Limited information is available on the transmission of Campylobacter jejuni subtypes in the beef production continuum and the foodborne risk posed to humans. Cattle were colonized by diverse subtypes of C. jejuni, and the densities of the bacterium shed in feces increased during the confined feeding period. Campylobacter jejuni was readily associated with the digesta, feces, and hides of cattle entering the abattoir, as well as the local environment. Moreover, C. jejuni cells were deposited on carcasses via direct contact and aerosols, but the bacterium was not detected in the ground beef generated from contaminated carcasses. We conclude that C. jejuni bacterial cells associated with beef cattle do not represent a significant risk through food consumption and suggest that clinically relevant subtypes are transmitted through alternate routes of exposure.
KEYWORDS: beef cattle, antimicrobial growth promoter, Campylobacter jejuni, longitudinal transmission, health risk
ABSTRACT
Increasing evidence exists for the role that cattle play in the epidemiology of campylobacteriosis. In this study, the prevalence and distribution of Campylobacter jejuni were longitudinally examined at the subspecies level in the beef cattle production continuum. Animals were subdivided into two groups: those that were not administered antibiotics and those that were administered the antimicrobial growth promoter chlortetracycline and sulfamethazine (AS700). Samples were longitudinally collected throughout the confined feeding operation (CFO) period and during the slaughter process, and C. jejuni was isolated and genotyped to assess subtype richness and to elucidate transmission dynamics from farm to fork. The bacterium was frequently isolated from cattle, and the bacterial densities shed in feces increased over the CFO period. Campylobacter jejuni was also isolated from digesta, hides, the abattoir environment, and carcasses. The administration of AS700 did not conspicuously reduce the C. jejuni densities in feces or within the intestine but significantly reduced the bacterial densities and the diversity of subtypes on abattoir samples. All cattle carried multiple subtypes, including clinically relevant subtypes known to represent a risk to human health. Instances of intra-animal longitudinal transmission were observed. Although clinically relevant subtypes were transmitted to carcasses via direct contact and aerosols, the bacterium could not be isolated nor could its DNA be detected in ground beef regardless of treatment. Although the evidence indicated that beef cattle represent a significant reservoir for C. jejuni, including high-risk subtypes strongly associated with the bovine host, they do not appear to represent a significant risk for direct foodborne transmission. This implicates alternate routes of human transmission.
IMPORTANCE Limited information is available on the transmission of Campylobacter jejuni subtypes in the beef production continuum and the foodborne risk posed to humans. Cattle were colonized by diverse subtypes of C. jejuni, and the densities of the bacterium shed in feces increased during the confined feeding period. Campylobacter jejuni was readily associated with the digesta, feces, and hides of cattle entering the abattoir, as well as the local environment. Moreover, C. jejuni cells were deposited on carcasses via direct contact and aerosols, but the bacterium was not detected in the ground beef generated from contaminated carcasses. We conclude that C. jejuni bacterial cells associated with beef cattle do not represent a significant risk through food consumption and suggest that clinically relevant subtypes are transmitted through alternate routes of exposure.
INTRODUCTION
Campylobacter jejuni commonly colonizes the intestinal tract of cattle and is frequently shed in large numbers in feces (1–6). Southwestern Alberta (SWA), Canada, possesses a high density of cattle, predominately beef cattle (≈1,166,000). As in the rest of North America, the majority of beef cattle in SWA (51%) are finished in confined feeding operations (CFOs) (7) and, until recently, were frequently administered antimicrobial agents in feeds as antimicrobial growth promoters (AGPs). Although their mechanisms of action are poorly understood at present (8), AGPs are administered at relatively low concentrations in the feed for prolonged periods to promote growth (9). It is currently unknown whether AGP administration has an effect on the C. jejuni populations in the cattle production continuum, including rates of carriage and subtype prevalence.
The rates of campylobacteriosis, caused primarily by C. jejuni (59.7 cases 100,000−1) in SWA, are substantially higher than the provincial (28.5 cases 100,000−1) and national (30.0 cases 100,000−1) averages (10–12). Although the epidemiology of campylobacteriosis in SWA is currently unresolved, some evidence suggests that direct contact with cattle (i.e., occupational risk) contributes to the burden of campylobacteriosis (13). A recent clinical study of campylobacteriosis conducted over a 1-year period in SWA indicated significant underreporting of clinical cases, with a majority of infections remaining undetected when using conventional microbiological culture methods (14). Moreover, this study highlighted that temporal clusters comprise a significant proportion of cases of campylobacteriosis and that a majority of infections (70.3%) are linked to subtypes associated with beef cattle. Our research and that of others have shown that only a subset of C. jejuni subtypes are associated with human infections, with a majority of subtypes being rarely, if ever, implicated in cases of campylobacteriosis. This emphasizes the necessity of examining the epidemiology of the bacterium using high-resolution subtyping data to more accurately ascertain human health risks that can be attributed to clinically relevant subtypes.
A number of studies have conducted snapshot examinations of C. jejuni associated with carcasses (15–18) and retail beef (19–22), but to date, few have longitudinally examined the transmission of C. jejuni throughout the beef production continuum or whether the consumption of beef contaminated with C. jejuni contributes to the burden of disease. We hypothesized (i) that the C. jejuni densities shed in beef cattle feces increase during the CFO period regardless of AGP administration, (ii) that C. jejuni subtypes are longitudinally transmitted to carcasses and ground beef, and (iii) that a subset of the C. jejuni subtypes associated with cattle represent an infection risk to humans. To test these hypotheses, the study objectives were to (i) conduct a replicated study using an experimental CFO in which beef cattle were administered no antibiotics or were administered the AGP chlortetracycline and sulfamethazine (AS700), (ii) temporally collect feces from animals throughout the CFO period and quantify the C. jejuni DNA within feces, (iii) longitudinally sample cattle throughout the slaughter process and isolate C. jejuni, (iv) perform high-resolution genotyping of C. jejuni isolates to ascertain the transmission dynamics of subtypes from farm to fork, and (v) determine the human health risk posed by the C. jejuni subtypes associated with beef cattle via food exposure.
RESULTS
Beef cattle regularly shed Campylobacter jejuni at high densities in their feces.
Cattle frequently shed C. jejuni in their feces throughout the CFO period (Fig. 1). The density of C. jejuni shed in feces increased over time (P < 0.001), but there was no difference in the density of the bacterium shed in feces between the cattle receiving the control treatment and the cattle receiving the AS700 treatment (P = 0.378).
FIG 1.
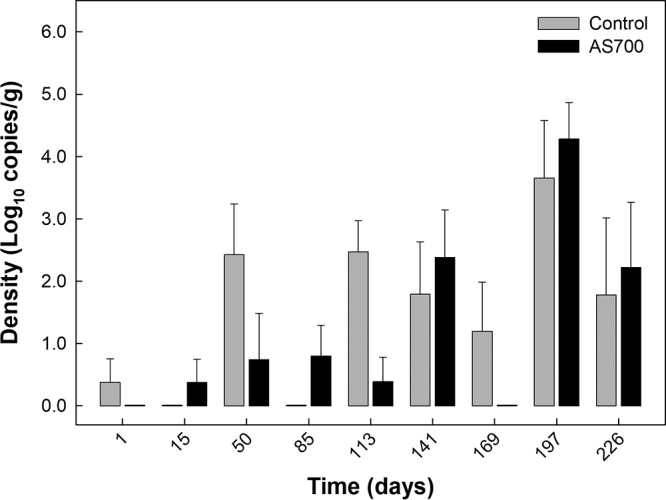
Densities of Campylobacter jejuni in feces (log10 number of copies per gram) from beef cattle administered chlortetracycline and sulfamethazine (AS700) or no antibiotics (control) in feed and housed in a beef cattle confined feeding operation for ca. 7.5 months. The AS700 treatment was withdrawn 28 days prior to slaughter (i.e., day 197). Vertical lines with histogram bars represent standard errors of the means (n = 5). There was no effect of AS700 administration (P = 0.378), and the densities of C. jejuni shed in the feces of the two groups increased equally over time (P < 0.001).
Campylobacter jejuni was detected throughout the intestinal tract of beef cattle.
The DNA of C. jejuni was frequently detected in association with the mucosa in the distal small and large intestines of beef cattle (Fig. 2). In some locations, the densities of C. jejuni cells were reduced (P ≤ 0.046) in animals administered AS700. The administration of AS700 did not affect the densities (P = 0.475) of C. jejuni in the digesta (see Fig. S1 in the supplemental material).
FIG 2.
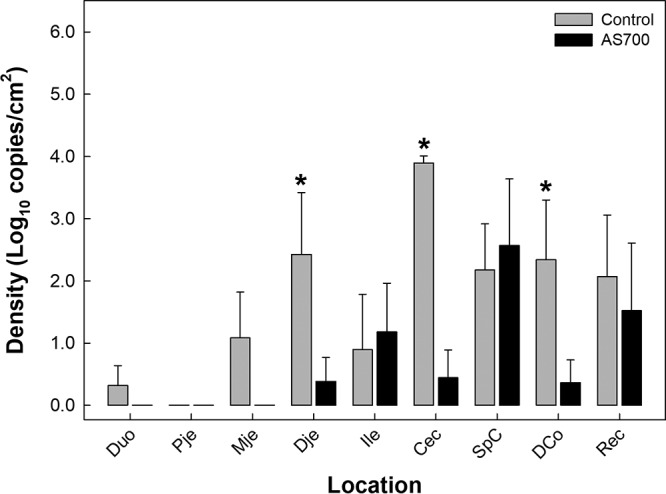
Densities of Campylobacter jejuni cells associated with intestinal mucosa in the duodenum (Duo), proximal jejunum (Pje), midjejunum (Mje), distal jejunum (Dje), ileum (Ile), cecum (Cec), spiral colon (SpC), descending colon (DCo), and rectum (Rec) of cattle administered chlortetracycline and sulfamethazine (AS700) or no antibiotics (control) during their time in a confined feeding operation. Vertical lines with histogram bars represent standard errors of the means (n = 5). Histogram bars with an asterisk indicate that the results for the controls differed (P < 0.050) from those for the AS700-treated animals at the corresponding location.
