Clavibacter spp. are economically important bacterial plant pathogens infecting a set of diverse agricultural crops, such as alfalfa, corn, pepper, potato, tomato, and wheat. A number of plant-pathogenic members of the genus (e.g., C. michiganensis sensu stricto and C. sepedonicus, infecting tomato and potato plants, respectively) are included in the A2 (high-risk) list of quarantine pathogens by the European and Mediterranean Plant Protection Organization (EPPO). Although tomato-associated members of Clavibacter spp. account for a significant portion of the genetic diversity in the genus, only the strains belonging to C. michiganensis sensu stricto (formerly C. michiganensis subsp. michiganensis) cause bacterial canker disease of tomato and are subjected to the quarantine inspections. Hence, discrimination between the pathogenic and nonpathogenic Clavibacter sp. strains associated with tomato seeds and transplants plays a pivotal role in the accurate detection and cost-efficient management of the disease. On the other hand, detailed information on the genetic contents of different lineages of the genus would lead to the development of genome-informed specific detection techniques. In this study, we have provided an overview of the phylogenetic and genomic differences between the pathogenic and nonpathogenic tomato-associated Clavibacter sp. strains. We also noted that the taxonomic status of newly introduced subspecies of C. michiganensis (i.e., C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli) should be reconsidered.
KEYWORDS: Actinobacteria, bacterial canker of tomato, bacterial taxonomy, Clavibacter michiganensis sensu stricto, quarantine pathogen
ABSTRACT
Members of the genus Clavibacter are economically important bacterial plant pathogens infecting a set of diverse agricultural crops (e.g., alfalfa, corn, potato, tomato, and wheat). Tomato-associated Clavibacter sp. strains account for a great portion of the genetic diversity of the genus, and C. michiganensis sensu stricto (formerly C. michiganensis subsp. michiganensis), causing bacterial canker disease, is considered one of the most destructive seed-borne agents for the crop worldwide. However, current taxonomic descriptions of the genus do not reflect the existing diversity of the strains, resulting in unsatisfactory results in quarantine surveys for the pathogens. In this study, we used all the available genome sequences of Clavibacter sp. strains, including the type strains of newly described subspecies, to provide precise insight into the diversity of tomato-associated members of the genus and further clarify the taxonomic status of the strains using genotypic and phenotypic features. The results of phylogenetic analyses revealed the existence of nine hypothetical new species among the investigated strains. None of the three new subspecies (i.e., C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli) is included within the tomato-pathogenic C. michiganensis sensu stricto lineage. Although comparative genomics revealed the lack of chp and tomA pathogenicity determinant gene clusters in the nonpathogenic strains, a number of pathogenicity-related genes were noted to be present in all the strains regardless of their pathogenicity characteristics. Altogether, our results indicate a need for a formal taxonomic reconsideration of tomato-associated Clavibacter sp. strains to facilitate differentiation of the lineages in quarantine inspections.
IMPORTANCE Clavibacter spp. are economically important bacterial plant pathogens infecting a set of diverse agricultural crops, such as alfalfa, corn, pepper, potato, tomato, and wheat. A number of plant-pathogenic members of the genus (e.g., C. michiganensis sensu stricto and C. sepedonicus, infecting tomato and potato plants, respectively) are included in the A2 (high-risk) list of quarantine pathogens by the European and Mediterranean Plant Protection Organization (EPPO). Although tomato-associated members of Clavibacter spp. account for a significant portion of the genetic diversity in the genus, only the strains belonging to C. michiganensis sensu stricto (formerly C. michiganensis subsp. michiganensis) cause bacterial canker disease of tomato and are subjected to the quarantine inspections. Hence, discrimination between the pathogenic and nonpathogenic Clavibacter sp. strains associated with tomato seeds and transplants plays a pivotal role in the accurate detection and cost-efficient management of the disease. On the other hand, detailed information on the genetic contents of different lineages of the genus would lead to the development of genome-informed specific detection techniques. In this study, we have provided an overview of the phylogenetic and genomic differences between the pathogenic and nonpathogenic tomato-associated Clavibacter sp. strains. We also noted that the taxonomic status of newly introduced subspecies of C. michiganensis (i.e., C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli) should be reconsidered.
INTRODUCTION
A number of plant-pathogenic bacterial species are reported to have nonpathogenic lineages which usually exist in the same ecological niche as their pathogenic counterparts (1–3). Nonpathogenic lineages or strains typically have genetic contents similar to those of their pathogenic relatives but lack some of the key pathogenicity determinants (e.g., pathogenicity islands, virulence genes, and plasmids) (4–6). As far as economically important quarantine plant-pathogenic bacteria are concerned, the presence of nonpathogenic strains in commercial seeds or propagative parts of plants will interfere in the accurate detection of the pathogens, leading to false-positive results in the quarantine inspections and unsatisfied seed producers and traders (7). This is due in part to the fact that most of the nonpathogenic bacterial strains are phenotypically similar to their pathogenic relatives; therefore, they are not differentiable from each other on the culture media (7). Furthermore, most of the molecular detection protocols (e.g., PCR primers, probes, and antibodies) are designed based on the general features of a given species/subspecies/pathovar rather than focused on the pathogenicity determinants of the pathogen (7). As a consequence, contradictions in the results of quarantine inspections will lead to economic losses for seed producers and will have negative impact on transportation of plant materials on a global scale (7, 8).
Clavibacter spp. are economically important Gram-positive bacterial plant pathogens infecting a set of diverse agricultural crops, e.g., alfalfa, corn, pepper, potato, tomato, and wheat (9). Until recently, the genus Clavibacter was considered to include only one species, C. michiganensis, comprising five plant-pathogenic subspecies, i.e., C. michiganensis subsp. insidiosus, C. michiganensis subsp. michiganensis, C. michiganensis subsp. nebraskensis, C. michiganensis subsp. sepedonicus, and C. michiganensis subsp. tessellarius (9, 10). Furthermore, all the tomato- and pepper-associated Clavibacter sp. strains were classified as members of C. michiganensis subsp. michiganensis regardless of whether they were pathogenic or nonpathogenic on the host of isolation. However, using multilocus sequence analysis (MLSA) and multilocus sequence typing (MLST), Jacques et al. (11) showed that tomato-associated nonpathogenic Clavibacter sp. strains are phylogenetically distinct from the pathogenic counterparts in the species. Differentiation of the pathogenic and nonpathogenic strains of C. michiganensis has always been an ongoing challenge for official sanitary agencies, quarantine inspectors, and seed providers (12), since false-positive results would lead to the rejection of seed/seedling lots on an economically significant scale (7, 8). This led to the belief that a comprehensive complete genome sequence-based reconsideration of C. michiganensis sensu lato (all the former members of C. michiganensis as described by Davis et al. [9]) is warranted to shed light on the genetic diversity, genomic repertories, and taxonomic status of the pathogenic and nonpathogenic tomato-associated strains of the species (11).
Following the emergence of high-throughput molecular-phylogenetic techniques, many Clavibacter sp. strains which were often previously misidentified based on phenotypic features were assigned to novel taxa. For instance, tomato-associated nonpathogenic members of C. michiganensis sensu lato were assigned to two new subspecies, C. michiganensis subsp. californiensis and C. michiganensis subsp. chilensis (13). Additionally, C. michiganensis subsp. phaseoli and C. michiganensis subsp. capsici were identified as the causal agents of bacterial bean leaf yellowing on common bean (Phaseolus vulgaris) and bacterial canker of pepper (Capsicum annuum), respectively (14, 15). Furthermore, nonpathogenic peach-colored strains isolated from the tomato phyllosphere were reported to be distinct from the tomato-pathogenic members of Clavibacter spp. (16).
Recently, a reclassification of Clavibacter spp. into five new species and a new combination was proposed based on genomic information, e.g., average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) indices (17–19). The original subspecies of C. michiganensis sensu lato were elevated to the species level and designated C. michiganensis (here referred to as C. michiganensis sensu stricto, formerly C. michiganensis subsp. michiganensis), C. tessellarius, C. insidiosus, C. nebraskensis, and C. capsici, as well as C. sepedonicus as new combination (17, 19). However, due to the lack of genomic information from the newly proposed subspecies (C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli) as well as several taxonomically undetermined strains, additional investigations are warranted to further clarify the taxonomy of the genus. Moreover, strains associated with solanaceous vegetables contain a large fraction of diversity within the Clavibacter sp. members, and much of the molecular, phylogenetic, and genomic information for these strains remains unexplored. For the tomato-associated strains of Clavibacter spp., comparative genomics on a wide collection of nonpathogenic and pathogenic strains would further elucidate the genetic diversity of these bacteria, resulting in the development of genome-informed specific molecular markers (e.g., specific conventional PCR and real-time PCR primers, as well as loop-mediated isothermal amplification) for the detection and differentiation of the pathogenic and nonpathogenic strains in the quarantine posts.
The objectives of the present study were to (i) investigate the genetic diversity of tomato-associated Clavibacter sp. strains using the genome sequences of all available nonpathogenic and pathogenic strains and (ii) provide a novel taxonomic overview into the status of tomato-pathogenic and nonpathogenic strains within the genus. For this aim, we used the genome sequences of 40 Clavibacter sp. strains, including the type strains of three newly described subspecies (C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli), as well as additional atypical nonpathogenic strains isolated from tomato plants around the globe (20). Draft genome sequence-based phylogenetic analyses revealed a higher diversity among the nonpathogenic strains of Clavibacter spp. than has previously been reported, delineating them into several new species. On the other hand, our data revealed that the two individual subspecies C. michiganensis subsp. chilensis and C. michiganensis subsp. phaseoli need to be considered members of one species, according to the 99% genome similarity among the type strains. Furthermore, comparative genomics among the pathogenic and nonpathogenic tomato-associated strains, as well as the type strains of the remaining species/subspecies within the genus, revealed several pathogenicity determinant genes presenting only in C. michiganensis sensu stricto, which could be considered suitable genomic targets for the development of specific detection methods for the tomato pathogen.
RESULTS
Pathogenicity and host range.
Tomato and pepper plants inoculated with the standard strain of C. michiganensis sensu stricto (ICMP 22049) showed the expected disease symptoms at 10 to 12 days postinoculation. Although tomato plants inoculated with strain ICMP 22049 showed wilting and plant death (see Fig. S1a in the supplemental material), pepper plants inoculated using the same strain showed only stem canker symptoms on the site of inoculation, with no wilting or plant death, in the same time frame (Fig. S1b). However, neither C. michiganensis subsp. phaseoli nor C. michiganensis subsp. chilensis induced symptoms on the inoculated plant species, i.e., common bean, cowpea, pepper, mung bean, and tomato (Table 1). Furthermore, we could not reisolate C. michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis from the stem, petiole, and leaf tissues 5 to 10 cm above the inoculation site on the stem. This could be an indication of the fact that C. michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis were unable to endophytically colonize the evaluated plant species. For the orange-pigmented tomato-associated strains CFBP 8615 and CFBP 8616, although no symptoms were observed on common bean, cowpea, pepper, mung bean, and tomato plants, bacterial colonies similar to those originally inoculated were consistently reisolated from the leaf tissues of common bean cultivar Navy plants inoculated with CFBP 8616 (Table 1). Furthermore, the standard strain ICMP 22049 was consistently reisolated from the symptomatic pepper and tomato plants on yeast extract-peptone-glucose agar (YPGA) medium, and their identity was confirmed using the genus-specific primer pair CMR16F1/CMR16R1 (data not shown). Similar results were obtained in both replications of the experiments, while the negative-control plants remained healthy.
TABLE 1.
Results of pathogenicity tests and host range assays of the type strains of Clavibacter michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis, as well as two atypical peach-colored Clavibacter sp. strains, on different annual crops under greenhouse conditionsa
| Taxon | Strain | Pathogenicityb
on: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Pepper |
Common bean |
Cowpea | Mung bean | Tomato | |||||
| cv. Sereno | cv. Aziz | cv. Red kidney | cv. Pinto | cv. Navy | |||||
| C. michiganensis subsp. phaseoli | CFBP 8627T | − | − | − | − | − | − | − | − |
| C. michiganensis subsp. chilensis | CFBP 8217T | − | − | − | − | − | − | − | − |
| Clavibacter sp. | CFBP 8615 | − | − | − | − | − | − | − | − |
| Clavibacter sp. | CFBP 8616 | − | − | − | − | −c | − | − | − |
| C. michiganensis sensu stricto | ICMP 22049 | +d | +d | − | − | − | − | − | + |
None of the evaluated bacterial strains was pathogenic on the tested plants, while the nonpathogenic strain CFBP 8616 was reisolated from common bean cv. Navy tissues.
