Abstract
Chloroplasts are photosynthetic plant organelles descended from a bacterial ancestor. The vast majority of chloroplast proteins are synthesized in the cytosol and then imported into the chloroplast post-translationally. Translocation complexes exist in the organelle's outer and inner envelope membranes (termed TOC and TIC, respectively) to facilitate protein import. These systems recognize chloroplast precursor proteins and mediate their import in an energy-dependent manner. However, many unanswered questions remain regarding mechanistic details of the import process and the participation and functions of individual components; for example, the cytosolic events that mediate protein delivery to chloroplasts, the composition of the TIC apparatus, and the nature of the protein import motor all require resolution. The flux of proteins through TOC and TIC varies greatly throughout development and in response to specific environmental cues. The import process is, therefore, tightly regulated, and it has emerged that the ubiquitin-proteasome system (UPS) plays a key role in this regard, acting at several different steps in the process. The UPS is involved in: the selective degradation of transcription factors that co-ordinate the expression of chloroplast precursor proteins; the removal of unimported chloroplast precursor proteins in the cytosol; the inhibition of chloroplast biogenesis pre-germination; and the reconfiguration of the TOC apparatus in response to developmental and environmental signals in a process termed chloroplast-associated protein degradation. In this review, we highlight recent advances in our understanding of protein import into chloroplasts and how this process is regulated by the UPS.
Keywords: chloroplasts, organelle biogenesis, plastid, protein import, protein translocation, ubiquitin-proteasome system
Introduction
Chloroplasts are organelles found within plants and algae which house a range of essential metabolic and biosynthetic functions, including photosynthesis [1,2]. Chloroplasts are endosymbiotically derived organelles, and consequently, contain a functional genome interpreted by eubacterial-type transcription and translation machineries, and possess a double-membraned envelope [3]. Over the course of plant evolution, roughly 98% of the endosymbiont's protein-coding sequences were either lost or relocated to the nuclear genome by the process of endosymbiotic gene transfer [4]. Approximately 100 genes are retained in the chloroplast, and these encode vital factors such as core genetic components [5]. The remaining 2000–3000 chloroplast proteins are encoded in the nucleus and imported post-translationally from the cytosol [6–8]. Chloroplasts are fully integrated cellular components that participate in bidirectional communication with the nucleus to co-ordinate gene expression for organelle biogenesis, photosynthesis, and plastid metabolism [9–11].
Chloroplast-localized proteins are typically synthesized as precursors, each with an N-terminal targeting signal called a transit peptide (TP), and delivered to the chloroplast surface by cytosolic chaperones. Entry into the chloroplast is achieved via translocon complexes in the outer (TOC) and inner (TIC) envelope membranes, which recognize the TP and facilitate transport into the stroma. The TPs of translocated precursor proteins (pre-proteins) are then cleaved off by the stromal processing peptidase (SPP) [12], before final folding and assembly in the stroma, or onward routing to the thylakoids [13,14] or the inner envelope membrane (IEM) [15–17].
It has been well established that the flux of protein import through TOC–TIC can vary greatly according to the developmental stage, environmental cues, or stress conditions [18–21]. The import process needs to be adequately regulated to maintain an optimally functioning chloroplast proteome [22]. The cytosolic ubiquitin-proteasome system (UPS) has emerged recently as an important regulator of chloroplast protein import [23]. The UPS is a eukaryotic protein-degrading mechanism that is involved in numerous protein homeostasis processes in the cell [24]. As a bacterially derived organelle, the chloroplast apparently contains no internal UPS machinery. Instead, ∼20 proteases of prokaryotic origin, notably Clp and FtsH enzymes, act to maintain internal protein homeostasis [18]. Alternatively, under conditions of stress or senescence, the entirety, or part, of the organelle may be degraded by autophagy [25–28]. However, autophagy and degradation by internal proteases are beyond the scope of this review, as indeed is the routing of proteins inside the chloroplast. Here, we will focus on the major events in the cytosol and at the chloroplast envelope membranes. The TOC–TIC system has been extensively reviewed elsewhere [2,6–8,29], and so our aim here is to summarize concisely recent advances. We endeavour to provide an overview of the mechanisms and components of the TOC–TIC apparatus, and to describe the several pathways through which protein import is regulated by the UPS.
Protein targeting to the chloroplast surface
Chloroplast TPs possess remarkably diverse amino acid sequences. This may reflect their need to accommodate varying domains for interaction with cytosolic, chloroplast membrane, and stromal chaperones and sorting factors [30], or with different plastid types [31]. Cytosolic chaperones may facilitate the navigation of pre-proteins to the organelle and maintain an unfolded conformation suitable for import [32]. Two cytosolic systems are reported to guide pre-proteins, though their mechanistic details and physiological significance both require clarification. Hsp90 has been proposed to operate with Hsp70/Hsp90-organizing protein (Hop) and the immunophilin FKBP73 to deliver pre-proteins to the outer envelope membrane (OEM) (Figure 1), where it may dock at Toc64 [33,34]; however, such delivery to Toc64 may not be necessary for protein import [35,36]. Alternatively, Hsp70 (Figure 1) has been shown to act with an undefined 14-3-3 protein to recognize phosphorylated TPs and deliver them to translocation complexes [37]; however, mutation of the relevant phosphorylation site did not impair chloroplast targeting [38].
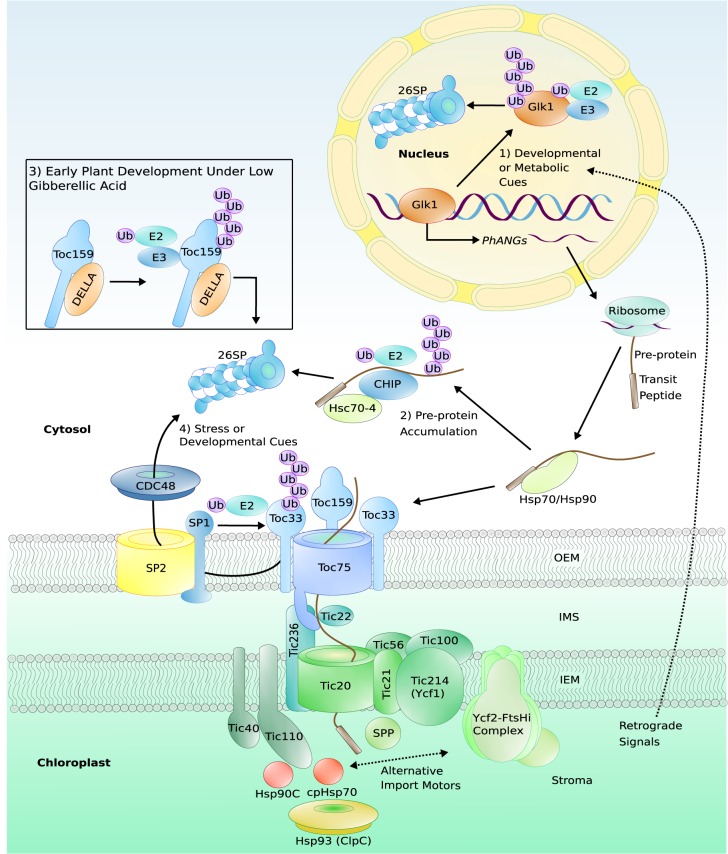
Figure 1. An overview of protein import into the chloroplast and its regulation by the UPS.