Campylobacter jejuni was commonly detected on the hides of cattle and transferred to carcasses.
Campylobacter jejuni isolates were recovered from the hides of cattle after euthanization and subsequently from the surfaces of the carcasses after hide removal (Fig. 3). DNA from C. jejuni cells with an intact membrane was detected from carcasses (brisket and rump) immediately after euthanization and after storage at 4°C for 24 h (Fig. 4A and B); the storage period had no effect (P ≥ 0.618) on C. jejuni DNA quantity. However, the amount of C. jejuni DNA on the carcasses was reduced (P ≤ 0.032) for animals administered AS700.
FIG 3.
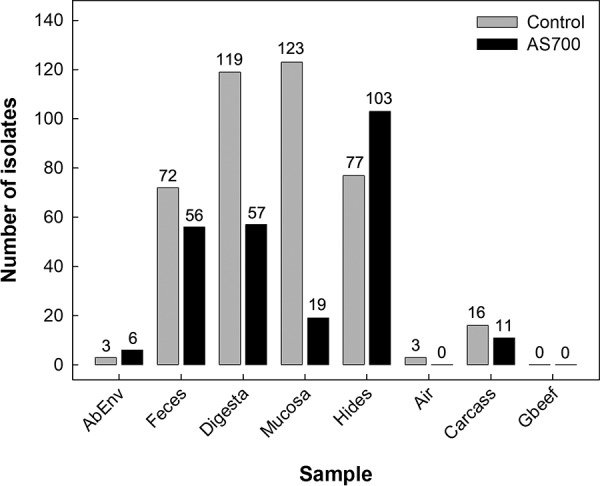
Total number of Campylobacter jejuni isolates recovered from the abattoir environment (AbEnv), feces, intestine (digesta and mucosa), hides, air, carcasses, and ground beef (Gbeef) from/of cattle administered chlortetracycline and sulfamethazine (AS700) or no antibiotics (control) in a beef cattle confined feeding operation.
FIG 4.
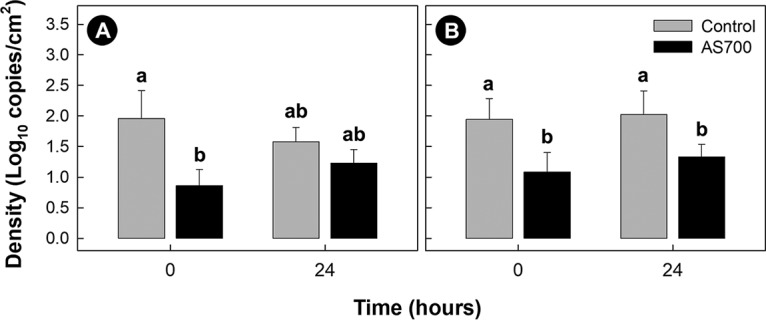
Densities of Campylobacter jejuni on the surfaces of carcasses after hide removal (time zero) and after 24 h in the chiller (time 24). Carcasses originated from cattle administered chlortetracycline and sulfamethazine (AS700) or no antibiotics (control) during their time in a confined feeding operation. (A) Brisket; (B) rump. Vertical lines with histogram bars represent standard errors of the means (n = 5), and the values for histogram bars not denoted with the same letter differ significantly (P < 0.050).
Multiple Campylobacter jejuni subtypes were recovered from the intestines of individual beef cattle.
A considerable diversity of C. jejuni subtypes was associated with cattle (Table 1; Fig. 5). Multiple comparative genomic fingerprinting (CGF) (in this case, CGF40, a genotyping method that targets 40 accessory genes located throughout the genome of C. jejuni) subtypes of C. jejuni were associated with individual steers (i.e., in the feces or digesta or in association with the mucosa) for most animals (Fig. S2). The number of C. jejuni subtypes recovered from individual animals ranged from 1 to 21 (i.e., in steer 476). The subtypes of C. jejuni associated with individual animals within and between pens differed (Fig. 5; Fig. S3).
TABLE 1.
CGF subtype clusters of Campylobacter jejuni isolates recovered throughout the beef cattle continuum from animals that were administered AS700 or no antibiotics (control) during the CFO period
| Steera | Treatment | Pen no. | CGF subtype cluster(s)b recovered from: |
|||
|---|---|---|---|---|---|---|
| Feces, digesta, mucosa | Hide | Air | Carcass | |||
| 577 | Control | 12 | 3, 5, 47, 65, 88, 89, 90, 94, 98, 101, 104, 105, 114 | 12, 18, 85, 96, 98 | ||
| 480 | Control | 2 | 114 | 18, 96, 108 | ||
| 573 | Control | 11 | 27 | 61, 87, 88 | ||
| 582 | Control | 12 | 88, 91, 109 | 9, 18, 68, 85 | ||
| 481 | Control | 2 | 58, 69, 77 | 4, 17, 26, 65, 69, 82 | 82, 97 | |
| 566 | Control | 11 | 76, 98 | 27, 69, 107 | 19, 60 | |
| 470 | Control | 1 | 1, 5, 17, 20, 27, 28, 30, 44, 46, 48, 49, 76, 88, 98, 103, 114, 118, 120, 121 | 18 | ||
| 578 | Control | 12 | 52, 88 | 15, 18, 68, 85 | ||
| 698 | Control | 24 | 56 | 56, 71, 85, 88 | ||
| 697 | Control | 24 | 14, 17, 18, 40, 54, 69, 70, 73, 78, 88, 98, 99, 100, 113, 114 | 88 | ||
| 472 | Control | 1 | 41, 49, 69, 88, 98 | 6, 56, 88 | 96 | |
| 703 | Control | 24 | 18, 69 | 56, 64 | ||
| 474 | Control | 1 | 43, 69, 88, 98 | 16, 18, 66, 67, 80 | 87 | |
| 572 | Control | 11 | 32, 69, 114 | 22, 65, 114 | 17 | 6, 65, 88 |
| 476 | Control | 2 | 2, 13, 17, 18, 35, 42, 69, 81, 98, 114, 115, 116, 117, 119, 122, 123, 124, 125, 126, 127, 128 | 62, 63 | ||
| 504 | AS700 | 4 | 18, 19, 21, 36, 37, 38, 39, 53, 59 | 53 | 82, 97 | |
| 499 | AS700 | 4 | 53, 58, 88, 106 | 53 | ||
| 643 | AS700 | 18 | 23, 24, 33 | 15, 17, 23, 53, 58, 82, 85, 110 | ||
| 632 | AS700 | 17 | 18, 22, 31, 69, 79 | 82 | ||
| 627 | AS700 | 17 | 88, 102 | 82 | ||
| 633 | AS700 | 17 | 3, 51, 82, 88, 91, 93 | 53, 82 | ||
| 490 | AS700 | 3 | 7, 8, 10, 11, 45, 50, 82, 84, 88, 92, 93, 110 | 53, 57, 82, 85, 110 | ||
| 492 | AS700 | 3 | 82, 88 | |||
| 669 | AS700 | 21 | 88, 94 | 53, 82, 86 | 15 | |
| 640 | AS700 | 18 | 17, 18, 55, 69, 72, 74, 75, 76, 86 | 21, 69, 82 | ||
| 644 | AS700 | 18 | 69, 112 | 17, 69, 82, 83 | 88 | |
| 666 | AS700 | 21 | 69, 82, 88 | |||
| 488 | AS700 | 3 | 18, 82 | 15, 25, 88 | ||
| 502 | AS700 | 4 | 82, 111 | |||
| 674 | AS700 | 21 | 29 | 82, 88, 95 | ||
Animals were processed in the order presented. Control animals were processed on day 1, and AS700 animals were processed on day 2.
CGF subtype clusters were determined at a 95% level of resolution. Digesta and mucosal samples were obtained from one animal per pen (i.e., steers 470, 476, 490, 504, 566, 577, 632, 640, 674, and 697) and processed. CGF subtype clusters 3, 69, and 77 were obtained on day 1 from the abattoir environment for control animals, and CGF subtype cluster 124 was obtained on day 2 from the abattoir environment for AS700 animals.
FIG 5.
