Abstract
Based on their identification as physiological nucleic acid carriers in humans and other organisms, extracellular vesicles (EVs) have been explored as therapeutic delivery vehicles for DNA, RNA, and other cargo. However, efficient loading and functional delivery of nucleic acids remain a challenge, largely because of potential sources of degradation and aggregation. Here, we report that protonation of EVs to generate a pH gradient across EV membranes can be utilized to enhance vesicle loading of nucleic acid cargo, specifically microRNA (miRNA), small interfering RNA (siRNA), and single-stranded DNA (ssDNA). The loading process did not impair cellular uptake of EVs, nor did it promote any significant EV-induced toxicity response in mice. Cargo functionality was verified by loading HEK293T EVs with either pro- or anti-inflammatory miRNAs and observing the effective regulation of corresponding cellular cytokine levels. Critically, this loading increase is comparable with what can be accomplished by methods such as sonication and electroporation, and is achievable without the introduction of energy associated with these methods that can potentially damage labile nucleic acid cargo.
Keywords: exosomes
Graphical Abstract
Jeyaram and colleagues report a pH gradient-based approach for loading cargo into extracellular vesicles (EVs). Using this method, functional delivery of small interfering RNA (siRNA) and microRNA (miRNA) was demonstrated, and no significant toxicity was observed in a mouse model. Thus, this approach could be employed to enhance EV-based therapies for a variety of applications.
Introduction
Extracellular vesicles (EVs) have emerged as potential drug carriers for a wide variety of therapeutic applications. These cell-derived lipid vesicles play critical roles in intercellular communication by acting as vehicles for biomolecule transfer between cells,1, 2, 3 and they may overcome biological hurdles that hinder the effectiveness of synthetic drug delivery systems.4 In particular, EVs have been identified as potential therapeutics via an RNA transfer mechanism,5,6 leading to their emergence as promising vehicles for therapeutic RNA delivery. However, intrinsic RNA levels in EVs are relatively low, raising questions about EV therapeutic potency and sparking investigations into methods to increase levels of specific RNAs in EVs.
To enhance therapeutic RNA levels in EVs, approaches have focused on modification of parent cells, as well as exogenous loading of fully formed EVs after isolation. The former has been successfully employed;7, 8, 9, 10, 11 however, this general strategy may hinder translation because of safety and scalability concerns, and also would not be applicable for EVs isolated from blood or biological fluids. Several exogenous loading techniques have been reported, including active methods such as electroporation and sonication. In particular, electroporation remains among the most commonly reported approaches to EV loading.12, 13, 14, 15, 16, 17 However, literature reports of successes with this method must be balanced against studies noting limitations such as induction of significant aggregation of RNA cargo and changes to EV morphology.18,19 Moreover, as with liposomes, electroporation can also induce aggregation and fusion of EVs.20 Our group has previously shown that sonication allows for small interfering RNA (siRNA) loading without significant aggregation, but we also demonstrated nucleic acid degradation with longer sonication times.21 Additionally, heat shock has been used to enable functional microRNA (miRNA) mimic loading,22 but this may also lead to nucleic acid destabilization and could impact EV membrane fluidity. Passive loading methods that do not involve exposing EVs and nucleic acid cargo to external force have also been explored. Notably, chemical modification of siRNAs23, 24, 25 has enabled high levels of functional loading. This approach is promising, but it requires specific modification of each prospective cargo molecule. Each of these strategies has strengths and weaknesses, but none has been established as universally applicable for unlocking the full therapeutic potential of nucleic acid delivery via EVs. Therefore, there remains a need to develop additional nucleic acid loading methods for EVs.
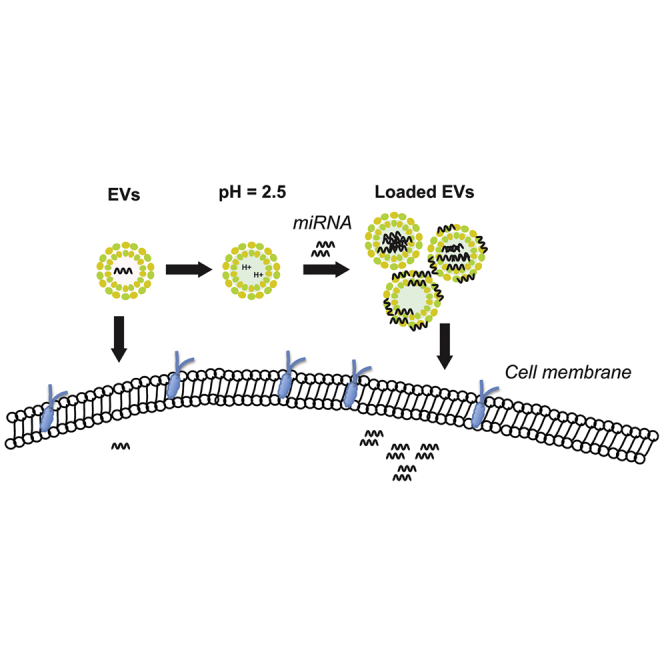
Approaches for loading cargo into EVs can be informed by research on liposomal systems, because EVs and liposomes share features of a phospholipid bilayer enclosing an aqueous lumen.26, 27, 28 Due to previous work with liposomes showing that a transmembrane pH gradient can be used to effectively encapsulate weakly basic or negatively charged small-molecule drugs,29, 30, 31, 32, 33 we hypothesized that this principle could be translated to loading negatively charged nucleic acids within EVs. Here, we report the parameter optimization of pH gradient-mediated loading of miRNAs to HEK293T-derived EVs. We further demonstrate the functionality of these loaded miRNAs via modulation of protein expression in cellular systems, as well as the ability to successfully administer pH gradient-modified EVs (pH-EVs) to animals.
Results
Preparation and Characterization of EVs
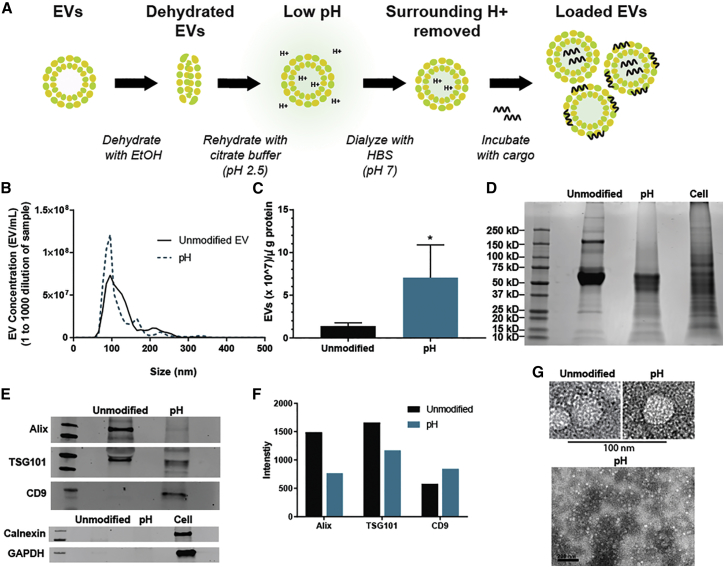
To promote the loading of negatively charged cargo, we used a pH gradient to render the interior of HEK293T-derived EVs acidic. This was accomplished by first dehydrating the EVs in 70% ethanol and then rehydrating them in an acidic citrate buffer (pH 2.5). To replace the surrounding acidic buffer with a more neutral buffer, we dialyzed EVs in HEPES-buffered saline (HBS; pH 7), resulting in a pH gradient between the intravesicular and extravesicular environments (Figure 1A). The effects of this process on EV yield and integrity were assessed by nanoparticle tracking analysis (NTA), bicinchoninic acid (BCA) assay, and immunoblotting. The results showed that both unmodified and pH-EVs fall within a similar size range (Figure 1B). Additionally, the total protein content associated with pH-EVs decreased in comparison with unmodified EVs, and thus the amount of EVs per microgram of protein increased (Figure 1C). Dialysis of EV samples that were not pH gradient modified led to similar effects as shown in experiments comparing filtration in place of dialysis in the procedure (Figures S1A–S1C), suggesting that dialysis steps during the pH gradient process may result in reduction of high molecular weight protein content in EV samples (Figure S1D). Alternatively, some EV-associated proteins may be degraded during the procedure. Supporting this latter possibility, a total protein gel showed some bands with decreased intensity (Figure 1D), and levels of EV-associated proteins Alix and TSG101 decreased after pH gradient modification (Figures 1E and 1F). However, the expression of EV-associated marker CD9 was enriched in the pH-EV sample (Figures 1E and 1F), indicating that some proteins may be enriched in pH-EVs, and thus that degradation is not solely responsible for any changes in protein content in these EVs. The absence of expression of the endoplasmic reticulum-associated protein calnexin further verified the absence of intracellular contaminants within the EV samples34 (Figures 1E and 1F). Transmission electron microscopy (TEM) (Figure 1G) revealed intact EVs after pH gradient modification.
