Abstract
Ethanol exposure during prenatal development causes fetal alcohol spectrum disorder (FASD), the most frequent preventable birth defect and neurodevelopmental disability syndrome. The molecular targets of ethanol toxicity during development are poorly understood. Developmental stages surrounding gastrulation are very sensitive to ethanol exposure. To understand the effects of ethanol on early transcripts during embryogenesis, we treated zebrafish embryos with ethanol during pre-gastrulation period and examined the transcripts by Affymetrix GeneChip microarray before gastrulation. We identified 521 significantly dysregulated genes, including 61 transcription factors in ethanol-exposed embryos. Sox2, the key regulator of pluripotency and early development was significantly reduced. Functional annotation analysis showed enrichment in transcription regulation, embryonic axes patterning, and signaling pathways, including Wnt, Notch and retinoic acid. We identified all potential genomic targets of 25 dysregulated transcription factors and compared their interactions with the ethanol-dysregulated genes. This analysis predicted that Sox2 targeted a large number of ethanol-dysregulated genes. A gene regulatory network analysis showed that many of the dysregulated genes are targeted by multiple transcription factors. Injection of sox2 mRNA partially rescued ethanol-induced gene expression, epiboly and gastrulation defects. Additional studies of this ethanol dysregulated network may identify therapeutic targets that coordinately regulate early development.
Subject terms: Gastrulation, Embryology
Introduction
Fetal alcohol spectrum disorder (FASD) is caused by the exposure to ethanol during prenatal developmental1–3. FASD patients display a range of morphological deformities and neurological deficits, including characteristic craniofacial dysmorphology, cognitive impairment, sensory defects, motor disabilities and organ deformities. A recent meta-analysis of FASD among children and youth showed the prevalence is approximately 0.8% globally, but it exceeds 1% in 76 countries4. The World Health Organization (WHO) European Region has the highest prevalence of FASD (1.98%) followed by the WHO Region of Americas (0.88%)4. Among all the countries studied to date, FASD is the most prevalent in South Africa where the prevalence is as high as 11.1%4. FASD prevalence is notably higher among special populations, for example, low socioeconomic status populations5,6, children in orphanages, people in psychiatric care etc.4.
Despite various proposed mechanisms to explain FASD etiology, the molecular targets of ethanol toxicity during development are poorly understood. Conception through gastrulation are sensitive periods for ethanol-induced defects7,8. During this period stem and progenitor cells transition from pluripotency to one of the three germ layers, and the cells undergo coordinated movements to organize the body plan9,10. These effects are regulated transcriptionally, for example, through the maternal to zygotic transition and the pluripotency transcriptional circuit. Since mammalian embryos develop inside their mother, it is difficult to study the effects of ethanol during gastrulation.
The zebrafish is an outstanding model to study early stages of embryogenesis because zebrafish produce hundreds of embryos synchronized at the same developmental stage and the embryos develop outside their mother9,11. There are morphological differences in early development stages between fish and humans. However, developmental gene expression networks are highly conserved from fish to human12. Zebrafish is an established model for the study of embryonic ethanol-exposure effects on development and its functional consequences, providing insights into the potential mechanisms of ethanol teratogenicity13–15.
Ethanol treatment of zebrafish embryos from 2 to 24 hours post-fertilization (hpf) reproducibly causes robust FASD-like defects, including craniofacial, cardiac, and neural defects14–21. This is a model for chronic ethanol exposure during early stages of pregnancy, when mothers may not know they are pregnant and may continue to drink alcohol. Our previous studies provided evidence of critical signaling dysregulation during organogenesis, including BMP, Notch, Wnt, and retinoic acid, which leads to heart and eye defects18,20. Phenotypic differences between ethanol-treated and untreated embryos were first detected during gastrulation, when ethanol-treated embryos displayed reduced epiboly progression22–24. Our studies examining changes in gene expression in ethanol-exposed embryos at mid-gastrulation (8 hpf) using microarray gene expression analysis identified various dysregulated genes including cell adhesion molecule Protocadherin-18a22. Defects in epiboly and gastrulation cell movements in ethanol-exposed embryos resembled the phenotype of embryos deficient in cadherin cell adhesion25. Protocadherin-18a was reduced after ethanol exposure, and injection of mRNA encoding protocadherin-18a partially rescued epiboly progression, cellular morphology of the enveloping layer cells during gastrulation, and convergence-extension of the anterior-posterior axes22. However, the ethanol dysregulated genes identified during mid-gastrulation could include indirect effects of ethanol exposure. We hypothesize that developmental signaling pathway defects seen during morphogenesis and organogenesis represent pleiotropic effects of ethanol on gene expression patterns that begin at the earliest stages of embryogenesis, when the embryo is first exposed to ethanol.
Gastrulation is a highly sensitive period for ethanol-induced defects7,8. In humans, gastrulation occurs at implantation and is the first time of exposure to maternal blood circulation and, hence, to maternal blood alcohol. Initial development is controlled by the maternally deposited proteins and mRNAs, until a burst of zygotic genes are transcribed in the early embryo prior to gastrulation that ultimately controls development26,27. In zebrafish, gastrulation starts around 5.25 hpf when the blastomere cells cover 50% of the yolk cells10,28. The initial zygotic transcriptional burst occurs ahead of gastrulation, during midblastula transition around 3 hpf27. The pluripotency factors Nanog, Pou5f1 (Oct4) and SoxB1, activate the zygotic program and pre-gastrulation development in zebrafish27. Zebrafish SoxB1 comprises of six sox genes: sox1a/1b/2/3/19a/19b29, of which, sox19b is supplied maternally30. These maternal factors play fundamental roles in activating transcription during early embryogenesis. Most of the initially activated genes guide early development27. The gene encoding the transcription factor Sox2 is one of the earliest zygotic genes activated (around 30% epiboly or 4.3 hpf)30. Sox2 plays critical roles in early vertebrate development by maintaining pluripotency and promoting differentiation later. This study examined the effects of ethanol on early transcripts before gastrulation. Affymetrix GeneChip microarray was performed and ethanol dysregulated genes that include critical transcription factors were identified. A gene regulatory network involving transcription factors and their target genes was identified. Experiments revealed significant reduction in sox2 transcripts and dysregulation of Sox2 target genes after ethanol exposure. Injecting sox2 mRNA partially rescued ethanol defects in early zebrafish embryos, showing an important role in FASD genesis.
Results
Ethanol exposure during blastula period perturbs gene expression patterns prior to gastrulation
Affymetrix GeneChip microarray analysis comparing control embryos to those treated with ethanol from 2 to 4.5 hpf (cleavage and pre-gastrulation stages) showed statistically significant changes of expression of many genes critical for embryogenesis (Supplementary Table S1) including sox genes, Notch ligands and Hairy/E(spl)-related (her) genes. To validate microarray results, a subset of genes were examined by either qPCR or by in situ hybridization. Downregulation of sox2 (array fold change −1.99, p < 0.0001), dlc (array fold change −1.82, p = 0.007) and her7 (array fold change −2.51, p < 0.001) genes at 4.5 hpf after ethanol exposure was confirmed by qPCR (Fig. 1A). In situ hybridization showed reduced staining for sox2, dlc and dld (array fold change −1.53, p < 0.001) at 4.5 hpf in ethanol-exposed embryos compared to control embryos (Fig. 1B–G).
Figure 1.