−, negative; +, positive.
The inoculated bacterial strain was reisolated from the asymptomatic leaf tissues of the test plants.
These plants showed only stem canker symptoms on the site of inoculation, with no systemic wilting and plant death.
Phylogenetic analyses.
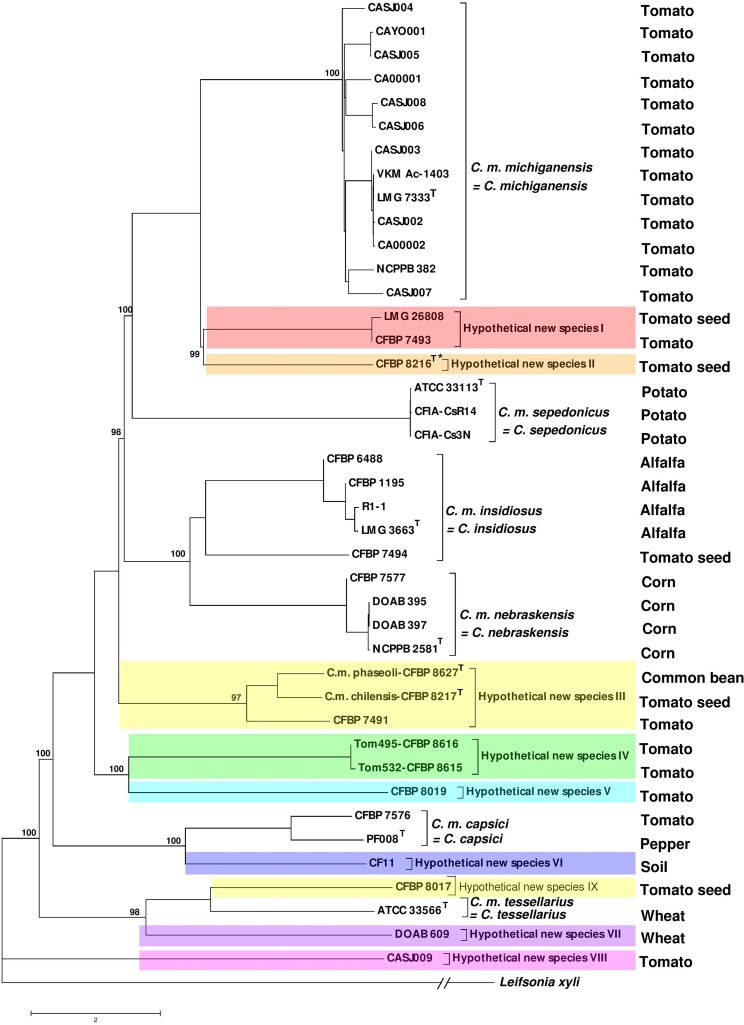
A neighbor-joining phylogenetic tree constructed using the genome sequences of 40 Clavibacter sp. strains (Table 2) via the ANI Calculator online service with the all-versus-all strategy revealed high genetic diversity among tomato-associated nonpathogenic strains of the genus (Fig. 1). ANI values between different pairs of strains varied from 87% to 100% among the Clavibacter sp. strains (Table 3). While all tomato-pathogenic strains of C. michiganensis sensu stricto clustered in a monophyletic clade showing 99 to 100% ANI with one another, nonpathogenic strains isolated from tomato were scattered in several clades, most of which had <96% ANI values with the other clades (Table 3). The closest nonpathogenic clade to the C. michiganensis sensu stricto group consisted of three strains, i.e., the type strain of C. michiganensis subsp. californiensis (CFBP 8216T), and the nonpathogenic strains LMG 26808 and CFBP 7493. the ANI value between the type strain of C. michiganensis sensu stricto (LMG 7333T) and the type strain of C. michiganensis subsp. californiensis was 95% in all the calculating strategies, retaining them at the threshold of species definition (21). Nevertheless, the dDDH value (57.70%) between the type strains of C. michiganensis sensu stricto and C. michiganensis subsp. californiensis was far below the threshold for species delineation (70%) with this method (Table 3). Altogether, given the differences in their pathogenicities and biochemical characteristics (13) as well as the below-threshold genomic similarity, the two taxa C. michiganensis sensu stricto and C. michiganensis subsp. californiensis could be considered separate species. Furthermore, the ANI between the type strains of C. michiganensis sensu stricto and C. michiganensis subsp. californiensis and the cluster which included the nonpathogenic strains LMG 26808 and CFBP 7493 was 94 to 95% (Table 3). The dDDH values between these type strains and LMG 26808 and CFBP 7493 were also 57 to 58%, indicating them as separate species (Fig. 1).
TABLE 2.
Clavibacter sp. genome sequences used for the comparative genomics and phylogenetic analysesa
| Previous nameb | New namec | Strain | Host of isolation | Pathogenicity | Date | Country of Isolation | GenBank accession no. | Reference |
|---|---|---|---|---|---|---|---|---|
| Clavibacter sp. | New species IV | CFBP 8615d | Solanum lycopersicum | NPe | 2015 | Iran | QWGT01000000 | 20 |
| Clavibacter sp. | new species IV | CFBP 8616d | Solanum lycopersicum | NP | 2015 | Iran | QWGU01000000 | 20 |
| C. michiganensis subsp. californiensis | New species II | CFBP 8216Td | Solanum lycopersicum | NP | 2000 | USA | QWEE01000000 | 20 |
| C. michiganensis subsp. chilensis | New species III | CFBP 8217Td | Solanum lycopersicum | NP | 2007 | Netherlands | QWGS01000000 | 20 |
| Clavibacter sp. | New species III | CFBP 7491d | Solanum lycopersicum | NP | NDf | ND | QWEB01000000 | 20 |
| Clavibacter sp. | New species I | CFBP 7493d | Solanum lycopersicum | NP | ND | ND | QWEC01000000 | 20 |
| C. michiganensis subsp. insidiosus | C. insidiosus | CFBP 1195 | Medicago sativa | Alfalfa | 1964 | UK | QWDZ01000000 | 20 |
| C. michiganensis subsp. insidiosus | C. insidiosus | CFBP 6488 | Medicago sativa | Alfalfa | 1998 | Czech Republic | QWEA01000000 | 20 |
| C. michiganensis subsp. nebraskensis | C. nebraskensis | CFBP 7577 | Zea mays | Corn | ND | ND | QWED01000000 | 20 |
| C. michiganensis subsp. phaseoli | New species III | CFBP 8627T | Phaseolus vulgaris | NP | 2009 | Spain | QWGV01000000 | 20 |
| Clavibacter sp. | New species VIII | CASJ009d | Solanum lycopersicum | NP | 2011 | USA | MDHJ00000000.1 | 22 |
| Clavibacter sp. | New species V | CFBP 8019d | Solanum lycopersicum | NP | 2011 | USA | NZ_MDJZ00000000.1 | 22 |
| Clavibacter sp. | New species IX | CFBP 8017d | Solanum lycopersicum | Wheat | 2006 | Netherlands | MDJY00000000 | 22 |
| Clavibacter sp. | New species VI | CF11 | soil | NP | 2011 | China | JROD00000000 | 23 |
| Clavibacter sp. | New species I | LMG 26808d | Solanum lycopersicum | NP | ND | Netherlands | AZQZ00000000 | 28 |
| Clavibacter sp. | C. insidiosus | CFBP 7494d | Solanum lycopersicum | Wheat | 1999 | Chile | MDJW00000000 | 22 |
| Clavibacter sp. | New species VII | DOAB 609 | Triticum aestivum | ND | 1976 | USA | LQXA00000000 | 50 |
| C. michiganensis subsp. michiganensis | C. michiganensis | LMG 7333T | Solanum lycopersicum | Tomato | 1957 | Hungary | NZ_MZMP00000000 | 51 |
| C. michiganensis subsp. michiganensis | C. michiganensis | NCPPB 382 | Solanum lycopersicum | Tomato | 1956 | UK | AM711867.1 | 26 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ004 | Solanum lycopersicum | Tomato | 1999 | USA | MDHE00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CAYO001 | Solanum lycopersicum | Tomato | 2001 | USA | MDHL00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ005 | Solanum lycopersicum | Tomato | 2001 | USA | MDHF00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CA00001 | Solanum lycopersicum | Tomato | 2000 | USA | MDHK00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ008 | Solanum lycopersicum | Tomato | 2002 | USA | MDHI00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ006 | Solanum lycopersicum | Tomato | 2002 | USA | MDHG00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ003 | Solanum lycopersicum | Tomato | 1999 | USA | MDHD00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | VKM Ac-1403 | Solanum lycopersicum | Tomato | 2017 | USA | FVZG00000000 | 52 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ002 | Solanum lycopersicum | Tomato | 1999 | USA | MDHC00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CA00002 | Solanum lycopersicum | Tomato | 2000 | USA | MDHM00000000 | 22 |
| C. michiganensis subsp. michiganensis | C. michiganensis | CASJ007 | Solanum lycopersicum | Tomato | 2011 | USA | MDHH00000000 | 22 |
| C. michiganensis subsp. capsici | C. capsici | PF008T | Capsicum sp. | Pepper | ND | South Korea | NZ_CP012573 | 15 |
| C. michiganensis subsp. capsici | C. capsici | CFBP 7576d | Solanum lycopersicum | Pepper | 1997 | ND | MDJX00000000 | 22 |
| C. michiganensis subsp. insidiosus | C. insidiosus | LMG 3663T | Medicago sativa | Alfalfa | 1955 | USA | MZMO00000000 | 51 |
| C. michiganensis subsp. insidiosus | C. insidiosus | R1-1 | Medicago truncatula | Alfalfa | 2009 | USA | NZ_CP011043 | 29 |
| C. michiganensis subsp. nebraskensis | C. nebraskensis | NCPPB 2581T | Zea mays | Corn | 1971 | USA | NC_020891.1 | 50 |
| C. michiganensis subsp. nebraskensis | C. nebraskensis | DOAB 395 | Zea mays | Corn | 2014 | Canada | LSOE00000000 | 50 |
| C. michiganensis subsp. nebraskensis | C. nebraskensis | DOAB 397 | Zea mays | Corn | 2014 | Canada | LAKL00000000 | 53 |
| C. michiganensis subsp. sepedonicus | C. sepedonicus | ATCC 33113T | Solanum tuberosum | Potato | ND | Canada | NC_010407.1 | 54 |
| C. michiganensis subsp. sepedonicus | C. sepedonicus | CFIA-CsR14 | Solanum tuberosum | Potato | ND | Canada | MZMN00000000 | 51 |
| C. michiganensis subsp. sepedonicus | C. sepedonicus | CFIA-Cs3N | Solanum tuberosum | Potato | ND | Canada | MZMM00000000 | 51 |
| C. michiganensis subsp. tessellarius | C. tessellarius | ATCC 33566T | Triticum aestivum | Wheat | 1978 | USA | MZMQ01000000 | 51 |
The first 10 sequences were obtained in this study and announced previously (20), while the remaining ones were retrieved from the NCBI GenBank database.
The original nomenclature of the taxon.
Either revised taxonomy of the strains (17) or their new taxonomic status as proposed in this study. Although new names of six species (i.e., C. capsici, C. insidiosus, C. michiganensis, C. nebraskensis, C. sepedonicus, and C. tessellarius) were formally described previously (17), a formal taxonomic description is needed for the hypothetical new species I to IX, which are based on the ANI/dDDH values (Table 3) and comparative genomics (Table 4).
The strain was isolated from tomato plant but was nonpathogenic on the host of isolation.
NP, nonpathogenic.
ND, not determined.
FIG 1.
Average nucleotide identity (ANI)-based neighbor-joining phylogenetic tree of 40 Clavibacter sp. strains constructed using the ANI calculator online service. Different colors represent hypothetical new species (I to IX). Seven hypothetical novel species were determined among tomato-associated nonpathogenic Clavibacter sp. strains. Furthermore, based on the ANI/dDDH indices, the type strains of C. michiganensis subsp. chilensis and C. michiganensis subsp. phaseoli belong to the same novel species. *, C. michiganensis subsp. californiensis.
TABLE 3.
