The RhlA specificity explains the observed differences in 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA) congeners. Whole-cell catalysts can now be designed for the synthesis of different congener mixtures of HAAs and rhamnolipids, thereby contributing to the envisaged synthesis of designer HAAs.
KEYWORDS: RhlA, HAA, rhamnolipids, glycolipids, chain length, 3-(3-hydroxyalkanoyloxy)alkanoic acid
ABSTRACT
While rhamnolipids of the Pseudomonas aeruginosa type are commercially available, the natural diversity of rhamnolipids and their origin have barely been investigated. Here, we collected known and identified new rhlA genes encoding the acyltransferase responsible for the synthesis of the lipophilic rhamnolipid precursor 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA). Generally, all homologs were found in Betaproteobacteria and Gammaproteobacteria. A likely horizontal gene transfer event into Actinobacteria is the only identified exception. The phylogeny of the RhlA homologs from Pseudomonas and Burkholderia species is consistent with the organism phylogeny, and genes involved in rhamnolipid synthesis are located in operons. In contrast, RhlA homologs from the Enterobacterales do not follow the organisms’ phylogeny but form their own branch. Furthermore, in many Enterobacterales and Halomonas from the Oceanospirillales, an isolated rhlA homolog can be found in the genome. The RhlAs from Pseudomonas aeruginosa PA01, Pseudomonas fluorescens LMG 05825, Pantoea ananatis LMG 20103, Burkholderia plantarii PG1, Burkholderia ambifaria LMG 19182, Halomonas sp. strain R57-5, Dickeya dadantii Ech586, and Serratia plymuthica PRI-2C were expressed in Escherichia coli and tested for HAA production. Indeed, except for the Serratia RhlA, HAAs were produced with the engineered strains. A detailed analysis of the produced HAA congeners by high-performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS) highlights the congener specificity of the RhlA proteins. The congener length varies from 4 to 18 carbon atoms, with the main congeners consisting of different combinations of saturated or monounsaturated C10, C12, and C14 fatty acids. The results are discussed in the context of the phylogeny of this unusual enzymatic activity.
IMPORTANCE The RhlA specificity explains the observed differences in 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA) congeners. Whole-cell catalysts can now be designed for the synthesis of different congener mixtures of HAAs and rhamnolipids, thereby contributing to the envisaged synthesis of designer HAAs.
INTRODUCTION
Surfactants are amphiphilic molecules that reduce surface and interfacial tensions, which allows them to accumulate at interfaces and form emulsions. These properties are of industrial interest and are exploited in multiple applications in such different fields as pharmaceuticals, agriculture, food, detergents, and cosmetics (1–3). Biosurfactants are surfactants of biological origin and are a promising alternative to synthetic surfactants, as they are nontoxic, biodegradable, and produced from renewable feedstocks. Their application window is extensive, as they might be effective in environments with extreme pH, temperature, or salinity (4–6).
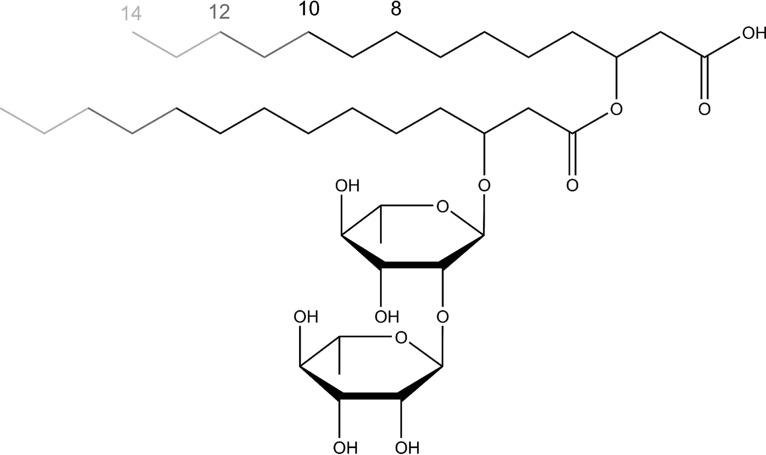
The biosurfactant 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA) is the hydrophobic moiety of rhamnolipids and most often consists of two hydroxy fatty acids linked by an ester bond (4, 7–10) (Fig. 1). Indeed, HAAs are not reported as typical products of microorganisms but, rather, were reported in trace amounts during rhamnolipid formation (11).
FIG 1.
Molecular structure of a rhamnolipid molecule. The chain lengths of the hydroxy fatty acids vary, resulting in different congeners. The main congener produced by P. aeruginosa contains 10 carbon atoms in both hydroxy fatty acid derivatives. Without the two rhamnose units, the molecule is a 3-(3-hydroxyalkanoyloxy)alkanoic acid (HAA). The synthesis of an HAA molecule is catalyzed by RhlA, which fuses two hydroxy fatty acids. RhlB links an activated dTDP-rhamnose to an HAA, resulting in a mono-rhamnolipid, which is the substrate that is transformed by RhlC, the second rhamnosyltransferase, into a di-rhamnolipid.
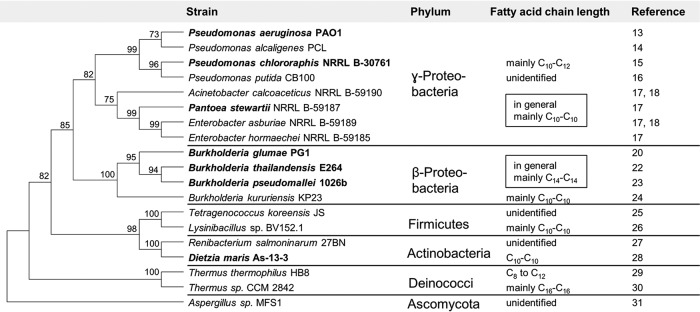
Jarvis and Johnson identified rhamnolipids in 1949 (12). Since then, these biosurfactants have been produced with different Pseudomonas species. Pseudomonas aeruginosa, a representative of the phylum Gammaproteobacteria, is one of the main producers in academia and industry. In the course of the past 2 decades, powerful analytical equipment such as Fourier transform infrared (FT-IR) spectroscopy and high-performance liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS) has enabled researchers to detect and verify the presence of rhamnolipids in culture supernatants. Rhamnolipid producers have been reported in the following phyla: Betaproteobacteria and Gammaproteobacteria (13–24), Firmicutes (25, 26), Actinobacteria (27, 28), Deinococcus-Thermus (29, 30), and Ascomycota (31) (Fig. 2).
FIG 2.
Phylogeny of published rhamnolipid producers based on 16S or 18S rRNA gene sequences. Strains in which rhl genes for rhamnolipid synthesis have been sequenced are marked in bold. The rRNA gene sequence of a reference strain was chosen to represent unsequenced rhamnolipid producers. The tree was constructed using the neighbor-joining method in MEGA7 with default settings. The numbers indicate bootstrap results.
The carbon chain lengths of HAAs determine their physical properties, such as their abilities to foam and emulsify, and their critical micelle concentration (CMC). Their chain lengths are strongly hinted to be determined by RhlA, an acyltransferase containing an α-/β-hydrolase domain that catalyzes the esterification of two activated hydroxy fatty acids to HAA (32). In in situ experiments, it has been shown that acyl-carrier protein (ACP)-activated hydroxy fatty acids are the preferred substrate for RhlA (8), while it has been shown in vivo in P. aeruginosa that CoA-activated hydroxyl fatty acids are incorporated preferably into the HAA molecule (33). Within the Gammaproteobacteria, Pseudomonas, Acinetobacter, Enterobacter (17, 18), and Pantoea (34) species produce mono- or diglycolipids. Their chain lengths vary, while the most common HAAs have 10 carbon atoms in both hydroxy fatty acids and are thus denoted C10-C10. In contrast, representatives of the Betaproteobacteria, namely, Burkholderia species, predominantly produce HAAs with chain lengths of 14 carbon atoms (Fig. 2). A few species do not follow this general categorization. Pseudomonas chlororaphis, e.g., produces rhamnolipids with one fatty acid chain of 10 carbon atoms and one of 12, resulting in the designation Rha-C10-C12 when these chains are fully saturated and Rha-C10-C12:1 when the C12 chain is unsaturated in one position (15, 35, 36). In contrast, Burkholderia kururiensis KP23 produces Gammaproteobacteria-like rhamnolipids containing mainly C10-C10 residues (24).
Rhamnolipid production has not been extensively explored in species of the phyla Firmicutes, Deinococcus-Thermus, Actinobacteria, and Ascomycota. Most promising are the results presented for Thermus species belonging to the phylum Deinococcus-Thermus. Pantazaki et al. (29) produced HAAs and rhamnolipids with chain lengths of 8 to 14 carbon atoms with Thermus thermophilus HB8. Rezanka et al. (30) reported the production of rhamnolipids by Thermus sp. strain CCM 2842, mainly containing the C16-C16 HAA congener, which has not been previously reported. Both groups used selective mass spectrometric methods.
A number of papers in the scientific literature report the synthesis of novel rhamnolipids with novel hosts, which we could not confirm, revealing the need for standardization and guidelines for determination of rhamnolipid and HAA structures. In contrast to rhamnolipids, only a few methods also cover HAAs. Again, HPLC-MS/MS is the method of choice to cover both rhamnolipids and HAAs (37, 38). The most comprehensive HPLC-MS/MS method focusing on HAA was presented by Lépine et al. (39). Therefore, our approach was to apply known and potential rhlA genes, express them recombinantly in Escherichia coli, and subject the resulting HAAs to a tailored HPLC-MS/MS analysis for confirmation.
The focus of our study was to explore the diversity of RhlAs and their potential to produce “designer HAAs.” The results are discussed in a phylogenetic context.
RESULTS
The natural diversity of RhlA, the acyltransferase of the rhamnolipid synthesis pathway, was investigated and exploited for the synthesis of the lipophilic intermediate HAA. We cloned eight rhlA homologs drawn from the full phylogenetic range of Proteobacteria into the Escherichia coli expression vector pET28a. Alternative RhlAs allowed the synthesis of different HAA congeners.
Phylogeny of RhlA.
