Abstract
Recent studies suggest that long noncoding RNAs (lncRNAs) play essential roles in tumor progression. However, the functional roles and underlying mechanisms of lncRNAs in neuroblastoma (NB), the most common malignant solid tumor in pediatric population, still remain elusive. Herein, through integrating analysis of a public RNA sequencing dataset, neuroblastoma highly expressed 1 (NHEG1) was identified as a risk-associated lncRNA, contributing to an unfavorable outcome of NB. Depletion of NHEG1 led to facilitated differentiation and decreased growth and aggressiveness of NB cells. Mechanistically, NHEG1 bound to and stabilized DEAD-box helicase 5 (DDX5) protein through repressing proteasome-mediated degradation, resulting in β-catenin transactivation that altered target gene expression associated with NB progression. We further determined a lymphoid enhancer binding factor 1 (LEF1)/transcription factor 7-like 2 (TCF7L2)/NHEG1/DDX5/β-catenin axis with a positive feedback loop and demonstrated that NHEG1 harbored oncogenic properties via its interplay with DDX5. Administration of small interfering RNAs against NHEG1 or DDX5 reduced tumor growth and prolonged survival of nude mice bearing xenografts. High NHEG1 or DDX5 expression was associated with poor survival of NB patients. These results indicate that lncRNA NHEG1 exhibits oncogenic activity that affects NB progression via stabilizing the DDX5 protein, which might serve as a potential therapeutic target for NB.
Keywords: neuroblastoma highly expressed 1, beta-catenin, DEAD-box helicase 5, neuroblastoma, tumor progression, tumorigenesis, long noncoding RNA
Graphical Abstract
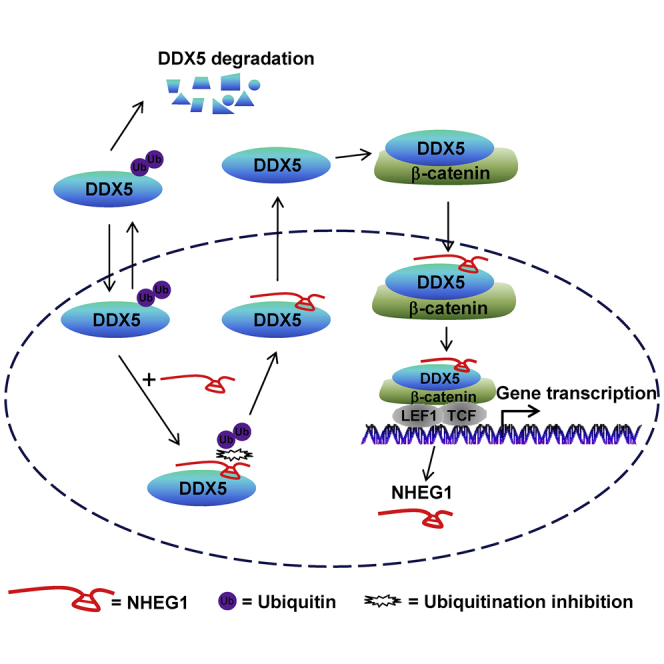
Emerging evidence indicates the roles of long noncoding RNAs (lncRNAs) in tumor progression. Zhao et al. show that a risk-associated lncRNA neuroblastoma highly expressed 1 (NHEG1) binds to and stabilizes DEAD-box helicase 5 (DDX5), resulting in β-catenin transactivation and tumor progression, which serves as a potential therapeutic target for neuroblastoma.
Introduction
Neuroblastoma (NB), a malignancy arising from the primitive neural crest, accounts for 15% of cancer-related mortality in childhood.1 Tumor invasion and metastasis are main death causes of high-risk NB patients.1 Recent evidence indicates that long noncoding RNAs (lncRNAs) play essential roles in NB progression.2, 3, 4, 5, 6 For example, high expression of cyclin-dependent kinase inhibitor 2A/alternate reading frame (ARF) intron 2 lncRNA (CAI2)2 or noncoding RNA expressed in aggressive neuroblastoma (ncRNA)3 is associated with poor survival of NB patients. A lncRNA transcribed from upstream of MYCN (lncUSMycN) promotes MYCN expression and proliferation of NB cells.4 lncRNA MYCN opposite strand (MYCNOS) cooperates with the CCCTC-binding factor to facilitate NB progression through increasing MYCN expression.5 Meanwhile, loss of neuroblastoma-associated transcript-1 (NBAT-1) increases the proliferation and reduces the differentiation of neuronal precursors via activating the RE1 silencing transcription factor.6 However, comprehensive discovery of lncRNAs associated with high risk, progression, and death of NB still remains to be determined.
In this study, through integrative analysis of a public RNA sequencing (RNA-seq) dataset, we discover neuroblastoma highly expressed 1 (NHEG1) as a lncRNA associated with poor prognosis of NB patients. We demonstrate that as a lymphoid enhancer binding factor 1 (LEF1)/transcription factor 7-like 2 (TCF7L2)-regulated lncRNA, NHEG1 is upregulated in NB. Knockdown of NHEG1 facilitates the differentiation and suppresses the tumorigenesis and aggressiveness of NB cell lines. In addition, NHEG1 interacts with and stabilizes protein partner DEAD-box helicase 5 (DDX5), resulting in transactivation of β-catenin, elevated NHEG1 levels, and altered expression of downstream genes. Moreover, administration of small interfering RNAs (siRNAs) against NHEG1 or DDX5 reduces the tumor growth and prolongs the survival of nude mice bearing xenografts, revealing the essential functions of NHEG1 and DDX5 in NB progression.
Results
Identification of NHEG1 as a lncRNA Essential for Tumor Progression
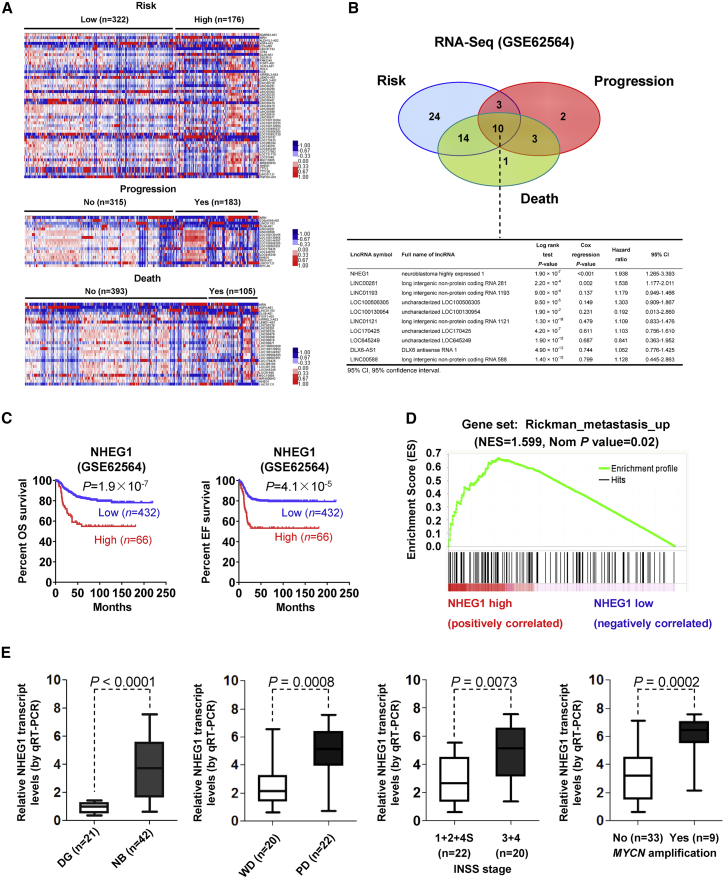
Mining of the RNA-seq dataset (Gene Expression Omnibus [GEO]: GSE62564)7 identified 51, 18, and 28 differentially expressed lncRNAs (fold change > 2.0, p < 0.05) in 498 NB specimens with different status of risk, clinical progression, or death, respectively (Figure 1A). Comprehensive analysis (p < 0.001) indicated that 10 lncRNAs were consistently associated with high risk, progression, and death of NB (Figure 1B). Among them, NHEG1, a conserved lncRNA among human and primate species (Figure S1A), was the most potential lncRNA (hazard ratio [HR] = 1.938, p < 0.001), contributing to poor outcome of NB patients (Figure 1B). Kaplan-Meier curve of these 498 NB patients (GEO: GSE62564) revealed significant difference in overall (p = 1.9 × 10−7) or event-free (p = 4.1 × 10−5) survival of cases with low or high NHEG1 expression (Figure 1C), which was in line with our findings in 42 NB patients (Table S1; Figure S1B). Notably, NHEG1 was of prognostic value for NB cases with low or high international neuroblastoma staging system (INSS) stages (Figure S1C). Gene set enrichment analysis (GSEA) on 3,999 NHEG1-correlated genes (p < 0.005; Table S2) in these 498 specimens revealed their association with cancer metastasis (Figure 1D). Higher NHEG1 transcript levels were validated in an independent cohort of 42 primary NB cases (Table S1) when compared to normal dorsal ganglia (p < 0.0001), especially in those with poor differentiation (p = 0.0008), advanced INSS stages (p = 0.0073), or MYCN amplification (p = 0.0002; Figure 1E). Consistently, in a public RNA-seq dataset (GEO: GSE62564), NHEG1 was highly expressed in NB specimens featured by advanced INSS stages (p = 0.0113), progression (p < 0.0001), high risk (p = 0.005), or death (p < 0.0001; Figure S1D). In public datasets, high NHEG1 expression was significantly associated with poor survival of bladder cancer, breast cancer, colon cancer, endometrioid carcinoma, Ewing sarcoma, gastric cancer, glioblastoma, liver cancer, lung cancer, lymphoma, ovarian cancer, and renal clear cell carcinoma (Figure S2). In addition, soft agar and Matrigel invasion assays indicated that stable ectopic expression of NHEG1 induced the anchorage-independent growth and invasiveness of nontransformed normal epithelial MCF 10A cells (Figures S3A–S3C). These results suggested that NHEG1 was a lncRNA associated with tumor progression.
Figure 1.
Identification of NHEG1 as a lncRNA Associated with NB Progression
(A) Cluster analysis and heatmap (fold change > 2.0, p < 0.05) of RNA-seq dataset (GEO: GSE62564) in 498 NB patients derived from the Gene Expression Omnibus (GEO) depicting the differentially expressed lncRNAs in tumors with various statuses of risk, progression, and death. (B) Venn diagram (top panel) and table of log-rank test or multivariate Cox regression analysis (bottom panel) indicating the comprehensive identification of lncRNAs with prognostic values for risk, progression, and death of NB. (C) Kaplan-Meier curves indicating overall survival (OS) and event-free (EF) survival of 498 well-defined NB patients (GEO: GSE62564) with high or low NHEG1 expression (cutoff value = 1.082). (D) Gene set enrichment analysis of NHEG1-correlated genes in 498 NB tissues (GEO: GSE62564). NES, normalized enrichment score; Nom, normalized. (E) Quantitative real-time RT-PCR assay revealing the levels of the NHEG1 transcript (normalized to GAPDH) in normal dorsal ganglia (DG; n = 21) and NB tissues (n = 42) with poor differentiation (PD) or well differentiation (WD), different INSS stages, or MYCN-amplification status. Fisher’s exact test for overlapping analysis in (B). Log-rank test for survival comparison in (C). Student’s t test compared gene expression levels in (E). Bars are means and whiskers (minimum [min] to maximum [max]) in (E).
