in animals fed HT115 bacteria carrying empty RNAi vector control; see Materials and Methods); and (C) day 1 adults with bus-8(e2882) loss-of-function background (quantification was relative to expression in wild-type animals; both groups were fed standard OP50 bacteria). (D) RT-qPCR was used to measure endogenous col gene expression in day 1 adult bus-8(e2882) animals fed with HT115 bacteria containing either lin-29(RNAi) vector or empty-vector control. In both groups, quantification was relative to wild type animals fed HT115 with empty-vector control. Error bars represent SEM. Unpaired t-test analyses were performed comparing to respective controls (* P < 0.05) or between the indicated groups (D; ** P < 0.05).
Genes encoding enzymes involved in fatty acid metabolism are enriched among LIN-29 downregulated targets
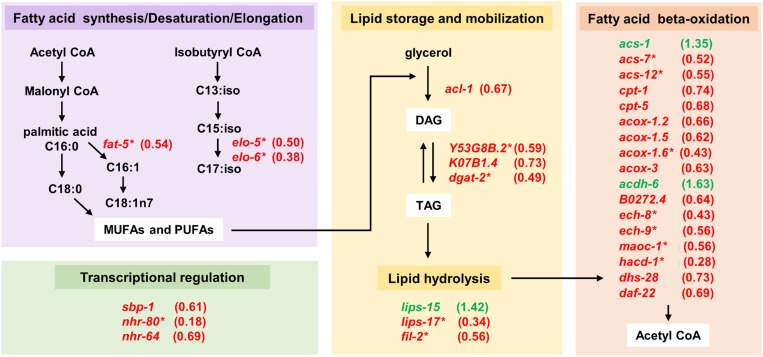
The most highly enriched category for the genes downregulated upon LIN-29 induction was “peroxisomal membrane”: this category contained three genes encoding peroxisomal assembly factors (prx-1, prx-5, prx-11), and two genes encoding enzymes acting within the peroxisome (ndx-8, maoc-1) (AbdelRaheim and McLennan 2002; Zhang et al. 2010). Surprisingly, the second highest overenriched category among the downregulated genes was “fatty acid metabolism.” To further explore the potential relevance of this result, we compared our gene target sets to a compendium of 471 C. elegans genes known to be involved in lipid metabolism (Y. Zhang et al. 2013). We found 25 of our 350 downregulated genes on this list, a number significantly higher than that expected by random sampling (hypergeometric P = 4.61E−7; Table 5 and Table S2). Likewise, when we searched the list of all 1101 LIN-29-regulated genes against the lipid metabolism gene list, we found 65 genes in total, 57 of which were downregulated upon LIN-29 misexpression (P = 1.72E−12; Table 5 and Table S2). These genes encode enzymes involved in a range of processes; however, many of them function in the synthesis of fatty acids, their storage, mobilization, and beta-oxidation (Figure 5). A potential link between the two enriched downregulated gene categories is the fact that many of the downregulated metabolic genes act in fatty acid beta-oxidation, a process which occurs in the peroxisome (and mitochondria) (Figure 5). Indeed, three lipid metabolic genes that are downregulated 30–50% by overexpression of LIN-29 function in peroxisomal beta-oxidation (maoc-1, dhs-28, and daf-22) (Figure S3), and reduction of their function by RNAi has been shown to cause an increase in lipid droplet size (Zhang et al. 2010). Similarly, reduction of function of the three peroxisomal assembly factor genes we identified (prx-1, prx-5, prx-11; Figure S3) also leads to an increase in lipid droplet size (Zhu et al. 2018), suggesting that both of these enriched categories of downregulated genes may impinge on lipid storage and utilization.
Figure 5.
LIN-29-regulated genes involved in lipid metabolism. Genes that were upregulated (green font) or downregulated (red font) upon misexpression of LIN-29 in the L3 stage are shown, with their respective fold change in parenthesis. Genes are grouped into broad categories (colored boxes) based on their gene product function in lipid metabolism. Note that there are more enzymes involved in these processes; however, only genes with a significant change (P < 0.05) upon LIN-29 overexpression are shown here. *LIN-29 target genes that were regulated ≥1.7-fold.
Also of note is the identification of genes encoding three transcription factors that regulate metabolic enzyme gene expression (Figure S4). The nuclear hormone receptor gene nhr-80 was the gene most downregulated upon lin-29 induction (more than fivefold decreased expression): NHR-80 physically interacts with NHR-49 to regulate genes involved in fatty acid metabolism, including fat-5 that was downregulated almost twofold upon LIN-29 overexpression (Van Gilst et al. 2005; Brock et al. 2006; Pathare et al. 2012). Likewise, sbp-1 encodes an SREBP (sterol regulatory element binding protein) homolog that is a major regulator of lipid metabolism genes (Lemieux and Ashrafi 2015). SBP-1 also regulates expression of fat-5 (Watts 2009) as well as several other genes that showed decreased expression upon LIN-29 induction: elo-5, elo-6, fil-2 (Kniazeva et al. 2004), and the nuclear hormone receptor gene nhr-64, which itself regulates lipid metabolism (Liang et al. 2010) (Figure S4).
The discovery of fatty acid metabolic enzyme genes among LIN-29-regulated target genes suggests the possibility that LIN-29 may act to regulate developmentally linked changes in metabolism that are part of the larval-to-adult transition. Indeed, LIN-29 activity from the hypodermis was shown to be required for intestinal expression of the vit genes, which encode lipid transport proteins necessary to move lipids from the intestine into the developing oocytes (Dowen et al. 2016). In this same work, the authors showed that lin-29(n333) adult animals had slightly reduced overall fat levels based on Oil Red O staining, although the cause for this decrease was not clear.
To test the idea that LIN-29 plays a broader role in regulating fatty acid metabolism in the larval-to-adult transition, we used GC-MS analysis to look for differences in the levels of various fatty acid species in young adult hs::lin-29 and hs::control animals that were subjected to heat-shock inductions in the L3 and L4 stages (see Materials and Methods). Consistent with the hypothesis, we found that levels of most individual fatty acid species, as well as total fatty acid levels, were decreased in adult animals subjected to early overexpression of LIN-29 (Table 7). Although the opposite of what may have been expected from the lin-29(n333) Oil Red O experiment (Dowen et al. 2016), this result supports the hypothesis that LIN-29 may normally regulate fatty acid metabolic gene expression in the L4 stage that affects fat content in the adult.
Table 7. Relative percent of fatty acid content in animals overexpressing lin-29 vs. control.
| Fatty acid (FA) | % FA in hs::lin-29 relative to hs::control | ||
|---|---|---|---|
| Run 1 | Run 2 | ||
| C14:0 | Myristic acid | 72 | 143 |
| C15:1 | 58 | 103 | |
| C15:0 | Pentadecanoic acid | 112 | 123 |
| C16:1 | Palmitoleic acid | 102 | 155 |
| C16:0 | Palmitic acid | 82 | 87 |
| C17:2 | 50 | 60 | |
| C17:1 | Heptadecanoic acid | 69 | 81 |
| C17:0 | Margaric acid | 80 | 66 |
| C18:3 | Linolenic acid | 72 | 92 |
| C18:2 | Linoleic acid | 90 | 85 |
| C18:1 | Oleic acid | 81 | 88 |
| C18:0 | Stearic acid | 79 | 76 |
| C19:1 | 147 | 108 | |
| C19:0 | Nonadecanoic acid | 58 | 60 |
| C20:5 | Eicosapentaenoic acid | 106 | 81 |
| C20:4 | Arachidonic acid | 91 | 65 |
| C20:3 | 77 | 51 | |
| C20:2 | 61 | 73 | |
| C20:0 | Eicosanoic acid | 67 | 65 |
| Total | 84 | 82 | |
Amounts of individual fatty acid in young adults were assessed by their esterification to fatty acid methyl esters (FAMEs) and quantification via gas chromatography–mass spectrometry in hs::lin-29 and hs::control animals after heat-shock treatment in the larva (see Materials and Methods). Each run included two independent biological trials. Shown is the percentage of each FAME in hs::lin-29 animals relative to the amount in hs::control animals. “Total” indicates the sum of all FAME species in hs::lin-29 animals relative to hs::control animals.
