Cells regulate α- and β-tubulin levels through a negative feedback loop which degrades tubulin mRNA upon detection of excess free tubulin protein. In a recent study inScience, Lin et al. discover a role for a novel factor, TTC5, in recognizing the N-terminal motif of tubulins as they emerge from the ribosome and in signaling co-translational mRNA decay.
Cells use translation-coupled mRNA decay for both quality control and general regulation of mRNA levels. A variety of known quality control pathways including Nonsense Mediated Decay (NMD), No-Go Decay (NGD), and Non-Stop Decay (NSD) specifically detect and degrade mRNAs encoding potentially toxic protein fragments or sequences which cause ribosomes to translate poorly or stall.1 More generally, canonical mRNA degradation is broadly thought to be translation dependent, though the mechanisms that drive these events are not completely understood.2,3
Tubulin mRNA levels and protein output are strictly controlled to allow for precisely timed alignment and segregation of chromosomes during cell division. While tubulin mRNAs have long been known to be degraded in a manner dependent on both their translation and the accessibility of the tubulin N-terminal motif, the mechanism responsible for this coupling remained unclear.4 In a recent study,5 Lin et al. reasoned that undiscovered protein factors might be critical for the regulated co-translational decay of α- and β-tubulins. To capture the putative factor, the authors translated tubulin in vitro to generate ribosome–nascent chain complexes (RNCs) containing a photoactivatable amino acid crosslinker positioned near the N-terminus. The protein TTC5 was identified by mass spectrometry as a factor closely associated with the tubulin N-terminal sequence. Cryo-electron microscopy structural studies and mutational analysis confirmed that TTC5 forms a bipartite interaction with the tubulin nascent chain and the 28S rRNA near the peptide exit tunnel, allowing for specific binding to ribosomes that are actively translating tubulin. Importantly, when Lin and colleagues knocked out TTC5 using CRISPR/Cas9 system, they observed that cells no longer degrade tubulin mRNAs after treatment with colchicine, a microtubule depolymerizing agent that induces an overabundance of free tubulin protein. Moreover, the lost decay phenotype was rescued by the addition of exogenous wild-type TTC5, but not by the addition of TTC5 in which key ribosome and nascent chain interacting residues were mutated. Lastly, TTC5-knockout cells exhibited increased numbers of chromosome segregation and alignment errors, consistent with the known requirement for tight regulation of tubulin expression for proper cell division.
Since TTC5 recognizes the N-terminus of actively translating tubulin mRNA, the authors wondered what normally limits TTC5-mediated mRNA decay to conditions where free tubulin is present. To address this, the authors mixed pre-formed TTC5–tubulin RNCs containing crosslinker with lysates from colchicine-treated or colchicine-untreated TTC5-knockout cells (either having or lacking abundant free tubulin, respectively). After irradiation, TTC5 only crosslinked to the RNC in lysates from cells that had previously been treated with colchicine; these data suggested to the authors that some other (unknown) factor may prevent TTC5 from binding under conditions of low free tubulin.
What are likely possibilities for how such coupling between translation and mRNA decay might occur? One example to consider is that of mRNA surveillance where extensive studies in yeast have identified a large group of proteins that recognize and resolve stalled RNCs found on problematic mRNAs and target those mRNAs for decay. Key among these factors are the E3 ligase Hel2 (ZNF598 in mammals) that recognizes colliding ribosomes, the Ribosome Quality Control Trigger Complex (RQT)6 that dissociates collided ribosomes, and Cue2 (N4BP2) and Xrn1 (XRN1) that promote endonucleolytic and exonucleolytic mRNA decay, respectively.7 While the precise mechanisms that lead to recruitment of factors involved in mRNA decay are unclear, a central role for ribosome collisions and for ubiquitination of these collided ribosomes seems likely. Studies in various systems have revealed that mRNA decay generally occurs co-translationally2 and correlates indirectly with codon optimality at a genome-wide level.3 Subsequent studies in yeast suggested a role for the DEAD-box protein Dhh1 in facilitating these differential rates of decay as a function of ribosome occupancy.8 More recent structural studies have shown that the CCR4–NOT complex binds to empty ribosomal E sites, potentially functioning as a read-out for slow ribosomes.9 Taken together, it seems clear that ribosome occupancy and activity are relevant not just to mRNA surveillance quality control pathways but also to canonical mRNA degradation.