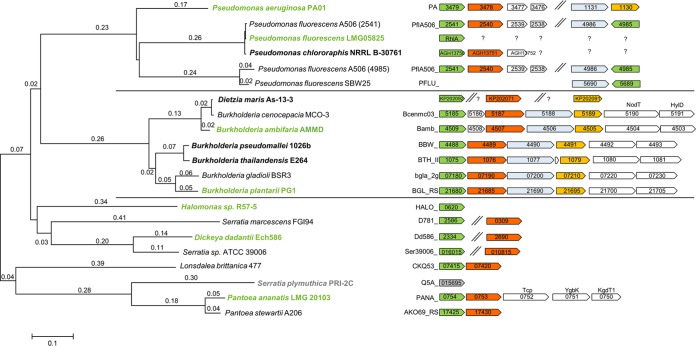
It has been shown that HAA synthesis in E. coli relies only on a recombinantly synthesized RhlA from P. aeruginosa (8, 32). Further, the experimental evidence strongly supports that RhlA selectively determines the β-hydroxy fatty acid chain lengths in HAAs (20). As a first step toward tailor-made HAAs, the natural genetic diversity of RhlA was investigated. Representative RhlA protein sequences for all phyla that were detectable by homology searches in GenBank and KEGG were collected. First, the RhlA of P. aeruginosa was used as a template. As the RhlAs from, for example, Pantoea species have limited homology with the protein from P. aeruginosa, homology searches with these sequences were also performed. All identified RhlA proteins are from the classes Betaproteobacteria and Gammaproteobacteria (Fig. 3). Strains from other phyla that are reported to produce rhamnolipids have not been sequenced, and the genes encoding their rhamnolipid synthesis pathways are not known, with two exceptions; an RhlA (GenBank accession number KP202092) was found in the Actinobacteria strain Dietzia maris As-13-3 (28), and the genome sequence of the Deinococcus-Thermus strain T. thermophilus HB8 (29) is known. However, in the latter genome, no rhlA homolog was found.
FIG 3.
Phylogenetic tree based on the amino acid sequences of RhlA. Operons associated with rhamnolipid formation are drawn next to the organism names, and genes are labeled with their gene locus or protein accession number. Organisms chosen for HAA production in this study are highlighted in green, while elsewhere-confirmed RhlAs are marked in bold. Others were chosen based on homology searches. S. plymuthica is marked in gray, as we could not confirm an RhlA activity. Double slashes depict independent genomic locations. In Dietzia maris, the synteny of rhlABC is not published. The strains P. fluorescens LMG05825 and P. chlororaphis NRRL-B-30761 are not genome sequenced; therefore, the putative homologous genes are indicated by question marks. The genes for rhamnolipid formation in P. aeruginosa are typically organized in two operons; rhlB (red) is located downstream of rhlA (green) and encodes rhamnosyltransferase I, which is necessary for mono-rhamnolipid formation. The genes rhlA and rhlB are colocalized with the regulator and inducer genes (rhlRI, white) that are involved in regulation via quorum sensing. In a second operon, rhlC (orange), the gene coding for rhamnosyltransferase II, is clustered with a putative transporter (light blue) gene. In the strains P. fluorescens LMG05835 and P. chlororaphis, only the genes shown are sequenced. In the P. ananatis LMG20103 operon containing rhlAB homologs, three genes are present that code for a methyl-accepting chemotaxis citrate transducer (tcp), a putative inner membrane protein (ygbK), and a 2-keto-3-deoxygluconate permease (kgdT1). In the Burkholderia species, the structural genes for di-rhamnolipid formation are organized in a single operon that further includes the genes nodT and hylD, which are potentially involved in the drug resistance systems of the cell. The tree was constructed using the neighbor-joining method in MEGA7 with default settings. Branch lengths shorter than 0.02 are omitted.
In general, the identified RhlAs can be divided into three main branches of a currently sparse phylogenetic tree (Fig. 3). In the first branch, the representatives of the genus Pseudomonas form a monophyletic lineage. In the P. aeruginosa strains, represented by strain PA01, two operons containing structural genes for rhamnolipid synthesis are known. In the first of these operons, rhlA and rhlB, the relevant genes for mono-rhamnolipid synthesis, are clustered with a regulator and inducer for quorum sensing, while rhlC, which enables the strain to produce di-rhamnolipids, is located in a different operon and is clustered with a putative transporter (40). Surprisingly, an analysis of the genetic environment of rhlA homologs detected using BLAST in the Pseudomonas fluorescens group showed two possible locations. Besides the colocalization with rhlB, an rhlA homolog is found in synteny with a putative transporter. In P. fluorescens strain A506, rhlA genes are present in both loci, while in P. fluorescens strain SBW25, only the latter location and no rhlB homolog can be found. Most P. fluorescens strains do not carry the genes for rhamnolipid synthesis (rhlA in synteny with rhlB, data not shown).
In the second branch, all representatives of the Burkholderia genus and the only Actinobacteria species, D. maris As-13-3 (28), are present. However, the RhlA of D. maris As-13-3 is reported to share 96% sequence identity with a Burkholderia cenocepacia protein, indicating that horizontal gene transfer is a probable explanation for its occurrence. In general, in Burkholderia, rhlAB are located on chromosome II and are in synteny with the putative transporter gene and rhlC. Furthermore, nodT and hylD, coding for enzymes related to efflux and secretion processes, are colocated. In B. cenocepacia and Burkholderia ambifaria, an open reading frame encoding a methyl transferase is placed between rhlA and rhlB. A second operon for rhamnolipid formation exists in Burkholderia pseudomallei and Burkholderia thailandensis on chromosome I (not shown).
The third branch includes homologous proteins from representatives of the orders Enterobacterales and Oceanospirillales, the latter with the only representative being Halomonas. In general, in this branch, the homology of the RhlA proteins is more divergent than in the Pseudomonas and Burkholderia branches. An rhlAB-like operon is found only in Pantoea strains (34) and Lonsdalea britanica, while rhlA homologs are found in Serratia and Dickeya strains but not in synteny with an rhlB homolog. No experimental evidence for HAA or rhamnolipid formation exists for the organisms in this branch, with the exception of Pantoea ananatis BRT175 (P. ananatis) producing the glucolipid ananatoside A, the hydrophobic part of which is an HAA molecule (34, 41). In P. ananatis LMG20103, the three genes tcp, ygbK, and kgdT1, which code for a putative methyl-accepting chemotaxis citrate transducer, an effector protein, and a 2-keto-3-deoxygluconate permease, respectively, are encoded in one common operon with the rhlAB homologs.
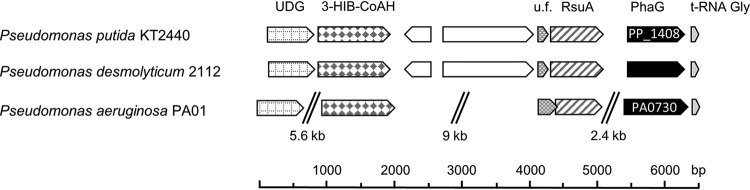
Determining the synteny of sequences identified by BLAST analyses using an RhlA query requires detailed analysis to distinguish RhlA from the transacylase PhaG, an enzyme that links de novo fatty acid and polyhydroxyalkanoate (PHA) biosynthesis (42–44) by catalyzing the reesterification from acyl carrier protein (ACP) to CoA. In P. aeruginosa, the protein sequences of RhlA and PhaG have a 44% sequence identity (44), which is similar to the 44 to 48% identity between Burkholderia RhlAs and RhlAs of P. aeruginosa. Fig. 3 shows that rhlA in Pseudomonas, Burkholderia, and Pantoea is part of a glycolipid synthesis operon. In contrast, phaG is located upstream of a tRNA gene, and furthermore, homologs of four of the six upstream genes of phaG in Pseudomonas putida can also be found upstream of phaG in P. aeruginosa (Fig. 4). We used this difference in the synteny of rhlA and phaG as a criterion for the identification of rhamnolipid genes in the reported rhamnolipid producer Pseudomonas desmolyticum NCIM-2112. We were especially interested in this strain, as it was reported to produce rhamnolipids with chain lengths of six to eight carbon atoms (45), a congener range not confirmed yet for an isolated RhlA. Full genome sequencing allowed a BLAST search for RhlA; however, only the transacylase-encoding phaG was identified. A gene encoding RhlB was not found in the genome of P. desmolyticum (data not shown). To improve the authoritative value of rhamnolipid literature, genetic evidence could be, besides high-quality analytics, a means to reduce or ideally avoid miscommunication of rhamnolipid-producing strains (46).
FIG 4.
Gene synteny of a phaG homolog in Pseudomonas desmolyticum. The gene synteny of the phaG homolog in P. desmolyticum is the same as in P. putida. Homolog genes coding for a uracil-DNA glycosylase (UDG), a 3-hydroxyisobutyryl-CoA hydrolase (3-HIB-CoAH), a protein of unknown function (u.f.), and a ribosomal small subunit pseudouridine A (RsuA) located upstream of phaG can also be found in the upstream region of phaG in P. aeruginosa. A tRNA homolog is placed downstream. This difference in synteny can be used as a criterion to distinguish rhlA from phaG in Pseudomonas species.
Considering gene synteny, the rhlA homologs identified by BLAST analysis of the Serratia, Dickeya, and Halomonas species are not well supported. Experimental evidence should confirm or disprove the RhlA activity.
HAA synthesis with recombinant E. coli.
E. coli strains BL21(DE3) and C43(DE3), each equipped with the rhlA gene from P. aeruginosa (pPA2), were grown in LB medium. Defined glucose pulses were given to provide an additional carbon source. When applying E. coli C43(DE3) as the host, glucose addition caused a steep increase in HAA titers 2 h after induction, which subsequently stagnated as glucose was depleted (6 h) (Fig. 5). The high HAA formation and growth rates were restored after the second glucose pulse at 20 h. While the growth rate slowed down 2 h later, the HAA production rate remained high, pointing to the fact that resources were efficiently allocated to the HAA synthesis pathway and diverted from supplying the growth machinery. With this strategy, an HAA titer of 1.7 g/liter 30 h after induction was reached, which is the highest concentration reported so far using recombinant microorganisms for HAA synthesis. Using E. coli BL21(DE3) as the host, the glucose supplementation had no enhancing effect on HAA formation at any time but was used for biomass formation. In this host, only 0.4 g/liter was achieved 20 h after induction.