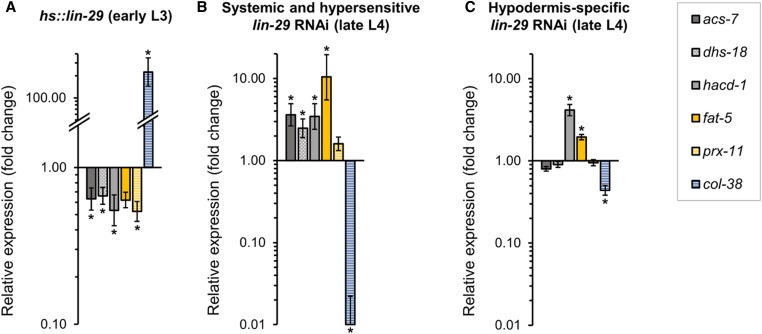
As showed above, repeated expression of LIN-29 in the adult life results in shorter mean life span and maximum life span. While we do not know the cause of this shortened life span, we consider a model in which repeated adult expression of lin-29 over time results in persistent downregulation of genes encoding metabolic enzymes that are required to keep metabolic homeostasis. To corroborate whether LIN-29 is capable of repressing metabolic targets in the adult context, we chose four downregulated genes encoding enzymes that function in lipid metabolism (acs-7, dhs-18, hacd-1, fat-5) and the peroxisome assembly factor gene prx-11, and examined their expression after LIN-29 induction in gravid adults. We found that three of these genes (dhs-18, hacd-1, and prx-11) were downregulated (Figure S5), suggesting that perhaps metabolic functions may be perturbed in these adult hs::lin-29 animals, contributing to their shortened life span.
The intersection of gain-of-function and loss-of-function transcriptomic data identifies a set of high-confidence LIN-29-regulated genes
We compared our list of genes differentially regulated by overexpression of wild-type LIN-29 in the L3 stage to data from Hunter et al. that examined gene expression in lin-29(n333) mutants vs. wild type in the L4 stage (Hunter et al. 2013). Although this mutation causes reduction-of-function phenotypes, the size and levels of lin-29 transcripts are not altered in n333 mutant animals (Rougvie and Ambros 1995). The n333 mutation causes a G > A mutation in the 3′ splice junction upstream of exon 5 of lin-29a (Blum et al. 2012), and mutations like this have been reported to retain some level of wild-type splicing (Blumenthal and Steward 1997), suggesting this is unlikely to be a true null allele. The intersection of these gene expression data sets gives a list of 21 strong candidates for LIN-29-activated genes: genes with increased expression in our gain-of-function/temporal misexpression approach and decreased expression in the Hunter et al. reduction-of-function data (hypergeometric P = 5.40E−06), and a list of 35 genes likely to be directly or indirectly repressed by LIN-29 (genes with decreased expression in our gain-of-function approach and increased expression in the Hunter et al. reduction-of-function data; hypergeometric P = 2.34E−20). We refer to the genes in common between these two transcriptomic data sets as genes for which LIN-29 is “necessary and sufficient” (N/S genes) and consider that these represent some of the best candidates for LIN-29 target genes (Table S2).
We were surprised by the small number of genes in this overlap, in particular the low number of cuticle collagen genes among the activated N/S genes (four genes). We previously showed that RNAi targeting lin-29 significantly reduced expression of the L4 cuticle col genes col-38, col-49, col-63, and col-138 in the L4 stage (Abete-Luzi and Eisenmann 2018), and col-49 was the most highly upregulated gene upon LIN-29 overexpression (48-fold; Table S2). Yet of these four genes, only col-38 was found as an activated N/S gene (Table S2). Examination of the lin-29(n333) data shows that col-49, col-63 and col-138 were downregulated 3- to 10-fold in the lin-29(n333) mutant; however, the data for these genes was slightly above the P < 0.05 cut-off. This suggests one (statistical) reason for the small overlap between the data sets.
Likewise, we were surprised that very few of our “metabolic” downregulated targets were in common with the lin-29(n333) data set for significantly upregulated genes. We first validated the RNA-seq result that some fatty acid metabolic genes are downregulated upon temporal misexpression of LIN-29 by performing qPCR on five of them (acs-7, dhs-18, hacd-1, fat-5, and prx-11) in a new set of biological replicates comparing hs::lin-29 and hs::control animals after early L3 induction: all five genes were downregulated (Figure 6A). We then determined whether reduction of lin-29 function by RNAi (which should unambiguously affect all lin-29 isoforms, unlike the n333 allele) caused increased expression of these metabolic genes in the L4 stage when LIN-29 protein normally accumulates. Consistent with our hs::lin-29 results, all five genes showed increased transcript levels at the L4 stage in lin-29 RNAi-treated animals (Figure 6B), suggesting that these genes are indeed repressed by LIN-29 activity at the L4 stage, yet for unknown reasons, the lin-29(n333) allele failed to significantly derepress them.
Figure 6.
LIN-29 represses intestinal genes involved in fatty acid metabolism and beta-oxidation in the L4 stage. Endogenous expression of intestinally expressed genes acs-7, dhs-18, hacd-1, fat-5, as well as peroxisome factor prx-11, was assessed by RT-qPCR in (A) hs::lin-29 animals 1 hr after induction in the early L3 stage (quantification relative to hs::control strain), and (B and C) L4 stage larvae after lin-29(RNAi) treatment (quantification relative to empty-vector control) in two different backgrounds: (B) a strain containing the RNAi-hypersensitive mutation rrf-3(pk1426), in which RNAi is stronger and effective in all tissues; and (C) a nonhypersensitive strain, where RNAi is only effective in the hypodermis (NR222; see Materials and Methods). In all cases, known LIN-29 upregulated gene col-38 (Abete-Luzi and Eisenmann 2018) was analyzed as a control for efficacy of lin-29 heat-shock induction and lin-29 RNAi. Error bars represent SEM. * P < 0.05 (unpaired t-test).
Temporal expression patterns of LIN-29-regulated genes
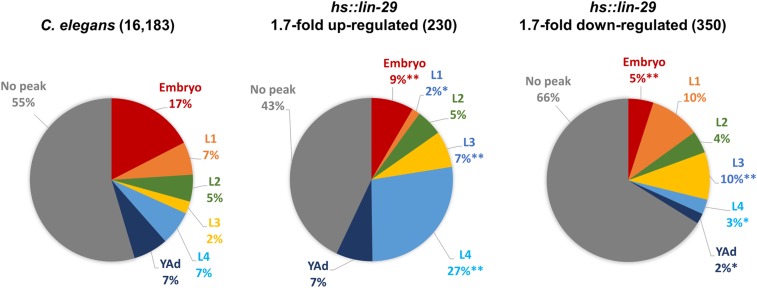
If the upregulated genes we identified upon early misexpression of LIN-29 are actual targets of LIN-29 regulation during the L4 stage, we would predict that these genes may show an increase in expression in the L4 stage during normal development; conversely, genes in our downregulated gene set would be predicted to decrease in expression at that time. To determine the pattern of temporal expression for our set of LIN-29 targets, we examined modENCODE developmental expression data for these genes (Gerstein et al. 2010), and categorized them based on whether they show a peak of expression in any particular stage of the worm life cycle. We then compared the pattern of temporal expression of our LIN-29 target genes to the pattern for all 16,183 C. elegans genes in the modENCODE data sets (Figure 7).
Figure 7.
Genes that normally peak in the L4 stage are overrepresented among LIN-29 upregulated targets. Temporal expression peaks were assessed for the indicated gene sets based on modENCODE RNA-seq data (Gerstein et al. 2010) using criteria from Jackson et al. (2014): genes showing ≥35% of their total developmental expression in one stage were identified as having a peak in that stage (color coded), the remainder are indicated as “no peak” (gray). Distributions for all genes in each set were calculated, displayed as percentages and compared to the genomic distribution (left). ** P < 0.0001 and * P < 0.01 (chi-square with Yates correction).
Notably, the proportion of our upregulated genes that show a peak of expression in the L4 stage during normal development is significantly larger than the percentage of L4-peak genes in the genome as a whole (27% vs. 7%; chi-square with Yates correction P < 0.0001; Figure 7). For the cuticle col genes specifically, we found that 85% (28 of 33) of the cuticle collagen genes upregulated upon LIN-29 misexpression show a single peak of expression in either the L4 or young adult stage during normal development, while 36% of all col genes show peak expression in those developmental times (Figure S6). We also looked at the distribution of temporal expression patterns for the smaller set of LIN-29-upregulated N/S genes and observed that genes that peak in the L4 stage are even more overrepresented (43% vs. 7%; chi-square with Yates correction P < 0.0001).