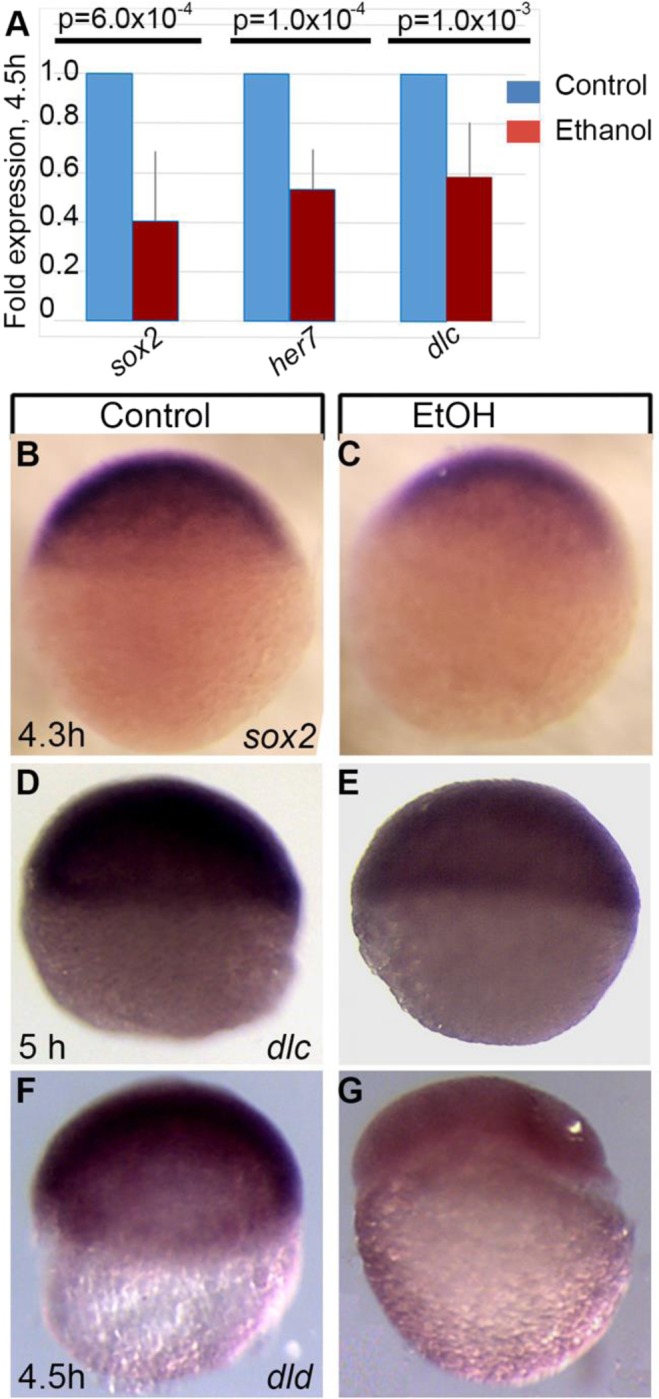
Ethanol exposure during mid-blastula transition changes the gene expression in zebrafish embryos. (A) Quantitative RT-PCR assays comparing transcript levels of sox2, her7, and dlc after ethanol treatment. Average fold change in expression was calculated from at least 3 independent experiments, with samples analyzed in triplicate. Samples were normalized to transcript levels for rsp15, and fold change for ethanol treated embryos was calculated by normalizing control levels to 1. (B–G) Whole mount in situ hybridization showed reduced expression of sox2 (B,C), dlc (D,E) and dld (F,G) in E100 embryos (C,E,G) compared to control (B,D,F).
There were significant changes in the expression of 651 probes, (absolute changes ≥ 1.25, FDR 0.15, p < 0.03) due to ethanol exposure (Supplementary Table S1). Out of those 651 probes, we were able to map Ensembl IDs for 534 probes, which correspond to 521 genes. Functional annotation analysis of ethanol dysregulated genes was done using DAVID that identified genes enriched in cellular processes, including transcription regulation and gene expression; DNA recombination; cell division and microtubule-based movement; cell-cell adhesion; and carbohydrate metabolic processes. Genes enriched in developmental processes, including dorso-ventral and anterior-posterior axes formation, cerebellum, somite, and optic fissure development were also detected in DAVID analysis. Dysregulated genes were enriched in Wnt, Notch, and retinoic acid signaling pathways (Table 1).
Table 1.
Gene ontology analysis of ethanol-dysregulated genes.
| Category | Term | Count | p-value |
|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0006355~regulation of transcription, DNA-templated | 45 | 0.016 |
| GOTERM_BP_DIRECT | GO:0001756~somitogenesis | 9 | 0.001 |
| GOTERM_BP_DIRECT | GO:0005975~carbohydrate metabolic process | 9 | 0.062 |
| GOTERM_BP_DIRECT | GO:0031101~fin regeneration | 8 | 0.001 |
| GOTERM_BP_DIRECT | GO:0009953~dorsal/ventral pattern formation | 7 | 0.050 |
| GOTERM_BP_DIRECT | GO:0016055~Wnt signaling pathway | 7 | 0.088 |
| GOTERM_BP_DIRECT | GO:0007219~Notch signaling pathway | 6 | 0.006 |
| GOTERM_BP_DIRECT | GO:0009952~anterior/posterior pattern specification | 6 | 0.007 |
| GOTERM_BP_DIRECT | GO:0007018~microtubule-based movement | 6 | 0.042 |
| GOTERM_BP_DIRECT | GO:0000278~mitotic cell cycle | 6 | 0.046 |
| GOTERM_BP_DIRECT | GO:0001889~liver development | 6 | 0.059 |
| GOTERM_BP_DIRECT | GO:0021549~cerebellum development | 5 | 0.001 |
| GOTERM_BP_DIRECT | GO:0016337~single organismal cell-cell adhesion | 5 | 0.030 |
| KEGG_PATHWAY | dre00561:Glycerolipid metabolism | 5 | 0.066 |
| KEGG_PATHWAY | dre03320:PPAR signaling pathway | 5 | 0.094 |
| GOTERM_BP_DIRECT | GO:0048384~retinoic acid receptor signaling pathway | 4 | 0.004 |
| GOTERM_BP_DIRECT | GO:0001878~response to yeast | 4 | 0.019 |
| GOTERM_BP_DIRECT | GO:0018279~protein N-linked glycosylation via asparagine | 4 | 0.041 |
| GOTERM_BP_DIRECT | GO:0006310~DNA recombination | 4 | 0.063 |
| GOTERM_BP_DIRECT | GO:0001757~somite specification | 3 | 0.038 |
| GOTERM_BP_DIRECT | GO:0061386~closure of optic fissure | 3 | 0.043 |
| GOTERM_BP_DIRECT | GO:0009948~anterior/posterior axis specification | 3 | 0.062 |
| GOTERM_BP_DIRECT | GO:0001574~ganglioside biosynthetic process | 3 | 0.076 |
| GOTERM_BP_DIRECT | GO:0006094~gluconeogenesis | 3 | 0.076 |
| GOTERM_BP_DIRECT | GO:0036342~post-anal tail morphogenesis | 3 | 0.098 |
| GOTERM_BP_DIRECT | GO:0071539~protein localization to centrosome | 2 | 0.070 |
| GOTERM_BP_DIRECT | GO:0045814~negative regulation of gene expression, epigenetic | 2 | 0.071 |
| GOTERM_BP_DIRECT | GO:0061056~sclerotome development | 2 | 0.093 |
| REACTOME_PATHWAY | Polyamine metabolic process:R-DRE-351202:R-DRE-351202 | 2 | 0.099 |
Category refers to the original database where the terms orient; term refers to the enriched term associated to the gene list; count refers to the total number of differentially expressed genes annotated to a given gene ontology term; the smaller the p-value, the more enriched the term.