FIG 5.
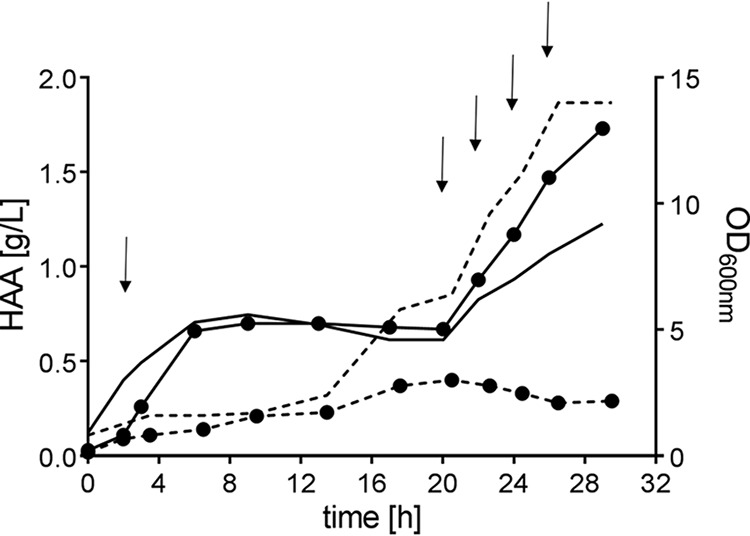
HAA formation (circles) by E. coli BL21(DE3) (dashed lines) and E. coli C43(DE3) (solid lines) transformed with plasmid pPA2 carrying rhlA of Pseudomonas aeruginosa PA01. Timepoint 0 indicates induction by IPTG. The OD is depicted without symbols. Glucose (0.2%, wt/vol) was supplemented after sampling to the time points marked by arrows. Each experiment was performed in duplicate.
The main HAA congeners synthesized by E. coli C43(DE3) pPA2 (Table 1) were the same as those produced by a recombinant P. putida KT2440 using the P. aeruginosa rhlA (11, 20) or the wild-type P. aeruginosa strain (12, 47–49). The HAA spectrum observed is, however, broader in E. coli, expanding to C14-containing congeners. The results support previous data showing that the RhlA enzyme is mainly responsible for HAA congener selectivity, while the host organisms play only a minor role.
TABLE 1.
Fractions of overall HAA congeners produced in E. coli C43(DE3) detected by HPLC-MS/MS
| Identity | m/z | Congenera | Fraction (%) forb: |
||||||
|---|---|---|---|---|---|---|---|---|---|
| pPA2 | pFLU | pDAD | pANA | pHAL | pAMB | pBUG | |||
| 14:0 | 273 | C4-C10* | 0.1 | ||||||
| 16:0 | 301 | C6-C10 | 0.1 | ||||||
| 18:0 | 329 | C8-C10 | 4.1 | 0.1 | 1.9 | 5.5 | |||
| 19:0 | 343 | C9-C10 | 0.1 | ||||||
| 20:0 | 357 | C8-C12 | 0.3 | 0.3 | 0.3 | ||||
| 20:0 | 357 | C10-C10 | 49.5 | 3.0 | 4.5 | 31.3 | |||
| 20:1 | 355 | C8-C12:1 | 0.1 | 0.3 | |||||
| 20:1 | 355 | C10-C10:1 | 0.3 | 0.2 | |||||
| 21:0 | 371 | C10-C11 | 0.1 | ||||||
| 22:0 | 385 | C8-C14* | 0.3 | 0.3 | 0.2 | ||||
| 22:0 | 399 | C10-C12 | 32.5 | 17.1 | 7.1 | 16.4 | 0.1 | 0.3 | 0.2 |
| 22:1 | 383 | C8-C14:1* | 0.4 | 0.1 | 0.1 | ||||
| 22:1 | 383 | C10-C12:1 | 4.2 | 21.3 | 5.6 | 10.7 | |||
| 23:0 | 399 | C10-C13* | 0.3 | 0.3 | 0.2 | 0.1 | |||
| 24:0 | 413 | C10-C14 | 4.1 | 23.8 | 25.8 | 10.0 | 2.0 | 2.3 | 4.5 |
| 24:0 | 413 | C12-C12 | 0.4 | 1.0 | 1.5 | 0.5 | 0.2 | 0.1 | |
| 24:1 | 411 | C10-C14:1 | 2.9 | 25.2 | 47.4 | 10.8 | 2.0 | 0.6 | 3.0 |
| 24:1 | 411 | C12-C12:1 | 1.5 | ||||||
| 24:2 | 409 | C10:1-C14:1 | 0.2 | ||||||
| 25:0 | 427 | C11-C14* | 0.3 | 0.1 | |||||
| 25:0 | 427 | C12-C13* | 0.1 | 0.6 | |||||
| 25:1 | 425 | C10-C15:1* | 0.1 | ||||||
| 25:1 | 425 | C11-C14:1* | 0.1 | ||||||
| 26:0 | 441 | C10-C16 | 0.2 | 0.1 | |||||
| 26:0 | 441 | C12-C14 | 1.2 | 1.2 | 1.5 | 10.1 | 12.1 | 5.1 | |
| 26:0 | 441 | C13-C13* | 0.2 | 1.2 | |||||
| 26:1 | 439 | C10-C16:1 | 4.1 | 2.6 | 3.3 | 0.1 | 0.1 | 0.4 | |
| 26:1 | 439 | C12-C14:1 | 1.2 | 1.6 | 2.2 | 8.8 | 4.9 | 3.9 | |
| 26:2 | 437 | C12:1-C14:1 | 0.3 | 0.6 | 0.7 | 0.1 | 0.1 | ||
| 27:0 | 455 | C12-C15* | 0.1 | ||||||
| 27:0 | 455 | C13-C14 | 0.4 | 6.3 | 0.6 | ||||
| 27:1 | 453 | C13-C14:1* | 0.3 | 4.0 | 0.4 | ||||
| 28:0 | 469 | C14-C14 | 0.1 | 22.1 | 12.2 | 24.4 | |||
| 28:1 | 467 | C10-C18:1* | 0.4 | 0.7 | |||||
| 28:1 | 467 | C12-C16:1* | 0.2 | 0.1 | 0.3 | 0.3 | 0.3 | ||
| 28:1 | 467 | C14-C14:1 | 0.1 | 0.4 | 30.8 | 36.5 | 28.3 | ||
| 28:2 | 465 | C12:1-C16:1* | 0.2 | ||||||
| 28:2 | 465 | C14:1-C14:1 | 0.2 | 0.2 | 14.9 | 10.3 | 11.7 | ||
| 29:0 | 483 | C13-C16* | 0.1 | ||||||
| 29:0 | 483 | C14-C15 | 0.3 | ||||||
| 29:1 | 481 | C13-C16:1* | 0.2 | ||||||
| 29:1 | 481 | C14-C15:1* | 0.1 | 0.5 | 0.2 | ||||
| 29:1 | 481 | C14:1-C15* | 0.5 | ||||||
| 29:2 | 479 | C14:1-C15:1 | 0.4 | ||||||
| 30:0 | 497 | C14-C16 | 0.5 | 1.2 | |||||
| 30:1 | 495 | C12-C18:1* | 0.1 | ||||||
| 30:1 | 495 | C14-C16:1 | 0.2 | 3.5 | 1.7 | 8.9 | |||
| 30:2 | 493 | C12:1-C18:1* | 0.1 | ||||||
| 30:2 | 493 | C14:1-C16:1* | 0.2 | 2.1 | 4.2 | 4.4 | |||
| 32:1 | 523 | C14-C18:1 | 0.5 | ||||||
| 32:1 | 523 | C16-C16:1 | 0.1 | 0.1 | |||||
| 32:2 | 521 | C14:1-C18:1* | 0.1 | 0.1 | 0.4 | ||||
| 32:2 | 521 | C16:1-C16:1 | 0.1 | 0.2 | |||||
| Average chain length | 10.5 | 11.6 | 11.8 | 10.8 | 13.8 | 13.7 | 14.0 | ||
New congeners are marked by an asterisk. Congeners include both variants for chain positions, e.g., C8-C10 and C10-C8.
The main congeners for each plasmid are in bold font. pPA2, P. aeruginosa PAO1; pFLU, P. fluorescens LMG 05825; pDAD, D. dadantii Dd586_2334; pANA, P. ananatis LMG 20103; pHAL, Halomonas sp. R57-5; pHAL, Halomonas sp. R57-5; pAMB, B. ambifaria AMMD; pBUG, B. plantarii (glumae) PG1.
Diversification of the HAA spectrum by exploiting natural genetic variance.
In order to increase HAA congener diversity, seven additional rhlAs of species representing the identified evolutionary space were used.
The first obvious choice from the Betaproteobacteria was RhlA of Burkholderia plantarii PG1 (formerly Burkholderia glumae PG1), which synthesizes mainly C14 rhamnolipids (20, 21). We also chose RhlA of B. ambifaria, which was of particular interest, as the protein shares 91% identity with RhlA of D. maris, which was reported to produce the C10-C10 congener (28). Our purpose was to verify this nontypical main congener for the Burkholderia genus with pAMB.
In contrast to the 16S rRNA phylogeny, in which Enterobacterales, Oceanospirillales, and Pseudomonas are classes of Gammaproteobacteria, the RhlA sequences of the Enterobacterales and the Oceanospirillales-representative Halomonas form a common third branch (Fig. 3). We thus selected RhlA from P. ananatis LMG20103 as the first representative from the Enterobacterales branch. This strain is fully genome sequenced, and the genes for glycolipid synthesis are present (34, 50). Rooney et al. (17) and Hošková et al. (18) reported that other Enterobacterales synthesize rhamnolipids with mainly C10-C10 HAAs (Fig. 2). The N terminus of RhlA in P. ananatis is longer than those of other RhlA proteins, which might be due to automated annotation. For this reason, two versions of rhlA were cloned, one representing a normal-sized rhlA and the long rhlA version. Both rhlAs led to HAA production (data for the long version not shown), suggesting that the normal-sized RhlA is the native protein. Additionally, sequencing indicated a frameshift in the published sequence that led to 13 incorrectly annotated amino acids. A comparison with RhlA from, e.g., Pantoea stewartii A206 confirms this finding, and a corrected sequence was submitted to GenBank (accession number MF671909). As mentioned above, the gene synteny in Dickeya dadantii Ech586, Halomonas sp. R57-5, and Serratia plymuthica PRI-2C does not show colocalization with genes related to glycolipid synthesis, but the rhlA homologs are isolated in the genome. To experimentally confirm the activity, we further investigated HAA formation using rhlA genes of these strains.
