Abstract
Background.
Inhaled corticosteroids (ICS) are the most widely prescribed and effective medication to control asthma symptoms and exacerbations. However, many children still have asthma exacerbations despite treatment, particularly in admixed populations, such as Puerto Ricans and African Americans. A few genome-wide association studies (GWAS) have been performed in European and Asian populations, and they have demonstrated the importance of the genetic component in ICS response.
Objective.
We aimed to identify genetic variants associated with asthma exacerbations in admixed children treated with ICS, and to validate previous GWAS findings.
Methods.
A meta-analysis of two GWAS of asthma exacerbations was performed in 1,347 admixed children treated with ICS (Hispanics/Latinos and African Americans), analyzing 8.7 million genetic variants. Those with p≤5×10−6 were followed up for replication in 1,697 asthmatic patients from six European studies. Associations of ICS response described in published GWAS were followed up for replication in the admixed populations.
Results.
A total of 15 independent variants were suggestively associated with asthma exacerbations in admixed populations (p≤5×10−6). One of them, located in the intergenic region of APOBEC3B and APOBEC3C, showed evidence of replication in Europeans (rs5995653, p = 7.52×10−3) and was also associated with change in lung function after treatment with ICS (p = 4.91×10−3). Additionally, the reported association of the L3MBTL4-ARHGAP28 genomic region was confirmed in admixed populations, although a different variant was identified.
Conclusions & Clinical Relevance.
This study revealed the novel association of APOBEC3B and APOBEC3C with asthma exacerbations in children treated with ICS and replicated previously identified genomic regions. This contributes to the current knowledge about the multiple genetic markers determining responsiveness to ICS which could lead in the future the clinical identification of those asthma patients who are not able to respond to such treatment.
Keywords: African American, childhood asthma, exacerbations, Latino, pharmacogenomics
INTRODUCTION
Asthma is the most common chronic condition in children and young adults. In addition to the direct impact of the illness on the individual, severe exacerbations of asthma generate considerable economic costs to healthcare systems, as well as work and/or school absenteeism [1].
Inhaled corticosteroids (ICS) are the most effective and commonly prescribed medications for symptom control and prevention of severe asthma exacerbations [1]. While most children using ICS experience a decrease in their asthma symptoms, 30–40% will continue to experience exacerbations, and of these non-responders, 10–15% may even have an increase in their exacerbations [2]. High variability in ICS response has been described also among ethnicities [3]. In addition to high asthma morbidity, exacerbations rates and mortality, admixed populations have reduced ICS response [4]. These strong ethnic differences suggest a substantial hereditary component in the ICS response [5]. In fact, approximately 40–60% of the variation in ICS response may be due to genetic factors [6].
For several decades, pharmacogenetic studies have utilized candidate-gene approaches, which only evaluate a small portion of the genetic variation. More recently, these have evolved towards hypothesis-free approaches by implementing genome-wide association studies (GWAS) [7]. Eight GWAS of ICS response have been performed to date [8–15], revealing an association between 14 genomic regions and this trait.
However, the polymorphisms identified by GWAS to date only represent a small proportion of the heritability of ICS response, and hence it is not possible to predict an individual’s response to this treatment [16]. The design of the GWAS performed to date may be the main reason, where analyses are statistically underpowered to detect genetic associations. Most GWAS of ICS response have included a relatively small number of individuals (N<1,000) of primarily European and, to a lesser extent, Asian ancestry, with poor representation of admixed populations [4], which include Hispanics/Latinos and African Americans. However, the increased asthma prevalence among admixed individuals with African ancestry, such as Puerto Ricans and African Americans, and the greater genetic diversity and specific genetic background of these populations present a unique opportunity to study the response to ICS treatment in asthma [3–4].
We hypothesized that a large pharmacogenetic study of ICS response in admixed individuals with asthma that exhaustively explores the association of genetic variants across the genome could reveal novel genes associated with this trait. We also attempted to evaluate whether the associations described in GWAS performed in European and Asian populations could be generalized to admixed populations.
METHODS
Study Populations
A total of eight independent studies participating in the Pharmacogenomics in Childhood of Asthma (PiCA) consortium [17] were analyzed as part of discovery and replication phases of this meta-GWAS. Individuals from two admixed populations were included in the discovery phase: the Genes-environments & Admixture in Latino Americans Study (GALA II) and the Study of African Americans, Asthma, Genes and Environments (SAGE). Samples from six European PiCA studies were used for replication. All studies have been approved by their local institutional review boards and all participants/parents provided written informed assent and consent, respectively. GALA II and SAGE were approved by the Human Research Protection Program Institutional Review Board of the University of California, San Francisco (San Francisco, United States) (ethics approval numbers: 217802 and 210362, respectively). PACMAN was approved by the Medical Ethics Committee of the University Medical Centre Utrecht (Utrecht, the Netherlands). The Tayside Committee on Medical Research Ethics (Dundee, United Kingdom) approved BREATHE. PASS was approved by the Liverpool Paediatric Research Ethics Committee (Liverpool, United Kingdom) (reference number: 08/H1002/56). SLOVENIA was approved by the Slovenian National Medical Ethics Committee (Ljubljana, Slovenia). ESTATe was approved by the Medische Ethische Toetsings Commissie, Erasmus Medical Center (Rotterdam, the Netherlands) (ethics approval number: MEC-2011–474). followMAGICS was approved by the Ethik-Kommission der Bayerischen Landesärztekammer (Munich, Germany) (ethics reference number: 01218).
Discovery phase
Patients from the GALA II and SAGE studies with a physician diagnosis of asthma who reported having active symptoms and asthma medication use within the last year were analyzed in the discovery phase. These are two independent studies focused on two different racial/ethnic groups based on the self-identified ethnicity of the four grandparents of each subject: Hispanics/Latinos (GALA II) and African Americans (SAGE). Both studies recruited unrelated children and young adults, aged 8 to 21 years old, using the same protocol and questionnaires from different areas in the United States. GALA II also recruited individuals in Puerto Rico [18].
Analyses were performed for a subset of 854 subjects from GALA II and 493 individuals from SAGE. Specifically, we assessed self-reported ICS use, age, gender, genome-wide genotypic data [19–20], and information regarding presence or absence of severe asthma exacerbations, as defined by the European Respiratory Society (ERS) and the American Thoracic Society (ATS) [21]. We examined exacerbations that occurred during the 12 months preceding the study enrollment (need to seek emergency asthma care, hospitalizations or the administration of oral corticosteroids).
