Pseudouridine, a frequent nucleoside modification in noncoding RNA and mRNA, is degraded in Arabidopsis peroxisomes to prevent the accumulation of the toxic intermediate pseudouridine monophosphate.
Abstract
Pseudouridine (Ψ) is a frequent nucleoside modification that occurs in both noncoding RNAs and mRNAs. In pseudouridine, C5 of uracil is attached to the Rib via an unusual C-glycosidic bond. This RNA modification is introduced on the RNA by site-specific transglycosylation of uridine (U), a process mediated by pseudouridine synthases. RNA is subject to constant turnover, releasing free pseudouridine, but the metabolic fate of pseudouridine in eukaryotes is unclear. Here, we show that in Arabidopsis (Arabidopsis thaliana), pseudouridine is catabolized in the peroxisome by (1) a pseudouridine kinase (PUKI) from the PfkB family that generates 5′-pseudouridine monophosphate (5′-ΨMP) and (2) a ΨMP glycosylase (PUMY) that hydrolyzes ΨMP to uracil and ribose-5-phosphate. Compromising pseudouridine catabolism leads to strong pseudouridine accumulation and increased ΨMP content. ΨMP is toxic, causing delayed germination and growth inhibition, but compromising pseudouridine catabolism does not affect the Ψ/U ratios in RNA. The bipartite peroxisomal PUKI and PUMY are conserved in plants and algae, whereas some fungi and most animals (except mammals) possess a PUMY-PUKI fusion protein, likely in mitochondria. We propose that vacuolar turnover of ribosomal RNA produces most of the pseudouridine pool via 3′-ΨMP, which is imported through the cytosol into the peroxisomes for degradation by PUKI and PUMY, a process involving a toxic 5′-ΨMP intermediate.
INTRODUCTION
To date, more than 150 distinct site-specific chemical modifications have been discovered in different eukaryotic RNA species. Most of these modifications are present in abundant noncoding RNAs (ncRNAs), including ribosomal RNAs (rRNAs) and tRNAs, and are important for promoting proper RNA folding and maintaining translational fidelity (Jack et al., 2011; Chou et al., 2017). Relatively few modified nucleosides, such as N6-methyladenosine (m6A; Zhong et al., 2008; Dominissini et al., 2012), 5-methylcytidine (Motorin et al., 2010; Yang et al., 2017), N1-methyladenosine (Dominissini et al., 2016), and pseudouridine (Ψ; Carlile et al., 2014), are present in mRNA, but these nucleosides can directly influence the metabolic fate and various functions of the respective transcripts (Frye et al., 2018), including mRNA decay, translation efficiency, and mRNA stability. Among these, m6A is the most prevalent mRNA modification in eukaryotes (0.4 to 0.6% of adenosine is modified; Dominissini et al., 2012). m6A, which is decoded by different reader proteins, influences pre-mRNA processing and promotes transcript stability, thereby regulating plant development and morphogenesis, such as the emergence of the first true leaves and trichome branching (Arribas-Hernández et al., 2018). Upon mRNA turnover, N6-methyl adenosine monophosphate (N6-mAMP) is released in the cytosol. An N6-mAMP deaminase that hydrolyzes N6-mAMP to IMP and methylamine in the cytosol was recently described in Arabidopsis (Arabidopsis thaliana) and human (Homo sapiens; Chen et al., 2018). IMP is an intermediate of AMP catabolism and GMP biosynthesis (Baccolini and Witte, 2019); thus, N6-mAMP deaminase introduces the modified nucleotide N6-mAMP into general nucleotide metabolism.
Pseudouridine (Ψ), a C5-glycoside isomer of uridine (U) is the most abundant modified nucleoside in ncRNA. Ψ is essential for rRNA folding and for regulating translational accuracy (Decatur and Fournier, 2002; Jack et al., 2011), and it is required for stabilizing the tRNA structure (Arnez and Steitz, 1994). In Arabidopsis, there are 187 Ψ sites in rRNAs and 232 Ψ sites in tRNAs (Sun et al., 2019). More than 2000 Ψ sites in more than 1900 human transcripts (Li et al., 2015) and 451 Ψ sites in 332 transcripts from Arabidopsis (Sun et al., 2019) were recently mapped. In Arabidopsis mRNAs, Ψ preferentially occurs in 5′ untranslated regions (UTRs) and coding sequences and is less abundant in 3′ UTRs (Sun et al., 2019), but the biological relevance of pseudouridine in mRNA remains unclear. Because rRNA and tRNA are more abundant than mRNA, the bulk of Ψ resides in ncRNA.
Pseudouridine synthases introduce Ψ through the isomerization of specific U residues in the RNA (Hamma and Ferré-D’Amaré, 2006). Interestingly in the yeast Saccharomyces cerevisiae, Ψ abundance at certain sites is dynamically regulated in response to stress conditions such as heat shock (Schwartz et al., 2014). Although Ψ is such a frequent RNA modification, the metabolic fate of pseudouridine monophosphate (ΨMP) upon degradation of Ψ-containing RNA in eukaryotes is unknown, and it is unclear whether and how ΨMP is connected to general nucleotide metabolism. Breitman (1970) discovered that in Escherichia coli, pseudouridine added to the growth medium could be used as the sole pyrimidine source by pyrimidine auxotrophic mutants. Almost 40 years later, E. coli was shown to use three enzymes to achieve this: (1) a pseudouridine kinase that generates ΨMP, (2) a ΨMP glycosidase that hydrolyzes ΨMP to uracil and ribose-5-phosphate, and (3) a uracil phosphoribosyltransferase that forms UMP from uracil and 5-phosphoribosyl-1-pyrophosphate (Preumont et al., 2008). Bioinformatic analyses suggested that orthologs of the pseudouridine kinase and the ΨMP glycosylase might also exist in eukaryotes (Preumont et al., 2008; Reumann, 2011). In Arabidopsis, these proteins have been detected in proteomic survey studies of isolated peroxisomes and their peroxisomal localization has been confirmed (Eubel et al., 2008; Reumann et al., 2009; Lingner et al., 2011). However, further experimental data about these proteins in eukaryotic organisms are lacking.
We are interested in the metabolic fate of noncanonical nucleotides in plants and have recently described how N6-mAMP is degraded in vivo. Here, we set out to elucidate the catabolic route for ΨMP in plants. We show that two peroxisomal enzymes similar to those from E. coli are required for pseudouridine degradation in vivo. We also investigated the phenotypic and molecular consequences of abrogated, mislocated, and partially enhanced pseudouridine catabolism for Arabidopsis. Based on these analyses, we propose a model for ΨMP catabolism.
RESULTS
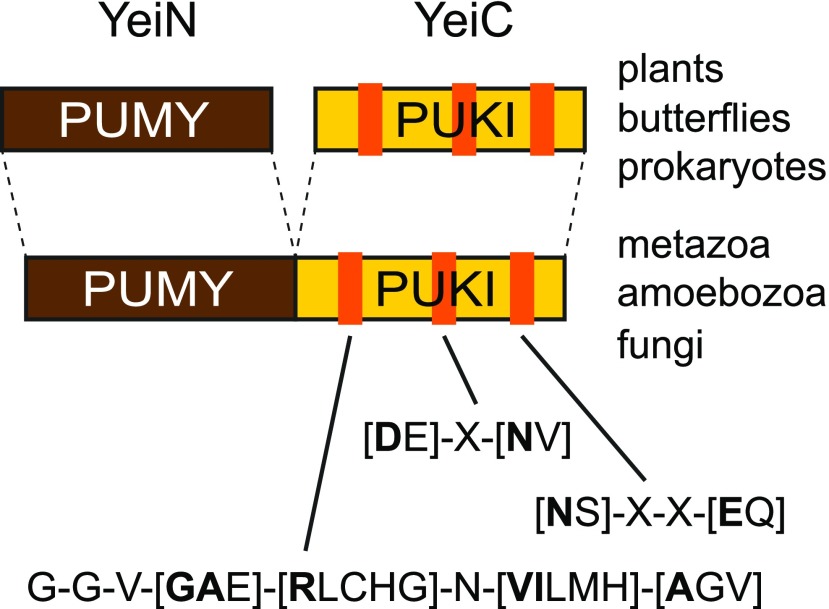
Identification of Enzymes Involved in Pseudouridine Catabolism
To investigate whether plants possess the pseudouridine-catabolizing enzymes identified and characterized so far only in E. coli, we used the protein sequence of ΨMP glycosylase (YeiN) from E. coli as a query in BLASTp searches against the Arabidopsis Araport11 protein database at The Arabidopsis Information Resource. Arabidopsis contains only a single protein homologous to YeiN. This protein, which is encoded at locus At1g50510, shares >46% identical and >62% similar amino acid residues with YeiN, making it a possible candidate for ΨMP glycosylase, although it is currently annotated as an indigoidine synthase A family protein. Similarly, we used the E. coli pseudouridine kinase (YeiC) as a query in a BLASTp search. Here, ribokinase from the fructokinase B family of proteins (PfkB kinases) had the highest score, but this enzyme is only ∼21% identical to YeiC. The substrate spectrum of ribokinase is limited to Rib (Riggs et al., 2016; Schroeder et al., 2018) and does not include nucleosides (R.Y. Schroeder and C.-P. Witte, unpublished results). Therefore, it is unlikely that ribokinase has pseudouridine kinase activity. In metazoa, homologs of YeiN and YeiC reside on a single polypeptide chain (Figure 1). Although the biochemical function of this fusion protein in eukaryotes is unknown, the relatively high conservation of YeiN homologs across the kingdoms of life suggests that this protein represents eukaryotic ΨMP glycosylase physically linked to pseudouridine kinase. Using the sequences of the kinase domains of such proteins as queries in protein homology searches in Arabidopsis, a PfkB kinase of unknown function encoded at the locus At1g49350 always had (by far) the highest BLASTp scores.
Figure 1.
Schematic Diagram of the Structures of PUKI (YeiC) and PUMY (YeiN) in Different Organisms.
Three conserved motifs characteristic of PUKI (YeiC) are shown. The diagram is based on data in Supplemental Figures 1 and 2 together with data from Schroeder et al. (2018). If several amino acids can occur at a certain position, the most frequently found amino acid is indicated in bold.
We performed extensive sequence database searches and compiled a multiple alignment of E. coli YeiN and YeiC with homologs from a wide evolutionary range of plants, metazoa, and fungi (Supplemental Figure 1). Peroxisomal targeting sequences (PTSs) were identified using previously described motifs (Kunze et al., 2011; Lingner et al., 2011). For YeiN of E. coli, a crystal structure with bound substrate is available (Huang et al., 2012). Side chains of residues involved in direct or water-mediated binding of the substrate and the coordination of a manganese ion at the active site are highly conserved in the aligned sequences (marked in red and blue, respectively, in Supplemental Figure 1), suggesting that these YeiN homologs might indeed encode ΨMP glycosylases.
The putative pseudouridine kinases are more divergent, and crystal structure data are currently not available. These kinases formed a well-supported clade in a phylogenetic analysis of the PfkB kinase family from five plant species (Schroeder et al., 2018). Three sequence motifs can be defined that clearly identify putative pseudouridine kinases among other PfkB kinases (Figure 1, absolutely conserved residues within these motifs are marked in yellow in Supplemental Figure 1). The motifs were validated and refined with bacterial YeiC sequences (Supplemental Figure 2) from an evolutionarily broad range of bacteria. To select only YeiC orthologs, which are likely functional, we only chose bacterial genomes in which YeiC is in the direct vicinity of YeiN. The first motif G-G-V-[GAE]-[RLCHG]-N-[VILMH]-[AGV] is a more defined variant of the PROSITE motif PS00583 used to identify PfkB kinases in general.
These in silico analyses suggest that in Arabidopsis, the locus At1g49350 encodes a pseudouridine kinase and At1g50510 encodes a ΨMP glycosylase, as had been hypothesized previously by Reumann (2011). It appears that like in E. coli, in Arabidopsis and many other eukaryotes, pseudouridine is phosphorylated, forming ΨMP that is then hydrolyzed to uracil and ribose 5-phosphate, using a pseudouridine kinase and a ΨMP glycosylase (Figure 2A). In metazoa, amoebozoa, and fungi, both activities generally appear to reside on the same polypeptide chain, whereas prokaryotes and plants have two independent enzymes (Figure 1). Interestingly, two independent enzymes are also found in butterflies. Mammals generally lack these enzymes, except for platypus (Ornithorhynchus anatinus).
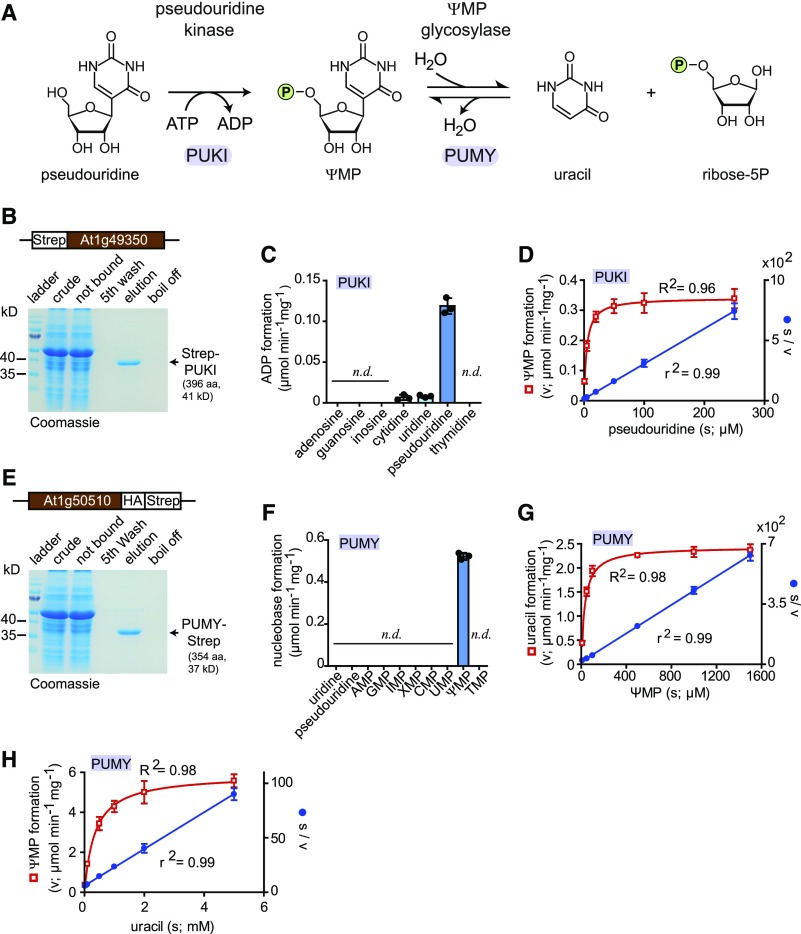
Figure 2.
