Abstract
Sex determination is remarkably variable among animals with examples of environmental sex determination, male heterogametic (XX/XY) and female heterogametic (ZZ/ZW) chromosomal sex determination, and other genetic mechanisms. The cephalochordate amphioxus occupies a key phylogenetic position as a basal chordate and outgroup to vertebrates, but its sex determination mechanism is unknown. During the course of generating Nodal mutants with transcription activator-like effector nucleases (TALENs) in amphioxus Branchiostoma floridae, serendipitously, we generated three mutant strains that reveal the sex determination mechanism of this animal. In one mutant strain, all heterozygous mutant offspring over three generations were female and all wild-type descendants were male. This pattern suggests the Nodal allele targeted is on a female-specific W chromosome. A second mutant showed the same W-linked inheritance pattern, with a female heterozygote passing the mutation only to daughters. In a third mutant strain, both male and female offspring could be heterozygous, but a female heterozygote passed the mutation only to sons. This pattern is consistent with the targeted allele being on a Z chromosome. We found an indel polymorphism linked to a Nodal allele present in most females, but no males in our cultured population. Together, these results indicate that Nodal is sex chromosome-linked in B. floridae, and that B. floridae has a ZZ/ZW sex chromosome system.
Keywords: Sex determination, ZZ/ZW, sex-linked marker, amphioxus, Genetics of Sex
SEXUAL reproduction (meiosis and syngamy of microgamete and macrogamete) is a fundamental character of metazoans. Remarkable mechanistic diversity underpins this shared character, with hermaphroditic species (individuals producing both microgametes or sperm and macrogametes or eggs) and gonochoristic species (with distinct males and females). Among the latter, determination of sex can occur through genetic sex-determining (GSD) or environmental sex-determining (ESD) systems (Beukeboom and Perrin 2014). Within vertebrates, mammals and birds utilize GSD systems and generally have specialized sex-determining chromosomes, denoted either X and Y (XX female, XY male) or Z and W (ZZ male, ZW female), while reptiles, amphibians, and fishes have a diversity of GSD systems (e.g., XX/XY, ZZ/ZW, X0/XX, Z0/ZZ) and ESD systems (for two recent reviews of sex determination, see Bachtrog et al. 2014; Capel 2017). Across invertebrates, many species have male or female heterogamety, but alternative chromosomal sex-determining systems are also known including haplodiploidy (Blackmon et al. 2015) and paternal genome loss (Beukeboom 2017; Hodson et al. 2017). It is not known what the sex-determination system was in the invertebrate ancestors of vertebrates.
The sex-determining system in GSD species can be determined experimentally via several approaches: chromosomal karyotype analysis (Dos Santos et al. 2016), gynogenesis, or breeding sex-reversed individuals (Wallace et al. 1999; Roco et al. 2015), and uncovering sex-linked markers (Gamble and Zarkower 2014). Obviously, karyotype analysis can be applied only in species with heteromorphic sex chromosomes. Gynogenesis is a method for producing animals in which all the genetic information originates from the female parent. Therefore, if gynogenesis were to yield only female offspring, it would reveal a female homogametic system (e.g., XX/XY), whereas if it were to yield both female and male offspring, it would suggest a male homogametic system (e.g., ZZ/ZW). However, inducing gynogenesis is generally difficult in most animals. In contrast to chromosomal karyotype analysis and gynogenesis, identifying sex-linked markers is universally applicable to GSD species, although, in practice, it is sometimes costly and labor-intensive. In a species, if a molecular marker is present only in males but not females, this indicates an XX/XY system (Gamble and Zarkower 2014). By contrast, when a marker is specifically present in females but not males, this would suggest a ZZ/ZW system (Wilson et al. 2014).
Amphioxus, or lancelets, belong to the Cephalochordata, a basally branching lineage of Chordata, and sister group to Tunicata and Vertebrata. In common with most vertebrates, but differing from most tunicates, amphioxus is gonochoristic with distinct male and female animals. The underlying mechanistic basis of sex determination has long been unclear, although investigations of wild populations reveal a 1:1 ratio of males and females (Stokes and Holland 1996; Henmi and Yamaguchi 2003; Kubokawa et al. 2003), consistent with a genetic mechanism. Nonetheless karyotype analysis of four well-studied amphioxus species have not revealed clear size differences among the two sets of homologous chromosomes, thus XY and ZW systems are both possible (Nogusa 1957; Wang et al. 2003, 2004). Here, we report a ZZ/ZW sex determination system in Branchiostoma floridae, and a female-linked genetic marker to distinguish male and female individuals.
