Abstract
A constant improvement in understanding of mitochondrial biology has provided new insights into mitochondrial dysfunction in human disease pathogenesis. Impaired mitochondrial dynamics caused by various stressors are characterized by structural abnormalities and leakage, compromised turnover, reactive oxygen species overproduction in mitochondria as well as increased mitochondrial DNA mutation frequency, which leads to impaired energy production and modified mitochondria-derived cell signaling. The mitochondrial dysfunction in airway epithelial, smooth muscle, and endothelial cells has been implicated in diseases including chronic obstructive lung diseases and acute lung injury. Increasing evidence indicates that the NRF2-antioxidant response element (ARE) pathway not only enhances redox defense but also facilitates mitochondrial homeostasis and bioenergetics. Identification of functional or potential AREs further supports the role for Nrf2 in mitochondrial dysfunction-associated airway disorders. While clinical reports indicate mixed efficacy, NRF2 agonists acting on respiratory mitochondrial dynamics are potentially beneficial. In lung cancer, growth advantage provided by sustained NRF2 activation is suggested to be through increased cellular antioxidant defense as well as mitochondria reinforcement and metabolic reprogramming to the preferred pathways to meet the increased energy demands of uncontrolled cell proliferation. Further studies are warranted to better understand NRF2 regulation of mitochondrial functions as therapeutic targets in airway disorders.
Keywords: lung, antioxidant response element, metabolism, sulforaphane
Introduction
Nuclear factor, erythroid 2 like 2 (NFE2L2) or NF-E2-related factor 2 (NRF2) is a transcriptional activator of antioxidant response element (ARE)-bearing genes encoding antioxidant, drug-metabolizing, glutathione-homeostasis enzymes as well as many other host defense proteins. For this, NRF2 dimerizes with other transcriptional activators including small Maf proteins (MafF, MafG, MafK). NRF2 homeostasis is regulated by a cytoplasmic inhibitor Kelch-like ECH-associated protein 1 (KEAP1) under housekeeping proteolytic demands or by exogenous stimuli such as oxidants, xenobiotics, carcinogens as well as antioxidants and chemopreventive agents (Taguchi et al. 2011). During the last two decades, in vitro and transgenic murine model (Cho 2013) studies revealed that NRF2 contributes to a broad spectrum of cellular functions including redox balance, cell cycle and death, immunity, metabolism, selective protein degradation, development, aging, and carcinogenesis.
Mitochondria are dynamic and multifunctional organelles that produce ATP and many biosynthetic intermediates by means of oxidative phosphorylation (OXPHOS) in response to cellular bioenergetic and biosynthetic demands. In this process, electrons liberated from reducing substrates are delivered to oxygen via respiratory proton (H+) pumps in complexes I-IV to establish a H+ gradient across the inner mitochondrial membrane, and electrochemical energy of this gradient is used to complete ATP synthesis by complex V (ATP synthases). Importantly, the mitochondrion is the only non-nuclear organelle bearing its own genome which encodes 13 polypeptides of the OXPHOS subunits and respiratory chain as well as two ribosomal RNAs and 22 transfer RNAs necessary for translation of polypeptides inside mitochondria in humans and mice. In addition, mitochondria are the major source of endogenous reactive oxygen species (ROS) production. Mitochondria also provide temporal storage of calcium ion (Ca2+) and are essential in cellular calcium homeostasis. Other important metabolic reactions that occur in mitochondria include steroid hormone and porphyrin synthesis, urea cycle, lipid metabolism, and interconversion of amino acids. Recent research has provided enormous insights into the molecular mechanisms underlying mitochondrial bioenergetics (ATP production) as well as biogenesis (increase in mitochondrial mass by de novo generation), fusion (mixing contents within a mitochondria population) and fission (binary division of mitochondria) in relation to mitophagy (lysosome-dependent selective degradation of defective mitochondria) as quality control programs. Maintenance of mitochondrial homeostasis and functions are critical components of mitochondrial health and interruption of these features are key determinants in aging and in the pathogenesis of neurodegenerative disorders, cardiac ischemia, type 2 diabetes, glaucoma, cancer, and other diseases (Ashar et al. 2017; Yue et al. 2018).
Mitochondria are constantly recycled as damaged or aging mitochondria undergo mitophagy for which PTEN-induced kinase 1 (PINK1), a mitochondrial serine/threonine-protein kinase, renders parkin (PARK2, E3 ubiquitin ligase) to bind to depolarized mitochondria to form autophagosome to be self-digested and replaced (Pickles et al. 2018). Mitophagy impacts various cellular process and diseases and may be either protective or deleterious. Biochemical changes in dysfunctional mitochondria include impaired OXPHOS subunits and energy production, diminished membrane potential, altered metabolic pathways such as glycolysis and fatty acid β-oxidation (FAO), disturbed calcium homeostasis, and increased ROS production. Dysfunctional mitochondria also change their physical morphology or lose their membrane integrity and may allow leakage of mitochondrial components including damage-associated molecular pattern (DAMP) molecules. DAMPs from mitochondria are mitochondrial DNA (mtDNA), ATP, mitochondrial transcription factor A (TFAM or mtTFA), succinate, cardiolipin, and N-formyl peptides [See review (Nakahira et al. 2015)]. Mitochondrial dysfunction can lead to necrotic and/or apoptotic cell death and interrupt cell proliferation (Ni et al. 2015). Dying cells (mostly necrotic) release DAMPs which alarm and activate the innate immune system (Venereau et al. 2015).
Mitochondrial numbers and morphology vary according to cell type and demand, whereby the balance between mitochondrial fusion/fission regulates mitochondrial distribution, morphology, and function. Depending on the energy substrates available, energy demands, or the redox state, the cell may increase or decrease the number and size of mitochondria, and altered copy number by damage can result in mitochondrial dysfunction (Clay Montier et al. 2009; Mishra and Chan 2016). Compared to genomic DNA, mtDNA is more susceptible to mutation and ROS damage possibly in part due to the lack of histones and less efficient DNA repair mechanisms (Alexeyev et al. 2013). mtDNA point mutations [including small insertion/deletion (indel) mutations] and mutations of nuclear genes encoding respiratory chain subunit proteins constitute a significant cause of human diseases such as mitochondrial encephalopathy (Schapira 2012). Currently, more than 250 pathogenic mtDNA mutations have been identified in various diseases (Alston et al. 2017) and a pathogenic mutation (e.g., m.3243A>G, m.8344A>G) is associated with multiple clinical features (phenotype heterogeneity), making disease prognosis extremely difficult to predict (Orsucci et al. 2018). In addition, non-repaired, damaged mtDNA under oxidative environments may be fragmented and released as a DAMP into the cytosol and extracellular space (Venereau et al. 2015) to trigger innate immune responses through toll-like receptor 9, cyclic guanosine monophosphate-adenosine monophosphate (GMP-AMP) synthase, and/or the nod-like receptor family, pyrin domain containing 3 (NLRP3) inflammasome (Banoth and Cassel 2018). As sources of ROS and reservoirs of antioxidant enzymes, mitochondria are critical organelles in ROS-dependent cellular redox pathways, which primarily connect mitochondria with NRF2.
Respiratory airways continuously interface a high oxygen environment for gas exchange and various environmental oxidants. In addition, the airway is a dynamic organ which constantly requires energy for various functions such as bronchial smooth muscle contraction, mucociliary clearance, and surfactant and mucus secretion. Therefore, airway cells place a large demand on mitochondrial bioenergetic functions. Recent studies have characterized the roles of mitochondrial dysfunction in diverse airway disorders. In this review, we profile the updated information on the role of NRF2 pathway in mitochondrial biology and discuss the association of NRF2 in airway pathogenesis.
NRF2 Regulation of Mitochondrial Function
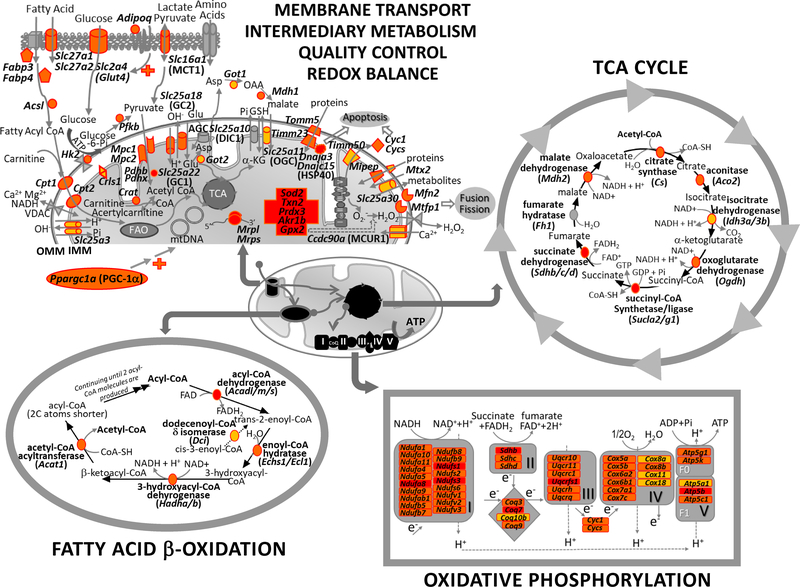
Recent studies indicated that loss or genetic mutation of NRF2 (or Nrf2) leads to oxidative stress-induced mitochondrial dysfunction and metabolic disorder (Mitsuishi et al. 2012; Ludtmann et al. 2014; Kovac et al. 2015). Accumulating evidence has determined that NRF2-mediated nuclear control of mitochondrial function is directly through ARE-mediated transcriptional activation of mitochondrial antioxidant defense, bioenergetic process, mitophagy, and biogenesis as well as mitochondria-associated intermediary metabolism (Piantadosi et al. 2008; Abdullah et al. 2012; Mitsuishi et al. 2012; Hayes and Dinkova-Kostova 2014; Cho et al. 2019) as summarized in Table 1. In addition, the NRF2-KEAP1 axis is known to interact with various proteins for direct and indirect influence of NRF2 on mitochondrial function (Fig. 1). An oncogenic role for overexpressed NRF2 is thought to be in part through facilitation of mitochondrial function and metabolic reprogramming to meet the increased energy demand of rapidly growing cancer cells.
Table 1.
NRF2 target proteins involved in mitochondrial biology and function
| Categories | Functions | Involved gene products |
|---|---|---|
| Nutrient Uptake/Transport | ↑ Glucose uptake | GLUT, ADIPO, ADIPOR |
| ↑ Fatty acid uptake | CD36, SLC27AI/2, ACSL, CPTI/2, FABP 3/4 | |
| ↑ Amino acid uptake | GOTI/2. GCI/2 | |
| ↑ Metabolites uptake | PDH. ACSL | |
| Intermediary Metabolism | ↑ Glycolysis | HK, PFK, PK, ME1, GSR |
| ↑ Amino acid degradation/Ketolysis | HIBCH. BCKDHA/B, BDHI, OAT | |
| ↓ Gluconeogenesis | G6PC,PEPC | |
| ↓ Lipogenesis | ACAC, ACLY, SCDI, FABP5, ELOVL | |
| Oxidative Phosphorylation | ↑ Complex I/II/III/IV/V subunits | |
| ↑ Electron transfer proteins/donors | CYCS, CYCI, CoQ, NADH, FADH2 | |
| TCA Cycle | ↑ Entry of metabolic substrates | Acetyl CoA, Malate, α-KG |
| ↑ NADH/FADH2 generation | MEI, G6PD, IDHI, ACAD | |
| ↑ Enzyme transcription | CS, ACO2, IDH3A/B, OGDH, SUCL, SDHA/B/C, MDH1/2. | |
| Lipid Metabolism | ↑ FAO substrates | FAD+/NAD+, FABP 3/4 |
| ↑ FAO enzyme transcription | ACSL, ACADL/M/S, ECHSI/ECHI, HADHA/B, ACATI | |
| ↑ Steroid hormone metabolism | AKRIBI5 | |
| Redox Homeostasis | ↓ ROS | SOD2, TRX2, TXNRD2, PRDXI/5, ALDH2/6/9/IBI, UCP3 |
| ↑ NADP/NAIY synthesis | MEI, G6PD, IDHI, PGD | |
| ↑ GSH synthesis | xCT, GG T, GST-AI/TI/PI, GS, GSR, GPXI/2/4, GCLC, GCLM | |
| ↑ GSH uptake | OGC, DIC, TTC | |
| ↑ Protein S-glutathionylation | GRX2, GST-P | |
| Quality Control | ↑ Biogenesis | PGC-Iα, NRF-I, NRF-2, T FAM, HO-I |
| ↑ Mitophagy | PINKI, PARK2, p62/SQSTRMI, LC3 | |
| ↑ Fusion/Fission | MFNI/2, DRPI | |
| ↑ Apoptosis | CYC, HSP40 | |
| ↑ Mobility | MIRO2 | |
| ↑ mtDNA repair | OGGI | |
| ↑ Calcium homeostasis | MCUI | |
| ↑ Membrane integrity | CRDI | |
| Cancer & Rapid Proliferating Cells | ↑ Pentose phosphate pathway | G6PD, PGD, MEI, TKT, TALDOI, PPAT, MTHFD2 |
| ↑ Lipid biosynthesis | ACACA, FASN, SCDI, ACLY | |
| ↑ Glutamate pathway | GLSI, GLUD |
↑, increase in function. ↓, decrease in function. ACAC, acetyl-CoA carboxylase; ACAD, acyl-CoA dehydrogenase; ACAT1, acetyl-CoA acetyltransferase, mitochondrial; ACLY, ATP-citrate lyase; ACSL, acyl-CoA synthetase long-chain family member; ACO2, aconitase 2; ADIPO, adiponectin, C1Q and collagen collagen domain containing; ADIPOR, adiponectin receptor; ALDH, aldehyde dehydrogenase; BCKDH, branched chain ketoacid dehydrogenase E1; BDH1, 3-hydroxybutyrate dehydrogenase, type 1 CoQ, coenzyme Q; CPT, carnitine O-palmitoyltransferase; CRD1, cardiolipin synthase; CS, citrate synthase; CYC1, cytochrome c1; CYCS, cytochrome c, somatic; DIC1, mitochondrial dicarboxylate transporter; DRP1, dystrophin related protein 1; ECH, enoyl Coenzyme A hydratase; ELOVL, ELOVL fatty acid elongase; FABP, fatty acid binding protein; FAO, fatty acid-β-oxidation; FASN, fatty acid synthase; FH, fumarate hydratase; G6PC, glucose-6-phosphate dehydrogenase; GC, mitochondrial glutamate carrier; GCLC, glutamate-cysteine ligase catalytic subunit; GCLM GCL modifier subunit; GGT, γ-glutamyltransferase; GLS, glutaminase; GLUD, glutamate dehydrogenase; GLUT, glucose transporter; GOT, glutamic-oxaloacetic transaminase; GPX, glutathione peroxidase; GRX2, glutaredoxin 2; GS, glutathione synthase; GSH, glutathione; GSR, glutathione reductase; GST, glutathione-S-transferase; HADH, hydroxyacyl-CoA dehydrogenase; HIBCH, 3-hydroxyisobutyryl-CoA hydrolase; HK, hexokinase; HO-1, heme oxygenase 1; HSP40, heat shock protein 40; IDH1, isocitrate dehydrogenase (NADP(+)) 1, cytosolic; KEAP1, Kelch-like ECH-associated protein 1; α-KG, α-ketoglutarate; LC3 (MAP1LC3), microtubule Associated Protein 1 Light Chain 3; MCU1, mitochondrial calcium uniporter; MDH, malate dehydrogenase; ME, malic enzyme; MFN, mitofusin; MIRO2, mitochondrial rho GTPase 2; MTHFD2, methylenetetrahydrofolate dehydrogenase (NADP+ Dependent) 2; NRF-1, nuclear respiratory factor-1; OAT, ornithine aminotransferase; OGC, oxoglutarate carrier; OGDH, oxoglutarate dehydrogenase; OGG1, 8-oxoguanine DNA glycosylase; PARK2, parkin; PDH, pyruvate dehydrogenase; PEPC, peptidase C; PFK, Phosphofructokinase; PGC-1α, peroxisome proliferative activated receptor, gamma, coactivator 1 α; PGD, phosphogluconate dehydrogenase; PINK1, PTEN-induced kinase 1; PK, pyruvate kinase; PPAT, phosphoribosyl pyrophosphate amidotransferase; PRDX, peroxiredoxin; SCD1, stearoyl-CoA desaturase; SDH, succinate dehydrogenase complex; SLC, solute carrier family; SOD2, superoxide dismutase 2; SQSTM1, sequestosome 1; SUCL, succinate-CoA ligase; TALDO1, transladolase 1; TCA, tricarboxylic acid; TFAM, transcription factor A, mitochondrial; TKT, transketolase; TRX2, thioredoxin 2; TTC, tricarboxylate carrier; TXNRD2, thioredoxin reductase 2; UCP3, uncoupling protein 3; xCT, cystine/glutamate transporter.