Biochemical Analyses of PUKI and PUMY.
(A) Reaction scheme.
(B) Purification of N-terminally Strep-tagged PUKI from leaf extracts of N. benthamiana via Strep-Tactin affinity chromatography. Crude, clarified extract; not bound, clarified extract after incubation with Strep-Tactin matrix; fifth wash, wash fluid after the fifth wash of the affinity matrix; elution, affinity eluate; boil off, supernatant after boiling the affinity matrix in SDS loading buffer. aa, amino acids.
(C) Enzymatic activity of Strep-PUKI with different nucleosides (each 1 mM) and 0.4 mM ATP. Error bars are sd (n = 3 independent reactions). n.d., not detectable.
(D) Determination of the kinetic constants for PUKI by fitting the kinetic data using the Michaelis–Menten equation (red curve). The data were also plotted as s/v over s (Hanes plot) and fitted by linear regression (blue line). Error bars are sd (n = 4 independent reactions). s, substrate concentration; v, enzymatic velocity.
(E) Purification of C-terminally Strep-tagged PUMY from leaf extracts of N. benthamiana via Strep-Tactin affinity chromatography. aa, amino acids.
(F) Enzymatic activity of PUMY-Strep with nucleosides and nucleotides (each 1 mM). Error bars are sd (n = 3 independent reactions). n.d., not detectable.
(G) Same as in (D) but for the forward reaction of PUMY, generating uracil and ribose-5-phosphate. s, substrate concentration; v, enzymatic velocity.
(H) Same as in (D) but for the reverse reaction of PUMY, generating ΨMP. s, substrate concentration; v, enzymatic velocity.
Biochemical Characterization
To validate the bioinformatic analyses, we cloned a cDNA of the putative pseudouridine kinase derived from the locus At1g49350 using 2-week-old Arabidopsis seedlings as starting material. The cDNA was transiently expressed in wild tobacco (Nicotiana benthamiana), generating an N-terminal Strep-tagged fusion protein, which was affinity purified to high homogeneity (Figure 2B), yielding an average of 15 µg of protein per gram of fresh leaf material.
We assessed the substrate spectrum of the enzyme by determining its activity with a range of nucleosides at a concentration of 1 mM. Pseudouridine was a good substrate, whereas cytidine and uridine were phosphorylated at an >16-fold lower rate than pseudouridine (Figure 2C). Purine nucleosides and thymidine were not accepted as substrates. Based on these results, we decided to name this enzyme PSEUDOURIDINE KINASE (PUKI). The catalytic efficiency (kcat/Km) of PUKI is 0.05 µM−1 s−1 for pseudouridine (Figure 2D; Tables 1 and 2), a value slightly lower than the catalytic efficiency of the E. coli ortholog YeiC (0.15 µM−1 s−1; Preumont et al., 2008). Interestingly, these values result from a 27-fold lower Km and an 80-fold lower kcat of PUKI versus YeiC (Tables 1 and 2), indicating that the plant enzyme has a much higher affinity for the substrate but markedly less catalytic activity than YeiC.
Table 1. Kinetic Constants of PUKI and PUMY from Arabidopsis.
| Enzymatic Reaction | Km (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) |
|---|---|---|---|
| PUKI | 4.4 ± 0.5 | 0.23 ± 0.01 | 0.0523 |
| PUMY, forward | 32.7 ± 2.6 | 1.41 ± 0.04 | 0.0431 |
| PUMY, reverse | 350.9 ± 38.8 | 3.41 ± 0.23 | 0.0097 |
Km for different substrates: pseudouridine at an ATP concentration of 0.4 mM (PUKI), uracil at a ribose-5-phosphate concentration of 4 mM (PUMY, reverse). Errors are sd (n = 4 independent enzymatic reactions).
Table 2. Kinetic Constants of PUKI and PUMY from E. coli as Reported by Preumont et al. (2008).
| Enzymatic Reaction | Km (µM) | kcat (s−1) | kcat/Km (µM−1 s−1) |
|---|---|---|---|
| PUKI (YeiC) | 120 | 18.58 | 0.15 |
| PUMY (YeiN), forward | 60 | 0.6 | 0.01 |
| PUMY (YeiN), reverse | n.d. | n.d. | n.d. |
Km for different substrates: pseudouridine at an ATP concentration of 0.4 mM (PUKI), uracil at a ribose-5-phosphate concentration of 4 mM (PUMY, reverse). n.d., not determined.
We also cloned a cDNA for the putative ΨMP glycosylase encoded at locus At1g50510 and expressed it in N. benthamiana, resulting in a C-terminal–tagged enzyme variant that we affinity purified to high homogeneity (Figure 2E). On average, 17 µg of protein per gram of fresh leaf material was obtained. We assessed the ability of the enzyme to release the nucleobase from eight nucleoside monophosphates and two nucleosides, each at a concentration of 1 mM. Catalytic activity was observed only for ΨMP (Figure 2F); therefore, this enzyme was called PSEUDOURIDINE MONOPHOSPHATE GLYCOSIDASE (PUMY). The catalytic efficiency of PUMY is 0.04 µM−1 s−1 for ΨMP (Figure 2G; Table 1s and 2), a value that is 4 times higher than that of the ortholog YeiN from E. coli (Tables 1 and 2). For YeiN, the reaction can run in reverse using ribose-5-phosphate and uracil as substrates to produce ΨMP, but the catalytic efficiency has not been reported (Preumont et al., 2008). We observed that PUMY from Arabidopsis also catalyzes the reverse reaction with a catalytic efficiency for urcil of 0.0097 µM−1 s−1 (Figure 2H; Tables 1 and 2), which is ∼4 times lower than the forward reaction.
Pseudouridine Catabolism Resides in Peroxisomes
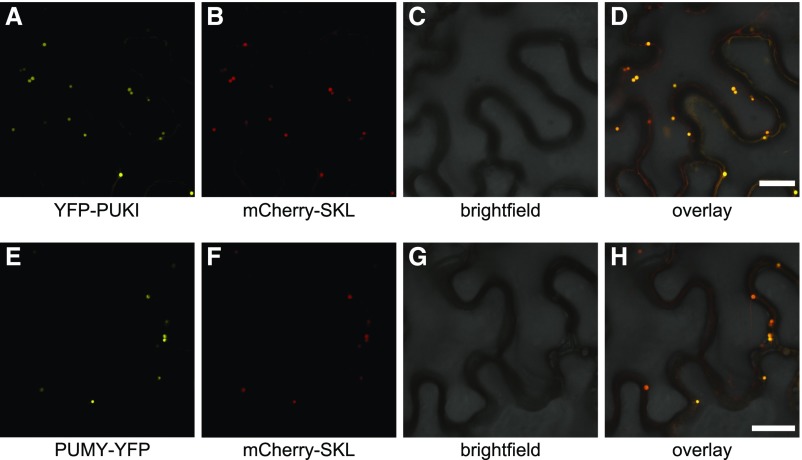
PUKI and PUMY have been detected in proteome studies of isolated peroxisomes, and their peroxisomal localizations have been confirmed by confocal laser scanning microscopy to validate the proteome data (Eubel et al., 2008; Reumann et al., 2009; Lingner et al., 2011). PUKI was called peroxisomal phosphofructokinase B family protein and PUMY was called indigoidine synthase A family protein in these studies. We aimed to confirm this subcellular localization of PUKI and PUMY by using fusion proteins comprising an N-terminal yellow fluorescent protein (YFP) tag for PUKI and a C-terminal YFP tag for PUMY transiently expressed from corresponding cDNA constructs in N. benthamiana leaves. As a positive control, a construct encoding mCherry fused to PTS1 (SKL) was coexpressed with these constructs (Nelson et al., 2007). Confocal microscopy confirmed that both proteins indeed reside in the peroxisomes (Figure 3). Interestingly, peroxisomal targeting signals are present in PUKI and PUMY sequences throughout the green linage, but the type of PTS (PTS1, PTS2, or both) varies between the enzymes of different species (Table 3; Supplemental Figure 1). By contrast, the PUMY-PUKI fusion protein found in the nongreen linages is generally predicted to reside in the mitochondria (Supplemental Table 1).
Figure 3.
Subcellular Localization of PUKI and PUMY.
(A) to (D) Confocal fluorescence microscopy images of N. benthamiana cells in the lower leaf epidermis transiently coproducing mCherry-SKL and PUKI fused to an N-terminal YFP tag (YFP-PUKI): YFP (A), mCherry (B), brightfield (C), and overlay of YFP, mCherry, and brightfield (D). All infiltrated cells exhibited the same pattern. Bar = 25 µm.
(E) to (H) Same as in (A) to (D) but for C-terminally YFP-tagged PUMY.
Table 3. Presence of Putative Peroxisomal Targeting Signals in PUKI and PUMY from Plants and Algae.
| Species | PUKI | PUMY | ||
|---|---|---|---|---|
| PTS1a | PTS2b | PTS1a | PTS2b | |
| Arabidopsis thaliana | ● | ● | ||
| Carica papaya | ● | ● | ||
| Gossypium raimondii | ● | ● | ||
| Manihot esculenta | ● | ● | ● | |
| Vitis vinifera | ● | ● | ● | |
| Cucumis sativus | ● | ● | ||
| Phaseolus vulgaris | ● | ● | ||
| Solanum lycopersicum | ● | ● | ||
| Mimulus guttatus | ● | ● | ||
| Zea mays | ● | ● | ||
| Sorghum bicolor | ● | ● | ||
| Serratia italica | ● | ● | ||
| Oryza sativa | ● | ● | ||
| Brachypodium distachyon | ● | ● | ||
| Zostera marina | ● | ● | ||
| Marchantia polymorpha | ● | ● | ||
| Spagnum phallax | ● | ● | ||
| Physcomitrella patens | ● | ● | ||
| Volvox cateri | ● | ● | ||
| Ostreococcus lucimarinus | ●c | ● | ||
Blank cells indicate the absense of a PTS.
S-[KMSA]-L was used as the motif for PTS1.
R-[VLIM]-x(5)-H-[VILMF] was used as the motif for PTS2.
Motif deviates from the consensus sequence.