Nucleus-encoded chloroplast proteins are synthesized in the cytosol as pre-proteins and post-translationally imported into the chloroplast. The pre-protein carries an N-terminal transit peptide which holds guidance information and initially allows interaction with cytosolic chaperones (e.g. Hsp70, Hsp90). The pre-protein is then recognized by receptor GTPases in the OEM, Toc33 and Toc159 (which exist in different isoforms with varying client specificity, and in Arabidopsis, these are termed Toc34 and Toc132/-120/-90, respectively). The receptors heterodimerize to allow the pre-protein to pass through the Toc75 pore into the IMS. Passage through the IEM is mediated by Tic20, which is reported to be part of the 1 MDa TIC complex containing Tic21, Tic56, Tic100, and Tic214 (Ycf1). Completion of translocation into the stroma is powered by an ATP-dependent import motor, which may be composed of stromal molecular chaperones (e.g. cpHsp70, Hsp90C) or a 2 MDa Ycf2–FtsHi complex. The Tic40 and Tic110 proteins are also involved in the import process, and may operate downstream in conjunction with stromal chaperones. Hsp93 (ClpC) has been proposed to perform a quality-control function at the point of import, or to act in the import motor. Upon arrival in the stroma, the transit peptide is cleaved from the pre-protein by the SPP. The UPS regulates protein import in a variety of ways: (1) The transcription factor Glk1, which regulates the expression of pre-protein-encoding PhaNGs, may be degraded by the UPS in response to unknown retrograde signals (from the chloroplast to the nucleus) that report on developmental or metabolic cues. (2) Accumulation of pre-proteins in the cytosol may trigger their UPS degradation to prevent the formation of cytotoxic aggregates, and this is mediated by the chaperone Hsc70-4 and the E3 ligase CHIP. (3) Before germination, DELLA factors repress chloroplast biogenesis under low gibberellic acid conditions by binding to Toc159 and triggering its UPS degradation. (4) During stress or particular phases of development, the CHLORAD system directly targets the TOC apparatus for proteolysis, with ubiquitination being mediated by the E3 ligase SP1. The targeted TOC proteins are retrotranslocated from the membrane via the channel protein SP2, using motive force provided by the cytosolic AAA+ ATPase CDC48. Note that the Toc33 and Toc159 receptor isoforms are depicted in the model here due to the known role of CHLORAD in suppressing the import of photosynthetic pre-proteins in response to abiotic stress [112], and because such photosynthetic pre-proteins are the primary clients of these isoforms [2]; however, all of the TOC receptor isoforms are likely substrates of SP1 at some stage, as revealed for example by the analysis of sp1 plants during de-etiolation [109]. Dashed lines indicate uncertainty. Abbreviations: E2, E2 conjugase; E3, E3 ligase; 26SP, 26S proteasome; Ub, ubiquitin; SP, suppressor of ppi1; CDC48, cell division cycle 48; TOC, translocon at the outer envelope membrane of chloroplasts; TIC, translocon at the inner envelope membrane of chloroplasts; OEM, outer envelope membrane; IMS, intermembrane space; IEM, inner envelope membrane; SPP, stromal processing peptidase; GLK1, Golden2-like1; CHIP, C-terminus of Hsc70-interacting protein; PhANGs, photosynthesis-associated nuclear genes.
Apart from the canonical, TP-mediated TOC–TIC import route, there are several further targeting pathways that deliver chloroplast proteins; these are less well understood and thought to serve only several hundred proteins [2]. Proteins of the OEM typically do not possess a TP, with targeting information instead residing within a transmembrane domain [39,40]. An interesting exception is the β-barrel protein Toc75, which is inserted with the aid of a bipartite targeting peptide, the first part of which is a canonical TP [41,42]. It has recently emerged that other β-barrel proteins may insert with the aid of a TP [43], and those that do not may be targeted to the chloroplast by their penultimate β-barrel strand [44] via components of the TOC apparatus [43]. Lastly, there exist at least two non-canonical pathways of chloroplast protein targeting, which deliver internal proteins lacking an N-terminal signal in TOC-independent fashion [45], or involve passage through the endomembrane system [46].
Protein import
The outer envelope membrane
In the TOC complex, Toc33 and Toc159 are receptors that possess cytosol-projecting GTPase domains that bind to the TPs of pre-proteins [2]. In higher plants, both receptors are encoded by gene families and so exist in multiple isoforms (the associated nomenclature is based on their molecular masses in kilodaltons). Members of the Toc33 family have a relatively simple architecture, with a single C-terminal membrane-spanning domain [47] and an N-terminal GTPase domain [8]. Members of the Toc159 family share a similar GTPase domain [48], located centrally, and have a large C-terminal membrane-anchoring domain [8]. Toc159 isoforms typically also have an N-terminal, intrinsically disordered acidic domain, which may act to convey recognition specificity [49,50].
Pre-proteins are translocated through the OEM via a channel made by the β-barrel protein Toc75 (Figure 1). Toc75 belongs to the bacterially-descended Omp85 protein superfamily [51]. A feature of this family is a soluble N-terminal polypeptide transport-associated (POTRA) domain [52], which in the case of Toc75 extends into the intermembrane space (IMS) and performs a proposed chaperone-like role [53]. The Toc75 channel has been found to reach a maximum diameter of 30 Å [54]. The stoichiometry of the Toc33 : Toc75 : Toc159 core complex has been determined to be in the range of 4 : 4 : 1 [55] to 3 : 3 : 1 [56], and so the channel may be formed by multiple copies of the Toc75 protein [57].
Initial TP interaction with the GTPase domains of Toc159 and Toc33 is transient and energy independent [58], potentially allowing for rapid and sequential interaction with pre-proteins [59]. Later, the TP interacts with the POTRA domain of Toc75 and the soluble IMS protein Tic22, before GTP hydrolysis [59]. While GTP hydrolysis at both receptors is not necessary for protein import in vivo [60,61], it is required for successful protein translocation in vitro [59], reinforcing the notion that the GTPase receptors are the first points of contact [7,8,62]. The TP may bind simultaneously to Toc33 and Toc159, as each preferentially binds to a distinct region of the peptide [63]. A bound TP may then encourage heterodimer formation between Toc33 and Toc159, as well as GTP hydrolysis [64], leading to an activated translocon conformation which the pre-protein can pass through [8].