Figure 1.
Preparation and Characterization of pH-EVs
(A) Schematic describing the steps to prepare and load pH-EVs. (B) The concentration of HEK293T EVs versus size was determined by nanoparticle tracking analysis via NanoSight. Samples were diluted 1,000-fold before analysis. (C) The amount of HEK293T EVs associated per microgram of protein with or without pH gradient modification was determined by the ratio of NTA and BCA results. Total protein (D) and immunoblotting (E) of unmodified and pH gradient-modified HEK293T EVs. (F) Densitometry of the immunoblot of EV-associated markers. (G) TEM at 200 kV showing EV morphology (lower scale bar, 200 nm). Statistical significance was evaluated using a two-tailed unpaired t test; *p < 0.05. Quantitative data were derived from three independent experiments with at least two technical replicates per experiment (n = 3); images are representative of three independent experiments.
Parameter Optimization to Enhance Cargo Incorporation
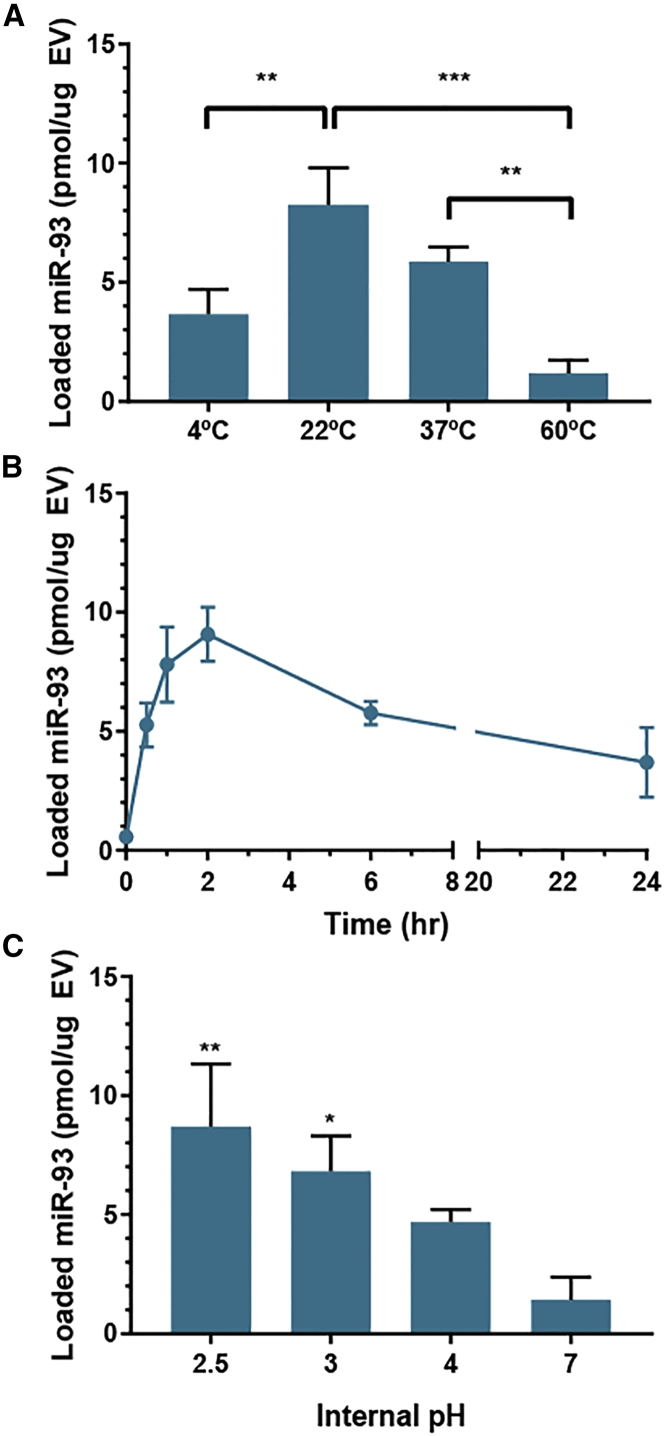
The process variables of incubation temperature, time, and pH were all investigated to promote the optimal loading of nucleic acid cargo. Comparing a range of temperatures from 4°C to 60°C, incubation at room temperature (22°C) was optimal for maximum observed incorporation of miRNA (Figure 2A). Cargo incorporation was found to be the lowest at 4°C or 60°C incubation. Varying the duration of EV incubation with cargo revealed that maximum observed incorporation was achieved after 2 h (Figure 2B). With longer incubation times, a decrease in cargo incorporation was observed, with a 57% decrease in total miRNA loaded between 2 and 6 h of incubation. This could be potentially caused by the degradation of miRNA in the buffer over time or by a loss of the pH gradient over time as negative cargo is entering the vesicles.
Figure 2.
Parameter Optimization for Nucleic Acid Loading to pH-EVs
Loading of pH gradient-modified HEK293T EVs with miR-93 was assessed by fluorescence quantification after (A) 1-h incubation at the indicated temperatures, (B) incubation at 22°C for the indicated times, and (C) 2-h incubation at 22°C with the indicated internal pH values. Two-way ANOVA (A) and one-way ANOVA (C) with Tukey’s multiple comparison test were used to determine significance; **p < 0.01, *p < 0.05 in comparison with pH 7. Data were derived from three independent experiments with at least two technical replicates per experiment (n = 3).
Based on liposomal studies,30 the degree of acidity of the intravesicular compartment was expected to significantly impact loading efficiency. Using citrate buffers of varying acidic strengths, we observed that miR-93 association with EVs increased with an increase in acidity of the internal pH, with the highest cargo loading achieved at pH 2.5 (Figure 2C). The optimal observed combination of loading parameters was thus a 2-h incubation time at room temperature with EVs that had an internal pH of 2.5. Using these optimized conditions, we then demonstrated that siRNA and single-stranded DNA (ssDNA) could also be loaded at similar levels via the pH gradient method (Figure 3).
Figure 3.