Pseudouridine Catabolism in Vivo
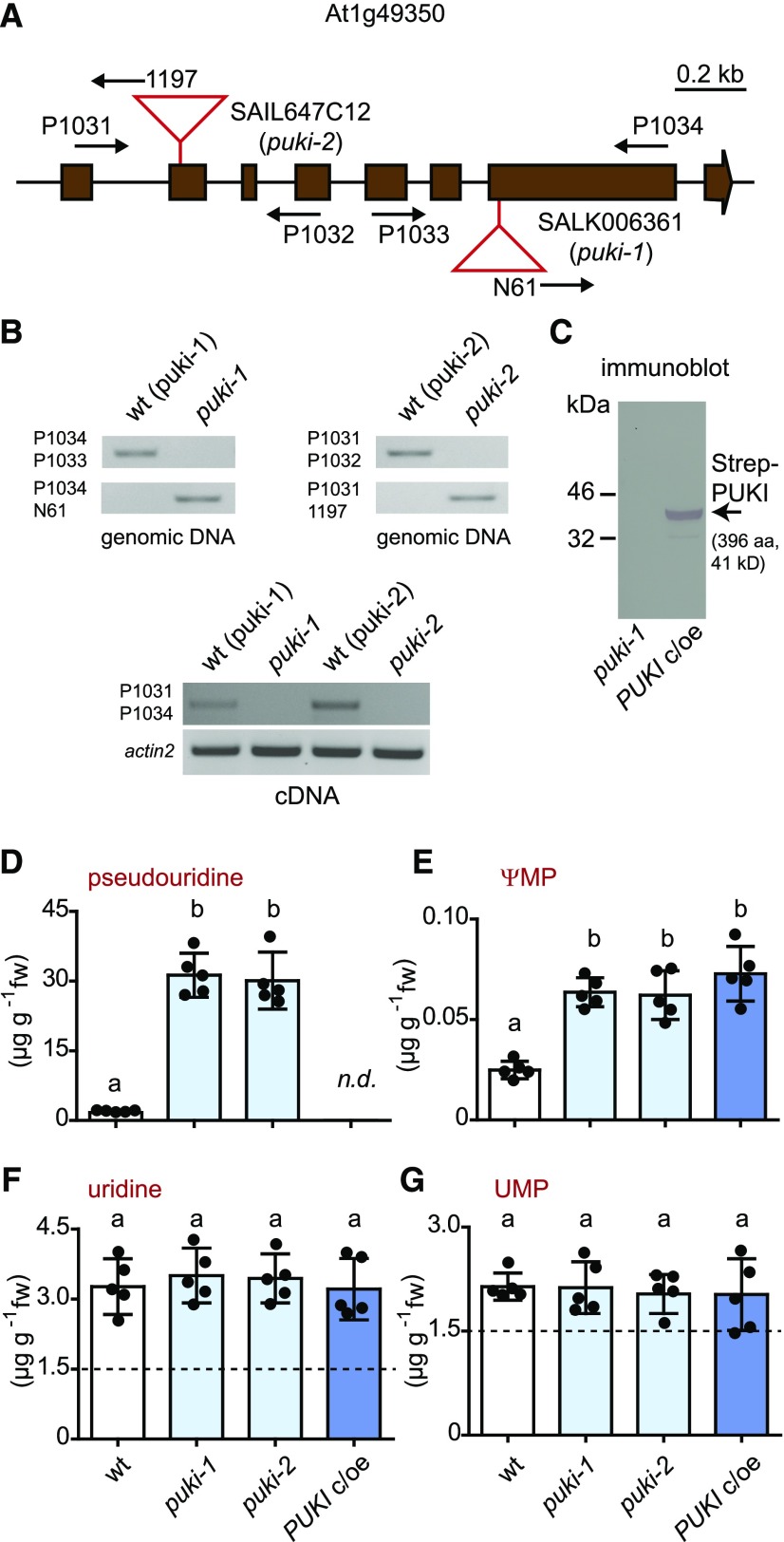
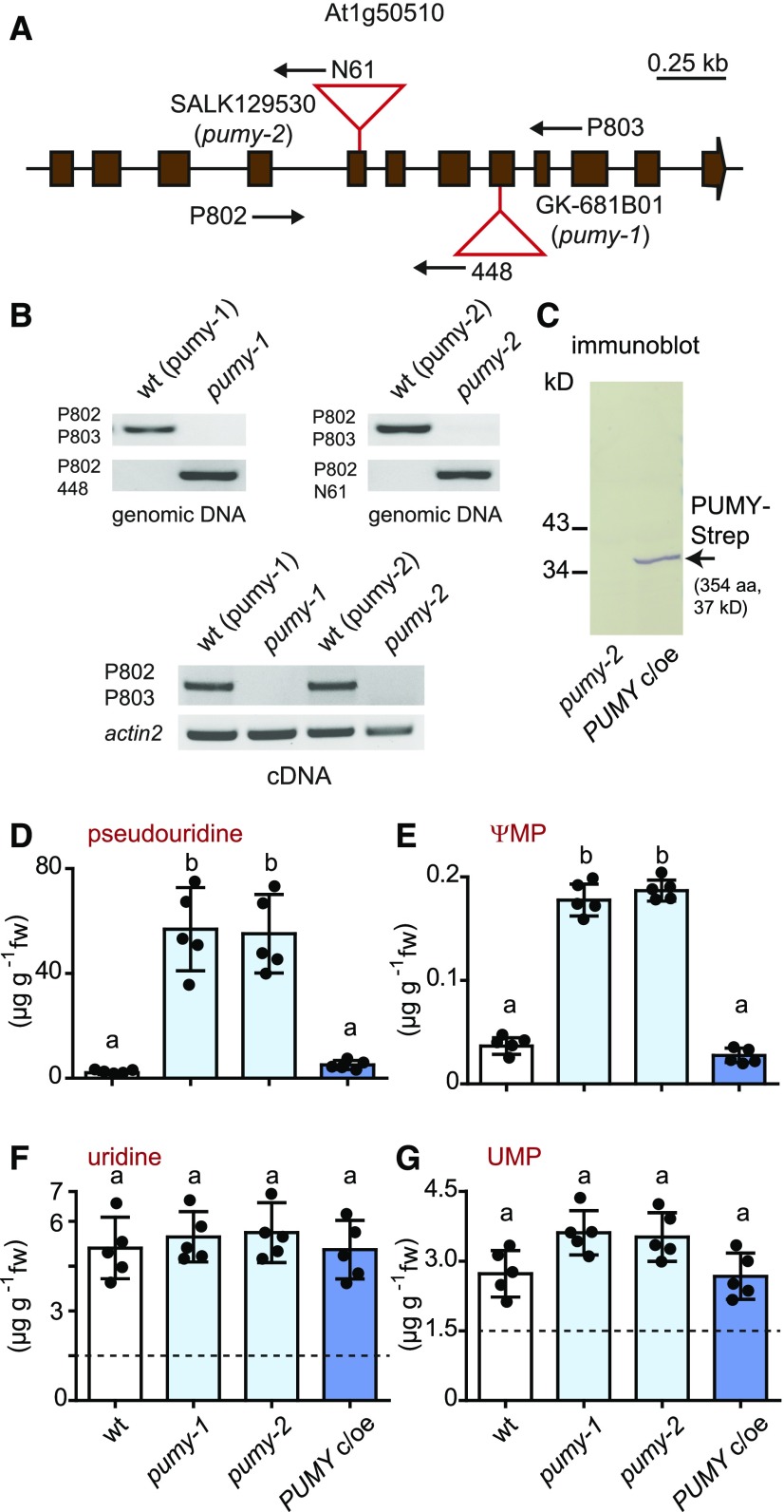
To assess the role of PUKI in vivo, we isolated two independent homozygous T-DNA insertion mutants of PUKI from segregating populations, including one from the SALK collection (Alonso et al., 2003) called puki-1 and one from the SAIL collection (Sessions et al., 2002) called puki-2 (Figures 4A and 4B). For each mutant, we also isolated the corresponding wild-type line to obtain a line genetically as close as possible to the respective mutant. The absence of intact PUKI mRNA was confirmed for both mutants (Figure 4B, bottom). Additionally, we generated a complementation and overexpression line producing N-terminal Strep-tagged PUKI in the puki-1 background called PUKI c/oe. The expression of the transgene for the tagged fusion protein was confirmed in transgenic seedlings (Figure 4C).
Figure 4.
Characterization and Metabolite Analysis of 2-Week-Old Seedlings Varying in PUKI Expression.
(A) Genomic organization of the At1g49350 locus encoding PUKI and positions of the T-DNA insertions (red triangles) in puki-1 (SALK006361) and puki-2 (SAIL647C12). The coding sequence (brown boxes) interrupted by noncoding regions (lines) is shown. Primer positions are indicated by arrows.
(B) PCR analysis of the wild type and T-DNA alleles (top panels) and RT-PCR to assess the presence of intact mRNA (bottom panel) in homozygous puki-1 and puki-2 lines and their corresponding wild types, wt (puki-1) and wt (puki-2), selected from the segregating mutant populations. aa, amino acids.
(C) Immunoblot analysis of seedling extracts from the Strep-PUKI complementation and overexpression line (PUKI c/oe) generated in the puki-1 background compared with the puki-1 line. An anti-Strep tag antibody was used for detection. aa, amino acids.
(D) to (G) pseudouridine, ΨMP, uridine, and UMP contents in 2-week-old seedlings of the wild type (wt, white), the two puki lines (light blue), and the PUKI c/oe line (dark blue). Error bars are sd (n = 5 biological replicates; lines were all grown together on one plate; for each line, material from five distinct plates was harvested.) The detection limit for pseudouridine was 0.4 ng (in matrix using a 20-μL sample). Different letters label data that are significantly different at P < 0.05. fw, fresh weight; n.d., not detectable.
Next, we quantified ΨMP and pseudouridine contents, as well as UMP and uridine contents, by performing liquid chromatography coupled to quadrupole mass spectrometry (LC-MS/MS) of extracts from 2-week-old seedlings of the wild type (derived from the segregating population of puki-1), the two PUKI mutants (puki-1 and puki-2), and the complementation and overexpression line (PUKI c/oe). The pseudouridine contents were ∼18 times higher in both PUKI mutants (mean, 30.7 μg g−1) compared to the wild type (mean, 1.7 μg g−1), whereas pseudouridine was not detected in the PUKI c/oe line (Figure 4D; Supplemental File). The data demonstrate that PUKI indeed functions as a pseudouridine kinase in vivo.
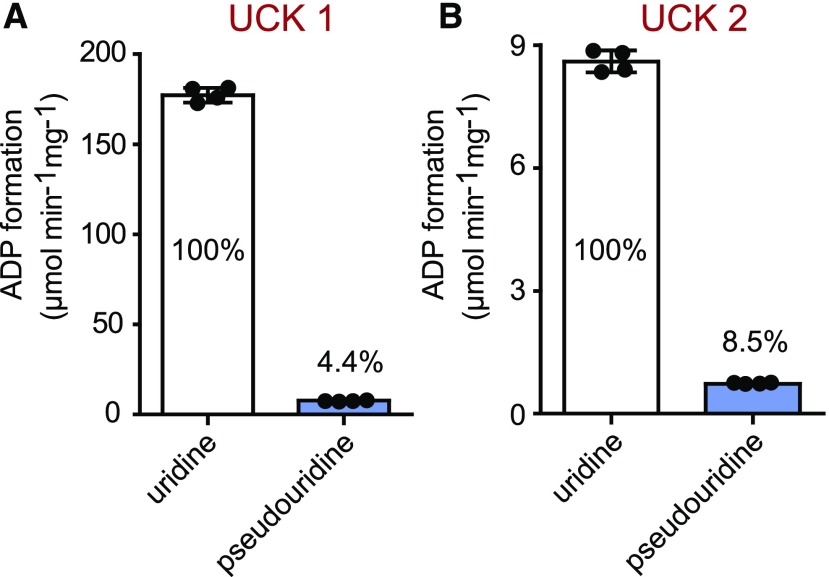
Surprisingly, both puki seedlings also contained ∼3 times more ΨMP than the wild type (Figure 4E). First, one should note that the pseudouridine concentration is generally by far higher than the ΨMP concentration, even in the wild type, which contained 1.7 μg g−1 pseudouridine but only ∼0.025 μg g−1 ΨMP (Figures 4D and 4E). In the puki background, the pseudouridine concentration is ∼10-fold higher than the uridine concentration (Figures 4D and 4F). We speculated that an unknown kinase, perhaps a cytosolic uridine cytidine kinase (UCK; Ohler et al., 2019), can phosphorylate pseudouridine under these extreme conditions, resulting in an increase in ΨMP contents in the puki background. We assessed whether Arabidopsis UCK1 and UCK2 accept pseudouridine as a substrate. Indeed, both kinases phosphorylated pseudouridine (Figure 5). At a substrate concentration of 1 mM, pseudouridine was converted at 4.4% of the rate measured for uridine by UCK1, which is responsible for most UCK activity in vivo (Ohler et al., 2019).
Figure 5.
Enzymatic Activity of UCK1 and UCK2 with Uridine and Pseudouridine.
(A) Specific activity of UCK1 purified after transient production in N. benthamiana leaves as C-terminal Strep-tagged variant, with 1 mM uridine or pseudouridine in the presence of 0.4 mM ATP. ADP formation was monitored for 10 min, and linear rates were observed in all cases. Error bars are sd (n = 4 independent enzyme assays). The rate for pseudouridine phosphorylation relative to uridine phosphorylation, which was set to 100%, is indicated.
(B) Same as in (A) but for UCK2.
The PUKI c/oe line contained ∼3 times more ΨMP (mean, 0.073 μg g−1) than the wild type (mean, 0.025 μg g−1; Figure 4E), likely due to the more pronounced phosphorylation of pseudouridine by the elevated PUKI levels in this line. In the wild type, the rate of pseudouridine-to-ΨMP conversion appears to be limited by the available catalytic capacity of PUKI.
The UMP and uridine contents were not significantly different among genotypes (Figures 4F and 4G), indicating that (1) PUKI does not act as a uridine kinase in vivo, even when overproduced, although it has limited UCK activity at high substrate concentrations in vitro (Figure 2C) and that (2) strong accumulation of pseudouridine does not interfere with the enzymes that maintain UMP and uridine homeostasis.
We also isolated two independent homozygous T-DNA insertion mutants for PUMY from segregating populations, including one from the GABI-Kat collection (Kleinboelting et al., 2012) called pumy-1 and one from the SALK collection (Alonso et al., 2003) called pumy-2 (Figures 6A and 6B). We isolated the corresponding wild-type lines as well. The absence of intact PUMY mRNA was confirmed for both mutants (Figure 6B, bottom). Additionally, we prepared a complementation and overexpression line producing C-terminal Strep-tagged PUMY in the pumy-2 background called PUMY c/oe. The expression of the transgene encoding the tagged fusion protein was confirmed in transgenic seedlings (Figure 6C).
Figure 6.
Characterization and Metabolite Analysis of 2-Week-Old Seedlings Varying in PUMY Expression.
(A) Genomic organization of the At1g50510 locus encoding PUMY and positions of the T-DNA insertions (red triangles) in pumy-1 (GK681B01) and pumy-2 (SALK129530). The coding sequence (brown boxes) interrupted by noncoding regions (lines) is shown. Primer positions are indicated by arrows.
(B) PCR analysis of the wild-type and T-DNA alleles (top panels) and RT-PCR to assess the presence of intact mRNA (bottom panel) in homozygous pumy-1 and pumy-2 lines and their corresponding wild types, wt (pumy-1) and wt (pumy-2), selected from the segregating mutant populations.
(C) Immunoblot analysis of seedling extracts from the PUMY-Strep complementation and overexpression line (PUMY c/oe) generated in the pumy-2 background compared with the pumy-2 line. An anti-Strep tag antibody was used for detection. aa, amino acids.
(D) to (G) pseudouridine, ΨMP, uridine, and UMP contents in 2-week-old seedlings of the wild type (wt, white), the two pumy lines (light blue), and the PUMY c/oe line (dark blue). Error bars are sd (n = 5 biological replicates; lines were all grown together on one plate; for each line, material from five distinct plates was harvested.) Different letters label data that are significantly different at P < 0.05. fw, fresh weight.
We detected 5 times more ΨMP (mean, 0.18 μg g−1) and 28 times more pseudouridine (mean, 56 μg g−1) in both pumy lines than in the wild type (mean ΨMP, 0.036 μg g−1; mean pseudouridine, 2 μg g−1) in 2-week-old seedlings (Figures 6D and 6E). In the PUMY c/oe line, the concentrations of both metabolites were nearly the same as in the wild type, demonstrating that the complementation was successful. These data demonstrate that PUMY is a ΨMP glycosylase in vivo, but the high accumulation of pseudouridine seems counterintuitive at first. There are several possible explanations for this finding: (1) PUKI is inhibited by accumulating ΨMP; (2) the putative activation of PUKI by interaction with PUMY is disrupted in the pumy background; and (3) ΨMP accumulation is prevented by a (pyrimidine) nucleotide phosphatase, which creates a futile cycle of pseudouridine phosphorylation by PUKI and dephosphorylation by this phosphatase. The first option is unlikely because the absolute concentration of ΨMP in vivo is quite low, and we did not observe substantial product inhibition of PUKI in our kinetic measurements. There is also no evidence for the second option because we did not detect an interaction between PUKI and PUMY (Supplemental Figure 3), and the enzymatic activity of PUKI was not influenced by the presence of PUMY in vitro (Supplemental Figure 4). However, there is some support for the third explanation because a slight increase in the UMP concentration was always observed in the pumy background (Figure 6G). This is potentially caused by the partial occupation of an unknown UMP phosphatase with ΨMP, reducing its capacity for UMP dephosphorylation. Because such a phosphatase likely resides outside the peroxisome, probably in the cytosol, this scenario requires the transport of ΨMP from the peroxisome to the cytosol. The peroxisomal membrane contains transporters that catalyze the import of ATP, NAD, and CoA in counter exchange for AMP (Charton et al., 2019). Perhaps ΨMP becomes a nonphysiological substrate of (one of) these transporters when it accumulates in mutants defective in ΨMP catabolism.
