The zygomycete Blakeslea trispora is an important strain for the production of carotenoids on a large scale. However, the regulation mechanism of carotenoid biosynthesis is still not well understood in this filamentous fungus. In the present study, we sought to investigate how crgA influences the expression of structural genes for carotenoids, carotenoid biosynthesis, and other anabolic phenotypes. This will lead to a better understanding of the global regulation mechanism of carotenoid biosynthesis and facilitate engineering this strain in the future for enhanced production of carotenoids.
KEYWORDS: Blakeslea trispora, crgA, carotenoids, biosynthesis, negative regulator, Mucorales
ABSTRACT
As an ideal carotenoid producer, Blakeslea trispora has gained much attention due to its large biomass and high production of β-carotene and lycopene. However, carotenogenesis regulation in B. trispora still needs to be clarified, as few investigations have been conducted at the molecular level in B. trispora. In this study, a gene homologous to carotenogenesis regulatory gene (crgA) was cloned from the mating type (−) of B. trispora, and the deduced CrgA protein was analyzed for its primary structure and domains. To clarify the crgA-mediated regulation in B. trispora, we used the strategies of gene knockout and complementation to investigate the effect of crgA expression on the phenotype of B. trispora. In contrast to the wild-type strain, the crgA null mutant (ΔcrgA) was defective in sporulation but accumulated much more β-carotene (31.2% improvement at the end) accompanied by enhanced transcription of three structural genes (hmgR, carB, and carRA) for carotenoids throughout the culture time. When the wild-type copy of crgA was complemented into the crgA null mutant, sporulation, transcription of structural genes, and carotenoid production were restored to those of the wild-type strain. A gas chromatography-mass spectrometry (GC-MS)-based metabolomic approach and multivariate statistical analyses were performed to investigate the intracellular metabolite profiles. The reduced levels of tricarboxylic acid (TCA) cycle components and some amino acids and enhanced levels of glycolysis intermediates and fatty acids indicate that more metabolic flux was driven into the mevalonate (MVA) pathway; thus, the increase of precursors and fat content contributes to the accumulation of carotenoids.
IMPORTANCE The zygomycete Blakeslea trispora is an important strain for the production of carotenoids on a large scale. However, the regulation mechanism of carotenoid biosynthesis is still not well understood in this filamentous fungus. In the present study, we sought to investigate how crgA influences the expression of structural genes for carotenoids, carotenoid biosynthesis, and other anabolic phenotypes. This will lead to a better understanding of the global regulation mechanism of carotenoid biosynthesis and facilitate engineering this strain in the future for enhanced production of carotenoids.
INTRODUCTION
As one of the most important natural pigments, carotenoids can quench singlet oxygen effectively, and their antioxidant capacity is considered to be helpful for alleviating cancer and heart disease (1, 2). A strain of the mating type (−) of the zygomycete Blakeslea trispora is able to accumulate large amounts of carotenoids, which drives interest for this filamentous fungus (3–5). In B. trispora, β-carotene is synthesized from the mevalonate (MVA) pathway. 3-Hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) catalyzed from acetyl coenzyme A (acetyl-CoA) is converted to mevalonate first by HMG-CoA reductase and to lycopene sequentially by several enzymes; then, lycopene is converted to β-carotene by lycopene cyclase. In the MVA pathway, HMG-CoA reductase encoded by hmgR is the first rate-limiting enzyme, because its activity decides the content of mevalonate (6). The phytoene synthase that converts geranylgeranyl pyrophosphate to phytoene and lycopene cyclase that controls lycopene cyclization are other important enzymes. They are encoded by the two domains of the carRA gene. Last but not least, phytoene dehydrogenase encoded by carB can convert phytoene to lycopene by dehydrogenation (7). The carotenogenesis pathway has been defined, but complexity in the regulation of carotenoid biosynthesis in B. trispora impedes the metabolic modification of B. trispora for enhanced carotenoid production.
Carotenogenesis regulatory gene (crgA) is a negative regulator of carotenoid biosynthesis detected in Mucor circinelloides (8, 9), Fusarium fujikuroi (10), and Fusarium oxysporum (11). Meanwhile, crgA plays an important role in the process of sporulation and carbon metabolism (12). The ortholog of crgA has been cloned from the mating type (+) of B. trispora and was then introduced into a crgA null mutant of M. circinelloides to investigate its function (13). The complemented strain restored the wild-type (WT) phenotype, which indicated that the function of this regulator was conserved in both filamentous fungi. Discovery of crgA provides a possibility to improve the carotenoid production in the crgA-containing carotenoid producer. As for B. trispora, the mating type (−) is the main contributor to carotenoid production, but there have been no investigations on the potential regulator of carotenoid biosynthesis in this strain. In this study, we present the isolation and characterization of the crgA gene from strain (−) of B. trispora. Effects of disruption and complementation of this gene on expression of structural genes for carotenoid and carbon metabolism provide evidence for a role of crgA as a negative regulator of carotenogenesis in B. trispora.
RESULTS
Cloning of a crgA-homologous gene from strain (−) of B. trispora.
The crgA-homologous sequence from strain (−) was cloned and sequenced. Figure S2A in the supplemental material shows that the nucleotide sequence was 4,245 bp in length (GenBank accession number MH345788.1) and it has an identity of 99.3% compared with that from strain (+) of B. trispora reported by Quiles-Rosillo et al. (13). Both sequences have a putative translation start GTG codon; the crgA gene in M. circinelloides begins at this GTG codon (18). Although this frequently occurs in bacterial genes (15), only a few well-documented examples have been reported in fungi (16). The upstream sequence of the GTG initiation codon of the B. trispora crgA gene contains pyrimidine-rich regions and a TATA box, which shows similarity with that in many other fungi, occurring in gene promoters and considered to contribute to the correct initiation of transcription (17).