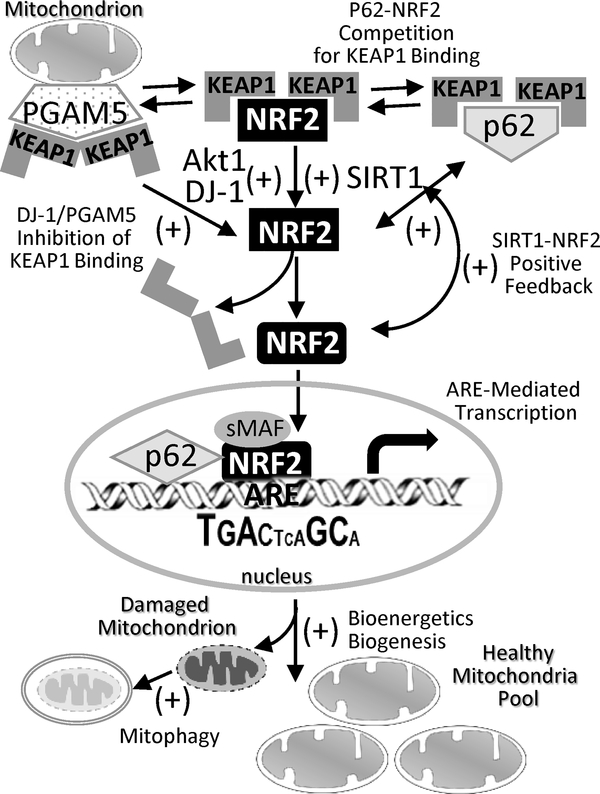
Fig 1. Interactions of NRF2-KEAP1 axis with other proteins in regulation of mitochondrial biology and functions.
Studies with transgenic animal and cell models or human samples demonstrated various proteins that may interact with NRF2-KEAP1 axis under normal physiologic and pathologic conditions to promote NRF2-mediated mitochondrial bioenergetics. NRF2-KEAP1 axis may communication or compete with SIRT1, PI3K/Akt, DJ-1, and/or p62 to liberate NRF2 from KEAP1 for antioxidant response element (ARE)-mediated transcriptional activation of genes involved in mitochondrial energy metabolism, biogenesis, and quality control as described in Table 1. Aktl, serine/threonine-specific protein kinase; DJ-1, protein deglycase (or Parkinson disease protein 7, PARK7); KEAP1, Kelch-like ECH-associated protein 1; p62, ubiquitin-binding protein (or sequestosome 1, SQSTM1); PGAM5, PGAM family member 5, mitochondrial serine/threonine protein phosphatase; PI3K, phosphoinositide 3-kinase; SIRT1, sirtuin 1; sMaf, small musculoaponeurotic fibrosarcoma (MafF, MafG, MafK).
Redox homeostasis
During consumption of oxygen for cellular ATP production, mitochondrial electron transport chain of the OXPHOS complex transfers single electrons to oxygen to form ROS, mainly superoxide hydrogen peroxide (H2O2) by complexes I and III. Mitochondria contain their own antioxidant defense system to detoxify and minimize ROS. This mitochondrial redox buffering capacity is controlled precisely to avoid mitochondrial dysfunction and cell death. However, mitochondrial ROS are increased under various pathophysiological conditions caused by hypoxia, ischemia/reperfusion injury, chemical stress, drug treatment, genetic defects, or metabolic fluctuations. NRF2 directly regulates mitochondrial ROS homeostasis through ARE-mediated induction of antioxidant enzymes localized in mitochondria. These enzymes include superoxide dismutase (SOD2 or MnSOD) neutralizing superoxide, glutathione (GSH) and multiple thiol-homeostasis enzymes such as glutathione reductase (GSR), glutathione peroxidases (e.g., GPX1, GPX4), glutathione-S-transferases (e.g., GST-A1, GST-T1, GST-P1), glutaredoxin 2 (GRX2, also called thioltransferase 2), thioredoxin 2 (TRX2), thioredoxin reductase 2 (TXNRD2), peroxiredoxins (e.g., PRDX3, PRDX5) (Ribas et al. 2014; Ryoo and Kwak 2018). PGAM family member 5, mitochondrial serine/threonine protein phosphatase (PGAM5) in the mitochondrial outer membrane is known to form a ternary complex with KEAP1 and NRF2, and thus an intact PGAM5-KEAP1-NRF2 complex constrains KEAP1 and allows NRF2 to sense and respond to mitochondrial ROS (Lo and Hannink 2008).
NRF2 transcriptionally induces all the enzymes involved in the synthesis of cellular reducing agents, nicotinamide adenine dinucleotide phosphate (NADPH) and GSH, in both normal and oxidant-stimulated tissues including lung (Cho 2005). GSH is transported to mitochondrial matrix by carriers, 2-oxoglutarate carrier (OGC or SLC25A11), dicarboxylate carrier (DIC or SLC25A10), and tricarboxylate carrier (TTC or SLC25A1) located in the mitochondrial inner membrane. Maintaining high mitochondrial GSH concentration (5–10 mM) is critical to prevent or repair oxidative damage generated during normal aerobic metabolism (Mailloux and Willmore 2014). In addition to ROS detoxification, recent studies indicated that mitochondrial GSH undergoes reversable S-glutathionylation by GST-P and GRX2 to prevent oxidation of mitochondrial proteins (Young et al. 2019). Protein S-glutathionylation is also known to be beneficial in mitochondrial metabolism processes, ATP production, ROS release, solute importation, permeability transition, protein uptake, and fission/fusion (Mailloux and Willmore 2014). OXPHOS complex I subunits (NADH:ubiquinone oxidoreductases or type I NADH dehydrogenases, e.g., MT-ND3, NDUFV1, NDUFS1) and uncoupling protein 3 as well as many other OXPHOS proteins are suggested to be targets of GRX2 for glutathionylation (Mailloux and Willmore 2014). Lack of murine Grx2 diminished ATP production and increased H2O2 and superoxide release from liver and cardiac mitochondria, and complex I and III proteins as well as mitochondrial redox sensor and tricarboxylic acid (TCA) cycle proteins including pyruvate dehydrogenase (PDH) and 2-oxoglutarate dehydrogenase complex (OGDC) are suggested to be S-glutathionylation targets in mouse mitochondria (Mailloux et al. 2014; O’Brien et al. 2017; Chalker et al. 2018). Overall, these studies support the importance of NRF2 in glutathione supply and usage for mitochondrial redox balance.
Bioenergetic process
Metabolic end-products of carbohydrates, fats, and proteins end up in mitochondrial FAO and TCA cycle and generate NADH (reduced form of NAD) and flavin adenine dinucleotide (FADH2, the reduced form FAD) for electron transport chains of OXPHOS complexes to produce ATP. Functional roles for NRF2 in mitochondrial metabolism and bioenergetics have been found from murine studies of high-fat diet-induced obesity and methionine-choline-deficient diet-induced fatty liver disease. In these studies, NRF2 negatively regulated gluconeogenic (e.g., phosphoenolpyruvate carboxylase, glucose-6-phosphatase) and lipogenic (e.g., acetyl-coA carboxylase 1) genes to protect against the metabolic disorders (Chartoumpekis et al. 2011; Kay et al. 2011; Uruno et al. 2013). The underlying molecular mechanisms are unclear. Furthermore, murine Nrf2 deficiency enhanced outer membrane permeability, reduced mitochondrial membrane potential, and suppressed oxygen consumption rate, and FAO in various tissues, cells, and isolated mitochondria (Ludtmann et al. 2014; Strom et al. 2016). Inhibition of OXPHOS complex II augmented neuron and astrocyte toxicity in Nrf2-deficient (Nrf2−/−) cells more than in wild-type cells, and Nrf2 overexpression reversed the inhibition effects in the cells (Calkins et al. 2004). Consistent with these findings, enhanced mitochondrial functions and upregulated FAO genes under caloric restriction were found in Keap1-knockdown mice (Kulkarni et al. 2013). Altered NAD and FAD homeostasis coupled with reduced ATP production were found in neurons from Nrf2−/− mice (Holmstrom et al. 2013).
As Nrf2 deficiency did not alter OXPHOS enzyme activities in this study, Holmstrom et al. (2013) proposed that Nrf2 deficiency mainly limited respiratory chain substrates availability. Supporting this hypothesis, limited substrates and impaired activity of complex I coincided with enhanced mitochondrial and total ROS production were found in cortical slice and glio-neuronal co-culture from Nrf2−/− mice (Kovac et al. 2015). In addition, functional AREs were determined in many genes generating the OXPHOS substrates. They include aldo-keto reductase family (e.g., AKR1B15) in the FAO process and pentose phosphate pathway (PPP) genes including glucose-6-phosphate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), malic enzyme 1 (ME1), in oxidative phase (generating a cellular reducing agent NADP), and transketolase (TKT) and transaldolase 1 (TALDO1) in non-oxidative phase (generating ribose 5-phosphate for nucleotide synthesis) (Nishinaka and Yabe-Nishimura 2005; Mitsuishi et al. 2012). However, potential AREs were identified in mitochondrial bioenergetic genes that were Nrf2-dependently activated by NRF2 agonists in mouse lung and liver (Abdullah et al. 2012; Cho et al. 2019). They included complex I-NADH dehydrogenases (e.g., Ndufs1), complex II-succinate dehydrogenases (e.g., Sdhb), complex III-ubiquinol-cytochrome c reductases (e.g., Uqcr11) and coenzyme Q9 (Coq9), complex IV-cytochrome c oxidases (e.g., Cox7a1), complex V-ATP synthases (e.g., Atp5a1, Atp5g1), acyl-Coenzyme A dehydrogenase, long-chain (Acadl), isocitrate dehydrogenase 3 (NAD+) alpha (Idh3a), solute carrier family 27 (fatty acid transporter), member 2 (Slc27a2), and ornithine aminotransferase (Oat) (Table 2). Some of these potential AREs have been functionally verified by chromatin immunoprecipitation followed by DNA sequencing (ChIP-Seq) analyses (Chorley et al. 2012; Mouse et al. 2012; Yue et al. 2014). Therefore, NRF2 may increase mitochondrial biogenesis by acting directly on OXPHOS enzyme transcription in addition to OXPHOS substrate production through induction of intermediary metabolism enzymes.
Table 2.
Potential antioxidant response elements (AREs) in mouse lung genes involved in mitochondrial function and energy metabolism.
| Gene Symbol | Gene Title | ARESequence/Orientation* | Distance to TSS§ | PWM† | MS‡ | Total AREs¶ | FI∞ |
|---|---|---|---|---|---|---|---|
| Acaa2 | acetyl-Coenzyme A acyltransferase 2 | gggtcTGCtatGTCACctggc/R | −4463 | 10.8 | 0.89 | 6 (5) | 1.48 |
| Acadl1 | acyl-Coenzyme A dehydrogenase, long-chain | gtggcTGCtgtGTCACaagtg/R | −3512 | 11.6 | 0.90 | 15 (7) | 1.75 |
| Acadm | acyl-Coenzyme A dehydrogenase, medium chain | gcaagATGACtctGACaacat/F | −4871 | 8.1 | 0.765 | 9 (7) | 1.66 |
| Acads | acyl-Coenzyme A dehydrogenase, short-chain | ccaggATGACtgaGCAcccgg/F | −4060 | 14.6 | 0.936 | 8 (3) | 1.41 |
| Acadvl | acyl-Coenzyme A dehydrogenase, very long chain | tagtaCTGACtgaGGAgttgg/F | −2518 | 8.8 | 0.796 | 10 (4) | 1.62 |
| Acat1 | acetyl-Coenzyme A acetyltransferase 1 | attttGGCtaaTTCAGctgga/R | −2339 | 8.8 | 0.816 | 6 (2) | 1.35 |
| Aco2 | aconitase 2, mitochondrial | gagggATGAGgaaGCGattac/F | −3839 | 6.9 | 0.784 | 9 (1) | 1.74 |
| Acsf2 | acyl-CoA synthetase family member 2 | tttctCTGACtcaGAAatgga/F | −1557 | 8.7 | 0.815 | 9 (5) | 1.30 |
| Acsl1 | acyl-CoA synthetase long-chain family member 1 | ttcatTGCtgtGTCATaacta/R | −2001 | 14.5 | 0.916 | 12 (8) | 1.66 |
| Adhfe1 | alcohol dehydrogenase, iron containing, 1 | aaataATTACtttGCAttcta/F | −3812 | 8.3 | 0.783 | 4 (3) | 1.34 |
| Adipoq1 | adiponectin, C1Q and collagen domain containing | agagaGTGAT acaGCTttgag/F | −1461 | 8.7 | 0.817 | 10 (6) | 8.73 |
| Adlh1l2 | aldehyde dehydrogenase 1 family, member L2 | acaccATGACcaaGGCagctc/F | −4967 | 8.1 | 0.764 | 12 (2) | 1.66 |
| Alox15 | arachidonate 15-lipoxygenase | tcactCTGACaatGCTcatgt/F | −4763 | 10.7 | 0.838 | 9 (5) | 2.33 |
| Atp2a21 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | ttgcaTTGAAtttGCAaatat/F | −1200 | 6.8 | 0.796 | 11 (1) | 2.07 |
| Atp5c11 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | cagaccTGACgctGCAgttta/F | −2442 | 10.1 | 0.855 | 9 (6) | 1.44 |
| Atp5g11 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 | tatgtTGCtatGTAACcccgg/R | −3067 | 11.4 | 0.807 | 9 (3) | 1.83 |
| Atp5k1 | ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit | aaaacCTGACaaaGCAtacat/F | −2805 | 12.3 | 0.895 | 16 (9) | 1.49 |
| Bckdha | branched chain ketoacid dehydrogenase E1, alpha polypeptide | tagatAGCatgGTCAGggcca/R | −3157 | 7.8 | 0.81 | 6 (3) | 1.26 |
| Bckshb | branched chain ketoacid dehydrogenase E1, beta polypeptide | gacaaATAACtcaGCAgggat/F | −118 | 8.7 | 0.825 | 9 (5) | 1.38 |
| Bdh1 | 3-hydroxybutyrate dehydrogenase, type 1 | tgtgaAGCtgtGTCACcaagg/R | −4175 | 9.6 | 0.877 | 14 (6) | 1.95 |
| Car5b | carbonic anhydrase 5b, mitochondrial | taagaATGAGcctGCAattga/F | −3847 | 10.8 | 0.808 | 6 (3) | 1.46 |
| Casq2 | calsequestrin 2 | aggacTGCttaGTCACacaca/R | −3816 | 13.5 | 0.923 | 8 (5) | 5.03 |
| ChkbCpt1b | carnitine palmitoyltransferase 1b, muscle | caaatCTCtgtGTCAGgagta/R | −2798 | 8.9 | 0.767 | 5 (4) | 3.98 |
| Cidea1 | cell death-inducing DNA fragmentation factor, alpha subunitlike effector A | atgacTTCtgaGTCAAggggc/R | −1960 | 11 | 0.832 | 7 (2) | 6.88 |
| Clybl1 | citrate lyase beta like | cccCTTGATtctGCAatcca/F | −3233 | 9.7 | 0.831 | 8 (5) | 1.76 |
| Coq31 | coenzyme Q3 homolog, metliyltransferase | tgacaGTGAT agaGCAgctgg/F | −1014 | 10.6 | 0.835 | 7 (5) | 1.37 |
| Coq7 | demethyl-Q 7 | caactTGCtgtGTCATccagg/R | −1516 | 16.1 | 0.927 | 17 (9) | 1.32 |
| Coq91 | coenzyme Q9 homolog | ctaccCTGACtcaGCAaaaag/F | −4217 | 17.4 | 0.96 | 10 (8) | 1.62 |
| Cox4i11 | cytochrome c oxidase subunit IV isol’onn 1 | aggccTGCgtaGTCAGcttca/R | −276 | 8.8 | 0.87 | 5 (3) | 1.30 |
| Cox5a1 | cytochrome c oxidase, subunit Va | gagagATGACttaGCTgctct F | −2459 | 9.4 | 0.878 | 11 (4) | 1.49 |
| Cox5b | cytochrome c oxidase, subunit Vb | tgctcCTGACtgaGCTcatga F | −2340 | 9.7 | 0.875 | 11 (7) | 1.52 |
| Cox6b1 | cytochrome c oxidase, subunit VIb polypeptide 1 | aaaccCTGACtaagCTGggtg/F | −4713 | 12.3 | 0.899 | 11 (8) | 1.34 |
| Cox7a1 | cytochrome c oxidase, subunit Vila 1 | agatcCTGACttgGCCctgag/F | −304 | 9.1 | 0.845 | 11 (7) | 4.06 |
| Cox7c | cytochrome c oxidase, subunit VIIc | tcatgGTGACtctgTCCttta/F | −3785 | 10.7 | 0.781 | 5 (4) | 1.30 |
| Cox8b | cytochrome c oxidase, subunit VIIIb | gttagTGCtgtGTTAT cctgc/R | −463 | 13.2 | 0.84 | 5 (3) | 3.70 |
| Cpt21 | carnitine palmitoyl transferase 2 | caaaaGTGACtgaGCCacaaa/F | −2058 | 11.3 | 0.90 | 9 (3) | 1.52 |
