The Pseudomonas syringae effector protein HopO1-1 targets and destabilizes plasmodesmata-located proteins to promote disease in plants.
Abstract
A hallmark of multicellular organisms is their ability to maintain physiological homeostasis by communicating among cells, tissues, and organs. In plants, intercellular communication is largely dependent on plasmodesmata (PD), which are membrane-lined channels connecting adjacent plant cells. Upon immune stimulation, plants close PD as part of their immune responses. Here, we show that the bacterial pathogen Pseudomonas syringae deploys an effector protein, HopO1-1, that modulates PD function. HopO1-1 is required for P. syringae to spread locally to neighboring tissues during infection. Expression of HopO1-1 in Arabidopsis (Arabidopsis thaliana) increases the distance of PD-dependent molecular flux between neighboring plant cells. Being a putative ribosyltransferase, the catalytic activity of HopO1-1 is required for regulation of PD. HopO1-1 physically interacts with and destabilizes the plant PD-located protein PDLP7 and possibly PDLP5. Both PDLPs are involved in bacterial immunity. Our findings reveal that a pathogenic bacterium utilizes an effector to manipulate PD-mediated host intercellular communication for maximizing the spread of bacterial infection.
INTRODUCTION
Multicellular organisms host a wide array of microorganisms. Although most microbes are beneficial or harmless to their hosts, infections caused by a few pathogenic microorganisms can lead to devastating diseases in animals and plants. Over the past three decades, progress has been made toward understanding how plants defend against pathogens at the molecular and cellular levels (Grant et al., 2006; Jones and Dangl, 2006; Nicaise et al., 2009). Plants detect the presence of microorganisms by recognizing microbial signatures such as bacterial flagellin and fungal chitin, collectively known as microbe-associated molecular patterns (Ranf, 2017). Recognition of microbe-associated molecular patterns by membrane-bound pattern recognition receptors on the plant cell surface initiates a cascade of signaling events, activating a form of plant innate immunity known as pattern-triggered immunity (Ranf, 2017; Saijo et al., 2018). To overcome host immunity, pathogenic microbes deliver virulence-intended microbial molecules, collectively called “effectors,” mostly into host cells as a major pathogenesis mechanism (Grant et al., 2006; Le Fevre et al., 2015; Toruño et al., 2016). To counter pathogen virulence, plants have evolved a second set of receptors, mainly intracellular NBS-LRR proteins, that recognize individual effectors and activate effector-triggered immunity (Cui et al., 2015). Current models suggest that pattern-triggered immunity and effector-triggered immunity constitute two major forms of cell-autonomous immunity in plants.
In addition to cell-autonomous immunity, uninfected host cells in an infected plant can exhibit immune responses (non-cell-autonomous immunity). Such immune responses in systemic tissues can limit subsequent infections by the same pathogen, a phenomenon known as systemic acquired resistance (Klessig et al., 2018). This process requires cell-to-cell communication. In plants, communication between cells is achieved through apoplastic and symplastic pathways. In the apoplastic pathway, signaling molecules exit signal-generating cells and enter into the apoplast (i.e., extracellular space). To enable intercellular communication, signaling molecules can enter signal-receiving cells through different means of trafficking (Lim et al., 2016). In the symplastic pathway, on the other hand, signaling molecules move from signal-generating cells to signal-receiving cells by passing through plasmodesmata (PD; Stahl and Simon, 2013; Lee, 2014, 2015; Cheval and Faulkner, 2018; Liu and Chen, 2018). PD are membrane-lined channels that span the cell walls of neighboring plant cells, providing cytoplasmic, endoplasmic reticulum (ER), and plasma membrane (PM) continuity between adjoining cells. The cytoplasmic sleeve between the two membranes, PM and ER, allows symplastic molecular movement between adjoining plant cells (Lucas et al., 2009). Three-dimensional ultrastructural analyses revealed that there are extensive ER-PM contact sites within the cytoplasmic sleeve (Nicolas et al., 2017).
Being a physical structure allowing the movement of molecules between plant cells, the aperture of PD, which determines the size exclusion limit, was known as a major determinant of PD function (Lucas and Lee, 2004). The PD aperture is controlled by dynamic deposition and degradation of callose, a plant polysaccharide, at PD within the cell walls. The accumulation and degradation of callose are mediated by callose synthases and β-1,3 glucanases, respectively (De Storme and Geelen, 2014). In addition, PD-localized proteins (PDLPs) are important regulators of callose homeostasis at PD (Lee et al., 2011; Cui and Lee, 2016). Expression of PDLP5 in Arabidopsis (Arabidopsis thaliana) is upregulated upon pathogen infection, coinciding with the accumulation of callose at PD, whereas the pdlp5 knockout mutant exhibits reduced callose deposition at PD (Lee et al., 2011). These findings suggest that PDLPs are required for pathogen-induced callose deposition at PD.
Pseudomonas syringae pv tomato (Pst) DC3000 is a Gram-negative bacterial pathogen that infects not only a crop plant, tomato (Solanum lycopersicum), but also the model plant Arabidopsis (Whalen et al., 1991). It injects 36 virulence-associated effector proteins into plant cells through the type III secretion system to modulate plant cellular processes (Xin and He, 2013, 2018; Wei et al., 2015). Using live-cell imaging, we discovered that one of the effectors, HopO1-1, is targeted to PD and increases the distance of PD-dependent molecular flux between cells in Arabidopsis. Furthermore, HopO1-1 physically interacts with PDLP7 and PDLP5 and destabilizes these two proteins. We found that this manipulation is linked to the ability of the bacterium to successfully colonize and maximize infection.
RESULTS
HopO1-1 Is Targeted to the PM and PD in Arabidopsis
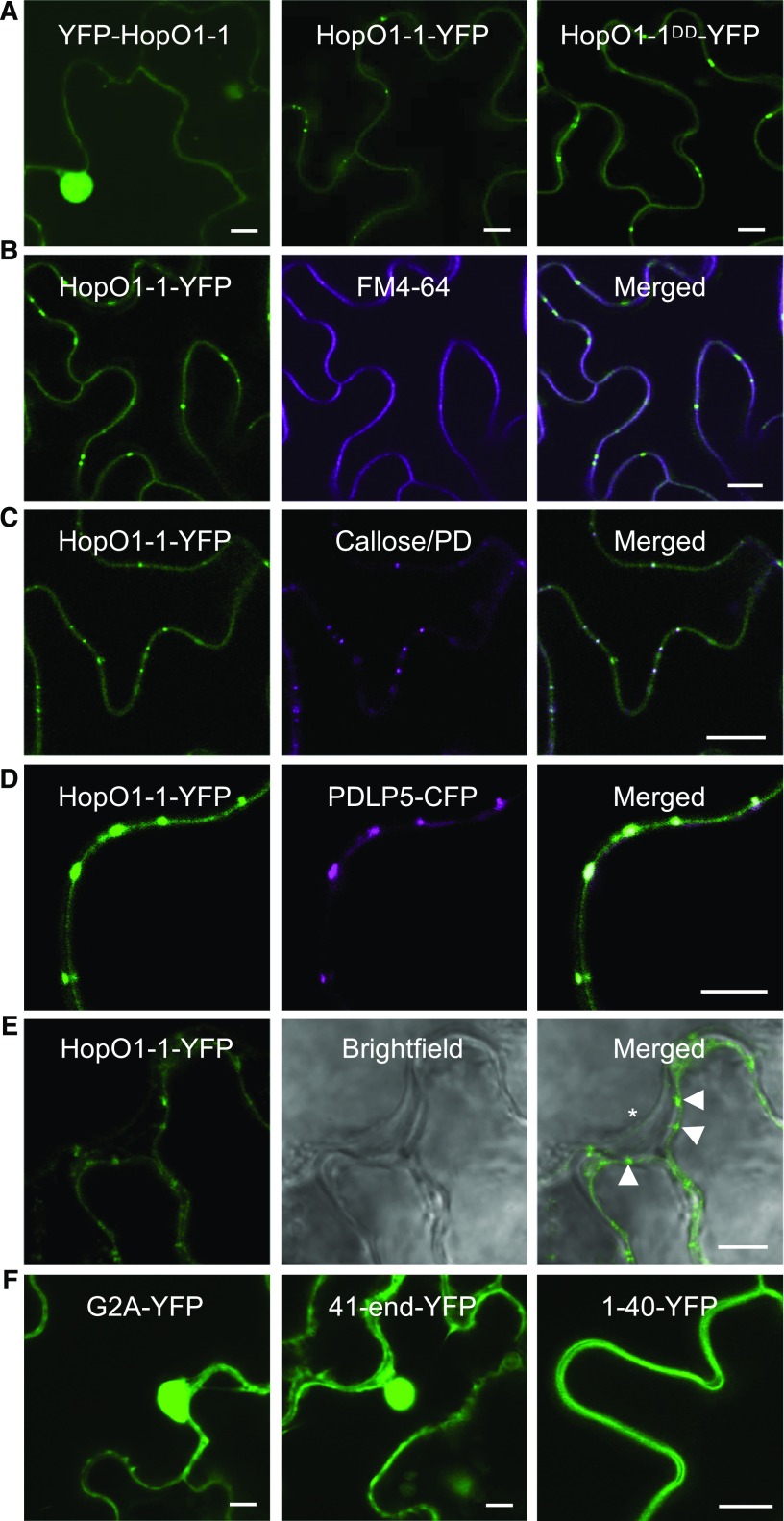
Our work on HopO1-1 was initiated following a systematic subcellular localization study of 32 Pst DC3000 effectors. Yellow fluorescent protein (YFP) fusions of effectors (both N- and C-terminal fusions) were generated and transiently expressed in tobacco (Nicotiana tabacum) leaves. Most interestingly to us, HopO1-1-YFP was observed as prominent, often symmetrical, punctate spots between two adjacent plant cells (Figure 1A). The distinct localization of HopO1-1 led us to speculate that HopO1-1 might be localized to PD.
Figure 1.
HopO1-1 Is Targeted to the PM and PD in Arabidopsis.
(A) Confocal images of leaf epidermal cells of Arabidopsis transgenic plants expressing fluorescent fusion protein of HopO1-1 variants. Bars = 10 µm.
(B) Confocal images show colocalization between HopO1-1-YFP and FM6-64-stained PM in Arabidopsis leaf epidermal cells. Bar = 10 µm.
(C) Confocal images show colocalization between HopO1-1-YFP and aniline blue fluorochrome-stained callose (PD) in Arabidopsis leaf epidermal cells. Bar = 10 µm.
(D) Confocal images show colocalization between HopO1-1-YFP and PDLP5-CFP. The fusion proteins were transiently expressed in N. benthamiana leaf epidermal cells. Bar = 5 µm.
(E) Confocal images of leaf epidermal cells of Arabidopsis transgenic plants expressing 35S-HopO1-1-YFP. Images were captured right after plasmolysis. The asterisk indicates a retracted plasma membrane. Arrowheads show HopO1-1-YFP retained on the cell wall. Bar = 5 µm.
(F) Subcellular localization of an N-myristoylation site mutant (HopO1-1G2A-YFP) and two truncated forms (HopO1-141-end-YFP and HopO1-11-40-YFP) of HopO1-1 stably expressed in Arabidopsis leaf epidermal cells. Bars = 10 µm.
