Abstract
Background
To date, responsiveness to tumor necrosis factor alpha inhibitors in ulcerative colitis (UC) patients is not predictable. This is partially due to a lack of understanding of the underlying inflammatory processes. The aim of this study was to identify immunological subgroups of patients with UC and to test responsiveness to adalimumab in these subgroups in the mouse model of ulcerative colitis (UC), which is based on NOD/scid IL-2Rγ null (NSG) mice reconstituted with peripheral blood mononuclear cells (PBMCs; NSG-UC).
Methods
The immunological profiles of 40 UC patients and 16 non-UC donors were determined by flow cytometric analysis of PBMCs in a snapshot and longitudinal study and analyzed by principal component, orthogonal partial least square discrimination (oPLS-DA), and hierarchical clustering analysis. NSG mice were reconstituted 5 times at consecutive time points with PBMCs from a single donor and were analyzed for frequencies of human leukocytes and histological phenotype. The response to adalimumab of 2 identified subgroups was tested in the NSG-UC model. We used the clinical, colon, and histological score, serum levels of glutamic and aspartic acid, and IL-6 and IL-1ß. Response was analyzed by oPLS-DA.
Results
Analysis revealed a distinction between UC and non-UC donors. Hierarchical clustering identified 2 major subgroups in UC patients. Group I was characterized by TH17 and M1 monocytes, group II by TH2/TH1, and switched B cells. These subgroups reflect the dynamics of inflammation as patients. NSG-UC mice achieved an immunological phenotype reflecting the patient’s immunological phenotype. oPLS-DA revealed that NSG-UC mice reconstituted with PBMCs from group II responded better to adalimumab.
Conclusions
The combination of profiling and testing of therapeutics in the NSG-UC model may lead to individualized and phase-dependent therapies.
Keywords: inflammatory bowel disease, ulcerative colitis, immunological profiling, NOD/scid IL-2Rγ null, NSG-UC, adalimumab
This study shows that ulcerative colitis (UC) patients display a TH2/TH1 or Th17/M1 monocyte-driven phenotype. These conditions are reflected in the NOD/scid IL-2Rγ null (NSG)–UC mouse model. The response to adalimumab in the NSG-UC model is higher in the TH2/TH1-driven group.
INTRODUCTION
Inflammation is a highly dynamic process that requires swift temporal and spatial responses of all cells involved and includes immune, epithelial, endothelial, and muscle cells and fibroblasts.1 These cells fulfill 3 major tasks: protection from invading pathogens, healing, and resolving inflammation. Although these 3 major processes are temporally harmonized in a healthy situation, they are not clearly distinct, and inducers of pro-inflammatory responses often simultaneously evoke immune-modulatory responses.2 This observation also implies that chronic inflammatory processes may not be distinguishable from inflammation in a healthy background, let alone that 2 different chronic inflammatory diseases, for example, ulcerative colitis (UC) and Crohn’s disease, can be discriminated based on patients’ immunological profiles. Indeed, to date no biological marker has been identified to discriminate reliably between these 2 diseases. However, this observation does not rule out that the dynamics of inflammatory processes cannot be reflected by immunological profiles of patients. A previous study has confirmed the assumption that inflammation in UC is characterized by pro-inflammatory and healing processes,3 but this is only a crude description and is insufficient to describe various manifestations of the disease. The delineation of underlying inflammatory processes, however, remains a prerequisite for individualized and phase-dependent therapies, which is the ultimate goal in all chronic inflammatory diseases.
Animal models are highly useful tools to examine patho-mechanisms underlying human diseases and to test the efficacy of novel therapeutics. However, the significance of these results is disputed, as most animal models poorly reflect human disease, and results obtained in these models are very often not translatable to clinical trials with patients. This especially applies to chronic inflammatory diseases like UC, which is still an umbrella diagnosis covering various subtypes of disease manifestations. Therefore, the value of results obtained from animal models remains questionable.
The NSG-UC mouse model is based on NOD/scid IL-2Rγ null (NSG) mice reconstituted with peripheral blood mononuclear cells (PBMCs) derived from patients with UC (NSG-UC).4, 5 It gives a better reflection of the inflammatory processes observed in humans. Most of the inflammatory cell types identified in UC patients have also been identified in the NSG-UC model to include M1- (CD14+ CD64+) and M2 monocytes (CD14+ CD163+ CD206+) and CD1a-expressing monocytes. Colonic inflammation is usually mild and characterized by a mixed infiltrate of immune cells into the mucosa, edema, crypt loss, and fibrosis; however, the observed pathological manifestations of the colon were highly donor dependent. Mice reconstituted with PBMCs from healthy donors developed no inflammation.6, 7 Even when PBMCs from patients in relapse were used for reconstitution, biological markers identified as hallmarks of inflammation displayed high variability5 and the pathological manifestations in the colon ranged from severe influx of inflammatory cells into the mucosa to severe fibrosis and almost no influx of inflammatory cells. These observations prompted us to speculate that the NSG-UC model reflects the ongoing inflammation in UC patients and that it can be used to predict responsiveness to tumor necrosis factor alpha (TNFα) inhibitors. To prove this hypothesis, first, the inflammatory profiles of UC patients and healthy donors were determined in snapshot and longitudinal studies. A clear distinction between healthy subjects and UC patients was achieved. Analysis of UC patients revealed 2 main inflammatory subtypes either driven by M1 monocytes/TH17 or by TH1/TH2 cells. Second, PBMCs from 1 donor profiled in the longitudinal study were used for reconstitution in the NSG-UC model. The results indicated that NSG-UC mice reflect the dynamics observed ex vivo. Third, we examined whether the 2 observed inflammatory processes might indicate response to adalimumab. The results suggest that mice reconstituted with PBMCs characterized by TH1/TH2 had a better response rate.
METHODS
Ethical Considerations
Written consent was given by all donors. The study was approved by the Institutional Review Board (IRB) of the Medical Faculty at the University of Munich (2015–22).
Animal studies were approved by the committees of the government of Upper Bavaria, Germany (55.2-2-1-54-2532-74-15), and performed in compliance with German Animal Welfare Laws.
Isolation of PBMC and Engraftment
Sixty milliliters of peripheral blood in trisodium citrate solution (S-Monovette, Sarstedt, Nürnberg, Germany) was collected from the arm vein of UC patients. The blood was diluted with Hank’s balanced salt solution (HBSS; Sigma Aldrich, Deisenhofen, Germany) in a 1:2 ratio. The suspension was loaded onto LeucoSep tubes (Greiner Bio One, Frickenhausen, Germany). Peripheral blood mononuclear cells (PBMCs) were separated by centrifugation at 400g for 30 minutes with no acceleration. The interphase was extracted and diluted with phosphate-buffered saline (PBS) to a final volume of 40 mL. Cells were counted and centrifuged at 1400g for 5 minutes. The cell pellet was resuspended in PBS at a concentration of 4×106 cells in 100 µL.
Six- to eight-week-old NOD.cg-PrkdcSCID Il2rgtm1Wjl/Szj mice (abbreviated as NOD-scid IL-2Rγ null [NSG]) were engrafted with 100-µL cell solution into the tail vein on day 1.