In another study that may share some parallels with tubulin mRNA regulation, it was found that association between the signal recognition particle (SRP) and the N-terminal signal sequence of translating secreted proteins is important for determining the stability of the corresponding mRNA.10 Typically, chaperones bind the N-termini of cytosolic proteins and the SRP binds the N-termini of secreted proteins as they emerge from the ribosome. The authors proposed that recognition of nascent peptides by the appropriate targeting machinery might be generally critical in determining the stability of the translating mRNA. It seems possible that the putative TTC5 inhibitory factor binds to the N-terminus of tubulin and stabilizes the mRNA in a similar manner.
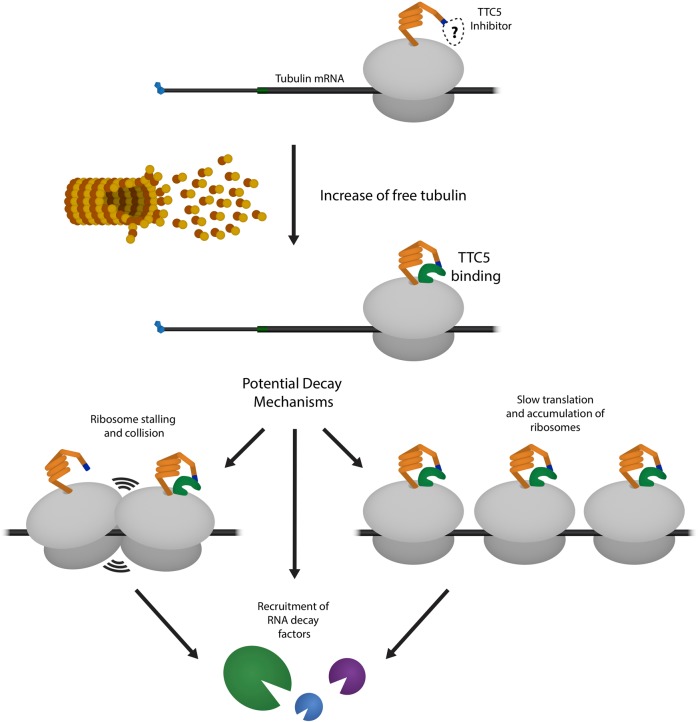
The recent work by Lin et al. provides new molecular insight into how coupling of tubulin translation to mRNA degradation takes place and suggests that there is more to learn. How, for example, does TTC5’s presence on tubulin nascent chains elicit decay of tubulin mRNA? One possibility is that TTC5 modulates the ribosome occupancy on the mRNA, or even leads to ribosome collisions, thus creating substrates for decay processes like NGD. Alternatively, TTC5 could directly facilitate recruitment of decapping and deadenylation factors or nucleases that directly target tubulin mRNAs for decay (Fig. 1). While the putative inhibitor of TTC5 binding remains unknown, its identity may also help dissect the mechanism of tubulin mRNA decay. Importantly, the tubulin example discussed here may provide insights into how ribosome function is broadly critical to mRNA decay.
Fig. 1. A model for TTC5’s role in tubulin mRNA decay and potential connections to known mRNA decay mechanisms.
TTC5 binds the ribosome and tubulin N-terminal motif upon increase of free tubulin in the cell. Translation-coupled decay of tubulin mRNA may happen via known mechanisms, such as induction of ribosome collisions or accumulation of slowly translating ribosomes, or TTC5 may help recruit RNA decay factors by other means.
References
- 1.Simms CL, Thomas EN, Zaher HS. Wiley Interdiscip. Rev. RNA. 2017;8:e1366. doi: 10.1002/wrna.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu W, et al. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Presnyak V, et al. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theodorakis NG, Cleveland DW. Mol. Cell. Biol. 1992;12:791–799. doi: 10.1128/MCB.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Z, et al. Science. 2019;4352:eaaz4352. [Google Scholar]
- 6.Matsuo Y, et al. Nat. Commun. 2017;8:159. doi: 10.1038/s41467-017-00188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Orazio KN, et al. Elife. 2019;8:e49117. doi: 10.7554/eLife.49117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radhakrishnan A, Green R. Biophys. J. 2015;108:391a. doi: 10.1016/j.bpj.2014.11.2144. [DOI] [Google Scholar]
- 9.Buschauer, R. et al. bioRxiv10.1101/854810 (2019)
- 10.Karamyshev ALL, et al. Cell. 2014;156:146–157. doi: 10.1016/j.cell.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]