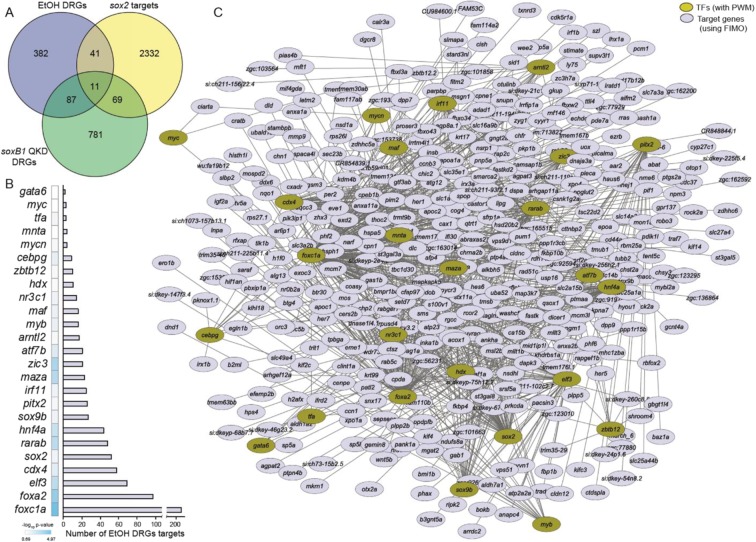
Among the 521 dysregulated genes, we identified 61 transcription factors (Table 2), including Sox2, a critical transcription factor. The expression of sox2 was significantly reduced after ethanol exposure. To identify Sox2 targets across the zebrafish genome, position weight matrixes for Sox2 were mapped within 2 kb upstream of transcription start sites of genes using find individual motif occurrences software31. Possible Sox2 targets were compared with the ethanol-dysregulated genes, which showed that 52 genes were common in both datasets (Fig. 2A). Transcriptome changes caused by SoxB1 knockdown (quadruple knockdown: sox2/3/19a/19b) at 30% epiboly (~4.7 hpf) were reported previously29. Results of this study were compared with ethanol dysregulated genes (4.5 hpf). We found 98 genes common in between SoxB1 knockdown dysregulated genes and ethanol dysregulated genes. Comparison of all three datasets showed 11 common genes in all these datasets (Fig. 2A). These data indicate that ethanol affects the expression of Sox2 and several Sox2 transcriptional targets.
Table 2.
Ethanol-dysregulated transcription factors.
| Gene Symbol | Fold change | p-value | PWM |
|---|---|---|---|
| tfa | −2.05 | 0.0016 | Available |
| sox2 | −1.99 | 0.0001 | Available |
| elf3 | −1.87 | 0.0003 | Available |
| si:ch211-222e23.7 | −1.78 | 0.0032 | Available |
| cdx4 | −1.62 | 0.0048 | Available |
| pitx2 | −1.62 | 0.0284 | Available |
| maf | −1.47 | 0.0015 | Available |
| foxc1a | −1.43 | 0.0002 | Available |
| zic3 | −1.32 | 0.0063 | Available |
| foxa2 | −1.31 | 0.0132 | Available |
| hnf4a | −1.30 | 0.0155 | Available |
| gata6 | −1.26 | 0.0052 | Available |
| LOC797948 | 1.28 | 0.0222 | Available |
| atf7b | 1.27 | 0.0082 | Available |
| mycn | 1.30 | 0.0100 | Available |
| arntl2 | 1.32 | 0.0022 | Available |
| rarab | 1.36 | 0.0101 | Available |
| mycb | 1.37 | 0.0076 | Available |
| cebpg | 1.41 | 0.0082 | Available |
| wu:fb82f02 | 1.46 | 0.0070 | Available |
| maza | 1.50 | 0.0219 | Available |
| myb | 1.57 | 0.0003 | Available |
| nr3c1 | 1.70 | 0.0053 | Available |
| sox9b | 1.72 | 0.0005 | Available |
| irf11 | 1.84 | 0.0030 | Available |
| her7 | −2.93 | 0.0003 | Not Available |
| msgn1 | −2.35 | 0.0012 | Not Available |
| her1 | −2.10 | 0.0126 | Not Available |
| zgc:136639 | −1.83 | 0.0026 | Not Available |
| sp5l | −1.82 | 0.0014 | Not Available |
| otx1a | −1.69 | 0.0138 | Not Available |
| irx1b | −1.56 | 0.0193 | Not Available |
| irx3a | −1.49 | 0.0119 | Not Available |
| nr0b2a | −1.46 | 0.0011 | Not Available |
| eve1 | −1.41 | 0.0004 | Not Available |
| pknox1.1 | −1.39 | 0.0005 | Not Available |
| hes6 | −1.35 | 0.0062 | Not Available |
| lhx1a | −1.34 | 0.0123 | Not Available |
| msxb | −1.34 | 0.0091 | Not Available |
| nr0b2a | −1.32 | 0.0045 | Not Available |
| sp5 | −1.27 | 0.0186 | Not Available |
| LOC407678 | 1.26 | 0.0141 | Not Available |
| etv5a | 1.26 | 0.0235 | Not Available |
| zgc:162349 | 1.27 | 0.0002 | Not Available |
| zgc:165515 | 1.27 | 0.0058 | Not Available |
| zgc:162349 | 1.27 | 0.0009 | Not Available |
| zorba | 1.28 | 0.0237 | Not Available |
| zhx3 | 1.30 | 0.0050 | Not Available |
| si:rp71-1g18.1 | 1.30 | 0.0134 | Not Available |
| znf277 | 1.30 | 0.0185 | Not Available |
| lrrfip1a | 1.33 | 0.0045 | Not Available |
| si:ch211-119o8.6 | 1.33 | 0.0086 | Not Available |
| lrrfip2 | 1.35 | 0.0099 | Not Available |
| her5 | 1.36 | 0.0023 | Not Available |
| rcor2 | 1.37 | 0.0075 | Not Available |
| pias4l | 1.38 | 0.0259 | Not Available |
| terf1 | 1.46 | 0.0108 | Not Available |
| klf2a | 1.46 | 0.0034 | Not Available |
| LOC100149164 | 1.48 | 0.0037 | Not Available |
| etv5a | 1.48 | 0.0261 | Not Available |
| si:dkeyp-68b7.7 | 1.50 | 0.0046 | Not Available |
| LOC797322 | 1.66 | 0.0283 | Not Available |
| tsc22d2 | 1.96 | 0.0028 | Not Available |
| zgc:77060 | 2.51 | 0.0109 | Not Available |
Figure 2.
Ethanol induced gene expression changes during zygotic genome activation. (A) Venn diagram shows overlapping of ethanol dysregulated genes identified in the Affymetrix GeneChip microarray analysis of control and ethanol treated embryos at 4.5 hpf with potential Sox2 targets and the genes differentially expressed in quadruple knockdown of SoxB1 factors, which includes Sox2 (Soxb1 QKD targets). (B) Twenty five of the ethanol-dysregulated transcription factors showing the enrichment of their targets in our ethanol-dysregulated gene set. (C) Transcription factor-target gene network visualizing using cytoscape shows interactions among the transcription factors (gold circles) and their target gene (blue circles) in our dataset. Ethanol dysregulated targets of Sox2 are co-shared by other dysregulated transcription factors.
To predict the possible binding sites of other ethanol-dysregulated transcription factors across the zebrafish genome, we explored the available position weight matrixes (TRANSFAC). Position weight matrixes were found for 24 of the 60 other ethanol-dysregulated transcription factors. Target genes of these 24 transcription factors were predicted by mapping position weight matrixes within 2 kb upstream of the start site of genes across zebrafish genome. Predicted targets of the dysregulated transcription factors show that many ethanol-dysregulated genes are targets of these transcription factors. We compared the predicted targets of these dysregulated transcription factors and examined for the enrichment of ethanol dysregulated genes by computing hypergeometric probability. This transcription factor-target gene interaction analysis identified 827 interactions that include 25 transcription factors targeting 423 dysregulated genes. Individual interaction counts for each transcription factor is listed in Table 3. The enrichment of ethanol-dysregulated targets over all possible genomic targets for a given transcription factor plotted as a bar graph is shown in Fig. 2B. A network analysis was done using cytoscape software to visualize the interactions between the dysregulated transcription factors and the dysregulated targets. This analysis showed that 25 transcription factors target many of the same ethanol dysregulated genes, which identifies a potential ethanol-induced transcription factor-target gene regulatory network in the early embryo (Fig. 2C). As many of the genes dysregulated by ethanol exposure were targeted by multiple dysregulated transcription factors, these factors could produce synergistic ethanol dysregulation effects during early zebrafish development.
