Diatoms, the world’s most diverse group of algae, are emerging as important model systems for studying eukaryotic phytoplankton biology and evolution.
Abstract
Diatoms are the world’s most diverse group of algae, comprising at least 100,000 species. Contributing ∼20% of annual global carbon fixation, they underpin major aquatic food webs and drive global biogeochemical cycles. Over the past two decades, Thalassiosira pseudonana and Phaeodactylum tricornutum have become the most important model systems for diatom molecular research, ranging from cell biology to ecophysiology, due to their rapid growth rates, small genomes, and the cumulative wealth of associated genetic resources. To explore the evolutionary divergence of diatoms, additional model species are emerging, such as Fragilariopsis cylindrus and Pseudo-nitzschia multistriata. Here, we describe how functional genomics and reverse genetics have contributed to our understanding of this important class of microalgae in the context of evolution, cell biology, and metabolic adaptations. Our review will also highlight promising areas of investigation into the diversity of these photosynthetic organisms, including the discovery of new molecular pathways governing the life of secondary plastid-bearing organisms in aquatic environments.
INTRODUCTION
Diatoms (Bacillariophyceae) are unicellular chlorophyll a/c-containing microalgae belonging to the division Heterokontophyta, also known as stramenopiles (e.g., Stiller et al., 2014). Diatoms represent a major class of phytoplankton in both marine and freshwater environments (Field et al., 1998; Pierella Karlusich et al., 2020) and are the most species-rich class of microalgae, with at least 100,000 extant species according to conservative estimates (Malviya et al., 2016). Diatoms are distributed worldwide, from tropical and subtropical regions to polar ecosystems (Figures 1 and 2). Thus, the diversity of diatom lifestyles and survival strategies can likely be attributed to their exceptional ability to adapt to highly dynamic aquatic environments. Diatoms and other aquatic protists contribute less than 1% to the Earth’s total biomass (550 gigatons of carbon [Gt C], 80 to 90% of which is derived from plants; (Bar-On et al., 2018; Pierella Karlusich et al., 2020), but diatoms alone contribute an estimated 20% to annual C fixation. This role in global C cycling is comparable to that of all the terrestrial rain forests combined (Field et al., 1998).
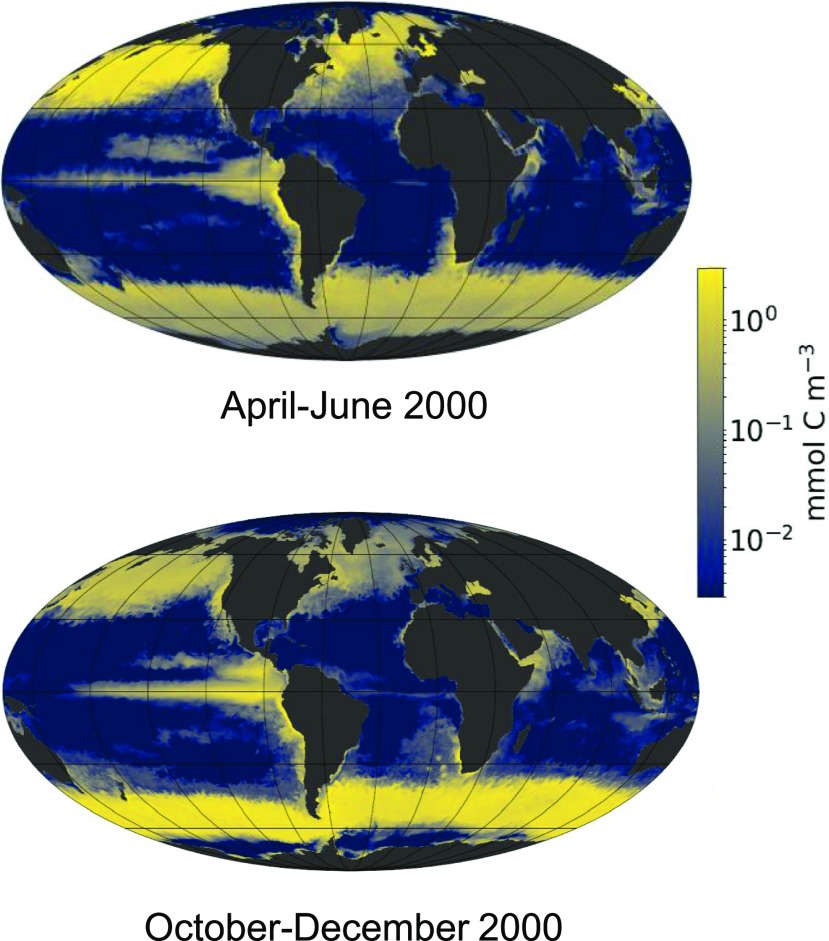
Figure 1.
Model of the Global Distribution of Diatom Biomass.
Model of the global distribution of diatom biomass in the ocean surface layer in April to June (left) and October to December (right) of 2000 (courtesy of Oliver Jahn, Stephanie Dutkiewicz, and Mick Follows). Biomass values, reported in log scale in units of mmol C m−3, were derived from the MIT ecosystem model, which simulates ocean circulation and key biogeochemical processes, e.g., nutrient fluxes, plankton growth, and death occurring in the ocean, at a global scale (Follows et al., 2007; Tréguer et al., 2018). Physiological data were derived from laboratory and field experiments. Because diatom biomass varies between ∼0.4 (P. tricornutum) and ∼7000 (Coscinodiscus wailesii) pmol C per cell, a biomass of 1 mmol C m−3 corresponds to a range of ∼150 cells L−1 to ∼2.5 106 cells L−1 (Marañón et al., 2013).
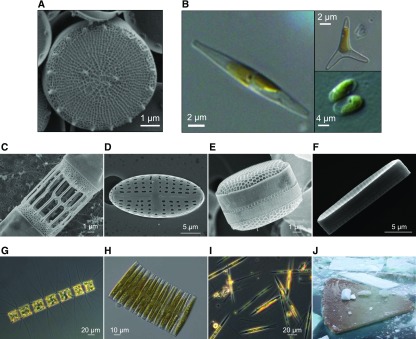
Figure 2.
Diatom Characteristics and Morphological Diversity.
(A) Scanning electron micrograph of the model centric species T. pseudonana with its characteristic cell wall of silica, viewed from the top.
(B) to (I) Light microscopy of the pennate model species P. tricornutum (B). This diatom exists in three interconvertible morphotypes: the fusiform, oval, and triradiate morphotypes, as shown in the figure. Scanning electron micrograph images of (C) Skeletonema tropicum, (D) a valve of a raphid pennate diatom, (E) Shionodiscus oestrupii var venrickiae, and (F) F. cylindrus. Light microscopy of (G) Chaetoceros sp, (H) Fragilariopsis kerguelensis, and (I) Pseudo-nitzschia sp from a natural phytoplankton sample collected at the Long Term Station MareChiara in the Gulf of Naples, Italy.
(J) Flipped ice floe in the Southern Ocean with dense population of sea-ice diatoms (e.g., F. cylindrus) at the ice-water interface. Images were kindly provided by Diana Sarno and Marina Montresor (Stazione Zoologica Anton Dohrn, Napoli, Italy) and James A. Raymond (Univeristy of Nevada, United States).
The most distinctive macroscopic feature of diatoms is the cell wall, which is made of hydrated silicon dioxide (silica; Figure 2) comprising two halves (thecae) that fit together like a Petri dish (Round et al., 1990). Subcellular features include a plastid surrounded by four membranes, loosely stacked thylakoids with no apparent subdomains, and a rough endoplasmic reticulum (ER) connected to the nucleus and outer plastid membrane. Unlike plants, diatoms store energy in the form of chrysolaminarin (β-1,3-glucan; Gruber and Kroth, 2017; Huang et al., 2018) and lipids (Armbrust et al., 2004). The metabolic strategies of different diatom species range from photoautotrophy to heterotrophy, including parasitism (Gruber and Kroth, 2017; Supplemental Table 1). Some diatoms live as free-floating single cells, whereas others form colonies (Figure 2) and participate in mutualistic or symbiotic relationships (Foster et al., 2011).
In the last 50 years, major advances in plant biology have been propelled by the development of genomic and genetic resources for experimentally controllable model systems such as Arabidopsis (Arabidopsis thaliana; Arabidopsis Genome Initiative, 2000; Koornneef and Meinke, 2010). In parallel, the green alga Chlamydomonas reinhardtii has emerged as a model system to study photosynthesis, chloroplast biology, sexual reproduction, flagellar motility, and micronutrient homeostasis (Merchant et al., 2007; Salomé and Merchant, 2019). Approximately 25 years ago, the diatom research community established the first genetic transformation protocols for microparticle bombardment of various species (Dunahay et al., 1995; Apt et al., 1996). This work prompted the nascent molecular marine community to start sequencing the genomes of phytoplankton organisms, first Thalassiosira pseudonana (Armbrust et al., 2004) and then Phaeodactylum tricornutum (Bowler et al., 2008), which are both diatom species.
In this review, we discuss the continuing rise of diatoms as model organisms. To date, T. pseudonana and P. tricornutum (Figures 2A and 2B) are the two species most amenable to laboratory studies, from simple growth experiments to the development of the latest genome-editing tools (Tables 1 to 4; Supplemental Tables 1 and 2). During the past 15 years, many genomic, metabolic, and cellular features of these algae have been discovered and are likely to be conserved in many diatom species. Nevertheless, many pertinent questions about the life cycles and environmental adaptation of ecologically relevant diatom species cannot be addressed with these two models alone, pointing to the pros and cons of having model species. This need has driven the implementation of new genome sequencing projects for Fragilariopsis cylindrus, Pseudo-nitzschia multistriata, and other species (Table 1; Supplemental Table 1). Information from these emerging models has extended our understanding of the evolution and diversification of diatom genomes. Thus, these diatoms may also represent the dawn of novel models that could be used to address biological questions that might have been missed by focusing on only a few models among the more than 100,000 extant diatom species.
Table 1. Genomic Features of Established and Emerging Diatom Model Species.
| Centric Diatoms | Pennate Diatoms | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Features | Thalassiosira pseudonanaa | Thalassiosira oceanica | Cyclotella cryptica | Skeletonema costatum | Phaeodactylum tricornutuma | Fragilariopsis cylindrus | Pseudo-nitzschia multiseries | Pseudo-nitzschia multistriata | Fistulifera solaris | |
| Strain | CCMP1335 | CCMP1005 | CCMP332 | Ariake8 | CCAP 1055/1 | CCMP 1102 | CLN-47 | B856 | JPCC DA0580 | |
| Ploidy | diploid | diploid | diploid | diploid | diploid, triploidy proposed | allodiploid | ||||
| Genome size (Mbp) | 32.1 | 81.6 | 161.7 | 46.9 | 27.4 | 61.1 | 218.7 | 59 | 24.9 | |
| Number of: | ||||||||||
| Chr. pairs | 24 | 34 | 42 | |||||||
| Scaffolds | 51,656 | 55 | 4890 | 297 | ||||||
| Haplotype divergence | 0.75% | 0.85% | up to 6% in 29% of genes | 14 to 38% | ||||||
| Number of genes | 11,776 | 34,642 | 21,121 | 16,449 | 12,233 | 21,066 | 19,703 | 12,008 | 11,448 | |
| Average gene size (bp) | 1,740.8 | 1,255 | 1,471 | 1,100 | 1,624 | 1,575 | 1,522 | 2,205 | ||
| Intron | ||||||||||
| Number/gene | 1.54 | 1.29 | 1.18 | 0.89 | 1.1 | 1.38 | 0.87 | |||
| Average length | 125.4 bp | 150 bp | 125 bp | 142 bp | 255.7 bp | 229 bp | 341 bp | |||
| Plastid | ||||||||||
| Genome size | 129 kbp | 141 kbp | 129 kbp | 117 kbp | 111. 5 kbp | 135 kbp | ||||
| Number of genes | 127 | 128 | 132 | 130 | 97 | 132 | ||||
| Mitochondria | ||||||||||
| Genome size | 43.8 kbp | 35.3 kbp | 58 kbp | 77.3 kbp | 46.3 kbp | >38.6 kbp | ||||
| Number of genes | 34 | 31 | 35 | 34 | 37 | 31 | ||||
| Annotation version | v3 | v1 | v3 | v1 | v1 | |||||
| References | (Armbrust et al., 2004) | (Lommer et al., 2012) | (Traller et al., 2016) | (Ogura et al., 2018) | (Bowler, 2008; Rastogi et al., 2018) | (Mock et al., 2017) | (Basu et al., 2017) | (Tanaka et al., 2015b) | ||
Genome portal: T. pseudonana, https://genome.jgi.doe.gov/Thaps3/Thaps3.home.html; T. oceanica, https://genome.jgi.doe.gov/Thaoce1/Thaoce1.info.html; C. cryptica, http://genomes.mcdb.ucla.edu/Cyclotella/download.html; S. costatum, https://ddbj.nig.ac.jp/DRASearch/study?acc=DRP004496; P. tricornutum, Phatr3:http://protists.ensembl.org/Phaeodactylum_tricornutum/Info/Index; F. cylindrus, https://genome.jgi.doe.gov/Fracy1/Fracy1.info.html; P. multiseries, https://genome.jgi.doe.gov/Psemu1/P semu1.home.html; P.multistriata, http://apollo.tgac.ac.uk/Pseudo-nitzschia_multistriata_V1_4_browser/sequences; F. solaris, https://trace.ddbj.nig.ac.jp/DRASearch/submission?acc=DRA002403. Chr., chromosome.
Blank cells indicate no data available.
Established diatom model species.
Table 4. Technologies and Genome-Enabled Tools available for T. pseudonana and P. tricornutum.
| Technologies | T. pseudonana | P. tricornutum | ||
|---|---|---|---|---|
| Techniques | ||||
| Nuclear Transformation Methods | ||||
| Biolistic | (Poulsen et al., 2006) | (Apt et al., 1996; Falciatore et al., 1999) | ||
| Conjugation | (Karas et al., 2015) | (Karas et al., 2015) | ||
| Electroporation | (Bugge, 2015) | (Niu et al., 2012) | ||
| Plastid Transformation | ||||
| Biolistic | (Materna et al., 2009) | |||
| Electroporation | (Xie et al., 2014) | |||
| Modular Cloning Technology | ||||
| Gateway | (Shrestha and Hildebrand, 2015) | (Siaut et al., 2007) | ||
| Golden Gate | (Hopes et al., 2016) | |||
| Gene Silencing | ||||
| Inverted repeat and antisense fragments | (Shrestha and Hildebrand, 2015) | (De Riso et al., 2009) | ||
| Genome Editing Methods | ||||
| CRISPR/Cas9 | (Hopes et al., 2016) | (Nymark et al., 2016) | ||
| Delivered as RNP | (Serif et al., 2018) | |||
| TALEN | (Daboussi et al., 2014) | |||
| Meganucleases | (Daboussi et al., 2014) | |||
| Molecular Tools | ||||
| Antibiotic Resistance/Selectable Markers | ||||
| Zeocin and Phleomycin/sh ble | (Apt et al., 1996) | |||
| Nourseothricin/nat | (Zaslavskaia et al., 2001) | |||
| Blasticidin-S/bsr | (Buck et al., 2018) | |||
| Streptothricin/sat | (Zaslavskaia et al., 2001) | |||
| Neomycin/nptII | (Zaslavskaia et al., 2001) | |||
| Reporter Genes | ||||
| eGFP | (Poulsen et al., 2006) | (Zaslavskaia et al., 2001) | ||
| YFP | (Karas et al., 2015) | (Siaut et al., 2007) | ||
| CFP | (Siaut et al., 2007) | |||
| uidA | (Sheppard et al., 2011) | (Zaslavskaia et al., 2001) | ||
| cat | (Apt et al., 1996) | |||
| Aequorin | (Falciatore et al., 2000) | |||
| Promoters | ||||
| Most Frequently Used | ||||
| Lhcf (Fcp), light responsive, strong | (Poulsen et al., 2006) | (Apt et al., 1996; Falciatore et al., 1999) | ||
| NR, NO3 inducible, strong | (Poulsen et al., 2006) | (Hempel et al., 2009) | ||
| H4, stable, mild | (Siaut et al., 2007) | |||
| Additionally Reported | ||||
| V-ATPase C, strong | (Watanabe et al., 2018) | |||
| Ef2, stable, strong | (Seo et al., 2015) | |||
| Ef-1α, 40SRPS8, γ-Tubulin, RBCMT, strong | (Slattery et al., 2018) | |||
| SIT1, Si-starvation inducible, strong | (Davis et al., 2017) | |||
| Thaps3_9619, Si-starvation inducible, strong | (Shrestha and Hildebrand, 2017) | |||
| AP1, strong, low P inducible | (Lin et al., 2017) | |||
| Fbp1, Fld, Isi1, iron-responsive | (Yoshinaga et al., 2014) | |||
| Ca1, Ca2, CO2-responsive | (Harada et al., 2005; Tanaka et al., 2016) | |||
| U6, RNA polymerase III transcribed | (Hopes et al., 2016) | (Nymark et al., 2016) | ||
| CMV, CaMV35S, RSV-LTR, PCMV, viral | (Sakaue et al., 2008) | |||
| CdP1, ClP1, ClP2, TnP1, TnP2, viral | (Kadono et al., 2015) | |||
AP1, alkaline phosphatase; bsr, blasticidin-S resistance gene; Ca1, carbonic anhydrase; CaMV, Cauliflower mosaic virus 35S; cat, chloramphenicol acetyl transferase conferring resistance to chloramphenicol; CdP, Chaetoceros debilis-infecting DNA virus; CFP, cyan fluorescent protein gene; ClP, Chaetoceros lorenzianus-infecting DNA virus; CMV, cytomegalovirus; CRISPR, clustered regularly interspaced short palindromic repeats; Ef-1α, Ef2, elongation factor 1α and 2; eGFP, enhanced green fluorescent protein gene; Fbp1, ferrichrome binding protein1; Fld, flavodoxin; γ-Tubulin, tubulin gamma chain; H4, histone H4; Isi1, iron-starvation-induced gene 1; Lhcf, diatom light harvesting complex; nat, nourseothricin acetyl transferase; nptII, neomycin phosphotransferase II; NR, nitrate reductase; P, phosphate; RBCMT, ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit N-methyltransferase I; RNP, ribonucleoprotein; RSV-LTR, Rous sarcomavirus long terminal repeat; sat, streptothricin acetyl transferase; sh ble, Streptoalloteichus hindustanus bleomycin resistance gene; SIT1, silicon transporter; TnP, Thalassionema nitzschioides-infecting DNA virus; U6, small nuclear RNA of the U6 complex; uidA, β-glucuronidase (GUS)-encoding gene; YFP, yellow fluorescent protein gene; 40SRPS8, 40S ribosomal protein S8.
Blank cells indicate no reference available.
In the following sections, we review diatom evolution and phylogeny and aspects of the cell and life cycle, discuss emerging areas of importance (“Selected Research Highlights” section) for diatom biology from genome organization to basic metabolism, and highlight the genomic and genetic resources and technologies that have been developed over the past decade (“Genomic and Genetic Resources” section). By placing current and future diatom research in the context of green algae and plants, we hope to make diatom research accessible to plant biologists, especially because most of the fascinating molecular secrets of diatoms await to be discovered.
