Salicylic acid antagonizes ethylene in apical hook development through NPR1-mediated repression of HLS1 expression and transcriptional activity of EIN3/EIL1.
Abstract
Salicylic acid (SA) and ethylene (ET) are important phytohormones that regulate numerous plant growth, development, and stress response processes. Previous studies have suggested functional interplay of SA and ET in defense responses, but precisely how these two hormones coregulate plant growth and development processes remains unclear. Our present work reveals antagonism between SA and ET in apical hook formation, which ensures successful soil emergence of etiolated dicotyledonous seedlings. Exogenous SA inhibited ET-induced expression of HOOKLESS1 (HLS1) in Arabidopsis (Arabidopsis thaliana) in a manner dependent on ETHYLENE INSENSITIVE3 (EIN3) and EIN3-LIKE1 (EIL1), the core transcription factors in the ET signaling pathway. SA-activated NONEXPRESSER OF PR GENES1 (NPR1) physically interacted with EIN3 and interfered with the binding of EIN3 to target gene promoters, including the HLS1 promoter. Transcriptomic analysis revealed that NPR1 and EIN3/EIL1 coordinately regulated subsets of genes that mediate plant growth and stress responses, suggesting that the interaction between NPR1 and EIN3/EIL1 is an important mechanism for integrating the SA and ET signaling pathways in multiple physiological processes. Taken together, our findings illuminate the molecular mechanism underlying SA regulation of apical hook formation as well as the antagonism between SA and ET in early seedling establishment and possibly other physiological processes.
INTRODUCTION
Shortly after the germination of dicot seeds in darkness, the apical part of the hypocotyl curves downward to form a transient structure known as the apical hook. The apical hook protects the shoot apical meristem from mechanical damage while pushing through the soil and is therefore essential for successful soil emergence (Harpham et al., 1991; Shen et al., 2016). Apical hook formation is mainly established by asymmetric growth on opposite sides of the apical hypocotyl that results from an asymmetric distribution of auxin (Li et al., 2004; Abbas et al., 2013). When treated with inhibitors of auxin polar transport or the auxin mimic 2,4-D, etiolated seedlings fail to form a hook (Lehman et al., 1996). The essential roles of some plant hormones in hook regulation are well characterized. Ethylene (ET) and gibberellins (GAs) are the two major phytohormones that promote and maintain hook curvature, while jasmonate (JA) inhibits hook formation (Raz and Ecker, 1999; Turner et al., 2002; Achard et al., 2003; An et al., 2012; Zhang et al., 2014; Wang and Guo, 2019). Light and high temperature are two environmental signals that negatively regulate hook formation (Liscum and Hangarter, 1993; Jin et al., 2018), and the involvement of multiple factors suggests the importance and complexity of hook development.
The ability of ET to promote apical hook formation is described as the ET-induced classic response, which results in an exaggerated hook curvature (Guzmán and Ecker, 1990; Raz and Ecker, 1999). ET is perceived by a family of endoplasmic reticulum–localized receptors (Hua and Meyerowitz, 1998; Chang and Stadler, 2001). In the absence of ET, the negative regulator CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), a Raf-like kinase (Kieber et al., 1993), phosphorylates ETHYLENE INSENSITIVE2 (EIN2; Ju et al., 2012; Qiao et al., 2012), resulting in the shutdown of ET signaling. EIN3/EIL1 are two downstream master transcription factors that are necessary and sufficient for the ET response and regulate the majority of ET-associated gene expression changes (Chao et al., 1997; Chang et al., 2013). ET perception by appropriate receptors eliminates the repressive action of CTR1, and the C-terminal fragment of dephosphorylated EIN2 is cleaved, after which the cleaved EIN2 C terminus translocates to the nucleus (Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012) and promotes transcriptional regulation by EIN3/EIL1 through the acetylation of histone H3 at Lys-14 and Lys-23 (Zhang et al., 2017). The EIN2 C terminus also functions in the cytosol and mediates the translational repression of two F-box proteins, EIN3 BINDING F-BOX1 (EBF1) and EBF2 (Guo and Ecker, 2003; Potuschak et al., 2003), which stabilize EIN3/EIL1 proteins (Li et al., 2015; Merchante et al., 2015). Acting together, EIN3/EIL1 are activated and accumulate upon ET induction, leading to the activation of the downstream cascade.
HOOKLESS1 (HLS1) encodes a putative N-acetyltransferase and is essential for hook formation (Lehman et al., 1996). Loss-of-function hls1 mutants display a complete hookless phenotype and are unresponsive to ET treatment in terms of hook formation (Guzmán and Ecker, 1990; Lehman et al., 1996). Studies have shown that ET-induced exaggeration of hook curvature is largely attributable to direct regulation of HLS1 transcription by EIN3/EIL1 (An et al., 2012). Interestingly, this EIN3-HLS1 signaling regime also mediates the effects of GA and JA on hook formation. GA promotes hook formation by eliminating DELLA repression of EIN3/EIL1 (An et al., 2012). JA inhibits hook formation by reducing EIN3/EIL1 abundance and hampering the binding of these proteins to the HLS1 promoter via MYC2 (Song et al., 2014; Zhang et al., 2014). Zhang et al. (2018) recently reported that the transcriptional coupling of EIN3/EIL1 and PIFs integrates the effects of multiple hormones and environmental factors on apical hook development by coregulating a subset of genes, including HLS1. This highlights the importance of HLS1 regulation by EIN3 for hook development.
Salicylic acid (SA) is critical for defense against plant pathogens, especially as a component of systemic acquired resistance (SAR; Malamy et al., 1990; Métraux et al., 1990). The core component of SA signaling, NONEXPRESSER OF PR GENES1 (NPR1), ensures SAR induction and the expression of a series of disease resistance genes (Cao et al., 1994, 1997). When uninduced, NPR1 is sequestered in its oxidized oligomeric form in the cytosol. When the cellular redox change is triggered by SA, NPR1 oligomers are reduced, and NPR1 monomers translocate to the nucleus, subsequently activating gene expression (Kinkema et al., 2000; Mou et al., 2003). NPR1 contains two identifiable domains associated with protein–protein interactions, the Broad-Complex, Tramtrack, and Bric-a-brac/pox virus and zinc finger (BTB/POZ) and ankyrin-repeat domains (Cao et al., 1997; Aravind and Koonin, 1999). However, NPR1 possesses no canonical DNA binding (DB) domain. Instead, it promotes transcriptional activation by interacting with TGACG sequence-specific binding protein (TGA) transcription factors, enhancing their transcriptional induction of pathogenesis-related genes (Zhang et al., 1999; Zhou et al., 2000; Després et al., 2003; Boyle et al., 2009). In addition to pathogen defense, SA also regulates numerous plant growth and development processes, including leaf senescence, flowering, cell growth, and root waving (Morris et al., 2000; Vanacker et al., 2001; Martínez et al., 2004; Zhao et al., 2015; Guo et al., 2017). However, the physiological significance of SA signaling in early seedling morphogenesis remains unclear. A recent transcriptomic analysis of seven developmental stages during the seed-to-seedling phase transition in Arabidopsis (Arabidopsis thaliana) identified a subset of genes whose expression levels increased after radicle protrusion and peaked when the cotyledons fully opened, a pattern tightly associated with early seedling establishment (Silva et al., 2016). These genes included a group of SA-responsive genes, suggesting SA involvement in early seedling morphogenesis.
In this study, we found that exogenous SA inhibited apical hook formation in an EIN3/EIL1-dependent manner and that NPR1 mediated the effect of SA on hook formation. The findings revealed direct interaction between the NPR1 N terminus and EIN3, uncovering a key mechanism in which NPR1 represses the transcriptional regulatory activity of EIN3/EIL1 by directly interfering with its binding to target gene promoters such as ProHLS1. Our transcriptomic analysis further indicated that the NPR1–EIN3 interaction is a critical module in the crosstalk between the SA and ET signaling pathways.
RESULTS
SA Inhibits Hook Formation in an EIN3/EIL1-Dependent Manner
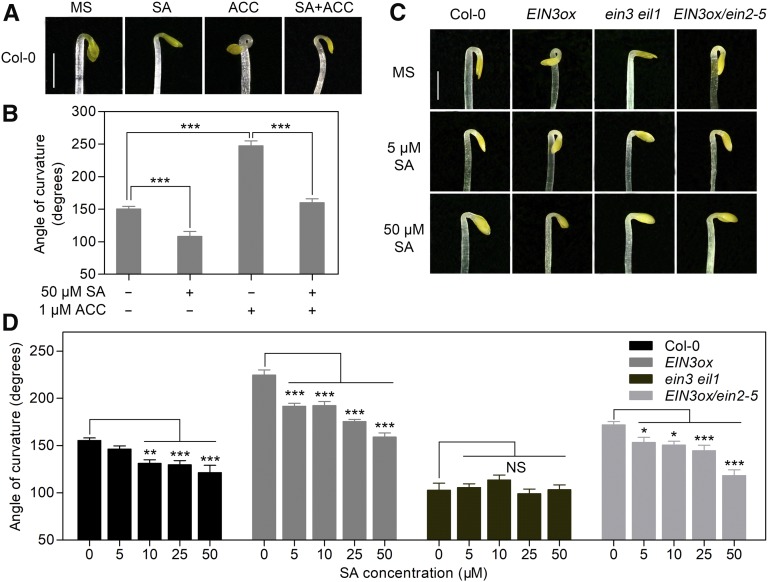
SA participates in a wide range of plant growth and development processes in addition to its pivotal function in the plant immune response (Rivas-San Vicente and Plasencia, 2011). To investigate the potential role of SA in early seedling establishment, we used apical hook formation in etiolated seedlings as a phenotypic feature, one in which the angle of hook curvature can be quantitatively measured (Supplemental Figure 1A). The hook curvature of wild-type (Columbia-0 [Col-0]) seedlings was reduced by SA treatment, indicating that SA negatively regulates hook formation (Figures 1A and 1B). ET is an important phytohormone in promoting hook formation (Guzmán and Ecker, 1990; Abbas et al., 2013), as evidenced by the dramatic exaggeration of hook curvature upon treatment with the ET biosynthesis precursor 1-aminocyclopropane-1-carboxylic acid (ACC; Figures 1A and 1B). To further evaluate the negative role of SA in hook formation, we applied ACC together with SA in the growth medium. SA significantly suppressed the positive effect of ACC on hook formation, suggesting an antagonism between SA and ET during this process (Figures 1A and 1B).
Figure 1.
Exogenous SA Represses Ethylene-Mediated Hook Formation in an EIN3/EIL1-Dependent Manner.
(A) Hook phenotypes of Col-0 seedlings treated with SA and ACC. Seedlings grown on MS medium for 3.5 d in the dark were either left untreated as controls (MS) or treated with 50 μM SA, 1 μM ACC, or 50 μM SA plus 1 μM ACC. Bar = 1 mm.
(B) Hook curvature of seedlings shown in (A). Values represent means ± se (n ≥ 15 seedlings). Statistical significance (***P < 0.001) was analyzed by one-way ANOVA along with Bonferroni’s comparison test at a significance level of 0.05 (Supplemental File 1; Supplemental Data Set 3).
(C) Hook phenotypes of 3.5-d-old etiolated Col-0, EIN3ox, ein3 eil1, and EIN3ox/ein2-5 seedlings grown on the indicated media. Bar = 1 mm.
(D) Hook curvature of seedlings from (C). Values represent means ± se (n ≥ 15 seedlings). Statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001) was analyzed by one-way ANOVA along with Bonferroni’s comparison test at a significance level of 0.05. NS, not significantly different.
