Abstract
Gastric cancer (GC) is one of the most common types of cancer worldwide. Patients must be identified at an early stage of tumor progression for treatment to be effective. The aim of the present study was to identify potential biomarkers with diagnostic value in patients with GC. To examine potential therapeutic targets for GC, four Gene Expression Omnibus (GEO) datasets were downloaded and screened for differentially expressed genes (DEGs). Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were subsequently performed to study the function and pathway enrichment of the identified DEGs. A protein-protein interaction (PPI) network was constructed. The CytoHubba plugin of Cytoscape was used to calculate the degree of connectivity of proteins in the PPI network, and the two genes with the highest degree of connectivity were selected for further analysis. Additionally, the two DEGs with the largest and smallest log Fold Change values were selected. These six key genes were further examined using Oncomine and the Kaplan-Meier plotter platform. A total of 99 upregulated and 172 downregulated genes common to all four GEO datasets were screened. The DEGs were primarily enriched in the Biological Process terms: ‘extracellular matrix organization’, ‘collagen catabolic process’ and ‘cell adhesion’. These three KEGG pathways were significantly enriched in the categories: ‘ECM-receptor interaction’, ‘protein digestion and absorption’, and ‘focal adhesion’. Based on Oncomine, expression of ATP4A and ATP4B were downregulated in GC, whereas expression of the other genes were all upregulated. The Kaplan-Meier plotter platform confirmed that upregulated expression of the identified key genes was significantly associated with worse overall survival of patients with GC. The results of the present study suggest that FN1, COL1A1, INHBA and CST1 may be potential biomarkers and therapeutic targets for GC. Additional studies are required to explore the potential value of ATP4A and ATP4B in the treatment of GC.
Keywords: gastric cancer, differentially expressed genes, key genes, bioinformatics analysis, diagnosis
Introduction
Gastric cancer (GC) is a malignant tumor that originates in the epithelium of the gastric mucosa and is one of the most common types of malignant tumors in the world (1). According to GLOBOCAN 2018, there were >1,000,000 new cases of GC and ~783,000 deaths in 2018, thus making it the cancer type with the fifth highest incidence rate and the third highest mortality in the world (2). The poor five-year survival rate of GC is primarily due the advanced stage of gastric tumors at the initial diagnosis in the majority of patients, and thus limits treatment opportunities (3). According to the Cancer Staging Manual, 8th edition, of the American Joint Committee on Cancer, only 30% of GC cases are diagnosed prior to metastasis, and the five-year survival for pathological Tumor-Node-Metastasis stage groups are between 68-80% for stage I, 46-60% for stage II, 8-30% for stage III and 5% for stage IV (4). Therefore, identifying potential biomarkers for patients with early GC is critical for improving patient outcomes.
In recent years, a variety of bioinformatics methods have contributed greatly to the discovery of biomarkers associated with tumor development, diagnosis and prognosis (5-8). The combined use of multiple databases of biological information for the analysis of cancer has also yielded certain breakthroughs. Yong et al (9) used Gene Expression Omnibus (GEO), Oncomine, Search Tool for Recurring Instances of Neighbouring Genes (STRING) and other databases for bioinformatic analysis, and concluded that PPP2CA may function as an oncogene and a prognostic biomarker or therapeutic target in the progression of colorectal cancer. Troiano et al (10) used the GEO database and Oncomine to examine the expression of BIRC5/Survivin in oral squamous cell carcinoma and showed that Survivin expression was upregulated compared with non-cancerous tissue. In addition, immunohistochemistry staining showed that cytoplasmic expression of Survivin was associated with poor overall survival in patients with oral squamous cell carcinoma. It may be beneficial to use multiple datasets and analysis tools to determine the potential mechanisms underlying development and progression of GC, and to identify potentially novel and specific diagnostic biomarkers for early detection of GC to improve the survival of patients.
In the present study, the expression profiles from four datasets (GSE13911, GSE19826, GSE54129 and GSE118916) in human GC and normal gastric tissue samples were obtained from the GEO database and analyzed to identify differentially expressed genes (DEGs). Gene Ontology (GO) and pathway enrichment analysis were performed to identify the biological functions and pathways of the DEGs. STRING and Cytoscape were used to construct a protein-protein interaction (PPI) network, and a total of six key genes were selected from the PPI network and DEGs. The value of the key genes was validated using the Oncomine and Kaplan-Meier platforms to further increase the reliability of the results and confirm the prognostic value of the key genes.
Materials and methods
Microarray data
The key word ‘gastric cancer’ was searched in the GEO database (ncbi.nlm.nih.gov/geo/), and a total of 9,224 datasets on human GC were retrieved. In the present study, four gene expression profiles from the GEO database were used, as they have not been studied together previously. The four datasets were: GSE13911(11), GSE19826(12), GSE54129 and GSE118916(13). Among these, GSE13911, GSE19826 and GSE54129 were based on the GPL570 platform [(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array]. GSE118916 was based on the GPL15207 platform [(PrimeView) Affymetrix Human Gene Expression Array].
Identification of DEGs
DEGs between GC samples and normal controls were identified using the GEO2R online analysis tool (ncbi.nlm.nih.gov/geo/geo2r); |log FC|≥1.0 and corrected P<0.05 were used as the cutoff criteria. The common DEGs of the four gene expression profiles were screened using Wayne analysis in Funrich (funrich.org/).
GO and KEGG enrichment analyses of DEGs
After obtaining the common DEGs, GO (14,15) and KEGG (16) analyses of the DEGs were performed using the Database for Annotation Visualization and Integrated Discovery (DAVID) online tool (17,18), with P<0.01 used as the threshold for significance. GO was used to identify the enrichment functions of three independent categories of genes; biological process (BP), cellular component (CC) and molecular function (MF). KEGG was used to search for the pathways associated with the identified genes (19). Only the top 10 BP, CC and MF terms, and the KEGG pathway with the smallest P-value were selected for further examination in the present study. The figures were generated using the OmicShare tools (omicshare.com/tools), a free online platform for data analysis.
PPI network construction
To explore the interaction between DEGs, the DEGs were analyzed using STRING (20) to generate a PPI network. PPI pairs with a combined score >0.4 were extracted, and disconnected nodes in the network were hidden. Subsequently, the PPI network was visualized using Cytoscape (21) and the degree of each protein node was calculated using the cytoHubba (22) plug-in in Cytoscape.
Identification of key genes
The two genes with the highest degree of connectivity in the PPI network, the two genes with the largest logFC values and the two genes with the smallest logFC among the shared DEGs were selected and considered key genes.
Analysis of key genes in Oncomine
The Oncomine database (oncomine.org/) was used to explore the mRNA expression differences of six key genes between GC and normal gastric tissue. Oncomine is a chip-based gene database and integrated data mining online cancer microarray database designed to facilitate the discovery of novel biomarkers from genome-wide expression analysis (23).
Survival analysis of key genes
The Kaplan-Meier plotter (24) is an online tool that can assess the effect of 54,000 genes on survival in 21 types of cancer. The largest datasets include breast (n=6,234), ovarian (n=2,190), lung (n=3,452) and gastric cancer (n=1,440) cancer. The primary purpose of the tool is to discover and validate biomarkers for survival. Online survival analysis of the selected key genes based on the GC database was performed using Kaplan-Meier Plotter. The hazard ratio (HR) with 95% confidence intervals (CIs) and log-rank P-values were calculated.
Results
Identification of DEGs
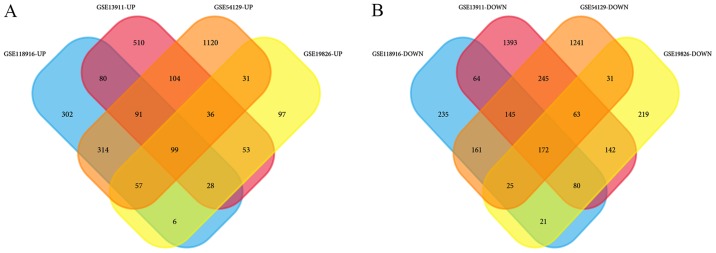
GSE13911 includes 38 GC samples and 31 normal samples, GSE19826 contains 12 GC samples and 15 normal samples, GSE54129 contains 111 GC samples and 21 normal samples, and GSE118916 contains 15 GC samples and 15 normal samples (Table I). In GSE13911, there are 26 intestinal, 4 mixed, 6 diffuse and 2 unclassified gastric carcinoma tissues, as well as 31 normal adjacent tissues. Unfortunately, information on the histological subtypes were not available in the other datasets. In the datasets, 1,001 upregulated and 2,304 downregulated DEGs were identified in GSE13911, 407 upregulated and 753 downregulated DEGs were identified in GSE19826, 1,852 upregulated and 2,083 downregulated DEGs were identified in GSE54129, and 977 upregulated and 903 downregulated DEGs were identified in GSE118916. Wayne analysis identified 99 common upregulated genes and 172 common downregulated genes were obtained from the 4 datasets (Table II; Fig. 1).