On the other hand, in the case of our LIN-29 downregulated targets, the proportion of genes that peak in the L4 stage during normal development was significantly smaller than that expected based on the known genomic distribution (3% vs. 7%; chi-square with Yates correction P = 0.0064; Figure 7). This was also true for genes with peak expression in the adult (2% vs. 7%; chi-square with Yates correction P = 0.0003; Figure 7). When we examined the temporal expression patterns of the N/S subset of LIN-29-downregulated genes, we noted that the proportion of genes with a peak of expression in stages before the L4 (63%) was much higher than that observed in the total genomic set (31%). Additionally, we examined postembryonic expression profiles generated from the modENCODE data for the 25 lipid metabolism genes downregulated ≥1.7-fold by early LIN-29 expression and found that 20 of 25 showed either a permanent (e.g., dhs-18; Figure S7) or temporary (e.g., elo-5, Figure S7) downregulation in the L4 stage during normal development.
Together, these trends are consistent with the hypothesis that many of the genes we identified as upregulated upon misexpression of LIN-29 in the early L3 are normally upregulated by the peak of LIN-29 protein in the L4 stage during development, and that many of the genes we identified as downregulated upon LIN-29 temporal misexpression may be expressed earlier in larval life and are normally downregulated in the L4 stage, when LIN-29 levels peak.
Spatial expression patterns of LIN-29-regulated genes are consistent with both cell-autonomous and -nonautonomous regulation
A major site of LIN-29 expression based on reporter gene expression and antibody staining is in hypodermal cells, with accumulation beginning in the early L4 in the seam cells, followed by expression in other hypodermal cells and the hypodermal syncytium, and remaining through adulthood (Bettinger et al. 1996; Harris and Horvitz 2011). However lin-29 expression is also seen earlier in the L3 stage in the hypodermal vulval cells, and the anchor cell and distal tip cells of the hermaphrodite gonad (Bettinger et al. 1996; Harris and Horvitz 2011). In males, LIN-29 is expressed in the linker cell—the cell that controls gonad migration in this sex—during the L3 stage and disappears in the late L4 stage, when this cell dies facilitating the connection of the gonad with the cloaca (Euling et al. 1999). Finally, LIN-29 shows steady expression in the pharynx of both males and hermaphrodites, beginning in the L1 stage and persisting through adulthood (Bettinger et al. 1996; Euling et al. 1999; Harris and Horvitz 2011); however, to our knowledge, a role of LIN-29 in pharyngeal cells remains to be determined.
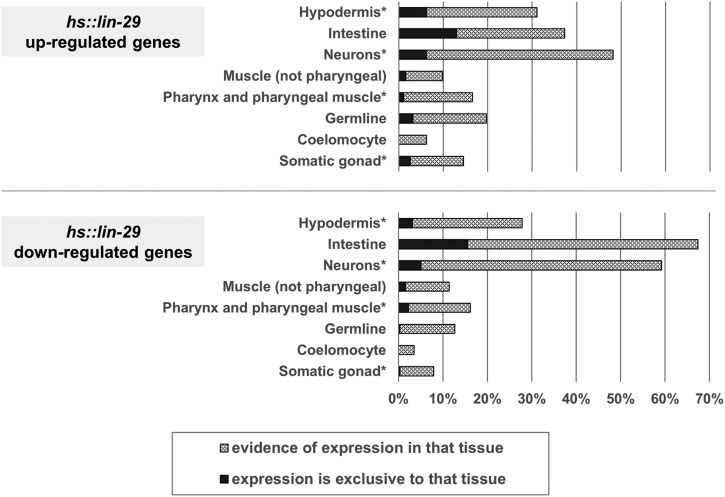
We examined the known spatial expression patterns of our LIN-29 differentially regulated genes in the C. elegans database (Table S2; see Materials and Methods). There is published gene expression data for 193 of the 230 upregulated genes, and based on this data, almost 75% of the upregulated genes show expression in at least one tissue known to express LIN-29 protein (Figure 8; Bettinger et al. 1996; Harris and Horvitz 2011). When the pattern of spatial expression for the upregulated genes is compared to the genome as a whole by tissue enrichment analysis (TEA) (Angeles-Albores et al. 2016), the most overrepresented expression site is the “epithelial system” with 74 of 193 of the upregulated genes (P = 1.0E−08; including all of the cuticle col genes with known expression). For the downregulated genes, there is spatial expression information for 316 of 350; 78% of the downregulated genes show expression in at least one tissue known to express LIN-29 (Figure 8). The fact that ≥75% of our up- and downregulated genes express in sites where LIN-29 is present is consistent with these genes being targets of LIN-29 during normal development.
Figure 8.
Spatial expression patterns of LIN-29 target genes. Spatial expression data available for 193 of 230 upregulated genes (top) and for 316 of 350 downregulated genes (bottom) was obtained (see Materials and Methods) and plotted as percentages of genes with expression in the indicated tissues. Tissues where LIN-29 is known to be expressed are denoted with an asterisk. Percentages sum to >100% because many genes are expressed in multiple tissues.
Nevertheless, for both the up- and downregulated gene sets, >20% of the genes show expression in tissues not known to express LIN-29. This site is most often the intestine, although this trend is much more prevalent for the downregulated genes: 213 out of 316 genes have intestinal expression, with 49 of these apparently expressing solely in this tissue (Figure 8 and Table S2). When we performed TEA for the downregulated gene set, the two overrepresented expression sites with the greatest numbers of genes are “intestine” (210 genes; TEA P = 1.5E−28) and “epithelial system” (100 genes; TEA P = 3.20E−06). Finally, among the 65 genes involved in lipid metabolism that we identified as possible LIN-29-regulated genes (from the full set of 1101 significant hs::lin-29-responsive genes), 75% of them show expression in the intestine, a major site of metabolic activity in the worm that is not known to express LIN-29.
One explanation for the identification of large numbers of intestinally expressed genes as potential LIN-29 targets is that expression from the heat-shock promoter led to the presence of LIN-29 in the intestine where it is not normally found, which bound to these genes and regulated their expression. Alternatively, LIN-29 expression in another tissue could have caused indirect (cell nonautonomous) regulation of these genes in the intestine. There is precedent for the idea of a signal from the hypodermis regulating intestinal gene expression. First, MacNeil et al. showed that hypodermis-specific transcription factors (e.g., LIN-26) can regulate the expression of reporters for intestinal genes (e.g., acdh-1) and proposed the existence of a signal that propagates regulatory information from one tissue to another (MacNeil et al. 2015). Second, Dowen et al. hypothesized that LIN-29 in the hypodermis activates expression of a secreted signal that mediates LIN-29-dependent regulation of vitellogenin gene expression in the intestine, acting through both insulin and mTORC2 signaling pathways (Dowen et al. 2016). Finally, Clark et al. suggest that activation of a BMP signaling pathway in the hypodermis leads to changes in fat accumulation in the intestine via insulin signaling (Clark et al. 2018). Supporting the idea that the downregulation of intestinal gene expression by ectopic LIN-29 may be indirect is the fact that at least some of these metabolic genes are expressed almost exclusively in the intestine (i.e., acs-7, dhs-18, and hacd-1; Table S2) and yet are derepressed by lin-29 RNAi treatment, a situation in which no ectopic/intestinal LIN-29 is involved (see above; Figure 6B). To bolster this result, we repeated the lin-29 RNAi treatment in a strain in which RNAi is only effective in the hypodermis (Figure 6C). Since this strain does not contain the rrf-3 mutation that renders animals hypersensitive to RNAi (Simmer et al. 2002), the lin-29 RNAi treatment was less effective (based on the smaller fold change observed for the hypodermal gene col-38; compare Figure 6B and Figure 6C). However, two of the lipid metabolic genes tested, hacd-1 and fat-5, which are expressed in the intestine but not the hypodermis, showed an increase in expression in L4 animals in which lin-29 function was compromised only in the hypodermis. We also performed fatty acid quantitation on these animals in which lin-29 function was reduced only in the hypodermis and found that levels of most fatty acid species, as well as total fatty acid content, were increased relative to control RNAi animals (Table 8), which is the opposite of the result obtained when we overexpressed lin-29 (Table 7). Together, these observations suggest that in addition to acting cell-autonomously to regulate gene expression in the hypodermis in the L4 stage, LIN-29 may also act cell-nonautonomously from the hypodermis to regulate expression of genes in the intestine, including many genes involved in lipid metabolism.