Table 3.
Ethanol-dysregulated transcription factors and the enrichment of their targets in the ethanol-dysregulated gene set.
| Transcription factors | Potential targets in the ethanol-dysregulated dataset | p-value | −log10 p-value |
|---|---|---|---|
| foxc1a | 207 | 0.0000 | 4.9689 |
| foxa2 | 97 | 0.0001 | 3.8315 |
| zic3 | 21 | 0.0006 | 3.2427 |
| maza | 23 | 0.0011 | 2.9524 |
| rarab | 48 | 0.0016 | 2.7962 |
| elf3 | 69 | 0.0019 | 2.7307 |
| hnf4a | 44 | 0.0034 | 2.4676 |
| atf7b | 21 | 0.0153 | 1.8144 |
| cdx4 | 58 | 0.0234 | 1.6299 |
| cebpg | 9 | 0.0312 | 1.5052 |
| myb | 16 | 0.0391 | 1.4078 |
| maf | 16 | 0.0454 | 1.3426 |
| irf11 | 25 | 0.0537 | 1.2701 |
| sox9b | 27 | 0.0540 | 1.2679 |
| nr3c1 | 14 | 0.0546 | 1.2631 |
| sox2 | 52 | 0.0567 | 1.2465 |
| tfa | 3 | 0.0739 | 1.1315 |
| pitx2 | 26 | 0.0793 | 1.1008 |
| arntl2 | 17 | 0.0952 | 1.0215 |
| mycn | 4 | 0.1035 | 0.9852 |
| zbtb12 | 10 | 0.1105 | 0.9568 |
| hdx | 11 | 0.1218 | 0.9143 |
| mnta | 4 | 0.1541 | 0.8121 |
Injection of sox2 mRNA partially rescues ethanol-induced epiboly, gastrulation and gene expression defects
Sox2 plays a critical role in pluiripotency and embryogenesis32,33. The pluripotency transcriptional circuit, which includes Sox2, regulates pre-gastrulation events in the zebrafish embryos27,29. Thus, Sox2 was selected for additional studies. To test the role of Sox2 activity in ethanol toxicity, 2–4 cell stage embryos were injected with 25 pg of synthetic sox2 mRNA to determine whether restoring this gene function remedies ethanol-induced defects. Epiboly progression was measured in the injected embryos after ethanol exposure and compared with controls. Untreated embryos reached 65.30 ± 5.5% epiboly at 8 hpf, but ethanol-treated embryos only reached 60.68 ± 4.2%, a significant delay (control vs. ethanol-treated, p < 0.05) (Fig. 3A,B,E). Average epiboly progression of sox2 mRNA injected embryos without ethanol treatment was 65.80 ± 3.6% at 8 hpf (Fig. 3C,E). Injection of sox2 mRNA with ethanol treatment rescued epiboly delay (64.39 ± 4.4%; control vs. sox2 mRNA + ethanol-treated, p = 0.72, ethanol-treated vs. sox2 mRNA + ethanol-treated, p < 0.05) (Fig. 3A–E).
Figure 3.
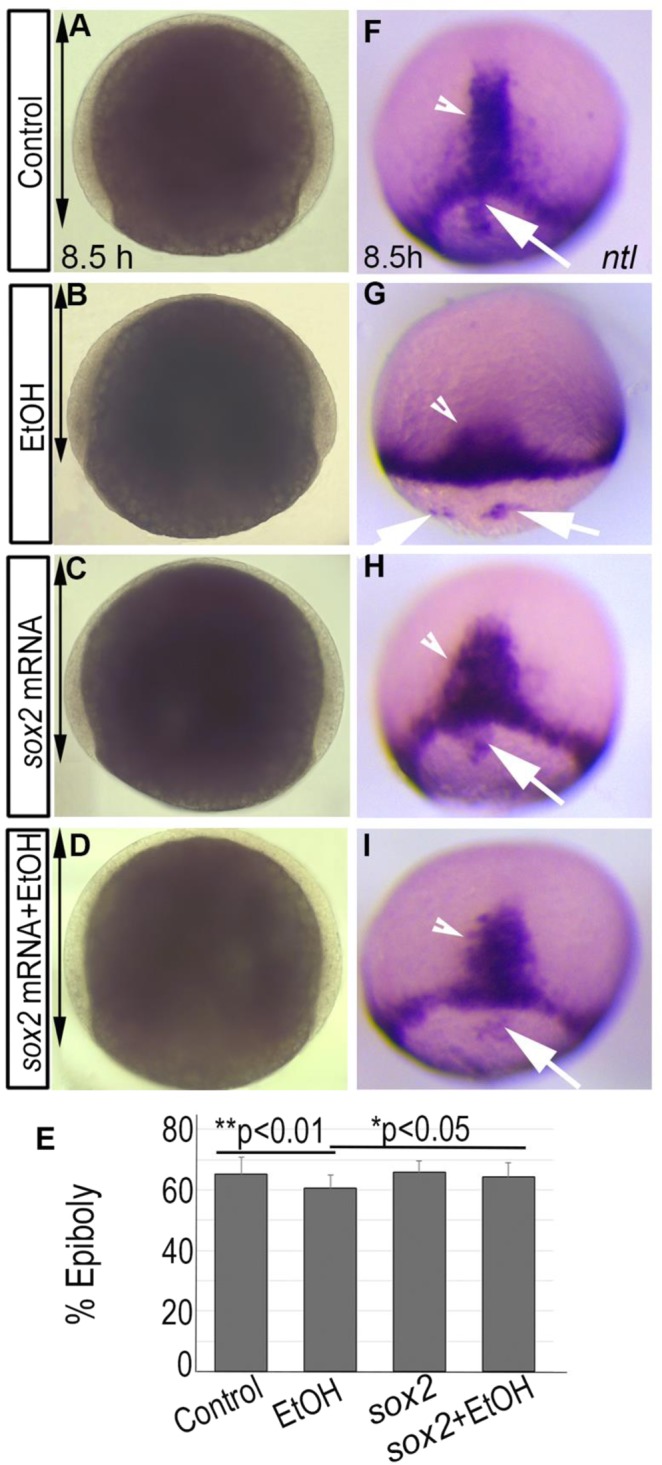
Ethanol induced epiboly and gastrulation defect was partially rescued by sox2 mRNA injection. (A–D) Bright field images showed reduced epiboly progression after ethanol exposure (B; double arrow), which was rescued by sox2 mRNA injection (D). (E) Graph shows the percentage of epiboly progression in control, ethanol-treated, sox2 mRNA injected, and sox2 mRNA + ethanol-treated injected embryos (see text for statistics). (F–I) In situ hybridization detecting ntl expression shows dorsal forerunner cells closely associated to the germ band in control, embryo (F), sox2 mRNA injected (H), and ethanol-treated + sox2 mRNA injected (I) embryos, and a dramatic separation and fragmentation of the dorsal forerunner cells from the germ band and from each other in the ethanol treated embryo (G).
