Abstract
Age-related macular degeneration (AMD) is a universal leading cause for irreversible blindness in the elderly population. Dedifferentiation of retinal pigment epithelium (RPE) cells initiates early pathological events in atrophic AMD. Herein, we aim to investigate effects of a circular RNA derived from the NR3C1 gene (circNR3C1) on regulating RPE function and AMD pathogenesis. circNR3C1 expression was consistently upregulated along with RPE differentiation and was downregulated in dysfunctional RPE and blood serum of AMD patients. Silencing of circNR3C1 reduced RPE characteristic transcripts and proteins, interrupted phagocytosis, accelerated intracellular reactive oxygen species (ROS) generation, and promoted RPE proliferation in vitro. circN3C1 silencing also decreased expressions of RPE characteristic markers and disturbed the ultrastructure of RPE in vivo, as shown by a thickened RPE with twisted basal infoldings and outer segments. Mechanistically, circNR3C1 acted as an endogenous microRNA-382-5p (miR-382-5p) sponge to sequester its activity, which increased phosphatase and tensin homolog on chromosome 10 (PTEN) expression and inhibited the protein kinase B/mammalian target of rapamycin (AKT/mTOR) pathway. miR-382-5p overexpression and PTEN silencing mimicked effects of circNR3C1 silencing on RPE phenotypes in vivo and in vitro. In conclusion, circNR3C1 prevents AMD progression and protects RPE by directly sponging miR-382-5p to block its interaction with PTEN and subsequently blocks the AKT/mTOR pathway. Pharmacological circNR3C1 supplementations are promising therapeutic options for atrophic AMD.
Keywords: circular RNA, age-related macular degeneration, retinal pigment epithelium, circNR3C1, miR-382-5p, PTEN, AKT/mTOR
Age-related macular degeneration (AMD) causes irreversible blindness in elderly population; however, roles of circular RNAs in AMD pathogenesis are not reported. Now in Molecular Therapy, Chen et al. (2019) show that circNR3C1 prevents AMD progression by sponging miR-382-5p to block its interaction with PTEN and subsequently blocks the AKT/mTOR pathway.
Introduction
Age-related macular degeneration (AMD) is a progressive disease affecting central retina and a universal leading cause for irreversible blindness in people aged over 55.1, 2, 3 Based on the presence or absence of choroidal vessels that disruptively invade the retina, the late stages of AMD, in which most vision loss occurs, can be categorized into the exudative and atrophic forms. Exudative AMD is characterized by choroidal neovascularization, and atrophic AMD is typified by atrophy of the retinal pigment epithelium (RPE), choriocapillaries, and photoreceptors.3 A detailed understanding of the molecular mechanisms of exudative AMD has led to several anti-vascular endothelial growth factor therapies, while no approved treatment has been developed for atrophic AMD. We have previously revealed that dedifferentiation of RPE cells is an early consequence of atrophic AMD.4, 5, 6, 7 As a monolayer of cuboidal, polarized, and pigmented cells, RPE is located in the outer retina between photoreceptors and choroidal blood vessels and forms a part of the blood/retina barrier.8, 9, 10 RPE is crucial in maintaining regular retinal functions, including absorbing light energy, transporting nutrients and metabolic end products, and secreting multiple growth factors.8 Elucidation of initiating events causing RPE abnormalities, especially RPE dedifferentiation, could help with the development of clinical preventions and therapies for atrophic AMD. However, the precise mechanism underlying RPE dedifferentiation is not clear yet.
Circular RNAs (circRNAs) have emerged as a novel group of noncoding RNAs presenting covalently connected loop structures without 5′ to 3′ polarity or a polyadenylated tail.11 They could derive from exons, introns, or both exons and introns of their parental genes.12,13 circRNAs are involved in many biological processes, like angiogenesis and tumorigenesis.14,15 They regulate gene expressions by acting as microRNA (miRNA) sponges,16 protein sponges,17,18 and nuclear transcriptional regulators.19,20 Evidence suggests that circRNAs are aberrantly expressed in several retinopathies, such as proliferative vitreoretinopathy and diabetic retinopathy.21, 22, 23 However, the role of circRNAs in atrophic AMD is still poorly understood. In this study, we investigate the expression and regulation of circNR3C1, a circRNA originating from the nuclear receptor subfamily 3 (NR3C1; OMIM: 138040) gene, in RPE cells and AMD pathogenesis. Our data suggest that circNR3C1 prevents AMD progression and protects RPE features by acting as miRNA sponges in vivo and in vitro.
Results
Identification of the Circular Structure and Clinical Features of circNR3C1
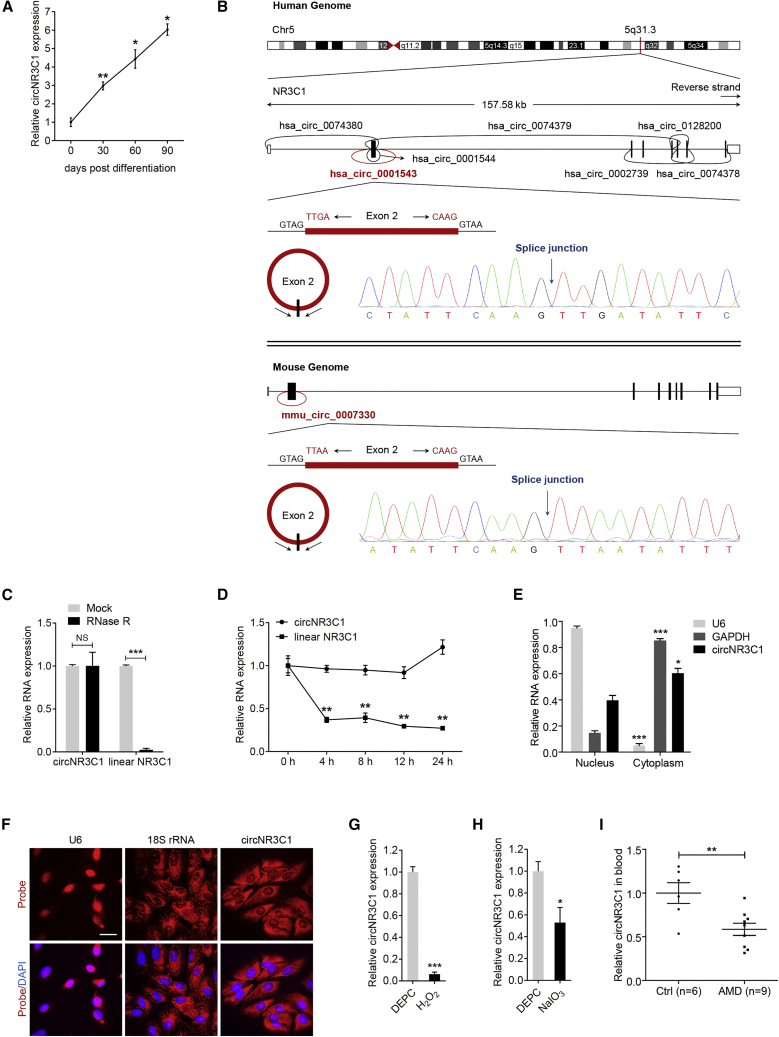
Many circRNAs have been found to be expressed in RPE.21 We have previously reported that dedifferentiation of RPE cells, typified by downregulation of RPE-specific proteins, is an early consequence of AMD.4 Herein, we applied a microarray to reveal circRNAs that consistently changed along with RPE differentiation (data not shown). We identified that hsa_circ_0001543, transcribed by the NR3C1 gene, was consistently upregulated along with RPE differentiation (Figure 1A). According to circBase (http://www.circbase.org/), NR3C1 host gene produces 7 circRNAs (hsa_circ_0074380, hsa_circ_0001543, hsa_circ_0001544, hsa_circ_0074379, hsa_circ_0002739, hsa_circ_0128200, and hsa_circ_0074378) in the human genome and one circRNA (mmu_circ_0007330) in the mouse genome (Figure 1B). hsa_circ_0001543, consisting of the head-to-tail splicing of exon 2, is located at chr5:142779220 to 142780417 in the human genome, and mmu_circ_0007330 is located at chr18:39645651 to 39646899 in the mouse genome. Both isoforms are highly conserved (Figure S1). We focused on these two isoforms for further functional analyses and named them circNR3C1. Amplified products of human and mouse circNR3C1 were sent for sequencing. Sequencing results were completely in accordance with the sequences shown in circBase (Figure 1B).
Figure 1.
