Recombinant protein overproduction often results in oxidative stress, causing deviations from the optimal redox cofactor regeneration balance. This becomes one of the limiting factors in obtaining high levels of heterologous protein production. Overexpression of redox-affecting enzymes has been explored in other organisms, such as Saccharomyces cerevisiae, as a means to fine tune the cofactor regeneration balance in order to obtain higher protein titers. In the present work, this strategy is explored in P. pastoris. In particular, one NADH kinase enzyme from S. cerevisiae (Pos5) is used, either in the cytosol or in mitochondria of P. pastoris, and its impact on the production of a model protein (antibody fragment) is evaluated. A significant improvement in the production of the model protein is observed when the kinase is directed to the cytosol. These results are significant in the field of heterologous protein production in general and in particular in the development of improved metabolic engineering strategies for P. pastoris.
KEYWORDS: NADH kinase, Pichia pastoris, Pos5, redox engineering, heterologous protein production
ABSTRACT
High-level expression and secretion of heterologous proteins in yeast cause an increased energy demand, which may result in altered metabolic flux distributions. Moreover, recombinant protein overproduction often results in endoplasmic reticulum (ER) stress and oxidative stress, causing deviations from the optimal NAD(P)H regeneration balance. In this context, overexpression of genes encoding enzymes catalyzing endogenous NADPH-producing reactions, such as the oxidative branch of the pentose phosphate pathway, has been previously shown to improve protein production in Pichia pastoris (syn. Komagataella spp.). In this study, we evaluate the overexpression of the Saccharomyces cerevisiae POS5-encoded NADH kinase in a recombinant P. pastoris strain as an alternative approach to overcome such redox constraints. Specifically, POS5 was cooverexpressed in a strain secreting an antibody fragment, either by directing Pos5 to the cytosol or to the mitochondria. The physiology of the resulting strains was evaluated in continuous cultivations with glycerol or glucose as the sole carbon source, as well as under hypoxia (on glucose). Cytosolic targeting of Pos5 NADH kinase resulted in lower biomass-substrate yields but allowed for a 2-fold increase in product specific productivity. In contrast, Pos5 NADH kinase targeting to the mitochondria did not affect growth physiology and recombinant protein production significantly. Growth physiological parameters were in silico evaluated using the recent upgraded version (v3.0) of the P. pastoris consensus genome-scale metabolic model iMT1026, providing insights on the impact of POS5 overexpression on metabolic flux distributions.
IMPORTANCE Recombinant protein overproduction often results in oxidative stress, causing deviations from the optimal redox cofactor regeneration balance. This becomes one of the limiting factors in obtaining high levels of heterologous protein production. Overexpression of redox-affecting enzymes has been explored in other organisms, such as Saccharomyces cerevisiae, as a means to fine tune the cofactor regeneration balance in order to obtain higher protein titers. In the present work, this strategy is explored in P. pastoris. In particular, one NADH kinase enzyme from S. cerevisiae (Pos5) is used, either in the cytosol or in mitochondria of P. pastoris, and its impact on the production of a model protein (antibody fragment) is evaluated. A significant improvement in the production of the model protein is observed when the kinase is directed to the cytosol. These results are significant in the field of heterologous protein production in general and in particular in the development of improved metabolic engineering strategies for P. pastoris.
INTRODUCTION
High-level expression and secretion of heterologous proteins in yeast have been reported to cause a metabolic burden that can significantly impact energy metabolism and alter the central carbon metabolism flux distribution (1, 2). Producing strains may not cope with the additional demand of ATP, NADPH, and precursors for de novo biosynthesis of amino acids, thereby leading to suboptimal cell fitness and reduced production yields (3). In addition, the folding, posttranslational modifications, and secretion processes of complex proteins demand many resources, particularly NADPH, which is required for disulfide bond formation and alleviation of endoplasmic reticulum (ER) oxidative stress (4). Thus, overproduction of recombinant proteins would result in altered redox cofactor state and, specifically, a reduction in NADPH availability. Such alterations in redox cofactor balance have a strong impact on cell metabolism (5). Therefore, strain engineering strategies targeting redox metabolism have been successfully applied to improve Escherichia coli (6, 7), Saccharomyces cerevisiae (8, 9), and Pichia pastoris (10–12) strains for a range of different applications.
NADPH availability is tightly related to biomass yields and recombinant protein production yields (13). Driouch et al. (14) reported that Aspergillus niger strains overproducing recombinant proteins show higher fluxes through the oxidative branch of the pentose phosphate pathway (PPP), which is the main cytosolic NADPH generation pathway. Also, Nocon et al. (12) found overexpressed genes coding for enzymes of the oxidative branch of the PPP, which led to higher productivity of recombinant cytosolic human superoxide dismutase (hSOD). Indeed, a previous study identified several metabolic engineering targets for improving recombinant protein production using a genome-scale metabolic model (11). Interestingly, about the 50% of the identified targets pointed toward NADPH supply reactions (15).
Based on the key metabolic role of NADPH on protein synthesis and secretion and the supporting evidence from previous studies pointing at the positive effect that its increased supply seems to have on recombinant protein production, we have investigated the impact of the overexpression of a heterologous gene encoding a NADH kinase on a recombinant P. pastoris strain secreting an antibody fragment (Fab). Previous studies in our research group reported an increase in Fab-specific productivity under reduced oxygen availability conditions (16). This was concomitant with a shift to a respirofermentative metabolism, as reflected in the generation of ethanol and arabitol for cofactor reoxidation (17). Furthermore, parallel metabolomics analyses revealed that the reduced oxygen availability for the electron transport chain leads to higher NADH/NAD+ ratios under such hypoxic conditions (18). In this context, we postulate that the NADH excess found under hypoxic conditions could be a potential source of electrons for NADPH regeneration; therefore, the physiological effects of the ectopic NADH kinase might be boosted under hypoxia in comparison to the reference normoxic condition. In order to test our hypothesis, redox-engineered strains were grown on glycerol and glucose under normoxic conditions as well as on glucose under hypoxic conditions. Overall, we aimed to investigate the combined effect of a process strategy (hypoxic conditions) and metabolic engineering strategy to improve the production of a secreted recombinant protein.
Moreover, we used the genome-scale metabolic model to evaluate the experimental physiological data sets obtained in chemostat cultivations and assist in the metabolic interpretation of the observed macroscopic changes.
(This research was conducted in partial fulfillment of the requirements for a Ph.D. from the Universitat Autònoma de Barcelona by M. Tomàs-Gamisans [19] and by S. Monforte [20].)
RESULTS
Cytosolic and mitochondrial overexpression of POS5 and its effect on recombinant Fab secretion.
The codon-optimized POS5 gene and its truncated version devoid of its native mitochondrial signal peptide-encoding sequence were overexpressed under the control of the constitutive GAP promoter into a P. pastoris X-33 strain expressing the genes encoding the 2F5 antibody fragment under the control of the same promoter, aiming for the targeted cooverexpression of NADH kinase in the mitochondria or in the cytosol (mPOS5 and cPOS5, respectively). A set of 12 independent clones of each cooverexpressing strain were verified for integration of mPOS5 or cPOS5 prior to the screening in baffled shake flasks.
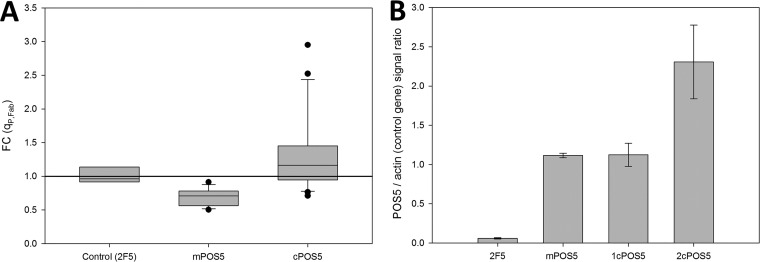
The two series of Fab-producing clones overexpressing each of the POS5 forms were first tested in a small-scale screening at a shake flask scale. Product titers and biomass were measured after 24 h of cultivation to check the effect of coexpression of mPOS5 or cPOS5 on recombinant Fab secretion. The average Fab yields of each series of 12 clones were normalized to those obtained from the reference strain X-33/2F5, as shown in Fig. 1A. Coexpression of the two POS5 gene forms demonstrated a largely unchanged recombinant protein secretion capacity. Interestingly, the average Fab yields of cPOS5 clones showed a high standard deviation as a result of two distinct clone populations, one with the same behavior as the control strain (fold change [FC], 1.07 ± 0.21) and another showing significantly higher product yields (FC, 2.34 ± 0.52). A plausible explanation for the observed clonal variation would be that isolated transformants differ in the dosage of the coexpressed cPOS5 gene. In order to test this hypothesis, the recombinant cPOS5 gene dosage was determined by droplet digital PCR (ddPCR) for a representative sample of each clone subpopulation, i.e., one giving significantly increased Fab yields and the other showing no significant effect on product yield compared to the reference strain. Notably, the cPOS5 clone population showing significantly increased product yields corresponded to those clones harboring 2 copies of the cPOS5 expression cassette (Fig. 1B).