Average nucleotide identity and digital DNA-DNA hybridization values among the type and/or representative strains of different lineages defined within the genus Clavibactera
| No. | Taxon | Strain | ANIb
(lower diagonal) or dDDH (upper diagonal) (%) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | |||
| 1 | C. michiganensis sensu stricto | LMG 7333T | 57.70 | 58.70 | 48.00 | 48.60 | 49.70 | 48.00 | 49.80 | 49.50 | 44.80 | 43.10 | 40.20 | 40.00 | 37.50 | 37.20 | 36.80 | 34.60 | |
| 2 | C. michiganensis subsp. californiensis | CFBP 8216T | 95/95/95 | 57.80 | 46.40 | 47.90 | 48.60 | 47.40 | 48.10 | 47.90 | 44.80 | 43.70 | 40.50 | 40.40 | 38.50 | 38.40 | 38.00 | 35.70 | |
| 3 | Clavibacter sp. | LMG 26808 | 95/94/95 | 94/95/95 | 45.80 | 47.80 | 48.40 | 47.10 | 47.90 | 47.70 | 44.30 | 42.60 | 39.60 | 39.50 | 37.10 | 37.00 | 36.40 | 34.50 | |
| 4 | C. sepedonicus | ATCC 33113T | 92/92/92 | 92/92/92 | 92/92/92 | 45.10 | 46.20 | 45.20 | 47.00 | 46.60 | 43.90 | 42.30 | 39.10 | 39.10 | 36.40 | 36.20 | 36.00 | 34.00 | |
| 5 | C. insidiosus | LMG 3663T | 93/93/93 | 93/93/93 | 92/92/93 | 92/92/92 | 64.60 | 59.90 | 51.20 | 51.00 | 47.10 | 43.30 | 40.50 | 40.20 | 37.90 | 37.40 | 37.20 | 34.80 | |
| 6 | Clavibacter sp. | CFBP 7494 | 93/93/93 | 92/93/93 | 93/92/93 | 92/92/92 | 96/96/96 | 60.20 | 52.20 | 52.10 | 46.00 | 44.10 | 40.70 | 40.60 | 38.10 | 37.60 | 37.50 | 34.90 | |
| 7 | C. nebraskensis | NCPPB 2581T | 92/93/92 | 92/93/92 | 92/92/92 | 92/92/92 | 95/95/95 | 95/95/95 | 51.40 | 51.10 | 45.30 | 43.30 | 40.70 | 40.70 | 37.70 | 37.20 | 36.90 | 34.90 | |
| 8 | C. michiganensis subsp. chilensis | CFBP 8217T | 93/93/93 | 93/93/93 | 92/93/93 | 92/93/92 | 93/93/93 | 93/93/94 | 93/93/93 | 87.50 | 47.00 | 45.80 | 43.40 | 42.90 | 40.20 | 39.50 | 39.20 | 37.10 | |
| 9 | C. michiganensis subsp. phaseoli | CFBP 8627T | 93/93/93 | 93/93/93 | 92/92/93 | 92/93/92 | 93/93/93 | 93/93/93 | 93/93/93 | 99/99/99 | 47.10 | 45.70 | 43.30 | 42.90 | 40.00 | 39.50 | 39.10 | 37.00 | |
| 10 | Clavibacter sp. | CFBP 8615 | 92/92/92 | 92/92/92 | 91/92/92 | 91/92/92 | 92/92/92 | 92/92/92 | 92/92/92 | 92/92/92 | 92/93/92 | 48.00 | 40.90 | 40.50 | 38.20 | 37.60 | 37.70 | 35.80 | |
| 11 | Clavibacter sp. | CFBP 8019 | 91/92/91 | 91/92/91 | 91/91/91 | 91/92/91 | 91/92/91 | 91/91/91 | 91/91/91 | 92/92/92 | 92/92/92 | 93/93/93 | 40.10 | 40.00 | 36.90 | 36.80 | 36.80 | 34.40 | |
| 12 | C. capsici | PF008T | 90/91/90 | 90/91/91 | 90/90/90 | 90/91/90 | 90/91/90 | 90/90/90 | 90/91/90 | 91/92/91 | 91/92/91 | 90/91/91 | 90/91/90 | 58.50 | 38.40 | 38.50 | 37.70 | 34.50 | |
| 13 | Clavibacter sp. | CF11 | 90/90/90 | 90/90/90 | 90/90/90 | 90/90/90 | 90/90/90 | 91/90/90 | 91/90/90 | 91/91/91 | 91/91/91 | 90/90/90 | 90/90/90 | 95/95/95 | 38.10 | 38.50 | 37.60 | 34.40 | |
| 14 | C. tessellarius | ATCC 33566T | 90/90/89 | 90/91/90 | 89/89/89 | 89/90/89 | 90/90/89 | 90/90/90 | 90/90/89 | 91/91/90 | 91/91/90 | 90/90/90 | 89/90/89 | 90/91/90 | 90/90/90 | 49.00 | 57.70 | 33.50 | |
| 15 | Clavibacter sp. | DOAB 609 | 89/89/89 | 89/90/90 | 89/89/89 | 89/89/89 | 89/89/89 | 90/90/89 | 89/89/89 | 90/90/90 | 90/90/90 | 89/90/89 | 89/89/89 | 90/90/90 | 90/90/90 | 93/93/93 | 47.20 | 33.60 | |
| 16 | Clavibacter sp. | CFBP 8017 | 89/89/89 | 89/90/90 | 89/89/89 | 89/89/89 | 89/89/89 | 90/90/89 | 89/89/89 | 90/90/90 | 90/90/90 | 89/90/89 | 89/89/89 | 90/90/89 | 90/90/89 | 95/95/95 | 92/92/92 | 33.30 | |
| 17 | Clavibacter sp. | CASJ009 | 88/89/89 | 89/90/88 | 88/88/88 | 88/89/88 | 88/89/88 | 88/88/88 | 88/89/88 | 89/90/89 | 89/90/90 | 89/90/89 | 88/89/88 | 88/89/89 | 88/88/88 | 88/89/88 | 88/88/87 | 87/88/87 | |
A combination of ANI and dDDH indices was used to designate a taxonomic status for a given phylogenetic clade, where the “new species” status was assigned to a clade only when both the ANI and dDDH values were below the accepted thresholds (≤95% and ≤70% for ANI and dDDH, respectively [21]).
ANI values were calculated using three different algorithms, i.e., JSpeciesWS, ANI calculator, and OrthoANIu, which are presented from left to right, respectively.
Clavibacter sepedonicus strains formed a monophyletic cluster separate from all the other lineages by ANI values of <93%, which is in coherence with its elevation at the species level (17, 18). Clavibacter insidiosus and C. nebraskensis strains clustered in a monophyletic group showing 95% ANI between the type strains of the species (Table 3). Here also, the dDDH value (59.90%) between the type strains of these two taxa was far below the threshold for species definition (70%) with this method (Table 3), supporting their elevation into separate species (17–19). The two taxa are also different in their hosts of isolation and pathogenicity patterns. While strain CFBP 7494 showed only a 64.60% dDDH value with the type strain of C. insidiosus, the 96% ANI (on the upper edge of species definition) prevents differentiation of this strain from the C. insidiosus species. Strain CFBP 7494 was isolated from tomato but was nonpathogenic on this plant species, while it has been shown that it induces disease symptoms on wheat plants under greenhouse conditions (22). Further evidence, including a comprehensive field survey and host range assay, is needed to elucidate the prevalence and exact taxonomic status of strain CFBP 7494. Surprisingly, the type strains of C. michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis shared 99% ANI with one another and 98% ANI with CFBP 7491, isolated from tomato seeds. These three strains had ANIs below 93% with all the remaining clades, suggesting a novel species within the genus. A high dDDH value (87.50%) also confirmed the close relationship between the type strains of C. michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis (Table 3).
Two peach-colored strains, CFBP 8615 and CFBP 8616, shared 100% ANI with one another, while they differed from all the remaining clades, with ANI values of <93%. Furthermore, nonpathogenic strain CFBP 8019 was determined to be the phylogenetically closest strain to the peach-colored strains, with 93% ANI values between the two clades. These ANI values are far below the accepted threshold (95 to 96%) for the definition of prokaryotic species (21), suggesting that strains CFBP 8615 and CFBP 8616 could be defined as forming a new species separated from CFBP 8019, while strain CFBP 8019 itself belongs to a new stand-alone species (Fig. 1). Low ANI values were also confirmed by dDDH values, which were <48% between the peach-colored strains and all the remaining clades (Table 3). Strain CF11, isolated from soil in a tomato-growing greenhouse (23), as well as the type strain of the pepper pathogen C. capsici (PF008T) clustered in a monophyletic clade, while they differed from one another with 95% ANI and 58.50% dDDH values. Hence, CF11 could be proposed as forming a new species within the genus, while the elevation of former C. michiganensis subsp. capsici to the species level (C. capsici) was confirmed, as proposed by Li et al. (17). The type strain of C. tessellarius showed <93% ANI with the type strains of all the other subspecies/species, confirming the wheat pathogen as a stand-alone species. However, neither strain CFBP 8017 nor strain DOAB 609, which were clustered in a shared clade with the type strain of C. tessellarius, could be included within this species. The ANIs of CFBP 8017 and DOAB 609 with the type strain of C. tessellarius were 95% and 93%, respectively, while the dDDH values between the same strains were 57.70% and 49.00%, respectively (Table 3). Thus, each of the CFBP 8017 and DOAB 609 strains could be defined as representing novel species. The nonpathogenic strain CASJ009 also had ANI values of <90% with all the Clavibacter sp. strains evaluated in this study, indicating that this strain also represents a novel species within the genus (Fig. 1; Table 3).
Comparative genomics.
Comparative genomics data obtained using the RAST online service revealed that the genome size among the Clavibacter sp. strains varied between 3,024 kbp in CFBP 8019 and 3,420 kbp in LMG 26808, with G+C contents ranging from 72.0% in LMG 26808 to 73.7% in C. tessellarius ATCC 33566T. Furthermore, the number of coding sequences (CDS) varied from 2,629 in C. michiganensis subsp. chilensis (CFBP 8217T) to 3,181 in DOAB 609. Genomic characteristics of Clavibacter spp. in a panel of 20 representative strains, which were selected on the basis of the phylogenetic analyses (as detailed above) to cover all lineages/clades of the genus, are shown in Table 4. The number of subsystems varied from 260 in CF11 to 345 in the reference strain of C. michiganensis sensu stricto, NCPB 382, and the type strain of C. sepedonicus, ATCC 33113T. Although the feature counts were similar in most of the subsystems among the pathogenic and nonpathogenic Clavibacter sp. strains, differences in the siderophore-producing subsystems were observed, and the siderophore assembly kit was detected only in the nonpathogenic strain CFBP 8616 (Table 4).
TABLE 4.