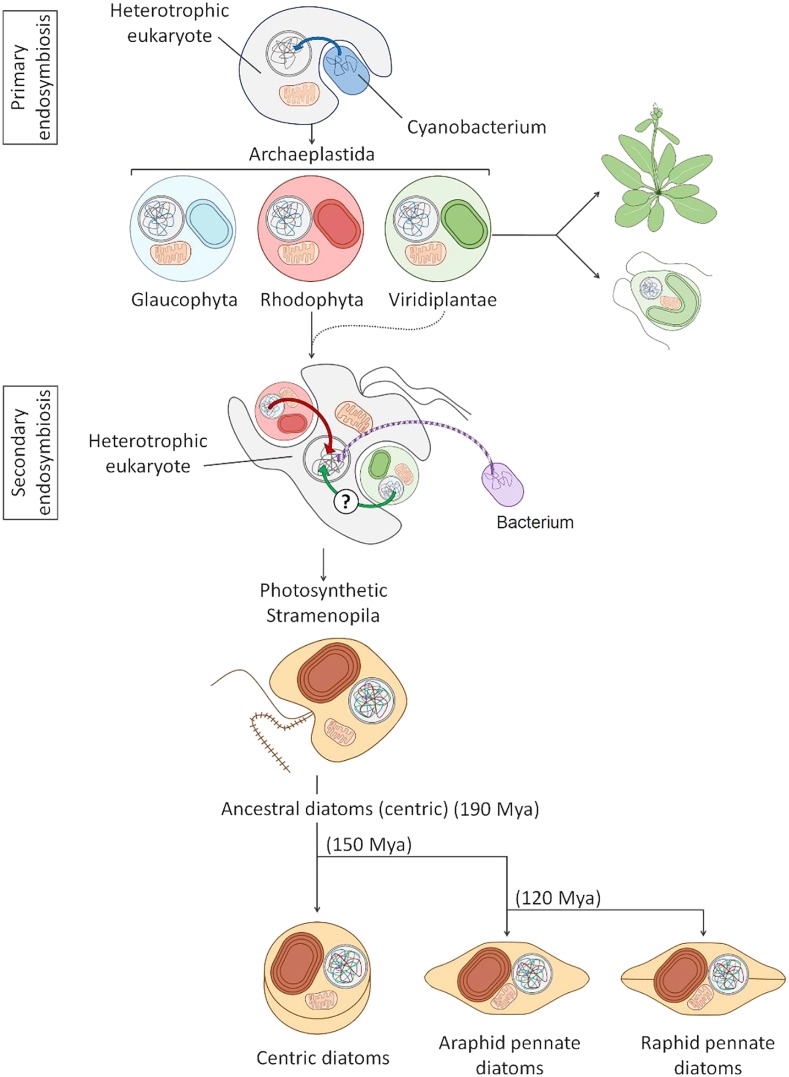
EVOLUTION AND PHYLOGENY
Photosynthetic eukaryotes evolved more than 1.5 billion years ago in the Proterozoic oceans. The progenitor of the plastid is considered to have been a free-living cyanobacterium that was engulfed by a heterotrophic host via primary endosymbiosis (Shih and Matzke, 2013). This event gave rise to the supergroup Archaeplastida, which diversified into three major lineages: the glaucophytes, rhodophytes, and Viridiplantae (Petersen et al., 2014). The rhodophytes gave rise to the stramenopiles (Figure 3) as well as the cryptophytes, alveolates, and haptophytes, together comprising the chromalveolate group (Stiller et al., 2014). Most chromalveolate species containing a red-algal-derived plastid or at least a remnant of it (e.g., Plasmodium falciparum) were thought to have originated from a single secondary endosymbiotic event (Cavalier-Smith, 1999), which is consistent with phylogenetic evidence based on plastid genes. However, phylogenomic studies based on nuclear genes do not support the proposed monophyly of chromalveolates (e.g., Stiller et al., 2014). Large-scale comparative genomic analyses have even suggested that a cryptic green-algal endosymbiont in chromalveolates predated the symbiosis with red alga (Moustafa et al., 2009). Based on the in silico prediction of 770 plastid-targeted proteins that are conserved across photosynthetic stramenopiles and on a smaller set of experimentally verified plastid-localized proteins, there is initial evidence that approximately 25% of the ancestral diatom plastid proteome consisted of nucleus-encoded proteins from green algae (Dorrell et al., 2017). Horizontal gene transfer (HGT) also contributed to the complex history of gene acquisitions in the chromalveolates (e.g., Nosenko and Bhattacharya, 2007; Chan et al., 2012; Basu et al., 2017).
Figure 3.
Simplified Scheme of the Major Events Leading to the Evolution of Diatoms through Primary and Secondary Endosymbiosis.
The initial primary endosymbiosis occurred when a heterotrophic host engulfed a cyanobacterium (represented in blue). Over time, a large proportion of the cyanobacterial genome was transferred to the nucleus of the host, as indicated by the blue arrow. The endosymbiotic process generated the plastids of the Archaeplastida, a major group including Glaucophyta (pale blue), Rhodophyta (pink), and the Viridiplantae (the model green alga C. reinhardtii and plant Arabidopsis are shown). During secondary endosymbiosis, a different heterotrophic cell acquired a red alga and potentially also a green alga. The algal endosymbiont became the plastid (in brown) of the Stramenopila, a group including diatoms, but also other algae such as pelagophytes or the multicellular kelps. Algal nuclear genomes were transferred to the heterotrophic nucleus, as represented by green and red curved arrows, while the algal nucleus and mitochondria were lost. Other bacterial genes in the Stramenopila genome were derived by horizontal gene transfer events from bacterial donors (violet arrow). The figure also shows the approximative dates of diatom evolution and separation between centric and pennate diatoms based on Nakov et al. (2018). Some pennate species further diversified and acquired the capacity to move by secreting mucilage through a longitudinal slit in the cell wall called raphe, hence the division between raphid (motile) and araphid (nonmotile) pennate diatoms. Mya, million years ago.
Despite the uncertain origins of many diatom genes, calibrated molecular clocks, multigene and taxon phylogeny, and the fossil record provide evidence that diatoms arose around the Triassic-Jurassic boundary ∼190 million years ago (Nakov et al., 2018). Diatoms can be divided into two major groups (Kooistra and Medlin, 1996): the radially symmetrical centric diatoms (e.g., T. pseudonana) and the pennate diatoms, with an elongated bilateral symmetry (e.g., P. tricornutum). Pennate diatoms can be subdivided into nonmotile araphid species and motile raphid species (Figures 2 and 3). Raphid species (e.g., P. tricornutum, F. cylindrus, P. multistriata) are mobile due to the presence of the raphe, a longitudinal slit in the cell wall through which mucilage containing actin-myosin protein complexes is secreted (Poulsen et al., 1999). Interestingly, the evolution of the raphe and the accompanying changes in life history traits (such as active motility of vegetative cells) approximately 120 million years ago appear to have facilitated range expansions or colonization of previously unavailable habitats (e.g., sediments) with varying rates of diversification. The raphid group outnumbers the species diversity estimated for both centric and araphid pennate diatoms, which might reflect the propensity of raphid diatoms to occupy and adapt to a variety of habitats, from sea ice in polar oceans to the epibiotic lifestyle in freshwater ecosystems. Most of the accelerated diversification took place in the Cenozoic over the past 75 million years (Nakov et al., 2018).
DIATOM CELL DIVISION AND LIFE CYCLE
Diatoms often display a “bloom and bust” life cycle, when rapid mitosis under favorable environmental conditions is followed by the demise of the bloom due to the depletion of nutrients, grazing of herbivores, sinking of cells, sexual reproduction, or even cell death if environmental conditions become unfavorable (Gross, 2012). Diatom cell division is complex and has unique aspects, as it resembles a patchwork of processes from plants and animals (Pickett-Heaps and Tippit, 1978; De Martino et al., 2009; Tanaka et al., 2015a). For instance, diatom mitosis commences at the microtubule-organizing center, similar to those in animal cells. Similar to most plants, diatoms do not have centrioles but rather possess a highly organized central spindle embedded in a matrix called the collar that enables chromosome attachment (Pickett-Heaps and Tippit, 1978). Diatom cytokinesis involves the centripetal development of a cleavage furrow, resembling cytokinesis in animals, but the process of centrifugal cell wall neosynthesis is more similar to plant cell wall deposition (Pickett-Heaps and Tippit, 1978). Cyclins, which regulate the cell cycle together with cyclin-dependent kinases, have expanded extensively in diatoms (Huysman et al., 2010). Some diatom-specific cyclins (dsCYCs) induced under specific growth conditions may control diatom growth in particular environments. dsCYC2 is a key regulator of the onset of cell division in P. tricornutum (Huysman et al., 2013). dsCYC2 controls the light-dependent G1-to-S cell cycle checkpoint, and its expression is induced by the blue-light sensor AUREOCHROME1a (described in more detail below).
The silica cell wall undergoes characteristic changes during cell division (Round et al., 1990). In general, the diatom cell wall (frustule) is composed of two identically shaped but slightly different sized thecae: the larger epitheca and the smaller hypotheca (Figure 4). The two thecae are fastened together by bands of silica, the girdle bands. When diatom cells divide, each daughter cell retains one theca from the original frustule, meaning that one new theca must be synthesized per daughter cell in addition to new girdle bands in order to keep the original and new thecae together. The synthesis of the silica structures is coordinated with the cell cycle, and when silicate is scarce in seawater, the cell cycle is halted (Brzezinski et al., 1990). It is generally assumed that intracellular silica deposition and patterning depend on both self-assembly processes and controlled silica polymerization in specialized silica deposition vesicles (Reimann et al., 1966), which deliver polymerized and macromolecule-complexed silica to specific cellular sites by exocytosis. Numerous organic molecules are involved in these processes, including soluble and transmembrane proteins (e.g., Kröger and Poulsen, 2008; Hildebrand et al., 2018). Proteins involved in cell wall biogenesis were initially identified by biochemical analysis and nanoscale resolution imaging. Putative novel regulators of silicon bioprocesses have been identified by whole-genome expression profiling of T. pseudonana grown under different conditions (Supplemental Table 2). Newly developed genome-editing tools (see section Genomic and Genetic Resources) have been used to define their functions in vivo. For instance, knockdown and knockout of T. pseudonana silacidin production revealed that this protein influences the size of thecae (Belshaw et al., 2017; Kirkham et al., 2017), whereas knockout of silicanin-1 suggested that this protein is important for ensuring that the diatom cell wall is mechanically robust (Görlich et al., 2019).
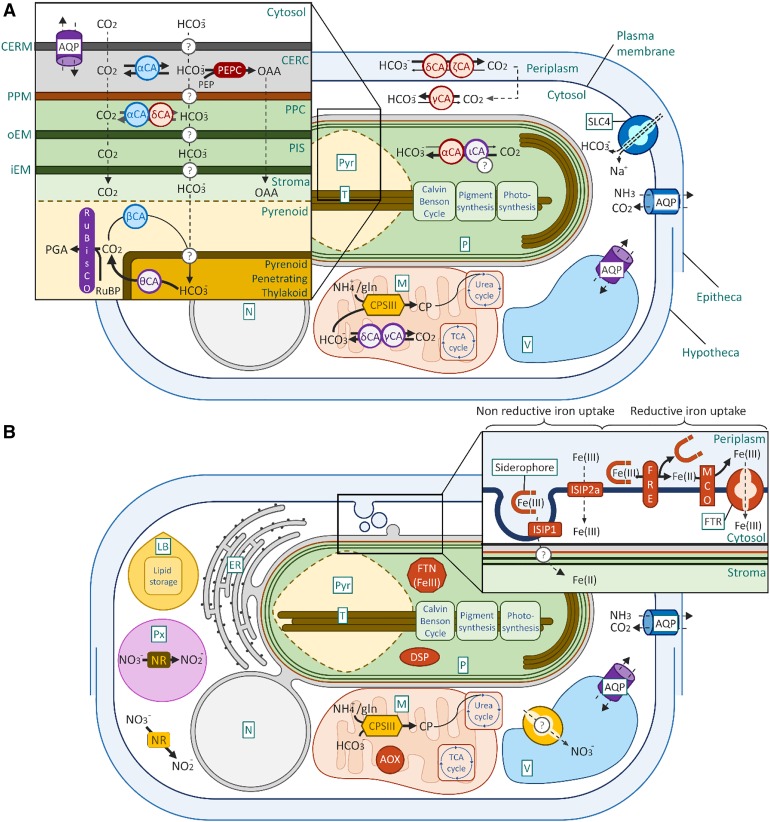
Figure 4.
Regulators of Carbon, Nitrogen, and Iron Metabolism Characterized in the Diatom Model Species.
(A) Regulators of carbon.
(B) Regulators of nitrogen and iron.
AOX, alternative oxidase; AQP, aquaporin; CA, carbonic anhydrases (characterized in P. tricornutum in blue, in T. pseudonana in red, in both in purple); CERC, chloroplast/endoplasmic reticulum compartment; CERM, chloroplast endoplasmic reticulum membrane; CP, carbamoyl phosphate; CPSIII, carbamoyl-phosphate synthase; DSP, death-specific protein; FRE, ferrireductase; FTN, ferritin; FTR, Fe(III) permease; iEM, internal plastid envelop membrane; ISIP1 and ISIP2a, iron-starvation-induced proteins; LB, lipid body; M, mitochrondrion; MCO, multicopper oxidase; N, nucleus; NR, nitrate reductase; oEM, outer plastid envelop membrane; P, plastid; PEPC, phosphoenolpyruvate carboxylase; PIS, plastid intermembrane space; PPC, periplastidial compartment; PPM, periplastidial membrane; Px, peroxysome; Pyr, pyrenoid; SCL4, bicarbonate solute carrier 4; T, thylakoids; TCA, tricarboxylic acid; V, vacuole. Question marks indicate still uncharacterized regualtors or uncertain cellular localisation.
For many diatoms, a progressive reduction in cell size occurs during each cell division because the hypotheca is synthesized inside the cells of the previous generation (Macdonald, 1869; Montresor et al., 2016). Restoration of the original cell size is thought to involve sexual recombination. For sexual reproduction in centric diatoms, eggs and flagellated sperm are usually produced from a clonal strain (homothallic), whereas pennate diatoms require strains of opposite mating types (heterothallic; Montresor et al., 2016). In many diatom species, sexual reproduction can only be induced below a specific cell size threshold. Several species (such as Seminavis robusta) emit pheromones as a signal to find appropriate mating types for sexual reproduction (Moeys et al., 2016). Little is known about how sex determination is controlled in diatoms, partly because this issue is not pertinent to the two most-studied models, T. pseudonana and P. tricornutum, as they appear to have lost the ability to reproduce sexually and do not go through cell size reduction.
Important information has recently been obtained from studies of the pennate diatom P. multistriata by taking advantage of novel reverse genetics tools developed for this species (Table 1; Supplemental Table 1). By comparing gene expression profiles of opposing mating types in P. multistriata, Russo et al. (2018) identified five genes with a mating-type-specific expression pattern, including MRP3, which is expressed exclusively in one mating type in a monoallelic manner. Overexpression of MRP3 in the opposite mating type induced sex reversal such that this transformant was able to mate with its own mating type. These findings indicate that a single allele of MRP3 is responsible for sex determination in P. multistriata (Russo et al., 2018). F. cylindrus contains an MRP3 homolog (Russo et al., 2018), but a population genomics study estimated that the recombination frequency in F. cylindrus is only approximately four times higher than the mutation rate, suggesting that sexual recombination is very infrequent or even lacking in this species (Mock et al., 2017). If MRP3 is part of a larger mating-type locus, techniques such as linkage mapping would reveal whether this locus is nonrecombining, as seen in sexually reproducing multicellular algae such as Ectocarpus siliculosus (Ahmed et al., 2014). Asexuality appears to have emerged several times in diatoms (Montresor et al., 2016), suggesting it might confer a selective advantage under certain conditions. In the open ocean where cell population density is low, the chance of encountering opposite mating types is very limited, so sex might not be the ideal way to recombine and diversify, as suggested for the coccolithophore Emiliania huxleyi (von Dassow et al., 2015). Other mechanisms such as allelic divergence and changes in ploidy might be as effective as sexual reproduction for diatom evolution and adaptation.
SELECTED RESEARCH HIGHLIGHTS
Genome Organization and Regulation of Gene Expression
T. pseudonana (Armbrust et al., 2004) and P. tricornutum (Bowler et al., 2008) were the first diatoms to have their genomes sequenced (Table 1). They were chosen as model systems because of their small genome sizes, rapid growth under controlled laboratory conditions (one division or more per day), their extensively characterized physiology, and the increasing availability of tools for reverse genetics (see section on “Genomic and Genetic Resources”). In the last decade, the hyperdiversity of diatoms, particularly the nested clade of pennate diatoms, has stimulated an interest in comparative genomics approaches to study the drivers of diversification and adaptation to a variety of aquatic habitats. New genome sequencing initiatives have been complemented by efforts to extend the existing molecular toolkit to a wider range of diatoms (Table 1; Supplemental Table 1). The rationale for deciding subsequent genome projects was either based on the importance of the habitats the diatom species occupy or their specific physiology: (1) F. cylindrus (Mock et al., 2017) to reveal adaptations to polar oceans, the preferred habitat of diatoms based on abundance; (2) Thalassiosira oceanica (Lommer et al., 2012) to study adaptation to iron-limited waters, which cover approximately 35% of the ocean surface; (3) P. multistriata (Basu et al., 2017) to reveal genes involved in sexual reproduction and recombination; (4) Pseudo-nitzschia multiseries (Basu et al., 2017) and Skeletonema costatum (Ogura et al., 2018) for the metabolic pathway underpinning the synthesis of a potent diatom toxin and to study harmful algal blooms; and (5) Fistulifera solaris (Tanaka et al., 2015b) and Cyclotella cryptica (Traller et al., 2016) for the production of biofuels.
The haploid size of the nuclear genomes of T. pseudonana (32.1 Mbp) and P. tricornutum (27.4 Mbp) are relatively small, with 11,766 and 12,233 predicted genes, respectively. At least 50% of the nuclear genome encodes proteins or smaller peptides, according to estimates based on P. tricornutum (e.g., Yang et al., 2018). The other sequenced species have larger genome sizes ranging from approximately 60 Mbp for P. multistriata and F. cylindrus up to over 200 Mbp for P. multiseries (Table 1).
All diatom species sequenced so far are thought to be diploid in their vegetative states. However, the results of several k-mer analyses suggest that F. cylindrus is either triploid or aneuploid (T.M., unpublished data). F. solaris has a confirmed allodiploid genome structure indicative of interspecies hybridization (Tanaka et al., 2015b, and below). Plastid and mitochondrial genomes have also been analyzed for most of the species listed in Table 1 in addition to the organellar genomes of other diatoms (Kowallik et al., 1995; Oudot-Le Secq et al., 2007; Lommer et al., 2010; Tanaka et al., 2011; Brembu et al., 2014). Organellar genomes are similarly compact, with few protein-coding genes (Table 1), and most organellar proteins are encoded in the nucleus, as in other eukaryotic phototrophs. Unlike the plastid genomes of green algae, diatom plastid genomes lack introns, except for S. robusta (Brembu et al., 2014). Overall, there appears to be no synteny among plastid genomes in diatoms.