Finally, we included the rhlA from P. fluorescens LMG 05825 (P. chlororaphis ATCC 17813), which is reported to be the same strain as P. chlororaphis NRRL B-30761 (35), a strain producing mainly C10-C12 and C10-C12:1 congeners (15). Solaiman et al. (36) found an operon containing rhlAB and the regulator gene rhlR (Fig. 3) in this strain. While we could confirm the previous results, the rhlA from strain LMG05825 carried two nucleotide changes resulting in one amino acid difference in RhlA.
E. coli strains were equipped with one of the eight rhlA genes and cultivated as described above. Glucose was fed 2 and 22 h after IPTG (isopropyl-β-d-thiogalactopyranoside) induction. Seven of the eight recombinant strains produced HAAs (Table 1); E. coli C43(DE3) pPLY was the exception. Again, while the main HAA congeners were highly similar to reported congener compositions of wild-type strains, the congener spectrum might be a bit wider, which however, could also be a result of the sensitive method used for identification in this study. By the combination of efficient chromatographic separation and structure informative tandem mass spectrometric detection, the resulting HPLC-MS/MS method enables selective and sensitive detection of HAAs. A limit of detection in the range of 0.1 mg/liter is achieved, and thus, HAA with a relative share of <0.1% can be detected (Table 1).
As expected from mono-rhamnolipids produced by the wild-type strain P. chlororaphis NRRL-B-30761 (15), our congener determination with plasmid pFLU revealed a different main congener spectrum than with pPA2 from P. aeruginosa. Accordingly, we detected C10-C12 and C10-C12:1 to be among the main congeners, but additionally, we identified C10-C14 and C10-C14:1 to be present in even slightly larger fractions. The C10-C14 congener was also detected by Gunther et al. (15), though in a smaller fraction. In contrast to pPA2, where the longest detected chain contained 14 carbon atoms, with pFLU, congeners containing C16, C16:1, or even C18:1 chains were present.
The two RhlA proteins from the Betaproteobacteria branch (pBUG and pAMB) showed 14 carbon atoms in both chains. For pBUG, this was expected due to the phylogenetic classification with other Burkholderia strains, for which C14-C14 rhamnolipid production has been reported in wild-type (19, 21–23) and recombinant strains (20). With pBUG, 16% of the HAAs incorporated at least one C16 or C16:1 fatty acid. In the phylogenetic tree shown in Fig. 3, pAMB is arranged with RhlA from Dietzia maris (28), shown to produce C10-C10-containing rhamnolipids. Therefore, the result with mainly C14 chain lengths was unexpected. Besides the main fraction of C14 chain lengths, we found with this plasmid the most significant fraction of unusual congeners containing chain lengths with odd numbers, namely, 12% containing at least one chain with 13 carbon atoms and in traces C15 or C15:1. To further confirm the presence of these odd-numbered hydroxy fatty acids, we conducted LC-MS/MS measurements applying high-resolution MS. Besides high resolution, the instrument used also delivers high mass accuracy (<5 ppm relative mass deviation compared to the theoretical value). Hence, elemental compositions can be deduced not only from the intact HAA molecule but also for the fragments in MS/MS mode. Exemplar data are presented in Fig. 6A, where the high-resolution MS/MS mass spectrum of an HAA molecule containing 27 carbon atoms with 1 unsaturation (HAA 27:1) after HPLC separation is depicted. Collision-induced dissociation of the precursor ion (measured m/z 453.3592, calculated m/z 453.3586, 1.3 ppm) gave rise to two complementary fragments (measured m/z 229.1810, calculated m/z 229.1809, 0.4 ppm and measured m/z 241.1811, calculated m/z 241.1809, 0.8 ppm). These fragments confirm the presence of C13:0 and C14:1 hydroxy fatty acids. The fragmentation is assigned according to Lépine et al. (39). Therefore, the detection of these two fragments also demonstrates that two congeners are contained, i.e., HAA C13:0-C14:1 and HAA C14:1-C13:0. Furthermore, the presence of odd-chain hydroxy fatty acids was confirmed using complementary gas chromatography-mass spectrometry (GC-MS) analysis. HAA samples were hydrolyzed and derivatized to yield the corresponding methyl ester. Additional trimethylsilylation of the hydroxy group facilitated the assignment of chain length as well as position of the hydroxy group in the mass spectrum obtained by electron ionization (Fig. 6B).
FIG 6.
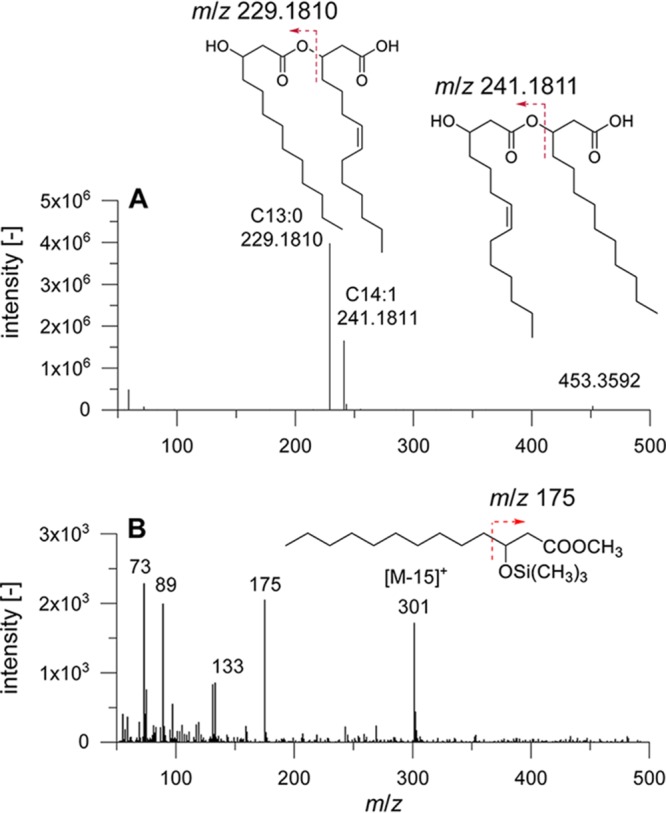
(A) HPLC high-resolution MS/MS spectrum of HAA 27:1. The fragments correspond to C13:0 and C14:1 hydroxy fatty acids as indicated by the shown structures. Please note that the double-bond position in C14:1 could not be determined using this method. (B) GC-MS spectrum of C13:0 hydroxy fatty acid after hydrolysis, methylation, and trimethylsilylation to confirm the presence of odd-chain hydroxy fatty acids. The chain length is determined by the characteristic [M-15]+ ion, while the diagnostic fragment (m/z 175) indicates the position of the hydroxy group.
Most surprising and divergent were the results we obtained with the plasmids from the Enterobacterales and Halomonas species. Though the RhlA homologs form their own branch (Fig. 3), the HAAs detected with the single plasmids do not show common characteristics. Again, for pPLY, HPLC-MS/MS did not confirm HAAs in the culture supernatant but did confirm other fatty acids. Notably, these free fatty acids experienced similar retention on the used HPLC column as HAAs. Using unspecific detection, such as charged aerosol detection, or by evaporative light scattering detection, false annotation cannot be ruled out. With plasmid pDAD, a comparable spectrum to pFLU was observed. Strikingly, the main fraction contained C10-C14 or C10-C14:1 (26 and 47%, respectively), indicating a high specificity for these congeners. With pANA, the main congeners contained C10-C10 (31%), C10-C12 or C10:C12:1 (27%), and C10-C14 or C10-C14:1 (21%), which is comparable to the congeners found with pPA2 and pFLU. Plasmid pHAL, in contrast, showed congeners like the Burkholderia strains with saturated and monounsaturated C14 chains.
Novel HAA congeners.
In total, we detected 53 congeners, 23 of which have not been reported before (see marked congeners in Table 1). In general, the new congeners can be categorized into three characteristics or combinations thereof. In the first, HAA molecules consist of two chains differing in four or more carbon atoms in length; in the second, chains with so far unknown monounsaturations can be found; and in the third, odd chain lengths occur. For the first category, we found C4-C10, C8-C14, C8-C14:1, C10-C15:1, C10-C18:1, and C12-C16:1. The chain lengths of the congeners C12-C18:1, C12:1-C18:1, and C14:1-C18:1 have been reported in rhamnolipids produced by Thermus sp. CCM 2842 (30), where the molecules were saturated. Other new congeners in the second category with monounsaturations are C8-C14:1, C12-C16:1, C12:1-C16:1, and combined with the third category including odd chain lengths, C10-C15:1, C11-C14:1, C13-C14:1, C13-C16:1, C14-C15:1, C14:1-C15, and C14:1-C15:1. More new odd chain lengths are C11-C14, C11-C14:1, C12-C13, C10-C15:1, C13-C13, C12-C15, C13-C14:1, and C13-C16. For the third category, it has to be mentioned that other odd chain lengths were detected earlier (30, 37, 51). In our results, it is striking that 7.7% of the overall congeners are new congeners with odd numbers synthesized by E. coli C43(DE3) pAMB.
The congeners that were produced covered the entire HAA spectrum known in wild-type Proteobacteria species. The congener C8-C8 produced by P. aeruginosa 57RP (49, 52) was found in some, but not all, experiments with pPA2 and hence is not listed in Table 1.
