Abstract
Increasing studies indicated that circular RNAs (circRNAs) play important roles in cancer progression. However, the roles of circRNAs in colorectal cancer (CRC) remain largely unknown. In this study, we determined the circRNA expression profile by next-generation RNA sequencing from eight CRC and paired non-cancerous matched tissues. circCAMSAP1 (originating from exon 2 to exon 3 of the CAMSAP1 gene, hsa_circ_0001900) was significantly upregulated in CRC tissues. Increased circCAMSAP1 expression was significantly correlated with advanced tumor/node/metastasis (TNM) stage and shortened overall survival. An elevation of circCAMSAP1 expression was detected via droplet digital PCR in the serum of CRC patients prior to surgery. Functionally, circCAMSAP1 promoted the malignant behavior of CRC. Mechanism study of upstream biogenesis of circCAMSAP1 indicated that circCAMSAP1 cyclization in CRC was mediated by splicing factor epithelial-splicing regulatory protein 1. Moreover, circCAMSAP1 acted as a sponge for miR-328-5p and abrogated its suppression on transcription factor E2F1. Taken together, our data indicated an essential role of the circCAMSAP1/miR-328-5p/E2F1 axis in the progression of CRC, which implied that circCAMSAP1 could serve as a diagnostic and prognostic biomarker as well as a potential therapeutic target for CRC.
Keywords: colorectal cancer, circCAMSAP1, miR-328-5p, E2F1, tumor growth
Graphical Abstract
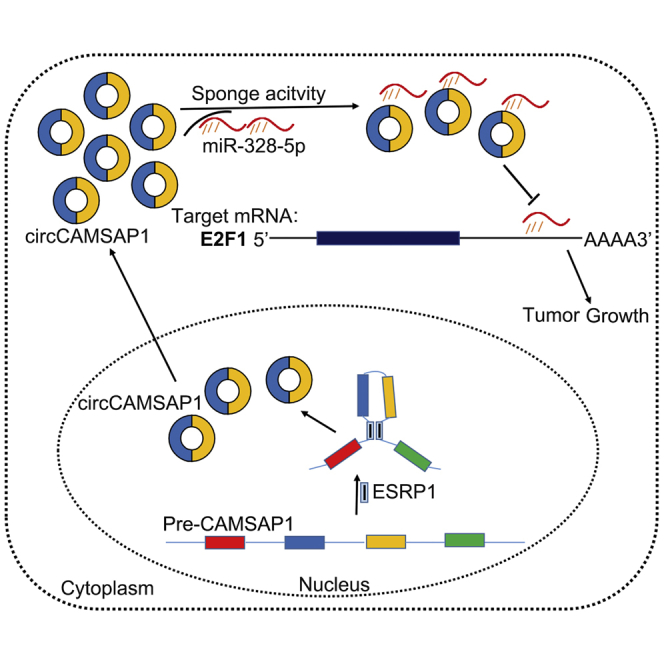
In this issue of Molecular Therapy, Lan and his colleagues profiled circRNA expression in colorectal cancer (CRC) tissues. They demonstrated that circCAMSAP1 could act as an oncogene in CRC and serve as a diagnostic and prognostic biomarker as well as a potential therapeutic target for CRC.
Introduction
Colorectal cancer (CRC) is the third most common cancer and one of the leading causes of cancer-related deaths in the world.1 Despite recent improvements in the diagnosis and therapies of CRC, its prognosis remains less than satisfactory, especially in patients with advanced stages.2 Hence, it is of great importance to seek new potential biomarkers and discover unknown mechanisms contributing to CRC pathogenesis.
Circular RNAs (circRNAs) are a subclass of endogenous non-coding RNAs that form covalently closed loops, without 5′ to 3′ polar or polyadenylation tails.3,4 This feature enables circRNAs to be resistant to RNA exonucleases and hence become more stable. circRNAs can be generated from precursor (pre-)mRNAs through alternative splicing, which have been detected extensively in various cell types in an evolutionarily conserved manner.5, 6, 7 Splicing factors are a group of RNA binding proteins that participate in the alternative splicing process of cells.8 Although several studies showed the importance of splicing factors in the formation of circRNAs,9, 10, 11 the regulatory roles of circRNAs biogenesis, especially in CRC, are not fully understood.
Given their stability, abundance, and evolutionary conservation, circRNAs could act as long-acting regulators of cellular behavior and robust potential biomarkers.12,13 Recently, circRNAs have been indicated to be involved in the natural history of several diseases, including neurodegenerative diseases such as Alzheimer’s disease,14 cardiovascular diseases,15 and various types of cancers.16, 17, 18, 19 Mechanistically, studies have shown that some circRNAs might serve as a microRNA (miRNA) sponge to inhibit miRNA functions.19,20 Some circRNAs could bind to proteins and regulate gene functions.15,16 Alternatively, some other circRNAs could encode functional proteins.21,22 Despite the improvement in the understanding of circRNAs, the potential correlation between circRNAs and CRC progression has not been fully elucidated.
In this study, we analyzed the expression profiles of circRNAs in CRC tissues and identified a circular RNA termed circCAMSAP1, which derives from the CAMSAP1 gene. circCAMSAP1 was significantly upregulated in CRC tissues and inversely associated with the prognosis of CRC. circCAMSAP1 expression in CRC tissues has a better prediction performance than carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) in the prognosis of CRC patients. Through detecting the circCAMSAP1 level in serum, we found that circCAMSAP1 expression was higher in the serum of CRC patients prior to surgery than that in patients after surgery or in healthy volunteers. Through a series of in vitro and in vivo experiments, we also revealed that circCAMSAP1 might function as the sponge of miR-328-5p to regulate the expression of transcription factor E2F1, thereby promoting CRC progression. Moreover, mechanism studies of the biogenesis of circCAMSAP1 indicated that epithelial-splicing regulatory protein 1 (ESRP1) induced the formation of circCAMSAP1. According to our results, circCAMSAP1 has the potential to act as a biomarker and therapeutic target of CRC.
Results
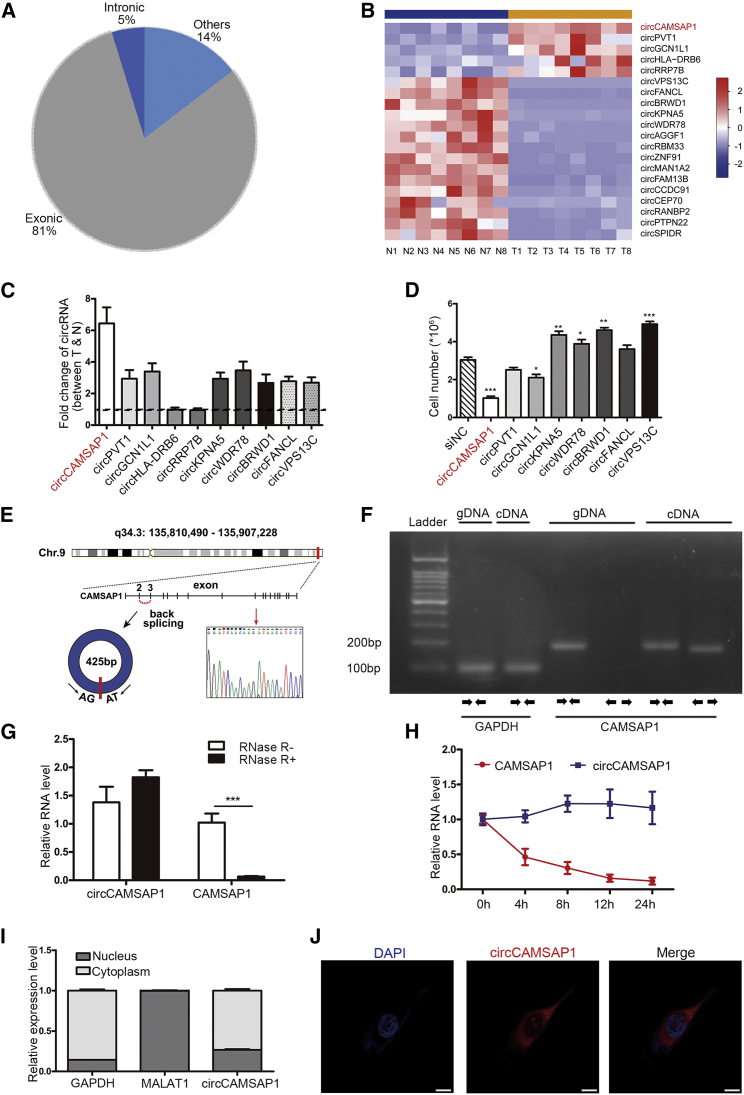
circRNA Expression Profiles in CRC
To screen the potential driver circRNAs in CRC pathogenesis, RNA-depleted total RNA sequence was performed to profile mRNA and circRNA expression in eight pairs of CRC tissues and their adjacent non-tumor tissue samples. 31,557 circRNAs were detected in the screening results, 81% of which were from the exon of genomic origin (Figure 1A). 46 circRNAs were differentially expressed between cancer and matched normal tissues after filtering differentially expressed circRNAs. Among these, five were significantly upregulated in CRC while 41 were downregulated (|log2(fold change)| > 1; Q value < 0.05; read count > 40; Figure 1B). We verified all upregulated circRNAs and the top five downregulated circRNAs in 20 pairs of fresh CRC tissues and corresponding non-tumor tissues by qRT-PCR (Figure 1C; Figure S1). We found that circCAMSAP1 (has_circ_0001900, named as circCAMSAP1) had abundant expression and was the most upregulated circRNA in CRC tissues (fold change = 6.4, p < 0.001). Moreover, the preliminary results showed that silencing of circCAMSAP1 using small interfering RNA (siRNA) had a significant inhibitory effect on CRC cell proliferation (Figure 1D). Therefore, we focused on circCAMSAP1 in this study.