To mark the axial mesendoderm, germ band (mesodermal cells at the leading-edge during epiboly progression) and the dorsal forerunner cells (a group of cells that migrate at the leading-edge of shield during gastrulation but do not involute) in control and treated embryos, ntl in situ hybridization was performed. Control embryos had dorsal forerunner cells closely associated with the germ ring. Dorsal forerunner cells were dissociated from one another and from the germ band in ethanol-treated embryos, which was partially rescued by sox2 mRNA injection (Fig. 3F–I). Functional annotation analysis detected dysregulation of genes involved in dorsal/ventral and anterior/posterior axes formation (Table 1). ntl staining confirmed that ethanol-exposed embryos had reduced convergence-extension of the axial mesendoderm cells compared to control embryos, producing shorter and wider axes. The convergence-extension defect was partially rescued by sox2 mRNA injection in the ethanol treated embryos (Fig. 3F–I).
The effects of sox2 mRNA injection on the expression of a few ethanol-dysregulated genes were analyzed. The expression level of dld was restored in sox2 mRNA injected, ethanol-exposed embryos as seen by dld in situ hybridization (Fig. 4A–D). Quantitative PCR was done to analyze the expression of sox2, her7, and dlc. The sox2 mRNA injection restored expression of sox2 that was downregulated in ethanol-treated embryos (Fig. 4E). The reduced expression of her7 and dlc in ethanol-treated embryos was significantly restored by sox2 mRNA injection in ethanol-treated embryos (ethanol-treated vs. sox2 mRNA + ethanol-treated: dlc; p < 0.01, her7; p < 0.01) (Fig. 4E). However, the expression levels of these genes in sox2 mRNA injected + ethanol-treated embryos were higher compared to control embryos (control vs. sox2 mRNA injected + ethanol-treated: dlc; p < 0.01, her7; p < 0.01) (Fig. 4E). Overall, sox2 mRNA injection partially rescued ethanol-induced early developmental defects.
Figure 4.
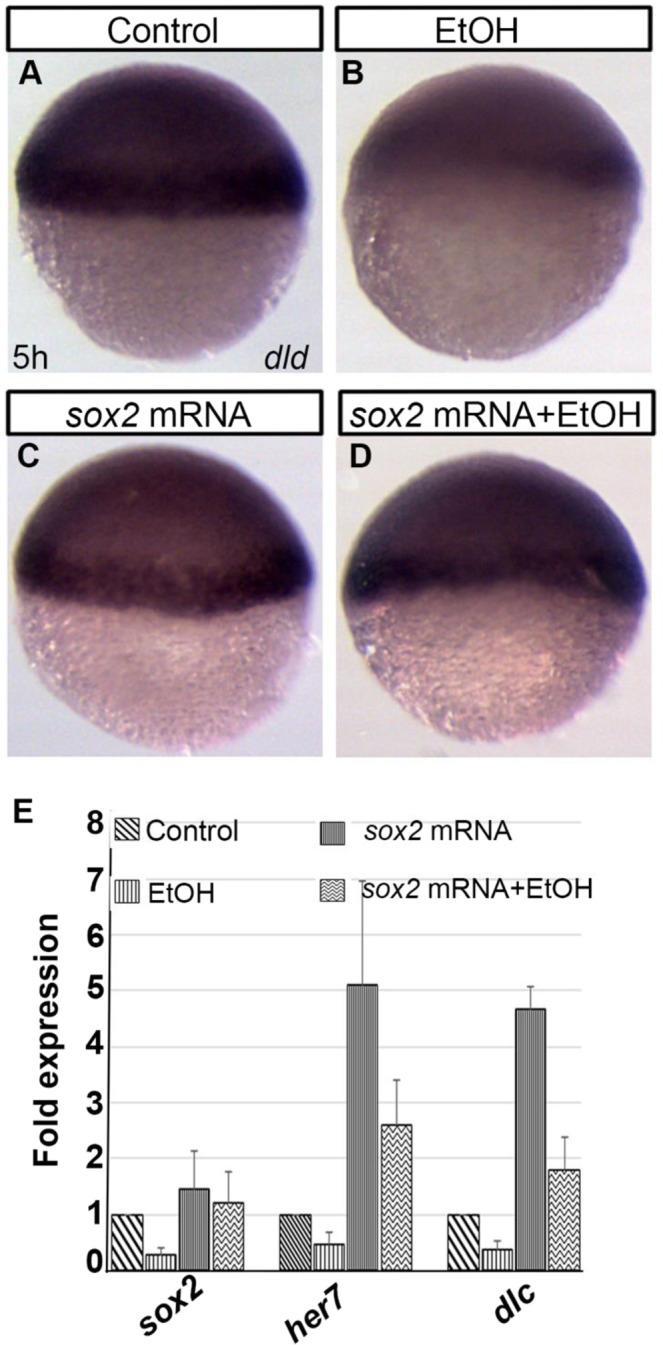
Ethanol induced gene expression changes was reversed by sox2 mRNA injection. (A–D) Whole mount ISH shows reduced dld expression after ethanol exposure, which was like control in sox2 mRNA injected and sox2 mRNA injected plus ethanol-treated embryos. (E) Quantitative PCR showed downregulation of sox2, her7, and dlc transcripts in the ethanol-treated embryos, which increased in the sox2 mRNA injected plus ethanol-treated embryos.
Discussion
Mouse and rat studies showed the association between prenatal ethanol exposure and gene expression changes in postnatal and adult stages34–37. This study examined ethanol-induced gene expression changes during embryogenesis before gastrulation. This is the first animal model study that identified the effects of ethanol on a master regulator, Sox2 that orchestrates embryogenesis, self-renewal, and pluripotency27,29,32. Ethanol exposure altered the expression of a large number of genes, which include other critical regulators of development. The differentially expressed genes are involved in various functions ranging from cellular processes, embryonic development to signaling pathways. This indicates possible multifactorial effects, which may include the alteration of epigenome by ethanol exposure, causing the changes in expression of critical genes.
Functional annotation analysis of the dysregulated genes identified enrichment of genes involved in mitotic nuclear division and microtubule-based movements. In fact, our previous study identified large multinucleated enveloping layer cells in 8 hpf ethanol-treated embryos and fragmentation of yolk microtubules18, which support current findings. The cell adhesion defects that was observed previously at 8 hpf18 were detected at 4.5 hpf ethanol-treated embryos, suggesting continuous defects in cell-to-cell communication and cell movements in those embryos. Interestingly, ethanol-sensitive signaling pathways detected during early embryogenesis were Wnt, Notch, and retinoic acid. Ethanol-induced dysregulation of retinoic acid signaling pathway was reported earlier16,17,19,38–41. Our studies examining the heart and the eye in ethanol-treated embryos identified disruption of Wnt, Notch, retinoic acid and Bmp signaling pathways during organogenesis17–20. This study showed that those signaling defects initiate early in ethanol-exposed embryos and continue to have their detrimental effects at later stages of development.
This microarray analysis detected a reduction of sox2 expression. Ethanol-induced effects on stoichiometry of Sox2 and Oct4 was reported during differentiation of mouse embryonic stem cell42. Sox2 and other Sox B1 type transcriptional regulators control a wide range of developmental effectors, including pcdh18a (gastrulation movement), neurog1, hesx1, and zic1 (neural differentiation), oep, and shh (neural patterning)29. Our previous Affymetrix GeneChip microarray study (GEO accession: GSE48380) comparing genes in control and ethanol-exposed embryos during mid-gastrulation identified significant dysregulation of all these genes after ethanol exposure at 8 hpf15,22. Reduction of Sox2 prior to gastrulation might be responsible for the dysregulation of these genes, interfering with gastrulation and other developmental events. Notably, convergence-extension defects seen in the ethanol-exposed embryos was also reported in the SoxB1 knockdown embryos, which display similar wedge-shaped ntl expression pattern. Presence of precise amount of sox2 transcripts is essential for normal development of the embryo. Injection of more than 30 pg of sox2 mRNA into the embryos caused delayed epiboly progression and other developmental defects, which caused the death of injected embryos (data not shown). Injection of 25 pg of sox2 mRNA partially rescued ethanol-induced epiboly defects. Injection of less than 25 pg of sox2 mRNA gave weaker rescue effects. sox2 mRNA injection raised the her7 and dlc transcript levels in ethanol-exposed embryos. However, the her7 and dlc transcript levels in sox2 mRNA-injected + ethanol-exposed embryos were significantly higher than the transcript levels in control embryos. Although sox2 mRNA injection did not fully rescue her7 and dlc expression in ethanol-exposed embryos, the results support their transcriptional regulation by Sox2 and ethanol. The transcription factor-target gene regulatory network (Fig. 2C) shows that her7 is regulated not only by Sox2, but also by Mnt, Cebpg3, and Atf7. Similarly, the expression of dlc is regulated by Sox2, Elf3, and Maz.