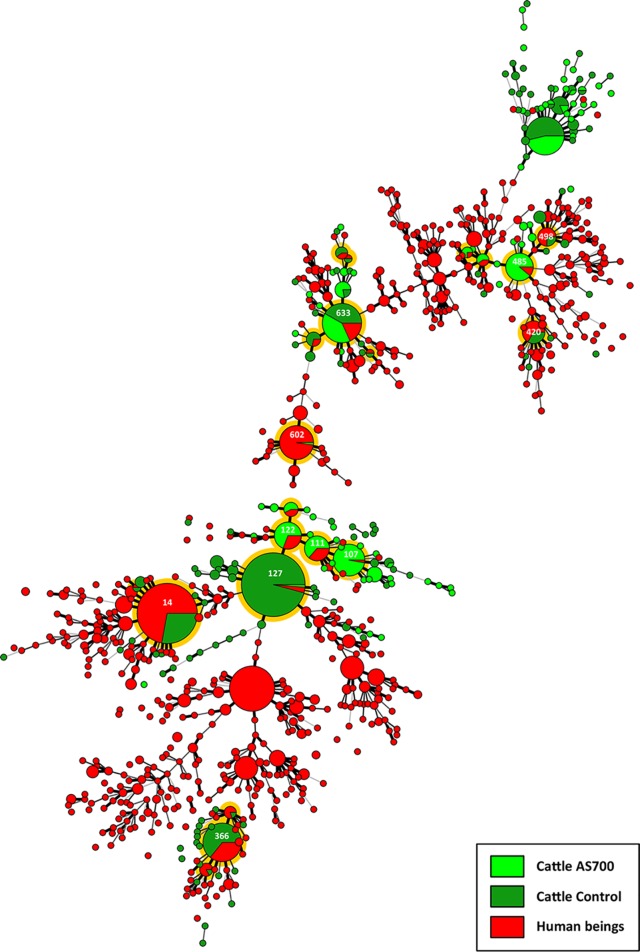
Campylobacter jejuni comparative genomic fingerprinting (CGF) subtype clusters at a 95% level of resolution (two mismatches or less) (n = 127) recovered from individual beef cattle throughout the production continuum. Cattle were administered chlortetracycline and sulfamethazine (i.e., AS700; light green) or no antibiotics (i.e., control; dark green). Intestinal digesta and mucosa were obtained from the steers indicated with an asterisk and processed. Black lines are subtypes recovered from feces or digesta/mucosa (sample type A), orange lines are subtypes recovered from hides (sample type B), green lines are subtypes recovered from carcasses (sample type C), and red lines are subtypes common to two or more sample types for individual animals. The red arrow points to C. jejuni subtype cluster 82, which was associated with the hides of a majority of the animals administered AS700. Black arrows point to C. jejuni subtype clusters 18, 69, and 88 (top to bottom, respectively).
The richness of Campylobacter jejuni subtypes isolated from cattle administered AS700 was reduced.
One hundred twenty-six C. jejuni subtypes were recovered from cattle, and the number of C. jejuni subtypes recovered from animals administered AS700 (n = 38) was less (P < 0.001) than the number recovered from control animals (n = 76) (Fig. 6).
FIG 6.
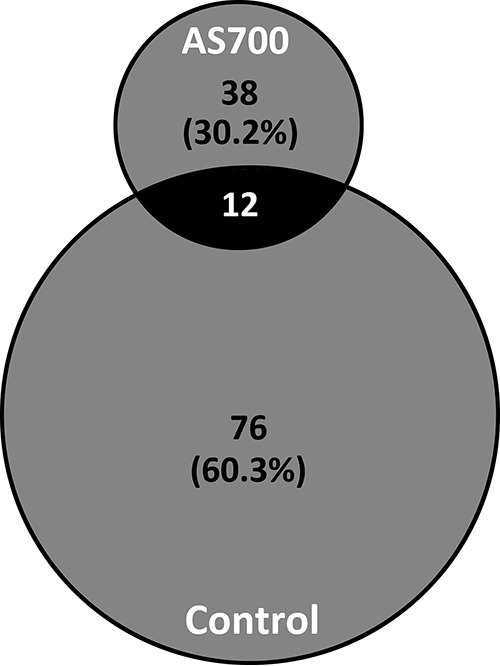
Two-way Venn diagram of Campylobacter jejuni comparative genomic fingerprinting (CGF) subtype clusters recovered from beef cattle throughout the production continuum (95% level of resolution). Subtypes were isolated from animals administered chlortetracycline and sulfamethazine (AS700) or no antibiotics (control). The total number of C. jejuni isolates examined and CGF subtypes obtained (95% level of resolution) were 664 and 126, respectively.
Campylobacter jejuni subtypes were longitudinally isolated from steers and carcasses but not from ground beef.
Multiple isolates were longitudinally recovered from beef cattle feces, intestines, hides, and carcasses but not from ground beef prepared from the briskets and rumps (Fig. 3). In addition, C. jejuni was recovered from the abattoir environment at the beginning of the day and from the air adjacent to the animals during the hide removal process. An examination of the C. jejuni subtypes revealed that the subtypes associated with an individual animal were also isolated from the surfaces of the hide and the carcass of the same animal (Table 1; Fig. 5; Fig. S3). Campylobacter jejuni subtype cluster 82 was recovered from the majority of beef cattle administered AS700 but was rarely recovered from the control animals (Table 2; Fig. 5). Notably, the subtypes within this cluster were primarily associated with beef cattle in Alberta and not with diarrheic humans or other livestock. In addition, subtype clusters 18, 69, and 88 were recovered from multiple animals and sample types and were primarily associated with cattle in Alberta. In the majority of instances the C. jejuni subtypes recovered from the hides and carcasses were not observed in the intestines of the same animal. Furthermore, the C. jejuni subtypes isolated from air adjacent to the animals during hide removal (subtype cluster 17) were not associated with that animal; rather, these subtypes were associated with other animals that were processed earlier in the day (i.e., within ca. ≥1.5 h). Moreover, the C. jejuni subtypes recovered from the abattoir environment before commencement of the processing of the animals were also observed in the feces and intestinal contents and on the hides of the animals, suggesting that the C. jejuni bacteria originating from the animals processed in the current study contaminated the abattoir environment.
TABLE 2.
Select CGF subtype clusters of Campylobacter jejuni longitudinally recovered from beef cattle in the production continuum with the corresponding subtype data within the C3GFdba
| Cluster identifier | C3GFdb identifier | Closest CGF identifier | Similarity | Rank | Size | % of isolates of the same cluster from the following: |
Primary source | % association with AB cattle | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Hu | Ca | Ch | ||||||||
| 18 | 0417.022.002 | 1.000 | 751 | 3 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 69 | Novel | 0735.005.001 | 0.975 | 12 | 227 | 18.5 | 52.4 | 14.1 | Bovine | 71.8 |
| 69 | Novel | 0735.004.002 | 0.975 | 3,121 | 1 | 0.0 | 0.0 | 0.0 | Environment | 100.0 |
| 82 | 0238.013.004 | 1.000 | 312 | 8 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 82 | 0238.015.001 | 1.000 | 3,121 | 1 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 82 | 0238.015.002 | 1.000 | 245 | 11 | 27.3 | 72.7 | 0.0 | Bovine | 90.9 | |
| 82 | 0238.016.002 | 1.000 | 95 | 38 | 2.6 | 92.1 | 2.6 | Bovine | 97.4 | |
| 82 | 0238.016.003 | 1.000 | 3,121 | 1 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 82 | 0238.016.004 | 1.000 | 751 | 3 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 88 | Novel | 0238.014.002 | 0.975 | 18 | 193 | 3.1 | 89.6 | 0.0 | Bovine | 96.9 |
| 88 | Novel | 0238.002.002 | 0.950 | 22 | 152 | 11.8 | 82.2 | 0.0 | Bovine | 84.9 |
Hu, humans; Ca, cattle; Ch, chickens; AB, Alberta.
A multitude of C. jejuni subtypes recovered from beef cattle represent a high health risk to people.
Twenty-two CGF clusters contained C. jejuni isolates that were recovered from beef cattle as well as from diarrheic humans (i.e., clinically relevant subtypes) within the study region of SWA (Fig. 7). Eleven of these clusters contained more than 10 isolates. Only one cluster (i.e., cluster 14) contained an identical match to isolates within the Canadian Campylobacter Comparative Genomic Fingerprinting database (C3GFdb; subtype 0169.001.002); this subtype is among the more prevalent within the C3GFdb and prominent in Alberta and contains isolates that were recovered from humans (30.8%), cattle (47.6%), and chickens (13.9%) (Table 3). The other 10 clusters contained subtypes that were novel to the C3GFdb but that were closely related to subtypes within the database and that were predominately associated with cattle as well as humans.
FIG 7.
Campylobacter jejuni comparative genomic fingerprinting (CGF) subtypes from beef cattle administered chlortetracycline and sulfamethazine (AS700; light green) or no antibiotics (control; dark green) and from diarrheic humans (red) within the study area (2004 to 2017). The minimum spanning tree was generated in BioNumerics software (version 6.6; Applied Maths). The size of the circle is proportional to the number of isolates within each CGF subtype (100% level of resolution), the thickness of the lines connecting the subtypes represents mismatched loci (i.e., one to three loci), and subtypes with no line represent four or more mismatched loci between the respective subtypes. Orange shading illustrates subtypes obtained from both cattle and diarrheic people, and clusters with more than 10 isolates are marked with a subtype cluster number (see Table 3 for specifics). The total numbers of isolates were 672 from cattle (238 from AS700 cattle, and 434 from control cattle) and 1,178 from diarrheic humans.
TABLE 3.