Materials and Methods
Animals and breeding
Wild type (WT) B. floridae juveniles were introduced from the laboratory of Jr-Kai Yu and bred in our aquarium as previously reported (Li et al. 2012). Adult females and males were selected randomly and crossed to generate progenies for laboratory population maintenance.
Generating amphioxus Nodal mutant
Two TALEN pairs, Nodal-Fw1/Rv1 and Nodal-Fw2/Rv2, targeting the exon I coding sequence of the Nodal gene (Figure 1), were assembled using the Goldy TALEN system as previously described (Li et al. 2014). The target sites were designed to span a PstI and a SacI restriction site, respectively. TALEN mRNAs were synthesized in vitro using T3 mMESSAGE mMACHINE kit (Ambion). Mutants were generated as previously reported (Li et al. 2017). In this study, we define animals injected with TALEN mRNAs as founders, progenies of crosses between founders and WT as F1 animals, progenies of crosses between F1 and WT as F2, and progenies of crosses between F2 and WT as F3.
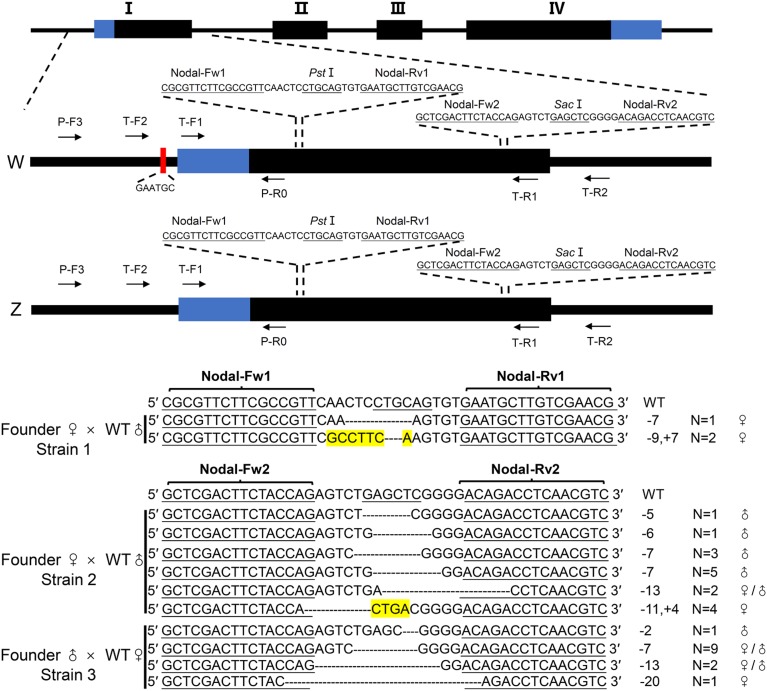
Figure 1.
Schematic diagram of Nodal gene alleles. Black boxes indicate exons (I, II, III and IV). Black lines indicate introns or flanking sequences of Nodal gene. Blue boxes represent 5′- or 3′- untranslated regions. A red box marks the 6-bp female-linked marker. Horizontal arrows show primer directions. The target sites of TALEN pair Nodal-Fw1/Rv1 and Nodal-Fw2/Rv2 are highlighted. W and Z indicate the Nodal allele on W and Z chromosome, respectively. The mutation types, relative to WT sequences carried by F1 heterozygotes from three strains, are listed, with dotted lines representing deletions and yellow-highlighted letters representing insertions. The sex and the number of F1 heterozygotes carrying an identical mutation type are also stated.