Study Protocol
NSG mice were obtained from Charles River Laboratories (Sulzfeld, Germany). Mice were kept under specific pathogen–free conditions in individually ventilated cages in a facility controlled according to Federation of Laboratory Animal Science Association (FELASA) guidelines. After engraftment on day 1, mice were presensitized by rectal application of 150 µL of 10% ethanol on day 8 using a 1-mm cat catheter (Henry Schein, Hamburg, Germany). The catheter was lubricated with Xylocain Gel 2% (AstraZeneca, Wedel, Germany). Rectal application was performed under general anesthesia using 4% isofluran. Postapplication, mice were kept at an angle of 30° to avoid ethanol dripping. On day 15, mice were challenged by rectal application of 50% ethanol following the protocol of day 8. On day 18, mice were killed. Adalimumab was provided by Sanofi-Aventis Deutschland GmbH (Frankfurt am Main, Germany) and was applied in PBS (30 mg/kg/d) on days 7 and 14. The control groups were injected with 30 mg/kg of the isotype antibody (Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany) on days 7 and 14.
Clinical Activity Score
Assessment of colitis severity was performed daily according to the following scoring system: loss of body weight: 0% (0), 0%–5% (1), 5%–10% (2), 10%–15% (3), 15%–20% (4); stool consistency: formed pellet (0), loose stool or unformed pellet (2), liquid stools (4); behavior: normal (0), reduced activity (1), apathy (4), and ruffled fur (1); body posture: intermediately hunched posture (1), permanently hunched posture (2). The scores were added daily into a total score, with a maximum of 12 points per day. Animals who suffered from weight loss >20%, rectal bleeding, rectal prolapse, self-isolation, or a severity score >7 were killed immediately and not taken into count. All scores were added for statistical analysis.
Colon Score
The colon was removed, a photograph was taken, and the colon was scored. Scoring was as follows: pellet: formed (0), soft (1), liquid (2); length of colon: >10 cm (0), 8–10 cm (1), <8 cm (2); dilation: no (0), minor (1), severe (2); hyperemia: no (0), yes (2); necrosis: no (0), yes (2).
Histological Analysis
At autopsy, samples from distal parts of the colon were fixed in 4% formaldehyde for 24 hours, before storage in 70% ethanol, and were routinely embedded in paraffin. Samples were cut into 3-µm sections and stained with hematoxylin and eosin (H&E) and Elastica-van-Gieson. Epithelial erosions were scored as follows: no lesions (1), focal lesions (2), multifocal lesions (3), major damage with involvement of basal membrane (4). Inflammation was scored as follows: infiltration of few inflammatory cells into the lamina propria (1), major infiltration of inflammatory cells into the lamina propria (2), confluent infiltration of inflammatory cells into the lamina propria (3), infiltration of inflammatory cells including tunica muscularis (4). Fibrosis was scored as follows: focal fibrosis (1), multifocal fibrosis and crypt atrophy (2). The presence of edema, hyperemia, and crypt abscess was scored with 1 additional point in each case. The scores for each criterion were added into a total score ranging from 0 to 12. Images were taken with a Zeiss AxioVert 40 CFL camera. Figures show representative longitudinal sections in the original magnification. In Adobe Photoshop CC, a tonal correction was used to enhance contrasts within the pictures.
Isolation of Human Leucocytes
To isolate human leucocytes, spleens were minced and cells filtrated through a 70-µL cell strainer, followed by centrifugation at 1400g for 5 minutes, and resuspended in fluorescence-activated cell sorting (FACS) buffer (1× PBS, 2 mM ethylene-diamine-tetra-acidic-acid [EDTA], 2% fetal calf serum [FCS]). For further purification, cell suspensions were filtrated using a 35-µm cell strainer and then labeled for flow cytometry analysis.
For isolation of lamina propria mononuclear cells (LPMCs), a protocol modified from Weigmann et al.8 was used. The washed and minced colon was predigested in an orbital shaker with slow rotation (40g) at 37°C for 1×20 minutes in predigestion solution containing 1× HBSS (Thermo Scientific, Darmstadt, Deutschland), 5 mM EDTA, 5% FCS, 100 U/mL pencillin-streptomycin (Sigma-Aldrich Co., St. Louis, MO, USA). Epithelial cells were removed by filtering through a nylon filter. After washing with Rossweel Park Memorial Istitute (RPMI), the remaining colon pieces were digested for 2×20 minutes in digestion solution containing 1× RPMI (Thermofisher Scientific, Darmstadt, Germany), 10% FCS, 1 mg/mL Collagenase A (Sigma-Aldrich Co., St. Louis, MO, USA), 10 KU/mL Dnase I (Sigma-Aldrich Co., St. Louis, MO, USA), 100 U/mL pencillin-streptomycin (Sigma-Aldrich Co., St. Louis, MO, USA) in an orbital shaker with slow rotation (40g) at 37°C.8
Isolated LPMC were centrifuged at 1400g for 5 minutes and resuspended in FACS buffer. Cell suspensions were filtrated 1 more time using a 35-µm cell strainer for further purification before labeling the cells for flow cytometry analysis.
Flow Cytometry Analysis
Labeling of human leucocytes was performed according to Supplementary Table 1.
All antibodies were purchased from Biolegend (San Diego, CA, USA) and used according to the manufacturer’s instructions. Flow cytometry was performed using a BD FACS Canto II and analyzed with FlowJo 10.1 software (FlowJo LLC, Ashland, OR, USA).
Detection of Cytokines in Sera and Colon
Whole blood was collected and allowed to clot at room temperature for 30 minutes. After a 10 minutes, centrifugation at 2000×g and 4°C, the supernatant was transferred to a fresh polypropylene tube and used immediately or stored at –80°C.
Approximately 10-mm-long sections of the terminal colon were dissected and cleaned of feces with ice-cold PBS; 500 µL of protease inhibitor cocktail (cOmplete, Roche, Penzberg, Germany) was added according to the manufacturer’s instructions. Samples were milled with a 5-mm stainless steel bead (Qiagen, Hilden, Germany), centrifuged for 5 minutes at 3000 rpm, and supernatants were shock-frozen and stored at –80°C.
Supernatants or sera were analyzed for cytokine content either with the MSD Mesoscale platform (Meso Scale Diagnostics, Rockville, MD, USA) using the V-PLEX Proinflammatory Panel 1 Mouse Kit or with the LUNARIS platform (AYOXXA Biosystems GmbH, Cologne, Germany) for multiplex protein analysis using the LUNARIS Mouse 12-Plex Th17 or the Human 11-Plex Cytokine Kit using the protocol provided by the respective manufacturers. Fluorescence signals of bound target proteins are recorded by high-resolution imaging for quantification by the proprietary LUNARIS Analysis Suite.
msCRP (Cat# EPX01A-26045–901, RRID:AB_2575963; Thermo Fisher Scientific, Darmstadt, Germany) and msTGFß (Cat# EPX01A-20608–901, RRID:AB_2575921; Thermo Fisher Scientific, Darmstadt, Germany) were detected by Luminex assay (MAGPIX; Luminex Corporation, Austin, TX, USA).
Detection of Amino Acids
Samples were prepared according to the manufacturer’s instructions. After incubation of 100 µL of serum with internal standards for 5 minutes, 25 µL of 15% 5-sulfosalicylic acid was added, and samples were centrifuged at 9000g for 15 minutes at 4°C. Supernatants were filtered through a 0.2-µm membrane, and 75 µL of lithium-loading buffer was added. Samples were analyzed with the amino acid analyzer Biochrom 30+ (Biochrom Ltd., Cambridge, UK).