Replication phase
Validation was carried out in European individuals from six independent studies participating in the PiCA consortium: the follow-up stage of the Multicenter Asthma Genetics in Childhood Study (followMAGICS); the Pharmacogenetics of Adrenal Suppression study (PASS); Pharmacogenetics of Asthma Medication in Children: Medication with Anti-inflammatory effects (PACMAN); Effectiveness and Safety of Treatment with Asthma Therapy in Children (ESTATe); BREATHE and SLOVENIA studies. Details for each study are described in the Supporting Information.
The use of ICS and availability of data related to the presence/absence of asthma exacerbations during the previous 12 or 6 months were also applied as inclusion criteria for the individuals from these studies analyzed in the current study, whereas non-availability of data related to ICS use, asthma exacerbations, age, gender and genotype data were considered as exclusion criteria. For those studies without data related to the events included in the ATS/ERS definition of asthma exacerbations, information regarding school absences, unscheduled general practitioner (GP) or respiratory system specialist visits was also considered.
Genome-wide genotyping, genetic ancestry assessment and imputation
Both GALA II and SAGE samples were genotyped using the Axiom® LAT1 array (Affymetrix Inc.), and quality control (QC) procedures were performed as described elsewhere [19–20]. Genotyping of the subjects included in the replication phase was performed on different genotyping platforms, as described in previous publications (see Supporting Information) (Table S1). In addition, four of the studies were genotyped for the purposes of the PiCA consortium and their QC is described in the Supporting Information.
Genetic ancestry was assessed by means of Principal Component (PC) analysis with EIGENSOFT 6.14 for the studies included in both discovery and replication phases [22]. Quantitative global genetic ancestry estimates were also obtained for the populations included in the discovery phase. An unsupervised model was applied using ADMIXTURE [23], assuming the European (CEU), African (YRI) and Native American (NAM) as the parental populations for the Hispanics/Latinos and YRI and CEU for African Americans. For that, reference haplotypes from CEU and YRI populations from the HapMap Project Phase III [24] were used. Moreover, haplotypes from individuals genotyped with Axiom® LAT1 array (Affymetrix Inc.) were considered as reference for NAM population, as described elsewhere [19, 25].
In all the studies, imputation was carried out by means of the Michigan Imputation Server (https://imputationserver.sph.umich.edu) using the second release of the Haplotype Reference Consortium (HRC) (r1.1 2016) as reference panel [26]. Haplotype reconstruction and imputation were performed with SHAPEIT v2.r790 [27] and Minimac3 [28], respectively.
Association testing and meta-analysis in the discovery phase
GWAS analyses were carried out separately for GALA II and SAGE. Logistic regressions were used to evaluate the association between genetic variants and ICS response by means of the binary Wald test implemented in EPACTS 3.2.6 [29]. The presence or absence of any asthma exacerbations during the last 12 or 6 months in patients treated with ICS was considered as a measure of ICS response, which was evaluated as a binary variable. Age, gender, and the first two PCs, obtained with EIGENSOFT 6.14 [22], were included as covariates in the regression models. The number of PCs included as covariates was chosen based on the comparison of different models that included up to 10 PCs, showing that results based on 2 PCs had the best fit with the expected values under the null hypothesis of no association.
Single nucleotide polymorphisms (SNPs) with a minor allele frequency (MAF)≥1% and with imputation quality (Rsq)≥0.3 in GALA II and SAGE, and shared among both populations were meta-analyzed using METASOFT [30]. Fixed-effects or random-effects models were selected for each variant depending on absence or presence of heterogeneity, respectively, which was assessed by means of the Cochran Q-test. A threshold of p-value≤5×10−6 was arbitrarily set to select variants suggestively associated with asthma exacerbations, since this threshold is commonly adopted in GWAS studies [31–35]. Among those variants, independent associations were detected by means of logistic regression analyses conditioned on the most significant SNP of each locus using R 3.4.3 [36]. This analysis provided a list of independent variants that were followed up for replication.
Association testing and meta-analysis in the replication phase
Statistical analyses were performed following the same methodology as in the discovery phase, except for the definition of asthma exacerbations available in each study and the number of PCs included as covariates in the association analyses (Table S1). Evidence of replication was considered for those SNPs that showed a combined p-value≤0.05 in a meta-analysis of all the European studies and consistent directions of effects in both discovery and validation populations.
Association with ICS response measured as change in FEV1
SNPs significantly associated with asthma exacerbations in both admixed and European populations, were evaluated for association with the change in the forced expiratory volume in 1 second (FEV1) after 6 weeks of treatment with ICS in 166 ICS users from the SLOVENIA study, the only cohort included in the analyses with this outcome measured. This variable was dichotomized to define responders and non-responders to ICS treatment using a cutoff of ≥8% improvement of FEV1, which has been established as a good predictor of asthma severity in children [37]. Logistic regression models were applied including age, gender, and the first two PCs as covariates.
Functional evaluation of variants associated with ICS response
Functional annotation and evidence of significant expression quantitative trait loci (eQTL) were searched with HaploReg v4.1[38] based on data provided by the Encyclopedia of DNA Elements (ENCODE) project [39]. This was performed for the SNP associated with ICS response in admixed and European populations and those in high linkage disequilibrium (LD) (r2>0.9) according to African populations from the 1000 Genomes Project (1KGP) data incorporated by HaploReg v4.1. Gene expression was inspected using the Portal for the Genotype-Tissue Expression (GTEx) [40] and the Gene Expression Atlas [41]. Moreover, evidence of association with enhancers was searched using the multiple sources available from GeneHancer [42].
Validation of previous associations in admixed populations
Since previous GWAS of ICS response have focused on European and Asian populations [8–15], we attempted to validate their results in admixed populations. A total of 25 SNPs near or within 14 genes declared as associated with ICS response [8–14], were followed up for replication in GALA II and SAGE.
Replication was attempted at the SNP level and also as genomic region, the latter considering variants located within 100 kilobases (kb) upstream or downstream from the gene where the variant was located or from the two closest genes in case the variant was intergenic. Evidence of replication was considered for SNPs nominally associated with ICS response (p≤0.05) that had the same direction of the effect as the published GWAS. For the replication at level of genomic region, a Bonferroni-like correction was applied to account for the number of independent variants tested within each genomic region, as estimated with empirical autocorrelations based on Markov Chain Monte Carlo (MCMC) simulations. For this, an autocorrelation matrix was obtained based on the −log10 p-value of each SNP analyzed using the effectiveSize function from the R package coda [43], as described elsewhere [44]. According to this, a Bonferroni-corrected significance threshold was estimated for each genomic region with α=0.05/number of independent variants.