Figure 1.
circRNA Expression Profile in CRC and Characterization of circCAMSAP1
(A) Genomic origin of circRNAs (n = 31,557) identified in human CRC tissues. 25,458 were derived from exons, 1,505 were derived from introns, and 4,594 were derived from the others. (B) Heatmap of the differentially expressed circRNAs in eight pairs of human CRC tissues and the matched non-tumor tissues. (C) Fold change of 10 indicated circRNAs expression between 20 CRC tumor tissues and adjacent non-tumor tissues validated by qPCR. N, non-tumor tissues; T, tumor tissues. (D) HCT15 cell numbers in indicated siRNA-treated groups compared to negative control (NC) control group on fifth day after siRNA treatment. (E) Genomic loci of the CAMSAP1 gene and circCAMSAP1. Red arrow indicates the back-splicing of CAMSAP1 exon 2 to exon 3 confirmed by Sanger sequencing. (F) RT-PCR for the analysis of the existence of circCAMSAP1 using the divergent primers and convergent primers in HCT15 cells. GAPDH was used as a control for a linear RNA transcript. (G) qRT-PCR analysis of circCAMSAP1 and CAMSAP1 linear mRNA with or without RNase R treatment. (H) qRT-PCR analysis of the abundance of circCAMSAP1 and CAMSAP1 linear mRNA in HCT15 cells treated with actinomycin D at the indicated time points. (I) Results of cytoplasmic and nuclear mRNA fractionation experiment. GADPH served as a marker of cytoplasmic location, while MALAT1 served as a marker of nuclear location. (J) Representative images for FISH circCAMSAP1 staining in HCT15 cells. The circCAMSAP1 probe was labeled with Cy3 (red); nuclei were stained with DAPI (blue). Scale bars, 5 μm. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SD.
Characterization of circCAMSAP1 in CRC
circCAMSAP1 is back-spliced from two exons (exon 2 and exon 3) of the CAMSAP1 gene (q34.3:135,810,490-135,907,228) and ultimately forms a length of 425 nt. Divergent primers were used to amplify the circular transcripts to confirm the circCAMSAP1 junction (Figure 1E), and the convergent primers were used to detect the linear transcripts. These two pairs of primers were used to amplify the circular and linear transcripts of CAMSAP1 in both complementary DNA (cDNA) and genomic DNA (gDNA) by PCR. The results showed that the circular transcript of CAMSAP1 could only be amplified using the divergent primers in cDNA (Figure 1F). Studies have indicated that circRNAs with internal ribosome entry sites (IREs) and an open reading frame (ORF) have protein translation potentials.21 circCAMSAP1 had no IREs (http://www.circbank.cn/), implicating a lower possibility of protein encoding (http://reprod.njmu.edu.cn/circrnadb). RNase R treatment and a half-life assay further showed that circCAMSAP1 was much more stable than CAMSAP1 linear mRNA (Figures 1G and 1H). Moreover, a nuclear mass separation assay showed that more than 70% of circCAMSAP1 localized in the cytoplasm (Figure 1I), which was further confirmed by fluorescence in situ hybridization (FISH) analysis (Figure 1J).
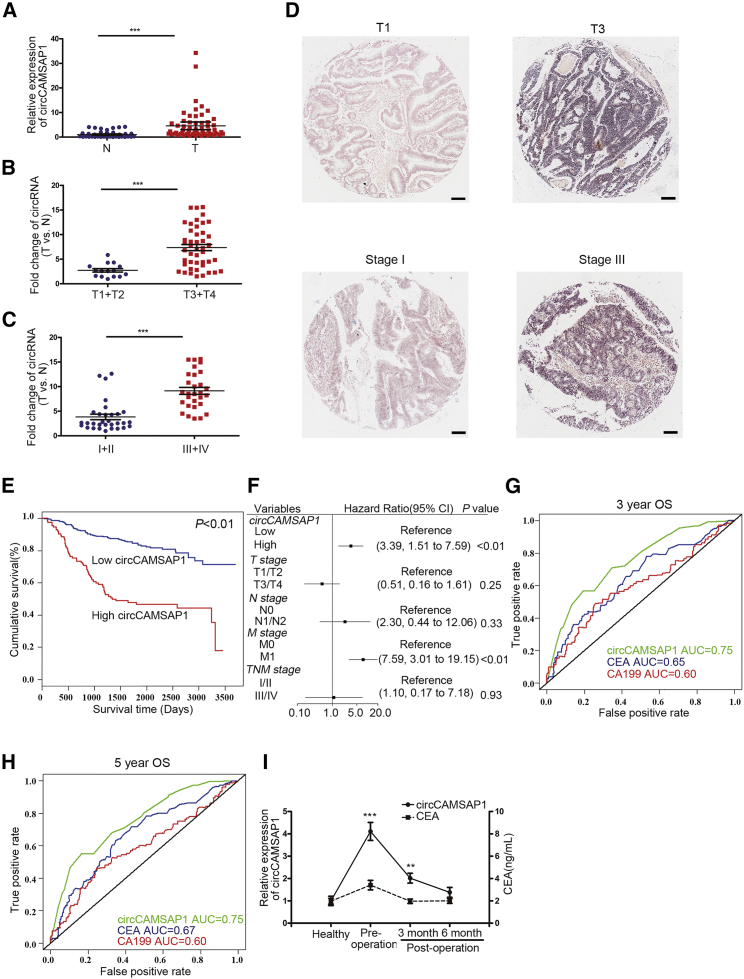
Clinical Implication of circCAMSAP1
We further explored the clinical significance of circCAMSAP1 expression in CRC. As shown in Figure 2A, circCAMSAP1 was significantly upregulated in 60 cases of CRC tumor tissue compared with paired non-tumor tissues (p < 0.001, Figure 2A). Patients with advanced T stage and clinical stage exhibited higher expression of circCAMSAP1 in the tumor tissues (Figures 2B and 2C). Tissue array slides with 424 cases of CRC were subjected to in situ hybridization (ISH) staining to further validate the expression of circCAMSAP1 in CRC tissues. Consistently, patients with advanced T stage and clinical stage exhibited higher circCAMSAP1 expression (Figure 2D; Table S1). More importantly, higher circCAMSAP1 expression in CRC tissues was closely correlated to poorer prognosis of CRC patients (Figure 2E). Multivariate analyses indicated that circCAMSAP1 expression was an independent negative prognostic parameter for CRC patients (hazard ratio [HR], 3.39; 95% confidence interval [CI], 1.51–7.59; p < 0.01; Figure 2F). Furthermore, we found that circCAMSAP1 expression in CRC tissues had a better prediction performance than did CEA and CA19-9 in the prognosis of CRC patients (Figures 2G and 2H). Therefore, we hypothesized that circCAMSAP1 might be detected in blood and has the potential to act as a tumor marker. circCAMSAP1 expression in patient serum was further detected to test the hypothesis. Serum samples from 20 patients with CRC before and after surgery and 20 healthy volunteers were subjected to droplet digital PCR (ddPCR). We found that circCAMSAP1 was enriched in the serum of CRC patients prior to surgery (Figure 2I; Figure S2). Collectively, these results suggested that circCAMSAP1 was potent to serve as a biomarker for CRC diagnosis and prognosis.
Figure 2.
Upregulated circCAMSAP1 Expression in CRC Tissues and Its Correlation with the Prognosis of Patients
(A) qRT-PCR for the expression of circCAMSAP1 in CRC and matched non-tumor tissues (n = 60). N, non-tumor tissues; T, tumor tissues. (B) Comparison of circCAMSAP1 expression between patients with T stage 3–4 (n = 45) and those with T stage 1–2 (n = 15), detected by qRT-PCR. (C) Comparison of circCAMSAP1 expression between patients with clinical stage III–IV (n = 31) and those with clinical stage I–II (n = 29), detected by qRT-PCR. (D) Representative images of in situ hybridization staining of circCAMSAP1 in CRC tissue of T stage T1 and T3 and clinical stage I and III. Scale bars, 100 μm. Original magnification, ×100. (E) Kaplan-Meier analysis of the correlation between circCAMSAP1 expression and overall survival. Kaplan-Meier survival curves for CRC patients with high (n = 113) and low (n = 311) expression of circCAMSAP1, determined by ISH. The optimal survival cut point was determined by X-Tile statistical software. (F) Multivariate regression analysis of overall survival (OS) in CRC patients (bars indicate 95% confidence intervals). (G and H) The receiver operating characteristic (ROC) curves for the prediction performance of circCAMSAP1, CEA, and CA19-9 in the 3-year (G) and 5-year (H) overall survival analysis of CRC patients. (I) The solid line shows the expression of circCAMSAP1 in the serum of healthy people (n = 20) and CRC patients (n = 20) before and after the operation, validated by RT-ddPCR. The dashed line shows the levels of CEA in the serum of healthy people (n = 20) and CRC patients before and after the operation (n = 20). *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SEM.