The antagonistic roles of SA and ET prompted us to assess whether SA hook curvature regulation is associated with the ET pathway. By analyzing the hook phenotypes of several ET pathway mutants under different dosages of SA treatment, we found that SA suppressed the hook curvature of an EIN3 overexpression (EIN3ox) line, the ethylene overproduction1 (eto1-2), and ctr1-1 mutants that showed a constitutive ET response, but not the hook curvature of ein2-5 and ein3 eil1, two ET-insensitive mutants (Figures 1C and 1D; Supplemental Figures 1B and 1C). Interestingly, SA treatments had little effect on the alteration of hypocotyl length in all these genotypes (Supplemental Figure 1D). Meanwhile, increased SA concentrations caused the progressive inhibition of root length in these genotypes with a similar pattern (Supplemental Figure 1E), implying that SA-induced repression of root elongation is independent of ET signaling.
We further observed that the exaggeration of the hook curvature of EIN3ox induced by ACC treatment could be largely repressed by SA, while the hook curvature of ein3 eil1 was not altered by either of these treatments (Supplemental Figures 1F and 1G). Because EIN3/EIL1 proteins do not accumulate in the ein2-5 mutant (Li et al., 2015), the SA-nonresponsive phenotype of ein2-5 and ein3 eil1 mutants suggests that EIN3/EIL1 are required for the negative effect of SA on hook formation and that EIN3/EIL1 may integrate SA and ET signaling in hook formation. This hypothesis was further supported by the hook curvature phenotype of EIN3ox/ein2-5 seedlings. Because of increased EIN3 accumulation, EIN3ox/ein2-5 exhibits a mildly enhanced hook curvature compared to ein2-5 (An et al., 2010). However, it is insensitive to ACC because of the ein2-5 background (Supplemental Figure 1H). Utilizing EIN3ox/ein2-5 seedlings, in which the upstream response of ACC is blocked, we found that SA could still reduce the hook curvature of EIN3ox/ein2-5 seedlings in a dosage-dependent manner, indicating a direct inhibitory effect on EIN3 (Figures 1C and 1D). Collectively, these findings suggest that SA negatively regulates hook formation and that the inhibitory effect of SA occurs in an EIN3/EIL1-dependent manner.
SA Suppresses HLS1 Expression by Inhibiting the Transcriptional Activity of EIN3/EIL1
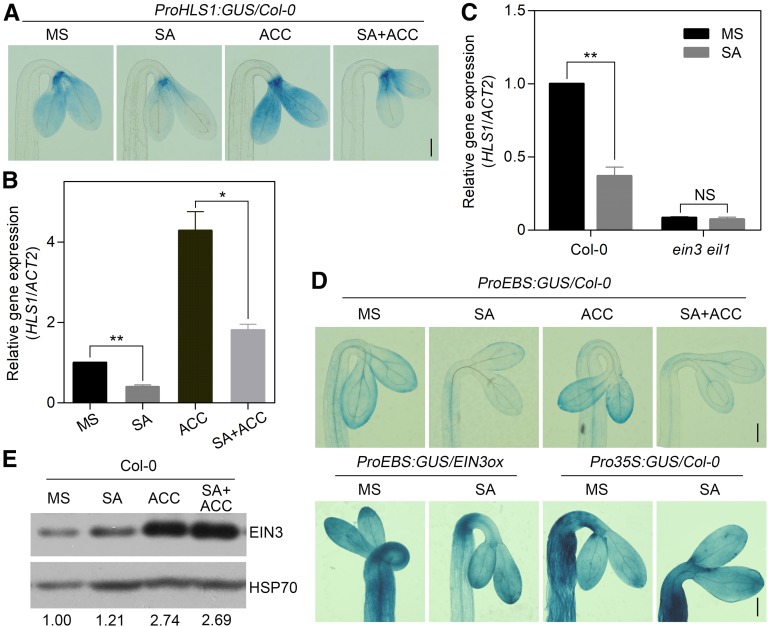
Our previous work showed that the transcriptional regulation of HLS1 gene expression by EIN3/EIL1 is a major regulatory mechanism of ET-mediated hook formation (An et al., 2012). Because the inhibitory effect of SA on hook formation was dependent on functional EIN3/EIL1, we hypothesized that SA might also regulate HLS1 gene expression to repress hook formation. Consistent with the significance of HLS1 regulation, the hook of the loss-of-function mutant hls1-1 was completely insensitive to ACC and SA, applied either individually or in combination (Supplemental Figure 2A). To further investigate the regulation of HLS1 by SA, we either monitored a β-glucuronidase (GUS) reporter line driven by the HLS1 promoter (ProHLS1:GUS/Col-0; Zhang et al., 2014) or conducted RT-qPCR to determine the endogenous HLS1 expression. SA significantly reduced HLS1 transcript levels and suppressed the positive effect of ACC on HLS1 transcription (Figures 2A and 2B). Moreover, the inhibitory effect of SA was EIN3/EIL1 dependent, as suggested by the unaltered HLS1 transcript levels in ein3 eil1 upon SA treatment (Figure 2C). These results suggest that SA negatively regulates hook formation by impairing EIN3/EIL1-dependent HLS1 gene expression.
Figure 2.
SA Suppresses HLS1 Gene Expression by Inhibiting the Transcriptional Activity of EIN3/EIL1.
(A) GUS staining of 3.5-d-old etiolated seedlings that were grown on MS medium and then left either untreated (MS) or treated with 500 μM SA and/or 50 μM ACC for 4 h. GUS was driven by a 1.6-kb HLS1 promoter in Col-0. Bar = 200 μm.
(B) Real-time PCR analysis of HLS1 transcript levels in etiolated Col-0 seedlings. Seedlings were grown on MS medium for 3.5 d and then treated with 500 μM SA and/or 50 μM ACC for 4 h. Three biological replicates were performed. Values represent means ± sd (n = 3). Statistical significance was analyzed using a two-tailed Student’s t test (*P < 0.05, **P < 0.01).
(C) Real-time PCR analysis of HLS1 gene expression in both Col-0 and the ein3 eil1 double mutant. Seedlings were grown on MS medium for 3.5 d and then treated with 500 μM SA for 4 h. Three biological replicates were performed. Values represent means ± sd (n = 3) relative to the ACTIN2 (ACT2) control. Statistical significance was analyzed using a two-tailed Student’s t test (**P < 0.01). NS, not significantly different.
(D) GUS staining of 3.5-d-old etiolated transgenic seedlings. Seedlings were grown on MS medium either unsupplemented (MS) or supplemented with 50 μM SA and/or 1 μM ACC. GUS was driven by a synthetic EBS promoter either in Col-0 (top) or in EIN3ox (bottom left). GUS driven by the constitutive cauliflower mosaic virus 35S promoter is shown as a control in Col-0 (bottom right). Bar = 200 μm.
(E) Total endogenous EIN3 protein levels in etiolated Col-0 seedlings. Seedlings were grown on MS medium for 3.5 d followed by either no treatment (MS) or treatment with 500 μM SA and/or 50 μM ACC for 4 h. Total protein was extracted with SDS buffer as described in Methods. Endogenous EIN3 and HSP70 were detected using the corresponding antibody. The numbers below the lanes represent the ratio of EIN3 to HSP70 based on gray-value analysis normalized to the control (MS, left lane). More than three biological replicates were performed with similar results.
To explore how SA influences EIN3/EIL1-dependent HLS1 gene expression, we compared the general function of EIN3/EIL1 between Murashige and Skoog (MS, no SA) and SA treatment conditions. For this analysis, we used a GUS reporter construct driven by a synthetic promoter containing five tandem-repeat EIN3 binding sites (ProEBS:GUS) as an indicator of the general transcriptional activity of EIN3/EIL1 (Stepanova et al., 2007). As expected, the GUS staining in Col-0 was enhanced by ACC treatment and was higher in both EIN3ox and ctr1-1 seedlings compared to Col-0. However, the enhanced reporter activity was greatly decreased by SA treatment (Figure 2D; Supplemental Figures 2B to 2E), indicating a repressive effect of SA on the general transcriptional activity of EIN3/EIL1. By contrast, the GUS staining in either the constitutive 35S promoter-GUS reporter line (Pro35S:GUS/Col-0) or the ProEBS:GUS/ein3 eil1 line was not affected by SA treatment (Figure 2D; Supplemental Figures 2C to 2E), further confirming that the effect of SA is EIN3/EIL1 dependent. Principally, the function of EIN3 can be regulated through two means: its protein abundance and its transcriptional regulatory activity. To dissect how SA repressed EIN3 function, we quantified endogenous EIN3 protein levels by immunoblotting. With or without ACC treatment, SA did not notably affect EIN3 protein abundance (Figure 2E), suggesting that SA instead inhibits the transcriptional regulatory activity of EIN3/EIL1.
NPR1 Is Partially Required for SA-Mediated Inhibition of Hook Formation
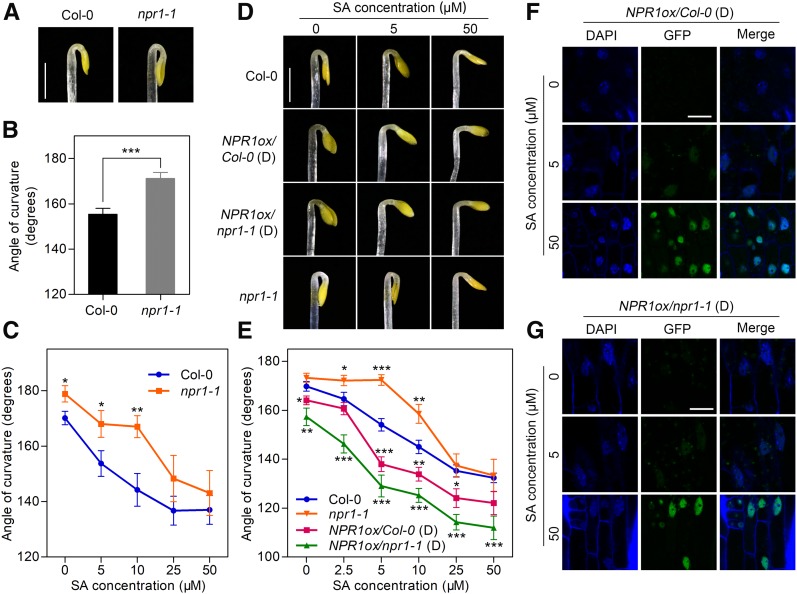
NPR1 is a key SA signaling component involved in defense responses and adaption to abiotic stress (Cao et al., 1994; Scott et al., 2004). We therefore hypothesized that NPR1 might also participate in the SA-mediated regulation of hook formation. To test this hypothesis, we first analyzed the npr1-1 hook phenotype and found that it was enhanced compared to Col-0 (Figures 3A and 3B), suggesting that NPR1 is a negative regulator of hook formation. The hook curvature responsiveness of three npr1 mutants (npr1-1, npr1-2, and npr1-3) was also compared to that of Col-0 under a variety of SA concentrations. With low SA, the hook curvature of these npr1 mutants showed relatively less change than that of Col-0 (Figure 3C; Supplemental Figure 3A), supporting our hypothesis that NPR1 is involved in SA-mediated hook repression. However, with higher SA concentrations, similar inhibition of hook formation was observed between npr1 mutants and Col-0 (Figure 3C; Supplemental Figure 3A), indicating the involvement of NPR1-independent factors in SA-mediated regulation of hook formation.
Figure 3.
SA Represses Hook Formation Partially through NPR1.