RESULTS
Characteristics of the study populations
The characteristics of the 1,347 admixed asthmatic patients from GALA II and SAGE analyzed in the discovery phase and the 1,697 Europeans subjects included in the replication are shown in Table 1 and Table S1, respectively. In terms of estimates of global ancestry in the admixed populations, Hispanics/Latinos had 13.6% African ancestry, 51.5% European ancestry and 34.9% Native American ancestry. In contrast, African Americans had 79.4% African admixture and 20.6% European ancestry. Hispanics/Latinos reported a higher proportion of asthma exacerbations in the 12 months preceding study enrollment (66.4%) than African Americans (51.9%). Although asthma exacerbations were differentially defined in the validation populations, similar proportions were found across the discovery and replication studies, except for PACMAN and SLOVENIA, with values of 11.0% and 34.1%, respectively (Table S1).
Table 1.
Clinical and demographic characteristics of the admixed populations analyzed in the discovery phase.
| GALA II (n = 854) | SAGE (n = 493) | |
|---|---|---|
| Gender (% male) | 57.3 | 54.2 |
| Mean age ± SD (years) | 12.1 ± 3.2 | 13.5 ± 3.4 |
| Ethnicity | Hispanic/Latino | African American |
| Mean genetic ancestry (%) | ||
| African | 13.6 | 79.4 |
| European | 51.5 | 20.6 |
| Native-American | 34.9 | NA |
| Asthma exacerbations in the last 12 months (%) | 66.4 | 51.9 |
| Emergency asthma care (%) | 56.6 | 43.2 |
| OCS use (%) | 40.2 | 29.4 |
| Hospitalizations (%) | 12.6 | 5.7 |
SD: standard deviation; OCS: oral corticosteroids; NA: not available.
Discovery phase
The meta-analysis of the GALA II and SAGE GWAS included 8.7 million SNPs that were shared among Hispanics/Latinos and African Americans and had MAF≥1% and Rsq≥0.3. The Q-Q plots of the association results for each individual study (Figure S1A and Figure S1B) and those obtained after combining both admixed populations did not reveal major genomic inflation due to population stratification (λGC = 1.04, Figure S1C). Although the genome-wide significant threshold (p-value≤5×10−8) was not reached by any of the variants, 27 SNPs with Rsq values ranging from 0.59–1.00 and located near or within 13 loci were suggestively associated with asthma exacerbations despite the use of ICS (p-value≤5×10−6) in admixed children and young adults (Figure 1 and Table S2).
Figure 1. Manhattan plot of association results of ICS response in the discovery phase.
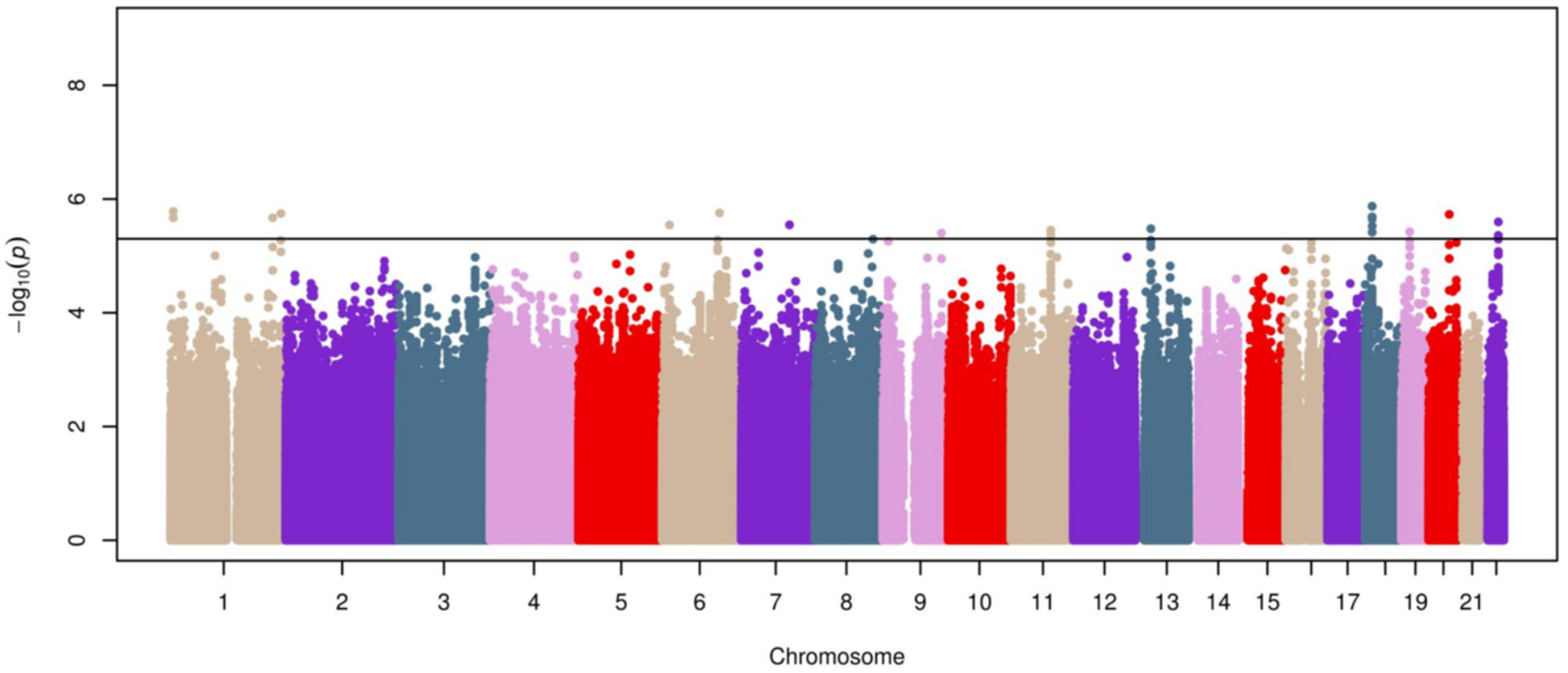
Association results are represented as −log10 p-value on the y-axis along the chromosomes (x-axis). The suggestive significance threshold for replication is indicated by the black line (p≤5×10−6).