Genotype identification and mutation type analysis
Animal genotyping was performed as previously reported (Li et al. 2017). In brief, by juvenile stage or adulthood, a tiny tail tip of each individual was cut and lysed with Animal Tissue Direct PCR Kit (Foregene). Lysed samples were amplified individually using primer pair T-F1/T-R1 (for TALEN pair Nodal-Fw1/Rv1) or T-F2 /T-R2 (for TALEN pair Nodal-Fw2/Rv2). PCR products were digested with PstI (for Nodal-Fw1/Rv1) or SacI (for Nodal-Fw2/Rv2) enzyme and separated on 1.2% agarose gel to identify their genotypes. To analyze the mutation type of each Nodal+/− heterozygous progeny, PCR products amplified using both primer pairs T-F1/T-R1 and T-F2/T-R2 were sequenced with primer T-F1. The primer sequences used in this study are listed in Supplemental Material, Table S1.
Hybrid experiments
Three amphioxus strains were generated in this study. Strain 1 is from a female founder injected with Nodal-Fw1/Rv1 mRNAs. This female was crossed with a WT male to generate F1 progeny. From the F1 population, two female heterozygotes, respectively carrying a 2 bp (−9 bp, +7 bp, mutation type I) and 7 bp deletion (mutation type II), were identified. Each one was crossed with a different WT male to generate F2 progenies. One randomly selected F2 female Nodal+/− carrying type I mutation was further crossed with a WT male to generate F3 progenies. Strains 2 and 3 are from female and male founders, respectively, injected with Nodal-Fw2/Rv2 mRNAs. From their F1 descendants, one female carrying a (−11 bp, +4 bp) mutation from strain 2 and one female carrying a 7 bp deletion (−7 bp) from strain 3 were identified, and each was crossed with a different WT male to generate their F2 progenies.
Survey of sex-linked markers
A female-specific 6-bp polymorphism identified in strains 2 and 3 was also surveyed in 11 females and 9 males from strain 1, and 15 female and 10 male WT animals from our laboratory population. The DNA fragment was amplified with primers T-F2/T-R2 and sequenced with primer P-R0, or amplified with primers P-F3/P-R0 and digested with BsmI. Sequences from other Nodal genomic regions were amplified with primers shown in Figure S3. Another 12 individuals (6 females and 6 males) randomly selected from our aquarium were included in this analysis. All PCR products except those amplified with G-F1/G-R1 were sequenced directly, and amplicons from G-F1/G-R1 were ligated to pGEM-T Easy vector (Promega) before sequencing due to its high polymorphism. Two to five colonies were sequenced for each ligation. Only sequences from an animal from which both haplotypes were detected were used for further analysis. The primer sequences are listed in Table S1.
Linkage analysis
To clarify if the mutation carried by strain 1 heterozygotes is linked with a 6-bp polymorphism on W chromosome (female heteromorphic sex chromosome), a DNA fragment spanning the marker and the two TALEN-targeted sites was amplified from the genome of one F1 female Nodal+/− heterozygote of strain 1 with primer pair T-F2/T-R1, and then cloned into pGEM-T Easy vector (Promega). A total of 24 clones were collected randomly for sequencing.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental material at figshare. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11689311.
Results
Identification of ZZ/ZW sex chromosome system for amphioxus by analyzing Nodal mutants
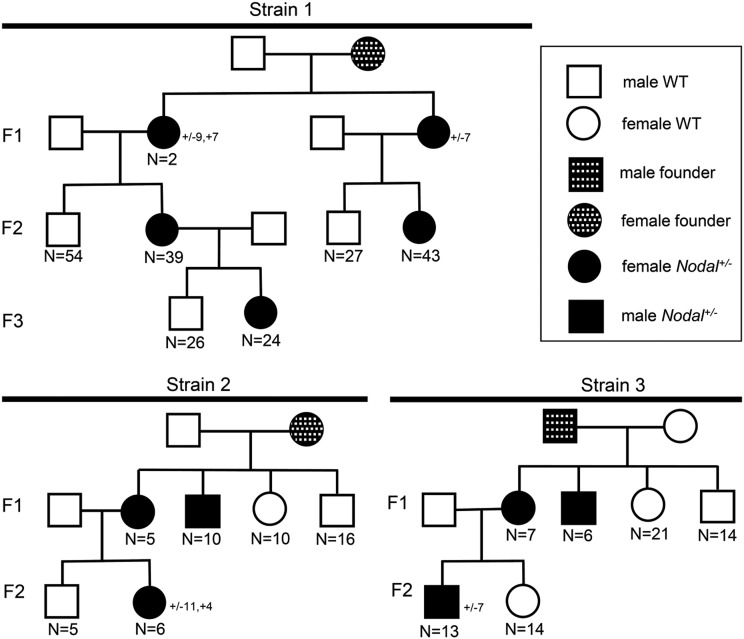