Table I.
Information for four gene expression profiles from Gene Expression Omnibus.
Table II.
The differentially expressed genes identified from the four gene expression profiles, between gastric cancer and normal tissues.
| Differentially expressed genes | Gene terms |
|---|---|
| Upregulated | INHBA CST1 COL11A1 FAP COL10A1 FNDC1 COL8A1 SERPINH1 CDH3 THBS2 CLDN1 TNFRSF11B SPP1 COL1A2 SFRP4 SULF1 CPXM1 BMP1 MFAP2 COL1A1 CTHRC1 BGN RARRES1 IGF2BP3 THBS4 COL6A3 SRPX2 OSR2 HOXB7 TIMP1 ASPN THY1 FKBP10 PRRX1 SDS APOE PMEPA1 COL12A1 GPNMB FBN1 ADAM12 C3 APOC1 COL5A1 SPARC EPHB2 NID2 CMTM3 PLEKHO1 TNFRSF10B EHD2 FN1 MMP11 COCH AMIGO2 COL5A2 OLFML2B KLHL23 SPOCK1 CDH11 TWIST1 RAB31 SULF2 FGD6 VCAN ITGBL1 PCOLCE HAVCR2 THBS1 DNM1 IGFBP7 PLAU TMEM158 COL3A1 FLNA EDNRA LEF1 LIPG FZD2 GXYLT2 S100A10 LGALS1 NRP2 SIRPA ANTXR1 CD9 LIF COL4A2 TGM2 COL6A1 PDPN KCNJ8 ACTN1 GPR161 ZAK RCN3 BAG2 BHLHE40 COL4A1 |
| Downregulated | ATP4A ATP4B KCNE2 AQP4 GIF LIPF GKN1 GKN2 DPCR1 PGC SOSTDC1 ESRRG MUC6 SST FBP2 CPA2 VSIG1 CXCL17 PDIA2 CCKBR TMED6 CHGA TFF2 PSCA FUT9 CA9 SCNN1G GUCA2B C16orf89 SLC26A9 KLK11 CWH43 DNER PSAPL1 CNTN3 ALDH3A1 GATA5 SCGB2A1 UGT2B15 RDH12 CLIC6 NRG4 CLDN18 CAPN9 SLC16A7 SSTR1 FBXL13 TCN1 VSIG2 AKR1B10 B3GNT6 FOLR1 MUM1L1 CHGB MAL TRIM50 AKR7A3 KIAA1324 PAIP2B SULT2A1 PTPRZ1 ARX LIFR ALDH1A1 HYAL1 BEX5 CA2 CYP2C18 ME1 SCNN1B ADH7 GCNT2 ACER2 FMO5 HPGD RASSF6 TFF1 TMEM171 CA4 KCNJ16 LDHD KCNJ15 GABRB3 HOMER2 TMPRSS2 LYPD6B KLHDC7A ARHGAP42 PLAC8 IGFBP2 CAPN13 SYTL5 PDGFD RNASE1 RORC CYP2C9 EPN3 PBLD METTL7A ZBTB7C UBL3 SH3RF2 RNASE4 ARHGEF37 ALDH6A1 RAB27B SULT1B1 PKIB PXMP2 GPRC5C RIMBP2 ATP8A1 FAM20A PIGR GOLM1 CYP3A5 FAM46C C9orf152 COBLL1 FA2H SORBS2 DGKD SGK2 TMEM220 ANG PLLP MYCN C1orf116 FGD4 SLC41A2 ADAM28 MAGI1 GRAMD1C IQGAP2 GULP1 SYTL2 DHRS7 OASL RNF128 DBT ELL2 RAB27A NOSTRIN NEDD4L PPFIBP2 AKR1C3 PELI2 SMPD3 PTPRN2 RASEF TMEM92 ABCC5 GALNT12 LMO4 NTN4 TMEM116 ID4 ELOVL6 ALDOB EPB41L4B CD36 GALNT5 SH3BGRL2 MAGI3 MICALL1 HIPK2 MAOA WWC1 SLC7A8 CDC14B FAM107B SUCLG2 |
Upregulated genes are listed from largest to smallest fold change values. Downregulated genes are listed from smallest to largest fold change values.
Figure 1.
Venn diagram of shared differentially expressed genes. (A) Upregulated and (B) downregulated genes from four gene expression profiles.
GO and KEGG pathway enrichment analyses of DEGs
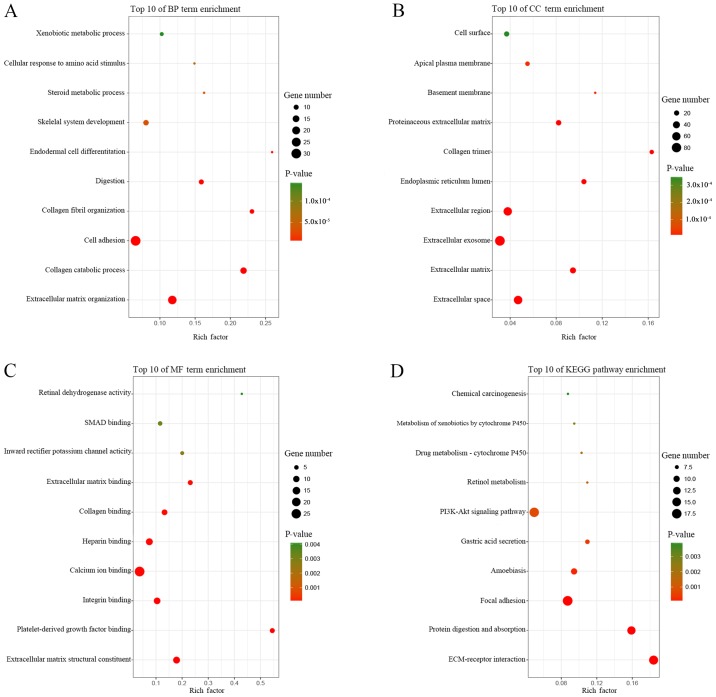
GO and KEGG pathway enrichment analyses of the DEGs was performed using the online tool DAVID, and the results are presented in Table III. GO analysis showed that in BP, the DEGs were primarily enriched for the GO terms: ‘extracellular matrix organization’, ‘collagen catabolic process’, ‘cell adhesion’, ‘collagen fibril organization’ and ‘digestion’ (Table III; Fig. 2A). CC analysis revealed that the DEGs were significantly enriched for the terms: ‘extracellular space’, ‘extracellular matrix’, ‘extracellular exosome’, ‘extracellular region’ and ‘endoplasmic reticulum lumen’ (Table III; Fig. 2B). For MF, the DEGs were enriched for the GO terms: ‘platelet-derived growth factor binding’, ‘collagen binding’, ‘extracellular matrix binding’, ‘inward rectifier potassium channel activity’ and ‘SMAD binding’ (Table III; Fig. 2C). According to KEGG pathway analysis, the DEGs were primarily enriched for the pathway terms: ‘ECM-receptor interaction’, ‘protein digestion and absorption’, ‘focal adhesion’, ‘amoebiasis’ and ‘gastric acid secretion’ (Table III; Fig. 2D).
Table III.
