Abstract
Heat Shock Protein 101 (HSP101), the homolog of Caseinolytic Protease B (CLPB) proteins, has functional conservation across species to play roles in heat acclimation and plant development. In wheat, several TaHSP101/CLPB genes were identified, but have not been comprehensively characterized. Given the complexity of a polyploid genome with its phenomena of homoeologous expression bias, detailed analysis on the whole TaCLPB family members is important to understand the genetic basis of heat tolerance in hexaploid wheat. In this study, a genome-wide analysis revealed thirteen members of TaCLPB gene family and their expression patterns in various tissues, developmental stages, and stress conditions. Detailed characterization of TaCLPB gene and protein structures suggested potential variations of the sub-cellular localization and their functional regulations. We revealed homoeologous specific variations among TaCLPB gene copies that have not been reported earlier. A study of the Chromosome 1 TaCLPB in four wheat genotypes demonstrated unique patterns of the homoeologous gene expression under moderate and extreme heat treatments. The results give insight into the strategies to improve heat tolerance by targeting one or some of the TaCLPB genes in wheat.
Subject terms: Gene expression profiling, Abiotic
Introduction
Increasing global temperature is a serious concern in agriculture as it affects crop productivity and food production. Heat stress often causes irreversible damages to plant physiological process and development. Wheat, one of the major cereal crops grown worldwide, is highly sensitive to heat stress. Frequent occurrence of days with super optimal temperatures during a wheat growing season directly affects yield1,2. At cellular level, high temperature damages membranes of different sub-cellular compartments and degrades proteins3,4. Kinetic activities of enzymes involved in photosynthesis, respiration, cell division, and many other vital processes are also affected by heat stress5–8.
Caseinolytic Protease B (CLPB) proteins play important roles in organisms, especially in stress response and at different developmental stages. These proteins are high molecular weight chaperones that are part of the Heat Shock Protein 100 (HSP100) family9. The initial study on one of the CLPB members in yeast discovered the role of Heat Shock Protein 104 (HSP104) in heat acclimation10. A process that involves protein disaggregation by HSP104 was later observed as the mechanism of heat tolerance in yeast11. The orthologous proteins were also identified in other organisms with similar functional characteristics to yeast HSP104. Bacterial CLPB12,13 and plant HSP101 or CLPB114–16 were characterized as the functional orthologs of yeast HSP104. In plants, beside HSP101 that is localized in the cytoplasm, the homologs of CLPB were also identified within the plastid and mitochondria17,18. Plastid localized CLPB in Arabidopsis thaliana (Arabidopsis) was known to play a role in plastid development and plant survival19,20, while the ortholog in tomato was known to be important for heat acclimation21.
Detailed structural features of CLPB protein were initially characterized by Lee et al. (2003) in the model bacterium Thermus thermophilus. The protein was described as a two-tiered ring of hexamers connected with coiled-coil linkers. The monomers are comprised of five conserved domains: the N-terminal domain; the D1-large domain (Nucleotide Binding Domain 1/NBD1); the D1-small domain including linker region; the D2-large domain (Nucleotide Binding Domain 2/NBD2); and the D2-small domain. CLPB was also classified as the member of the AAA+ (ATPases associated with diverse cellular activities) superfamily of ATPases22. The C-terminal domain of the NBD2 was predicted to be critical for CLPB oligomerization while the interaction of ATP with the NBD1 stabilizes the CLPB oligomer23,24. The N-terminal domain was not found to be essential for oligomerization and disaggregation activity, but was important for CLPB binding with specific substrates such as casein25.
To be active, CLPB requires energy from ATP hydrolysis that is triggered by the interaction of coiled-coil linkers of the middle domain with the DnaK-DnaJ complex26. During protein disaggregation, DnaK-DnaJ (HSP70-HSP40) exposes the peptide segment of damaged proteins to the central pore of the CLPB hexamer which will progressively pull and unfold it27. The long coiled-coil structure of the linker region has been known as a characteristic feature that distinguishes CLPB from the other AAA+ or the HSP100 family members28. In Thermus thermophillus, the CLPB linker forms a two-bladed propeller with two motifs that is similar to that of leucine zippers in eukaryotic transcription factors29,30. It was predicted that HSP70 triggers the active state of CLPB by its interaction with the CLPB linker region which acts as a molecular toggle31,32. A species-specific characteristic is also possibly present in the middle domain structure as shown by a specific interaction of the linker region from E. coli CLPB and the yeast HSP104 with DnaK and HSP70, respectively33.
In hexaploid wheat Triticum aestivum, several studies revealed the presence of HSP101 gene copies and their expression under high temperature or other types of stress treatments. The first wheat ortholog of HSP101 was identified as a 102 kDa Ω-binding protein that can complement the thermotolerance defect in yeast hsp10434. The protein was also shown to act as a translational regulator of Ferredoxin-1 (Fed-1)34–36. The other two genes of wheat HSP101 were cloned later and named as TaHSP101B and TaHSP101C, while the first HSP101 was renamed as TaHSP101A37. An in-silico study of the Caseinolytic Protease Class I family has predicted five members of the CLPB family in wheat: three of them are cytoplasmic copies and one copy each of the other two is targeted to the chloroplast and mitochondria38. These genes were shown to be differentially expressed at different tissues and stress conditions37,38. Cytoplasmic CLPBs were up-regulated in leaves under heat, salt and oxidative stress38. The increased expression of wheat CLPBs was also observed under drought stress, but not observed under chilling and wounding treatments37,38.
The wheat genome has its own complexity due to polypoidy. Genus Triticum, with 7 as the monoploid number of chromosomes [1x = 7], consists of diploid [2n = 2x = 14], tetraploid [2n = 4x = 28] and hexaploid wheat [2n = 6x = 42] species39. During the evolution of hexaploid wheat, the A genome came from Triticum urartu [AA]40, which is similar to Triticum monococcum; however, the B genome donor, Aegilops speltoides, is still controversial41,42. The A and the B genomes then combined to form Triticum turgidum [AABB]43 and the Allohexaploid Triticum aestivum [AABBDD] arose from a spontaneous hybridization of T. turgidum with the donor of the D genome Aegilops tauschii44. The term homoeolog or homeolog refers to genes or chromosomes that are inherited from different progenitors through interspecific hybridization, resulted in allopolyploidization39,45. It is distinguishable with homolog which refers to the genes or genomes that share similarities which are inherited from common ancestors45.
Differential expression of homoeologous genes are common phenomena in polyploids. Reconciliation of genomes gave consequences to the anomaly of gene expression patterns and phenotypes by the presence of changes at the genetic and epigenetic levels46–48. Homoeolog expression bias is unequal expression among the homoeologs at different tissues or developmental stages; or as anomaly of their expression level relative to their diploid progenitors47,49. Subgenomic preferences have been reported in octoploid strawberry with a single subgenome exhibited significant dominance in gene expression and control of certain metabolomic and disease resistance traits50. Contribution of homoeolog expression dominance in facilitating selection of glucosinolate and lipid metabolism genes was also reported in the vegetable-use and oil-use sub-varieties of Brassica juncea51. In cotton and wheat, alterations of expression patterns among homoeologs under variation in stress conditions, tissues, and developmental stages have also been documented52–57. Given the consequences of unequal expression to the natural gene selections in polyploids, it is important to understand the genetic basis of valuable traits with respect to the homology and homoeology perspectives for the success of selective breeding programs.
Previous studies have identified HSP101/CLPB copies in wheat without a clear map of the whole gene family in the genome of this polyploid species (Wells et al. 1998; Campbell et al. 2001; Muthusamy et al. 2016). In tetraploid wheat Triticum turgidum subsp. durum, four copies of TdHSP101 were cloned and physically mapped on the two homoeologous chromosomes of groups 1 and 358. Orthologs of the two genes were also placed on the corresponding chromosomes of the A genome progenitor, Triticum monococcum58. The corresponding gene copies are not known in the hexaploid wheat. Since more than one sequence were reported as the putative HSP101/CLPB homologs in wheat, there has been confusion on how many genes exactly present in the genome and functional, which copies are mainly playing role and potential to be targeted for crop improvement. Besides, lack of thorough observation on the entire gene family will potentially introduce bias in gene expression analyses due to high similarities among homologous or homoeologous genes. The bias might lead to inaccurate predictions about gene responses and functions. Specificity in gene targeting even more crucial if genome editing is the choice for genetic modification as currently has become a trend in today’s methods59,60. Hence, detailed analysis on all the HSP101/CLPB family members is required.