Genomic characteristics of Clavibacter sp. strains used in this studya
| Parameter or subsystem feature | C. michiganensis sensu stricto (NCPPB 382) | C. capsici (PF008T) | Clavibacter sp. (CFBP 8615) | Clavibacter sp. (CFBP 8616) | C. michiganensis subsp. californiensis (CFBP 8216T) | Clavibacter sp. (CFBP 7491) | Clavibacter sp. (CFBP 7493) | Clavibacter sp. (CFBP 7494) | Clavibacter sp. (CFBP 8019) | Clavibacter sp. (CFBP 8017) | Clavibacter sp. (DOAB 609) | Clavibacter sp. (CF11) | Clavibacter sp. (LMG 26808) | C. michiganensis subsp. phaseoli (CFBP 8627T) | C. michiganensis subsp. chilensis (CFBP 8217T) | Clavibacter sp. (CASJ009) | C. insidiosus (LMG 3663T) | C. nebraskensis (NCPPB 2581T) | C. sepedonicus (ATCC 33113T) | C. tessellarius (ATCC 33566T) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (bp) | 3,297 | 3,056 | 3,129 | 3,094 | 3,193 | 3,288 | 3,275 | 3,313 | 3,024 | 3,172 | 3,296 | 3,118 | 3,420 | 3,052 | 3,044 | 3,268 | 3,387 | 3,063 | 3,258 | 3,318 |
| GC content (%) | 72.7 | 73.6 | 73.2 | 73.2 | 72.7 | 73.0 | 72.9 | 73.3 | 73.5 | 73.5 | 73.2 | 73.6 | 72.0 | 73.5 | 73.5 | 73.6 | 72.7 | 73.0 | 72.6 | 73.7 |
| No. of CDS | 2,979 | 2,725 | 2,807 | 2,730 | 2,784 | 2,917 | 2,897 | 2,956 | 2,676 | 3,014 | 3,181 | 3,002 | 3,097 | 2,642 | 2,629 | 3,054 | 3,091 | 2,739 | 3,047 | 2,956 |
| No. of subsystems | 345 | 326 | 323 | 319 | 338 | 316 | 342 | 330 | 326 | 263 | 266 | 260 | 340 | 315 | 311 | 341 | 332 | 325 | 345 | 317 |
| No. of RNAs | 51 | 51 | 47 | 47 | 48 | 50 | 48 | 50 | 48 | 48 | 53 | 49 | 57 | 50 | 49 | 51 | 52 | 51 | 51 | 52 |
| No. of CDS for: | ||||||||||||||||||||
| Cofactors, vitamins, pigments | 177 | 158 | 169 | 157 | 165 | 139 | 173 | 161 | 158 | 107 | 116 | 99 | 175 | 167 | 130 | 181 | 165 | 139 | 169 | 176 |
| Virulence, disease, defense | 34 | 38 | 37 | 36 | 38 | 36 | 36 | 32 | 38 | 26 | 25 | 25 | 40 | 35 | 34 | 42 | 36 | 30 | 33 | 22 |
| Resistance to antibiotics/toxic compounds | 19 | 23 | 22 | 21 | 23 | 21 | 21 | 17 | 24 | 17 | 16 | 15 | 25 | 21 | 19 | 27 | 21 | 17 | 20 | 18 |
| Invasion and intracellular resistance | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 14 | 9 | 9 | 10 | 15 | 14 | 15 | 15 | 15 | 13 | 13 | 4 |
| Potassium metabolism | 13 | 12 | 12 | 12 | 13 | 12 | 13 | 13 | 16 | 5 | 5 | 5 | 9 | 9 | 13 | 8 | 13 | 9 | 10 | 11 |
| Phages, prophages, transposable elements | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Iron acquisition and metabolism | 16 | 17 | 13 | 25 | 16 | 8 | 19 | 18 | 13 | 5 | 10 | 5 | 17 | 10 | 10 | 12 | 15 | 14 | 17 | 16 |
| Siderophoresb | ||||||||||||||||||||
| Yersiniabactin | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 | 5 | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 0 |
| Aerobactin | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 3 | 3 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Nucleosides and nucleotides | 87 | 72 | 93 | 92 | 89 | 76 | 100 | 76 | 81 | 80 | 79 | 75 | 74 | 98 | 68 | 90 | 79 | 80 | 99 | 65 |
| Protein metabolism | 211 | 201 | 209 | 192 | 189 | 185 | 209 | 204 | 208 | 166 | 162 | 167 | 205 | 178 | 198 | 192 | 205 | 178 | 213 | 185 |
| Cell division and cell cycle | 26 | 21 | 8 | 8 | 23 | 24 | 24 | 23 | 21 | 0 | 0 | 0 | 8 | 23 | 8 | 22 | 24 | 20 | 22 | 23 |
| Motility and chemotaxis | 1 | 2 | 2 | 2 | 2 | 3 | 1 | 2 | 2 | 0 | 0 | 0 | 3 | 3 | 3 | 3 | 2 | 1 | 2 | 2 |
| Regulation and cell signaling | 33 | 30 | 19 | 20 | 22 | 22 | 25 | 30 | 33 | 14 | 17 | 18 | 36 | 28 | 17 | 29 | 31 | 33 | 35 | 24 |
| Secondary metabolism | 6 | 18 | 5 | 5 | 4 | 5 | 11 | 12 | 11 | 9 | 9 | 9 | 12 | 0 | 1 | 11 | 5 | 5 | 13 | 11 |
| Fatty acids, lipids, and isoprenoids | 88 | 78 | 84 | 103 | 82 | 108 | 78 | 88 | 59 | 44 | 44 | 42 | 79 | 85 | 76 | 82 | 65 | 65 | 80 | 101 |
| Nitrogen metabolism | 7 | 10 | 6 | 6 | 10 | 6 | 7 | 6 | 6 | 6 | 6 | 6 | 7 | 6 | 6 | 11 | 7 | 7 | 6 | 11 |
| Stress response | 67 | 73 | 76 | 69 | 74 | 67 | 64 | 70 | 78 | 27 | 27 | 28 | 68 | 71 | 61 | 82 | 74 | 71 | 78 | 64 |
| Metabolism of aromatic compounds | 10 | 8 | 9 | 8 | 9 | 14 | 10 | 10 | 8 | 4 | 3 | 3 | 10 | 10 | 10 | 18 | 10 | 9 | 10 | 10 |
| Amino acids and derivatives | 224 | 190 | 223 | 212 | 233 | 221 | 210 | 192 | 199 | 217 | 221 | 203 | 236 | 193 | 195 | 224 | 219 | 229 | 232 | 220 |
| Sulfur metabolism | 20 | 16 | 16 | 11 | 9 | 12 | 10 | 16 | 17 | 6 | 5 | 5 | 13 | 10 | 11 | 26 | 17 | 9 | 22 | 18 |
| Phosphorus metabolism | 38 | 26 | 25 | 27 | 28 | 26 | 27 | 23 | 25 | 28 | 27 | 27 | 36 | 25 | 26 | 25 | 23 | 23 | 25 | 24 |
| Carbohydrates | 285 | 266 | 255 | 268 | 267 | 283 | 247 | 268 | 237 | 181 | 194 | 175 | 278 | 263 | 261 | 382 | 266 | 266 | 269 | 281 |
Individual genomes were analyzed using the online annotating service RAST, and protein-encoding sequences (CDS), functions of the genes, and subsystems represented in the genomes were determined for each genome using the SEED-Viewer comparative environment. Features corresponding to cell wall and capsule, DNA metabolism, dormancy and sporulation, membrane transport, respiration, RNA metabolism, and miscellaneous groups are not included since they were common among all the evaluated taxa.
The siderophore assembly kit was detected only in Clavibacter sp. strain CFBP 8616.
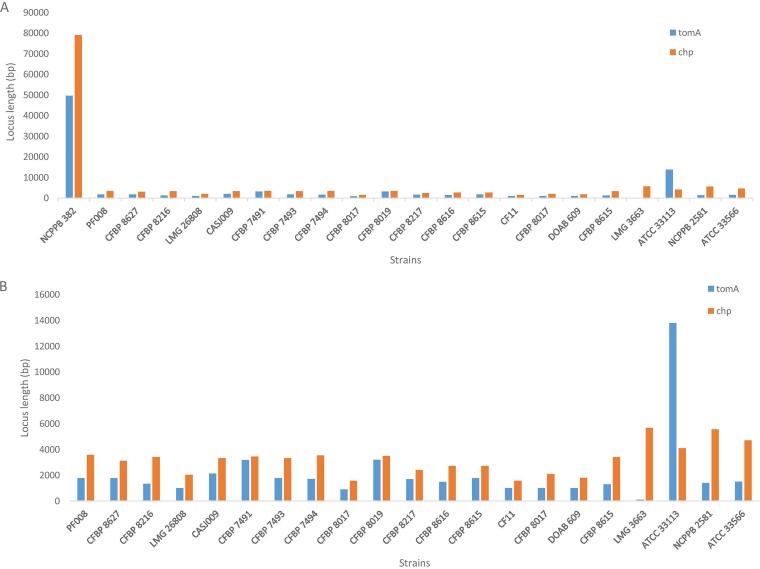
One-versus-one BLASTn- and BLASTp-based explorations using the complete genome sequence of C. michiganensis sensu stricto NCPPB 382 as the reference genome versus the individual Clavibacter sp. strains revealed the lack of pathogenicity determinant genes/clusters in all the tomato-associated nonpathogenic strains evaluated in this study (Table 5). For the chp gene cluster (i.e., loci CMM_0034 to CMM_0077 in the NCPPB 382 genome sequence [accession no. AM711867.1]), only a fraction of the genes were detected in the nonpathogenic strains (Fig. 2). For instance, a sugar phosphate isomerase (CMM_0034) was present in all the nonpathogenic strains. A putative phosphotransferase (CMM_0065) and ATPase (parX = CMM_0066) were found in CFBP 7491, CFBP 7493, and LMG 26808. A hypothetical protein produced by the CMM_0054 locus and a transcriptional regulator protein secreted by CMM_0055 were found only in the type strain of C. michiganensis subsp. californiensis. A serine protease (ppaD = CMM_0075) and a putative ATPase (CMM_0067) were found only in CASJ009 and CFBP 7491, respectively, while a putative DNA invertase (CMM_PS_07) were found in both of the last strains. Among the pathogenicity determinant genes inside the chp gene cluster, ppaA (CMM_0041), pelA1 (CMM_0043), pelA2 (CMM_0051), chpC (CMM_0052), and chpG (CMM_0059) were found in none of the evaluated nonpathogenic strains. Non-chp pathogenicity determinant genes clvG (CMM_1963), clvF (CMM_1964), clvA (micA = CMM_1967), and perF (CMM_2382) were also not detected in the evaluated nonpathogenic bacterial strains. Interestingly, a subtilisin-like serine protease (sbtA = CMM_0070) was found in a number of nonpathogenic strains (Table 5), while nucleotides 1 to 600 were missing in all the nonpathogenic members. The expansin-encoding gene expA (CMM_1480) was found in CFBP 8017, DOAB 609, CFBP 7493, and LMG 26808.
TABLE 5.
Results of one-versus-one BLASTn/BLASTp searches using the genome sequence of Clavibacter michiganensis sensu stricto NCPPB 382 (accession no. AM711867.1) against all other genome sequences shown in Table 2a
| Locus tag on NCPPB 382 genome | Gene or cluster | Length (bp) | Query coverage and sequence similarity (%)b
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. michiganensis sensu stricto (NCPPB 382) | Clavibacter sp. (CFBP 8615) | Clavibacter sp. (CF11) | Clavibacter sp. (CFBP 8017) | Clavibacter sp. (DOAB 609) | C. michiganensis subsp. californiensis (CFBP 8216T) | C. michiganensis subsp. chilensis (CFBP 8217T) | C. capsici (PF008T) | Clavibacter sp. (CFBP 7491) | Clavibacter sp. (CFBP 7493) | Clavibacter sp. (LMG 26808) | Clavibacter sp. (CFBP 7494) | Clavibacter sp. (CFBP 8019) | Clavibacter sp. (CASJ009) | C. michiganensis subsp. phaseoli (CFBP 8627T) | C. insidiosus (LMG 3663T) | C. nebraskensis (NCPPB 2581T) | C. sepedonicus (ATCC 33113T) | C. tessellarius (ATCC 33566T) | Reference | |||
| CMM_0070 | sbtA | 3,102 | [100]100 | [75]75 | [98]82 | ND | [97]81 | [82]82 | [76]94 | [97]82 | [97]89 | ND | ND | ND | [98]82 | [77]82 | [96]89 | ND | [99]80 | ND | ND | 43 |
| CMM_0013 | srtA | 801 | [100]100 | [100]92 | [100]87 | [100]87 | [100]87 | [100]97 | [100]91 | [100]87 | [100]91 | [100]97 | [100]97 | [100]90 | [100]89 | [100]89 | [100]91 | [100]92 | [100]92 | [100]93 | [100]88 | 43 |
| CMM_1480 | expA | 762 | [100]100 | ND | ND | [77]81 | [77]82 | ND | ND | ND | ND | [100]99 | [100]99 | ND | ND | ND | ND | ND | ND | ND | NDc | 26 |
| CMM_1673 | xysA | 1,296 | [100]100 | [98]90 | [99]87 | [99]92 | [97]91 | [99]95 | [81]76 | [100]88 | [83]76 | [99]94 | [99]94 | [99]93 | [99]89 | [48]80 | [81]75 | [99]95 | [99]92 | ND | [99]92 | 43 |
| CMM_1674 | xysB | 2,010 | [100]100 | [92]93 | [100]89 | [100]89 | [54]75 | [100]93 | [100]93 | [100]90 | [100]93 | [94]95 | [99]94 | [100]92 | [100]92 | [95]90 | [93]93 | [100]90 | [100]91 | ND | [99]89 | 43 |
| CMM_2443 | celB | 1,608 | [100]100 | ND | ND | [100]89 | [100]87 | ND | ND | [100]88 | ND | [100]91 | [100]91 | [100]91 | ND | ND | ND | [100]92 | [64]85 | [100]91 | [100]85 | 26 |
| CMM_2691 | NAd | 1,002 | [100]100 | [99]89 | [100]89 | [100]89 | [99]90 | [100]91 | [100]89 | [99]89 | [100]89 | [100]94 | [100]94 | [89]92 | [99]90 | [97]83 | [100]88 | [100]94 | [98]92 | [99]89 | [100]90 | 26 |
| CMM_2692 | NA | 1,410 | [100]100 | [90]93 | [100]86 | [100]86 | [100]86 | [91]96 | [77]90 | [100]89 | [30]86 | [100]91 | [100]91 | [100]90 | [100]87 | ND | [90]90 | [100]92 | [100]91 | [100]86 | [100]88 | 26 |
| CMM_2871 | NA | 1,491 | [100]100 | ND | ND | ND | ND | ND | ND | ND | ND | [100]90 | [100]90 | ND | ND | ND | ND | [92]88 | [98]92 | ND | [100]92 | 26 |
| CMM_2645 | vatr1 | 624 | [100]100 | [100]94 | [100]92 | [100]92 | [100]91 | [85]98 | [100]93 | [100]92 | [100]94 | [100]98 | [100]98 | [100]96 | [100]93 | [100]89 | [100]94 | [100]96 | [100]96 | [100]94 | [100]92 | 25 |
| CMM_2969 | vatr2 | 1,323 | [100]100 | [100]92 | [100]92 | [99]92 | [100]92 | [100]96 | [100]94 | [100]92 | [100]94 | [100]95 | [100]95 | [100]94 | [100]93 | [100]90 | [100]94 | [100]93 | [100]93 | [100]93 | [100]91 | 25 |
| pCM1_0020 | celA | 2,241 | ND | ND | [46]84 | [46]75 | [47]75 | ND | ND | [45]77 | ND | ND | ND | ND | ND | ND | ND | [97]90 | [76]84 | ND | [45]75 | 26 |
Putative pathogenicity determinant genes/regions described in the literature were subjected to the analyses. While nine chromosomal genes (i.e., chpC, chpG, clvA [micA], clvF, clvG, pelA1, pelA2, perF, and ppaA) as well as the pCM2 plasmid-borne pathogenicity-associated gene pat-1 were determined as present exclusively in C. michiganensis sensu stricto, the remaining genes were detected in different phylogenetic lineages regardless of their pathogenicity and host range.