| Crat | carnitine acetyltransferase | ccaatTGCagaCTCAGgctga/R | −3648 | 14.6 | 0.86 | 5 (3) | 1.29 |
| Cs1 | citrate synthase | gagagATGACtcaGCAgttaa/F | 2727 | 16.2 | 0.947 | 11(0) | 1.43 |
| Cycl | cytochrome c-1 | actccAGCtgtGTCAAgttga/R | −4786 | 12 | 0.892 | 8 (3) | 1.68 |
| Cycs1 | cytochrome c, somatic | acccaGGCttgGTCAAggctl/R | −421 | 6.7 | 0.84 | 8 (3) | 1.70 |
| Cyp2e1 | cytochrome P450, family 2, subfamily e, polypeptide 1 | taagaTTGACtcaGCCtgagc/F | −2573 | 12.3 | 0.911 | 6 (5) | 3.52 |
| Decrl1 | 2,4-dienoyl CoA reductase 1, mitochondrial | ggagcTGCtgaATCAGgtcct/R | −4367 | 9.5 | 0.854 | 8 (5) | 1.58 |
| Dgat2 | diacylglycerol O-acyltransferase 2 | gcagcATCAGtcaGCAtgtct/F | −1343 | 9.8 | 0.788 | 10 (5) | 2.47 |
| Dhrs7c | dihydrolipoamide S-acetyltransferase | gactcATAACtcaGCAgttcc/F | −2314 | 10.8 | 0.838 | 10 (4) | 3.51 |
| Dlat1 | dihydrolipoamide S-acetyltransferase | ctagtGTCACtgaGCAgcatc/F | −1643 | 7.9 | 0.819 | 7 (1) | 1.49 |
| Dut | deoxyuridine triphosphatase, mitochondrial | aataaTGCcctGTCATgtgta/R | −3266 | 10.8 | 0.85 | 8 (4) | 1.30 |
| Echl | enoyl coenzyme A hydratase 1, peroxisomal | tcttgTGCtgcCTCATggagg/R | −4260 | 11.2 | 0.847 | 8 (6) | 1.56 |
| Echsl | enoyl Coenzyme A hydratase, short chain, 1, mitochondrial | caaccCTGACacaGCAggaca/F | −852 | 15.2 | 0.925 | 9 (7) | 1.43 |
| Eci1 | dodecenoyl-Coenzyme A delta isomerase | agaagCTGACtctGCCtgaga/F | −643 | 9.2 | 0.859 | 7 (4) | 1.34 |
| Endog | endonuclease G | tggctGTGATcctGCGcttgt F | −2877 | 7.4 | 0.768 | 8 (2) | 1.23 |
| Etfa | electron transferring llavoprotein, alpha polypeptide | actctTGCtgaGTCATctggt R | −2545 | 16.4 | 0.951 | 10 (6) | 1.52 |
| Etfb | electron transferring flavoprotein, beta polypeptide | atatcTGCtgcCTCACagaga/R | −3260 | 11.3 | 0.849 | 7 (3) | 1.48 |
| Fabp31 | fatly acid binding protein 3, muscle and heart | tgagtGTGACcatGCCctgaa/F | −1115 | 8.1 | 0.82 | 13 (6) | 3.10 |
| Fh1 | fumarate hydratase 1 | gcagtCTGACacaGTAaacat F | −4197 | 10.5 | 0.808 | 7 (4) | 1.35 |
| Gatd3a | Glutamine amidotransferase-like class 1 domain-containing protein 3A, mitochondrial | tcagcATGAGttgGCAcctgt/F | −1503 | 12.5 | 0.833 | 10 (5) | 1.48 |
| Gpd1 | glycerol-3-phosphate dehydrogenase 1 | taaccTGCtgtCTCAGtgtcc R | −4600 | 12 | 0.851 | 7 (5) | 2.19 |
| Gpx21 | glutathione peroxidase 2 | ccgggATGACttaGCAaaaaa F | 59 | 16.6 | 0.937 | 13 (11) | 1.78 |
| Gstm41 | glutathione S-transferase, mu 4 | tacctGTGACtcaGCAtcttc F | −2627 | 19 | 0.992 | 1 1 (3) | 1.30 |
| Gsto11 | glutathione S-transferase omega 1 | tggatAGCtgaGTCACtgccc/R | −2726 | 13.1 | 0.916 | 9 (6) | 2.22 |
| Hadhb | hydroxyacyl-Coenzyme A dehydrogenase | taaagGTGAAttaGCAcccag/F | −1398 | 10.3 | 0.853 | 10 (6) | 1.50 |
| Hibadh | 3-hydroxyisobutyrate dehydrogenase | caaagGGCtaaG’l CATagttt R | −2325 | 14.7 | 0.914 | 6 (4) | 1.27 |
| Hibch | 3-hydroxyisobutyryl-Coenzyme A hydrolase | ccacaATGACacaGTTcctct F | −2170 | 9.4 | 0.787 | 9 (4) | 1.45 |
| Hk2 | hexokinase 2 | gccacCGCcgcGTCAGgctca R | −265 | 9.3 | 0.836 | 7 (4) | 1.38 |
| Hrc | histidine rich calcium binding protein | ctcagGGCtctGTCACtgata R | −4140 | 10.1 | 0.862 | 11 (7) | 3.75 |
| Idh3a1 | isocitrate dehydrogenase 3 (NAD+) alpha | tttctCTGACtcaGCActttg/F | −2163 | 14.9 | 0.942 | 12 (6) | 1.56 |
| Ifi27l2a | interferon, alpha-inducible protein 27 like 2A | tatttTTCtgtGTCATccata/R | −3889 | 11.4 | 0.803 | 7 (5) | 5.07 |
| Ldhb1 | lactate dehydrogenase B | gccacTGCaaaGTCAGcaggc/R | 65 | 10.1 | 0.869 | 13 (8) | 1.43 |
| Macrodl1 | MACRO domain containing 1 | ttaacTGCtgaGTCATctctc/R | −1327 | 16.2 | 0.947 | 8 (5) | 1.58 |
| Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | ttgctTTCtctGTCACtgtcc/R | −1518 | 7.6 | 0.782 | 13 (3) | 1.37 |
| Me3 | malic enzyme 3, NADP(+)-dependent, mitochondrial | actatTGCaaaGTCAActagg/R | −4844 | 10.4 | 0.873 | 7 (2) | 1.92 |
| Mfn2 | mitofusin 2 | ctctgTGCtgaTTCAGgtcca R | 1472 | 11.2 | 0.861 | 4 (2) | 1.42 |
| Mpcl1 | mitochondrial pyruvate carrier 1 | tgtagTGCtgaTTAATgatta R | −3305 | 11.1 | 0.788 | 11 (7) | 1.41 |
| Mrpl12 | mitochondrial ribosomal protein L12 | gattcTGCagaCTCATctggt R | −559 | 10.9 | 0.833 | 4 (3) | 1.4 |
| Mybpc3 | myosin binding protein C, cardiac | tgcacATGACttaGGAgcagg/F | −899 | 9.3 | 0.801 | 10 (3) | 4.90 |
| Myh7 | myosin, heavy polypeptide 7, cardiac muscle, beta | tcaagCTGACttaGACaattc/F | −4150 | 9.4 | 0.789 | 12 (4) | 3.63 |
| Myl3 | myosin, light polypeptide 3 | gagtcTGCtgtGTCAAggggt/R | −2096 | 14.1 | 0.92 | 10 (1) | 5.90 |
| Mylk3 | myosin light chain kinase 3 | acacaGTAACccaGCAcatta/F | −2004 | 12.5 | 0.834 | 9 (8) | 4.25 |
| Mylk4 | myosin light chain kinase family, member 4 | tcttgGGCagtGTCACcatag/R | −1053 | 9.7 | 0.843 | 10 (7) | 2.49 |
| Myom2 | myomesin 2 | cagatTGCtatGTCACactga/R | −3988 | 16.3 | 0.924 | 17 (10) | 4.29 |
| Ndrg41 | N-myc downstream regulated gene 4 | cggcaATGAGtgtGCAgaaag/F | −2645 | 8.7 | 0.805 | 17 (11) | 5.77 |
| Ndufa11 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1 | gtgagATGACtcaGCGggtaa/F | −2810 | 11.1 | 0.901 | 11 (6) | 1.31 |
| Ndufa101 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | cacaaATGAAtcaGCAcaaat/F | −2392 | 12.1 | 0.867 | 7 (3) | 1.47 |
| Ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | aagctGTGACaaaGTAtcata/F | −4369 | 7.9 | 0.778 | 4 (3) | 1.35 |
| Ndufa51 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 | gcttcTGCttaGTAAT cgtgt/R | −3593 | 14.8 | 0.855 | 6 (4) | 1.59 |
| Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | tcaggCTGATcttGCAaataa/F | −1464 | 8 | 0.781 | 7 (2) | 1.34 |
| Ndufa91 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | agtttGGCggaGTCATtctca/R | −2217 | 12.5 | 0.879 | 10 (6) | 1.48 |
| Ndufab11 | NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 | cattcTGCcttGTCATggtca/R | −3775 | 15.3 | 0.88 | 8 (4) | 1.61 |
| Ndufb51 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5 | gcgacTGCaagGTCACgctct/R | −531 | 10.4 | 0.852 | 11 (7) | 1.37 |
| Ndufb71 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 | aggagGGCtcaGTCATtactg/R | −776 | 9.9 | 0.881 | 11 (4) | 1.43 |
| Ndufb8 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8 | gagttGCCttgGTCATggtgt/R | −1792 | 9 | 0.768 | 9 (5) | 1.50 |
| Ndufb91 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | ccacaATAACtcaGCActccg/F | −3274 | 13.7 | 0.866 | 13 (5) | 1.46 |
| Ndufs11 | NADH dehydrogenase (ubiquinone) Fe-S protein 1 | ctggcCGCtaaGTCAAtatgg/R | 171 | 9.7 | 0.889 | 11 (6) | 1.63 |
| Ndufs21 | NADH dehydrogenase (ubiquinone) Fe-S protein 2 | ttcacTGCttaGTCACactct/R | −1449 | 15.3 | 0.937 | 14 (5) | 1.44 |
| Ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | gctttTGCcgaGTCACtaagg/R | −2898 | 14.2 | 0.907 | 7 (4) | 1.44 |
| Ndufs61 | NADH dehydrogenase (ubiquinone) Fe-S protein 6 | tctgaTGCcaaGTAACcttga/R | −2794 | 9.5 | 0.794 | 7 (4) | 1.43 |
| Ndufv11 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | ccttcTGCtttGTCAGacaga/R | −846 | 12.7 | 0.894 | 11 (5) | 1.41 |
| Ndufv2 | NADH dehydrogenase (ubiquinone) flavoprotein 2 | gcttcGGCtgtGTAAGagtca/R | −4523 | 10 | 0.793 | 6 (3) | 1.44 |
| Ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | gcatgGTGAGactGCCttaaa/F | −3718 | 8.5 | 0.786 | 3 (2) | 1.50 |
| Nnmt | nicotinamide N-methyltransferase | catttTGCtgaATGAT catgc/R | −2776 | 10.8 | 0.79 | 7 (4) | 1.74 |
| Nppa | natriuretic peptide type A | atattTGCagtGTGACtcgta/R | −2299 | 9.5 | 0.772 | 14 (6) | 31.7 |
| Nudt8 | nudix (nucleoside diphosphate linked moiety X)-type motif 8 | tcacaGTCACacaGCAaagta/F | −878 | 14.4 | 0.846 | 7 (1) | 2.12 |
| Oxct1 | 3-oxoacid CoA transferase 1 | tgaggGTCAGtcaGCAcgctg/F | −279 | 9.5 | 0.78 | 6 (2) | 1.31 |
| Pdha1 | pyruvate dehydrogenase E1 alpha 1 | gaagaGTGACtgaGGAagact/F | −1276 | 10 | 0.814 | 6 (4) | 1.34 |
| Pdhb1 | pyruvate dehydrogenase beta | ctgatTGCtgaGTCATctaag/R | −1822 | 13.3 | 0.934 | 9 (5) | 1.44 |
| Pfkfb1 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 | tcttgTGCtgtGTAAGgaggg/R | −4051 | 11.2 | 0.815 | 10 (5) | 1.55 |
| Phgdh | 3-phosphoglycerate dehydrogenase | ctccgGTGACtggGCAgggtt/F | 59 | 9.8 | 0.871 | 11 (3) | 1.98 |
| Phyh | phytanoyl-CoA hydroxylase | catttTGCtgtCTCACaggtg/R | −2798 | 12.9 | 0.853 | 8 (5) | 1.79 |
| Pkm1 | pyruvate kinase, muscle | tctgcTGCtgaGTCATtactg/R | −2757 | 15.1 | 0.939 | 10 (4) | 1.24 |
| Pla2g16 | phospholipase A2, group XVI | gggagGTGACtgaGCAgagcc/F | −4218 | 10.9 | 0.909 | 8 (5) | 1.61 |
| Plin4 | perilipin 4 | tctggGTGACacaGCAgctcg/F | 94 | 14.9 | 0.924 | 5 (3) | 2.74 |
| Plin5 | perilipin 5 | taatcTGCtgtTTAAT cctga/R | −1 119 | 12.1 | 0.773 | 12 (9) | 2.80 |
| Pln | phospholamban | taaagTGCtgaATCATaatgc R | −4526 | 14.8 | 0.895 | 9 (4) | 10.1 |
| Pnpla3 | patatin-like phospholipase domain containing 3 | gtccaGGCtgaGTCACtgaag R | −813 | 10.3 | 0.901 | 4 (3) | 3.03 |
| Ppal | pyrophosphatase (inorganic) 1 | aaaccATAACcaaGCActaga/F | −2110 | 10.3 | 0.804 | 9 (4) | 1.85 |
| Ppargcla | peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | actagATAACtctGCAttatt/F | −2195 | 9.5 | 0.802 | 8 (6) | 1.76 |
| Ppargcab | peroxisome proliferative activated receptor, gamma, coactivator 1 beta | gctgcTGCtgaATCAActtgg/R | −2602 | 11 | 0.874 | 10 (6) | 1.50 |
| Prdx31 | peroxiredoxin 3 | ccaacATGAAgcaGCAtatga/F | −4476 | 10.9 | 0.851 | 11 (8) | 1.34 |
| Pxmp21 | peroxisomal membrane protein 2 | acaaaCGCttaGTCAGcgcgg R | −3633 | 14.7 | 0.909 | 7 (5) | 1.50 |
| Qsox11 | quiescin Q6 sufhydryl oxidase 1 | atagtTGCttgGTCAGgtgcc R | −2189 | 9.9 | 0.872 | 10 (1) | 1.56 |
| Sdha | succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | tatcgGTGACttaGAGataaa F | −1070 | 9.4 | 0.785 | 10 (3) | 1.31 |
| Sdhb1 | succinate dehydrogenase complex, subunit B, iron sulfur (Ip) | gaaagCTGACgcaGCCcagtg/F | −3662 | 9.1 | 0.868 | 12 (6) | 1.58 |
| Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein | aaaaaGTAACagaGCAaatgt/F | −1373 | 8.8 | 0.797 | 7 (5) | 1.34 |
| Slc16a11 | solute carrier family 16 (monocarboxylic acid transporters), member 1 | cgcggCGCcggGTCACgtggc/R | −212 | 6.5 | 0.825 | 10 (6) | 1.33 |
| Slc25a18 | solute carrier family 16 (monocarboxylic acid transporters), member | aaataCGCtgtGTCACtttat/R | −1773 | 12.5 | 0.894 | 15 (6) | 1.44 |
| Slc27a11 | solute carrier family 27 (fatty acid transporter), member 1 | gagcaCTGACtgtGCTagctt/F | −1175 | 7 | 0.831 | 5 (4) | 1.57 |
| Slc27a2 | solute carrier family 27 (fatty acid transporter), member 2 | ttcctGTTACtcaGCAggctt/F | −4469 | 11.3 | 0.844 | 14 (8) | 3.50 |
| Slc2a4 | solute carrier family 2 (facilitated glucose transporter), member 4 | gagaaTGCctaTTCATcctcc/R | −2266 | 9.5 | 0.817 | 4 (3) | 2.18 |
| Slc47a1 | solute carrier family 47, member 1 | ccctgATGACtaaGCAttcca/F | −1687 | 13.3 | 0.925 | 9 (4) | 4.02 |
| Sucla2 | succinate-Coenzyme A ligase, ADP-forming, beta subunit | gaagtAGCagcGTCACagttc/R | −220 | 8.7 | 0.841 | 7 (2) | 1.35 |
| Tecrl | trans-2,3-enoyl-CoA reductase-like | gaatcATAACgctGCAaaata/F | −213 | 7.5 | 0.768 | 5 (1) | 2.28 |
| Thrsp | thyroid hormone responsive SPOT14 | atcagGGCtaaGTCAGactca/R | −3275 | 12.4 | 0.896 | 8 (5) | 4.11 |
| Tnnc1 | troponin C, cardiac slow skeletal | gcccaGTGAGgcaGCAccagt F | −3469 | 10.4 | 0.843 | 9 (6) | 4.85 |
| Tnni3 | troponin I, cardiac 3 | accttTACtgaGTCATctctc R | −3438 | 10.8 | 0.823 | 9 (5) | 6.94 |
| Tomm51 | translocase of outer mitochondrial membrane 5 | ccaggGTGACacaGAAaaatc F | −906 | 14.3 | 0.83 | 5 (2) | 1.36 |
| Tmp1 | tropomyosin 1, alpha | tggggGTGACtggGCAtcctc/F | −2579 | 10.4 | 0.872 | 13 (8) | 1.69 |
| Trf | transferrin | ccaatTGCccaATCACcccgc/R | −71 | 9.9 | 0.816 | 9 (1) | 1.29 |
| Uox1 | urate oxidase | taacgGGCtttGTCATccctt/R | −4600 | 11.2 | 0.863 | 14 (7) | 1.64 |
| Uqcr10 | ubiquinol-cytochrome c reductase, complex III subunit X | ttaatTACttaGTCAAtttaa/R | −2159 | 7.9 | 0.804 | 3 (2) | 1.44 |
| Uqcr11 | ubiquinol-cytochrome c reductase, complex III subunit XI | ttttcTGCttgGTCACtgggg/R | −3126 | 13.7 | 0.901 | 10 (4) | 1.50 |
| Uqcrc1 | ubiquinol-cytochrome c reductase core protein 1 | tttttTGCttaGTTATtttat/R | −593 | 9.1 | 0.822 | 4 (3) | 1.55 |
| Uqcrfs1 | ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 | tctacTGCagaGTGATgcttc/R | −2866 | 11.6 | 0.814 | 6 (2) | 1.58 |
| Uqcrh | ubiquinol-cytochrome c reductase hinge protein | gtgtgTGCtgaCTCACacctg/R | −3276 | 11.9 | 0.869 | 7 (4) | 1.38 |
| Uqcrq | ubiquinol-cytochrome c reductase, complex III subunit VII | tctccTGCtcaGTCACtctag/R | −3503 | 12.9 | 0.925 | 11 (4) | 1.50 |
| Usp13 | ubiquitin specific peptidase 13 | cccgcCTGACgctGCAgctgg/F | −106 | 9.9 | 0.857 | 9 (3) | 1.36 |
Selected putative AREs in the upstream of each gene are shown. Full list of potential AREs and details are in the supplemental Table 3 of previous publication (Cho et al. 2019). Bioinformatic analysis (Wang et al. 2007) done for Nrf2-dependent, sulforaphane-induced lung genes in ICR mice (Cho et al. 2019). Mouse genome build mm9 used.