Table 8. Relative percent of fatty acid content in animals treated with hypodermis-specific lin-29(RNAi) vs. control RNAi.
| Fatty acid (FA) | % of FA in hypodermis-specific lin-29(RNAi) animals relative to control | |
|---|---|---|
| C14:0 | Myristic acid | 134 |
| C15:1 | 110 | |
| C15:0 | Pentadecanoic acid | 118 |
| C16:1 | Palmitoleic acid | 145 |
| C16:0 | Palmitic acid | 127 |
| C17:2 | 98 | |
| C17:1 | Heptadecanoic acid | 109 |
| C17:0 | Margaric acid | 117 |
| C18:3 | Linolenic acid | 135 |
| C18:2 | Linoleic acid | 119 |
| C18:1 | Oleic acid | 143 |
| C18:0 | Stearic acid | 126 |
| C19:1 | 81 | |
| C19:0 | Nonadecanoic acid | 116 |
| C20:5 | Eicosapentaenoic acid | 182 |
| C20:4 | Arachidonic acid | 159 |
| C20:3 | 180 | |
| C20:2 | 185 | |
| C20:0 | Eicosanoic acid | 147 |
| Total | 132 |
Amounts of individual fatty acid in young adults were assessed by their esterification to fatty acid methyl esters (FAMEs) and quantification via gas chromatography–mass spectrometry in NR222 animals fed with lin-29(RNAi) and empty RNAi vector control (see Materials and Methods). A single run was performed on three independent biological trials per treatment.
LIN-29-regulated extracellular signaling proteins are required for the regulation of LIN-29 intestinal targets
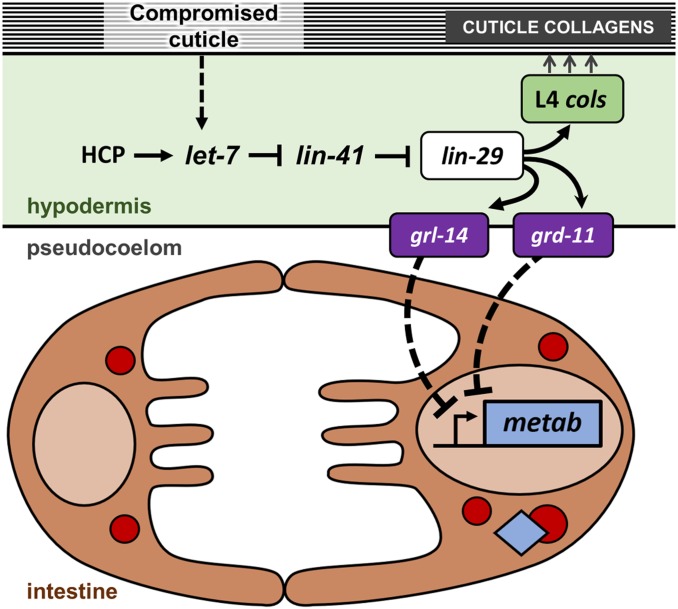
Our results on LIN-29-dependent downregulation of intestinal gene expression, combined with the fatty acid analysis in LIN-29 overexpressing and reduction-of-function animals, suggest a possible role for LIN-29 in coordinating intestinal metabolic activity at the larval to adult transition from the hypodermis.
We identified four genes encoding signaling molecules among our LIN-29 upregulated genes: three encoding C. elegans proteins related to the Hedgehog family of signals (grd-11, grl-14, wrt-6) and one encoding an insulin-like peptide (ins-37). When examining the wild-type postembryonic expression data (modENCODE) for these genes, we observed that grd-11 and grl-14 show low expression during larval life, but both wrt-6 and ins-37 show a marked peak of expression in the L4 stage, when hypodermal LIN-29 protein is active (Figure S7). Expression of three of these signal genes goes down in the lin-29(n333) L4 data set (Hunter et al. 2013) [grl-14 (0.57, P = 0.065); wrt-6 (0.17, P = 0.046); ins-37 (0.51, P = 0.058)]. Moreover, a wrt-6 reporter is expressed only in the hypodermis and in the socket cells of the amphids (Aspöck et al. 1999).
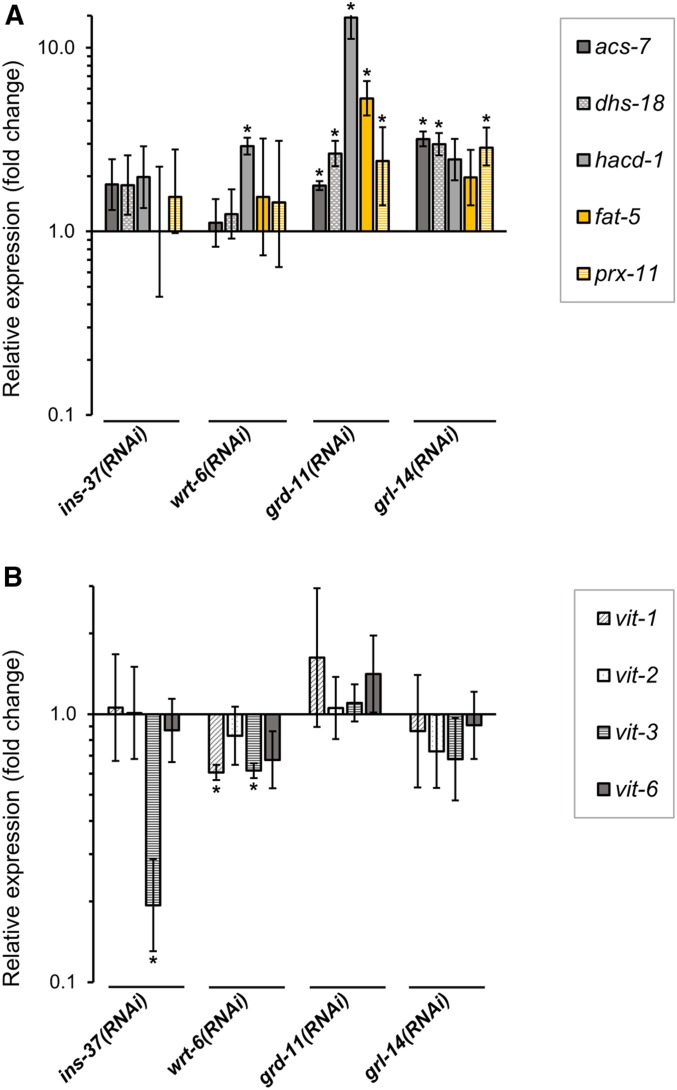
We individually tested the requirement for each of these four signaling genes both for repression of LIN-29-downregulated intestinal metabolic gene targets and for activation of vit genes in the late L4 stage using RNAi and qPCR. For the metabolic genes tested (acs-7, dhs-18, hacd-1, fat-5, and prx-11), at least one of the genes showed significantly increased expression in wrt-6, grd-11, or grl-14 RNAi-treated animals (Figure 9A). In the case of grd-11(RNAi), four of the metabolic genes showed significant increases, and for grl-14(RNAi), three of five genes did so. The gene hacd-1 showed an increase in expression in all three treatments of reduction-of-function for a Hedgehog-related factor. In the case of ins-37(RNAi) animals, four of five genes showed an almost twofold increase in expression, but the results were not significant at P < 0.05, so it is unclear if ins-37 functions to regulate these genes in the L4 stage.
Figure 9.
Four lin-29 target genes that encode signaling molecules regulate expression of LIN-29 intestinal targets in the L4 stage. Endogenous expression of five metabolic genes downregulated upon LIN-29 expression (acs-7, dhs-18, hacd-1, fat-5, and prx-11; A), and four vitellogenin genes previously shown to require lin-29 for their expression (vit-1, vit-2, vit-3 and vit-6; Dowen et al. 2016; B) was evaluated by RT-qPCR in late L4 stage ins-37(RNAi), wrt-6(RNAi), grd-11(RNAi)l or grl-14(RNAi) animals. Quantifications were relative to expression in animals treated with empty-vector RNAi control. Error bars represent SEM. * P < 0.05 (unpaired t-test).
