ONAC054 promotes ABA-induced leaf senescence, and its activity is regulated through alternative 3′ splice site selection and cleavage of a C-terminal transmembrane domain.
Abstract
In most plants, abscisic acid (ABA) induces premature leaf senescence; however, the mechanisms of ABA signaling during leaf senescence remain largely unknown. Here, we show that the rice (Oryza sativa) NAM/ATAF1/2/CUC2 (NAC) transcription factor ONAC054 plays an important role in ABA-induced leaf senescence. The onac054 knockout mutants maintained green leaves, while ONAC054-overexpressing lines showed early leaf yellowing under dark- and ABA-induced senescence conditions. Genome-wide microarray analysis showed that ABA signaling-associated genes, including ABA INSENSITIVE5 (OsABI5) and senescence-associated genes, including STAY-GREEN and NON-YELLOW COLORING1 (NYC1), were significantly down-regulated in onac054 mutants. Chromatin immunoprecipitation and protoplast transient assays showed that ONAC054 directly activates OsABI5 and NYC1 by binding to the mitochondrial dysfunction motif in their promoters. ONAC054 activity is regulated by proteolytic processing of the C-terminal transmembrane domain (TMD). We found that nuclear import of ONAC054 requires cleavage of the putative C-terminal TMD. Furthermore, the ONAC054 transcript (termed ONAC054α) has an alternatively spliced form (ONAC054β), with seven nucleotides inserted between intron 5 and exon 6, truncating ONAC054α protein at a premature stop codon. ONAC054β lacks the TMD and thus localizes to the nucleus. These findings demonstrate that the activity of ONAC054, which is important for ABA-induced leaf senescence in rice, is precisely controlled by multilayered regulatory processes.
INTRODUCTION
Leaf senescence is the final stage of leaf development, in which intracellular organelles and macromolecules are actively destabilized to relocate nutrients into developing tissues or storage organs (Lim et al., 2007). The initiation of leaf senescence is tightly controlled by endogenous factors, such as the state of phytohormones and metabolites (Moore et al., 2003; Sakuraba et al., 2012; Kusaba et al., 2013), and external stimuli, such as light, drought, high salinity, and pathogens (Quirino et al., 1999; Gepstein and Glick, 2013; Sakuraba et al., 2014, 2018). One informative approach for understanding leaf senescence mechanisms is the isolation and analysis of stay-green mutants, which retain greenness during the senescence phase, and early leaf-yellowing mutants, which senesce earlier than the wild type. To date, several stay-green and early leaf-yellowing mutants have been isolated in Arabidopsis (Arabidopsis thaliana) and crop plants, leading to the identification of many senescence-associated genes (SAGs) and senescence signaling cascades (Kusaba et al., 2013; Woo et al., 2019).
The NAC genes (NAM, ATAF1/2, and CUC2) encode plant-specific transcription factors (TFs) and are one of the largest TF families in plants, with 106 NAC genes in Arabidopsis and 149 genes in rice (Oryza sativa; Gong et al., 2004; Xiong et al., 2005; Nuruzzaman et al., 2010). The NAC proteins commonly harbor a highly conserved DNA binding domain in their N-terminal regions (Aida et al., 1997) but have diverse C-terminal regions, suggesting that the C-terminal regions may play a critical role in determining the specific cis elements that characterize the respective target genes of each NAC TF (Tran et al., 2010). NAC TFs are involved in multiple developmental processes, such as embryo and shoot meristem development (Souer et al., 1996), lateral root formation (Xie et al., 2000), phytohormone signaling (Zhu et al., 2015), and abiotic stress responses (Tran et al., 2004; Wu et al., 2009).
In addition to these roles, many of the NAC TFs in Arabidopsis and crop plants have been identified as positive or negative regulators of leaf senescence (Kim et al., 2016; Li et al., 2018). In Arabidopsis, ANAC002/ATAF1, ANAC016, ANAC019, ANAC029/NAC-LIKE, ACTIVATED BY AP3/PI (NAP), ANAC032, ANAC046, ANAC092/ORESARA1 (ORE1), ANAC057/ORE1 SISTER1 (ORS1), ANAC055, and ANAC072 promote senescence, and ANAC042/JUNGBRUNNEN1 (JUB1) and ANAC083/VND-INTERACTING2 (VNI2) inhibit senescence (Garapati et al., 2015; Takasaki et al., 2015; Kim et al., 2016; Oda-Yamamizo et al., 2016).
Among these senescence-associated NAC (senNAC) TFs, the TF activity of ANAC016 is posttranslationally regulated by the C-terminal transmembrane domain (TMD). Under normal growth conditions, ANAC016 localizes in the cytoplasm. However, under senescence-inducing conditions, such as light deprivation and drought stress, the cleavage of the TMD allows ANAC016 to translocate to the nucleus (Sakuraba et al., 2015a). There, ANAC016 activates its target genes associated with leaf senescence, ANAC029/NAP and STAY-GREEN1 (AtSGR1; Kim et al., 2013; Sakuraba et al., 2016a), and represses genes associated with drought stress response, such as ABA RESPONSIVE ELEMENTS BINDING PROTEIN1 (AREB1; Sakuraba et al., 2015a). Like ANAC016, the TF activities of some Arabidopsis TMD-containing NACs, ANAC053/NAC WITH TRANSMEMBRANE MOTIF1-LIKE4 (NTL4) and ANAC062/NTL6, are posttranslationally regulated by their C-terminal TMD, and they play important roles in leaf senescence and/or abiotic stress responses (Kim et al., 2012; Lee et al., 2012).
Considering the number of Arabidopsis senNACs identified, other crop plant species, such as rice and barley (Hordeum vulgare), also should have more than 10 senNACs, but there have not been many studies of senNACs in crop plants. OsNAP is one of the few characterized senNACs in rice. OsNAP-overexpressing transgenic rice showed an early leaf-yellowing phenotype, and an RNA interference line targeting OsNAP had delayed leaf senescence (Liang et al., 2014), indicating that OsNAP functions as a senescence-promoting TF. OsNAC2 was recently identified as another senescence-promoting TF, and it directly activates the expression of chlorophyll (Chl)-degradation genes, SGR and NON-YELLOW COLORING3 (NYC3), and abscisic acid (ABA) biosynthetic genes, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3 (OsNCED3) and ZEP1/ABA1 (encodes zeaxanthin epoxidase 1, which catalyzes epoxidation of zeaxanthin to yield violaxanthin, a substrate of NCED; Mao et al., 2017). By contrast, ONAC106 has been identified as a senescence-inhibiting TF. ONAC106 directly binds to the promoters of the Chl-degradation genes SGR and NYC1, and the expression of these genes is significantly down-regulated by ONAC106 overexpression (Sakuraba et al., 2015b). However, further identification and characterization of senNAC TFs in rice will be necessary to understand the regulatory mechanisms of leaf senescence.
In this study, we identified and characterized a rice senNAC TF, ONAC054, which is a TMD-containing NAC. ONAC054 was previously termed RICE DWARF VIRUS MULTIPLICATION1 (RIM1) and was shown to be a regulator of jasmonate signaling; its knockout mutants show semidwarf and root growth inhibition phenotypes in young seedlings (Yoshii et al., 2010). The regulatory functions of ONAC054 in leaf development are not well understood. Here, we characterize the expression of ONAC054 and report that ONAC054 activates genes associated with ABA signaling, indicating that ONAC054 plays an important role in ABA-induced leaf senescence in rice. Moreover, we examine the regulation of ONAC054 by cleavage of its C-terminal TMD and reveal a role for alternative splicing in regulation of ONAC054. Finally, we discuss the significance of this multilayered regulation of ONAC054 for ABA-induced leaf senescence in rice.
RESULTS
A Null Mutation of ONAC054 Delays Leaf Yellowing during Dark-Induced Senescence
Most of the senNAC genes are dramatically up-regulated during natural senescence (NS) and artificially induced leaf senescence, including dark-induced senescence (DIS; Kim et al., 2009, 2013; Sakuraba et al., 2015b). Like the typical senNACs, ONAC054 transcripts accumulated in the yellowing sectors (regions c and d; see Supplemental Figure 1A) of senescing leaves compared to the green sectors (regions a and b; Supplemental Figure 1A). ONAC054 expression significantly increased until 4 d of dark incubation (DDI; Supplemental Figure 1B), indicating that ONAC054 is a senNAC gene.
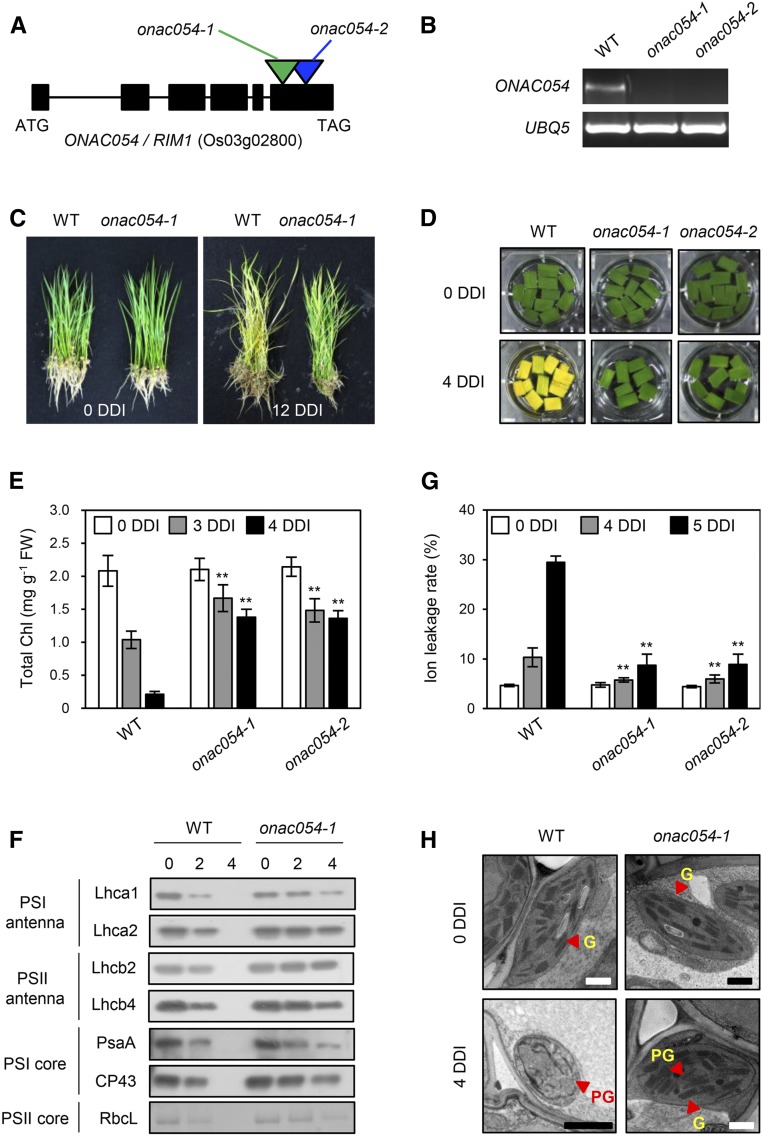
To investigate the function of ONAC054, we searched for a rice mutant in the RiceGE database (http://signal.salk.edu/cgi-bin/RiceGE). We found two independent T-DNA insertion lines, PFG_3A-07241 (designated as onac054-1) and PFG_3A-07240 (onac054-2), in which a single T-DNA fragment is inserted into different regions of the sixth exon of ONAC054 (Figure 1A). Reverse transcription-PCR analysis revealed that the ONAC054 transcript was absent in the leaves of the two T-DNA insertion lines (Figure 1B), indicating that they are both knockout mutants. A previous study reported that the onac054 knockout mutants (Japanese japonica cultivar ‘Nipponbare’ background) exhibited both semidwarf and root growth retardation phenotypes at the young seedling stage (Yoshii et al., 2010). As expected, we observed similar root phenotypes in the onac054-1 and onac054-2 mutants (hereafter referred to jointly as onac054 mutants; Supplemental Figure 2).
Figure 1.
The onac054 Knockout Mutants Show a Delayed Senescence Phenotype during Dark-Induced Senescence.
(A) Gene structure of ONAC054 and T-DNA insertion sites of two independent knockout lines of ONAC054.
(B) Knockout of ONAC054 in the onac054-1 and onac054-2 mutants was confirmed by RT-PCR analysis. UBQ5 was used as an internal control. WT, wild type.