Identification of the Circular Structure and Clinical Features of circNR3C1
(A) Quantitative real-time PCR assay was utilized to reveal circNR3C1 expression in hiPSC and hiPSC-RPE at 30, 60, and 90 days post differentiation (one-way ANOVA, Bonferroni’s test). (B) The head-to-tail splicing of circNR3C1 in human (upper) and mouse (below) genome were confirmed by Sanger sequencing, which was consistent with the sequence reported in CircBase database. (C) circNR3C1 and linear NR3C1 expressions were detected by quantitative real-time PCR in total RNAs in the presence or absence of RNase R supplementation (two-tailed Student’s t test). (D) Quantitative real-time PCR was applied to determine expressions of circNR3C1 and linear NR3C1 in ARPE-19 cells after actinomycin D treatment (one-way ANOVA, Bonferroni’s test). (E) Expressions of nuclear control transcript (U6), cytoplasm control transcript (GAPDH), and circNR3C1 were detected by quantitative real-time PCR in the nuclear and cytoplasm fractions of ARPE-19 cells (one-way ANOVA, Bonferroni’s test). (F) RNA-FISH assays were performed to reveal the expression pattern of circNR3C1 in ARPE-19 cells using Cy3-labeled antisense probes (U6, 18S rRNA, and circNR3C1). Scale bar represents 20 μm. (G) circNR3C1 expression was monitored in ARPE-19 cells incubated with culture medium containing H2O2 (100 μM) for 24 h by quantitative real-time PCR. DEPC, diethylpyrocarbonate (two-tailed Student’s t test). (H) We used quantitative real-time PCR to determine circNR3C1 expression in ARPE-19 cells maintained in culture medium supplemented with NaIO3 (1.25 mmol/L) for 96 h (two-tailed Student’s t test). DEPC, diethylpyrocarbonate. (I) Quantitative real-time PCR was utilized to show circNR3C1 expressions in blood serum of AMD patients (n = 9) and non-AMD controls (n = 6; two-tailed Student’s t test). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
circRNAs are expected to be resistant to ribonuclease R (RNase R), while linear RNA will degrade upon RNase R treatment.24 Our data revealed that circNR3C1, rather than linear NR3C1 mRNA, could resist digestion by RNase R (Figure 1C). We further examined circNR3C1 stability in human retinal pigmented epithelial cells (ARPE-19) cells under treatment with actinomycin D, a transcription inhibitor. We found that circNR3C1 was highly stable with a half-life of over 24 h (Figure 1D). However, the linear transcript was easily degraded with a half-life of less than 4 h (Figure 1D). Quantitative real-time PCR analysis of nuclear and cytoplasmic RNA and fluorescent in situ hybridization (FISH) analysis indicated that circNR3C1 was mainly expressed in the cytoplasm of RPE cells (Figures 1E and 1F).
We next analyzed whether circNR3C1 expression was altered under abnormal RPE conditions and in AMD patients. Our data revealed that circNR3C1 expression was significantly reduced in RPE cells treated with H2O2 or sodium iodate (NaIO3) when compared to the control group treated with diethylpyrocarbonate (DEPC) (Figures 1G and 1H). Furthermore, we found that circNR3C1 expression was downregulated in the blood serum of AMD patients than that in non-AMD controls (Figure 1I).
circNR3C1 Protects RPE Function In Vitro
We next performed transfection assays to modulate expression levels of circNR3C1 in RPE cells to investigate whether it participates in mediating RPE features. Endogenous circNR3C1 expression was downregulated in ARPE-19 cells transfected with circNR3C1-small interfering RNA (siRNA; Figure S2A), while mRNA levels of linear NR3C1 was not affected (Figure S2B). Three pairs of siRNA oligos targeting circNR3C1 were initially designed and tested, and siRNA-1 with best efficiency was selected for further analyses (Figures S2A and S2B).
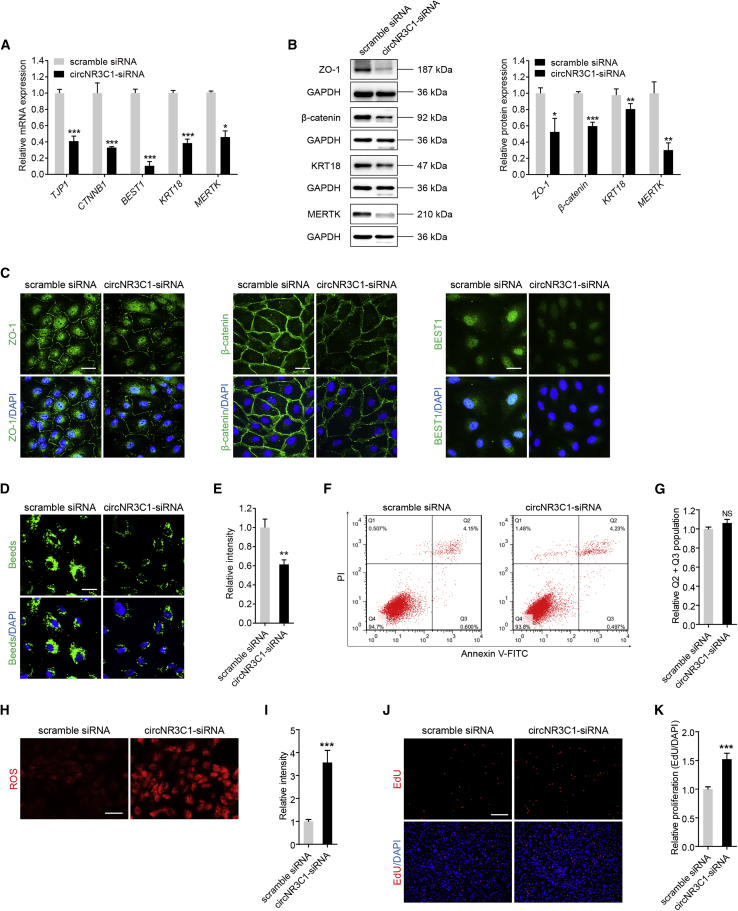
We initially monitored mRNA expression levels of RPE characteristic markers, including tight junction protein ZO-1 (TJP1; NM_003257), catenin beta-1 (CTNNB1; NM_0010904), bestrophin-1 (BEST1; NM_001139443), cytokeratin-18 (KRT18; NM_000224), and tyrosine-protein kinase Mer (MERTK; NM_006343), by quantitative real-time PCR. In ARPE-19 cells, decreased endogenous circNR3C1 expression downregulated expressions of those RPE markers (Figure 2A). Immunoblotting and immunofluorescent staining was then applied to test the intracellular expression and localization of ZO-1 (protein encoded by TJP1), β-catenin (protein encoded by CTNNB1), KRT18 (protein encoded by KRT18), MERTK (protein encoded by MERTK), and BEST1 (protein encoded by BEST1). Consistent with the mRNA data, decreased expression of all three proteins was revealed in cells transfected with circNR3C1-siRNA (Figures 2B and 2C). No obvious mislocalization of the three proteins was identified in all transfected groups.
Figure 2.
circNR3C1 Protects RPE Function In Vitro
(A) Quantitative real-time PCR was utilized to determine expressions of RPE-specific markers, including TJP1, CTNNB1, BEST1, KRT18, and MERTK, in ARPE-19 cells transfected with scramble siRNA or circNR3C1-siRNA (two-tailed Student’s t test). (B) Immunoblotting was used to show expression levels of RPE characteristic markers, including ZO-1, β-catenin, KRT18, and MERTK, in ARPE-19 cells transfected with scramble siRNA or circNR3C1-siRNA. Representative images along with the quantification results were shown (two-tailed Student’s t test). (C) We used immunofluorescence staining to visualize expression patterns of ZO-1, β-catenin, and BEST1 in ARPE-19 cells transfected with scramble siRNA or circNR3C1-siRNA. Scale bar represents 20 μm. (D and E) Phagocytic ability was tested using carboxylate-modified polystyrene latex beads with yellow-green fluorescence. Phagocytic ability was reduced in ARPE-19 cells transfected with circNR3C1-siRNA compared to cells transfected with scramble siRNA (two-tailed Student’s t test). Scale bar represents 20 μm. (F and G) Apoptosis rates were measured using flow cytometric analyses. No difference was detected between ARPE-19 cells transfected with circNR3C1-siRNA and scramble siRNA (two-tailed Student’s t test). (H and I) Mitochondrial ROS, reflected by red signals, were more accumulated in ARPE-19 cells transfected with circNR3C1-siRNA when compared to cells transfected with scramble siRNA (two-tailed Student’s t test). Scale bar represents 50 μm. (J and K) Proliferation of ARPE-19 cells transfected with scramble siRNA or circNR3C1-siRNA was analyzed by calculating the incorporation of EdU during DNA synthesis. Proliferation was promoted in ARPE-19 cells transfected with circNR3C1-siRNA compared to cells transfected with scramble siRNA (two-tailed Student’s t test). Scale bar represents 200 μm. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; NS, no significant difference.
A crucial function of RPE is the phagocytosis of daily shed photoreceptor outer segments, which is essential to maintain retinal homeostasis.25 Impairment of RPE phagocytic ability contributes to AMD pathogenesis.26, 27, 28 We next tested whether circNR3C1 affects RPE phagocytosis. Phagocytic ability was suppressed in ARPE-19 cells transfected with circNR3C1-siRNA when compared to cells transfected with scramble siRNA (Figures 2D and 2E). To further tell whether circNR3C1 has a direct effect on RPE phagocytosis independent of RPE cell death, we next used the Annexin-V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis assay to measure RPE apoptosis in different transfected groups. Our data revealed no detectable effect of circNR3C1 on RPE apoptosis (Figures 2F and 2G). Taken together, our results indicated that circNR3C1 promotes RPE phagocytosis.