The complex evolutionary history of diatoms is the main reason their genomes are considered to be chimeric (Armbrust et al., 2004; Bowler et al., 2008). The extant gene repertoire common to all diatom genomes was recruited from (1) the ancient heterotrophic host, (2) a red algal (and potentially also a green algal) endosymbiont, and (3) diverse bacteria via HGT. As green and red algae are subgroups of the Archaeplastida that arose from the primary endosymbiosis event, diatom genomes also contain cyanobacterial genes and genes from their hosts (Figure 3; Petersen et al., 2014).
A comparison of several diatom genomes with those of other taxonomic groups confirmed that they include a significant proportion of species-specific genes (e.g., 26% of P. tricornutum genes) among smaller proportions of “core diatom-specific genes” (not present in nondiatom genomes, 15.8%) and genes shared with diatoms and other stramenopiles (5.3%; Rastogi et al., 2018). Comparative genomics in the green lineage revealed similar proportions of genes across different species. For instance, ∼15% of genes of the model chlorophyte C. reinhardtii are considered to be species specific (Merchant et al., 2007).
Repetitive elements such as transposable elements (TEs) are some of the main evolutionary drivers affecting genome structure and gene expression in diatoms. For instance, TEs make up less than 3% of the T. pseudonana genome but more than 30% of the F. cylindrus genome (Mock et al., 2017) and 73% of the P. multiseries genome (Basu et al., 2017), possibly reflecting differences in diversification and adaptation. Chlorophyte genomes show less variability in TE content, which varies between 6 and 7% (Philippsen et al. 2016). In diatoms, retrotransposons flanked with long terminal repeats appear to be the most abundant TEs in diatom genomes (Maumus et al., 2009). Some P. tricornutum-specific Ty1/copia-like retrotransposons are strongly induced when nitrate is depleted (Maumus et al., 2009), and cytosine methylation is thought to control the mobility of these TEs, as they are hypomethylated when expressed (Veluchamy et al., 2015). Long terminal repeat retrotransposons might induce homologous recombination between highly similar elements, leading to homologous and nonhomologous recombinant genomic loci, including gene-containing flanking regions. The detection of de novo insertions of these elements into genomic loci in different P. tricornutum accessions supports the hypothesis that transposition contributes to rearrangements and diversification of diatom genomes, which might be adaptive, as there is conditional activation of TEs (Maumus et al., 2009). How TE-mediated recombination contributes to the evolution of genomes in terms of the accumulation and spread of de novo single nucleotide polymorphisms in a population depends on the selective advantage conferred by the mutation. Depending on the strength of selection and the population structure, this might lead to selective sweeps. Experimental evolution under different types of selection pressure combined with population genomics based on single-cell genomes will shed light on how mutations come to prominence in diatom populations and therefore become established in the genome. Such work will also reveal the temporal dynamics of the fixation of mutations and provide fundamental insights into the pace of genetic adaptation (e.g., Krasovec et al., 2019) under changing environmental conditions in the ocean.
An additional influence on diatom genomes is the acquisition of genes via HGT (Bowler et al., 2008). Recent data have estimated that 2 to 6% of diatom genes are of bacterial origin (Basu et al., 2017). However, long-read sequencing technology, which was not applied to T. pseudonana or P. tricornutum, will reveal how many of these genes were derived from contaminating bacteria. A recent genome project with 10 red algal species based on long-read sequencing technology revealed that only ∼1% of genes were horizontally transferred (Rossoni et al., 2019). However, based on the currently available HGT data from diatoms, ∼50% of these genes were acquired before the separation of the main lineages ∼150 million years ago. How genes are transferred into diatom genomes is still unknown. Foreign DNA with a lower GC content can be maintained in P. tricornutum as episomes. Since diatom centromeres have a low GC content (Diner et al., 2017), perhaps low GC foreign DNA in diatoms could recruit specific chromosome maintenance proteins such as histones. This hijacking mechanism might thus favor the acquisition of foreign DNA (Diner et al., 2017). Diatoms have a long history of entangled relationships with bacteria for the acquisition of essential nutrients including vitamins, and bacterial conjugation is effective in transferring genetic material by direct cell-to-cell contact between bacteria and diatoms. Therefore, it is reasonable to assume that their inter-kingdom co-evolution contributed to shaping the structure and organization of diatom genomes.
In addition to TEs and bacterial genes acquired via HGT, gene duplication accompanied by neofunctionalization significantly contributes to the level of innovation in diatom genomes. Recent comparative analyses (Basu et al., 2017) have provided the most comprehensive insights into the role of gene family expansions in the evolution of diatom genomes. In generally, diatoms contain significantly more genes encoding high mobility group boxes, heat-shock transcription factors, and cyclins compared to other phototrophs (Bowler et al., 2008; Basu et al., 2017). Species-specific gene family expansions have also been detected. For example, the expansion of ice binding protein family genes in F. cylindrus is thought to help these diatoms cope with frequent freezing in polar oceans (Mock et al., 2017). Whole-genome duplication appears to play a role in diatom evolution, diversification, and adaptation, especially in Thalassiosirales and several pennate diatoms, such as those in the genus Gyrosigma (Parks et al., 2018). Genome duplication also plays a significant role in plant evolution, but not in the evolution of green algae, as they appear to be haploid or diploid depending on their life cycle stage (Qiao et al., 2019). The mechanisms inducing whole-genome duplication are less clear. The pennate diatom F. solaris appears to be allodiploid, which may be a common mode of polyploidization based on hybridization events in distant parental lineages (Tanaka et al., 2015b). However, for other pennate lineages, low hybrid viability is commonly observed, which minimizes the evolutionary success of allopolyploidy (Amato and Orsini, 2015). Much higher hybrid viability is seen with autopolyploidy, which is often caused by meiotic nonreduction (Mann, 1994; von Dassow et al., 2008). However, there is insufficient genomic evidence to reliably assess the ploidy levels in different diatom species. We currently know little about the occurrence and potential significance of aneuploidy in diatoms. If it occurs in addition to polyploidy, it could potentially generate exceptional allelic diversity, which could be exploited experimentally to test the potential fitness effects of mutations. To reveal the significance of any type of ploidy, it will be crucial to investigate how the different alleles are expressed and if specific conditions drive the evolution of ploidy. The recent sequencing of the F. cylindrus genome showed that ∼25% of genetic loci have highly divergent alleles, which are differentially expressed under various environmental conditions (Mock et al., 2017). Alleles with the highest ratios of nonsynonymous to synonymous substitutions showed the most pronounced levels of differential expression, which suggests they resulted from adaptive evolution. Results from recent genome projects with several Picochlorum (Chlorophyta) species (Foflonker et al., 2018) suggest that significant haplotype divergence is not restricted to polar diatoms. It may therefore have evolved independently in some algal lineages as a mechanism to adapt to variable environments.
The mechanisms controlling gene expression in diatoms are still largely unknown. Although diverse transcription factor (TF) families have been identified from diatom gene sequence analysis (Rayko et al., 2010), to date, only a few studies have confirmed TF activity and TF DNA binding sites in diatoms (Banerjee et al., 2016; Matthijs et al., 2017). More generally, cis- and trans-acting regulatory elements controlling gene expression are still largely unknown in diatoms due to poor sequence conservation. Some novel information is currently emerging from computational analyses of large transcriptome data sets (Ashworth et al., 2016).
In plants and green algae, epigenetic mechanisms regulating gene expression, including DNA methylation and interfering small non-coding RNAs (sRNAs), significantly contribute to acclimation and evolution (Law and Jacobsen, 2010; Lopez et al., 2015; Kronholm et al., 2017). Although diatom epigenetics is still in its infancy, there is preliminary evidence that cytosine methylation contributes to the regulation of gene expression and silencing of repetitive sequences such as TEs (Tirichine et al., 2017). Canonical microRNAs (miRNAs) that control gene expression in plants (Law and Jacobsen, 2010) and the green alga Chlamydomonas (Molnár et al., 2007) have not been found in diatoms (Lopez-Gomollon et al., 2014; Rogato et al., 2014). However, a large and diverse group of canonical miRNAs has been discovered in the multicellular brown alga E. siliculosus (Tarver et al., 2015), suggesting that miRNAs independently evolved in different heterokont lineages. However, various classes of 21- to 30-nucleotide sRNAs have been characterized in both P. tricornutum and T. pseudonana (Rogato et al., 2014; Lopez-Gomollon et al., 2014). These include known noncoding RNA classes such as very abundant tRNA-derived sRNAs, splicing sRNAs (e.g., U2 small nuclear RNA), and highly expressed transcripts of undefined origin. In P. tricornutum, most sRNAs are 25 to 30 nucleotides long and map to repetitive and silenced TEs marked by DNA methylation. This suggests that epigenetic mechanisms drive RNA-dependent DNA methylation of TEs, as observed in plants and metazoans. However, unlike in plants, sRNAs in diatoms target DNA methylated protein-coding genes (Rogato et al., 2014). A multitude of long intergenic noncoding RNAs have also been identified in P. tricornutum that respond to phosphorus depletion (Cruz de Carvalho et al., 2016).
The mechanisms underpinning the biogenesis and regulation of noncoding RNAs and the dynamic control of chromatin marks in diatoms are still largely unknown. Studies aimed at characterizing key components of plant epigenetic pathways identified in diatom genomes such as the Dicer-, Argonaute-, and Polycomb-based systems (De Riso et al. 2009; Tirichine et al., 2017) should help to reveal how epigenetics influences the adaptation and evolution of diatoms in their aquatic environment.
Photosynthesis and Energy Production
Diatoms usually dominate algal blooms (Boyd et al., 2007), and their photosynthesis is highly efficient under dynamic light regimes (Wagner et al., 2006). Although the general biochemical structures and functions of the complexes involved in oxygenic photosynthesis are conserved in most phototrophs, important differences have been identified in diatoms, including differences in pigment composition, the spatial organization of photosynthetic complexes, the response to high-light conditions, and the integration of photosynthetic activity with cellular metabolism. We are only starting to discover unique features of the photosynthetic process in diatoms in terms of how they acclimate to the dynamic physicochemical environment of aquatic systems. On the other hand, the shared photosynthetic features between diatoms and plants or green algae tell us about the universal constraints that apply to the photosynthetic electron transfer chain.
Due to their origins from secondary endosymbiosis (Cavalier-Smith, 2003), diatom plastids are enclosed by four membranes, unlike plastids in the green lineage, which are surrounded by only two membranes. The outermost chloroplast membrane in diatoms is connected to the ER (Figure 4). Diatom plastids contain stacks of three thylakoids that run through the entire organelle. Immunolocalization, three-dimensional plastid reconstruction, and in vivo spectroscopy in P. tricornutum have revealed the segregation of PSI and PSII: PSI is found mostly within the peripheral membranes facing the stroma, and PSII is located within the core membranes at the junction between two thylakoid membranes (Pyszniak and Gibbs, 1992; Flori et al., 2017). This structural organization, in contrast to that found in plants (with grana stacks interspaced by stroma lamellae), has important implications for how light capture and photosynthetic electron flow are regulated in these microalgae. The distance between the two photosystems is short, as they face each other in the two external thylakoids, and there are physical connections between thylakoids (Flori et al., 2017). Therefore, the diffusion of mobile electron carriers from PSII to PSI does not limit photosynthetic electron flow even under high-light irradiance, which is not the case in plants (Kirchhoff et al., 2004). This unusual segregation of the two photosystems prevents the loss of efficiency of the photosystems via possible spillover of excitons from PSII to PSI (Flori et al., 2017), as found in cyanobacteria and red algae (Biggins and Bruce, 1989). This organization might also explain why state transitions, which are observed in the green lineage when light harvesting complexes move from PSII to PSI, are absent in diatoms (Owens, 1986).
In terms of pigments, diatoms use chlorophyll (Chl) c and Chl a, and the main carotenoids are β-carotene, fucoxanthins (Fx), and diadinoxanthin/diatoxanthin. The fucoxanthin Chl a/c binding protein (FCP), which belongs to the family of the Light Harvesting Complex (LHC) proteins, binds seven Chl a, two Chl c, seven Fx, and likely one diadinoxanthin within the protein scaffold (Wang et al., 2019). FCPs are divided into three groups (Gundermann and Büchel, 2014): LHCF (the main light-harvesting antenna), LHCR (specific to PSI), and LHCX (involved in photoprotection, see below). A specific member of the FCP family (LHCF15) is involved in acclimation to far-red light conditions, at least in P. tricornutum (Herbstová et al., 2015, 2017). PSII-FCP forms a homodimer, with two FCP homotetramers and three FCP monomers per monomer, forming a complicated protein-pigment network that is different from its counterpart in the green lineage (Nagao et al., 2019; Pi et al., 2019). The structural arrangement between pigments in FCPs ensures efficient energy transfer from Fx to Chl, optimizing the use of the blue-green light spectrum, which is enriched in the aquatic environment and absorbed by Fx (Wang et al., 2019). This arrangement might also allow for the rapid conversion to a dissipative state under high light stress.
Indeed, diatoms continuously cope with highly dynamic light environments and the risk of PSII inactivation under excessive light. Diatoms respond to PSII photoinactivation by using a PSII repair cycle mediated by FtsH protease complexes (Campbell et al., 2013). Diatoms accumulate significant pools of PSII repair cycle intermediates (Wu et al., 2012) even in darkness (Li et al., 2017). In addition, diatoms can rapidly adjust PSII activity to light conditions by dissipating the energy from excess photons as heat (Lavaud et al., 2002; Wilhelm et al., 2006; Lepetit et al., 2012). This process is usually visualized in vivo as a quenching of Chl a fluorescence, hereafter called nonphotochemical quenching (NPQ), which is up to five times more efficient in diatoms than in land plants (Ruban et al., 2004). Less is known about the regulation of NPQ in diatoms than in green algae and land plants, but there have been some interesting findings (Goss and Lepetit, 2015). NPQ capacity in diatoms is dependent on a reversible xanthophyll cycle, which converts diadinoxanthin to diatoxanthin under high light in a single de-epoxidation step, as opposed to the two steps in plants (Hager and Stransky, 1970; Stransky and Hager, 1970). Specialized members of the LHC family, LHCX proteins, are required for NPQ in diatoms (Bailleul et al., 2010; Lepetit et al., 2017; Buck et al., 2019), as demonstrated for the PsbS protein in Arabidopsis (Li et al., 2000) and LHCSR (light harvesting complex stress related) proteins in Chlamydomonas (Peers et al., 2009). LHCX1 is essential for responses to high-light conditions (Bailleul et al., 2010; Buck et al., 2019), acting as a modulator of NPQ, and it contributes to the phenotypic plasticity in diatoms underpinning adaptive evolution (Bailleul et al., 2010). The constitutive expression of LHCX1 enables a high basal NPQ capacity in P. tricornutum, as also observed in T. pseudonana and C. cryptica/meneghiniana (Zhu and Green, 2010). In addition to LHCX1, diatoms possess high-light-induced LHCX2 and LHCX3 proteins, which, like Chlamydomonas LHCSRs (Peers et al., 2009), help regulate NPQ under high-light stress (Buck et al., 2019). The quenching of Chl fluorescence (the dissipation of excess energy in the form of heat) is thought to occur in two different locations within PSII (Chukhutsina et al., 2014; Kuzminov and Gorbunov, 2016; Taddei et al., 2018). The various LHCX proteins might contribute differently to these two quenching sites. Diatom LHCX family members are differentially expressed in response to nutrient stress, and more intriguingly, under prolonged periods of darkness (Taddei et al., 2016; Lepetit et al., 2017). Thus, perhaps the expansion of the LHCX family, especially in the polar diatom F. cylindrus (containing 11 LHCXs; Mock et al., 2017), reflects functional diversification, which could help diatoms respond to variable environmental conditions, such as strong seasonal changes (Taddei et al., 2016). The pioneering work on diatom LHCXs lays the foundation for more general studies investigating the roles of these proteins in the ecophysiological adaptation of marine microalgae to dynamic environments, as LHCX or LHCX-like genes have been identified in other clades with secondary plastids, such as haptophytes and alveolates (Giovagnetti and Ruban, 2018).
Light-dependent photosynthetic reactions lead to the biosynthesis of ATP and NADPH at a ratio lower than that required for C-fixation, resulting in an ATP deficit. In the green lineage, this deficit is mostly overcome by the production of additional ATP through cyclic electron transport around PSI (Allen, 2002). Diatoms cope with this deficit by “meta-cyclic” electron flow, where the ATP/NADPH ratio is increased by two processes: (1) the export of NADPH to mitochondria and (2) the subsequent import of ATP from mitochondria to plastids (Bailleul et al., 2015). While exchanges of NADPH and ATP between the two organelles have also been observed in the green lineage (e.g., Lemaire et al., 1988; Yoshida et al., 2007; Joliot and Joliot 2008), the observation that this process is constitutive in diatoms is unprecedented. One recent hypothesis suggests that this energetic coupling involves a strong functional connection between mitochondrial glycolysis, an unusual metabolic configuration in stramenopiles (Liaud et al., 2000; Kroth et al., 2008; Río Bártulos et al., 2018), and the Calvin-Benson-Bassham cycle (Smith et al., 2016). Additional characterization of the mitochondrial alternative oxidase (AOX) knock-down lines also showed an hypersensitivity to stress, indicating that the coupling between the two organelles is essential for diatom sensitivity to changing environments (Murik et al., 2018). Regardless of its exact mechanism, the coupling between mitochondria and plastids requires the displacement of metabolites between the two organelles. How this exchange takes place despite the presence of four membranes surrounding the plastid remains an enigma.
Another fundamental issue is the role of luminal pH in regulating photosynthetic electron flow in diatoms. In the green lineage, the proton concentration in the lumen is a master regulator that adjusts photosynthetic electron transfer based on the availability of light and metabolic requirements. An acidic lumen can slow electron transport at the cytochrome b6f level through a process called photosynthetic control (Foyer et al., 1990). In the green lineage, luminal pH also controls PSII activity by regulating the two main actors of NPQ: the xanthophyll cycle, which regulates the concentration of the quencher diatoxanthin, and PSII subunits PsbS and LHCSR, whose protonation induce conformational changes (Horton, 2012). In diatoms like in plants, de-epoxidase activity (and therefore NPQ) is modulated by the proton gradient (Caron et al., 1987). But a role for pH-induced conformational changes to PSII subunits in NPQ is unlikely given the linear relationship between diatoxanthin and NPQ (Lavaud et al., 2002; Goss et al., 2006; Barnett et al., 2015).
The biogenesis of the diatom photosynthetic apparatus is yet to be unexplored. The proteins of the photosynthetic apparatus are composed of subunits encoded in the plastid and nucleus, like those of the green lineage. It is well established that tight cooperation between the nuclear and chloroplast genetic systems is critical for the stoichiometric biogenesis and complex assembly of the photosynthetic machinery in green algae and plants (Woodson and Chory, 2008; Choquet and Wollman, 2009). No such information is currently available for secondary endosymbionts such as diatoms. Genomic analysis has suggested that plastid-targeting systems function in the delivery of nuclear-encoded plastid proteins in diatoms, first through the ER and then across the four plastid membranes (Gruber et al., 2015). Plastid proteins are transported from the ER to the periplastidial compartment via an ER-associated protein degradation-derived translocon (Hempel et al., 2009) and to the stroma via systems similar to that of the Toc and Tic plant translocon (reviewed in Gruber and Kroth, 2017; Maier et al., 2015). Plastid-targeted proteins possess N-terminal bipartite presequences consisting of a signal peptide and transit peptide, with a conserved ASAFAP motif at the signal peptide cleavage site (Table 2; Gruber et al., 2015), which are relatively well conserved in diatoms compared to the corresponding plant presequences. However, our knowledge of the mature N-terminal sequences of these plastid-targeted proteins is far less advanced (Huesgen et al., 2013). The protein import mechanisms to different plastid compartments are still largely undefined, and novel systems regulating the photosynthetic process are expected to be found.