(A) Hook phenotypes of Col-0 and npr1-1 on MS medium. Seedlings were grown in the dark for 3.5 d. Representative phenotypes are shown. Bar = 1 mm.
(B) Quantification of hook curvature in Col-0 and npr1-1 seedlings grown on MS medium for 3.5 d. Values represent means ± se (n ≥ 15 seedlings). Statistical significance was analyzed by a two-tailed Student’s t test (***P < 0.001).
(C) Hook curvature of 3.5-d-old etiolated seedlings as a function of SA concentration. Values represent means ± se (n ≥ 15 seedlings). Statistical significance of npr1-1 compared with Col-0 under different concentrations was analyzed by a two-tailed Student’s t test (*P < 0.05, **P < 0.01).
(D) Hook phenotypes of Col-0, npr1-1, NPR1ox/Col-0 (D), and NPR1ox/npr1-1 (D; overexpression of NPR1-GFP driven by the 35S promoter) etiolated seedlings grown on MS medium supplemented with 0, 5, or 50 μM SA for 3.5 d. Bar = 1 mm.
(E) Hook curvature of 3.5-d-old NPR1ox/Col-0 (D) and NPR1ox/npr1-1 (D) etiolated seedlings as a function of SA concentration. Values represent means ± se (n ≥ 15 seedlings). Statistical significance was analyzed by a two-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001).
(F) and (G) NPR1-GFP fluorescence detection in the cotyledons of NPR1ox/Col-0 (D) seedlings (F) and NPR1ox/npr1-1 (D) seedlings (G). Etiolated seedlings were grown on MS medium supplemented with the indicated concentrations of SA for 3.5 d before confocal observation. DAPI staining was used to indicate the nucleus position. Bar = 10 μm.
To verify the negative effect of NPR1 on hook formation, we analyzed two previously published NPR1 overexpression (NPR1ox) lines referred to as NPR1ox/Col-0 (D) and NPR1ox/npr1-1 (D; Mou et al., 2003). In these lines, the expression of a NPR1-GFP fusion protein driven by the 35S promoter was detected in the hook and cotyledon regions (Supplemental Figures 3B to 3D). Compared with the respective nontransgenic controls (Col-0 and npr1-1), the NPR1ox transgenic plants [NPR1ox/Col-0 (D) and NPR1ox/npr1-1 (D)] displayed a significant hypersensitive response to SA treatment in terms of hook curvature inhibition, especially at lower concentrations of SA (Figures 3D and 3E). Previous studies found that SA induces the translocation of NPR1 from the cytosol to the nucleus in light-grown plants (Kinkema et al., 2000), so we examined whether this phenomenon also occurs in the etiolated NPR1ox seedlings. Whereas limited GFP signal was detected in the nuclei of NPR1ox/Col-0 (D) and NPR1ox/npr1-1 (D) seedlings grown on MS medium, the NPR1-GFP fusion protein gradually accumulated in the nucleus as the SA concentration increased (Figures 3F and 3G; Supplemental Figures 3C and 3D), resembling the expression pattern of NPR1 in green seedlings. Taken together, these results indicate that NPR1 acts as a negative regulator and partly mediates SA regulation of hook formation.
NPR1 Acts Upstream of EIN3/EIL1 and Inhibits EIN3/EIL1-Induced Hook Formation
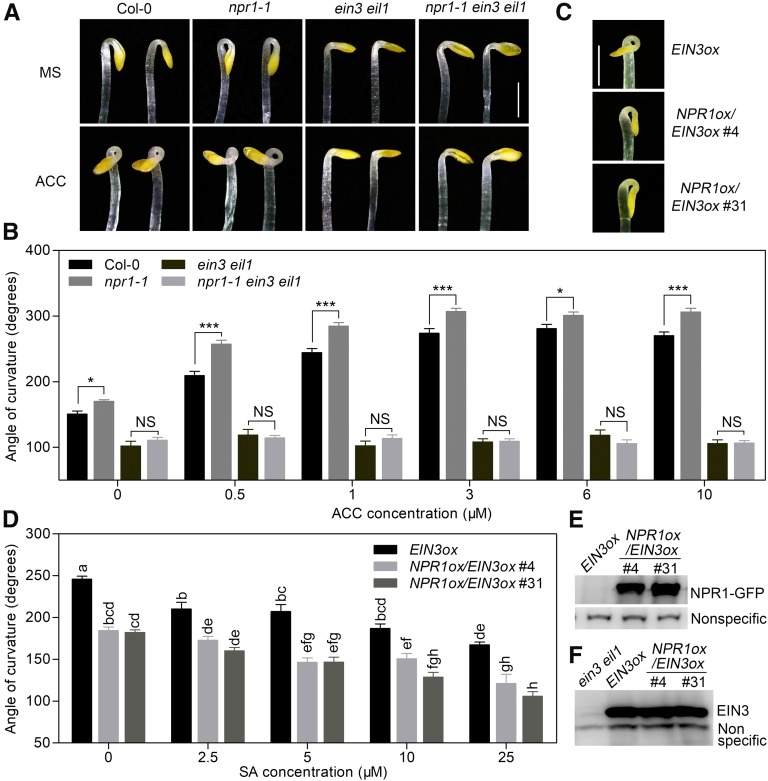
The involvement of both EIN3/EIL1 and NPR1 in SA regulation of hook formation prompted us to dissect their genetic relationship in this process. Phenotypic analysis revealed a suppression of the enhanced hook formation of npr1-1 by the mutations in EIN3/EIL1 (Figures 4A and 4B), indicating that EIN3/EIL1 function downstream of NPR1 in regulating hook curvature. Moreover, npr1-1 exhibited a mild hypersensitivity of hook curvature to low concentrations of ACC compared to Col-0 (Figures 4A and 4B), reflecting a negative effect of endogenous NPR1 on ET-induced hook curvature exaggeration. As expected, the responsiveness of npr1-1 to ACC treatment was fully blocked in npr1-1 ein3 eil1 triple mutant (Figures 4A and 4B), confirming that NPR1 acts upstream of EIN3/EIL1 during hook curvature regulation.
Figure 4.
NPR1 Acts Upstream of EIN3/EIL1 and Inhibits Hook Formation through EIN3/EIL1.
(A) Hook phenotypes of Col-0, npr1-1, ein3 eil1, and npr1-1 ein3 eil1 etiolated seedlings grown on MS with (ACC) or without (MS) 1 μM ACC medium for 3.5 d. Bar = 1 mm.
(B) Hook curvature angles of Col-0, npr1-1, ein3 eil1, and npr1-1 ein3 eil1 grown on MS containing a variety of ACC concentrations in the dark for 3.5 d. Values represent means ± se (n ≥ 15 seedlings). Statistical significance among Col-0, npr1-1, ein3 eil1, and npr1-1 ein3 eil1 was analyzed by one-way ANOVA with Bonferroni’s comparison test at a significance level of 0.05 (*P < 0.05, ***P < 0.001). NS, not significantly different.
(C) Hook phenotypes of EIN3ox and two independent NPR1ox/EIN3ox transgenic seedlings (#4 and #31) grown on MS medium for 3.5 d. Bar = 1 mm.
(D) Hook curvature in EIN3ox, NPR1ox/EIN3ox #4, and NPR1ox/EIN3ox #31 seedlings grown for 3.5 d on MS containing a variety of SA concentrations. Values represent means ± se (n ≥ 15 seedlings). Statistical significance was analyzed by one-way ANOVA along with Bonferroni correction at a significance level of 0.05. Different lowercase letters above the bars indicate a significant difference.
(E) Detection of NPR1-GFP in NPR1ox/EIN3ox transgenic seedlings using an anti-GFP antibody. A nonspecific band (bottom lanes) detected by the antibody represents a loading control.
(F) Detection of EIN3 in EIN3ox and NPR1ox/EIN3ox transgenic seedlings using an anti-EIN3 antibody. A nonspecific band (bottom bands) detected by the antibody represents a loading control.
To further examine the negative effect of NPR1 on EIN3/EIL1-induced hook formation, we overexpressed NPR1-GFP in the EIN3ox and ein3 eil1 backgrounds (Figure 4E; Supplemental Figures 4A to 4C). We found that the exaggerated hook curvature of EIN3ox was dramatically suppressed by NPR1ox (Figures 4C and 4D), whereas the hook curvature of ein3 eil1 was not affected (Supplemental Figure 4E). Thus, the negative regulation of hook formation by NPR1 was dependent on EIN3/EIL1. Nuclear NPR1 signal was also detected in the apical hypocotyl and cotyledons of these etiolated seedlings (Supplemental Figures 4B and 4C). Consistent with the action of SA, NPR1ox did not alter the abundance of EIN3 (Figure 4F; Supplemental Figure 4D), reinforcing the likelihood that NPR1 impairs EIN3/EIL1 transcriptional regulatory activity. In addition, we found that low-dose ACC treatments further augmented the hook bending of EIN3ox, while high-dose ACC treatments of EIN3ox failed to result in a typical hook structure, probably due to the extremely shortened hypocotyl induced by overactivated EIN3 activity (Supplemental Figures 4F and 4G). The hook curvature of NPR1ox/EIN3ox seedlings was reduced compared with that of EIN3ox without or with low-dose ACC treatments (Supplemental Figures 4F and 4G). Upon high-dose ACC treatments, the NPR1ox/EIN3ox seedlings were instead able to develop a hook structure (Supplemental Figures 4F and 4G), suggesting a repression of NPR1 for the excess EIN3 function. Taken together, these results indicate that NPR1 inhibits EIN3/EIL1-induced hook formation.
NPR1 Physically Interacts with the EIN3 N-Terminal DB Domain
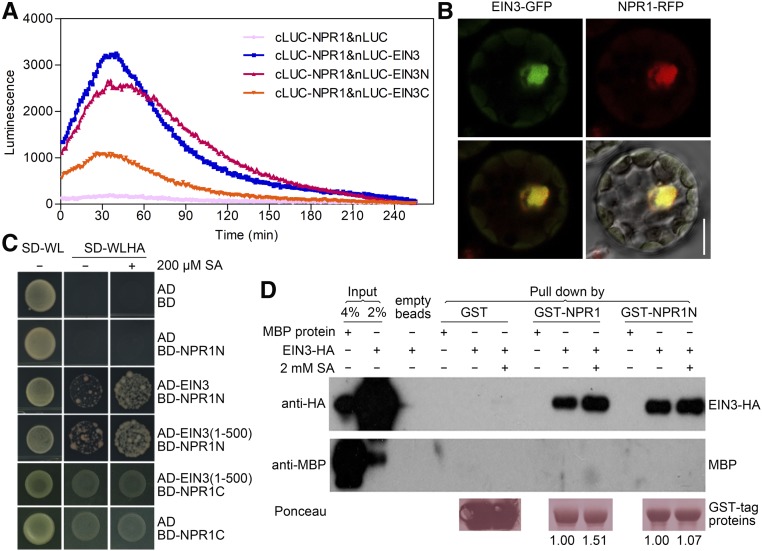
Considering the role of NPR1 in promoting the transcriptional activation of disease resistance genes through its interaction with basic leucine zipper (bZIP) transcription factors such as TGAs (Zhang et al., 1999), we hypothesized that NPR1 might influence EIN3 function through physical interaction. To test this hypothesis, we conducted split-luciferase (LUC) complementation assays in Arabidopsis protoplasts and tobacco (Nicotiana benthamiana) leaves. For these experiments, full-length EIN3 (EIN3FL), the N-terminal portion of EIN3 containing the DB domain (EIN3N, 1 to 384 amino acids), and the C-terminal portion of EIN3 containing the putative transcriptional activation domain (EIN3C, 385 to 628 amino acids) were individually fused with the LUC N terminus (nLUC), while full-length NPR1 was fused with the LUC C terminus (cLUC; Figure 5A; Supplemental Figure 5). Compared to the negative control, strong LUC signals were detected when NPR1 was coexpressed with each EIN3 fragment, and especially high activity was observed with EIN3FL and EIN3N (Figure 5A; Supplemental Figure 5). These results indicated in vivo interaction of NPR1 with EIN3 and the N-terminal portion of EIN3 in particular. The in vivo interaction was further supported by the nuclear colocalization of NPR1 and EIN3 when EIN3-GFP and NPR1-red fluorescent protein (RFP) were coexpressed in Arabidopsis protoplasts (Figure 5B).