To confirm the PD localization, we generated stable transgenic plants expressing HopO1-1 tagged with YFP in Arabidopsis. Using immunoblot analyses, we detected the expression of full-length fusion proteins (Supplemental Figure 1B). The T2 generation of HopO1-1-YFP transgenic plants was subjected to subcellular localization using confocal laser scanning microscopy. Consistent with transient expression results, HopO1-1-YFP signals were detected in the periphery (with puncta) of Arabidopsis cells, suggesting that the fusion protein targets to both the PM and PD. YFP-HopO1-1 signals, on the other hand, were detected in both the nucleus and the cytoplasm (Figure 1A). HopO1-1-1 contains a putative myristoylation site (N-terminal Gly: G2), which targets a protein to the PM. We thus reasoned that tagging YFP to the C terminus of HopO1-1 (HopO1-1-YFP fusion protein) is correctly targeted to the right cellular compartment in Arabidopsis, whereas tagging YFP to the N terminus of HopO1-1 (i.e., YFP-HopO1-1) resulted in its mislocalization to the nucleus and the cytoplasm. Consistent with this hypothesis, we observed that expression of HopO1-1-YFP resulted in slower plant growth compared with that of wild-type Col-0 or YFP-HopO1-1 (Supplemental Figure 1C). A similar growth defect was also observed in transgenic plants expressing wild-type HopO1-1 without any fusion (Supplemental Figures 1D and 1E), suggesting that HopO1-1-YFP is functionally similar to nontagged wild-type HopO1-1, whereas YFP-HopO1-1 is likely nonfunctional.
To further validate the PM/PD localization of HopO1-1, we stained the PM of Arabidopsis transgenic plant 35S-HopO1-1-YFP with FM 4-64 (Speth et al., 2009). HopO1-1-YFP signals overlapped with FM 4-64-stained PM (Figure 1B), verifying the PM localization of HopO1-1-YFP. To confirm the PD localization, we labeled callose deposited at PD of 35S-HopO1-1-YFP transgenic plants using aniline blue fluorochrome (Guseman et al., 2010). As expected, HopO1-1-YFP fusion proteins were colocalized with aniline blue-stained PD (Figure 1C). The PD localization of HopO1-1 was further tested by transiently coexpressing HopO1-1-YFP with PDLP5-cyan fluorescent protein (CFP) in Nicotiana benthamiana leaves. HopO1-1-YFP signals were found to overlap with PDLP5-CFP signals at PD (Figure 1D). To further confirm the PD association of HopO1-1, we performed plasmolysis with leaves of Arabidopsis transgenic plants expressing HopO1-1-YFP or PDLP7-YFP. These fusion proteins were detected in the periphery with puncta in between plant cells (Supplemental Figures 2A and 2B). After plasmolysis, punctate signals of HopO1-1-YFP and PDLP7-YFP fusion proteins were retained on the cell wall (Figure 1E; Supplemental Figure 1C). These findings confirmed that HopO1-1 is targeted to both the PM and PD in Arabidopsis.
Role of the ADP-Ribosyltransferase Domain in HopO1-1 Localization to PD in Arabidopsis
Although several plant and viral proteins are localized to PD, a consensus PD-targeting signal has not emerged and remains largely unknown (Thomas et al., 2008; Caillaud et al., 2014; Yuan et al., 2016). We next examined the domain/sequence of HopO1-1 required for PD localization. As HopO1-1 contains a putative N-myristolation site (G2), we first looked at its role in PD localization. Consistent with its predicted role in membrane association, the G2A mutant abolishes the PM localization as well as the PD localization of HopO1-1 (Figure 1F), suggesting that N-myristolation is essential for PM/PD localization. We next searched for putative functional domains of HopO1-1 using NCBI Conserved Domain Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Amino acids 41 to 283 (C-terminal end residue) are predicted to encode an ADP-ribosyltransferase (ADP-RT; Supplemental Figure 2A); other than an ADP-RT domain, none of other known targeting signals or transmembrane domains were detected. To determine whether the ADP-RT domain is important for the PM/PD localization, we generated two deletion forms: deletion of the ADP-RT domain (HopO1-11-40-YFP) and deletion of the first 40 amino acids (HopO1-141-end-YFP). Confocal analyses showed that, while amino acids 1 to 40 of HopO1-1 are sufficient to localize the fusion protein to the PM, there is no detectable PD signal. HopO1-141-end-YFP, on the other hand, was mainly detected in the nucleus and the cytoplasm (Figure 1F). We further investigated whether the putative catalytic residues of HopO1-1 are involved in the correct subcellular localization by generating a YFP-tagged catalytic mutant of HopO1-1 (HopO1-1DD-YFP), in which two catalytic residues, E247 and E249, were mutated to D. Unlike HopO1-1-YFP plants, HopO1-1DD-YFP transgenic plants lost the ability to slow plant growth (Supplemental Figure 1C), suggesting that the predicted catalytic site residues are required for HopO1-1 function in planta. However, HopO1-1DD-YFP was found in the periphery (with puncta) of Arabidopsis cells, similar to HopO1-1-YFP (Figure 1A). Taken together, these results indicate that the PM/PD localization of HopO1-1 likely requires two signals: amino acids 1 to 40 of HopO1-1 contains the first signal that is needed for localization to the PM, whereas the ADP-RT domain (but not catalytic site residues) is necessary for PD localization.
HopO1-1 Exhibits Some Mono-ADP-Riobsyltransferase Activity in Vitro
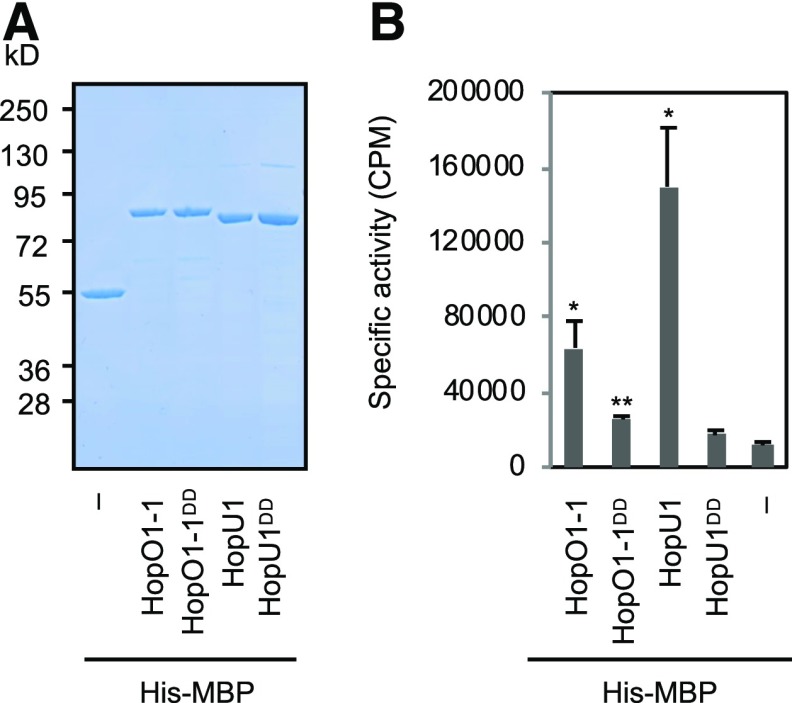
We next performed experiments to directly test whether HopO1-1 is indeed a mono-ADP-ribosyltransferase (mADP-RT), as previously predicted along with HopU1 (Fu et al., 2007). His-MBP-HopO1-1, His-MBP-HopO1-1DD, and His-MBP (a negative control) were expressed in Escherichia coli and purified. Additionally, we included His-MBP-HopU1 and His-MBP-HopU1DD as positive and negative controls, respectively (Fu et al., 2007; Figure 2A). Purified proteins were incubated together with a generic substrate, poly-l-Arg, to perform an in vitro ADP-ribosylation assay as described (Fu et al., 2007). Consistent with a previous report (Fu et al., 2007), His-MBP-HopU1 ribosylates the generic substrate in vitro. His-MBP-HopO1-1 produced a significantly higher amount of ADP-ribosylated poly-l-Arg compared with His-MBP. Furthermore, the catalytic mutant, His-MBP-HopO1-1DD, has a reduced activity (Figure 2B). However, we noticed that the activity of HopO1-1 is significantly lower than that of HopU1 in this assay. Together, these results suggest that HopO1-1 is likely an active mADP-RT, although we cannot exclude the possibility that it also has another enzymatic activity.
Figure 2.
HopO1-1 Exhibits mADP-RT Activity.
(A) SDS-PAGE analysis. Recombinant proteins of wild types (HopO1-1 and HopU1), catalytic mutants (HopO1-1DD and HopU1DD), and His-MBP were purified with Ni-NTA resin. Purified proteins (2 mM) were separated by SDS-PAGE and stained with SimplyBlue SafeStain. Minus (–) indicates His-MBP only. Numbers on the left indicate molecular mass in kilodaltons.
(B) In vitro ADP-ribosylation assay. Recombinant proteins were incubated with poly-l-Arg to examine their ribosyltransferase activity. His-MBP served as a negative control. Error bars represent se from four reactions. Statistical differences between different recombinant proteins and His-MBP were analyzed with a two-tailed t test (*, P < 5 × 10−2; **, P < 5 × 10−3). Minus (–) indicates His-MBP only.
HopO1-1 Contributes to Bacterial Virulence
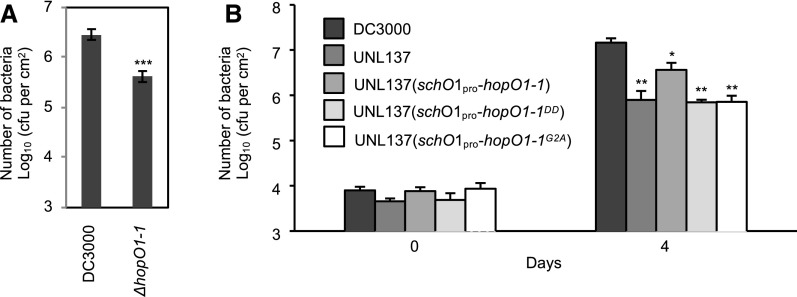
To investigate the virulence function of HopO1-1 in the context of bacterial infection, we generated a ΔhopO1-1 deletion strain of Pst DC3000. The leaves of 5-week-old Arabidopsis plants (wild-type accession Col-0) were infected with 2 × 108 cfu/mL Pst DC3000 and the ΔhopO1-1 mutant using a dip inoculation method. The ΔhopO1-1 mutant was significantly compromised in virulence compared with wild-type Pst DC3000 (Figure 3A). In parallel, we also tested the virulence activity of an independent mutant, UNL137, in which hopO1-1 and the adjacent hopT1-1 genes are deleted (Guo et al., 2005). As shown in Figure 3B, the double mutant was also compromised in virulence. To investigate whether the PM/PD localization and catalytic activity of HopO1-1 are required for the function of HopO1-1, we conducted complementation experiments with HopO1-1G2A (defective in PM/PD localization) and HopO1-1DD (defective in ADP-RT activity), respectively. Whereas wild-type hopO1-1 partially complemented UNL137, hopO1-1G2A and hopO1-1DD failed to rescue the pathogenicity of UNL137 (Figure 3B). These results are consistent with the hypothesis that HopO1-1 is targeted to the PM/PD in plants to exert its virulence through its putative ADP-RT activity.
Figure 3.
HopO1-1 Is Required for Full Virulence of P. syringae.