(C) Plants at different DDI. The wild-type (WT) and onac054-1 mutants grown for 1 month under LD (14 h light per day) conditions were transferred to darkness at 28°C for 12 d.
(D) to (H) The changes in leaf color (D), total Chl level (E), photosystem protein levels (F), ion leakage rate (G), and chloroplast structure (H) in the leaf discs of 1-month-old wild type (WT) and onac054 mutants during dark incubation. Detached leaf discs were incubated on 3 mM MES (pH 5.8) buffer with the abaxial side up at 28°C in darkness and sampled at the specified DDI for each experiment. (F) Antibodies against PSI antenna (Lhca1 and Lhca2), PSII antenna (Lhcb2 and Lhcb4), PSI core (PsaA), and PSII core (CP43) proteins were used for immunoblot analysis. RbcL was detected by Coomassie Brilliant Blue staining.
(E) and (G) The mean and SD were obtained from more than five biological samples (one leaf disc each). Asterisks indicate a significant difference compared to the WT (Student’s t test, **P < 0.01).
(G) G, Grana thylakoid; PG, plastoglobule. Scale bars, 1 μm. These experiments were repeated twice with similar results.
We subsequently examined the phenotype of the onac054 mutants during DIS. For DIS, 4-week-old plants were transferred to complete darkness. At 12 DDI, the leaves of the wild type (the parental japonica cv Dongjin) turned yellow, while those of the onac054-1 mutant retained their green color (Figure 1C). The senescence phenotype of the onac054 mutants was also investigated using detached leaf discs. At 4 DDI, the leaf discs of onac054 mutants retained their green color, while those of the wild type turned completely yellow (Figure 1D). Consistent with the visible phenotype, total Chl levels remained high in the onac054 mutants during DIS (Figure 1E).
We further examined the stability of photosystem proteins during DIS by immunoblot analysis. At 4 DDI, all the photosystem proteins, including PSI antenna proteins (Lhca1 and Lhca2), PSII antenna proteins (Lhcb2 and Lhcb4), PSI core protein (PsaA), and PSII core protein (CP43), remained more stable in the onac054-1 leaves (Figure 1F). Moreover, the membrane ion leakage rate, an indicator of membrane disintegration, was much lower in the leaves of both onac054 mutants during DIS (Figure 1G). At 0 DDI, the chloroplasts of wild-type and onac054-1 leaves were intact, with highly stacked grana thylakoids, but at 4 DDI, the grana structure remained intact in the onac054-1 leaves, whereas it was hardly detected in the wild-type leaves (Figure 1H). Therefore, the null mutation of ONAC054 results in delayed leaf yellowing under DIS conditions.
onac054 Mutants Showed a Functional Stay-Green Phenotype during NS
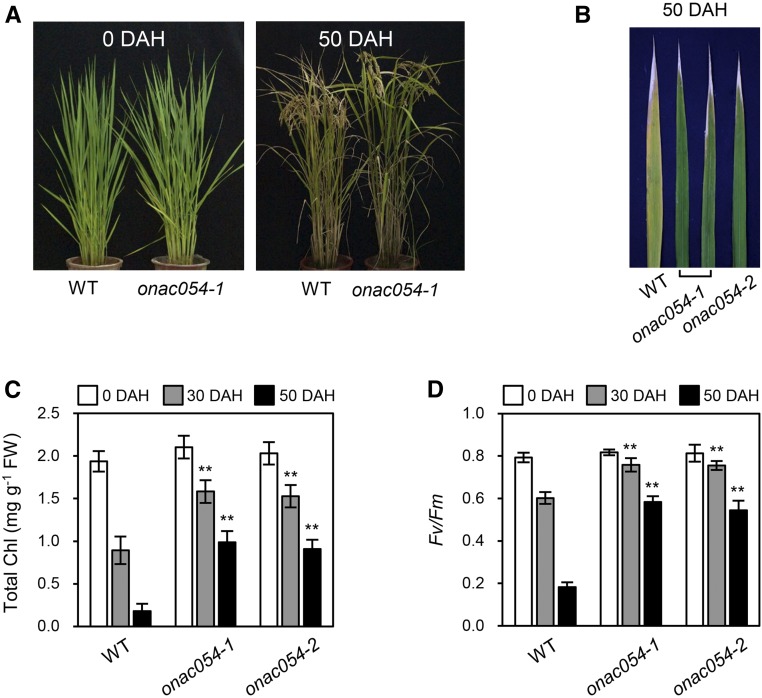
We subsequently examined the NS phenotype of onac054 mutants in the paddy field under natural long-day (NLD) conditions (≥14 h light/day at 37°N latitude, Suwon, South Korea). Throughout the vegetative stage, the growth rate and plant size of the onac054-1 mutant were not significantly different from those of the wild type (Figure 2A, left), and the heading dates of onac054 mutants were also very similar to the wild type (Supplemental Figure 3). During grain filling, however, the onac054-1 mutant had delayed leaf yellowing (Figure 2A, right), which was also observed in the onac054-2 leaves (Figure 2B). Consistent with their color, the leaves of onac054 mutants retained much higher total Chl levels, while the wild-type leaves showed a significant decrease in Chl levels (Figure 2C). To examine the efficiency of photosynthesis, we measured the Fv/Fm ratio (efficiency of PSII) after heading. The Fv/Fm ratio in the wild type drastically decreased at 50 d after heading, but much higher ratios were sustained in the onac054 mutants (Figure 2D), indicating that the mutation in ONAC054 results in delayed leaf senescence under NLD conditions in the field.
Figure 2.
onac054 Knockout Mutants Show a Delayed Senescence Phenotype in Natural Paddy Field Conditions.
(A) Phenotypes of the wild type (WT; the parental japonica rice cv Dongjin) and the onac054-1 mutant at 0 and 50 d after heading (DAH).
(B) Leaf colors of the wild type (WT) and onac054 mutants at 50 DAH. The third leaves of the main culm were used.
(C) and (D) Changes in total Chl level (C) and Fv/Fm ratio (D) in the leaves of the wild type (WT) and onac054 mutants during grain filling (0, 30, and 50 DAH). FW, Fresh weight. The means and SD were obtained from six biological replicates (one leaf disc each). These experiments were repeated twice with similar results. Asterisks indicate a significant difference compared to the wild type (Student’s t test, **P < 0.01).
Plants that retain the capacity to photosynthesize and thus to produce fixed carbon may produce greater yields of grain or biomass compared with plants that senesce earlier. Indeed, previous reports showed that some stay-green rice mutants show increased crop yield (Liang et al., 2014; Sakuraba et al., 2015b). To test whether this holds true for onac054, we examined several agronomic traits of onac054 mutants at 70 d after heading. We found that the number of panicles per plant and the number of grains per panicle in onac054 mutants were significantly higher than those in the wild type (Supplemental Figure 4). However, spikelet fertility of onac054 mutants was lower than the wild type (Supplemental Figure 4E). Thus, grain yield per plant of onac054 plants was almost the same as that of the wild type (Supplemental Figure 4F).
Nuclear Localization of ONAC054 Is Controlled by Cleavage of the C-Terminal TMD
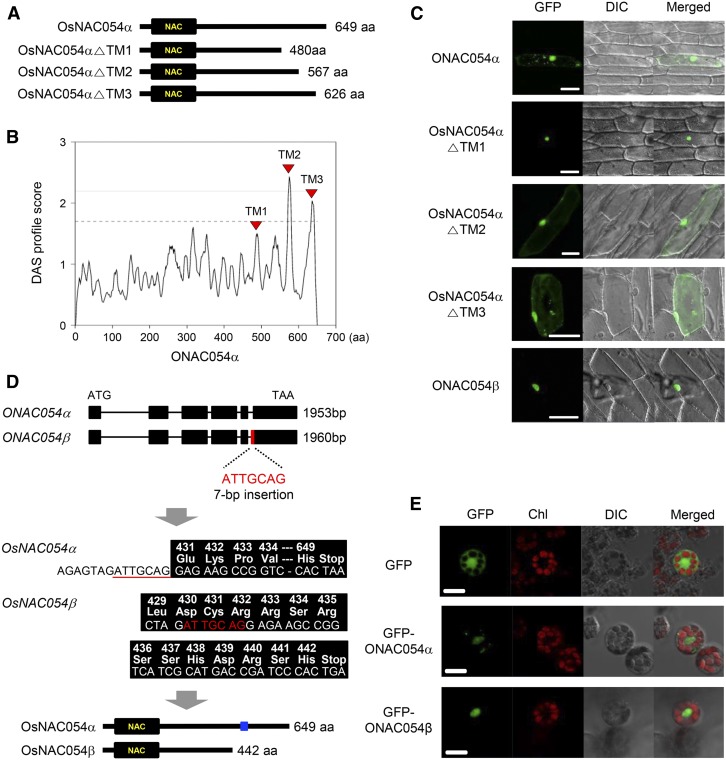
In many cases, the TMD-containing NAC TFs are much larger than the NAC TFs without a TMD (Kim et al., 2013). ONAC054 is a large protein (649 amino acids) compared with other ONAC TFs (Figure 3A); therefore, we examined whether the ONAC054 TF contains a TMD, which is important for the regulation of subcellular localization. First, we searched for a possible TMD using the Dense Alignment Surface transmembrane prediction server (https://tmdas.bioinfo.se/DAS/index.html). This identified three putative TMD regions, TM1 (496 to 502), TM2 (573 to 578), and TM3 (634 to 641; Figure 3B).
Figure 3.
The C-Terminal TMD Determines the Subcellular Localization of ONAC054 Proteins.
(A) Amino acid (aa) length of mature ONAC054 (ONAC054α), ONAC054△TM1(481-649), ONAC054△TM2(568-649), and ONAC054△TM3(627-649) proteins.
(B) Putative regions of the TMD in ONAC054 predicted by the Dense Alignment Surface (DAS) transmembrane prediction server (https://tmdas.bioinfo.se/DAS/index.html). aa, amino acid.
(C) Subcellular localization of GFP-ONAC054α, GFP-ONAC054△TM1, GFP-ONAC054△TM2, GFP-ONAC054△TM3, and GFP-ONAC054β examined by transient expression in onion epidermal cells. DIC, differential interference contrast. Scale bar, 10 μm.
(D) Gene structures of ONAC054α and ONAC054β. The insertion of 7 bp in the fifth intron of ONAC054β caused alternative 3′ splice site selection and a premature stop codon. Asterisks indicate the position of the premature stop codon. The blue box indicates the TM1 region of ONAC054α. aa, amino acid.
(E) Subcellular localization of GFP (control), GFP-ONAC054α, and GFP-ONAC054β examined by transient expression using protoplasts isolated from 10-d-old seedlings of rice. Scale bar, 10 μm. These experiments were repeated twice with similar results. Chl, chlorophyll.
To test which putative TMD is involved in the translocation of ONAC054, the subcellular localization of green fluorescent protein (GFP)-ONAC054△TM1(481-649), GFP-ONAC054△TM2(568-649), and GFP-ONAC054△TM3(627-649) fusion proteins was examined by transient expression assays in onion (Allium cepa) epidermal layer cells (Figure 3C). The full-length ONAC054 protein fused with GFP at the N terminus (GFP-ONAC054α) was localized both inside and outside of the nucleus (Figure 3C). The fluorescence outside of the nucleus overlapped with the fluorescence of ER-Tracker Red (Supplemental Figure 5), which specifically stains the endoplasmic reticulum (ER), indicating that some portion of intact ONAC054 proteins localized in the ER, similar to Arabidopsis TMD-containing NAC proteins (Ng et al., 2013). Similar to GFP-ONAC054, GFP-ONAC054△TM2 and GFP-ONAC054△TM3 were localized both inside and outside of the nucleus, but GFP-ONAC054△TM1 exclusively located in the nucleus (Figure 3C), indicating that cleavage of the TM1 region is required for the localization of ONAC054 in the nucleus.
In this study, we also found a variant ONAC054 transcript, in which seven nucleotides (ATTGCAG) are inserted between the fifth intron and the sixth exon (Figure 3D) by alternative 3′ splice site selection, one type of alternative splicing event (Syed et al., 2012). This insertion caused a frame shift in the protein, leading to a premature stop codon (Figure 3D). Thus, the protein encoded by the alternative 3′ splice site selection form of ONAC054 (hereafter referred to as ONAC054β) is much smaller (443 amino acids) than that of the standard ONAC054 (649 amino acids, hereafter referred to as ONAC054α; Figure 3D). Notably, ONAC054β lacks the TM1 region. Transient expression assays using onion epidermal layer cells showed that ONAC054β localized only in the nucleus, while ONAC054α localized both inside and outside of the nucleus (Figure 3C). Similar results were obtained from transient expression assays using protoplasts isolated from 10-d-old rice seedlings (Figure 3E). It is probable that the activity of ONAC054 depends on its nuclear localization, which is regulated by alternative 3′ splice site selection as well as external signal-induced TM1 cleavage of ONAC054α.