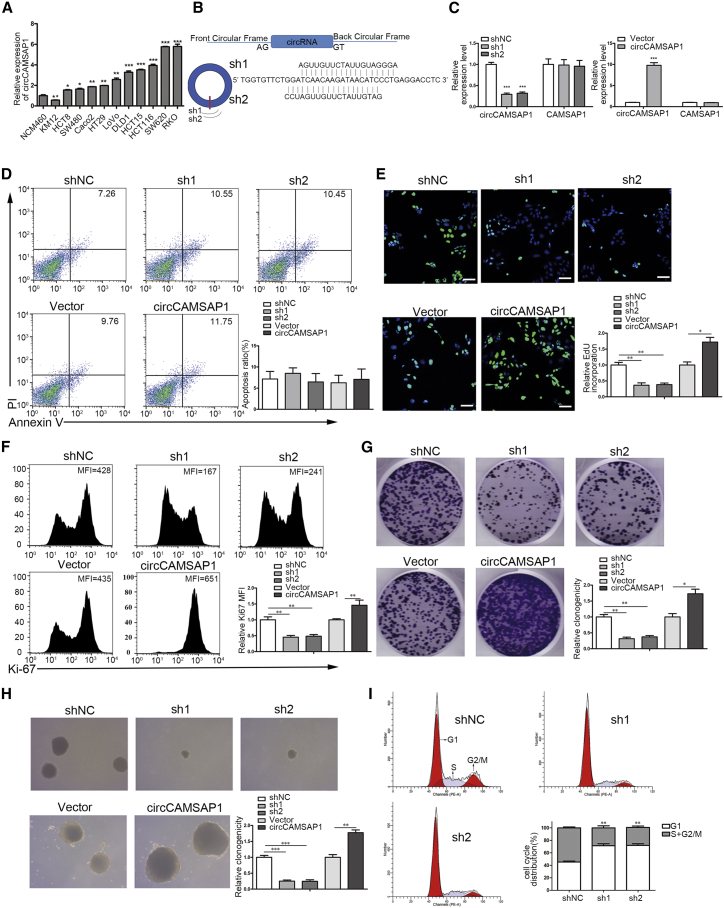
circCAMSAP1 Promotes CRC Cell Proliferation
Next, we investigated the biological functions of circCAMSAP1 in CRC. To choose the optimal cell lines used for silencing and overexpression of circCAMSAP1, we first checked the expression of circCAMSAP1 in various CRC cell lines. HCT15 and DLD1 were chosen as target cell lines due to the moderate expression of circCAMSAP1 (Figure 3A). We constructed circCAMSAP1 knockdown stable cell lines using short hairpin RNAs (shRNAs), including sh1 and sh2, that specifically target the back-spliced junction site of circCAMSAP1 to downregulate the expression of circCAMSAP1, but not the linear CAMSAP1 (Figures 3B and 3C). Additionally, we constructed ectopic circCAMSAP1 overexpression stable cell lines transfected with the full-length cDNA of the circCAMSAP1-specific vector (Figures 3B and 3C). The results of flow cytometry analysis indicated that neither silencing nor overexpressing circCAMSAP1 would affect tumor cell apoptosis (Figure 3D). As shown in Figure 1D, the silencing of circCAMSAP1 had a strong effect on inhibiting CRC cell proliferation (Figure 1D). EdU (Figure 3E), FACS analysis of Ki-67 staining (Figure 3F), plate colony formation (Figure 3G), and soft agar colony formation (Figure 3H) assays were further performed to confirm the effect of circCAMSAP1 on CRC cell proliferation. All of these results showed that knockdown of circCAMSAP1 could remarkably impair cell proliferation of HCT15, while the ectopic overexpression of circCAMSAP1 substantially increased cell proliferation of HCT15. Furthermore, the results of cell cycle progression showed that knockdown of circCAMSAP1 could induce G1 phase cell cycle arrest (Figure 3I). Similar results can also be seen in DLD1 cells (Figure S3). Collectively, the results above suggested that circCAMSAP1 played an important role in the proliferation of CRC cells.
Figure 3.
circCAMSAP1 Promotes the Proliferation of CRC Cells
(A) qRT-PCR analysis for the expression of circCAMSAP1 in normal colonic epithelial cell line (NCM460) and CRC cell lines. (B) Schematic illustration showing shRNAs and circCAMSAP1 overexpression construct. shRNAs including sh1 and sh2 target the back-splice junction of circCAMSAP1. (C) qRT-PCR analysis of circCAMSAP1 and CAMSAP1 RNA expression of circCAMSAP1 knockdown or overexpression of stable HCT15 cell lines, compared to negative control shRNA (shNC) or vector control, respectively. (D) Propidium iodide (PI)/annexin V double staining analysis of apoptosis of HCT15 cells with circCAMSAP1 knockdown or overexpression, compared to shNC or vector control, respectively. Numerical valves denote percentage of positive cells in annexin V+ populations. (E–H) Effect of circCAMSAP1 knockdown or overexpression on the proliferation of HCT15 cells was assessed by EdU (E), FACS analysis of Ki-67 staining (F), plate colony formation (G), and soft agar colony formation (H) assays. Scale bars, 50 μm. (I) FACS cell cycle analysis of HCT15 cells with circCAMSAP1 knockdown, compared to shNC-treated HCT15 cells, using PI DNA staining. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SD.
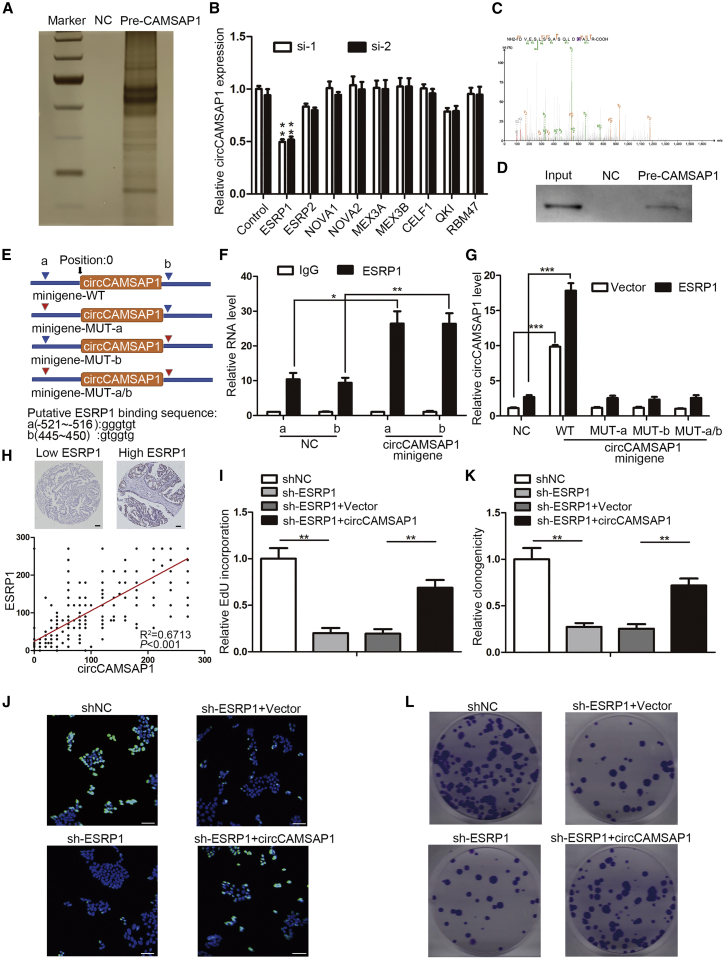
Splicing Factor ESRP1 Promotes circCAMSAP1 Production by Binding to the Flanked Intron Regions of circCAMSAP1
We next explored the mechanism by which circCAMSAP1 is generated. Recent studies showed that the generation of circRNAs is related to alternative splicing, while splicing factors are capable of regulating the formation of circRNA via binding to the flanked intron regions.9,11,23 Therefore, we designed probes targeting flanked intron regions of circCAMSAP1 within pre-CAMSAP1 and performed RNA pull-down assays to detect proteins binding to pre-CAMSAP1. Silver staining showed numerous proteins interacting with pre-CAMSAP1 (Figure 4A). The proteins interacting with pre-CAMSAP1 were then identified using mass spectrometry (MS). Based on the results of MS and previous published studies about RNA splicing factors associated with alternative splicing, tumorigenesis, or circRNAs formation, we focused on the following RNA-binding proteins: ESRP1,11,24 ESRP2,25 NOVA1,26 NOVA2,27 MEX3A,28 MEX3B,28 CELF1,28 QKI,9 and RBM47.29 A series of siRNAs was designed to target the candidate genes above. The results of qRT-PCR showed that only silencing ESRP1 of these genes could noticeably decrease circCAMSAP1 expression, suggesting that ESRP1 could serve as an upstream splicing factor for circCAMSAP1 biogenesis (Figure 4B). MS and western blotting of purified protein confirmed that ESRP1 protein could bind to pre-CAMSAP1 (Figures 4C and 4D). We further tested whether ESRP1-binding sequences (GGT-rich)11,30, 31, 32 on the flanked intron regions were required for the formation of circCAMSAP1. Two putative ESRP1 binding sites were identified, as shown in Figure 4E. Next, we constructed the circCAMSAP1 minigene to conduct an RNA immunoprecipitation (RIP) assay (Figure 4E). As shown in Figure 4F, the enrichment of the two putative ESRP1 binding sites was higher in the anti-ESRP1 group than in the anti-immunoglobulin G (IgG) group, while the transfection of the circCAMSAP1 minigene could still further improve the enrichment (Figure 4F). These results confirmed that ESRP1 could bind to these two putative binding sites on the flanked intron regions. Next, individual mutation minigenes were constructed. Then, the CRC cells with or without ESRP1 overexpression were transfected with wild-type (WT) or individual mutation circCAMSAP1 minigenes. Only the WT circCAMSAP1 minigene could enhance the expression of circCAMSAP1 in both control cells and ESRP1-overexpressed