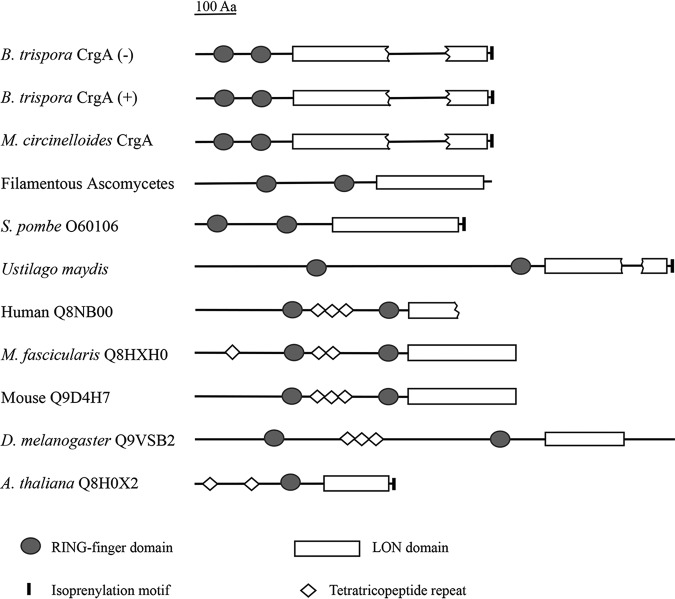
Although nucleotide sequences of crgA from (−) and (+) strains are not identical, the amino acid sequences of CrgA protein deduced from the coding sequences are almost the same (Fig. S2B). Both 611-amino-acid-long polypeptides contain several domains, including two RING fingers (RFs), one disrupted LON domain, and one isoprenylation motif (Fig. S2B). No CrgA homologous protein was found in the SWALL database, though several putative eukaryotic proteins presenting similarity in domain architecture to that of the CrgA proteins obtained by searching against a translation of the fungal genome were compared (Fig. 1). The domain architectures of two RING finger domains at the amino-terminal region followed by a LON domain are widespread in eukaryotes from fungi to animals and humans, suggesting this type of protein is conserved in function. Another obvious characteristic of CrgA proteins from (−) and (+) strains of B. trispora is the existence of an isoprenylation motif at the carboxy terminus (Fig. S2B), which has also been observed in M. circinelloides, Schizosaccharomyces pombe, Ustilago maydis, and Arabidopsis thaliana. In addition, CrgA proteins contain five repetitive fragments of heptapeptide PASMMAR (Fig. S2B), much like that of CrgA protein from M. circinelloides, with unknown function.
FIG 1.
Comparison of domain architecture of CrgA with similar proteins from different species. Filamentous Ascomycetes (Aspergillus nidulans, GenBank accession number [no.] AACD01000101.1; Gibberella zeae, GenBank accession no. AACM01000228.1), Schizosaccharomyces pombe (SWALL accession no. 060106), Ustilago maydis (GenBank accession no. AACP01000182.1), human (SWALL accession no. Q8NBOO), Macaca fascicularis (SWALL accession no. Q8HXHO), mouse (SWALL accession no. Q9D4H7), Drosophila melanogaster (SWALL accession no. Q9VSB2), and Arabidopsis thaliana (SWALL accession no. Q8HOX2).
Construction of the crgA null mutants and gene complementary strains.
The WT strain of B. trispora did not grow when cultured on wort agar containing hygromycin B. In contrast, transformants containing the resistance gene were able to grow on the resistance medium (see Fig. S3A). Although the transformants had resistance to hygromycin B, it was necessary to confirm whether the hph gene was integrated in the correct location. The confirmation was performed by PCR analysis with specific primers (see Fig. S3B). The amplicon for the P1/P2 primer pair amplified only in PCRs from the wild-type strain, while amplicons for P3/P4, P5/P6, and hR/hF primer pairs were amplified from the ΔcrgA strain only. Gel electrophoresis analyses (Fig. S3C) indicated that crgA was disrupted successfully. crgA complementation was also performed with a split-marker strategy, and the integrated fragment sequence in the transformant was in accordance with the crgA sequence of the wild-type strain, which indicated that a complemented (C-ΔcrgA) strain was obtained. The crgA-complemented strains reverted to being hygromycin sensitive, which was confirmed by the fact that they were not able to grow on the wort agar medium containing hygromycin B while demonstrating vigorous growth on the same medium without supplementation of hygromycin B (data not shown).
Effect of crgA on sporulation.
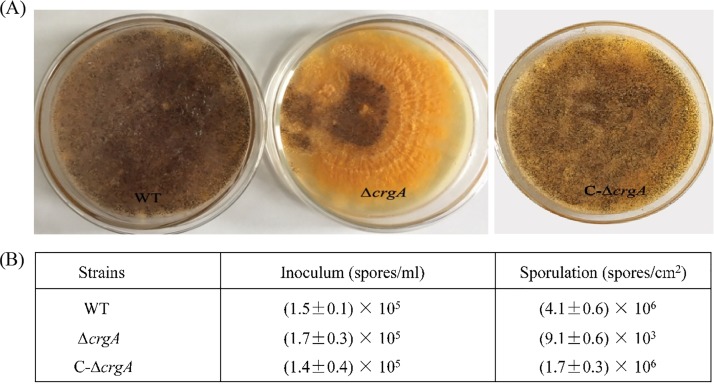
One hundred microliters of spore suspensions from the wild-type strain, ΔcrgA strain, and C-ΔcrgA strain were inoculated on wort agar plates. Growth was observed and sporulation was analyzed with a hemacytometer counting method after the strains were cultured at 25°C for 5 days. Wild-type strains sporulated soon under suitable conditions, but the ΔcrgA strain formed thick yellow fungus membranella with a small amount of spores (Fig. 2A). Amounts of ΔcrgA strain spores were reduced by approximately three orders of magnitude compared with that of the wild-type strain, while amounts of C-ΔcrgA strain spores were similar to that of the wild-type strain (Fig. 2B), which indicated that crgA influenced the sporulation.
FIG 2.
Effect of crgA on sporulation in B. trispora. (A) Sporulation of different B. trispora strains (wild-type [WT], crgA null strain [ΔcrgA], and crgA-complemented strain [C-ΔcrgA]) on wort agar plates. (B) Generated spore amounts of each strain from the nearly same inoculum amount.
Effect of crgA on the transcription of structural genes.