The ONAC054 TF Binds to the Mitochondrial Dysfunction Motif and Its Variants
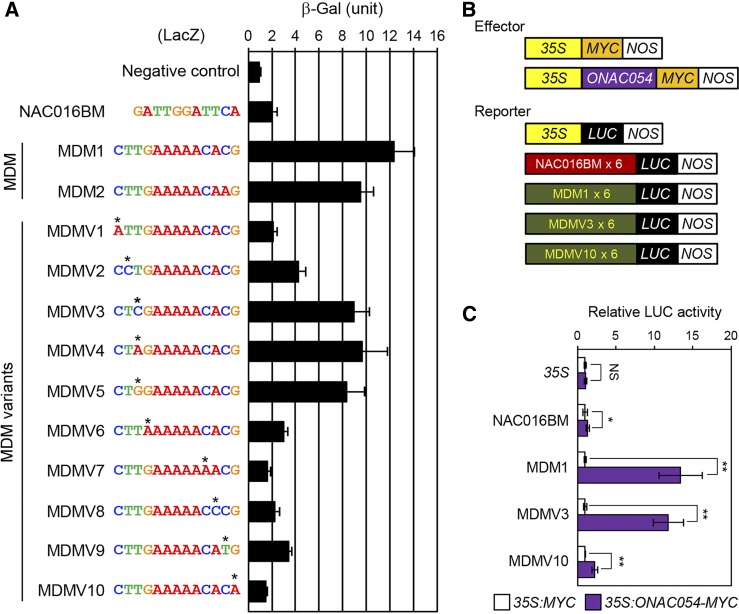
A previous study defined the 4-bp core sequence of the NAC binding motif as CACG (Olsen et al., 2005). However, some Arabidopsis TMD-containing NAC TFs specifically interact with the sequence CTTGXXXXXCA[C/A]G, named the mitochondrial dysfunction motif (MDM) (De Clercq et al., 2013). Arabidopsis NAC016 specifically binds to the NAC016 binding motif (NAC016BM) GATTGGAT[A/T]CA, which is similar to the MDM sequence (Sakuraba et al., 2015a). Therefore, it is possible that ONAC054 also binds the MDM, NAC016BM, or their variants.
To examine this hypothesis, we first determined whether ONAC054 interacts with the NAC016BM or MDM by yeast one-hybrid (Y1H) assays. The sequences of the NAC016BM and MDM were fused to the promoter of TUBULIN BETA CHAIN2 (TUB2), which does not contain those motifs, and ONAC054α was used as a bait. ONAC054 does not bind to the TUB2 promoter (Figure 4A, negative control). We found that ONAC054 strongly binds to the two types of MDMs, MDM1 (CTTGAAAAACACG) and MDM2 (CTTGAAAAACAAG), but not to the NAC016BM (Figure 4A). We subsequently examined whether ONAC054 also interacts with the MDM variants (MDMV) by point mutation analysis of the MDM in Y1H assays. We found that point mutations in the first (C), fourth (G), and 10th to 13th (CACG) nucleotides of the MDM significantly decreased the binding capacity of ONAC054, while mutations in the second (T) and third (T) nucleotides did not significantly reduce the binding efficiency, indicating that ONAC054 has binding capacity for the MDM, but the second and third nucleotides of the MDM are relatively flexible for interacting with ONAC054.
Figure 4.
ONAC054 Binds to the MDM and Its Variants.
(A) Interaction of ONAC054 with the NAC16BM, MDM, and its substituted sequences (MDMV) in Y1H assays. The sequences of interest were fused to a promoter fragment of TUB2 that does not contain a NAC016BM and MDM. β-Galactosidase (β-Gal) activity was measured by liquid assays (see “Methods”).
(B) and (C) The effect of ONAC054 expression on the activity of NAC016BM, MDM, and MDM variants in rice protoplasts. LUC was fused to the 6× tandem NAC016BM, MDM1, MDMV3, and MDMV10, and the ONAC054 overexpression vector or empty vector was cotransfected in the protoplasts. The 35S promoter was used as a negative control.
(A) and (C) The mean and SD values were obtained from more than four reaction mixtures. Asterisks indicate a significant difference compared to the wild type (Student’s t test, *P < 0.05 and **P < 0.01). These experiments were repeated twice with similar results.
To evaluate whether binding to the MDM is sufficient for direct regulation of the genes targeted by ONAC054, we performed transient expression assays using rice protoplasts. In this experiment, the reporter plasmid contained the six tandem repeats of the NAC016BM, MDM1, MDMV3, and MDMV10 fused with the Cauliflower mosaic virus 35S promoter located upstream of the luciferase (LUC) reporter gene (Figure 4B). The experiment revealed that ONAC054 (ONAC054α) strongly activates the MDM1 and MDMV3, in which the third nucleotide of the MDM is substituted, but not the NAC016BM, MDMV10, or the 35S control promoter (Figure 4C). Thus, it is possible that ONAC054 acts as a transcriptional activator by binding to the MDM or MDM variants in the promoters of its target genes.
SAGs Are Differentially Expressed in onac054 Mutants during Leaf Senescence
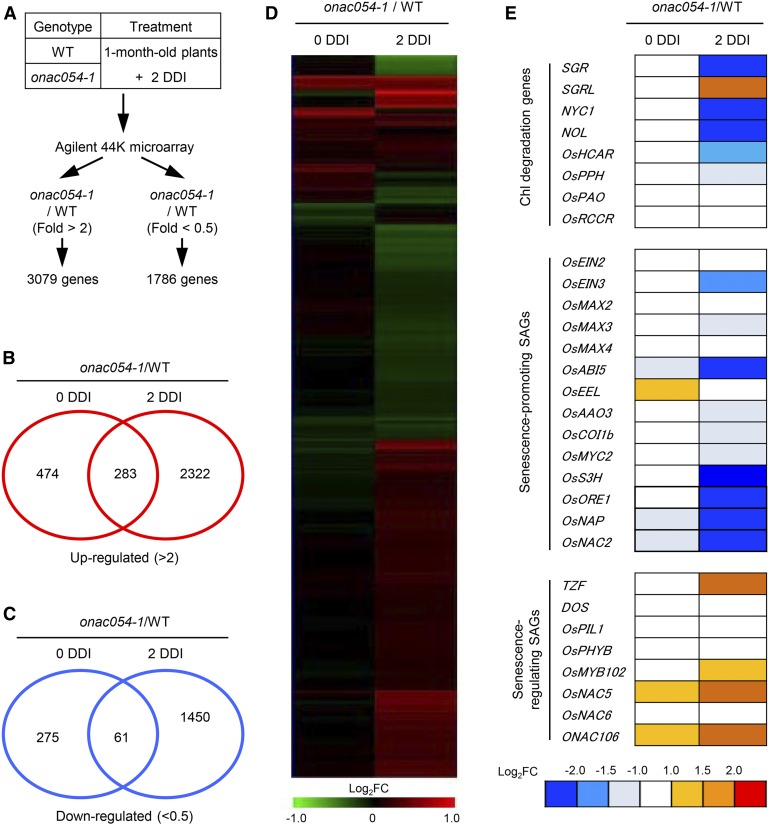
To identify differentially expressed genes (DEGs) in onac054 mutants compared with the wild type and the gene regulatory network of ONAC054-mediated leaf senescence, we performed a genome-wide microarray analysis. Detached leaf discs from 1-month-old wild-type and onac054-1 plants were incubated in the dark for 2 d (2 DDI), and their RNAs were extracted for microarray analysis (Figure 5A). We identified 757 and 2605 genes that were significantly up-regulated (onac054-1/WT; >2-fold) and 336 and 1511 genes that were down-regulated (onac054-1/WT; <0.5-fold) at 0 and 2 DDI, respectively (Figures 5B and 5C). The up- and down-regulated genes included in the Venn diagrams are listed in Supplemental Data Sets 1 and 2. The hierarchical average linkage cluster analysis showed that more DEGs were detected at 2 DDI (Figure 5D), suggesting a significant regulatory role of ONAC054 during leaf senescence.
Figure 5.
Altered Expression of SAGs in the onac054-1 Mutant during DIS.
(A) A summary of the microarray analysis. A filter for Student’s t test P value of < 0.05 was applied to DEGs. WT, wild type.
(B) and (C) Venn diagrams of the number of DEGs in the onac054-1 mutant compared to the wild type (WT). The numbers of up-regulated (B) and down-regulated genes (C) in the onac054-1 mutant at 0 and 2 DDI are shown.
(D) Hierarchical average linkage clustering of genes differentially expressed between wild-type (WT) and onac054-1 plants.
(E) The ratios of expression levels (onac054-1/wild type [WT]) for known or putative SAGs at 0 and 2 DDI are illustrated. The relative expression value was normalized to the WT expression level.
In the microarray data set, we first examined whether SAGs are differentially expressed in the onac054-1 mutant compared with the wild type (onac054-1/WT). We found that genes related to Chl degradation, such as SGR, NYC1, and NYC1-LIKE(NOL), were down-regulated in the onac054-1 mutant (Figure 5E). Among the SAGs, OsABI5, encoding a basic LEUCINE-ZIPPER)-type (bZIP) TF in ABA signaling (Zou et al., 2008), and SALICYLIC ACID 3-HYDROXYLASE (OsS3H), encoding a salicylic acid synthesis enzyme (Zhang et al., 2013), were down-regulated (Figure 5E). Among the NAC TFs, OsNAP and OsNAC2, which promote leaf senescence (Liang et al., 2014; Mao et al., 2017), were down-regulated, while ONAC106, encoding a senescence-inhibiting NAC TF (Sakuraba et al., 2015b), was up-regulated in the onac054-1 mutant (Figure 5E). However, the expression of some SAGs, such as PHYTOCHROME INTERACTING FACTOR 3-LIKE1 (OsPIL1; Sakuraba et al., 2017) and Chl-degradation genes (OsPAO and OsRCCR), was not altered in the onac054-1 mutant, even under DIS conditions (Figure 5E). Our findings suggest that the lack of ONAC054 activity caused large-scale changes of SAG expression during leaf senescence.
ONAC054 Up-regulates Genes Associated with ABA-Induced Leaf Senescence
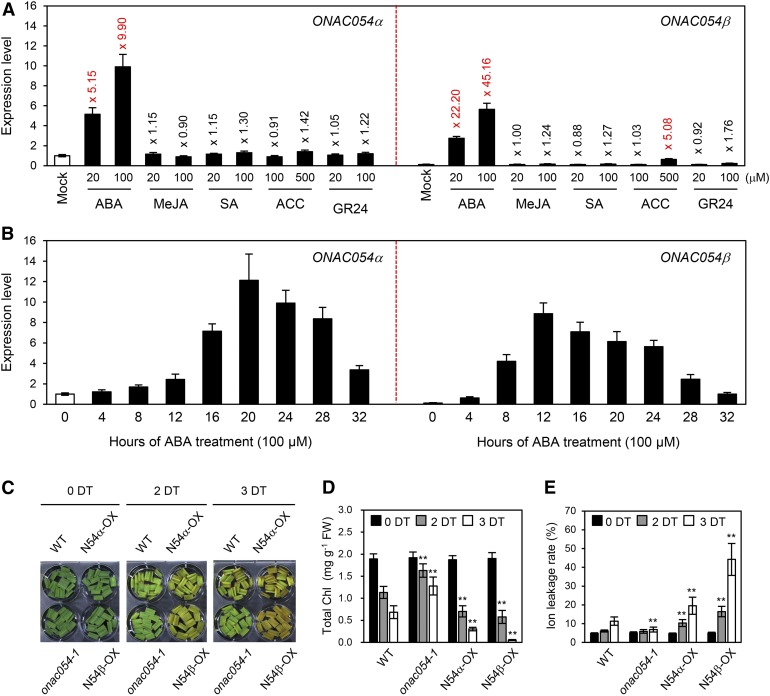
In the microarray analysis, we further found that many genes associated with phytohormone signaling and metabolism were differentially expressed in the onac054-1 mutant, especially at 2 DDI (Supplemental Figure 6). Thus, we examined the expression of ONAC054 in response to treatments with the senescence-promoting phytohormones ABA, methyl jasmonate, salicylic acid, 1-aminocyclopropane-1-carboxylic acid (ACC; an ethylene intermediate), and GR24 (a strigolactone analog). In this experiment, we checked the levels of both the ONAC054α and ONAC054β transcripts to determine if the expression of the two forms differs in response to phytohormone treatments. At 24 h of ABA treatment, the levels of both ONAC054α and ONAC054β were significantly increased compared with the untreated control (Figure 6A). ONAC054β was induced much more strongly by ABA; ONAC054β increased to around 45-fold, but ONAC054α increased to around 10-fold at 24 h of ABA treatment (100 μM; Figure 6A). Notably, ONAC054β was also induced by a high concentration of ACC (500 μM), whereas ONAC054α was not induced by any of the other phytohormones (Figure 6A). To examine the ABA-induced expression of ONAC054α and ONAC054β in more detail, we checked the levels of the ONAC054α and ONAC054β transcripts during ABA treatment (100 μM) at 4-h intervals for a total of 32 h of ABA treatment. Interestingly, ONAC054β levels increased much faster than those of ONAC054α; ONAC054β levels peaked at 12 h, but ONAC054α peaked at 20 h of ABA treatment (Figure 6B), indicating that ONAC054β is closely associated with the early ABA response. We further checked the levels of the ONAC054α and ONAC054β transcripts in senescing leaves. Compared with their levels in green sectors of leaves (regions a and b), the levels of both ONAC054α and ONAC054β significantly increased in the yellowing sectors (regions c and d), but the ONAC054β levels increased faster and were the highest in the region c, while the ONAC054α levels were the highest in region d (Supplemental Figure 7).