cells (Figure 4G). Within these groups, the ESRP1-overexpressed cells transfected with the WT circCAMSAP1 minigene showed the highest circCAMSAP1 expression level (Figure 4G). These results indicated that both of these two binding sites on upstream and downstream flanked intron regions of circCAMSAP1 were necessary for ESRP1-mediated circCAMSAP1 circularization. Furthermore, we detected the ESRP1 expression on CRC tissues using immunohistochemistry (IHC) staining. The results showed that the expression of circCAMSAP1 was strongly associated with ESRP1 in CRC patients (Figure 4H). Additionally, the sequence of si-1 of ESRP1 was selected to construct shRNA for generating ESRP1 knockdown-stable cell lines. Knockdown of ESRP1 inhibited the proliferation of CRC cells while the effect could be partly rescued by circCAMSAP1 overexpression, as shown by EdU and plate colony formation assays (Figures 4I–4L; Figure S4). Taken together, the results above indicated that ESRP1 could regulate the biogenesis of circCAMSAP1 in CRC cells through binding to the flanked intron regions of circCAMSAP1.
Figure 4.
Splicing Factor ESRP1 Promotes circCAMSAP1 Production by Binding to the Flanked Intron Regions of circCAMSAP1
(A) Silver staining of the purified proteins in pre-CAMSAP1 RNA pull-down experiment. NC, scramble RNA control. Pre-CAMSAP1, proteins obtained by RNA pull-down using probes targeting flanked intron regions of circCAMSAP1 within pre-CAMSAP1. (B) qRT-PCR analysis of circCAMSAP1 expression in HCT15 cells transfected with indicated siRNAs. (C) Mass spectrogram of ESRP1 protein. (D) Immunoblotting of ESRP1 in indicated groups. NC, scramble RNA control. Pre-CAMSAP1, proteins obtained by RNA pull-down using probes targeting flanked intron regions of circCAMSAP1 within pre-CAMSAP1. Input, total protein. (E) Schematic of circCAMSAP1 minigenes with two wild-type (WT) or mutant (MUT) ESRP1 binding sites on the flanked intron regions of circCAMSAP1. (F) ESRP1 RIP-qPCR results using primers targeting the sequences around the putative binding site of ESRP1 in HCT15 cells transfected with or without WT circCAMSAP1 minigene. Values were normalized to the level of background RIP, as detected by an IgG isotype control. (G) qRT-PCR analysis of the circCAMSAP1 expression in HCT15 cells. Cells with or without ESRP1 overexpression were transfection with WT or various MUT circCAMSAP1 minigenes. (H) Correlation of the ESRP1 IHC score and circCAMSAP1 ISH score in the clinical samples of patients with CRC (n = 424, Pearson’s correlation coefficient R and p value are shown). (I–L) Proliferation of ESRP1-silenced HCT15 cells transfected with circCAMSAP1 vector, assessed by EdU (I and J) and plate colony formation (K and L) assays. Scale bars, 50 μm. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SD.
circCAMSAP1 Serves as a Sponge for miR-328-5p
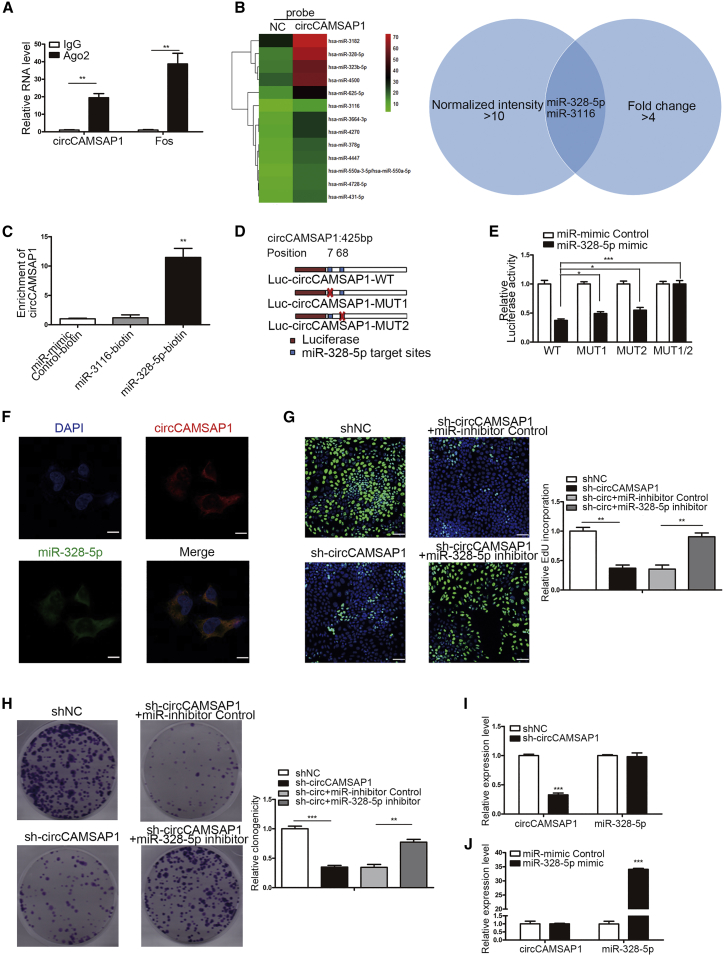
We further explored the underlying molecular mechanism responsible for the effect of circCAMSAP1 on CRC cells. Given that circRNAs have been shown to act as a miRNA sponge in the cytoplasm,19 and circCAMSAP1 is mainly located in the cytoplasm, we hypothesized that circCAMSAP1 might exert its function through sponging certain miRNAs.
It is well known that miRNAs inhibit translation and degrade mRNA in an Ago2-dependent manner by binding to their targets.33 Therefore, to test the hypothesis, we performed an anti-Ago2 RIP assay to pull down RNA transcripts that bind to Ago2. As shown in Figure 5A, the enrichment of circCAMSAP1 was much higher in the anti-Ago2 group compared with the anti-IgG group (Figure 5A), suggesting that circCAMSAP1 was involved in the regulation process of miRNA (RNA-induced silencing complex [RISC] complex). We subsequently conducted a pull-down assay using biotin-labeled probes targeting the circCAMSAP1 junction site to enrich the potential directly binding miRNAs, which was followed by a miRNA microarray. The results of the miRNA microarray showed that miR-328-5p and miR-3116 were significantly more abundant compared with control (fold change > 4; normalized intensity > 10; Table S3; Figure 5B). To further validate whether circCAMSAP1 could sponge these miRNAs, a miRNA pull-down assay was performed to purify the miRNA binding RNA, using biotinylated miRNA mimics (miR-328-5p and miR-3116). The results showed that miR-328-5p rather than miR-3116 could significantly enrich circCAMSAP1 (Figure 5C). Moreover, we generated luciferase constructs harboring the potential target sites and miR-328-5p binding site mutant constructs of circCAMSAP1 (Figure 5D). Overexpression of miR-328-5p by miRNA mimics could remarkably reduce the luciferase activities of a WT reporter but not the mutant ones (Figure 5E). Furthermore, the results of FISH analysis showed that circCAMSAP1 was co-localized with miR-328-5p in the cytoplasm (Figure 5F). Previous studies have reported the tumor suppressive effect of miR-328-5p in different kinds of malignancies.34,35 Similarly, we also found that miR-328-5p could inhibit the proliferation of CRC cells and induce G1 phase arrest (Figure S5). We next performed an in vitro rescue assay to confirm the interaction of circCAMSAP1 and miR-328-5p. As revealed by EdU and plate colony formation assays, the miR-328-5p inhibitor could significantly rescue the proliferation-inhibitory effect induced by circCAMSAP1 knockdown (Figures 5G and 5H; Figure S6). Additionally, circCAMSAP1 and miR-328-5p did not affect the expression of one another (Figures 5I and 5J). Collectively, these results indicated that circCAMSAP1 could enhance the proliferation and tumor growth of CRC through serving as a miR-328-5p sponge.