GO term and KEGG pathway enrichment analyses of the 271 differentially expressed genes.
| Category | Term | Description | Count | P-Value |
|---|---|---|---|---|
| BP term | GO:0030198 | Extracellular matrix organization | 23 | 1.28x10-13 |
| BP term | GO:0030574 | Collagen catabolic process | 14 | 7.06x10-12 |
| BP term | GO:0007155 | cell adhesion | 30 | 3.59x10-11 |
| BP term | GO:0030199 | Collagen fibril organization | 9 | 7.87x10-08 |
| BP term | GO:0007586 | Digestion | 10 | 3.19x10-07 |
| BP term | GO:0035987 | Endodermal cell differentiation | 7 | 2.13x10-06 |
| BP term | GO:0001501 | Skeletal system development | 11 | 3.42x10-05 |
| BP term | GO:0008202 | Steroid metabolic process | 7 | 3.60x10-05 |
| BP term | GO:0071230 | Cellular response to amino acid stimulus | 7 | 6.04x10-05 |
| BP term | GO:0006805 | Xenobiotic metabolic process | 8 | 1.45x10-04 |
| BP term | GO:0042060 | Wound healing | 8 | 1.70x10-04 |
| BP term | GO:0006081 | Cellular aldehyde metabolic process | 4 | 4.70x10-04 |
| BP term | GO:0030277 | Maintenance of gastrointestinal epithelium | 4 | 6.20x10-04 |
| BP term | GO:0010107 | Potassium ion import | 5 | 6.98x10-04 |
| BP term | GO:0007584 | Response to nutrient | 7 | 7.50x10-04 |
| BP term | GO:0002576 | Platelet degranulation | 8 | 7.99x10-04 |
| BP term | GO:0060021 | Palate development | 7 | 8.64x10-04 |
| BP term | GO:0010812 | Negative regulation of cell-substrate adhesion | 4 | 0.001003 |
| BP term | GO:0001503 | Ossification | 7 | 0.001131 |
| BP term | GO:0030168 | Platelet activation | 8 | 0.001523 |
| BP term | GO:0051216 | Cartilage development | 6 | 0.001703 |
| BP term | GO:0010628 | Positive regulation of gene expression | 12 | 0.001721 |
| BP term | GO:0001523 | Retinoid metabolic process | 6 | 0.001977 |
| BP term | GO:0016525 | Negative regulation of angiogenesis | 6 | 0.002125 |
| BP term | GO:0055114 | Oxidation-reduction process | 19 | 0.002857 |
| BP term | GO:0032964 | Collagen biosynthetic process | 3 | 0.003084 |
| BP term | GO:0008284 | Positive regulation of cell proliferation | 16 | 0.003752 |
| BP term | GO:0001649 | Osteoblast differentiation | 7 | 0.004274 |
| BP term | GO:0022617 | Extracellular matrix disassembly | 6 | 0.005144 |
| BP term | GO:0071711 | Basement membrane organization | 3 | 0.005647 |
| BP term | GO:0050891 | Multicellular organismal water homeostasis | 3 | 0.005647 |
| BP term | GO:0001525 | Angiogenesis | 10 | 0.005716 |
| BP term | GO:0042476 | Odontogenesis | 4 | 0.007007 |
| BP term | GO:0010575 | Positive regulation of vascular endothelial growth factor production | 4 | 0.007007 |
| BP term | GO:0050909 | Sensory perception of taste | 4 | 0.008568 |
| BP term | GO:0001937 | Negative regulation of endothelial cell proliferation | 4 | 0.008568 |
| BP term | GO:0040037 | Negative regulation of fibroblast growth factor receptor signaling pathway | 3 | 0.008901 |
| BP term | GO:0042572 | Retinol metabolic process | 4 | 0.009418 |
| CC term | GO:0005615 | Extracellular space | 63 | 9.65x10-17 |
| CC term | GO:0031012 | Extracellular matrix | 28 | 2.46x10-14 |
| CC term | GO:0070062 | Extracellular exosome | 87 | 1.68x10-12 |
| CC term | GO:0005576 | Extracellular region | 61 | 4.86x10-12 |
| CC term | GO:0005788 | Endoplasmic reticulum lumen | 20 | 4.73x10-11 |
| CC term | GO:0005581 | Collagen trimer | 15 | 5.56x10-11 |
| CC term | GO:0005604 | Basement membrane | 9 | 1.82x10-05 |
| CC term | GO:0005578 | Proteinaceous extracellular matrix | 22 | 3.57x10-10 |
| CC term | GO:0016324 | Apical plasma membrane | 16 | 2.29x10-05 |
| CC term | GO:0009986 | Cell surface | 20 | 3.51x10-04 |
| CC term | GO:0005887 | Integral component of plasma membrane | 34 | 0.004256 |
| CC term | GO:0005886 | Plasma membrane | 79 | 0.004569 |
| CC term | GO:0030141 | Secretory granule | 6 | 0.004319 |
| CC term | GO:0031093 | Platelet alpha granule lumen | 5 | 0.008125 |
| CC term | GO:0031090 | Organelle membrane | 6 | 0.008522 |
| MF term | GO:0048407 | Platelet-derived growth factor binding | 6 | 2.55x10-07 |
| MF term | GO:0005518 | Collagen binding | 8 | 2.37x10-05 |
| MF term | GO:0050840 | Extracellular matrix binding | 6 | 3.05x10-05 |
| MF term | GO:0005242 | Inward rectifier potassium channel activity | 4 | 0.002802 |
| MF term | GO:0046332 | SMAD binding | 5 | 0.003328 |
| MF term | GO:0005201 | Extracellular matrix structural constituent | 12 | 2.77x10-09 |
| MF term | GO:0001758 | Retinal dehydrogenase activity | 3 | 0.004132 |
| MF term | GO:0005178 | Integrin binding | 11 | 2.77x10-06 |
| MF term | GO:0005509 | Calcium ion binding | 27 | 1.47x10-05 |
| MF term | GO:0008201 | Heparin binding | 12 | 2.07x10-05 |
| MF term | GO:0016491 | Oxidoreductase activity | 9 | 0.008547 |
| MF term | GO:0008083 | Growth factor activity | 8 | 0.009105 |
| KEGG pathway | hsa04512 | ECM-receptor interaction | 16 | 5.16x10-11 |
| KEGG pathway | hsa04974 | Protein digestion and absorption | 14 | 7.73x10-09 |
| KEGG pathway | hsa04510 | Focal adhesion | 18 | 2.67x10-07 |
| KEGG pathway | hsa05146 | Amoebiasis | 10 | 1.63x10-04 |
| KEGG pathway | hsa04971 | Gastric acid secretion | 8 | 4.23x10-04 |
| KEGG pathway | hsa04151 | PI3K-Akt signaling pathway | 17 | 7.35x10-04 |
| KEGG pathway | hsa00830 | Retinol metabolism | 7 | 0.00124 |
| KEGG pathway | hsa00982 | Drug metabolism-cytochrome P450 | 7 | 0.001703 |
| KEGG pathway | hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 7 | 0.002628 |
| KEGG pathway | hsa05204 | Chemical carcinogenesis | 7 | 0.003889 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological processes; CC, cellular component; MF, molecular function.
Figure 2.
Gene Ontology terms and KEGG pathway enrichment analyses of 271 differentially expressed genes. Top 10 terms of enrichment for (A) BP, (B) CC and (C) MF. (D) Top 10 enriched KEGG pathways. KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; CC, cellular component; MF, molecular function.
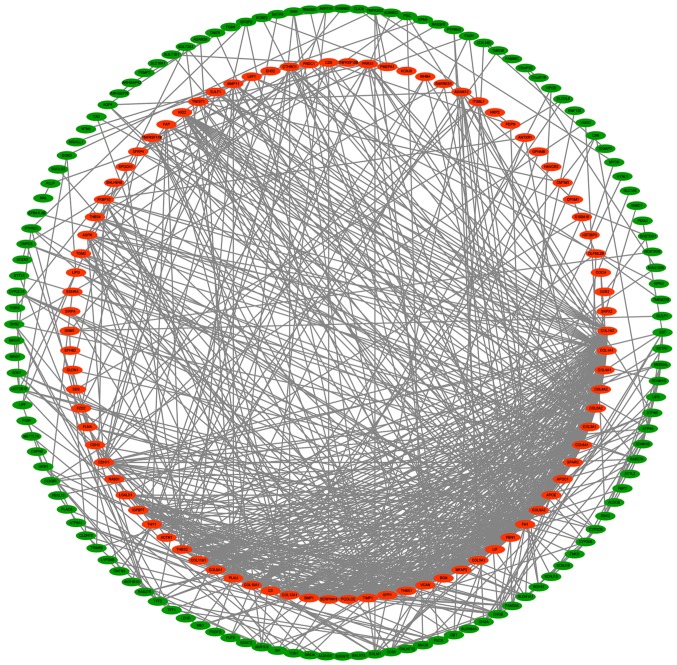
PPI network construction
Based on the STRING prediction results, a PPI network with 211 nodes and 741 sides was constructed in Cytoscape (Fig. 3), and the number of segments connected to each gene in the figure represents its degree.
Figure 3.
Protein-protein interaction network of differentially expressed genes. Red indicates upregulated genes, and green represents downregulated genes.