Oxidative stress, which increases mitochondrial damage and generates reactive oxygen species (ROS), is a generally recognized risk factor for AMD.29,30 We thus tested whether circNR3C1 modulates oxidative stress of RPE cells. Based on our data, ROS generation was remarkably accelerated in ARPE-19 cells with circNR3C1 knocked down when compared to the control group (Figures 2H and 2I). Because dedifferentiation of post-mitotic tissues, including RPE cells, can be followed by cell proliferation,31 we further hypothesized that circNR3C1 could promote RPE proliferation. According to our result, 5-ethynyl-2′-deoxyuridine (EdU)-positive RPE cells increased in ARPE-19 cells transfected with circNR3C1-siRNA when compared with the control group (Figures 2J and 2K). Therefore, our results indicated that circNR3C1 reduced ROS generation in RPE and restrained RPE proliferation.
circNR3C1 Serves as a miRNA Sponge of miR-382-5p in RPE Cells
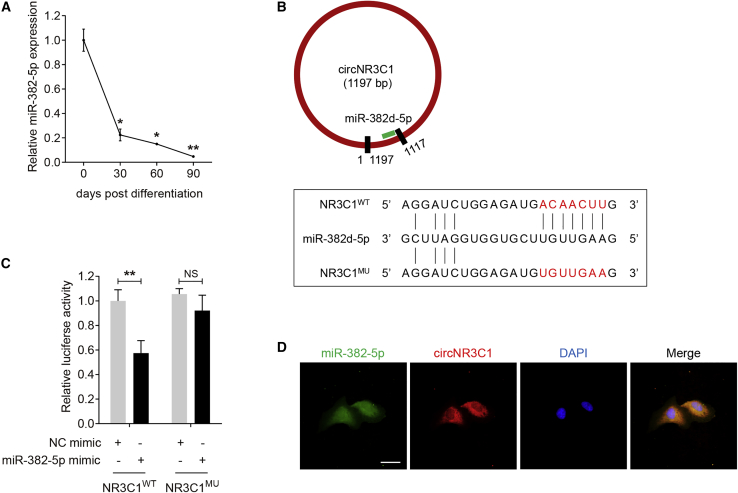
circRNAs with miRNA-binding sites might act as miRNA sponges, and circRNA-miRNAs-mRNA network is reported to participate in lots of biological pathways and disease etiologies.32,33 circNR3C1 was found mainly localized in the cytoplasm of RPE cells (Figures 1E and 1F), we therefore speculated that circNR3C1 might function as a miRNA sponge in RPE cells. Online prediction software Circular RNA Interactome (https://circinteractome.nia.nih.gov/) suggested miR-382-5p as a potential miRNA target of circNR3C1.34 miR-382-5p was highly conserved between human and murine genome (Figure S3). miR-382-5p expression was consistently downregulated along with RPE differentiation (Figure 3A), suggesting its potential inhibitory role on RPE function. The mutant NR3C1 (NR3C1MU) plasmid covered 7 mutated nucleotides located in the core binding region with miR-382-5p as displayed in Figure 3B. Luciferase activity was reduced in ARPE-19 cells co-transfected with wild-type NR3C1 (NR3C1WT) and miR-382-5p mimic compared to cells co-transfected with NR3C1MU and miR-382-5p mimic (Figure 3C). Introduction of the 7 mutated nucleotides abolished the ability of circNR3C1 to bind miR-382-5p. If circNR3C1 indeed bound with miR-382-5p in RPE cells, they should be co-expressed. Herein, FISH assay showed a large degree of overlap between circNR3C1 and miR-382-3p (Figure 3D). These results suggested that circNR3C1 served as a binding platform for miR-382-5p and might act as a miRNA sponge of miR-382-5p in RPE cells.
Figure 3.
circNR3C1 Serves as a miRNA Sponge of miR-382-5p in RPE Cells
(A) Quantitative real-time PCR assay was applied to reveal miR-382-5p expression in hiPSC and hiPSC-RPE at 30, 60, and 90 days post differentiation (one-way ANOVA, Bonferroni’s test). (B) Schematic diagram of the interaction between miR-382d-5p and circNR3C1. (C) Entire circNR3C1 sequence was synthesized, amplified, and inserted into the pGL3-Promoter Vector to generate the recombinant plasmids NR3C1WT and NR3C1MU. ARPE-19 cells were co-transfected with NR3C1WT/NR3C1MU with NC mimic/miR-382-5p mimic. Relative luciferase activity was measured using the dual luciferase assay (two-tailed Student’s t test). Renilla luciferase activity was taken as an indicator for transfection efficiency. (D) RNA-FISH assays were performed to reveal the expression pattern of miR-382-5p (FITC-labeled antisense probe) and circNR3C1 (Cy3-labeled antisense probes) in ARPE-19 cells. Scale bar represents 20 μm. *p < 0.05; **p < 0.01; NS, no significant difference.
circNR3C1-miR-382-5p-PTEN Network Moderates RPE Features In Vitro
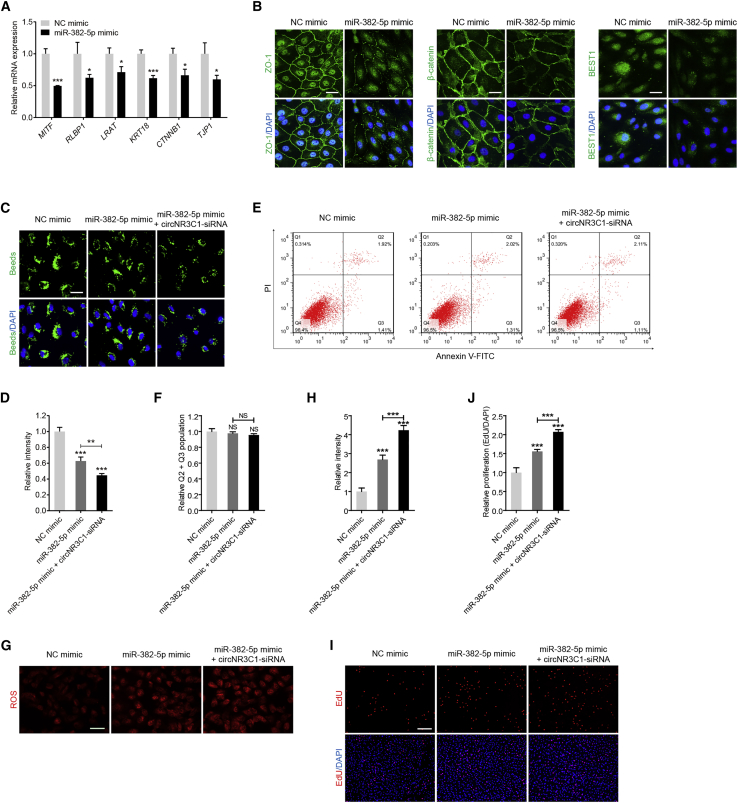
miR-382-5p mimic transfection in RPE cells downregulated mRNA expression levels of RPE characteristic markers, including microphthalmia-associated transcription factor (MITF; NM_198159), retinaldehyde binding protein 1 (RLBP1; NM_000326), lecithin retinol acyltransferase (LRAT; NM_004744), KRT18, CTNNB1, and TJP1 (Figure 4A). Consistent with quantitative real-time PCR results, immunofluorescent staining indicated that ZO-1, β-catenin, and BEST1 expression was reduced in cells overexpressing miR-382-5p (Figure 4B). In addition, miR-382-5p overexpression also decreased phagocytic ability of RPE cells (Figures 4C and 4D) without affecting their apoptosis rate (Figures 4E and 4F), accelerated ROS generation in RPE cells (Figures 4G and 4H), and promoted RPE proliferation (Figures 4I and 4J). The above findings indicated that overexpressing miR-382-5p mimicked the effects of circNR3C1 silencing in RPE cells.
Figure 4.
miR-382-5p Moderates RPE Features In Vitro
(A) mRNA levels of RPE-specific markers, including MITF, RLBP1, LRAT, KRT18, CTNNB1, and TJP1 were detected by quantitative real-time PCR in ARPE-19 cells transfected with NC mimic or miR-382-5p mimic (two-tailed Student’s t test). (B) Expression patterns of ZO-1, β-catenin, and BEST1 were observed in ARPE-19 cells transfected with NC mimic or miR-382-5p mimic using immunofluorescent staining. Scale bar represents 20 μm. (C and D) Phagocytic ability was disturbed in ARPE-19 cells transfected with miR-382-5p mimic compared to cells transfected with NC mimic, and was more reduced in cells co-transfected with mir-382-5p mimic and circNR3C1-siRNA (one-way ANOVA, Bonferroni’s test). Beads representing ability of phagocytosis were reflected by green signals. Scale bar represents 20 μm. (E and F) As revealed by flow cytometric analyses, no difference in apoptosis rates was detected among ARPE-19 cells transfected with NC mimic, miR-382-5p mimic, or mir-382-5p mimic together with circNR3C1-siRNA (one-way ANOVA, Bonferroni’s test). (G and H) Mitochondrial ROS were increased in ARPE-19 cells transfected with miR-382-5p mimic compared with cells transfected with NC mimic, and were more accumulated in cells co-transfected with mir-382-5p mimic together with circNR3C1-siRNA (one-way ANOVA, Bonferroni’s test). Scale bar represents 50 μm. (I and J) Proliferation of ARPE-19 cells was promoted in the miR-382-5p mimic transfected group when compared with the NC mimic transfected group, and was more promoted in cells co-transfected with mir-382-5p mimic and circNR3C1-siRNA (one-way ANOVA, Bonferroni’s test). Scale bar represents 200 μm. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; NS, no significant difference.
We next tested whether circNR3C1 knockdown and miR-382-5p overexpression at the same time would show double hit effects on RPE cells. According to our results, circNR3C1 silencing aggravated miR-382-5p-mediated suppressive effects on RPE cells. RPE phagocytosis was more disturbed (Figures 4C and 4D), intracellular ROS was more accumulated (Figures 4G and 4H), and RPE proliferation was more stimulated (Figures 4I and 4J) in cells co-transfected with circNR3C1 siRNA and miR-382-5p mimic when compared to cells transfected with miR-382-5p mimic alone.