Transcription factor-target gene interaction analysis showed that the number of potential targets among the ethanol-induced dysregulated genes are more for Foxc1a and Foxa2 (members of forkhead transcription factors), Elf3 (a member of the E26 transformation specific family of transcription factors), and Cdx4 (a member of caudal gene family) than for Sox2. However, ethanol-induced gene expression change for sox2 was higher (and possibly playing a more significant role) than foxc1a, foxa2, elf3 and cdx4. Foxc1a plays crucial role in somitogenesis, cardiovascular and retina development during embryogenesis43–48. Foxa transcription factors are essential for developmental of dorsal axis structures including prechordal plate, notochord, hypochord, and floor plate49–51. A growing body of research shows that Elf3 plays significant roles in the development of cancer52–56. Although poorly understood, there is evidence that Elf3 is important during development. A null mutation of Elf3 caused the death of about 30% of mice in utero57,58. Another Sox family gene sox9b was also detected as the ethanol-dysregulated gene in our study. Sox9 is involved in many developmental processes including craniofacial, heart, brain and retinal development in mammal59. Zebrafish has two homologues of sox9, sox9a and sox9b. Both Sox9a and Sox9b play roles on neural, cardiac, and cartilage development Zebrafish60–64. Network analysis showed that many of these dysregulated transcription factors interact with each other, which suggests that the ethanol-induced developmental defects were the combined and perhaps synergistic effects of multiple regulators and that manipulation of one gene would not lead to complete rescue of ethanol-induced defects. Future study is needed to analyze the potential interaction between combinations of genes coordinately disrupt early development, contributing to ethanol-induced defects.
Methods
Zebrafish husbandry
Zebrafish (Danio rerio) ABTL strain was raised and maintained under standard laboratory conditions65 following Indiana University Policy on Animal Care and Use. The use of zebrafish adults for breeding, embryo collection and embryo experiments were approved by the campus animal care and use ethics committee: IUPUI School of Science Institutional Animal Care and Use Committee (IACUC).
Ethanol treatment
Embryos were kept in embryo medium after fertilization. At 2 hpf, embryos were divided into two groups. One group was transferred to embryo medium containing 100 mM ethanol (E100) and the other group was kept in embryo medium without ethanol (control).
RNA isolation, microarray analysis, and bioinformatics approach
Ethanol-treated and untreated embryos were incubated until 4.5 hpf. At this point, total RNA was extracted from pools of 20 control and 20 E100 embryos using TRIzol reagent (Sigma, St Louis, MO, USA). Seven independent experiments were done. RNA samples were purified by passing through the Qiagen RNAeasy column (Cat. No. 74104). The RNA quality was examined by Agilent Bioanalyzer RNA Nanochip (Agilent Technologies, Santa Clara, CA, USA). The RNA integrity number (RIN) for one of the ethanol samples was 2.5, so that experiment (both treated and control) was not further analyzed. For the remaining 12 samples (from 6 experiments) RIN ≥ 8.2. Standard protocol for the Affymetrix 3′IVT Express kit (Affymetrix, Santa Clara, CA, USA) was followed to label the samples starting with 100 ng of total RNA. The 12 samples were each hybridized to a Zebrafish Genome Array (Affymetrix) for 17 h, washed, stained and scanned following the standard protocol; all 12 were handled in parallel as a single batch. Arrays were visually scanned for abnormalities or defects; none were found.
Affymetrix gene expression console software was used to generate MAS566 signals and detection calls; arrays were scaled to a target of 1000. Only those probe sets that had a MAS5 signal fraction present ≥ 0.50 in at least one of the two treatments were analyzed67. MAS5 signals were imported into Partek Genomics Suite (Partek, Inc., St Louis, MO, USA) and log2 transformed. These log2 transformed signals were used for principal components analysis, hierarchical clustering and signal histograms to determine if there were any outlier arrays, and no outliers were detected. A 2-way ANOVA with factors for treatment (ethanol vs., control) and independent experiment (random effect) was used to analyze log2 transformed signals. This analysis indicated that the embryo batch was indeed significant. The False Discovery Rate (FDR) was calculated using the Storey qvalue method68. Microarray data were deposited in the NCBI GEO database, accession number GSE145574.
A subset of the differentially expressed probes with absolute changes ≥ 1.25 were included in downstream analysis (FDR 0.15; p < 0.03). We performed function annotation analysis of these ethanol dysregulated probes using DAVID69. Additionally, we annotated the transcription factors for these differentially expressed probe IDs using AnimalTFDB database70. For these annotated ethanol-dysregulated transcription factors, we obtained the position weight matrices available in TRANSFAC71 and searched for their occurrence using “find individual motif occurrences” algorithm31 to predict the target site within 2 kb upstream of start site of every gene in zebrafish genome (Zv11). Find individual motif occurrences computes a log-likelihood ratio score for the occurrence of each motif in a specific input sequence and hence enables the discovery of recognition sequences of transcription factors in the upstream regions of gene starts. Find individual motif occurrences computes converts these scores into p-values using dynamic programming generated at random genomic loci with user defined background frequencies for genomic alphabet (A, T, G, C) with false discovery rates. It provides a ranked list of motif occurrences per position weight matrices, each with an associated log-likelihood ratio score, p-value and other statistical metrics. Find individual motif occurrences computes based predicted targets of these dysregulated transcription factors were compared and examined for the enrichment of ethanol induced dysregulated targets by computing hypergeometric probability for the gene set enrichment. Resulting data was represented as networks using cytoscape72 network visualization software.
Quantitative PCR analysis
One µg of total RNA extracted from control and E100 embryos was reverse transcribed to cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA), and cDNA was diluted tenfold with RNase free water. Each 20 µl PCR reaction was performed with 1–4 µl of cDNA using Power SYBR Green PCR mix (Applied Biosystems/Life Technologies, Inc.) and 0.5 µM of each primer. Primer sets used are listed in the Supplementary Table S2. Three independent experiments were performed on the 7300 Real Time PCR System (Applied Biosystems), each in triplicate, using rsp15 as internal control. Relative expression was calculated as described73. Fold changes in gene expression was calculated using comparative CT method (ΔΔCT)73. Unpaired two-tailed student’s t-test was used for comparisons between control and ethanol treated groups using GraphPad software (GraphPad Software, La Jolla, CA, USA).