The results for the vit genes were more variable (Figure 9B), with a significant decrease in expression for vit-3 in ins-37(RNAi) animals, and for vit-1 and vit-3 in wrt-6(RNAi) animals. Unlike the case for repression of the metabolic genes, the gene grd-11 appears to not be required for the activation of the vit genes in the intestine at the L4 stage, and while the results with grl-14 RNAi were consistent with a role, they were not significant. Nonetheless, it is interesting to note that the L4 expression of these vit genes is affected when the function is reduced for the two signaling genes that show a normal peak of expression at the L4 stage.
Together, these data suggest that several genes encoding signaling molecules that we identified as upregulated targets of LIN-29 may function in the repression of intestinal metabolic gene expression in the L4 stage when LIN-29 levels peak. These results support the hypothesis that LIN-29 may play a role in mediating the cell-nonautonomous regulation of at least some of the intestinal targets of LIN-29 through activation of signaling molecules (this work; Dowen et al. 2016).
Discussion
The LIN-29 transcription factor is the terminal effector of the heterochronic pathway in C. elegans and is necessary for the execution of a number of developmental processes at the larval-to-adult transition; however, the sufficiency of lin-29 for these processes has not be assessed. Here, we utilize a gain-of-function reagent to examine the phenotypic and gene regulatory effects caused by direct expression of lin-29 at earlier (L3) or later (adult) time points. We show that early expression of LIN-29 causes Dumpy, Egg-laying defective, and Small body size phenotypes, as well as precocious fusion of seam cells, all phenotypes displayed by lin-41(lf) mutant and lin-41(RNAi) animals (Slack et al. 2000; Tocchini et al. 2014). This result, together with our previous demonstration that early and late misexpression of LIN-29 can induce expression of endogenous col genes and a col-38 reporter (Abete-Luzi and Eisenmann 2018), indicates that LIN-29 alone is sufficient to initiate a number of processes occurring at the larval-to-adult transition. This is consistent with recent work indicating that there are only four relevant targets downstream of the heterochronic miRNA let-7, two of which are lin-29 and mab-10, which encodes a LIN-29-interacting protein (Aeschimann et al. 2019). We also found that early expression of lin-29 caused a curious “L4 vulva in an adult body” phenotype, which we hypothesize may be the result of the lin-29 causing precocious differentiation of the vulval cells; interestingly, vertebrate homologs of LIN-29 also have known prodifferentiation activities (Pagel and Deindl 2011). Finally, overexpression of LIN-29 in the adult was found to shorten life span. Taken together, these phenotypic effects suggest the importance of keeping lin-29 levels properly restrained until the correct time for the important transition to adulthood has been attained.
To further characterize the role of this transcription factor, we identified target genes regulated by LIN-29 using a gain-of-function approach involving temporal misexpression of LIN-29 at an earlier than normal time in development, followed by analysis of transcriptional changes an hour later. We believe this approach may more closely reflect the biological situation—a rapid increase in LIN-29 levels from a low starting point—and does not suffer from the caveat that observed changes in gene expression could be due to changes in cell fate earlier in development caused by a mutant background. One caveat of the gain-of-function approach is that overexpression of LIN-29 in tissues where it is not normally expressed may lead to the identification of spurious targets. However, it is worth pointing out that temporal misexpression of LIN-29 both earlier and later in development leads to col gene reporter expression only in the tissues in which it is normally detected, simply at an earlier or later time (Abete-Luzi and Eisenmann 2018). This suggests that lack of necessary cofactors, or a nonpermissive chromatin state, may prevent LIN-29 from activating targets genes in inappropriate tissues. This is consistent with work showing that after heat-shock expression of a neuronal transcription factor, targets genes only showed ectopic spatial expression when activity of a chromatin factor was compromised (Tursun et al. 2011). Although we cannot rule out that ectopically expressed LIN-29 may be directly affecting gene expression due to its presence in tissues where it is not normally expressed or due to higher levels of expression than normal, the fact that many of the putative LIN-29 target genes we identified in our gain-of-function approach were regulated in the opposite manner when lin-29 function was reduced by RNAi, and that many of them naturally show changes of expression in the L4 stage, strongly suggests that LIN-29 is likely to participate in the regulation of expression of these genes during normal development.
The major category of known genes upregulated by LIN-29 are those encoding cuticle collagen (col) genes (14% of known 1.7-fold upregulated genes). We identified 33 col genes, and the majority of them have a known peak of expression in the L4 or adult stage, after LIN-29 levels peak in the hypodermis. Although LIN-29 was previously shown to regulate several col genes (Liu et al. 1995; Rougvie and Ambros 1995; Abete-Luzi and Eisenmann 2018), this result suggests a much more pervasive role for LIN-29 in regulating the formation of the adult cuticle. The large number of L4 and adult col genes regulated by LIN-29 may reflect the fact that the adult cuticle is the most structurally complex of the cuticles synthesized by the worm (Page and Johnstone 2007), and unlike other cuticles, which need to last for less than a day, the adult lifelong cuticle may require more robustness.
Interestingly, we found that several col gene targets of LIN-29 in the L4 stage also show increased expression in the adult when cuticle integrity is compromised in both bli-1(RNAi) and bus-8(e2882) animals. As it was previously shown that bli-1(RNAi) treatment alters transcript levels for the heterochronic genes let-7, hbl-1, and lin-41, this suggests that the regulation of col genes by LIN-29 may also occur in response to cuticle damage, via a signal that acts through upstream heterochronic regulators of LIN-29 expression (Zhang et al. 2015; Zhao et al. 2019). This suggests that a mechanism to repair the cuticle during adult life may exist, and that this mechanism acts via the same molecular pathway that was utilized to synthesize the adult cuticle initially (Figure 10). Additional work will be needed to establish the mechanism by which the presence of cuticle damage in the adult leads to regulation of heterochronic pathway components to impinge on LIN-29 activity.
Figure 10.
Model of the roles of LIN-29 and its target genes in the hypodermis and intestine. In the hypodermis, lin-29 expression is regulated by the heterochronic pathway (HPC) acting through let-7 and lin-41. LIN-29 activates expression of many L4 and adult specific cuticle collagen (col) genes which contribute to the adult cuticle. Cuticle damage in the adult [e.g., in bli-1(RNAi) animals] signals through let-7 to increase expression of lin-29 and col genes. Dashed lines indicate that a mechanism is not yet known. Hypodermal LIN-29 also activates expression of signaling genes (grd-11 and grl-14 are shown as examples), which act to reduce expression of genes involved in fat metabolism in the intestine.
Besides the col genes, several other genes previously shown to depend on lin-29 activity for their expression were not identified in our list of hs::lin-29 upregulated genes. Several explanations for this result are possible: (1) a target could be expressed in too few cells to be identified by our global approach, (2) a target could require a different lin-29 isoform than lin-29a, (3) LIN-29 may be necessary but not sufficient for target gene regulation, or (4) a target gene was regulated in the expected direction but did not rise to statistical significance. However, two examples of putative LIN-29 targets bear commenting on. First, the vit genes (vit-1–6) have been shown to be positively regulated in the intestine by lin-29 activity in the hypodermis (Dowen et al. 2016), yet only two vit genes show increased expression in our hs::lin-29 animals (vit-2 and vit-6). Recently it was shown that expression of the vit genes in the intestine is regulated by the conserved transcription factors CEH-60 and UNC-62 (Dowen 2019). Neither ceh-60 nor unc-62 showed a significant change in expression in our gain-of-function data (Table S2) or in the lin-29(n333) reduction-of-function data (Hunter et al. 2013), suggesting they may not function directly downstream of LIN-29. Therefore, the fact that we did not observe significant effects on most vit genes upon LIN-29 overexpression could be an example of genes for which lin-29 is necessary but not sufficient for expression. Second, the transcription factor MAB-10 is known to physically interact with LIN-29 and participate in a number of lin-29-mediated processes (Harris and Horvitz 2011). In previous work, it was suggested that expression of mab-10 was not regulated by lin-29 (Harris and Horvitz 2011); however, we find mab-10 transcripts levels increase 2.3-fold upon overexpression of LIN-29 (Table S2), and mab-10 levels also decreased in both lin-29(n333) and let-7(lf) mutants (Hunter et al. 2013). This transcriptional regulation may be evolutionarily conserved, as the LIN-29 orthologs EGR1, EGR2, and EGR3 all regulate expression of the MAB-10 ortholog NAB2 through binding sites in the NAB2 gene promoter (Kumbrink et al. 2005, 2010). These results suggest that mab-10 is likely to be a transcriptional target of LIN-29, and that lin-29 and mab-10 may have complicated effects on each other’s expression and activity.