Statistical Analysis
Statistical analysis was performed with R (R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/). Variables are represented by mean, standard deviation, and median. A 2-sided Student t test and a confidence level of 0.95 were used to compare binary groups and >2 groups. Analysis of variance (ANOVA) followed by Tukey’s honestly significant difference (HSD) was conducted. For correlation analysis, Pearson’s product–moment correlation was performed, and a 95% confidence interval was applied. Heatmap was performed using R (default), then 2-groups ANOVA followed by Tukey’s HSD was conducted. Principal component analysis (PCA) was performed using the plyr, ChemometricsWithR, maptools, car, and rgeos packages. Orthogonal partial least square discrimination analysis (oPLS-DA) was performed using the ropls package.9
RESULTS
Immunological Profiles of Patients With UC
As inflammatory processes are not unique to UC and are also highly dynamic to exercise all functions from protection to wound healing and resolving, to date it has not been possible to completely delineate these processes. In a previous study, we used a panel of immunological cells and cytokines and growth factors to prove the hypothesis that known components driving wound healing processes can also be detected in UC patients.3 To further explore the dynamics of inflammation and the driving components in UC, 40 profiles of UC patients and 16 profiles of non-UC individuals were determined based on flow cytometric analysis of PBMCs. Basic patient demographics are presented in Table 1.
TABLE 1.
Basic Demographics
| UC (n = 40) | Non-UC (n = 16) | |
|---|---|---|
| Age, y | ||
| Mean (SD) | 37.38 (15.43) | 40 (17.1) |
| Range | 21–77 | 21–66 |
| Gender, male, No. (%) | 23 (57.5) | 5 (31.1) |
| Duration of UC, y | ||
| Mean (SD) | 11 (7.9) | |
| Range | 2–37 | |
| SCCAI | ||
| Mean (SD) | 4.70 (3.35) | |
| Range | 0–1 | |
| Treatment (current) | ||
| Infliximab | 11 | |
| Adalimumab | 3 | |
| Glucocorticoids | 5 | |
| Mesalazine | 17 | |
| Azothioprine | 3 | |
| No | 4 |
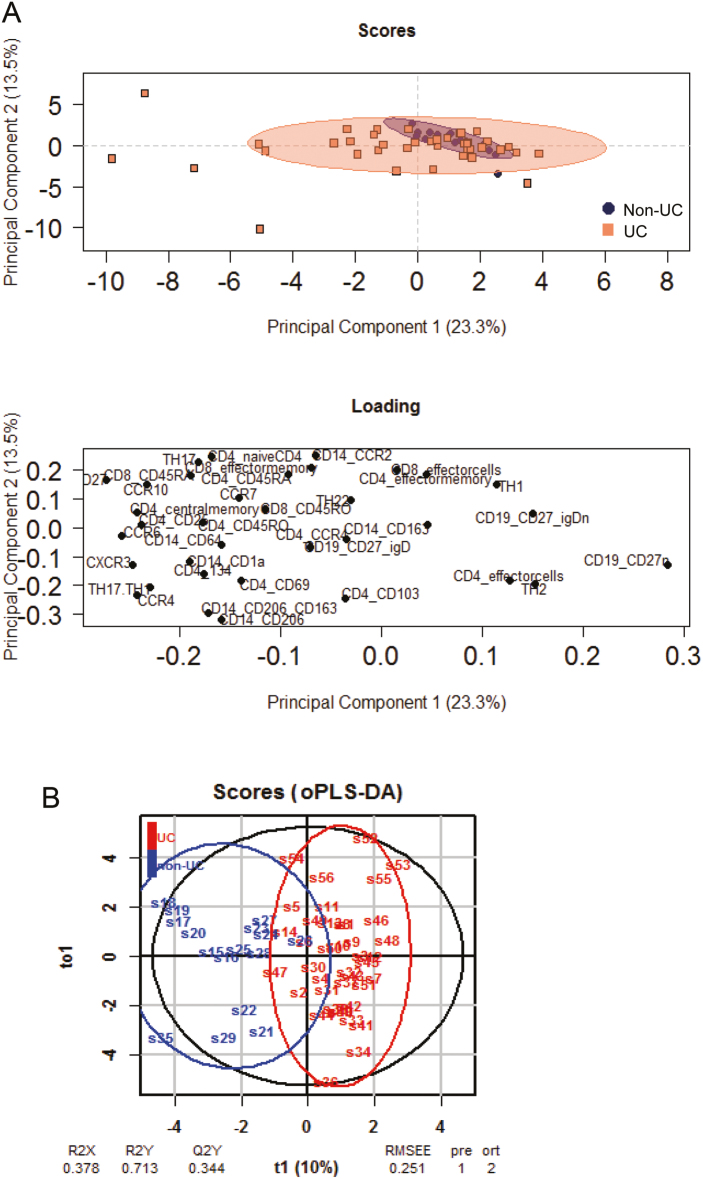
The panel included surface markers of subgroups of CD4+ and CD8+ T cells, CD14+ monocytes, and B cells and chemokine receptors (Supplementary Table 2). For gating strategy, the complete raw data set, and differences in frequencies observed in UC vs non-UC, see Supplementary Figure 1 and Supplementary Tables 3 and 4. The capacity of these surface markers to discriminate between the non-UC and UC groups was evaluated by 2 different methods. First, principal component analysis was used to analyze the correlation between all variables (for component loadings, component variances, and importance of components, see Supplementary Table 5). This analysis revealed that 10 principal components shaped the inflammation. As depicted in Figure 1A, 2 components described merely 37.1% of the variance and were not sufficient to discriminate between the 2 groups; however, it became apparent that the non-UC group clustered more closely and that UC patients were more heterogeneous. Therefore, an oPLS-DA was performed. This method has been used in IBD to detect differences in metabolic profiles10 and is advantageous when a comparably high number of variables are included in the analysis and differences are subtle. Although this method (predicted component = 1, orthogonal component = 2) also depicted an overlap between the 2 groups, a discrimination between profiles of non-UC and UC donors was achieved (Fig. 1B). The values of R2X, R2Y, and R2Q of 0.378, 0.713, and 0.344, respectively, and the low value of the root mean square error of estimation (RMSEE) of 0.251 indicate the validity of the model. For significance diagnostics, R2Y and R2Q values were compared with values after random permutation of the y response. The obtained values of pR2Y = 0.05 and pR2Q = 0.05 further corroborated the validity of the oPLS model. To test the predictive performance of this model, cross-validation with training and testing subsets (28 samples, 12 variables, and 1 response for each subset) was performed. In the training subset, 1 false negative was detected (non-UC: 8 non-UC and 1 UC; UC: 0 non-UC and 20 UC). As expected, the predictions in the testing subset (28 samples, 12 variables, and 1 response) were lower (non-UC: 3 non-UC and 5 UC: UC: 15 UC and 5 non-UC). Thus, this classifier achieves 71% of correct predictions.
FIGURE 1.
Subgroups of immune cells lead to discrimination of non-UC and UC patients. Profiles of 40 UC and 16 non-UC donors were compared using (A) PCA analysis and (B) oPLS-DA analysis. Abbreviations: R2X, fraction of the variation of the X variables explained by the model; R2Y, fraction of the variation of the Y variables explained by the model; Q2Y, fraction of variation of the y variables predicted by the model.
As the PCA analysis suggested a high heterogeneity of the patient group, the profiles of 40 patients were analyzed by cluster analysis depicted as a heatmap. Among the patients were 3 who had repeatedly donated blood and who experienced high clinical activity under different treatment regimens (Supplementary Table 6).