(A) P. syringae infection assay. Leaves of Col-0 were dip-inoculated with Pst DC3000 wild-type and ΔhopO1-1 mutant strains at 2 × 108 cfu/mL. Bacterial multiplication was determined 3 d after infection by counting bacterial numbers (cfu/cm2 leaf area). Six replicates were analyzed. Error bars represent se. Statistical differences between DC3000 and the ΔhopO1-1 mutant were analyzed with a two-tailed t test (***, P < 5 × 10−5).
(B) Sequence motifs associated with membrane targeting and catalytic activity of HopO1-1 contribute to full virulence of P. syringae. Arabidopsis plants (Col-0) were spray-inoculated with 5 × 107 cfu/mL of the following DC3000 strains: wild-type DC3000, UNL137, UNL137 (schO1pro-hopO1-1), UNL137 (schO1pro-hopO1-1DD), and UNL137 (schO1pro-hopO1-1G2A). Bacterial population was measured at 0 and 4 d postinoculation. Statistical differences between DC3000 and the mutants were analyzed with a two-tailed t test (*, P < 5 × 10−2; **, P < 5 × 10−3).
HopO1-1 Alters PD-Mediated Cell-to-Cell Molecular Trafficking in Arabidopsis
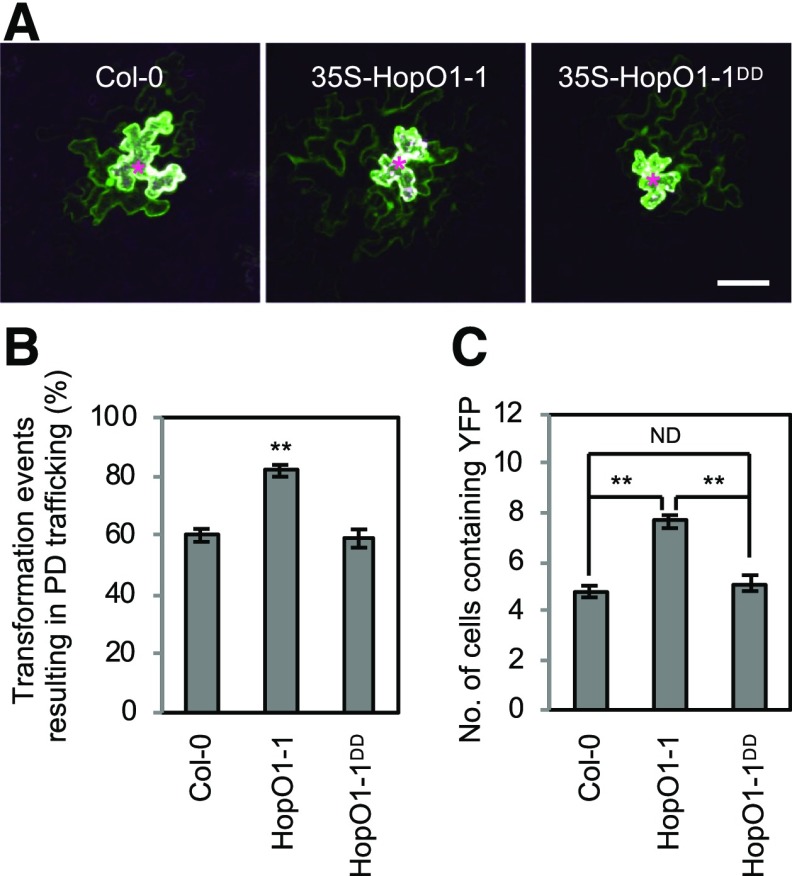
Given the PD localization of HopO1-1, we hypothesized that HopO1-1 modulates PD-dependent molecular flux between plant cells. To test this hypothesis, we adopted a microparticle bombardment approach. Gold particles were coated with plasmids that express YFP (which can move from transformed cells to adjacent cells through PD in Arabidopsis epidermal cells) and ER-trapped CFP (ER-CFP; which cannot move beyond transformed cells). Plasmid-coated particles were bombarded into leaves of wild-type Col-0, 35S-HopO1-1, and 35S-HopO1-1DD transgenic plants following the protocol described previously (Thomas et al., 2008; Faulkner et al., 2013; Aung et al., 2017). Fluorescent signals were detected at ∼20 h after bombardment using confocal microscopy. The degree of diffusion of YFP proteins was used to determine PD-dependent molecular flux between Arabidopsis abaxial epidermis cells, whereas transformed cells were marked by nondiffusible ER-CFP. We observed a greater movement of YFP molecules in transgenic plants expressing HopO1-1 compared with wild-type Col-0 plants or transgenic plants expressing the HopO1-1DD mutant (Figure 4A). Overall, around 60% of transformed cells led to PD-dependent trafficking of YFP molecules in the wild type and HopO1-1DD, whereas expression of HopO1-1 resulted in PD-dependent trafficking in over 80% of transformed cells (Figure 4B). More strikingly, transgenic expression of HopO1-1 promoted the movement of YFP molecules to more surrounding plant cells (Figures 4A and 4C).
Figure 4.
HopO1-1 Promotes PD Permeability in Arabidopsis.
(A) Confocal images show the diffusion of YFP. Images were taken of leaf epidermal cells of Col-0 and transgenic plants expressing HopO1-1 variants. YFP and ER-CFP were cobombarded into the leaves of wild-type Col-0, 35S-HopO1-1, and 35S-HopO1-1DD. PD-dependent diffusion of YFP molecules was examined. Asterisks indicate the bombarded sites expressing both YFP and ER-CFP. Bar = 50 µm.
(B) Quantitative data show the percentage of transformation events resulting in PD trafficking of YFP in wild-type Col-0, 35S-HopO1-1, and 35S-HopO1-1DD. The degree of trafficking is scored by counting the number of transformation events yielding the diffusion of YFP to surrounding cells versus the total transformation events per experiment. Error bars represent se from three biological replicates. Statistical differences between wild-type Col-0 and the transgenic plants were analyzed with a two-tailed t test (**, P < 0.005).
(C) Quantitative data show the average number of cells containing YFP in wild-type Col-0, 35S-HopO1-1, and 35S-HopO1-1DD. Transformation events resulting in PD trafficking from all three independent experiments are combined for the analysis. Error bars represent se. Statistical differences among wild-type Col-0 and the transgenic plants were analyzed with a Mann-Whitney U test (**, P < 0.0001). ND, no statistical difference.
Because HopO1-1 increases the cell-to-cell movement of YFP, we examined whether it also enlarges the size exclusion limit of PD. We built a YFP cancatemer with two or three YFP molecules to increase the size of YFP, resulting in 2xYFP (∼54 kD) or 3xYFP (∼81 kD), respectively (Supplemental Figure 3A). We did not observe an enhanced movement of 2xYFP molecules in transgenic plants expressing HopO1-1 compared with wild-type Col-0 (Supplemental Figure 3B). In addition, there was no movement of 3xYFP in either Col-0 or 35S-HopO1-1 leaves (Supplemental Figure 3C). Together, these results suggest that transgenic expression of HopO1-1 increases the distance of PD-mediated molecular flux without drastically increasing the size exclusion limit of PD, as detected by incremental 27-kD size increases. However, our data cannot exclude the possibility that a small increase of the PD aperture in HopO1-1 transgenic plants might contribute to increasing PD-mediated trafficking of 1xYFP.
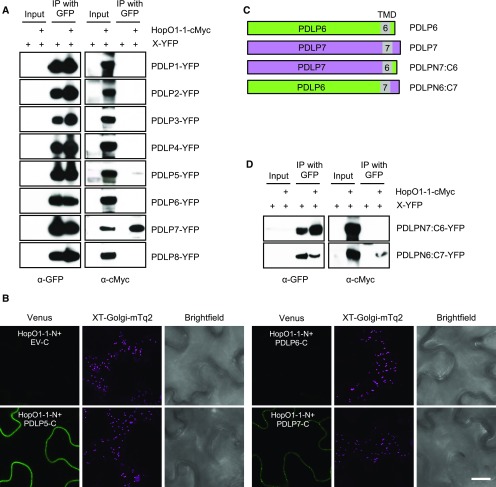
HopO1-1 Physically Associates with PDLPs
To modulate PD-dependent molecular flux, we hypothesized that HopO1-1 might manipulate one or more PD regulators. We noticed that PDLP5 is involved in bacterial immunity (Lee et al., 2011) through maintaining callose homeostasis at PD (Cui and Lee, 2016). PDLP5 belongs to the PDLP family, which has eight members (PDLP1–PDLP8), in Arabidopsis (Thomas et al., 2008). To examine whether HopO1-1 targets PDLPs, we tested the physical interaction between HopO1-1 and all eight PDLPs using coimmunoprecipitation (co-IP) analyses in planta. PDLP-YFP with or without HopO1-1-cMyc was transiently expressed in tobacco leaves. Co-IP followed by immunoblot analyses showed that HopO1-1 interacts with PDLP5 and PDLP7 (Figure 5A). We further confirmed these interactions using bimolecular fluorescence complementation (BiFC) assays. Confocal images showed that when HopO1-1:NVen210 was coexpressed together with PDLP5:CVen210 or PDLP7:CVen210, the fluorescent signals could be reconstituted (Figure 5B). PLDP6 and a cytosolic CVen peptide (X:CVen210), on the other hand, do not complement the fluorescent signals when expressed together with HopO1-1 (Figure 5B). Intriguingly, the complemented fluorescent signals between HopO1-1 and PDLP5 were mainly detected on the PM, whereas PD-like puncta were observed when HopO1-1 and PDLP7 were coexpressed. Together, the BiFC results agree with the co-IP results, validating that HopO1-1 interacts with PDLP5 and PDLP7 in planta.
Figure 5.
HopO1-1 Physically Associates with PDLPs.
(A) Co-IP analysis of the interaction between HopO1-1 and PDLPs. Various combinations of HopO1-1-cMyc and PDLP-YFP fusion proteins as indicated were transiently expressed in tobacco leaves followed by immunoprecipitation (IP) using GFP-Trap_A. A GFP or cMyc antibody was used to detect the fusion proteins. Three biological replicates were performed for each sample.
(B) BiFC assay of the interaction between HopO1-1 and PDLPs. Various combinations of HopO1-1:NVen210 and PDLPs:CVen210 were transiently expressed in tobacco leaves. The infiltrated leaves were subjected to confocal imaging 2 d postinfiltration. At least 10 images were captured from randomly chosen regions of infiltrated leaves for each experiment. Three biological replicates were performed for each sample. C, CVen210; EV, empty vector; N, NVen210; XT-Golgi-mTq2, an integrated mTurquoise2 marker labeling Golgi. Bar = 10 µm.
(C) Schematic representations of the sequences of wild-type PDLPs and chimeric PDLPs. TMD, transmembrane domain.
(D) Co-IP analysis of the interaction between HopO1-1 and the C-terminal tail of PDLP7. Various combinations of HopO1-1-cMyc and different variants of PDLP-YFP fusion proteins as indicated were transiently expressed in tobacco leaves followed by IP using GFP-Trap_A. A GFP or cMyc antibody was used to detect the protein.