In this study, we identified all the members of HSP101/CLPB gene family in hexaploid wheat and located their position on wheat chromosomes. The sequences were characterized based on their predicted protein structures as compared to the known HSP101/CLPB sequences from two model species, rice and Arabidopsis. Unique conserved domains and motifs were analyzed throughout the linker regions to study variation of the proteins at the functional level. Gene expression patterns were characterized in silico and in real time PCR with respect to plant developmental stages and stress treatments. TaCLPBs of the group 1 chromosomes, of which a member was shown to complemet the hsp10434, were found to be more responsive to drought and heat stress. We specifically cloned and studied this group members for their homoeologous expression patterns in four wheat genotypes under moderate and extreme high temperatures.
Materials and Methods
Identification of CLPB gene copies and their mapping to the wheat genome
Using a tblastx tool61,62, the rice sequences (Os05g0519700, Os03g0426900, Os02g0181900) were used as references and queries to retrieve the orthologs from the wheat sequence databases in NCBI (https://www.ncbi.nlm.nih.gov), Swissprot/Uniprot (http://www.uniprot.org), and EnsemblPlants (http://plants.ensembl.org/Triticum_aestivum). The retrieved sequences were then confirmed as orthologs of CLPB following the criteria developed by Dhaliwal et al.63. Briefly, orthologous sequences have to meet four criteria: the highest level of sequence identities and query coverage, the presence of domains and motifs of CLPB at the protein level, the relative size and distance among domains and motifs to be similar to the query, and that orthologs must retrieve the reference sequences at the first place when the basic local alignment (BLAST) against nucleotide or protein databases are performed. Ensemble Plants database was used to confirm the ancestral relationship of the putative sequences with the orthologous genes from other species.
The TaCLPBs gene family were mapped on wheat chromosomes using the BLAST (tblastn) tool in the Wheat Chromosome Survey Sequence (https://wheat-urgi.versailles.inra.fr/Seq-Repository) database generated by the International Wheat Genome Sequencing Consortium (https://www.wheatgenome.org/About). In this early wheat database, individual chromosome arms were derived and sequenced from double ditelosomic stocks of the hexaploid wheat cultivar Chinese Spring62. Partial sequences retrieved during the analysis were recovered through a sequence search in: (1) the NCBI EST database of bread wheat (https://www.ncbi.nlm.nih.gov/dbEST/); (2) the draft assembly of gene rich regions of Chinese Spring wheat in the Cereal Database (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB); the genome database of wheat progenitors, including Triticum urartu, Aegilops speltoides, Aegilops tauschii, and Triticum turgidum subsp. durum (https://urgi.versailles.inra.fr/download/iwgsc/TGAC_WGS_assemblies_of_ other_wheat_species/). Full length sequence contigs were synthesized by a manual assembly of the partial sequences through their overlapping regions using the DNA alignment tool in Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). Recent updates with the release of IWGSC RefSeq assembly v1.1 (https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Assemblies/v1.0/), were incorporated later in the analysis and mostly confirmed the manual assembly and annotation in the previous analysis.
Analysis of CLPB genes and proteins
TaCLPB putative genes were aligned with the known cDNA/EST sequences by using a DNA alignment tool in Clustal Omega to manually identify the exon and intron junctions. Predicted CDS sequences were translated into protein sequences using the EMBOSS Transeq translation tool (https://www.ebi.ac.uk/Tools/st/emboss_transeq/). The translated sequences were then used to analyze homology among TaCLPB proteins under multiple sequence alignment using a protein alignment tool in Clustal Omega. Protein conserved domains were identified using the NCBI’s CD-Search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) with SMART and Pfam databases as references. To predict subcellular localization of CLPB proteins, the ChloroP 1.1 Server64 and the TargetP 1.1 Server (http://www.cbs.dtu.dk/services)65 were used to identify signatures of signal peptides. Default parameters were used for all the analyses. Phylogenetic analysis was done using Maximum Likelihood method by RAxML v.8.2.12 on the CIPRES Science Gateway with the GTR + Γ model of evolution66. Bootstrap analyses of 1000 replicates were used as the support for the optimum maximum likelihood tree. The phylogenetic tree was visualized using Dendroscope v.3.5.967.
The 3D structure of CLPB proteins was predicted using the I-TASSER software68. The predicted models with the highest C-score values were used to identify the ligand binding residues using the COACH program69. Comparisons among protein models were performed in the PDBeFold (http://pdbe.org/fold/). CLPB protein sequences of OsHSP101, OsCLPB-C, OsCLPB-M (UniProt ID: Q6F2Y7, Q75GT3, Q0E3C8); AtCLPB-1, AtCLPB-3, AtCLPB-4 (UniProt ID: P42730, Q9LF37, Q8VYJ7); and TCLPB (UniProt ID: TTHA1487) were included in the protein analysis as references to represent the models of functionally characterized CLPBs from rice, Arabidopsis, and Thermus thermophilus.
In silico RNA-seq expression analysis
The manually annotated and mapped TaCLPB sequences were compared to latest version of the gene models (Wheat RefSeq v1.1) in EnsemblPlants. The expVIP tool was used to analyze the expression of TaCLPB copies in silico by retrieving the RNA-seq expression data of TaCLPB transcripts in polyploid wheat56. Two datasets were selected for the analysis: 1) the wheat development time course (ENA: ERP004714) and 2) the drought and heat stress (SRA: SRP045409). The expression values were visualized as the unit of transcript per kilobase exons per million reads (TPM). Differential expression analysis was done on the transcript raw-count data by the EdgeR package version 3.24.2 in the R program70.
Real-time expression analyses
Real-time gene expression for five chromosomal group of TaCLPB members was analyzed in PBW343 variety under normal temperature (22 °C) at three developmental stages: seedling stage, anthesis stage, and grain filling stage (7DAA). In three biological replicates, four types of tissue collected were seedling leaves, mature leaves (second leaves), flag leaves, and spikes. Five primer pairs were designed as common primers to amplify TaCLPB members of each chromosomal group with 150–200 bp expected amplicon size.
The expression study of the homoeologous TaCLPB members of the chromosome 1 was done under control and heat treatments on four wheat genotypes: Chinese Spring, Red Fife, Giza 168, and PBW 343. Chinese Spring was chosen as the reference accession for wheat as its genome was sequenced. The other three genotypes are varieties originated from three different regions that are considered to pose unique temperature or climate regime. Giza 168 is a variety from Egypt, Red Fife is originated from Canada, and PBW 343 from India. Leaf samples were collected from three biological replicates of 12-day old seedlings following the treatments of (1) 2 h at 37 °C; (2) 4 h at 37 °C; and (3) 2 h at 37 °C plus 4 h at 42 °C. Homoeologous specific primers were designed to amplify around 200–300 bp amplicons and the specificity of each was tested using Chinese Spring Nullisomic-tetrasomic lines for the group I chromosomes by PCR amplification. All primers were listed in the Supplement 1.
Total RNA was isolated using a modified Hot Phenol Extraction method71 and the cDNA was synthesized using M-MLV Reverse Transcriptase enzyme kit (Promega, WI, USA). Relative transcript abundance was measured by Real-time qPCR using the SYBR Green I detection system from Kapa Biosystem for Roche LightCycler 480. PCR mixtures were composed of 50x dilution of cDNA samples (synthesized from 1 µg RNA), 0.2 pmol/µl primers, 1.2x Kapa Sybr Fast LC480 (Kapa Biosystems, USA). As the amplicons were expected to have high GC contents, 2.5% DMSO was added into the reaction. The cycling conditions were 95 °C/3 min pre-incubation; 32 cycles of 95 °C/10 sec denaturation, 62 °C/20 sec annealing, 72 °C/1 sec extension. Data analyses were done using the LinReg PCR program72. The expression levels are shown as the means of normalized ratios of the target gene to the actin gene expression along with the standard deviation of three biological replicates73. The fold change values represent the ratio of the target gene expression as compared to the control with the error bars showing relative standard deviations (rsd) of three biological replicates73. Statistical analysis of the expression data was done by the analysis of variance (anova) for multiway-treatment structure, followed by a post-hoc multiple comparison using Tukey’s test in the R program.