Identification of six key genes
The two genes with the most nodes were FN1 and COL1A1. In the PPI network, FN1 was the most prominent, with the highest degree of connectivity at 52. The degree of connectivity of COL1A1 is 43 (Table IV). Expression of these two genes is upregulated in GC tissues. Additionally, of those DEGs shared among the four gene expression profiles, the two DEGs with the largest logFC and the two DEGs with the smallest logFC values were selected. The higher the logFC in the upregulated DEGs, the greater the increase in expression of the gene. Similarly, the lower the logFC values in the downregulated DEGs, the greater the decrease in expression of the gene. When sorting DEGs according to logFC, the logFC of GSE19826 was used as the standard, as chip GSE19826 represented a homogenous cancer tissue population at each Tumor-Node-Metastasis stage (25), which increases the accuracy of the expression profile (Table V). The two DEGs with the largest logFC values were INHBA (logFC=4.35) and CST1 (logFC=4.18) (Table VI). The two DEGs with the smallest logFC values were ATP4A (logFC=-6.46) and ATP4B (logFC=-5.91) (Table VII). Therefore, these six genes were selected as key genes.
Table IV.
The 10 genes with the largest degree of connectivity in the protein-protein-interaction network.
| Rank | Gene | Degree |
|---|---|---|
| 1 | FN1 | 52 |
| 2 | COL1A1 | 43 |
| 3 | COL1A2 | 38 |
| 4 | COL3A1 | 37 |
| 5 | FBN1 | 35 |
| 6 | BGN | 32 |
| 6 | COL5A2 | 32 |
| 8 | TIMP1 | 31 |
| 9 | SPARC | 30 |
| 10 | THBS2 | 28 |
Table V.
The expression data from GSE19826 in gastric cancer.
| Tissue type | Accession no. | Title | Stage |
|---|---|---|---|
| Noncancer tissue | GSM495051 | CB2008210-1N | n/a |
| Gastric cancer tissue | GSM495052 | CB2008210-1T | II |
| Noncancer tissue | GSM495053 | CB2008210-2N | n/a |
| Gastric cancer tissue | GSM495054 | CB2008210-2T | IV |
| Noncancer tissue | GSM495055 | CB2008210-3N | n/a |
| Gastric cancer tissue | GSM495056 | CB2008210-3T | I |
| Noncancer tissue | GSM495057 | CB2008210-4N | n/a |
| Gastric cancer tissue | GSM495058 | CB2008210-4T | II |
| Noncancer tissue | GSM495059 | CB2008210-5N | n/a |
| Gastric cancer tissue | GSM495060 | CB2008210-5T | III |
| Noncancer tissue | GSM495061 | CB2008210-6N | n/a |
| Gastric cancer tissue | GSM495062 | CB2008210-6T | IV |
| Noncancer tissue | GSM495063 | CB2008210-7N | n/a |
| Gastric cancer tissue | GSM495064 | CB2008210-7T | IV |
| Noncancer tissue | GSM495065 | CB2008210-9N | n/a |
| Gastric cancer tissue | GSM495066 | CB2008210-9T | III |
| Noncancer tissue | GSM495067 | CB2008210-12N | n/a |
| Gastric cancer tissue | GSM495068 | CB2008210-12T | II |
| Noncancer tissue | GSM495069 | CB2008210-13N | n/a |
| Gastric cancer tissue | GSM495070 | CB2008210-13T | I |
| Noncancer tissue | GSM495071 | CB2008210-14N | n/a |
| Gastric cancer tissue | GSM495072 | CB2008210-14T | III |
| Noncancer tissue | GSM495073 | CB2008210-15N | n/a |
| Gastric cancer tissue | GSM495074 | CB2008210-15T | I |
| Normal gastric tissue | GSM495075 | CB2008210-3C | n/a |
| Normal gastric tissue | GSM495076 | CB2008210-5C | n/a |
| Normal gastric tissue | GSM495077 | CB2008210-9C | n/a |
Table VI.
The 10 genes with the largest logFC values in GSE19826.
| Rank | Name | LogFC |
|---|---|---|
| 1 | INHBA | 4.35 |
| 2 | CST1 | 4.18 |
| 3 | COL11A1 | 4.11 |
| 4 | FAP | 3.91 |
| 5 | COL10A1 | 3.72 |
| 6 | FNDC1 | 3.27 |
| 6 | COL8A1 | 3.17 |
| 8 | SERPINH1 | 2.97 |
| 9 | CDH3 | 2.95 |
| 10 | THBS2 | 2.94 |
FC, fold change.
Table VII.
The 10 genes with the smallest logFC values in GSE19826.
| Rank | Name | LogFC |
|---|---|---|
| 1 | ATP4A | -6.46 |
| 2 | ATP4B | -5.91 |
| 3 | KCNE2 | -5.88 |
| 4 | AQP4 | -5.81 |
| 5 | GIF | -5.75 |
| 6 | LIPF | -5.53 |
| 6 | CHIA | -5.51 |
| 8 | GKN1 | -5.49 |
| 9 | GKN2 | -5.44 |
| 10 | DPCR1 | -4.83 |
FC, fold change.
Analysis of the six key genes in Oncomine
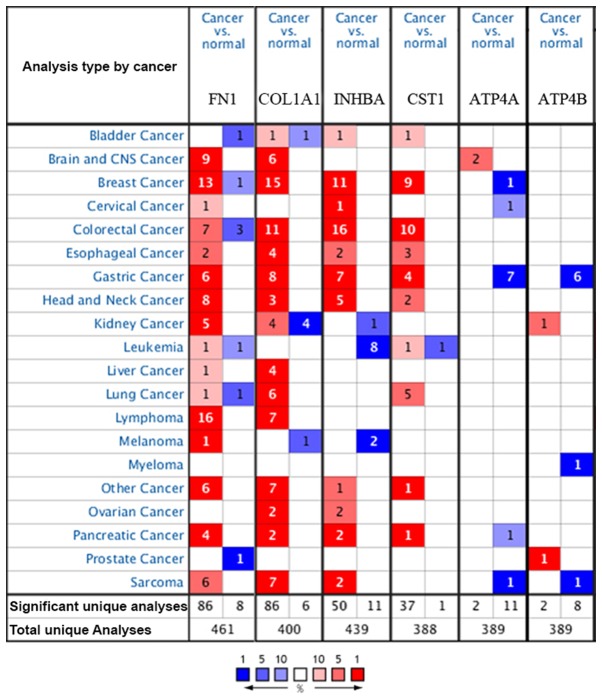
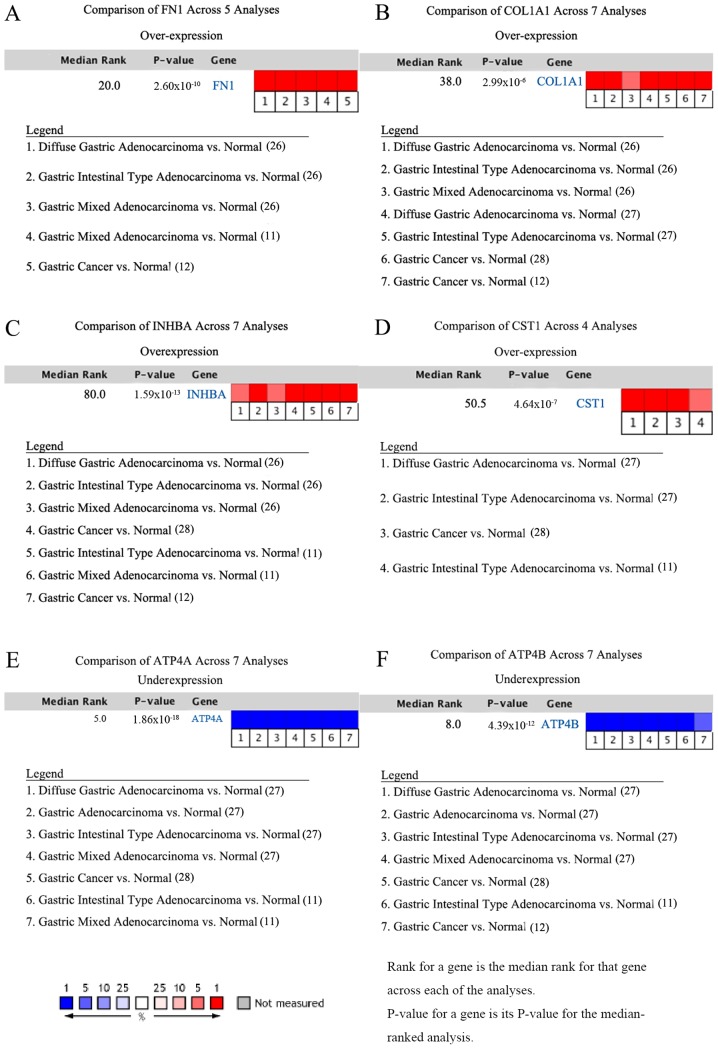
The Oncomine database was used to confirm the expression of the six key genes in 20 different types of cancer. The results showed that there were statistically significant differences in their expression. In the Oncomine database, there were no studies showing low expression of FN1, COL1A1, INHBA or CST1 in GC, but there were six, eight, seven and four studies showing increased expression, respectively. For ATP4A and ATP4B, the reverse was observed with no studies showing high expression, but seven and six studies, respectively, showing decreased expression (Fig. 4).