In situ hybridization
Whole-mount in situ hybridization of zebrafish embryos was performed using digoxigenin-labeled riboprobes for ntl, dlc, and dld. The riboprobes were synthesized using DIG RNA Labeling Kit (Roche, Indianapolis, IN, USA) according to manufacturer’s recommendations. For sox2 riboprobe, pCBA3-zf-sox2 plasmid was cut using BamHI restriction enzyme, and the Dig RNA probe was synthesized using T7 RNA polymerase. Images were collected using a Leica MZ12 microscope equipped with Leica DFC290 camera.
sox2 rescue experiments
mRNA was synthesized from a pCBA3-zf-sox2 vector29 using a SP6 mMessage mMachine kit (Ambion, Austin, TX, USA). Synthetic mRNA (25 pg/embryo) was injected into the embryos at the 2-cell stage. Injected and uninjected embryos were treated with or without 100 mM ethanol until analyzed. For epiboly measurement, embryos were fixed at 8 hpf, dechorionated and imaged focusing on enveloping cell layer at the embryo margin. Percent epiboly progression was calculated using Image J software. For gen expression analyses, embryos were dechorionated and total RNA was extracted at 4.3 hpf, and quantitative PCR was performed. One-way ANOVA and post hoc Tukey HSD for individual comparisons were used for analyses in rescue experiments.
Supplementary information
Acknowledgements
This work was supported by NIH/NIAAA R21 AA026711. Microarray study was carried out in the Center for Medical Genomics at Indiana University School of Medicine. We thank Dr. Yusuke Kamchi for providing pCBA3-zf-sox2 plasmid. We thank Drs. Ela K. Knapik and Lilianna Solnica-Krezel for providing us the probes for in situ experiments. We thank Jessica Howard, Saratalai Tinubu for helping us in preliminary experiments and the members of Marrs lab for helpful discussion.
Author contributions
S.S., H.J.E. and J.A.M. conceived of and designed the study. S.S. and J.N.M. acquired data. S.S., R.S. J.N.M., S.C.J., H.J.E. and J.A.M. analyzed and interpreted data. The manuscript was written and edited by S.S., R.S., S.C.J., H.J.E. and J.A.M.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-59043-x.
References
- 1.May PA, et al. Prevalence and characteristics of fetal alcohol spectrum disorders. Pediatrics. 2014;134:855–866. doi: 10.1542/peds.2013-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.May PA, et al. Prevalence of Fetal Alcohol Spectrum Disorders in 4 US Communities. Jama. 2018;319:474–482. doi: 10.1001/jama.2017.21896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.May PA, et al. Prevalence and epidemiologic characteristics of FASD from various research methods with an emphasis on recent in-school studies. Dev. Disabil. Res. Rev. 2009;15:176–192. doi: 10.1002/ddrr.68. [DOI] [PubMed] [Google Scholar]
- 4.Lange S, et al. Global Prevalence of Fetal Alcohol Spectrum Disorder Among Children and Youth: A Systematic Review and Meta-analysis. JAMA pediatrics. 2017;171:948–956. doi: 10.1001/jamapediatrics.2017.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abel EL. An update on incidence of FAS: FAS is not an equal opportunity birth defect. Neurotoxicology teratology. 1995;17:437–443. doi: 10.1016/0892-0362(95)00005-C. [DOI] [PubMed] [Google Scholar]
- 6.Abel, E. L. Fetal alcohol abuse syndrome. Plenum Press, New York (1998).
- 7.Haycock PC. Fetal alcohol spectrum disorders: the epigenetic perspective. Biol. Reprod. 2009;81:607–617. doi: 10.1095/biolreprod.108.074690. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert-Barness E. Teratogenic causes of malformations. Ann. Clin. laboratory Sci. 2010;40:99–114. [PubMed] [Google Scholar]
- 9.Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu. Rev. Cell developmental Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- 10.Solnica-Krezel L. Gastrulation in zebrafish–all just about adhesion? Curr. Opin. Genet. Dev. 2006;16:433–441. doi: 10.1016/j.gde.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Sarmah, S. & Marrs, J. A. Zebrafish as a Vertebrate Model System to Evaluate Effects of Environmental Toxicants on Cardiac Development and Function. International journal of molecular sciences17, 10.3390/ijms17122123 (2016). [DOI] [PMC free article] [PubMed]
- 12.Kalinka AT, Tomancak P. The evolution of early animal embryos: conservation or divergence? Trends Ecol. evolution. 2012;27:385–393. doi: 10.1016/j.tree.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes Y, Buckley DM, Eberhart JK. Diving into the world of alcohol teratogenesis: a review of zebrafish models of fetal alcohol spectrum disorder. Biochem. Cell Biol. = Biochim. et. biologie cellulaire. 2018;96:88–97. doi: 10.1139/bcb-2017-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lovely CB, Fernandes Y, Eberhart JK. Fishing for Fetal Alcohol Spectrum Disorders: Zebrafish as a Model for Ethanol Teratogenesis. Zebrafish. 2016;13:391–398. doi: 10.1089/zeb.2016.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muralidharan P, Sarmah S, Zhou FC, Marrs JA. Fetal Alcohol Spectrum Disorder (FASD) Associated Neural Defects: Complex Mechanisms and Potential Therapeutic Targets. Brain Sci. 2013;3:964–991. doi: 10.3390/brainsci3020964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrs JA, et al. Zebrafish fetal alcohol syndrome model: effects of ethanol are rescued by retinoic acid supplement. Alcohol. 2010;44:707–715. doi: 10.1016/j.alcohol.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarmah S, Marrs JA. Complex cardiac defects after ethanol exposure during discrete cardiogenic events in zebrafish: prevention with folic acid. Dev. Dyn. 2013;242:1184–1201. doi: 10.1002/dvdy.24015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarmah S, Muralidharan P, Marrs JA. Embryonic Ethanol Exposure Dysregulates BMP and Notch Signaling, Leading to Persistent Atrio-Ventricular Valve Defects in Zebrafish. PLoS One. 2016;11:e0161205. doi: 10.1371/journal.pone.0161205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muralidharan P, Sarmah S, Marrs JA. Zebrafish retinal defects induced by ethanol exposure are rescued by retinoic acid and folic acid supplement. Alcohol. 2015;49:149–163. doi: 10.1016/j.alcohol.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muralidharan P, Sarmah S, Marrs JA. Retinal Wnt signaling defect in a zebrafish fetal alcohol spectrum disorder model. PLoS One. 2018;13:e0201659. doi: 10.1371/journal.pone.0201659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarmah, S. & Marrs, J. A. Embryonic Ethanol Exposure Affects Early- and Late-Added Cardiac Precursors and Produces Long-Lasting Heart Chamber Defects in Zebrafish. Toxics5, 10.3390/toxics5040035 (2017). [DOI] [PMC free article] [PubMed]
- 22.Sarmah S, et al. Ethanol exposure disrupts extraembryonic microtubule cytoskeleton and embryonic blastomere cell adhesion, producing epiboly and gastrulation defects. Biol. Open. 2013;2:1013–1021. doi: 10.1242/bio.20135546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blader P, Strahle U. Ethanol impairs migration of the prechordal plate in the zebrafish embryo. Developmental Biol. 1998;201:185–201. doi: 10.1006/dbio.1998.8995. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Ethanol exposure affects cell movement during gastrulation and induces split axes in zebrafish embryos. Int. J. developmental neuroscience: Off. J. Int. Soc. Developmental Neurosci. 2010;28:283–288. doi: 10.1016/j.ijdevneu.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Babb SG, Marrs JA. E-cadherin regulates cell movements and tissue formation in early zebrafish embryos. Developmental dynamics: an. Off. Publ. Am. Assoc. Anatomists. 2004;230:263–277. doi: 10.1002/dvdy.20057. [DOI] [PubMed] [Google Scholar]
- 26.Schulz, K. N. & Harrison, M. M. Mechanisms regulating zygotic genome activation. Nature reviews. Genetics, 10.1038/s41576-018-0087-x (2018). [DOI] [PMC free article] [PubMed]
- 27.Lee MT, et al. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nat. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Dev. 1990;108:569–580. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- 29.Okuda Y, Ogura E, Kondoh H, Kamachi Y. B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet. 2010;6:e1000936. doi: 10.1371/journal.pgen.1000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda Y, et al. Comparative genomic and expression analysis of group B1 sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev. Dyn. 2006;235:811–825. doi: 10.1002/dvdy.20678. [DOI] [PubMed] [Google Scholar]
