Lactobacillus rhamnosus GG is one of the best-studied probiotic strains. One of the well-characterized features of the strain is the pili encoded by the spaCBA-srtC1 gene cluster. These pili are involved in persistence in the gastrointestinal tract and are important for the probiotic properties of L. rhamnosus GG. Previous studies demonstrated that the L. rhamnosus GG genome can be unstable under certain conditions and can lose the spaCBA-srtC1 gene cluster. Since in vitro studies have shown that the loss of the spaCBA-srtC1 gene cluster decreases certain L. rhamnosus GG probiotic properties, we assessed both the genomic stability and phenotypic properties of L. rhamnosus GG throughout an industrial production process. We found that neither genomic nor phenotypic changes occurred in the samples. Therefore, we demonstrate that L. rhamnosus GG retains the spaCBA-srtC1 cluster and exhibits excellent genomic and phenotypic stability in the specific industrial process examined here.
KEYWORDS: Lactobacillus rhamnosus GG, genomic instability, industrial fermentation, probiotics
ABSTRACT
Lactobacillus rhamnosus GG is one of the most widely marketed and studied probiotic strains. In L. rhamnosus GG, the spaCBA-srtC1 gene cluster encodes pili, which are important for some of the probiotic properties of the strain. A previous study showed that the DNA sequence of the spaCBA-srtC1 gene cluster was not present in some L. rhamnosus GG variants isolated from liquid dairy products. To examine the stability of the L. rhamnosus GG genome in an industrial production process, we sequenced the genome of samples of L. rhamnosus GG (DSM 33156) collected at specific steps of the industrial production process, including the culture collection stock, intermediate fermentations, and final freeze-dried products. We found that the L. rhamnosus GG genome sequence was unchanged throughout the production process. Consequently, the spaCBA-srtC1 gene locus was intact and fully conserved in all 31 samples examined. In addition, different production batches of L. rhamnosus GG exhibited consistent phenotypes, including the presence of pili in final freeze-dried products, and consistent characteristics in in vitro assays of probiotic properties. Our data show that L. rhamnosus GG is highly stable in this industrial production process.
IMPORTANCE Lactobacillus rhamnosus GG is one of the best-studied probiotic strains. One of the well-characterized features of the strain is the pili encoded by the spaCBA-srtC1 gene cluster. These pili are involved in persistence in the gastrointestinal tract and are important for the probiotic properties of L. rhamnosus GG. Previous studies demonstrated that the L. rhamnosus GG genome can be unstable under certain conditions and can lose the spaCBA-srtC1 gene cluster. Since in vitro studies have shown that the loss of the spaCBA-srtC1 gene cluster decreases certain L. rhamnosus GG probiotic properties, we assessed both the genomic stability and phenotypic properties of L. rhamnosus GG throughout an industrial production process. We found that neither genomic nor phenotypic changes occurred in the samples. Therefore, we demonstrate that L. rhamnosus GG retains the spaCBA-srtC1 cluster and exhibits excellent genomic and phenotypic stability in the specific industrial process examined here.
INTRODUCTION
Lactobacillus rhamnosus GG was originally isolated from a human fecal sample by Sherwood Gorbach and Barry Goldin in 1985 and was proposed to be a potential probiotic strain based on its high acid and bile tolerance, adhesion capacity, and growth characteristics (1, 2). Since its introduction into the market in 1990, L. rhamnosus GG has become one of the most widely marketed and studied probiotic strains. L. rhamnosus GG has been tested in several hundred clinical trials and has shown beneficial effects in a variety of areas, including shortening the duration of acute infectious diarrhea (3, 4), preventing the development of atopic dermatitis (5, 6), increasing tolerance to cow’s milk in allergic infants (7), and eliminating colonization by vancomycin-resistant enterococci (VRE), which are a leading cause of nosocomial infections (8). To understand the mechanisms of action underlying these benefits, many genomic and in vitro studies have been conducted on L. rhamnosus GG, and several genes important for probiotic function have been identified, including pilus-encoding genes.
Pili are hair-like appendages on the surface of some bacteria that can facilitate adhesion, conjugation, and immune modulation. While structure-function studies of pili have long focused on their role as virulence factors in Gram-negative pathogens (9), the presence and functions of pili in Gram-positive commensal bacteria such as L. rhamnosus GG have begun to be appreciated and characterized more recently (10). The pili on L. rhamnosus GG, which are composed of the pilin subunits SpaC, SpaB, and SpaA and are encoded by the spaCBA-srtC1 gene cluster (11), are important for several functions in vitro. First, pili play a key role in mucus adhesion, demonstrated by the fact that wild-type L. rhamnosus GG adheres strongly to mucus in vitro, whereas mutants of L. rhamnosus GG that lack pili, as well as the closely related Lactobacillus rhamnosus strain Lc705 that naturally lacks pili, adhere poorly (12). A human intervention study showed that L. rhamnosus GG persists longer in the gastrointestinal tract than Lc705 (11), suggesting that pili may facilitate transient colonization. Second, pili may be important for immunomodulatory effects. Purified pili from L. rhamnosus GG interact with dendritic cells (DCs) via CD209 (commonly known as dendritic cell-specific ICAM-grabbing nonintegrin, or DC-SIGN) to stimulate the production of the Th1-related cytokine interleukin-12 (IL-12) (13), thus implicating pili in an immunomodulatory function that would be consistent with observed clinical effects in preventing and treating infections and allergic conditions (14). Third, the pili of L. rhamnosus GG have been proposed to facilitate the elimination of the opportunistic pathogens VRE in the gastrointestinal tract via a competitive exclusion mechanism. The SpaCBA pili of L. rhamnosus GG exhibit high sequence similarity to the PGC-3 pili of the clinical VRE isolate Enterococcus faecium E1165, and binding of E1165 to mucus could be prevented by coincubation with antibodies raised against SpaC or L. rhamnosus GG (15).
Although most of the evidence for the functional importance of pili has been shown in vitro and needs to be substantiated in vivo, the in vitro evidence suggests that the loss of the spaCBA-srtC1 gene cluster might eliminate some probiotic functions of L. rhamnosus GG. Thus, conservation of the spaCBA-srtC1 genomic region may be of great importance for the efficacy of the strain. Genomic stability can be influenced by genome composition in certain regions. For example, insertion sequence (IS) elements can mediate larger genomic changes (16–18). The L. rhamnosus GG genome has 69 annotated IS-like elements, many of which are located around the spaCBA-srtC1 gene cluster region (11). Recent studies have examined the genomic stability of L. rhamnosus GG (16, 19). One study isolated and genome-sequenced L. rhamnosus GG from three different liquid dairy products and found that the products contained variants of L. rhamnosus GG, where two out of the three variants featured large genomic deletions, resulting in the loss of the spaCBA-srtC1 region (19). Based on this finding, Sybesma et al. (19) proposed a quality assurance approach in the production process. Genomic stability can also be influenced by growth conditions. Exposure to stressful conditions can increase the frequency of genomic changes, as shown in experimental evolution studies (20–22). The genomic stability of L. rhamnosus GG grown under different stress conditions for approximately 1,000 generations was recently investigated (16). That study showed that the L. rhamnosus GG genome is generally highly stable, but in some samples that were continuously exposed to bile stress, a mutation that led to larger genomic changes emerged after approximately 900 generations.