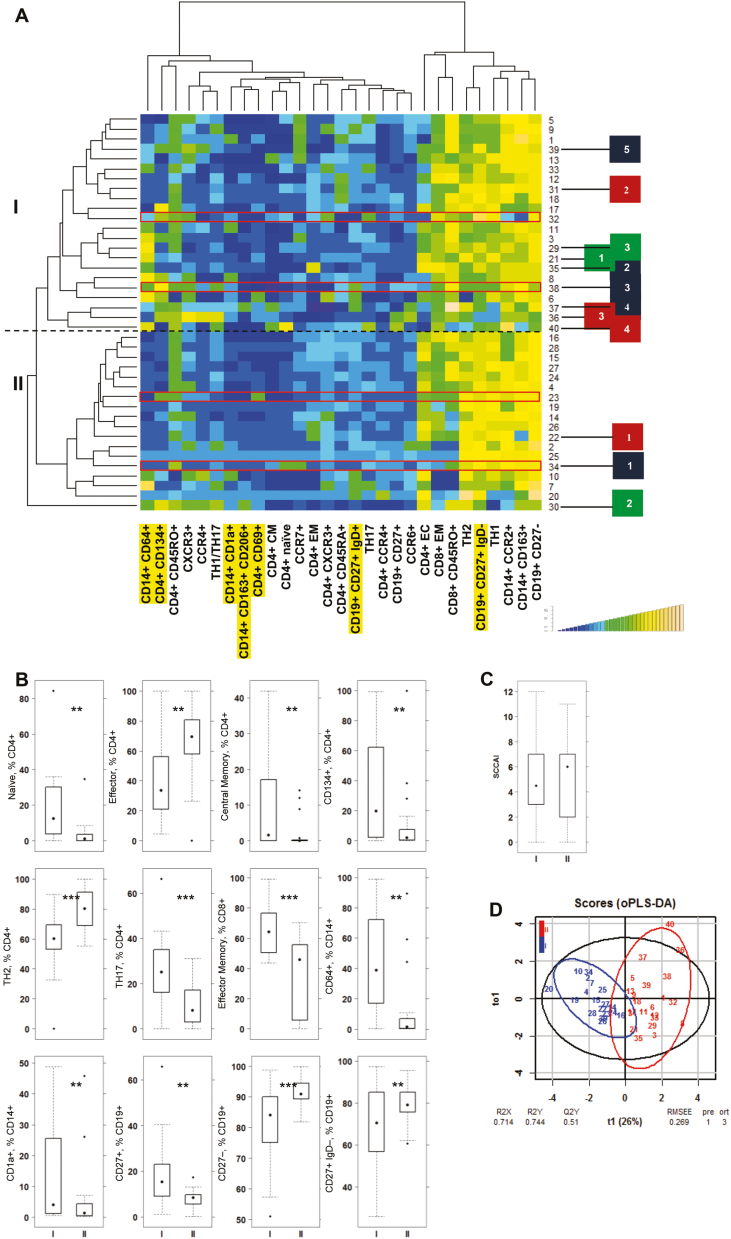
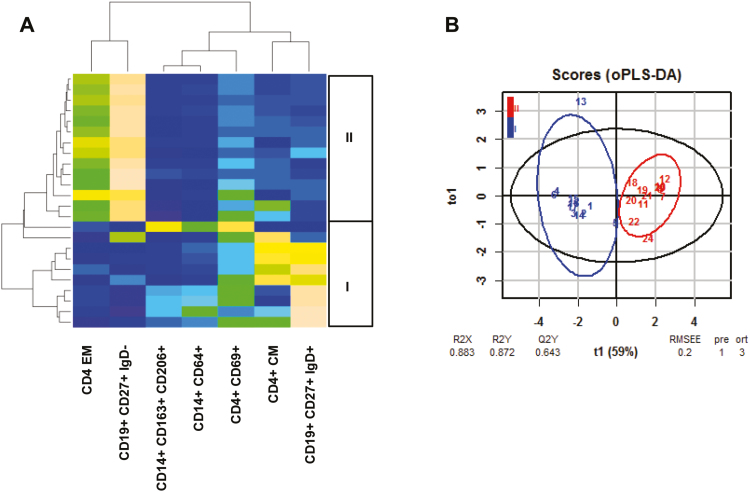
As shown in Figure 2A, 2 main subgroups of UC patients were detected. The patient group I was characterized by increased frequencies of pro-inflammatory monocytes, naïve and central memory (CM) CD4+ cells, OX40-expressing CD4+ T cells (CD4+ CD134+), TH17 cells, pro-inflammatory M1 monocytes (CD14+ CD64+ and CD14+ CD1a+), and antigen-experienced B cells (CD19+ CD27+), whereas group II was signified by increased frequencies of effector CD4+ and TH2 cells, switched B cells (CD19+ CD27+ IgD-), and antigen-inexperienced B cells (CD19+ CD27-). No significance was observed with regard to the Simple Clinical Colitis Activity Index (SCCAI), exhibiting similar mean ± SD values of 5.18 ± 3.40 in patient group I and 4.88 ± 3.53 in patient group II (Fig. 2C). Boxplot analysis followed by the Student t test identified naïve and effector CD4+, CD4+ CD134+, TH2, TH17, effector CD8+ cells, CD14+ CD64+ and CD14+ CD1a+ monocytes, and CD19+ CD27+ IgD-, CD19+ CD27-, and CD19+ CD27+ to be significantly different in both groups (Fig. 2B; for the complete data set, see Supplementary Table 7). To examine whether these variables have the capacity to discriminate between groups I and II, oPLS-DA was performed. As depicted in Figure 2D, both groups were separated, with high R2X, R2Y, and Q2Y values of 0.71, 0.74, and 0.51, respectively. The low RMSEE value of 0.26 indicates a high probability of classifying patients correctly based on these variables. For significance diagnostics, R2Y and R2Q values were compared with values after random permutation of the y response. The obtained values of pR2Y = 0.05 and pR2Q = 0.05 further corroborated the validity of the oPLS model. To further evaluate the predictive performance of this model, cross-validation with training and testing subsets (20 samples, 12 variables, and 1 response for each subset) was performed. In the training subset, no false negative was detected (I: 9 I and 0 II; II: 0 I and 11 II). As expected, the predictions in the testing subset were lower (I: 8 I and 3 II; II: 1 I and 8 II). Thus, this classifier achieves 80% of correct predictions.
FIGURE 2.
Patient profiling leads to identification of subgroups of UC. A, Hierarchically clustered heatmap of 40 patients. Subgroups I and II are indicated on the left side. Patients participating in the longitudinal study are indicated as boxes on the right side (blue: patient A, time points 1–5; red: patient B, time points 1–4; green: patient C, time points 1–3). B, Boxplot analysis of frequencies of subtypes of CD4+ and CD8+ T cells, monocytes, and B cells. C, Boxplot analysis of SCCAI score. Boxes represent upper and lower quartiles, whiskers represent variability, and outliers are plotted as individual points. Labels given on x-axes on the bottom and top row apply to all charts. For comparison of groups, a Student t test was conducted (*P = 0.05; **P = 0.01; ***P = 0.001). D, oPLS-DA analysis of patient group I and II. The parameters used were identical to those used for hierarchical clustering. PBMCs selected for testing of responsiveness to adalimumab in the NSG-UC are boxed in red. Labels highlighted in yellow refer to markers used in the animal studies for discriminating groups. Abbreviations: R2X, fraction of the variation of the X variables explained by the model; R2Y, fraction of the variation of the Y variables explained by the model; Q2Y, fraction of variation of the y variables predicted by the model.
