A protein phosphatase positively regulates the trafficking of phosphate transporters in response to phosphate levels.
Abstract
Phosphate (Pi) uptake in plants depends on plasma membrane (PM)-localized phosphate transporters (PTs). OsCK2 phosphorylates PTs and inhibits their trafficking from the endoplasmic reticulum (ER) to the PM in rice (Oryza sativa), but how PTs are dephosphorylated is unknown. We demonstrate that the protein phosphatase type 2C (PP2C) protein phosphatase OsPP95 interacts with OsPT2 and OsPT8 and dephosphorylates OsPT8 at Ser-517. Rice plants overexpressing OsPP95 reduced OsPT8 phosphorylation and promoted OsPT2 and OsPT8 trafficking from the ER to the PM, resulting in Pi accumulation. Under Pi-sufficient conditions, Pi levels were lower in young leaves and higher in old leaves in ospp95 mutants than in those of the wild type, even though the overall shoot Pi levels were the same in the mutant and the wild type. In the wild type, OsPP95 accumulated under Pi starvation but was rapidly degraded under Pi-sufficient conditions. We show that OsPHO2 interacts with and induces the degradation of OsPP95. We conclude that OsPP95, a protein phosphatase negatively regulated by OsPHO2, positively regulates Pi homeostasis and remobilization by dephosphorylating PTs and affecting their trafficking to the PM, a reversible process required for adaptation to variable Pi conditions.
INTRODUCTION
Phosphorus (P) is an essential mineral nutrient for plant development and reproduction and an integral component of biomacromolecules such as phospholipids and nucleic acids. The levels of phosphate (Pi), the only form of P that can be absorbed by plants, are commonly limited in soil due to chemical fixation and microbial activity (Raghothama, 1999). Plants have developed a series of adaptive responses that allow them to withstand suboptimal Pi conditions, such as enhancing Pi-scavenging capacity from the external environment by modifying root system architecture, secreting acid phosphatases, inducing Pi transport and symbiosis with mycorrhizal fungi, and recycling and remobilizing internal Pi via RNase activity, and lipid remodeling (Raghothama, 1999; Lin et al., 2009).
Pi uptake in plants is largely mediated by plasma membrane (PM)-localized phosphate transporters (PTs) that belong to the PHOSPHATE TRANSPORTER1 (PHT1) symporter family. A sequence similarity comparison with the high-affinity yeast (Saccharomyces cerevisiae) phosphate transporter PHO84 revealed that 13 and 9 PHT1 genes have been identified in rice (Oryza sativa) and Arabidopsis (Arabidopsis thaliana), respectively (Goff et al., 2002; Karthikeyan et al., 2002). Rice PHT1 genes show different tissue-specific expression patterns and responses to Pi starvation and encode enzymes with different affinities for Pi. For instance, the high-affinity Pi transporter OsPT8 (PHOSPHATE TRANSPORTER8) is not responsive to Pi starvation at the transcriptional level but is universally expressed in rice tissues and plays an important role in Pi uptake (Chen et al., 2011; Jia et al., 2011). Knockdown of OsPT8 in shoots inhibits the redistribution of Pi from source to sink organs (Li et al., 2015). OsPT1 and OsPT4 are also constitutively expressed in rice and function in Pi uptake and redistribution (Sun et al., 2012; Zhang et al., 2015). The low-Pi-induced transporter OsPT2, which is localized in the stele of roots, plays important roles in Pi uptake and root-to-shoot translocation under Pi-deficient conditions (Ai et al., 2009). Other functionally characterized PHT1 genes including OsPT3/6/9/10 are also induced by low Pi and play diverse roles in Pi uptake and translocation (Ai et al., 2009; Sun et al., 2012; Wang et al., 2014; Chang et al., 2019).
Although most PHT1 genes in rice are induced at the transcript level by Pi starvation or mycorrhizal symbiosis (Yang et al., 2012; Secco et al., 2013), posttranslational regulation of PHT1 family proteins is also important for their activities (Wang et al., 2018). In Arabidopsis, several PHT1 members are degraded by the ubiquitin E2 conjugase PHOSPHATE2 (AtPHO2) and the ubiquitin E3 ligase NITROGEN LIMITATION ADAPTATION (AtNLA; Huang et al., 2013; Lin et al., 2013; Park et al., 2014). Although rice OsPHO2 does not interact with PHT1 family members (at least not OsPT2/6/8), it does interact with PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (OsPHF1; Ying et al., 2017). PHF1 is localized to the endoplasmic reticulum (ER) and plays an important role in regulating the exit of PTs from the ER and their trafficking to the PM (González et al., 2005; Bayle et al., 2011; Chen et al., 2011). Notably, the phosphorylation of PHT1 family transporters affects their ER exit (Bayle et al., 2011; Chen et al., 2015). We previously revealed that OsPT2 and OsPT8 can be phosphorylated by CASEIN KINASE2 (CK2) under Pi-sufficient conditions and that phosphorylated PTs cannot interact with OsPHF1, resulting in the ER retention of PTs, allowing fewer PTs to target the PM to absorb Pi from the rhizosphere (Chen et al., 2015).
Protein phosphorylation is a reversible reaction catalyzed by two types of antagonistic enzymes: protein kinases and protein phosphatases (Uhrig et al., 2013). Although PTs are phosphorylated by CK2 in response to Pi levels in rice, how PTs are dephosphorylated in plants is currently unknown. Here, using yeast two-hybrid (Y2H) screening, we identified a PP2C protein phosphatase, OsPP95, that interacts with OsPT2 and OsPT8. OsPP95 dephosphorylates OsPT8, promoting its ER exit and trafficking to the PM. OsPP95 plays an important role in Pi uptake and redistribution. In addition, OsPP95 is targeted by OsPHO2 under Pi-sufficient conditions, resulting in its more rapid degradation under Pi-sufficient versus Pi-starvation conditions. These results provide a mechanistic understanding of a pathway in which OsPP95 acts antagonistically with CK2 to regulate the reversible phosphorylation of PTs, thereby modulating their ER exit and trafficking to the PM, ultimately regulating plant Pi homeostasis and distribution.
RESULTS
OsPP95 Physically Interacts with PTs
To investigate whether protein phosphatase is responsible for the dephosphorylation of PTs and affects their ER exit and trafficking to the PM, we investigated the subcellular localizations of GFP-tagged OsPT2 and OsPT8 (driven by the 35S promoter) treated with or without a general protein phosphatase inhibitor (cocktail II; Sigma-Aldrich; Supplemental Figure 1). When transiently expressed in rice protoplasts, both OsPT2-GFP and OsPT8-GFP were mainly detected in the PM (Supplemental Figure 1A), as previously reported (Chen et al., 2011). However, both OsPT2-GFP and OsPT8-GFP were retained in the ER when treated with protein phosphatase inhibitor cocktail II (Supplemental Figure 1A). To further test the effects of cocktail II on PT localization, we examined the ProPT8:PT8-GFP line (Supplemental Figure 1B). In contrast to untreated plants, in which most OsPT8-GFP was localized to the PM, OsPT8-GFP was also detected in the ER after treatment with protein phosphatase inhibitor, resembling the localization of OsPT8-GFP in the OsCK2α3 overexpression line. These results suggest that protein phosphatase is required for the dephosphorylation of PTs for their trafficking from the ER to the PM. OsPT8-GFP was mainly localized to the PM of root epidermal cells in ProPT8:PT8-GFP treated with or without okadaic acid (Supplemental Figure 1B), an inhibitor of PPP but not PP2C protein phosphatases, suggesting that the protein phosphatase(s) that specifically dephosphorylate PTs belong to the PP2C family.
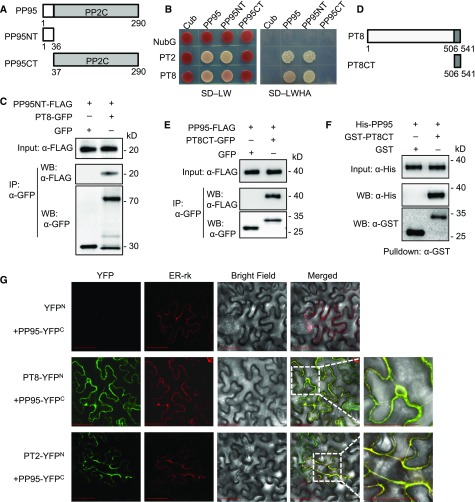
To narrow down the potential candidate PP2Cs, we examined the transcript abundance of PP2C family genes (90 members) in rice root according to previous data reported by Secco et al. (2013). Fifty-nine out of the 90 PP2C family genes were filtered out due to low transcript abundances (fragments per kilobase of exon per million fragments mapped < 10). The remaining 31 candidates were tested for interaction with OsPT8 using a split-ubiquitin membrane-based Y2H assay. OsPP95 was identified as the candidate protein phosphatase interacting with OsPT8, using the experimental workflow presented in Supplemental Figure 2. OsPP95 interacted with both OsPT8 and OsPT2 in a Y2H assay (Figures 1A and 1B). To identify the PT-interacting domain of OsPP95, the OsPP95 protein was divided into OsPP95NT (amino acids 1–36, containing a unique OsPP95 sequence) and OsPP95CT (amino acids 37 to 290, including the conserved PP2C catalytic domain), as shown in Figure 1A and Supplemental Figure 3. Both OsPT2 and OsPT8 interacted with the N terminus but not the C terminus of OsPP95 (Figure 1B). Further coimmunoprecipitation (co-IP) assays using rice protoplasts transiently expressing OsPP95-NT-FLAG together with GFP or OsPT8-GFP confirmed that OsPP95NT-FLAG could be coimmunoprecipitated by OsPT8-GFP fusion protein but not the GFP control (Figure 1C). Because the C terminus of OsPT8 can be phosphorylated by CK2, which impairs its interaction with OsPHF1, resulting in the retention of OsPT8 in the ER (Chen et al., 2015), we speculated that OsPP95 also interacts with the C terminus of OsPT8 (OsPT8CT; Figure 1D). To test this interaction, we performed co-IP assays using tobacco (Nicotiana Benthamiana) leaves transiently expressing OsPP95-FLAG together with GFP or OsPT8CT-GFP. OsPP95-FLAG was detected in OsPT8CT-GFP immunoprecipitated proteins but not in the GFP control (Figure 1E). Further GST pulldown assay confirmed that OsPT8CT interacts with OsPP95 in vitro (Figure 1F), suggesting that OsPP95 interacts with the C terminus of OsPT8. All these results suggest that OsPP95 plays a specific role in regulating PTs among PP2C proteins.
Figure 1.
Rice PP95 Physically Interacts with PTs.
(A) Schematic representation of the N-terminal (NT) and C-terminal (CT) structures of PP95 used for Y2H or co-IP analysis. White box and gray fill represent the NT and CT structures of PP95, respectively.
(B) Split-ubiquitin Y2H analysis of the interaction between PP95 and PT8 or PT2. Cub represents C-terminal ubiquitin and NubG represents the mutated N-terminal fragment of ubiquitin. SD-LW, SD/-Leu-Trp; SD-LWHA, SD/-Leu-Trp-His-Ade.
(C) Co-IP assay of PT8 with PP95-NT in planta. Plasmids containing Pro35S:PT8-GFP or Pro35S:GFP and Pro35S:PP95NT-FLAG were cotransformed into N. benthamiana leaves. Anti-GFP magnetic beads were used to immunoprecipitate the proteins, which were further analyzed by immunoblotting with anti-FLAG and anti-GFP antibodies. IP, immunoprecipitate; WB, western blot.
(D) Schematic representation of the N-terminal (NT) and C-terminal (CT) structures of PT8 used for pulldown or co-IP analysis. White box and gray fill represent the NT and CT structures of PT8, respectively.
(E) Co-IP assay of PT8CT with PP95 in planta. Plasmids containing Pro35S:PT8CT-GFP or Pro35S:GFP and Pro35S:PP95-FLAG were cotransformed into tobacco leaves. Anti-GFP magnetic beads were used to immunoprecipitate the proteins, which were further analyzed by immunoblotting with anti-FLAG and anti-GFP antibodies.
(F) Pulldown assay of His-PP95 by GST-PT8CT. Fusion proteins were expressed in E. coli and purified for pulldown assay. Immunoblots were detected using anti-His and anti-GST antibodies.