Figure 5.
NPR1 N terminus Directly Interacts with EIN3.
(A) Split-LUC complementation assay in Arabidopsis protoplasts. Plasmids of an nLUC-EIN3 fusion (nLUC alone was used as the negative control) and a cLUC-NPR1 fusion were cotransformed into Col-0 protoplasts. EIN3N indicates the EIN3 N terminus (fragment 1 to 384 amino acids), and EIN3C indicates the EIN3 C terminus (fragment 385 to 628 amino acids). Luminescence was measured after culturing the protoplasts under low light (2.5 μmol/m2/s) for 16 h. Three biological replicates were performed with similar results.
(B) Colocalization of NPR1 and EIN3 in Col-0 protoplasts. The EIN3 and NPR1 proteins were fused with GFP and RFP, respectively. Top panels show localization of EIN3 (left, GFP signal) and NPR1 (right, RFP signal). Bottom left panel shows the merged signals, bottom right panel shows these signals merged with a differential interference contrast image. Bar = 10 μm.
(C) Y2H assay to detect interaction between NPR1 and EIN3. AD and BD indicate the empty vectors pGADT7 and pGBKT7, respectively. NPR1N indicates the NPR1 N terminus (fragment 1 to 194 amino acids), and NPR1C indicates the NPR1 C terminus (fragment 178 to 593 amino acids). EIN3(1-500) represents the N terminus of EIN3. SA (200 μM) was (+) or was not (−) added to the media. Cells were plated on SD medium either lacking Trp (W) and Leu (L; SD-WL) or lacking Trp, Leu, His (H), and Ade (A; SD-WLHA). Three biological replicates were performed.
(D) Pull-down assay to test the direct interaction between NPR1 and EIN3 in vitro. Purified GST-NPR1 and GST-NPR1N were incubated with glutathione Sepharose 4B for 2 h, followed by the addition of either EIN3-HA or MBP to the reaction. SA (2 mM) was supplied. Anti-HA and anti-MBP antibodies were used for protein detection. Ponceau staining indicates the quantity of GST-tagged proteins pulled down. The numbers below the lanes represent the ratio of EIN3 relative to GST-tagged proteins.
To identify the NPR1 and EIN3 segments critical for their interaction, yeast two-hybrid (Y2H) analysis was performed, with the segmentation based on the functional domains contained therein. We split NPR1 into the N-terminal portion containing the BTB/POZ domain (NPR1N, 1 to 194 amino acids) and the C-terminal portion containing the ankyrin-repeat domain (NPR1C, 178 to 593 amino acids). Both domains are important for NPR1 function in SA signal transduction, including the interaction between NPR1 and TGA proteins (Zhang et al., 1999; Zhou et al., 2000). As shown in Figure 5C, NPR1N, but not NPR1C, interacted with both EIN3FL and the EIN3 fragment containing the DB domain (1 to 500 amino acids; Figure 5C), suggesting that the BTB/POZ domain of NPR1 is important for its interaction with EIN3. This hypothesis was further supported by an in vitro pull-down assay (Figure 5D). Interestingly, we found that SA application noticeably enhanced the NPR1N–EIN3 interaction in Y2H as well as the NPR1–EIN3 interaction in the pull-down assay (Figures 5C and 5D). This enhancement may be attributable to SA-promoted monomerization of NPR1 that occurs at its N terminus or NPR1 conformational change upon SA binding in its C terminus (Mou et al., 2003; Wu et al., 2012), but further investigation is needed. Based on these findings, we conclude that the NPR1 N terminus physically interacts with the DNA binding region of EIN3. The BTB/POZ domain of NPR1 may therefore be critical for regulating the DNA binding activity of EIN3.
NPR1 Impairs the Binding of EIN3 to Its Target Promoters
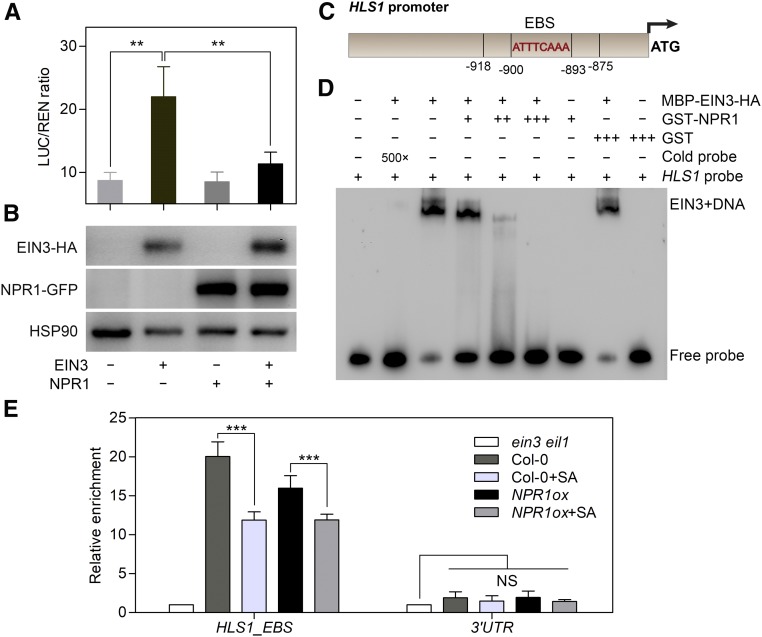
Given the direct interaction of NPR1 and EIN3 and their regulatory relationship in the context of hook formation, we tested whether NPR1 influences the transcriptional activation of HLS1 by EIN3 in a dual-LUC assay (Hellens et al., 2005). Different combinations of effector constructs were coexpressed, followed by sequential recording of luminescence from firefly (Photinus pyralis) LUC driven by the HLS1 promoter (ProHLS1:LUC) and Renilla (Renilla reniformis) LUC driven by the 35S promoter (Pro35S:REN). The ratio of LUC-to-REN activity was used as an indicator of the level of transcriptional regulation of the HLS1 gene. Consistent with a previous report (Zhang et al., 2014), EIN3 greatly enhanced the transcriptional activation of the HLS1 promoter (Figure 6A). However, coexpression of NPR1 and EIN3 significantly repressed this enhancement by EIN3 (Figure 6A), wherein the protein levels of EIN3 and NPR1 were monitored by immunoblotting to ensure comparable expression in each group (Figure 6B). These results indicate that NPR1 interferes with the transcriptional activation of the HLS1 promoter by EIN3.
Figure 6.
NPR1 Represses the Binding of EIN3 to the HLS1 Promoter.
(A) A transient dual-LUC reporter assay illustrates the repressive effect of NPR1 on EIN3-induced HLS1 transcription. The ProHLS1:LUC/Pro35S:REN ratio indicates the relative transcriptional activity of EIN3 at the HLS1 promoter. Values represent means ± sd (n = 3 biological replicates). Statistical significance was analyzed by one-way ANOVA along with Bonferroni’s comparison test at a significance level of 0.05 (**P < 0.01).
(B) Detection of expressed EIN3-HA and NPR1-GFP protein in transformed protoplast cells from (A) using an anti-HA or anti-GFP antibody, respectively. Detection of HSP90 was used as a loading control. The label below the figures indicates the expressed effector proteins in each group.
(C) Schematic illustration of EBS in the HLS1 promoter used in our experiment. The 44-bp HLS1 probe used for EMSA contained sequences that harbored the EBS motif (from −918 bp to −875 bp upstream of the ATG codon). ATTTCAAA represents the core nucleic acid sequence of the EBS motif.
(D) Binding of EIN3 to the HLS1 promoter was repressed by NPR1 in vitro. Biotin-labeled HLS1 probe (10 fmol) was used in each reaction. Cold probe indicates unlabeled HLS1 probe, and 5 pmol of cold probe was used for competition with biotin-labeled HLS1 probe. GST-NPR1 was mixed with MBP-EIN3-HA protein at 1:1, 2:1, and 3:1 molar ratio. GST was used as a negative control. − indicates no protein supplemented; +, ++, and +++ indicate NPR1:EIN3 at molar ratios of 1:1, 2:1, and 3:1, respectively; 500× indicates that cold probe is a 500-fold excess of labeled HLS1 probe.
(E) ChIP-qPCR assays indicate the enrichment of EIN3 protein in the HLS1 promoter in vivo. An anti-EIN3 polyclonal antibody was used for EIN3-DNA immunoprecipitation from 3.5-d-old etiolated Col-0 and NPR1ox/Col-0 (D) seedlings treated with 50 μM ACC and/or 500 μM SA for 4 h. ein3 eil1 seedlings were regarded as the negative control. Values represent means ± sd (n = 3 biological replicates). Statistical significance (***P < 0.001) was analyzed by one-way ANOVA with Bonferroni’s comparison test at a significance level of 0.05. NS, not significantly different; UTR, untranslated region.
Together with the finding that NPR1 interacts with the DB domain of EIN3, we hypothesized that NPR1 may directly interfere with the binding of EIN3 to the HLS1 promoter. To test this hypothesis, we first confirmed the binding of purified maltose binding protein (MBP)–tagged EIN3 to the HLS1 promoter by electrophoretic mobility shift assay (EMSA). A specific band denoting the binding of EIN3 to biotin-labeled HLS1 promoter probe was verified by the competition assay with the wild-type or mutated unlabeled probes (Figure 6C; Supplemental Figure 6A). We then purified glutathione S-transferase (GST)–tagged NPR1 protein and made use of this well-established system to investigate the effect of NPR1. With increasing amounts of NPR1 protein added to the reaction, the binding of EIN3 protein to the HLS1 promoter was gradually reduced until undetectable (Figure 6D), supporting that NPR1 directly interferes with EIN3 binding to the HLS1 promoter. Considering the weak association between NPR1 and the C-terminal portion of EIN3 in Arabidopsis protoplasts (Figure 5A), we further investigated whether the EIN3 C-terminal portion was required for the inhibitory action of NPR1. We used a fragment of EIN3 corresponding to amino acids 1 to 314, a region that contains the DB domain of EIN3 (Song et al., 2015). This N-terminal portion of EIN3 bound to the HLS1 promoter effectively in vitro (Supplemental Figure 6B). While NPR1 inhibited the binding of this truncated protein to the HLS1 promoter, a relatively higher amount of NPR1 was required (Supplemental Figure 6B). Therefore, the interaction between NPR1 and the N-terminal portion of EIN3 is sufficient for the inhibitory action of NPR1 on EIN3, but a possible contribution of EIN3 C-terminal portion also exists.
To address whether NPR1 interference is found with a wide range of EIN3 target genes, we conducted EMSA and dual-LUC assays using the synthetic EBS promoter in place of the HLS1 promoter. Similar results were observed, with NPR1 inhibiting the transcriptional activation of EBS by EIN3, although seemingly to a lesser extent compared with the HLS1 promoter (Supplemental Figures 6C to 6E). Thus, NPR1 may influence a subset of EIN3-regulated genes through the inhibitory interaction effect.