NHEG1 Is a LEF1/TCF7L2-Regulated Nuclear lncRNA in NB
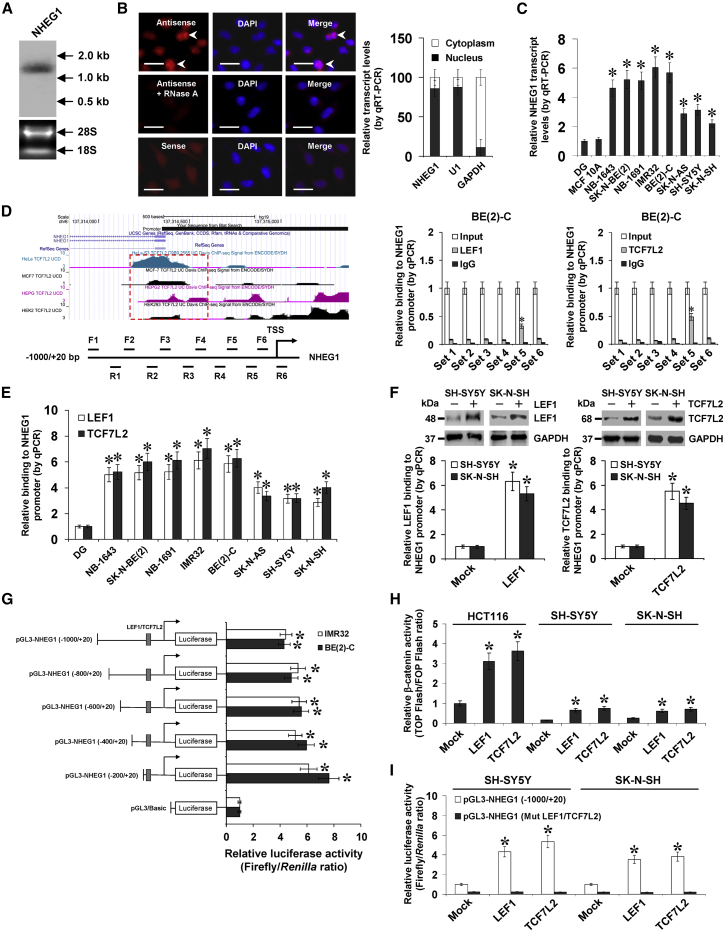
The intergenic lncRNA NHEG1, consisting of three exons, is located at chromosome 6q23.3. In BE(2)-C cells, the existence of the 1,360-bp NHEG1 transcript (GenBank: KX069230) was validated by rapid amplification of cDNA ends (RACE) and northern blot assays (Figures S3D and 2A), with nuclear localization and enrichment revealed by subcellular fractionation and RNA fluorescence in situ hybridization (RNA-FISH) assays (Figure 2B). The Coding-Potential Assessment Tool8 indicated no coding probability of NHEG1 (value = 0.0107). High NHEG1 expression was observed in embryonic brain, spleen, kidney, and muscle tissues (Figure S3E). Mining of public datasets revealed low deletion or amplification frequency of the NHEG1 gene in most cancers (Figure S4A) and revealed no copy number alteration of NHEG1 in 83 NB cases (Figure S4B). In The Cancer Genome Atlas (TCGA) datasets, NHEG1 was upregulated in lung cancer, cervical cancer, prostate cancer, and neuroblastic tumors (Figure S4C). Higher NHEG1 levels were detected in NB cell lines than those of normal dorsal ganglia or MCF 10A cells (Figure 2C). The chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) datasets derived from the University of California Santa Cruz (UCSC) Genome Browser indicated the presence of the TCF7L2 peak in the region adjacent to the transcription start site of NHEG1 (Figure 2D). Notably, endogenous enrichment of TCF7L2 and LEF1 (another TCF/LEF family member) on the proximal NHEG1 promoter was observed in NB cells (Figures 2D and 2E). In addition, in SH-SY5Y and SK-N-SH cells transfected with LEF1 or TCF7L2, there was an increased NHEG1 promoter fragment enriched by LEF1 or TCF7L2 antibody, respectively (Figure 2F). Dual-luciferase array indicated that NHEG1 transcription was predominantly controlled by a region spanning −200 to +20 nucleotides relative to the transcription start site, which contained a putative binding site for LEF1/TCF7L2 (Figure 2G). Meanwhile, transfection of LEF1 or TCF7L2 led to an increase of β-catenin activity in SH-SY5Y and SK-N-SH cells, with response tendency similar to that in colon cancer HCT116 cells (Figure 2H). The activity of the NHEG1 promoter reporter pGL3-NHEG1 (−1,000/+20) was significantly elevated in SH-SY5Y and SK-N-SH cells transfected by LEF1 or TCF7L2, whereas mutation of the LEF1/TCF7L2 binding site abolished this alteration (Figure 2I). Accordingly, transfection of LEF1 or TCF7L2, but not of MYCN, increased the NHEG1 transcript levels in these NB cells (Figures S5A and S5B). These data indicated that NHEG1 was a LEF1/TCF7L2-regulated nuclear lncRNA.
Figure 2.
NHEG1 Is a LEF1/TCF7L2-Regulated Nuclear lncRNA
(A) Northern blot assay with a 268-bp specific probe indicating the existence of 1.3 kb NHEG1 transcript in BE(2)-C cells. (B) RNA fluorescence in situ hybridization images (left panel) showing the nuclear localization of NHEG1 in BE(2)-C cells using a 268-bp antisense probe (red), with nuclei staining with DAPI (blue). The sense probe and antisense probe with RNase A (20 μg) treatment were used as negative controls. Scale bars, 10 μm. Quantitative real-time RT–PCR (right panel) showing the distribution of NHEG1, U1, and GAPDH in the nuclear and cytoplasmic fractions (n = 4). (C) Quantitative real-time RT-PCR assay indicating the NHEG1 transcript levels (normalized to GAPDH) in normal DG (n = 21), MCF 10A cells, and NB cell lines. (D) UCSC Genome Browser view (top left panel) and ChIP and qPCR assays (middle and right panels) with tiled primer sets (bottom left panel), indicating the endogenous binding of TCF7L2 and LEF1 to the NHEG1 promoter (normalized to input DNA) in NB cells (n = 6). (E) ChIP and qPCR assays with primer set 5 showing the enrichment of LEF1 and TCF7L2 on the NHEG1 promoter (normalized to input DNA) in normal DG (n = 10) and NB cell lines (n = 5). (F) ChIP and qPCR assays with primer set 5 (lower panel) and western blot (upper panel) revealing the enrichment of LEF1 and TCF7L2 on the NHEG1 promoter (normalized to input DNA) in NB cells transfected with empty vector (Mock), LEF1, or TCF7L2 (n = 5). (G) Dual-luciferase assay indicating the activity of NHEG1 promoter reporters (normalized to pGL3-Basic) in NB cells (n = 6). (H) Dual-luciferase assay showing the relative activity of β-catenin in colon cancer HCT116 cells and NB cell lines transfected with mock, LEF1, or TCF7L2 (n = 5). (I) Dual-luciferase assay indicating the relative activity of the NHEG1 promoter reporter pGL3-NHEG1 (−1,000/+20) with the wild-type (WT) or mutant (Mut) LEF1/TCF7L2 binding site in NB cells transfected with mock, LEF1, or TCF7L2 (n = 5), with normalization to activity of WT reporter in mock cells. ANOVA and Student’s t test compared the difference in (C)–(I). *p < 0.01 versus DG, IgG, mock, or pGL3-Basic. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (B)–(I).
Knockdown of NHEG1 Suppresses the Progression of NB
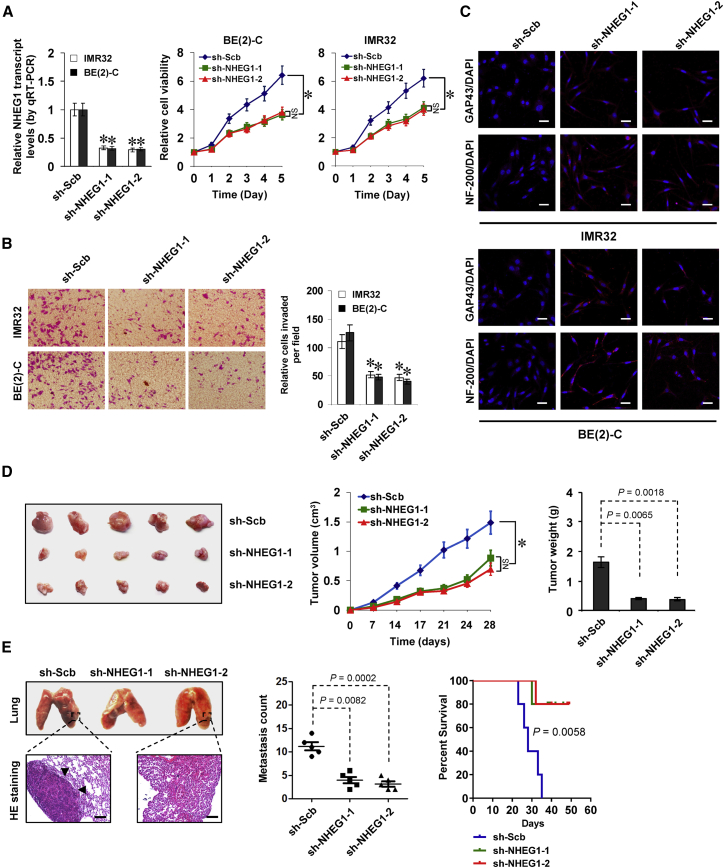
Then, the effects of NHEG1 knockdown were explored in IMR32 and BE(2)-C cells, representing high expression levels. Transfection of two independent short hairpin RNAs (shRNAs), sh-NHEG1-1 and sh-NHEG1-2, resulted in depletion of NHEG1 and a decrease of viability in NB cells (Figure 3A). In addition, decreased invasiveness was observed in these viable NB cells with stable depletion of NHEG1 (Figure 3B). Meanwhile, elevated levels of growth-associated protein 43 (GAP43) and neurofilament protein 200 (NF-200), two established neuronal differentiation markers, were observed in NB cells following stable NHEG1 knockdown (Figure 3C). Consistent with these findings, stable NHEG1 knockdown into BE(2)-C cells resulted in decreased growth and weight of formed subcutaneous xenograft tumors in nude mice (Figure 3D). Moreover, fewer lung metastatic colonies and longer survival were noted in nude mice that received tail-vein administration of BE(2)-C cells stably transfected by sh-NHEG1-1 or sh-NHEG1-2 (Figure 3E). These results indicated that knockdown of NHEG1 inhibited NB progression.
Figure 3.