(G) BiFC analysis of the interaction between PP95 and PT8 or PT2 in tobacco leaves. The N-terminal fragment of YFP (YFPN) was fused to the C terminus of PT8 or PT2. The C-terminal fragment of YFP (YFPC) was fused to the C terminus of PP95. Combinations of YFPN or YFPC with the corresponding PP95 and PT8 or PT2 fusion constructs were used as negative controls. The two right-most images are amplified images of the two boxed regions. The localization of the ER is indicated by the expression of an ER marker (ER-rk). Bars = 50 μm.
To validate the interaction between OsPP95 and PTs and to investigate the subcellular location of their interaction in planta, we conducted bimolecular fluorescence complementation (BiFC) assays with OsPT8 or OsPT2 fused to the N terminus of YFP (OsPT8-YFPN or OsPT2-YFPN) and OsPP95 fused to the C terminus of YFP (OsPP95-YFPC). When OsPT8-YFPN or OsPT2-YFPN was coexpressed with OsPP95-YFPC in tobacco leaves, YFP fluorescence was well aligned with the red fluorescence from the ER marker (Figure 1G), suggesting that OsPP95 interacts with PTs in the ER. Together, these results demonstrate that OsPP95 interacts with PTs in vivo and in vitro.
OsPP95 Is Expressed in Roots and Shoots, and Its Protein Is Localized in All Subcellular Compartments
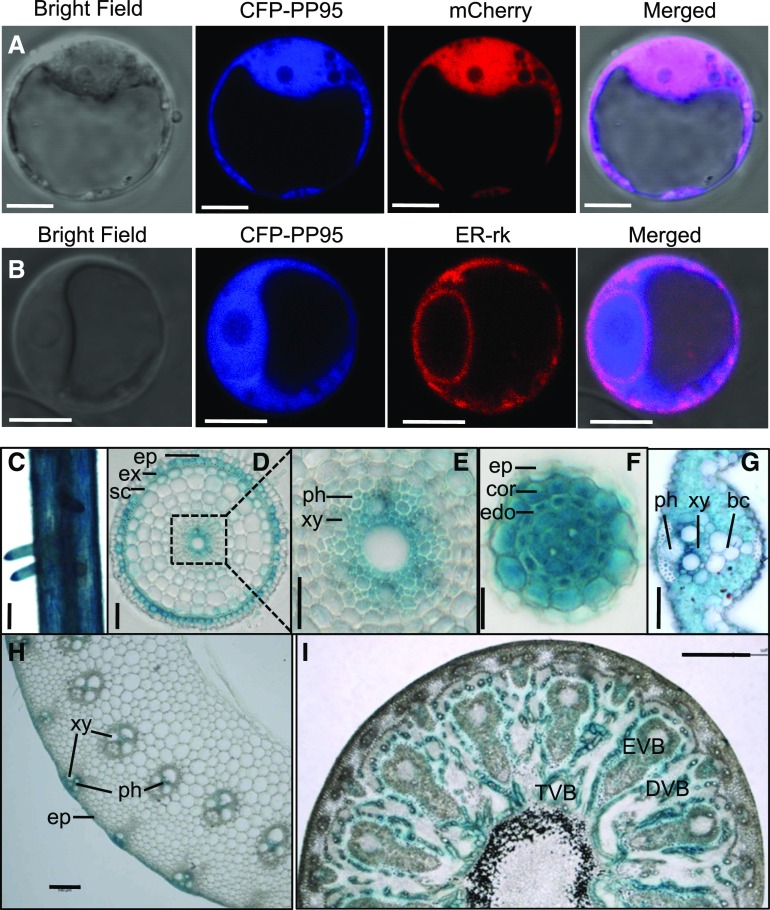
We examined the subcellular localization of OsPP95 by transiently expressing OsPP95 fused with GFP or cyan fluorescent protein (CFP) in rice protoplasts (Figures 2A and 2B; Supplemental Figure 4). Unlike the ER localization of the interaction between OsPP95 and PTs, OsPP95-GFP and CFP-OsPP95 fluorescent signals were ubiquitously expressed in all subcellular components, including the nucleus, cytoplasm, and ER, as the signals aligned with Pro35S:mCherry fluorescence and partially also with that of an ER marker (ER-rk; Figures 2A and 2B; Supplemental Figure 4).
Figure 2.
Subcellular Localization and Tissue-Specific Expression of PP95.
(A) and (B) Subcellular localization of CFP-PP95 fusion proteins. Pro35S:CFP-PP95 constructs together with Pro35S:mCherry (A) or ER marker ER-rk (B) were expressed in rice protoplasts. Bar = 5 μm.
(C) to (I) Tissue-specific expression of PP95. GUS staining in the root maturation zone (C), cross section of a primary root (D), amplified image of the root central cylinder shown in the boxed region in (D) [(E)], lateral root (F), leaf blade (G), stem (H), and node (I) of ProPP95:gPP95-GUS transgenic plants. bc, bulliform cells; DVB, diffuse vascular bundles; edo, endodermis; ep, epidermis; EVB, enlarged vascular bundles; ex, exodermis; ph, phloem; sc, sclerenchyma; TVB, transit vascular bundles; xy, xylem. Bars = 100 μm in (C) and (H), 25 μm in (D) and (G), 15 μm in (E) and (F), and 200 μm in (I).
To determine the tissue-specific expression patterns of OsPP95 in rice, we generated transgenic plants harboring the β-glucuronidase (GUS) reporter gene driven by a putative OsPP95 promoter (2334-bp region immediately upstream of the ATG translation initiation codon) and performed histochemical staining. OsPP95 was expressed in roots, nodes, and leaves (Figures 2C–2I). In roots, GUS activity was detected in the primary root epidermis, exodermis, xylem, and both the cortex and stele of the lateral root (Figures 2C–2F). In leaf blades, GUS staining was observed in all cell types except bulliform cells (Figure 2G). In the stem, GUS staining was detected in vascular tissue (Figure 2H). OsPP95 was expressed in transit vascular bundles and diffuse vascular bundles but not in enlarged vascular bundles (Figure 2I).
OsPP95 Regulates Pi Homeostasis
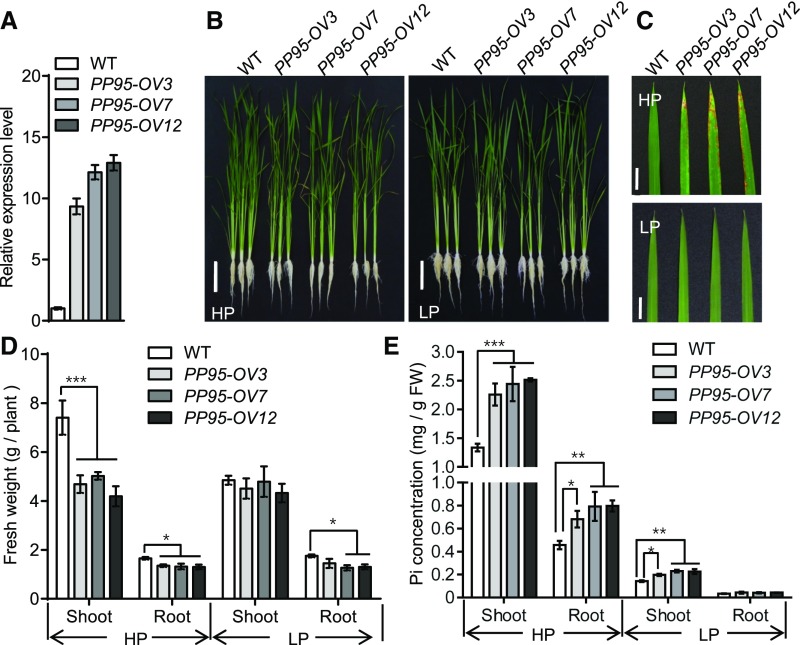
To examine the role of OsPP95 in regulating Pi homeostasis in rice, we generated OsPP95 overexpression transgenic lines in the wild-type background (cv Nipponbare). Three independent representative overexpression lines (OsPP95-OV3, OsPP95-OV7, and OsPP95-OV12) were chosen for further analysis (Figure 3A). For phenotyping, 10-d-old seedlings were transferred into Pi-sufficient (200 μM Pi) and Pi-deficient (10 μM Pi) solutions and grown for another 20 d. Under Pi-sufficient conditions, both the shoots and roots were smaller in OsPP95 overexpression plants than in wild-type plants (Figures 3B and 3D). The OsPP95-OV plants displayed leaf tip necrosis in old leaves, a typical Pi toxicity symptom in rice (Figure 3C). In addition, the Pi concentrations in OsPP95-OV plants were ∼1.6-fold that of the wild type in roots and 1.8-fold that in shoots under Pi-sufficient conditions (Figure 3E). However, under Pi-deficient conditions, both shoot and root growth in OsPP95-OV plants were comparable to that of the wild type, although two of the overexpression lines showed lower root fresh weight than the wild type (Figures 3B and 3D). Under Pi-deficient conditions, leaf tip necrosis was not observed in old leaves of OsPP95-OV (Figure 3C), although Pi levels were 0.5-fold greater in OsPP95-OV shoots than in wild-type shoots (Figure 3E). In addition, Pi levels in various leaves were higher in OsPP95-OV than in the wild type under both Pi-sufficient and Pi-deficient conditions (Supplemental Figure 5).
Figure 3.
Overexpression of PP95 Results in Pi Accumulation in Rice.
(A) Expression levels of PP95 in PP95 overexpression lines grown under Pi-sufficient conditions (200 μM). Error bars represent sd (n = 3).
(B) and (C) Phenotypes of the wild type and three PP95 overexpression transgenic lines (PP95-OV3, PP95-OV7, and PP95-OV12). Photographs of whole seedlings (B) and leaf blades (C) were taken of plants grown under Pi-sufficient (HP; 200 μM) or Pi-deficient (LP; 10 μM) conditions for 20 d (10-d-old plants were used for treatment). Bars = 10 cm in (B) and 1 cm in (C).
(D) Shoot and root fresh weight of the plants described in (B). Error bars represent sd (n = 9 independent plants). Asterisks indicate significant differences from the wild-type control (*, P < 0.05 and ***, P < 0.001; Student’s t test).
(E) Pi levels in shoots and roots of wild-type plants and three PP95 overexpression transgenic lines (PP95-OV3, PP95-OV7, and PP95-OV12) grown under HP (200 μM) or LP (10 μM) conditions for 20 d (10-d-old plants were used for treatment). Error bars represent sd (n = 3 independent plants). FW, fresh weight. Asterisks indicate significant differences from the wild-type control (*, P < 0.05, **, P < 0.01, and ***, P < 0.001; Student’s t test).
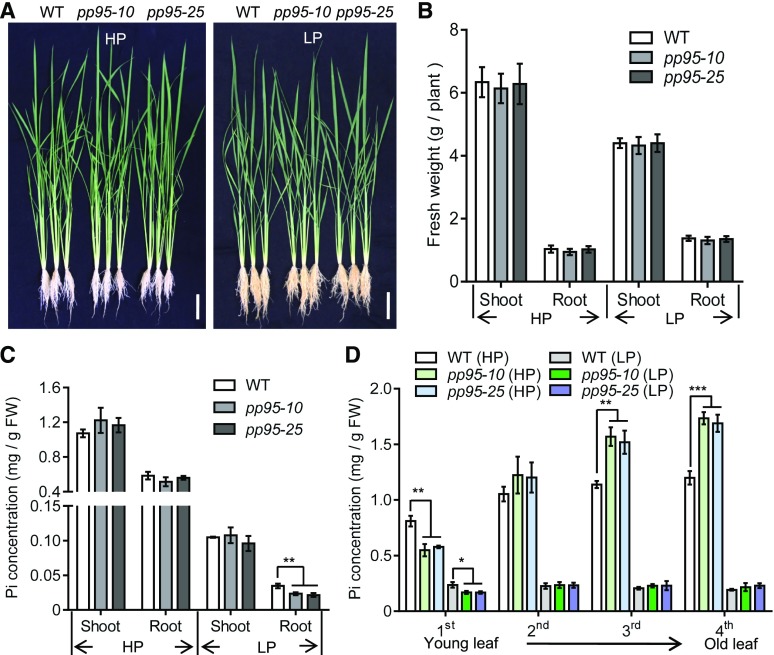
To further investigate the physiological function of OsPP95 in rice, we created ospp95 knockout mutants in the cv Nipponbare background using the CRISPR-Cas9 system. Two independent mutants, pp95-10 and pp95-25, were identified by sequencing analysis, which contained a 1-bp insertion and a 10-bp deletion in the second exon of OsPP95, respectively (Supplemental Figure 6). To rule out the possibility of off-target effects of CRISPR/Cas9, we backcrossed the pp95 mutants to wild-type plants and used homozygous pp95 plants from the F3 populations for further analysis. The growth of the pp95 mutants was similar to that of wild-type plants under both Pi-sufficient and Pi-deficient conditions (Figures 4A and 4B). Under Pi-sufficient conditions, the Pi concentrations in pp95 shoot and roots were not significantly different from the wild type (Figure 4C). However, under Pi-deficient conditions, Pi levels in pp95 roots decreased by 30% compared with the wild type, whereas Pi levels in shoots did not significantly differ among lines (Figure 4C).