Figure 5.
circCAMSAP1 Serves as a Sponge for miR-328-5p
(A) RIP assay using an antibody against Ago2, followed by detection of circCAMSAP1 and Fos (positive control). Values were normalized to the level of background RIP, as detected by an IgG isotype control. (B) Results of miRNA array of circCAMSAP1 pull-down products. (C) miRNA biotin pull-down assay using biotin-coupled miR-328-5p or miR-3116 mimics, compared to control miR-mimics. circCAMSAP1 expression levels were determined by qRT-PCR. (D) Schematic of luciferase reporter vectors containing wild-type (WT) or mutant (MUT) putative miR-328-5p binding sites of circCAMSAP1. (E) The luciferase activities of HCT15 co-transfected with WT or MUT circCAMSAP1 luciferase reporter vector and miR-328-5p mimics or control miR mimics. (F) Co-localization of circCAMSAP1 and miR-328-5p in HCT15 cells was evaluated by a FISH assay. Scale bars, 5 μm. (G and H) The sh1-mediated circCAMSAP1 knockdown HCT15 cells were transfected with miR-328-5p inhibitor or control inhibitor. The proliferation of HCT15 cells in indicated groups was assessed by EdU (G) and plate colony formation (H) assays. Scale bars, 50 μm. (I) The expression of miR-328-5p after circCAMSAP1 silencing using sh1. (J) The expression of circCAMSAP1 after treatment with miR-328-5p mimics. **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SD.
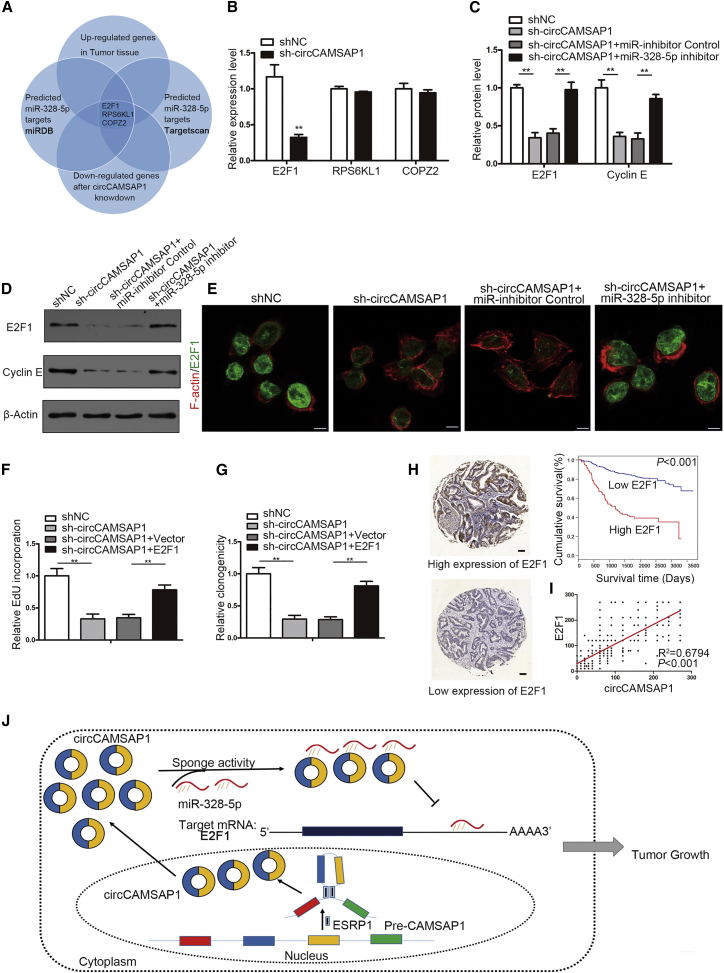
miR-328-5p Suppresses CRC Cell Proliferation through Targeting E2F1
To identify the targets of miR-328-5p, we performed whole-transcriptome deep sequencing on circCAMSAP1 knockdown and control cells. 125 mRNAs were downregulated by circCAMSAP1 knockdown (fold change ≥ 2.00; false discovery rate [FDR] ≤ 0.001; Table S4). Furthermore, through filtering the genes that were upregulated in our sequencing results of CRC tissue (Table S5) and predicted target genes of miR-328-5p using miRDB (http://mirdb.org/cgi-bin/search.cgi) and TargetScan (http://www.targetscan.org/), we identified three candidate target genes of miR-328-5p (E2F1, RPS6KL1, and COPZ2; Figure 6A). The results of qRT-PCR showed that only E2F1 was downregulated after circCAMSAP1 knockdown (Figure 6B). Therefore, we further focused on verifying whether E2F1 was the direct target of miR-328-5p. An anti-Ago2 RIP assay showed that mRNA levels of E2F1 enriched in the anti-Ago2 group were significantly increased compared with the IgG group (Figure S7A). The miRNA biotin pull-down assay revealed that miR-328-5p could combine with the 3′ UTR of E2F1 mRNA (Figure S7B). Furthermore, luciferase assays showed a reduction of luciferase activities when the constructs containing miR-328-5p binding sites were co-transfected with miR-328-5p mimics. Mutations of the binding sites abolished the repression effect (Figure S7C). Moreover, the protein levels of E2F1 were significantly decreased after transfection of miR-328-5p mimics but increased after transfection of miR-328-5p inhibitor (Figure S7D). The similar changes of the expression of Cyclin E, one of the downstream genes of E2F1, can be obtained following transfection of miR-328-5p mimics or inhibitor (Figure S7D). Furthermore, the overexpression of E2F1 could partly rescue the inhibition of miR-328-5p upon cell proliferation (Figures S7E and S7F). Collectively, the results above suggested that miR-328-5p could suppress proliferation of CRC cells through directly suppressing the expression of E2F1.
Figure 6.
circCAMSAP1 Promotes Cell Proliferation via the circCAMSAP1/miR-328-5p/E2F1 Axis
(A) Schematic drawing of the screening procedure of candidate genes. (B) qRT-PCR analysis of the expression of three candidate genes after silencing of circCAMSAP1 using sh1 in HCT15 cells. (C–E) The sh1-mediated circCAMSAP1 silencing HCT15 cells were transfected with control miR inhibitor or miR-328-5p inhibitor. (C and D) The quantitative results (C) of immunoblotting (D) for E2F1, Cyclin E, and β-actin in HCT15 cells as indicated (n = 3). (E) Representative immunofluorescence (IF) images of E2F1 staining in control cells, circCAMSAP1 silencing cells, and circCAMSAP1 silencing cells transfected with miR-328-5p inhibitor or control miR inhibitor (n = 3). Scale bars, 5 μm. (F and G). sh1-mediated circCAMSAP1 knockdown HCT15 cells were transfected with E2F1 overexpression construct or control construct. The proliferation of HCT15 cells was assessed by EdU (F) and plate colony formation (G) assays. (H) Kaplan-Meier analysis of the correlation between E2F1 expression and overall survival. Kaplan-Meier survival curves for CRC patients with high (n = 88) and low (n = 336) expression of E2F1, determined by IHC staining, are shown. The optimal survival cut point was determined by X-Tile statistical software. Scale bars, 100 μm. Original magnification, ×100. (I) Correlation of the E2F1 IHC score and circCAMSAP1 ISH score in the clinical samples of CRC patients (n = 424, Pearson’s correlation coefficient R and p value are shown). (J) Graphical abstract. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. Error bars indicate SD.
circCAMSAP1 Promotes Cell Proliferation via the circCAMSAP1/miR-328-5p/E2F1 Axis
To further validate whether circCAMSAP1 exerts its function via the circCAMSAP1/miR-328-5p/E2F1 axis, we next examined the expression of E2F1 and its downstream gene Cyclin E after knockdown of circCAMSAP1. As shown in Figures 6C–6E, silencing of circCAMSAP1 significantly reduced the expression levels of E2F1 in CRC cells (Figures 6C–6E). In addition, both immunofluorescence (IF) and western blotting revealed that the miR-328-5p inhibitor could significantly reverse the downregulation of E2F1 after silencing of circCAMSAP1 (Figures 6C–6E). More importantly, overexpression of E2F1 restored proliferation-inhibitory effect induced by circCAMSAP1 knockdown, confirming functionally that circCAMSAP1 could promote CRC progression via the miR-328-5p/E2F1 axis (Figures 6F and 6G; Figure S9). Next, we detected the E2F1 expression using IHC staining on tissue array slides of 424 CRC patients. The results showed that higher expression of E2F1 was associated with poorer overall survival (Figure 6H). Moreover, the expression of circCAMSAP1 was strongly associated with E2F1 in CRC patients (Figure 6I), which further supported the circCAMSAP1/miR-328-5p/E2F1 axis. Taken together, these results convincingly indicated that circCAMSAP1 could promote CRC cell proliferation as a sponge of miR-328-5p and functions via the circCAMSAP1/miR-328-5p/E2F1 axis (Figure 6J).