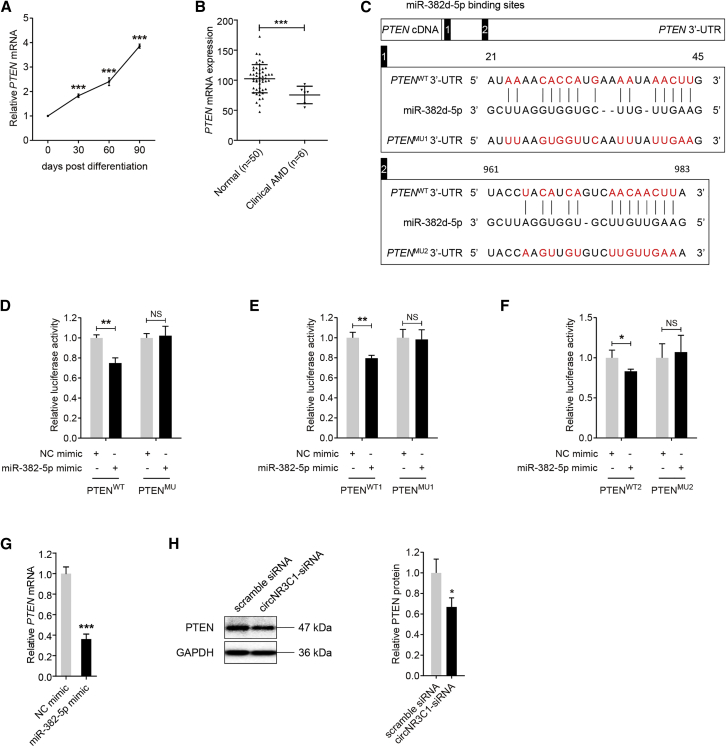
Phosphatase and tensin homolog on chromosome 10 (PTEN; NM_000314) was previously identified as a target gene of miR-382-5p in several cells, including human gastric cancer cells,35 human epithelial kidney cells,36 human hemangioma-derived endothelial cells,37 and mouse liver cells,38 but not in retinal cells. Inactivation of PTEN has also been reported to be involved in retinal degeneration.39 Herein, we found that PTEN mRNA expression was consistently downregulated along with RPE differentiation (Figure 5A). To further tell the role of PTEN in AMD pathogenesis, we compared its expression between macular RPE-choroid samples of 6 AMD patients and 50 healthy controls. Clinical diagnosis and personal particulars for each participant were detailed previously.40 We found that PTEN mRNA expression was decreased in the macular RPE-choroid tissue of patients diagnosed with clinical AMD when compared to the control group (Figure 5B), supporting its protective role in AMD etiology. We therefore tested whether PTEN is a target gene of miR-382-5p in RPE cells. We first investigated whether miR-382-5p directly binds to PTEN 3′ untranslated region (3′ UTR) by luciferase reporter assay. Two potential binding sites were revealed (Figure 5C). As shown in Figure 5C, the PTENMU plasmid covered 15 and 13 mutated nucleotides in the two binding spots, respectively. Reduction in luciferase activity was detected in ARPE-19 cells co-transfected with PTENWT and miR-382-5p compared to cells transfected with PTENMU and miR-382-5p, while introduction of mutated nucleotides abolished this reduction (Figure 5D). To further tell which one was the exact binding site, the PTENMU1 plasmid covering 15 mutated nucleotides in the first binding spot and PTENMU2 plasmid covering 13 mutated nucleotides in the second binding spot were applied. Our data suggested that luciferase activity was reduced in cells transfected with PTENWT1/2 and miR-382-5p compared to cells transfected with PTENMU1/2 and miR-382-5p (Figures 5E and 5F). Thus, our data suggested that both sites acted as miR-382-5p binding sites. Further assessment revealed that overexpression of miR-382-5p in ARPE-19 cells suppressed PTEN expression (Figure 5G), indicating that PTEN expression was inversely correlated with miR-382-5p. Thus, our findings supported PTEN as a target gene of miR-382-5p in RPE cells. We also revealed that circNR3C1 silencing reduced PTEN protein expression in RPE (Figure 5H).
Figure 5.
PTEN Is a Direct Target of miR-382-5p in RPE Cells
(A) We utilized quantitative real-time PCR assay to reveal PTEN expression in hiPSC and hiPSC-RPE at 30, 60, and 90 days post differentiation (one-way ANOVA, Bonferroni’s test). (B) PTEN mRNA expressions in the macular RPE-choroid tissue of AMD patients (n = 6) and non-AMD controls (n = 50) as indicated by the microarray data (two-tailed Student’s t test). (C) Schematic diagram of the interaction between miR-382d-5p and PTEN 3′ UTR. A 986-bp fragment in PTEN 3′ UTR covering the two binding sites was synthesized, amplified, and inserted into the pGL3-Promoter Vector to construct the recombinant plasmids PTENWT and PTENMU. Another two 200-bp fragments in PTEN 3′ UTR covering the first and the second binding sites, respectively, were applied to construct recombinant plasmids PTENWT1/PTENMU1 and PTENWT2/PTENMU2. (D-F) ARPE-19 cells were co-transfected with PTENWT/PTENMU/PTENWT1/PTENMU1/PTENWT2/PTENMU2 with NC mimic/miR-382-5p mimic. Relative luciferase activity was measured using the dual luciferase assay. Renilla luciferase activity was taken as an indicator for transfection efficiency. Luciferase activity was reduced in cells co-transfected with miR-382-5p mimic and PTENWT/PTENWT1/PTENWT2 (two-tailed Student’s t test). (G) Quantitative real-time PCR assay revealed mRNA expression of PTEN in ARPE-19 cells transfected with NC mimic or miR-382-5p mimic (two-tailed Student’s t test). (H) PTEN protein expressions in ARPE-19 cells transfected with scramble siRNA or circNR3C1-siRNA were displayed by immunoblotting. A representative image along with the quantification result was shown (two-tailed Student’s t test). *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significant difference.
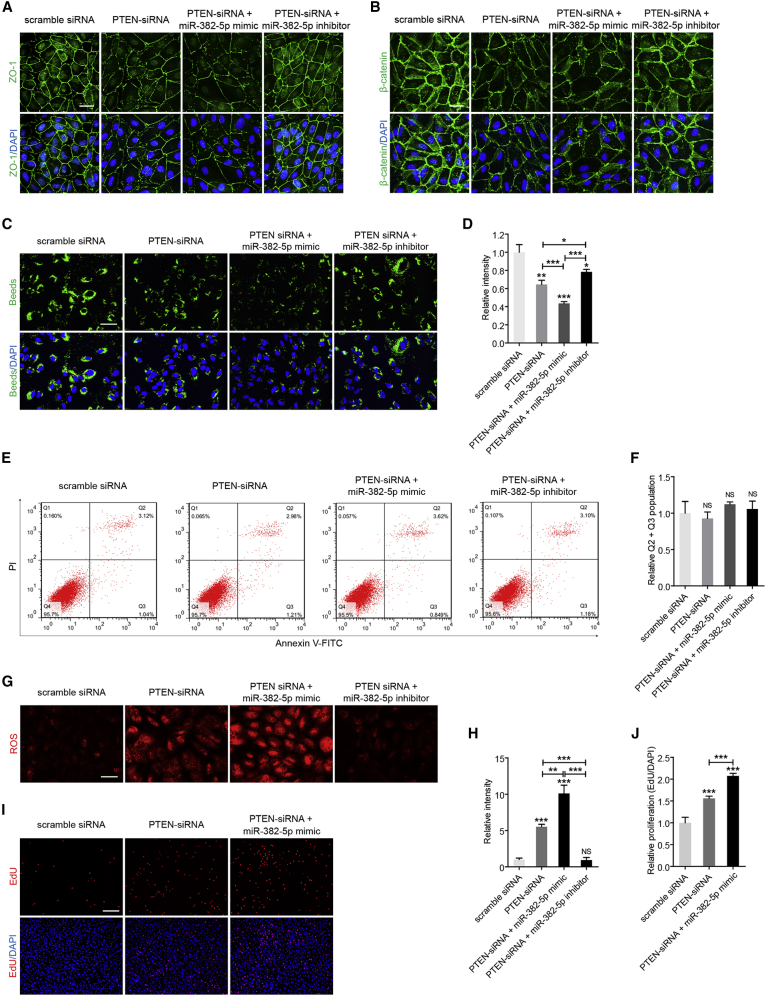
We then determined whether miR-382-5p-PTEN interaction mediated RPE function. We designed three pairs of siRNA oligos targeting PTEN and selected siRNA-1 with best efficiency for further assessment (Figures S4A and S4B). Our results implied that, similar to knockdown of circNR3C1 and overexpression of miR-382-5p, silencing of PTEN in RPE cells reduced ZO-1 and β-catenin expression (Figures 6A and 6B), interfered with the phagocytic ability of RPE cells (Figures 6C and 6D) without affecting their apoptosis rate (Figures 6E and 6F), increased ROS production (Figures 6G and 6H), and accelerated proliferation of RPE cells (Figures 6I and 6J). Meanwhile, miR-382-5p mimic aggravated effects induced by PTEN silencing in RPE cells (Figures 6A–6J), while co-transfection of miR-382-5p inhibitor abrogated those effects (Figures 6A–6H). Collectively, our data suggested that circNR3C1-miR-382-5p-PTEN crosstalk moderated RPE phenotypes in vitro.