Knockdown of NHEG1 Leads to Suppression of NB Progression
(A) Quantitative real-time RT-PCR (left panel) and MTT colorimetric assay (middle and right panels) depicting the changes in NHEG1 expression (normalized to GAPDH levels) and cell viability of NB cells stably transfected with two independent shRNAs against NHEG1 (sh-NHEG1) compared with scramble shRNA (sh-Scb; n = 5). (B) Representative images (left panel) and quantification (right panel) of Transwell Matrigel invasion assay indicating the invasion capability of NB cells stably transfected with sh-Scb or sh-NHEG1 (n = 5). (C) Immunofluorescent confocal showing the neuronal differentiation of IMR32 and BE(2)-C cells stably transfected with sh-Scb or sh-NHEG1, using the antibodies specific to neuronal markers GAP43 and NF-200. Scale bars, 10 μm. (D) Representative images (left panel), in vivo growth curve (middle panel), and weight at the end points (right panel) of subcutaneous xenograft tumors formed by BE(2)-C cells stably transfected with sh-Scb or sh-NHEG1 in nude mice (n = 5 for each group). (E) Representative images (top left panel), hematoxylin and eosin (H&E) staining (bottom left panel), quantification of lung metastatic colonization (middle panel), and Kaplan-Meier curves (right panel) of nude mice treated with tail-vein injection of BE(2)-C cells stably transfected with sh-Scb or sh-NHEG1 (n = 5 for each group). ANOVA compared the difference in (A), (B), (D), and (E). Log-rank test for survival comparison in (E). *p < 0.01 versus sh-Scb; NS, not significant. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (A)–(C).
NHEG1 Stabilizes DDX5 Protein through Physical Interaction
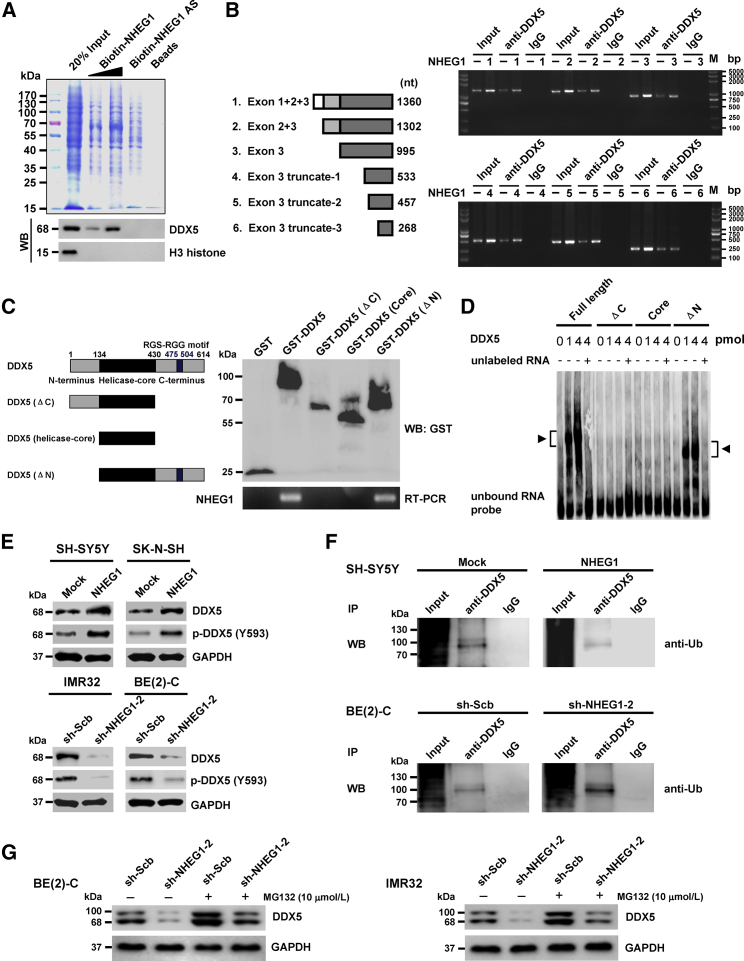
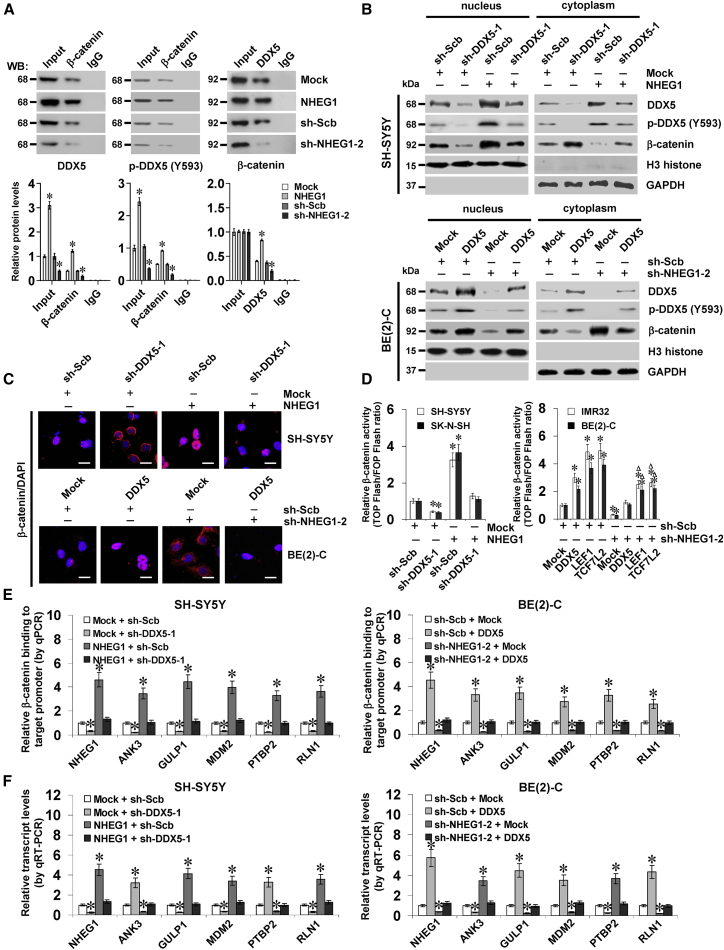
To investigate the protein partner of NHEG1, biotin-labeled RNA pull-down was carried out, followed by a nonquantitative proteomic analysis of the RNA-associated protein complex. Mass spectrometry revealed six and nine differential proteins pulled by NHEG1, but not by NHEG1 antisense transcript, from BE(2)-C and IMR32 cells, respectively, whereas DDX5 was the only protein (with 85 detectable peptides) consistently pulled down by NHEG1 in both cell lines (Figure 4A; Table S3). Western blot further validated that DDX5 was detected in the NHEG1 pull-down complex but not in the control samples pulled down by the NHEG1 antisense transcript or beads only (Figure 4A). RNA immunoprecipitation (RIP) assay indicated endogenous interaction of NHEG1 with both DDX5 and tyrosine 593-phosphorylated DDX5 (p-DDX5 (Y593)) in SH-SY5Y and SK-N-SH cells, which was increased by stable transfection of NHEG1 but not of empty vector (Figure S6A). In addition, through establishing truncation constructs, exon 3 (especially the 904–1,092 nt) of NHEG1 was found to be crucial for its binding to DDX5 (Figure 4B), whereas carboxyl-terminus (431–614 amino acids [aa]), but not amino-terminus (1–134 aa) or helicase-core domain (135–430 aa), of glutathione S-transferase (GST)- or hemagglutinin (HA)-tagged DDX5 was essential for the binding to NHEG1 (Figuress 4C, 4D, and S6B). Notably, overexpression of NHEG1 decreased the degradation of DDX5 in cycloheximide-treated BE(2)-C cells and markedly increased the half-life of DDX5 degradation from 17.5 h to 33.2 h (Figure S6C). Ectopic expression or depletion of NHEG1 facilitated and reduced the protein levels of DDX5 and p-DDX5 (Y593) in NB cells, without impact on transcript levels (Figures 4E and S6D). Endogenous ubiquitination of DDX5 (predominant band at 100 kDa) was observed in NB cells, which was repressed and facilitated by overexpression or depletion of NHEG1 (Figure 4F). Pretreatment of BE(2)-C and IMR32 cells with a proteasome inhibitor (MG132) prevented the degradation of DDX5 protein induced by knockdown of NHEG1 (Figure 4G). These results suggested that NHEG1 interacted with and stabilized DDX5 protein in NB cells.
Figure 4.
NHEG1 Interacts with and Stabilizes DDX5 Protein in NB Cells
(A) Biotin-labeled RNA pull-down, Coomassie blue staining (top panel) and validating western blot (bottom) assays revealing the interaction between NHEG1 and DDX5 protein in BE(2)-C cells. NHEG1 antisense (AS)- and bead-bound protein served as negative controls. (B) RIP assay using DDX5 antibody indicating the interaction between NHEG1 and DDX5 protein in SH-SY5Y cells transfected with a series of NHEG1 truncations. The immunoglobulin G (IgG)-bound RNA was taken as negative control. (C) In vitro binding assay depicting the recovered NHEG1 levels by RIP (right lower panel) after incubation with full-length (1–614 amino acids), ΔC (1–430 amino acids), helicase-core (135–430 amino acids), or ΔN (135–614 amino acids) of GST-tagged recombinant DDX5 protein (left panel) validated by western blot (right upper panel). (D) RNA EMSA assay determining the interaction between recombinant DDX5 protein and biotin-labeled RNA probe for NHEG1 (arrowheads), with or without competition, using an excess of the unlabeled homologous RNA probe. (E) Western blot assay showing the total and phosphorylated (at tyrosine 593) DDX5 in NB cells stably transfected with empty vector (Mock), NHEG1, scramble shRNA (sh-Scb), or sh-NHEG1-2. (F) coIP and western blot assays indicating the endogenous interaction between DDX5 and ubiquitin (Ub) in SH-SY5Y and BE(2)-C cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2. (G) Western blot assay showing the expression of DDX5 in NB cells stably transfected with sh-Scb or sh-NHEG1-2 and those treated with proteasome inhibitor MG132 for 48 hr. Data are representative of three independent experiments in (A)–(G).