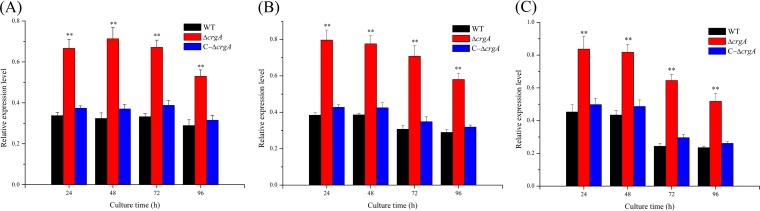
In the MVA pathway, there are several structural genes for carotenoid biosynthesis, and transcript levels of these genes are related to carotenoid accumulation (14). To verify whether the carotenogenesis pathway was regulated by the crgA gene, we explored the transcription of structural genes for carotenoid biosynthesis in different strains. Total RNA was isolated from B. trispora wild-type, ΔcrgA, and C-ΔcrgA strains, and then transcription of hmgR, carRA, and carB was analyzed. It was found that all of these genes were expressed at comparable levels in the WT strains and the C-ΔcrgA strains (Fig. 3A to C), despite the substantially enhanced expression of these genes in the crgA null strains. Transcript levels of hmgR, carB, and carRA in the ΔcrgA strain were upregulated by 90% to 120%, 100% to 130%, and 80% to 160%, respectively, compared with those in the wild-type strain. When crgA was complemented into the ΔcrgA strain, transcription of these genes (hmgR, carB, and carRA) was inhibited (Fig. 3A to C). This result suggests that the carotenogenesis pathway is inhibited by the crgA gene in the WT strain; therefore, the knockout of crgA induces transcriptional activation of the structural genes for carotenoid biosynthesis.
FIG 3.
Transcriptional analysis of structural genes for carotenoid biosynthesis, hmgR (A), carB (B), and carRA (C), in WT, ΔcrgA, and C-ΔcrgA strains. The relative transcription levels of these genes were normalized using the internal reference gene tefl. Data represent means ± standard deviations from three independent experiments. **, P < 0.01 (ΔcrgA versus WT).
Effect of crgA on carotenoid biosynthesis in strain (−) of B. trispora.
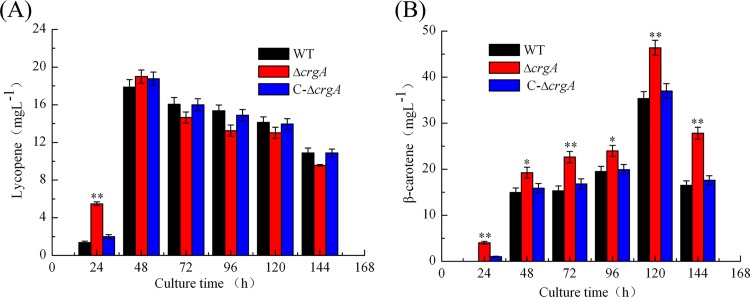
To analyze the effect of crgA on carotenoid biosynthesis in strain (−) of B. trispora, the time course of lycopene and β-carotene production was determined. As shown in Fig. 4A, lycopene content in all strains increased sharply from the beginning to 48 h and decreased slowly with incubation time. Especially, lycopene content in the ΔcrgA strain was much higher than in wild-type and C-ΔcrgA strains at 24 h of incubation, but this difference disappeared at longer incubation times, indicating that crgA has a weak influence on lycopene production. As for β-carotene production, the accumulation trends in all the strains were almost the same, i.e., the contents were low at early culture stage, increased step by step, and reached the maximum at 120 h (Fig. 4B). However, the ΔcrgA strain accumulated more β-carotene than the other strains across the incubation time. The maximum content in the ΔcrgA strain increased by 31.2% at 120 h of incubation compared with that in wild-type strain, while the carotenoid level in the C-ΔcrgA strain was similar to that in the wild-type strain.
FIG 4.
Time course of carotenoid production. (A) Lycopene production profile in WT, ΔcrgA, and C-ΔcrgA strains. (B) β-Carotene production profile in WT, ΔcrgA, and C-ΔcrgA strains. *, P < 0.05; **, P < 0.01 (ΔcrgA versus WT).
Effect of crgA on metabolite profiles in B. trispora.
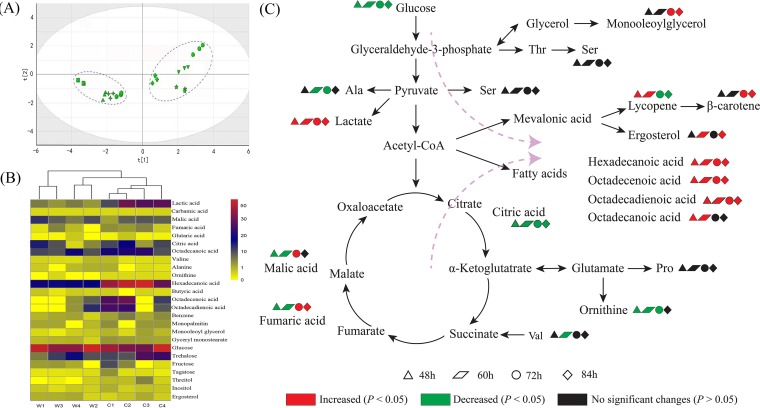
Metabolomics technology was used to investigate the effect of crgA on B. trispora metabolism. Metabolites in ΔcrgA and wild-type strains were measured with gas chromatography-mass spectrometry (GC-MS), and more than 100 peaks were detected. To establish a proper model to show differences among the samples, 25 metabolites whose signal/noise ratios were higher than 500 were selected, and a principal-component analysis was performed with SIMCA-P (versions 13.0). As shown in Fig. 5A, samples of the ΔcrgA strain (C1, C2, C3, and C4) and wild-type strain (W1, W2, W3, and W4) clustered together, which indicated that there was a notable difference between the two strains. Metabolites with the greatest changes, including amino acids, fatty acids, organic acids, saccharides, and polyols, are shown in Fig. 5B. Compared with those in the wild-type strain, the contents of amino acids (alanine, valine, and ornithine) in the ΔcrgA strain were much lower and the contents of fatty acids (hexadecanoic acid, octadecadienoic acid, octadecenoic acid, and octadecanoic acid) were much higher. The contents of organic acids (malic acid, fumaric acid, and citric acid) in the ΔcrgA strain were lower than those in the wild-type strain at first, but then the contents of malic acid and fumaric acid increased. In terms of saccharides, the content of glucose in the ΔcrgA strain was much lower, but the content of trehalose was higher. At the same time, the contents of inositol and ergosterol in the ΔcrgA strain were higher. Changes of the metabolites relevant to carotenoid biosynthesis are shown in Fig. 5C.