Table 2. Centralized Genome-Enabled Resources and Databases for T. pseudonana and P. tricornutum.
We propose two research directions that might advance our understanding of diatom photosynthesis. First, setting up appropriate genetic resources in diatom strains that can be grown both autotrophically and heterotrophically, such as C. cryptica and Cylindrotheca fusiformis, could be a powerful way to dissect plastid regulation, as has been achieved in C. reinhardtii (Salomé and Merchant, 2019). Genetic investigations into photosynthetic processes have been somewhat limited to the obligate phototrophy of P. tricornutum and T. pseudonana, where photosynthetic mutants are lethal, so characterizing them is unmanageable. Second, the increasing amount of genomic information from diverse diatom species and from other phytoplankton can be used to identify proteins associated with photosynthetic functions in these organisms (Dorrell et al., 2017). Inspired by the GreenCut inventory that used the Chlamydomonas genome as a reference to identify proteins conserved in green lineage organisms (Merchant et al., 2007), a catalog using diatoms as the reference could be exploited to reconstruct the plastid proteomes of stramenopiles. This would in turn help us identify regulators of diatom photosynthesis, thereby extending the current view of photosynthetic constraints in eukaryotes.
Nutrient Metabolism
Nutrient uptake and assimilation by prominent primary producers such as diatoms are particularly relevant because they influence the entire marine food web and global biogeochemical cycling of key elements such as carbon and nitrogen (Falkowski, 2015). Furthermore, due to their silicified cell walls, diatoms are also major players in the silicon cycle, and silicon is one of the most abundant elements in the Earth’s crust. Despite its relevance, the regulation of silica metabolism is still largely uncharacterized. Diatoms adapt to changes in silicic acid availability in part by altering the expression of genes encoding silicon transporter proteins (Hildebrand et al., 1997; Thamatrakoln and Hildebrand, 2008). Novel putative regulators of this important process identified by whole-genome expression profiling of T. pseudonana (Supplemental Table 2). Common sets of genes are induced by both silicate and nutrient limitation (Supplemental Table 2; Mock et al., 2008), suggesting that the pathways involved in cell cycle regulation, silica deposition, and nutrient metabolism are strongly coupled. In this section, we focus on the regulation of carbon, nitrogen, and iron metabolism, as recent studies on these topics have helped unveil some peculiar adaptations of diatoms to the marine environment (Figure 4).
Carbon Fixation
Like other aquatic phototrophs, diatoms must overcome the problem of low CO2 concentrations in water (10 to 30 μM); these concentrations are much lower than that required to saturate the CO2-fixing enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) in diatoms (Reinfelder, 2011). Physiological and molecular investigations have shown that the carbon concentration mechanism (CCM) in diatoms is critical for the efficient uptake and assimilation of inorganic Carbon (Ci; both CO2 and HCO3−) from the environment (see Hopkinson et al., 2011; Tsuji et al., 2017; Schoefs et al., 2017, for a detailed overview on this topic; and Figure 4). Except for some highly conserved proteins, the CCM in diatoms is different from that of cyanobacteria and green algae (Wang et al., 2015; Kaplan, 2017; Tomar et al., 2017), supporting the hypothesis that the many proteins and metabolic pathways of the CCM independently evolved in different lineages of unicellular photosynthetic organisms. Diatoms do not contain homologs of Ci transporters from green algae (e.g., the Chlamydomonas LCI1 and HLA3; Ohnishi et al., 2010; Yamano et al., 2015). However, 10 putative HCO3− transporters phylogenetically related to those from metazoans are encoded in the genomes of model diatom species, including the solute carrier SLC4, a major plasmalemma bicarbonate transporter of functional relevance under low-CO2 conditions (Nakajima et al., 2013; Shen et al., 2017). The existence of additional plastid-localized HCO3− transporters has been hypothesized based on the CCM model (Hopkinson, 2014), but no such transporters have thus far been characterized.
Like other photosynthetic organisms, diatoms possess multiple carbonic anhydrase (CA) subclasses that convert (1) HCO3− to CO2 at the cell surface to facilitate diffusive uptake, (2) CO2 to HCO3− in the cytosol to limit CO2 diffusion out of the cells and to facilitate its transport to the plastid, and (3) HCO3− in the stroma to CO2 close to Rubisco in the pyrenoid. CA variants have also been found in some diatom species (Matsuda et al., 2017; Young and Hopkinson, 2017; e.g., Figure 4 for P. tricornutum and T. pseudonana), an indication of how the CCM process has further diversified in these microalgae. The θ-type CA in P. tricornutum (Kikutani et al., 2016), which is targeted to the lumen of the pyrenoid-penetrating thylakoid, is essential for efficient photosynthesis and growth. Interestingly, in green algae, the final conversion of HCO3− to CO2 by the α-type CAH3 CA also occurs within the thylakoids that penetrate the pyrenoid (Mitra et al., 2004), suggesting that the tight association between CCM in the pyrenoid and thylakoid membrane function was a driving force in the independent evolution of photosynthesis in distant aquatic phototrophs. New CA metallovariants that use cadmium or cobalt as cofactors, rather than the commonly used zinc, have also been described in diatoms (Lane et al., 2005; Lionetto et al., 2016). The ability to use different metal cofactors may facilitate diatom growth in the ocean, where large areas contain low levels of trace metals. This hypothesis is supported by the recent discovery of the diatom ιCA that uses manganese as a cofactor (Jensen et al., 2019). The sequence of this CA variant shares only low identity with known CAs, but its ecological relevance is supported by its wide distribution in diatoms and other phytoplanktonic algae, bacteria, and archaea and its prevalence in metagenomic samples from marine environments (Jensen et al., 2019).
Overall, comparative analysis of diatom genomes has revealed that diatoms have more genes that take part in C metabolism than green algae (Kroth et al., 2008; Smith et al., 2012). Moreover, diatom species differ considerably in terms of the intracellular localization, gene duplication, or deletion of some enzymes of the C-partitioning pathway (e.g., some pyruvate hub enzymes; Smith et al., 2012). It is likely that these enzymes acquired specialized metabolic functions in different species as diatoms diversified. Diatoms also contain genes that are potentially involved in C4-like photosynthesis (Kroth et al., 2008). Well described in land and aquatic plants, this pathway favors the formation of organic molecules with four C atoms, which increase local CO2 concentrations when decarboxylated near Rubisco. The existence of the C4 pathway in diatoms was originally proposed in Thalassiosira weissflogii (Reinfelder et al., 2000) and then in T. pseudonana (Kustka et al., 2014) based on physiological and biochemical analyses, but functional characterization of the putative C4 genes does not support these hypotheses. The silencing of the essential pyruvate-orthophosphate dikinase (PPDK) gene of C4 metabolism (Haimovich-Dayan et al., 2013) did not alter Ci acquisition in P. tricornutum. However, perhaps this pathway aids in the dissipation of excess energy and pH homeostasis. The intracellular localization of all putative C4-like proteins in P. tricornutum (Ewe et al., 2018) and T. pseudonana (Tanaka et al., 2014) is quite different from the distribution of the core C4 pathway enzymes in plants. In particular, the lack of decarboxylases in the diatom plastid would prevent a functional C4 pathway from operating as a CCM. The roles of diatom C4-like genes and many other genes putatively involved in C acquisition and metabolism are still poorly understood. We anticipate that their functional characterization in different model species will reveal novel regulatory processes that explain how diatoms became such successful primary producers in the contemporary ocean.
Nitrogen Assimilation
Diatoms quickly assimilate NO3−, the most stable, abundant ionic form of inorganic N in the ocean (Garcia et al., 2010). They also quickly assimilate NO2, NH4+, and organic forms of N such as urea and diverse amino acids, making them highly competitive in nitrate-rich environments such as coastal upwelling systems and the Southern Ocean.
Diatoms contain many genes encoding N transporters, including high- and low-affinity nitrate (NRT2/NPF) and ammonium (AMT) transporters and a urea transporter that is not found in green algae or plants (Rogato et al., 2015). Meta-omics analysis has unveiled complex genetic diversification of these transporters, which is likely shaped by environmental N abundance (Busseni et al., 2019). Recent characterization of plasma membrane-type aquaporins (PtAQP1 and PtAQP2) from P. tricornutum also defined novel CO2/NH3 channels (Figures 4A and 4B) (Matsui et al., 2018), which control the cell’s permeability to these nutrients. PtAQP1 and PtAQP2 also participate in regulating NPQ under high light, likely mitigating the potentially toxic effects of high concentrations of ammonia on the photosynthetic apparatus by facilitating NH3 efflux.
As observed in other algal lineages (Breuer et al., 2012; Merchant et al., 2012), N starvation triggers intracellular N recycling in diatoms and redirects C from storage carbohydrates to C-rich compounds such as lipids. A negative effect of N depletion is the degradation of pigments, which is detrimental to photosynthetic capacity, C fixation, and ultimately cell division and growth (Kolber et al., 1988; Allen et al., 2008; Levitan et al., 2015a). Transcriptomic, proteomic, and metabolic profiling of diatoms responding to N limitation revealed an extremely pronounced remodeling of lipid metabolism compared to other phototrophs (Supplemental Table 2). Functional characterization of key regulators of N metabolism such as the nitrate reductase gene (NR) helped confirm this finding. Initial analysis of P. tricornutum NR knockdown lines revealed that under N starvation, 40% more C is redirected toward the biosynthesis of storage triacylglycerols in the mutants compared to wild-type lines (Levitan et al., 2015b), suggesting that these important metabolic changes are regulated by the N assimilation pathway. NR knockout lines cannot grow in medium with NO3− as the sole N source because N assimilation is abolished (McCarthy et al., 2017), but they continue to import NO3− despite growth arrest, leading to NO3− accumulation inside the vacuole, likely via the action of recently identified vacuolar NO3− transporters (McCarthy et al., 2017). NO3− accumulation in the vacuole is thought to help diatoms cope with periods of nitrate scarcity and even suboptimal light conditions.
The identification of a metazoan-type ornithine-urea cycle in diatoms, previously only known for its role in N excretion, is one of the most surprising discoveries from diatom genomic analysis (Armbrust et al., 2004). The study of one of these proteins, carbamoyl phosphate synthase III (unCPS; Allen et al., 2011), which is targeted to the mitochondria in P. tricornutum, has elucidated some of the functions of the urea cycle in these algae. Metabolic analysis of CPSIII knockdown cell lines showed that intermediate products of the urea cycle are particularly depleted in these lines. In particular, CPSIII regulates the incorporation of NH4+ and HCO3− into carbamoyl phosphate, which is subsequently converted into other urea cycle intermediates and products, such as Arg and the signaling molecule nitric oxide (NO). Down-regulation of CPSIII also impairs the response of N-limited diatoms to N addition, suggesting that the urea cycle contributes to the rapid recovery from prolonged N limitation. It has therefore been proposed that the urea cycle plays a key role in controlling and redistributing C and N fluxes into the cell based on N availability. This pathway is strongly linked to the tricarboxylic acid (TCA) and glutamine synthase/glutamate synthase cycles due to the shared pools of precursor metabolites (Allen et al., 2011). Since their discovery in diatoms, CPS genes have been found in multicellular brown algae and haptophytes (Allen et al., 2011), supporting the hypothesis that a urea cycle was inherited from the common exosymbiont of the cryptophyte, alveolate, stramenopile, and haptophyte lineage. In diatoms, CPSIII down-regulation also affects the biosynthesis of the major osmolyte Pro and the precursors of long-chain polyamines required for cell wall formation (Kröger et al., 2000). Interestingly, some of these metabolites are generated from ornithine-urea cycle intermediates by the products of genes of bacterial origin. From an evolutionary perspective, the coupling of proteins derived from bacteria and exosymbionts likely allowed the functionality of the urea cycle to expand in diatoms toward processes that are critical for their growth and physiological acclimation to variable environments (Allen et al., 2011). It is important to mention that homologs of the ornithine-urea cycle such as CPS and Orn carbamoyltransferase are also found in green algae and plants. However, these proteins are localized to the plastid as part of the Arg biosynthesis pathway and are phylogenetically distinct from the diatom enzymes (Winter et al., 2015).
Iron
Iron, which functions as an electron carrier in iron-sulfur proteins and hemoproteins, plays essential roles in photosynthesis (PSI, ferredoxin, cytochrome b6f) and other metabolic processes (TCA cycle, respiration, nutrient assimilation, and so on) in all phototrophs. In marine ecosystems, iron is the prime limiting factor for phytoplankton growth, as large phytoplankton blooms have been induced in high-nutrient low-chlorophyll regions in several iron fertilization experiments (Boyd et al., 2007). Diatoms often dominated these blooms, suggesting they have efficient strategies for iron uptake and a subsequent growth response that enable them to outcompete other algal groups.
Different coastal and oceanic diatom species employ a wide variety of physiological strategies to deal with iron availability. Diatoms possess diverse high-affinity reductive uptake systems, and like vascular plants, they can reduce ferric iron, the predominant form of iron in the environment, before or during uptake into the cell (Allen et al., 2008; Marchetti et al., 2009; Groussman et al., 2015). Many diatom species contain high-affinity ferric reductases (FREs) that dissociate iron III from ligands, multicopper oxidases that oxidize Fe2+ to Fe3+, and permease (FTR) systems that receive iron (II) for translocation across membranes (Figure 4B; Groussman et al., 2015), as observed in green algae such as Chlamydomonas (Glaesener et al., 2013). Several species can also take up ferric iron without reducing it. The direct intracellular uptake of siderophores was recently demonstrated in several different diatom species (Kazamia et al., 2018), as Lesuisse et al. (1998) and Chu et al. (2010) previously described in bacteria and fungi. In P. tricornutum, siderophores are taken up by interacting with the cell surface, followed by endocytosis and delivery to the plastid. This siderophore uptake system is highly efficient, with an affinity thought to be higher than for any other eukaryotes (Kazamia et al., 2018). Therefore, it is likely that this system strongly contributes to the assimilation of iron, especially in regions where iron is very scarce.
Once iron enters the cytosol of diatoms, it is allocated to iron-based metabolism, but its intracellular content must be tightly regulated because free iron can trigger the production of harmful radical oxygen species (ROS). ROS can irreversibly damage essential Fe-S-containing proteins such as those that function in photosynthesis. Ferritin is thought to be crucial for maintaining iron homeostasis in diatoms (Groussman et al., 2015) by sequestering iron and providing an efficient iron storage system. For instance, ferritin in the pennate diatom Pseudo-nitzschia granii was thought to confer a competitive advantage over non-ferritin-containing algae, as the stored iron in P. granii corresponded with a prolonged growth phase and extended diatom blooms (Marchetti et al., 2009). However, recent data indicate that diatom ferritin is optimized for initial Fe2+ oxidation, pointing to an additional role for this protein in buffering iron availability (Pfaffen et al., 2015).
When iron is scarce in the environment, diatoms adjust their metabolism by reducing their cellular requirements for iron. For instance, as observed in most cyanobacteria and other algae (Pierella Karlusich and Carrillo, 2017), diatoms can replace the plastid-localized ferredoxin with flavodoxin, as the latter uses flavin as a cofactor instead of iron (La Roche et al., 1996; Groussman et al., 2015). Unlike most diatoms, green algae also possess other interchangeable soluble electron carriers: the copper-containing plastocyanin, which is abundant in copper-supplemented medium and remains expressed under iron limitation; and the iron-cytochrome c6, which replaces plastocyanin under copper deficiency (Merchant and Bogorad, 1986). So far, the plastocyanin has been reported only in T. oceanica (Peers and Price, 2006), which appears to have adapted to low-iron environments. The constitutive expression of plastocyanin observed in this species may reduce the requirement for iron, which is less abundant than copper in the open ocean.
Novel candidate proteins specific to diatoms and other phytoplanktonic organisms have been put forward as regulators of acclimation to iron limitation. These include a protein previously annotated as “death-specific”, which regulates photosynthesis under limiting iron and light conditions (Thamatrakoln et al., 2013), and the iron-starvation-induced proteins (ISIPs), whose expression is induced by iron limitation (Allen et al., 2008). ISIP1 was originally identified as a putative iron receptor on the cell surface (Lommer et al., 2012), but more recent studies in P. tricornutum showed that ISIP1 plays a key role in the endocytic uptake of siderophores (Kazamia et al., 2018). Functional characterization of ISIP2a revealed its influence on ferric-iron concentration and high-affinity uptake (Morrissey et al., 2015). ISIP2a is an algal transferrin-like proteins named phytotransferrin that is functionally equivalent to its human homologue (McQuaid et al., 2018), as iron uptake was recovered in a P. tricornutum ISIP2a mutant complemented with the human transferrin gene. These findings, together with the characterization of a transferritin-like protein in the green alga Dunaliella salina (Fisher et al., 1997), highlight the importance of this iron acquisition mechanism for a wide variety of microalgae in aquatic environments (Morrissey et al., 2015; McQuaid et al., 2018). Moreover, the discovery that carbonate ions control iron uptake by ISIP2 uncovered a relationship between carbonate availability and iron uptake in diatoms. Regulators of iron metabolism can therefore be used as molecular markers for assessing iron status in natural diatom populations. For example, ISIP proteins are highly expressed in iron-limited oceanic regions (Bertrand et al., 2015; Carradec et al., 2018), which is consistent with the results of laboratory-based experiments examining iron uptake (Kazamia et al., 2018). Furthermore, the wide occurrence of carbonate-sensitive phytotransferrin sequences in metagenomic data sets suggests that this system is of ecological importance and raises the possibility that ocean acidification has a negative impact on iron uptake by diatoms (McQuaid et al., 2018).
Perception and Responses to Environmental Signals
Massive diatom blooms in the marine environment are controlled by light and nutrient availability, as well as interactions with other organisms, including partners, competitors, or predators (Brodie et al., 2017). Studies of diatom responses to specific abiotic and biotic signals or stresses are revealing how diatoms acclimate to complex environments and survive adverse conditions.
Diatoms possess a palette of photoreceptors that use information on the changing light spectrum and intensity to modulate critical photophysiological responses (Jaubert et al., 2017). As expected from the abundance of blue light wavelengths through the entire water column, diatoms have a multitude of blue light sensors (Jaubert et al., 2017). The animal-like protein PtCPF1, a member of the expanded cryptochrome/photolyase family (CPF) in P. tricornutum, shows both blue light-dependent transcriptional regulation and DNA repair activity (Coesel et al., 2009). This double function is thought to allow PtCPF1 to regulate different cell cycle phases, which are strongly synchronized with periodic light/dark variations. Following their discovery in diatoms, bifunctional cryptochromes have been also found in green algae (Heijde et al., 2010; Franz et al., 2018). Diatoms also possess a protein distantly related to plant-type cryptochrome (CryP) that regulates transcript abundance in the light as well as in darkness (Juhas et al., 2014; König et al., 2017). Diatoms do not possess blue-light-absorbing phototropins, which control responses such as phototropism and chloroplast movement in plants (Briggs, 2014) and photoprotection in green algae (Petroutsos et al., 2016). However, multiple aureochromes (AUREOs) have been found in diatoms. AUREOs are light-induced TFs specific to stramenopiles (Kroth et al., 2017). Like phototropins, AUREOs contain a light-oxygen-voltage-sensing domain for blue light perception in a novel combination with a b-ZIP-domain responsible for DNA binding. PtAUREO1a functions synergistically with the basic leucine zipper TF bZIP10 to control the expression dsCYC2, thus rendering the regulation of cell cycle progression dependent on blue light (Huysman et al., 2013). Both PtAUREO1a and AUREO1b contribute to photoacclimation in diatoms, as their corresponding knockout mutants show less photosynthetic activity than the wild type (Mann et al., 2017). The NPQ process was only modified in the AUREO1a mutant, indicating that these gene family members have diversified functions.