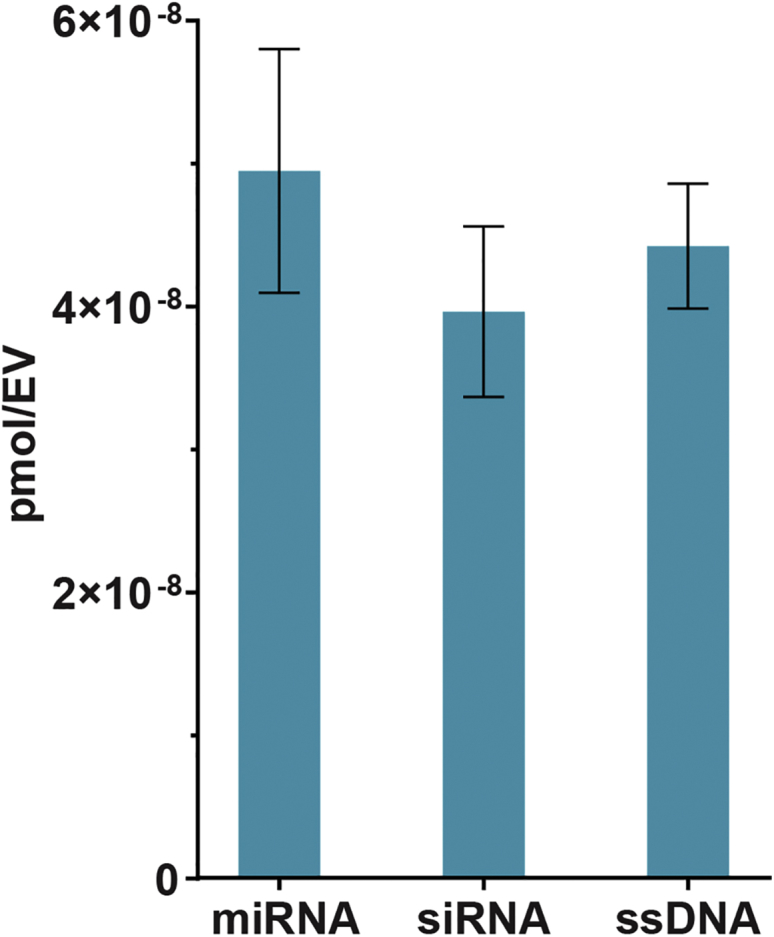
Loading of Multiple Cargo Classes into EVs via pH Gradient
Loading of miRNA (miR-93), siRNA (to GAPDH), and ssDNA (custom sequence) into EVs by the pH gradient method was evaluated via fluorescent quantification of labeled nucleic acid cargo. Data were derived from three independent experiments with at least two technical replicates per experiment (n = 3).
Reusability of Excess Nucleic Acid Cargo
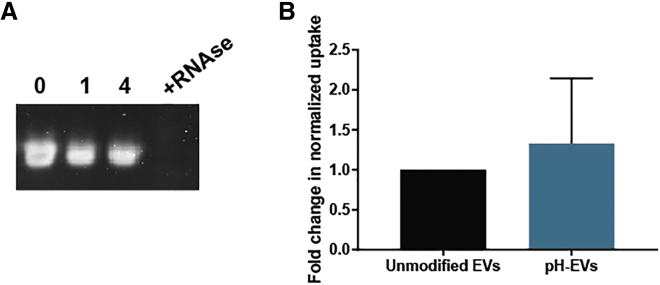
EV loading methods, such as electroporation, sonication, and heat shock, that ultimately rely on cargo diffusion are inherently limited in terms of efficiency. In these cases, the maximum possible loading efficiency is dictated by the volumetric ratio between the cargo-containing solution and the total intravesicular volume of EVs in suspension. Solubility considerations and practical working volume limitations often lead to this ratio being 20:1 or greater in favor of the cargo solution volume, meaning that a maximum of 5% or less of the total cargo in solution would be expected to associate with or be encapsulated by EVs, even assuming perfect conditions for diffusion. Previous studies have shown that loading by electroporation or sonication can damage nucleic acid cargo through aggregation and/or degradation,18,21,35 rendering any non-EV-associated cargo useless. In contrast, we hypothesized that the pH gradient method may allow for non-EV-associated cargo to be reused in additional loading processes, thus increasing overall cargo utilization efficiency. Testing this hypothesis with siRNA, we isolated non-EV-associated cargo by filtration with 300-kDa molecular weight cut off (MWCO) filters after 2-h incubation with pH-EVs. The recovered siRNA was then added to fresh pH-EVs, and successful loading was confirmed by fluorescence measurements of labeled siRNA (Table 1). Iterations of this process were continued until we could no longer detect RNA after washing steps, showing that we could reuse the same RNA to load four additional sets of EVs. Cargo miRNA integrity was evaluated after four cycles of loading in pH buffer (Figure 4A) by visualization after gel electrophoresis.
Table 1.
Reusability of Nucleic Acids after Cargo Loading with pH-EVs
| Amount of siRNA in Loading Buffer (pmol) | Amount of pH-EVs Used (μg) | Amount of siRNA Loaded in EVs (pmol) | % of siRNA Loaded | Theoretical Unused siRNA (pmol) | siRNA Recovered after Washing (pmol) |
|---|---|---|---|---|---|
| 1,000 | 10 | 65 | 6.5 | 935 | 720 (filtrate 1) |
| 720 (filtrate 1) | 10 | 41 | 5.6 | 679 | 543 (filtrate 2) |
| 543 (filtrate 2) | 10 | 32 | 5.8 | 511 | 420 (filtrate 3) |
| 420 (filtrate 3) | 10 | 23 | 5.4 | 397 | 280 (filtrate 4) |
| 280 (filtrate 4) | 10 | 7 | 3.2 | 273 | 198 (filtrate 5) |
| 198 (filtrate 5) | 10 | ∼0 | ∼0 | ∼198 | 125 (filtrate 6) |
After incubation with pH-EVs, the amount of siRNA was calculated via fluorescence measurements. The remaining loading buffer (filtrate after washing loaded EVs) was then used to load a fresh set of EVs, and the measurements were repeated until RNA could no longer be loaded. Data were derived from three independent experiments with at least two technical replicates per experiment (n = 3).
Figure 4.
pH Gradient-Modified EV Uptake and Cargo Integrity
(A) Cargo integrity in pH-modified EVs was evaluated by subjecting miR-93 (RNA) to 0, 1, or 4 cycles of loading by pH gradient process; RNase I treatment was used as a control. (B) Uptake of BODIPY TR Ceramide-labeled unmodified and pH gradient-modified HEK293T EVs by HUVECs was assessed by fluorescence quantification. Uptake is shown as fold change in comparison with unmodified EVs and was normalized to measurements of labeled EV fluorescence before cell treatment. Data were derived from three independent experiments with at least two technical replicates per experiment (n = 3). Statistical significance was evaluated using a two-tailed unpaired t test.
pH-EVs Are Effectively Taken up and Exhibit No Significant Toxicity
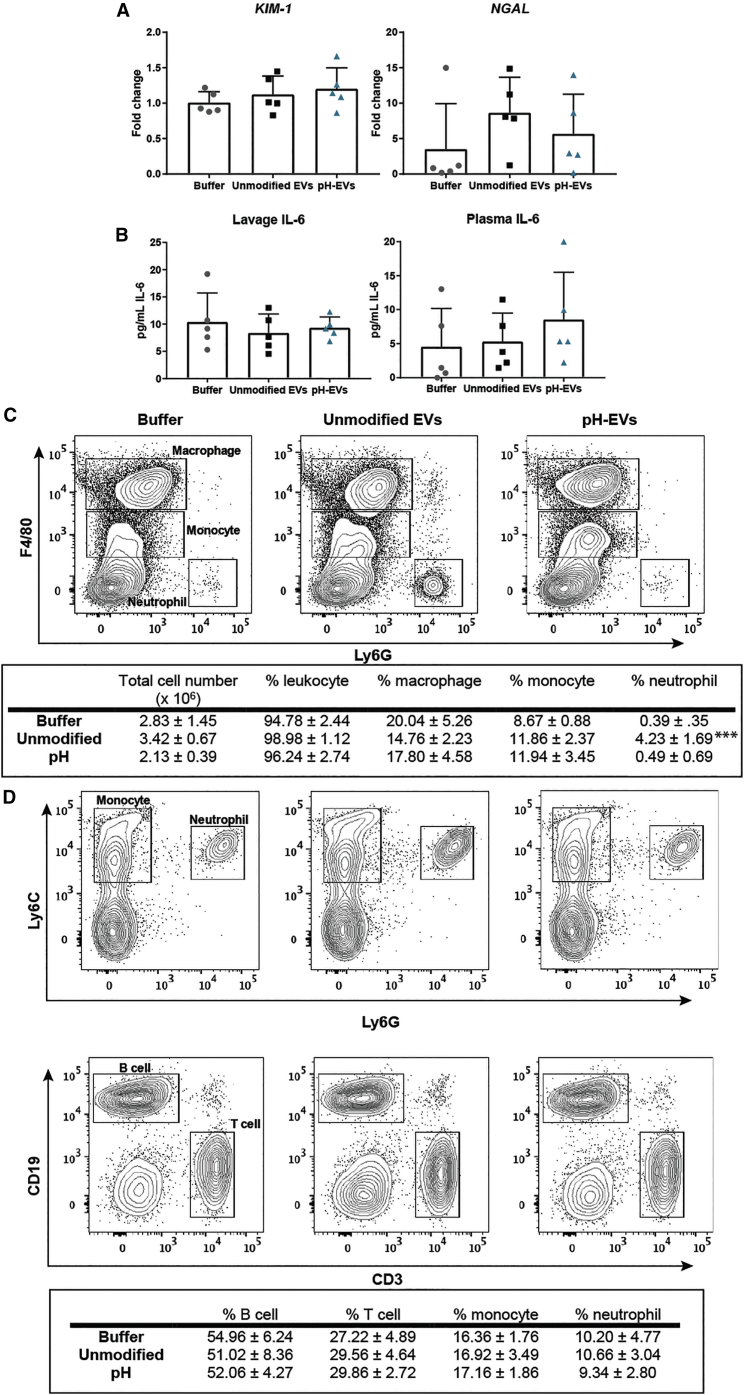
One potential pitfall of the pH gradient method was the possibility of inhibited EV uptake via damage to surface proteins or lipids during the dehydration/rehydration process. However, no significant difference in uptake between pH gradient-modified and unmodified HEK293T EVs by HUVECs in culture was observed (Figure 4B); data are shown after normalization to measurements of EV fluorescence before cell treatment. Toxicity was also evaluated in vivo by assessing lavage fluid in the peritoneal cavity and plasma after multiple EV injections into C57BL/6J mice. A total of 4.5 × 109 EVs of either unmodified or pH-modified HEK293T-derived EVs were injected i.p. daily for 3 days, and kidney injury, inflammation, and peritoneal/blood cell population were