Figure 6.
ONAC054 Promotes ABA-Induced Leaf Senescence.
(A) Expression patterns of ONAC054α and ONAC054β during treatment with phytohormones. Detached leaf discs from 1-month-old wild-type plants were floated on 3 mM MES buffer (pH 5.8) containing ABA (20 or 100 μM), methyl jasmonate (MeJA; 20 or 100 μM), SA (20 or 100 μM), ACC (500 or 1000 μM), or GR24 (20 or 100 μM) and incubated for 24 h under continuous light. The mean and SD values were obtained from four biological samples (three leaf discs each).
(B) Expression patterns of ONAC054α and ONAC054β during ABA treatment (100 μM) were checked every 4 h for 32 h after the start of ABA treatment. The mean and SD values were obtained from four biological samples (three leaf discs each).
(C) to (E) Changes of leaf color (C), total Chl level (D), and ion leakage rate (E) in wild-type (WT), onac054-1, 35S:GFP-ONAC054α (N54α-OX in figures), and 35S:GFP-ONAC054β (N54β-OX in figures) plants after 2 and 3 d of treatment (DT) with 100 μM ABA.
(D) and (E) Black, gray, and white bars indicate 0, 2, and 3 d of treatment with ABA, respectively. The mean and SD values were obtained from six biological samples (four leaf discs each). Asterisks indicate a significant difference compared to the WT (Student’s t test, **P < 0.01). These experiments were repeated at least twice with similar results.
To understand the biological roles of ONAC054α and ONAC054β, we generated rice transgenic plants overexpressing ONAC054α or ONAC054β fused with GFP at the N terminus (hereafter referred to as the ONAC054α-OX and ONAC054β-OX lines, respectively). The level of GFP-ONAC054α in the ONAC054α-OX transgenic lines was more than 15-fold higher than that of ONAC054α in the wild type (Supplemental Figure 8A), and that of GFP-ONAC054β in the ONAC054β-OX transgenic lines was more than 25-fold higher than that of ONAC054β in the wild type (Supplemental Figure 8B). We also found by RT-qPCR analysis that the expression of ONAC054β in the two onac054 mutants is considerably lower than that in the wild type (Supplemental Figure 9). We examined ABA-induced leaf senescence in the leaves of wild-type, onac054-1, ONAC054α-OX, and ONAC054β-OX plants. Rosette leaf discs from 1-month-old wild-type, onac054-1, ONAC054α-OX, and ONAC054β-OX plants were all green before ABA treatment; however, the leaves of the ONAC054α-OX and ONAC054β-OX lines turned yellow much faster, with a dramatic decrease in Chl levels, during ABA treatment (100 μM), while those of the onac054-1 mutant retained their green color and Chl levels (Figures 6C and 6D). Consistent with the changes in color, the ion leakage rates of leaf discs dramatically increased in both ONAC054α-OX and ONAC054β-OX lines but was much lower in the onac054-1 mutant under ABA treatment (Figure 6E). Notably, leaf yellowing of the ONAC054β-OX line was faster than that of the ONAC054α-OX line (Figures 6C to 6E), probably due to differences in ONAC054α and ONAC054β transcript levels. Similar early leaf-yellowing phenotypes of ONAC054α-OX and ONAC054β-OX plants were observed during DIS (Supplemental Figure 10), indicating that ONAC054 up-regulates ABA-induced leaf senescence.
We subsequently examined whether the C-terminal TMD of ONAC054 is cleaved in response to ABA treatment. First, we confirmed the accumulation of constitutively expressed GFP-ONAC54 proteins in leaf tissues under normal conditions, differentiating the GFP-ONAC54 proteins by their respective molecular weights (Supplemental Figure 11). Under ABA treatment or dark incubation, the molecular weight of GFP-ONAC54α was reduced to a similar molecular weight as that of GFP-ONAC54β. This indicates that the C-terminal TMD of ONAC54α is cleaved by proteolytic processing under ABA treatment and dark incubation.
ONAC054 Up-regulates OsABI5 and NYC1
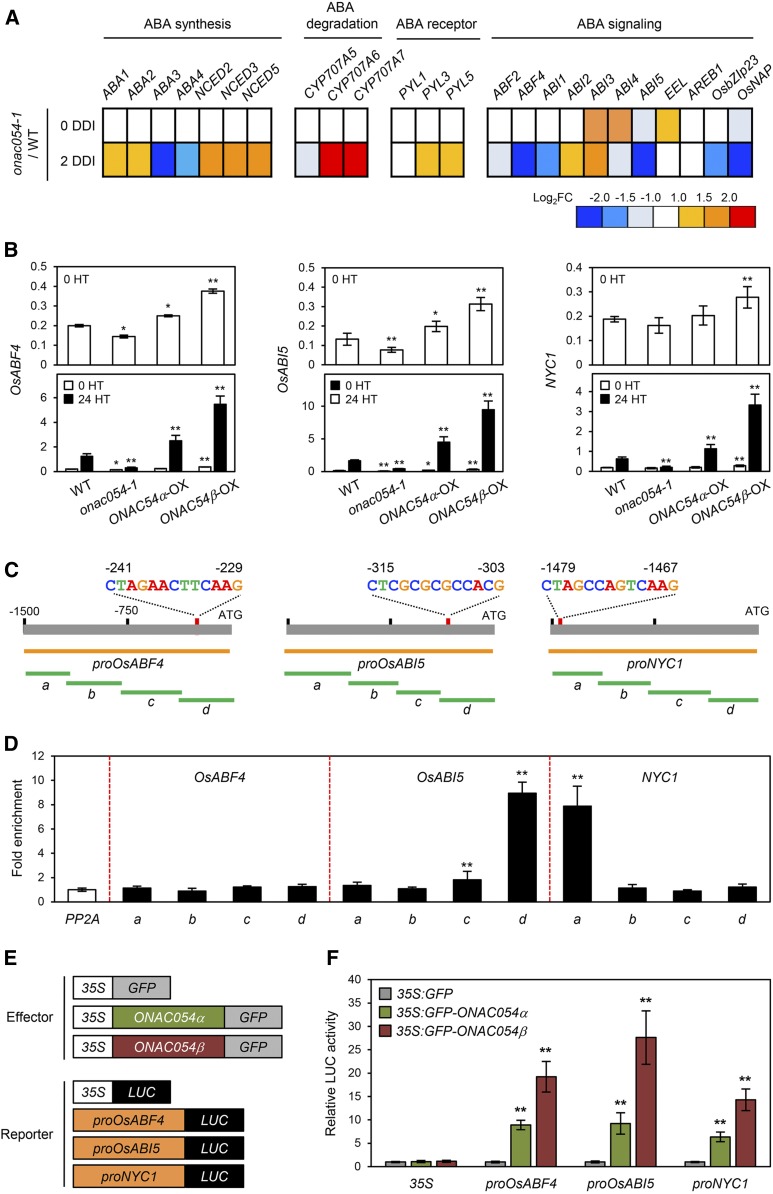
To examine how ONAC054 promotes ABA-induced leaf senescence, we first checked the expression patterns of genes associated with ABA metabolism and ABA signaling in the onac054-1 mutant during DIS, obtained from the microarray analysis. Among the genes associated with ABA metabolism, ABA DEFICIENT3 (OsABA3) and OsABA4, encoding ABA biosynthesis enzymes (Schwartz et al., 1997; North et al., 2007), were down-regulated in the onac054-1 mutant at 2 DDI (Figure 7A). By contrast, among the genes associated with ABA signaling, we found that in addition to OsNAP, several bZIP TFs (OsABF4, OsABI5, and OsbZIP23) were significantly down-regulated in the onac054-1 mutant during DIS (Figure 7A). As shown in Figure 6C, onac054-1 leaves retained their green color, while the leaves of ONAC054α-OX and ONAC054β-OX showed an early-senescence phenotype during ABA treatment. In addition, we measured ABA contents in wild-type, onac054-1, and onac054-2 leaves and found that ABA contents of onac054 mutants were almost the same as the wild type at 0 DDI, although the ABA content of the wild type at 3 DDI was higher than that of onac054-1 (Supplemental Figure 12). Therefore, we focused on the relationship between ONAC054 and ABA signaling-related genes. To this end, we checked the expression patterns of ABA signaling-associated genes (OsABF4, OsABI5, OsbZIP23, and OsNAP) in the leaves of wild-type, onac054-1, ONAC054α-OX, and ONAC054β-OX plants before and after 24 h of treatment with ABA (100 μM). In addition, we checked the expression patterns of several SAGs (SGR, NYC1, NOL, OsORE1, and OsNAC2) that were significantly down-regulated in onac054-1 leaves during DIS. Before ABA treatment, the expression levels of OsABF4, OsABI5, and OsNAP were down-regulated in the onac054-1 mutant, and several genes examined were slightly up-regulated in the ONAC054β-OX line (Figure 7B; Supplemental Figure 13). After 24 h of ABA treatment, all the genes examined were down-regulated in the onac054-1 mutant, and they were up-regulated in the ONAC054α-OX and ONAC054β-OX lines, although more strongly in ONAC054β-OX. Consistent with this, we found that these genes were down-regulated in the onac054-2 mutant during ABA treatment (Supplemental Figure 14), indicating a strong association between expression levels of ONAC054 and the downstream genes examined.
Figure 7.
ONAC054 Directly or Indirectly Activates OsABF4, OsABI5, and NYC1.
(A) The ratios of expression levels (onac054-1/wild type [WT]) for genes associated with ABA metabolism and signaling at 0 and 2 DDI obtained from the microarray analysis are illustrated. The relative expression value was normalized to the WT expression level.
(B) Expression patterns of OsABF4, OsABI5, and NYC1 in wild-type (WT), onac054-1, ONAC054α-OX (N54α-OX), and ONAC054β-OX (N54β-OX) plants before and after 24 h of treatment with ABA. Detached leaf discs from 1-month-old rice seedlings were used for RT-qPCR analysis. The mean and SD values were obtained from four biological samples (three leaf discs each). HT, hour(s) of treatment.
(C) The positions of the MDM variants in the promoters of OsABF4, OsABI5, and NYC1, and promoter fragments used for the ChIP assay (green horizontal lines) and transactivation assay (orange horizontal lines).
(D) ONAC054β binding affinity to the promoter regions of OsABF4, OsABI5, and NYC1 in planta examined by ChIP assays. GFP-ONAC054β was transiently expressed in protoplasts isolated from 10-d-old wild-type rice seedlings. Fold enrichment of the promoter fragments was measured by immunoprecipitation with an anti-GFP antibody (see Methods). Enrichment of a promoter region of an unrelated gene, PP2A, was used as a negative control. The mean and SD values were obtained from four reaction mixtures.