The intermembrane space
In the IMS, Tic22 is suggested to act as a chaperone [65,66] and facilitate pre-protein delivery to the TIC complex at the IEM [67] (Figure 1). A recent study identified the IEM protein Tic236 as part of a 1.25 MDa TOC–TIC supercomplex [68]. Tic236 was suggested to provide a physical link between the TOC and TIC complexes, through its anchorage in the IEM, where it associates with Tic20, and its interaction with the POTRA domain of Toc75 [68] (Figure 1). However, a study of its maize orthologue, defective kernel5 (DEK5), identified functions in envelope biogenesis [69]. DEK5 was suggested to mediate the insertion of β-barrel proteins involved in protein import and metabolite transport, in accordance with the fact that it shares homology with the bacterial TamB protein [69]. A distinct role for Tic236 in protein import was proposed based on the observation that tic236 mutant chloroplasts show reduced protein import but no change in the abundance of TOC proteins [68]. In contrast, dek5 mutants displayed a reduction in TOC protein abundance, and thus the impact on protein import was interpreted to be a secondary effect [69]. Ultrastructure analysis revealed a reduction in the proportion of envelope relative to other chloroplast compartments in dek5 [69], but a similar analysis was not done for tic236 plants.
The inner envelope membrane
The recent discovery of a 1 MDa TIC complex [70] has led to reconsideration of previously proposed models for the operation of the TIC machinery [6]. The 1 MDa complex consists of Tic214 (encoded by the chloroplast gene ycf1), Tic100, Tic56, Tic20, and Tic21 [70] (Figure 1). It is expected that the translocon channel is formed by Tic20, which has four membrane-spanning α-helical domains [71]. Three copies of Tic20 could theoretically exist within the 1 MDa complex [70], so a pore size of up to 30 Å has been predicted [57]. Tic21 has a similar structure to Tic20 and may function in a complementary way [72].
The originally identified components of the TIC import machinery include the proteins Tic110, Tic40, Tic20, and Tic21 (Figure 1). The functional relationship between Tic110 and Tic40 and the 1 MDa complex is unclear, but it may be that the former are recruited to import complexes during later stages of import to co-ordinate chaperone functions [6] and/or are required for the import of only some pre-proteins [73]. Stromal chaperones co-operate with Tic110 and Tic40 to facilitate pre-protein import and subsequent folding [74,75]. Translocation into the stroma is driven by an energy-dependent process [76] and several chaperones, including cpHsp70, Hsp90C, and Hsp93, have been implicated in the provision of the motive force [8] (Figure 1). However, the exact roles of these chaperones require clarification. While cpHsp70 has been strongly linked to the role of the main protein import motor [77–79], Hsp90C was also found to be essential for protein translocation [80]. A motor function has also been proposed for Hsp93 (ClpC) [81,82], although recent data showing that it interacts with the ClpP proteolytic subunit at the envelope (an interaction long known to occur in other contexts [83]) imply that it works in a protein quality-control process at the point of import [84,85].
The situation was further complicated by the recent identification of a 2 MDa Ycf2–FtsHi protein complex that was proposed to act as an import motor associated with the 1 MDa TIC complex [86] (Figure 1). In view of the evidence supporting 30 Å pore diameters for the import channels [57], it was suggested that such a powerful ATPase might be specifically recruited to handle the import of recalcitrant or tightly-folded proteins [87]. It seems plausible that cpHsp70 is the general import motor [77–79], with the energetically more expensive Ycf2–FtsHi complex being utilized only under specific conditions.
The 1 MDa TIC complex remains an enigmatic discovery in need of further research. It was the first import complex to contain a chloroplast-encoded protein (Tic214/Ycf1), and this component is notably absent from the family Poaceae [2]. The complex has been proposed to act as a general TIC translocon at the IEM, and in support of this notion, it co-purifies with TOC proteins [70], an interaction that could in principle be mediated by Tic236. Knock-out mutants of ycf1 are embryo lethal, and tic100 and tic56 mutants display chlorotic phenotypes [70] typical of impaired chloroplast protein import. However, analysis of tic56 mutants attributed the observed phenotypes to defects in ribosome assembly, and revealed no impairment of protein import [88–91], a result shared when ycf1 translation was repressed by the specific plastid ribosomal inhibitor spectinomycin [92]. Tic214 has also been found to have functions in the biogenesis of photosynthetic complexes in thylakoid membranes [93], which may contribute to the severity of its knock-out phenotype. It is plausible that the 1 MDa TIC components act in multiple processes, including protein import, with additional machinery operating either in series or in parallel. The extent of involvement of the 1 MDa complex in import may depend on the nature of the client proteins, or on specific developmental or environmental conditions.
Ubiquitin-dependent regulation of protein import
The UPS is a major regulatory system involved in the targeting of misfolded or unnecessary proteins for degradation. It functions within many biological processes, such as hormone signalling [94]. Ubiquitination (or ubiquitylation) is a post-translational modification involving the addition of one or more copies of the 8.5 kDa ubiquitin protein to lysine residues of target proteins [24]. The addition of polyubiquitin chains identifies the protein for degradation by the nucleocytosolic 26S proteasome (26SP) [24]. The 26SP is an ATP-dependent proteolytic complex formed from a cylindrical 20S core particle and a 19S regulatory particle; ubiquitinated proteins are recognized by the regulatory particle which guides them to the core where they are degraded [95]. Importantly, this cytosolic machinery also targets organelles, most famously the endoplasmic reticulum (ER) in ER-associated protein degradation (ERAD) [96], but also the endosymbiotically derived mitochondria and chloroplasts [97]. Indeed, proteomic studies of plant ubiquitinomes have identified many chloroplast proteins which may be UPS targets, although it remains to be determined whether these are processed as precursors in the cytosol or later [98–100].
Ubiquitination requires an enzyme cascade involving the activation and targeted conjugation of ubiquitin. An E1 ubiquitin activase first forms a thioester bond with ubiquitin in an ATP-dependent reaction [24]. The ubiquitin moiety is then transferred to an E2 ubiquitin conjugase. Finally, an E3 ubiquitin ligase conveys substrate specificity through a selective interaction with its targets, catalysing the transfer of ubiquitin to the target from the E2 enzyme [24]. Reiterative rounds of conjugation onto ubiquitin lysine residues result in the formation of a polyubiquitin chain degradation signal. Upon degradation, the ubiquitin moieties are recycled through the action of deubiquitinating enzymes. The E3 ligases are necessarily numerous and diverse, given their role in specificity, with roughly 1400 E3 proteins in Arabidopsis, far outnumbering the ∼40 E2 and two E1 enzymes [23]. In plants, there are four classes of E3 ligase: homologous to the E6-AP carboxyl terminus (HECT), really interesting new gene (RING), U-box, and cullin-RING ligase (CRL) [23]. Each class has a different mechanism of action and subunit composition, with particular diversity in the substrate-interacting domain or component [24].
Degradation of precursor proteins
Initial evidence for the UPS regulation of chloroplast proteins came from observations of cytosolic E3 activity targeting pre-proteins [101]. The C-terminus of Hsc70-interacting protein (CHIP) E3 ligase was initially shown to direct the degradation of Clp and FtsH precursors under high-light conditions [102,103]. A subsequent study revealed that in the Arabidopsis Toc159 mutant plastid protein import2 (ppi2), pre-proteins had accumulated in the cytosol and the expression of the cytosolic chaperone Hsc70-4 (an Hsp70 isoform) was elevated [101]. The Hsc70-4 protein was shown to interact with the TPs of pre-proteins, recruiting them to CHIP for ubiquitination and degradation by the 26SP [101] (Figure 1). This system is suggested to function as a quality-control process to degrade mis-targeted proteins and/or prevent the accumulation of pre-proteins in the cytosol, as unfolded proteins may accumulate into cytotoxic aggregates [101]. A recent study in wheat identified another cytosolic E3 ligase, stress-associated protein 5 (SAP5), which interacts with the pre-protein of Hsp90C to trigger its degradation [104].