carefully examined. qPCR results indicated no effect of the pH gradient process on EV-induced expression of acute kidney injury markers kidney injury molecule-1 (KIM-1) and neutrophil gelatinase-associated lipocalin (NGAL) (Figure 5A), while levels of pro-inflammatory cytokine interleukin-6 (IL-6) in plasma and peritoneal lavage supernatant were also unaffected (Figure 5B). To investigate whether pH-EV injection would impact host immune cell composition, we performed immunophenotyping experiments both locally in the peritoneal cavity where the EVs were delivered and systemically in the blood circulation. No significant differences between EV treatments were observed in total cell numbers, resident macrophages (F4/80high/ Ly6G−), or monocytes (F4/80middle/ Ly6G−), whereas a slight increase of neutrophil population (F4/80−/Ly6G+) in the peritoneal cavity was seen following administration of unmodified EVs compared with control (Figure 5C). These data suggest that pH-EVs did not significantly affect immune cell composition in the peritoneal cavity. Further, the percentages of monocytes (Ly6C+/Ly6G−), neutrophils (Ly6C+/Ly6G+), T cells (CD3+/CD19−), and B cells (CD3−/CD19+) in the blood as measured by flow cytometry analysis suggested no significant systemic immunogenicity following repeated administration of unmodified or pH-EVs (Figure 5D). EV treatment also had little effect on hemoglobin levels, as well as leukocyte and platelet counts (Table 2).
Figure 5.
Assessment of Toxicity and Immunogenicity of pH Gradient-Modified EVs In Vivo
Unmodified or pH gradient-modified HEK293T EVs were injected i.p. into 9- to 10-week-old male wild-type (C57BL/6J) mice at 4.5 × 109 EVs/mouse/day for 3 days. PBS was used as the buffer control. (A) Acute kidney injury markers KIM-1 and NGAL were measured in the kidney using qRT-PCR. (B) Plasma and peritoneal lavage IL-6 levels were assessed by ELISA. (C) Representative flow cytometry of gated macrophage (F4/80high/Ly6G−), monocyte (F4/80middle/Ly6G−), and neutrophil (F4/80−/Ly6G+) in the peritoneal cavity. The percentage of each cell population over leukocyte (CD45+) was listed in the table. (D) Representative flow cytometry of gated monocyte (Ly6C+/Ly6G−), neutrophil (Ly6C+/Ly6G+), B cell (CD19+/CD3−), and T cell (CD19−/CD3+) in the blood. Percentage to total leukocytes (CD45+) was calculated. For all experiments, five animals per group were analyzed (n = 5). One-way ANOVA with Tukey’s multiple comparison test was used to determine statistical significance (none observed).
Table 2.
Blood Cell Count
| White Blood Cells (×103/μL) | Hemoglobin (g/dL) | Platelets (×103/μL) | |
|---|---|---|---|
| Buffer | 3.76 ± 1.14 | 12.78 ± 0.31 | 1,111 ± 49.87 |
| Unmodified EVs | 3.18 ± 0.36 | 13.04 ± 0.50 | 1,121 ± 47.64 |
| pH-EVs | 3.90 ± 1.15 | 12.94 ± 0.46 | 1,096 ± 58.41 |
Whole blood was collected following 3 days of EV administration and transferred to EDTA-coated blood collection tube. Leukocyte, platelet, and hemoglobin levels were analyzed using an automated cell counter (n = 5 animals/group).
pH-EVs Deliver Functionally Active RNA Cargo
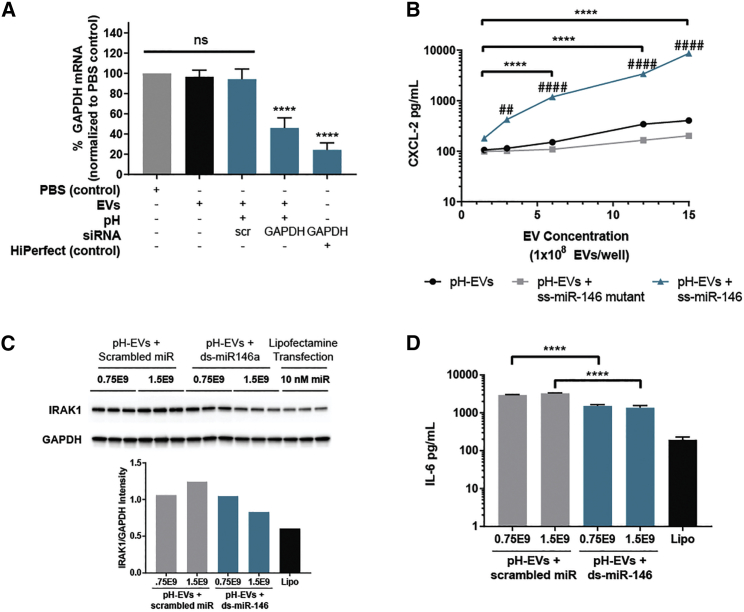
After evaluating that the pH gradient process did not induce significant effects counter to productive therapeutic cargo delivery, we next assessed the ability of pH-EVs to deliver functionally active RNA to cells. Delivery by pH-EVs of siRNA targeted to the protein glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in HEK293T cells resulted in 54% knockdown compared with scrambled RNA or PBS controls (Figure 6A), whereas unmodified EVs did not induce significant knockdown.
Figure 6.
Delivery of Functionally Active Cargo by pH Gradient-Modified EVs
(A) GAPDH mRNA in cells after 48-h incubation with pH gradient-modified HEK293T EVs or controls as indicated was evaluated by qPCR; statistical significance is shown with respect to the PBS control. (B) MIP-2 release by BMDMs after 16-h incubation with the indicated EV groups was determined by ELISA; asterisk (*) indicates statistical significance as shown by the bars; number sign (#) indicates statistical significance with respect to the corresponding EV-only sample for each corresponding time point. Effects of scramble control of ds-miR-146a delivery by pH gradient-modified HEK293T EVs at the indicated doses on (C) IRAK1 (by immunoblot and densitometry quantification) and (D) IL-6 (by ELISA) production by BMDMs were assessed. Asterisk (*) indicates significance with respect to 0.75 × 109 EVs + scramble; number sign (#) indicates significance with respect to Lipofectamine treatment. Data were derived from three independent experiments with at least two technical replicates per experiment (n = 3). Statistical significance was evaluated using one-way ANOVA with Tukey’s multiple comparison test.