FIG 5.
(A) PCA score plot of metabolites in B. trispora WT and ΔcrgA strains at different time points. C l (squares)/W l (circles), C2 (triangles)/W2 (diamonds), C3 (plus signs)/W3 (inverted triangles), and C4 (hexagons)/W4 (stars) represent samples of ΔcrgA/WT strains after culture for 48 h, 60 h, 72 h, and 84 h, respectively. (B) Cluster analysis of B. trispora WT and ΔcrgA strains. Cl/W l, C2/W2, C3/W3, and C4/W4 represent samples of ΔcrgA/WT strains after culture for 48 h, 60 h, 72 h, and 84 h, respectively. (C) Effects of crgA on B. trispora at the metabolism level. Red symbols denote significant increase (P < 0.05), and green symbols denote significant decrease (P < 0.05), while black symbols denote no significant change in metabolite levels (P > 0.05). Ala, alanine; Ser, serine; Thr, threonine; Val, valine; Pro, proline.
DISCUSSION
A gene homologous to M. circinelloides crgA has been found in strain (+) of B. trispora, which is involved in the photoinduction of the carotenoid biosynthesis (13). In this study, the homologous gene was also isolated from strain (−) of B. trispora, which is usually used in the industrial production of carotenoids. The deduced product equally corresponds to the putative CrgA protein of the strain (+) of B. trispora and shows high similarity to the counterpart of M. circinelloides, indicating that the function of CrgA may be conserved between these zygomycetes. Although the RING-finger and LON domains of CrgA from strain (−) from other reports are essential for the negative regulation in carotenogenesis (10), the function of CrgA remains unclear among the zygomycetes. Even so, the comparison and functional analyses of similar domain architectures in different species can still provide some clues. The RING-finger and LON domains appear in organisms from fungi to mammals (Fig. 1), suggesting that the function of these proteins containing the above-mentioned domains is universal and that these proteins appeared before the differentiation of major eukaryotic groups. The RING-finger domains usually exist in a class of E3 ligases involved with recognition and degradation of target proteins (38), while LON domains are found in ATP-dependent Lon proteases (39). The putative isoprenylation motif has been regarded as related to posttranslation by the attachment of either a farnesyl or a geranyl-geranyl group to a cysteine residue (40). These similar domain architectures give us reason to believe that they play a role in the regulation of specific cellular metabolic functions, although there is little known of the above-described proteins. The above-mentioned discussion suggests that the CrgA protein of B. trispora strain (−) may act as a regulatory protein rather than a biosynthetic enzyme. The lack of similarity with the previously described carotenogenic enzymes (data not shown) reinforces this regulatory role proposed for the crgA gene.
To gain insight into this gene in strain (−) of B. trispora, the gene was disrupted to investigate phenotypic changes. The split-marker strategy is a frequently used method for gene disruption in fungi due to their high transformation rate and low level of ectopic integration (19). Ectopic insertion occurs with high frequency in filamentous fungi because of nonhomologous end joining (20); therefore, specific primers were designed to verify the disruption (see Fig. S3B in the supplemental material). The expected PCR amplicons indicated that crgA was replaced by hph accurately (Fig. S3C). Then, gene complementation of the ΔcrgA strain was conducted by transforming crgA into the ΔcrgA strain with the same method.
Sporulation was inhibited when crgA was disrupted, and this phenotype was restored after the mutant was complemented (Fig. 2B), which suggests that crgA is able to control sporulation. It has been reported that sporulation is under the control of crgA in M. circinelloides (21), which is taxonomically close to B. trispora. As for carotenoid production, the content of β-carotene in ΔcrgA strain increased 31.2% compared with that in the wild-type strain, while the C-ΔcrgA and wild-type strains accumulated similar β-carotene levels (Fig. 4B). The results indicated that crgA functions as a negative regulator of carotenoid biosynthesis. For lycopene production, it seems that the three strains had almost the same content across the culture process except at the 24-h incubation point. Although the exact reason is unknown, this may be related to the fact that lycopene is an intermediate metabolite. A previous study reported that crgA from strain (+) of B. trispora is able to restore the wild-type phenotype of a crgA null mutant of M. circinelloides (13), which suggests that the regulation role of crgA on carotenogenesis is conserved between B. trispora and M. circinelloides.
At the RNA level, transcription of structural genes (hmgR, carRA, and carB) in the ΔcrgA strain increased compared to that in wild-type and C-ΔcrgA strains (Fig. 3A and B). HMG-CoA reductase encoded by hmgR can regulate the content of mevalonate and its derivatives, such as carotenoids (6). Besides, the phytoene dehydrogenase encoded by carB can convert phytoene to lycopene by dehydrogenation. carRA is a bifunctional gene that encodes phytoene synthase and lycopene cyclase. The phytoene synthase catalyzes the conversion of geranylgeranyl pyrophosphate, and lycopene cyclase can catalyze lycopene cyclization (7). It can be deduced that enhanced transcript levels of carB and carRA made the ΔcrgA strain accumulate more carotenoids (Fig. 4A and B), which indicates that crgA regulates the accumulation of carotenoids by controlling the transcription of structural genes. Furthermore, the maximum expression levels of carotenoid structural genes in the ΔcrgA strain appeared at different culture times, which may be related to the location of these genes in the metabolic pathway.