Strikingly, the pseudouridine concentration was 2 times higher in 2-week-old seedlings in the pumy (Figure 6D) versus the puki background (Figure 4D), indicating that even in the absence of PUKI, there is still some pseudouridine degradation. A bypass reaction at high pseudouridine concentration may be operative here: pseudouridine might be phosphorylated by the UCKs (Figure 5) in the cytosol, followed by the import of ΨMP into the peroxisome and degradation by PUMY. It appears that depending on the chemical gradient, ΨMP might be transported in both directions over the peroxisomal membrane.
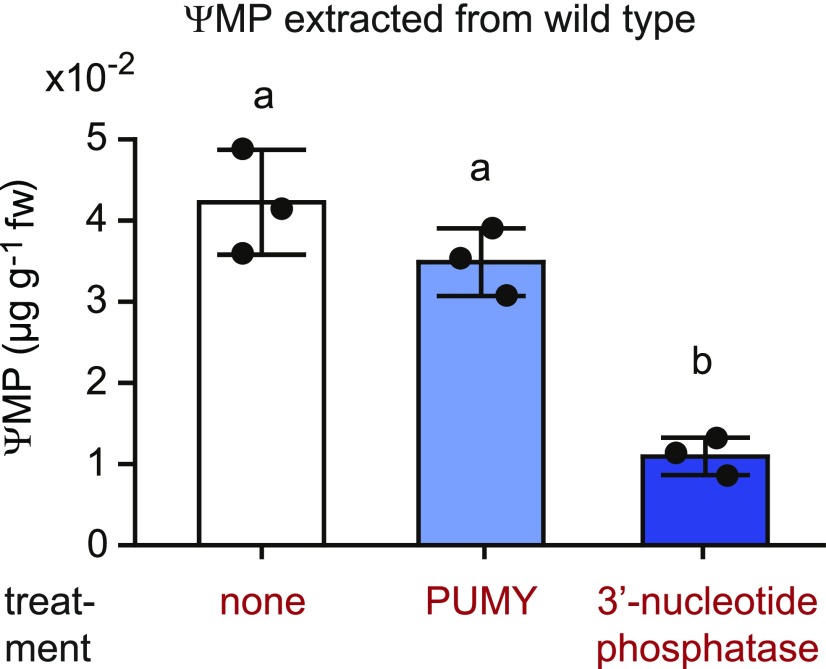
Interestingly, ΨMP was not completely consumed in the PUMY c/oe line but was present at roughly the same concentration as in the wild type (Figure 6E). This indicates that the ΨMP detectable in both lines is not located in the peroxisome, because otherwise it would be hydrolyzed by PUMY, at least in the PUMY c/oe line. ΨMP extracted from the wild type was highly resistant to added PUMY in vitro but could be dephosphorylated to large extent by a 3′-nucleoside monophosphate–specific phosphatase (Figure 7; Supplemental Figure 5), demonstrating that most of the total ΨMP is actually 3′-ΨMP rather than 5′-ΨMP. Vacuolar RNA degradation generates 3′ nucleotides as intermediates before they are dephosphorylated to nucleosides for export into the cytosol via the equilibrative nucleoside exporter1 (ENT1; Bernard et al., 2011). Therefore, it is likely that this 3′-ΨMP resides in the vacuole, representing most of the ΨMP detected in the wild type, whereas the concentration of 5′-ΨMP (the PUMY substrate) is very low. It appears that cells need PUKI for the following reasons: (1) 5′-ΨMP is not an intermediate of RNA degradation in the vacuole, where most Ψ-containing RNAs (rRNA and tRNA) are degraded; thus, RNA degradation does not generate the PUMY substrate directly; and (2) the vacuole provides nucleosides and likely not nucleotides to the cytosol.
Figure 7.
Most of the Detectable ΨMP in the Wild Type Is 3′-ΨMP.
ΨMP extracted from 2-week-old wild-type seedlings was incubated for 15 min with PUMY (5′-ΨMP glycosylase), a 3′-nucleotide phosphatase (nuclease P1 from P. citrinum), or without enzyme addition as a negative control. In a separate positive control experiment, the added enzymes were confirmed to be specific and sufficiently active for full substrate conversion under the chosen conditions (Supplemental Figure 5). Error bars are sd (n = 3 independent enzymatic reactions).
We also investigated the metabolic profiles of the PUKI and PUMY variants later during development using leaves of different ages from 37-d-old, soil-grown plants. The results are very similar to those obtained from 2-week-old seedlings grown on agar medium (Supplemental Figures 6 and 7). However, unlike in seedlings, it appears that only slightly more pseudouridine was produced in the pumy background compared to the puki background. This may be explained by the two- to fourfold lower pseudouridine levels in these older plants compared with 2-week-old seedlings (Figures 4D and 6D), resulting in less PUKI-independent degradation initiated (possibly) via the cytosolic UCKs (Figure 5).
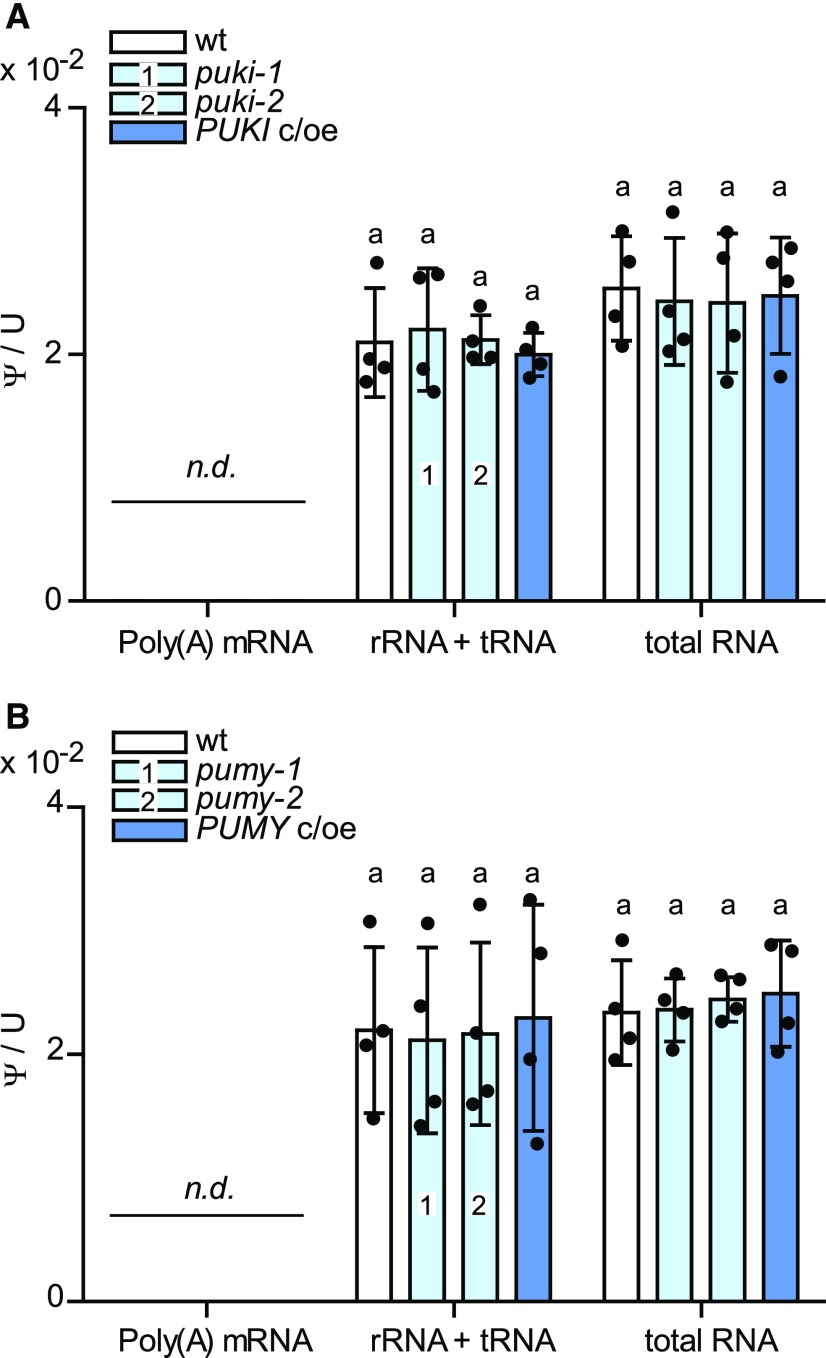
Is the Ψ/U Ratio in RNA Influenced by Defects in Pseudouridine Catabolism?
Pseudouridine and ΨMP accumulate in plants lacking PUKI or PUMY. Perhaps ΨMP becomes phosphorylated to the corresponding triphosphate and is then randomly incorporated into RNA species by RNA polymerases. A major reason plants and other organisms possess ΨMP catabolism might be to prevent such misincorporation into RNA.
We tested this hypothesis by quantifying Ψ/U ratios in mRNA, rRNA + tRNA, and total RNA pools from the puki and pumy lines and the corresponding wild types and complementation lines. The rRNA + tRNA pool showed no significant changes in the Ψ/U ratio in any of the investigated genotypes (Figure 8). There are only 451 Ψ sites in mRNA in the Arabidopsis transcriptome (Sun et al., 2019), and our mass spectrometry method was not sensitive enough to detect this low degree of pseudouridylation. We conclude that plants do not require ΨMP catabolism to protect rRNA and tRNA from erroneous incorporation of Ψ, but whether mRNA might be affected remains an open question. The insensitivity of Ψ abundance in bulk RNA to different pseudouridine and ΨMP concentrations in the respective genotypes indicates that the UMP kinases (uridylate kinases) and the RNA polymerases are sufficiently selective against ΨMP and pseudouridine 5'-triphophate (ΨTP), respectively. A similar scenario has been described for the catabolic pathway of N6-mAMP in Arabidopsis. The abrogation of N6-mAMP catabolism did not alter the m6A/A ratio in different RNA species, suggesting that adenylate kinases function as strong molecular filters that prevent the phosphorylation of most N6-mAMP to the triphosphate (Chen et al., 2018).
Figure 8.
Ψ/U Ratios for Different RNA Species in Seedlings Varying in PUKI or PUMY Expression.
(A) Ψ frequency relative to U for total RNA, nonpolyadenylated RNA (rRNA and tRNA), and mRNA from 2-week-old wild-type seedlings derived from the segregating population of puki-1 (wt; white), the two PUKI mutants (light blue), and the complementation and overexpression line (PUKI c/oe; dark blue). Error bars are sd (n = 4; two independent RNA extractions and RNA pool isolations, each RNA digested twice). The detection limit of pseudouridine was 150 pg (in water using 15-μL sample). n.d., not detectable.
(B) Same as in (A) but for 2-week-old seedlings varying in PUMY expression.
Compromising Pseudouridine Catabolism Delays Seed Germination and Root Growth
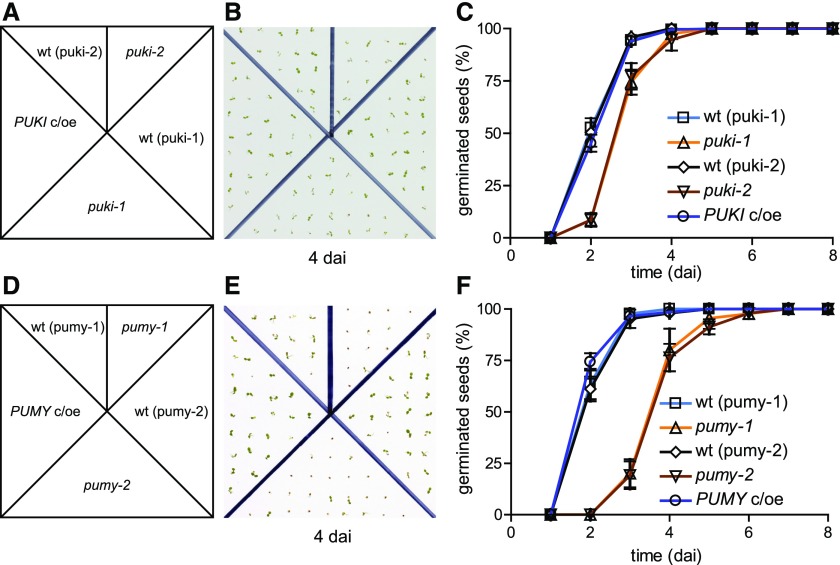
We assessed the physiological performance of the PUKI and PUMY mutants and the respective complementation and overexpression lines (PUKI c/oe and PUMY c/oe). The PUKI mutants, but not the PUKI c/oe line, reproducibly showed a slight delay in germination of ∼0.5 d compared to the wild type (Figures 9A to 9C). The PUMY mutants displayed an even more severe germination delay of ∼2 d, which was not observed in the complementation line PUMY c/oe (Figures 9D to 9F). No other phenotypic alterations occurred in any of the lines during the later stages of development when the plants were grown on soil.
Figure 9.
Comparison of Seed Germination in Plants Varying in PUKI or PUMY Expression.
(A) Scheme for genotype distribution on test plates for PUKI variants. wt (puki-1), wild type derived from the segregating puki-1 population; wt (puki-2), wild type derived from the segregating puki-2 population.
(B) Image of a test plate 4 dai. The plates were incubated under a 16-h-light/8-h-dark cycle.
(C) Germination frequency of freshly harvested seeds over time. Error bars are sd (n = 4; Each test plate contained all genotypes. Seeds from the same genotype were derived from a different mother plant on each test plate.) Mother plants for the seeds had been grown side by side in a randomized setup.
(D) Scheme for genotype distribution on test plates for the PUMY variants.
(E) Image of a test plate 4 dai.
(F) Same as in (C) but for the PUMY expression variants.