FIG 1.
Representation of the results of clone screening and selection. (A) Specific Fab production (qP,Fab) was measured and normalized to the reference strain X-33/2F5 and expressed as fold changes (FC). Dots indicate individual extreme clones, boxes indicate quartiles, and whiskers indicate variability outside the upper and lower quartiles. (B) A representative clone of each population was analyzed for determining the POS5 gene copy number. Error bars show the standard deviation of the average from at least three different clones.
Physiological characterization of the POS5-cooverexpressing strains growing in chemostat cultures.
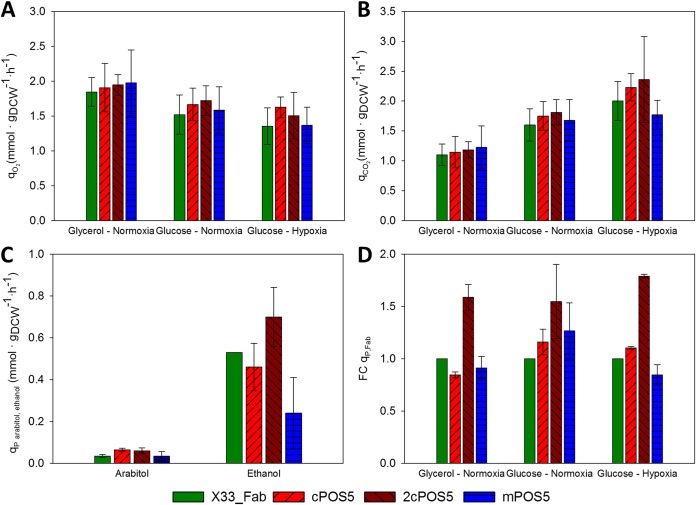
In order to study the effects of altering NADPH generation on cell physiology and Fab extracellular production under different environmental conditions, the reference strain X-33/2F5, as well as representative clones of the mPOS5 strain (mitochondrial expression of POS5), the cPOS5 and 2cPOS5 strains (cytosolic expression with one and two copies of POS5, respectively), were cultivated in carbon-limited chemostat bioreactors using glycerol or glucose as the carbon source under normoxic conditions, as well as under hypoxia when using glucose (see Table S2 in the supplemental material for a summary of physiological growth parameters). Since Pos5p catalyzes an NADPH-generating reaction, an alteration of the redox cofactor balance can be expected. In order to check whether POS5 overexpression has a significant impact on the redox cofactor balance, the NADPH/NADP+ ratios were measured and calculated for all strains under the tested growth conditions (Fig. 2). The reference strain X-33/2F5 showed the lowest ratios under all three growth conditions. Strains cooverexpressing mPOS5 and cPOS5 had NADPH/NADP+ values comparable to those of the reference strain when grown on glycerol, whereas this value was 2-fold higher in the 2cPOS5 strain. Cooverexpression of POS5 had a higher impact in cells grown on glucose than on glycerol, as shown in Fig. 2A and B. In this case, cPOS5 and mPOS5 strains showed a trend of increasing NADPH/NADP+ ratios of about 20% and 30% compared to the reference strain, respectively, even though this effect was not statistically significant. The 2cPOS5 strain also had a higher impact on the NADPH/NADP+ ratio in glucose-grown cells than in glycerol-grown cells. Notably, when the oxygen supply was reduced, this adaptation pattern was further accentuated, i.e., NADPH/NADP+ ratios were comparatively higher for all four strains (Fig. 2C). Such increase in the NADPH/NADP+ ratio observed under hypoxic conditions may be a consequence of the NADH excess or accumulation observed under these conditions (18), which is generally converted to ethanol or other by-products such as arabitol (16, 21). Thus, an increased availability of NADH under hypoxic conditions would allow higher conversions to NADPH by Pos5p and, consequently, increased NADPH/NADP+ ratios.
FIG 2.
(A to C) Representation of NADPH/NADP+ molar ratios in all the strains for growth on glycerol (A) or glucose (B) under normoxic conditions and under hypoxic conditions (C).
Such cofactor balance alteration due to POS5 overexpression had a further impact on the physiological growth profile of the POS5-engineered strains (Fig. 3). Although such an impact was not significant in all strains and under all growth conditions, some tendencies can be appreciated (Fig. 3). In particular, the specific oxygen uptake rate (qO2) showed a tendency to increase under all growth conditions in both cytosol- and mitochondrion-directed POS5-overexpressing strains. Notably, 2cPOS5 showed a higher qO2 than the other strains under most of the tested conditions, probably reflecting an increased activity of the respiratory chain due to the additional ATP demand for consumption in the NADH kinase reaction (see Discussion for further discussion). Similarly, the specific CO2 production rate (qCO2) showed an increasing trend as a result of higher cytosolic NADH kinase activity levels, both under normoxic and hypoxic conditions, whereas the mPOS5 strain seemed to show an opposite trend, i.e., reduced qCO2 compared to that of the reference strain (Fig. 3). Notably, the variation in CO2-specific production rates, although not statistically significant, appears to be at the expense of biomass yields (YXS), which show the opposite pattern, a statistically significant reduction for 2cPOS5 and an increase in mPOS5 under hypoxic conditions (Table S2).
FIG 3.
Main macroscopic growth parameters for all strains and chemostat cultivation conditions. (A) Specific O2 consumption rate. (B) Specific CO2 production rate. (C) Specific by-product generation rate under glucose-hypoxic conditions. (D) Fold change of Fab productivity of the generated strains in comparison to the X-33/2F5 strain. Each measure shown is the mean ± standard deviation of the results from two different experiments.
As expected, by-product formation was detected only under respirofermentative conditions attained under hypoxia. In particular, while both the cPOS5 and 2cPOS5 strains showed similar or higher arabitol- and ethanol-specific production rates, ethanol formation decreased in the mPOS5 strain compared to that in the reference strain (Fig. 3C).
POS5 overexpression also had a clear impact on Fab production (Fig. 3D). When grown on glycerol, only 2cPOS5 improved Fab productivity (qP,Fab) significantly with respect to the reference strain. However, all three redox-engineered strains showed significantly enhanced qP,Fab when grown on glucose under normoxic conditions, with 2cPOS5 being the strain with the best performance (1.55-fold increase compared to the reference strain). Under oxygen-limiting conditions, although mPOS5 showed a decrease in productivity, the cytosolic POS5-overexpressing strains improved Fab production up to 1.8-fold compared to the reference strain grown under the same conditions, i.e., showing an additive effect of hypoxia and cPOS5 overexpression.
Cytosolic POS5 overexpression further enhances glycolysis upregulation caused by hypoxia.