Moreover, metabolic profile change was investigated by GC-MS and multivariate statistical analysis, as was done previously for the related species Phycomyces blakesleeanus (22). Principal-component analysis (PCA) is a statistical procedure that uses an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of linearly uncorrelated variables called principal components. Figure 5A shows that samples of the ΔcrgA strain and wild-type strain were separated significantly, which indicates that metabolic fluxes changed after crgA was disrupted. Metabolites with great changes can be divided into five categories, including saccharides, polyols, fatty acids, amino acids, and organic acids (Fig. 5B). At each time point, the content of glucose in the ΔcrgA strain was much lower than in the wild-type strain, which indicates that carbohydrate metabolism was altered after crgA was disrupted. Surprisingly, the content of trehalose increased notably (Fig. 5B). Since an important function of trehalose is to protect cell proteins from attack due to intracellular and extracellular changes (23), crgA disruption might induce the stress response, leading to an increase of trehalose secretion to protect cells. Meanwhile, the level of ergosterol was positively correlated with that of β-carotene. This may be ascribed to the fact that ergosterol and β-carotene share the same precursor (mevalonic acid), driving the metabolic flux to the mevalonate pathway. In addition, ergosterol plays a crucial part in cell permeability and can further enhance the accumulation of β-carotene (24). Contents of fatty acids (hexadecanoic acid, octadecanoic acid, octadecenoic acid, and octadecadienoic acid) in the ΔcrgA strain were much higher than in the wild-type strain. This phenomenon could be explained by the following aspects. On one hand, fatty acids and carotenoids were converted from the common precursor acetyl-CoA; therefore, levels of fatty acids were enhanced along with carotenoids. At the same time, fatty acids can stimulate more conversion of acetyl-CoA to carotenoids because of feedback inhibition (25). On the other hand, fatty acids could be used as a cosubstrate of glucose and take part in cell growth and cytomembrane or lipid biosynthesis (26, 27) and thus could activate B. trispora to synthesize carotenoids. crgA disruption also induced a content change of intermediates involved in the tricarboxylic acid (TCA) cycle. The citric acid content in the ΔcrgA strain was much lower than in the wild-type strain at each time point. Meanwhile, malic acid and fumaric acid contents decreased at first. The decrease might suggest that metabolic flux to the TCA cycle was inhibited and this metabolic pressure would drive acetyl-CoA to flow to the MVA and fatty acid pathway. However, malic acid and fumaric acid contents increased later, which might be related to the decreased amounts of amino acids (valine and ornithine). Valine can be degraded to succinate directly (28), and ornithine participates in the TCA cycle after transamination and oxidation (29, 30), which suggests that amino acid metabolism is influenced by crgA disruption. These results provide a potential mechanism of crgA regulating carotenoid biosynthesis in B. trispora. Moreover, many other metabolites are under the control of crgA, and further study needs to be performed to understand how CrgA impacts the biology of B. trispora and other fungi.
MATERIALS AND METHODS
Strains and culture conditions.
Strain (−) of B. trispora NRRL2896 was used in this study, which was cultivated under the conditions described in a previous report (31). Briefly, to produce mycelia, a 100-μl spore suspension with a density of 1.0 × 106 spores per ml was inoculated onto wort agar medium and then transferred to 50 ml seed medium (30 g/liter corn starch, 50 g/liter soybean meal, 1.5 g/liter KH2PO4, 0.5 g/liter MgSO4·7H2O, pH 6.5) in 250-ml Erlenmeyer flasks and cultivated at 25°C at 180 rpm for 48 h.
Cloning and analyses of crgA from strain (−) of B. trispora.
The mycelia were triturated with liquid nitrogen, and genomic DNA was isolated with the Plant Genomic DNA kit (Tiangen, China). The crgA gene was cloned with primers (Table 1) that were designed according to the crgA gene sequence of strain (+) (EMBL accession number AJ585199). The gene was amplified in 50-μl reaction mixtures containing pfu DNA polymerase (Sangon, China). PCR products were ligated into the pMD-18T vector, transformed into Escherichia coli DH5α, and sequenced (Sangon, China).
TABLE 1.
Primers used in this study
| Primer | Sequence, 5′→3′a | Purpose |
|---|---|---|
| Fu/Cu | GGAAATTAAGCTATGCACCGCAGTATAGTC | crgA cloning/complementation |
| Fd/Cr | TATTTTCATATGGAACAAGATTTGTCTATA | |
| Cd | CGTGGATCCATTGTCGAACGACAAGGCAGT | |
| Cf | TCAAAAATATTAAATGCTAAAAATGGAGAA | |
| hF | GTCGGAGACAGAAGATGATATTGAAGGAG | hph cloning |
| hR | GTTGGAGATTTCAGTAACGTTAAGTGGAT | |
| 5f | GGAAATTAAGCTATGCACCGCAGTATAGTCA | Cloning of 5′ flank region of crgA |
| 5r | ATCCACTTAACGTTACTGAAATCTCCAACAGFGAAGGTTTGAACAGAAAACTCTTGTAGC | |
| 3f | CTCCTTCAATATCATCTTCTGTCTCCGACACAGACGACTGAAGAGATGATTGATGAACT | Cloning of 3′ flank region of crgA |
| 3r | TATTTTCATATGGAACAAGATTTGTCTATACTG | |
| hf | GCGAAGAATCTCGTGCTTTCA | Cloning of split marker |
| hr | TCCAGAAGAAGATGTTGGCGAC | |
| P1 | AGCCTACGTTTTGAGTAGCTCGATC | Confirming mutants |
| P2 | ATACATTGTTGTGATGAAGCCACAC | |
| P3 | ATGGGCATGTTTTTGGGCTAGCAGT | |
| P4 | CGCGCAGGCTCTCGATGAGCTGAT | |
| P5 | CTCCTACATCGAAGCTGAAAGCACG | |
| P6 | ACTCCTCTTCCAAGAGCACTAGGTA | |
| hmgR-F | AAACGATGGATTGAACAAGAGGG | RT-qPCR |
| hmgR-R | TAGACTAGACGACCGGCAAGAGC | |
| carB-F | TATTGGCGGAACTGCTACTGC | |
| carB-R | CCCTGATCAAAGCGATGACC | |
| carRA-F | TCTTGAGCGTCGTCCTATCC | |
| carRA-R | GCACGGTCAATTATCCAAGC | |
| tef1-F | AACTCGGTAAGGGTTCCTTCAAG | |
| tef1-R | CGGGAGCATCAATAACGGTAAC |
Sequences in boldface are the reverse complement of hR and hF, respectively. The hph was inserted in the direction of crgA antisense.
Construction of gene deletion and complementation cassettes.