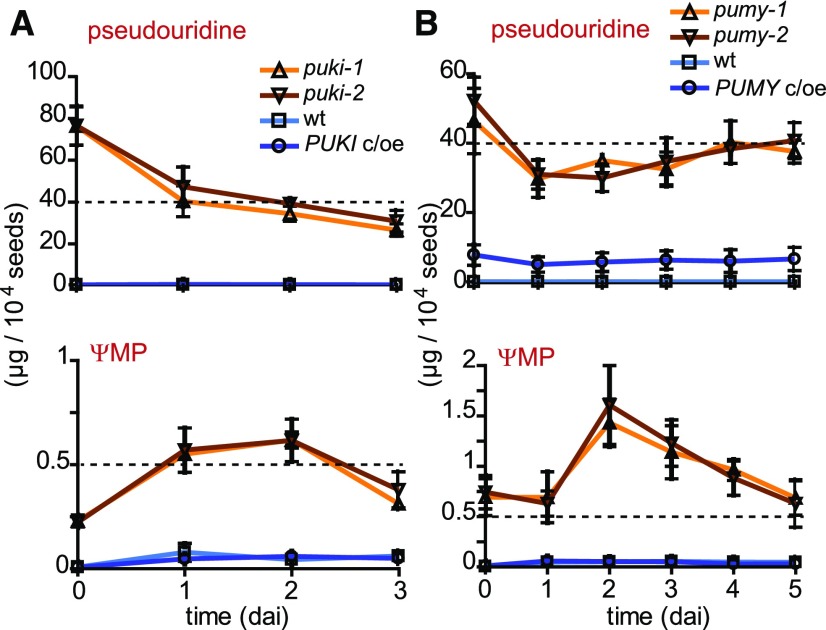
We wondered whether the degree to which germination is delayed might be correlated to the level of pseudouridine or ΨMP accumulation. Therefore, we quantified these metabolites in seeds of the PUKI and PUMY variants, respectively, during the first 3 or 5 days after imbibition (dai), covering the duration of the germination process in these genotypes. The pseudouridine content in the germinating seeds appeared slightly higher in puki compared with pumy lines (Figures 10A and 10B, top), whereas ΨMP was always more abundant in the pumy seeds (Figures 10A and 10B, bottom). The ΨMP contents of all genotypes increased during the first 2 dai, but the effect was small in the wild types and the complementation lines, where ΨMP eventually reached ∼0.05 µg per 104 seeds at 2 dai. At that time, many seeds of the wild types and the c/oe lines had already germinated (Figures 9C and 9F). By contrast, less than 10% of the puki seeds and none of the pumy seeds had germinated at 2 dai, when 0.7 and 1.5 µg of ΨMP per 104 seeds had accumulated in these genotypes, respectively (Figures 10A and 10B, bottom). Perhaps a ΨMP precursor (such as Ψ-containing RNA) that is stored in seeds releases ΨMP during the early days after imbibition, thereby delaying germination.
Figure 10.
Pseudouridine and ΨMP Contents in Lines Varying in PUKI or PUMY Expression during Seed Germination.
(A) Pseudouridine (top) and ΨMP (bottom) contents in germinating seeds of the different PUKI lines. Error bars are sd (n = 3 biological replicates; For each genotype, the seeds were derived from three distinct mother plants.)
(B) Same as in (A) but for germinating seeds of different PUMY expression variants.
UMP concentrations did not differ among genotypes during germination, whereas uridine contents were always significantly lower in germinating seeds of the puki and pumy backgrounds (Supplemental Figure 8).
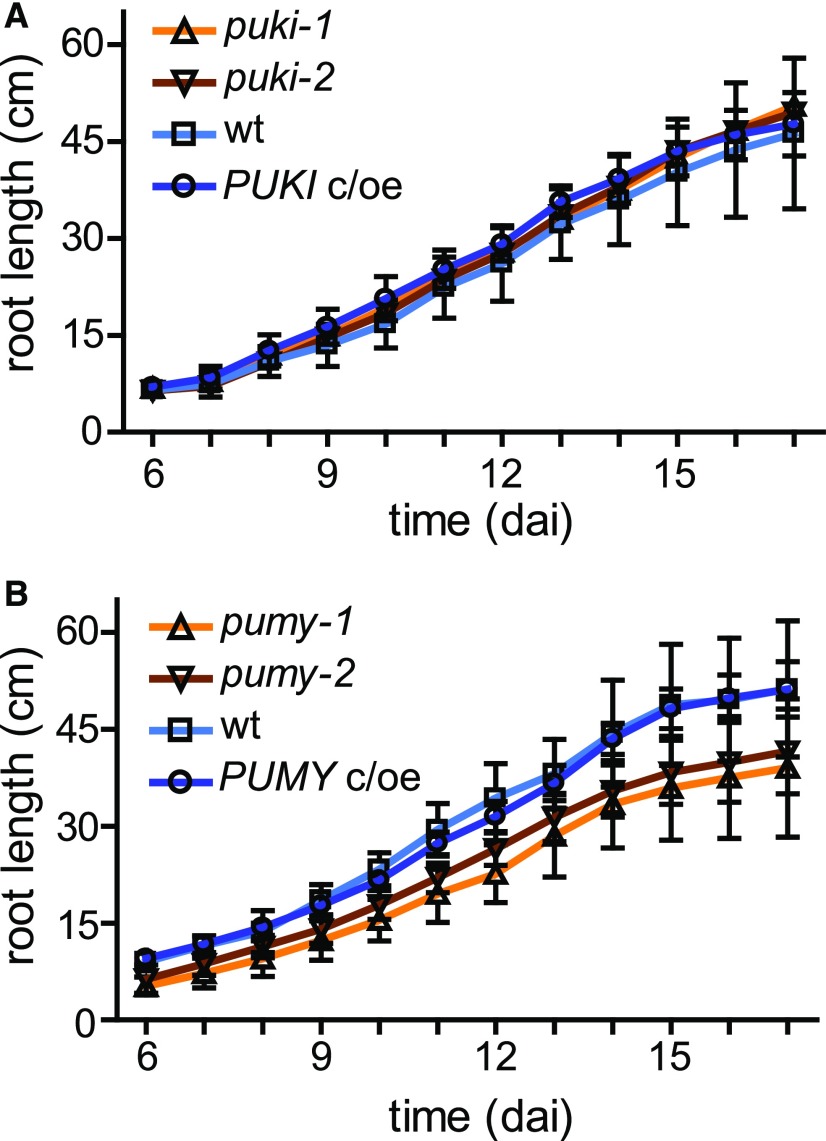
Ψ was recently mapped in different RNA species of Arabidopsis (Sun et al., 2019). Ψ was found at 451 sites in mRNA, frequently occurring in transcripts of genes responding to salt stress. Therefore, we investigated whether our genetic variants of PUKI and PUMY exhibit phenotypic differences in response to salt stress. This was not the case (Supplemental Figure 9), but the root growth of the PUMY mutants was compromised in both the presence and absence of salt, which was not observed in the PUMY c/oe line (Figure 11; Supplemental Figure 9). We also analyzed the pseudouridine and ΨMP contents of 17-d-old seedlings grown in the presence of 125 mM NaCl compared with seedlings grown on control medium without additional salt. Although pseudouridine and ΨMP contents increased in the PUKI variant genotypes by ∼2.0- and 1.2-fold, respectively, and in PUMY variant genotypes by ∼2.7- and 1.2-fold, respectively, in the presence of NaCl, there were no significant differences between genotypes. Control metabolites such as adenosine and AMP showed similar increases in concentration in the presence of salt stress. We therefore attributed these differences to the smaller seedling size of stressed plants rather than to any specific effects of salt stress on pseudouridine or ΨMP accumulation.
Figure 11.
Root growth of PUKI and PUMY expression variants.
(A) Comparison of root growth in the wild type, PUKI mutants, and PUKI c/oe. At 6 dai, germinated seedlings were placed on horizontal plates containing half-strength MS medium (without salt) and root lengths were determined daily up to 17 dai. Error bars are sd (n = 9 seedlings per genotype in groups of three on three different plates).
(B) Same as in (A) but for different PUMY expression variants.
ΨMP Is Toxic to Plants
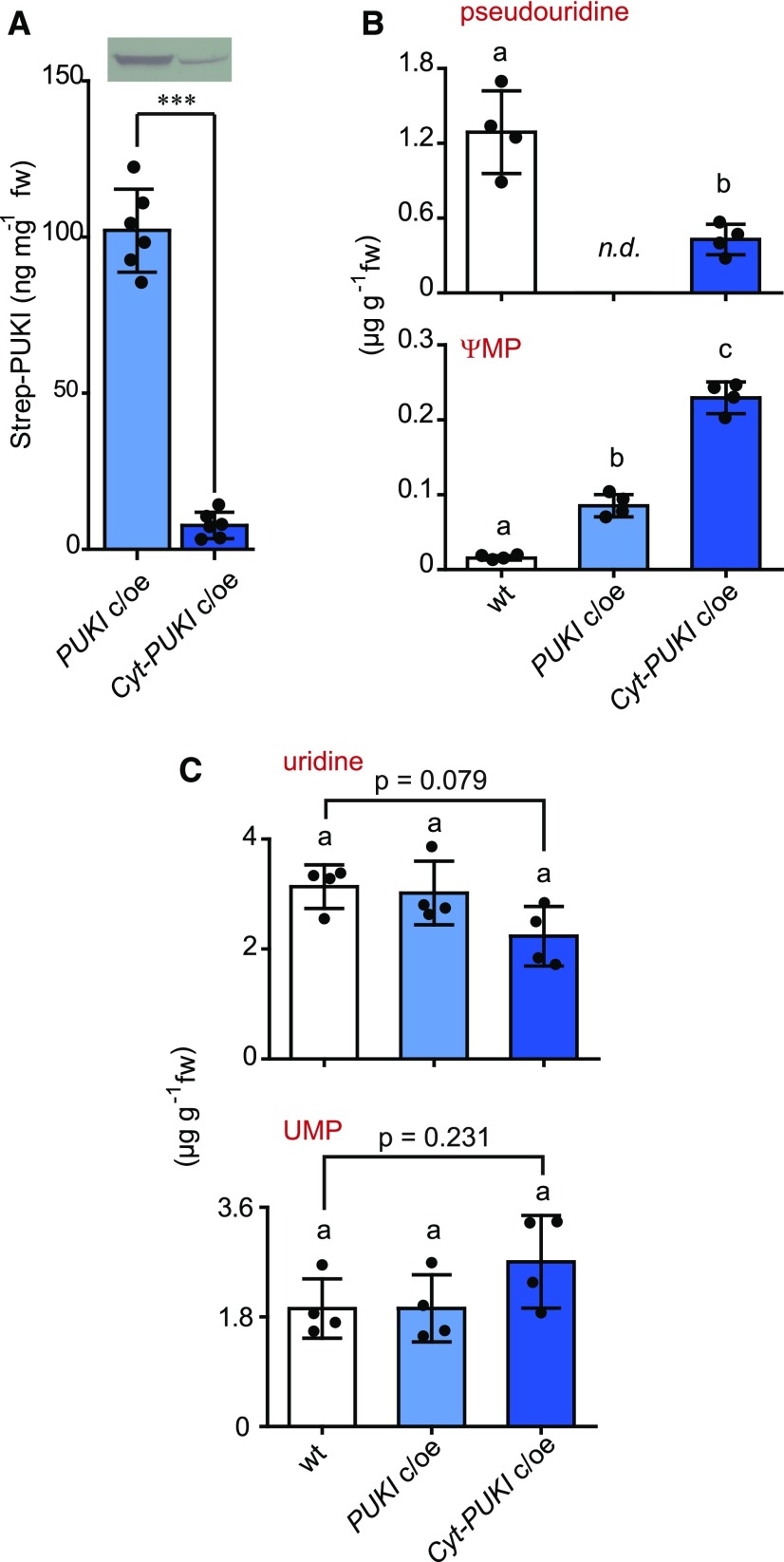
The reason that pseudouridine phosphorylation coupled to ΨMP degradation is confined to the peroxisome might lie in the toxicity of ΨMP. In the puki lines, peroxisomal pseudouridine phosphorylation is abrogated and only bypass phosphorylation takes place, likely via the activity of cytosolic UCKs, resulting in aberrant ΨMP generation in the cytosol. According to this model, the steady-state concentration of ΨMP in puki is the result of (1) pseudouridine phosphorylation by UCKs, (2) ΨMP dephosphorylation by a cytosolic (UMP) phosphatase, and (3) ΨMP import into the peroxisome coupled to degradation by PUMY. We reasoned that it might be possible to boost the ΨMP concentration to test its toxicity by increasing the rate of pseudouridine phosphorylation in the cytosol. Therefore, we generated transgenic Arabidopsis plants overproducing a cytosolic PUKI variant in the puki background designated cytosolic (Cyt)-PUKI c/oe. Only plant lines with a relatively low degree of Cyt-PUKI overproduction were recovered after selection, indicating that strong overproducers did not survive. In the Cyt-PUKI c/oe line used for further experiments, the enzyme is 13-fold less abundant than in the peroxisomal PUKI c/oe line used throughout this study (Figure 12A). Despite the relatively low abundance of Cyt-PUKI, the accumulation of ∼30 µg g−1 pseudouridine normally observed in seedlings in the puki background (Figure 4D) was efficiently suppressed in the Cyt-PUKI c/oe line to less than 0.5 µg g−1, which is lower than in the wild type (Figure 12B, top). The ΨMP concentration in the Cyt-PUKI c/oe line (mean, 0.229 μg g−1) was higher than in any other line used in this study, that is, >2-fold higher than in PUKI c/oe (mean, 0.085 μg g−1) and ∼14-fold higher than in the wild type (mean, 0.016 μg g−1; Figure 12B, bottom). Uridine and UMP concentrations were not significantly altered in this line, although it tended to exhibit a reduction in uridine levels and an increase in UMP levels (Figure 12C). These results demonstrate that pseudouridine degradation is operational even if PUKI is misplaced in the cytosol, confirming the previous notion that cytosolic ΨMP can be imported into the peroxisome to reach PUMY. Interestingly, we repeatedly failed to introduce Cyt-PUKI into the pumy background despite several transformation attempts. In the pumy background, the escape route of cytosolic ΨMP into the peroxisome to the PUMY sink is not functional, which apparently cannot be tolerated, supporting the idea that ΨMP becomes toxic at more elevated concentrations.
Figure 12.
Comparison of the Peroxisomal and Cyt-PUKI Overproduction Lines and the Wild Type.