Rhodopsins (RHOs), a diverse group of light-dependent reactive membrane proteins with retinal as the chromophore, have been identified in several diatom species. The first diatom RHO gene discovered was found in F. cylindrus (Strauss, 2012). Preliminary data suggest that RHOs from F. cylindrus and P. granii are likely xanthorhodopsins acquired from bacteria via HGT. Xanthorhodopsins are light-driven proton pumps that generate a proton motive force for ATP biosynthesis (Strauss, 2012). F. cylindrus RHO is targeted to the plastid (Strauss, 2012), suggesting it contributes to ATP biosynthesis in this organelle. Alternatively, the proton motive force generated by RHOs might enhance iron uptake when iron is limiting. This might explain why RHO transcripts are abundant in the metatranscriptomes of diatom communities from iron-limited ocean surfaces (Marchetti et al., 2012).
Light sensing in diatoms is further augmented by bacterial-like phytochrome photoreceptors. Diatom phytochromes (DPHs) absorb longer red and far-red wavebands than plants, green algae, and glaucophyte algae (Rockwell et al., 2014) and can trigger far-red light signaling (Fortunato et al., 2016). These solar radiation wavelengths are quickly absorbed in the water column, and DPH might act as ocean surface light sensors. However, since DPH genes have been found in the genomes of many diatom species and in metatranscriptomic data from subsurface layer samples (M.J. and A.F., unpublished data), their roles in light sensing in deeper waters cannot be excluded. Most far-red-light-induced genes regulated by DPH encode proteins of unknown function. Characterizing these genes and proteins should reveal novel far-red-light-sensing processes and their importance for diatom biology.
Diatoms also experience the daily light-dark cycle, and basic biological processes such as the cell cycle (Huysman, et al., 2010), gene expression (Ashworth et al., 2013; Chauton et al., 2013; Smith et al., 2016), and pigment biosynthesis (Ragni and d’Alcalà, 2007) show daily oscillations. In P. tricornutum, these rhythms persist under constant light conditions, providing evidence that they are regulated by an endogenous circadian clock (Annunziata et al., 2019). The bHLH-PAS protein RITMO1 was the first regulator of circadian rhythms identified in diatoms, as deregulating its expression strongly altered cellular rhythmicity (Annunziata et al., 2019). No obvious homologs of bacterial, green algae, plant, or animal circadian clock components have thus far been identified in diatoms. Therefore, RITMO1 is a valuable entry point for characterizing circadian regulation in diatoms and in many marine microalgae possessing RITMO1-like proteins.
Diatoms also perceive and respond to changes in nutrient availability, which can occur rapidly in turbulent waters, as evidenced by the massive changes in gene expression that occur under different nutrient stresses (Supplemental Table 2). Based on the important metabolic changes observed in NR mutants compared to wild-type cells under N stress, as mentioned above (Levitan et al., 2015b; McCarthy et al., 2017), NR might function in sensing cellular N fluxes and remodeling intermediate metabolism (Levitan et al., 2015b). Regarding C sensing, CO2 itself might elicit the modifications in cell physiology observed in response to changes in external CO2 levels. CO2 activates a signaling cascade involving cAMP as a second messenger and the TF PtbZIP11 by binding to CO2/cAMP-responsive elements in CO2-regulated genes such as PtCA1 and PtCA2. Interestingly, light-driven signals and metabolic signals act on the same pathway (Kikutani et al., 2012; Tanaka et al., 2016). Therefore, a complex interconnection of regulatory networks controls diatom physiology under dynamic environmental conditions (Supplemental Table 2).
Diatoms also respond to intracellular metabolic signals that are strongly dependent on environmental changes. A retrograde plastid-to-nucleus signaling pathway was identified in P. tricornutum by artificially modifying the redox state of the plastoquinone pool (Lepetit et al., 2013). As shown in plants and green algae, plastid signals induce changes in the expression of nuclear genes, such as LHCX genes, and the biosynthesis of photoprotective pigments (Lepetit et al., 2013; Taddei et al., 2016). ROS normally produced by oxygen-based metabolism (photosynthesis, photorespiration, and oxidative phosphorylation) can act as central secondary messengers in diatoms, as in many other organisms including plants and green algae (Mittler et al., 2011). A compartmentalized redox-sensitive signaling network (redoxome) was characterized in P. tricornutum by analyzing the redox proteome and redox-sensitive biosensors localized to different cellular compartments (e.g., roGFP; green fluorescent protein) (Rosenwasser et al., 2014) and by monitoring phenotypic variability within diatom populations at the single-cell level (Mizrachi et al., 2019). When oxidative stress is induced by other stresses (e.g., high light), the redoxome regulates redox metabolism and cell fate decisions toward acclimation or cell death.
The life of diatoms in the ocean is also strongly influenced by surrounding organisms. Some diatoms can assimilate N via symbiosis with N2-fixing cyanobacteria (Foster et al., 2011). As observed in a variety of marine and freshwater microalgae (Croft et al., 2005), some bacteria can provide diatoms with vitamin B12, whose deficiency would otherwise limit growth in certain environments (Bertrand et al., 2015). P. multiseries and diverse co-occurring bacteria interact via complex communication systems (Amin et al., 2015) involving the exchange of hormones, macro and micronutrients, and signaling molecules released by both the diatoms and bacteria. Diatoms and their grazers also interact in an elaborate way. In the presence of copepods, some diatoms produce unsaturated aldehyde oxylipins that inhibit the growth of the predator (Ianora et al., 2004). These aldehydes can act as sensing molecules to control the diatom population itself by activating a signaling cascade involving calcium, NO, and oxidation of the glutathione pool (Vardi et al., 2006; van Creveld et al., 2015). NO production was initially thought to be controlled by a calcium-dependent NO synthase-like protein (NOA; Vardi, 2008), but subsequent studies revealed that NO is likely produced by a nitrite-dependent pathway catalyzed by the side activity of NR. Consequently, NO appears to play a role in a nitrite-sensing and acclimation system (Dolch et al., 2017), as previously demonstrated in Chlamydomonas (Wei et al., 2014). In Chlamydomonas, however, NO is also produced in NR-defective strains by other pathways, such as the polyamine pathway, from Arg by an unknown NO synthase-like pathway or from nitrite by molybdenum cofactor-containing enzymes (De Mia et al., 2019). Considering the interconnectivity of signaling networks in diatoms, it is conceivable that NO produced by different N sources participates in both cell acclimation and cell fate regulation, depending on the environmental context and the physiological state of the cells. A recent study in Skeletonema marinoi supports this scenario. In this species, another pathway activated by intracellular sterol sulfates triggers programmed cell death by inducing ROS and NO production during cell aging (Gallo et al., 2017).
Currently, information about diatom signaling pathways, from stimulus to response, remains largely fragmentary, and many aspects of diatom interaction networks remain to be clarified. None of the signaling components involved in light sensing downstream of photoreceptors of plants and green algae have thus far been identified in diatom genomes. As in other eukaryotes, calcium appears to be a critical second messenger for detecting fluid motion, osmotic stress, iron, and infochemicals in diatoms (Falciatore et al., 2000; Vardi et al., 2006). A Ca2+ signaling mechanism was recently discovered in diatoms when EukCats, a novel class of single-domain voltage-gated channels, was characterized (Helliwell et al., 2019). EukCats, which are widely distributed in eukaryote phytoplankton, are involved in regulating cellular motility in P. tricornutum.
GENOMIC AND GENETIC RESOURCES
Genomic Resources
P. tricornutum and T. pseudonana are the most advanced diatom model species for molecular studies. Strains of both P. tricornutum and T. pseudonana have been isolated from locations worldwide and are curated in various culture collections (Supplemental Figure 1). The ecological relevance of T. pseudonana and its obligate requirement for silica were strong considerations when the CCMP1335 strain was chosen for the first sequencing project of a centric diatom (Armbrust et al., 2004). P. tricornutum Pt1.1 8.6 Bohlin (CCMP2561) was then chosen as a representative diatom of the pennate lineage (Bowler et al., 2008). The availability of many transcriptomic and proteomic data sets has greatly facilitated genome annotation efforts and therefore gene discovery in both species (Table 2; Supplemental Table 2). Resequencing of seven additional T. pseudonana strains (Koester et al., 2018) and 10 P. tricornutum accessions (Supplemental Figure 1; Rastogi et al., 2019) has revealed polymorphisms, providing evidence for genome variation, evolution, and adaptation. The following diatom genome sequences have recently become available, as previously mentioned (Table 1; Supplemental Table 1): F. cylindrus, T. oceanica, P. multistriata, P. multiseries, F. solaris, C. cryptica, and S. costatum. Most diatom genome projects are accompanied by the production of extensive RNA-sequencing data sets and other omics data obtained under a variety of growth conditions relevant to the individual species (see Table 2 and Supplemental Table 2 for P. tricornutum and T. pseudonana). However, a database centralizing all of these omic resources is still lacking.
The availability of genomic information from P. tricornutum has made it possible to begin reconstructing the metabolic networks of diatoms, as done in other models of the green lineage (e.g., Dal’Molin et al., 2011). Diatom metabolism databases (Table 2) include DiatomCyc and the genome scale metabolic model based on improved genome annotation and protein subcellular localization predictions (Fabris et al., 2012; Levering et al., 2016). These resources are instrumental in charting intracellular metabolic fluxes in diatoms grown under different sources of energy and C (Kim et al., 2016) to explore the roles of novel pathways (e.g., lower glycolysis in mitochondria) or to predict the existence of new regulators of energy fluxes between organelles in relation to diatom acclimation (Kim et al., 2016; Broddrick et al., 2019). Although the biotechnological exploitation of diatoms is beyond the scope of this review, it is important to mention that these genomic resources and genetic tools (Tables 1 to 4) also provide the basis for metabolic engineering of these algae (e.g., Daboussi et al., 2014; Kim et al., 2016; Levering et al., 2016).
Large amounts of metagenomics and metatranscriptomics data are now being generated from the sequencing of natural phytoplankton populations (Table 3). These data represent powerful resources for exploring the distribution of diatom genes and species in an environmental context.
Table 3. Large Omics Data for Phytoplankton Including Diatoms.
| Resource | Data Types | Website |
|---|---|---|
| The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP) | 678 transcriptomes from 411 different strains, among which 92 different diatoms | https://www.imicrobe.us/#/projects/104 |
| The Tara Ocean | MetaTranscriptomic MetaGenomic | http://tara-oceans.mio.osupytheas.fr/ocean-gene- atlas/ |
| https://doi.org/10.1594/PANGAEA.875582 | ||
| Sea of Change project | MetaTranscriptomic MetaGenomic | https://genome.jgi.doe.gov/portal/AntOcenscriptome_8_FD/AntOcenscriptome_8_FD.info.html |
Genetic Resources
The breakthrough that made reverse genetics in diatoms possible was the biolistic delivery of transgenes to C. cryptica and Navicula saprophila cells (Dunahay et al., 1995). Many transformation experiments in other diatom species, including P. tricornutum and T. pseudonana (Table 4; Supplemental Table 1), were subsequently performed using the original protocol from 1995 with minor modifications, which gives reproducible results even though the transformation efficiency is relatively low (∼10−6 to 10−8 transformants per μg of DNA). DNA can be delivered to P. tricornutum via electroporation (Table 4), but the transformation efficiency is more variable. A drawback of both approaches is that the transgene is randomly integrated into the genome, with multiple integration events. Chromosomal position effects and variable transgene copy numbers cause significant variability in transgene expression levels in independent lines. Biolistic gene transfer affects genome integrity due to the physical forces that produce the double-strand breaks that are repaired by nonhomologous end joining (Liu et al., 2019). To overcome these issues, new vectors have been designed containing a yeast-derived sequence, which can be replicated as episomes in diatom cells (Karas et al., 2015). These episomes can be delivered to P. tricornutum and T. pseudonana through bacterial conjugation using Escherichia coli (Table 4). This transformation strategy is more efficient than previously described methods (∼10−4 transformants per μg of DNA for P. tricornutum and T. pseudonana) but is less stable, as episomes are eventually lost in the absence of selective pressure. On the positive side, this feature can be exploited to achieve transient expression, which could be particularly useful for potentially deleterious transgenes.
Vectors have also been constructed to express foreign genes in P. tricornutum plastids transformed by electroporation (Table 4). There is also evidence that plastid mutagenesis by homologous recombination is feasible in this species (Table 4).
In parallel, vectors have been designed for high-throughput protein tagging and promoter-reporter/target transgenes to study gene expression patterns and functions (Table 4). In P. tricornutum and T. pseudonana, Lhcf promoters are most commonly used to drive strong expression of the target gene, while the Nitrate reductase promoter allows gene expression to be induced by shifting the transgenic cells from nitrate-deficient to nitrate-sufficient medium. Examples of reporter genes include the beta-glucuronidase gene uidA for monitoring gene expression and GFP variants for subcellular localization of proteins (Table 4). Down-regulation of gene expression has been achieved in P. tricornutum and T. pseudonana by expressing antisense or inverted repeat sequences of target genes (Table 4). This strategy often results in the reduction of mRNA and protein levels (De Riso et al., 2009), but it is not yet known how gene silencing works in diatoms. A recent major achievement was the development of genome-editing tools such as TALEN and CRISPR/Cas9 in P. tricornutum and T. pseudonana (Table 4; Kroth et al., 2018). Nucleotide insertions or deletions resulting from nonhomologous end joining have been described, as well as gene replacement mediated by homology-directed repair in the presence of template DNA (Table 4). More recently, transgene-free editing of the P. tricornutum genome was achieved by delivering Cas9/single guide RNA ribonucleoprotein complexes (Serif et al., 2018). With ribonucleoprotein delivery, endogenous counter-selectable markers have been used to facilitate the identification of mutated genes.
PERSPECTIVES
In the last two decades, major international initiatives such as Marine Genomics Europe, the European Marine Biological Resource Centre, and the Marine Microbiology Initiative have promoted diatom genetics and genomics research. Since diatoms include the most advanced molecular model systems among phytoplankton organisms, previously unknown cellular and metabolic processes have been discovered in these species, broadening our awareness of the diversity of photosynthetic organisms.
Molecular insights into the life of diatoms obtained from studies of established and emerging model species can be now transferred to the ∼100 nonmodel diatom species whose transcriptomes have been sequenced in the framework of the Marine Microbial Eukaryote Transcriptome Sequencing Project (Keeling et al., 2014). Diatom models also provide a reference system for genomic and phylogenomic comparisons (Blaby-Haas and Merchant, 2019) with other microalgae and macroalgae such as E. siliculosus (Cock et al., 2010) of the heterokont clade that are still recalcitrant to genetic investigations.
Many molecular secrets of diatoms remain to be revealed. The unveiling of the chimeric nature of diatom genomes was a step forward, but it prompted the following question: How did regulators derived from different ancestors coevolve and integrate their functionalities to cooperatively fine-tune diatom genome expression and metabolism while responding to environmental changes? Further characterization of genetic and epigenetic processes in diatoms will undoubtedly uncover novel regulatory pathways, their significance for diatom success in contemporary oceans, and their evolutionary past.
The lack of functional information for roughly half of the diatom gene repertoire greatly limits our understanding of their biology, as is the case for many other algae (Blaby-Haas and Merchant, 2019). The discovery of gene products with no known protein domain hints at functional capabilities unique to diatom species that cannot be surmised by comparative genomics. In other model organisms, such as Arabidopsis and Chlamydomonas, forward genetics has been among the most successful strategies to determine gene functions by inducing and hybridizing mutants and carrying out large-scale phenotypic screens. These approaches are precluded in diatoms that do not reproduce sexually in a way that can be controlled in the laboratory. Forward genetics could possibly be established in sexually reproducing diatom species. However, in the coming years, the generation of CRISPR-Cas9 knockout libraries targeting all genes in diatom genomes may allow us to establish loss-of-function screens in any diatom species, paving the way for major breakthroughs.
One of the biggest challenges for the future will be to assess the significance of diatom biology in the marine environment. Being the most advanced phytoplanktonic model species, diatoms offer unique opportunities to address fundamental questions through integrative gene-to-ecosystem approaches (e.g., Guidi et al., 2016; Carradec et al., 2018). Functional investigations of diatom model species have enabled us to identify genes with strong phenotypic impacts that might be targets for selection and therefore drive the evolution and adaptation of diatoms to their highly variable environment. More links between diatom ecology, genomics, and genetics will intensify in the future due to the tangible potential to learn more about the factors shaping phytoplanktonic communities and the capacity of marine ecosystems to tolerate, adapt to, and contribute to a changing climate.
Supplemental Data
Supplemental Figure 1. Map of the globe indicating the geographical origin of T. pseudonana and P. tricornutum isolates used for molecular and cellular investigations.
Supplemental Table 1. Characteristics of the established and emerging model species.
Supplemental Table 2. Available omics data recently generated in T. pseudonana and P. tricornutum under different conditions.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. We thank Alessandro Manzotti and Carole Duchêne for fruitful discussions and support in preparing the figures and Bernard Lepetit, Francis-André Wollman, Chris Bowler, Bianca Sclavi, Mariella Ferrante, and Alessandra Rogato for critical suggestions. Research was supported by funding from the European H2020 EMBRIC (European Marine Biological Research Infrastructure Cluster, grant GA 654008 to A.F.), the European H2020 ASSEMBLE Plus (Association of European Marine Biological Research Laboratories Expanded, grant GA 730984 H2020-INFRAIA-1-2016-2017 to A.F.), the Gordon and Betty Moore Foundation Marine Microbial Initiative (grants GBMF 4966 to A.F. and GBMF 4961 to T.M.), the Fondation Bettencourt-Schueller (Coups d'élan pour la recherche francaise-2018 to A.F.), the grant “DYNAMO” (ANR-11-LABX-0011-01- to A.F. and B.B.), the European Research Council under the EU Horizon 2020 research and innovation program (ERC starting grant PhotoPHYTOMICS, grant 715579- to B.B.), the Natural Environment Research Council (NERC grant NE/R000883/1 to T.M.), and the Leverhulme Trust (grant RPG-2017364 to T.M.). T.M. also acknowledges support from the School of Environmental Sciences, University of East Anglia, Norwich, United Kingdom.
Footnotes
Articles can be viewed without a subscription.