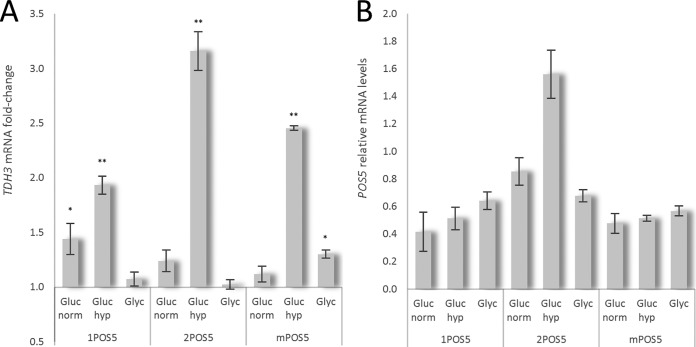
To gain further insight into the additive effect of hypoxia and cytosolic POS5 overexpression observed in chemostat cultures, the relative transcriptional levels of TDH3 (as marker gene for glycolysis) and POS5 were determined by ddPCR for all strains and growth conditions tested (Fig. 4). Previous studies demonstrate that, in contrast to S. cerevisiae, glycolysis is transcriptionally upregulated by hypoxia in P. pastoris (17, 22). Glucose-grown cells showed an increase in TDH3 transcripts when POS5 was overexpressed compared to reference strain. Specifically, TDH3 was upregulated 1.44- and 1.24-fold in the 1-copy and 2-copy cPOS5 strains, respectively, under normoxic conditions, whereas the mPOS5 strain showed only a slight increase (1.12-fold) in TDH3 mRNA levels. Such upregulation of TDH3 was further enhanced by hypoxic conditions, with 1.93-, 3.16-, and 2.46-fold increases observed in the 1cPOS5, 2cPOS5, and mPOS5 strains, respectively, compared to the reference strain. Notably, such results correlate with the increased Fab secretion observed under this condition. In contrast, no significant changes were observed in TDH3 transcriptional levels when glycerol was used as a carbon source in the 1cPOS5 and 2cPOS5 strains compared to the reference strain, except for the mPOS5 strain, which showed a 1.3-fold increase.
FIG 4.
Transcriptional analysis of chemostat cultivations. (A) Relative changes of TDH3 mRNA levels in POS5 strains are expressed as fold change from the TDH3 levels in the reference strain. (B) POS5 mRNA levels are expressed as relative expression levels to ACT1 (POS5/ACT1 signal) for each individual strain and condition (*, P < 0.05; **, P < 0.01) compared to the control strain. Error bars show the standard deviation of the results from three technical replicates. Gluc, glucose; norm, normoxia; Glyc, glycerol; hyp, hypoxia.
Concerning POS5 transcription, the 2cPOS5 strain showed the highest POS5 transcriptional levels under all tested conditions. As expected, POS5 mRNA levels in glucose-grown 2cPOS5 cells were at least 2-fold higher than in the 1cPOS5 strain. Moreover, as POS5 was expressed under the control of the GAP promoter, i.e., the TDH3 promoter, POS5 expression was 1.8-fold higher in 2cPOS5 cells grown under glucose/hypoxia than that in glucose/normoxia-grown cells. Such a hypoxic boost effect was not observed in glucose-grown 1cPOS5 cells, suggesting that ectopic POS5 mRNA levels are too low to cause a significant metabolic effect in these cells. Similarly, POS5 expression levels in glucose/normoxia-grown mPOS5 cells did not show significant differences between cultivation conditions.
Notably, POS5 transcriptional levels in glycerol-grown cells appeared to be on the same range as in glucose/normoxia-grown cells. This is in contradiction to previous studies reporting that TDH3 promoter expression level is around two-thirds of that in glucose (23, 24). Such differences could be related to the different growth conditions used in previous studies (shake flask cultures) and the different analytical method employed in our study (ddPCR).
In silico interpretation of POS5 overexpression physiological impact.
One of the applications of genome-scale metabolic models (GSMM) is to assist in the interpretation of biological data to reinforce or discard hypotheses (25). We used the iMT1026 v3.0 GSMM for P. pastoris (25), which enables improved prediction capabilities for growth on glycerol compared to the original version (26).
The effect of Fab overproduction on flux distribution was tested by performing a series of simulations successively increasing Fab production constraints (employing flux scanning based on enforced objective function [FSEOF]). NADPH turnover ratios were calculated for the resulting predicted fluxes. A clear correlation between Fab production and this cofactor turnover was obtained due to the increased demand of amino acid biosynthesis for Fab production (Fig. S1).
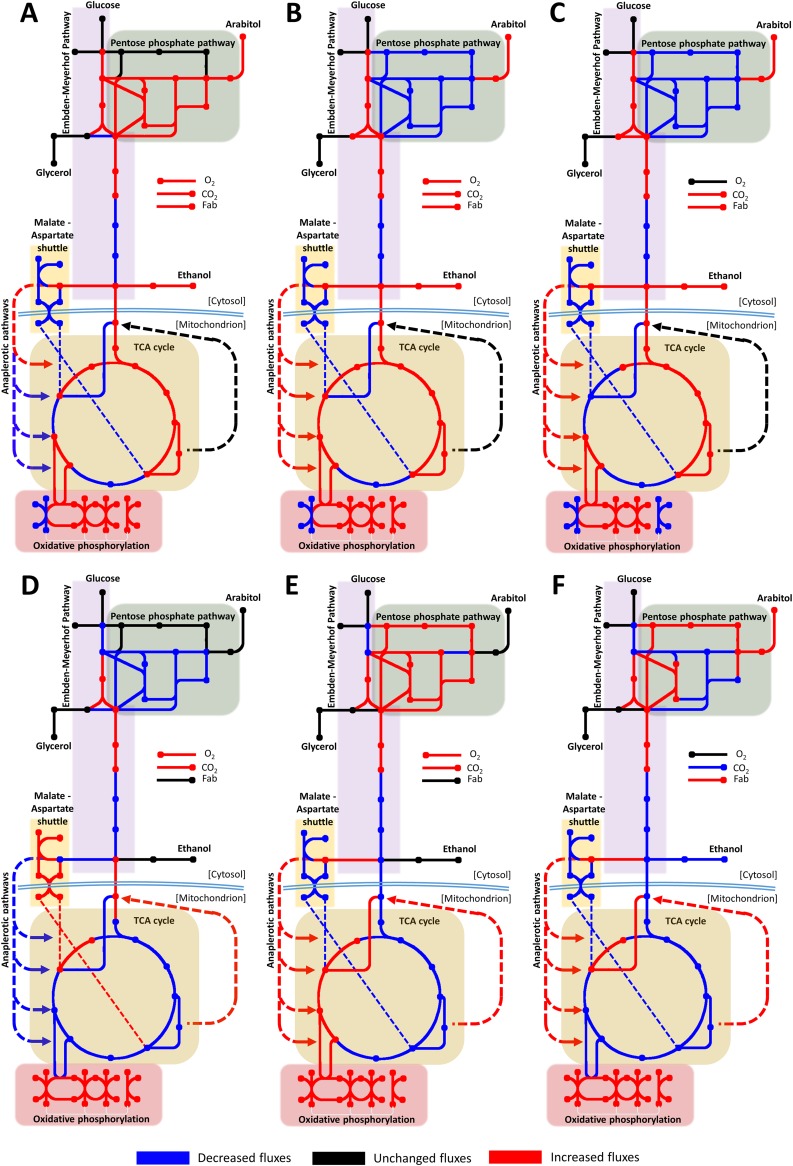
The metabolic impact of POS5 overexpression was also modeled by constraining successively increasing fluxes through the specific NADH kinase reaction (cytosolic or mitochondrial) and using minimization of metabolic adjustment (MOMA) for assessing the new distribution. As a result, metabolic fluxes through some pathways are predicted to be up- or downshifted concomitantly with an increased flux through an NADH kinase reaction. Mitochondrion- or cytosol-directed POS5 overexpression impacts differently on metabolic flux distribution, as it is affecting different compartmentalized NAD(P)H pools. For example, cytosolic Pos5p synthesis supplies an important fraction of NADPH under glucose-growing conditions (Fig. 5B and C), and consequently, the flux through the oxidative branch of the pentose phosphate pathway decreased. In contrast, mitochondrial NADPH kinase synthesis would not be able to supply enough cytosolic NADPH, and therefore, the flux of the NADPH-generating reactions of the pentose phosphate pathway is enhanced (Fig. 5D and E). Another predicted and important difference between cytosolic and mitochondrial Pos5p overexpression is the increased activity of the glycolysis pathway (coherent with experimental ddPCR data) and tricarboxylic acid (TCA) cycle when the cytosolic NADPH kinase is overexpressed, while mitochondrial overexpression is predicted to cause a decreased flux through this pathway. However, some predicted changes are common between cPOS5 and mPOS5 strains, as in both cases, there is an increase in the activity of the oxidative phosphorylation that supplies the additional ATP demanded in the NADPH kinase reaction. The increased electron transfer to the respiratory chain is predicted to be through NADH oxidation in the mPOS5 strain, but for the cPOS5 strain, succinate oxidation to fumarate would be the additional electron supply. Overall, increases in oxygen consumption, CO2 production, by-product formation, and Fab production are predicted for all the conditions tested in simulations for the cPOS5 strain. Similarly, an increase in qO2 and qCO2 during growth on glycerol and glucose-normoxia is predicted for the mPOS5 strain, whereas no significant changes are predicted in terms of by-product formation or Fab production for this strain. Conversely, mitochondrial overexpression of NADH kinase leads to a reduction in CO2, ethanol, and Fab production under hypoxic conditions, which is in line with the experimentally observed behavior.