Figure 6.
miR-382-5p-PTEN Interaction Moderates RPE Functions In Vitro
(A and B) Expression patterns of ZO-1 (A) and β-catenin (B) were visualized by immunofluorescence staining in ARPE-19 cells transfected with scramble siRNA, PTEN-siRNA, PTEN-siRNA together with miR-382-5p mimic, or PTEN-siRNA together with miR-382-5p inhibitor. (C and D) Phagocytosis was disturbed in ARPE-19 cells transfected with PTEN-siRNA compared to cells transfected with scramble siRNA. This negative effect was aggravated by co-transfection of miR-382-5p mimic, and was rescued by co-transfection of miR-382-5p inhibitor (one-way ANOVA, Bonferroni’s test). Scale bar represents 20 μm. (E and F) No difference was identified in apoptosis rates of ARPE-19 cells transfected with distinct oligonucleotides (one-way ANOVA, Bonferroni’s test). (G and H) Mitochondrial ROS was accumulated in ARPE-19 cells transfected with PTEN-siRNA compared to cells transfected with scramble siRNA. Co-transfection of miR-382-5p mimic aggravated this effect, while co-transfection of miR-382-5p inhibitor abrogated this effect (one-way ANOVA, Bonferroni’s test). Scale bar represents 50 μm. (I and J) Proliferation was increased in ARPE-19 cells transfected with PTEN-siRNA compared to cells transfected with scramble siRNA. Proliferation was more promoted by co-transfection of miR-382-5p mimic, and was reduced by co-transfection of miR-382-5p inhibitor (one-way ANOVA, Bonferroni’s test). Scale bar represents 200 μm. *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significant difference.
circNR3C1-miR-382-5p-PTEN Network Regulates AKT/mTOR Pathway
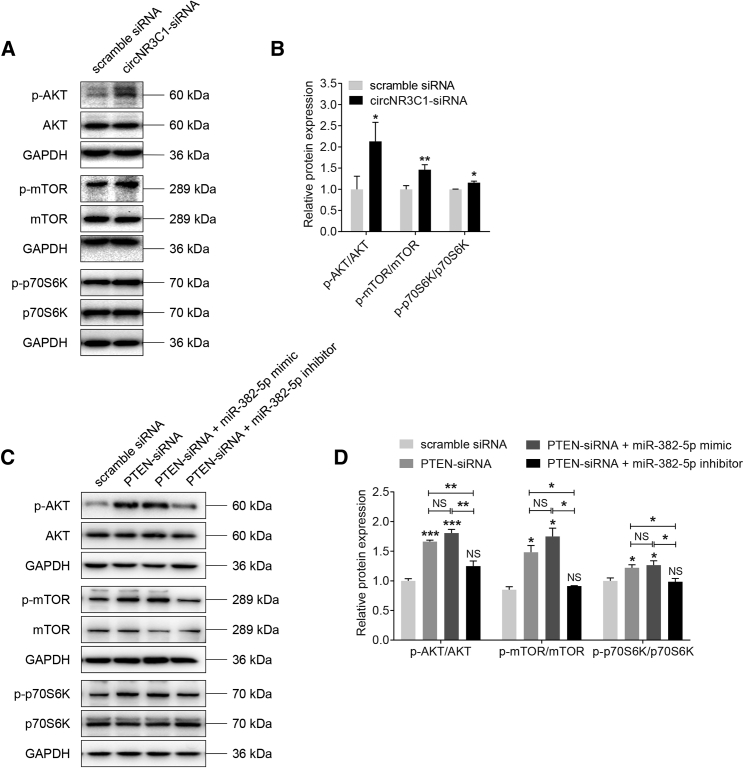
Because PTEN is an upstream regulator of the protein kinase B/mammalian target of rapamycin (AKT/mTOR) signaling pathway,41 and the AKT/mTOR pathway is involved in AMD etiology,4,6 we thus tried to determine whether circNR3C1-miR-382-5p-PTEN network regulated AKT/mTOR pathway. As shown in Figures 7A and 7B, western blot analysis showed increased phosphorylation of AKTSer473, mTORSer2488, and p70S6KThr389/412 in RPE cells with circNR3C1 knocked down when compared to the control group, suggesting that circNR3C1 could block the AKT/mTOR pathway. Our data also implied that the AKT/mTOR pathway was activated in RPE cells transfected with PTEN siRNA (Figures 7C and 7D), and co-transfection of miR-382-5p mimic with PTEN siRNA would promote such activation to a greater extent. By contrast, miR-382-5p inhibitor could block activation of AKT/mTOR pathway (Figures 7C and 7D). Thus, our data implied that the circNR3C1-miR-382-5p-PTEN network might moderate RPE features via regulating the AKT/mTOR pathway.
Figure 7.
circNR3C1-miR-382-5p-PTEN Network Regulates AKT/mTOR Pathway
(A-D) Immunoblotting was applied to show expression levels of proteins involved in the AKT/mTOR pathway, including AKT, phosphorylated AKT, mTOR, phosphorylated mTOR, p70S6K, and phosphorylated p70S6K, in ARPE-19 cells transfected with distinct oligonucleotides. Based on our findings, the AKT/mTOR pathway was activated in cells transfected with circNR3C1-siRNA compared to cells transfected with scramble siRNA (A and B; two-tailed Student’s t test). In addition, silencing of PTEN activated the AKT/mTOR pathway. Overexpression of miR-382-5p aggravated this effect, while co-transfected of miR-382-5p inhibitor abrogated this effect (one-way ANOVA, Bonferroni’s test). *p < 0.05; **p < 0.01; ***p < 0.001; NS, no significant difference.
circNR3C1-miR-382-5p-PTEN Network Mediates RPE Phenotypes In Vivo
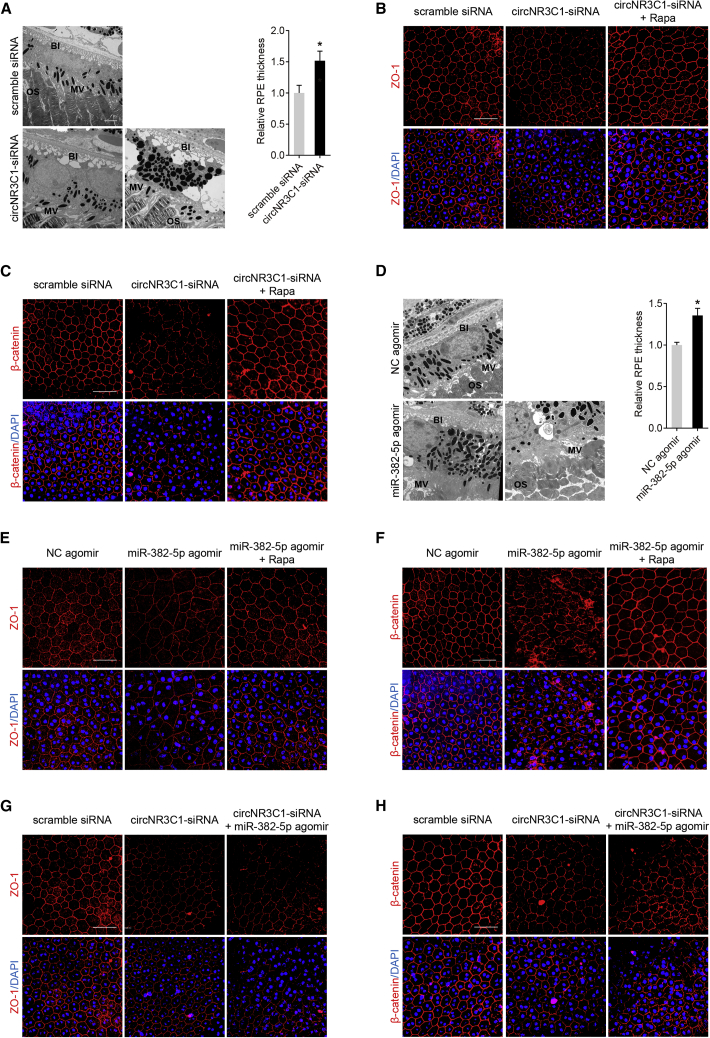
We then determined whether circNR3C1 was also involved in promoting RPE function in vivo. Three pairs of siRNAs for circNR3C1 (mmu_circ_0007330) silencing in mice were designed (Figure 1B). circNR3C1 siRNA-3 injection reduced retinal circNR3C1 expression but not linear NR3C1 mRNA expression (Figures S5A–S5B), and was selected for further investigations. In comparison with the scramble siRNA-injected eye, transmission electron microscopy (TEM) showed a thickened RPE layer with twisted basal infoldings (BIs) and outer segments (OSs) in the circNR3C1 siRNA-injected eye (n = 6; Figure 8A). Abnormalities in RPE morphology and loss of RPE characteristic markers, ZO-1 and β-catenin, at cell junctions were evident by immunofluorescent staining in RPE flat mounts of circNR3C1 siRNA-injected mice (n = 6; Figures 8B and 8C).
Figure 8.
circNR3C1-miR-382-5p-PTEN Network Mediates RPE Phenotypes In Vivo
(A) Ultrastructure of mice retina overexpressing scramble siRNA or circNR3C1-siRNA was visualized by TEM. A representative image along with the quantification results is shown (two-tailed Student’s t test). Scale bar represents 2 μm. BI, basal infoldings; MV, microvilli; OS, outer segments. (B and C) Immunofluorescence staining was applied to observed expression patterns of ZO-1 and β-catenin in RPE flat mounts of mice eyes injected with scramble siRNA, circNR3C1-siRNA, or circNR3C1-siRNA together with rapamycin (intraperitoneally injected). ZO-1 and β-catenin expressions were decreased in mice eyes injected with circNR3C1-siRNA when compared to eyes injected with scramble siRNA. However, intraperitoneal injection of rapamycin could rescue the negative effect of circNR3C1 silencing. Scale bar represents 50 μm. (D) TEM was utilized to compare the ultrastructure of mice retina overexpressing NC agomir or circNR3C1 agomir. A representative image along with the quantification results were shown (two-tailed Student’s t test). Scale bar represents 2 μm. BI, basal infoldings; MV, microvilli; OS, outer segments. (E and F) We used immunofluorescent staining to see expression patterns of ZO-1 and β-catenin in RPE flat mounts of mice eyes injected with NC agomir, miR-382-5p agomir, or miR-382-5p together with rapamycin (intraperitoneally injected). ZO-1 and β-catenin expressions were decreased in mice eyes injected with miR-382-5p agomir when compared to eyes injected with NC agomir. However, intraperitoneal injection of rapamycin could rescue the negative effect of miR-382-5p overexpression. Scale bar represents 50 μm. (G and H) Immunofluorescence was utilized to visualize ZO-1 and β-catenin in RPE flat mounts of mice eyes injected with scramble siRNA, circNR3C1-siRNA, or circNR3C1-siRNA together with miR-382-5p agomir. ZO-1 and β-catenin expressions were more decreased in mice eyes co-injected with circNR3C1-siRNA and miR-382-5p agomir when compared to eyes injected with circNR3C1-siRNA alone. Scale bar represents 50 μm. *p < 0.05.