We next examined the potential of pH-EVs to modulate the production of inflammatory cytokines known to be important in sepsis, a condition for which EVs have been both proposed as a treatment36, 37, 38, 39 and implicated as a contributing factor.40,41 In one experiment, pH-EVs were loaded with the pro-inflammatory ss-miR-14640 and exposed to bone marrow-derived macrophage (BMDM) cells. In this case, loaded pH-EVs induced dose-dependent cellular production of MIP-2 (macrophage inflammatory protein 2; CXCL2), a classical inflammatory cytokine, compared with a minimal response from pH-EVs alone or pH-EVs with a mutant sequence (Figure 6B). In another experiment, pH-EVs were loaded with anti-inflammatory double-stranded (ds)-miR-146.36,42 Here, loaded pH-EVs induced a decrease in inflammatory protein expression, as evidenced by immunoblotting for IRAK1 (Figure 6C), a proinflammatory enzyme. ELISA-based detection also revealed loaded pH-EV-induced decrease in levels of IL-6 (Figure 6D), a pro-inflammatory cytokine whose inhibition has previously been shown to improve survival in sepsis models.43, 44, 45 Thus, these results suggest therapeutic potential for utilizing pH-EVs in sepsis treatment.
Discussion
In this study, we demonstrated versatile loading potential and functional delivery of miRNA cargo using a pH gradient-based method. Using this approach, we were able to load thousands of copies of miRNA per EV as calculated from a bulk population (Figure 3), comparable with what has been reported for other EV loading methods, such as sonication,21 electroporation,19 chemical modification of cargo,23,24 and others.7,20 Additionally, this strategy enabled preservation of unloaded nucleic acid cargo such that it could be reused in subsequent loading operations, resulting in more efficient use of valuable biological therapeutic molecules (Table 1).
Despite exposing EVs to dehydration by ethanol during the loading process, cell uptake and functional cargo delivery were still achieved (Figures 4 and 6). This suggests that any proteins and lipids critical to cell uptake of EVs were not critically negatively impacted by the pH gradient process. This finding echoes prior work that showed that exposure of EVs to Proteinase K, which would be expected to degrade surface proteins, has little effect on EV physicochemical properties.46 This same study found that after injection in mice, the half-life and mean residence time of EVs did not change based on Proteinase K exposure. However, the area under the curve increased significantly for Proteinase K-treated EVs, indicating an increased percent of EV dose in the blood and decreased clearance. In our study, levels of Alix and TSG101 decreased after pH modification, whereas CD9 was increased (Figures 1E and 1F). Because TEM confirmed the presence of intact EVs, it is possible that the pH gradient process results in rearrangement and differential exposure of surface proteins and lipids on EVs, which could impact uptake in unpredictable ways. Overall, further dedicated study of EV uptake mechanisms would shed light on how this and other methods might be optimized to reduce any detrimental effects on EV cargo delivery.
The possibility of different loading levels in different EV subsets should also be considered. Studies have shown that the composition and function of small and large EVs can differ in morphology, as well as protein and lipid content;47,48 these differences in the surface composition of EV subtypes may make them more or less conducive to loading via the pH method. Further, the roles of EV-associated proteins differ depending on the parent cell type, and thus the method described here may not be universally appropriate for EV loading.
The results of this study also have implications for the field with regard to normalization of EV amount for functional studies, a subject on which there is not yet a consensus best practice.34 Many studies determine EV dose based on total protein content of EVs; however, as shown here, processing can dramatically impact the relationship between EVs and their associated proteins. Thus, in cases where EVs undergo downstream modification, dosage on the basis of number of EVs may be more accurate. Of course, because proteins could be the active cargo of interest, dosing by protein amount may be more relevant for some studies.
Ultimately, the utility of the method described here was shown by functional delivery of both siRNA and miRNA cargoes. Treating BMDM cells with pro- or anti-inflammatory miRNA led to regulation of corresponding cytokine levels. Specifically, we observed that delivery of pro-inflammatory ss-miR-146a resulted in increased MIP-2 secretion by BMDM cells. This suggests that pH-EVs can be employed in further studies exploring the roles of miRNAs in inflammatory diseases, such as sepsis. Further, we showed that delivery of ds-miR-146a reduced IL-6 production by the same cells, a potentially anti-inflammatory outcome, because endogenous EV-associated miR-146 has been shown to reduce inflammatory gene expression and inhibit endotoxin-induced inflammation in mice.36 This same study showed that there was approximately one copy of miR-146 per EV and an average of 370 copies per cell after treatment. It has been reported that at least 100 copies of miRNA per cell are necessary for regulatory capacity.49 These results suggest that pH gradient-based loading of EVs may be a promising approach to achieve functional therapeutic effects of miRNA delivery.
Materials and Methods
Cell Culture
Human embryonic kidney cells (HEK293T; CRL-3216; ATCC, Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) [+] 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Corning 10-013-CV; Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS) in T175 tissue culture polystyrene flasks in the presence of EV-depleted serum. Human umbilical vein endothelial cells (HUVECs; Lonza C2519A) were cultured in endothelial cell growth media (C-22121; PromoCell, Heidelberg, Germany) in flasks coated with 0.1% gelatin. Bone marrow cells were harvested from mouse tibias and femurs, cultured, and differentiated into macrophages in the presence of macrophage colony-stimulating factor (M-CSF; 10 ng/mL). In brief, bone marrow cells were cultured in RPMI 1640 culture medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% FBS (Thermo Fisher Scientific), 5% horse serum (Thermo Fisher Scientific), and penicillin (100 U/mL)/streptomycin (100 μg/mL) (Thermo Fisher Scientific) in a CO2 incubator at 37°C. Three days later, culture media were changed, and macrophages were ready for experiments at day 4.
EV Isolation and Characterization
Conditioned media from HEK293T cells grown in EV-depleted media were collected and subjected to differential ultracentrifugation with a 118,000 × g final centrifugation step as previously described.19,21 Pelleted EVs were resuspended in 1X PBS and washed using Nanosep 300-kDa MWCO spin columns (OD300C35; Pall, Port Washington, NY, USA). The washed EVs were resuspended in 1X PBS and filtered using an 0.2-μm syringe filter. EV size distribution and concentration were evaluated by NTA via a NanoSight LM10 using NTA analytical software version 2.4. Each sample was analyzed in triplicate using different fields of view with a 30-s video acquisition time. The camera level and threshold were set consistently at 13 and 3, respectively, for all samples. Total EV protein concentration was measured using a BCA assay following the manufacturer’s protocol. Relative levels of EV-associated proteins were assessed via total protein and western blotting. The membrane was imaged using a LI-COR Odyssey CLX Imager. Specific EV-associated protein marker levels and control proteins were quantified using immunoblot analysis for Alix (ab186429; Abcam, Cambridge, MA, USA), TSG101 (ab125011; Abcam), CD9 (ab92726; Abcam), Calnexin (2679S; Cell Signaling Technology, Danvers, MA, USA), and GAPDH (2118L; Cell Signaling Technology). Primary antibodies were added at 1:1,000 dilution, besides anti-GAPDH, which was added at 1:2,000 dilution. Secondary antibody IRDye 800CW anti-Mouse and anti-Rabbit (926-32210 and 926-32211; LI-COR Biosciences, Lincoln, NE, USA) were added at 1:10,000 dilutions.
TEM
Samples were fixed in in 2% EM-grade paraformaldehyde for 1 h before adsorption to formvar-coated copper grids stabilized with evaporated carbon film and type 200 mesh (FCF200-Cu; Electron Microscopy Sciences, Hatfield, PA, USA). After adsorption, grids were stained with uranyl acetate replacement (22405; Electron Microscopy Services), air-dried, and imaged at 200 kV.
Preparation and Cargo Loading of pH-EVs
To create a pH gradient, we first dehydrated EVs in 70% ethanol at a concentration of 300 μg/mL. The dehydration was allowed to proceed for 12 h in a 24-well plate with 1 mL of the dehydration solution per well. Once most of the liquid evaporated, the EVs were rehydrated in 1 mL of citrate buffer (150 mM [pH 2.5] for 1 h). After rehydration, EVs were resuspended and transferred to 300-kDa Spectra/Por Float-A-Lyzer G2 dialysis tubes (G235072; Spectrum Labs, Waltham, MA, USA) and dialyzed in 1X HBS at pH 7.0. EVs were dialyzed for 24 h with buffer changes every 2 h for the first 6 h and then left overnight. After dialysis, the volume of EV solution was concentrated down using 100-kDa MWCO centrifuge tubes and filtered through 0.2-μm syringe filters. EVs were then characterized as above.