Figure 4.
Mutation of PP95 Alters Pi Homeostasis in Rice.
(A) Phenotypes of the wild type and two independent pp95 mutants. Photographs of whole seedlings were taken of plants grown under Pi-sufficient (HP; 200 μM) or Pi-deficient (LP; 10 μM) conditions for 20 d (10-d-old plants were used for treatment). Bars = 10 cm.
(B) Shoot and root fresh weight of plants described in (A). Error bars represent sd (n = 9).
(C) Pi levels in shoots and roots of the wild type and two independent pp95 mutants grown under HP or LP conditions. FW, fresh weight.
(D) Pi levels in different leaves of the wild type and two independent pp95 mutants under HP or LP conditions.
Error bars represent sd (n = 3). Asterisks indicate significant differences from the wild-type control (*, P < 0.05, **, P < 0.01, and ***, P < 0.001; Student’s t-test).
To examine whether the mutation of OsPP95 affects the distribution of Pi among leaves in pp95, we measured Pi levels in different leaves. Under Pi-sufficient conditions, Pi levels were decreased in the youngest leaf of pp95 compared with the wild type, while Pi levels were increased in older leaves (leaf 3 and leaf 4). However, under Pi-deficient conditions, Pi levels were decreased in the youngest leaf but not in old leaves (leaf 2 to leaf 4) of pp95 compared with the wild type (Figure 4D). No significant differences in the dry weights of different tissues were detected between the wild type and pp95 under Pi-sufficient conditions. However, the dry weight of the youngest leaf was lower in pp95 than in the wild type under Pi-deficient conditions (Supplemental Figure 7). These results are consistent with those of a previous report showing that the spdt mutant has a reduced Pi concentration and inhibited growth of the youngest leaf, but not of the whole rice plant, under Pi-deficient conditions (Yamaji et al., 2017). These results indicate that OsPP95 is involved in regulating Pi homeostasis and distribution under different Pi conditions in rice.
OsPP95 Acts Antagonistically with CK2 to Regulate Pi Homeostasis
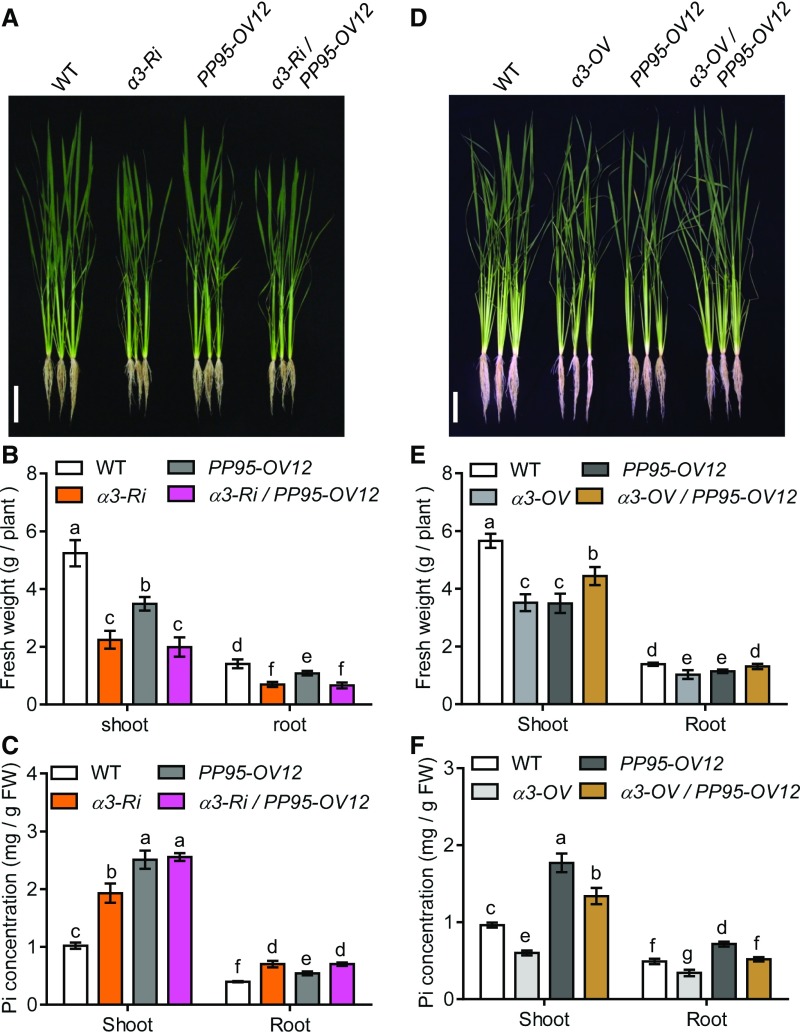
As both OsCK2 and OsPP95 interacted with the C termini of PTs, we reasoned that OsPP95 might act antagonistically with OsCK2. To test the genetic interaction between OsPP95 and OsCK2, we crossed OsCK2α3-RNAi (α3-Ri) and OsCK2α3 overexpression (α3-OV) plants with OsPP95 overexpression (PP95-OV12) plants, generating α3-Ri/OsPP95-OV and α3-OV/OsPP95-OV plants, respectively. Pi levels were higher in roots and shoots of a3-Ri than in the wild type under Pi-sufficient conditions. However, the α3-Ri/OsPP95-OV line did not exhibit an additive effect on Pi levels compared with a3-Ri (Figures 5A–5C), suggesting that OsPP95 and OsCK2α3 function in the same genetic pathway to regulate Pi homeostasis. Furthermore, the α3-OV/OsPP95-OV12 double overexpression plants showed better shoot and root growth than α3-OV and OsPP95-OV12 (Figures 5D and 5E). Whereas α3-OV showed lower Pi levels than the wild type under Pi-sufficient conditions in both shoots and roots, OsPP95-OV12 showed higher Pi levels than the wild type in both organs (Figure 5F). Furthermore, the α3-OV/OsPP95-OV12 double overexpression plants showed lower Pi levels than OsPP95-OV12 and higher Pi levels than α3-OV and wild-type plants (Figure 5F). These results suggest that overexpression of OsPP95 reverses the effects of OsCK2α3 overexpression.
Figure 5.
Genetic Interaction between CK2 and PP95 in Rice.
(A) Phenotypes of wild-type, CK2α3-RNAi (α3-Ri), PP95-OV12, and α3-Ri/PP95OV-12 whole seedlings grown under Pi-sufficient (HP, 200 μM) conditions for 20 d (10-d-old plants were used for treatment). Bar = 10 cm.
(B) Shoot and root fresh weight of plants described in (A). Error bars represent sd (n = 9).
(C) Pi concentrations in shoots and roots of plants described in (A). FW, fresh weight. Error bars represent sd (n = 3).
Different letters in (B) and (C) indicate significant differences (Duncan’s multiple range test, P < 0.05).
(D) Phenotypes of wild-type, CK2α3 overexpression line (α3-OV), PP95-OV12, and α3-OV/PP95-OV12 whole seedlings grown under HP conditions for 20 d (10-d-old plants were used for treatment). Bar = 10 cm.
(E) Shoot and root fresh weight of plants described in (D). Error bars represent sd (n = 9).
(F) Pi concentrations in shoots and roots of plants described in (D). Error bars represent sd (n = 3).
Different letters in (E) and (F) indicate significant differences (Duncan’s multiple range test, P < 0.05).
As mutation of OsPP95 affects the Pi distribution among different leaves, we reasoned that OsCK2 might also regulate Pi distribution. To test this hypothesis, we analyzed the Pi levels in different leaves of α3-Ri and β3-Ri (OsCK2β3-RNAi line) plants under Pi-sufficient and Pi-deficient conditions (Supplemental Figure 8). Under Pi-sufficient conditions, knockdown of OsCK2α3 or OsCK2β3 led to Pi accumulation in all leaves examined (Supplemental Figure 8A). However, under Pi-deficient conditions, knockdown of OsCK2α3 led to Pi accumulation in all leaves, whereas knockdown of OsCK2β3 led to increased levels of Pi in young leaves (first leaf and second leaf) but decreased levels in old leaves (third leaf and fourth leaf; Supplemental Figure 8B). These results indicate that OsPP95 acts antagonistically with OsCK2 to regulate Pi distribution among different leaves in rice.
OsPP95 Directly Dephosphorylates PTs
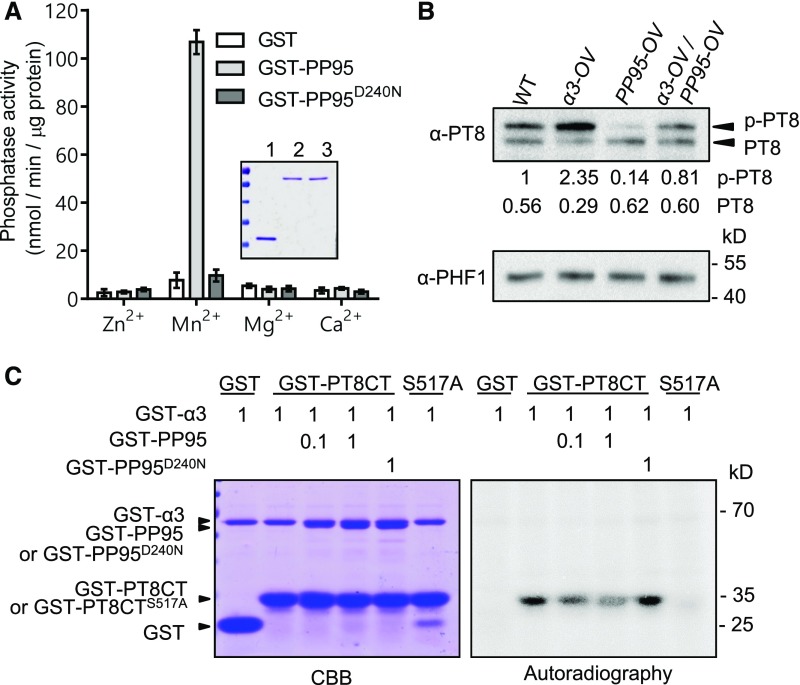
OsPP95 is grouped into the F1 clade of PP2Cs, which belongs to the Mn2+/Mg2+-dependent PPM family (Xue et al., 2008). To determine whether OsPP95 has phosphatase activity, we performed an in vitro phosphatase assay using the chromogenic substrate p-nitrophenylphosphate (pNPP). The activity of GST-OsPP95 purified from Escherichia coli cells was tested in solutions containing different metal ions. GST-OsPP95 had phosphatase activity in the presence of Mn2+ (Figure 6A). However, GST-OsPP95D240N, in which the highly conserved metal binding site (Asp) was mutated to an inactive form (Asn), lost its phosphatase activity in all solutions examined (Figure 6A). These results indicate that OsPP95 is an Mn2+-dependent protein phosphatase.
Figure 6.
PP95 Directly Dephosphorylates PTs.
(A) Analysis of the phosphatase activity of PP95 and its mutated version. GST fusion proteins were expressed in E. coli and purified for pNPP-based phosphatase assays. Dephosphorylated pNPP levels were calculated based on absorbance at 405 nm after the dephosphorylation reaction was complete. The inset shows purified GST (lane 1), GST-PP95 (lane 2), and GST-PP95D240N (lane 3) on a Coomassie Brilliant Blue-stained SDS-PAGE gel.