We recently revealed an essential role for Nodal signaling in left–right axis specification of amphioxus (Li et al. 2017). To define the function of the Nodal gene in the process, we constructed a TALEN pair Nodal-Fw1/Rv1 (Figure 1) and used it to generate Nodal mutants. We identified one female founder carrying germline mutations and crossed it with a WT male (named strain 1). Among the F1 progeny, three individuals carrying two types of frame-shift mutations (−9 bp, +7 bp, type I mutation; and −7 bp, type II mutation) in one allele of Nodal locus were identified, which were later found to be all females (Figure 1 and Figure 2). To culture this Nodal+/− population, females with the two different mutations were crossed with two WT males each. In all, 39 F2 heterozygotes and 54 WT siblings were identified from the brood inheriting the type I mutation (Figure 2). Remarkably, when these animals became sexually mature, we found that all identified heterozygotes are female, and all their WT siblings are male (Figure 2). We genotyped the offspring from the brood carrying the type II mutation and found a similar result: all 43 identified Nodal+/− animals are female and all 27 identified WT siblings are male (Figure 2). To examine if the sex-linked pattern is inherited further, we crossed one female F2 Nodal+/− carrying the type I mutation with a WT male. Again, among F3 offspring, all identified Nodal+/− (24) are female, and all identified WT siblings (26) are male (Figure 2).
Figure 2.
Breeding experiments of Nodal mutant strains. The mutation types carried in Nodal+/− heterozygotes of this figure are frame-shift only. Strain 1: a female founder injected with TALEN pair Nodal-Fw1/Rv1 was crossed with a WT male. One F1 female Nodal+/− heterozygote carrying type I mutation (−9 bp, +7 bp) and one carrying the type II mutation (−7 bp) were crossed with two different WT males. Further, one F2 female Nodal+/− heterozygote carrying the type I mutation was crossed with another WT male. Strain 2: a female founder injected with TALEN pair Nodal-Fw2/Rv2 was crossed with a WT male. One F1 female Nodal+/− heterozygote carrying mutation (−11 bp, +4 bp) was crossed a WT male. Strain 3: a male founder injected with TALEN pair Nodal-Fw2/Rv2 was crossed with a WT female. One F1 female Nodal+/− heterozygote carrying mutation (−7 bp) was crossed with a WT male. The animal numbers analyzed (if >2) are noted.
We hypothesized two alternative mechanisms to explain those observations. Either the Nodal gene is involved in sex determination by an unknown mechanism, or the Nodal gene is sex chromosome-linked with the two mutations from the founders of strain 1 occurring on a female-specific chromosome. To distinguish between these two possibilities, we generated another batch of F0 founders by injection with a new TALEN mRNA pair (Nodal-Fw2/Rv2) (Figure 1). We collected one female founder (which generated strain 2) and one male founder (which generated strain 3) with germline mutations and crossed each with a male and a female WT animal, respectively (Figure 2). After gonads developed, enabling visual separation of sexes, we genotyped animals by restriction enzyme assay and DNA sequencing. From strain 2, we identified 15 individuals carrying frame-shift mutations: 5 females and 10 males (Figure 1 and Figure 2). Males and females do not necessarily inherit the same mutations. From strain 3, we identified seven females and six males carrying frame-shift mutations at one allele of the Nodal gene (Figure 1 and Figure 2). The mixture of sexes in these mutant animals demonstrate that the Nodal gene is not sex determining. We further crossed an F1 female from strain 2 (carrying an 11 bp deletion plus 4 bp insertion in one allele of Nodal) with a WT male. All six identified Nodal+/− progeny were female and all five identified WT siblings were male (Figure 2). This is consistent with W-chromosome linkage, as in mutant strain 1. In contrast, when a female F1 animal from founder 3 (carrying a 7 bp Nodal deletion) was crossed with a wild type male, all 13 identified Nodal+/− F2 animals were male and all 14 identified WT siblings were female (Figure 2). This indicates that the mutant allele is Z-linked. Together, these results demonstrate that B. floridae has distinct Z and W chromosomes, indicative of a ZZ/ZW sex chromosome system. The Nodal gene has alleles on the W and Z chromosomes, but does not control sex determination.