Figure 4.
mRNA expression of the six key genes in 20 different types of cancer. Cell color is determined by the best gene rank percentile for the analyses within the cell.
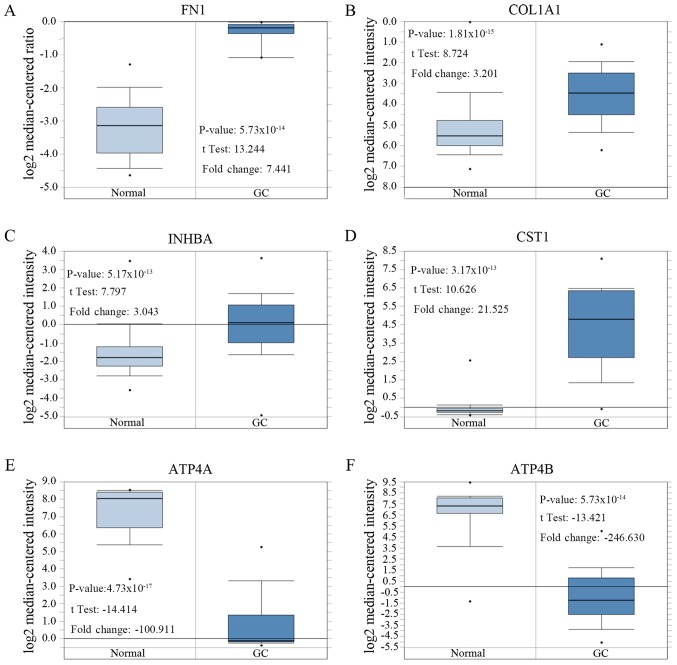
After comparing the expression levels of these six genes in cancerous and normal gastric tissue, the expression levels of FN1, COL1A1, INHBA and CST1 in GC tissues were significantly higher compared with the control group, and the expression levels of ATP4A and ATP4B in GC tissues were significantly lower compared with the control group (Table VIII; Fig. 5).
Table VIII.
Additional information for the six key genes shown in Figure 5.
| Author, year | Gene | Normal tissue samples | Gastric cancer samples | P-value | Fold Change | Published journal | (Refs.) |
|---|---|---|---|---|---|---|---|
| Chen et al, 2003 | FN1 | 28 | 8 | 5.73x10-14 | 7.441 | Molecular Biology of The Cell | (26) |
| Cui et al, 2011 | COL1A1 | 80 | 80 | 1.81x10-15 | 3.201 | Nucleic Acids Research | (28) |
| Cui et al, 2011 | INHBA | 80 | 80 | 5.17x10-13 | 3.043 | Nucleic Acids Research | (28) |
| Cho et al, 2011 | CST1 | 19 | 31 | 3.17x10-13 | 21.525 | Clinical Cancer Research | (27) |
| Cho et al, 2011 | ATP4A | 19 | 20 | 4.73x10-17 | -100.911 | Clinical Cancer Research | (27) |
| D'Errico et al, 2009 | ATP4B | 31 | 26 | 6.15x10-19 | -246.630 | European Journal of Cancer | (11) |
Figure 5.
Expression of six key genes in different gastric cancer gene chips in Oncomine. P<0.0001 and a |fold change|>2 were used as the threshold. Comparison of mRNA expression in cancerous vs. normal gastric tissue. (A) FN1, (B) COL1A1, (C) INHBA, (D) CST1, (E) ATP4A and (F) ATP4B.
In addition, meta-analyses of the six key genes in GC in the Oncomine database also supported the findings that expression of FN1, COL1A1, INHBA and CST1 is upregulated in GC, whereas expression of ATP4A and ATP4B is downregulated in GC (11,12,26-28). The studies and references involved are shown in Fig. 6. In the meta-analyses, P=-0.000, FC≥2.0 and gene rank ≤300 were selected as the cutoff criteria.
Figure 6.
Meta-analyses of the six key genes in gastric cancer in Oncomine. (A) FN1, (B) COL1A1, (C) INHBA, (D) CST1, (E) ATP4A and (F) ATP4B.
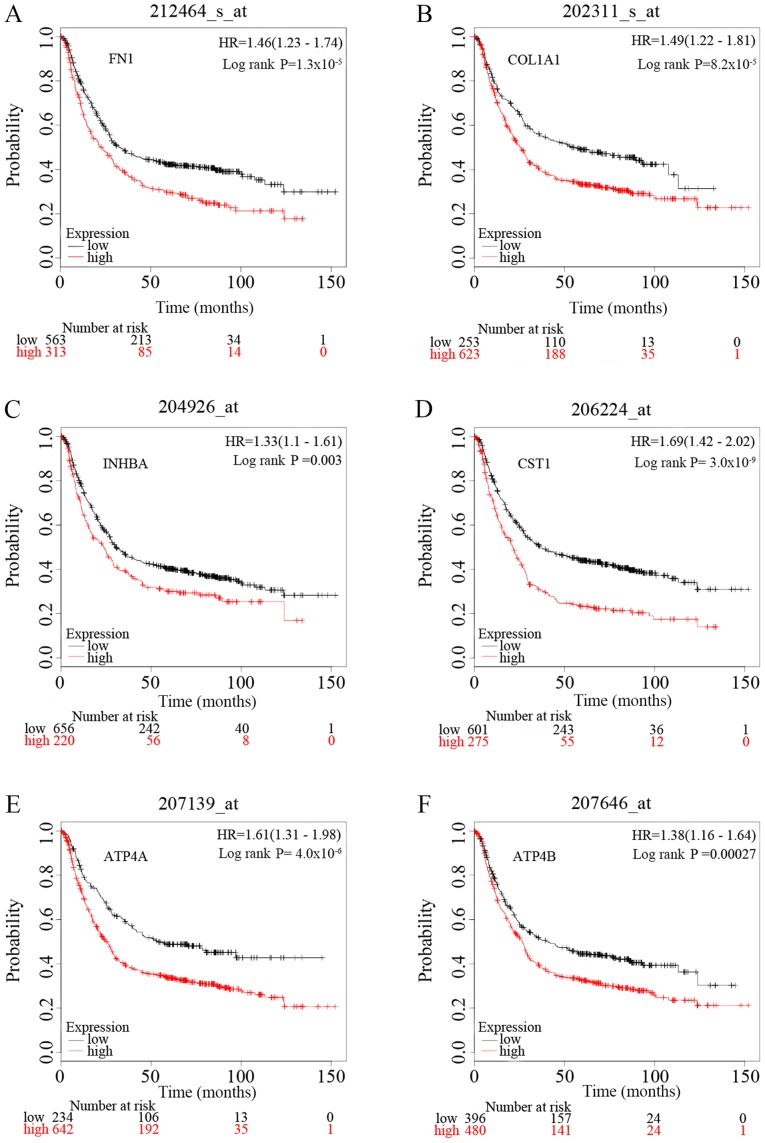
Survival analysis of the six key genes
To identify the prognostic value of the six potential key genes, overall survival curves based on differential expression of the six key genes were plotted using Kaplan-Meier plotter (Fig. 7). There were 1,440 patients with GC on the Kaplan-Meier plotter platform who were suitable for the analysis of overall survival. The curves indicate that overexpression of the six key genes is significantly associated with decreased overall survival times of patients with GC. However, it is worth noting that ATP4A and ATP4B were significantly downregulated in GC samples in the present study.
Figure 7.
Kaplan-Meier overall survival analyses of patients with gastric cancer based on expression of the six key genes. (A) FN1, (B) COL1A1, (C) INHBA, (D) CST1, (E) ATP4A, (F) ATP4B. HR, hazard ratio.