After performing pairwise regression models conditioned on the most significant variant for each locus with at least two suggestive associations, one independent variant was detected per locus, except for APOBEC3B-APOBEC3C and ANKRD30B, where two SNPs remained significant after conditioning on each gene’s most significant variant (Table S3). As a result, 15 SNPs were identified as independently associated with ICS response in admixed populations (Table S3) and were followed up for replication.
Replication phase
Of the 15 SNPs selected for replication in Europeans, 11 SNPs had a MAF≥1% and Rsq≥0.3 (ranging from 0.36–1.00) in Europeans and were forwarded for replication (Table 2). Of those, rs5995653, located within the intergenic region of APOBEC3B and APOBEC3C (Figure 2), showed evidence of nominal replication after combining the European studies. To check that the association of this SNP in the admixed populations was not confounded by unaccounted components of ancestry, different regression models were tested including estimates of genetic ancestry, different number of PCs, or following the method described by Conomos et al. [45], which provided similar results (Table S4). The direction of effect for this SNP was the same in Europeans (OR for A allele: 0.76, 95% CI: 0.62–0.93, p = 7.52×10−3) as in the admixed samples (OR for A allele = 0.66, 95% CI: 0.56–0.79, p = 4.80×10−6) (Table 2). A meta-analysis of this SNP across the two phases resulted in a suggestive genome-wide significant association (OR for A allele = 0.70, 95% CI: 0.61–0.81, p = 3.31×10−7, Figure 3).
Table 2.
Association results for the suggestive SNPs followed up for replication in European populations.
| Admixed populations (n=1,347) | European populations (n=1,697) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr.a | Positionb | Nearest gene(s) | A1/A2 | Freq.c | OR (95% CI)d | p-value | Freq.c | OR (95% CI)d | p-value |
| rs11121611 | 1 | 6367219 | ACOT7 | G/T | 0.201 | 0.55 (0.43–0.70) | 1.65 × 10−6 | 0.062 | 0.97 (0.61–1.56) | 0.247e |
| rs35514893 | 6 | 15909525 | DTNBP1-MYLIP | T/C | 0.020 | 0.36 (0.23–0.55) | 2.86 × 10−6 | 0.082 | 0.73 (0.22–2.46) | 0.613 |
| rs4897302 | 6 | 123886231 | TRDN | T/C | 0.505 | 1.58 (1.31–1.91) | 1.75 × 10−6 | 0.221 | 0.96 (0.81–1.13) | 0.637 |
| rs61585310 | 7 | 104006510 | LHFPL3 | G/T | 0.796 | 0.61 (0.49–0.75) | 2.85 × 10−6 | 0.763 | 0.91 (0.74–1.11) | 0.352 |
| rs7851998 | 9 | 126828514 | LHX2-NEK6 | A/G | 0.191 | 0.56 (0.44–0.72) | 3.97 × 10−6 | 0.046 | 0.83 (0.65–1.06) | 0.132 |
| rs2125362 | 11 | 86167136 | ME3 | A/G | 0.684 | 1.31 (0.68–2.56) | 3.53 × 10−6 e | 0.750 | 0.97 (0.82–1.16) | 0.764 |
| rs450789 | 13 | 33578233 | KL | G/A | 0.334 | 0.64 (0.53–0.77) | 3.33 × 10−6 | 0.271 | 0.97 (0.83–1.15) | 0.756 |
| rs12959468 | 18 | 15182381 | ANKRD30B-ROCK1 | A/G | 0.039 | 0.39 (0.26–0.58) | 2.99 × 10−6 | 0.077 | 1.39 (0.74–2.62) | 0.309 |
| rs2278992 | 19 | 18095769 | KCNN1 | C/T | 0.176 | 0.59 (0.47–0.74) | 3.76 × 10−6 | 0.151 | 1.00 (0.81–1.24) | 0.991 |
| rs6001366 | 22 | 39399941 | APOBEC3B-APOBEC3C | T/C | 0.079 | 0.47 (0.35–0.65) | 2.53 × 10−6 | 0.064 | 1.00 (0.72–1.38) | 0.995 |
| rs5995653 | 22 | 39404249 | APOBEC3B-APOBEC3C | A/G | 0.285 | 0.66 (0.56–0.79) | 4.80 × 10−6 | 0.508 | 0.76 (0.62–0.93) | 7.52 ×10−3 |
Chromosome;
Positions based on GRCh37/hg19 build;
Frequency of the effect allele;
Odds ratio for the effect alleles (additive model);
Random-effect model was applied since heterogeneity was found between admixed/European populations.
A1: Effect allele; A2: Non-effect allele; CI: Confidence Interval.
Figure 2. Regional plot of association results in the discovery phase for the APOBEC3B-APOBEC3C intergenic region, which represents a novel association with ICS response.