References
- Ahmed S., et al. (2014). A haploid system of sex determination in the brown alga Ectocarpus sp. Curr. Biol. 24: 1945–1957. [DOI] [PubMed] [Google Scholar]
- Allen A.E., Dupont C.L., Oborník M., Horák A., Nunes-Nesi A., McCrow J.P., Zheng H., Johnson D.A., Hu H., Fernie A.R., Bowler C. (2011). Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473: 203–207. [DOI] [PubMed] [Google Scholar]
- Allen A.E., Laroche J., Maheswari U., Lommer M., Schauer N., Lopez P.J., Finazzi G., Fernie A.R., Bowler C. (2008). Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proc. Natl. Acad. Sci. USA 105: 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. (2002). Photosynthesis of ATP-electrons, proton pumps, rotors, and poise. Cell 110: 273–276. [DOI] [PubMed] [Google Scholar]
- Amato A., Orsini L. (2015). Rare interspecific breeding in Pseudo-nitzschia (Bacillariophyceae). Phytotaxa 217: 145–154. [Google Scholar]
- Amin S.A., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Annunziata R., et al. (2019). bHLH-PAS protein RITMO1 regulates diel biological rhythms in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 116: 13137–13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt K.E., Kroth-Pancic P.G., Grossman A.R. (1996). Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. 252: 572–579. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Armbrust E.V., et al. (2004). The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306: 79–86. [DOI] [PubMed] [Google Scholar]
- Ashworth J., Coesel S., Lee A., Armbrust E.V., Orellana M.V., Baliga N.S. (2013). Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc. Natl. Acad. Sci. USA 110: 7518–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth J., Turkarslan S., Harris M., Orellana M.V., Baliga N.S. (2016). Pan-transcriptomic analysis identifies coordinated and orthologous functional modules in the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum. Mar. Genomics 26: 21–28. [DOI] [PubMed] [Google Scholar]
- Bailleul B., et al. (2015). Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524: 366–369. [DOI] [PubMed] [Google Scholar]
- Bailleul B., Cardol P., Breyton C., Finazzi G. (2010). Electrochromism: A useful probe to study algal photosynthesis. Photosynth. Res. 106: 179–189. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Herman E., Serif M., Maestre-Reyna M., Hepp S., Pokorny R., Kroth P.G., Essen L.-O., Kottke T. (2016). Allosteric communication between DNA-binding and light-responsive domains of diatom class I aureochromes. Nucleic Acids Res. 44: 5957–5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett A., Méléder V., Blommaert L., Lepetit B., Gaudin P., Vyverman W., Sabbe K., Dupuy C., Lavaud J. (2015). Growth form defines physiological photoprotective capacity in intertidal benthic diatoms. ISME J. 9: 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Phillips R., Milo R. (2018). The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 115: 6506–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S., et al. (2017). Finding a partner in the ocean: molecular and evolutionary bases of the response to sexual cues in a planktonic diatom. New Phytol. 215: 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw N., Grouneva I., Aram L., Gal A., Hopes A., Mock T. (2017). Efficient CRISPR/Cas-mediated homologous recombination in the model diatom Thalassiosira pseudonana. bioRxiv 215582. [DOI] [PubMed] [Google Scholar]
- Bertrand E.M., et al. (2015). Phytoplankton-bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc. Natl. Acad. Sci. USA 112: 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins J., Bruce D. (1989). Regulation of excitation energy transfer in organisms containing phycobilins. Photosynth. Res. 20: 1–34. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas C.E., Merchant S.S. (2019). Comparative and functional algal genomics. Annu. Rev. Plant Biol. 70: 605–638. [DOI] [PubMed] [Google Scholar]
- Bowler C., et al. (2008). The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456: 239–244. [DOI] [PubMed] [Google Scholar]
- Boyd P.W., et al. (2007). Mesoscale iron enrichment experiments 1993-2005: Synthesis and future directions. Science 315: 612–617. [DOI] [PubMed] [Google Scholar]
- Brembu T., Winge P., Tooming-Klunderud A., Nederbragt A.J., Jakobsen K.S., Bones A.M. (2014). The chloroplast genome of the diatom Seminavis robusta: New features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genomics 16: 17–27. [DOI] [PubMed] [Google Scholar]
- Breuer G., Lamers P.P., Martens D.E., Draaisma R.B., Wijffels R.H. (2012). The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 124: 217–226. [DOI] [PubMed] [Google Scholar]
- Briggs W.R. (2014). Phototropism: Some history, some puzzles, and a look ahead. Plant Physiol. 164: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broddrick J.T., Du N., Smith S.R., Tsuji Y., Jallet D., Ware M.A., Peers G., Matsuda Y., Dupont C.L., Mitchell B.G., Palsson B.O., Allen A.E. (2019). Cross-compartment metabolic coupling enables flexible photoprotective mechanisms in the diatom Phaeodactylum tricornutum. New Phytol. 222: 1364–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie J., Chan C.X., De Clerck O., Cock J.M., Coelho S.M., Gachon C., Grossman A.R., Mock T., Raven J.A., Smith A.G., Yoon H.S., Bhattacharya D. (2017). The algal revolution. Trends Plant Sci. 22: 726–738. [DOI] [PubMed] [Google Scholar]
- Brzezinski M., Olson R., Chisholm S. (1990). Silicon availability and cell-cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 67: 83–96. [Google Scholar]
- Buck J.M., Sherman J., Bártulos C.R., Serif M., Halder M., Henkel J., Falciatore A., Lavaud J., Gorbunov M.Y., Kroth P.G., Falkowski P.G., Lepetit B. (2019). Lhcx proteins provide photoprotection via thermal dissipation of absorbed light in the diatom Phaeodactylum tricornutum. Nat. Commun. 10: 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J.M., Río Bártulos C., Gruber A., Kroth P.G. (2018). Blasticidin-S deaminase, a new selection marker for genetic transformation of the diatom Phaeodactylum tricornutum. PeerJ 6: e5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge J. (2015). Electroporation-mediated transformation and post-transcriptional gene regulation of nitrate reductase in the marine diatom Thalassiosira pseudonana Master’s thesis (Worcester, MA: Clark University; ). [Google Scholar]
- Busseni G., et al. (2019). Meta-omics reveals genetic flexibility of diatom nitrogen transporters in response to environmental changes. Mol. Biol. Evol. 36: msz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D.A., Hossain Z., Cockshutt A.M., Zhaxybayeva O., Wu H., Li G. (2013). Photosystem II protein clearance and FtsH function in the diatom Thalassiosira pseudonana. Photosynth. Res. 115: 43–54. [DOI] [PubMed] [Google Scholar]
- Caron L., Berkaloff C., Duval J.C., Jupin H. (1987). Chlorophyll fluorescence transients from the diatom Phaeodactylum tricornutum: Relative rates of cyclic phosphorylation and chlororespiration. Photosynth. Res. 11: 131–139. [DOI] [PubMed] [Google Scholar]
- Carradec Q., et al. ; Tara Oceans Coordinators (2018). A global ocean atlas of eukaryotic genes. Nat. Commun. 9: 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. (1999). Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46: 347–366. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. (2003). Genomic reduction and evolution of novel genetic membranes and protein-targeting machinery in eukaryote-eukaryote chimaeras (meta-algae). Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 109–133, NaN–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.X., Bhattacharya D., Reyes-Prieto A. (2012). Endosymbiotic and horizontal gene transfer in microbial eukaryotes: Impacts on cell evolution and the tree of life. Mob. Genet. Elements 2: 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauton M.S., Winge P., Brembu T., Vadstein O., Bones A.M. (2013). Gene regulation of carbon fixation, storage, and utilization in the diatom Phaeodactylum tricornutum acclimated to light/dark cycles. Plant Physiol. 161: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y., Wollman F.-A. (2009). The CES process In The Chlamydomonas Sourcebook, 2nd ed, Chapter 29, Harris E.H., Stern D.B., Witman G.B., eds (London: Academic Press; ), pp. 1027–1063. [Google Scholar]
- Chu B.C., Garcia-Herrero A., Johanson T.H., Krewulak K.D., Lau C.K., Peacock R.S., Slavinskaya Z., Vogel H.J. (2010). Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23: 601–611. [DOI] [PubMed] [Google Scholar]
- Chukhutsina V.U., Büchel C., van Amerongen H. (2014). Disentangling two non-photochemical quenching processes in Cyclotella meneghiniana by spectrally-resolved picosecond fluorescence at 77K. Biochim. Biophys. Acta 1837: 899–907. [DOI] [PubMed] [Google Scholar]
- Cock J.M., et al. (2010). The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465: 617–621. [DOI] [PubMed] [Google Scholar]
- Coesel S., Mangogna M., Ishikawa T., Heijde M., Rogato A., Finazzi G., Todo T., Bowler C., Falciatore A. (2009). Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Rep. 10: 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M.T., Lawrence A.D., Raux-Deery E., Warren M.J., Smith A.G. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93. [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho M.H., Sun H.-X., Bowler C., Chua N.-H. (2016). Noncoding and coding transcriptome responses of a marine diatom to phosphate fluctuations. New Phytol. 210: 497–510. [DOI] [PubMed] [Google Scholar]
- Daboussi F., et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5: 3831. [DOI] [PubMed] [Google Scholar]
- Dal’Molin C.G., Quek L.E., Palfreyman R.W., Nielsen L.K. (2011). AlgaGEM--a genome-scale metabolic reconstruction of algae based on the Chlamydomonas reinhardtii genome. BMC Genomics 12 (Suppl 4): S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow P., Petersen T.W., Chepurnov V.A., Virginia Armbrust E. (2008). Inter- and intraspecific relationships between nuclear DNA content and cell sirze in selected members of the centric diatom genus Thalassiosira (Bacillariophyceae). J. Phycol. 44: 335–349. [DOI] [PubMed] [Google Scholar]
- Davis A., Crum L.T., Corbeil L.B., Hildebrand M. (2017). Expression of Histophilus somni IbpA DR2 protective antigen in the diatom Thalassiosira pseudonana. Appl Microbiol Biotechnol 101: 5313–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martino A., Amato A., Bowler C. (2009). Mitosis in diatoms: Rediscovering an old model for cell division. BioEssays 31: 874–884. [DOI] [PubMed] [Google Scholar]
- De Mia M., Lemaire S.D., Choquet Y., Wollman F.-A. (2019). Nitric oxide remodels the photosynthetic apparatus upon S-starvation in Chlamydomonas reinhardtii. Plant Physiol. 179: 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. (2009). Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 37: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner R.E., et al. (2017). Diatom centromeres suggest a mechanism for nuclear DNA acquisition. Proc. Natl. Acad. Sci. USA 114: E6015–E6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolch L.-J., et al. (2017). Nitric oxide mediates nitrite-sensing and acclimation and triggers a remodeling of lipids. Plant Physiol. 175: 1407–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell R.G., Gile G., McCallum G., Méheust R., Bapteste E.P., Klinger C.M., Brillet-Guéguen L., Freeman K.D., Richter D.J., Bowler C. (2017). Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife 6: e23717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunahay T.G., Jarvis E.E., Roessler P.G. (1995). Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J. Phycol. 31: 1004–1012. [Google Scholar]
- Ewe D., Tachibana M., Kikutani S., Gruber A., Río Bártulos C., Konert G., Kaplan A., Matsuda Y., Kroth P.G. (2018). The intracellular distribution of inorganic carbon fixing enzymes does not support the presence of a C4 pathway in the diatom Phaeodactylum tricornutum. Photosynth. Res. 137: 263–280. [DOI] [PubMed] [Google Scholar]
- Fabris M., Matthijs M., Rombauts S., Vyverman W., Goossens A., Baart G.J.E. (2012). The metabolic blueprint of Phaeodactylum tricornutum reveals a eukaryotic Entner-Doudoroff glycolytic pathway. Plant J. 70: 1004–1014. [DOI] [PubMed] [Google Scholar]
- Falciatore A., d’Alcalà M.R., Croot P., Bowler C. (2000). Perception of environmental signals by a marine diatom. Science 288: 2363–2366. [DOI] [PubMed] [Google Scholar]
- Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. (1999). Transformation of nonselectable reporter genes in marine diatoms. Mar. Biotechnol. 1: 239–251. [DOI] [PubMed] [Google Scholar]
- Falkowski P.G. (2015). Life’s Engines: How Microbes Made Earth Habitable. (Princeton, NJ: Princeton University Press; ). [Google Scholar]
- Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P. (1998). Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 281: 237–240. [DOI] [PubMed] [Google Scholar]
- Fisher M., Gokhman I., Pick U., Zamir A. (1997). A structurally novel transferrin-like protein accumulates in the plasma membrane of the unicellular green alga Dunaliella salina grown in high salinities. J. Biol. Chem. 272: 1565–1570. [DOI] [PubMed] [Google Scholar]
- Flori S., et al. (2017). Plastid thylakoid architecture optimizes photosynthesis in diatoms. Nat. Commun. 8: 15885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foflonker F., Mollegard D., Ong M., Yoon H.S., Bhattacharya D. (2018). Genomic analysis of Picochlorum species reveals how microalgae may adapt to variable environments. Mol. Biol. Evol. 35: 2702–2711. [DOI] [PubMed] [Google Scholar]
- Follows M.J., Dutkiewicz S., Grant S., Chisholm S.W. (2007). Emergent biogeography of microbial communities in a model ocean. Science 315: 1843–1846. [DOI] [PubMed] [Google Scholar]
- Fortunato A.E., et al. (2016). Diatom phytochromes reveal the existence of far-red-light-based sensing in the ocean. Plant Cell 28: 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R.A., Kuypers M.M.M., Vagner T., Paerl R.W., Musat N., Zehr J.P. (2011). Nitrogen fixation and transfer in open ocean diatom-cyanobacterial symbioses. ISME J. 5: 1484–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C., Furbank R., Harbinson J., Horton P. (1990). The mechanisms contributing to photosynthetic control of electron transport by carbon assimilation in leaves. Photosynth. Res. 25: 83–100. [DOI] [PubMed] [Google Scholar]
- Franz S., Ignatz E., Wenzel S., Zielosko H., Putu E.P.G.N., Maestre-Reyna M., Tsai M.-D., Yamamoto J., Mittag M., Essen L.-O. (2018). Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii. Nucleic Acids Res. 46: 8010–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo C., d’Ippolito G., Nuzzo G., Sardo A., Fontana A. (2017). Autoinhibitory sterol sulfates mediate programmed cell death in a bloom-forming marine diatom. Nat. Commun. 8: 1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H.E., Locarnini R.A., Boyer T.P., Antonov J.I., Zweng M.M., Baranova O.K., Johnson D.R. (2010). World Ocean Atlas 2009, Levitus S., ed, Volume Vol. 4: Nutrients (phosphate, nitrate, and silicate) (Washington, DC: NOAA Atlas NESDIS 71, U.S. Government Printing Office; ). [Google Scholar]
- Giovagnetti V., Ruban A.V. (2018). The evolution of the photoprotective antenna proteins in oxygenic photosynthetic eukaryotes. Biochem. Soc. Trans. 46: 1263–1277. [DOI] [PubMed] [Google Scholar]
- Glaesener A.G., Merchant S.S., Blaby-Haas C.E. (2013). Iron economy in Chlamydomonas reinhardtii. Front. Plant Sci. 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich S., Pawolski D., Zlotnikov I., Kröger N. (2019). Control of biosilica morphology and mechanical performance by the conserved diatom gene Silicanin-1. Commun Biol 2: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss R., Lepetit B. (2015). Biodiversity of NPQ. J. Plant Physiol. 172: 13–32. [DOI] [PubMed] [Google Scholar]
- Goss R., Lepetit B., Wilhelm C. (2006). Evidence for a rebinding of antheraxanthin to the light-harvesting complex during the epoxidation reaction of the violaxanthin cycle. J. Plant Physiol. 163: 585–590. [DOI] [PubMed] [Google Scholar]
- Gross M. (2012). The mysteries of the diatoms. Curr. Biol. 22: R581–R585. [DOI] [PubMed] [Google Scholar]
- Groussman R.D., Parker M.S., Armbrust E.V. (2015). Diversity and evolutionary history of iron metabolism genes in diatoms. PLoS One 10: e0129081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A., Kroth P.G. (2017). Intracellular metabolic pathway distribution in diatoms and tools for genome-enabled experimental diatom research. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A., Rocap G., Kroth P.G., Armbrust E.V., Mock T. (2015). Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 81: 519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi L., et al. ; Tara Oceans coordinators (2016). Plankton networks driving carbon export in the oligotrophic ocean. Nature 532: 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundermann K., Büchel C. (2014). Structure and functional heterogeneity of fucoxanthin-chlorophyll proteins in diatoms In The Structural Basis of Biological Energy Generation, Hohmann-Marriott M.F., ed (Dordrecht, the Netherlands: Springer Netherlands; ), pp. 21–37. [Google Scholar]
- Hager A., Stransky H. (1970). [The carotenoid pattern and the occurrence of the light-induced xanthophyll cycle in various classes of algae. V. A few members of Cryptophyceae]. Arch. Mikrobiol. 73: 77–89. [PubMed] [Google Scholar]
- Haimovich-Dayan M., Garfinkel N., Ewe D., Marcus Y., Gruber A., Wagner H., Kroth P.G., Kaplan A. (2013). The role of C4 metabolism in the marine diatom Phaeodactylum tricornutum. New Phytol. 197: 177–185. [DOI] [PubMed] [Google Scholar]
- Harada H., Nakatsuma D., Ishida M., Matsuda Y. (2005). Regulation of the expression of intracellular β-Carbonic Anhydrase in response to CO2 and light in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 139: 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijde M., et al. (2010). Characterization of two members of the cryptochrome/photolyase family from Ostreococcus tauri provides insights into the origin and evolution of cryptochromes. Plant Cell Environ. 33: 1614–1626. [DOI] [PubMed] [Google Scholar]
- Helliwell K.E., Chrachri A., Koester J.A., Wharam S., Verret F., Taylor A.R., Wheeler G.L., Brownlee C. (2019). Alternative mechanisms for fast Na+/Ca2+ signaling in eukaryotes via a novel class of single-domain voltage-gated channels. Curr. Biol. 29: 1503–1511.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel F., Bullmann L., Lau J., Zauner S., Maier U.G. (2009). ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol. Biol. Evol. 26: 1781–1790. [DOI] [PubMed] [Google Scholar]
- Herbstová M., Bína D., Kaňa R., Vácha F., Litvín R. (2017). Red-light phenotype in a marine diatom involves a specialized oligomeric red-shifted antenna and altered cell morphology. Sci. Rep. 7: 11976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbstová, M., Bína, D., Koník, P., Gardian, Z., Vácha, F., Litvín, R. (2015). Molecular basis of chromatic adaptation in pennate diatom Phaeodactylum tricornutum. Biochim. Biophys. Acta 1847: 534–543. [DOI] [PubMed]
- Hildebrand M., Lerch S.J.L., Shrestha R.P. (2018). Understanding diatom cell wall silicification—Moving forward. Front. Mar. Sci. 125. [Google Scholar]
- Hildebrand M., Volcani B.E., Gassmann W., Schroeder J.I. (1997). A gene family of silicon transporters. Nature 385: 688–689. [DOI] [PubMed] [Google Scholar]
- Hopes A., Nekrasov V., Kamoun S., Mock T. (2016). Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana. Plant Methods 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkinson B.M. (2014). A chloroplast pump model for the CO2 concentrating mechanism in the diatom Phaeodactylum tricornutum. Photosynth. Res. 121: 223–233. [DOI] [PubMed] [Google Scholar]