Results from laboratory-scale genomic stability studies cannot necessarily be extrapolated to industrial-scale production, where bacteria may be subjected to certain stressful conditions that may affect genomic stability. Batch fermentation is a common method used in the industrial production of bacteria. In a closed system, a fermentation tank is filled with a sterilized nutrient solution and inoculated with a selected strain. Throughout the fermentation process, parameters such as pH and temperature are continuously controlled. When nutrients are exhausted, growth declines, and bacteria are harvested with the aim of combining the shortest production time with the highest yield. Next, bacteria are concentrated, and cryoprotective reagents are added to protect the bacteria from detrimental conditions during subsequent freezing and freeze-drying processes (23). Despite measures taken to optimize for fast growth and high survival rates, bacteria may still encounter periods of acid stress, nutrient deprivation, and cold shock. Because of the observed loss of the spaCBA-srtC1 region in some L. rhamnosus GG variants (19), it is important to assess both the genome stability and phenotype of L. rhamnosus GG in a large-scale production process.
RESULTS
The genome integrity of L. rhamnosus GG is conserved throughout the industrial production process.
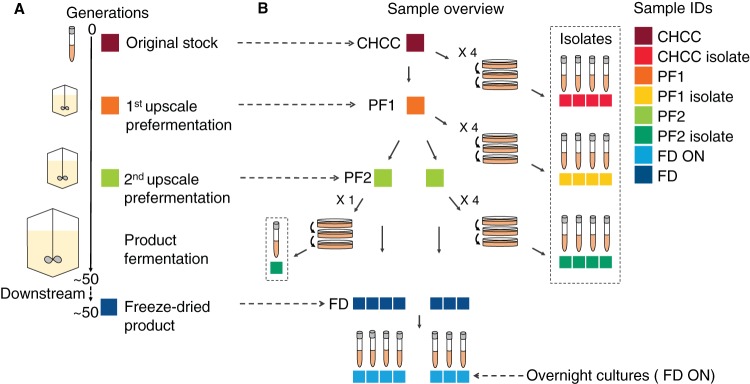
To analyze the genome stability of industrially produced L. rhamnosus GG, we collected and whole-genome sequenced 31 samples from different steps throughout the production process at Chr. Hansen A/S (Fig. 1). These included the final freeze-dried products from seven different production batches, their respective prefermentations, and the stock from the Chr. Hansen Culture Collection (CHCC) (24) used in the production process. In addition, cultures grown overnight were prepared from the freeze-dried products, and single-colony isolates were prepared from prefermentations. As assessed by the optical density (OD), approximately 50 generations occurred in the production process from the CHCC stock to the freeze-dried product.
FIG 1.
Overview of sampling from the production process. Samples were taken from the CHCC stock, the intermediate growth steps in the production process (PF1 and PF2), and the freeze-dried product from the final fermentation of the product (FD). From each of these samples, single-colony isolates (isolates) or cultures grown overnight (ON) were prepared. (A) Industrial process from which samples were taken. (B) Specific samples taken, including isolates. Sample identifications and colors, as defined in the key, are consistently used in all figures.
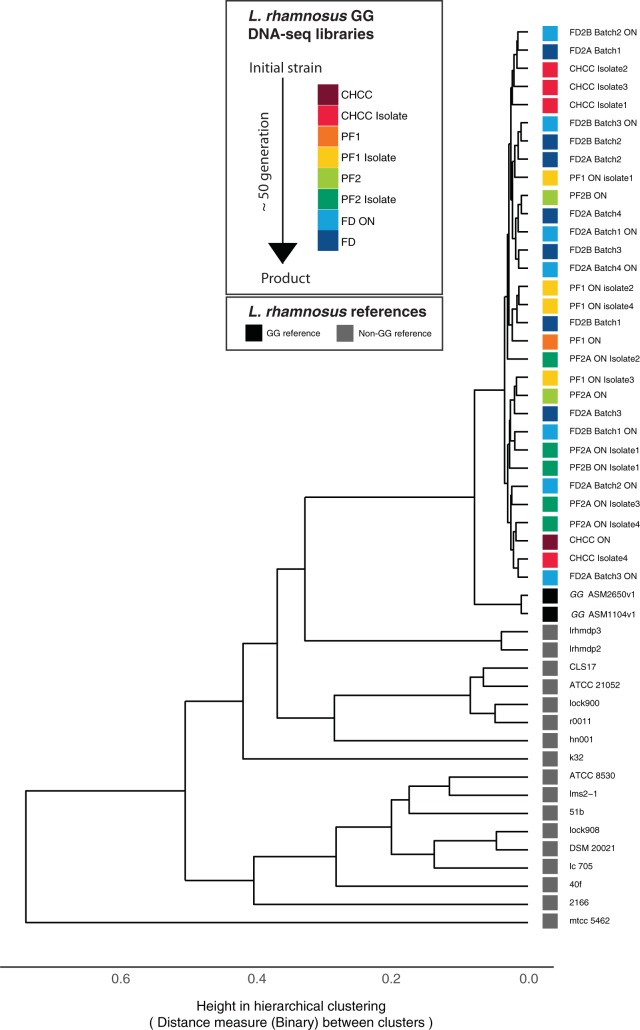
Genome sequence analysis resulted in high-quality contigs with an average read coverage of 357× from de novo-assembled genomes (see Materials and Methods). First, we wanted to ensure the purity of our sequenced genomes. We started by filtering away short contigs of low coverage (see Materials and Methods). Next, we performed hierarchical clustering of the 31 genomes together with all published L. rhamnosus genomes, based on gene presence (Fig. 2). This showed that all 31 samples clustered tightly with the L. rhamnosus GG reference genome, indicating that all samples are the same strain, which allows for comparative genome analysis and variant analysis within the strain.
FIG 2.
Genome similarity between samples and available public genomes for L. rhamnosus GG and non-GG strains. Shown is hierarchical clustering of genome similarities. The distance matrix was calculated based on gene presence and absence in the genomes using a binary method. The bottom scale indicates the distance between clusters. The 31 production samples are color-coded as in Fig. 1.
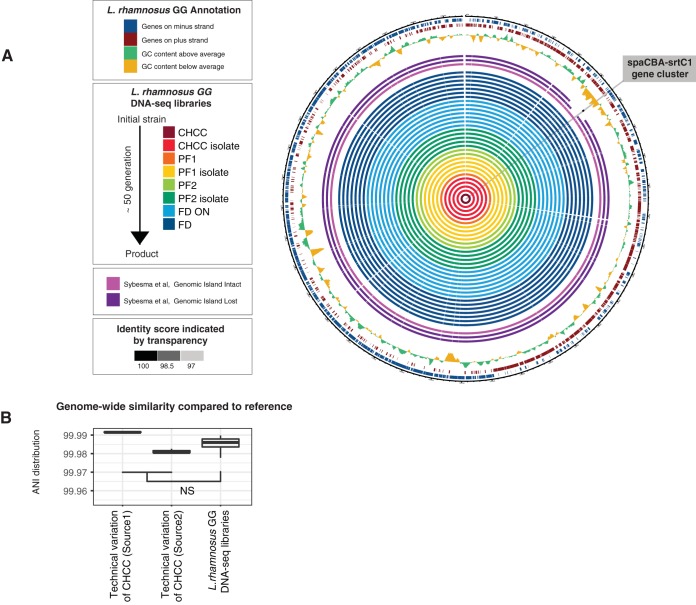
Alignment of contigs derived from the 31 production samples to the reference genome showed high consistency in genome coverage across all libraries and high identity scores compared to the reference genome (Fig. 3A). Importantly, the spaCBA-srtC1 region that was lost in the isolates examined by Sybesma et al. (19) was retained in all 31 samples in this study (Fig. 3A), confirming the genomic stability of Lactobacillus rhamnosus GG in the production process. In addition, the multiple-sequence alignment of the spaCBA-srtC1 region was 100% identical across all 31 samples and the reference genome (see Fig. S1 in the supplemental material).
FIG 3.