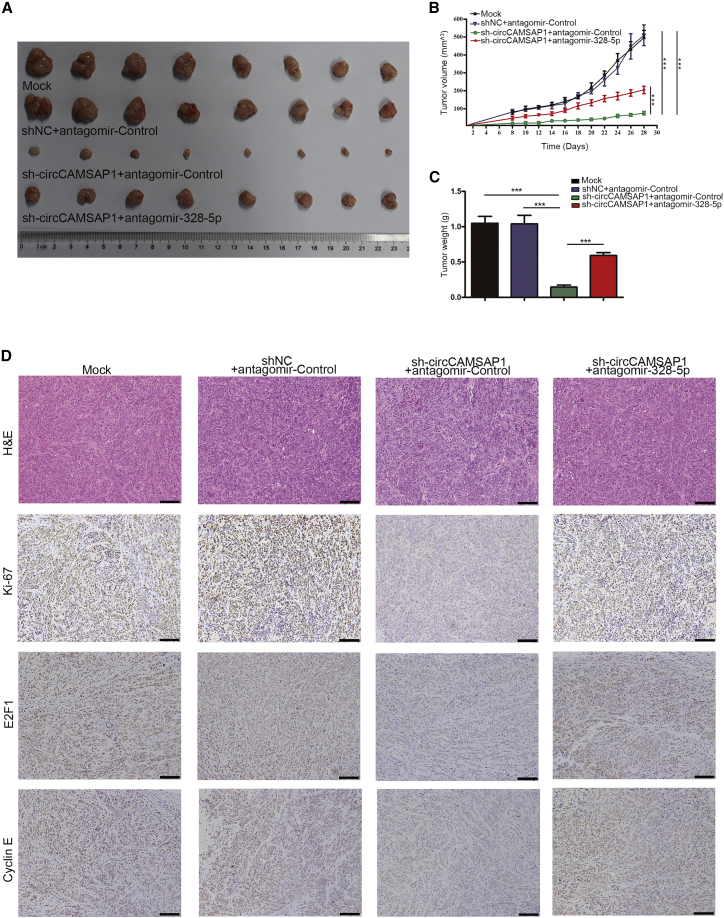
The circCAMSAP1/miR-328-5p/E2F1 Axis Is Essential for CRC Progress In Vivo
To further assess and confirm whether the oncogenic potential of circCAMSAP1 was through the miR-328-5p/E2F1 axis in vivo, we established a xenograft tumor model in nude mice. The results showed that circCAMSAP1 silencing tumors with miR-328-5p inhibition had a significantly larger size and higher weight compared to circCAMSAP1 silencing counterparts, highlighting the ability of inhibition of miR-328-5p to neutralize the tumor-suppressive effect caused by circCAMSAP1 silencing (Figures 7A–7C). Likewise, the results of IHC revealed that tumor tissues collected from the circCAMSAP1 silencing group had lower expression of Ki-67, E2F1, and Cyclin E when compared with the control group, which could be partly counteracted by miR-328-5p inhibition (Figure 7D). Taken together, these results demonstrated that circCAMSAP1 played an essential role in CRC progression in vivo via the miR-328-5p/E2F1 axis.
Figure 7.
The circCAMSAP1/miR-328-5p/E2F1 Axis Is Essential for CRC Progress In Vivo
The sh1-mediated circCAMSAP1 silencing HCT15 cells were treated with antagomir-control or antagomir-328-5p. (A) Images of tumors of indicated groups (n = 8). (B) Tumor growth curves of nude mice with indicated treatments. Tumor volumes were monitored with digital calipers every other day during the time course of 4 weeks. (C) Tumor weights were measured at day 28. (D) H&E staining and IHC staining of Ki-67, E2F1, and Cyclin E in tumors with indicated treatments. Scare bars, 100 μm. Original magnification, ×200. ***p < 0.001 by Student’s t test. Error bars indicate SEM.
Discussion
In the present study, we identified circRNA expression profile in CRC tissues via RNA sequencing (RNA-seq). We demonstrated that circCAMSAP1 was significantly upregulated in CRC tissue and negatively correlated with patients’ survival. We found that circCAMSAP1 expression in CRC tissues had a better prediction performance than did CEA and CA19-9 in the prognosis of CRC patients. Interestingly, circCAMSAP1 was more abundant in the serum of CRC patients prior to surgery, compared to the healthy volunteers and in the patients after surgery. Functionally, circCAMSAP1 promoted tumor growth both in vitro and in vivo. Mechanistically, circCAMSAP1 acted as the sponge of miR-328-5p and upregulated E2F1 expression. More importantly, we found that ESRP1 mediated circCAMSAP1 biogenesis via interaction with its flanked intron regions. Thus, our findings revealed that circCAMSAP1 acts as an oncogene of CRC and could serve as a diagnostic and prognostic biomarker as well as a therapeutic target for CRC.
circRNAs were initially considered as junk products of aberrant RNA splicing.36 This view is now being challenged since increasing evidence shows that circRNAs are abundant in various cell types in an evolutionarily conserved manner.5 In our study, by comparing the sequence of circCAMSAP1 between humans and Mus musculus, we observed 90% of commonality, which revealed that circCAMSAP1 was highly conserved and might possess many important biological functions. Presently, a great number of circRNAs have been reported to play an important role in the progression of different cancer types.19,32 For instance, higher expression of circPRKCI can promote tumor growth in lung adenocarcinoma,19 while upregulated circANKS1B promotes metastasis of breast cancer.32 In the present study, both in vitro and in vivo results clearly demonstrated that circCAMSAP1 was an oncogene in CRC, promoting tumor growth.
The longer half-lives and specific expression patterns enable circRNAs to be robust biomarkers.37,38 Until now, several circRNAs have been reported to be diagnostic and prognostic biomarkers in CRC,12 glioma,39 lung cancer,19 gastric cancer,17 and other malignancies.33,40 In this study, we found that circCAMSAP1 was more abundant in CRC tissues than the adjacent normal tissues, and it correlated with T stage and clinical stage. Patients with higher levels of circCAMSAP1 had shortened overall survival. Additionally, higher expression of circCAMSAP1 was an independent factor for a more unsatisfactory outcome. More interestingly, we found that circCAMSAP1 was abundant in the serum of CRC patients before surgery, while its expression was low in CRC patients after surgery and in normal donors. Studies have reported that circular RNAs could be secreted from tumor cells by exosomes.41,42 Based on RNA-seq and qRT-PCR, one study has reported several circRNAs enriched in exosomes. Within these, circCAMSAP1 was one of the circRNAs that had a higher expression level and circRNA/mRNA ratio in cell-free exosomes than in cells.42 Therefore, it is quite possible that CRC cells could secrete circCAMSAP1 into serum by exosomes, so that a higher level of circCAMSAP1 in the serum of CRC patients before surgery can be detected. However, this still needs further experiments to confirm.
Previous studies revealed that circRNA generation is related to alternative splicing while abnormal splicing was shown to be involved in CRC carcinogenesis.9,11,43 However, the relationships between circRNAs and alternative splicing in CRC remain largely unknown. Previous studies suggested that splicing factors play an essential role in the formation of circRNAs via binding to the flanked intron regions of circRNAs.9,11 In the present study, we identified a splicing factor, ESRP1, which could mediate the biogenesis of circCAMSAP1 via interacting with “GGT-rich” motifs on the flanked intron regions of circCAMSAP1-forming exons. ESRP1 has been reported to stimulate CRC cells growth and promote CRC progression.44 Similarly, we also found that ESRP1 acted importantly in the proliferation of CRC cells, at least partly through promoting circCAMSAP1 biogenesis. Although ESRP1 served as an upstream splicing factor for circCAMSAP1 biogenesis, ESRP1 silence only reduced circCAMSAP1 expression level by about 50%. Therefore, other splicing factors might also participate in the circCAMSAP1 biogenesis process. Previous studies demonstrated that the silence of QKI affected the expression of a large number of circRNAs.9 In accordance with this, we also found that knockdown of QKI reduced circCAMSAP1 expression level by about 20%. Therefore, these findings indicated that the formation of circRNA was a complicated process and might be simultaneously controlled by multiple splicing factors. As the previous study showed that ESRP1 could regulate the expression of circBIRC6 in human embryonic stem cells,11 ESRP1 might regulate the expression of different circRNAs in several disease entities, which needs further investigation.