Identification of a female marker on amphioxus chromosome W
During our analysis on the progenies of strains 2 and 3, we found a 6-bp (GAATGC) polymorphism upstream of one Nodal allele in all 41 examined females, but not in any of the 45 F1 males (Figure 1 and Table 1). This 6-bp polymorphism was also detected in one allele of Nodal gene in the 11 heterozygous females analyzed, but not in the 9 WT (male) siblings examined, from strain 1 (Table 2). Linkage analysis shows that the 6-bp polymorphism is present upstream of the mutated Nodal allele on chromosome W, but not the normal Nodal allele on chromosome Z, in the F1 progenies of strain 1 (Figure S1). This result is consistent with the 6-bp marker being a female-specific marker on chromosome W in strains 1, 2, and 3. To define if the 6-bp polymorphism is a universal female-specific marker, another 15 female and 10 male WT animals selected randomly from our laboratory population were surveyed. The 6-bp marker was detected in one haplotype of 12 females analyzed, but in none of the 10 males analyzed. This is consistent with the 6-bp polymorphism being a female-specific marker, although it is not present in all females analyzed.
Table 1. Genotyping and marker detection in Nodal mutant strain 2 and 3.
| Broods | Generation | Genotype | Sex | Analyzed number | Marker |
|---|---|---|---|---|---|
| Founder ♀ × WT ♂ (strain 2) | F1 | Nodal+/− | ♀ | 5 | 5 |
| ♂ | 11 | 0 | |||
| WT | ♀ | 3 | 3 | ||
| ♂ | 4 | 0 | |||
| Nodal+/− ♀ × WT♂ | F2 | Nodal+/− | ♀ | 6 | 6 |
| WT | ♂ | 5 | 0 | ||
| Founder♂ × WT ♀ (strain 3) | F1 | Nodal+/− | ♀ | 7 | 7 |
| ♂ | 6 | 0 | |||
| WT | ♀ | 6 | 6 | ||
| ♂ | 6 | 0 | |||
| Nodal+/− ♀ × WT♂ | F2 | WT | ♀ | 14 | 14 |
| Nodal+/− | ♂ | 13 | 0 | ||
| Total | ♀ | 41 | 41 | ||
| ♂ | 45 | 0 |
Table 2. Genotyping and marker detection in Nodal mutant strain 1.
| Broods | Generation | Genotype | Sex | Analyzed number | Marker |
|---|---|---|---|---|---|
| Founder ♀ × WT ♂ | F1 | Nodal+/− | ♀ | 3 | 3 |
| WT | ♂/♀ | NA | NA | ||
| Nodal+/− ♀ × WT♂ | F2 | Nodal+/− | ♀ | 6 | 6 |
| WT | ♂ | 5 | 0 | ||
| Nodal+/− ♀ × WT ♂ | F3 | Nodal+/− | ♀ | 2 | 2 |
| WT | ♂ | 4 | 0 | ||
| Total | Nodal+/− | ♀ | 11 | 11 | |
| WT | ♂ | 9 | 0 |
The 6-bp is a BsmI restriction site unique to the amplicon of P-F3/P-R0. This provides the possibility to detect the 6-bp with BsmI digestion. We tested this with the 25 WT animals mentioned above and the F2 animals from strains 2 and 3. As expected, the PCR products from either WT (12) or F2 (20) females carrying the 6-bp marker were partially cut (∼50%), and those from the three WT females and all WT males (10) and F2 males (18), which do not carry the 6-bp marker, remain intact after being digested with the BsmI enzyme (Figure S2).
To identify a more reliable sex-linked marker, we further screened other Nodal genomic regions spanning from −2.7 kb upstream of the start codon to the stop codon (Figure S3). We selected the Nodal gene locus for this analysis because our breeding experiments in all three mutant strains show that the Nodal gene seems to be closely linked to the sex-determining locus. We detected hundreds of polymorphisms (including base pair replacement and small indels) within this 6.9 kb region. None of them is female-specific and many of them are shared between the Z and the W, indicating ongoing gene conversion between Z-linked and W-linked sequences (Figure S4). We then extended our survey to 238 kb upstream and 254 kb downstream of the Nodal gene using a binary search method. Again, none of the detected polymorphisms is female-specific, and many of them are shared between males and females (Figure S5). These results implied that the Nodal gene is in the pseudoautosomal region, which is close to the female-specific region of the W (∼0–1.2 cM, because there were no recombinants in 251 meioses).