For cargo loading, unless otherwise described, 3 × 109 EVs were incubated with 1,000 pmol of labeled nucleic acid cargo for 2 h before washing in a 300-kDa MWCO filter with 1X HBS. Nucleic acids were pre-labeled for detection by mixing 1,000 pmol of nucleic acids with 10 μL dye reagent at room temperature for 5 min according to the manufacturer’s instructions for Quant-iT PicoGreen Assay Kits (catalog [cat.] #P11496; Thermo Fisher Scientific). EV samples were subsequently washed using 300-kDa MWCO filter tubes to remove unincorporated nucleic acids. The specifications of the cargo used are as follows: miRNA = hsa-mir-93-5p (CTM-548920; Dharmacon; sequence: 5′-CAA AGU GCU GUU CGU GCA GGU AG-3′); siRNA = Invitrogen Silencer Select GAPDH Positive Control siRNA (43-908-50; Fisher Scientific); and ssDNA = custom sequence: 5′-ATA TGT TCA TCT TAG CAT TCG AGT ATA A-3′ (IDT).
To optimize maximum loading efficiency, we incubated pH-EVs or unmodified EVs with equal amounts of miR-93 labeled immediately prior to each set of experiments. To determine the optimal temperature for miRNA incubation, we incubated miRNA with EVs at 4°C, 22°C, 37°C, and 60°C. Unincorporated excess cargo was removed by washing with 1X HBS buffer after 1 h of incubation. To determine the optimal length of incubation with cargo, we incubated the labeled miRNA with the EVs for 0 min, 30 min, 1 h, 2 h, 6 h, and 24 h. After each time point, excess miRNA was removed by washing, as described previously. To evaluate the effect of the citrate buffer pH, we prepared the buffer at pH 2.5, 3, 4, and 7. For all optimization experiments, the unincorporated excess cargo was removed by washing with 1X HBS. Fluorescence was quantified by comparing the values measured at 480 excitation and 520 emission with a standard curve.
EV Cargo Integrity and Uptake
To evaluate the integrity of loaded miRNA cargo, we incubated miR-93 for 0, 1, or 4 pH loading cycles in 1X HBS or incubated it with RNase I (EN0601; Thermo Fisher Scientific) at 37°C for 45 min. RNA was subsequently incubated at 70°C for 10 min after 1:1 dilution in RNA loading buffer (1 mM EDTA, 96% formamide) before running on a polyacrylamide gel at 100 V for 10 min. After staining with SYBR Gold Nucleic Acid Gel Stain, gels were imaged using a FluorChem E System Gel Imaging System.
To assess EV uptake, we plated HUVECs in black-walled 96-well plates coated with 0.1% gelatin at a seeding density of 6,000 cells/well the day before the experiment. A total of 200 μg EVs were labeled with BODIPY TR Ceramide (D7540; Thermo Fisher) at a 10 μM concentration in a total volume of 100 μL. The solution was incubated at 37°C for 20 min, protected from light. Excess unincorporated dye was removed by washing in 300-kDa MWCO filter tubes. Fluorescence of the labeled EVs was measured at excitation 592 nm and emission 618 nm before treatment. HUVECs were then treated with 3 × 109 labeled EVs and incubated at 37°C for 24 h. Cells were washed, and fluorescence of the cells was measured and normalized to the initial fluorescence intensity. Fold change of uptake is shown with respect to unmodified EVs.
In Vivo Cytotoxicity
The in vivo study was performed in 9- to 10-week-old male wild-type (C57BL/6J) mice (The Jackson Laboratories, Bar Harbor, ME, USA). Mice were housed in an animal facility of University of Maryland School of Medicine (Baltimore, MD, USA) for 1 week before the study under specific pathogen-free environment. They were fed with autoclaved bacteria-free diet, and the housing facilities were temperature controlled and air-conditioned with 12-h/12-h light/dark cycles. All animal protocols were approved by the Institutional Animal Care and Use Committees of University of Maryland School of Medicine. For the in vivo experiments, mice were randomly assigned into three groups (five mice/group) and intraperitoneally (i.p.) injected with either buffer (HBS), unmodified EV, or pH-EV (4.5 × 109 EVs/mouse/day in 250 μL volume) for 3 days.
All animals were sacrificed on day 4. Peritoneal lavage was collected following anesthetizing by ketamine (100 mg/kg) and xylazine (4 mg/kg) subcutaneous injection and centrifuged to prepare cell-free peritoneal lavage supernatant and peritoneal cell pellets. Whole blood was collected by cardiac puncture using a 26G needle and transferred to an EDTA-coated blood collection tube (Beckton Dickinson), and a 100 μL aliquot was used for complete blood count analysis (Ac-T diff Analyzer; Beckman Coulter, Brea, CA, USA). Plasma was harvested by centrifugation and stored at −80°C for cytokine analysis. Kidneys were collected, snap frozen in liquid nitrogen, and stored at −80°C for future analysis. Peritoneal and blood cells were further analyzed by flow cytometry as described below.
IL-6 in the plasma and peritoneal lavage supernatant was measured using commercially available DuoSet ELISA kits (R&D Systems, Minneapolis, MN, USA) following the manufacturer’s instructions.
Total RNA was extracted from the kidney using TRIzol (Sigma, St. Louis, MO, USA). Reverse transcription was performed using the M-MLV reverse transcriptase (Promega, Madison, WI, USA). qPCR was performed on a QuantStudio 3 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using the GoTaq qPCR master mix (Promega, Madison, WI, USA). The transcripts of NGAL and KIM-1 were quantified with GAPDH as the internal control. The PCR primer sequences are listed as follows: GAPDH (forward: 5′-AACTTT GGCATTGTGGAAGG-3′, reverse: 5′-GGATGCAGGGATGATGTTCT-3′); NGAL (forward: 5′-CTCAGAACTTGATCCCTGCC-3′, reverse: 5′-TCCTTGAGGCCCAGAGACTT-3′); and KIM-1 (forward: 5′-CATTTAGGCCTCATACTGC-3′, reverse: 5′-ACAAGCAGAAGATGGGCATT-3′). Transcript expression was calculated using the comparative cycle threshold (Ct) method normalized to GAPDH (2−ΔΔCt) and expressed as the fold change in the EV-injected group over the buffer (HBS)-treated group.
Peritoneal cells were manually counted using a hemocytometer, and a fraction of cells (1 × 106) was stained with the surface marker CD45-phycoerythrin (PE) (BD Biosciences, San Jose, CA, USA), F4/80-Alexa Fluro 647 (BD Biosciences, San Jose, CA, USA), and Ly6G-BV421 (BD Biosciences, San Jose, CA, USA). After red blood cell lysis, cells from a 100-μL blood sample were stained with CD45-PE (BD Biosciences, San Jose, CA, USA), Ly6C-BV510 (BioLegend, San Diego, CA, USA), CD3-fluorescein isothiocyanate (FITC) (BioLegend, San Diego, CA, USA), and CD19-PE-Cy7 (BioLegend, San Diego, CA, USA). Cells were incubated with antibodies in the dark at 4°C for 30 min and then washed once with 4 mL PBS. Cell pellets were resuspended in PBS containing 5% FBS and analyzed using a BD LSR II flow cytometer. Data analysis was performed with FlowJo software.