(E) Reporter and effector constructs used in the protoplast transient assay. Each construct also contained the NOS terminator.
(F) The activation of OsABF4, OsABI5, and NYC1 promoters (−1500 to −1, relative to the start codon) by GFP-ONAC054α and GFP-ONAC054β expression in protoplasts that were incubated in darkness. The 35S promoter and an empty expression vector (35S:GFP) served as negative controls. The mean and SD values were obtained from five reaction mixtures. Asterisks indicate a significant difference compared to the wild type or negative controls (Student’s t test, *P < 0.05 and **P < 0.01). These experiments were repeated at least twice with similar results.
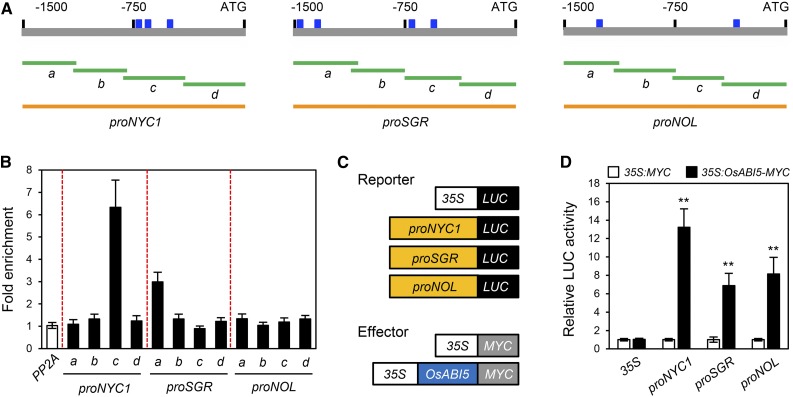
We subsequently checked whether the promoters (−1500 to −1 bp from the ATG start site) of the nine genes examined above (OsABF4, OsABI5, OsbZIP23, OsNAP, SGR, NYC1, NOL, OsORE1, and OsNAC2) harbor the MDM or MDM variants. Among these promoters, the promoters of OsABF4, OsABI5, and NYC1 harbor the MDM variants CTAGAACTTCAAG, CTCGCGCGCCACG, and CTAGCCAGTCAAG, respectively (Figure 7C), while the other promoters do not contain the MDM or MDM variants. Therefore, we further analyzed the relationship between ONAC054 and OsABF4, OsABI5, and NYC1 expression. First, we examined direct binding of ONAC054 to the promoter regions of three genes in vivo by a chromatin immunoprecipitation (ChIP) assay using rice protoplasts transiently expressing GFP-ONAC054α. Region “d” of OsABI5 and region “a” of NYC1, which include the MDM variants (see Figure 7C), were highly enriched in the immunoprecipitate, while other regions showed no enrichment (Figure 7D).
We subsequently performed protoplast transient assays to determine the effect of ONAC054 on the transcript levels of OsABF4, OsABI5, and NYC1. For the reporter constructs, the promoters of OsABF4, OsABI5, and NYC1 were fused with the LUC reporter, and for the effector constructs, both GFP-ONAC054α and GFP-ONAC054β were used (Figure 7E). We found that both forms of ONAC054 activated the promoters of OsABF4, OsABI5, and NYC1, and ONAC054β had a stronger effect. However, neither form of ONAC054 activated the 35S control promoter (Figure 7F). These results indicate that ONAC054 directly up-regulates the expression of OsABI5 and NYC1 and indirectly up-regulates OsABF4.
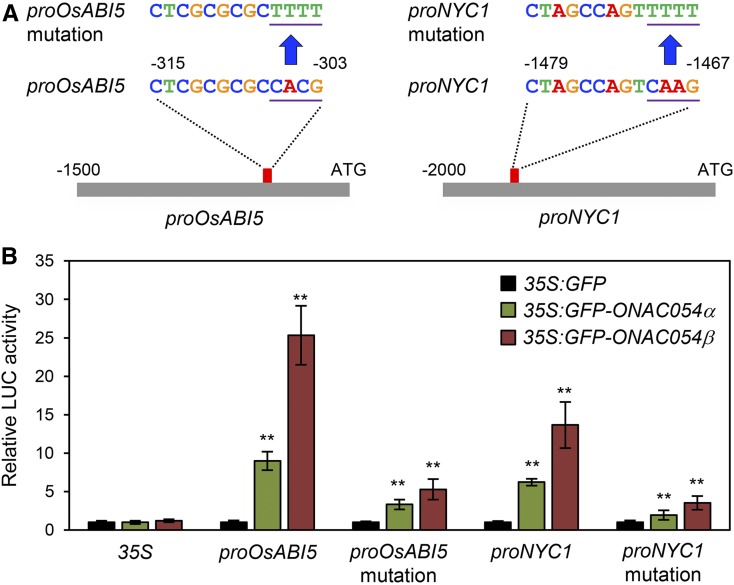
To determine the exact binding site of ONAC054 in the promoters of OsABI5 and NYC1, we used protoplast transient assays to examine whether ONAC054 could activate the promoters of OsABI5 and NYC1 in which the MDM variants (CTCGCGCGCCACG and CTAGCCAGTCAAG) were substituted to CTCGCGCGTTTT and CTAGCCAGTTTTT, respectively (Figure 8A). We found that the mutated promoters of OsABI5 and NYC1 were not strongly activated by cotransfection with GFP-ONAC054α or GFP-ONAC054β (Figure 8B), indicating that ONAC054 activates the transcription of OsABI5 and NYC1 by directly binding to the MDM sequences in their promoter regions.
Figure 8.
The MDM Variants in the Promoters of OsABI5 and NYC1 Are Essential for Transcriptional Activation by ONAC054.
(A) The 10th to 13th nucleotides of OsABI5 and NYC1 were substituted by TTTT.
(B) The activation of OsABI5 and NYC1 promoters (−1500 to −1, relative to the start codon) and their variants, in which the 10th to 13th nucleotides of the MDM were substituted by TTTT, by GFP-ONAC054α and GFP-ONAC054β expression in protoplasts that were incubated in darkness. The 35S promoter and an empty expression vector (35S:GFP) served as negative controls. The mean and SD values were obtained from four reaction mixtures. Asterisks indicate a significant difference compared to the negative controls (Student’s t test, **P < 0.01). These experiments were repeated at least twice with similar results.
OsABI5 Also Directly Up-regulates SGR and NYC1
The Arabidopsis ABI5 TF promotes leaf senescence by directly up-regulating AtSGR1 and AtNYC1 (Sakuraba et al., 2014). Additionally, we recently revealed that OsABF4 directly up-regulates SGR and NYC1 (Piao et al., 2019). Thus, it is highly possible that OsABI5 directly activates other Chl-degradation genes. In our microarray analysis, three rice Chl-degradation genes (NYC1, SGR, and NOL) were down-regulated in the onac054-1 mutant during DIS and ABA treatment (Figure 5D; Supplemental Figure 9), and we found that the promoters of the three genes contain an ABA-responsive element (ABRE) consensus sequence (ACGTG; Figure 9A) in many ABA-inducible genes (Busk and Pagès, 1998). Therefore, we focused on the transcriptional relationship between ONAC054 and these three Chl-degradation genes.
Figure 9.
OsABI5 Directly Activates the Transcription of NYC1 and SGR.
(A) The positions of the ABRE binding motif (ACGTG) in the promoters of NYC1, SGR, and NOL and promoter fragments used for the ChIP assay (green horizontal lines) and transactivation assay (orange horizontal lines).
(B) Binding affinity of OsABI5 to the promoters of NYC1, SGR, and NOL in planta examined by ChIP assays. OsABI5-MYC was transiently expressed in protoplasts isolated from 10-d-old wild-type rice seedlings. Fold enrichment of the promoter fragments was measured by immunoprecipitation with an anti-MYC antibody (see “Methods”). Enrichment of a promoter region of an unrelated gene, PP2A, was used as a negative control. The mean and SD values were obtained from four biological samples.
(C) Reporter and effector constructs used in the protoplast transient assay. Each construct also contained the NOS terminator.
(D) The activation of NYC1, SGR, and NOL promoters (−1500 to −1, relative to the start codon) by OsABI5-MYC expression in protoplasts that were incubated in darkness. The 35S promoter and an empty expression vector (35S:MYC) served as negative controls. The mean and SD values were obtained from five reaction mixtures. Asterisks indicate a significant difference compared to the negative control (Student’s t test, **P < 0.01). These experiments were repeated twice with similar results.
First, we examined whether OsABI5 directly interacts with the promoters of SGR, NYC1, and NOL using ChIP assays with rice protoplasts transiently expressing OsABI5-MYC. We found that region “a” of SGR and region “c” of NYC1, which include ABRE sequences (see Figure 9A), were highly enriched in the immunoprecipitates, while the other regions showed no significant enrichment (Figure 9B). We next performed protoplast transient assays to determine the activity of OsABI5 for the transcription of NYC1, SGR, and NOL. For the reporter constructs, we used SGR, NYC1, and NOL fused with the LUC reporter gene (Figure 9C). We found that the LUC activities from the proSGR:LUC, proNYC1:LUC, and proNOL:LUC transgenes significantly increased when cotransfected with 35S:OsABI5-MYC (Figure 9D). In conclusion, it is probable that OsABI5 participates in the promotion of leaf senescence by directly up-regulating SGR and NYC1 and indirectly up-regulating NOL.
DISCUSSION
Multilayered Mechanisms Regulating ONAC054 Activity
In this study, we found that ONAC054, a senNAC TF in rice, plays an important role in the promotion of leaf senescence by up-regulating ABA signaling: onac054 mutants showed a delayed leaf-yellowing phenotype and ONAC054-OX plants showed an early leaf-yellowing phenotype during DIS (Figure 1; Supplemental Figure 10) and NS (Figure 2) and under ABA treatment (Figure 6). In the phylogenetic tree of 139 NACs in rice and 105 NACs in Arabidopsis, the amino acid sequence of ONAC054 is phylogenetically distant from those of other rice senNACs, such as OsNAP/ONAC058, OsNAC2, and ONAC106, and Arabidopsis senNACs (Supplemental Figure 15; Supplemental Data Set 3). This phylogeny suggests that ONAC054 represents a different class of senNACs from known senNACs.
Our observations indicate that the activity of ONAC054 is regulated at multiple steps. First, ONAC054 has a TMD at the C terminus, which determines its subcellular localization (Figure 3). We found that under normal growth conditions, GFP-fused ONAC054 proteins localize in the ER and the nucleus (Supplemental Figure 5). Under ABA treatment, however, the TMD of the native form of ONAC054, ONAC054α, is removed by currently unidentified proteases and then most of the processed ONAC054 proteins are transported into the nucleus (Supplemental Figure 11). Similarly, the cleavage of the TMD and translocation of an Arabidopsis NAC TF, NTL6/ANAC062, to the nucleus are regulated by Snf1-related protein kinase (SnRK2.8)-mediated phosphorylation. Since SnRK2.8 is activated by ABA treatment or under dehydration conditions, ABA or abiotic stress may induce the proteolysis of the C-terminal TMD of NTL6 (Seo and Park, 2010; Kim et al., 2012). Similar to the case of ONAC054 and NTL6, the activity of many TMD-containing TFs is regulated by proteolytic processing in response to specific environmental cues; thus, many plant TMD-containing TFs play important roles in the response to environmental stresses (Liu et al., 2007; Kim et al., 2012; Lee et al., 2012; Ng et al., 2013; Sakuraba et al., 2015a).
Splicing is a second step at which ONAC054 activity is regulated. ONAC054 has an alternative splice form, ONAC054β, in which selection of an alternative 3′ splice site causes the insertion of a 7-bp sequence, ATTGCAG, in the sixth exon, thus producing a premature stop codon. As a result, ONAC054β is much smaller (443 amino acids) than ONAC054α (649 amino acids) and lacks the C-terminal TMD (Figure 3D). Our transient expression assays using onion epidermal layer cells and protoplasts from rice seedlings revealed that all expressed GFP-ONAC054β localizes exclusively in the nucleus (Figures 3C and 3E) since it lacks the TMD at the C terminus. Previous studies have reported similar mechanisms of TMD-containing TF activity mediated by alternative splicing. For example, the Arabidopsis TMD-containing bZIP TF, bZIP60, has an alternative splice form, in which 23 nucleotides are absent, which causes a frameshift that results in a short C-terminal region without a TMD (Nagashima et al., 2011). However, studies of alternative splicing-mediated regulation of TMD-containing TFs are still limited, and the importance of such mechanisms is not well understood.