Chloroplast biogenesis
A further influence of the UPS on cytosolic events controlling chloroplast biogenesis was reported to occur during early plant development, pre-germination. DELLA proteins inhibit seed germination in processes regulated by UPS-mediated degradation [105]. Germination depends on the accumulation of the hormone gibberellic acid, which down-regulates DELLA factor accumulation to enable germination and, in turn, chloroplast biogenesis [106]. It was reported that all DELLA factors can interact with cytosolic Toc159, prior to its assembly into the TOC complex, and initiate its degradation by the 26SP [106] (Figure 1). Low gibberellic acid conditions also resulted in the UPS-dependent down-regulation of pre-proteins [106].
The UPS also exerts indirect effects on chloroplast development through nuclear activities [23,25,97,107]. In Arabidopsis, the two golden2-like (Glk) transcription factors function redundantly to promote the expression of photosynthetic proteins, thereby promoting chloroplast biogenesis [108]. Glk1 itself is regulated at the transcriptional level through plastid-to-nucleus signals mediated by genomes uncoupled1 (GUN1) in response to the developmental state of the organelle [9]. Interestingly, Glk1 has also been found to be regulated at the posttranslational level by the UPS in response to an as yet unknown, GUN1-independent plastid signal [108] (Figure 1). This signal may derive from an environmental or developmental source to control chloroplast biogenesis [108].
Chloroplast-associated protein degradation
The first evidence for direct interaction between the UPS and chloroplast proteins in situ came from the discovery of the ubiquitin-dependent degradation of TOC complexes. A forward-genetic screen identified the RING-type E3 ligase, suppressor of ppi1 locus 1 (SP1) [109]; ppi1 is a Toc33 knock-out mutant with a pale yellow phenotype [110]. Located in the OEM [111], SP1 possesses two transmembrane domains separated by an IMS domain, which acts in target recognition, and a C-terminal cytosolic RING domain [109]. SP1 directly interacts with all TOC proteins, mediating their ubiquitination and degradation (Figure 1). Thus, there is an increase in TOC protein abundance when SP1 is lost, which, when in a chlorotic ppi1 background, causes enhanced greening (or suppression of ppi1), a phenotype mirrored by UPS inhibition [109]. Further experiments revealed that such degradation of TOC complexes provides for important regulation of the import machinery, and may act to alter the proteome, functions and developmental fate of the organelle [112]. This regulation can also help to promote the plant's tolerance of abiotic stress, by attenuating the import of photosynthetic proteins and thus suppressing photosynthesis and the tendency to overproduce reactive oxygen species during stress [112].
To be degraded by the cytosolic 26SP, polyubiquitinated TOC proteins first need to overcome the physical and energetic barriers to their removal from the OEM. Degradation of ER membrane and mitochondrial outer membrane proteins involves retrotranslocation across the membrane before degradation by the 26SP [96,113], and it was thought that protein degradation at the chloroplast OEM may involve an analogous process. Strikingly, an additional product of the suppressor screen that identified SP1 was the Omp85 protein SP2, and this OEM protein was shown to assist TOC retrotranslocation [114]. As an Omp85 family member, SP2 shares homology with Toc75 and is capable of forming a channel, although it lacks a POTRA domain. Like SP1, SP2 physically interacts with TOC proteins, and it is hypothesized to form the retrotranslocon channel [114] (Figure 1). The entry of substrates into the channel may occur by lateral gating in the membrane, as with other Omp85 proteins [57].
The motive force for extraction through SP2 is provided by a cytosolic factor: cell division cycle 48 (CDC48) is a homohexameric ATPase and a member of the ATPases associated with various cellular activities (AAA+) family of proteins [115]. Conformational changes in CDC48 induced by ATP hydrolysis create a motive force, allowing for the extraction and denaturation of bound substrates through the central pore [116,117] (Figure 1). CDC48 functions in a variety of cellular activities beyond protein homeostasis, such as cell cycle regulation and autophagy, but it is especially well known as the core motor component in ERAD [118]. Of the five reported homologues of CDC48 in Arabidopsis, CDC48A was found to associate with Toc33 [114], and has recently been identified by proteomic analysis of the chloroplast envelope [119]. Reconstitution experiments demonstrated that CDC48 operates as a cytosolic motor to retrotranslocate ubiquitinated TOC proteins prior to their degradation, a process in which SP2 was also shown to be critical [114] (Figure 1). This pathway of TOC degradation by the UPS involving SP1, SP2, and CDC48 has been named CHLORAD [114].
Perspectives
Importance of the field: The chloroplast is a dynamic plant organelle with indispensable metabolic functions including photosynthesis. The import of chloroplast proteins is, therefore, an essential and tightly controlled process, defects in which result in severe phenotypes. The UPS-mediated regulation of chloroplast protein import enables the plant to respond to the fluctuating protein demands of the organelle.
Summary of current thinking: Despite concerted efforts to unravel the mechanisms of protein import into chloroplasts over many years, there remain substantial gaps in our understanding. Passage through the TOC apparatus is a reasonably well-defined process involving initial docking at receptor GTPases and subsequent translocation through the β-barrel pore of Toc75. However, precisely how pre-proteins are delivered across the IEM is unclear: two TIC systems, involving Tic110/Tic40 and Tic214 (the 1 MDa complex), have been implicated, but whether and/or how they co-operate is unknown. Moreover, while it is well established that an ATP-dependent motor drives protein import, whether this comprises stromal chaperones (including cpHsp70) or a 2 MDa Ycf2–FtsHi complex is debated. The UPS is known to regulate the import process in a variety of ways: it degrades pre-proteins in the cytosol; it down-regulates chloroplast biogenesis in early development through the destruction of Toc159, and the transcription factor Glk1; and it reconfigures the TOC apparatus during developmental transitions and periods of stress.
Future directions: It is clear that there is a need to reconcile the conflicting reports on the cytosolic guidance systems, the TIC apparatus, and the stromal import motor. With regard to UPS involvement in chloroplast protein import, our understanding is still in its infancy. It is apparent from proteomic studies that a greater number of chloroplast proteins may be regulated by the UPS than is currently appreciated. Processing of many of these additional proteins may occur through the degradation of pre-proteins in the cytosol, and so additional cytosolic E3 ligases may be identified. Moreover, how the UPS regulation of chloroplasts acts in a physiological context requires further elucidation: we have seen that pre-germination conditions promote pre-protein degradation, and that CHLORAD can mediate stress responses, and so it seems likely that additional integration with developmental and environmental cues will be revealed. The discovery of the CHLORAD pathway offers much promise for future studies. In addition to identifying additional components and cofactors of the system, it will be important to determine the full range of its substrates. As things stand, only TOC proteins (and SP1) are established CHLORAD substrates, but there does exist the potential for other substrates to be found, and indeed for the identification of additional E3 ligases. Lastly, how the activity of CHLORAD is co-ordinated by intracellular signalling networks is completely unknown, and this will be very interesting to explore.