CGF subtype clusters of Campylobacter jejuni predominantly associated with beef cattle in the production continuum with the corresponding subtype data within the C3GFdba
| Cluster identifier | Size | C3GFdb identifier | Closest CGF identifier | Similarity | Rank | Size | % of isolates of the same cluster from the following: |
Primary host | % association with AB cattle | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hu | Ca | Ch | |||||||||
| 14 | 129 | 0169.001.002 | 1.000 | 1 | 775 | 30.8 | 47.6 | 13.9 | Bovine | 76.5 | |
| 107 | 36 | Novel | 0238.007.004 | 0.950 | 3,121 | 1 | 100.0 | 0.0 | 0.0 | Human | 100.0 |
| 107 | Novel | 0238.007.002 | 0.925 | 55 | 70 | 20.0 | 65.7 | 8.6 | Bovine | 67.1 | |
| 111 | 22 | Novel | 0238.007.004 | 0.975 | 3,121 | 1 | 100.0 | 0.0 | 0.0 | Human | 100.0 |
| 111 | Novel | 0238.007.002 | 0.950 | 55 | 70 | 20.0 | 65.7 | 8.6 | Bovine | 67.1 | |
| 122 | 26 | Novel | 0238.002.008 | 0.975 | 751 | 3 | 0.0 | 100.0 | 0.0 | Bovine | 66.7 |
| 122 | Novel | 0238.002.002 | 0.950 | 22 | 152 | 11.8 | 82.2 | 2.6 | Bovine | 84.9 | |
| 127 | 144 | Novel | 0238.014.002 | 0.950 | 18 | 193 | 3.1 | 89.6 | 2.6 | Bovine | 96.9 |
| 127 | Novel | 0238.002.008 | 0.950 | 751 | 3 | 0.0 | 100.0 | 0.0 | Bovine | 66.7 | |
| 366 | 53 | Novel | 0982.005.001 | 0.975 | 179 | 16 | 93.8 | 0.0 | 0.0 | Human | 100.0 |
| 366 | Novel | 0982.001.007 | 0.975 | 3,121 | 1 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 420 | 18 | Novel | 0853.012.004 | 0.925 | 245 | 11 | 90.9 | 9.1 | 0.0 | Human | 100.0 |
| 420 | Novel | 0853.011.006 | 0.925 | 3,121 | 1 | 0.0 | 100.0 | 0.0 | Bovine | 100.0 | |
| 485 | 26 | Novel | 0949.001.003 | 0.875 | 554 | 4 | 50.0 | 50.0 | 0.0 | Bovine | 50.0 |
| 485 | Novel | 0949.007.001 | 0.875 | 554 | 4 | 100.0 | 0.0 | 0.0 | Human | 100.0 | |
| 498 | 12 | Novel | 0949.007.001 | 0.875 | 554 | 4 | 100.0 | 0.0 | 0.0 | Human | 100.0 |
| 498 | Novel | 0949.004.001 | 0.875 | 1,160 | 2 | 0.0 | 100.0 | 0.0 | Bovine | 50.0 | |
| 602 | 41 | Novel | 0695.006.005 | 0.875 | 554 | 4 | 25.0 | 50.0 | 0.0 | Bovine | 75.0 |
| 602 | Novel | 0695.006.003 | 0.875 | 554 | 4 | 25.0 | 50.0 | 0.0 | Bovine | 75.0 | |
| 633 | 57 | Novel | 0904.001.001 | 0.875 | 152 | 20 | 15.0 | 5.0 | 60.0 | Poultry | 15.0 |
| 633 | Novel | 0730.002.001 | 0.875 | 312 | 8 | 12.5 | 12.5 | 0.0 | Bovine | 25.0 | |
Hu, humans; Ca, cattle; Ch, chickens; AB, Alberta.
DISCUSSION
In North America, beef calves born in the spring on pasture are typically transferred to CFOs in the fall, and they are backgrounded and finished in the CFO over a ca. 7-month period, at which point they are transported to an abattoir for processing. Within the CFO, cattle are commonly colonized by C. jejuni, and large densities of the bacterium are frequently shed in their feces (2–5, 23–25). Our research and that of others has also shown that not all subtypes of C. jejuni represent a high risk to humans. There are subtypes of the bacterium commonly associated with beef cattle that are frequently recovered from diarrheic people in SWA (14), suggesting that cattle are an important reservoir of C. jejuni subtypes infecting people. However, the degree and the mechanisms by which these subtypes are longitudinally transmitted within the beef production continuum have not been extensively studied. Moreover, there is a paucity of data on the primary transmission routes of exposure through which cattle-associated C. jejuni isolates are transmitted to humans, including the potential foodborne risk from beef consumption.
We conducted an empirical study in which beef cattle housed in an experimental feedlot were administered an industry standard AGP (i.e., AS700) or no antimicrobials within a replicated experimental design (n = 5). Ten steers were housed in individual pens within the feedlot at stocking densities that are representative of those of industry standards in western Canada. Individual animals were monitored throughout the CFO period (feces) and during processing in the abattoir (abattoir environment, hides, carcass surfaces, intestines, and ground beef generated from the carcasses). Our results indicate that all cattle housed in the CFO that we examined were colonized by C. jejuni and shed the bacterium in large densities exceeding 104 cells/g of feces. This is consistent with previous reports that showed that beef cattle chronically shed C. jejuni in large quantities in their feces (3). With the exception of the final sample time (i.e., day 226), we did not conduct isolations of C. jejuni from feces, and we restricted characterization of the shedding dynamics of the bacterium to quantification of DNA. Quantification of C. jejuni using quantitative PCR (qPCR) has a number of limitations, including sensitivity, specificity, and the inclusiveness of primers (14), as well as the potential for the amplification of DNA from nonviable cells, which can lead to misleading conclusions on viable cell densities. Although no selective media currently exist for C. jejuni, the application of the most-probable-number strategy to enumerate the bacterium, in combination with subtyping, could be applied in future studies to fully elucidate the temporal shedding dynamics of C. jejuni in beef cattle.
Prior to 1 December 2018, the administration of medically important AGPs to enhance feed efficiency and health was a common husbandry practice in North America. AGPs are administered at relatively low concentrations in feed for prolonged periods to promote growth (9). The mechanisms by which AGPs function are not well understood at present, and a proposed mechanism of AGP action is via direct or indirect modulation of the enteric immune system, as opposed to via a direct impact on pathogenic bacteria (8). It has previously been shown that beef cattle in CFOs administered AGPs collectively shed diverse C. jejuni subtypes in their feces (25), but it is currently unknown whether AGP administration has an effect on the rates of C. jejuni carriage and subtype prevalence. In the current study, AGPs were administered at low doses throughout the background period and the majority of the finishing period. We observed that although the densities of C. jejuni cells shed in feces increased over the feedlot period, they were not influenced by AS700 administration. However, the diversity of subtypes was reduced among the C. jejuni bacteria recovered from animals administered AS700. The practice of AGP administration to beef cattle in Canada ended in December 2018 (26), and it remains to be determined if this will affect the prevalence and diversity of the C. jejuni subtypes in beef cattle and the subsequent rates of campylobacteriosis in people. It is noteworthy that the administration of medically important antimicrobials to beef cattle is still permitted as therapeutic treatments under prescription from a licensed veterinarian.
The distribution of C. jejuni throughout the intestinal tract was examined by isolation and quantitative PCR (qPCR). The DNA of the bacterium was prominently associated with the mucosa in the large intestine (cecum, spiral colon, descending colon, and rectum). This is consistent with the findings of a previous study that showed that C. jejuni bacteria in the digesta were distributed throughout the intestinal tract of beef steers but that C. jejuni bacteria associated with the mucosa were concentrated in the distal small intestine and large intestine (5). In a study in which calves were inoculated with C. jejuni, the bacterium was subsequently isolated throughout the intestinal tract, but the highest densities of cells were observed in the large intestine (27). Some evidence exists that Escherichia coli O157:H7 is associated with lymphoid follicle dense mucosa at the terminal rectum (28). Thus, the rectal mucosa was examined for C. jejuni, and we observed that the DNA of the bacterium was prominent in the rectum. Campylobacter jejuni was also detected at high levels in the digesta of the rectum in inoculated calves (27). The densities of C. jejuni within the digesta of the intestinal lumen were not affected by AS700 administration, and cells associated with the mucosa also were not conspicuously reduced due to AGP administration. Consistent with the large numbers of C. jejuni subtypes shed in feces, the bacterium was readily detected throughout the intestinal tract of beef cattle.
Although diverse subtypes of C. jejuni are collectively associated with beef cattle housed in CFOs (25, 29), to our knowledge, the diversity of subtypes colonizing individual animals has not previously been examined. We observed that the majority of the 30 animals examined (n = 22) were colonized with more than one C. jejuni subtype; from 10 individual animals, greater than 5 subtypes were recovered, and from 1 animal, 21 subtypes were recovered. Although it has not been extensively studied, current evidence indicates that C. jejuni subtype diversity is limited in individual diarrheic people. For example, the majority of diarrheic people were observed to be infected by a single strain of C. jejuni (14). Coinfection of humans with more than one C. jejuni strain has been documented in 5% to 10% of sporadic cases (14, 30) and approximately one-half of cases of campylobacteriosis linked to outbreaks in the United Kingdom (31, 32). With the possible exception of young calves (27), C. jejuni is not thought to adversely affect the health of beef cattle in CFOs (i.e., they are asymptomatic), and our evidence indicates that they are colonized by multiple subtypes of C. jejuni and shed these subtypes in large quantities in their feces. The niches occupied by the different subtypes of C. jejuni within the intestinal tract of cattle are currently unknown, but the large quantities of diverse subtypes released from cattle feces and the consequences of this on the epidemiology of campylobacteriosis in people warrant additional study. It is noteworthy that beef cattle are prolific producers of feces, and in areas with high densities of cattle, such as SWA, beef cattle produce the vast majority of the manure (i.e., relative to other livestock and humans).