PDLPs are type I membrane proteins, which contain a short fragment (7–19 amino acids) of the C-terminal cytoplasmic tail (C-tail; Figure 5C). As HopO1-1 is secreted into plant cells, the interaction between HopO1-1 and PDLPs would likely be mediated in part by the C-tail of PDLPs. To examine whether the C-tail of PDLP7 confers specificity in the PDLP interaction with HopO1-1, we generated two different chimeric forms between PDLP6 and PDLP7 by swapping the transmembrane domain plus the C-tail (Figure 5C). Using the above-mentioned co-IP approach, we found that PDLPN7:C6 failed to interact with HopO1-1. PDLPN6:C7, on the other hand, became competent as a HopO1-1-interacting protein (Figure 5D). Together, these results show that the interaction between HopO1-1 and PDLP7 is mediated through the C-tail of PDLP7.
HopO1-1 Affects the Stability of PDLP5 and PDLP7
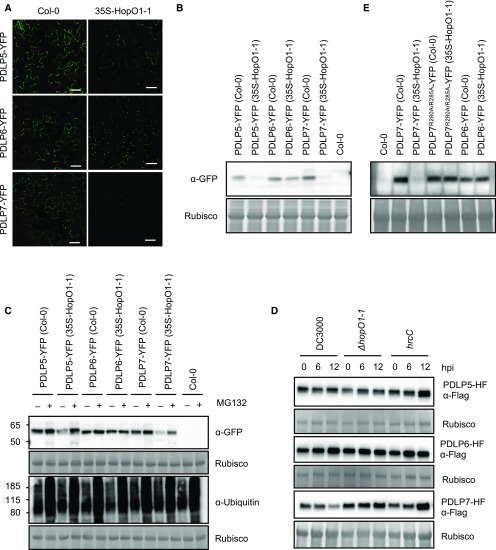
Having found that HopO1-1 physically associates with PDLP5 and PDLP7, we next investigated whether HopO1-1 affects the levels or molecular weights of the PDLPs in planta. We generated transgenic plants stably expressing PDLP-YFP fusion proteins in the wild-type Col-0 or 35S-HopO1-1 background. As the transgenic plants were generated in different backgrounds, we analyzed three independent lines for each construct. The transgenic plants were subjected to confocal imaging to examine levels of PDLP-YFP fusion proteins. As shown in Figure 6A, we noticed that YFP signals from PDLP5-YFP and PDLP7-YFP plants were much dimmer in the 35S-HopO1-1 background, whereas YFP signals in PDLP6-YFP plants were not affected by the expression of HopO1-1. This raised the possibility that HopO1-1 might destabilize PDLP5 and PDLP7. To determine the levels of PDLPs more qualitatively, we performed immunoblot analyses to detect YFP fusion proteins using a GFP antibody. In line with confocal images, we found that expression of HopO1-1 affects the levels of PDLP5 and PDLP7, while the level of PDLP6 was not drastically affected (Figure 6B; Supplemental Figure 4). However, HopO1-1 does not affect the molecular weights of the PDLPs (Figure 6C).
Figure 6.
HopO1-1 Affects PDLP Protein Stability in Arabidopsis.
(A) Confocal images of 2-week-old Arabidopsis transgenic plants expressing 35S-PDLPs-YFP in wild-type Col-0 or 35S-HopO1-1. Bars = 20 µm.
(B) Immunoblot analysis shows the expression of 35S-PDLP-YFP fusion proteins. The transgenic plants were generated in the wild-type Col-0 or 35S-HopO1-1 background. A GFP antibody was used to detect YFP fusion proteins. Rubisco served as an internal control.
(C) Immunoblot analysis shows that PDLP5 and PDLP7 are degraded through a proteasome-dependent pathway. Arabidopsis transgenic plants expressing 35S-PDLP-YFP fusion proteins were grown in 0.5× Linsmaier and Skoog (LS) liquid medium. Ten-day-old seedlings were treated with mock (–; 1% DMSO) and 50 µM MG132 (+). The samples were collected 24 h after the treatment and subjected to immunoblot analyses. A GFP antibody was used to detect YFP fusion proteins and a ubiquitin antibody was used to detect ubiquitinated proteins. Rubisco served as an internal control.
(D) Immunoblot analysis shows the expression of 35S-PDLPs-HF upon bacterial infection. Five-week-old Arabidopsis transgenic plants were infiltrated with 2 × 108 cfu/mL Pst DC3000, ΔhopO1-1, or hrcC. The infected leaves were collected at different time points as indicated (hpi, hours postinfection). Expression of the PDLPs was detected using a Flag antibody. Rubisco served as a loading control.
(E) Immunoblot analysis detects the expression of 35S-PDLP-YFP fusion proteins in transgenic plants in the wild-type Col-0 or 35S-HopO1-1 background. A GFP antibody was used to detect YFP fusion proteins. Rubisco served as a loading control.
Given that HopO1-1 affects the protein stability of PDLPs, we next examined whether the PDLPs are degraded through a proteasome-dependent pathway. Arabidopsis transgenic seedlings expressing PDLP-YFP in wild-type Col-0 or 35S-HopO1-1 were treated with MG132, a proteasome inhibitor, and subjected to immunoblot analyses. As shown in Figure 6C, MG132 blocked the degradation of PDLP5-YFP and PDLP7-YFP fusion proteins in the 35S-HopO1-1 background. We further showed that HopO1-1 did not affect the transcript levels of PDLP5, PDLP6, and PDLP7 (Supplemental Figure 5A). Collectively, our results suggest that HopO1-1 affects the stability of PDLP5 and PDLP7 through a proteasome-dependent mechanism without affecting the transcript levels.
To directly test the possible degradation of PDLP5 and/or PDLP7 in the context of bacterial infection, we infected PDLP-HF (His and Flag epitopes) transgenic plants with different bacterial strains: Pst DC3000, ΔhopO1-1, or hrcC. Infected leaves were harvested at 0, 6, and 12 h postinfection and subjected to immunoblot analyses. As shown in Figure 6D, the level of PDLP7 is lower at 12 h postinfection with Pst DC3000, whereas ΔhopO1-1 and hrcC mutants do not destabilize PDLP7. By contrast, Pst DC3000 does not degrade PDLP5 (Figure 6D). These findings suggest that PDLP7, but not PDLP5, is a biologically relevant host target of HopO1-1 during infection. As a negative control, the PDLP6 level was not changed in response to Pst DC3000, ΔhopO1-1, or hrcC infection (Figure 6D).
Putative Ribosylation Sites Are Crucial for the Degradation of PDLPs
mADP-RT can modify target proteins by ribosylating Arg (Hassa et al., 2006). PDLP5 and PDLP6 contain one Arg, whereas PDLP7 has two Arg residues, at their C-terminal ends (Supplemental Figure 5B). To examine the role of putative ribosylation sites on PDLP7 protein stability, we mutated Arg-280 and Arg-285 into Ala (PDLP7R280A/R285A) and generated transgenic plants stably expressing PDLP7R280A/R285A-YFP in the wild-type Col-0 or 35S-HopO1-1 background. Unlike HopO1-1’s effect on wild-type PDLP7, the expression of HopO1-1 does not affect the level of PDLP7R280A/R285A (Figure 6E). Similarly, the expression of PDLP transcripts was not affected by HopO1-1 (Supplemental Figure 5C). This result suggests that the Arg residues at the C terminus of PDLP7 are important for HopO1-1-dependent protein degradation.
We next performed experiments to determine whether HopO1-1 directly ribosylates PDLP7 to modulate its stability. To test mADP-RT activity of HopO1-1 in planta, we incubated total proteins extracted from Arabidopsis or tobacco with the recombinant His-MBP fusion protein purified from E. coli or the YFP fusion protein enriched from N. benthamiana. Consistent with a previous report, HopU1 ribosylated both Arabidopsis and tobacco proteins (Fu et al., 2007), whereas HopO1-1 did not ribosylate any plant proteins (Supplemental Figures 6A and 6B). We next enriched PDLP-YFP fusion proteins from Arabidopsis transgenic plants stably expressing PDLP-YPF in wild-type Col-0 or 35S-HopO1-1 to detect possible HopO1-1-mediated ribosylation of PDLPs using an anti-pan-ADP-ribose binding reagent (Redditt et al., 2019). Although ribosylated plant proteins were detected in transgenic plants expressing HopU1 (DEX-HopU1), we did not observe any ribosylation on PDLPs (Supplemental Figure 6C). Given that PDLPs are destabilized through a proteasome-dependent pathway (Figure 6C), we treated the transgenic plants with MG132 and subjected them to immunoblot analyses. Again, we did not detect HopO1-1-dependent ribosylation of PDLP7 in Arabidopsis (Supplemental Figure 6D).
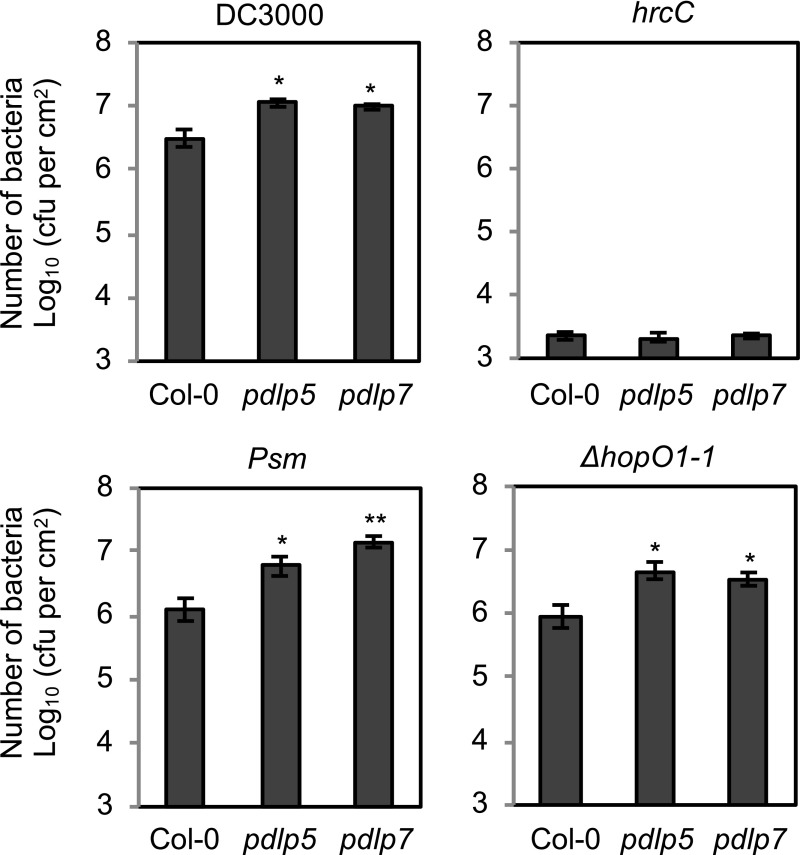
PDLP7 Is Required for Bacterial Immunity
It was previously reported that the pdlp5 mutant is more susceptible to P. syringae pv maculicola (Psm) ES4326 (Lee et al., 2011). Because HopO1-1 physically interacts with and destabilizes PDLP7, we speculated that PDLP7 might also play a role in plant immunity. To investigate the role of PDLP7 in plant immunity against bacterial pathogens, we identified and characterized a pdlp7 mutant that carries a T-DNA in the first exon of PDLP7 (Supplemental Figure 7A). An RT-PCR assay showed that the pdlp7 mutant is a knockout, as there are no PDLP7 transcripts in the mutant (Supplemental Figure 7B). The pdlp7 mutant displays normal plant morphology compared with the wild type (Supplemental Figure 7C), suggesting that PDLP7 does not make major contributions to plant growth and development. Consistent with a previous report (Lee et al., 2011), we observed that pdlp5 was more susceptible to Psm ES4326 (Figure 7). In addition, pdlp5 showed enhanced disease susceptibility to Pst DC3000 but not to the hrcC mutant. We observed that the pdlp7 mutant was also more susceptible to both Psm ES4326 and Pst DC3000 but not to the hrcC mutant (Figure 7). A double pdlp5 pdlp7 mutant was generated by crossing. The double mutant displayed similar susceptibility to bacterial infection compared with the single mutants (Supplemental Figure 7D), suggesting that PDLP5 and PDLP7 likely function together in plant immunity. PDLP5 and PDLP7 uniquely contribute to the bacterial immunity among the PDLP family, as a pdlp1 pdlp2 pdlp3 (pdlp1/2/3) triple mutant and a pdlp4 single mutant were not compromised in bacterial defense (Supplemental Figure 7D). We next infected the pdlp5 and pdlp7 mutants with the ΔhopO1-1 mutant. Both mutants were more susceptible to the ΔhopO1-1 mutant (Figure 7), suggesting that genetic removal of PDLP5 and PDLP7 from the plant is sufficient to substitute for the loss of HopO1-1 in the bacterium.