Discussion
GC is a complex heterogeneous disease with high incidence and mortality rates, and poses a serious threat to afflicted patients. Therefore, it is important to identify biomarkers that are meaningful for both diagnostic and prognostic assessment (29).
In the present study, 271 DEGs were screened, including 99 upregulated and 172 downregulated genes, by analyzing four gene expression profiles containing a combined 176 GC tissue samples and 82 normal gastric tissue samples. Of the causes of cancer-associated deaths, 90% are the result of metastasis (30). In the present study, GO enrichment results showed that the occurrence and development of GC was closely associated with metastasis. GO analysis indicated that DEGs were primarily associated with extracellular matrix organization, collagen catabolic process and cell adhesion. Collagen is the primary component of the extracellular matrix and of the interstitial microenvironment. Collagen can provide a scaffold for tumor cell growth and induce migration of tumor cells (31,32). There is evidence that collagen synthesis increases in the presence of a gastric tumor (33). Zhou et al (32) reported that collagen components are quantitatively and qualitatively reorganized in the tumor microenvironment of GC, and collagen width was identified as a useful prognostic indicator for GC (32). In addition, studies have shown that changes in cell-cell adhesion and cell-matrix adhesion can promote cancer cell metastasis (34). MF analysis showed that the DEGs were significantly enriched in platelet-derived growth factor binding. It has been demonstrated that inhibition of platelet-derived growth factor receptor-a can reduce the proliferation of gastrointestinal stromal tumor cells with mutant v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT) by affecting the KIT-dependent transcription factor ETV1(35).
KEGG pathway analysis showed that the DEGs were primarily enriched in ECM-receptor interaction, protein digestion and absorption, and focal adhesion. ECM-receptor interaction serves a vital role in several types of cancer (36-38). The interaction between membrane receptors of tumor cells and ECM proteins serve an important role in tumor invasion and metastasis (39), and ECM-receptor interaction serve a crucial role in the process of tumor shedding, adhesion, degradation, movement and hyperplasia (38). In addition, the non-steroidal anti-inflammatory drug celecoxib may exhibit anti-GC effects by inhibiting the expression of various proteins and inhibiting leukocyte transendothelial migration and focal adhesion (40), which provides a possible mechanism for future investigations of the role of focal adhesion in GC and developing new anti-GC drugs.
The degree of connectivity of a gene in a PPI network reflects its association with GC. The greater the connectivity, the closer a gene is to the disease mechanism. The logFC values of DEGs reflects the level of up or downregulation of the gene. The higher the logFC values in the upregulated DEGs, the greater the degree of upregulation of the gene, and the lower the logFC values in the downregulated DEGs, the greater the degree of downregulation (41-43). Thus it was hypothesized that the DEGs with the highest and lowest logFC values would be the genes most closely associated with disease mechanisms.
In the present study, the two genes with the highest degree of connectivity in the PPI network, and the two DEGs with the largest and smallest logFC values, were all selected as key genes. These were FN1, COL1A1, INHBA, CST1, ATP4A and ATP4B. These six key genes were verified in the Oncomine database. Expression of FN1, COL1A1, INHBA and CST1 were upregulated in GC, and expression of ATP4A and ATP4B were downregulated, consistent with the results obtained from analysis of the GEO datasets. Furthermore, survival analysis showed that upregulation of the six key genes was significantly associated with worse overall survival, and downregulation of ATP4A and ATP4B expression predicted a more favorable prognosis for patients with GC, providing novel insights into potential GC treatment strategies.
FN1 was the gene with the highest degree of connectivity. It is expressed in a wide range of healthy plasmalemmas, lamina propria mucosae and smooth-muscle cell layers, and it is involved in a variety of cellular processes including embryogenesis, blood coagulation, wound healing, host defense and metastasis (44). As a glycoprotein involved in cell adhesion and migratory processes, FN1 is hypothesized to be associated with signaling pathways associated with cancer (13). Expression of FN1 is significantly increased in anti-chemotherapy osteosarcoma cell lines and tissues, and is associated with a poor prognosis (45). Knockdown of FN1 gene expression results in reduced cell proliferation, increased cellular senescence and apoptosis, and reduced migration and invasion, by blocking the activation of the PI3K/AKT signaling pathway (46). Furthermore, downregulation of FN1 inhibits proliferation, migration and invasion, and thus reduces progression of colorectal cancer (47). The results of the present study suggest that FN1 may be a potential biomarker and therapeutic target for diagnosis and treatment of GC, consistent with previous studies (13,48,49), and thus further confirming the significance of FN1 in GC.
COL1A1 is one of the most important components of the ECM, and it is highly expressed in most connective tissues and various human solid tumors (50). It is also the primary component of type I collagen, which serves a key role in tumor cell adhesion and invasion (51). A mechanistic study revealed that COL1A1 and COL1A2 affects angiogenesis in GC, and their expression is also significantly associated with progression of GC (52). In addition, Zhang et al (53) further confirmed that overexpression of COL1A1 promoted GC cell proliferation in vitro. These previous studies support the use of COL1A1 as a key potential GC biomarker in the present study.
INHBA is a member of the transforming growth factor-β (TGF-β) superfamily, which is closely associated with tumor proliferation and expression is upregulated in lung cancer (54), GC (12) and colon cancer (55), where INHBA expression is closely associated with their prognosis. In a study of GC, Chen et al (56) found that INHBA gene silencing reduced migration and invasion of GC cells by blocking the activation of the TGF-β signaling pathway. They suggested that INHBA was a potential target for GC therapy (56). Another study showed that INHBA mRNA expression in GC may be a useful prognostic biomarker for patients with stage II or III GC who receive adjuvant chemotherapy with S-1(57). The results of the present study support the conclusions drawn in these previous studies.
Cystatin SN (CST1) is a member of the type 2 cystatin superfamily, the primary role of which is to limit the proteolytic activity of cysteine proteases (58). The dysregulated expression of CST1 is hypothesized to be involved in several types of cancer, including cholangiocarcinoma (59), breast cancer (58), GC (60) and colorectal cancer (61). CST1 prevents cell aging and promotes cancer development by affecting the activity of cathepsin B (62). However, CST1 has not been analyzed using bioinformatics for survival prognosis in GC, to the best of our knowledge. Using multiple databases, the present study is the first to validate CST1 as a novel prognostic biomarker and a potential therapeutic target for treatment of GC.
ATP4A encodes the α subunit and ATP4B encodes the β subunit of the gastric H+, K+-ATPase, respectively. They regulate gastric acid secretion and, as a result, are targets for acid reduction (63). Fei et al (64) found that expression of ATP4A and ATP4B were significantly downregulated in patients with GC, but their expression was not significantly correlated with overall survival (64). In the present study, downregulation of ATP4A and ATP4B expression was associated with favorable overall survival in patients with GC. Downregulation of ATP4A and ATP4B mRNA expression in GC tissue is associated with the development of GC (65). Correa's Cascade is inversely associated with gastric acid secretion rate, the downregulation of ATP4A and ATP4B mRNA expression begins in the early stages of gastric mucosal lesions, and the expression of both is gradually decreased as Correa's cascade progresses (66). In addition, Helicobacter pylori (H. pylori) inhibits parietal acid secretion by downregulating the expression of ATP4A and ATP4B in gastric parietal cells prior to the formation of GC, suggesting that H. pylori is closely associated with the development of GC (67). Thus, it was hypothesized that ATP4A and ATP4B may inhibit the formation of GC. Survival analysis showed that ATP4A and ATP4B in GC are adverse prognostic factors for patients with GC, suggesting that upregulation is associated with progression of GC. However, studies have reported that the expression of ATP4A and ATP4B is not regulated by H. pylori in GC (68-70). Other studies have shown significant decreases in the abundance of Helicobacter and Neisseria, and significant increases in Achromobacter, Citrobacter, Phyllobacterium, Clostridium, Rhodococcus and Lactobacillus in gastric carcinoma in comparison with chronic gastritis (71,72). Additionally, the gastric microbiota composition in patients with gastric carcinoma is significantly different compared with patients with chronic gastritis (71). Therefore, it was hypothesized that the formation of an altered gastric microbiota composition may result in the expression of ATP4A and ATP4B to be passively upregulated as GC progresses. Further research is required to more accurately determine the biological function of ATP4A and ATP4B in GC.