Genome similarity of samples versus the L. rhamnosus GG reference genome. (A) Circular representation of alignments of DNA sequence contigs derived from the 31 production samples to the reference genome. Each solid circle indicates one sample, color-coded as in Fig. 1, where the opacity indicates the alignment identity score. For comparison, contigs from three samples reported previously by Sybesma et al. (19) are also included. The three outer circles represent GC content and annotated genes on both strands, as indicated in the key. (B) Analysis of genome-wide similarity of selected samples versus the reference genome. The y axis shows the distribution of average nucleotide identity (ANI) scores versus the reference genome for a given set of samples as box plots (two sets of technical replicates for CHCC samples sequenced at two different locations [see Materials and Methods] and all L. rhamnosus GG samples from Fig. 1). NS, not significant (P > 0.05 by a two-sided Mann-Whitney test).
Next, we wanted to assess nucleotide differences on the whole-genome level between samples. To distinguish true biological differences from technical sequencing noise (25), we measured the pairwise average nucleotide identities (ANIs) of the 31 genomes to the reference and those of 6 additional CHCC stock samples sequenced at two different places to the reference (Fig. 3B). Because the CHCC samples are identical, they serve as technical controls: ANIs between these samples and the reference represent technical variations in the analysis. We observed that the ANI score distributions of the 31 genomes were not significantly different (P > 0.05 by a two-sided Mann-Whitney test) from the ANI scores of all the technical replicates (Fig. 3B) (see Materials and Methods). In fact, sequencing the same sample at different locations could induce larger variations than those existing in the 31 genomes. Therefore, the differences between the 31 genomes were not greater than what would be expected due to technical noise, and this also allowed for further analysis examining minor genomic variants.
L. rhamnosus GG does not accumulate single nucleotide polymorphisms during an industrial production process.
Since single nucleotide polymorphisms (SNPs) were previously identified in marketed L. rhamnosus GG isolates (26), we complemented the whole-genome integrity analysis by assessing the potential accumulation of small mutations (27). To ensure a robust analysis, we used seven different approaches for variant calling, and the intersections of the outputs of these approaches were used as the final variant results. First, we analyzed samples of the CHCC stock, prefermentations, and final freeze-dried products, which represent bacterial populations. No significant SNPs were called in any of these production samples, except for two bases (positions 615483 and 1883242) that matched known sequencing errors present in the L. rhamnosus GG reference genome (26). Indels were not detected in any of the samples.
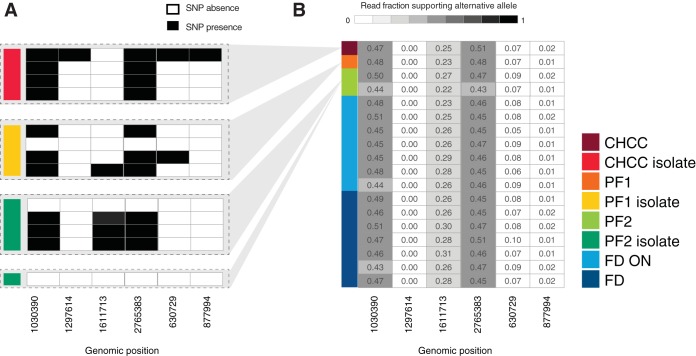
Next, we examined single-colony isolates that we purified from the CHCC stock and production samples. In contrast to the population analysis, when we analyzed the genomes of single-colony isolates, we identified six SNPs that occurred consistently across isolates regardless of the sample type from which they originated (Fig. 4A). To analyze whether these SNPs may represent minor alleles in the production samples, we calculated the fraction of reads in the production samples that supported the alternative alleles identified in isolates. The majority of SNPs identified in the isolates had some read support in the production samples, ranging from 1% to 51% of reads (Fig. 4B). Importantly, the read support of a given allele was demonstrated to be consistent in all production samples from the CHCC stock to the freeze-dried products and among the seven production batches (Fig. 4B). In addition, the fraction of read support was invariant when comparing the CHCC stock with the original stock from 1994, from which the CHCC stock was made (Fig. S2A, left). Coupling this with an analysis comparing genome-wide ANI scores between the assemblies (Fig. S2A, right), we demonstrated that the CHCC stock that is currently used for the production of the LGG® strain and the original stock from 1994 are identical.
FIG 4.
Analysis of SNPs in isolates. (A) Binary table of SNP calls in CHCC, PF1, and PF2 isolates. Each row represents an isolate sample, colored as in Fig. 1. Gray areas indicate the production samples from which the isolates are derived. Columns represent genome locations. Black cells represent calls of significant SNPs compared to the reference genome (note that no SNPs were called in the production sample genomes, only in isolates). (B) Fraction of read support in production samples for SNPs identified in panel A. Rows indicate each production sample, colored as in Fig. 1. Columns indicate the same genome locations as in panel A. Cell numbers and shading indicate the fraction of reads in each sample that support the alternative allele.
To examine whether SNPs identified in the isolates could change the strain phenotype, we overlapped them with gene annotation and predicted the consequence of amino acid substitution using the PROVEAN (Protein Variation Effect Analyzer) method (28) based on alignments and chemical properties of amino acids. This analysis predicted the effects of all SNPs to be neutral (Fig. S2B). It is worth noting that no marginal mutations were found in the spaCBA-srtC1 region (average coverage of 751×), confirming the stability of the pilus-related genes.
Pili are present on L. rhamnosus GG freeze-dried products.
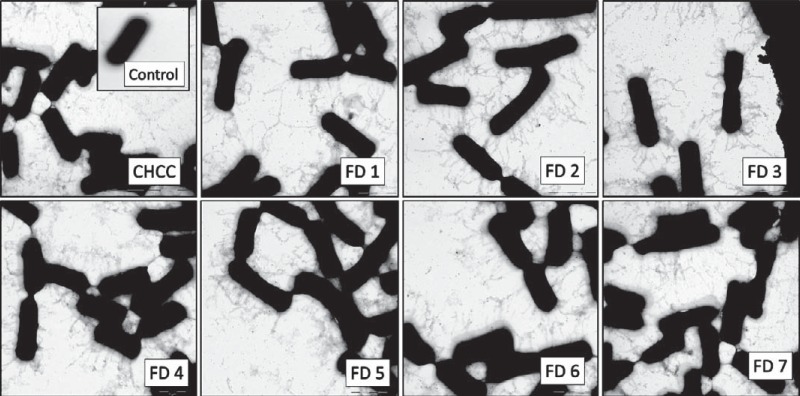
As described above, we demonstrated that the spaCBA-srtC1 genomic region is intact and conserved throughout the industrial production process. In the L. rhamnosus GG genome, 69 IS-like elements have been annotated, with many located around the spaCBA-srtC1 gene cluster region (11). A high frequency of IS elements has been shown to impact gene expression (29). Furthermore, mechanical forces in a production process could disrupt pili. Hence, we investigated the preservation of the phenotypic expression of pili throughout the production process. Using electron microscopy (EM), we investigated L. rhamnosus GG from the CHCC stock and from the freeze-dried final product from each of the seven production batches. Immunolabeling with polyclonal anti-SpaA rabbit antibodies highlighted the pilus structure and enabled visualization of pili. Protein A-gold particles were retained on the grids only in the presence of anti-SpaA antibodies, which demonstrated that protein A-gold was specifically binding to the anti-SpaA antibodies. A control sample not labeled with anti-SpaA rabbit antibodies demonstrated that the observed structures were pili (Fig. 5).
FIG 5.
Electron microscopy images of pili on L. rhamnosus GG. EM was performed on samples from the CHCC stock and the seven freeze-dried industrial production batches (FD1 to FD7) of L. rhamnosus GG labeled with polyclonal anti-SpaA rabbit antibodies and protein A-gold particles to visualize pili. For the control, L. rhamnosus GG from the CHCC stock without polyclonal anti-SpaA rabbit antibodies was used.
We found that pili were present in all samples, and there was no visible difference in pilus phenotypes among the samples. This analysis confirmed that pilus structures, which are important for host interaction and probiotic functionality, were maintained in the production process.