Increasing evidence showed that circRNAs play an important role in the regulation of gene expression. Due to their abundance and stability, circRNAs can exert biological functions through a miRNA sponge for a longer period of time than long non-coding RNAs (lncRNAs).40 In this study, we showed that circCAMSAP1, one highly expressed oncogene in CRC cells, is a stable circularized transcript, resistant to RNase R digestion, and with a longer half-time. In addition, circCAMSAP1 incorporated into the Argonaute (AGO)-RISC complex in CRC cells. Therefore, we inferred that circCAMSAP1 functions as a miRNA sponge. To date, miRNAs targeted by circRNAs are mostly predicted and selected by bioinformatics analysis, which seems to be blindness. In this study, we performed circRNA pull-down experiments to pull down circCAMSAP1 and used miRNA microarrays to detect the miRNAs that bind to circCAMSAP1. Finally, we found out that miR-328-5p was the circCAMSAP1-binding miRNA. miR-328-5p has been proposed to be a potential tumor suppressor in many cancer types, including nasopharyngeal cancer35 and breast cancer.34 In the present study, we showed that miR-328-5p could inhibit proliferation of CRC cells, in accordance with the previous reports.
miRNAs could bind to the 3′ UTR of mRNA and inhibit the gene function. To better identify the downstream gene regulated by miR-328-5p in CRC cells, we used whole-transcriptome deep sequencing of circCAMSAP1-silenced cells to detect the differential gene expression. While considering the upregulated genes in CRC tissue and predicted targets of miR-328-5p in the online database, we finally found that E2F1 was the target of miR-328-5p in CRC cells. As a cell cycle driver, E2F1 is an important transcription factor and plays an essential role in tumor progression.45,46 Previous studies reported that E2F1 promotes the aggressiveness of human CRC cells.47,48 In our present study, we found that E2F1 silence using siRNA could significantly inhibit cell proliferation of CRC cells (Figure S8). Moreover, we found that the expression of E2F1 positively correlated with circCAMSAP1 while overexpression of E2F1 could reverse the tumor suppressive effect of miR-328-5p.
There are several limitations to this study. First, although we found out the dynamic tendency of circCAMSAP1 expression in the serum of CRC patients before and after surgery, the sample size is relatively small. A larger patient population for future validation is needed. Second, since the circCAMSAP1 could be secreted by CRC cells, it would be interesting to further investigate the role of circCAMSAP1 in the tumor microenvironment. Third, it is also possible that circCAMSAP1 may regulate the development of CRC through other mechanisms such as protein binding, which requires further investigation.
Conclusions
In summary, we showed that circCAMSAP1 biogenesis was mediated by ESRP1 in CRC. circCAMSAP1 could serve as an oncogenic circRNA and promote tumor growth of CRC via the circCAMSAP1/miR-328-5p/E2F1 axis. Our data suggested that circCAMSAP1 could be a diagnostic and prognostic biomarker as well as a potential treatment target for CRC.
Materials and Methods
Additional materials and methods can be found in Supplemental Materials and Methods.
Human Tissue and Cell Lines
The 60 pairs of freshly frozen CRC and adjacent normal tissues and 424 samples of CRC formalin-fixed, paraffin-embedded (FFPE) tissues were obtained from patients who underwent operations at The Sixth Affiliated Hospital of Sun Yat-sen University. Serum from 20 CRC patients before surgery and 3 and 6 months after surgery, and from 20 healthy volunteers was collected for ddPCR detection. The study was approved by the Institutional Review Board of The Sixth Affiliated Hospital of Sun Yat-sen University.
CRC cancer cell lines HT29, SW480, HCT116, SW620, Lovo, SW480, DLD1, Caco2, KM12, RKO, and HCT15 and human normal colonic epithelial NCM460 cells were cultured in DMEM medium (Gibco, NY, USA) supplemented with 10% fetal bovine serum (Gibco, NY, USA). All of the cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
RNA Sequencing, Identification, and Quantification of Human circRNAs
Total RNA was extracted from eight pairs of freshly frozen CRC tissues. The RNA quality was checked by Agilent 2200 (Agilent Technologies, USA). RNA was treated with a RiboMinus eukaryote kit (QIAGEN, Valencia, CA, USA) to delete ribosomal RNA, followed by the cDNA library construction. Next, deep sequencing was performed with an Illumina HiSeq 3000 (Illumina, San Diego, CA, USA). The clean reads were aligned to the reference genome (GRCH37.p13 NCBI). Unmapped reads were collected to identify the circRNAs. Reads that mapped to the circRNA junction (with an overhang of at least 6 nt) were counted for each candidate.
FISH Assay
The FISH assay was performed to detect the location of circCAMSAP1, using a FISH kit (c10910, RiboBio, Guangzhou, China) according to the manufacturer’s protocol. The hybridization was performed with Cy3-labeled circCAMSAP1 probes and FAM-labeled miR-328-5p probes (GenePharma, Shanghai, China). Briefly, cells were seeded in a glass-bottom dish overnight, after which the cells were incubated with pre-hybridization solution at 37°C for 30 min. Probes were dissolved in hybridization solution at a concentration of 20 μM and added to slides and hybridized overnight. The slides were washed by 4× SSC (saline sodium citrate) in 0.1% Tween 20 for 5 min three times, 2× SSC for 5 min once, and 1× SSC for 5 min once. Then, all slides were incubated with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min at room temperature. The results were analyzed by using a confocal microscopy. The probe sequences are shown in Table S2.
RNA Immunoprecipitation
Anti-Ago2 (#03-110, Millipore) and anti-ESRP1 (#21045-1-AP, Proteintech) were used for the RIP assay for Ago2 and ESRP1, respectively. RIP was performed using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Merck, Germany) according to the manufacturer’s guidelines. The isolated RNA was purified, followed by qRT-PCR. The enrichment values were normalized to the level of background RIP, as detected by IgG isotype control.
circRNA Pull-Down
Biotin-labeled circCAMSAP1 probes and control probes were synthesized by Sangon Biotech (Shanghai, China). The biotin-labeled probes were used for circRNA pull-down as mentioned previously, with some modifications.49 In brief, 1 × 107 cells were cross-linked by 1% formaldehyde for 30 min and then lysed in co-immunoprecipitation (coIP) buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM ethylenediaminetetraacetate acid, 0.5% Nonidet P-40, and 5 g/mL aprotinin).50 The mixture was sonicated at high amplitude for 30 cycles of 30 s (on/off) pulses. The cell lysis was incubated with a circCAMSAP1 probe-streptavidin beads (Life Technologies, CA, USA) mixture overnight at 37°C. Then, the beads were washed and incubated with proteinase K. Finally, TRIzol reagent was added to the bead mixture for RNA extraction, followed by miRNA microarray (ArrayStar, Rockville, MD, USA) or qRT-PCR. The probe sequences are shown in Table S2.
Western Blotting
The CRC cells were collected, washed, and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer. Then, the concentration of protein was detected by a bicinchoninic acid (BCA) kit (CWBio, Beijing, China). After that, the equal amounts of protein were separated on 10% SDS-PAGE and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Schwalbach, Germany). The membrane was then blocked with Tris-buffered saline with Tween 20 (TBST) buffer containing 5% skim milk powder and incubated with corresponding primary antibodies at 4°C overnight. The primary antibodies used were anti-E2F1 (1:1,000, #ab179445, Abcam), anti-Cyclin E (1:500, #sc-247, Santa Cruz Biotechnology), anti-ESRP1 (1:1,000, #ab107278, Abcam), and anti-β-actin (1:25,000, #ab49900, Abcam). Then, the membrane was washed with TBST three times and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (anti-rabbit, 7074S; anti-mouse, 7076S; Cell Signaling Technology, 1:5,000) for 1 h at room temperature, after which the bands were visualized using Pierce enhanced chemiluminescence (ECL) western blotting substrate (Thermo Fisher Scientific, MA, USA).
IHC
IHC was performed on tissue microarray (TMA) or FFPE tissue sections as described previously.51 The primary antibodies used were anti-ESRP1 (1:100, #ab107278, Abcam), anti-E2F1 (1:100, #ab179445, Abcam), anti-Cyclin E (1:100, #sc-247, Santa Cruz Biotechnology), and anti-Ki-67(1:50, #ab833, Abcam). Tissue sections were incubated with primary antibodies at 4°C overnight and then incubated with secondary antibody. DAB complex was used as the chromogen. The nuclei were counterstained with hematoxylin. All sections were scored as follows: proportion score: 0–100 (1%–100%); intensity score: 0, negative; 1, weak; 2, intermediate; 3, strong; total score = proportion score × intensity score.51
Xenograft Tumor Model
The animal assay was approved by the Committee for Animal Care and Use of Sun Yat-sen University. For the xenograft tumor model, 4-week-old BALB/c nude mice were randomly divided into four groups (n = 8 for each group): HCT15 cells, HCT15 cells with negative control shRNA (shNC) and antagomir-control, HCT15 cells with circCAMSAP1 knockdown and antagomir-control, and HCT15 cells with circCAMSAP1 knockdown and antagomir-328-5p. Cells were inoculated subcutaneously into nude mice (5 × 106/0.2 mL of PBS/each). Tumor volumes were measure every 2 days by digital calipers. After 4 weeks, the mice were sacrificed and the weight of the tumors was measured. The tumor tissues were harvested, followed by hematoxylin and eosin (H&E) and IHC staining.