ARE core-like sequence 5’-RTKAYnnnGCR-3’ (R=A or G, K=G or T, Y=C or T, n=A, C, G, or T), forward (F) or reverse complementary (R) orientation.
transcription start site.
position weight matrix score. The minimal PWM of functional ARE=6.4 and the maximal PWM of functional ARE=21.8 (the median PWM of all known functional AREs= 15) in the current bioinformatic model (Wang et al. 2007).
matrix similarity score.
Total number of ARE-like sequences determined in the gene loci (including up to 5 kb of upstream and downstream sequences). Number of the potential AREs in the upstream region including 5’-UTR is in the parenthesis.
Fold increase over PBS-pretreatment control in Nrf2 wild-type (ICR) mouse lung at day 9 after sulforaphane treatment (oral, 9 μmol) on days 1, 3, and 5 (Cho et al. 2019).
Genes with Nrf2/sMaf-bound functional AREs based on mouse ENCODE ChIP-seq data (Mouse et al. 2012; Yue et al. 2014).
Biogenesis, mitophagy, and quality control
Mitochondrial biogenesis is known to be cooperatively regulated by several transcription factors. Peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC-1α, gene is PPARGC1A) is a master co-regulator (Austin and St-Pierre 2012; Johri et al. 2013), which forms heteromeric complexes with nuclear respiratory factors, NRF-1 (also called alpha palindromic-binding protein, α-PAL) and NRF-2 (also called GA binding protein transcription factor subunit alpha, GABPA), and the nuclear receptors (e.g., peroxisome proliferator-activated receptors such as PPARα, PPARδ, and PPARγ). These transcription factors, in turn, regulate the expression of OXPHOS complex genes including cytochrome c (CYCS) as well as TFAM and families (TFB1M, TFB2M) that are essential in transcription of respiratory chain subunits and translational components (Hock and Kralli 2009). PGC-1α also induces the expression of genes involved in heme biosynthesis, ion transport, mitochondrial protein translation, and protein importation and also stimulates respiratory function (Austin and St-Pierre 2012). Importantly, NRF2 is reported to control mitochondrial quality characterized by copy number, mitophagy, and biogenesis. Piantadosi et. al. (2008) first demonstrated that NRF2 induces NRF-1 through functional AREs via the heme oxygenase 1 (HO-1)-carbon monoxide (CO) system in mice. Overexpression of HO-1 caused CO production to activate phosphoinositide 3-kinase (PI3K)-serine/threonine-specific protein kinase (Akt) pathway, which in turn liberated NRF2 from Keap1 sequestration to induce NRF-1 and enhanced mitochondria copy number in murine cardiomyocytes (Piantadosi et al. 2008). A similar observation for CO- and NRF2-mediated mitochondrial biogenesis was demonstrated in rodent liver, neuron, and lung (Athale et al. 2012; Hota et al. 2012; MacGarvey et al. 2012). The mouse PGC-1α gene (Ppargc1a) also possesses multiple putative AREs (Cho et al. 2019) and a functional ARE was determined in PPARγ-encoding gene (Cho et al. 2010). In mice trained with treadmill exercise, Nrf2 deficiency reduced whole body energy expenditure accompanying decreased skeletal muscle mitochondria mass and antioxidant activities (Merry and Ristow 2016), supporting further the role for NRF2 in mitochondrial biogenesis.
Mitochondria constantly undergo fission and fusion, and the rates of fusion and fission are tightly controlled. When cells experience metabolic or environmental stresses, mitochondrial fusion and fission work to maintain the balance. An increase in fusion activity leads to mitochondrial elongation, whereas an increase in fission activity results in mitochondrial fragmentation (Chan 2012). Mitofusin-1 and −2 (MFN1, MFN2) mediate fusion. Complex of dynamin-1-like protein (DRP1) and mitochondrial fission factor (MFF) promote fission and it is coordinated with DNA replication (biogenesis) as fission requires mtDNA for their function. Mitochondrial fission has significant implications in stress response and apoptosis (Chan 2012). KEAP1 was identified as a negative regulator for this process, and KEAP1 inhibition/NRF2 activation degraded the mitochondrial fission protein DRP1 (Sabouny et al. 2017). ARE motifs were identified in the promoters of mouse Mfn2 (Cho et al. 2019). When damaged mitochondria are separated from the mitochondrial network by fission, mitophagy subsequently degrades them to maintain a healthy pool of mitochondria. Mitophagy is also accelerated by oxidative stress and monitored and regulated by redox sensors; PINK1 recruits and phosphorylates PARK2 in mitochondria to ubiquitinate MFNs and other proteins (e.g., voltage-dependent anion-selective channel 1, VDAC1) to induce mitophagy. The ubiquitinated mitochondrial proteins are then recognized by autophagy proteins, p62/sequestosome 1 (SQSTM1) and microtubule-associated proteins 1A/1B light chain 3B (LC3B), to form an autophagosome for lysosomal degradation. NRF2 increased PINK1 transcription in damaged mitochondria under various stress conditions (Murata et al. 2015). The p62/SQSTM1 promoter also possesses functional AREs (Jain et al. 2010). Nrf2 activated p62/SQSTM1 following mitophagy induction in neuroblastoma cells, and Nrf2 knockdown impaired increase in p62 (Ivankovic et al. 2016). In addition, p62 (KEAP1-interacting region) competed with NRF2 (DLG motif) for KEAP1 binding (in the DC pocket) and thus accumulation of p62 caused persistent NRF2 activation (Kageyama et al. 2018) (Fig. 1). NRF2 and p62 are also known to cooperate in suppressing aberrant KEAP1 activity for mitochondrial motility (Bertrand et al. 2015). Mitochondria are anchored to microtubule motor proteins by attachments with the small mitochondrial Rho GTPases (MIRO1, MIRO2) in the mitochondrial outer membrane and they primarily traffic on microtubules in the cell. NRF2 or p62 binding to KEAP1 within the PGAM5 complex on the mitochondrial outer membrane suppressed KEAP1-CUL3 ligase-mediated degradation of MIRO2 and preserved mitochondrial motility (O’Mealey et al. 2017).
Protein deglycase DJ-1 (also called Parkinson disease protein 7, PARK7) is essential in mitochondria quality control. DJ-1 is linked to recessively inherited Parkinson’s disease when mutated, i.e. human carriers of E64D mutation (c.926G>A) develop early-onset recessive Parkinsonism accompanying selective dopamine-generating cell death and oxidative stress (Bonifati et al. 2003). Oxidation of the Cys106 residue caused mitochondrial localization of DJ-1 (Canet-Aviles et al. 2004). Loss of DJ-1 caused impaired mitochondrial respiration, increased mitochondrial ROS, mitochondrial membrane depolarization, mitochondrial fragmentation, and alterations of mitochondrial shape in human neuronal cells and mouse embryonic fibroblasts (Krebiehl et al. 2010; Thomas et al. 2011). Defective mitochondrial complex I activity in PARK7 null dopaminergic neuronal cells was thought to be due to the aberrant formation of the supercomplex, which impaired the flow of electrons through the channels between respiratory chain complexes, resulting in mitochondrial dysfunction (Heo et al. 2012). Importantly, DJ-1 was known as an upstream regulator for NRF2 to prevent its KEAP1 binding and subsequent ubiquitination (Clements et al. 2006) (Fig. 1). Knockout of Park7 in mice or rodent neuronal cells significantly reduced ARE-mediated TRX1 and glutamate cysteine ligase catalytic expression and increased susceptibility to oxidants (Zhou and Freed 2005; Im et al. 2012). DJ-1 was also proposed to regulate redox and mitochondrial homeostasis independent of Nrf2 [See review (Ariga 2015)]. In summary, these studies implied that DJ-1-NRF2 axis contributes to mitochondrial dynamics and quality control through maintenance of redox balance.
NRF2 in Mitochondrial Dysfunction of Airway Disorders
Mitochondrial biology has been extensively investigated in airway disorders during the last decade. Mitochondrial energy provision is an important function of the airway cells. For example, alveolar type 2 cells in the parenchyma are enriched with mitochondria to provide ATP for surfactant synthesis, secretion, and recycling as well as differentiation into type 1 pneumocytes (Schumacker et al. 2014). Mitochondrial [Ca2+] and ATP production are also critical for ion transport, maintaining airway surface hydration, ciliary beat, and mucin secretion in bronchial epithelium (Lazarowski and Boucher 2009). In addition, mitochondria have a vital role in bronchial airway smooth muscle (ASM) contraction. The mitochondrial genome has been known as a cellular target under oxidative stresses such as hyperoxia, acute lung injury (ALI), or pneumonia (Schumacker et al. 2014). To compensate for damaged mitochondria in airway disorders, promotion of mitochondrial biogenesis or mitochondria transfer has been suggested (Schumacker et al. 2014). In the case of mitochondrial transfer, donation of healthy mitochondria by mesenchymal stem cell (MSC)-conducted mitochondrial transfer to the mouse airway epithelial or ASM cells increased alveolar ATP concentration and protected against experimental lung inflammation and hyperresponsiveness (Islam et al. 2012; Li et al. 2018). These findings indicated transfer may be a therapeutic strategy for diseases linked to genetic mitochondrial defects. In airway pathogenesis, there is relatively little evidence of a role for NRF2 in mitochondrial functions. However, studies discussed below have implicated NRF2 as an important component to support airway mitochondrial structural and functional integrity against oxidative stress and inflammation.
Chronic obstructive pulmonary disease (COPD)
Cigarette smoke (CS) contains over 500 chemical compounds and is a well-known source of ROS (e.g., H2O2, superoxide). COPD and lung cancer are the two major CS-related lung diseases. COPD is a complex, debilitating lung disease that encompasses a variety of phenotypes including chronic bronchitis, oxidative stress, loss of alveolar surface area (emphysema), and remodeling of small airways (Barnes et al. 2003). Diagnosis of COPD is difficult until it is clinically apparent, and the prognosis remains poor. Moreover, current treatment options do not substantially alter the course of the disease. Patients with COPD are at increased risk (2–5 fold) for both the development of lung cancer and poor outcome after lung cancer diagnosis and treatment (de Torres et al. 2007).
Many in vitro studies have reported CS-induced mitochondrial defects in primary airway cells from patients or airway epithelial cell lines. CS increased ROS production from mitochondria and decreased mitochondrial membrane potential, released mitochondrial DAMPs, apoptosis, and mitophagy leading to airway injury [See review (Ryter et al. 2018)]. Evidence suggests that mitophagy is in general deleterious in the lungs of COPD patients, and LC3B and other autophagic proteins have a pro-pathogenic role in COPD. Relative to wild-type littermate mice, significantly decreased lung apoptosis and emphysema were found in LC3B gene (Map1lc3b)-deficient mice after CS exposure (Chen et al. 2010; Lam et al. 2013). Consistent with this observation, Pink1-knockout mice were resistant to mitochondrial dysfunction and COPD phenotypes development by CS exposure (Mizumura et al. 2014). Alveolar macrophages from COPD patients were found to have increased mitochondrial ROS and reduced bactericidal activity against Streptococcus peumoniae (Bewley et al. 2017). Necroptosis is a programmed form of necrosis regulated by receptor-interacting protein kinase 1 (RIPK1), RIPK3, and mixed lineage kinase domain-like pseudokinase (MLKL) in chronic lung diseases including COPD (Mizumura et al. 2014). Increased RIPK3 level coincided with increased amount of PINK1 in macrophages of COPD lung cells (Mizumura et al. 2014), indicating mitophagy may induce the necroptosis pathway and contribute to COPD phenotypes. Therefore, mitophagy and necroptosis are potential targets for COPD treatment. Differential susceptibility to CS-induced neutrophilia of inbred mouse strains was related to the release of mtDNA into bronchoalveolar lavage fluids (BALFs) (Pouwels et al. 2017). Inhibition of mitochondrial iron chelation and FAO was also suggested as a therapeutic approach based on a genome-wide association study (GWAS) which determined iron-responsive element-binding protein 2 (IREB2 or IRP2) as a susceptibility gene for COPD (DeMeo et al. 2009). IRP2 increases cellular iron uptake by iron responsive element-mediated transcriptional activation under iron depletion conditions. Excess buildup of iron in mitochondria by IRP2 was thought to lead to mitochondrial dysfunction due to inefficient oxygen consumption and energy metabolism. Consistent with this finding, inhibited iron (non-heme) loading and complex IV (COX) was found in Irp2-deficient mice and these mice were resistant to CS-caused experimental COPD (Cloonan et al. 2016). COPD is also a multiorgan systemic disease including locomotor muscle dysfunction (i.e., skeletal muscle weakness, cachexia). COPD patients stratified by cachexia had reduced expression of PPARα/PPARδ, PGC-1α and TFAM in skeletal muscle, relative to healthy subjects (Remels et al. 2007). These findings indicated that disrupted mitochondrial biogenesis may underlie skeletal muscle dysfunction in COPD.
Supporting a role for NRF2 in COPD, genetic disruption of Nrf2 caused early onset and severe emphysema in mice (Rangasamy et al. 2004). In cultured bronchial epithelial cells or primary lung cells from COPD patients, air pollution-derived particulate matter (PM2.5) exposure partially inactivated the NRF2 pathway and critically impaired mitochondrial redox homeostasis and functions (Leclercq et al. 2018). Most COPD patients are elderly, and many features of COPD including oxidant/antioxidant, protease/antiprotease, and proliferative/antiproliferative balance as well as control of inflammatory response are shared with those in aged lung (the aging hypothesis for COPD) (Ito and Barnes 2009). Silent mating type information regulation 2 homolog 1 (sirtuin 1, SIRT1) is a NAD-dependent deacetylase that modulates oxidative stress response and premature cellular senescence and aging. Recent studies have associated onset/progression of COPD with deacetylation of many transcription factors by SIRT1 [see review (Conti et al. 2015)]. SIRT1 was decreased in the lungs of smokers and COPD patients compared with nonsmokers or healthy subjects, and knockdown of SIRT1 increased NF-κB acetylation and interleukin (IL)-8 release (Rajendrasozhan et al. 2008). Consistent with this finding, Sirt1-heterozygous knockout mice developed an emphysematous phenotype at 1 year of age and were susceptible to CS-induced COPD symptoms (Yao et al. 2012). SIRT1 has a number of beneficial roles in mitochondria including PGC-1α deacetylation for nuclear translocation to induce mitochondrial biogenesis (Canto et al. 2009). Sirt1 significantly enhanced the NRF2 pathway by KEAP1 depression and/or NRF2 stabilization (deacetylation, reduction of ubiquitination) in rat renal cells (Huang et al. 2017) (Fig. 1). NRF2 also positively regulated protein expression and deacetylation activity of SIRT1 (Huang et al. 2017). In addition, NRF2 and SIRT1 were recruited to AREs to induce multidrug resistance-associated protein 2 (MRP2 or ABCC2) (Kulkarni et al. 2014) which has a role in glutathione excretion and xenobiotic conjugate formation (Vollrath et al. 2006). Taken together, a positive feedback loop by crosstalk between SIRT1 and the NRF2 is potentially beneficial against COPD pathogenesis. Well-known COPD susceptibility genes identified from multiple GWAS includes family with sequence similarity 13 member (FAM13A) in the FAO pathway (Hancock et al. 2012). CS-induced experimental COPD indicated that FAM13A shapes the cellular metabolic response to CS by promoting the FAO, and Fam13a deletion attenuated CS-induced mitochondrial respiration interruption and provided resistance to emphysema in mice (Jiang et al. 2017). A recent transcriptome analysis found increased lung ARE-responsive genes (e.g., Akr1b8, Nqo1, Gpx2) in CS-resistant Fam13a knockout mice relative to wild-type mice (Yun et al. 2017), which linked FAM13A and NRF2 in model COPD.
Idiopathic pulmonary fibrosis (IPF)
IPF is a fatal disease of the lower respiratory tract characterized by inflammation and fibrosis of the interstitium, leading to destruction of alveolar structure. Development of IPF is significantly associated with age, and people older than 75 years have 50 times higher prevalence than people 35 years old or younger (Selman et al. 2010). Mitochondria are particularly susceptible to aging. Structural and functional abnormality of mitochondria have been found with aging and aging-associated mtDNA mutations, likely related to mtDNA replication errors (Bratic and Larsson 2013).
Elevated mtDNA levels in the lung tissue, BALF, and plasma were found in IPF patients, and plasma mtDNA was strongly associated with disease progression and mortality (Ryu et al. 2017). Accumulation of mtDNA and endoplasmic reticulum (ER) stress markers was found in the lung and in type 2 pneumocytes in severe fibrotic areas from IPF patients compared to controls (Bueno et al. 2015; Patel et al. 2015). The IPF type 2 cell mitochondria were dysmorphic, dysfunctional, and defective in LC3-mediated autophagy (Bueno et al. 2015). Old mice (> 18 months) with diminished PINK1 levels were more susceptible to ER stress-induced fibrosis than young mice (2–3 months), and Pink1-deficient mouse lungs displayed more swollen mitochondria, lower mtDNA copy number, and higher collagen accumulation than wild-type mouse lungs (Bueno et al. 2015; Patel et al. 2015). Pink1−/− and Park2−/− mice were susceptible to the fibrogens bleomycin, murine gammaherpesvirus 68 than their wild-type controls (Bueno et al. 2015; Patel et al. 2015; Kobayashi et al. 2016). ATP release from healthy cells is tightly controlled by extracellular ATP/ADPases, and ATP released from injured cells is also a DAMP that initiates NLRP3-mediated immune responses through the purinergic receptor (P2X7, P2RX7) signaling (Mariathasan et al. 2006). Increased ATP levels were found in BALF from IPF patients compared to controls (Riteau et al. 2010). Extracellular ATP level was also associated with bleomycin-induced lung fibrosis, and reduced bleomycin-induced lung inflammation and fibrosis markers were found in P2rx7-deficient mice compared to wild-type controls (Riteau et al. 2010).