DISCUSSION
The esterification of (hydroxy-) fatty acids as it is catalyzed by RhlA is a rare enzyme activity. A similar activity can be found in the black yeast fungus Aureobasidium pullulans. In this strain, liamocin, a glycolipid consisting of mannitol linked with three or four 3,5-dihydroxydecanoic ester groups, is produced (53). Our survey for RhlA in microorganisms showed its presence mainly in the Betaproteobacteria and Gammaproteobacteria phyla, with little evidence in other phyla. We exploited the natural diversity of RhlA, allowing the synthesis of distinct HAA congener mixtures using E. coli as a host. The confirmed substrate specificity of RhlA opens the door for the production of tailor-made HAAs.
Fig. 2 shows that rhamnolipid producers are not restricted to representatives of the Betaproteobacteria and Gammaproteobacteria phyla. However, we and others (46, 54) have experienced difficulties in reproducing and confirming previous studies showing rhamnolipid synthesis by bacteria of different phyla. In many cases, we did not detect rhamnolipid production and/or genetic evidence for rhamnolipid synthesis despite having cultivated and/or sequenced the reported rhamnolipid producers, respectively. Having had similar experiences, Irorere et al. (46) ascertained that unequivocal analytical techniques to determine rhamnolipid production were not used and concluded that particular reports might be erroneous. As mentioned above, Jadhav et al. (45), for example, reported that P. desmolyticum NCIM-2112 produced mono-rhamnolipids with fatty acid chain lengths of from six to eight carbon atoms. Our efforts to identify an rhlA homolog after genome sequencing failed. We cultivated the organism as described by the authors but detected no rhamnolipids using HPLC-MS/MS (data not shown). The question of whether P. desmolyticum encodes an enzyme with RhlA activity but of a different phylogeny remains, which is consistent with the observations of Kügler et al. (54) finding no evidence of reports of rhamnolipid production by Actinobacteria. Indeed, we found no RhlA with homology searches in non-Proteobacteria species, with the exception of the actinobacterium Dietzia maris AS-13-3. A detailed survey of the analytical methods applied for the identification of novel rhamnolipid-producing strains is necessary. In this regard, the reports of rhamnolipid production in Renibacterium salmoninarum 27BN (27), Tetragenococcus koreensis JS (25), and Aspergillus sp. strain MFS1 (31) do not fulfill the criteria for unequivocal rhamnolipid identification proposed by Irorere et al. (46).
The diversity of HAA congeners might be broadened by identifying RhlAs from Betaproteobacteria and Gammaproteobacteria.
The evolutionary relationships between the known RhlAs (Fig. 3) are to a large extent consistent with the species phylogeny based on 16S rRNA gene sequences (Fig. 2). However, it is striking that the RhlA proteins within the genus Pseudomonas do not form a monophyletic lineage with the RhlAs from the Enterobacterales (Serratia, Dickeya, Lonsdalea, and Pantoea) as the 16S rRNA genes do. While pseudomonads and Enterobacterales are both Gammaproteobacteria, the RhlA proteins of the Enterobacterales are outgrouped, forming a separate branch.
Within the pseudomonads, species-specific HAA congeners could be produced. While pPA2 is the most prominent C10-C10 producer, we detected with pFLU a C12 or C14 chain combined with a C10 chain, which confirms the findings of Gunther et al. (15). However, Gunther found C10-C12 or C10-C12:1 as the main congener; in our study, C10-C14 and C10-C14:1 turned out to be even more prominent.
The Burkholderia species B. plantarii, B. thailandensis, and B. mallei synthesize mainly C14-C14 rhamnolipids (19, 21–23, 55). This was confirmed in this study. Using the B. plantarii RhlA, the average carbon chain was determined to have 14.0 carbon atoms. However, in terms of RhlA diversity, the phylogeny of the known RhlA proteins indicates that other Burkholderiaceae might produce HAAs with shorter fatty acids. B. kururiensis, belonging to the genus Paraburkholderia (24), is reported to mainly produce the C10-C10 rhamnolipid congeners. Two explanations for this finding are possible. On the one hand, a C10-specific protein might have been transferred to B. kururiensis from, e.g., P. aeruginosa via horizontal gene transfer. On the other hand, the rhlA might have evolved from the original Burkholderia type rhlA to be more promiscuous toward shorter fatty acid chain lengths.
Most new congeners with odd chain lengths were produced when RhlA from B. ambifaria was applied, which we confirmed with GC analytics (Fig. 6B). It was shown that in contrast to acetyl-CoA, propionyl-CoA can be accepted by the enzyme FabH as a precursor to chain elongation, resulting in odd-chain-length fatty acids (56, 57). FabH varies depending on its bacterial origin and accepts acetyl-CoA or propionyl-CoA in bacteria synthesizing straight-chain fatty acids, while in branched-chain fatty acid-producing bacteria, branched-chain acyl-CoAs serve as precursors for chain elongation (57). Our results showing straight C13 chains when using pAMB in E. coli indicate that FabH of E. coli is of the straight-chain type delivering the substrate for the B. ambifaria RhlA.
Most interesting and representing the group with the most potential toward novel HAA congeners are the results we obtained with RhlA proteins from the Enterobacterales and Halomonas. Except for Pantoea and Lonsdalea, the genes coding for RhlAs in this group are not colocalized with other genes related to glycolipid formation and thus are difficult to distinguish from phaG. In contrast to the Pseudomonas and Burkholderia branches shown in Fig. 3, RhlA homologs from five genera are combined in the third branch. With only four RhlAs tested, we found a diversity within this group ranging from no HAAs with pPLY over similar congeners like in the pseudomonads (pDAD and pANA) to a Burkholderia-like spectrum with pHAL. So far, few results from Enterobacterales have been presented in the literature. Reports about rhamnolipid formation by the wild-type strains Enterobacter asburiae (17, 18), Enterobacter hormaechei, P. stewartii (17) (Fig. 1), and P. ananatis BRT175, a strain producing a glycolipid with a sugar moiety other than rhamnose (34, 41), show similar fatty acid chain lengths to those we detected with pANA and pDAD. Though the HAA spectrum from, e.g., pDAD and pFLU or pHAL and pBUG are similar, the RhlAs are only distantly related and not arranged in the same phylogenetic lineage. The diversity of HAAs within the Enterobacterales indicated by long branch lengths hints at the existence of more proteins with RhlA activity in this and other orders of the Gammaproteobacteria, such as the Oceanospirillales. With confirmed RhlA activity from numerous species, the sparse tree depicted in Fig. 3 might develop toward distinct branches related to genera. A tendency can already be seen with our data obtained with pHAL, pDAD, pPLY, and pANA. Eliminating the unconfirmed S. plymuthica strain, to date, three strains from Lonsdalea and Pantoea form their own lineage. In these species, a colocalization of the rhlA homologs with an rhlB homolog are found.
Our results indicate that the rhlA genes are conserved within microbial genera. Because RhlA mainly determines substrate specificity in the rhamnolipid synthesis pathway, the main fatty acid congener of HAAs and rhamnolipids can be inferred from knowledge of the species of the producing organism. Insights into the correlation between microbial and RhlA phylogeny and RhlA specificity may be fostered by additional genomic and production data from rhamnolipid producers, ideally increasing the number of HAA-producing species and genera for which genetic evidence for rhlA genes exists.
HAAs synthesized by non-Proteobacteria.
Toribio et al. (58) argued in 2010, when hundreds of genomes were already available in databases, that the rare occurrence of rhlA homologs outside of the Betaproteobacteria and Gammaproteobacteria species suggested that horizontal gene transfer occurs only in rare circumstances. This conclusion agrees with the results for D. maris presented here (28). Although 360,000 genomes are currently available in the Genomes On Line Database (GOLD), and many more are available in others, the early observation by Toribio et al. (58) is still valid. Although no evidence is presented here, a massive gene loss in most other phyla and genera cannot be excluded. With BLAST searches, RhlAs cannot be detected in, e.g., Thermus. Despite having a common ancestor, it is possible that the phylogenetic distance increased during evolution. This hypothesis is supported by the fact that the Betaproteobacteria RhlA proteins from P. aeruginosa and P. ananatis, which show a similar HAA spectrum, share a mere 35% identity or, to name another example, proteins from B. plantarii PG1 and Halomonas spp. show only 50% identical positions (data not shown). Alternatively, rhamnolipids might be synthesized by proteins that do not share an evolutionary origin with RhlA.
Some evidence exists for alternative genes, especially for strains of the genus Thermus, which is encouraging. Pantazaki et al. (29) reported the production of HAAs and rhamnolipids with chain lengths of 8 to 14 carbon atoms using the fully sequenced strain T. thermophilus HB8. No homologs of rhlA and rhlB were found in the genome using conventional BLAST approaches. The main congener detected in Thermus sp. CCM 2842 was Rha-C16-C16, and fatty acid chain lengths of up to 24 occur in small fractions (30), indicating that an RhlA with different substrate specificity exists; again, no genetic evidence is available. The numerous reports of HAA and rhamnolipid synthesis by species not belonging to Betaproteobacteria and Gammaproteobacteria remain something of a mystery, with explanations as divergent as erroneous analytics, horizontal gene transfer, massive gene loss, and parallel evolution. The challenge to identifying the genetic origin of rhamnolipid synthesis in phyla such as Firmicutes, Actinobacteria, and Deinococcus-Thermus thus remains.
MATERIALS AND METHODS
Bacterial strains, plasmids, and synthetic genes.
P. ananatis LMG 20103, P. fluorescens LMG 05825, and B. ambifaria LMG 19182 (= B. ambifaria AMMD) were purchased from the BCCM/LMG Bacteria Collection, and P. desmolyticum NCIM-2112 (45) was purchased from the National Collection of Industrial Microorganisms (NCIM). Plasmid pVLT33_rhlABBG, containing rhlA from B. plantarii PG1, was kindly provided by A. Wittgens (20). The native gene from Dickeya and codon-optimized genes for the rhlA homologs from Halomonas and Serratia were synthesized by IDT (Coralville, IA, USA).
Construction of expression plasmids.