Data Availability
RNA-seq data have been submitted to the GEO database (GEO: GSE138202).
Statistical Analysis
Statistical analysis was conducted using SPSS 19.0 (IBM, SPSS, Chicago, IL, USA). The differences among groups were tested using Student’s t test or one-way ANOVA. Kaplan-Meier plots and log-rank tests were used for survival analysis. The univariate and multivariate analyses were analyzed by the Cox proportional hazards model. The correlations were analyzed using Pearson’s correlation coefficients. p values <0.05 were used as significance criteria.
Author Contributions
C.Z., X.-r.W., H.-s.L., and F.-w.W. contributed to study design and drafting of the manuscript. T.H., Z.-x.L., N.L, X.-w.H., and X.-b.Z. performed the experiments. X.-j.W. and D.X. contributed to the manuscript review. X.-r.W. and P.L. supervised the study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Key R&D Program of China (no. 2017YFC1308800), the National Natural Science Foundation of China (no. 81870383), the Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (No. 2018GZR0201005), the Guangdong Natural Science Foundation (no. 2017A030313785), the Science and Technology Planning Project of Guangzhou City (no. 201804010014), and the Science and Technology Planning Project of (no. 2015B020229001 and no. 20160916). We thank Novel Bioinformatics Ltd., Co. for the support of bioinformatics analysis.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.12.008.
Contributor Information
Xian-rui Wu, Email: wuxianr5@mail.sysu.edu.cn.
Ping Lan, Email: lanping@mail.sysu.edu.cn.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Jeon J., Du M., Schoen R.E., Hoffmeister M., Newcomb P.A., Berndt S.I., Caan B., Campbell P.T., Chan A.T., Chang-Claude J. Determining risk of colorectal cancer and starting age of screening based on lifestyle, environmental, and genetic factors. Gastroenterology. 2018;154:2152–2164.e19. doi: 10.1053/j.gastro.2018.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 8.Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H., Huang X., Wang J., Yang L., Kong Y., Gao G., Zhang L., Chen Z.S., Xie X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer. 2019;18:23. doi: 10.1186/s12943-019-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Floris G., Zhang L., Follesa P., Sun T. Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 2017;54:5156–5165. doi: 10.1007/s12035-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 16.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rong D., Lu C., Zhang B., Fu K., Zhao S., Tang W., Cao H. circPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol. Cancer. 2019;18:25. doi: 10.1186/s12943-019-0958-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Yang F., Fang E., Mei H., Chen Y., Li H., Li D., Song H., Wang J., Hong M., Xiao W. Cis-acting circ-CTNNB1 promotes β-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 2019;79:557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 19.Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., Xu W., Zhang E., Wang J., Fang T. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 20.Chen S., Huang V., Xu X., Livingstone J., Soares F., Jeon J., Zeng Y., Hua J.T., Petricca J., Guo H. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843.e22. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong W., Dai Z.H., Liu F.C., Guo X.G., Ge C.M., Ding J., Liu H., Yang F. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167. doi: 10.1016/j.ebiom.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2019 doi: 10.1038/s41418-019-0423-5. Published online September 30, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayakawa A., Saitoh M., Miyazawa K. Dual roles for epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) in cancer progression. Adv. Exp. Med. Biol. 2017;925:33–40. doi: 10.1007/5584_2016_50. [DOI] [PubMed] [Google Scholar]
- 26.Ludlow A.T., Wong M.S., Robin J.D., Batten K., Yuan L., Lai T.P., Dahlson N., Zhang L., Mender I., Tedone E. NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer. Nat. Commun. 2018;9:3112. doi: 10.1038/s41467-018-05582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek S., Oh T.G., Secker G., Sutton D.L., Okuda K.S., Paterson S., Bower N.I., Toubia J., Koltowska K., Capon S.J. The alternative splicing regulator Nova2 constrains vascular Erk signaling to limit specification of the lymphatic lineage. Dev. Cell. 2019;49:279–292.e5. doi: 10.1016/j.devcel.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Chatterji P., Rustgi A.K. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol. Med. 2018;24:490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rokavec M., Kaller M., Horst D., Hermeking H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci. Rep. 2017;7:4687. doi: 10.1038/s41598-017-04234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warzecha C.C., Jiang P., Amirikian K., Dittmar K.A., Lu H., Shen S., Guo W., Xing Y., Carstens R.P. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dittmar K.A., Jiang P., Park J.W., Amirikian K., Wan J., Shen S., Xing Y., Carstens R.P. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng K., He B., Yang B.B., Xu T., Chen X., Xu M., Liu X., Sun H., Pan Y., Wang S. The pro-metastasis effect of circANKS1B in breast cancer. Mol. Cancer. 2018;17:160. doi: 10.1186/s12943-018-0914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 34.Luo T., Yan Y., He Q., Ma X., Wang W. miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by targeting RAGE. Oncol. Rep. 2018;39:2906–2914. doi: 10.3892/or.2018.6353. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S., Li P., Zhao L., Xu L. LINC00210 as a miR-328-5p sponge promotes nasopharyngeal carcinoma tumorigenesis by activating NOTCH3 pathway. Biosci. Rep. 2018;38 doi: 10.1042/BSR20181168. BSR20181168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cocquerelle C., Mascrez B., Hétuin D., Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 37.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hang D., Zhou J., Qin N., Zhou W., Ma H., Jin G., Hu Z., Dai J., Shen H. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7:2783–2791. doi: 10.1002/cam4.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J., Ye J., Zhang L., Xia L., Hu H., Jiang H., Wan Z., Sheng F., Ma Y., Li W. Differential expression of circular RNAs in glioblastoma multiforme and its correlation with prognosis. Transl. Oncol. 2017;10:271–279. doi: 10.1016/j.tranon.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Chen R.X., Wei W.S., Li Y.H., Feng Z.H., Tan L., Chen J.W., Yuan G.J., Chen S.L., Guo S.J. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 2018;24:6319–6330. doi: 10.1158/1078-0432.CCR-18-1270. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Li Z., Jiang P., Peng M., Zhang X., Chen K., Liu H., Bi H., Liu X., Li X. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J. Exp. Clin. Cancer Res. 2018;37:177. doi: 10.1186/s13046-018-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J., Li H., Shen S., Sun L., Yuan Y., Xing C. Alternative splicing events implicated in carcinogenesis and prognosis of colorectal cancer. J. Cancer. 2018;9:1754–1764. doi: 10.7150/jca.24569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fagoonee S., Picco G., Orso F., Arrigoni A., Longo D.L., Forni M., Scarfò I., Cassenti A., Piva R., Cassoni P. The RNA-binding protein ESRP1 promotes human colorectal cancer progression. Oncotarget. 2017;8:10007–10024. doi: 10.18632/oncotarget.14318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedele M., Visone R., De Martino I., Troncone G., Palmieri D., Battista S., Ciarmiello A., Pallante P., Arra C., Melillo R.M. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 46.Sun C.C., Li S.J., Hu W., Zhang J., Zhou Q., Liu C., Li L.L., Songyang Y.Y., Zhang F., Chen Z.L. Comprehensive analysis of the expression and prognosis for E2Fs in human breast cancer. Mol. Ther. 2019;27:1153–1165. doi: 10.1016/j.ymthe.2019.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Fang Z., Gong C., Liu H., Zhang X., Mei L., Song M., Qiu L., Luo S., Zhu Z., Zhang R. E2F1 promote the aggressiveness of human colorectal cancer by activating the ribonucleotide reductase small subunit M2. Biochem. Biophys. Res. Commun. 2015;464:407–415. doi: 10.1016/j.bbrc.2015.06.103. [DOI] [PubMed] [Google Scholar]
- 48.Fang Z., Gong C., Yu S., Zhou W., Hassan W., Li H., Wang X., Hu Y., Gu K., Chen X. NFYB-induced high expression of E2F1 contributes to oxaliplatin resistance in colorectal cancer via the enhancement of CHK1 signaling. Cancer Lett. 2018;415:58–72. doi: 10.1016/j.canlet.2017.11.040. [DOI] [PubMed] [Google Scholar]
- 49.Zeng K., Chen X., Xu M., Liu X., Hu X., Xu T., Sun H., Pan Y., He B., Wang S. circHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng K., Wang Z., Ohshima K., Liu Y., Zhang W., Wang L., Fan L., Li M., Li X., Wang Y. BRAF V600E mutation correlates with suppressive tumor immune microenvironment and reduced disease-free survival in Langerhans cell histiocytosis. OncoImmunology. 2016;5:e1185582. doi: 10.1080/2162402X.2016.1185582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been submitted to the GEO database (GEO: GSE138202).