Abbreviations
- 26SP
26S proteasome
- AAA+
ATPases associated with various cellular activities
- CDC48
cell division cycle 48
- CHIP
C-terminus of Hsc70-interacting protein
- CHLORAD
chloroplast-associated protein degradation
- CRL
cullin-RING ligase
- DEK5
defective kernel5
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- FKBP73
FK506-binding protein 73
- Glk1
golden2-like 1
- HECT
homologous to the E6-AP carboxyl terminus
- Hop
Hsp70/Hsp90-organizing protein
- IEM
inner envelope membrane
- IMS
intermembrane space
- OEM
outer envelope membrane
- POTRA
polypeptide transport-associated
- ppi
plastid protein import
- RING
really interesting new gene
- SP
suppressor of ppi1
- TIC
translocon at the inner envelope membrane of chloroplasts
- TOC
translocon at the outer envelope membrane of chloroplasts
- TP
transit peptide
- UPS
ubiquitin-proteasome system
Author Contribution
S.M.T. wrote the initial draft and revisions of the manuscript. P.P. and R.P.J. edited the manuscript and provided critical guidance.
Funding
The authors’ research is supported by the Oxford Interdisciplinary Bioscience Doctoral Training Partnership (DTP) and the Biotechnology and Biological Sciences Research Council (BBSRC) [grant numbers BB/M011224/1, BB/R016984/1, and BB/R009333/1].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Rolland N., Curien G., Finazzi G., Kuntz M., Marechal E., Matringe M. et al. (2012) The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu. Rev. Genet. 46, 233–264 10.1146/annurev-genet-110410-132544 [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P. and López-Juez E. (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802 10.1038/nrm3702 [DOI] [PubMed] [Google Scholar]
- 3.McFadden G.I. (2014) Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 6, a016105 10.1101/cshperspect.a016105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Timmis J.N., Ayliffe M.A., Huang C.Y. and Martin W. (2004) Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 10.1038/nrg1271 [DOI] [PubMed] [Google Scholar]
- 5.Leister D. (2003) Chloroplast research in the genomic age. Trends Genet. 19, 47–56 10.1016/S0168-9525(02)00003-3 [DOI] [PubMed] [Google Scholar]
- 6.Nakai M. (2018) New perspectives on chloroplast protein import. Plant Cell Physiol. 59, 1111–1119 10.1093/pcp/pcy083 [DOI] [PubMed] [Google Scholar]
- 7.Sjuts I., Soll J. and Bölter B. (2017) Import of soluble proteins into chloroplasts and potential regulatory mechanisms. Front. Plant Sci. 8, 168 10.3389/fpls.2017.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paila Y.D., Richardson L.G.L. and Schnell D.J. (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 427, 1038–1060 10.1016/j.jmb.2014.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kakizaki T., Matsumura H., Nakayama K., Che F.S., Terauchi R. and Inaba T. (2009) Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353 10.1104/pp.109.145987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodson J.D., Perez-Ruiz J.M., Schmitz R.J., Ecker J.R. and Chory J. (2013) Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13 10.1111/tpj.12011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bräutigam K., Dietzel L., Kleine T., Stroher E., Wormuth D., Dietz K.J. et al. (2009) Dynamic plastid redox signals integrate gene expression and metabolism to induce distinct metabolic states in photosynthetic acclimation in Arabidopsis. Plant Cell 21, 2715–2732 10.1105/tpc.108.062018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trösch R. and Jarvis P. (2011) The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS ONE 6, e23039 10.1371/journal.pone.0023039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schünemann D. (2007) Mechanisms of protein import into thylakoids of chloroplasts. Biol. Chem. 388, 907–915 10.1515/BC.2007.111 [DOI] [PubMed] [Google Scholar]
- 14.Celedon J.M. and Cline K. (2013) Intra-plastid protein trafficking: how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta 1833, 341–351 10.1016/j.bbamcr.2012.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viana A.A., Li M. and Schnell D.J. (2010) Determinants for stop-transfer and post-import pathways for protein targeting to the chloroplast inner envelope membrane. J. Biol. Chem. 285, 12948–12960 10.1074/jbc.M110.109744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Martin J.R., Aldama G.A., Fernandez D.E. and Cline K. (2017) Identification of putative substrates of SEC2, a chloroplast inner envelope translocase. Plant Physiol. 173, 2121–2137 10.1104/pp.17.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Singhal R., Taylor I.W., McMinn P.H., Chua X.Y., Cline K. et al. (2015) The Sec2 translocase of the chloroplast inner envelope contains a unique and dedicated SECE2 component. Plant J. 84, 647–658 10.1111/tpj.13028 [DOI] [PubMed] [Google Scholar]
- 18.Watson S.J., Sowden R.G. and Jarvis P. (2018) Abiotic stress-induced chloroplast proteome remodelling: a mechanistic overview. J. Exp. Bot. 69, 2773–2781 10.1093/jxb/ery053 [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Yu Q., Xiong H., Wang J., Chen S., Yang Z. et al. (2016) Proteomic insight into the response of Arabidopsis chloroplasts to darkness. PLoS ONE 11, e0154235 10.1371/journal.pone.0154235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M., Takahashi S., Kondo T., Dohra H., Ito Y., Kiriiwa Y. et al. (2015) Plastid proteomic analysis in tomato fruit development. PLoS ONE 10, e0137266 10.1371/journal.pone.0137266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu C.C. and Li H.M. (2018) Developmental regulation of protein import into plastids. Photosynth. Res. 138, 327–334 10.1007/s11120-018-0546-4 [DOI] [PubMed] [Google Scholar]
- 22.Yang X., Li Y., Qi M., Liu Y. and Li T. (2019) Targeted control of chloroplast quality to improve plant acclimation: from protein import to degradation. Front. Plant Sci. 10, 958 10.3389/fpls.2019.00958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling Q. and Jarvis P. (2015) Functions of plastid protein import and the ubiquitin-proteasome system in plastid development. Biochim. Biophys. Acta 1847, 939–948 10.1016/j.bbabio.2015.02.017 [DOI] [PubMed] [Google Scholar]
- 24.Vierstra R.D. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397 10.1038/nrm2688 [DOI] [PubMed] [Google Scholar]
- 25.Rochaix J.D. and Ramundo S. (2018) Chloroplast signaling and quality control. Essays Biochem. 62, 13–20 10.1042/EBC20170048 [DOI] [PubMed] [Google Scholar]
- 26.Nishimura K., Kato Y. and Sakamoto W. (2017) Essentials of proteolytic machineries in chloroplasts. Mol. Plant 10, 4–19 10.1016/j.molp.2016.08.005 [DOI] [PubMed] [Google Scholar]