Cloning of TaCLPB homoeologous copies from the group 1 chromosome
Based on our sequence analysis, the TaCLPBs of the chromosome 1 are the functional orthologs of HSP101. TaCLPB in the chromosome 1A has been shown to complement yeast hsp104 mutant34. It became of interest to identify all the homoeologs in this group by sequence cloning and characterize the homoeologous expression patterns. The putative full-length sequences of the chromosome 1 TaCLPB members were used as references for designing homoeologous specific primers (Supplement 1). The PCR reactions were performed on the Chinese Spring genomic DNA and cDNA templates. Amplicons with the expected size were cloned using the Gateway cloning system (Invitrogen, USA) and sequenced (at least three colonies per clone).
Results and Discussions
TaCLPB gene family members and their chromosomal locations in polyploid wheat
The rice CLPB proteins that have been annotated and functionally characterized are: (1) cytoplasm targeted CLPB (CLPB-c) that is also known as rice HEAT SHOCK PROTEIN 101 (OsHSP101); (2) plastid targeted CLASS I CLP ATPASE B-C (OsCLPB-C); and (3) mitochondria targeted CLASS I CLP ATPASE B-M (OsCLPB-M). Using these three proteins as references for sequence search and annotation, 13 wheat sequences were identified as the members of the wheat CLPB family; seven of which were predicted to be targeted to the cytoplasm and physically mapped to the long arm of chromosomes 1A, 1B, 1D, 3A, 3B, 3D, and 4B (1AL, 1BL, 1DL, 3AL, 3BL, 3DL, 4BL), respectively; three putative plastid targeted sequences are mapped to the long arms of the chromosome 5A, 4B, and 4D (5AL, 4BL, 4DL); three putative mitochondria targeted sequences mapped to the short arms of the chromosome 6A, 6B, and 6D (6AS, 6BS, 6DS), respectively. The alignment of TaCLPB sequences with their orthologs in rice are given in the Supplements 2–4. Table 1 listed the information of TaCLPB sequences by the genome analysis. The sequence names are symbols to differentiate the sequences based on the chromosomal location (with the addition of “1” in “TaCLPB-4B1” for an extra copy at the chromosome 4B). Information about the synteny of these sequences with the orthologs from other species was provided in the Supplement 5.
Table 1.
TaCLPB sequences with respect to their corresponding ortholog in rice, chromosomal mapping and subcellular target locations.
| No. | TaCLPB | Map | EnsemblPlants ID (RefSeq. 1.1 gene model) |
Gene length | Aa # | Predicted sub-cellular target | Transit peptide length |
|---|---|---|---|---|---|---|---|
| 1 | TaCLPB-1A | 1AL | TraesCS1A02G340100 | 2757 | 918 | cytoplasm | NA |
| 2 | TaCLPB-1B | 1BL | TraesCS1B02G352400 | 2754 | 917 | cytoplasm | NA |
| 3 | TaCLPB-1D | 1DL | TraesCS1D02G342100 | 2757 | 918 | cytoplasm | NA |
| 4 | TaCLPB-3A | 3AL | TraesCS3A02G274400 | 2742 | 913 | cytoplasm | NA |
| 5 | TaCLPB-3B | 3BL | TraesCS3B02G308100 | 2742 | 913 | cytoplasm | NA |
| 6 | TaCLPB-3D | 3DL | TraesCS3D02G273600 | 2742 | 913 | cytoplasm | NA |
| 7 | TaCLPB-4B1 | 4BL | TraesCS4B02G393100 | 2712 | 903 | cytoplasm | NA |
| 8 | TaCLPB-5A | 5AL | TraesCS5A02G547300 | 2925 | 974 | plastid | 70 |
| 9 | TaCLPB-4B | 4BL | TraesCS4B02G380800 | 2928 | 975 | plastid | 71 |
| 10 | TaCLPB-4D | 4DL | TraesCSU02G131300 | 2917 | 971 | plastid | 67 |
| 11 | TaCLPB-6A | 6AS | TraesCS6A02G146400 | 2976 | 991 | mitochondria | 85 |
| 12 | TaCLPB-6B | 6BS | TraesCS6B02G174500 | 2976 | 991 | mitochondria | 85 |
| 13 | TaCLPB-6D | 6DS | TraesCS6D02G135600 | 2976 | 991 | mitochondria | 85 |
Results from the genome analysis support our hypothesis that at least six family members of CLPB sequences are present in hexaploid wheat, as the diploid and tetraploid progenitors have two and four copies of the gene, respectively58. Based on the survey mapping of chromosomal locations, followed by the prediction of subcellular localizations, we found that the three previously reported TaHSP10I genes34,37 are cytoplasmic CLPBs on the chromosome group1 and 3. We are reporting that homoeologous sequences from the group 1 and 3 are present on A, B, and D chromosomes. Based on our in silico mapping, we found that the HSP101 (AF083344.2) that was originally isolated from wheat and functionally characterized by Wells et al. (1998) is present on chromosome 1AL. The HSP101 gene previously known as TaHSP101B (AF097363.1) is present on chromosome 1DL and TaHSP101C (AF174433.1) is present on the chromosome 3DL.
Congruent with the five wheat CLPB copies reported by Muthusamy et al. (2016), our analysis showed the presence of eight additional copies clustered into three groups of the cytoplasmic CLPB, one group of the plastid targeted CLPB, and one group of the mitochondria targeted CLPB. Each group consists of three genes that are present on the corresponding three homoeologous chromosomes. Only one of the sequences that was reported (GenBank ID: AK330787) does not match with our sequence annotation. This sequence was previously predicted as a plastid targeted CLPB. This partial sequence is actually located on chromosome 3D, while our predicted sequences of the plastid targeted CLPB are mapped on chromosome 5A, 4B, and 4D.
A TaCLPB gene that is present on 4BL does not have any homoeologs on the chromosome 4. Although the wheat genome sequence coverage is good, it is still not clear if the lack of homoeologs for this sequence is real or is simply because of the lack of corresponding sequences in the database. The same results were obtained after we reanalyzed the sequence with the newly released IWGSC RefSeq assembly v 1.0. We did however find a sequence in Triticum urartu (EnsemblePlants ID: TRIUR3_09779) that appears to be an ortholog of this copy suggesting the presence of the copy A-homoeolog in the progenitor species. Interestingly, the copy in T. urartu has a unique insertion in the 5’ end of the mRNA, giving an additional start codon to the sequence. This additional sequence encodes 160 amino acids that contains a transposon domain motif (Supplement 6).
Structural features of TaCLPBs
Structural comparisons of TaCLPB genes showed variations in their intron number and size (Fig. 1). While the organellar copies have a higher number of introns than the cytoplasmic ones, no intron is present in TraesCS4B02G393100 (TaCLPB-4B1) sequence. At the protein level, amino acid similarities among TaCLPBs range between 45.4–98.8% (Supplement 7). High similarities were observed among sequences of the same sub-cellular target. There are less sequence similarities between cytoplasmic and organellar CLPBs (46–50%). TraesCS4B02G393100 (TaCLPB-4B1) protein uniquely has the least sequence similarity with the other CLPB members (46–78%). A phylogenetic tree constructed by the maximum likelihood method showed sequence clusters that followed the predicted groups of sub-cellular localizations (Fig. 2).
Figure 1.
Exon-intron organization of TaCLPB genes. The gene structures are drawn from the start codon to the stop codon. Exons and introns are shown in blue and grey, respectively.
Figure 2.

Phylogenetic tree showing relationship among CLPB family in Arabidopsis, rice, and wheat. The tree was constructed using a maximum likelihood method in RAxML v.8.2.12 based on the alignment of amino acid sequences.
The 3D structures were compared among TaCLPBs and with their orthologs in rice, Arabidopsis, and Thermus thermophilus. The results showed that structural similarities among CLPB proteins are high (78–93%) regardless of the sequence similarities (Table 2). Interestingly, there are higher structural similarities between TaCLPBs with their corresponding orthologs in Arabidopsis although their sequence identities are higher with the orthologs in rice. As an example, TraesCS1A02G340100 (TaCLPB-1A) has 92% identity with the rice ortholog and 84% identity with AtCLPB1, but its 3D structure similarity is 84% with the OsHSP101 and 91% 3D similarity with the AtCLPB1.
Table 2.