Phenotypic characteristics are consistent for different production batches of L. rhamnosus GG.
To examine the consistency in phenotype among different production batches of L. rhamnosus GG, we compared the seven freeze-dried production batches in four in vitro assays that represent important characteristics for probiotic bacteria: acid tolerance, bile tolerance, the ability to stimulate an increase in the transepithelial electrical resistance (TEER) of Caco-2 cells, and the ability to stimulate the release of the cytokines IL-12p70 and tumor necrosis factor alpha (TNF-α) from human dendritic cells.
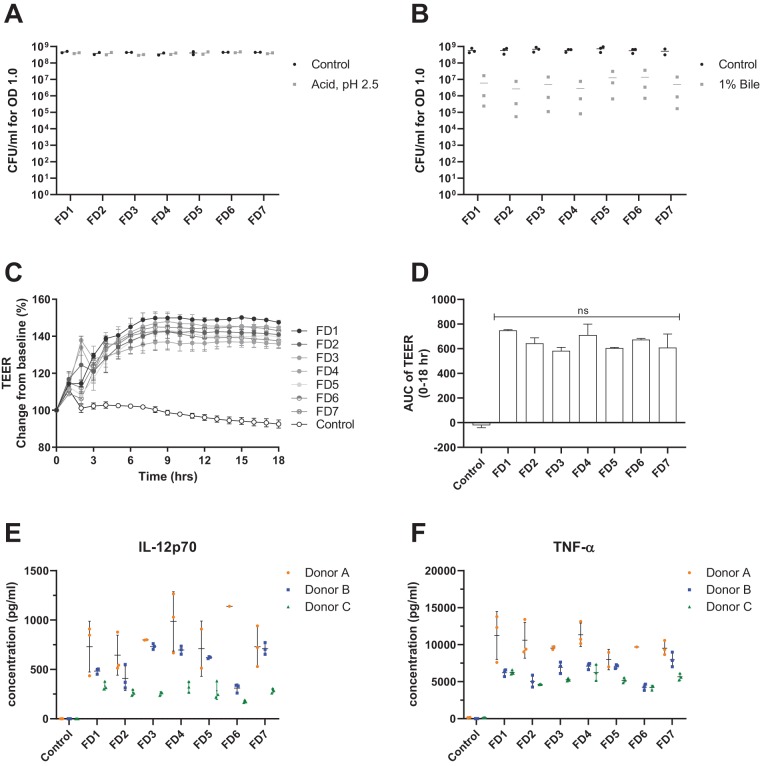
All seven production batches (FD1 to FD7) exhibited high tolerance to acid shock, showing no significant decrease in CFU after a 1-h incubation at pH 2.5 compared with their respective controls. In addition, there were no significant differences in CFU counts among any of the acid-treated samples (Fig. 6A). In contrast, the L. rhamnosus GG samples were somewhat sensitive to bile, losing an average of 2-log viability after a 1-h incubation. However, there were no significant differences in CFU counts among any of the bile-treated samples (Fig. 6B), indicating that freeze-dried L. rhamnosus GG bacteria from different production batches exhibit consistent phenotypes for both acid and bile tolerance.
FIG 6.
Phenotypic comparisons of seven different production batches of L. rhamnosus GG. (A) CFU on MRS agar for 1 ml of the sample at an OD of 1.0 after a 1-h incubation in gastric acid solution (pH 2.5) (Acid) or 10% MRS (pH 6.5) (Control) (y axis) for production batches (FD1 to FD7) (x axis). Data points represent the means for technical triplicates from two independent experiments. (B) CFU on MRS agar for 1 ml of the sample at an OD of 1.0 after a 1-h incubation in 1% porcine bile solution (Bile) or 10% MRS (pH 6.5) (y axis) for production batches (FD1 to FD7) (x axis). Data points represent the means for technical triplicates from three independent experiments, and horizontal lines represent the means of data from the three experiments. (C) Transepithelial electrical resistance (TEER) of Caco-2 cell monolayers stimulated with L. rhamnosus GG from one of the seven freeze-dried industrial production batches measured for 18 h. The y axis shows TEER percent changes relative to the baseline. Each line shows TEER measurements using production batches (FD1 to FD7) and the control (cell culture medium only), where data points represent the means ± SD of results from technical duplicates. The x axis shows the time in hours. (D) Area under the curve (AUC) of TEER measurements from 0 to 18 h relative to the baseline (100%) (y axis) for production batches (FD1 to FD7) and the control (x axis), as described above for panel C. Bars represent the means ± SD for technical duplicates. ns, no statistical difference (by one-way ANOVA). (E) Concentration of IL-12p70 secreted by monocyte-derived dendritic cells (DCs) from three different donors (y axis) that were stimulated with L. rhamnosus GG from one of seven freeze-dried industrial production batches (FD1 to FD7) and the control (x axis). Controls were incubated in cell culture medium only. Data points represent the results of biological replicates. Color represents the donor. Black bars represent the means ± SD for each donor. (F) Concentration of TNF-α secreted by monocyte-derived DCs from three different donors (y axis) that were stimulated with L. rhamnosus GG from one of seven freeze-dried industrial production batches (FD1 to FD7) (x axis). The plot follows the same conventions as the ones described above for panel E.
In an in vitro assay of intestinal barrier function, all production batches stimulated a similar increase in the TEER of Caco-2 cell monolayers (Fig. 6C), indicating that L. rhamnosus GG stimulates a tightening of the Caco-2 cell tight junctions in vitro. Similarly, analysis of the area under the curve (AUC) over an 18-h TEER time course showed that there were no significant differences in the TEER responses among the stimulated FD1 to FD7 (Fig. 6D).
In in vitro assays of monocyte-derived DCs, FD1 to FD7 all stimulated the secretion of the cytokines IL-12p70 and TNF-α in cells derived from three different donors (Fig. 6E and F). Donor variation is typically observed for in vitro DC assays (30), and therefore, statistical analysis was performed separately for each donor. For any given donor, there were no statistically significant differences in either IL-12p70 or TNF-α secretion levels stimulated by the different production batches.
Altogether, the data from multiple assays support that different production batches of L. rhamnosus GG exhibit consistent genotypes and phenotypes.
DISCUSSION
Our study demonstrates the high genomic stability of L. rhamnosus GG during an industrial production process. In particular, the spaCBA-srtC1 locus, which is important for the probiotic properties of L. rhamnosus GG, is unchanged during the production process. In addition, the high genomic stability of L. rhamnosus GG implies that other genes important for probiotic properties are intact and conserved, such as the genes encoding bile salt hydrolase (LGG_00501) (31) and the major secreted proteins Msp1 and Msp2 (also known as p75 and p40, respectively) (32, 33) and gene clusters involved in the biosynthesis of exopolysaccharides (11) and lipoteichoic acid (34).
Microbial evolution studies have shown that an increased mutational rate is associated with nutrient deficiency and stressful conditions (16, 21, 35–38). In an industrial production process using batch fermentation, bacteria may be subjected to stress due to, for example, the production of acidic metabolites and depletion of nutrients, which can lead to increased mutation and recombination rates. However, industrial fermentations are highly regulated to obtain maximum yield in minimal time. Therefore, the high genomic stability observed in the large-scale fermentation could be a result of optimal growth conditions. Importantly, in the specific production process examined here, the number of generations from the CHCC stock to the final product is estimated to be around 50, which limits the opportunity for mutations to occur.
The mutational rate is one method to calculate genomic stability and to assess the influence of different growth conditions on the stability of the genome (16, 39). Douillard et al. (16) reported a mutation rate of 2.65 · 10−9 polymorphisms per nucleotide per generation for L. rhamnosus GG under stress-free growth conditions, defined as classical anaerobic growth conditions in De Man-Rogosa-Sharpe (MRS) broth at 37°C. Given this mutation rate and the approximately 50 generations from the CHCC stock to the freeze-dried product, approximately 4 mutational events per batch would be estimated to occur. In the present study, no SNPs were detected in any of the production samples of L. rhamnosus GG, indicating that no elevated mutation rate exists. In the rare instance that a mutation did occur, there would not be enough generations for a mutated variant to establish a noteworthy proportion of the population.