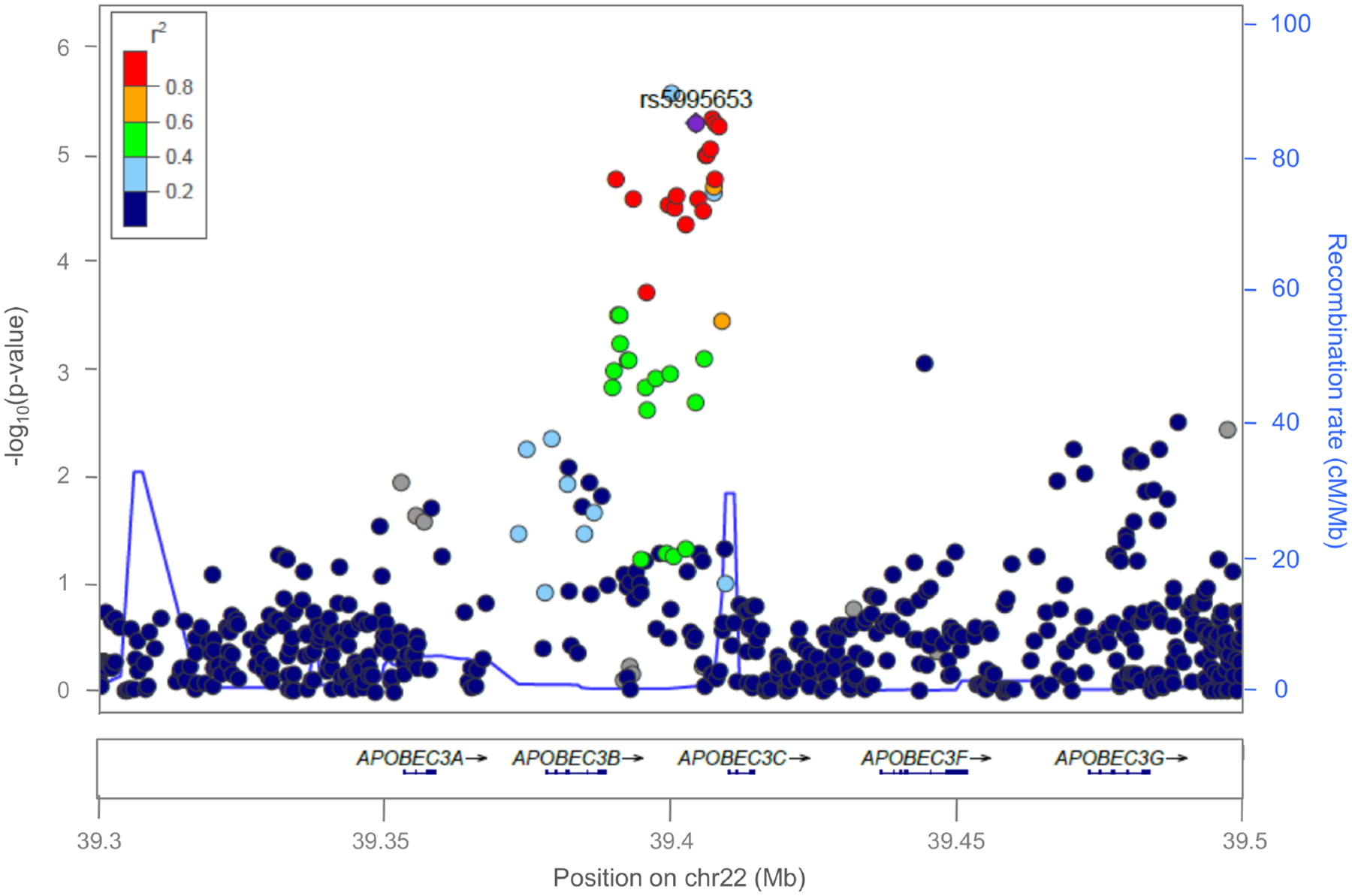
Statistical significance of association results (−log10 p-value) (y-axis) is represented by chromosome position (x-axis) for each SNP as a dot. A diamond represents the independent association signal with evidence of replication in Europeans (rs5995653) and the remaining SNPs are color-coded based on their LD with this SNP, indicated by pairwise r2 values for American populations from the 1KGP.
Figure 3. Forest plot of association effect of rs5995653 across studies.
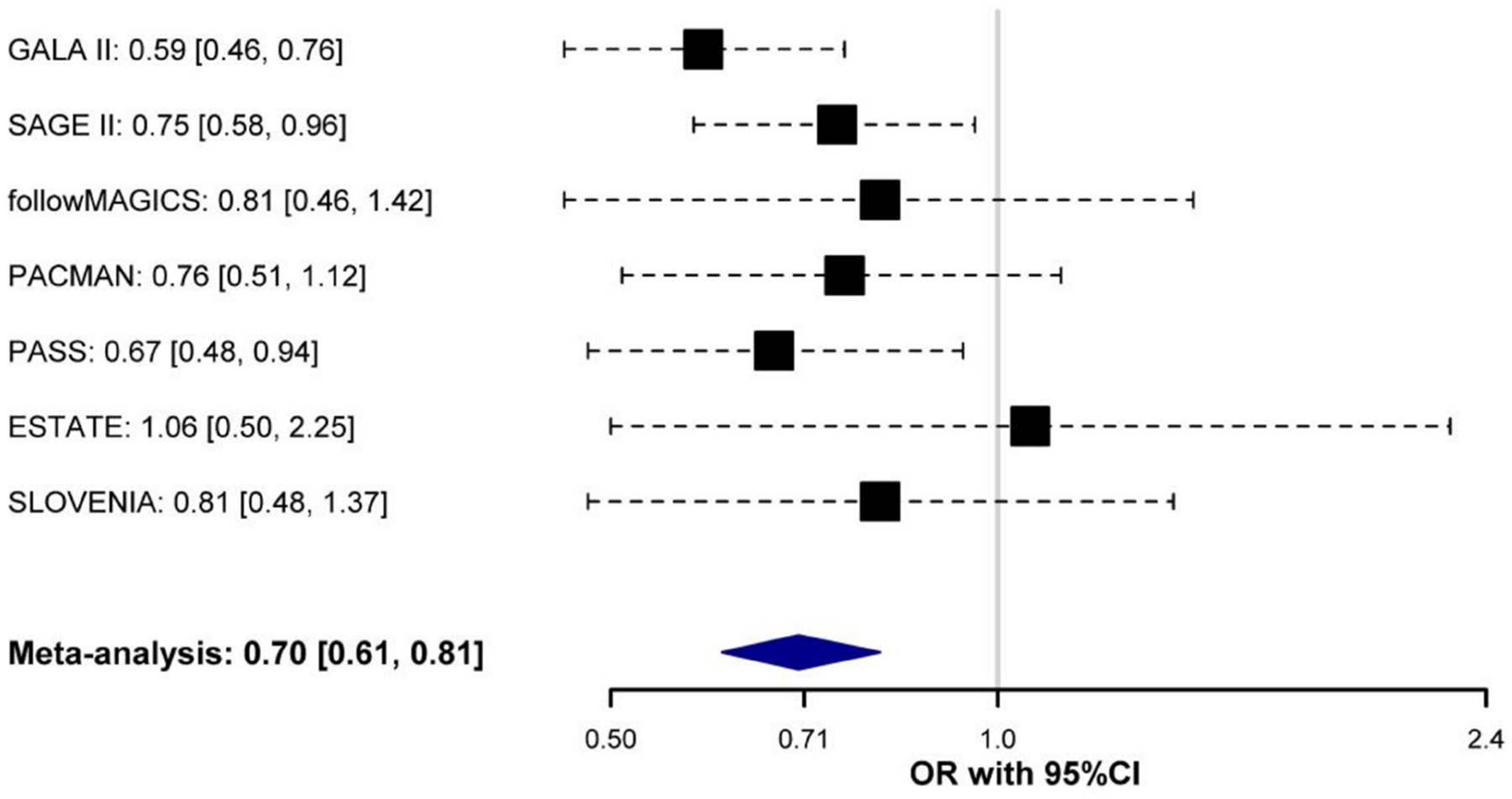
Odds ratio (OR) for the effect allele (A) is shown for each study and after combining them by black boxes and a blue diamond. Black dash lines indicate the corresponding 95% Confidence Intervals (95% CI) for each individual study.