Although several genes were identified as promising diagnostic and prognostic biomarkers for GC, the present study has the following limitations. First, the present study lacked experimental and clinical validation. Second, the possibility that different histological types may affect the accuracy of results cannot be eliminated. Thus, future bioinformatics analysis should be designed such that samples can be stratified by histological type. Finally, the sample size was relatively small for the RNA-Seq experiments, which may result in inaccuracies or results which are not completely representative of the wider populace. Therefore, it is necessary to use larger samples to perform bioinformatics analysis, and further experimental and clinical studies are required.
In conclusion, the present study used bioinformatics to analyze biological processes and signaling pathways closely associated with GC occurrence and development and identified FN1, COL1A1, INHBA and CST1 as promising diagnostic and prognostic biomarkers for GC patients. Additionally, the results of the survival analysis of ATP4A and ATP4B were inconsistent with other international studies. Therefore, further studies are required to assess the effects of ATP4A and ATP4B on GC initiation and development. Furthermore, experimental and clinical studies are required to validate the findings of the present study and determine the potential clinical value of these potential biomarkers.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Key R&D Program of China (grant nos. 2018YFC1704100 and 2018YFC1704106).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
WW and YH conceived of and designed the study. YH and QZ performed the bioinformatics analysis and analyzed the data. WW and QZ wrote the manuscript. WW and ZL revised the manuscript. XZ contributed to the design of the study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Shi J, Qu YP, Hou P. Pathogenetic mechanisms in gastric cancer. World J Gastroenterol. 2014;20:13804–13819. doi: 10.3748/wjg.v20.i38.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–2664. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 4.In H, Solsky I, Palis B, Langdon-Embry M, Ajani J, Sano T. Validation of the 8th Edition of the AJCC TNM staging system for gastric cancer using the national cancer database. Ann Surg Oncol. 2017;24:3683–3691. doi: 10.1245/s10434-017-6078-x. [DOI] [PubMed] [Google Scholar]
- 5.Peng H, Deng Y, Wang L, Cheng Y, Xu Y, Liao J, Wu H. Identification of potential biomarkers with diagnostic value in pituitary adenomas using prediction analysis for microarrays method. J Mol Neurosci. 2019;69:399–410. doi: 10.1007/s12031-019-01369-x. [DOI] [PubMed] [Google Scholar]
- 6.Wu Y, Jamal M, Xie T, Sun J, Song T, Yin Q, Li J, Pan S, Zeng X, Xie S, Zhang Q. Uridine-cytidine kinase 2 (UCK2): A potential diagnostic and prognostic biomarker for lung cancer. Cancer Sci. 2019;110:2734–2747. doi: 10.1111/cas.14125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Zhou Y, Luo R, Liu K, Chen Z. Trophinin-associated protein expression is an independent prognostic biomarker in lung adenocarcinoma. J Thorac Dis. 2019;11:2043–2050. doi: 10.21037/jtd.2019.04.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Lin C, Li N, Du Y, Yang C, Bai Y, Feng Z, Su C, Wu R, Song S, et al. PLAGL2 and POFUT1 are regulated by an evolutionarily conserved bidirectional promoter and are collaboratively involved in colorectal cancer by maintaining stemness. EBioMedicine. 2019;45:124–138. doi: 10.1016/j.ebiom.2019.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yong L, YuFeng Z, Guang B. Association between PPP2CA expression and colorectal cancer prognosis tumor marker prognostic study. Int J Surg. 2018;59:80–89. doi: 10.1016/j.ijsu.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Troiano G, Guida A, Aquino G, Botti G, Losito NS, Papagerakis S, Pedicillo MC, Ionna F, Longo F, Cantile M, et al. Integrative histologic and bioinformatics analysis of BIRC5/Survivin expression in oral squamous cell carcinoma. Int J Mol Sci. 2018;19(E2664) doi: 10.3390/ijms19092664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Wen YG, Li DP, Xia J, Zhou CZ, Yan DW, Tang HM, Peng ZH. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29:77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Zhu Z, Zhao Y, Zhang Q, Wu X, Miao B, Cao J, Fei S. FN1, SPARC, and SERPINE1 are highly expressed and significantly related to a poor prognosis of gastric adenocarcinoma revealed by microarray and bioinformatics. Sci Rep. 2019;9(7827) doi: 10.1038/s41598-019-43924-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. doi: 10.1093/nar/gky1055. The Gene Ontology Consortium: The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 47: D330.D338, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M. ‘Post-genome Informatics’, Oxford University Press (2000). https://www.kanehisa.jp/docs/archive/PGI-contents.html. [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu W, Wang Y, Wang Y, Lv S, Xu X, Dong X. Screening of differentially expressed genes and identification of NUF2 as a prognostic marker in breast cancer. Int J Mol Med. 2019;44:390–404. doi: 10.3892/ijmm.2019.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. doi: 10.1186/1752-0509-8-S4-S11. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 8 (Suppl 4): S11, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Cui S, Li W, Zhao Y, Yan X, Xu J. PAX3 is a biomarker and prognostic factor in melanoma: Database mining. Oncol Lett. 2019;17:4985–4993. doi: 10.3892/ol.2019.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(9227) doi: 10.1038/s41598-018-27521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rausei S, Ruspi L, Galli F, Pappalardo V, Di Rocco G, Martignoni F, Frattini F, Rovera F, Boni L, Dionigi G. Seventh tumor-node-metastasis staging of gastric cancer: Five-year follow-up. World J Gastroenterol. 2016;22:7748–7753. doi: 10.3748/wjg.v22.i34.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.e02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, Ni Z, Zhang M, Kong X, Hoffman LL, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serra O, Galán M, Ginesta MM, Calvo M, Sala N, Salazar R. Comparison and applicability of molecular classifications for gastric cancer. Cancer Treat Rev. 2019;77:29–34. doi: 10.1016/j.ctrv.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 30.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Climent M, Pera M, Aymar I, Ramón JM, Grande L, Nogués X. Bone health in long-term gastric cancer survivors: A prospective study of high-dose vitamin D supplementation using an easy administration scheme. J Bone Miner Metab. 2018;36:462–469. doi: 10.1007/s00774-017-0856-1. [DOI] [PubMed] [Google Scholar]
- 32.Zhou ZH, Ji CD, Xiao HL, Zhao HB, Cui YH, Bian XW. Reorganized collagen in the tumor microenvironment of gastric cancer and its association with prognosis. J Cancer. 2017;8:1466–1476. doi: 10.7150/jca.18466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang M, Koh I, Lee SJ, Cheong JH, Kim P. Droplet-based microtumor model to assess cell-ECM interactions and drug resistance of gastric cancer cells. Sci Rep. 2017;7(41541) doi: 10.1038/srep41541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Chu KM. E-cadherin and gastric cancer: Cause, consequence, and applications. Biomed Res Int. 2014;2014(637308) doi: 10.1155/2014/637308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi Y, Bardsley MR, Toyomasu Y, Milosavljevic S, Gajdos GB, Choi KM, Reid-Lombardo KM, Kendrick ML, Bingener-Casey J, Tang CM, et al. Platelet-derived growth factor receptor-α regulates proliferation of gastrointestinal stromal tumor cells with mutations in KIT by stabilizing ETV1. Gastroenterology. 2015;149:420–432.e16. doi: 10.1053/j.gastro.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahbari NN, Kedrin D, Incio J, Liu H, Ho WW, Nia HT, Edrich CM, Jung K, Daubriac J, Chen I, et al. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci Transl Med. 2016;8(360ra135) doi: 10.1126/scitranslmed.aaf5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen MK, Rise K, Giskeødegård GF, Richardsen E, Bertilsson H, Størkersen Ø, Bathen TF, Rye M, Tessem MB. Integrative metabolic and transcriptomic profiling of prostate cancer tissue containing reactive stroma. Sci Rep. 2018;8(14269) doi: 10.1038/s41598-018-32549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, Chen C, Ren Y, Jia Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett. 2019;24(38) doi: 10.1186/s11658-019-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, Chen H, Wang J, Zhou L, Liu S. Collagen stiffness promoted non-muscle-invasive bladder cancer progression to muscle-invasive bladder cancer. Onco Targets Ther. 2019;12:3441–3457. doi: 10.2147/OTT.S194568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin GH, Xu W, Shi Y, Wang LB. Celecoxib exhibits an anti-gastric cancer effect by targeting focal adhesion and leukocyte transendothelial migration-associated genes. Oncol Lett. 2016;12:2345–2350. doi: 10.3892/ol.2016.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He WQ, Gu JW, Li CY, Kuang YQ, Kong B, Cheng L, Zhang JH, Cheng JM, Ma Y. The PPI network and clusters analysis in glioblastoma. Eur Rev Med Pharmacol Sci. 2015;19:4784–4790. [PubMed] [Google Scholar]