Functional Evaluation of Loaded RNA Cargo
To assess the function of loaded siRNA, we incubated pH gradient or unmodified EVs at room temperature for 1 h with GAPDH siRNA, washed and isolated them as described above, and then added them to HEK293T cells (plated the day before in a six-well plate with 3 × 105 cells/well). A total of 100 μg of each EV sample (unmodified EVs, pH-EVs, pH-EVs + scrambled siRNA, pH-EVs + GAPDH siRNA) was added to the cells dropwise. As a positive control, 20 nM GAPDH siRNA was applied to cells with HiPerFect transfection reagent (301705; QIAGEN, Germantown, MD, USA) and a PBS-only negative control. After 2 days of incubation, total RNA was isolated (RNeasy Kit 74104; QIAGEN) from the cells. One microgram of total RNA from each type of sample was converted into cDNA, and mRNA level of GAPDH was confirmed by qPCR. The change in mRNA level was normalized by using Ct value from B-actin mRNA.50
GAPDH siRNA: Dharmacon ON-TARGETplus GAPDH Control siRNA D-001830-01-20; Scrambled siRNA: Dharmacon; 5′-GGUGCCAGUUCUCCAAGAUUdTdT-3′.
To assess cytokine regulation by loaded miRNAs, we treated BMDMs seeded in a 96-well plate (2 × 105 cells/well) with ss-miR-146a-5p mimic-loaded EVs (doses ranging from 1.5 × 108 to 1.5 × 109 EVs/mL) overnight. Cells were incubated in serum-free culture medium containing 0.05% BSA for 1 h before treatment with EVs. Medium was collected for detection of MIP-2 using ELISA kits (R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. Final cytokine concentrations were calculated based on a standard curve. The primer sequences were as follows: ss-miR-146-5p: IDT; 5′-UGAGAACUGAAUUCCAUGGGUU-3′; and ss-miR146 (U→A) mutant: IDT; 5′-AGAGAACAGAAAACCAAGGGAA-3′.
Additionally, BMDMs seeded in a 12-well plate (4 × 106 cells/well) were treated with ds-miR-146a-mimic-loaded EVs (100 μg/mL) for 2 days. Cell culture media were collected from cultures and stored at −80°C for ELISA analysis. Cells were lysed using Nonidet P-40 (NP-40) lysis buffer, and protein concentration was determined using Bradford assay (Bio-Rad). A total of 20 μg of protein from each sample was separated using 4%–12% gradient gel, transferred to polyvinylidene fluoride (PVDF) membrane, and blotted with primary Abs (anti-IRAK1, anti-GAPDH, 1:1,000; Cell Signaling Technology) overnight at 4°C. After incubation with secondary Abs, protein bands were visualized using Luminata Forte Western HRP substrate (Millipore) and imaged in a ChemiDoc system (Bio-Rad Laboratories, Hercules, CA, USA). The band intensity was quantified using ImageJ. IL-6 levels in the media were measured via ELISA (R&D Systems) according to the manufacturer’s instructions. The primer sequences were as follows: ds-miR-146a-5p mimic: QIAGEN Syn-mmu-miR-146a-5p miScript miRNA Mimic 219600, 5′-UGAGAACUGAAUUCCAUGGGUU-3′; and ds-miR-146a scramble: IDT, 5′-ACGAGUUACGUGGUACGUUAAU-3′.
Statistical Analysis
Data were analyzed using GraphPad Prism software version 7.04. Analysis of variance (one- or two-way as appropriate) was used with Tukey’s post hoc test unless otherwise stated. Statistical significance was shown with the following notation: nsp > 0.05 (not significant); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; #p ≤ 0.05, ##p ≤ 0.01, ###p ≤ 0.001, ####p ≤ 0.0001. Data are plotted as mean ± standard error.
Author Contributions
Conceptualization: A.J., T.N.L., E.D., and S.M.J. Methodology: A.J., T.N.L., S.W., and L.Z. Investigation: A.J., T.N.L., S.W., L.Z., E.D., S.M.K., D.L., B.P., and D.R.K. Writing: A.J. and S.M.J. Supervision: W.C. and S.M.J.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by a PhRMA Foundation Predoctoral Fellowship in Pharmaceutics (to A.J.), by the National Institutes of Health (grant HL141611 to S.M.J., grant GM097259 to W.C., and grant GM122908 to W.C.), and the National Science Foundation (grant 1750542 to S.M.J.). The authors appreciate the help and support of Maryland NanoCenter and AIMLab staff including Dr. Wen-An Chiou and Dr. Sz-Chian Liou. The authors thank Dr. Xiaoxuan Fan for the technical support from the Flow Cytometry Core, University of Maryland School of Medicine, Center for Innovative Biomedical Resources (Baltimore, MD, USA).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.12.007.
Supplemental Information
References
- 1.Lamichhane T.N., Sokic S., Schardt J.S., Raiker R.S., Lin J.W., Jay S.M. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng. Part B Rev. 2015;21:45–54. doi: 10.1089/ten.teb.2014.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EL Andaloussi S., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 3.Vader P., Mol E.A., Pasterkamp G., Schiffelers R.M. Extracellular vesicles for drug delivery. Adv. Drug Deliv. Rev. 2016;106(Pt A):148–156. doi: 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong J.P.K., Stevens M.M. Strategic design of extracellular vesicle drug delivery systems. Adv. Drug Deliv. Rev. 2018;130:12–16. doi: 10.1016/j.addr.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deregibus M.C., Cantaluppi V., Calogero R., Lo Iacono M., Tetta C., Biancone L., Bruno S., Bussolati B., Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 6.Hergenreider E., Heydt S., Tréguer K., Boettger T., Horrevoets A.J.G., Zeiher A.M., Scheffer M.P., Frangakis A.S., Yin X., Mayr M. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012;14:249–256. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 7.Ferguson S.W., Wang J., Lee C.J., Liu M., Neelamegham S., Canty J.M., Nguyen J. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci. Rep. 2018;8:1419. doi: 10.1038/s41598-018-19581-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shimbo K., Miyaki S., Ishitobi H., Kato Y., Kubo T., Shimose S., Ochi M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014;445:381–387. doi: 10.1016/j.bbrc.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Mizrak A., Bolukbasi M.F., Ozdener G.B., Brenner G.J., Madlener S., Erkan E.P., Ströbel T., Breakefield X.O., Saydam O. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol. Ther. 2013;21:101–108. doi: 10.1038/mt.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yim N., Ryu S.-W., Choi K., Lee K.R., Lee S., Choi H., Kim J., Shaker M.R., Sun W., Park J.H. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein-protein interaction module. Nat. Commun. 2016;7:12277. doi: 10.1038/ncomms12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J., Lee H., Goh U., Kim J., Jeong M., Lee J., Park J.H. Cellular Engineering with Membrane Fusogenic Liposomes to Produce Functionalized Extracellular Vesicles. ACS Appl. Mater. Interfaces. 2016;8:6790–6795. doi: 10.1021/acsami.6b01315. [DOI] [PubMed] [Google Scholar]
- 12.El-Andaloussi S., Lee Y., Lakhal-Littleton S., Li J., Seow Y., Gardiner C., Alvarez-Erviti L., Sargent I.L., Wood M.J. Exosome-mediated delivery of siRNA in vitro and in vivo. Nat. Protoc. 2012;7:2112–2126. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 13.Wahlgren J., De L Karlson T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., Valadi H. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 15.Mendt M., Kamerkar S., Sugimoto H., McAndrews K.M., Wu C.-C., Gagea M., Yang S., Blanko E.V.R., Peng Q., Ma X. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3:e99263. doi: 10.1172/jci.insight.99263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao C.Y., Papoutsakis E.T. Engineering human megakaryocytic microparticles for targeted delivery of nucleic acids to hematopoietic stem and progenitor cells. Sci. Adv. 2018;4:eaau6762. doi: 10.1126/sciadv.aau6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooijmans S.A.A., Stremersch S., Braeckmans K., de Smedt S.C., Hendrix A., Wood M.J.A., Schiffelers R.M., Raemdonck K., Vader P. Electroporation-induced siRNA precipitation obscures the efficiency of siRNA loading into extracellular vesicles. J. Control. Release. 2013;172:229–238. doi: 10.1016/j.jconrel.