NHEG1 Regulates Target Gene Expression by a LEF1/TCF7L2/NHEG1/DDX5/β-Catenin Positive Feedback Loop
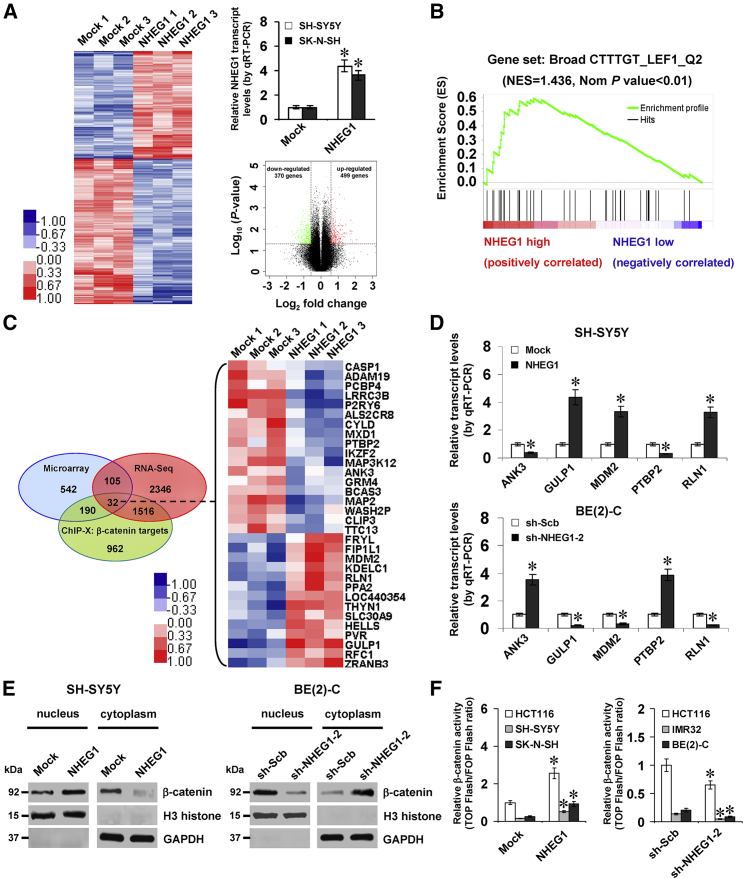
To explore the target genes of NHEG1, microarray analysis was performed in SH-SY5Y cells stably overexpressing NHEG1, which indicated 499 upregulated (Table S4) and 370 downregulated (Table S5) genes upon NHEG1 overexpression (fold change > 1.5, p < 0.05; Figure 5A). The GSEA on these genes revealed significant association with β-catenin signaling (normalized enrichment score [NES] = 1.436, normalized p < 0.01; Figure 5B). Further overlapping analysis (p < 0.001) with genes positively or negatively correlated with NHEG1 (p < 0.005, false discovery rate < 0.05; Table S2) in the RNA-seq dataset (GEO: GSE62564) and β-catenin target genes in the ChIP-X database9 indicated that thirty-two downstream genes were significantly regulated by NHEG1 (Figure 5C). Among them, the expression of ankyrin 3 (ANK3), engulfment adaptor phosphotyrosine-binding (PTB) domain containing 1 (GULP1), murine double minute 2 (MDM2), polypyrimidine tract binding protein 2 (PTBP2), and relaxin 1 (RLN1) was significantly correlated with NHEG1 levels in a public NB dataset (GEO: GSE62564; Figure S7A) and associated with outcome of patients (Figure S7B). Stable overexpression or knockdown of NHEG1 altered the transcript levels of these genes (Figure 5D) and increased or decreased the nuclear translocation and transactivation of β-catenin in NB cells (Figures 5E and 5F), respectively.
Figure 5.
NHEG1 Regulates the Expression of β-Catenin Target Genes in NB Cells
(A) Microarray heatmap (left, fold change > 1.5, p < 0.05), quantitative real-time RT-PCR (top right panel, n = 3), and volcano plots (bottom right panel) revealing the alteration of gene expression in SH-SY5Y cells stably transfected with empty vector (Mock) or NHEG1. (B) Gene set enrichment analysis of altered genes in SH-SY5Y cells stably transfected with NHEG1, compared with mock-transfected control. NES, normalized enrichment score; Nom, normalized. (C) Venn diagram (left panel) and heatmap (right panel, fold change > 1.5, p < 0.05) showing the NHEG1-altered expression of β-catenin target genes that were intersected with NHEG1-correlated transcripts in the RNA-seq dataset (GEO: GSE62564) and ChIP-X database. (D) Quantitative real-time RT-PCR assay indicating the transcript levels of β-catenin target genes ANK3, GULP1, MDM2, PTBP2, and RLN1 (normalized to GAPDH) in NB cells stably transfected with mock, NHEG1, scramble shRNA (sh-Scb), or sh-NHEG1-2 (n = 5). (E) Western blot assay showing the nuclear and cytoplasmic expression of β-catenin and H3 histone in NB cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2. (F) Dual-luciferase assay indicating the relative activity of β-catenin in colon cancer HCT116 and NB cells transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 (n = 5). Student’s t test analyzed the difference in (A), (D), and (F). Fisher’s exact test for overlapping analysis in (C). *p < 0.01 versus mock or sh-Scb. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (D)–(F).
Since previous studies indicate the coactivator role of DDX5 in regulating β-catenin activity,10 we further investigated their interplay in NB. Colocalization of DDX5 and β-catenin was observed in NB cells (Figure S7C). Depletion of DDX5 prevented the increased nuclear translocation of β-catenin in NB cells transfected with β-catenin (Figures S8A and S8B). Conversely, transfection of DDX5 into NB cells facilitated the nuclear translocation of β-catenin, whereas knockdown of β-catenin abolished these effects (Figures S8C and S8D). The β-catenin activity was significantly decreased and increased by stable knockdown or overexpression of DDX5 in NB cells, which was rescued by overexpression or depletion of β-catenin, respectively (Figure S8E). Consistently, the expression of β-catenin downstream genes was significantly changed in NB cells (Figure S8F). Notably, overexpression or depletion of NHEG1 increased and reduced the levels of DDX5 and p-DDX5 (Y593) and their interaction with β-catenin in NB cells (Figure 6A). In addition, ectopic expression or knockdown of NHEG1 led to an increase and decrease in the cytoplasmic and nuclear levels of DDX5 and p-DDX5 (Y593) and nuclear translocation and transactivation of β-catenin, which was prevented by depletion or overexpression of DDX5, respectively (Figures 6B–6D). Depletion of NHEG1 attenuated the LEF1- or TCF7L2-facilitated β-catenin activity in NB cells (Figure 6D). Importantly, the enrichment of β-catenin on the promoters of NHEG1 and its downstream genes ANK3, GULP1, MDM2, PTBP2, and RLN1 was significantly increased and decreased in NB cells with stable transfection of NHEG1 or sh-NHEG1-2, which was rescued by knockdown and ectopic expression of DDX5, respectively (Figure 6E). Moreover, the expression of these β-catenin target genes was correspondingly altered in these NB cells (Figure 6F). Meanwhile, individual or combinatorial knockdown of DDX5 and NHEG1 exerted similar effects on β-catenin enrichment and expression of these target genes in SH-SY5Y cells (Figures S9A and S9B) and repressed the viability of MYCN-nonamplified (SH-SY5Y and SK-N-SH) and MYCN-amplified (BE(2)-C and IMR32) NB cells to the similar extent (Figures S9C and S9D). These results demonstrated that NHEG1 regulated the target gene expression through a LEF1/TCF7L2/NHEG1/DDX5/β-catenin positive feedback loop in NB cells.
Figure 6.
NHEG1 Regulates β-Catenin Target Gene Expression by a LEF1/NHEG1/ DDX5/β-Catenin Positive Feedback Loop in NB Cells
(A) Representative images (top panel) and quantification (bottom panel) of coIP and western blot assays revealing the interaction among DDX5, p-DDX5 (Y593), and β-catenin in SH-SY5Y cells stably transfected with empty vector (Mock), NHEG1, scramble shRNA (sh-Scb), or sh-NHEG1-2, with normalization to input of mock or sh-Scb. (B) Western blot assay indicating the nuclear and cytoplasmic expression of DDX5, p-DDX5 (Y593), β-catenin, and H3 histone in NB cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 and those cotransfected with sh-DDX5-1 or DDX5. (C) Fluorescence immunocytochemical staining showing the localization of β-catenin in SH-SY5Y and BE(2)-C cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 and those cotransfected with sh-DDX5-1 or DDX5. Scale bars, 10 μm. (D) Dual-luciferase assay indicating the relative activity of β-catenin in NB cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 and those cotransfected with sh-DDX5-1, DDX5, LEF1, or TCF7L2 (n = 5). (E) ChIP and qPCR assays showing the enrichment of β-catenin on target gene promoters (normalized to input DNA) in SH-SY5Y and BE(2)-C cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 and those cotransfected with sh-DDX5-1 or DDX5 (n = 5). (F) Quantitative real-time RT-PCR assay revealing the transcript levels of target genes (normalized to GAPDH) in SH-SY5Y and BE(2)-C cells stably transfected with mock, NHEG1, sh-Scb, or sh-NHEG1-2 and those cotransfected with sh-DDX5-1 or DDX5 (n = 5). Student’s t test and ANOVA analyzed the difference in (A) and (D)–(F). *p < 0.01 versus mock, sh-Scb, or mock + sh-Scb. Δp < 0.01 versus sh-Scb + LEF1 or sh-Scb + TCF7L2. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (A)–(F).
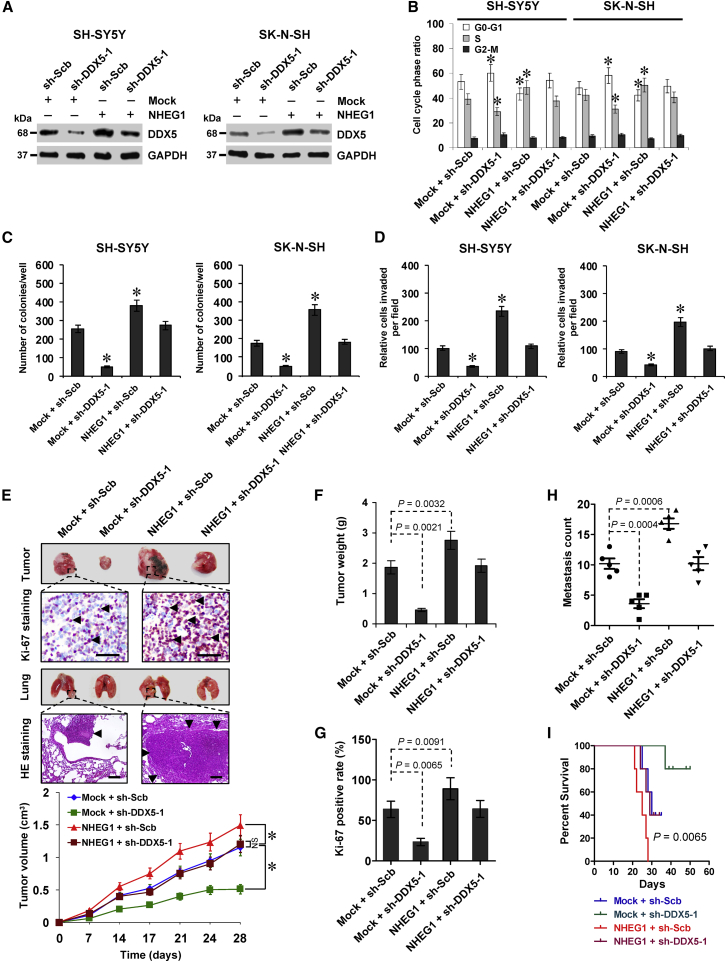
NHEG1 Exerts Oncogenic Roles via Interacting with DDX5
To explore the functional interplay of NHEG1 and DDX5 during the aggressiveness of NB cells, rescue studies were performed in SH-SY5Y and SK-N-SH cells with moderate NHEG1 levels. Stable overexpression of NHEG1 into these NB cells led to an increase in DDX5 expression, which was restored by transfection of sh-DDX5-1 (Figure 7A). The cell-cycle progression into S phase, viability, and growth of NB cells was facilitated by stable overexpression of NHEG1 (Figures 7B, 7C, S9E, and S9F). In addition, ectopic expression of NHEG1 reduced the levels of neuronal differentiation markers GAP43 and NF-200 (Figure S9G). Stable overexpression of NHEG1 resulted in increased invasiveness of viable NB cells (Figures 7D and S9H). Restoration of DDX5 levels via transfection of sh-DDX5-1 rescued the changes in cell-cycle progression, viability, growth, differentiation, and invasion of NB cells following stable transfection of NHEG1 (Figures 7B–7D and S9E–S9H). Moreover, stable transfection of NHEG1 in SH-SY5Y cells led to an increase in the tumorigenesis, weight, and Ki-67 proliferation index of subcutaneous xenograft tumors (Figures 7E–7G) and led to statistically more lung metastatic colonies and lower survival probability in nude mice (Figures 7E, 7H, and 7I). Meanwhile, depletion of DDX5 abolished the impacts of NHEG1 on in vivo tumorigenesis and aggressiveness of NB cells (Figures 7E–7I). These results suggested that NHEG1 exerted oncogenic roles via interacting with DDX5.