Figure 7.
PDLP5 and PDLP7 Are Involved in Bacterial Immunity.
Leaves of Col-0 and pdlp mutants were syringe-infiltrated with Pst DC3000, the ΔhopO1-1 mutant, the hrcC mutant, or Psm ES4326 at 2 × 105 cfu/mL. Bacterial multiplication was determined 2 d after infection by counting bacterial number (cfu/cm2 leaf area). Error bars represent se from six biological replicates. Statistical differences between wild-type Col-0 and mutants were analyzed with a two-tailed t test (*, P < 0.05; **, P < 0.005).
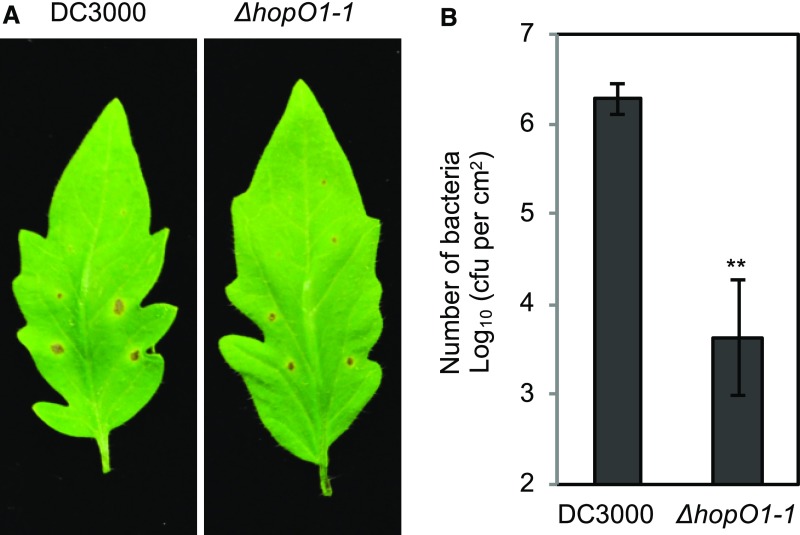
HopO1-1 Is Important for Bacterial Colonization and Invasion of Tissues Surrounding Infection Sites
The finding that Pst DC3000 injects HopO1-1 to manipulate PD prompted us to test the role of HopO1-1 in allowing bacteria to colonize surrounding tissues. For this purpose, we locally infected Pst DC3000 and the ΔhopO1-1 mutant on tomato (‘Castlemart’) leaves using a leaf-stab assay and counted bacterial numbers in tissues surrounding the infection sites. The ΔhopO1-1 mutant caused smaller halo spots (Figure 8A) and had many fewer bacteria in the surrounding tissues (Figure 8B). This result supports the hypothesis that Pst DC3000 delivers HopO1-1 to promote bacterial colonization and invade host tissues around infection sites.
Figure 8.
HopO1-1 Is Crucial for Colonization of P. syringae Surrounding Infection Sites.
(A) Disease phenotypes of tomato leaves after local infection with Pst DC3000 and the ΔhopO1-1 mutant using a needle. The images were taken 7 d after infection.
(B) Bacterial multiplication was determined 7 d after infection by counting bacterial number (cfu/cm2 leaf area). The needle infection sites were removed using a biopsy punch (2 mm radius), and the distal tissues were collected to determine bacterial growth. Error bars represent se from six samples. Statistical differences between DC3000 and ΔhopO1-1 are analyzed with a two-tailed t test (**, P < 0.005).
DISCUSSION
Bacterial effectors have been detected in different cellular compartments within plant cells, including the PM (Shan et al., 2000; Robert-Seilaniantz et al., 2006; Xin et al., 2015), ER (Block et al., 2014), trans-Golgi network/early endosome (Nomura et al., 2011), chloroplast (Jelenska et al., 2007; Li et al., 2014), mitochondrion (Block et al., 2010), and nucleocytoplasm (Fu et al., 2007; Giska et al., 2013). Determination of the subcellular localization of effectors within plant cells is important for explaining the biologically relevant functions of pathogen effectors. In this study, we found that Pst DC3000 HopO1-1 is targeted to PD. This finding strongly suggests that pathogenic bacteria manipulate not only cell-autonomous host functions but also PD-mediated non-cell-autonomous host processes to spread infection.
Recent findings began to reinforce the notion that PD are important battlegrounds during plant-pathogen interactions (Kankanala et al., 2007; Lee, 2014; Cheval and Faulkner, 2018; Sakulkoo et al., 2018; Ganusova and Burch-Smith, 2019). In particular, different members of PDLPs have roles not only in regulating basic PD function but also in plant immunity. For example, PDLP1 is a receptor for plant viral movement proteins and plays an important role in promoting viral movement (Amari et al., 2010). PDLP1 is also required to resist Hyaloperonospora arabidopsidis infection by depositing callose at haustoria, a feeding structure of the pathogen (Caillaud et al., 2014). We found no evidence that PDLP1 is involved in Pst DC3000 infection of Arabidopsis. Instead, HopO1-1 selectively targets PDLP7 and possibly PDLP5, which are involved in bacterial immunity (Figures 5 and 7). We further demonstrated that the short intracellular C-tail of PDLP7 determines the specific interaction between PDLP and HopO1-1 (Figures 5C and 5D). In fact, changing two amino acids at the C-tail of PDLP7 was sufficient to prevent the protein from being degraded by HopO1-1 (Figure 6E). If many PD-manipulating pathogen effectors target the short C-tails of PDLPs, future efforts to edit the C-tails of PDLPs might provide a broadly applicable novel means of engineering plants with enhanced plasmodesmal immunity against pathogens.
At this point, we favor the hypothesis that HopO1-1 targets immunity-associated PDLP7 for degradation by ribosylating the PDLP protein based on the following observations: HopO1-1 ribosylates a generic substrate (Figure 2), the catalytic activity of HopO1-1 is required for its virulence function (Figure 3B), and the putative ribosylation sites of PDLP7 are required for HopO1-1-dependent protein degradation (Figure 6E). However, we have been unable to detect HopO1-1-dependent ADP-ribosylation of PDLPs or other plant proteins using various methodologies (Supplemental Figure 6). It is possible that the sensitivity of the tested approaches is not high enough to detect the ribosylated signals, although we could robustly detect the ADP-RT activity of HopU1 in all these assays. Alternatively, an unknown biochemical activity of HopO1-1 might regulate the protein stability of PDLP7 in Arabidopsis. Interestingly, a recent report demonstrated that a putative mADP-RT, Legionella pneumophila effector SdeA, functions as a ubiquitin-conjugating enzyme in an E1- and E2-independent manner (Qiu et al., 2016). However, HopO1-1 does not share sequence similarity with SdeA. Further study is necessary to elucidate how HopO1-1 precisely destabilizes PDLP7.
Here, we found that expression of HopO1-1 facilitates the movement of YFP molecules between plant cells without drastically changing the apparent PD aperture using the YFP diffusion assays (Figure 4; Supplemental Figure 3). A recent report showed that PD-dependent trafficking is independent of PD aperture, PD density, or callose deposition at PD (Yan et al., 2019). Those authors reported that PHLOEM UNLOADING MODULATOR (PLM) is required for the formation of the ER-PM contact sites within the cytoplasmic sleeve of PD. PD in the plm mutant have no visible cytoplasmic sleeve between the ER and the PM (Nicolas et al., 2017) but exhibit enhanced PD-dependent trafficking. Moreover, in the plm mutant, apparent PD aperture (as observed by transmission electron microscopy analyses), PD density, and PD-associated callose accumulation are comparable to those in wild-type plants (Yan et al., 2019). These findings suggest that, besides apparent PD aperture and density, there are other aspects of PD that might play a crucial role in determining PD function. In this regard, future examination of PD in 35S-HopO1-1 transgenic plants or during Pst DC3000 infection may shed light on an aspect of PD regulation that may have evaded discovery so far.
It has long been established that viral pathogens exploit PD to move between plant cells (Benitez-Alfonso et al., 2010; Heinlein, 2015; Kumar et al., 2015). Recent studies showed that fungal pathogens also exploit the function of PD to spread in plants (Khang et al., 2010; Cao et al., 2018). Our finding that a bacterial effector protein, HopO1-1, targets the host PD suggests that diverse pathogenic microbes have evolved virulence factors to modulate PD-mediated cell-to-cell communication in plants. Understanding how pathogenic microbes modulate PD at the molecular level represents a promising area of research that has potential to substantially advance our understanding of fundamental PD biology and novel disease control strategies in plants.
METHODS
Plant Materials, Growth Conditions, Transformation, Plant Selection, and Chemical Treatment
Arabidopsis (Arabidopsis thaliana), tobacco (Nicotiana tabacum), and tomato (Solanum lycopersicum cv Castlemart) plants were grown at 22°C with 50% humidity and irradiated with ∼100 μmol m−2 s−1 white light. Arabidopsis T-DNA insertion mutants pdlp4 (SALK_028613), pdlp5 (SAIL_46_E06.v1), and pdlp7 (SALK_015341) were obtained from the ABRC. pdlp1/2/3 (Caillaud et al., 2014) was a gift from Christine Faulkner’s lab. The presence of the T-DNAs and the homozygosity of mutants were identified by genomic PCR using the following primers: pdlp4-1 (PDLP4-LP, PDLP4-RP, and LBb1.3), pdlp5 (PDLP5-LP, PDLP5-RP, and SAIL-LB2), and pdlp7 (PDLP7-LP, PDLP7-RP, and LBb1.3). The absence of PDLP4, PDLP5, and PDLP7 transcripts was determined by RT-PCR using primers PDLP4-Fwd + PDLP4-Rev, PDLP5-Fwd + PDLP5-Rev, and PDLP7-Fwd + PDLP7-Rev, respectively. All primers used in this study are listed in Supplemental Data Set 1. Arabidopsis transgenic plants were generated using the simplified transformation method (https://plantpath.wisc.edu/simplified-arabidopsis-transformation-protocol/). For transgenic plants harboring resistance to kanamycin, T0 seeds were selected on 0.5× LS medium with 50 µg/mL kanamycin. For basta resistance transgenic plants, T0 seeds were germinated on soil and 1-week-old seedlings were sprayed with 0.1% (v/v) Finale Herbicide (Bayer) and 0.05% (v/v) Silwet L-77 (PhytoTech). The T2 and T3 plants were screened on 0.5× LS medium with 10 µg/mL glufosinate-ammonium. For MG132 treatment, Arabidopsis seeds were germinated and grown in half strength LS liquid medium with 1% (w/v) Suc. Ten-day-old seedlings were treated with 1% (v/v) DMSO (; mock) or 50 μM MG132. Samples were collected 24 h after the treatment.