SNPs were detected only in single-colony isolates. Principally, such SNPs could (i) be induced during the three purification steps used to prepare the single-colony isolates (the purification process to prepare isolates would likely be more stressful than the production conditions, as bacteria encounter prolonged exposure to low pH and nutrient exhaustion when grown on agar plates) or (ii) be due to the fact that subpopulations carrying different alleles are present in the production samples.
Importantly, our analysis shows that most SNPs detected in individual isolates had a consistent minor allele frequency (as defined by the fraction of read support) in the production samples, regardless of whether they were taken early or late in the production process. This finding was corroborated by the high read coverage at these sites (average coverage of 552×). This could indicate that the one SNP identified only in an isolate sample (genomic position 1297614) was likely induced by purification steps, whereas the remaining isolate samples represent the allele variation seen in the full bacterial population, which does not change throughout the production process and is consistent with the allele variation present in the original stock from 1994. This may be because the SNPs detected were predicted to have no functional consequence and therefore do not affect fitness. Thus, we demonstrate that this production process does not induce measurable genetic drift and that the genome of L. rhamnosus GG in the original stock from 1994 is identical to the genome in the products currently produced.
While genome stability is important to ensure the retention of all the genes that may be important for strain functionality, the conditions of the production process may influence how these genes are expressed and, hence, how the strain behaves phenotypically. The phenotypic assays performed are commonly used to examine probiotic functionality and demonstrated consistent characteristics in acid and bile tolerance, TEER, and the cytokine response, with no significant differences among any of the production batches.
An important specific question related to genome stability and probiotic functionality was whether the production process impacts the propensity of L. rhamnosus GG to express and retain pili, which are important for its probiotic properties. Some strains of Lactobacillus casei contain an intact spaCBA-srtC1 cluster but do not express pili under any of the conditions tested so far (26). In addition, studies have shown that the spaCBA-srtC1 genes in L. rhamnosus GG are differentially expressed in different growth phases (40) and that strong centrifugal forces may shear off pili from L. rhamnosus GG (41). If the pili of L. rhamnosus GG are either not expressed or not retained in the industrial production process, probiotic efficacy in the gastrointestinal tract may be impaired. Our study shows that the spaCBA-srtC1 gene cluster is fully conserved during the production process and that pili are present in the CHCC stock and all freeze-dried samples, with no visible difference in pilus phenotype. Thus, neither the genotype nor the pilus phenotype was modified throughout the production process. Sybesma et al. (19) previously isolated two L. rhamnosus GG variants from dairy products lacking the genomic region encoding pili and hypothesized that this loss may occur as a result of the production process. Our results strongly indicate that this explanation is unlikely, as neither the genotype nor the phenotype changed throughout the process. A more likely scenario is that the L. rhamnosus GG variants identified by Sybesma et al. (19) represent variants existing among different producers that are distinct from the L. rhamnosus GG samples examined here, which are identical to the reference genome. However, the exact mechanism underlying the pilus gene loss reported by Sybesma et al. (19) remains to be investigated.
In conclusion, our results demonstrate high genome stability and consistent phenotypes of pili and probiotic properties in the examined production process for L. rhamnosus GG.
MATERIALS AND METHODS
Sampling of the bacterial strain and growth conditions.
The Chr. Hansen Culture Collection (CHCC) holds the original ampoules of L. rhamnosus GG, commercially known as LGG® (LGG® is a trademark of Chr. Hansen A/S), received from Valio Ltd. in 1994. CHCC samples of L. rhamnosus GG have been deposited in the German Collection of Microorganisms and Cell Cultures (DSM 33156).
From the stock in the CHCC used for the production of LGG® strain, intermediate fermentations are performed to prepare stocks to be used as inoculation material in the large-scale fermentation producing the final product (38). The media and cryoprotectants used in the manufacturing process are based primarily on carbohydrates, amino acids, vitamins, and minerals, the exact compositions of which are proprietary information of Chr. Hansen A/S. Upon the completion of fermentation, bacterial cells are harvested and concentrated by centrifugation using a separator. The concentrated bacterial cells are then mixed with cryoprotectants and frozen into pellets. The frozen pellets are lyophilized, resulting in very low water activity, which ensures the viability of the freeze-dried bacteria.
Samples of the strain L. rhamnosus GG were collected from both intermediate fermentations and the final fermentation of large-scale industrial productions to cover the different steps in the production pipeline (Fig. 1). Samples from the intermediate fermentation were collected as frozen pellets, whereas final batches were collected as a freeze-dried product. Samples were collected over a 6-month period and stored at −80°C until use. Samples included seven final batches originating from two different prefermentations. All batches originated from the L. rhamnosus GG stock in the CHCC. In addition, CHCC samples used for assessing technical variations and the original stock from 1994 were included in the study.
Isolates were prepared from the CHCC stock and intermediate fermentations by growing a culture overnight and streaking it on MRS agar (Difco) plates. After 3 days of anaerobic growth at 37°C, single colonies were selected and streaked onto new plates. After three successive streaking procedures to ensure the purification of a single colony, one colony was inoculated into a broth culture grown overnight to ensure that there was enough material for extraction. A culture grown overnight was also prepared from each of the seven final batches (Fig. 1). In total, 31 samples were included in the study.
DNA extraction.
DNA was extracted directly from the seven freeze-dried batches. To extract DNA directly from industrial production samples, the freeze-dried pellets were weighed to an amount corresponding to 5 · 109 CFU/g based on CFU counts conducted on each of the batches. Samples from prefermentations (including isolates), the original stock from 1994, and CHCC samples used for assessing technical variations were anaerobically incubated overnight in MRS broth (Difco) at 37°C prior to DNA extraction.
DNA was extracted from samples using a DNeasy blood and tissue minikit (Qiagen). Frozen pellets were thawed and adjusted to an OD at 600 nm (OD600) of 5 in TES (50 mM Tris, 1 mM EDTA, 6.7% sucrose) buffer. Two milliliters of each sample was transferred to an Eppendorf tube, centrifuged at 6,000 × g, and washed twice by dissolving the pellet in TES buffer. Extraction was carried out using the DNeasy blood and tissue minikit according to the manufacturer’s protocol. In addition to the standard protocol, an extra washing step was included after enzymatic lysis. The three samples from the original stock from 1994 and the three CHCC samples used for comparison were prepared in a single batch and extracted at Novogene.
Generation and processing of shotgun sequencing data.
DNA from the 31 samples collected from the industrial pipeline were whole-genome sequenced at Eurofins in paired-end reads (2 by 101 bp) using the Illumina HiSeq 2500 system. The additional CHCC samples used for evaluating technical variation were sequenced at two different locations: source 1 triplicates were sequenced at Novogene using Illumina PE150, and source 2 triplicates were sequenced at Baseclear using the paired-end (2- by 101-bp) Illumina HiSeq 2500 system. The three samples from the original 1994 stock were sequenced in a single batch with the source 1 CHCC triplicates.
Trim Galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) was used to trim low-quality base pairs from the reads. The first 5 bp from the 5′ ends of both paired reads were also trimmed due to the observation of biased quality scores and nucleotide composition.
L. rhamnosus GG reference genome.
The reference genome for L. rhamnosus GG used for all genomic analyses was described previously by Kankainen et al. (11) (ASM2650v1), where L. rhamnosus GG was obtained from the Valio culture collection (Valio Ltd.). For Fig. 2, ASM1104v1 and available Ensembl (42) genomes for L. rhamnosus strains other than GG were also used.