- 27.Woodson J.D., Joens M.S., Sinson A.B., Gilkerson J., Salome P.A., Weigel D. et al. (2015) Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350, 450–454 10.1126/science.aac7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Izumi M., Ishida H., Nakamura S. and Hidema J. (2017) Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29, 377–394 10.1105/tpc.16.00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnell D.J. (2019) The TOC GTPase receptors: regulators of the fidelity, specificity and substrate profiles of the general protein import machinery of chloroplasts. Protein J. 38, 343–350 10.1007/s10930-019-09846-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D.W. and Hwang I. (2018) Evolution and design principles of the diverse chloroplast transit peptides. Mol. Cells 41, 161–167 10.14348/molcells.2018.0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H.M. and Teng Y.S. (2013) Transit peptide design and plastid import regulation. Trends Plant Sci. 18, 360–366 10.1016/j.tplants.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 32.Flores-Pérez U. and Jarvis P. (2013) Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta 1833, 332–340 10.1016/j.bbamcr.2012.03.019 [DOI] [PubMed] [Google Scholar]
- 33.Fellerer C., Schweiger R., Schöngruber K., Soll J. and Schwenkert S. (2011) Cytosolic HSP90 cochaperones HOP and FKBP interact with freshly synthesized chloroplast preproteins of Arabidopsis. Mol. Plant 4, 1133–1145 10.1093/mp/ssr037 [DOI] [PubMed] [Google Scholar]
- 34.Panigrahi R., Adina-Zada A., Whelan J. and Vrielink A. (2013) Ligand recognition by the TPR domain of the import factor Toc64 from Arabidopsis thaliana. PLoS ONE 8, e83461 10.1371/journal.pone.0083461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronsson H., Boij P., Patel R., Wardle A., Topel M. and Jarvis P. (2007) Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 52, 53–68 10.1111/j.1365-313X.2007.03207.x [DOI] [PubMed] [Google Scholar]
- 36.Hofmann N.R. and Theg S.M. (2005) Toc64 is not required for import of proteins into chloroplasts in the moss Physcomitrella patens. Plant J. 43, 675–687 10.1111/j.1365-313X.2005.02483.x [DOI] [PubMed] [Google Scholar]
- 37.May T. and Soll J. (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell 12, 53–64 10.1105/tpc.12.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakrieko K.A., Mould R.M. and Smith A.G. (2004) Fidelity of targeting to chloroplasts is not affected by removal of the phosphorylation site from the transit peptide. Eur. J. Biochem. 271, 509–516 10.1046/j.1432-1033.2003.03950.x [DOI] [PubMed] [Google Scholar]
- 39.Lee D.W., Lee J. and Hwang I. (2017) Sorting of nuclear-encoded chloroplast membrane proteins. Curr. Opin. Plant Biol. 40, 1–7 10.1016/j.pbi.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 40.Kim J., Na Y.J., Park S.J., Baek S.H. and Kim D.H. (2019) Biogenesis of chloroplast outer envelope membrane proteins. Plant Cell Rep. 38, 783–792 10.1007/s00299-019-02381-6 [DOI] [PubMed] [Google Scholar]
- 41.Inoue K. and Keegstra K. (2003) A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J. 34, 661–669 10.1046/j.1365-313X.2003.01755.x [DOI] [PubMed] [Google Scholar]
- 42.Day P.M., Potter D. and Inoue K. (2014) Evolution and targeting of Omp85 homologs in the chloroplast outer envelope membrane. Front. Plant Sci. 5, 535 10.3389/fpls.2014.00535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Day P.M., Inoue K. and Theg S.M. (2019) Chloroplast outer membrane β-barrel proteins use components of the general import apparatus. Plant Cell 31, 1845–1855 10.1105/tpc.19.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klinger A., Gosch V., Bodensohn U., Ladig R. and Schleiff E. (2019) The signal distinguishing between targeting of outer membrane beta-barrel protein to plastids and mitochondria in plants. Biochim. Biophys. Acta Mol. Cell Res. 1866, 663–672 10.1016/j.bbamcr.2019.01.004 [DOI] [PubMed] [Google Scholar]
- 45.Jarvis P. (2004) Organellar proteomics: chloroplasts in the spotlight. Curr. Biol. 14, R317–R319 10.1016/j.cub.2004.03.054 [DOI] [PubMed] [Google Scholar]
- 46.Baslam M., Oikawa K., Kitajima-Koga A., Kaneko K. and Mitsui T. (2016) Golgi-to-plastid trafficking of proteins through secretory pathway: insights into vesicle-mediated import toward the plastids. Plant Signal. Behav. 11, e1221558 10.1080/15592324.2016.1221558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H. and Chen L.J. (1997) A novel chloroplastic outer membrane-targeting signal that functions at both termini of passenger polypeptides. J. Biol. Chem. 272, 10968–10974 10.1074/jbc.272.16.10968 [DOI] [PubMed] [Google Scholar]
- 48.Leipe D.D., Wolf Y.I., Koonin E.V. and Aravind L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 10.1006/jmbi.2001.5378 [DOI] [PubMed] [Google Scholar]
- 49.Agne B., Andrès C., Montandon C., Christ B., Ertan A., Jung F. et al. (2010) The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 153, 1016–1030 10.1104/pp.110.158048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue H., Rounds C. and Schnell D.J. (2010) The molecular basis for distinct pathways for protein import into Arabidopsis chloroplasts. Plant Cell 22, 1947–1960 10.1105/tpc.110.074328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schleiff E. and Becker T. (2011) Common ground for protein translocation: access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 12, 48–59 10.1038/nrm3027 [DOI] [PubMed] [Google Scholar]
- 52.Koenig P., Mirus O., Haarmann R., Sommer M.S., Sinning I., Schleiff E. et al. (2010) Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J. Biol. Chem. 285, 18016–18024 10.1074/jbc.M110.112649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Neil P.K., Richardson L.G.L., Paila Y.D., Piszczek G., Chakravarthy S., Noinaj N. et al. (2017) The POTRA domains of Toc75 exhibit chaperone-like function to facilitate import into chloroplasts. Proc. Natl Acad. Sci. U.S.A. 114, E4868–E4876 10.1073/pnas.1621179114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganesan I., Shi L.X., Labs M. and Theg S.M. (2018) Evaluating the functional pore size of chloroplast TOC and TIC protein translocons: import of folded proteins. Plant Cell 30, 2161–2173 10.1105/tpc.18.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schleiff E., Soll J., Kuchler M., Kuhlbrandt W. and Harrer R. (2003) Characterization of the translocon of the outer envelope of chloroplasts. J. Cell Biol. 160, 541–551 10.1083/jcb.200210060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kikuchi S., Hirohashi T. and Nakai M. (2006) Characterization of the preprotein translocon at the outer envelope membrane of chloroplasts by blue native PAGE. Plant Cell Physiol. 47, 363–371 10.1093/pcp/pcj002 [DOI] [PubMed] [Google Scholar]
- 57.Ganesan I. and Theg S.M. (2019) Structural considerations of folded protein import through the chloroplast TOC/TIC translocons. FEBS Lett. 593, 565–572 10.1002/1873-3468.13342 [DOI] [PubMed] [Google Scholar]