A major category of genes showing downregulation upon LIN-29 misexpression are genes involved in lipid metabolism (7% of 1.7× downregulated genes, n = 25). Many of the lipid metabolic genes we identified show temporary or permanent downregulation of expression in the L4 stage when LIN-29 levels peak. We also showed that lin-29 reduction of function by RNAi causes an increase in expression of several of these genes at the L4 stage. LIN-29 overexpression also caused a decrease in expression of nhr-80 and sbp-1, two transcription factors that regulate downstream metabolic genes (McKay et al. 2003; Kniazeva et al. 2004; Van Gilst et al. 2005; Brock et al. 2006; Nomura et al. 2010; Pathare et al. 2012; MacNeil et al. 2015), including fat-5 (Watts and Browse 2000; Brock et al. 2006). Thus, our target gene identification suggests a broader role for LIN-29 in regulating fat metabolism at the adult transition than previously suggested. The demonstration that both increased expression of lin-29 (hs::lin-29 animals) and decreased expression of lin-29 [lin-29(RNAi) animals] during larval life lead to opposite changes in fatty acid content in young adult animals is supportive of the hypothesis that lin-29 regulates genes involved in fat metabolism during normal development.
Many of the metabolic genes we identified have known expression in the intestine, a major site of metabolic activity that does not express lin-29, suggesting that lin-29 may regulate metabolic gene expression cell-nonautonomously. We identified four LIN-29 upregulated target genes that encode extracellular signaling proteins belonging to the insulin-like peptide and hedgehog-related families and showed that they negatively regulate expression of several LIN-29 metabolic gene targets in the L4 stage. While RNAi of these individual genes in a sensitized background was sufficient to cause an effect on metabolic gene expression, we were unable to show that downregulation of those metabolic gene targets by hs::lin-29 was significantly dependent on activity of the signaling genes (data not shown). Whether this is due to the increased level of signal gene transcript caused by the higher levels of LIN-29 induced by heat shock, or to other reasons, is currently unknown. Regardless, the identification of these signaling genes as activated targets of LIN-29 that can regulate fatty acid metabolic genes expressed in the intestine, combined with the demonstration that reduction of lin-29 activity in only the hypodermal tissue can alter expression of these same fatty acid metabolic genes and increase fatty acid levels, supports a model that lin-29 activity in the hypodermis cell-nonautonomously coordinates fat metabolic gene expression in the intestine during normal development (Figure 10).
Three of the consequences of the transition from larval life to adulthood are (1) the period of rapid growth in length and volume that occurs during larval life comes to an end (Knight et al. 2002); (2) consistent with this, the active metabolism seen in larval life slows down (Braeckman et al. 2009); and (3) worms become reproductive and mobilize fat from the intestine into developing oocytes via movement of yolk (the major protein component of which are vitellogenins) (Dowen et al. 2016; Watts and Ristow 2017). The identification of genes involved in fatty acid synthesis and beta-oxidation as downregulated targets of LIN-29 (this work), combined with the previous identification of vit genes as activated targets (Dowen et al. 2016), suggests that LIN-29 may play a major role in effecting these changes. For example, if the adult worm needs to slow its growth and mobilize fat stores from somatic tissues to germ cells, it must slow down the burning of fat, which is consistent with the decrease in expression of many genes involved in fatty acid beta-oxidation (Figure 5). Likewise, a decrease in the storage of fat in somatic tissues (to make it available for mobilization to oocytes) is consistent with a decrease in expression of Y53G8B.2, K07B1.4, and dgat-2, which encode three of four C. elegans DGAT enzymes that function in triacylglycerol synthesis (Figure 5) (Watts and Ristow 2017). Thus, the changes in fatty acid metabolic gene expression we observed on early overexpression of LIN-29 can be rationalized based on a normal role for LIN-29 in coordinating metabolic changes at the larval-to-adult transition. Intriguingly, one vertebrate homolog of LIN-29, EGR-1 (Harris and Horvitz 2011), is expressed in adipose tissue, and misexpression of EGR-1 correlates with dietary-induced obesity and insulin resistance in both mice and humans (J. Zhang et al. 2013). Conversely, loss of Egr-1 function protects mice from dietary-induced pathologies such as insulin resistance and hyperlipidemia, most likely due to an altered balance between energy expenditure and storage (J. Zhang et al. 2013). Therefore, it is possible that the regulation of energy metabolism by this type of zinc finger transcriptional regulator may have been conserved during evolution.
Acknowledgments
We thank Andy Golden for providing plasmids, Helge Großhans for providing lin-29 strains, Joshua Wilhide from the University of Maryland Baltimore County (UMBC) Molecular Characterization and Analysis Complex for technical assistance with the GC-MS analysis, and Belinda Jackson for assistance with bli-1 RNAi. We thank the WormBase consortium and its developers for their achievements and their assistance. We also thank members of the Eisenmann laboratory for advice and support. Lastly, we thank the Caenorhabditis Genetics Center, which provided some of the strains. This work was financially supported by the Department of Biological Sciences, UMBC (biology.umbc.edu); the National Institute of Child Health and Human Development (nichd.nih.gov; award R03 HD096313 to D.M.E.); and the intramural program of the National Institute of Diabetes and Digestive and Kidney Diseases (to M.W.K.).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11663883.
Communicating editor: S. Kennedy
Literature Cited
- Abbott A. L., Alvarez-Saavedra E., Miska E. A., Lau N. C., Bartel D. P. et al. , 2005. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell 9: 403–414. 10.1016/j.devcel.2005.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbdelRaheim S. R., and McLennan A. G., 2002. The Caenorhabditis elegans Y87G2A.14 Nudix hydrolase is a peroxisomal coenzyme A diphosphatase. BMC Biochem. 3: 5 10.1186/1471-2091-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abete-Luzi P., and Eisenmann D. M., 2018. Regulation of C. elegans L4 cuticle collagen genes by the heterochronic protein LIN-29. Genesis 56: 1–8. 10.1002/dvg.23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. C., Lu Y., and Shaham S., 2007. A morphologically conserved nonapoptotic program promotes linker cell death in Caenorhabditis elegans. Dev. Cell 12: 73–86. 10.1016/j.devcel.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Abrahante J. E., Miller E. A., and Rougvie A. E., 1998. Identification of heterochronic mutants in Caenorhabditis elegans: temporal misexpression of a collagen:green fluorescent protein fusion gene. Genetics 149: 1335–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante J. E., Daul A. L., Li M., Volk M. L., Tennessen J. M. et al. , 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. 10.1016/S1534-5807(03)00127-8 [DOI] [PubMed] [Google Scholar]
- Aeschimann F., Kumari P., Bartake H., Gaidatzis D., Xu L. et al. , 2017. LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell 65: 476–489.e4. 10.1016/j.molcel.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Aeschimann F., Neagu A., Rausch M., and Großhans H., 2019. let-7 coordinates the transition to adulthood through a single primary and four secondary targets. Life Sci. Alliance 2: e201900335 10.26508/lsa.201900335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altun Z. F., and Hall D. H., 2009. Introduction. In WormAtlas. 10.3908/wormatlas.1.1 [DOI] [Google Scholar]
- Ambros V., and Horvitz H., 1984. Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226: 409–416. 10.1126/science.6494891 [DOI] [PubMed] [Google Scholar]
- Angeles-Albores D., N Lee R. Y., Chan J., and Sternberg P. W., 2016. Tissue enrichment analysis for C. elegans genomics. BMC Bioinformatics 17: 366 10.1186/s12859-016-1229-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspöck G., Kagoshima H., Niklaus G., and Bürglin T. R., 1999. Caenorhabditis elegans has scores of hedgehog-related genes: sequence and expression analysis. Genome Res. 9: 909–923. 10.1101/gr.9.10.909 [DOI] [PubMed] [Google Scholar]
- Bettinger J. C., Lee K., and Rougvie A. E., 1996. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122: 2517–2527. [DOI] [PubMed] [Google Scholar]