To assess whether the interference of NPR1 on the binding of EIN3 to the HLS1 promoter represents the action mode of SA on HLS1 gene expression, we performed chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) analysis. Because the low abundance of endogenous EIN3 makes it difficult to precipitate using the anti-EIN3 antibody, we first induced EIN3 accumulation by ACC treatment. The relative enrichment of the EBS-containing HLS1 promoter fragment by anti-EIN3 antibody was significantly reduced by SA treatment in both Col-0 and NPR1ox seedlings (Figure 6E). Moreover, compared with Col-0, NPR1ox seedlings showed an obvious decrease in EIN3 binding, and SA treatment further reduced EIN3 binding in NPR1ox (Figure 6E). By contrast, no effect was observed using the HLS1 3′ untranslated region negative control fragment. Because SA did not alter EIN3 protein abundance (Figure 2E), the reduction reflected a decrease of EIN3 binding to the HLS1 promoter. Taken together, these results indicate that NPR1 represses the transcriptional regulatory activity of EIN3 by directly interfering with the binding of EIN3 to its target promoters, including the HLS1 promoter.
SA, NPR1, and EIN3/EIL1 Coordinately Regulated a Subset of Genes
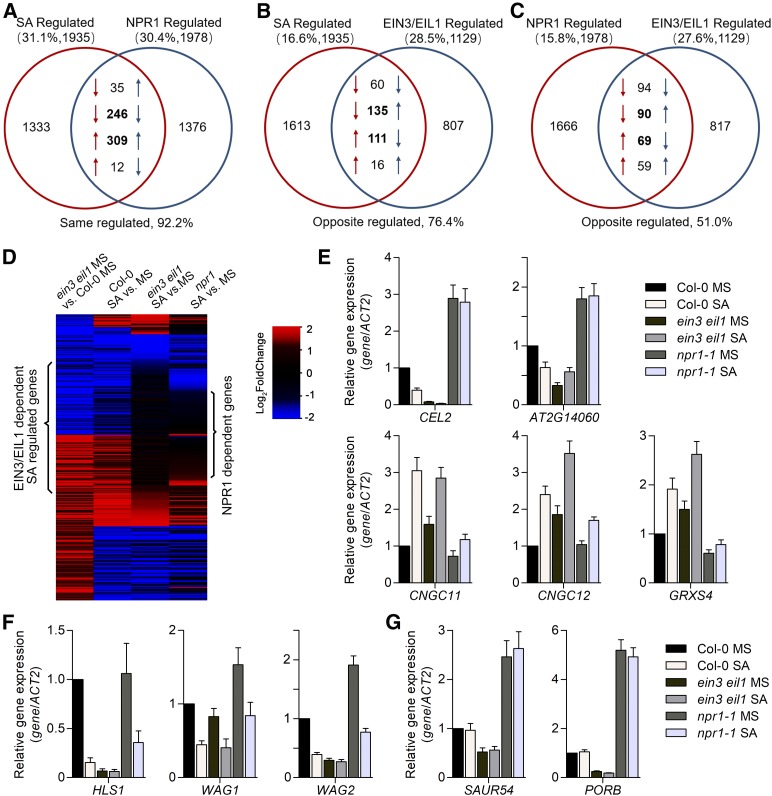
The findings from the dual-LUC and EMSA assays using the EBS promoter revealed that the interference effect of NPR1 on the transcriptional activity of EIN3 affects a wide range of genes in addition to HLS1. To investigate potential overlap in the gene regulatory networks of NPR1 and EIN3/EIL1 in hook development and other physiological processes, we conducted transcriptome profiling analysis using Col-0, ein3 eil1, and npr1-1 etiolated seedlings with and without SA treatment. Differentially expressed genes between Col-0 (+SA) and Col-0 (−SA) were considered to be the SA-regulated gene set, while genes differentially expressed between ein3 eil1 (−SA) and Col-0 (−SA) were defined as EIN3/EIL1-regulated genes. Differentially expressed genes between npr1 (+SA) and Col-0 (+SA) composed the NPR1-regulated gene set (listed in Supplemental Data Set 1). Among the 1935 SA-responsive genes in the Col-0 background, 31.1% were also identified in the NPR1-regulated gene set and 92.2% of the shared genes were regulated by SA or NPR1 in the same direction of expression (Figure 7A). The largely same directional expression patterns between SA and NPR1 coregulated genes are consistent with the role of NPR1 as a positive regulator of SA-mediated changes in etiolated seedlings. Of the SA-regulated genes, 16.6% overlapped with the EIN3/EIL1-regulated set (322 genes in total), which accounted for 28.5% of EIN3/EIL1-regulated genes (Figure 7B). Among these 322 genes, 246 genes (76.4%) had opposite expression patterns upon SA treatment and EIN3/EIL1 induction (Figure 7B), suggesting a predominant antagonism between SA treatment and EIN3/EIL1 function in the etiolated seedlings. Among NPR1 and EIN3/EIL1 coordinately regulated genes, 51.0% (159 of 312 genes) had opposite expression patterns in the two gene sets (Figure 7C), suggesting a comparable degree of antagonism and synergy between NPR1 and EIN3/EIL1.
Figure 7.
SA and NPR1 Coordinately Regulate a Subset of Genes Dependent on EIN3/EIL1.
(A) to (C) Venn diagrams showing the pairwise overlap between SA- and NPR1-regulated genes (A), SA- and EIN3/EIL1-regulated genes (B), and NPR1- and EIN3/EIL1-regulated genes (C). Up arrows (↑) indicate activated genes with an increased transcript level regulated by SA, EIN3/EIL1, or NPR1. Down arrows (↓) indicate repressed genes with a decreased transcript level regulated by SA, EIN3/EIL1, or NPR1. The expression patterns of a given gene in the left and right sets are indicated in red and blue, respectively. Percentages above the diagrams indicate the proportion of overlapping genes among total regulated genes. Percentages below the diagrams indicate the proportion of genes marked in bold (i.e., oppositely regulated and similarly regulated) among total overlapping genes.
(D) Heatmap showing transcriptomic differences in the expression levels of overlapping genes in (B) for Col-0, ein3 eil1, and npr1-1 plants following SA treatment versus MS treatment.
(E) to (G) Representative expression of class 1 genes (E), class 2 genes (F), and class 3 genes (G), as shown in Supplemental Figure 7. Expression levels were measured by quantitative RT-PCR analysis and normalized to the ACTIN2 (ACT2) expression level. Values represent means ± sd (n = 3 biological replicates).
Given that HLS1 is a primary target of EIN3/EIL1 involved in SA-mediated hook development, we screened for EIN3/EIL1-dependent SA-regulated genes with expression patterns similar to that of HLS1 to identify additional hook-regulated genes. To this end, we analyzed the expression of EIN3/EIL1-regulated genes in Col-0, ein3 eil1, and npr1-1 plants with SA induction (Figure 7D). Genes that met the following two criteria were defined as EIN3/EIL1-dependent SA-regulated genes: (i) exhibited opposite expression patterns between SA-regulated gene set and EIN3/EIL1-regulated gene set (135+111 genes; Figure 7B), or showed same directional patterns in Col-0 (+SA) and ein3 eil1 (−SA) compared to Col-0 (−SA) and (ii) not affected by SA treatment in ein3 eil1 seedlings (Figure 7D). We found that the majority of these genes (87 of the total of 145 genes) relied on NPR1 function, as their expression was not altered in npr1-1 upon SA induction (Figure 7D). Among the EIN3/EIL1-dependent SA-downregulated genes including HLS1, WAG1, WAG2, and several auxin-responsive genes (Figures 7E and 7F; Supplemental Figure 7B; Supplemental Data Set 2), WAG2 was previously shown to prevent the opening of the apical hook by maintaining the asymmetric auxin response (Willige et al., 2012). Therefore, this subset of genes might participate in the antagonistic regulation of SA and ET in the regulation of hook curvature.
We further analyzed the functional categories in three classes of genes: SA-, EIN3/EIL1-, and NPR1-regulated genes (class 1); SA- and EIN3/EIL1-regulated genes (class 2); and EIN3/EIL1- and NPR1-regulated genes (class 3; Figures 7E to 7G; Supplemental Figure 7A). We performed gene ontology analysis for each gene class and summarized the biological processes and corresponding genes (Supplemental Figure 7B). Genes in class 1 were mainly associated with defense responses and several abiotic stress responses including salt and cold stress. Class 1 genes were also associated with the cell wall organization process, which is relevant to cell growth, raising the possibility that some genes involved in plant growth might also function in hook development mediated by NPR1 and EIN3/EIL1. Some genes regulated by NPR1 and EIN3/EIL1, but not by SA treatment (class 3), were associated with auxin and JA response as well as the oxidation-reduction process (Figure 7G). Genes regulated by SA and EIN3/EIL1, but not by NPR1 (class 2), were widely involved in defense, water deprivation, auxin response, and ET-associated processes, further indicating the importance of EIN3/EIL1 regulation by SA. This functional analysis indicates that SA, NPR1, and EIN3/EIL1 both coordinately and differentially regulate a subset of genes involved in myriad aspects of plant growth and stress responses.
DISCUSSION
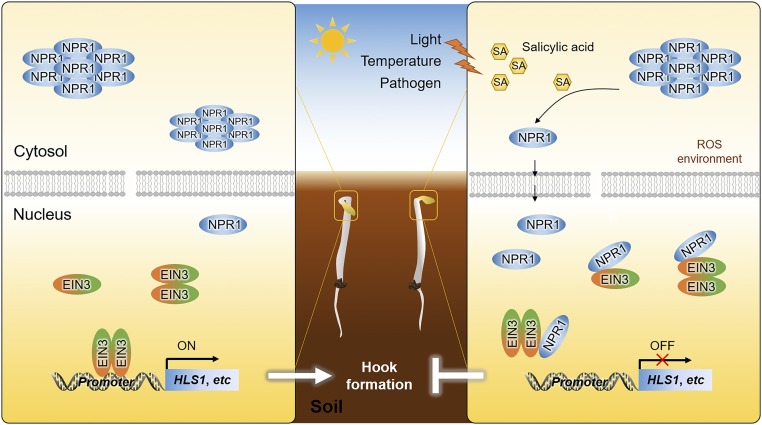
SA is a well-known inducer of SAR and plays crucial roles in plant pathogen defense (Malamy et al., 1990; Métraux et al., 1990; Durrant and Dong, 2004). SA has also been implicated in various growth and development processes, such as leaf senescence, flowering time, and cell growth (Morris et al., 2000; Vanacker et al., 2001; Martínez et al., 2004; Rivas-San Vicente and Plasencia, 2011). Here, we showed that SA inhibited formation of the apical hook, a structure associated with skotomorphogenesis that protects dicotyledonous seedlings from mechanical damage while pushing through the soil. We found that SA inhibition of hook formation was dependent on EIN3/EIL1, two ET-activated transcription factors. NPR1, the core component in SA signaling, was involved in SA-regulated hook formation. Specifically, NPR1 interfered with EIN3 binding to its target genes including HLS1 through a direct interaction between the N-terminal BTB/POZ domain of NPR1 and the N-terminal DB domain of EIN3. Beyond hook formation, this interaction likely represents an important regulatory node between the SA and ET signaling pathways, as transcriptomic analysis revealed that SA, NPR1, and EIN3/EIL1 both cooperatively and independently regulated distinctive classes of genes associated with various physiological processes. Based on these findings, we propose a working model in which SA activates NPR1 that translocates into the nucleus and impedes the transcriptional regulatory activity of EIN3, inhibiting hook formation through reduced expression of numerous hook-regulated genes, such as HLS1 and WAG2 (Figure 8).