- 31.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinforma. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang S, Cui W. Sox2, a key factor in the regulation of pluripotency and neural differentiation. World J. Stem Cell. 2014;6:305–311. doi: 10.4252/wjsc.v6.i3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paranjpe SS, Veenstra GJ. Establishing pluripotency in early development. Biochim. Biophys. Acta. 2015;1849:626–636. doi: 10.1016/j.bbagrm.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal C, Park KS, Jung KH, Chai YG. Ethanol-related alterations in gene expression patterns in the developing murine hippocampus. Acta biochimica et. biophysica Sin. 2015;47:581–587. doi: 10.1093/abbs/gmv050. [DOI] [PubMed] [Google Scholar]
- 35.Lussier AA, et al. Prenatal Alcohol Exposure: Profiling Developmental DNA Methylation Patterns in Central and Peripheral Tissues. Front. Genet. 2018;9:610. doi: 10.3389/fgene.2018.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camargo Moreno M, Mooney SM, Middleton FA. Heterogeneity of p53 dependent genomic responses following ethanol exposure in a developmental mouse model of fetal alcohol spectrum disorder. PLoS one. 2017;12:e0180873. doi: 10.1371/journal.pone.0180873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleiber ML, Laufer BI, Wright E, Diehl EJ, Singh SM. Long-term alterations to the brain transcriptome in a maternal voluntary consumption model of fetal alcohol spectrum disorders. Brain Res. 2012;1458:18–33. doi: 10.1016/j.brainres.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 38.Yelin R, et al. Ethanol exposure affects gene expression in the embryonic organizer and reduces retinoic acid levels. Dev. Biol. 2005;279:193–204. doi: 10.1016/j.ydbio.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A, Singh CK, DiPette DD, Singh US. Ethanol impairs activation of retinoic acid receptors in cerebellar granule cells in a rodent model of fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 2010;34:928–937. doi: 10.1111/j.1530-0277.2010.01166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrelli B, Bendelac L, Hicks GG, Fainsod A. Insights into retinoic acid deficiency and the induction of craniofacial malformations and microcephaly in fetal alcohol spectrum disorder. Genes. 2019;57:e23278. doi: 10.1002/dvg.23278. [DOI] [PubMed] [Google Scholar]
- 41.Shabtai Y, Fainsod A. Competition between ethanol clearance and retinoic acid biosynthesis in the induction of fetal alcohol syndrome. Biochem. Cell Biol. 2018;96:148–160. doi: 10.1139/bcb-2017-0132. [DOI] [PubMed] [Google Scholar]
- 42.Ogony JW, Malahias E, Vadigepalli R, Anni H. Ethanol alters the balance of Sox2, Oct4, and Nanog expression in distinct subpopulations during differentiation of embryonic stem cells. Stem Cell Dev. 2013;22:2196–2210. doi: 10.1089/scd.2012.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, et al. Zebrafish foxc1a plays a crucial role in early somitogenesis by restricting the expression of aldh1a2 directly. J. Biol. Chem. 2015;290:10216–10228. doi: 10.1074/jbc.M114.612572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skarie JM, Link BA. FoxC1 is essential for vascular basement membrane integrity and hyaloid vessel morphogenesis. Invest. Ophthalmol. Vis. Sci. 2009;50:5026–5034. doi: 10.1167/iovs.09-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topczewska JM, et al. The winged helix transcription factor Foxc1a is essential for somitogenesis in zebrafish. Genes. Dev. 2001;15:2483–2493. doi: 10.1101/gad.907401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umali J, Hawkey-Noble A, French CR. Loss of foxc1 in zebrafish reduces optic nerve size and cell number in the retinal ganglion cell layer. Vis. Res. 2019;156:66–72. doi: 10.1016/j.visres.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Veldman MB, Lin S. Etsrp/Etv2 is directly regulated by Foxc1a/b in the zebrafish angioblast. Circ. Res. 2012;110:220–229. doi: 10.1161/circresaha.111.251298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yue Y, et al. The transcription factor Foxc1a in zebrafish directly regulates expression of nkx2.5, encoding a transcriptional regulator of cardiac progenitor cells. J. Biol. Chem. 2018;293:638–650. doi: 10.1074/jbc.RA117.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Norton WH, et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Dev. 2005;132:645–658. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamplin OJ, Cox BJ, Rossant J. Integrated microarray and ChIP analysis identifies multiple Foxa2 dependent target genes in the notochord. Dev. Biol. 2011;360:415–425. doi: 10.1016/j.ydbio.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Dal-Pra S, Thisse C, Thisse B. FoxA transcription factors are essential for the development of dorsal axial structures. Dev. Biol. 2011;350:484–495. doi: 10.1016/j.ydbio.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Kaplan MH, Wang XP, Xu HP, Dosik MH. Partially unspliced and fully spliced ELF3 mRNA, including a new Alu element in human breast cancer. Breast cancer Res. Treat. 2004;83:171–187. doi: 10.1023/B:BREA.0000010710.51614.2d. [DOI] [PubMed] [Google Scholar]
- 53.Luk, I. Y., Reehorst, C. M. & Mariadason, J. M. ELF3, ELF5, EHF and SPDEF Transcription Factors in Tissue Homeostasis and Cancer. Molecules23, 10.3390/molecules23092191 (2018). [DOI] [PMC free article] [PubMed]
- 54.Prescott JD, Koto KS, Singh M, Gutierrez-Hartmann A. The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol. Cell. Biol. 2004;24:5548–5564. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CH, et al. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–1622. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 56.Chang J, et al. Over-expression of ERT(ESX/ESE-1/ELF3), an ets-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19:151–154. doi: 10.1038/sj.onc.1203252. [DOI] [PubMed] [Google Scholar]
- 57.Ng AY, et al. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- 58.Oliver JR, Kushwah R, Hu J. Multiple roles of the epithelium-specific ETS transcription factor, ESE-1, in development and disease. Laboratory investigation; a J. technical methods Pathol. 2012;92:320–330. doi: 10.1038/labinvest.2011.186. [DOI] [PubMed] [Google Scholar]
- 59.Plavicki JS, et al. Construction and characterization of a sox9b transgenic reporter line. Int. J. Dev. Biol. 2014;58:693–699. doi: 10.1387/ijdb.140288jp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, et al. Fgf-signaling-dependent Sox9a and Atoh1a regulate otic neural development in zebrafish. J. Neurosci. 2015;35:234–244. doi: 10.1523/JNEUROSCI.3353-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim YI, et al. Cartilage development requires the function of Estrogen-related receptor alpha that directly regulates sox9 expression in zebrafish. Sci. Rep. 2015;5:18011. doi: 10.1038/srep18011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dalcq J, et al. RUNX3, EGR1 and SOX9B form a regulatory cascade required to modulate BMP-signaling during cranial cartilage development in zebrafish. PLoS one. 2012;7:e50140. doi: 10.1371/journal.pone.0050140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor Sox9. Dev. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 64.Gawdzik JC, et al. sox9b is required in cardiomyocytes for cardiac morphogenesis and function. Sci. Rep. 2018;8:13906. doi: 10.1038/s41598-018-32125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Westerfield, M. The zebrafish book. Eugene, OR: The University of Oregon Press (2000).
- 66.Pepper SD, Saunders EK, Edwards LE, Wilson CL, Miller CJ. The utility of MAS5 expression summary and detection call algorithms. BMC Bioinforma. 2007;8:273. doi: 10.1186/1471-2105-8-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinforma. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. U S Am. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 70.Hu H, et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinforma. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 72.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinforma. 2014;47(8 13):11–24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.