- Bettinger J. C., Euling S., and Rougvie A. E., 1997. The terminal differentiation factor LIN-29 is required for proper vulval morphogenesis and egg laying in Caenorhabditis elegans. Development 124: 4333–4342. [DOI] [PubMed] [Google Scholar]
- Blum E. S., Abraham M. C., Yoshimura S., Lu Y., and Shaham S., 2012. Control of nonapoptotic developmental cell death in Caenorhabditis elegans by a polyglutamine-repeat protein. Science 335: 970–973. 10.1126/science.1215156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., and Steward K., 1997. RNA processing and gene structure, pp. 117–145 in C. elegans II, edited by Riddle D. L. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Boehm M., and Slack F., 2005. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957. 10.1126/science.1115596 [DOI] [PubMed] [Google Scholar]
- Braeckman B. P., Houthoofd K., and Vanfleteren J. R., 2009. Intermediary metabolism (February 16, 2009), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.146.1, http://www.wormbook.org. 10.1895/wormbook.1.146.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock T. J., Browse J., and Watts J. L., 2006. Genetic regulation of unsaturated fatty acid composition in C. elegans. PLoS Genet 2: e108 10.1371/journal.pgen.0020108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos V., and Partridge L., 2017. Good Ol’ fat: links between lipid signaling and longevity. Trends Biochem. Sci. 42: 812–823. 10.1016/j.tibs.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B. et al. , 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. F., Meade M., Ranepura G., Hall D. H., and Savage-Dunn C., 2018. Caenorhabditis elegans DBL-1/BMP regulates lipid accumulation via interaction with insulin signaling. G3 (Bethesda) 8: 343–351. 10.1534/g3.117.300416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen R. H., 2019. CEH-60/PBX and UNC-62/MEIS coordinate a metabolic switch that supports reproduction in C. elegans. Dev. Cell 49: 235–250.e7. 10.1016/j.devcel.2019.03.002 [DOI] [PubMed] [Google Scholar]
- Dowen R. H., Breen P. C., Tullius T., Conery A. L., and Ruvkun G., 2016. A microRNA program in the C. elegans hypodermis couples to intestinal mTORC2/PQM-1 signaling to modulate fat transport. Genes Dev. 30: 1515–1528. 10.1101/gad.283895.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J., Mutlu A. S., and Wang M. C., 2016. Lipid metabolism, lipid signalling and longevity, pp. 307–329 in Ageing: Lessons from C. elegans, Springer, Cham, Switzerland. [Google Scholar]
- Ecsedi M., and Großhans H., 2013. Lin-41/Trim71: emancipation of a miRNA target. Genes Dev. 27: 581–589. 10.1101/gad.207266.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euling S., Bettinger J. C., and Rougvie A. E., 1999. The LIN-29 transcription factor is required for proper morphogenesis of the Caenorhabditis elegans male tail. Dev. Biol. 206: 142–156. 10.1006/dbio.1998.9063 [DOI] [PubMed] [Google Scholar]
- Ewald C. Y., Landis J. N., Abate J. P., Murphy C. T., and Blackwell T. K., 2015. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature 519: 97–101. 10.1038/nature14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., Guardavaccaro D., Neubert K., Chan T., Li D. et al. , 2007. DRE-1: an evolutionarily conserved F Box protein that regulates C. elegans developmental age. Dev. Cell 12: 443–455. 10.1016/j.devcel.2007.01.018 [DOI] [PubMed] [Google Scholar]
- Friedlander-Shani L., and Podbilewicz B., 2011. Heterochronic control of AFF-1-mediated cell-to-cell fusion in C. elegans, pp. 5–11 in Cell Fusion in Health and Disease, Vol. 713 Springer, Dordrecht, The Netherlands: 10.1007/978-94-007-0763-4_2 [DOI] [PubMed] [Google Scholar]
- Garigan D., Hsu A. L., Fraser A. G., Kamath R. S., Abringet J. et al. , 2002. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics 161: 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. 10.1126/science.1196914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrepati L., Krause M. W., Chen W., Brodigan T. M., Correa-Mendez M., et al. , 2015. Identification of Wnt pathway target genes regulating the division and differentiation of larval seam cells and vulval precursor cells in Caenorhabditis elegans. G3 (Bethesda) 5: 1551–1566. 10.1534/g3.115.017715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, B. P., W. Hanna-Rose, and P. W. Sternberg, 2012 Morphogenesis of the vulva and the vulval-uterine connection (November 30, 2012), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.152.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. K., Lee D., Lee H., Kim D., Son H. G. et al. , 2016. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7: 56147–56152. 10.18632/oncotarget.11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris D. T., and Horvitz H. R., 2011. MAB-10/NAB acts with LIN-29/EGR to regulate terminal differentiation and the transition from larva to adult in C. elegans. Development 138: 4051–4062. 10.1242/dev.065417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes G. D., Frand A. R., and Ruvkun G., 2006. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development 133: 4631–4641. 10.1242/dev.02655 [DOI] [PubMed] [Google Scholar]
- Hibbs M. A., Hess D. C., Myers C. L., Huttenhower C., Li K. et al. , 2007. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23: 2692–2699. 10.1093/bioinformatics/btm403 [DOI] [PubMed] [Google Scholar]
- Hong Y., Roy R., and Ambros V., 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- Houbaviy H. B., Murray M. F., and Sharp P. A., 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5: 351–358. 10.1016/S1534-5807(03)00227-2 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., and Lempicki R. A., 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. F., Cho C. Y., Cheng Y. T., Huang J. W., Wu Y. Z. et al. , 2014. BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 10: e1004428 10.1371/journal.pgen.1004428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S. E., Finnegan E. F., Zisoulis D. G., Lovci M. T., Melnik-Martinez K. V. et al. , 2013. Functional genomic analysis of the let-7 regulatory network in Caenorhabditis elegans. PLoS Genet. 9: e1003353 10.1371/journal.pgen.1003353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Wang M., Ririe T. O., Fernandes J. S., and Sternberg P. W., 2005. Transcriptional network underlying Caenorhabditis elegans vulval development. Proc. Natl. Acad. Sci. USA 102: 4972–4977. 10.1073/pnas.0408122102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson B. M., Abete-Luzi P., Krause M. W., and Eisenmann D. M., 2014. Use of an activated beta-catenin to identify Wnt pathway target genes in Caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3 (Bethesda) 4: 733–747. 10.1534/g3.113.009522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., and Ahringer J., 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. 10.1016/S1046-2023(03)00050-1 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., and Ahringer J., 2000. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: RESEARCH0002 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M., Crawford Q. T., Seiber M., Wang C. Y., and Han M., 2004. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2: E257 10.1371/journal.pbio.0020257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. G., Patel M. N., Azevedo R. B. R., and Leroi A. M., 2002. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol. Dev. 4: 16–27. 10.1046/j.1525-142x.2002.01058.x [DOI] [PubMed] [Google Scholar]
- Kumbrink J., Gerlinger M., and Johnson J. P., 2005. Egr-1 induces the expression of its corepressor Nab2 by activation of the Nab2 promoter thereby establishing a negative feedback loop. J. Biol. Chem. 280: 42785–42793. 10.1074/jbc.M511079200 [DOI] [PubMed] [Google Scholar]
- Kumbrink J., Kirsch K. H., and Johnson J. P., 2010. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J. Cell. Biochem. 111: 207–217. 10.1002/jcb.22690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. Y. N., Howe K. L., Harris T. W., Arnaboldi V., Cain S. et al. , 2018. WormBase 2017: molting into a new stage. Nucleic Acids Res. 46: D869–D874. 10.1093/nar/gkx998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux G. A., and Ashrafi K., 2015. Insights and challenges in using C. elegans for investigation of fat metabolism. Crit. Rev. Biochem. Mol. Biol. 50: 69–84. 10.3109/10409238.2014.959890 [DOI] [PubMed] [Google Scholar]
- Liang B., Ferguson K., Kadyk L., and Watts J. L., 2010. The role of nuclear receptor NHR-64 in fat storage regulation in caenorhabditis elegans. PLoS One 5: e9869 10.1371/journal.pone.0009869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-Y., Johnson S. M., Abraham M., Vella M. C., Pasquinelli A. et al. , 2003. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. 10.1016/S1534-5807(03)00124-2 [DOI] [PubMed] [Google Scholar]
- Lints R., and Hall D. H., 2009. The cuticle, in WormAtlas. 10.3908/wormatlas.1.12 [DOI] [Google Scholar]