NRF2 has a protective role in pulmonary fibrosis in mice (Cho et al. 2004; Traver et al. 2017). In fibrotic airways, mitochondrial levels of SOD2 were markedly heightened in Nrf2−/− mice compared to Nrf2+/+ controls (Carraway et al. 2008). NRF2 also improved mitochondrial functions by inhibition of NADP oxidase 4 (NOX4) which generates ROS (superoxide) in the inner mitochondrial membrane (Bernard et al. 2017); lung fibroblasts from Nox4-deficient mice had improved mitochondrial respiration and biogenesis (determined by mtDNA/nDNA ratio, TFAM, NRF-1) compared to normal mouse cells. In addition, silencing NRF2 in human lung fibroblast cells abrogated the NOX4-knockout effect on mitochondrial bioenergetics and biogenesis (Bernard et al. 2017). These data imply that the protective role of NRF2 in lung fibrogenesis is likely through control of mitochondrial functions.
Asthma
Asthma affects more than 300 million people worldwide and the incidence and severity of the disease increased between 2001 and 2010 (Moorman et al. 2012). Asthma is characterized by reversible airflow obstruction due to ASM hyperresponsiveness and mucus overproduction. ASM resides in respiratory airways from trachea to terminal bronchioles, and ASM plasticity is critical to lung development as well as the pathogenesis of asthma, chronic bronchitis, and emphysema.
Mitochondria sense intracellular [Ca2+] for calcium homeostasis and energy production, which is particularly critical in metabolically active, energy-consuming contractile responses in myocardium, ASM, and vascular smooth muscle for stable and persistent force generation. Increased energy demand of the contractile response must be matched by equivalent increase in energy supply and [Ca2+] in ASM. Increased cytosolic [Ca2+] signals the contractile response through excitation-contraction coupling and elevated mitochondrial [Ca2+] stimulates ATP production. While the major calcium storage in ASM is sarcoplasmic reticulum (SR) or ER, mitochondria have temporal calcium buffering storage. The mitochondrial outer membrane is relatively permeable to most ions and small molecules although Ca2+ transport is facilitated by a nonselective VDAC1 (Rapizzi et al. 2002). However, the inner membrane strictly regulates Ca2+ transport through highly selective mitochondrial calcium uniport (MCU) and sodium-calcium exchanger (NCX or SLC8A1) for influx and release, respectively (Kirichok et al. 2004; Palty et al. 2010). Mitochondria-ER/SR coupling is therefore essential for activation of MCU for ASM contraction and relaxation (Zhao et al. 2018). In addition to calcium homeostasis, excess ROS production is also a key determinant of ASM hypercontractility (Sutcliffe et al. 2012).
Asthmatics have increased oxygen consumption and impaired calcium homeostasis during inflammation which leads to ROS production and ER/SR stress as well as enhanced mitochondrial biogenesis and cell proliferation (Trian et al. 2007; Delmotte and Sieck 2015). Increased release of extracellular ATP found in BALF from asthmatics indicated mitochondrial damage in an experimental asthma model (Idzko et al. 2007). Dendritic cell-driven T helper 2 (Th2) cytokine production and bronchial hyperreactivity are suppressed by ATP neutralization (by ATPase or P2 receptor antagonist) (Idzko et al. 2007). Similarly, airborne allergen-treated mouse bronchial cells induced extracellular ATP release accompanying increased intracellular [Ca2+] and IL-33-mediated Th2 cell responses (Kouzaki et al. 2011).
In mouse models of allergic asthma, mitochondrial dysfunction and oxidative stress were key factors in the pathogenesis. Compared to wild-type controls, mice deficient in Nrf2 or its downstream genes Gpx2 and Gsto1 are more susceptible to asthma-like symptoms including elevated oxidative stress, inflammation, mucus, and airway hyperresponsiveness (Rangasamy et al. 2005; Dittrich et al. 2010). Moreover, reduced NRF2 activity was found in ASM cells from severe asthmatic patients (Michaeloudes et al. 2011). NRF2 activation by an agonist (sulforaphane) increased ARE-responsive SOD2 and HO-1 expression and reduced ASM proliferation in cultured ASM cells (Michaeloudes et al. 2011). In peripheral blood monocytes from asthmatics, hyperoxidation rate of NRF2-dependent peroxiredoxins including mitochondrial PRDX3 were related to the severity of asthma (Kwon et al. 2012). These investigations warrant further research on NRF2 in the pathogenesis of asthma.
Pulmonary arterial hypertension (PAH)
PAH is a vascular disease caused by hyperproliferation of vascular cells leading to ventricular failure and premature death. Mitochondrial compromise has been found to be a characteristic of PAH. Mitochondrial ROS and diffusible metabolites and DAMPs activate NLRP3-inflammasome and TLR9, which lead to vascular remodeling and PAH (Sutendra and Michelakis 2014). In a persistent pulmonary hypertension model using newborn lambs, reduced SOD2 expression and activity caused an increase of mitochondrial superoxide, and it depleted endothelial nitric oxide synthase (eNOS) leading to pulmonary artery endothelial dysfunction (Afolayan et al. 2012). Exogenous administration of SOD2 improved eNOS function and alleviated PAH of newborn lambs (Afolayan et al. 2012). In a rat model of PAH and in pulmonary artery smooth muscle cells from PAH patients, mitochondrial fragmentation was associated with decreased MFN2 and PGC1-α levels (Ryan et al. 2013), suggesting a potential MFN2 therapy for PAH in combination with inhibition of mitochondrial fission. Mitochondrial FAO generating substrates for OXPHOS were found to be decreased in association with intracellular lipid accumulation in PAH patients (Talati and Hemnes 2015). In addition, 3-fold greater glycolytic rate was found in pulmonary artery endothelial cells from PAH patients compared to healthy controls, indicating a metabolic switch to anaerobic glycolysis (Xu et al. 2007). In summary, vascular mitochondrial dysfunction and metabolic disturbance may at least partly underlie the PAH pathogenesis and a beneficial role for NRF2 in PAH through modulation of SOD2, PGC-1α, and intermediary metabolism is postulated.
Pneumonia, acute lung injury and other airway disorders
In a murine model of Staphylococcus aureus sepsis, mitochondrial biogenesis was enhanced to rescue damaged type 2 pneumocytes and lung function (Athale et al. 2012; Suliman et al. 2017). Nrf2−/− mice are highly susceptible to bacterial sepsis and lung inflammation (MacGarvey et al. 2012). NRF-1 and TFAM mRNA induction was evident in wild-type mouse lungs, but not in Nrf2−/− mouse lungs with pneumonia caused by S. aureus (Athale et al. 2012). Inhaled CO increased survival after sepsis, which was accompanied by activation of Akt1-NRF2 axis-induced mitochondrial biogenesis factors including NRF-1, TFAM, and PGC-1α. Sepsis-induced loss of hepatic mtDNA copy number was also greater in Nrf2−/− and Akt−/− mice than in corresponding wild-type mice (MacGarvey et al. 2012). Nrf2-mediated mitochondrial quality control also affected lung resolution from septic pneumonia as reduced mitophagy determined by lowered LC3 levels and elevation of p62 expression was detected in Nrf2−/− type 2 pneumocytes and alveolar macrophages compared to the cells from wild-type mice (Chang et al. 2015).
Circulating mtDNA has been associated with severe trauma and sepsis patients, indicating that this DAMP could be a potential biomarker or proinflammatory alarm signal for acute respiratory distress syndrome (ARDS) or sepsis (Kung et al. 2012; Simmons et al. 2013). Another mitochondrial DAMP, cardiolipin, is a rare mitochondrial-specific phospholipid in the inner membrane and is an apoptotic cell surface marker vital for overall mitochondrial function and membrane integrity. Increased lung injury and disrupted surfactant activity and lung function were found in mice treated with cardiolipin (Ray et al. 2010). Markedly elevated cardiolipin was also evident in tracheal aspirates from pneumonia patients and BALF from bacteria-infected mice (Ray et al. 2010). Increased airway cardiolipin secretion and development of pneumonia were detected in type 2 cells of mice with a functional mutation (G308V) in a cardiolipin transporter (ATPase, aminophospholipid transporter, class I, type 8B, member 1, ATP8B1) (Ray et al. 2010). In addition, decreased lung NRF2-ARE responses were found in the aged Atp8b1 mutant mice (14 month) compared to aged wild-type mice (Soundararajan et al. 2016). Overall, these studies indicated that cardiolipin homeostasis may be protective in age-related lung disorders.
Experimental hyperoxia exposure is a model of supplemental oxygen therapy in ALI/ARDS of adults and bronchopulmonary dysplasia (BPD) of preterm infants. Although supplemental oxygen is clinically important in these clinical settings, oxygen paradoxically causes significant toxicity in the lung. While multiple factors contribute to the risk of the disease, the strongest predictor is lower gestational age which places the most premature infants at greatest risk to BPD. Supporting this notion, hyperoxia exposure exaggerated mitochondrial oxidation in late saccular/early alveolar stage of rodent lung pneumocytes (postnatal days P5-P7) than in matured rodent lung cells (Berkelhamer et al. 2013). The magnitude of hyperoxia-caused arrest in alveolar development was associated with declined mitochondrial respiration and complex I activity in a murine model of BPD (Ratner et al. 2009). Exposure of immature (canalicular/saccular stage) rat fetal lung explants to hyperoxia caused mtDNA damage, impaired branching morphogenesis, and diminished surfactant expression, indicating modulation of mtDNA repair as a potential novel strategy for treatment of oxidant-induced lung disease in the preterm infant (Gebb et al. 2013). Adult type 2 cells and mitochondria isolated from mice exposed to hyperoxia also had significant decrease in mitochondrial respiration and oxygen consumption rate (via complexes I and II) as well as glycolysis, indicating hyperoxia also impairs mitochondrial energy metabolism (Das 2013). NRF2 protects lungs against ALI caused by LPS, hyperoxia, and other insults (Cho et al. 2002; Thimmulappa et al. 2006; Marzec et al. 2007; Cho et al. 2012). In models of ALI induced by LPS and hyperoxia, mitochondrial localization of SOD2 was increased, supporting the NRF2-mediated increase of mitochondrial redox processes (Carraway et al. 2008). In experimental BPD, mitochondria-associated genes such as Akr1b8, Sod2, and GSH synthesis genes (GSH synthase, Gss; cystine/glutamate transporter, Slc7a11) as well as complex V (Atp6v1d) were significantly suppressed in hyperoxia-susceptible Nrf2−/− neonate lungs relative to Nrf2+/+ neonate lungs, which indicated significant arrest in alveolarization by Nrf2 deficiency (Cho et al. 2012). In addition, mtDNA damage was significantly greater in Nrf2−/− neonate lungs than in Nrf2+/+ neonate lungs under hyperoxic and normoxic conditions (Cho et al. 2012). These results supported the potential role for NRF2 in pathogenesis and mitochondrial dysfunction of BPD.
Cystic fibrosis, a lethal inflammatory disease accompanied by abnormal mucus secretion, is caused by mutations in cystic fibrosis transmembrane conductance regulator (CFTR). Many studies have demonstrated decreased function or expression of OXPHOS subunits (e.g., complex I) in cystic fibrosis patients [see review (Sureshbabu and Bhandari 2013)]. Bacterial flagellin (a TLR5 ligand) exacerbated pro-inflammatory responses of cystic fibrosis airway epithelial cells driven by Pseudomonas aeruginosa and MCU had a role in TLR-NLRP3 inflammasome signaling (Rimessi et al. 2015). These findings indicated that mitochondrial [Ca2+] contributes to the regulation of inflammatory cystic fibrosis (Rimessi et al. 2015). A functional ARE was identified in the far upstream (−44kb) enhancer region of CFTR (Zhang et al. 2015), which is consistent with a role for NRF2 in mitochondrial dysfunction in this disease.
Lung Cancer
Lung cancer is the leading cause of cancer-related deaths worldwide. In particular, non-small cell lung cancer (NSCLC) including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma accounts for 85% of the cases and is the major cigarette smoking-related lung disease (Tan et al. 2009). Although environmental (e.g., cigarette smoking, COPD with airflow obstruction) and genetic (e.g., epidermal growth factor receptor, EGFR) factors are important contributors to predisposition and progress, lung cancer is also characterized by anti-apoptosis and uncontrolled cell proliferation and angiogenesis processes. While two EGFR tyrosine kinase inhibitors (TKIs, gefitinib and erlotinib) are representative chemotherapeutics for NSCLC, many patients eventually experience resistance to EGFR-TKI due to EGFR somatic mutation development in cancer cells. Overexpression of mitochondria-localized gene clusters was evident in squamous cell carcinoma regardless of presence or absence of COPD (Boelens et al. 2011). TFAM levels were also correlated with lung function indices in NSCLC (Peng et al. 2013).
Cancer cells generate elevated levels of ROS and become highly vulnerable to death by oxidative stress. Different from non-neoplastic disorders, the paradoxically pathogenic role of enhanced NRF2 expression and activity in carcinogenesis is well documented and thus inhibition of NRF2 has been developed as a strategy for tumor suppression. Somatic mutations in NRF2 or in the repressor KEAP1 (Cho et al. 2015), likely due to mis-incorporation during DNA replication or by exposure to mutagens, are the ‘driver’ mutations causing persistent activation of NRF2-ARE to provide growth advantage and are causally implicated in cancer development. A recent study indicated that disrupted KEAP1-NRF2 pathway in NCSLC also contributes to the EGFR-TKI resistance, indicating crosstalk between the pathways (Park et al. 2018).
Increased mtDNA copy number is correlated with increased risk of lung cancer and mtDNA instability has been associated with lung cancer. Chronic inflammation during carcinogenesis causes ROS overproduction and implicates mtDNA damage. In NSCLC, there are frequent alterations in mtDNA D-loop which regulates mtDNA replication and expression and the D-loop SNPs (e.g., 16390A) are prognostic markers for NSCLC outcome (Ding et al. 2012). The majority of the coding mtDNA mutations in lung cancers targeted complex I, and a mutation in the ND5 (G13289A) of the complex I forced its overexpression and led to lung cancer cell proliferation (Dasgupta et al. 2012). Reduced mitochondrial fusion (MFN2) and increased fission (DRP1 and mitochondrial dynamin like GTPase or optic atrophy 1, OPA1) proteins were evident in lung adenocarcinoma tissues or cell lines and increased DRP1 was related to poor prognosis (Fang et al. 2012; Rehman et al. 2012) [Also see review (Nam et al. 2017)].
While normal cells and cancer cells utilize the same metabolic pathways, cancer cells have different preference for metabolic pathways (Chartoumpekis et al. 2015). Cancer cells require more energy and nutrients than normal cells to meet the increased demands of uncontrolled cell growth and proliferation. The most prominent adaptive changes for cancer cells to accomplish this demand is to increase glucose uptake and favor aerobic glycolysis (fermentation). This ‘Warburg Effect’ is used rather than the efficient OXPHOS pathway (Warburg 1956). Other anabolic pathways which provide cancer cells the necessary energy and substrates to synthesize lipids, proteins, and nucleic acids are the glutamate pathway, the PPP, and lipid biosynthesis (Chartoumpekis et al. 2015).
Studies in A549 cells, a human lung adenocarcinoma cell line bearing a KEAP1 somatic mutation and hypermethylation which drives constitutive NRF2 activation, indicated that NRF2 induces genes encoding PPP enzymes (G6PD, PGD, TKT, TALDO1) as well as NADPH generating ME1 and isocitrate dehydrogenase 1 (Mitsuishi et al. 2012; Hayes and Dinkova-Kostova 2014). These genes are either direct NRF2 effectors or indirectly regulated by NRF2 (Mitsuishi et al. 2012). This reprogramming of glucose metabolism by NRF2 from the aerobic pyruvate/TCA cycle into anabolic PPP was also found in animals with high glucose availability (Chartoumpekis et al. 2015). PI3K-Akt signaling is involved in nuclear accumulation of NRF2 to redirect glucose and glutamate into the adaptive, anabolic pathways (Mitsuishi et al. 2012). Fumarate, an oncogenic metabolite, was also connected with KEAP1-NRF2 pathway in cancer. In the TCA cycle, fumarate hydratase (FH) catalyzes the hydration of fumarate into malate, and germline heterozygous FH mutations have been associated with renal cancer (Tomlinson et al. 2002). In FH-deficient cells and mouse tumors, ARE-responsive antioxidant enzymes were markedly increased due to the stabilization of NRF2 via succination of critical cysteine residues in KEAP1 with accumulated fumarate (Adam et al. 2011). Other mitochondrial events linked with oncogenic NRF2 is dysregulation of the p62-KEAP1-NRF2 axis (Inami et al. 2011). Aberrant accumulation of p62 (which competes with NRF2 for KEAP1 binding) elevated NRF2 translocation and GSH synthesis leading to chemoprevention tolerance of hepatic cancer cells (Saito et al. 2016). Increased NRF2 levels in NCSLC tissues with the poor prognosis was also linked with enhanced autophagy proteins (e.g., LC3, Beclin 1) and autophagosome formation was dependent on NRF2 levels in NCSLC cells (Wang et al. 2017b). NRF2 showed oncogenic effects in colon and breast cancer cells by inhibition of a microRNA miR-181c which reduces abundance of mitochondria-encoded COX (e.g., MT-CO1) in complex IV (Jung et al. 2017).