Comparison of TaCLPB protein structures with rice, Arabidopsis, and Thermus thermophilus CLPB proteins.
| Sequence No |
Name | OsHSPI01 | AtCLPB1 | TCLPB | |||
|---|---|---|---|---|---|---|---|
| % seq | % sse | % seq | % sse | % seq | % sse | ||
| 1 | TaCLPB-1A | 92 | 91 | 83 | 91 | 51 | 84 |
| 2 | TaCLPB-1B | 93 | 84 | 84 | 91 | 52 | 86 |
| 3 | TaCLPB-1D | 92 | 86 | 83 | 93 | 52 | 84 |
| 4 | TaCLPB-3A | 92 | 93 | 84 | 91 | 51 | 86 |
| 5 | TaCLPB-3B | 91 | 84 | 84 | 93 | 52 | 84 |
| 6 | TaCLPB-3D | 91 | 87 | 85 | 87 | 51 | 82 |
| 7 | TaCLPB-4B1 | 74 | 85 | 69 | 93 | 49 | 85 |
|
Sequence No |
Name | OsCLPB-C | AtCLPB3 | TCLPB | |||
| % seq | % sse | % seq | % sse | % seq | % sse | ||
| 8 | TaCLPB-5A | 93 | 84 | 82 | 82 | 56 | 80 |
| 9 | TaCLPB-4B | 93 | 80 | 84 | 86 | 56 | 78 |
| 10 | TaCLPB-4D | 93 | 85 | 83 | 83 | 56 | 85 |
|
Sequence No |
Name | OsCLPB-M | AtCLPB4 | TCLPB | |||
| % seq | % sse | % seq | % sse | % seq | % sse | ||
| 11 | TaCLPB-6A | 91 | 85 | 78 | 89 | 55 | 85 |
| 12 | TaCLPB-6B | 92 | 84 | 78 | 80 | 54 | 82 |
| 13 | TaCLPB-6D | 90 | 83 | 78 | 87 | 55 | 83 |
% seq = the percentage of sequence identities; % sse = the percentage of protein structure identities.
We referred to Thermus thermophilus protein structure29 and identified domain conservation across the sequences. Three domain clusters were observed in all the TaCLPB proteins (Fig. 3): (1) Clp_N (Clp amino terminal domain); (2) P-Loop_NTPase (P-Loop containing Nucleoside Triphosphate Hydrolase); and 3) Clp_D2-Small (C-terminal, D2-small domain, of CLPB protein). Two nucleotide binding domains (P-Loop_NTPase) and one C-terminal domain (Clp_D2-Small) were identified in all CLPB proteins. Only one N-domain motif is present in the cytoplasmic chromosome group 1 and group 3 TaCLPB while the other members have two motifs.
Figure 3.
Conserved domain motifs present in CLPB proteins of Arabidopsis, rice and wheat.
In the middle region, which is a coiled coil structure, domain motifs were identified to be varied among the CLPB proteins (Fig. 3). Some motifs are related to the autophagy protein (APG6), and several other motifs are related to the proteins involved in cell divisions (TACC, Spc7, SMC_N, Mnd1). In general, these domain motifs reflect variation that could be present at the functional level, determined by the linker region. The importance of a middle domain for the specificity of CLPB activities has been well studied in yeast and bacterial systems33. While other domains were interchangeable in the chimeras of yeast HSP104 and bacterial CLPB, exchanging the middle domain led to a failure in protein function. The middle domain of yeast HSP104 was not able to interact with bacterial DnaK and the middle domain of CLPB could not interact with yeast HSP70. The regions within helix 2 and helix 3 of the middle regions were identified to be responsible for this specificity33.
Some studies have also shown the role of middle domains as a molecular toggle that triggers different functions31,32. The loop regions, that were marked as motif 1 and motif 2 in the middle domain, were found to be essential for the interaction with trigger factors such as HSP70. We then looked at the area of the CLPB middle domains in Arabidopsis, rice, and wheat (Fig. 4). We specifically marked the regions that are aligned with motif 1 and motif 2 of TCLPB from Thermus thermophillus. These two motifs were known to be essential for the functionality of CLPB in the bacteria29. Higher sequence conservations in the regions were observed among CLPB members of the same subcellular target locations. Only TraesCS4B02G393100 (TaCLPB-4B1) showed less similarity with the other cytoplasmic CLPBs. Some residues were identified to be unique for different plant species with some minor variations being observed among wheat homoeologs. Looking at the data, it is possible that types of molecules or proteins that interact with CLPBs are unique for different subcellular locations, different chromosomal copies, or even different plant species.
Figure 4.
Variation in the residues of middle domain motifs of CLPB proteins in Arabidopsis, rice, and wheat. Secondary structures along with motif locations were characterized through protein sequence alignments with the 3D model of Thermus thermophilus CLPB using the I-TASSER program. Conserved residues are shown in white color with grey shade, semi-conserved residues are shown in black color with grey shade, non-conserved residues are shown in different colors without shade.
At the sequence level there are signature residues that have been identified in the previous studies and used to differentiate between CLPB with the other ATPase family members28. We confirmed the presence of these signatures in all TaCLPBs with some minor variations (Fig. 5 and Supplement 8); xKFTxxxxxALAxAxxLAxxxxHxxhxPhHLAxALh at the N-terminus; Gx4GKT of Walker A, Kx6–10H4D of Walker B1, and Rx6AIDLHD of Walker B2 at the NBD1; RWTGIPVxKH at the middle domain; GxGKT of Walker A and Rx6h4D of Walker B at the NBD2; FRPEFLNRLDEIIVFxxL at the C-terminus. We also observed some motifs of KYRG of pore 1, GYVG of pore 2, and GARPHxRxHx of sensor and substrate determination (SSD) that are important for the activity of CLPB29,74. Additionally, three unique signatures that are specific for the CLPB proteins were identified at the N-terminus and used to distinguish sequences of different subcellular localizations, they are: MNPxx for cytoplasmic targeted sequences; HTQQE for the plastid targeted sequences; and HSPDx for mitochondria targeted sequences. Since the N-domain is considered to function as a substrate recognition element of the protein25, the motifs may indicate variation in the substrates that interact with CLPB members.
Figure 5.
Sequence signatures identified in CLPB sequences of Arabidopsis, rice, and wheat. Consensus sequences show signature residues of cytoplasm, plastid and mitochondria targeted CLPBs identified at different conserved regions, including: N-terminal domain, middle domain, C-terminal domain, Sensor and Substrate Discrimination (SSD) motif, Nucleotide binding domain I and II. Consensus residues shown in grey shade share similarity among the three different CLPB proteins. Conserved residues are shown in white color with green shade, semi-conserved residues are shown in black color with green shade, and non-conserved residues are shown in different colors without shade. Positions of residues in the sequences are given in the Supplement 8.
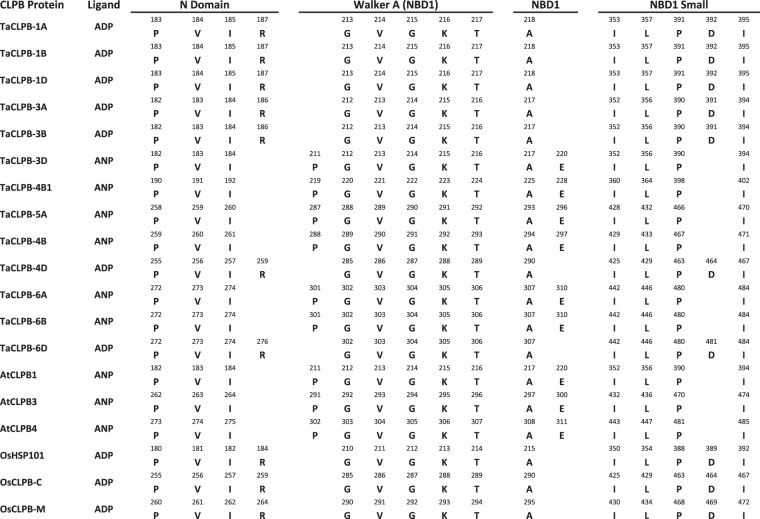
To detect interactions of TaCLPB with other molecules, a ligand binding prediction was performed using the COACH program. Several ligand binding sites were identified in the wheat CLPB sequences as shown in the Table 3. Since this prediction relied on the database of conserved ligand binding across species, unique binding sites might not be identified through the analysis. High confidence scores (C-score) were shown by the binding of wheat CLPB proteins to ANP (the analog of ATP) and ADP. We mapped residues of these two binding sites with respect to the position of conserved domains of CLPBs in the Fig. 6. The major binding sites were located at the N-terminal and the first nucleotide-binding domain (NBD1). In yeast, the type of nucleotide ligand was found to regulate the affinity of HSP104 toward polypeptides75. It will be interesting to see whether the tendency to bind ADP or ANP ligands control the affinity of plant CLPBs to their substrate polypeptides. Some minor binding sites with a lower C-score were also observed for the CLPB members, including ATP, AF3 (aluminum fluoride), MG (magnesium), and GAI (Guanidine), that have also been reported earlier76,77.