We further investigated the role of miR-382-5p in regulating RPE features in vivo. Similar to the above findings, TEM also revealed that miR-382-5p upregulation by agomir-injection-induced RPE thickening, together with BI and OS twisting (n = 6; Figure 8D). Immunofluorescent staining indicated disturbed RPE structure and reduced ZO-1 and β-catenin expressions in miR-382-5p agomir-injected eye when compared to the negative control (NC) agomir-injected eye (n = 6; Figures 8E and 8F).
We also determined whether exogenous miR-382-5p addition could aggravate the negative effect of circNR3C1 silencing on RPE cells. circNR3C1 silencing could increase the release of miR-382-5p and induce RPE degeneration. According to our findings, miR-382-5p upregulation by exogenous agomir injection could exacerbate the phenotype of circNR3C1 silencing on RPE degeneration in vivo (n = 6; Figures 8G and 8H). Since we have revealed that the circNR3C1-miR-382-5p-PTEN network might moderate RPE features via regulating the AKT/mTOR pathway, we next tested whether rapamycin, an inhibitor of the AKT/mTOR pathway, could rescue the negative effects of circNR3C1-siRNA and miR-382-5p agomir on RPE cells. Immunofluorescence staining suggested that treatment with rapamycin could abrogated the repressive effects mediated by circNR3C1-siRNA (n = 6; Figures 8B and 8C) and miR-382-5p (n = 6; Figures 8E and 8F). Collectively, our data implied that circNR3C1-miR-382-5p-PTEN network mediated RPE phenotypes in vivo.
Discussion
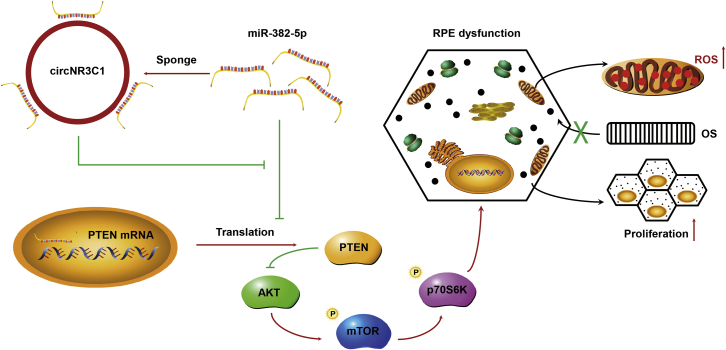
RPE dedifferentiation is a crucial contributing factor to atrophic AMD.4, 5, 6, 7 Thus, blocking RPE dedifferentiation is a promising approach to treat such retinal degenerative diseases. Previously, our group has demonstrated the involvement of a series of non-coding RNAs, including miRNAs and long noncoding RNAs (lncRNAs), in regulating RPE differentiation. We found that miR-184 regulated the AKT/mTOR signaling pathway and suppressed RPE dedifferentiation,6 miR-302d-3p mediated by c-Jun induced RPE dedifferentiation by targeting cycling protein p21Waf1/Cip1,7 and lncRNA ZNF503-AS1 inhibited RPE dedifferentiation through reducing ZNF503 expression.5 circRNAs are important regulators in gene expression. Increasing evidence has shown that circRNA dysregulation leads to ocular diseases.21, 22, 23 However, whether circRNAs are implicated in RPE differentiation or AMD pathology has never been illustrated. In the present study, we found that circNR3C1 prevented AMD progression and protected regular RPE function both in vivo and in vitro. circNR3C1 directly sponged miR-382-5p to block its interaction with PTEN and further suppressed its downstream AKT/mTOR signal pathway (Figure 9).
Figure 9.
Schematic Diagram of the circNR3C1-Regulated Pathway in RPE Cells
Red arrows indicate promotive function and green lines indicate suppressive function.
Roles of circNR3C1 are poorly understood. circNR3C1 is reported to restrain bladder cancer,42 while its impact on AMD or other diseases is not clear. Herein, we confirmed the circular structures of circNR3C1 in both human and mice, which consisted of the head-to-tail splicing of exon 2 in the NR3C1 gene. We found that circNR3C1 expression was suppressed in blood samples of AMD patients. In addition, circNR3C1 level was increased along with RPE differentiation and was decreased in H2O2 or NaIO3 treated RPE cells, further supporting its potential involvement in AMD pathogenesis. Insufficient endogenous circN3C1 expression disturbed RPE ultrastructure, reduced RPE characteristic transcripts and proteins, interrupted phagocytosis, generated mitochondrial ROS, and promoted RPE proliferation. Noteworthy, a previous study identified a suppressive effect of circNR3C1 on the proliferation of bladder cancer cells,42 which was similar to our findings in RPE cells. circNR3C1 was found located in the cytoplasm of RPE cells with multiple predicted miRNA binding sites, indicating its potential role as a miRNA sponge.32,33 Former findings suggested that circNR3C1 sponged miR-27a-3p, thus increasing cyclin D1 expression in bladder cancer cells.42 Our experimental data clarified that circNR3C1 harbored a binding site for miR-382-5p and could sponge miR-382-5p as competing endogenous RNA in RPE cells. However, whether circNR3C1 may target other miRNAs and signal pathways, or even possess other functions such as protein sponges and nuclear transcriptional regulators, is not fully elucidated.
The role of miR-382-5p in mediating retinal function has never been illustrated before. PTEN was previously found as a target gene of miR-382-5p,35, 36, 37, 38 while their interaction in RPE remains inclusive. In the present study, we first revealed that miR-382-5p overexpression triggered RPE dysfunction both in vivo and in vitro, and identified PTEN as a direct target of miR-382-5p in RPE cells. PTEN is essential in maintaining RPE cell function and the response of RPE cells to oxidative stress, and inactivation of PTEN associates with retinal degeneration and disrupts intercellular adhesion in RPE cells.39,43, 44, 45, 46 Reportedly, RPE-specific deletion of mouse PTEN gene results in RPE cells that fail to maintain basolateral adhesions, undergo epithelial-mesenchymal transition, and migrate out of the retina entirely.39 PTEN is an upstream suppressive regulator of the AKT/mTOR signaling pathway. Stimulation of the AKT/mTOR pathway is reported to induce RPE dedifferentiation, proliferation, migration, and hypertrophy and is supposed as a crucial pathological process for atrophic AMD.4,6,47 In this study, our data supported that reduced endogenous PTEN expression interrupted regular function of RPE cells through both in vivo and in vitro studies. We also found that circNR3C1-miR-382-5p-PTEN network regulated RPE features through the AKT/mTOR pathway.
Although both in vitro and in vivo studies support that circNR3C1 inhibited AMD progression through sponging miR-382d-5p and regulating PTEN/AKT/mTOR pathway, it is still not clear whether circNR3C1 may sponge other miRNAs, target distinct signaling pathways, or function as protein sponges or nuclear transcriptional regulators. In addition, other circRNAs involved in RPE degeneration and AMD etiology have not been revealed yet. Thus, more investigations in this area are still warranted.
In conclusion, our study reveals the protective role of circNR3C1 in the etiology of atrophic AMD. Silencing of circNR3C1 disturbs RPE structure, reduces expression of RPE characteristic transcripts and proteins, interrupts RPE phagocytosis, increases ROS generation, and promotes RPE proliferation. We show that circNR3C1 prevents AMD by acting as a miR-382-5p sponge and a PTEN/AKT/mTOR signaling pathway regulator (Figure 9). circNR3C1 is a promising target for treating atrophic AMD.
Materials and Methods
Samples
Blood samples of 9 AMD patients and 6 healthy controls were collected in The First Affiliated Hospital of Nanjing Medical University. All procedures followed standard procedures of blood donation for research and were approved by the local institutional ethical committees conformed to Declaration of Helsinki. Written informed consent was obtained from all donors before their donation. In addition, sample information and microarray data (GEO: GSE29801) of 56 independent macular RPE-choroid samples were downloaded from Gene Expression Omnibus datasets and analyzed as described previously.5,40 Sample information is detailed in Table S1.
Cell Culture and Treatments
Human induced pluripotent stem cells (hiPSCs, IMR90-57) were cultured on mouse embryonic fibroblasts (SiDan-Sai Biotechnology, Shanghai, China) in 6-well tissue culture plates according to a previously described protocol.6 The hiPSCs were then differentiated into RPE cells per the SFEB/CS method using low-molecular-weight compounds CKI-7 (5 μM) and SB-431542 (5 μM).48 ARPE-19 cells, purchased from American Type Culture Collection, were maintained in DMEM/F12 medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), penicillin (100 U/mL), and streptomycin (100 U/mL) at 37°C, 5% CO2. We used complete medium short for supplemented culture medium in the following text. For actinomycin D assay, ARPE-19 cells were cultured in complete medium added with actinomycin D (1 mg/mL) and were harvested at 4, 8, 12, and 24 h post treatment, respectively. For H2O2 assay, cells were treated with H2O2 (100 μM) for 24 h before collection. For NaIO3 assay, ARPE-19 cells were maintained in complete medium supplemented with NaIO3 (1.25 mmol/L) for 96 h. Medium was changed every 24 h with NaIO3 added.
RNA Preparation, RNase R Treatment, PCR, and Sanger Sequencing
Total RNA was extracted from cell lysates and peripheral blood samples using TRIzol reagent (Invitrogen). ARPE-19 cells were collected at 48 h post transfection for RNA isolation. Nuclear and cytoplasmic fractions were isolated with NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Waltham, MA, USA). RNA concentration and quality were tested using Nano-Drop ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). For RNase R treatment, 2 μg of extracted RNA was incubated with RNase R (3 U/μg) at 37°C for 30 min and then purified with an RNasey MinElute cleaning kit (QIAGEN, Hilden, Germany). cDNA was generated for circRNA and mRNA with a PrimeScript RT Kit (Takara, Otsu, Shiga, Japan), and was synthesized for miRNA by stem-loop reverse transcription with oligo-dT primers (RiboBio, Guangzhou, China). RNA amounts were determined by quantitative real-time PCR using FastStart Universal SYBR Green Master (ROX; Roche, Basel, Switzerland) with StepOne Plus Real-Time PCR System (Applied Biosystems, Darmstadt, Germany). Human 18S ribosomal RNA (18S rRNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 gene expressions were analyzed in parallel for normalization of circRNA, mRNA, and miRNA expressions, respectively. Standard protocol for Sanger sequencing had been detailed previously.49 Primers information is provided in Table S2.