NRF2-dependent Modulation of Lung Mitochondria Transcriptome by Sulforaphane
There is great interest to develop new therapeutic solutions for human diseases related to mitochondrial dysfunction. Pharmacological agents that act as NRF2 agonists, including sulforaphane, resveratrol, 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole (CDDO-Im), and curcumin (Cho and Kleeberger 2015), have been highlighted in studies on mitochondrial biology (Negrette-Guzman et al. 2013; Dinkova-Kostova and Abramov 2015; Denzer et al. 2016). Sulforaphane is a special form of the phytochemical isothiocyanates, and Talalay and Fahey initially recognized its efficacy in induction of ARE-bearing phase 2 enzymes for cytoprotection and cancer prevention (Fahey et al. 1997). Sulforaphane was then found to act mainly through NRF2 stabilization from KEAP1 inhibition (Kensler et al. 2013). Additional investigations found that sulforaphane has an anti-proliferative property (cell cycle arrest) and induces intrinsic apoptosis in cancer cells (Gamet-Payrastre et al. 2000), which provided potential for sulforaphane as a tumor suppressing agent, not a blocking agent, possibly effective in post-initiation stage (Myzak and Dashwood 2006). Further studies identified a role for sulforaphane in mitochondria protection and preservation against xenobiotics while it drove mitochondrial dysfunction to bring the tumor cell to an apoptotic death as an anti-carcinogenetic mechanism (Negrette-Guzman et al. 2013). Through either mechanism, sulforaphane has had a beneficial agent in human disorders.
In airways, NRF2-dependent effects of sulforaphane include attenuation of lung inflammation and phagocytosis against bacterial infection following emphysema, RSV infection, and inhaled arsenic (Cho and Kleeberger 2015). Sulforaphane pre-treatment markedly inhibited PM2.5-induced type 2 cell damage by inhibition of the mitochondrial apoptotic pathway (Wang et al. 2017a). Sulforaphane administration also restored benzo(a)pyrene-induced decline of mitochondrial bioenergetic enzyme activities and increased BCL2 levels during lung cancer initiation in mice (Priya et al. 2011). In NCSLC, an active sulforaphane metabolite (sulforaphane-cysteine) inhibited cell viability and activated the ERK1/2-mediated mitochondrial signaling pathway leading to apoptosis (Lin et al. 2017). However, clinical reports have indicated mixed or negative efficacy of sulforaphane (Egner et al. 2014; Wise et al. 2016).
In a murine ALI model, we determined that orally dosing with sulforaphane significantly prevented hyperoxia-induced lung inflammation and injury in a NRF2-dependent manner (Cho et al. 2019). Importantly, lung cDNA microarray analysis elucidated that sulforaphane markedly induced the transcriptome of mitochondrial energy metabolism in wild-type mice, but not in Nrf2−/− mice (Cho et al. 2019), consistent with Nrf2-mediated increase of mitochondrial functions. Expression of many of these mitochondria-related genes were consistently higher in Nrf2+/+ lungs than in Nrf2−/− lungs after hyperoxia exposure. Genes encoding OXPHOS complex subunits and assembly factors/ancillary proteins, TCA cycle enzymes, mitochondrial transporters/symporters, and mitochondrial antioxidants as well as biogenesis and fusion/fission proteins were coordinately increased with those encoding enzymes in FAO, glycolysis, and amino acid degradation pathways (Fig. 2). These results suggested that sulforaphane-NRF2 pathways supply metabolic substrates and electrons for mitochondrial functions and enhance mitochondrial mass to enable the lung to defend against subsequent oxidant attack while Nrf2 deficiency limits intermediary metabolic substrates and mitochondrial function to impair the mitochondria-mediated cellular processes.
Fig 2. Pulmonary NRF2 effectors known to be involved in mitochondrial bioenergetics and biogenesis.
NRF2-dependently regulated lung genes involved in mitochondrial biogenesis and bioenergetics were determined in a murine study with a NRF2 agonist sulforaphane (Cho et al. 2019). Highlighted proteins in color are encoded from sulforaphane-induced genes in Nrf2-deficient mice but not in Nrf2-sufficient mice at normal physiological condition (in orange), after hyperoxia exposure (an experimental model of acute lung injury, in yellow), or under both conditions (in red). Most of these protein-encoding genes are found to possess potential antioxidant response elements in their upstream region (Selected list in Table 2). In addition to mitochondrial redox balance through reactive oxygen species quenching and protein S-glutathionylation, NRF2 may enhance cellular entry of nutrients and mitochondrial entry of intermediary metabolites by transcriptional activation of plasma and mitochondrial membrane transporters to increase substrates for oxidative phosphorylation and ATP production. NRF2 may also maintain the healthy pool of mitochondria through quality control by induction of genes involved in biogenesis, fusion, fission, mitophagy, and apoptosis. Most importantly, NRF2 may directly induce expression of the genes encoding enzymes for fatty acid β-oxidation (FAO), tricarboxylic acid (TCA) cycle, and oxidative phosphorylation. Overall, NRF2 may exert its action in various cells such as type 2 pneumocytes, bronchial epithelial cells, smooth muscle cells, and endothelial cells to defend airway from pathogenesis. IMM=inner mitochondrial membrane. OMM=outer mitochondrial membrane. Acsl, acyl-CoA synthetase long-chain family member; adipoq, adiponectin, C1Q and collagen domain containing; ADIPOR, adiponectin receptor; AGC, aspartate-glutamate carrier; Aldh, aldehyde dehydrogenase; Asp, aspartic acid; Atp, ATP synthase, H+ transporting, mitochondrial; Ccdc90a, coiled-coil domain containing 90A or mitochondrial calcium uniporter regulator 1 (Mcur1); CoA, coenzyme A; Coq, coenzyme Q; Cox, cytochrome c oxidase subunit; Cpt, carnitine palmitoyltransferase; Crat, carnitine O-acetyltransferase; Crd1, cardiolipin synthase; CYC1, cytochrome cl; CYCS, cytochrome c, somatic; Dic1, mitochondrial dicarboxylate transporter; Dnaj, DnaJ heat shock protein family; Fabp, fatty acid binding protein; GC, glutamate carrier; Glu, glutamic acid; Glut4, glucose transporter type 4; Got, glutamic-oxaloacetic transaminase; GSH, glutathione; Gpx2, glutathione peroxidase 2; Hk2, hexokinase 2; HSP40, heat shock protein 40; α-KG, α-ketoglutarate; MCT1, monocarboxylate transporter 1; MCUR1, mitochondrial calcium uniporter regulator 1; Mdh1, malate dehydrogenase 1; Mfn2, mitofusin 2; Mipep, mitochondrial intermediate peptidase; Mpc, mitochondrial pyruvate carrier; Mrpl, mitochondrial ribosomal protein L; Mrps, mitochondrial ribosomal protein S; mtDNA, mitochondrial DNA; Mtfp1, mitochondrial fission process 1; Mtx2, Metaxin 2; Nduf, NADH dehydrogenase (ubiquinone); OGC, oxoglutarate carrier; OAA, oxaloacetate; Pdh, pyruvate dehydrogenase; Pfkb, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase; Ppargc1a, peroxisome proliferative activated receptor, gamma, coactivator 1 α; Prdx3, peroxiredoxin 3; Sdh, succinate dehydrogenase complex; Slc, solute carrier family; Sod2, superoxide dismutase 2; Timm, translocase of IMM; Tomm5, translocase of outer mitochondrial membrane 5; Txn2, thioredoxin 2; Uqcr, ubiquinol-cytochrome c reductase, complex. VDAC, voltage dependent anion channel.
Using a position weight matrix (PWM) statistical model (Wang et al. 2007), we found potential ARE sequences in the NRF2-dependent genes associated to the mitochondrial functions (Cho et al. 2019). The multiple ARE-like motifs (PWM score of ≥ 6.4) were enriched in these genes (Table 2). A similar approach was used to define potential AREs from NRF2-dependently regulated mitochondrial genes by butylated hydroxyanisole treatment in mouse liver (Abdullah et al. 2012). Some potential AREs have been functionally verified by ChIP assays (Mouse et al. 2012; Yue et al. 2014) and further supported the role for the NRF2 pathway in facilitating mitochondrial biology to protect the airway against oxidant disorders.
Conclusions
Healthy maintenance of mitochondrial function is essential to prevent various human diseases. It is becoming clear that regulation of mitochondrial function constitutes an important part of cytoprotection by NRF2 under normal physiology as well as in pathological conditions. Many recent investigations have also investigated the novel transcriptional targets of NRF2 in mitochondrial dynamics as well as the crosstalk between mitochondrial proteins/metabolites and the KEAP1-NRF2 axis. NRF2 activates intermediary metabolism pathways which facilitate the flow of carbon into the FAO and TCA cycle for mitochondrial respiration and ATP production. Emerging studies have also linked mitochondrial dysfunction with ROS- and inflammation-associated lung diseases and studies have underscored the role of airway NRF2 in management of mitochondrial dysfunctions in COPD, asthma, IPF, PAH, bacterial pneumonia, and ALI. In lung cancer which has heightened NRF2 activity, an oncogenic role of NRF2 may be attributed to the improvement of mitochondrial quality and function which provides adaptive behaviors and growth advantage of cancer cells. Overall, further accumulation of the insights into the novel roles for NRF2 in modulation of mitochondrial homeostasis and function should lead to new therapeutic or preventive strategies in both neoplastic and non-neoplastic airway disorders.
Acknowledgement
The research involved in the current review article was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (NIEHS). The authors thank Drs. Michael Fessler and Xuting Wang at the NIEHS for providing excellent review of the manuscript.
Footnotes
Conflict of Interest
All authors declare that they have no conflict of interest.
Special Issue of Archives of Pharmacal Research:
Redox-based therapeutic target Nrf2 in human diseases
References
- Abdullah A, Kitteringham NR, Jenkins RE, Goldring C, Higgins L, Yamamoto M, Hayes J, Park BK (2012). Analysis of the role of Nrf2 in the expression of liver proteins in mice using two-dimensional gel-based proteomics. Pharmacol Rep 64(3):680–697 [DOI] [PubMed] [Google Scholar]
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, Stevens M, Fischer R, Carmeliet P, Maxwell PH, Pugh CW, Frizzell N, Soga T, Kessler BM, El-Bahrawy M, Ratcliffe PJ, Pollard PJ (2011). Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 20:524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afolayan AJ, Eis A, Teng RJ, Bakhutashvili I, Kaul S, Davis JM, Konduri GG (2012). Decreases in manganese superoxide dismutase expression and activity contribute to oxidative stress in persistent pulmonary hypertension of the newborn. Am J Physiol Lung Cell Mol Physiol 303:L870–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev M, Shokolenko I, Wilson G, LeDoux S (2013). The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb Perspect Biol 5:a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW (2017). The genetics and pathology of mitochondrial disease. J Pathol 241:236–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga H (2015). Common mechanisms of onset of cancer and neurodegenerative diseases. Biol Pharm Bull 38:795–808 [DOI] [PubMed] [Google Scholar]
- Ashar FN, Zhang Y, Longchamps RJ, Lane J, Moes A, Grove ML, Mychaleckyj JC, Taylor KD, Coresh J, Rotter JI, Boerwinkle E, Pankratz N, Guallar E, Arking DE (2017). Association of Mitochondrial DNA Copy Number With Cardiovascular Disease. JAMA Cardiol 2:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athale J, Ulrich A, Chou Macgarvey N, Bartz RR, Welty-Wolf KE, Suliman HB, Piantadosi CA (2012). Nrf2 promotes alveolar mitochondrial biogenesis and resolution of lung injury in Staphylococcus aureus pneumonia in mice. Free Radic Biol Med 53:1584–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin S, St-Pierre J (2012). PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. Journal Cell Sci 125(Pt 21):4963–71 [DOI] [PubMed] [Google Scholar]
- Banoth B, Cassel SL (2018). Mitochondria in innate immune signaling. Transl Res 202:52–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ, Shapiro SD, Pauwels RA (2003). Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J 22:672–688 [DOI] [PubMed] [Google Scholar]
- Berkelhamer SK, Kim GA, Radder JE, Wedgwood S, Czech L, Steinhorn RH, Schumacker PT (2013). Developmental differences in hyperoxia-induced oxidative stress and cellular responses in the murine lung. Free Radic Biol Med 61:51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard K, Logsdon NJ, Miguel V, Benavides GA, Zhang J, Carter AB, Darley-Usmar VM, Thannickal VJ (2017). NADPH Oxidase 4 (Nox4) Suppresses Mitochondrial Biogenesis and Bioenergetics in Lung Fibroblasts via a Nuclear Factor Erythroid-derived 2-like 2 (Nrf2)-dependent Pathway. J Biol Chem 292:3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand HC, Schaap M, Baird L, Georgakopoulos ND, Fowkes A, Thiollier C, Kachi H, Dinkova-Kostova AT, Wells G (2015). Design, Synthesis, and Evaluation of Triazole Derivatives That Induce Nrf2 Dependent Gene Products and Inhibit the Keap1-Nrf2 Protein-Protein Interaction. J Med Chem 58:7186–7194 [DOI] [PubMed] [Google Scholar]
- Bewley MA, Preston JA, Mohasin M, Marriott HM, Budd RC, Swales J, Collini P, Greaves DR, Craig RW, Brightling CE, Donnelly LE, Barnes PJ, Singh D, Shapiro SD, Whyte MKB, Dockrell DH (2017). Impaired Mitochondrial Microbicidal Responses in Chronic Obstructive Pulmonary Disease Macrophages. Am J Respir Crit Care Med 196:845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boelens MC, Gustafson AM, Postma DS, Kok K, van der Vries G, van der Vlies P, Spira A, Lenburg ME, Geerlings M, Sietsma H, Timens W, van den Berg A, Groen HJ (2011). A chronic obstructive pulmonary disease related signature in squamous cell lung cancer. Lung Cancer 72:177–183 [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P (2003). Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299:256–259 [DOI] [PubMed] [Google Scholar]
- Bratic A, Larsson NG (2013). The role of mitochondria in aging. J Clin Invest 123:951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, Duncan SR, Rojas M, Shiva S, Chu CT, Mora AL (2015). PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest 125:521–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA (2004). Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc Natl Acad Sci USA 102:244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR (2004). The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA 101:9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway MS, Suliman HB, Kliment C, Welty-Wolf KE, Oury TD, Piantadosi CA (2008). Mitochondrial biogenesis in the pulmonary vasculature during inhalational lung injury and fibrosis. Antioxid Redox Signal 10:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker J, Gardiner D, Kuksal N, Mailloux RJ (2018). Characterization of the impact of glutaredoxin-2 (GRX2) deficiency on superoxide/hydrogen peroxide release from cardiac and liver mitochondria. Redox Biol 15:216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC (2012). Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46:265–287 [DOI] [PubMed] [Google Scholar]
- Chang AL, Ulrich A, Suliman HB, Piantadosi CA (2015). Redox regulation of mitophagy in the lung during murine Staphylococcus aureus sepsis. Free Radic Biol Med 78:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartoumpekis DV, Wakabayashi N, Kensler TW (2015). Keap1/Nrf2 pathway in the frontiers of cancer and non-cancer cell metabolism. Biochem Soc Trans 43:639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartoumpekis DV, Ziros PG, Psyrogiannis AI, Papavassiliou AG, Kyriazopoulou VE, Sykiotis GP, Habeos IG (2011). Nrf2 represses FGF21 during long-term high-fat diet-induced obesity in mice. Diabetes 60:2465–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, Ifedigbo E, Parameswaran H, Ryter SW, Choi AM (2010). Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci USA 107:18880–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-Y, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR (2005). Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic Biol Med 38:325–343 [DOI] [PubMed] [Google Scholar]
- Cho HY (2013) Genomic structure and variation of nuclear factor (erythroid-derived 2)-like 2. Oxid Med Cell Longev 2013:286524 doi: 10.1155/2013/286524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Gladwell W, Wang X, Chorley B, Bell D, Reddy SP, Kleeberger SR (2010). Nrf2-regulated PPARγ expression is critical to protection against acute lung injury in mice. Am J Respir Crit Care Med 182:170–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Jedlicka AE, Reddy SP, Kensler TW, Yamamoto M, Zhang LY, Kleeberger SR (2002). Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol 26:175–182 [DOI] [PubMed] [Google Scholar]
- Cho HY, Kleeberger SR (2015). Association of Nrf2 with airway pathogenesis: lessons learned from genetic mouse models. Arch Toxicol 89:1931–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Marzec J, Kleeberger SR (2015). Functional polymorphisms in NRF2: implications for human disease. Free Radic Biol Med 88:362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Miller-DeGraff L, Blankenship-Paris T, Wang X, Bell DA, Lih F, Deterding L, Panduri V, Morgan DL, Yamamoto M, Reddy AJ, Talalay P, Kleeberger SR (2019). Sulforaphane enriched transcriptome of lung mitochondrial energy metabolism and provided pulmonary injury protection via Nrf2 in mice. Toxicol Appl Pharmacol 364:29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Reddy SP, Yamamoto M, Kleeberger SR (2004). The transcription factor NRF2 protects against pulmonary fibrosis. FASEB J 18:1258–1260 [DOI] [PubMed] [Google Scholar]