FIG 5.
(A to F) Graphical representation of the predicted metabolic flux redistribution when overexpressing the NADH kinase in the cytosol (A to C) or mitochondria (D to F). Cells were grown in glycerol (A and D), glucose under normoxic conditions (B and E), and glucose under hypoxia (C and F). Blue, black, and red lines denote reactions with decreased, unchanged, and increased metabolic fluxes, respectively.
DISCUSSION
POS5 overexpression increases NADPH availability and recombinant protein production.
Several studies have addressed the impact of heterologous protein expression on the metabolism and cell physiology of P. pastoris (27) and how environmental conditions can modulate the product yields (17, 28). Heyland et al. (13) postulated and Nocon et al. (11, 12) provided strong evidence that increasing flux through the PPP allows the supply of additional NADPH, thereby compensating for the extra demand caused by biosynthetic processes involved in heterologous protein production. In fact, our in silico simulations indicate that an increase in Fab extracellular production correlates with higher NADPH turnover rates. Redox-engineered strains are able to generate additional NADPH compared to the reference X-33/2F5 strain, resulting in increased NADPH/NADP+ ratios. Moreover, a higher POS5 gene dosage further increased POS5 transcriptional levels, correlating with higher NADPH/NADP+ ratios and Fab protein production. Higher NADPH/NADP+ ratios may reflect more NADPH available for cellular processes, not only for biosynthesis of recombinant protein, but also for its potential use in protein folding and ER oxidative stress response processes involved in protein secretion (14). Therefore, although POS5 overexpression leads to a drain in cell energy resources (i.e., ATP consumption), it ensures a nonlimiting supply of NADPH, which has been demonstrated to allow for increased recombinant protein production yields (12). Our in silico calculations show increased Fab secretion when the ectopic Pos5p NADH kinase is targeted to the cytosol. This may indicate that Fab biosynthesis and secretion could compensate for the cofactor perturbation (boosted NADPH levels) by draining such NADPH excess, thereby restoring the redox cofactor original state.
Metabolic impact of POS5 overexpression.
Our results strongly suggest that Pos5p NADH kinase overexpression perturbs cofactor balance. This is further reflected in the differences observed in growth profiles of POS5-overexpressing strains compared to the reference strain, pointing to an impact in the distribution of fluxes through the cell’s metabolic network. Previous studies in other yeast and fungi have reported that the overexpression of NADH kinases has a strong effect on metabolic flux distribution (29–31), as also predicted by the simulations performed with iMT1026 v3.0. Noteworthy, the impermeability of organelle membranes to NAD(P)H leads to cofactor reoxidation in the same compartment where they are reduced (32). Thus, different effects of the cofactor perturbation are expected in the cPOS5 and mPOS5 strains. POS5 overexpression provides a source of NADPH in addition to the oxidative branch of the PPP, which is the main cytosolic NADPH-generating pathway in yeast (33). As extra NADPH is supplied by the NADH kinase, flux through the oxidative branch of the PPP is predicted to decrease, as observed when POS5 overexpression is targeted to the cytosol of S. cerevisiae (30). Indeed, in S. cerevisiae, NADPH inhibits ZWF1, the first step in the oxidative PPP (34). Therefore, the increased levels of NADPH in cPOS5 (and 2cPOS5) would be coherent with the predicted reduction of flux through the oxidative branch of the PPP. Conversely, mitochondrial overexpression of POS5 would result in an increased flux through the PPP. Part of the carbon flux would be redirected to the mitochondria for generating the required NADH surplus at the expense of cytosolic NADH yields. This reduction in cytosolic NADH generation would be compensated for by the additional NADPH synthesized. Thus, enzymes able to use both NADH and NADPH would turn their specificity to NADPH. Therefore, despite the production of additional mitochondrial NADPH, the impermeability of mitochondrial membrane to redox cofactors would force cytosolic NADPH generation pathways to supply the required NADPH for compartment-specific biosynthetic processes. The correlation between the flux through the oxidative branch of the pentose phosphate pathway and biomass yields has been widely reported (33, 35). Accordingly, the predicted flux increase and decrease through the PPP as a consequence of mPOS5 and cPOS5 overexpression, respectively, are in agreement with the biomass yields observed in chemostat cultivations, being an increase in YXS yield in mPOS5 strains and a reduction in cPOS5 strains (Table S2). Similarly, in other species, while mitochondrial POS5-overexpressing strains show an increase in YXS (5, 29, 31), the cytosolic overexpression of the NADH kinase results in reduced biomass yields. It is noteworthy to mention that the scaled reduced costs of Fab production over biomass generation are narrow (1.01 × 10−4), and therefore, differences in biomass yields are mainly a consequence of POS5 overexpression and the concomitant redistribution of metabolic fluxes and energy consumption, rather than increased Fab yields.
Redox cofactor balance and energy metabolism are very closely linked in the respiratory chain. It is therefore plausible that a perturbation in redox cofactor levels would cause a metabolic flux redistribution to restore the energy supply capacity of the cells. Moreover, in addition to the redox cofactor imbalance created by a surplus of NADPH at the expense of NADH, the Pos5p-catalyzed NADH kinase reaction is ATP consuming. According to the performed simulations, higher fluxes in oxidative phosphorylation would compensate for this ATP drain. These predictions are also in agreement with the increased oxygen consumption rates of the mutant strains during the chemostat cultivations. Nevertheless, in the cPOS5 strain, part of the cytosolic NADH generated is consumed in the NADH kinase reaction and cannot be used for biosynthetic purposes nor transported by mitochondrial redox shuttles to further deliver its electrons into the respiratory chain. Despite the higher TCA cycle activity and consequent increase in mitochondrial NADH generation, it would not provide enough reducing power for the electron transport chain, and a reduction in the NADH electron transfer and an increase in alternative electron delivery mechanisms (i.e., succinate oxidation to fumarate) are predicted for the cPOS5 strain. Since the NADH demand is located in mitochondria in mPOS5, the compensation of the drain would rely on flux readjustments in mitochondrially located reactions. In this case, an activation of the malic enzyme as well as an increase in the anaplerotic feed of TCA cycle intermediates would supply the additional NADH, enabling increased electron transfer to the respiratory chain. As with S. cerevisiae and Aspergillus nidulans, we do not observe a reduction in biomass yields (29, 30).
When grown under hypoxic conditions, the additional supply of ATP required by Pos5p is limited due to the restricted oxygen availability constraining the respiratory chain activity. Simulations predict that when cPOS5 strain cells grow under hypoxia, the ATP drain caused by Pos5p activity cannot be completely compensated, leading to a decrease in cell fitness and increased hypoxic effects. Accordingly, experimental data show an increase in by-product formation when cPOS5 is overexpressed. These results are also supported by the ddPCR analyses performed, showing a transcriptional adaptation of the glycolytic pathway under hypoxic conditions positively correlated with the gene copy number (and transcriptional levels) of POS5. TDH3 has been reported to increase its transcription levels in hypoxia (Baumann et al. [17]). Our results strongly support that increasing NADPH availability by overexpressing POS5 enhances this hypoxic effect. Consequently, since the ectopic expression of POS5 is also under the transcriptional control of the TDH3 promoter, POS5 positively feeds back its own overexpression under low-oxygen-availability conditions.