Figure 7.
NHEG1 Exerts Oncogenic Roles via Interacting with DDX5
(A) Western blot assay showing the expression of DDX5 in SH-SY5Y and SK-N-SH cells stably transfected with empty vector (Mock) or NHEG1 and those cotransfected with scramble shRNA (sh-Scb) or sh-DDX5-1. (B–D) Quantification of flow cytometric (B), soft agar (C), and Transwell Matrigel invasion (D) assays indicating the cell-cycle progression, anchorage-independent growth, and invasion capability of NB cells stably transfected with mock or NHEG1 and those cotransfected with sh-Scb or sh-DDX5-1 (n = 6). (E–H) Representative images (E, top panel), in vivo growth curve (E, bottom panel), tumor weight (F), and Ki-67 positive rate (G) at the end points of xenograft tumors in athymic nude mice formed by hypodermic injection of SH-SY5Y cells stably transfected with mock, NHEG1, sh-Scb, and sh-DDX5-1 (n = 5 for each group). Representative images (E, top panel), H&E staining (E, arrowheads), and quantification (H) of lung metastatic colonies in athymic nude mice (n = 5 for each group) after tail-vein injection of SH-SY5Y cells stably transfected with mock, NHEG1, sh-Scb, or sh-DDX5-1. Scale bars, 100 μm. (I) Kaplan-Meier curves of nude mice treated with tail-vein injection of SH-SY5Y cells stably transfected with mock or NHEG1 and those cotransfected with sh-Scb or sh-DDX5-1 (n = 5 for each group). ANOVA analyzed the difference in (B)–(H). Log-rank test for survival comparison in (I). *p < 0.01 versus mock + sh-Scb. NS, not significant. Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (A)–(D).
Therapeutic Efficiencies of siRNAs for NHEG1 and DDX5 In Vivo
We further explored the therapeutic efficiencies of NHEG1- and DDX5-specific siRNAs, si-NHEG1 and si-DDX5, on the tumor growth and survival of athymic nude mice bearing xenografts. Intravenous administration of si-NHEG1 or si-DDX5 significantly reduced the volume and weight of xenograft tumors formed by hypodermic injection of BE(2)-C cells when compared to those treated with scramble siRNA (si-Scb; Figures S10A and S10B). Meanwhile, the body weight of nude mice, an indicator of developing tumor burden, was improved in si-NHEG1 or si-DDX5 treatment groups than that of the si-Scb treatment group (Figure S10C). In addition, Kaplan-Meier survival analysis revealed that intravenous administration of si-NHEG1 or si-DDX5 obviously prolonged the survival time of nude mice (Figure S10D). Administration of si-NHEG1 or si-DDX5 decreased the expression of NHEG1 and DDX5 within tumors, respectively, with corresponding changes in downstream gene expression levels (Figures S10E and S10F). These results indicated the therapeutic efficiencies of si-NHEG1 and si-DDX5 in nude mice bearing xenograft tumors.
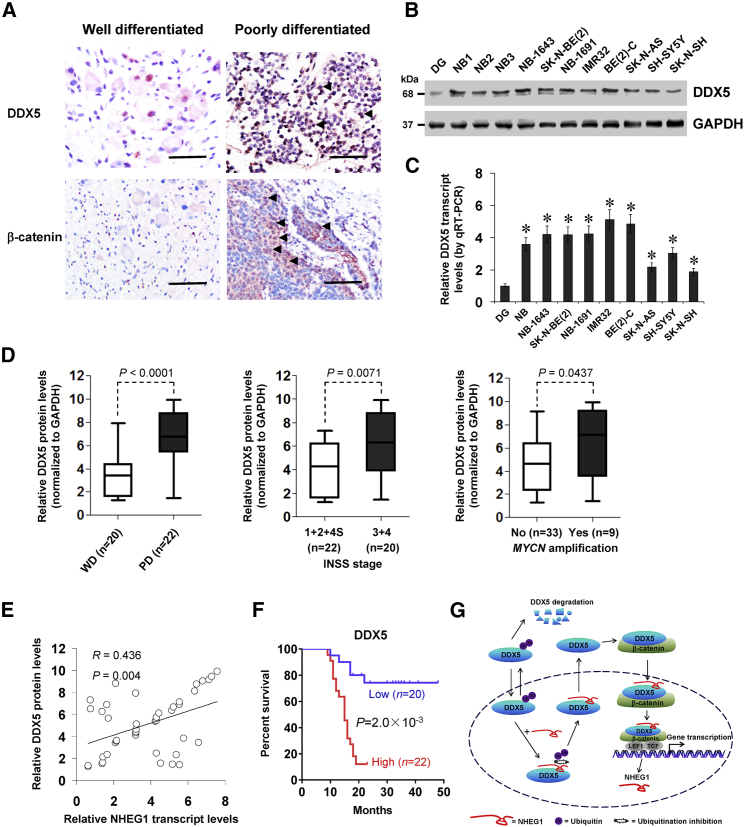
High DDX5 Expression Is Associated with an Unfavorable Outcome of NB
We further investigated the expression of DDX5 in NB, using fresh and paraffin-embedded specimens from 42 primary cases. Immunohistochemical staining indicated that DDX5 and β-catenin were observed in the nuclei and cytoplasm of tumor cells (Figure 8A). The DDX5 immunostaining was observed in 23/42 (54.8%) NB specimens and higher in those with poor differentiation (p = 0.041), higher mitosis karyorrhexis index (MKI; p = 0.045), advanced INSS stages (p = 0.007), or MYCN amplification (p = 0.027; Table S6). Elevated DDX5 levels were detected in NB tissues and cell lines when compared to normal dorsal ganglia (Figures 8B and 8C), especially in NB specimens featured by poor differentiation (p < 0.0001), advanced INSS stages (p = 0.0071), or MYCN amplification (p = 0.0437; Figure 8D). The NHEG1 expression was positively correlated with that of DDX5 protein (R = 0.436, p = 0.004) in these NB specimens (Figure 8E). The Kaplan-Meier survival curve of 42 NB patients revealed that high DDX5 levels had lower survival probability (p = 0.002) (Figure 8F). These results indicated that high DDX5 expression was associated with an unfavorable outcome of NB.
Figure 8.
High DDX5 Expression Is Associated with Unfavorable Outcome of NB Patients
(A) Representative immunohistochemical staining showing the nuclear and cytoplasmic expression pattern of DDX5 and β-catenin in the tumor cells of NB specimens (arrowheads, brown). Scale bars. 100 μm. (B and C) Western blot (B) and quantitative real-time RT-PCR (C) assays indicating the levels of DDX5 (normalized to GAPDH) in normal DG (n = 10 and 21, respectively), NB tissues (n = 42), and cultured NB cell lines. (D) Quantification of western blot assay revealing the DDX5 levels (normalized to GAPDH) in NB tissues with different differentiation, INSS stages, or MYCN-amplification status. (E) Pearson’s coefficient correlation analysis depicting the positive correlation between NHEG1 transcript and DDX5 protein levels in NB tissues (n = 42). (F) Kaplan-Meier survival curve of 42 NB patients with high or low expression of DDX5 (cutoff value = 5.0). (G) The mechanisms underlying NHEG1-promoted NB progression: as a LEF1/TCF7L2-regulated lncRNA, NHEG1 interacts with and stabilizes DDX5 protein through repressing the ubiquitination and proteasomal degradation, which in turn, leads to nuclear translocation and activation of β-catenin, elevated NHEG1 levels, and altered transcription of downstream genes associated with NB progression. ANOVA and Student’s t test analyzed the difference in (C) and (D). Pearson’s correlation coefficient analysis for gene expression in (E). Log-rank test for survival comparison in (F). *p < 0.01 versus DG. Bars are means and whiskers (min to max) in (D). Data are shown as mean ± SEM (error bars) and representative of three independent experiments in (A)–(C).
Discussion
In our previous studies, we discover that Ets-1 promoter-associated noncoding RNA (pancEts-1) binds to heterogeneous nuclear ribonucleoprotein K (hnRNPK) to facilitate its physical interaction with β-catenin, resulting in β-catenin stabilization and NB progression.11 However, whether other lncRNAs contribute to β-catenin transactivation during NB progression still remains elusive. In this study, we discover that NHEG1, a lncRNA locating at chromosome 6q23.3, is associated with progression and poor outcome of NB. Recent studies indicate that loss of distal 6q (6q27) is associated with poor survival of high-risk NB patients.12 However, the mining of public dataset shows no copy number alteration of NHEG1 in NB cases. Our evidence shows that NHEG1 does not affect the levels of β-catenin in NB cells. Instead, we demonstrate the crucial roles of LEF1/TCF7L2/β-catenin in controlling NHEG1 transcription and that NHEG1 interacts with and stabilizes DDX5 protein to increase the nuclear translocation of β-catenin (Figure 8G), which subsequently associates with LEF1 to regulate gene transcription,13 suggesting the LEF1/TCF7L2/NHEG1/DDX5/β-catenin positive feedback loop in NB. Our evidence from in vitro and in vivo studies clearly demonstrates that NHEG1 exerts oncogenic properties via interplay with DDX5. The discovery of such a regulatory loop indicates a promising step toward harnessing this lncRNA for therapeutic intervention against NB.