(B) In vivo dephosphorylation of PT8 by PP95. Phosphorylated and dephosphorylated PT8 proteins extracted from ER in α3-OV, PP95-OV, α3-OV/PP95-OV, and wild-type plants grown under Pi-sufficient conditions were detected by immunoblotting after Phos-Tag SDS-PAGE using an anti-OsPT8 antibody. PHF1 detected using anti-PHF1 antibody was used as an ER protein control. Values represent relative quantification of PT8 proteins in the indicated plants.
(C) In vitro dephosphorylation of GST-PT8CT by GST-PP95. GST-PT8CT proteins were phosphorylated by GST-CK2α3 in the presence of [γ-32P]ATP, followed by the addition of GST-PP95 or GST-PP95D240N and 1 h of incubation. Proteins were visualized by Coomassie Brilliant Blue (CBB) staining (left panel), and phosphorylated proteins were visualized by autoradiography (right panel). GST and GST-PT8CTS517A were used as controls. GST-CK2α3, GST-PP95, and GST-PP95D240N protein levels (in μg) used in the assays are indicated by numbers (0.1 and 1).
Since OsPP95 directly interacts with PTs at the ER and functions as a protein phosphatase, we reasoned that OsPP95 directly dephosphorylates PTs. To test this hypothesis, we used an OsPT8-specific antibody to detect the phosphorylation of OsPT8 in the ER of OsPP95-OV12 and α3-OV (Figure 6B). Two bands were detected on Phos-Tag immunoblots, including phosphorylated and nonphosphorylated OsPT8 under Pi-sufficient conditions (Figure 6B). More phosphorylated OsPT8 was detected using the ER extracts from α3-OV versus the wild type, but significantly less was detected in OsPP95-OV extracts. Furthermore, less phosphorylated OsPT8 was detected in the α3-OV/OsPP95-OV double overexpression line than in α3-OV (Figure 6B). These findings support the notion that OsPP95 plays a critical role in dephosphorylating OsPT8.
We previously reported that Ser-517 at the C terminus of OsPT8 is phosphorylated by OsCK2α3 (Chen et al., 2015). We therefore examined whether OsPP95 is involved in dephosphorylating OsPT8 at Ser-517 using the C terminus of OsPT8 (OsPT8CT) as the substrate in an in vitro phosphorylation assay. OsPT8CT, but not the nonphosphorylatable form (PT8CTS517A), was phosphorylated by OsCK2α3 (Figure 6C), which is consistent with previous results (Chen et al., 2015). However, a striking reduction in PT8CT phosphorylation was detected in samples treated with OsPP95 but not the mutated version, OsPP95D240N (Figure 6C), suggesting that OsPP95 dephosphorylates OsPT8 at Ser-517.
OsPP95 Acts Antagonistically with CK2 to Regulate the ER Exit of PTs
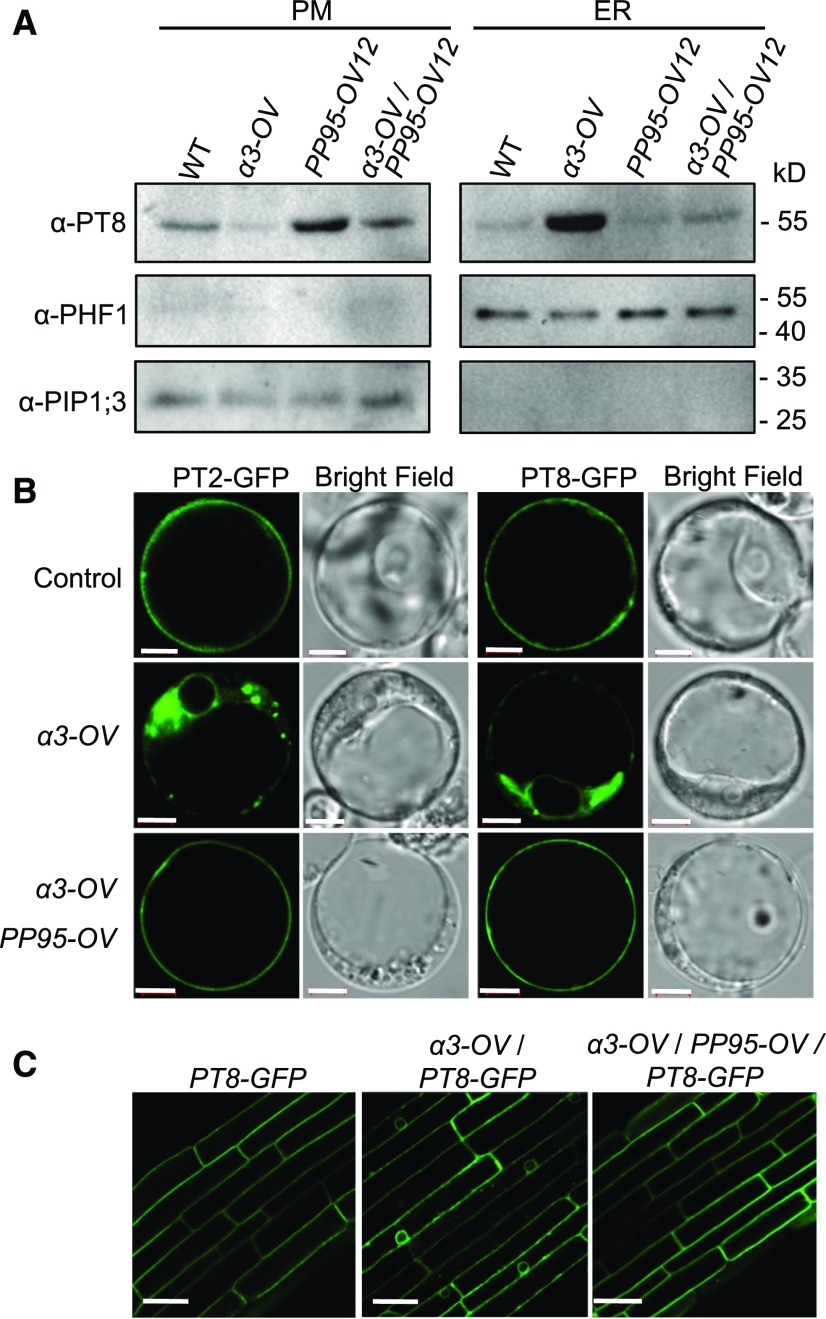
Since OsPP95 acts antagonistically with CK2 to regulate the phosphorylation status of OsPT8 at Ser-517, we reasoned that OsPP95 might regulate the ER exit of PTs. To test this possibility, we measured PT8 protein levels in the ER and PM of the OsPP95-OV lines and pp95 mutants. Consistent with the Pi accumulation patterns in the OsPP95-OV lines (Figure 3E), immunoblotting revealed that more OsPT8 accumulated at the PM, but less accumulated in the ER, in OsPP95-OV versus the wild type under Pi-sufficient conditions (Supplemental Figures 9A and 9B). Immunoblot analysis further indicated that less OsPT8 accumulated in the PM in the shoot base of pp95 compared with the wild type under both Pi-sufficient and Pi-deficient conditions (Supplemental Figure 9C); however, the OsPT8 levels in the ER of the pp95 mutants were increased in Pi-sufficient conditions, but not in Pi-deficient conditions, compared with the wild type (Supplemental Figure 9D). Consistently, less OsPT8 accumulated in the PM but more accumulated in the ER in α3-OV versus the wild type; more OsPT8 accumulated at the PM but less accumulated in the ER in OsPP95-OV12 versus the wild type; and the levels of both PM- and ER-localized OsPT8 in α3-OV/OsPP95-OV12 were between those of α3-OV and PP95-OV12 (Figure 7A).
Figure 7.
PP95 Acts Antagonistically with CK2 to Modulate the ER Exit of PTs.
(A) PT8 protein levels at the PM and the ER in plants. Proteins were isolated from plants grown under Pi-sufficient conditions. Anti-PT8 antibody was used to detect PT8 protein levels. PHF1 and PIP1;3 were used as ER and PM protein controls, respectively.
(B) Subcellular localization of PT2-GFP and PT8-GFP fusion proteins in protoplasts. Pro35S:PT2/8-GFP (control), Pro35S:PT2-GFP, or Pro35S:PT8-GFP constructs together with Pro35S:CK2α3 (a3-OV) or Pro35S:CK2α3 and Pro35S:PP95 (a3-OV/PP95-OV) were transformed into rice protoplasts. Bars = 5 μm.
(C) Subcellular localization of PT8-GFP in root epidermal cells of 7-d-old ProPT8:PT8-GFP, ProPT8:PT8-GFP/a3-OV, and ProPT8:PT8-GFP/a3-OV/PP95-OV transgenic plants. Bars = 20 μm.
We examined the subcellular localization of GFP-tagged OsPT2/OsPT8 in rice protoplasts cotransformed with or without OsPP95 and OsCK2α3 overexpression constructs. OsPT2-GFP and OsPT8-GFP fusion proteins were mainly observed on the PM in control and OsPP95-overexpressing protoplasts; overexpressing OsCK2α3 resulted in the ER retention of OsPT2-GFP and OsPT8-GFP (Figure 7B; Supplemental Figures 10A to 10D), which is consistent with a previous report (Chen et al., 2015). When OsCK2α3 and OsPP95 were coexpressed in protoplasts, OsPT2-GFP and OsPT8-GFP were mainly detected in the PM (Figure 7B). By contrast, PT8S517D-GFP (phosphorylation-mimic version of OsPT8) and PT8S517A-GFP (mimicking the nonphosphorylatable form of OsPT8) were mainly detected in the ER and PM, respectively, regardless of coexpression with OsPP95 (Supplemental Figures 10E to 10L).
To further confirm this result in planta, we generated transgenic plants overexpressing OsPP95 in the α3-OV/OsPT8-GFP background. Whereas OsPT8-GFP was detected in the ER in α3-OV/OsPT8-GFP plants, it was mainly observed in the PM of root epidermal cells in OsPP95-OV/α3-OV/OsPT8-GFP plants, as in OsPT8-GFP plants (Figure 7C). These results indicate that OsPP95 plays an important role in modulating the ER exit of PTs by acting antagonistically with CK2.
OsPP95 Is More Stable under Pi-Deficient Versus Pi-Sufficient Conditions
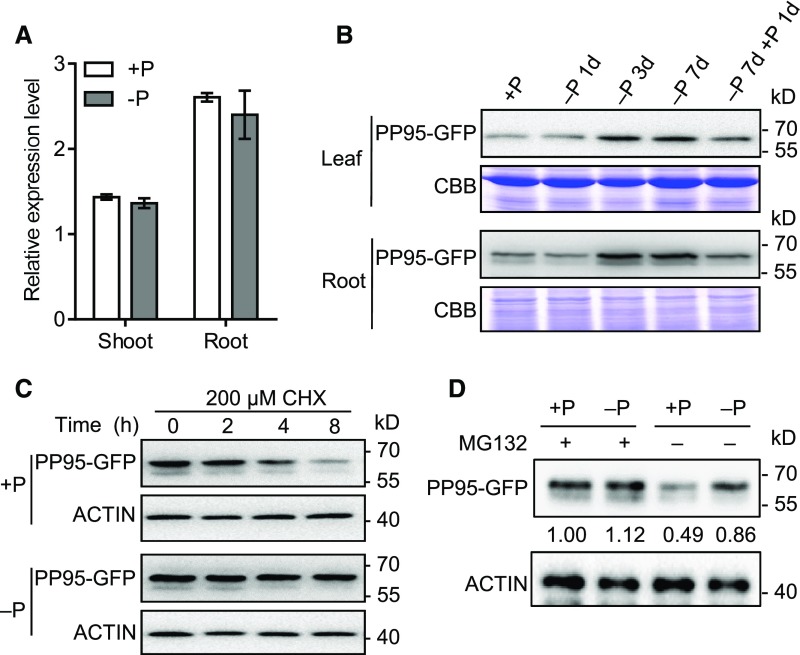
OsPP95 transcript levels were similar under both Pi-sufficient and Pi-deficient conditions, as determined by qRT-PCR (Figure 8A). To investigate whether OsPP95 protein levels vary under different Pi conditions, we generated ProPP95:gPP95-GFP/pp95-10 transgenic plants (hereafter named PP95-GFP) harboring ProPP95:gPP95-GFP in the pp95-10 mutant background (Supplemental Figure 11). Pi levels in the youngest leaf and roots were significantly higher in the transgenic plants than in pp95-10 and comparable to those of the wild type, indicating that the PP95-GFP fusion protein was functional (Supplemental Figures 11A and 11B). PP95-GFP signals were observed in the nuclei and cytoplasm of root cells (Supplemental Figure 11C), which is consistent with its localization pattern in protoplasts (Figures 2A and 2B; Supplemental Figure 4). However, PP95-GFP signals were weaker in plants grown under Pi-sufficient versus Pi-deficient conditions (Supplemental Figure 11C). Consistently, immunoblot analysis using ProPP95:gPP95-GFP/pp95-10 transgenic plants indicated that OsPP95 levels gradually increased under Pi-deprived (without Pi) conditions in a time-dependent manner and rapidly decreased in all the leaves and roots after Pi resupply (Figure 8B). These results suggest that OsPP95 is more stable under Pi-deficient conditions.