The values in brackets refer to the query coverage of the locus, while the other values refer to the sequence similarity between the reference strain (NCPPB 382T) and the strain in question. Query coverage of <50% was not considered reliable data. ND, not detected (absence of the target gene/cluster).
A putative expansin protein was found in this species (GenBank accession no. OQJ63896.1), in which the nucleotide sequence was different from that of CMM_1480 expA.
NA, not assigned.
FIG 2.
Results of comparative genomics on the two main pathogenicity determinant regions of Clavibacter michiganensis sensu stricto NCPPB 382 (i.e., chp and tomA) against the nonpathogenic tomato-associated strains, as well as the type strains of five species that are pathogenic on other plants. While only a fraction (<10%) of the clusters were detected in the nonpathogenic strains (A), variations were observed in the patterns of the genes (B).
For the tomA gene cluster (CMM_0078 to CMM_0112 in the genome sequence of NCPPB 382), a β-glucosidase-related gene (bglC = CMM_0083) was found in the type strains of C. michiganensis subsp. chilensis, C. capsici, and C. michiganensis subsp. phaseoli, as well as strains CFBP 8615, CFBP 8616, CFBP 7493, LMG 26808, and CFBP 7494. A putative alpha-glucosidase gene (aglA = CMM_0106) was found in CFBP 7491, CFBP 8019, and CASJ009. Furthermore, a putative ABC-type sugar transport permease gene (CMM_0108) was found in CFBP 7491 and CASJ009, while the srtA gene (CMM_0013), which encodes a putative sortase enzyme, was found in all the evaluated nonpathogenic strains in this study (Table 5).
We also assessed the presence of a set of eight virulence genes, i.e., celB (CMM_2443), pelA1 (CMM_0043), pelA2 (CMM_0051), xysA (CMM_1673), xysB (CMM_1674), CMM_2691, CMM_2692, and CMM_2871, which are responsible for cell wall degradation at the later stages of tomato infection by C. michiganensis sensu stricto. The polygalacturonase-encoding locus CMM_2871 was found in tomato-associated strains CFBP 7493 and LMG 26808, as well as the type strains of C. insidiosus (LMG 3663T), C. nebraskensis (NCPPB 2581T), and C. tessellarius (ATCC 33566T). Furthermore, celB (CMM_2443), which is a homolog of the plasmid-born celA gene (24), was present in tomato-associated nonpathogenic strains CFBP 8017, DOAB 609, CFBP 7493, LMG 26808, and CFBP 7494. The four loci CMM_1673, CMM_1674, CMM_2691, and CMM_2692 were present in all tomato-associated strains, except for CMM_2692, which was absent in CASJ009, while the query coverage varied between 30 and 100% and the sequence similarity varied from 75 to 100% among the strains. Surprisingly, the virulence-associated transcriptional regulator genes vatr1 (CMM_2645) and vatr2 (CMM_2969), which regulate C. michiganensis sensu stricto virulence during infection (25), were present in all the strains evaluated in this study regardless of their pathogenicity status (Table 5).
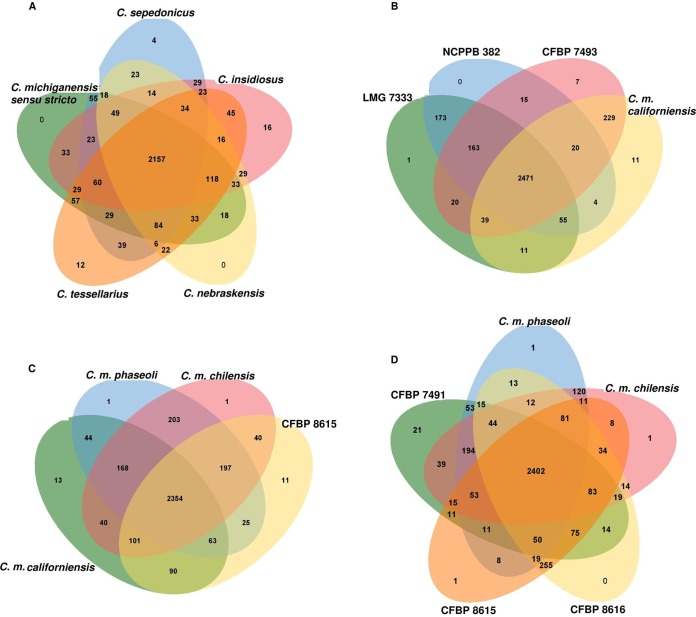
Orthologous gene clusters were determined using the OrthoVenn online service through four-versus-four and five-versus-five designations of the representative strains from different phylogenetic lineages (Fig. 3A to D). Type strains of the five former C. michiganensis sensu lato subspecies shared 2,157 proteins in their genome sequences (Fig. 3A). Although none of the type strains of C. michiganensis sensu stricto and C. nebraskensis showed unique proteins in their sequences, the type strains of C. sepedonicus, C. tessellarius, and C. insidiosus showed 4, 12, and 16 unique proteins among their genome sequences, respectively. Furthermore, when the two phylogenetic neighboring clades of C. michiganensis sensu stricto (i.e., C. michiganensis subsp. californiensis and CFBP 7493) were compared with the type/reference strains of C. michiganensis sensu stricto (LMG 7333T and NCPPB 382), 7 and 11 unique proteins were detected in the genome sequences of CFBP 7493 and C. michiganensis subsp. californiensis strain CFBP 8216T, respectively (Fig. 3B). Unique and shared proteins in the type strains of C. michiganensis subsp. californiensis, C. michiganensis subsp. phaseoli, and C. michiganensis subsp. chilensis as well as the atypical peach-colored strain CFBP 8615 are shown in Fig. 3C and D.
FIG 3.
Venn diagrams constructed using the OrthoVenn online service, showing the distribution of shared gene families (orthologous clusters) among different sets of Clavibacter sp. strains.
Plasmids, phages, and bacteriocins.
No integrative plasmid (episome) was detected using the PlasmidFinder online service in the draft genome sequences of bacterial strains investigated in this study, except for LMG 26808, in which two Enterobacteriaceae plasmids, IncL/M(pOXA-48) and IncR, were identified. Surprisingly, sequences homologous to the plasmid-born celA gene were detected in the sequences of the type strains of C. insidiosus (LMG 3663T) and C. nebraskensis (NCPPB 2581T), with query coverage of 97% and 76%, respectively, and sequence similarity of 90% and 84% to the sequence of the reference strain NCPPB 382 (Table 5). On the other hand, plasmid profiling detected the two expected plasmids pCM1 (≈27 kb) and pCM2 (≈70 kb) in the tomato-pathogenic strain C. michiganensis sensu stricto ICMP 22049 (data not shown). However, type strains of C. michiganensis subsp. phaseoli and C. michiganensis subsp. chilensis, as well as the two peach-colored strains CFBP 8615 and CFBP 8616, did not carry any detectable plasmid (data not shown).
The PHASTER online service was used to detect prophage sequences within the bacterial genomes. Altogether, five hypothetical prophage groups, i.e., Gordon_Schwabeltier (accession no. NC_031255), Gordon_Smoothie (NC_030696), N15 (NC_001901), P1 (NC_005856), and Phi92 (NC_023693), were detected in the Clavibacter sp. strains investigated in this study (Table 6). Prophages were detected in tomato-associated strains CFBP 8615, CFBP 7491, LMG 26808, and CASJ009, as shown in Table 6. While strain LMG 26808 contained three prophages, only one prophage per strain was detected in strains CFBP 8615, CFBP 7491, and CASJ009. The phage Phi92, which was originally isolated from a pathogenic Escherichia coli strain, was detected in CFBP 8615, LMG 26808, CASJ009, C. sepedonicus (ATCC 33113T), and C. tessellarius (ATCC 33566T), while each of the remaining four prophages was detected in only one strain (Table 6).
TABLE 6.
Prophages within the genome sequences of Clavibacter sp. strains detected using the online service PHASTERa
| Taxon | Strain | Region length (kb) | Completeness | No. of proteins | Position (contig: bp) | Phage (GenBank accession no.) | G+C content (%) |
|---|---|---|---|---|---|---|---|
| Clavibacter sp. | CFBP 8615 | 6.9 | Incomplete | 8 | 38: 3643–10586 | Phi92 (NC_023693) | 71.28 |
| Clavibacter sp. | CFBP 7491 | 6.8 | Incomplete | 14 | 4: 13543–20391 | Gordon_Schwabeltier (NC_031255) | 66.29 |
| Clavibacter sp. | LMG 26808 | 9.9 | Incomplete | 10 | 13: 128032–137979 | Phi92 (NC_023693) | 71.79 |
| 7 | Incomplete | 11 | 15: 712–7762 | N15 (NC_001901) | 46.49 | ||
| 23.7 | Incomplete | 10 | 15: 5546–29310 | P1 (NC_005856) | 48.19 | ||
| Clavibacter sp. | CASJ009 | 7.7 | Incomplete | 9 | 1009722–1017492 | Phi92 (NC_023693) | 71.29 |
| C. insidiosus | LMG 3663T | 6.7 | Incomplete | 16 | scaffold2: 5055–11844 | Gordon_Smoothie (NC_030696) | 66.95 |
| C. sepedonicus | ATCC 33113T | 7.1 | Incomplete | 10 | 42745–49937 | Phi92 (NC_023693) | 70.71 |
| 7.7 | Incomplete | 8 | 660037–667813 | Phi92 (NC_023693) | 71.20 |
Five prophage groups, i.e., Gordon_Schwabeltier, Gordon_Smoothie, N15, P1, and Phi92, were detected in strains CFBP 8615, CFBP 7491, LMG 26808, CASJ009, LMG 3663, and ATCC 33113. No prophage was detected within the genome sequences of the strains ATCC 33566T, CF11, CFBP 7493, CFBP 7494, CFBP 8017, CFBP 8019, CFBP 8216T, CFBP 8217T, CFBP 8616, CFBP 8627T, DOAB 609, NCPPB 2581T, NCPPB 382, and PF008T.
In silico screening for bacteriocins and antibiotic peptides revealed distinct differences between the pathogenic and nonpathogenic tomato-associated Clavibacter sp. strains. The lantibiotic Michiganin A was detected in all 12 C. michiganensis sensu stricto strains (data not shown) but was not detected in the nonpathogenic tomato-associated strains or in the pathogenic strains on other plant species. Furthermore, sactipeptides (peptides with cysteine sulfur-to-α-carbon cross-links) were the most common group of bacteriocins among all the Clavibacter sp. strains. Indeed, except for tomato-pathogenic C. michiganensis sensu stricto strains, all the strains which contained bacteriocins had at least one type of sactipeptides (Table 7). Linear azol(in)e-containing peptides (LAPs) were detected in both the pathogenic and nonpathogenic strains, while thiopeptides which are commonly produced by Actinobacteria were found in the two phylogenetically closely related nonpathogenic strains CFBP 7493 and LMG 26808. Enterocin_AS_48, a circular bacteriocin produced by Enterococcus spp., was detected exclusively in strain CF11, which was originally isolated from soil in a tomato-growing greenhouse (23).