A split-marker strategy was used to construct a deletion cassette (see Fig. S1A in the supplemental material), which was PCR-based and needed two rounds of PCRs (32). In round 1, primers were designed by Primer Premier 5.0 for cloning of 5′ and 3′ flanks of the crgA sequence and hph fragment. The selectable marker hph from plasmid pCSN44 containing the trpC promoter confers hygromycin resistance. In round 2, 5′ and 3′ flanks were fused to the marker by overlap extension PCR. A complementation cassette was also constructed with the split-marker strategy (Fig. S1B). Two fragments of crgA were amplified with the primers listed in Table 1, and part of each fragment was complementary to the other.
PEG-mediated transformation of B. trispora protoplasts.
The mycelia were dispersed with a magnetic stirrer and washed twice with osmotic stabilizer (0.6 M NaCl). Then, ∼1 g of mycelia was suspended in 5 ml solution supplemented with mixed enzymes combined with 2% lysozyme, 3% cellulose, 3% snailase, and osmotic stabilizer (33). The mixture was incubated at 25°C with an agitation rate of 75 rpm for 12 h in the dark. To remove remnant mycelia, the solution was then filtered with four pieces of lens wiping paper, and protoplasts were obtained. The transformation of B. trispora protoplasts was performed according to the previous protocol (33, 34) with some modifications. The protoplasts were pelleted by centrifuging at 3,000 rpm for 10 min and washed twice with ST solution (188.2 g/liter sucrose, 1.2 g/liter Tris, pH 7.5). The solution was centrifuged at 3,000 rpm for 10 min, and then the protoplasts were resuspended with STC solution (188.2 g/liter sucrose, 1.2 g/liter Tris, 2.78 g/liter CaCl2, pH 7.5) at a final density of 1 × 106 protoplasts/ml. Approximately 5 μg deletion cassette was mixed with 200 μl of protoplasts and incubated in ice water for 20 min. Then PTC solution (600 g/liter polyethylene glycol [PEG] 6000, 1.2 g/liter Tris, 2.78 g/liter CaCl2, pH 7.5) was added gently and incubated at 25°C for 20 min. The transformed protoplasts were washed with osmotic stabilizer and cultivated initially on wort agar medium for 12 h, and then a layer of wort agar medium containing 200 μg/ml hygromycin B and 0.1% Triton X-100 was poured onto the plates to select the resistant mutants. PCR analyses of transformants were performed with specific primers after subculturing several times (35), and the crgA null transformants were named ΔcrgA strains. ΔcrgA strains were complemented with the wild-type copy of crgA, transformed in the same way described above, and crgA-complemented transformants were named C-ΔcrgA strains.
Transcription analyses of carotenoid structural genes.
Total RNA was isolated by TRIzol (Invitrogen, USA) from the mycelia cultured at 25°C at 180 rpm for a certain time in fermentation medium (50 g/liter corn starch, 25 g/liter soybean meal, 1.5 g/liter KH2PO4, 0.5 g/liter MgSO4·7H2O, pH 6.5). After that, reverse transcription was performed immediately using PrimeScript RT master mix (TaKaRa, Japan) with the primers for carotenoid structural genes (hmgR, carRA, and carB) listed in Table 1. Sequentially, fluorogenic quantitative PCR was performed with SYBR Premix Ex-TaqII (TaKaRa, Japan) to analyze the transcript levels of the three genes. tef1 was used as the reference gene, because its transcript level remained relatively steady in different environments (14); the primers used to amplify tef1 are listed in Table 1. The amplification program was as follows: 95°C for 5 min followed by 40 cycles at 95°C for 5 s and 60°C for 20 s. The method to present relative gene expression is the comparative threshold cycle (CT) method, in which the expression level of tef1 is used as the reference. Since accurate quantification requires primer sets that facilitate maximum amplification efficiency, primers are best evaluated using SYBR green fluorescence and melting curve analysis. The primer sets referred to in the previous report generate no primer dimers and result in near 100% amplification efficiency for each gene (14).
Analyses of carotenoids.
Strains were cultivated in 25 ml fermentation medium at 25°C and 180 rpm for a certain time to obtain mycelia. Mycelia were washed and centrifuged at 12,000 × g for 5 min, dried in vacuum oven at 40°C for 24 h, and triturated with a mortar and pestle in liquid nitrogen. Carotenoids were extracted by petroleum ether until the mycelia were colorless, and then the extracts supplemented with 5% butylated hydroxytoluene were concentrated by rotary evaporator. Sediments were resuspended with acetonitrile and analyzed with a high-performance liquid chromatograph (HPLC) equipped with an Agilent TC C18 column at 28°C. Acetonitrile (80%) and methanol (20%) were used as mobile phase with a flow rate of 1.0 ml/min. Lycopene and β-carotene were detected at 502 nm and 450 nm, respectively.
Preparation of metabolome samples.
One hundred microliters spore suspension with the density of 1.0 × 106 spores per ml was inoculated into 50 ml SUP medium (36) (50 g/liter glucose, 5 g/liter yeast extract, 4 g/liter KH2PO4, 0.9 g/liter K2HPO4, 1 g/liter NH4Cl, 0.25 g/liter MgSO4·7H2O, pH 6.5) and cultivated at 25°C at 180 rpm for various times (48 h, 60 h, 72 h, and 84 h). The fermentation broth was harvested, and mycelia were obtained after centrifugation at 12,000 × g at 4°C for 5 min. Cell metabolism was quenched with liquid nitrogen, and then the mycelia were ground to powder with mortar and pestle (25). The powder was transferred to a tube, and metabolites were extracted with 0.75 ml prechilled (−20°C) methanol. The supernatant was collected after extracts were centrifugated at 12,000 × g at 4°C for 10 min. The pellet was resuspended with prechilled methanol, and metabolites were extracted once again as described above. Supernatants were mixed as samples for derivatization. Mixtures of 0.5 ml supernatant and 10 μl internal standards (ribitol in water, 1 mg/ml) were dried in a Termovap sample concentrator. Samples were derivatized in two steps. First, the solutions of methoxylamine hydrochloride in pyridine (175 μl, 20 mg/ml) were added to the samples and incubated at 30°C for 2 h under agitation. Then, 175 μl solutions of N-methyl-N-(trimethylsilyl) trifluoroacetamide (MSTFA) and trimethychlorosilane (99:1 [vol/vol]) were added to the samples and incubated at 30°C for 1 h.