- 58.Ma Y., Kouranov A., LaSala S.E. and Schnell D.J. (1996) Two components of the chloroplast protein import apparatus, IAP86 and IAP75, interact with the transit sequence during the recognition and translocation of precursor proteins at the outer envelope. J. Cell Biol. 134, 315–327 10.1083/jcb.134.2.315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson L.G.L., Small E.L., Inoue H. and Schnell D.J. (2018) Molecular topology of the transit peptide during chloroplast protein import. Plant Cell 30, 1789–1806 10.1105/tpc.18.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Agne B., Infanger S., Wang F., Hofstetter V., Rahim G., Martin M. et al. (2009) A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J. Biol. Chem. 284, 8670–8679 10.1074/jbc.M804235200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aronsson H., Combe J., Patel R., Agne B., Martin M., Kessler F. et al. (2010) Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J. 63, 297–311 10.1111/j.1365-313X.2010.04242.x [DOI] [PubMed] [Google Scholar]
- 62.Andrès C., Agne B. and Kessler F. (2010) The TOC complex: preprotein gateway to the chloroplast. Biochim. Biophys. Acta 1803, 715–723 10.1016/j.bbamcr.2010.03.004 [DOI] [PubMed] [Google Scholar]
- 63.Wiesemann K., Simm S., Mirus O., Ladig R. and Schleiff E. (2019) Regulation of two GTPases Toc159 and Toc34 in the translocon of the outer envelope of chloroplasts. Biochim. Biophys. Acta Proteins Proteom. 1867, 627–636 10.1016/j.bbapap.2019.01.002 [DOI] [PubMed] [Google Scholar]
- 64.Lumme C., Altan-Martin H., Dastvan R., Sommer M.S., Oreb M., Schuetz D. et al. (2014) Nucleotides and substrates trigger the dynamics of the Toc34 GTPase homodimer involved in chloroplast preprotein translocation. Structure 22, 526–538 10.1016/j.str.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 65.Glaser S., van Dooren G.G., Agrawal S., Brooks C.F., McFadden G.I., Striepen B. et al. (2012) Tic22 is an essential chaperone required for protein import into the apicoplast. J. Biol. Chem. 287, 39505–39512 10.1074/jbc.M112.405100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripp J., Hahn A., Koenig P., Flinner N., Bublak D., Brouwer E.M. et al. (2012) Structure and conservation of the periplasmic targeting factor Tic22 protein from plants and cyanobacteria. J. Biol. Chem. 287, 24164–24173 10.1074/jbc.M112.341644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kouranov A., Chen X., Fuks B. and Schnell D.J. (1998) Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002 10.1083/jcb.143.4.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y.L., Chen L.J., Chu C.C., Huang P.K., Wen J.R. and Li H.M. (2018) TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129 10.1038/s41586-018-0713-y [DOI] [PubMed] [Google Scholar]
- 69.Zhang J., Wu S., Boehlein S.K., McCarty D.R., Song G., Walley J.W. et al. (2019) Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. J. Cell Biol. 218, 2638–2658 10.1083/jcb.201807166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M. et al. (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574 10.1126/science.1229262 [DOI] [PubMed] [Google Scholar]
- 71.Kovács-Bogdan E., Benz J.P., Soll J. and Bölter B. (2011) Tic20 forms a channel independent of Tic110 in chloroplasts. BMC Plant Biol. 11, 133 10.1186/1471-2229-11-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teng Y.S., Su Y.S., Chen L.J., Lee Y.J., Hwang I. and Li H.M. (2006) Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18, 2247–2257 10.1105/tpc.106.044305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee D.W. and Hwang I. (2019) Protein import into chloroplasts via the Tic40-dependent and -independent pathways depends on the amino acid composition of the transit peptide. Biochem. Biophys. Res. Commun. 518, 66–71 10.1016/j.bbrc.2019.08.009 [DOI] [PubMed] [Google Scholar]
- 74.Inaba T., Li M., Alvarez-Huerta M., Kessler F. and Schnell D.J. (2003) Attic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627 10.1074/jbc.M306367200 [DOI] [PubMed] [Google Scholar]
- 75.Chou M.L., Fitzpatrick L.M., Tu S.L., Budziszewski G., Potter-Lewis S., Akita M. et al. (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22, 2970–2980 10.1093/emboj/cdg281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi L.X. and Theg S.M. (2013) Energetic cost of protein import across the envelope membranes of chloroplasts. Proc. Natl Acad. Sci. U.S.A. 110, 930–935 10.1073/pnas.1115886110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu L., McNeilage R.T., Shi L.X. and Theg S.M. (2014) ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26, 1246–1255 10.1105/tpc.113.121822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi L.X. and Theg S.M. (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22, 205–220 10.1105/tpc.109.071464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Su P.H. and Li H.M. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531 10.1105/tpc.109.071415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inoue H., Li M. and Schnell D.J. (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl Acad. Sci. U.S.A. 110, 3173–3178 10.1073/pnas.1219229110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akita M., Nielsen E. and Keegstra K. (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J. Cell Biol. 136, 983–994 10.1083/jcb.136.5.983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen E., Akita M., Davila-Aponte J. and Keegstra K. (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J. 16, 935–946 10.1093/emboj/16.5.935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halperin T., Ostersetzer O. and Adam Z. (2001) ATP-dependent association between subunits of Clp protease in pea chloroplasts. Planta 213, 614–619 10.1007/s004250100527 [DOI] [PubMed] [Google Scholar]
- 84.Flores-Pérez U., Bédard J., Tanabe N., Lymperopoulos P., Clarke A.K. and Jarvis P. (2016) Functional analysis of the Hsp93/ClpC chaperone at the chloroplast envelope. Plant Physiol. 170, 147–162 10.1104/pp.15.01538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sjögren L.L., Tanabe N., Lymperopoulos P., Khan N.Z., Rodermel S.R., Aronsson H. et al. (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 289, 11318–11330 10.1074/jbc.M113.534552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kikuchi S., Asakura Y., Imai M., Nakahira Y., Kotani Y., Hashiguchi Y. et al. (2018) A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell 30, 2677–2703 10.1105/tpc.18.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herrmann J.M. (2018) A force-generating machine in the plant's powerhouse: a pulling AAA-ATPase motor drives protein translocation into chloroplasts. Plant Cell 30, 2646–2647 10.1105/tpc.18.00751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Köhler D., Montandon C., Hause G., Majovsky P., Kessler F., Baginsky S. et al. (2015) Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol. 167, 972–990 10.1104/pp.114.255562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Köhler D., Helm S., Agne B. and Baginsky S. (2016) Importance of translocon subunit Tic56 for rRNA processing and chloroplast ribosome assembly. Plant Physiol. 172, 2429–2444 10.1104/pp.16.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Agne B., Köhler D. and Baginsky S. (2017) Protein import-independent functions of Tic56, a component of the 1-MDa translocase at the inner chloroplast envelope membrane. Plant Signal. Behav. 12, e1284726 10.1080/15592324.2017.1284726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schäfer P., Helm S., Kohler D., Agne B. and Baginsky S. (2019) Consequences of impaired 1-MDa TIC complex assembly for the abundance and composition of chloroplast high-molecular mass protein complexes. PLoS ONE 14, e0213364 10.1371/journal.pone.0213364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bölter B. and Soll J. (2017) Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol. Plant 10, 219–221 10.1016/j.molp.2016.10.012 [DOI] [PubMed] [Google Scholar]