Conversely, despite the oxygen limitations, the mPOS5 strain would be able to compensate for the drained ATP by supplying additional mitochondrial NADH to the respiratory chain, thereby increasing ATP production. This strain, similarly to A. nidulans, is able to overcome the ATP drain and increase biomass yields with respect to the control strain, even under hypoxic conditions (29). Although P. pastoris is commonly classified as a Crabtree-negative yeast, it can produce a certain amount of ethanol and other by-products (35), particularly under hypoxic conditions (17). By-product formation is a consequence of limitations in TCA cycle and oxidative phosphorylation capacities, leading to an excess of reduced NAD(P)H that the cell is not able to reoxidize by the respiratory pathway (35, 36). The mPOS5 strain showed a decrease in ethanol secretion due to the reduction in available mitochondrial NADH, while arabitol production remained comparable to that in the reference X-33/2F5 strain. The cPOS5 strains showed increased arabitol and ethanol production both in experimental data and simulations. Under hypoxic conditions, increased NADH kinase levels would convert the excess of NADH to NADPH (as reflected in the experimentally determined increased NADPH/NADP+ ratio); this NADPH surplus would be subsequently reoxidized through the generation of arabitol. In addition, simulations indicate an increase in TCA cycle flux leading to enhanced NADH generation. Due to the reduced capacity of oxidative phosphorylation caused by the oxygen limitation, the additional NADH generated has to be reoxidized, forming ethanol, in agreement with the experimental observation.
To conclude, in this study, the S. cerevisiae POS5 gene, encoding an NADH kinase, was overexpressed in P. pastoris, either directed to the cytosol or to the mitochondria. The physiological characterization of these strains in chemostat cultivations showed a clear effect of POS5 overexpression on the redox cofactor balance. Indeed, POS5 overexpression increased the NADPH/NADP+ ratio in all the strains and under all conditions tested. Furthermore, the strain containing two copies of POS5 integrated in the cell’s genome (2cPOS5) showed the highest increase in NADPH/NADP+ ratio compared to the reference strain. This strongly supports a positive correlation between POS5 gene dosage and NADPH availability. Moreover, this correlation can also be observed in a comparison of the strain performance, where 2cPOS5 strain cells showed the greatest fold change increase in Fab productivity. These results are also in agreement with the performed simulations, which show a positive correlation between NADPH turnover and Fab production as well as increased Fab productivity when flux through the cytosolic NADH kinase reaction is increased.
The different behaviors of the mPOS5 and cPOS5 strains indicate the complexity of cell metabolism with organelle membranes impermeable to redox cofactors and highlight the importance of directing enzymes to the appropriate compartment when designing metabolic engineering strategies.
As a result of POS5 overexpression, metabolic fluxes through the central carbon metabolism redistribute. Notably, redox-engineered strains showed higher oxygen requirements concomitant with increased oxidative phosphorylation in order to replenish the ATP pools drained in the reaction catalyzed by the NADH kinase. Consequently, these strains, particularly the cPOS5 strain, are more sensitive to O2 (i.e., show a lower threshold for the onset of respirofermentative metabolism) and showed more extreme/drastic hypoxic effects (increased by-product formation). In fact, the effects of POS5 overexpression are boosted under hypoxic conditions, and redox-engineered strains show higher NADPH/NADP+ ratios and Fab productivity. Moreover, this effect is particularly notorious when the gene copy number of cPOS5 is increased, remarking how important gene dosage is while performing strain modifications at a metabolic level. The gene dosage effect is supported by the ddPCR results, with 2cPOS5 showing the highest fold change in TDH3 transcription (the glycolytic marker) and the highest POS5 transcription level. Notably, both in silico flux distributions and macroscopic growth parameters of the POS5-engineered strains were coherent in terms of increased demand of oxygen, CO2, and by-product generation profiles, as well as Fab productivity, revealing iMT1026 v3.0 as a useful tool for consistently assessing the interpretation of the cultivation results, as well as taking into account the effect of growth conditions on the metabolic phenotype of the engineered strains.
Overall, we demonstrated the impact of redox cofactor perturbation in cell metabolism and provided further evidence of NADPH metabolism as a key cell engineering target for improved recombinant protein production. Nonetheless, further studies are needed in order to dissect the actual contributions of protein folding and secretion (versus protein synthesis) to the increased NADPH demand. In this respect, future comparative studies of the impact of NADPH metabolism perturbation on the production of the same model protein produced in the cytosol or secreted should bring further insights. In addition, future development of production processes (i.e., high-cell-density fed-batch cultivations) using POS5-engineered strains would also benefit from regulated POS5 expression (e.g., be coinduced with the recombinant product gene, expressed under the control of an easily tunable promoter) to minimize the potential metabolic burden associated with the GAP-driven constitutive expression of this gene.
MATERIALS AND METHODS
Strain generation.
A Pichia pastoris X-33 (Thermo Fisher Scientific)-derived strain expressing multiple copies of the genes encoding the human antigen-binding fragment (Fab) 2F5 under the transcriptional control of the constitutive GAP promoter and with the α-mating factor secretion signal sequence from Saccharomyces cerevisiae (37) was used in this study.
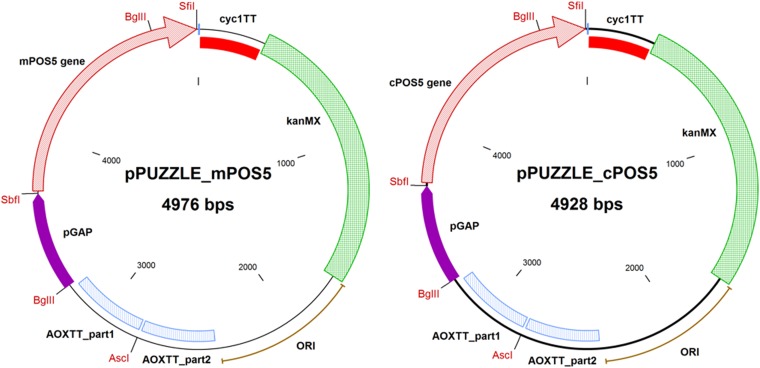
The S. cerevisiae POS5 gene, encoding the mitochondrial NADH kinase Pos5p (38, 39), was codon optimized for heterologous expression in P. pastoris, synthesized by GeneArt (Thermo Fisher Scientific), and cloned into a pPUZZLE vector (40) under the control of GAP promoter, thereby generating vector pPUZZLE_mPOS5 (Fig. 6). Similarly, an analogous construction, pPUZZLE_cPOS5, was constructed expressing a 5′-truncated POS5 excluding the first 48 bp coding for the N-terminal 16 amino acids, allowing for cytosolic Pos5p localization. Escherichia coli DH5α was used for plasmid propagation.
FIG 6.
Plasmid maps for pPUZZLE_mPOS5 and pPUZZLE_cPOS5. In red, the restriction enzymes used were SbfI and SfiI for cloning mPOS5 and cPOS5, AscI for plasmid linearization, and BglII for plasmid ligation verification. pPUZZLE contains the kanMX gene encoding kanamycin resistance (E. coli) and Geneticin G418 resistance (P. pastoris). ORI, origin of replication.
P. pastoris X-33/2F5 strain transformation and recombinant clone isolation were performed as described in reference 40. The presence of an integrated expression cassette into the host genome was confirmed by colony PCR (41) using the primer pairs described in Table S1.
Clone screening at a small scale.
A set of 12 recombinant clones for each strain construct were screened for growth and Fab 2F5 production in triplicate baffled shake flask cultures using glucose minimal medium, as described by Baumann et al. (40).
Chemostat cultivations.