To make sure that the Nodal gene is also sex-linked in females that do not share the 6-bp insertion polymorphism, we examined polymorphisms of a WT female founder without the 6-bp polymorphism, and its three female and two male offspring. The result showed that the detected polymorphism specific to the female parent are passed on to either all male or all female offspring (Figure S6). This result, together with the finding of a ZZ/ZW sex chromosome system in amphioxus, indicated that the Nodal gene is also sex-linked in females without the 6-bp insertion.
Discussion
Sex-determining mechanisms are remarkably labile across metazoan lineages. Especially, it is considered that GSD systems have evolved independently and repeatedly during metazoan evolution (Beukeboom and Perrin 2014; Mignerot and Coelho 2016). For example, among vertebrates, mammals generally have XX/XY systems, birds use ZZ/ZW, but fishes and frogs use either (or neither) of the two systems to determine sex. The cephalochordates, commonly known as amphioxus, hold a key phylogenetic position for studying the evolution of vertebrates. The ratio of male to female is 1:1 in all amphioxus species examined so far, suggesting that sex is probably genetically controlled. Consistent with this, Nogusa (1957) claimed a XX/XY sex chromosome system for B. japonicum on the basis of karyotype; however, amphioxus chromosomes are small and of very similar size, which made the above conclusion difficult to verify (Wang et al. 2003; Zhang et al. 2009). In the present study, we provide molecular evidence that B. floridae utilize a ZZ/ZW system for sex determination, and we identify a female-linked marker in this species.
No morphological or genetic markers are currently available to identify female and male amphioxus apart before maturation of the gonads. This has hampered studies of sex determination. Indeed, in our mutant strains, in which Nodal mutations are inherited by a single sex, we have not observed detectable morphological differences between sexes at early developmental stages. However, our finding that the Nodal gene is located on sex chromosomes provides a new tool for investigating sex determination, even though we suggest the gene is not involved in sex determination itself. Since the Nodal gene is closely linked to sex, this afforded a means to identify a naturally occurring sex-linked polymorphism. Specifically, we identified a 6-bp indel specifically inserted on the W chromosome of some, but not all, females. This fragment can transmit stably from F0 females to F1, F2, and F3 female offspring, and thus can be used as a molecular marker to identify male or female progenies if a laboratory population has the insertion.
Acknowledgments
We thank Peter Holland (University of Oxford, UK) for his critical reading and language editing and the two anonymous reviewers for their constructive comments on the manuscript. We also thank Luohao Xu (University of Vienna, Austria), Qi Zhou (Zhejiang University, China), Zhen Huang (Fujian Normal University, China) and Tokiharu Takahashi (University of Manchester, UK) for their helpful discussions. The present work is supported by grants from the National Natural Science Foundation of China (No. 31872186, 31672246 and 31471986).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.11689311.