Limited research has examined the mechanisms of C. jejuni transmission within beef cattle CFOs and elsewhere in the production continuum. It is currently thought that a small number of calves entering the feedlot are colonized by C. jejuni and that the bacterium is rapidly transmitted among animals within the CFO, facilitated by the close proximity of animals to one another within CFOs (23–25). A limitation of our study design was that some replicate pens were situated adjacent to one another, including pens containing cattle exposed to AS700 and cattle not administered antimicrobials (e.g., pens 2 and 3); this treatment assignment was necessitated by the logistics of administering the AGP and control treatment feed. Examination of the subtypes, however, indicated that there was limited transmission of subtypes between cattle housed within adjacent pens separated by a wood slat fence. Although the cattle sampled within pens contained common C. jejuni subtypes, they also carried subtypes that were unique to each individual. We observed that the densities of C. jejuni shed in feces increased over the feedlot period, but AGP administration did not conspicuously affect the densities of C. jejuni in feces. That the diversity of C. jejuni subtypes recovered from cattle administered AS700 was reduced suggests that selection occurs at the strain level. The Campylobacter jejuni subtypes present in the intestinal tract and shed in feces were present on the hides of individual animals at the time of euthanization, and they were transferred to the surfaces of the carcasses during the dressing process and subsequently survived on the carcasses at 4°C for 24 h. The degree to which C. jejuni persists at low temperatures on dressed carcasses has not been extensively examined, but C. jejuni has been shown to survive to different degrees on uncooked beef at refrigeration temperatures (33, 34). To our knowledge, examination of survival by C. jejuni has not been done at the subtype level of resolution. As subtypes show different survivabilities under adverse conditions, such as oxygen exposure (35), studies to ascertain the survival of C. jejuni on beef and other meats should be performed at the strain level. In the current study, the C. jejuni subtypes associated with the cattle were present in the abattoir environment and were recovered from the air during hide removal. Thus, the evidence obtained indicates that C. jejuni strains originating from incoming animals are directly and indirectly transmitted to the carcasses during processing and survive on the carcasses for 24 h at 4°C. From surveillance investigations, the low-prevalence presence of C. jejuni on beef carcasses in abattoirs has been previously observed in snapshot examinations (16–18, 36, 37) but, to our knowledge, has not been observed in longitudinal studies or at the subtype level of resolution.
The vast majority of studies examining the contamination of beef carcasses and meat by C. jejuni have been survey based. We examined ground beef generated from the carcasses that were contaminated with C. jejuni, albeit at low densities, and were unable to isolate any C. jejuni bacterial cells or detect the DNA of the bacterium. As we did not detect the DNA of viable or nonviable cells using PCR, this would suggest that the dilution of the already low densities of C. jejuni on carcass surfaces (<500 cells/cm2) in ground beef was responsible, at least in part. Our findings support the findings of a multitude of surveys that have concluded that viable C. jejuni cells are uncommon in retail beef in Canada and elsewhere (19–21, 38–43). This contrasts with the high prevalence of contamination of retail poultry by C. jejuni (44). Besides the obvious differences in how beef and chicken carcasses are processed, it has been proposed that chilling reduces the surface humidity of red meat more than that of poultry meat, which may contribute to the low prevalence of C. jejuni in beef compared to that in poultry (38). Collectively, however, our longitudinal data, coupled with food surveillance findings, indicate that the C. jejuni bacteria associated with beef meat and meat products do not represent a significant foodborne risk.
Although our results allow the conclusion that the consumption of beef contaminated with C. jejuni is not a foodborne risk factor, accumulated evidence indicates that C. jejuni bacteria associated with cattle are associated with campylobacteriosis. At present, the degree of this risk and the mechanisms by which clinically relevant subtypes are transmitted to people remain enigmatic. In the current study, SWA was used as model agroecosystem, and the C. jejuni subtypes associated with cattle and diarrheic people living in the study region during the same time period were observed to be similar. This is in agreement with the findings of a recently published study that showed that temporal case clusters of C. jejuni subtypes associated with cattle contribute to the high rates of campylobacteriosis in SWA (14). To discern host associations, the data within the C3GFdb were utilized; the C3GFdb currently includes data for >25,000 C. jejuni isolates that have been subtyped by CGF, including ≈10,000 isolates from SWA. The host breakdown within the C3GFdb is ≈4,000 C. jejuni isolates from diarrheic people, ≈4,400 isolates from water, ≈6,000 isolates from cattle, ≈7,000 isolates from poultry, and ≈3,500 isolates from other sources. Examination of the C. jejuni isolates recovered from cattle in the current study further implicates beef cattle as a primary reservoir of C. jejuni infecting people in SWA, a region with a high density of cattle and with a high number of animals in CFOs (7). In a recent snapshot examination, Thepault et al. (6) collected C. jejuni isolates from the intestines of cattle (beef and dairy) in France, subtyped them using CGF40, and compared the subtypes to those of a set of clinical C. jejuni isolates that were recovered from people throughout France and that had been subtyped. Similar to the findings of the current study, they observed that genetic diversity was significantly lower among C. jejuni isolates recovered from cattle than among those recovered from people and that there was overlap between the genotypes from both origins. They did not ascertain the mechanisms by which people may have come into contact with the high-risk subtypes of C. jejuni. Although chickens are thought to be the primary reservoir of clinically relevant C. jejuni subtypes (45), this does not appear to be the case in all locations. For example, evidence suggests that in Finland, cattle are as important a reservoir of C. jejuni subtypes infecting people as poultry (46). In addition to the commonality of the C. jejuni subtypes associated with cattle and diarrheic people in SWA, the results of an epidemiological study conducted in the region indicated that occupational contact with cattle is a significant risk factor for infection by the bacterium (13). Another possibility is that beef cattle serve as a primary reservoir of C. jejuni subtypes infecting poultry flocks and that C. jejuni bacteria originating from beef cattle subsequently infect people via consumption of inappropriately handled/cooked poultry. This is consistent with information in the C3GFdb that shows that the subtypes isolated from beef cattle and poultry are also frequently recovered from diarrheic humans. This hypothesis is under investigation by our research team.
In summary, we completed a longitudinal transmission study of C. jejuni throughout the beef cattle production continuum using SWA as a model agroecosystem. Cattle were exposed to the AGP AS700 or no antibiotics. We showed that beef cattle were colonized by diverse C. jejuni subtypes, including a large number of different subtypes within some individuals. The administration of AS700 did not affect the quantities of C. jejuni bacteria shed in feces but did reduce the subtype diversity. Furthermore, evidence for the horizontal transmission of C. jejuni subtypes from cattle to carcasses was observed, but no C. jejuni bacteria were detected in ground beef generated from the contaminated carcasses. Although the subtypes of C. jejuni associated with cattle were also isolated from diarrheic people during the study period, the evidence indicated that the foodborne risk posed by clinically relevant C. jejuni subtypes is negligible. Thus, alternate mechanisms to foodborne transmission from beef cattle are likely responsible for the high rates of campylobacteriosis observed in SWA.
MATERIALS AND METHODS
Ethics approvals.
Before commencement of the study, the experiment was approved by the Agriculture and Agri-Food Canada Lethbridge Research and Development Centre (LeRDC) Animal Care Committee. Approval to transfer Campylobacter isolates from the Chinook Regional Hospital to LeRDC was obtained from the University of Lethbridge Office of Research Ethics (Certificate of Human Subject Research number 715) and the University of Alberta Research Ethics Office (Health Research Ethics Board number Pro00094238). The information that was transferred with the isolates was restricted to the isolate identifier, the year of isolation, and whether the bacterium was isolated from a stool or blood sample. No information that could reveal the identity of the infected individual was disclosed to the research personnel.
Animals and husbandry.
One hundred Angus-cross beef cattle were housed in an experimental CFO located at the Lethbridge Research and Development Centre. Calves originated from a common location and did not receive antimicrobial agents before the initiation of the experiment. Upon arrival at the CFO, calves were arbitrarily assigned to one of two treatments: (i) no antimicrobial agents (i.e., control) and (ii) 350 mg head (hd)−1 day−1 chlortetracycline and 350 mg hd−1 day−1 sulfamethazine (AS700; catalog number Aureo S 700 G; Alpharma Inc., NJ), as recommended by the manufacturer. AS700 was selected, as it was commonly used in the Canadian beef industry prior to the ban on AGP administration in late 2018. Each treatment was replicated five times and was arranged as a randomized complete block design; each block consisted of a separate pen containing 10 steers. Water troughs were shared between adjacent pens in some instances, but treatments were arranged in a manner so that only cattle that received the same antimicrobial agent could drink from the same trough.
All of the animals involved in this study were housed according to the guidelines set out by the Canadian Council on Animal Care (47). Steers entering the CFO were fed for the first 84 days a forage-based diet consisting of 70% barley silage, 25% barley grain, and 5% (dry matter basis) supplement containing vitamins and minerals (i.e., backgrounding period). The cattle were subsequently transitioned from the forage-based diet to a grain-based diet (85% barley, 10% barley silage, 5% supplement) over a 21-day period, and then they were maintained on the grain-based diet for an additional 126 days (i.e., finishing period). The cattle were fed once daily in a manner to ensure that all feed allotted to a pen was consumed. For the AS700-treated animals, the antibiotics were first introduced into the feed 5 days after the cattle arrived at the CFO and were continually applied thereafter until 28 days before shipment of the animals to the abattoir (i.e., to meet the requisite withdrawal period for animals entering the human food chain). To avoid cross contamination, antimicrobial agents were mixed with 5 kg of a supplement containing minerals and vitamins, and the mixture was manually spread over the surface of the feed within each of the appropriate pens during the morning feeding. All animals in the pen were capable of feeding at the feed trough within each pen at the same time. For the control group, cattle were fed supplement without antimicrobial agents. Besides AS700, none of the study cattle were administered other antimicrobial agents (i.e., therapeutically or metaphylactically).
Confined feeding operation sampling.
It was not possible to longitudinally sample all 100 steers enrolled in the study. Thus, 3 of the 10 steers per pen per treatment (AS700 or control) were selected for detailed longitudinal examination (i.e., 15 steers per treatment); the animals were identified by ear tags. Feces were collected from the 30 selected animals at ca. 28-day intervals throughout the feeding period. Fecal samples were obtained per rectum as described previously (23). The final fecal samples were obtained upon transport to the abattoir. After collection, the samples were placed on ice and processed within 4 h.