Association of rs5995653 with ICS response measured as change in FEV1
The SNP rs5995653 was significantly associated with a positive response to the ICS treatment in SLOVENIA, measured as an increase of FEV1 (OR for A allele = 2.16, 95% CI: 1.26–3.70, p = 4.91×10−3), which is concordant with the protective effect of this SNP with asthma exacerbations in both discovery and validation studies.
In silico functional role of the novel association detected
The experimental data provided by the ENCODE project shows that the SNP rs5995653 is located within a histone H3 lysine 4 mono-methylation (H3K4me1) mark of an active gene enhancer and a DNAse hypersensitivity site in blood cells [39]. This is concordant with the GeneHancer evidence that APOBEC3B has been associated with enhancers that regulate multiple transcription factor binding sites, indicating its involvement in the regulation of gene expression in different cell types, including lung fibroblasts [42]. Moreover, this variant is also in high LD with several eQTL in blood cells associated with the expression of APOBEC3A (rs9607601: p=1.80×10−13 and rs5995654: p=9.10×10−14), APOBEC3G (rs9607601: p=0.003), and CBX6 (rs9607601: p=3.94×10−4 and rs5995654: p=4.00×10−4) [38–39, 46]. In addition, previous functional studies have evidenced high levels of gene expression of both APOBEC3B and APOBEC3C in pulmonary cells (GTEx) [40–41].
Validation of previous associations of ICS response
None of the 25 SNPs previously associated with ICS response was consistently associated with asthma exacerbations in admixed populations (Table S5). To assess whether the lack of replication of previous GWAS hits could be due to the association of alternative genetic variants among different populations, a replication analysis was also performed at genomic region level. A total of 36,261 variants located within 100 kb upstream and downstream from 14 loci previously associated with ICS response were evaluated. After applying a Bonferroni-like correction for the number of variants analyzed within each genomic region, suggestive associations were observed for nine SNPs near three genomic regions: ALLC (min p-value = 4.69×10−4 for the SNP rs113903375), L3MBTL4-ARHGAP28 (min p-value = 1.57×10−5 for the SNP rs62081416), and ELMO2-ZNF334 (min p-value = 3.56×10−4 for the SNP rs2425845) (Table S6). However, applying a more restrictive correction for the total number of independent variants across all genomic regions (p≤1.71×10−5 for 2,916 independent variants tested), only the association of rs62081416, located within the intergenic region of L3MBTL4 and ARHGAP28, was significantly associated with ICS response in admixed individuals (OR for A allele = 2.44, 95% CI: 1.63–3.65, p = 1.57×10−5).
DISCUSSION
In this study, we carried out the first GWAS of ICS response in Hispanic/Latino and African American children and young adults with asthma. After combining the association results from these two populations, 15 independent suggestive association signals were associated with asthma exacerbations despite use of ICS, and one of them showed evidence of nominal replication in Europeans. This SNP was also significantly associated with an increase in FEV1 after 6 weeks of treatment with ICS in one of the European studies where this outcome was measured. These results revealed for the first time the association of APOBEC3B and APOBEC3C genes with ICS response in asthmatic children and young adults. Additionally, we validated the association of the L3MBTL4-ARHGAP28 genomic region in admixed populations, which was previously described in a GWAS of ICS response in subjects of European descent.
The APOBEC3B and APOBEC3C genes encode two members of the apolipoprotein B mRNA-editing catalytic polypeptide 3 (APOBEC3) family. APOBEC3 proteins are involved in RNA editing through the deamination of cytidine to uracil [47]. Their main function is related to innate immunity and are considered important restriction factors against a broad range of viruses [48]. However, APOBEC3 proteins are also involved in cellular processes related to mutagenic activity [49], including the development of several types of cancer, while APOBEC3B specifically has been associated with an increased risk of lung cancer [50].
We found that the A allele of rs5995653, located 5.8 kb from the 3’UTR of APOBEC3C, showed a protective effect against asthma exacerbations and was associated with improvement on FEV1 in patients treated with ICS. While no asthma-related functions have been attributed to any of the APOBEC3 flanking genes, evidence of high levels of RNA expression has been found in pulmonary fibroblasts for both genes [40–41]. Furthermore, the functional evidence found for rs5995653 suggests that this SNP plays a key role in regulating the expression of genes involved in several cellular processes in the lung. Interestingly, respiratory viral infections are important risk factors for exacerbations in asthmatic children [51]. This fact is concordant with the consistent function of APOBEC3B and APOBEC3C as restrictors of viral infections, suggesting that the expression of these genes in pulmonary tissues could be involved in fighting against viral-induced asthma exacerbations in patients treated with ICS.
Our study has several strengths. First, this is the largest meta-GWAS of ICS response with a discovery phase specifically focused on Hispanic/Latino and African American asthma patients, the minority ethnic groups most affected by asthma in the United States [4]. Admixed populations with African and Native American have been underrepresented in the asthma pharmacogenomic studies of ICS response [4]. Secondly, we identified a novel association shared among admixed and European populations, which could be also influential in other populations. Third, we validated the association of three genomic regions previously described in GWAS of ICS response in European and Asian populations [11, 13] and one of them was associated with an improvement in FEV1 after treatment with ICS in adults [11]. This evidence reinforces the validity of asthma exacerbations as a good measure of response to the asthma treatment with ICS. Finally, the fact that the intergenic region of L3MBTL4 and ARHGAP28 has been previously identified in adults could suggest the existence of common genetic markers of ICS response among adulthood and childhood asthma [13].