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Lamichhane T.N., Raiker R.S., Jay S.M. Exogenous DNA Loading into Extracellular Vesicles via Electroporation is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharm. 2015;12:3650–3657. doi: 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sutaria D.S., Badawi M., Phelps M.A., Schmittgen T.D. Achieving the Promise of Therapeutic Extracellular Vesicles: The Devil is in Details of Therapeutic Loading. Pharm. Res. 2017;34:1053–1066. doi: 10.1007/s11095-017-2123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamichhane T.N., Jeyaram A., Patel D.B., Parajuli B., Livingston N.K., Arumugasaamy N., Schardt J.S., Jay S.M. Oncogene Knockdown via Active Loading of Small RNAs into Extracellular Vesicles by Sonication. Cell. Mol. Bioeng. 2016;9:315–324. doi: 10.1007/s12195-016-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D., Lee H., Zhu Z., Minhas J.K., Jin Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017;312:L110–L121. doi: 10.1152/ajplung.00423.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Didiot M.-C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R. Exosome-mediated Delivery of Hydrophobically Modified siRNA for Huntingtin mRNA Silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haraszti R.A., Miller R., Didiot M.-C., Biscans A., Alterman J.F., Hassler M.R., Roux L., Echeverria D., Sapp E., DiFiglia M. Optimized Cholesterol-siRNA Chemistry Improves Productive Loading onto Extracellular Vesicles. Mol. Ther. 2018;26:1973–1982. doi: 10.1016/j.ymthe.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Didiot M.-C., Haraszti R.A., Aronin N., Khvorova A. Loading of Extracellular Vesicles with Hydrophobically Modified siRNAs. Methods Mol Biol. 2018;1740:199–214. doi: 10.1007/978-1-4939-7652-2_16. [DOI] [PubMed] [Google Scholar]
- 26.Stremersch S., Vandenbroucke R.E., Van Wonterghem E., Hendrix A., De Smedt S.C., Raemdonck K. Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release. 2016;232:51–61. doi: 10.1016/j.jconrel.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Antimisiaris S., Mourtas S., Papadia K. Targeted si-RNA with liposomes and exosomes (extracellular vesicles): How to unlock the potential. Int. J. Pharm. 2017;525:293–312. doi: 10.1016/j.ijpharm.2017.01.056. [DOI] [PubMed] [Google Scholar]
- 28.Lane R.E., Korbie D., Anderson W., Vaidyanathan R., Trau M. Analysis of exosome purification methods using a model liposome system and tunable-resistive pulse sensing. Sci. Rep. 2015;5:7639. doi: 10.1038/srep07639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurer-Spurej E., Wong K.F., Maurer N., Fenske D.B., Cullis P.R. Factors influencing uptake and retention of amino-containing drugs in large unilamellar vesicles exhibiting transmembrane pH gradients. Biochim. Biophys. Acta. 1999;1416:1–10. doi: 10.1016/s0005-2736(98)00204-1. [DOI] [PubMed] [Google Scholar]
- 30.Bertrand N., Bouvet C., Moreau P., Leroux J.-C. Transmembrane pH-gradient liposomes to treat cardiovascular drug intoxication. ACS Nano. 2010;4:7552–7558. doi: 10.1021/nn101924a. [DOI] [PubMed] [Google Scholar]
- 31.Li X., Hirsh D.J., Cabral-Lilly D., Zirkel A., Gruner S.M., Janoff A.S., Perkins W.R. Doxorubicin physical state in solution and inside liposomes loaded via a pH gradient. Biochim. Biophys. Acta. 1998;1415:23–40. doi: 10.1016/s0005-2736(98)00175-8. [DOI] [PubMed] [Google Scholar]
- 32.Harrigan P.R., Wong K.F., Redelmeier T.E., Wheeler J.J., Cullis P.R. Accumulation of doxorubicin and other lipophilic amines into large unilamellar vesicles in response to transmembrane pH gradients. Biochim. Biophys. Acta. 1993;1149:329–338. doi: 10.1016/0005-2736(93)90218-o. [DOI] [PubMed] [Google Scholar]
- 33.Qiu L., Jing N., Jin Y. Preparation and in vitro evaluation of liposomal chloroquine diphosphate loaded by a transmembrane pH-gradient method. Int. J. Pharm. 2008;361:56–63. doi: 10.1016/j.ijpharm.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnsen K.B., Gudbergsson J.M., Skov M.N., Christiansen G., Gurevich L., Moos T., Duroux M. Evaluation of electroporation-induced adverse effects on adipose-derived stem cell exosomes. Cytotechnology. 2016;68:2125–2138. doi: 10.1007/s10616-016-9952-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander M., Hu R., Runtsch M.C., Kagele D.A., Mosbruger T.L., Tolmachova T., Seabra M.C., Round J.L., Ward D.M., O’Connell R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015;6:7321. doi: 10.1038/ncomms8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Gu H., Qin D., Yang L., Huang W., Essandoh K., Wang Y., Caldwell C.C., Peng T., Zingarelli B., Fan G.C. Exosomal miR-223 Contributes to Mesenchymal Stem Cell-Elicited Cardioprotection in Polymicrobial Sepsis. Sci. Rep. 2015;5:13721. doi: 10.1038/srep13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J., Wang Y., Li L. Functional significance of exosomes applied in sepsis: A novel approach to therapy. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:292–297. doi: 10.1016/j.bbadis.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Miksa M., Wu R., Dong W., Komura H., Amin D., Ji Y., Wang Z., Wang H., Ravikumar T.S., Tracey K.J., Wang P. Immature dendritic cell-derived exosomes rescue septic animals via milk fat globule epidermal growth factor-factor VIII [corrected] J. Immunol. 2009;183:5983–5990. doi: 10.4049/jimmunol.0802994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J., Feng Y., Jeyaram A., Jay S.M., Zou L., Chao W. Circulating Plasma Extracellular Vesicles from Septic Mice Induce Inflammation via MicroRNA- and TLR7-Dependent Mechanisms. J. Immunol. 2018;201:3392–3400. doi: 10.4049/jimmunol.1801008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Essandoh K., Yang L., Wang X., Huang W., Qin D., Hao J., Wang Y., Zingarelli B., Peng T., Fan G.C. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim. Biophys. Acta. 2015;1852:2362–2371. doi: 10.1016/j.bbadis.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boldin M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damas P., Ledoux D., Nys M., Vrindts Y., De Groote D., Franchimont P., Lamy M. Cytokine serum level during severe sepsis in human IL-6 as a marker of severity. Ann. Surg. 1992;215:356–362. doi: 10.1097/00000658-199204000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shalova I.N., Lim J.Y., Chittezhath M., Zinkernagel A.S., Beasley F., Hernández-Jiménez E., Toledano V., Cubillos-Zapata C., Rapisarda A., Chen J. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Kalbitz M., Fattahi F., Herron T.J., Grailer J.J., Jajou L., Lu H., Huber-Lang M., Zetoune F.S., Sarma J.V., Day S.M. Complement Destabilizes Cardiomyocyte Function In Vivo after Polymicrobial Sepsis and In Vitro. J. Immunol. 2016;197:2353–2361. doi: 10.4049/jimmunol.1600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charoenviriyakul C., Takahashi Y., Morishita M., Nishikawa M., Takakura Y. Role of Extracellular Vesicle Surface Proteins in the Pharmacokinetics of Extracellular Vesicles. Mol. Pharm. 2018;15:1073–1080. doi: 10.1021/acs.molpharmaceut.7b00950. [DOI] [PubMed] [Google Scholar]
- 47.Tkach M., Kowal J., Théry C. Why the need and how to approach the functional diversity of extracellular vesicles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20160479. doi: 10.1098/rstb.2016.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Durcin M., Fleury A., Taillebois E., Hilairet G., Krupova Z., Henry C., Truchet S., Trötzmüller M., Köfeler H., Mabilleau G. Characterisation of adipocyte-derived extracellular vesicle subtypes identifies distinct protein and lipid signatures for large and small extracellular vesicles. J. Extracell. Vesicles. 2017;6:1305677. doi: 10.1080/20013078.2017.1305677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullokandov G., Baccarini A., Ruzo A., Jayaprakash A.D., Tung N., Israelow B., Evans M.J., Sachidanandam R., Brown B.D. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat. Methods. 2012;9:840–846. doi: 10.1038/nmeth.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witwer K.W., Buzás E.I., Bemis L.T., Bora A., Lässer C., Lötvall J., Nolte-’t Hoen E.N., Piper M.G., Sivaraman S., Skog J. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2:20360. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.