TABLE 7.
In silico screening for bacteriocins and antibiotic peptides among the Clavibacter sp. genome sequences analyzed in this studya
| Taxon | Strain | Contig no. | Start codon | End codon | Class |
|---|---|---|---|---|---|
| C. michiganensis sensu stricto | NCPPB 382 | 1 | 2211104 | 2233501 | 63.1; Michiganin A (Lantibiotic) |
| C. capsici | PF008T | 1 | 2406476 | 2426476 | Sactipeptides |
| Clavibacter sp. | CFBP 8615 | 14 | 7079 | 27079 | LAPs |
| 91 | 1570 | 18430 | Sactipeptides | ||
| Clavibacter sp. | CFBP 8616 | 166 | 4888 | 15112 | Sactipeptides |
| C. michiganensis subsp. californiensis | CFBP 8216T | 201 | 9178 | 10822 | Sactipeptides |
| Clavibacter sp. | CFBP 7491 | 224 | 8131 | 11869 | Sactipeptides |
| Clavibacter sp. | CFBP 7493 | 1 | 25925 | 45925 | Thiopeptide, LAPs |
| 226 | -8455 | 11545 | Sactipeptides | ||
| Clavibacter sp. | CFBP 7494 | 9 | 267758 | 287758 | LAPs |
| 15 | 89075 | 109075 | Sactipeptides | ||
| Clavibacter sp. | CFBP 8019 | 11 | 186443 | 206443 | Sactipeptides |
| 17 | 190604 | 210604 | LAPs | ||
| Clavibacter sp. | LMG 26808 | 6 | 43154 | 63154 | Thiopeptide, LAPs |
| 10 | 130865 | 150865 | Sactipeptides | ||
| Clavibacter sp. | CF11 | 17 | 47765 | 67765 | Sactipeptides |
| 2 | 247508 | 267508 | LAPs | ||
| 13 | 138056 | 158368 | 150.1, Enterocin_AS_48 | ||
| Clavibacter sp. | DOAB 609 | 20 | 4781 | 24781 | Sactipeptides |
| 30 | 31400 | 51400 | LAPs | ||
| Clavibacter sp. | CFBP 8017 | 33 | 39236 | 59236 | LAPs |
| 46 | 11546 | 31546 | Sactipeptides | ||
| C. michiganensis subsp. phaseoli | CFBP 8627T | 212 | −9916 | 10084 | Sactipeptides |
| C. michiganensis subsp. chilensis | CFBP 8217T | NDb | ND | ND | ND |
| Clavibacter sp. | CASJ009 | ND | ND | ND | ND |
| C. insidiosus | LMG 3663T | 1 | 2292719 | 2312719 | Sactipeptides |
| C. nebraskensis | NCPPB 2581T | 1 | 2422475 | 2442475 | Sactipeptides |
| C. sepedonicus | ATCC 33113T | 1 | 2663669 | 2683669 | Sactipeptides |
| 1 | 3119744 | 3139861 | 294.1, Plantathiazolicin (Plantazolicin) | ||
| C. tessellarius | ATCC 33566T | 1 | 1820699 | 1840699 | Sactipeptides |
The lantibiotic Michiganin A was detected in all 12 C. michiganensis sensu stricto strains (Fig. 1) but was not found in nonpathogenic tomato-associated strains or in the pathogenic strains on other plant species.
ND, not detected.
DISCUSSION
In this study, using phylogenetic analyses, comparative genomics, and pathogenicity assays, we provide novel insight into the diversity of Clavibacter sp. strains, with a special focus on tomato-associated members of the genus. Phylogenetic analyses accomplished with ANI and dDDH calculations revealed a higher genetic diversity of Clavibacter sp. strains than has so far been assumed (17). We also aimed to decipher the phylogenetic positions of the three newly described subspecies of C. michiganensis sensu lato (i.e., C. michiganensis subsp. californiensis, C. michiganensis subsp. chilensis, and C. michiganensis subsp. phaseoli). Although our results confirm that these three subspecies are no longer included in C. michiganensis sensu stricto, we still used their original names in this study to avoid confusion. A formal taxonomic study would provide appropriate epithets for these taxa. On the other hand, BLAST-based comparative genomics revealed that several genes (i.e., vatr1, vatr2, xysA, xysB, and srtA) which had previously been identified as pathogenicity determinants were present in all the pathogenic and nonpathogenic tomato-associated strains, indicating further complexities in the functions of these genes (25, 26). Nonpathogenic counterparts of actinobacterial plant pathogens have frequently been isolated from a set of taxonomically diverse plant species which were distant from the main host of the pathogen. For instance, both the pathogenic and nonpathogenic Curtobacterium flaccumfaciens strains phylogenetically closely related to the common bean pathogen C. flaccumfaciens pv. flaccumfaciens were isolated from solanaceous annual crops, i.e., eggplant, pepper, and tomato (2). While the pathogenic and nonpathogenic strains of C. flaccumfaciens are not differentiable using routine molecular techniques, e.g., MLSA (27), all the nonpathogenic members of C. michiganensis sensu lato could be differentiated from the tomato-pathogenic C. michiganensis sensu stricto strains (11).
Nonpathogenic strains of Clavibacter spp. were consistently reported to associate with seeds, transplants, and aerial portions of tomato plants (11, 13, 22, 28). However, until recently, only a few genome sequences from nonpathogenic Clavibacter sp. strains were available (22, 28), limiting our understanding of the putative role of these bacteria on the host plants and their environment. In a preliminary complete genome sequence-based comparative study, Zaluga et al. (28) investigated the genome of tomato-associated nonpathogenic strain LMG 26808 and provided initial insights into the genetic bases of differences between the pathogenic and nonpathogenic members of C. michiganensis sensu lato. However, it has been noted that LMG 26808 is phylogenetically very close to the C. michiganensis sensu stricto clade, leaving a greater portion of nonpathogenic Clavibacter sp. diversity uninvestigated (28). Our results revealed that strain LMG 26808 as well as two other strains, i.e., CFBP 7493 and CFBP 8216T, are phylogenetically closely related and fall into a monophyletic clade along with the pathogenic members of C. michiganensis sensu stricto (Fig. 1). The genomic contents of the two clades represented by LMG 26808/CFBP 7493 as “hypothetical new species I” and CFBP 8216T as “hypothetical new species II” varied in the pathogenicity-related genes sbtA, expA, and celB and the locus CMM_2871, while there was no difference between the strains LMG 26808 and CFBP 7493 in the evaluated genomic areas (Table 5). More specifically, the expA gene (CMM_1480), which is responsible for expansin production (29), and a polygalacturonase encoded by the CMM_2871 locus at the final stages of infection were found in the CFBP 7493 and LMG 26808 strains but not in the type strain of C. michiganensis subsp. californiensis (Table 5).
Recently, Li and colleagues (17) reevaluated the taxonomy of C. michiganensis sensu lato and proposed the reclassification of each of the former C. michiganensis subspecies into species status. Since only two genome sequences of C. insidiosus were available at that time (both isolated in the United States) (29), we sequenced two further “Old World” strains (i.e., CFBP 1195 and CFBP 6488, isolated in the United Kingdom and the Czech Republic, respectively) to gain a precise vision of the intraspecies diversity of the alfalfa pathogen (20). Our analyses confirm the existence of C. capsici, C. insidiosus, C. michiganensis sensu stricto, C. nebraskensis, C. sepedonicus, and C. tessellarius as stand-alone species. While the 95% ANI did not solely support the separation of the alfalfa and maize pathogens, the 59.90% dDDH between the type strains of the two taxa, as well as their distinct host plants, could be considered evidence for the separation of C. insidiosus and C. nebraskensis (Tables 2 and 3). Strain CFBP 7494, which was isolated from tomato seeds and causes disease symptoms on wheat plants under greenhouse conditions (22), clustered in a monophyletic clade with the alfalfa-pathogenic strains and still fell into the C. insidiosus species, with 96% ANI and 64.60% dDDH. While the phylogenetic position of strain CFBP 7494 was clarified in these analyses, only further investigations using a larger collection of strains will shed a light on the genetic content, biological characteristics, and taxonomic status of tomato-associated wheat-pathogenic members of Clavibacter spp.
Draft genome sequences of the three new subspecies of C. michiganensis sensu lato revealed their phylogenetic position, highlighting inaccuracy in the nomenclature of C. michiganensis subsp. chilensis and C. michiganensis subsp. phaseoli. Type strains of these two subspecies shared 99% ANI and 87.50% dDDH with one another, indicating a synonymy and orientating toward the proposal of a novel unique new species (Fig. 1 and Table 3). The type strain of C. michiganensis subsp. phaseoli (CFBP 8627T) was isolated from common bean seeds in Spain and reported to cause bacterial bean leaf-yellowing disease in greenhouse assays (14). However, we could not observe any symptoms on inoculated common bean plants even when three different cultivars were evaluated in the pathogenicity tests (Table 1). This could be attributed to differences in the environmental conditions between the two assays and probably also differences in the susceptibility of common bean cultivars used in the two studies. Further field surveys are needed to decipher potential natural occurrence under the field conditions and the putative frequency and prevalence of the common bean-associated Clavibacter sp. strains.
Comparative genomics revealed that tomato-associated C. michiganensis sensu lato strains are adapted to a nonpathogenic lifestyle, which is reflected by the lack of pathogenicity gene clusters present in the pathogenic members (Table 5; Fig. 2). Although the absence of almost all of the 129-kb chp/tomA region was common among all the nonpathogenic strains, some of the putative virulence factors were present in the nonpathogenic strains, suggesting contributions of these genes in the endophytic lifestyle of the bacteria. An in-depth comparative analysis with newly sequenced Clavibacter sp. genomes allowed us to illustrate a more precise insight underlying genetic contents of these bacteria. For instance, the expA gene was detected in the nonpathogenic strains CFBP 7493 and LMG 26808 as well as the wheat-pathogenic strains CFBP 8017 and DOAB 609 but not in the type strain of C. tessellarius (Table 5). Microbial expansins are found in the genomes of several plant-pathogenic bacteria, and it is assumed that they provide particular advantages to xylem-dwelling phytopathogens (29, 30). Expansin enhances cellulose breakdown by cellulase enzymes in the later stages of pathogen invasion (31). These observations correlate with the initial assumptions that nonpathogenic Clavibacter sp. strains must have lost or never contained prominent virulence determinants (e.g., the 129-kb chp/tomA region) responsible for disease induction in tomato plants. With the availability of genome sequences covering a broader diversity of nonpathogenic Clavibacter sp. strains, one would assume that the gene flow and evolutionary pathways of pathogenicity determinants in the genus are similar to those that have previously been estimated for plant-pathogenic xanthomonads (4).
In conclusion, our results obtained from the analyses of 40 genome sequences provide a comprehensive insight into the genetic diversity of Clavibacter spp. and confirm the recent taxonomic revision of the genus. However, phylogenetic analyses suggest that the recently described subspecies C. michiganensis subsp. chilensis and C. michiganensis subsp. phaseoli should be classified as members of the same novel species (13, 14). Taking together all the phylogenetic, genomic, and pathogenicity data, nine hypothetical novel species could be identified within Clavibacter spp., seven of which (i.e., hypothetical new species I, II, III, IV, V, VIII, and IX as shown in Fig. 1) were isolated from asymptomatic tomato tissues or seed lots. These findings raise the question whether the current taxonomy of tomato-associated Clavibacter sp. strains is technically applicable for quarantine purposes and emphasize at the same time the need for more detailed taxonomic investigations among the phylogenetically diverse tomato-associated Clavibacter sp. strains. Indeed, the only pathogenic lineage of tomato-associated strains is C. michiganensis sensu stricto, while the seven nonpathogenic lineages need to be designated novel formal taxa. This would help the plant pathology agencies and tomato seed industry inspectors to specifically target the enemy and neglect the nonpathogenic lineages. Only a formal taxonomic study would address this issue, with delineation of appropriate epithets and species descriptions for these new taxa. On the other hand, the nine pathogenicity determinant genes (Table 5) would be appropriate targets for the development of novel genome-informed detection methods for differentiation of tomato-pathogenic and nonpathogenic strains.
MATERIALS AND METHODS
Bacterial strains and genome sequences.