Abattoir.
The 30 steers selected for longitudinal sample acquisition in the CFO were further sampled at a provincial inspected medium-capacity plant (Ben’s Quality Meats, Picture Butte, AB, Canada). Control animals were transported in the late afternoon of day 1 and euthanized on the morning of day 2. AS700-treated animals were transported on the afternoon of day 2 and euthanized on the morning of day 3. Following transport of the control animals, the stock trailer was thoroughly cleaned using a pressure washer. At the abattoir, the animals were maintained on a barley silage diet and were euthanized humanely according to the Canadian Council on Animal Care (47).
Abattoir samples were acquired on the slaughter floor and within the carcass chiller room. Standard sanitation procedures for the abattoir at the end of the day included (i) removal of all edible offals and trimmings; (ii) prerinsing of all equipment, knock box, walls, leg bench, viscera tub, inspection tray, cradle, scale, splitting saw, brisket saw, gutting stand, doors, door handles, and head rack with warm (i.e., >32°C) water; (iii) removal and rinsing of floor gates; (iv) pumping of the blood pit; (v) scraping and discarding of excess material from the floor; (vi) treatment of all equipment and surfaces with HydroChem foam (HydroChem Industrial Services, Inc., Deer Park, TX) (Pinnacle Distribution Inc., Saskatoon, SK, Canada) for a minimum of 15 min; (vii) further hand washing of all foamed equipment, inspection tray, and brisket saw; (viii) rinsing of all foamed equipment and surfaces with hot water (starting at the top and working down and washing the floors toward the center floor drain); (ix) application of a 12% bleach solution (0.6% sodium hypochlorite) to all equipment and surfaces with a minimum exposure time of 5 min before a final rinsing with hot water; and (x) application of the quaternary ammonium product Quatromyicide (Dustbane Products Ltd., Ottawa, ON, Canada) at a rate of 500:1 to all surfaces and equipment.
Abattoir sampling.
Five environmental samples were obtained from the abattoir prior to the commencement of and immediately after the slaughter of the 15 animals processed on each day. Samples were obtained from (i) the hydraulic gate of the knock box, (ii) the blade of the splitting saw, (iii) the wall immediately behind the inspection area, (iv) the viscera tub, and (v) the wall adjacent to the rendering room entrance. Within 5 min after euthanization, samples from the surface of the hide of all animals (a 900-cm2 area of the brisket and rump) were obtained with a sterile 2- by 4-cm cellulose acetate sponge (Nasco Canada, Newmarket, ON, Canada) moistened with Columbia broth (CB; Difco, BD Biosciences, Mississauga, ON, Canada); the area sampled (i.e., 30 × 30 cm) was delimited using a sanitized wire frame. Air samples were obtained during the hide removal process for each animal. An inertial air sampler (catalog number MAS-100; EMD Chemicals Inc., Gibbstown, NJ) operated for 2 min (100 liters/min) was used; the sampler was situated at a height of 1 m adjacent to the carcass during hide removal; Karmali agar (KA; Oxoid Inc., Nepean, ON, Canada) containing selective supplement SR167 (KSA; Oxoid Inc.) was used. Following hide removal, evisceration, and breaking, the carcass was sprayed with warm water (38 to 43°C) according to the standard operating procedures of the plant. After the carcass wash, the surface of one side of each carcass was sampled; like the hides, a 900-cm2 area of the brisket and rump was sponge sampled. The surface of the other side of each carcass was sampled in the same manner after 24 h in the carcass chiller (6°C). All sponges were maintained in sterile bags on ice until they were processed (ca. 2 to 6 h). After 7 days in the chiller room, the carcasses were graded.
Meat trimmings (20% fat) from the brisket and rump of the right side of the carcass of each animal were obtained after 7 days in the chiller, placed in a plastic bag, and transported to the Lethbridge Research and Development Centre on ice. Meat from each sample was processed separately by grinding (Porkert number 32 bolt-down manual meat grinder) through a 5-mm-diameter plate. The grinder was dismantled between each sample, and all parts were washed in hot water (85°C), followed by surface sanitation with 95% ethanol. In a preliminary assessment, this treatment was found to result in no discernible carryover of campylobacteria or fecal coliforms (data not presented). A subsample of ground beef (150 g) was placed in a sterile filtered stomacher bag (model 400 bags; Seward Laboratory Systems Inc., Bohemia, NY) and maintained on ice until it was processed. The remaining ground beef sample was placed at 5°C for 7 days, and a subsample was similarly placed in a stomacher bag on ice. Samples were processed ≈2 h after generating ground beef.
The intestinal tract of five animals per antimicrobial treatment (one animal per pen) was removed approximately 10 min after death and placed on a clean sheet of plastic on a cool cement floor within the abattoir. Nine intestinal sections (20 cm in length) were obtained from each animal at the following locations: (i) descending portion of the duodenum (i.e., following the cranial flexure), (ii) proximal jejunum, (iii) central jejunum, (iv) distal jejunum, (v) ileum (10 cm before the ileal-cecal junction), (vi) free end of the cecum, (vii) central flexure of the ascending colon, (viii) descending colon (20 cm before the sigmoid colon), and (ix) rectum (48). Before excision of the intestinal sections, bilateral ligatures were applied adjacent to the excision site to minimize external contamination of the tissues with digesta. Tissue samples were placed in individual bags on ice, transported to the laboratory, and processed within ≈4 h.
Isolation.
At the laboratory, intestinal sections were aseptically excised longitudinally and the digesta was removed. Mucosal surfaces were washed gently with chilled phosphate buffer (0.01 M, pH 7.2) to remove the residual digesta but retain the mucus, and a section of the intestinal wall (3 by 3 cm) was removed with a DNA-free scalpel blade. Feces and digesta (1 g) were placed in a 10 times volume of CB, and the sample was vortexed on the high setting for 45 s. The sponges and mucosa were homogenized in 20 ml of CB for 2 min on the high setting using a Stomacher 80 lab blender (Seward Laboratory Systems Inc.), the sponges were removed, the suspension was centrifuged at 14,900 × g for 10 min, the supernatant was removed, and the pellet was resuspended in 1 ml of CB. The ground beef was homogenized in 200 ml of CB for 2 min on the high setting using a Stomacher 400 circulator (Seward Laboratory Systems Inc.), and the suspension was concentrated by centrifugation as described above for the sponge samples. An aliquot (25 μl) of the homogenate/stomached suspension was spread in duplicate onto Campylobacter blood-free selective medium (modified CCDA-Preston medium) with CCDA selective supplement CM739 (CSA; Oxoid Inc.) and KSA in 60-mm-diameter petri dishes. The cultures were maintained at 42°C in a microaerobic atmosphere (10% CO2, 3% H2, 5% O2, 82% N2). In addition, samples from the ground beef were enriched in Bolton broth (Oxoid Inc.) with selective supplement SR0183 (BBS; Oxoid Inc.). An aliquot of 25 μl of the original sample was added to 5.0 ml of each enrichment broth sample in 100- by 16-mm tubes. Tubes containing BBS were incubated at 175 rpm for 3 h at 30°C, 2 h at 37°C, and 24 h at 42°C; 10 μl of the enrichment broth was then removed and streaked in duplicate onto CSA and KSA in 60-mm-diameter petri dishes, and the cultures were maintained in the microaerobic atmosphere as described above.
The cultures were examined after 48 h and 72 h, and where applicable, the biomass from each of two colonies per colony morphology characteristic of Campylobacter was streaked onto KA and maintained at 37°C in a microaerobic atmosphere (i.e., 5% O2, 3% H2, 10% CO2, and 82% N2). When necessary, cells from the colonies were examined for size, shape, and characteristic motility for Campylobacter-like bacteria using phase-contrast microscopy. All presumptive Campylobacter-like isolates were streaked for purity, the biomass was transferred to CB containing 40% glycerol, and the cultures were maintained frozen at −80°C until the bacteria were identified.
Identification.
Genomic DNA of presumptive Campylobacter isolates was extracted using an AutoGen 740 robot (Holliston, MA) according to the manufacturer’s protocol for Gram-negative bacteria. All isolates were identified by PCR using a Campylobacter genus-specific PCR targeting the 16S rRNA gene (49) and a C. jejuni-specific PCR targeting the hipO gene (50).
Genotyping.
All recovered C. jejuni isolates were fingerprinted using the CGF40 method (51). Briefly, eight-multiplex PCRs were performed for each C. jejuni isolate; each five-multiplex reaction mix contained 1 U Taq DNA polymerase (MP Biomedicals, Irvine, CA, USA), 1× buffer, 2.5 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (dNTP), 0.12 to 0.74 μM the 10 primers (of note, the primer concentration was optimized to produce a strong amplicon for each primer set in the multiplex), and 1 μl of DNA template (20 to 100 ng) in a 25-μl reaction mix. An EP Gradient master cycler (Eppendorf, Mississauga, ON, Canada) was used, and PCR conditions were an initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and a final extension step at 72°C for 5 min. Amplicons were resolved using a QIAxcel high-throughput capillary electrophoresis system with DNA screening cartridges (Qiagen Inc.), the AM320 separation method, and a 20-s injection time. The 15- to 3,000-bp alignment marker and a 100-bp to 2.5-kbp size ladder were used as size standards (Qiagen Inc.).
Preparation and DNA extraction from samples.