Gene Cloning and Plasmid Construction
Plasmid DNAs were constructed using a Gateway cloning system (Life Technologies; Thermo Fisher Scientific) or restriction enzyme (RE) digestion. In this study, we reported the following constructs: 35S-HopO1-1, 35S-HopO1-1DD, 35S-HopO1-1-cMyc, 35S-YFP-HopO1-1, 35S-HopO1-1-YFP, 35S-HopO1-1DD-YFP, 35S-HopO1-1G2A-YFP, 35S-HopO1-141-end-YFP, 35S-HopO1-11-40-YFP, 35S-PDLP5-YFP, 35S-PDLP6-YFP, 35S-PDLP7-YFP, 35S-PDLPN6:C7-YFP, 35S-PDLPN7:C6-YFP, 35S-PDLP7R280A/R285A-YFP, 35S-PDLP5-HF, 35S-PDLP6-HF, 35S-PDLP7-HF, 35S-1xYFP, 35S-2xYFP, 35S-3xYFP, 35S-ER-CFP, HopO1-1:NVen210-X:CVen210, HopO1-1:NVen210-PDLP5:CVen210, HopO1-1:NVen210-PDLP6:CVen210, HopO1-1:NVen210-PDLP7:CVen210, HopO1-1-His-MBP, HopO1-1DD-His-MBP, HopU1-His-MBP, and HopU1DD-His-MBP.
The genes of interest were amplified from Pseudomonas syringae pv tomato DC3000 genomic DNA or the cDNA synthesized from total RNA of wild-type (Col-0) Arabidopsis seedlings using Phusion High-Fidelity DNA polymerase (New England Biolabs). For Gateway cloning (Life Technologies), the genes of interest were amplified with Gateway-compatible primers. The PCR fragments were cloned into a donor vector (pDONR 207) and different destination vectors using a standard Gateway cloning system (Life Technologies). For plasmids cloned by RE digestion, the genes of interest were amplified with gene-specific primers containing the chosen RE recognition sites (see Supplemental Data Set 1 for details). The PCR fragments were digested with the REs and ligated into the RE-digested destination vector using T4 ligase (Thermo Fisher Scientific). The vectors used in this study are listed in Supplemental Data Set 2.
To generate the HopO1-1G2A mutation, the mutation site was introduced in the forward primer. To create catalytic mutants (HopO1-1DD and HopU1DD) and chimeric fusion proteins (PDLPN6:C7 and PDLPN7:C6), an overlapping PCR approach (https://gfp.dpb.carnegiescience.edu/protocol/index4.html) was adopted using the overlapping primers. The mutation sites of PDLP7 R280A/R285A-YFP were introduced in the reverse primer used for PCR amplification. For building a single-vector BiFC plasmid, HopO1-1 was amplified with primers containing restriction sites NcoI and BamHI on the end of forward and reverse primers, respectively. The amplified fragment was digested with the REs and ligated into an enzyme-digested recipient plasmid, pDOE-05 (Gookin and Assmann, 2014), to generate HopO1-1-NVen210. The resulting plasmid, HopO1-1:NVen210-X:CVen210, was used as a negative control as well as a vector to introduce PDLPs. PDLP5 was amplified with primers containing AvaII and BspEI cutting sites, whereas PDLP6 and PDLP7 were amplified with SanDI and BspEI cutting sites. The amplified and digested PCR products were ligated into the enzyme-digested recipient plasmid, HopO1-1:NVen210-X:CVen210. ER-CFP and EYFP were amplified using ER-CFP (CD3-953) and ER-YFP (CD3-957), respectively, as templates (Nelson et al., 2007). For cloning 2xYFP, the two fragments of EYFP coding sequences were ligated using EcoRI. 3xYFP was built by ligating the third coding sequence using BamHI at the 3′ end of 2xYFP. The amplified products were then cloned into pDnor 207 and pEarley Gate 100 (Earley et al., 2006). To express recombinant proteins in Escherichia coli, HopO1-1 and HopU1 variants were amplified with primers containing restriction sites NdeI and BamHI on the end of forward and reverse primers, respectively. The amplified fragments were digested with the REs and ligated into enzyme-digested recipient plasmid, pET17b HMR, to generate His-MPB fusion protein.
Transient Expression
For Agrobacterium tumefaciens-mediated transient expression, A. tumefaciens strains GV3101 (pMP90) harboring the plasmid of interest (cell density at A600 of 0.1) were suspended in sterile distilled water and infiltrated into the leaves of 5-week-old tobacco or Nicotiana benthamiana plants. The infiltrated leaves were subjected to live-cell imaging 2 d after infiltration. A similar transient expression method was used for BiFC assays. The two candidate genes for testing the interaction were cloned into a double open reading frame expression BiFC system with an XT-Golgi, mTurquoise2 (mTq2) marker (Gookin and Assmann, 2014). The A. tumefaciens-infected cells were identified by locating the plant cells with mTq2 expression, and the complementation of Venus signals was examined 2 d after infiltration using confocal microscopy.
To examine PD-dependent molecular flux, a microprojectile bombardment approach combined with confocal imaging was adopted as previously described (Thomas et al., 2008; Faulkner et al., 2013; Aung et al., 2017). In short, 20 mg of 1.0-μm gold particles (Bio-Rad) was soaked in 70% (v/v) ethanol for 15 min and rinsed with 1 mL of sterile distilled water three times. The rinsed particles were stored in 333 μL of 50% (v/v) glycerol. To coat plasmid DNA on the particles, 25 μL of rinsed particles was mixed with 5 μL (1 μg/μL) each of plasmid DNAs (ER-CFP and YFPs), 25 μL of 2.5 M CaCl2, and 5 μL of 0.2 M spermidine. The mixture was vortexed at maximum speed for 3 min, settled for 1 min, and centrifuged at 3000g for 5 s, and the supernatant was removed. Then, the pellet was resuspended in 100 μL of 100% ethanol. The ethanol-rinsed coated particles were then spun down at 3000g for 5 s and subjected to two more rounds of ethanol washes. The particles were suspended in 25 μL of 100% ethanol. Eight-microliter particles were loaded on a macrocarrier disc. The disc was assembled into a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad) following the manufacturer’s instructions. The vacuum chamber was set at 27 inches of Hg, and the particles were delivered into the abaxial side of 5-week-old Arabidopsis leaves at 1100 p.s.i. The whole procedure was performed at room temperature. The bombarded leaves were kept in the same growth chamber at high humidity for 16 to 20 h before imaging. To quantitatively compare the PD-dependent diffusion efficiency of 1xYFP, we collected 518, 402, and 375 images of Col-0, 35S-HopO1-1, and 35S-HopO1-1DD, respectively, from three biological replicates. To determine the PD-dependent trafficking, we first calculated the ratios between transformation events/cells resulting in PD trafficking and total transformation events/cells per experiment. The values from three biological replicates were averaged, and se was calculated. To quantify the number of cells containing YFP, we pooled all images that show PD-dependent movement from three biological replicates. Totals of 311, 330, and 221 images from Col-0, 35S-HopO1-1, and 35S-HopO1-1DD, respectively, were used for analysis. The number of cells containing YFP signals was averaged, and se was calculated. A Mann-Whitney U (https://www.socscistatistics.com/tests/mannwhitney/default2.aspx) test was used to determine the statistical difference between different genotypes. Mann-Whitney U test results are shown in Supplemental Data Set 3. To determine the PD-dependent trafficking of 2xYFP, we calculated the ratios between transformation events/cells resulting in PD trafficking and total transformation events/cells per experiment. At least 50 images were collected from Col-0 and 35S-HopO1-1 from each biological replicate. Values from three biological replicates were averaged, and se was calculated.
Confocal Imaging and Chemical Staining Analyses
A Zeiss Laser Scanning Microscope 510 was used to image fluorescent signals. A small piece (∼4 mm2) of leaf tissue was mounted with water on a glass slide with the abaxial side facing upward. Different fluorescent signals are excited with the following laser lines: callose (405 nm), CFP and mTq (458 nm), YFP and Venus (514 nm), and FM 4-64 (595 nm). The signals were then collected using the following settings: callose (Bandpass [BP] 420–480 nm), CFP and mTq (BP 460–510 nm), YFP and Venus (BP 530–600 nm), and FM 4-64 (590–630 nm). Callose staining of live tissues was performed as described previously by Guseman et al. (2010). In brief, leaves of 4-week-old Arabidopsis plants were infiltrated with 0.1 mg/mL aniline blue fluorochrome (Biosupplies Australia). The callose signals were collected ∼30 min after infiltration for imaging. Cotyledons of 2-week-old Arabidopsis seedlings were stained with 2 μM FM 4-64 (Life Technologies)for 5 min before imaging.
Plasmolysis
Leaves of 2-week-old Arabidopsis transgenic plants expressing 35S-HopO1-1-YFP or 35S-PDLP7-YFP were infiltrated with 1 M NaCl and imaged immediately using confocal microscopy as mentioned above.
Generation of P. syringae Deletion Mutant and Complementation Strains
A ΔhopO1-1 deletion strain was generated in the Pst DC3000 background as previously described (Kvitko and Collmer, 2011) with minor changes. In brief, 1.1- and 1.5-kb genomic DNA fragments flanking hopO1-1 were amplified using the primers listed in Supplemental Data Set 1. The amplified fragments were digested with SalI and ligated with T4 ligase. The ligated 2.6-kb product was gel purified and digested with EcoRI and HindIII and cloned into EcoRI- and HindIII-digested pK18mobsacB using T4 ligase (Thermo Fisher Scientific). The ligated product was then transformed into E. coli RHO5. Both Pst DC3000 and E. coli RHO5 carrying pK18mobsacB plasmid were mixed and spotted on a sterile nitrocellulose filter square on LM medium with 400 μg/mL diaminopimelic acid for conjugation. Transconjugated Pst DC3000 merodiploids were screened with LM medium containing rifampicin and kanamycin. Merodiploids were then selected on LM medium containing rifampicin and 10% (w/v) Suc to counterselect the integration. The Suc-resistant and kanamycin-sensitive colonies were then genotyped by PCR using primers listed in Supplemental Data Set 1 to confirm the deletion.
To complement the UNL137 mutant, wild-type hopO1-1 (pLN1622), catalytic mutant hopO1-1DD (pLN4191), and G2A mutant hopO1-1G2A (pLN5543) were fused with their native promoter (schO1pro) and cloned into a pML123 vector. The constructs were then transformed into the UNL137 mutant.