Assembly and Circos visualization.
Spades (43) with the “—careful” setting was used for performing de novo assembly. Using the assembled contigs, DNA contamination was examined in quality control plots made by plotting contig coverage as a function of contig length (data not shown). Contigs with low coverage were considered of low quality and removed from downstream analysis. Cutoffs differed based on the observations in the quality control plots: 2- to 5-fold coverage was used for all the samples from this study, while 2- to 2.5-fold coverage was used for samples described previously by Sybesma et al. (19), which were of a lower sequencing depth. The majority of the filtered contigs were short contigs. The remaining contigs were used for checking gene presence and absence across all 31 samples and the reference genome (ASM2650v1). Assembly statistics were performed by using quast (44). The Circlize package (45) was used to perform the circular genome plot.
Gene calling.
Prokka version 1.12 (46) was used for gene annotation of both de novo assemblies and public reference genomes. The gene annotation outputs were then used in Roary version 3.8.0 (47) to generate a gene presence-and-absence matrix for all the genomes analyzed.
Pairwise average nucleotide identity.
Average nucleotide identity (ANI) scores against the reference genome were calculated for the 31 samples from this study, the 2 dairy isolates, and the 2 sets of CHCC technical triplicates. For each genome X, one ANI score was generated using X as a query genome and the reference genome as a subject genome. Pairwise ANI scores were calculated to compare the differences between the original 1994 stock triplicates and the source 1 CHCC triplicates from the same batch. For each pair of genomes X and Y, two ANI scores were generated: one ANI score was calculated using X as a query genome and Y as a subject genome, and the other ANI score was calculated by switching X to a subject genome and Y to a query genome. An online Perl script (https://github.com/jhbadger/scripts/blob/master/ANI.pl) was used to calculate all ANIs.
Mapping and variant calling.
Initially, Smalt (0.7.6; http://www.sanger.ac.uk/science/tools/smalt-0) and Bowtie2 version 2.2.3 (48) were applied to map all sequencing libraries to the reference genome (ASM2650v1). Base quality score recalibration (BQSR) with the gatk tool (49) was then performed to eliminate potential biases of the base quality score from the sequencer. After BQSR, mutation analysis was done by using three different tools: gatk, SAMtools (50), and freebayes (51) in haploid mode. Coupled with the two above-described mapping tools, this resulted in six ways of variant calling. In addition, breseq (52) was used to validate the outputs from the above-described methods. The intersects from all seven approaches were used as final results in this study. SAMtools was used for quantifying read support for the reference and alternative alleles. The impact of amino acid substitutions was predicted using PROVEAN (28).
Immunoelectron microscopy.
Selected samples were visualized by transmission electron microscopy. The immunogold labeling method was modified from a protocol described previously by Reunanen et al. (53). Polyclonal anti-SpaA rabbit antibodies and protein A-gold particles (10-nm diameter; Nano Biosols) were used for the detection of pili on carbon-coated copper grids (pure C, 200-mesh Cu, catalog number 01840-F). Copper grids were discharged, and one drop of the bacterial sample containing approximately 109 CFU/ml was added to the carbon side for 30 min. The grids were washed three times with phosphate-buffered saline (PBS). Pili were labeled for 1 h with a 1:100-diluted antibodies in 1% bovine serum albumin (BSA) in PBS. Pilus-antibody complexes were formed by incubation for 20 min on drops with 1:55-diluted protein A-gold label in 1% BSA in PBS. Grids were then stained for 30 s with 2% phosphotungstic acid and subsequently visualized using a Leica CM 100 transmission electron microscope.
Preparation of bacteria for phenotypic assays.
Freeze-dried samples from the different production batches (FD1 to FD7) were prepared by dissolving 0.1 g freeze-dried bacteria in 10 ml Hanks’ balanced salt solution (HBSS). Samples were washed twice with HBSS and centrifuged to remove the cryogen (first wash for 10 min at 10,000 × g and second wash for 5 min at 6,000 × g). Samples were subsequently resuspended in the appropriate media for the assay.
Acid and bile tolerance assays.
All samples were adjusted to an OD of 1.0 in 10% MRS (10% [vol/vol] dilution of MRS medium in demineralized water, adjusted to pH 6.5). For each sample, the culture at an OD of 1.0 was divided into three separate tubes and centrifuged for 5 min at 6,000 × g, and the supernatants were carefully removed. The three bacterial pellets were resuspended in three different media: (i) control medium (10% MRS, pH 6.5), (ii) gastric acid medium (10% MRS containing 3% [wt/vol] pepsin [catalog number P7000; Sigma-Aldrich] and 2% [wt/vol] sodium chloride [catalog number S7653; Sigma-Aldrich], adjusted to pH 2.5 with hydrochloric acid), and (iii) bile medium (10% MRS containing 1% [wt/vol] porcine bile [catalog number B8631; Sigma-Aldrich] [pH 6.5]). The cultures were incubated for 1 h at 37°C and analyzed for CFU directly thereafter.
To measure CFU per milliliter, 1 ml of the culture was diluted in 9 ml of peptone water (Dilucup; Holm & Halby), and a 10-fold dilution series was prepared using Dilucups in the Dilushaker system. For each dilution, triplicate MRS agar plates were prepared by adding 1 ml of the sample from the Dilucup and pour plating melted MRS agar (pH 6.5) (catalog number 288210; Difco, UK). Plates were incubated for 2 days at 37°C, and colonies were counted. Only plates with colony counts of between 20 and 300 colonies were used for calculating CFU per milliliter. The means of data for technical triplicates from 2 to 3 independent experiments are shown.
Statistical analysis for bile and acid tolerance assays.
Statistical analysis for bile and acid tolerance assays was performed with GraphPad Prism 8 software (GraphPad Software, La Jolla, CA, USA). Mean values from 2 to 3 independent experiments were compared for each sample (control versus acid or bile treatment) and among all samples (FD1 to FD7) within each treatment group using two-way analysis of variance (ANOVA). P values were adjusted for multiple testing by using the Sidak method.
Transepithelial electrical resistance assay.
Caco-2 cells (ACC 169; DSMZ GmbH, Germany) were cultured at 37°C in an atmosphere of 5% CO2 in Gibco DMEM with GlutaMAX (Dulbecco’s modified Eagle medium formulated with low glucose and sodium pyruvate; Gibco by Life Technologies, UK) supplemented with 20% FBS (fetal bovine serum) (South American origin; Gibco by Life Technologies), 1% minimum essential medium (MEM) nonessential amino acid solution (Sigma-Aldrich), and 100 U/ml penicillin G, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (pen-strep-ampB solution; Biological Industries [BI], Israel). Caco-2 cells were seeded onto transwell membrane inserts in 12-well cell culture plates (1.12 cm2, polyethylene terephthalate membranes with 0.4-μm pores; Corning) at an initial cell density of 105 cells/well and cultured for 21 days in Gibco DMEM with GlutaMAX containing supplements. Media in both the apical and the basolateral compartments were replaced every 3 to 4 days.
Transwell membrane inserts with Caco-2 monolayers were transferred to wells of a CellZscope2 instrument (nanoAnalytics GmbH, Germany) and replenished with antibiotic-free cell culture medium in both the apical and basolateral compartments 1 day prior to bacterial stimulation to allow the cells to adapt to the CellZscope. Transepithelial electrical resistance (TEER) was measured every hour prior to stimulation to establish baseline TEER. For bacterial stimulation, 100 μl antibiotic-free cell culture medium was removed from apical compartments and replaced with either 100 μl of fresh antibiotic-free cell culture medium (unstimulated controls) or 100 μl of bacterial cultures prepared in antibiotic-free cell culture medium at an OD of 3.8, corresponding to a final OD600 of 0.5 and approximately 5 × 107 bacteria in the wells. Under each condition, duplicate wells were analyzed. Cells were stimulated for 18 h at 37°C with 5% CO2, and TEER was measured every hour. The TEER measure of each well was normalized to the baseline TEER of that well before stimulation (t = 0 h), and TEER is expressed as a percent increase relative to the baseline, where the baseline is equal to 100%. The averages and standard deviations (SD) for the duplicates are displayed for each time point and each condition. Area under the curve (AUC) values were calculated for each well using GraphPad Prism 8, and the averages and standard deviations for the duplicates are shown.