Aliquots of feces and digesta (200 ± 5 mg) were collected and stored at −20°C until they were processed. Biopsy specimens (5 mm in diameter) of mucosa were frozen at −20°C until they were processed. For sponge samples concentrated by centrifugation (see “Isolation” above), 200-μl aliquots were frozen at −20°C until they were processed. Aliquots (200 μl) from the carcasses and ground beef were additionally treated with ethidium monoazide before freezer storage as described previously (52). Genomic DNA was extracted from feces, digesta, and sponge washes using a QIAamp DNA stool minikit (Qiagen Inc.) according to the manufacturer’s specifications for pathogen detection. For mucosal tissues, a Qiagen DNeasy tissue kit (Qiagen Inc.) was used according to the manufacturer’s recommendation. An internal amplification control (IAC) was added to all substrates before they were extracted (2).
Quantification of Campylobacter jejuni.
The Campylobacter jejuni bacteria in/on the feces, digesta, carcasses, and ground beef were quantified by PCR as described previously (53). Briefly, the mapA gene was targeted, and the PCR efficiency, the optimum primer concentration, and the dynamic range were determined in advance. The SYBR green-based standard curve method for quantification of DNA was carried out using QuantiTect SYBR green (Qiagen Inc.) Each 20-μl PCR mixture contained 2 μl of DNA (20 to 50 ng), 10 μl of the 2× QuantiTect SYBR green master mix, 1.0 μl of each of the forward and reverse primers (10 μM QCjmapANF and QCjmapANR, respectively), 0.1 μg μl−1 bovine serum albumin, and 3 μl nuclease-free water (Qiagen Inc.). Samples were amplified as follows: 1 cycle at 95°C for 15 min and 40 cycles at 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. A Stratagene Mx3005 qPCR system (Stratagene Products, La Jolla, CA) was used. All reactions were run in duplicate, and the mean value of the observations was used for analysis. The number of bacteria was expressed as the log10 copy number gram−1 of feces or digesta and log10 copy number centimeter−2 of mucosa, hides, or carcasses. Standard curves were established using genomic DNA from known concentrations of C. jejuni NCTC 11168. For all reactions, melt curve analysis was conducted to confirm the amplification specificity.
Human health risk determination.
To ascertain health risk, the CGF40 profiles of the C. jejuni isolates recovered from beef cattle in the current study were compared to the CGF40 profiles of C. jejuni isolates recovered from 1,171 diarrheic humans in the study region from 2004 to 2017. In addition, the CGF40 profiles of prominent subtypes associated with both cattle and humans were queried against the profiles of the subtypes within the C3GFdb; the database includes >25,000 fingerprinted C. jejuni isolates from humans, livestock, wildlife, and environmental matrices.
Analyses.
Most analyses were conducted using the mixed procedure of the Statistical Analysis System software (SAS Institute Inc., Cary, NC). In instances in which variables were not independent, such as fecal collection time and intestinal location, they were treated as repeated measures; the appropriate covariance structure was utilized according to the lowest Akaike’s information criterion. In the event of a significant main effect, the least-squares means test was used to evaluate differences among means of interest. Isolates were assigned to CGF subtype clusters at 100% and 95% levels of resolution using the simple matching analysis coefficient with the unweighted pair group method with arithmetic mean (UPGMA) clustering in BioNumerics software (version 6.6; Applied Maths, Austin, TX). Population structures were visualized as minimum spanning trees (MSTs) using BioNumerics software (version 6.6; Applied Maths). Venn diagrams of subtypes were generated using pivot tables at a 95% level of resolution. Fisher’s exact test was used to compare the C. jejuni subtype counts between the AS700 and control treatments.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the assistance of the following individuals at Agriculture and Agri-Food Canada, Lethbridge Research and Development Centre: Wendi Smart, Brant Baker, and Fred Van Herk for collection of fecal samples and for transporting cattle from the CFO to the abattoir; Jennilee Jamison and Cindy Barkley for abattoir sampling; Ryan Lammertsen and Christa King for processing samples; and Daniel Hymus for genotyping C. jejuni isolates. We also thank Tim McAllister (Agriculture and Agri-Food Canada, LeRDC) for facilitating access to the animals, the staff and management of Ben’s Quality Meats (Picture Butte, AB, Canada) for allowing us to obtain samples from within their facility, and the many contributors to the C3GFdb.
Financial support for this study was provided by grants from the Alberta Livestock and Meat Agency (grant 2012F034R), the Canada-Alberta Beef Industry Development Fund (CABIDF), and Agriculture and Agri-Food Canada (a peer review grant).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Inglis GD, Kalischuk LD. 2004. Direct quantification of Campylobacter jejuni and Campylobacter lanienae in feces of cattle by real-time quantitative PCR. Appl Environ Microbiol 70:2296–2306. doi: 10.1128/aem.70.4.2296-2306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inglis GD, Kalischuk LD. 2003. Use of PCR for direct detection of Campylobacter species in bovine feces. Appl Environ Microbiol 69:3435–3447. doi: 10.1128/aem.69.6.3435-3447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglis GD, Kalischuk LD, Busz HW. 2004. Chronic shedding of Campylobacter species in beef cattle. J Appl Microbiol 97:410–420. doi: 10.1111/j.1365-2672.2004.02313.x. [DOI] [PubMed] [Google Scholar]
- 4.Inglis GD, Kalischuk LD, Busz HW. 2003. A survey of Campylobacter species shed in faeces of beef cattle using polymerase chain reaction. Can J Microbiol 49:655–661. doi: 10.1139/w03-087. [DOI] [PubMed] [Google Scholar]
- 5.Inglis GD, Kalischuk LD, Busz HW, Kastelic JP. 2005. Colonization of cattle intestines by Campylobacter jejuni and Campylobacter lanienae. Appl Environ Microbiol 71:5145–5153. doi: 10.1128/AEM.71.9.5145-5153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thepault A, Poezevara T, Quesne S, Rose V, Chemaly M, Rivoal K. 2018. Prevalence of thermophilic Campylobacter in cattle production at slaughterhouse level in France and link between C. jejuni bovine strains and campylobacteriosis. Front Microbiol 9:471. doi: 10.3389/fmicb.2018.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberta Government. 2011. Census of agriculture for Alberta. Alberta Agriculture and Rural Development, Edmonton, AB, Canada: https://www1.agric.gov.ab.ca/$Department/deptdocs.nsf/all/agdex4091/$FILE/852-6.pdf. [Google Scholar]
- 8.Brown K, Uwiera RR, Kalmokoff ML, Brooks SP, Inglis GD. 2017. Antimicrobial growth promoter use in livestock: a requirement to understand their modes of action to develop effective alternatives. Int J Antimicrob Agents 49:12–24. doi: 10.1016/j.ijantimicag.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Cromwell GL. 2002. Why and how antibiotics are used in swine production. Anim Biotechnol 13:7–27. doi: 10.1081/ABIO-120005767. [DOI] [PubMed] [Google Scholar]
- 10.Alberta Government. 2004. Notifiable diseases in Alberta annual report. Alberta Health and Wellness, Calgary, AB, Canada. [Google Scholar]
- 11.Public Health Agency of Canada. 2018. Notifiable diseases online. https://diseases.canada.ca/notifiable/.
- 12.Inglis GD, Boras VF, Houde A. 2011. Enteric campylobacteria and RNA viruses associated with healthy and diarrheic humans in the Chinook Health Region of southwestern Alberta, Canada. J Clin Microbiol 49:209–219. doi: 10.1128/JCM.01220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasselback P. 2002. Feedlot alley and enteric illness: are they related or is southern Alberta just a wonderful place for humans, cattle and bugs to live? Canadian Laboratory Medicine Congress, Calgary, AB, Canada. [Google Scholar]
- 14.Inglis GD, Boras VF, Webb AL, Suttorp VV, Hodgkinson P, Taboada EN. 2019. Enhanced microbiological surveillance reveals that temporal case clusters contribute to the high rates of campylobacteriosis in a model agroecosystem. Int J Med Microbiol 309:232–244. doi: 10.1016/j.ijmm.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Wysok B, Uradziński J, Wojtacka J. 2015. Determination of the cytotoxic activity of Campylobacter strains isolated from bovine and swine carcasses in north-eastern Poland. Pol J Vet Sci 18:579–586. doi: 10.1515/pjvs-2015-0075. [DOI] [PubMed] [Google Scholar]
- 16.Wieczorek K, Denis E, Lynch O, Osek J. 2013. Molecular characterization and antibiotic resistance profiling of Campylobacter isolated from cattle in Polish slaughterhouses. Food Microbiol 34:130–136. doi: 10.1016/j.fm.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Van Donkersgoed J, Bohaychuk V, Besser T, Song XM, Wagner B, Hancock D, Renter D, Dargatz D. 2009. Occurrence of foodborne bacteria in Alberta feedlots. Can Vet J 50:166–172. [PMC free article] [PubMed] [Google Scholar]
- 18.Hakkinen M, Heiska H, Hanninen ML. 2007. Prevalence of Campylobacter spp. in cattle in Finland and antimicrobial susceptibilities of bovine Campylobacter jejuni strains. Appl Environ Microbiol 73:3232–3238. doi: 10.1128/AEM.02579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narvaez-Bravo C, Taboada EN, Mutschall SK, Aslam M. 2017. Epidemiology of antimicrobial resistant Campylobacter spp. isolated from retail meats in Canada. Int J Food Microbiol 253:43–47. doi: 10.1016/j.ijfoodmicro.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 20.Llarena AK, Sivonen K, Hanninen ML. 2014. Campylobacter jejuni prevalence and hygienic quality of retail bovine ground meat in Finland. Lett Appl Microbiol 58:408–413. doi: 10.1111/lam.12206. [DOI] [PubMed] [Google Scholar]
- 21.Zhao S, Young SR, Tong E, Abbott JW, Womack N, Friedman SL, McDermott PF. 2010. Antimicrobial resistance of Campylobacter isolates from retail meat in the United States between 2002 and 2007. Appl Environ Microbiol 76:7949–7956. doi: 10.1128/AEM.01297-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannon SJ, Inglis GD, Allan B, Waldner C, Russell ML, Potter A, Babiuk LA, Townsend H. 2009. Prevalence and risk factor investigation of Campylobacter species in retail ground beef from Alberta, Canada. Food Prot Trends 29:780–786. [Google Scholar]
- 23.Inglis GD, McAllister TA, Busz HW, Yanke LJ, Morck DW, Olson ME, Read RR. 2005. Effects of subtherapeutic administration of antimicrobial agents to beef cattle on the prevalence of antimicrobial resistance in Campylobacter jejuni and Campylobacter hyointestinalis. Appl Environ Microbiol 71:3872–3881. doi: 10.1128/AEM.71.7.3872-3881.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inglis GD, Morck DW, McAllister TA, Entz T, Olson ME, Yanke LJ, Read RR. 2006. Temporal prevalence of antimicrobial resistance in Campylobacter spp. from beef cattle in Alberta feedlots. Appl Environ Microbiol 72:4088–4095. doi: 10.1128/AEM.02830-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb AL, Selinger LB, Taboada EN, Inglis GD. 2018. Subtype-specific selection for resistance to fluoroquinolones but not to tetracyclines is evident in Campylobacter jejuni isolates from beef cattle in confined feeding operations in southern Alberta. Appl Environ Microbiol 84:e02713-17. doi: 10.1128/AEM.02713-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Public Health Agency of Canada. 2019. Responsible use of medically important antimicrobials in animals. https://www.canada.ca/en/public-health/services/antibiotic-antimicrobial-resistance/animals/actions/responsible-use-antimicrobials.html.