- Hopkinson B.M., Dupont C.L., Allen A.E., Morel F.M.M. (2011). Efficiency of the CO2-concentrating mechanism of diatoms. Proc. Natl. Acad. Sci. USA 108: 3830–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P. (2012). Optimization of light harvesting and photoprotection: molecular mechanisms and physiological consequences. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 3455–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Haferkamp I., Lepetit B., Molchanova M., Hou S., Jeblick W., Río Bártulos C., Kroth P.G. (2018). Reduced vacuolar β-1,3-glucan synthesis affects carbohydrate metabolism as well as plastid homeostasis and structure in Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 115: 4791–4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen P.F., Alami M., Lange P.F., Foster L.J., Schröder W.P., Overall C.M., Green B.R. (2013). Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS One 8: e74483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman M.J.J., et al. (2013). AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). Plant Cell 25: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman M.J.J., Martens C., Vandepoele K., Gillard J., Rayko E., Heijde M., Bowler C., Inzé D., Van de Peer Y., De Veylder L., Vyverman W. (2010). Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 11: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianora A., Miralto A., Poulet S.A., Carotenuto Y., Buttino I., Romano G., Casotti R., Pohnert G., Wichard T., Colucci-D’Amato L., Terrazzano G., Smetacek V. (2004). Aldehyde suppression of copepod recruitment in blooms of a ubiquitous planktonic diatom. Nature 429: 403–407. [DOI] [PubMed] [Google Scholar]
- Jaubert M., Bouly J.-P., Ribera d’Alcalà M., Falciatore A. (2017). Light sensing and responses in marine microalgae. Curr. Opin. Plant Biol. 37: 70–77. [DOI] [PubMed] [Google Scholar]
- Jensen E.L., Clement R., Kosta A., Maberly S.C., Gontero B. (2019). A new widespread subclass of carbonic anhydrase in marine phytoplankton. ISME J. 13: 2094–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot, P., Joliot, A. (2008). Quantification of the electrochemical proton gradient and activation of ATP synthase in leaves. Biochim. Biophys. Acta 1777: 676–683. [DOI] [PubMed]
- Juhas M., von Zadow A., Spexard M., Schmidt M., Kottke T., Büchel C. (2014). A novel cryptochrome in the diatom Phaeodactylum tricornutum influences the regulation of light-harvesting protein levels. FEBS J. 281: 2299–2311. [DOI] [PubMed] [Google Scholar]
- Kadono T., Miyagawa-Yamaguchi A., Kira N., Tomaru Y., Okami T., Yoshimatsu T., Hou L., Ohama T., Fukunaga K., Okauchi M., Yamaguchi H., Ohnishi K., et al. (2015). Characterization of marine diatom-infecting virus promoters in the model diatom Phaeodactylum tricornutum. Sci Rep 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan A. (2017). On the cradle of CCM research: Discovery, development, and challenges ahead. J. Exp. Bot. 68: 3785–3796. [DOI] [PubMed] [Google Scholar]
- Karas B.J., et al. (2015). Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 6: 6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazamia, E. et al. (2018). Endocytosis-mediated siderophore uptake as a strategy for Fe acquisition in diatoms. Sci. Adv. 4: eaar4536. [DOI] [PMC free article] [PubMed]
- Keeling P.J., et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikutani S., Nakajima K., Nagasato C., Tsuji Y., Miyatake A., Matsuda Y. (2016). Thylakoid luminal θ-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc. Natl. Acad. Sci. USA 113: 9828–9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikutani S., Tanaka R., Yamazaki Y., Hara S., Hisabori T., Kroth P.G., Matsuda Y. (2012). Redox regulation of carbonic anhydrases via thioredoxin in chloroplast of the marine diatom Phaeodactylum tricornutum. J. Biol. Chem. 287: 20689–20700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Fabris M., Baart G., Kim M.K., Goossens A., Vyverman W., Falkowski P.G., Lun D.S. (2016). Flux balance analysis of primary metabolism in the diatom Phaeodactylum tricornutum. Plant J. 85: 161–176. [DOI] [PubMed] [Google Scholar]
- Kirchhoff H., Tremmel I., Haase W., Kubitscheck U. (2004). Supramolecular photosystem II organization in grana thylakoid membranes: Evidence for a structured arrangement. Biochemistry 43: 9204–9213. [DOI] [PubMed] [Google Scholar]
- Kirkham A.R., Richthammer P., Schmidt K., Wustmann M., Maeda Y., Hedrich R., Brunner E., Tanaka T., van Pée K.-H., Falciatore A., Mock T. (2017). A role for the cell-wall protein silacidin in cell size of the diatom Thalassiosira pseudonana. ISME J. 11: 2452–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester J.A., Berthiaume C.T., Hiranuma N., Parker M.S., Iverson V., Morales R., Ruzzo W.L., Armbrust E.V. (2018). Sexual ancestors generated an obligate asexual and globally dispersed clone within the model diatom species Thalassiosira pseudonana. Sci. Rep. 8: 10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolber Z., Zehr J., Falkowski P. (1988). Effects of growth irradiance and nitrogen limitation on photosynthetic energy conversion in photosystem II. Plant Physiol. 88: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S., Juhas M., Jäger S., Kottke T., Büchel C. (2017). The cryptochrome-photolyase protein family in diatoms. J. Plant Physiol. 217: 15–19. [DOI] [PubMed] [Google Scholar]
- Kooistra W.H., Medlin L.K. (1996). Evolution of the diatoms (Bacillariophyta). IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Mol. Phylogenet. Evol. 6: 391–407. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Meinke D. (2010). The development of Arabidopsis as a model plant. Plant J. 61: 909–921. [DOI] [PubMed] [Google Scholar]
- Kowallik K.V., Stoebe B., Schaffran I., Kroth-Pancic P., Freier U. (1995). The chloroplast genome of a chlorophyll a+c-containing alga, Odontella sinensis. Plant Mol. Biol. Rep. 13: 336–342. [Google Scholar]
- Krasovec M., Sanchez-Brosseau S., Piganeau G. (2019). First estimation of the spontaneous mutation rate in diatoms. Genome Biol. Evol. 11: 1829–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger N., Deutzmann R., Bergsdorf C., Sumper M. (2000). Species-specific polyamines from diatoms control silica morphology. Proc. Natl. Acad. Sci. USA 97: 14133–14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger N., Poulsen N. (2008). Diatoms-from cell wall biogenesis to nanotechnology. Annu. Rev. Genet. 42: 83–107. [DOI] [PubMed] [Google Scholar]
- Kronholm I., Bassett A., Baulcombe D., Collins S. (2017). Epigenetic and genetic contributions to adaptation in Chlamydomonas. Mol. Biol. Evol. 34: 2285–2306. [DOI] [PubMed] [Google Scholar]
- Kroth P.G., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PLoS One 3: e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroth P.G., Wilhelm C., Kottke T. (2017). An update on aureochromes: Phylogeny - mechanism - function. J. Plant Physiol. 217: 20–26. [DOI] [PubMed] [Google Scholar]
- Kroth P.G., Bones A.M., Daboussi F., Ferrante M.I., Jaubert M., Kolot M., Nymark M., Río Bártulos C., Ritter A., Russo M.T., Serif M., Winge P., et al. (2018). Genome editing in diatoms: achievements and goals. Plant Cell Rep 37: 1401–1408. [DOI] [PubMed] [Google Scholar]
- Kustka A.B., Milligan A.J., Zheng H., New A.M., Gates C., Bidle K.D., Reinfelder J.R. (2014). Low CO2 results in a rearrangement of carbon metabolism to support C4 photosynthetic carbon assimilation in Thalassiosira pseudonana. New Phytol. 204: 507–520. [DOI] [PubMed] [Google Scholar]
- Kuzminov F.I., Gorbunov M.Y. (2016). Energy dissipation pathways in Photosystem 2 of the diatom, Phaeodactylum tricornutum, under high-light conditions. Photosynth. Res. 127: 219–235. [DOI] [PubMed] [Google Scholar]
- Lane T.W., Saito M.A., George G.N., Pickering I.J., Prince R.C., Morel F.M.M. (2005). Biochemistry: A cadmium enzyme from a marine diatom. Nature 435: 42. [DOI] [PubMed] [Google Scholar]
- La Roche J., Boyd P.W., McKay R.M.L., Geider R.J. (1996). Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382: 802–805. [Google Scholar]
- Lavaud J., Rousseau B., Etienne A.-L. (2002). In diatoms, a transthylakoid proton gradient alone is not sufficient to induce a non-photochemical fluorescence quenching. FEBS Lett. 523: 163–166. [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire C., Wollman F.A., Bennoun P. (1988). Restoration of phototrophic growth in a mutant of Chlamydomonas reinhardtii in which the chloroplast atpB gene of the ATP synthase has a deletion: An example of mitochondria-dependent photosynthesis. Proc. Natl. Acad. Sci. USA 85: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepetit B., Gélin G., Lepetit M., Sturm S., Vugrinec S., Rogato A., Kroth P.G., Falciatore A., Lavaud J. (2017). The diatom Phaeodactylum tricornutum adjusts nonphotochemical fluorescence quenching capacity in response to dynamic light via fine-tuned Lhcx and xanthophyll cycle pigment synthesis. New Phytol. 214: 205–218. [DOI] [PubMed] [Google Scholar]
- Lepetit B., Goss R., Jakob T., Wilhelm C. (2012). Molecular dynamics of the diatom thylakoid membrane under different light conditions. Photosynth. Res. 111: 245–257. [DOI] [PubMed] [Google Scholar]
- Lepetit B., Sturm S., Rogato A., Gruber A., Sachse M., Falciatore A., Kroth P.G., Lavaud J. (2013). High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiol. 161: 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesuisse E., Simon-Casteras M., Labbe P. (1998). Siderophore-mediated iron uptake in Saccharomyces cerevisiae: The SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144: 3455–3462. [DOI] [PubMed] [Google Scholar]
- Levering J., Broddrick J., Dupont C.L., Peers G., Beeri K., Mayers J., Gallina A.A., Allen A.E., Palsson B.O., Zengler K. (2016). Genome-scale model reveals metabolic basis of biomass partitioning in a model diatom. PLoS One 11: e0155038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan O., Dinamarca J., Zelzion E., Gorbunov M.Y., Falkowski P.G. (2015a). An RNA interference knock-down of nitrate reductase enhances lipid biosynthesis in the diatom Phaeodactylum tricornutum. Plant J. 84: 963–973. [DOI] [PubMed] [Google Scholar]
- Levitan O., Dinamarca J., Zelzion E., Lun D.S., Guerra L.T., Kim M.K., Kim J., Van Mooy B.A.S., Bhattacharya D., Falkowski P.G. (2015b). Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc. Natl. Acad. Sci. USA 112: 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Beardall J., Collins S., Gao K. (2017). Decreased photosynthesis and growth with reduced respiration in the model diatom Phaeodactylum tricornutum grown under elevated CO2 over 1800 generations. Glob. Change Biol. 23: 127–137. [DOI] [PubMed] [Google Scholar]
- Li X.P., Björkman O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395. [DOI] [PubMed] [Google Scholar]
- Liaud M.-F., Lichtlé C., Apt K., Martin W., Cerff R. (2000). Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: Evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 17: 213–223. [DOI] [PubMed] [Google Scholar]
- Lin H.-Y., Yen S.-C., Kuo P.-C., Chung C.-Y., Yeh K.-L., Huang C.-H., Chang J., Lin H.-J. (2017). Alkaline phosphatase promoter as an efficient driving element for exogenic recombinant in the marine diatom Phaeodactylum tricornutum. Algal Research 23: 58–65. [Google Scholar]
- Lionetto M.G., Caricato R., Giordano M.E., Schettino T. (2016). The complex relationship between metals and carbonic anhydrase: new insights and perspectives. Int. J. Mol. Sci. 17: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Nannas N.J., Fu F.F., Shi J., Aspinwall B., Parrott W.A., Dawe R.K. (2019). Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 31: 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommer M., Roy A.-S., Schilhabel M., Schreiber S., Rosenstiel P., LaRoche J. (2010). Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genomics 11: 718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommer M., et al. (2012). Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 13: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D.A., Hamaji T., Kropat J., De Hoff P., Morselli M., Rubbi L., Fitz-Gibbon S.T., Gallaher S.D., Merchant S.S., Umen J.G., Pellegrini M. (2015). Dynamic changes in the transcriptome and methylome of Chlamydomonas reinhardtii throughout its life cycle In Plant Physiol., Volume 169, pp. 2730–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gomollon S., Beckers M., Rathjen T., Moxon S., Maumus F., Mohorianu I., Moulton V., Dalmay T., Mock T. (2014). Global discovery and characterization of small non-coding RNAs in marine microalgae. BMC Genomics 15: 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J.D. (1869). I.— On the structure of the Diatomaceous frustule, and its genetic cycle. Ann. Mag. Nat. Hist. 3: 1–8. [Google Scholar]
- Maier U.G., Zauner S., Hempel F. (2015). Protein import into complex plastids: Cellular organization of higher complexity. Eur. J. Cell Biol. 94: 340–348. [DOI] [PubMed] [Google Scholar]
- Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., Wincker P., Iudicone D., de Vargas C., Bittner L., Zingone A., Bowler C. (2016). Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 113: E1516–E1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann D.G. (1994). Auxospore formation, reproductive plasticity and cell structure in Navicula ulvacea and the resurrection of the genus Dickieia (Bacillariophyta). Eur. J. Phycol. 29: 141–157. [Google Scholar]
- Mann M., Serif M., Jakob T., Kroth P.G., Wilhelm C. (2017). PtAUREO1a and PtAUREO1b knockout mutants of the diatom Phaeodactylum tricornutum are blocked in photoacclimation to blue light. J. Plant Physiol. 217: 44–48. [DOI] [PubMed] [Google Scholar]
- Marañón E., Cermeño P., López-Sandoval D.C., Rodríguez-Ramos T., Sobrino C., Huete-Ortega M., Blanco J.M., Rodríguez J. (2013). Unimodal size scaling of phytoplankton growth and the size dependence of nutrient uptake and use. Ecol. Lett. 16: 371–379. [DOI] [PubMed] [Google Scholar]
- Marchetti A., Parker M.S., Moccia L.P., Lin E.O., Arrieta A.L., Ribalet F., Murphy M.E.P., Maldonado M.T., Armbrust E.V. (2009). Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature 457: 467–470. [DOI] [PubMed] [Google Scholar]
- Marchetti A., Schruth D.M., Durkin C.A., Parker M.S., Kodner R.B., Berthiaume C.T., Morales R., Allen A.E., Armbrust E.V. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl. Acad. Sci. USA 109: E317–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna A.C., Sturm S., Kroth P.G., Lavaud J. (2009). First induced plastid genome mutations in an alga with secondary plastids: psbA mutations in the diatom Phaeodactylum tricornutum (BACILLARIOPHYCEAE) reveal consequences on the regulation of photosynthesis. Journal of Phycology 45: 838–846. [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Hopkinson B.M., Nakajima K., Dupont C.L., Tsuji Y. (2017). Mechanisms of carbon dioxide acquisition and CO2 sensing in marine diatoms: a gateway to carbon metabolism. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Hopkinson B.M., Nakajima K., Matsuda Y. (2018). Plasma membrane-type aquaporins from marine diatoms function as CO2/NH3 channels and provide photoprotection In Plant Physiol., Volume 178, pp. 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijs M., Fabris M., Obata T., Foubert I., Franco-Zorrilla J.M., Solano R., Fernie A.R., Vyverman W., Goossens A. (2017). The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 36: 1559–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F., Allen A.E., Mhiri C., Hu H., Jabbari K., Vardi A., Grandbastien M.-A., Bowler C. (2009). Potential impact of stress activated retrotransposons on genome evolution in a marine diatom. BMC Genomics 10: 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.K., Smith S.R., McCrow J.P., Tan M., Zheng H., Beeri K., Roth R., Lichtle C., Goodenough U., Bowler C.P., Dupont C.L., Allen A.E. (2017). Nitrate reductase knockout uncouples nitrate transport from nitrate assimilation and drives repartitioning of carbon flux in a model pennate diatom. Plant Cell 29: 2047–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaid J.B., Kustka A.B., Oborník M., Horák A., McCrow J.P., Karas B.J., Zheng H., Kindeberg T., Andersson A.J., Barbeau K.A., Allen A.E. (2018). Carbonate-sensitive phytotransferrin controls high-affinity iron uptake in diatoms. Nature 555: 534–537. [DOI] [PubMed] [Google Scholar]
- Merchant S., Bogorad L. (1986). Regulation by copper of the expression of plastocyanin and cytochrome c552 in Chlamydomonas reinhardi. Mol. Cell. Biol. 6: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S.S., Kropat J., Liu B., Shaw J., Warakanont J. (2012). TAG, you’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr. Opin. Biotechnol. 23: 352–363. [DOI] [PubMed] [Google Scholar]
- Merchant S.S., et al. (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra M., Lato S.M., Ynalvez R.A., Xiao Y., Moroney J.V. (2004). Identification of a new chloroplast carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol. 135: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. (2011). ROS signaling: The new wave? Trends Plant Sci. 16: 300–309. [DOI] [PubMed] [Google Scholar]
- Mizrachi A., Graff van Creveld S., Shapiro O.H., Rosenwasser S., Vardi A. (2019). Light-dependent single-cell heterogeneity in the chloroplast redox state regulates cell fate in a marine diatom. eLife 8: e47732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mock T., et al. (2017). Evolutionary genomics of the cold-adapted diatom Fragilariopsis cylindrus. Nature 541: 536–540. [DOI] [PubMed] [Google Scholar]
- Mock T., et al. (2008). Whole-genome expression profiling of the marine diatom Thalassiosira pseudonana identifies genes involved in silicon bioprocesses. Proc. Natl. Acad. Sci. USA 105: 1579–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeys S., et al. (2016). A sex-inducing pheromone triggers cell cycle arrest and mate attraction in the diatom Seminavis robusta. Sci. Rep. 6: 19252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A., Schwach F., Studholme D.J., Thuenemann E.C., Baulcombe D.C. (2007). miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447: 1126–1129. [DOI] [PubMed] [Google Scholar]
- Montresor M., Vitale L., D’Alelio D., Ferrante M.I. (2016). Sex in marine planktonic diatoms: insights and challenges. Perspect. Phycol. 3: 61–75. [Google Scholar]
- Morrissey J., et al. (2015). A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Curr. Biol. 25: 364–371. [DOI] [PubMed] [Google Scholar]
- Moustafa A., Beszteri B., Maier U.G., Bowler C., Valentin K., Bhattacharya D. (2009). Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324: 1724–1726. [DOI] [PubMed] [Google Scholar]
- Murik O., Tirichine L., Prihoda J., Thomas Y., Araújo W.L., Allen A.E., Fernie A.R., Bowler C. (2018). Downregulation of mitochondrial alternative oxidase affects chloroplast function, redox status and stress response in a marine diatom. New Phytol 221: 1303–1316. [DOI] [PubMed] [Google Scholar]
- Nagao R., Kato K., Suzuki T., Ifuku K., Uchiyama I., Kashino Y., Dohmae N., Akimoto S., Shen J.-R., Miyazaki N., Akita F. (2019). Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex. Nat. Plants 5: 890–901. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Tanaka A., Matsuda Y. (2013). SLC4 family transporters in a marine diatom directly pump bicarbonate from seawater. Proc. Natl. Acad. Sci. USA 110: 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakov T., Beaulieu J.M., Alverson A.J. (2018). Insights into global planktonic diatom diversity: The importance of comparisons between phylogenetically equivalent units that account for time. ISME J. 12: 2807–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y.-F., Yang Z.-K., Zhang M.-H., Zhu C.-C., Yang W.-D., Liu J.-S., Li H.-Y. (2012). Transformation of diatom Phaeodactylum tricornutum by electroporation and establishment of inducible selection marker. Biotech. [DOI] [PubMed]