- Cho HY, van Houten B, Wang X, Miller-Degraff L, Fostel J, Gladwell W, Perrow L, Panduri V, Kobzik L, Yamamoto M, Bell DA, Kleeberger SR (2012). Targeted Deletion of Nrf2 Impairs Lung Development and Oxidant Injury in Neonatal Mice. Antioxid Redox Signal 17:1066–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA (2012). Identification of novel NRF2-regulated genes by ChlP-Seq: influence on retinoid X receptor alpha. Nuc Acid Res 40: 7416–7429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay Montier LL, Deng JJ, Bai Y (2009). Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomics 36:125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP (2006). DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA 103:15091–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabon MA, Konrad C, Polverino F, Siempos II, Perez E, Mizumura K, Ghosh MC, Parameswaran H, Williams NC, Rooney KT, Chen ZH, Goldklang MP, Yuan GC, Moore SC, Demeo DL, Rouault TA, D’Armiento JM, Schon EA, Manfredi G, Quackenbush J, Mahmood A, Silverman EK, Owen CA, Choi AM (2016). Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med 22:163–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti V, Corbi G, Manzo V, Pelaia G, Filippelli A, Vatrella A (2015). Sirtuin 1 and aging theory for chronic obstructive pulmonary disease. Anal Cell Pathol 2015:897327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das KC (2013). Hyperoxia decreases glycolytic capacity, glycolytic reserve and oxidative phosphorylation in MLE-12 cells and inhibits complex I and II function, but not complex IV in isolated mouse lung mitochondria. PLoS One 8:e73358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Soudry E, Mukhopadhyay N, Shao C, Yee J, Lam S, Lam W, Zhang W, Gazdar AF, Fisher PB, Sidransky D (2012). Mitochondrial DNA mutations in respiratory complex-I in never-smoker lung cancer patients contribute to lung cancer progression and associated with EGFR gene mutation. J Cell Physiol 227:2451–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres JP, Bastarrika G, Wisnivesky JP, Alcaide AB, Campo A, Seijo LM, Pueyo JC, Villanueva A, Lozano MD, Montes U, Montuenga L, Zulueta JJ (2007). Assessing the relationship between lung cancer risk and emphysema detected on low-dose CT of the chest. Chest 132:1932–1938 [DOI] [PubMed] [Google Scholar]
- Delmotte P, Sieck GC (2015). Interaction between endoplasmic/sarcoplasmic reticulum stress (ER/SR stress), mitochondrial signaling and Ca(2+) regulation in airway smooth muscle (ASM). Can J Physiol Pharmacol 93:97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, Litonjua A, Bueno R, Pillai SG, Lomas DA, Sparrow D, Shapiro SD, Criner GJ, Kim HP, Chen Z, Choi AM, Reilly J, Silverman EK (2009). Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet 85:493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer I, Munch G, Friedland K (2016). Modulation of mitochondrial dysfunction in neurodegenerative diseases via activation of nuclear factor erythroid-2-related factor 2 by food-derived compounds. Pharmacol Res 103:80–94 [DOI] [PubMed] [Google Scholar]
- Ding C, Li R, Wang P, Fan H, Guo Z (2012). Sequence polymorphisms of the mitochondrial displacement loop and outcome of non-small cell lung cancer. Exp Ther Med 3(5):861–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Abramov AY (2015) The emerging role of Nrf2 in mitochondrial function. Free Rad Biol Med 88:179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich AM, Meyer HA, Krokowski M, Quarcoo D, Ahrens B, Kube SM, Witzenrath M, Esworthy RS, Chu FF, Hamelmann E (2010). Glutathione peroxidase-2 protects from allergen-induced airway inflammation in mice. Eur Respir J 35(5):1148–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner PA, Chen JG, Zarth AT, Ng DK, Wang JB, Kensler KH, Jaconson LP, Munoz A, Johnson JL, Groopman JD, Fahey JW, Talalay P, Zhu J, Chen TY, Qian GS, Carmella SG, Hecht SS, Kensler TW (2014). Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prev Res 7:813–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey JW, Zhang Y, Talalay P (1997). Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci 94:10367–10372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang HY, Chen CY, Chiou SH, Wang YT, Lin TY, Chang HW, Chiang IP, Lan KJ, Chow KC (2012). Overexpression of optic atrophy 1 protein increases cisplatin resistance via inactivation of caspase-dependent apoptosis in lung adenocarcinoma cells. Hum Pathol 43:105–11 [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li P, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Terce F (2000). Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer Res 60:1426–1433 [PubMed] [Google Scholar]
- Gebb SA, Decoux A, Waggoner A, Wilson GL, Gillespie MN (2013). Mitochondrial DNA damage mediates hyperoxic dysmorphogenesis in rat fetal lung explants. Neonatology 103:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock DB, Soler Artigas M, Gharib SA, Henry A, Manichaikul A, Ramasamy A, Loth DW, Imboden M, Koch B, McArdle WL, Smith AV, Smolonska J, Sood A, Tang W, Wilk JB, Zhai G, Zhao JH, Aschard H, Burkart KM, Curjuric I, Eijgelsheim M, Elliott P, Gu X, Harris TB, Janson C, Homuth G, Hysi PG, Liu JZ, Loehr LR, Lohman K, Loos RJ, Manning AK, Marciante KD, Obeidat M, Postma DS, Aldrich MC, Brusselle GG, Chen TH, Eiriksdottir G, Franceschini N, Heinrich J, Rotter JI, Wijmenga C, Williams OD, Bentley AR, Hofman A, Laurie CC, Lumley T, Morrison AC, Joubert BR, Rivadeneira F, Couper DJ, Kritchevsky SB, Liu Y, Wjst M, Wain LV, Vonk JM, Uitterlinden AG, Rochat T, Rich SS, Psaty BM, O’Connor GT, North KE, Mirel DB, Meibohm B, Launer LJ, Khaw KT, Hartikainen AL, Hammond CJ, Glaser S, Marchini J, Kraft P, Wareham NJ, Volzke H, Stricker BH, Spector TD, Probst-Hensch NM, Jarvis D, Jarvelin MR, Heckbert SR, Gudnason V, Boezen HM, Barr RG, Cassano PA, Strachan DP, Fornage M, Hall IP, Dupuis J, Tobin MD, London SJ (2012). Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet 8:e1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Dinkova-Kostova AT (2014). The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39:199–218 [DOI] [PubMed] [Google Scholar]
- Heo JY, Park JH, Kim SJ, Seo KS, Han JS, Lee SH, Kim JM, Park JI, Park SK, Lim K, Hwang BD, Shong M, Kweon GR (2012). DJ-1 null dopaminergic neuronal cells exhibit defects in mitochondrial function and structure: involvement of mitochondrial complex I assembly. PLoS One 7:e32629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A (2009).Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol 71:177–203 [DOI] [PubMed] [Google Scholar]
- Holmstrom KM, Baird L, Zhang Y, Hargreaves I, Chalasani A, Land JM, Stanyer L, Yamamoto M, Dinkova-Kostova AT, Abramov AY (2013). Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol Open 2:761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota KB, Hota SK, Chaurasia OP, Singh SB (2012). Acetyl-L-carnitine-mediated neuroprotection during hypoxia is attributed to ERK1/2-Nrf2-regulated mitochondrial biosynthesis. Hippocampus 22:723–736 [DOI] [PubMed] [Google Scholar]
- Huang K, Gao X, Wei W (2017). The crosstalk between Sirt1 and Keap1/Nrf2/ARE anti-oxidative pathway forms a positive feedback loop to inhibit FN and TGF-beta1 expressions in rat glomerular mesangial cells. Exp Cell Res 361:63–72 [DOI] [PubMed] [Google Scholar]
- Idzko M, Hammad H, van Nimwegen M, Kool M, Willart MA, Muskens F, Hoogsteden HC, Luttmann W, Ferrari D, Di Virgilio F, Virchow JC Jr., Lambrecht BN (2007). Extracellular ATP triggers and maintains asthmatic airway inflammation by activating dendritic cells. Nat Med 13:913–919 [DOI] [PubMed] [Google Scholar]
- Im JY, Lee KW, Woo JM, Junn E, Mouradian MM (2012). DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum Mol Genet 21:3013–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, Lee MS, Tanaka K, Komatsu M (2011). Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193(2):275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Das SR, Emin MT, Wei M, Sun L, Westphalen K, Rowlands DJ, Quadri SK, Bhattacharya S, Bhattacharya J (2012). Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat Med 18:759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, COPD as a disease of accelerated lung aging. Chest 135(1):173–180 [DOI] [PubMed] [Google Scholar]
- Ivankovic D, Chau KY, Schapira AH, Gegg ME (2016). Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy. J Neurochem 136:388–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T (2010). p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285:22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Knudsen NH, Wang G, Qiu W, Naing ZZC, Bai Y, Ai X, Lee CH, Zhou X (2017). Genetic Control of Fatty Acid beta-Oxidation in Chronic Obstructive Pulmonary Disease. Am J Respir Cell Mol Biol 56:738–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri A, Chandra A, Flint Beal M (2013). PGC-1alpha, mitochondrial dysfunction, and Huntington’s disease. Free Radic Biol Med 62:37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KA, Lee S, Kwak MK (2017). NFE2L2/NRF2 Activity Is Linked to Mitochondria and AMP-Activated Protein Kinase Signaling in Cancers Through miR-181c/Mitochondria-Encoded Cytochrome c Oxidase Regulation. Antioxid Redox Signal 27:945–961 [DOI] [PubMed] [Google Scholar]
- Kageyama S, Saito T, Obata M, Koide RH, Ichimura Y, Komatsu M (2018). Negative Regulation of the Keap1-Nrf2 Pathway by a p62/Sqstm1 Splicing Variant. Mol Cell Biol 38:pii: e00642–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay HY, Kim WD, Hwang SJ, Choi HS, Gilroy RK, Wan YJ, Kim SG (2011). Nrf2 inhibits LXRalpha-dependent hepatic lipogenesis by competing with FXR for acetylase binding. Antioxid Redox Signal 15:2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Egner PA, Agyeman AS, Visvanathan K, Groopman JD, Chen JG, Chen TY, Fahey JW, Talalay P (2013). Keap1-nrf2 signaling: a target for cancer prevention by sulforaphane. Top Curr Chem 329:163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichok Y, Krapivinsky G, Clapham DE (2004). The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427:360–364 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Araya J, Minagawa S, Hara H, Saito N, Kadota T, Sato N, Yoshida M, Tsubouchi K, Kurita Y, Ito S, Fujita Y, Takasaka N, Utsumi H, Yanagisawa H, Hashimoto M, Wakui H, Kojima J, Shimizu K, Numata T, Kawaishi M, Kaneko Y, Asano H, Yamashita M, Odaka M, Morikawa T, Nakayama K, Kuwano K (2016). Involvement of PARK2-Mediated Mitophagy in Idiopathic Pulmonary Fibrosis Pathogenesis. J Immunol 197:504–516 [DOI] [PubMed] [Google Scholar]
- Kouzaki H, Iijima K, Kobayashi T, O’Grady SM, Kita H (2011). The danger signal, extracellular ATP, is a sensor for an airborne allergen and triggers IL-33 release and innate Th2-type responses. J Immunol 186:4375–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovac S, Angelova PR, Holmstrom KM, Zhang Y, Dinkova-Kostova AT, Abramov AY (2015). Nrf2 regulates ROS production by mitochondria and NADPH oxidase. Biochim Biophys Acta 1850:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebiehl G, Ruckerbauer S, Burbulla LF, Kieper N, Maurer B, Waak J, Wolburg H, Gizatullina Z, Gellerich FN, Woitalla D, Riess O, Kahle PJ, Proikas-Cezanne T, Kruger R (2010). Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One 5:e9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SR, Armstrong LE, Slitt AL (2013). Caloric restriction-mediated induction of lipid metabolism gene expression in liver is enhanced by Keap1-knockdown. Pharm Res 30:2221–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SR, Donepudi AC, Xu J, Wei W, Cheng QC, Driscoll MV, Johnson DA, Johnson JA, Li X, Slitt AL (2014). Fasting induces nuclear factor E2-related factor 2 and ATP-binding Cassette transporters via protein kinase A and Sirtuin-1 in mouse and human. Antioxid Redox Signal 20:15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung CT, Hsiao SY, Tsai TC, Su CM, Chang WN, Huang CR, Wang HC, Lin WC, Chang HW, Lin YJ, Cheng BC, Su BY, Tsai NW, Lu CH (2012). Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J Transl Med 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Bae YJ, Moon KA, Lee YS, Lee T, Lee KY, Kim TB, Park CS, Moon HB, Cho YS (2012). Hyperoxidized peroxiredoxins in peripheral blood mononuclear cells of asthma patients is associated with asthma severity. Life Sci 90:502–508 [DOI] [PubMed] [Google Scholar]
- Lam HC, Cloonan SM, Bhashyam AR, Haspel JA, Singh A, Sathirapongsasuti JF, Cervo M, Yao H, Chung AL, Mizumura K, An CH, Shan B, Franks JM, Haley KJ, Owen CA, Tesfaigzi Y, Washko GR, Quackenbush J, Silverman EK, Rahman I, Kim HP, Mahmood A, Biswal SS, Ryter SW, Choi AM (2013). Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest 123(12):5212–5230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowski ER, Boucher RC (2009). Purinergic receptors in airway epithelia. Curr Opin Pharmacol 9:262–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq B, Kluza J, Antherieu S, Sotty J, Alleman LY, Perdrix E, Loyens A, Coddeville P, Lo Guidice JM, Marchetti P, Garcon G (2018). Air pollution-derived PM2.5 impairs mitochondrial function in healthy and chronic obstructive pulmonary diseased human bronchial epithelial cells. Environ Pollut 243:1434–1449 [DOI] [PubMed] [Google Scholar]
- Li X, Michaeloudes C, Zhang Y, Wiegman CH, Adcock IM, Lian Q, Mak JCW, Bhavsar PK, Chung KF (2018). Mesenchymal stem cells alleviate oxidative stress-induced mitochondrial dysfunction in the airways. J Allergy Clin Immunol 141:1634–1645 [DOI] [PubMed] [Google Scholar]
- Lin K, Yang R, Zheng Z, Zhou Y, Geng Y, Hu Y, Wu S, Wu W (2017). Sulforaphane-cysteine-induced apoptosis via phosphorylated ERK1/2-mediated maspin pathway in human non-small cell lung cancer cells. Cell Death Discov 3:17025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SC, Hannink M (2008). PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res 314:1789–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtmann MH, Angelova PR, Zhang Y, Abramov AY, Dinkova-Kostova AT (2014). Nrf2 affects the efficiency of mitochondrial fatty acid oxidation. Biochem J 457:415–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGarvey NC, Suliman HB, Bartz RR, et al. (2012). Activation of mitochondrial biogenesis by heme oxygenase-1-mediated NF-E2-related factor-2 induction rescues mice from lethal Staphylococcus aureus sepsis. Am J Respir Crit Care Med 185:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux RJ, Willmore WG (2014). S-glutathionylation reactions in mitochondrial function and disease. Front Cell Dev Biol 2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailloux RJ, Xuan JY, McBride S, Maharsy W, Thorn S, Holterman CE, Kennedy CR, Rippstein P, deKemp R, da Silva J, Nemer M, Lou M, Harper ME (2014). Glutaredoxin-2 is required to control oxidative phosphorylation in cardiac muscle by mediating deglutathionylation reactions. J Biol Chem 289:14812–14828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM (2006). Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440:228–232 [DOI] [PubMed] [Google Scholar]
- Marzec JM, Christie JD, Reddy SP, Jedlicka AE, Vuong H, Lanken PN, Aplenc R, Yamamoto T, Yamamoto M, Cho HY, Kleeberger SR (2007). Functional polymorphisms in the transcription factor NRF2 in humans increase the risk of acute lung injury. FASEB J 21:2237–2246 [DOI] [PubMed] [Google Scholar]
- Merry TL, Ristow M (2016). Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J Physiol 594:5195–5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeloudes C, Chang PJ, Petrou M, Chung KF (2011). Transforming growth factor-beta and nuclear factor E2-related factor 2 regulate antioxidant responses in airway smooth muscle cells: role in asthma. Am J Respir Crit Care Med 184:894–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC (2016). Metabolic regulation of mitochondrial dynamics. J Cell Biol 212:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H (2012). Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22:66–79 [DOI] [PubMed] [Google Scholar]
- Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, Glass K, Owen CA, Mahmood A, Washko GR, Hashimoto S, Ryter SW, Choi AM (2014). Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest 124:3987–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman JE, Akinbami LJ, Bailey CM, Zahran HS, King ME, Johnson CA, Liu X (2012). National surveillance of asthma: United States, 2001–2010. Vital Health Stat 3:1–58 [PubMed] [Google Scholar]
