It has been known for several decades, and suggested as early as the 1930s (Caldwell, 1931), that viruses move their proteins and other molecules from one plant cell to the next by altering and regulating plasmodesmata (PD), the tiny doors connecting the cytoplasm between plant cells. And, only in the past decade (Khang et al., 2010) have scientists discovered that fungi are doing the same. But, apparently, pathogenic viruses and fungi are not the only ones who know how to modulate PD for spreading virulence signals between cells. Aung et al. (2020) demonstrate, in a groundbreaking study, that pathogenic bacteria have also found a way.
HopO1-1, a putative mono-ADP-ribosyltransferase effector injected into host cells by the bacterium Pseudomonas syringae pv tomato (Pst) DC3000, was found to increase virulence by interacting with and causing proteasomal degradation of two of the eight Arabidopsis (Arabidopsis thaliana) PD-localized proteins (PDLPs). These two PDLPs, PDLP5 and PDLP7, were found to interact with HopO1-1 via their C-terminal cytoplasmic tails. However, PDLP7 appears to be the main target of HopO1-1: it was degraded to a greater extent than was PLDP5, and only PDLP7 levels were reduced following Pst DC3000 infection. Degradation of PDLP7 was found to require Arg residues, known targets of ribosylation, in its cytoplasmic tail. A strain of Pst DC3000 lacking HopO1-1 (ΔhopO1-1) exhibited compromised virulence and failed to reduce PLDP7 levels upon infection. Additionally, both single-knockout pdlp5 and pdlp7 mutant plants were more susceptible to Pst DC3000 than were the wild-type plants and single-knockout pdlp4 and triple-knockout pdlp1/2/3 mutant plants.
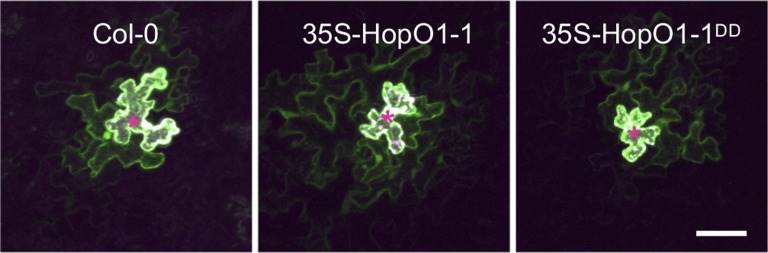
Strikingly, the downstream effect of HopO1-1–induced PDLP7 degradation appears to be an increase in intercellular molecular flux, which very well may serve as a general mechanism whereby pathogenic bacteria spread virulence-promoting signals through plants. Via an elegant experiment using particle-bombardment delivery of transgenes into leaf tissue, it was determined that soluble yellow fluorescent protein (YFP) transiently expressed in single cells traveled more often and further to neighboring cells in Arabidopsis plants overexpressing HopO1-1 than in both the wild-type plants and plants overexpressing a version of HopO1-1 that is mutated at its two putative catalytic-site residues (see figure). HopO1-1 seems to exert its influence on molecular flux without drastically affecting the size exclusion limit of PD, as the movement of 2×YFP into neighboring cells was significantly reduced to a similar level in both transgenic HopO1-1 and the wild-type plants, and 3×YFP did not move at all into neighboring cells.
Soluble YFP Moves through More Plant Cells in the Presence of a Functional HopO1-1 Bacterial Effector
A transgene encoding soluble YFP capable of intercellular movement through PD and (as a positive control for transformation) a transgene encoding an endoplasmic reticulum–localized cyan fluorescent protein incapable of intercellular movement were cobombarded into the leaf epidermal cells of the Arabidopsis wild-type Columbia (Col-0) plants, transgenic plants overexpressing HopO1-1 (35S-HopO1-1), and transgenic plants overexpressing a putative catalytic-mutant form of HopO1-1 (35S-HopO1-1DD). Green, soluble YFP; asterisks, transformed cells. Scale bars = 50 μm. (Reprinted from Aung et al. [2020], Figure 4A.)
Many questions remain to be answered regarding how, exactly, HopO1-1 influences signaling through PD and alters the architecture of PD themselves. It would be interesting to determine, for example, whether and how endoplasmic reticulum–plasma membrane contact sites, endoplasmic reticulum–derived desmotubules, and their associated actin proteins, all found within the PD channel (Nicolas et al., 2018), are regulated by HopO1-1. But what is clear is that just as pathogenic bacteria are opening the doors between plant cells, this study by Aung et al. (2020) is likewise opening its own doors to a whole new field of research into bacterial effector-mediated plasmodesmatal modulation.
Footnotes
Articles can be viewed without a subscription.
References
- Aung K., Kim P., Li Z., Joe A., Kvitko B., Alfano J.R., He S.Y. (2020). Pathogenic bacteria target plant plasmodesmata to colonize and invade surrounding tissues. Plant Cell 32: 595–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell J. (1931). The physiology of virus diseases in plants: II. Further studies on the movement of mosaic in the tomato plant. Ann. Appl. Biol. 18: 279–298. [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.-Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas W.J., Grison M.S., Bayer E.M. (2018). Shaping intercellular channels of plasmodesmata: The structure-to-function missing link. J. Exp. Bot. 69: 91–103. [DOI] [PubMed] [Google Scholar]