Figure 8.
A Working Model of NPR1-Mediated Inhibition of Hook Formation through the Repression of EIN3 Activity.
When seedlings protrude through the soil, mechanical compression induces ethylene production and thus EIN3 accumulation in the nucleus. EIN3 can form homodimers, which activate the transcription of HLS1 and other hook formation genes, promoting apical hook formation. Under low SA concentrations, NPR1 protein is mostly located in the cytoplasm in the oligomeric form. Upon exposure to environmental stress signals such as light, heat, cold, and pathogen infection, the level of SA increases in etiolated seedlings, wherein NPR1 oligomers are converted into monomers and translocate into the nucleus. Activated NPR1 interacts with EIN3 through its N-terminal BTB domain and impedes the binding of EIN3 to its target genes, including HLS1. Hook formation is therefore repressed in SA-induced seedlings.
In disease resistance against biotrophic pathogens, SA is considered to act antagonistically with ET (Thomma et al., 1999; Gu et al., 2000; Berrocal-Lobo et al., 2002; Díaz et al., 2002). However, the molecular mechanism underlying the SA–ET antagonism is largely unclear. Previous studies reported that EIN3/EIL1 reduces SA levels by suppressing the transcription of SID2, which encodes a key enzyme in the SA biosynthesis pathway, and SA can also inhibit ET biogenesis (Leslie and Romani, 1988; Wildermuth et al., 2001; Chen et al., 2009). The NPR1-EIN3 interference model illustrated in this study provides an alternative explanation for the SA–ET antagonism, which may not only occur in the skotomorphogenesis process but also in the defense and other developmental processes. In support of this notion, our transcriptomic data revealed a subset of defense genes that are oppositely regulated by EIN3/EIL1 and SA or NPR1 (Figures 7B, 7C and 7E; Supplemental Data Set 1). Of them, CNGC11 and CNGC12, which were shown to induce multiple defense responses (Yoshioka et al., 2006), might represent a downstream regulatory node integrating SA and ET signaling, although the physiological relevance of this node needs to be further confirmed.
In canonical SA-mediated immune signaling, NPR1 is a transcriptional coactivator with two conserved domains, the N-terminal BTB/POZ domain and the C-terminal ankyrin-repeat domain (Cao et al., 1997; Aravind and Koonin, 1999). Both domains contribute to the activation of NPR1-interacting TGA transcription factors, wherein the BTB/POZ domain of NPR1 was shown to associate with TGA2 to form an enhanceosome (Zhang et al., 1999; Rochon et al., 2006). By contrast, our study demonstrated that NPR1 acts as a transcriptional corepressor of EIN3 by interfering with its DNA binding activity in regulating hook development. Consistent with this finding, NPR1 was recently reported to function as a repressor of bZIP28 and bZIP60, two master transcription factors mediating the unfolded protein response (Lai et al., 2018). It is not clear how NPR1 confers its effect on different groups of transcription factors as either a coactivator or a corepressor. Direct binding between the N-terminal BTB/POZ domain of NPR1 and the DB domain of EIN3 was sufficient to disrupt the DNA binding activity of EIN3 (Supplemental Figure 6B). This finding suggests that NPR1 negates the transcriptional activity of EIN3 probably through a steric hindrance effect to EIN3-target DNA binding. Additionally, the EIN3 proteins can form homodimers via their N-terminal domains, and this enhances their DNA binding ability (Solano et al., 1998; Song et al., 2015). Thus, it is also likely that the BTB/POZ domain interferes with the dimerization of EIN3 and reduces its DNA binding activity (Figure 8). Furthermore, NPR1 can also interact with the C terminus of EIN3 (Figure 5A). Such interaction might recruit some NPR1-associated repressive factors, such as NIMINs (Weigel et al., 2005), to form a transcriptional repressive complex with EIN3. Future investigation is required to delineate the exact regulatory mechanism of NPR1 repression on EIN3 activity.
NPR1 participates in the repression of hook formation, especially when EIN3 activity is high (Figures 4C and 4D). However, the negative regulation of hook formation by SA is not fully dependent on NPR1 (Figure 3C), implying the existence of NPR1-independent factors. NPR3 and NPR4, two NPR1 paralogs, have a domain structure similar to that of NPR1 but function as negative regulators of SA signaling (Zhang et al., 2006). As reported in a recent study, NPR3 and NPR4 act oppositely to NPR1 in the transcriptional regulation of immune genes via directly interacting with and repressing TGAs in an NPR1-independent manner (Ding et al., 2018). It is therefore worth investigating whether NPR3 and NPR4 also interact with EIN3 and regulate EIN3 activity in SA-mediated hook development. Meanwhile, SA modulates the redox states of NPR1 to facilitate an oligomer-to-monomer transition and consequently its nuclear translocation (Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008). As aforementioned, EIN3 proteins can form homodimers that facilitate their DNA binding activity (Song et al., 2015), so whether SA modulates the state of EIN3 dimer/monomer through redox regulation warrants further investigation. Given that SA regulation of hook formation is fully dependent on EIN3/EIL1 but partly on NPR1 (Figures 1C and 1D), it is conceivable that SA influences the functions of EIN3/EIL1 in a variety of ways.
HLS1 is a direct target of EIN3/EIL1 and a major regulator of apical hook formation. In SA regulation of hook formation, the repression of EIN3/EIL1-mediated HLS1 transcription seems to be a key mechanism. Our transcriptomic analysis had identified 145 genes that are regulated by SA in an EIN3/EIL1-dependent manner (Figure 7D). Eighty-seven of them are NPR1 dependent, further supporting the presence of NPR1-independent pathways. Besides HLS1, many other genes in this list may also participate in the regulation of hook development, including WAG1, WAG2, and CEL2 (Figures 7D to 7G). Particularly interesting is WAG2 that encodes a protein kinase that regulates auxin transport and that was shown to prevent hook opening (Willige et al., 2012). The CEL2 gene is associated with cell wall organization (Wieczorek et al., 2008) and is probably associated with differential growth in the apical hook region. Therefore, SA inhibition of hook formation involves the transcriptional regulation of a subset of growth-associated genes that are targets of EIN3/EIL1. These findings reveal a mechanism in which the NPR1–EIN3 interaction partially mediates the SA/ET antagonism in the apical hook development. Our transcriptomic analysis suggests that the NPR1–EIN3 interaction represents a key regulatory module that mediates other physiological processes coregulated by SA and ET.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) mutants used in this study are Col-0 ecotype. ein2-5 (Alonso et al., 1999), eto1-2 (Wang et al., 2004), ctr1-1 (Kieber et al., 1993), EIN3ox (Chao et al., 1997), ein3-1 eil1-1 (Alonso et al., 2003), hls1-1 (Guzmán and Ecker, 1990), ProHLS1:GUS/Col-0 (Zhang et al., 2014), ProEBS:GUS/Col-0, ProEBS:GUS/EIN3ox, ProEBS:GUS/ctr1-1, ProEBS:GUS/ein3 eil1 (He et al., 2011), and npr1-1 and npr1-3 (Cao et al., 1994) were lab stocks and were reported in previous studies. NPR1ox/Col-0 (D) and NPR1ox/npr1-1 (D) were previously described by Mou et al. (2003). npr1-2 (CS3801) was purchased from Arabidopsis Biological Resource Center. Surface-sterilized seeds were spread on MS medium (4.4 g/L MS salts [PhytoTech], 1% [w/v] Suc, pH 5.7 to 5.8, and 0.8% [w/v] agar). After stratification at 4°C in darkness for 3 d, the plates were placed under white light for 5 h and then cultured in darkness at 22°C for 3.5 d to observe the hook phenotype.
Generation of Transgenic Plants and Mutants
To generate NPR1ox/EIN3ox and NPR1ox/ein3-1 eil1-1 plants, the NPR1 coding sequence linked with the GFP coding sequence at the C terminus was inserted into the pCAMBIA1307 (Li et al., 2015) vector using SacI and SalI sites. The target sequence in this vector was verified, and the vector was introduced into Agrobacterium tumefaciens GV3101 by electroporation at 2.2 V using a MicroPulser (Bio-Rad). The floral dip method (Clough and Bent, 1998) was used for plant transformation.
To construct the npr1-1 ein3-1 eil1-1 triple mutant, npr1-1 was crossed with the ein3-1 eil1-1 double mutant, followed by PCR-based genotyping in the F2 population. Pro35S:GUS/Col-0 was constructed by fusing the Cauliflower mosaic virus 35S promoter with the GUS reporter gene in the pBI101 vector (Jefferson et al., 1987). The fused vector was transformed into Col-0 plants, which were used as a control for the SA treatment.
Preparation of Chemical Solutions
ACC and sodium salicylic acid were purchased from Sigma-Aldrich. ACC and sodium salicylic acid were dissolved in distilled water to make 10 mM and 1 M stock solutions. Working solutions were diluted from the stock solution.
Hook Curvature Measurement
A Canon camera (EOS 760D) with Canon macro lens (EF 100mm f/2.8L IS USM) was used to photograph the hooks of individual seedlings. Supplemental Figure 1A depicts the method of hook curvature angles determination. ImageJ (http://rsbweb.nih.gov/ij/) was used to measure the hook curvature angles between the hypocotyl and cotyledons.
Statistical Analysis
For multiple pairwise comparison, significance analysis was performed by one-way ANOVA along with Bonferroni’s multiple comparison test (*0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001). For comparison in all groups (Figure 4D; Supplemental Figure 4E), statistical significance was analyzed by one-way ANOVA with Bonferroni correction. Different lowercase letters above the bars indicate a significant difference. All of the ANOVA analysis were performed at a significance level of 0.05. For other statistical analysis, two-tailed Student’s t test was used to analyze the significance between two noted samples at a significance level of 0.05 (*0.01 < P < 0.05; **0.001 < P < 0.01; ***P < 0.001). Detailed descriptions of statistical analyses are presented in Supplemental Data Set 3 and Supplemental File 1.
RNA Extraction and Real-Time PCR
Total RNA of whole etiolated seedlings was extracted using the TRIzol reagent (Invitrogen). RT of total RNA was performed using Moloney murine leukemia virus reverse transcriptase (Promega) at 42°C for 60 min. Real-time PCR was performed on the Light Cycler 480 system (Roche) with SYBR Premix ExTaq reagents (Takara). Each sample was detected in three technical replicates, and three biological replicates were performed. ACTIN2 was used as the reference gene. The relative expression of target genes was calculated by the 2−ΔΔCt method (Udvardi et al., 2008). Sequences of oligonucleotides used in this study are listed in Supplemental Table 1.
Protein Extraction and Immunoblotting
Samples treated with ACC or SA were immediately frozen and ground in liquid nitrogen. Equivalent volumes of ground powder were suspended in protein extraction buffer (100 mM Tris-HCl, pH 6.8, 4% [w/v] SDS, 10% [v/v] glycerol, 2% [v/v] β-mercaptoethanol, 100 mM DTT, and 0.02% [w/v] bromophenol blue). The extracts were thoroughly mixed, maintained on ice for 15 min, and then heated at 75°C for 10 min. After 13,000g centrifugation for 10 min, the supernatant was collected for detection. An anti-GFP antibody (ABclonal, AE012; 1:5000 dilution) was used together with an anti-mouse IgG horseradish peroxidase conjugate (Promega, W4028; 1:10,000 dilution) to detect target NPR1 proteins with GFP tag. Endogenous EIN3 antibody was from rabbit (1:5000 dilution; Guo and Ecker, 2003) and was used for EIN3 protein detection in combination with anti-rabbit IgG horseradish peroxidase conjugate (Promega, W4018; 1:10,000 dilution).