Figure 8.
PP95 Is More Stable under Pi-Deficient Versus Pi-Sufficient Conditions.
(A) Relative PP95 expression levels in the shoots and roots of plants grown in solution with (+P; 200 μM) or without (–P; 0 μM) Pi.
(B) PP95 protein levels in plants under different durations of Pi starvation. Proteins were extracted from ProPP95:gPP95-GFP/PP95-10 transgenic plants grown under +P conditions for 10 d and transferred to –P conditions for 1 to 7 d, followed by Pi resupply for 1 d. PP95-GFP proteins were detected by immunoblotting after SDS-PAGE using an anti-GFP antibody. Coomassie Brilliant Blue (CBB) staining was used as a loading control.
(C) Effect of CHX treatment on PP95 in rice. Fourteen-day-old ProPP95:gPP95-GFP/PP95-10 transgenic plants grown under +P and –P conditions were treated with the protein synthesis inhibitor CHX for the indicated durations. PP95-GFP extracted from roots was detected by immunoblotting using an anti-GFP antibody. ACTIN detected using anti-ACTIN antibody was used as a control.
(D) Effect of MG132 treatment on PP95 degradation in rice. ProPP95:gPP95-GFP/pp95-10 transgenic plants grown under +P and –P conditions were treated with or without MG132 for 24 h. PP95-GFP extracted from roots was detected by immunoblotting using an anti-GFP antibody. ACTIN was used as a control. Values represent relative quantification of PP95-GFP proteins in related samples.
Immunoblot analysis showed that PP95-GFP levels started to decrease after a 4-h treatment with the protein synthesis inhibitor cycloheximide (CHX). This fusion protein was almost completely degraded after an 8-h treatment in ProPP95:gPP95-GFP/pp95-10 seedlings grown under Pi-sufficient conditions but not under Pi-deprived conditions (Figure 8C). No difference in OsPP95 protein level was detected in ProPP95:gPP95-GFP/pp95-10 transgenic plants grown under Pi-sufficient versus Pi-deprived conditions when treated with MG132, a 26S proteasome inhibitor (Figure 8D), suggesting that OsPP95 degradation is dependent on the ubiquitin/26S proteasome pathway. Together, these results suggest that Pi starvation promotes OsPP95 stability.
Degradation of OsPP95 Partially Relies on OsPHO2
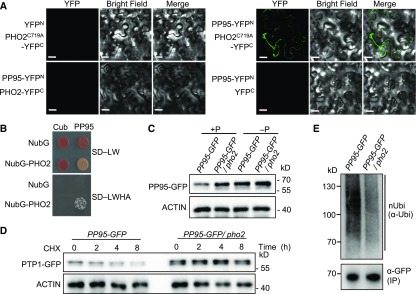
To investigate whether known Pi-homeostasis-related ubiquitin ligases OsPHO2 (Hu et al., 2011), OsNLA1 (Yang et al., 2017; Yue et al., 2017), OsUPS (Hur et al., 2012), and OsPIE1 (Yang et al., 2018) are responsible for the degradation of OsPP95 in rice, we performed Y2H assays using OsPP95 as bait. Only OsPHO2 interacted with OsPP95 in yeast, which was confirmed by BiFC assays (Figures 9A and 9B). No YFP signal was detected when OsPP95-YFPN and OsPHO2-YFPC were coexpressed in tobacco leaves (Figure 9A). However, YFP signals were detected when OsPP95-YFPN was coexpressed with OsPHO2C719A-YFPC (the catalytic residue Cys-719 of OsPHO2 was substituted with Ala; Figure 9A). These results indicate that OsPHO2 interacts with OsPP95, and they suggest that OsPHO2 might degrade OsPP95.
Figure 9.
The Degradation of OsPP95 Is Partially Dependent on OsPHO2.
(A) BiFC analysis of the interaction between PP95 and PHO2 or PHO2C719A in tobacco leaves. The N-terminal fragment of YFP (YFPN) was fused to the C terminus of PP95. The C-terminal fragment of YFP (YFPC) was fused to the C terminus of PHO2 or PHO2C719A. Combinations of YFPN or YFPC with the corresponding PHO2 or PHO2C719A and PP95 constructs were used as negative controls. Bars = 20 μm.
(B) Split-ubiquitin Y2H analysis of the interaction between PP95 and PHO2. Cub, C-terminal ubiquitin; NubG, the mutated N-terminal fragment of ubiquitin; SD-LW, SD/-Leu-Trp; SD-LWHA, SD/-Leu-Trp-His-Ade.
(C) Protein levels of PP95 in plants. Roots of ProPP95:gPP95-GFP/pp95-10 (PP95-GFP) and ProPP95:gPP95-GFP/pp95-10/pho2 (PP95-GFP/pho2) transgenic plants grown with (+P; 200 μM) or without (–P; 0 μM) Pi were sampled for protein extraction. PP95-GFP protein was detected by immunoblotting using an anti-GFP antibody. ACTIN was used as the loading control.
(D) PP95 is more stable in pho2 than in the wild type under Pi-sufficient conditions. PP95-GFP and PP95-GFP/pho2 transgenic plants grown under Pi-sufficient conditions were treated with CHX for the indicated durations. PP95-GFP extracted from roots was detected by immunoblotting using an anti-GFP antibody. ACTIN was used as the loading control.
(E) In vivo ubiquitination assay of PP95. The roots of 14-d-old wild-type and pho2 plants treated with MG132 under Pi-sufficient conditions were sampled for protein extraction. PP95-GFP was immunoprecipitated using α-GFP antibody. Ubiquitinated PP95-GFP protein was detected using α-Ubi antibody. The levels of immunoprecipitated (IP) proteins are shown below.
To determine whether OsPHO2 degrades OsPP95, we crossed PP95-GFP plants with pho2 to obtain PP95-GFP/pho2 plants. Immunoblot analysis showed that under Pi-sufficient conditions, OsPP95 protein levels were higher in PP95-GFP/pho2 than in PP95-GFP. However, under Pi-deprived conditions, OsPP95 levels were similar in PP95-GFP and PP95-GFP/pho2 plants (Figure 9C). After CHX treatment, PP95-GFP was degraded more rapidly in PP95-GFP than in PP95-GFP/pho2 under Pi-sufficient conditions (Figure 9D). Finally, we examined the levels of ubiquitin-conjugated OsPP95 in PP95-GFP and PP95-GFP/pho2 plants using anti-ubiquitin antibody and found that the levels of ubiquitinated PP95-GFP were greatly reduced in PP95-GFP/pho2 compared with PP95-GFP (Figure 9E). These results indicate that OsPHO2 is involved in degrading OsPP95 through the ubiquitination-mediated degradation pathway in rice.
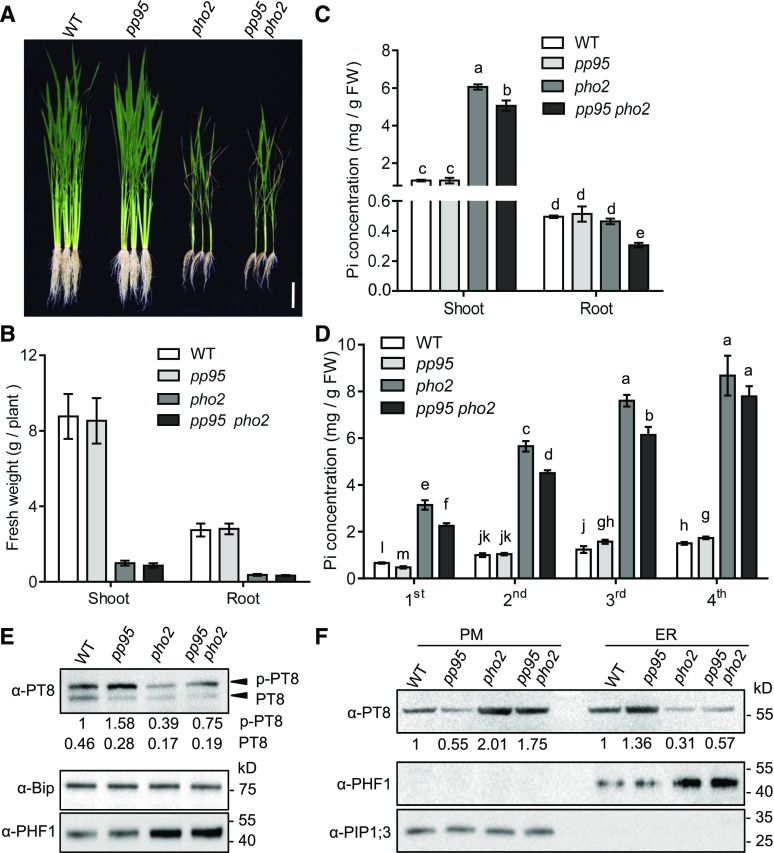
To investigate the genetic interaction between pp95 and pho2, we produced the pp95 pho2 double mutant by crossing pp95-10 with pho2 (Figures 10A and 10B). Under Pi-sufficient conditions, Pi levels in the shoots and roots of pp95 pho2 plants were reduced 17 and 35%, respectively, compared with those of pho2 (Figure 10C). We measured Pi levels in various leaves of pp95 pho2 plants. In contrast to pp95-10, whose Pi levels were only reduced in the youngest leaf compared with the wild type under Pi-sufficient conditions (Figure 4D), Pi levels in the three youngest leaves were significantly reduced in pp95 pho2 plants (Figure 10D).
Figure 10.
Genetic Interaction between PHO2 and PP95 in Rice.
(A) Phenotypes of wild-type, pp95-10, pho2, and pp95 pho2 double mutant whole seedlings after plants were grown under Pi-sufficient (200 μM) condition for 45 d. Bar = 10 cm.
(B) Shoot and root fresh weight of plants described in (A). Error bars represent sd (n = 9).
(C) and (D) Pi concentrations in shoot, root (C), and different leaves (D) of wild-type, pp95 mutant, pho2 mutant, and pp95 pho2 double mutant plants grown under Pi-sufficient conditions. FW, fresh weight. Error bars represent sd (n = 3). Different letters indicate significant differences (Duncan’s multiple range test, P < 0.05).
(E) Phosphorylated and dephosphorylated PT8 proteins extracted from the ER in wild-type, pp95 mutant, pho2 mutant, and pp95 pho2 double mutant plants grown in Pi-sufficient conditions. PT8 proteins were detected by immunoblotting in Phos-Tag SDS-PAGE using an anti-OsPT8 antibody. Bip and PHF1 detected using anti-Bip and anti-PHF1 antibody were used as controls. Values represent relative quantifications of phosphorylated PT8 and dephosphorylated PT8 proteins in related plants.
(F) PT8 protein levels at the PM and the ER in related plants. Proteins were isolated from plants grown under Pi-sufficient (200 μM) conditions. Anti-PHF1 antibody and anti-PIP1;3 antibody were used as ER and PM protein controls, respectively. Values represent relative quantifications of PT8 proteins in the indicated plants. The relative intensity of PM OsPT8 proteins or ER OsPT8 proteins in the wild type was set to 1.