In this study, we found that both ONAC054α and ONAC054β significantly increased in response to ABA treatment, but their expression patterns were somewhat different, as the transcript levels of ONAC054β increased much faster than those of ONAC054α (Figures 6A and 6B). It is not clear why the expression patterns of ONAC054α and ONAC054β differ under ABA treatment. One possibility is that the different 3′ untranslated region (UTR) sequence of ONAC054β, caused by the frameshift and following the insertion of a premature stop codon, results in a different expression pattern. The cis elements responsible for gene expression can be located downstream of a coding sequence. For example, the sequence downstream of the stop codon of NIA1, encoding a nitrate reductase, contains nitrate-responsive cis elements that are required for the nitrate-dependent induction of NIA1 (Konishi and Yanagisawa, 2011). The 3′ UTR sequence is sometimes important for mRNA stability. Various sequence elements are required for mRNA degradation and are enriched among short- and long-lived transcripts (Narsai et al., 2007). THIAMINC (THIC), encoding a protein involved in thiamine biosynthesis, has two splicing forms with different lengths of 3′ UTR caused by alternative splicing, and their expression levels significantly differ as the 3′ UTR extension produced by alternative splicing leads to a decrease of mRNA stability (Wachter et al., 2007). Based on these results, we speculate that the stability of ONAC054α and ONAC054β mRNAs is regulated at the posttranscriptional level.
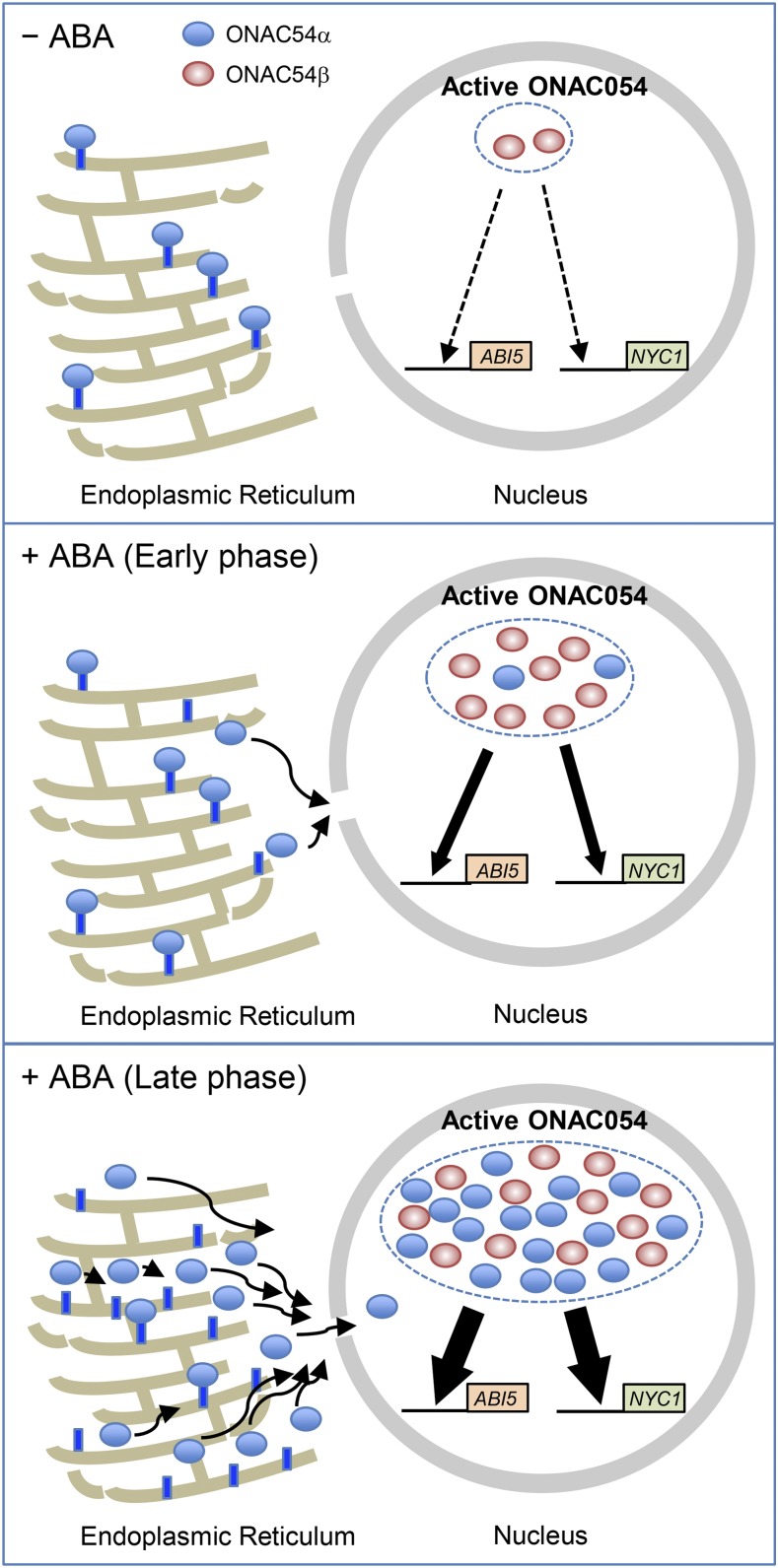
Based on our findings, we propose a tentative regulatory model of ONAC054 activity (Figure 10). Under normal growth conditions, the expression levels of ONAC054α and ONAC054β are low (Supplemental Figure 1; Figure 6A), which probably causes low accumulation of ONAC054 proteins. In addition, the TMD of ONAC054α is not cleaved under non-stress conditions. Therefore, in this situation, the accumulation of the active form of ONAC054 in the nucleus is low (Figure 10, top). At the beginning of the ABA treatment, ONAC054β expression dramatically increases, but ONAC054α increases more slowly (Figure 6B). Thus, in the early phase of ABA treatment, the majority of the active form of ONAC054 is probably ONAC054β. In contrast with ONAC054α, ONAC054β can be transported into the nucleus as soon as it is translated. Therefore, the induction of ONAC054β is probably necessary for the prompt response to increasing ABA concentration. In the late phase of ABA treatment, the levels of both ONAC054α and ONAC054β increase dramatically (Figure 6B). Furthermore, the TMD of most ONAC054α proteins is cleaved after ABA treatment (Supplemental Figure 11); thus, the active ONAC054 pool now includes both ONAC054β and ONAC054α△TM at high levels, leading to the strong induction of ABA-responsive genes, such as OsABI5 and OsABF4 (Figure 7). Collectively, the activity of ONAC054 is regulated at the transcriptional, posttranscriptional, and posttranslational levels. This multilayered regulation of TMD-containing TFs will be a model case for studying the rapid and proper expression of downstream genes in response to environmental stimuli. It is still unclear how ABA induces the transcription of ONAC054α and ONAC054β, although the promoter of ONAC054 contains an ABRE motif. Elucidation of the regulatory mechanism for the induction of ONAC054α and ONAC054β in the presence of ABA is necessary for further understanding ONAC054-mediated ABA-induced leaf senescence.
Figure 10.
Tentative Model of ONAC054-Mediated Leaf Senescence through ABA Signaling.
Considering the low expression of ONAC054α and ONAC054β and the fact that the TMD is not cleaved in the absence of ABA, the accumulation of active ONAC054 proteins in the nucleus (ONAC054β and ONAC054α△TM) is low (top) under normal growth conditions. In the early phase of ABA treatment, ONAC054β dramatically accumulates in the nucleus (middle). In the late phase of ABA treatment, ONAC054α accumulates in addition to ONAC054β (bottom). Furthermore, the TMD of ONAC054α is cleaved, leading to the high accumulation of active ONAC054 proteins in the nucleus, which promotes leaf senescence through direct activation of OsABI5 and NYC1 transcription.
The Regulatory Networks of ABA-Induced Leaf Senescence Mediated by ONAC054
In this study, we found that ONAC054 is involved in the promotion of leaf senescence by modulating the ABA signaling response. In ABA signaling, ONAC054 directly activates OsABI5, while it also affects the expression levels of other genes associated with the ABA signaling response through as-yet-unidentified routes (Figure 7). ONAC054 also directly activates NYC1 (Figures 7 and 8), encoding a Chl b reductase that catalyzes the first step of Chl degradation (Kusaba et al., 2007). Furthermore, OsABI5 directly activates the transcription of NYC1 and SGR, in addition to indirectly activating NOL (Figure 9), which encodes another Chl b reductase (Sato et al., 2009). Thus, ONAC054 and OsABI5 may form coherent feed-forward loops that strongly activate NYC1 expression during leaf senescence. Such coherent feed-forward loops have been suggested to increase the robustness of biological signaling processes (Mangan and Alon, 2003). Since NYC1 homologs catalyze the first reaction of the Chl degradation pathway from Chl b to 7-hydroxymethyl Chl a, nyc1 knockout mutants in Arabidopsis and rice showed delayed leaf yellowing while retaining Chl b (Kusaba et al., 2007; Horie et al., 2009). In this study, the Chl a/b ratio in the onac054-1 mutant gradually decreased during ABA treatment (Supplemental Figure 16), indicating that Chl b was predominantly retained in onac054-1 leaves during ABA-induced leaf senescence, which was probably caused by strong repression of NYC1.
The transcriptional regulatory network of NYC1 has been well studied in Arabidopsis. NYC1 expression is activated by ABA-responsive bZIP TFs, including ABI5, EEL, and ABF4 (Sakuraba et al., 2014; Gao et al., 2016), and ABA-responsive NAC TFs, including ANAC019 and ANAC092/ORE1 (Qiu et al., 2015; Zhu et al., 2015), in addition to other TFs, such as EIN3, MYC2, and PIF5 (Qiu et al., 2015; Zhang et al., 2015; Zhu et al., 2015). Similarly, the expression of rice NYC1 is directly regulated by ABA-responsive NAC TFs, such as ONAC106 and OsNAP (Liang et al., 2014; Sakuraba et al., 2015b). The ABA-responsive TFs OsABF4 and OsABI5 bind to the same ABRE-containing region of the NYC1 promoter and directly activate its transcription (Figure 9; Piao et al., 2019). Furthermore, ONAC054 binds to the promoter region of NYC1 that contains the MDM sequence (Figure 7). Since these TFs bind different sequences, they bind to different regions in the promoter of NYC1, although it is still unclear whether their binding can be differentially regulated. Furthermore, NYC1 was shown to be strongly induced by ABA treatment (Figure 7B; Kusaba et al., 2007), and thus it is probable that NYC1 has a key role in ABA-induced leaf yellowing, which is regulated by multiple TFs, including ABA-responsive bZIP and NAC TFs. Characterization of the response of nyc1 mutants to ABA treatment will be necessary to clarifying the role of NYC1 in ABA responses.
In this study, we found that mutation of ONAC054 greatly affects the expression of other senNAC genes: OsNAP and OsNAC2, encoding senescence-promoting NAC TFs (Liang et al., 2014; Mao et al., 2017) were significantly down-regulated, while ONAC106, encoding a senescence-inhibiting NAC TF (Sakuraba et al., 2015b), was up-regulated in the onac054-1 mutant during ABA treatment and/or DIS (Figure 5D; Supplemental Figure 13). Although ONAC054 and the other three senNAC TFs (OsNAC2, OsNAP, and ONAC106) are involved in the regulation of leaf senescence, their target genes are different. For instance, OsNAC054 directly promotes the expression of NCED3 and ZEP1/ABA1, encoding ABA synthesis enzymes, and thus, ABA content in OsNAC2-OX plants was significantly higher than in the wild type (Mao et al., 2017). By contrast, ONAC054 does not affect ABA accumulation: the levels of ABA biosynthesis genes and ABA content in the onac054-1 mutant were similar to those of the wild type before the induction of senescence (Figure 7; Supplemental Figure 12). It is probable that such differences in downstream target genes lead to the robustness of ABA-induced leaf senescence mediated by ONAC054 and other senNAC TFs. To date, the binding sequences of OsNAC2, OsNAP, and ONAC106 have not been determined, although ChIP or Y1H assays showed that OsNAC2 and ONAC106 bind promoter fragments of their target genes that contain the “CACG” core sequence of the NAC binding motif (Sakuraba et al., 2015b; Mao et al., 2017). Here, we showed that ONAC054 can bind to MDM (Figure 4), the binding sequence of some Arabidopsis TMD-containing NAC TFs (De Clercq et al., 2013). Identification of the binding sites of OsNAC2, OsNAP, and ONAC106 will be required for further understanding the gene regulatory network of ABA-induced leaf senescence in rice.