GUS Staining
For GUS staining, 3.5-d-old etiolated seedlings grown on the different media were washed three times with PBS buffer (100 mM Na3PO4, pH 7.0) and then incubated with GUS staining buffer (100 mM Na3PO4, pH 7.0, 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 1 mM Na2⋅EDTA, 1% [v/v] Triton X-100, and 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronide; Jefferson et al., 1987) at 37°C for 2 to 4 h or longer in the dark. After staining, the seedlings were washed three times with PBS, followed by decolorization using 95% ethanol. Finally, the seedlings were placed in 75% ethanol, and the staining of individual seedlings was observed with a Zeiss microscope.
Quantitative Analysis of GUS Activity
Etiolated 3.5-d-old seedlings grown on 50 μM SA and/or 1 μM ACC media were collected and ground into powder. Proteins were extracted with GUS extraction buffer (100 mM PBS, pH 7.0, 10 mM Na2⋅EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] SDS, and 10 mM β-mercaptoethanol). Enzyme reactions were performed in GUS extraction buffer containing 2 mM 4-methylumbelliferyl-β-d-glucuronide at 37°C for 30 min. Production of 4-methylumbelliferon was detected using a SynergyHTX multi-mode reader (BioTek) with fluorescence measurement at 460 nm following excitation at 365 nm. The total protein concentration was quantified using a BCA Protein Assay Kit (Pierce) with the microplate procedure. GUS activity was calculated as nanomole per liter 4-methylumbelliferon per minute and per microgram of total soluble proteins.
Protein Expression, Purification, and In Vitro Pull-Down
The NPR1 full-length coding sequence and the NPR1 fragment (1 to 194 amino acids) were digested with BamHI and SalI and inserted into the pGEX-5X-1 vector (GE Healthcare). To generate the MBP-EIN3-HA construct, the EIN3 coding sequence with a C-terminal hemagglutinin (HA) tag was digested with BamHI and SalI and inserted into the pMAL-p2X vector (GE Healthcare). All constructs were transformed into Escherichia coli BL21 (DE3) competent cells. Expression of the target proteins was induced by 0.3 mM isopropyl-β-d-thiogalactopyranoside, and cells were cultured at 22°C for 3 h before collection for purification. Protein purification was performed using the ÄKTA pure system (GE Healthcare) according to the manufacturer’s instructions.
Purified GST-NPR1, GST-NPR1 (1 to 194 amino acids), and empty GST proteins were incubated with glutathione Sepharose 4B (GE Healthcare) in pull-down buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 10% [v/v] glycerol, 0.1% [v/v] Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 0.1% [v/v] β-mercaptoethanol, and 1× protease inhibitor cocktail [Roche, 04693132001]). The reaction was incubated at 4°C for 2 h. MBP-EIN3-HA protein was added to the mixture, followed by incubation for another 3 h at 4°C. After gently washing the complex five to six times, protein-bound beads were collected. The proteins were released from the beads using SDS buffer (detailed in the immunoblotting section) at 90°C for 10 min. Interaction was analyzed either by using anti-GST (TIANGEN, AB101), anti-MBP (NEB, E8032S), and anti-HA (Roche, 12013819001) antibodies or by Ponceau staining.
Split-LUC Complementation
The NPR1 coding sequence was inserted into the pCAMBIA1300-cLUC vector at the BamHI-SalI site (Chen et al., 2008). Segments of the EIN3 coding sequence were amplified and inserted into the pCAMBIA1300-nLUC vector at the SacI-SalI site for EIN3FL and at KpnI-SalI for both EIN3N (1 to 384 amino acids) and EIN3C (385 to 628 amino acids).
Ten-day-old Arabidopsis seedlings were treated with 1.5% (w/v) cellulose R10 and 0.4% (w/v) macerozyme R10 (YAKULT) for 5 h for protoplast extraction. The nLUC and cLUC plasmids were cotransformed into protoplasts using the polyethylene glycol standard protocol (Yoo et al., 2007). After culturing the protoplasts for 12 to 16 h under low light, the interaction was analyzed by reading the kinetic curve of luciferin signal on the Centro LB 960 system (Berthold Technologies).
For the assays using Nicotiana benthamiana leaves, the constructs were first electroporated into Agrobacterium tumefaciens GV3101 cells. The bacteria were cultured, collected, and suspended in IFB buffer (0.5% [w/v] Glc, 10 mM MgCl2, 10 mM MES, and 150 μM acetosyringone, pH 5.7) at 0.5 OD600, before mixing pairwise in a 1:1 OD600 ratio. The bacterial mixture was then infiltrated into tobacco leaves using a syringe. The injected plants were cultured for 3 d and detected using the LB 985 NightSHADE system (Berthold Technologies).
Y2H Analyses
The pGADT7 and pGBKT7 vectors were used, and polyethylene glycol–induced transformations were performed according to the instructions in the Yeast Protocols Handbook (Clontech). An N-terminal fragment of the NPR1 coding sequence (1 to 194 amino acids) and a fragment lacking the N-terminal region (178 to 593 amino acids) were separately cloned into the pGBKT7 vector. Similarly, EIN3 coding sequence fragments (1 to 500 amino acids and full length) were inserted into the pGADT7 vector. Plasmid pairs were cotransformed into AH109 cells that were incubated at 30°C on selective dropout (SD)-Trp-Leu medium (Clontech). Finally, the AH109 cells were selected on SD medium lacking Trp, Leu, His, and adenine (Ade; SD-Trp-Leu-His-Ade).
Microscopy
For protein localization, the NPR1ox etiolated seedlings expressing NPR1-GFP protein were grown on media with varying concentrations of SA for 3.5 d and then observed using a confocal microscope (Zeiss LSM880). More than 10 random regions in cotyledons and hook were observed for each treatment. The 4′,6-diamidino-2-phenylindole (DAPI) staining indicates the position of the nucleus. For colocalization of EIN3 and NPR1, the EIN3 coding sequence was inserted into the pCHF3-GFP vector (Yin et al., 2002), and the NPR1 coding sequence linked with RFP at the C terminus was inserted into the pCAMBIA1307 vector (Li et al., 2015). The plasmids were used for transformation of Arabidopsis protoplasts to observe their localization. The nuclei of cells were clearly visible under confocal microscope. More than 10 randomly chosen regions were observed, and similar colocalization was found in the cells with both EIN3-GFP and NPR1-RFP expression. GFP and RFP were excited at 488 and 561 nm, respectively, and detected at 493 to 590 and 582 to 642 nm wavelength, respectively. DAPI was excited at 405 nm and detected at 415 to 515 nm.
Dual-LUC Reporter System
The pGreen II 0800-LUC vector carrying the 1.5-kb HLS1 promoter was used as a reporter plasmid as described previously (Zhang et al., 2014). For the effector plasmids, the EIN3 and NPR1 coding sequences that linked with HA or GFP tag at C terminus, respectively, were amplified and then digested with BamHI-XhoI and BamHI-ApaI before being inserted into the pGreen II 62-SK vector (Hellens et al., 2005). Arabidopsis protoplasts were extracted from 10-d-old seedlings as described above. The two effector plasmids were combined equally and transformed into protoplasts together with the reporter plasmid at a 2:1 ratio. Cells were cultured at 22°C for 16 h. In the control group, empty pGreen II 62-SK vector replaced the effector plasmid at the same concentration.
We used the dual-LUC reporter assay system (Promega) to sequentially measure the activities of firefly (Photinus pyralis) and Renilla (Renilla reniformis) LUCs from a single sample. After collection of the cultured cells, cells were broken using passive lysis buffer (Promega, E194A). Next, the suitable volume of cell lysate was mixed with 5 times volume of LUC assay buffer (Promega, E195A) to detect the activity of firefly LUC. After that, the same volume of Stop and Glo buffer (Promega, E641A) equal to LUC assay buffer was added to the reaction to detect the Renilla LUC activity. During detection, a GLO-MAX 20/20 luminometer was used for recording the value (Promega). The ratio of firefly relative to Renilla LUC activity was used as an indicator for the transcriptional efficiency of the HLS1 and EBS promoters. More than three biological replicates were performed.
EMSA Method
An oligonucleotide probe was synthesized for HLS1 with the sequence information described in An et al. (2012), and EBS sequence information was described in Song et al. (2015). The 3′ hydroxyl end of the HLS1 or EBS sense strand was labeled with biotin along with nucleic acid synthesis by company. EMSA was performed using the LightShift Chemiluminescent EMSA Kit (Pierce). In the binding system, 10 or 20 fmol of biotin-labeled probe was incubated with the proteins in binding buffer (2.5% [v/v] glycerol, 50 mM KCl, 5 mM MgCl2, 10 mM EDTA, and 1 μg of poly dI⋅dC) on ice for 1 h. For the unlabeled probe (cold probe) competition group, a 500-fold excess of cold probe was added to the reaction system. Each reaction product was electrophoresed on a 6% polyacrylamide gel in TBE buffer (45 mM Tris, 45 mM boric acid, and 1 mM Na2⋅EDTA, pH 8.3) for 40 min to 1 h. The oligonucleotide sequences used in our study are listed in Supplemental Table 1.
ChIP-qPCR Assays
The ChIP assays were performed according to protocols previously described by Gendrel et al. (2005), with minor modifications. Col-0 and NPR1ox/Col-0 (D) seedlings (2 g) treated with ACC or ACC plus SA were collected. The samples were cross-linked with 1% (v/v) formaldehyde, and the reactions were terminated with 0.125 M Gly, with all steps performed under vacuum. After washing the seedling samples with 500 mL of deionized water, the water was removed and the samples were immediately frozen in liquid nitrogen. The samples were ground into powder for chromatin DNA extraction and sonication. The chromatin DNA mixture was diluted with ChIP dilution buffer (16.7 mM Tris-HCl, pH 8.0, 167 mM NaCl, 1.1% [v/v] Triton, and 1.2 mM EDTA) and incubated with Protein G Sepharose 4B (Invitrogen) at 4°C for 2 h for preclearing. The supernatant was collected and transferred to a fresh tube, followed by the addition of an anti-EIN3 antibody at 4°C overnight. Protein G Sepharose 4B was used to immunoprecipitate the protein-DNA complex. After washing the beads, chromatin DNA was eluted with TE (10 mM Tris-HCl, pH 8.0, 1 mM Na2⋅EDTA) buffer, followed by protein digestion with 20 mg/mL Proteinase K (Merck, 539480) and reverse cross-linking with 200 mM NaCl. The DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1, [v/v/v]) and dissolved in deionized water thoroughly. The DNA was stored at −20°C and used for real-time PCR detection.
mRNA-Sequencing Analysis
Etiolated 3.5-d-old seedlings of Col-0, ein3-1 eil1-1, and npr1-1 were treated for 4 h with MS liquid medium supplemented with or without 500 μM SA before tissue collection. Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen). mRNA sequencing was performed by the Beijing Genomics Institute using the Illumina HiSeq X Ten system. Specifically, after the total RNA extraction and DNase I treatment, magnetic beads with oligo(dT; NEB, E7530S) was used to isolate mRNA. Mixed with the fragmentation buffer, the mRNA was fragmented into short fragments. cDNA was synthesized using the mRNA fragments as templates. After purification of fragments, end reparation and single nucleotide A (Ade) addition, the short fragments were connected with adapters (GATCGGAAGAGCACACGTCTGAACTCCAGTCACTGAATCATATCTCGTAT). The suitable fragments were selected for the PCR amplification as templates with several quality control steps. Finally, the library was sequenced using the Illumina HiSeq X Ten system. The Arabidopsis Information Resource 10 genome was used as the Arabidopsis reference genome (www.arabidopsis.org). Reads were mapped to the genome using TopHat2 (https://ccb.jhu.edu/software/tophat/index.shtml) software with two mismatches allowed. Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/) was used for differential expression analysis. Differentially expressed genes were identified based on a fold change > 2 and q-value < 0.05 when comparing the test group sample to the control group sample. Venn diagram analysis was conducted using the Bioinformatics and Evolutionary Genomics online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). Gene ontology enrichment analysis was performed using the BiNGO app (https://www.psb.ugent.be/cbd/papers/BiNGO/Home.html) in the Cytoscape software package (http://cytoscape.org) and DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/summary.jsp).