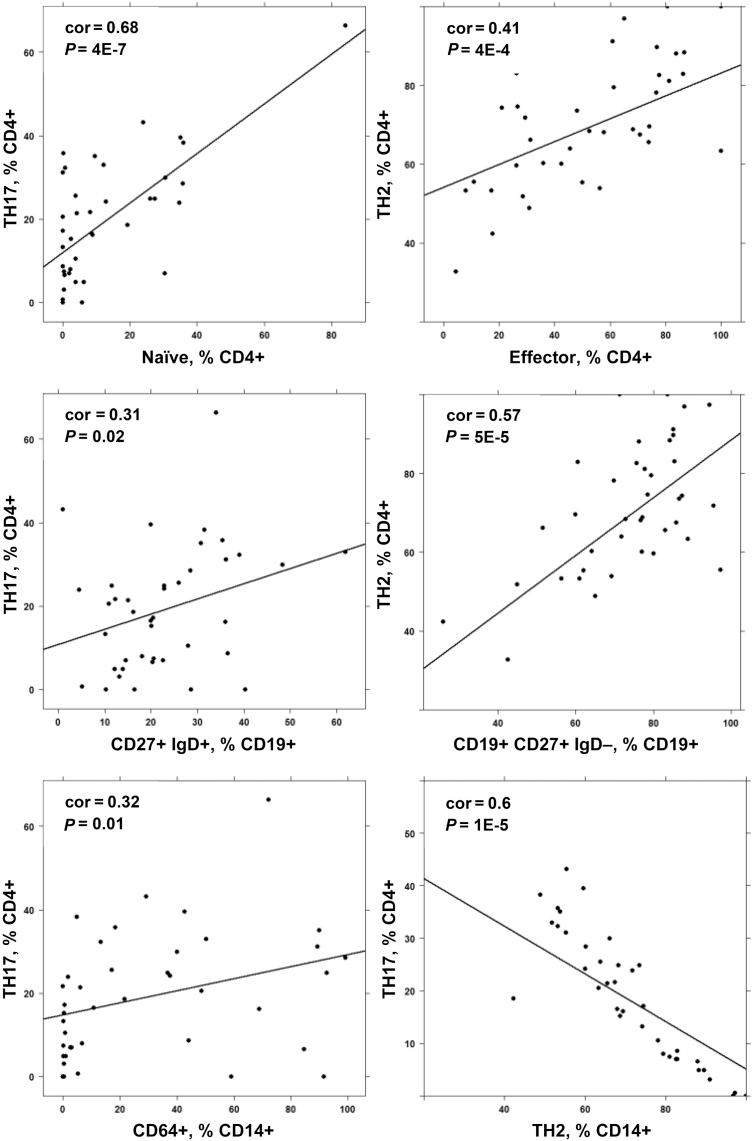
Pearson correlation analysis of the 2 TH17 and TH2 cellular populations that signified patient groups I and II, respectively, and that had previously been identified as drivers in UC11, 12 revealed partially opposing correlations. As shown in Figure 3, TH17 correlated positively with naïve CD4+ T cells, unswitched B cells, and M1 monocytes and CD14+ CD64+, whereas TH2 positively correlated with CD4+ effector cells and switched B cells (CD19+ CD27+ IgD-). Both cell types strongly correlated negatively, suggesting that both pathways evoke distinct responses (for the complete correlation matrix, see Supplementary Table 8).
FIGURE 3.
Correlation analysis of subtypes of immune cells identifies TH17 and TH2 as opposing immunological pathways. Method = Pearson; numbers display Pearson’s product–moment correlation values (cor) and P values (P).
Patients who had repeatedly donated blood switched from group I to group II or vice versa during the course of the analysis, indicating that classification of the groups was not based on clearly defined inflammatory subgroups but rather reflects the dynamic of the inflammation (Fig. 2A).
The Immunological Phenotype of the Patient Is Preserved in the NSG-UC Mouse Model
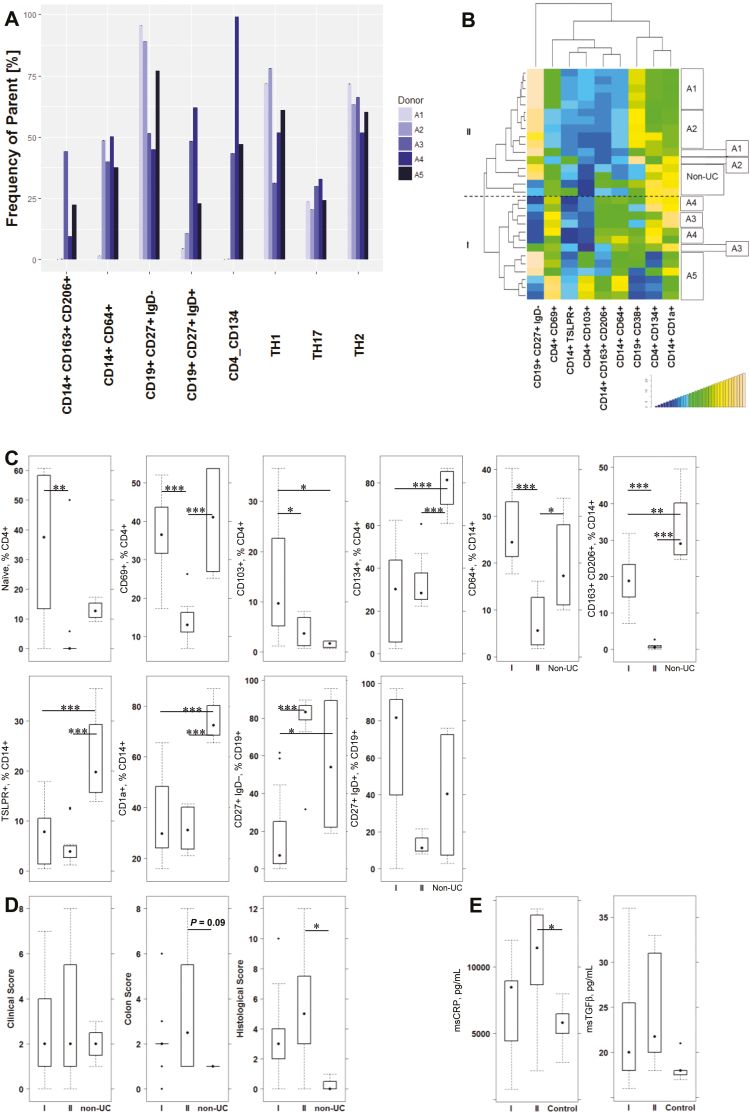
As we have previously observed various pathological phenotypes in the NSG-UC mouse model to include a high influx of inflammatory cells and severe damage of the colon architecture or highly fibrotic alterations (Roswitha Gropp, the pathological phenotype in the NSG-UC mouse model), we wanted to examine whether the inflammation in the mouse corresponds to the observed inflammatory profile of patients. Therefore, mice were reconstituted with PBMCs from donor A 5 times at 5 consecutive time points (Fig. 2A1–A5; Supplementary Table 6). These groups were compared with a group of mice reconstituted with a non-UC donor. Before reconstitution, the immunological profile of the donor was assessed. As shown in Figures 2A and 4A, the profile of the donor underwent a shift. Although at the first time point the profile was that of the TH1/2- switched B cell type, the type shifted to the Th17/monocyte-driven inflammation throughout the course of the study. At time point 5, M1 and M2 monocytes were increased, and this correlated with increased Th17 cells and unswitched B cells. Six NSG mice were reconstituted with PBMCs from donor A in consecutive experiments, and 4 NSG mice were reconstituted with PBMCs from a non-UC donor. All mice were challenged with ethanol according to a standard protocol as described in the “Methods” section. As usual, the levels of reconstitution, as reflected by hCD45+ cells, varied; however, no significant differences were found between the groups.