Draft genome sequences of 10 Clavibacter sp. strains (Table 2) were prepared using the shotgun genome sequencing facility of the Illumina HiSeq X platform. The culture media, bacterial growth conditions, genomic DNA preparation, sequencing procedure, and genome annotation were described previously (20). In this framework, we investigated the type strains of C. michiganensis subsp. californiensis (CFBP 8216T), C. michiganensis subsp. chilensis (CFBP 8217T), and C. michiganensis subsp. phaseoli (CFBP 8627T), two C. insidiosus strains (CFBP 1195 and CFBP 6488), one C. nebraskensis strain (CFBP 7577), and the nonpathogenic peach-colored (i.e., CFBP 8615 and CFBP 8616) and yellow-pigmented (i.e., CFBP 7491 and CFBP 7493) strains. Furthermore, all the publicly available genome sequences of Clavibacter spp., until April 2019, were retrieved from the NCBI GenBank database and included in the phylogenetic analysis and comparative genomics. Table 2 describes the 40 Clavibacter sp. strains used in this study, their origins of isolation, and their pathogenicity features.
Pathogenicity tests and host range.
Due to the close phylogenetic relationships between the type strains of C. michiganensis subsp. phaseoli (CFBP 8627T) and C. michiganensis subsp. chilensis (CFBP 8217T), these two strains, as well as the atypical peach-colored strains recently isolated from tomato (16), were subjected to the pathogenicity tests and host range assays under greenhouse conditions. Pathogenicity tests were performed on bell pepper (cv. Sereno), chili pepper (cv. Aziz), common bean (cv. Red kidney, Pinto, and Navy), cowpea (Vigna unguiculata cv. Partow), mung bean (Vigna radiata cv. Mashhad), and tomato (cv. Sunseed 6189) plants. The plant growth conditions, inoculation procedure, and incubation environment were the same as detailed previously (32, 33). Inoculated plants were periodically monitored for the appearance of disease symptoms up to 30 days postinoculation. Positive- and negative-control plants were treated in the same manner using the standard strain of C. michiganensis sensu stricto (ICMP 22049, isolated from a symptomatic tomato plant in Iran in 2015 [16]) and sterile distilled water, respectively. Koch’s postulates were satisfied by reisolating the inoculated strains on yeast extract-peptone-glucose agar (YPGA) medium from all inoculated plants. Confirmation of the identity of the reisolated bacteria was made by determining Gram reaction and colony characteristics on yeast extract-dextrose-calcium carbonate (YDC) agar medium as well as by using the genus-specific primer pair CMR16F1/CMR16R1 (34) as described previously (16). The pathogenicity tests were conducted twice.
Phylogenetic analyses.
Average nucleotide identity (ANI) was calculated among all the Clavibacter sp. genome sequences included in this study. The ANI was estimated using both one-versus-one and all-versus-all strategies via different algorithms, i.e., JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/) (35), ANI calculator (http://enve-omics.ce.gatech.edu/g-matrix/) (36), and OrthoANIu (https://www.ezbiocloud.net/tools/orthoaniu) (37). An ANI-based neighbor-joining phylogenetic tree was constructed using the ANI calculator online service, and the genome sequence of Leifsonia xyli subsp. cynodontis (DSM 46306; accession no. NC_022438.1) was used as an outgroup in the tree. Additionally, the Genome-to-Genome Distance Calculator online service (http://ggdc.dsmz.de/distcalc2.php) was used to calculate digital DNA-DNA hybridization (dDDH) values, which infer the genome-to-genome distances between pairs of genomes based on the Genome BLAST Distance Phylogeny (38). A combination of ANI and dDDH indices was used to designate a taxonomic status to a given phylogenetic clade, where the “new species” status was assigned to a clade only when both ANI and dDDH values were below the accepted threshold (≤95% and ≤70% for ANI and dDDH, respectively) (21).
Comparative genomics.
Twenty strains representing the entire genetic diversity of Clavibacter spp. based on the ANI/dDDH data, host of isolation, and pathogenicity characteristics were subjected to the comparative genomics analyses. Type strains of all the C. michiganensis sensu lato species/subspecies, as well as all the individual strains sharing ≤95% and ≤70% ANI and dDDH values, respectively, with the other taxa were selected for comparative genomics analyses. Genome length (bp), G+C content (%), and total numbers of protein-coding sequences (CDS), RNA genes, and pseudogenes were determined for all the genomes.
The online annotating service RAST (Rapid Annotations using Subsystems Technology) (http://rast.nmpdr.org/) (39) was used for fully automated annotation of the bacterial genomes, and the obtained information was used to reconstruct metabolic networks and subsystems. A subsystem is a set of functional roles that the annotator considers related categories. Subsystems represent a collection of functionally related protein families that make up a metabolic pathway (e.g., iron acquisition and metabolism), a complex (e.g., the ribosome), or a class of proteins (e.g., bacteriocins) (40). Subsequently, the genomes were transferred to the comparative environment of the SEED-Viewer (http://www.theseed.org/wiki/Main_Page) (41) for comparative genomics analyses. The SEED-Viewer was used for the identification of protein-encoding sequences (CDS), assigning functions to the genes, and prediction of represented gene clusters in the genomes. The distribution of the genes among various clusters and specific protein-encoding genes within each cluster were estimated using the same service. Furthermore, BLASTn/BLASTp-based investigation was performed to decipher whether the pathogenicity determinant genes/clusters are present in the genomes (26). Using the complete genome of C. michiganensis sensu stricto NCPPB 382, one-versus-one BLASTn/BLASTp searches were done against the sequences of the pathogenicity island (a 129-kb low-G+C region which includes chp and tomA clusters) as well as several individual genes proposed to have effective contributions to the virulence of C. michiganensis sensu stricto (22, 25, 26, 28, 42, 43). Proteins with amino acid sequence similarities higher than 50% and with a query coverage higher than 70% were considered homologs (28).
We also screened the genome sequences for the presence of hypothetical bacteriocin-encoding genes/clusters using the web-based tool BAGEL4 (http://bagel4.molgenrug.nl/) (44). BAGEL4 combines direct mining for the structural genes with indirect mining for bacteriocin-associated genes. Furthermore, the online service PlasmidFinder 2.0 (https://cge.cbs.dtu.dk/services/PlasmidFinder/) (45) was used for the screening of all the genomic sequences for the presence of integrative plasmids/episomes. Identification and annotation of prophage sequences within bacterial genomes were performed using the online service PHASTER (PHAge Search Tool Enhanced Release) (http://phaster.ca/) (46). Given the fact that identification of overlaps among the orthologous clusters can enable us to elucidate the function and evolution of proteins across multiple species, genome-wide comparisons and visualization of orthologous clusters were performed using the online service OrthoVenn (http://www.bioinfogenome.net/OrthoVenn/) (47). The analyses were conducted on the “bacteria” section of the platform using default settings (E value, 1e−5; inflation value, 1.5). Regarding the numeric limitation of OrthoVenn in the handling of bacterial genomes (up to six genomes per run), different series of the strains were evaluated using the same parameters.
Plasmid profiling.
In order to further investigate the genetic contents of nonpathogenic tomato-associated Clavibacter sp. strains, we evaluated the plasmid profiles of the type strains of C. michiganensis subsp. chilensis (CFBP 8217T) and C. michiganensis subsp. phaseoli (CFBP 8627T) as well as the two peach-colored strains CFBP 8615 and CFBP 8616 isolated from tomato. Plasmids were isolated according to the procedure described by Kotchoni et al. (48) with minor modifications. The tomato-pathogenic C. michiganensis sensu stricto strain ICMP 22049 was used as positive control (49). Bacterial strains were grown in 50 ml Luria-Bertani (LB) medium on a shaker at 150 rpm and 27°C for 48 h. Bacterial cells were harvested by centrifugation at 13,000 × g for 1 min at room temperature, and the pellet was resuspended in 200 μl of “solution I” of the protocol of Kotchoni et al. (48), mixed well, and incubated at 37°C for 20 min. Subsequently, 400 μl of freshly prepared “solution II” was added to the microtubes and mixed well by inverting gently four to six times to avoid breaking the plasmid(s), and then 200 μl of “solution III” was immediately added, mixed very gently, and incubated at 4°C for 15 min without any intervention. The mixture was centrifuged at 10,000 × g for 5 min, the supernatant was transferred to a new microtube, and a 0.6 volume of isopropanol was added to the supernatant, mixed gently by inverting four to six times, kept at room temperature for 10 min, and then centrifuged at 10,000 × g for 5 min, and the supernatant was discarded. The pellet containing precipitated plasmid DNA was washed with 400 μl of 70% (vol/vol) ethanol and centrifuged at 10,000 × g for 3 min at room temperature. The supernatant was removed, and the pellet was air dried. Finally, plasmid DNA was resuspended in 50 μl sterile distilled water containing 10 mg/ml RNase A. The presence of plasmids was analyzed on a 0.6% agarose gel as described previously (27).
Data availability.
The data sets generated for this study were previously published (20) and can be found in the NCBI GenBank Database (see Table 2).
Supplementary Material
ACKNOWLEDGMENTS
We thank the French Collection of Plant-Associated Bacteria (CIRM-CFBP) (http://www6.inra.fr/cirm_eng/CFBP-Plant-Associated-Bacteria) for strain conservation and supply, CATI BBRIC for bioinformatics facilities, and the French Network on Xanthomonads (FNX) (https://www.reseau-xantho.org/) for recurrent scientific exchanges. We benefited from interactions promoted by COST Action CA16107 EuroXanth (https://euroxanth.eu/).
We declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
E.O. and M.-A.J. conceived and designed the study, with assistance from T.R. E.O., S.M.T., M.A., and S.Z. carried out the experiments. M.B. and P.P. performed the genome sequencing and annotation. E.O. analyzed and interpreted the data with assistance from T.R., M.B., and M.-A.J. E.O. prepared the manuscript with assistance from M.-A.J. All authors revised the final manuscript.