Table 3.
Predicted ligand binding properties of CLPB proteins in wheat, rice, and Arabidopsis.
| CLPB protein | Ligand binding prediction in C-score | |||||
|---|---|---|---|---|---|---|
| ADP | ATP | ANP | AF3 | MG | GAI | |
| TaCLPB-1A |
0.75 0.21 |
0.05 | ||||
| TaCLPB-1B |
0.77 0.21 |
0.03 0.02 |
0.05 | |||
| TaCLPB-1D |
0.77 0.21 |
0.01 |
0.03 0.02 |
0.05 0.01 |
||
| TaCLPB-3A |
0.75 0.21 |
0.01 | 0.05 | |||
| TaCLPB-3B |
0.77 0.21 |
0.01 | 0.03 | 0.05 | ||
| TaCLPB-3D |
0.78 0.20 |
0.01 | 0.05 | |||
| TaCLPB-4B1 | 0.22 | 0.01 | 0.77 |
0.05 0.01 |
||
| TaCLPB-5A | 0.19 | 0.01 | 0.75 | 0.05 | ||
| TaCLPB-4B | 0.33 | 0.01 | 0.98 | 0.02 | 0.05 | |
| TaCLPB-4D |
0.74 0.17 |
0.03 | 0.05 | |||
| TaCLPB-6A | 0.20 | 0.73 | 0.05 | 0.02 | ||
| TaCLPB-6B | 0.19 | 0.76 | 0.03 |
0.05 0.02 |
0.02 | |
| TaCLPB-6D |
0.76 0.21 |
0.01 | 0.05 | 0.02 | ||
| AtCLPB1 |
0.93 0.37 |
0.05 0.01 |
||||
| AtCLPB3 |
0.93 0.32 |
0.01 | 0.02 | 0.05 | 0.02 | |
| AtCLPB4 |
0.91 0.33 |
0.05 | ||||
| OsHSP101 | 0.36 | 0.01 | 0.94 | 0.02 |
0.04 0.01 |
|
| OsCLPB-C | 0.32 | 0.92 |
0.05 0.01 |
|||
| OsCLPB-M | 0.33 | 0.01 | 0.90 | 0.02 | 0.05 | 0.02 |
ADP = Adenosine Diphosphate; ATP = Adenosine Triphosphate; ANP = Phosphoaminephosponic Acid-Adenylate Ester; AF3 = Aluminium Fluoride; MG = Magnesium; GAI = Guanidine.
Figure 6.
Residues in the predicted ligand binding sites were mapped to the CLPB conserved domains of Arabidopsis, rice, and wheat. The single letter amino acids are labelled with their positions in the protein.
Expression analyses of TaCLPB genes
Initial expression study of TaCLPB genes was performed in silico by using publicly available RNA-seq databases. The expression patterns of the family members were studied at different developmental stages, tissues, and abiotic stress (drought and heat) conditions. The TaCLPB gene model IDs obtained from Ensemble database were listed in the Table 1.
Figure 7 shows TaCLPB gene expression in roots, stems, leaves, spikes, and grains at three different life stages following the Zadok’s growth scale. Low expression of TaCLPBs were shown in roots and stems. In the leaves, expression increases were observed from the three-tiller stage to 2DAA. Cytoplasmic TaCLPBs showed lower expression compared to the organellar members at the vegetative stages but increased significantly after meiosis until the early grain filling stage in leaves and reproductive tissues. TraesCS4B02G393100 (TaCLPB-4B1) expression was observed in mature leaves at the grain filling stage. Meanwhile, organellar targeted genes showed relatively stable expression at all stages, with decreases in reproductive tissues at the later stages. In general, at 30DAA, the expression levels of all TaCLPBs were decreased four to eight folds (Supplement 10).
Figure 7.
The expression of TaCLPB genes in different tissues and life stages. Digital analysis was done using the expVIP wheat expression browser against an RNAseq database of the wheat development time course (T. aestivum cv. Chinese Spring; ENA: ERP00471 4). Transcript per million (TPM) is the RNAseq expression unit. Differential expression was analyzed statistically in the EdgeR program. Stars are representing significant difference at one stage in comparison with the earliest stage of each tissue in the analysis (Significance codes: <0.0001 ‘***’; <0.001 ‘**’; <0.01 ‘*’; <0.05 ‘ . ’).
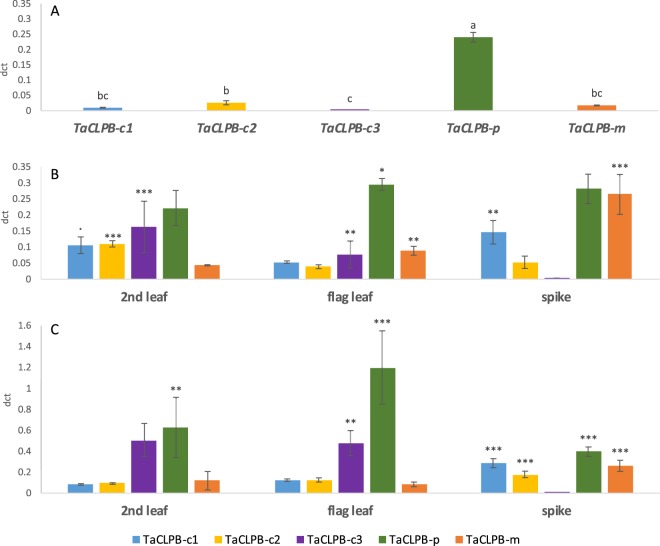
Using real-time PCR, we confirmed the expression patterns of TaCLPB genes in a group-wise. In total, five primers were designed to amplify TaCLPBs of the cytoplasmic (TaCLPB-c) groups from the group 1 chromosomes (TaCLPB-c1), the group 3 chromosomes (TaCLPB-c2), and the group 4 chromosomes (TaCLPB-c3); the plastid targeted group (TaCLPB-p); and the mitochondria targeted group (TaCLPB-m). Variation in expression levels were observed among the TaCLPB groups at the seedling, anthesis, and grain filling (7DAA) stages in different tissues including young leaves, mature leaves (second leaves), flag leaves, and spikes (Fig. 8). Similar patterns of group expression were observed between the real-time PCR and the in silico analyses. At the seedling stage, except the plastid group, all the TaCLPB members showed low expression. Cytoplasmic groups of the chromosome 1 and 3 were more expressed in leaves and reproductive tissues (spike) in the beginning of the reproductive stages (anthesis-7DAA). At these stages, TraesCS4B02G393100 (TaCLPB-4B1) was significantly expressed in the second leaves and flag leaves. Plastid targeted TaCLPB group showed highest expression level in all stages, except in the spike at the 7DAA which tend to be lower. Meanwhile, expression of mitochondrial group was significantly increased in spikes at the anthesis and 7DAA.
Figure 8.
Real time expression of TaCLPB genes in different tissues at the seedling (A), the anthesis (B) and & the 7DAA grain filling (C) stages. Five primers were used to amplify transcripts from the cytoplasmic (TaCLPB-c1, TaCLPB-c2, TaCLPB-c3), the plastid (TaCLPB-p), and the mitochondria (TaCLPB-m) groups respectively. Statistics were done using the analysis of variance (anova) for multiway-treatment structure, followed by a post-hoc multiple comparison using Tukey’s test. Letters are indicating statistical variation among genes at the control. Stars are representing significant differences of expression in comparison with the control (Significance codes: <0.0001 ‘***’; <0.001 ‘**’; <0.01 ‘*’; <0.05 ‘ . ’).
Previous studies on the developing plant organs of maize and wheat have revealed HSP101 expression during plant growth and development. Without stress treatments, cytoplasmic HSP101 proteins were abundant in tassels (at the pre-meiosis stage), ears, silks, endosperms, and the embryos of both plants. During kernel imbibition, maize HSP101 decreased and finally disappeared within 3 days78. Very little HSP101 protein was present in the leaves and roots under this non-stress condition. However, in maize the level of HSP101 protein and transcript were increased after heat treatments in the vegetative and floral meristematic regions, fully expanded foliar leaves, young ears and roots; but not in anthers at the anthesis, mature pollens, and in the developing endosperm or embryos79. We observed similarities between the previous studies with our observation on the cytoplasmic TaCLPBs of the chromosomes 1 and 3. It seems the proteins are produced and accumulated during seed formation but not required in the vegetative stage, unless there is a stress.