P. syringae Infection Assays
For dip inoculation, Pst DC3000 and ΔhopO1-1 were grown at 30°C overnight in LM medium (Kvitko and Collmer, 2011). The overnight cultures were then resuspended with water supplemented with 0.02% (v/v) Silwet L-77 to a final concentration of 2 × 108 cfu/mL. The entire rosette of 5-week-old Arabidopsis plants (Col-0) was dipped into the bacterial suspension with gentle swirling for ∼20 s. The dipped plants were then placed under a plastic dome to maintain high humidity (∼80%). Bacterial multiplication was determined 3 d after infection by counting cfu/cm2 leaf disc extracts.
For spray inoculation, Pst DC3000 and derivative strains were grown at 30°C overnight on Suc containing King’s B (King et al., 1954) agar plates and resuspended in 10 mM MgCl2 containing Silwet L-77 (Lehle Seeds) to 5 × 107 cells/mL. The cell suspensions were sprayed onto Arabidopsis plants (Col-0), and the plant leaves were sampled at 0 and 4 d postinoculation. For each treatment, four leaf discs (0.4 cm2) were crushed in 250 μL of sterilized water, and the serial dilutions were plated onto King’s B agar plates containing rifampicin (100 mg/L). The plates were incubated at 30°C for 2 or 3 d until the bacterial colonies appeared. The following strains were used in the pathogenicity assay: Pst DC3000, UNL137, UNL137 (schO1pro-hopO1-1), UNL137 (schO1pro-hopO1-1DD), and UNL137 (schO1pro-hopO1-1G2A).
For the syringe infiltration assay, bacteria were grown as mentioned above for dip inoculation. Different P. syringae strains at 2 × 105 cfu/mL were infiltrated into the leaves of 5-week-old Arabidopsis plants. The infiltrated plants were dried under low humidity (∼20%) for 1 h to let water evaporate and covered with a plastic dome to maintain high humidity (∼80%). Bacterial multiplication was determined 2 d after infection by counting cfu/cm2 leaf disc extracts.
For the leaf-stab assay, the bacteria were grown at 30°C overnight on LM agar plates. Bacteria were picked and inoculated on tomato leaves using a 30G PrecisionGlide needle (BD). The inoculated plants were fully covered with plastic wrap. The distal spreading of bacteria was determined 7 d after infection. During the sampling, the infected sites were removed with a 2-mm biopsy punch. The surrounding tissues were then collected using a 4-mm biopsy punch (nine punches for each sample) and counted as cfu/cm2 leaf disc extracts.
Immunoblot Analyses
Fresh tissues were frozen with liquid nitrogen and homogenized with TissueLyser II (Qiagen). SDS-containing extraction buffer (60 mM Tris-HCl, pH 8.8, 2% [v/v] SDS, 2.5% [v/v] glycerol, 0.13 mM EDTA, pH 8.0, and 1× protease inhibitor cocktail complete from Roche) was added to the homogenized tissues (100 μL/10 mg). The samples were vortexed for 30 s, heated at 70°C for 10 min, and centrifuged at 13,000g for 5 min at room temperature. The supernatants were then transferred to new tubes. For SDS-PAGE analysis, 5 μL of the extract in 1× NuPAGE LDS sample buffer (Life Technologies) was separated on 4–12% NuPage (Life Technologies). The separated proteins were transferred to a polyvinylidene fluoride (PVDF; EMD Millipore) membrane. The membrane was incubated in a blocking buffer (3% [w/v] BSA, 50 mM Tris base, 150 mM NaCl, and 0.05% [v/v] Tween 20, pH 8.0) at room temperature for 1 h. Then it was incubated with an antibody prepared in the blocking buffer at 4°C overnight. The antibodies used are as follows: 1:20,000 α-GFP (Abcam catalog No. ab290), 1:20,000 α-streptavidin-Horseradish Peroxidase (HRP; Abcam catalog No. ab7403), 1:10,000 α-cMyc (Abcam catalog No. ab9106), 1:100 α-ubiquitin (Sigma-Aldrich catalog No. U5379), 1:10,000 α-Flag-HRP (Sigma-Aldrich catalog No. A8592), and 1:1,000 α-pan-ADP-ribose binding reagent (EMD Millipore catalog No. MABE1016). The probed membranes were washed three times with 1× TBST (50 mM Tris base, 150 mM NaCl, and 0.05% [v/v] Tween 20, pH 8.0) for 5 min before being incubated with a secondary antibody at room temperature for 1 h except for α-streptavidin-HRP and α-Flag-HRP. The secondary antibodies used were 1:20,000 goat anti-rabbit IgG (Thermo Fisher Scientific catalog No. 31,460). Finally, the membranes were washed four times with 1× TBST for 10 min before the signals were visualized with SuperSignal West Dura Extended Duration Substrate (Pierce Biotechnology).
Co-IP Assays
PDLP-YFP and HopO1-1-cMyc proteins were transiently expressed in tobacco, and co-IP assays were performed as previously described (Aung and Hu, 2011) with minor modifications. One gram fresh weight of infiltrated leaf was collected 2 d after infiltration. The tissues were ground in 3 mL of RIPA buffer (Thermo Fisher Scientific) with 1× complete protease inhibitor cocktail (Roche) and lysed on a rotator at 4°C for 1 h. The samples were centrifuged at 13,000g for 10 min at 4°C to remove cell debris. Twenty microliters of the supernatants (total proteins) served as the input controls. The remaining supernatants were then incubated with 20 μL of GFP-Trap_A (ChromoTek) on a rotator for 1 h to pull down the YFP fusion proteins. The agarose beads were then spun down at 3000g for 15 s and washed four times with RIPA buffer. Proteins associated with the YFP fusion protein were eluted by adding 50 μL of 1× NuPAGE LDS sample buffer (Life Technologies) and heating at 70°C for 10 min. The eluted proteins were analyzed by immunoblot assay as mentioned above.
Expression and Purification of Recombinant Proteins in E. coli
The plasmids containing HopO1-1 or HopU1 variants were transformed into E. coli Rosetta. The transformants were inoculated in 2 mL of Luria-Bertani (LB) medium with 100 mg/mL ampicillin and 10% (v/v) Glc and incubated in a 37°C shaking incubator. The overnight culture were refreshed in Luria-Bertani medium (1:20 ratio) containing 10% (w/v) Glc at 37°C for another 2 h. Expression of the proteins was induced by adding 300 μM IPTG and incubation for 3 h at 28°C. Soluble recombinant proteins were purified using Ni-NTA resin as recommended by the manufacturer (Qiagen). In short, bacterial pellet from 200 mL of induced culture was pelleted and suspended with 40 mL of Native Binding buffer (50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole, pH 8.0). The suspension was sonicated and centrifuged at 13,000g at 4°C for 10 min to remove cell debris. The supernatant was filtered through a 0.22-μm sterile filter and incubated with 2 mL of Ni-NTA agarose at 4°C for 1 h. The agarose beads were then rinsed with Native Wash buffer (50 mM NaH2PO4, 300 mM NaCl, and 20 mM imidazole, pH 8.0) and eluted with Elution buffer (50 mM NaH2PO4, 300 mM NaCl, and 250 mM imidazole, pH 8.0). The purity and enrichment of the fusion proteins were determined by separating the proteins with SDS-PAGE and staining with SimplyBlue SafeStain (Life Technologies).
mADP-RT Activity Assays
Poly-l-Arg-ADP-RT assay was performed as previously described (Fu et al., 2007). In brief, 2 mM purified proteins (HopO1-1-His-MBP, HopO1-1DD-His-MBP, HopU1-His-MBP, HopU1DD-His-MBP, and His-MBP) were incubated with 0.5 mg of a generic substrate, poly-l-Arg (Sigma-Aldrich; 80 μL of 10 mg/mL in 0.1 M dimethyl glutaric acid buffer, pH 7.0) and 0.25 mM [32P]NAD (Perkin Elmer; radiolabeled on the ADP-ribose moiety) at room temperature for 1 h. Recombinant protein His-MBP was used as a negative control. The reaction was stopped by adding 1 mL of 0.1 M phosphate buffer. The substrate was centrifuged at 3000g at room temperature and rinsed with 1 mL of PBS three times. The substrate was then resuspended with 250 μL of 0.1 M HCl and 500 μL of 0.1 M dimethyl glutaric acid buffer, pH 7.0. Specific incorporated radioactivity was quantified using liquid scintillation (Beckman LS 5000TD).
To test for mADP-RT activity using plant extracts as substrates, 10 mg fresh weight of Arabidopsis or tobacco was isolated using 100 μL of protein extraction buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, and 1 mM DTT). Total proteins were centrifuged at 700g for 10 min at 4°C to remove tissue debris. Ten microliters of the total proteins was mixed with 1.25 mM Biotinylated-NAD+ (Trevigen) and 2 μg of recombinant proteins purified from E. coli. The reaction was incubated at room temperature for 1 h and stopped by adding NuPAGE LDS sample buffer (Life Technologies). To use proteins transiently expressed in tobacco as enzymes, the proteins were pulled down using 20 μL of GFP-Trap_A (ChromoTek) as mentioned above. The beads were then incubated with 1.25 mM Biotinylated-NAD+ (Trevigen) and total proteins extracted from Arabidopsis or tobacco. The ribosylated proteins were detected using α-streptavidin-HRP as mentioned above.
Detection of ADP-Ribosylated Proteins in Planta
To detect ADP-ribosylated proteins in planta, YFP fusion proteins were enriched from Arabidopsis transgenic plants expressing 35S-PDLP5-YFP, 35S-PDLP6-YFP, or 35S-PDLP7-YFP in the wild-type Col-0 or 35S-HopO1-1 background using 20 μL of GFP-Trap_A (ChromoTek) as mentioned above. Total proteins of wild-type Col-0 and DEX-HopU1 (4 h after 30 μM dexamethasone [DEX; Sigma-Aldrich] treatment) were isolated as described above and served as negative and positive controls, respectively. Ribosylated proteins were detected using an anti-pan-ADP-ribose binding reagent.
RNA Extraction and RT-PCR Analyses
Total RNA from leaves of 2- or 5-week-old Arabidopsis plants was purified as previously described by Chen et al. (2014). Total RNA (0.32 μg) was used to make cDNA using SuperScript VILO Master Mix (Life Technologies). For RT-PCR, gene-specific primers for hopO1-1, UBQ10, PDLP5, PDLP6, and PDLP7 were used to amplify the target genes (Supplemental Data Set 1). UBQ10 was used as an internal control and amplified for 25 cycles. The rest of the genes were amplified for 35 cycles. PCR products were separated on a 1% agarose gel.
Experimental Repeats and Data Analyses
At least three independent experimental repeats were performed for all experiments. The statistical method and sample size for each experiment are listed in the relevant figures and figure legends.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this articles are as follows: PDLP1 (At5g43980), PDLP2 (At1g04520), PDLP3 (At2g33330), PDLP4 (At3g04370), PDLP5 (At1g70690), PDLP6 (At2g01660), PDLP7 (At5g37660), and UBQ10 (At4g05320). Germplasm identification numbers mentioned in this work are as follows: pdlp4 (SALK_028613), pdlp5 (SAIL_46_E06.v1), and pdlp7 (SALK_015341).
Supplemental Data
Supplemental Figure 1. Expression of HopO1-1 variants in Arabidopsis.
Supplemental Figure 2. Subcellular localization of HopO1-1 and PDLP7 in Arabidopsis.
Supplemental Figure 3. HopO1-1 promotes PD permeability in Arabidopsis.
Supplemental Figure 4. HopO1-1 affects PDLP protein stability in Arabidopsis.
Supplemental Figure 5. Expression of PDLP transcripts in Arabidopsis.