- 42.Miryala SK, Anbarasu A, Ramaiah S. Discerning molecular interactions: A comprehensive review on biomolecular interaction databases and network analysis tools. Gene. 2018;642:84–94. doi: 10.1016/j.gene.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 43.Yan H, Zheng G, Qu J, Liu Y, Huang X, Zhang E, Cai Z. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. J Cell Physiol. 2019;234:23785–23797. doi: 10.1002/jcp.28947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H, Sun Z, Li Y, Fan D, Jiang H. MicroRNA-200c binding to FN1 suppresses the proliferation, migration and invasion of gastric cancer cells. Biomed Pharmacother. 2017;88:285–292. doi: 10.1016/j.biopha.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 45.Kun-Peng Z, Chun-Lin Z, Xiao-Long M, Lei Z. Fibronectin-1 modulated by the long noncoding RNA OIP5-AS1/miR-200b-3p axis contributes to doxorubicin resistance of osteosarcoma cells. J Cell Physiol. 2019;234:6927–6939. doi: 10.1002/jcp.27435. [DOI] [PubMed] [Google Scholar]
- 46.Liao YX, Zhang ZP, Zhao J, Liu JP. Effects of fibronectin 1 on cell proliferation, senescence and apoptosis of human glioma cells through the PI3K/AKT signaling pathway. Cell Physiol Biochem. 2018;48:1382–1396. doi: 10.1159/000492096. [DOI] [PubMed] [Google Scholar]
- 47.Cai X, Liu C, Zhang TN, Zhu YW, Dong X, Xue P. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J Cell Biochem. 2018;119:4717–4728. doi: 10.1002/jcb.26651. [DOI] [PubMed] [Google Scholar]
- 48.Yan P, He Y, Xie K, Kong S, Zhao W. In silico analyses for potential key genes associated with gastric cancer. PeerJ. 2018;6(e6092) doi: 10.7717/peerj.6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang K, Liu H, Xie D, Xiao Q. Differentially expressed genes ASPN, COL1A1, FN1, VCAN and MUC5AC are potential prognostic biomarkers for gastric cancer. Oncol Lett. 2019;17:3191–3202. doi: 10.3892/ol.2019.9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun S, Wang Y, Wu Y, Gao Y, Li Q, Abdulrahman AA, Liu XF, Ji GQ, Gao J, Li L, et al. Identification of COL1A1 as an invasion-related gene in malignant astrocytoma. Int J Oncol. 2018;53:2542–2554. doi: 10.3892/ijo.2018.4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW, Zhang GJ. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25:211–223. [PubMed] [Google Scholar]
- 52.Huang C, Yang X, Han L, Fan Z, Liu B, Zhang C, Lu T. The prognostic potential of alpha-1 type I collagen expression in papillary thyroid cancer. Biochem Biophys Res Commun. 2019;515:125–132. doi: 10.1016/j.bbrc.2019.04.119. [DOI] [PubMed] [Google Scholar]
- 53.Zhang QN, Zhu HL, Xia MT, Liao J, Huang XT, Xiao JW, Yuan C. A panel of collagen genes are associated with prognosis of patients with gastric cancer and regulated by microRNA-29c-3p: An integrated bioinformatics analysis and experimental validation. Cancer Manag Res. 2019;11:4757–4772. doi: 10.2147/CMAR.S198331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seder CW, Hartojo W, Lin L, Silvers AL, Wang Z, Thomas DG, Giordano TJ, Chen G, Chang AC, Orringer MB, et al. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia. 2009;11:388–396. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Wu J, Zhang J, Yang Z, Jin W, Li Y, Jin L, Yin L, Liu H, Wang Z. Integrated bioinformatics analysis of key genes involved in progress of colon cancer. Mol Genet Genomic Med. 2019;7(e00588) doi: 10.1002/mgg3.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen ZL, Qin L, Peng XB, Hu Y, Liu B. INHBA gene silencing inhibits gastric cancer cell migration and invasion by impeding activation of the TGF-β signaling pathway. J Cell Physiol. 2019;234:18065–18074. doi: 10.1002/jcp.28439. [DOI] [PubMed] [Google Scholar]
- 57.Katayama Y, Oshima T, Sakamaki K, Aoyama T, Sato T, Masudo K, Shiozawa M, Yoshikawa T, Rino Y, Imada T, Masuda M. Clinical significance of INHBA gene expression in patients with gastric cancer who receive curative resection followed by adjuvant S-1 chemotherapy. In Vivo. 2017;31:565–571. doi: 10.21873/invivo.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai DN, Li Y, Chen B, Du Y, Li SB, Lu SX, Zhao ZP, Zhou AJ, Xue N, Xia TL, et al. Elevated expression of CST1 promotes breast cancer progression and predicts a poor prognosis. J Mol Med (Berl) 2017;95:873–886. doi: 10.1007/s00109-017-1537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tian A, Pu K, Li B, Li M, Liu X, Gao L, Mao X. Weighted gene coexpression network analysis reveals hub genes involved in cholangiocarcinoma progression and prognosis. Hepatol Res. 2019;49:1195–1206. doi: 10.1111/hepr.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J, Bae DH, Kim JH, Song KS, Kim YS, Kim SY. HOXC10 overexpression promotes cell proliferation and migration in gastric cancer. Oncol Rep. 2019;42:202–212. doi: 10.3892/or.2019.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang J, Liu HL, Tao L, Lin XY, Yang YD, Tan SW, Wu B. Let-7d inhibits colorectal cancer cell proliferation through the CST1/p65 pathway. Int J Oncol. 2018;53:781–790. doi: 10.3892/ijo.2018.4419. [DOI] [PubMed] [Google Scholar]
- 62.Oh SS, Park S, Lee KW, Madhi H, Park SG, Lee HG, Cho YY, Yoo J, Dong Kim K. Extracellular cystatin SN and cathepsin B prevent cellular senescence by inhibiting abnormal glycogen accumulation. Cell Death Dis. 2017;8(e2729) doi: 10.1038/cddis.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin S, Lin B, Wang X, Pan Y, Xu Q, He JS, Gong W, Xing R, He Y, Guo L, et al. Silencing of ATP4B of ATPase H+/K+ transporting beta subunit by intragenic epigenetic alteration in human gastric cancer cells. Oncol Res. 2017;25:317–329. doi: 10.3727/096504016X14734735156265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fei HJ, Chen SC, Zhang JY, Li SY, Zhang LL, Chen YY, Chang CX, Xu CM. Identification of significant biomarkers and pathways associated with gastric carcinogenesis by whole genome-wide expression profiling analysis. Int J Oncol. 2018;52:955–966. doi: 10.3892/ijo.2018.4243. [DOI] [PubMed] [Google Scholar]
- 65.Lozano-Pope I, Sharma A, Matthias M, Doran KS, Obonyo M. Effect of myeloid differentiation primary response gene 88 on expression profiles of genes during the development and progression of Helicobacter-induced gastric cancer. BMC Cancer. 2017;17(133) doi: 10.1186/s12885-017-3114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Di Mario F, Goni E. Gastric acid secretion: Changes during a century. Best Pract Res Clin Gastroenterol. 2014;28:953–65. doi: 10.1016/j.bpg.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Saha A, Hammond CE, Beeson C, Peek RM Jr, Smolka AJ. Helicobacter pylori represses proton pump expression and inhibits acid secretion in human gastric mucosa. Gut. 2010;59:874–881. doi: 10.1136/gut.2009.194795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Friis-Hansen L. Achlorhydria is associated with gastric microbial overgrowth and development of cancer: Lessons learned from the gastrin knockout mouse. Scand J Clin Lab Invest. 2006;66:607–621. doi: 10.1080/00365510600873894. [DOI] [PubMed] [Google Scholar]
- 69.Sáenz JB, Mills JC. Acid and the basis for cellular plasticity and reprogramming in gastric repair and cancer. Nat Rev Gastroenterol Hepatol. 2018;15:257–273. doi: 10.1038/nrgastro.2018.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vinasco K, Mitchell HM, Kaakoush NO, Castaño-Rodríguez N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim Biophys Acta Rev Cancer. 2019;1872(188309) doi: 10.1016/j.bbcan.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 71.Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang LL, Liu JX, Yu XJ, Si JL, Zhai YX, Dong QJ. Microbial community reshaped in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22:7257–7264. doi: 10.26355/eurrev_201811_16260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.