Statistical analysis for the TEER assay.
Statistical analysis for the TEER assay was performed with GraphPad Prism 8 software. To compare the AUCs of the unstimulated control and the different bacterial samples (FD1 to FD7), one-way ANOVA followed by Tukey’s multiple-comparison test was performed.
Cytokine secretion from human dendritic cells.
The in vitro generation of human dendritic cells (DCs) from monocytes was slightly modified from a protocol described previously by Zeuthen et al. (54). Briefly, human peripheral blood mononuclear cells (PBMCs) were separated from buffy coats of three healthy donors by density gradient separation using Ficoll-Paque Plus (GE Healthcare, Freiburg, Germany). Monocytes were isolated by positive selection for CD14 using magnetically activated cell sorting with CD14 microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and cultured at a density of 2 × 106 cells/ml in complete DC medium (RPMI 1640 supplemented with 10 mM HEPES [Sigma-Aldrich, Schnelldorf, Germany], 50 μM 2-mercaptoethanol [2-ME; Sigma-Aldrich, Schnelldorf, Germany], 2 mM l-glutamine [Life Technologies Ltd., Paisley, UK], 10% heat-inactivated fetal bovine serum [Invitrogen, Paisley, UK], 100 U/ml penicillin [Biological Industries, Kibbutz Beit-Haemek, Israel], and 100 μg/ml streptomycin [Biological Industries, Kibbutz Beit-Haemek, Israel]). The DC medium also contained 30 ng/ml human recombinant IL-4 and 20 ng/ml human recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) (both from Sigma-Aldrich, St. Louis, MO, USA). Cells were kept at 37°C with 5% CO2. Fresh complete DC medium containing full doses of IL-4 and GM-CSF was added after 3 days of culture. At day 6, differentiation to immature DCs was verified by surface marker expression analysis (CD14, <3%; CD11c, >97% expression; CD1a, >95%).
Immature DCs were resuspended in fresh complete DC medium containing no antibiotics, seeded into 96-well plates at 1 × 105 cells/well, and allowed to acclimate at 37°C in 5% CO2 for at least 1 h before bacterial stimulation. DCs were stimulated for 20 h with the different production batches (FD1 to FD7) at a final concentration of an OD of 0.01. For the control, DCs were left unstimulated. Cells were kept at 37°C with 5% CO2. After stimulation, DC supernatants were sterile filtered through a 0.2-μm Acro-Prep Advance 96-well filter plate (Pall Corporation, Ann Arbor, MI, USA) and stored at −80°C until cytokine quantification.
Secreted levels of IL-12p70 were quantified using V-plex human proinflammatory panel 1 from Meso Scale Discovery (MSD) (catalog number K15049D), and secreted levels of TNF-α were quantified using a customized U-plex panel (MSD, Rockville, MD, USA) according to the manufacturer’s instructions. The means of the lower detection limits were 0.05 pg/ml for IL-12p70 and 0.26 pg/ml for TNF-α.
Statistical analysis for cytokine secretion.
Statistical analysis for cytokine secretion was performed with GraphPad Prism 8 software. Donor variation is typically observed for in vitro DC assays (30), and therefore, statistical analysis was performed separately for each donor. For each donor, cytokine secretion levels stimulated by the different bacterial samples (FD1 to FD7) were compared using a Kruskal-Wallis test followed by Dunn’s adjustment for multiple testing. Data are expressed in picograms per milliliter. Values below the detection limit or below the fitting curve were replaced by the half-limit of detection.
Data availability.
All DNA sequence data are stored at the Gene Expression Omnibus (GEO) database under series accession number GSE123727.
Supplementary Material
ACKNOWLEDGMENTS
The present study was supported by the Innovation Foundation Denmark (M.S., A.S., and A.B. by grants 5139-00024B and Y.C., A.S., and A.B. by 5158-00023B). M.J. was supported by the Højteknologifonden (grant 159-2013-6).
We thank William de Vos for kindly providing polyclonal anti-SpaA rabbit antibodies. We thank Eric Johansen, Johan van Hylckama Vlieg, and Willem de Vos for valuable comments on the manuscript.
M.S., A.S., A.B., M.J., and A.W. designed the experiments. M.S., N.I.V.-J., and M.W. performed the experiments. Y.C. performed bioinformatics analysis. M.S. performed electron microscopy. M.J. performed exploratory bioinformatics analysis. M.S., Y.C., A.W., N.I.V.-J., and A.S. wrote the paper. D.S.N. and A.B. revised the paper.
M.S., A.W., N.I.V.-J., M.W., and A.B. are employed by Chr. Hansen A/S, which produces and markets the LGG® strain. M.J. and Y.C. were employed by Chr. Hansen A/S until August 2017 and February 2019, respectively.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Silva M, Jacobus NV, Deneke C, Gorbach SL. 1987. Antimicrobial substance from a human Lactobacillus strain. Antimicrob Agents Chemother 31:1231–1233. doi: 10.1128/aac.31.8.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldin BR, Gorbach SL, Saxelin M, Barakat S, Gualtieri L, Salminen S. 1992. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci 37:121–128. doi: 10.1007/bf01308354. [DOI] [PubMed] [Google Scholar]
- 3.Isolauri E, Juntunen M, Rautanen T, Sillanaukee P, Koivula T. 1991. A human Lactobacillus strain (Lactobacillus casei sp strain GG) promotes recovery from acute diarrhea in children. Pediatrics 88:90–97. [PubMed] [Google Scholar]
- 4.Aggarwal S, Upadhyay A, Shah D, Teotia N, Agarwal A, Jaiswal V. 2014. Lactobacillus GG for treatment of acute childhood diarrhoea: an open labelled, randomized controlled trial. Indian J Med Res 139:379–385. [PMC free article] [PubMed] [Google Scholar]
- 5.Kalliomäki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 6.Kalliomäki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. 2003. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 7.Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, Cosenza L, Passariello A, Leone L, Granata V, Di Costanzo M, Pezzella V, Troncone R. 2013. Formula selection for management of children with cow’s milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. J Pediatr 163:771–777. doi: 10.1016/j.jpeds.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Manley KJ, Fraenkel MB, Mayall BC, Power DA. 2007. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust 186:454–457. doi: 10.5694/j.1326-5377.2007.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 9.Hospenthal MK, Costa TRD, Waksman G. 2017. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat Rev Microbiol 15:365–379. doi: 10.1038/nrmicro.2017.40. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan V. 2015. Pilins in gram-positive bacteria: a structural perspective. IUBMB Life 67:533–543. doi: 10.1002/iub.1400. [DOI] [PubMed] [Google Scholar]