(A) Protein quantification of Strep-PUKI produced in 2-week-old PUKI c/oe and Cyt-PUKI c/oe seedlings. PUKI c/oe, peroxisomal PUKI complementation and overexpression line in the puki-1 background; Cyt-PUKI c/oe, Cyt-PUKI complementation and overexpression line in the puki-1 background. (Top) Immunoblot with anti-Strep tag detection. Error bars are sd (n = 6 biological replicates; plants derived from six different growth plates analyzed in six immunoblots as shown top). ***, t test indicates significant difference at P < 0.001. fw, fresh weight.
(B) Pseudouridine and ΨMP contents in 2-week-old wild type, PUKI c/oe, and Cyt-PUKI c/oe seedlings. Error bars are sd (n = 4 biological replicates). Different letters label data that are significantly different at P < 0.05. fw, fresh weight; n.d., not detectable.
(C) Same as in (B) but showing uridine and UMP contents. P values of ANOVA followed by Tukey’s post test are given where indicated. fw, fresh weight.
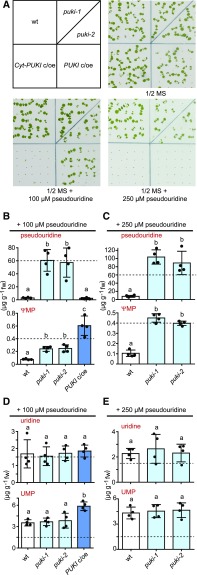
To further assess the toxicity of ΨMP, we compared the susceptibility of PUKI c/oe and Cyt-PUKI c/oe seedlings to externally supplemented pseudouridine. The wild type and PUKI mutants were also included in this experiment. The Cyt-PUKI c/oe line failed to grow on medium supplemented with 100 µM pseudouridine, whereas all other genotypes grew (Figure 13A). At 250 µM pseudouridine, the PUKI c/oe line also stopped growing. The pseudouridine concentration in 2-week-old PUKI mutant seedlings increased from ∼30 µg g−1 (Figure 4D) to 60 µg g−1 when supplemented with 100 µM pseudouridine (Figure 13B) and to almost 100 µg g−1 when supplemented with 250 µM pseudouridine (Figure 13C), whereas the ΨMP concentration increased from ∼0.06 to 0.24 µg g−1 and then to >0.44 µg g−1 under these conditions. However, the highest ΨMP concentration (∼0.6 µg g−1) was reached in the PUKI c/oe line supplemented with 100 µM pseudouridine (Figure 13B, bottom), that is, sixfold more than without pseudouridine addition (Figure 12B, bottom), whereas this line never contained more pseudouridine than the wild type on normal medium or in the presence of external pseudouridine (Figures 12B and 13B).
Figure 13.
Phenotypes and Metabolite Contents of Seedlings Varying in PUKI Expression Grown in the Presence of Pseudouridine.
(A) Growth responses of the wild type (wt), the puki-1 and puki-2 lines, the PUKI c/oe line, and the Cyt-PUKI c/oe line to 100 or 250 µM pseudouridine in the growth medium. Images were taken 7 dai. All seeds germinated, but plant growth was inhibited, depending on the genotype and the pseudouridine content of the medium. ½ MS, half-strength MS.
(B) Quantification of pseudouridine (top) and ΨMP (bottom) in 2-week-old PUKI variant seedlings in the presence of 100 µM pseudouridine. Error bars are sd (n = 4 biological replicates; Four plates containing all genotypes. Seeds from the same genotype were derived from a different mother plant on each plate.) Different letters label data that are significantly different at P < 0.05. fw, fresh weight.
(C) Same as in (B) but for growth medium containing 250 µM pseudouridine. fw, fresh weight.
(D) and (E) Same as in (B) and (C) but showing uridine (top) and UMP (bottom) levels. fw, fresh weight.
These results support the hypothesis that ΨMP becomes toxic at relatively low concentrations, because plants with the highest capacity for ΨMP generation (PUKI c/oe and Cyt-PUKI c/oe) were the least resistant to external pseudouridine addition (Figure 13A), although they did not accumulate any pseudouridine (Figures 12B and 13B). By contrast, a high pseudouridine content does not appear to strongly influence plant growth, although pseudouridine accumulated in the PUKI mutants to more than two orders of magnitude higher concentrations than ΨMP, exceeding the cellular uridine concentration by more than 30-fold (Figures 13B to 13E).
In 2-week-old pumy seedlings, growth in the presence of external pseudouridine also led to a strong increase in ΨMP contents (to ∼1 µg g−1). Internal pseudouridine levels even reached 200 µg g−1 (Supplemental Figure 10). Although the PUMY mutants appeared smaller in response to 250 µM pseudouridine treatment, the plants still survived, indicating that these metabolite concentrations are still tolerable.
Uridine concentrations were generally reduced by pseudouridine supplementation but did not differ between the respective genotypes (compare Figure 4F with Figures 13D and 13E, top; and Figure 6F with Supplemental Figure 10D and 10E, top), whereas UMP concentrations increased in response to added pseudouridine (comapre Figure 4G with Figures 13D and 13E, bottom; and compare Figure 6G with Supplemental Figure 8E, bottom; and Supplemental Figure 10D). A particularly strong increase in UMP levels was observed in PUKI c/oe in the presence of 100 µM external pseudouridine (Figure 13D, bottom). As mentioned above, PUKI c/oe also had a very high ΨMP content under these conditions (Figure 13B, bottom). Elevated UMP concentrations were consistently observed in lines with high ΨMP contents (Figure 6G; Figure 12C, bottom; Figure 13D, bottom; Supplemental Figures 10D and 10E, bottom). It is therefore quite possible that ΨMP interferes with a pyrimidine phosphatase required for UMP homeostasis. ΨMP is also likely a substrate of this phosphatase, as indicated by the high pseudouridine but low ΨMP concentrations in the PUMY mutants (see above). By contrast, the UMP content was not correlated with the pseudouridine concentration, as clearly observed in 2-week-old PUKI c/oe seedlings supplemented with 100 µM pseudouridine: under these conditions, the PUKI c/oe line contained the same amount of pseudouridine (Figure 13B, top) but more UMP (Figure 13D, bottom) than the wild type. These data again support the notion that ΨMP can also leave the peroxisome, because peroxisomal ΨMP generation increased in the PUKI c/oe line, but UMP homeostasis is maintained in the cytosol (Ohler et al., 2019).
Because the ΨMP concentrations were strongly elevated in the Cyt-PUKI c/oe line and in response to the addition of pseudouridine to the puki lines, we investigated whether the Ψ contents of different RNA pools were affected under these more extreme conditions. However, again, no differences from the wild type were observed (Supplemental Figure 11). These findings confirm the notion that Ψ catabolism is not required to protect the bulk RNA, and they demonstrate that the observed phenotypes are not caused by alterations in the Ψ/U ratios of rRNA and tRNA.
Interestingly, seeds of the Cyt-PUKI c/oe and PUKI c/oe lines showed no germination delay, even in the presence of 100 µM external pseudouridine (Supplemental Figures 12A and 12B). According to our hypothesis that ΨMP is responsible for the germination delay, these lines should contain relatively low levels of ΨMP during this early developmental stage. This was indeed the case (Supplemental Figures 12C and 12D, bottom). Cyt-PUKI c/oe (no external pseudouridine) and PUKI c/oe (in the presence of 100 µM external pseudouridine) contained both only ∼0.1 µg of ΨMP in 104 seeds at 2 dai, a level ∼6-fold and 15-fold lower than that of the puki and pumy lines, respectively (Figure 10). It appears that the strong accumulation of ΨMP in the PUKI overexpression lines occurs later in development and that external pseudouridine is not taken up and metabolically converted during germination.
Taken together, these findings strongly suggest that ΨMP at a relative low concentration of ∼1 µg g−1 is toxic for seedlings and interferes with UMP homeostasis, whereas pseudouridine is tolerated at a level of up to (at least) 200 µg g−1. Similarly, higher ΨMP concentrations also appear to interfere with seed germination.
DISCUSSION
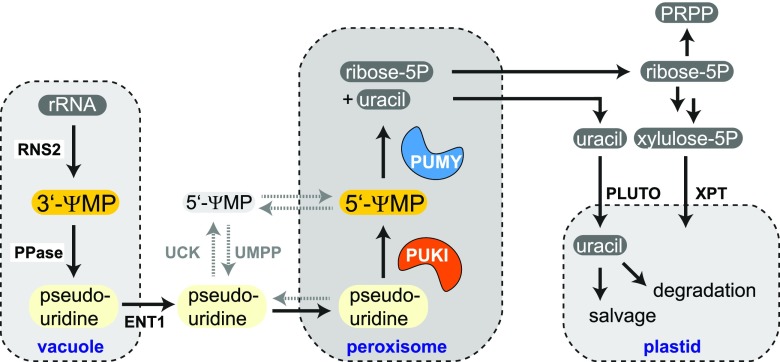
We constructed a model based on this data by integrating all findings and speculating about the metabolic fate of the pseudouridine degradation products (Figure 14). Most pseudouridine is probably derived from the vacuolar turnover of ncRNA, particularly rRNA and tRNA (Floyd et al., 2015), first releasing 3′-ΨMP, that is then dephosphorylated while still in the vacuole. The export into the cytosol is probably mediated by the ENT1 (Bernard et al., 2011). Most reactions of nucleoside metabolism in plants are located in the cytosol, including guanosine and cytidine deamination (Dahncke and Witte, 2013; Chen et al., 2016), nucleoside hydrolysis (Jung et al., 2009; Jung et al., 2011; Riegler et al., 2011; Baccolini and Witte, 2019), but also nucleoside phosphorylation of adenosine by the adenosine kinases (Moffatt et al., 2000) and of uridine by the UCKs (Ohler et al., 2019). Pseudouridine is a substrate of the UCKs (Figure 5), but the catalytic rates in the wild type are likely negligible because the pseudouridine concentration is too low. By contrast, in the PUKI and PUMY mutants, which strongly accumulate pseudouridine, the UCK reaction might gain some relevance for pseudouridine (Figure 14, reactions/transport occurring only in the mutant backgrounds are depicted by gray dashed arrows).
Figure 14.
Model of the Arabidopsis Pseudouridine Catabolic Pathway in the Context of General Plant Metabolism.
Enzymatic reactions and metabolite transport occurring only in the PUKI or PUMY variant background are shown by gray dashed arrows. Note that this model contains several speculative elements, in particular the generation of 3′-ΨMP in the vacuole and the details of pseudouridine transport over the vacuole and peroxisome membranes. PPase, vacuolar phosphatase; PLUTO, plastidic nucleobase transporter; PRPP, 5-phosphoribosyl-1-pyrophosphate; ribose-5P, ribose-5-phosphate; RNS2, ribonuclease 2; UMPP, UMP phosphatase; XPT, xylulose-5-phosphate/phosphate translocator; xylulose-5P, xylulose-5-phosphate.
Several lines of evidence indicate that ΨMP accumulation delays germination and is toxic for plants at relatively low concentrations. Because 5′-ΨMP is an intermediate of pseudouridine catabolism, the toxicity of this compound might explain why the pseudouridine catabolic route is confined to the peroxisome, an organelle that does not have general nucleotide metabolism of its own. The small size of the peroxisome guarantees that the diffusion path for ΨMP from generation to degradation is short, resulting in little accumulation of ΨMP. The short diffusion distance might be of particular importance in plants, because unlike in metazoa, PUKI and PUMY in plants do not reside on the same polypeptide (Figure 1), and the enzymes do not interact physically (Supplemental Figures 3 and 4). Perhaps metazoa can afford to place the PUMY-PUKI fusion protein into the mitochondria, because ΨMP might be efficiently channeled between both enzyme domains. The accumulation of ΨMP will also be avoided due to the kinetic characteristics of the enzymes (Tables 1 and 2). Although PUKI binds pseudouridine with high affinity (low Km), it does not rapidly catalyze the generation of ΨMP (low kcat). In situations of high rRNA and tRNA turnover, these characteristics will likely prevent ΨMP accumulation, instead leading to higher pseudouridine concentrations, which is less harmful. Consistent with this notion, pseudouridine accumulates in the wild-type seedlings to a level of 1.7 μg g−1, whereas the ΨMP concentration is only 0.025 μg g−1 (Figures 4D and 4E). In addition, most of the ΨMP is 3′-ΨMP (Figure 7), which is likely sequestered in the vacuole. Additionally, pseudouridine accumulation can be abolished by the overproduction of PUKI in the PUKI c/oe line (Figure 4D), indicating that the catalytic capacity of PUKI is limited in the wild type to prevent ΨMP toxicity.
Avoiding ΨMP accumulation is important because our data indicate that ΨMP is potentially mobile over the peroxisomal membrane. We hypothesize that ΨMP is quickly turned over by an unknown phosphatase in the cytosol (UMP phosphatase in Figure 14), but neither this reaction nor ΨMP transport over the peroxisomal membrane is likely of high relevance in the wild type, where PUMY represents an efficient peroxisomal sink of ΨMP.
The products of the PUMY reaction, uracil and ribose-5-phosphate, probably leave the peroxisome (Figure 14). Uracil is imported into the plastids (Witz et al., 2012) to either undergo degradation (Zrenner et al., 2009) or salvage to UMP catalyzed by the plastidic uracil phosphoribosyltransferase (Mainguet et al., 2009; Ohler et al., 2019). Ribose-5-phosphate can be used in the cytosol to generate activated Rib, 5-phosphoribosyl-1-pyrophosphate, for use in nucleobase salvage reactions and for de novo biosynthesis of cytosolic pyrimidine (Howles et al., 2006; Witz et al., 2012). Alternatively, ribose-5-phosphate can be converted to xylulose-5-phosphate in the cytosol by cytosolic forms of ribose-5-phosphate isomerase together with ribulose-5-phosphate epimerase. The xylulose-5-phosphate is imported into the plastids via the xylulose-5-phosphate/phosphate translocator (Eicks et al., 2002).
One may speculate that the efficient removal of modified nucleotides/nucleosides is necessary to protect RNA species from random incorporation of modified nucleotides by RNA polymerases. In support of this idea, N6-mATP is a substrate of RNA polymerase II (Chen et al., 2018). However, neither the abrogation of N6-mAMP catabolism (Chen et al., 2018) nor the mutation or overexpression of genes or coding sequences for the pseudouridine catabolic enzymes (Figure 8; Supplemental Figure 11) altered the respective ratios of modified-to-unmodified bases in several RNA species. Apparently, the primary role of modified base catabolism is not the protection of RNA. It is still unclear which selective pressure has maintained the catabolic pathway for N6-mAMP degradation in many eukaryotes. However, our results strongly suggest that avoiding toxic ΨMP accumulation is an important function of the catabolic pathway for pseudouridine degradation.