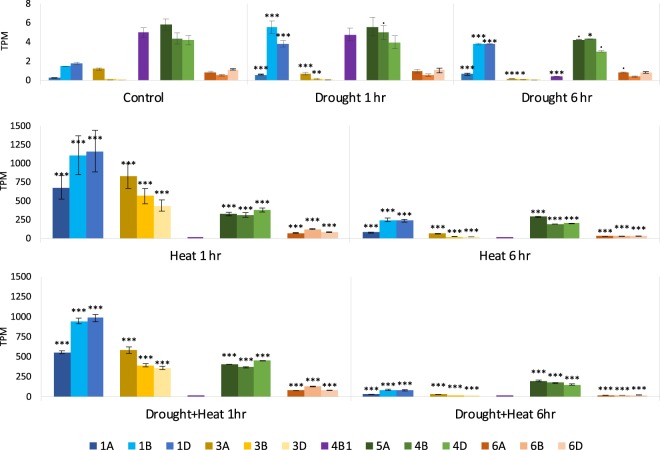
In silico expression of TaCLPBs under drought and heat treatments were shown in the Fig. 9. Under drought treatment. TaCLPBs of the chromosome 1 increased expression 2–4 folds without significant differences between 1 h and 6 h treatments (Supplement 11). Expression decreases were observed in TraesCS3A02G274400 (TaCLPB-3A) and TraesCS4B02G393100 (TaCLPB-4B1) by 6 h drought stress. Under heat stress, expression increases were observed in all the TaCLPB members, except TraesCS4B02G393100 (TaCLPB-4B1). Five-hour extension of heat stress resulted in lower level of gene expression. Similar patterns were observed under the combination of heat and drought treatments.
Figure 9.
The expression of TaCLPB genes under drought and heat treatments. Digital analysis was done using the expVIP wheat expression browser against an RNAseq database of the heat and drought stress (T, aestivum cv. TAM107; SRA: SRX25791 5). The RNA libraries were made from leaves of 1-week old seedlings at a control condition (A); one-hour and six-hour drought treatments using 20% PEG6000 (A); one-hour and six-hour 40 °C heat treatments (B); and the combinations of drought and heat treatments (C). Transcript per million (TPM) is the RNAseq expression unit. Differential expression was analyzed statistically in the EdgeR program. Stars are representing significant changes by treatments in comparison with the control (Significance codes: <0.0001 ‘***’; <0.001 ‘**’; <0.01 ‘*’; <0.05 ‘ . ’).
The roles of organellar CLPBs in heat tolerance have not been explored in as much detail as the cytoplasmic one. In Arabidopsis and rice, potency of the proteins to confer thermotolerance was revealed by the ability of the CLPB genes to complement yeast hsp104 mutant14,16,80. Over expression of cytoplasmic CLPB could improve thermotolerance to 45–50 °C heat stress in rice81. In Arabidopsis, besides its ability to confer thermotolerance82, cytoplasmic CLPB or known as HSP101 was found to have pleiotropic effects which affect plant fitness83. Studies on organellar CLPB have been reported in Arabidopsis and tomato. Plastid target CLPB was predicted to play a role in chloroplast formation and conferring thermotolerance in this organelle19. Silencing of a plastid targeted CLPB caused impaired acquisition of thermotolerance in tomato21. In silico, we observed increases of expression of the organellar CLPBs under heat treatments in leaves at the seeding stage. This increased expression indicates a potential role of organellar CLPBs under the stress, perhaps in collaboration with the cytoplasmic CLPBs.
Homoeologous specific copies of the Chromosome group 1 TaCLPBs
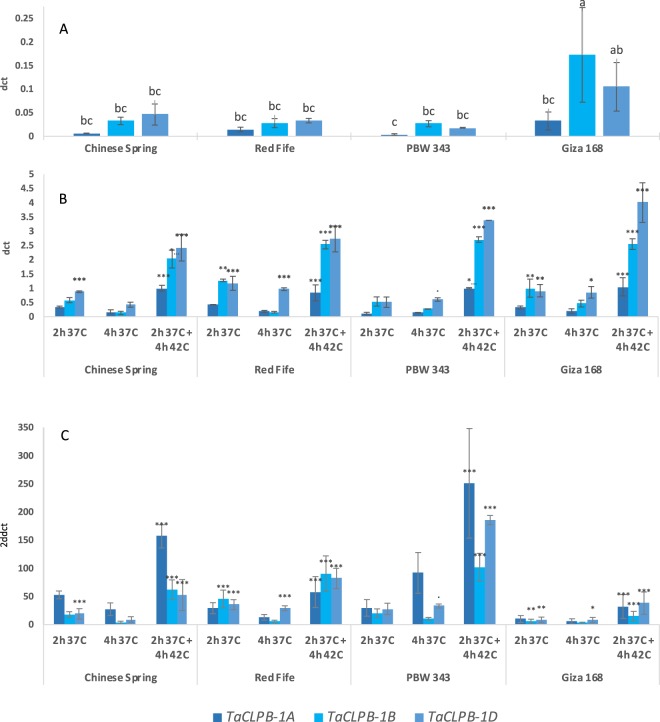
The cytoplasmic members of CLPBs, that are typically known as HSP101, have been well studied in several plant species, including Arabidopsis, maize, soybean, and rice14–16,84. Most of these studies characterized the gene as a functional ortholog through a yeast hsp104 complementation, and or by showing its positive effect on thermotolerance. In wheat, a cytoplasmic TaCLPB from the chromosome 1A was known to play a role as an mRNA binding protein that activates protein translations34. Though it is still not clear how this role is related to thermotolerance, the ability of the copy to complement yeast hsp104 indicated its similar function with HSP104. B and D copies, of the chromosome group 1 (TraesCS1B01G352400 and TraesCS1D01G342100) that have not been well characterized, were expected to also share similar functions with yeast HSP104. We confirmed the presence of the A, B, and D homoeologs through sequence cloning and compared their expression under different heat treatments in four wheat genotypes (Fig. 10).
Figure 10.
Homoeologous-specific expression of the chromosome I cytoplasmic TaCLPBs in four wheat genotypes under different heat treatments. Transcript analysis was done by qPCR using a relative quantification in three biological replicates. 2dct represents a normalized expression ratio (in comparison with actin reference), in the leaves of 12-day old seedlings under 22 °C control (A), and heat treatments of 2 h at 37 °C; 4 h at 37 °C; 2 h 37 °C plus 4 h at 42 °C (B). 2ddct represents a calibrated expression ratio of transcripts between a treatment and the control. Statistics were done using the analysis of variance (anova) for multiway-treatment structure, followed by a post-hoc multiple comparison using Tukey’s test. Letters are indicating statistical variation among genes at the control. Stars are representing significant changes of expression in comparison with the control (Significance codes: <0.0001 ‘***’; <0.001 ‘**’; <0.01 ‘*’; <0.05 ‘ . ’).
The genomic clones of TaCLPBs from the group 1 chromosomes were sequenced and used to confirm the full-length sequences identified through the bioinformatics analysis. To confirm the intron-exon junctions, the three homoeologous copies were aligned with a cDNA sequence of chromosome 1D copy. Minor mistakes observed in the sequences from the database were corrected using these cloned sequences. Homoeologous specific primers were then designed and optimized through a PCR on the Chinese Spring nulli-tetra lines as shown in the Supplement 9. Nulli-tetra lines are the lines that are missing one chromosome pair which is replaced by another homoeologous chromosome pair (nulli 1A means missing a pair of the A homoeologs of the chromosome 1). Using the primers that are designed specifically to amplify the chromosome 1A copy in the PCR, there should be no amplification detected on the nulli 1A (nulli-tetra N1AT1B) template. The same principle is applied for 1B and 1D copy-specific primers, there were no amplification in the PCR on the nulli 1B (nulli-tetra N1BT1D) and nulli 1D (nulli-tetra N1DT1A) templates, respectively.
In silico analysis has shown interesting responses of the chromosome 1 TaCLPBs under drought and heat treatments. Using real-time PCR, we confirmed this group expression in four wheat genotypes originated from different climate regions (Fig. 10). We found the 1A homoeolog showed significant increase of expression in four genotypes by 2 h 37 °C followed by 4 h 42 °C treatment. Significant increases of 1B homoeolog expression were observed under 2 h 37 °C and the combination of 2 h 37 °C followed by 4 h 42 °C. The 1D homoeolog showed increased expression in all the three treatments in Giza168 and Red Fife. Extended 37 °C heat treatment up to 4 h exposure resulted in a lower expression of TaCLPB copies when compared to the shorter 2 h exposure. This is similar with the results from the in-silico analysis of the SRA: SRP045409 dataset for the 2 h vs 6 h under 40 °C heat treatments (see Fig. 9), longer exposure to the heat stress decreased TaCLPB expression. Decreased gene expression after a long-term heat stress might be related to the optimum level of the CLPB proteins that created a negative feedback loop to the transcript expression.