Two independent carbon-limited chemostat cultivations were performed for each strain under three different growing conditions, using as a carbon source either glycerol, glucose under normoxic conditions (100% air in the inlet gas composition), or glucose under hypoxic conditions (25:75 of air/N2 in the inlet gas composition), as previously described (42). Cultivations were performed at a working volume of 1 liter in a 2-liter benchtop Biostat B (B. Braun Biotech International) bioreactor. Operational conditions were set to 25°C, 700 rpm, 1 vol/vol/min inlet gas flow, 20,000 Pa overpressure, 0.1 h−1 dilution rate (D), and pH 5.0 controlled by the addition of 15% (vol/vol) NH4. Samples were taken at the 3rd, 4th, and 5th residence times for cell density monitoring, Fab titers, extracellular metabolites, and dry cell weight (DCW) analyses. The off-gases were cooled dawn in a condenser at 4°C and further desiccated in two silica gel columns. The off-gas O2 and CO2 concentrations were measured using BCP-O2 (zirconium dioxide) and BCP-CO2 (infrared) BlueSens gas analyzers, respectively.
For reactor inoculation, strains were cultivated in a 1-liter baffled Erlenmeyer flask containing 150 ml YPG broth (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 1% [wt/vol] glycerol) and antibiotic (100 μg·liter−1 zeocin for the control strain or 500 μg·liter−1 Geneticin G418 for NADH kinase recombinant clone selection) at an optical density at 600 nm (OD600) between 0.15 and 0.30. Precultures were incubated at 25°C under 130 rpm for 16 to 24 h.
Batch medium composition was as previously described (16). Briefly, it contained 40 g·liter−1 glycerol, 1.8 g·liter−1 citric acid, 12.6 g·liter−1 (NH4)2HPO4, 0.5 g·liter−1 MgSO4·7H2O, 0.9 g·liter−1 KCl, 0.02 g·liter−1 CaCl2·2H2O, 4.6 ml·liter−1 trace salt stock solution, 2 ml·liter−1 biotin solution (0.2 g·liter−1), and 250 μl·liter−1 Glanapon 2000 antifoam (Bussetti). The chemostat medium was also adapted from reference 16. Briefly, it contained 50 g·liter−1 carbon source (glycerol or glucose), 0.92 g·liter−1 monohydrate citric acid, 4.35 g·liter−1 (NH4)2HPO4, 0.65 g·liter−1 MgSO4·7H2O, 1.7 g·liter−1 KCl, 0.01 g·liter−1 CaCl2·2H2O, 1.6 ml trace salt solution, 1 ml biotin solution (0.2 g·liter−1), and 200 μl·liter−1 Glanapon antifoam. The trace salt solution was composed of 6.0 g·liter−1 CuSO4·5H2O, 0.08 g·liter−1 NaI, 3.36 g·liter−1 MnSO4·H2O, 0.2 g·liter−1 Na2MoO4·2H2O, 0.02 g·liter−1 H3BO3, 0.82 g·liter−1 CoCl2·6H2O, 20 g·liter−1 ZnCl2, 65 g·liter−1 FeSO4·7H2O, and 5.0 ml H2SO4 (95 to 98% [wt/wt]). The medium pH was adjusted to 5.0 with 6 N HCl.
Analytical methods.
(i) Biomass concentration. Cell density was assessed by the optical density at 600 nm measured in a DR3900 spectrophotometer (Hach Lange GMBH). Dry cell weight (DCW) was measured by gravimetric methods, as follows: 2 to 10 ml of sample was filtered in glass fiber prefilters (Merck Millipore) preweighed after drying at 105°C for 24 h. Each filter was washed twice with 10 ml of distilled water, dried at 105°C for 24 h, cooled in a desiccator, and weighed.
(ii) Fermentation product analysis.
Citric acid, glucose, glycerol, arabitol, succinic acid, acetic acid, and ethanol were analyzed by high-performance liquid chromatography (HPLC) in an UltiMate 3000 liquid chromatography system (Dionex) using an ICSep ICE-Coregel 87H3 ion exchange column (Transgenomic) and a Waters 2410 refraction index detector. Sulfuric acid (6 mM) was used as continuous phase at 0.5 ml/min flow and 20-μl sample injection volume. Data were analyzed using the CROMELEON software (Dionex).
(iii) Quantification of Fab. Fab 2F5 was quantified by enzyme-linked immunosorbent assay (ELISA) in 96-well Immuno Plates (Nunc; Thermo Scientific), as described by Gasser and coworkers (37). Briefly, plates were subjected to an overnight precoating of Fab-specific anti-human IgG primary antibody (I5260; Sigma) in phosphate-buffered saline (PBS) buffer (1:1,000). Then, plates were washed three times with PBS–1% Tween 20, and samples and Fab standard (Bethyl, Inc.) were diluted in PBS buffer containing 10% (wt/vol) bovine serum albumin (BSA; Sigma) and 0.1% (vol/vol)–Tween 80. Plates were incubated for 2 h, washed again with PBS–1% Tween 20 three times, and incubated for 1 h after the addition of anti-human kappa light-chain (bound)-alkaline phosphatase (Sigma) secondary antibody. Plates were washed with PBS–1% Tween 20 three times and treated with para-nitrophenylphosphate (pNPP) phosphatase substrate (Sigma), and the absorbance at 405 nm was measured using a Multiskan FC microplate reader (Thermo Scientific).
(iv) Droplet digital PCR analysis. ddPCR was used to determine the recombinant POS5 gene copy number and transcriptional levels of TDH3 and POS5.
For gene dosage determination, genomic DNA was purified using the Wizard genomic DNA purification kit (Promega), according to the manufacturer’s instructions, and quantified in a NanoDrop 2000 spectrophotometer (Thermo Scientific). Half a microgram of DNA was digested by the EcoRI and BamHI FastDigest enzymes (Thermo Scientific) to produce DNA fragments smaller than 5 kb and purified with the Wizard SV gel and PCR clean-up system (Promega). Reaction conditions (1× Supermix ddPCR TaqMan, 300 nM each primer, 200 nM each probe, and 0.02 ng·μl−1 digested genomic DNA) and operational conditions were performed as suggested by Bio-Rad and optimized for P. pastoris, as described in reference 43. The annealing temperature was set to 57°C, after temperature gradient determination. The primers used for ddPCR are listed in Table 1.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′–3′) | Tm (°C)a |
|---|---|---|
| POS5-Fb ,c | ATCCGCGCCTGCAGGAATGTTTGTTAGAGTTAAGTTGAACAAGCCAGTTAA | 67 |
| POS5_cyt-Fb ,c | ATCCGCGCCTGCAGGAATAATGTCCACTTTGGACTCCCATTCCTTGAA | 68 |
| POS5-Rb ,c | ATGACTAGGCCGAGGCGGCCTTAGTCGTTGTCAGTCTGTCTC | 68 |
| POS5_int-Rb ,c | AGCAACACCGTCAGCAGTAG | |
| POS5_amp-Fd | GGAGTGTCACTTGAAGAA | 38.6 |
| POS5_amp-Rd | CGTCAGCAGTAGTTCTAG | 36.6 |
| POS5 probed | ACTCCAACTCCTCCATCGTTACTCA (5′, 6-FAM/3′, BHQ-1)e | 57.1 |
Tm, melting temperature.
Primer used for cloning POS5 into pPUZZLE.
Primer used for clone verification.
Primer used for gene copy number determination.
FAM, 6-carboxyfluorescein; BHQ, black hole quencher.
The expression levels of TDH3 and POS5 were quantified by ddPCR, as described in reference 44, with minor modifications. Briefly, 1 ng of cDNA was used for the reaction mixture instead of 0.4 ng, and the annealing temperature was set to 56.5°C in the PCR. The housekeeping gene β-actin (ACT1) was selected to normalize the data. The primers used are described in Table 1.
(v) NADPH/NADP+ ratio determination. Samples for NADPH and NADP+ quantification were taken and rapidly quenched with cold 60% (vol/vol) methanol (45, 46). Cell suspensions were centrifuged and washed twice with quenching solution, as described by Ortmayr et al. (45) (4,000 × g, −10°C, 10 min in a 5804 R centrifuge; Eppendorf). Finally, pellets were stored at −80°C. NADPH and NADP+ concentrations were determined using the EnzyChrom NADP+/NADPH assay kit (BioAssay Systems), and the optical densities at a wavelength of 595 nm were measured using a Multiskan FC microplate reader (Thermo Scientific). Analyses were performed in duplicate. The relative standard deviation (RSD) of the analytical method was 20%.
Statistical analysis.