- Mouse EC, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, Giste E, Johnson A, Zhang M, Balasundaram G, Byron R, Roach V, Sabo PJ, Sandstrom R, Stehling AS, Thurman RE, Weissman SM, Cayting P, Hariharan M, Lian J, Cheng Y, Landt SG, Ma Z, Wold BJ, Dekker J, Crawford GE, Keller CA, Wu W, Morrissey C, Kumar SA, Mishra T, Jain D, Byrska-Bishop M, Blankenberg D, Lajoie BR, Jain G, Sanyal A, Chen KB, Denas O, Taylor J, Blobel GA, Weiss MJ, Pimkin M, Deng W, Marinov GK, Williams BA, Fisher-Aylor KI, Desalvo G, Kiralusha A, Trout D, Amrhein H, Mortazavi A, Edsall L, McCleary D, Kuan S, Shen Y, Yue F, Ye Z, Davis CA, Zaleski C, Jha S, Xue C, Dobin A, Lin W, Fastuca M, Wang H, Guigo R, Djebali S, Lagarde J, Ryba T, Sasaki T, Malladi VS, Cline MS, Kirkup VM, Learned K, Rosenbloom KR, Kent WJ, Feingold EA, Good PJ, Pazin M, Lowdon RF, Adams LB (2012). An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Takamatsu H, Liu S, Kataoka K, Huh NH, Sakaguchi M (2015). NRF2 Regulates PINK1 Expression under Oxidative Stress Conditions. PLoS One 10:e0142438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myzak MC, Dashwood RH (2006). Chemoprotection by sulforaphane: keep one eye beyond Keap1. Cancer Lett 233:208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K, Hisata S, Choi AM (2015). The Roles of Mitochondrial Damage-Associated Molecular Patterns in Diseases. Antioxid Redox Signal 23:1329–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam HS, Izumchenko E, Dasgupta S, Hoque MO (2017). Mitochondria in chronic obstructive pulmonary disease and lung cancer: where are we now? Biomark Med 11:475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrette-Guzman M, Huerta-Yepez S, Tapia E, Pedraza-Chaverri J (2013). Modulation of mitochondrial functions by the indirect antioxidant sulforaphane: a seemingly contradictory dual role and an integrative hypothesis. Free Radic Biol Med 65:1078–1089 [DOI] [PubMed] [Google Scholar]
- Ni HM, Williams JA, Ding WX (2015). Mitochondrial dynamics and mitochondrial quality control. Redox Biol 4:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishinaka T, Yabe-Nishimura C (2005). Transcription factor Nrf2 regulates promoter activity of mouse aldose reductase (AKR1B3) gene. J Pharmacol Sci 97:43–51 [DOI] [PubMed] [Google Scholar]
- O’Brien M, Chalker J, Slade L, Gardiner D, Mailloux RJ (2017). Protein S-glutathionylation alters superoxide/hydrogen peroxide emission from pyruvate dehydrogenase complex. Free radic Biol Med 106:302–314 [DOI] [PubMed] [Google Scholar]
- O’Mealey GB, Plafker KS, Berry WL, Janknecht R, Chan JY, Plafker SM (2017). A PGAM5-KEAP1-Nrf2 complex is required for stress-induced mitochondrial retrograde trafficking. J Cell Sci 130:3467–3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsucci D, Siciliano G, Mancuso M (2018) Revealing the Complexity of Mitochondrial DNA-Related Disorders. EBioMedicine 30:3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan-Barmatz V, Herrmann S, Khananshvili D, Sekler I (2010). NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci USA 107:436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Kim JH, Ko E, Kim JY, Park MJ, Kim MJ, Seo H, Li S, Lee JY (2018). Resistance to gefitinib and cross-resistance to irreversible EGFR-TKIs mediated by disruption of the Keap1-Nrf2 pathway in human lung cancer cells. FASEB J 32:5862–5873 [DOI] [PubMed] [Google Scholar]
- Patel AS, Song JW, Chu SG, Mizumura K, Osorio JC, Shi Y, El-Chemaly S, Lee CG, Rosas IO, Elias JA, Choi AM, Morse D (2015). Epithelial cell mitochondrial dysfunction and PINK1 are induced by transforming growth factor-beta1 in pulmonary fibrosis. PLoS One 10:e0121246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Yang M, Chen ZY, Chen P, Guan CX, Xiang XD, Cai S, Chen Y, Fang X (2013). Expression and methylation of mitochondrial transcription factor a in chronic obstructive pulmonary disease patients with lung cancer. PLoS One 8:e82739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piantadosi CA, Carraway MS, Babiker A, Suliman HB (2008). Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res 103:1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Vigie P, Youle RJ (2018). Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr Biol 28:R170–R185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouwels SD, Faiz A, den Boef LE, Gras R, van den Berge M, Boezen HM, Korstanje R, Ten Hacken NHT (2017). Genetic variance is associated with susceptibility for cigarette smoke-induced DAMP release in mice. AmJ Physiol Lung Cell Mol Biol 313:L559–L580 [DOI] [PubMed] [Google Scholar]
- Priya DK, Gayathri R, Gunassekaran G, Murugan S, Sakthisekaran D (2011). Chemopreventive role of sulforaphane by upholding the GSH redox cycle in pre- and post-initiation phases of experimental lung carcinogenesis. Asian Pac J Cancer Prevent 12:103–110 [PubMed] [Google Scholar]
- Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I (2008). SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 177:861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S (2004). Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest 114:1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangasamy T, Guo J, Mitzner WA, Roman J, Singh A, Fryer AD, Yamamoto M, Kensler TW, Tuder RM, Georas SN, Biswal S (2005). Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J Exp Med 202:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapizzi E, Pinton P, Szabadkai G, Wieckowski MR, Vandecasteele G, Baird G, Tuft RA, Fogarty KE, Rizzuto R (2002). Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J Cell Biol 159:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner V, Starkov A, Matsiukevich D, Polin RA, Ten VS (2009). Mitochondrial dysfunction contributes to alveolar developmental arrest in hyperoxia-exposed mice. Am J Respir Cell Mol Biol 40:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray NB, Durairaj L, Chen BB, McVerry BJ, Ryan AJ, Donahoe M, Waltenbaugh AK, O’Donnell CP, Henderson FC, Etscheidt CA, McCoy DM, Agassandian M, Hayes-Rowan EC, Coon TA, Butler PL, Gakhar L, Mathur SN, Sieren JC, Tyurina YY, Kagan VE, McLennan G, Mallampalli RK (2010). Dynamic regulation of cardiolipin by the lipid pump Atp8b1 determines the severity of lung injury in experimental pneumonia. Nat Med 16:1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL (2012). Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 26:2175–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remels AH, Schrauwen P, Broekhuizen R, Willems J, Kersten S, Gosker HR, Schols AM (2007). Peroxisome proliferator-activated receptor expression is reduced in skeletal muscle in COPD. Eur Respir J 30:245–252 [DOI] [PubMed] [Google Scholar]
- Ribas V, Garcia-Ruiz C, Fernandez-Checa JC (2014). Glutathione and mitochondria. Front Pharmacol 5:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimessi A, Bezzerri V, Patergnani S, Marchi S, Cabrini G, Pinton P (2015). Mitochondrial Ca2+-dependent NLRP3 activation exacerbates the Pseudomonas aeruginosa-driven inflammatory response in cystic fibrosis. Nat Comm 6:6201. [DOI] [PubMed] [Google Scholar]
- Riteau N, Gasse P, Fauconnier L, Gombault A, Couegnat M, Fick L, Kanellopoulos J, Quesniaux VF, Marchand-Adam S, Crestani B, Ryffel B (2010). Extracellular ATP is a danger signal activating P2X7 receptor in lung inflammation and fibrosis. Am J Respir Crit Care Med 182:774–783 [DOI] [PubMed] [Google Scholar]
- Ryan JJ, Marsboom G, Fang YH, Toth PT, Morrow E, Luo N, Piao L, Hong Z, Ericson K, Zhang HJ, Han M, Haney CR, Chen CT, Sharp WW, Archer SL (2013). PGC1alpha-mediated mitofusin-2 deficiency in female rats and humans with pulmonary arterial hypertension. Am J Respir Crit Care Med 187:865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo IG, Kwak MK (2018). Regulatory crosstalk between the oxidative stress-related transcription factor Nfe2l2/Nrf2 and mitochondria. Toxicol App Pharmacol 359:24–33 [DOI] [PubMed] [Google Scholar]
- Ryter SW, Rosas IO, Owen CA, Martinez FJ, Choi ME, Lee CG, Elias JA, Choi AMK (2018). Mitochondrial Dysfunction as a Pathogenic Mediator of Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. Ann Am Thorac Soc 15(Supplement_4):S266–S272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C, Sun H, Gulati M, Herazo-Maya JD, Chen Y, Osafo-Addo A, Brandsdorfer C, Winkler J, Blaul C, Faunce J, Pan H, Woolard T, Tzouvelekis A, Antin-Ozerkis DE, Puchalski JT, Slade M, Gonzalez AL, Bogenhagen DF, Kirillov V, Feghali-Bostwick C, Gibson K, Lindell K, Herzog RI, Dela Cruz CS, Mehal W, Kaminski N, Herzog EL, Trujillo G (2017). Extracellular Mitochondrial DNA Is Generated by Fibroblasts and Predicts Death in Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med 196:1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabouny R, Fraunberger E, Geoffrion M, Ng AC, Baird SD, Screaton RA, Milne R, McBride HM, Shutt TE (2017). The Keap1-Nrf2 Stress Response Pathway Promotes Mitochondrial Hyperfusion Through Degradation of the Mitochondrial Fission Protein Drp1. Antioxid Redox Signal 27:1447–1459 [DOI] [PubMed] [Google Scholar]
- Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, Ohishi M, Endo K, Uemura T, Nishito Y, Okuda S, Obata M, Kouno T, Imamura R, Tada Y, Obata R, Yasuda D, Takahashi K, Fujimura T, Pi J, Lee MS, Ueno T, Ohe T, Mashino T, Wakai T, Kojima H, Okabe T, Nagano T, Motohashi H, Waguri S, Soga T, Yamamoto M, Tanaka K, Komatsu M. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming (2016). p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun 7:12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapira AH (2012). Mitochondrial diseases. Lancet 379:1825–1834 [DOI] [PubMed] [Google Scholar]
- Schumacker PT, Gillespie MN, Nakahira K, Choi AM, Crouser ED, Piantadosi CA, Bhattacharya J (2014). Mitochondria in lung biology and pathology: more than just a powerhouse. Am J Physiol Lung Cell Mol Physiol 306:L962–L974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman M, Rojas M, Mora AL, Pardo A (2010). Aging and interstitial lung diseases: unraveling an old forgotten player in the pathogenesis of lung fibrosis. Semin Respir Crit Care Med 31:607–617 [DOI] [PubMed] [Google Scholar]
- Simmons JD, Lee YL, Mulekar S, Kuck JL, Brevard SB, Gonzalez RP, Gillespie MN, Richards WO (2013). Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg 258:591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan R, Stearns TM, Czachor A, Fukumoto J, Turn C, Westermann-Clark E, Breitzig M, Tan L, Lockey RF, King BL, Kolliputi N (2016). Global gene profiling of aging lungs in Atp8b1 mutant mice. Aging 8:2232–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom J, Xu B, Tian X, Chen QM (2016). Nrf2 protects mitochondrial decay by oxidative stress. FASEB J 30:66–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suliman HB, Kraft B, Bartz R, Chen L, Welty-Wolf KE, Piantadosi CA (2017). Mitochondrial quality control in alveolar epithelial cells damaged by S. aureus pneumonia in mice. Am J Physiol Lung Cell Mol Physiol 313:L699–L709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshbabu A, Bhandari V (2013). Targeting mitochondrial dysfunction in lung diseases: emphasis on mitophagy. Front Physiol 4:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe A, Hollins F, Gomez E, Saunders R, Doe C, Cooke M, Challiss RA, Brightling CE (2012). Increased nicotinamide adenine dinucleotide phosphate oxidase 4 expression mediates intrinsic airway smooth muscle hypercontractility in asthma. Am J Respir Crit Care Med 185:267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Michelakis ED (2014). The metabolic basis of pulmonary arterial hypertension. Cell Metab 19:558–573 [DOI] [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M (2011). Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16:123–140 [DOI] [PubMed] [Google Scholar]
- Talati M, Hemnes A (2015). Fatty acid metabolism in pulmonary arterial hypertension: role in right ventricular dysfunction and hypertrophy. Pulm Circ 5:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan XL, Moslehi R, Han W, Spivack SD (2009). Haplotype-tagging single nucleotide polymorphisms in the GSTP1 gene promoter and susceptibility to lung cancer. Cancer Detect Prev 32:403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Scollick C, Traore K, Yates M, Trush MA, Liby KT, Sporn MB, Yamamoto M, Kensler TW, Biswal S (2006). Nrf2-dependent protection from LPS induced inflammatory response and mortality by CDDO-Imidazolide. Biochem Biophys Res Commun 351:883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, Sandebring A, Miller D, Maric D, Cedazo-Minguez A, Cookson MR (2011). DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet 20:40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, Roylance RR, Olpin S, Bevan S, Barker K, Hearle N, Houlston RS, Kiuru M, Lehtonen R, Karhu A, Vilkki S, Laiho P, Eklund C, Vierimaa O, Aittomaki K, Hietala M, Sistonen P, Paetau A, Salovaara R, Herva R, Launonen V, Aaltonen LA, Multiple Leiomyoma C (2002). Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 30:406–410 [DOI] [PubMed] [Google Scholar]
- Traver G, Mont S, Gius D, Lawson WE, Ding GX, Sekhar KR, Freeman ML (2017). Loss of Nrf2 promotes alveolar type 2 cell loss in irradiated, fibrotic lung. Free rRadic Biol Med 112:578–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trian T, Benard G, Begueret H, Rossignol R, Girodet PO, Ghosh D, Ousova O, Vernejoux JM, Marthan R, Tunon-de-Lara JM, Berger P (2007). Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J Exp Med 204:3173–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uruno A, Furusawa Y, Yagishita Y, et al. (2013). The Keap1-Nrf2 system prevents onset of diabetes mellitus. Mol Cell Biol 33:2996–3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venereau E, Ceriotti C, Bianchi ME (2015). DAMPs from Cell Death to New Life. Front Immunol 6:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J (2006). Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J 395:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AS, Xu Y, Zhang ZW, Lu BB, Yin X, Yao AJ, Han LY, Zou ZQ, Li Z, Zhang XH (2017a). Sulforaphane protects MLE-12 lung epithelial cells against oxidative damage caused by ambient air particulate matter. Food Funct 8:4555–4562 [DOI] [PubMed] [Google Scholar]
- Wang J, Liu Z, Hu T, Han L, Yu S, Yao Y, Ruan Z, Tian T, Huang T, Wang M, Jing L, Nan K, Liang X (2017b). Nrf2 promotes progression of non-small cell lung cancer through activating autophagy. Cell Cycle 16:1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA (2007). Identification of polymorphic antioxidant response elements in the human genome. Hum Mol Genet 16:1188–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O (1956). On respiratory impairment in cancer cells. Science 124:269–270 [PubMed] [Google Scholar]
- Wise RA, Holbrook JT, Criner G, Sethi S, Rayapudi S, Sudini KR, Sugar EA, Burke A, Thimmulappa R, Singh A, Talalay P, Fahey JW, Berenson CS, Jacobs MR, Biswal S, Broccoli Sprout Extract Trial Research G (2016). Lack of Effect of Oral Sulforaphane Administration on Nrf2 Expression in COPD: A Randomized, Double-Blind, Placebo Controlled Trial. PLoS One 11:e0163716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Koeck T, Lara AR, Neumann D, DiFilippo FP, Koo M, Janocha AJ, Masri FA, Arroliga AC, Jennings C, Dweik RA, Tuder RM, Stuehr DJ, Erzurum SC (2007). Alterations of cellular bioenergetics in pulmonary artery endothelial cells. Proc Natl Acad Sci USA 104:1342–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Chung S, Hwang JW, Rajendrasozhan S, Sundar IK, Dean DA, McBurney MW, Guarente L, Gu W, Ronty M, Kinnula VL, Rahman I (2012). SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest 122:2032–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A, Gill R, Mailloux RJ (2019). Protein S-glutathionylation: The linchpin for the transmission of regulatory information on redox buffering capacity in mitochondria. Chem Biol Interact 299:151–162 [DOI] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, Shen Y, Pervouchine DD, Djebali S, Thurman RE, Kaul R, Rynes E, Kirilusha A, Marinov GK, Williams BA, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See LH, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K, Bender MA, Zhang M, Byron R, Groudine MT, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu YC, Rasmussen MD, Bansal MS, Kellis M, Keller CA, Morrissey CS, Mishra T, Jain D, Dogan N, Harris RS, Cayting P, Kawli T, Boyle AP, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi VS, Cline MS, Erickson DT, Kirkup VM, Learned K, Sloan CA, Rosenbloom KR, Lacerda de Sousa B, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent WJ, Ramalho Santos M, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo PJ, Wilken MS, Reh TA, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds AP, Neph S, Humbert R, Hansen RS, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler EE, Orkin SH, Levasseur D, Papayannopoulou T, Chang KH, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y, Weiss MJ, Blobel GA, Cao X, Zhong S, Wang T, Good PJ, Lowdon RF, Adams LB, Zhou XQ, Pazin MJ, Feingold EA, Wold B, Taylor J, Mortazavi A, Weissman SM, Stamatoyannopoulos JA, Snyder MP, Guigo R, Gingeras TR, Gilbert DM, Hardison RC, Beer MA, Ren B, Mouse EC (2014). A comparative encyclopedia of DNA elements in the mouse genome. Nature 515:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Yang X, Ning P, Xi X, Yu H, Feng Y, Shao R, Meng X (2018). A mitochondria-targeted ratiometric two-photon fluorescent probe for detecting intracellular cysteine and homocysteine. Talanta 178:24–30 [DOI] [PubMed] [Google Scholar]
- Yun JH, Morrow J, Owen CA, Qiu W, Glass K, Lao T, Jiang Z, Perrella MA (2017). Transcriptomic Analysis of Lung Tissue from Cigarette Smoke-Induced Emphysema Murine Models and Human Chronic Obstructive Pulmonary Disease Show Shared and Distinct Pathways. Am J Respir Cell Mol Biol 57:47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Leir SH, Harris A (2015). Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am J Respir Cell Mol Biol 52:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Wu AY, Yu X, et al. (2018). Microdomain elements of airway smooth muscle in calcium regulation and cell proliferation. J Physiol Pharmacol 69:151–163 [DOI] [PubMed] [Google Scholar]
- Zhou W, Freed CR (2005). DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem 280:43150–43158 [DOI] [PubMed] [Google Scholar]