Eight expression plasmids carrying rhlA from either P. aeruginosa PA01, P. ananatis LMG 20103, B. plantarii PG1, P. fluorescens LMG 05825, B. ambifaria LMG 19182, Halomonas sp. R57-5, D. dadantii Ech586, or S. plymuthica PRI-2C were constructed. The rhlA gene from P. aeruginosa PA01 was amplified from plasmid pPS05 (59) and cloned into pET28 as described previously (9). The rhlA genes of P. ananatis LMG20103, P. fluorescens LMG 05825, and B. ambifaria LMG 19182, were amplified from genomic DNA and cloned into pET28a, and the B. plantarii PG1 rhlA was amplified from plasmid pVLT33_rhlABBG. For the construction of plasmids pPLY, pHAL, and pDAD, rhlA was amplified from G-strings. Purified PCR products and digested pET28a were used for subsequent Gibson assembly using the respective cloning kits (NEB, Ipswich, MA, USA) following the supplier’s instructions. The plasmids used are listed in Table 2.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Characteristics/purpose | Reference |
|---|---|---|
| Strains | ||
| P. aeruginosa PA01 | Wild-type, cloning of rhlA | 13 |
| P. fluorescens LMG 05825 | Wild-type, cloning of rhlA, strain used instead of P. chlororaphis NRRL B-30761 | 15 |
| P. desmolyticum NCIM-2112 | Wild-type, identification of phaG | 45 |
| P. ananatis LMG 20103 | Wild-type, cloning of rhlA | 34, 41, 50 |
| B. ambifaria LMG 19182 | Wild-type, cloning of rhlA, strain used instead of P. ambifaria AMMD | 64 |
| E. coli DH5α | Cloning procedures | 65 |
| E. coli BL21(DE3) | Expression host | 66 |
| E. coli C43(DE3) | Expression host | 67 |
| Plasmids | ||
| pPS05 | Plasmid harboring rhlA from Pseudomonas aeruginosa PA01, SynPro16, TetR | 59 |
| pVLT33_rhlABBG | Plasmid harboring rhlA from Burkholderia plantarii (glumae) PG1, lacI, Ptac KmR | 20 |
| pET28a | Expression vector, T7 promoter, His-Tag, lacI, pBR322 ori, f1 ori, KmR | 68 |
| pPA2 | rhlA from P. aeruginosa PA01 cloned into pET28a (NdeI/SalI), KmR | 9 |
| pANA | rhlA from P. ananatis LMG 20103 cloned into pET28a (NdeI/BamHI), KmR | This work |
| pBUG | rhlA from B. plantarii PG1 cloned into pET28a (NdeI/BamHI), KmR | This work |
| pFLU | rhlA from P. fluorescens 05825 cloned into pET28a (NdeI/SalI), KmR | This work |
| pAMB | rhlA from B. ambifaria 19182 cloned into pET28a (NdeI/BamHI), KmR | This work |
| pHAL | Codon optimized rhlA from Halomonas sp. R57-5 cloned into pET28a (NdeI/BamHI), KmR | This work |
| pDAD | rhlA from D. dadantii Ech586 cloned into pET28a (NdeI/BamHI), KmR | This work |
| pPLY | Codon optimized rhlA from S. plymuthica cloned into pET28a (NdeI/BamHI), KmR | This work |
Culture conditions for HAA production.
Plasmids pPA2, pANA, pBUG, pFLU, pAMB, pDAD, pPLY, and pHAL were separately transformed in E. coli. For HAA production, a 100-ml shake flask with 10 ml of LB containing 50 μg/ml kanamycin was inoculated with a freshly transformed E. coli strain. The cells were grown overnight at 37°C in a Multitron shaker (Infors HT, Bottmingen, Switzerland) at 200 rpm with a throw of 25 mm and humidity of 80%. HAA synthesis was conducted in 50 ml of the same medium in a 500-ml shake flask without baffles. The culture was inoculated to an optical density at 600 nm (OD600) of 0.1, and cells were grown at 37°C until the OD600 was between 0.5 and 0.9. Expression of rhlA was induced by the addition of 0.5 mM IPTG, at which point the temperature was lowered to 30°C and the shaking speed raised to 300 rpm to ensure that sufficient oxygen was supplied. Next, 2 and 20 h after the cultures were induced, 200 μl of 50 g/liter glucose was added, and the supernatant for HPLC-MS/MS was harvested 28 h after induction. In the experiment shown in Fig. 5, the cultures received 0.2% (wt/vol) glucose 2, 20, 22, 24, and 26 h after induction.
Quantification of HAAs.
For HAA quantification, components of the samples were separated using reverse-phase chromatography on a C18 column and detected with a charged aerosol detector (RP-HPLC-CAD) as described by Tiso et al. (60), based on the method established by Behrens et al. (61). Harvesting of cells and cell residue was accomplished by centrifugation at 13,000 × g for 3 min. Next, 400 μl of the supernatant was mixed with 400 μl of acetonitrile, and the resulting mixture was vortexed. Samples were subsequently incubated overnight at 4°C to facilitate precipitation of any residual material that might clog the column. Prior to HPLC analysis, the samples were centrifuged again, and the supernatant was filtered with a Phenex-RC (regenerated cellulose) syringe filter (diameter, 4 mm; pore size, 0.2 μm; Phenomenex, Torrance, CA, USA).
An UltiMate3000 series HPLC system was used. The instrument is composed of an LPG-3400SD pump, a WPS-3000 (RS) autosampler, a TCC-3000 (RS) column oven, and a Corona charged aerosol detector (CAD) (all Thermo Fisher Scientific, Inc., Waltham, MA, USA). The Corona CAD was supplied with a continuous nitrogen stream by a Parker Balston NitroVap-1LV nitrogen generator (Parker Hannifin GmbH, Kaarst, Germany).
A Nucleodur C18 gravity column from Macherey-Nagel GmbH and Co. KG (Düren, Germany) with a precolumn cartridge of 4 mm length, a particle size of 3 μm, and dimensions of 150 × 4.6 mm was utilized to separate the HAAs. The gradient program started with 70% acetonitrile and 30% purified water containing 0.2% formic acid. After this, composition was kept constant for 1 min, and the acetonitrile percentage began to increase and continued until it reached 100% at 8 min. After 11 min of total running time, the acetonitrile percentage was decreased to 70% within 1 min. The total analysis time was 15 min. The column oven temperature was set to 40°C, and 5 μl of the sample was injected. The flow rate was set to 1 ml/min.
HAA composition analysis by HPLC-MS/MS.
For HAAs produced by E. coli C43(DE3) pPA2, BL21(DE3) pPA2, C43(DE3) pANA, C43(DE3) pFLU, and C43(DE3) pBUG, sample preparation and chromatographic separation were carried out as described by Behrens et al. (37). In short, 2 ml of the sample material [4 ml for the less concentrated E. coli C43(DE3) pBUG sample] was subjected to solid-phase extraction on a strong quaternary ammonium-modified polymeric anion-exchange material (Chromabond HR-XA, 3 ml volume, 200 mg adsorbent weight; Macherey-Nagel GmbH and Co. KG, Düren, Germany). Then, 10 μl of solid-phase extraction (SPE) extract [20 μl for the E. coli C43(DE3) pBUG sample] was analyzed with HPLC using a Nucleodur Sphinx RP column (150 × 2 mm, 3 μm; Macherey-Nagel, Düren, Germany). A mobile phase gradient consisting of 5 mmol/liter aqueous ammonium formate buffer (pH 3.3) containing 5% acetonitrile (vol/vol) (A) and acetonitrile (B) was used. Mass spectrometric detection was carried out with electrospray ionization in negative ionization mode. Structural information was provided by performing additional MS/MS experiments on two different mass spectrometers as follows. Samples of E. coli C43(DE3) pPA2 and C43(DE3) pBUG were analyzed on a Micromass Quattro micro triple quadrupole mass spectrometer (product ion scans) as detailed previously (37). MS/MS characterization of extracts from E. coli C43(DE3) pANA, C43(DE3) pFLU, and BL21(DE3) pPA2 was carried out on a linear ion trap mass spectrometer (LTQ XL; Thermo Fisher Scientific, Inc., San Jose, CA, USA) under the conditions described by Behrens et al. (38).
Additional confirmatory experiments were conducted using high-resolution MS. The analytes were identified by their accurate masses detected on a QExactive hybrid quadrupole Orbitrap (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer. The instrument was operated in negative electrospray ionization mode with the following parameters: spray voltage, 3.0 kV; sheath gas, 40 arbitrary units (AU); auxiliary gas, 10 AU; sweep gas, 1 AU; resolution, 140,000× (full width at half maximum [FWHM] at m/z 200); and mass range, m/z 200 to 1,000.
The intact HAAs were detected as deprotonated molecules ([M-H]–); e.g., a peak at m/z 301 was observed for C8-C8 HAA. MS/MS product ion spectra were dominated by the cleavage of the ester bond between the two β-hydroxy fatty acids as described by Lépine et al. (39). The product ion spectrum of the parent ion at m/z 301 showed a major fragment at m/z 159, which corresponds to a C8 fatty acid moiety, thus confirming the assignment of the parent as C8-C8 HAA. Fragments with m/z 131 and 187 were also present. These ions indicate the presence of C6 and C10 fatty acid moieties, therefore confirming by LC-MS/MS that not only C8-C8 HAA but also C6-C10 and C10-C6 HAAs were present.
Confirmation of hydroxy fatty acids by GC-MS.
The HAAs were analyzed using gas chromatography-mass spectrometry (GC-MS). Therefore, an aliquot of each sample was dried under a gentle stream of nitrogen and hydrolyzed with 0.5 M NaOH in MeOH-H2O solution (9:1 [vol/vol], 2 ml, 70°C, 1 h). Afterward, the solution was acidified to pH 3 with 1 M HCl, and the fatty acids were extracted with chloroform (3 × 3 ml). After removal of the solvent, fatty acid methyl esters (FAMEs) were prepared by adding 100 μl of BF3-MeOH (14%, wt/vol) and heating (75°C, 1 h). Then, 2 ml of H2O was added, and the FAMEs were extracted with chloroform (3 × 2 ml). The solvent was evaporated using a gentle stream of nitrogen. The residue was redissolved in 25 μl of pyridine and 50 μl of the silylating agent (BSTFA:TMCS [99:1, vol/vol]) and then heated (70°C, 1 h). Finally, the silylating agent was removed under a gentle stream of nitrogen, and the residue was rediluted in 0.2 ml n-hexane and used for GC-MS analysis.