To further confirm the effect of OsPHO2 on OsPP95, we assessed the phosphorylation status of OsPT8 in the ER of pho2 plants (Figure 10E). Phosphorylated OsPT8 accumulated to a lesser extent in the ER of pho2 than in that of the wild type, whereas more phosphorylated OsPT8 was detected in the ER of pp95 pho2 plants than in that of pho2 (Figure 10E). Correspondingly, more OsPT8 accumulated in the PM of pho2 plants than in that of pp95 pho2, and less OsPT8 accumulated in the ER of pho2 plants than in that of pp95 pho2 (Figure 10F). Together, these results indicate that OsPHO2 regulates Pi homeostasis and distribution partially through OsPP95.
DISCUSSION
Protein phosphorylation is a reversible posttranslational modification mediated by protein kinases and protein phosphatases that plays critical roles in protein localization, stability, and activity (Uhrig et al., 2013). Several protein kinases are involved in Pi homeostasis (Fragoso et al., 2009; Lei et al., 2014; Chen et al., 2015; Zhang et al., 2016), but to date, no protein phosphatase was shown to function in Pi acquisition and homeostasis in plants. Here, we identified the PP2C protein phosphatase OsPP95, which regulates Pi acquisition and homeostasis via dephosphorylating PTs. The identification of this reversible PT phosphorylation mechanism for plant adaptation to variable Pi environments should facilitate the genetic improvement of Pi-efficient crops.
OsPP95 Is a PP2C That Positively Regulates Pi Homeostasis
The rice and Arabidopsis genomes contain 90 and 80 PP2C genes, respectively, which are divided into 11 clades, with OsPP95 grouped into the F1 clade (Xue et al., 2008; Singh et al., 2010; Fuchs et al., 2013). PP2C activity relies on the binding of two metal ions, Mg2+ and Mn2+ (Shi, 2009). In this study, the protein phosphatase OsPP95 was activated in vitro by Mn2+ but not by Mg2+, suggesting that OsPP95 is slightly different from other PP2Cs, although it contains a conserved PP2C domain (Supplemental Figure 3). A version of OsPP95 mutated in the conserved metal binding site (Asp-240 to Asn) lacked phosphatase activity, indicating that OsPP95 is a typical PP2C protein phosphatase (Figure 6A). In addition to having a conserved PP2C catalytic domain, many PP2C proteins also contain N- and/or C-terminal domains, which may contribute to substrate specificity, cellular localization, and/or stability (Shi, 2009). In this study, OsPP95 interacted with OsPT8 through its nonconserved N-terminal sequence (Figures 1A to 1C). The rice genome encodes four F1 clade PP2Cs (Singh et al., 2010). Only OsPP95 interacted with OsPT8 in our Y2H screen, which is consistent with the finding that the N termini of these PP2Cs are quite different from that of OsPP95 (Supplemental Figure 3). However, we cannot exclude the possibility that other PP2C proteins interact with other PTs.
PP2Cs are involved in regulating plant growth and development, plant immunity, biotic and abiotic stress responses, and potassium nutrient signaling (Schweighofer et al., 2007; Ma et al., 2009; Spartz et al., 2014; Sun et al., 2016). Despite this, no PP2C protein was previously shown to regulate plant Pi acquisition or Pi starvation responses. We determined that OsPP95 is degraded more rapidly under Pi-sufficient versus Pi-deprived conditions, although OsPP95 transcription is not responsive to Pi starvation, resulting in OsPP95 accumulation under Pi starvation (Figure 8). In addition, our results suggest that OsPP95 is degraded by OsPHO2 via the 26S proteasome pathway under Pi-sufficient conditions, suggesting that OsPP95 is involved in the response to Pi starvation in rice.
Pi levels were higher in roots (∼1.6-fold) and shoots (1.8-fold) of the OsPP95-OV lines compared with the wild type under Pi-sufficient conditions, and Pi levels in shoots were also higher (0.5-fold) in these lines than in the wild type under Pi-deficient conditions (Figure 3E). By contrast, the Pi concentration in roots was 30% lower in ospp95 compared with the wild type, although there was no significant difference in Pi concentration in the shoots of ospp95 versus the wild type under Pi-deficient conditions (Figure 4C). These results suggest that OsPP95 positively regulates Pi homeostasis.
OsPP95 Acts Antagonistically with CK2 to Regulate PT Trafficking
Pi acquisition in plants is mainly mediated by PM-localized PTs. PTs are initially targeted to the ER to ensure their correct folding and modification, followed by vesicle trafficking to the PM (González et al., 2005; Bayle et al., 2011). Trafficking of PTs from the ER to the PM requires PHF1, a Sec12-related protein that facilitates the exit of PTs from the ER (Bayle et al., 2011; Chen et al., 2011). PTs are subject to phosphorylation, which affects their trafficking from the ER to the PM in response to Pi status in Arabidopsis and rice (Bayle et al., 2011; Chen et al., 2015). Rice CK2 phosphorylates OsPT2 and OsPT8, impairing their interaction with OsPHF1 and resulting in their retention in the ER (Chen et al., 2015). In this study, we identified OsPP95, an Mn2+-dependent PP2C phosphatase, which interacts with OsPT2 and OsPT8 and affects their ER and PM localization (Figures 1 and 7). An in vitro kinase assay indicated that OsPP95 dephosphorylates OsPT8 at Ser-517 (Figure 6), the same amino acid that is phosphorylated by OsCK2 (Chen et al., 2015). The phosphorylation-mimic OsPT8 was retained in the ER, whereas nonphosphorylatable OsPT8 was localized to the PM (Supplemental Figure 10). Furthermore, the phosphorylation level of OsPT8 was lower in OsPP95-OV plants (Figure 6B), and more OsPT8 in the PM and less in the ER was detected in these plants than in the wild type (Supplemental Figures 9A and 9B). These findings suggest that OsPP95 dephosphorylates PTs and affects their ER exit and trafficking to the PM. In addition, overexpressing OsPP95 reversed the ER retention of PTs caused by the overexpression of OsCK2 (Figure 7), suggesting that OsPP95 functions antagonistically with OsCK2 to regulate PT trafficking and therefore Pi homeostasis.
Notably, overexpressing OsPP95 led to Pi accumulation in shoots, even under Pi-deficient conditions (Figure 3), suggesting that PTs might also be phosphorylated under Pi-deficient conditions. The phosphorylation and protein levels of OsCK2β3 decrease in response to Pi deficiency (Chen et al., 2015). Although OsCK2β3 protein levels were lower under Pi-deficient versus Pi-sufficient conditions, OsCK2β3 was detectable by immunoblotting under Pi-deficient conditions (Supplemental Figure 12), suggesting that CK2 might also function under Pi-deficient conditions. Moreover, RNAi lines of both OsCK2α3 and OsCK2β3 accumulated Pi under both Pi-sufficient and Pi-deficient conditions (Supplemental Figure 8), further supporting the notion that CK2 also functions under Pi-deficient conditions to maintain Pi homeostasis.
OsPP95 Regulates Pi Translocation and Distribution
Plants tend to remobilize P from senescing leaves, representing an important P source for plant growth, especially during the later stages of development and in situations where soil P is limited (Veneklaas et al., 2012). However, how Pi remobilization is regulated between old leaves and young leaves at the molecular level is unknown.
Under Pi-sufficient conditions, more Pi accumulated in old versus young leaves, and Pi was remobilized to young leaves under Pi-starvation conditions in the wild type. However, in the pp95 mutants, Pi levels were lower in the youngest leaf and higher in older leaves (third and fourth leaves) than in the wild type under Pi-sufficient conditions (Figure 4D). Moreover, Pi levels were lower in pp95 pho2 than in pho2 in young leaves (first to third leaves) but not in older leaves (Figure 10D). These findings support the notion that OsPP95 is involved in Pi remobilization from old to young leaves. By contrast, under Pi-deficient conditions, β3-Ri showed higher Pi concentrations in young leaves, but lower Pi concentrations in old leaves, compared with the wild type (Supplemental Figure 8B). Consistently, OsCK2β3 levels were higher in young versus old leaves (Supplemental Figure 12).
OsPT3 and OsPT8 regulate Pi remobilization between old and young leaves (Gu et al., 2016; Wang et al., 2018). Under Pi-deficient conditions, less Pi accumulated in the youngest leaf in ospt3 and ospt8 compared with the wild type (Li et al., 2015; Chang et al., 2019). Furthermore, our results indicate that OsPP95 acts antagonistically with CK2 to regulate OsPT8 trafficking, although whether OsPP95 regulates OsPT3 trafficking requires further investigation. These observations indicate that the regulation of the phosphorylation levels of PTs by OsCK2 and OsPP95 in leaves is essential for proper Pi distribution and remobilization. As the transcripts of OsPP95 and its protein levels in different leaves are stable (Supplemental Figure 13), whereas the protein levels of OsCK2β3 decreases in the old leaves (Supplemental Figure 12), the balance between OsCK2 and OsPP95 affects Pi distribution and remobilization. Interestingly, the Pi concentration in α3-Ri was higher in older leaves than in young leaves under both Pi-sufficient and Pi-deficient conditions, which is not consistent with that of β3-Ri under Pi-deficient conditions (Supplemental Figure 8B), perhaps because PT phosphorylation requires a CK2 holoenzyme, and OsCK2β3 but not OsCK2α3 is unstable under Pi-deprived conditions (Chen et al., 2015). The rice genome contains two CK2β subunits, OsCK2β1 and OsCK2β3. OsCK2β3 was found to be necessary for the interaction between OsCK2α3 and PTs (Chen et al., 2015). However, OsCK2β1 did not interact with PTs (Chen et al., 2015), suggesting that only OsCK2β3 functioned together with OsCK2α3 to regulate PT trafficking. Recently, results from phosphoproteomic profiling under Pi-sufficient versus Pi-deficient conditions revealed that OsCK2 might also phosphorylate other substrates, such as OsSIZ1 (SAP and Miz1), to regulate Pi homeostasis in rice (Yang et al., 2019). Although OsCK2α3 functioned together with OsCK2β3, not OsCK2β1, to regulate PT trafficking, it remains to be determined whether OsCK2α3 functions with OsCK2β1 to regulate Pi homeostasis through other substrates.
The node is a well-organized vascular system in graminaceous plants that distributes mineral nutrients, including Pi (Yamaji and Ma, 2014; Clemens and Ma, 2016; Yamaji et al., 2017). OsPP95 was expressed in the transit vascular bundles and diffuse vascular bundles of nodes (Figure 2I), suggesting that OsPP95 is involved in regulating Pi distribution in nodes. OsPT3 is expressed in nodes under Pi deficiency, and OsPT8 is expressed throughout the plant (Li et al., 2015; Chang et al., 2019), suggesting that OsPP95 regulates the phosphorylation status of OsPT3, OsPT8, or other PTs in nodes to affect Pi distribution.
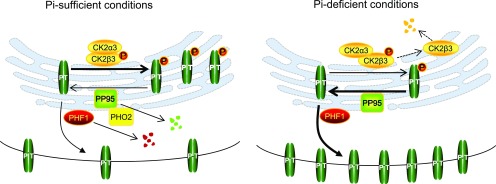
In conclusion, we identified the PT-interacting protein phosphatase OsPP95, which positively regulates Pi homeostasis and distribution by acting antagonistically with OsCK2 to regulate the phosphorylation status of PTs and their trafficking to the PM. As summarized in our working model (Figure 11), under Pi-sufficient conditions, OsPP95 protein is degraded by OsPHO2 and PTs are phosphorylated by the OsCK2 holoenzyme. These processes weaken the interaction between OsPHF1 and PTs and enhance the ER retention of PTs, allowing less PT to traffic to the PM to absorb Pi. Under Pi-deficient conditions, OsCK2β3 is degraded, while OsPP95 and OsPHF1 are more stable, and OsPHO2 is cleaved by OsmiR399. Therefore, more dephosphorylated PTs exit from the ER with the help of OsPHF1 and traffic to the PM to enhance Pi absorption.
Figure 11.
Working Model for the Role of PP95 in Regulating PT Trafficking from the ER to the PM.
Under Pi-sufficient conditions, CK2 holoenzyme (CK2α3 together with phosphorylated CK2β3) phosphorylates the PTs, which inhibits their ER exit by inhibiting their interaction with the trafficking facilitator PHF1. Furthermore, PHO2 degrades PP95, preventing it from dephosphorylating the PTs. Consequently, few PTs are targeted to the PM. Under Pi-deficient conditions, CK2β3 is degraded. PHO2 is downregulated, allowing PP95 to be stable and to dephosphorylate the PTs. The dephosphorylated PTs exit the ER with the help of PHF1 and are trafficked to the PM. Thus, more PTs are targeted to the PM, which facilitates increased Pi absorption.