We recognize some limitations of our study. First, the most significant variant associated with ICS response in admixed and European populations did not reach genome-wide significance. This result was replicated in independent samples at nominal level, although it would not still be significant after a multiple comparison correction. Second, this study did not include a considerable larger sample size compared to the largest GWAS of ICS response published to date [17]. Third, even though the HRC reference panel is the largest catalogue of variants from the whole genome available to date [26], admixed populations with African and Native American ancestries are not well represented. Fourth, asthma exacerbations were differentially defined in the European populations included in the replication phase. Nevertheless, this outcome was homogeneously defined in the studies included in the discovery phase, suggesting that the identified locus is robustly associated with asthma exacerbation across a range of definitions. Fifth, ICS response was evaluated as the presence or absence of asthma exacerbations in asthmatic patients with a self-reported use of ICS, which might not correspond to compliance or changes with the asthma control therapy. For this reason, the association signal detected was followed up for replication using a quantitative measurement of ICS response, which was only available in one of the European populations. Additional studies should seek to validate the association signal when using change in FEV1 after the treatment with ICS as the response variable. Finally, functional evidence relating the intergenic region of APOBEC3B and APOBEC3C with ICS response in asthma patients was not directly assessed in this current study, since only experimental data available in public databases was queried. Therefore, in vitro experiments in relevant tissues and cell types for ICS response are needed to evaluate the functional roles of these loci in order to confirm their implication in this trait.
In summary, our meta-GWAS in admixed children and young adults identified a novel association of genetic variants from the intergenic region of APOBEC3B and APOBEC3C as with ICS response in subjects with asthma. We also validated the association of one genomic region previously associated with ICS response. Our study demonstrates the advantages of including admixed populations in asthma pharmacogenomic studies of ICS response.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the patients, families, recruiters, health care providers and community clinics for their participation in all the studies involved in the PiCA consortium (http://pica-consortium.org/). The authors also thank Sandra Salazar for her support as the GALA II and SAGE study coordinator and the contribution of Teide High-Performance Computing facilities (http://teidehpc.iter.es) provided by the Instituto Tecnológico y de Energías Renovables (ITER, S.A.) to the results of this research.
This work was supported by the award number AC15/00015 by the Instituto de Salud Carlos III (ISCIII) through Strategic Action for Health Research (AES) and European Community (EC) within the Active and Assisted Living (AAL) Programme framework (MP-Y), and the SysPharmPedia grant from the ERACoSysMed 1st Joint Transnational Call from the European Union under the Horizon 2020. N.H-P was funded by a fellowship (FI16/00136) from Instituto de Salud Carlos III (ISCIII) and co-funded by the European Social Funds from the European Union (ESF) “ESF invests in your future” and MP-Y was supported by the Ramón y Cajal Program (RYC-2015-17205) by the Spanish Ministry of Economy, Industry and Competitiveness. The GALA II and SAGE studies were funded by the Sandler Family Foundation, the American Asthma Foundation, the RWJF Amos Medical Faculty Development Program, Harry Wm. and Diana V. Hind Distinguished Professor in Pharmaceutical Sciences II, National Institutes of Health (1R01HL117004, R01Hl128439, R01HL135156, and 1X01HL134589), National Institute of Health and Environmental Health Sciences (R01ES015794 and R21ES24844), the National Institute on Minority Health and Health Disparities (1P60MD006902, RL5GM118984, and 1R01MD010443), and the Tobacco-Related Disease Research Program under Award Number 24RT-0025 to E.G.B. The PACMAN cohort study was funded by a strategic alliance between GlaxoSmithKline and Utrecht Institute for Pharmaceutical Sciences. The SLOVENIA study was financially supported by the Slovenian Research Agency (research core funding No. P3-0067).
Footnotes
CONFLICT OF INTEREST STATEMENT
NH-P declares funding from Instituto de Salud Carlos III (ISCIII) and the European Social Funds. SSO and HF report funding from the National Institutes of Health (NIH). KV declares funding from ZonMw. MK reports funding from the European Union, the German Ministry of Education and Research, German Research Foundation and other sources. A-HM declares funding from GlaxoSmithKline, Boehringer Ingelheim and Astra Zeneca. EGB reports funding from NIH, National Institute of Health and Environmental Health Sciences, National Institute on Minority Health and Health Disparities and the Tobacco-Related Disease Research Program. MP-Y declares funding from ISCIII and Spanish Ministry of Economy, Industry and Competitiveness.
The rest of authors have no conflict of interest.
REFERENCES
- 1.Global strategy for asthma management and prevention. Global Initiative for Asthma (GINA) 2017. http://ginasthma.org/. Accesed January 15, 2018.