- 11.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hämäläinen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjärvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasinkangas P, Reunanen J, Douillard FP, Ritari J, Uotinen V, Palva A, de Vos WM. 2014. Genomic characterization of non-mucus-adherent derivatives of Lactobacillus rhamnosus GG reveals genes affecting pilus biogenesis. Appl Environ Microbiol 80:7001–7009. doi: 10.1128/AEM.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tytgat HLP, van Teijlingen NH, Sullan RMA, Douillard FP, Rasinkangas P, Messing M, Reunanen J, Satokari R, Vanderleyden J, Dufrêne YF, Geijtenbeek TBH, de Vos WM, Lebeer S. 2016. Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated heterotrimeric pili. PLoS One 11:e0151824. doi: 10.1371/journal.pone.0151824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segers ME, Lebeer S. 2014. Towards a better understanding of Lactobacillus rhamnosus GG-host interactions. Microb Cell Fact 13(Suppl 1):S7. doi: 10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tytgat HLP, Douillard FP, Reunanen J, Rasinkangas P, Hendrickx APA, Laine PK, Paulin L, Satokari R, de Vos WM. 2016. Lactobacillus rhamnosus GG outcompetes Enterococcus faecium via mucus-binding pili: evidence for a novel and heterospecific probiotic mechanism. Appl Environ Microbiol 82:5756–5762. doi: 10.1128/AEM.01243-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douillard FP, Ribbera A, Xiao K, Ritari J, Rasinkangas P, Paulin L, Palva A, Hao Y, de Vos WM. 2016. Polymorphisms, chromosomal rearrangements, and mutator phenotype development during experimental evolution of Lactobacillus rhamnosus GG. Appl Environ Microbiol 82:3783–3792. doi: 10.1128/AEM.00255-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider D, Lenski RE. 2004. Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol 155:319–327. doi: 10.1016/j.resmic.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Thomas CM, Nielsen KM. 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 19.Sybesma W, Molenaar D, van IJcken W, Venema K, Kort R. 2013. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol 79:2233–2239. doi: 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 21.Finkel SE, Kolter R. 1999. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci U S A 96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jerison ER, Desai MM. 2015. Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments. Curr Opin Genet Dev 35:33–39. doi: 10.1016/j.gde.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacroix C, Yildirim S. 2007. Fermentation technologies for the production of probiotics with high viability and functionality. Curr Opin Biotechnol 18:176–183. doi: 10.1016/j.copbio.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Johansen E, Øregaard G, Sørensen KI, Derkx PMF. 2015. Modern approaches for isolation, selection, and improvement of bacterial strains for fermentation applications, p 227–248. In Holzapfel W. (ed), Advances in fermented foods and beverages: improving quality, technologies and health benefits. Elsevier, Cambridge, United Kingdom. [Google Scholar]
- 25.Qi Y, Liu X, Liu C-G, Wang B, Hess KR, Symmans WF, Shi W, Pusztai L. 2015. Reproducibility of variant calls in replicate next generation sequencing experiments. PLoS One 10:e0119230. doi: 10.1371/journal.pone.0119230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J, Caggia C, Lähteinen T, Brouns SJJ, Satokari R, von Ossowski I, Reunanen J, Palva A, de Vos WM. 2013. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9:e1003683. doi: 10.1371/journal.pgen.1003683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Skoog T, Einarsdottir E, Kaartokallio T, Laivuori H, Grauers A, Gerdhem P, Hytönen M, Lohi H, Kere J, Jiao H. 2016. Investigation of rare and low-frequency variants using high-throughput sequencing with pooled DNA samples. Sci Rep 6:33256. doi: 10.1038/srep33256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi Y, Chan AP. 2015. PROVEAN Web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saedler H, Reif HJ, Hu S, Davidson N. 1974. IS2, a genetic element for turn-off and turn-on of gene activity in E. coli. Mol Gen Genet 132:265–289. doi: 10.1007/bf00268569. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Oosting M, Deelen P, Ricaño-Ponce I, Smeekens S, Jaeger M, Matzaraki V, Swertz MA, Xavier RJ, Franke L, Wijmenga C, Joosten LAB, Kumar V, Netea MG. 2016. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nat Med 22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, Savijoki K, Nyman TA, Surakka A, Salusjärvi T, de Vos WM, Tynkkynen S, Kalkkinen N, Varmanen P. 2011. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics 10:M110.002741. doi: 10.1074/mcp.M110.002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth A, Yan F, Polk DB, Rao RK. 2008. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 294:G1060–G1069. doi: 10.1152/ajpgi.00202.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan F, Cao H, Cover TL, Washington MK, Shi Y, Liu L, Chaturvedi R, Peek RM, Wilson KT, Polk DB. 2011. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest 121:2242–2253. doi: 10.1172/JCI44031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claes IJJ, Segers ME, Verhoeven TLA, Dusselier M, Sels BF, De Keersmaecker SCJ, Vanderleyden J, Lebeer S. 2012. Lipoteichoic acid is an important microbe-associated molecular pattern of Lactobacillus rhamnosus GG. Microb Cell Fact 11:161. doi: 10.1186/1475-2859-11-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL. 2009. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 1:239–257. doi: 10.1093/gbe/evp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Péter G, Reichart O. 2001. The effect of growth phase, cryoprotectants and freezing rates on the survival of selected micro-organisms during freezing and thawing. Acta Aliment 30:89–97. doi: 10.1556/AAlim.30.2001.1.10. [DOI] [Google Scholar]
- 37.Saarela M, Rantala M, Hallamaa K, Nohynek L, Virkajärvi I, Mättö J. 2004. Stationary-phase acid and heat treatments for improvement of the viability of probiotic lactobacilli and bifidobacteria. J Appl Microbiol 96:1205–1214. doi: 10.1111/j.1365-2672.2004.02286.x. [DOI] [PubMed] [Google Scholar]
- 38.Saxelin M, Grenov B, Svensson U, Fondén R, Reniero R, Mattila-Sandholm T. 1999. The technology of probiotics. Trends Food Sci Technol 10:387–392. doi: 10.1016/S0924-2244(00)00027-3. [DOI] [Google Scholar]
- 39.Drake JW. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci U S A 88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laakso K, Koskenniemi K, Koponen J, Kankainen M, Surakka A, Salusjärvi T, Auvinen P, Savijoki K, Nyman TA, Kalkkinen N, Tynkkynen S, Varmanen P. 2011. Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microb Biotechnol 4:746–766. doi: 10.1111/j.1751-7915.2011.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tripathi P, Dupres V, Beaussart A, Lebeer S, Claes IJJ, Vanderleyden J, Dufrêne YF. 2012. Deciphering the nanometer-scale organization and assembly of Lactobacillus rhamnosus GG pili using atomic force microscopy. Langmuir 28:2211–2216. doi: 10.1021/la203834d. [DOI] [PubMed] [Google Scholar]
- 42.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, Kumar N, Liu Z, Maurel T, Moore B, McDowall MD, Maheswari U, Naamati G, Newman V, Ong CK, Paulini M, Pedro H, Perry E, Russell M, Sparrow H, Tapanari E, Taylor K, Vullo A, Williams G, Zadissia A, Olson A, Stein J, Wei S, Tello-Ruiz M, Ware D, Luciani A, Potter S, Finn RD, Urban M, Hammond-Kosack KE, Bolser DM, De Silva N, Howe KL, Langridge N, Maslen G, Staines DM, Yates A. 2018. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res 46:D802–D808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mikheenko A, Valin G, Prjibelski A, Saveliev V, Gurevich A. 2016. Icarus: visualizer for de novo assembly evaluation. Bioinformatics 32:3321–3323. doi: 10.1093/bioinformatics/btw379. [DOI] [PubMed] [Google Scholar]
- 45.Gu Z, Gu L, Eils R, Schlesner M, Brors B. 2014. circlize implements and enhances circular visualization in R. Bioinformatics 30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 46.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 47.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv 1207.3907 [q-bio.GN].
- 52.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reunanen J, von Ossowski I, Hendrickx APA, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78:2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeuthen LH, Fink LN, Frøkiaer H. 2008. Toll-like receptor 2 and nucleotide-binding oligomerization domain-2 play divergent roles in the recognition of gut-derived lactobacilli and bifidobacteria in dendritic cells. Immunology 124:489–502. doi: 10.1111/j.1365-2567.2007.02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All DNA sequence data are stored at the Gene Expression Omnibus (GEO) database under series accession number GSE123727.