FISH
U6, 18S rRNA, circNR3C1, and miR-382-5p probes were synthesized by RiboBio. FISH was performed using a FISH kit according to the manufacturer’s instructions (RiboBio). Briefly, ARPE19 cells were fixed with 4% paraformaldehyde, permeabilized in 0.5% Triton X-100 on ice, and then hybridized with Cy3-labeled RNA of U6, 18S rRNA and circNR3C1 probe mixes, and FITC-labeled miR-382-5p probes in moist chambers, respectively. Cell nuclei were counterstained by 4’,6-diamidino-2-phenylindole (DAPI; Sigma, St. Louis, MO, USA). Images were taken with a LSM 510 confocal microscope (Carl Zeiss, Jena, Germany).
Reagents and Cell Transfection
Oligonucleotides used in both human and mouse species models were purchased from RiboBio (RiboBio, Guangzhou, China), including scramble siRNA (human and mouse), circNR3C1-siRNA (human and mouse), PTEN-siRNA (human), NC mimic (human), miR-382-5p mimic (human), miR-382-5p inhibitor (2′-O-methyl modification; human), NC agomir (mouse), and miR-382-5p agomir (mouse). Sequences of these oligonucleotides were detailed in Table S3. Entire circNR3C1 sequence was synthesized, amplified, and inserted into the pGL3-Promoter Vector (Promega, Madison, WI, USA) using the Xba I restriction site to generate the recombinant plasmids NR3C1WT and NR3C1MU. In addition, a 986-bp fragment in PTEN 3′ UTR covering the two binding sites were also synthesized, amplified, and inserted into the pGL3-Promoter Vector using the Xba I restriction site to construct the recombinant plasmids PTENWT and PTENMU. Another two 200-bp fragments in PTEN 3′ UTR covering the first and the second binding sites, respectively, were applied to construct recombinant plasmids PTENWT1/PTENMU1 and PTENWT2/PTENMU2. Constructed plasmids were sequenced and confirmed using Sanger sequencing. For transfection assay, ARPE-19 cells were seeded into 6-well templates, and then transfected with 100 pmol siRNA/mimic/inhibitor and/or 4 μg plasmids at 50%–60% confluence using Lipofectamine 3000 transfection reagent (Invitrogen) per the manufacturers’ protocol.
Immunofluorescence
Immunofluorescence was conducted according to a previously described protocol.6,50 Briefly, ARPE-19 cells were planted into 8-well chamber slides (Millipore, Billerica, MA, USA), and were harvested at 72 h post transfection. Cells were then incubated with primary antibodies (Table S4) at 4°C overnight, and corresponding fluorescence-conjugated secondary antibodies (1:1,000 diluted in 1X phosphate buffered saline [PBS]; Invitrogen) at room temperature for 1 h. Cell nuclei were counterstained by DAPI. A LSM 510 confocal microscope was applied for image collection.
Analysis of Phagocytosis
Phagocytic ability was analyzed per a previously defined protocol.7 Briefly, ARPE-19 cells were grown on 8-well chamber slides and were harvested at 72 h post transfection. Cells were then incubated with carboxylate-modified polystyrene latex beads (1 μm in diameter; Sigma) with yellow-green fluorescence (emission maximum: 515 nm; 70 beads per cell) at 37°C for 12 h. After incubation, cells were washed with 1X PBS to stop phagocytosis and treated with 0.2% trypan blue to quench extracellular fluorescence. Cell nuclei were counterstained by DAPI. We then used a LSM 510 confocal microscope to collect images. ImageJ software (http://rsb.info.nih.gov/ij/index.html) was applied to quantify fluorescence.
Apoptosis Assay
At 72 h post transfection, ARPE-19 cells were collected and incubated with Annexin V-FITC (R&D, NJ, USA) and PI (R&D) according to the manufacturers’ protocol. Annexin-V positive cells were recognized as apoptotic cells, and were identified by a gallios flow cytometry (Beckman Coulter, Brea, USA). A total of 10,000 living cells were counted for each sample. Three additional groups of ARPE-19 cells without treatment were included for scatter gating as follows: (1) unstained cells for cell selection and adjustment of photomultiplier voltage, (2) Annexin V-FITC only stained cells for adjustment of the FITC channel, and (3) PI only stained cells for adjustment of the phycoerythrin channel. Data were presented as two-color dot plot with Annexin V-FITC (x axis) versus PI (y axis).
Mitochondrial ROS Measurement
Mitochondrial ROS levels were measured using the MitoSOX Red mitochondrial superoxide indicator (Invitrogen) per the manufacturer’s protocols. ARPE-19 cells were planted into 12-well plates and were collected at 72 h post transfection. Cells were incubated with 5 μM MitoSOX reagent at 37°C for 10 min in the dark. We then used a Leica DM4000 B LED microscope (Leica, Wetzlar, Germany) to observe mitochondrial ROS, which was reflected by red signals.
EdU Incorporation Assay
An EdU assay kit (Invitrogen) was utilized to detect cell proliferation per the manufacturer’s protocols. ARPE-19 cells were seeded into 12-well templates and harvested at 72 h post transfection. ARPE-19 cells were then incubated with 50 mM EdU for 2 h, fixed with 4% paraformaldehyde, and incubated with Apollo Dye Solution to label proliferating cells. Cell nuclei were counterstained by DAPI. Proliferating cells with red signals were visualized by a Leica DM4000 B LED microscope.
Luciferase Reporter Assay
Luciferase reporter assay was performed according to a previously defined protocol.5,6 ARPE-19 cells, grown on 24-well plates, were transfected with 16 ng cytomegalovirus-Renilla (Promega), 20 pmol miR-382-5p mimic or NC mimic, and 800 ng NR3C1WT/NR3C1MU/PTENWT/PTENMU. At 72 h post transfection, cells were collected for luciferase activities measurement using the dual luciferase system (Promega) with a GloMax-96 luminometer. Firefly luciferase activities were normalized to Renilla luciferase activities, which were taken as internal standard indicators for transfection efficiency.
Immunoblotting
Immunoblotting was performed using a previously described protocol.6,50 ARPE-19 cells were harvested at 72 h post transfection in ice-cold protein lysis buffer (Beyotime, Shanghai, China) containing protease inhibitors cocktail (Roche) for protein extraction. Extracted proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore). Membranes were incubated with primary antibodies (Table S4) at 4°C overnight and probed with corresponding horseradish peroxidase-conjugated secondary antibodies (1:10,000 diluted in 1X PBS; ICL, Newberg, OR) at room temperature for 1 h. Blots were developed by the ECL-western blotting system (BioRad, Hercules, CA, USA). Protein expression was quantified using ImageJ software (http://rsb.info.nih.gov/ij/index.html).
Mouse Breeding, Intravitreal Injection, and Tissue Preparation
C57BL/6 mice were housed in a specific pathogen-free facility in Nanjing Medical University and conformed to the guidelines of the Care and Use of Laboratory Animals (published by the NIH publication no. 86–23, revised 1996). Embryo rearing was maintained at 28.5°C with a 12 h light/dark cycle. All embryos were produced by natural mating. Animal experiments were approved by ethical review board of Nanjing Medical University and conformed to the Guide for the Care and Use of Laboratory Animals. For intravitreal injection, 6-week-old C57BL/6 mice were anesthetized by intraperitoneal injection of ketamine (80 mg/kg) and xylazine (4 mg/kg). 1 μL (1 nmol/μL) of circNR3C1 siRNA/scramble siRNA/miR-382-5p agomir/NC agomir was delivered into the vitreous of mice using a 33-gauge needle. To maximize siRNA/agomir delivery efficiency, we administered an additional intravitreal injection 2 weeks after the initial injection. Mice were sacrificed for tissue preparation 1 month after the initial injection. For rapamycin administration, 6-week-old C57BL/6 mice were injected daily with rapamycin (3 mg/kg) intraperitoneally until sacrifice. For preparation of eyecups, C57BL/6 mice eyes were enucleated and rinsed with 1X PBS. After trimming the connective tissues and removing the anterior portion, the remaining posterior eyecup was sent for TEM. For immunofluorescent staining, we further separated neural retina from the posterior eyecup and used whole mounts of RPE/choroid/sclera.
TEM
Posterior eyecups were fixed with 2.5% glutaradehyde at 4°C overnight, and dehydrated with ethanol and propylene oxide. The dehydrated samples were then cut into slide sections and stained with 0.3% lead citrate. A JEM-1010 electron microscope (JEOL, Tokyo, Japan) was further utilized to visualize the ultrastructure of retina.
Statistics
For statistical analyses, we used GraphPad Prism (version 4.0; GraphPad Software, San Diego, CA, USA). Student’s t test was applied for comparisons between two different groups, and one-way analysis of variance (ANOVA) followed by the Bonferroni’s post hoc test was used in comparisons among three or more groups. Data were displayed as mean ± standard error of the mean, and p < 0.05 was considered as statistically significant. All experiments were conducted in both biological and technical triplicates with data averaged.
Author Contributions
X.C. and Q.L. conceived and designed the study. X.C., C.J., R.S., and D.Y. conducted experiments. X.C. interpreted the data and drafted the manuscript. Q.L. revised the manuscript. D.Y. and C.J. coordinated and analyzed the data. All the authors contributed to, read, and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank all donors for sample donations. This study was supported by National Natural Science Foundation of China (81770973 to Q.L. and 81700877 to X.C.); National Key Research and Development Program of China (2017YFA0104100 to Q.L.); Natural Science Foundation of Jiangsu Province (BK20171087 to X.C.); Six Talent Peaks Project in Jiangsu Province (WSW-004 to X.C.); and a project funded by the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.01.010.