FIGURE 4.
The immunological phenotype of the donor is partially preserved in the NSG-UC model. NSG mice were reconstituted with PBMCs from patient A isolated at 5 consecutive time points (A1–A5; n = 6 for each experiment) and with PBMCs from a non-UC donor (n = 4). Mice were challenged with 10% ethanol at day 8 and 50% ethanol at day 15. A, Comparison of frequencies of subtypes of T, B, and monocytes in PBMCs from patient A at time points of reconstitution. B, Hierarchical cluster analysis of frequencies of human leukocytes isolated from the spleen of reconstituted mice, analyzed by flow cytometry, and depicted as a heatmap. Boxes on the right side indicate mice reconstituted with PBMCs from patient A at different time points and those of the non-UC donor. I and II indicate the main groups. C, Frequencies of human leukocytes isolated from mouse spleen of groups I and II and the non-UC donor are depicted as boxplots. D, Clinical, colon, and histological scores of mice from groups I and II and the non-UC donor are depicted as boxplots. E, Mouse CRP levels in colons of mice from groups I and II in comparison with levels in unrecontituted mice (control, n = 7). Boxes represent upper and lower quartiles, whiskers represent variability, and outliers are plotted as individual points. Labels given on x-axes on the bottom row apply to all charts. For comparison of groups, a Student t test was conducted (0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05).
To compare the immunological profiles of these mice, human leukocytes were isolated from mouse spleen during autopsy and subjected to flow cytometric analysis using a similar, albeit more restricted, panel, as in the profiling of UC patients (for the complete data set, see Supplementary Table 9). Figure 4B shows the hierarchical clustering of the leukocytes as a heatmap, which revealed 2 main groups. In the first 2 experiments (A1 and A2), mice exhibited fewer M1 and M2 monocytes, whereas in the following experiments (A3–A5), frequencies of M1 and M2 increased. As observed in the correlation analysis, in UC these increased frequencies correlated with increased frequencies of CD4+ CD69+ and CD4+ CD134+ T cells. Thus, in analogy to the distinction in UC patients, the mouse groups were labeled mouse group I and mouse group II. The group of mice reconstituted with a non-UC donor clustered between group I and II. To analyze whether the discrimination of the 2 groups also referred to naïve and effector T cells, switched and unswitched B cells, and M1 monocytes, leukocytes were separated into groups I and II. As shown in Figure 4C, the pattern was similar to the one in PBMCs from UC patients. The non-UC-derived leukocytes exhibited a distinct pattern. It is noteworthy that the frequencies of M2 monocytes and TSLPR-expressing monocytes were increased as compared with groups I and II, indicating that the protective effect of PBMCs might be due to these immune-modulatory cells.
To analyze whether PBMCs from groups I and II also affect symptoms and pathological phenotype, mice were monitored throughout the experiment and classified according to a clinical, colon, and histological score, as described in the “Methods” section. All scores seemed more elevated in mouse group II; however, differences were not significant (Fig. 4D). To corroborate the observation that inflammation declined in group I as compared with group II, proteins were extracted from colon, and mouse calprotectin (CRP) levels were determined by luminex assay. In this experiment, the control samples were collected from nonreconstituted mice (n = 7). As shown in Figure 4E, CRP levels were highest in group II, and the difference was significant when group II was compared with the control group. As observed in previous studies by reverse transcription polymerase chain reaction analysis from mRNA colon extracts,6 msTGFß levels increased upon challenge and were not significantly different in both groups.
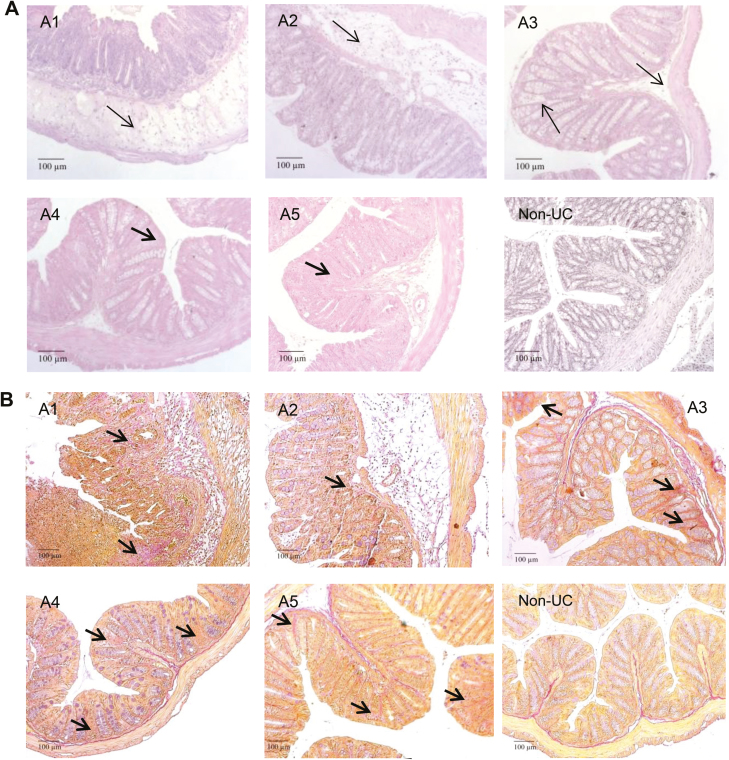
Major differences were observed when histopathological manifestations of the colon were analyzed (Fig. 5A). Histological analysis revealed more severe inflammation characterized by a pronounced influx of inflammatory cells into the mucosa and severe edema in the first 2 experiments, whereas the influx of inflammatory cells and the development of edema were less severe in the last 3 experiments. However, loss of goblet cells and fibrotic alterations prevailed. Staining of collagen deposition by Elastica-van-Gieson corroborated fibrotic alterations in the mucosa (Fig. 5B) This observation was also supported by the TGFß levels (Fig. 4E).
FIGURE 5.
Histological analysis of a section of the distal part of a mouse colon. Mice were treated as described in Figure 4. A1–A5 refers to 5 consecutive studies with PBMCs from donor A, and non-UC refers to a study with PBMC from a non-UC donor. A, Photomicrographs of H&E-stained sections. B, Photomicrographs of Elastica-van-Gieson-stained sections. Arrow indicates edema and influx of inflammatory cells; bold arrow indicates fibrosis.
Response to Adalimumab in the NSG-UC Mouse Model
As the previous experiment had shown that the immunological phenotype is partially preserved in the NSG-UC mouse model, we further explored whether response to treatment with adalimumab was different in both inflammatory subgroups. As in previous studies, outcomes to evaluate the efficacy of adalimumab were the clinical, colon, and histological score and serum levels of glutamic and aspartic acid.4 In addition, the serum levels of IL-6 and IL-1ß were included in the analysis.