- Liu Z., Kirch S., and Ambros V., 1995. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development 121: 2471–2478. [DOI] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- MacNeil L. T., Pons C., Arda H. E., Giese G. E., Myers C. L. et al. , 2015. Transcription factor activity mapping of a tissue-specific in vivo gene regulatory network. Cell Syst. 1: 152–162. 10.1016/j.cels.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R. M., McKay J. P., Avery L., and Graff J. M., 2003. C elegans: a model for exploring the genetics of fat storage. Dev. Cell 4: 131–142. 10.1016/S1534-5807(02)00411-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss E. G., 2007. Heterochronic genes and the nature of developmental time. Curr. Biol. 17: R425–R434. 10.1016/j.cub.2007.03.043 [DOI] [PubMed] [Google Scholar]
- Moss E. G., and Romer-Seibert J., 2014. Cell-intrinsic timing in animal development. Wiley Interdiscip. Rev. Dev. Biol. 3: 365–377. 10.1002/wdev.145 [DOI] [PubMed] [Google Scholar]
- Munoz-Torres M., and Carbon S., 2017. Get GO! retrieving GO data using AmiGO, QuickGO, API, files, and tools, pp. 149–160 in Methods in Molecular Biology, Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- Newman A. P., Inoue T., Wang M., and Sternberg P. W., 2000. The Caenorhabditis elegans heterochronic gene lin-29 coordinates the vulval-uterine-epidermal connections. Curr. Biol. 10: 1479–1488. 10.1016/S0960-9822(00)00827-7 [DOI] [PubMed] [Google Scholar]
- Nimmo R. A., and Slack F. J., 2009. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma 118: 405–418. 10.1007/s00412-009-0210-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Horikawa M., Shimamura S., Hashimoto T., and Sakamoto K., 2010. Fat accumulation in Caenorhabditis elegans is mediated by SREBP homolog SBP-1. Genes Nutr. 5: 17–27. 10.1007/s12263-009-0157-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. P., and Johnstone I. L., 2007. The cuticle, WormBook, ed. The C. elegans Research Community WormBook, http://www.wormbook.org, doi/10.1895/wormbook.1.138.1 [Google Scholar]
- Pagel J.-I., and Deindl E., 2011. Early growth response 1–a transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 48: 226–235. [PubMed] [Google Scholar]
- Partridge F. A., Tearle A. W., Gravato-Nobre M. J., Schafer W. R., and Hodgkin J., 2008. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 317: 549–559. 10.1016/j.ydbio.2008.02.060 [DOI] [PubMed] [Google Scholar]
- Pathare P. P., Lin A., Bornfeldt K. E., Taubert S., and van Gilst M. R., 2012. Coordinate regulation of lipid metabolism by novel nuclear receptor partnerships. PLoS Genet. 8: e1002645 10.1371/journal.pgen.1002645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H., Inoue M., Hikita T., Köppen M., Hardin J. D. et al. , 2007. Establishment of a tissue-specific RNAi system in C. elegans. Gene 400: 166–173. 10.1016/j.gene.2007.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch M., Ecsedi M., Bartake H., Müllner A., and Großhans H., 2015. A genetic interactome of the let-7 microRNA in C. elegans. Dev. Biol. 401: 276–286. 10.1016/j.ydbio.2015.02.013 [DOI] [PubMed] [Google Scholar]
- Ririe T. O., Fernandes J. S., and Sternberg P. W., 2008. The Caenorhabditis elegans vulva: a post-embryonic gene regulatory network controlling organogenesis. Proc. Natl. Acad. Sci. USA 105: 20095–20099. 10.1073/pnas.0806377105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie A. E., and Ambros V., 1995. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development 121: 2491–2500. [DOI] [PubMed] [Google Scholar]
- Rougvie A. E., and Moss E. G., 2013. Developmental transitions in C. elegans larval stages. Curr. Top Dev. Biol. 105: 153–180. 10.1016/B978-0-12-396968-2.00006-3 [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A.-S. S. et al. , 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14: 2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L. et al. , 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. 10.1016/S0960-9822(02)01041-2 [DOI] [PubMed] [Google Scholar]
- Slack F. J., Basson M., Liu Z., Ambros V., Horvitz H. R. et al. , 2000. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell 5: 659–669. 10.1016/S1097-2765(00)80245-2 [DOI] [PubMed] [Google Scholar]
- Sternberg P. W., Vulval development . 2005. WormBook, ed. The C. elegans Research Community, WormBook, 10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Su J.-L., Chen P.-S., Johansson G., and Kuo M.-L., 2012. Function and regulation of let-7 family microRNAs. MicroRNA 1: 34–39. 10.2174/2211536611201010034 [DOI] [PubMed] [Google Scholar]
- Sutphin G. L., and Kaeberlein M., 2009. Measuring Caenorhabditis elegans life span on solid media. J. Vis. Exp. (27): 1152 10.3791/1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini C., Keusch J. J., Miller S. B., Finger S., Gut H. et al. , 2014. The TRIM-NHL protein LIN-41 controls the onset of developmental plasticity in Caenorhabditis elegans. PLoS Genet. 10: e1004533 10.1371/journal.pgen.1004533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsialikas J., and Romer-Seibert J., 2015. LIN28: roles and regulation in development and beyond. Development 142: 2397–2404. 10.1242/dev.117580 [DOI] [PubMed] [Google Scholar]
- Tursun B., Patel T., Kratsios P., and Hobert O., 2011. Direct conversion of C. elegans germ cells into specific neuron types. Science 331: 304–308. 10.1126/science.1199082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Jolly A., and Yamamoto K. R., 2005. Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53 10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., 2009. Fat synthesis and adiposity regulation in Caenorhabditis elegans. Trends Endocrinol. Metab. 20: 58–65. 10.1016/j.tem.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., and Browse J., 2000. A palmitoyl-CoA-specific Δ9 fatty acid desaturase from Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 272: 263–269. 10.1006/bbrc.2000.2772 [DOI] [PubMed] [Google Scholar]
- Watts J. L., and Browse J., 2002. Genetic dissection of polyunsaturated fatty acid synthesis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 99: 5854–5859. 10.1073/pnas.092064799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts J. L., and Ristow M., 2017. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 207: 413–446. 10.1534/genetics.117.300106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildwater M., Sander N., de Vreede G., and van den Heuvel S., 2011. Cell shape and Wnt signaling redundantly control the division axis of C. elegans epithelial stem cells. Development 138: 4375–4385. 10.1242/dev.066431 [DOI] [PubMed] [Google Scholar]
- Worringer K. A., Rand T. A., Hayashi Y., Sami S., Takahashi K. et al. , 2014. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 14: 40–52. 10.1016/j.stem.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zhang Y., Sun T., Guo F., Huang S. et al. , 2013. Dietary obesity-induced Egr-1 in adipocytes facilitates energy storage via suppression of FOXC2. Sci. Rep. 3: 1–10. 10.1038/srep01476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. O., Box A. C., Xu N., Le Men J., Yu J. et al. , 2010. Genetic and dietary regulation of lipid droplet expansion in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107: 4640–4645. 10.1073/pnas.0912308107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zou X., Ding Y., Wang H., Wu X. et al. , 2013. Comparative genomics and functional study of lipid metabolic genes in Caenorhabditis elegans. BMC Genomics 14: 164 10.1186/1471-2164-14-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li W., Li L., Li Y., Fu R. et al. , 2015. Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 42: 309–320. 10.1016/j.immuni.2015.01.014 [DOI] [PubMed] [Google Scholar]
- Zhao L., Dong S., Zhao Y., Shao H., Krasteva N. et al. , 2019. Dysregulation of let-7 by PEG modified graphene oxide in nematodes with deficit in epidermal barrier. Ecotoxicol. Environ. Saf. 169: 1–7. 10.1016/j.ecoenv.2018.10.106 [DOI] [PubMed] [Google Scholar]
- Zhu X., Liu Y., Zhang H., and Liu P., 2018. Whole-genome RNAi screen identifies methylation-related genes influencing lipid metabolism in Caenorhabditis elegans. J. Genet. Genomics 45: 259–272. 10.1016/j.jgg.2018.03.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Original gene expression data underlying this work are fully available without restriction from the Gene Expression Omnibus archive (www.ncbi.nlm.nih.gov/geo/) under accession number GSE118433. After publication, some of the data from this paper will be available in the publicly accessible, curated database WormBase (wormbase.org). Any reagents and strains utilized in this work will either be available from a publicly accessible strain repository (the Caenorhabditis Genetic Center) or freely available upon request from the corresponding author. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11663883.