METHODS
Plant Materials and Growth Conditions
The onac054 T-DNA insertion knockout lines, ONAC054-overexpressing transgenic lines, and the parental wild-type japonica rice (Oryza sativa) cv Dongjin were grown in a growth chamber under long-day (14.5 h cool-white fluorescent light, 30°C/9.5 h dark, 24°C, 500 μmol m−2 s−1) conditions or in a paddy field under NLD (>14 h light per day) conditions in Suwon, South Korea (37°N latitude). The seeds were sown on seedbeds in a greenhouse, and 1-month-old seedlings were transplanted to the paddy field. Two independent T-DNA insertion knockout lines of ONAC054 (onac054-1, PFG_3A-07241; onac054-2, PFG_3A-07240) were obtained from the Crop Biotech Institute at Kyung Hee University, South Korea. For DIS, detached leaf discs from 1-month-old or 3-week-old whole plants were floated on 3 mM MES buffer (pH 5.8) and incubated in complete darkness.
Plasmid Construction and Transformation
The cDNAs of ONAC054α and ONAC054β were amplified by RT-PCR using gene-specific primers (Supplemental Table), subcloned into the pCR8/GW/TOPO vector, and ligated into the pMDC43 Gateway binary vectors containing the 35S promoter (Curtis and Grossniklaus, 2003). 35S:GFP-ONAC054α and 35S:GFP-ONAC054β in the pMDC43 plasmids were introduced into calli generated from the mature embryos of Dongjin seeds by Agrobacterium tumefaciens (strain LBA4404)-mediated transformation (Lee et al., 2006). The transgenic rice plants were selected on 2N6 medium containing hygromycin (50 mg L−1) and confirmed by genomic PCR using specific primers (Supplemental Table).
Chl Quantification
To measure total Chl concentrations, frozen leaf tissues were homogenized by zirconia beads, and pigments were extracted from leaf homogenates with 80% (v/v) ice-cold acetone. Chl concentrations were determined by spectrophotometry using an ABU6300 Pro (Amersham Biosciences) as previously described by Porra et al. (1989).
Ion Leakage Rates
Ion leakage rates were measured as described previously (Lee et al., 2015). In brief, membrane leakage was determined by measuring electrolytes (or ions) leaking from the rice leaf discs (1 cm2). Three leaf discs from each treatment were immersed in 6 mL of 0.4 M mannitol at room temperature with gentle shaking for 3 h, and initial conductivity of the solution was measured with a conductivity meter (CON6METER). Total conductivity was determined after incubation of the samples at 85°C for 20 min. The ion leakage rate is expressed as a percentage (initial conductivity divided by total conductivity).
SDS-PAGE and Immunoblot Analysis
Total protein extracts were prepared from leaf tissues using detached leaf discs from 1-month-old rice plants before and after 2 or 4 d of dark treatment. Leaf tissues were ground in liquid nitrogen, and 10-mg aliquots were homogenized with 100 μL of sample buffer (50 mM Tris, pH 6.8, 2 mM EDTA, 10% [v/v] glycerol, 2% [w/v] SDS, and 6% [v/v] 2-mercaptoethanol). The homogenates were centrifuged at 10,000g for 3 min, and the supernatants were denatured at 80°C for 5 min. A 4-μL aliquot of each sample was subjected to 12% (w/v) SDS-PAGE, followed by electroblotting onto a Hybond P membrane (GE Healthcare). Antibodies against photosystem proteins, including Lhca1 (Agrisera, no. AS01005), Lhcb2 (Agrisera, no. AS01003), Lhcb4 (Agrisera, no. AS04045), PsaA (Agrisera, no. AS06172), CP43 (Agrisera, no. AS0111787), and α-tubulin (Agrisera, no. AS10680), and anti-rabbit IgG horseradish peroxidase-linked antibody (Cell Signaling Technology, no. 7074) were used for immunoblot analysis, and RbcL was visualized by Coomassie Brilliant Blue staining. The level of each protein was examined using the enhanced chemiluminescence system with WESTSAVE (AbFRONTIER) according to the manufacturer’s protocol.
Transmission Electron Microscopy
Transmission electron microscopy was performed using a method previously described by Inada et al., 1998 with some modifications. The middle part of the first leaf in the main culm was used for the experiments. Small leaf pieces were fixed in modified Karnovsky’s fixative (2% [v/v] paraformaldehyde, 2% [v/v] glutaraldehyde, and 50 mM sodium cacodylate buffer, pH 7.2), followed by three washes with 50 mM sodium cacodylate buffer, pH 7.2, at 4°C for 10 min. The samples were postfixed at 4°C for 2 h with 1% osmium tetroxide in 50 mM sodium cacodylate buffer, pH 7.2, and washed twice with distilled water at room temperature. The samples were stained en bloc in 0.5% (w/v) uranyl acetate at 4°C overnight and dehydrated in an ethanol gradient solution with propylene oxide, followed by infiltration with Spurr’s resin. The samples were polymerized at 70°C for 24 h and sectioned with an ultramicrotome MT-X (Boeckeler Instruments). The sections were mounted on copper grids and stained with 2% (w/v) uranyl acetate for 7 min and with Reynolds’ lead citrate for 7 min. Micrographs were obtained with a LIBRA 120 transmission electron microscope (Carl Zeiss).
Fv/Fm Ratios
The Fv/Fm ratios of flag leaf tissues in the wild type (Dongjin) and onac054 mutants grown in the paddy field were measured using an OS-30p instrument (OptiSciences) as previously described by Sakuraba et al. (2016b). The middle section of each flag leaf was adapted in the dark for 5 min to complete oxidation of QA (a bound plastoquinone). After the dark treatment, the Fv/Fm ratio was measured in the paddy field. More than three experimental replicates per plant were conducted.
Subcellular Localization of ONAC054α and ONAC054β
The cDNAs of ONAC054α and ONAC054β were amplified by RT-PCR and then ligated into the pCR8/GW/TOPO plasmid (Invitrogen). ONAC054 cDNA was inserted into the pMDC43 vector with two copies of the Cauliflower mosaic virus 35S promoter and the GFP coding sequence by LR recombination reaction (Invitrogen). For subcellular localization analysis, the vectors containing GFP-ONAC054α and GFP-ONAC054β were bombarded into onion (Allium cepa) epidermal layer cells using a DNA particle delivery system (Biolistic PDS-1000/He, Bio-Rad). At 16 h after bombardment, GFP fluorescence was analyzed in epidermal cells using a confocal laser scanning microscope II (LSM710, Carl Zeiss).
Y1H Assays
For the Y1H assays, the ONAC054α coding sequence was inserted into the pGAD424 vector (Clontech) as a prey. For mutation analysis of the MDM, we prepared a series of one-nucleotide substituted sequences, which were fused to a fragment of the TUBULIN2 (TUB2) promoter (−465 to −214 from the start codon). Primers used for cloning are listed in the Supplemental Table. The yeast strain YM4271 was used for bait and prey clones, and β-galactosidase activity was measured by liquid assay using chlorophenol red-β-D-galactopyranoside (Roche Applied Science) according to the Yeast Protocol Handbook (Clontech).
RNA Isolation and RT-qPCR Analysis
For the reverse transcription reactions, total RNA was extracted from rice leaf blades and other tissues using an RNA extraction kit (Macrogen). First-strand cDNA was prepared with 2 μg of total RNA using M-MLV reverse transcriptase and oligo(dT)15 primer (Promega) in a total volume of 25 μL and diluted with 75 μL of distilled water. For qPCR, a 20-μL mixture was prepared, including first-strand cDNA equivalent to 2 μL total RNA, 10 μL 2× GoTaq master mix (Promega), 6 μL distilled water, and gene-specific forward and reverse primers (Supplemental Table). The qPCR was performed using a LightCycler 480 (Roche Diagnostics). Rice UBIQUITIN5 (OsUBQ5) was used as an internal control. The relative expression level of each gene was calculated using the 2−ΔΔCT method as previously described by Livak and Schmittgen (2001).
Microarray Analysis
The 1-month-old wild-type (Dongjin) and onac054-1 plants grown under long-day conditions were incubated in darkness for 2 d. Total RNA was extracted from the second leaves using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA quality was checked using a 2100 Bioanalyzer (Agilent Technologies). Microarray analysis was performed using the Rice Gene Expression Microarray, design identifier 015241 (Agilent Technologies), containing 43,803 rice genes. Total RNA (150 ng) was used to prepare Cy3-labeled probes, using the low-RNA-input linear amplification kit PLUS (Agilent Technologies). Labeled RNA probes were fragmented using the Gene Expression Hybridization buffer kit (Agilent Technologies). All microarray experiments, including data analysis, were performed according to the manufacturer’s manual. The arrays were air-dried and scanned using a high-resolution array scanner (Agilent Technologies) with the appropriate settings for two-color gene expression arrays. GeneSpring GX 7.3 (Agilent Technologies) was used to calculate the intensity ratio and fold changes. Microarray analysis was performed in two experimental replicates with two different biological replicates of the wild-type and onac054-1 plants. For evaluating the statistical significance and obtaining the P value, one-sample t tests were performed using GeneSpring GX 7.3 (Agilent Technologies). DEGs were selected that had P < 0.05 and normalized onac054-1/wild type ratios > 2 or < 0.5. These DEGs were used for the Venn diagram. The information on phytohormone-associated genes was obtained from Oryzabase (www.shigen.nig.ac.jp/rice/oryzabase).
Phytohormone Treatments
Detached leaf discs from 1-month-old wild-type plants were floated on 3 mM MES buffer (pH 5.8) containing ABA (20 or 100 μM), methyl jasmonate (20 or 100 μM), salicylic acid (20 or 100 μM), ACC (500 or 1000 μM), and GR24 (20 or 100 μM) and incubated for 24 h under continuous light. Leaves were then collected for RT-qPCR analysis. For leaf senescence induction by ABA, detached leaf discs from 1-month-old wild-type, onac054 mutants, and OANC054-OX plants were floated on 3 mM MES buffer (pH 5.8) containing 100 μM ABA and incubated for the indicated periods. The leaves were then frozen in liquid nitrogen for further analyses.
Quantification of ABA Contents
To determine the ABA contents, leaf discs from 1-month-old plants were collected and weighed. To extract ABA, the leaves were ground in liquid nitrogen and were resuspended with 80% (v/v) methanol containing 1 mM butylated hydroxytoluene (1 mL for 20 mg leaf tissue) overnight at 4°C. The ABA contents were analyzed using the ABA ELISA kit according to the manufacturer’s instructions (MyBiosource).
ChIP Assays
For the ChIP assay, the 35S:GFP-ONAC054α and 35S:GFP-ONAC054β constructs in the pMDC43 binary vector (Earley et al., 2006) were transfected into rice protoplasts as previously described by Zhang et al. (2011). The protoplasts were then subjected to cross-linking for 20 min with 1% (v/v) formaldehyde under vacuum. The chromatin complexes were isolated and sonicated as previously described (Saleh et al., 2008) with slight modifications. An anti-GFP polyclonal antibody (Abcam, no. ab290) and protein A agarose/salmon sperm DNA (Millipore, no. 11719408001) were used for immunoprecipitation. After reverse cross-linking and protein digestion, the DNA was purified using a QIAquick PCR purification kit (Qiagen). Quantitative PCR was performed using 2× GoTaq master mix (Promega) and gene-specific primers listed in Supplemental Table.
Protoplast Transient Assays
To construct reporter plasmids containing the LUC reporter gene under the control of various promoters, promoter fragments of OsABI5 (−1501 to −1), OsABF4 (−1516 to −1), NOL (−1510 to −1), SGR (−1514 to −1), and NYC1 (−1508 to −1) were cloned into the pGreenII-0579 vector, which contains the LUC reporter gene at the C terminus. For the effector plasmids, the cDNAs of ONAC054α, ONAC054β, and OsABI5 were cloned into the pCR8/GW/TOPO Gateway vector (Invitrogen). These cDNAs were cloned upstream of a sequence encoding four copies of a MYC epitope tag in the pEarley203 vector (Earley et al., 2006). The reporter (4 μg) and effector plasmids (8 μg) were cotransfected into 5 × 104 rice protoplasts by the polyethylene glycol (PEG)-mediated transfection method (Yoo et al., 2007). Transfected protoplasts were then suspended in protoplast culture medium (0.4 mM mannitol, 4 mM MES buffer, and 15 mM MgCl2, pH 5.8) and kept in darkness for 16 h. The LUC activity in each cell lysate was determined using the luciferase assay system kit (Promega).