Previous studies show upregulation of β-catenin in high-risk NB tissues and cell lines without MYCN amplification, whereas the mechanisms regulating activation of β-catenin remain elusive.14 Subsequent studies indicate that activation of β-catenin signaling is associated with increased luciferase activity of the TOP/FOP reporter in MYCN nonamplified SK-N-SH cells.15 Transfection of sialidase NEU4 long (NEU4L) leads to hyperactivation of β-catenin signaling and stem-like growth in MYCN-amplified SK-N-BE cells.16 Gain- and loss-of-function studies indicate that β-catenin enhances cell viability of NB cells17 and confers resistance to crizotinib in MYCN-amplified and nonamplified NB cells.18 In this study, we detected endogenous β-catenin activity in NB cell lines. As β-catenin-interacting proteins, LEF1 serves as an activator of gene transcription, whereas TCF7L2 displays active or repressive transcriptional effects in a cell-context-dependent manner.19 Transfection of LEF1 or TCF7L2 led to increased β-catenin activity in NB cells, with response tendency similar to that in colon cancer cells. In addition, we found high expression of NHEG1 in NB tissues and cell lines. As a lncRNA was regulated by LEF1/TCF7L2, but not by MYCN, NHEG1 facilitated the transactivation of β-catenin in both MYCN-amplified and nonamplified NB cells, indicating the crucial roles of NHEG1 in activating β-catenin signaling in NB.
As a nuclear prototypic member of the DEAD box family, DDX5 is overexpressed in a variety of malignancies, including breast cancer, prostate cancer, and colorectal cancer, and is associated with higher tumor grade and poor prognosis.20 DDX5 is essential for proliferation of breast cancer cells via controlling the transcription of DNA replication genes.21 Through interacting with calcium/calmodulin, DDX5 facilitates the migration and metastasis of tumor cells,22 indicating the important roles of DDX5 in cancer progression. In the current study, we demonstrated that high DDX5 levels were associated with an unfavorable outcome of NB, and DDX5 exerted oncogenic functions in NB progression. We found that DDX5 promoted the nuclear translocation and transactivation of β-catenin via physical interaction, resulting in altered expression of downstream target genes associated with tumor progression, such as ANK3,23 GULP1,24 MDM2,25 PTBP2,26 and RLN1.27 Previous studies show conflicting findings in the impact of c-Abl kinase-mediated Tyr593 phosphorylation on the roles of DDX5 in stimulating β-catenin-dependent transcription in colon cancer cells.28,29 Our data indicated that NHEG1 facilitated the levels of wild-type and Tyr593-phosphorylated DDX5 protein, resulting in β-catenin transactivation in NB cells. Since knockdown of DDX5 rescued the NB cells from alteration in biological behaviors upon NHEG1 overexpression, our evidence suggests that the oncogenic roles of NHEG1 are exerted, at least in part, by stabilizing DDX5 protein.
Recent evidence shows that DDX5 also forms an RNA-protein complex with HOX transcript antisense RNA (HOTAIR) to regulate the stability of suppressor of zeste 12 homolog and PRC2-mediated gene repression in hepatocarcinogenesis.30 In the current study, we identified NHEG1 as a lncRNA binding to the DDX5 protein. Mechanistically, NHEG1 stabilizes DDX5 protein to facilitate its binding to β-catenin and enhances β-catenin-mediated transcriptional activation, suggesting a function of NHEG1 in recruiting critical factors to assemble active LEF1/TCF7L2/β-catenin transcription complexes. We demonstrate that NHEG1 physically binds to the carboxyl-terminus of the DDX5 protein to prevent its degradation. The ubiquitylated proteins are usually degraded via the 26S proteasome.31 However, DDX5 is aberrantly modified by polyubiquitylation in colon adenomas and carcinomas, and there is a possible defect in proteasome-mediated degradation of DDX5 in these tumors.32 Our evidence shows that endogenous ubiquitination of DDX5 is responsive to the proteasome inhibitor, and NHEG1 represses the ubiquitination and proteasomal degradation of DDX5 in NB cells, whereas the involved ubiquitination sites warrant further investigation.
In summary, we have demonstrated that NHEG1, a nuclear lncRNA, is upregulated and associated with an unfavorable outcome of NB. NHEG1 directly interacts with and stabilizes DDX5 protein to increase its expression and β-catenin activity. NHEG1 expression is essential for promoting the tumorigenesis and aggressiveness of NB cells. Treatment with siRNAs against NHEG1 or DDX5 reduces the tumor growth and prolongs the survival time of nude mice bearing xenografts. Due to a limited number of specimens, the association of NHEG1 or DDX5 levels with MYCN amplification warrants further investigation with a larger cohort of NB cases. Mutagenic studies are needed to reveal the exact roles of p-DDX5 (Y593) in NHEG1-facilitated transactivation of β-catenin. In addition, since potential interplay exists between the hemostatic system and malignancy,33 further studies are warranted to explore the roles of NHEG1 in homeostasis and oncogenesis using transgenic or knockout animal models. Due to the limitation of the nonquantitative proteomic approach in identifying proteins of relatively low abundance,34 further quantitative proteomics are needed to explore the potential protein partners of NHEG1 and underlying mechanisms in progression of other cancers. The current study extends our understanding about the genes contributing to NB progression and indicates that NHEG1 and DDX5 are potential targets for the therapeutics of NB.
Materials and Methods
Cell Lines
Human nontransformed mammary epithelial MCF 10A (CRL-10317) cells, NB cell lines SK-N-BE(2) (CRL-2271), IMR32 (CCL-127), BE(2)-C (CRL-2268), NB-1643, NB-1691, SH-SY5Y (CRL-2266), SK-N-SH (HTB-11), and SK-N-AS (CRL-2137) and colon cancer HCT116 (CCL-247) cells were purchased from American Type Culture Collection (Rockville, MD, USA) and Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). Cells were authenticated by short tandem repeat profiling and used within 6 months after resuscitation. Mycoplasma contamination was regularly examined using the Lookout Mycoplasma PCR Detection Kit (Sigma, St. Louis, MO, USA). Cells were grown in RPMI-1640 medium, supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), and incubated with cycloheximide (CHX) or MG132 (Sigma).
Northern Blotting
Northern blotting was performed, as previously described,35 using a 268-bp probe prepared by the PCR DIG Probe Synthesis Kit (Roche, Indianapolis, IN, USA) and primer sets (Table S7). For northern blot, 20 μg of total RNA was separated on 3-(N-morpholino) propanesulfonic acid-buffered 2% (w/v) agarose gel containing 1.2% (v/v) formaldehyde under denaturing conditions for 4 h at 80 V and transferred to Hybond-N+ membrane (Pall, Port Washington, NY, USA). Prehybridization was carried out at 65°C for 30 min in DIG Easy Hyb solution (Roche). Hybridization was performed at 65°C for 16–18 h. Blots were washed stringently, detected by anti-digoxigenin (DIG) antibody, and recorded on X-ray films with chemiluminescence substrate CSPD (Roche).
Access and Analysis of Public Data
Public datasets of RNA-seq, microarray, and copy number alteration were downloaded from GEO, TCGA, or cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Data processing and quantile normalization were performed using R software (version 3.4.1). The DESeq2 package was used to compare gene expression in different tumor tissues characterized by clinical features using default parameters. Statistical significance of differential gene expression was indicated by log2 values of fold change (greater than 1 or less than −1) and p value (<0.05). Then, overlapping analysis was performed to identify genes consistently associated with clinical features of tumors, while their expression correlation and survival significance were also assessed.
GSEA Assay
GSEA was undertaken, as described previously.36 The gene sets from publication were applied as indicated. Datasets were prepared from either the public RNA-seq (GEO: GSE62564) assay or NHEG1 overexpression experiments.
RACE Assay
Total RNA was isolated from BE(2)-C cells to prepare RACE-ready cDNA using the SMARTer RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA), which was further amplified by PCR primers and nested PCR primers (Table S7).
RNA-FISH
Biotin-labeled NHEG1 probes were transcribed and purified by using the T7 RNA polymerase, Biotin RNA Labeling Mix (Roche), and RNeasy Mini Kit (QIAGEN, Redwood City, CA, USA). Cells were seeded on coverslips and fixed with 4% paraformaldehyde. Hybridization was undertaken in a humidified chamber at 37°C for 16 h, with or without RNase A (20 μg) treatment. Cells were treated by streptavidin-conjugated Cy3, with nuclei staining by 4′,6-diamidino-2-phenylindole (DAPI).
Gene Overexpression and Knockdown
Full-length and truncations of NHEG1 cDNA (1360 bp) were obtained from NB tissues (Table S8) and inserted into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). Human β-catenin and MYCN expression vectors were kind gifts from Dr. Ralf Janknecht29 and Dr. Arturo Sala.37 Human LEF1 cDNA (1,200 bp) and TCF7L2 cDNA (1,809 bp; Genechem, Shanghai, China) were inserted into pcDNA3.1 and pCMV-3Tag-1A (Addgene, Cambridge, MA, USA), respectively (Table S8). Oligonucleotides encoding shRNAs against NHEG1, DDX5, or β-catenin (Table S8) were inserted into GV102 (Genechem). Stable cells were established by treatment with puromycin or neomycin (Invitrogen).
Microarray and Gene Ontology Assays
High-throughput mRNA microarray analysis was applied for determining gene expression, using Affymetrix Human Transcriptome Array 2.0 (Shanghai Technology, Shanghai, China). Raw data were deposited in the Gene Expression Omnibus database (GEO: GSE80393).
Quantitative Real-Time RT-PCR
The RNA Subcellular Isolation Kit (Active Motif, Carlsbad, CA, USA) or RNeasy Mini Kit (QIAGEN) was used to prepare nuclear, cytoplasmic, or total RNA. The Transcriptor First Strand cDNA Synthesis Kit (Roche), SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), and primers (Table S7) were applied for reverse transcription reactions and quantitative real-time RT-PCR. The transcript levels were normalized to those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and analyzed by the 2−ΔΔCt method.
Western Blotting
Cytoplasmic or nuclear fractions were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific). Protein was prepared using 1 times cell lysis buffer (Promega, Madison, WI, USA). Western blotting was carried our as described previously,5,38, 39, 40 using specific antibodies for LEF1 (ab53293), TCF7L2 (ab76151), DDX5 (ab126730), p-DDX5 (Y593; ab62255), β-catenin (ab32572), HA (ab9110), and ubiquitin (Ub; ab7254; Abcam) and GST (sc-53909), histone H3 (sc-24516), and GAPDH (sc-365062; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
CoIP Assay
Co-immunoprecipitation (coIP) was performed as described previously,41 using specific antibodies against DDX5 (ab126730) or β-catenin (ab32572; Abcam). Bead-bound proteins were released and analyzed by western blot analysis.
Fluorescence Immunocytochemical Staining
Cells were plated on coverslips, fixed with a solution of 95% ethanol and 5% glacial acetic acid, permeabilized with 0.3% Triton X-100, and blocked with 5% milk for 1 h. Cells were incubated at 4°C overnight with antibodies specific for DDX5 (ab126730; 1:100 dilution), β-catenin (ab32572; 1:200 dilution), and GAP43 (ab75810; Abcam; 1:100 dilution) or NF-200 (sc-32729; Santa Cruz Biotechnology; 1:100 dilution) and treated with goat anti-rabbit immunoglobulin G (IgG; 1:500 dilution), with nuclei staining with DAPI (300 nM).5
Luciferase Reporter Assay
Human NHEG1 promoter (−1,000/+20) and its truncations were amplified by PCR using genomic DNA (Table S8) and inserted into pGL3-Basic (Promega). The mutating binding site of LEF1/TCF7L2 was established using the GeneTailor Site-Directed Mutagenesis System (Invitrogen) and PCR primers (Table S8). The TOP-FLASH and FOP-FLASH reporters were purchased from Millipore (Bedford, MA). Dual-luciferase assay was performed according to the manufacturer’s instructions (Promega). Luciferase activity was measured with a luminometer (Lumat LB9507; Berthold Technologies, Bad Wildbad, Germany).5,41, 42, 43, 44, 45
Rescue of Gene Expression
The truncations of HA-tagged DDX5 were generated using its cDNA template, provided by Dr. Ralf Janknecht,29 and PCR primers (Table S8). To restore downregulation or upregulation of DDX5 induced by NHEG1 knockdown or overexpression, stable cells were transfected by the DDX5 expression construct or shRNAs specific for DDX5 (Table S8).