- 27.Terzolo HR, Lawson GH, Angus KW, Snodgrass DR. 1987. Enteric campylobacter infection in gnotobiotic calves and lambs. Res Vet Sci 43:72–77. doi: 10.1016/S0034-5288(18)30745-8. [DOI] [PubMed] [Google Scholar]
- 28.Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DG, Gally DL. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun 71:1505–1512. doi: 10.1128/iai.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannon SJ, Taboada EN, Russell ML, Allan B, Waldner C, Wilson HL, Potter A, Babiuk L, Townsend HG. 2009. Genomics-based molecular epidemiology of Campylobacter jejuni isolates from feedlot cattle and from people in Alberta, Canada. J Clin Microbiol 47:410–420. doi: 10.1128/JCM.01432-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson JF, Frost JA, Kramer JM, Thwaites RT, Bolton FJ, Wareing DR, Gordon JA. 2001. Coinfection with Campylobacter species: an epidemiological problem? J Appl Microbiol 91:206–211. doi: 10.1046/j.1365-2672.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 31.Frost JA, Gillespie IA, O'Brien SJ. 2002. Public health implications of campylobacter outbreaks in England and Wales, 1995-9: epidemiological and microbiological investigations. Epidemiol Infect 128:111–118. doi: 10.1017/s0950268802006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forbes KJ, Gormley FJ, Dallas JF, Labovitiadi O, MacRae M, Owen RJ, Richardson J, Strachan NJ, Cowden JM, Ogden ID, McGuigan CC. 2009. Campylobacter immunity and coinfection following a large outbreak in a farming community. J Clin Microbiol 47:111–116. doi: 10.1128/JCM.01731-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balamurugan S, Nattress FM, Baker LP, Dilts BD. 2011. Survival of Campylobacter jejuni on beef and pork under vacuum packaged and retail storage conditions: examination of the role of natural meat microflora on C. jejuni survival. Food Microbiol 28:1003–1010. doi: 10.1016/j.fm.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Bostan K, Aksu H, Ozgen O, Ugur M. 2001. Effects of refrigerated and frozen storage on the survival of Campylobacter jejuni in ground and cubed beef. Pakistan J Biol Sci 4:888–890. doi: 10.3923/pjbs.2001.888.890. [DOI] [Google Scholar]
- 35.Oh E, McMullen LM, Chui L, Jeon B. 2017. Differential survival of hyper-aerotolerant Campylobacter jejuni under different gas conditions. Front Microbiol 8:954. doi: 10.3389/fmicb.2017.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohaychuk VM, Gensler GE, Barrios PR. 2011. Microbiological baseline study of beef and pork carcasses from provincially inspected abattoirs in Alberta, Canada. Can Vet J 52:1095–1100. [PMC free article] [PubMed] [Google Scholar]
- 37.Wieczorek K. 2009. Relationship between the molecular typing of Campylobacter strains and the prevalence of their virulence genes. Bull Vet Inst Pulawy 53:193–198. [Google Scholar]
- 38.Premarathne J, Anuar AS, Thung TY, Satharasinghe DA, Jambari NN, Abdul-Mutalib NA, Huat JTY, Basri DF, Rukayadi Y, Nakaguchi Y, Nishibuchi M, Radu S. 2017. Prevalence and antibiotic resistance against tetracycline in Campylobacter jejuni and C. coli in cattle and beef meat from Selangor. Front Microbiol 8:2254. doi: 10.3389/fmicb.2017.02254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bosilevac JM, Guerini MN, Brichta-Harhay DM, Arthur TM, Koohmaraie M. 2007. Microbiological characterization of imported and domestic boneless beef trim used for ground beef. J Food Prot 70:440–449. doi: 10.4315/0362-028x-70.2.440. [DOI] [PubMed] [Google Scholar]
- 40.Phillips D, Jordan D, Morris S, Jenson I, Sumner J. 2008. A national survey of the microbiological quality of retail raw meats in Australia. J Food Prot 71:1232–1236. doi: 10.4315/0362-028x-71.6.1232. [DOI] [PubMed] [Google Scholar]
- 41.Wong TL, Hollis L, Cornelius A, Nicol C, Cook R, Hudson JA. 2007. Prevalence, numbers, and subtypes of Campylobacter jejuni and Campylobacter coli in uncooked retail meat samples. J Food Prot 70:566–573. doi: 10.4315/0362-028x-70.3.566. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Ge B, De Villena J, Sudler R, Yeh E, Zhao S, White DG, Wagner D, Meng J. 2001. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the greater Washington, D.C., area. Appl Environ Microbiol 67:5431–5436. doi: 10.1128/AEM.67.12.5431-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong J, Kim JM, Jung WK, Kim SH, Bae W, Koo HC, Gil J, Kim M, Ser J, Park YH. 2007. Prevalence and antibiotic resistance of Campylobacter spp. isolated from chicken meat, pork, and beef in Korea, from 2001 to 2006. J Food Prot 70:860–866. doi: 10.4315/0362-028x-70.4.860. [DOI] [PubMed] [Google Scholar]
- 44.Silva J, Leite D, Fernandes M, Mena C, Gibbs PA, Teixeira P. 2011. Campylobacter spp. as a foodborne pathogen: a review. Front Microbiol 2:200. doi: 10.3389/fmicb.2011.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thibodeau A, Fravalo P, Taboada EN, Laurent-Lewandowski S, Guevremont E, Quessy S, Letellier A. 2015. Extensive characterization of Campylobacter jejuni chicken isolates to uncover genes involved in the ability to compete for gut colonization. BMC Microbiol 15:97. doi: 10.1186/s12866-015-0433-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Haan CPA, Kivistö RI, Hakkinen M, Corander J, Hänninen M-L. 2010. Multilocus sequence types of Finnish bovine Campylobacter jejuni isolates and their attribution to human infections. BMC Microbiol 10:200. doi: 10.1186/1471-2180-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olfert ED, Cross BM, McWilliam AA (ed). 2017. Guide to the care and use of experimental animals. Canadian Council on Animal Care, Ottawa, ON, Canada: https://www.ccac.ca/Documents/Standards/Guidelines/Experimental_Animals_Vol1.pdf. [Google Scholar]
- 48.Reti KL, Thomas MC, Yanke LJ, Selinger LB, Inglis GD. 2013. Effect of antimicrobial growth promoter administration on the intestinal microbiota of beef cattle. Gut Pathog 5:8. doi: 10.1186/1757-4749-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linton D, Owen RJ, Stanley J. 1996. Rapid identification by PCR of the genus Campylobacter and of five Campylobacter species enteropathogenic for man and animals. Res Microbiol 147:707–718. doi: 10.1016/s0923-2508(97)85118-2. [DOI] [PubMed] [Google Scholar]
- 50.Inglis GD, Zaytsoff SJM, Selinger LB, Taboada EN, Uwiera R. 2018. Therapeutic administration of enrofloxacin in mice does not select for fluoroquinolone resistance in Campylobacter jejuni. Can J Microbiol 64:681–694. doi: 10.1139/cjm-2017-0741. [DOI] [PubMed] [Google Scholar]
- 51.Taboada EN, Ross SL, Mutschall SK, MacKinnon JM, Roberts MJ, Buchanan CJ, Kruczkiewicz P, Jokinen CC, Thomas JE, Nash JH, Gannon VP, Marshall B, Pollari F, Clark CG. 2012. Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni. J Clin Microbiol 50:788–797. doi: 10.1128/JCM.00669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inglis GD, McAllister TA, Larney FJ, Topp E. 2010. Prolonged survival of Campylobacter species in bovine manure compost. Appl Environ Microbiol 76:1110–1119. doi: 10.1128/AEM.01902-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lone AG, Selinger LB, Uwiera RR, Xu Y, Inglis GD. 2013. Campylobacter jejuni colonization is associated with a dysbiosis in the cecal microbiota of mice in the absence of prominent inflammation. PLoS One 8:e75325. doi: 10.1371/journal.pone.0075325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.