During the first 2 d of germination, the ΨMP content in Arabidopsis seeds increased (Figure 10), which was also observed during germination in common bean (Phaseolus vulgaris; Al-Baldawi and Brown, 1983). Seeds acquire DNA and RNA damage during storage, which contributes to the loss of germination vigor over time. Extensive rRNA degradation upon imbibition has been observed in carrot seeds (Brocklehurst and Fraser, 1980). Damaged rRNA might be degraded during the early stages of germination and be replaced by new rRNA. This rRNA turnover might give rise to the increased ΨMP concentrations in seeds after imbibition.
ΨMP catabolism is highly conserved in vascular plants and mosses but also in algae. Organisms always need to avoid ΨMP toxicity, but they might also solve this problem by excreting pseudouridine. For example, mammals lack PUKI and PUMY orthologs, but mammalian urine is known to contain pseudouridine (Sekula et al., 2017), indicating that these animals utilize the excretion strategy. The human pathogen E. coli might use the pseudouridine catabolic pathway primarily to utilize Ψ supplied by the host as carbon and uracil sources.
Many other modified nucleosides in addition to pseudouridine occur in RNA, such as 5-methylcytidine and 7-methyguanosine. These give rise to numerous modified nucleotides or nucleosides upon RNA turnover, but how most of these are metabolized is currently unknown. Elucidating these catabolic pathways represents an interesting challenge for the future.
METHODS
Plant Material and Cultivation
Segregating T-DNA insertion mutants of Arabidopsis (Arabidopsis thaliana) from the SALK collection (SALK_006361, puki-1; and SALK_129530, pumy-2; Alonso et al., 2003), the SAIL collection (SAIL_647C12, puki-2; Sessions et al., 2002), and the GABI-Kat collection (GK-681B01, pumy-1; Kleinboelting et al., 2012) were obtained from the Nottingham Arabidopsis Stock Centre. The corresponding wild types and the homozygous mutants were screened from the offspring of the segregating populations.
Arabidopsis and Nicotiana benthamiana plants were cultivated as previously described by Witte et al. (2005). Agar plates were prepared with half-strength Murashige and Skoog (MS) nutrients (product no. MO255, Duchefa Biochemie) including vitamins, but without sugar. For the pseudouridine feeding experiments, the plants were grown on this agar medium supplemented with 100 or 250 µM pseudouridine. For the NaCl treatment experiments, 6-d-old seedlings were transferred from half-strength MS medium to new half-strength MS medium supplied with 0, 125, and 200 mM NaCl and the root lengths were recorded daily. The plates were incubated under long-day conditions (Binder KBFW 720 with Osram Lumilux lights; 16 h of light at 55 µmol m−2 s−1, 22°C day, 20°C night). For metabolic profiling of 2-week-old plants, seedlings grown on agar were used, whereas the metabolic profiling of 37-d-old plants was performed with soil-grown material. Germination trials were performed on half-strength MS medium. For metabolite analysis during germination, the seeds were germinated in a thin layer of water (or Ψ when indicated).
RNA Extraction, Cloning, and Quantitative RT-PCR Analyses
Total RNA was isolated from 2-week-old Arabidopsis seedlings, and cDNA was prepared as described previously (Chen et al., 2016). The following primers were used for cloning: for PUKI (At1g49350), P1009 and P1011; for PUMY (At1g50510), P804 and P805; for PUKI (cytosolic), P1009 and P1235; for UCK1 (At5g40870), P107 and P108; and for UCK2 (At3g27190), P109 and P110 (Supplemental Table 2). By introducing EcoRI and XmaI sites via the primers, PUKI and PUMY were cloned into pXNS1cpmv-Strep (V89) and pXCScpmv-hemagglutinin (HA)-Strep (V69), respectively, for protein production (Myrach et al., 2017). The vector V89 was generated by cloning a 523-bp XhoI SalI fragment from V69 into the XhoI site of pXNS1pat-Strep (V41; Cao et al., 2010). A 189-bp fragment from V69 amplified with primers N0004 and N0005 and digested with SpeI and XbaI was inserted into the XbaI site of the modified V41. The resulting vector V89 contains 5′ and 3′ UTR enhancer sequences of Cowpea mosaic virus (CPMV) RNA-2, which improve protein production. V89 differs from the very similar V90 (Baccolini and Witte, 2019) in that it contains an NdeI site instead of an NcoI site for cloning. Both vectors facilitate the enhanced production of proteins with N-terminal Strep tag in plants. PUKI and PUMY were also cloned via EcoRI and XmaI into pXCS-YFP (V36; Witte et al., 2004) and pXNS1pat-YFP (V99), respectively, for subcellular localization analysis. V99 was generated by cloning a 1455-bp AscI SfoI fragment of pXNS2pat-YFP (V63; Dahncke and Witte, 2013) into pXNPpat-Strep (V41; Cao et al., 2010) digested with the same enzymes. V99 differs from the very similar V63 (Dahncke and Witte, 2013) in that it contains an NdeI site instead of an NcoI site for cloning. Both vectors facilitate the production of proteins with N-terminal YFP tag in plants. UCK1 and UCK2 were cloned via PstI and XmaI sites, introduced by the primers during amplification, into pXCS-HA-Strep (V13; Witte et al., 2004) for the production of proteins with a C-terminal HA-Strep tag in planta. For the PUKI and PUMY interaction experiment with one untagged enzyme, the untagged PUKI was generated by cloning the PUKI fragment produced with primers P1009 and P1011 (see above) into V69. To generate untagged PUMY, the PUMY fragment was amplified with primers P804 and P1824 and cloned into V69.
Arabidopsis homozygous mutant and the corresponding wild-type lines were screened from the segregating mutant populations by PCR using the following primer pairs for the wild type and the respective mutant alleles: P1033 and P1034, N61 and P1034 for puki-1; P1031 and P1032, 1197 and P1031 for puki-2; P802 and P803, 448 and P802 for pumy-1; and P802 and P803, N61 and P802 for pumy-2. The PCR products from the mutants were cloned and sequenced. The T-DNA insertions were positioned exactly as annotated on the Signal website (http://signal.salk.edu/cgi-bin/tdnaexpress). To measure gene-specific mRNA levels in the mutants, cDNAs were prepared from seedlings of each genotype. The PCR used primer pairs P1031 + P1034 and P802 + P803, giving rise to a product of 1749 bp and 1251 bp from the wild-type alleles of puki and pumy, respectively. The primers flanked the T-DNA insertions in each case. To amplify ACTIN2 as a control, primers 1033 and 1034 were used. The PCR (Analytik Jena) was performed with 40 cycles for PUKI, PUMY, and ACTIN2 detection at annealing temperatures of 65, 60, and 60°C, respectively. All primer sequences are listed in Supplemental Table 2.
mRNA and nonpolyadenylated RNAs were isolated from total RNA for digestion using the PolyATract mRNA Isolation System III (Promega) as described by Chen et al. (2018).
Protein Purification, Enzymatic Assays, and Immunoblot Analyses
N-Terminal Strep-tagged PUKI, C-terminal Strep-tagged PUMY, C-terminal Strep-tagged UCK1, and C-terminal Strep-tagged UCK2 were affinity purified after 5 d of transient protein production in N. benthamiana as described previously (Werner et al., 2008). In brief, 1.5-mL samples of clarified extracts were generated from 0.75 g of leaf material and incubated with 40 μL of Streptactin-Macroprep (50% [w/v] slurry, IBA). The matrix was washed five times with 1 mL of washing buffer and eluted in 150 μL of elution buffer. Purified proteins were quantified using the Bradford reagent (Serva) with BSA as a standard.
For the kinase assay, the phosphorylation activity of PUKI was assessed by quantifying the conversion of pseudouridine to ΨMP and the phosphorylation activity of UCK1 and UCK2 was assessed by quantifying the conversion of uridine to UMP. An Agilent HPLC 1200 system equipped with a Polaris 5 C18-A column (150 × 4.6 mm, particle size 5 µm; Agilent) column operated as described by Schroeder et al. 2018) was used to quantify the reaction products. The enzymatic reaction was conducted at 30°C in 0.3-mL reaction buffer containing 40 mM Tris-HCl buffer, pH 7.5, 2 mM MgCl2, 10 mM KCl, 0.4 mM ATP, 1 mM DTT, and varying concentrations of the substrates. ΨMP dissolved in reaction buffer was used as a standard. 5′-ΨMP was custom synthesized by Jena Bioscience (Jena, Germany), and its identity was confirmed by the company using 13C NMR. Additionally, we confirmed that 3′-specific nuclease P1 was unable to dephosphorylate the custom synthesized 5′-ΨMP, showing the absence of contamination with any 3′-ΨMP. The kinase reaction was started by adding 10 μL (3 µg) of purified enzyme and incubated for 0, 1, and 3 min. The kinetic constants were determined at a pseudouridine concentration of 1, 5, 20, 50, 100, and 250 µM. The reaction (70 µL) was stopped by adding 33 μL of HClO4 (1.2 M), followed by 3 μL of alkaline solution (5 M KOH and 2 M K2CO3). After centrifugation at 20,000g for 15 min, 20 μL of supernatant was injected into the HPLC machine. The kinetic measurements were conducted in four replicates. The kinetic constants were determined by fitting the data to the Michaelis–Menten equation using Prism 4 (GraphPad Software). To determine the enzymatic activity of UCK1 and UCK2 with uridine and pseudouridine, 1 mM uridine was incubated for 0, 4, and 10 min with UCK1 (0.64 ng µL−1) and UCK2 (5 ng µL−1), respectively, and 1 mM pseudouridine was incubated for 0, 4, and 10 min with UCK1 (3.2 ng µL−1) and UCK2 (25 ng µL−1), respectively, in the kinase reaction mixture described above. The activity was determined by quantifying ADP production over time via HPLC.
The enzymatic activity of PUMY was assessed by quantifying the generation of uracil over time via HPLC analysis as described for PUKI. Uracil and ΨMP were used as standards for the forward and reverse reactions, respectively. The enzymatic reaction was conducted at 30°C in 0.3-mL reaction buffer containing 40 mM Tris-HCl buffer, pH 7.5, 2 mM MgCl2, 10 mM KCl, 1 mM DTT, and varying concentrations of the substrates. The reaction was started by adding 10 μL (3 µg) of purified enzyme. The kinetic constants for the ΨMP glycosidase activity of PUMY were determined using ΨMP at concentrations of 10, 50, 100, 500, 1000, and 1500 µM. The kinetic constants for the reverse reaction of PUMY were determined using 4 mM ribose-5-phosphate and uracil at concentrations of 20, 100, 500, 1000, 2000, and 5000 µM. The reaction was incubated for 0, 1, and 3 min. Each enzymatic reaction was repeated four times.
For the enzymatic assays of leaf extracts, 1.5 mL of clarified extracts were desalted using Zeba spin desalting columns (Thermo Scientific). The desalted extracts were used for the enzymatic assays as described above and for Streptactin affinity purification starting with a volume of 1 mL of extract and eluting in 100 μL of elution buffer.
Monoclonal anti-Strep-tag (1:5000 dilution, clone 7G8, Antibody Facility, University of Braunschweig, Germany; http://antibody-facility.com/) or anti-myc antibody (1:400 dilution; Roche) from mouse were used together with anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich) to detect Strep-tagged or myc-tagged proteins on immunoblots. Peroxisomal and Cyt-PUKI proteins in the complementation lines were quantified from images of immunoblots using a concentration series (5, 10, 25, 50, 100, and 200 ng/µL) of Strep-PUKI purified from N. benthamiana as a standard with ImageJ 1.47 software.
Subcellular Localization
YFP-PUKI or PUMY-YFP was transiently coexpressed with the peroxisomal marker protein mCherry-SKL in N. benthamiana leaves for 5 d. The samples were analyzed using a True Confocal Scanner SP8 microscope equipped with an HC PL APO CS2 40 × 1.10 water immersion objective (Leica Microsystems). The acquired images were processed using Leica Application Suite Advanced Fluorescence software (Leica Microsystems).
Metabolite Analyses
Metabolites were quantified using an Agilent 1200 HPLC system coupled to an Agilent 6460 triple-quadrupole mass spectrometer as previously described by Chen et al. (2018), with minor modifications. Nucleotides and nucleosides were separated on a Polaris 5 C18A column (50 × 4.6 mm, particle size 5 µm; Agilent) at a flow rate of 0.65 mL min−1 with the following gradient: 0 min, 5% (v/v) B; 2 min, 5% B; 5.5 min, 15% B; 9.5 min, 85% B; 11 min, 85% B; 11.1 min, 5% B; 20 min, 5% B. Solvent A was 10 mM ammonium acetate, pH 9.5, and solvent B was methanol.
The plant material was frozen in liquid nitrogen and ground in centrifuge vials with steal beads in a mixer mill (Mixer Mill MM400, Retsch) for 3 min. For seedlings and rosette leaves, 100 mg of homogenized plant material was extracted with 1 mL of methanol:acetonitrile:water at a ratio of 2:2:1 containing freshly added internal standards (1 µg mL−1 15N2-UMP and 0.5 µg mL−1 15N2-U; Cambridge Isotope Laboratories). For seeds, 10 mg of homogenized material was used for extraction. The extracts were vortexed, incubated on ice for 10 min, and centrifuged at 15,000g at 4°C for 15 min. The supernatant (0.5 mL) was concentrated by freeze drying and resuspended in 0.05 mL of 5% (v/v) acetonitrile, 95% (v/v) water. Directly after resuspension, 20-μL samples were injected for analysis. The following mass transitions were monitored: m/z 325.01 to 212.9 (UMP), m/z 327.18 to 212.19 (15N2-UMP), m/z 325.01 to 125 (ΨMP), m/z 245.21 to 133 (U), m/z 247.2 to 115 (15N2-U), and m/z 245.1 to 125 (pseudouridine). UMP and uridine were quantified against their internal standards. The other compounds were quantified using external standards added to the wild-type extracts from the corresponding growth conditions as a matrix. ΨMP (0, 4, 8, 40, 80, and 160 ng mL−1) and pseudouridine (0, 250, 500, 2500, 5000, 10,000, and 25,000 ng mL−1) were used as external standards.