Accession Numbers
Sequence information from this study can be found in the Arabidopsis Genome Initiative and GenBank/EMBL databases, and the accession numbers are listed in Supplemental Table 2. Raw data of the mRNA sequence are available at the Gene Expression Omnibus database with the accession number GSE137212.
Supplemental Data
Supplemental Figure 1. SA-ET antagonism in the regulation of hook formation but not hypocotyl or root elongation. Supports Figure 1.
Supplemental Figure 2. SA inhibits the EIN3/EIL1-activated transcription of an EBS-containing promoter. Supports Figure 2.
Supplemental Figure 3. NPR1 overexpression seedlings exhibit a hypersensitive response to SA. Supports Figure 3.
Supplemental Figure 4. Hook curvature phenotypes of NPR1ox/ein3 eil1 and NPR1ox/EIN3ox seedlings. Supports Figure 4.
Supplemental Figure 5. NPR1 interacts with EIN3 in tobacco leaves. Supports Figure 5.
Supplemental Figure 6. NPR1 interferes with EIN3 binding to its targets. Supports Figure 6.
Supplemental Figure 7. GO analysis of genes coordinately and differentially regulated by SA, NPR1, and EIN3/EIL1. Supports Figure 7.
Supplemental Table 1. Sequences of oligonucleotides used in this study.
Supplemental Table 2. Accession numbers.
Supplemental File 1. ANOVA and t test tables.
Supplemental Data Set 1. Differentially expressed genes in three gene sets from transcriptomic analysis.
Supplemental Data Set 2. EIN3/EIL1-dependent SA-regulated genes from heat map analysis.
Supplemental Data Set 3. ANOVA statistical analysis tables.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Xinnian Dong (Duke University) and Shunping Yan (Huazhong Agricultural University) for kindly providing NPR1ox/Col-0 (D), NPR1ox/npr1-1 (D), and npr1 mutant seeds. We also thank all members of the Guo lab for helpful discussions and suggestions. This work was funded by the National Natural Science Foundation of China (grants 31570286 and 91740203 to H.G.), the National Key Research and Development Program of China (grant 2018YFA0507101 to H.G.), and the Peking-Tsinghua Center for Life Sciences (to H.G.)
AUTHOR CONTRIBUTIONS
H.G. and P.H. conceived the project and designed the experiments; P.H. performed most of the experiments and data organization; P.H. and Z.D. performed the Y2H assay; P.H. and P.G. performed the ChIP-qPCR and dual-LUC assays; X.Z. constructed several plasmids for the split-LUC complementation assay and provided several genetic materials; P.H. and Y.Q. performed fluorescence detection using confocal microscopy; P.H. and B.L. analyzed the transcriptomic data; P.H., Y.W., and H.G. wrote the article. All authors analyzed and discussed the data in this article.
References
- Abbas M., Alabadí D., Blázquez M.A. (2013). Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 4: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Vriezen W.H., Van Der Straeten D., Harberd N.P. (2003). Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhang X., Zhu Z., Ji Y., He W., Jiang Z., Li M., Guo H. (2012). Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., Zhang C., Han Y., He W., Liu Y., Zhang S., Ecker J.R., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22: 2384–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Koonin E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285: 1353–1361. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M., Molina A., Solano R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29: 23–32. [DOI] [PubMed] [Google Scholar]
- Boyle P., Le Su E., Rochon A., Shearer H.L., Murmu J., Chu J.Y., Fobert P.R., Després C. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Chang C., Stadler R. (2001). Ethylene hormone receptor action in Arabidopsis. BioEssays 23: 619–627. [DOI] [PubMed] [Google Scholar]
- Chang K.N., et al. (2013). Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2: e00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. (1997). Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89: 1133–1144. [DOI] [PubMed] [Google Scholar]
- Chen H., Xue L., Chintamanani S., Germain H., Lin H., Cui H., Cai R., Zuo J., Tang X., Li X., Guo H., Zhou J.M. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz J., ten Have A., van Kan J.A.L. (2002). The role of ethylene and wound signaling in resistance of tomato to Botrytis cinerea. Plant Physiol. 129: 1341–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Sun T., Ao K., Peng Y., Zhang Y., Li X., Zhang Y. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467.e15. [DOI] [PubMed] [Google Scholar]
- Durrant W.E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42: 185–209. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2: 213–218. [DOI] [PubMed] [Google Scholar]
- Gu Y.Q., Yang C., Thara V.K., Zhou J., Martin G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12: 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. (2003). Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677. [DOI] [PubMed] [Google Scholar]
- Guo P., Li Z., Huang P., Li B., Fang S., Chu J., Guo H. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Ecker J.R. (1990). Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpham N.V.J., Berry A.W., Knee E.M., Rovedahoyos G., Raskin I., Sanders I.O., Smith A.R., Wood C.K., Hall M.A. (1991). The effect of ethylene on the growth and development of wild-type and mutant Arabidopsis thaliana (L). Heynh. Ann. Bot. (Lond.) 68: 55–61. [Google Scholar]
- He W., et al. (2011). A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Meyerowitz E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Pang L., Fang S., Chu J., Li R., Zhu Z. (2018). High ambient temperature antagonizes ethylene-induced exaggerated apical hook formation in etiolated Arabidopsis seedlings. Plant Cell Environ. 41: 2858–2868. [DOI] [PubMed] [Google Scholar]
- Ju C., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.S., Renna L., Yarema J., Ruberti C., He S.Y., Brandizzi F. (2018). Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 115: E5203–E5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Leslie C.A., Romani R.J. (1988). Inhibition of ethylene biosynthesis by salicylic acid. Plant Physiol. 88: 833–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Johnson P., Stepanova A., Alonso J.M., Ecker J.R. (2004). Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., Guo H. (2015). EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell 163: 670–683. [DOI] [PubMed] [Google Scholar]
- Liscum E., Hangarter R.P. (1993). Light-stimulated apical hook opening in wild-type Arabidopsis thaliana seedlings. Plant Physiol. 101: 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J., Carr J.P., Klessig D.F., Raskin I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004. [DOI] [PubMed] [Google Scholar]
- Martínez C., Pons E., Prats G., León J. (2004). Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 37: 209–217. [DOI] [PubMed] [Google Scholar]
- Merchante C., Brumos J., Yun J., Hu Q., Spencer K.R., Enríquez P., Binder B.M., Heber S., Stepanova A.N., Alonso J.M. (2015). Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell 163: 684–697. [DOI] [PubMed] [Google Scholar]
- Métraux J.P., Signer H., Ryals J., Ward E., Wyss-Benz M., Gaudin J., Raschdorf K., Schmid E., Blum W., Inverardi B. (1990). Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250: 1004–1006. [DOI] [PubMed] [Google Scholar]
- Morris K., MacKerness S.A.H., Page T., John C.F., Murphy A.M., Carr J.P., Buchanan-Wollaston V. (2000). Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. 23: 677–685. [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. (2003). EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 115: 679–689. [DOI] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V., Ecker J.R. (1999). Regulation of differential growth in the apical hook of Arabidopsis. Development 126: 3661–3668. [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: Its role in plant growth and development. J. Exp. Bot. 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Rochon A., Boyle P., Wignes T., Fobert P.R., Després C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18: 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A.J. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Li Y., Pan Y., Zhong S. (2016). Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front. Plant Sci. 7: 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.T., Ribone P.A., Chan R.L., Ligterink W., Hilhorst H.W.M. (2016). A predictive coexpression network identifies novel genes controlling the seed-to-seedling phase transition in Arabidopsis thaliana. Plant Physiol. 170: 2218–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12: 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Zhu C., Zhang X., Wen X., Liu L., Peng J., Guo H., Yi C. (2015). Biochemical and structural insights into the mechanism of DNA recognition by Arabidopsis ETHYLENE INSENSITIVE3. PLoS One 10: e0137439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Huang H., Gao H., Wang J., Wu D., Liu X., Yang S., Zhai Q., Li C., Qi T., Xie D. (2014). Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P.H.J., Eggermont K., Tierens K.F.M., Broekaert W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121: 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.G., Ellis C., Devoto A. (2002). The jasmonate signal pathway. Plant Cell 14 (Suppl): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H., Lu H., Rate D.N., Greenberg J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J. 28: 209–216. [DOI] [PubMed] [Google Scholar]
- Wang K.L.C., Yoshida H., Lurin C., Ecker J.R. (2004). Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950. [DOI] [PubMed] [Google Scholar]
- Wang Y., Guo H. (2019). On hormonal regulation of the dynamic apical hook development. New Phytol. 222: 1230–1234. [DOI] [PubMed] [Google Scholar]
- Weigel R.R., Pfitzner U.M., Gatz C. (2005). Interaction of NIMIN1 with NPR1 modulates PR gene expression in Arabidopsis. Plant Cell 17: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek K., Hofmann J., Blöchl A., Szakasits D., Bohlmann H., Grundler F.M.W. (2008). Arabidopsis endo-1,4-β-glucanases are involved in the formation of root syncytia induced by Heterodera schachtii. Plant J. 53: 336–351. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Willige B.C., Ogiso-Tanaka E., Zourelidou M., Schwechheimer C. (2012). WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development 139: 4020–4028. [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J.Y., Boyle P., Wang Y., Brindle I.D., De Luca V., Després C. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Reports 1: 639–647. [DOI] [PubMed] [Google Scholar]
- Yin Y., Wang Z.Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Moeder W., Kang H.G., Kachroo P., Masmoudi K., Berkowitz G., Klessig D.F. (2006). The chimeric Arabidopsis CYCLIC NUCLEOTIDE-GATED ION CHANNEL11/12 activates multiple pathogen resistance responses. Plant Cell 18: 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang L., Qi B., Zhao B., Ko E.E., Riggan N.D., Chin K., Qiao H. (2017). EIN2 mediates direct regulation of histone acetylation in the ethylene response. Proc. Natl. Acad. Sci. USA 114: 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Ji Y., Xue C., Ma H., Xi Y., Huang P., Wang H., An F., Li B., Wang Y., Guo H. (2018). Integrated regulation of apical hook development by transcriptional coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 30: 1971–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhu Z., An F., Hao D., Li P., Song J., Yi C., Guo H. (2014). Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 26: 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y.T., Qu N., Zhao Q., Bi D., Li X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48: 647–656. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96: 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Wang J., Yuan J., Wang X.L., Zhao Q.P., Kong P.T., Zhang X. (2015). NITRIC OXIDE-ASSOCIATED PROTEIN1 (AtNOA1) is essential for salicylic acid-induced root waving in Arabidopsis thaliana. New Phytol. 207: 211–224. [DOI] [PubMed] [Google Scholar]
- Zhou J.M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., Klessig D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13: 191–202. [DOI] [PubMed] [Google Scholar]