- Nosenko T., Bhattacharya D. (2007). Horizontal gene transfer in chromalveolates. BMC Evol. Biol. 7: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M., Sharma A.K., Sparstad T., Bones A.M., Winge P. (2016). A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci Rep 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura A., Akizuki Y., Imoda H., Mineta K., Gojobori T., Nagai S. (2018). Comparative genome and transcriptome analysis of diatom, Skeletonema costatum, reveals evolution of genes for harmful algal bloom. BMC Genomics 19: 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi N., Mukherjee B., Tsujikawa T., Yanase M., Nakano H., Moroney J.V., Fukuzawa H. (2010). Expression of a low CO2-inducible protein, LCI1, increases inorganic carbon uptake in the green alga Chlamydomonas reinhardtii. Plant Cell 22: 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudot-Le Secq M.-P., Grimwood J., Shapiro H., Armbrust E.V., Bowler C., Green B.R. (2007). Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: Comparison with other plastid genomes of the red lineage. Mol. Genet. Genomics 277: 427–439. [DOI] [PubMed] [Google Scholar]
- Owens T.G. (1986). Light-harvesting function in the diatom Phaeodactylum tricornutum: II. Distribution of excitation energy between the photosystems. Plant Physiol. 80: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks M.B., Nakov T., Ruck E.C., Wickett N.J., Alverson A.J. (2018). Phylogenomics reveals an extensive history of genome duplication in diatoms (Bacillariophyta). Am. J. Bot. 105: 330–347. [DOI] [PubMed] [Google Scholar]
- Peers G., Price N.M. (2006). Copper-containing plastocyanin used for electron transport by an oceanic diatom. Nature 441: 341–344. [DOI] [PubMed] [Google Scholar]
- Peers G., Truong T.B., Ostendorf E., Busch A., Elrad D., Grossman A.R., Hippler M., Niyogi K.K. (2009). An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462: 518–521. [DOI] [PubMed] [Google Scholar]
- Petersen J., Ludewig A.-K., Michael V., Bunk B., Jarek M., Baurain D., Brinkmann H. (2014). Chromera velia, endosymbioses and the rhodoplex hypothesis--plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol. Evol. 6: 666–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroutsos D., Tokutsu R., Maruyama S., Flori S., Greiner A., Magneschi L., Cusant L., Kottke T., Mittag M., Hegemann P., Finazzi G., Minagawa J. (2016). A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537: 563–566. [DOI] [PubMed] [Google Scholar]
- Pfaffen S., Bradley J.M., Abdulqadir R., Firme M.R., Moore G.R., Le Brun N.E., Murphy M.E.P. (2015). A diatom ferritin optimized for iron oxidation but not iron storage. J. Biol. Chem. 290: 28416–28427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen G.S., Avaca-Crusca J.S., Araujo A.P.U., DeMarco R. (2016). Distribution patterns and impact of transposable elements in genes of green algae. Gene 594: 151–159. [DOI] [PubMed] [Google Scholar]
- Pi X., Zhao S., Wang W., Liu D., Xu C., Han G., Kuang T., Sui S.F., Shen J.R. (2019). The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex. Science 365: eaax4406. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H. (1978). The diatom spindle in perspective. Cell 14: 455–467. [DOI] [PubMed] [Google Scholar]
- Pierella Karlusich J.J., Carrillo N. (2017). Evolution of the acceptor side of photosystem I: Ferredoxin, flavodoxin, and ferredoxin-NADP+ oxidoreductase. Photosynth. Res. 134: 235–250. [DOI] [PubMed] [Google Scholar]
- Pierella Karlusich J.J., Ibarbalz F.M., Bowler C. (2020). Phytoplankton in the Tara Ocean. Annu. Rev. Mar. Sci. 12: 233–265. [DOI] [PubMed] [Google Scholar]
- Poulsen N.C., Spector I., Spurck T.P., Schultz T.F., Wetherbee R. (1999). Diatom gliding is the result of an actin-myosin motility system. Cell Motil. Cytoskeleton 44: 23–33. [DOI] [PubMed] [Google Scholar]
- Poulsen N., Chesley P.M., Kröger N. (2006). Molecular genetic manipulation of the diatom Thalassiosira pseudonana (BACILLARIOPHYCEAE). J Phycol 42: 1059–1065. [Google Scholar]
- Pyszniak A.M., Gibbs S.P. (1992). Immunocytochemical localization of photosystem I and the fucoxanthin-chlorophyll a/c light-harvesting complex in the diatom Phaeodactylum tricornutum. Protoplasma 166: 208–217. [Google Scholar]
- Qiao X., Li Q., Yin H., Qi K., Li L., Wang R., Zhang S., Paterson A.H. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni M., d’Alcalà M.R. (2007). Circadian variability in the photobiology of Phaeodactylum tricornutum: pigment content. J. Plankton Res. 29: 141–156. [Google Scholar]
- Rastogi A., Maheswari U., Dorrell R.G., Vieira F.R.J., Maumus F., Kustka A., McCarthy J., Allen A.E., Kersey P., Bowler C., Tirichine L. (2018). Integrative analysis of large scale transcriptome data draws a comprehensive landscape of Phaeodactylum tricornutum genome and evolutionary origin of diatoms. Sci. Rep. 8: 4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi A., Vieira F.R.J., Deton-Cabanillas A.-F., Veluchamy A., Cantrel C., Wang G., Vanormelingen P., Bowler C., Piganeau G., Hu H., Tirichine L. (2019). A genomics approach reveals the global genetic polymorphism, structure, and functional diversity of ten accessions of the marine model diatom Phaeodactylum tricornutum. ISME J.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayko E., Maumus F., Maheswari U., Jabbari K., Bowler C. (2010). Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytol. 188: 52–66. [DOI] [PubMed] [Google Scholar]
- Reimann B.E.F., Leivin J.C., Volcani B.E. (1966). Studies on the biochemistry and fine structure of silica shell formation in diatoms. II. The structure of the cell wall of Navicula pelliculosa (Bréb.) Hilse. J. Phycol. 2: 74–84. [DOI] [PubMed] [Google Scholar]
- Reinfelder J.R. (2011). Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Annu. Rev. Mar. Sci. 3: 291–315. [DOI] [PubMed] [Google Scholar]
- Reinfelder J.R., Kraepiel A.M., Morel F.M. (2000). Unicellular C4 photosynthesis in a marine diatom. Nature 407: 996–999. [DOI] [PubMed] [Google Scholar]
- Río Bártulos C., Rogers M.B., Williams T.A., Gentekaki E., Brinkmann H., Cerff R., Liaud M.-F., Hehl A.B., Yarlett N.R., Gruber A., Kroth P.G., van der Giezen M. (2018). Mitochondrial glycolysis in a major lineage of eukaryotes. Genome Biol. Evol. 10: 2310–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell N.C., Duanmu D., Martin S.S., Bachy C., Price D.C., Bhattacharya D., Worden A.Z., Lagarias J.C. (2014). Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. USA 111: 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogato A., Amato A., Iudicone D., Chiurazzi M., Ferrante M.I., d’Alcalà M.R. (2015). The diatom molecular toolkit to handle nitrogen uptake. Mar. Genomics 24: 95–108. [DOI] [PubMed] [Google Scholar]
- Rogato A., Richard H., Sarazin A., Voss B., Cheminant Navarro S., Champeimont R., Navarro L., Carbone A., Hess W.R., Falciatore A. (2014). The diversity of small non-coding RNAs in the diatom Phaeodactylum tricornutum. BMC Genomics 15: 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwasser S., Graff van Creveld S., Schatz D., Malitsky S., Tzfadia O., Aharoni A., Levin Y., Gabashvili A., Feldmesser E., Vardi A. (2014). Mapping the diatom redox-sensitive proteome provides insight into response to nitrogen stress in the marine environment. Proc. Natl. Acad. Sci. USA 111: 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoni A.W., Price D.C., Seger M., Lyska D., Lammers P., Bhattacharya D., Weber A.P. (2019). The genomes of polyextremophilic cyanidiales contain 1% horizontally transferred genes with diverse adaptive functions. eLife 8: e45017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round F.E., Crawford R.M., Mann D.G. (1990). The Diatoms: Biology & Morphology of the Genera. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Ruban A., Lavaud J., Rousseau B., Guglielmi G., Horton P., Etienne A.-L. (2004). The super-excess energy dissipation in diatom algae: comparative analysis with higher plants. Photosynth. Res. 82: 165–175. [DOI] [PubMed] [Google Scholar]
- Russo M.T., et al. (2018). MRP3 is a sex determining gene in the diatom Pseudo-nitzschia multistriata. Nat. Commun. 9: 5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue K., Harada H., Matsuda Y. (2008). Development of gene expression system in a marine diatom using viral promoters of a wide variety of origin. Physiol Plant 133: 59–67. [DOI] [PubMed] [Google Scholar]
- Salomé P.A., Merchant S.S. (2019). A series of fortunate events: Introducing Chlamydomonas as a reference organism. Plant Cell 31: 1682–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoefs B., Hu H., Kroth P.G. (2017). The peculiar carbon metabolism in diatoms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S., Jeon H., Hwang S., Jin E., Chang K.S. (2015). Development of a new constitutive expression system for the transformation of the diatom Phaeodactylum tricornutum. Algal Research 11: 50–54. [Google Scholar]
- Serif M., Dubois G., Finoux A.-L., Teste M.-A., Jallet D., Daboussi F. (2018). One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat. Commun. 9: 3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Dupont C.L., Hopkinson B.M. (2017). The diversity of CO2-concentrating mechanisms in marine diatoms as inferred from their genetic content. J. Exp. Bot. 68: 3937–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard V.C., Scheffel A., Poulsen N., Kröger N. (2011). Live diatom silica immobilization of multimeric and redox-active enzymes. Appl. Environ. Microbiol. 78: 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.M., Matzke N.J. (2013). Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc. Natl. Acad. Sci. USA 110: 12355–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R.P., Hildebrand M. (2015). Evidence for a regulatory role of diatom silicon transporters in cellular silicon responses. Eukaryotic Cell 14: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R.P., Hildebrand M. (2017). Development of a silicon limitation inducible expression system for recombinant protein production in the centric diatoms Thalassiosira pseudonana and Cyclotella cryptica. Microb Cell Fact 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C. (2007). Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene 406: 23–35. [DOI] [PubMed] [Google Scholar]
- Slattery S.S., Diamond A., Wang H., Therrien J.A., Lant J.T., Jazey T., Lee K., Klassen Z., Desgagné-Penix I., Karas B.J., Edgell D.R. (2018). An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum. ACS Synth. Biol. 7: 328–338. [DOI] [PubMed] [Google Scholar]
- Smith S.R., Abbriano R.M., Hildebrand M. (2012). Comparative analysis of diatom genomes reveals substantial differences in the organization of carbon partitioning pathways. Algal Res. 1: 2–16. [Google Scholar]
- Smith S.R., Gillard J.T.F., Kustka A.B., McCrow J.P., Badger J.H., Zheng H., New A.M., Dupont C.L., Obata T., Fernie A.R., Allen A.E. (2016). Transcriptional orchestration of the global cellular response of a model pennate diatom to diel light cycling under iron limitation. PLoS Genet. 12: e1006490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J.W., Schreiber J., Yue J., Guo H., Ding Q., Huang J. (2014). The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5: 5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky H., Hager A. (1970). [A carotenoid pattern and the occurence of the light-induced xanthophyll cycle in various classes of algae. II. Xanthophyceae]. Arch. Mikrobiol. 71: 164–190. [PubMed] [Google Scholar]
- Strauss, J. (2012). A genomic analysis using RNA-seq to investigate the adaptation of the psychrophilic diatom Fragilariopsis cylindrus to the polar environment. PhD thesis. (Norwich, UK: University of East Anglia).
- Taddei L., Chukhutsina V.U., Lepetit B., Stella G.R., Bassi R., van Amerongen H., Bouly J.-P., Jaubert M., Finazzi G., Falciatore A. (2018). Dynamic changes between two LHCX-related energy quenching sites control diatom photoacclimation. Plant Physiol. 177: 953–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei L., et al. (2016). Multisignal control of expression of the LHCX protein family in the marine diatom Phaeodactylum tricornutum. J. Exp. Bot. 67: 3939–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., De Martino A., Amato A., Montsant A., Mathieu B., Rostaing P., Tirichine L., Bowler C. (2015a). Ultrastructure and membrane traffic during cell division in the marine pennate diatom Phaeodactylum tricornutum. Protist 166: 506–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Ohno N., Nakajima K., Matsuda Y. (2016). Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic β-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum. Plant Physiol. 170: 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Kikutani S., Mahardika A., Matsuda Y. (2014). Localization of enzymes relating to C4 organic acid metabolisms in the marine diatom, Thalassiosira pseudonana. Photosynth. Res. 121: 251–263. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fukuda Y., Yoshino T., Maeda Y., Muto M., Matsumoto M., Mayama S., Matsunaga T. (2011). High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth. Res. 109: 223–229. [DOI] [PubMed] [Google Scholar]
- Tanaka T., et al. (2015b). Oil accumulation by the oleaginous diatom Fistulifera solaris as revealed by the genome and transcriptome. Plant Cell 27: 162–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarver J.E., Cormier A., Pinzón N., Taylor R.S., Carré W., Strittmatter M., Seitz H., Coelho S.M., Cock J.M. (2015). microRNAs and the evolution of complex multicellularity: identification of a large, diverse complement of microRNAs in the brown alga Ectocarpus. Nucleic Acids Res. 43: 6384–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K., Bailleul B., Brown C.M., Gorbunov M.Y., Kustka A.B., Frada M., Joliot P.A., Falkowski P.G., Bidle K.D. (2013). Death-specific protein in a marine diatom regulates photosynthetic responses to iron and light availability. Proc. Natl. Acad. Sci. USA 110: 20123–20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K., Hildebrand M. (2008). Silicon uptake in diatoms revisited: a model for saturable and nonsaturable uptake kinetics and the role of silicon transporters. Plant Physiol. 146: 1397–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., Rastogi A., Bowler C. (2017). Recent progress in diatom genomics and epigenomics. Curr. Opin. Plant Biol. 36: 46–55. [DOI] [PubMed] [Google Scholar]
- Tomar V., Sidhu G.K., Nogia P., Mehrotra R., Mehrotra S. (2017). Regulatory components of carbon concentrating mechanisms in aquatic unicellular photosynthetic organisms. Plant Cell Rep. 36: 1671–1688. [DOI] [PubMed] [Google Scholar]
- Traller J.C., et al. (2016). Genome and methylome of the oleaginous diatom Cyclotella cryptica reveal genetic flexibility toward a high lipid phenotype. Biotechnol. Biofuels 9: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tréguer P., et al. (2018). Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 11: 27–37. [Google Scholar]
- Tsuji Y., Nakajima K., Matsuda Y. (2017). Molecular aspects of the biophysical CO2-concentrating mechanism and its regulation in marine diatoms. J. Exp. Bot. 68: 3763–3772. [DOI] [PubMed] [Google Scholar]
- van Creveld S.G., Rosenwasser S., Schatz D., Koren I., Vardi A. (2015). Early perturbation in mitochondria redox homeostasis in response to environmental stress predicts cell fate in diatoms. ISME J. 9: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A. (2008). Cell signaling in marine diatoms. Commun. Integr. Biol. 1: 134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi A., Formiggini F., Casotti R., De Martino A., Ribalet F., Miralto A., Bowler C. (2006). A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 4: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy A., et al. (2015). An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 16: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow P., et al. (2015). Life-cycle modification in open oceans accounts for genome variability in a cosmopolitan phytoplankton. ISME J. 9: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner H., Jakob T., Wilhelm C. (2006). Balancing the energy flow from captured light to biomass under fluctuating light conditions. New Phytol. 169: 95–108. [DOI] [PubMed] [Google Scholar]
- Wang, W., et al. (2019). Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science 363: eaav0365. [DOI] [PubMed]
- Wang Y., Stessman D.J., Spalding M.H. (2015). The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: How Chlamydomonas works against the gradient. Plant J. 82: 429–448. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Kadono T., Kira N., Suzuki K., Iwata O., Ohnishi K., Yamaguchi H., Adachi M. (2018). Development of endogenous promoters that drive high-level expression of introduced genes in the model diatom Phaeodactylum tricornutum. Marine Genomics 42: 41–48. [DOI] [PubMed] [Google Scholar]
- Wei L., Derrien B., Gautier A., Houille-Vernes L., Boulouis A., Saint-Marcoux D., Malnoë A., Rappaport F., de Vitry C., Vallon O., Choquet Y., Wollman F.-A. (2014). Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm C., Büchel C., Fisahn J., Goss R., Jakob T., Laroche J., Lavaud J., Lohr M., Riebesell U., Stehfest K., Valentin K., Kroth P.G. (2006). The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157: 91–124. [DOI] [PubMed] [Google Scholar]
- Winter G., Todd C.D., Trovato M., Forlani G., Funck D. (2015). Physiological implications of arginine metabolism in plants. Front. Plant Sci. 6: 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson J.D., Chory J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9: 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Roy S., Alami M., Green B.R., Campbell D.A. (2012). Photosystem II photoinactivation, repair, and protection in marine centric diatoms. Plant Physiol. 160: 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W.-H., Zhu C.-C., Zhang N.-S., Li D.-W., Yang W.-D., Liu J.-S., Sathishkumar R., Li H.-Y. (2014). Construction of novel chloroplast expression vector and development of an efficient transformation system for the diatom Phaeodactylum tricornutum. Mar Biotechnol 16: 538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Sato E., Iguchi H., Fukuda Y., Fukuzawa H. (2015). Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 112: 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Lin X., Liu X., Zhang J., Ge F. (2018). Genome annotation of a model diatom Phaeodactylum tricornutum using an integrated proteogenomic pipeline. Mol. Plant 11: 1292–1307. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Terashima I., Noguchi K. (2007). Up-regulation of mitochondrial alternative oxidase concomitant with chloroplast over-reduction by excess light. Plant Cell Physiol. 48: 606–614. [DOI] [PubMed] [Google Scholar]
- Yoshinaga R., Niwa-Kubota M., Matsui H., Matsuda Y. (2014). Characterization of iron-responsive promoters in the marine diatom Phaeodactylum tricornutum. Marine Genomics 16: 55–62. [DOI] [PubMed] [Google Scholar]
- Young J.N., Hopkinson B.M. (2017). The potential for co-evolution of CO2-concentrating mechanisms and Rubisco in diatoms. J. Exp. Bot. 68: 3751–3762. [DOI] [PubMed] [Google Scholar]
- Zaslavskaia L.A., Lippmeier J.C., Kroth P.G., Grossman A.R., Apt K.E. (2001). Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. Journal of Phycology 36: 379–386. [Google Scholar]
- Zhu S.-H., Green B.R. (2010). Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochim. Biophys. Acta 1797: 1449–1457. [DOI] [PubMed] [Google Scholar]