Chemostat cultivation data were checked for consistency, and standard reconciliation procedures were applied (47). A statistical consistency test, based on the h-index as described in reference 47, was passed with a confidence level of 95%. Consequently, there was no evidence of gross measurement errors. A statistical comparison of the macroscopic growth profiles of the different strains was performed using the Microsoft Excel 2-tailed Student t test.
Metabolic modeling.
The iMT1026 v3.0 metabolic model (BioModels database MODEL1612130000) of P. pastoris (26) was used in the COBRA Toolbox v2.0.6 (48) under Matlab 2014 (Mathworks, USA) with SBML toolbox v4.1.0 (49) and libSBML library v5.12.0 (50) and with IBM ILOG CPLEX optimization studio 12.7 as the solver. The prediction of flux redistribution due to Fab overexpression was performed employing flux scanning based on enforced objective function (FSEOF) (51) by maximizing the biomass production at a constrained range of Fab secretion (0 to 0.12 mg·gDCW−1·h−1). Particularly, redox cofactor turnover rates were calculated using the flux-sum analysis (52) on each resulting flux distribution. A cytosolic NADH kinase reaction was incorporated into the model (the corresponding mitochondrial reaction was not added, as iMT1026 v3.0 already contained the endogenous mitochondrial NADH kinase reaction). The perturbation of the NADH kinase activity on flux distribution was calculated by minimization of metabolic adjustment (MOMA) (53) performing a series of simulations enforcing a minimal flux through the NADH kinase reaction (cytosolic or mitochondrial) constraining the uptake of the carbon source to the control strain experimental values in the case of normoxia (glycerol and glucose) and additionally constraining the oxygen uptake rate for the simulations under hypoxic conditions. The resulting flux distributions at different NADH kinase reaction fluxes (0 to 2 mmol·gDCW−1·h−1) were compared against the control strain (X-33/2F5), in which the NADH kinase reaction flux is 0.
Data availability.
Data corresponding to this study can be found in the supplemental material files and additional data in the UAB data repository database (54).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by projects CTQ2013-42391-R and CTQ2016-74959-R (AEI/FEDER, UE) of the Spanish Ministry of Economy, Industry and Competitiveness; 2014-SGR-452 from the Reference Network in Biotechnology (XRB; Generalitat de Catalunya); grants FPU FPU12/06185 (to M.T.-G.) and FPI BES-2014-067935 (to S.M.) of the Spanish Ministry of Education, Culture and Sport; and a postdoctoral grant from the Ciência sem Fronteiras Program (CNPq, Brazil, process 249872/2013-7) to C.C.P.A.
We thank Master of Science student Ane Quesada for supporting small-scale clone screening experiments.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ferrer P, Albiol J. 2014. 13C-based metabolic flux analysis in yeast: the Pichia pastoris case, p 209–232. In Mapelli V. (ed), Methods in molecular biology. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 2.Klein T, Niklas J, Heinzle E. 2015. Engineering the supply chain for protein production/secretion in yeasts and mammalian cells. J Ind Microbiol Biotechnol 42:453–464. doi: 10.1007/s10295-014-1569-2. [DOI] [PubMed] [Google Scholar]
- 3.Wu G, Yan Q, Jones JA, Tang YJ, Fong SS, Koffas M. 2016. Metabolic burden: cornerstones in synthetic biology and metabolic engineering applications. Trends Biotechnol 34:652–664. doi: 10.1016/j.tibtech.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Delic M, Rebnegger C, Wanka F, Puxbaum V, Haberhauer-Troyer C, Hann S, Köllensperger G, Mattanovich D, Gasser B. 2012. Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast. Free Radic Biol Med 52:2000–2012. doi: 10.1016/j.freeradbiomed.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 5.Hou J, Lages NF, Oldiges M, Vemuri GN. 2009. Metabolic impact of redox cofactor perturbations in Saccharomyces cerevisiae. Metab Eng 11:253–261. doi: 10.1016/j.ymben.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Lee HC, Kim JS, Jang W, Kim SY. 2010. High NADPH/NADP+ ratio improves thymidine production by a metabolically engineered Escherichia coli strain. J Biotechnol 149:24–32. doi: 10.1016/j.jbiotec.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Siedler S, Bringer S, Bott M. 2011. Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl Microbiol Biotechnol 92:929–937. doi: 10.1007/s00253-011-3374-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim S, Hahn JS. 2015. Efficient production of 2,3-butanediol in Saccharomyces cerevisiae by eliminating ethanol and glycerol production and redox rebalancing. Metab Eng 31:94–101. doi: 10.1016/j.ymben.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Geertman J-M, van Maris AJA, van Dijken JP, Pronk JT. 2006. Physiological and genetic engineering of cytosolic redox metabolism in Saccharomyces cerevisiae for improved glycerol production. Metab Eng 8:532–542. doi: 10.1016/j.ymben.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Krainer FW, Dietzsch C, Hajek T, Herwig C, Spadiut O, Glieder A. 2012. Recombinant protein expression in Pichia pastoris strains with an engineered methanol utilization pathway. Microb Cell Fact 11:22. doi: 10.1186/1475-2859-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nocon J, Steiger MG, Pfeffer M, Sohn SB, Kim TY, Maurer M, Rußmayer H, Pflügl S, Ask M, Haberhauer-Troyer C, Ortmayr K, Hann S, Koellensperger G, Gasser B, Lee SY, Mattanovich D. 2014. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng 24:129–138. doi: 10.1016/j.ymben.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nocon J, Steiger M, Mairinger T, Hohlweg J, Rußmayer H, Hann S, Gasser B, Mattanovich D. 2016. Increasing pentose phosphate pathway flux enhances recombinant protein production in Pichia pastoris. Appl Microbiol Biotechnol 100:5955–5963. doi: 10.1007/s00253-016-7363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyland J, Fu J, Blank LM, Schmid A. 2010. Quantitative physiology of Pichia pastoris during glucose-limited high-cell density fed-batch cultivation for recombinant protein production. Biotechnol Bioeng 107:357–368. doi: 10.1002/bit.22836. [DOI] [PubMed] [Google Scholar]
- 14.Driouch H, Melzer G, Wittmann C. 2012. Integration of in vivo and in silico metabolic fluxes for improvement of recombinant protein production. Metab Eng 14:47–58. doi: 10.1016/j.ymben.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Mattanovich D, Sauer M, Gasser B. 2016. Industrial microorganisms: Pichia pastoris, p 687–714. In Wittmann C, Liao JC (ed), Industrial biotechnology. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 16.Baumann K, Maurer M, Dragosits M, Cos O, Ferrer P, Mattanovich D. 2008. Hypoxic fed-batch cultivation of Pichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol Bioeng 100:177–183. doi: 10.1002/bit.21763. [DOI] [PubMed] [Google Scholar]
- 17.Baumann K, Carnicer M, Dragosits M, Graf AB, Stadlmann J, Jouhten P, Maaheimo H, Gasser B, Albiol J, Mattanovich D, Ferrer P. 2010. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst Biol 4:141. doi: 10.1186/1752-0509-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carnicer Heras M. 2012. Systematic metabolic analysis of recombinant Pichia pastoris under different oxygen conditions: a metabolome and fluxome based study. PhD thesis. Universitat Autònoma de Barcelona, Barcelona, Spain. [Google Scholar]
- 19.Tomàs-Gamisans M. 2017. Developing strategies for systems metabolic engineering of Pichia pastoris. PhD thesis. Universitat Autònoma de Barcelona, Barcelona, Spain: http://hdl.handle.net/10803/458538. [Google Scholar]
- 20.Monforte S. 2019. Systems metabolic engineering for recombinant protein production in Pichia pastoris. PhD thesis. Universitat Autònoma de Barcelona, Barcelona, Spain. [Google Scholar]