The variation in expression among homoeologous copies indicates necessity to identify all the homoeologs before studying the gene expression in polyploids. A bias in the expression level could be easily introduced to the analysis by using the primers that are not specific to only one copy, or otherwise common to represent all the homoeologs. This aspect is critical especially if one need to compare the gene expression among genotypes.
In conclusions, complexity of the wheat genome creates special challenges to study gene function and its potential use in breeding programs. In this study, a systematic approach was taken to understand the role of HSP101/CLPB genes in heat tolerance through a genome-wide bioinformatics analysis, followed by real-time expression studies. Thirteen copies of CLPB genes were identified and characterized for their structural variations and differential expression patterns. The results suggest possible different functions of TaCLPBs with respect to their chromosomal and subcellular localizations. The expression analysis of TaCLPBs of the group 1 chromosomes revealed variation among homoeologous copies with respect to different temperature treatments. In this experiment, the variation of TaCLPB expression among four genotypes would not be enough to show the link between TaCLPB with thermotolerance in varieties. Nevertheless, it provides basic information for further studies to reveal the potentials of TaCLPB for variety improvement, including development of gene-based markers.
Supplementary information
Author contributions
E.E., R.N., N.G. and K.S.G. conceived the original screening and research plans; E.E. as the main contributor of the manuscript designed and performed most of the experiments, conceived the project and wrote the article with contributions of all the authors; R.N. contributed for some technical assistance, experimental design and complemented the writing; N.G. contributed for some preliminary experiment; K.S.G. supervised and complemented the writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60673-4.
References
- 1.Farooq M, Bramley H, Palta JA, Siddique KHM. Heat Stress in Wheat during Reproductive and Grain-Filling Phases. CRC. Crit. Rev. Plant Sci. 2011;30:491–507. doi: 10.1080/07352689.2011.615687. [DOI] [Google Scholar]
- 2.Cossani CM, et al. Physiological traits for improving heat tolerance in wheat. Front. Plant Sci. 2017;2:1–18. doi: 10.1104/pp.112.207753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wahid A, Gelani S, Ashraf M, Foolad MR. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007;61:199–223. doi: 10.1016/j.envexpbot.2007.05.011. [DOI] [Google Scholar]
- 4.Gupta NK, et al. Effect of short-term heat stress on growth, physiology and antioxidative defence system in wheat seedlings. Acta Physiol. Plant. 2013;35:1837–1842. doi: 10.1007/s11738-013-1221-1. [DOI] [Google Scholar]
- 5.Su Xiaoyu, Wu Si, Yang Li, Xue Ruili, Li Hua, Wang Yuexia, Zhao Huijie. Exogenous progesterone alleviates heat and high light stress-induced inactivation of photosystem II in wheat by enhancing antioxidant defense and D1 protein stability. Plant Growth Regulation. 2014;74(3):311–318. doi: 10.1007/s10725-014-9920-1. [DOI] [Google Scholar]
- 6.Law RD, Crafts-Brandner SJ. Inhibition and acclimation of photosynthesis to heat stress is closely correlated with activation of ribulose-1,5-bisphosphate Carboxylase/Oxygenase. Plant Physiol. 1999;120:173–82. doi: 10.1104/pp.120.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammed Abdul-Razack, Tarpley Lee. Impact of High Nighttime Temperature on Respiration, Membrane Stability, Antioxidant Capacity, and Yield of Rice Plants. Crop Science. 2009;49(1):313–322. doi: 10.2135/cropsci2008.03.0161. [DOI] [Google Scholar]
- 8.SMERTENKO A., DRABER 1,2 P., VIKLICKY V., OPATRNY Z. Heat stress affects the organization of microtubules and cell division in Nicotiana tabacum cells. Plant, Cell and Environment. 1997;20(12):1534–1542. doi: 10.1046/j.1365-3040.1997.d01-44.x. [DOI] [Google Scholar]
- 9.Schirmer E, Glover J, Singer M, Lindquist S. {HSP100/Clp} proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 1996;21:289–296. doi: 10.1016/S0968-0004(96)10038-4. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science (80-.). 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 11.Parsell DA, Lindquist S. the Function of Heat Shock Proteins in Stress Tolerance: Degradation and Reactivation of Damaged Proteins. Annu. Rev. Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 12.Squires CL, Pedersen S, Ross BM, Squires C. C1pB Is the Escherichia coli Heat Shock Protein F84. 1. J. Bacteriol. 1991;173:4254–4262. doi: 10.1128/JB.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson MJ, Clarke AK. The Escherichia coli heat shock protein ClpB restores acquired thermotolerance to a cyanobacterial clpB deletion mutant. Cell. Stress Chaperones. 2000;5:255–64. doi: 10.1379/1466-1268(2000)005<0255:TECHSP>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schirmer EC, Lindquist S, Vierling E. An Arabidopsis Heat Shock Protein Complements a Thermotolerance Defect in Yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee YR, et al. A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell. 1994;6:1889–97. doi: 10.1105/tpc.6.12.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal M, et al. Molecular characterization of rice hsp101: Complementation of yeast hsp104 mutation by disaggregation of protein granules and differential expression in indica and japonica rice types. Plant Mol. Biol. 2003;51:543–553. doi: 10.1023/A:1022324920316. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt M, Neupert W, Langer T. The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J. Cell. Biol. 1996;134:1375–1386. doi: 10.1083/jcb.134.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keeler SJ, et al. Acquired thermotolerance and expression of the HSP100/ClpB genes of lima bean. Plant Physiol. 2000;123:1121–32. doi: 10.1104/pp.123.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myouga F, Motohashi R, Kuromori T, Nagata N, Shinozaki K. An Arabidopsis chloroplast-targeted Hsp101 homologue, APG6, has an essential role in chloroplast development as well as heat-stress response. Plant J. 2006;48:249–260. doi: 10.1111/j.1365-313X.2006.02873.x. [DOI] [PubMed] [Google Scholar]
- 20.Lee U, et al. The Arabidopsis ClpB/Hsp100 family of proteins: Chaperones for stress and chloroplast development. Plant J. 2007;49:115–127. doi: 10.1111/j.1365-313X.2006.02940.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang JY, et al. The involvement of chloroplast HSP100/ClpB in the acquired thermotolerance in tomato. Plant Mol. Biol. 2006;62:385–395. doi: 10.1007/s11103-006-9027-9. [DOI] [PubMed] [Google Scholar]
- 22.Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: Common structure-diverse function. Genes to Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- 23.Barnett ME, Zolkiewska A, Zolkiewski M. Structure and activity of ClpB from Escherichia coli. Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 2000;275:37565–37571. doi: 10.1074/jbc.M005211200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 25.Beinker P, Schlee S, Groemping Y, Seidel R, Reinstein J. The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 2002;277:47160–47166. doi: 10.1074/jbc.M207853200. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, et al. Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc. Natl. Acad. Sci. USA. 2013;110:8513–8518. doi: 10.1073/pnas.1217988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the Mechanism of Protein Disaggregation Through a ClpB-DnaK Interaction. Science (80-.). 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 28.Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP lO0/Clp proteins: a common mechanism explains diverse functions. Trends Biochem. Sci. 1996;21:289–296. doi: 10.1016/S0968-0004(96)10038-4. [DOI] [PubMed] [Google Scholar]
- 29.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/S0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Sielaff B, Lee J, Tsai FTF. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl. Acad. Sci. USA. 2010;107:8135–40. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat. Struct. Mol. Biol. 2012;19:1338–46. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 32.Carroni M, et al. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife. 2014;2014:1–22. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miot M, et al. Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc. Natl. Acad. Sci. USA. 2011;108:6915–6920. doi: 10.1073/pnas.1102828108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells DR, Tanguay RL, Le H, Gallie DR. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 1998;12:3236–51. doi: 10.1101/gad.12.20.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallie DR, Kado CI. A translational enhancer derived from tobacco mosaic virus is functionally equivalent to a Shine-Dalgarno sequence. Proc. Natl. Acad. Sci. USA. 1989;86:129–32. doi: 10.1073/pnas.86.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling J, et al. Heat Shock Protein HSP101 Binds to the Fed-1 Internal Light Regulatory Element and Mediates Its High Translational Activity. Plant Cell. 2000;12:1213–1227. doi: 10.1105/tpc.12.7.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell JL, et al. Cloning of new members of heat shock protein HSP101 gene family in wheat (Triticum aestivum (L.) Moench) inducible by heat, dehydration, and ABA. Biochim. Biophys. Acta. 2001;1517:270–277. doi: 10.1016/S0167-4781(00)00292-X. [DOI] [PubMed] [Google Scholar]