After derivatization, samples were analyzed using a GCMS-QP-2020 equipped with a Nexis GC-2030 gas chromatograph (both Shimadzu, Kyoto, Japan). A 30-m, 0.25-mm-inside-diameter (i.d.), 0.25-μm-film-thickness DB-5MS column (J&W Scientific, Folsom, CA, USA) was used for the separation. Samples (1 μl) were injected using an AOC-20i Plus autosampler (Shimadzu, Kyoto, Japan) and a programmed temperature vaporization (PTV) inlet (250°C) in splitless mode. Helium (5.0) was used as a carrier gas with a flow rate of 1.22 ml/min. The column oven was programmed as follows: starting at 50°C, the temperature was increased at a rate of 10°C/min to 300°C, which was held for 10 min. Mass spectra were obtained by electron ionization (EI; 70 eV). The temperatures of the ion source and interface were set to 250°C. Data were recorded from m/z 50 to 500 with a rate of 10 scans/s. For comparison of retention times and fragmentation patterns, a bacterial acid methyl ester standard solution (BAME) (47080-U; Sigma-Aldrich, Steinheim, Germany) was used (10-fold diluted with methyl tert‐butyl ether).
Computational methods.
The evolutionary history was inferred using the neighbor-joining method (62). Evolutionary analyses were conducted in MEGA7 (63).
Accession numbers.
The corrected sequence of the rhlA gene in P. ananatis LMG 20103 and the sequence containing PhaG in P. desmolyticum were deposited under the GenBank accession numbers MF671909 and MG099922, respectively. The codon-optimized rhlA homologs for the construction of pHAL and pPLY are accessible under MN369027 and MN369028.
ACKNOWLEDGMENTS
We received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement number 633962 for project P4SB. This work was partially funded by the Cluster of Excellence “Tailor-Made Fuels from Biomass” (TMFB), which is financed by the Excellence Initiative of the German federal and state governments to promote science and research at German universities, and by the Federal Ministry of Education and Research (BMBF, Germany) as part of the project NemaContAnt (FKZ 031B0352B). Further funding was received by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—Exzellenzcluster 2186, “The Fuel Science Center ID: 390919832.”
REFERENCES
- 1.Randhawa KKS, Rahman PKSM. 2014. Rhamnolipid biosurfactants: past, present, and future scenario of global market. Front Microbiol 5:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banat IM, Makkar RS, Cameotra SS. 2000. Potential commercial applications of microbial surfactants. Appl Microbiol Biotechnol 53:495–508. doi: 10.1007/s002530051648. [DOI] [PubMed] [Google Scholar]
- 3.Singh A, Van Hamme JD, Ward OP. 2007. Surfactants in microbiology and biotechnology. Part 2. Application aspects. Biotechnol Adv 25:99–121. doi: 10.1016/j.biotechadv.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Mawgoud AM, Lépine F, Déziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzoigwe C, Burgess JG, Ennis CJ, Rahman PKSM. 2015. Bioemulsifiers are not biosurfactants and require different screening approaches. Front Microbiol 6:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahman PKSM, Gakpe E. 2008. Production, characterization and applications of biosurfactants: review. Biotechnology 7:360–370. doi: 10.3923/biotech.2008.360.370. [DOI] [Google Scholar]
- 7.Wittgens A, Tiso T, Arndt TT, Wenk P, Hemmerich J, Müller C, Wichmann R, Küpper B, Zwick M, Wilhelm S, Hausmann R, Syldatk C, Rosenau F, Blank LM. 2011. Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact 10:80. doi: 10.1186/1475-2859-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu K, Rock CO. 2008. RhlA converts beta-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the beta-hydroxydecanoyl-beta-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J Bacteriol 190:3147–3154. doi: 10.1128/JB.00080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blank LM, Tiso T, Germer A. January 2017. Extracellular production of designer hydroxyalkanoyloxy alkanoic acids with recombinant bacteria. World patent WO2017006252A1.
- 10.Déziel E, Lépine F, Milot S, Villemur R. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3–(3-hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005–2013. doi: 10.1099/mic.0.26154-0. [DOI] [PubMed] [Google Scholar]
- 11.Tiso T, Zauter R, Tulke H, Leuchtle B, Li WJ, Behrens B, Wittgens A, Rosenau F, Hayen H, Blank LM. 2017. Designer rhamnolipids by reduction of congener diversity: production and characterization. Microb Cell Fact 16:225. doi: 10.1186/s12934-017-0838-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis FG, Johnson MJ. 1949. A glyco-lipide produced by Pseudomonas-aeruginosa. J Am Chem Soc 71:4124–4126. doi: 10.1021/ja01180a073. [DOI] [Google Scholar]
- 13.Müller MM, Hörmann B, Syldatk C, Hausmann R. 2010. Pseudomonas aeruginosa PAO1 as a model for rhamnolipid production in bioreactor systems. Appl Microbiol Biotechnol 87:167–174. doi: 10.1007/s00253-010-2513-7. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira FJS, Vazquez L, de Campos NP, de França FP. 2009. Production of rhamnolipids by a Pseudomonas alcaligenes strain. Process Biochem 44:383–389. doi: 10.1016/j.procbio.2008.11.014. [DOI] [Google Scholar]
- 15.Gunther NW IV, Nunez A, Fett W, Solaiman DK. 2005. Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl Environ Microbiol 71:2288–2293. doi: 10.1128/AEM.71.5.2288-2293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martinez-Toledo A, Rios-Leal E, Vazquez-Duhalt R, Gonzalez-Chavez Mdel C, Esparza-Garcia JF, Rodriguez-Vazquez R. 2006. Role of phenanthrene in rhamnolipid production by P. putida in different media. Environ Technol 27:137–142. doi: 10.1080/09593332708618628. [DOI] [PubMed] [Google Scholar]
- 17.Rooney AP, Price NP, Ray KJ, Kuo TM. 2009. Isolation and characterization of rhamnolipid-producing bacterial strains from a biodiesel facility. FEMS Microbiol Lett 295:82–87. doi: 10.1111/j.1574-6968.2009.01581.x. [DOI] [PubMed] [Google Scholar]
- 18.Hošková M, Ježdík R, Schreiberová O, Chudoba J, Šír M, Čejková A, Masák J, Jirků V, Řezanka T. 2015. Structural and physiochemical characterization of rhamnolipids produced by Acinetobacter calcoaceticus, Enterobacter asburiae and Pseudomonas aeruginosa in single strain and mixed cultures. J Biotechnol 193:45–51. doi: 10.1016/j.jbiotec.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 19.Hörmann B, Müller MM, Syldatk C, Hausmann R. 2010. Rhamnolipid production by Burkholderia plantarii DSM 9509(T). Eur J Lipid Sci Technol 112:674–680. doi: 10.1002/ejlt.201000030. [DOI] [Google Scholar]
- 20.Wittgens A, Santiago-Schuebel B, Henkel M, Tiso T, Blank LM, Hausmann R, Hofmann D, Wilhelm S, Jaeger KE, Rosenau F. 2018. Heterologous production of long-chain rhamnolipids from Burkholderia glumae in Pseudomonas putida: a step forward to tailor-made rhamnolipids. Appl Microbiol Biotechnol 102:1229–1239. doi: 10.1007/s00253-017-8702-x. [DOI] [PubMed] [Google Scholar]
- 21.Costa SG, Déziel E, Lépine F. 2011. Characterization of rhamnolipid production by Burkholderia glumae. Lett Appl Microbiol 53:620–627. doi: 10.1111/j.1472-765X.2011.03154.x. [DOI] [PubMed] [Google Scholar]
- 22.Dubeau D, Déziel E, Woods DE, Lépine F. 2009. Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol 9:263. doi: 10.1186/1471-2180-9-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Häussler S, Nimtz M, Domke T, Wray V, Steinmetz I. 1998. Purification and characterization of a cytotoxic exolipid of Burkholderia pseudomallei. Infect Immun 66:1588–1593. doi: 10.1128/IAI.66.4.1588-1593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tavares LF, Silva PM, Junqueira M, Mariano DC, Nogueira FC, Domont GB, Freire DM, Neves BC. 2013. Characterization of rhamnolipids produced by wild-type and engineered Burkholderia kururiensis. Appl Microbiol Biotechnol 97:1909–1921. doi: 10.1007/s00253-012-4454-9. [DOI] [PubMed] [Google Scholar]
- 25.Lee M, Kim MK, Vancanneyt M, Swings J, Kim SH, Kang MS, Lee ST. 2005. Tetragenococcus koreensis sp. nov., a novel rhamnolipid-producing bacterium. Int J Syst Evol Microbiol 55:1409–1413. doi: 10.1099/ijs.0.63448-0. [DOI] [PubMed] [Google Scholar]
- 26.Aleksic I, Petkovic M, Jovanovic M, Milivojevic D, Vasiljevic B, Nikodinovic-Runic J, Senerovic L. 2017. Anti-biofilm properties of bacterial di-rhamnolipids and their semi-synthetic amide derivatives. Front Microbiol 8:2454. doi: 10.3389/fmicb.2017.02454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christova N, Tuleva B, Lalchev Z, Jordanova A, Jordanov B. 2004. Rhamnolipid biosurfactants produced by Renibacterium salmoninarum 27BN during growth on n-hexadecane. Z Naturforsch C J Biosci 59:70–74. doi: 10.1515/znc-2004-1-215. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Cai B, Shao Z. 2014. Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris As-13-3. Front Microbiol 5:711. doi: 10.3389/fmicb.2014.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pantazaki AA, Dimopoulou MI, Simou OM, Pritsa AA. 2010. Sunflower seed oil and oleic acid utilization for the production of rhamnolipids by Thermus thermophilus HB8. Appl Microbiol Biotechnol 88:939–951. doi: 10.1007/s00253-010-2802-1. [DOI] [PubMed] [Google Scholar]
- 30.Rezanka T, Siristova L, Sigler K. 2011. Rhamnolipid-producing thermophilic bacteria of species Thermus and Meiothermus. Extremophiles 15:697–709. doi: 10.1007/s00792-011-0400-5. [DOI] [PubMed] [Google Scholar]
- 31.Seghal Kiran G, Thajuddin N, Hema TA, Idhayadhulla A, Surendar Kumar R, Selvin J. 2010. Optimization and characterization of rhamnolipid biosurfactant from sponge associated marine fungi Aspergillus sp. MSF1. Desalination Water Treat 24:257–265. doi: 10.5004/dwt.2010.1569. [DOI] [Google Scholar]
- 32.Cabrera-Valladares N, Richardson A-P, Olvera C, Treviño LG, Déziel E, Lépine F, Soberón-Chávez G. 2006. Monorhamnolipids and 3–(3-hydroxyalkanoyloxy)alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl Microbiol Biotechnol 73:187–194. doi: 10.1007/s00253-006-0468-5. [DOI] [PubMed] [Google Scholar]
- 33.Abdel-Mawgoud AM, Lépine F, Déziel E. 2014. A stereospecific pathway diverts beta-oxidation intermediates to the biosynthesis of rhamnolipid biosurfactants. Chem Biol 21:156–164. doi: 10.1016/j.chembiol.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Smith DD, Nickzad A, Déziel E, Stavrinides J. 2016. A novel glycolipid biosurfactant confers grazing resistance upon Pantoea ananatis BRT175 against the social amoeba Dictyostelium discoideum. mSphere 1:e00075-15. doi: 10.1128/mSphere.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunther NW, Solaiman DKY, Fett WF. April 2007. Processes for the production of rhamnolipids. US patent 7202063.