PBMCs from 2 different donors each from patient groups I and II (Fig. 2A) were reconstituted, and 7 days postreconstitution, mice were divided into 2 groups: mice challenged with ethanol and treated with isotype (isotype; mouse group I reconstituted with PBMCs from patient group I: donor n = 2; mice n = 12, mouse group II reconstituted with PBMCs from patient group II: donor n = 2; mice n = 12) and mice challenged with ethanol and treated with adalimumab (adalimumab; mouse group I: donor n = 2; mice n = 12, mouse group II: donor n = 2; mice n = 12). Challenge was performed a described in the “Methods” section; 30 mg/kg of isotype or adalimumab was applied intraperitoneally on days 6 and 13.
To validate the previous results, a cluster analysis was performed of frequencies of human leukocytes isolated from the spleen and depicted as a heatmap. For this analysis, only mice from the challenged control group were included. As shown in Figure 6A, mice clustered according to the group of, patients, and oPLS-DA analysis revealed a clear distinction between both groups, as indicated by the high R2X, R2Y, and R2Q values of 0.83, 0.872, and 0.643, respectively, and by the low RMSEE value of 0.2 (Fig. 6B).
FIGURE 6.
The NSG-UC model reflects the immunological profile of patients. NSG-UC mice were engrafted with PBMCs derived from UC patients of patient group I (donor n = 2; samples n = 11) or II (donor n = 2; samples n = 13) challenged with 10% ethanol at day 8 and 50% ethanol at day 15 and treated with isotype (isotype n = 12). A, Heatmap of the frequencies of human leukocytes isolated from the spleen of mice treated with isotype. B, oPLS-DA analysis of mouse groups I and II using the same parameters as shown in the heatmap.
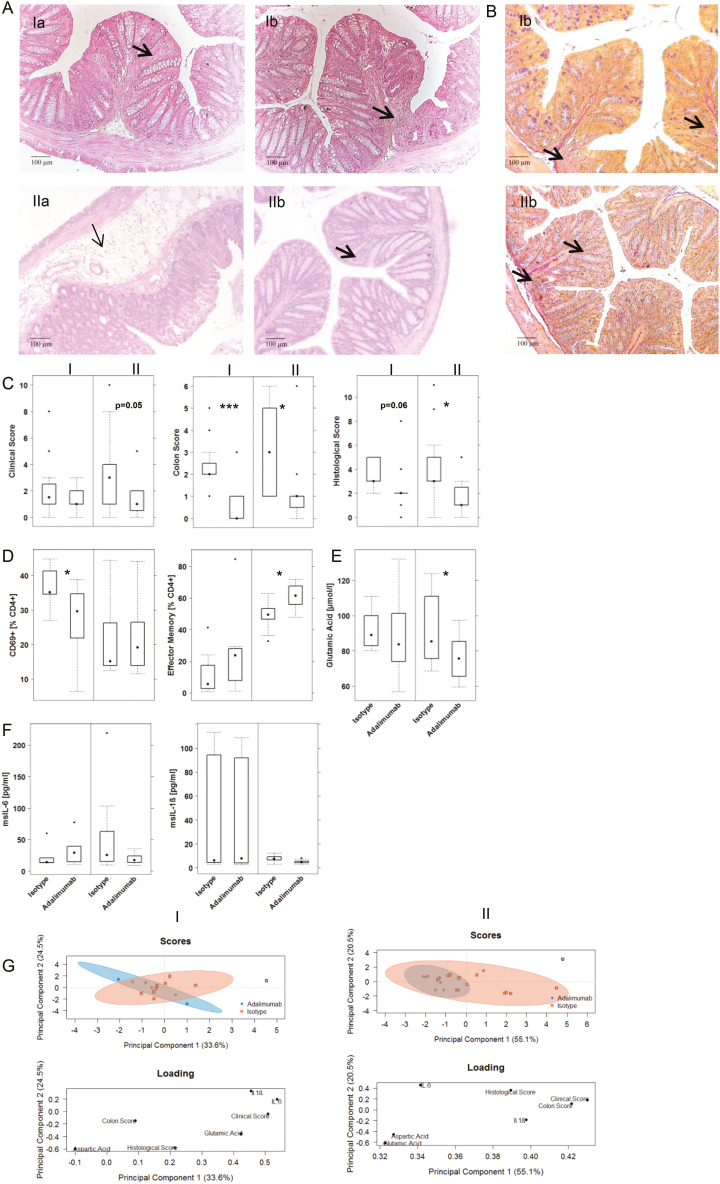
Second, the response to ethanol was compared in mouse groups I and II. As indicated by the clinical score (Fig. 7C), mice from patient groups I and II responded differently to challenge with ethanol (for the complete data set, see Supplementary Table 10). The clinical score was less pronounced in patient group I, which was mostly due to less severe diarrhea. No difference was observed when the colon score was analyzed and the histological score was not profoundly different. As observed in the previous experiment, the influx of inflammatory cells into the mucosa was less pronounced in mouse group I (Fig. 7A). In some cases, however, loss of goblet cells and fibrosis were detected in these colons. The response to adalimumab also differed in the 2 mouse groups. In mouse group II, the differences in clinical and histological scores were significantly different when the isotype and adalimumab groups were compared, as opposed to mouse group I, where only the differences in the colon score were significant (Fig. 7C). Histological analysis revealed what we have observed in a clinical study3 and in mouse studies using infliximab,6 namely that the remodeling of inflammation is not affected by anti-TNFα inhibitors (Fig. 7A and B).
FIGURE 7.
NSG mice reconstituted with PMBCs derived from patient groups I and II responded differently to treatment with adalimumab. Mice were engrafted and treated as described in Figure 4. A, Photomicrographs of H&E-stained sections of distal parts of the colon from mice. B, Photomicrographs of Elastica-van-Gieson-stained sections: (a) isotype, (b) adalimumab. Arrow indicates edema and influx of inflammatory cells; bold arrow indicates fibrosis. C, Clinical, colon, and histological scores. D, Frequencies of human leukocytes isolated from the spleen. E, Serum levels of glutamic acid. F, Serum levels of mouse cytokines. Results are depicted as boxplots. Boxes represent upper and lower quartiles, whiskers represent variability, and outliers are plotted as individual points. Labels given on x-axes on the bottom and top row apply to all charts. For comparison of groups, a Student t test was conducted (0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05). G, PCA analysis.
Comparison of human leukocytes isolated from the spleen and subjected to flow cytometric analysis revealed different responses to adalimumab (Fig. 7D). Adalimumab induced a significant decline of frequencies of activated CD4+ CD69+ T cells in mouse group I, whereas CD4+ effector cells were significantly affected in mouse group II. A difference was also observed when serum levels of glutamic acid were analyzed (Fig. 7E). Only in mouse group II did the levels of glutamic acids decrease significantly in response to adalimumab. The difference between the 2 groups was also apparent when the serum levels of IL-6 and IL-1ß were compared (Fig. 7F). Mice from group II displayed higher levels of IL-6, which decreased on treatment with adalimumab, whereas levels of IL-6 were unaffected by treatment with adalimumab in mice from patient group I. As the variability of IL-1ß expression in group I was high, one cannot conclude that IL-1ß is generally higher in patient group I. The response to adalimumab was marginal in both groups. To compare the response to adalimumab in both groups of mice, the parameters were used which had previously been identified as inflammatory markers responses as all scores, IL-6, IL-1ß, and aspartic and glutamic acid to perform PCA. As shown in Figure 7G, both groups responded, but in group II mice clustered more closely together, indicating a higher response rate than group I.