GC-MS determination and data analyses.
After derivatization, metabolites of the samples were detected on a gas chromatograph-mass spectrometer (TSQ8000; Thermo Scientific, USA) equipped with DB-5 capillary, and the detection was performed according to the protocol described in reference 37, with slight modifications. One microliter of sample was injected by a Thermo Scientific autoinjector at 300°C with a split ratio of 1:1. Helium was used as carrier gas, and the flow rate was 1 ml/min. GC oven temperature was raised to 80°C and maintained for 1 min and then raised to 160°C at a constant speed in 10 min, raised to 300°C at a constant speed in 30 min, and maintained for 5 min at 300°C. Electron impact ionization was used at 40 to 800 m/z. Metabolites in samples were identified by searching the NIST mass spectral library and reference compounds, and the content was calculated according to the peak area of every metabolite and internal standard. Principal-component analysis was applied to the data with SIMCA (version 13.0), and cluster analysis was performed with R software.
Statistical analyses.
All experiments were performed in triplicates, and values are expressed as the means with standard deviations from three independent measurements. The significance of differences between groups was assessed by a one-way analysis of variance. A P value of <0.05 indicates the presence of a statistically significant difference, and a P value of <0.01 was considered highly significant.
Data availability.
The nucleotide sequence for crgA from B. trispora is available from GenBank (accession number MH345788.1).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants 21606105 and 21878123), the Open Funding Project of the State Key Laboratory of Bioreactor Engineering (grant number 2018OPEN17), the Program of the Key Laboratory of Industrial Biotechnology, Ministry of Education, China (grant number KLIB-KF201604), the Project Funded by China Postdoctoral Science Foundation (grant number 2018M630525), the Fundamental Research Funds for the Central Universities (grant number JUSRP51504), and the 111 Project (grant number 111-2-06).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Srivastava S, Srivastava AK. 2013. Lycopene; chemistry, biosynthesis, metabolism and degradation under various abiotic parameters. J Food Sci Technol 52:41–53. doi: 10.1007/s13197-012-0918-2. [DOI] [Google Scholar]
- 2.Wang Q, Feng LR, Luo W, Li HG, Wang L, Zhou Y, Yu XB. 2014. Mutation breeding of lycopene-producing strain Blakeslea trispora by a novel atmospheric and room temperature plasma (ARTP). Appl Biochem Biotechnol 174:452–460. doi: 10.1007/s12010-014-0998-8. [DOI] [PubMed] [Google Scholar]
- 3.Papaioannou EH, Liakopoulou-Kyriakides M. 2010. Substrate contribution on carotenoids production in Blakeslea trispora cultivations. Food Bioprod Process 88:305–311. doi: 10.1016/j.fbp.2009.03.001. [DOI] [Google Scholar]
- 4.Pegklidou K, Mantzouridou F, Tsimidou MZ. 2008. Lycopene production using Blakeslea trispora in the presence of 2-methyl imidazole: yield, selectivity, and safety aspects. J Agr Food Chem 56:4482–4490. doi: 10.1021/jf800272k. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, Feng LR, Luo W, Li HG, Zhou Y, Yu XB. 2015. Effect of inoculation process on lycopene production by Blakeslea trispora in a stirred-tank reactor. Appl Biochem Biotechnol 175:770–779. doi: 10.1007/s12010-014-1327-y. [DOI] [PubMed] [Google Scholar]
- 6.Yan GL, Wen KR, Duan CQ. 2012. Enhancement of beta-carotene production by over-expression of HMG-CoA reductase coupled with addition of ergosterol biosynthesis inhibitors in recombinant Saccharomyces cerevisiae. Curr Microbiol 64:159–163. doi: 10.1007/s00284-011-0044-9. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Saiz M, Paz B, De La Fuente JL, Lopez-Nieto MJ, Cabri W, Barredo JL. 2004. Blakeslea trispora genes for carotene biosynthesis. Appl Environ Microbiol 70:5589–5594. doi: 10.1128/AEM.70.9.5589-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro E, Ruiz Pérez VL, Torres-Martínez S. 2000. Overexpression of the crgA gene abolishes light requirement for carotenoid biosynthesis in Mucor circinelloides. Eur J Biochem 267:800–807. doi: 10.1046/j.1432-1327.2000.01058.x. [DOI] [PubMed] [Google Scholar]
- 9.Navarro E, Lorca-Pascual J, Quiles-Rosillo M, Nicolás F, Garre V, Torres-Martínez S, Ruiz-Vázquez R. 2001. A negative regulator of light-inducible carotenogenesis in Mucor circinelloides. Mol Genet Genomics 266:463–470. doi: 10.1007/s004380100558. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Ortiz R, Limón MC, Avalos J. 2013. Functional analysis of the carS gene of Fusarium fujikuroi. Mol Genet Genomics 288:157–173. doi: 10.1007/s00438-013-0739-7. [DOI] [PubMed] [Google Scholar]
- 11.Rodríguez-Ortiz R, Michielse C, Rep M, Limón MC, Avalos J. 2012. Genetic basis of carotenoid overproduction in Fusarium oxysporum. Fungal Genet Biol 49:684–696. doi: 10.1016/j.fgb.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Navarro E, Peñaranda A, Hansberg W, Torres-Martínez S, Garre V. 2013. A white collar 1-like protein mediates opposite regulatory functions in Mucor circinelloides. Fungal Genet Biol 52:42–52. doi: 10.1016/j.fgb.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Quiles-Rosillo MD, Ruiz-Vazquez RM, Torres-Martinez S, Garre V. 2005. Light induction of the carotenoid biosynthesis pathway in Blakeslea trispora. Fungal Genet Biol 42:141–153. doi: 10.1016/j.fgb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt AD, Heinekamp T, Matuschek M, Liebmann B, Bollschweiler C, Brakhage AA. 2005. Analysis of mating-dependent transcription of Blakeslea trispora carotenoid biosynthesis genes carB and carRA by quantitative real-time PCR. Appl Microbiol Biotechnol 67:549–555. doi: 10.1007/s00253-005-1941-2. [DOI] [PubMed] [Google Scholar]