- 36.Solaiman DK, Ashby RD, Gunther NW, Zerkowski JA. 2015. Dirhamnose-lipid production by recombinant nonpathogenic bacterium Pseudomonas chlororaphis. Appl Microbiol Biotechnol 99:4333–4342. doi: 10.1007/s00253-015-6433-4. [DOI] [PubMed] [Google Scholar]
- 37.Behrens B, Engelen J, Tiso T, Blank LM, Hayen H. 2016. Characterization of rhamnolipids by liquid chromatography/mass spectrometry after solid-phase extraction. Anal Bioanal Chem 408:2505–2514. doi: 10.1007/s00216-016-9353-y. [DOI] [PubMed] [Google Scholar]
- 38.Behrens B, Helmer PO, Tiso T, Blank LM, Hayen H. 2016. Rhamnolipid biosurfactant analysis using online turbulent flow chromatography-liquid chromatography-tandem mass spectrometry. J Chromatogr A 1465:90–97. doi: 10.1016/j.chroma.2016.08.044. [DOI] [PubMed] [Google Scholar]
- 39.Lépine F, Déziel E, Milot S, Villemur R. 2002. Liquid chromatographic/mass spectrometric detection of the 3–(3-hydroxyalkanoyloxy) alkanoic acid precursors of rhamnolipids in Pseudomonas aeruginosa cultures. J Mass Spectrom 37:41–46. doi: 10.1002/jms.244. [DOI] [PubMed] [Google Scholar]
- 40.Blank L, Rosenau F, Wilhelm S, Wittgens A, Tiso T. January 2018. Means and methods for rhamnolipid production. US patent 9854799.
- 41.Gauthier C, Lavoie S, Piochon M, Martinez S, Milot S, Déziel E. 2019. Structural determination of ananatoside A: an unprecedented 15-membered macrodilactone-containing glycolipid from Pantoea ananatis. Carbohydr Res 471:13–18. doi: 10.1016/j.carres.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Rehm BH, Mitsky TA, Steinbüchel A. 2001. Role of fatty acid de novo biosynthesis in polyhydroxyalkanoic acid (PHA) and rhamnolipid synthesis by pseudomonads: establishment of the transacylase (PhaG)-mediated pathway for PHA biosynthesis in Escherichia coli. Appl Environ Microbiol 67:3102–3109. doi: 10.1128/AEM.67.7.3102-3109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gutierrez M, Choi MH, Tian B, Xu J, Rho JK, Kim MO, Cho YH, Yoon SC. 2013. Simultaneous inhibition of rhamnolipid and polyhydroxyalkanoic acid synthesis and biofilm formation in Pseudomonas aeruginosa by 2-bromoalkanoic acids: effect of inhibitor alkyl-chain-length. PLoS One 8:e73986. doi: 10.1371/journal.pone.0073986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehm BH, Krüger N, Steinbüchel A. 1998. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. The phaG gene from Pseudomonas putida KT2440 encodes a 3-hydroxyacyl-acyl carrier protein-coenzyme a transferase. J Biol Chem 273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 45.Jadhav M, Kalme S, Tamboli D, Govindwar S. 2011. Rhamnolipid from Pseudomonas desmolyticum NCIM-2112 and its role in the degradation of Brown 3REL. J Basic Microbiol 51:385–396. doi: 10.1002/jobm.201000364. [DOI] [PubMed] [Google Scholar]
- 46.Irorere VU, Tripathi L, Marchant R, McClean S, Banat IM. 2017. Microbial rhamnolipid production: a critical re-evaluation of published data and suggested future publication criteria. Appl Microbiol Biotechnol 101:3941–3951. doi: 10.1007/s00253-017-8262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller MM, Hörmann B, Kugel M, Syldatk C, Hausmann R. 2011. Evaluation of rhamnolipid production capacity of Pseudomonas aeruginosa PAO1 in comparison to the rhamnolipid over-producer strains DSM 7108 and DSM 2874. Appl Microbiol Biotechnol 89:585–592. doi: 10.1007/s00253-010-2901-z. [DOI] [PubMed] [Google Scholar]
- 48.Burger MM, Glaser L, Burton RM. 1963. The enzymatic synthesis of a rhamnose-containing glycolipid by extracts of Pseudomonas aeruginosa. J Biol Chem 238:2595–2602. [PubMed] [Google Scholar]
- 49.Déziel E, Lépine F, Dennie D, Boismenu D, Mamer OA, Villemur R. 1999. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim Biophys Acta 1440:244–252. doi: 10.1016/s1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 50.De Maayer P, Chan WY, Venter SN, Toth IK, Birch PR, Joubert F, Coutinho TA. 2010. Genome sequence of Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J Bacteriol 192:2936–2937. doi: 10.1128/JB.00060-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irorere VU, Smyth TJ, Cobice D, McClean S, Marchant R, Banat IM. 2018. Fatty acid synthesis pathway provides lipid precursors for rhamnolipid biosynthesis in Burkholderia thailandensis E264. Appl Microbiol Biotechnol 102:6163–6174. doi: 10.1007/s00253-018-9059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Déziel E, Lépine F, Milot S, Villemur R. 2000. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim Biophys Acta 1485:145–152. doi: 10.1016/s1388-1981(00)00039-1. [DOI] [PubMed] [Google Scholar]
- 53.Saur KM, Brumhard O, Scholz K, Hayen H, Tiso T. 2019. A pH shift induces high-titer liamocin production in Aureobasidium pullulans. Appl Microbiol Biotechnol 103:4741–4752. doi: 10.1007/s00253-019-09677-3. [DOI] [PubMed] [Google Scholar]
- 54.Kügler JH, Le Roes-Hill M, Syldatk C, Hausmann R. 2015. Surfactants tailored by the class Actinobacteria. Front Microbiol 6:212. doi: 10.3389/fmicb.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Häussler S, Rohde M, von Neuhoff N, Nimtz M, Steinmetz I. 2003. Structural and functional cellular changes induced by Burkholderia pseudomallei rhamnolipid. Infect Immun 71:2970–2975. doi: 10.1128/iai.71.5.2970-2975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heath RJ, Rock CO. 1996. Inhibition of beta-ketoacyl-acyl carrier protein synthase III (FabH) by acyl-acyl carrier protein in Escherichia coli. J Biol Chem 271:10996–11000. doi: 10.1074/jbc.271.18.10996. [DOI] [PubMed] [Google Scholar]
- 57.Rock CO, Jackowski S. 2002. Forty years of bacterial fatty acid synthesis. Biochem Biophys Res Commun 292:1155–1166. doi: 10.1006/bbrc.2001.2022. [DOI] [PubMed] [Google Scholar]
- 58.Toribio J, Escalante AE, Soberón-Chávez G. 2010. Rhamnolipids: production in bacteria other than Pseudomonas aeruginosa. Eur J Lipid Sci Technol 112:1082–1087. doi: 10.1002/ejlt.200900256. [DOI] [Google Scholar]
- 59.Tiso T, Sabelhaus P, Behrens B, Wittgens A, Rosenau F, Hayen H, Blank LM. 2016. Creating metabolic demand as an engineering strategy in Pseudomonas putida: rhamnolipid synthesis as an example. Metab Eng Commun 3:234–244. doi: 10.1016/j.meteno.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiso T, Germer A, Küpper B, Wichmann R, Blank LM. 2016. Methods for recombinant rhamnolipid production, p 65–94. In McGenity TJ, Timmis KN, Nogales Fernández B (ed), Hydrocarbon and lipid microbiology protocols: synthetic and systems biology - applications. Springer, Berlin, Germany. [Google Scholar]
- 61.Behrens B, Baune M, Jungkeit J, Tiso T, Blank LM, Hayen H. 2016. High performance liquid chromatography-charged aerosol detection applying an inverse gradient for quantification of rhamnolipid biosurfactants. J Chromatogr A 1455:125–132. doi: 10.1016/j.chroma.2016.05.079. [DOI] [PubMed] [Google Scholar]
- 62.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 63.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnson SL, Bishop-Lilly KA, Ladner JT, Daligault HE, Davenport KW, Jaissle J, Frey KG, Koroleva GI, Bruce DC, Coyne SR, Broomall SM, Li P-E, Teshima H, Gibbons HS, Palacios GF, Rosenzweig CN, Redden CL, Xu Y, Minogue TD, Chain PS. 2015. Complete genome sequences for 59 Burkholderia isolates, both pathogenic and near neighbor. Genome Announc 3:e00159-15. doi: 10.1128/genomeA.00159-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci U S A 87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 67.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol 260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 68.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol 185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]