DISCUSSION
The Holy Grail in chronic inflammatory diseases is the identification of biological markers that might explain the heterogeneity of disease manifestations and predict responses to treatment. Recent studies have identified Oncostatin M as a driver of inflammation and a predictive marker of response to therapy with anti-TNFα antibodies.13 In this study, we show that a broad panel of subtypes of CD4+ T cells, CD14+ monocytes, and B cells led to the discrimination of 2 main inflammatory conditions in UC patients. Patients in group I were characterized by increased levels of M1 monocytes (CD14+ CD64+), CD4+ CD134+ activated T cells, and unswitched B cells (CD19+ CD27+ IgD+). Correlation analysis revealed that this condition may be driven by TH17 CD4+ T cells. In contrast, patients from group II were signified by elevated TH2 CD4+ T cells, and these cells correlated negatively with M1 monocytes and M2 monocytes, respectively, and unswitched B cells. These conditions do not represent distinct subtypes of the disease but rather reflect the dynamics of inflammation, as patient profiles analyzed in longitudinal studies switched from group I to group II and vice versa during the course of the study. Although we are far from understanding which factors are responsible for the switch, our study suggests that the monocyte-driven arm of the inflammation is not addressed by adalimumab, and suppressing inflammation in UC may require targeting of both processes. Patients who respond poorly to treatment with anti-TNFα antibodies may need additional therapy to gain remission, and therapeutically naïve patients experiencing predominantly monocyte-driven inflammation may be spared from treatment with anti-TNFα antibodies. Recently, specific JAK1 inhibitors and the pan JAK inhibitor tofacitinib have been shown to inhibit the activation of M1 macrophages in vitro and induce M2 polarization.14 It is noteworthy that tofacitinib lacks efficacy in acute dextran-sulfate sodium (DSS)–induced colitis but ameliorates disease activity in the acute DSS rescue model. Our own unpublished results have also shown efficacy in the NSG-UC model without affecting TGFß levels, suggesting the suppression of M1 but not M2 monocytes. Thus, tofacitinib might be the first approved inhibitor addressing M1 monocytes, and defining subtypes of patients according to the scheme presented in this study may be the first step to individualized and phase-dependent therapies.
These 2 inflammatory conditions were also observed in the NSG-UC mouse model, where PBMCs derived from patient group I achieved a phenotype corresponding to the inflammatory profile of patient group I. Conversely, PBMCs from patient group II resulted in a phenotype of patient group II. Thus, the inflammatory profile in the NSG-UC mouse model was preserved. This observation was corroborated by analysis of the phenotype of mice that had been reconstituted with PBMCs from a patient who took part in the longitudinal study and whose PBMCs were simultaneously used for reconstitution. The switch (transition from group I to group II) observed in the immunological profile was also reflected in the corresponding mice.
Next, we assessed whether the 2 different inflammatory conditions were prone to respond to the TNFα inhibitor adalimumab and whether a difference in efficacy could be observed. Analysis revealed that mice were responsive to adalimumab, but that mice reconstituted with PBMCs from patient group II were more responsive as compared with mice reconstituted with PBMCs from patient group I. The results presented in this study show that the combination of immunological profiling of patients and studies in the NSG-UC model leads to a better understanding of the inflammatory processes; however, for several reasons, the approach presented in this study is still limited and needs to be improved. First, although the panel of subtypes of inflammatory cells leads to distinction of inflammatory states, it still needs to be expanded, as it, for example, neglects important cell types such as basophils, mast cells, and eosinophils. In this regard, the NSG-UC model is chimeric, and it is not known how well mouse signals conveyed by basophils, for example, are received by human cells or whether basophils derived from patients have a different impact on inflammation than mouse basophils. Second, the onset of inflammation in the model requires a challenge with ethanol, and it is not known whether inflammation in humans also requires a trigger or is perpetuating due to the lack of mechanisms resolving inflammation. Third, it is a short-lived inflammation, and repeated challenge with ethanol leads to increased wound healing processes (Roswitha Gropp, repeated challenge with ethanol leads to increased frequencies of M2 monocytes); thus it does not have the capacity to reproduce inflammation in humans who endure lifelong inflammation and may experience phases of remission and relapses. Even in light of these limitations, however, the NSG-UC model presents an improvement as compared with conventional mouse models, and results obtained by the combination of patient profiling and preclinical tests in this model may have a higher translatability to future clinical trials.
Supplementary Material
ACKNOWLEDGMENTS
Our special thanks go to the donors. Without their commitment, this work would not have been possible. We thank the team in the animal facility for their excellent work and their enduring friendliness in stressful situations and Paula Winkelmann for excellent technical assistance.
Supported by: This work was funded by Sanofi-Aventis Deutschland GmbH, Frankfurt am Main, Germany.
Conflicts of interest: The authors declare no competing interests. Sanofi-Aventis GmbH, Frankfurt am Main, provided support in the form of salaries for author E.B., but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. All other authors declare no conflict of interest.
REFERENCES
- 1. Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. [DOI] [PubMed] [Google Scholar]
- 2. Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. [DOI] [PubMed] [Google Scholar]
- 3. Föhlinger M, Palamides P, Mansmann U, et al. Immunological profiling of patients with ulcerative colitis leads to identification of two inflammatory conditions and CD1a as a disease marker. J Transl Med. 2016;14:310:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jodeleit H, Al-Amodi O, Caesar J, et al. Targeting ulcerative colitis by suppressing glucose uptake with ritonavir. Dis Model Mech. 2018;11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jodeleit H, Palamides P, Beigel F, et al. Design and validation of a disease network of inflammatory processes in the NSG-UC mouse model. J Transl Med. 2017;15:265:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palamides P, Jodeleit H, Föhlinger M, et al. A mouse model for ulcerative colitis based on NOD-scid IL2R γnull mice reconstituted with peripheral blood mononuclear cells from affected individuals. Dis Model Mech. 2016;9:985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Amodi O, Jodeleit H, Beigel F, et al. CD1a-expressing monocytes as mediators of inflammation in ulcerative colitis. Inflamm Bowel Dis. 2018; 24:1225–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weigmann B, Tubbe I, Seidel D, et al. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2:2307–2311. [DOI] [PubMed] [Google Scholar]
- 9. Thévenot EA, Roux A, Xu Y, et al. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J Proteome Res. 2015;14:3322–3335. [DOI] [PubMed] [Google Scholar]
- 10. Williams HR, Willsmore JD, Cox IJ, et al. Serum metabolic profiling in inflammatory bowel disease. Dig Dis Sci. 2012;57:2157–2165. [DOI] [PubMed] [Google Scholar]
- 11. Fukaura K, Iboshi Y, Ogino H, et al. Mucosal profiles of immune molecules related to t helper and regulatory t cells predict future relapse in patients with quiescent ulcerative colitis. Inflamm Bowel Dis. 2019;25:1019–1027. [DOI] [PubMed] [Google Scholar]
- 12. Heller F, Florian P, Bojarski C, et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology. 2005;129:550–564. [DOI] [PubMed] [Google Scholar]
- 13. West NR, Hegazy AN, Owens BMJ, et al. ; Oxford IBD Cohort Investigators Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Vries LCS, Duarte JM, De Krijger M, et al. A JAK1 selective kinase inhibitor and tofacitinib affect macrophage activation and function. Inflamm Bowel Dis. 2019;25:647–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.