Supplemental Information
References
- 1.Bressler N.M. Age-related macular degeneration is the leading cause of blindness. JAMA. 2004;291:1900–1901. doi: 10.1001/jama.291.15.1900. [DOI] [PubMed] [Google Scholar]
- 2.Jager R.D., Mieler W.F., Miller J.W. Age-related macular degeneration. N. Engl. J. Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 3.Lim L.S., Mitchell P., Seddon J.M., Holz F.G., Wong T.Y. Age-related macular degeneration. Lancet. 2012;379:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 4.Zhao C., Yasumura D., Li X., Matthes M., Lloyd M., Nielsen G., Ahern K., Snyder M., Bok D., Dunaief J.L. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X., Jiang C., Qin B., Liu G., Ji J., Sun X., Xu M., Ding S., Zhu M., Huang G. LncRNA ZNF503-AS1 promotes RPE differentiation by downregulating ZNF503 expression. Cell Death Dis. 2017;8:e3046. doi: 10.1038/cddis.2017.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang C., Qin B., Liu G., Sun X., Shi H., Ding S., Liu Y., Zhu M., Chen X., Zhao C. MicroRNA-184 promotes differentiation of the retinal pigment epithelium by targeting the AKT2/mTOR signaling pathway. Oncotarget. 2016;7:52340–52353. doi: 10.18632/oncotarget.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang C., Xie P., Sun R., Sun X., Liu G., Ding S., Zhu M., Yan B., Liu Q., Chen X., Zhao C. c-Jun-mediated microRNA-302d-3p induces RPE dedifferentiation by targeting p21Waf1/Cip1. Cell Death Dis. 2018;9:451. doi: 10.1038/s41419-018-0481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005;85:845–881. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 9.Bok D. The retinal pigment epithelium: a versatile partner in vision. J. Cell Sci. Suppl. 1993;17:189–195. doi: 10.1242/jcs.1993.supplement_17.27. [DOI] [PubMed] [Google Scholar]
- 10.Marmorstein A.D. The polarity of the retinal pigment epithelium. Traffic. 2001;2:867–872. doi: 10.1034/j.1600-0854.2001.21202.x. [DOI] [PubMed] [Google Scholar]
- 11.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H.D., Jiang L.H., Sun D.W., Hou J.C., Ji Z.L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 15.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 16.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 17.Qu S., Zhong Y., Shang R., Zhang X., Song W., Kjems J., Li H. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14:992–999. doi: 10.1080/15476286.2016.1220473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37 e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yao J., Hu L.L., Li X.M., Shan K., Zhou R.M., Ge H.M., Yao M.D., Jiang Q., Zhao C., Yan B. Comprehensive circular RNA profiling of proliferative vitreoretinopathy and its clinical significance. Biomed. Pharmacother. 2019;111:548–554. doi: 10.1016/j.biopha.2018.12.044. [DOI] [PubMed] [Google Scholar]
- 22.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 23.Liu C., Ge H.M., Liu B.H., Dong R., Shan K., Chen X., Yao M.D., Li X.M., Yao J., Zhou R.M. Targeting pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A inhibition aggravates diabetes-induced microvascular dysfunction. Proc. Natl. Acad. Sci. USA. 2019;116:7455–7464. doi: 10.1073/pnas.1814874116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 25.Tian B., Al-Moujahed A., Bouzika P., Hu Y., Notomi S., Tsoka P., Miller J.W., Lin H., Vavvas D.G. Atorvastatin Promotes Phagocytosis and Attenuates Pro-Inflammatory Response in Human Retinal Pigment Epithelial Cells. Sci. Rep. 2017;7:2329. doi: 10.1038/s41598-017-02407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W. Phagocyte dysfunction, tissue aging and degeneration. Ageing Res. Rev. 2013;12:1005–1012. doi: 10.1016/j.arr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.Y., Zhao H., Martinez J., Doggett T.A., Kolesnikov A.V., Tang P.H., Ablonczy Z., Chan C.C., Zhou Z., Green D.R., Ferguson T.A. Noncanonical autophagy promotes the visual cycle. Cell. 2013;154:365–376. doi: 10.1016/j.cell.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishibashi T., Sorgente N., Patterson R., Ryan S.J. Pathogenesis of drusen in the primate. Invest. Ophthalmol. Vis. Sci. 1986;27:184–193. [PubMed] [Google Scholar]
- 29.Jarrett S.G., Boulton M.E. Consequences of oxidative stress in age-related macular degeneration. Mol. Aspects Med. 2012;33:399–417. doi: 10.1016/j.mam.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karunadharma P.P., Nordgaard C.L., Olsen T.W., Ferrington D.A. Mitochondrial DNA damage as a potential mechanism for age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2010;51:5470–5479. doi: 10.1167/iovs.10-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Xin Y., Ye F., Wang W., Lu Q., Kaplan H.J., Dean D.C. Taz-tead1 links cell-cell contact to zeb1 expression, proliferation, and dedifferentiation in retinal pigment epithelial cells. Invest. Ophthalmol. Vis. Sci. 2010;51:3372–3378. doi: 10.1167/iovs.09-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panda A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 33.Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seok J.K., Lee S.H., Kim M.J., Lee Y.M. MicroRNA-382 induced by HIF-1α is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42:8062–8072. doi: 10.1093/nar/gku515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D., Zhong L., Yuan Z., Yao J., Zhong P., Liu J., Yao S., Zhao Y., Liu L., Chen M. miR-382-5p modulates the ATRA-induced differentiation of acute promyelocytic leukemia by targeting tumor suppressor PTEN. Cell. Signal. 2019;54:1–9. doi: 10.1016/j.cellsig.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Li D., Li P., Guo Z., Wang H., Pan W. Downregulation of miR-382 by propranolol inhibits the progression of infantile hemangioma via the PTEN-mediated AKT/mTOR pathway. Int. J. Mol. Med. 2017;39:757–763. doi: 10.3892/ijmm.2017.2863. [DOI] [PubMed] [Google Scholar]
- 38.Bei Y., Song Y., Wang F., Dimitrova-Shumkovska J., Xiang Y., Zhao Y., Liu J., Xiao J., Yang C. miR-382 targeting PTEN-Akt axis promotes liver regeneration. Oncotarget. 2016;7:1584–1597. doi: 10.18632/oncotarget.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J.W., Kang K.H., Burrola P., Mak T.W., Lemke G. Retinal degeneration triggered by inactivation of PTEN in the retinal pigment epithelium. Genes Dev. 2008;22:3147–3157. doi: 10.1101/gad.1700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman A.M., Gallo N.B., Hancox L.S., Miller N.J., Radeke C.M., Maloney M.A., Cooper J.B., Hageman G.S., Anderson D.H., Johnson L.V., Radeke M.J. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corradetti M.N., Guan K.L. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- 42.Zheng F., Wang M., Li Y., Huang C., Tao D., Xie F., Zhang H., Sun J., Zhang C., Gu C. CircNR3C1 inhibits proliferation of bladder cancer cells by sponging miR-27a-3p and downregulating cyclin D1 expression. Cancer Lett. 2019;460:139–151. doi: 10.1016/j.canlet.2019.06.018. [DOI] [PubMed] [Google Scholar]
- 43.He J., Long C., Huang Z., Zhou X., Kuang X., Liu L., Liu H., Tang Y., Fan Y., Ning J. PTEN Reduced UVB-Mediated Apoptosis in Retinal Pigment Epithelium Cells. BioMed Res. Int. 2017;2017:3681707. doi: 10.1155/2017/3681707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang K.H., Lemke G., Kim J.W. The PI3K-PTEN tug-of-war, oxidative stress and retinal degeneration. Trends Mol. Med. 2009;15:191–198. doi: 10.1016/j.molmed.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee E.J., Kim N., Kang K.H., Kim J.W. Phosphorylation/inactivation of PTEN by Akt-independent PI3K signaling in retinal pigment epithelium. Biochem. Biophys. Res. Commun. 2011;414:384–389. doi: 10.1016/j.bbrc.2011.09.083. [DOI] [PubMed] [Google Scholar]
- 46.Lin X., Zhou X., Liu D., Yun L., Zhang L., Chen X., Chai Q., Li L. MicroRNA-29 regulates high-glucose-induced apoptosis in human retinal pigment epithelial cells through PTEN. In Vitro Cell. Dev. Biol. Anim. 2016;52:419–426. doi: 10.1007/s11626-015-9990-z. [DOI] [PubMed] [Google Scholar]
- 47.Huang J., Gu S., Chen M., Zhang S.J., Jiang Z., Chen X., Jiang C., Liu G., Radu R.A., Sun X. Abnormal mTORC1 signaling leads to retinal pigment epithelium degeneration. Theranostics. 2019;9:1170–1180. doi: 10.7150/thno.26281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osakada F., Jin Z.B., Hirami Y., Ikeda H., Danjyo T., Watanabe K., Sasai Y., Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J. Cell Sci. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- 49.Zhao C., Lu S., Zhou X., Zhang X., Zhao K., Larsson C. A novel locus (RP33) for autosomal dominant retinitis pigmentosa mapping to chromosomal region 2cen-q12.1. Hum. Genet. 2006;119:617–623. doi: 10.1007/s00439-006-0168-3. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y., Chen X., Xu Q., Gao X., Tam P.O., Zhao K., Zhang X., Chen L.J., Jia W., Zhao Q. SPP2 Mutations Cause Autosomal Dominant Retinitis Pigmentosa. Sci. Rep. 2015;5:14867. doi: 10.1038/srep14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.