Construction of Phylogenetic Tree
To examine the phylogenetic relationship among ONAC054 and other NAC proteins in rice and Arabidopsis (Arabidopsis thaliana), full-length NAC protein sequences were retrieved from the Rice Annotation Project Database (RAP-DB, https://rapdb.dna.affrc.go.jp) and The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/index.jsp), respectively. The phylogenetic tree was constructed using the MEGA7 software (Kumar et al., 2016) by the neighbor-joining method with 1000 bootstrap replication support. The amino acid sequences of Arabidopsis and rice NAC TFs used for the construction of the phylogenetic tree are listed in Supplemental Data Set 3.
Accession Numbers
Sequence data generated in this study can be found in the National Center for Biotechnology Information (NCBI): NYC1, Os01g0227100; NOL, 0s03g0654600; ONAC054, Os03g0119966; OsABF4, Os09g0456200; OsABI5, Os01g0859300; OsbZIP23, Os02g0766700; OsNAC2, Os04g0460600; OsNAP, Os03g0327800; OsUBQ5, Os01g0328400; SGR, Os09g0532000.
Supplemental Data
Supplemental Figure 1. Expression patterns of ONAC054 during leaf senescence
Supplemental Figure 2. onac054 mutants show semidwarf and root growth retardation phenotypes
Supplemental Figure 3. Heading dates of the onac054 mutants
Supplemental Figure 4. Agronomic traits of onac054 mutants
Supplemental Figure 5. ONAC054α localizes in the ER and the nucleus
Supplemental Figure 6. Phytohormone biosynthesis and signaling genes were differentially expressed in onac054-1 leaf discs during DIS
Supplemental Figure 7. Expression patterns of ONAC054α and ONAC054β in senescent leaves
Supplemental Figure 8. Expression levels of ONAC054α and ONAC054β in the ONAC54α-OX and ONAC054β-OX lines, respectively
Supplemental Figure 9. Expression level of ONAC054β in wild-type and onac054 mutants
Supplemental Figure 10. The ONAC054α-OX and ONAC054β-OX lines showed an early leaf-yellowing phenotype during DIS
Supplemental Figure 11. The TMD of ONAC054α is cleaved in response to ABA and dark treatments
Supplemental Figure 12. Relative ABA contents in the leaves of onac054 mutants compared with the wild type during DIS
Supplemental Figure 13. ONAC054α and ONAC054β up-regulate the expression of genes associated with leaf senescence and ABA signaling
Supplemental Figure 14. OsABI5 and OsABF4 are down-regulated in onac054 mutants
Supplemental Figure 15. Phylogenetic tree of the NAC TFs of rice and Arabidopsis
Supplemental Figure 16. The change of the Chl a/b ratio in onac054-1, ONAC054α-OX, and ONAC054β-OX plants during ABA treatment
Supplemental Table.. Primers used in this study
Supplemental Data Set 1. List of genes up-regulated or down-regulated in onac054-1 mutant at 0 DDI
Supplemental Data Set 2. List of genes up-regulated or down-regulated in onac054-1 mutant at 2 DDI
Supplemental Data Set 3. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 15
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This work was carried out with the support of the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2017R1A2B3003310). The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Y.S. and N.-C.P. designed experiments. Y.S., D.K., S.-H.H., and W.P. carried out experiments. Y.S., D.K., and S.-H.K. analyzed data. S.Y. provided molecular biology reagents and analytical tools. G.A. developed plant materials. S.Y. and G.A. provided advice about the article. Y.S., D.K., and N.-C.P. wrote the article.
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Pagès M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37: 425–435. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq I., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Gao S., Gao J., Zhu X., Song Y., Li Z., Ren G., Zhou X., Kuai B. (2016). ABF2, ABF3, and ABF4 promote ABA-mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 9: 1272–1285. [DOI] [PubMed] [Google Scholar]
- Garapati P., Xue G.P., Munné-Bosch S., Balazadeh S. (2015). Transcription factor ATAF1 in Arabidopsis promotes senescence by direct regulation of key chloroplast maintenance and senescence transcriptional cascades. Plant Physiol. 168: 1122–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S., Glick B.R. (2013). Strategies to ameliorate abiotic stress-induced plant senescence. Plant Mol. Biol. 82: 623–633. [DOI] [PubMed] [Google Scholar]
- Gong W., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y., Ito H., Kusaba M., Tanaka R., Tanaka A. (2009). Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J. Biol. Chem. 284: 17449–17456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada N., Sakai A., Kuroiwa H., Kuroiwa T. (1998). Three-dimensional analysis of the senescence program in rice (Oryza sativa L.) coleoptiles. Planta 205: 153–164. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Nam H.G., Lim P.O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33: 48–56. [DOI] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Park M.J., Seo P.J., Song J.S., Kim H.J., Park C.M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448: 353–363. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Sakuraba Y., Han S.H., Yoo S.C., Paek N.C. (2013). Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54: 1660–1672. [DOI] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. (2011). The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant Cell Physiol. 52: 824–836. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Ito H., Morita R., Iida S., Sato Y., Fujimoto M., Kawasaki S., Tanaka R., Hirochika H., Nishimura M., Tanaka A. (2007). Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba M., Tanaka A., Tanaka R. (2013). Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosynth. Res. 117: 221–234. [DOI] [PubMed] [Google Scholar]
- Lee S., Seo P.J., Lee H.J., Park C.M. (2012). A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J. 70: 831–844. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Sakuraba Y., Lee T., Kim K.W., An G., Lee H.Y., Paek N.C. (2015). Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 57: 562–576. [DOI] [PubMed] [Google Scholar]
- Lee S.M., Kang K., Chung H., Yoo S.H., Xu X.M., Lee S.B., Cheong J.J., Daniell H., Kim M. (2006). Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol. Cells 21: 401–410. [PMC free article] [PubMed] [Google Scholar]
- Li Z., Woo H.R., Guo H. (2018). Genetic redundancy of senescence-associated transcription factors in Arabidopsis. J. Exp. Bot. 69: 811–823. [DOI] [PubMed] [Google Scholar]
- Liang C., Wang Y., Zhu Y., Tang J., Hu B., Liu L., Ou S., Wu H., Sun X., Chu J., Chu C. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 111: 10013–10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136. [DOI] [PubMed] [Google Scholar]
- Liu J.X., Srivastava R., Che P., Howell S.H. (2007). An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19: 4111–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Mangan S., Alon U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. USA 100: 11980–11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C., Lu S., Lv B., Zhang B., Shen J., He J., Luo L., Xi D., Chen X., Ming F. (2017). A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 174: 1747–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B., Zhou L., Rolland F., Hall Q., Cheng W.H., Liu Y.X., Hwang I., Jones T., Sheen J. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336. [DOI] [PubMed] [Google Scholar]
- Nagashima Y., Mishiba K., Suzuki E., Shimada Y., Iwata Y., Koizumi N. (2011). Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep. 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Howell K.A., Millar A.H., O’Toole N., Small I., Whelan J. (2007). Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19: 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North H.M., De Almeida A., Boutin J.P., Frey A., To A., Botran L., Sotta B., Marion-Poll A. (2007). The Arabidopsis ABA-deficient mutant aba4 demonstrates that the major route for stress-induced ABA accumulation is via neoxanthin isomers. Plant J. 50: 810–824. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A.M., Satoh K., Kondoh H., Ooka H., Kikuchi S. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465: 30–44. [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C., Mitsuda N., Sakamoto S., Ogawa D., Ohme-Takagi M., Ohmiya A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 6: 23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169: 785–797. [Google Scholar]
- Piao W., Kim S.H., Lee B.D., An G., Sakuraba Y., Paek N.C. (2019). Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 70: 2699–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Qiu K., Li Z., Yang Z., Chen J., Wu S., Zhu X., Gao S., Gao J., Ren G., Kuai B., Zhou X. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in Arabidopsis. PLoS Genet. 11: e1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino B.F., Normanly J., Amasino R.M. (1999). Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol. Biol. 40: 267–278. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Balazadeh S., Tanaka R., Mueller-Roeber B., Tanaka A. (2012). Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol. 53: 505–517. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Han S.H., Lee S.H., Hörtensteiner S., Paek N.C. (2016a). Arabidopsis NAC016 promotes chlorophyll breakdown by directly up-regulating STAYGREEN1 transcription. Plant Cell Rep. 35: 155–166. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Han S.H., Yang H.J., Piao W., Paek N.C. (2016b). Mutation of Rice Early Flowering3.1 (OsELF3.1) delays leaf senescence in rice. Plant Mol. Biol. 92: 223–234. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Jeong J., Kang M.Y., Kim J., Paek N.C., Choi G. (2014). Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat. Commun. 5: 4636. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Kim D., Paek N.C. (2018). Salt treatments and induction of senescence. Methods Mol. Biol. 1744: 141–149. [DOI] [PubMed] [Google Scholar]
- Sakuraba Y., Kim E.Y., Paek N.C. (2017). Roles of rice PHYTOCHROME-INTERACTING FACTOR-LIKE1 (OsPIL1) in leaf senescence. Plant Signal. Behav. 12: e1362522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Kim Y.S., Han S.H., Lee B.D., Paek N.C. (2015a). The Arabidopsis transcription factor NAC016 promotes drought stress responses by repressing AREB1 transcription through a trifurcate feed-forward regulatory loop involving NAP. Plant Cell 27: 1771–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y., Piao W., Lim J.H., Han S.H., Kim Y.S., An G., Paek N.C. (2015b). Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 56: 2325–2339. [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025. [DOI] [PubMed] [Google Scholar]
- Sato Y., Morita R., Katsuma S., Nishimura M., Tanaka A., Kusaba M. (2009). Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 57: 120–131. [DOI] [PubMed] [Google Scholar]
- Schwartz S.H., Léon-Kloosterziel K.M., Koornneef M., Zeevaart J.A.D. (1997). Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 114: 161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Park C.M. (2010). A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav. 5: 481–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E., van Houwelingen A., Kloos D., Mol J., Koes R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85: 159–170. [DOI] [PubMed] [Google Scholar]
- Syed N.H., Kalyna M., Marquez Y., Barta A., Brown J.W.S. (2012). Alternative splicing in plants—coming of age. Trends Plant Sci. 17: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki H., Maruyama K., Takahashi F., Fujita M., Yoshida T., Nakashima K., Myouga F., Toyooka K., Yamaguchi-Shinozaki K., Shinozaki K. (2015). SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 84: 1114–1123. [DOI] [PubMed] [Google Scholar]
- Tran L.-S.P., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S.P., Nishiyama R., Yamaguchi-Shinozaki K., Shinozaki K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1: 32–39. [DOI] [PubMed] [Google Scholar]
- Wachter A., Tunc-Ozdemir M., Grove B.C., Green P.J., Shintani D.K., Breaker R.R. (2007). Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell 19: 3437–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H.R., Kim H.J., Lim P.O., Nam H.G. (2019). Leaf senescence: Systems and dynamics aspects. Annu. Rev. Plant Biol. 70: 347–376. [DOI] [PubMed] [Google Scholar]
- Wu Y., Deng Z., Lai J., Zhang Y., Yang C., Yin B., Zhao Q., Zhang L., Li Y., Yang C., Xie Q. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19: 1279–1290. [DOI] [PubMed] [Google Scholar]
- Xie Q., Frugis G., Colgan D., Chua N.H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14: 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Liu T., Tian C., Sun S., Li J., Chen M. (2005). Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 59: 191–203. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshii M., Yamazaki M., Rakwal R., Kishi-Kaboshi M., Miyao A., Hirochika H. (2010). The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J. 61: 804–815. [DOI] [PubMed] [Google Scholar]
- Zhang K., Halitschke R., Yin C., Liu C.J., Gan S.S. (2013). Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc. Natl. Acad. Sci. USA 110: 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Z., Chen Y., He J.X., Bi Y. (2015). PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) positively regulates dark-induced senescence and chlorophyll degradation in Arabidopsis. Plant Sci. 237: 57–68. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Chen J., Xie Z., Gao J., Ren G., Gao S., Zhou X., Kuai B. (2015). Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84: 597–610. [DOI] [PubMed] [Google Scholar]
- Zou M., Guan Y., Ren H., Zhang F., Chen F. (2008). A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 66: 675–683. [DOI] [PubMed] [Google Scholar]