- 38.Muthusamy SK, Dalal M, Chinnusamy V, Bansal KC. Differential Regulation of Genes Coding for Organelle and Cytosolic ClpATPases under Biotic and Abiotic Stresses in Wheat. Front. Plant Sci. 2016;7:929. doi: 10.3389/fpls.2016.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldman M, Levy AA. Genome evolution due to allopolyploidization in wheat. Genetics. 2012;192:763–774. doi: 10.1534/genetics.112.146316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dvořák J, Terlizzi P, di, Zhang H-B, Resta P. The evolution of polyploid wheats: identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 41.McFadden ES, Sears ER. The Origin of Triticum spelta and Its Free-Threshing Hexaploid Relatives. J. Hered. 1946;37:81–89. doi: 10.1093/oxfordjournals.jhered.a105590. [DOI] [PubMed] [Google Scholar]
- 42.Riley R, Unrau J, Chapman V. Evidence on the Origin of the B Genome of Wheat. J. Hered. 1958;49:91–98. doi: 10.1093/oxfordjournals.jhered.a106784. [DOI] [Google Scholar]
- 43.Sarkar P, Stebbins GL. Morphological Evidence Concerning the Origin of the B Genome in Wheat. Am. J. Bot. 1956;43:297–304. doi: 10.1002/j.1537-2197.1956.tb10494.x. [DOI] [Google Scholar]
- 44.Kihara H. Discovery of the DD-Analyser, One of the Ancestors of Triticum vulgare. Agric. Hortic. 1944;19:13–14. [Google Scholar]
- 45.Glover NM, Redestig H, Dessimoz C. Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016;21:609–621. doi: 10.1016/j.tplants.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doyle JJ, et al. Evolutionary Genetics of Genome Merger and Doubling in Plants. Annu. Rev. Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- 47.Yoo M, Szadkowski E, Wendel JF. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity (Edinb). 2013;110:171–180. doi: 10.1038/hdy.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu G, et al. Evolutionary conservation and divergence of gene coexpression networks in gossypium (Cotton) seeds. Genome Biol. Evol. 2016;8:3765–3783. doi: 10.1093/gbe/evw280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grover CE, et al. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012;196:966–971. doi: 10.1111/j.1469-8137.2012.04365.x. [DOI] [PubMed] [Google Scholar]
- 50.Nomaguchi T, et al. Homoeolog expression bias in allopolyploid oleaginous marine diatom Fistulifera solaris. BMC Genomics. 2018;19:1–17. doi: 10.1186/s12864-018-4691-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016;48:1225–1235. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- 52.Dong S, Adams KL, Adams KL. Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol. 2011;190:1045–1057. doi: 10.1111/j.1469-8137.2011.03650.x. [DOI] [PubMed] [Google Scholar]
- 53.Leach LJ, et al. Patterns of homoeologous gene expression shown by RNA sequencing in hexaploid bread wheat. BMC Genomics. 2014;15:276. doi: 10.1186/1471-2164-15-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, Z. et al. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15 (2015). [DOI] [PMC free article] [PubMed]
- 55.Mutti JS, Bhullar RK, Gill KS. Evolution of Gene Expression Balance Among Homeologs of Natural Polyploids. G3 (Bethesda). 2017;7:1225–1237. doi: 10.1534/g3.116.038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramírez-González RH, et al. The transcriptional landscape of polyploid wheat. Science (80-.). 2018;361:1–12. doi: 10.1126/science.aar6089. [DOI] [PubMed] [Google Scholar]
- 57.Yue H, et al. Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Wheat. Genes (Basel). 2018;9:70. doi: 10.3390/genes9020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gullì M, Corradi M, Rampino P, Marmiroli N, Perrotta C. Four members of the HSP101 gene family are differently regulated in Triticum durum Desf. FEBS Lett. 2007;581:4841–4849. doi: 10.1016/j.febslet.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 59.Borrill P, Harrington SA, Uauy C. Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J. 2019;97:56–72. doi: 10.1111/tpj.14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaman QU, Li C, Cheng H, Hu. Q. Genome editing opens a new era of genetic improvement in polyploid crops | Elsevier Enhanced Reader.pdf. Crop J. 2019;7:141–150. doi: 10.1016/j.cj.2018.07.004. [DOI] [Google Scholar]
- 61.Pertsemlidis A, Fondon JW, Fondon JW., III Having a BLAST with bioinformatics (and avoiding BLASTphemy) Genome biology. 2001;2:1–10. doi: 10.1186/gb-2001-2-10-reviews2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The International Wheat Genome Sequencing Consortium, (IWGSC) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 63.Dhaliwal, A. K., Mohan, A. & Gill, K. S. Comparative analysis of ABCB1 reveals novel structural and functional conservation between monocots and dicots. Front. Plant Sci.5, 657 (2014). [DOI] [PMC free article] [PubMed]
- 64.Emanuelsson O, Nielsen H, Heijne G. Von. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 66.Stamatakis A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics. 2014;30:8–10. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huson DH, Scornavacca CS. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 68.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang J, Roy A, Zhang Y. Structural bioinformatics Protein – ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics. 2013;29:2588–2595. doi: 10.1093/bioinformatics/btt447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sambrook, J., Fristch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor Laboratory press, 1989).
- 72.Ruijter JM, et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:1–12. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Livak, K. J. & S, D. Analysis of Relative Gene Expression Data Using Real- Time Quantitative PCR and the 2 Ϫ ⌬⌬ C T Method. 408, 402–408 (2001). [DOI] [PubMed]
- 74.Smith CK, Baker TA, Sauer RT. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bösl B, Grimminger V, Walter S. Substrate binding to the molecular chaperone Hsp104 and its regulation by nucleotides. J. Biol. Chem. 2005;280:38170–38176. doi: 10.1074/jbc.M506149200. [DOI] [PubMed] [Google Scholar]
- 76.Zeymer C, Barends TRM, Werbeck ND, Schlichting I, Reinstein J. Elements in nucleotide sensing and hydrolysis of the AAA+ disaggregation machine ClpB: A structure-based mechanistic dissection of a molecular motor. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014;70:582–595. doi: 10.1107/S1399004713030629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kummer E, Oguchi Y, Seyffer F, Bukau B, Mogk A. Mechanism of Hsp104/ClpB inhibition by prion curing Guanidinium hydrochloride. FEBS Lett. 2013;587:810–817. doi: 10.1016/j.febslet.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 78.Nieto-Sotelo J, et al. Maize HSP101 Plays Important Roles in Both Induced and Basal Thermotolerance and Primary Root Growth. Plant Cell. 2002;14:1621–1633. doi: 10.1105/tpc.010487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Young TE, et al. Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol. 2001;127:777–791. doi: 10.1104/pp.010160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh A, Grover A. Plant Hsp100/ClpB-like proteins: Poorly-analyzed cousins of yeast ClpB machine. Plant Mol. Biol. 2010;74:395–404. doi: 10.1007/s11103-010-9682-8. [DOI] [PubMed] [Google Scholar]
- 81.Katiyar-Agarwal S, Agarwal M, Grover A. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol. 2003;51:677–686. doi: 10.1023/A:1022561926676. [DOI] [PubMed] [Google Scholar]
- 82.Queitsch C. Heat Shock Protein 101 Plays a Crucial Role in Thermotolerance in Arabidopsis. Plant Cell Online. 2000;12:479–492. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.TONSOR S. J., SCOTT C., BOUMAZA I., LISS T. R., BRODSKY J. L., VIERLING E. Heat shock protein 101 effects in A. thaliana: genetic variation, fitness and pleiotropy in controlled temperature conditions. Molecular Ecology. 2008;17(6):1614–1626. doi: 10.1111/j.1365-294X.2008.03690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nieto-Sotelo J, Kannan KB, Martínez LM, Segal C. Characterization of a maize heat-shock protein 101 gene, HSP101, encoding a ClpB/Hsp 100 protein homologue. Gene. 1999;230:187–195. doi: 10.1016/S0378-1119(99)00060-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.