- 2.Szefler SJ, Phillips BR, Martinez FD, Chinchilli VM, Lemanske RF, Strunk RC, et al. Characterization of within-subject responses to fluticasone and montelukast in childhood asthma. J Allergy Clin Immunol 2005;115:233–242. [DOI] [PubMed] [Google Scholar]
- 3.Mersha TB. Mapping asthma-associated variants in admixed populations. Front Genet 2015;6:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega VE, Meyers DA. Pharmacogenetics: implications of race and ethnicity on defining genetic profiles for personalized medicine. J Allergy Clin Immunol 2014;133:16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss ST. New approaches to personalized medicine for asthma: where are we? J Allergy Clin Immunol 2012;129:327–334. [DOI] [PubMed] [Google Scholar]
- 6.Lemiere C, Bai T, Balter M, Bayliff C, Becker A, Boulet LP, et al. Adult Asthma Consensus Guidelines update 2003. Can Respir J 2004;11 Suppl A:9A–18A. [DOI] [PubMed] [Google Scholar]
- 7.Vijverberg SJH, Farzan N, Slob EMA, Neerincx AH, Maitland-van der Zee AH. Treatment response heterogeneity in asthma: the role of genetic variation. Expert Rev Respir Med 2018;12:55–65. [DOI] [PubMed] [Google Scholar]
- 8.Tantisira KG, Lasky-Su J, Harada M, Murphy A, Litonjua AA, Himes BE, et al. Genomewide association between GLCCI1 and response to glucocorticoid therapy in asthma. N Engl J Med 2011;365:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tantisira KG, Damask A, Szefler SJ, Schuemann B, Markezich A, Su J, et al. Genome-wide association identifies the T gene as a novel asthma pharmacogenetic locus. Am J Respir Crit Care Med 2012;185:1286–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu AC, Himes BE, Lasky-Su J, Litonjua A, Peters SP, Lima J, et al. Inhaled corticosteroid treatment modulates ZNF432 gene variant’s effect on bronchodilator response in asthmatics. J Allergy Clin Immunol 2014;133:723–728 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park TJ, Park JS, Cheong HS, Park BL, Kim LH, Heo JS, et al. Genome-wide association study identifies ALLC polymorphisms correlated with FEV(1) change by corticosteroid. Clin Chim Acta 2014;436:20–26. [DOI] [PubMed] [Google Scholar]
- 12.Park HW, Dahlin A, Tse S, Duan QL, Schuemann B, Martinez FD, et al. Genetic predictors associated with improvement of asthma symptoms in response to inhaled corticosteroids. J Allergy Clin Immunol. 2014;133:664–9 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlin A, Denny J, Roden DM, Brilliant MH, Ingram C, Kitchner TE, et al. CMTR1 is associated with increased asthma exacerbations in patients taking inhaled corticosteroids. Immun Inflamm Dis 2015;3:350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Tong C, Wang Z, Mauger D, Tantisira KG, Israel E, et al. Pharmacodynamic genome-wide association study identifies new responsive loci for glucocorticoid intervention in asthma. Pharmacogenomics J 2015;15:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosteller M, Hosking L, Murphy K, Shen J, Song K, Nelson M, et al. No evidence of large genetic effects on steroid response in asthma patients. J Allergy Clin Immunol 2017;139:797–803 e7. [DOI] [PubMed] [Google Scholar]
- 16.Ortega VE, Meyers DA, Bleecker ER. Asthma pharmacogenetics and the development of genetic profiles for personalized medicine. Pharmgenomics Pers Med 2015;8:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farzan N, Vijverberg SJ, Andiappan AK, Arianto L, Berce V, Blanca-Lopez N, et al. Rationale and design of the multiethnic Pharmacogenomics in Childhood Asthma consortium. Pharmacogenomics 2017;18:931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura KK, Galanter JM, Roth LA, Oh SS, Thakur N, Nguyen EA, et al. Early-life air pollution and asthma risk in minority children. The GALA II and SAGE II studies. Am J Respir Crit Care Med 2013;188:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pino-Yanes M, Thakur N, Gignoux CR, Galanter JM, Roth LA, Eng C, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol 2015;135:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White MJ, Risse-Adams O, Goddard P, Contreras MG, Adams J, Hu D, et al. Novel genetic risk factors for asthma in African American children: Precision Medicine and the SAGE II Study. Immunogenetics 2016;68:391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reddel HK, Taylor DR, Bateman ED, Boulet LP, Boushey HA, Busse WW, et al. An official American Thoracic Society/European Respiratory Society statement: asthma control and exacerbations: standardizing endpoints for clinical asthma trials and clinical practice. Am J Respir Crit Care Med 2009;180:59–99. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 23.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature 2007;449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernandez-Pacheco N, Flores C, Alonso S, Eng C, Mak ACY, Hunstman S, et al. Identification of a novel locus associated with skin colour in African-admixed populations. Scientific Reports 2017;7:44548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaneau O, Coulonges C, Zagury JF. Shape-IT: new rapid and accurate algorithm for haplotype inference. BMC Bioinformatics 2008;9:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics 2015;31:782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HM. EPACTS (Efficient and Parallelizable Association Container Toolbox). http://genome.sph.umich.edu/wiki/EPACTS (2016).
- 30.Han B, Eskin E. Random-effects model aimed at discovering associations in meta-analysis of genome-wide association studies. Am J Hum Genet 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed E, Nunez S, Kulp D, Qian J, Reilly MP, Foulkes AS. A guide to genome-wide association analysis and post-analytic interrogation. Stat Med 2015;34:3769–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oikkonen J, Kuusi T, Peltonen P, Raijas P, Ukkola-Vuoti L, Karma K, et al. Creative Activities in Music--A Genome-Wide Linkage Analysis. PLoS One 2016;11:e0148679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanders AE, Sofer T, Wong Q, Kerr KF, Agler C, Shaffer JR, et al. Chronic Periodontitis Genome-wide Association Study in the Hispanic Community Health Study / Study of Latinos. J Dent Res 2017;96:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roosenboom J, Lee MK, Hecht JT, Heike CL, Wehby GL, Christensen K, et al. Mapping genetic variants for cranial vault shape in humans. PLoS One 2018;13:e0196148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medina-Gomez C, Kemp JP, Trajanoska K, Luan J, Chesi A, Ahluwalia TS, et al. Life-Course Genome-wide Association Study Meta-analysis of Total Body BMD and Assessment of Age-Specific Effects. Am J Hum Genet 2018;102:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R Development Core Team. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ (2013). [Google Scholar]
- 37.Tse SM, Gold DR, Sordillo JE, Hoffman EB, Gillman MW, Rifas-Shiman SL, et al. Diagnostic accuracy of the bronchodilator response in children. J Allergy Clin Immunol 2013;132:554–9 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016;44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, et al. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res 2010;38:D690–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards 2017. [DOI] [PMC free article] [PubMed]
- 43.Plummer M, Best N, Cowles K, Vines K. CODA: Convergence Diagnosis and Output Analysis for MCMC. R News 2006;6:7–11. [Google Scholar]
- 44.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS Comput Biol 2011;7:e1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conomos MP, Laurie CA, Stilp AM, Gogarten SM, McHugh CP, Nelson SC, et al. Genetic Diversity and Association Studies in US Hispanic/Latino Populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2016;98:165–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desimmie BA, Delviks-Frankenberrry KA, Burdick RC, Qi D, Izumi T, Pathak VK. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 2014;426:1220–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janahi EM, McGarvey MJ. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J Viral Hepat 2013;20:821–828. [DOI] [PubMed] [Google Scholar]
- 49.Kanu N, Cerone MA, Goh G, Zalmas LP, Bartkova J, Dietzen M, et al. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol 2016;17:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gansmo LB, Romundstad P, Hveem K, Vatten L, Nik-Zainal S, Lonning PE, et al. APOBEC3A/B deletion polymorphism and cancer risk. Carcinogenesis 2018;39:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duenas Meza E, Jaramillo CA, Correa E, Torres-Duque CA, Garcia C, Gonzalez M, et al. Virus and Mycoplasma pneumoniae prevalence in a selected pediatric population with acute asthma exacerbation. J Asthma 2016;53:253–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