METHODS
Plant Materials and Growth Conditions
All rice (Oryza sativa) materials used in the study were generated in the cv Nipponbare (japonica rice) background. OsPP95 overexpression lines, pp95 mutants (generated via CRISPR/Cas9), ProPP95:gPP95-GUS, and ProPP95:gPP95-GFP/pp95-10 were generated by Agrobacterium tumefaciens (strain EHA105)-mediated transformation by transforming callus induced from mature cv Nipponbare or pp95-10 embryos with the corresponding vectors (described below) as described by Zhou et al. (2008). Other plant materials, including the ProPT8:PT8-GFP line, α3-OV, α3-Ri, β3-Ri, and pho2, were previously described by Liu et al. (2010) and Chen et al. (2015). The α3-OV line was crossed with PP95-OV12 to generate the α3-OV/PP95-OV double overexpression line. The ospp95-10 line was crossed with pho2 to generate the pp95 pho2 double mutant. The α3-Ri line was crossed with OsPP95-OV12 to generate the α3-Ri/OsPP95-OV line.
Pi-sufficient (HP or +P), Pi-deficient (LP), and Pi-deprived (–P) treatments were performed via hydroponics using rice nutrient solution (Yang et al., 2018) containing 200 μM Pi, 10 μM Pi, and 0 μM Pi, respectively. The nutrient solution was adjusted to pH 5.5 with 1 M HCl or NaOH. Plants were grown in a growth chamber at 30/22°C (day/night) and 60 to 70% humidity, bulb-type light with a photon density of ∼300 μmol m–2 s–1, and a photoperiod of 14 h as previously described by Shao et al. (2019).
Construction of Vectors to Produce Transgenic Plants
For the OsPP95 overexpression vector construct, the OsPP95 coding region (870 bp) was amplified by PCR and introduced into the binary expression vector pF3PZPY122 using BamHI and SmaI enzyme recognition sites downstream of the 35S promoter. To generate the ProOsPP95:gOsPP95-GUS vector, a 2519-bp fragment, including the promoter and 5′ untranslated region, and the 3889-bp genomic sequence of OsPP95 without the stop codon were amplified by PCR from cv Nipponbare DNA and inserted into the GUS-pBI101.3 vector between the SalI and BamHI sites and fused in-frame to GUS. The GUS fragment of the ProOsPP95:gPP95-GUS vector was replaced with GFP to generate the ProOsPP95:gPP95-GFP vector. The CRISPR/Cas9-OsPP95 vector was constructed as previously described (Ma et al., 2015). All constructs were sequenced to confirm the presence of the correct expression cassette prior to transformation.
Y2H Assays
A DUAL Membrane Pairwise Interaction Kit (Dualsystems Biotech) was used for the Y2H assays. The full length (1−873 bp), N-terminal region (1−108 bp), and C-terminal region (109−873 bp) of OsPP95 were cloned into the pDHB1 vectors to generate Cub-OsPP95, Cub-OsPP95NT, and Cub-OsPP95CT, respectively. The full-length OsPT2 and OsPT8 sequences were cloned into the pPR3-STE vectors to generate NubG-OsPT2 and NubG-OsPT8, respectively. The full-length OsPHO2 sequence was cloned into the pPR3-N vector to generate NubG-OsPHO2. Primers are listed in Supplemental Table. The constructs were cotransformed into yeast strain NMY51 and plated onto medium without Trp and Leu to select positive clones. Protein-protein interactions were indicated by the growth of yeast colonies on medium without Leu, Trp, His, and Ade.
BiFC Assays
The BiFC assays were performed using a split YFP system as previously described by Lv et al. (2014). The coding sequence of OsPP95 was cloned into the N-terminal and C-terminal fragments of YFP vectors to generate OsPP95-YFPN and OsPP95-YFPC. The coding sequences of OsPT2 and OsPT8 were cloned into the N-terminal fragments of YFP vectors to generate OsPT2-YFPN and OsPT8-YFPN. The coding sequences of OsPHO2 and OsPHO2C719A (Cys-719 changed to Ala) were cloned into the C-terminal fragments of YFP vectors to generate OsPHO2-YFPC and OsPHO2C719A-YFPC, respectively. Primers are listed in Supplemental Table. The constructs and ER marker gene ER-rk were transiently expressed in Nicotiana benthamiana leaves. YFP fluorescence in N. benthamiana leaves was imaged 3 d after infiltration using a Zeiss LSM710 laser scanning confocal microscope.
Co-IP Assays
The co-IP assays were performed as previously described (Lv et al., 2014). The full-length and the bp 1516 to 1626 coding sequences of OsPT8 were amplified by PCR and cloned in-frame into the modified binary vector pCAMBIA1300-GFP (Pro35S:GFP) to generate Pro35S:OsPT8-GFP and Pro35S:OsPT8CT-GFP. The full-length and the bp 1 to 108 coding sequences of OsPP95 were amplified by PCR and introduced into the binary expression vector pF3PZPY122 to generate Pro35S:OsPP95-FLAG and Pro35S:OsPP95NT-FLAG. The related primers are listed in Supplemental Table. To assess the interaction between PP95NT and OsPT8, the Pro35S:OsPP95NT-FLAG vector was cotransformed with Pro35S:OsPT8-GFP or Pro35S:GFP into rice protoplasts as previously described by Chen et al. (2011). To assess the interaction between PP95 and OsPT8CT, the Pro35S:OsPP95-FLAG vector was transiently expressed in N. benthamiana leaves together with Pro35S:OsPT8CT-GFP or Pro35S:GFP. GFP-Trap Magnetic Agarose (Chromotek) was used to immunoprecipitate proteins, which was further analyzed by immunoblotting with anti-FLAG produced in mouse (Sigma-Aldrich, F1804; 1:5000) and anti-GFP antibodies produced in rabbit (Sigma-Aldrich, G1544; 1:5000). Proteins were resolved by 12% (w/v) SDS-PAGE, transferred to polyvinylidene fluoride membranes, and detected by immunoblot analysis using an ECL reagent (Millipore) and the ChemDoc XRS system (Bio-Rad).
Pulldown Assays
Full-length OsPP95 and the bp 1516 to 1626 coding sequence of OsPT8 were cloned into the C terminus of 6xHis and GST in pET28a (Invitrogen) and pGEX-4T-1 (GE Healthcare), respectively. Primers used are listed in Supplemental Table. The GST-OsPT8CT and His-OsPP95 proteins were expressed in Escherichia coli TransB (DE3) (Transgen) and purified by Ni-NTA Agarose (Qiagen) and GST-affinity chromatography (GE Healthcare), respectively. GST pulldown was conducted as previously reported by Lv et al. (2014). Proteins were further analyzed by immunoblotting with anti-His produced in mouse (Abcam, ab18184; 1:5000) and anti-GST produced in mouse (TransGen Biotech, HT601-01; 1:5000) antibodies.
Protein Expression and Phosphatase Assays
OsGST-PP95 and OsGST-PP95D240N fusion constructs were generated by inserting the PCR-amplified coding sequence (OsPP95 or OsPP95D240N) in-frame into pGEX-4T-1. The coding sequence of OsPP95D240N was generated with specific primers using gene splicing via overlap extension PCR. Primers are listed in Supplemental Table. GST-OsCK2α3, GST-OsPT8CT, and GST-OsPT8CTS517A construction was previously described (Chen et al., 2015). The GST-tagged proteins were expressed in E. coli TransB (DE3) (Transgen) and purified via GST-affinity chromatography (GE Healthcare).
The phosphatase assay was performed as described (Spartz et al., 2014). Briefly, 0.2 μg of OsPP95 was incubated in 100 μL of assay buffer containing 75 mM Tris, pH 7.6, 0.5 mM EDTA, 100 mM NaCl, and 5 mM pNPP supplemented with 10 mM MnCl2, ZnCl2, CaCl2, or MgCl2. Absorbance at 405 nm was recorded after 20 min using a Spectroquant NOVA 60 spectrophotometer (Merck). A standard curve was generated using 4-nitrophenol to calculate phosphatase activity.
For the in vitro dephosphorylation assays, GST-OsPT8CT was labeled with 32P by the catalytic subunit OsCK2α3 in the presence of [γ-32P]ATP at 30°C for 30 min as previously reported (Chen et al., 2015). Following the addition of 1 μg of GST-OsPP95 or GST-OsPP95D240N and 50 μM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (a CK2 kinase inhibitor), the reactions were incubated at 30°C for 1 h. The samples were separated by electrophoresis on a 10%(w/v) SDS-PAGE gel, followed by staining and visualization via exposure of x-ray film.
For the in vivo dephosphorylation assays, microsomal fractions were isolated from the shoot bases of 14-d-old plants grown under Pi-sufficient conditions using a Plant Microsomal Membrane Extraction Kit (Invent Biotechnologies) according to the manufacturer’s instructions. Protein samples were separated on 12% (w/v) acrylamide gels with or without Phos-Tag and analyzed by immunoblotting with anti-OsPT8 antibody (Chen et al., 2015). The relative intensity value of each band was analyzed using ImageJ software (https://imagej.en.softonic.com/).
Subcellular Localization of OsPT2 and OsPT8 in Rice Protoplasts
Rice protoplasts were isolated and transformed as previously described (Chen et al., 2011). For drug treatment, 1× phosphatase inhibitor cocktail II (Sigma-Aldrich) or 20 μM okadaic acid was added to the samples, followed by incubation for 3 h. Fluorescence in the protoplasts was imaged using a confocal laser scanning microscope (Zeiss LSM710). Excitation/emission wavelengths were 488 nm/495 to 540 nm for GFP, 458 nm/465 to 500 nm for CFP, 488 nm/515 to 545 nm for YFP, and 543 nm/575 to 630 nm for mCherry.
Isolation of ER Proteins and PM Proteins
ER proteins were isolated from the shoot bases of plants grown under Pi-sufficient or Pi-deficient conditions for 14 d with an Endoplasmic Reticulum Isolation Kit (Sigma-Aldrich) with some modifications. Briefly, the materials were ground to a powder and resuspended in extraction buffer (10 mM HEPES, pH 7.8, 1 mM EGTA, 25 mM KCl, and 250 mM sucrose supplemented with 1 mM PMSF, 20 μM MG132, and 1× protease inhibitor cocktail [Roche]). The resulting homogenate was centrifuged at 12,000g for 15 min at 4°C, and 7.5× volume of 8 mM CaCl2 was added dropwise to the resulting supernatant. The mixture was stirred for 15 min and then centrifuged at 8000g for 10 min at 4°C. The pellet (containing enriched ER proteins) was resuspended in extraction buffer. PM proteins were isolated from the shoot bases of plants grown under Pi-sufficient and Pi-deficient conditions as previously described by Chen et al. (2015). The proteins were analyzed by immunoblotting with anti-OsPHF1 produced in rabbit (Chen et al., 2015), anti-OsPIP1;3 produced in rabbit (PM marker antibody; Agrisera, AS09504; 1:5000), anti-OsBip produced in rabbit (ER marker antibody; Agrisera, AS09481; 1:5000), and anti-OsPT8 (Chen et al., 2015) antibodies produced in rabbit.
Histochemical Localization of GUS Expression
T2 ProOsPP95:gOsPP95-GUS seedlings were grown under Pi-sufficient conditions. The leaves and roots of 14-d-old seedlings and the stems (internode II/III) and node I of ∼60-d-old seedlings were collected for histochemical GUS staining as described by Chen et al. (2011). Briefly, the leaves were stained with GUS for 8 h and then washed with 80% (v/v) ethanol to remove the chlorophyll. All materials were embedded in ∼3% (w/v) agar and cut into 40- to 60-μm sections using a vibrating microtome (Leica). Cross sections of roots, leaves, stems, and nodes were photographed with a microscope (Nikon).