- 21.Carnicer M, Baumann K, Töplitz I, Sánchez-Ferrando F, Mattanovich D, Ferrer P, Albiol J. 2009. Macromolecular and elemental composition analysis and extracellular metabolite balances of Pichia pastoris growing at different oxygen levels. Microb Cell Fact 8:65. doi: 10.1186/1475-2859-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann K, Dato L, Graf AB, Frascotti G, Dragosits M, Porro D, Mattanovich D, Ferrer P, Branduardi P. 2011. The impact of oxygen on the transcriptome of recombinant S. cerevisiae and P pastoris–a comparative analysis. BMC Genomics 12:218. doi: 10.1186/1471-2164-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterham HR, Digan ME, Koutz PJ, Lair SV, Cregg JM. 1997. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- 24.Prielhofer R, Cartwright SP, Graf AB, Valli M, Bill RM, Mattanovich D, Gasser B. 2015. Pichia pastoris regulates its gene-specific response to different carbon sources at the transcriptional, rather than the translational, level. BMC Genomics 16:167. doi: 10.1186/s12864-015-1393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomàs-Gamisans M, Ferrer P, Albiol J. 2018. Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb Biotechnol 11:224–237. doi: 10.1111/1751-7915.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomàs-Gamisans M, Ferrer P, Albiol J. 2016. Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PLoS One 11:e0148031. doi: 10.1371/journal.pone.0148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordà J, Jouhten P, Cámara E, Maaheimo H, Albiol J, Ferrer P. 2012. Metabolic flux profiling of recombinant protein secreting Pichia pastoris growing on glucose:methanol mixtures. Microb Cell Fact 11:57. doi: 10.1186/1475-2859-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dragosits M, Stadlmann J, Albiol J, Baumann K, Maurer M, Gasser B, Sauer M, Altmann F, Ferrer P, Mattanovich D. 2009. The effect of temperature on the proteome of recombinant Pichia pastoris. J Proteome Res 8:1380–1392. doi: 10.1021/pr8007623. [DOI] [PubMed] [Google Scholar]
- 29.Panagiotou G, Grotkjaer T, Hofmann G, Bapat PM, Olsson L. 2009. Overexpression of a novel endogenous NADH kinase in Aspergillus nidulans enhances growth. Metab Eng 11:31–39. doi: 10.1016/j.ymben.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Hou J, Vemuri GN, Bao X, Olsson L. 2009. Impact of overexpressing NADH kinase on glucose and xylose metabolism in recombinant xylose-utilizing Saccharomyces cerevisiae. Appl Microbiol Biotechnol 82:909–919. doi: 10.1007/s00253-009-1900-4. [DOI] [PubMed] [Google Scholar]
- 31.Qiao K, Wasylenko TM, Zhou K, Xu P, Stephanopoulos G. 2017. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol 35:173–177. doi: 10.1038/nbt.3763. [DOI] [PubMed] [Google Scholar]
- 32.Bakker BM, Overkamp KM, van Maris AJ, Kötter P, Luttik M A H, van Dijken JP, Pronk JT. 2001. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37. doi: 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- 33.Blank LM, Lehmbeck F, Sauer U. 2005. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res 5:545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Llobell A, Lopez-Ruiz A, Peinado J, Lopez-Barea J. 1988. Glutathione reductase directly mediates the stimulation of yeast glucose-6-phosphate dehydrogenase by GSSG. Biochem J 249:293–296. doi: 10.1042/bj2490293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyland J, Fu J, Blank LM, Schmid A. 2011. Carbon metabolism limits recombinant protein production in Pichia pastoris. Biotechnol Bioeng 108:1942–1953. doi: 10.1002/bit.23114. [DOI] [PubMed] [Google Scholar]
- 36.Vemuri GN, Eiteman M, McEwen JE, Olsson L, Nielsen J. 2007. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104:2402–2407. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasser B, Maurer M, Gach J, Kunert R, Mattanovich D. 2006. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol Bioeng 94:353–361. doi: 10.1002/bit.20851. [DOI] [PubMed] [Google Scholar]
- 38.Outten CE, Culotta VC. 2003. A novel NADH kinase is the mitochondrial source of NADPH in Saccharomyces cerevisiae. EMBO J 22:2015–2024. doi: 10.1093/emboj/cdg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strand MK, Stuart GR, Longley MJ, Graziewicz MA, Dominick OC, Copeland WC. 2003. POS5 gene of Saccharomyces cerevisiae encodes a mitochondrial NADH kinase required for stability of mitochondrial DNA. Eukaryot Cell 2:809–820. doi: 10.1128/ec.2.4.809-820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baumann K, Adelantado N, Lang C, Mattanovich D, Ferrer P. 2011. Protein trafficking, ergosterol biosynthesis and membrane physics impact recombinant protein secretion in Pichia pastoris. Microb Cell Fact 10:93. doi: 10.1186/1475-2859-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergkessel M, Guthrie C. 2013. Colony PCR, p 299–309. In Abelson JN, Simon MI (ed), Laboratory methods in enzymology. Elsevier, Waltham, MA. [DOI] [PubMed] [Google Scholar]
- 42.Adelantado N, Tarazona P, Grillitsch K, García-Ortega X, Monforte S, Valero F, Feussner I, Daum G, Ferrer P. 2017. The effect of hypoxia on the lipidome of recombinant Pichia pastoris. Microb Cell Fact 16:86. doi: 10.1186/s12934-017-0699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cámara E, Albiol J, Ferrer P. 2016. Droplet digital PCR-aided screening and characterization of Pichia pastoris multiple gene copy strains. Biotechnol Bioeng 113:1542–1551. doi: 10.1002/bit.25916. [DOI] [PubMed] [Google Scholar]
- 44.Cámara E, Landes N, Albiol J, Gasser B, Mattanovich D, Ferrer P. 2017. Increased dosage of AOX1 promoter-regulated expression cassettes leads to transcription attenuation of the methanol metabolism in Pichia pastoris. Sci Rep 7:44302. doi: 10.1038/srep44302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortmayr K, Nocon J, Gasser B, Mattanovich D, Hann S, Koellensperger G. 2014. Sample preparation workflow for the liquid chromatography tandem mass spectrometry based analysis of nicotinamide adenine dinucleotide phosphate cofactors in yeast. J Sep Sci 37:2185–2191. doi: 10.1002/jssc.201400290. [DOI] [PubMed] [Google Scholar]
- 46.Carnicer M, Canelas AB, Ten Pierick A, Zeng Z, van Dam J, Albiol J, Ferrer P, Heijnen JJ, van Gulik W. 2012. Development of quantitative metabolomics for Pichia pastoris. Metabolomics 8:284–298. doi: 10.1007/s11306-011-0308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noorman HJ, Romein B, Luyben KCAM, Heijnen JJ. 2000. Classification, error detection, and reconciliation of process information in complex biochemical systems. Biotechnol Bioeng 49:364–376. doi:. [DOI] [PubMed] [Google Scholar]
- 48.Schellenberger J, Que R, Fleming RMT, Thiele I, Orth JD, Feist AM, Zielinski DC, Bordbar A, Lewis NE, Rahmanian S, Kang J, Hyduke DR, Palsson B. 2011. Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0. Nat Protoc 6:1290–1307. doi: 10.1038/nprot.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keating SM, Bornstein BJ, Finney A, Hucka M. 2006. SBMLToolbox: an SBML toolbox for MATLAB users. Bioinformatics 22:1275–1277. doi: 10.1093/bioinformatics/btl111. [DOI] [PubMed] [Google Scholar]
- 50.Bornstein BJ, Keating SM, Jouraku A, Hucka M. 2008. LibSBML: an API library for SBML. Bioinformatics 24:880–881. doi: 10.1093/bioinformatics/btn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi HS, Lee SY, Kim TY, Woo HM. 2010. In silico identification of gene amplification targets for improvement of lycopene production. Appl Environ Microbiol 76:3097–3105. doi: 10.1128/AEM.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung B, Lee D-Y. 2009. Flux-sum analysis: a metabolite-centric approach for understanding the metabolic network. BMC Syst Biol 3:117. doi: 10.1186/1752-0509-3-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segrè D, Vitkup D, Church GM. 2002. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A 99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Albiol J. 2019. Dataset from redox engineering by ectopic overexpression of NADH kinase in recombinant Pichia pastoris (Komagataella phaffii). Universitat Autònoma de Barcelona Digital Data Repository, Barcelona, Spain: https://ddd.uab.cat/record/216543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data corresponding to this study can be found in the supplemental material files and additional data in the UAB data repository database (54).