Financial support for this study was provided by Shiraz University (Iran) and INRA, Agrocampus-Ouest, Université d’Angers (France).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Jacques MA, Arlat M, Boulanger A, Boureau T, Carrère S, Cesbron S, Chen NW, Cociancich S, Darrasse A, Denancé N, Fischer-Le Saux M, Gagnevin L, Koebnik R, Lauber E, Noël LD, Pieretti I, Portier P, Pruvost O, Rieux A, Robène I, Royer M, Szurek B, Verdier V, Vernière C. 2016. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu Rev Phytopathol 54:163–187. doi: 10.1146/annurev-phyto-080615-100147. [DOI] [PubMed] [Google Scholar]
- 2.Osdaghi E, Taghavi SM, Hamzehzarghani H, Fazliarab A, Harveson RM, Tegli S, Lamichhane JR. 2018. Epiphytic Curtobacterium flaccumfaciens strains isolated from symptomless solanaceous vegetables are pathogenic on leguminous but not on solanaceous plants. Plant Pathol 67:388–398. doi: 10.1111/ppa.12730. [DOI] [Google Scholar]
- 3.Mafakheri H, Taghavi SM, Puławska J, de Lajudie P, Lassalle F, Osdaghi E. 2019. Two novel genomospecies in the Agrobacterium tumefaciens species complex associated with rose crown gall. Phytopathology 109:1859–1868. doi: 10.1094/PHYTO-05-19-0178-R. [DOI] [PubMed] [Google Scholar]
- 4.Merda D, Briand M, Bosis E, Rousseau C, Portier P, Barret M, Jacques MA, Fischer-Le Saux M. 2017. Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Mol Ecol 26:5939–5952. doi: 10.1111/mec.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Triplett LR, Verdier V, Campillo T, Van Malderghem C, Cleenwerck I, Maes M, Deblais L, Corral R, Koita O, Cottyn B, Leach JE. 2015. Characterization of a novel clade of Xanthomonas isolated from rice leaves in Mali and proposal of Xanthomonas maliensis sp. nov. Antonie Van Leeuwenhoek 107:869–881. doi: 10.1007/s10482-015-0379-5. [DOI] [PubMed] [Google Scholar]
- 6.Eichenlaub R, Gartemann KH. 2011. The Clavibacter michiganensis subspecies: molecular investigation of Gram-positive bacterial plant pathogens. Annu Rev Phytopathol 49:445–464. doi: 10.1146/annurev-phyto-072910-095258. [DOI] [PubMed] [Google Scholar]
- 7.Gitaitis R, Walcott R. 2007. The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol 45:371–397. doi: 10.1146/annurev.phyto.45.062806.094321. [DOI] [PubMed] [Google Scholar]
- 8.Schaad NW, Abrams J, Madden LV, Frederick RD, Luster DG, Damsteegt VD, Vidaver AK. 2006. An assessment model for rating high-threat crop pathogens. Phytopathology 96:616–621. doi: 10.1094/PHYTO-96-0616. [DOI] [PubMed] [Google Scholar]
- 9.Davis MJ, Gillaspie AG, Vidaver AK, Harris RW. 1984. Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and Bermudagrass stunting disease. Int J Syst Bacteriol 34:107–117. doi: 10.1099/00207713-34-2-107. [DOI] [Google Scholar]
- 10.Dye DW, Kemp WJ. 1977. A taxonomic study of plant pathogenic Corynebacterium species. New Zeal J Agric Res 20:563–582. doi: 10.1080/00288233.1977.10427375. [DOI] [Google Scholar]
- 11.Jacques MA, Durand K, Orgeur G, Balidas S, Fricot C, Bonneau S, Quillévéré A, Audusseau C, Olivier V, Grimault V, Mathis R. 2012. Phylogenetic analysis and polyphasic characterization of Clavibacter michiganensis strains isolated from tomato seeds reveal that nonpathogenic strains are distinct from C. michiganensis subsp. michiganensis. Appl Environ Microbiol 78:8388–8402. doi: 10.1128/AEM.02158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.EPPO. 2016. PM 7/42 (3) Clavibacter michiganensis subsp. michiganensis. Bull OEPP/EPPO Bull 46:202–225. doi: 10.1111/epp.12302. [DOI] [Google Scholar]
- 13.Yasuhara-Bell J, Alvarez AM. 2015. Seed-associated subspecies of the genus Clavibacter are clearly distinguishable from Clavibacter michiganensis subsp. michiganensis. Int J Syst Evol Microbiol 65:811–826. doi: 10.1099/ijs.0.000022. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez AJ, Trapiello E. 2014. Clavibacter michiganensis subsp. phaseoli subsp. nov., pathogenic in bean. Int J Syst Evol Microbiol 64:1752–1755. doi: 10.1099/ijs.0.058099-0. [DOI] [PubMed] [Google Scholar]
- 15.Oh EJ, Bae C, Lee HB, Hwang IS, Lee HI, Yea MC, Yim KO, Lee S, Heu S, Cha JS, Oh CS. 2016. Clavibacter michiganensis subsp. capsici subsp. nov., causing bacterial canker disease in pepper. Int J Syst Evol Microbiol 66:4065–4070. doi: 10.1099/ijsem.0.001311. [DOI] [PubMed] [Google Scholar]
- 16.Osdaghi E, Ansari M, Taghavi SM, Zarei S, Koebnik R, Lamichhane JR. 2018. Pathogenicity and phylogenetic analysis of Clavibacter michiganensis strains associated with tomato plants in Iran. Plant Pathol 67:957–970. doi: 10.1111/ppa.12801. [DOI] [Google Scholar]
- 17.Li X, Tambong J, Yuan KX, Chen W, Xu H, Lévesque CA, De Boer SH. 2018. Re-classification of Clavibacter michiganensis subspecies on the basis of whole-genome and multi-locus sequence analyses. Int J Syst Evol Microbiol 68:234–240. doi: 10.1099/ijsem.0.002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk H-P, Goodfellow M, Göker M. 2018. Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007. doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, De Boer SH. 2019. Descriptions of Clavibacter insidiosus sp. nov. and Clavibacter tessellarius sp. nov. Int J Syst Evol Microbiol 69:2069. doi: 10.1099/ijsem.0.003439. [DOI] [PubMed] [Google Scholar]
- 20.Osdaghi E, Portier P, Briand M, Taghouti G, Jacques M-A, Osdaghi E, Portier P, Briand M, Taghouti G, Jacques M-A. 2018. Draft genome sequences of the type strains of three Clavibacter subspecies, and atypical peach-colored strains isolated from tomato. Microbiol Resour Announc 7:e01357-18. doi: 10.1128/MRA.01357-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M, Oh HS, Park SC, Chun J. 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 22.Thapa SP, Pattathil S, Hahn MG, Jacques MA, Gilbertson RL, Coaker G. 2017. Genomic analysis of Clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a Gram-positive bacterial pathogen. Mol Plant Microbe Interact 30:786–802. doi: 10.1094/MPMI-06-17-0146-R. [DOI] [PubMed] [Google Scholar]
- 23.Du Y, Yuan B, Zeng Y, Meng J, Li H, Wang R, Li G, Feng F. 2015. Draft genome sequence of the cellulolytic bacterium Clavibacter sp. CF11, a strain producing cold-active cellulase. Genome Announc 3:e01304-14. doi: 10.1128/genomeA.01304-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahr H, Dreier J, Meletzus D, Bahro R, Eichenlaub R. 2000. The endo-β-1,4-glucanase CelA of Clavibacter michiganensis subsp. michiganensis is a pathogenicity determinant required for induction of bacterial wilt of tomato. Mol Plant Microbe Interact 13:703–714. doi: 10.1094/MPMI.2000.13.7.703. [DOI] [PubMed] [Google Scholar]
- 25.Savidor A, Chalupowicz L, Teper D, Gartemann KH, Eichenlaub R, Manulis-Sasson S, Barash I, Sessa G. 2014. Clavibacter michiganensis subsp. michiganensis Vatr1 and Vatr2 transcriptional regulators are required for virulence in tomato. Mol Plant Microbe Interact 27:1035–1047. doi: 10.1094/MPMI-02-14-0061-R. [DOI] [PubMed] [Google Scholar]
- 26.Gartemann KH, Abt B, Beke T, Burger A, Engemann J, Flugel M, Gaigalat L, Goesmann A, Grafen I, Kalinowski J, Kaup O, Kirchner O, Krause L, Linke B, McHardy A, Meyer F, Pohle S, Ruckert C, Schneiker S, Zellermann E, Puhler A, Eichenlaub R, Kaiser O, Bartels D. 2008. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp michiganensis NCPPB 382 reveals a large island involved in pathogenicity. J Bacteriol 190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osdaghi E, Taghavi SM, Calamai S, Biancalani C, Cerboneschi M, Tegli S, Harveson RM. 2018. Phenotypic and molecular-phylogenetic analysis provide novel insights into the diversity of Curtobacterium flaccumfaciens. Phytopathology 108:1154–1164. doi: 10.1094/PHYTO-12-17-0420-R. [DOI] [PubMed] [Google Scholar]
- 28.Załuga J, Stragier P, Baeyen S, Haegeman A, Van Vaerenbergh J, Maes M, De Vos P. 2014. Comparative genome analysis of pathogenic and non-pathogenic Clavibacter strains reveals adaptations to their lifestyle. BMC Genomics 15:392. doi: 10.1186/1471-2164-15-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Samac DA, Glazebrook J, Ishimaru CA. 2015. Complete genome sequence of Clavibacter michiganensis subsp. insidiosus R1-1 using PacBio single-molecule real-time technology. Genome Announc 3:e00396-15. doi: 10.1128/genomeA.00396-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tancos MA, Lowe-Power TM, Peritore-Galve FC, Tran TM, Allen C, Smart CD. 2018. Plant-like bacterial expansins play contrasting roles in two tomato vascular pathogens. Mol Plant Pathol 19:1210–1221. doi: 10.1111/mpp.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgelis N, Nikolaidis N, Cosgrove DJ. 2015. Bacterial expansins and related proteins from the world of microbes. Appl Microbiol Biotechnol 99:3807–3823. doi: 10.1007/s00253-015-6534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yaripour Z, Taghavi SM, Osdaghi E, Lamichhane JR. 2018. Host range and phylogenetic analysis of Xanthomonas alfalfae causing bacterial leaf spot of alfalfa in Iran. Eur J Plant Pathol 150:267–274. doi: 10.1007/s10658-017-1271-0. [DOI] [Google Scholar]
- 33.Osdaghi E, Taghavi SM, Hamzehzarghani H, Fazliarab A, Harveson RM, Lamichhane JR. 2016. Occurrence and characterization of a new red-pigmented variant of Curtobacterium flaccumfaciens, the causal agent of bacterial wilt of edible dry beans in Iran. Eur J Plant Pathol 146:129–145. doi: 10.1007/s10658-016-0900-3. [DOI] [Google Scholar]
- 34.Lee IM, Bartoszyk IM, Gundersen DE, Mogen B, Davis RE. 1997. Nested PCR for ultrasensitive detection of the potato ring rot bacterium, Clavibacter michiganensis subsp. sepedonicus. Appl Environ Microbiol 63:2625–2630. doi: 10.1128/AEM.63.7.2625-2630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez-R LM, Konstantinidis KT. 2016. The Enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Preprints 4:e1900v1. doi: 10.7287/peerj.preprints.1900v1. [DOI] [Google Scholar]
- 37.Yoon SH, Ha SM, Lim JM, Kwon SJ, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 38.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yasuhara-Bell J, Marrero G, Alvarez AM. 2014. Genes clvA, clvF and clvG are unique to Clavibacter michiganensis subsp. michiganensis and highly conserved. Eur J Plant Pathol 140:655–664. doi: 10.1007/s10658-014-0495-5. [DOI] [Google Scholar]
- 43.Chalupowicz L, Cohen-Kandli M, Dror O, Eichenlaub R, Gartemann KH, Sessa G, Barash I, Manulis-Sasson S. 2010. Sequential expression of bacterial virulence and plant defense genes during infection of tomato with C. michiganensis subsp. michiganensis. Phytopathology 100:252–261. doi: 10.1094/PHYTO-100-3-0252. [DOI] [PubMed] [Google Scholar]
- 44.de Jong A, van Hijum SA, Bijlsma JJ, Kok J, Kuipers OP. 2006. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res 34:W273–W279. doi: 10.1093/nar/gkl237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Coleman-Derr D, Chen G, Gu YQ. 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–w84. doi: 10.1093/nar/gkv487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotchoni SO, Gachomo EW, Betiku E, Shonukan OO. 2003. A home made kit for plasmid DNA mini-preparation. Afr J Biotechnol 2:88–90. [Google Scholar]
- 49.Ansari M, Taghavi SM, Hamzehzarghani H, Valenzuela M, Siri MI, Osdaghi E, Ansari M, Taghavi SM, Hamzehzarghani H, Valenzuela M, Siri MI, Osdaghi E. 2019. Multiple introductions of tomato pathogen Clavibacter michiganensis subsp. michiganensis into Iran as revealed by a global-scale phylogeographic analysis. Appl Environ Microbiol 85:e02098-19. doi: 10.1128/AEM.02098-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tambong JT, Xu R, Daayf F, Brière S, Bilodeau GJ, Tropiano R, Hartke A, Reid LM, Cott M, Cote T, Agarkova I. 2016. Genome analysis and development of a multiplex TaqMan real-time PCR for specific identification and detection of Clavibacter michiganensis subsp. nebraskensis. Phytopathology 106:1473–1485. doi: 10.1094/PHYTO-05-16-0188-R. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Yuan X. 2017. Genome sequences for multiple Clavibacter strains from different subspecies. Genome Announc 5:e00721-17. doi: 10.1128/genomeA.00721-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vasilenko OV, Starodumova IP, Dorofeeva LV, Tarlachkov SV, Prisyazhnaya NV, Chizhov VN, Subbotin SA, Huntemann M, Clum A, Duffy K, Pillay M, Palaniappan K, Varghese N, Chen I-MA, Stamatis D, Reddy TBK, O'Malley R, Daum C, Shapiro N, Ivanova N, Kyrpides NC, Woyke T, Whitman WB, Evtushenko LI, Vasilenko OV, Starodumova IP, Dorofeeva LV, Tarlachkov SV, Prisyazhnaya NV, Chizhov VN, Subbotin SA, Huntemann M, Clum A, Duffy K, Pillay M, Palaniappan K, Varghese N, Chen I-MA, Stamatis D, Reddy TBK, O’Malley R, Daum C, Shapiro N, Ivanova N, Kyrpides NC, Woyke T, Whitman WB, Evtushenko LI. 2018. Draft genome sequences of new isolates and the known species of the family Microbacteriaceae associated with plants. Microbiol Resour Announc 7:e01051-18. doi: 10.1128/MRA.01051-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tambong JT, Xu R, Adam Z, Cott M, Rose K, Reid LM, Daayf F, Brière S, Bilodeau GJ. 2015. Draft genome sequence of Clavibacter michiganensis subsp. nebraskensis strain DOAB 397, isolated from an infected field corn plant in Manitoba, Canada. Genome Announc 3:e00768-15. doi: 10.1128/genomeA.00768-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bentley SD, Corton C, Brown SE, Barron A, Clark L, Doggett J, Harris B, Ormond D, Quail MA, May G, Francis D, Knudson D, Parkhill J, Ishimaru CA. 2008. Genome of the actinomycete plant pathogen Clavibacter michiganensis subsp. sepedonicus suggests recent niche adaptation. J Bacteriol 190:2150–2160. doi: 10.1128/JB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated for this study were previously published (20) and can be found in the NCBI GenBank Database (see Table 2).