Biotin-Labeled RNA Pull-Down and Mass Spectrometry Analysis
Biotin-labeled NHEG1 was in vitro transcribed, as mentioned above. Biotin-labeled RNA pull-down was performed as described previously.5 Briefly, nuclear protein was extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) and incubated with biotin-labeled RNAs and streptavidin agarose beads (Invitrogen). Retrieved protein was detected by western blot or mass spectrometry analysis (Wuhan Institute of Biotechnology, Wuhan, China).
Crosslinking RIP Assay
Cells were ultraviolet-light crosslinked at 254 nm (200 J/cm2) in phosphate buffer saline and collected by scraping. RIP assay was performed according to the instructions of the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore), using antibodies against DDX5 (ab126730 [Abcam] and sc-166167 [Santa Cruz Biotechnology]). Co-precipitated RNA was measured by RT-PCR with primer sets (Table S7). Total RNAs (input) and isotype antibody (IgG) were applied as controls.
In Vitro Binding Assay
The GST-tagged DDX5 proteins were established by preparing truncations of GST-tagged DDX5 using its cDNA template, provided by Dr. Ralf Janknecht,29 and PCR primer sets (Table S8) and transforming into E. coli,5 which were incubated with NHEG1 cRNA, prepared by in vitro transcription using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific). DDX5-RNA complexes were pulled down using GST beads (Sigma). The SDS-PAGE and western blot were applied in detection of protein, and RT-PCR, using primer sets (Table S7), was used to detect the levels of RNA.
RNA EMSA
The biotin-labeled NHEG1 probe was generated as mentioned above. RNA electrophoretic mobility shift assay (EMSA) was undertaken using the LightShift Chemiluminescent RNA EMSA Kit (Thermo Fisher Scientific) and recombinant DDX5 protein.
ChIP Assay
ChIP and quantitative real-time PCR were carried out using the EZ-ChIP kit (Upstate Biotechnology, Temecula, CA), SYBR Green PCR Master Mix (Applied Biosystems), and primer sets (Table S7). The amount of immunoprecipitated DNA was calculated in reference to a standard curve and normalized to input DNA, while DNA immunoprecipitated with an unspecific antibody (isotype IgG) served as a negative control.41,44,46,47
In Vitro Growth, Cell Cycle, and Invasion Assays
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma) colorimetric,41,43,48 soft agar,5,38,44,49,50 flow cytometric,39 and Transwell Matrigel invasion5,38, 39, 40,46,51, 52, 53 assays were applied for detecting the viability, growth, cell-cycle progression, and invasiveness of tumor cells.
In Vivo Growth, Metastasis, and Therapeutic Assays
All animal studies were approved by the Animal Care Committee of Tongji Medical College (approval number: Y20080290) and carried out in accordance with NIH Guidelines for the Care and Use of Laboratory Animals. For in vivo tumor growth studies, blindly randomized 4-week-old female BALB/c nude mice (n = 5 per group) were injected subcutaneously in the dorsal flanks with 1 × 106 tumor cells. 1 Month later, mice were sacrificed and examined for tumor weight and corresponding indicators. Experimental metastasis studies were performed by tail-vein injection of tumor cells (0.4 × 106 per mouse, n = 5 per group) into blindly randomized 4-week-old female BALB/c nude mice, as previously described.5,38,44,52 For in vivo therapeutic studies, tumor cells (1 × 106) were injected subcutaneously into the dorsal flanks of nude mice. 2 Weeks after tumor cell inoculation with confirmation of successful maturation of tumors, mice were blindly randomized and treated by intravenous injection of 21 nucleotide-synthesized siRNAs (Table S8; RiboBio) using Invivofectamine 3.0 Reagent (Life Technologies) as indicated. The tumor volume, body weight, and survival time of each mouse were monitored and recorded.
Patient Tissue Samples
Human tissue study was approved by the Institutional Review Board of Tongji Medical College (approval number: 2011-S085) and carried out in accordance with the guidelines set forth by the Declaration of Helsinki. Written, informed consents were obtained from all legal guardians of patients. All cases received no preoperative chemotherapy or other treatment. Human normal dorsal root ganglia or embryonic tissues (at day 50 of gestation) were, respectively, collected from interrupted pregnancies and therapeutic abortion. All fresh specimens were frozen in liquid nitrogen, validated by pathological diagnosis, and stored at −80°C until use.
Immunohistochemistry
Immunohistochemistry was undertaken, as described previously,5,38,44,52 using antibodies against Ki-67 (ab92742; Abcam; 1:100 dilution), DDX5 (ab126730; Abcam; 1:100 dilution), or β-catenin (sc-59737; Santa Cruz Biotechnology; 1:200 dilution). The staining intensity was evaluated on a range from 0 to 3 (0, negative; 1, weakly positive; 2, moderately positive; 3, strongly positive), whereas percentage of positive cells was evaluated ranging from 0 to 4 (0, negative; 1, positive in 1%–25%; 2, positive in 26%–50%; 3, positive in 51%–75%; 4, positive in 76%–100%). Based on the products of staining intensity, multiplied by percentage of positive cells, the results of immunohistochemistry were classified into negative (−, 0–1), mildly (+, 2–3), moderately (++, 4–8), and strongly positive (+++, 9–12). The moderate (++) or strong (+++) reactivity was defined as high expression, whereas negative (−) or mild-positive (+) reactivity was defined as low expression.
Data Availability
Microarray data supporting the results of this study have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/), under GEO: GSE80393. The public datasets are available from the GEO (GEO: GSE62564, GSE31684, GSE20685, GSE17536, GSE17679, GSE7696, GSE31210, GSE10846, and GSE18520) or TCGA database. All remaining data are presented within the article and Supplemental information and available from the corresponding author upon request.
Statistical Analysis
All results were presented as mean ± standard error of the mean (SEM). Cutoff values were defined by average levels of gene expression. The significant difference of tumor tissues or cell lines was determined by analysis of variance (ANOVA), χ2 analysis, and Student’s t test. The significance of overlap analysis was evaluated by Fisher’s exact test. Gene expression relationship was analyzed by Pearson’s correlation coefficient method. Survival difference was assessed by log-rank test and Cox regression models. All statistical tests were two sided and considered statistically significant when p values were less than 0.05.
Author Contributions
X.Z., D.L., and F.Y. conceived and performed most of the experiments. H. Lian, J.W., X.W., E.F., H.S., and A.H. accomplished some of the in vitro experiments. Y.G., Y.L., H. Li., and Y.C. accomplished the in vivo studies. F.Y. and E.F. undertook the mining of publicly available datasets. K.H. critically reviewed the manuscript. Q.T. and L.Z. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We are grateful for Drs. Ralf Janknecht and Arturo Sala in providing vectors. This work was granted by the National Natural Science Foundation of China (81272779, 81372667, 81472363, 81402301, 81402408, 81572423, 81672500, 81773094, 81772967, 81874085, 81874066, 81802925, 81903011, and 81903008); Fundamental Research Funds for the Central Universities of China (2019kfyRCPY032, 2012QN224, 2013ZHYX003, 01-18-530112, and 01-18-530115); and Natural Science Foundation of Hubei Province of China (2014CFA012).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.12.013.
Contributor Information
Liduan Zheng, Email: ld_zheng@hotmail.com.
Qiangsong Tong, Email: qs_tong@hotmail.com.