- 93.Yang X.F., Wang Y.T., Chen S.T., Li J.K., Shen H.T. and Guo F.Q. (2016) PBR1 selectively controls biogenesis of photosynthetic complexes by modulating translation of the large chloroplast gene Ycf1 in Arabidopsis. Cell Discov. 2, 16003 10.1038/celldisc.2016.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Christians M.J., Gingerich D.J., Hansen M., Binder B.M., Kieber J.J. and Vierstra R.D. (2009) The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 57, 332–345 10.1111/j.1365-313X.2008.03693.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sako K., Yanagawa Y., Kanai T., Sato T., Seki M., Fujiwara M. et al. (2014) Proteomic analysis of the 26S proteasome reveals its direct interaction with transit peptides of plastid protein precursors for their degradation. J. Proteome Res. 13, 3223–3230 10.1021/pr401245g [DOI] [PubMed] [Google Scholar]
- 96.Liu Y. and Li J. (2014) Endoplasmic reticulum-mediated protein quality control in Arabidopsis. Front. Plant Sci. 5, 162 10.3389/fpls.2014.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hua Z. and Vierstra R.D. (2016) Ubiquitin goes green. Trends Cell Biol. 26, 3–5 10.1016/j.tcb.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 98.Kim D.Y., Scalf M., Smith L.M. and Vierstra R.D. (2013) Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 25, 1523–1540 10.1105/tpc.112.108613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xie H., Wang Y., Ding Y., Qiu C., Sun L., Gai Z. et al. (2019) Global ubiquitome profiling revealed the roles of ubiquitinated proteins in metabolic pathways of tea leaves in responding to drought stress. Sci. Rep. 9, 4286 10.1038/s41598-019-41041-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Svozil J., Hirsch-Hoffmann M., Dudler R., Gruissem W. and Baerenfaller K. (2014) Protein abundance changes and ubiquitylation targets identified after inhibition of the proteasome with syringolin A. Mol. Cell Proteomics 13, 1523–1536 10.1074/mcp.M113.036269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee S., Lee D.W., Lee Y., Mayer U., Stierhof Y.D., Lee S. et al. (2009) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21, 3984–4001 10.1105/tpc.109.071548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shen G., Adam Z. and Zhang H. (2007) The E3 ligase AtCHIP ubiquitylates FtsH1, a component of the chloroplast FtsH protease, and affects protein degradation in chloroplasts. Plant J. 52, 309–321 10.1111/j.1365-313X.2007.03239.x [DOI] [PubMed] [Google Scholar]
- 103.Shen G., Yan J., Pasapula V., Luo J., He C., Clarke A.K. et al. (2007) The chloroplast protease subunit ClpP4 is a substrate of the E3 ligase AtCHIP and plays an important role in chloroplast function. Plant J. 49, 228–237 10.1111/j.1365-313X.2006.02963.x [DOI] [PubMed] [Google Scholar]
- 104.Zhang N., Xu J., Liu X., Liang W., Xin M., Du J. et al. (2019) Identification of HSP90C as a substrate of E3 ligase TaSAP5 through ubiquitylome profiling. Plant Sci. 287, 110170 10.1016/j.plantsci.2019.110170 [DOI] [PubMed] [Google Scholar]
- 105.Li K., Yu R., Fan L.M., Wei N., Chen H. and Deng X.W. (2016) DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 7, 11868 10.1038/ncomms11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shanmugabalaji V., Chahtane H., Accossato S., Rahire M., Gouzerh G., Lopez-Molina L. et al. (2018) Chloroplast biogenesis controlled by DELLA-TOC159 interaction in early plant development. Curr. Biol. 28, 2616–2623.e5 10.1016/j.cub.2018.06.006 [DOI] [PubMed] [Google Scholar]
- 107.Hirosawa Y., Ito-Inaba Y. and Inaba T. (2017) Ubiquitin-proteasome-dependent regulation of bidirectional communication between plastids and the nucleus. Front. Plant Sci. 8, 310 10.3389/fpls.2017.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tokumaru M., Adachi F., Toda M., Ito-Inaba Y., Yazu F., Hirosawa Y. et al. (2017) Ubiquitin-proteasome dependent regulation of the GOLDEN2-LIKE 1 transcription factor in response to plastid signals. Plant Physiol. 173, 524–535 10.1104/pp.16.01546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ling Q., Huang W., Baldwin A. and Jarvis P. (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338, 655–659 10.1126/science.1225053 [DOI] [PubMed] [Google Scholar]
- 110.Jarvis P., Chen L.-J., Li H.-m., Peto C., Fankhauser C. and Chory J. (1998) An Arabidopsis mutant defective in the chloroplast general protein import apparatus. Science 282, 100–103 https://www.ncbi.nlm.nih.gov/pubmed/9756470 [DOI] [PubMed] [Google Scholar]
- 111.Ling Q., Li N. and Jarvis P. (2017) Chloroplast ubiquitin E3 ligase SP1: does it really function in peroxisomes? Plant Physiol. 175, 586–588 10.1104/pp.17.00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ling Q. and Jarvis P. (2015) Regulation of chloroplast protein import by the ubiquitin E3 ligase SP1 is important for stress tolerance in plants. Curr. Biol. 25, 2527–2534 10.1016/j.cub.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu X., Li L. and Jiang H. (2016) Doa1 targets ubiquitinated substrates for mitochondria-associated degradation. J. Cell Biol. 213, 49–63 10.1083/jcb.201510098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ling Q., Broad W., Trösch R., Topel M., Demiral Sert T., Lymperopoulos P. et al. (2019) Ubiquitin-dependent chloroplast-associated protein degradation in plants. Science 363, eaav4467 10.1126/science.aav4467 [DOI] [PubMed] [Google Scholar]
- 115.Ye Y., Tang W.K., Zhang T. and Xia D. (2017) A mighty “protein extractor” of the cell: structure and function of the p97/CDC48 ATPase. Front. Mol. Biosci. 4, 39 10.3389/fmolb.2017.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bodnar N.O. and Rapoport T.A. (2017) Molecular mechanism of substrate processing by the Cdc48 ATPase complex. Cell 169, 722–735 10.1016/j.cell.2017.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Twomey E.C., Ji Z., Wales T.E., Bodnar N.O., Ficarro S.B., Marto J.A. et al. (2019) Substrate processing by the Cdc48 ATPase complex is initiated by ubiquitin unfolding. Science 365, eaax1033 10.1126/science.aax1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bègue H., Jeandroz S., Blanchard C., Wendehenne D. and Rosnoblet C. (2017) Structure and functions of the chaperone-like p97/CDC48 in plants. Biochim. Biophys. Acta 1861, 3053–3060 10.1016/j.bbagen.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 119.Bouchnak I., Brugiere S., Moyet L., Le Gall S., Salvi D., Kuntz M. et al. (2019) Unraveling hidden components of the chloroplast envelope proteome: opportunities and limits of better MS sensitivity. Mol. Cell Proteomics 18, 1285–1306 10.1074/mcp.RA118.000988 [DOI] [PMC free article] [PubMed] [Google Scholar]