Supplemental Figure 6. HopO1-1 does not ribosylate plant proteins.
Supplemental Figure 7. Characterization of pdlp mutants.
Supplemental Data Set 1. Primers used in this study.
Supplemental Data Set 2. Vectors used in this study.
Supplemental Data Set 3. Summary of statistical tests.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the ABRC for providing the T-DNA insertion mutants, Honggao Yan (Michigan State University) for sharing the pET17b HMR plasmid, and Christine Faulkner (John Innes Centre) for sharing the pdlp1/2/3 mutant. We also thank Terra Livingston, Katie Walicki, and Deliana May (Michigan State University) for providing technical support. This work was supported by the National Institute of General Medical Sciences (Grant 4R00GM115766-02 to K.A. and Grant GM109928 to S.Y.H.), the Gordon and Betty Moore Foundation (Grant GBMF3037 to S.Y.H.), and the National Science Foundation Division of Integrative Organismal Systems (Grant 1508504 to J.R.A.). During the resubmission of this manuscript, we lost our friend and collaborator, James Robert Alfano, a senior coauthor of this article. Dr. Alfano made many important contributions to the fields of plant pathology and bacterial effector biology. His legacy will live on.
AUTHOR CONTRIBUTIONS
K.A. and S.Y.H. designed the research; K.A. performed most experiments; P.K., A.J., and J.R.A. designed and performed the disease assay shown in Figure 3B; B.K. helped generate the ΔhopO1-1 mutant; Z.P.L. performed the immunoblot analyses shown in Figure 6D; K.A. and S.Y.H. analyzed data and wrote the article with input from all authors.
Footnotes
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Amari K., et al. (2010). A family of plasmodesmal proteins with receptor-like properties for plant viral movement proteins. PLoS Pathog. 6: e1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Hu J. (2011). The Arabidopsis tail-anchored protein PEROXISOMAL AND MITOCHONDRIAL DIVISION FACTOR1 is involved in the morphogenesis and proliferation of peroxisomes and mitochondria. Plant Cell 23: 4446–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K., Xin X., Mecey C., He S.Y. (2017). Subcellular localization of Pseudomonas syringae pv. tomato effector proteins in plants. Methods Mol. Biol. 1531: 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y., Faulkner C., Ritzenthaler C., Maule A.J. (2010). Plasmodesmata: Gateways to local and systemic virus infection. Mol. Plant Microbe Interact. 23: 1403–1412. [DOI] [PubMed] [Google Scholar]
- Block A., Guo M., Li G., Elowsky C., Clemente T.E., Alfano J.R. (2010). The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell. Microbiol. 12: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A., Toruño T.Y., Elowsky C.G., Zhang C., Steinbrenner J., Beynon J., Alfano J.R. (2014). The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 201: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Caillaud M.C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S.J., Jones A.M., Robatzek S., Jones J.D., Faulkner C. (2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Blekemolen M.C., Tintor N., Cornelissen B.J.C., Takken F.L.W. (2018). The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol. Plant 11: 691–705. [DOI] [PubMed] [Google Scholar]
- Chen Y., Aung K., Rolčík J., Walicki K., Friml J., Brandizzi F. (2014). Inter-regulation of the unfolded protein response and auxin signaling. Plant J. 77: 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheval C., Faulkner C. (2018). Plasmodesmal regulation during plant-pathogen interactions. New Phytol. 217: 62–67. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. (2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66: 487–511. [DOI] [PubMed] [Google Scholar]
- Cui W., Lee J.Y. (2016). Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants 2: 16034. [DOI] [PubMed] [Google Scholar]
- De Storme N., Geelen D. (2014). Callose homeostasis at plasmodesmata: Molecular regulators and developmental relevance. Front. Plant Sci. 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Faulkner C., Petutschnig E., Benitez-Alfonso Y., Beck M., Robatzek S., Lipka V., Maule A.J. (2013). LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc. Natl. Acad. Sci. USA 110: 9166–9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Guo M., Jeong B.R., Tian F., Elthon T.E., Cerny R.L., Staiger D., Alfano J.R. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288. [DOI] [PubMed] [Google Scholar]
- Ganusova E.E., Burch-Smith T.M. (2019). Review: Plant-pathogen interactions through the plasmodesma prism. Plant Sci. 279: 70–80. [DOI] [PubMed] [Google Scholar]
- Giska F., Lichocka M., Piechocki M., Dadlez M., Schmelzer E., Hennig J., Krzymowska M. (2013). Phosphorylation of HopQ1, a type III effector from Pseudomonas syringae, creates a binding site for host 14-3-3 proteins. Plant Physiol. 161: 2049–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin T.E., Assmann S.M. (2014). Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J. 80: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.R., Fisher E.J., Chang J.H., Mole B.M., Dangl J.L. (2006). Subterfuge and manipulation: Type III effector proteins of phytopathogenic bacteria. Annu. Rev. Microbiol. 60: 425–449. [DOI] [PubMed] [Google Scholar]
- Guo M., Chancey S.T., Tian F., Ge Z., Jamir Y., Alfano J.R. (2005). Pseudomonas syringae type III chaperones ShcO1, ShcS1, and ShcS2 facilitate translocation of their cognate effectors and can substitute for each other in the secretion of HopO1-1. J. Bacteriol. 187: 4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guseman J.M., Lee J.S., Bogenschutz N.L., Peterson K.M., Virata R.E., Xie B., Kanaoka M.M., Hong Z., Torii K.U. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137: 1731–1741. [DOI] [PubMed] [Google Scholar]
- Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. (2006). Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70: 789–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M. (2015). Plasmodesmata: Channels for viruses on the move. Methods Mol. Biol. 1217: 25–52. [DOI] [PubMed] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B.A., Wright C.M., Brodsky J.L., Greenberg J.T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E.O., Ward M.K., Raney D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44: 301–307. [PubMed] [Google Scholar]
- Klessig D.F., Choi H.W., Dempsey D.A. (2018). Systemic acquired resistance and salicylic acid: Past, present, and future. Mol. Plant Microbe Interact. 31: 871–888. [DOI] [PubMed] [Google Scholar]
- Kumar D., Kumar R., Hyun T.K., Kim J.Y. (2015). Cell-to-cell movement of viruses via plasmodesmata. J. Plant Res. 128: 37–47. [DOI] [PubMed] [Google Scholar]
- Kvitko B.H., Collmer A. (2011). Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Methods Mol. Biol. 712: 109–128. [DOI] [PubMed] [Google Scholar]
- Lee J.Y. (2014). New and old roles of plasmodesmata in immunity and parallels to tunneling nanotubes. Plant Sci. 221–222: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y. (2015). Plasmodesmata: A signaling hub at the cellular boundary. Curr. Opin. Plant Biol. 27: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., Zybaliov B., van Wijk K., Zhang C., Lu H., Lakshmanan V. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23: 3353–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Fevre R., Evangelisti E., Rey T., Schornack S. (2015). Modulation of host cell biology by plant pathogenic microbes. Annu. Rev. Cell Dev. Biol. 31: 201–229. [DOI] [PubMed] [Google Scholar]
- Li G., Froehlich J.E., Elowsky C., Msanne J., Ostosh A.C., Zhang C., Awada T., Alfano J.R. (2014). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77: 310–321. [DOI] [PubMed] [Google Scholar]
- Lim G.H., Shine M.B., de Lorenzo L., Yu K., Cui W., Navarre D., Hunt A.G., Lee J.Y., Kachroo A., Kachroo P. (2016). Plasmodesmata localizing proteins regulate transport and signaling during systemic acquired immunity in plants. Cell Host Microbe 19: 541–549. [DOI] [PubMed] [Google Scholar]
- Liu L., Chen X. (2018). Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas W.J., Ham B.K., Kim J.Y. (2009). Plasmodesmata: Bridging the gap between neighboring plant cells. Trends Cell Biol. 19: 495–503. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Lee J.Y. (2004). Plasmodesmata as a supracellular control network in plants. Nat. Rev. Mol. Cell Biol. 5: 712–726. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nicaise V., Roux M., Zipfel C. (2009). Recent advances in PAMP-triggered immunity against bacteria: Pattern recognition receptors watch over and raise the alarm. Plant Physiol. 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas W.J., Grison M.S., Trépout S., Gaston A., Fouché M., Cordelières F.P., Oparka K., Tilsner J., Brocard L., Bayer E.M. (2017). Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants 3: 17082. [DOI] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Sheedlo M.J., Yu K., Tan Y., Nakayasu E.S., Das C., Liu X., Luo Z.Q. (2016). Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533: 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S. (2017). Sensing of molecular patterns through cell surface immune receptors. Curr. Opin. Plant Biol. 38: 68–77. [DOI] [PubMed] [Google Scholar]
- Redditt T.J., Chung E.H., Zand Karimi H., Rodibaugh N., Zhang Y., Trinidad J.C., Kim J.H., Zhou Q., Shen M., Dangl J.L., Mackey D.M., Innes R.W. (2019). AvrRpm1 functions as an ADP-ribosyl transferase to modify NOI-domain containing proteins, including Arabidopsis and soybean RPM1-interaction protein 4. Plant Cell 31: 2664–2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Shan L., Zhou J.M., Tang X. (2006). The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant Microbe Interact. 19: 130–138. [DOI] [PubMed] [Google Scholar]
- Saijo Y., Loo E.P., Yasuda S. (2018). Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 93: 592–613. [DOI] [PubMed] [Google Scholar]
- Sakulkoo W., Osés-Ruiz M., Oliveira Garcia E., Soanes D.M., Littlejohn G.R., Hacker C., Correia A., Valent B., Talbot N.J. (2018). A single fungal MAP kinase controls plant cell-to-cell invasion by the rice blast fungus. Science 359: 1399–1403. [DOI] [PubMed] [Google Scholar]
- Shan L., He P., Zhou J.M., Tang X. (2000). A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant Microbe Interact. 13: 592–598. [DOI] [PubMed] [Google Scholar]
- Speth E.B., Imboden L., Hauck P., He S.Y., et al. (2009). Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiology 149: 1824–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. (2013). Gated communities: Apoplastic and symplastic signals converge at plasmodesmata to control cell fates. J. Exp. Bot. 64: 5237–5241. [DOI] [PubMed] [Google Scholar]
- Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño T.Y., Stergiopoulos I., Coaker G. (2016). Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54: 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H.L., Chakravarthy S., Mathieu J., Helmann T.C., Stodghill P., Swingle B., Martin G.B., Collmer A. (2015). Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe 17: 752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen M.C., Innes R.W., Bent A.F., Staskawicz B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.F., He S.Y. (2013). Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 51: 473–498. [DOI] [PubMed] [Google Scholar]
- Xin X.F., Kvitko B., He S.Y. (2018). Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 16: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.F., Nomura K., Ding X., Chen X., Wang K., Aung K., Uribe F., Rosa B., Yao J., Chen J., He S.Y. (2015). Pseudomonas syringae effector avirulence protein E localizes to the host plasma membrane and down-regulates the expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 gene required for antibacterial immunity in Arabidopsis. Plant Physiol. 169: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., et al. (2019). Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat. Plants 5: 604–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Lazarowitz S.G., Citovsky V. (2016). Identification of a functional plasmodesmal localization signal in a plant viral cell-to-cell-movement protein. MBio 7: e02052-15. [DOI] [PMC free article] [PubMed] [Google Scholar]