Supplemental Information
References
- 1.Westermann F., Schwab M. Genetic parameters of neuroblastomas. Cancer Lett. 2002;184:127–147. doi: 10.1016/s0304-3835(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 2.Barnhill L.M., Williams R.T., Cohen O., Kim Y., Batova A., Mielke J.A., Messer K., Pu M., Bao L., Yu A.L., Diccianni M.B. High expression of CAI2, a 9p21-embedded long noncoding RNA, contributes to advanced-stage neuroblastoma. Cancer Res. 2014;74:3753–3763. doi: 10.1158/0008-5472.CAN-13-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu M., Ohira M., Li Y., Niizuma H., Oo M.L., Zhu Y., Ozaki T., Isogai E., Nakamura Y., Koda T. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. Int. J. Oncol. 2009;34:931–938. doi: 10.3892/ijo_00000219. [DOI] [PubMed] [Google Scholar]
- 4.Liu P.Y., Erriquez D., Marshall G.M., Tee A.E., Polly P., Wong M., Liu B., Bell J.L., Zhang X.D., Milazzo G. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J. Natl. Cancer Inst. 2014;106:dju113. doi: 10.1093/jnci/dju113. [DOI] [PubMed] [Google Scholar]
- 5.Zhao X., Li D., Pu J., Mei H., Yang D., Xiang X., Qu H., Huang K., Zheng L., Tong Q. CTCF cooperates with noncoding RNA MYCNOS to promote neuroblastoma progression through facilitating MYCN expression. Oncogene. 2016;35:3565–3576. doi: 10.1038/onc.2015.422. [DOI] [PubMed] [Google Scholar]
- 6.Pandey G.K., Mitra S., Subhash S., Hertwig F., Kanduri M., Mishra K., Fransson S., Ganeshram A., Mondal T., Bandaru S. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 7.Su Z., Fang H., Hong H., Shi L., Zhang W., Zhang W., Zhang Y., Dong Z., Lancashire L.J., Bessarabova M. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z., Luo Z., Zhou L., Li X., Jiang T., Fu E. DDX5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating β-catenin signaling pathway. Cancer Sci. 2015;106:1303–1312. doi: 10.1111/cas.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., Wang X., Mei H., Fang E., Ye L., Song H., Yang F., Li H., Huang K., Zheng L., Tong Q. Long Noncoding RNA pancEts-1 Promotes Neuroblastoma Progression through hnRNPK-Mediated β-Catenin Stabilization. Cancer Res. 2018;78:1169–1183. doi: 10.1158/0008-5472.CAN-17-2295. [DOI] [PubMed] [Google Scholar]
- 12.Depuydt P., Boeva V., Hocking T.D., Cannoodt R., Ambros I.M., Ambros P.F., Asgharzadeh S., Attiyeh E.F., Combaret V., Defferrari R. Genomic amplifications and distal 6q loss: novel markers for poor survival in high-risk neuroblastoma patients. J. Natl. Cancer Inst. 2018;110:1084–1093. doi: 10.1093/jnci/djy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morin P.J. β-catenin signaling and cancer. BioEssays. 1999;21:1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Liu X., Mazanek P., Dam V., Wang Q., Zhao H., Guo R., Jagannathan J., Cnaan A., Maris J.M., Hogarty M.D. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene. 2008;27:1478–1488. doi: 10.1038/sj.onc.1210769. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Zhou B., Liu Y., Chen K., Bao P., Wang Y., Wang J., Zhou Z., Sun X., Li Y. Wnt inhibitory factor-1 functions as a tumor suppressor through modulating Wnt/β-catenin signaling in neuroblastoma. Cancer Lett. 2014;348:12–19. doi: 10.1016/j.canlet.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Tringali C., Cirillo F., Lamorte G., Papini N., Anastasia L., Lupo B., Silvestri I., Tettamanti G., Venerando B. NEU4L sialidase overexpression promotes β-catenin signaling in neuroblastoma cells, enhancing stem-like malignant cell growth. Int. J. Cancer. 2012;131:1768–1778. doi: 10.1002/ijc.27450. [DOI] [PubMed] [Google Scholar]
- 17.Jansen S.R., Holman R., Hedemann I., Frankes E., Elzinga C.R.S., Timens W., Gosens R., de Bont E.S., Schmidt M. Prostaglandin E2 promotes MYCN non-amplified neuroblastoma cell survival via β-catenin stabilization. J. Cell. Mol. Med. 2015;19:210–226. doi: 10.1111/jcmm.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alshareef A., Gupta N., Zhang H.F., Wu C., Haque M., Lai R. High expression of β-catenin contributes to the crizotinib resistant phenotype in the stem-like cell population in neuroblastoma. Sci. Rep. 2017;7:16863. doi: 10.1038/s41598-017-17319-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cadigan K.M., Waterman M.L. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb. Perspect. Biol. 2012;4:a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuller-Pace F.V., Moore H.C. RNA helicases p68 and p72: multifunctional proteins with important implications for cancer development. Future Oncol. 2011;7:239–251. doi: 10.2217/fon.11.1. [DOI] [PubMed] [Google Scholar]
- 21.Mazurek A., Luo W., Krasnitz A., Hicks J., Powers R.S., Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer Discov. 2012;2:812–825. doi: 10.1158/2159-8290.CD-12-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., Gao X., Yang J.J., Liu Z.R. Interaction between p68 RNA helicase and Ca2+-calmodulin promotes cell migration and metastasis. Nat. Commun. 2013;4:1354. doi: 10.1038/ncomms2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang T., Abou-Ouf H., Hegazy S.A., Alshalalfa M., Stoletov K., Lewis J., Donnelly B., Bismar T.A. Ankyrin G expression is associated with androgen receptor stability, invasiveness, and lethal outcome in prostate cancer patients. J. Mol. Med. (Berl.) 2016;94:1411–1422. doi: 10.1007/s00109-016-1458-4. [DOI] [PubMed] [Google Scholar]
- 24.Ma C.I., Martin C., Ma Z., Hafiane A., Dai M., Lebrun J.J., Kiss R.S. Engulfment protein GULP is regulator of transforming growth factor-β response in ovarian cells. J. Biol. Chem. 2012;287:20636–20651. doi: 10.1074/jbc.M111.314997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slack A.D., Chen Z., Ludwig A.D., Hicks J., Shohet J.M. MYCN-directed centrosome amplification requires MDM2-mediated suppression of p53 activity in neuroblastoma cells. Cancer Res. 2007;67:2448–2455. doi: 10.1158/0008-5472.CAN-06-1661. [DOI] [PubMed] [Google Scholar]
- 26.Guo J., Jia J., Jia R. PTBP1 and PTBP2 impaired autoregulation of SRSF3 in cancer cells. Sci. Rep. 2015;5:14548. doi: 10.1038/srep14548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng S., Agoulnik I.U., Bogatcheva N.V., Kamat A.A., Kwabi-Addo B., Li R., Ayala G., Ittmann M.M., Agoulnik A.I. Relaxin promotes prostate cancer progression. Clin. Cancer Res. 2007;13:1695–1702. doi: 10.1158/1078-0432.CCR-06-2492. [DOI] [PubMed] [Google Scholar]
- 28.Yang L., Lin C., Liu Z.R. P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 29.Shin S., Rossow K.L., Grande J.P., Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67:7572–7578. doi: 10.1158/0008-5472.CAN-06-4652. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Xing Z., Mani S.K.K., Bancel B., Durantel D., Zoulim F., Tran E.J., Merle P., Andrisani O. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64:1033–1048. doi: 10.1002/hep.28698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciechanover A., Orian A., Schwartz A.L. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 32.Causevic M., Hislop R.G., Kernohan N.M., Carey F.A., Kay R.A., Steele R.J., Fuller-Pace F.V. Overexpression and poly-ubiquitylation of the DEAD-box RNA helicase p68 in colorectal tumours. Oncogene. 2001;20:7734–7743. doi: 10.1038/sj.onc.1204976. [DOI] [PubMed] [Google Scholar]
- 33.Yapijakis C., Bramos A., Nixon A.M., Ragos V., Vairaktaris E. The interplay between hemostasis and malignancy: the oral cancer paradigm. Anticancer Res. 2012;32:1791–1800. [PubMed] [Google Scholar]
- 34.Jazurek M., Ciesiolka A., Starega-Roslan J., Bilinska K., Krzyzosiak W.J. Identifying proteins that bind to specific RNAs - focus on simple repeat expansion diseases. Nucleic Acids Res. 2016;44:9050–9070. doi: 10.1093/nar/gkw803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu C., Tong Q., Zheng L., Liang Z., Pu J., Mei H., Hu T., Du Z., Tian F., Zeng F. TSEG-1, a novel member of histone H2A variants, participates in spermatogenesis via promoting apoptosis of spermatogenic cells. Genomics. 2010;95:278–289. doi: 10.1016/j.ygeno.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chayka O., Corvetta D., Dews M., Caccamo A.E., Piotrowska I., Santilli G., Gibson S., Sebire N.J., Himoudi N., Hogarty M.D. Clusterin, a haploinsufficient tumor suppressor gene in neuroblastomas. J. Natl. Cancer Inst. 2009;101:663–677. doi: 10.1093/jnci/djp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., Pu J., Qi T., Qi M., Yang C., Li S., Huang K., Zheng L., Tong Q. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene. 2014;33:387–397. doi: 10.1038/onc.2012.574. [DOI] [PubMed] [Google Scholar]
- 39.Zheng L., Qi T., Yang D., Qi M., Li D., Xiang X., Huang K., Tong Q. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS ONE. 2013;8:e55719. doi: 10.1371/journal.pone.0055719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng L., Pu J., Qi T., Qi M., Li D., Xiang X., Huang K., Tong Q. miRNA-145 targets v-ets erythroblastosis virus E26 oncogene homolog 1 to suppress the invasion, metastasis, and angiogenesis of gastric cancer cells. Mol. Cancer Res. 2013;11:182–193. doi: 10.1158/1541-7786.MCR-12-0534. [DOI] [PubMed] [Google Scholar]
- 41.Jiang G., Zheng L., Pu J., Mei H., Zhao J., Huang K., Zeng F., Tong Q. Small RNAs targeting transcription start site induce heparanase silencing through interference with transcription initiation in human cancer cells. PLoS ONE. 2012;7:e31379. doi: 10.1371/journal.pone.0031379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D., Zhao X., Xiao Y., Mei H., Pu J., Xiang X., Jiao W., Song H., Qu H., Huang K. Intelectin 1 suppresses tumor progression and is associated with improved survival in gastric cancer. Oncotarget. 2015;6:16168–16182. doi: 10.18632/oncotarget.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li D., Mei H., Pu J., Xiang X., Zhao X., Qu H., Huang K., Zheng L., Tong Q. Intelectin 1 suppresses the growth, invasion and metastasis of neuroblastoma cells through up-regulation of N-myc downstream regulated gene 2. Mol. Cancer. 2015;14:47. doi: 10.1186/s12943-015-0320-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D., Mei H., Qi M., Yang D., Zhao X., Xiang X., Pu J., Huang K., Zheng L., Tong Q. FOXD3 is a novel tumor suppressor that affects growth, invasion, metastasis and angiogenesis of neuroblastoma. Oncotarget. 2013;4:2021–2044. doi: 10.18632/oncotarget.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang X., Zhao X., Qu H., Li D., Yang D., Pu J., Mei H., Zhao J., Huang K., Zheng L., Tong Q. Hepatocyte nuclear factor 4 alpha promotes the invasion, metastasis and angiogenesis of neuroblastoma cells via targeting matrix metalloproteinase 14. Cancer Lett. 2015;359:187–197. doi: 10.1016/j.canlet.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Zheng L., Li D., Xiang X., Tong L., Qi M., Pu J., Huang K., Tong Q. Methyl jasmonate abolishes the migration, invasion and angiogenesis of gastric cancer cells through down-regulation of matrix metalloproteinase 14. BMC Cancer. 2013;13:74. doi: 10.1186/1471-2407-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gade P., Kalvakolanu D.V. Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity. Methods Mol. Biol. 2012;809:85–104. doi: 10.1007/978-1-61779-376-9_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng L., Jiao W., Song H., Qu H., Li D., Mei H., Chen Y., Yang F., Li H., Huang K., Tong Q. miRNA-558 promotes gastric cancer progression through attenuating Smad4-mediated repression of heparanase expression. Cell Death Dis. 2016;7:e2382. doi: 10.1038/cddis.2016.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D., Song H., Mei H., Fang E., Wang X., Yang F., Li H., Chen Y., Huang K., Zheng L., Tong Q. Armadillo repeat containing 12 promotes neuroblastoma progression through interaction with retinoblastoma binding protein 4. Nat. Commun. 2018;9:2829. doi: 10.1038/s41467-018-05286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiao W., Chen Y., Song H., Li D., Mei H., Yang F., Fang E., Wang X., Huang K., Zheng L., Tong Q. HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 2018;37:2728–2745. doi: 10.1038/s41388-018-0128-0. [DOI] [PubMed] [Google Scholar]
- 51.Marshall J. Transwell(®) invasion assays. Methods Mol. Biol. 2011;769:97–110. doi: 10.1007/978-1-61779-207-6_8. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H., Qi M., Li S., Qi T., Mei H., Huang K., Zheng L., Tong Q. microRNA-9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol. Cancer Ther. 2012;11:1454–1466. doi: 10.1158/1535-7163.MCT-12-0001. [DOI] [PubMed] [Google Scholar]
- 53.Zheng L., Jiang G., Mei H., Pu J., Dong J., Hou X., Tong Q. Small RNA interference-mediated gene silencing of heparanase abolishes the invasion, metastasis and angiogenesis of gastric cancer cells. BMC Cancer. 2010;10:33. doi: 10.1186/1471-2407-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data supporting the results of this study have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/), under GEO: GSE80393. The public datasets are available from the GEO (GEO: GSE62564, GSE31684, GSE20685, GSE17536, GSE17679, GSE7696, GSE31210, GSE10846, and GSE18520) or TCGA database. All remaining data are presented within the article and Supplemental information and available from the corresponding author upon request.