In Vivo Degradation Assay
Fourteen-day-old ProOsPP95:gOsPP95-GFP/ospp95-10 and ProOsPP95:gOsPP95-GFP/ospho2 plants grown under +P and/or –P conditions were treated with 200 µM CHX for 2, 4, and 8 h. Proteins were extracted from roots using extraction buffer containing 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% (v/v) Triton X-100, and 5% (v/v) glycerol plus 1 mM PMSF, 20 mM MG132, and 1× protease inhibitor cocktail (Roche). Proteins were separated by 12% (v/v) SDS-PAGE, transferred to polyvinylidene fluoride membranes, and detected by immunoblot analysis using an ECL reagent (Millipore) and the ChemDoc XRS system (Bio-Rad). Anti-GFP antibody was used to detect PP95-GFP fusion proteins, and anti-actin antibody (Sigma-Aldrich, A3853; 1:5000) was used as a loading control.
RNA Isolation and qRT-PCR Analysis
Total RNA was isolated from the samples using an RNA Extraction Kit (NucleoSpin RNA Plant; Macherey-Nagel). cDNA was synthesized from 1 μg of total RNA using a SuperScript II Reverse Transcriptase Kit (Invitrogen) with oligo(dT) primer, and qRT-PCR was performed as described (Yang et al., 2017). The primers are listed in Supplemental Table.
In Vivo Ubiquitination Assays
Seventeen-day-old ProOsPP95:gOsPP95-GFP/ospp95-10 (OsPP95-GFP) and ProOsPP95-gOsPP95-GFP/ospp95-10/ospho2 (OsPP95-GFP/ospho2) plants grown under Pi-sufficient conditions were treated with 20 µM MG132 for 7 d. Proteins were extracted from roots in extraction buffer containing 25 mM Tris, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, and 5% glycerol plus 1 mM PMSF, 20 µM MG132, 20 mM DTT, and 1× protease inhibitor cocktail (Roche).
GFP-Trap Magnetic Agarose (Chromotek) was used to immunoprecipitate GFP fusion proteins. The immunoprecipitation assays were performed as previously described (Zhang et al., 2017). The ubiquitin modifications of immunoprecipitated proteins were detected using the P4D1 mouse monoclonal anti-ubiquitin antibody produced in mouse (Santa Cruz Biotechnology, sc-8017; 1:500). Anti-GFP antibody produced in rabbit (Sigma-Aldrich, G1544; 1:5000) was used as an internal control.
Measurement of Pi Concentrations
Pi levels in shoots, leaves, and roots were measured as described previously (Chen et al., 2011).
Statistical Analysis
Statistical analysis was conducted using SPSS Statistics v. 22.0 (IBM). Data were analyzed by Duncan’s multiple range test (P < 0.05 as the level of significance) or by Student’s t test (Supplemental File 1).
Accession Numbers
Sequence data in this article can be found in the GenBank/EMBL databases under the following accession numbers: OsPP95 (Os07g0507000), OsPT2 (Os03g0150800), OsPT8 (Os10g0444700), OsPHO2 (Os05g0557700), OsCK2α3 (Os03g0207300), and OsCK2β3 (Os07g0495100).
Supplemental Data
Supplemental Figure 1. Protein phosphatase inhibitor affects the subcellular localization of PT2 and PT8.
Supplemental Figure 2. Schematic representation of the experimental workflow for identifying protein phosphatase that interacts with PTs.
Supplemental Figure 3. Alignment of PP2C F1-clade protein homologs in rice and Arabidopsis.
Supplemental Figure 4. Subcellular localization of the PP95-GFP fusion protein in rice protoplast.
Supplemental Figure 5. PP95 overexpression transgenic lines showed Pi accumulation in different leaves under HP (A) or LP (B) conditions.
Supplemental Figure 6. Identification of pp95 mutants.
Supplemental Figure 7. Growth phenotype of pp95 mutant under HP and LP conditions.
Supplemental Figure 8. Pi concentration in different leaves of Ck2α3-RNAi (α3-Ri) and Ck2β3-RNAi (β3-Ri) plants.
Supplemental Figure 9. PT8 protein levels in the PM and ER of PP95-OV and pp95 plants.
Supplemental Figure 10. OsPP95 affects the subcellular localization of PT8 but not PT8S517A or PT8S517D.
Supplemental Figure 11. Pi and PP95 levels in wild type, pp95-10, and ProPP95:gPP95-GFP/pp95-10 transgenic plants under different Pi conditions.
Supplemental Figure 12. Immunoblot analysis of CK2β3 protein levels in different leaves.
Supplemental Figure 13. PP95 distribution in different leaves under HP or LP conditions.
Supplemental Table. Primers used in this study.
Supplemental File. The results of statistical analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We dedicate this work to Ping Wu. This work was supported by the National Key Research and Development Program of China (grant 2016YFD0100700), the National Natural Science Foundation of China (grants 31701984 and 31570244), the Ministry of Agriculture of China (grant 2016ZX08001003-009), and the Ministry of Education and Bureau of Foreign Experts of China (grant B14027).
AUTHOR CONTRIBUTIONS
C.M., J.Y., and Z.Y. conceived and designed the experiments; J.Y., Z.Y., Y.W., F.W., W.M., Q.H., and J.X. performed the experiments; C.M., J.Y., Z.Y., and Z.W. analyzed the data; J.Y., C.M., and Z.Y. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809. [DOI] [PubMed] [Google Scholar]
- Bayle V., Arrighi J.F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M.X., Gu M., Xia Y.W., Dai X.L., Dai C.R., Zhang J., Wang S.C., Qu H.Y., Yamaji N., Feng Ma J., Xu G.H. (2019). OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol. 179: 656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Liu Y., Ni J., Wang Y., Bai Y., Shi J., Gan J., Wu Z., Wu P. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157: 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., et al. (2015). The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S., Ma J.F. (2016). Toxic heavy metal and metalloid accumulation in crop plants and foods. Annu. Rev. Plant Biol. 67: 489–512. [DOI] [PubMed] [Google Scholar]
- Fragoso S., Espíndola L., Páez-Valencia J., Gamboa A., Camacho Y., Martínez-Barajas E., Coello P. (2009). SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 149: 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S., Grill E., Meskiene I., Schweighofer A. (2013). Type 2C protein phosphatases in plants. FEBS J. 280: 681–693. [DOI] [PubMed] [Google Scholar]
- Goff S.A., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M., Chen A., Sun S., Xu G. (2016). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 9: 396–416. [DOI] [PubMed] [Google Scholar]
- Hu B., Zhu C., Li F., Tang J., Wang Y., Lin A., Liu L., Che R., Chu C. (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156: 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.K., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur Y.J., Yi Y.B., Lee J.H., Chung Y.S., Jung H.W., Yun D.J., Kim K.M., Park D.S., Kim D.H. (2012). Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Mol. Biol. Rep. 39: 5883–5888. [DOI] [PubMed] [Google Scholar]
- Jia H., Ren H., Gu M., Zhao J., Sun S., Zhang X., Chen J., Wu P., Xu G. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156: 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A.S., Varadarajan D.K., Mukatira U.T., D’Urzo M.P., Damsz B., Raghothama K.G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L., Li Y., Wang Q., Xu J., Chen Y., Yang H., Ren D. (2014). Activation of MKK9-MPK3/MPK6 enhances phosphate acquisition in Arabidopsis thaliana. New Phytol. 203: 1146–1160. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang J., Zhang X., Fan H., Gu M., Qu H., Xu G. (2015). Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 230: 23–32. [DOI] [PubMed] [Google Scholar]
- Lin W.Y., Huang T.K., Chiou T.J. (2013). Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.Y., Lin S.I., Chiou T.J. (2009). Molecular regulators of phosphate homeostasis in plants. J. Exp. Bot. 60: 1427–1438. [DOI] [PubMed] [Google Scholar]
- Liu F., Wang Z., Ren H., Shen C., Li Y., Ling H.Q., Wu C., Lian X., Wu P. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62: 508–517. [DOI] [PubMed] [Google Scholar]
- Lv Q., Zhong Y., Wang Y., Wang Z., Zhang L., Shi J., Wu Z., Liu Y., Mao C., Yi K., Wu P. (2014). SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Park B.S., Seo J.S., Chua N.H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26: 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693. [DOI] [PubMed] [Google Scholar]
- Schweighofer A., et al. (2007). The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 19: 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Jabnoune M., Walker H., Shou H., Wu P., Poirier Y., Whelan J. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Zhou H.Z., Wu Y., Zhang H., Lin J., Jiang X., He Q., Zhu J., Li Y., Yu H., Mao C. (2019). OsSPL3, a SBP-domain protein, regulates crown root development in rice. Plant Cell 31: 1257–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. (2009). Serine/threonine phosphatases: Mechanism through structure. Cell 139: 468–484. [DOI] [PubMed] [Google Scholar]
- Singh A., Giri J., Kapoor S., Tyagi A.K., Pandey G.K. (2010). Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz A.K., Ren H., Park M.Y., Grandt K.N., Lee S.H., Murphy A.S., Sussman M.R., Overvoorde P.J., Gray W.M. (2014). SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N., Wang J., Gao Z., Dong J., He H., Terzaghi W., Wei N., Deng X.W., Chen H. (2016). Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Natl. Acad. Sci. USA 113: 6071–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu M., Cao Y., Huang X., Zhang X., Ai P., Zhao J., Fan X., Xu G. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159: 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig R.G., Labandera A.M., Moorhead G.B. (2013). Arabidopsis PPP family of serine/threonine protein phosphatases: Many targets but few engines. Trends Plant Sci. 18: 505–513. [DOI] [PubMed] [Google Scholar]
- Veneklaas E.J., Lambers H., Bragg J., Finnegan P.M., Lovelock C.E., Plaxton W.C., Price C.A., Scheible W.R., Shane M.W., White P.J., Raven J.A. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195: 306–320. [DOI] [PubMed] [Google Scholar]
- Wang F., Deng M., Xu J., Zhu X., Mao C. (2018). Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 74: 114–122. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Y., Piñeros M.A., Wang Z., Wang W., Li C., Wu Z., Kochian L.V., Wu P. (2014). Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37: 1159–1170. [DOI] [PubMed] [Google Scholar]
- Xue T., Wang D., Zhang S., Ehlting J., Ni F., Jakab S., Zheng C., Zhong Y. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N., Ma J.F. (2014). The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 19: 556–563. [DOI] [PubMed] [Google Scholar]
- Yamaji N., Takemoto Y., Miyaji T., Mitani-Ueno N., Yoshida K.T., Ma J.F. (2017). Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541: 92–95. [DOI] [PubMed] [Google Scholar]
- Yang J., Wang L., Mao C., Lin H. (2017). Characterization of the rice NLA family reveals a key role for OsNLA1 in phosphate homeostasis. Rice (N. Y.) 10: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Xie M.Y., Wang L., Yang Z.L., Tian Z.H., Wang Z.Y., Xu J.M., Liu B.H., Deng L.W., Mao C.Z., Lin H.H. (2018). A phosphate-starvation induced RING-type E3 ligase maintains phosphate homeostasis partially through OsSPX2 in rice. Plant Cell Physiol. 59: 2564–2575. [DOI] [PubMed] [Google Scholar]
- Yang J., Xie M.Y., Yang X.L., Liu B.H., Lin H.H. (2019). Phosphoproteomic profiling reveals the importance of CK2, MAPKs and CDPKs in response to phosphate starvation in rice. Plant Cell Physiol. 60: 2785–2796. [DOI] [PubMed] [Google Scholar]
- Yang S.Y., et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24: 4236–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y., Yue W., Wang S., Li S., Wang M., Zhao Y., Wang C., Mao C., Whelan J., Shou H. (2017). Two h-type thioredoxins interact with the E2 ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in rice. Plant Physiol. 173: 812–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W., Ying Y., Wang C., Zhao Y., Dong C., Whelan J., Shou H. (2017). OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. 90: 1040–1051. [DOI] [PubMed] [Google Scholar]
- Zhang F., et al. (2015). Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82: 556–569. [DOI] [PubMed] [Google Scholar]
- Zhang H., Huang L., Hong Y., Song F. (2016). BOTRYTIS-INDUCED KINASE1, a plasma membrane-localized receptor-like protein kinase, is a negative regulator of phosphate homeostasis in Arabidopsis thaliana. BMC Plant Biol. 16: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wei B., Yuan R., Wang J., Ding M., Chen Z., Yu H., Qin G. (2017). The Arabidopsis RING-type E3 ligase TEAR1 controls leaf development by targeting the TIE1 transcriptional repressor for degradation. Plant Cell 29: 243–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Jiao F., Wu Z., Li Y., Wang X., He X., Zhong W., Wu P. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]