- 15.Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 16.Conlon H, Zadra I, Haas H, Arst HN Jr, Jones MG, Caddick MX. 2001. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol Microbiol 40:361–375. doi: 10.1046/j.1365-2958.2001.02399.x. [DOI] [PubMed] [Google Scholar]
- 17.Punt PJ, Dingemanse MA, Kuyvenhoven A, Soede RD, Pouwels PH, van den Hondel CA. 1990. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene 93:101–109. doi: 10.1016/0378-1119(90)90142-E. [DOI] [PubMed] [Google Scholar]
- 18.Murcia-Flores L, Lorca-Pascual JM, Garre V, Torres-Martínez S, Ruiz-Vázquez RM. 2007. Non-AUG translation initiation of a fungal RING finger repressor involved in photocarotenogenesis. J Biol Chem 282:15394–15403. doi: 10.1074/jbc.M610366200. [DOI] [PubMed] [Google Scholar]
- 19.Catlett NL, Lee BN, Yoder OC, Turgeon BG. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Rep 50:9–11. doi: 10.4148/1941-4765.1150. [DOI] [Google Scholar]
- 20.Krappmann S, Sasse C, Braus GH. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 5:212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas FE, Calo S, Murcia-Flores L, Garre V, Ruiz-Vazquez RM, Torres-Martinez S. 2008. A RING-finger photocarotenogenic repressor involved in asexual sporulation in Mucor circinelloides. FEMS Microbiol Lett 280:81–88. doi: 10.1111/j.1574-6968.2007.01044.x. [DOI] [PubMed] [Google Scholar]
- 22.Alcalde E, Fraser PD. 2016. Metabolite profiling of Phycomyces blakesleeanus carotene mutants reveals global changes across intermediary metabolism. Microbiology 162:1963–1971. doi: 10.1099/mic.0.000376. [DOI] [PubMed] [Google Scholar]
- 23.Leslie SB, Israeli E, Lighthart B, Crowe JH, Crowe LM. 1995. Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl Environ Microbiol 61:3592–3597. doi: 10.1128/AEM.61.10.3592-3597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young LY, Hull CM, Heitman J. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob Agents Chemother 47:2717–2724. doi: 10.1128/aac.47.9.2717-2724.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jie S, Hao L, Yuan Q. 2012. Metabolic regulation of trisporic acid on Blakeslea trispora revealed by a GC-MS-based metabolomic approach. PLoS One 7:e46110. doi: 10.1371/journal.pone.0046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantzouridou F, Tsimidou MZ, Roukas T. 2006. Performance of crude olive pomace oil and soybean oil during carotenoid production by Blakeslea trispora in submerged fermentation. J Agric Food Chem 54:2575–2581. doi: 10.1021/jf0526339. [DOI] [PubMed] [Google Scholar]
- 27.Calder PC, Yaqoob P, Harvey DJ, Watts A, Newsholme EA. 1994. Incorporation of fatty acids by concanavalin A-stimulated lymphocytes and the effect on fatty acid composition and membrane fluidity. Biochem J 300:509–518. doi: 10.1042/bj3000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murín R, Mohammadi G, Leibfritz D, Hamprecht B. 2009. Glial metabolism of valine. Neurochem Res 34:1195–1203. doi: 10.1007/s11064-008-9895-2. [DOI] [PubMed] [Google Scholar]
- 29.Boyle SM, Markham GD, Hafner EW, Wright JM, Tabor H, Tabor CW. 1984. Expression of the cloned genes encoding the putrescine biosynthetic enzymes and methionine adenosyltransferase of Escherichia coli (speA, speB, speC and metK). Gene 30:129–136. doi: 10.1016/0378-1119(84)90113-6. [DOI] [PubMed] [Google Scholar]
- 30.Shaibe E, Metzer E, Halpern YS. 1985. Control of utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol 163:938–942. doi: 10.1128/JB.163.3.938-942.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Luo W, Gu QY, Feng LR, Li HG, Yu XB. 2013. Enhanced lycopene content in Blakeslea trispora by effective mutation-screening method. Appl Biochem Biotechnol 171:1692–1700. doi: 10.1007/s12010-013-0468-8. [DOI] [PubMed] [Google Scholar]
- 32.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. 1996. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast 12:1439–1457. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Yuan Q, Du X. 2008. Protoplast from β-carotene-producing fungus Blakeslea trispora: preparation, regeneration and validation. Korean J Chem Eng 25:1416–1421. doi: 10.1007/s11814-008-0232-x. [DOI] [Google Scholar]
- 34.Turgeon BG, Garber RC, Yoder OC. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol Cell Biol 7:3297–3305. doi: 10.1128/mcb.7.9.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, DiGuistini S, Wang TC, Bohlmann J, Breuil C. 2010. Agrobacterium-meditated gene disruption using split-marker in Grosmannia clavigera, a mountain pine beetle associated pathogen. Curr Genet 56:297–307. doi: 10.1007/s00294-010-0294-2. [DOI] [PubMed] [Google Scholar]
- 36.Wöstemeyer J. 1985. Strain-dependent variation in ribosomal DNA arrangement in Absidia glauca. Eur J Biochem 146:443–448. doi: 10.1111/j.1432-1033.1985.tb08671.x. [DOI] [PubMed] [Google Scholar]
- 37.Hu X, Li H, Tang P, Sun J, Yuan Q, Li C. 2013. GC-MS-based metabolomics study of the responses to arachidonic acid in Blakeslea trispora. Fungal Genet Biol 57:33–41. doi: 10.1016/j.fgb.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Joazeiro CA, Weissman AM. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552. doi: 10.1016/S0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- 39.Roudiak SG, Shrader TE. 1998. Functional role of the N-terminal region of the Lon protease from Mycobacterium smegmatis. Biochemistry 37:11255–11263. doi: 10.1021/bi980945h. [DOI] [PubMed] [Google Scholar]
- 40.Zhang FL, Casey PJ. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem 65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence for crgA from B. trispora is available from GenBank (accession number MH345788.1).