Communicating editor: J. Birchler
Literature Cited
- Bachtrog D., Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P. et al. , 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12: e1001899 10.1371/journal.pbio.1001899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L. W., 2017. An extraordinary sex determination mechanism in a book louse. Genetics 206: 751–753. 10.1534/genetics.117.201236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom L. W., and Perrin N., 2014. The Evolution of Sex Determination. Oxford University Press, Oxford, UK: 10.1093/acprof:oso/9780199657148.001.0001 [DOI] [Google Scholar]
- Blackmon H., Hardy N. B., and Ross L. J. E., 2015. The evolutionary dynamics of haplodiploidy: genome architecture and haploid viability. Evolution 69: 2971–2978. 10.1111/evo.12792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B., 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 18: 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Dos Santos S. M., Pompolo S. D., Goncalves T. C. M., de Freitas S. P. C., Rangel E. F. et al. , 2016. New sex-determination system in the genus Panstrongylus (Hemiptera: Reduviidae) revealed by chromosomal analysis of Panstrongylus lutzi. Parasit. Vectors 9: 295 10.1186/s13071-016-1574-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T., and Zarkower D., 2014. Identification of sex-specific molecular markers using restriction site-associated DNA sequencing. Mol. Ecol. Resour. 14: 902–913. [DOI] [PubMed] [Google Scholar]
- Henmi Y., and Yamaguchi T., 2003. Biology of the amphioxus, Branchiostoma belcheri in the Ariake Sea, Japan I. Population structure and growth. Zool. Sci. 20: 897–906. 10.2108/zsj.20.897 [DOI] [PubMed] [Google Scholar]
- Hodson C. N., Hamilton P. T., Dilworth D., Nelson C. J., Curtis C. I. et al. , 2017. Paternal genome elimination in Liposcelis booklice (Insecta: Psocodea). Genetics 206: 1091–1100. 10.1534/genetics.117.199786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubokawa K., Mizuta T., Morisawa M., and Azuma N., 2003. Gonadal state of wild amphioxus populations and spawning success in captive conditions during the breeding period in Japan. Zool. Sci. 20: 889–895. 10.2108/zsj.20.889 [DOI] [PubMed] [Google Scholar]
- Li G., Yang X., Shu Z., Chen X., and Wang Y., 2012. Consecutive spawnings of Chinese amphioxus, Branchiostoma belcheri, in captivity. PLoS One 7: e50838 10.1371/journal.pone.0050838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Feng J., Lei Y., Wang J., Wang H. et al. , 2014. Mutagenesis at specific genomic loci of amphioxus Branchiostoma belcheri using TALEN method. J. Genet. Genomics 41: 215–219. 10.1016/j.jgg.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Liu X., Xing C. F., Zhang H. Y., Shimeld S. M. et al. , 2017. Cerberus-Nodal-Lefty-Pitx signaling cascade controls left-right asymmetry in amphioxus. Proc. Natl. Acad. Sci. USA 114: 3684–3689. 10.1073/pnas.1620519114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignerot L., and Coelho S. M., 2016. The origin and evolution of the sexes: novel insights from a distant eukaryotic linage. C. R. Biol. 339: 252–257. 10.1016/j.crvi.2016.04.012 [DOI] [PubMed] [Google Scholar]
- Nogusa S., 1957. The chromosomes of the Japanese lancelet, Branchiostoma belcheri (gray),with special reference to the sex-chromosomes. Annot. Zool. Jpn. 30: 42–46. [Google Scholar]
- Roco A. S., Olmstead A. W., Degitz S. J., Amano T., Zimmerman L. B. et al. , 2015. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc. Natl. Acad. Sci. USA 112: E4752–E4761. 10.1073/pnas.1505291112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes M. D., and Holland N. D., 1996. Reproduction of the Florida lancelet (Branchiostoma floridae): spawning patterns and fluctuations in gonad indexes and nutritional reserves. Invertebr. Biol. 115: 349–359. 10.2307/3227024 [DOI] [Google Scholar]
- Wallace H., Badawy G. M. I., and Wallace B. M. N., 1999. Amphibian sex determination and sex reversal. Cell. Mol. Life Sci. 55: 901–909. 10.1007/s000180050343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. L., Zhang S. C., and Zhang Y. Z., 2003. The karyotype of amphioxus Branchiostoma belcheri tsingtauense (Cephalochordata). J. Mar. Biol. Assoc. U. K. 83: 189–191. 10.1017/S0025315403006969h [DOI] [Google Scholar]
- Wang C. L., Zhang S. C., and Chu J. S., 2004. G-banding patterns of the chromosomes of amphioxus Branchiostoma belcheri tsingtauense. Hereditas 141: 2–7. 10.1111/j.1601-5223.2004.01750.x [DOI] [PubMed] [Google Scholar]
- Wilson C. A., High S. K., McCluskey B. M., Amores A., Yan Y. L. et al. , 2014. Wild sex in zebrafish: loss of the natural sex determinant in domesticated strains. Genetics 198: 1291–1308. 10.1534/genetics.114.169284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. J., Li G., Sun Y., and Wang Y. Q., 2009. Chromosome preparation and preliminary observation of two amphioxus species in Xiamen. Zool. Res. 30: 131–136. 10.3724/SP.J.1141.2009.02131 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, and supplemental material at figshare. Supplemental material available at figshare: https://doi.org/10.25386/genetics.11689311.