To determine the susceptibility of ΨMP extracted from the wild type to enzymatic degradation, freeze-dried metabolite extract was resuspended in 95% (v/v) water, 5% (v/v) acetonitrile and incubated for 15 min with (1) no further additions, (2) PUMY at 30°C, or (3) nuclease P1 from Penicillium citrinum (New England Biolabs) at 37°C. Nuclease P1 has 3′-, but not 5′-, nucleotidase activity, and the incubation time and the added enzyme quantities were sufficient to fully degrade any available nucleotides (Supplemental Figure 5). Samples were directly analyzed by LC-MS/MS without further processing.
Quantitative Analysis of the Ψ/U Ratio in RNA by LC-MS/MS
RNA samples were fully digested to nucleosides as described previously (Chen et al., 2018), with minor modifications. Next, 100 ng of mRNA or 800 ng of total RNA or nonpolyadenylated RNA was digested by benzonase (1 unit; Sigma-Aldrich) and phosphodiesterase I (4 × 10−3 units; Sigma-Aldrich) and dephosphorylated by shrimp alkaline phosphatase (1 unit; New England Biolabs) in 50 μL of buffer containing 10 mM Tris-HCl, pH 7.9, 1 mM MgCl2, and 0.1 mg mL−1 BSA. After an incubation at 37°C for 8 h, the samples were filtered in ultra-filtration tubes (3-kD cutoff; Pall), and 15-μL aliquots were analyzed by LC-MS/MS. The nucleosides were separated and identified as described above. Standard solutions of uridine (5, 10, 50, 100, 500, 1000, and 5000 ng µL−1) and pseudouridine (50, 100, 500, 1000, 5000, and 10,000 ng µL−1) were used for quantification.
Sequence Analyses
PUKI and PUMY sequences were identified by BLASTp searches using the sequences from Arabidopsis as queries at the Phytozome v12.1.6 web server for plants and algae (phytozome.jgi.doe.gov/pz/portal.html) and at the National Center for Biotechnology Information for all nonplant organisms. Multiple alignments were generated with Muscle hosted by European Bioinformatics Institute (www.ebi.ac.uk/Tools/msa/muscle/) and shaded with Boxshade (https://embnet.vital-it.ch/software/BOX_form.html).
Quantification and Statistical Analysis
Analysis of variance (ANOVA) followed by Tukey’s honestly significant difference test was performed in R for statistical evaluation. Different letters represent differences at the P < 0.05 significance level.
Accession Numbers
Sequence data from this article can be found under the following accession/locus numbers: At1g49350 (PUKI), At1g50510 (PUMY), At5g40870 (UCK1), and At3g27190 (UCK2).
Supplemental Data
Supplemental Figure 1. Comparative sequence analysis of plant YeiC and YeiN homologs (PUKI and PUMY) with the corresponding sequences from metazoa and fungi as well as Escherichia coli.
Supplemental Figure 2. Multiple alignment of putative YeiC (PUKI) sequences from a wide phylogenetic range of bacteria.
Supplemental Figure 3. PUKI does not interact with PUMY.
Supplemental Figure 4. PUKI activity is not stimulated by the presence of PUMY.
Supplemental Figure 5. Confirmation of nuclease P1 and PUMY enzymatic activities.
Supplemental Figure 6. Metabolite contents in rosette leaves of 37-day-old Arabidopsis plants varying in PUKI expression.
Supplemental Figure 7. Metabolite contents in rosette leaves of 37-day-old Arabidopsis plants varying in PUMY expression.
Supplemental Figure 8. Uridine and UMP contents in germinating seeds of variants with different PUKI and PUMY expression levels.
Supplemental Figure 9. Phenotypes and metabolite contents of seedlings varying in PUMY expression grown in the presence of pseudouridine.
Supplemental Figure 10. Phenotypes of seedlings varying in PUKI and PUMY expression grown in the presence of 0, 125, and 200 mM NaCl.
Supplemental Figure 11. Ψ/U ratios for different RNA species in Cyt-PUKI c/oe seedlings grown on standard medium and seedlings varying in PUKI expression grown in the presence of 100 µM pseudouridine.
Supplemental Figure 12. Germination and metabolite contents of Cyt-PUKI c/oe Seeds germinated on standard medium and seeds varying in PUKI expression germinated in the presence of 100 µM pseudouridine.
Supplemental Table 1. Predicted subcellular localization of the PUMY-PUKI fusion enzyme from metazoa and fungi.
Supplemental Table 2. Primers used in this study.
Supplemental File. ANOVA analyses.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Anting Zhu for preparing vector V89, André Specht, Hildegard Thölke for technical support, Katharina J. Heinemann and Xiaoguang Chen for help with plant cultivation and harvesting, Xiaoye Liu for assistance with the statistical analyses, Wenlei Wang for help with salt stress analyses, and Nieves Medina Escobar for revising the article. This work was financially supported by the National Natural Science Foundation of China (grant 31900907), the Natural Science Foundation of Jiangsu Province (grant BK20190528), the Leibniz University Hannover (Wege in die Forschung II to M.C.), and Deutsche Forschungsgemeinschaft (grant CH2292/1-1 to M.C. and grant WI3411/4-1 to C.-P.W.)
AUTHOR CONTRIBUTIONS
M.C. and C.-P.W. conceived the research; M.C. performed the experiments; C.-P.W. carried out the bioinformatic analyses; M.C. and C.-P.W. analyzed the data and wrote the article.
References
- Al-Baldawi N.F., Brown E.G. (1983). Accumulation of 5-ribosyluracil (pseudouridine) within the tissues of Phaseolus vulgaris. Phytochemistry 22: 419–421. [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Arnez J.G., Steitz T.A. (1994). Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Arribas-Hernández L., Bressendorff S., Hansen M.H., Poulsen C., Erdmann S., Brodersen P. (2018). An m6A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30: 952–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccolini C., Witte C.-P. (2019). AMP and GMP catabolism in Arabidopsis converge on xanthosine, which is degraded by a nucleoside hydrolase heterocomplex. Plant Cell 31: 734–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C., Traub M., Kunz H.-H., Hach S., Trentmann O., Möhlmann T. (2011). Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J. Exp. Bot. 62: 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman T.R. (1970). Pseudouridulate synthetase of Escherichia coli: Correlation of its activity with utilization of pseudouridine for growth. J. Bacteriol. 103: 263–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst P.A., Fraser R.S. (1980). Ribosomal RNA integrity and rate of seed germination. Planta 148: 417–421. [DOI] [PubMed] [Google Scholar]
- Cao F.-Q., Werner A.K., Dahncke K., Romeis T., Liu L.-H., Witte C.-P. (2010). Identification and characterization of proteins involved in rice urea and arginine catabolism. Plant Physiol. 154: 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charton L., Plett A., Linka N. (2019). Plant peroxisomal solute transporter proteins. J. Integr. Plant Biol. 61: 817–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Herde M., Witte C.-P. (2016). Of the nine cytidine deaminase-like genes in Arabidopsis, eight are pseudogenes and only one is required to maintain pyrimidine homeostasis in vivo. Plant Physiol. 171: 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Urs M.J., Sánchez-González I., Olayioye M.A., Herde M., Witte C.-P. (2018). m6A RNA degradation products are catabolized by an evolutionarily conserved N6-methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30: 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-J., Donnard E., Gustafsson H.T., Garber M., Rando O.J. (2017). Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell 68: 978–992.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahncke K., Witte C.-P. (2013). Plant purine nucleoside catabolism employs a guanosine deaminase required for the generation of xanthosine in Arabidopsis. Plant Cell 25: 4101–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur W.A., Fournier M.J. (2002). rRNA modifications and ribosome function. Trends Biochem. Sci. 27: 344–351. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Dominissini D., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicks M., Maurino V., Knappe S., Flügge U.-I., Fischer K. (2002). The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 128: 512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H., Meyer E.H., Taylor N.L., Bussell J.D., O’Toole N., Heazlewood J.L., Castleden I., Small I.D., Smith S.M., Millar A.H. (2008). Novel proteins, putative membrane transporters, and an integrated metabolic network are revealed by quantitative proteomic analysis of Arabidopsis cell culture peroxisomes. Plant Physiol. 148: 1809–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd B.E., Morriss S.C., MacIntosh G.C., Bassham D.C. (2015). Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11: 2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Harada B.T., Behm M., He C. (2018). RNA modifications modulate gene expression during development. Science 361: 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T., Ferré-D’Amaré A.R. (2006). Pseudouridine synthases. Chem. Biol. 13: 1125–1135. [DOI] [PubMed] [Google Scholar]
- Howles P.A., Birch R.J., Collings D.A., Gebbie L.K., Hurley U.A., Hocart C.H., Arioli T., Williamson R.E. (2006). A mutation in an Arabidopsis ribose 5-phosphate isomerase reduces cellulose synthesis and is rescued by exogenous uridine. Plant J. 48: 606–618. [DOI] [PubMed] [Google Scholar]
- Huang S., Mahanta N., Begley T.P., Ealick S.E. (2012). Pseudouridine monophosphate glycosidase: A new glycosidase mechanism. Biochemistry 51: 9245–9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K., Bellodi C., Landry D.M., Niederer R.O., Meskauskas A., Musalgaonkar S., Kopmar N., Krasnykh O., Dean A.M., Thompson S.R., Ruggero D., Dinman J.D. (2011). rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B., Flörchinger M., Kunz H.-H., Traub M., Wartenberg R., Jeblick W., Neuhaus H.E., Möhlmann T. (2009). Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. Plant Cell 21: 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B., Hoffmann C., Möhlmann T. (2011). Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of purines. Plant J. 65: 703–711. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N., Huep G., Kloetgen A., Viehoever P., Weisshaar B. (2012). GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40: D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze M., Neuberger G., Maurer-Stroh S., Ma J., Eck T., Braverman N., Schmid J.A., Eisenhaber F., Berger J. (2011). Structural requirements for interaction of peroxisomal targeting signal 2 and its receptor PEX7. J. Biol. Chem. 286: 45048–45062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. (2015). Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11: 592–597. [DOI] [PubMed] [Google Scholar]
- Lingner T., Kataya A.R., Antonicelli G.E., Benichou A., Nilssen K., Chen X.-Y., Siemsen T., Morgenstern B., Meinicke P., Reumann S. (2011). Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell 23: 1556–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainguet S.E., Gakière B., Majira A., Pelletier S., Bringel F., Guérard F., Caboche M., Berthomé R., Renou J.P. (2009). Uracil salvage is necessary for early Arabidopsis development. Plant J. 60: 280–291. [DOI] [PubMed] [Google Scholar]
- Moffatt B.A., Wang L., Allen M.S., Stevens Y.Y., Qin W., Snider J., von Schwartzenberg K. (2000). Adenosine kinase of Arabidopsis. Kinetic properties and gene expression. Plant Physiol. 124: 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y., Lyko F., Helm M. (2010). 5-Methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res. 38: 1415–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrach T., Zhu A., Witte C.-P. (2017). The assembly of the plant urease activation complex and the essential role of the urease accessory protein G (UreG) in delivery of nickel to urease. J. Biol. Chem. 292: 14556–14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Ohler L., Niopek-Witz S., Mainguet S.E., Möhlmann T. (2019). Pyrimidine salvage: Physiological functions and interaction with chloroplast biogenesis. Plant Physiol. 180: 1816–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preumont A., Snoussi K., Stroobant V., Collet J.-F., Van Schaftingen E. (2008). Molecular identification of pseudouridine-metabolizing enzymes. J. Biol. Chem. 283: 25238–25246. [DOI] [PubMed] [Google Scholar]
- Reumann S. (2011). Toward a definition of the complete proteome of plant peroxisomes: Where experimental proteomics must be complemented by bioinformatics. Proteomics 11: 1764–1779. [DOI] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P.M., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riegler H., Geserick C., Zrenner R. (2011). Arabidopsis thaliana nucleosidase mutants provide new insights into nucleoside degradation. New Phytol. 191: 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs J.W., Rockwell N.C., Cavales P.C., Callis J. (2016). Identification of the plant ribokinase and discovery of a role for Arabidopsis ribokinase in nucleoside metabolism. J. Biol. Chem. 291: 22572–22582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R.Y., Zhu A., Eubel H., Dahncke K., Witte C.-P. (2018). The ribokinases of Arabidopsis thaliana and Saccharomyces cerevisiae are required for ribose recycling from nucleotide catabolism, which in plants is not essential to survive prolonged dark stress. New Phytol. 217: 233–244. [DOI] [PubMed] [Google Scholar]
- Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., Fink G., Regev A. (2014). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekula P., Dettmer K., Vogl F.C., Gronwald W., Ellmann L., Mohney R.P., Eckardt K.-U., Suhre K., Kastenmüller G., Oefner P.J., Köttgen A. (2017). From discovery to translation: Characterization of C-mannosyltryptophan and pseudouridine as markers of kidney function. Sci. Rep. 7: 17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xu Y., Bai S., Bai X., Zhu H., Dong H., Wang W., Zhu X., Hao F., Song C.-P. (2019). Transcriptome-wide analysis of pseudouridylation of mRNA and non-coding RNAs in Arabidopsis. J. Exp. Bot. 70: 5089–5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A.K., Sparkes I.A., Romeis T., Witte C.-P. (2008). Identification, biochemical characterization, and subcellular localization of allantoate amidohydrolases from Arabidopsis and soybean. Plant Physiol. 146: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte C.-P., Noël L.D., Gielbert J., Parker J.E., Romeis T. (2004). Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. Plant Mol. Biol. 55: 135–147. [DOI] [PubMed] [Google Scholar]
- Witte C.P., Rosso M.G., Romeis T. (2005). Identification of three urease accessory proteins that are required for urease activation in Arabidopsis. Plant Physiol. 139: 1155–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witz S., Jung B., Fürst S., Möhlmann T. (2012). De novo pyrimidine nucleotide synthesis mainly occurs outside of plastids, but a previously undiscovered nucleobase importer provides substrates for the essential salvage pathway in Arabidopsis. Plant Cell 24: 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., et al. (2017). 5-Methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 27: 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20: 1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R., Riegler H., Marquard C.R., Lange P.R., Geserick C., Bartosz C.E., Chen C.T., Slocum R.D. (2009). A functional analysis of the pyrimidine catabolic pathway in Arabidopsis. New Phytol. 183: 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]